
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 18 February 2022
Sec. Food Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.842268
This article is part of the Research TopicFood Safety and Public HealthView all 33 articles
Alternaria alternata is a principal plant pathogen responsible for the biosynthesis of mycotoxins, including tenuazonic acid (TeA), alternariol (AOH), and alternariol monomethyl ether (AME). The velvet gene VeA is involved in fungal growth, development, and secondary metabolism, including mycotoxin biosynthesis via light regulation. In this study, the detailed regulatory roles of AaVeA in A. alternata with various light sources were investigated from the comparative analyses between the wild type and the gene knockout strains. In fungal growth and conidiation, mycelial extension was independent of light regulation in A. alternata. Red light favored conidiation, but blue light repressed it. The absence of AaVeA caused the marked reduction of hyphae extension and conidiophore formation even though red light could not induce more spores in ΔAaVeA mutant. The differentially expressed genes (DEGs) enriched in hyphal growth and conidiation were drastically transcribed from the comparatively transcriptomic profile between the wild type and ΔAaVeA mutant strains with or without light. In mycotoxin production, TeA biosynthesis seems no obvious effect by light regulation, but AOH and AME formation was significantly stimulated by blue light. Nevertheless, the disruption of AaVeA resulted in a marked decrease in mycotoxin production and the action of the stimulation was lost via blue light for the abundant accumulation of AOH and AME in the ΔAaVeA strain. From DEG expression and further verification by RT-qPCR, the loss of AaVeA caused the discontinuous supply of the substrates for mycotoxin biosynthesis and the drastic decline of biosynthetic gene expression. In addition, pathogenicity depends on AaVeA regulation in tomato infected by A. alternata in vivo. These findings provide a distinct understanding of the roles of AaVeA in fungal growth, development, mycotoxin biosynthesis, and pathogenicity in response to various light sources.
The genus Alternaria forms colonies worldwide as saprophytes in plant residue, soil, and air, as well as in pathogens of pre- and post-harvest crops (Thomma, 2003). Alternaria alternata is the most common species contaminating a wide range of plants, including wheat, sorghum, tomato, apple, and their products (Logrieco et al., 2009), which produces a variety of mycotoxins treating human and animal health. Generally, of the Alternaria mycotoxins, tenuazonic acid (TeA), alternariol (AOH), and alternariol monomethyl ether (AME) are the most serious and frequent toxin contaminants (Ostry, 2008; Patriarca, 2016). TeA is the most toxic of Alternaria mycotoxins and exhibits acute toxicity and cytotoxicity by suppressing protein biosynthesis on ribosomes and has phytotoxic activity by blocking photosystem II electron transport (Logrieco et al., 2009; Chen and Qiang, 2017). AOH and AME possess cytotoxicity, genotoxic, and mutagenic properties (Patriarca, 2016; Wenderoth et al., 2019). Additionally, AOH also plays as a pathogenicity factor during plant infection (Wenderoth et al., 2019). Furthermore, data on dietary exposure have been recorded to estimate its prevalence in Europe [EFSA on Contaminants in the Food Chain (CONTAM), 2011; Arcella et al., 2016]. However, no regulations have been developed worldwide, except by the Bavarian Health and Food Safety Authority, who set a TeA maximum limit at 500 μg/kg in sorghum/millet-based infant food (Solfrizzo, 2017).
TeA is regarded the hybrid of an isoleucine and two acetates from the skeleton structure. It was identified to be biosynthesized by TeA synthetase 1 (TAS1), a nonclassical type of non-ribosomal peptide synthetase and polyketide synthase (NRPS-PKS) hybrid enzyme (Yun et al., 2015). TAS1 catalyzes cyclization from an isoleucine with an acetoacetyl-CoA. Furthermore, TAS1 expression is specifically modulated by a Zn(II)2-Cys6-type transcriptional factor encoded by TAS2, located on the neighboring TAS1 in M. oryzae (Yun et al., 2017). However, based on the Basic Local Alignment Search Tool (BLAST) results, no homologous proteins (similarity <30%) were found after searching the genome of A. alternata. AOH and its methyl derivative AME were confirmed to be biosynthesized by a PKS gene cluster, including pksI and omtI responsible for their formation (Wenderoth et al., 2019). A Gal4-like transcription factor aohR positively modulated the expression of the gene cluster. In addition, a pksI orthologue SnPKS19 was validated to be responsible for AOH production in the wheat pathogen Parastagonospora nodorum (Chooi et al., 2015).
Fungal growth, development, and secondary metabolism are regulated by light signals via light sensing systems, including photoreceptor-like white collar (WC-1 and 2) orthologs and phytochrome FphA (Blumenstein et al., 2005; Idnurm and Heitman, 2005). In response to light regulation, the heterotrimeric velvet complex VelB/VeA/LaeA controls secondary metabolites in Aspergillus nidulans (Bayram et al., 2008). These components of the velvet complex exert diverse functions in different fungal species. Several scientists have separately characterized the functions of LaeA, VeA, and VelB in mycotoxin producing fungi (Amaike and Keller, 2009; Wiemann et al., 2010; Chang et al., 2013; Estiarte et al., 2016; Takao et al., 2016; Eom et al., 2018). VeA is regarded as an important light regulator, influencing fungal growth, development, and secondary metabolite biosynthesis (Wiemann et al., 2010; Merhej et al., 2012). LaeA was identified as the main regulator of secondary metabolites and as a global regulator for asexual and sexual development, growth, and virulence (Wiemann et al., 2010). PoLAE1 positively regulates TeA biosynthesis via TAS1 and TAS2 expression in M. oryzae (Yun et al., 2017). Moreover, the regulatory function of LaeA and VeA for AOH and AME production was characterized in a previous study (Estiarte et al., 2016). Both regulators could modulate the expression of genes responsible for their biosynthesis, including pksJ and altR (Saha et al., 2012; Wenderoth et al., 2019). In addition, growth morphology and conidiation were markedly altered via comparative analysis between the wild type and mutant of LaeA or VeA loss in response to light or dark conditions. However, light was characterized by no obvious differences in AOH and AME formation from the comparison of mycotoxin contents in both strains of A. alternata cultured under either dark or light conditions. On the contrary, in the study on the roles of blue-light receptor LreA for mycotoxin production, AOH formation was stimulated by light, especially blue light (Pruß et al., 2014). The mechanism of the contradictive outcomes on the roles of AOH production by light remains unclear from these two studies. Additionally, the effects on the critically important mycotoxin TeA production in response to light have not yet been characterized.
To our knowledge, this is the first report on the roles and potential mechanisms of AaVeA on TeA production with various light sources. In this study, the deletion of AaVeA was carried out, and the analyses of fungal growth, development, and mycotoxin biosynthesis were compared between the wild-type and ΔAaVeA strains under diverse light conditions. Furthermore, the comparatively transcriptomic profile was performed to uncover the regulatory mechanisms for mycotoxin production. Blue light was unfavorable to sporulation but did not obviously affect hyphal extension in A. alternata. In contrast, red light had no significant effect on sporulation, whose in vitro morphology was similar to that of white-light treatment. TeA biosynthesis depended on the regulation of AaVeA, but light caused no marked alteration on its production, which was distinct from the stimulation of AOH and AME biosynthesis by blue light.
A. alternata P3 and the mutant strains were cultured on potato dextrose agar (PDA) at 25°C. The spores were then washed with 0.1% Tween-80 water, collected from the culture of A. alternata, and diluted to 1 × 105 spores/ml (Jiang et al., 2019; Wang et al., 2019). The spore suspension was cultured on PDA at 25°C for 10 days to monitor hyphal extension, colony morphology, and conidial formation (Wang et al., 2019). The spores were inoculated and incubated on potato dextrose broth (PDB) under dark, white, blue, and red-light conditions at 25°C for 10 days for the following analysis on mycotoxin determination and gene expression. For the continuous light experiment, white, blue, or red LED lamps (18 W/lamp) were used, and the distance between the light source and the medium was 30 cm. The average illumination intensities of white, blue, and red light were separately measured at 5200, 4350, and 2,270 lux, respectively.
