- 1Vaccine and Infectious Disease Organization, Saskatoon, SK, Canada
- 2Department of Biochemistry, Microbiology and Immunology, University of Saskatchewan, Saskatoon, SK, Canada
- 3Department of Veterinary Microbiology, University of Saskatchewan, Saskatoon, SK, Canada
The global poultry industry has grown to the extent that the number of chickens now well exceeds the number of humans on Earth. Escherichia coli infections in poultry cause significant morbidity and economic losses for producers each year. We obtained 94 E. coli isolates from 12 colibacillosis outbreaks on Saskatchewan farms and screened them for antimicrobial resistance and biofilm formation. Fifty-six isolates were from broilers with confirmed colibacillosis, and 38 isolates were from healthy broilers in the same flocks (cecal E. coli). Resistance to penicillins, tetracyclines, and aminoglycosides was common in isolates from all 12 outbreaks, while cephalosporin resistance varied by outbreak. Most E. coli were able to form biofilms in at least one of three growth media (1/2 TSB, M63, and BHI broth). There was an overall trend that disease-causing E. coli had more antibiotic resistance and were more likely to form biofilms in nutrient-rich media (BHI) as compared to cecal strains. However, on an individual strain basis, there was no correlation between antimicrobial resistance and biofilm formation. The 21 strongest biofilm forming strains consisted of both disease-causing and cecal isolates that were either drug resistant or susceptible. Draft whole genome sequencing indicated that many known antimicrobial resistance genes were present on plasmids, with disease-causing E. coli having more plasmids on average than their cecal counterparts. We tested four common disinfectants for their ability to kill 12 of the best biofilm forming strains. All disinfectants killed single cells effectively, but biofilm cells were more resistant, although the difference was less pronounced for the disinfectants that have multiple modes of action. Our results indicate that there is significant diversity and complexity in E. coli poultry isolates, with different lifestyle pressures affecting disease-causing and cecal isolates.
Introduction
Colibacillosis is a broad term for infections caused by pathogenic strains of Escherichia coli, and it is known to affect wide range of farm animals including poultry, pigs, and calves (Foster and Smith, 2009; Guabiraba and Schouler, 2015; Renzhammer et al., 2020). Avian colibacillosis is an infectious disease of poultry caused by Avian pathogenic E. coli (APEC) (Dziva and Stevens, 2008; Kemmett et al., 2013; Guabiraba and Schouler, 2015). It is associated with collection of extraintestinal infections including acute fatal septicemia, air sacculitis, chronic respiratory diseases, cellulitis, pericarditis, peritonitis and salphingitis (Nolan et al., 2020). Outbreaks among flocks cause significant economic losses in the poultry industry due to carcass condemnation, reduced egg laying, morbidity, and mortality, as well as costs incurred for disinfection and antimicrobial treatment (Dziva and Stevens, 2008; Mellata, 2013; Graveline et al., 2014; Guabiraba and Schouler, 2015). Colibacillosis is thought to begin as a respiratory tract infection in broilers that inhale fecal dust contaminated with APEC (Dziva and Stevens, 2008; Guabiraba and Schouler, 2015). Due to a lack of consistent phenotypic and genotypic characteristics, the gold standard for APEC classification is the chicken lethality assay (Wooley et al., 2000; Collingwood et al., 2014; Mageiros et al., 2021). APEC strains, which encompass a large number of E. coli sequence types, are one of the subpathotypes of ExPEC (Extra intestinal pathogenic E. coli) together with uropathogenic, sepsis-associated and neonatal meningitis E. coli (Johnson et al., 2008). Several studies have suggested that APEC and human ExPEC strains share similar virulence-associated genes despite their sources of isolation, indicating the increased potential for zoonotic E. coli infections in humans (Johnson et al., 2008; Jakobsen et al., 2011). For example, a human pathogenic E. coli strain belonging to the O25b:H4-ST131 pandemic clonal group has been isolated from poultry, pigs, and cattle (Mora et al., 2010; Nicolas-Chanoine et al., 2014). Although they cause significant damage to broiler flocks, little is known about the ecological niche of the APEC strains. Do they live within the barn setting or do they colonize and later escape the intestinal niche? Many of the details surrounding colibacillosis infections remain a mystery.
The primary treatment option for colibacillosis is antimicrobial therapy (Sargeant et al., 2019). Many antimicrobials that are critical for human medicine are used in the global poultry industry to prevent and treat bacterial diseases (Landoni and Albarellos, 2015). Antimicrobials had been used for decades to enhance growth and prevent infection in food animals (Xiong et al., 2018), but this overuse of antimicrobials lead to the emergence of antimicrobial resistance (AMR) among bacterial pathogens and was thought to be a significant contributor to the AMR crisis in human clinical settings. Hence, the usage of antimicrobials as a growth promotor has now been banned in many countries (Maron et al., 2013; Government of Canada, 2018; Xiong et al., 2018; McMillan et al., 2019). E. coli are resident microbes in the intestinal flora of most food animals (Partridge et al., 2018). In the gut, they are exposed to billions of other bacteria and viruses which may harbor antimicrobial resistant mobile genetic elements. E. coli are highly promiscuous and may be a significant reservoir of AMR (Lugsomya et al., 2018; Chuppava et al., 2019). For example, E. coli that possess extended-spectrum β-lactamases (ESBL) are resistant to penicillins and certain cephalosporins (i.e., 3rd and 4th generations) and because many of the resistance genes are located on mobile genetic elements, ESBL E. coli are now globally disseminated (Thaden et al., 2016). Infections caused by ESBL E. coli are associated with higher economic costs, longer hospital stays, and increased mortality compared to non-ESBL E. coli (Tumbarello et al., 2007; Malande et al., 2019). Poultry are a known reservoir of ESBL E. coli, and transmission from livestock to humans has been proven via multi-locus sequence typing and whole genome sequencing (Leverstein-van Hall et al., 2011; Overdevest et al., 2011; Nagy et al., 2021). This all becomes more prominent since E. coli were recently identified as the #1 AMR-associated bacterial species causing morbidity and mortality in humans worldwide (Antimicrobial Resistance Collaborators, 2022).
Most APEC strains are also likely to form biofilms, an important factor contributing to increased persistence and survival of APEC within broiler farms (Reisner et al., 2006; Rodrigues et al., 2019). Biofilms are a physiological state where bacterial cells become irreversibly attached to surfaces and enclosed in a self-secreted extracellular polymeric matrix composed of proteins, polysaccharides, and nucleic acids (Hung et al., 2013; Sanchez-Vizuete et al., 2015). Growth in biofilms offers certain advantages to APEC including facilitating the exchange of AMR and virulence-associated genes via horizontal gene transfer between bacteria of same or different species (van den Bogaard and Stobberingh, 2000). In addition, the biofilm matrix provides protection for the embedded cells, resisting detrimental effects of antibiotics, disinfectants, and the host immune system, all of which makes it difficult to eradicate biofilm-related infections in clinical and food production settings (Reisner et al., 2006; Kester and Fortune, 2014; Rodrigues et al., 2019). Despite the increasing occurrence of AMR, bacteria seem to remain susceptible to disinfectants; the presence of multiple active ingredients with different mechanisms of action means that resistance is more unlikely to arise (Davin-Regli, 2012). However, disinfectants are often used incorrectly in practice; the use of inappropriate concentrations, expired products, or the inactivation of compounds due to the presence of inorganic matter on insufficiently cleaned surfaces are common errors. The exposure of bacteria to sub-inhibitory concentrations of disinfectants may result in reduced susceptibility (Soumet et al., 2016), which can cause significant problems for producers at multiple steps in the food production pipeline. Further, insufficient cleaning of barn surfaces or in meat packaging facilities may allow biofilm formation to occur on equipment. We feel it is important to understand the lifestyle of APEC strains to prevent dissemination of disease in birds and potentially to humans.
We isolated 94 E. coli from 12 different colibacillosis outbreaks on Saskatchewan farms and analyzed these isolates for biofilm formation, and resistance to 27 antimicrobial compounds and four disinfectants. We wanted to determine if biofilm formation and AMR could be correlated and if E. coli from diseased birds were different than E. coli from healthy birds.
Materials and Methods
Broiler Sampling
Four- to six-week-old broiler birds that were suspected of having colibacillosis were submitted to the Poultry Extension Team (PEX) at the Western College of Veterinary Medicine, University of Saskatchewan, as part of a routine analysis for the Saskatchewan broiler industry. In each case, the birds originated from local producers that had farms within a 2-h driving distance of Saskatoon. Members of the PEX performed necropsy on each bird and submitted organ samples for microbial isolation and diagnosis by MALDI-TOF mass spectrometry (Prairie Diagnostic Services1). If E. coli infection in the birds was confirmed, the PEX sent samples of the heart, liver, and spleen, previously stored at 4°C, to the White lab. At the same time, the PEX requested from the same producers that they submit 3–4 healthy broilers from the flocks where the diseased birds had originated. The cecal contents from these birds were subsequently sent to the White lab.
Escherichia coli Isolation and Identification
Samples of the heart, liver, and spleen (diseased birds) or cecal contents (presumed healthy birds) were placed in 2 ml Eppendorf Safe-Lock tubes containing 1 ml of phosphate-buffered saline (PBS) and a 5-mm steel bead (Qiagen #69989) and were homogenized for 5 min at 30 Hz using a mixer mill (Retsch; MM400). Aliquots of the organ homogenates were spread on MacConkey Agar and colonies allowed to grow overnight at 37°C. Suspected E. coli colonies were confirmed via positive indole and negative Simmons-citrate biochemical tests. In total, we analyzed 56 disease-causing or “systemic” E. coli isolates and 38 healthy or “cecal” E. coli isolates, collected from 12 outbreaks over a 2-year period (Supplementary Table 1). E. coli isolates with visually distinct colony morphologies were selected for further analysis.
Bacterial Growth and Media Conditions
Bacterial isolates were stored in 20% glycerol at –80°C for long-term preservation. Isolates were inoculated from frozen stocks onto Tryptic soy agar (TSA; Becton Dickinson) and were incubated for 18 h at 37°C. A single colony was inoculated into 5 mL of LB broth and grown overnight at 37°C with shaking at 200 rpm. Overnight cultures were used for all subsequent experiments.
Curli and Cellulose Testing
Overnight cultures were standardized to an optical density of 1.0 at 600 nm. Four microliter aliquots were spotted on Congo Red Agar (20 μg/mL of Congo Red dye in 1% tryptone, 0.5% yeast extract, 1.5% agar; pH 7.2–7.4), to visualize the rdar morphotype and curli production (Collinson et al., 1991) and onto Calcofluor White agar [200 mg/L fluorescent brightener #28; (Sigma-Aldrich, St. Louis, MO, United States) in 1% tryptone, 0.5% yeast extract, 1.5% agar] to visualize cellulose production (Solano et al., 2002).
Crystal Violet Assay
Crystal violet assays were performed using the method of Merritt et al. (2011), with details outlined previously (Liu et al., 2018). For each isolate, 20 μL of standardized inoculum was used to inoculate wells of different 96-well plates, containing 180 μL of three biofilm media: (1) 1/2 tryptic soy broth (TSB), (2) M63 media (ammonium sulfate, potassium monophosphate, and ferrous sulfate), and (3) brain heart infusion (BHI) broth. Each isolate was inoculated into six replicate wells and grown at 28°C for 24 h; negative control wells contained 180 μL of broth and were not inoculated. After 24 h, liquid was removed and each plate was washed twice with 200 μL of PBS. Bacterial cells and extracellular biofilm matrix were fixed by adding 200 μL of methanol to each well and incubating at room temperature for 15 min. The methanol was removed, and plates were left to air dry for 30 min, prior to staining with 200 μL of 0.2% crystal violet for 5 min. Excess crystal violet was rinsed from each well using distilled water. Plates were left to air dry before addition of 160 μL of 33% glacial acetic acid. The absorbance of the resulting solution was measured at 570 nm using a spectrophotometer. The classification system developed by Stepanović et al. (2000) was used to describe biofilm formation by each strain. The cut-off OD (ODc) was calculated as three standard deviations above the average OD of the negative control wells; this value was compared to the mean OD for each strain. The biofilm categories were strong (mean OD > 4*ODc); moderate (4*ODc > mean OD > 2* ODc); weak (2* ODc > mean OD > ODc); or none (ODc > mean OD).
