
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 07 April 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.840520
This article is part of the Research TopicAntimicrobial Resistance in Foodborne Pathogens: ecology, epidemiology, and mechanismsView all 12 articles
 Jufang Hu1,2†
Jufang Hu1,2† Jie Yang1,2†
Jie Yang1,2† Wenxin Chen1,2†
Wenxin Chen1,2† Zhihong Liu1,2
Zhihong Liu1,2 Qin Zhao3
Qin Zhao3 Hui Yang3
Hui Yang3 Zhiliang Sun1,2
Zhiliang Sun1,2 Xiaojun Chen1,2*
Xiaojun Chen1,2* Jiyun Li1,2*
Jiyun Li1,2*
Colistin is one of the last-line drugs against difficult to treat and multidrug-resistant Gram-negative bacteria. The emergence of mobile colistin resistance gene mcr-1 increased worldwide attention on colistin resistance. mcr-1 is the dominant gene that caused resistance to colistin in chicken-derived Escherichia coli (E. coli) in China; it has a broad resistance spectrum and causes multiple drug resistance problems. There are only few studies on mcr-positive E. coli (MCRPEC) from laying ducks and quails in China. Here, the molecular and epidemiological characteristics of MCRPEC from three kinds of poultry farms (laying duck, quail, and broiler) were investigated in Changsha, China. A total of 17 mcr-positive E. coli (MCRPEC) strains were screened in 690 samples from the three kinds of poultry farms. This is the first report on MCRPEC, to our best knowledge, derived from quail. All the MCRPEC strains were resistant to colistin, sulfamethoxazole-trimethoprim, florfenicol, tetracycline, and ciprofloxacin, and mildly resistant to tigecycline, gentamicin, piperacillin/tazobactam, cefotaxime, and ceftiofur. All the strains were sensitive to meropenem and amikacin. By bioinformatics analysis, 17 MCRPEC strains belonging to 11 MLST types were distributed in phylogroups A (58.8%), B1 (23.5%), and phylogroup D (17.6%). mcr-1 was located in IncI2 plasmid with typical plasmid conjugation transfer part, type IV secretory system, and tellurium-resistant protein, increasing transmission capacity to other bacteria. Monitoring of colistin-resistant bacteria in poultry farms should be strengthened.
The use of colistin as an antibiotic for the treatment of Gram-negative infections has been gradually reduced because of its nephrotoxicity and neurotoxicity (Koch-Weser et al., 1970). With the emergence of multi-drug-resistant bacteria, especially carbapenem-resistant bacteria, lack of new antibiotics against Gram-negative pathogens has forced the reuse of traditional antibiotic (colistin) (Falagas and Kasiakou, 2005). However, it will be decreasing effectiveness of colistin in clinical after the emergence of mobile colistin resistance gene, mcr-1(Liu et al., 2016), and its variants (mcr-2 to mcr-10) (Xavier et al., 2016; AbuOun et al., 2017; Borowiak et al., 2017; Carattoli et al., 2017; Yin et al., 2017; Wang X. et al., 2018; Yang et al., 2018; Carroll et al., 2019; Wang C. et al., 2020). Colistin-resistant isolates harboring these plasmid-mediated colistin resistance genes had been increasing on human clinical (Li et al., 2021), livestock and poultry farms (Hassen et al., 2020), sewage treatment systems (Yang et al., 2019), meat products retail (Sadek et al., 2021), waterfowl breeding (Shen et al., 2019), and home water purifiers (Chen et al., 2021). mcr genes have been mainly found in Escherichia coli, Klebsiella pneumoniae, Salmonella enterica, Enterobacter spp., and Aeromonas spp. (Ling et al., 2020). mcr-1 is the most widely disseminated plasmid-mediated colistin-resistant gene and is mainly carried by the IncI2 and IncX4 types of plasmid (Tang et al., 2021).
