
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 10 February 2022
Sec. Food Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.853263
This article is part of the Research Topic New Knowledge of Food Microbiology in Asia, Volume II View all 15 articles
In this study, we investigated the species composition and diversity of psychrotrophic bacteria in raw milk from Heilongjiang, Inner Mongolia, Gansu, Henan, Anhui, Jiangsu, Chongqing, and Hunan provinces in China using traditional cultivation and PacBio Single Molecule Real-Time sequencing methods. The isolated psychrotrophic bacteria were highly diverse, which composed of 21 genera and 59 species. Pseudomonas accounted for 58.9% of the total genera while Stenotrophomonas and Enterococcus were also highly represented (above 5.0%). In particular, P. azotoformans occurred at a level of 16.9% and P. paralactis, P. lactis, E. faecalis, and P. marginalis were present in relatively high proportions (above 4.0%). Regional differences were found significantly among the test regions except samples from Heilongjiang and Inner Mongolia were similar. Additionally, differences were observed between days in Henan, Anhui, and Jiangsu samples. Therefore, control strategies must be implemented on regional and season basis.
Milk is an indispensable food for humans and its high nutrient content plays a key role in maintaining health especially for the elderly and children. The quality and safety of dairy products is deeply trusted by consumers (America Department of Health and Human Services, 2019) and the high level of safety procedures in the dairy industry have all but eliminated any physical and chemical dangers of milk consumption (Wang et al., 2018; Du et al., 2019). In contrast, milk contamination by microorganisms is still a constant threat and raw milk may be exposed to microorganisms during milking, storage, transportation, and processing. This risk is offset by the use of rapid cooling equipment and cold chain systems in the dairy industry. The temperature of the raw milk after extrusion is rapidly reduced to <6°C and kept at this level during storage and transportation. Interestingly, the presence of psychrotrophic bacteria can result in contamination of the milk even at these storage temperatures (Ahmed et al., 2014; Fusco and Quero, 2014; Hahne et al., 2019; Du et al., 2020).
Psychrophilic bacteria are widely distributed in the environment and can contaminate cow udders, milking equipment and storage tanks. Cold storage conditions provide ideal conditions for their growth (Yuan et al., 2019). These organisms also secrete heat-resistant proteases and lipases that can significantly alter the quality of dairy products. For example, lipid hydrolysis by contaminating lipases can lead to rancidity while released proteases hydrolyze casein and produces peculiar smells and cause aging and gelation thereby shortening the shelf life (Xin et al., 2017; Meng et al., 2018; Yang et al., 2020; Bu et al., 2022).
The primary species of psychrotrophic bacteria in raw milk have been identified in the genera Pseudomonas, Chromobacterium, Clostridium, Lactobacillus, Alcaligenes, Flarobacterium, Micrococcus, Corynebacterium, Streptococcus, and Enterobacterium (Wang et al., 2018). This diversity was further cataloged in raw milk samples from Heilongjiang Province in Northern China and psychrophilic milk bacteria were primarily found for the genera Chryseobacterium, Pseudomonas, Staphylococcus, Acinetobacter, Clostridium, Flavobacterium, Lactococcus, Kocuria, Bacillus, Serratia, Sphingobacterium, and Aerococcus (Yang et al., 2020). However, the diversity of psychrotrophic bacteria in raw milk for different regions of China was not clear. Therefore, this study was focused on the identification of psychrophilic bacterial community structures in raw milk from different regions of China through a combination of traditional culture and PacBio single-molecule real-time sequencing. We also examined whether regional differences in the community structure of psychrophiles were present. These results provide suggestions for the prevention and control of psychrotrophic bacteria in raw milk.
From December 2018 to 2020, eight farms were selected from eight regions in China for raw milk collection. The regions included Hunan, Henan, Heilongjiang, Chongqing, Inner Mongolia, Gansu, Anhui, and Jiangsu. At each ranch, one batch of samples was collected every day for 3 consecutive days except for Henan when sample collection was 4 consecutive days. The milk was immediately placed in 200 mL sterile bottles and refrigeration at 4°C for pending analysis.
Psychrophiles were cultured using 25 mL raw milk that was diluted 10 times and 200 μL of three suitable dilutions were spread on Psychrophilic bacterial selective agar plates (Qingdao Hope Bio-Technology Co., Ltd., Qingdao, China). The plates were incubated at 6.5°C incubator for 10 d according to IDF method (IDF 101:2005) (International Dairy Federation, 2005). Single colonies were then streak-purified until pure cultures were obtained. Randomly selected colonies were used for identification in triplicate.
DNA was extracted from individual bacterial colonies on agar plates and directly lysed in PCR tubes containing 50 μL lysis buffer (Takara Biomedical Technology, Beijing, China) that was heated at 80°C for 15 min. Bacterial species were identified by PCR using 16S rRNA universal primers (Table 1). The PCR reactions were carried out using Emerald Amp Max PCR Master Mix (Takara Biomedical Technology, Beijing, China) using 2 μL DNA template and other conditions as specified by the manufacturer. The PCR amplification program for 16S rRNA was as follows: 94°C 4 min, 30 cycles of 94°C for 30 s, 57°C for 30 s and 72°C for 90 s and a final step of 72°C for 10 min.
Amplicons were electrophoresed through 1.5% agarose gels to determine whether a target band was present. The PCR products of 16S rRNA amplification were purified and sequenced by a commercial company [BGI Tech Solutions (Beijing Liuhe), Beijing, China].