Biofilm Formation in the MBEC Assay®
Escherichia coli biofilm formation and the measurement of disinfectant susceptibility was performed using the MBEC Assay® biofilm inoculator 96-well plates (Innovotech Inc., Edmonton, AB, Canada) according to previously described method (Harrison et al., 2010). A standardized inoculum of 1.5 × 107 CFU/mL was added to biofilm growth media to a final volume of 150 μL per well. The MBEC plate lid containing 96 polystyrene pegs was placed into the 96-well microtitre plate base, sealed with parafilm and incubated at 37°C for 24 h with slight rocking on a tilting platform shaker. After incubation, peg lids were removed from the base plate and washed twice with sterile PBS to remove loosely attached cells. Biofilm formation on pegs was quantified by viable cell counts following disruption of the biofilm by sonication for 30 min with a bath sonicator (Branson #3510, Canada). Dislodged biofilm cells were serially diluted and grown on Mueller Hinton (MH) agar plates (Liu et al., 2018; Sivaranjani et al., 2021).
Determining the Minimum Inhibitory Concentration and Minimum Bactericidal Concentration for Disinfectants
The minimum inhibitory concentration (MIC) was determined by broth microdilution according to the Clinical and Laboratory Standards Institute (CLSI) standard for antimicrobial susceptibility testing (CLSI, 2020). Serial twofold dilutions of each disinfectant (Supplementary Table 2) were prepared in MH broth to a final volume of 90 μL per well. Ten μL aliquots of culture, representing 1.5 × 107 bacterial cells, were added to each well, the plates were covered and incubated at 37°C with slight rocking. After 24 h of growth, the OD at 600 nm of each well was measured by an xMarkTM Microplate Absorbance Spectrophotometer (Bio-Rad, Mississauga, ON, Canada). The MIC was recorded as the concentration in the wells where bacterial growth was visibly inhibited. To determine the Minimum Bactericidal Concentration (MBC), 100 μl of aliquots from MIC wells were sub-cultured on MH agar and incubated for 24 h. The lowest concentration that resulted in no bacterial growth on MH agar plates was determined as the MBC.
Disinfectant Susceptibility Testing
Planktonic Cells
Cells from overnight cultures were washed twice with sterile PBS and resuspended in 5 mL of fresh MH broth. The MH broth cultures were normalized to an optical density of 1.0 at 600 nm and the starting concentration of bacteria (CFU/mL) for each culture before disinfectant challenge was determined by viable cell counts. Serial twofold dilutions of each disinfectant were prepared in MH broth with a final volume of 90 μL in each well of a 96-well microtitre plate. Ten μL of inoculum was added to 90 μL of MH broth containing disinfectant and incubated at 37°C for 30 min. To test the viability of cells within each well, 10 μL aliquots were removed and sub-cultured into fresh MH broth and the remaining 90 μL was spread on a fresh MH agar plate. Both the sub-cultures were incubated at 37°C for 24 h. The lowest concentration resulting in no growth in liquid and on agar plates was recorded as the bactericidal concentration of disinfectant.
Biofilm Cells
After 24 h growth, biofilms formed on polystyrene pegs were transferred to a 96-well microtitre plate in which serial double dilutions of disinfectants were prepared in MH broth. Prior to disinfectant challenge, 6 control pegs were broken off from each plate, sonicated for 30 min in recovery media and the resuspended cells were serially diluted to determine the average starting number of biofilm cells for each strain. The remaining pegs were exposed to disinfectant for 30 min, rinsed twice in sterile PBS and cells were dislodged by bath sonication as described above. For each disinfectant concentration, cells from six replicate pegs were serially diluted and grown on MH agar plates to determine the number of viable cells remaining. The killing efficacy of disinfectants on biofilm cells was determined as the lowest concentration of disinfectant resulting in viable cell counts at or below the detection limit of 125 CFU per mL.
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility of all strains was determined via broth microdilution using the Gram-negative panel for the MicroScan system (Beckman Coulter, Mississauga, ON, Canada), testing the following 26 agents: amikacin, amoxicillin-clavulanic acid, ampicillin-sulbactam, ampicillin, aztreonam, cefazolin, cefepime, cefotaxime, cefoxitin, ceftazidime, ceftriaxone, cefuroxime, cephalothin, ciprofloxacin, doripenem, ertapenem, gentamicin, imipenem, levofloxacin, meropenem, nitrofurantoin, piperacillin-tazobactam, tetracycline, tigecycline, tobramycin and trimethoprim-sulfamethoxazole. Chloramphenicol, colistin, and nalidixic acid testing was performed using agar dilution (Wiegand et al., 2008). For each antibiotic, petri plates were prepared containing LB agar (lysogeny broth, 1.5% agar) plus antibiotic ranging from 0.5 μg/mL to 32 μg/mL. Isolates were streaked on blood agar (Oxoid #CM0055) and incubated for 18 h at 37°C. A 0.5 MacFarland standard was made for each isolate and isolates were spotted onto antibiotic plates using a 96-pin microplate replicator. Plates were incubated at 35°C for 24 h before reading MIC values. For quality control, E. coli ATCC 25922, Staphylococcus aureus ATCC 29213 and Pseudomonas aeruginosa ATCC 27853 were included as appropriate.
The results were interpreted according to the CLSI Guidelines (CLSI, 2020). MIC breakpoints were as follows: ampicillin (≥32 μg/mL), cefazolin (≥8 μg/mL), cephalothin (≥32 μg/mL), ceftazidime (≥16 μg/mL), ceftriaxone (≥4 μg/mL), cefuroxime (≥32 μg/mL), cefotaxime (≥4 μg/mL), cefepime (≥16 μg/mL), amoxicillin + clavulanic acid (≥32 μg/mL), ampicillin + sulbactam (≥32 μg/mL), cefoxitin (≥32 μg/mL), aztreonam (≥16 μg/mL), gentamicin (≥8 μg/mL), tobramycin (≥16 μg/mL), nalidixic acid (≥64 μg/mL), tetracycline (≥16 μg/mL), colistin (≥4 μg/mL), chloramphenicol (≥32 μg/mL), ciprofloxacin (≥1 μg/mL), doripenem (≥4 μg/mL), ertapenem (≥2 μg/mL), imipenem (≥4 μg/mL), meropenem (≥4 μg/mL), nitrofurantoin (≥128 μg/mL).
Statistics
Graphing of data and statistical analyses were performed using GraphPad Prism software v.8.0.2 (San Diego, CA, United States). Data from the disinfectant susceptibility assays were logarithmically transformed and analyzed using the Kruskal–Wallis test with post hoc analysis via Dunn’s multiple tests. Statistical differences in biofilm biomass, as measured by crystal violet staining, were determined using an ordinary-one way ANOVA test [F = 14.28, p > 0.0001]. Distribution of systemic isolates was analyzed using a chi-square test to determine whether the frequency of positive biofilm formation in each media was statistically significant [chi-square statistic: 15.74, df = 2, p > 0.0001].
Genomic DNA Extraction
Genomic DNA was extracted from each E. coli isolate using the GenElute Bacterial Genomic DNA Extraction Kit (Sigma-Aldrich, St. Louis, MO, United States), according to the manufacturer’s instructions. Purity of the DNA was assessed using a NanoDrop spectrophotometer (Thermo Fisher, Ottawa, ON, Canada) and quantified using a Qubit Fluorometer (Thermo Fisher Scientific, Ottawa, ON, Canada).
Nanopore Sequencing
All E. coli isolates were sequenced on a Nanopore MinION according to the protocol developed by Lohman et al. (2018), with the amount of ligase added to each step calculated based on the expected number of DNA molecules available for sequencing. Small fragments were removed from the genomic DNA eluate by adding 0.4x magnetic beads to each sample (NucleoMag NGS beads, Macherey-Nagel, Allentown, PA, United States). DNA fragments larger than 500 bp were bound, washed in 80% ethanol and eluted in 1 mM Tris buffer, pH 8.0. DNA was stored at –20°C for up to 1 week before sequencing.
Up to 200 fmol of purified and size-selected DNA was added to a Lobind Eppendorf tube. Distilled water was added to a final volume of 30 μL. Nicks and gaps in the DNA were repaired using FFPE DNA Repair Mix (#M6630; New England Biolabs, Whitby, ON, Canada) and polyA-tails were added to DNA fragments using the Ultra II End Repair/dA-Tailing Module (#M7645; New England Biolabs). Nanopore barcodes (#NBD-104; Oxford Nanopore Technologies, Oxford, United Kingdom) were ligated to DNA fragments using Blunt/TA Master Mix (New England Biolabs). Multiplexed samples were pooled into a single Eppendorf tube and the reaction was adjusted to 1 M NaCl. DNA repair enzymes and excess DNA barcodes were removed using a 0.2X magnetic bead clean-up. Beads were washed twice with 80% EtOH and the DNA was eluted in 10 mM Tris, pH 8.0. Sequencing adapters (AMII; Oxford Nanopore Technologies EXP-NBD104) were ligated to the ends of DNA fragments using Quick T4 DNA ligase (New England Biolabs) before a final bead clean-up (1:1 ratio of beads to sample) was performed to remove T4 DNA ligase and excess sequencing adapters from the final purified DNA library. Beads were washed by incubating two times in long chain fragment buffer and resuspending by flicking. DNA was eluted in 1mM Tris Buffer, pH 8.0. The final library was prepared and loaded onto the sequencer as described in the SQK-LSK109 protocol. Sequencing was performed using R.9.4.1 MinION flow cells. Sequencing runs were terminated after approximately 24 h.
Base-calling and de-barcoding of Nanopore reads was performed using MinKNOW (version 2.0) and Guppy (version 2.1.1) software (Oxford Nanopore Technologies). Read quality and lengths for each sequencing run were determined and visualized using the NanoPack software (version 1.35.4) developed by De Coster et al. (2018). Additional read trimming was performed using Porechop to remove any barcodes missed by Guppy (Wick, 2017). Reads were filtered for quality using Nanofilt to ensure that only reads with a Q score of 10 or greater were included in draft assemblies (–q 10) (De Coster et al., 2018).
Rough draft genomes were assembled using Unicycler (version 0.4.8) (Wick et al., 2017). A summary of the characteristics of the 96 E. coli strains selected for draft genome assembly, as well as sequencing information can be found in Supplementary Table 1. Each isolate tested was considered distinct due to having a unique combination of biofilm and AMR profiles.
Phylogroup Determination, Plasmid Identification, and Antimicrobial Resistance Gene Localization
The phylogroup of each E. coli strain was determined using the Clermontyping tool2, a web-based platform that predicts phylogroups based on an in silico quadruplex PCR to screen the whole genome FASTA sequence of an isolate for the arpA, chuA, yjaA, TspE4.C2 and TrpAgpC genes (Beghain et al., 2018).
We screened E. coli draft assemblies for plasmid sequences using PlasFlow, a program that uses a neural network to differentiate plasmid from chromosome sequences and has a higher accuracy than BLAST-based programs such as Plasmid Finder (Krawczyk et al., 2018). The draft chromosome and plasmid sequences were screened for antimicrobial resistance genes using the ResFinder database (ResFinder 4.0) from the Centre for Disease Epidemiology (Bortolaia et al., 2020). Genes were identified and used to generate presence and absence data for all 94 E. coli isolates. In the end, 28 known AMR genes were identified as being present in at least one isolate, encoding for resistance to tetracycline (tetA, tetB), sulfonamides (sul1, sul2), trimethoprim (dfraS), aminoglycosides [aph(6)-Id, aph(3′′), aph(4)-I1a, aph (3′), aadA1, aac(2), aac(3)-IV, aac(3)-VI, aac(3)-2d], and beta-lactam antibiotics (blaTEM–18, blaTEM–176, blaTEM–105, blaCMY–2, blaCMY–138, blaCMY–14, blaCMY–149, blaCMY–15, blaCMY–16, blaCMY–155, blaCMY–122, blaCMY–4, blaOXA–1). The mdfA gene encodes a multi-drug efflux pump.
Results
Disease-Causing Escherichia coli Are More Drug-Resistant Than Cecal Escherichia coli
A total of 94 E. coli strains were isolated from birds with colibacillosis (n = 56; systemic E. coli) and from presumed healthy birds of the same flock (n = 38; cecal E. coli). These E. coli strains represented 12 outbreaks on Saskatchewan farms between 2019 and 2020. Antimicrobial susceptibility testing was performed on all 94 isolates against 27 different antimicrobials: the results are summarized in Table 1. Resistance to Tetracycline (29.8%), Ampicillin (24.6%), Gentamicin (23.4%), and Tobramycin (19.1%) was the most observed. In contrast, there was no resistance detected to colistin, chloramphenicol, or the carbapenems. 43 isolates were susceptible to all 27 tested antimicrobials, while 32 were resistant to three or more drugs, and nine isolates were resistant to drugs from six different classes (Supplementary Table 1). Overall, the systemic isolates were significantly more drug resistant than the cecal isolates; 71% of systemic E. coli were resistant to at least one drug compared to only 29% of cecal E. coli (Supplementary Table 1). Tetracycline resistance was common in both groups, but resistance to ampicillin, gentamycin, and tobramycin was 20–30% higher in systemic isolates (Table 1). Systemic isolates also had higher MIC values for first and third generation cephalosporins, compared to cecal isolates (Table 1). Resistance to cefoxitin was found exclusively in systemic isolates, while Cefepime resistance was exclusively identified in cecal isolates.
