- 1School of Life Sciences, Henan University, Kaifeng, China
- 2College of Geography and Environmental Science, Henan University, Kaifeng, China
Climate models predict significant changes in precipitation magnitude in semi-arid grasslands, so it is vital to improve our understanding of how changing precipitation affects microbial communities in grassland ecosystems. Using a long-term field manipulation experiment, we evaluated the responses of microbial communities to a decrease (DP) and an increase (IP) in precipitation on a semi-arid grassland in northern China. The results showed that bacterial species richness decreased significantly with DP but remained stable under IP. Relative abundance of oligotrophic, slow-growing bacterial phyla (e.g., Chloroflexi and Acidobacteria) increased with DP but decreased with IP, whereas the relative abundance of copiotrophic, fast-growing bacterial phyla (e.g., Proteobacteria and Bacteroidetes) decreased with DP but increased with IP. In contrast, diversity, species composition, and relative abundance of different fungal phyla change little with DP or IP. These results indicate a greater sensitivity of bacteria to precipitation changes than fungi, and the sensitivity of bacteria to DP was higher than IP. Our findings are important for understanding soil microbial dynamics under future climate change scenarios.
Introduction
Climate change is affecting precipitation patterns and causing changes in the amount of precipitation (Huntington, 2006). Changes in the amount of precipitation greatly affect soil microbes and the biogeochemical pathways they control, especially in semi-arid grasslands where water availability is increasingly important for regulating microbial community dynamics (Nielsen and Ball, 2015). However, limited knowledge is available on how altering soil water and nutrient availability induced by precipitation changes regulating microbial communities (Evans and Wallenstein, 2014).
The effects of change in the amount of precipitation on microbial communities have been well-reported (Clark et al., 2009; Landesman and Dighton, 2010; Nielsen and Ball, 2015; Gao et al., 2016; Zhou et al., 2018). However, experimental results have been highly variable. Some studies have found that drought shifts the bacterial community toward Gram-positive bacteria (with strong, thick, interlinked peptidoglycan cell walls) because they are inherently resistant to dry conditions (Uhlirová et al., 2005; Zhou et al., 2018). In contrast, no changes in species composition, either in bacterial or fungi communities after decreases in precipitation, have been demonstrated in tallgrass prairie (Williams, 2007). Fungi tend to be drought-tolerant due to their ability to accumulate osmoregulatory solutes to protect their metabolism and filamentous structure; these properties allow fungi to exploit substrates in very dry soils (Schimel et al., 2007; Nielsen and Ball, 2015). Therefore, it is necessary to examine how changes in precipitation amount affect bacterial groups, fungal groups, and different phyla of bacteria and fungi that display contrasting water-related life strategies.
Abiotic factors such as soil temperature (Feng and Simpson, 2009), soil moisture (Brockett et al., 2012), and soil pH (Fierer and Jackson, 2006; Lauber et al., 2009) are major drivers of microbial communities. Changes in precipitation could directly influence soil microbes by altering soil water availability or indirectly by regulating nutrient availability and soil temperature (Brockett et al., 2012; Koyama et al., 2017). Plant communities can also affect microbial communities by regulating microclimatic variability, resource availability, and habitat complexity (Waldrop et al., 2006; Koyama et al., 2017). Maestre et al. (2015) showed that drought reduced diversity and abundance of soil bacteria and fungi by decreasing plant cover and consequent organic carbon inputs into the soil. Changes in plant diversity can lead to changes in plant products and diversity of litter organic components, thus affecting the composition and function of soil microbial communities (Spehn et al., 2000). However, Meier and Bowman (2008) have found that soil microbial diversity is positively correlated with the diversity of chemical components produced by plant litters while there is no direct correlation with the diversity of plant species.
Semi-arid grasslands are very sensitive to changes in soil water availability, and changes in precipitation have been reported to affected plant species diversity and community composition (Zhang et al., 2020a). However, how changes in precipitation shift microbial communities and the drivers of microbial communities are still unclear due to the complexity of soil ecosystem and the limitation of soil microbial research techniques. Here, we performed a manipulation experiment in a semi-arid steppe in northern China to answer the following two questions: (1) how do increases (IP) and decreases (DP) in precipitation change the bacterial and fungal groups? (2) which factors drive the alterations in microbial communities under changing precipitation?
Materials and Methods
Study Site
The study site is located in a typical temperate steppe at the Duolun Restoration Ecology Station (42°02′N, 116°16′E, 1324 m a.s.l.) in Inner Mongolia, China. Mean annual precipitation from 1961 to 2014 in the area is ∼383 mm with more than 90% occurring in the growing season (from May to October). Mean annual temperature is 2.1°C, with a monthly mean temperature of −17.5°C in January and 18.9°C in July (Yang et al., 2020).
Experimental Design
We used a randomized block design with three treatments: ambient precipitation (C), a 60% decrease in growing season precipitation (from April to October, DP), and a 60% increase in growing season precipitation (from April to October, IP). Each treatment had five replicates giving 15 plots in total. Each plot had an area of 16 m2 (4 m × 4 m), and was randomly distributed in each block. Buffer zones between plots were 1.5 m wide (Zhang et al., 2020a). Precipitation was blocked using shelters in the DP plots when IP plots received additional 60% precipitation with a handheld irrigation system. The experiment was established on April 15, 2015.
Vegetation Monitoring
The above-ground vegetation was sampled in 1 m × 1 m quadrat in each plot at the end of September when plant biomass reached its peak level (Sagar et al., 2017). Species richness and abundance were estimated, and individual plants in each quadrat were clipped to the soil surface. Biomass was then sorted by species, and number of the species was also recorded for each quadrat. All shoot samples were dried at 80°C for 48 h, and weighed (Liang et al., 2013).
Soil Moisture and Soil Temperature Measurement
The soil temperature (ST, 0–10 cm) was measured using a temperature probe, an accessory to the Li-8100 (Li-Cor, Inc., Lincoln, NE, United States) on September 10, 2018 (Sagar et al., 2017). Volumetric soil water content (SM, 0–10 cm) was measured with a portable device (Diviner 2000, Sentek Pty Ltd., Balmain, NSW, Australia).
