
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 18 March 2022
Sec. Terrestrial Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.846359
 Yubin Zhao1
Yubin Zhao1 Dawei Guan1
Dawei Guan1 Xu Liu2
Xu Liu2 Gui-Feng Gao2
Gui-Feng Gao2 Fangang Meng3
Fangang Meng3 Bingqiang Liu4
Bingqiang Liu4 Pengfei Xing1
Pengfei Xing1 Xin Jiang1
Xin Jiang1 Mingchao Ma1
Mingchao Ma1 Fengming Cao1
Fengming Cao1 Li Li1
Li Li1 Jun Li1*
Jun Li1*
Rhizosphere microbial communities are vital for plant growth and soil sustainability; however, the composition of rhizobacterial communities, especially the assembly process and co-occurrence pattern among microbiota after the inoculation of some beneficial bacteria, remains considerably unclear. In this study, we investigated the structure of rhizomicrobial communities, their assembly process, and interactions contrasting when Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 are co-inoculated or Bradyrhizobium japonicum 5038 mono-inoculated in black and cinnamon soils of soybean fields. The obtained results indicated that the Chao and Shannon indices were all higher in cinnamon soil than that in black soil. In black soil, the co-inoculation increased the Shannon indices of bacteria comparing with that of the mono-inoculation. In cinnamon soil, the co-inoculation decreased the Chao indices of fungi comparing with that of mono-inoculation. Compared with the mono-inoculation, the interactions of microorganisms of co-inoculation in the co-occurrence pattern increased in complexity, and the nodes and edges of co-inoculation increased by 10.94, 40.18 and 4.82, 16.91% for bacteria and fungi, respectively. The co-inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 increased the contribution of stochastic processes comparing with Bradyrhizobium japonicum 5038 inoculation in the assembly process of soil microorganisms, and owing to the limitation of species diffusion might restrict the direction of pathogenic microorganism movement. These findings support the feasibility of rebuilding the rhizosphere microbial system via specific microbial strain inoculation and provide evidence that the co-inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 can be adopted as an excellent compound rhizobia agent resource for the sustainable development of agriculture.
Rhizobia have been identified as symbiotic nitrogen-fixing bacteria that fix atmospheric nitrogen to the soil, thus forming an important nitrogen input for the sustainability of the agricultural ecosystem (Stewart, 1967; Kumar et al., 2019); in addition, rhizobial inoculants can be the biocontrol for a number plant diseases and minimize the prevalence of some pathogenic bacteria (Nimnoi et al., 2011; Neda, 2021), which is an effective approach to reducing the application of chemical fertilizers and pesticides. The close association of Rhizobium and soybean provide sufficient mutual benefits. However, owing to the specific symbiotic matching characteristics between different rhizobia and plants host (Wang et al., 2009), environmental stress (Thilakarathna and Raizada, 2017), and the competition of native rhizobial strains (Schumpp and Deakin, 2010), the multivariate mechanism of Rhizobium inoculation on nodulation of soybean is unclear. Several studies indicated that the co-inoculation of some functional plant growth-promoting rhizobacteria (PGPR) with rhizobia could increase the colonization ability of rhizobia and promote the nodulation and nitrogen fixation of soybean (Srinivasan et al., 1997; Moretti et al., 2020; Roper et al., 2020). Concurrently, various microbial inoculants can dissolve phosphorus and potassium, increase the content of soluble phosphorus, potassium, and other mineral elements in the soil, and further increase soybean yield (Jun et al., 2019; Iturralde et al., 2020; Ju et al., 2020). Although the positive effects of PGPR on plant growth have been well known for decades, the underlying molecular mechanisms of plant growth promotion, especially in soil microbial communities, remain unexplored.
To date, Bacillus has been the most extensively studied PGPR. Bacillus amyloliquefaciens or B. subtilis added in wheat (Mahmud et al., 2021), cucumber (Bi et al., 2019; Wang et al., 2021), tomato (Qin et al., 2017; Wan et al., 2018), and Populus alba L. (Chen et al., 2020) all exhibited significant impact on bacterial diversity and its composition. These reports verify several effects of growth promotion in microbial communities via the inoculation of common and well-researched Bacillus, such as B. amyloliquefaciens and B. subtilis. However, the structure of microbial communities when exogenous B. aryabhattai and the underlying mechanisms are introduced in agricultural soil remain unclear. B. aryabhattai strain B8W22 was initially isolated from cryotubes used to collect air samples from the Earth’s stratosphere at an altitude between 27 and 41 km (Shivaji et al., 2009), and strains of this species was later isolated from rhizosphere soil in several parts of the world (Lee et al., 2012; Pailan et al., 2015; Yan et al., 2016). Some studies indicated that B. aryabhattai can be a valuable organism for its incorporation into biofertilizers and other soil amendments to improve crop productivity and tolerance toward pesticides by regulating the homeostasis of several phytohormones and producing different gibberellic acids (Park et al., 2017). In addition, the effects of the changes in B. aryabhattai on soil microorganisms have also been studied. B. aryabhattai broth has been supplemented with insoluble zinc compounds. In another study, further inoculation substantially decreased the rhizosphere soil pH and increased dehydrogenase, β-glucosidase, auxin production, microbial respiration, and microbial biomass in the rhizosphere soils of soybean and wheat crops (Ramesh et al., 2014). When B. aryabhattai was added in Chinese cabbages, it changed soil fungal community structure considerably, and the control efficiency of clubroot was 76.92% (Liu et al., 2018). The inoculation of B. aryabhattai could alter the soil microbial community; however, deeper microbial mechanisms when Rhizobium and B. aryabhattai co-inoculate and interact lack in-depth research.
The assembly process and co-occurrence pattern are two advanced and prevalent approaches currently adopted in ecology for exploring microbial mechanisms, and they have obtained extensive research results (Zhou et al., 2017a; Tripathi et al., 2018; Chen et al., 2019). It is generally accepted that deterministic and stochastic processes were simultaneously presented for community assembly in various ecosystem types (Feng et al., 2018; Shi et al., 2018; Sengupta et al., 2019; Zhao et al., 2019). Furthermore, the co-occurrence pattern indicates that soil microbial microorganisms interact with each other through the links of nodes and edges (Faust and Raes, 2012). To elucidate the links of community assembly processes and the ecological strategies of co-occurring species, two different but complementary category theories, the niche-based theory and the neutral theory, were raised to perform quantitative analysis and examine the contributions of deterministic and stochastic processes in the microbial community assembly. The niche-based theory is based on the differences in ecological niches of co-occurring species (Hubbell, 2011), and the neutral theory relies on dispersal and stochastic demographic processes (Silvertown, 2004). A recent study demonstrated that in soybean fields, deterministic processes play a dominant role in bacterial community assembly (Zhang et al., 2018). Moreover, in different soybean cultivars, the assembly process and co-occurrence networks of rhizobacterial communities were determined to be different (Zhong et al., 2019). Soil type can also alter the co-occurrence of microbial communities (Liu et al., 2019). Previous studies also indicate that the changes in the assembly process and co-occurrence pattern are related to the downstream impacts on the function of the system (Tedersoo et al., 2014; Maynard et al., 2019). Hence, revealing the details of the complex interactions and assembly process is potentially crucial in developing novel strategies for sustainable agriculture and comprehending the ecological mechanisms of the introduced Rhizobium and Bacillus.
Considering spatial heterogeneity, the effect of different species of functional PGPR inoculated on soil bacterial and fungal communities was conducted in two soil types for two cultivars in a major soybean production region in China. Soil microbial diversity, co-occurrence pattern, and assembly process of co-inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 were collectively investigated to verify the growth-promoting effect of co-inoculating Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 for the first time.
To explore the effects of co-inoculating Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 on soybean, no-inoculation (100% chemical fertilizer), one-inoculation (75% chemical fertilizer + Bradyrhizobium japonicum 5038), and two inoculation (75% chemical fertilizer + Bradyrhizobium japonicum 5038 + Bacillus aryabhattai MB35-5) treatments were conducted in black and cinnamon soils with a total of six samples (B0, B1, B2, C0, C1, and C2) marked in this study. The doses of chemical fertilizers were 100 kg N/ha, 100 kg P2O5/ha, and 100 kg K2O/ha for no-inoculation treatment, and 75 kg N/ha, 75 kg P2O5/ha, and 75 kg K2O/ha for one-inoculation and two inoculation treatments. The chemical fertilizer used in this study was composed of urea (CON2H4), Ca(H2PO4)2⋅H2O and K2SO4.
The Bradyrhizobium japonicum 5038 (CGMCC16747) and Bacillus aryabhattai MB35-5 used in this study were screened in our laboratory. Bradyrhizobium japonicum 5038, a nitrogen-fixing strain that can tolerate water stress, was screened from 160 purified strains of Bradyrhizobium that had previously been isolated from the root nodules of different Asiatic soybean cultivars. This research was published by Zhu et al. (2022). Bacillus aryabhattai MB35-5 was screened from 204 purified strains of bacteria, which had previously been isolated from soils in different rice and wheat fields, that were mostly located in 16 Chinese cities. This strain is resistant to salt, alkali, acid and anaerobic conditions. It increased the root dry matter of corn and maize by 24.02 and 16.21%, respectively, compared with the control Silicate bacteria 3016. These results have already been used to obtain a Chinese invention patent (CN112772678B).
Single colonies of Bradyrhizobium japonicum 5038 were cultured in 100 ml liquid yeast extract mannitol broth (YMB) containing 10.0 g mannitol, 0.2 g MgSO4⋅7H2O, 0.1 g NaCl, 0.5 g K2HPO4, 0.5 g yeast extract, 1000 ml H2O (pH = 7.0). Single colonies of B. aryabhattai MB35-5 were cultured in 100 ml liquid R2A containing 0.5 g yeast extract, 0.5 g tryptone, 0.5 g glucose, 0.3g K2HPO4, 5.0 g MgO3Si, 1000 ml H2O (pH = 7.0). The cells were incubated at 30°C with shaking at 180 rpm until reached to the value of OD600 for 0.6–0.8. The cells were incubated at 30°C with shaking at 180 rpm until they reached OD600 levels of 0.6–0.8. For the one-inoculation treatment, 3 ml of active culture of Br. japonicum 5038 was mixed with 1 kg of soybean seeds. When both strains were co-inoculated simultaneously, 1.5 ml of each activated bacterial liquid was mixed with 1 kg of soybean seeds. All the inoculated seeds were air-dried 2–3 h in the shade before planting.
