- 1Tianjin Academy of Agricultural Sciences, The Institute of Agriculture Resources and Environmental Sciences, Tianjin, China
- 2Tianjin Key Laboratory of Indoor Air Environmental Quality Control, School of Environmental Science and Engineering, Tianjin University, Tianjin, China
Antibiotic resistance is one of the most important environmental challenges. Microalgae has been considered as a promising green media for environmental purification. In this work, sulfadimethoxine (SDM) biodegradation potential of Chlorella sp. L38 and Phaeodactylum tricornutum MASCC-0025 is investigated. Experimental results indicated that the tested freshwater and marine microalgae strains presented stress response to SDM addition. For Chlorella sp. L38, it has a good adaptability to SDM condition via antioxidant enzyme secretion (SOD, MDA, and CAT up to 23.27 U/mg, 21.99 μmol/g, and 0.31 nmol/min/mg) with removal rate around 88%. P. tricornutum MASCC-0025 exhibited 100% removal of 0.5 mg/L SDM. With increasing salinity (adding a certain amount of NaCl) of cultivation media, the removal rate of SDM by microalgae increased. Although its adaptive process was slower than Chlorella sp. L38, the salinity advantage would facilitate enzyme accumulation. It indicated that microalgae could be used to remove SDM from freshwater and marine environment via suitable microalgae strain screening.
Introduction
Sulfonamides are one of the most frequently used antibiotics for therapeutic purposes and are also used as feed additives in certain intensive farming operations (Peiris et al., 2017). Due to the continuous consumption, sulfonamides, such as sulfadimethoxine (SDM), sulfamethoxazole (SMX), and sulfamethazine (SMZ), have been widely found in wastewater, freshwater, and groundwater (Mulla et al., 2018, 2021). In the last decades, antibiotics utilization in veterinary and human medicine was widespread (0.1–0.2 million tons per annum of antibiotics have been utilized all around the world), which increased the risk of environmental contamination (Homem and Santos, 2011; Kumar and Pal, 2018). For example, being the largest country of antibiotics consumption and production in the world, China consumed 92,700 tons of antibiotics in which 48% are consumed by humans and the remaining 52% are by animals (Zhang et al., 2015; Qiao et al., 2018).
Associated with extensive utilization, they pose potential hazards both environmentally and health wise since they are not easily biodegradable and can cause numerous ecological impacts (e.g., promoting the growth of antibiotic-resistant genes and antibiotic-resistant bacteria) (Shao et al., 2005; Zhou et al., 2009). Until now, the dominant antibiotics removal technologies include chemical oxidation, adsorption, membrane and biodegradation, etc. (Ahmed et al., 2015; Yu et al., 2016; Anjali and Shanthakumar, 2019; Bilal et al., 2019). The operation and maintenance cost of chemical oxidation technology is high. Adsorption method and membrane separation method do not realize the harmlessness of pollutants, and there is a lack of reasonable disposal technology for the separated antibiotics. The growth and metabolism of microorganisms will be affected by antibiotics, resulting in the low treatment efficiency of antibiotics by traditional sewage treatment technology (Ali et al., 2018; Sutherland and Ralph, 2019). In addition, there is also a risk of secondary pollution due to the substantial dosage requirements on sorbents, solvent, flocculants, and other agents (Li et al., 2019; Cheng et al., 2020; Zhang and Li, 2020).
As a green and cost-effective alternative of conventional antibiotics removal techniques, microalgae-based processes have attracted more and more attention in the recent years (Cheng et al., 2017; Leng et al., 2020). Compared with the conventional chemical and physical technologies, microalgae-based processes have presented the advantage of lower capital and operational costs, natural disinfection, and high efficiency (Craggs et al., 2012; Cheng et al., 2017). Meanwhile, application of microalgae for bioremediation purpose provides an opportunity to reuse the contaminants as nutrition via photosynthesis process, and produce potential value-added biomass (namely polysaccharides, protein, pigment, and lipid) (Sutherland et al., 2018; Song et al., 2020). From the existing results, some microalgae strains have the capacity to resist to antibiotics in the natural environment and can remove them by various mechanisms (such as biosorption, bioaccumulation, and intracellular/extracellular biodegradation) (Nagarajan et al., 2019; Song et al., 2019, 2020).
It should be noted that the antibiotics biodegradation mechanism would be different for different microalgae species. The objective of this work is to investigate the biodegradation of SDM by two different microalgae, derived from freshwater (Chlorella sp. L38) and marine (Phaeodactylum tricornutum MASCC-0025) environment. To intensify the biodegradation performance, algae cultivation condition was studied and optimized under different SDM initial concentrations. Meanwhile, the antibiotic removal rate is further intensified via increasing the salinity of cultivation medium.
Materials and Methods
Microalgae Species
Two microalgae (Chlorella sp. L38 and P. tricornutum MASCC-0025) were used as target strains, since existing literatures have indicated that Chlorella sp. has an excellent capacity on environmental purification. Meanwhile, marine environment is also an important contaminator via antibiotics. Therefore, P. tricornutum MASCC-0025 is also investigated for antibiotic removal. Chlorella sp. L38 and P. tricornutum MASCC-025 were obtained from the algae collection of Applied Microalgae Biology Laboratory of Ocean University of China and the Algae Collection of Institute of Oceanography, Chinese Academy of Sciences. SDM is selected as the representative antibiotic for all the experiments, and it was purchased from Aladdin Chemistry Co., Ltd. (Shanghai, China).
Cultivation Conditions
The cultivation media for Chlorella sp. L38 and P. tricornutum MASCC-0025 are BG11 and f/2, respectively. Different SDM concentrations (0.5, 1, and 3 mg/L) were added to the cultivation media. To investigate the influence of high salinity on SDM removal performance, different concentration of NaCl solution is added into freshwater microalgae cultivation media. Both SDM and NaCl were added into media after filtration (0.45 μm polytetrafluoroethylene/PTFE) for bacteria removal. The temperature and illumination conditions were 25 ± 1°C and 5,000 Lux under 24 h. Three parallel samples were set in each group to eliminate the interference of other factors.
