- 1State Key Laboratory of Microbial Technology, Marine Biotechnology Research Center, Shandong University, Qingdao, China
- 2College of Marine Life Sciences, Frontiers Science Center for Deep Ocean Multispheres and Earth System, Ocean University of China, Qingdao, China
- 3Laboratory for Marine Biology and Biotechnology, Pilot National Laboratory for Marine Science and Technology, Qingdao, China
- 4School of Bioengineering, Qilu University of Technology, Jinan, China
- 5Engineering Research Center of Sustainable Development and Utilization of Biomass Energy, Ministry of Education, Yunnan Normal University, Kunming, China
Trimethylamine N-oxide (TMAO), which was detected at nanomolar concentrations in surface seawaters, is an important carbon, nitrogen and/or energy source for marine bacteria. It can be metabolized by marine bacteria into volatile methylated amines, the second largest source of nitrogen after N2 gas in the oceans. The SAR11 bacteria are the most abundant oligotrophic plankton in the oceans, which represents approximately 30% of the bacterial cells in marine surface waters. Genomic analysis suggested that most SAR11 bacteria possess an ATP-binding cassette transporter TmoXWV that may be responsible for importing TMAO. However, it was still unclear whether SAR11 bacteria can utilize TMAO as the sole nitrogen source and how they import TMAO. Here, our results showed that Pelagibacter strain HTCC1062, a SAR11 bacterium, can grow with TMAO as the sole nitrogen source. TmoXWV from strain HTCC1062 (TmoXWV1062) was verified to be a functional TMAO importer. Furthermore, TmoX1062, the periplasmic substrate binding protein of TmoXWV1062, was shown to have high binding affinities toward TMAO at 4°C (Kd = 920 nM), 10°C (Kd = 500 nM) and 25°C (Kd = 520 nM). The high TMAO binding affinity and strong temperature adaptability of TmoX1062 reveal a possible oligotrophic niche adaptation strategy of strain HTCC1062, which may help it gain a competitive advantage over other bacteria. Structure comparison and mutational analysis indicated that the TMAO binding mechanism of TmoX1062 may have differences from the previously reported mechanism of TmoX of Ruegeria pomeroyi DSS-3. This study provides new insight into TMAO utilization by the widespread SAR11 bacteria.
Introduction
Marine phytoplankton generate approximate one-half of the global primary production in the oceans, with a large fraction turning into dissolved organic matter (DOM) by various mechanisms (Falkowski et al., 1998; Azam and Malfatti, 2007). Trimethylamine N-oxide (TMAO) is an important component of marine DOM and a compatible osmolyte for a variety of marine biota (Gibb and Hatton, 2004; Carpenter et al., 2012). It is also a nitrogen and/or carbon source for marine heterotrophic bacteria (Lidbury et al., 2015). The concentrations of TMAO range from low nanomolar (nM) in coastal and open ocean surface waters to low micromolar (μM) in deep sea (Gibb et al., 1999; Gibb and Hatton, 2004). TMAO participates in various physiological processes in marine organisms (Seibel and Walsh, 2002). In deep-sea organisms, TMAO can act as a potent protein stabilizer, playing a central role in counteracting the protein-denaturing effect of urea (Ma et al., 2014; Liao et al., 2017; Ganguly et al., 2020). TMAO can also serve as a piezolyte, which can be accumulated in bacteria and fish to improve the survival of organisms at high hydrostatic pressure (Yancey et al., 2014; Yin et al., 2018; Qin et al., 2021). Furthermore, TMAO can be catabolized by marine bacteria to small, volatile, methylated amines (MAs), such as trimethylamine (TMA), dimethylamine (DMA) and monomethylamine (MMA), which are precursors of the greenhouse gas nitrous oxide (Dos Santos et al., 1998; Lidbury et al., 2017).
SAR11 bacteria are the most abundant oligotrophic bacteria in ocean surface waters, and play an important role in mineralizing marine DOM (Morris et al., 2002). Pelagibacter strain HTCC1062, the first cultivable SAR11 bacterium, can utilize TMAO to generate ATP (Giovannoni et al., 2005; Sun et al., 2011). The marine Roseobacter clade (MRC) bacterium Ruegeria pomeroyi DSS-3, which can grow with TMAO as the sole nitrogen source, can also utilize TMAO to produce intracellular ATP (Lidbury et al., 2015). The gene cluster encoding proteins for TMAO transport and metabolism has been identified in R. pomeroyi DSS-3 (Figure 1A; Lidbury et al., 2014, 2015, 2017). In strain DSS-3, TMAO can either be imported from marine environment by TmoXWV, an ATP-binding cassette (ABC) importer specific for TMAO, or be converted in vivo from TMA through the catalysis of TMA monooxygenase Tmm (Chen et al., 2011; Lidbury et al., 2014; Li et al., 2017). Then, TMAO in the cells is catabolized to DMA by TMAO demethylase Tdm (Lidbury et al., 2014; Sun et al., 2019). With the catalysis of DMA monooxygenase DmmDABC, DMA is further catabolized to MMA (Lidbury et al., 2017; Sun et al., 2019), which can be converted to γ-glutamylmethylamide by γ-glutamylmethylamide synthetase GmaS (Chen Y. et al., 2010; Wischer et al., 2015; Wang et al., 2021). Bioinformatic analysis indicated that tmoXWV homologs are prevalent in SAR11 bacteria (Lidbury et al., 2014). However, genomic analysis suggested that SAR11 bacteria lack dmmDABC (Figure 1B; Lidbury et al., 2017), which is essential for TMAO utilization as the nitrogen source in R. pomeroyi DSS-3. So far, it is still unknown whether SAR11 bacteria can utilize TMAO as a nitrogen source.