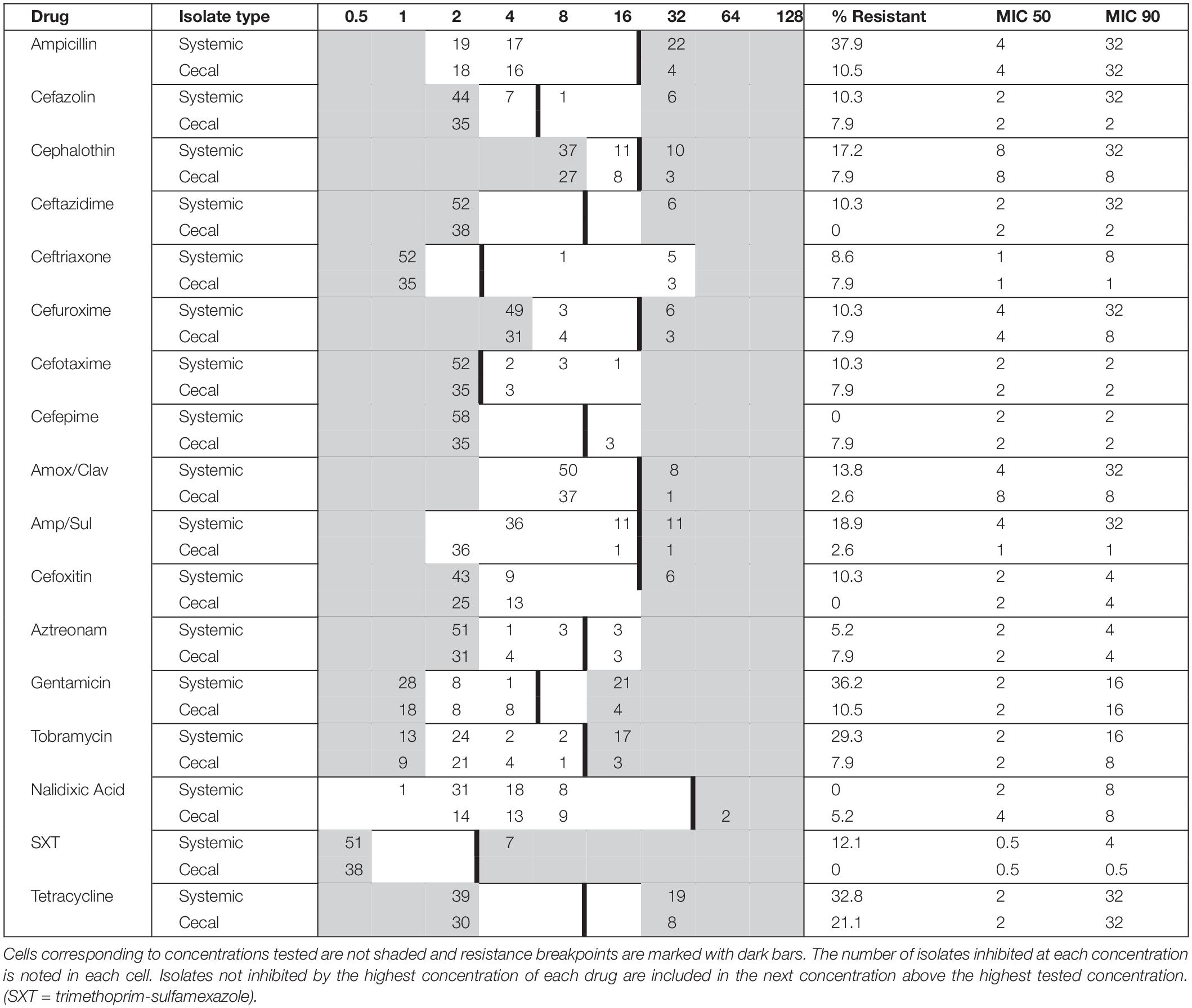
Table 1. Minimum inhibitory concentration (MIC) distribution for systemic (n = 56) and cecal (n = 38) E. coli isolates cultured from Saskatchewan broilers.
Systemic Isolates Form Greater Biofilms in Rich Media
Biofilm formation is associated with bacterial persistence, and we wanted to determine if this physiological state was correlated with antibiotic resistance. All E. coli isolates were screened for biofilm-forming ability in three different liquid media: nutrient rich BHI broth versus relatively nutrient poor ½ TSB and M63 media. Crystal violet staining was used to quantify and classify the level of biofilm. Overall, the systemic isolates generated significantly more biomass in nutrient rich BHI broth, as compared to the other two media (Figure 1A), whereas the cecal isolates did not show a clear preference for any one media type (Figure 1B).
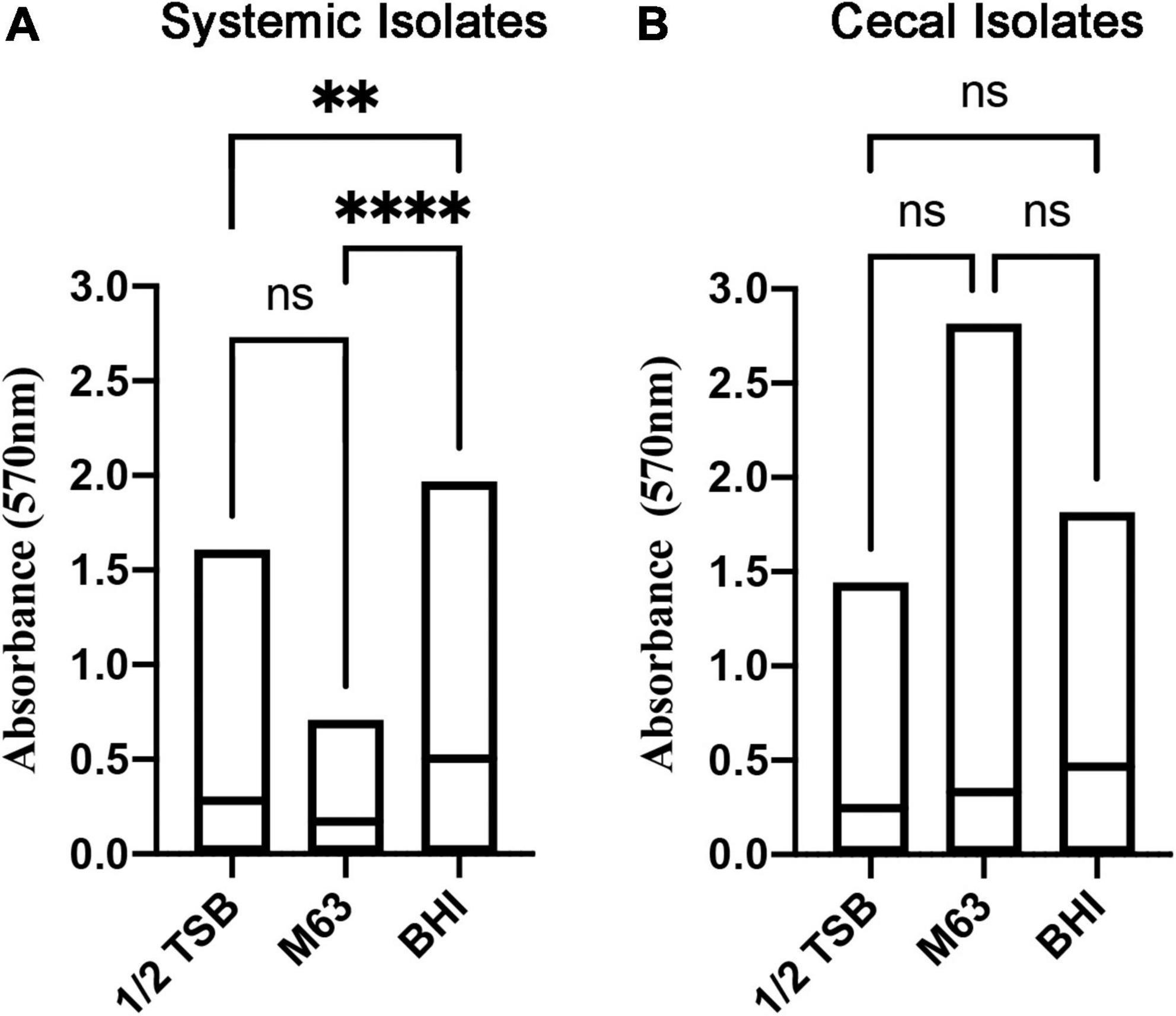
Figure 1. Quantitation of biofilm biomass for systemic and cecal E. coli isolates grown in three different growth media. Bars represent the mean absorbance of dissolved crystal violet that was used to stain biofilm cells and extracellular matrix from 56 systemic E. coli (A) or 38 cecal E. coli (B). Statistical significance between different growth media was tested by one-way ANOVA (****p < 0.0001; **p < 0.01; ns, p > 0.05).
To classify the level of biofilm formation for individual isolates, we used the formula described by Stepanović et al. (2000) (see section “Materials and Methods”). Isolates were categorized as positive if they formed strong or moderate biofilms, while isolates that produced none or weak biofilms were categorized as negative. 72% of systemic isolates were positive for biofilms in BHI broth, as compared to only 55% of cecal isolates (Figure 2). In contrast, 53% of cecal isolates were biofilm positive in M63 media, as compared to only 23% of systemic isolates (Figure 2). In general, systemic isolates were significantly worse at forming biofilms in nutrient poor media, with 28% positive in ½ TSB and 23% positive in M63 media (X2, p < 0.005) (Figure 2). In contrast, a roughly equal proportion of cecal isolates formed biofilms in M63 (53%) as BHI broth (55%), suggesting that these strains are equally capable of forming biofilms in nutrient-poor and nutrient-rich conditions (Figure 2). Approximately 30% of cecal isolates formed no biofilm in each media tested (Figure 2).
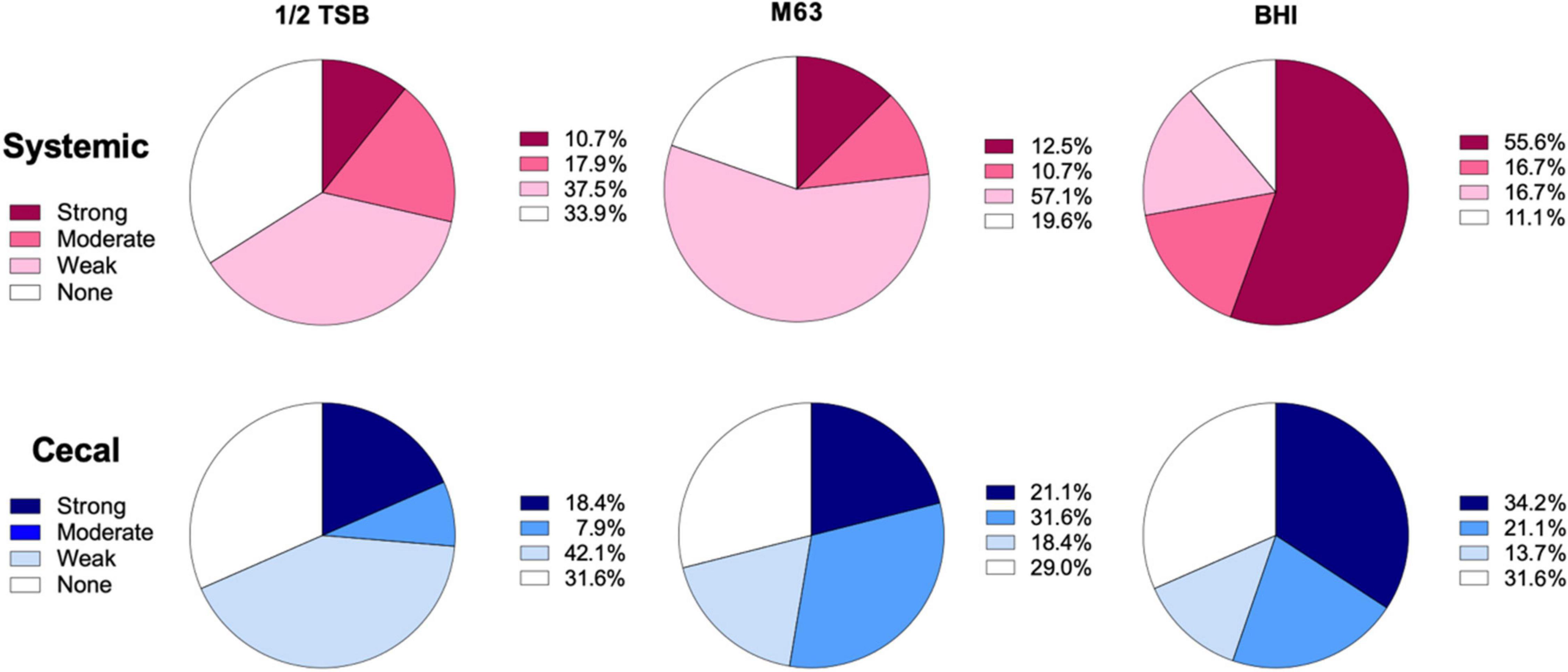
Figure 2. Classification of biofilm formation by systemic and cecal E. coli isolates grown in three different growth media. Biofilm formation in ½ TSB, M63, and BHI broth was classified using a formula where the cut-off OD (ODc) was set as three standard deviations above the average OD of the negative control wells: Strong = mean OD > 4*ODc; Moderate = 4*ODc > mean OD > 2* ODc; Weak = 2* ODc > mean OD > ODc; or None = ODc > mean OD. Systemic isolates (n = 56) are represented by shades of red/pink while cecal isolates (n = 38) are represented by shades of blue.