Parameter Determination
The algal biomass variation can be evaluated via optical density (OD) value and specific growth rate determination, and the detailed determination and calculation procedures of Chlorella sp. L38 have been described in our previous work (Song et al., 2020). The specific growth rate (μ) was measured by fitting the dry cell weight (DCW, μg/L) to an exponential function using the equation proposed by Kabra et al. (2014). The relationship between OD450 and DCW of P. tricornutum (PT) MASCC-0025 was evaluated by the following equation:
In addition, pH of cultivation media was measured every 2 days. Chlorophyll (a and b) and carotenoids were detected every 4 days in the whole cultivation period. The total chlorophyll content was measured by Kurade et al. (2016). In addition, the superoxide dismutase (SOD), malondialdehyde (MDA), and catalase (CAT) enzyme contents were determined via corresponding methods. The potential degradation product of SDM is analyzed via LC-MS (e2695 system; Waters Co., Ltd., United States).
Statistical Analysis
One-way ANOVA was used for statistical analysis. Results were expressed as means ± SEM based on parallel experiments and were considered at 95% CIs.
Results and Discussion
Microalgae Growth
Microalgae growth performance (OD, specific growth rate, and pH) of Chlorella sp. L38 and P. tricornutum MASCC-0025 under different SDM concentrations is shown in Figure 1. From Figure 1A, it can be seen that when the concentration was lower than 3 mg/L, SDM presented a certain stimulating effect on the growth of Chlorella sp. L38. Especially, when the initial concentration of SDM was set at 0.5 mg/L, the growth rate of Chlorella sp. L38 was significantly higher than that of the control group (without SDM addition). It could also be observed that SDM had a significant inhibitory effect on the growth of Chlorella sp. L38 on the 6th day, and the inhibitory effect increased with SDM concentration. Associated with growth, from the 8th to the 14th days, Chlorella sp. L38 gradually adapted the cultivation condition with adding SDM, and the OD value presented an increased trend and specific growth rate also became higher (as shown in Figure 2A). According to Figures 1B, 2B, for different concentration gradients of SDM with and without adding NaCl solution, it can be seen that the growth rate of Chlorella sp. 38 increased with the increase of salt concentration. When the addition dosage of NaCl increased to a certain extent, the growth promotion effect on Chlorella sp. L38 would decrease with the continuous increase of salt concentration. It indicated that a certain amount of NaCl can promote Chlorella sp. L38 growth, and it could be also seen that the facilitation effect of the salt on Chlorella sp. L38 growth would vary with initial SDM concentration.
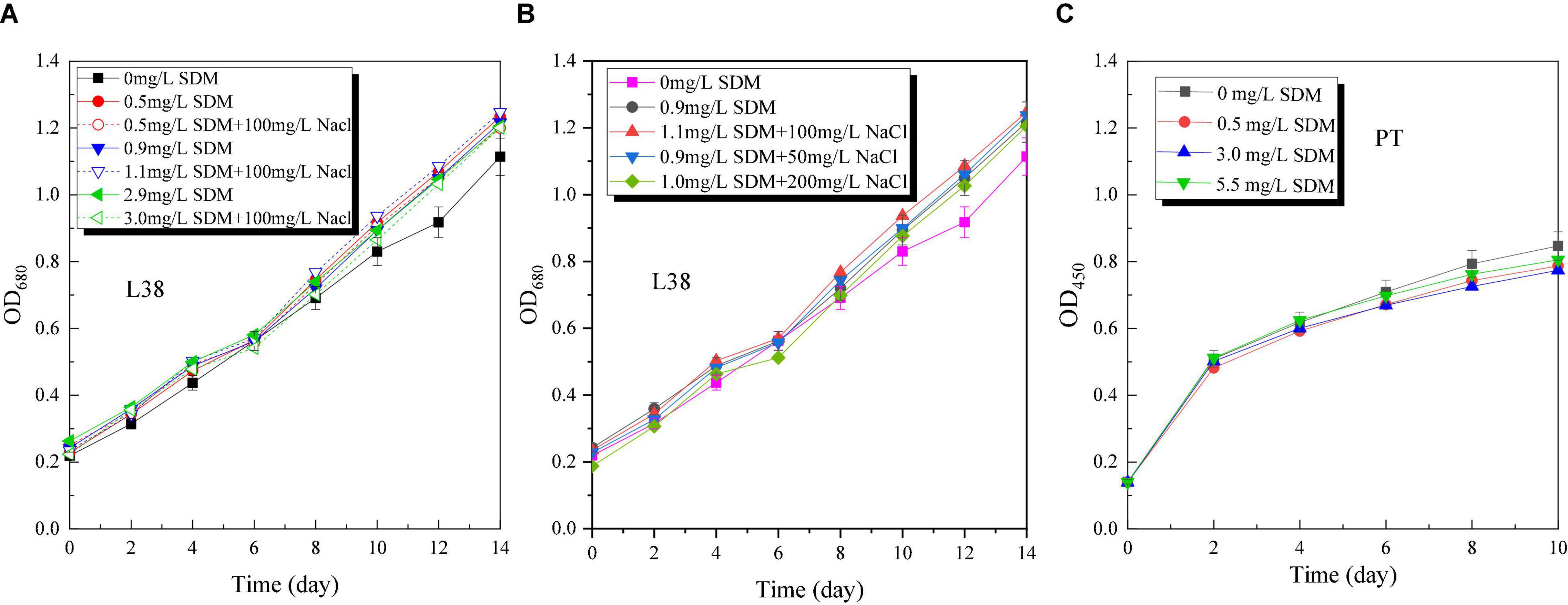
Figure 1. Optical density (OD) variation of Chlorella sp. L38 and Phaeodactylum tricornutum MASCC-0025 under different sulfadimethoxine (SDM) concentrations.