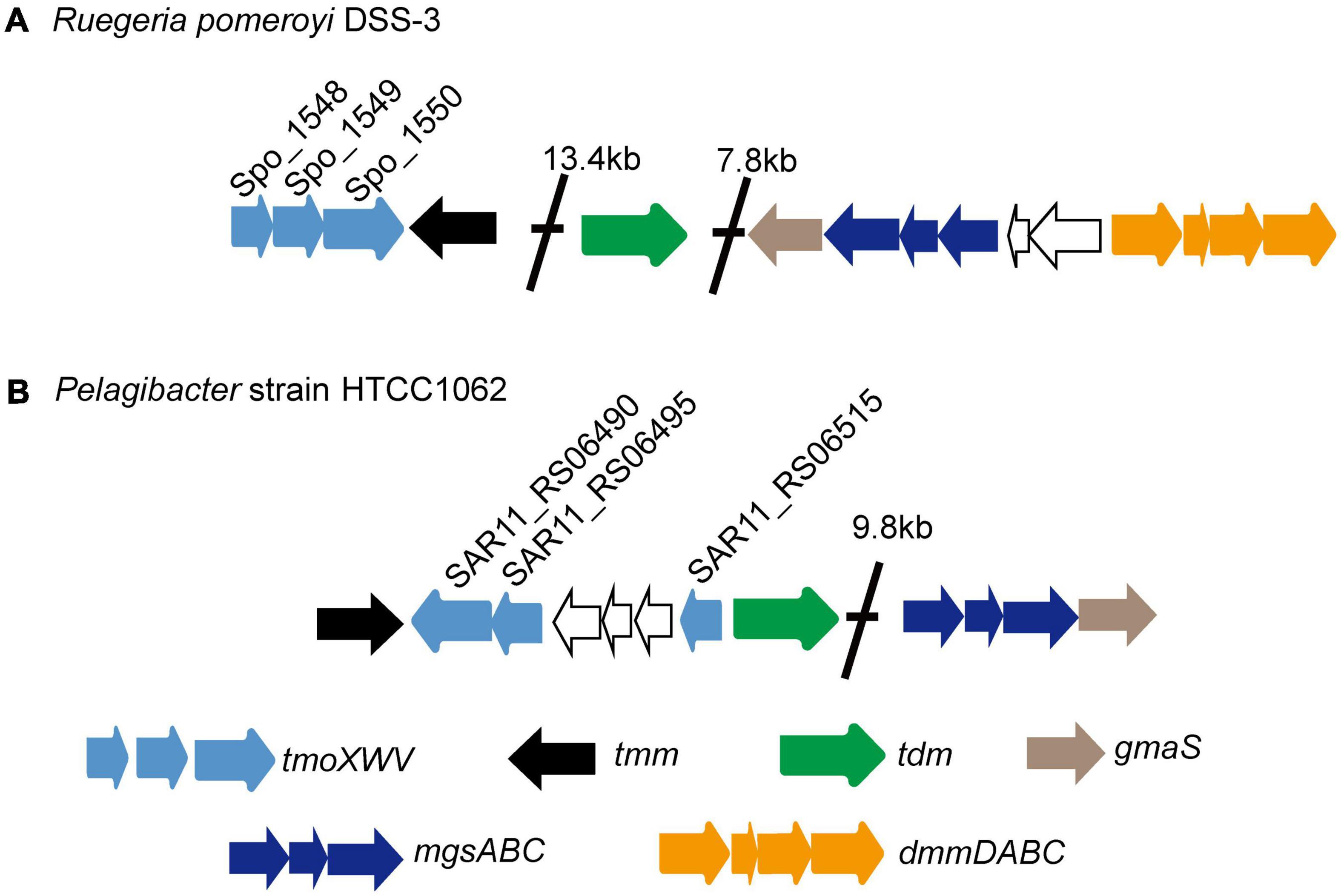
Figure 1. The gene clusters involved in TMAO transport and metabolism in the MRC bacterium R. pomeroyi DSS-3 (A) and in the SAR11 bacterium HTCC1062 (B). Tdm, trimethylamine N-oxide demethylase; Tmm, trimethylamine monooxygenase; TmoXVW, ATP-dependent TMAO transporter; DmmDABC, DMA monooxygenase; MgsABC, N-methylglutamate synthase; GmaS, γ-glutamylmethylamide synthetase.
In the TMAO transporter TmoXWV, TmoX is the periplasmic TMAO binding protein, the TMAO binding mechanism of which in R. pomeroyi DSS-3 has been revealed on the basis of structural and biochemical analyses (Li et al., 2015). Phylogenetic analysis indicated that TmoXWV belongs to the glycine betaine/proline betaine-type ABC transporter family, and TmoX belongs to the cluster F III of the ABC transporter superfamily (Lidbury et al., 2014). Cluster F III consists of substrate binding proteins specific for different compatible osmolytes, including betaine, carnitine, choline and TMAO (Berntsson et al., 2010; Lidbury et al., 2014; Rice et al., 2014; Beis, 2015). It has been found that TmoX homologs from MRC and those from SAR11 bacteria form two different branches in the phylogenetic tree (Lidbury et al., 2014). Therefore, the TMAO binding mechanism of SAR11 TmoX may have differences from that of R. pomeroyi DSS-3 TmoX.
This study aimed to investigate whether SAR11 bacteria can utilize TMAO as a nitrogen source and how they import TMAO with strain HTCC1062 as a model. We found that strain HTCC1062 can grow with TMAO as the sole nitrogen source. Genetic work demonstrated that TmoXWV1062, the TmoXWV homolog in HTCC1062, is a functional TMAO importer. TmoX1062, the periplasmic substrate binding protein of TmoXWV1062, was shown to have high binding affinities toward TMAO at 4–25°C by biochemical studies. The TMAO binding mechanism of TmoX1062 was further analyzed by structural modeling and mutational analysis.
Materials and Methods
Bacterial Strains and Growth Conditions
Strain HTCC1062 was cultured in AMS1 medium amended with 25 μM glycine, 10 μM methionine and 50 μM pyruvate at 16°C according to the reported protocol (Rappe et al., 2002; Tripp, 2013). AMS1 was sparged with CO2 for 5 h followed by sparging with air for 10 h. The pH of the resulting AMS1 typically ranged from 7.5 to 7.7. Cells of strain HTCC1062 were stained with SYBR Green I (Molecular Probes, America) and counted with a Guava Technologies flow cytometer (Millipore, America). The E. coli strains DH5α, BL21(DE3) and WM3064 were grown in the Lysogeny Broth (LB) medium at 37°C. Diaminopimelic acid (0.3 mM) was added into the LB medium to culture E. coli WM3064. R. pomeroyi DSS-3 was purchased from the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures and was cultured in 974 medium at 30°C according to the protocol provided1.
Real-Time qPCR Analysis
Strain HTCC1062 was firstly cultured in AMS1 medium amended with 25 μM glycine, 10 μM methionine and 50 μM pyruvate. When the concentration of cells reached 2 × 107 cells/ml, TMAO was added into the medium with a final concentration of 0.8 mM. The group without the addition of TMAO was set up as a control. After 0.5 or 2 h incubation, RNA was extracted from the cells using the RNeasy mini kit (Qiagen, America), and was subsequently reverse-transcribed to cDNA using Goldenstar™RT6 cDNA Synthesis Kit (TsingKe, China). The qPCR experiments were performed using a Light Cycler II 480 System (Roche, Switzerland) following the instructions of SYBR® Premix Ex TaqTM (TaKaRa, Japan) with the following cycling conditions: 95°C for 5 min, 45 cycles of 95°C for 10 s and 60°C for 30 s. The recA gene was used as an internal reference gene.
Genetic Manipulations
Deletion of the tmoW gene of R. pomeroyi DSS-3 was performed by pK18mobsacB-Ery based homolog recombination (Wang et al., 2015). The upstream and downstream sequences of the tmoW gene were amplified with primer sets tmoW-UP-F/tmoW-UP-R and tmoW-Down-F/tmoW-Down-R (Supplementary Table 1 and Supplementary Figure 1). Then, the PCR fragments were inserted to the vector pK18mobsacB-Ery with HindIII/BamHI as the restriction sites to generate pK18Ery-tmoW, which was transferred into E. coli WM3064. Next, the plasmid pK18Ery-tmoW was mobilized into R. pomeroyi DSS-3 by intergeneric conjugation with E. coli WM3064. To select for colonies in which the pK18Ery-tmoW had integrated into the R. pomeroyi DSS-3 genome by a single crossover event, cells were plated on the marine 2,216 agar plates containing erythromycin (25 μg/ml). Subsequently, the resultant mutant was cultured in the marine broth 2,216 medium and plated on the marine 2,216 agar plates containing 10% (w/v) sucrose to select for colonies in which the second recombination event occurred. For complementation of the ΔtmoW mutant, the tmoXWV1062 gene cluster with its native promoter was amplified from the genomic DNA of HTCC1062 with primer sets tmoXWV1062-350Up-F/tmoXWV1062-Down-R (Supplementary Table 1). The PCR fragments were digested with BamHI and EcoRI, and then inserted into the vector pHG101 to generate pHG101-tmoXWV1062. This plasmid was then transformed into E. coli WM3064, and mobilized into the ΔtmoW mutant of R. pomeroyi DSS-3 by conjugation.
