
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 19 May 2022
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.850807
Malic acid is a component of the rhizosphere exudate and is vital for crop growth. However, little information is available about the effects of external applications of malic acid on the nutrient absorption and quality of grape fruit, and few studies have been performed on the relationship between the changes in the rhizosphere microbial community and nutrient absorption and fruit quality of grapes after adding malic acid. Here, the LM (low concentration of malic acid) and HM (high concentration of malic acid) treatments comprised 5% and 10% malic acid (the ratio of acid to the total weight of the fertilizer) combined with NPK fertilizer, respectively. Applying malic acid changed the grape rhizosphere microbial community structure and community-level physiological profile (CLPP) significantly, and HM had a positive effect on the utilization of substrates. The microbial community structure in the rhizosphere of the grapes with added malic acid was closely related to the CLPP. The N and P content in the leaves and fruits increased after applying malic acid compared to the control, while K content in the fruits increased significantly. In addition, malic acid significantly reduced the weight per fruit, significantly increased soluble sugar content (SSC) and vitamin C content of the fruit, and significantly improved the fruit sugar-acid ratio and grape tasting score. Moreover, the principal component analysis and grape nutrient and fruit quality scores showed that grape nutrients and fruit quality were significantly affected by malic acid and ranked as 5% malic acid > 10% malic acid > control. Pearson’s correlation heatmap of microbial composition, nutrient absorption and fruit quality of the grapes showed that the grape microbial community was closely related to grape nutrients and fruit quality. Adding malic acid was positively correlated to Planococcaceae, Bacillaceae, Woeseiaceae and Rhodobacteraceae. Furthermore, Planococcaceae, Bacillaceae, Woeseiaceae and Rhodobacteraceae were closely related to grape nutrient absorption and fruit quality. Bacillaceae and Woeseiaceae were positively correlated with total soluble sugar, while Planococcaceae and Rhodobacteraceae were positively correlated with titratable acid. Hence, Bacillaceae and Woeseiaceae were the key bacteria that played a major role in grape fruit quality and nutrient absorption after applying malic acid water-soluble fertilizer.
Malic acid (2-hydroxybutanedioic acid) is a four-carbon dicarboxylic acid (Dai et al., 2018) used as an enzyme substrate (Bassham and Calvin, 1980; Buser-Suter et al., 1982; Casati et al., 1999; Edwards et al., 2001) and a carrier of carbon-reducing power to transfer carbon and reducing power between the cytoplasm and organelles (Drincovich et al., 2001; Scheibe, 2004; Chen et al., 2019). Malic acid often links various metabolic pathways in different organelles and participates in the regulation of various metabolic reactions in plant cells (Outlaw and Oliver, 1977; Song et al., 2009). Malic acid in leaves is involved in the regulation of stomatal opening and closing, which provides a large number of counter ions to open the stomata and take up K+. When the stomata are open, the concentration of malic acid in the guard cells is six times higher than that when the stomata are closed, while the K+ concentration increases two to four times (Yao et al., 2020). In particular, malic acid has a biological “phytohormone” effect, which promotes the growth and cold resistance of seedlings (Lou et al., 1993; Lasa et al., 2002; Guo et al., 2017).
Malic acid is a low molecular weight organic acid (LMWOA) that is closely related to soil nutrient content and is a link between carbon and nitrogen metabolism. The difference in the ratio of added nitrate-nitrogen to ammonium-nitrogen causes changes in the type and content of organic acids in the rhizosphere (Lasa et al., 2002; Dong et al., 2004). It is an important energy source for bacterial respiration located in the nodules of nitrogen-fixing bacteria (Driscoll and Finan, 2010). This acid provides most of the carbon skeleton for nitrogen fixation (Rosendahl et al., 1990; Galvez, 2000; Schulze et al., 2002; Vance and Heichel, 2003) and participates in the binding of the oxygen diffusion barrier through an osmotic electrical mechanism (Denison, 1998). Malic acid exchanges and chelates with Fe and Al ligands, thereby reducing the adsorption of P in the soil (Wang et al., 2016), resulting in a larger pool of P in the soil solution that is available for plant uptake (Bolan et al., 1994). Malic acid is secreted by potassium-dissolving bacteria to dissolve potassium from aluminum potassium silicate (Prajapati and Modi, 2012) and drives the surface chemical reactions of acid hydrolysis and complex dissolution and promotes the release of mineral potassium and soil potassium, which increases the effective potassium content in the soil (Wang and Wang, 2009).
As a plant rhizosphere exudate, malic acid has a screening effect on the plant rhizosphere microbial community (Jones, 1998; Rudrappa et al., 2008; Berendsen et al., 2012; Chen et al., 2012; Lakshmanan et al., 2012; Beauregard et al., 2013; Tan et al., 2013; Yuan et al., 2018). When Arabidopsis was infected with Pseudomonas syringae, the secretion of malic acid into the rhizosphere increased, contributing to the proliferation of Bacillus subtilis FB17 in the rhizosphere. Hence, the formation of a B. subtilis biofilm is closely related to the presence of malic acid (Berendsen et al., 2012; Chen et al., 2012). Bacillus amyloliquefaciens T-5 is significantly induced by malic acid in a chemotactic reaction and cluster movement but has no significant effect on the formation of the biofilm (Tan et al., 2013). Moreover, malic acid, citric acid, and oxalic acid are common rhizosphere exudates of watermelon that induce the biocontrol bacterium Paenibacillus polymyxa SQR-21 to drive toward the root. Malic acid has a strong driving ability (Ling et al., 2011).
Therefore, malic acid has the potential to act as a synergist of NPK water-soluble fertilizer. Research on exogenous malic acid has mainly focused on its mitigation effect in response to heavy metal stress (Ebrahimian and Bybordi, 2014; Chen et al., 2020; Yao et al., 2020), and several studies have investigated the preservation of fresh-cut Lilium cv. Brunello as well as growth and flowering in Gazania and the uptake of K by tobacco (Darandeh and Hadavi, 2012; Talebi et al., 2014; Han et al., 2016). A previous study showed that malic acid combined with NPK fertilizer significantly improved pear fruit quality and nutrient uptake (Shao et al., 2022). However, the role of the microbial community in this process is not clear.
In this study, Shine Muscat grapes were used as experimental materials, conventional NPK fertilizer was used as the control, and 5% and 10% malic acid combined with NPK fertilizer were applied as treatments. We explored the relationship between the grape rhizosphere microbial community, nutrient uptake and fruit quality after adding malic acid to evaluate how malic acid-driven microbial communities affect grape nutrients and fruit quality and the prospect of applying malic acid as a synergist for fruit quality improvement.
The experiment was carried out in Kangcun, Xinxiang City, Henan Province, China (35°9′28″N, 113°42′17″E) from April to September 2019. The physical and chemical properties of the 0–20 cm soil layers were measured according to Lu (2000) and were as follows: organic matter 0.62%, nitrate-nitrogen 93.25 mg/kg, ammonium-nitrogen 59.20 mg/kg, available P 102.92 mg/kg, available K 213.4 mg/kg, pH 6.7 and electrical conductivity 152.47 us/cm.
Shine Muscat grapes (Vitis labrusca × V. vinifera) were planted in 2014 and arranged for the trials, and the spacing between the rows was 2.5 m × 1.5 m. Malic acid and NPK fertilizer were formulated into water-soluble fertilizer solutions in the proportions shown in Table 1. The fertilizers were applied at the flowering, young fruit, fruit expansion and 20-days-before-harvest stages during the grape growing period. Each treatment had three repeated plots, and each plot had nine grape trees. Each plot was arranged randomly. The fruits matured, the grapes were sampled after they matured (16 bunches of grapes from each treatment) and were brought back to the laboratory on September 20, 2019, for testing of various indicators. The rhizosphere soil samples were collected 1 month after the last fertilization, and three grape trees in each plot were randomly collected and mixed into one sample. Then, three soil samples from each treatment were processed, and a portion of each sample was dried naturally, with the rest stored at −80°C for the determination of microbial indicators. KNO3, urea (NH4N2O) and KH2PO4 were supplied by Sinopharm Chemical Reagent Beijing Co., Ltd. (Beijing, China).
According to Si et al. (2018), a community-level physiological profile (CLPP) was constructed using the Biolog EcoPlate (Biolog Inc., Hayward, CA, United States). Three soil samples from each treatment were analyzed for the experiment. Briefly, 1 g of soil and 99 mL of 0.85% sterilized NaCl solution were added to an autoclaved triangular flask, and the flask was shaken at 120 rpm for 30 min and then stored at 4°C for 30 min. A total of 150 μL of the resulting suspension was placed in each well, and the mixture was incubated at 25°C for 192 h. Then, the plates were read every 24 h using a Biolog MicroStation TM reader at both 590 and 750 nm (Guanghua et al., 2008) (Biolog Inc.).
The CLPP was constructed using Biolog Ecoplate (Biolog Inc., Hayward, CA, United States). The 120 h data collected during the exponential phase were used to construct the CLPPs for the Shine Muscat grape rhizosphere soil. Principal component analysis (PCA) was used to assess differences relating to the different amounts of malic acid added for the CLPPs, after normalizing the absorbance associated with each substrate (Kolton et al., 2017). Six C source groups were calculated to assess catabolic activity with the different malic acid treatments (Jiang et al., 2013; Wu et al., 2013).
Nitrate (NO3-N) and ammonium (NH4-N) were measured according to Lu (2000) by extracting with 1.0 M KCl at a 1:10 soil-to-solution ratio, followed by measurements using an automated discrete analyser (CleverChem 380, DeChem-Tech Inc., Hamburg, Germany) (Si et al., 2018). According to the method described by Lu (2000), available K was extracted in 1 M ammonium acetate using atomic absorption spectrophotometry (AAS; ZEEnit 700P; German Jena Analytical Instrument Co., Ltd., Jena, Germany), and available P was extracted from the soil samples with 0.5 M NaHCO3 (pH 8.5) and measured spectrophotometrically (Tu-1901; Persee Inc., Beijing, China). The pH was measured using a pH meter (DPH-2; ATAGO, Tokyo, Japan) at a 1:2.5 (w/v) ratio of soil to distilled water. The electrical conductivity of the soil was measured using a conductometer (DEC-2; ATAGO) at a 1:5 (w/v) ratio of soil to distilled water (Lu, 2000). Total carbon (TC) and inorganic carbon (IC) in the soil were determined using a carbon and nitrogen analyser (Primacs100, Skalar, Breda, Netherlands). Soil organic matter (SOM) = 1.724 × (TC − IC).
Leaf photosynthesis was measured using a CIRAS-3 instrument (PP systems, Amesbury, MA, United States) at 1 week before harvest.
Soluble solid content (SSC) was measured with a handheld digital refractometer (PR-101, Atago, Tokyo, Japan). Vitamin C (Vc) content was measured using the 2,6-dichlorophenol indophenol method (Cao et al., 2007). Total soluble sugar (TSS) content was determined by the anthrone method (Wang, 2006). Titratable acid (TA) content was determined by the NaOH titration method (Lu, 2000). The solid acid ratio (SAR) was calculated as TSS/TA. The grape tasting score (TS) was evaluated using a 10-point sensory evaluation according to a previous method (Lin et al., 2020).
N, P, and K contents of the leaves and fruits were determined by digestion with H2SO4-H2O2 (Lu, 2000). An automatic discontinuous chemical analyser (Clever Chem 380) was used to determine the N and P contents in leaves and fruit, and the K content was determined using an AAS device (AAS ZEEnit 700P, Jena, Germany).
The DNA extracted from three independent soil samples served as a template to amplify the 16S rRNA gene and the internal transcribed spacer (ITS) region. The V3-V4 hypervariable region of the bacterial 16S rRNA gene (Wan et al., 2018) was amplified with the primer pairs 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). The fungal-specific primers (Jamil et al., 2020) ITS3F (5′-GATGAAGAACGYAGYRAA-3′) and ITS4R (5′-TCCTCCG CYYATTGATATGC-3′) were employed to amplify the fungal ITS region. Polymerase chain reaction (PCR) amplification of the 16S rRNA gene was performed as follows: initial denaturation at 95°C for 3 min, followed by 27 cycles of denaturing at 95°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 45 s, a single extension at 72°C for 10 min and ending at 4°C. The PCR mixtures contained 5× TransStart FastPfu buffer 4 μL, 2.5 mM dNTPs 2 μL, forward primer (5 μM) 0.8 μL, reverse primer (5 μM) 0.8 μL, TransStart FastPfu DNA Polymerase 0.4 μL, template DNA 10 ng and ddH2O up to 20 μL. The PCR reactions were performed in triplicate. The PCR product was extracted after 2% agarose gel electrophoresis and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States) according to the manufacturer’s instructions and quantified using the Quantus™ Fluorometer (Promega, Madison, WI, United States).
