- Department of Zoology, Faculty of Natural Sciences, Comenius University in Bratislava, Bratislava, Slovakia
Like many other aquatic animals, freshwater planarians have also become partners of symbiotic ciliates from the class Oligohymenophorea. In the present study, we explored the hidden diversity and addressed the questionable systematic position of mouthless obligatory gut endosymbionts of freshwater planarians, using the nuclear and mitochondrial SSU rRNA genes. Although all isolated ciliates morphologically corresponded to a single species, molecular analyses suggested the existence of three genetically distinct entities: Haptophrya planariarum, Haptophrya dugesiarum nov. spec., and Haptophrya schmidtearum nov. spec. The two former species share the same planarian host, which indicates a speciation model involving one duplication event without host switching. Such a diversification pattern was recognized also in astome ciliates inhabiting megascolecid and glossoscolecid earthworms. The present multi-gene phylogenies along with the secondary structure of the mitochondrial 16S rRNA molecule, however, challenge the traditional classification of Haptophrya within the subclass Astomatia. Haptophrya very likely evolved from an orphan scuticociliate lineage by the loss of oral apparatus and by the transformation of the thigmotactic field into an adhesive sucker. Since astomy evolved multiple times independently within the Oligohymenophorea, the loss of cell mouth cannot be used as a sole argument for the assignment of Haptophrya to the Astomatia anymore.
Introduction
Freshwater triclad planarians (order Tricladida Lang, 1884) represent a relatively derived monophyletic lineage of free-living flatworms. The age of their origin was mostly inferred by biogeographical events, as the proper fossil record of triclads is extremely limited due to their fragile bodies. Diversification of the family Dugesiidae Ball, 1974 might date back even to the epoch of the supercontinent Pangaea (~300 Mya). Molecular clock estimates, however, suggest that triclads could be a way more ancient (for a review, see Vila-Farré and Rink, 2018 and references cited therein). Interestingly, their divergence time estimates meet those of their ciliate endosymbionts, which indicates that the ciliate-planarian associations could be very ancient (Rataj and Vďačný, 2018; Zhang and Vďačný, 2022). Despite the very long evolutionary history, the bauplan of these dorso-ventrally flattened worms remained highly uniform. The limbless narrowly leaf-shaped body covered with mucus and carrying a protrusive pharynx has ensured them a position of successful predators over millions of years. The conservativeness of the planarian bauplan might have also lowered the pressure on the morphological evolution of their ciliate symbionts, which in turn could lead to their cryptic speciation. As concerns ecological demands, planarians inhabit all kinds of freshwater habitats all over the globe. For instance, they live in fishponds, lakes of various sizes and depths, in temporary water bodies such as puddles, swamps, cave ponds, but also in springs, creeks, streams, and shallow sections of rivers (Reynoldson, 1981). All these environments are commonly inhabited by ciliates as well (Lynn, 2008), a fact enabling encounters of planarians with ciliates.
Like many other aquatic animals, freshwater planarians have also become partners of symbiotic ciliates. Even though only a small fraction of triclads has been properly examined for the presence of symbiotic ciliates, at least three different types of associations were detected. The character of these symbiotic systems and the impact on planarians differ significantly, however. All ciliates so far isolated either from the gut, the body cavity, or from the surface of planarians are members of the highly diverse class Oligohymenophorea de Puytorac et al., 1974. The first description of a symbiotic ciliate from freshwater planarians comes from von Siebold (1839). He allegedly found a harmless endocommensal carrying an adhesive sucker without any additional spine-like structures, currently known as Haptophrya planariarum (von Siebold, 1839) Stein, 1867, in the gut of Planaria torva Müller, 1773. The morpho-molecular characterization of H. planariarum and its host range, which accounts for seven triclad species living in the Holarctis and Palaearctic, were reviewed elsewhere (Rataj and Vďačný, 2018). Another endozoic ciliate described from freshwater triclads is Annelophrya sphaeronucleata (Georgévitch, 1950) Lom, 1959. It is easily distinguished from H. planariarum by the presence of numerous hooks in the thigmotactic area (Georgévitch, 1950; Lom, 1959). There are, unfortunately, no recent records of this species and no DNA samples are available from the initial discovery. Haptophrya Stein, 1867 and Annelophrya Lom, 1959 are traditionally classified in the subclass Astomatia Schewiakoff, 1896 in compendium of Lynn (2008).