Black soil samples were collected from a trial site located in Songyuan, Jilin Province (N 45°13′, E 124°82′), whereas cinnamon soil samples were collected from a trial site located in Shijiazhuang, Hebei Province (N 38°05′, E 114°50′). These sites are two experimental stations belonging to China’s agricultural research system. Soybean cultivars were Jiyu47 and Jidou12 for black soil and cinnamon soil, respectively. Soil samples were collected in September 7 and August 22 of 2020 at soybean early pod stage. Rhizosphere soil at a depth of 10–20 cm (plow layer) was collected, gently shaking off the soil which loosely adhered to the roots, retaining the soil tightly adhered to roots, collecting the soil by brush, and mixing to obtain one composite soil sample per plot. Then, we randomly selected five samples from each plot for the different treatments, and a total of 30 samples were collected. After the collection, all soil samples were stored in an icebox and transferred to a laboratory for examination. Each sample was separated into two parts: one part was stored at −80°C for biological analysis and the other part was air-dried in the shade for physical and chemical analyses.
The soil nitrate-nitrogen (NN) and ammonium-nitrogen (AN) contents were determined using samples of fresh soil, whereas the other soil properties were analyzed after drying soil samples at 25 ± 5°C and passing them through a 1.0 mm mesh. The soil pH was measured using a pH meter with soil to water ratio of 1:1 (Li et al., 2013). The NN and AN contents were determined via flow injection analysis (Mulvaney, 1996), whereas the soil total nitrogen (TN) and organic carbon (TC) levels were determined according to the procedure presented by Strickland and Sollins (1987). The alkali-hydrolyzable nitrogen (HN) was measured by an alkaline extraction method (alkaline potassium permanganate method). The available phosphorus (AP) was quantified using the NaHCO3– extraction method, whereas the available potassium (AK) was quantified using the NH4OAc extraction method and a flame photometer (FP640, Shanghai, China) (Mulvaney, 1996). In addition, the soil metallic elements (Na, Ca, and Mg) were measured according to Acero et al. (2006).
When the soil sample was collected, at the same time, five completely plant roots of each treatment was collected and transported to the lab. Then, we selected the relatively full and suitable size nodules for counting and then weighted the dry weight of the nodules at 80°C until we reached a constant weight. The increasing numbers and dry matter of nodules that enable the inoculation of different PGPR species are illustrated in Supplementary Figure S1.
We extracted total DNA from rhizosphere soil with a Power Soil DNA Isolation Kit (MOBIO Laboratories Inc., Carlsbad, CA, United States) according to the manufacturer’s instructions. The DNA quality and concentration (A260/A280) were estimated using a NanoDrop ND-1000 UV-Vis Spectrophotometer (Thermo Scientific, Rockwood, TN, United States). Each DNA sample was stored at −80°C until further analysis. Subsequently, bacteria were analyzed by sequencing the V3-V4 hypervariable region of the 16S ribosomal RNA (rRNA) gene. The V3-V4 region was amplified using universal primers, 338F and 806R (338F: 5′-ACTCCTACGGGAGGCAGCA-3′ and 806R: 5′-GGACTACHVGGGTWTCTAAT-3′) (Xu et al., 2016). The PCR mixtures contain 5 × TransStart FastPfu buffer 4 μL, 2.5 mM dNTPs 2 μL, forward primer (5 μM) 0.8 μL, reverse primer (5 μM) 0.8 μL, TransStart FastPfu DNA Polymerase 0.4 μL, BSA 0.2 μL, template DNA 10 ng, and finally ddH2O up to 20 μL. The hypervariable regions of the fungal 18S rRNA gene were amplified with the 817F and 1196R primers (817F: 5′-TTAGCATGGAATAATRRAATAGGA-3′ and 1196R: 5′-TCTGGACCTGGTGAGTTTCC-3′) using a thermocycler polymerase chain reaction (PCR) system (GeneAmp 9700, ABI, California, United States) (Rousk et al., 2010). The PCR mixtures contain 10 × buffer 2 μL, 2.5 mM dNTPs 2 μL, forward primer (5 μM) 0.8 μL, reverse primer (5 μM) 0.8 μL, rTaq Polymerase 0.2 μL, BSA 0.2 μL, template DNA 10 ng, and finally ddH2O up to 20 μL. The PCR amplification of 16S rRNA gene was performed as follows: initial denaturation at 95°C for 3 min, followed by 30 cycles of denaturing at 95°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 45 s, and single extension at 72°C for 10 min, and end at 4°C.
The PCR products were utilized to purify and generate the amplicon libraries; subsequently, the obtained data were analyzed using QIIME pipeline version 1.8.0. We excluded low-quality reads in an initial quality-filtering step. Chimeric sequences were then identified using UCHIME. Operational taxonomic units (OTUs) were defined via clustering at 97% similarity. The rarefaction and diversity indices were calculated after clustering the OTUs. The taxonomy of each 16S rRNA gene sequence was analyzed by the ribosomal database project classifier1 against the silva138/16s_bacteria 16S rRNA database and Unite8.0/its_fungi as references with a confidence threshold of 70% (Amato et al., 2013). Data were analyzed using a free online platform, Majorbio I-Sanger Cloud Platform2. The raw data were deposited in the short reads archive database of the National Center for Biotechnology Information (SRA accession: PRJNA788980, PRJNA788360).
The network analysis was designed based on all the 30 samples at the phylum and class levels of bacteria and fungi. To explore all the pairwise associations, correlation scores (Spearman’s correlation), OTUs with relative abundances less than 0.005% or in less than five soil samples were discarded. The relationship among OTUs was examined by Pearson’s correlation, Spearman’s correlation, Bray–Curtis dissimilarity, and Kullback–Leibler dissimilarity using R (version 3.6.3). The P-values of the five methods were integrated using the Brown method, and the correlations | P| > 0.7 were retained for the downstream procedure. The resulting correlations were imported into the Gephi platform and then visualized by the Fruchterman–Reingold algorithms.
To determine the potential importance of stochastic processes on community assembly, we adopted a neutral community model (NCM) to predict the relationship between OTU detection frequencies and their relative abundance across the wider metacommunity, which were performed using R (version 3.6.3) and the program presented by Chen et al. (2019). The assembly processes of bacterial and fungal communities were evaluated by calculating the nearest taxon index and beta nearest taxon index (betaNTI) using the “ses.mntd” function in a picante package (Stegen et al., 2012; Xun et al., 2019). When | betaNTI| < 2, the contribution was considered a stochastic process, and when | betaNTI| > 2 the shifts in community composition were deterministic processes.
Soil physicochemical property data were analyzed using Microsoft Excel 2010 pro and SPSS version 20. A Mantel test was adopted to determine the significance of the relationship between soil physicochemical properties and community structure using QIIME. A random forest model was used to identify the bacterial and fungal taxa on the order level that accurately predict the co-inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 by adopting the random forest package (importance = TRUE, proximity = TRUE, ntree = 1000). We further adopted the variation partitioning analysis (VPA) with adjusted R2 coefficients based on the redundancy analysis (RDA) and partial Mantel test to quantify the relative effects of environmental and spatial factors in shaping community composition (R version 3.3.1).
An analysis of similarities indicated the inoculation treatments that produced substantial effects on the diversity of bacterial (R2 = 0.76, p < 0.01) and fungal (R2 = 0.78, p < 0.01) communities relative to the OTU level (Figure 1). The Chao and Shannon indices of the three treatments were all higher in cinnamon soil than that in black soil, respectively. In black soil, the co-inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 increased the Shannon indices of bacteria comparing with that of the Bradyrhizobium japonicum 5038 mono-inoculated treatment. In cinnamon soil, the co-inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 decreased the Chao indices of fungi comparing with that in Bradyrhizobium japonicum 5038 mono-inoculated treatment (Supplementary Figure S3).
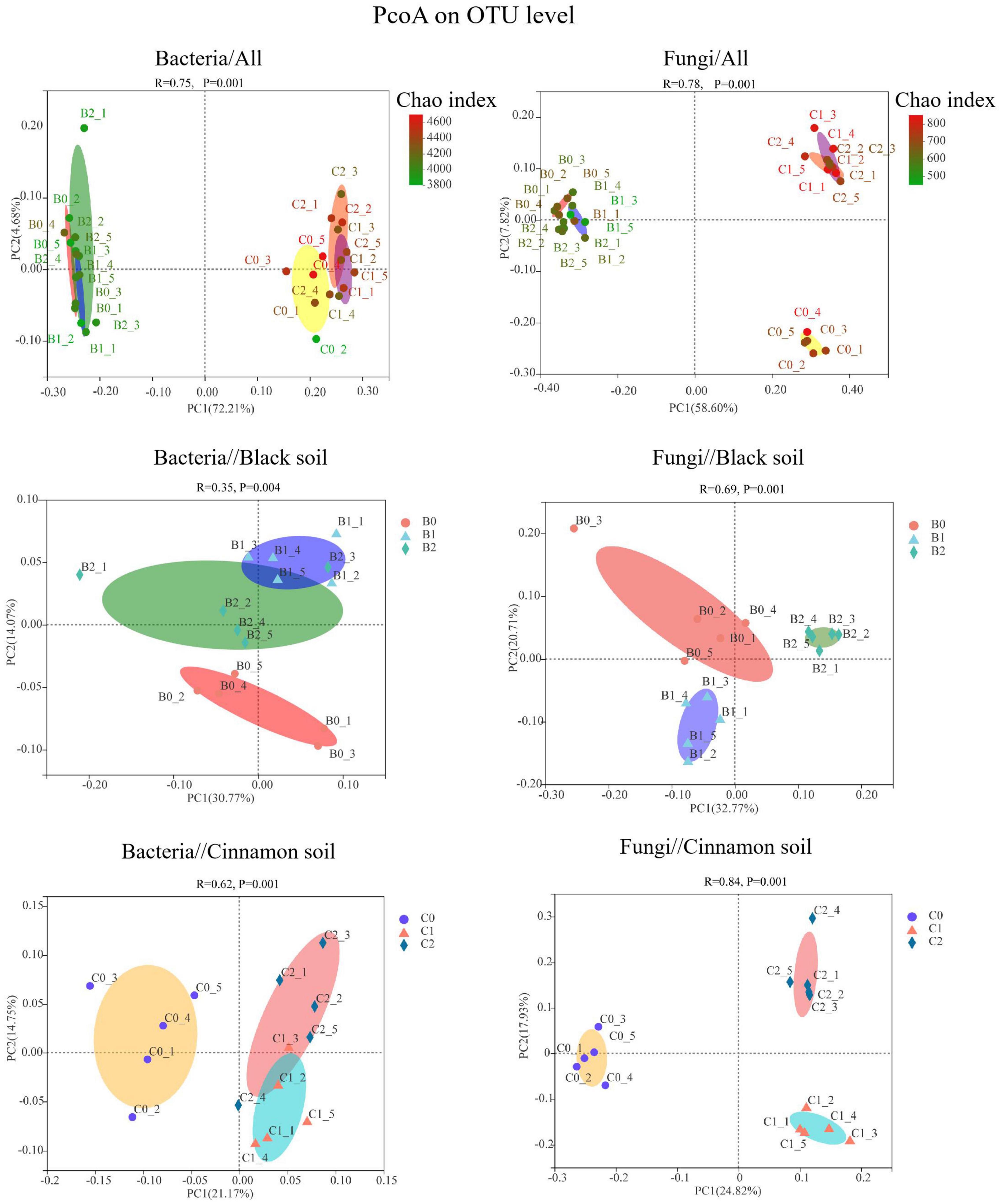
Figure 1. PCoA based on Bray–Curtis dissimilarities showing the difference in bacterial and fungal community in black soil and cinnamon soil. B0, B1, and B2 means no-inoculation, inoculation of Bradyrhizobium japonicum 5038, and co-inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 in the black soil, and C0, C1, and C2 means no-inoculation, inoculation of Bradyrhizobium japonicum 5038, and co-inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 in the cinnamon soils.