We also screened all E. coli isolates for curli and cellulose production since these polymers are central to biofilm formation in the host and the environment (White et al., 2011; MacKenzie et al., 2017; Chuppava et al., 2019). Sixty-two isolates produced both curli and cellulose, distributed as 73% (41/56) of systemic isolates and 55% (21/38) of cecal isolates (Supplementary Table 1). Twenty isolates produced only curli, consisting of 10 systemic and 10 cecal isolates. Nine isolates produced only cellulose, consisting of five systemic and four cecal isolates, and five cecal isolates were the only strains negative for both curli and cellulose. Taken together, the results indicated that systemic isolates had a slight increase in biofilm phenotypes as compared to cecal isolates.
Identifying the Strongest Biofilm Forming Escherichia coli Isolates and Testing Their Resistance to Commercial Disinfectants
We performed additional screening of 55 isolates that were classified as strong biofilm formers in BHI, M63 and ½ TSB media (Supplementary Table 1). We tested the ability of strains to adhere and form biofilms on polystyrene pegs in MBEC biofilm inoculator plates. Rather than measure biomass by staining, we focused on live cell counts, looking for strains with cell counts greater than 107 CFU, which we considered an appropriate cut off for good biofilm formation, based on previous MBEC assays (Liu et al., 2018). In total, 21 isolates formed biofilms with cell counts above 107 CFU (Figure 3; stars). consisting of 11 systemic and 10 cecal E. coli.
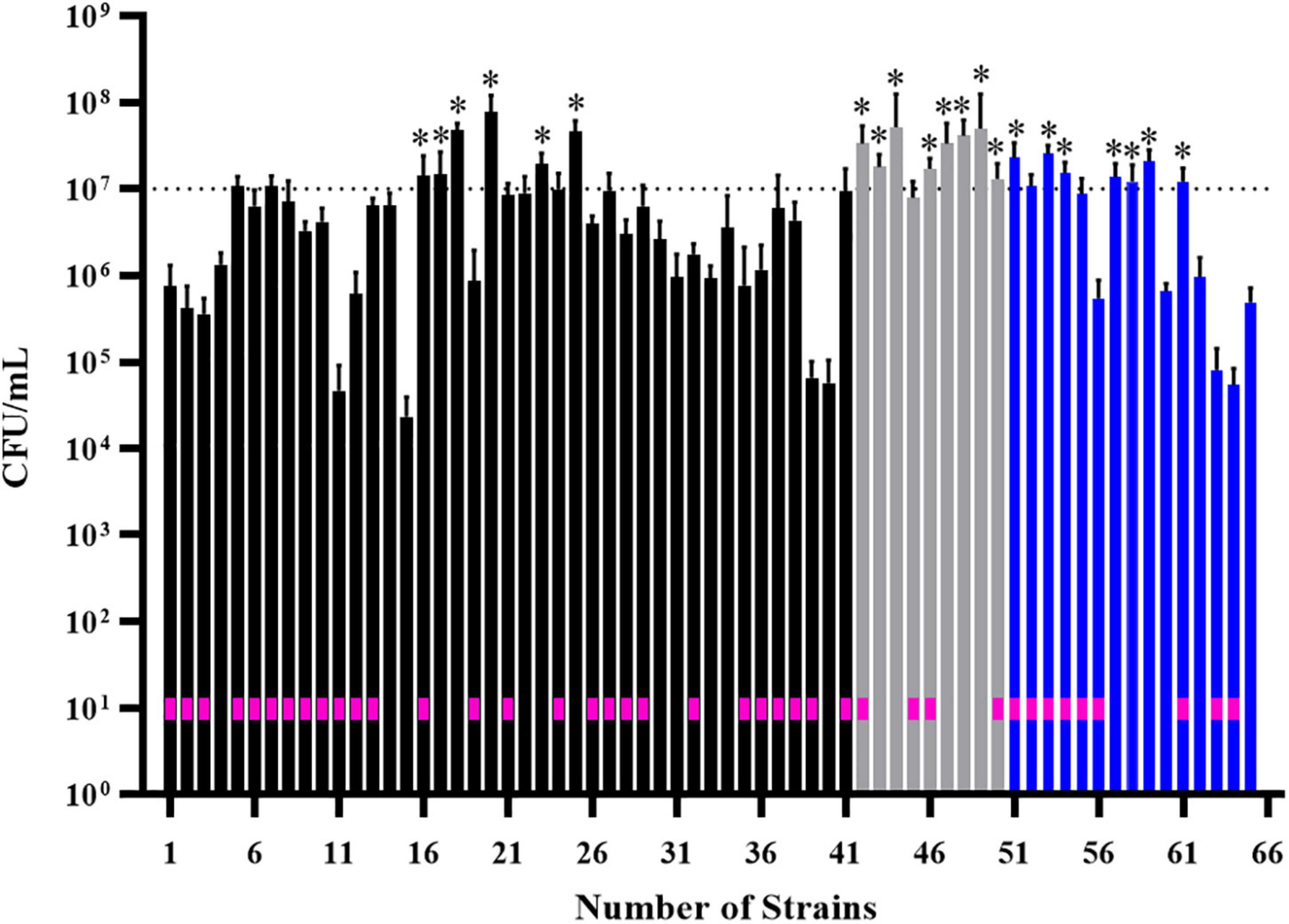
Figure 3. Screening of 55 biofilm-positive E. coli isolates for their ability to form biofilms on polystyrene pegs. After growth for 24 h at 37°C in BHI (black bars), M63 (gray bars) or ½ TSB (blue bars) media, cells were dislodged and enumerated from each peg (n = 6); histogram bars represent the average CFU/mL values and error bars represent the standard deviations. Some isolates formed strong biofilms in more than one media, which is why 65 bars are shown on the graph. Stars above the bars denote the isolates with cell densities greater than 107 CFU/mL; these were considered the strongest biofilm forming isolates (n = 21). The E. coli strains isolated from diseased birds are designated by a pink box within the corresponding histogram bar.
We further screened 12 of these isolates (7 systemic + 5 cecal) for resistance to four commercial disinfectants (Supplementary Table 2). Each disinfectant was able to kill planktonic cells of all E. coli isolates at low concentrations, ranging from 0.0016-0.0031% for Virocid, 0.125-0.25% for Virkon, 1.9–3.8 ppm for the quaternary ammonium disinfectant DDAC and 0.031–0.063% for H2O2 (Supplementary Table 3). The MBC values of all four disinfectants were either equal or one serial dilution higher than the corresponding MIC values. We deemed these results to be somewhat misleading because MIC and MBC determination requires exposure to test antimicrobials for 24 h, whereas most common disinfectants require contact times of only minutes to achieve bactericidal effects. Therefore, we repeated disinfectant screening on planktonic and biofilm cells of each of the 12 E. coli strains with a 30 min exposure. The starting cell numbers for each strain ranged between 106 and 108 CFU (Supplementary Table 4). Virkon and Virocid disinfectants exhibited effective bactericidal activity against both cell types of all 12 isolates even at low concentrations (Table 2 and Supplementary Figures 1, 2). The concentration of Virocid required to achieve full bactericidal effect was 1/2- to 1/32- fold lower than the suppliers’ recommended concentration, and for Virkon it was reduced approximately by 1/2- to 1/8- fold (Table 2 and Supplementary Table 2). DDAC was effective at killing planktonic cells of the 12 E. coli isolates with bactericidal concentrations ranging from 4.7 to 9.4 ppm (Table 2). However, biofilm cells of all 12 isolates had 2- to 64-fold increased resistance to DDAC, and one of the systemic isolates was not killed at the manufacturer recommended concentration of 300 ppm (Table 2 and Supplementary Figure 3). Higher concentrations of DDAC were required to eradicate the biofilms of three systemic isolates as compared to the five cecal isolates (Table 2). The results with H2O2 were similar to DDAC, as planktonic cells of all tested isolates were susceptible to low concentrations, but biofilms cells had 4- to 64-fold increased resistance (Table 2 and Supplementary Figure 4). Again, 4 systemic isolates were more resistant and required 4% H2O2 for the complete eradication of biofilms, whereas all five cecal isolates were susceptible to concentrations of 2% or less (Table 2 and Supplementary Figure 4). To determine if these trends based on isolate source were consistent, we would need to analyze more E. coli strains.
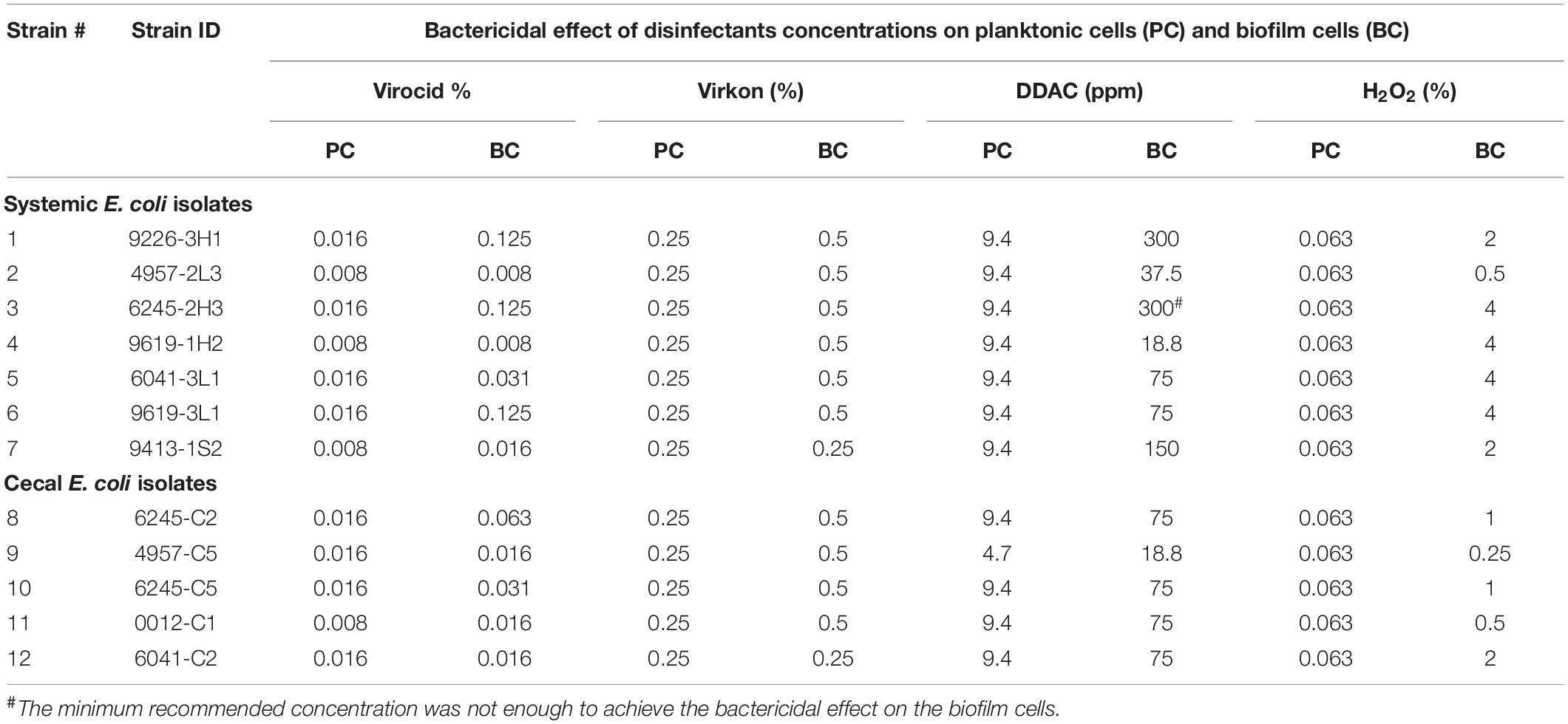
Table 2. Bactericidal effect of disinfectants on planktonic and biofilm cells within 30 min of contact time.