Milk samples (10 mL each) were centrifuged at 14,000 × g for 5 min and the precipitate was used for DNA extraction using the PowerSoil DNA Isolation kit (Mobio, Carlsbad, CA, United States) according to the manufacturer’s instructions. Extracted DNA was amplified using the following PCR cycling program: 95°C 5 min, followed by 30 cycles of 95°C 30 s, 50°C 30 s, 72°C 90 s and a final extension at 72°C for 7 min. The PCR primers (5′- 3′) specific for the 16S rDNA were 27F (GAGAGTTTGATCCTGGCTCAG) and 1492R (AAGGAGGTGATCCAGCCGCA) (Wang et al., 2018). The amplified products were purified using MagicPure Size Selection DNA Beads (TransGen Biotech, Beijing, China). Electrophoresis results were quantified using Image J software1 and a Sequel II kit (PacBio, Menlo Park, CA, United States) was used for sequence analysis following the instructions of the manufacturer.
The raw data reads were corrected to obtain the CCS (Circular Consensus Sequencing) sequence using SMRT Link (version 8.0) and lima (v1.7.0) software to identify the CCS sequence through the barcode sequence. Chimeras were removed using UCHIME version 8.1 to obtain the final high-quality CCS sequences (Edgar et al., 2011). The composition of psychrotrophic bacterial microbiota was investigated by submitting the psychrotrophic bacteria database to the raw milk microbiota database. Using the sequence similarity relationship between valid data, different data clusters were grouped in operational taxonomic units (OTU) at a 97% level and annotated based on OTU abundance and sequence information (Dixon, 2003). The relative abundance map of microbial community composition was carried out in R ggplot2 (version 2.2.1) (Wickham, 2016), the diversity index was calculated using https://view.qiime2.org, and the principal coordinate analysis (PCoA) was generated in the R project Vegan package version 2.5.3 (Dixon, 2003), Heat map, Venny, and linear discriminant analysis were generated using the online software at https://www.genescloud.cn.
In the current study we examined 248 different psychrophilic bacterial colonies that were isolated from 25 raw milk samples, which identified as 59 species in 21 genera. The dominant genus was Pseudomonas (58.9%) with Stenotrophomonas (7.3%), Enterococcus (6.5%), Chryseobacterium (4.8%), and Serratia (3.6%) as secondary dominants. P. azotoformans accounted for 16.9% of the total species followed by P. paralactis (7.7%), P. lactis (5.2%), E. faecalis (4.8%), and P. marginalis (4.0%) (Table 2).
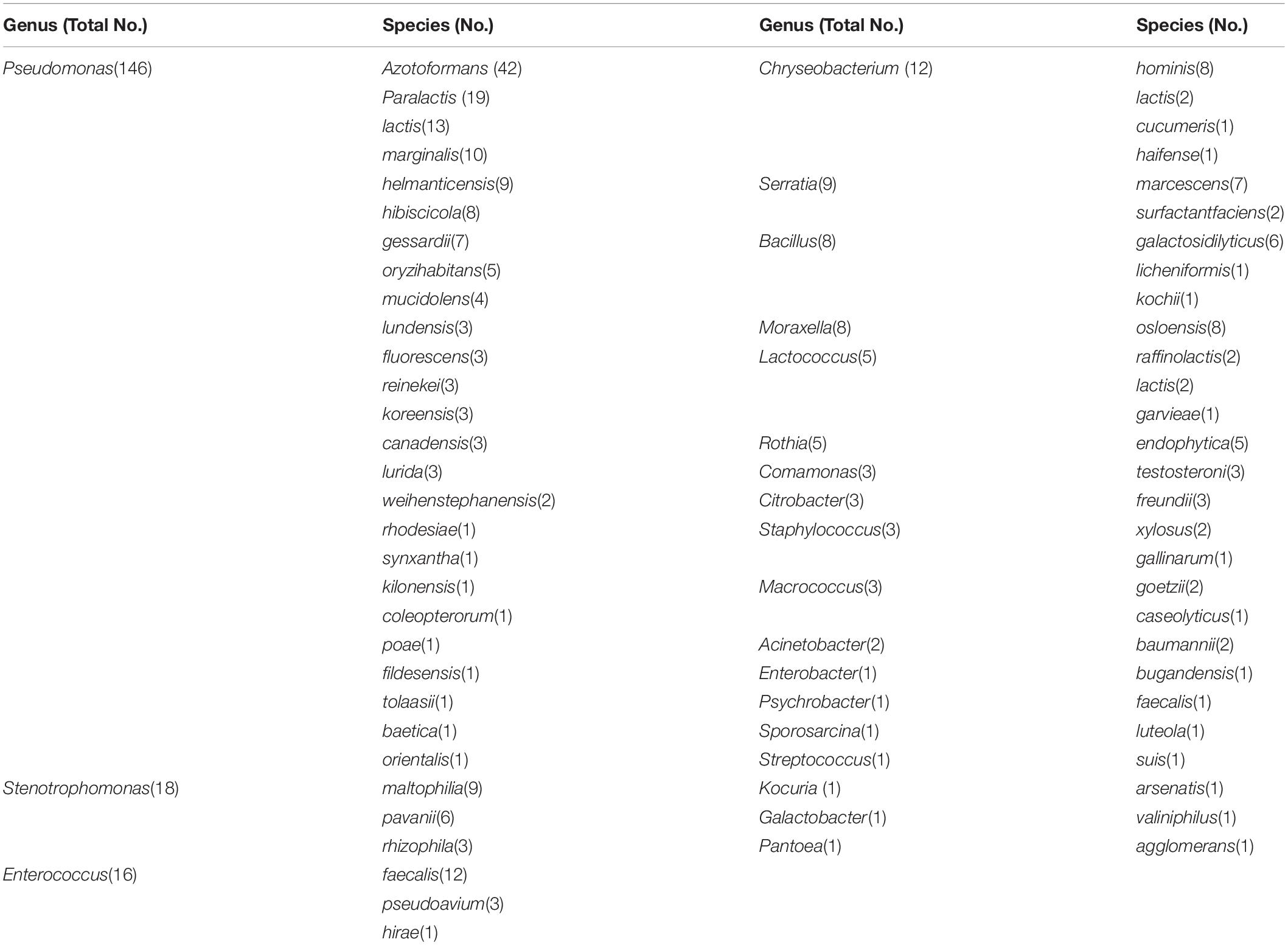
Table 2. Identification and distribution of psychrotrophic bacteria isolated from raw milk samples collected from different regions in China.