A total of 7592 rarefied OTUs from 40 phyla were identified for bacteria. The top 10 most abundant phyla were Actinobacteria (37.37, 15.84%), Proteobacteria (21.03, 25.30%), Acidobacteria (12.68, 26.82%), Chloroflexi (14.31, 7.14%), Bacteroidetes (2.34, 6.71%), Gemmatimonadetes (3.15, 3.53%), Myxococcota (1.77, 4.07%), Firmicutes (2.61, 1.52%), Patescibacteria (1.13, 1.59%), and Methylomirabilota (0.59, 1.49%) in the black and cinnamon soils, respectively (Figure 2). To identify the most important predictors for shaping bacterial communities, 13 soil physicochemical properties were measured (Supplementary Table S1). The Mantel tests demonstrated that the bacterial communities were significantly (p < 0.05) correlated with TC, HN, and Ca for black soil and TC, TN, NN, AN, Ca, and pH for the cinnamon soil (Supplementary Table S2). Among these variables, HN (Mantel r = 0.36, p < 0.05) and NN (Mantel r = 0.58, p < 0.05) exhibited the strongest correlation with the bacterial community structure for black and cinnamon soil, respectively. Principal coordinates analysis (PCoA) results indicated that the bacterial communities were divided along the soil type and number of bacteria inoculation in axes 1 and 2, which could explain 72.21 and 4.68% of the total variations, respectively (Figure 1). The relative abundance of most of the top 10 phyla was significantly (p < 0.05) correlated with the main soil physicochemical properties. Among the soil physicochemical properties, pH and AK exhibit the significant correlation (p < 0.05) with more bacterial communities in black soil (Supplementary Table S4); TN and Mg exhibit the significant correlation (p < 0.05) with more bacterial communities in the cinnamon soil (Supplementary Table S5). Random forest analysis was adopted to identify biomarkers at the family level that are strongly associated with the PGPR species (Supplementary Figure S4). We determined that several of them were enriched by the increase of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5, including Pezizaceae, Aspergillaceae, Ascodesmidaceae, and Rhizophlyctidaceae.
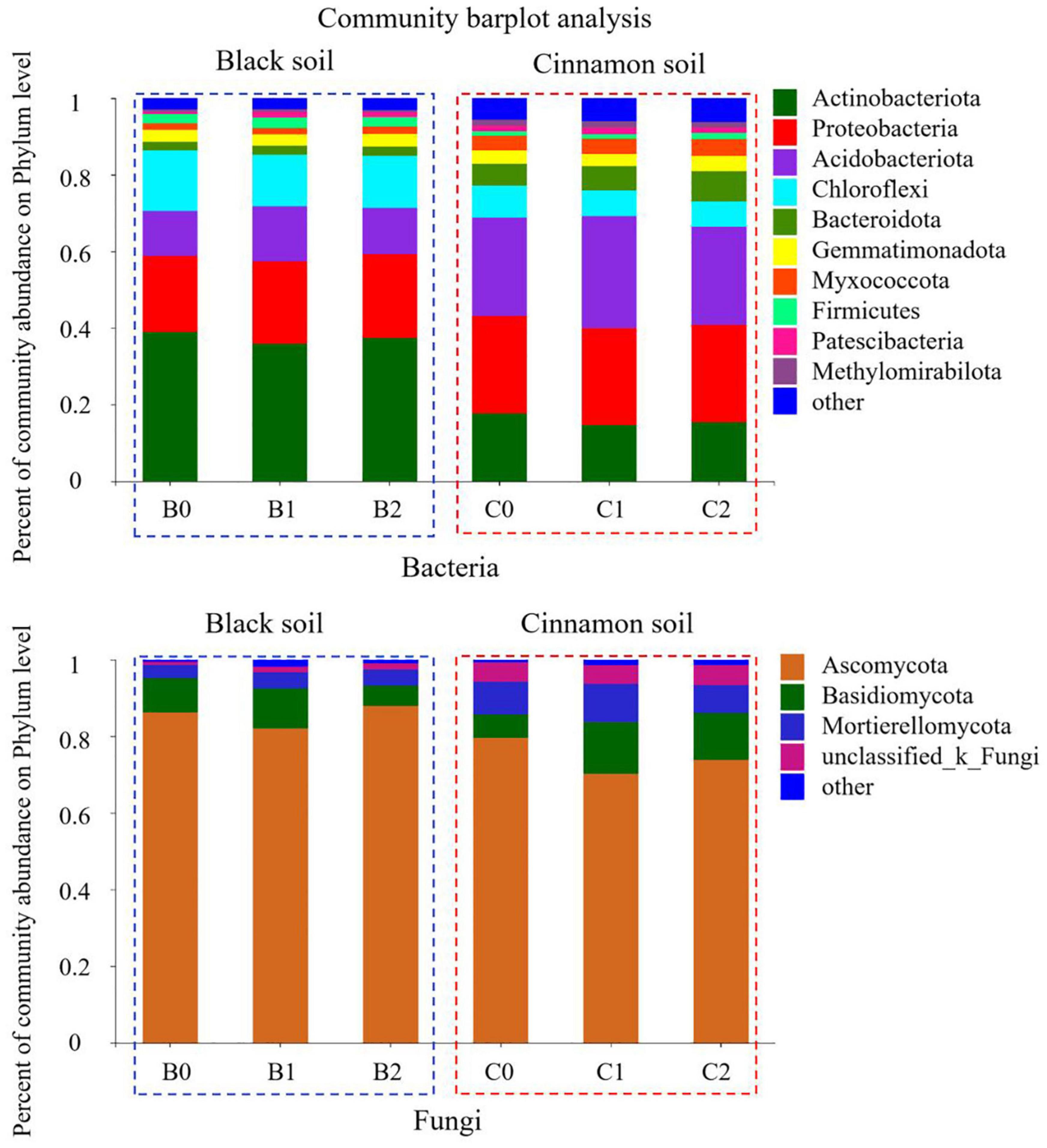
Figure 2. The relative abundance of top 10 phyla for each three treatments in black and cinnamon soil.
A total of 2405 rarefied OTUs from 16 phyla were identified for fungi. The top three most abundant phyla were Ascomycota (85.38, 74.54%), Basidiomycota (8.23, 10.54%), and Mortierellomycota (3.97, 8.63%) in the black and cinnamon soils, respectively (Figure 2). Moreover, the same phenomenon emerged in the two soil types, such that with the increase in the species of inoculated bacteria, Ascomycota first decreased and then increased, whereas Basidiomycota increased first before decreasing. In other words, the inoculation with Bradyrhizobium japonicum 5038 reduced Ascomycota and increased Basidiomycota, and then the addition of Bacillus increased Ascomycota and decreased Basidiomycota. Mantel tests demonstrated that the fungal communities were significantly (p < 0.05) correlated with TC, NN, AN, AP, TK, Na, Ga, and Mg for the black soil and with TC, TN, NN, AP, AK, Na, Ca, and pH for the cinnamon soil (Supplementary Table S2). Among these variables, NN (Mantel r = 0.48, p < 0.05; r = 0.58, p < 0.05) exhibited the strongest correlation with the fungal community structure for both black and cinnamon soils. PCoA results showed that the fungal communities were divided along the soil type and number of inoculation of bacteria in axes 1 and 2, which could explain 58.60 and 7.82% of the total variations, respectively (Figure 1). The relative abundance of most of the top three phyla was significantly (p < 0.05) correlated with the main soil physicochemical properties. Among the soil physicochemical properties, HN and TN are more significantly correlated with the microbial communities in the black soil (Supplementary Table S4); pH and Na have a more significant correlation with the microbial communities in the cinnamon soil (Supplementary Table S5). Rhizophlyctidales, Auriculariales, Diversisporales, Capnodiales, and other fungi in the order level were analyzed as biomarkers that were strongly associated with the co-inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 (Supplementary Figure S4).
In the co-occurrence network constructed by all 30 soil samples, 124 nodes and 1231 edges were identified for soil bacteria, whereas 80 nodes and 830 edges were identified for soil fungi (Figures 3, 4). In total, 12 sub-network analyses were also performed, as illustrated in Figures 3, 4 for bacteria and fungi, respectively, which showed that as the Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 inoculated, the nodes and edges increase. In other words, the inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 could increase the complexity of the co-occurrence pattern for both bacteria and fungi. The rich microorganisms in the sub-network were different in the two soil types, and the complexity of the co-occurrence pattern in the cinnamon soil was higher than that in the black soil, which is especially reflected in the increase in the amount of Proteobacteria and Dothideomycetes.
