
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 27 April 2022
Sec. Microbial Physiology and Metabolism
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.838042
Iron acquisition and modulation of its intracellular concentration are critical for the development of all living organisms. So far, several proteins have been described to be involved in iron homeostasis. Among them, ferritins act as the major iron storage proteins, sequestering internalized iron and modulating its concentration inside bacterial cells. We previously described that the deletion of the 3’-untranslated region (3’UTR) of the ftnA gene, which codes for ferritin in Staphylococcus aureus, increased the ftnA mRNA and ferritin levels. Here, we show that the ferritin levels are affected by RNase III and PNPase, which target the ftnA 3’UTR. Rifampicin mRNA stability experiments revealed that the half-life of the ftnA mRNA is affected by both RNase III and the ftnA 3’UTR. A transcriptional fusion of the ftnA 3’UTR to the gfp reporter gene decreased green fluorescent protein (GFP) expression, indicating that the ftnA 3’UTR could work as an independent module. Additionally, a chromosomal deletion of the ftnA 3’UTR impaired S. aureus growth under conditions of iron starvation. Overall, this work highlights the biological relevance of the ftnA 3’UTR for iron homeostasis in S. aureus.
Iron is an essential micronutrient for several biological processes in living cells such as oxygen transport, methanogenesis, the tricarboxylic cycle (TCA), gene regulation, and DNA biosynthesis (Andrews et al., 2003; Price and Boyd, 2020). When infecting the host, pathogenic bacteria like Staphylococcus aureus need to overcome the restricted availability of free iron, a process known as nutritional immunity (Hood and Skaar, 2012; Marchetti et al., 2020). Staphylococcus aureus, one of the most relevant nosocomial bacteria worldwide (Tong et al., 2015), has developed diverse strategies to obtain iron by secreting siderophores and hemophores that chelate iron and heme molecules, respectively, as well as expressing iron and heme uptake systems (Wandersman and Delepelaire, 2004; Haley and Skaar, 2012).
Although iron is essential for life, at high cytosolic concentrations, it can be toxic for the bacterium. This is due to the formation of reactive oxygen species (ROS) that cause oxidative stress and damage DNA as well as other essential molecules (Meneghini, 1997; Price and Boyd, 2020). For this reason, bacteria dedicate a significant amount of resources to regulating intracellular iron concentration. In S. aureus, the ferric uptake regulator (Fur) and peroxide stress transcriptional regulator (PerR) are the two main transcriptional regulators that modulate the excess of iron and oxidative stress. When high levels of iron are sensed inside the cell, Fur inhibits the transcription of several genes involved in iron and heme uptake and promotes the expression of efflux pumps (Horsburgh et al., 2001; Friedman et al., 2006; Troxell and Hassan, 2013). Alternatively, the iron surplus is dealt with by the storage ferritin protein, FtnA, whose levels are increased under iron-rich conditions. However, when iron is scarce, ftnA expression is transcriptionally repressed by PerR (Morrissey et al., 2004; Zühlke et al., 2016).
3’-untranslated regions (3’UTRs) have recently emerged as impactful post-transcriptional regulatory elements that regulate the levels of the mRNAs in which they are encoded through different mechanisms (Miyakoshi et al., 2015; Zhao et al., 2018; Menendez-Gil and Toledo-Arana, 2021). For example, the 3’UTR of the icaR mRNA modulates the production of IcaR, the main repressor of PIA-PNAG exopolysaccharide biosynthesis in S. aureus. This 3’UTR contains a UCCCC motif that binds to the Shine-Dalgarno (SD) sequence at the 5’UTR of the same mRNA and inhibits ribosome binding and translation. This interaction results in the formation of a double-stranded RNA (dsRNA) substrate for endoribonuclease III (RNase III) to process (de Los Mozos et al., 2013). The 3’UTR of the Bacillus subtilis hbs mRNA follows a similar pattern, interacting with its own 5’UTR, but in this case to prevent RNase Y cleavage (Braun et al., 2017). Another way in which 3’UTRs may influence RNA stability and protein expression is by carrying AU-rich motifs that are processed by RNase E and/or PNPase. This has been described for the Yersinia pestis hmsT, Salmonella enterica hilD, and Corynebacterium glutamicum aceA mRNAs, among others (Maeda and Wachi, 2012; López-Garrido et al., 2014; Zhu et al., 2016; Zhao et al., 2018). 3’UTRs can also interact with RNA-binding proteins (RBPs) and small RNAs (sRNAs) to regulate specific biological processes. In Escherichia coli, under iron deprivation conditions, the apo-AcnB protein binds to its own mRNA at a loop located in its 3’UTR to protect it against the degradosome (Benjamin and Massé, 2014). The Salmonella hilD and S. aureus icaR 3’UTRs are targeted by the Spot42 and RsaI sRNAs, respectively, promoting protein expression (El-Mouali et al., 2018; Bronesky et al., 2019). Additionally, 3’UTRs can be reservoirs of trans-acting sRNAs that can be generated either by an internal promoter (type I) or by an mRNA processing site (type II) located at the 3’UTR (Miyakoshi et al., 2015).
In a previous study, we unveiled that the S. aureus ftnA gene expression was modulated by its own 3’UTR. Deletion of the ftnA 3’UTR increased ferritin expression, portraying said 3’UTR as a putative post-transcriptional regulatory element (Menendez-Gil et al., 2020). Here, we show that the ftnA 3’UTR could act as an independent regulatory module that is targeted by RNase III and PNPase to decrease ferritin expression. Deletion of the ftnA 3’UTR impaired bacterial growth under iron starvation conditions. In this scenario, the ftnA 3’UTR-mediated regulation would play an essential role in achieving a tightly regulated iron homeostasis in S. aureus.
Bacterial strains, plasmids, and oligonucleotides used in this study are listed in Tables 1–3, respectively. Staphylococcus aureus strains were grown in Tryptic Soy Broth (Pronadisa) supplemented with 0.25% glucose (TSBg) or, when indicated, in modified chemically defined medium (Toledo-Arana et al., 2005). To prepare S. aureus and E. coli competent cells, B2 (casein hydrolysate, 10 g L−1; yeast extract, 25 g L−1; NaCl, 25 g L−1; K2HPO4, 1 g L−1; glucose, and 5 g L−1; pH 7.5), and SuperBroth (tryptone, 30 g L−1; yeast extract, 20 g L−1; and MOPS, 10 g L−1; pH 7) media were used, respectively. For selective growth, media were supplemented with the appropriate antibiotics at the following concentrations: ampicillin (Amp), 100 μg ml−1 for E. coli transformants; erythromycin (Erm), 1,5 μg ml−1 or 10 μg ml−1 for S. aureus cells harboring the pMAD or pCN plasmids, respectively.
Most of the plasmids used in this study were engineered as previously described (Caballero et al., 2018; Menendez-Gil et al., 2020). pMAD plasmids used for chromosomal deletions were constructed by amplifying flanking sequences (AB and CD) of the target regions using primers A/B and C/D (Table 3). PCR fragments were digested and ligated into pMAD through a double-fragment ligation process using BamHI, EcoRI, and KpnI or NheI (Tables 2, 3).
The green fluorescent protein (GFP) reporter plasmids were constructed using the Listeria monocytogenes pAD-cGFP plasmid as a template (Balestrino et al., 2010). To build pGFP, the hly 5’UTR and GFP sequences were amplified with primers Sal-GFP-fw and BcuI-TT-BamHI-GFP-rvs (Table 3) and the resulting PCR fragment was cloned into the pEW plasmid. The 3’UTR of ftnA was amplified using primers BamHI-EcoRI-3UTR-ftn-fw and SmaI-3UTR-ftn-rvs (Table 3) and inserted downstream of the gfp gene using restriction sites BamHI and SmaI. The pGFP-Δ3’UTRftnA was constructed using the pGFP-3’UTRftnA as a template and primers SalI-GFP-fw and KpnI-D3UTR-term-ftn (Table 3). The amplification product was ligated into the pGFP plasmid using SalI and KpnI.
The plasmids expressing 3XFFtnAΔ3’UTR19-56 and 3XFFtnAΔ3’UTR57-93 were constructed using the p3xFFtnA plasmid backbone. For the 3XFFtnAΔ3’UTR19-56 plasmid, an overlapping PCR was performed using oligonucleotide pairs +1-ftn and 3’UTR-ftn-19-56-fw and 3’UTR-ftn-19-56-rv and term-ftn. Analogously, oligonucleotides +1-ftn and 3UTR-ftn-term-1/2 were used for the 3XFFtnAΔ3’UTR57-93 plasmid (Table 3). The resulting amplicons were then inserted into pEW using BamHI and KpnI (Table 2).
