
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 18 April 2022
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.835647
This article is part of the Research Topic Emergence and re-emergence of plant diseases caused by Xanthomonas species View all 5 articles
 Mustafa Ojonuba Jibrin1,2*
Mustafa Ojonuba Jibrin1,2* Sujan Timilsina3
Sujan Timilsina3 Gerald V. Minsavage3
Gerald V. Minsavage3 Garry E. Vallad3,4
Garry E. Vallad3,4 Pamela D. Roberts3,5*
Pamela D. Roberts3,5* Erica M. Goss3,6*
Erica M. Goss3,6* Jeffrey B. Jones3*
Jeffrey B. Jones3*
Bacterial spot disease was first reported from South Africa by Ethel M. Doidge in 1920. In the ensuing century after the initial discovery, the pathogen has gained global attention in plant pathology research, providing insights into host–pathogen interactions, pathogen evolution, and effector discovery, such as the first discovery of transcription activation-like effectors, among many others. Four distinct genetic groups, including Xanthomonas euvesicatoria (proposed name: X. euvesicatoria pv. euvesicatoria), Xanthomonas perforans (proposed name: X. euvesicatoria pv. perforans), Xanthomonas gardneri (proposed name: Xanthomonas hortorum pv. gardneri), and Xanthomonas vesicatoria, are known to cause bacterial spot disease. Recently, a new race of a bacterial spot pathogen, race T5, which is a product of recombination between at least two Xanthomonas species, was reported in Nigeria. In this review, our focus is on the progress made on the African continent, vis-à-vis progress made in the global bacterial spot research community to provide a body of information useful for researchers in understanding the diversity, evolutionary changes, and management of the disease in Africa.
Tomato (Solanum lycopersicum L.) and pepper (Capsicum species) are two of the most important vegetable crops cultivated in Africa and worldwide (FAOSTAT, 2020). Egypt and Nigeria, which, respectively, rank fifth and fifteenth globally, are the largest producers of tomatoes in Africa (FAOSTAT, 2020). FAOSTAT reports statistics on pepper production in two different categories: chilies and green pepper. Ethiopia and Ghana lead the continent in the production of dry chilies and pepper; Nigeria and Egypt lead in green pepper production (FAOSTAT, 2020). Tomato and pepper production in Africa is hampered by many constraints, including pests (insect pests, nematodes, and diseases due to fungal, bacterial, and viral pathogens), low soil fertility, low input utilization, and low market prices. One of the most important constraints in production results from attack by pathogens, some of which could lead to total crop failures, where control measures are not implemented (Jones et al., 2016).
Bacterial diseases constitute a major component of the disease challenges reducing tomato and pepper yields in all production areas (Wai et al., 2015; Jones et al., 2016). Bacterial diseases such as bacterial spot (caused by several species belonging to the genus Xanthomonas), bacterial speck (caused by Pseudomonas syringae pv. tomato; Young et al., 1978), bacterial canker (caused by Clavibacter michiganensis; Davis et al., 1984), and bacterial wilt (caused by Ralstonia solanacearum) are among the most devastating tomato and pepper diseases (Jones et al., 2016). When they occur, the severity or level of infection of these diseases is frequently accelerated under humid conditions and could lead to severe losses when not adequately controlled.
In this review, we focus on Africa and review efforts in understanding the bacterial spot disease in this region of the world where little information is available, but recent studies suggest important findings. The bacterial spot pathogen was first isolated from tomatoes purchased at the Pretoria market in South Africa in 1914 (Doidge, 1920). Since its first report, the disease has caused devastating yield reduction globally, with yield losses up to 45 and 50% reported in Africa and the United States, respectively (Pohronezny and Volin, 1983; Kaaya and Massomo, 2003). We trace important milestones from its first published report in 1920 to the recent report of a new race in Nigeria (Jibrin et al., 2018b). Although there are recent reviews on the bacterial spot disease and its pathogens (Potnis et al., 2015; Abrahamian et al., 2021; Osdaghi et al., 2021; Potnis, 2021), we provide an African context for research progress, especially highlighting the progress that has not been emphasized in previous reviews, to provide a useful resource for future studies and management of the disease on the African continent. We hope this information provides the platform for future research on the disease and pathogens that cause bacterial spot disease on the continent and around the world.
The period 1850–1915 is generally regarded as the Golden Age of Microbiology, especially for the discovery of bacteria as pathogenic agents (Blevins and Bronze, 2010; Arguelles, 2013). This period witnessed in 1876 the discovery of the first bacterial pathogen, Bacillus anthracis, a causal agent of anthrax in cattle and sheep. Shortly after the discovery of anthrax, fire blight of apple and pear caused by Erwinia amylovora was discovered by T.J. Burrill in 1877 (Glawe, 1992). In 1882, Mycobacterium tuberculosis was discovered by Robert Koch. The pathogen-causing bacterial spot of tomato, initially described as “canker of tomato,” was also discovered within this golden period by Ethel M. Doidge in South Africa in 1914 (Doidge, 1920). She named the bacterium Bacterium vesicatorium, which was rod-shaped, aerobic, gram-negative, and contained a single polar flagellum (Doidge, 1920; Stall et al., 2009). Shortly after the publication by Doidge, the disease was identified in the United States by Gardner and Kendrick, and they named the causal organism Bacterium exitiosum (Gardner and Kendrick, 1921). Thus, the bacterial spot pathogen was subject to international focus very early on in discovery. Based on the reports from both studies, the strains described by Doidge were feebly amylolytic, whereas the strains described by Gardner and Kendrick were strongly amylolytic (Jones et al., 1998). Soon afterward, Gardner and Kendrick established that B. vesicatorium also caused bacterial spot on pepper (Gardner and Kendrick, 1923). The bacterial spot disease of pepper was also described within the same period by Sherbakoff (1918). Attempts by Higgins in 1922 and Gardner and Kendrick in 1924 to differentiate these two differently named pathogens were unsuccessful (Higgins, 1922; Gardner and Kendrick, 1924). The result was the retention of the name proposed by Doidge, B. vesicatorium, because of the priority rule in nomenclature (Gardner and Kendrick, 1923; Stall et al., 2009). From this point onward, the taxonomic journey of the bacterial spot pathogen took different routes.
Bacterium vesicatorium appeared to be recognized as the sole causal agent of bacterial spot of tomato and pepper for a long time. However, the nomenclature was changed several times to Pseudomonas vesicatoria in 1925, Phytomonas vesicatoria in 1930, Xanthomonas vesicatoria in 1939, and Xanthomonas campestris pv. vesicatoria in 1980 (Stevens, 1925; Burkholder and Li, 1941; Hayward and Waterston, 1964; Dye et al., 1980; Stall et al., 2009; Potnis et al., 2015). It was realized early in the 1990s that two distinct phenotypic and genotypic groups, named A and B, existed in X. campestris pv. vesicatoria (Vauterin et al., 1990; Stall et al., 1994). Group A strains do not hydrolyze starch or pectin, whereas group B strains do hydrolyze starch and degrade pectin (Bouzar et al., 1994c). Additionally, two genetically distinct strains identified in tomatoes from the former Yugoslavia and Florida were determined to belong to groups D and C, respectively (Šutic, 1957; Jones et al., 1995; Jones et al., 2000). In an extensive genome-based study using DNA–DNA hybridization, 16S ribosomal RNA (rRNA), and restriction fragment analyses, X. campestris pv. vesicatoria was reclassified into two species (Jones et al., 2004). These species included X. euvesicatoria (group A) and X. perforans (group C) (Jones et al., 2004). In summary, four species were designated as the causal organisms of the bacterial spot disease, including X. euvesicatoria, X. vesicatoria, X. perforans, and X. gardneri (Jones et al., 2004; Potnis et al., 2015). Recently, X. euvesicatoria and X. perforans were described as pathovars of the same species, and the names X. euvesicatoria pv. euvesicatoria and X. euvesicatoria pv. perforans were proposed (Constantin et al., 2016). Similarly, X. gardneri was recently reclassified as Xanthomonas cynarae pv. gardneri and then as X. hortorum pv. gardneri (Timilsina et al., 2019a; Morinière et al., 2020).
Symptoms of the bacterial spot disease on tomato and pepper often occur as coalesced dark-brown necrotic lesions visible on all plant parts depending on the stage of infection (Doidge, 1920; Stall et al., 2009; Osdaghi et al., 2021; Figures 1A–C). Pith necrosis on tomatoes caused by X. perforans has also been described (Aiello et al., 2013; Figure 1C). On nutrient agar, the bacterium appears as yellow mucoid colonies within 24–48 h (Figures 1D,E). Pathogenicity tests often involve inoculating a bacterial suspension adjusted to 108 cfu/ml from a 24-h-old culture on young plants of susceptible tomato and pepper cultivars, incubating under high humidity and at warm temperatures (28°C) for 48–72 h and observing for development of symptoms after an incubation period. Subsequently, biochemical, serological, and molecular tests have been used to help in differentiating the bacterial spot pathogens. Serological assays such as dot-immunobinding assay, immunofluorescence, and double diffusion have been used to diagnose bacterial spot of tomatoes in Egypt (Tawfik et al., 2009). However, biochemical tests are used more commonly in Africa (Shenge et al., 2007; Opara and Odibo, 2009). Such biochemical tests often include Gram-negative reaction (KOH test), positive motility test, positive xanthan gum production, hydrogen sulfide production, and unique patterns of carbon utilization patterns on BIOLOG plates (BIOLOG, Hayward, CA, United States). Sometimes, as with the first discovery, bacterial spot may be recovered from tomatoes that are designated for sale (Figure 1F shows typical packing in markets in Nigeria).
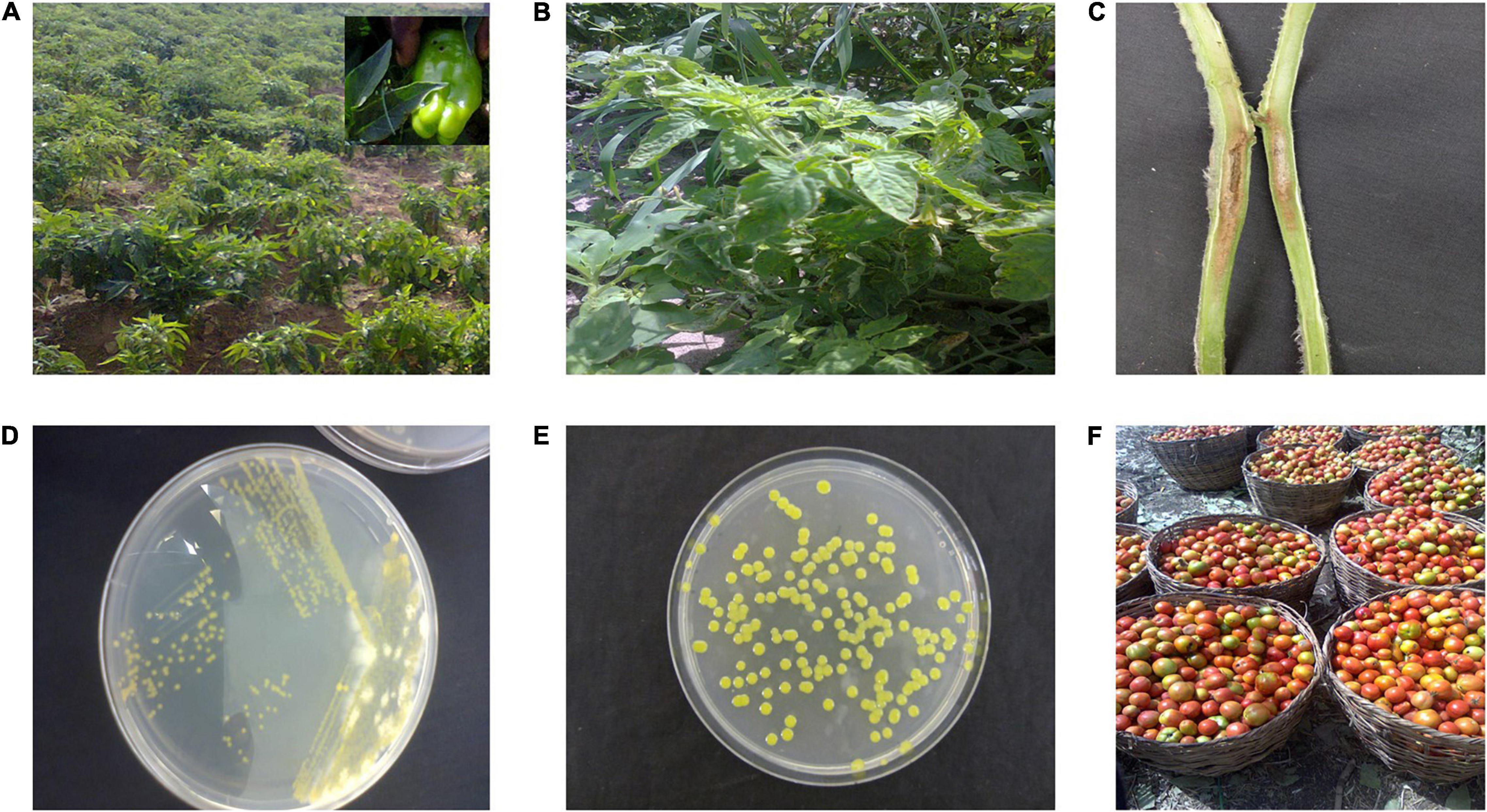
Figure 1. (A) Pepper field in northwestern Nigeria infected with bacterial spot disease. Most pepper race 6 Nigerian strains (Jibrin et al., 2014) were isolated from this field. (B) Tomato field in Tanzania infected with bacteria spot disease. (C) Pith necrosis observed in tomato stem artificially inoculated with Xanthomonas perforans strain Xp91-118. (D) Quadrant streaking to isolate pure culture of bacterial spot strains from field samples of tomato. (E) Pure culture obtained after sub-culturing strain from (D). (F) Typical harvest of tomato in Nigeria. Diseased fruits are separated from healthy fruits before marketing.
Several species-specific primers have been designed for rapid polymerase chain reaction (PCR) and quantitative PCR (qPCR) identification of bacterial spot pathogens (Koenraadt et al., 2009; Strayer et al., 2016). Although current primers are largely successful in distinguishing between the four species causing bacterial spot of tomato, it may be important to develop unique primers based on local strains in Africa because of the possibility of genetic variation. For example, the qPCR protocol based on hrcN primers utilized by Strayer et al. (2016) was accurate in diagnosing tomato X. perforans strains but not X. euvesicatoria strains from Nigeria (Jibrin et al., 2018b). This was because the tomato X. euvesicatoria strains from Nigeria had an identical hrcN sequence to X. perforans, which may have been acquired through horizontal gene transfer (Jibrin et al., 2018b). Similarly, Xanthomonas arboricola was identified as one of the pathogens isolated from tomatoes in Tanzania based on gumD and fyuA primers (Mbega et al., 2012a). An improved PCR technique was developed for differentiating members of pathogenic and nonpathogenic Xanthomonas species (Adriko et al., 2014). Such techniques could similarly be improved using available genomic resources to improve further the accuracy of diagnosing bacterial spot strains.
