- Department of Agriculture, Agribusiness, and Environmental Sciences, Citrus Center, Texas A&M University-Kingsville, Weslaco, TX, United States
Citrus Huanglongbing (HLB) or citrus greening, is the most destructive disease for citrus worldwide. It is caused by the psyllid-transmitted, phloem-limited bacteria “Candidatus Liberibacter asiaticus” (CLas). To date, there are still no effective practical strategies for curing citrus HLB. Understanding the mechanisms against CLas can contribute to the development of effective approaches for combatting HLB. However, the unculturable nature of CLas has hindered elucidating mechanisms against CLas. In this review, we summarize the main aspects that contribute to the understanding about the mechanisms against CLas, including (1) CLas virulence targets, focusing on inhibition of virulence genes; (2) activation of citrus host defense genes and metabolites of HLB-tolerant citrus triggered by CLas, and by agents; and (3) we also review the role of citrus microbiome in combatting CLas. Finally, we discuss novel strategies to continue studying mechanisms against CLas and the relationship of above aspects.
Introduction
Citrus Huanglongbing (HLB), or citrus greening, is the most destructive citrus disease worldwide. It is associated with three species of fastidious, phloem-restricted α-proteobacteria: “Candidatus Liberibacter asiaticus” (CLas), “Candidatus Liberibacter americanus”(CLam), and “Candidatus Liberibacter africanus” (CLaf), which are transmitted by the psyllids Diaphorina citri or Trioza erytreae (Jagoueix et al., 1994; Bové, 2006; Gottwald, 2010). CLas is the most prevalent species found in commercial citrus production regions, including the United States, China, and Brazil (Jagoueix et al., 1994; Bové, 2006; Gottwald, 2010; Bassanezi et al., 2020; Zhou, 2020). HLB symptomology include yellowing of shoots, blotchy mottled leaves, corky veins, malformed and discolored fruits, premature fruit drop, root loss, and eventually tree death (Wang and Trivedi, 2013; Blaustein et al., 2018). Unfortunately, no commercial citrus varieties are resistant to HLB.
The HLB epidemic has affected all major citrus growing regions in the world (Hodges and Spreen, 2012; Kumagai et al., 2013; da Graça et al., 2015; Graham et al., 2020). In the United States, Florida has been the most affected citrus producing state. Since HLB arrival in 2005 (Bové, 2006), citrus production in Florida has decreased by 74% (Singerman and Rogers, 2020). Production losses due to HLB have resulted in the reduction of citrus growers from 7,389 in 2002 to 2,775 in 2017, juice processing facilities from 41 in 2003/2004 to 14 in 2016/2017, and packinghouses from 79 to 26 during the same period (Singerman and Rogers, 2020). In China, HLB was first reported in Guangdong province nearly a century ago (Reinking, 1919). To date, HLB has occurred in 10 provinces in China, including Guangdong, Guangxi, Fujian, Zhejiang, Jiangxi, Hunan, Guizhou, Hainan, and Sichuan. Especially, citrus production in Guangdong, Guangxi, and Fujian have been affected by HLB for a long time (Zhou, 2020). In Brazil, HLB was first reported in São Paulo State in 2004 (Coletta-Filho et al., 2004). After the first HLB outbreak, the disease spread to the States of Minas Gerais, Paraná and Mato Grosso do Sul, causing reduction of citrus production (Bassanezi et al., 2020).
Currently, many strategies have been developed for HLB mitigation, including application of antimicrobials (Zhang et al., 2011a, 2012, 2014, 2021; Hu and Wang, 2016; Hu et al., 2018; Yang et al., 2018), thermotherapy (Hoffman et al., 2013; Fan et al., 2016; Yang et al., 2016b; Doud et al., 2017; Ghatrehsamani et al., 2019; Vincent et al., 2019), macro-and micronutrients (Spann and Schumann, 2009; Gottwald et al., 2012; Rouse et al., 2017; Mattos-Jr et al., 2020; Dong et al., 2021; Zhou et al., 2021), plant defense inducers (Canales et al., 2016; Li et al., 2016, 2019, 2021a; Hu et al., 2018; Wang, 2021), control of the insect vector (Grafton-Cardwell et al., 2013; Boina and Bloomquist, 2015; Cocuzza et al., 2017; Pierre et al., 2021), biocontrol (Trivedi et al., 2011; Hopkins and Wall, 2021; Nan et al., 2021; Poveda et al., 2021), and eradication of HLB symptomatic citrus trees (Bassanezi et al., 2013; Yuan et al., 2020). However, these strategies have shown limited success in field applications and effective HLB management remains a challenge. Three-pronged approach including control of the psyllid vector, aggressive removal of infected trees to reduce sources of the disease, and planting with HLB-free nursery stock, has proven successful in China and Brazil, and has resulted in drastic reductions in the proportion of symptomatic trees (Bové, 2006). While this approach was advocated early on in Florida’s HLB outbreak, it was deemed to be too expensive by most producers, who instead decided to maintain symptomatic trees as long as they were bearing usable fruit (Hall and Gottwald, 2011). In addition, non-uniform distribution of CLas within citrus tree (Tatineni et al., 2008; Li et al., 2009) makes early detection of CLas very difficult, which is crucial for the management of citrus HLB. Thus, breeding for HLB disease-resistance may provide the most effective and sustainable solution to combat HLB (Bové, 2006).
In order to develop novel and effective strategies to suppress HLB, it is important to understand the virulence mechanisms employed by CLas to be able to elucidate potential targets against the pathogen. In this review, we describe the different virulence mechanisms of CLas and strategies used to identify virulence inhibitors. We also discuss the role of plant defenses in conferring HLB tolerance and the potential role of the citrus microbiome against CLas. We conclude with a discussion about the new pathways for studying this uncultured bacterial pathogen.
CLas Virulence Targets
Most insights of CLas virulence and biological processes are derived from the genome sequence of CLas (Duan et al., 2009), and other related Liberibacters (Coyle et al., 2018). Many putative virulence factors have been identified by utilizing surrogate models, and several strategies also have been developed for targeting these virulence genes associated with CLas pathogenicity and survival.
Secretion Systems and Effectors
Systems capable of secreting bacterial proteins, called effectors, into host cells are among the most important virulence factors of bacterial pathogens. Protein effectors often suppress plant defenses or manipulate developmental processes within the host to benefit the pathogen (Jones and Dangl, 2006). CLas encodes type I secretion systems (T1SS), a complete general secretory pathway (Sec), and an autotransporter type V secretion system (T5SS), but lacks other secretion systems (Duan et al., 2009; Fagen et al., 2014; Wulff et al., 2014; Wang et al., 2017). The Sec machinery facilitates the majority of proteins transport across the cytoplasmic membrane and is essential for bacterial viability (Segers and Anné, 2011). The Sec apparatus also secretes important virulence factors in some plant-pathogenic bacteria. It has been reported that CLas has at least 86 proteins with functional Sec-dependent secretion signals (Prasad et al., 2016). Many of these proteins, also called Sec-delivered effectors (SDEs) are highly conserved in CLas genomes and exhibit differential expression patterns in the citrus host and the psyllid vector (Thapa et al., 2020). CLas Sec-delivered effector 1 (SDE1, CLIBASIA_05315), is conserved across CLas isolates with a typical Sec-dependent secretion signal (Pitino et al., 2016; Prasad et al., 2016; Pagliaccia et al., 2017). SDE1 is highly expressed in citrus relative to psyllid, indicating a plausible role in CLas colonization of citrus and HLB disease progression (Yan et al., 2013). SDE1 inhibits the enzymatic activity of citrus papain-like cysteine proteases (PLCPs), which regulate multiple processes in plants, including defense against microbial pathogens (Clark et al., 2018). Other studies also suggested that SDE1 contributes to CLas colonization and the development of leaf yellowing symptoms, possibly by promoting premature senescence in citrus (Pitino et al., 2016; Clark et al., 2020). Although, there is no evidence that targeting effectors would lead to CLas suppression, targeting the Sec system could inhibit protein translocation and have a significant effect on CLas virulence and survival. The SecA ATPase drives protein translocation when it is bound to the SecYEG complex (Economou and Wickner, 1994; Van den Berg et al., 2004). Based on characteristics of SecA, 20 small molecules against CLas were identified by molecular docking in silico, and five of these compounds were confirmed to have antimicrobial activity in vitro using Agrobacterium tumefaciens as culturable model (Akula et al., 2012). Using a similarity search methodology, 11 compounds were identified based on the five SecA inhibitors (Hu et al., 2016). Although these 11 compounds had poor aqueous solubility, they were coupled in a micro-emulsion to assess their antimicrobial activities on eight bacteria phylogenetically related to CLas (A. tumefaciens, Liberibacter crescens, Rhizobium etli, Bradyrhizobium japonicum, Mesorhizobium loti, and Sinorhizobium meliloti). The inhibitions obtained from these compounds were similar to those described for streptomycin (Hu et al., 2016). Thus, the compounds targeting SecA, could also inhibit protein translocation in CLas and have a significant effect on HLB suppression.
Transcriptional Regulators
The reduced genome of CLas has a small number of transcriptional regulators that if targeted by high affinity inhibitors could result in strong reduction of CLas fitness and survival (Table 1). For instance, the transcriptional regulator PrbP was identified and the genome of CLas and was shown to bind to specific promoter regions of CLas DNA as well as to interact with RpoB, the β subunit of RNA polymerase (Gardner et al., 2016). In vitro screening of chemical compounds that target this gene identified one compound, tolfenamic acid, that inhibited PrbP/RpoB interaction and PrbP DNA binding. Further evaluation showed that tolfenamic acid inhibited in vitro growth of L. crescens, affected viability of CLas in citrus leaf-soaking assays, and reduced CLas titers in infected seedlings causing the recovery of roots and canopy tissues (Gardner et al., 2016). The antimicrobial activity of Tolfenamic acid against CLas might be the result of targeting key regulatory components that inhibit multiple pathways for bacterial survival.
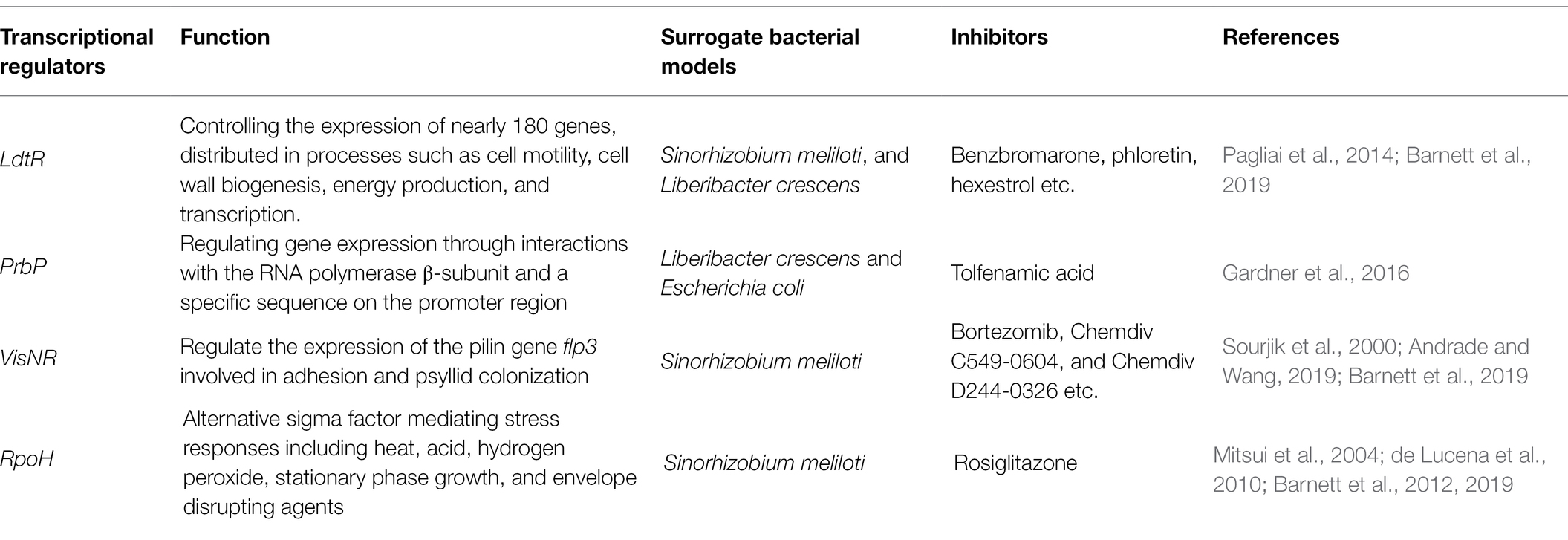
Table 1. Transcriptional regulators in uncultured bacteria Candidatus Liberibacter and inhibitors found in surrogate models for screening chemicals targeted the gene.
LdtR belongs to the MarR family transcription regulator and it has been linked to the regulation of more than 180 genes in Liberibacter species (Pagliai et al., 2017). In S. meliloti, mutation of LdtR resulted in morphological changes and reduced tolerance to osmotic stress. Small molecules including benzbromarone that targeted at LdtR were identified that caused a phenotype in S. meliloti and L. crescens similar with the insertional mutants (Pagliai et al., 2014). These small molecules were then assessed via a citrus shoot assay and shown to decrease the expression of LdtR and a gene regulated by LdtR potentially involved in cell wall biosynthesis. Therefore, application of small molecules that target LdtR, as a potential treatment option against citrus HLB.