The World Organization for Animal Health (OIE) recommends that organizations urgently prohibit colistin use as a growth promoter because of increasing colistin resistance.1 At the same time, governments began to issue policies to limit colistin in livestock and poultry. The Chinese government formally banned colistin as an animal growth promoter on April 30, 2017 (Walsh and Wu, 2016). Other countries, including India, Japan, Malaysia, and Thailand, have banned or agreed to ban colistin as a feed additive for animal growth (Olaitan et al., 2021). The withdrawal of colistin from animal feed in China has significantly reduced colistin resistance and the prevalence of mcr-1 in both animals and humans (Wang Y. et al., 2020). It proved that the intervention policies effectively reduced the use of colistin and reduced colistin resistance in animals.
mcr-1 and its variants have been found in various countries and regions but mainly focused on livestock breeding, especially pig and broiler chickens. The prevalence of mcr-1 in other kinds of poultry is rare. Here, we investigated the prevalence and characteristics of mcr-positive Escherichia coli (MCRPEC) isolate from three types of poultry farms, namely, quail farm, laying duck farm, and broiler farm, in Changsha, China.
From May 2019 to May 2020, animal cloacal swabs and surrounding environmental samples were collected from three kinds of poultry farms in Changsha as previously described (Wang et al., 2017; Shi et al., 2021; Zhao et al., 2021). Briefly, 50 animal cloacal swabs per farm were collected using sterile swabs and then suspended in 1 ml phosphate-buffered saline (PBS). Flies were caught using fly glue boards (Green Leaf Co., China) and transferred to the 1 mL PBS with sterile tweezers. Four ml of drinking water and sewage were collected in a clean bottle. Around the farming area, surface soil of 5–10 cm was removed with a shovel, and 5-g soil samples were collected with sterile bags. Five samples from within one square meter were mixed. All the samples were stored in an icebox and transported to the laboratory. The samples were seeded in MacConkey (Solarbio) containing 2 mg/L of colistin and incubated at 37°C overnight; Then, a single pink clone was picked up.
In all the bacteria, we detected the presence of mcr-1 to mcr-10 by polymerase chain reaction (PCR) (Rebelo et al., 2018; Borowiak et al., 2020; Wang C. et al., 2020). Species identification of MCRPEC was confirmed with the 16S rRNA gene (Srivastava et al., 2008). PCR products were sequenced by Tsingke Biological Technology (Changsha, China) using the sanger sequencing method and compared to the GenBank database.
As recommended by Clinical & Laboratory Standards Institute (CLSI) M100-S30,2 CLSI VET01-A4 (see text footnote 2), and the European Committee on Antimicrobial Susceptibility Testing (EUCAST),3 minimum inhibitory concentrations (MICs) of colistin were evaluated by broth microdilution, and 11 antibiotics (meropenem, amikacin, florfenicol, sulfamethoxazole-trimethoprim, piperacillin-tazobactam, tetracycline, ciprofloxacin, gentamicin, tigecycline, cefotaxime, and ceftiofur) were determined with the agar dilution method. E. coli ATCC 25922 was used as a quality control strain. The transferability of mcr-1 was determined by plasmid conjugation assay; streptomycin-resistant E. coli C600 serves as the recipient and MCRPEC as the donor strains. Their transconjugants were selected on MacConkey agar with colistin (2 mg/L) and streptomycin (500 mg/L), and confirmed by ERIC-PCR fingerprinting (Amin et al., 2020) for E. coli C600 and PCR analysis for the mcr-1 gene.
Genomic DNA was extracted from the MCRPEC strains using a TIANamp Bacteria DNA kit (Tiangen Biotech Company, China) according to the manufacturer’s instructions, and 150-bp paired-end reads were generated with a HiSeq X Ten platform (Illumina). Bacterial genome assembly was performed with the SPAdes software (Bankevich et al., 2012).
Multilocus sequence typing (MLST) was performed, antimicrobial-resistant genes were determined by SRST2 (Inouye et al., 2014), and E. coli phylogroups were identified with the ClermonTyping scheme (Beghain et al., 2018). The clonal relationship of MCRPEC was evaluated by core genome alignments and with phylogenetic trees, which were constructed with Parsnp (Treangen et al., 2014) and visualized with the online tool iTOL.4 Plasmid comparisons of the isolates were performed with BLAST Ring Image Generator (BRIG) (Alikhan et al., 2011), and EasyFig 2.2.5 was used for gene-environment analysis (Sullivan et al., 2011).