Polymerase chain reaction amplification of the ITS region was performed using the KAPA HiFiHot Start ReadyMix PCR Kit in a GeneAmp PCR System 9700 instrument (Life Technologies, Carlsbad, CA, United States). The PCR reactions were conducted in 25 μL total volume reaction cocktails consisting of 12.5 μL of KAPA HiFi HotStart ReadyMix (2×), 0.25 μmol L-1 of each primer and 10 ng of the DNA template. Amplification was performed with the following thermal profile: 3 min of initial denaturation at 95°C followed by 27 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 30 s and a final extension at 72°C for 10 min. After purification, the PCR products were quantified using the 2100 Bioanalyses System (Agilent Technologies Inc., Santa Clara, CA, United States) (Mueller et al., 2000) and pooled at equal concentrations.
The purified amplicons were pooled in equimolar concentrations and paired-end sequenced on an Illumina MiSeq PE300 platform (Illumina, San Diego, CA, United States) according to the standard protocols of Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China). The raw reads were deposited into the NCBI Sequence Read Archive database (Accession Number: PRJNA786655).
The raw 16S rRNA gene and ITS region sequencing reads were demultiplexed and quality-filtered using fastp version 0.20.0 (Chen et al., 2018) and merged with FLASH version 1.2.7 (Magoč and Salzberg, 2011) using the following criteria: (i) the 300 bp reads were truncated at any site receiving an average quality score < 20 over a 50 bp sliding window, and truncated reads < 50 bp were discarded; reads containing ambiguous characters were also discarded; (ii) only overlapping sequences > 10 bp were assembled according to their overlapped sequence. The maximum mismatch ratio of the overlap region was 0.2. Reads that could not be assembled were discarded; (iii) samples were distinguished according to the barcode (Supplementary Table 1) and primers, and the sequence direction was adjusted, the exact barcode was matched and two nucleotide mismatches were used for primer matching.
Operational taxonomic units (OTUs) with a 97% similarity cut-off (Stackebrandt and Goebel, 1999; Edgar, 2013) were clustered using UPARSE version 7.1 (Edgar, 2013) and chimeric sequences were identified and removed. The taxonomy of each representative OTU sequence was analyzed using RDP Classifier version 2.2 (Wang et al., 2007) against 16S rRNA and the ITS database (e.g., Silva v138) with a confidence threshold of 0.7.
Experiments were performed using a completely randomized design. Statistical analysis was performed using SPSS Statistics 22 software (SPSS Inc., Chicago, IL, United States). All data are expressed as the mean ± standard error (SE). One-way analysis of variance and Duncan’s test were used to detect differences. A P-value < 0.05 was considered significant. The PCA was performed using Canoco 4.5 (Microcomputer Power, Ithaca, NY, United States). Non-metric multidimensional scaling (NMDS) was conducted and a Pearson’s correlation heatmap was produced using an R package.
The number of OTUs in the grape rhizosphere increased, while the OTUs of specific bacteria and fungi decreased after adding malic acid (Supplementary Figure 1). NMDS analysis and the percentage of microbial composition were used to evaluate the effect of applying malic acid on the microbial community structure of the grape rhizosphere (Figure 1 and Supplementary Figure 2). The NMDS of the microbial community revealed that each treatment formed its own cluster, and the control cluster was separated from the malic acid samples (LM and HM clusters). Additionally, the LM cluster was close to the HM cluster in the bacterial and fungal communities. These results demonstrate that the grape rhizosphere microbial community structure changed significantly after applying malic acid.
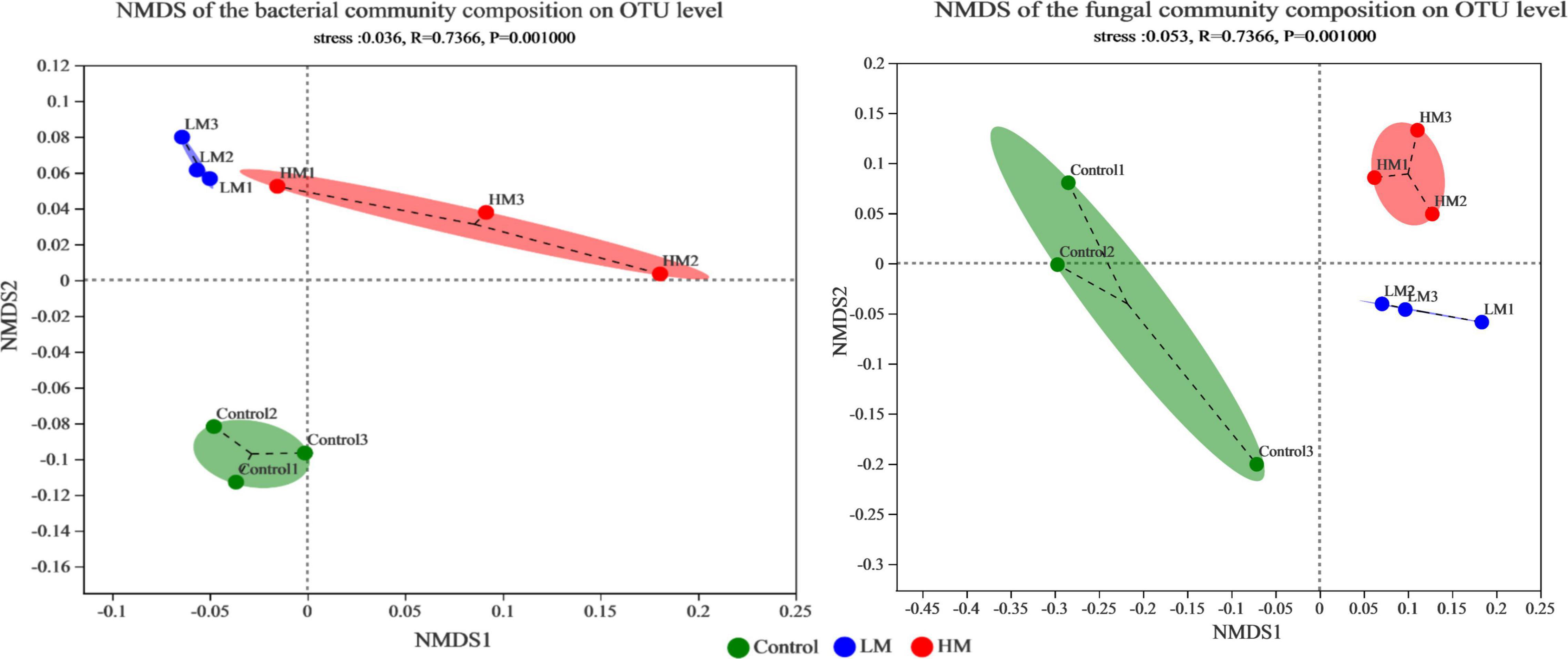
Figure 1. NMDS plots showing differences based on Bray-Curtis distance in the grape rhizosphere microbial community structure after applying different amounts of malic acid. Left: bacterial community. Right: fungal community.
The PCA of the soil microbial CLPP showed that the malic acid treatments affected the functional structure of the soil microbial community (Figure 2). Two PCs accounted for 85% of the total variation and each treatment formed its own cluster. The control cluster was close to the LM cluster, and distributed on the negative axis of PC1, whereas the HM cluster was farther away and distributed on the positive axis of PC1. However, this was significantly different from the NMDS analysis of the microbial OTUs by ANOSIM (p = 0.001).
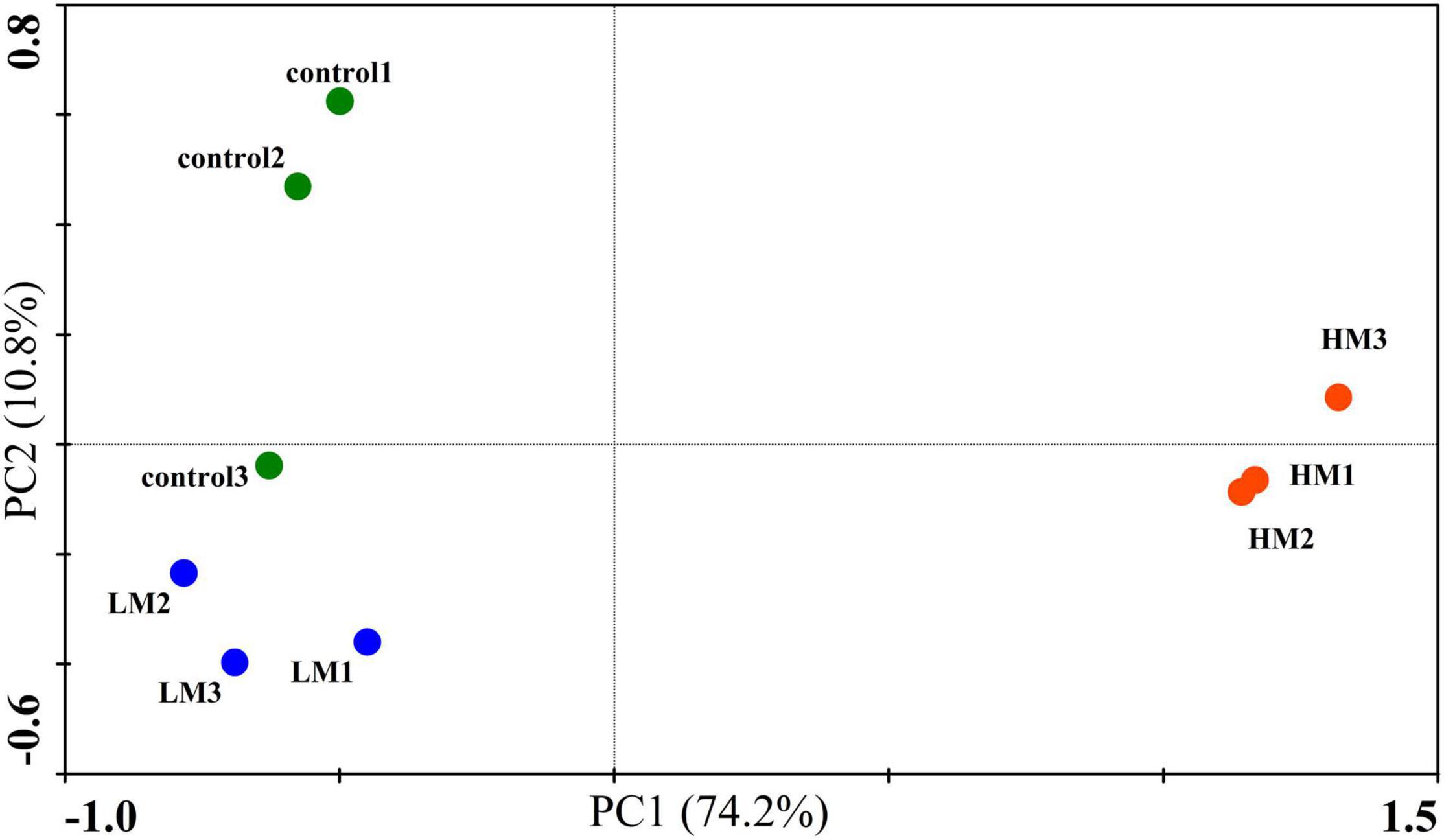
Figure 2. Effect of malic acid on carbon utilization by the grape rhizosphere microbial community. PCA plot of carbon substrate utilization patterns after the addition of malic acid.
The use of six substrate types (polymers, carbohydrates, phenolic compounds, carboxylic acids, amino acids and amines) with different malic acid applications is shown in Supplementary Figure 3. The results show that utilization of the six substrate types by the microbial community changed significantly under the HM treatment (p < 0.05). Utilization of the six substrate types by the grape rhizosphere microbial community increased with an increase in the amount of malic acid added. The utilization intensity of HM for the six major carbon sources was significantly higher than that of LM and the control, of which the utilization intensity for phenols was the most significant. Moreover, HM had a positive effect on substrate utilization.
Pearson’s correlation heatmap between bacterial composition and microbial carbon shows that Gemmatimonadetes and Firmicutes were positively correlated with utilization of the six carbon sources, among which Gemmatimonadetes, Firmicutes and amines were extremely significantly positively correlated, with correlation coefficients of 0.85 (p = 0.003) and 0.81 (p = 0.008), respectively (Supplementary Table 2). Patescibacteria and Sumerlaeota were significantly negatively correlated (p < 0.05) with the utilization of the six carbon sources. The correlations between Patescibacteria and polymers and amines were −0.7 (p = 0.035) and −0.67 (p = 0.048), while the correlations between Sumerlaeota and phenols, amines, and carbohydrates were −0.68 (p = 0.046), −0.7 (p = 0.037) and −0.69 (p = 0.039), respectively (Supplementary Table 2).