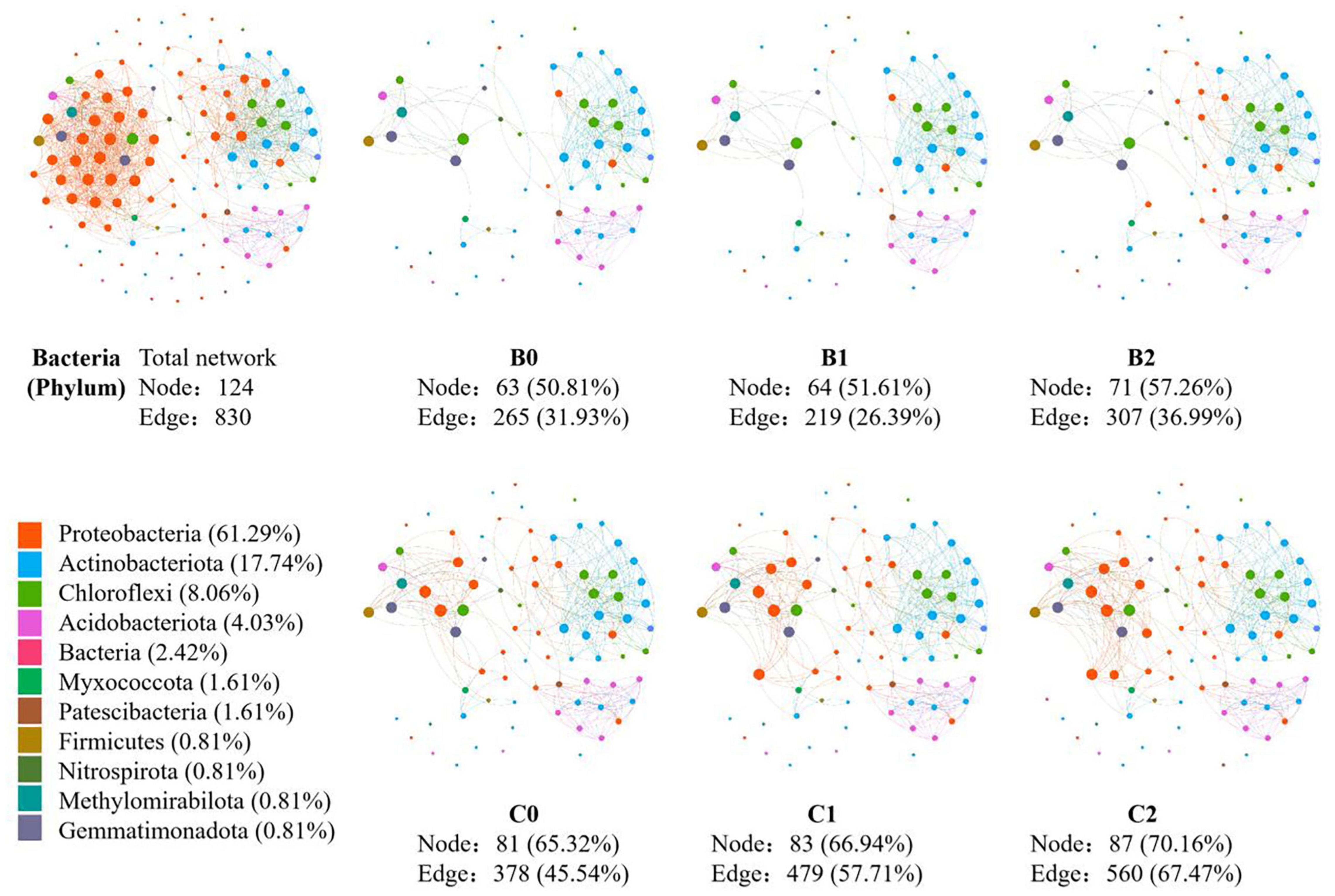
Figure 3. The co-occurrence pattern of total bacterial community and each sub-network fallowing co-inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 in black and cinnamon soil.
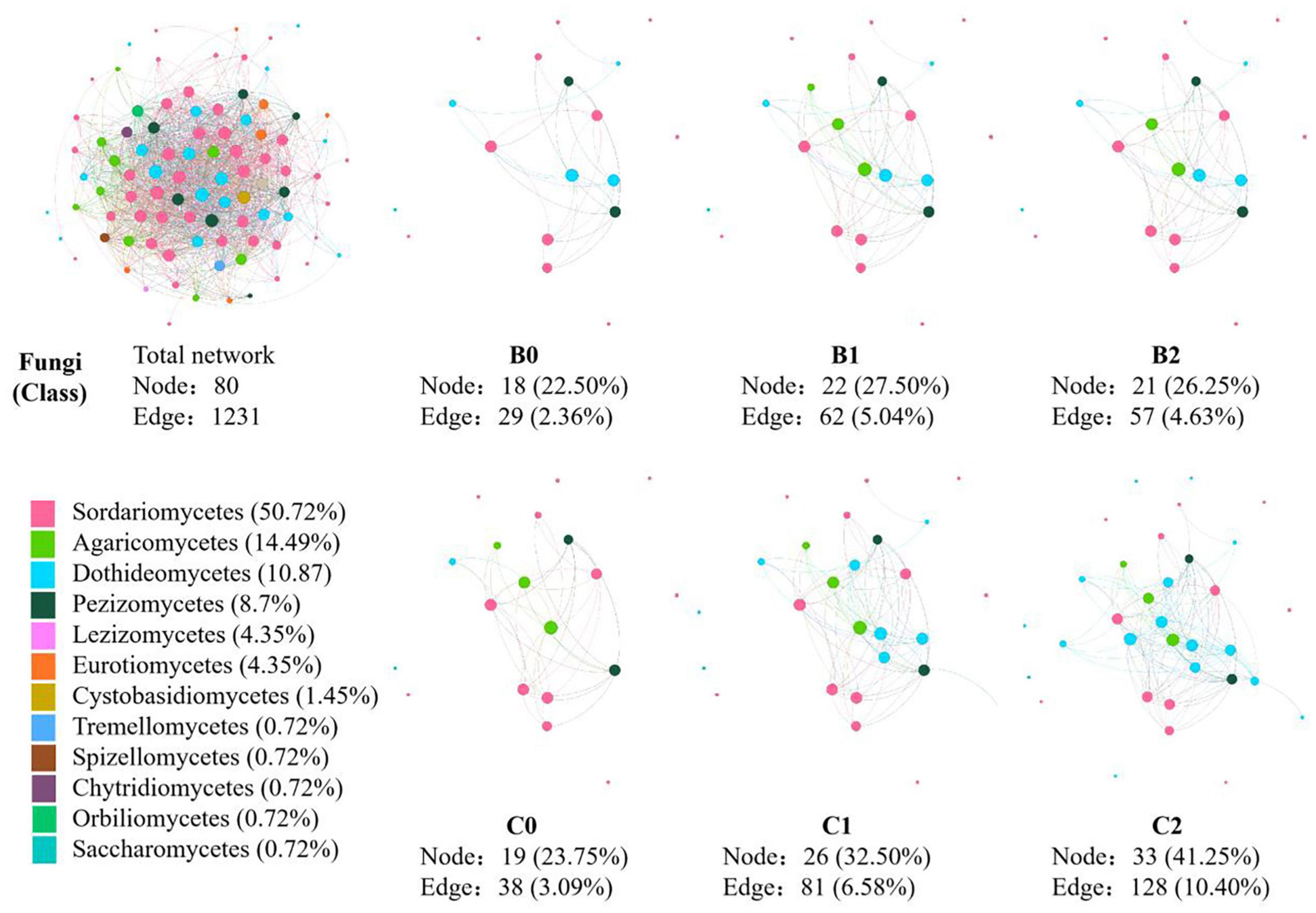
Figure 4. The co-occurrence pattern of total fungal community and each sub-network fallowing co-inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 in black and cinnamon soil.
In total, 21 and 3 key species of bacteria and fungi within the network were identified (Supplementary Figure S5), which included Gymnoascus, Cyphellophora, Azospirillales, and several other microorganisms (Supplementary Table S6). Moreover, we identified several key species as biomarkers, Cyphellophora (R2 = 0.484, P = 0.002) in the black soil and Solirubrobacteraceae (R2 = 0.379, P = 0.009) in the cinnamon soil, which was exhibited a strong negative association with PGPR numbers, and Leucoagaricus (R2 = 0.425, P = 0.005) in the cinnamon soil, which exhibited a strong positive correlation with PGPR species (Supplementary Figures S6, S7).
The relationship between network characteristics and the physical and chemical properties of soil is illustrated in Supplementary Figure S8. HN, TN, and TK were significantly positive with the number of nodes in 12 sub-networks, whereas the Ca and Mg contents were also important factors with a significant negative relationship with the number of bacteria and fungi nodes, respectively. In addition, environmental factors via the degree of bacteria network and average path length of the fungi network affected the co-occurrence pattern.
The NCM successfully estimated a large fraction of the relationship between the occurrence frequency of OTUs and their relative abundance variations (Figure 5), with 73, 71, and 72% of explained bacterial community variance for non-inoculation, one-inoculation, and two co-inoculation treatments, respectively. The obtained results indicated that stochastic processes were crucial in shaping the bacterial community assembly, and the co-inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 can increase the stochastic processes for driving the microbial community assembly. The contribution of the stochastic and deterministic processes with the inoculation of the Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 was also in agreement with these results (Figure 6). Although the relationships between the occurrence frequency of OTUs and their relative abundance variations were 35, 30, and 37%, we acknowledge that the co-inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5increased the stochastic processes for the fungi community assembly. The contribution of the deterministic processes in fungi was higher than those in bacteria, which also indicates that the deterministic process plays a more significant role in driving the fungal community assembly (Figure 6). The Nm-value was lower for bacterial taxa with the one-inoculation treatment of Bradyrhizobium japonicum 5038 (Nm = 26 170) and two co-inoculation treatment of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 (Nm = 26 858) than the non-inoculation treatment (Nm = 27 825). This phenomenon was more significant for the fungi community, and the Nm-value decreased to the one-inoculation treatment of Bradyrhizobium japonicum 5038 (Nm = 3961) and two co-inoculation treatments of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 (Nm = 3514) from non-inoculation (Nm = 5902). These results indicate that the species diffusion decreased after inoculating PGPR.