Soil Sampling and Analysis
The soil samples were collected on September 10, 2018, at depth 0–20 cm. Six soil cores (approximately 5 cm diameter and 20 cm in depth) were taken from each plot and mixed to form one composite sample. Each sample was placed in a sterile plastic bag, sealed, and placed on ice when transported to the laboratory. Soil samples were sieved through a 2.0 mm mesh and stored at 4°C for analysis of soil characteristics or subsamples at -80°C for DNA extraction. The concentrations of NH4+ and NO3– were detected using a flow injection analyser (Tecator Inc., Hoganas, Sweden) (Miao et al., 2019). Soil total C (TC) and N (TN) contents were determined on a Vario MICRO cube elemental analyzer (Elementar, Germany). The soil total P (phosphorus, TP) was analyzed using the Olsen methods (Long et al., 2020).
DNA Extraction and Sequencing
DNA was extracted from 0.3 g fresh soil of each sample using MoBio Power soil TMDNA isolation kits (San Diego, CA, United States) according to the manufacturer’s instructions. Primer sets ITS1F (CTTGGTCATTTAGAGGAAGTAA)/ITS2R (GCTGCGTTCTTCATCGATGC) and 338F (ACTCCTACG GGAGGCAGCAG)/806R (GGACTACHVGGGTWTCTAAT) were selected to amplify the genes of fungi ITS and bacterial 16SrRNA, respectively. The PCR products for fungal and bacterial genes were gel purified and further quantified using PicoGreen Kits (Invitrogen, Shanghai, China). Sequencing was performed using the Illumina MiSeq platform at Majorbio Company (Shanghai, China). The raw high-throughput sequencing data were first processed using the Quantitative Insights Into Microbial Ecology (QIIME) toolkit. The potentially similar sequences were clustered into operational taxonomic units (OTUs) at a similarity level of 97%. Taxonomic assignment was performed by blasting the representative sequences against the MaarjAM (fungi) and SILVA (bacteria) database. Finally, this resulted in a total of 991,215 bacterial sequences and 1,132,095 fungal sequences for all samples, respectively. The rarefaction curves for the observed OTUs for both bacteria and fungi reached saturation for each treatment (Supplementary Figure 1), suggesting that the analyzed reads were sufficient to detect most of sequence types. All sequences have been submitted to the GenBank Sequence Read Archive with accession number MN834158–MN836334, KDPW01000000.
Data Analysis
The sequence data was analyzed on the Majorbio I-Sanger Cloud Platform.1 In order to examine how DP and IP affect community composition of plant, bacteria, and fungi, dissimilarity in compositions between treatments were calculated with the non-metric multidimensional scaling (NMDS) analysis using the Bray-Curtis distance matricesbased on plant species, bacterial and fungal OTUs. NMDS values were computed using the “metaMDS” function in the R package vegan v. 2.2–0 (Oksanen et al., 2014). A mixed-effects model analysis was used to test effects of DP and IP on community composition, species richness, and relative abundances of bacterial and fungal, where DP and IP were viewed as fixed between-subjects effect, and block was viewed as random variable. LSD post hoc tests were used to test for significant differences in these variables among C, DP, and IP treatments. The analyses were conducted using SPSS 16.0 (SPSS, Inc., Chicago, IL, United States). In order to explore the drivers of different phyla, a heat map analysis was used to examine potential correlations between ST, SM, plant species richness, aboveground net primary productivity (ANPP), NH4+ and NO3– concentration, total nitrogen (TN), total carbon (TC), soil pH, ratio of carbon to nitrogen (C/N), soil TP, and plant community composition with the observed bacterial group (abundance) patterns. The heat map was constructed using the pheatma package of R software 4.0.0.
Results
Soil and Plant Attributes
Mean soil moisture was 6.4% in the control plots. DP significantly reduced soil moisture by an average of 1.3%, whereas IP significantly increased soil moisture by an average of 1.3% (Table 1). Both TN and TC were significantly reduced by IP, but remained stable under the DP treatment. DP significantly increased NO3– concentration, whereas IP had little effect on it. Neither DP nor IP affected the NH4+ concentration compared with control (Table 1).
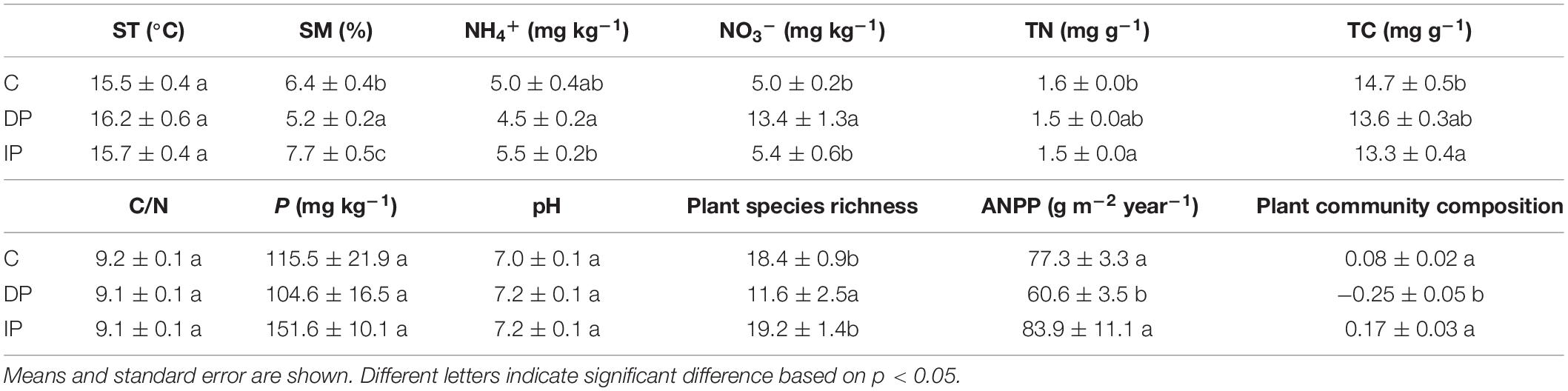
Table 1. Results (F-values) of one-way ANOVAs on effects of decreasing precipitation (DP) and increasing precipitation (IP) on soil temperature (ST, °C), soil moisture (SM, %), NH4+ (mg kg–1), NO3– (mg kg–1), total nitrogen (TN, mg kg–1), total carbon (TC, mg kg–1), the ratio of carbon to nitrogen (C/N), phosphorus content (P, mg°kg–1), soil pH, plant species richness (m–2), aboveground net primary productivity (ANPP, g m–2 year–1), and plant community composition (The scores of X axis in NMDS).