Pearson’s correlation heatmap between fungal composition and microbial carbon metabolism showed that Mortierellomycota was negatively correlated with microbial carbon metabolism, and significantly negatively correlated with polymers, carbohydrates, carboxylic acids and amino acids, with correlation coefficients of −0.85 (very significant, p = 0.004), −0.7 (p = 0.036), −0.71 (p = 0.034) and −0.72 (p = 0.029), respectively (Figure 3 and Supplementary Table 3). Monoblepharomycota was significantly positively correlated with polymers, carbohydrates and amino acids, with coefficients of 0.71 (p = 0.033), 0.67 (p = 0.048) and 0.76 (p = 0.018), respectively (Figure 3 and Supplementary Table 3). Blastocladiomycota was very significantly positively correlated with phenols (0.82, p = 0.007) and amines (0.84, p = 0.005), and significantly positively correlated with carbohydrates (0.77, p = 0.016), carboxylic acids (0.75, p = 0.02) and amino acids (0.67, p = 0.047) (Figure 3 and Supplementary Table 3).
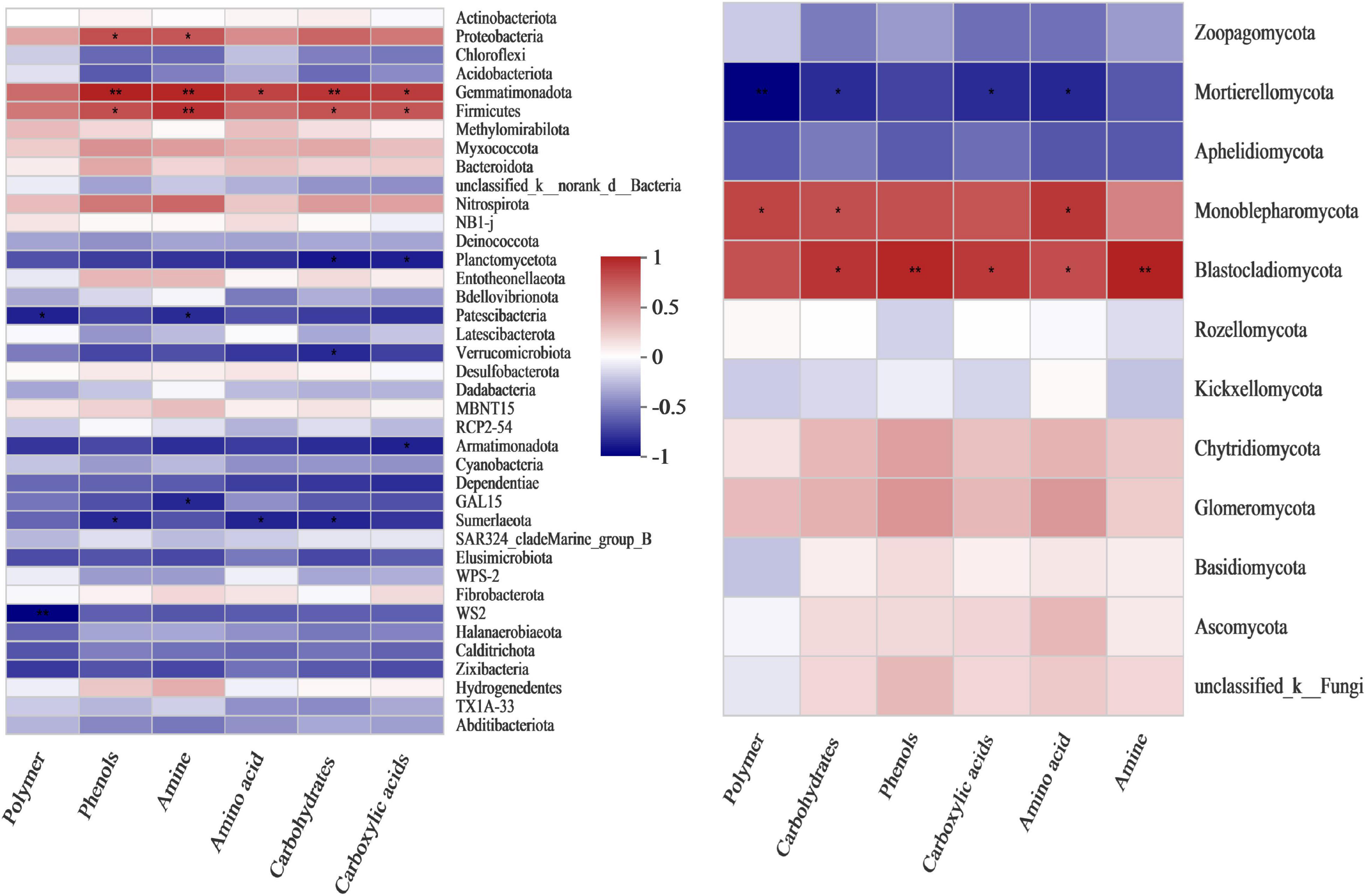
Figure 3. Pearson’s correlation heatmap showing the relationship between microbial composition and metabolism at the phylum level. Left: bacterial community. Right: fungal community. **Correlation is significant at the 0.01 level. *Correlation is significant at the 0.05 level.
As shown in Table 2, the rhizosphere soil NO3-N trended downward with increased application of malic acid, and that under the LM treatment was significantly lower than the control. In contrast, available P content trended upward, but no significant difference was observed between the treatments. NH4-N content increased first and then decreased, and that under the LM treatment was significantly higher than that under the HM treatment and in the control. No significant difference in available K content was observed among the treatments.
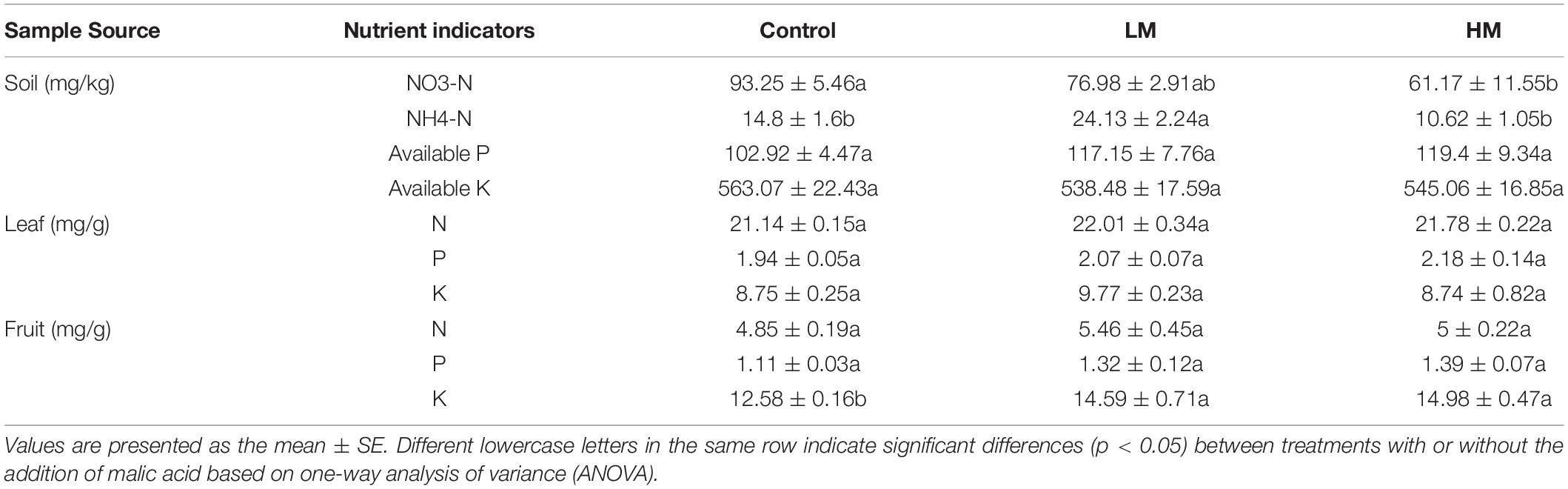
Table 2. Nutrient contents of the grape rhizosphere soil, leaves, and fruits with different amounts of added malic acid.
There were no significant differences in grape soil pH between the LM, HM, and control samples. The SOM content and EC value were significantly higher in the LM samples than in the control and HM samples (with SOM being 132.84% higher under LM vs. the control; Supplementary Table 4). The LM and HM treatments significantly increased the net photosynthetic rate (Pn) of leaves. The LM treatment significantly increased stomatal conductivity (Gs) and the water utilization rate (WUE) compared to the control (Supplementary Table 5).
The leaf nutrient analysis showed that although there was no significant difference between the treatments, the N and P contents of the LM and HM leaves (except the K content of HM leaves) tended to be higher than those of the control. In addition, NPK content in the fruit trended upward with increased addition of malic acid, and the K content in the LM and HM fruits was significantly higher than that of the control.
As shown in Table 3 and Figure 4, the quality of grape fruit was obviously affected by malic acid. Weight per fruit (WPF) decreased significantly as the amount of malic acid added was increased. However, the TSS, Vc, and SSC in the fruit increased significantly after adding malic acid, but no significant differences were observed between LM and HM. In addition, TA of the fruits with HM was significantly higher than that of LM and the control. The solid acid ratio (SAR) and TS of the fruits with LM were significantly higher than those of HM and the control.
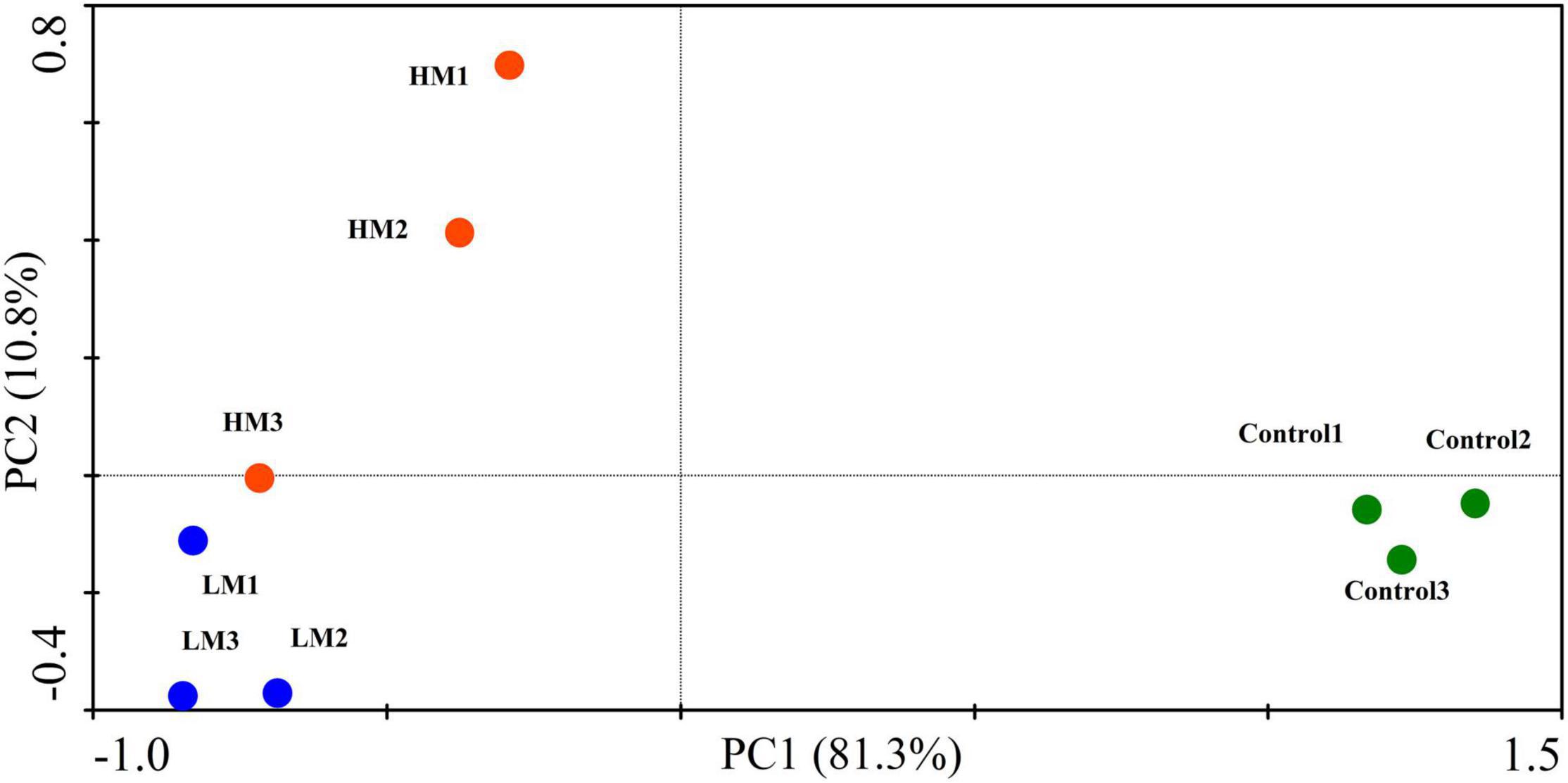
Figure 4. PCA plot showing the effect of adding different amounts of malic acid on grape nutrient absorption and fruit quality.