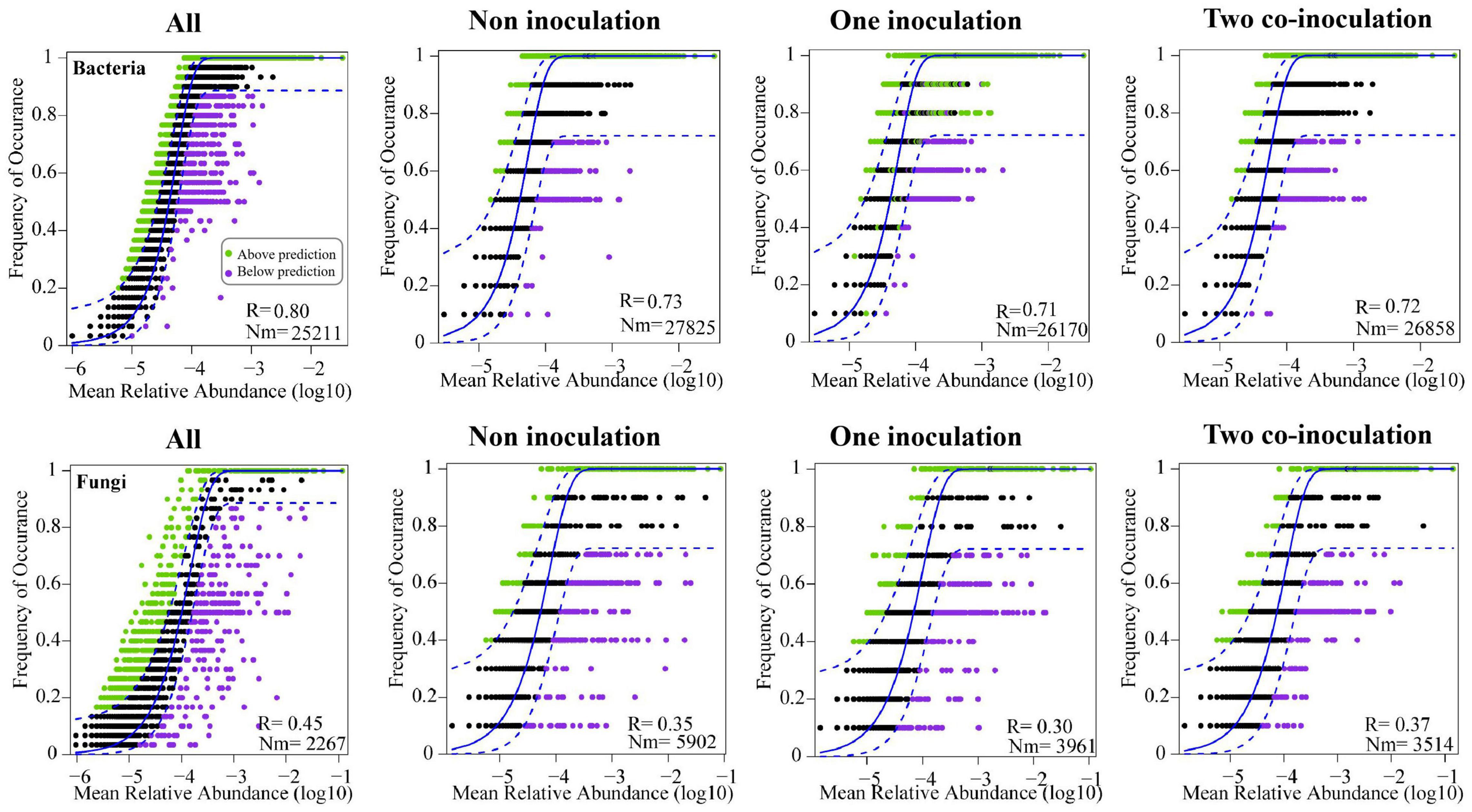
Figure 5. Fit of the neutral community model (NCM) of community assembly. The predicted occurrence frequencies for non-inoculation, one inoculation, two co-inoculation, and all representing soil bacterial and fungal communities. The solid blue lines indicate the best fit to the NCM, and the dashed blue lines represent 95% confidence intervals around the model prediction. OTUs that occur more or less frequently than predicted by the NCM are shown in different colors. Nm indicates the metacommunity size times immigration, R indicates the fit to this model.
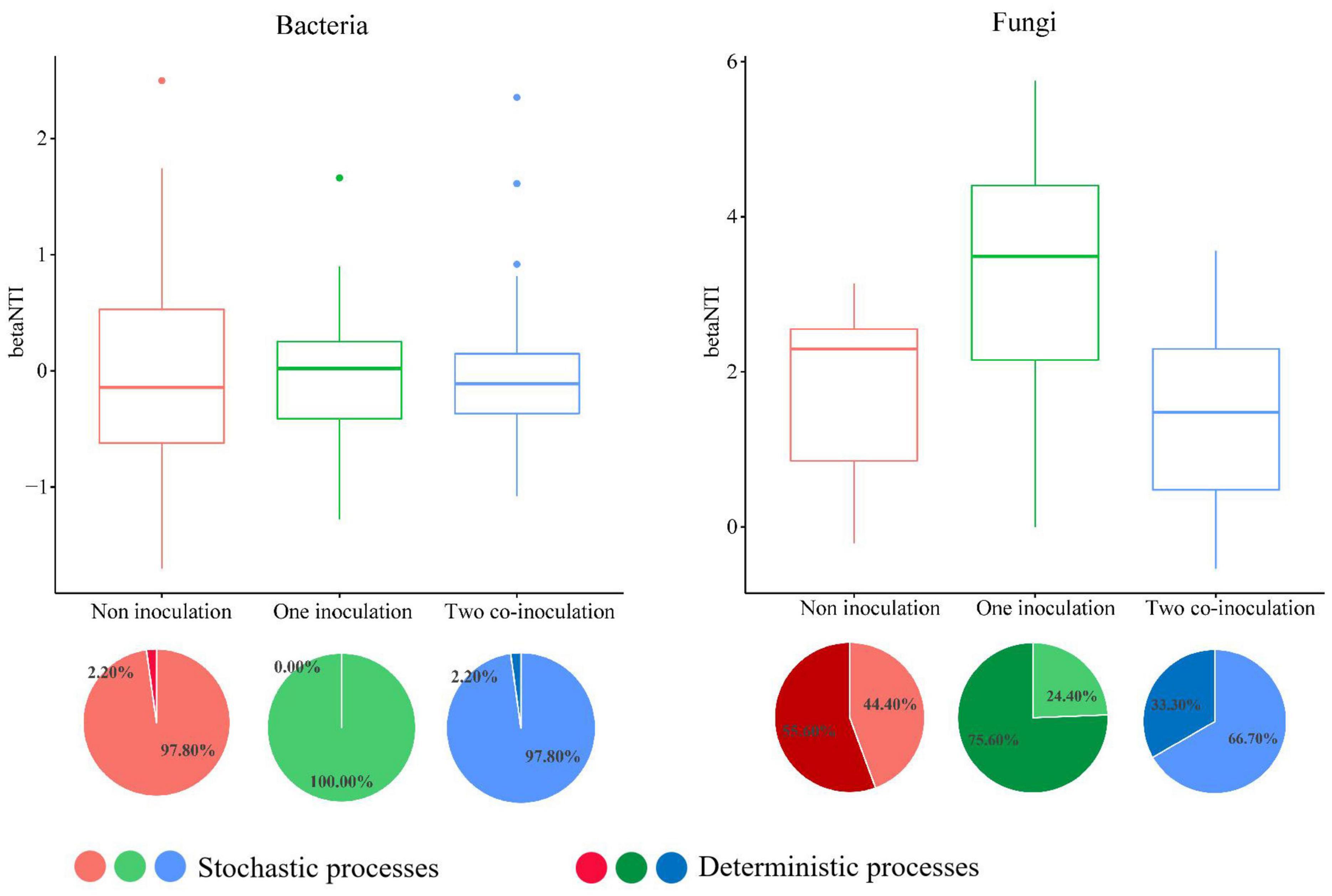
Figure 6. The community assembly processes by fitting niche-based theories. The relationship between inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 and betaNTI. The top 50 and 100 abundance OTU were chose for constructing phylogenetic tree. Pie plots showing the relative contribution of each ecological process in community assembly response along the inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5.unity model of community assembly.
Recently, the co-occurrence pattern in ecology has been widely adopted to explore the ecological interactions among microbial communities across various ecosystems (Williams et al., 2014; Fan et al., 2021; Gao et al., 2021). In this study, the obtained results indicated that the network of the microbial community in two soil types after inoculation of Bradyrhizobium japonicum 5038 became complex, and some functional microorganisms increased, e.g., the nitrogen-fixing Azospirillales (Supplementary Table S6). Several previous studies also presented similar results, which indicated that in addition to simulating the proliferation of potential beneficial microbes, Bradyrhizobium japonicum 5038 inoculation also increased connections in rhizobacterial networks and altered the hub taxa (Zhong et al., 2019; Shang et al., 2021). In another study, the results indicated that compared to grass treatments, legumes increased soil bacterial diversity and the abundance of nitrogen-fixing Nitrospirae groups (Zhou et al., 2017b). For fungi, Bradyrhizobium japonicum 5038 inoculation in the soybean also facilitated an increase in the number of connections between fungi, including the hub fungi shift to Phaeosphaeria, Sporobolomyces, Septoria, Edenia, and Leptospora (Xu et al., 2020). Moreover, by building mutants with a defective Nod factor, previous research confirmed that the symbiosis compromised NoeI mutant-inoculation could reduce microbial diversity and co-occurrence interactions, and decrease the abundance of beneficial microbes (Liu et al., 2020). This could be because the inoculated Bradyrhizobium japonicum 5038 forms a more symbiotic system with the host plant, which was changed by altering plant root exudates such as flavonoid (White et al., 2015), Catechin (Mommer et al., 2016), and some volatile organic compounds to change the soil physical and chemical environment and influence the prevalence of rhizosphere microorganisms (Vishwakarma et al., 2020).
It is widely acknowledged that Bacillus produces highly resistant spores that can survive for prolonged periods in the soil, which are very promising as potential inoculants in agriculture (Kumar et al., 2016; Ferreira et al., 2020; Oleńska et al., 2020). In this study, the nodes and edges of the co-occurrence networks increased with the PGPR species, thus indicating that the co-inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 leads to networks with more complexity than the single inoculated Bradyrhizobium japonicum 5038 (Figures 3, 4). Previous studies indicated with an increase in the diversity of microorganisms, the interaction relationship becomes stronger, which increases plant tolerance to a range of other abiotic stresses such as heavy metals contamination, water stress (flooding and drought), salinity, and cold (Gerhardt et al., 2009; Ju et al., 2020). After introducing Bacillus, pathogenic Fusarium certainly decreased to 0.89% from 1.24% in the individually inoculated Bradyrhizobium japonicum 5038 treatment in our study. Several studies verified that Bacillus spp. could produce 1-aminocyclopropane-1-carboxylate (ACC) to inhibit the stress-related hormone production in the plants and increase biotic stress resistance (Saleem et al., 2007; Hayat et al., 2010). Given the important role of environmental factors in driving microbial communities, the increase in the number of PGPR species can specifically provide microbial populations with more interactions owing to niche differentiation. In addition, keystone species are highly connected with other species within the networks, and they potentially exert a considerable influence on the entire microbial community (Gao et al., 2021). The primary reason for such effect is that different co-inoculated PGPR species can secrete different enzyme systems, such as iron transporter IRT1 (Zhang et al., 2009) and cell wall degrading enzyme (Bell et al., 2019), to increase the nutrient utilization rate. Increasing the supply of nutrients will increase the ecological niche of microorganisms, reduce competition, and allow more microorganisms to participate in the biochemical process. From the random forest analysis, our study also verified that Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 are the main microorganisms that alter the community structure.