Gene Cloning, Point Mutation, and Protein Expression and Purification
The full-length tmoX1062 gene was amplified from the genomic DNA of HTCC1062 by PCR using FastPfu DNA polymerase (TransGen Biotech, China), and was subcloned into the NdeI/XhoI restriction sites of the pET22b (Novagen, America) vector with a C-terminal His-tag. All of the point mutations in tmoX1062 were performed with the QuikChange® mutagenesis kit II (Agilent, America). The wild-type (WT) TmoX1062 protein and all of the mutants were expressed in E. coli strain BL21(DE3). The recombinant E. coli strains were cultured at 37°C in LB medium to an OD600 of 0.8–1.0 and then incubated at 16°C for 16 h with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) as an inducer for recombinant protein expression. The recombinant proteins were purified first with Ni-affinity column (GE Healthcare, America), and then with gel filtration on a Superdex-75 column (GE Healthcare, America) eluted with the buffer containing 10 mM Tris–HCl (pH 8.0) and 100 mM NaCl. Approximately 2 mg recombinant TmoX1062 protein was obtained from 1 liter of culture.
Isothermal Titration Calorimetry Measurements
Isothermal titration calorimetry (ITC) measurements were performed using a PEAQ-ITC system (Malvern, Britain). The sample cell was loaded with 250 μl of protein sample (30 μM), and the reference cell contained distilled water. The syringe was filled with 70 μl of TMAO (200 μM). The proteins and TMAO were kept in the same buffer containing 10 mM Tris–HCl (pH 8.0) and 100 mM NaCl. Titrations were carried out by adding 0.4 μl of TMAO for the first injection and 1.5 μl for the following 12 injections, with stirring at 750 rpm/min.
Circular-Dichroism Spectroscopic Assays
Wild-type TmoX1062 and all of the mutants were subjected to circular-dichroism (CD) spectroscopic assays at 20°C on a J-1500 spectropolarimeter (Jasco, Japan). CD spectra of the samples at a final concentration of approximately 10 μM were collected from 250 nm to 200 nm at a scan speed of 200 nm/min with a bandwidth of 1 nm. All of the samples were in a buffer containing 10 mM Tris–HCl (pH 8.0) and 100 mM NaCl. To determine the Tm of TmoX1062, the temperature was raised from 20 to 80°C in 1 h.
Results and Discussion
Pelagibacter Strain HTCC1062 Can Grow With Trimethylamine N-Oxide as the Sole Nitrogen Source
To investigate whether strain HTCC1062 can grow with TMAO as a nitrogen source, we replaced (NH4)2SO4 in AMS1 medium by TMAO. Methionine, which is usually used as the reduced sulfur, was also replaced by dimethylsulfoniopropionate (DMSP) to avoid the possible interference of its nitrogen atom. As shown in Figure 2A, strain HTCC1062 showed noticeable growth in the medium with TMAO as the sole nitrogen source, although its growth on TMAO was much weaker compared to that on (NH4)2SO4. This result suggests that strain HTCC1062 should contain TMAO transporter and enzymes involved in TMAO metabolism.
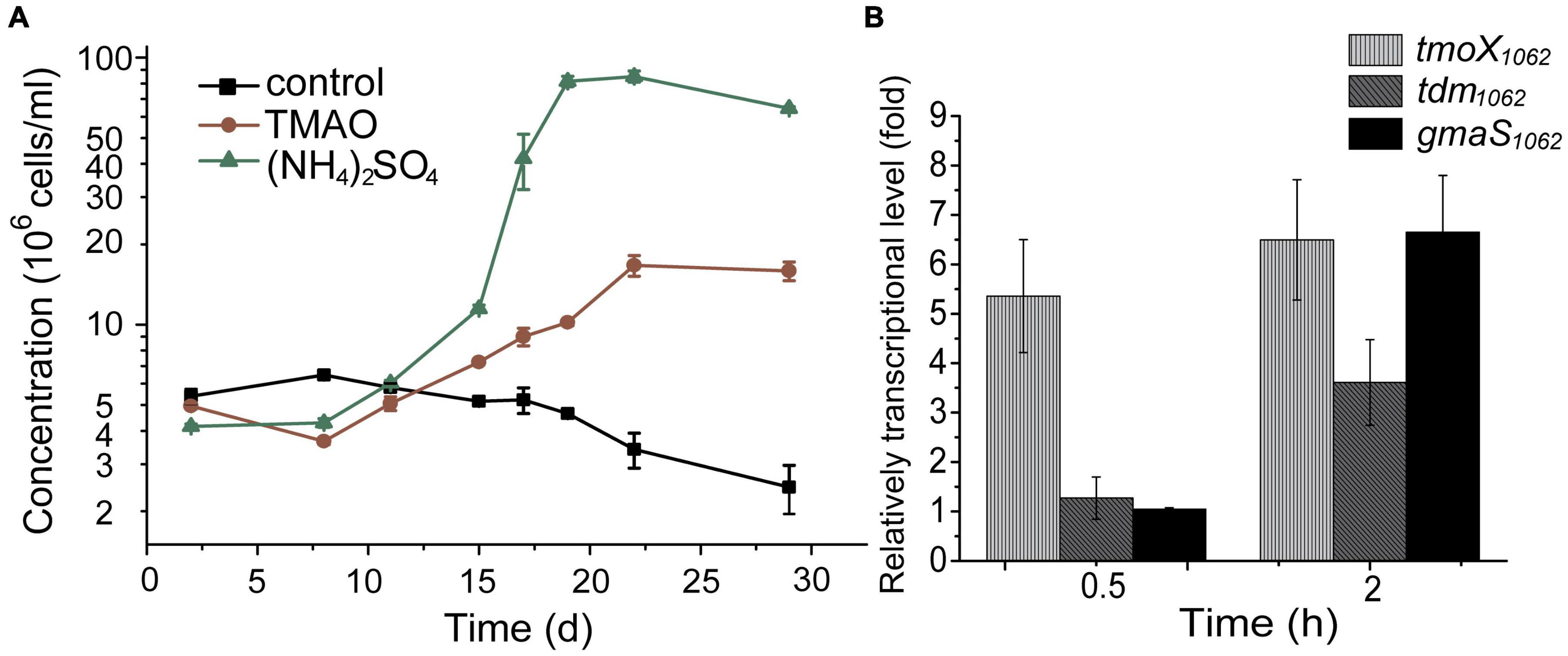
Figure 2. TMAO utilization by strain HTCC1062. (A) The growth curves of HTCC1062 with 0.8 mM TMAO or 0.4 mM (NH4)2SO4 as the sole nitrogen source. DMSP was used as the sulfur source. Bacterial cells cultured without nitrogen source were used as the control. The concentration of bacteria was determined by flow cytometry. (B) RT-qPCR assay of the transcriptions of genes possibly involved in the TMAO transport and metabolism in HTCC1062. The bacterium cultured without TMAO in the same medium was used as the control. The recA gene was used as an internal reference gene. The error bar represents standard deviation of triplicate experiments.
In R. pomeroyi DSS-3, TMAO can be transported into the cell through a TMAO specific transporter TmoXWV (Lidbury et al., 2014), and is then utilized as a nitrogen and energy source with the catalysis of several enzymes, including Tdm, DmmDABC and GmaS (Lidbury et al., 2015). Genomic analysis suggests that strain HTCC1062 possesses tmoXWV, tdm and gmaS homologs (tmoXWV1062, tdm1062 and gmaS1062, respectively). However, no dmmDABC homolog was identified from the genome of strain HTCC1062 (Lidbury et al., 2017; Figure 1B). RT-qPCR analysis showed that the transcriptions of tmoX1062, tdm1062 and gmaS1062 were all up-regulated by TMAO (Figure 2B), suggesting that these genes may be functional in TMAO import and metabolism. Thus, the SAR11 bacterial strain HTCC1062 may import and metabolize TMAO via a pathway generally similar to that of the MRC bacterial strain DSS-3, except that strain HTCC1062 may recruit an isoenzyme of DmmDABC to convert DMA to MMA. Next, we characterized TmoXWV1062 of strain HTCC1062 to investigate how SAR11 bacteria import TMAO in this study.