Precultures were grown in 5 ml of TSBg supplemented with Erm (TSBg+Erm) and incubated overnight (ON) at 37°C and 200 rpm. Bacterial concentrations of the precultures were estimated by measuring their optical density (OD600). Erlermeyer flasks containing fresh TSBg+Erm were inoculated with precultures to reach starting OD600 of 0.02. For rifampicin mRNA stability assays, test cultures were grown at 37°C and 200 rpm until an OD600 of 0.5 (exponential phase). Six aliquots of 20 ml of the culture were transferred to 50 ml Falcon tubes containing 300 μg ml−1 of rifampicin and incubated at 37°C for 0, 2, 4, 8, 15, and 30 min. Then, 5 ml of stop solution (95% ethanol and 5% phenol) were added to the samples and centrifuged for 2 min at 4,400 g. Pellets were frozen in liquid nitrogen and stored at −80°C. RNA extraction and Northern blot analysis were performed as described in Toledo-Arana et al. (2009) and Menendez-Gil et al. (2020). Radiolabeled riboprobes were synthesized from a PCR carrying the T7 promoter (Table 3) using the MAXIscript T7 transcription kit (Ambion) and [α32P]-UTP, following the manufacturer’s recommendations. These riboprobes were designed to target the ftnA or the 3XFftnA mRNAs. The mRNA levels were quantified by densitometry of Northern blot autoradiographies using ImageJ.1 Each of the mRNA levels was normalized to the levels of the sample at 0 min of rifampicin incubation.
The mutants generated in this study (Table 1) were obtained as previously described (Valle et al., 2003) by a two-step homologous recombination that exchanges a specific chromosomic region by the mutant allele present in the pMAD plasmid (Arnaud et al., 2004). The marker-less mutants were verified by PCR using oligonucleotides E and F (Table 3) and Sanger sequencing.
Bacteria were grown as described above. At OD600 0.5, 30 ml samples were taken for protein extraction as described by Menendez-Gil et al. (2020). Western blotting was performed as previously described (Caballero et al., 2018). The 3xFLAG tagged protein samples were incubated with mouse monoclonal anti-FLAG M2-Peroxidase (HRP) antibodies (Sigma) diluted 1:1,000, whereas the GFP samples were incubated with mouse monoclonal anti-GFP antibodies 1:5,000 (Living Colors, Clontech) and peroxidase-conjugated goat anti-mouse immunoglobulin G and M antibodies 1:2,500 (Pierce-Thermo Scientific). Membranes were developed using the SuperSignal West Pico Chemiluminiscent Substrate kit (Thermo Scientific). Mean intensities of developed protein bands were quantified by densitometry of Western blot images using ImageJ and plotted as arbitrary units (A.U.). Statistical significances were calculated by running a paired t-test in GraphPad Prim; asterisks (*) indicate p-values lower than 0.05 (p < 0.05) while ns indicate not significant differences.
Precultures were grown in 5 ml of modified chemically defined medium without iron (MMwo/Fe) overnight at 37°C and 200 rpm (Toledo-Arana et al., 2005). Since all glass material contains iron traces, bacteria can grow in this medium. In order to eliminate the remaining free iron, we used 2′2-dipyridil (DIP; Sigma) as an iron chelator. Precultures were normalized to OD600 0,1 and 5 μl of these aliquots were diluted in 195 μl of modified chemically defined medium containing different concentrations of DIP in 96-well microtiter plates. The growth curve was monitored using the SpectraMax 340 PC Microplate Reader (Molecular Devices). OD650 measurements were performed every 30 min at 37°C for a period of 20 h.
In a previous study, we showed that deletion of the ftnA 3’UTR increased both the ftnA mRNA and ferritin protein levels in S. aureus (Menendez-Gil et al., 2020). To evaluate whether the ftnA 3’UTR deletion affected ftnA mRNA stability, we performed rifampicin mRNA stability assays and half-life determinations. To that end, we transformed the S. aureus 15981 ΔftnA strain with the p3xFFtnA and p3xFFtnAΔ3’UTR plasmids, which expressed the WT and Δ3’UTR ftnA mRNAs under the control of the PblaZ constitutive promoter, respectively. This allowed us to exclusively monitor the plasmidic ftnA gene using a strand-specific riboprobe. The resulting strains were grown until exponential phase and their total RNAs extracted at different time points after rifampicin addition. Northern blots revealed that the half-life of the ftnA mRNA was higher in the p3xFFtnAΔ3’UTR strain (2.5 min) when compared to the p3xFFtnA strain (0.8 min; Figure 1). Note that since the decrease in concentration of the two mRNAs was not exponential for all time points, we used only the first three time points (0–4 min) for the WT mRNA and the first four time points (0–8 min) for the Δ3’UTRftnA deletion mutant to calculate their half-life (Figure 1B). These results indicated that the ftnA 3’UTR may be targeted by unknown RNases to modulate FtnA expression.
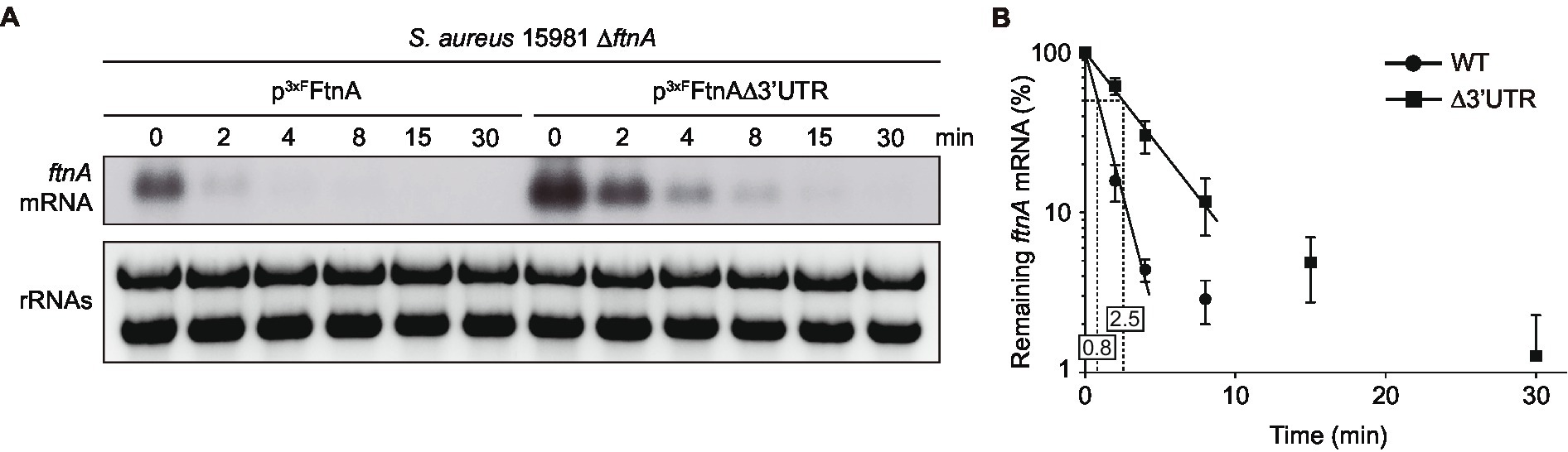
Figure 1. The ftnA ∆3’UTR mRNA has a longer half-life than the ftnA WT mRNA. (A) Half-life measurements of ftnA WT and ∆3’UTR mRNAs expressed from a constitutive promoter. Strains carrying the constructs were grown in TSBg at 37°C until exponential phase when 300 μg/ml of rifampicin was added. Samples were taken at the indicated time points (min). Ribosomal RNAs (rRNAs) stained with Midori Green are shown as loading controls. A representative image of the experiment, which was repeated three times, is shown. (B) The mRNA levels were quantified by densitometry of Northern blot images using Image J (http://rsbweb.nih.gov/ij/) and normalized using time 0 as a reference. The mean of the mRNA levels was plotted in function of time. Error bars represent the SD from three independent replicates.