There was 18.4 plant species m–2 in the control plots. DP significantly reduced plant species richness by an average of 6.8 species m–2, whereas IP had no significant effect. ANPP was 71.6 g m–2 year–1 in the control plots. DP decreased ANPP by 16.7 g m–2 year–1, whereas IP had no effect on ANPP. The scores of X axis in NMDS in 2018 data showed that plant species composition in the DP plots diverged from the control plots, whereas composition did not differ in the IP communities from the control plots (Table 1 and Supplementary Figure 2).
Bacterial and Fungal Diversity
High throughput sequencing yielded a total of 1,177,241 bacterial and 1,315,445 fungal DNA sequences, with 3,444 and 2,142 operational taxonomic units, average read lengths of 439 and 267 bp, and average library coverage of 97.7 and 94.9%, respectively. Species richness of the bacterial communities decreased with DP [F(1,9) = 11.9, P = 0.009], whereas IP had no significant effect compared to ambient precipitation [F(1,9) = 2.7, P > 0.05]. Neither DP nor IP affected fungal species richness (Figure 1).
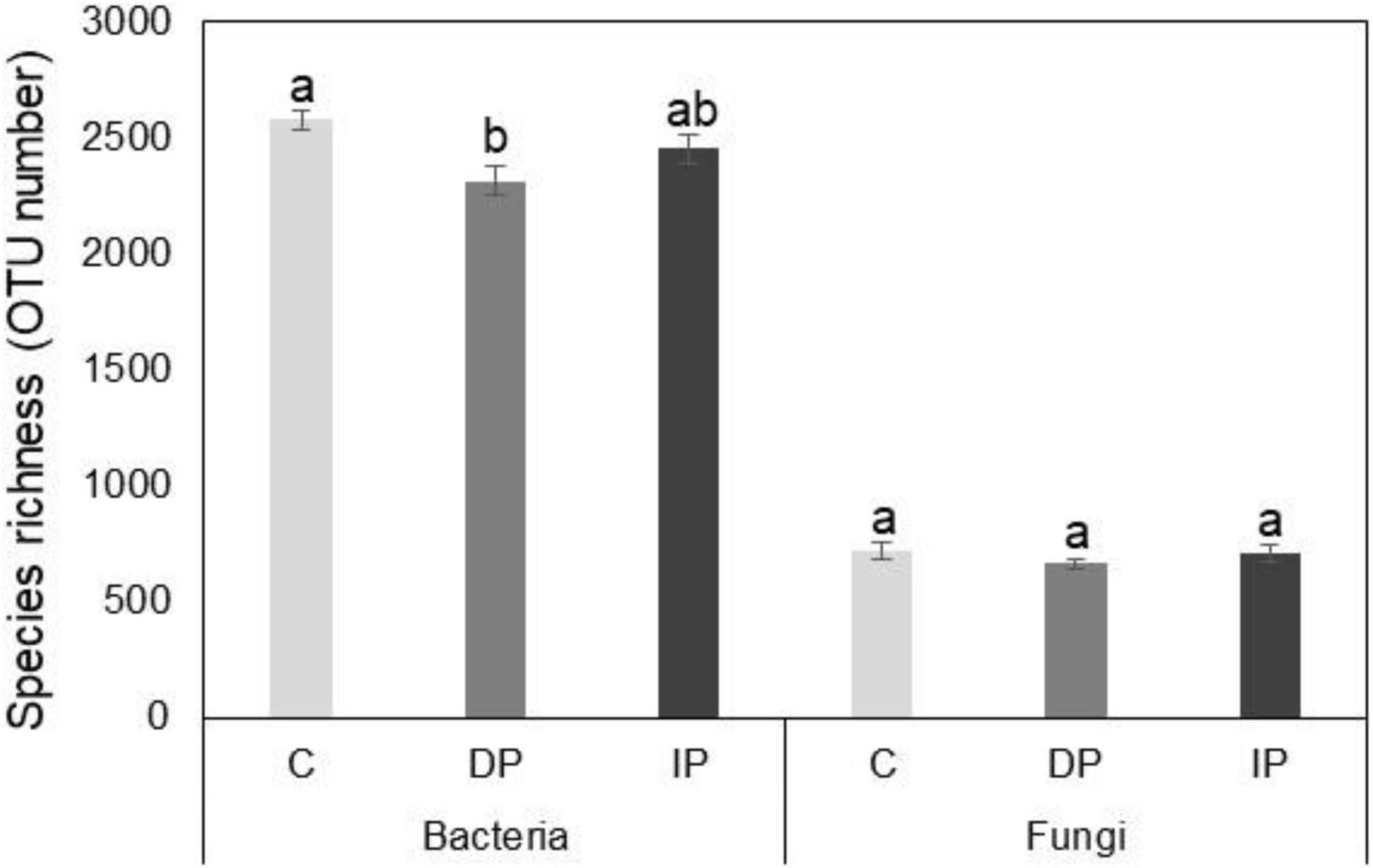
Figure 1. Effect of increasing precipitation (IP) and decreasing precipitation (DP) on species richness of soil bacteria and fungi. C represents control. Different letters indicate significant differences based on p < 0.05.