As inhibition of transcriptional regulators provide an alternative method for mitigating CLas and HLB, a synthetic, high-throughput screening system to identify molecules that target CLas transcriptional regulators was developed (Barnett et al., 2019). This system used the closely related model bacterium, S. meliloti, as a heterologous host for expression of the CLas transcriptional activator, the activity of which was detected through expression of an enhanced green fluorescent protein (EGFP) gene fused to a target promoter. Around 120,000 compounds were screened by this system to target regulators including LdtR, RpoH, and VisNR and compounds that inhibited regulator activity were selected as candidate compound for combating HLB (Barnett et al., 2019). CLas sigma factor RpoH is most similar to RpoH1 in S. meliloti (72% identity), which mediate response to various stressors, including heat, acid, hydrogen peroxide, stationary phase growth, and envelope disrupting agents (Mitsui et al., 2004; de Lucena et al., 2010; Barnett et al., 2012). VisN and VisR, members of the LuxR transcriptional factor family, negatively regulate the expression of the CLas pilin gene flp3, which is associated with bacterial adherence and psyllid colonization (Andrade and Wang, 2019). Thus, targeting transcriptional regulators is a potential strategy for reducing CLas fitness and HLB mitigation.
Role of Prophage in CLas Survival
A prophage, also considered as a temperate phage, can integrate into the circular bacterial DNA chromosome, continuing this lysogenic cycle for as long as host physiology remains stable. However, stresses such as heat, UV light, starvation, or chemicals like antibiotics, which cause DNA damage to bacterial cells, activate the “SOS” stress response inducing the excision of phage DNA from the host (Oppenheim et al., 2005). Three prophage regions have been identified in CLas and have been classified as SC1, SC2, and SC3, based on genomic data (Zhang et al., 2011b; Zheng et al., 2016, 2018). SC1 carries putative lytic cycle genes, as phage particles in the phloem of infected periwinkle have been observed by transmission electron microscopy, although phage particles have not been observed in citrus (Fleites et al., 2014). SC2 lacks lytic cycle genes and can be integrated in the CLas genome or replicate as an excision plasmid prophage (Zhang et al., 2011b). Study of Zhang (2011b) indicated that SC1 and SC2 also encode multiple virulence factors that might contribute to the pathogenicity of CLas. Two predicated peroxidases are encoded by SC1 and SC2, which might detoxify CLas against reactive oxygen species (ROS), including superoxide radicals, hydrogen peroxide, and hydroxyl radicals. SC1 and SC2 also encode two predicated adhesins, which might be useful in transmission by psyllid (Zhang et al., 2011b). SC3 is not capable of reproduction via the lytic cycle. A restriction-modification (R-M) system of SC3 was speculated to play a role against Type 1 prophage-phage invasion (Zheng et al., 2018). The involvement of SC3 in survive of CLas still needs to be investigated.
Study of Ding et al. (2018) demonstrated that the relative copy number of both prophage SC1 and SC2 increased in HLB-affected host plants (citrus and periwinkle), in response to heat and antibiotic (tetracycline) treatments. These results suggest a potential mechanism for the activity of heat treatment and antibiotics against HLB through induction of CLas prophages causes lysis of CLas bacteria, reducing CLas population and mitigating HLB symptoms in citrus trees (Ding et al., 2018). Therefore, understanding the factors that trigger the lytic cycle in CLas prophages can provide a potential control strategy of citrus HLB.
Mechanisms of HLB-Tolerant Citrus to CLas
Citrus Huanglongbing affects all commercial citrus varieties, citrus species, and relatives (Bové, 2006). Nevertheless, several citrus cultivars and relatives have shown tolerance to CLas, and many studies have deciphered the mechanism of these tolerance to HLB (Table 2). Here, we would discuss host defense genes and metabolites against CLas (Figure 1).
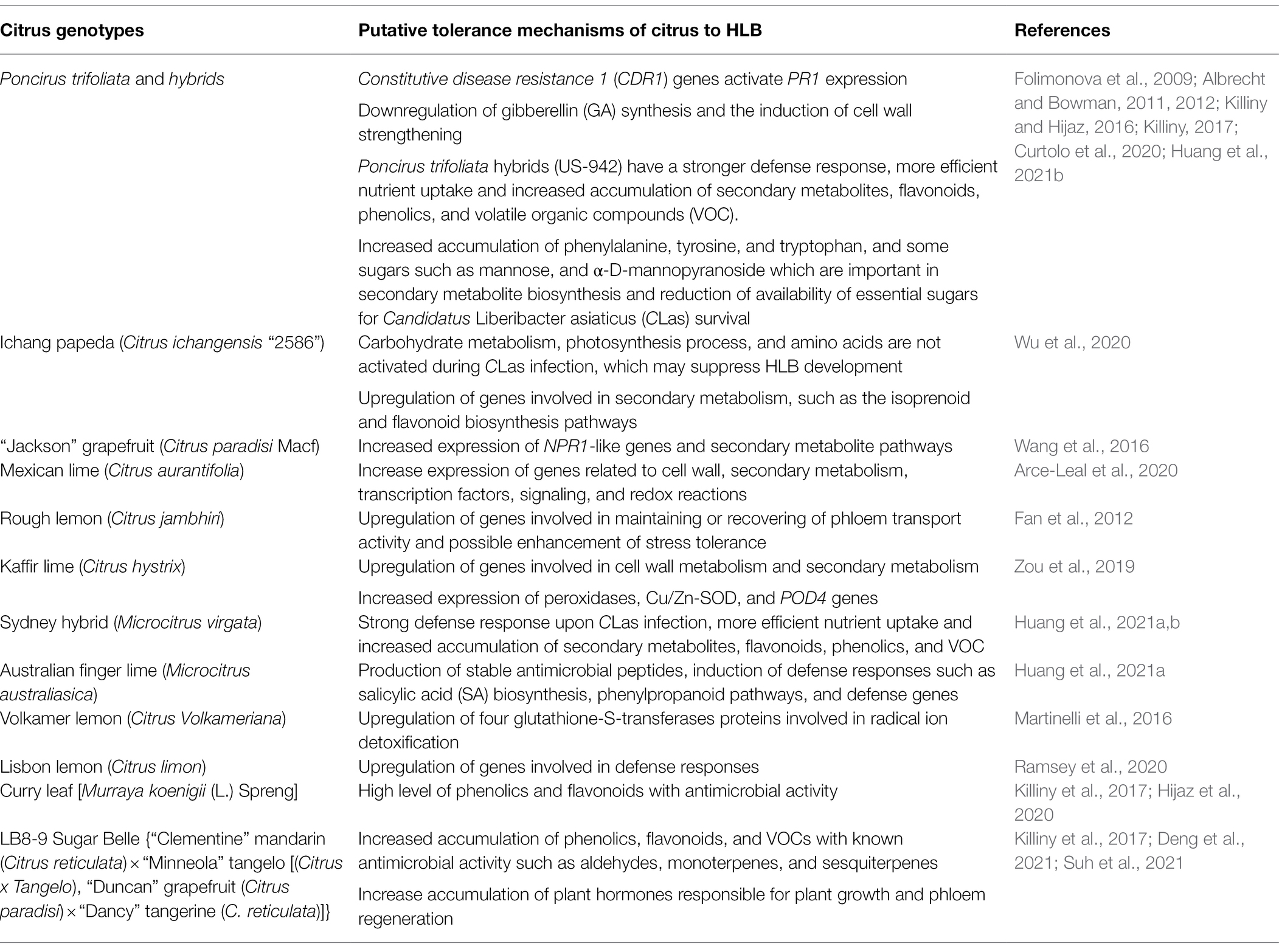
Table 2. The mechanisms of Citrus Huanglongbing (HLB)-tolerant citrus elucidated by multi-omics approaches.
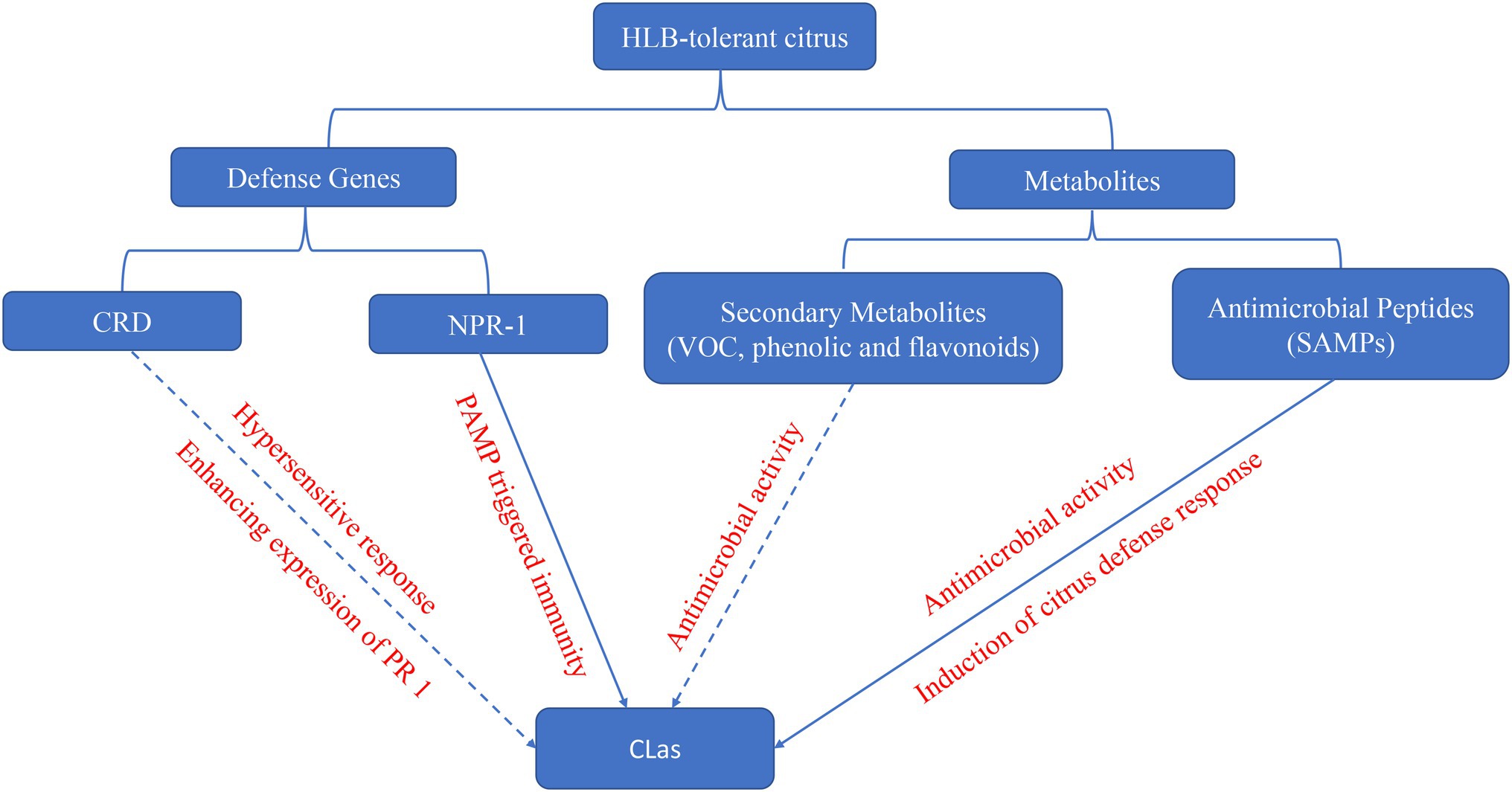
Figure 1. The potential mechanism of defense genes and metabolites in HLB-tolerant citrus against CLas. Solid line indicates that the functions were confirmed in citrus, and dash line indicates that the function were just confirmed in other species.
Citrus Defense Genes Involved in Combating CLas
Multiple defense genes in HLB-tolerant citrus have been identified by multi-omics approaches (Table 2), although just the function of Constitutive disease resistance (CDR) and Non-expressor of Pathogenesis Related genes 1 (NPR1) was confirmed in surrogate models or citrus.
Constitutive disease resistance genes belong to the plant aspartic proteinase (APs) gene family. CDR1 was first identified and cloned in Arabidopsis. Its product has been implicated in disease resistance signaling (Xia et al., 2004). Overexpression of a rice (Oryza sativa L) CDR 1 gene, led to constitutive activation of defense response and enhanced resistance in rice and Arabidopsis against bacterial and fungal pathogens (Prasad et al., 2009). Several studies have demonstrated that CDR1 as potential candidate genes for HLB tolerance in Poncirus (Albrecht and Bowman, 2012; Du et al., 2015; Rawat et al., 2015). A study was undertaken to mine and characterize the CDR gene family in Citrus and Poncirus and to understand its association with HLB tolerance in Poncirus. It found that PtCDR2 and PtCDR8 were high abundance in Poncirus leaf transcriptomes. The expression of PtCDR2 and PtCDR8 genes responded to CLas infection differently in HLB-tolerant and susceptible genotypes (Rawat et al., 2017). The role of PtCDR2 and PtCDR8 in disease resistance was confirmed in Arabidopsis mutants that showed that transformation of PtCDR2 and PtCDR8 into Arabidopsis cdr1 mutant induced PR1 expression and recovered the hypersensitive response to Pseudomonas syringae pv. tomato strain DC3000 (Ying et al., 2020). Therefore, PtCDR2 and PtCDR8 play a key role in plant defense responses and serve as strong candidate genes for engineering citrus for HLB disease tolerance.