Functional Analysis of TmoXWV in HTCC1062
It has been reported that the tmoW-deleted mutation in R. pomeroyi DSS-3 disables its capacity to grow with TMAO as the sole nitrogen source (Lidbury et al., 2014). TmoXWV1062 of strain HTCC1062 shares ∼41% sequence identity to the functional TmoXWV of R. pomeryi DSS-3. Because currently genetic manipulation cannot be performed in SAR11 bacteria, we tried to demonstrate the TMAO-importing function of TmoXWV1062 in a tmoW-deleted mutant of R. pomeroyi DSS-3. We constructed the mutant ΔtmoWDSS–3 by deleting the majority of gene tmoW from the R. pomeroyi DSS-3 genome, and then complemented this mutant with tmoXWV1062 to generate the complemented strain ΔtmoWDSS–3-tmoXWV1062 that contains the tmoXWV1062 cluster from strain HTCC1062. As shown in Figure 3, the ΔtmoWDSS–3 mutant was unable to grow on TMAO, consistent with that previously reported (Lidbury et al., 2014). In contrast, the growth of the complemented strain ΔtmoWDSS–3-tmoXWV1062 on TMAO was comparable to that of strain R. pomeroyi DSS-3, suggesting that the tmoXWV1062 cluster was involved in TMAO transport in strain ΔtmoWDSS–3-tmoXWV1062. Considering that strain HTCC1062 can grow on TMAO (Figure 2A), this result indicates that tmoXWV1062 is most likely to encode a functional TMAO importer in strain HTCC1062.
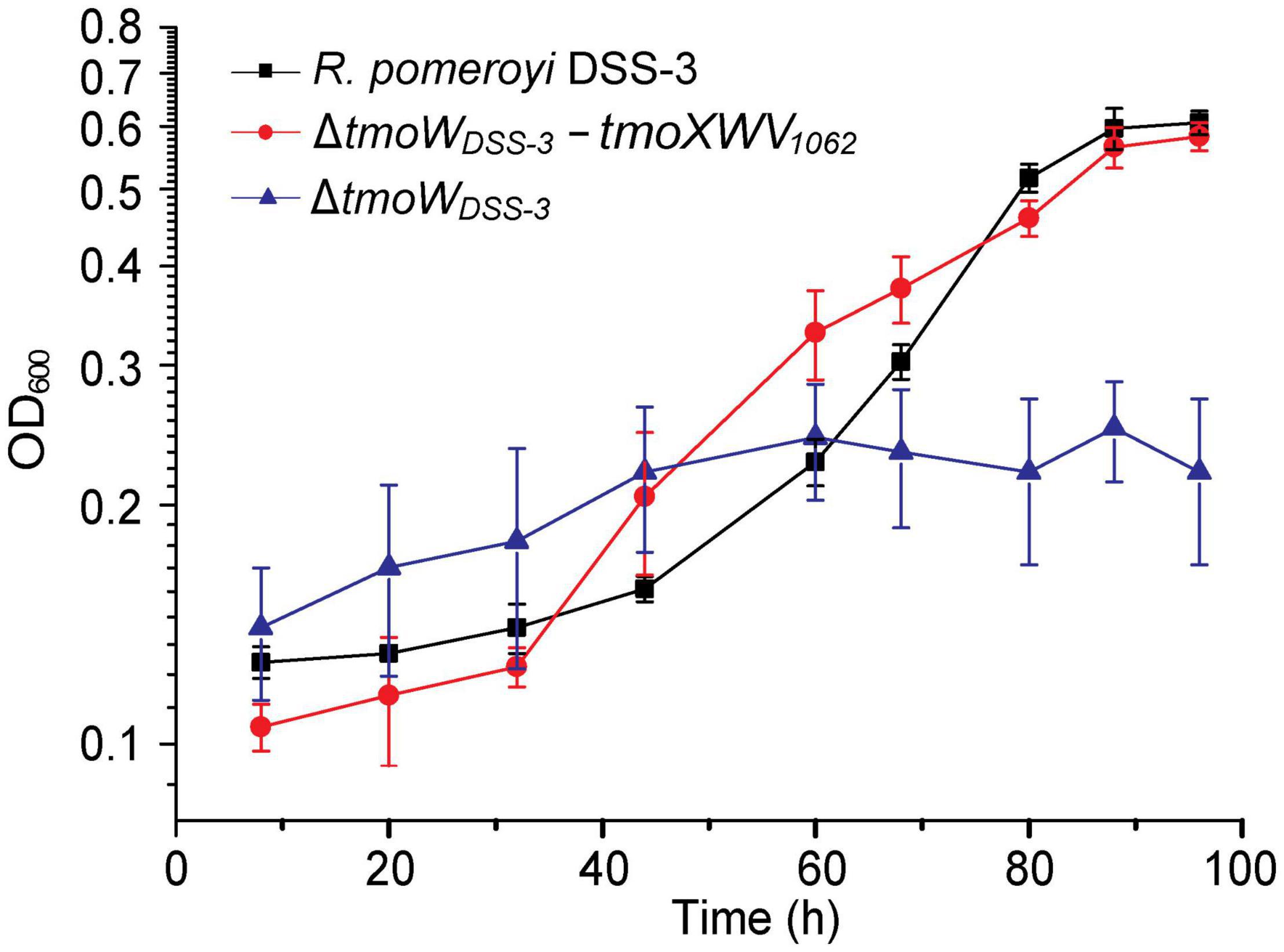
Figure 3. Growth curves of the WT R. pomeroyi DSS-3, the ΔtmoWDSS–3 mutant, and the complemented mutant ΔtmoWDSS–3-tmoXWV1062. All strains were cultivated with TMAO (2 mM) as the sole nitrogen source. The error bar represents standard deviation of triplicate experiments.
Characterization of TmoX1062
The periplasmic substrate binding protein of an ABC transporter is usually responsible for the first-step recognition of substrate, and can bind a given ligand with high affinity (Albers et al., 1999; Chen C. L. et al., 2010). To characterize TmoX1062, the substrate binding protein of TmoXWV1062, the full-length tmoX1062 gene containing 934 nucleotides was amplified from the genome of strain HTCC1062 and was expressed in E. coli BL21(DE3) cells. To analyze the substrate specificity of TmoX1062, the binding affinities of the recombinant TmoX1062 toward TMAO, betaine, choline, TMA, DMA and carnitine were determined by ITC measurements. Among the tested substrates, TmoX1062 possessed a high binding affinity toward TMAO, with a Kd (dissociation constant) of 520 nM (Figure 4A), but presented little binding affinity toward betaine, carnitine, TMA or DMA (Figure 4 and Table 1). Compared to TmoXDSS–3 of R. pomeroyi DSS-3, which exhibited a Kd of 1.6 μM toward TMAO (Li et al., 2015), TmoX1062 possessed a higher binding affinity toward TMAO. Considering the concentrations of TMAO range from nanomolar to low micromolar in marine environments (Gibb et al., 1999; Gibb and Hatton, 2004), the higher binding affinity of TmoX1062 toward TMAO would help strain HTCC1062 gain a competitive advantage over other bacteria at low TMAO concentrations. Surprisingly, the recombinant TmoX1062 also presented binding affinity toward choline, with a Kd of 2.5 μM (Figure 4B and Table 1). A similar phenomenon was also observed in TmoXDSS–3 (Li et al., 2015). RT-qPCR results indicated that choline did not induce the transcription of tmoX1062 in strain HTCC1062 (Supplementary Figure 2), suggesting that the binding of recombinant TmoX1062 toward choline may not make physiological sense. Alternatively, strain HTCC1062 may utilize TmoXWV1062 as a multifunctional transporter to import both TMAO and choline, as this strain possesses a highly streamlined genome (Giovannoni et al., 2005; Sun et al., 2011; Noell and Giovannoni, 2019).