The deletion of AaVeA was used to investigate the functional roles for the growth, development, and secondary metabolite biosynthesis. AaVeA was disrupted by homologous recombination using the hygromycin resistance gene as a marker gene for antibiotic resistance screening (Supplementary Figure S1). Flanking sequences of 1,458-bp upstream and 1,485-bp downstream were separately amplified by polymerase chain reaction (PCR) using genomic DNA of A. alternata P3 as a template with the primer pairs AaVeA-up-F/R and AaVeA-down-F/R (Supplementary Table S1). The hygromycin resistance gene encoding hygromycin phosphotransferase (hph) was amplified using the primer pair hph-F/R. The reaction mixture (50 μl) consisted of 2 μl of the template DNA, 1 μl of each forward and backward primer (10 μM), 25 μl of GoTaq Master Mix (Promega, Madison, WI, United States), and 21 μl of nuclease-free water. The PCR program was set as follows: initial denaturation at 95°C for 5 min, 35 cycles of the amplification at 95°C for 30 s, 54°C for 30 s and 72°C for 90 s, and finally extension for 5 min. The PCR product was extracted using a Gel Extraction Kit (Sangon Biotech, Shanghai, China). Three fragments were assembled by overlapping extension PCR in combination with nested PCR using the primer pair AaVeA-knock-F/R. The assembled fragment was also purified for transformation. The protoplast of A. alternata was obtained by enzymatic hydrolysis of fungal cell wall with 30 mg/ml of snailase, 5 mg/ml of driselase, 2 mg/ml of lyticase, 8 mg/ml of β-D-dextranase, and 20 mg/ml of cellulase. Then, the PCR product was transformed into the protoplast by polyethylene glycol (PEG)-CaCl2 mediated genetic transformation. The transformants were screened by 100 μg/ml hygromycin B (Roche Diagnostics, Mannheim, Germany) and identified by PCR and quantitative PCR according to the method of Zhang et al. (2016). The mutant of AaVeA deletion was finally obtained for the following study (Supplementary Figure S1).
The spore suspension of A. alternata P3 and the mutant was acquired (1 × 105 spores/ml). A 5-μl aliquot of the suspension was inoculated in a PDA medium and grown for 10 days at 25°C. The characteristics of the colony were observed, and then the conidiation was compared between the wild type and ΔAaVeA mutant under white, red, blue, and dark conditions. The spore counts were determined using the hemocytometer after they were washed completely from the culture using 0.1% Tween-80.
The microscopical structure was evaluated by scanning electron microscopy (SEM). The samples were harvested and fixed in 2.5% glutaraldehyde. Then, the samples were washed, dehydrated, and vacuum-dried following the processes by Wang et al. (2019).
The 1%(v/v) spore suspension of A. alternata P3 and the AaVeA null mutant were inoculated and cultured in PDB. The strains were incubated for 10 days at 25°C with or without various light treatments. The supernatant was centrifuged and used to determine the concentration of mycotoxin. The determination of mycotoxin production was assayed under the direction of Jiang et al. (2019) using LC–MS/MS.
The mycelial samples were collected after culture under various light conditions and ground with liquid nitrogen. Total RNA was extracted from the mycelia with three biological replications cultivated in PDB medium with or without light using TRIzol reagent according to the Cold Spring Harbor Laboratory (Rio et al., 2010). The quality and purity of the extracted RNA were evaluated using the Bioanalyzer 2,100 and RNA 6000 Nano LabChip Kit (Agilent, CA, United States) with RIN number > 7.0. cDNA library construction and RNA-seq were carried out by Hangzhou Lianchuan Biotechnology Co., Ltd. (Hangzhou, China). Using the Illumina paired-end RNA-seq approach, the transcriptome was sequenced, and millions of paired-end reads were generated. After removing the low-quality reads, we mapped the reads to the reference genome of A. alternata using the HISAT package (Kim et al., 2015). HISAT allows multiple alignments per read (up to 20 by default) and a maximum of two mismatches when mapping the reads to the reference. The remaining reads, after trimmed from the raw reads, were then bioinformatically analyzed based on the genome annotation of A. alternata (genome assembly: GCF_001642055.1; Dang et al., 2015).
Differentially expressed genes (DEGs) were selected with a |log2 (fold change) | > 1 and statistical significance of value of p < 0.05 by the R package. Gene Ontology (GO) functional annotation (Young et al., 2010) and Kyoto Encyclopedia of Genes and Genomes (KEGG; Kanehisa et al., 2008) pathway enrichment analysis were conducted with the DEGs to further understand their functions.
The relative expression levels of TeA biosynthetic genes and velvet genes were further confirmed and quantified by reverse transcription quantitative real-time PCR (RT-qPCR). Total RNA was separately extracted by the EasyPure Plant RNA Kit (TransGen Biotech, Beijing, China) containing DNase I digestion for the genomic DNA residue. The RNA samples were diluted to an equal concentration, and then cDNA was reverse transcribed by EasyScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). Quantitative PCR was carried out by TransStart Top Green qPCR SuperMix (+Dye I), using cDNA as the template. The 20-μL reaction mixture comprised the following: 2 μl of cDNA, 0.4 μl of forward and backward primer (10 μM; Supplementary Table S2), 10 of 2 × TransStart Top Green qPCR SuperMix (+Dye I), and 7.2 μl of nuclease-free water. The transcriptomic level of clustered biosynthetic and regulatory genes was separately normalized to the expression of β-tubulin and evaluated by the 2-ΔΔCt calculation from the results of StepOnePlus Real-Time PCR Systems (Applied Biosystems, Foster City, CA, United States).
To evaluate the pathogenicity and mycelial extension in vivo for the wild and VeA-disrupted strains of A. alternata, an equal volume (5 μl) of conidial suspension (105/ml) was separately inoculated into the man-made wound on the surface of tomatoes after they were sterilized with 10% sodium hypochlorite and 75% alcohol. Tomatoes were treated with or without light at 25°C for 10 days.
Fungal growth was measured, and the infected area on tomatoes was used to extract and determine mycotoxin production. The samples were first dried by vacuum freezing and drying technology. Then, they were weighed and ground into powder, of which 0.2 g was used to determine the mycotoxin content. The final mycotoxin concentration was expressed as mycotoxin content per unit area.
All the experiments were carried out with at least three replicates. The data of fungal extension diameters, spore counts, and mycotoxins concentrations were analyzed by IBM SPSS statistics 23.0 (IBM Inc., Armonk, NY, United States) and compared by the one-way analysis of variance (ANOVA) using Tukey’s test. Significance was considered as p < 0.05. The figures were generated by Graphpad Prism 8.02 (GraphPad Software Inc., San Diego, CA, United States).
To understand the effects of the AaVeA gene on fungal growth and conidiation, the wild type and AaVeA knockout strains were used to characterize the properties with white, red, or blue light or without light. The wild type and the ΔAaVeA mutant strain were separately grown on PDA for 10 days. The colonies and the microscopic morphology are shown in Figure 1. Light seemed to hardly influence the mycelial extension of A. alternata from the colonies with light illumination (Figure 2A). However, lacking the gene AaVeA resulted in the inhibition of hyphal growth by 24.2% in the ΔAaVeA mutant under dark conditions (p < 0.001). Similarly, the mycelia of the mutant extended 25.5%, 23.0%, and 18.9% more slowly than the wild type with white, red, or blue light, respectively. There was no significant difference in the inhibition rate under the different light conditions due to the gene deletion (p > 0.05).