Trends in Antimicrobial Resistance by Outbreak
As a final comparison of the E. coli strains analyzed, we divided each group of isolates from outbreak to outbreak. This perhaps has more biological relevance than comparing the large groups of systemic and cecal strains to each other, since in almost all cases outbreaks occurred on different poultry farms. We identified 20 unique antibiotic resistance profiles across 12 Saskatchewan colibacillosis outbreaks (Supplementary Table 1). The most common resistance pattern was ampicillin + gentamicin + tobramycin, which was identified in isolates from 7 of 12 outbreaks. In all but three outbreaks, we identified systemic E. coli isolates that were resistant to at least four drugs. While the cecal isolates were generally susceptible, we identified at least one drug resistant cecal isolate in half of the outbreaks. In two outbreaks, cecal isolates were identified that were resistant to more than four drugs. This comparison again revealed an overall trend for systemic E. coli isolates to be more antibiotic resistant than cecal isolates, but there was a lot of diversity and no absolute patterns.
Draft Genome Sequencing and Phylogeny of Systemic and Cecal Escherichia coli Isolates
For each outbreak, disease-causing E. coli were isolated from up to three diseased birds and cecal E. coli were isolated from up to three healthy birds from the same flocks. Strain profiles based on the combination of curli, cellulose and AMR phenotype screening, indicated that the 94 isolates did not consist of groups of identical clones (Supplementary Table 1).
We determined the phylogroup of each E. coli strain using the Clermontyping tool (see footnote 2). Systemic isolates were primarily distributed into phylogroups G (32.8%), A (20.7%), and D (18.9%) (Supplementary Table 1). This was consistent with extraintestinal pathogenic E. coli, with phylogroups G and D known to contain many pathogenic E. coli strains (Clermont et al., 2000, 2019). In contrast, the majority of Saskatchewan cecal strains were distributed in phylogroups A (60.5%) and B1 (15.8%) (Supplementary Table 1), which are phylogroups known to typically contain commensal and environmental E. coli isolates (Duriez et al., 2001). The wide genetic diversity identified in the pool of systemic isolates suggests that transmissible elements, such as plasmids, may play a role in infection (Sola-Gines et al., 2015).
Known Antimicrobial Resistance Genes Appear to Be Concentrated on Plasmids of Systemic and Cecal Escherichia coli
The E. coli Nanopore draft assemblies were analyzed for plasmid sequences using PlasFlow (Krawczyk et al., 2018). We found that the 56 systemic isolates possessed an average of four plasmids (range from 0 to 9), while 38 cecal isolates possessed an average of 1 plasmid (range from 0 to 4) (Supplementary Figure 5). Next, we screened draft chromosome and plasmid sequences for antimicrobial resistance genes to see if we could identify genes unique to either group of isolates. We used the publicly available ResFinder 4.0 database from the Centre for Disease Epidemiology (Bortolaia et al., 2020). Due of the high error rates of Nanopore sequencing, and a lack of any polishing or consensus step in the Unicycler assembly program, we did not search for AMR acquired via point mutations.
The chromosomal and plasmid DNA sequences for each isolate were screened for the presence of 28 known AMR genes. We identified few chromosomal antimicrobial resistance genes in our E. coli population, except for mdfA, an outer membrane transporter that confers resistance to quaternary ammonium compounds, the active ingredient in a common commercial barn disinfectant. A small number of systemic isolates had chromosomal TetA genes, and four cecal isolates had chromosomal aminoglycoside or sulfonamide genes (Figures 4A,B). The remaining AMR genes identified in both groups were present on plasmids (Figures 4C,D). Over 25% of systemic isolates possessed plasmid-encoded tetracycline [tet(A)], beta-lactam (blaTEM-1B), and aminoglycoside [aac(3)-2d, aph(3”)] resistance genes (Figure 4C). Plasmid-mediated aminoglycoside resistance [aph(6)-Id, aac(3)-IV] was the most common type found in cecal isolates, at 9% abundance (Figure 4D). Almost half (41%) of cecal isolates lacked any plasmid-encoded AMR genes, compared to only 10% of systemic isolates.
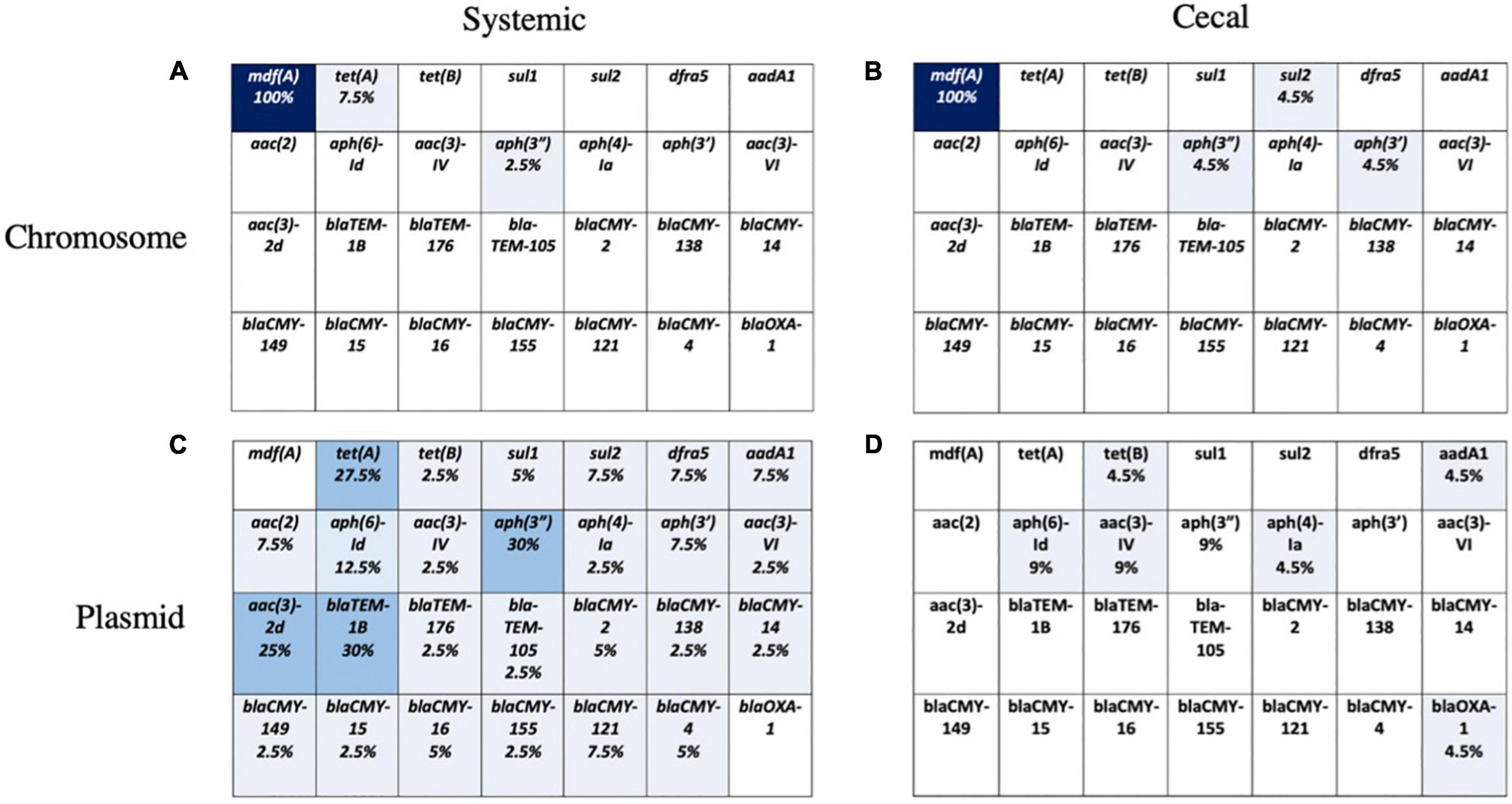
Figure 4. Presence of 28 known AMR genes on chromosome and plasmid for systemic and cecal E. coli strains. Percent abundance of AMR genes on the chromosome of systemic E. coli (A) and cecal E. coli (B) or the plasmids of systemic E. coli (C) and cecal E. coli (D). Light blue represents a proportion greater than 10%, medium blue = 10–30%, and dark blue represents greater than 30%.
Discussion
In this study, we analyzed E. coli isolates from colibacillosis outbreaks on Saskatchewan broiler farms. The comparison between 56 systemic or disease-causing E. coli strains and 38 cecal isolates from presumed healthy birds in the same flocks revealed an overall trend where the systemic isolates had a greater degree of AMR and were more likely to form biofilms than cecal isolates. This trend was not absolute and there were exceptions depending on the outbreak. The patterns of AMR and biofilm formation tended to be unique within each outbreak and between systemic and cecal isolates within each outbreak. Overall, there was a lot of phenotypic diversity, as reported by others (Schouler et al., 2012; Clermont et al., 2013; Riley, 2014; Cordoni et al., 2016). The differences observed between the systemic and cecal E. coli isolates indicated that they likely represent distinct groups of E. coli within the broiler farm environment. If the cecal E. coli were the source of disease, we would have expected to identify conserved AMR patterns within each outbreak, but we did not. Draft genome sequencing indicated that there were differences in plasmid content between the two groups of strains analyzed. Understanding the biological reasons that systemic and cecal E. coli are different is difficult without knowing the exact pressures that these groups of E. coli are subject to (Reisner et al., 2006; Brzuszkiewicz et al., 2009; Coombes, 2009; Azevedo et al., 2016; O’Boyle and Roe, 2021).
One of the goals for performing phenotypic analysis was to determine if there was a correlation between the degree of AMR and the biofilm-forming ability of individual E. coli strains, regardless of source. Despite the trend for systemic isolates having higher levels of antibiotic resistance and slightly more biofilm formation, there was no statistically significant relationship between AMR status and biofilm production in either the systemic or cecal group of E. coli strains analyzed. A roughly equal proportion of systemic and cecal isolates formed strong biofilms despite systemic isolates being more drug resistant. Nine of 11 systemic isolates that formed the strongest biofilms in 96-well plates and on polystyrene pegs were multi-drug resistant; the remaining two systemic isolates and 10 cecal isolates that formed strong biofilms were susceptible to all tested antimicrobials. This indicated that there was no direct correlation between the two phenotypes. Curli and cellulose production were both prevalent, even though there were a group of isolates (cecal) (roughly 30%) that did not form biofilms in any media.
Despite biofilm formation not being correlated with increased AMR, it is well established that this growth state is important for other aspects of persistence and survival (Harms et al., 2016; Jung et al., 2019; Yan and Bassler, 2019). The biofilm environment offers bacteria a source of nutrients as well as protection from external stressors such as desiccation, antimicrobials, and disinfectants. In our study, systemic isolates formed significantly more biofilms in BHI broth than in any other media, with 72% of isolates forming a positive biofilm. In contrast, less than 25% of systemic strains formed a positive biofilm in relatively nutrient-poor media. The significance of the media type is not fully understood, but is a little surprising since previous studies have shown that APEC strains form negligible biofilms in most media (Skyberg et al., 2007; Nielsen et al., 2018). However, the strains characterized in those studies were obtained from geographically diverse collections of APEC strains isolated from birds with various manifestations of disease. To our knowledge, our study is the first to analyze presumed APEC strains isolated exclusively from birds with confirmed sepsis in a small geographic area. Perhaps the E. coli strains responsible for a majority of systemic colibacillosis cases in Saskatchewan outbreaks possess genetic characteristics that favor biofilm formation in nutrient-rich media (Mellata et al., 2010, 2012). In contrast, for cecal isolates, there was no significant preference for the type of biofilm media, with approximately 50% of cecal isolates producing a biofilm in two media: M63 and BHI broth. This is consistent with previous reports that E. coli isolates of fecal origin can form biofilms in both minimal and nutrient-rich media (Skyberg et al., 2007; Nielsen et al., 2018).