Single-molecule real-time sequencing was used to determine the identities of microorganisms in the raw milk samples. The richness, uniformity and coverage of the sample sequencing were sufficient to predict the diversity within each sample. Interestingly, the samples from Heilongjiang possessed significantly lower Chao 1 species richness scores than samples from the other regions. However, the Simpson and Pielou evenness indices were not significantly different (Figure 1). These data indicated that the microbial diversity between the sampling locations was similar.

Figure 1. Alpha analysis metrics for microbiota present in milk samples collected in eight locations for this study. A. Heilongjiang, B. Inner Mongolia, C. Gansu, D. Henan, E. Anhui, F. Jiangsu, G. Chongqing, and H. Hunan. *Significant difference.
Milk samples were also used to culture psychrotrophic bacteria that were then identified using 16S rDNA sequencing from pure cultures. We then combined this data with the single-molecule sequencing data to analyze the diversity of psychrotrophic bacteria in raw milk. The most abundant genera in our samples were Enterococcus, Kaistella, Commonas, Pseudomonas, Sporosarcina, Epilithonimonas, Rothia, Stenotrophomonas, Cytobacillus, Chryseobacterium, Macrococcus, Streptococcus, Lactococcus, Enterobacter, Bacillus, Serratia, Acinetobacter, Moraxella, Psychrobacter, and Staphylococcus. In particular, Sporosarcina was the most abundant psychrophilic genus (23.8%) in raw milk in Heilongjiang. Comamonas was the most abundant (18.8%) in samples from Hunan and Pseudomonas (20.6%) from Anhui. Overall, Pseudomonas accounted for 9.0, 3.1, 7.4, 11.4, 20.6, 11.9, 6.3, and 5.7% from Heilongjiang, Inner Mongolia, Gansu, Henan, Jiangsu, Hunan, and Chongqing, respectively (Figure 2A).

Figure 2. Relative OTU abundance at the (A) genus and (B) species levels from raw milk samples used for this study. A. Heilongjiang, B. Inner Mongolia, C. Gansu, D. Henan, E. Anhui, F. Jiangsu, G. Chongqing, and H. Hunan.