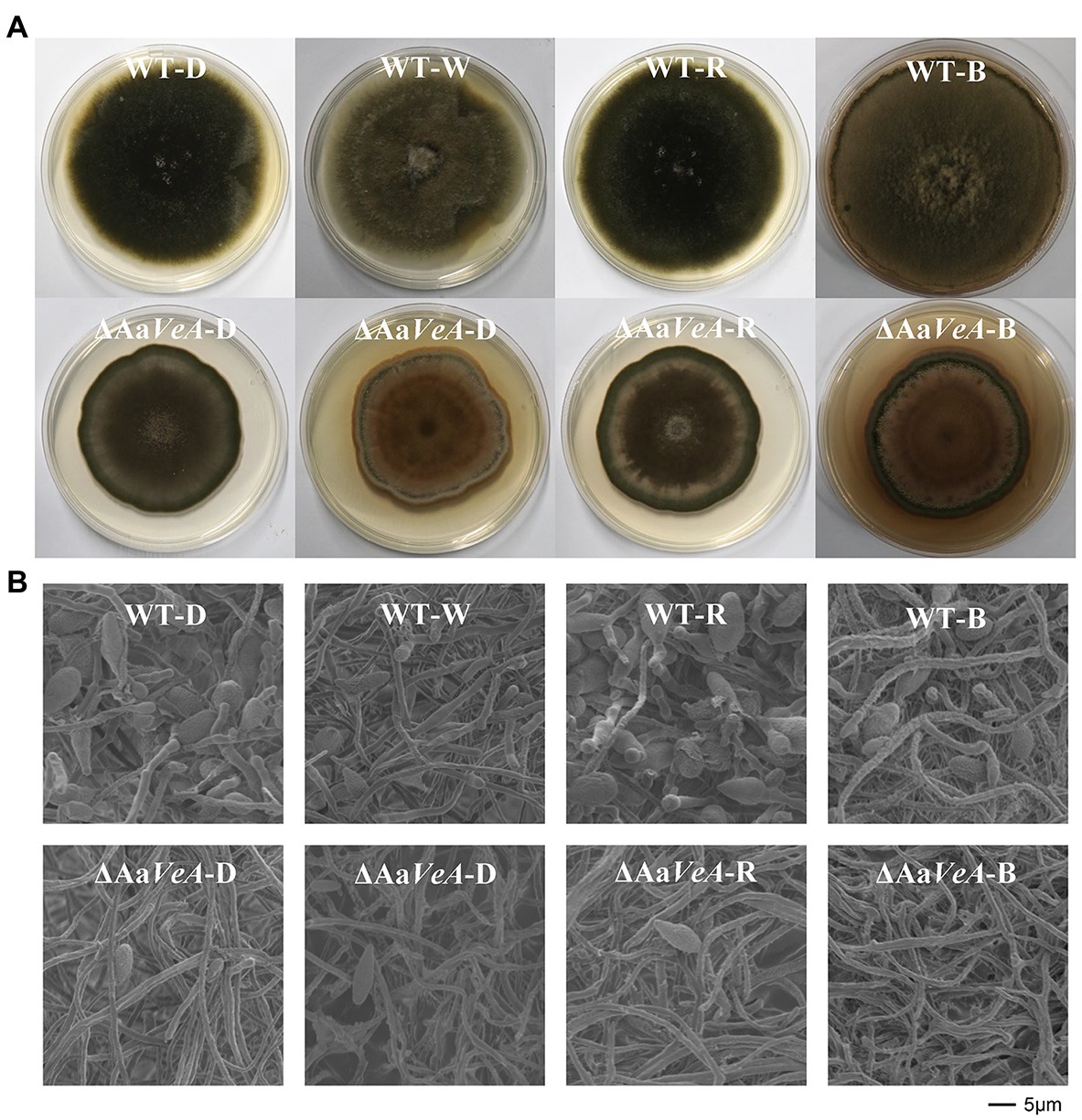
Figure 1. Colony and morphology characters of the wild type (WT) and AaVeA disrupted mutant (ΔAaVeA) strains with various light sources in Alternaria alternata. (A) The front diagram of WT and ΔAaVeA strains cultured on potato dextrose agar (PDA) for 10 days at 25°C in darkness (D) or with white (W), red (R) and blue (B) light. The colony of the wild strain extended at a similar rate while the mutant strain grew slower. The similar morphology was observed in the colony of the wild strain in darkness and with red light. However, the density of the mycelia became thicker in the wild strain with white and blue light. (B) Microscopic morphology of mycelia and spores of WT and ΔAaVeA strains by scanning electron microscopy (SEM). The microscopic morphology of mycelial and spore structure was observed with no marked alteration in dark and light conditions. However, the fungal spores were much more abundant under dark and red-light conditions.
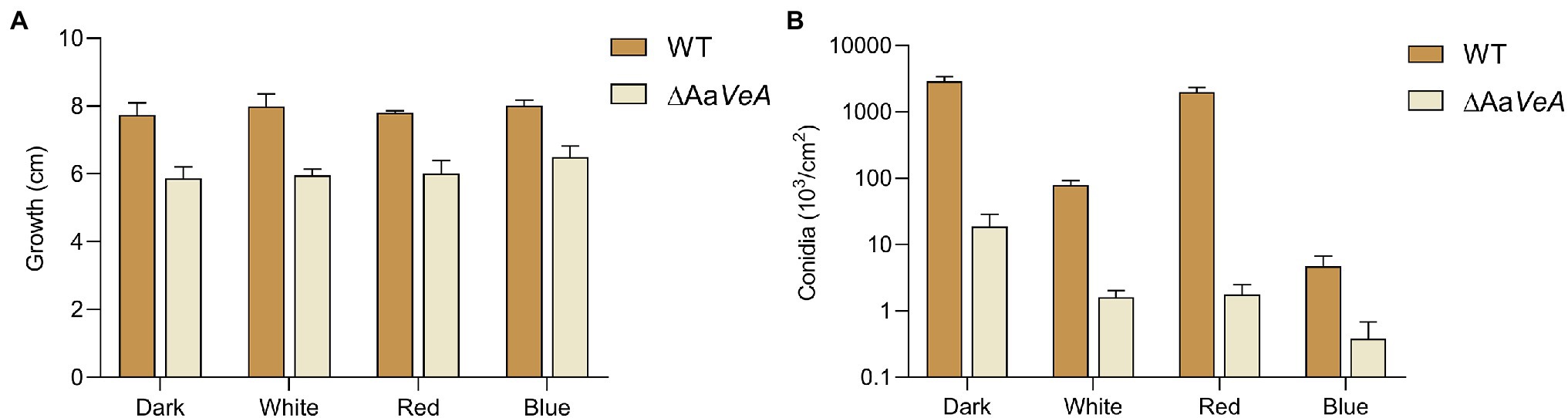
Figure 2. Mycelial growth (A) and conidia formation (B) of WT and ΔAaVeA strains. The colony diameter of WT and ΔAaVeA strains cultured on PDA medium was separately measured. Accordingly, the conidia were washed and determined using the hemocytometer.
Although no obvious difference and a similar trend was found on the hyphal extension, the conidiation of A. alternata was markedly changed under different conditions (Figures 1B, 2B). White light illumination caused a 97.2% decrease in spores (p < 0.001). Red light stimulated conidiation, while blue light had a negative effect on conidial formation. Compared to the dark condition, red light recovered about 68.7% of conidial formation, but under blue light, the formation of conidia was repressed by 99.8%. Different from mycelial extension, the disruption of AaVeA led to a significant reduction in conidia under both dark and light conditions. Interestingly, unlike the wild strain, red light hardly stimulated asexual sporulation when the AaVeA gene was inactivated. Blue light also resulted in the lack of sporulation in both wild and ΔAaVeA strains.
In general, light hardly affected fungal growth, while red light favored spore formation and blue light did the opposite.
To evaluate the AaVeA roles in TeA biosynthesis, mycotoxin extraction and quantitation were performed, and the changes in mycotoxin production were compared between the wild-type and ΔAaVeA strains under various light sources. The TeA concentration reached up to 48.8 μg/ml in darkness, and no significant change was observed with white light illumination (Figure 3A). Furthermore, mycotoxin production also seemed no more marked fluctuation under red and blue light conditions. TeA production became much weaker after the gene, AaVeA, was disrupted under dark or light conditions. Similarly, no obviously positive or negative effect on TeA biosynthesis was detected in the mutant strain in response to various light sources (p > 0.05).
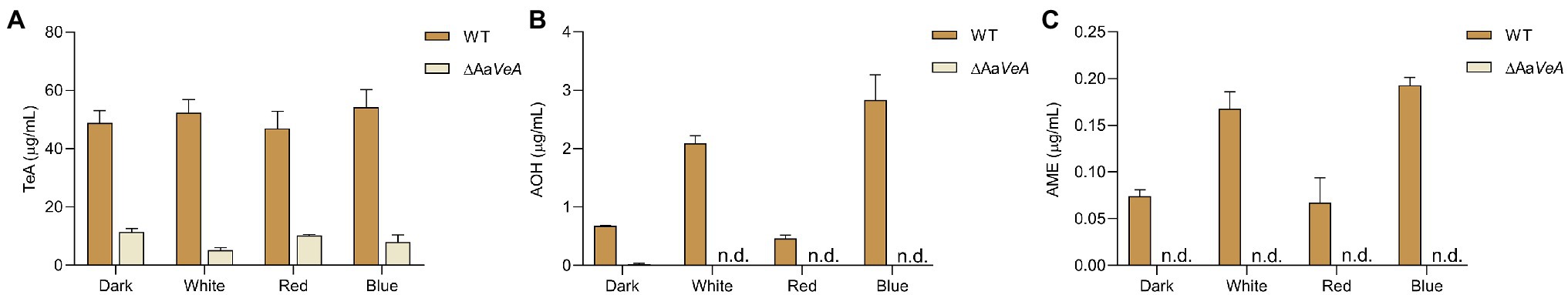
Figure 3. Mycotoxin biosynthesis produced by the WT and AaVeA disrupted mutant (ΔAaVeA) strain of Alternaria alternata under various light sources. The concentrations of mycotoxins, including tenuazonic acid (TeA; A), alternariol (AOH; B), and alternariol monomethyl ether (AME; C), were, respectively, extracted and determined by LC–MS.
In a previous study, light promoted AOH and AME production (Pruß et al., 2014; Igbalajobi et al., 2019), which was also confirmed in this study (Figures 3B,C), although it could not result in more accumulation in TeA biosynthesis. AOH and AME concentrations were extremely stimulated by light illumination, especially blue light. AOH and AME contents were separately promoted 2.08- and 1.26-times with white light higher than its production in the dark (p < 0.001). Furthermore, blue light exposure caused much more AOH and AME accumulation while red light had no positive stimulation. When AaVeA was deleted, AOH and AME production was significantly repressed by 96.4% and 100% in the dark (p < 0.001). Light could no longer stimulate more abundant AOH and AME biosynthesis, even blue light.