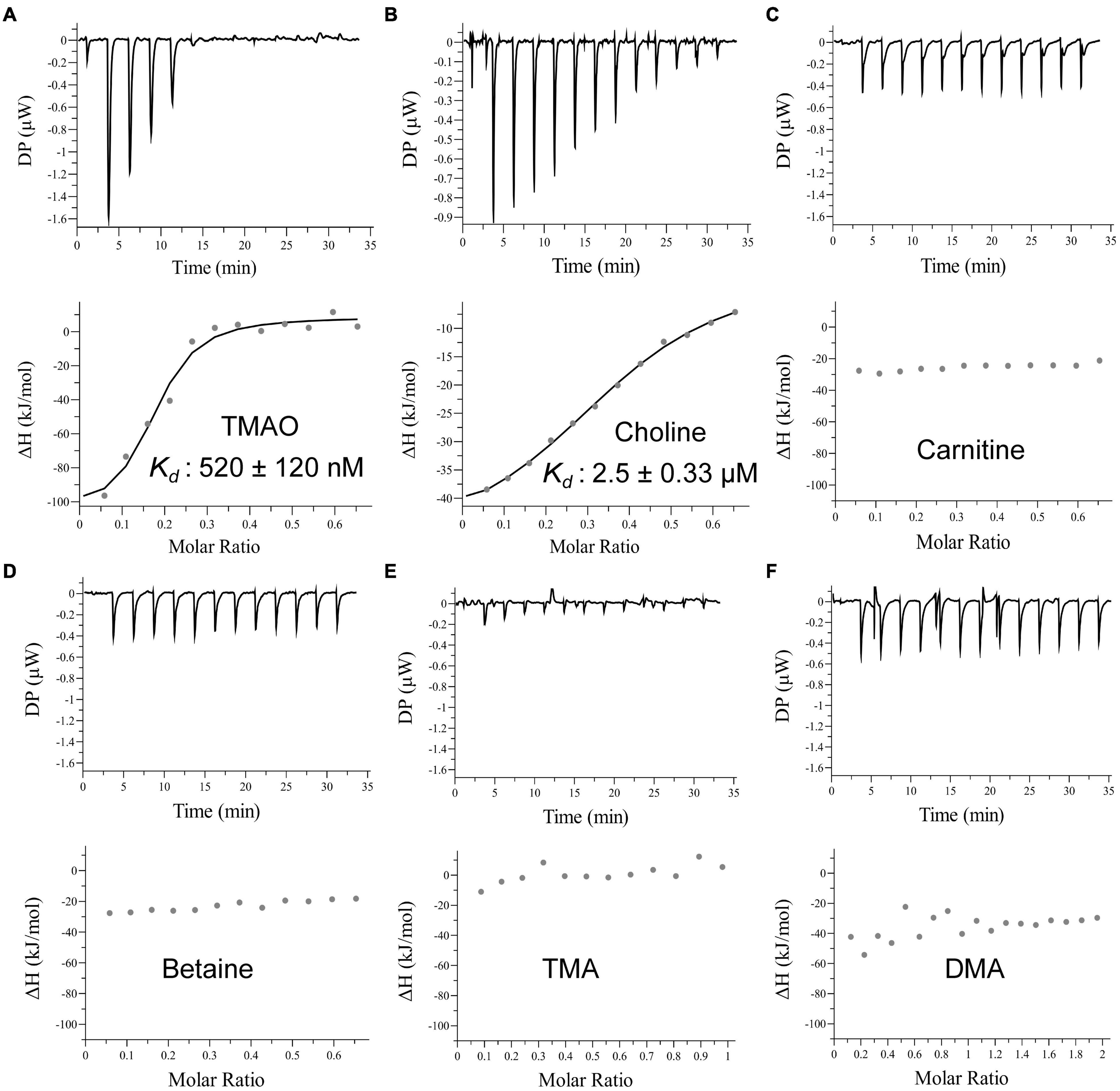
Figure 4. ITC data for titrations of different substrates into TmoX1062. ITC traces (top) and integrated binding isotherms (bottom) are shown. Substrates are shown in each integrated binding isotherm.
The tmoX gene is widespread in divergent marine bacteria, especially in SAR11 bacteria (Lidbury et al., 2014). The seawater temperatures are different at different depths and change with the seasons regularly, especially for surface seawaters (Malmstrom et al., 2010). Therefore, marine bacteria need to adapt different temperatures. To investigate the thermostability of TmoX1062, we measured the melting temperature (Tm) of TmoX1062. The Tm of TmoX1062 is 62.5°C (Figure 5A), which is higher than that of TmoXDSS–3 (Tm = 54.5°C) (Li et al., 2015), suggesting that TmoX1062 has higher thermostability than TmoXDSS–3. The binding affinities of TmoX1062 toward TMAO at different temperatures were also detected. TmoX1062 exhibited high binding affinities toward TMAO at 4°C (Figure 5B), 10°C (Figure 5C) and 25°C (Figure 5D), indicating that TmoXWV1062 should be able to import TMAO into cells of strain HTCC1062 efficiently at different temperatures. The nanomolar-level TMAO binding affinity, the high thermostability and the strong temperature adaptability of TmoX1062 may reflect the niche adaptation of HTCC1062 to the volatile marine environment, especially to the oligotrophic environment.
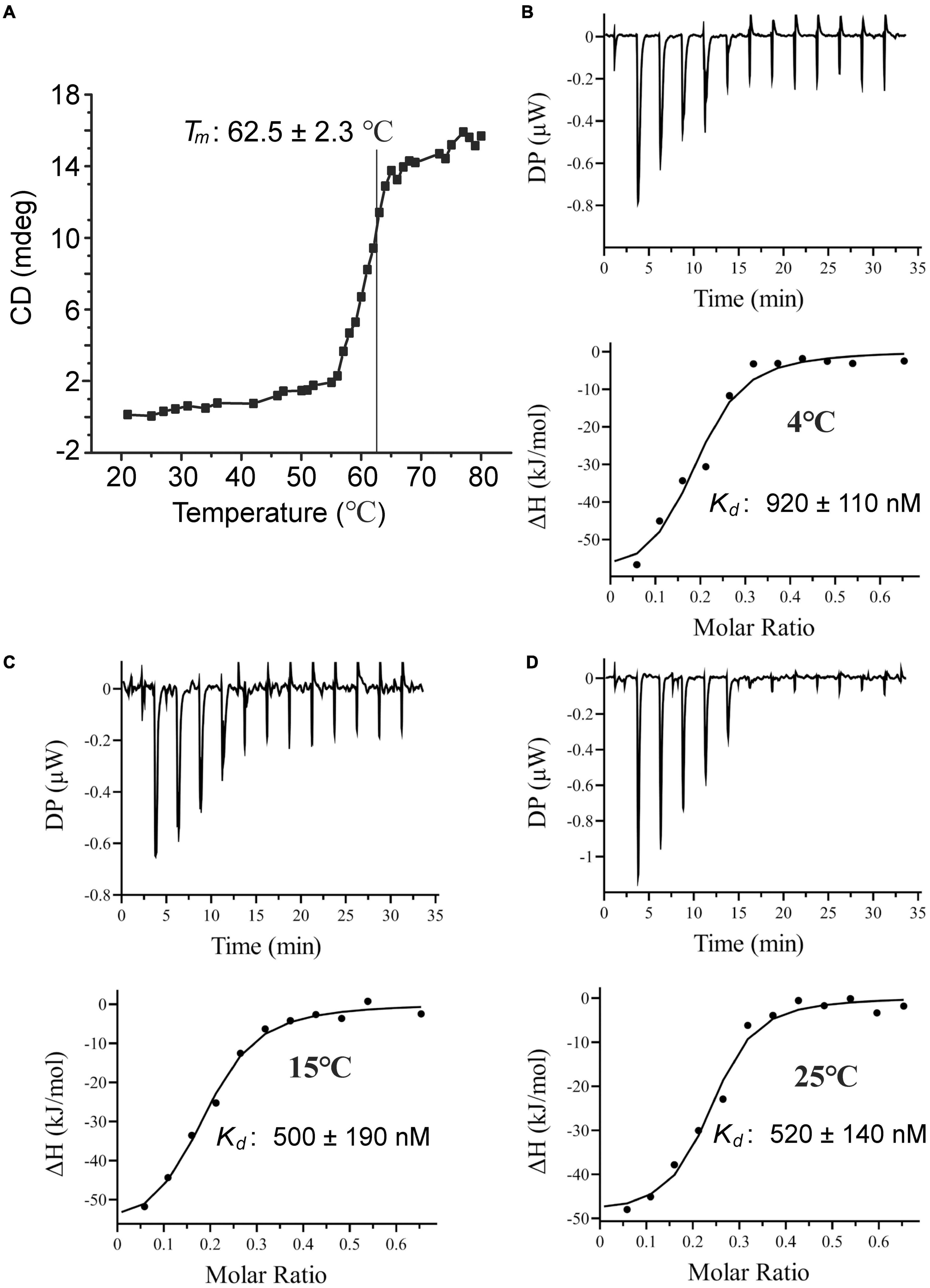
Figure 5. Characterization of TmoX1062. (A) The Tm of TmoX1062 determined by CD. (B–D) ITC data for titrations of TMAO into TmoX1062 at 4°C (B), 10°C (C), and 25°C (D).