The most abundant species were Chryseobacterium haifense, Comamonas testosteroni, Sporosarcina luteola, Enterococcus faecalis, Epilithonimonas hominis, Rothia endophytica, Cytobacillus kochii, Chryseobacterium cucumeris, Enterococcus pseudoavium, Macrococcus goetzii, Streptococcus suis, Stenotrophomonas maltophilia, Stenotrophomonas rhizophila, Pseudomonas lundensis, Enterobacter bugandensis, Bacillus licheniformis, Serratia marcescens, Acinetobacter baumannii, Pseudomonas azotoformans, and Moraxella osloensis (Figure 2B). S. luteola was the most abundant in Heilongjiang (23.8%) and C. testosteroni in Henan (17.2%) and P. lundii in Anhui (14.5%) raw milk samples (Figure 2B).
We analyzed the correlations between community structures of the psychrophilic bacteria in raw milk samples from different regions using PCoA. The Heilongjiang and Inner Mongolia raw milk samples clustered indicating similar communities. Interestingly, these two groups were clearly separated from the remaining sample regions indicating significant differences in community structures (Figure 3).
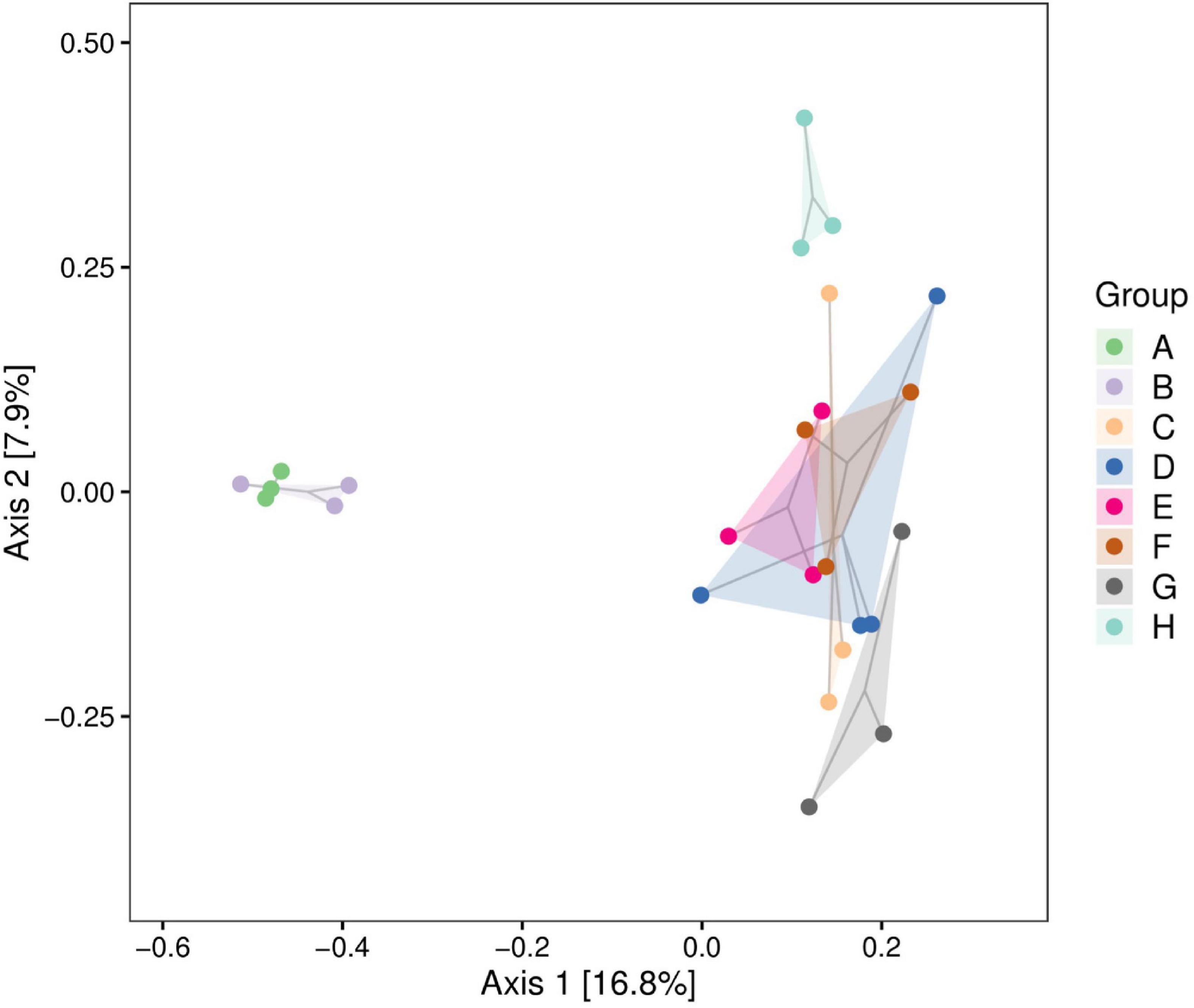
Figure 3. Principal coordinates analysis (PCoA) plot of raw milk samples used in this study. A. Heilongjiang, B. Inner Mongolia, C. Gansu, D. Henan, E. Anhui, F. Jiangsu, G. Chongqing, and H. Hunan.
We used clustering heat map analysis to further explore the differences in the community structure of psychrotrophic bacteria using the top 50 relative abundance scores. This analysis utilized the Euclidean distance of the microbial composition data and species were clustered according to Pearson correlation coefficient data. We found that Heilongjiang and Inner Mongolia samples were clustered together and separated from the other groups due to the presence of S. luteola, C. kochii, and E. faecalis. The clustering of Henan, Anhui, and Jiangsu raw milk samples were due to the presence of P. lurida, P. gessardii, L. galactosidilyticus, G. valiniphilus, and C. freundii (Figure 4).
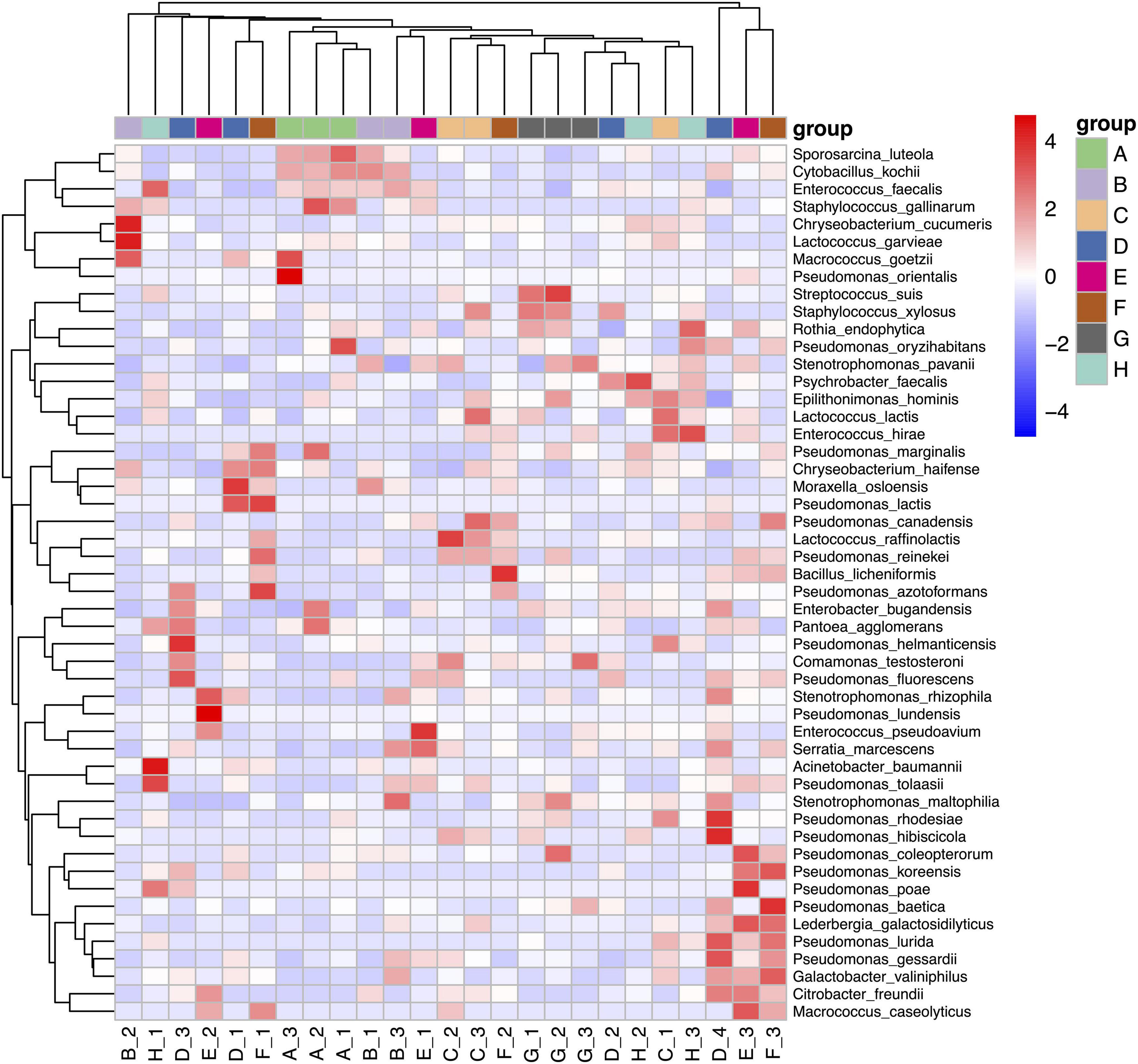
Figure 4. Heatmap analysis of raw milk samples used in this study. A. Heilongjiang, B. Inner Mongolia, C. Gansu, D. Henan, E. Anhui, F. Jiangsu, G. Chongqing, and H. Hunan.