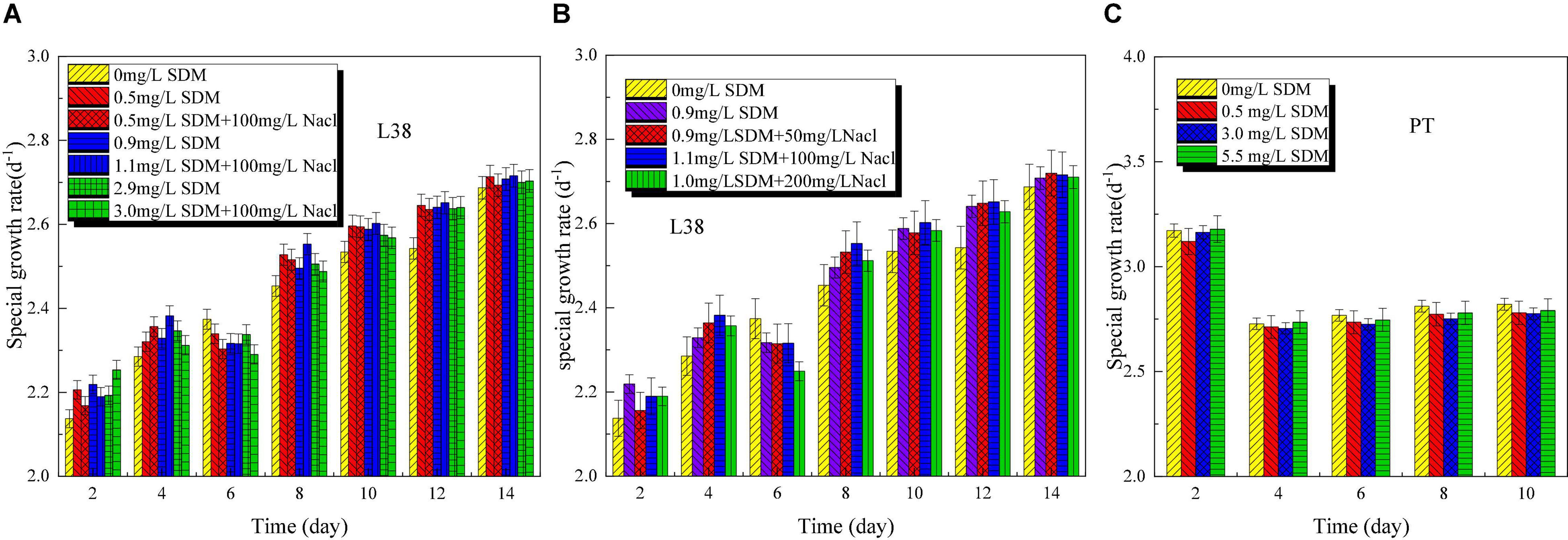
Figure 2. Specific growth rate variation of Chlorella sp. L38 and Phaeodactylum tricornutum MASCC-0025 under different sulfadimethoxine (SDM) concentrations.
For P. tricornutum MASCC-0025, the OD and specific growth rate variation are shown in Figures 1C, 2C. It could be found that the presence of SDM had a slight inhibitory effect on the growth of algae, and the inhibitory effect was the strongest at the SDM concentration of 3 mg/L. However, when the concentration of SDM increased to 5.5 mg/L, the inhibitory effect on microalgae slowed down. It could be observed that for the different microalgae species, the SDM effect was also different. Therefore, to select suitable microalgae strain for antibiotics degradation, more efforts should be paid on the investigation of the metabolism mechanism and genetic modification of microalgae.
The pH of Chlorella sp. L38 increased rapidly from about 7.5 to 11 in the initial 2 days of cultivation, and then varied slowly from 4 to 14 days (maintained at 10.5–11.0), as shown in Figure 3A. It also indicated that Chlorella sp. L38 has a good adaptive capacity to SDM. For P. tricornutum MASCC-0025, the pH value of all the groups varied around 9.5, and the control group was slightly higher than that of the other groups with adding SDM, as illustrated in Figure 3B. It might be due to the inhibitory effect of antibiotics on the growth of P. tricornutum MASCC-0025, leading to the growth state of the experimental group that was slightly slower than that of the control group. Overall, the pH variation trend was consistent with algae biomass accumulation.
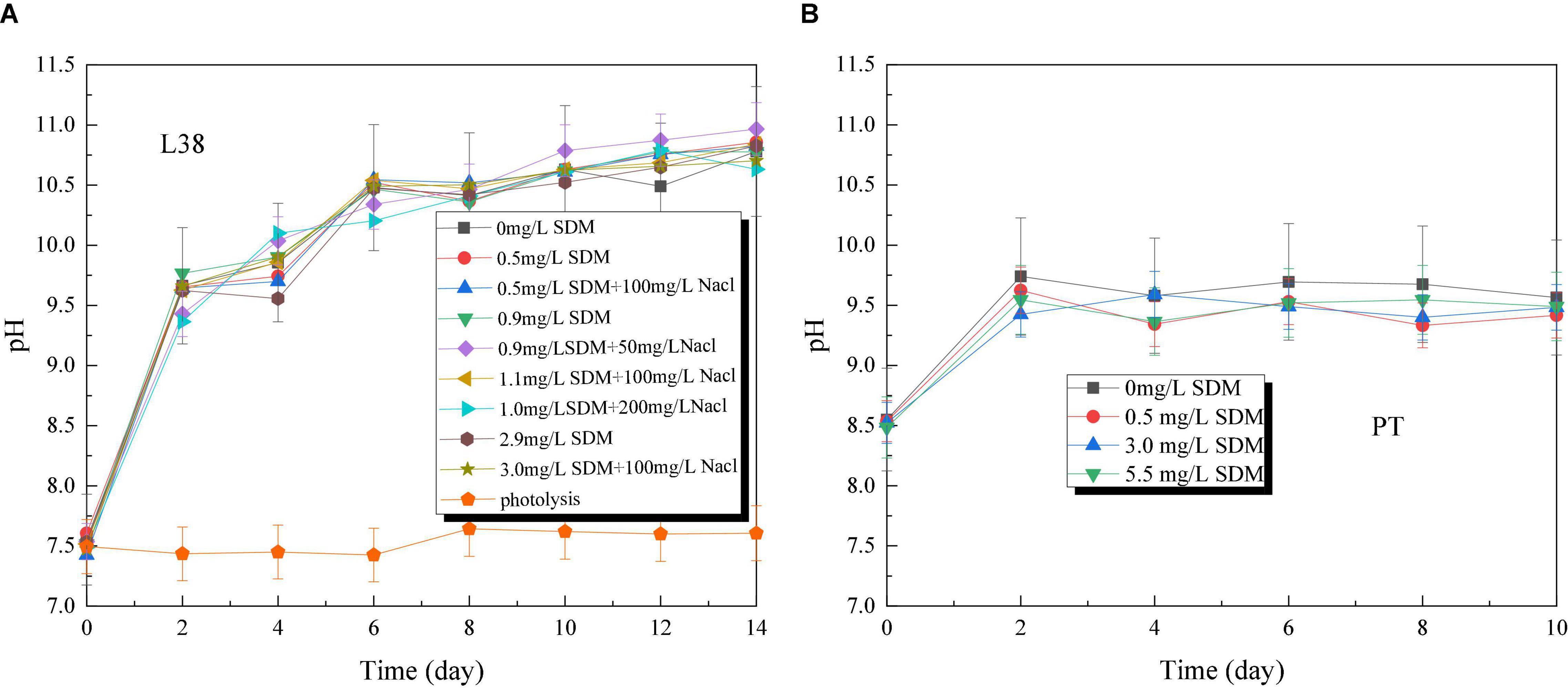
Figure 3. pH variation of Chlorella sp. L38 and Phaeodactylum tricornutum MASCC-0025 under different sulfadimethoxine (SDM) concentrations.