To uncover the regulatory roles of AaVeA, the detailed transcriptomic profile of A. alternata with or without light illumination was separately analyzed and shown in Supplementary Tables S3 and S4. In total, gene expression at the transcriptional level exhibited the most dramatic changes from the number of DEGs in wild type strain compared to the ΔAaVeA mutant grown in the dark (Supplementary Figure S2). There were 1,496 DEGs, including 614 upregulated and 882 downregulated DEGs. Nevertheless, the comparative transcriptomic profile of the mutant had fewer fluctuations between the dark and white light conditions, as there were 660 DEGs in total (350 upregulated and 310 downregulated genes). This demonstrated that AaVeA is a critically important regulator for the physiological-biochemical processes in A. alternata.
From the analysis of GO enrichment, some biological processes, including conidium formation, pathogenicity, and secondary metabolite biosynthetic process, were distinctly enriched from the comparatively expressed DEGs (Figure 4). The biological process related to conidial formation and development was enriched from the DEGs between the wild and AaVeA null strains under white light and dark conditions (Figure 5). The secondary metabolite biosynthetic process was markedly influenced by AaVeA regulation. The biosynthesis of mycotoxins, including TeA, AOH, and AME, were affected to various degrees under light regulation via the regulator.
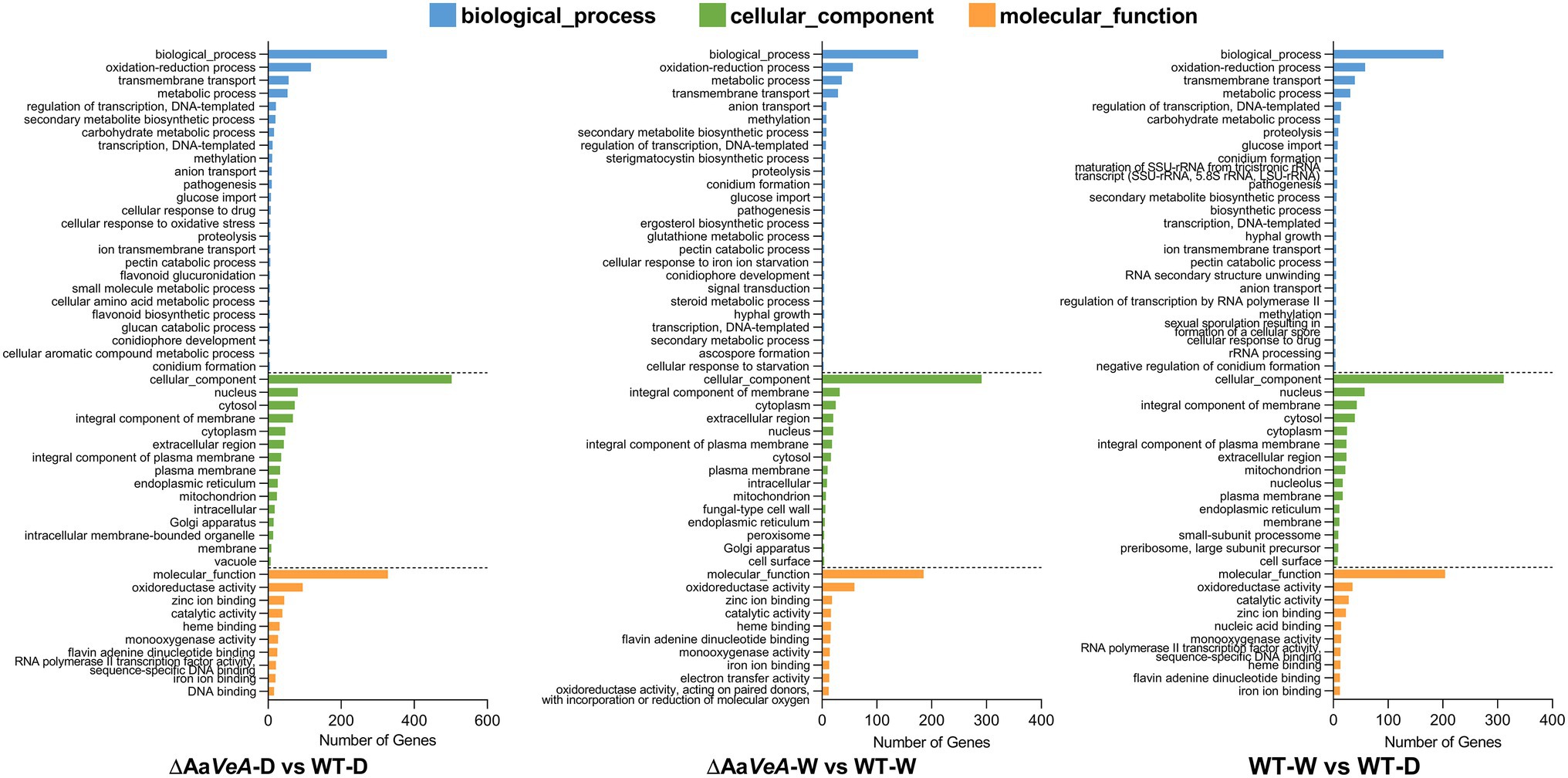
Figure 4. Gene Ontology (GO) enrichment analysis of differentially expressed genes (DEGs). The comparative DEGs were employed to carry out GO analysis in both groups of ΔAaVeA-D/WT-D (darkness), ΔAaVeA-W/WT-W (white light illumination), and WT-D/WT-W. The number of distribution of DEGs in GO terms enriched in biological process, cellular component, and molecular function was reflected as the histogram.

Figure 5. The heat map of the transcriptional level of DEGs enriched in the biological process of hyphal growth, conidium formation, and pathogenesis. The transcriptional level of DEGs was expressed as the value of log2 FPKM (Fragments Per Kilobase per Million) from the detailed transcriptomic profile.
Many DEGs partly enriched in the process of hyphal growth (Figure 5). This explains why the mycelial extension became slower when AaVeA was disrupted in A. alternata. In addition, the DEGs responsible for pathogenesis were enriched and hence AaVeA was probaly involved in the pathogenicity of fungal infection. To investigate whether AaVeA was responsible for pathogenicity, the trials were carried out in tomato infected by A. alternata with or without AaVeA.
From KEGG pathway enrichment analysis, the metabolism, biosynthesis, and degradation of amino acids, including valine, leucine, and isoleucine were found to be enriched (Supplementary Figure S3). The substrate of TeA biosynthesis comprises isoleucine and acetoacetyl-CoA. The fluctuation of gene expression related to amino acid metabolism could lead to the alteration of the supply of isoleucine. Acetoacetyl-CoA could be formed by carbohydrate metabolism, including fatty acid biosynthesis and degradation, which were also enriched in KEGG pathway enrichment analysis.
From the prediction of the secondary metabolite biosynthesis gene clusters, the TeA biosynthetic gene cluster contains two genes encoding a biosynthetic enzyme and a transporter. The enzymatic protein has a NRPS portion and a ketosynthase (KS) domain for the condensation and cyclization of the mycotoxin, which is similar to the homologous protein encoded by the TAS1 (MGG_07803) gene in Magnaporthe oryzae (Yun et al., 2015). Additionally, another TAS2 gene encoding a Zn(II)2-Cys6-type transcription factor harbored the neighboring TAS1 and was identified to be responsible for the regulation of TAS1 expression and mycotoxin biosynthesis (Yun et al., 2017). However, we searched for the homologous gene throughout the genome of A. alternata, but no matched protein was found. The transporter gene may be responsible for the export of TeA from the cytoplasm to the extracellular matrix. Generally, the trend of these two genes expression was similar to the production of the mycotoxin (Figure 6A). Accordingly, AOH/AME biosynthetic gene cluster has been identified, which consists of four biosynthetic enzymatic genes (pksI, omtI, moxI, sdrI, and doxI) and a transcriptional factor gene (aohR; Chooi et al., 2015; Wenderoth et al., 2019). Among them, pksI is responsible for AOH formation. AOH is then converted to AME by an O-methyltransferase encoded by omtI. Both the transcription of pksI and omtI became lower but the AaLaeA gene transcript had no downtrend in the AaVeA knockout strain (Figure 6B). This demonstrates that AaVeA could positively affect TeA biosynthesis by regulating the transcription of the clustered biosynthetic genes, but it did not modulate AaLaeA expression.