We used Venny and linear discriminant analyses to categorize the different raw milk bacterial species that we identified from our eight study areas. We found that E. faecalis and R. endophytica were common species identified in samples from all regions. In addition, the relative abundance of Bacillus in Jiangsu samples was significantly greater than for the other regions while Streptococcus and Comamonas in Hunan samples were significantly greater than other regions (Figures 5, 6).

Figure 5. Venny analysis of raw milk samples used in this study. A. Heilongjiang, B. Inner Mongolia, C. Gansu, D. Henan, E. Anhui, F. Jiangsu, G. Chongqing, and H. Hunan.
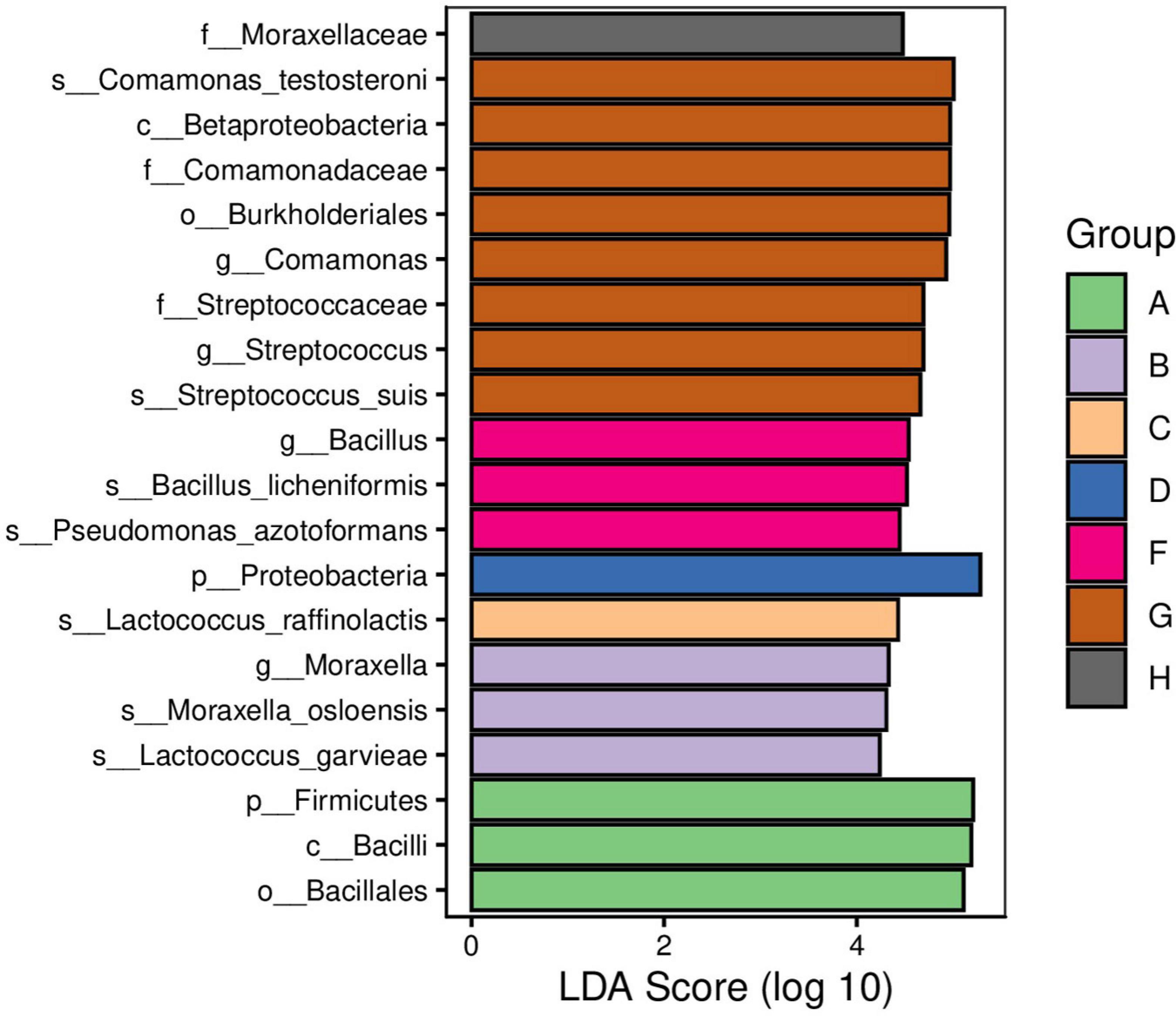
Figure 6. Linear discriminant analysis (LDA) of raw milk samples used in this study. A. Heilongjiang, B. Inner Mongolia, C. Gansu, D. Henan, E. Anhui, F. Jiangsu, G. Chongqing, and H. Hunan.