Six hundred ninety samples (250 broiler chicken cloacal swabs, 150 laying duck cloacal swabs, 150 quail cloacal swabs, and 140 environmental samples) were collected from 10 poultry farms in Changsha (Supplementary Table 1). After screening, 17 strains were positive for mcr-1 (Table 1), but no other mcr variants were detected in this study, indicating that mcr-1 was the dominant mcr gene in poultry farms in Changsha. Among the three kinds of poultry breeding, the isolation rate of mcr-1-positive MCRPE was highest in broiler (5.6%, 14/250), followed by quail (0.7%, 1/150), and laying duck (0.7%, 1/150) (Table 1). In addition, MCRPEC was detected in the surrounding environment (flies, 1.8%, 1/56) (Table 1). MCRPEC was present in five broiler farms, and detection rate was higher than 2% (2, 4, 4, 8, and 10%, respectively), indicating that urgent attention is needed in broiler breeding.
The MIC range of poultry MCRPEC against colistin was 4 to >128 mg/L. All the MCRPEC isolates were resistant to sulfamethoxazole, florfenicol, tetracycline, and ciprofloxacin, and sensitive to meropenem and amikacin. We found that some of the MCRPEC strains were tetracycline-resistant, but no tetracycline-resistant gene was detected (Figure 1), which may be because of overexpression of the efflux pump gene. In addition, most of the strains were mildly resistant to tigecycline, gentamicin, piperacillin-tazobactam, cefotaxime, and ceftiofur (Table 2 and Supplementary Table 2). Interestingly, different β-lactam-resistant genes (blaCTX, blaOXA, and blaTEM) were found in the 17 MCRPEC strains.
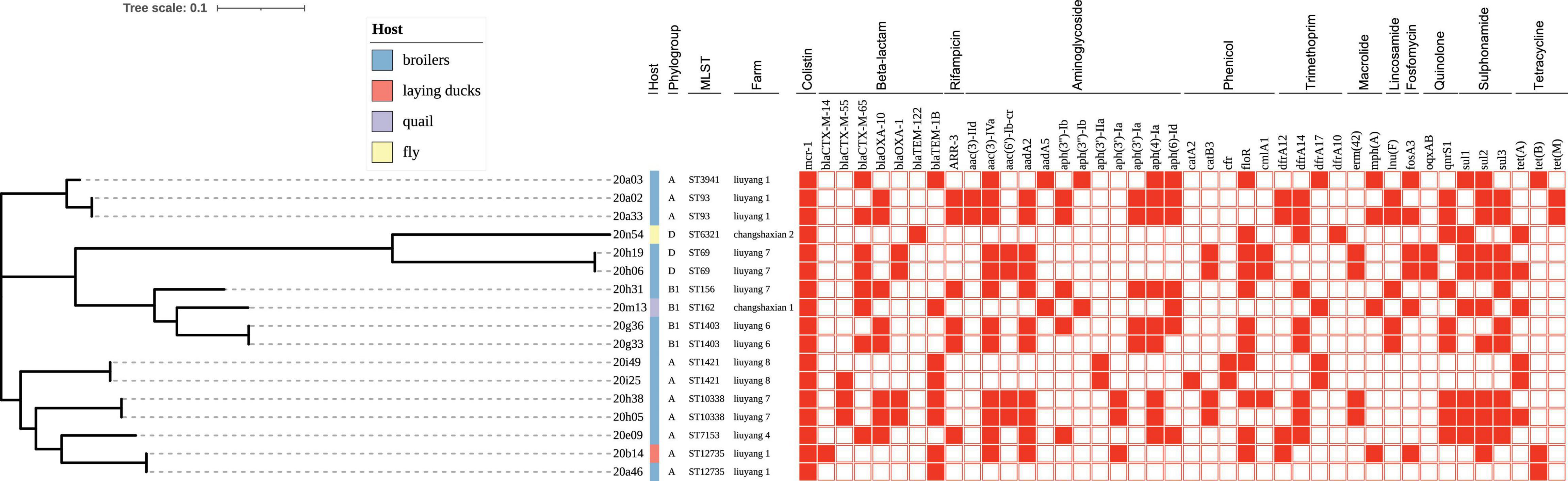
Figure 1. Phylogenetic tree of the 17 mcr-positive Escherichia coli (MCRPEC) isolates. Phylogroup, multilocus sequence type, and farm origin are shown in text after the corresponding isolate name. Sources of the isolates are differentiated by color. Blue, broiler; Orange, laying duck; Purple, quail; Yellow, fly. Filled squares denote the resistance genes for presence and empty squares for absence.