Non-expressor of Pathogenesis Related genes 1 gene is a key regulator in the signal transduction pathway that leads to SAR response. The NPR1 gene may act as a regulator of the transcription factor/s that controls PR gene expression (Kinkema et al., 2000) and mediates the salicylic acid (SA) induced expression of PR genes and SAR (Clarke et al., 1998). Plants over expressing NPR1 display enhanced resistance to several pathogens (Cao et al., 1998). For instance, transcriptome profiling of HLB-tolerant “Jackson” (grapefruit hybrid) and HLB-susceptible “Marsh” grapefruit found that four NPR1-like genes were significantly upregulated in HLB tolerant citrus trees (Wang et al., 2016). Furthermore, transgenic sweet orange cultivars “Hamlin” and “Valencia” expressing an A. thaliana npr1 gene under the control of a constitutive CaMV 35S promoter or a phloem specific Arabidopsis SUC2 (AtSUC2) promoter resulted in trees with normal phenotypes that exhibited enhanced resistance to HLB. Additionally, the transgenic trees exhibited reduced diseased severity and a few lines remained disease-free even after 36 months of planting in a high-disease pressure field site (Dutt et al., 2015). AtNPR1 can enhance expression of transcription of genes encoding pathogen-associated molecular patterns (PAMPs), transcription factors, leucine-rich repeat receptor kinases (LRR-RKs), and putative ankyrin repeat-containing proteins, in AtNPR1 transgenic line compared to the control plant (Qiu et al., 2020). These results suggested that NPR1 positively regulates the innate defense mechanisms in citrus, contributing to enhance tolerance to citrus HLB.
Activation of Antimicrobial Metabolites
Plants have a number of unique defense mechanisms including physical barriers to pathogen invasion as well as a wide range of secondary metabolites and antimicrobial peptides (AMP). Secondary metabolites have long been suggested to interact with pathogen (Hartmann, 2008). Several studies have revealed a vast number of secondary metabolites with proven or putative functions in plant responses to pathogen microorganisms (Piasecka et al., 2015). In several HLB-tolerance citrus cultivars, the transcriptomic analysis reveals that most differentially expressed genes (DEGs) increase in secondary metabolites pathways from HLB-tolerant citrus including Poncirus trifoliata and its hybrids (Albrecht and Bowman, 2012), “Jackson” (grapefruit hybrid; Wang et al., 2016), Mexican lime (Citrus aurantifolia; Arce-Leal et al., 2020), and Kaffir lime (Citrus hystrix; Zou et al., 2019). The secondary metabolites are higher in HLB-affected tolerant citrus cultivars, indicating a strong relationship between HLB-tolerance and secondary metabolites accumulation (Rao et al., 2018). In addition, amino acids including phenylalanine, tyrosine, and tryptophan were accumulated on HLB-tolerant citrus relative P. trifoliata (Killiny and Hijaz, 2016), which are involved in synthesis of many secondary metabolites. Furthermore, several studies demonstrated that HLB-tolerant citrus including US-942 (P. trifoliata×Citrus reticulata), Curry leaf [Murraya koenigii (L.) Spreng], and LB8-9 Sugar Belle contained high level of secondary metabolites such as volatile organic compounds (VOC), phenolics, and flavonoids (Killiny et al., 2017; Hijaz et al., 2020; Deng et al., 2021; Huang et al., 2021b).
Volatile organic compounds play a key role in protecting plants under insect and pathogen attack. VOCs, including aldehydes, monoterpenes, sesquiterpenes, thymol, b-elemene, and (E)-b-caryophyllene, have antimicrobial activities against pathogens, and accumulate in HLB-tolerant LB8-9 Sugar Belle (Killiny, 2017; Deng et al., 2021). Phenolics are a group of secondary metabolites, which are produced via the shikimic acid pathway through the phenylpropanoid pathway (Lin et al., 2016). It has been demonstrated that the accumulation of phenolic compounds at the infection site could result in pathogen restriction and prevention of their spread to other plant’s tissues (Nicholson and Hammerschmidt, 1992). Flavonoids are widely distributed in plants and they are synthesized in the cytosol through the phenylpropanoid pathway by a set of enzymes (Treutter, 2006; Petrussa et al., 2013). Flavonoids could exhibit their resistance to pathogens by inhibition and crosslinking of the microbial enzymes, chelation of metals necessary for enzyme activity, and formation of physical barrier (Treutter, 2006). The HLB-tolerant Curry leaf [M. koenigii (L.) Spreng] and LB8-9 Sugar Belle contain high level of phenolics and flavonoids, which correlate with their enhanced tolerance to CLas (Killiny et al., 2017; Hijaz et al., 2020). Therefore, increased levels of VOCs, phenolics, and flavonoids in citrus may contribute to HLB tolerance.
Antimicrobial peptides stand out as one of the most prominent components of the plant immune system. These small and usually basic peptides are deployed as a generalist defense strategy that grants direct and durable resistance against plant pathogens. A recent study identified a novel class of heat stable antimicrobial peptides (SAMPs), from HLB-tolerant citrus Australian finger lime (Microcitrus australiasica). SAMPs not only effectively reduced CLas titer and disease symptoms in HLB-positive trees but also prevented and inhibited infections by induction of defense response genes such as PR1 and PR2, an enzyme of SA biosynthesis, phenylpropanoid pathways, and phenylalanine ammonia-lyase 1(PAL; Huang et al., 2021a). Thus, HLB-tolerant citrus can also be a source of defense peptides against CLas.
Host Defense Triggered by Agents
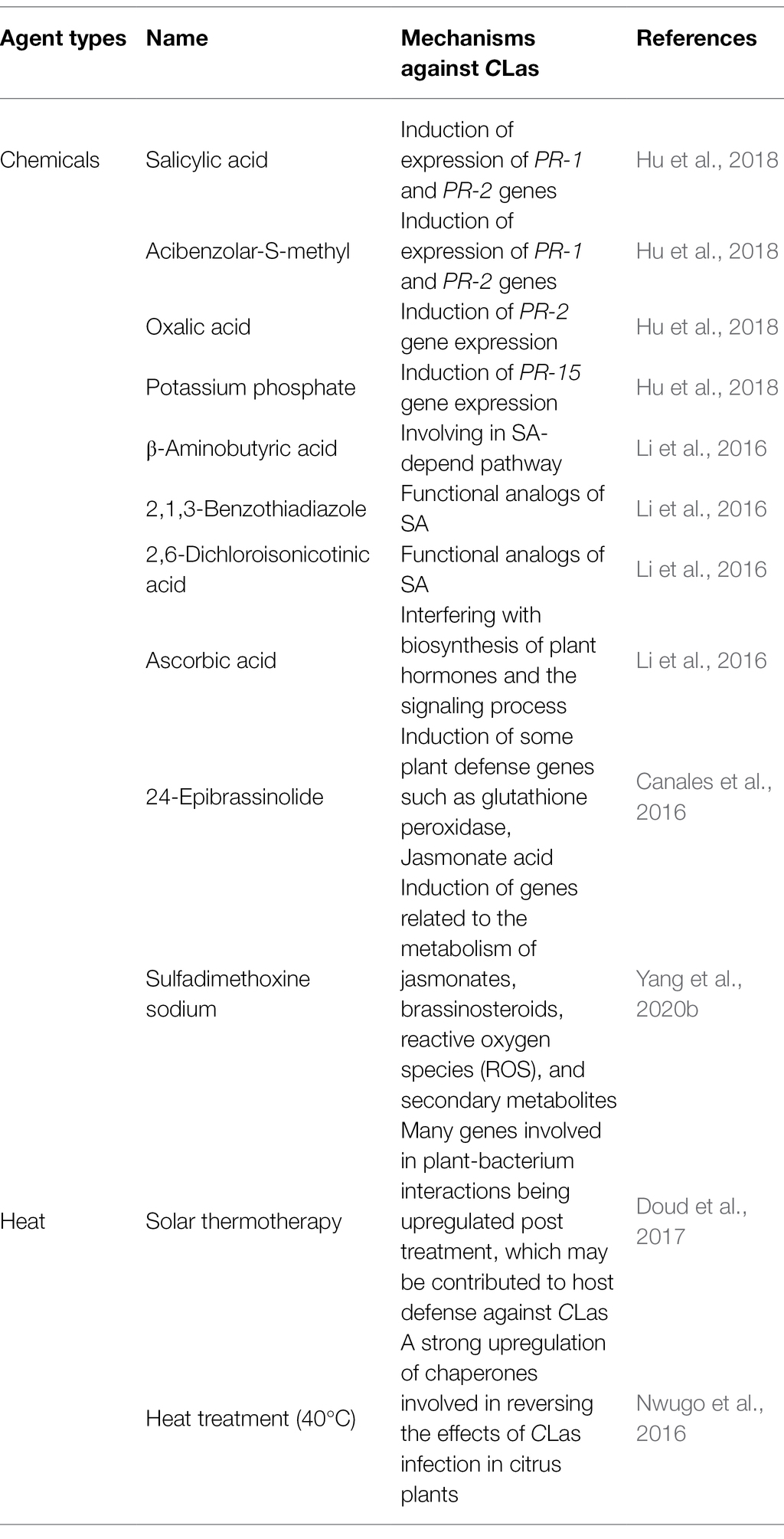
Citrus defense mechanisms not only can be activated by pathogens, but also induced by agents such as chemical compounds, heat and nutrients. Systemic acquired resistance (SAR) can be useful to control of several plant diseases (Ryals et al., 1996; Sticher et al., 1997; Durrant and Dong, 2004). SAR involves in a specific defense signaling pathway that required SA and is associated with accumulation of pathogenesis-related proteins (PR). Several chemical compounds can activate SAR in plant. Four SAR activators including SA, oxalic acid, acibenzolar-S-methyl, and potassium phosphate, provided significant control of HLB by suppressing CLas titer and disease progress when applied by trunk injection (Hu et al., 2018). Furthermore, both SA and acibenzolar-S-methyl significantly induced expression of PR-1 and PR-2 genes, and oxalic acid and potassium phosphate resulted in significant induction of PR-2 and PR-15 gene expression, respectively (Hu et al., 2018). In addition, foliar spray application of several plant defense inducers [i.e., β-aminobutyric acid (BABA), 2,1,3-benzothiadiazole (BTH), 2,6-dichloroisonicotinic acid (INA), and ascorbic acid (AA)] were reported to suppress progress of HLB in the field. BTH and INA, which are functional analogs of SA, can induce plant defenses in citrus. The effect control of BABA on citrus HLB may be in SA-depend pathway. Furthermore, AA may alleviate HLB symptoms by interfering with biosynthesis of plant hormones (including salicylic acid and jasmonic acid; Li et al., 2016). Other plant hormones, such as brassinosteroids, can induce plant defenses against a wide range of pathogens including CLas. Foliar spray of brassinosteroid (24-epibrassinolide) in greenhouse and field experiments of HLB-affected citrus showed CLas titer was reduction after treatment under both conditions (Canales et al., 2016). Moreover, several chemical compounds have antimicrobial activities against CLas, and can also induce plant defense against the pathogen. Sulphonamide antibiotics such as sulfadimethoxine sodium (SDX) and sulfathiazole sodium (STZ) have been proved to be effective against CLas (Zhang et al., 2014; Yang et al., 2016a). Transcriptomic analysis of citrus plants revealed that SDX can induce genes related to the metabolism of jasmonates, brassinosteroids, ROS, and secondary metabolites, which are beneficial for resistance against HLB (Yang et al., 2020b; Table 3).
Candidatus Liberibacter asiaticus is a heat-tolerant bacterium and can thrive under high temperature conditions extending to 35°C (Lopes et al., 2009). Many studies demonstrated that heat treatment (temperature ranged from 40 to 50°C) can eliminate or suppress CLas titer in HLB-affected citrus (Hoffman et al., 2013; Fan et al., 2016; Yang et al., 2016a,b). Moreover, the heat treatment also can enhance vigor of HLB-affected citrus and promote new flush (Hoffman et al., 2013; Yang et al., 2016b; Armstrong et al., 2021). Transcriptome analysis has shown that the gene expression profiles of HLB-affected trees post solar-heat treatment more closely modeled healthy trees than their gene profiles prior to treatment, with many genes involved in plant-bacterium interactions being upregulated post treatment, which may contribute to host defense against CLas (Doud et al., 2017). In addition, proteomics analysis indicated that a strong upregulation of chaperones including small (23.6, 18.5, and 17.9 kDa) heat shock proteins, a HSP70-like protein and a ribulose-1,5-bisphosphate carboxylase oxygenase (RuBisCO)-binding 60 kDa chaperonin, in response to heat treatment (40°C), which has been involved in reversing the effects of CLas infection in citrus plants (Nwugo et al., 2016; Table 3).
For several years, it has been reported that the application of enhanced nutritional products can extend the vigor of HLB-affected citrus and trigger citrus defense against CLas (Spann and Schumann, 2009; Shen et al., 2013; da Silva et al., 2020; Shahzad et al., 2020; Dong et al., 2021). Although nutrient treatments have no effect on reducing CLas titer and cannot enhance yield of HLB-affected citrus in the field (Gottwald et al., 2012; da Silva et al., 2020; Phuyal et al., 2020), the application of macro-and micronutrients have been adopted worldwide as they induce host defenses and help maintain production of HLB-affected trees. For instance, a field study in Florida showed application of phosphorus (P) oxyanion solutions to HLB-affected citrus mitigated disease symptom severity during a 3-year field trial (Zhao et al., 2013). It is known that phosphite has a direct action on plant defense mechanisms by the activation of the PAL activity and the biosynthesis of phytoalexins (Saindrenan and Guest, 1994), which may be involved in citrus defense induced by P. HLB-affected citrus display interveinal chlorotic leaves due to iron (Fe) deficiency caused by CLas (Masaoka et al., 2011) and foliar application of Fe2+ have shown to alleviate symptoms of HLB-affected citrus trees (Inoue et al., 2020). In other pathosystems, such as in rice-Magnoporthe interactions, rice plants growing at high Fe levels have enhanced resistance against the fungus. Although this has not been evaluated, application of Fe may induce host defense against CLas (Peris-Peris et al., 2017). Recent research indicates that elevated levels of manganese (Mn) promote better tree response to the effects of HLB increasing citrus tree lifespan (Morgan et al., 2016; Zambon et al., 2019). Sufficient Mn in the rhizosphere is critical for scavenging ROS (Alscher et al., 2002), which is known to be produced extensively in CLas-damaged cells (Ma et al., 2022). Although many nutrients can mitigate symptoms of HLB-affected trees, the mechanisms of how these nutrients trigger citrus defenses are still unclear and warrant investigation.