Community Composition of Bacteria and Fungi
The composition of bacterial communities were significantly changed by both DP and IP (Figure 2), and the top 1% of OTUs loadings on the axis1 in MNDS were shown in Supplementary Table 1. In contrast, neither DP nor IP affected the composition of the fungal community, the top 2% of OTUs loadings on the axis1 in MNDS were shown in Supplementary Table 2. Based on the number of OTUs, we categorized microbal groups at the phyla level. Among the bacteria, the most abundant phyla were Actinobacteria, with 27.2–30.8% of the sequences, followed by Proteobacteria (20.3–27.8%), Acidobacteria (16.2–21.4%), and Chloroflexi (8.2–13.1%). Actinobacteria was stable regardless of DP or IP. DP increased Chloroflexi and Nitrospirae by 59.39% [F(1,9) = 50.45, P < 0.001] and 33.41% [F(1,9) = 9.21, P < 0.05], respectively, whereas IP had no effect on either Chloroflexi [F(1,9) = 0.7, P > 0.05] or Nitrospirae [F(1,9) = 0.8, P > 0.05]. Acidobacteria decreased 23.40% [F(1,9) = 9.84, P = 0.01] by IP but was not affected by DP [F(1,9) = 0.14, P > 0.05]. Bacteroidetes decreased 43.06% [F(1,9) = 30.65, P = 0.001] by DP but remained stable under IP [F(1,9) = 1.50, P > 0.05]. In contrast, Proteobacteria increased 21.96% [F(1,9) = 14.17, P = 0.006] by IP but had little response to DP [F(1,9) = 1.89, P > 0.05]. Soil fungal communities were dominated by Ascomycota (66.5–73.7%), Zygomycota (9.9–11.3%), and Basidiomycota (7.1–9.8%). However, the relative abundance of the fungal phyla remains stable under either DP or IP (Figure 3).
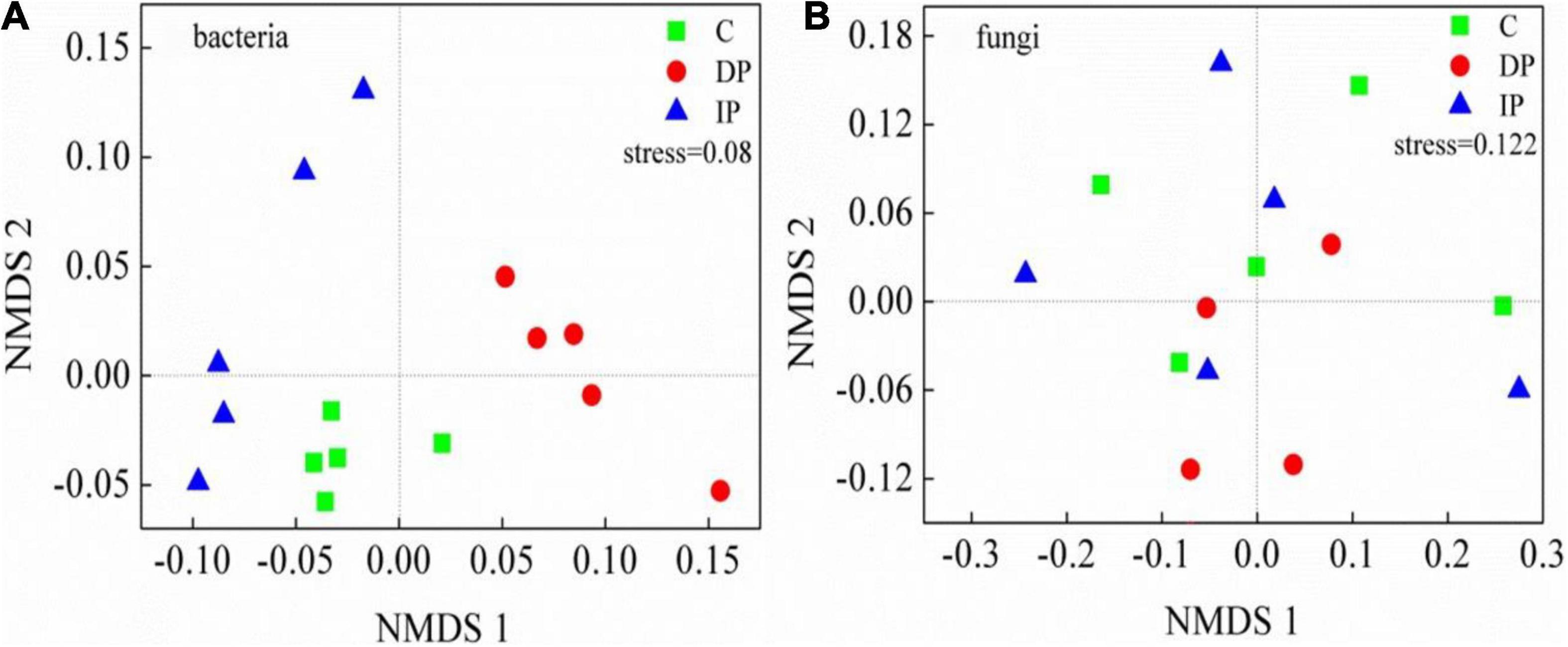
Figure 2. Non-metric multidimensional scaling analysis (NMDS) of soil bacterial and fungal communities under decreasing precipitation (DP) and increasing precipitation (IP). See Figure 1 for abbreviations.