Key Residues of TmoX1062 Involved in Binding Trimethylamine N-Oxide
The TMAO binding mechanism of TmoXDSS–3 has been proposed based on structural and mutational analyses (Li et al., 2015). In TmoXDSS–3, the TMAO binding pocket is composed of Trp55, Trp102, Phe106, Glu131, Trp177, Phe220, and Trp222 (Figure 6A), among which Glu131 forms a hydrogen bond with the oxygen atom of TMAO, and four tryptophan residues (Trp55, Trp102, Trp177, and Trp222) form a rectangular aromatic box and interact with TMAO by cation-π interactions (Li et al., 2015; Figure 6A). The aromatic rings of two phenylalanine residues (Phe106 and Phe220) also participate in forming the hydrophobic cage to accommodate TMAO (Li et al., 2015; Figure 6A). TmoX1062 shares ∼51% sequence identity with TmoXDSS–3, and the TMAO binding mechanism of TmoX1062 is still unclear.
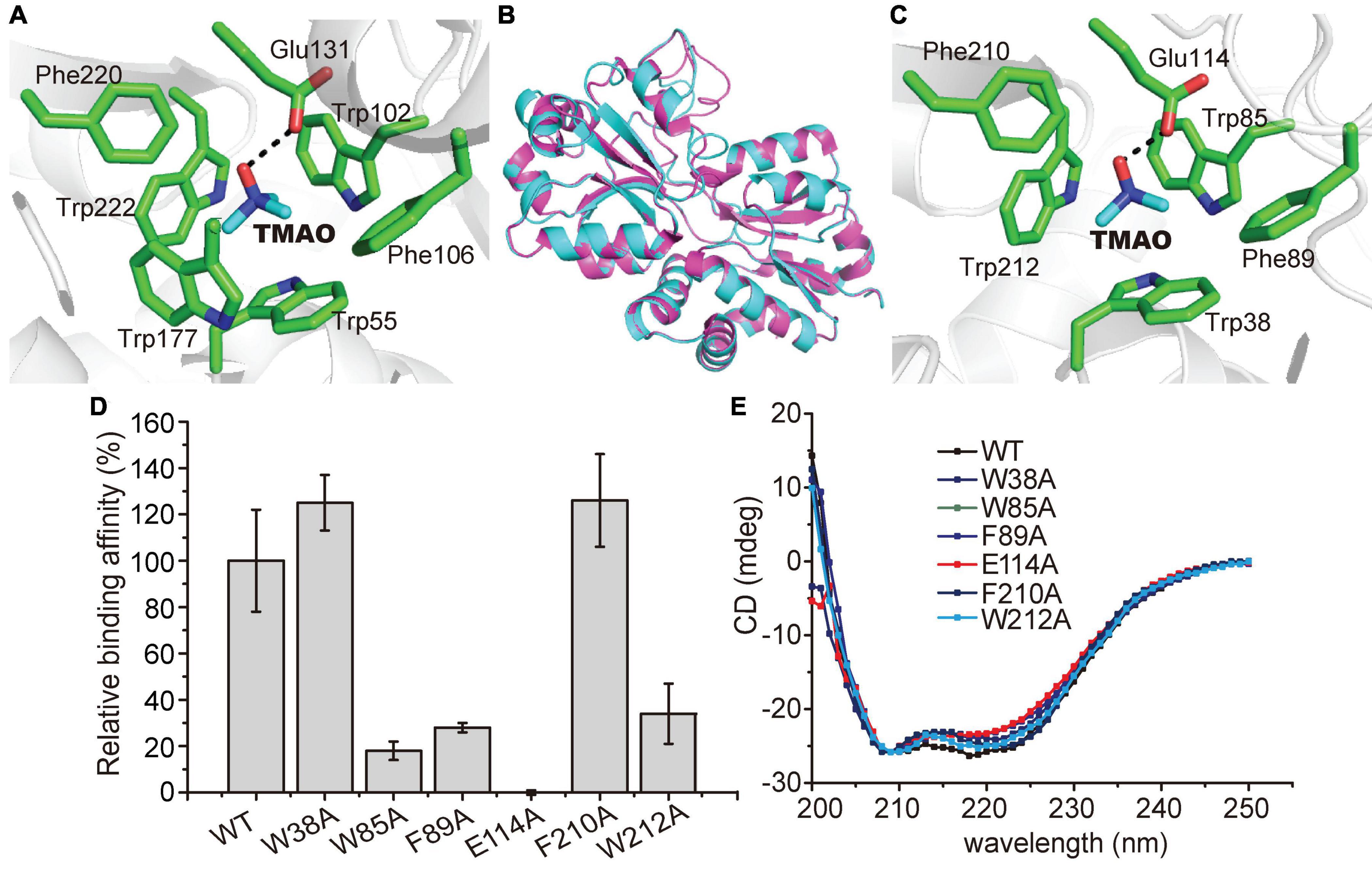
Figure 6. Possible residues involved in the binding of TMAO in TmoX1062. (A) Residues composing the TMAO binding pocket in TmoXDSS–3 (PDB code: 4XZ6). The TMAO molecule is colored in blue, and TmoXDSS–3 residues are colored in green. The possible hydrogen bond is represented by the dashed line. (B) Superimposed structures of TmoXDSS–3 (purple) and TmoX1062 (cyan). (C) Residues composing the TMAO binding pocket based on modeling structure of TmoX1062. The TMAO molecule is colored in blue, and TmoX1062 residues are colored in green. The possible hydrogen bond is represented by the dashed line. (D) Binding affinities of WT TmoX1062 and its mutants toward TMAO. The binding affinity of WT TmoX1062 was defined as 100%. The error bar represents standard deviation of triplicate experiments. (E) CD spectra of WT TmoX1062 and its mutants.
To probe the TMAO binding mechanism of TmoX1062, we tried to co-crystallize TmoX1062 and TMAO and solve the crystal structure of TmoX1062. However, all the attempts failed. We then modeled the structure of TmoX1062 via Swiss-model2 (Waterhouse et al., 2018), with the crystal structure of TmoXDSS–3 (PDB code: 4XZ6) as the template. The overall structure of TmoX1062 is similar to that of TmoXDSS–3 (Figure 6B), with a root mean square deviation (RMSD) between these two structures of 0.1 Å over 226 Cα atoms.