In order to identify the RNases that could potentially target the ftnA 3’UTR, we transformed the most relevant non-essential RNase mutants (Bonnin and Bouloc, 2015) of the S. aureus 15981 WT strain with the p3xFFtnA and p3xFFtnAΔ3’UTR plasmids, which expressed the 3xFLAG-tagged ferritin protein (3xFFtnA) from the WT and Δ3’UTR ftnA mRNAs, respectively. The selected RNase mutants included ∆rnc (RNase III, a dsRNA endonuclease; Lasa et al., 2011; Lioliou et al., 2012), ∆mrnc (mini-RNase III, a double-stranded RNA endonuclease paralog to RNase III, which was initially identified in B. subtilis; Olmedo and Guzmán, 2008, with an uncharacterized ortholog in S. aureus), ∆pnpA (PNPase, 3′-5′ exonuclease; Anderson and Dunman, 2009), ∆rnr (RNase R, 3′-5′ exonuclease; Oussenko et al., 2002), ∆rny (RNase Y, a single-stranded RNA endonuclease; Marincola et al., 2012), and ∆rnjA (RNase J1, a bifunctional RNase with endonuclease and 5′ to 3′ exonuclease activities; Linder et al., 2014). We then determined their 3xFFtnA protein levels by Western blot using anti-FLAG antibodies (Figure 2A). As expected, the S. aureus 15981 WT strain that carried the p3xFFtnAΔ3’UTR plasmid expressed higher 3xFFtnA protein levels than the strain expressing the whole ftnA mRNA. Such protein increase was also obtained when ferritin was expressed from the p3xFFtnAΔ3’UTR plasmid in the ∆rnr, ∆mrnc, and ∆rny mutant strains. Since 3xFFtnA could not be detected in the ∆rnjA strain, we performed the same experiment but loading a higher amount of total protein (Figure 2B). The ∆rnjA mutant strains also showed an increase in ferritin expression when the 3’UTR was deleted (Figures 2A,B). These results suggested that RNase R, mini-RNase III, RNase Y, and RNase J1 were not involved in the 3’UTR-mediated processing of the ftnA mRNA. In contrast, the Western blots revealed that the ∆rnc mutant strains carrying the p3xFFtnA and p3xFFtnAΔ3’UTR plasmids expressed similar levels of the 3xFFtnA protein regardless of the 3’UTR deletion from the 3xFLAG-ftnA mRNA. Similar results were obtained in the ∆pnpA mutant strains (Figure 2A). This suggests that RNase III and PNPase could be targeting the ftnA 3’UTR to process the ftnA mRNA.
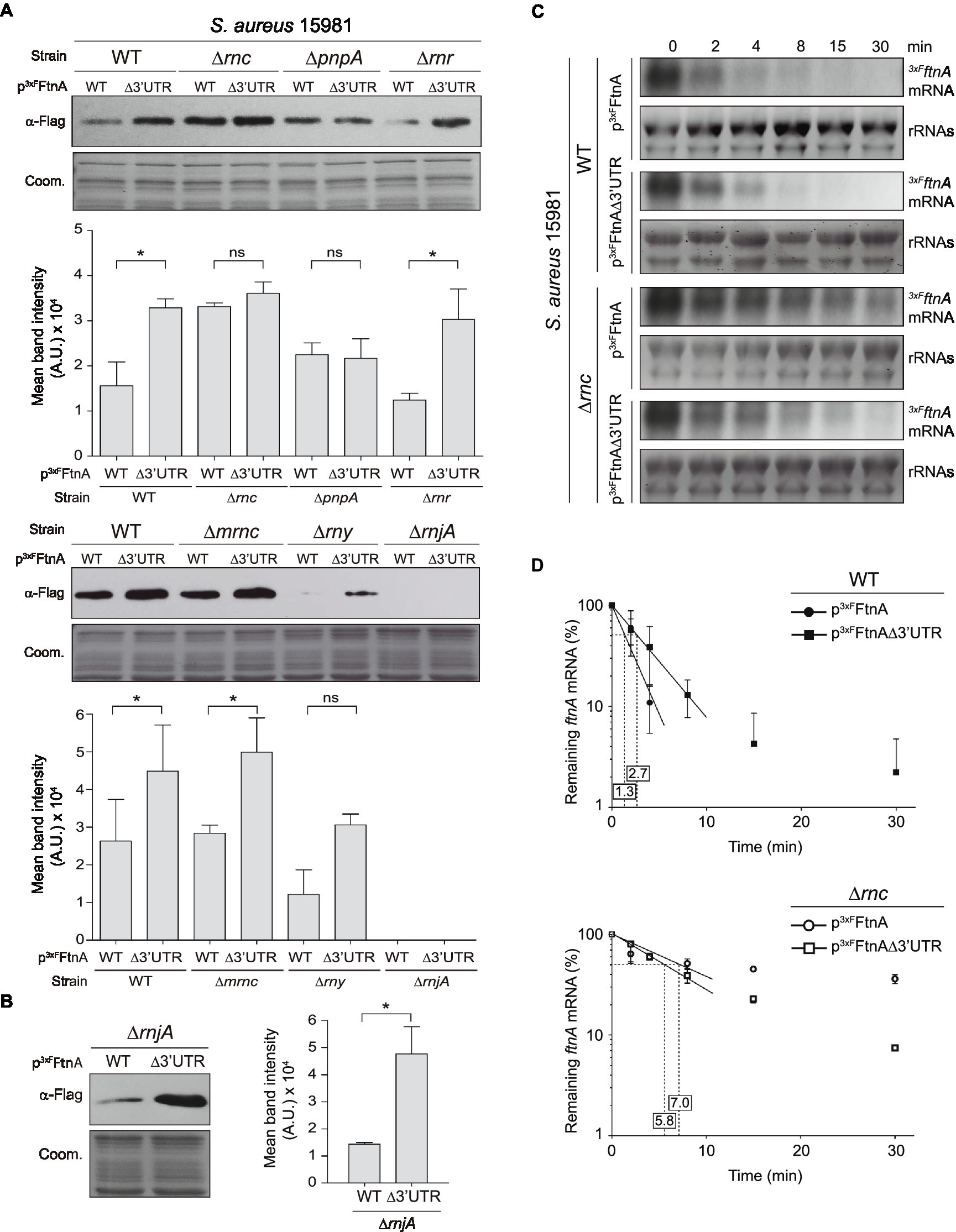
Figure 2. Role of RNases in ftnA 3’UTR-mediated regulation. Western blots showing: (A) 3xFFtnA levels from different Staphylococcus aureus 15981 RNase mutants harboring the p3xFFtnA and p3xFFtnAΔ3’UTR constructs. (B) 3xFFtnA levels in the ∆rnjA mutant increasing the protein load used in (A). Western blots were developed using peroxidase-conjugated anti-FLAG antibodies. Coomassie (Coom.) stained gel portions are shown as loading controls. Western blot images show representative results from at least three independent replicates. The mean intensity of the bands was estimated by densitometry of blot images using ImageJ (A.U., arbitrary units). Statistical significances were determined through paired t-tests in GraphPad Prim; *p < 0.05; and ns, not significant. (C) Rifampicin half-life assays of 3xFLAG ftnA WT and ∆3’UTR mRNAs expressed from a constitutive promoter in the S. aureus 15981 WT strain and its isogenic ∆rnc mutant. Strains were grown in TSBg at 37°C until exponential phase when 300 μg/ml of rifampicin was added. Samples were taken at the indicated time points (min). rRNAs stained with Midori Green are shown as loading control. Representative images of the experiment, which was repeated twice, are shown. (D) The mRNA levels were quantified by densitometry of Northern blot images in Image J (http://rsbweb.nih.gov/ij/) and normalized using time 0 as a reference. The mean of the mRNA levels was plotted in function of time. Error bars represent the SD from two independent replicates.
mRNA decay is often initiated by endoribonucleases, including RNase Y and RNase III (Durand et al., 2015). To confirm the implication of RNase III in the ftnA mRNA decay, we evaluated the half-life of the WT and Δ3’UTRftnA mRNAs in S. aureus 15981 WT and its isogenic ∆rnc mutant strain. The rifampicin mRNA stability assays revealed that the ftnA mRNA half-life increased from 1.3 min in the WT strain to 7.0 min in the ∆rnc mutant (Figures 2C,D). This was higher than the half-life observed for the Δ3’UTRftnA mRNA mutant, indicating that the RNase III might target the ftnA mRNA through additional mechanisms. Note that the half-life of the ftnA mRNA and its Δ3’UTR mutant were similar when expressed from the ∆rnc mutant (7.0 vs. 5.8 min, respectively; Figures 2C,D). Taken together, these results indicate that RNase III promotes ftnA mRNA decay in a process in which the ftnA 3’UTR plays a critical role.