Based on routine pathogenicity and biochemical tests, the bacterial spot pathogens of tomato and pepper have been reported from a considerable number of countries in Africa and the associated Islands. Alongside South Africa, these countries include Egypt, Ethiopia, Ghana, Kenya, Malawi, Morocco, Mozambique, Niger, Nigeria, Reunion, Senegal, Seychelles, Sudan, Togo, Tunisia, Zambia, and Zimbabwe (OEPP/EPPO, 1988; Okoli and Erinle, 1989; Bouzar et al., 1994a,b; Black et al., 2001; Abd-Alla and Bashandy, 2008; Tawfik et al., 2009; Hamza et al., 2010, 2012; Opara and Odibo, 2009; Jibrin et al., 2014, 2015; Kebede et al., 2014; Lamini et al., 2015; Algam et al., 2018; Ashmawy et al., 2020). The specific species of the pathogen responsible for the disease are yet to be determined for a considerable number of countries. Figure 2 shows the map of Africa and associated Southwest Indian Ocean (SWIO) islands where bacterial spot pathogen(s) has been reported.
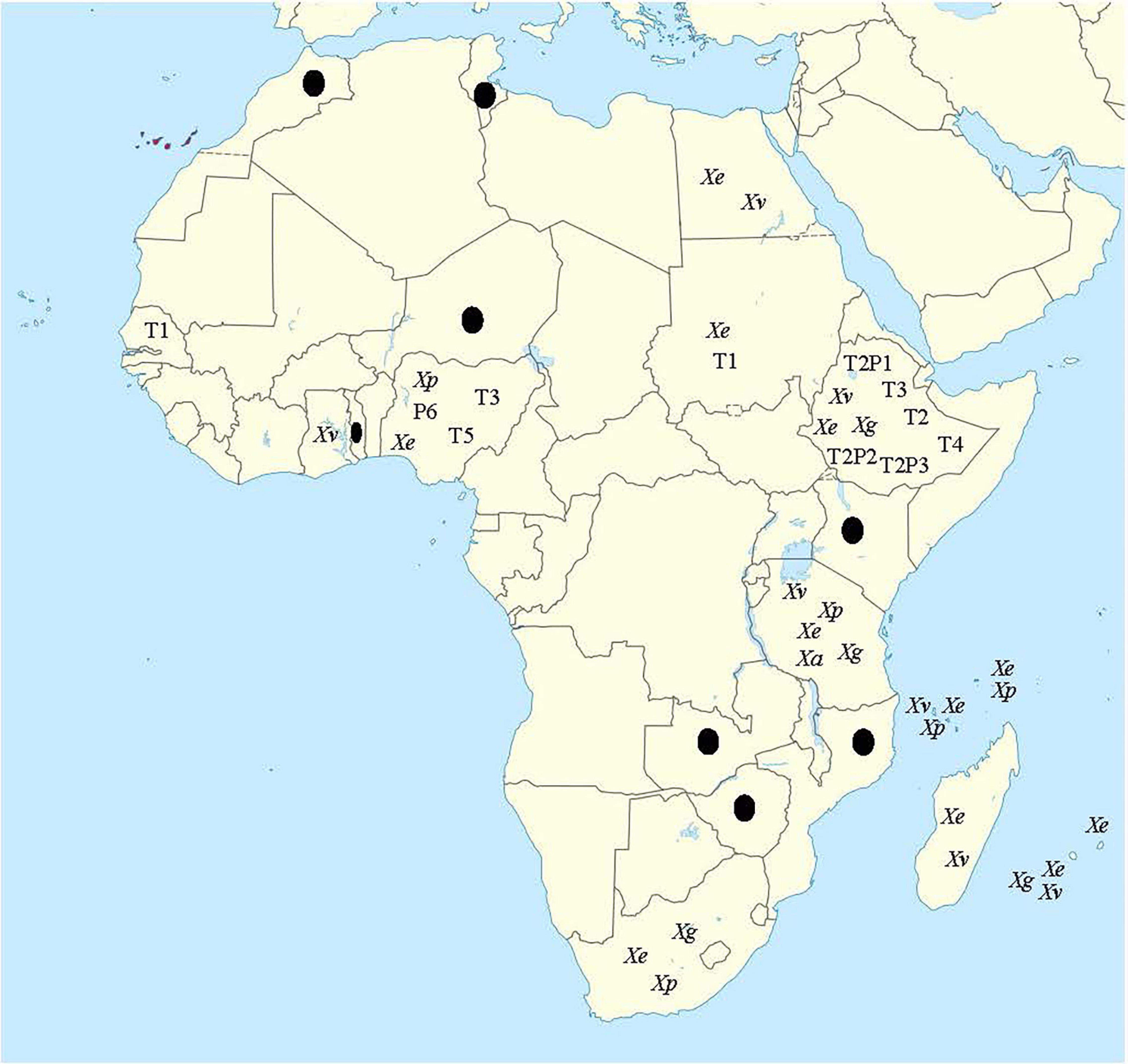
Figure 2. Map of Africa showing countries where bacterial spot disease has been reported on tomato and/or pepper. Xe, Xp, Xg, and Xv represent X. euvesicatoria (X. euvesicatoria pv. euvesicatoria), X. perforans (X. euvesicatoria pv. perforans), X. hortorum pv. gardneri, and X. vesicatoria, respectively. Xa represents X. arboricola reported only in Tanzania. Races include T1, T2, T3, T5, T2P1, T2P2, and T2P3. Countries with black dots are those for which bacterial spot have been reported, but neither species nor race was identified. Map for this figure was obtained from https://commons.wikimedia.org/wiki/File:Canary_ Islands_in_Africa_(-mini_map_-rivers).svg (date of access: 6 September 2021).
Identification of species of Xanthomonas on tomato and pepper in Africa has routinely followed the four species nomenclature. Based on this classification, all four bacterial spot Xanthomonas spp. have been identified in Africa. In a study of the bacterial spot pathogen in the Southwest Indian Ocean, X. euvesicatoria, X. perforans, X. vesicatoria, and X. gardneri were identified from recovered strains (Hamza et al., 2010). X. euvesicatoria was found in Seychelles, Mauritius, and Reunion on tomato and pepper, with three distinct groups described. Strains in the first group, which were isolated from tomatoes in Reunion and Comoros Island over a 6-year period, showed a higher population in lesions when artificially inoculated over a longer period than other strains. These strains also produced avirulent reactions on pepper cv. Aiguille. Strains in the second group were isolated from tomatoes in Seychelles and Comoros Islands and were also pathogenic on pepper cv. Aiguille. Strains in the third group were isolated from pepper in Reunion, Mauritius, Seychelles, and the Comoros Islands and were pathogenic on tomato cv. Marmande but were genetically heterogeneous based on amplified fragment length polymorphism (Hamza et al., 2010). Most strains in the third group produced a low population when inoculated on tomato cv. Marmande (Hamza et al., 2010). X. perforans was reported in Seychelles and Mauritius, whereas X. vesicatoria was found in Madagascar and Reunion. All recovered X. perforans and X. vesicatoria strains were from tomato and were pathogenic on tomato but not pepper (Hamza et al., 2010). X. gardneri was isolated from tomato and pepper but was found only in Reunion (Hamza et al., 2010). In a study covering tomato-growing regions of northern, central, and southern Tanzania, all four species, in addition to X. arboricola, were reported to cause bacterial spot of tomato (Mbega et al., 2012a). The report of X. arboricola causing bacterial spot of tomato in Tanzania was the first of its kind, and no other studies have identified this species as a similar bacterial spot pathogen. In Nigeria, X. euvesicatoria was reported in pepper in northwestern Nigeria (Jibrin, 2014; Jibrin et al., 2014). From the same region in Nigeria, an atypical X. perforans strain was obtained from tomato that contains alleles of genes in X. euvesicatoria and other unknown xanthomonads, suggesting extensive recombination (Jibrin et al., 2015; Timilsina et al., 2015). In Ghana, a survey of tomato fields in Ashanti, Brong Ahafo, and Upper East regions identified X. vesicatoria as the causal organism of bacterial spot disease (Lamini et al., 2015). In Sudan, X. euvesicatoria was reported from tomato and pepper in Blue Nile State and central Sudan (Bouzar et al., 1994a; Algam et al., 2018). X. perforans, X. vesicatoria, and X. gardneri are associated with bacterial spot of tomato in Ethiopia (Kebede et al., 2014). Alongside the previously described X. euvesicatoria by Doidge, X. perforans and X. gardneri have also been associated with bacterial spot disease in South Africa (Jibrin et al., 2018a; Jibrin, 2019).
Africa is the most tropical of all continents, and temperatures are expected to rise in the coming years if adequate measures are not taken (Engelbrecht et al., 2015; Nangombe et al., 2018). Although bacterial spot pathogens are known to cause disease globally, they often prefer warm and humid conditions for optimum disease epidemics. Early in the discovery of the disease, it was shown that the incidence of the disease was directly related to rainfall, with moist, humid conditions favorable for disease development (Doidge, 1920). Tomato and pepper cultivation in Africa is done under rain-fed and irrigation conditions and protected agriculture (Gordon, 2009; Abolmaaty and Abd El-Ghafar, 2010; NAERLS., 2012; Shenge et al., 2018). The climate of Africa coupled with the irrigation or rainfall under which tomato and pepper are cultivated inadvertently provide optimum conditions for severe disease epidemics. However, variation in epidemics due to seasons, hosts, seed source, and agronomic practices are known to occur (Abolmaaty and Abd El-Ghafar, 2010; Shenge et al., 2018). In a trial in Egypt to compare disease severity in protected agriculture (plastic house) and open field, the disease severity was significantly higher under irrigated protected agriculture than in open fields (Abolmaaty and Abd El-Ghafar, 2010). The trial also recorded that disease severity was positively correlated with average temperature and relative humidity over a 3-year period (Abolmaaty and Abd El-Ghafar, 2010). In a trial in Nigeria, the incidence and severity of the bacterial spot pathogen were more severe under rain-fed production than irrigation during the dry season (Shenge et al., 2018).
Solanaceous weeds have been reported to be hosts for the bacterial spot pathogen and may be important in disease epidemiology (Potnis et al., 2015). A recent study in Egypt identified X. vesicatoria associated with solanaceous plants based on pathogenicity tests and 16S rRNA analysis (Ashmawy et al., 2020). In field surveys, it was found that field sanitation influenced bacterial spot disease severity in Tanzania, with abandoned fields with no weeding having high disease severity (Shenge et al., 2010). In a recent study in Brazil, artificial inoculation of weeds with X. euvesicatoria, X. perforans, and X. gardneri resulted in the development of lesions typical of bacterial spot (Araújo et al., 2015; Santos et al., 2020). This is indirect evidence that the bacteria can survive on weeds. Recently, X. perforans strains were isolated from forest seedlings of Eucalyptus pellita in Indonesia (Bophela et al., 2019), suggesting that the bacteria could survive as epiphytes in forest trees.
Seeds are important in bacterial spot disease epidemiology, as they often serve as sources of inoculum for the pathogens. In Africa, the bacterial spot pathogens have been routinely isolated from seeds (Abdalla, 2000; Black et al., 2001; Opara and Odibo, 2009; Shenge et al., 2010; Mbega et al., 2012b). Isolation of pathogens from fruits is also common in Africa. Bacterial spot pathogens have been routinely isolated from fruits in Tanzania and Nigeria, with some of the strains in Nigeria being identified as X. euvesicatoria (Opara and Odibo, 2009; Shenge et al., 2010; Jibrin, 2014).
Wind and rain are known to disperse bacterial spot xanthomonads (McInnes et al., 1988; Bernal and Berger, 1996). However, the impact of wind-driven events such as the harmattan, which spread dry and dusty northeasterly winds from the Sahara Desert over much of West Africa into the Gulf of Guinea, have not been evaluated. Africa has also experienced recent histories of climate extremes such as cyclones, floods, droughts, and heatwaves (Reason and Keibel, 2004; Russo et al., 2016; Barbier et al., 2018). These events, which are becoming sustained in several parts of Africa, could consequently affect microbial epidemiology, including those of plant pathogens, including bacterial spot xanthomonads, as has been shown, for example, for pathogens of animals (Bannor and Ogunsan, 1988).
Race determination of strains is based on the hypersensitive reaction (HR) in response to effector proteins in Xanthomonas strains delivered via the type III secretion system into host cells and recognition by specific resistance proteins in tomato or pepper (Stall et al., 2009). Table 1 shows the reactions of different bacterial spot pathogen races on the differential lines for tomato and pepper. Resistance genes in tomato include the nondominant genes rx1, rx2, and rx3 in Hawaiian accession line H7998 and dominant Xv3 gene in Fl216 and Xv4 gene in Solanum pennellii that were introgressed into S. lycopersicum (Stall et al., 2009). Strains that produce HR on H7998 only are referred to as T1 strains. Only X. euvesicatoria strains have been identified as T1 strains to date (Stall et al., 2009; Timilsina et al., 2016). Race T2 represents strains that do not produce an HR on any of the differential lines. This includes all strains that did not contain avrXv3, avrXv4, or avrRxv. All X. vesicatoria strains and some X. gardneri strains belong to this race (Stall et al., 2009; Timilsina et al., 2016). Race T3 initially included strains that produced a susceptible reaction on H7998 and carried the avrXv3 gene when expressed elicited an HR in FL216 or H7981 (Jones et al., 1988; Stall et al., 2009). Race T3 strains were determined to be X. perforans, as they were genetically different from X. euvesicatoria (Stall et al., 2009). Later, it was shown that T3 strains contain functional xopJ4 (avrXv4) and elicit an HR on tomato lines with Xv4 resistance gene (Astua-Monge et al., 2000; Stall et al., 2009). Strains that have functional xopJ4 but nonfunctional avrXv3 were later designated as race T4 (Timilsina et al., 2016). Pepper races are determined by the reactions of avrBs1, avrBs2, avrBs3, and avrBs4c genes in pathogens upon recognition by Bs1, Bs2, Bs3, and Bs4c resistance genes in pepper, respectively (Stall et al., 2009). The different lines used for characterization of pepper races include PI163192, PI260435, PI271322, and PI235047, which, respectively, contain the Bs1, Bs2, Bs3, and Bs4 resistance genes (Stall et al., 2009; Table 1).