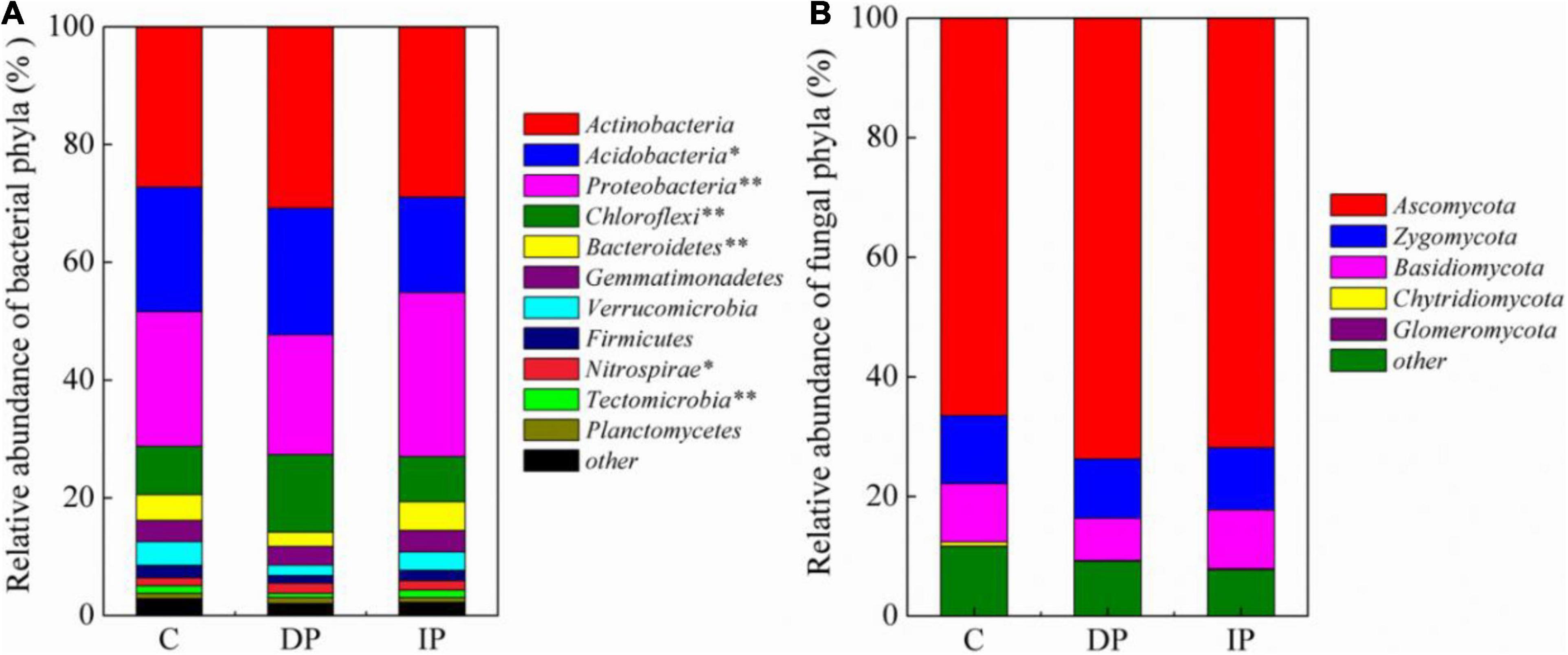
Figure 3. The relative abundances of bacterial (A) and fungal (B) phyla in the soil samples. See Figure 1 for abbreviations. The value of p < 0.05 are marked “*”, the value of p < 0.001 are marked “**”.
Factors Influencing Relative Abundance of Bacterial Phyla
The correlation heatmap showed that the relative abundances of drought-tolerant bacterial phyla (e.g., Acidobacteria and Chloroflexi) showed a significant negative correlation with SM, plant species richness, and ANPP but positively correlated with plant species composition. The relative abundances of drought-sensitive bacterial phyla (e.g., Proteobacteria and Bacteroidetes) were positively correlated with SM, plant species richness, and ANPP but negative correlation with plant species composition (Figure 4).
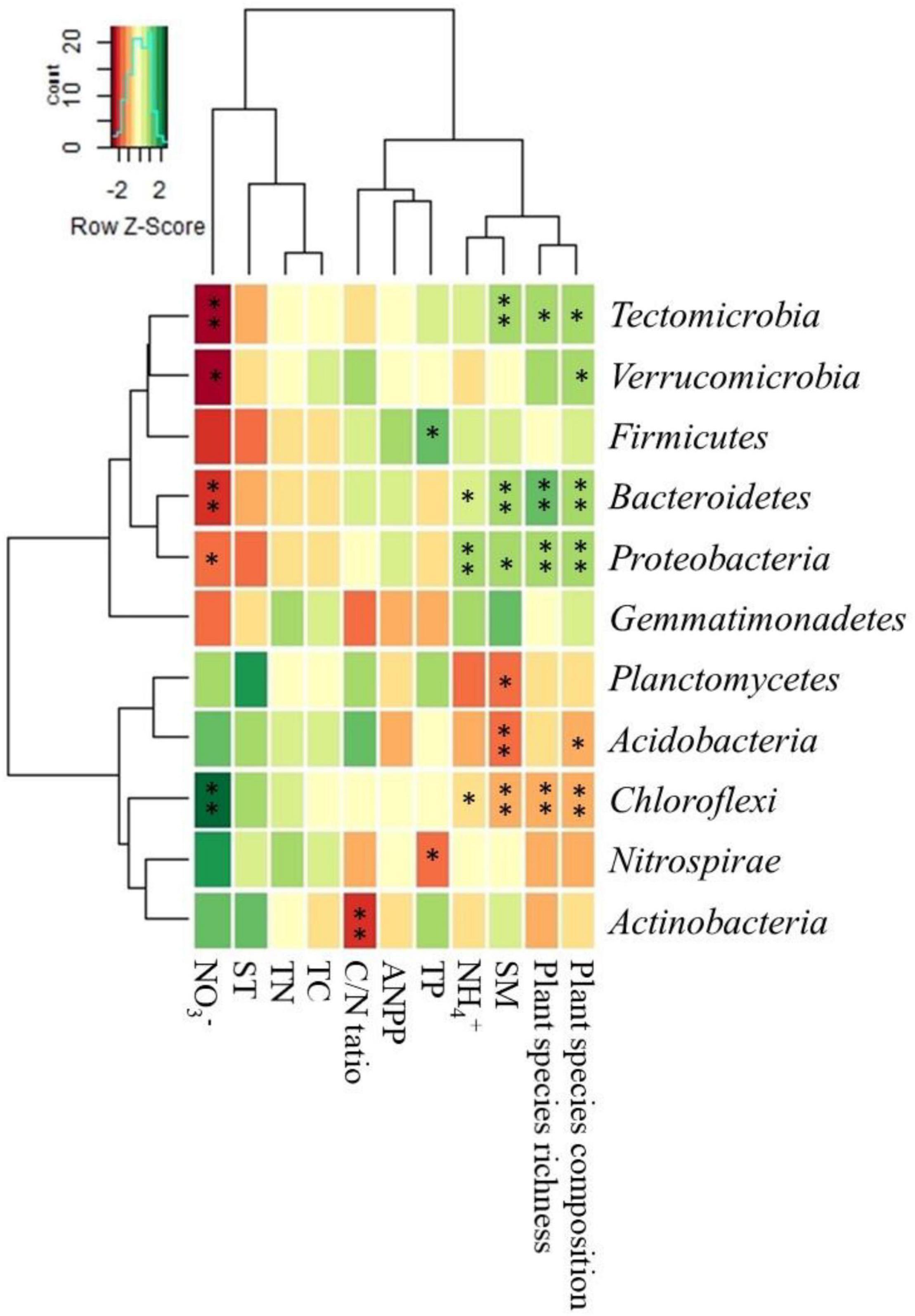
Figure 4. Correlation heat map of soil and vegetation properties with relative abundance of the top eleven bacterial phyla. The right side of the legend is the color range of R-values. See Table 1 for abbreviations. The values of p < 0.05 are marked with “*.” The values of p < 0.001 are marked with “**”.