Structural analysis of the model of TmoX1062 showed that the binding pocket of TmoX1062 may be composed of Trp38, Trp85, Phe89, Glu114, Phe210, and Trp212 (Figure 6C), and Trp164, the corresponding residue of Trp177 in TmoXDSS–3 (Figure 6A), may not participate in binding TMAO. The residue Glu114, corresponding to Glu131 in TmoXDSS–3, may form a hydrogen bond with TMAO (Figure 6C). Mutation of Glu114 to alanine abolished the binding affinity of TmoX1062 toward TMAO (Figure 6D), indicating the important role of Glu114 in binding TMAO. The side chains of Trp38, Trp85 and Trp212 (corresponding to Trp55, Trp102 and Trp222 in TmoXDSS–3, respectively) form an aromatic box (Figure 6C). Together with the side chains of Phe89 and Phe210 (corresponding to Phe106 and Phe220 in TmoXDSS–3), this box forms a hydrophobic cage to accommodate the quaternary amine of TMAO (Figure 6C). Mutations of Trp85, Phe89 and Trp212 to alanine severely deceased the binding affinity of TmoX1062 toward TMAO, suggesting the important roles of these three residues in substrate binding (Figure 6D). However, mutants W38A and F210A still maintained a relatively high TMAO binding affinity (Figure 6D), indicating that these two residues may not participate in TMAO binding, or the other residues of TmoX1062 may compensate the function of Trp38 and Phe210. In TmoXDSS–3, mutations of the corresponding residues composing the TMAO binding pocket all decreased its TMAO binding affinity to a large extent (Li et al., 2015). Therefore, our biochemical results suggested that several residues of TmoX1062 participating in TMAO binding may be different from those of TmoXDSS–3, although sequence analysis showed that the residues comprising the binding pocket of TmoX are all highly conserved in the MRC and the SAR11 clade (Li et al., 2015). CD spectroscopy assays showed that the secondary structures of the mutants are similar to that of WT TmoX1062 (Figure 6E), indicating that the decrease in the binding affinities of the mutants is a result of residue replacement rather than structural changes.
Conclusion
Trimethylamine N-oxide is widespread in the oceans, and can be utilized by marine bacteria as carbon, nitrogen and/or energy source (Lidbury et al., 2015, 2017). The SAR11 bacteria are widespread in marine environment (Brown et al., 2012). Here, our results showed that the SAR11 bacterium HTCC1062 is capable of utilizing TMAO as a nitrogen source for growth, which likely absorbs TMAO via the ABC transporter TmoXWV1062. The periplasmic substrate binding protein TmoX1062 of this transporter has high binding affinity toward TMAO, and exhibits a relatively high thermostability and strong temperature adaptability, which may reflect the niche adaptation of HTCC1062 to the oligotrophic marine environment. Mutational analysis indicated that the TMAO binding mechanism of TmoX1062 may have differences from the previously reported mechanism of TmoXDSS–3 of MRC bacteria. This study provides insights into how SAR11 bacteria utilize TMAO and offers a better understanding of marine nitrogen cycling.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
C-YL and Y-ZZ designed the research. X-LC and J-MD directed the research. CG performed the experiments. NZ and X-YH helped in experiments. NW, X-YZ, and PW helped in data analysis. CG, C-YL, and X-LC wrote the manuscript. C-YL edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key R&D Program of China (2021YFA0909600 and 2018YFC1406700), the National Science Foundation of China (grants 91851205, 42076229, 31961133016, 31630012, U1706207 and 31870052), the Fundamental Research Funds for the Central Universities (202041011), the Major Scientific and Technological Innovation Project (MSTIP) of Shandong Province (2019JZZY010817), the Program of Shandong for Taishan Scholars (tspd20181203), AoShan Talents Cultivation Program Supported by Qingdao National Laboratory for Marine Science and Technology (2017ASTCP-OS14 and QNLM2016ORP0310), the grant of Laboratory for Marine Biology and Biotechnology (OF2019NO02), and Pilot National Laboratory for Marine Science and Technology (Qingdao).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Zhifeng Li from State Key laboratory of Microbial Technology of Shandong University for her help in ITC experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.838608/full#supplementary-material
Footnotes
References
Albers, S. V., Elferink, M. G. L., Charlebois, R. L., Sensen, C. W., Driessen, A. J. M., and Konings, W. N. (1999). Glucose transport in the extremely thermoacidophilic Sulfolobus solfataricus involves a high-affinity membrane-integrated binding protein. J. Bacteriol. 181, 4285–4291. doi: 10.1128/Jb.181.14.4285-4291.1999
Azam, F., and Malfatti, F. (2007). Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 5, 782–791. doi: 10.1038/nrmicro1747
Beis, K. (2015). Structural basis for the mechanism of ABC transporters. Biochem. Soc. Trans. 43, 889–893. doi: 10.1042/Bst20150047
Berntsson, R. P. A., Smits, S. H. J., Schmitt, L., Slotboom, D. J., and Poolman, B. (2010). A structural classification of substrate-binding proteins. FEBS Lett. 584, 2606–2617. doi: 10.1016/j.febslet.2010.04.043
Brown, M. V., Lauro, F. M., DeMaere, M. Z., Muir, L., Wilkins, D., Thomas, T., et al. (2012). Global biogeography of SAR11 marine bacteria. Mol. Syst. Biol. 8:595. doi: 10.1038/msb.2012.28
Carpenter, L. J., Archer, S. D., and Beale, R. (2012). Ocean-atmosphere trace gas exchange. Chem. Soc. Rev. 41, 6473–6506. doi: 10.1039/c2cs35121h
Chen, C. L., Malek, A. A., Wargo, M. J., Hogan, D. A., and Beattie, G. A. (2010). The ATP-binding cassette transporter Cbc (choline/betaine/carnitine) recruits multiple substrate-binding proteins with strong specificity for distinct quaternary ammonium compounds. Mol. Microbiol. 75, 29–45. doi: 10.1111/j.1365-2958.2009.06962.x
Chen, Y., Patel, N. A., Crombie, A., Scrivens, J. H., and Murrell, J. C. (2011). Bacterial flavin-containing monooxygenase is trimethylamine monooxygenase. Proc. Natl. Acad. Sci. U.S.A. 108, 17791–17796. doi: 10.1073/pnas.1112928108
Chen, Y., Scanlan, J., Song, L. J., Crombie, A., Rahman, M. T., Schafer, H., et al. (2010). Gamma-Glutamylmethylamide is an essential intermediate in the metabolism of methylamine by Methylocella silvestris. Appl. Environ. Microbiol. 76, 4530–4537. doi: 10.1128/Aem.00739-10
Dos Santos, J. P., Iobbi-Nivol, C., Couillault, C., Giordano, G., and Mejean, V. (1998). Molecular analysis of the trimethylamine N-oxide (TMAO) reductase respiratory system from a Shewanella species. J. Mol. Biol. 284, 421–433. doi: 10.1006/jmbi.1998.2155