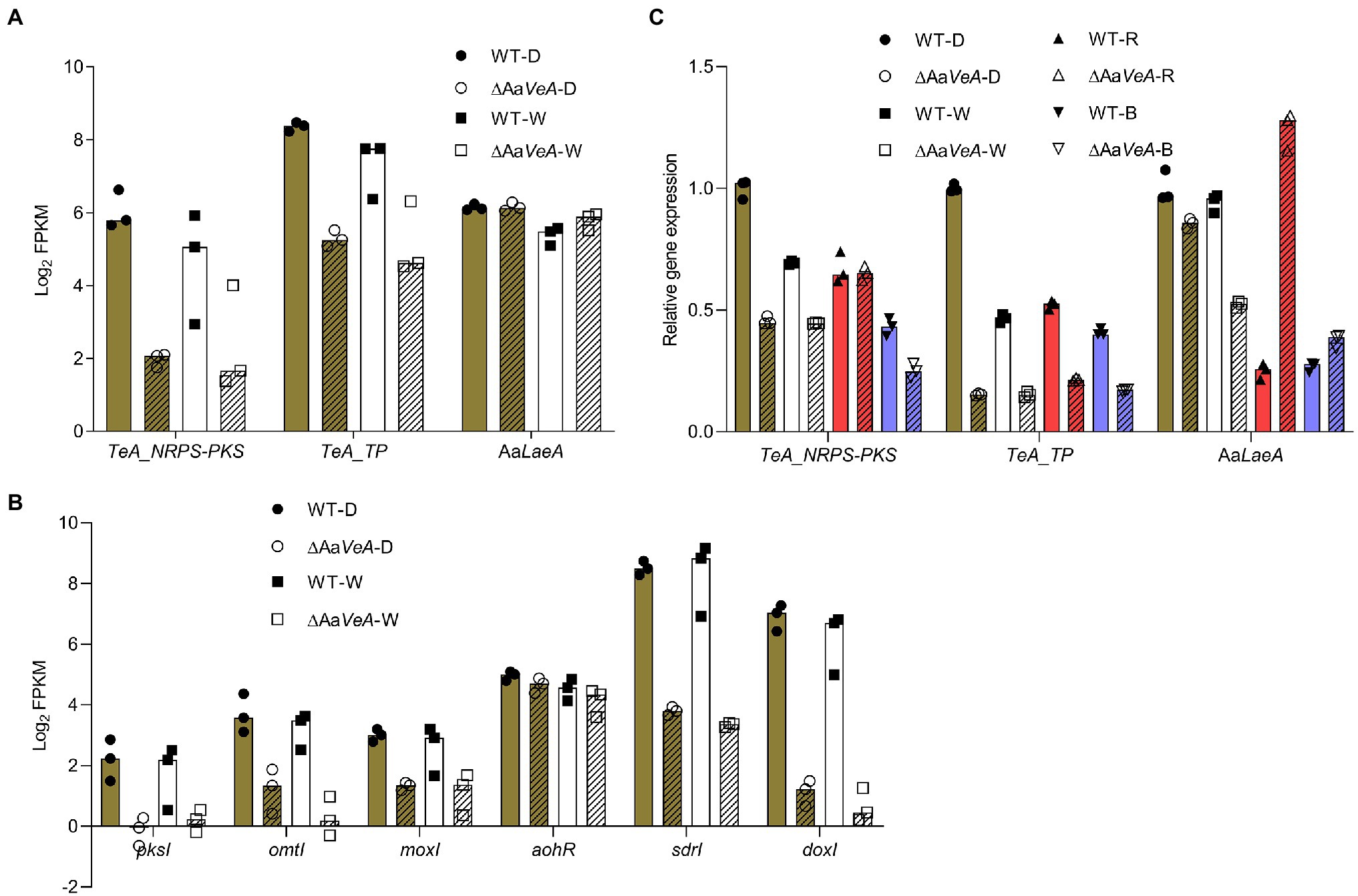
Figure 6. The expression analyses of biosynthetic and regulatory genes involved in mycotoxin biosynthesis. The biosynthetic and regulatory genes contain the biosynthetic clustered genes of TeA and AOH/AME, and the master regulator gene of secondary metabolites AaLaeA in Alternaria alternata. The results of the gene expression analyses were plotted using the median with range and separately expressed as the scatter dots. (A) The expression level of TeA biosynthetic clustered genes and AaLaeA was expressed as the value of log2 FPKM from the transcriptomic analysis. (B) The expression level of biosynthetic clustered genes for AOH and AME biosynthesis was expressed as the value of log2 FPKM from the transcriptomic analysis. (C) The relative gene expression was confirmed by reverse transcription quantitative real-time PCR (RT-qPCR) and calculated by the 2-ΔΔCt method.
To validate the data of RNA-seq, RT-qPCR was carried out for the analysis of fungal gene expression (Figure 6C). The trend of the clustered genes and LaeA transcription was similar to the analysis from RNA-seq. In addition, the red and blue light treatments had similar effects in the wild type and ΔAaVeA strains.
The wild and ΔAaVeA mutant strains of A. alternata were separately used to inoculate and colonize the surface of sterilized tomatoes. For 10 days culture, the disruption of AaVeA resulted in a reduction of 34.6% or 24.7% in lesion size under dark or light conditions, respectively (Figure 7). The hyphae of the mutant had weakness to be extending. Light seemed to have no significant influence on hyphal growth in the wild strain, which was consistent with the results of medium culture.
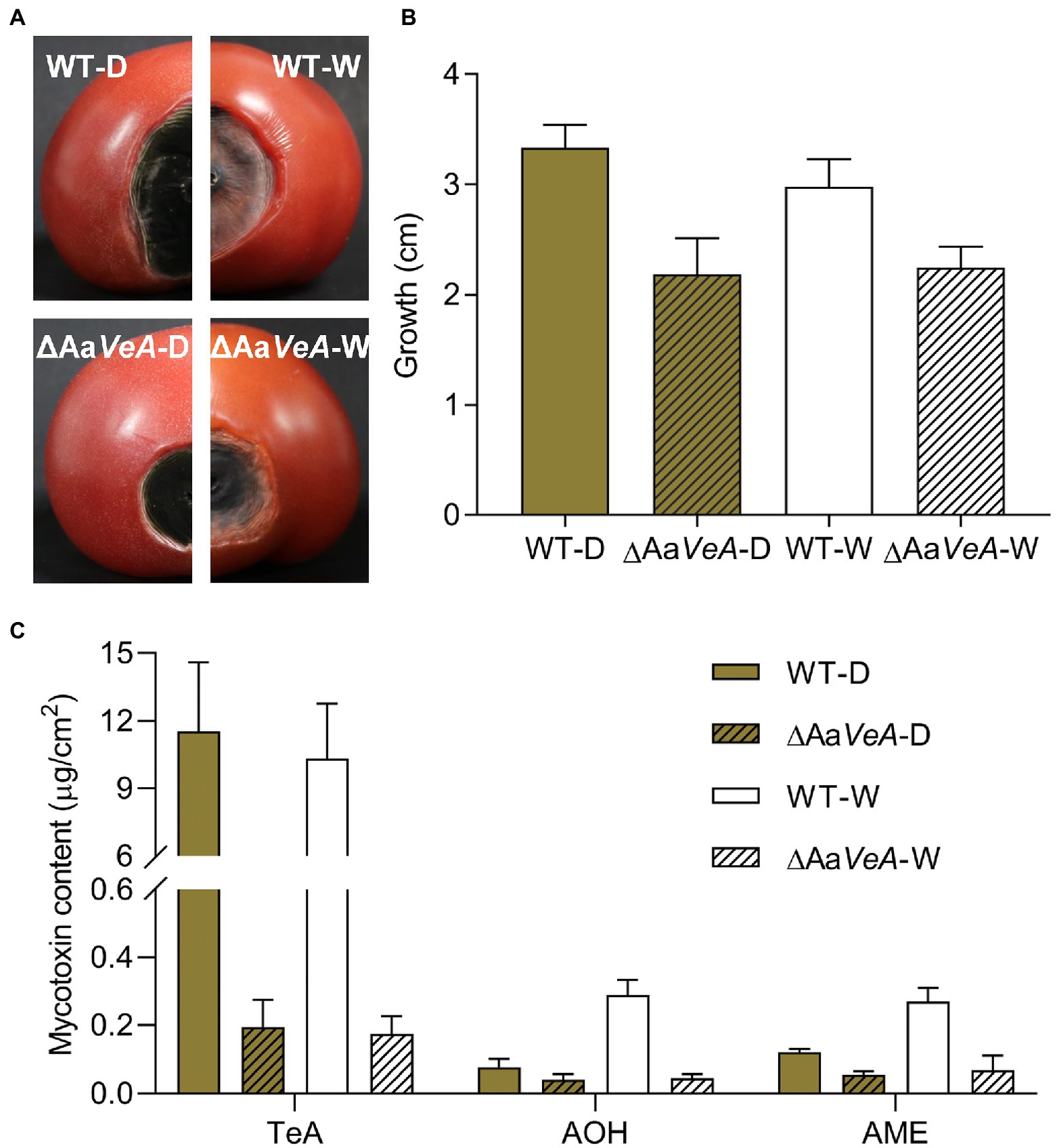
Figure 7. The infection and mycotoxin production of ΔAaVeA compared to the wild type (WT) strain on tomatoes with or without light treatment. The spore suspension of WT and ΔAaVeA strains was separately inoculated onto tomatoes (1 × 105 spores/ml; 5 μl) and cultured for 10 days at 25°C under dark or white light condition. (A) The diagram of tomatoes infected by WT and ΔAaVeA strains of Alternaria alternata. (B) The growth diameter of fungal infection. (C) The mycotoxin production of Alternaria alternata on the tomatoes.