Discussion
Effects of Changing Precipitation on Microbial Communities
Bacteria are more sensitive to changes in precipitation than fungi, particularly to decreases in precipitation (Maestre et al., 2015). Our results show that changes in precipitation influenced bacterial communities rather than fungal communities. Phyla within soil bacterial groups respond differently to changing precipitation due to different evolutionary adaptations and physiological acclimation mechanisms (Cregger et al., 2012). Our study demonstrates that bacteria within the phyla Acidobacteria, Chloroflexi, and Nitrospirae were more abundant under precipitation decreases. These groups are characterized by slow growth rates and mini-colony formation, which are typical characteristics of oligotrophic organisms. These bacterial taxa can maintain their abundance better under dry environments due to a greater tolerance to water stress (Evans and Wallenstein, 2014; Ochoa-Hueso et al., 2018). In contrast, Proteobacteria and Bacteroidetes, which are water-sensitive bacteria, increase under precipitation increases and decrease under precipitation decreases. These groups are generally considered to be copiotrophic, fast-growing bacteria and tend to respond to the altered availability of labile carbon induced by changes in precipitation (Fierer et al., 2009).
Fungi are characterized by a resistant response pattern, with generally stable abundance, species richness, and community composition under both precipitation increases and precipitation decreases. A possible explanation is that fungi have a relatively broad water optimum (compared to bacteria) without significant inhibition of growth (de Vries et al., 2018). Fungi are more drought tolerant than bacteria (except actinomycetes) because their hyphae can transfer water and nutrients from water-filled micropores (de Boer et al., 2005). However, some fungi grow well in wetter conditions. Abundant precipitation generally stimulates plant growth (Zhang et al., 2020b), leading to a large amount of organic matter supplied to the soil. Some fungi are copiotrophs and play an important role in decomposing organic matter, such as cellulose and chitin (Fierer et al., 2009).
Factors Affecting Microbial Communities
Soil water availability plays important role in structuring bacterial communities (Zhou et al., 2018). Limited soil water availability induced by drought decreases bacterial diversity by decreasing solute mobility and constraining the substrate supply to the decomposers (Ilstedt et al., 2000; Manzoni et al., 2014). In this way, decreases in precipitation directly inhibit Gram-negative bacteria (e.g., Proteobacteria) that are highly sensitive to soil moisture (Fierer et al., 2007; Bardgett et al., 2008; Manzoni et al., 2012). In contrast, Acidobacteria, which belongs to Gram-positive bacteria, are much more water-tolerant (Schimel et al., 2007) because of their stronger cell wall and a potentially more advanced osmoregulatory strategy than Gram-negative bacteria (Harris, 1981). Moreover, microbes provide plant nutrients only when their nutrient needs are met in the process of decomposition of organic matter, thus soil available nitrogen regulates the composition of microbial communities (Hodge et al., 2000). However, increased nitrate-nitrogen under drought stress induced by reduced plant and microbial uptake (Zhou et al., 2018) and/or reduction in denitrification and leaching losses in exposed dry soil (Gómez et al., 2012) suggests that soil available nitrogen has a weak effect on bacterial diversity in the current study. The changes in plant species identity and composition related to drought stress tolerance alter microbial communities by favoring plant associations with mycorrhizal fungi and mutualistic soil bacteria (Mariotte et al., 2017; Rubin et al., 2017). For example, mismatches between plant-microbe partners induced by the change in plant communities under drought create fitness differences among microbial species that reorder their relative abundances in the community by altering rates of population growth (Rudgers et al., 2020).
Conclusion
Our study shows that fungi were insensitive to either increases or decreases in precipitation. In contrast, changes in precipitation shifted the soil bacterial communities. The relative abundance of oligotrophic, slow-growing bacterial phyla increased in response to DP but decreased under IP. In contrast, the relative abundance of copiotrophic, fast-growing bacterial phyla decreased in response to DP but increased under IP. Effects of water availability, nitrate-nitrogen content, and plant-mediated effects are critical for driving the changes in soil microbial communities under changing precipitation. Our study suggests that changes in the amount of precipitation in the semi-arid grassland play an important role in determining soil bacterial communities under future climate change scenarios.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author Contributions
RX and ZY proposed the scientific hypotheses and supervised the project. JL, GB, and DW collected data. RX and JL performed data analyses and wrote the draft of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This project was financially supported by Natural Science Foundation of Henan Province (202300410084) and National Natural Science Foundation of China (31570429 and 31300363).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.842446/full#supplementary-material
Abbreviations
C, ambient precipitation; DP, a 60% decrease in growing season precipitation; IP, a 60% increase in growing season precipitation; OTUs, operational taxonomic units; NMDS, non-metric multidimensional scaling; ST, soil temperature; SM, soil moisture; TN, total nitrogen; TC, total carbon; C/N, the ratio of carbon to nitrogen; ANPP, above ground net primary productivity.