Soil is a natural medium for cultivating soil microorganisms, and changes in its physical and chemical properties will trigger changes in the structure and interaction of the microbial community. The physical and chemical properties of different soil types will vary significantly, including the microbial communities (Gowda et al., 2021; Zhu et al., 2021). In the black soil, HN and Ca are the main influencing factors of the bacterial community, and NN and Na are the main influencing factors of the fungal community; in the cinnamon soil, the effects of TC, and NN are more significant in determining the microbial community (Supplementary Tables S2, S3). Interestingly, the same phenomenon emerged after co-inoculating Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 in the two soil types. In other words, the complexity of the network increased after inoculating the PGPR, and as the species of PGPR increased, the interaction between microorganisms also increased. Hence, the co-inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 has universal and positive effects on the soil microbial interaction. Collectively, the inoculation of beneficial microorganisms plays a crucial role in maintaining soil ecological health and increasing its resistance. Compound rhizobia agent positively influences plant growth and represents a promising sustainable trend to increase plant production.
In this study, we examined bacterial and fungal community assembly in agroecosystems across two major soybean production areas throughout north China. Elucidating the relationship between community assembly and species coexistence is fundamental and crucial for understanding ecosystem diversity and functioning. Previous reports indicated that microbial co-occurrence associations tended to be higher when communities were primarily driven by the dispersal limitation relative to species sorting (Jiao et al., 2020). Our findings are in agreement with the finding that the co-inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 increased the diversity of bacterial and fungal communities and the complexity of the co-occurrence pattern, at the same time, species diffusion in the community assembly process decreased after inoculating the PGPR species.
The deterministic processes refer to local environmental factors and biotic interactions that are responsible for shaping the microbial community, whereas stochastic processes consider birth, death, speciation, and immigration, which play a key role in shaping the microbial community assembly (Sloan et al., 2006; Zhou and Ning, 2017). In our study, the changes in the stochastic and deterministic processes in the bacterial and fungal communities differed within the inoculation of PGPR. The main assembly processes were stochastic processes for bacteria in the two ecological models, whereas the deterministic process kept increasing for fungi. These results could be attributed to the diversity of the microbial community. Xun et al. (2019) demonstrated that deterministic and stochastic assembly processes are dominant in low- and high-diversity communities, respectively. The bacterial communities exhibit more diversity (Chao index of 4002-4401) than fungi communities (Chao index of 604–814); thus, stochastic processes are dominant for bacteria. Although a previous study demonstrated that deterministic processes play a dominant role in bacterial community assembly in soybean fields (Zhang et al., 2018), the assembly processes of soybean rhizobacterial communities differed in different soybean cultivars and soil types (Liu et al., 2019; Zhong et al., 2019). The inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5s also exerts less impact on the assembly process, because bacteria have more species in sufficient ecological niches. Hence, although the interaction of bacteria increased with the PGPR species (Figure 3), the contribution of the biotic factor was limited. The environmental factor might be the main driving factor for the changes in the bacterial community. In addition, these speculations are well supported by the VPA, which indicated that the environmental factor could explain 86% of the changes in the bacterial community (Supplementary Figure S9).
The contributions of the deterministic assembly process were higher for fungi than bacteria. The VPA demonstrated that the environment factor explains 48% of the changes in the fungal communities, and the 52% residuals, probably owing to the increasing interaction following the co-inoculation of PGPR (Supplementary Figure S9). The inoculation of Bradyrhizobium japonicum 5038, due to the increase in the deterministic assembly process (75.60%) (Figure 6), indicated that the environmental factor could exert a more significant effect; thus, the resistance of the fungal community may decrease. When Bacillus was added, the contribution of the deterministic assembly processes decreased to 33.30%, whereas the contribution of the stochastic processes increased to 66.70% (Figure 6). Bacillus further increased the diversity of fungi, could replenish the ecological niche in time, and competitively prevent the invasion of pathogenic microorganisms (Ongena and Jacques, 2008; Cawoy et al., 2011). Furthermore, the added Bacillus further limited the species diffusion and restricted the direction of movement of pathogenic microorganisms. The shift of balance between the deterministic and stochastic processes was also due to different ecological responses of special taxa (e.g., habitat specialists) to the environmental changes under a specific situation (Leibold et al., 2017; Gad et al., 2020). For example, the abundance of Cyphellophora with a significant relationship with the PGPR species decreased after the inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 in black soil (Supplementary Figure S6). Previous research showed that several Cyphellophora species have been associated with potential pathogens (Decock et al., 2003). Consequently, the co-inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 could form a protective microdomain in the soybean rhizosphere to defense against soil-borne fungal pathogens, especially Fusarium (wilt disease) (Fu et al., 2017; Mendes et al., 2018).
The VPA (Supplementary Table S7) and Mantel test (Supplementary Tables S2, S3) all demonstrated that nitrogen content was a vital environment factor for altering the microbial community, and by altering HN and AN contents, the co-inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 could change the microbial community and further drive the assembly process in the black and cinnamon soil, respectively. This consistency may be attributed to the increase in the abundance of Azospirillales with the co-inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 (Supplementary Table S6). According to the above analysis, although the change in the microbial diversity is not significant, the co-inoculation of Rhizobium and Bacillus can substantially influence the interaction and community construction of soil microorganisms to a great extent via changes in the nitrogen content.
In conclusion, the co-inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 could structure the bacterial and fungi communities, and with an increase in the number of PGPR species, the bacterial and fungal diversity and complexity of the networks increased. The co-inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 negligibly influenced the assembly process of the soil bacterial community; however, it exerted a significant impact on the assembly process of the fungal community. Moreover, our results suggest that the inoculation of Bradyrhizobium japonicum 5038 increases the deterministic process for fungi, whereas the addition of Bacillus aryabhattai MB35-5 to Bradyrhizobium japonicum 5038 increases the stochastic process for fungi. The co-inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 could limit the process of species dispersal diffusion, which might be one reason for controlling the pathogen invasion. In general, these findings have broadened our understanding of the response of the bacterial and fungi communities on the co-inoculation of Rhizobium and Bacillus in farmland, which provides a certain reference for the development and application of Rhizobium compound inoculants.
The original contributions presented in the study are publicly available. This data can be found here: NCBI, under accession numbers: PRJNA788980, PRJNA788360.
YZ, DG, and JL designed the experiment. YZ analyzed all data and wrote the first draft of the manuscript. XL and G-FG contributed new reagents or analytical tools. DG, FM, BL, and PX performed the field experiment. XJ, MM, FC, LL, and JL edited the manuscript and checked the language. All the authors contributed to manuscript revisions.
This work was supported by National Key Research and Development Program of China (2021YFD1700200), the China Postdoctoral Science Foundation (2021M693450), and the earmarked fund for Modern Agro-industry Technology Research System (CARS-04).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Prof. Haiyan Chu of the Institute of Soil Science, Chinese Academy of Sciences for his help in microbial data analysis, and thank Dr. Yaowei Kang (Life Sciences Collage of Zhaoqing University) for his guidance on the study of Bacillus aryabhattai MB35-5.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.846359/full#supplementary-material
Acero, P., Ayora, C., Torrentó, C., and Nieto, J. M. (2006). The behavior of trace elements during schwertmannite precipitation and subsequent transformation into goethite and jarosite. Geochim. Cosmochim. Ac. 70, 4130–4139. doi: 10.1016/j.gca.2006.06.1367
Amato, K. R., Yeoman, C. J., Kent, A., Righini, N., Carbonero, F., Estrada, A., et al. (2013). Habitat degradation impacts black howler monkey (alouatta pigra) gastrointestinal microbiomes. ISME J. 7, 1344–1353. doi: 10.1038/ismej.2013.16
Bell, C. A., Lilley, C. J., McCarthy, J., Atkinson, H. J., and Urwin, P. E. (2019). Plant-parasitic nematodes respond to root exudate signals with host-specific gene expression patterns. PLoS Pathog. 15:e1007503.1007503 doi: 10.1371/journal.ppat
Bi, X. D., Dai, W., Wang, X. Y., Dong, S. J., Zhang, S. L., Zhang, D. J., et al. (2019). Effects of Bacillus subtilis on the growth, colony maintenance, and attached bacterial community composition of colonial cyanobacteria. Environ. Sci. Pollut. R. 26, 14977–14987. doi: 10.1007/s11356-019-04902-y
Cawoy, H., Bettiol, W., Fickers, P., and Ongena, M. (2011). “Bacillus-based biological control of plant diseases,” in Pesticides in the Modern World-pesticides Use and Management, ed. Stoytcheva (InTech: Academic Press).