As shown in Figure 4, the PCA of nutrient absorption and fruit quality showed that the PC1 and PC2 scores were 81.3% and 10.8%, respectively, and each treatment formed its own cluster. Adding malic acid obviously changed the nutrient absorption and fruit quality of the grapes. The control and malic acid treatments (LM and HM) were located on the positive and negative axes of PC1, respectively. However, the difference in malic acid was mainly reflected in PC2; HM was distributed on the positive axis of PC2, and LM was distributed on the negative axis of PC2. Additionally, the comprehensive score results of grape nutrient absorption and fruit quality showed that LM > HM > control (Table 4).
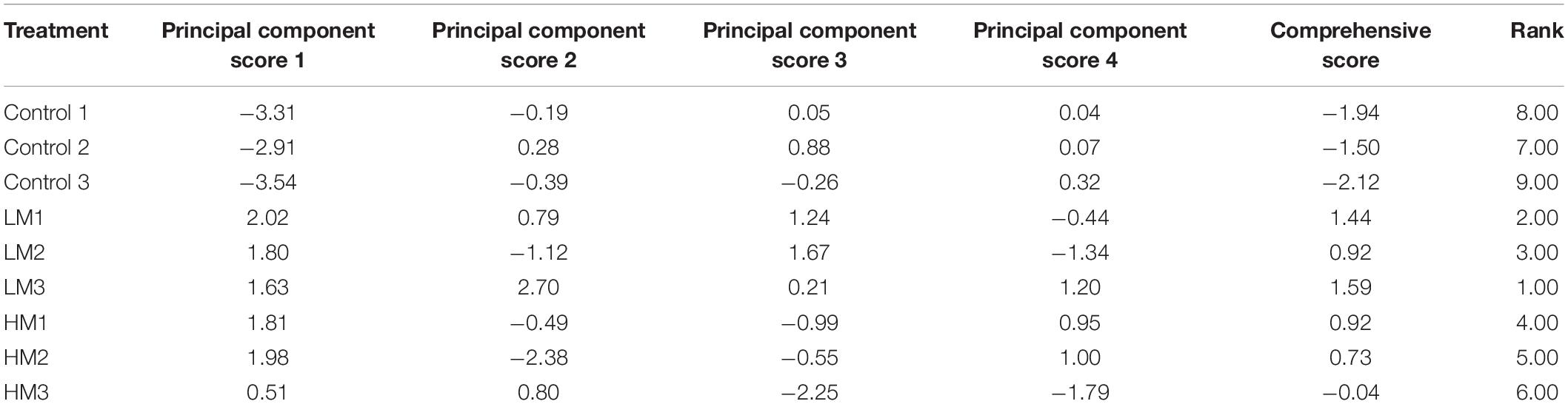
Table 4. Comprehensive results of PCA on the effect of adding different amounts of malic acid on nutrient absorption and fruit quality in grape.
In summary, although the malic acid treatment reduced WPF, and the high-concentration treatment (HM) risked a reduction in yield, the low-concentration (LM) treatment improved nutrient absorption capacity and fruit quality, resulting in the best taste and the highest nutrient and fruit quality scores.
Pearson’s correlation heatmap (Figure 5) shows that the environmental factors of EC, pH, SOM, and added malic acid were closely related to the composition of the bacterial and fungal communities. The amount of added malic acid was very significantly positively correlated with Firmicutes and Blastocladiomycota, with coefficients of 0.798 (p = 0.01) and 0.836 (p = 0.005) (Supplementary Tables 6, 7), respectively. In particular, the amount of added malic acid was very significantly negatively correlated with Patescibacteria, with a correlation coefficient of −0.87 (p = 0.002) (Figure 4 and Supplementary Table 6). Soil OM was significantly positively correlated with Deinococcota, Verrucomicrobiota, Sumerlaeota and Abditibacteriota, and had a very significant relationship with Deinococcota, with a correlation coefficient of 0.822 (p = 0.006), and a significant negative correlation with Myxococcota (Figure 4 and Supplementary Table 6). EC had a closer relationship with the bacterial communities than pH and a very significant positive correlation with Deinococcota, with a coefficient of 0.808 (p = 0.008), and a very significant negative correlation with Entotheonellaeota, with a correlation coefficient of −0.815 (p = 0.007). The fungus Rozellomycota was significantly positively correlated with OM, with a coefficient of 0.775 (p = 0.014).
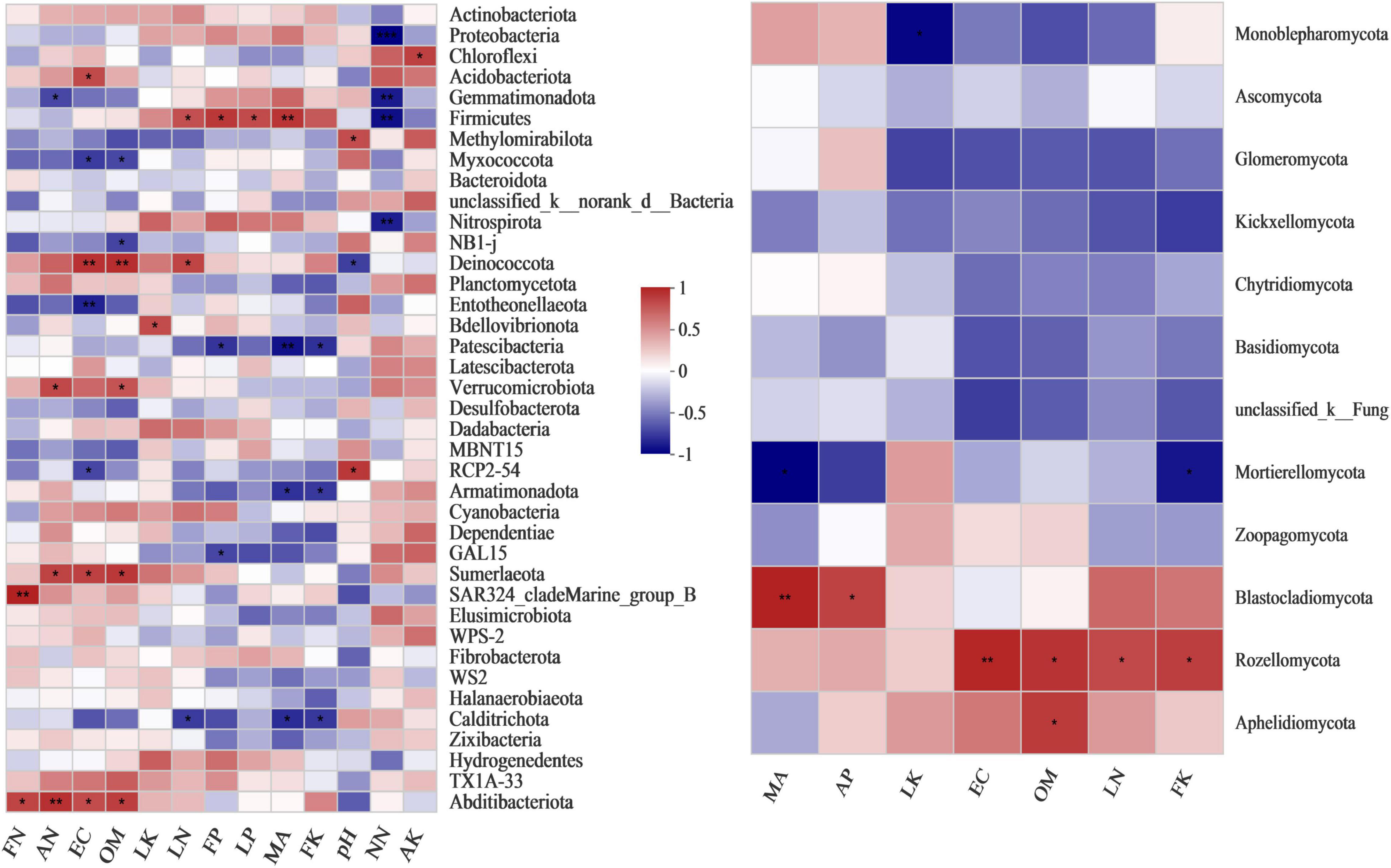
Figure 5. Pearson’s correlation heatmap showing the relationship between microbial composition, environmental factors and nutrient content at the phylum level. Left: bacterial community. Right: fungal community. MA, malic acid; OM, organic matter; EC, electrical conductivity; LN, leaf nitrogen; LP, leaf phosphorus; LK, leaf potassium; FN, fruit nitrogen; FP, fruit phosphorus; FK, fruit potassium. **Correlation is significant at the 0.01 level. *Correlation is significant at the 0.05 level.
As shown in Figure 5, the bacterial community components were closely related to rhizosphere soil nitrogen content (ammonium-nitrogen and nitrate-nitrogen). In particular, Proteobacteria, Gemmatimonadota, Firmicutes and Nitrospirota were very significantly negatively correlated with nitrate-nitrogen content, with coefficients of −0.934 (p = 0), −0.837 (p = 0.005), −0.881 (p = 0.002) and −0.829 (p = 0.006), respectively (Supplementary Table 6). Studies on the nutrient and microbial community composition of grape leaves and fruits have shown that grapes are closely related to absorption of the nutrient element potassium. Leaf K content was negatively correlated with Monoblepharomycota (Supplementary Table 7, r = −0.754, and p = 0.019) and positively correlated with Bdellovibrionota, with a correlation coefficient of 0.703 (p = 0.035). Fruit K content was negatively correlated with Patescibacteria and Calditrichota, with correlation coefficients of −0.764 (p = 0.017) and −0.738 (p = 0.023), respectively.
Pearson’s correlation heatmap analysis of the fruit quality bacterial community showed that the SAR, TS and WPF were closely related to the bacterial community (Figure 6 and Supplementary Table 8). In particular, TS was closely related to the bacterial and fungal communities, with a very significant negative correlation with Gemmatimonadota and Myxococcota, and correlation coefficients of −0.82 (p = 0.007) and −0.863 (p = 0.003), respectively, in the bacterial community, and a very significant positive correlation with Sumerlaeota, with a correlation coefficient of 0.836 (p = 0.005). TS was very significantly negatively correlated with Chytridiomycota and Glomeromycota in the fungal community with correlation coefficients of −0.86 (p = 0.003) and −0.86 (p = 0.003), respectively.