A total of 11 MLST types (ST10338, ST1403, ST1421, ST156, ST162, ST3941, ST6321, ST69, ST7153, ST93, and ST12735) were identified among the 17 MCRPEC strains (Figure 1). MCRPEC isolates predominantly belonged to phylogroup A (58.8%, 10/17), followed by phylogroup B1 (23.5%, 4/17) and phylogroup D (17.6%, 3/17) (Figure 1). Strain 20m13, belonging to ST162, was isolated from a quail farm. To the best of our knowledge, this is the first report on detection of mcr-1 from quail. In another quail farm, one MCRPEC strain (20n54) from a fly belongs to ST6321 ClermontTypingD E. coli, which contains seven resistant genes, namely, β-lactam-resistant gene (blaTEM–122), tetracycline-resistant gene [tet(A)], colistin-resistant gene (mcr-1), phenicol-resistant gene (floR), fluoroquinolone-resistant gene (qnrS1), trimethoprim-resistant gene (dfrA10, dfrA14), and sulfonamide-resistant gene (sul1), which pose a potential risk to quail’s health by spreading to quails.
By constructing a phylogenetic tree, Five groups, with two strains per group isolated from the same farm and closely related, possessed a similar ST type, phylogroup (Figure 1). In addition, two isolates (20a46 and 20b14) that we found in the proximity of the duck and broiler farms had a close genetic relationship (Figure 1).
mcr-1-bearing plasmid of most strains (82.4%, 14/17) could transfer to streptomycin-resistant E. coli C600 by conjugation assay. By bioinformatics analysis, type IV secretion systems (virB1, virB2, virB4, virB8, virB10, and virB11) and plasmid conjugation elements (pilM, pilV, and pilL) were found in almost all the isolates, which are related to the transferability of bacteria and contribute mcr-1 to other bacteria (Figure 2). Besides, 70.6% (12/17) mcr-1-bearing plasmids belonged to Incl2 except for the additional five contigs whose plasmid type was difficult to distinguish because of the short sequence. Therefore, we speculated that the Incl2 plasmid was prevalent in local poultry farms. Studies show that ISApl1 has been deemed to be the critical element mediating the translocation of mcr-1 into various plasmid backbones (Sun et al., 2017). However, ISApl1 was not found in the 17 isolates, showing a more stable mcr-1 genetic environment (Supplementary Figure 1). In addition, this study also found that some of the strains carry tellurium-resistant proteins (TerA, TerB, and TerD), which are involved in tellurite resistance, colicine resistance, phage inhibition, and pathogenicity (Turkovicova et al., 2016).