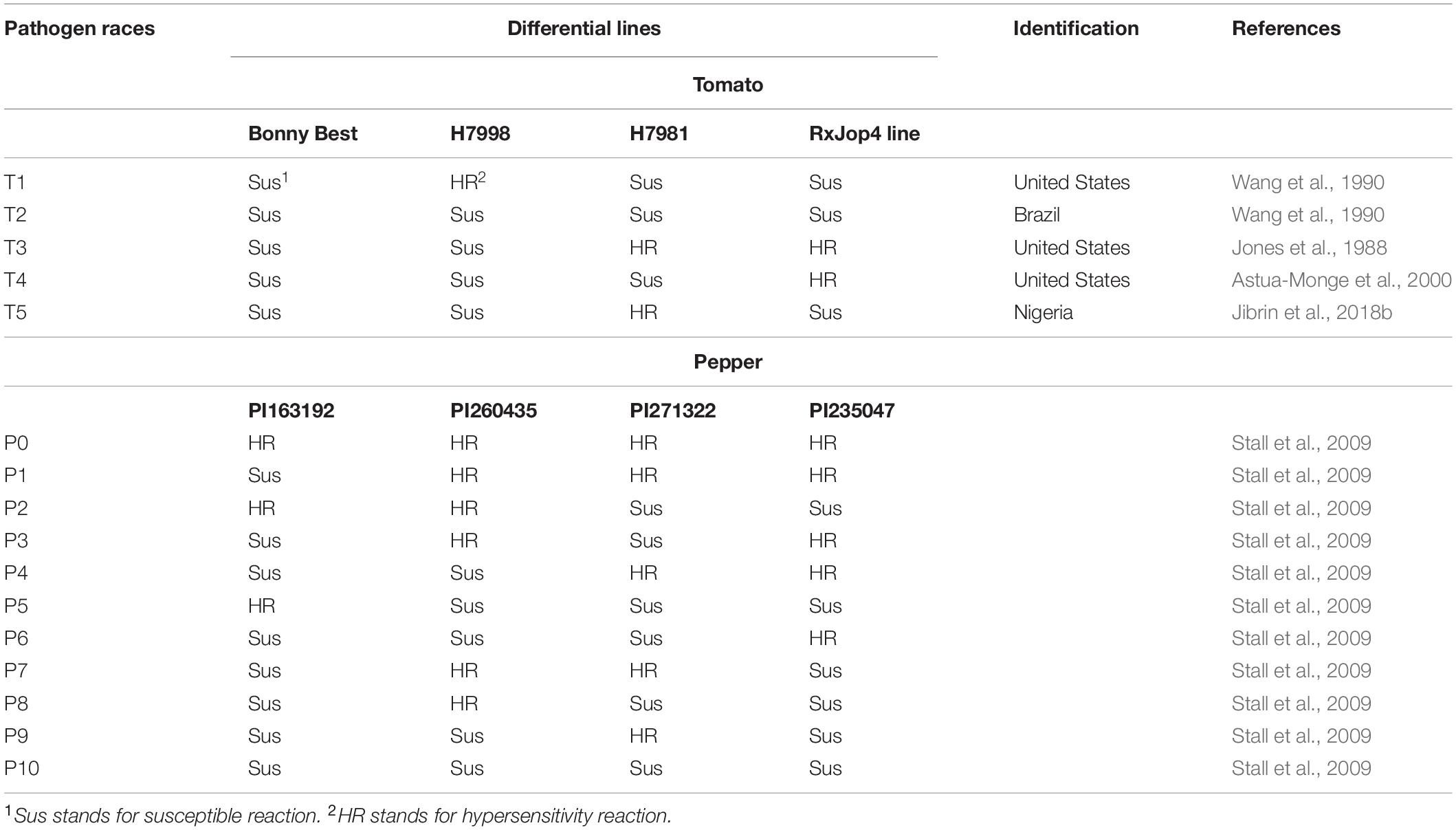
Table 1. Races of bacterial spot xanthomonads following Stall et al. (2009) and emergence of race T5 in Africa.
Earlier studies of race distribution of bacterial spot of tomato and pepper strains from the Sudan and Senegal identified T1 races (Bouzar et al., 1994a,b; Jones et al., 1998). Pepper race 6 and tomato race T3 were previously identified in Nigeria (Jibrin et al., 2014, 2015). The initial race test did not include the Xv4 (also referred to as RXopJ4 differential line), and those strains were originally designated as T3 strains (Jibrin, 2014; Jibrin et al., 2015). Subsequently, it was shown that these strains, with a representative NI1 strain whose genome was sequenced, lacked xopJ4 and would not elicit an HR on the RXopJ4 differential line (Jibrin et al., 2018b). These strains produce a new set of reactions on the available differential lines, and we propose that they should be designated as T5 race (Table 1). The highest diversity in races was reported in Ethiopia, where races T2, T2P1, T2P2, T2P3, T3, and T4 have been reported (Kebede et al., 2014). The races of bacterial spot xanthomonads that occur in other African countries are yet to be determined probably because race determination requires the use of race differential lines that are not widely available on the continent. The emergence of NI1 race T5 in northwestern Nigeria and multiple races within the central region of Ethiopia may point to unique and unsampled diversity in Africa. For example, Hamza et al. (2010) reported differential pathogenicity when X. euvesicatoria strains from countries in the SWIO were inoculated on pepper cultivar Aiguille as opposed to the similar pathogenicity reaction on the cultivar Marmande (Hamza et al., 2010). Future studies may yet uncover unknown races and possible novel pathogen–host interaction dynamics.
Natural environments and geographies do have an impact on bacterial evolution through various interactions that lead to mutations, gain and loss of genes, and recombination (Martinez, 2009; Pearson et al., 2009; Li et al., 2014). Understanding how bacteria evolve in different environments and their effects on population structure is important from an ecological perspective and can impact disease management (Allen and Banfield, 2005; Potnis, 2021). In several studies, multilocus sequence analyses indicated a unique population from Africa (Hamza et al., 2012; Kebede et al., 2014; Timilsina et al., 2015). In an analysis utilizing four housekeeping genes (atpD, dnaK, efp, and gyrB), strains from countries in the SWIO showed multiple sequence types (STs) in each species (Hamza et al., 2010). The type strains used in the study included X. euvesicatoria NCPPB2968 from the United States, X. vesicatoria LMG911 from New Zealand, X. perforans NCPPB4321 from the United States, and X. gardneri NCPPB881 (ATCC19865) from the former Yugoslavia. There were two (E1 and E3), three (P1, P2, and P3), one (G1), and two (V1 and V2) STs of X. euvesicatoria, X. perforans, X. gardneri, and X. vesicatoria strains, respectively, from countries in the SWIO (Hamza et al., 2010). Although most X. euvesicatoria strains were ST E1 (which included the type strain), ST E3 was only identified in some strains from Mauritius and Reunion (Hamza et al., 2010). ST P1 included X. perforans type strain and was comprised of most of the strains from Seychelles, except for a single strain that was assigned to ST P2 (Hamza et al., 2010). A clonal population was inferred for X. gardneri, as there was no observed diversity in the sequenced genes. Most X. vesicatoria strains belonged to ST V1, including the reference strain. However, strains from Madagascar and Reunion belonged to ST V2 and were identical to strains originating from Europe (Hamza et al., 2010). Interestingly too, there was evidence of recombination events in the atpD gene of X. euvesicatoria strains (Hamza et al., 2010). In a separate multilocus sequence analysis (MLSA) study based on the same housekeeping gene set, a tomato strain from Zambia (LMG927) and two tomato strains originating from the United States (ICMP110 and LMG10429) were shown to be more closely related to X. campestris pv. raphani and were pathogenic on solanaceous species and radish (Hamza et al., 2012).
Following the proposal for a standardized and uniform set of housekeeping genes for MLSAs to aid the continued study of populations of xanthomonads, subsequent studies utilized partial sequences of six genes, including fusA, gapA, gltA, gyrB, lacF, and lepA (Almeida et al., 2010). MLSAs utilizing this scheme identified a diverse population of bacterial spot pathogens consisting of X. vesicatoria, X. perforans, and X. gardneri in Central Ethiopia (Kebede et al., 2014). The reference strains used in the studies included X. euvesicatoria 85-10 from the United States, X. perforans 91-118 from the United States, X. gardneri ATCC19865 from the former Yugoslavia, and X. vesicatoria ATCC35937 from New Zealand. Two multilocus haplotypes were identified in X. vesicatoria strains from Ethiopia, whereas a third haplotype for the species was identified in Reunion and Madagascar (Kebede et al., 2014; Timilsina et al., 2015). In Ethiopia, there were no sequence differences between the X. perforans and X. gardneri strains and the reference strains of both species, suggesting that the X. perforans strains were similar to X. perforans strains from the United States. There was limited variation in X. gardneri in general (Kebede et al., 2014). Although the diversity of the bacterial spot pathogen recovered from samples within a short distance in Central Ethiopia was interesting, as three species were identified, there was no observed recombination in multilocus sequences of the analyzed strains (Kebede et al., 2014). Studies based on partial sequences of six housekeeping genes showed that bacterial spot of tomato strains from Nigeria gave unique results (Jibrin et al., 2015; Timilsina et al., 2015). The pepper strains were easily assigned to X. euvesicatoria based on MLSA (Jibrin et al., 2014); however, although the tomato strains were similar to X. perforans based on the routinely used hrcN gene sequence, they were phylogenetically difficult to assign to X. euvesicatoria or X. perforans based on MLSA (Jibrin et al., 2015; Timilsina et al., 2015). Specifically, an unusual recombination event was identified in the sequenced genes of the tomato strains from Nigeria. The partial sequences of gltA and lacF genes were identical to X. euvesicatoria, whereas the fusA and gyrB genes were identical to X. perforans. However, gapA and, more significantly, lepA genes were unique. The lepA gene was not related to any bacterial spot xanthomonad lepA gene (Timilsina et al., 2015). MLSAs failed to resolve the taxonomy of the tomato strains from Nigeria, and they were termed “atypical X. euvesicatoria” strains (Jibrin et al., 2015; Timilsina et al., 2015).
Population structure of African strains based on these three studies in the SWIO, Ethiopia, and Nigeria suggests unique events that drive the type and population of bacterial spot pathogens in each country. In all three studies, sampling was done within a limited area of the countries surveyed, and there is an opportunity for more studies. These types of studies utilizing multilocus analyses may be an important front for studying bacterial spot populations in Africa.
The reference genomes of the four species causing bacterial spot disease have been sequenced, and none of the reference strains were isolated in Africa (Thieme et al., 2005; Potnis et al., 2011). Analyses of the sequenced genomes provided important insights into types of secretion systems, effectors, lipopolysaccharide (LPS) clusters, copper resistance genes, and species-specific genes that underpin pathogen evolution and host–pathogen interaction (Thieme et al., 2005; Potnis et al., 2011).
Genomes of X. euvesicatoria, X. perforans, and X. gardneri from Africa have been sequenced (Richard et al., 2017a; Jibrin et al., 2018a,b; Jibrin, 2019). Figure 3 shows the comparison of representative African strains with the reference strains of the bacterial spot xanthomonads. The first complete genomes of bacterial spot strains from Africa were those of X. euvesicatoria strain LH3 and X. gardneri strain JS749-3 from Mauritius and Reunion, respectively (Richard et al., 2017a). The genome of strain NI1 from Nigeria was the first X. perforans genome from Africa that was sequenced and, alongside the genome of X. euvesicatoria strain NI38, were also the first genomes of African bacterial spot xanthomonads that were described in detail (Jibrin et al., 2018b). Recombination based on whole-genome sequencing among the bacterial spot pathogens was initially described in the genome of the Nigerian X. perforans strain, NI1 (Jibrin et al., 2018b). Subsequently, recombination was also shown in the genomes of X. perforans strains from Florida and Alabama in the United States (Newberry et al., 2019; Timilsina et al., 2019b). Notably, the core genome phylogeny and pangenome matrix suggest that the NI1 strain was more like Xanthomonas axonopodis pv. allii (now X. euvesicatoria pv. allii) and was intermediate between previously described X. euvesicatoria and X. perforans strains (Jibrin et al., 2018b). It was, therefore, initially thought that the Nigerian tomato strains may have been a result of the host jump between onion (Allium species) and tomato. The genome of strain JS749-3 from Reunion in 1997 was the first sequenced X. gardneri strain that was described to have the copper resistance copLAB gene cluster (Richard et al., 2017a). Similarly, the plasmids of both LH3 and JS749-3 contain the cusAB-smmD, czcABCD, and arsBHCR resistance gene clusters, except that LH3 lacks the latter gene cluster (Richard et al., 2017a). Interestingly, genome mining showed that the spread of these mobile element-associated copper resistance genes and the plasmid carrying them is restricted among bacteria within the Xanthomonas genus (Richard et al., 2017b). These gene clusters are important in the survival of the bacteria under heavy metal conditions (Richard et al., 2017a,b). We further provide a detailed description of variation in the secretion systems, LPS cluster, and effectors of African strains compared with the reference strains for each species.
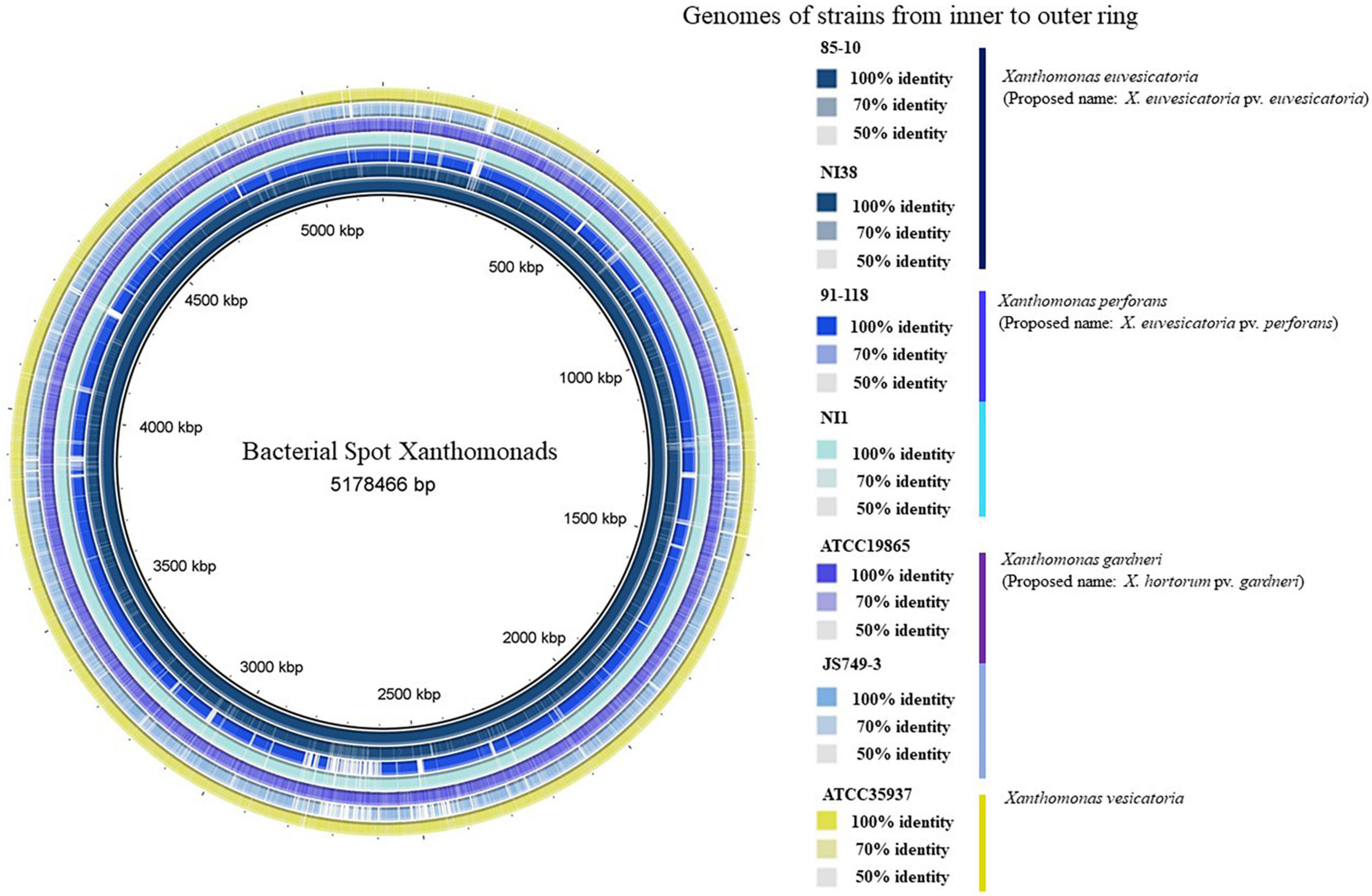
Figure 3. Comparisons of reference strains of each bacterial spot species against sequenced representative strains from Africa. From innermost to outermost ring: Xanthomonas euvesicatoria 85-10, X. euvesicatoria NI38, X. perforans 91-118, X. perforans NI1, X. gardneri ATCC19865, X. gardneri JS749-3, and X. vesicatoria ATCC35937. There was no representative African X. vesicatoria strain, as no known strain has been sequenced. Varying color gradients in percentage identities are based on BLAST match of a minimum of 50% percentage identity (1e-5) of genomic regions against genome of strain 85-10 as shown in key. Because BLAST matches are calculated using 85-10 as reference sequences, regions that are absent from genome of strain 85-10 but present in one or more of other genomes are not displayed. Vertical lines in key are, respectively, colored according to color of each genome in ring. Circular genome was produced using BLAST Ring Image Generator (BRIG; Alikhan et al., 2011).