In order to investigate whether the ftnA 3’UTR had functional capacities on its own, we fused the ftnA 3’UTR downstream of the gfp gene, which encodes the GFP, thus, generating the pGFP-3’UTRftnA plasmid. As a control, we constructed a plasmid that included the transcriptional terminator (TT) of the ftnA mRNA downstream of the gfp gene (pGFP-∆3’UTRftnA; Figure 3A). Then, we transformed the S. aureus 15981 WT strain with these plasmids and determined the GFP levels by Western blot analysis. The results revealed that the pGFP-3’UTRftnA plasmid expressed lower GFP levels when compared to the pGFP-∆3’UTRftnA plasmid (Figure 3B). This confirmed that the ftnA 3’UTR alone worked as an independent module able to reduce the expression of a heterologous gene like gfp. To analyze whether such GFP expression reduction was mediated by RNase III and PNPase, we introduced the pGFP-3’UTRftnA plasmid into the ∆rnc and ∆pnpA mutant strains. Western blot results showed that the ∆rnc and ∆pnpA mutants expressed similar GFP levels to the ones produced by the strain carrying the pGFP-∆3’UTRftnA plasmid, indicating that the 3’UTR would still be targeted and processed by RNase III and PNPase regardless of the CDS (Figure 3C).
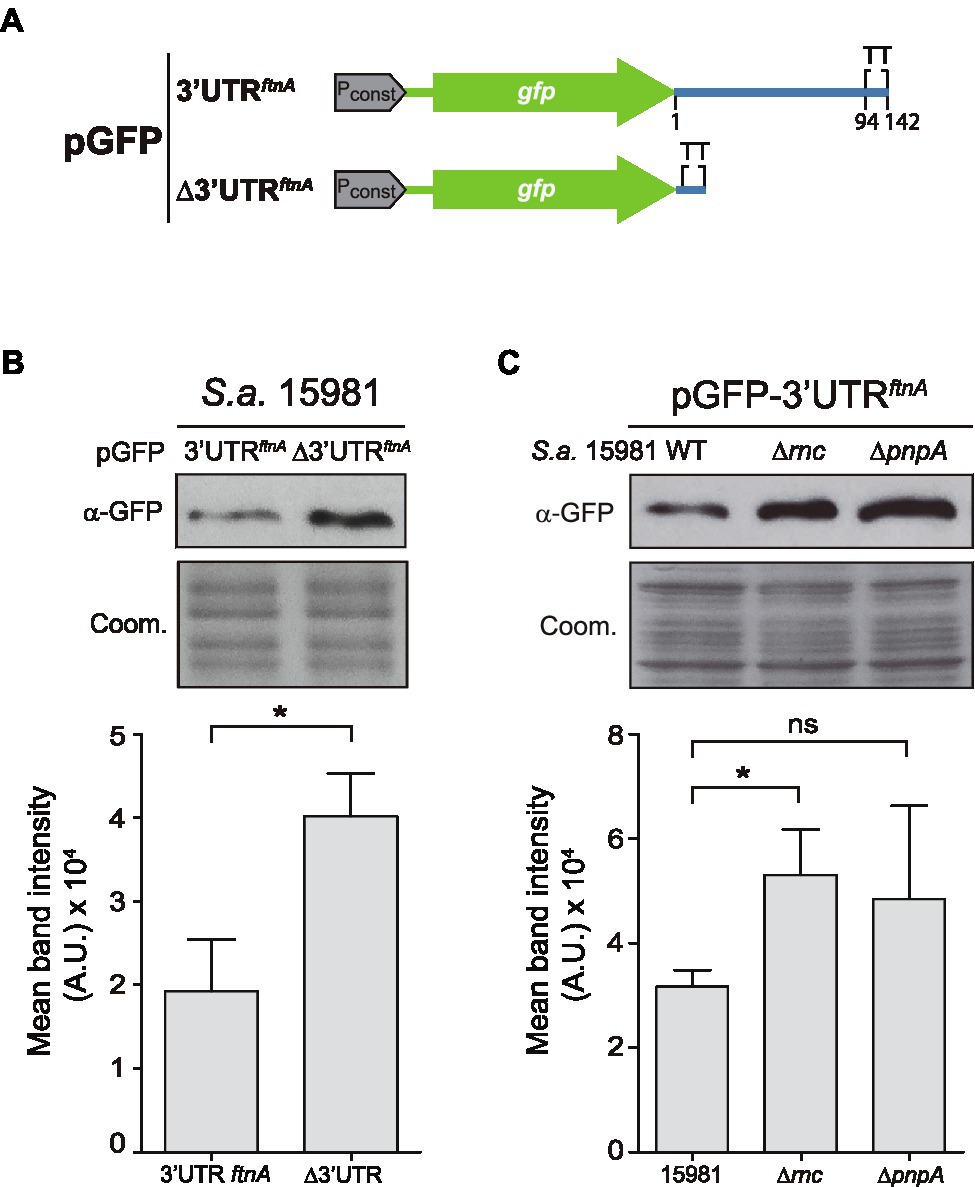
Figure 3. The ftnA 3’UTR can act as an independent module. (A) Schematic representation of the green fluorescent protein (GFP) constructs generated. Pconst: constitutive promoter; TT: transcriptional terminator. (B) Western blot showing the GFP levels of the Staphylococcus aureus 15981 WT strain carrying either pGFP-3’UTRftnA or pGFP-∆3’UTRftnA. (C) Western blot showing the GFP levels of the 15981 ∆rnc and ∆pnpA strains carrying pGFP-3’UTRftnA. Western blots were developed using monoclonal anti-GFP antibodies and peroxidase-conjugated goat anti-mouse immunoglobulin G and M antibodies. Coomassie (Coom.) stained gel portions are shown as loading controls. Western blot images show the representative results from at least three independent replicates. Mean intensity bands were quantified by densitometry of blot images in ImageJ (A.U., arbitrary units). Statistical significances were determined by running paired t-tests using the GraphPad Prim software; *p < 0.05; and ns, not significant.
Since RNase III is a double-stranded endoribonuclease, we looked for putative double-stranded RNA regions within the ftnA 3’UTR secondary structure. We used the RNAstructure version 6.2 software to predict the ftnA mRNA conformation.2 We could not find any evident secondary structures susceptible to RNase III within the ftnA 3’UTR, nor a hypothetical 5’UTR-3’UTR interaction, as previously described for the icaR mRNA (de Los Mozos et al., 2013). A plausible alternative would be for the ftnA 3’UTR to be targeted by a trans-acting small RNA that, upon interaction, generated a double-stranded substrate for RNase III to process. This idea would require the putative paring region to be conserved among S. aureus strains and close relatives. Previous multiple sequence alignment analyses revealed that 8,193 out of the 10,000 S. aureus genomes available at the NCBI database presented an ftnA 3’UTR with a 100% of identity. The remaining ones showed just few nucleotide differences (Menendez-Gil et al., 2020). Although, such a high degree of conservation suggested an important role for the ftnA 3’UTR, it prevented us from identifying a putative functional region. Our previous analyses also showed that among other Staphylococcus species carrying the ftnA gene, the ftnA 3’UTR conservation only applied to S. argenteus (Menendez-Gil et al., 2020). Further nucleotide comparison analysis between the S. aureus and S. argenteus ftnA 3’UTRs revealed two conserved regions comprised between nucleotides 18–56 (conserved region I) and 68–89 (conserved region II), respectively, besides the putative TT (Figure 4A). To determine whether one of these regions could be involved in the ftnA mRNA processing, we constructed two plasmids expressing the 3xFLAG tagged ftnA mRNA carrying either a deletion between nucleotides 19 and 56 (p3xFFtnA∆3’UTR19-56), or 57 and 93 (p3xFFtnA∆3’UTR57-93), which selectively eliminated conserved regions I and II, respectively (Figure 4B). We used such plasmids to transform the S. aureus 15981 WT strain and evaluated their 3xFFtnA protein expression. Western blot analyses revealed that both mutations produced similar 3xFFtnA protein levels when compared to the full-length ftnA mRNA, suggesting that deletions of a few nucleotides are not enough to reproduce the effect generated by the ∆3’UTR mutant (Figure 4C). At the same time, it indicated that both conserved regions contribute to RNase III action to modulate ferritin expression.