Cleaning with disinfectants is one the most practical intervention strategies to eliminate bacterial contamination in the poultry barn setting. Many disinfectants are used in the poultry industry, with active ingredients belonging to aldehydes (e.g., formaldehyde), halogens (e.g., sodium hypochlorite), surface active agents [e.g., quaternary ammonium compounds (QAC)], oxidizing agents (e.g., H2O2), phenols and alcohols (Sagripanti and Bonifacino, 2000). The continuous exposure of residual disinfectants at lower concentrations may increase bacterial tolerance (Wieland et al., 2017). Any increase in tolerance could increase the bacterial adaptive resistance to antibiotics and enhance their survival fitness against various environmental stresses (Morente et al., 2013). We used the Calgary biofilm device to evaluate the killing efficacy of four commonly used disinfectants on the adherent population of cells from 12 of the strongest biofilm forming isolates. Each of the selected disinfectants is formulated with single or multiple active ingredients. Virkon is a solution formulated with multiple active ingredients including peroxygens and surfactants. It kills bacteria by oxidizing sulfur bonds in proteins and enzymes, resulting in cell wall rupture. Virocid is also a broad-spectrum disinfectant, formulated with synergistic blend of two different QAC (i.e., single chain and twin chain quaternary ammonia) along with glutaraldehyde and isopropanol. QACs interact with negatively charged cell membranes to elicit bactericidal action, while glutaraldehyde synergistically interacts with functional thiol and amine groups of proteins (Gilbert and Moore, 2005; Masadeh et al., 2013). It is believed that the interaction of active components with matrix proteins may facilitate the effective penetration of biofilm and synergistically kills the embedded bacterial cells (Osland et al., 2020). Our results showed that Virocid and Virkon had excellent killing at low concentrations against the planktonic and biofilm cells of all test strains, presumably due to the synergistic blend of multiple ingredients with non-specific bactericidal modes of action. In contrast, DDAC and H2O2 showed a large gap in killing where up to 64-fold higher concentrations were necessary to eradicate biofilm cells. The germicidal action of DDAC depends on numerous factors including the length of the N-alkyl chain, their combination with other active ingredients, pH, concentration used and the different target pathogens (Gilbert and Moore, 2005; He et al., 2013). H2O2 is a strong oxidizing agent that has been shown to damage bacterial DNA, proteins, and cellular membranes (Bell et al., 1997). It is well-known that bacteria residing in the inner matrix of a biofilm are protected from bactericidal concentrations of chemical agents, providing prime opportunities to develop resistance. Several studies have implicated that the sub-inhibitory concentrations of QACs may have a potential to select the emergence of AMR among pathogens, which could be raised from cross-resistance between QACs and other medically important antibiotics such as ampicillin, ceftazidime, cefotaxime, and ciprofloxacin (Nhung et al., 2015; Soumet et al., 2016; Nasr et al., 2018). Wieland and coworkers reported that the regular use of DDAC at low residual concentration would be able to select antibiotic resistance among E. coli and Enterococcus spp., which was confirmed by correlating the increased MIC values of DDAC with increased MIC values for several antibiotics, and high-level resistance was observed against aminoglycoside in enterococci (Wieland et al., 2017). These reports are in line with our results, that most of our isolates showed relatively common resistance to aminoglycoside and several other antibiotics. There was also a clear difference in susceptibility between systemic and cecal biofilms, as systemic isolates required higher concentration of DDAC and H2O2 for complete killing of biofilm cells. Our results suggest that the disinfectants with multiple active ingredients with different modes of action (i.e., Virocid and Virkon) are more effective against both systemic and cecal biofilms than the disinfectants with single mode of action (DDAC and H2O2).
Resistance to therapeutic antibiotics among E. coli isolated from poultry is of serious concern for both human and veterinary medicine, especially since it was just recognized that E. coli is the #1 cause of bacterial deaths due to resistant infections (Antimicrobial Resistance Collaborators, 2022). Recently, Canada has restricted the use of medically important antibiotics in food animals from being used as a growth promoter and requiring producers to obtain a veterinary prescription to treat infections in their flocks (Government of Canada, 2018). However, studies suggest that resistant bacteria can persist in the environment and spread AMR genes to commensal bacteria or between premises even with declining or no antimicrobial usage (Ge et al., 2005; Agunos et al., 2013). Reasons for this fitness advantage are likely to be complex but not limited to horizontal transmission of AMR genes between bacteria, improper cleaning and disinfection, biofilm formation and co-selection of resistance to certain antimicrobials. 71% of systemic isolates were resistant to between one and nine tested antimicrobial compounds. In contrast, only 29% of E. coli recovered from the cecal contents of uninfected birds were drug resistant or multi-drug resistant (MDR). Previous studies have demonstrated that resistance to specific drugs is correlated with the presence of ExPEC virulence genes (Johnson et al., 2006, 2012). Resistance to trimethoprim-sulfamexazole was exclusive to systemic isolates – this drug is significantly associated with the possession of the afa gene (Johnson et al., 2012). Afa is a fimbrial adhesin which is found in uropathogenic E. coli (UPEC) and is associated with recurrent infection (Bien et al., 2012). Most E. coli strains tested in our study demonstrated resistance to Ampicillin, Tetracycline, Gentamycin and Tobramycin, and displayed MDR as classified by resistance to as many as three different antimicrobial classes. These resistance trends are comparable to those of previous reports, with most clinical E. coli isolates recovered from either diseased or uninfected chickens exhibiting resistance to aminoglycosides, tetracyclines and sulfa drugs (Sáenz et al., 2003; Yang et al., 2004; Zhao et al., 2005; Varga et al., 2019).
Despite the presence of E. coli from diverse phylogroups, similar AMR profiles appeared in multiple outbreaks, suggesting that mobile genetic elements could be playing a role in resistance. To this end, draft genome sequencing revealed that numerous antimicrobial resistance genes were carried on plasmids. E. coli isolates with the same 9 drug resistance profiles were isolated from four outbreaks, while resistance to ampicillin, gentamicin, and tobramycin was present in over 50% of outbreaks. This may be indicative of transmission between farms, though further genetic characterization would be required to confirm this. Given the small geographic location of our study population and only two hatchling suppliers servicing central Saskatchewan, vertical transmission of APEC may also play a role in some of the similarities observed between outbreaks (Giovanardi et al., 2005). These systemic isolates may establish biofilms in the broiler barn environment and act as reservoirs for virulence and AMR genes in future outbreaks. E. coli can survive for years in the barn environment, particularly in dust (Schulz et al., 2016), perhaps supporting the prevailing hypothesis that colibacillosis infections occur via the inhalation of contaminated dust (Kabir, 2010). We also found that E. coli from Saskatchewan colibacillosis outbreaks had outbreak-specific media preferences for biofilm formation, suggesting that outbreak isolates may share properties which influence biofilm formation in a particular media type (Skyberg et al., 2007; Nielsen et al., 2018). In some cases, systemic and cecal isolates from a single outbreak exclusively formed biofilms in only a single media type. However, analysis of more outbreaks and a greater number of isolates is required to see if any trends are consistent. While biofilm preference was not correlated with any particular AMR profile, previous studies have identified biofilm formation as a key tool for the persistence of AMR genes in broiler barns (Zhai et al., 2020). The genetic components that promote biofilm formation in different media types may be linked to survival in the presence of different disinfecting agents (Zhai et al., 2020). A survey of the cleaning and disinfecting strategies used by Saskatchewan producers would help us to understand the role that biofilm formation plays in the persistence of outbreak-specific E. coli populations.
Proper biosecurity is critical for the management of colibacillosis on broiler farms. While biofilm formation and AMR profiles are not directly correlated, both likely play a role in the survival and pathogenicity of systemic isolates on farms. Outbreak tracking may provide a better information about the population of E. coli causing colibacillosis in a particular area. Bacterial persistence in the environment plays a key role in other recurrent bacterial infections, and tracking outbreaks may make it possible to identify E. coli reservoirs (Da Silva and De Martinis, 2013; Liao et al., 2021).
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
MS, MCM, JRD, JER, and APW: conceptualization, formal analysis, and writing—review and editing. MS, MCM, MKS, and LW: investigation. JER and APW: resources. MS, MCM, and APW: data curation and writing—original draft preparation. All authors: read and agreed to the published version of the manuscript.
Funding
This research was supported by the following grants: Natural Sciences and Engineering Research Council (NSERC) Collaborative Research and Development (Grant #543702-19), Saskatchewan Agriculture Development Fund (#20190173), and Saskatchewan Chicken Industry Development Fund to APW, JER, and JRD. MCM was a recipient of a COMGRAD scholarship from the University of Saskatchewan. VIDO receives operational funding from the Government of Saskatchewan through Innovation Saskatchewan and the Ministry of Agriculture and from the Canada Foundation for Innovation through the Major Science Initiatives for its CL3 facility. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank Tyra Dickson, Tennille Knezacek, and Nikki Storbakken, who form the Poultry Extension Group at the University of Saskatchewan, for project planning, collecting bird and tissue samples, and many helpful discussions; Neil Rawlyk at VIDO for general technical assistance; the Chicken Farmers of Saskatchewan and Saskatchewan poultry producers for supporting our colibacillosis project. Published as VIDO manuscript series no. 965.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.841516/full#supplementary-material
Footnotes
References
Agunos, A., Leger, D., Avery, B. P., Parmley, E. J., Deckert, A., Carson, C. A., et al. (2013). Ciprofloxacin-resistant Campylobacter spp. in retail chicken, Western Canada. Emerg. Infect. Dis. 19, 1121–1124. doi: 10.3201/eid1907.111417
Antimicrobial Resistance Collaborators (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
Azevedo, M., Sousa, A., Moura de Sousa, J., Thompson, J. A., Proenca, J. T., and Gordo, I. (2016). Trade-Offs of Escherichia coli Adaptation to an Intracellular Lifestyle in Macrophages. PLoS One 11:e0146123. doi: 10.1371/journal.pone.0146123
Beghain, J., Bridier-Nahimas, A., Nagard, H., Denamur, E., and Clermont, O. (2018). Clermontyping: an easy-to-use and accurate in silico method for Escherichia genus strain typing. Microb. Genom. 4:e000192. doi: 10.1099/mgen.0.000192
Bell, K. Y., Cutter, C. N., and Sumner, S. S. (1997). Reduction of foodborne micro-organisms on beef carcass tissue using acetic acid, sodium bicarbonate, and hydrogen peroxide spray washes. Food Microbiol. 14, 439–448. doi: 10.1006/fmic.1997.0108
Bien, J., Sokolova, O., and Bozko, P. (2012). Role of uropathogenic Escherichia coli virulence factors in development of urinary tract infection and kidney damage. Int. J. Nephrol. 2012:681473. doi: 10.1155/2012/681473
Bortolaia, V., Kaas, R. F., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Brzuszkiewicz, E., Gottschalk, G., Ron, E., Hacker, J., and Dobrindt, U. (2009). Adaptation of pathogenic E. coli to various niches: genome flexibility is the key. Microb. Pathogenomics 6, 110–125. doi: 10.1159/000235766
Chuppava, B., Keller, B., El-Wahab, A., Sürie, C., and Visscher, C. (2019). Resistance reservoirs and multi-drug resistance of commensal Escherichia coli from excreta and manure isolated in broiler houses with different flooring designs. Front. Microbiol. 10:2633. doi: 10.3389/fmicb.2019.02633
Clermont, O., Bonacorsi, S., and Bingen, E. (2000). Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66, 4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000
Clermont, O., Christenson, J. K., Denamur, E., and Gordon, D. M. (2013). The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5, 58–65. doi: 10.1111/1758-2229.12019
Clermont, O., Dixit, O. V. A., Vangchhia, B., Condamine, B., Dion, S., Bridier-Nahmias, A., et al. (2019). Characterization and rapid identification of phylogroup G in Escherichia coli, a lineage with high virulence and antibiotic resistance potential. Environ. Microbiol. 21, 3107–3117. doi: 10.1111/1462-2920.14713
CLSI (2020). Performance Standards for Antimicrobial Susceptibility Testing. In CLSI Document M100-30. Wayne, PA: CLSI.