Antioxidant Enzyme Variation
Usually, reactive oxygen species (ROS), namely superoxide anions (O2–) and hydrogen peroxide (H2O2), are important signaling molecules that control cell metabolism. They play a key role in the development, growth, differentiation, and proliferation of multicellular organisms, which are usually stimulated by organic pollutants (Vaahtera et al., 2014; Zandalinas and Mittler, 2018). On the other hand, excessive ROS has a risk to damage the microbial membrane system and ultimately impedes growth. To protect against and eliminate the toxicity caused by ROS, microalgae could secrete various antioxidant enzymes through antioxidant defense mechanisms, such as SOD and CAT (Xiong et al., 2017a). For example, as an important antioxidant enzyme, SOD can transfer O2– to H2O2 and O2 to avoid its accumulation. When algae are stressed by low concentrations of exogenous pollutants, it can avoid or reduce oxidative damage by increasing the activity of these antioxidant enzymes in microalgae cells. However, when the tolerable threshold is exceeded, serious oxidative damage to cell structure and even death may result. As the main product of lipid peroxidation, MDA content can represent the damage degree of cell membrane.
Figure 4 shows SOD, MDA, and CAT concentration variation of Chlorella sp. L38 and P. tricornutum MASCC-0025 under different SDM concentrations. According to Figure 4A, due to the presence of SDM, O2– increased rapidly associated with the growth of Chlorella sp. L38, resulting in the corresponding increase of SOD in the algal cell. The amount of O2– was positively correlated to the increase of SDM concentration. In Figure 4B, it could be also seen that the addition of NaCl could promote the generation of SOD and CAT enzymes in Chlorella sp. L38. Thus, increasing salinity of growth condition facilitated to alleviate the toxicity of SDM to Chlorella sp. L38. It should be noted that when the NaCl concentration increased to 200 mg/L, the CAT enzyme content would be decreased and lower than that of 100 mg/L NaCl. It might be because the high salt concentration adversely affected the transport of substances. Therefore, under the action of SOD enzyme, O2– would be transferred into H2O2 but not decomposed into H2O and O2 in time. In Figure 4A, when the concentration of SDM was set at 0.5 mg/L, the SOD and CAT enzymatic reactions of algal cells were intensified and the stress resistance was enhanced. When the concentration of SDM varied from 1 to 3 mg/L, with the increase of the concentration of antibiotics, the equilibrium reaction was more destructive, and thus the content of MDA increased. However, in the group of adding NaCl, it was observed that salt presented a certain maintenance effect on the balance of the reaction, and the content of MDA in algal cells was much lower than that in the group without adding NaCl.
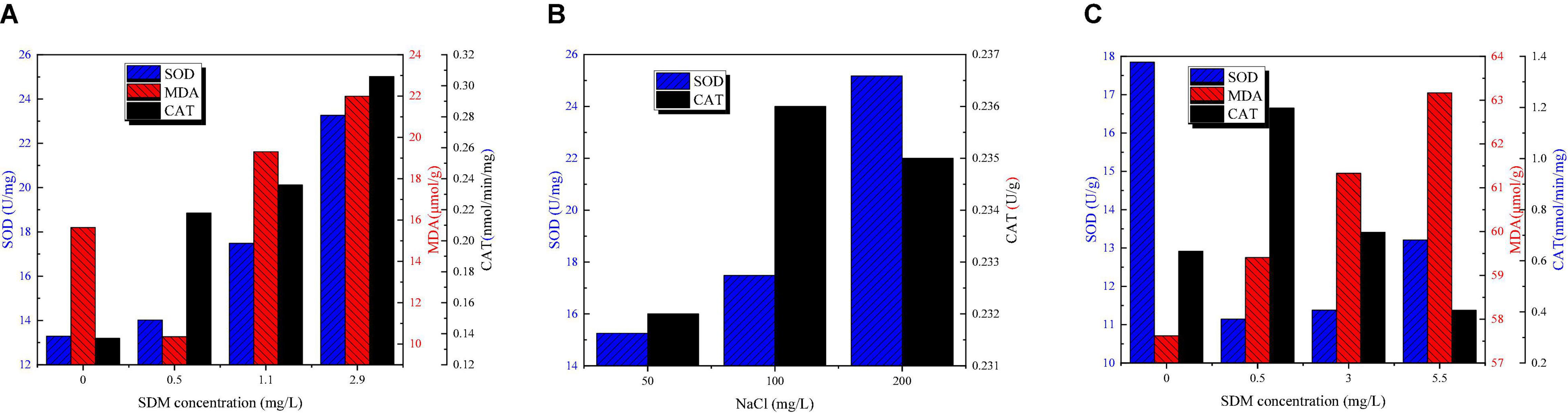
Figure 4. Superoxide dismutase (SOD), malondialdehyde (MDA), and catalase (CAT) content variation of Chlorella sp. L38 and Phaeodactylum tricornutum MASCC-0025 under different sulfadimethoxine (SDM) concentrations.
For P. tricornutum MASCC-0025, as shown in Figure 4C, it can be seen that with the increase of the concentration of antibiotics, the content of MDA and SOD also increased, indicating that the degree of algal cell damage increased. However, when SDM concentration was set at 1 mg/L, it could be seen that SOD enzyme concentration was significantly lower than that in the control group, while CAT enzyme concentration was the highest. That might be since at the initial stage of P. tricornutum MASCC-0025 cultivation, part of O2– has been converted into H2O2. Thus, due to the reduction of O2–, the content of SOD decreased. Meanwhile, the increased H2O2 content would stimulates the production of CAT enzymes in algae cells to continuously convert H2O2 into H2O and O2.