Light had no obvious influence as an activator or inhibitor for TeA biosynthesis. Accordingly, TeA production was obviously decreased by 98.3% in the ΔAaVeA strain grown in the dark (p < 0.001). A similar reduction occurred under white light illumination. However, light stimulated the accumulation of AOH and AME by 2.81- and 1.24-times more than that of mycotoxin production in the dark (p < 0.001). Moreover, AOH and AME contents were separately suppressed by 45.9% and 53.7%, respectively, due to the deletion of AaVeA under dark condition. Similarly, the loss of AaVeA led to a decrease in the AOH and AME concentration by 84.3% and 74.7% in ΔAaVeA with light illumination. This showed that AOH and AME biosynthesis was regulated by light via AaVeA.
In total, these results from the pathogenic infection on tomato in vivo were similar to the outcome in vitro, in which AaVeA positively played vital roles in hyphal growth, pathogenicity, and mycotoxin biosynthesis.
Light modulates fungal growth, development, and virulence (Idnurm and Heitman, 2005; Purschwitz et al., 2006; Losi and Gaertner, 2021). The mode of action of various filamentous fungi has different responses under light conditions (Losi and Gaertner, 2021). Members of the fungal kingdom have developed light sensing systems, such as phytochrome FphA as a red-light photosensor (Blumenstein et al., 2005) and white collar (WC-1 and 2) homologs as blue-light photoreceptors (Idnurm and Heitman, 2005). Accordingly, FphA has been suggested positive function for sporulation while LreA, the WC-1 orthologue, is required for the spore formation independently of light in A. alternata (Pruß et al., 2014; Igbalajobi et al., 2019). The velvet complex, including VeA, LaeA, and VelB proteins, has been found to be involved in fungal growth, development, and virulence in response to light (Bayram et al., 2008; Amaike and Keller, 2009). In this study, the disruption of AaVeA enabled weaker growth and pathogenesis independent of light stimulation in A. alternata. Red light stimulates conidiospore formation, and blue light represses it, which is consistent with a previous study, in which blue light acted as a suppressor in A. alternata and A. cichorii (Vakalounakis and Christias, 1985, 1986; Pruß et al., 2014). Nevertheless, continuous light favors sporulation in A. eichhorniae (Shabana et al., 2000). The distinct results for conidiation in light conditions may be due to the diversity of species in the genus Alternaria and culture conditions (Varma et al., 2015). The loss of AaVeA results in the marked reduction of spore formation in A. alternata. Even red light could not stimulate more spores in the mutant strain and thus AaVeA was required for sporulation.
Light modulation could affect fungal metabolism as well, including primary and secondary metabolism (Tisch and Schmoll, 2010; Fanelli et al., 2015). Light regulates fungal primary metabolism, such as the production of carotenoids in the previous studies in the genus Fusarium (Estrada and Avalos, 2008; Tang et al., 2020), as well as Neurospora crassa (Nelson et al., 1989). In addition to the influence of primary metabolites, light was confirmed to be an important signal for secondary metabolism, including mycotoxin production (Fanelli et al., 2015). Furthermore, the effects of light were distinctly different on the biosynthesis of various secondary metabolites. For instance, light played no significant role in penicillin production in Penicillium chrysogenum (Kopke et al., 2013). Unlike P. chrysogenum, penicillin biosynthesis seemed to be positively influenced by light in A. nidulans (Kato et al., 2003; Spröte and Brakhage, 2007; Röhrig et al., 2017). In the research of mycotoxins, the production of aflatoxin B1 by A. flavus and ochratoxin A by A. ochraceus was considerably higher with light stimulation (Aziz and Moussa, 1997). However, in other reports, light exposure, especially blue light, led to the drastic reduction of ochratoxin A formation in the producing Penicillium and Aspergillus species (Schmidt-Heydt et al., 2011, 2012). In addition, sterigmatocystin production levels were differently altered under dark or light conditions in A. nidulans (Kato et al., 2003; Bayram et al., 2010; Röhrig et al., 2017). Sterigmatocystin biosynthesis was stimulated with the alteration from light to darkness in A. nidulans FGSC A4 (Kato et al., 2003; Bayram et al., 2010). Nevertheless, Light illumination preferred to the elevation of sterigmatocystin level in A. nidulans SRJ7 (Röhrig et al., 2017). In A. alternata, light was suggested distinct roles in AOH production in the previous studies (Pruß et al., 2014; Estiarte et al., 2016; Igbalajobi et al., 2019). Hence, we further performed the function of various light sources on the production of AOH and AME as well as TeA. In our study, AOH and AME biosynthesis were confirmed to be stimulated by light, especially blue light. However, TeA production was independently of light in that no stimulative impact occurred with light illumination. This demonstrates the distinct roles in the biosynthesis of various secondary metabolites in response to various light sources.
The heterotrimeric velvet complex, including VeA, LaeA, and VelB, is involved in secondary metabolism by coordinating light signals. In the velvet complex, VeA plays important roles in fungal growth and secondary metabolism in response to various light sources (Wiemann et al., 2010; Merhej et al., 2012; Eom et al., 2018). LaeA is responsible for the biosynthesis of secondary metabolites as the master regulator (Perrin et al., 2007; Bayram et al., 2008; Kosalková et al., 2009). Interestingly, in the study of the velvet complex for penicillin biosynthesis, veA exhibited opposing roles in penicillin biosynthesis in A. nidulans (Kato et al., 2003; Spröte and Brakhage, 2007). Nevertheless, VeA acts similar role as a positive regulator for sterigmatocystin production in A. nidulans (Kato et al., 2003; Bayram et al., 2010). The function of LaeA and VeA on AOH and AME production had been performed with or without white light in A. alternata (Estiarte et al., 2016). Based on the previous studies, the regulatory impacts of VeA were carried out for the production of AOH, AME, and TeA in A. alternata in response to various light sources. The marked reduction of mycotoxins occurred, and light had no stimulative impact on AOH and AME biosynthesis in the AaVeA null strain. Thus, all the mycotoxins were partly regulated by the velvet gene, AaVeA.
Mycotoxins are involved in fungal pathogenicity, as they facilitate mycotoxin-producing fungi to colonize plant tissues or utilize their nutrients (Graf et al., 2012; Wenderoth et al., 2019). TeA exerts phytotoxic activity and induces plant necrosis by blocking electron transport (Chen and Qiang, 2017). In addition, AOH was confirmed as a virulence factor for its contribution to the infection and extension of fungal hosts. In this study, the deletion of AaVeA led to the marked reduction of TeA, AOH, and AME. Therefore, the infection of the mutant became weaker on tomato. In total, this above illustrates that the regulator AaVeA coordinates light signals for mycelial growth, sporulation, pathogenesis, and mycotoxin production.
The continuous supply of substrates is a prerequisite of mycotoxin biosynthesis. Accordingly, TeA has been confirmed to be formed by the condensation of an isoleucine and an acetoacetyl-CoA (Yun et al., 2015). The biosynthesis of AOH and AME initially starts from acetyl-CoA (Saha et al., 2012; Wenderoth et al., 2019). Hence, the influence of metabolic processes may lead to an obvious change in mycotoxin biosynthesis. The loss of AaVeA could cause an obvious reduction in mycotoxin production possibly due to the drastic fluctuation in amino acid metabolic process, carbohydrate metabolic process, secondary metabolites biosynthetic process, etc., which were enriched in the comparatively transcriptomic profile of wild type and AaVeA null strains with or without light.
Generally, secondary metabolites of biosynthetic genes are clustered and harbored closely in mold. Not only TeA but also AOH and AME have been identified. The TeA biosynthetic gene cluster contains at least one enzyme catalyzing the cyclization reaction and a pathway-specific Zn(II)2-Cys6-type transcriptional factor in M. oryzae (Yun et al., 2015, 2017). In fact, there is a homologous enzyme responsible for the catalytic reaction, but no specific regulatory gene is harbored near the enzymatic gene. However, there is a neighboring transporter gene, which may be connected to the export of the mycotoxin avoiding toxicity to it. AOH and AME were biosynthesized by a gene cluster consisting of a polyketide synthase gene (pksI), an O-methyltransferase (omtI), and other enzymatic genes, as well as a transcriptional factor gene, aohR (Wenderoth et al., 2019). The disruption of AaVeA led to a drastic decline on the expression level of biosynthetic genes in A. alternata under both light and dark conditions, which agrees with the trend of mycotoxin production.