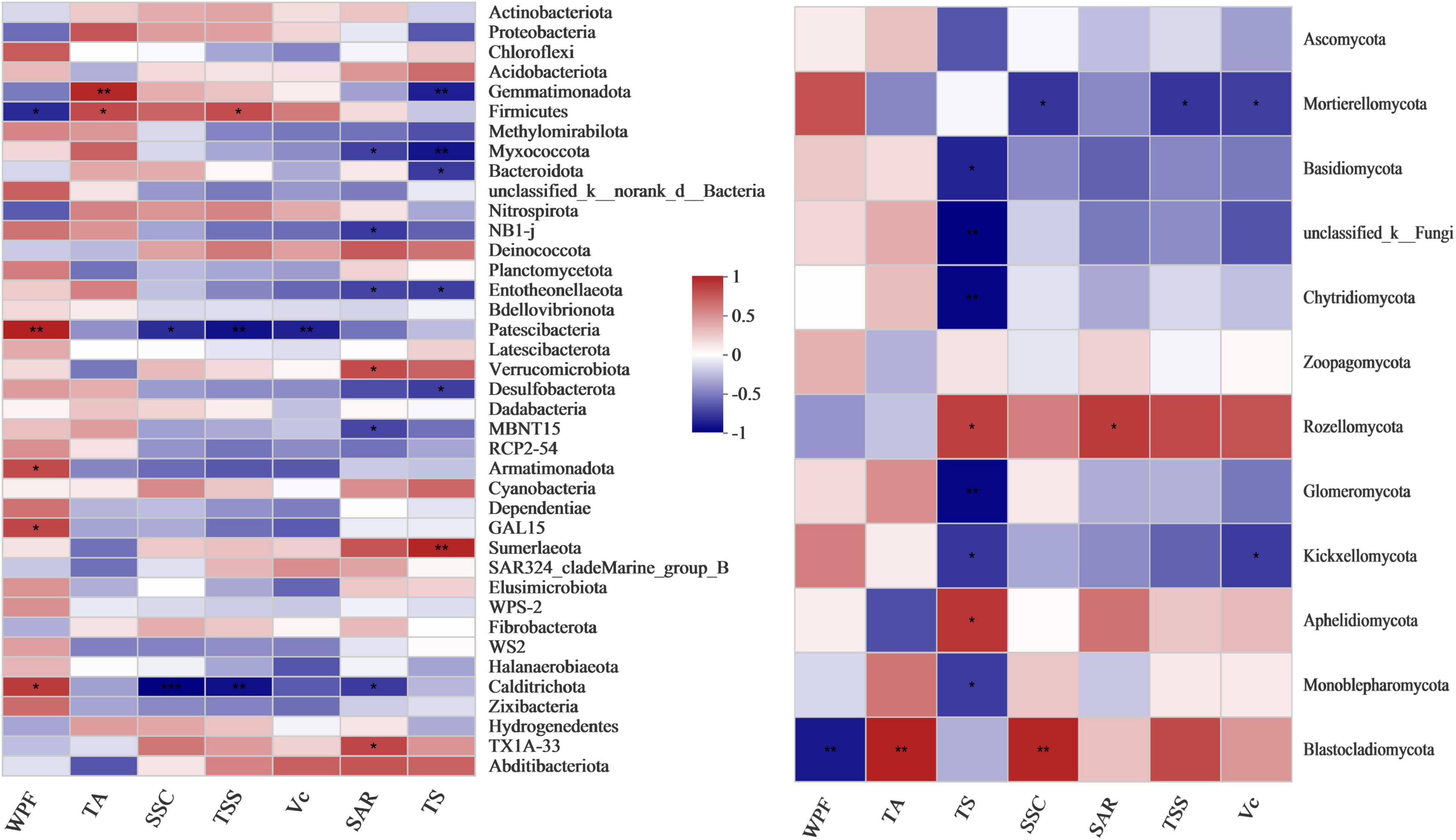
Figure 6. Pearson’s correlation heatmap showing the relationship between microbial composition and grape fruit quality at the phylum level. WPF, weight per fruit; SSC, soluble solid content; Vc, vitamin C; TSS, total soluble sugar; TA, titratable acid; TS, grape tasting score; SAR, solid-acid ratio (TSS/TA). **Correlation is significant at the 0.01 level. *Correlation is significant at the 0.05 level.
Firmicutes had a significant positive correlation with TA and TSS and a significant negative correlation with WPF (Figure 6). Patescibacteria was negatively correlated with TSS, SSC and Vc, with correlation coefficients of −0.867 (p = 0.002), −0.773 (p = 0.015) and −0.813 (p = 0.008), respectively, and a very significant positive correlation with WPF, with a correlation coefficient of 0.847 (p = 0.004). Calditrichota was significantly negatively correlated with TSS and SAR (r = −0.872 and −0.723, p = 0.002 and 0.028, respectively), and significantly positively correlated with WPF (r = 0.748, p = 0.02).
The increase in the ratio of the fungi Mortierellomycota and Blastocladiomycota reduced fruit quality. Mortierellomycota had a significant negative correlation with TSS, SSC and Vc, while Blastocladiomycota had a very significant positive correlation with TA, and a very significant negative correlation with WPF, with correlation coefficients of 0.814 (p = 0.008) and −0.802 (p = 0.009), respectively. The increase in the Rozellomycota ratio had the potential to improve fruit quality, which was significantly positively correlated with SAR and TS.
The nutrient absorption and fruit quality of the grapes were significantly affected by the bacterial community at the family level (Figure 7 and Supplementary Tables 8, 9). Adding malic acid was significantly positively correlated with Planococcaceae (p = 0.008), Bacillaceae (p = 0.047), Woeseiaceae (p = 0.012) and Rhodobacteraceae (p = 0.01), and the correlation coefficients were 0.78, 0.84, 0.79 and 0.8, respectively. In contrast to the malic acid-added treatments, nitrate-nitrogen was extremely significantly negatively correlated with Planococcaceae, Bacillaceae, Woeseiaceae and Rhodobacteraceae with correlation coefficients of −0.88 (p = 0.002), −0.9 (p = 0.001), −0.82 (p = 0.006) and −0.95 (p = 0), respectively. Similar to the malic acid-added treatments, the P contents of leaves and fruits were significantly positively correlated with Planococcaceae (r = 0.68 and 0.81, and p = 0.045 and 0.008, respectively) and Bacillaceae (r = 0.75 and 0.77, and p = 0.021 and 0.016), and soil available P content was significantly positively correlated with Woeseiaceae (r = 0.77, and p = 0.015) and Rhodobacteraceae (r = 0.73, and p = 0.026). Fruit K content was significantly positively correlated with Bacillaceae (r = 0.67, and p = 0.049) and Woeseiaceae (r = 0.76, and p = 0.018). Therefore, Planococcaceae, Bacillaceae, Woeseiaceae and Rhodobacteraceae were related to the malic acid treatments and played an important role in the nutrient absorption of grapes.
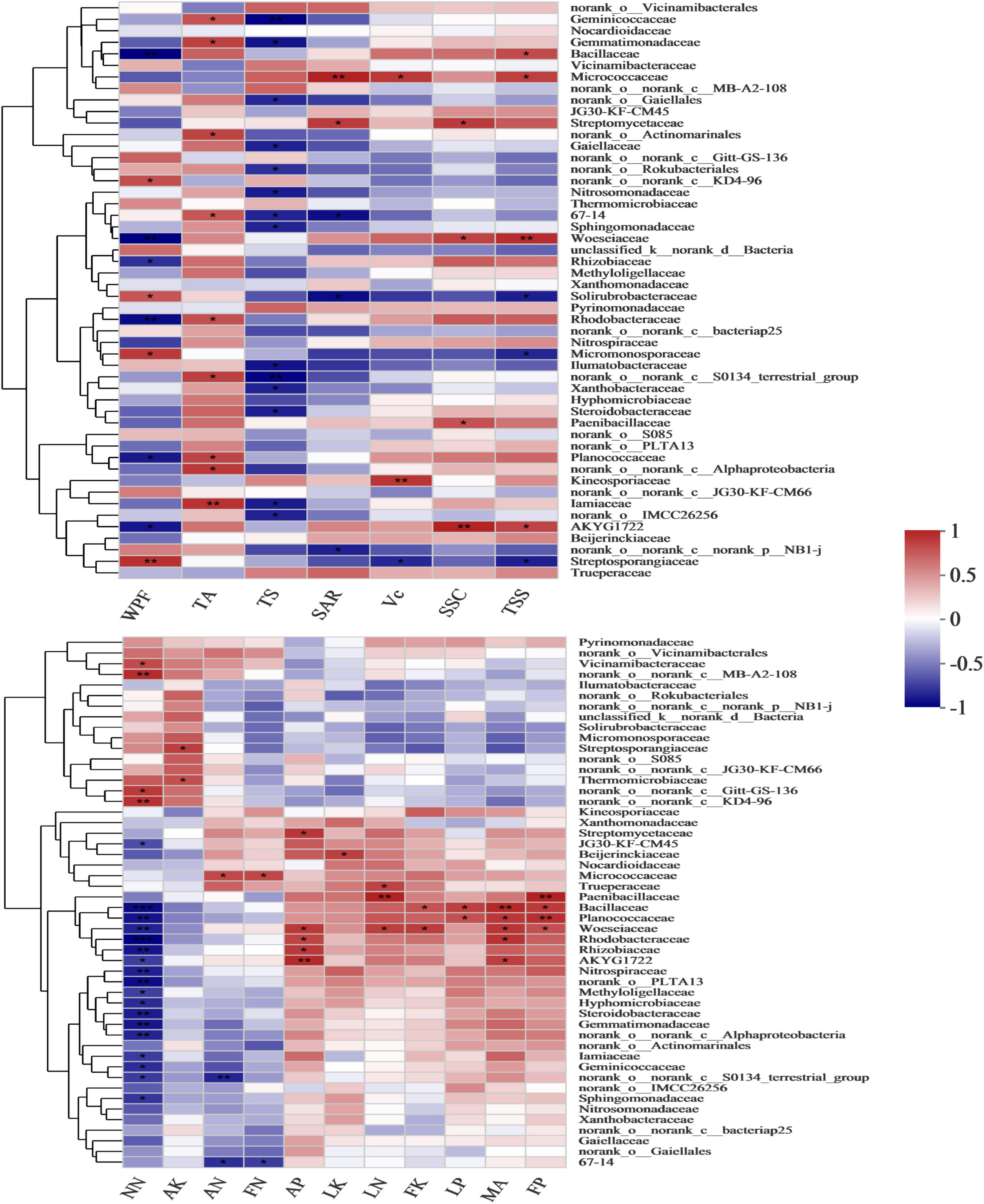
Figure 7. Pearson’s correlation heatmap showing the relationships between bacterial composition, fruit quality and nutrient content at the family level. MA, malic acid; OM, organic matter; EC, electrical conductivity; LN, leaf nitrogen; LP, leaf phosphorus; LK, leaf potassium; FN, fruit nitrogen; FP, fruit phosphorus; FK, fruit potassium; WPF, weight per fruit; SSC, soluble solid content; Vc, vitamin C; TSS, total soluble sugar; TA, titratable acid; TS, grape tasting score; SAR, solid-acid ratio. **Correlation is significant at the 0.01 level. *Correlation is significant at the 0.05 level.
Furthermore, contrary to malic acid, Planococcaceae, Bacillaceae, Woeseiaceae and Rhodobacteraceae all had significant negative correlations with WPF, with correlation coefficients of −0.75 (p = 0.019), −0.84 (p = 0.005), −0.84 (p = 0.005) and −0.8 (p = 0.009), respectively. Bacillaceae and Woeseiaceae were significantly positively correlated with TSS, with correlation coefficients of 0.72 (p = 0.028) and 0.84 (p = 0.005), while Planococcaceae and Rhodobacteraceae were significantly positively correlated with TA (r = 0.75 and 0.71, p = 0.021 and 0.033, respectively), and the proliferation of Bacillaceae and Woeseiaceae increased the TSS of fruit exposed to added malic acid.
As an important intermediate product of many metabolic processes in plants, malic acid links multiple metabolic pathways in cells (Fernie and Martinoia, 2009) and plays an important physiological function during plant growth. In addition, malic acid, as one of the main exudates of the plant rhizosphere, affects the composition of the rhizosphere microbial community and soil nutrient cycling.
Adding malic acid affected the pH of the soil, which, in turn, affected the soil microbial community. In addition, it served as a carbon source to stimulate and screen the soil microbial communities. However, the effects of 5% and 10% malic acid combined with NPK fertilizer on soil pH were not significant, and this was consistent with research on peach and pear rhizosphere soil after adding malic acid (Shao et al., 2022). Therefore, malic acid as a carbon source stimulates and screens the soil microbial community, which is the main reason why it affected the grape rhizosphere microbial community.
Malic acid, a major organic acid in root plant secretions, is selectively secreted and effectively signaled to beneficial rhizosphere bacteria, regulating root metabolites during the recruitment of beneficial microorganisms, which emphasizes the breadth and sophistication of plant-microbe interactions (Rudrappa et al., 2008). The secretion of malic acid into the rhizosphere of three emergent plant species has a significant negative correlation with ammonia-oxidizing bacterial activity (Chen et al., 2021). Malic acid induces a stronger chemotactic response and swarming motility of Bacillus amyloliquefaciens than citric acid, succinic acid or fumaric acid, which produces a variety of antibiotics with broad-spectrum activity against different plant pathogens, thereby inducing plant host system resistance (Basi et al., 2006; Chen et al., 2009; Arrebola et al., 2010; Ramarathnam et al., 2010; Tan et al., 2013). Additionally, malic acid in the presence of a pathogen recruits the beneficial bacterium Bacillus subtilis FB17 to Arabidopsis roots (Rudrappa et al., 2008). Similarly, we revealed that the amount of added malic acid was extremely significantly positively correlated with Bacillaceae in the grape rhizosphere. Furthermore, malic acid and citric acid in watermelon root exudates, which are intermediate products of the tricarboxylic acid (TCA) cycle, i.e., also significantly induce Paenibacillus polymyxa SQR-21 motility (Ling et al., 2011). Hence, malic acid, as the main organic acid in the rhizosphere exudate, is the second most preferred carbon source for organisms, such as Bacillus subtilis and Azospirillum brasilense (Bashan and de-Bashan, 2002; Meyer et al., 2011; Rekha et al., 2018). Malic acid and citric acid released from tomato roots attract Pseudomonas fluorescens strains (Weert et al., 2002; Liu et al., 2020).
The bacterial community was more sensitive to malic acid than the fungal community. In structuring rhizosphere microbial communities with different root exudates, differences in fungal community structure have been attributed to citric acid and differences in bacterial community structure have been attributed to cisaconitic, citric and malic acids (Dennis et al., 2010).