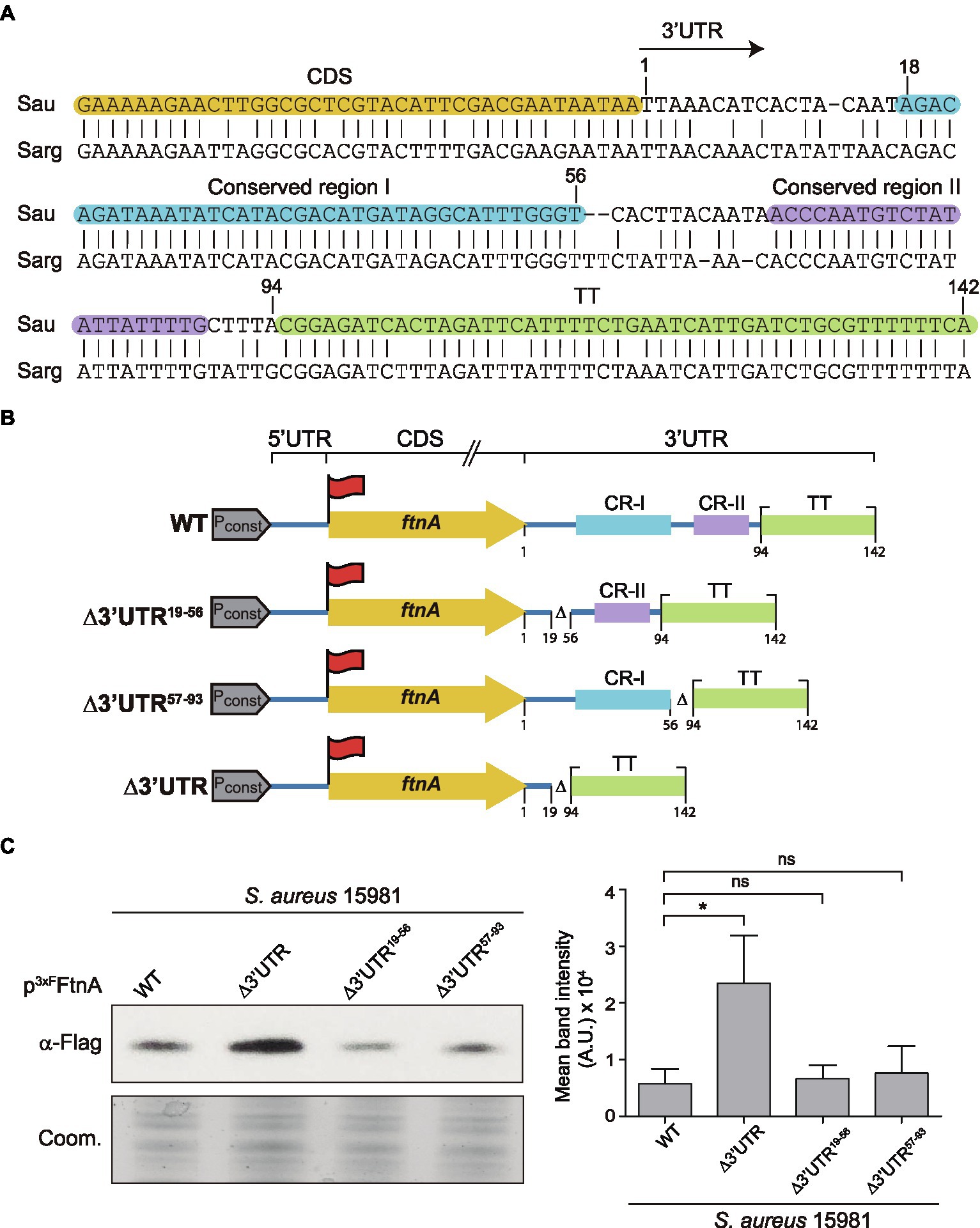
Figure 4. The ftnA 3’UTR sequence is conserved in Staphylococcus aureus and Staphylococcus argenteus. (A) Blastn alignment of the ftnA 3’UTR from Staphylococcus aureus (Sau) and Staphylococcus argenteus (Sarg). Nucleotides corresponding to the CDS, conserved regions I and II (CR-I and CR-II), and the TT are highlighted in orange, blue, purple, and green, respectively. The arrow indicates the start of the 3’UTR. (B) Schematic representation of the generated constructs to identify the functional region of the ftnA 3’UTR. 3xFlag is represented with a red flag, Pconst: constitutive promoter. (C) Western blot showing the levels of 3xFFtnA when expressed from the constructs shown in section (B), introduced in the S. aureus 15981 WT strain. A Coomassie (Coom.) stained gel portion is shown as a loading control. Western blot images show the representative results from at least three independent replicates. Mean intensity bands were quantified by densitometry of blot images in ImageJ (A.U., arbitrary units). Statistical significances were determined through paired t-tests using the GraphPad Prim software; *p < 0.05; and ns, not significant.
To evaluate the biological relevance of the ftnA 3’UTR-mediated control of ferritin production, we constructed a chromosomal ftnA∆3’UTR mutant in the S. aureus 15981 genetic background. First, to control that the chromosomal mutant behaved as the plasmidic one, we performed Northern blot analyses to monitor the ftnA mRNA levels. Total RNAs were extracted from the WT and ∆3’UTR mutant strains grown until exponential phase in a rich medium (TSBg). Northern blot results showed that the chromosomal ftnA∆3’UTR mutant expressed higher ftnA mRNA levels than the WT strain as it occurred with the plasmidic mutant (Figure 5A).
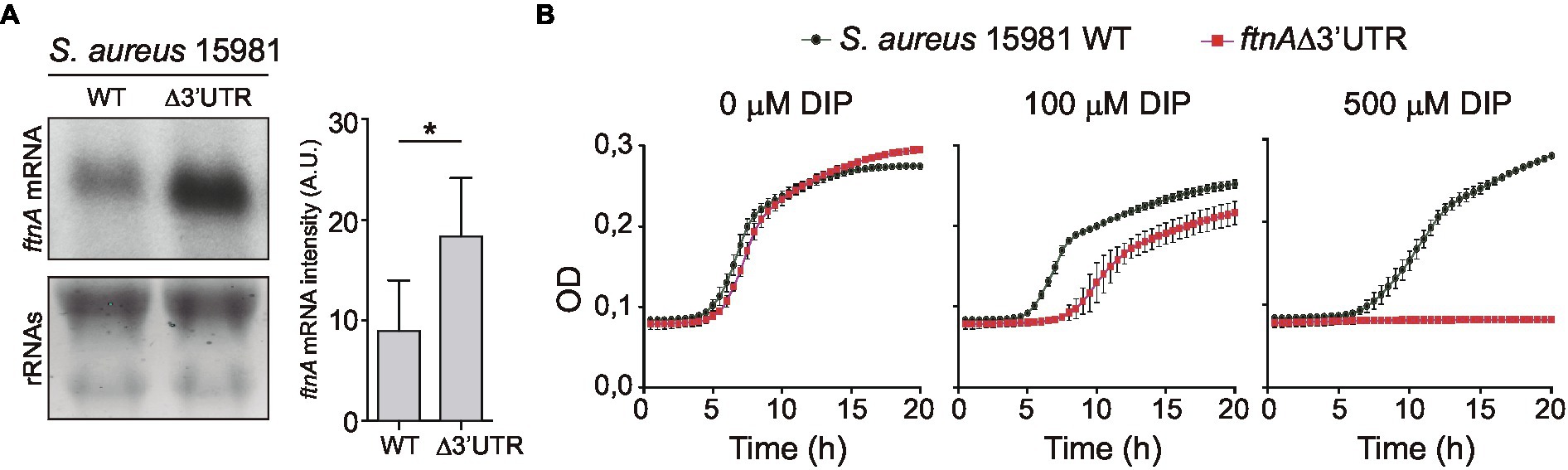
Figure 5. The 3’UTR of ftnA is essential for Staphylococcus aureus growth under iron starvation conditions. (A) Northern blot showing the ftnA mRNAs from S. aureus 15981 WT and chromosomal ftnAΔ3’UTR mutant strains grown in TSBg at 37°C until exponential phase. The lower panel shows ribosomal RNAs (rRNAs) stained with Midori Green as a loading control. A representative image of the experiment, which was repeated twice, is shown. Mean intensity bands were quantified by densitometry of blot images in ImageJ (A.U., arbitrary units). Statistical significances were determined by running paired t-tests in GraphPad Prim; *p < 0.05. (B) The S. aureus 15981 WT and ftnA∆3’UTR strains were grown for 20 h in a modified chemically defined medium without iron (MMwo/Fe; Toledo-Arana et al., 2005) and increasing concentrations of the iron chelator 2′2-dipyridil (DIP). Optical density (OD) measurements were registered every half an hour. Error bars represent the SD from three independent replicates.