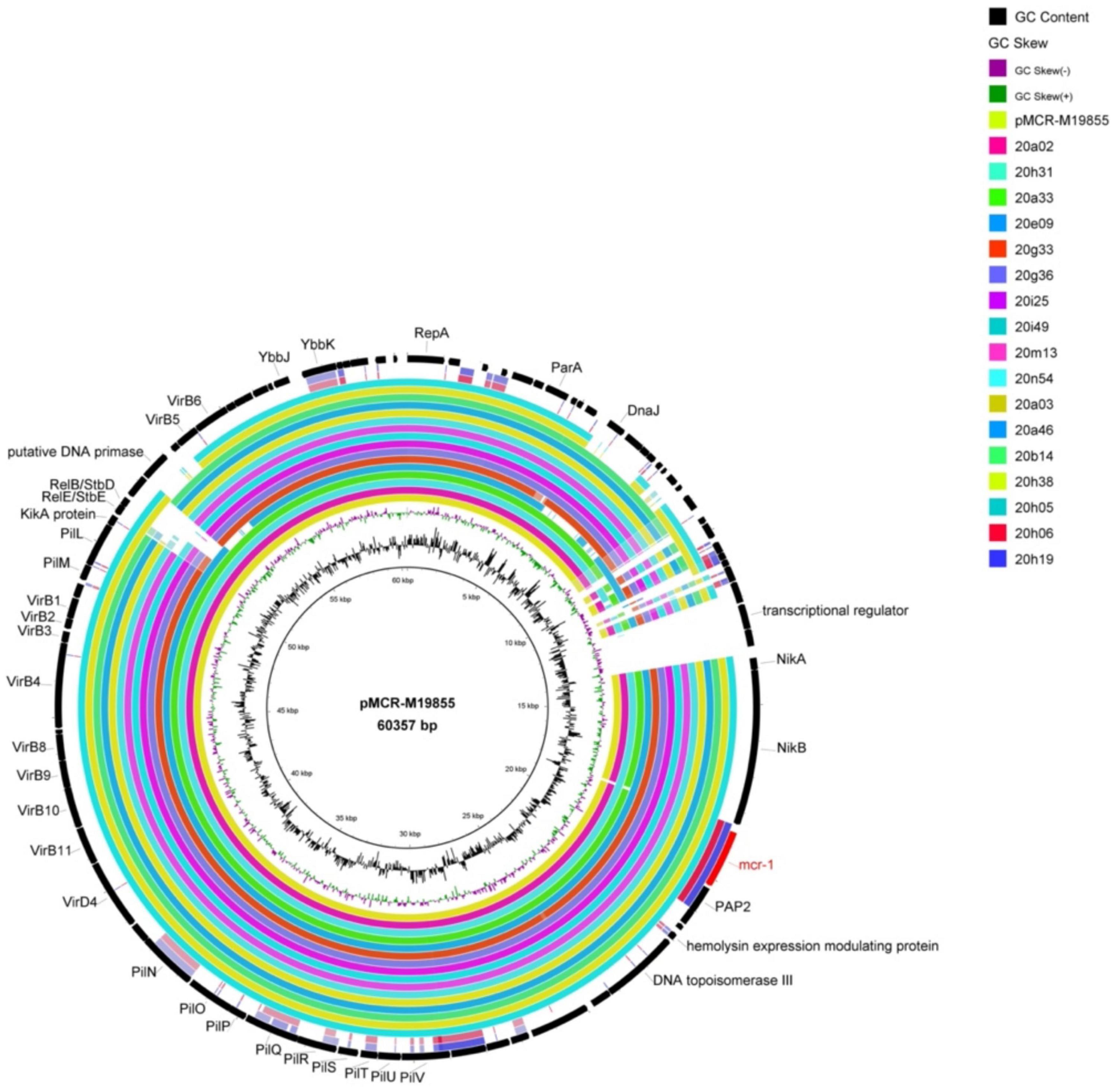
Figure 2. Comparison of whole-genome sequences of MCRPEC against the sequence of mcr-1-carrying plasmid pMCR-M19855 (GenBank accession no. KY471315). The first ring from the inside and the coordinates correspond to pMCR-M19855. Each ring represents one of the genomes, and the outside ring represents the regions from the 17 MCRPEC strains in this study.
In this study, colistin was not used in the poultry farms, but the detection rate of the MCRPEC isolates in the poultry farms, especially in the broiler farms, was relatively high. By inquiry, antibiotics are used extensively to promote growth in the broiler; on the contrary, laying ducks are prohibited from using antibiotics during the egg-producing period, because the addition of antibiotics will give rise to veterinary drug residues and reduce the output of egg production. In addition, co-carrier essential resistance genes (sul, floR, tet(M), bla, aph, etc.) may be related to using other antibiotics (florfenicol, enrofloxacin, doxycycline), along with mcr-1 which had the potential risk of multi-drug resistant bacteria. Another study showed that mcr-1could co-transfer with the blaCTX–M, fosA3, oqxAB, and floR genes (Sun et al., 2017). Therefore, local regulatory authorities should strengthen the supervision of antibiotic usage and monitor bacterial resistance, especially to MCRPEC strains.
Only a strain from laying ducks (farm liuyang 1) had a close relationship with one MCRPEC strain and a high mcr-1 detection rate (8%, 4/50) in the broiler farm. Two farms are located in different parts of the same land and managed by the same breeder, which is a potential for transmission from broilers to laying ducks. MCRPEC strains with Incl2 plasmid carrying the type IV secretion system and plasmid conjugation elements promote their mcr-1 transfer to streptomycin-resistant E. coli C600. In this way, it makes the Incl2 plasmid become the dominant carrier of mcr-1.