The second group of ciliates associated with planarians includes surface-dwelling mobilid peritrichs (subclass Peritrichia Stein, 1859) belonging to the genera Urceolaria Stein, 1867 and Trichodina Ehrenberg, 1830. Mobilids thus entered into relationships with planarians at least two times independently. For a long time, only a single species from each genus was known to inhabit planarians (Haider, 1964). Recently, we have recognized that Trichodina steinii Claparède and Lachmann, 1859 is a complex of three morphologically cryptic species, whose occurrence perfectly correlates with that of their different planarian hosts. Urceolaria mitra (von Siebold, 1848) Stein, 1867 very likely also covers two cryptic but co-occurring species (Rataj and Vďačný, 2021). Interestingly, Urceolaria and Trichodina can either occupy their hosts separately or they can co-occur as well (Reynoldson, 1947). Both attach to their hosts with the aid of a skeletal ring, leaving a visible mark on the host surface (Bowen and Ryder, 1994). The extensive work on ecology, dispersal, and population dynamics of U. mitra showed that mobilids only use the host-produced water currents for feeding purposes and cause no direct harm to their hosts (Reynoldson, 1950, 1951, 1955, 1956).
Unlike mobilids, representatives of the third group of ciliates detected in freshwater triclads are histophagous hymenostomes (subclass Hymenostomatia Delage and Hérouard, 1896). They are capable of causing severe damage to their hosts by feeding on their parenchyma tissues, not seldomly leading even to death (Wright, 1981). Members of only two hymenostome genera have been recognized to infect planarians. More specifically, three Ophryoglena Ehrenberg, 1830 species (O. intestinalis Rossolimo, 1926, O. parasitica André, 1909 and O. pyriformis Rossolimo, 1926) and six Tetrahymena Furgason, 1940 species were isolated from the gut and body cavity of at least seven different freshwater triclads (André, 1909; Rossolimo, 1926; Wright, 1981; Rataj and Vďačný, 2020). However, only four Tetrahymena species were identified by a means of molecular methods (Rataj and Vďačný, 2020). The identity of the two remaining Tetrahymena species remains questionable due to their substantial morphological uniformity.
Symbiotic relationships are in the center of interest of a broad variety of ecological, physiological, parasitological, cell biological, and environmental studies (e.g., Bignell, 2000; Dimijian, 2000a,b; Sapp, 2004). The development of molecular methods enabled the examination of symbionts and their hosts also from the viewpoint of molecular taxonomy, phylogenetics, and biogeography. The recent multi-gene analyses brought serious concerns about the alleged omnipresence and non-specificity of ciliate symbionts (c.f. Lynn et al., 2014; Moon-van der Staay et al., 2014; Rataj and Vďačný, 2020, 2021; Obert et al., 2021). Although the species concept and approaches used to discriminate cryptic species have a paramount effect on the diversity recognized (Caron, 2013; Struck et al., 2018), the very deep divergences in mitochondrial genes of symbiotic ciliates isolated from different hosts cannot be ignored any more (e.g., Lynn et al., 2014; Obert et al., 2021; Rataj and Vďačný, 2021). In the present paper, we studied nuclear and mitochondrial genes encoding for the small subunit rRNA molecules, including their secondary and tertiary structures, to address the real diversity of the morphologically uniform, gut endosymbiont of freshwater planarians, H. planariarum. The integrative approach significantly confronted the current taxonomic concept of H. planariarum and challenged its traditional systematic position in the subclass Astomatia as well.