At least six (6) secretion systems were described in the sequenced reference strains of the bacterial spot xanthomonads with varying organizations (Potnis et al., 2011). These include type 1 secretion system (T1SS), type 2 secretion system (T2SS), type 3 secretion system (T3SS), type 4 secretion system (T4SS), type 5 secretion system (T5SS), and type 6 secretion system (T6SS). We review the organization of these secretion systems in NI38, NI1, and JS749-3 compared with their reference strains, which are, respectively, X. euvesicatoria 85-10, X. perforans Xp91-118, and X. gardneri ATCC19865. A schematic diagram showing the organization of the six secretion systems and LPS cluster is shown in Figure 4.
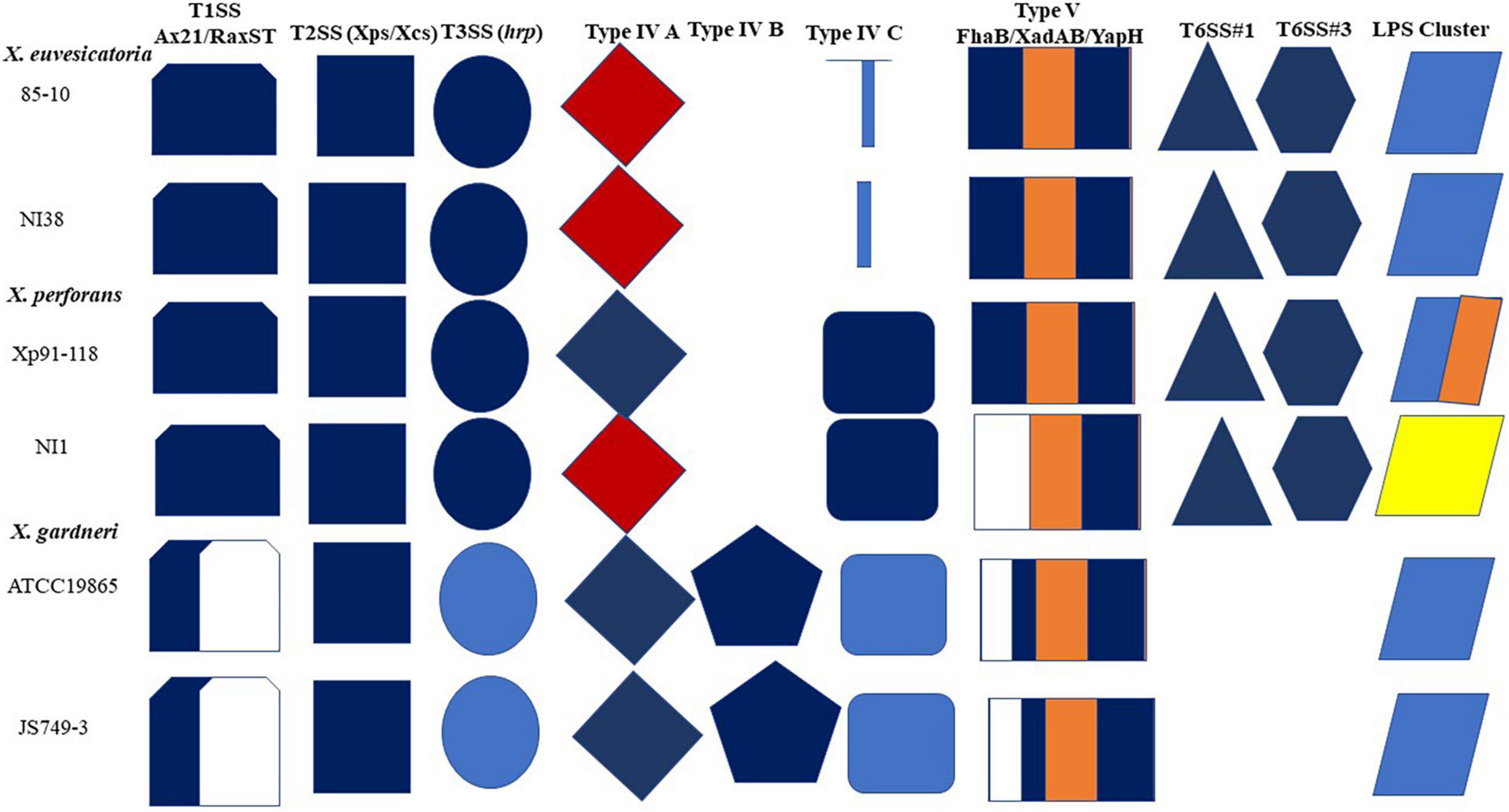
Figure 4. A diagrammatic representation of secretion systems and LPS clusters in reference strains compared with sequenced African strains. Shapes and color codes follow methodology of Potnis et al. (2011) with modifications. For T1SS, dark blue color in shape shows presence of homologs of Ax21 and RaxST; white space indicates absence of RaxST. T2SS systems show presence of both Xps and Xcs systems in reference strains and African strains. T3SS shows similarity in hrp cluster. Unique X. gardneri hrp cluster is in faint blue. Type IVA shows Dot/Icm cluster in red and Vir system in deep blue. Unique type IVB cluster in X. gardneri is also shown, whereas type IVC is shown. T5SS shows that NI1 lacks homologs of genes encoding FhaB protein, whereas one copy is present in X. gardneri. X. gardneri lacks T6SS. LPS cluster of NI1 is most unique and is similar to that of X. translucence pv. translucence (Jibrin et al., 2018b).
Type 1 secretion system: An ax21 (activator of XA21-mediated immunity) gene was reported to be conserved in all bacterial spot xanthomonads (Potnis et al., 2011). The ax21 gene was predicted to encode a type 1 secreted protein that is involved in quorum sensing signaling (Lee et al., 2008). Homologs of the ax21 gene are also found in NI38, NI1, and JS749-3 and are like the copies found in their reference strains. However, there is variability in the raxST gene, which encodes the putative sulfotransferase raxST gene necessary for recognition of XA21, which is consistent with previous findings (da Silva et al., 2004; Potnis et al., 2011). Although a different homolog of the raxST gene is found in both NI38 and NI1 compared with the reference strains of X. euvesicatoria and X. perforans, respectively, no raxST gene was found in JS749-3, which is consistent with the absence of the gene in reference X. gardneri genome.
Type 2 secretion system: Two T2SS clusters, the Xps and Xcs systems, are found in the reference strains of the bacterial spot xanthomonads (Potnis et al., 2011). The Xps system is activated by HrpG and HrpX and represses xylanases and proteases (Szczesny et al., 2010). However, the function of the Xcs system in bacterial virulence is unclear (Szczesny et al., 2010). Both the Xps and Xcs systems are present in all the sequenced African strains, including the reference strains.
Type 3 secretion system: A hrp (hypersensitivity, response, and pathogenicity) gene cluster of the T3SS is found in all sequenced reference strains (Potnis et al., 2011). The hrp cluster of X. euvesicatoria is almost identical to that of X. perforans, with differences in the organization of the hpaG and hpaF, which are fused in X. perforans but occur as separate genes in X. euvesicatoria (Potnis et al., 2011). Previous analyses of the T3SS gene clusters in NI1 and NI38 showed that genes in the hrp and hrc (hrp conserved) are similar to their reference strains, with evidence of recombination in some genes (Jibrin et al., 2018b). It was also shown that although the core and pangenome of NI1 were similar to X. axonopodis pv. allii, the T3SS genes of NI1 were more like X. perforans (Jibrin et al., 2018b). The hrcN gene of NI38 was notably similar to the hrcN gene of X. perforans strain 4P1S2 isolated from Italy (Jibrin et al., 2018b). The organization of the hrp cluster in strain JS749-3 is similar to the X. gardneri reference strain.
Type 4 secretion system: Two T4SS clusters, Vir and Dot/Icm types, are found in bacterial spot xanthomonads. All reference strains contained more than one copy of the T4SS cluster, but only 85-10 contain both the Vir and Dot/Icm clusters, which are found on the plasmid (Thieme et al., 2005; Potnis et al., 2011). The reference strains of X. perforans, X. vesicatoria, and X. gardneri lack the Dot/Icm type. Additionally, two clusters of T4SS are found in Xp91-118, with one on the plasmid and the second on the chromosome. In X. gardneri, two unique T4SSs are found on the plasmids, whereas one cluster is present on the chromosome (Potnis et al., 2011). Based on this information, the T4SSs in bacterial spot xanthomonads were classified as type IV A (which include the Dot/Icm type in X. euvesicatoria and other plasmid-borne clusters), type IV B (representing the unique clusters in X. gardneri), and type IV C (representing the chromosomal T4SS clusters) (Potnis et al., 2011). Like their reference strains, NI38 has both the Vir and Dot/Icm clusters, whereas JS749-3 has the sets of Vir clusters described for the reference X. gardneri strain. However, race T5 Nigerian X. perforans strain NI1 T4SS cluster is different from the X. perforans reference strain and has both the Vir and Dot/Icm clusters, such as X. euvesicatoria strain 85-10 (Figure 4).
Type 5 secretion system: The T5SS secreted proteins described in the reference genomes include FhaB, XadA, XadB, and YapH (Potnis et al., 2011). FhaB hemagglutinin is present in all the reference genomes of the bacterial spot pathogen, which is possibly inactivated by an internal stop codon (Potnis et al., 2011). Homologs of the two separate open reading frames for fhaB are present in NI38 and JS749-3 but absent in NI1. Homologs of Xanthomonas adhesin-like proteins XadA and XadB, which enhance bacterial colonization of the leaf surface and entry into hydathodes (Das et al., 2009), are also found in all the reference strains and the strains NI1, NI38, and JS743-9. YapH1 and YapH2, shown to be involved in virulence in Xanthomonas oryzae pv. oryzae at later stages of growth, are present in 85-10, NI38, Xp91-118, and NI1. However, only YapH2 but not YapH1 is present in JS749-3 and the X. gardneri reference strain ATCC19865.
Type 6 secretion system: Three different clusters of T6SS, named types 1, 2, and 3, are reported in xanthomonads and have been shown to be involved in host–pathogen interactions (Potnis et al., 2011). The reference strains of X. euvesicatoria and X. perforans both possess the types 1 and 3 T6SS, whereas X. vesicatoria has the type 3 T6SS only. No T6SS cluster was reported in the reference strain of X. gardneri (Potnis et al., 2011). Similarly, only types 1 and 3 T6SSs are found in NI1 and NI38. No T6SS was found in strain JS743-9.
The genes in the LPS biosynthetic locus in the bacterial spot xanthomonads are positioned between two highly conserved housekeeping genes, cystathionine gamma-lyase (metB) and electron transport flavoprotein (etfA; Patil et al., 2007; Potnis et al., 2011). The LPS cluster of X. gardneri reference strain is identical to the LPS cluster of X. vesicatoria reference strain and differs from that of X. euvesicatoria reference strain in the functions of the two glycosyl transferases found in the cluster (Molinaro et al., 2003; Thieme et al., 2005; Potnis et al., 2011). However, X. perforans evolved a novel LPS cluster (Potnis et al., 2011). The X. euvesicatoria strain NI38 was reported to have identical LPS cluster to 85-10, but NI1 has evolved a different LPS cluster not found in other sequenced bacterial spot strains (Jibrin et al., 2018b). The LPS cluster of strain NI1 was identical to the LPS cluster of Xanthomonas translucens pv. translucens, a pathogen that causes bacterial wilt and black chaff of barley (Hordeum vulgare) (Jibrin et al., 2018b; Figure 4). The LPS cluster of strain JS749-3 is identical to that of the X. gardneri reference strain.
A diverse array of effectors governs the interaction of the bacterial spot xanthomonads with their host, with 11 core effectors initially described as shared among all bacterial spot species (Potnis et al., 2011). These effectors include AvrBs2, XopD, XopF1, XopK, XopL, XopN, XopQ, XopR, XopX, XopZ1, and XopAD. Subsequent studies identified XopE2 as a core bacterial spot Xanthomonas effector (Schwartz et al., 2015). Comparison of the genomes of African strains of bacterial spot pathogens with the reference strains and other published genomes showed that the core effectors are conserved with few notable differences (Jibrin et al., 2018b; Table 2). For X. euvesicatoria, all the core effectors in NI38 have the same allele as the reference strain 85-10 except for XopQ, XopAD, and XopE2, which are different alleles. There is an early termination in XopQ of NI38, resulting in 42 amino acids in the protein sequence compared with 464 amino acids in the protein sequence of XopQ of strain 85-10, suggesting that it may be nonfunctional. In X. perforans, there is a difference in alleles of XopD, XopK, XopL, XopQ, XopZ1, and XopAD. XopE2, which was found to be conserved in Florida, United States field strains of X. perforans (Schwartz et al., 2015), is absent in NI1 and Xp91-118 (Jibrin et al., 2018b). In X. gardneri, there are differences in alleles for XopD, XopL, XopN, XopQ, XopAD, and XopE2 in strains JS749-3 and ATCC19865.