We isolated 248 psychrophilic bacterial colonies using traditional culture methods and found 21 genera and 59 species that were identified by 16 rDNA gene sequencing. At the genus level, Pseudomonas, Stenotrophomonas, Enterococcus, Chryseobacterium, and Serratia were the present in the highest proportions and Pseudomonas accounted for 58.9% of the total. At the species level, P. azotoformans, P. paralactis, P. lactis, E. faecalis, and P. marginalis were the present in the highest proportions and P. azotoformans accounted for to 16.9% of the total species numbers in our samples. These results were consistent with a previous study where Pseudomonas, Lactococcus, and Acinetobacter accounted for 62% of the total (Von Neubeck et al., 2015). Another study found that the Pseudomonas was the most prevalent genus (74.79%) in raw milk and was represented by 25 species, the most common of these species were P. fragi (10.92%), P. lundensis (6.72%), and P. fluorescens (6.72%) (Zhang et al., 2020). Additionally, a study utilizing 595 strains of psychrotrophic bacteria from raw milk samples indicated 90% of the samples contained Pseudomonas at proportions as high as 25.7% (Yang et al., 2020).
The traditional culture of microorganisms has numerous advantageous over other methods and is an indispensable technical means for the in-depth exploration of microbial resources in raw milk. The composition of psychrotrophic bacterial microbiota was investigated by submitting the psychrotrophic bacteria database to the raw milk microbiota database. We used single-molecule real-time sequencing to also identify un-culturable microorganisms to understand the total diversity of microorganisms in raw milk. These combined data indicated that Enterococcus, Kaistella, Comamonas, Pseudomonas, Sporosarcina, Epilithonimonas, Rothia, Stenotrophomonas, Cytobacillus, Chryseobacterium, Macrococcus, Streptococcus, Lactococcus, Enterobacter, Bacillus, Serratia, Acinetobacter, Moraxella, Psychrobacter, and Staphylococcus were the 20 most abundant genus in all samples. This was consistent with the results of previous studies that found Pseudomonas, Acinetobacter, Lactococcus, Corynebacterium, and Streptococcus as the most abundant genus in raw milk (McHugh et al., 2020). An additional study indicated that Lactococcus, Bacillus, Streptococcus, Sporosarcina, Pseudomonas, and Acinetobacter were the primary psychrophilic bacteria at the genus level in raw milk (Hou et al., 2015). Raw milk samples analyzed in another study indicated and Pseudomonas, Lactococcus, and Acinetobacter were the most common genera (Li et al., 2018). In our study we found that Enterococcus posed a high risk for contamination and a previous study of 1,584 batches of raw milk samples from four dairy factories found that Enterococcus was presence at levels ranging from 40.6 to 79.7% (Yoon and Lee, 2021). This agreed with another study where the relative abundance of Enterococcus in raw yak milk reached levels as high as 36% (Qazalbash et al., 2021).
At the species level we found that the most abundant organisms were C. haifense, C. testosterone, S. luteola, E. faecalis, E. hominis, R. endophytica, C. kochii, C. cucumeris, E. pseudoavium, M. goetzii, S. suis, S. maltophilia, S. rhizophila, P. lundensis, E, bugandensis, B. licheniformis, S. marcescens, A. baumannii, P. azotoformans, and M. osloensis. A previous study indicated that E. faecalis, S. rhizophila, P. lundensis, and M. osloensi were the primary psychrophilic bacteria at the species level in raw milk (Yang et al., 2020). The most common source for A. baumannii is environmental pollution and this bacterium can cause pneumonia, meningitis, and wound infections (Engür et al., 2014). We found that the relative abundance of A. baumannii in Chongqing was relatively high and this was consistent with previous studies implicating raw milk and commercial infant formula milk powder as sources (Tamang et al., 2014). Streptococcus originates primarily in the farm environment and is a common cause of dairy cow mastitis and most animals have no obvious clinical symptoms at the time of infection (Moroni et al., 2006). S. suis has been also identified in raw milk in cows with mastitis (Ma et al., 2021). We found S. suis in Hunan samples and this may be related to farm management practices.
China is a vast country with many farms, we also analyzed the influence of geographic regions factors on the community structure of psychrotrophic bacteria in raw milk, and the results showed that geographic regions factors significantly influenced the community structure of psychrotrophic bacteria in raw milk, with Heilongjiang and Inner Mongolia clustered together in both principal coordinates analysis and cluster heat map analysis due to their location in the north of China, while Henan, Anhui, and Jiangsu clustered together due to their proximity. This is consistent with previous findings that geographic factors were significant factors influencing the milk microbiota (Guo et al., 2021), while studies on psychrotrophic bacteria had also shown that geographic factors were determining factors attributing to significant influence in the raw milk psychrotrophic bacteria (Yap et al., 2021). In addition, differences in the community structure of psychrotrophic bacteria in raw milk between different geographic regions may be influenced by storage temperature (Vithanage et al., 2016; Hahne et al., 2019; Quinto et al., 2020). The low-temperature storage resulted in an increasing relative abundance of psychrotrophic bacteria (Yang et al., 2020), the psychotropic bacterial community structure diversity in the summer was higher than that in the winter (Vithanage et al., 2016). Cold storage tests using raw water buffalo milk indicated that the microbial community structure changed with storage time. The first 24 h were dominated by Lactococcus and Streptococcus while at 72 h Pseudomonas and Acinetobacter dominated and latter achieved levels more than 60% (Li et al., 2016). A comparison of the effects of different cold storage conditions on the microorganism content of raw milk indicated that Pseudomonas isolates also dominated all temperatures and were more than 80% at 2 and 4°C (Vithanage et al., 2017), however, only 8–10% at 10 and 30°C (Hahne et al., 2019), however, another study found that all Pseudomonas can grow at 2–25°C (Meng et al., 2019), 67% Pseudomonas can grow at 40°C (Caldera et al., 2016). Differences were observed between days in Henan, Anhui, and Jiangsu samples that may be related to farm management or disinfection procedures. Disinfection of farm milking equipment and acid or alkaline disinfection are usually performed every other day.