In conclusion, light stimulation had no marked alteration on mycelial growth but caused significant changes in sporulation in A. alternata. Although red light favored the formation of spores, blue light weakened the sporulation. However, the lack of AaVeA led to slower fungal growth and the marked reduction of conidiospore formation in A. alternata despite light stimulation. The deletion of AaVeA resulted in drastic transcriptional fluctuation of DEGs enriched in hyphal growth, conidiation, and pathogenicity in the wild type and ΔAaVeA strains under dark and light conditions. Light, especially blue light stimulated AOH and AME accumulation but had no obvious effects on TeA production. The biosynthesis of these mycotoxins was dependent on the modulation of the velvet gene, AaVeA, in A. alternata. The regulator positively modulated mycotoxin production via the continuous supply of substrates and the activation of biosynthetic gene expression. From this study, the application of visible light on the prevention and control of the mycotoxins seems inappropriate due to the stimulus for AOH and AME biosynthesis by light, especially blue light. In combination with our previous study, UV-C irradiation could probably be a promising tactic for the mycotoxin control. In addition, it would be more effective to avoid the continuously visual light when some other control measures are taken for reducing the mycotoxins contamination in agricultural products.
The original contributions presented in the study are publicly available. This data can be found at: NGDC, CRA005946, https://ngdc.cncb.ac.cn/gsa/browse/CRA005946.
LW and MW conceptualized and designed the manuscript and wrote and revised the manuscript. LW carried out the experiments. HL and JJ gave the technical supports and revised the manuscript. All authors read and approved the manuscript.
This research was supported by the National Natural Science Foundation of China (grant no. 31801648), the National Project for Quality and Safety Risk Assessment of Agricultural Products of China (grant no. GJFP2019002), and the Beijing Natural Science Foundation (grant nos. 6184038 and 7192026).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.842268/full#supplementary-material
TeA, Tenuazonic acid; AOH, Alternariol; AME, Alternariol monomethyl ether; TAS1, TeA synthetase 1; NRPS, Non-ribosomal peptide synthetase; PKS, Polyketide synthase; BLAST, Basic Local Alignment Search Tool; WC, White collar; PDA, Potato dextrose agar; PDB, Potato dextrose broth; PCR, Polymerase chain reaction; Hph, Hygromycin phosphotransferase; PEG, Polyethylene glycol; SEM, Scanning electron microscopy; DEGs, Differentially expressed genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; RT-qPCR, Reverse transcription quantitative real-time PCR; ANOVA, Analysis of variance; KS, Ketosynthase.
Amaike, S., and Keller, N. P. (2009). Distinct roles for VeA and LaeA in development and pathogenesis of Aspergillus flavus. Eukaryot. Cell 8, 1051–1060. doi: 10.1128/EC.00088-09
Arcella, D., Eskola, M., and Ruiz, J. A. G. (2016). Dietary exposure assessment to Alternaria toxins in the European population. EFSA J. 14:4654. doi: 10.2903/j.efsa.2016.4654
Aziz, N. H., and Moussa, L. A. (1997). Influence of white light, near-UV irradiation and other environmental conditions on production of aflatoxin B1 by Aspergillus flavus and ochratoxin A by Aspergillus ochraceus. Nahrung 41, 150–154. doi: 10.1002/food.19970410307
Bayram, Ö. S., Bayram, Ö., Valerius, O., Park, H. S., Irniger, S., Gerke, J., et al. (2010). LaeA control of velvet family regulatory proteins for light-dependent development and fungal cell-type specificity. PLoS Genet. 6:e1001226. doi: 10.1371/journal.pgen.1001226
Bayram, Ö., Krappmann, S., Ni, M., Jin, W. B., Helmstaedt, K., Valerius, O., et al. (2008). VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320, 1504–1506. doi: 10.1126/science.1155888
Blumenstein, A., Vienken, K., Tasler, R., Purschwitz, J., Veith, D., Frankenberg-Dinkel, N., et al. (2005). The Aspergillus nidulans phytochrome FphA represses sexual development in red light. Curr. Biol. 15, 1833–1838. doi: 10.1016/j.cub.2005.08.061
Chang, P.-K., Scharfenstein, L. L., Li, P., and Ehrlich, K. C. (2013). Aspergillus flavus VelB acts distinctly from VeA in conidiation and may coordinate with FluG to modulate sclerotial production. Fungal Genet. Biol. 58–59, 71–79. doi: 10.1016/j.fgb.2013.08.009
Chen, S., and Qiang, S. (2017). Recent advances in tenuazonic acid as a potential herbicide. Pestic. Biochem. Physiol. 143, 252–257. doi: 10.1016/j.pestbp.2017.01.003
Chooi, Y.-H. H., Muria-Gonzalez, M. J., Mead, O. L., and Solomon, P. S. (2015). SnPKS19 encodes the polyketide synthase for alternariol mycotoxin biosynthesis in the wheat pathogen Parastagonospora nodorum. Appl. Environ. Microbiol. 81, 5309–5317. doi: 10.1128/aem.00278-15
Dang, H. X., Pryor, B., Peever, T., and Lawrence, C. B. (2015). The Alternaria genomes database: a comprehensive resource for a fungal genus comprised of saprophytes, plant pathogens, and allergenic species. BMC Genomics 16:239. doi: 10.1186/s12864-015-1430-7
EFSA on Contaminants in the Food Chain (CONTAM) (2011). Scientific opinion on the risks for animal and public health related to the presence of Alternaria toxins in feed and food. EFSA J. 9, 1–97. doi: 10.2903/j.efsa.2011.2407
Eom, T.-J., Moon, H., Yu, J.-H., and Park, H.-S. (2018). Characterization of the velvet regulators in Aspergillus flavus. J. Microbiol. 56, 893–901. doi: 10.1007/s12275-018-8417-4
Estiarte, N., Lawrence, C. B., Sanchis, V., Ramos, A. J., and Crespo-Sempere, A. (2016). LaeA and VeA are involved in growth morphology, asexual development, and mycotoxin production in Alternaria alternata. Int. J. Food Microbiol. 238, 153–164. doi: 10.1016/j.ijfoodmicro.2016.09.003
Estrada, A. F., and Avalos, J. (2008). The white collar protein WcoA of Fusarium fujikuroi is not essential for photocarotenogenesis, but is involved in the regulation of secondary metabolism and conidiation. Fungal Genet. Biol. 45, 705–718. doi: 10.1016/j.fgb.2007.12.003
Fanelli, F., Geisen, R., Schmidt-Heydt, M., Logrieco, A. F., and Mulè, G. (2015). Light regulation of mycotoxin biosynthesis: new perspectives for food safety. World Mycotoxin J. 9, 129–146. doi: 10.3920/wmj2014.1860
Graf, E., Schmidt-Heydt, M., and Geisen, R. (2012). HOG MAP kinase regulation of alternariol biosynthesis in Alternaria alternata is important for substrate colonization. Int. J. Food Microbiol. 157, 353–359. doi: 10.1016/j.ijfoodmicro.2012.06.004
Idnurm, A., and Heitman, J. (2005). Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 3:e95. doi: 10.1371/journal.pbio.0030095
Igbalajobi, O., Yu, Z., and Fischer, R. (2019). Red- and blue-light sensing in the plant pathogen Alternaria alternata depends on phytochrome and the white-collar protein LreA. mBio 10, 1–17. doi: 10.1128/mbio.00371-19
Jiang, N., Li, Z., Wang, L., Li, H., Zhu, X., Feng, X., et al. (2019). Effects of ultraviolet-c treatment on growth and mycotoxin production by Alternaria strains isolated from tomato fruits. Int. J. Food Microbiol. 311:108333. doi: 10.1016/j.ijfoodmicro.2019.108333
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. doi: 10.1093/nar/gkm882
Kato, N., Brooks, W., and Calvo, A. M. (2003). The expression of sterigmatocystin and penicillin genes in Aspergillus nidulans is controlled by veA, a gene required for sexual development. Eukaryot. Cell 2, 1178–1186. doi: 10.1128/EC.2.6.1178-1186.2003