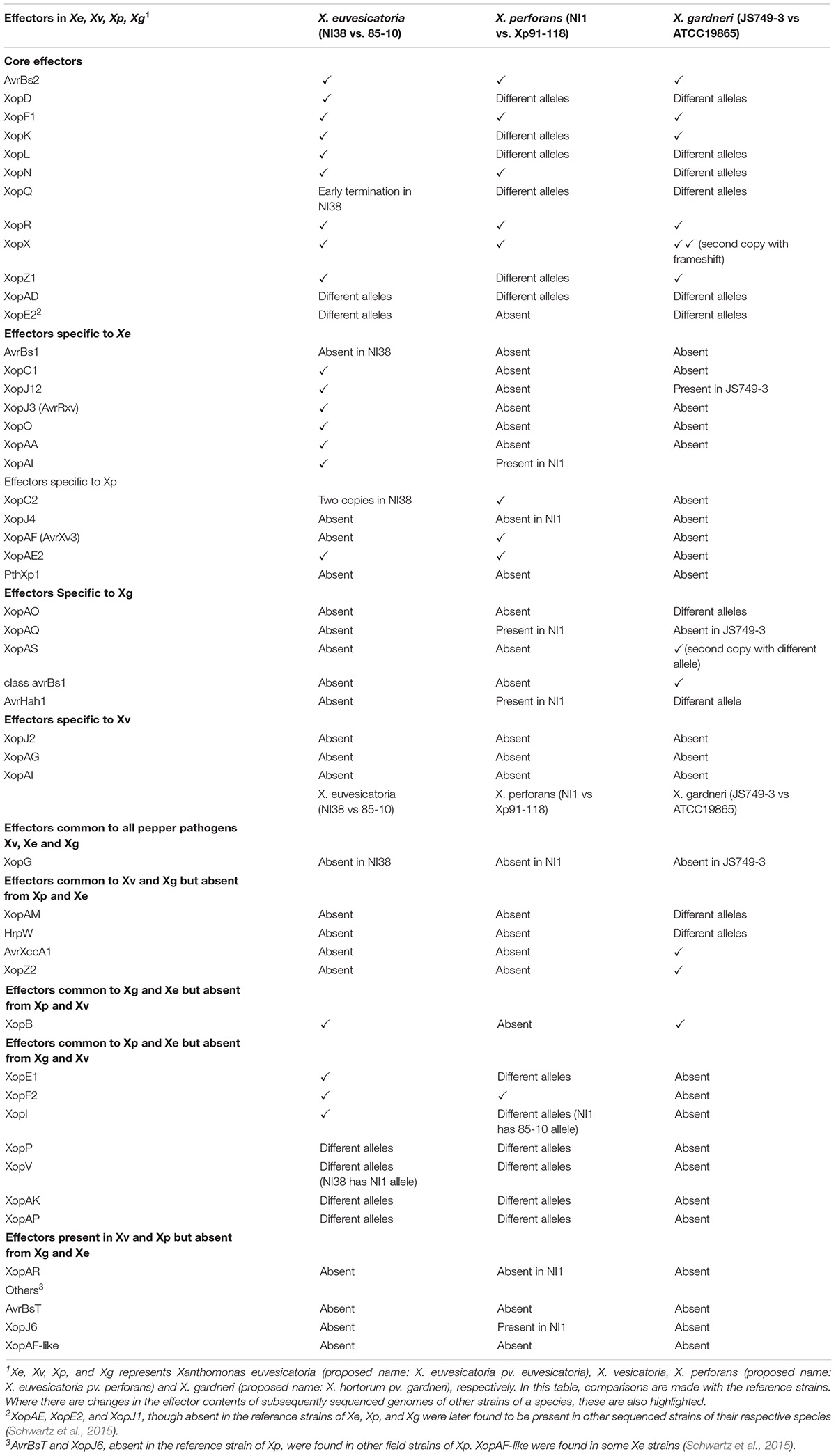
Table 2. Effector diversity in reference strains of X. euvesicatoria, X. perforans, and X. gardneri compared with African strains of same species.
Among effectors that are specific to X. euvesicatoria, NI38 has a similar allele with 85-10 except for AvrBs1, which is not present in NI38. All X. euvesicatoria-specific effectors are absent in the X. perforans reference strain and NI1 except for XopAI, which is present in NI1. Similarly, the reference strain of X. gardneri lacks all the initially described X. euvesicatoria-specific effectors, but XopJ1 was later found in subsequently sequenced strains (Potnis et al., 2011; Schwartz et al., 2015). X. gardneri strain JS749-3 possesses XopJ1 but lacks all other X. euvesicatoria-specific effectors (Table 2). XopAE and XopC2, initially described as specific to X. perforans (Potnis et al., 2011), are present in NI38 (Jibrin et al., 2018b). Although XopAE is not present in 85-10, some sequenced strains of X. euvesicatoria have XopAE (Schwartz et al., 2015). Interestingly, NI1 lacks XopJ4, the effector that led to the discovery of race 4 in X. perforans. The absence of this effector in NI1 and NI-1-like strains defines the new race T5. Recently, a new AvrBs3-type transcription activation-like (TAL) effector, pthXp1, which enhances symptom development in tomatoes and delays pustule formation in citrus, was reported in X. perforans strains from Italy and the United States (Jibrin et al., 2019; Newberry et al., 2019). This TAL effector is absent in all the reference strains and the African strains (Table 2). None of the effectors that were originally described to be specific to X. perforans are present in all sequenced strains of X. gardneri, including strain JS749-3. All the originally described X. gardneri-specific effectors are absent in sequenced strains of X. euvesicatoria, including NI38. However, XopAQ and AvrHah1 are present in the X. perforans race T5 strain NI1, representing the first time these effectors were reported in X. perforans (Jibrin et al., 2018b). Subsequently, AvrHah1 was also reported in some X. perforans strains in the United States (Newberry et al., 2019). Interestingly, X. gardneri strain JS749-3 lacks XopAQ and has a different allele of AvrHah1 compared with the reference strain. None of the originally described X. vesicatoria-specific effectors were reported in the three strains representing the three species from Africa.
XopG, an effector present in the reference strains of X. euvesicatoria, X. perforans, and X. gardneri, is absent in NI1, NI38, and JS749-3. XopAM, HrpW, AvrXccA1, and XopZ2 were only found in JS749-3, with the first two effectors having different alleles from the reference strain ATCC19865. XopB is a common effector found in the reference strains 85-10 and ATCC19865, and similar alleles are found in NI38 and JS749-3, respectively. The effectors that are common in 85-10 and Xp91-118 but absent in the reference strains for X. vesicatoria and X. gardneri include XopE1, XopF2, XopI, XopP, XopV, XopAK, and XopAP (Potnis et al., 2011). These effectors are absent in JS749-3, but a different mix of alleles of the effectors is present in NI1 and NI38 (Table 2). XopAR, an effector present in the reference strains of X. perforans and X. vesicatoria but absent in X. euvesicatoria and X. gardneri, is absent in NI1. Effector AvrBsT was shown to be responsible for HR on pepper (Potnis et al., 2011). Although this effector was not present in X. perforans reference strain Xp91-118, multiple sequenced field strains were shown to possess a copy of AvrBsT. NI1 has a different allele of AvrBsT, which was subsequently characterized and renamed as XopJ6 (Iruegas-Bocardo et al., 2018; Jibrin et al., 2018b).
Evidently, strains from Africa possess genomic profiles that have some genetic differences from the reference strains and strains in other studies, which contributed novel insights to the bacterial spot pathogens. The sequencing and study of additional strains from Africa should provide additional information into the evolution and diversity of bacterial spot xanthomonads.
Seeds play central roles in vegetable production in Africa (Abdalla, 2000; Opara and Odibo, 2009). Much of the tomato and pepper production in Africa results from transplants on nurseries raised by smallholder farmers themselves (Perez et al., 2017). The seeds are often sourced from commercial outlets or research institutes, grown on farmer-made beds, and transplanted to small plots (Perez et al., 2017). Open-pollinated varieties developed by national research institutes are very common, although hybrid varieties are becoming popular (Ugonna et al., 2015; Schreinemachers et al., 2021). Hybrid seeds are imported in most countries (Ugonna et al., 2015; Schreinemachers et al., 2021). Seeds, therefore, play a significant role in disease spread in Africa.
Bacterial spot pathogens can be seed-borne, and pathogen contamination of seeds and seedlings is a major factor in disease spread (McGuire and Jones, 1989; Dutta et al., 2016; Ignjatov et al., 2017; Giovanardi et al., 2018; Abrahamian et al., 2019; Abrahamian et al., 2021). In Africa, bacterial spot pathogens have frequently been isolated from commercial seeds. In a survey of farmer-saved seeds in Tanzania, bacterial spot pathogens were isolated from seeds saved from different commercial cultivars (Black et al., 2001; Shenge et al., 2007; Mbega et al., 2012c). The 16S rRNA sequences of recovered strains were identical to X. euvesicatoria and X. gardneri sequences (Mbega et al., 2012b). In Egypt, bacterial spot pathogens have also been isolated from imported seeds (Abdalla, 2000; Ashmawy et al., 2020). One of the identified species from the imported seeds was designated as X. vesicatoria based on 16S rRNA (Ashmawy et al., 2020).
Obviously, 16S rRNA sequences are insufficient in confirming the species of strains, and sequencing of additional loci is necessary. However, these results, which are also accompanied by pathogenicity tests, are confirmatory that bacterial spot pathogens are associated with seed trade in Africa. In the studies by Hamza et al. (2010), there was a strong hypothesis for linking bacterial spot disease in the SWIO to the importation of contaminated tomato and pepper seeds because X. vesicatoria strains from Madagascar and Reunion clustered closely with strains from France and Italy (Hamza et al., 2010).
Commercially produced transplants are rare in Africa, and farmers produce their own seedlings (Perez et al., 2017). It is therefore difficult to assess the role of seedlings in the spread of bacterial spot disease. However, the bacterial spot pathogen has been isolated from tomato seedlings in Egypt (El-Hendawy et al., 2005). The role of seedlings may become important in the future if large-scale tomato and pepper production gains a foothold on the continent.
Globally, bacterial spot disease continues to cause significant yield losses of tomato and pepper (Potnis et al., 2015). Generally, efforts to improve management of the disease have focused on different strategies, including deployment of host resistance, use of antibiotics such as streptomycin, copper bactericides, systemic acquired resistance inducers, nano-based formulations of promising bactericides, small molecules, and improvement with surfactants (Stall et al., 2009; Araújo et al., 2015; Paret et al., 2013; Griffin et al., 2017; Hassan and Zyton, 2017; Liu et al., 2019; Awad-Allah et al., 2021; Jibrin et al., 2021a; Qiao et al., 2021).
Extensive use of streptomycin and copper-based bactericides can lead to the development of streptomycin and copper-resistant strains as recorded in Florida, United States (Stall and Thayer, 1962; Ritchie and Dittapongpitch, 1991; Araújo et al., 2015). Antibiotic and copper-sensitive and -resistant strains have been reported from laboratory assays in Africa. Tomato and pepper xanthomonad strains from Sudan and Ethiopia were determined to be sensitive to streptomycin and copper (Bouzar et al., 1994a,b, c; Kebede et al., 2014; Kebede and Yesuf, 2013). In Tanzania, strains isolated from tomato from three agroecological zones were sensitive to gentamycin, streptomycin, and ampicillin (Shenge et al., 2007). Similarly, strains evaluated from tomato and pepper fields in northwestern Nigeria were sensitive to copper (Jibrin, 2014). However, X. euvesicatoria strain LH3 and X. gardneri strain JS749-3 from Reunion are copper-resistant and carry the copper resistance cluster of genes on its plasmid (Richard et al., 2017a). These results may point to local management strategies deployed in individual countries influencing the composition of copper and antibiotic resistance traits of strains or to strains being imported and spread via seed trade. Although several studies have demonstrated the recovery of bacterial pathogens from imported seeds, there are no reports on copper resistance profiles of strains isolated from seeds, and such a study might provide market-door information on the presence or absence of copper resistance in strains imported with seeds.
Biological control strategies have also been implemented for managing bacterial spot of tomatoes. Biological control aims to prevent pathogen infection of plants or their establishment on plants (O’Brien, 2017). However, many of the experiments with biological control carried out in Africa were on already infected plant parts. In Egypt, treatment of seeds, seedlings, and foliar sprays with Rahnella aquatilis was shown to be effective in reducing deleterious effects of bacterial spot disease (El-Hendawy et al., 2005). Similarly, treatment of seeds and seedlings with Pseudomonas fluorescens alone and in combination with acibenzolar-S-methyl reduced bacterial spot disease severity (Abo-Elyousr and El-Hendawy, 2008).
Plant extracts have also shown potential in managing bacterial spot on seeds and seedlings. Extracts of ginger rhizome, mustard, and a local Ethiopian beer have been shown to inactivate a Xanthomonas strain of bacterial spot of tomato (Kebede and Yesuf, 2013). In Tanzania, oils of eucalyptus (Eucalyptus globules) and rosemary (Rosmarinus officinalis) and niaouli (Melaleuca viridiflora) at 2% concentration were shown to inhibit X. perforans growth in in vitro assays and in planta evaluations (Mbega et al., 2012b). Similarly, extracts of several plants, including Aloe vera, Coffea arabica, and Yucca schidigera, were shown to inhibit seed-borne X. perforans (Mbega et al., 2012c).
In general, studies involving field trials for the management of bacterial spot disease are limited. Perhaps, the absence of streptomycin and copper-resistant strains in some countries in Africa provides a window for the careful deployment of antibiotics and copper-based formulations in bacterial spot disease management. Recent advances in small molecules and nano-formulations may provide additional opportunities for the management of bacterial spot disease in the future (Liu et al., 2019; Liao et al., 2021; Qiao et al., 2021; Srivastava et al., 2021).
We present progress made on the African continent on the bacterial spot of tomato and pepper disease since its discovery in the early 20th Century. Although appreciable progress has been made, gaps still exist. The identity and variation of the bacterial spot pathogen are yet to be determined in many countries in Africa. In many countries where the disease has been reported, there is a paucity of information on the species and races causing the disease. As molecular diagnostics and sequencing tools become accessible on the continent, it is expected that this situation will improve. MLSAs have proved to be a great technique in the identification and subsequent population studies of recovered isolates (Hamza et al., 2012; Kebede et al., 2014; Timilsina et al., 2015; Osdaghi et al., 2018). In as much as Sanger sequencing remains accessible, this will be an important tool for studying the bacterial spot pathogen populations throughout Africa to provide insights into population structure. Such preliminary knowledge is important for species identification and laying a benchmark for subsequent whole-genome sequence studies.