Soil OM is a dynamic nutrient storage medium that provides macronutrients to produce protein in plants through soil biota activities (Garcia-Pausas and Paterson, 2011; Wood et al., 2018). A low malic acid treatment stimulates the microorganisms and primes the soil organic carbon in a nutrient-poor system (Chowdhury et al., 2014). Hence, the SOM content was significantly increased by 5% malic acid compared with the control.
Malic acid was negatively correlated with NO3-N in the emergent plant rhizosphere of a constructed wetland in northern China (Tan et al., 2013). As the amount of malic acid added increased in the current study, the NO3-N content in the grape rhizosphere decreased and was significantly reduced by 10% malic acid combined with the NPK fertilizer. Additionally, absorption of NH4-N was closely related to ammonium-nitrogen content. Soil NH4-N content increased significantly after adding 5% malic acid combined with the NPK fertilizer. The absorption of NH4-N upregulates the synthesis of malic acid and oxaloacetic acid by promoting the activities of malate dehydrogenase and phosphoenolpyruvate carboxykinase (Britto and Kronzucker, 2005; Wang et al., 2021). Thus, root cytosol alkalinization induced by NH4-N uptake distinctly enhanced the activities of phosphoenolpyruvate carboxylase and malate dehydrogenase but reduced malic enzyme activities (Xu et al., 2021).
Malic acid, as one of the LMWOAs, increases plant-available P fractions by solubilizing inorganic P fractions, which are virtually insoluble, retarding the reaction of fertilizer P with soil components and decreasing the relative saturation of metal ions in solution (Harrold and Tabatabai, 2006; Pavinato et al., 2008; Miller and Fox, 2011; Oral and Uygur, 2018). In addition, plants produce a series of protective mechanisms when exposed to a phosphorus deficiency by secreting small molecules, such as malic acid, into the rhizosphere (Ozawa et al., 1995; Ascencio, 1997; McGrail et al., 2021). Although the ability of malic acid to complex with metal ions is weaker than that of dicarboxylic acid and TCA (McGrail et al., 2021), malic acid combined with NPK fertilizer increased soil available phosphorus content, thereby increasing the phosphorus content of leaves and fruits, and ultimately increasing the absorption of phosphorus by grapes.
K is an essential macronutrient for plant growth that plays important roles in various metabolic processes involving protein synthesis, photosynthesis, enzymes and resistance to pests and diseases (Prajapati and Modi, 2012). Potassium is solubilized from potassium-aluminum silicate minerals through the secretion of different organic acids, such as malic acid and citric acid, by potassium-dissolving bacteria (Prajapati and Modi, 2012). Although no significant difference was observed in the results, the available K content of the grape rhizosphere soil with added malic acid was lower than that of the control, while leaf K content in the 5% malic acid treatment was higher than that in the other treatments. However, regardless of the 5% and 10% malic acid combined with the NPK fertilizer, the K content of fruits was significantly higher than that of the control, indicating that malic acid promoted the absorption of potassium by grapes and contributed to the accumulation of potassium in fruits. Secretion of malic acid into the rhizosphere is strongly affected by potassium status (Freeman, 1967). Moreover, the combination of potassium nutrition and exogenous organic acids improves the absorption of iron by monocots and dicots and mediates iron-biofortified crops (Awad-Allah and Elsokkary, 2020).
Malic acid is stored in vacuoles, constituting a major carbon pool and a potential substrate for respiration (Blanke and Lenz, 1989), but is also the predominant organic acid associated with taste, flavor and juice quality in fruit (Yao et al., 2020). Malic acid promotes plant growth by increasing chlorophyll content and mitigating stress damage to photosynthetic structures, thereby significantly increasing plant biomass (Chen et al., 2020). Photosynthetic assimilates are mainly used for fruit growth during the early stage of fruit development, and the sugar in the fruit accumulates 2 weeks after fruit expansion stops, leading to an increase in SSC (Long et al., 2006). Adding malic acid potentially improved the photosynthetic capacity of grape leaves. We speculate that the photosynthetic rate and water use efficiency of grape leaves would increase after adding malic acid, which facilitates the accumulation of soluble solids in the fruit. Similar results were found in pears when applying malic acid combined with NPK (Shao et al., 2022). A study of organic acids and potassium fertilizer in fruits reported that applying potassium fertilizer increases TA of fruits, particularly malic acid content (Cummings and Reeves, 1971; Du, 1985; Biaiłczyk and Lechowski, 1989), and malic acid content is usually positively correlated with ash alkalinity during fruit ripening, while ash content alkalinity is closely related to potassium content (Genevois and Peynaud, 1947; Souty et al., 1967; Lobit et al., 2006).
Malic acid as a rhizosphere exudate secreted by plants drives microorganisms to participate in OM mineralization that indirectly mediates nutrient uptake and indirectly mediates nutrient absorption through dissolution and chelation of nutrients (McGrail et al., 2021). Malic acid was significantly positively correlated with Planococcaceae, Bacillaceae, Woeseiaceae and Rhodobacteracea.
It was revealed that the 5% malic acid treatment increased soil ammonium-nitrogen content and decreased soil nitrate-nitrogen content, while excessive malic acid reduced the available nitrogen content in the soil. However, most ammonia-oxidizing bacteria OTUs were negatively correlated with malic acid content (Fang et al., 2019). Contrary to adding malic acid, nitrate-nitrogen was significantly negatively correlated with Planococcaceae, Bacillaceae, Woeseiaceae and Rhodobacteraceae. Planococcaceae is composed mainly of the genus Planococcus, and Planococcus includes denitrifying bacteria (Chen et al., 2016; Ismail et al., 2021). Bacillaceae, particularly the genus Bacillus, is involved in denitrification and dissimilatory nitrogen reduction to ammonium in several strains, and various members of Bacillus have flexible physiological functions during the process of dissimilated nitrate reduction and its intermediates or by-products (Verbaendert, 2014). Woeseiaceae is an abundant core member of the microbial community in global marine sediments that are involved in the incomplete denitrification pathway, including subunits of nitrite reduction (nirS) and NO reduction (norB) to the ozone-depleting greenhouse gas N2O (Hinger et al., 2019). However, Rhodobacteraceae, which oxidize NH4-N to nitrate or nitrite, is significantly negatively correlated with nitrate-nitrogen (Liu et al., 2018).
Similar to the added malic acid treatments, the P content of leaves and fruits was significantly positively correlated with Planococcaceae and Bacillaceae, and soil available P content was significantly positively correlated with Woeseiaceae and Rhodobacteraceae. B. subtilis, isolated from mangrove soil in Chollangi, East Godavari, exhibits a phosphate solubilizing ability in the range of 80–100 mg/l (Anzuay et al., 2015). Research on the role of P limitations in shaping soil bacterial communities has revealed that Firmicutes, including Planococcaceae and Bacillaceae, are enriched in high P soils, and Planococcaceae is relatively more abundant than Bacillaceae (Oliverio et al., 2020). Notably, the abundance of Planococcaceae and Bacillacea related to functions of carbon degradation and P cycling increase sugarcane yield (Silva et al., 2021). Rhodobacteraceae is a family in Alphaproteobacteria that is involved in C, N and S cycling processes in the marine environment (Zheng et al., 2015; Zhang et al., 2019). Non-marine Rhodobacteriaceae gained high-affinity transporters in response to much lower sulfate concentrations and lost genes associated with reduced sodium chloride and organohalogen concentrations in their habitats (Simon et al., 2017). The bacterial carbon-phosphorus lyase pathway, an enzyme complex that evolved to extract phosphate from phosphonates, is prevalent in a considerable proportion of Rhodobacteraceae bacteria (11–40% of organisms) across all ocean regions in the mesopelagic zone (Sosa et al., 2019).
Fruit K content was significantly positively correlated with Bacillaceae and Woeseiaceae. Bacillaceae is a family of potassium-dissolving bacteria (KSB) microorganism that secrete organic acids from insoluble potassium-containing minerals that directly dissolve rock K or chelated silicon (Meena et al., 2014, 2016; Zhang and Kong, 2014). Hence, both the malic acid and the increase in the abundance of Bacillaceae with added malic acid stimulated the absorption of potassium by grapes; thus, the proliferation of Planococcaceae, Bacillaceae, Woeseiaceae and Rhodobacteraceae stimulated by malic acid has the potential to enhance nutrient absorption of grapes. Planococcaceae, Bacillaceae, Woeseiaceae and Rhodobacteraceae were significantly negatively correlated with WPF. Bacillaceae and Woeseiaceae were significantly positively correlated with TSS, while Planococcaceae and Rhodobacteraceae were significantly positively correlated with TA. However, SSC and TSS of grape fruit increased after adding malic acid. In addition, the nutrient content of leaves and fruits also increased after the malic acid treatment. Bacillaceae, involved in plant rhizosphere growth, and Woeseiaceae, involved in the nitrogen cycle, have the potential to improve fruit quality. Therefore, Bacillaceae and Woeseiaceae were the key bacteria playing a major role in grape fruit quality and nutrient absorption after applying the malic acid water-soluble fertilizer.
Nutrient absorption and fruit quality of grapes were improved after adding malic acid, and the best formula was 5% malic acid combined with NPK fertilizer. In addition, the structure and carbon metabolism of the soil microbial community were affected significantly by applying malic acid, and the composition of the microbial community was closely related to nutrient absorption and the quality of the grapes. Adding malic acid was significantly positively correlated with Planococcaceae, Bacillaceae, Woeseiaceae, and Rhodobacteraceae with correlation coefficients of 0.78, 0.84, 0.79 and 0.8, respectively. The proliferation of Planococcaceae, Bacillaceae, Woeseiaceae, and Rhodobacteraceae stimulated by malic acid has the potential to enhance nutrient absorption of grapes. Bacillaceae and Woeseiaceae were significantly positively correlated with the TSS of grape fruit with correlation coefficients of 0.72 and 0.84, respectively, while Planococcaceae and Rhodobacteraceae were significantly positively correlated with the TA content of grape fruit (0.75 and 0.71, respectively). Hence, Bacillaceae and Woeseiaceae are the key bacteria that play a major role in grape fruit quality and nutrient absorption after applying malic acid water-soluble fertilizer.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
WS, GX, and HY: conceptualization. WS and GX: methodology. WS: software, formal analysis, and data curation. PS, WS, HY, GX, and GD: validation. WS and HY: investigation. PS: resources and visualization. WS and PS: writing—original draft preparation and writing—review and editing. PS and GD: supervision, project administration, and funding acquisition. All authors have read and agreed to the published version of the manuscript.
This research was funded by a Major Scientific and Technological Project of Xinjiang Corps (2019AA004) and an earmarked Fund for Hebei Agriculture Research System Grant (HBCT2018100204).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank the Henan Key Laboratory of Fruit and Cucurbit Biology and the Laboratory of Fruit Breeding Technology of the Ministry of Agriculture and Rural Affairs for providing equipment and technical support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.850807/full#supplementary-material
Anzuay, M. S., Ludueña, L. M., Angelini, J. G., Fabra, A., and Taurian, T. (2015). Beneficial effects of native phosphate solubilizing bacteria on peanut (Arachis hypogaea L) growth and phosphorus acquisition. Symbiosis 66, 89–97. doi: 10.1007/s13199-015-0337-z
Arrebola, E., Jacobs, R., and Korsten, L. (2010). Iturin a is the principal inhibitor in the biocontrol activity of Bacillus amyloliquefaciens PPCB004 against postharvest fungal pathogens. J. Appl. Microbiol 108, 386–395. doi: 10.1111/j.1365-2672.2009.04438.x
Ascencio, J. (1997). Root secreted acid phosphatase kinetics as a physiological marker for phosphorus deficiency. J. Plant Nutr. 20, 9–26. doi: 10.1080/01904169709365230
Awad-Allah, E. F., and Elsokkary, I. H. (2020). Influence of potassium nutrition and exogenous organic acids on iron uptake by monocot and dicot plants. Open J. Soil Sci. 10, 486–500. doi: 10.4236/ojss.2020.1010025
Basi, H. P., Weir, T. L., Perry, L. G., Gilroy, S., and Vivanco, J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol 57, 233–266. doi: 10.1146/annurev-plant-57-033010-200001
Bashan, Y., and de-Bashan, L. E. (2002). Protection of tomato seedlings against infection by Pseudomonas syringae pv. tomato by using the plant growth-promoting bacterium Azospirillum brasilense. Appl. Environ. Microbiol 68, 2637–2643. doi: 10.1128/AEM.68.6.2637-2643.2002
Bassham, J. A., and Calvin, M. (1980). The Path of Carbon in Photosynthesis. In Die CO2-Assimilation/The Assimilation of Carbon Dioxide. Berlin, Heidelberg: Springer, 884–922. doi: 10.1007/978-3-642-94798-8_30
Beauregard, P. B., Chai, Y., Vlamakis, H., Losick, R., and Kolter, R. (2013). Bacillus subtilis biofilm induction by plant polysaccharides. Proc. Natl. Acad. Sci. 110, E1621–E1630. doi: 10.1073/pnas.1218984110
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Biaiłczyk, J., and Lechowski, Z. (1989). Malic acid synthesis in relation to K+ and CI- availability in Phaseolus coccineus L. pulvini. Biochemie Physiol. Pflanzen 184, 79–86. doi: 10.1016/S0015-3796(89)80125-8
Blanke, M. M., and Lenz, F. (1989). Fruit photosynthesis. Plant Cell Environ. 12, 31–46. doi: 10.1111/j.1365-3040.1989.tb01914.x
Bolan, N. S., Naidu, R., Mahimairaja, S., and Baskaran, S. (1994). Influence of low-molecular-weight organic acids on the solubilization of phosphates. Biol. Fert. Soils 18, 311–319. doi: 10.1007/BF00570634
Britto, D. T., and Kronzucker, H. J. (2005). Nitrogen acquisition, PEP carboxylase, and cellular pH homeostasis: new views on old paradigms. Plant Cell Environ. 28, 1396–1409. doi: 10.1111/j.1365-3040.2005.01372.x
Buser-Suter, C., Andres, W., and Philippe, M. (1982). A malic acid permease in isolated vacuoles of a crassulacean acid metabolism plant. Plant Physiol. 69, 456–459. doi: 10.1104/pp.69.2.456
Cao, J. K., Jiang, W. B., and Zhao, Y. M. (2007). Physiological and Biochemical Experiment Guidance After Fruit and Vegetable Harvest. Beijing: China Light Industry Press.