Role of Citrus Microbiome in Combatting CLas
The plant microbiome is an important contributor to plant health and defense against pathogens. Plant-associated microbiota can suppress pathogens through direct competition, producing antimicrobial compounds or stimulating plant immunity to resist or tolerate pathogen infection (Saikkonen et al., 2004; Kaul et al., 2016; Brader et al., 2017). To date, a plethora of studies have focused on deciphering the role of the citrus microbiome with the goal of identifying members of the microbial community associated with HLB and CLas suppression. However, comparison of microbiomes from healthy and HLB-affected citrus have shown that CLas affects the microbial community structure and reduce the putative beneficial microbe associations within citrus leaves and roots (Trivedi et al., 2011; Zhang et al., 2017; Ginnan et al., 2020; Yan et al., 2021).
For example, CLas infection in mandarin leaves (C. reticulata cv. Shatangju) causes reduction of several beneficial bacteria genera including Variovorax, Novosphingobium, Methylobacillus, Methylotenera, and Lysobacters, which are known to be involved in promoting plant growth and antibiotic production (Yan et al., 2021). Study of Blaustein (2017)identified citrus-health-associated endophytes of leaves (such as Methylpbacterium, Burkholderia, and Sphingomonas) and roots (Bradyrhizobiaceae) based on increased relative abundances in healthy vs. HLB-diseased citrus trees. These potential beneficial microbes are known to be involved in competing with pathogens for nutrients, antagonize pathogens through antibiosis, assist the host with nutrient acquisition, and induce host defense responses (Compant et al., 2005; Madhaiyan et al., 2006; Enya et al., 2007; Lugtenberg and Kamilova, 2009; Verma et al., 2010; Innerebner et al., 2011; Ardanov et al., 2012). However, their reduction in HLB-affected citrus provides insights into the role of the microbial community into HLB progression.
Interestingly, inoculations of Burkholderia stains isolated from the rhizosphere of healthy citrus roots can induce the expression of genes involved in activation of citrus defenses and SA mediated induced systemic resistance (Zhang et al., 2017). Other studies have shown that Bacillus sp. can also induce host defense responses against CLas through enhancing expression of several transcription factors involved in disease resistance (Tang et al., 2018; Munir et al., 2020). Moreover, when the biocontrol agent Xylella fastidiosa strain EB92-1 was applied to HLB-affected citrus plants, the results indicated that it could reduce the incidence of HLB symptoms in mature trees through 18 months after inoculation and the incidence of severe symptoms up to 3 years (Hopkins and Wall, 2021). Although the mechanism of HLB suppression by X. fastidiosa EB92-1 and the other bacteria remain to be studied in depth, these studies show that beneficial bacteria can be used to suppress CLas and improve plant health by induction of plant defenses that confer broad-spectrum resistance against pathogens.
Manipulation of the citrus microbiome to enrich the populations of beneficial microbes in HLB-affected trees could aid in disease suppression. Actually, nutrients play a role in activating plant immunity system by altering the microbial community structure and the metabolism (Sugimoto et al., 2010; Shi et al., 2012; Huber and Jones, 2013). A recent study has reported that application of calcium, magnesium, and boron to the soil can alter microbial structure and communities in phyllosphere and rhizosphere of HLB-affected citrus and promoted beneficial microorganism (Burkholderiaceae, Xanthomonas, and Stenotrophomonas) enrichment, which may have contributed to the reduced HLB incidence, and CLas titers (Zhou et al., 2021).
Chemotherapy is another method that can shape the citrus microbiome. Antimicrobial activity of the effective antibiotics against CLas have been associated with shifts in endophytic microbial structure and communities in HLB-affected citrus after treatment (Zhang et al., 2014; Yang et al., 2015, 2020a; Li et al., 2019). Foliar application of penicillin and oxytetracycline to HLB-affected citrus, caused an increase in the relative abundance of beneficial bacterial species, including Streptomyces avermitilis and Bradyrhizobium, compared to those treated with water control (Yang et al., 2020a). Moreover, the relative abundance of the bacterial species associated with CLas survival, such as Propionibacterium acnes and Synechocystis sp. PCC 6803, was lower in penicillin and oxytetracycline treated plants compared to the control (Yang et al., 2020a). Other studies have shown that the endophytic microbiome was altered in HLB-affected scion treated with ampicillin, and 10 abundant operational taxonomic units (OTUs) from antibiotic producing Stenotrophomonas spp. were only detected in the ampicillin-treatment (Zhang et al., 2013). Study of Ascunce et al. (2019) also showed that Bacilli, involved in the elicitation of plant defenses against pests and pathogens, were relatively more abundant in petioles and roots from penicillin treated HLB-affected citrus. Moreover, it was also found that the endophytic microbiome was changed in HLB-affected citrus plants under heat and sulfonamide (sulfathiazole sodium–STZ, and sulfadimethoxine sodium—SDX) treatments (Yang et al., 2016a). Following antibiotic treatment with SDX and STZ, there was enhanced abundance of OTUs belonging to the families Streptomycetaceae, Desulfobacteraceae, Chitinophagaceae, and Xanthomonadaceae, which are beneficial for control of plant pathogens and promoting plant growth (Hell, 1997; Kim and Jung, 2007; Mhedbi-Hajri et al., 2011; Mendes et al., 2013). Therefore, the enrichment of beneficial bacteria in these antibiotic treatments, may be contributed to their antimicrobial activity against CLas.
It is clear that the citrus microbiome plays a key role in citrus health. Whether some bacteria have an effect in survival of CLas is still unclear. The enrichment of beneficial bacteria in healthy citrus or in response to effective chemical compounds, may be involved in combating CLas. However, more studies are needed to validate the role of beneficial bacteria in citrus, and identify antagonistic bacteria against CLas (Trivedi et al., 2011; Riera et al., 2017; Zhang et al., 2017). Therefore, isolation and identification of the enriched beneficial bacteria can provide more insight into the role of citrus microbiome in HLB mitigation.
New Approaches for Studying Mechanisms Against Uncultured Bacterial Pathogens
Despite the advances in uncovering virulence mechanisms of CLas, identification of genes conferring disease tolerance, discovering potential antagonistic bacteria, and identifying many small molecules that inhibit CLas, we are still far from deploying sustainable solutions to the HLB epidemic.
Establishing CLas in culture can provide an extended vision in mechanism of agents against CLas. Although several reports of transient CLas cultures have been published, most of these attempts have only been able to maintain CLas in coculture (Davis et al., 2008; Parker et al., 2014; Fujiwara et al., 2018; Ha et al., 2019; Merfa et al., 2019). These studies partially fulfilled Koch’s postulates and could potentially be used to unravel the complex relationships of CLas with other citrus endophytes; however, no follow-up research using these approaches to obtain a pure CLas culture has been published. Currently, the methods employed to study the mode of action of small molecules with antimicrobial activity have been elucidated in vitro using as culturable surrogate models such as L. crescens and S. meliloti (Pagliai et al., 2014; Barnett et al., 2019). However, all sequenced CLas strains have reduced genome size of about 1.2 Mb (Duan et al., 2009; Thapa et al., 2020), compared with the slightly larger 1.5 Mb of L. crescens BT-1 (Leonard et al., 2012), and the about 6.7 Mb genome of the phylogenetically related S. meliloti (Sugawara et al., 2013) which could cause differences in biosynthetic pathways, metabolic enzymes, and secretion systems. Therefore, novel approaches are needed to uncover how chemicals, nutrition, beneficial microorganisms or hosts, directly or indirectly affect CLas. One such approach is the recent development of a plant hairy root system that mimics the host environment and supports the growth of CLas (Irigoyen et al., 2020). This system was developed as a tool for high throughput screening of antimicrobials against CLas and Candidatus Liberibacter solanacearum (CLso), which is faster and more reliable compared to conventional compound screening approaches (Irigoyen et al., 2020). Thus, this system could also be used as a model to study the mode of action of antimicrobials against CLas inside the citrus host.
Uncovering the complex interactions of CLas and the host, is key to discover pathways that can be exploited for disease suppression. However, genome-wide transcriptome profiling of a phloem-restricted pathogen in planta is very difficult, since the bacterial mRNA constitutes a minor fraction of the total mRNA. Thus, most research has focused on gene expression of the citrus host, and smaller number of studies describe global gene expression profiles of the pathogen. To examine the expression profiles of CLas in the host, most studies rely on quantitative reverse transcription-polymerase chain reaction approaches which only address targeted genes. In vivo transcriptomic analyses are required to understand the active pathways in CLas. A recent study identified the regions in the citrus fruit pith with higher bacterial titers which was used to conduct RNA-seq analysis after rRNA removal (Fang et al., 2021). This study compared the gene expression profiles of the fruit pit vs. leaf midribs and found different gene expression profiles related to virulence genes; however, the resolution of the transcriptome profile was lower in midribs compared to fruit pit mainly due to the lower bacterial titers (Fang et al., 2021).
Because the main limitation of conducting transcriptomic profiles of CLas is bacterial titers, different enrichment approaches are being developed. A CLas enrichment system using dodder showed about 419-fold CLas titer increase in dodder system as compared to the corresponding citrus hosts, and the dual RNA-seq data indicated that similar CLas gene expression profiles in dodder and citrus samples, yet dodder samples generated a higher solution than those obtained in citrus host (Li et al., 2021b). Although the CLas-enrichment dodder system could be used as surrogate model for studying interaction of CLas and host, dodder defense system against CLas is very different from citrus. To overcome the limitation of surrogate systems, a bacterial cell enrichment procedure has been developed for transcriptome profiling of CLas in citrus in which bacteria is isolated from citrus samples prior to RNA extraction, reaching detectable expression to 84% of the CLas genome coverage (De Francesco et al., 2022). This CLas-enrichment method will be useful for mechanisms of CLas within the citrus host and for elucidating potential targets for CLas suppression.
Conclusion
The mechanisms to study phloem-limited and uncultured plant bacterial pathogens are a complicated process. The development of novel approaches to understand the virulence mechanisms of the pathogen, the mode of action of antimicrobial therapies, the interactions with host and other endophytic microbes will aid in the search of effective and sustainable methods to combat CLas and ultimately HLB.
Until we unravel the mechanistic black box in the interactions between citrus phloem and CLas, the combination of effective agents including chemicals, nutrition, and plant defense activators will continue to be the only path to combat HLB (Figure 2). To effectively combat HLB, multiple strategies need to be applied against CLas: (1) the use of antimicrobial agents that directly disturb the biological processes of CLas, thus affecting bacterial survival; (2) the use of chemical agents that suppress CLas by inducing citrus host defense systems; (3) modifying the environment by agents to promote the enrichment of beneficial bacteria to antagonize CLas; and (4) the enrichment of beneficial bacteria that trigger citrus defense system against CLas. These pathways may work separately or together to promote tree health, mitigate HLB and recover tree productivity. Therefore, the relationship of virulence targets, citrus defenses and microbiome plays a key role in elucidating mechanisms against CLas.
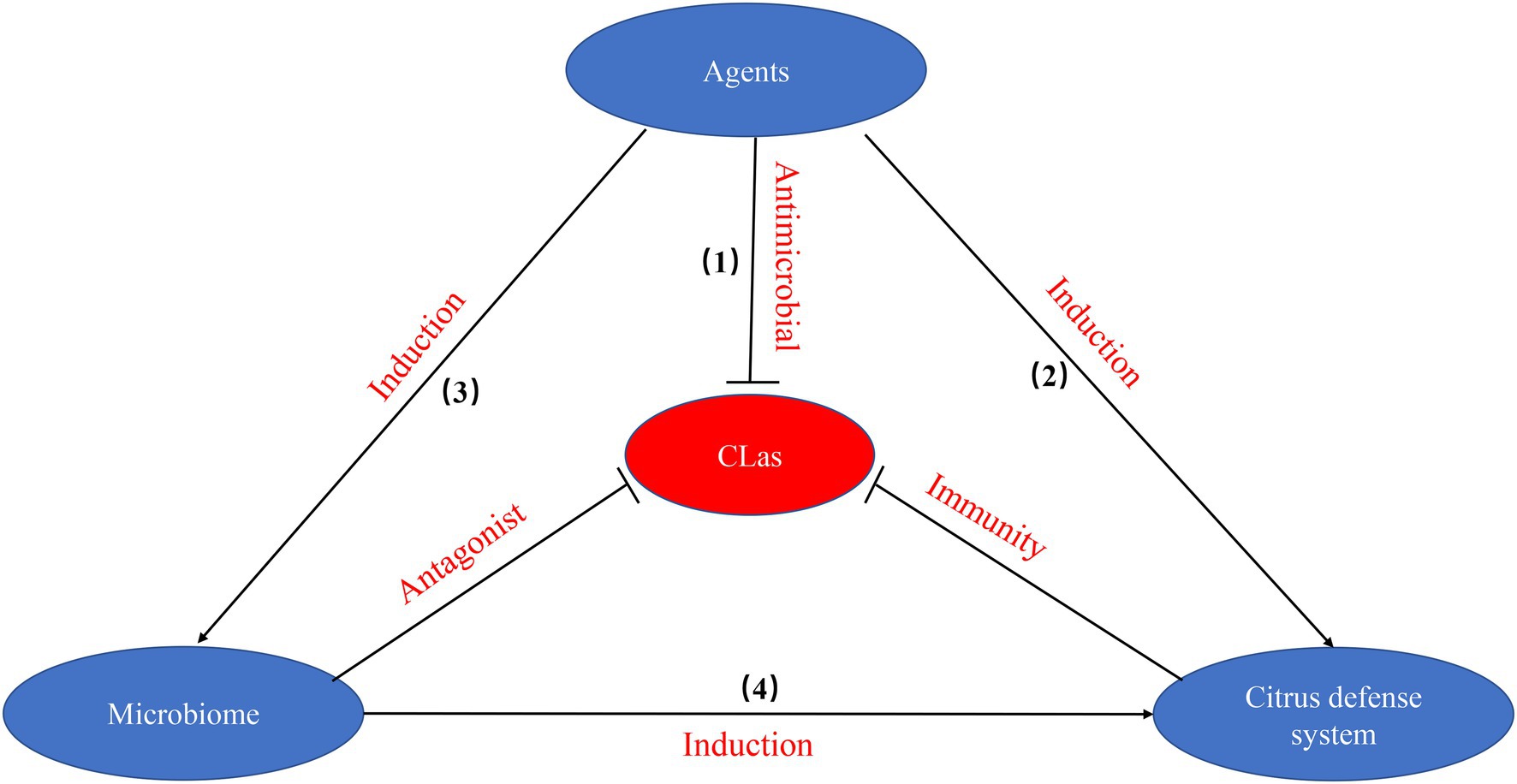
Figure 2. The pathways involved in effective agents including chemicals, nutrition, and plant defense activators against CLas.