Materials and Methods
Material Collection and Processing
Planarians were collected from a variety of freshwater bodies at 28 localities in Slovakia (Central Europe). Detailed descriptions of collection sites were provided by Rataj and Vďačný (2020) and are summarized in Supplementary Table S1. Identification of planarians was based on morphological criteria, and the identity of some taxa was confirmed also by sequencing their cytochrome c oxidase subunit I (COI). Primers and PCR conditions used for amplification of the planarian COI gene are listed in Supplementary Tables S2, S3. The molecular assignment of the examined planarians to species is shown in Supplementary Figure S1. Planarians were processed and dissected as described by Rataj and Vďačný (2018, 2019). Haptophryans were isolated from the pharynx plicatus and gut content of their planarian hosts with Pasteur micropipettes. They were investigated at low (50–400×) and high (1,000×) magnifications with bright field and differential interference contrast under a Leica DM2500 optical microscope. Their ciliary pattern and nuclear apparatus were revealed with protargol impregnation (Wilbert, 1975). Habitus and taxonomically important features were captured on microphotographs by a Canon EOS 80D camera. Measurements were taken in ImageJ ver. 1.49.
Single cells of ciliates were collected for molecular analyses as described by Rataj and Vďačný (2018, 2019). More specifically, each specimen was separately placed in 180 μl of cell lysis buffer (Promega, Fitchburg, Wisconsin, United States) and its genomic DNA was extracted with the ReliaPrep™ Blood gDNA Miniprep System (Promega, Fitchburg, Wisconsin, United States). The mitochondrial 16S rRNA gene (16S henceforth) and the nuclear 18S rRNA gene (18S henceforth) were consequently PCR amplified. Primers and PCR conditions are provided in Supplementary Tables S2, S3. Unfortunately, all attempts to amplify the nuclear ITS1-5.8S-ITS2 region and the mitochondrial COI gene failed. The list of tested primers is provided in Supplementary Table S2. PCRs were carried out with the GoTaq® Long PCR Master Mix (Promega, Fitchburg, Wisconsin, United States), following the protocol described in Rataj and Vďačný (2021). Sequencing was conducted in Macrogen Europe B.V. (Amsterdam, The Netherlands) on an ABI 3730 automatic sequencer. Newly acquired sequences were examined in Chromas ver. 2.6.6 (Technelysium Pty Ltd., South Brisbane, Australia) and only high-quality sequence fragments were assembled into contigs in BioEdit ver. 7.2.5 (Hall, 1999). More specifically, the Phred score was ≥20 and the length of fragments typically exceeded 900 nt. The assemble parameters were as follows: a minimum of 150 base overlap and a minimum match of 95%.
Predicting Secondary Structures of rRNA Molecules
The secondary structure of 16S and 18S rRNA molecules was predicted using R2DT (Sweeney et al., 2021). The 16S secondary structure model proposed for Paramecium tetraurelia and Tetrahymena pyriformis (Cannone et al., 2002)1 as well as the 18S model proposed for Tetrahymena thermophila by Lee and Gutell (2012) were also taken into account. These models were based on ribosomal crystal structures and hence are consistent with 3D ribosomal structures, accounting for non-canonical base pairs on parity with Watson-Crick base pairs. Since helices 39 and 40 of 16S are highly variable and much longer in Haptophrya than in other ciliates, the RNAstructure web server (Mathews, 2004)2 was employed to find their common secondary structure motif. The secondary structure of the divergent helices 21, 33, 39, 40, and 44 of 16S was also analyzed using the free-energy minimization approach on the Mfold web server ver. 3.0 (Zuker, 2003).3
The incorporation of non-canonical base pairs in 16S helices modeled on the RNAstructure and Mfold web servers followed the RC/Rp pipeline proposed by Rybarczyk et al. (2015). More specifically, RNA 3D structures were predicted from the putative secondary structures using RNAComposer (Popenda et al., 2012).4 Resulting PDB files with tertiary structure information served as input for RNApdbee (Antczak et al., 2014; Zok et al., 2018).5 Canonical and non-canonical base pairs were identified from the tertiary structures with the 3DNA/DSSR option, secondary structures were resolved using the hybrid algorithm and visualized using the VARNA-based procedure. Finally, the recognized non-canonical base pairs were included in the 16S rRNA models. Secondary structures were plotted either in Varna ver. 3.93 (Darty et al., 2009) or with the help of R2DT (Sweeney et al., 2021) and TRAVeLer (Elias and Hoksza, 2017). The helix number system of rRNA molecules was according to Lee and Gutell (2012) and Petrov et al. (2014).