To the best of our knowledge, this study is the first to report on the detection of mcr-1 on quails. Before this, Salmonella enterica carrying tetA, tetB, and tetG (Bacci et al., 2012), and multidrug-resistant methicillin-resistant Staphylococcus aureus (LA-MRSA) CC398 carrying the blaZ, mecA, ermB, and ermC genes (Silva et al., 2021) were found on quails. There was no significant relationship between the E. coli isolated from quail farms and that from other poultry farms (Figure 1). At the same time, MCRPEC from flies in the quail farms was determined, increasing the transmission probability of mcr-1 in quails (Wang et al., 2017).
We demonstrated the prevalence of MCRPEC isolates in poultry farms in Changsha, but only mcr-1 gene was found. Poultry MCRPEC shows multi-drug resistance when co-harboring with other essential resistant genes. Likewise, comprehensive genomic analyses revealed that the mcr-1 genes located in the IncI2 plasmid might transfer to a different host, with the transfer element posing a potential risk to humans. Monitoring of colistin-resistant bacteria in poultry farms should be strengthened.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
JL and XC designed the research. JH, QZ, ZL, and HY collected the data. JH, JY, and WC analyzed and interpreted the data. JH drafted the manuscript. JL, JH, ZS, and XC revised the manuscript. All the authors read and approved the final version of the manuscript.
This research was supported by the Hunan Provincial Natural Science Foundation of China (2021JJ40234) and the China Postdoctoral Science Foundation (2020M682569).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.840520/full#supplementary-material
Supplementary Figure 1 | Genetic environment of mcr-1 in the 17 mcr-positive Escherichia coli (MCRPEC) strains. The direction of arrows indicates the direction of transcription.
AbuOun, M., Stubberfield, E. J., Duggett, N. A., Kirchner, M., Dormer, L., Nunez-Garcia, J., et al. (2017). Mcr-1 and mcr-2 (mcr-6.1) variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J. Antimicrob. Chemother. 72, 2745–2749. doi: 10.1093/jac/dkx286
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Amin, M. B., Sraboni, A. S., Hossain, M. I., Roy, S., Mozmader, T. A. U., Unicomb, L., et al. (2020). Occurrence and genetic characteristics of mcr-1-positive colistin-resistant E. coli from poultry environments in Bangladesh. J. Glob. Antimicrob. Resist. 22, 546–552. doi: 10.1016/j.jgar.2020.03.028
Bacci, C., Boni, E., Alpigiani, I., Lanzoni, E., Bonardi, S., and Brindani, F. (2012). Phenotypic and genotypic features of antibiotic resistance in Salmonella enterica isolated from chicken meat and chicken and quail carcasses. Int. J. Food Microbiol. 160, 16–23. doi: 10.1016/j.ijfoodmicro.2012.09.014
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Beghain, J., Bridier-Nahmias, A., Le Nagard, H., Denamur, E., and Clermont, O. (2018). ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 4:e000192. doi: 10.1099/mgen.0.000192
Borowiak, M., Baumann, B., Fischer, J., Thomas, K., Deneke, C., Hammerl, J. A., et al. (2020). Development of a novel mcr-6 to mcr-9 multiplex PCR and assessment of mcr-1 to mcr-9 occurrence in colistin-resistant Salmonella enterica isolates from environment, feed, animals and food (2011-2018) in Germany. Front. Microbiol. 11:80. doi: 10.3389/fmicb.2020.00080
Borowiak, M., Fischer, J., Hammerl, J. A., Hendriksen, R. S., Szabo, I., and Malorny, B. (2017). Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J. Antimicrob. Chemother. 72, 3317–3324. doi: 10.1093/jac/dkx327
Carattoli, A., Villa, L., Feudi, C., Curcio, L., Orsini, S., Luppi, A., et al. (2017). Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill 22:30589. doi: 10.2807/1560-7917.ES.2017.22.31.30589