Casati, P., Drincovich, M. F., Edwards, G. E., and Andreo, C. S. (1999). Malate metabolism by NADP-malic enzyme in plant defense. Photosyn. Res. 1999, 99–105. doi: 10.1023/A:1006209003096
Chen, D., Wei, L., Zou, Z., Yang, K., and Wang, H. (2016). Bacterial communities in a novel three-dimensional bioelectrochemical denitrification system: the effects of pH. Appl. Microbiol. Biotechnol 100, 6805–6813. doi: 10.1007/s00253-016-7499-3
Chen, H. C., Zhang, S. L., Wu, K. J., Li, R., He, X. R., He, D. N., et al. (2020). The effects of exogenous organic acids on the growth, photosynthesis and cellular ultrastructure of Salix variegata franch. under Cd stress. Ecotoxicol. Environ. Safety 187:109790. doi: 10.1016/j.ecoenv.2019.109790
Chen, J., Dong, J., Li, C., Chen, H., Wang, L., Lyu, T., et al. (2021). Response of microbial community composition and function to emergent plant rhizosphere of a constructed wetland in northern China. Appl. Soil Ecol. 168:104141. doi: 10.1016/j.apsoil.2021.104141
Chen, Q., Wang, B., Ding, H., Zhang, J., and Li, S. (2019). The role of NADP-malic enzyme in plants under stress. Plant Sci. 281, 206–212.
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884-i890. doi: 10.1093/bioinformatics/bty560
Chen, X., Koumoutsi, A., Scholz, R., Schneider, K., Vater, J., Sussmuth, R., et al. (2009). Genome analysis of Bacillus amyloliquefaciens FZB42 reveals its potential for biocontrol of plant pathogens. J. Biotechnol 140, 27–37. doi: 10.1016/j.jbiotec.2008.10.011
Chen, Y., Cao, S., Chai, Y., Clardy, J., Kolter, R., Guo, J. H., et al. (2012). A Bacillus subtilis sensor kinase involved in triggering biofilm formation on the roots of tomato plants. Mol. Microbiol. 85, 418–430. doi: 10.1111/j.1365-2958.2012.08109.x
Chowdhury, S., Farrell, M., and Bolan, N. (2014). Priming of soil organic carbon by malic acid addition is differentially affected by nutrient availability. Soil Biol. Biochem. 77, 158–169. doi: 10.1016/j.soilbio.2014.06.027
Cummings, G. A., and Reeves, J. (1971). Factors influencing chemical characteristics of peaches. J. Am. Soc. Hortic. Sci. 96, 320–322. doi: 10.1007/s00442-019-04405-0
Dai, Z., Zhou, H., Zhang, S., Gu, H., Yang, Q., Zhang, W., et al. (2018). Current advance in biological production of malic acid using wild type and metabolic engineered strains. Bioresour. Technol. 258, 345–353. doi: 10.1016/j.biortech.2018.03.001
Darandeh, N., and Hadavi, E. (2012). Effect of pre-harvest foliar application of citric acid and malic acid on chlorophyll content and post-harvest vase life of Lilium cv. brunello. Front. Plant Sci. 2:106. doi: 10.3389/fpls.2011.00106
Denison, R. F. (1998). Decreased oxygen permeability: an universal stress response in legume root nodules. Plant Biol. 111, 191–192. doi: 10.1111/j.1438-8677.1998.tb00694.x
Dennis, P. G., Miller, A. J., and Hirsch, P. R. (2010). Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol. Ecol. 72, 313–327. doi: 10.1111/j.1574-6941.2010.00860.x
Dong, C., Shen, Q., and Wang, G. (2004). Tomato growth and organic acid changes in response to partial replacement of NO3–N by NH4+-N. Pedosphere 14, 159–164.
Drincovich, M. F., Casati, P., and Andreo, C. S. (2001). NADP-malic enzyme from plants: a ubiquitous enzyme involved in different metabolic pathways. FEBS Lett. 490, 1–6. doi: 10.1016/S0014-5793(00)02331-0
Driscoll, B., and Finan, T. (2010). NAD (+)-dependent malic enzyme of rhizobium meliloti is required for symbiotic nitrogen fixation. Mol. Microbiol. 7, 865–873. doi: 10.1111/j.1365-2958.1993.tb01177.x
Ebrahimian, E., and Bybordi, A. (2014). Effect of organic acids on heavy-metal uptake and growth of canola grown in contaminated soil. Commun. Soil Sci. Plant Anal. 45, 1715–1725. doi: 10.1080/00103624.2013.875206
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edwards, G. E., Furbank, R. T., Hatch, M. D., and Osmond, C. B. (2001). What does it take to be C4? Lessons from the evolution of C4 photosynthesis. Plant Physiol. 125, 46–49. doi: 10.1104/pp.125.1.46
Fang, J., Zhao, R., Cao, Q., Quan, Q., Sun, R., and Liu, J. (2019). Effects of emergent aquatic plants on nitrogen transformation processes and related microorganisms in a constructed wetland in northern China. Plant Soil 443, 473–492. doi: 10.1007/s11104-019-04249-w
Fernie, A. R., and Martinoia, E. (2009). Malate. jack of all trades or master of a few? Phytochemistry 70, 828–832. doi: 10.1016/j.phytochem.2009.04.023
Freeman, G. G. (1967). Studies on potassium nutrition of plants. II.—some effects of potassium deficiency on the organic acids of leaves. J. Sci. Food Agric. 18, 569–576. doi: 10.1002/jsfa.2740181205
Galvez, S. (2000). Oxygen regulation of a nodule-located carbonic anhydrase in alfalfa. Plant Physiol. 124, 1059–1068. doi: 10.1104/pp.124.3.1059
Garcia-Pausas, J., and Paterson, E. (2011). Microbial community abundance and structure are determinants of soil organic matter mineralisation in the presence of labile carbon. Soil Biol. Biochem. 43, 1705–1713. doi: 10.1016/j.soilbio.2011.04.016
Genevois, L., and Peynaud, E. (1947). Composition de neuf variétés de prunes. Revue Horticole 1947, 317–318.
Guanghua, W., Junjie, L., Xiaoning, Q., Jian, J., Yang, W., and Xiaobing, L. (2008). Effects of fertilization on bacterial community structure and function in a black soil of Dehui region estimated by Biolog and PCR-DGGE methods. Acta Ecol. Sin. 28, 220–226. doi: 10.1016/S1872-2032(08)60023-2
Guo, H., Chen, H., Hong, C., Jiang, D., and Zheng, B. (2017). Exogenous malic acid alleviates cadmium toxicity in Miscanthus sacchariflorus through enhancing photosynthetic capacity and restraining ROS accumulation. Ecotoxicol. Environ. Safety 141, 119–128. doi: 10.1016/j.ecoenv.2017.03.018
Han, Z. J., Xu, J., Wei, X. Z., He, B., Xue, G., and Yang, T. Z. (2016). Effect of exogenous organic acids on tobacco physiology index and soil potassium content. J. Agric. Sci. Technol. 18:109.
Harrold, S. A., and Tabatabai, M. A. (2006). Release of inorganic phosphorus from soils by low-molecular-weight organic acids. Commun. Soil Sci. Plant Anal. 37, 1233–1245. doi: 10.1080/00103620600623558
Hinger, I., Pelikan, C., and Mußmann, M. (2019). Role of the ubiquitous bacterial family woeseiaceae for N2O production in marine sediments. Geophys. Res. Abstr. 21:17441.
Ismail, S., Elreedy, A., Fiji, M., Ni, S. Q., Tawfik, A., and Esiason, M. (2021). Fatigue of anammox consortia under long-term 1, 4-dioxane exposure and recovery potential: N-kinetics and microbial dynamics. J. Hazardous Materials 414:125533. doi: 10.1016/j.jhazmat.2021.125533
Jamil, A., Yang, J. Y., Su, D. F., Tong, J. Y., Chen, S. Y., Luo, Z. W., et al. (2020). Rhizospheric soil fungal community patterns of Duchesnea indica in response to altitude gradient in Yunnan, southwest China. Can. J. Microbiol. 66, 359–367. doi: 10.1139/cjm-2019-0589
Jiang, Z., Li, P., Wang, Y., Li, B., and Wang, Y. (2013). Effects of roxarsone on the functional diversity of soil microbial community. Int. Biodeter. Biodegrad. 76, 32–35. doi: 10.1016/j.ibiod.2012.06.010
Jones, D. L. (1998). Organic acids in the rhizosphere–a critical review. Plant Soil 205, 25–44. doi: 10.1023/A:1004356007312
Kolton, M., Graber, E. R., Tsehansky, L., Elad, Y., and Cytryn, E. (2017). Biochar-stimulated plant performance is strongly linked to microbial diversity and metabolic potential in the rhizosphere. New Phytol. 213, 1393–1404. doi: 10.1111/nph.14253
Lakshmanan, V., Kitto, S. L., Caplan, J. L., Hsueh, Y. H., Kearns, D. B., Wu, Y. S., et al. (2012). Microbe-associated molecular patterns-triggered root responses mediate beneficial rhizobacterial recruitment in Arabidopsis. Plant Physiol. 160, 1642–1661. doi: 10.1104/pp.112.200386
Lasa, B., Frechilla, S., Aparicio-Tejo, P. M., and Lamsfus, C. (2002). Alternative pathway respiration is associated with ammonium ion sensitivity in spinach and pea plants. Plant Growth Regul. 37, 49–55. doi: 10.1023/A:1020312806239
Lin, C. C., He, Z. D., and Shan, W. L. (2020). Comprehensive evaluation of fruit quality of 12 red table grape cultivars cultivated in yangling area based on principal component and cluster analyses. J. Fruit Sci. 37, 520–532.
Ling, N., Raza, W., Ma, J., Huang, Q., and Shen, Q. (2011). Identification and role of organic acids in watermelon root exudates for recruiting Paenibacillus polymyxa SQR-21 in the rhizosphere. Eur. J. Soil Biol. 47, 374–379. doi: 10.1016/j.ejsobi.2011.08.009
Liu, J., Zhang, P., Li, H., Tian, Y., Wang, S., Song, Y., et al. (2018). Denitrification of landfill leachate under different hydraulic retention time in a two-stage anoxic/oxic combined membrane bioreactor process: Performances and bacterial community. Bioresour. Technol. 250, 110–116. doi: 10.1016/j.biortech.2017.11.026
Liu, W., Zhao, Q., Zhang, Z., Li, Y., Xu, N., Qu, Q., et al. (2020). Enantioselective effects of imazethapyr on Arabidopsis thaliana root exudates and rhizosphere microbes. Sci. Total Environ. 716:137121. doi: 10.1016/j.scitotenv.2020.137121
Lobit, P., Genard, M., Soing, P., and Habib, R. (2006). Modelling malic acid accumulation in fruits: relationships with organic acids, potassium, and temperature. J. Exp. Bot. 57, 1471–1483. doi: 10.1093/jxb/erj128
Long, R. L., Walsh, K. B., Midmore, D. J., and Rogers, G. (2006). Irrigation scheduling to increase muskmelon fruit biomass and soluble solids concentration. Hortscience 41, 367–369. doi: 10.21273/HORTSCI.41.2.367
Lou, X., Xu, W., and Wu, H. (1993). Study of effects of DI-malic acid on the performance of rice seedling roots. J. Zhen. Agric. Univ. 19, 388–388.