Then, we aimed at comparing the capacities of the S. aureus 15981 WT and the chromosomal ftnA∆3’UTR mutant strains to grow under iron starvation conditions. We incubated microplates containing minimal medium lacking iron (MMwo/Fe) at 37°C and measured bacterial growth by registering the optical density every 30 min. However, no growth differences were observed between the WT and ∆3’UTR mutant in the MMwo/Fe (Figure 5B). Since iron traces could be still present in this medium, iron starvation might be difficult to achieve under laboratory conditions without the use of chelating agents (Pi and Helmann, 2017). Therefore, the MMwo/Fe was complemented with increasing concentrations of 2,2′-dipyridyl (DIP), a strong iron chelator. Figure 5B shows that the addition of the DIP chelator at a concentration of 100 μM significantly affected the growth of the ftnA∆3’UTR mutant, while adding 500 μM of DIP completely impaired it. Altogether, these data portrayed the ftnA 3’UTR as an essential module to control the ftnA mRNA expression and maintain proper iron levels for S. aureus growth under iron starvation conditions.
The right amount of iron concentration inside the cells is essential for bacterial growth since it is utilized as a cofactor for a wide variety of enzymes. However, intracellular iron excess can lead to oxidative stress and, ultimately, cell damage (Andrews et al., 2003; Hood and Skaar, 2012). For this purpose, the existence of regulating agents such as the ferritin, which removes free intracellular iron, is paramount for protecting cells from its potential toxic effects (Zühlke et al., 2016). As a consequence, the levels of ferritin must also be tightly regulated and in accordance with iron availability (Morrissey et al., 2004). In this study, we showed that ferritin expression is controlled at the post-transcriptional level by the ftnA 3’UTR, which is mainly targeted by RNase III and PNPase (Figure 2). The ftnA 3’UTR seems to work as an independent cis-regulatory module since its fusion to the heterologous gfp reporter gene also decreased GFP expression with the participation of RNase III and PNPase (Figure 3). PNPase is a 3′-5′ exoribonuclease whose activity is inhibited by the presence of strong RNA secondary structures (Spickler and Mackie, 2000; Dar and Sorek, 2018; Ingle et al., 2021). The 3′ end of the ftnA mRNA contains a putative intrinsic Rho-independent terminator that should avoid PNPase processing. Therefore, one would expect the action of RNase III to trigger ftnA mRNA processing, which would provide an mRNA carrying now a 3′ end accessible for PNPase cleavage. This is in agreement with the canonical mechanism of RNA degradation found in the majority of Gram-positive bacteria, which it is initiated by either RNase Y or RNase III and followed by the action of 3′-5′ exoribonucleases such as PNPase and RNase R (Broglia et al., 2020; Mediati et al., 2021). Our data indicated that RNase III would process the ftnA mRNA, at least in part, through its 3’UTR (Figure 3). Note that the rifampicin mRNA stability assays also showed that RNase III affects the ftnA mRNA independently of the 3’UTR (Figure 2). This mechanism would require further investigations.
RNase III cleaves dsRNAs. However, no internal double-stranded RNA structures were predicted in the 3’UTR that could provide a dsRNA substrate for RNase III as previously described (de Los Mozos et al., 2013). This suggested that either a cis-antisense RNA or a trans-acting sRNA may be required to create such RNA substrate for RNase III to cleave (Lasa et al., 2011; Lioliou et al., 2012). Although, we predicted some putative interactions between the ftnA 3’UTR and previously identified sRNAs in S. aureus (Geissmann et al., 2009; Bohn et al., 2010; Carroll et al., 2016), we failed to validate such interactions in vivo (data not shown). Whether other sRNAs and/or asRNAs interact with the ftnA 3’UTR remains to be explored. Moreover, knowing that global regulatory RNA chaperones have been already shown to bind 3’UTRs (Holmqvist et al., 2016, 2018; Potts et al., 2017) and iron-sensing proteins like aconitase bind their own mRNA (Benjamin and Massé, 2014), it would be interesting to evaluate whether RNA-binding proteins or even the ferritin itself could interact with the ftnA mRNA through the 3’UTR to control ferritin expression.
It is noteworthy that the whole ftnA 3’UTR was highly conserved in S. aureus and S. argenteus. This suggested that the ftnA 3’UTR sequence may be relevant for both species, which preserve throughout evolution a similar post-transcriptional control of ferritin production. It is also interesting that the corresponding 3’UTR sequences from other Staphylococcus species were completely different (both in length and sequence) despite the ftnA CDS being conserved (Figure 6). We previously showed that constructs of chimeric mRNAs including the S. aureus ftnA CDS and the ftnA 3’UTRs from Staphylococcus simiae, Staphylococcus epidermidis, and Staphylococcus capitis were unable to decrease 3xFFtnA expression (Menendez-Gil et al., 2020), indicating the presence of species-specific 3’UTR-mediated regulatory mechanisms. How the ftnA 3’UTRs from different Staphylococcus species participate in the modulation of ferritin production remains to be investigated.
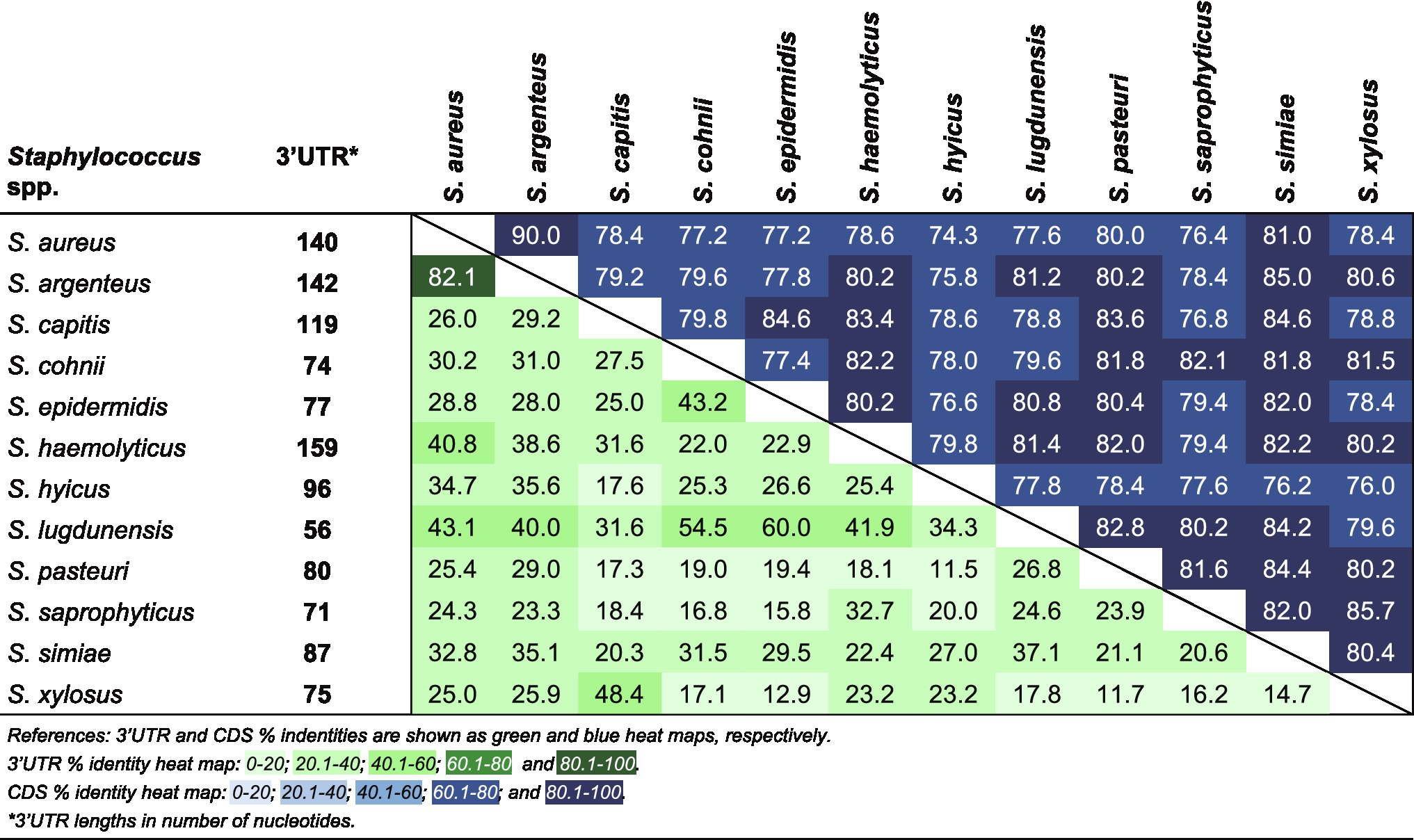
Figure 6. Percent identity matrix of a multiple sequence alignment for the ftnA 3’UTRs and ftnA CDSs from several Staphylococcus species.