Footnotes
References
Bardgett, R. D., Freeman, C., and Ostle, N. J. (2008). Microbial contributions to climate change through carbon cycle feedbacks. ISME J. 2, 805–814. doi: 10.1038/ismej.2008.58
Brockett, B. F. T., Prescott, C. E., and Grayston, S. J. (2012). Soil moisture is the major factor influencing microbial community structure and enzyme activities across seven biogeoclimatic zones in western Canada. Soil Biol. Biochem. 44, 9–20.
Clark, J., Campbell, J., Grizzle, H., Acosta-Martinez, V., and Zak, J. (2009). Soil microbial community response to drought and precipitation variability in the Chihuahuan Desert. Microb. Ecol. 57, 248–260. doi: 10.1007/s00248-008-9475-7
Cregger, M. A., Schadt, C. W., McDowell, N. G., Pockman, W. T., and Classen, A. T. (2012). Response of the soil microbial community to changes in precipitation in a semiarid ecosystem. Appl. Environ. Microbiol. 78, 8587–8594. doi: 10.1128/AEM.02050-12
de Boer, W., Folman, L. B., Summerbell, R. C., and Boddy, L. (2005). Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 29, 795–811. doi: 10.1016/j.femsre.2004.11.005
de Vries, F. T., Griffiths, R. I., Bailey, M. J., Craig, H., Girlanda, M., Gweon, H. S., et al. (2018). Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 9:3033. doi: 10.1038/s41467-018-05516-7
Evans, S. E., and Wallenstein, M. D. (2014). Climate change alters ecological strategies of soil bacteria. Ecol. Lett. 17, 155–164. doi: 10.1111/ele.12206
Feng, X., and Simpson, M. J. (2009). Temperature and substrate controls on microbial phospholipid fatty acid composition during incubation of grassland soils contrasting in organic matter quality. Soil Biol. Biochem. 41, 804–812. doi: 10.1016/j.soilbio.2009.01.020
Fierer, N., Bradford, M. A., and Jackson, R. B. (2007). Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364. doi: 10.1890/05-1839
Fierer, N., and Jackson, R. B. (2006). The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. 103, 626–631. doi: 10.1073/pnas.0507535103
Fierer, N., Strickland, M. S., Liptzin, D., Bradford, M. A., and Cleveland, C. C. (2009). Global patterns in belowground communities. Ecol. Lett. 12, 1238–1249. doi: 10.1111/j.1461-0248.2009.01360.x
Gao, C., Kim, Y., Zheng, Y., Yang, W., Chen, L., Ji, N., et al. (2016). Increased precipitation, rather than warming, exerts a strong influence on arbuscular mycorrhizal fungal community in a semiarid steppe ecosystem. Botany 94, 459–469. doi: 10.1139/cjb-2015-0210
Gómez, R., Arce, M. I., Sánchez, J. J., and Sanchez-Montoya, M. M. (2012). The effects of drying on sediment nitrogen content in a Mediterranean intermittent stream: a microcosms study. Hydrobiologia 679, 43–59. doi: 10.1007/s10750-011-0854-6
Harris, R. F. (1981). “Effect of water potential on microbial growth and activity,” in Water Potential Relations in Soil Microbiology, eds J. F. Parr, W. R. Gardner, and L. F. Elliott (Madison: American Society of Agronomy), 23–95.
Hodge, A., Robinson, D., and Fitter, A. (2000). Are microorganisms more effective than plants at competing for nitrogen? Trends Plant Sci. 5, 304–308. doi: 10.1016/S1360-1385(00)01656-3
Huntington, T. G. (2006). Evidence for intensification of the global water cycle: review and synthesis. J. Hydrol. 319, 83–95. doi: 10.1016/j.jhydrol.2005.07.003
Ilstedt, U., Nordgren, A., and Malmer, A. (2000). Optimum soil water for soil respiration before and after amendment with glucose in humid tropical acrisols and a boreal mor layer. Soil Biol. Biochem. 32, 1591–1599. doi: 10.1016/S0038-0717(00)00073-0
Koyama, A., Steinweg, J. M., Haddix, M. L., Dukes, J. S., and Wallenstein, M. D. (2017). Soil bacterial community responses to altered precipitation and temperature regimes in an old field grassland are mediated by plants. FEMS Microbiol. Ecol. 94, 1–15. doi: 10.1093/femsec/fix156
Landesman, W. J., and Dighton, J. (2010). Response of soil microbial communities and the production of plant-available nitrogen to a two-year rainfall manipulation in the New Jersey Pinelands. Soil Biol. Biochem. 42, 1751–1758. doi: 10.1016/j.soilbio.2010.06.012
Lauber, C. L., Hamady, M., Knight, R., and Fierer, N. (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120. doi: 10.1128/AEM.00335-09
Liang, J., Xia, J., Liu, L., and Wan, S. (2013). Global patterns of the responses of leaf-level photosynthesis and respiration in terrestrial plants to experimental warming. J. Plant Ecol. 6, 437–447.