Only genomes of isolates from Mauritius, Reunion, Nigeria, Ethiopia and South Africa have been sequenced (Richard et al., 2017a; Jibrin et al., 2018a,b; Jibrin, 2019). Sequencing of these genomes produced important “firsts” in understanding the bacterial spot pathogens. It is therefore expected that studies of additional genomes of the bacterial spot pathogen in Africa should provide significant insights into the evolution and biology of the pathogens. This could provide foundations for genome-informed host–pathogen and ecological interaction studies that are unique to production systems in Africa. Additional ecological studies of these pathogens are certainly needed. In vitro assays in studies conducted in Tanzania showed that P. syringae pv. tomato outgrows bacterial spot xanthomonads when grown together in nutrient yeast dextrose broth (Shenge et al., 2008). The importance of bacteriocins in the emergence of X. perforans and subsequent displacement of X. euvesicatoria have also been studied in Florida populations (Hert et al., 2005; Marutani-Hert et al., 2020). Studies on bacteriocin evolution among strains are lacking in Africa and could provide important insights into the evolution of the bacterial spot pathogens on the continent. Transcriptomics can improve understanding of the interaction of bacterial spot pathogens and their hosts (Strauss et al., 2012; Du et al., 2015; Shi and Panthee, 2020). Recently, known and novel metabolites from nonbacterial spot systems were reported in X. perforans (Jibrin et al., 2021b). Combining metabolomics and transcriptomics in studying isolates of the bacterial spot pathogens from Africa could provide additional insights into the diversity of molecules that are important in host–pathogen or other ecological interactions and their pathways and expressed genes.
Although much progress has been achieved with the management of the pathogen on pepper, managing the disease on tomatoes is still very difficult. Often, clean seeds and good cultural practices are advocated (Jones et al., 2016). However, as has been demonstrated in this review in studies in Tanzania and Egypt, imported and farmer-saved seeds are already often contaminated with pathogens (El-Hendawy et al., 2005; Shenge et al., 2010). Seed treatment with appropriate chemicals may therefore provide a frontline in disease management. The deployment of durable resistance is still to be achieved for bacterial spot on tomatoes. The Bs2 gene from pepper was initially touted as a possible resistance gene against bacterial spot disease on tomatoes (Tai et al., 1999; Horvath et al., 2015). However, resistance broke down, and efforts at deploying other genes have not been successful (Horvath et al., 2012, 2015). Screening tomato accessions in Africa against the bacterial spot pathogens could reveal novel resistance genes against bacterial spot.
Communicating research findings to farmers is important to help them improve disease management. Although there may be other offline resources, only one extension fact sheet was found online that targeted Africa in this review (Massawe et al., 2012). Development of management resources and training of farmers would be key to improving the management of the disease in Africa.
MJ wrote the original draft. All authors read and accepted the manuscript.
This research was supported by the United States Department of Agriculture (USDA), National Institute of Food and Agriculture (NIFA), and Specialty Crop Research Initiative Grants Program Grant Numbers: 2015-51181-24312 and 2019-51181-30010.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Debra Jones for critically reading the manuscript and providing helpful suggestions. MJ is grateful to all members of the P.D. Roberts Lab, J.B. Jones Lab, and E.M. Goss Lab at the University of Florida for all-round support and mentorship in understanding the bacterial spot of tomato and pepper pathosystem. MJ was initially supported for his M.Sc by a Borlaug Leadership Enhancement in Agricultural (Borlaug LEAP) fellowship award by the University of California—Davis, which included study visits to the University of Florida, United States, and International Institute for Tropical Agriculture, Tanzania. MJ was further supported for his Ph.D. by a generous graduate assistantship by P.D. Roberts. MJ also thanks Ahmadu Bello University, Zaria, for an award of university study fellowship. Special thanks to members and staff of the Department of Crop Protection, Ahmadu Bello University, Nigeria; International Institute for Tropical Agriculture, Tanzania; University of Florida, United States; and all others who aided in field surveys, laboratory analyses, and all other forms of assistance during the course of the studies.
Abdalla, M. E. (2000). Detection and identification of seed-borne pathogenic bacteria of imported tomato seeds in Egypt. EPPO Bulletin 30, 327–331. doi: 10.1111/j.1365-2338.2000.tb00904.x
Abd-Alla, M. H., and Bashandy, S. R. (2008). Bacterial wilt and spot of tomato caused by Xanthomonas vesicatoria and Ralstonia solanacearum in Egypt. World Journal of Microbiology and Biotechnology 24, 291–292. doi: 10.1007/s11274-007-9385-8
Abo-Elyousr, K. A. M., and El-Hendawy, H. H. (2008). Integration of Pseudomonas fluorescens and acibenzolar-S-methyl to control bacterial spot disease of tomato. Crop Protection 27, 1118–1124. doi: 10.1016/j.cropro.2008.01.011
Abolmaaty, S. M., and Abd El-Ghafar, N. Y. (2010). The relationship between the incidence of bacterial spot disease of pepper and some environmental factors. Journal of Plant Protection and Pathology 1, 157–164. doi: 10.21608/jppp.2010.86711
Abrahamian, P., Klein-Gordon, J. M., Jones, J. B., and Vallad, G. E. (2021). Epidemiology, diversity, and management of bacterial spot of tomato caused by Xanthomonas perforans. Applied microbiology and biotechnology 105, 6143–6158. doi: 10.1007/s00253-021-11459-9
Abrahamian, P., Timilsina, S., Minsavage, G. V., Kc, S., Goss, E. M., Jones, J. B., et al., (2019). The Type III Effector AvrBsT Enhances Xanthomonas perforans Fitness in Field-Grown Tomato. Phytopathology® 108, 1355–1362. doi: 10.1094/PHYTO-02-18-0052-R
Adriko, J., Mbega, E. R., Mortensen, C. N., Wulff, E. G., Tushemereirwe, W. K., Kubiriba, J., et al., (2014). Improved PCR for identification of members of the genus Xanthomonas. Eur J Plant Pathol. 138, 293–306. doi: 10.1007/s10658-013-0329-x
Aiello, D., Scuderi, G., Vitale, A., Firrao, G., Polizzi, G., and Cirvilleri, G. (2013). A pith necrosis caused by Xanthomonas perforans on tomato plants. European Journal of Plant Pathology 137, 29–41. doi: 10.1007/s10658-013-0214-7
Algam, S., Eltayeb, A., Mohammed, F. E. I., Teia, F. K., Elwagia, M. E. A., Abdalbagi, S., et al., (2018). First Report of Xanthomonas euvesicatoria Causing Bacterial Spot Disease in Tomato in Blue Nile State, Sudan. J. Agric. Sci. 26, 97–106.
Alikhan, N. F., Petty, N. K., and Ben Zakour, N. L. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Allen, E. E., and Banfield, J. F. (2005). Community genomics in microbial ecology and evolution. Nature Reviews Microbiology 3, 489–498. doi: 10.1038/nrmicro1157
Almeida, N. F., Yan, S., Cai, R., Clarke, C. R., Morris, C. E., Schaad, N. W., et al., (2010). PAMDB, A Multilocus Sequence Typing and Analysis Database and Website for Plant-Associated Microbes. Phytopathology§ 100, 208–215. doi: 10.1094/PHYTO-100-3-0208
Araújo, E. R., Costa, J. R., and Pontes, N. C. (2015). Xanthomonas perforans and X. gardneri associated with bacterial leaf spot on weeds in Brazilian tomato fields. Eur J Plant Pathol 143, 543–548. doi: 10.1007/s10658-015-0705-9
Arguelles, J. (2013). The early days of the Nobel Prize and Golden Age of Microbiology. Highlighted in Frontispiece 5, *page,
Ashmawy, N. A., Shoeib, A. A., Youssef, H. F. B., and Mahmoud, S. M. (2020). Pathological, biochemical and molecular characterization of the seed-borne bacteria “Pantoea spp., Xanthomonas spp. and Pseudomonas spp.” from solanaceous plants in Egypt. Journal of microbiology, biotechnology and food sciences 10, 289–295. doi: 10.15414/jmbfs.2020.10.2.289-295
Astua-Monge, G., Minsavage, G. V., Stall, R. E., Vallejos, C. E., Davis, M. J., and Jones, J. B. (2000). Xv4-vrxv4: A new gene-for-gene interaction identified between Xanthomonas campestris pv. vesicatoria race T3 and the wild tomato relative Lycopersicon pennellii. MOLECULAR PLANT-MICROBE INTERACTIONS 13, 1346–1355. doi: 10.1094/MPMI.2000.13.12.1346
Awad-Allah, E. F. A., Shams, A. H. M., and Helaly, A. A. (2021). Suppression of Bacterial Leaf Spot by Green Synthesized Silica Nanoparticles and Antagonistic Yeast Improves Growth, Productivity and Quality of Sweet Pepper. Plants 10, 689. doi: 10.3390/plants10081689
Bannor, T. T., and Ogunsan, E. A. (1988). Harmattan—A predisposing factor in the spread of viral diseases in domestic animals. Tropical Animal Health and Production 20, 211–211. doi: 10.1007/BF02239984
Barbier, J., Guichard, F., Bouniol, D., Couvreux, F., and Roehrig, R. (2018). Detection of Intraseasonal Large-Scale Heat Waves: Characteristics and Historical Trends during the Sahelian Spring. Journal of Climate 31, 61–80. doi: 10.1175/JCLI-D-17-0244.1
Bernal, R. F., and Berger, R. D. (1996). The spread of epiphytic populations of Xanthomonas campestris pv. vesicatoria on pepper in the field. J. Phytopathol. 144, 479–484. doi: 10.1111/j.1439-0434.1996.tb00328.x
Black, R., Seal, S., Abubakar, Z., Nono-Womdim, R., and Swai, I. (2001). Bacterial spot (Xanthomonas campestris pv. vesicatoria) of tomato and sweet pepper in Tanzania. Plant Pathology 50, 810–810. doi: 10.1046/j.1365-3059.2001.00633.x
Blevins, S. M., and Bronze, M. S. (2010). Robert Koch and the ‘golden age’ of bacteriology. International Journal of Infectious Diseases 14, e744–e751. doi: 10.1016/j.ijid.2009.12.003
Bophela, K. N., Venter, S. N., and Wingfield, M. J. (2019). Xanthomonas perforans: a tomato and pepper pathogen associated with bacterial blight and dieback of Eucalyptus pellita seedlings in Indonesia. Australasian Plant Pathol. 48, 543–551. doi: 10.1007/s13313-019-00657-9
Bouzar, H., Ahmed, N. E., Somodi, G. C., Jones, J. B., and Stall, R. (1994a). Characterization of Xanthomonas campestris pv.vesicatoria strains from tomato and pepper grown in Sudan. Plant Disease 78, 12–19.
Bouzar, H., Jones, J., Minsavage, G., Stall, R., and Scott, J. (1994b). Proteins unique to phenotypically distinct groups of Xanthomonas campestris pv. vesicatoria revealed by silver staining. Phytopathology 84, 39–44. doi: 10.1094/phyto-84-39
Bouzar, H., Jones, J. B., Stall, R. E., Hodge, N. C., and Minsavage, V. G. (1994c). Physiological, chemical, serological, and pathogenic analyses of a worldwide collection of Xanthomonas campestris pv. vesicatoria strains. Phytopathology 84, 663–671. doi: 10.1094/phyto-84-663
Burkholder, W. H., and Li, C. (1941). Variations in Phytomonas vesicatoria. Phytopathology 31, 753–755.
Constantin, E. C., Cleenwerck, I., Maes, M., Baeyen, S., Van Malderghem, C., De Vos, P., et al., (2016). Genetic characterization of strains named as Xanthomonas axonopodis pv. dieffenbachiae leads to a taxonomic revision of the X. axonopodis species complex. Plant Pathology 65, 792–806. doi: 10.1111/ppa.12461
da Silva, F. G., Shen, Y., Dardick, C., Burdman, S., Yadav, R. C., de Leon, A. L., et al., (2004). Bacterial genes involved in type I secretion and sulfation are required to elicit the rice Xa21-mediated innate immune response. Molecular plant-microbe interactions : MPMI 17, 593–601. doi: 10.1094/MPMI.2004.17.6.593
Das, A., Rangaraj, N., and Sonti, R. V. (2009). Multiple adhesin-like functions of Xanthomonas oryzae pv. oryzae are involved in promoting leaf attachment, entry, and virulence on rice. Molecular plant-microbe interactions : MPMI 22, 73–85. doi: 10.1094/MPMI-22-1-0073
Doidge, E. M. (1920). A tomato canker. Annals of Applied Biology 7, 407–430. doi: 10.1111/j.1744-7348.1921.tb05528.x
Du, H., Wang, Y., Yang, J., and Yang, W. (2015). Comparative Transcriptome Analysis of Resistant and Susceptible Tomato Lines in Response to Infection by Xanthomonas perforans Race T3. Front. Plant Sci. 6:1173. doi: 10.3389/fpls.2015.01173
Dutta, B., Gitaitis, R., Sanders, H., Booth, C., Smith, S., and Langston, D. B. Jr. (2016). Role of blossom colonization in pepper seed infestation by Xanthomonas euvesicatoria. Phytopathology 104, 232–239. doi: 10.1094/PHYTO-05-13-0138-R
Dye, D., Bradbury, J., Goto, M., Hayward, A., Lelliott, R., and Schroth, M. N. (1980). International standards for naming pathovars of phytopathogenic bacteria and a list of pathovar names and pathotype strains. Review of Plant pathology 59, 153–168.
El-Hendawy, H. H., Osman, M. E., and Sorour, N. M. (2005). Biological control of bacterial spot of tomato caused by Xanthomonas campestris pv. vesicatoria by Rahnella aquatilis. Microbiological Research 160, 343–352. doi: 10.1016/j.micres.2005.02.008
Engelbrecht, F., Adegoke, J., Bopape, M.-J., Naidoo, M., Garland, R., Thatcher, M., et al., (2015). Projections of rapidly rising surface temperatures over Africa under low mitigation. Environmental Research Letters 10, 085004. doi: 10.1088/1748-9326/10/8/085004
FAOSTAT (2020). FAOSTAT. Available online at: https://www.fao.org/faostat/en/ *accessed qu
Gardner, M. W., and Kendrick, J. B. (1923). Bacterial spot of tomato and pepper. Phytopathology 13, 307–315.
Giovanardi, D., Biondi, E., Ignjatov, M., Jevtić, R., and Stefani, E. (2018). Impact of bacterial spot outbreaks on the phytosanitary quality of tomato and pepper seeds. Plant Pathology 1168–1176. *vol, doi: 10.1111/ppa.12839
Glawe, D. A. (1992). Thomas J. Burrill, Pioneer in Plant Pathology. Annual Review of Phytopathology 30, 17–25. doi: 10.1146/annurev.py.30.090192.000313
Gordon, J. S. (2009). Reseeding tomato production in West Africa: Identification and deployment of high yielding cultivars resistant to tomato yellow leaf curl disease.: A Dissertation presented to the faculty of the graduate school of Cornell University in partial fulfillment of the requirements for the Degree of Doctor of Philosophy. *city pub.