Chlorophyll Variation
The photosynthetic processes of microalgae include the photosystem I (PSI) and the photosystem II (PSII) (Mirkovic et al., 2017). For PSI, the light collection efficiency of photosynthesis is related to chlorophyll a. The content of chlorophyll a is important for assessing the adaptability of microalgae to environmental stresses such as salinity. Chlorophyll a, b and carotenoid content variation of Chlorella sp. L38 under different SDM concentrations is shown in Figure 5. It should be pointed out that the chlorophyll content of P. tricornutum MASCC-0025 is too low to be detected. From Figures 5A,B, it can be seen that SDM had a potential to promote the synthesis of chlorophyll a at 8–14 days. By contrast, the addition of SDM would inhibit the synthesis of chlorophyll b within 0–4 days, and then promoted the synthesis of chlorophyll b after the later adaptation. Under the same SDM concentration, with the increase of NaCl concentration, the synthesis of chlorophyll a in Chlorella sp. L38 cells was inhibited. Compared with chlorophyll a, chlorophyll b showed no obvious inhibition or promotion at the initial stage of culture, as shown in Figures 5B,C. With the increase of time, the synthesis of chlorophyll b was promoted after 8 days. Meanwhile, from 12 days of cultivation, the presence of SDM significantly promoted the synthesis of carotenoids, as illustrated in Figures 5E,F. Within 0–8 days, the presence of NaCl had a slight stimulating effect on the synthesis of carotenoids. However, with the increase of salt concentration, the synthesis of carotenoids decreased. At 12–14 days, the presence of salt turned to inhibit the synthesis of carotenoids. The aforementioned results indicated that Chlorella sp. L38 could adapt to the cultivation environment containing SDM and NaCl. It should be noted that the synthesis of chlorophyll and carotenoids in microalgae might be also inhibited if the NaCl concentration was too high. The reason might be that except Na+ absorbed by microalgae, Cl– could not be utilized, and which had a certain toxic effect on microalgae, thus inhibiting the synthesis of chlorophyll and carotenoids.
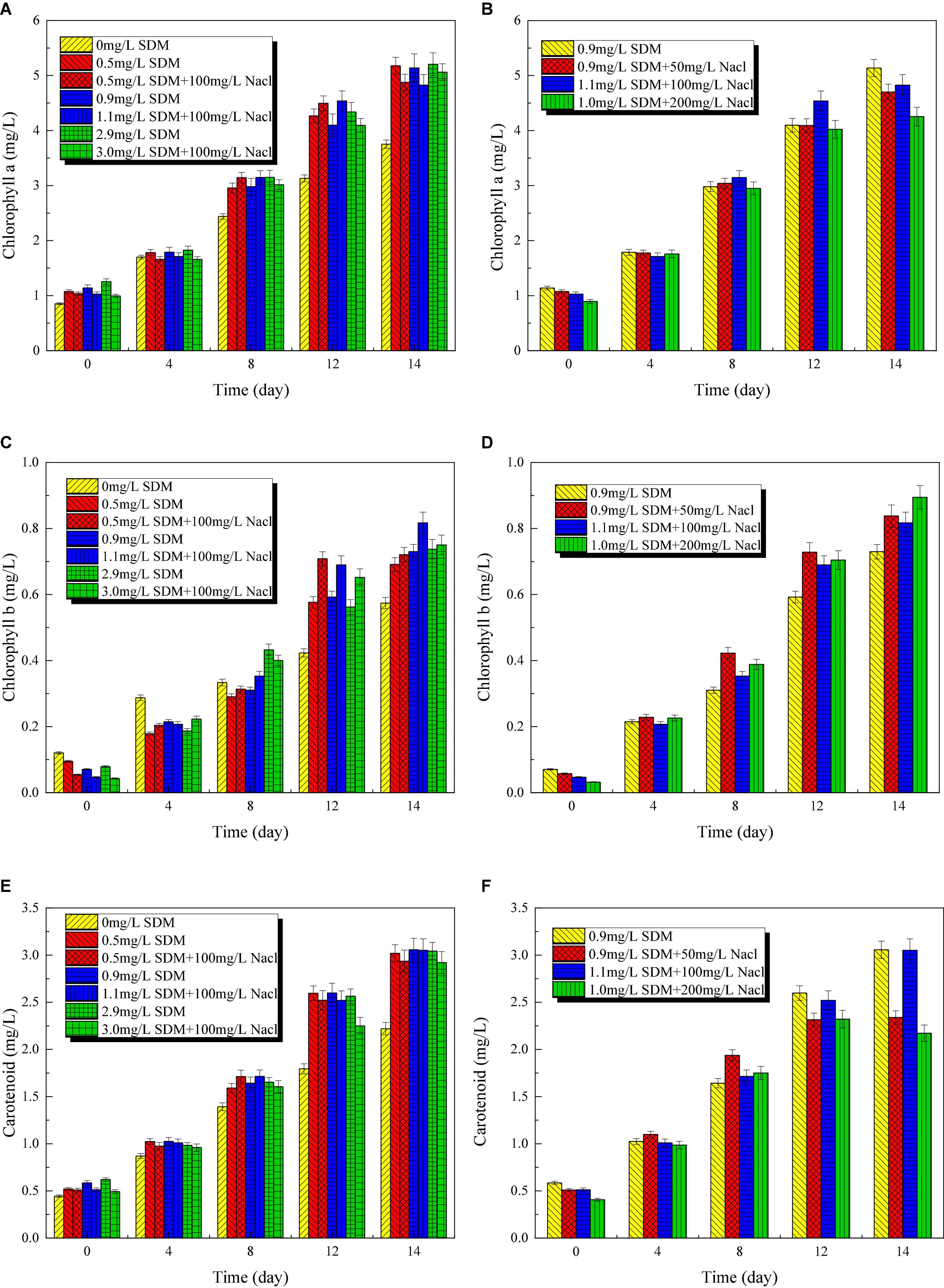
Figure 5. Chlorophyll a (A,B), b (C,D), and carotenoid (E,F) content variation of Chlorella sp. L38 and Phaeodactylum tricornutum MASCC-0025 under different sulfadimethoxine (SDM) concentrations.