Author Contributions
CY and VA contributed to writing and editing this manuscript. All authors read and approved the final version of the manuscript.
Funding
This study was supported by funds from the USDA-NIFA-SCRI (2019-70016-29096, 2018-70016-28198, and 2016-70016-24833) to VA.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Akula, N., Trivedi, P., Han, F. Q., and Wang, N. (2012). Identification of small molecule inhibitors against SecA of Candidatus Liberibacter asiaticus by structure based design. Eur. J. Med. Chem. 54, 919–924. doi: 10.1016/j.ejmech.2012.05.035
Albrecht, U., and Bowman, K. D. (2011). Tolerance of the trifoliate citrus hybrid US-897 (Citrus reticulata Blanco× Poncirus trifoliata L. Raf.) to huanglongbing. HortScience 46, 16–22. doi: 10.21273/HORTSCI.46.1.16
Albrecht, U., and Bowman, K. D. (2012). Transcriptional response of susceptible and tolerant citrus to infection with Candidatus Liberibacter asiaticus. Plant Sci. 185–186, 118–130. doi: 10.1016/j.plantsci.2011.09.008
Alscher, R. G., Erturk, N., and Heath, L. S. (2002). Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 53, 1331–1341. doi: 10.1093/jexbot/53.372.1331
Andrade, M., and Wang, N. (2019). The tad pilus apparatus of ‘Candidatus Liberibacter asiaticus’ and its regulation by VisNR. Mol. Plant-Microbe Interact. 32, 1175–1187. doi: 10.1094/MPMI-02-19-0052-R
Arce-Leal, Á. P., Bautista, R., Rodríguez-Negrete, E. A., Manzanilla-Ramírez, M. Á., Velázquez-Monreal, J. J., Santos-Cervantes, M. E., et al. (2020). Gene expression profile of Mexican lime (Citrus aurantifolia) trees in response to huanglongbing disease caused by Candidatus Liberibacter asiaticus. Microorganisms 8:528. doi: 10.3390/microorganisms8040528
Ardanov, P., Sessitsch, A., Häggman, H., Kozyrovska, N., and Pirttilä, A. M. (2012). Methylobacterium-induced endophyte community changes correspond with protection of plants against pathogen attack. PLoS One 7:e46802. doi: 10.1371/journal.pone.0046802
Armstrong, C. M., Doud, M. S., Luo, W., Raithore, S., Baldwin, E. A., Zhao, W., et al. (2021). Beneficial horticultural responses from the application of solar thermotherapy to mature huanglongbing-affected citrus trees. Horticult. Plant J. 7, 411–422. doi: 10.1016/j.hpj.2021.04.008
Ascunce, M. S., Shin, K., Huguet-Tapia, J. C., Poudel, R., Garrett, K. A., Van Bruggen, A. H., et al. (2019). Penicillin trunk injection affects bacterial community structure in citrus trees. Microb. Ecol. 78, 457–469. doi: 10.1007/s00248-018-1302-1
Barnett, M. J., Bittner, A. N., Toman, C. J., Oke, V., and Long, S. R. (2012). Dual RpoH sigma factors and transcriptional plasticity in a symbiotic bacterium. J. Bacteriol. 194, 4983–4994. doi: 10.1128/JB.00449-12
Barnett, M. J., Solow-Cordero, D. E., and Long, S. R. (2019). A high-throughput system to identify inhibitors of Candidatus Liberibacter asiaticus transcription regulators. Proc. Natl. Acad. Sci. U. S. A. 116, 18009–18014. doi: 10.1073/pnas.1905149116
Bassanezi, R., Belasque, J., and Montesino, L. (2013). Frequency of symptomatic trees removal in small citrus blocks on citrus huanglongbing epidemics. Crop Prot. 52, 72–77. doi: 10.1016/j.cropro.2013.05.012
Bassanezi, R. B., Lopes, S. A., De Miranda, M. P., Wulff, N. A., Volpe, H. X. L., and Ayres, A. J. (2020). Overview of citrus huanglongbing spread and management strategies in Brazil. Trop. Plant Pathol. 45, 251–264. doi: 10.1007/s40858-020-00343-y
Blaustein, R. A., Lorca, G. L., Meyer, J. L., Gonzalez, C. F., and Teplitski, M. (2017). Defining the core citrus leaf-and root-associated microbiota: Factors associated with community structure and implications for managing huanglongbing (citrus greening) disease. Appl. Environ. Microbiol. 83, e00210–00217. doi: 10.1128/AEM.00210-17
Blaustein, R. A., Lorca, G. L., and Teplitski, M. (2018). Challenges for managing Candidatus Liberibacter spp. (huanglongbing disease pathogen): current control measures and future directions. Phytopathology 108, 424–435. doi: 10.1094/PHYTO-07-17-0260-RVW
Boina, D. R., and Bloomquist, J. R. (2015). Chemical control of the Asian citrus psyllid and of huanglongbing disease in citrus. Pest Manag. Sci. 71, 808–823. doi: 10.1002/ps.3957
Bové, J. M. (2006). Huanglongbing: a destructive, newly-emerging, century-old disease of citrus. J. Plant Pathol. 88, 7–37. doi: 10.4454/jpp.v88i1.828
Brader, G., Compant, S., Vescio, K., Mitter, B., Trognitz, F., Ma, L.-J., et al. (2017). Ecology and genomic insights into plant-pathogenic and plant-nonpathogenic endophytes. Annu. Rev. Phytopathol. 55, 61–83. doi: 10.1146/annurev-phyto-080516-035641
Canales, E., Coll, Y., Hernández, I., Portieles, R., García, M. R., López, Y., et al. (2016). ‘Candidatus Liberibacter asiaticus’, causal agent of citrus huanglongbing, is reduced by treatment with brassinosteroids. PLoS One 11:e0146223. doi: 10.1371/journal.pone.0146223
Cao, H., Li, X., and Dong, X. (1998). Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc. Natl. Acad. Sci. U. S. A. 95, 6531–6536. doi: 10.1073/pnas.95.11.6531
Clark, K., Franco, J. Y., Schwizer, S., Pang, Z., Hawara, E., Liebrand, T. W., et al. (2018). An effector from the huanglongbing-associated pathogen targets citrus proteases. Nat. Commun. 9, 1–11. doi: 10.1038/s41467-018-04140-9
Clark, K. J., Pang, Z., Trinh, J., Wang, N., and Ma, W. (2020). Sec-delivered effector 1 (SDE1) of ‘Candidatus Liberibacter asiaticus’ promotes citrus huanglongbing. Mol. Plant-Microbe Interact. 33, 1394–1404. doi: 10.1094/MPMI-05-20-0123-R
Clarke, J. D., Liu, Y., Klessig, D. F., and Dong, X. (1998). Uncoupling PR gene expression from NPR1 and bacterial resistance: characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell 10, 557–569. doi: 10.1105/tpc.10.4.557
Cocuzza, G. E. M., Alberto, U., Hernández-Suárez, E., Siverio, F., Di Silvestro, S., Tena, A., et al. (2017). A review on Trioza erytreae (African citrus psyllid), now in mainland Europe, and its potential risk as vector of huanglongbing (HLB) in citrus. J. Pest. Sci. 90, 1–17. doi: 10.1007/s10340-016-0804-1
Coletta-Filho, H., Targon, M., Takita, M., De Negri, J., Pompeu, J., Machado, M., et al. (2004). First report of the causal agent of huanglongbing (“Candidatus Liberibacter asiaticus”) in Brazil. Plant Dis. 88:1382. doi: 10.1094/PDIS.2004.88.12.1382C
Compant, S., Reiter, B., Sessitsch, A., Nowak, J., Clément, C., and Barka, E. D. A. (2005). Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. strain PsJN. Appl. Environ. Microbiol. 71, 1685–1693. doi: 10.1128/AEM.71.4.1685-1693.2005
Coyle, J. F., Lorca, G. L., and Gonzalez, C. F. (2018). Understanding the physiology of Liberibacter asiaticus: an overview of the demonstrated molecular mechanisms. J. Mol. Microbiol. Biotechnol. 28, 116–127. doi: 10.1159/000492386
Curtolo, M., De Souza Pacheco, I., Boava, L. P., Takita, M. A., Granato, L. M., Galdeano, D. M., et al. (2020). Wide-ranging transcriptomic analysis of Poncirus trifoliata, Citrus sunki, Citrus sinensis and contrasting hybrids reveals HLB tolerance mechanisms. Sci. Rep. 10, 1–14. doi: 10.1038/s41598-020-77840-2
Da Graça, J., Kunta, M., Sétamou, M., Rascoe, J., Li, W., Nakhla, M., et al. (2015). Huanglongbing in Texas: report on the first detections in commercial citrus. J. Citrus Pathol. 2
Da Silva, J. R., De Alvarenga, F. V., Boaretto, R. M., Lopes, J. R. S., Quaggio, J. A., Coletta Filho, H. D., et al. (2020). Following the effects of micronutrient supply in HLB-infected trees: plant responses and ‘Candidatus Liberibacter asiaticus’ acquisition by the Asian citrus psyllid. Trop. Plant Pathol. 45, 597–610. doi: 10.1007/s40858-020-00370-9
Davis, M. J., Mondal, S. N., Chen, H., Rogers, M. E., and Brlansky, R. H. (2008). Co-cultivation of ‘Candidatus Liberibacter asiaticus’ with actinobacteria from citrus with huanglongbing. Plant Dis. 92, 1547–1550. doi: 10.1094/PDIS-92-11-1547
De Francesco, A., Lovelace, A. H., Shaw, D., Qiu, M., Wang, Y., Gurung, F., et al. (2022). Transcriptome profiling of ‘Candidatus Liberibacter asiaticus’ in citrus and psyllids. Phytopathology 112, 116–130. doi: 10.1094/PHYTO-08-21-0327-FI
De Lucena, D. K., Pühler, A., and Weidner, S. (2010). The role of sigma factor RpoH1 in the pH stress response of Sinorhizobium meliloti. BMC Microbiol. 10, 1–17. doi: 10.1186/1471-2180-10-265
Deng, H., Zhang, Y., Reuss, L., Suh, J. H., Yu, Q., Liang, G., et al. (2021). Comparative leaf volatile profiles of two contrasting mandarin cultivars against Candidatus Liberibacter asiaticus infection illustrate huanglongbing tolerance mechanisms. J. Agric. Food Chem. 69, 10869–10884. doi: 10.1021/acs.jafc.1c02875
Ding, F., Allen, V., Luo, W., Zhang, S., and Duan, Y. (2018). Molecular mechanisms underlying heat or tetracycline treatments for citrus HLB control. Hortic. Res. 5, 1–11. doi: 10.1038/s41438-018-0038-x
Dong, Z., Srivastava, A. K., Liu, X., Riaz, M., Gao, Y., Liang, X., et al. (2021). Interactions between nutrient and huanglongbing pathogen in citrus: an overview and implications. Sci. Hortic. 290:110511. doi: 10.1016/j.scienta.2021.110511
Doud, M. M., Wang, Y., Hoffman, M. T., Latza, C. L., Luo, W., Armstrong, C. M., et al. (2017). Solar thermotherapy reduces the titer of Candidatus Liberibacter asiaticus and enhances canopy growth by altering gene expression profiles in HLB-affected citrus plants. Hortic. Res. 4, 1–9. doi: 10.1038/hortres.2017.54
Du, D., Rawat, N., Deng, Z., and Gmitter, F. G. (2015). Construction of citrus gene coexpression networks from microarray data using random matrix theory. Hortic. Res. 2, 1–8. doi: 10.1038/hortres.2015.26
Duan, Y., Zhou, L., Hall, D. G., Li, W., Doddapaneni, H., Lin, H., et al. (2009). Complete genome sequence of citrus huanglongbing bacterium, ‘Candidatus Liberibacter asiaticus’ obtained through metagenomics. Mol. Plant-Microbe Interact. 22, 1011–1020. doi: 10.1094/MPMI-22-8-1011
Durrant, W. E., and Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. doi: 10.1146/annurev.phyto.42.040803.140421
Dutt, M., Barthe, G., Irey, M., and Grosser, J. (2015). Transgenic citrus expressing an Arabidopsis NPR1 gene exhibit enhanced resistance against huanglongbing (HLB; citrus greening). PLoS One 10:e0137134. doi: 10.1371/journal.pone.0137134
Economou, A., and Wickner, W. (1994). SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell 78, 835–843. doi: 10.1016/S0092-8674(94)90582-7
Enya, J., Shinohara, H., Yoshida, S., Tsukiboshi, T., Negishi, H., Suyama, K., et al. (2007). Culturable leaf-associated bacteria on tomato plants and their potential as biological control agents. Microb. Ecol. 53, 524–536. doi: 10.1007/s00248-006-9085-1