This is the first study to analyze the diversity of psychrophilic bacteria in raw milk in different regions of China by a combination of traditional culture and PacBio single-molecule real-time sequencing. The community structure of psychrotrophic bacteria in raw milk was very different across regions. In this study, we found that the community structure and diversity of psychrotrophic bacteria in raw milk in Heilongjiang and Inner Mongolia were similar although they differed significantly from the other study regions. Additionally, differences were observed between days in Henan, Anhui, and Jiangsu samples. These procedures assisted in a more accurate understanding of the community structure of psychrophilic bacteria in raw milk from different regions and can be used to develop timely individual control strategies on a case or a reginal basis in order to ensure milk safety.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://bigd.big.ac.cn, CRA005869.
BD: conceptualization, methodology, investigation, and writing – original draft. LM: methodology. HL: data curation. NZ: conceptualization and supervision. YZ and SZ: writing – review, supervision, and funding acquisition. JW: conceptualization and funding acquisition. All authors contributed to the article and approved the submitted version.
This study was funded by the Scientific Research Project for Major Achievements of Agricultural Science and Technology Innovation Program (CAAS-ZDXT2019004), the Agricultural Science and Technology Innovation Program (ASTIP-IAS12), and the Modern Agro-Industry Technology Research System of the PR China (CARS-36).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ahmed, M. M. M., Hafez, E. E., Mona, A. M., Abdelrrassoul, H. A., and Mabrouk, Y. M. (2014). Detection of baby milk powder contamination by microorganisms. World Appl. Sci. J. 30, 93–98. doi: 10.5829/idosi.wasj.2014.30.01.81255
America Department of Health and Human Services (2019). Grade “A” Pasteurized Milk Ordinance. Washington, DC: America Department of Health and Human Services.
Bu, Y., Qiao, W., Zhai, Z., Liu, T., Gong, P., Zhang, L., et al. (2022). Establishment and evaluation of a loop-mediated isothermal amplification assay for rapid detection of Pseudomonas fluorescens in raw milk. Front. Microbiol. 12:810511. doi: 10.3389/fmicb.2021.810511
Caldera, L., Franzetti, L., Van Coillie, E., De Vos, P., Stragier, P., De Block, J., et al. (2016). Identification, enzymatic spoilage characterization and proteolytic activity quantification of Pseudomonas spp. isolated from different foods. Food Microbiol. 54, 142–153. doi: 10.1016/j.fm.2015.10.004
Dixon, P. (2003). VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Du, B., Meng, L., Liu, H., Zheng, N., Zhang, Y., Guo, X., et al. (2020). Impacts of milking and housing environment on milk microbiota. Animals 10, 1–12. doi: 10.3390/ani10122339
Du, B., Wen, F., Zhang, Y., Zheng, N., Li, S., Li, F., et al. (2019). Presence of tetracyclines, quinolones, lincomycin and streptomycin in milk. Food Control 100, 171–175. doi: 10.1016/j.foodcont.2019.01.005
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Engür, D., Çakmak, B. Ç, Kaynak Türkmen, M., Telli, M., Eyigör, M., and Güzünler, M. (2014). A milk pump as a source for spreading Acinetobacter baumannii in a neonatal intensive care unit. Breastfeed. Med. 9, 551–554. doi: 10.1089/bfm.2014.0054
Fusco, V., and Quero, G. M. (2014). Culture-dependent and culture-independent nucleic-acid-based methods used in the microbial safety assessment of milk and dairy products. Compr. Rev. Food Sci. Food Safety 13, 493–537. doi: 10.1111/1541-4337.12074
Guo, X., Yu, Z., Zhao, F., Sun, Z., Kwok, L.-Y., and Li, S. (2021). Both sampling seasonality and geographic origin contribute significantly to variations in raw milk microbiota, but sampling seasonality is the more determining factor. J. Dairy Sci. 104, 10609–10627. doi: 10.3168/jds.2021-20480
Hahne, J., Isele, D., Berning, J., and Lipski, A. (2019). The contribution of fast growing, psychrotrophic microorganisms on biodiversity of refrigerated raw cow’s milk with high bacterial counts and their food spoilage potential. Food Microbiol. 79, 11–19. doi: 10.1016/j.fm.2018.10.019
Hou, Q., Xu, H., Zheng, Y., Xi, X., Kwok, L., Sun, Z., et al. (2015). Evaluation of bacterial contamination in raw milk, ultra-high temperature milk and infant formula using single molecule, real-time sequencing technology. J. Dairy Sci. 98, 8464–8472. doi: 10.3168/jds.2015-9886
International Dairy Federation (2005). IDF 101:2005, Milk-Enumeration of Colony-Forming Units of Psychrotrophic Microorganisms-Colony-Count Technique at 6.5°C. Brussels: International Dairy Federation.