Genetic Distances, Networks, and Molecular Diagnostic Characters
16S and 18S sequences were aligned according to the primary and predicted secondary structures (see above) using the package 4SALE ver. 1.7.1 (Seibel et al., 2006). Subsequently, the number of compensatory base changes (CBC) was identified with the CBCAnalyzer option (Wolf et al., 2005) supplied with 4SALE. Pairwise p-distances were calculated for each rRNA gene separately with a custom Python script, using a pairwise deletion option to exclude alignment gaps. Parsimony networks were constructed from the secondary structure-based alignments in PopART v.1.7 (Leigh and Bryant, 2015) using the TCS method (Clement et al., 2000). Nucleotide positions suitable for species diagnoses (i.e., molecular autapomorphies) were identified within the secondary structure-based alignments, using a custom Python script.
Phylogenetic Analyses
To determine the phylogenetic position of Haptophrya within the class Oligohymenophorea, two datasets were assembled following mostly our previous study (Zhang and Vďačný, 2022) and taking into account newly published sequences of oligohymenophorean ciliates isolated from a variety of invertebrates (Rataj and Vďačný, 2020, 2021; Obert et al., 2021). The first dataset contained 18S sequences sampled across almost the whole class Oligohymenophorea. However, only taxa having also 16S sequences were selected for this dataset. The single exceptions were Conchophthirus and Dexiotricha, which were included as they are the nearest relatives of Haptophrya according to the BLAST search and previous single-gene analyses (e.g., Rataj and Vďačný, 2018, 2019; Antipa et al., 2020; Obert et al., 2021; Zhang and Vďačný, 2022). The second dataset contained 18S and corresponding 16S sequences. Taxon sampling and GenBank accession numbers of both datasets are collated in Supplementary Table S4.
Individual molecular markers were aligned on the MAFFT ver. 7 web server,6 using the iterative G-INS-i method as well as the progressive G-INS-1 method with an accurate guide tree (Katoh et al., 2019). Both alignment strategies were coupled with a gap opening penalty of 1.53 and the 200PAM/κ = 2 scoring matrix. Ambiguous nucleotide alignment positions were masked on the Gblocks ver. 0.91b server (Talavera and Castresana, 2007)7 with a less stringent option, allowing smaller final blocks, gap positions within the final blocks, and less strict flanking positions. Phylogenetic trees were constructed with the maximum likelihood (ML) approach as implemented in the program IQTREE ver. 1.6.10 (Nguyen et al., 2015) on the IQTREE web server (Trifinopoulos et al., 2016)8 and with Bayesian (BI) inference as implemented in the program MrBayes ver. 3.2.7 (Ronquist et al., 2012). Each molecular partition was assigned the best evolutionary substitution model, as selected under the BI information criterion in IQTREE. The ML search started from a BioNJ tree and the branching pattern of ML trees was assessed with 1,000 ultrafast bootstrap pseudoreplicates. All other parameters were left default. Settings in Bayesian analyses and convergence diagnostics followed our previous protocols (Rataj and Vďačný, 2021). Prior parameters of evolutionary models in Bayesian analyses were estimated in IQTREE and implemented with the “prset” command. Trees were rooted in FigTree ver. 1.2.39 by placing the root on a branch separating members of the class Oligohymenophorea from those of the class Colpodea, which served as an outgroup.
Results
Alpha-Diversity, Prevalence, and Intensity of Haptophryans in Planarians
Two ecological groups of planarians were examined for the presence of haptophryans during the years 2016 and 2019. The first group comprised stream- and river-dwelling planarians (169 specimens of Dugesia gonocephala, 166 individuals of Polycelis felina, and four exemplars of Crenobia alpina), while the second group contained pond planarians (426 specimens of Girardia tigrina, eight individuals of Dendrocoelum lacteum, six exemplars of Schmidtea lugubris, six specimens of Schmidtea polychroa, and a single individual of Polycelis nigra). Haptophrya was detected only in two out of the eight planarian species examined, namely, in D. gonocephala collected from a variety of running waters and in S. polychroa living in stagnant waters. Although the abundance of haptophryans was generally very low (one to dozen of specimens per host), these endosymbiotic ciliates were noted at 12 out of the 28 localities studied.