Fagen, J. R., Leonard, M. T., Mccullough, C. M., Edirisinghe, J. N., Henry, C. S., Davis, M. J., et al. (2014). Comparative genomics of cultured and uncultured strains suggests genes essential for free-living growth of Liberibacter. PLoS One 9:e84469. doi: 10.1371/journal.pone.0084469
Fan, J., Chen, C., Yu, Q., Khalaf, A., Achor, D. S., Brlansky, R. H., et al. (2012). Comparative transcriptional and anatomical analyses of tolerant rough lemon and susceptible sweet orange in response to ‘Candidatus Liberibacter asiaticus’ infection. Mol. Plant-Microbe Interact. 25, 1396–1407. doi: 10.1094/MPMI-06-12-0150-R
Fan, G.-C., Xia, Y.-L., Lin, X.-J., Hu, H.-Q., Wang, X.-D., Ruan, C.-Q., et al. (2016). Evaluation of thermotherapy against huanglongbing (citrus greening) in the greenhouse. J. Integr. Agric. 15, 111–119. doi: 10.1016/S2095-3119(15)61085-1
Fang, F., Guo, H., Zhao, A., Li, T., Liao, H., Deng, X., et al. (2021). A significantly high abundance of “Candidatus Liberibacter asiaticus” in citrus fruit pith: in planta transcriptome and anatomical analyses. Front. Microbiol. 12:681251. doi: 10.3389/fmicb.2021.681251
Fleites, L. A., Jain, M., Zhang, S., and Gabriel, D. W. (2014). “Candidatus Liberibacter asiaticus” prophage late genes may limit host range and culturability. Appl. Environ. Microbiol. 80, 6023–6030. doi: 10.1128/AEM.01958-14
Folimonova, S. Y., Robertson, C. J., Garnsey, S. M., Gowda, S., and Dawson, W. O. (2009). Examination of the responses of different genotypes of citrus to huanglongbing (citrus greening) under different conditions. Phytopathology 99, 1346–1354. doi: 10.1094/PHYTO-99-12-1346
Fujiwara, K., Iwanami, T., and Fujikawa, T. (2018). Alterations of Candidatus Liberibacter asiaticus-associated microbiota decrease survival of Ca. L. asiaticus in in vitro assays. Front. Microbiol. 9:3089. doi: 10.3389/fmicb.2018.03089
Gardner, C. L., Pagliai, F. A., Pan, L., Bojilova, L., Torino, M. I., Lorca, G. L., et al. (2016). Drug repurposing: tolfenamic acid inactivates PrbP, a transcriptional accessory protein in Liberibacter asiaticus. Front. Microbiol. 7:1630. doi: 10.3389/fmicb.2016.01630
Ghatrehsamani, S., Czarnecka, E., Verner, F. L., Gurley, W. B., Ehsani, R., and Ampatzidis, Y. (2019). Evaluation of mobile heat treatment system for treating in-field HLB-affected trees by analyzing survival rate of surrogate bacteria. Agronomy 9:540. doi: 10.3390/agronomy9090540
Ginnan, N. A., Dang, T., Bodaghi, S., Ruegger, P. M., Mccollum, G., England, G., et al. (2020). Disease-induced microbial shifts in citrus indicate microbiome-derived responses to huanglongbing across the disease severity spectrum. Phytobiomes J. 4, 375–387. doi: 10.1094/PBIOMES-04-20-0027-R
Gottwald, T. R. (2010). Current epidemiological understanding of citrus huanglongbing. Annu. Rev. Phytopathol. 48, 119–139. doi: 10.1146/annurev-phyto-073009-114418
Gottwald, T., Graham, J., Irey, M., Mccollum, T., and Wood, B. W. (2012). Inconsequential effect of nutritional treatments on huanglongbing control, fruit quality, bacterial titer and disease progress. Crop Prot. 36, 73–82. doi: 10.1016/j.cropro.2012.01.004
Grafton-Cardwell, E. E., Stelinski, L. L., and Stansly, P. A. (2013). Biology and management of Asian citrus psyllid, vector of the huanglongbing pathogens. Annu. Rev. Entomol. 58, 413–432. doi: 10.1146/annurev-ento-120811-153542
Graham, J., Gottwald, T., and Setamou, M. (2020). Status of huanglongbing (HLB) outbreaks in Florida, California and Texas. Trop. Plant Pathol. 45, 265–278. doi: 10.1007/s40858-020-00335-y
Ha, P. T., He, R., Killiny, N., Brown, J. K., Omsland, A., Gang, D. R., et al. (2019). Host-free biofilm culture of “Candidatus Liberibacter asiaticus,” the bacterium associated with huanglongbing. Biofilms 1:100005. doi: 10.1016/j.bioflm.2019.100005
Hall, D. G., and Gottwald, T. R. (2011). Pest management practices aimed at curtailing citrus huanglongbing disease. Outlooks Pest Manag. 22:189. doi: 10.1564/22aug11
Hartmann, T. (2008). The lost origin of chemical ecology in the late 19th century. Proc. Natl. Acad. Sci. U. S. A. 105, 4541–4546. doi: 10.1073/pnas.0709231105
Hell, R. (1997). Molecular physiology of plant sulfur metabolism. Planta 202, 138–148. doi: 10.1007/s004250050112
Hijaz, F., Al-Rimawi, F., Manthey, J. A., and Killiny, N. (2020). Phenolics, flavonoids and antioxidant capacities in citrus species with different degree of tolerance to huanglongbing. Plant Signal. Behav. 15:1752447. doi: 10.1080/15592324.2020.1752447
Hodges, A. W., and Spreen, T. H. (2012). “Economic impacts of citrus greening (HLB) in Florida, 2006/07-2010/11,” in Electronic Data Information Source (EDIS) FE903. Gainseville, FL: University of Florida.
Hoffman, M. T., Doud, M. S., Williams, L., Zhang, M.-Q., Ding, F., Stover, E., et al. (2013). Heat treatment eliminates ‘Candidatus Liberibacter asiaticus’ from infected citrus trees under controlled conditions. Phytopathology 103, 15–22. doi: 10.1094/PHYTO-06-12-0138-R
Hopkins, D., and Wall, K. (2021). Biological control of citrus huanglongbing with EB92-1, a benign strain of Xylella fastidiosa. Plant Dis. 105, 2914–2918. doi: 10.1094/PDIS-02-21-0362-RE
Hu, J., Akula, N., and Wang, N. (2016). Development of a microemulsion formulation for antimicrobial SecA inhibitors. PLoS One 11:e0150433. doi: 10.1371/journal.pone.0167792
Hu, J., Jiang, J., and Wang, N. (2018). Control of citrus huanglongbing via trunk injection of plant defense activators and antibiotics. Phytopathology 108, 186–195. doi: 10.1094/PHYTO-05-17-0175-R
Hu, J., and Wang, N. (2016). Evaluation of the spatiotemporal dynamics of oxytetracycline and its control effect against citrus huanglongbing via trunk injection. Phytopathology 106, 1495–1503. doi: 10.1094/PHYTO-02-16-0114-R
Huang, C.-Y., Araujo, K., Sánchez, J. N., Kund, G., Trumble, J., Roper, C., et al. (2021a). A stable antimicrobial peptide with dual functions of treating and preventing citrus huanglongbing. Proc. Natl. Acad. Sci. U. S. A. 118:e2019628118. doi: 10.1073/pnas.2019628118
Huang, C. Y., Niu, D., Kund, G., Jones, M., Albrecht, U., Nguyen, L., et al. (2021b). Identification of citrus immune regulators involved in defence against huanglongbing using a new functional screening system. Plant Biotechnol. J. 19, 757–766. doi: 10.1111/pbi.13502
Huber, D. M., and Jones, J. B. (2013). The role of magnesium in plant disease. Plant Soil 368, 73–85. doi: 10.1007/s11104-012-1476-0
Innerebner, G., Knief, C., and Vorholt, J. A. (2011). Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl. Environ. Microbiol. 77, 3202–3210. doi: 10.1128/AEM.00133-11
Inoue, H., Yamashita-Muraki, S., Fujiwara, K., Honda, K., Ono, H., Nonaka, T., et al. (2020). Fe2+ ions alleviate the symptom of citrus greening disease. Int. J. Mol. Sci. 21:4033. doi: 10.3390/ijms21114033
Irigoyen, S., Ramasamy, M., Pant, S., Niraula, P., Bedre, R., Gurung, M., et al. (2020). Plant hairy roots enable high throughput identification of antimicrobials against Candidatus Liberibacter spp. Nat. Commun. 11, 1–14. doi: 10.1038/s41467-020-19631-x
Jagoueix, S., Bove, J.-M., and Garnier, M. (1994). The phloem-limited bacterium of greening disease of citrus is a member of the α subdivision of the Proteobacteria. Int. J. Syst. Evol. Microbiol. 44, 379–386. doi: 10.1099/00207713-44-3-379
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kaul, S., Sharma, T., and Dhar, M. K. (2016). “Omics” tools for better understanding the plant–endophyte interactions. Front. Plant Sci. 7:955. doi: 10.3389/fpls.2016.00955
Killiny, N. (2017). Metabolite signature of the phloem sap of fourteen citrus varieties with different degrees of tolerance to Candidatus Liberibacter asiaticus. Physiol. Mol. Plant Pathol. 97, 20–29. doi: 10.1016/j.pmpp.2016.11.004
Killiny, N., and Hijaz, F. (2016). Amino acids implicated in plant defense are higher in Candidatus Liberibacter asiaticus-tolerant citrus varieties. Plant Signal. Behav. 11:e1171449. doi: 10.1080/15592324.2016.1171449
Killiny, N., Valim, M. F., Jones, S. E., Omar, A. A., Hijaz, F., Gmitter, F. G., et al. (2017). Metabolically speaking: possible reasons behind the tolerance of ‘Sugar Belle’mandarin hybrid to huanglongbing. Plant Physiol. Biochem. 116, 36–47. doi: 10.1016/j.plaphy.2017.05.001
Kim, M. K., and Jung, H.-Y. (2007). Chitinophaga terrae sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 57, 1721–1724. doi: 10.1099/ijs.0.64964-0
Kinkema, M., Fan, W., and Dong, X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12, 2339–2350. doi: 10.1105/tpc.12.12.2339
Kumagai, L., Levesque, C., Blomquist, C., Madishetty, K., Guo, Y., Woods, P., et al. (2013). First report of Candidatus Liberibacter asiaticus associated with citrus huanglongbing in California. Plant Dis. 97:283. doi: 10.1094/PDIS-09-12-0845-PDN
Leonard, M. T., Fagen, J. R., Davis-Richardson, A. G., Davis, M. J., and Triplett, E. W. (2012). Complete genome sequence of Liberibacter crescens BT-1. Stand. Genomic Sci. 7, 271–283. doi: 10.4056/sigs.3326772
Li, J., Kolbasov, V. G., Pang, Z., Duan, S., Lee, D., Huang, Y., et al. (2021a). Evaluation of the control effect of SAR inducers against citrus huanglongbing applied by foliar spray, soil drench or trunk injection. Phytopathol. Res. 3, 1–15. doi: 10.1186/s42483-020-00079-2
Li, W., Levy, L., and Hartung, J. S. (2009). Quantitative distribution of ‘Candidatus Liberibacter asiaticus’ in citrus plants with citrus huanglongbing. Phytopathology 99, 139–144. doi: 10.1094/PHYTO-99-2-0139
Li, J., Li, L., Pang, Z., Kolbasov, V. G., Ehsani, R., Carter, E. W., et al. (2019). Developing citrus huanglongbing (HLB) management strategies based on the severity of symptoms in HLB-endemic citrus-producing regions. Phytopathology 109, 582–592. doi: 10.1094/PHYTO-08-18-0287-R
Li, J., Trivedi, P., and Wang, N. (2016). Field evaluation of plant defense inducers for the control of citrus huanglongbing. Phytopathology 106, 37–46. doi: 10.1094/PHYTO-08-15-0196-R
Li, T., Zhang, L., Deng, Y., Deng, X., and Zheng, Z. (2021b). Establishment of a Cuscuta campestris-mediated enrichment system for genomic and transcriptomic analyses of ‘Candidatus Liberibacter asiaticus’. Microb. Biotechnol. 14, 737–751. doi: 10.1111/1751-7915.13773
Lin, D., Xiao, M., Zhao, J., Li, Z., Xing, B., Li, X., et al. (2016). An overview of plant phenolic compounds and their importance in human nutrition and management of type 2 diabetes. Molecules 21:1374. doi: 10.3390/molecules21101374
Lopes, S., Frare, G., Bertolini, E., Cambra, M., Fernandes, N., Ayres, A., et al. (2009). Liberibacters associated with citrus huanglongbing in Brazil: ‘Candidatus Liberibacter asiaticus’ is heat tolerant, ‘Ca. L. americanus’ is heat sensitive. Plant Dis. 93, 257–262. doi: 10.1094/PDIS-93-3-0257