Li, L. Jr., Renye, J. A., Feng, L., Zeng, Q., Tang, Y., Huang, L., et al. (2016). Characterization of the indigenous microflora in raw and pasteurized buffalo milk during storage at refrigeration temperature by high throughput sequencing. J. Dairy Sci. 99, 7016–7024. doi: 10.3168/jds.2016-11041
Li, N., Wang, Y., You, C., Ren, J., Chen, W., Zheng, H., et al. (2018). Variation in raw milk microbiota throughout 12 months and the impact of weather conditions. Sci. Rep. 8, 1–10. doi: 10.1038/s41598-018-20862-8
Ma, T., Shen, L., Wen, Q., Lv, R., Hou, Q., Kwok, L. Y., et al. (2021). PacBio sequencing revealed variation in the microbiota diversity, species richness and composition between milk collected from healthy and mastitis cows. Microbiology 167, 1–10. doi: 10.1099/mic.0.000968
McHugh, A. J., Feehily, C., Fenelon, M. A., Gleeson, D., Hill, C., and Cotter, P. D. (2020). Tracking the dairy microbiota from farm bulk tank to skimmed milk powder. MSystems 5, 1–16. doi: 10.1128/mSystems.00226-20
Meng, L., Liu, H., Dong, L., Zheng, N., Xing, M., Zhang, Y., et al. (2018). Identification and proteolytic activity quantification of Pseudomonas spp. isolated from different raw milks at storage temperatures. J. Dairy Sci. 101, 2897–2905. doi: 10.3168/jds.2017-13617
Meng, L., Zhang, Y., Liu, H., Zhao, S., Wang, J., and Zheng, N. (2019). Characterization of Pseudomonas spp. and associated proteolytic properties in raw milk stored at low temperatures. Front. Microbiol. 8:2158. doi: 10.3389/fmicb.2017.02158
Moroni, P., Sgoifo Rossi, C., Pisoni, G., Bronzo, V., Castiglioni, B., and Boettcher, P. J. (2006). Relationships between somatic cell count and intramammary infection in buffaloes. J. Dairy Sci. 89, 998–1003. doi: 10.3168/jds.S0022-0302(06)72165-8
Qazalbash, M., Masud, T., Ahmad, A., Hayat, R., Ibrahim, M., Mujtaba, A., et al. (2021). Diversity of lactic acid bacteria associated with raw yak (Bos grunniens) milk produced in Pakistan. J. Anim. Feed Sci. 30, 42–51. doi: 10.22358/jafs/133201/2021
Quinto, E. J., Marín, J. M., Caro, I., Mateo, J., and Schaffner, D. W. (2020). Modelling growth and decline in a two-species model system: pathogenic Escherichia coli O157:H7 and psychrotrophic spoilage bacteria in milk. Foods 9, 1–15. doi: 10.3390/foods9030331
Scarpellini, M., Franzetti, L., and Galli, A. (2004). Development of PCR assay to identify Pseudomonas fluorescens and its biotype. FEMS Microbiol. Lett. 236, 257–260. doi: 10.1016/j.femsle.2004.05.043
Tamang, M. D., Gurung, M., Nam, H. M., Kim, S. R., Jang, G. C., Jung, S. C., et al. (2014). Short communication: genetic characterization of antimicrobial resistance in Acinetobacter isolates recovered from bulk tank milk. J. Dairy Sci. 97, 704–709. doi: 10.3168/jds.2013-7403
Vithanage, N. R., Dissanayake, M., Bolge, G., Palombo, E. A., Yeager, T. R., and Datta, N. (2016). Biodiversity of culturable psychrotrophic microbiota in raw milk attributable to refrigeration conditions, seasonality and their spoilage potential. Int. Dairy J. 57, 80–90. doi: 10.1016/j.idairyj.2016.02.042
Vithanage, N. R., Dissanayake, M., Bolge, G., Palombo, E. A., Yeager, T. R., and Datta, N. (2017). Microbiological quality of raw milk attributable to prolonged refrigeration conditions. J. Dairy Res. 84, 92–101. doi: 10.1017/s0022029916000728
Von Neubeck, M., Baur, C., Krewinkel, M., Stoeckel, M., Kranz, B., Stressler, T., et al. (2015). Biodiversity of refrigerated raw milk microbiota and their enzymatic spoilage potential. Int. J. Food Microbiol. 211, 57–65. doi: 10.1016/j.ijfoodmicro.2015.07.001
Wang, J., Zheng, Y., Xi, X., Hou, Q., Xu, H., Zhao, J., et al. (2018). Application of PacBio Single Molecule Real-Time (SMRT) sequencing in bacterial source tracking analysis during milk powder production. Food Control 93, 226–234. doi: 10.1016/j.foodcont.2018.05.044
Xin, L., Meng, Z., Zhang, L., Cui, Y., Han, X., and Yi, H. (2017). The diversity and proteolytic properties of psychrotrophic bacteria in raw cows’ milk from North China. Int. Dairy J. 66, 34–41. doi: 10.1016/j.idairyj.2016.10.014
Yang, X., Guo, X., Liu, W., Tian, Y., Gao, P., Ren, Y., et al. (2020). The complex community structures and seasonal variations of psychrotrophic bacteria in raw milk in Heilongjiang Province, China. LWT Food Sci. Technol. 134, 1–12. doi: 10.1016/j.lwt.2020.110218
Yap, M., Gleeson, D., O’Toole, P. W., O’Sullivan, O., Cotter, P. D., and Björkroth, J. (2021). Seasonality and geography have a greater influence than the use of chlorine-based cleaning agents on the microbiota of bulk tank raw milk. Appl. Environ. Microbiol. 87, 1–11. doi: 10.1128/AEM.01081-21
Yoon, S., and Lee, Y. J. (2021). Molecular characteristics of enterococcus faecalis and enterococcus faecium from bulk tank milk in Korea. Animals 11, 1–9. doi: 10.3390/ani11030661
Yuan, L., Sadiq, F., Burmolle, M., Wang, N., and He, G. (2019). Insights into psychrotrophic bacteria in raw milk: a review. J. Food Prot. 82, 1148–1159. doi: 10.4315/0362-028x.Jfp-19-032
Keywords: milk, PacBio single molecule real-time sequencing, psychrophilic bacteria, community structures, regional
Citation: Du B, Meng L, Liu H, Zheng N, Zhang Y, Zhao S and Wang J (2022) Single Molecule Real-Time Sequencing and Traditional Cultivation Techniques Reveal Complex Community Structures and Regional Variations of Psychrotrophic Bacteria in Raw Milk. Front. Microbiol. 13:853263. doi: 10.3389/fmicb.2022.853263
Received: 12 January 2022; Accepted: 20 January 2022;
Published: 10 February 2022.
Edited by:
Qingli Dong, University of Shanghai for Science and Technology, ChinaReviewed by:
Steve Flint, Massey University, New ZealandCopyright © 2022 Du, Meng, Liu, Zheng, Zhang, Zhao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiaqi Wang, amlhcWl3YW5nQHZpcC4xNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.