Chen, W. D., Ren, K. X., Isabwe, A., Chen, H. H., Liu, M., and Yang, J. (2019). Stochastic processes shape microeukaryotic community assembly in a subtropical river across wet and dry seasons. Microbiome 7, 1–16. doi: 10.1186/s40168-019-0749-8
Chen, Y. F., Ye, J. R., and Kong, Q. Q. (2020). Potassium-solubilizing activity of Bacillus aryabhattai SK1-7 and its growth-promoting effect on Populus alba L. Forests 11:1348. doi: 10.3390/f11121348
Decock, C., Delgado-Rodríguez, G., Buchet, S., and Seng, J. M. (2003). A new species and three new combinations in Cyphellophora, with a note on the taxonomic affinities of the genus, and its relation to Kumbhamaya and Pseudomicrodochium. Antonie Van Leeuwenhoek 84, 209–216. doi: 10.1023/A:102601503185
Fan, K., Delgado-Baquerizo, M., Guo, X., Wang, D., and Zhu, Y. G. (2021). Biodiversity of key-stone phylotypes determines crop production in a 4-decade fertilization experiment. ISME J. 15, 550–561. doi: 10.1038/s41396-020-00796-8
Faust, K., and Raes, J. (2012). Microbial interactions: from networks to models. Nat. Rev. Microbiol. 10, 538–550. doi: 10.1038/nrmicro2832
Feng, Y., Chen, R., Stegen, J. C., Guo, Z., Zhang, J., Li, Z., et al. (2018). Two key features influencing community assembly processes at regional scale: initial state and degree of change in environmental conditions. Mol. Ecol. 27, 5238–5251. doi: 10.1111/mec.14914
Ferreira, L. D. V. M., De Carvalho, F., Andrade, J. F. C., Oliveira, D. P., De Medeiros, F. H. V., and Moreira, F. M. D. S. (2020). Co-inoculation of selected nodule endophytic rhizobacterial strains with Rhizobium tropici promotes plant growth and controls damping off in common bean. Pedosphere 30, 98–108. doi: 10.1016/S1002-0160(19)60825-8
Fu, L., Penton, C. R., Ruan, Y., Shen, Z. Z., Xue, C., Li, R., et al. (2017). Inducing the rhizosphere microbiome by biofertilizer application to suppress banana Fusarium wilt disease. Soil Boil. Biochem. 104, 39–48. doi: 10.1016/j.soilbio.2016.10.008
Gad, M., Hou, L. Y., Li, J. W., Wu, Y., Rashid, A., Chen, N. W., et al. (2020). Distinct mechanisms underlying the assembly of microeukaryotic generalists and specialists in an anthropogenically impacted river. Sci. Total Environ. 748:141434. doi: 10.1016/j.scitotenv.2020.141434
Gao, G. F., Peng, D., Zhang, Y., Li, Y., Fan, K., Tripathi, B. M., et al. (2021). Dramatic change of bacterial assembly process and co-occurrence pattern in Spartina alterniflora salt marsh along an inundation frequency gradient. Sci. Total. Environ. 755:142546. doi: 10.1016/j.scitotenv.2020.142546
Gerhardt, K. E., Huang, X. D., Glick, B. R., and Greenberg, B. M. (2009). Phytoremediation and rhizoremediation of organic soil contaminants: potential and challenges. Plant Sci. 176, 20–30. doi: 10.1016/j.plantsci.2008.09.014
Gowda, S., Swarnalakshmi, K., Sharma, M., Reddy, K., Bhoumik, A., Suman, A., et al. (2021). Soil Type Influence Nutrient Availability, Microbial Metabolic Diversity, Eubacterial and Diazotroph Abundance in Chickpea Rhizosphere. Bethesda, MD: ORCID
Hayat, R., Ali, S., Amara, U., Khalid, R., and Ahmed, I. (2010). Soil beneficial bacteria and their role in plant growth promotion: a review. Ann. Microbiol. 60, 579–598. doi: 10.1007/s13213-010-0117-1
Hubbell, S. P. (2011). The Unified Neutral Theory of Biodiversity and Biogeography (MPB-32). Princeton, NJ: Princeton University Press, doi: 10.1515/9781400837526
Iturralde, E. T., Stocco, M. C., Faura, A., Mónaco, C. I., Cordo, C., Pérez-Giménez, J., et al. (2020). Coinoculation of soybean plants with Bradyrhizobium japonicum and Trichoderma harzianum: coexistence of both microbes and relief of nitrate inhibition of nodulation. Biotechnol. Rep. 26:e00461. doi: 10.1016/j.btre.2020.e00461
Jiao, S., Yang, Y. F., Xu, Y. Q., Zhang, J., and Lu, Y. H. (2020). Balance between community assembly processes mediates species coexistence in agricultural soil microbiomes across eastern China. ISME J. 14, 202–216. doi: 10.1038/s41396-019-0522-9
Ju, W., Jin, X., Liu, L., Shen, G., Zhao, W., Duan, C., et al. (2020). Rhizobacteria inoculation benefits nutrient availability for phytostabilization in copper contaminated soil: drivers from bacterial community structures in rhizosphere. Appl. Soil. Ecol. 150:103450. doi: 10.1016/j.apsoil.2019.103450
Jun, Y. A. N., Xiaozeng, H. A. N., Xu, C. H. E. N., Xinchun, L. U., Wenfeng, C. H. E. N., Entao, W. A. N. G., et al. (2019). Effects of long-term fertilization strategies on soil productivity and soybean rhizobial diversity in a Chinese Mollisol. Pedosphere 29, 784–793. doi: 10.1016/S1002-0160(17)60470-3
Kumar, A., Meena, V. S., Roy, P., and Vandana Kumari, R. (2019). “Role of rhizobia for sustainable agriculture: lab to land,” in Plant Growth Promoting Rhizobacteria for Agricultural Sustainability, eds A. Kumar and V. Meena (Singapore: Springer), 129–149. doi: 10.1007/978-981-13-7553-8_7
Kumar, P., Pandey, P., Dubey, R. C., and Maheshwari, D. K. (2016). Bacteria consortium optimization improves nutrient uptake, nodulation, disease suppression and growth of the common bean (Phaseolus vulgaris) in both pot and field studies. Rhizosphere 2, 13–23. doi: 10.1016/j.rhisph.2016.09.002
Lee, S., Ka, J. O., and Song, H. G. (2012). Growth promotion of Xanthium italicum by application of rhizobacterial isolates of Bacillus aryabhattai in microcosm Soil. J. Microbiol. 50, 45–49. doi: 10.1007/s12275-012-1415-z
Leibold, M. A., Chase, J. M., and Ernest, S. M. (2017). Community assembly and the functioning of ecosystems: how metacommunity processes alter ecosystems attributes. Ecology 98, 909–919. doi: 10.1002/ecy.1697
Li, X. Y., Deng, Y., Li, Q., Lu, C. Y., Wang, J. J., Zhang, H. W., et al. (2013). Shifts of functional gene representation in wheat rhizosphere microbial communities under elevated ozone. ISME J. 7, 660–671. doi: 10.1038/ismej.2012.120
Liu, C. M., Yang, Z. F., He, P. F., Munir, S., Wu, Y. X., Ho, H. H., et al. (2018). Deciphering the bacterial and fungal communities in clubroot-affected cabbage rhizosphere treated with Bacillus subtilis XF-1. Agr. Ecosyst. Environ. 256, 12–22. doi: 10.1016/j.agee.2018.01.001
Liu, F., Hewezi, T., Lebeis, S. L., Pantalone, V., Grewal, P. S., and Staton, M. E. (2019). Soil indigenous microbiome and plant genotypes cooperatively modify soybean rhizosphere microbiome assembly. BMC Microbiol. 19:201. doi: 10.1186/s12866-019-1572-x
Liu, Y., Ma, B., Chen, W., Schlaeppi, K., Erb, M., Stirling, E., et al. (2020). The Symbiotic Capacity of Rhizobium Shapes Root-associated Microbiomes. Bethesda, MD: ORCID
Mahmud, K., Missaoui, A., Lee, K., Ghimire, B., Presley, H. W., and Makaju, S. (2021). Rhizosphere microbiome manipulation for sustainable crop production. Curr. Plant Biol. 27:100210. doi: 10.1016/j.cpb.2021.100210
Maynard, D. S., Bradford, M. A., Covey, K. R., Lindner, D., Glaeser, J., Talbert, D. A., et al. (2019). Consistent trade-offs in fungal trait expression across broad spatial scales. Nat. Microbiol. 4, 846–853. doi: 10.1038/s41564-019-0361-365
Mendes, L. W., Raaijmakers, J. M., de Hollander, M., Mendes, R., and Tsai, S. M. (2018). Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. ISME J. 12, 212–224. doi: 10.1038/ismej.2017.158
Mommer, L., Hinsinger, P., Prigent-Combaret, C., and Visser, E. J. W. (2016). Advances in the rhizosphere: stretching the interface of life. Plant Soil. 407, 1–8. doi: 10.1007/s11104-016-3040-9
Moretti, L. G., Crusciol, C. A., Kuramae, E. E., Bossolani, J. W., Moreira, A., Costa, N. R., et al. (2020). Effects of growth-promoting bacteria on soybean root activity, plant development, and yield. Agron. J. 112, 418–428. doi: 10.1002/agj2.20010
Mulvaney, R. L. (1996). “Nitrogen-inorganic forms,” in Method of Soil Analysis. Part 3. Chemical Methods, ed. D. L. Sparks (Madison, WI: American Society of Agronomy and Soil Science Society of America).