Collingwood, C., Kemmett, K., Williams, N., and Wigley, P. (2014). Is the concept of avian pathogenic Escherichia coli as a single pathotype fundamentally flawed? Front. Vet. Sci. 1:5. doi: 10.3389/fvets.2014.00005
Collinson, S., Emödy, L., Müller, K., Trust, T., and Kay, W. (1991). Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J. Bacteriol. 173, 4773–4781. doi: 10.1128/jb.173.15.4773-4781.1991
Coombes, B. K. (2009). Type III secretion systems in symbiotic adaptation of pathogenic and non-pathogenic bacteria. Trends Microbiol. 17, 89–94. doi: 10.1016/j.tim.2008.11.006
Cordoni, G., Woodward, M. J., Wu, H., Alanazi, M., Wallis, T., and La Ragione, R. M. (2016). Comparative genomics of European avian pathogenic E. coli (APEC). BMC Genomics 17:960. doi: 10.1186/s12864-016-3289-7
Da Silva, E. P., and De Martinis, E. C. P. (2013). Current knowledge and perspectives on biofilm formation: the case of Listeria monocytogenes. Appl. Microbiol. Biotechnol. 97, 957–968. doi: 10.1007/s00253-012-4611-1
Davin-Regli, A. (2012). Cross-resistance between biocides and antimicrobials: an emerging question. Rev. Sci. Techn. 31, 89–104. doi: 10.20506/rst.31.1.2099
De Coster, W., D’Hert, S., Schultz, D. T., Cruts, M., and Van Broeckhoven, C. (2018). NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34, 2666–2669. doi: 10.1093/bioinformatics/bty149
Duriez, P., Clermont, O., Bonacorsi, S., Bingen, E., Chanventre, A., Elion, J., et al. (2001). Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiol. Soc. 147(Pt 6), 1671–1676. doi: 10.1099/00221287-147-6-1671
Dziva, F., and Stevens, M. P. (2008). Colibacillosis in poultry: unravelling the molecular basis of virulence of avian pathogenic Escherichia coli in their natural hosts. Avian Pathol. 37, 355–366. doi: 10.1080/03079450802216652
Foster, D. M., and Smith, G. W. (2009). Pathophysiology of diarrhea in calves. Vet. Clin. North Am. Food Anim. Pract. 25, 13–36. doi: 10.1016/j.cvfa.2008.10.013
Ge, B., McDermott, P. F., White, D. G., and Meng, J. (2005). Role of efflux pumps and topoisomerase mutations in fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob. Agents Chemother. 49, 3347–3354. doi: 10.1128/AAC.49.8.3347-3354.2005
Gilbert, P., and Moore, L. (2005). Cationic antiseptics: diversity of action under a common epithet. J. Appl. Microbiol. 99, 703–715. doi: 10.1111/j.1365-2672.2005.02664.x
Giovanardi, D., Campagnari, E., Ruffoni, L. S., Pesente, P., Ortali, G., and Furlattini, V. (2005). Avian pathogenic Escherichia coli transmission from broiler breeders to their progeny in an integrated poultry production chain. Avian Pathol. 34, 313–318. doi: 10.1080/03079450500179046
Government of Canada (2018). Responsible Use of Medically Important Antimicrobials in Animals. Ottawa, ON: Government of Canada.
Graveline, R., Garneau, P., Martin, C., Mourez, M., Hancock, M. A., Lavoie, R., et al. (2014). Leucine-responsive regulatory protein Lrp and PapI homologues influence phase variation of CS31A fimbriae. J. Bacteriol. 196, 2944–2953. doi: 10.1128/JB.01622-14
Guabiraba, R., and Schouler, C. (2015). Avian colibacillosis: still many black holes. FEMS Microbiol. Lett. 362:fnv118. doi: 10.1093/femsle/fnv118
Harms, A., Maisonneuve, E., and Gerdes, K. (2016). Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354:aaf4268. doi: 10.1126/science.aaf4268
Harrison, J. J., Stremick, C. A., Turner, R. J., Allan, N. D., Olson, M. E., and Ceri, H. (2010). Microtiter susceptibility testing of microbes growing on peg lids: a miniaturized biofilm model for high-throughput screening. Nat. Prot. 5, 1236–1254. doi: 10.1038/nprot.2010.71
He, J., Söderling, E., Vallittu, P. K., and Lassila, L. V. (2013). Investigation of double bond conversion, mechanical properties, and antibacterial activity of dental resins with different alkyl chain length quaternary ammonium methacrylate monomers (QAM). J. Biomater. Sci. Polym. Ed. 24, 565–573. doi: 10.1080/09205063.2012.699709
Hung, C., Zhou, Y., Pinkner, J. S., Dodson, K. W., Crowley, J. R., Heuser, J., et al. (2013). Escherichia coli biofilms have an organized and complex extracellular matrix structure. mBio 4:e00645-13. doi: 10.1128/mBio.00645-13
Jakobsen, L., Garneau, P., Kurbasic, A., Bruant, G., Stegger, M., Harel, J., et al. (2011). Microarray-based detection of extended virulence and antimicrobial resistance gene profiles in phylogroup B2 Escherichia coli of human, meat and animal origin. J. Med. Microbiol. 60, 1502–1511. doi: 10.1099/jmm.0.033993-0
Johnson, J. R., Porter, S. B., Zhanel, G., Kuskowski, M. A., and Denamur, E. (2012). Virulence of Escherichia coli clinical isolates in a murine sepsis model in relation to sequence type ST131 status, fluoroquinolone resistance, and virulence genotype. Infect. Immun. 80, 1554–1562. doi: 10.1128/IAI.06388-11
Johnson, T. J., Siek, K. E., Johnson, S. J., and Nolan, L. K. (2006). DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. J. Bacteriol. 188, 745–758. doi: 10.1128/JB.188.2.745-758.2006
Johnson, T. J., Wannemuehler, Y., Johnson, S. J., Stell, A. L., Doetkott, C., Johnson, J. R., et al. (2008). Comparison of extraintestinal pathogenic Escherichia coli strains from human and avian sources reveals a mixed subset representing potential zoonotic pathogens. Appl. Environ. Microbiol. 74, 7043–7050. doi: 10.1128/AEM.01395-08
Jung, S.-H., Ryu, C.-M., and Kim, J.-S. (2019). Bacterial persistence: fundamentals and clinical importance. J. Microbiol. 57, 829–835. doi: 10.1007/s12275-019-9218-0
Kabir, S. (2010). Avian colibacillosis and salmonellosis: a closer look at epidemiology, pathogenesis, diagnosis, control and public health concerns. Int. J. Environ. Res. Pub. Health 7, 89–114. doi: 10.3390/ijerph7010089
Kemmett, K., Humphrey, T., Rushton, S., Close, A., Wigley, P., and Williams, N. J. (2013). A longitudinal study simultaneously exploring the carriage of APEC virulence associated genes and the molecular epidemiology of faecal and systemic E. coli in commercial broiler chickens. PLoS One 8:e67749. doi: 10.1371/journal.pone.0067749
Kester, J. C., and Fortune, S. M. (2014). Persisters and beyond: mechanisms of phenotypic drug resistance and drug tolerance in bacteria. Crit. Rev. Biochem. Mol. Biol. 49, 91–101. doi: 10.3109/10409238.2013.869543
Krawczyk, P. S., Lipinski, L., and Dziembowski, A. (2018). PlasFlow: predicting plasmid sequences in metagenomic data using genome signatures. Nucleic Acid Res. 46:e35. doi: 10.1093/nar/gkx1321
Landoni, M. F., and Albarellos, G. (2015). The use of antimicrobial agents in broiler chickens. Vet. J. 205, 21–27. doi: 10.1016/j.tvjl.2015.04.016
Leverstein-van Hall, M., Dierikx, C., Cohen Stuart, J., Voets, G., Van Den Munckhof, M., van Essen-Zandbergen, A., et al. (2011). Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin. Microbiol. Infect. 17, 873–880. doi: 10.1111/j.1469-0691.2011.03497.x
Liao, J., Bergholz, P., and Wiedmann, M. (2021). Adjacent terrestrial landscapes impact the biogeographical pattern of soil Escherichia coli strains in produce fields by modifying the importance of environmental selection and dispersal. Appl. Environ. Microbiol. 87, e02516–e02520. doi: 10.1128/AEM.02516-20
Liu, F., Hansra, S., Crockford, G., Köster, W., Allan, B. J., Blondeau, J. M., et al. (2018). Tetrasodium EDTA is effective at eradicating biofilms formed by clinically relevant microorganisms from patients’ central venous catheters. mSphere 3:e00525-18. doi: 10.1128/mSphere.00525-18
Lohman, N., Watson, M., and Loose, M. (2018). A Training Bootcamp Based on the Oxford Nanopore MinION. Available online at: http://porecamp.github.io/2015/handbook.html (accessed April, 2021).
Lugsomya, K., Yindee, J., Niyomtham, W., Tribuddharat, C., Tummaruk, P., Hampson, D. J., et al. (2018). Antimicrobial resistance in commensal Escherichia coli isolated from pigs and pork derived from farms either routinely using or not using in-feed antimicrobials. Microb. Drug Resist. 24, 1054–1066. doi: 10.1089/mdr.2018.0154
MacKenzie, K. D., Palmer, M. B., Köster, W. L., and White, A. P. (2017). Examining the link between biofilm formation and the ability of pathogenic Salmonella strains to colonize multiple host species. Front. Vet. Sci. 4:138. doi: 10.3389/fvets.2017.00138
Mageiros, L., Méric, G., Bayliss, S. C., Pensar, J., Pascoe, B., Mourkas, E., et al. (2021). Genome evolution and the emergence of pathogenicity in avian Escherichia coli. Nat. Commun. 12, 1–13.
Malande, O. O., Nuttall, J., Pillay, V., Bamford, C., and Eley, B. (2019). A ten-year review of ESBL and non-ESBL Escherichia coli bloodstream infections among children at a tertiary referral hospital in South Africa. PLoS One 14:e0222675. doi: 10.1371/journal.pone.0222675
Maron, D. F., Smith, T. J., and Nachman, K. E. (2013). Restrictions on antimicrobial use in food animal production: an International regulatory and economic survey. Global. Health. 9, 1–11. doi: 10.1186/1744-8603-9-48
Masadeh, M. M., Gharaibeh, S. F., Alzoubi, K. H., Al-Azzam, S. I., and Obeidat, W. M. (2013). Antimicrobial activity of common mouthwash solutions on multidrug-resistance bacterial biofilms. J. Clin. Med. Res. 5:389. doi: 10.4021/jocmr1535w
McMillan, E. A., Gupta, S. K., Williams, L. E., Jové, T., Hiott, L. M., Woodley, T. A., et al. (2019). Antimicrobial resistance genes, cassettes, and plasmids present in Salmonella enterica associated with United States food animals. Front. Microbiol. 10:832. doi: 10.3389/fmicb.2019.00832
Mellata, M. (2013). Human and avian extraintestinal pathogenic Escherichia coli: infections, zoonotic risks, and antibiotic resistance trends. Foodborne Pathog. Dis. 10, 916–932. doi: 10.1089/fpd.2013.1533
Mellata, M., Ameiss, K., Mo, H., and Curtiss, R. III (2010). Characterization of the contribution to virulence of three large plasmids of avian pathogenic Escherichia coliχ7122 (O78: K80: H9). Infect. Immun. 78, 1528–1541. doi: 10.1128/IAI.00981-09
Mellata, M., Maddux, J. T., Nam, T., Thomson, N., Hauser, H., Stevens, M. P., et al. (2012). New insights into the bacterial fitness-associated mechanisms revealed by the characterization of large plasmids of an avian pathogenic E. coli. PLoS One 7:e29481. doi: 10.1371/journal.pone.0029481
Merritt, J. H., Kadouri, D. E., and O’Toole, G. A. (2011). Growing and analyzing static biofilms. Curr. Prot. Microbiol. 22, 1–17. doi: 10.1002/9780471729259.mc01b01s00
Mora, A., Herrera, A., Mamani, R., López, C., Alonso, M. P., Blanco, J. E., et al. (2010). Recent emergence of clonal group O25b: K1: H4-B2-ST131 ibeA strains among Escherichia coli poultry isolates, including CTX-M-9-producing strains, and comparison with clinical human isolates. Appl. Environ. Microbiol. 76, 6991–6997. doi: 10.1128/AEM.01112-10
Morente, E. O., Fernández-Fuentes, M. A., Burgos, M. J. G., Abriouel, H., Pulido, R. P., and Galvez, A. (2013). Biocide tolerance in bacteria. Int. J. Food Microbiol. 162, 13–25. doi: 10.1016/j.ijfoodmicro.2012.12.028
Nagy, B. J., Balázs, B., Benmazouz, I., Gyüre, P., Kövér, L., Kaszab, E., et al. (2021). Comparison of extended-spectrum beta-lactamase-producing Escherichia coli Isolates from Rooks (Corvus frugilegus) and contemporary human-derived strains: a one health perspective. Front. Microbiol. 12:785411–785411. doi: 10.3389/fmicb.2021.785411
Nasr, A. M., Mostafa, M. S., Arnaout, H. H., and Elshimy, A. A. A. (2018). The effect of exposure to sub-inhibitory concentrations of hypochlorite and quaternary ammonium compounds on antimicrobial susceptibility of Pseudomonas aeruginosa. Am. J. Infect. Cont. 46, e57–e63. doi: 10.1016/j.ajic.2018.04.201
Nhung, N. T., Thuy, C. T., Trung, N. V., Campbell, J., Baker, S., Thwaites, G., et al. (2015). Induction of antimicrobial resistance in Escherichia coli and non-typhoidal Salmonella strains after adaptation to disinfectant commonly used on farms in Vietnam. Antibiotics. 4, 480–494. doi: 10.3390/antibiotics4040480
Nicolas-Chanoine, M. H., Bertrand, X., and Madec, J. Y. (2014). Escherichia coli ST131, an intriguing clonal group. Clin. Microbiol. Rev. 27, 543–574. doi: 10.1128/CMR.00125-13
Nielsen, D. W., Klimavicz, J. S., Cavender, T., Wannemuehler, Y., Barbieri, N. L., Nolan, L. K., et al. (2018). The impact of media, phylogenetic classification, and E. coli pathotypes on biofilm formation in extraintestinal and commensal E. coli from humans and animals. Front. Microbiol. 9:902. doi: 10.3389/fmicb.2018.00902
Nolan, L. K., Vaillancourt, J. P., Barbieri, N. L., and Logue, C. M. (2020). Colibacillosis in: Diseases of Poultry. Ames, IA: Wiley Online Library.