Lu, R. (2000). Method of Soil Agrochemical Analysis. Beijing: Chinese Agricultural Science and Technology Press.
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957-2963. doi: 10.1093/bioinformatics/btr507
McGrail, R. K., Van Sanford, D. A., and McNear, D. H. (2021). Semidwarf winter wheat roots contain fewer organic acids than wild-type varieties under phosphorus stress. Crop Sci. 61, 3586–3597. doi: 10.1002/csc2.20470
Meena, V. S., Maurya, B. R., and Verma, J. P. (2014). Does a rhizospheric microorganism enhance K+ availability in agricultural soils? Microbiol. Res. 169, 337–347. doi: 10.1016/j.micres.2013.09.003
Meena, V. S., Maurya, B. R., Verma, J. P., and Meena, R. S. (2016). Potassium Solubilizing Microorganisms for Sustainable Agriculture. New Delhi: Springer, 1–20. doi: 10.1007/978-81-322-2776-2
Meyer, F. M., Jules, M., Mehne, F. M. P., Le Coq, D., Landmann, J. J., Görke, B., et al. (2011). Malate-mediated carbon catabolite repression in Bacillus subtilis involves the HPrK/CcpA pathway. J. Bacteriol. 193, 6939–6949. doi: 10.1128/JB.06197-11
Miller, B. W., and Fox, T. R. (2011). Long-term fertilizer effects on oxalate-desorbable phosphorus pools in a typic paleaquult. Soil Sci. Soc. Am. J. 2011, 1110–1116. doi: 10.2136/sssaj2010.0037
Mueller, O., Hahnenberger, K., Dittmann, M., Yee, H., Dubrow, R., Nagle, R., et al. (2000). A microfluidic system for high-speed reproducible DNA sizing and quantitation. Electrophoresis 21, 128–134. doi: 10.1002/(SICI)1522-2683(20000101)21:1<128::AID-ELPS128>3.0.CO;2-M
Oliverio, A. M., Bissett, A., McGuire, K., Saltonstall, K., Turner, B. L., and Fierer, N. (2020). The role of phosphorus limitation in shaping soil bacterial communities and their metabolic capabilities. MBio 11, e1718–e1720. doi: 10.1128/mBio.01718-20
Oral, A., and Uygur, V. (2018). Effects of low-molecular-mass organic acids on P nutrition and some plant properties of Hordeum vulgare. J. Plant Nutr. 41, 1482–1490. doi: 10.1080/01904167.2018.1458866
Outlaw, W. H., and Oliver, H. L. (1977). Organic acid and potassium accumulation in guard cells during stomatal opening. Proc. Natl. Acad. Sci. U.S.A. 1977, 4434–4438. doi: 10.1073/pnas.74.10.4434
Ozawa, K., Osaki, M., Matsui, H., Honma, M., and Tadano, T. (1995). Purification and properties of acid phosphatase secreted from lupin roots under phosphorus-deficiency conditions. Soil Sci. Plant Nutr. 41, 461–469. doi: 10.1080/00380768.1995.10419608
Pavinato, P. S., Merlin, A., and Rosolem, C. A. (2008). Organic compounds from plant extracts and their effect on soil phosphorus availability. Pesquisa Agropecuária Brasileira 43, 1379–1388. doi: 10.1590/S0100-204X2008001000017
Prajapati, K. B., and Modi, H. A. (2012). Isolation and characterization of potassium solubilizing bacteria from ceramic industry soil. CIBTech J. Microbiol. 1, 8–14.
Ramarathnam, R., Fernando, W. G. D., and Kievit, T. (2010). The role of antibiosis and induced systemic resistance, mediated by strains of Pseudomonas chlororaphis, Bacillus cereus and B. amyloliquefaciens, in controlling blackleg disease of canola. BioControl 56, 225–235. doi: 10.1007/s10526-010-9324-8
Rekha, K., Baskar, B., Srinath, S., and Usha, B. (2018). Plant-growth-promoting rhizobacteria Bacillus subtilis RR4 isolated from rice rhizosphere induces malic acid biosynthesis in rice roots. Can. J. Microbiol. 64, 20–27. doi: 10.1139/cjm-2017-0409
Rosendahl, L., Vance, C. P., and Pedersen, W. B. (1990). Products of dark CO2 fixation in pea root nodules support bacteroid metabolism. Plant Physiol. 93, 12–19. doi: 10.1104/pp.93.1.12
Rudrappa, T., Czymmek, K. J., Pare, P. W., and Bais, H. P. (2008). Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 148, 1547–1556. doi: 10.1104/pp.108.127613
Scheibe, R. (2004). Malate valves to balance cellular energy supply. Physiol. Plant. 120, 21–26. doi: 10.1111/j.0031-9317.2004.0222.x
Schulze, J., Tesfaye, M., Litjens, R., Bucciarelli, B., Trepp, G., and Miller, S. (2002). Malate plays a central role in plant nutrition. Plant Soil 247, 133–139. doi: 10.1023/A:1021171417525
Shao, W., Xu, G. Y., Yu, H. L., Gao, D. T., Liu, Y., and Si, P. (2022). Low molecular weight organic acid water-soluble fertilizer improves leaf photosynthesis and fruit quality of pear. J. Fruit Sci. 2022, 1–16. doi: 10.13925/j.cnki.gsxb.20210523
Si, P., Shao, W., Yu, H., Yang, X., Gao, D., Qiao, X., et al. (2018). Rhizosphere microenvironments of eight common deciduous fruit trees were shaped by microbes in northern China. Front. Microbiol. 9:3147. doi: 10.3389/fmicb.2018.03147
Silva, A. M. M., Estrada-Bonilla, G. A., Lopes, C. M., Matteoli, F. P., Cotta, S. R., Feiler, H. P., et al. (2021). Does organomineral fertilizer combined with phosphate-solubilizing bacteria in sugarcane modulate soil microbial community and functions? Microbial. Ecol. 2021, 1–17. doi: 10.1007/s00248-021-01855-z
Simon, M., Scheuner, C., Meier-Kolthoff, J. P., Brinkhoff, T., Wagner-Döbler, I., Ulbrich, M., et al. (2017). Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J. 11, 1483–1499. doi: 10.1038/ismej.2016.198
Song, C., Zhang, L., Jia, Y., Cui, G., Cui, Z., and Zhu, Y. (2009). Malate metabolism and transport in plants. Plant Physiol. J. 5, 419–428.
Sosa, O. A., Repeta, D. J., DeLong, E. F., Ashkezari, M. D., and Karl, D. M. (2019). Phosphate-limited ocean regions select for bacterial populations enriched in the carbon-phosphorus lyase pathway for phosphonate degradation. Environ. Microbiol. 21, 2402–2414. doi: 10.1111/1462-2920.14628
Souty, M., Perret, A., and André, P. (1967). Premières observations sur quelques variétés de pêches destinées à la conserve. Ann. Technol. Agric. 6, 775–791.
Stackebrandt, E., and Goebel, B. M. (1999). Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44, 846–849. doi: 10.1099/00207713-44-4-846
Talebi, M., Hadavi, E., and Jaafari, N. (2014). Foliar sprays of citric acid and malic acid modify growth, flowering, and root to shoot ratio of gazania (Gazania rigens L.): a comparative analysis by ANOVA and structural equations modeling. Adv. Agric. 2014:147278. doi: 10.1155/2014/147278
Tan, S., Yang, C., Mei, X., Shen, S., Raza, W., Shen, Q., et al. (2013). The effect of organic acids from tomato root exudates on rhizosphere colonization of Bacillus amyloliquefaciens T-5. Appl. Soil Ecol. 64, 15–22. doi: 10.1016/j.apsoil.2012.10.011
Vance, C. P., and Heichel, G. H. (2003). Carbon in N2 fixation: limitation or exquisite adaptation. Ann. Rev. Plant Biol. 42, 373–390. doi: 10.1146/annurev.pp.42.060191.002105
Verbaendert, I. (2014). Denitrification in Gram-Positive Bacteria, With Focus on Members of the Bacillaceae Ph. D, Thesis.
Wan, Y., Zhou, L., Wang, S., Liao, C., Li, N., Liu, W., et al. (2018). Syntrophic growth of geobacter sulfurreducens accelerates anaerobic denitrification. Front. Microbiol. 9:1572. doi: 10.3389/fmicb.2018.01572
Wang, D., and Wang, J. (2009). Mechanism of soil mineral potassium release extracted by low-molecular-weigh organic acids. J. Liaoning Tech. Univ. 28, 259–261.
Wang, J. F., Li, W. L., Li, Q. S., Wang, L. L., He, T., Wang, F. P., et al. (2021). Nitrogen fertilizer management affects remobilization of the immobilized cadmium in soil and its accumulation in crop tissues. Environ. Sci. Pollut. Res. 2021, 1–13. doi: 10.1007/s11356-021-12868-z
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261-5267. doi: 10.1128/AEM.00062-07
Wang, X. (2006). Experimental Principles and Techniques of Plant Physiology and Biochemistry. Beijing: Higher Education Press.
Wang, Y., Whalen, J. K., Chen, X., Cao, Y., Huang, B., Lu, C., et al. (2016). Mechanisms for altering phosphorus sorption characteristics induced by low-molecular-weight organic acids. Can. J. Soil Sci. 96, 289–298. doi: 10.1139/cjss-2015-0068
Weert, S., Vermeiren, H., Mulders, I. H., Kuiper, I., Hendrickx, N., Bloemberg, G. V., et al. (2002). Flagella-drive chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol. Plant Microbe Int. 15, 1173–1180. doi: 10.1094/MPMI.2002.15.11.1173
Wood, S. A., Tirfessa, D., and Baudron, F. (2018). Soil organic matter underlies crop nutritional quality and productivity in smallholder agriculture. Agric. Ecosyst. Environ. 266, 100–108. doi: 10.1016/j.agee.2018.07.025
Wu, L., Li, Z., Li, J., Khan, M. A., Huang, W., Zhang, Z., et al. (2013). Assessment of shifts in microbial community structure and catabolic diversity in response to Rehmannia glutinosa monoculture. Appl. Soil Ecol. 67, 1–9. doi: 10.1016/j.apsoil.2013.02.008
Xu, Z. M., Wang, J. F., Li, W. L., Wang, Y. F., He, T., Wang, F. P., et al. (2021). Nitrogen fertilizer affects rhizosphere Cd re-mobilization by mediating gene AmALM2 and AmALMT7 expression in edible amaranth roots. J. Hazardous Mater. 2021:126310. doi: 10.1016/j.jhazmat.2021.126310
Yao, H., Zhang, S., Zhou, W., Liu, Y., Liu, Y., and Wu, Y. (2020). The effects of exogenous malic acid in relieving aluminum toxicity in Pinus massoniana. Int. J. Phytoremed. 22, 669–678. doi: 10.1080/15226514.2019.1707162
Yuan, J., Raza, W., and Shen, Q. (2018). Root Exudates Dominate the Colonization of Pathogen and Plant Growth-Promoting Rhizobacteria. Berlin: Springer, 167–180. doi: 10.1007/978-3-319-75910-4_6
Zhang, C., and Kong, F. (2014). Isolation and identification of potassium-solubilizing bacteria from tobacco rhizospheric soil and their effect on tobacco plants. Appl. Soil Ecol. 82, 18–25. doi: 10.1016/j.apsoil.2014.05.002
Zhang, M., Pan, L., Huang, F., Gao, S., Su, C., Zhang, M., et al. (2019). Metagenomic analysis of composition, function and cycling processes of microbial community in water, sediment and effluent of Litopenaeus vannamei farming environments under different culture modes. Aquaculture 506, 280–293. doi: 10.1016/j.aquaculture.2019.03.038
Keywords: malic acid, CLPP, Illumina MiSeq sequencing, nutrient absorption, total soluble sugar, titratable acid, Bacillaceae, Woeseiaceae
Citation: Si P, Shao W, Yu H, Xu G and Du G (2022) Differences in Microbial Communities Stimulated by Malic Acid Have the Potential to Improve Nutrient Absorption and Fruit Quality of Grapes. Front. Microbiol. 13:850807. doi: 10.3389/fmicb.2022.850807
Received: 08 January 2022; Accepted: 31 March 2022;
Published: 19 May 2022.
Edited by:
Bernardo González, Adolfo Ibáñez University, ChileReviewed by:
Vlad Stoian, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaCopyright © 2022 Si, Shao, Yu, Xu and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Si, c2lwZW5nQGNhYXMuY24=; Guoqiang Du, Z2R1QGhlYmF1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.