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Ma, W., Pang, Z., Huang, X., Xu, J., Pandey, S. S., Li, J., et al. (2022). Citrus huanglongbing is a pathogen-triggered immune disease that can be mitigated with antioxidants and gibberellin. Nat. Commun. 13, 1–13. doi: 10.1038/s41467-022-28189-9
Madhaiyan, M., Reddy, B. S., Anandham, R., Senthilkumar, M., Poonguzhali, S., Sundaram, S., et al. (2006). Plant growth–promoting methylobacterium induces defense responses in groundnut (Arachis hypogaea L.) compared with rot pathogens. Curr. Microbiol. 53, 270–276. doi: 10.1007/s00284-005-0452-9
Martinelli, F., Reagan, R. L., Dolan, D., Fileccia, V., and Dandekar, A. M. (2016). Proteomic analysis highlights the role of detoxification pathways in increased tolerance to huanglongbing disease. BMC Plant Biol. 16, 1–14. doi: 10.1186/s12870-016-0858-5
Masaoka, Y., Pustika, A., Subandiyah, S., Okada, A., Hanundin, E., Purwanto, B., et al. (2011). Lower concentrations of microelements in leaves of citrus infected with ‘Candidatus Liberibacter asiaticus’. Jpn. Agr. Res. Q. 45, 269–275. doi: 10.6090/jarq.45.269
Mattos-Jr, D., Kadyampakeni, D. M., Da Silva, J. R., Vashisth, T., and Boaretto, R. M. (2020). Reciprocal effects of huanglongbing infection and nutritional status of citrus trees: a review. Trop. Plant Pathol. 45, 586–596. doi: 10.1007/s40858-020-00389-y
Mendes, R., Garbeva, P., and Raaijmakers, J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. doi: 10.1111/1574-6976.12028
Merfa, M. V., Pérez-López, E., Naranjo, E., Jain, M., Gabriel, D. W., and De La Fuente, L. (2019). Progress and obstacles in culturing ‘Candidatus Liberibacter asiaticus’, the bacterium associated with huanglongbing. Phytopathology 109, 1092–1101. doi: 10.1094/PHYTO-02-19-0051-RVW
Mhedbi-Hajri, N., Jacques, M.-A., and Koebnik, R. (2011). Adhesion mechanisms of plant-pathogenic Xanthomonadaceae. Adv. Exp. Med. Biol. 715, 71–89. doi: 10.1007/978-94-007-0940-9_5
Mitsui, H., Sato, T., Sato, Y., Ito, N., and Minamisawa, K. (2004). Sinorhizobium meliloti RpoH 1 is required for effective nitrogen-fixing symbiosis with alfalfa. Mol. Gen. Genomics. 271, 416–425. doi: 10.1007/s00438-004-0992-x
Morgan, K. T., Rouse, R. E., and Ebel, R. C. (2016). Foliar applications of essential nutrients on growth and yield of ‘Valencia’sweet orange infected with huanglongbing. HortScience 51, 1482–1493. doi: 10.21273/HORTSCI11026-16
Munir, S., Li, Y., He, P., He, P., Ahmed, A., Wu, Y., et al. (2020). Unraveling the metabolite signature of citrus showing defense response towards Candidatus Liberibacter asiaticus after application of endophyte Bacillus subtilis L1-21. Microbiol. Res. 234:126425. doi: 10.1016/j.micres.2020.126425
Nan, J., Zhang, S., and Jiang, L. (2021). Antibacterial potential of Bacillus amyloliquefaciens GJ1 against citrus huanglongbing. Plants 10:261. doi: 10.3390/plants10020261
Nicholson, R. L., and Hammerschmidt, R. (1992). Phenolic compounds and their role in disease resistance. Annu. Rev. Phytopathol. 30, 369–389. doi: 10.1146/annurev.py.30.090192.002101
Nwugo, C. C., Doud, M. S., Duan, Y.-P., and Lin, H. (2016). Proteomics analysis reveals novel host molecular mechanisms associated with thermotherapy of ‘Ca. Liberibacter asiaticus’-infected citrus plants. BMC Plant Biol. 16, 1–15. doi: 10.1186/s12870-016-0942-x
Oppenheim, A. B., Kobiler, O., Stavans, J., Court, D. L., and Adhya, S. (2005). Switches in bacteriophage lambda development. Annu. Rev. Genet. 39, 409–429. doi: 10.1146/annurev.genet.39.073003.113656
Pagliaccia, D., Shi, J., Pang, Z., Hawara, E., Clark, K., Thapa, S. P., et al. (2017). A pathogen secreted protein as a detection marker for citrus huanglongbing. Front. Microbiol. 8:2041. doi: 10.3389/fmicb.2017.02041
Pagliai, F. A., Coyle, J. F., Kapoor, S., Gonzalez, C. F., and Lorca, G. L. (2017). LdtR is a master regulator of gene expression in Liberibacter asiaticus. Microb. Biotechnol. 10, 896–909. doi: 10.1111/1751-7915.12728
Pagliai, F. A., Gardner, C. L., Bojilova, L., Sarnegrim, A., Tamayo, C., Potts, A. H., et al. (2014). The transcriptional activator LdtR from ‘Candidatus Liberibacter asiaticus’ mediates osmotic stress tolerance. PLoS Pathog. 10:e1004101. doi: 10.1371/journal.ppat.1004101
Parker, J. K., Wisotsky, S. R., Johnson, E. G., Hijaz, F. M., Killiny, N., Hilf, M. E., et al. (2014). Viability of ‘Candidatus Liberibacter asiaticus’ prolonged by addition of citrus juice to culture medium. Phytopathology 104, 15–26. doi: 10.1094/PHYTO-05-13-0119-R
Peris-Peris, C., Serra-Cardona, A., Sánchez-Sanuy, F., Campo, S., Ariño, J., and San Segundo, B. (2017). Two NRAMP6 isoforms function as iron and manganese transporters and contribute to disease resistance in rice. Mol. Plant-Microbe Interact. 30, 385–398. doi: 10.1094/MPMI-01-17-0005-R
Petrussa, E., Braidot, E., Zancani, M., Peresson, C., Bertolini, A., Patui, S., et al. (2013). Plant flavonoids—biosynthesis, transport and involvement in stress responses. Int. J. Mol. Sci. 14, 14950–14973. doi: 10.3390/ijms140714950
Phuyal, D., Nogueira, T. A. R., Jani, A. D., Kadyampakeni, D. M., Morgan, K. T., and Ferrarezi, R. S. (2020). ‘Ray Ruby’grapefruit affected by huanglongbing II. Planting density, soil, and foliar nutrient management. HortScience 55, 1420–1432. doi: 10.21273/HORTSCI15255-20
Piasecka, A., Jedrzejczak-Rey, N., and Bednarek, P. (2015). Secondary metabolites in plant innate immunity: conserved function of divergent chemicals. New Phytol. 206, 948–964. doi: 10.1111/nph.13325
Pierre, M. O., Salvatierra-Miranda, J., Rivera, M. J., Etxeberria, E., Gonzalez, P., and Vincent, C. I. (2021). White and red-dyed kaolin particle films reduce Asian citrus psyllid populations, delay huanglongbing infection, and increase citrus growth. Crop Prot. 150:105792. doi: 10.1016/j.cropro.2021.105792
Pitino, M., Armstrong, C. M., Cano, L. M., and Duan, Y. (2016). Transient expression of Candidatus Liberibacter asiaticus effector induces cell death in Nicotiana benthamiana. Front. Plant Sci. 7:982. doi: 10.3389/fpls.2016.00982
Poveda, J., Roeschlin, R. A., Marano, M. R., and Favaro, M. A. (2021). Microorganisms as biocontrol agents against bacterial citrus diseases. Biol. Control 158:104602. doi: 10.1016/j.biocontrol.2021.104602
Prasad, B. D., Creissen, G., Lamb, C., and Chattoo, B. B. (2009). Overexpression of rice (Oryza sativa L.) OsCDR1 leads to constitutive activation of defense responses in rice and Arabidopsis. Mol. Plant-Microbe Interact. 22, 1635–1644. doi: 10.1094/MPMI-22-12-1635
Prasad, S., Xu, J., Zhang, Y., and Wang, N. (2016). SEC-translocon dependent extracytoplasmic proteins of Candidatus Liberibacter asiaticus. Front. Microbiol. 7:1989. doi: 10.3389/fmicb.2016.01989
Qiu, W., Soares, J., Pang, Z., Huang, Y., Sun, Z., Wang, N., et al. (2020). Potential mechanisms of AtNPR1 mediated resistance against huanglongbing (HLB) in citrus. Int. J. Mol. Sci. 21:2009. doi: 10.3390/ijms21062009
Ramsey, J. S., Chin, E. L., Chavez, J. D., Saha, S., Mischuk, D., Mahoney, J., et al. (2020). Longitudinal transcriptomic, proteomic, and metabolomic analysis of citrus limon response to graft inoculation by Candidatus Liberibacter asiaticus. J. Proteome Res. 19, 2247–2263. doi: 10.1021/acs.jproteome.9b00802
Rao, M. J., Ding, F., Wang, N., Deng, X., and Xu, Q. (2018). Metabolic mechanisms of host species against citrus huanglongbing (greening disease). Crit. Rev. Plant Sci. 37, 496–511. doi: 10.1080/07352689.2018.1544843
Rawat, N., Kiran, S. P., Du, D., Gmitter, F. G., and Deng, Z. (2015). Comprehensive meta-analysis, co-expression, and miRNA nested network analysis identifies gene candidates in citrus against huanglongbing disease. BMC Plant Biol. 15, 1–21. doi: 10.1186/s12870-015-0568-4
Rawat, N., Kumar, B., Albrecht, U., Du, D., Huang, M., Yu, Q., et al. (2017). Genome resequencing and transcriptome profiling reveal structural diversity and expression patterns of constitutive disease resistance genes in huanglongbing-tolerant Poncirus trifoliata and its hybrids. Hortic. Res. 4, 1–8. doi: 10.1038/hortres.2017.64
Riera, N., Handique, U., Zhang, Y., Dewdney, M. M., and Wang, N. (2017). Characterization of antimicrobial-producing beneficial bacteria isolated from huanglongbing escape citrus trees. Front. Microbiol. 8:2415. doi: 10.3389/fmicb.2017.02415
Rouse, R. E., Ozores-Hampton, M., Roka, F. M., and Roberts, P. (2017). Rehabilitation of huanglongbing-affected citrus trees using severe pruning and enhanced foliar nutritional treatments. HortScience 52, 972–978. doi: 10.21273/HORTSCI11105-16
Ryals, J. A., Neuenschwander, U. H., Willits, M. G., Molina, A., Steiner, H.-Y., and Hunt, M. D. (1996). Systemic acquired resistance. Plant Cell 8, 1809–1819. doi: 10.1105/tpc.8.10.1809
Saikkonen, K., Wäli, P., Helander, M., and Faeth, S. H. (2004). Evolution of endophyte–plant symbioses. Trends Plant Sci. 9, 275–280. doi: 10.1016/j.tplants.2004.04.005
Saindrenan, P., and Guest, D. I. (1994). “Involvement of phytoalexins in the response of phosphonate-treated plants to infection by Phytophthora species.” in Handbook of Phytoalexin Metabolism and Action. eds. Daniel, M., and Purkayasthsa, R. P. (New York: CRC Press), 375–390.
Segers, K., and Anné, J. (2011). Traffic jam at the bacterial sec translocase: targeting the SecA nanomotor by small-molecule inhibitors. Chem. Biol. 18, 685–698. doi: 10.1016/j.chembiol.2011.04.007
Shahzad, F., Chun, C., Schumann, A., and Vashisth, T. (2020). Nutrient uptake in huanglongbing-affected sweet orange: transcriptomic and physiological analysis. J. Am. Soc. Hortic. Sci. 145, 349–362. doi: 10.21273/JASHS04929-20
Shen, W., Cevallos-Cevallos, J. M., Nunes Da Rocha, U., Arevalo, H. A., Stansly, P. A., Roberts, P. D., et al. (2013). Relation between plant nutrition, hormones, insecticide applications, bacterial endophytes, and Candidatus Liberibacter Ct values in citrus trees infected with huanglongbing. Eur. J. Plant Pathol. 137, 727–742. doi: 10.1007/s10658-013-0283-7
Shi, X., Li, B., Qin, G., and Tian, S. (2012). Mechanism of antifungal action of borate against Colletotrichum gloeosporioides related to mitochondrial degradation in spores. Postharvest Biol. Technol. 67, 138–143. doi: 10.1016/j.postharvbio.2012.01.003
Singerman, A., and Rogers, M. E. (2020). The economic challenges of dealing with citrus greening: the case of Florida. J. Integr. Pest Manag. 11:3. doi: 10.1093/jipm/pmz037
Sourjik, V., Muschler, P., Scharf, B., and Schmitt, R. D. (2000). VisN and VisR are global regulators of chemotaxis, flagellar, and motility genes in Sinorhizobium (Rhizobium) meliloti. J. Bacteriol. 182, 782–788. doi: 10.1128/JB.182.3.782-788.2000
Spann, T. M., and Schumann, A. W. (2009). The role of plant nutrients in disease development with emphasis on citrus and huanglongbing. Proc. Fla. State Hort. Soc. 122, 169–171.