Falkowski, P. G., Barber, R. T., and Smetacek, V. V. (1998). Biogeochemical controls and feedbacks on ocean primary production. Science 281, 200–207. doi: 10.1126/science.281.5374.200
Ganguly, P., Polak, J., van der Vegt, N. F. A., Heyda, J., and Shea, J. E. (2020). Protein stability in TMAO and mixed Urea-TMAO solutions. J. Phys. Chem. B 124, 6181–6197. doi: 10.1021/acs.jpcb.0c04357
Gibb, S. W., and Hatton, A. D. (2004). The occurrence and distribution of trimethylamine N-oxide in Antarctic coastal waters. Mar. Chem. 91, 65–75. doi: 10.1016/j.marchem.2004.04.005
Gibb, S. W., Mantoura, R. F. C., Liss, P. S., and Barlow, R. G. (1999). Distributions and biogeochemistries of methylamines and ammonium in the Arabian Sea. Deep Sea Res. 2 Top. Stud. Oceanogr. 46, 593–615. doi: 10.1016/S0967-0645(98)00119-2
Giovannoni, S. J., Tripp, H. J., Givan, S., Podar, M., Vergin, K. L., Baptista, D., et al. (2005). Genome streamlining in a cosmopolitan oceanic bacterium. Science 309, 1242–1245. doi: 10.1126/science.1114057
Li, C. Y., Chen, X. L., Shao, X., Wei, T. D., Wang, P., Xie, B. B., et al. (2015). Mechanistic insight into trimethylamine N-Oxide recognition by the marine bacterium Ruegeria pomeroyi DSS-3. J. Bacteriol. 197, 3378–3387. doi: 10.1128/Jb.00542-15
Li, C. Y., Chen, X. L., Zhang, D., Wang, P., Sheng, Q., Peng, M., et al. (2017). Structural mechanism for bacterial oxidation of oceanic trimethylamine into trimethylamine N-oxide. Mol. Microbiol. 103, 992–1003. doi: 10.1111/mmi.13605
Liao, Y. T., Manson, A. C., DeLyser, M. R., Noid, W. G., and Cremer, P. S. (2017). Trimethylamine N-oxide stabilizes proteins via a distinct mechanism compared with betaine and glycine. Proc. Natl. Acad. Sci. U.S.A. 114, 2479–2484. doi: 10.1073/pnas.1614609114
Lidbury, I., Mausz, M. A., Scanlan, D. J., and Chen, Y. (2017). Identification of dimethylamine monooxygenase in marine bacteria reveals a metabolic bottleneck in the methylated amine degradation pathway. ISME J. 11, 1592–1601. doi: 10.1038/ismej.2017.31
Lidbury, I., Murrell, J. C., and Chen, Y. (2014). Trimethylamine N-oxide metabolism by abundant marine heterotrophic bacteria. Proc. Natl. Acad. Sci. U.S.A. 111, 2710–2715. doi: 10.1073/pnas.1317834111
Lidbury, I. D. E. A., Murrell, J. C., and Chen, Y. (2015). Trimethylamine and trimethylamine N-oxide are supplementary energy sources for a marine heterotrophic bacterium: implications for marine carbon and nitrogen cycling. ISME J. 9, 760–769. doi: 10.1038/ismej.2014.149
Ma, J. Q., Pazos, I. M., and Gai, F. (2014). Microscopic insights into the protein-stabilizing effect of trimethylamine N-oxide (TMAO). Proc. Natl. Acad. Sci. U.S.A. 111, 8476–8481. doi: 10.1073/pnas.1403224111
Malmstrom, R. R., Coe, A., Kettler, G. C., Martiny, A. C., Frias-Lopez, J., Zinser, E. R., et al. (2010). Temporal dynamics of Prochlorococcus ecotypes in the Atlantic and Pacific oceans. ISME J. 4, 1252–1264. doi: 10.1038/ismej.2010.60
Morris, R. M., Rappe, M. S., Connon, S. A., Vergin, K. L., Siebold, W. A., Carlson, C. A., et al. (2002). SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420, 806–810. doi: 10.1038/nature01240
Noell, S. E., and Giovannoni, S. J. (2019). SAR11 bacteria have a high affinity and multifunctional glycine betaine transporter. Environ. Microbiol. 21, 2559–2575. doi: 10.1111/1462-2920.14649
Qin, Q. L., Wang, Z. B., Su, H. N., Chen, X. L., Miao, J., Wang, X. J., et al. (2021). Oxidation of trimethylamine to trimethylamine N-oxide facilitates high hydrostatic pressure tolerance in a generalist bacterial lineage. Sci. Adv. 7:eabf9941. doi: 10.1126/sciadv.abf9941
Rappe, M. S., Connon, S. A., Vergin, K. L., and Giovannoni, S. J. (2002). Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418, 630–633. doi: 10.1038/nature00917
Rice, A. J., Park, A., and Pinkett, H. W. (2014). Diversity in ABC transporters: type I, II and III importers. Crit. Rev. Biochem. Mol. 49, 426–437. doi: 10.3109/10409238.2014.953626
Seibel, B. A., and Walsh, P. J. (2002). Trimethylamine N-oxide accumulation in marine animals: relationship to acylglycerol storage. J. Exp. Biol. 205, 297–306.
Sun, J., Mausz, M. A., Chen, Y., and Giovannoni, S. J. (2019). Microbial trimethylamine metabolism in marine environments. Environ. Microbiol. 21, 513–520. doi: 10.1111/1462-2920.14461
Sun, J., Steindler, L., Thrash, J. C., Halsey, K. H., Smith, D. P., Carter, A. E., et al. (2011). One carbon metabolism in SAR11 pelagic marine bacteria. PLoS One 6:e23973. doi: 10.1371/journal.pone.0023973
Tripp, H. J. (2013). The unique metabolism of SAR11 aquatic bacteria. J. Microbiol. 51, 147–153. doi: 10.1007/s12275-013-2671-2
Wang, N., Chen, X. L., Gao, C., Peng, M., Wang, P., Zhang, N., et al. (2021). Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine. J. Biol. Chem. 296:100081. doi: 10.1074/Jbc.Ra120.015952
Wang, P. X., Yu, Z. C., Li, B. Y., Cai, X. S., Zeng, Z. S., Chen, X. L., et al. (2015). Development of an efficient conjugation-based genetic manipulation system for Pseudoalteromonas. Microb. Cell Fact. 14:11. doi: 10.1186/s12934-015-0194-8
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Wischer, D., Kumaresan, D., Johnston, A., El Khawand, M., Stephenson, J., Hillebrand-Voiculescu, A. M., et al. (2015). Bacterial metabolism of methylated amines and identification of novel methylotrophs in Movile Cave. ISME J. 9, 195–206. doi: 10.1038/ismej.2014.102
Yancey, P. H., Gerringer, M. E., Drazen, J. C., Rowden, A. A., and Jamieson, A. (2014). Marine fish may be biochemically constrained from inhabiting the deepest ocean depths. Proc. Natl. Acad. Sci. U.S.A. 111, 4461–4465. doi: 10.1073/pnas.1322003111
Keywords: TMAO, ABC transporter, substrate binding protein, SAR11 bacteria, niche adaptation
Citation: Gao C, Zhang N, He X-Y, Wang N, Zhang X-Y, Wang P, Chen X-L, Zhang Y-Z, Ding J-M and Li C-Y (2022) Characterization of the Trimethylamine N-Oxide Transporter From Pelagibacter Strain HTCC1062 Reveals Its Oligotrophic Niche Adaption. Front. Microbiol. 13:838608. doi: 10.3389/fmicb.2022.838608
Received: 18 December 2021; Accepted: 07 February 2022;
Published: 28 February 2022.
Edited by:
Mohamed Jebbar, Université de Bretagne Occidentale, FranceReviewed by:
Luis Manuel Bolaños, University of Exeter, United KingdomKaoru Nakasone, Kindai University, Japan
Copyright © 2022 Gao, Zhang, He, Wang, Zhang, Wang, Chen, Zhang, Ding and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun-Mei Ding, ZGptMzQxN0AxNjMuY29t; Chun-Yang Li, TGN5QG91Yy5lZHUuY24=
 Chao Gao
Chao Gao Nan Zhang4
Nan Zhang4 Xi-Ying Zhang
Xi-Ying Zhang Peng Wang
Peng Wang Xiu-Lan Chen
Xiu-Lan Chen Yu-Zhong Zhang
Yu-Zhong Zhang Jun-Mei Ding
Jun-Mei Ding Chun-Yang Li
Chun-Yang Li