Long, M., Zhang, J. J., Liu, Z. Y., Zhou, L. Y., Su, F. L., Xiao, R., et al. (2020). Can the scaling of plant nitrogen to phosphorus be altered by global change? An empirical test. J. Plant Ecol. 13, 442–449. doi: 10.1093/jpe/rtaa032
Maestre, F. T., Delgado-Baquerizo, M., Jeffries, T. C., Eldridge, D. J., and Singh, B. K. (2015). Increasing aridity reduces soil microbial diversity and abundance in global drylands. Proc. Natl. Acad. Sci. U.S.A. 112, 15684–15689. doi: 10.1073/pnas.1516684112
Manzoni, S., Schaeffer, S. M., Katul, G., Porporato, A., and Schimel, J. P. (2014). A theoretical analysis of microbial eco-physiological and diffusion limitations to carbon cycling in drying soils. Soil Biol. Biochem. 73, 69–83. doi: 10.1016/j.soilbio.2014.02.008
Manzoni, S., Schimel, J. P., and Porporato, A. (2012). Responses of soil microbial communities to water stress: results from a meta-analysis. Ecology 93, 930–938. doi: 10.1890/11-0026.1
Mariotte, P., Canarini, A., and Dijkstra, F. A. (2017). Stoichiometric N:P flexibility and mycorrhizal symbiosis favour plant resistance against drought. J. Ecol. 105, 958–967. doi: 10.1111/1365-2745.12731
Meier, C. L., and Bowman, W. D. (2008). Links between plant litter chemistry, species diversity, and below-ground ecosystem function. Proc. Natl. Acad. Sci. U.S.A. 105, 19780–19785. doi: 10.1073/pnas.0805600105
Miao, R., Ma, J., Liu, Y. Z., Liu, Y. C., and Yang, Z. L. (2019). Variability of aboveground litter inputs alters soil carbon and nitrogen in a Coniferous-Broadleaf Mixed Forest of central China. Forests 10:188. doi: 10.3390/f10020188
Nielsen, U., and Ball, B. A. (2015). Impacts of altered precipitation regimes on soil communities and biogeochemistry in arid and semi-arid ecosystems. Glob. Change Biol. 21, 1407–1421. doi: 10.1111/gcb.12789
Ochoa-Hueso, R., Collins, S. L., Delgado-Baquerizo, M., Hamonts, K., Pockman, W. T., Sinsabaugh, R. L., et al. (2018). Drought consistently alters the composition of soil fungal and bacterial communities in grasslands from two continents. Glob. Change Biol. 24, 2818–2827. doi: 10.1111/gcb.14113
Oksanen, J., Blanchet, F. G., Friendly, M., Legendre, P., O’Hara, R. G., Simpson, G. L., et al. (2014). Vegan: Community Ecology Package. R Package version 2.2-0. https://cran.r-project.org/web/packages/vegan/index.html. (Accessed Mar 20, 2018).
Rubin, R. L., van Groenigen, K. J., and Hungate, B. A. (2017). Plant growth promoting rhizobacteria are more effective under drought: a meta-analysis. Plant Soil 416, 309–323. doi: 10.1007/s11104-017-3199-8
Rudgers, J. A., Afkhami, M. E., Bell-Dereske, L., Chung, Y. A., and Nuez, M. A. (2020). Climate Disruption of Plant-Microbe Interactions. Annu. Rev. Ecol. Evol. Syst. 51, 561–586. doi: 10.1146/annurev-ecolsys-011720-090819
Sagar, R., Li, G., Singh, J. S., and Wan, S. (2017). Carbon fluxes and species diversity in grazed and fenced typical steppe grassland of Inner Mongolia, China. J. Plant Ecol. 12, 10–22.
Schimel, J., Balser, T. C., and Wallenstein, M. D. (2007). Microbial stress-response physiology and its implications for ecosystem function. Ecology 88, 1386–1394. doi: 10.1890/06-0219
Spehn, E. M., Joshi, J., Schmid, B., Alphei, J., and Körnor, C. (2000). Plant diversity effects on soil heterotrophic activity in experimental grassland ecosystems. Plant Soil 224, 217–230. doi: 10.1007/s00442-012-2585-4
Uhlirová, E., Elhottova, D., Triska, J., and Santruckova, H. (2005). Physiology and microbial community structure in soil at extreme water content. Folia Microbiol. 50, 161–166. doi: 10.1007/BF02931466
Waldrop, M. P., Zak, D. R., Blackwood, C. B., Curtis, C. D., and Tilman, D. (2006). Resource availability controls fungal diversity across a plant diversity gradient. Ecol. Lett. 9, 1127–1135. doi: 10.1111/j.1461-0248.2006.00965.x
Williams, M. A. (2007). Response of microbial communities to water stress in irrigated and drought-prone tallgrass prairie soils. Soil Biol. Biochem. 39, 2750–2757. doi: 10.1016/j.soilbio.2007.05.025
Yang, Z., Wei, Y., Fu, G., Song, H., Li, G., and Xiao, R. (2020). Asymmetric effect of increased and decreased precipitation in different periods on soil and heterotrophic respiration in a semiarid grassland. Agric. For. Meteorol. 291:108039. doi: 10.1016/j.agrformet.2020.108039
Zhang, J., Miao, Y., Zhang, T., Wei, Y., Qiao, X., Miao, R., et al. (2020a). Drought timing and primary productivity in a semiarid grassland. Land Degrad. Dev. 31, 2185–2195. doi: 10.1002/ldr.3603
Zhang, J., Li, J., Xiao, R., Zhang, J., Wang, D., Miao, R., et al. (2020b). The response of productivity and its sensitivity to changes in precipitation: a meta-analysis of field manipulation experiment. J. Veg. Sci. 32:e12954. doi: 10.1111/jvs.12954
Keywords: bacterial and fungal communities, community composition, changing precipitation, diversity, grasslands
Citation: Li J, Benti G, Wang D, Yang Z and Xiao R (2022) Effect of Alteration in Precipitation Amount on Soil Microbial Community in a Semi-Arid Grassland. Front. Microbiol. 13:842446. doi: 10.3389/fmicb.2022.842446
Received: 23 December 2021; Accepted: 14 February 2022;
Published: 17 March 2022.
Edited by:
Leonardo Erijman, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaReviewed by:
Natasja Van Gestel, Texas Tech University, United StatesEric G. Lamb, University of Saskatchewan, Canada
Copyright © 2022 Li, Benti, Wang, Yang and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rui Xiao, eGlhb3IxMTMwQDE2My5jb20=
 Junyong Li
Junyong Li Girmaye Benti
Girmaye Benti Dong Wang
Dong Wang Zhongling Yang
Zhongling Yang Rui Xiao
Rui Xiao