These putative regulatory differences are not restricted to the post-transcriptional level. It was also shown that the transcriptional regulation of ferritin expression in response to metals in S. epidermidis was significantly different from S. aureus (Morrissey et al., 2004), suggesting that members of the Staphylococcus genus have developed different strategies to regulate iron homeostasis in species-specific manners.
In addition to the ftnA 3’UTR, we recently found 3’UTR sequence variability in several staphylococcal genes, indicating that this phenomenon may be widespread among bacteria. Asides from iron homeostasis, long 3’UTRs with evolutionary variability (Menendez-Gil et al., 2020) also affect relevant biological processes such as metabolism (Maeda and Wachi, 2012), biofilm formation (de Los Mozos et al., 2013; Zhu et al., 2016), and hemolysin production (Menendez-Gil et al., 2020). We proposed that these regions may be prone to changes that reflect in bacterial diversity in a similar way as it occurred for eukaryotes, promoting the diversification of species (Menendez-Gil and Toledo-Arana, 2021).
Another relevant observation in this study was the impaired growth of S. aureus under iron starvation conditions upon chromosomal deletion of the ftnA 3’UTR (Figure 5). Considering that deletion of the ftnA 3’UTR increased ferritin concentration (Figure 2; Menendez-Gil et al., 2020), it could be speculated that higher ferritin levels would sequester the scarce iron available inside the cells. As a result, the essential functions carried out by enzymes requiring iron as a cofactor would be affected, leading to bacterial growth arrest.
In summary, our study highlights the relevance of 3’UTRs to fine-tune the expression of genes involved in relevant processes such as iron homeostasis.
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
PM-G and AT-A conceived and designed the experiments. PM-G and AC-M performed the experiments. PM-G, AC-M, CC, and AT-A analyzed the data and contributed to the interpretation of results. PM-G, CC, and AT-A wrote the manuscript. All authors contributed to the article and approved the submitted version.
AT-A was supported by the European Research Council under the European Union’s Horizon 2020 research and innovation program (ERC-CoG-2014-646869); and the Spanish Ministry of Science and Innovation (PID2019-105216GB-I00) grants.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
Anderson, K. L., and Dunman, P. M. (2009). Messenger RNA turnover processes in Escherichia coli, Bacillus subtilis, and emerging studies in Staphylococcus aureus. Int. J. Microbiol. 2009, 1–15. doi: 10.1155/2009/525491
Andrews, S. C., Robinson, A. K., and Rodríguez-Quiñones, F. (2003). Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237. doi: 10.1016/S0168-6445(03)00055-X
Arnaud, M., Chastanet, A., and Debarbouille, M. (2004). New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70, 6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004
Balestrino, D., Hamon, M. A., Dortet, L., Nahori, M.-A., Pizarro-Cerda, J., Alignani, D., et al. (2010). Single-cell techniques using chromosomally tagged fluorescent bacteria to study Listeria monocytogenes infection processes. Appl. Environ. Microbiol. 76, 3625–3636. doi: 10.1128/AEM.02612-09
Benjamin, J.-A. M., and Massé, E. (2014). The iron-sensing aconitase B binds its own mRNA to prevent sRNA-induced mRNA cleavage. Nucleic Acids Res. 42, 10023–10036. doi: 10.1093/nar/gku649
Bohn, C., Rigoulay, C., Chabelskaya, S., Sharma, C. M., Marchais, A., Skorski, P., et al. (2010). Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res. 38, 6620–6636. doi: 10.1093/nar/gkq462
Bonnin, R. A., and Bouloc, P. (2015). RNA degradation in Staphylococcus aureus: diversity of ribonucleases and their impact. Int. J. Genomics 2015:395753. doi: 10.1155/2015/395753
Braun, F., Durand, S., and Condon, C. (2017). Initiating ribosomes and a 5′/3′-UTR interaction control ribonuclease action to tightly couple B. subtilis hbs mRNA stability with translation. Nucleic Acids Res. 45, 11386–11400. doi: 10.1093/nar/gkx793
Broglia, L., Lécrivain, A.-L., Renault, T. T., Hahnke, K., Ahmed-Begrich, R., Le Rhun, A., et al. (2020). An RNA-seq based comparative approach reveals the transcriptome-wide interplay between 3′-to-5′ exoRNases and RNase Y. Nat. Commun. 11, 1587–1512. doi: 10.1038/s41467-020-15387-6
Bronesky, D., Desgranges, E., Corvaglia, A., Francois, P., Caballero, C. J., Prado, L., et al. (2019). A multifaceted small RNA modulates gene expression upon glucose limitation in Staphylococcus aureus. EMBO J. 38:e99363. doi: 10.15252/embj.201899363
Caballero, C. J., Menendez-Gil, P., Catalan-Moreno, A., Vergara-Irigaray, M., García, B., Segura, V., et al. (2018). The regulon of the RNA chaperone CspA and its auto-regulation in Staphylococcus aureus. Nucleic Acids Res. 46, 1345–1361. doi: 10.1093/nar/gkx1284
Carroll, R. K., Weiss, A., Broach, W. H., Wiemels, R. E., Mogen, A. B., Rice, K. C., et al. (2016). Genome-wide annotation, identification, and global transcriptomic analysis of regulatory or small RNA gene expression in Staphylococcus aureus. mBio 7, e01990–e01915. doi: 10.1128/mBio.01990-15
Dar, D., and Sorek, R. (2018). Extensive reshaping of bacterial operons by programmed mRNA decay. PLoS Genet. 14:e1007354. doi: 10.1371/journal.pgen.1007354
de Los Mozos, I. R., Vergara-Irigaray, M., Segura, V., Villanueva, M., Bitarte, N., Saramago, M., et al. (2013). Base pairing interaction between 5′- and 3′-UTRs controls icaR mRNA translation in Staphylococcus aureus. PLoS Genet. 9:e1004001. doi: 10.1371/journal.pgen.1004001
Durand, S., Tomasini, A., Braun, F., Condon, C., and Romby, P. (2015). sRNA and mRNA turnover in gram-positive bacteria. FEMS Microbiol. Rev. 39, 316–330. doi: 10.1093/femsre/fuv007
El-Mouali, Y., Gaviria-Cantin, T., Sánchez-Romero, M. A., Gibert, M., Westermann, A. J., Vogel, J., et al. (2018). CRP-cAMP mediates silencing of Salmonella virulence at the post-transcriptional level. PLoS Genet. 14:e1007401. doi: 10.1371/journal.pgen.1007401
Friedman, D. B., Stauff, D. L., Pishchany, G., Whitwell, C. W., Torres, V. J., and Skaar, E. P. (2006). Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog. 2:e87. doi: 10.1371/journal.ppat.0020087
Geissmann, T., Chevalier, C., Cros, M.-J., Boisset, S., Fechter, P., Noirot, C., et al. (2009). A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res. 37, 7239–7257. doi: 10.1093/nar/gkp668
Haley, K. P., and Skaar, E. P. (2012). A battle for iron: host sequestration and Staphylococcus aureus acquisition. Microbes Infect. 14, 217–227. doi: 10.1016/j.micinf.2011.11.001
Holmqvist, E., Li, L., Bischler, T., Barquist, L., and Vogel, J. (2018). Global maps of ProQ binding in vivo reveal target recognition via RNA structure and stability control at mRNA 3′ ends. Mol. Cell 70, 971.e6–982.e6. doi: 10.1016/j.molcel.2018.04.017
Holmqvist, E., Wright, P. R., Li, L., Bischler, T., Barquist, L., Reinhardt, R., et al. (2016). Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 35, 991–1011. doi: 10.15252/embj.201593360
Hood, M. I., and Skaar, E. P. (2012). Nutritional immunity: transition metals at the pathogen–host interface. Nat. Rev. Microbiol. 10, 525–537. doi: 10.1038/nrmicro2836