Griffin, K., Gambley, C., Brown, P., and Li, Y. (2017). Copper-tolerance in Pseudomonas syringae pv. tomato and Xanthomonas spp. and the control of diseases associated with these pathogens in tomato and pepper. A systematic literature review. Crop Protection 96, 144–150. doi: 10.1016/j.cropro.2017.02.008
Hamza, A. A., Robène-Soustrade, I., Jouen, E., Gagnevin, L., Lefeuvre, P., Chiroleu, F., et al., (2010). Genetic and Pathological Diversity Among Xanthomonas Strains Responsible for Bacterial Spot on Tomato and Pepper in the Southwest Indian Ocean Region. Plant Disease 94, 993–999. doi: 10.1094/PDIS-94-8-0993
Hamza, A. A., Robene-Soustrade, I., Jouen, E., Lefeuvre, P., Chiroleu, F., Fisher-Le Saux, M., et al., (2012). MultiLocus Sequence Analysis- and Amplified Fragment Length Polymorphism-based characterization of xanthomonads associated with bacterial spot of tomato and pepper and their relatedness to Xanthomonas species. Systematic and Applied Microbiology 35, 183–190. doi: 10.1016/j.syapm.2011.12.005
Hassan, E. O., and Zyton, M. A. (2017). Management of Bacterial Spot of Pepper Caused by Xanthomonas campestris pv. vesicatoria. American Journal of Bioscience and Bioengineering 5, 41–49. doi: 10.11648/j.bio.20170501.17
Hayward, A., and Waterston, J. (1964). Xanthomonas vesicatoria. CMI descriptions of pathogenic fungi and bacteria. 20. *city pub.
Hert, A. P., Roberts, P. D., Momol, M. T., Minsavage, G. V., Tudor-Nelson, S. M., and Jones, J. B. (2005). Relative importance of bacteriocin-like genes in antagonism of Xanthomonas perforans tomato race 3 to Xanthomonas euvesicatoria tomato race 1 strains. Applied and environmental microbiology 71, 3581–3588. doi: 10.1128/AEM.71.7.3581-3588.2005
Horvath, D. M., Pauly, M. H., Hutton, S. F., Vallad, G. E., Scott, J. W., Jones, J. B., et al., (2015). “The pepper bs2 gene confers effective field resistance to bacterial leaf spot and yield enhancement in Florida tomatoes,” in Proceedings of the IVth International Symposium on Tomato Diseases, 1069.
Horvath, D. M., Stall, R. E., Jones, J. B., Pauly, M. H., Vallad, G. E., Dahlbeck, D., et al., (2012). Transgenic resistance confers effective field level control of bacterial spot disease in tomato. PloS one 7:e42036. doi: 10.1371/journal.pone.0042036
Ignjatov, M., Gašić, K., Ivanović, M., Šević, M., Obradović, A., and Milošević, M. (2017). Characterization of Xanthomonas euvesicatoria strains pathogens of pepper in Serbia. Pesticidi i fitomedicina 25, 139–149. doi: 10.2298/pif1002139i
Iruegas-Bocardo, F., Abrahamian, P., Minsavage, G. V., Potnis, N., Vallad, G. E., Jones, J. B., et al., (2018). XopJ6, a new member of the XopJ family of type III effectors, in Xanthomonas perforans. PHYTOPATHOLOGY 108, 37–37.
Jibrin, M. O. (2014). Race classification and molecular characterization of xanthomonads causing bacterial spot of tomato (Solanum lycopersicum L.) In north-western Nigeria: A Thesis submitted to the School of Post Graduate Studies, Ahmadu Bello University, Zaria, i n partial fulfillment of the requirements for the Award of Masters Degree in Crop Protection. *city pub.
Jibrin, M. O. (2019). Global and regional studies on population genomics, recombination, spread and diversification of bacterial spot xanthomonads: A Dissertation presented to the graduate school of the University of Florida in partial fulfillment of the requirements for the degree of Doctor of Philosophy. *city pub.
Jibrin, M. O., Liu, Q., Jones, J. B., and Zhang, S. (2021a). Surfactants in plant disease management: A brief review and case studies. Plant Pathol. 70, 495–510.
Jibrin, M. O., Liu, Q., Guingab-Cagmat, J., Jones, J. B., Garrett, T. J., and Zhang, S. (2021b). Metabolomics Insights into Chemical Convergence in Xanthomonas perforans and Metabolic Changes Following Treatment with the Small Molecule Carvacrol. Metabolites. 2021 11, 879. doi: 10.3390/metabo11120879
Jibrin, M. O., Minsavage, G. E., Jones, J. B., and Roberts, P. D. (2019). “Identification and Characterization of a Novel avrBs3-like TAL Effector, pthXp1, in Xanthomonas perforans which Delays Pustule Formation in Citrus,” in Proceeding of the 2019 Florida Phytopathological Society Biennial Meeting, Lake Alfred, Florida.
Jibrin, M. O., Potnis, N., Timilsina, S., Minsavage, G. V., Osdaghi, E., Vou, S., et al., (2018a). A global outlook on the evolution of type three effectors in Xanthomonads causing bacterial spot on tomato and pepper. International Congress of Plant Pathology (ICPP) 2018: Plant Health in A Global Economy *vol page,
Jibrin, M. O., Potnis, N., Timilsina, S., Minsavage, G. V., Vallad, G. E., Roberts, P. D., et al., (2018b). Genomic Inference of Recombination-Mediated Evolution in Xanthomonas euvesicatoria and X. perforans. Applied and environmental microbiology 84, e136–e118. doi: 10.1128/AEM.00136-18
Jibrin, M. O., Timilsina, S., Potnis, N., Minsavage, G. V., Shenge, K. C., Akpa, A. D., et al., (2015). First Report of Atypical Xanthomonas euvesicatoria Strains Causing Bacterial Spot of Tomato in Nigeria. Plant Disease 99, 415–415. doi: 10.1094/PDIS-09-14-0952-PDN
Jibrin, M. O., Timilsina, S., Potnis, N., Minsavage, G. V., Shenge, K. C., Akpa, A. D., et al., (2014). First Report of Xanthomonas euvesicatoria Causing Bacterial Spot Disease in Pepper in Northwestern Nigeria. Plant Disease 98, 1426–1426. doi: 10.1094/PDIS-06-14-0586-PDN
Jones, J. B., Bouzar, H., Somodi, G. C., Stall, R. E., Pernezney, K., El-Morsy, G., et al. (1988). Evidence for the preemptive nature of tomato race 3 of Xanthomonas campestris pv. vesicatoria in Florida. Phytopathology 88, 33–38.
Jones, J. B., Bouzar, H., Stall, R., Almira, E., Roberts, P., Bowen, B., et al., (2000). Systematic analysis of xanthomonads (Xanthomonas spp.) associated with pepper and tomato lesions. Int. J. Syst. Evol. Microbiol. 50, 1211–1219. doi: 10.1099/00207713-50-3-1211
Jones, J. B., Lacy, G. H., Bouzar, H., Stall, R. E., and Schaad, N. W. (2004). Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Syst Appl Microbiol 27, 755–762. doi: 10.1078/0723202042369884
Jones, J. B., Stall, R. E., and Bouzar, H. (1998). Diversity among Xanthomonads pathogenic on pepper and tomato. Annual Review of Phytopathology 36, 41–58. doi: 10.1146/annurev.phyto.36.1.41
Jones, J. B., Stall, R. E., Somodi, G. C., Bouzar, H., and Hodge, N. C. (1995). A third tomato race of Xanthomonas campestris pv. vesicatoria. Plant Dis 79, 395–398.
Jones, J. B., Zitter, T. A., Momol, T. M., and Miller, S. A. (2016). Compendium of Tomato Diseases and Pests, Second Edition: The American Phytopathological Society. *city pub.
Kaaya, N. K. F., and Massomo, S. M. S. (2003). A guide on seed-borne bacterial diseases of tomato in Tanzania. Copenhagen, Denmark.: Technical Bulletin, Danish Government Institute of Seed Pathology for Developing Countries (DGISP). *city pub.
Kebede, M. A., and Yesuf, M. (2013). Efficacy of plant extracts, traditional materials and antibacterial chemicals against Xanthomonas campestris pv. vesicatoria on tomato seed. African Journal of Microbiology Research 7, 2395–2400.
Kebede, M., Timilsina, S., Ayalew, A., Admassu, B., Potnis, N., Minsavage, G. V., et al., (2014). Molecular characterization of Xanthomonas strains responsible for bacterial spot of tomato in Ethiopia. European Journal of Plant Pathology 140, 677–688. doi: 10.1007/s10658-014-0497-3
Koenraadt, H., van Betteray, B., Germain, R., Hiddink, G., Jones, J. B., Oosterhof, J., et al., (2009). Development of specific primers for the molecular detection of bacterial spot of pepper and tomato. Acta Horticulturae 808, 99–102. doi: 10.1038/s41598-018-32295-4
Lamini, S., Cornelius, E. W., Nutsugah, S. K., Asiedu-Coffie, S., Miller, S., and Gilbertson, R. (2015). Molecular Identification of Bacterial Spot of Tomato incited by Xanthomonas campestris pv. vesicatoria from Fields in the Ashanti, Brong Ahafo and the Upper East Regions of Ghana. International Journal of Research in Agricultural Sciences 2, 2348–3997.
Lee, S. W., Jeong, K. S., Han, S. W., Lee, S. E., Phee, B. K., Hahn, T. R., et al., (2008). The Xanthomonas oryzae pv. oryzae PhoPQ two-component system is required for AvrXA21 activity, hrpG expression, and virulence. Journal of bacteriology 190, 2183–2197. doi: 10.1128/JB.01406-07
Li, S.-J., Hua, Z.-S., Huang, L.-N., Li, J., Shi, S.-H., Chen, L.-X., et al., (2014). Microbial communities evolve faster in extreme environments. Scientific Reports 4, 6205. doi: 10.1038/srep06205
Liao, Y. Y., Huang, Y., Carvalho, R., Choudhary, M., Da Silva, S., Colee, J., et al., (2021). Magnesium Oxide Nanomaterial, an Alternative for Commercial Copper Bactericides: Field-Scale Tomato Bacterial Spot Disease Management and Total and Bioavailable Metal Accumulation in Soil. Environmental science & technology 55, 13561–13570. doi: 10.1021/acs.est.1c00804
Liu, Q., Qiao, K., and Zhang, S. (2019). Potential of a Small Molecule Carvacrol in Management of Vegetable Diseases. Molecules (Basel, Switzerland) 24, 1932. doi: 10.3390/molecules24101932
Martinez, J. L. (2009). The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proceedings of the Royal Society B: Biological Sciences 276, 2521–2530. doi: 10.1098/rspb.2009.0320
Marutani-Hert, M., Hert, A. P., Tudor-Nelson, S. M., Preston, J. F., Minsavage, G. V., Stall, R. E., et al., (2020). Characterization of three novel genetic loci encoding bacteriocins associated with Xanthomonas perforans. PloS one 15:e0233301–e0233301. doi: 10.1371/journal.pone.0233301
Massawe, A., Tilya, M., Kimani, M., Musebe, R., and Tengeru, H. (2012). Factsheet for farmers: Bacterial Spot disease management in tomatoes. Available online at https://www.plantwise.org/KnowledgeBank/factsheetforfarmers/20137803392. (accessed August 08, 2021)
Mbega, E. R., Mabagala, R. B., Adriko, J., Lund, O. S., Wulff, E. G., and Mortensen, C. N. (2012a). Five Species of Xanthomonads Associated with Bacterial Leaf Spot Symptoms in Tomato from Tanzania. Plant Disease 96, 760–760. doi: 10.1094/PDIS-01-12-0105-PDN
Mbega, E. R., Mabagala, R. B., Mortensen, C. N., and Wulff, E. G. (2012b). Evaluation of essential oils as seed treatment for the control of Xanthomonas spp. associated with the bacterial leaf spot of tomato in Tanzania. Journal of Plant Pathology 94, 273–281.
Mbega, E. R., Mortensen, C. N., Mabagala, R. B., and Wulff, E. G. (2012c). The effect of plant extracts as seed treatments to control bacterial leaf spot of tomato in Tanzania. Journal of General Plant Pathology 78, 277–286. doi: 10.1007/s10327-012-0380-z
McGuire, R. G., and Jones, J. B. (1989). “Detection of Xanthomonas campestris pv. vesicatoria in tomato,” in Detection of bacteria in seed and other planting material, eds A. W. Saettler, N. W. Schaad, and D. A. Roth 59–62.
McInnes, T., Gitaitis, R., McCarter, S., Jaworski, C., and Phatak, S. (1988). Airborne dispersal of bacteria in tomato and pepper transplant fields. Plant Disease 72, 575–579.
Molinaro, A., Silipo, A., Lanzetta, R., Newman, M. A., Dow, J. M., and Parrilli, M. (2003). Structural elucidation of the O-chain of the lipopolysaccharide from Xanthomonas campestris strain 8004. Carbohydrate research 338, 277–281. doi: 10.1016/s0008-6215(02)00433-0
Morinière, L., Burlet, A., Rosenthal, E. R., Nesme, X., Portier, P., Bull, C. T., et al., (2020). Clarifying the taxonomy of the causal agent of bacterial leaf spot of lettuce through a polyphasic approach reveals that Xanthomonas cynarae Trébaol et al., 2000 emend. Timilsina et al., 2019 is a later heterotypic synonym of Xanthomonas hortorum Vauterin et al., 1995. Systematic and Applied Microbiology 43, 126087. doi: 10.1016/j.syapm.2020.126087
NAERLS. (2012). Agricultural Performance Survey Report of Nigeria of 2012 Wet Season in Nigeria, National Agricultural Extension Research Liaison Service (NAERLS), Ahmadu Bello University. NAERLS Press. 9, 126.
Nangombe, S., Zhou, T., Zhang, W., Wu, B., Hu, S., Zou, L., et al., (2018). Record-breaking climate extremes in Africa under stabilized 1.5 °C and 2 °C global warming scenarios. Nature Climate Change 8, 375–380. doi: 10.1038/s41558-018-0145-6
Newberry, E. A., Bhandari, R., Minsavage, G. V., Timilsina, S., Jibrin, M. O., Kemble, J., et al., (2019). Independent Evolution with the Gene Flux Originating from Multiple Xanthomonas Species Explains Genomic Heterogeneity in Xanthomonas perforans. Applied and environmental microbiology 85, e885–e819. doi: 10.1128/AEM.00885-19
O’Brien, P. A. (2017). Biological control of plant diseases. Australasian Plant Pathology 46, 293–304. doi: 10.1007/s13313-017-0481-4
OEPP/EPPO (1988). Data sheets on quarantine organisms No. 157, Xanthomonas campestris pv. vesicatoria. OEPP/EPPO Bulletin 18, 521–526.