Neda, E. K. (2021). Competency of rhizobial inoculation in sustainable agricultural production and biocontrol of plant diseases. Front. Sustain. Food S. 5:728014. doi: 10.3389/fsufs.2021.728014
Nimnoi, P., Lumyong, S., and Pongsilp, N. (2011). Impact of rhizobial inoculants on rhizosphere bacterial communities of three medicinal legumes assessed by denaturing gradient gel electrophoresis (DGGE). Ann. Microbiol. 61, 237–245. doi: 10.1007/s13213-010-0128-y
Oleńska, E., Małek, W., Wójcik, M., Swiecicka, I., Thijs, S., and Vangronsveld, J. (2020). Beneficial features of plant growth-promoting rhizobacteria for improving plant growth and health in challenging conditions: a methodical review. Sci. Total Environ. 743:140682. doi: 10.1016/j.scitotenv.2020.140682
Ongena, M., and Jacques, P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–125. doi: 10.1016/j.tim.2007.12.009
Pailan, S., Gupta, D., Apte, S., Krishnamurthi, S., and Saha, P. (2015). Degradation of organophosphate insecticide by a novel Bacillus aryabhattai strain SanPS1, isolated from soil of agricultural field in Burdwan, West Bengal, India. Int. Biodeterior. Biodegrad. 103, 191–195. doi: 10.1016/j.ibiod.2015.05.006
Park, Y. G., Mun, B. G., Kang, S. M., Hussain, A., and Yun, B. W. (2017). Bacillus aryabhattai srb02 tolerates oxidative and nitrosative stress and promotes the growth of soybean by modulating the production of phytohormones. PLoS One 12:e0173203. doi: 10.1371/journal.pone.0173203
Qin, Y. X., Shang, Q. M., Zhang, Y., Li, P. L., and Chai, Y. R. (2017). Bacillus amyloliquefaciens L-S60 reforms the rhizosphere bacterial community and improves growth conditions in cucumber plug seedling. Front. Microbiol. 8:2620. doi: 10.3389/fmicb.2017.02620
Ramesh, A., Sharma, S. K., Sharma, M. P., Yadav, N., and Joshi, O. P. (2014). Inoculation of zinc solubilizing Bacillus aryabhattai strains for improved growth, mobilization and biofortification of zinc in soybean and wheat cultivated in Vertisols of central India. Appl. Soil Ecol. 73, 87–96. doi: 10.1016/j.apsoil.2013.08.009
Roper, W. R., Duckworth, O. W., Grossman, J. M., and Israel, D. W. (2020). Rhizobium leguminosarum strain combination effects on nodulation and biological nitrogen fixation with Vicia villosa. Appl. Soil Ecol. 156:103703. doi: 10.1016/j.apsoil.2020.103703
Rousk, J., Bååth, E., Brookes, P. C., Lauber, C. L., Lozupone, C., Caporaso, J. G., et al. (2010). Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4, 1340–1351. doi: 10.1038/ismej.2010.58
Saleem, M., Arshad, M., Hussain, S., and Bhatti, A. S. (2007). Perspective of plant growth promoting rhizo-bacteria (PGPR) containing ACC deaminase in stress agriculture. J. Ind. Microbiol. Biotechnol. 34, 635–648. doi: 10.1007/s10295-007-0240-6
Schumpp, O., and Deakin, W. J. (2010). How inefficient rhizobia prolong their existence within nodules. Trends. Plant Sci. 15, 189–195. doi: 10.1016/j.tplants.2010.01.001
Sengupta, A., Indivero, J., Gunn, C., Tfaily, M. M., Chu, R. K., Toyoda, J., et al. (2019). Spatial gradients in the characteristics of soil-carbon fractions are associated with abiotic features but not microbial communities. Biogeosciences 16, 3911–3928. doi: 10.5194/bg-16-3911-2019
Shang, J. Y., Wu, Y., Huo, B., Chen, L., Wang, E. T., Sui, Y., et al. (2021). Potential of bradyrhizobia inoculation to promote peanut growth and beneficial rhizobacteria abundance. J. Appl. Microbiol. 131, 2500–2515. doi: 10.1111/jam.15128
Shi, Y., Li, Y., Xiang, X., Sun, R., Yang, T., He, D., et al. (2018). Spatial scale affects the relative role of stochasticity versus determinism in soil bacterial communities in wheat fifields across the North China Plain. Microbiome 6:27. doi: 10.1186/s40168-018-0409-404
Shivaji, S., Chaturvedi, P., Begum, Z., Pindi, P. K., Manorama, R., Padmanaban, D. A., et al. (2009). Janibacter hoylei sp nov., Bacillus isronensis sp nov and Bacillus aryabhattai sp nov., isolated from cryotubes used for collecting air from the upper atmosphere. Int. J. Syst. Evol. Microbiol. 59, 2977–2986. doi: 10.1099/ijs.0.002527-0
Silvertown, J. (2004). Plant coexistence and the niche. Trends. Ecol. Evol. 19, 605–611. doi: 10.1016/j.tree.2004.09.003
Sloan, W. T., Lunn, M., Woodcock, S., Head, I. M., Nee, S., and Curtis, T. P. (2006). Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ. Microbiol. 8, 732–740. doi: 10.1111/j.1462-2920.2005.00956.x
Srinivasan, M., Holl, F. B., and Petersen, D. J. (1997). Nodulation of Phaseolus vulgaris by Rhizobium etli is enhanced by the presence of Bacillus. Can. J. Microbiol. 43, 1–8. doi: 10.1139/m97-001
Stegen, J. C., Lin, X. J., Konopka, A. E., and Fredrickson, J. K. (2012). Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 6, 1653–1664. doi: 10.1038/ismej.2012.22
Stewart, W. D. P. (1967). Nitrogen-fixing plants: the role of biological agents as providers of combined nitrogen is discussed. Science 158:1426. doi: 10.1126/science.158.3807.1426
Strickland, T. C., and Sollins, P. (1987). Improved method for separating light-and heavy-fraction organic material from soil. Soil Sci. Soc. Am. J. 51, 1390–1393. doi: 10.2136/sssaj1987.03615995005100050056x
Tedersoo, L., Bahram, M., Põlme, S., Kõljalg, U., Yorou, N. S., Wijesundera, R., et al. (2014). Global diversity and geography of soil fungi. Science 346:1256688. doi: 10.1126/science.1256688
Thilakarathna, M. S., and Raizada, M. N. (2017). A meta-analysis of the effectiveness of diverse rhizobia inoculants on soybean traits under field conditions. Soil Bio. Biochem. 105, 177–196. doi: 10.1016/j.soilbio.2016.11.022
Tripathi, B. M., Stegen, J. C., Kim, M., Dong, K., Adams, J. M., and Lee, Y. K. (2018). Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. ISME J. 12, 1072–1083. doi: 10.1038/s41396-018-0082-4
Vishwakarma, K., Kumar, N., Shandilya, C., Mohapatra, S., Bhayana, S., and Varma, A. (2020). Revisiting plant–microbe interactions and microbial consortia application for enhancing sustainable agriculture: a review. Front. Microbiol. 11:3195. doi: 10.3389/fmicb.2020.560406
Wan, T. T., Zhao, H. H., and Wang, W. (2018). Effects of the biocontrol agent Bacillus amyloliquefaciens SN16-1 on the rhizosphere bacterial community and growth of tomato. J. Phytopathol. 166, 324–332. doi: 10.1111/jph.12690
Wang, H., Man, C. X., Wang, E. T., and Chen, W. X. (2009). Diversity of rhizobia and interactions among the host legumes and rhizobial genotypes in an agricultural-forestry ecosystem. Plant Soil 314, 169–182. doi: 10.1007/s11104-008-9716-z
Wang, J. J., Xu, S., Yang, R., Zhao, W., Zhu, D., Zhang, X. X., et al. (2021). Bacillus amyloliquefaciens FH-1 significantly affects cucumber seedlings and the rhizosphere bacterial community but not soil. Sci. Rep. 11:12055. doi: 10.1038/s41598-021-91399-6
White, L. J., Jothibasu, K., Reese, R. N., Brözel, V. S., and Subramanian, S. (2015). Spatio temporal influence of isoflavonoids on bacterial diversity in the soybean rhizosphere. Mol. Plant Microbe Interact. 28, 22–29. doi: 10.1094/MPMI-08-14-0247-R
Williams, R. J., Howe, A., and Hofmockel, K. S. (2014). Demonstrating microbial co-occurrence pattern analyses within and between ecosystems. Front. Microbiol. 5:358. doi: 10.3389/fmicb.2014.00358
Xu, H., Yang, Y., Tian, Y., Xu, R., Zhong, Y., and Liao, H. (2020). Rhizobium inoculation drives the shifting of rhizosphere fungal community in a host genotype dependent manner. Front. Microbiol. 10:3135. doi: 10.3389/fmicb.2019.03135
Xu, N., Tan, G. G., Wang, H. Y., and Gai, X. P. (2016). Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil Biol. 74, 1–8. doi: 10.1016/j.ejsobi.2016.02.004
Xun, W. B., Li, W., Xiong, W., Ren, Y., Liu, Y. P., Miao, Y. Z., et al. (2019). Diversity-triggered deterministic bacterial assembly constrains community functions. Nat. Commun. 10:3833. doi: 10.1038/s41467-019-11787-5
Yan, Y., Zhang, L., Yu, M. Y., Wang, J., Tang, H., and Yang, Z. W. (2016). The genome of Bacillus aryabhattai T61 reveals its adaptation to Tibetan Plateau environment. Genes Genom. 38, 293–301. doi: 10.1007/s13258-015-0366-2
Zhang, B. G., Zhang, J., Liu, Y., Guo, Y. Q., Shi, P., and Wei, G. H. (2018). Biogeography and ecological processes affecting root-associated bacterial communities in soybean fields across China. Sci. Total Environ. 627, 20–27. doi: 10.1016/j.scitotenv.2018.01.230
Zhang, H., Sun, Y., Xie, X., Kim, M. S., Dowd, S. E., and Paré, P. W. (2009). A soil bacterium regulates plant acquisition of iron via deficiency-inducible mechanisms. Plant J. 58, 568–577. doi: 10.1111/j.1365-313X.2009.03803.x
Zhao, P., Bao, J., Wang, X., Liu, Y., Li, C., and Chai, B. (2019). Deterministic processes dominate soil microbial community assembly in subalpine coniferous forests on the Loess Plateau. Peer J. 7:e6746. doi: 10.7717/peerj.6746
Zhong, Y. J., Yang, Y. Q., Liu, P., Xu, R. N., Rensing, C., and Fu, X. D. (2019). Genotype and rhizobium inoculation modulate the assembly of soybean rhizobacterial communities. Plant Cell Environ. 42, 2028–2044. doi: 10.1111/pce.13519
Zhou, J., and Ning, D. (2017). Stochastic community assembly: does it matter in microbial ecology? Microbiol. Mol. Biol. Rev. 81:e00002-17. doi: 10.1128/MMBR.00002-17
Zhou, Y., Zhu, H., Fu, S., and Yao, Q. (2017a). Metagenomic evidence of stronger effect of stylo (legume) than bahiagrass (grass) on taxonomic and functional profiles of the soil microbial community. Sci. Rep. 7:10195. doi: 10.1038/s41598-017-10613-6
Zhu, J., Jiang, X., Guan, D. W., Kang, Y. W., Li, L., Cao, F. M., et al. (2022). Effects of rehydration on physiological and transcriptional responses of a water-stressed rhizobium. J. Microbiol. 60, 31–46. doi: 10.1007/s12275-022-1325-7
Zhou, Y., Zhu, H. Z., Fu, S. L., and Yao, Q. (2017b). Variation in soil microbial community structure associated with different legume species is greater than that associated with different grass species. Front. Microbiol. 8:1007. doi: 10.3389/fmicb.2017.01007
Zhu, Y., Guo, B., Liu, C., Lin, Y. C., Fu, Q. L., Li, N. Y., et al. (2021). Soil fertility, enzyme activity, and microbial community structure diversity among different soil textures under different land use types in coastal saline soil. J. Soil Sediment. 21, 2240–2252. doi: 10.1007/s11368-021-02916-z
Keywords: Rhizobium, Bacillus, co-inoculation, microbial assembly process, co-occurrence pattern
Citation: Zhao Y, Guan D, Liu X, Gao G-F, Meng F, Liu B, Xing P, Jiang X, Ma M, Cao F, Li L and Li J (2022) Profound Change in Soil Microbial Assembly Process and Co-occurrence Pattern in Co-inoculation of Bradyrhizobium japonicum 5038 and Bacillus aryabhattai MB35-5 on Soybean. Front. Microbiol. 13:846359. doi: 10.3389/fmicb.2022.846359
Received: 31 December 2021; Accepted: 18 February 2022;
Published: 18 March 2022.
Edited by:
Andreas Ulrich, Leibniz Centre for Agricultural Landscape Research (ZALF), GermanyReviewed by:
Sushil K. Sharma, National Institute of Biotic Stress Management, IndiaCopyright © 2022 Zhao, Guan, Liu, Gao, Meng, Liu, Xing, Jiang, Ma, Cao, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Li, bGlqdW4wMUBjYWFzLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.