Horsburgh, M. J., Ingham, E., and Foster, S. J. (2001). In Staphylococcus aureus, fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183, 468–475. doi: 10.1128/JB.183.2.468-475.2001
Ingle, S., Chhabra, S., Laspina, D., Salvo, E., Liu, B., and Bechhofer, D. H. (2021). Polynucleotide phosphorylase and RNA helicase CshA cooperate in Bacillus subtilis mRNA decay. RNA Biol. 18, 1692–1701. doi: 10.1080/15476286.2020.1864183
Lasa, I., Toledo-Arana, A., Dobin, A., Villanueva, M., de los Mozos, I. R., Vergara-Irigaray, M., et al. (2011). Genome-wide antisense transcription drives mRNA processing in bacteria. Proc. Natl. Acad. Sci. U. S. A. 108, 20172–20177. doi: 10.1073/pnas.1113521108
Linder, P., Lemeille, S., and Redder, P. (2014). Transcriptome-wide analyses of 5′-ends in RNase J mutants of a gram-positive pathogen reveal a role in RNA maturation, regulation and degradation. PLoS Genet. 10:e1004207. doi: 10.1371/journal.pgen.1004207
Lioliou, E., Sharma, C. M., Caldelari, I., Helfer, A. C., Fechter, P., Vandenesch, F., et al. (2012). Global regulatory functions of the Staphylococcus aureus endoribonuclease III in gene expression. PLoS Genet. 8:e1002782. doi: 10.1371/journal.pgen.1002782
López-Garrido, J., Puerta-Fernández, E., and Casadesús, J. (2014). A eukaryotic-like 3′ untranslated region in Salmonella enterica hilD mRNA. Nucleic Acids Res. 42, 5894–5906. doi: 10.1093/nar/gku222
Maeda, T., and Wachi, M. (2012). 3′ untranslated region-dependent degradation of the aceA mRNA, encoding the glyoxylate cycle enzyme isocitrate lyase, by RNase E/G in Corynebacterium glutamicum. Appl. Environ. Microbiol. 78, 8753–8761. doi: 10.1128/AEM.02304-12
Marchetti, M., De Bei, O., Bettati, S., Campanini, B., Kovachka, S., Gianquinto, E., et al. (2020). Iron metabolism at the interface between host and pathogen: from nutritional immunity to antibacterial development. Int. J. Mol. Sci. 21:2145. doi: 10.3390/ijms21062145
Marincola, G., Schäfer, T., Behler, J., Bernhardt, J., Ohlsen, K., Goerke, C., et al. (2012). RNase Y of Staphylococcus aureus and its role in the activation of virulence genes. Mol. Microbiol. 85, 817–832. doi: 10.1111/j.1365-2958.2012.08144.x
Mediati, D. G., Lalaouna, D., and Tree, J. J. (2021). Burning the candle at both ends: have exoribonucleases driven divergence of regulatory RNA mechanisms in bacteria? mBio 12:e0104121. doi: 10.1128/mBio.01041-21
Meneghini, R. (1997). Iron homeostasis, oxidative stress, and DNA damage. Free Radic. Biol. Med. 23, 783–792. doi: 10.1016/s0891-5849(97)00016-6
Menendez-Gil, P., Caballero, C. J., Catalan-Moreno, A., Irurzun, N., Barrio-Hernandez, I., Caldelari, I., et al. (2020). Differential evolution in 3'UTRs leads to specific gene expression in Staphylococcus. Nucleic Acids Res. 48, 2544–2563. doi: 10.1093/nar/gkaa047
Menendez-Gil, P., and Toledo-Arana, A. (2021). Bacterial 3′UTRs: a useful resource in post-transcriptional regulation. Front. Mol. Biosci. 7:617633. doi: 10.3389/fmolb.2020.617633
Miyakoshi, M., Chao, Y., and Vogel, J. (2015). Cross talk between ABC transporter mRNAs via a target mRNA-derived sponge of the GcvB small RNA. EMBO J. 34, 1478–1492. doi: 10.15252/embj.201490546
Morrissey, J. A., Cockayne, A., Brummell, K., and Williams, P. (2004). The staphylococcal ferritins are differentially regulated in response to iron and manganese and via PerR and Fur. Infect. Immun. 72, 972–979. doi: 10.1128/IAI.72.2.972-979.2004
Olmedo, G., and Guzmán, P. (2008). Mini-III, a fourth class of RNase III catalyses maturation of the Bacillus subtilis 23S ribosomal RNA. Mol. Microbiol. 68, 1073–1076. doi: 10.1111/j.1365-2958.2008.06203.x
Oussenko, I. A., Sanchez, R., and Bechhofer, D. H. (2002). Bacillus subtilis YhaM, a member of a new family of 3′-to-5′ exonucleases in gram-positive bacteria. J. Bacteriol. 184, 6250–6259. doi: 10.1128/JB.184.22.6250-6259.2002
Pi, H., and Helmann, J. D. (2017). Sequential induction of Fur-regulated genes in response to iron limitation in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 114, 12785–12790. doi: 10.1073/pnas.1713008114
Potts, A. H., Vakulskas, C. A., Pannuri, A., Yakhnin, H., Babitzke, P., and Romeo, T. (2017). Global role of the bacterial post-transcriptional regulator CsrA revealed by integrated transcriptomics. Nat. Commun. 8:1596. doi: 10.1038/s41467-017-01613-1
Price, E. E., and Boyd, J. M. (2020). Genetic regulation of metal ion homeostasis in Staphylococcus aureus. Trends Microbiol. 28, 821–831. doi: 10.1016/j.tim.2020.04.004
Spickler, C., and Mackie, G. A. (2000). Action of RNase II and polynucleotide phosphorylase against RNAs containing stem-loops of defined structure. J. Bacteriol. 182, 2422–2427. doi: 10.1128/JB.182.9.2422-2427.2000
Toledo-Arana, A., Dussurget, O., Nikitas, G., Sesto, N., Guet-Revillet, H., Balestrino, D., et al. (2009). The Listeria transcriptional landscape from saprophytism to virulence. Nature 459, 950–956. doi: 10.1038/nature08080
Toledo-Arana, A., Merino, N., Vergara-Irigaray, M., Débarbouillé, M., Penadés, J. R., and Lasa, I. (2005). Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J. Bacteriol. 187, 5318–5329. doi: 10.1128/JB.187.15.5318-5329.2005
Tong, S. Y. C., Davis, J. S., Eichenberger, E., Holland, T. L., and Fowler, V. G. (2015). Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28, 603–661. doi: 10.1128/CMR.00134-14
Troxell, B., and Hassan, H. M. (2013). Transcriptional regulation by ferric uptake regulator (Fur) in pathogenic bacteria. Front. Cell. Infect. Microbiol. 3:59. doi: 10.3389/fcimb.2013.00059
Valle, J., Toledo-Arana, A., Berasain, C., Ghigo, J.-M., Amorena, B., Penadés, J. R., et al. (2003). SarA and not sigma B is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48, 1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x
Wandersman, C., and Delepelaire, P. (2004). Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58, 611–647. doi: 10.1146/annurev.micro.58.030603.123811
Zhao, J.-P., Zhu, H., Guo, X.-P., and Sun, Y.-C. (2018). AU-rich long 3′ untranslated region regulates gene expression in bacteria. Front. Microbiol. 9:3080. doi: 10.3389/fmicb.2018.03080
Zhu, H., Mao, X.-J., Guo, X.-P., and Sun, Y.-C. (2016). The hmsT 3′ untranslated region mediates c-di-GMP metabolism and biofilm formation in Yersinia pestis. Mol. Microbiol. 99, 1167–1178. doi: 10.1111/mmi.13301
Keywords: Staphylococcus aureus, 3’UTRs, post-transcriptional regulation, RNase III, PNPase, mRNA decay, ferritin, iron homeostasis
Citation: Menendez-Gil P, Catalan-Moreno A, Caballero CJ and Toledo-Arana A (2022) Staphylococcus aureus ftnA 3’-Untranslated Region Modulates Ferritin Production Facilitating Growth Under Iron Starvation Conditions. Front. Microbiol. 13:838042. doi: 10.3389/fmicb.2022.838042
Received: 17 December 2021; Accepted: 31 March 2022;
Published: 27 April 2022.
Edited by:
Harold J. Schreier, University of Maryland, Baltimore County, United StatesReviewed by:
Franz Narberhaus, Ruhr University Bochum, GermanyCopyright © 2022 Menendez-Gil, Catalan-Moreno, Caballero and Toledo-Arana. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alejandro Toledo-Arana, YS50b2xlZG8uYXJhbmFAY3NpYy5lcw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.