Okoli, C. A. N., and Erinle, J. D. (1989). Factors responsible for market losses of tomato fruit in the Zaria area of Nigeria. Journal of Horticultural Science 64, 69–71.
Opara, E. U., and Odibo, F. J. C. (2009). Studies and Characterization of Bacterial Spot Pathogen of Tomato Xanthomonas campestris pv. Vesicatoria. Journal of Molecular Genetics 1, 35–43.
Osdaghi, E., Jones, J. B., Sharma, A., Goss, E. M., Abrahamian, P., Newberry, E. A., et al., (2021). A centenary for bacterial spot of tomato and pepper. Molecular Plant Pathology *vol page, doi: 10.1111/mpp.13125
Osdaghi, E., Taghavi, S. M., Koebnik, R., and Lamichhane, J. R. (2018). Multilocus sequence analysis reveals a novel phylogroup of Xanthomonas euvesicatoria pv. perforans causing bacterial spot of tomato in Iran. Plant Pathology 67, 1601–1611. doi: 10.1111/ppa.12864
Paret, M. L., Vallad, G. E., Averett, D. R., Jones, J. B., and Olson, S. M. (2013). Photocatalysis: Effect of Light-Activated Nanoscale Formulations of TiO2 on Xanthomonas perforans and Control of Bacterial Spot of Tomato. Phytopathology® 103, 228–236. doi: 10.1094/PHYTO-08-12-0183-R
Patil, P. B., Bogdanove, A. J., and Sonti, R. V. (2007). The role of horizontal transfer in the evolution of a highly variable lipopolysaccharide biosynthesis locus in xanthomonads that infect rice, citrus and crucifers. BMC evolutionary biology 7:243. doi: 10.1186/1471-2148-7-243
Pearson, T., Giffard, P., Beckstrom-Sternberg, S., Auerbach, R., Hornstra, H., Tuanyok, A., et al., (2009). Phylogeographic reconstruction of a bacterial species with high levels of lateral gene transfer. BMC Biology 7:78. doi: 10.1186/1741-7007-7-78
Perez, K., Froikin-Gordon, J. S., Abdourhamane, I. K., Levasseur, V., Alfari, A. A., Mensah, A., et al., (2017). Connecting smallholder tomato producers to improved seed in West Africa. Agriculture & Food Security 6, 42. doi: 10.1186/s40066-017-0118-4
Pohronezny, K., and Volin, R. B. (1983). The effect of bacterial spot on yield and quality of fresh market tomatoes. HortScience 18, 69–70.
Potnis, N. (2021). Harnessing Eco-Evolutionary Dynamics of Xanthomonads on Tomato and Pepper to Tackle New Problems of an Old Disease. Annual Review of Phytopathology 59, 289–310. doi: 10.1146/annurev-phyto-020620-101612
Potnis, N., Krasileva, K., Chow, V., Almeida, N. F., Patil, P. B., Ryan, R. P., et al., (2011). Comparative genomics reveals diversity among xanthomonads infecting tomato and pepper. BMC Genomics 12:146. doi: 10.1186/1471-2164-12-146
Potnis, N., Timilsina, S., Strayer, A., Shantharaj, D., Barak, J. D., Paret, M. L., et al., (2015). Bacterial spot of tomato and pepper: diverse Xanthomonas species with a wide variety of virulence factors posing a worldwide challenge. Molecular plant pathology 16, 907–920. doi: 10.1111/mpp.12244
Qiao, K., Liu, Q., Xia, Y., and Zhang, S. (2021). Evaluation of a Small-Molecule Compound, N-Acetylcysteine, for the Management of Bacterial Spot of Tomato Caused by Copper-Resistant Xanthomonas perforans. Plant disease 105, 108–113. doi: 10.1094/PDIS-05-20-0928-RE
Reason, C. J. C., and Keibel, A. (2004). Tropical Cyclone Eline and Its Unusual Penetration and Impacts over the Southern African Mainland. Weather and Forecasting 19, 789–805. doi: 10.1175/1520-04342004019<0789:TCEAIU<2.0.CO;2
Richard, D., Boyer, C., Lefeuvre, P., Canteros, B. I., Beni-Madhu, S., Portier, P., et al., (2017a). Complete Genome Sequences of Six Copper-Resistant Xanthomonas Strains Causing Bacterial Spot of Solaneous Plants, Belonging to X. gardneri, X. euvesicatoria, and X. vesicatoria, Using Long-Read Technology. Genome announcements 5, e1693–e1616. *e1693–16 doi: 10.1128/genomeA.01693-16
Richard, D., Ravigné, V., Rieux, A., Facon, B., Boyer, C., Boyer, K., et al., (2017b). Adaptation of genetically monomorphic bacteria: evolution of copper resistance through multiple horizontal gene transfers of complex and versatile mobile genetic elements. Molecular Ecology 26, 2131–2149. doi: 10.1111/mec.14007
Ritchie, D. F., and Dittapongpitch, V. (1991). Copper-and streptomycin-resistant strains and host differentiated races of Xanthomonas campestris pv. vesicatoria in North Carolina. Plant Dis 75, 733–736.
Russo, S., Marchese, A. F., Sillmann, J., and Immé, G. (2016). When will unusual heat waves become normal in a warming Africa? Environmental Research Letters 11, 054016. doi: 10.1088/1748-9326/11/5/054016
Santos, L. V. S., Melo, E. A., and Silva, A. M. F. (2020). Weeds as alternate hosts of Xanthomonas euvesicatoria pv. euvesicatoria and X. campestris pv. campestris in vegetable-growing fields in the state of Pernambuco, Brazil. Trop. plant pathol. 45, 484–492. doi: 10.1007/s40858-020-00350-z
Schreinemachers, P., Howard, J., Turner, M., Groot, S. N., Dubey, B., Mwadzingeni, L., et al., (2021). Africa’s evolving vegetable seed sector: status, policy options and lessons from Asia. Food Security 13, 511–523. doi: 10.1007/s12571-021-01146-y
Schwartz, A. R., Potnis, N., Timilsina, S., Wilson, M., Patané, J., Martins, J., et al., (2015). Phylogenomics of Xanthomonas field strains infecting pepper and tomato reveals diversity in effector repertoires and identifies determinants of host specificity. Frontiers in Microbiology 6:535.
Shenge, K. C., Jimoh, R. O., Akpa, A. D., Chindo, P. S., and Ajene, I. J. (2018). Seasonal variations in incidence and severity of bacterial spot and bacterial speck of tomato (Solanum lycopersicum L.) under rain-fed and irrigated conditions in Samaru Zaria, Nigeria. Nigerian Journal of Plant Protection 31, *page,
Shenge, K. C., Mabagala, R. B., and Mortensen, C. N. (2007). Identification and characterization of strains of Xanthomonas campestris pv. vesicatoria from Tanzania by Biolog system and sensitivity to antibiotics. African Journal of Biotechnology 6, 15–22.
Shenge, K. C., Mabagala, R. B., and Mortensen, C. N. (2008). Coexistence between neighbours: Pseudomonas syringae pv. tomato and Xanthomonas campestris pv. vesicatoria, incitants of bacterial speck and spot diseases of tomato. Archives of Phytopathology and Plant Protection 41, 559–571. doi: 10.1080/03235400600881752
Shenge, K. C., Mabagala, R. B., and Mortensen, C. N. (2010). Current status of bacterial -speck and -spot diseases of tomato in three tomato-growing regions of Tanzania. Journal Agricultural Extension and Rural Development 2, 84–88.
Sherbakoff, C. D. (1918). Report of the associate plant pathologist. Florida Agricultural Experimental Station Repertoire 1916–1917, 66R–86R.
Shi, R., and Panthee, D. R. (2020). Transcriptome-Based Analysis of Tomato Genotypes Resistant to Bacterial Spot (Xanthomonas perforans) Race T4. International journal of molecular sciences 21, 4070. doi: 10.3390/ijms21114070
Srivastava, V., Deblais, L., Kathayat, D., Rotondo, F., Helmy, Y. A., Miller, S. A., et al., (2021). Novel Small Molecule Growth Inhibitors of Xanthomonas spp. Causing Bacterial Spot of Tomato. Phytopathology 111, 940–953. doi: 10.1094/PHYTO-08-20-0341-R
Stall, R. E., and Thayer, P. L. (1962). Streptomycin resistance of the bacterial spot pathogen and control with streptomycin. Plant Dis. Rep. 46, 389–392.
Stall, R. E., Beaulieu, C., Egel, D., Hodge, N. C., Leite, R. P., Minsavage, G. V., et al. (1994). Two genetically diverse groups of strains are included in Xanthomonas campestris pv. vesicatoria. Int. J. Syst. Evol. Microbiol. 44, 47–53.
Stall, R. E., Jones, J. B., and Minsavage, G. V. (2009). Durability of Resistance in Tomato and Pepper to Xanthomonads Causing Bacterial Spot. Annual Review of Phytopathology 47, 265–284. doi: 10.1146/annurev-phyto-080508-081752
Stevens, F. L. (1925). Plant disease fungi In: Plant disease fungi. New York: The Macmillan company; Print, 1871–1934.
Strauss, T., van Poecke, R. M., Strauss, A., Römer, P., Minsavage, G. V., Singh, S., et al., (2012). RNA-seq pinpoints a Xanthomonas TAL-effector activated resistance gene in a large-crop genome. Proceedings of the National Academy of Sciences of the United States of America 109, 19480–19485. doi: 10.1073/pnas.1212415109
Strayer, A. L., Jeyaprakash, A., Minsavage, G. V., Timilsina, S., Vallad, G. E., Jones, J. B., et al., (2016). A Multiplex Real-Time PCR Assay Differentiates Four Xanthomonas Species Associated with Bacterial Spot of Tomato. PLANT DISEASE 100, 1660–1668. doi: 10.1094/PDIS-09-15-1085-RE
Šutic, D. (1957). Tomato bacteriosis. Posebna Izd. Inst. Zasht. Bilja, Beograd [Spec. Edit. Inst. Plant Prot., Beograd]. English Summary. Rev. Appl. Mycol 36, 734–735.
Szczesny, R., Jordan, M., Schramm, C., Schulz, S., Cogez, V., Bonas, U., et al., (2010). Functional characterization of the Xcs and Xps type II secretion systems from the plant pathogenic bacterium Xanthomonas campestris pv vesicatoria. The New phytologist 187, 983–1002. doi: 10.1111/j.1469-8137.2010.03312.x
Tai, T. H., Dahlbeck, D., Clark, E. T., Gajiwala, P., Pasion, R., Whalen, M. C., et al., (1999). Expression of the <em>Bs2</em> pepper gene confers resistance to bacterial spot disease in tomato. Proceedings of the National Academy of Sciences 96, 14153. doi: 10.1073/pnas.96.24.14153
Tawfik, A. E., El Shaimaa, M. M., Barakat, M. I. E., and Shalaby, O. Y. (2009). The Occurrence of bacterial spot of tomato in Egypt. Proc. 2nd Intl. Symposium on Tomato Diseases. Acta Hort 808, 295–300. doi: 10.17660/ActaHortic.2009.808.46
Thieme, F., Koebnik, R., Bekel, T., Berger, C., Boch, J., Buttner, D., et al., (2005). Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. JOURNAL OF BACTERIOLOGY 187, 7254–7266. doi: 10.1128/JB.187.21.7254-7266.2005
Timilsina, S., Abrahamian, P., Potnis, N., Minsavage, G. V., White, F. F., Staskawicz, B. J., et al., (2016). Analysis of Sequenced Genomes of Xanthomonas perforans Identifies Candidate Targets for Resistance Breeding in Tomato. PHYTOPATHOLOGY 106, 1097–1104. doi: 10.1094/PHYTO-03-16-0119-FI
Timilsina, S., Jibrin, M. O., Potnis, N., Minsavage, G. V., Kebede, M., Schwartz, A., et al., (2015). Multilocus sequence analysis of xanthomonads causing bacterial spot of tomato and pepper plants reveals strains generated by recombination among species and recent global spread of Xanthomonas gardneri. Applied and environmental microbiology 81, 1520–1529. doi: 10.1128/AEM.03000-14
Timilsina, S., Kara, S., Jacques, M. A., Potnis, N., Minsavage, G. V., Vallad, G. E., et al., (2019a). Reclassification of Xanthomonas gardneri (ex Šutič 1957) Jones et al., 2006 as a later heterotypic synonym of Xanthomonas cynarae Trébaol et al., 2000 and description of X. cynarae pv. cynarae and X. cynarae pv. gardneri based on whole genome analyses. International journal of systematic and evolutionary microbiology 69, 343–349. doi: 10.1099/ijsem.0.003104
Timilsina, S., Pereira-Martin, J. A., Minsavage, G. V., Iruegas-Bocardo, F., Abrahamian, P., Potnis, N., et al., (2019b). Multiple Recombination Events Drive the Current Genetic Structure of Xanthomonas perforans in Florida. FRONTIERS IN MICROBIOLOGY 10:448. doi: 10.3389/fmicb.2019.00448
Ugonna, C. U., Jolaoso, M. A., and Onwualu, A. P. (2015). Tomato Value Chain in Nigeria: Issues, Challenges and Strategies. Journal of Scientific Research & Reports 7, 501–515. doi: 10.9734/jsrr/2015/16921
Vauterin, L., Swings, J., Kersters, K., Gillis, M., Mew, T. W., Schroth, M. N., et al. (1990). Towards an improved taxonomy of Xanthomonas. Int. J. Syst. Evol. Microbiol. 40, 312–316.
Wai, K. P. P., Siddique, M. I., Mo, H.-S., Yoo, H. J., Byeon, S.-E., Jegal, Y., et al., (2015). Pathotypes of Bacterial Spot Pathogen Infecting Capsicum Peppers in Korea. The plant pathology journal 31, 428–432. doi: 10.5423/PPJ.NT.05.2015.0074
Keywords: bacterial spot, tomato, pepper (Capsicum annum L.), Africa, Xanthomonas, Management
Citation: Jibrin MO, Timilsina S, Minsavage GV, Vallad GE, Roberts PD, Goss EM and Jones JB (2022) Bacterial Spot of Tomato and Pepper in Africa: Diversity, Emergence of T5 Race, and Management. Front. Microbiol. 13:835647. doi: 10.3389/fmicb.2022.835647
Received: 14 December 2021; Accepted: 21 January 2022;
Published: 18 April 2022.
Edited by:
Mohammad Arif, University of Hawai’i at Mãnoa, United StatesReviewed by:
Toni A. Chapman, New South Wales Department of Primary Industries, AustraliaCopyright © 2022 Jibrin, Timilsina, Minsavage, Vallad, Roberts, Goss and Jones. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mustafa Ojonuba Jibrin, amlicmlubW9AZ21haWwuY29t; Pamela D. Roberts, cGRyQHVmbC5lZHU=; Erica M. Goss, ZW1nb3NzQHVmbC5lZHU=; Jeffrey B. Jones, amJqb25lc0B1ZmwuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.