Sulfadimethoxine Removal Performance
Figure 6 presents SDM removal rate of Chlorella sp. L38 and P. tricornutum MASCC-0025 under different initial concentrations. The experiment proved that SDM almost could not be degraded under the condition of light alone. Also, the maximum degradation rate reached about 88% after Chlorella sp. L38 were introduced, as shown in Figure 6A. Yu et al. (2011) proposed that SDM could be removed by 52% through biological action in the immobilized cell system. By comparing different NaCl concentration gradients under the same SDM initial dosage, it could be seen that certain salt concentrations could stimulate Chlorella sp. L38 to degrade SDM. On the 14th day of cultivation, the degradation times achieved up to 3.3 times compared with the group without adding NaCl. For example, when the initial SDM concentration was set at 1 mg/L, the degradation rate of microalgae with adding 100 mg/L NaCl reached 50% after 14 days of cultivation. This was because under salt-treated conditions, microalgae could regulate their growth at low NaCl concentrations by altering their biochemical properties, such as enzyme systems and extracellular polymers (EPS). Xiong et al. (2017b) also indicated that microalgae could present an increased photosynthetic activity (enhanced CO2 assimilation by transferring carbon and energy resource into algal biomass) and adaptive mechanism under salt stress.
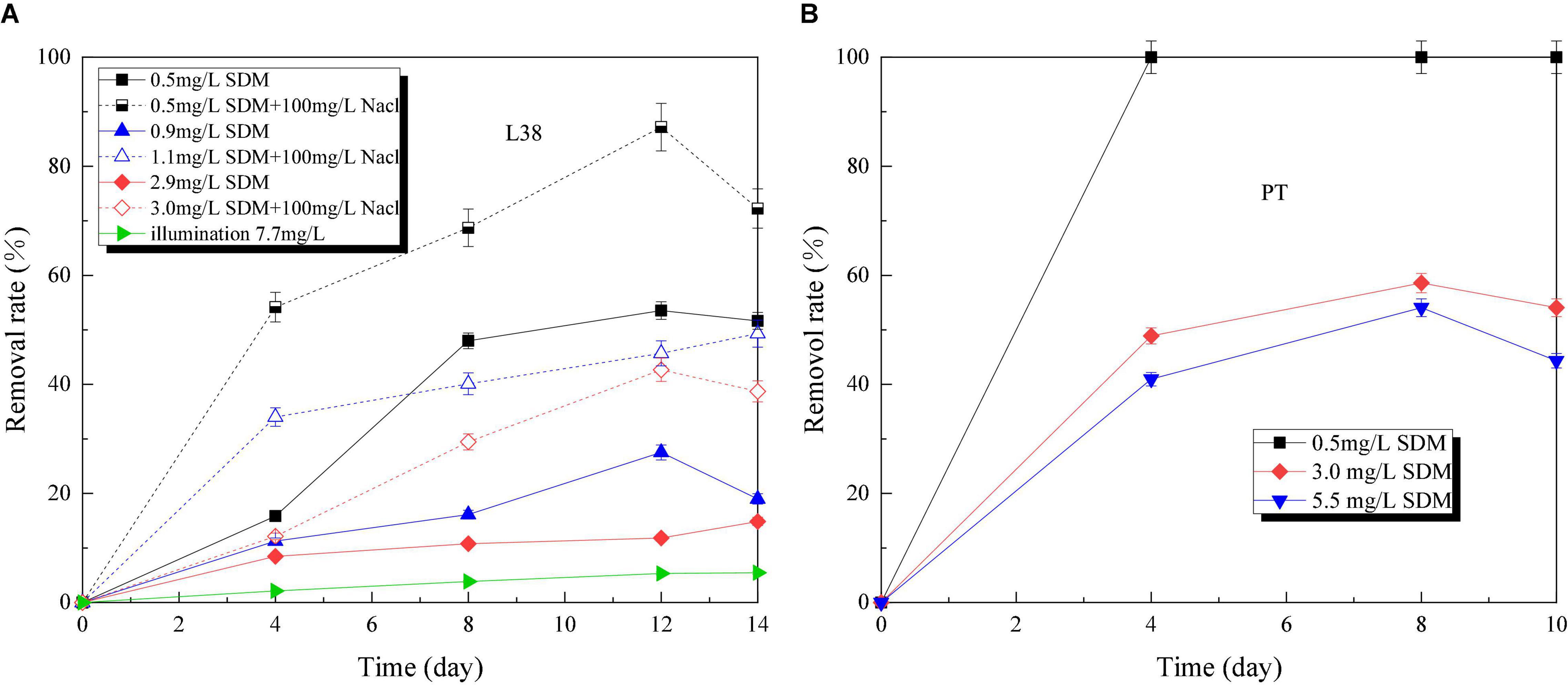
Figure 6. Sulfadimethoxine (SDM) removal rate variation of Chlorella sp. L38 and Phaeodactylum tricornutum MASCC-0025 under different initial concentrations.
Compared with Chlorella sp. L38, P. tricornutum MASCC-0025 showed a good SDM removal performance. Under the SDM concentration at 3.0 mg/L, the SDM removal rates of Chlorella sp. L38 (adding NaCl) and P. tricornutum MASCC-0025 were 29.45 and 58.59% (1.98 times higher than Chlorella sp. L38), respectively. It could be seen that marine algae might have a superiority to be a potential option to remove SDM.
Metabolic Product Analysis
To further investigate biodegradation pathway of SDM, its microalgal metabolic products were also investigated. Kiki et al. (2020) proposed that microalgal biodegradation could reduce the toxicity of antibiotics and then transform them into less toxic intermediates. Xiong et al. (2019) found that the major bioconversion pathways of SMZ were hydrolysis, methylation, dechlorination, and hydroxylation, and the main pathways of SMX degradation were nitrosation, deamination, methylation, and hydroxylation. As another typical member of sulfonamide antibiotics, Sun et al. (2018) proposed that due to the similarity of SDM and SMX structures (side chain groups of amino substituted benzene ring), their degradation process and degree might be similar. According to the identification of metabolites via LC-MS, metabolic pathway of SDM via Chlorella sp. L38 was mainly through deamination reaction to generate hydroxyl and amino compounds.