O’Boyle, N., and Roe, A. J. (2021). Heterogeneity in populations of enterohaemorrhagic Escherichia coli undergoing d-serine adaptation. Curr. Gen. 67, 221–224. doi: 10.1007/s00294-020-01130-7
Osland, A. M., Vestby, L. K., and Nesse, L. L. (2020). The effect of disinfectants on Quinolone Resistant E. coli (QREC) in biofilm. Microorganisms 8:1831. doi: 10.3390/microorganisms8111831
Overdevest, I., Willemsen, I., Rijnsburger, M., Eustace, A., Xu, L., Hawkey, P., et al. (2011). Extended-spectrum β-lactamase genes of Escherichia coli in chicken meat and humans, The Netherlands. Emerg. Infect. Dis. 17:1216. doi: 10.3201/eid1707.110209
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31:e00088-e17.
Reisner, A., Krogfelt, K. A., Klein, B. M., Zechner, E. L., and Molin, S. (2006). In vitro biofilm formation of commensal and pathogenic Escherichia coli strains: impact of environmental and genetic factors. J. Bacteriol. 188, 3572–3581. doi: 10.1128/JB.188.10.3572-3581.2006
Renzhammer, R., Loncaric, I., Roch, F. F., Pinior, B., Käsbohrer, A., Spergser, J., et al. (2020). Prevalence of virulence genes and antimicrobial resistances in E. coli associated with neonatal diarrhea, postweaning diarrhea, and edema disease in pigs from Austria. Antibiotics 9:208. doi: 10.3390/antibiotics9040208
Riley, L. (2014). Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin. Microbiol. Infect. 20, 380–390. doi: 10.1111/1469-0691.12646
Rodrigues, S. V., Laviniki, V., Borges, K. A., Furian, T. Q., Moraes, H. L., Nascimento, V. P., et al. (2019). Biofilm formation by avian pathogenic Escherichia coli is not related to in vivo pathogenicity. Curr. Microbiol. 76, 194–199. doi: 10.1007/s00284-018-1608-8
Sáenz, Y., Zarazaga, M., Briñas, L., Ruiz-Larrea, F., and Torres, C. (2003). Mutations in gyrA and parC genes in nalidixic acid-resistant Escherichia coli strains from food products, humans and animals. J. Antimicrob. Chemother. 51, 1001–1005. doi: 10.1093/jac/dkg168
Sagripanti, J.-L., and Bonifacino, A. (2000). Resistance of Pseudomonas aeruginosa to liquid disinfectants on contaminated surfaces before formation of biofilms. J. AOAC Int. 83, 1415–1422. doi: 10.1093/jaoac/83.6.1415
Sanchez-Vizuete, P., Orgaz, B., Aymerich, S., Le Coq, D., and Briandet, R. (2015). Pathogens protection against the action of disinfectants in multispecies biofilms. Front. Microbiol. 6:705. doi: 10.3389/fmicb.2015.00705
Sargeant, J. M., Bergevin, M. D., Churchill, K., Dawkins, K., Deb, B., Dunn, J., et al. (2019). The efficacy of antibiotics to control colibacillosis in broiler poultry: a systematic review. Anim. Health Res. Rev. 20, 263–273. doi: 10.1017/S1466252319000264
Schouler, C., Schaeffer, B., Brée, A., Mora, A., Dahbi, G., Biet, F., et al. (2012). Diagnostic strategy for identifying avian pathogenic Escherichia coli based on four patterns of virulence genes. J. Clin. Microbiol. 50, 1673–1678. doi: 10.1128/JCM.05057-11
Schulz, J., Ruddat, I., Hartung, J., Hamscher, G., Kemper, N., and Ewers, C. (2016). Antimicrobial-resistant Escherichia coli survived in dust samples for more than 20 years. Front. Microbiol. 7:866. doi: 10.3389/fmicb.2016.00866
Sivaranjani, M., Liu, F., and White, A. P. (2021). Synergistic activity of tetrasodium EDTA, ethanol and chlorhexidine hydrochloride against planktonic and biofilm cells of clinically relevant pathogens. J. Glob. Antimicrob. Resist. 24, 148–157. doi: 10.1016/j.jgar.2020.12.002
Skyberg, J., Siek, K., Doetkott, C., and Nolan, L. (2007). Biofilm formation by avian Escherichia coli in relation to media, source and phylogeny. J. Appl. Microbiol. 102, 548–554. doi: 10.1111/j.1365-2672.2006.03076.x
Sola-Gines, M., Cameron-Veas, K., Badiola, I., Dolz, R., Majo, N., Dahbi, G., et al. (2015). Diversity of multi-drug resistant avian pathogenic Escherichia coli (APEC) causing outbreaks of colibacillosis in broilers during 2012 in Spain. PLoS One 10:e0143191. doi: 10.1371/journal.pone.1043191
Solano, C., García, B., Valle, J., Berasain, C., Ghigo, J. M., Gamazo, C., et al. (2002). Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43, 793–808. doi: 10.1046/j.1365-2958.2002.02802.x
Soumet, C., Méheust, D., Pissavin, C., Le Grandois, P., Frémaux, B., Feurer, C., et al. (2016). Reduced susceptibilities to biocides and resistance to antibiotics in food-associated bacteria following exposure to quaternary ammonium compounds. J. Appl. Microbiol. 121, 1275–1281. doi: 10.1111/jam.13247
Stepanović, S., Vuković, D., Dakić, I., Savić, B., and Švabić-Vlahović, M. (2000). A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Met. 40, 175–179. doi: 10.1016/s0167-7012(00)00122-6
Thaden, J. T., Fowler, V. G., Sexton, D. J., and Anderson, D. J. (2016). Increasing incidence of extended-spectrum β-lactamase-producing Escherichia coli in community hospitals throughout the southeastern United States. Infect. Cont. Hosp. Epidemiol. 37, 49–54. doi: 10.1017/ice.2015.239
Tumbarello, M., Sanguinetti, M., Montuori, E., Trecarichi, E. M., Posteraro, B., Fiori, B., et al. (2007). Predictors of mortality in patients with bloodstream infections caused by extended-spectrum-β-lactamase-producing Enterobacteriaceae: importance of inadequate initial antimicrobial treatment. Antimicrob. Agents Chemother. 51, 1987–1994. doi: 10.1128/AAC.01509-06
van den Bogaard, A. E., and Stobberingh, E. E. (2000). Epidemiology of resistance to antibiotics: links between animals and humans. Int. J. Antimicrob. Agent 14, 327–335. doi: 10.1016/s0924-8579(00)00145-x
Varga, C., Guerin, M. T., Brash, M. L., Slavic, D., Boerlin, P., and Susta, L. (2019). Antimicrobial resistance in fecal Escherichia coli and Salmonella enterica isolates: a two-year prospective study of small poultry flocks in Ontario, Canada. BMC Vet. Res. 15, 1–10. doi: 10.1186/s12917-019-2187-z
White, A., Sibley, K., Sibley, C., Wasmuth, J., Schaefer, R., Surette, M., et al. (2011). Intergenic sequence comparison of Escherichia coli isolates reveals lifestyle adaptations but not host specificity. Appl. Environ. Microbiol. 77, 7620–7632. doi: 10.1128/AEM.05909-11
Wick, R. R. (2017). Porechop: Adapter Trimmer for Oxford Nanopore Reads. Available online at: https://github.com/rrwick/Porechop (accessed June, 2021).
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wiegand, I., Hilpert, K., and Hancock, R. E. (2008). Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Prot. 3, 163–175. doi: 10.1038/nprot.2007.521
Wieland, N., Boss, J., Lettmann, S., Fritz, B., Schwaiger, K., Bauer, J., et al. (2017). Susceptibility to disinfectants in antimicrobial-resistant and-susceptible isolates of Escherichia coli, Enterococcus faecalis and Enterococcus faecium from poultry–ESBL/AmpC-phenotype of E. coli is not associated with resistance to a quaternary ammonium compound, DDAC. J. Appl. Microbiol. 122, 1508–1517. doi: 10.1111/jam.13440
Wooley, R., Gibbs, P., Brown, T., and Maurer, J. (2000). Chicken embryo lethality assay for determining the virulence of avian Escherichia coli isolates. Avian Dis. 44, 318–324. doi: 10.2307/1592546
Xiong, W., Sun, Y., and Zeng, Z. (2018). Antimicrobial use and antimicrobial resistance in food animals. Environ. Sci. Pol. Res. 25, 18377–18384. doi: 10.1007/s11356-018-1852-2
Yan, J., and Bassler, B. L. (2019). Surviving as a community: antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe 26, 15–21. doi: 10.1016/j.chom.2019.06.002
Yang, H., Chen, S., White, D. G., Zhao, S., McDermott, P., Walker, R., et al. (2004). Characterization of multiple-antimicrobial-resistant Escherichia coli isolates from diseased chickens and swine in China. J. Clin. Microbiol. 42, 3483–3489. doi: 10.1128/JCM.42.8.3483-3489.2004
Zhai, R., Fu, B., Shi, X., Sun, C., Liu, Z., Wang, S., et al. (2020). Contaminated in-house environment contributes to the persistence and transmission of NDM-producing bacteria in a Chinese poultry farm. Environ. Int. 139:105715. doi: 10.1016/j.envint.2020.105715
Keywords: antibiotics, antimicrobial resistance (AMR), Avian pathogenic E. coli (APEC), biofilm, disinfectant, colibacillosis, poultry
Citation: Sivaranjani M, McCarthy MC, Sniatynski MK, Wu L, Dillon JR, Rubin JE and White AP (2022) Biofilm Formation and Antimicrobial Susceptibility of E. coli Associated With Colibacillosis Outbreaks in Broiler Chickens From Saskatchewan. Front. Microbiol. 13:841516. doi: 10.3389/fmicb.2022.841516
Received: 22 December 2021; Accepted: 19 April 2022;
Published: 17 June 2022.
Edited by:
Hans P. Steenackers, KU Leuven, BelgiumReviewed by:
Catherine M. Logue, University of Georgia, United StatesIsidro García Meniño, University of Santiago de Compostela, Spain
Copyright © 2022 Sivaranjani, McCarthy, Sniatynski, Wu, Dillon, Rubin and White. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aaron P. White, YWFyb24ud2hpdGVAdXNhc2suY2E=
†These authors have contributed equally to this work
 Murugesan Sivaranjani
Murugesan Sivaranjani Madeline C. McCarthy
Madeline C. McCarthy Michelle K. Sniatynski
Michelle K. Sniatynski Linzhi Wu
Linzhi Wu Jo-Anne R. Dillon
Jo-Anne R. Dillon Joseph E. Rubin
Joseph E. Rubin Aaron P. White
Aaron P. White