Sticher, L., Mauch-Mani, B., and Métraux, J. P. (1997). Systemic acquired resistance. Annu. Rev. Phytopathol. 35, 235–270. doi: 10.1146/annurev.phyto.35.1.235
Sugawara, M., Epstein, B., Badgley, B. D., Unno, T., Xu, L., Reese, J., et al. (2013). Comparative genomics of the core and accessory genomes of 48 Sinorhizobium strains comprising five genospecies. Genome Biol. 14, R17–R20. doi: 10.1186/gb-2013-14-2-r17
Sugimoto, T., Watanabe, K., Yoshida, S., Aino, M., Furiki, M., Shiono, M., et al. (2010). Field application of calcium to reduce Phytophthora stem rot of soybean, and calcium distribution in plants. Plant Dis. 94, 812–819. doi: 10.1094/PDIS-94-7-0812
Suh, J. H., Tang, X., Zhang, Y., Gmitter, F. G., and Wang, Y. (2021). Metabolomic analysis provides new insight into tolerance of huanglongbing in citrus. Front. Plant Sci. 12:710598. doi: 10.3389/fpls.2021.710598
Tang, J., Ding, Y., Nan, J., Yang, X., Sun, L., Zhao, X., et al. (2018). Transcriptome sequencing and ITRAQ reveal the detoxification mechanism of Bacillus GJ1, a potential biocontrol agent for huanglongbing. PLoS One 13:e0200427. doi: 10.1371/journal.pone.0200427
Tatineni, S., Sagaram, U. S., Gowda, S., Robertson, C. J., Dawson, W. O., Iwanami, T., et al. (2008). In planta distribution of ‘Candidatus Liberibacter asiaticus’ as revealed by polymerase chain reaction (PCR) and real-time PCR. Phytopathology 98, 592–599. doi: 10.1094/PHYTO-98-5-0592
Thapa, S. P., De Francesco, A., Trinh, J., Gurung, F. B., Pang, Z., Vidalakis, G., et al. (2020). Genome-wide analyses of Liberibacter species provides insights into evolution, phylogenetic relationships, and virulence factors. Mol. Plant Pathol. 21, 716–731. doi: 10.1111/mpp.12925
Treutter, D. (2006). Significance of flavonoids in plant resistance: a review. Environ. Chem. Lett. 4, 147–157. doi: 10.1007/s10311-006-0068-8
Trivedi, P., Spann, T., and Wang, N. (2011). Isolation and characterization of beneficial bacteria associated with citrus roots in Florida. Microb. Ecol. 62, 324–336. doi: 10.1007/s00248-011-9822-y
Van Den Berg, B., Clemons, W. M., Collinson, I., Modis, Y., Hartmann, E., Harrison, S. C., et al. (2004). X-ray structure of a protein-conducting channel. Nature 427, 36–44. doi: 10.1038/nature02218
Verma, J., Yadav, J., Tiwari, K., Lavakush, S., and Singh, V. (2010). Impact of plant growth promoting rhizobacteria on crop production. Int. J. Agric. Res. 5, 954–983. doi: 10.3923/ijar.2010.954.983
Vincent, C., Pierre, M., Li, J., and Wang, N. (2019). Implications of heat treatment and systemic delivery of foliar-applied oxytetracycline on citrus physiological management and therapy delivery. Front. Plant Sci. 10:41. doi: 10.3389/fpls.2019.00041
Wang, N. (2021). A promising plant defense peptide against citrus huanglongbing disease. Proc. Natl. Acad. Sci. U. S. A. 118:e2026483118. doi: 10.1073/pnas.2026483118
Wang, N., Pierson, E. A., Setubal, J. C., Xu, J., Levy, J. G., Zhang, Y., et al. (2017). The Candidatus Liberibacter–host interface: insights into pathogenesis mechanisms and disease control. Annu. Rev. Phytopathol. 55, 451–482. doi: 10.1146/annurev-phyto-080516-035513
Wang, N., and Trivedi, P. (2013). Citrus huanglongbing: a newly relevant disease presents unprecedented challenges. Phytopathology 103, 652–665. doi: 10.1094/PHYTO-12-12-0331-RVW
Wang, Y., Zhou, L., Yu, X., Stover, E., Luo, F., and Duan, Y. (2016). Transcriptome profiling of huanglongbing (HLB) tolerant and susceptible citrus plants reveals the role of basal resistance in HLB tolerance. Front. Plant Sci. 7:933. doi: 10.3389/fpls.2016.00933
Wu, H., Hu, Y., Fu, S., Zhou, C., and Wang, X. (2020). Coordination of multiple regulation pathways contributes to the tolerance of a wild citrus species (Citrus ichangensis ‘2586’) against huanglongbing. Physiol. Mol. Plant Pathol. 109:101457. doi: 10.1016/j.pmpp.2019.101457
Wulff, N. A., Zhang, S., Setubal, J. C., Almeida, N. F., Martins, E. C., Harakava, R., et al. (2014). The complete genome sequence of ‘Candidatus Liberibacter americanus’, associated with citrus huanglongbing. Mol. Plant-Microbe Interact. 27, 163–176. doi: 10.1094/MPMI-09-13-0292-R
Xia, Y., Suzuki, H., Borevitz, J., Blount, J., Guo, Z., Patel, K., et al. (2004). An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J. 23, 980–988. doi: 10.1038/sj.emboj.7600086
Yan, Q., Sreedharan, A., Wei, S., Wang, J., Pelz-Stelinski, K., Folimonova, S., et al. (2013). Global gene expression changes in Candidatus Liberibacter asiaticus during the transmission in distinct hosts between plant and insect. Mol. Plant Pathol. 14, 391–404. doi: 10.1111/mpp.12015
Yan, H., Zhou, B., Jiang, B., Lv, Y., Moniruzzaman, M., Zhong, G., et al. (2021). Comparative analysis of bacterial and fungal endophytes responses to Candidatus Liberibacter asiaticus infection in leaf midribs of Citrus reticulata cv. Shatangju. Physiol. Mol. Plant Pathol. 113:101590. doi: 10.1016/j.pmpp.2020.101590
Yang, C., Hu, H., Wu, Y., Lin, X., Fan, G., Duan, Y., et al. (2020a). Metagenomic analysis reveals the mechanism for the observed increase in antibacterial activity of penicillin against uncultured bacteria Candidatus Liberibacter asiaticus relative to oxytetracycline in planta. Antibiotics 9:874. doi: 10.3390/antibiotics9120874
Yang, C., Powell, C. A., Duan, Y., Ancona, V., Huang, J., and Zhang, M. (2020b). Transcriptomic analysis reveals root metabolic alteration and induction of huanglongbing resistance by sulphonamide antibiotics in huanglongbing-affected citrus plants. Plant Pathol. 69, 733–743. doi: 10.1111/ppa.13154
Yang, C., Powell, C. A., Duan, Y., Shatters, R., Fang, J., and Zhang, M. (2016a). Deciphering the bacterial microbiome in huanglongbing-affected citrus treated with thermotherapy and sulfonamide antibiotics. PLoS One 11:e0155472. doi: 10.1371/journal.pone.0155472
Yang, C., Powell, C. A., Duan, Y., Shatters, R. G., Lin, Y., and Zhang, M. (2016b). Mitigating citrus huanglongbing via effective application of antimicrobial compounds and thermotherapy. Crop Prot. 84, 150–158. doi: 10.1016/j.cropro.2016.03.013
Yang, C., Powell, C. A., Duan, Y., Shatters, R., and Zhang, M. (2015). Antimicrobial nanoemulsion formulation with improved penetration of foliar spray through citrus leaf cuticles to control citrus huanglongbing. PloS one 10:e0133826. doi: 10.1371/journal.pone.0133826
Yang, C., Zhong, Y., Powell, C. A., Doud, M. S., Duan, Y., Huang, Y., et al. (2018). Antimicrobial compounds effective against Candidatus Liberibacter asiaticus discovered via graft-based assay in citrus. Sci. Rep. 8, 1–11. doi: 10.1038/s41598-018-35461-w
Ying, X., Redfern, B., Gmitter, F. G., and Deng, Z. (2020). Heterologous expression of the constitutive disease resistance 2 and 8 genes from Poncirus trifoliata restored the hypersensitive response and resistance of Arabidopsis cdr1 mutant to bacterial pathogen Pseudomonas syringae. Plants 9:821. doi: 10.3390/plants9070821
Yuan, X., Chen, C., Bassanezi, R., Wu, F., Feng, Z., Shi, D., et al. (2020). Region-wide comprehensive implementation of roguing infected trees, tree replacement, and insecticide applications successfully controls citrus HLB. Phytopathology 111, 1361–1368. doi: 10.1094/PHYTO-09-20-0436-R
Zambon, F. T., Kadyampakeni, D. M., and Grosser, J. W. (2019). Ground application of overdoses of manganese have a therapeutic effect on sweet orange trees infected with Candidatus Liberibacter asiaticus. HortScience 54, 1077–1086. doi: 10.21273/HORTSCI13635-18
Zhang, S., Flores-Cruz, Z., Zhou, L., Kang, B.-H., Fleites, L. A., Gooch, M. D., et al. (2011b). ‘Ca. Liberibacter asiaticus’ carries an excision plasmid prophage and a chromosomally integrated prophage that becomes lytic in plant infections. Mol. Plant-Microbe Interact. 24, 458–468. doi: 10.1094/MPMI-11-10-0256
Zhang, M., Guo, Y., Powell, C. A., Doud, M. S., Yang, C., and Duan, Y. (2014). Effective antibiotics against ‘Candidatus Liberibacter asiaticus’ in HLB-affected citrus plants identified via the graft-based evaluation. PLoS One 9:e111032. doi: 10.1371/journal.pone.0116056
Zhang, M., Karuppaiya, P., Zheng, D., Sun, X., Bai, J., Ferrarezi, R. S., et al. (2021). Field evaluation of chemotherapy on HLB-affected citrus trees with emphasis on fruit yield and quality. Front. Plant Sci. 12:611287. doi: 10.3389/fpls.2021.611287
Zhang, M., Powell, C. A., Benyon, L. S., Zhou, H., and Duan, Y. (2013). Deciphering the bacterial microbiome of citrus plants in response to ‘Candidatus Liberibacter asiaticus’-infection and antibiotic treatments. PLoS One 8:e76331. doi: 10.1371/journal.pone.0085170
Zhang, M., Powell, C. A., Guo, Y., Doud, M. S., and Duan, Y. (2012). A graft-based chemotherapy method for screening effective molecules and rescuing huanglongbing-affected citrus plants. Phytopathology 102, 567–574. doi: 10.1094/PHYTO-09-11-0265
Zhang, M., Powell, C. A., Zhou, L., He, Z., Stover, E., and Duan, Y. (2011a). Chemical compounds effective against the citrus huanglongbing bacterium ‘Candidatus Liberibacter asiaticus’ in planta. Phytopathology 101, 1097–1103. doi: 10.1094/PHYTO-09-10-0262
Zhang, Y., Xu, J., Riera, N., Jin, T., Li, J., and Wang, N. (2017). Huanglongbing impairs the rhizosphere-to-rhizoplane enrichment process of the citrus root-associated microbiome. Microbiome 5, 1–17. doi: 10.1186/s40168-017-0304-4
Zhao, H., Sun, R., Albrecht, U., Padmanabhan, C., Wang, A., Coffey, M. D., et al. (2013). Small RNA profiling reveals phosphorus deficiency as a contributing factor in symptom expression for citrus huanglongbing disease. Mol. Plant 6, 301–310. doi: 10.1093/mp/sst002
Zheng, Z., Bao, M., Wu, F., Chen, J., and Deng, X. (2016). Predominance of single prophage carrying a CRISPR/cas system in “Candidatus Liberibacter asiaticus” strains in southern China. PLoS One 11:e0146422. doi: 10.1371/journal.pone.0169138
Zheng, Z., Bao, M., Wu, F., Van Horn, C., Chen, J., and Deng, X. (2018). A type 3 prophage of ‘Candidatus Liberibacter asiaticus’ carrying a restriction-modification system. Phytopathology 108, 454–461. doi: 10.1094/PHYTO-08-17-0282-R
Zhou, C. (2020). The status of citrus huanglongbing in China. Trop. Plant Pathol. 45, 279–284. doi: 10.1007/s40858-020-00363-8
Zhou, Y., Tang, Y., Hu, C., Zhan, T., Zhang, S., Cai, M., et al. (2021). Soil applied Ca, Mg and B altered phyllosphere and rhizosphere bacterial microbiome and reduced huanglongbing incidence in gannan navel orange. Sci. Total Environ. 791:148046. doi: 10.1016/j.scitotenv.2021.148046
Keywords: HLB, unculturable bacteria, CLas, citrus defenses, HLB-tolerance
Citation: Yang C and Ancona V (2022) An Overview of the Mechanisms Against “Candidatus Liberibacter asiaticus”: Virulence Targets, Citrus Defenses, and Microbiome. Front. Microbiol. 13:850588. doi: 10.3389/fmicb.2022.850588
Edited by:
Changyong Zhou, Southwest University, ChinaReviewed by:
Xuefeng Wang, Citrus Research Institute, ChinaOrlando Borras-Hidalgo, Qilu University of Technology, China
Copyright © 2022 Yang and Ancona. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuanyu Yang, Y2h1YW55dS55YW5nQHRhbXVrLmVkdQ==; Veronica Ancona, dmVyb25pY2EuYW5jb25hLWNvbnRyZXJhc0B0YW11ay5lZHU=
 Chuanyu Yang
Chuanyu Yang Veronica Ancona
Veronica Ancona