Conclusion
The biodegradation fate of SDM via Chlorella sp. L38 and P. tricornutum MASCC-0025 was investigated and the biodegradation performance was optimized via cultivation condition variation. The experimental results indicated that Chlorella sp. L38 could better adapt SDM addition via antioxidant enzyme secretion. In addition, moderate salinity condition could stimulate SOD (25.18 U/mg) and CAT (0.24 nmol/min/mg) enzyme generation. By contrast, P. tricornutum MASCC-0025 has the advantage of high salinity to stimulate SDM degradation rate (up to 100%). It could be observed that microalgae have the potential for antibiotics removal from freshwater and marine environment under the optimized species and cultivation conditions.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
BL: data curation, original draft preparation, revising and software. DW: writing, reviewing, data curation, original draft preparation, and software. YL: original draft preparation, reviewing and editing. YS: reviewing and editing. CW and JS: data curation. CS: conceptualization, methodology, writing, reviewing, editing and supervision. All authors contributed to the article and approved the submitted version.
Funding
This research was financially supported by the innovative research and experiment project of young researchers of Tianjin Academy of Agricultural Sciences (201919), the innovation team construction project of Tianjin dairy cattle (mutton sheep) Industry Technology System (ITTCRS2021000), the sub-project of the National Key Research and Development Program (2017YFD0801404-1), and International Cooperation Research Centre of Carbon Capture in Ultra-low Energy-consumption (Tianjin).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.840562/full#supplementary-material
References
Ahmed, M. B., Zhou, J. L., Ngo, H. H., and Guo, W. (2015). Adsorptive removal of antibiotics from water and wastewater: progress and challenges. Sci. Total Environ. 532, 112–126. doi: 10.1016/j.scitotenv.2015.05.130
Ali, M. E., El-Aty, A. M. A., Badawy, M. I., and Ali, R. K. (2018). Removal of pharmaceutical pollutants from synthetic wastewater using chemically modified biomass of green alga Scenedesmus obliquus. Ecotoxicol. Environ. Saf. 151, 144–152. doi: 10.1016/j.ecoenv.2018.01.012
Anjali, R., and Shanthakumar, S. (2019). Insights on the current status of occurrence and removal of antibiotics in wastewater by advanced oxidation processes. J. Environ. Manag. 246, 51–62. doi: 10.1016/j.jenvman.2019.05.090
Bilal, M., Ashraf, S. S., Barceló, D., and Iqbal, H. M. N. (2019). Biocatalytic degradation/redefining “removal” fate of pharmaceutically active compounds and antibiotics in the aquatic environment. Sci. Total Environ. 691, 1190–1211. doi: 10.1016/j.scitotenv.2019.07.224
Cheng, D., Ngo, H. H., Guo, W., Chang, S. W., Nguyen, D. D., Liu, Y., et al. (2020). Removal process of antibiotics during anaerobic treatment of swine wastewater. Bioresour. Technol. 300:122707. doi: 10.1016/j.biortech.2019.122707
Cheng, J., Ye, Q., Yang, Z., Yang, W., Zhou, J., and Cen, K. (2017). Microstructure and antioxidative capacity of the microalgae mutant Chlorella PY-ZU1 during tilmicosin removal from wastewater under 15% CO2. J. Hazard. Mater. 324, 414–419. doi: 10.1016/j.jhazmat.2016.11.006
Craggs, R., Sutherland, D., and Campbell, H. (2012). Hectare-scale demonstration of high rate algal ponds for enhanced wastewater treatment and biofuel production. J. Appl. Phycol. 24, 329–337. doi: 10.1007/s10811-012-9810-8
Homem, V., and Santos, L. (2011). Degradation and removal methods of antibiotics from aqueous matrices – a review. J. Environ. Manag. 92, 2304–2347. doi: 10.1016/j.jenvman.2011.05.023
Kabra, A. N., Ji, M. K., Choi, J., Kim, J. R., Govindwar, S. P., and Jeon, B. H. (2014). Toxicity of atrazine and its bioaccumulation and biodegradation in a green microalga, Chlamydomonas Mexicana. Environ. Sci. Pollut. Res. 21, 12270–12278. doi: 10.1007/s11356-014-3157-4
Kiki, C., Rashid, A., Wang, Y., Li, Y., Zeng, Q., Yu, C. P., et al. (2020). Dissipation of antibiotics by microalgae: kinetics, identification of transformation products and pathways. J. Hazard. Mater. 387:121985. doi: 10.1016/j.jhazmat.2019.121985
Kumar, A., and Pal, D. (2018). Antibiotic resistance and wastewater: correlation, impact and critical human health challenges. J. Environ. Chem. Eng. 6, 52–58. doi: 10.1016/j.jece.2017.11.059
Kurade, M. B., Kim, J. R., Govindwar, S. P., and Jeon, B. H. (2016). Insights into microalgae mediated biodegradation of diazinon by Chlorella vulgaris: microalgal tolerance to xenobiotic pollutants and metabolism. Algal Res. 20, 126–134. doi: 10.1016/j.algal.2016.10.003
Leng, L., Wei, L., Xiong, Q., Xu, S., Li, W., Lv, S., et al. (2020). Use of microalgae based technology for the removal of antibiotics from wastewater: a review. Chemosphere 238:124680. doi: 10.1016/j.chemosphere.2019.124680
Li, M., Liu, Y., Zeng, G., Liu, N., and Liu, S. (2019). Graphene and graphene-based nanocomposites used for antibiotics removal in water treatment: a review. Chemosphere 226, 360–380. doi: 10.1016/j.chemosphere.2019.03.117
Mirkovic, T., Ostroumov, E. E., Anna, J. M., van Grondelle, R., and Govindjee Scholes, G. D. (2017). Light absorption and energy transfer in the antenna complexes of photosynthetic organisms. Chem. Rev. 117, 249–293. doi: 10.1021/acs.chemrev.6b00002