Carroll, L. M., Gaballa, A., Guldimann, C., Sullivan, G., Henderson, L. O., and Wiedmann, M. (2019). Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. mBio 10, e00853–e00919. doi: 10.1128/mBio.00853-19
Chen, Z., Zhu, S., Zhao, L., Wang, X., Liang, M., Mao, D., et al. (2021). Occurrence of high-risk mcr-1 gene and blaNDM-1 positive superbug in the reverse osmosis filter cartridges of the household water purifiers. J. Hazard. Mater. Lett. 2:100011. doi: 10.1016/j.hazl.2020.100011
Falagas, M. E., and Kasiakou, S. K. (2005). Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 40, 1333–1341. doi: 10.1086/429323
Hassen, B., Abbassi, M. S., Ruiz-Ripa, L., Mama, O. M., Hassen, A., Torres, C., et al. (2020). High prevalence of mcr-1 encoding colistin resistance and first identification of blaCTX-M-55 in ESBL/CMY-2-producing Escherichia coli isolated from chicken faeces and retail meat in Tunisia. Int. J. Food Microbiol. 318:108478. doi: 10.1016/j.ijfoodmicro.2019.108478
Inouye, M., Dashnow, H., Raven, L. A., Schultz, M. B., Pope, B. J., Tomita, T., et al. (2014). SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 6:90. doi: 10.1186/s13073-014-0090-6
Koch-Weser, J., Sidel, V. W., Federman, E. B., Kanarek, P., Finer, D. C., and Eaton, A. E. (1970). Adverse effects of sodium colistimethate. Manifestations and specific reaction rates during 317 courses of therapy. Ann. Intern. Med. 72, 857–868. doi: 10.7326/0003-4819-72-6-857
Li, Y., Zhang, Y., Chen, M., Hu, J., Zhang, H., Xiang, Y., et al. (2021). Plasmid-borne colistin resistance gene mcr-1 in a multidrug resistant Salmonella enterica serovar Typhimurium isolate from an infant with acute diarrhea in China. Int. J. Infect. Dis. 103, 13–18. doi: 10.1016/j.ijid.2020.11.150
Ling, Z., Yin, W., Shen, Z., Wang, Y., Shen, J., and Walsh, T. R. (2020). Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J. Antimicrob. Chemother. 75, 3087–3095. doi: 10.1093/jac/dkaa205
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Olaitan, A. O., Dandachi, I., Baron, S. A., Daoud, Z., Morand, S., and Rolain, J.-M. (2021). Banning colistin in feed additives: a small step in the right direction. Lancet Infect. Dis. 21, 29–30. doi: 10.1016/s1473-3099(20)30915-4
Rebelo, A. R., Bortolaia, V., Kjeldgaard, J. S., Pedersen, S. K., Leekitcharoenphon, P., Hansen, I. M., et al. (2018). Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. 23:00672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672
Sadek, M., Ortiz de la Rosa, J. M., Abdelfattah Maky, M., Korashe Dandrawy, M., Nordmann, P., and Poirel, L. (2021). Genomic features of MCR-1 and extended-spectrum beta-lactamase-producing enterobacterales from retail raw chicken in Egypt. Microorganisms 9:195. doi: 10.3390/microorganisms9010195
Shen, Y., Lv, Z., Yang, L., Liu, D., Ou, Y., Xu, C., et al. (2019). Integrated aquaculture contributes to the transfer of mcr-1 between animals and humans via the aquaculture supply chain. Environ. Int. 130:104708. doi: 10.1016/j.envint.2019.03.056
Shi, X., Li, Y., Yang, Y., Shen, Z., Cai, C., Wang, Y., et al. (2021). High prevalence and persistence of carbapenem and colistin resistance in livestock farm environments in China. J. Hazard. Mater. 406:124298. doi: 10.1016/j.jhazmat.2020.124298
Silva, V., Vieira-Pinto, M., Saraiva, C., Manageiro, V., Reis, L., Ferreira, E., et al. (2021). Prevalence and characteristics of multidrug-resistant livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) CC398 Isolated from quails (Coturnix Coturnix Japonica) slaughtered for human consumption. Animals (Basel) 11:2038. doi: 10.3390/ani11072038
Srivastava, S., Singh, V., Kumar, V., Verma, P. C., Srivastava, R., Basu, V., et al. (2008). Identification of regulatory elements in 16S rRNA gene of Acinetobacter species isolated from water sample. Bioinformation 3, 173–176. doi: 10.6026/97320630003173