All isolated ciliates morphologically corresponded to H. planariarum (see below). However, according to the present molecular analyses, the morphospecies H. planariarum is a complex of three genetically distinct and morphologically cryptic species (see below). The most common species is assigned to H. planariarum, while the two rare species are endowed here with names: H. dugesiarum nov. spec. and H. schmidtearum nov. spec. The former species was detected exclusively in D. gonocephala at 11 localities and co-occurred with H. dugesiarum at two localities (in a shallow section of the river Váh near the village of Bystrá, Veľká Fatra Mts. and in a stream running through the Fončorda residential area, Banská Bystrica, Zvolenská kotlina basin). Haptophrya dugesiarum was recorded only at these two localities and was also restricted to D. gonocephala. Finally, H. schmidtearum was found only at a single locality (Jurské jazierko pond, an urban oak-hornbeam forest, district of the village of Svätý Jur, Malé Karpaty Mts.). Although two planarians co-occurred in the Jurské jazierko pond, H. schmidtearum was consistently and multiple times isolated only from S. polychroa and never from P. nigra.
The prevalence of H. planariarum and H. dugesiarum in D. gonocephala was 59.76 and 13.61%, respectively. However, the prevalence of H. schmidtearum in S. polychroa was 33.33%. The intensity of all three Haptophrya species was consistently very low. Namely, there were one to 14 specimens of H. planariarum per host, 1–7 individuals of H. dugesiarum, and only up to four exemplars of H. schmidtearum per host.
Molecular Identification of Haptophryans
In total, 96 new sequences were obtained from three morphologically cryptic Haptophrya species isolated from freshwater planarians (Table 1). They can be unambiguously distinguished by rRNA gene sequences, whereby the nuclear 18S rRNA gene can serve as a pre-barcode and the mitochondrial 16S rRNA gene as a DNA barcode.