Mulla, S. I., Bagewadi, Z. K., Faniband, B., Bilal, M., Chae, J. C., Bankole, P. O., et al. (2021). Various strategies applied for the removal of emerging micropollutant sulfamethazine: a systematic review. Environ. Sci. Pollut. Res. doi: 10.1007/s11356-021-14259-w
Mulla, S. I., Hu, A., Sun, Q., Li, J., Suanon, F., Ashfaq, M., et al. (2018). Biodegradation of sulfamethoxazole in bacteria from three different origins. J. Environ. Manage. 206, 93–102. doi: 10.1016/j.jenvman.2017.10.029
Nagarajan, D., Kusmayadi, A., Yen, H. W., Dong, C. D., Lee, D. J., and Chang, J. S. (2019). Current advances in biological swine wastewater treatment using microalgae-based processes. Bioresour. Technol. 289:121718. doi: 10.1016/j.biortech.2019.121718
Peiris, C., Gunatilake, S. R., Mlsna, T. E., Mohan, D., and Vithanage, M. (2017). Biochar based removal of antibiotic sulfonamides and tetracyclines in aquatic environments: a critical review. Bioresour. Technol. 246, 150–159. doi: 10.1016/j.biortech.2017.07.150
Qiao, M., Ying, G. G., Singer, A. C., and Zhu, Y. G. (2018). Review of antibiotic resistance in China and its environment. Environ. Int. 110, 160–172. doi: 10.1016/j.envint.2017.10.016
Shao, B., Dong, D., Wu, Y., Hu, J., Meng, J., Tu, X., et al. (2005). Simultaneous determination of 17 sulfonamide residues in porcine meat, kidney and liver by solidphase extraction and liquid chromatography–tandem mass spectrometry. Anal. Chim. Acta 546, 174–181. doi: 10.1016/j.aca.2005.05.007
Song, C., Wei, Y., Qiu, Y., Qi, Y., Li, Y., and Kitamura, Y. (2019). Biodegradability and mechanism of florfenicol via Chlorella sp. UTEX1602 and L38: experimental study. Bioresour. Technol. 272, 529–534. doi: 10.1016/j.biortech.2018.10.080
Song, C., Wei, Y., Sun, J., Song, Y., Li, S., and Kitamura, Y. (2020). Biodegradation and metabolic fate of thiamphenicol via Chlorella sp. UTEX1602 and L38. Bioresour. Technol. 296:122320. doi: 10.1016/j.biortech.2019.122320
Sun, P., Li, Y., Meng, T., Zhang, R., Song, M., and Ren, J. (2018). Removal of sulfonamide antibiotics and human metabolite by biochar and biochar/H2O2 in synthetic urine. Water Res. 147, 91–100. doi: 10.1016/j.watres.2018.09.051
Sutherland, D. L., and Ralph, P. J. (2019). Microalgal bioremediation of emerging contaminants – opportunities and challenges. Water Res. 164:114921. doi: 10.1016/j.watres.2019.114921
Sutherland, D. L., Heubeck, S., Park, J., Turnbull, M. H., and Craggs, R. J. (2018). Seasonal performance of a full-scale wastewater treatment enhanced pond system. Water Res. 136, 150–159. doi: 10.1016/j.watres.2018.02.046
Vaahtera, L., Brosché, M., Wrzaczek, M., and Kangasjärvi, J. (2014). Specificity in ROS signaling and transcript signatures. Antioxid. Redox Signal. 21, 1422–1441. doi: 10.1089/ars.2013.5662
Xiong, J., Kim, S., Kurade, M. B., Govindwar, S., Abou-Shanab, R. A. I., Kim, J. R., et al. (2019). Combined effects of sulfamethazine and sulfamethoxazole on a freshwater microalga, Scenedesmus obliquus: toxicity, biodegradation, and metabolic fate. J. Hazard. Mater. 370, 138–146. doi: 10.1016/j.jhazmat.2018.07.049
Xiong, J., Kurade, M. B., and Jeon, B. (2017a). Ecotoxicological effects of enrofloxacin and its removal by monoculture of microalgal species and their consortium. Environ. Pollut. 226, 486–493. doi: 10.1016/j.envpol.2017.04.044
Xiong, J., Kurade, M. B., Patil, D. V., Jang, M., Paeng, K. J., and Jeon, B. H. (2017b). Biodegradation and metabolic fate of levofloxacin via a freshwater green alga, Scenedesmus obliquus in synthetic saline wastewater. Algal Res. 25, 54–61. doi: 10.1016/j.algal.2017.04.012
Yu, F., Li, Y., Han, S., and Ma, J. (2016). Adsorptive removal of antibiotics from aqueous solution using carbon materials. Chemosphere 153, 365–385. doi: 10.1016/j.chemosphere.2016.03.083
Yu, T. H., Lin, A. Y., Panchangam, S. C., Hong, P. K., Yang, P. Y., and Lin, C. F. (2011). Biodegradation and bio-sorption of antibiotics and non-steroidal anti-inflammatory drugs using immobilized cell process. Chemosphere 84, 1216–1222. doi: 10.1016/j.chemosphere.2011.05.045
Zandalinas, S. I., and Mittler, R. (2018). ROS-induced ROS release in plant and animal cells. Free Radic. Biol. Med. 122, 21–27. doi: 10.1016/j.freeradbiomed.2017.11.028
Zhang, Q. Q., Ying, G. G., Pan, C. G., Liu, Y. S., and Zhao, J. L. (2015). Comprehensive evaluation of antibiotics emission and fate in the river basins of China: source analysis, multimedia modeling, and linkage to bacterial resistance. Environ. Sci. Technol. 49, 6772–6782. doi: 10.1021/acs.est.5b00729
Zhang, X., and Li, R. (2020). Electrodes bioaugmentation promotes the removal of antibiotics from concentrated sludge in microbial electrolysis cells. Sci. Total Environ. 715:136997. doi: 10.1016/j.scitotenv.2020.136997
Keywords: sulfadimethoxine, Chlorella sp., Phaeodactylum tricornutum, microalgae, NaCl, biodegradation
Citation: Li B, Wu D, Li Y, Shi Y, Wang C, Sun J and Song C (2022) Metabolic Mechanism of Sulfadimethoxine Biodegradation by Chlorella sp. L38 and Phaeodactylum tricornutum MASCC-0025. Front. Microbiol. 13:840562. doi: 10.3389/fmicb.2022.840562
Received: 22 December 2021; Accepted: 26 January 2022;
Published: 18 March 2022.
Edited by:
Qiang Wang, Henan University, ChinaReviewed by:
Sandhya Mishra, Key Laboratory of Tropical Forest Ecology, Xishuangbanna Tropical Botanical Garden (CAS), ChinaSikandar I. Mulla, REVA University, India
Copyright © 2022 Li, Wu, Li, Shi, Wang, Sun and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunfeng Song, Y2h1bmZlbmcuc29uZ0B0anUuZWR1LmNu
 Bing Li
Bing Li Di Wu1
Di Wu1 Chunfeng Song
Chunfeng Song