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Sun, J., Fang, L. X., Wu, Z., Deng, H., Yang, R. S., Li, X. P., et al. (2017). Genetic analysis of the IncX4 plasmids: implications for a unique pattern in the mcr-1 acquisition. Sci. Rep. 7:424. doi: 10.1038/s41598-017-00095-x
Tang, B., Chang, J., Hu, J., Qian, M., Xia, X., and Yang, H. (2021). Structures and characteristics of 103 plasmids carrying mcr gene. Acta Agric. Zhejiangensis 33, 43–51. doi: 10.3969/j.issn.1004-1524.2021.01.06
Treangen, T. J., Ondov, B. D., Koren, S., and Phillippy, A. M. (2014). The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15:524. doi: 10.1186/s13059-014-0524-x
Turkovicova, L., Smidak, R., Jung, G., Turna, J., Lubec, G., and Aradska, J. (2016). Proteomic analysis of the TerC interactome: novel links to tellurite resistance and pathogenicity. J. Proteomics 136, 167–173. doi: 10.1016/j.jprot.2016.01.003
Walsh, T. R., and Wu, Y. (2016). China bans colistin as a feed additive for animals. Lancet Infect. Dis. 16, 1102–1103. doi: 10.1016/s1473-3099(16)30329-2
Wang, C., Feng, Y., Liu, L., Wei, L., Kang, M., and Zong, Z. (2020). Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 9, 508–516. doi: 10.1080/22221751.2020.1732231
Wang, X., Wang, Y., Zhou, Y., Li, J., Yin, W., Wang, S., et al. (2018). Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microbes Infect. 7:122. doi: 10.1038/s41426-018-0124-z
Wang, Y., Xu, C., Zhang, R., Chen, Y., Shen, Y., Hu, F., et al. (2020). Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: an epidemiological comparative study. Lancet Infect. Dis. 20, 1161–1171. doi: 10.1016/S1473-3099(20)30149-3
Wang, Y., Zhang, R. M., Li, J. Y., Wu, Z. W., Yin, W. J., Schwarz, S., et al. (2017). Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat. Microbiol. 2:16260. doi: 10.1038/nmicrobiol.2016.260
Xavier, B. B., Lammens, C., Ruhal, R., Kumar-Singh, S., Butaye, P., Goossens, H., et al. (2016). Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium. Euro Surveill 21:27. doi: 10.2807/1560-7917.ES.2016.21.27.30280
Yang, F., Gu, Y., Zhou, J., and Zhang, K. (2019). Swine waste: a reservoir of high-risk blaNDM and mcr-1. Sci. Total Environ. 683, 308–316. doi: 10.1016/j.scitotenv.2019.05.251
Yang, Y. Q., Li, Y. X., Lei, C. W., Zhang, A. Y., and Wang, H. N. (2018). Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 73, 1791–1795. doi: 10.1093/jac/dky111
Yin, W., Li, H., Shen, Y., Liu, Z., Wang, S., Shen, Z., et al. (2017). Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio 8:3. doi: 10.1128/mBio.00543-17
Zhao, Q., Berglund, B., Zou, H., Zhou, Z., Xia, H., Zhao, L., et al. (2021). Dissemination of blaNDM-5 via IncX3 plasmids in carbapenem-resistant Enterobacteriaceae among humans and in the environment in an intensive vegetable cultivation area in eastern China. Environ. Pollut. 273:116370. doi: 10.1016/j.envpol.2020.116370
Keywords: poultry, quail, laying duck, broiler, colistin, mcr-1, Escherichia coli
Citation: Hu J, Yang J, Chen W, Liu Z, Zhao Q, Yang H, Sun Z, Chen X and Li J (2022) Prevalence and Characteristics of mcr-1-Producing Escherichia coli in Three Kinds of Poultry in Changsha, China. Front. Microbiol. 13:840520. doi: 10.3389/fmicb.2022.840520
Received: 21 December 2021; Accepted: 03 March 2022;
Published: 07 April 2022.
Edited by:
Bao-Tao Liu, Qingdao Agricultural University, ChinaReviewed by:
Rongmin Zhang, South China Agricultural University, ChinaCopyright © 2022 Hu, Yang, Chen, Liu, Zhao, Yang, Sun, Chen and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojun Chen, eGo5MmUyQDE2My5jb20=; Jiyun Li, bGlqaXl1bnlAZm94bWFpbC5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.