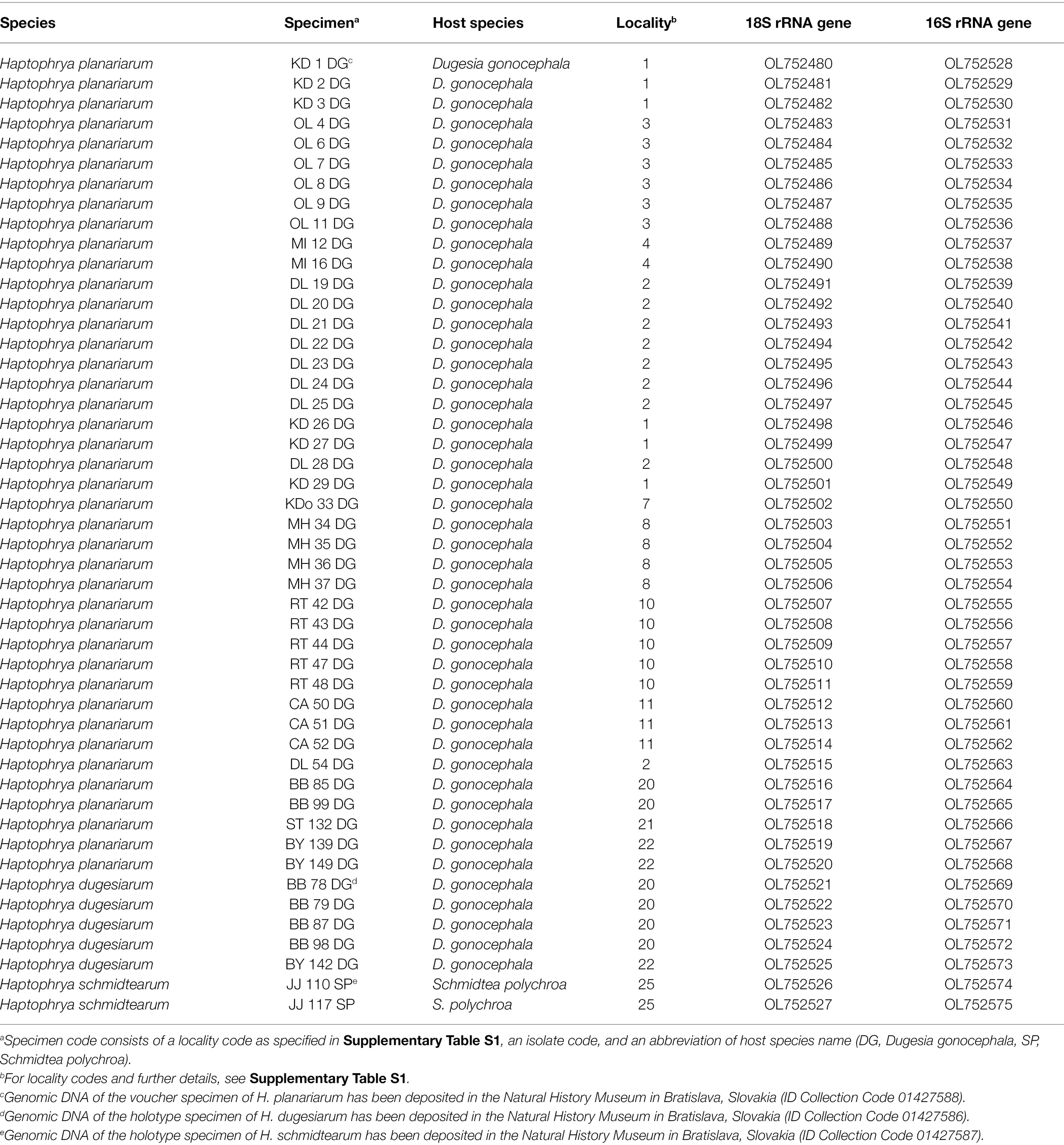
Table 1. Characterization and origin of nuclear and mitochondrial SSU rRNA gene sequences of Haptophrya species analyzed in the present study.
The 18S rRNA gene is 1761 nucleotides (nt) long in H. planariarum and H. dugesiarum, while 1760 nt long in H. schmidtearum. The guanosine–cytosine (GC) content ranges from 42.22% in H. schmidtearum to 42.48% in H. planariarum. The 18S sequences of H. planariarum and H. dugesiarum differ by only two or three nucleotide positions (Figure 1A), which corresponds to a p-distance of 0.11–0.17%. As evident from the parsimony network, there are two ribotypes in both Haptophrya species isolated from D. gonocephala. Interestingly, individual ribotypes of H. planariarum and H. dugesiarum are distinguished by the same site (position 975) in helix 24 where is either uracil or guanine. This nucleotide position is very likely ancestrally polymorphic and both nucleotide states retain the RNA helical structure. On the other hand, H. schmidtearum is fairly distant from H. planariarum and H. dugesiarum, as it differs from them by as many as 16–19 nucleotides (Figure 1A). The p-distance between H. schmidtearum and the two other Haptophrya species ranges from 0.97 to 1.14%, indicating a comparatively deep divergence. The 18S secondary structure models of the three Haptophrya species are shown in Figure 2 and Supplementary Figures S2, S3. The fraction of Watson–Crick pairs (AU and CG) ranges from 72.53% in H. schmidtearum to 73.12% in H. planariarum, wobble pairs (GU or UG) from 12.54% in H. planariarum to 12.93% in H. schmidtearum, and non-canonical pairs from 14.34% in H. planariarum to 14.54% in H. schmidtearum. The molecular diagnostic characters of these three species are accumulated in helices 21, 21es6b, and 21es6c of the V4 region, helices 38–40 of the V7 region, and helix 44 of the V9 region. The distinctness of H. schmidtearum from H. planariarum and H. dugesiarum is also strengthened by three CBCs.