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kopke, K., Hoff, B., Bloemendal, S., Katschorowski, A., Kamerewerd, J., and Kück, U. (2013). Members of the Penicillium chrysogenum velvet complex play functionally opposing roles in the regulation of penicillin biosynthesis and conidiation. Eukaryot. Cell 12, 299–310. doi: 10.1128/EC.00272-12
Kosalková, K., García-Estrada, C., Ullán, R. V., Godio, R. P., Feltrer, R., Teijeira, F., et al. (2009). The global regulator LaeA controls penicillin biosynthesis, pigmentation and sporulation, but not roquefortine C synthesis in Penicillium chrysogenum. Biochimie 91, 214–225. doi: 10.1016/j.biochi.2008.09.004
Logrieco, A., Moretti, A., and Solfrizzo, M. (2009). Alternaria toxins and plant diseases: An overview of origin, occurrence and risks. World Mycotoxin J. 2, 129–140. doi: 10.3920/WMJ2009.1145
Losi, A., and Gaertner, W. (2021). A light life together: photosensing in the plant microbiota. Photochem. Photobiol. Sci. 20, 451–473. doi: 10.1007/s43630-021-00029-7
Merhej, J., Urban, M., Dufresne, M., Hammond-Kosack, K. E., Richard-Forget, F., and Barreau, C. (2012). The velvet gene, FgVe1, affects fungal development and positively regulates trichothecene biosynthesis and pathogenicity in Fusarium graminearum. Mol. Plant Pathol. 13, 363–374. doi: 10.1111/j.1364-3703.2011.00755.x
Nelson, M. A., Morelli, G., Carattoli, A., Romano, N., and Macino, G. (1989). Molecular cloning of a Neurospora crassa carotenoid biosynthetic gene (Albino-3) regulated by blue light and the products of the white collar genes. Mol. Cell. Biol. 9, 1271–1276. doi: 10.1128/mcb.9.3.1271-1276.1989
Ostry, V. (2008). Alternaria mycotoxins: an overview of chemical characterization, producers, toxicity, analysis and occurrence in foodstuffs. World Mycotoxin J. 1, 175–188. doi: 10.3920/wmj2008.x013
Patriarca, A. (2016). Alternaria in food products. Curr. Opin. Food Sci. 11, 1–9. doi: 10.1016/j.cofs.2016.08.007
Perrin, R. M., Fedorova, N. D., Jin, W. B., Cramer, R. A., Wortman, J. R., Kim, H. S., et al. (2007). Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 3:e50. doi: 10.1371/journal.ppat.0030050
Pruß, S., Fetzner, R., Seither, K., Herr, A., Pfeiffer, E., Metzler, M., et al. (2014). Role of the Alternaria alternata blue-light receptor LreA (white-collar 1) in spore formation and secondary metabolism. Appl. Environ. Microbiol. 80, 2582–2591. doi: 10.1128/aem.00327-14
Purschwitz, J., Müller, S., Kastner, C., and Fischer, R. (2006). Seeing the rainbow: light sensing in fungi. Curr. Opin. Microbiol. 9, 566–571. doi: 10.1016/j.mib.2006.10.011
Rio, D. C., Ares, M., Hannon, G. J., and Nilsen, T. W. (2010). Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010, 1–4. doi: 10.1101/pdb.prot5439
Röhrig, J., Yu, Z., Chae, K.-S., Kim, J.-H., Han, K.-H., and Fischer, R. (2017). The Aspergillus nidulans velvet-interacting protein, VipA, is involved in light-stimulated heme biosynthesis: A. nidulans heme biosynthesis. Mol. Microbiol. 105, 825–838. doi: 10.1111/mmi.13739
Saha, D., Fetzner, R., Burkhardt, B., Podlech, J., Metzler, M., Dang, H., et al. (2012). Identification of a polyketide synthase required for alternariol (AOH) and alternariol-9-methyl ether (AME) formation in Alternaria alternata. PLoS One 7:e40564. doi: 10.1371/journal.pone.0040564
Schmidt-Heydt, M., Cramer, B., Graf, I., Lerch, S., Humpf, H. U., and Geisen, R. (2012). Wavelength-dependent degradation of ochratoxin and citrinin by light in vitro and in vivo and its implications on Penicillium. Toxins 4, 1535–1551. doi: 10.3390/toxins4121535
Schmidt-Heydt, M., Rüfer, C., Raupp, F., Bruchmann, A., Perrone, G., and Geisen, R. (2011). Influence of light on food relevant fungi with emphasis on ochratoxin producing species. Int. J. Food Microbiol. 145, 229–237. doi: 10.1016/j.ijfoodmicro.2010.12.022
Shabana, Y. M., Elwakil, M. A., and Charudattan, R. (2000). Effect of media, light and pH on growth and spore production by Alternaria eichhorniae, a mycoherbicide agent for waterhyacinth. J. Plant Dis. Prot. 107, 617–626.
Solfrizzo, M. (2017). Recent advances on Alternaria mycotoxins. Curr. Opin. Food Sci. 17, 57–61. doi: 10.1016/j.cofs.2017.09.012
Spröte, P., and Brakhage, A. A. (2007). The light-dependent regulator velvet A of Aspergillus nidulans acts as a repressor of the penicillin biosynthesis. Arch. Microbiol. 188, 69–79. doi: 10.1007/s00203-007-0224-y
Takao, K., Akagi, Y., Tsuge, T., Harimoto, Y., Yamamoto, M., and Kodama, M. (2016). The global regulator LaeA controls biosynthesis of host-specific toxins, pathogenicity and development of Alternaria alternata pathotypes. J. Gen. Plant Pathol. 82, 121–131. doi: 10.1007/s10327-016-0656-9
Tang, Y., Zhu, P., Lu, Z., Qu, Y., Huang, L., Zheng, N., et al. (2020). The photoreceptor components FaWC1 and FaWC2 of Fusarium asiaticum cooperatively regulate light responses but play independent roles in virulence expression. Microorganisms 8:365. doi: 10.3390/microorganisms8030365
Thomma, B. P. H. J. (2003). Alternaria spp.: From general saprophyte to specific parasite. Mol. Plant Pathol. 4, 225–236. doi: 10.1046/j.1364-3703.2003.00173.x
Tisch, D., and Schmoll, M. (2010). Light regulation of metabolic pathways in fungi. Appl. Microbiol. Biotechnol. 85, 1259–1277. doi: 10.1007/s00253-009-2320-1
Vakalounakis, D. J., and Christias, C. (1985). Blue-light inhibition of conidiation in Alternaria cichorii. Trans. Br. Mycol. Soc. 85, 285–289. doi: 10.1016/S0007-1536(85)80190-X
Vakalounakis, D. J., and Christias, C. (1986). Dark reversion of blue-light inhibition of conidiation in Alternaria cichorii. Can. J. Bot. 64, 1016–1017. doi: 10.1139/b86-138
Varma, P. K., Yamuna, C., and Mangala, U. N. (2015). The effect of media and light on in vitro sporulation of Alternaria solani. Int. J. Appl. Biol. Pharm. Technol. 6, 97–102.
Wang, L., Jiang, N., Wang, D., and Wang, M. (2019). Effects of essential oil citral on the growth, mycotoxin biosynthesis and transcriptomic profile of Alternaria alternata. Toxins 11:553. doi: 10.3390/toxins11100553
Wenderoth, M., Garganese, F., Schmidt-Heydt, M., Soukup, S. T., Ippolito, A., Sanzani, S. M., et al. (2019). Alternariol as virulence and colonization factor of Alternaria alternata during plant infection. Mol. Microbiol. 112, 131–146. doi: 10.1111/mmi.14258
Wiemann, P., Brown, D. W., Kleigrewe, K., Bok, J. W., Keller, N. P., Humpf, H. U., et al. (2010). FfVel1 and FfLae1, components of a velvet-like complex in Fusarium fujikuroi, affect differentiation, secondary metabolism and virulence. Mol. Microbiol. 77, 972–994. doi: 10.1111/j.1365-2958.2010.07263.x
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. doi: 10.1186/gb-2010-11-2-r14
Yun, C. S., Motoyama, T., and Osada, H. (2015). Biosynthesis of the mycotoxin tenuazonic acid by a fungal NRPS-PKS hybrid enzyme. Nat. Commun. 6, 1–9. doi: 10.1038/ncomms9758
Yun, C. S., Motoyama, T., and Osada, H. (2017). Regulatory mechanism of mycotoxin tenuazonic acid production in Pyricularia oryzae. ACS Chem. Biol. 12, 2270–2274. doi: 10.1021/acschembio.7b00353
Keywords: Alternaria alternata, mycotoxin, tenuazonic acid, alternariol, light regulation, velvet complex
Citation: Wang L, Wang M, Jiao J and Liu H (2022) Roles of AaVeA on Mycotoxin Production via Light in Alternaria alternata. Front. Microbiol. 13:842268. doi: 10.3389/fmicb.2022.842268
Received: 23 December 2021; Accepted: 27 January 2022;
Published: 18 February 2022.
Edited by:
Hosny El-Adawy, Friedrich Loeffler Institut, GermanyReviewed by:
Mustafa Shukry Atta, Kafrelsheikh University, EgyptCopyright © 2022 Wang, Wang, Jiao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meng Wang, d2FuZ21AYnJjYXN0Lm9yZy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.