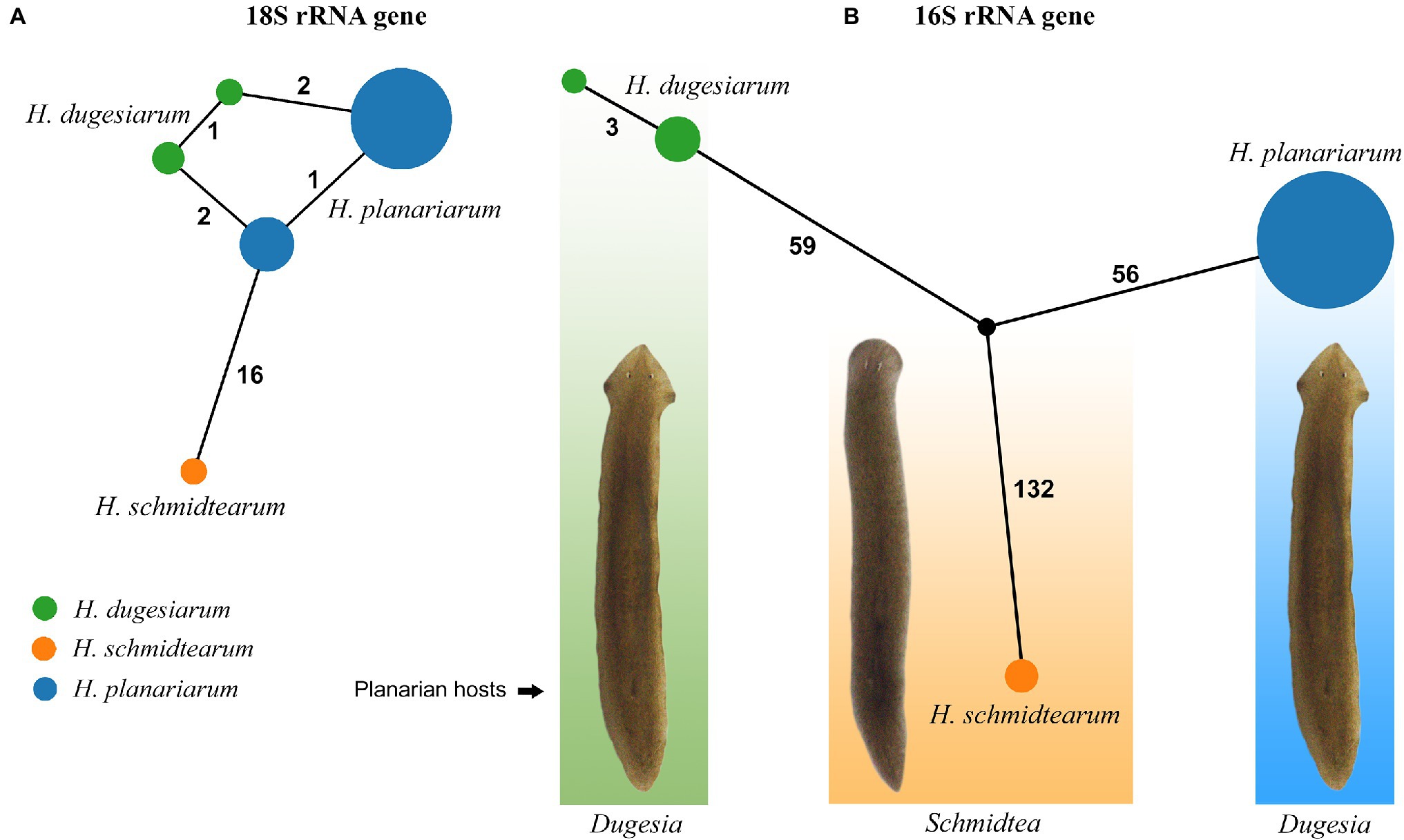
Figure 1. TCS networks of the nuclear 18S (A) and the mitochondrial 16S (B) rRNA gene sequences of three Haptophrya species isolated from freshwater planarians. Circle size corresponds to the haplotype frequency and color refers to individual Haptophrya species. Numbers along edges represent mutational steps.
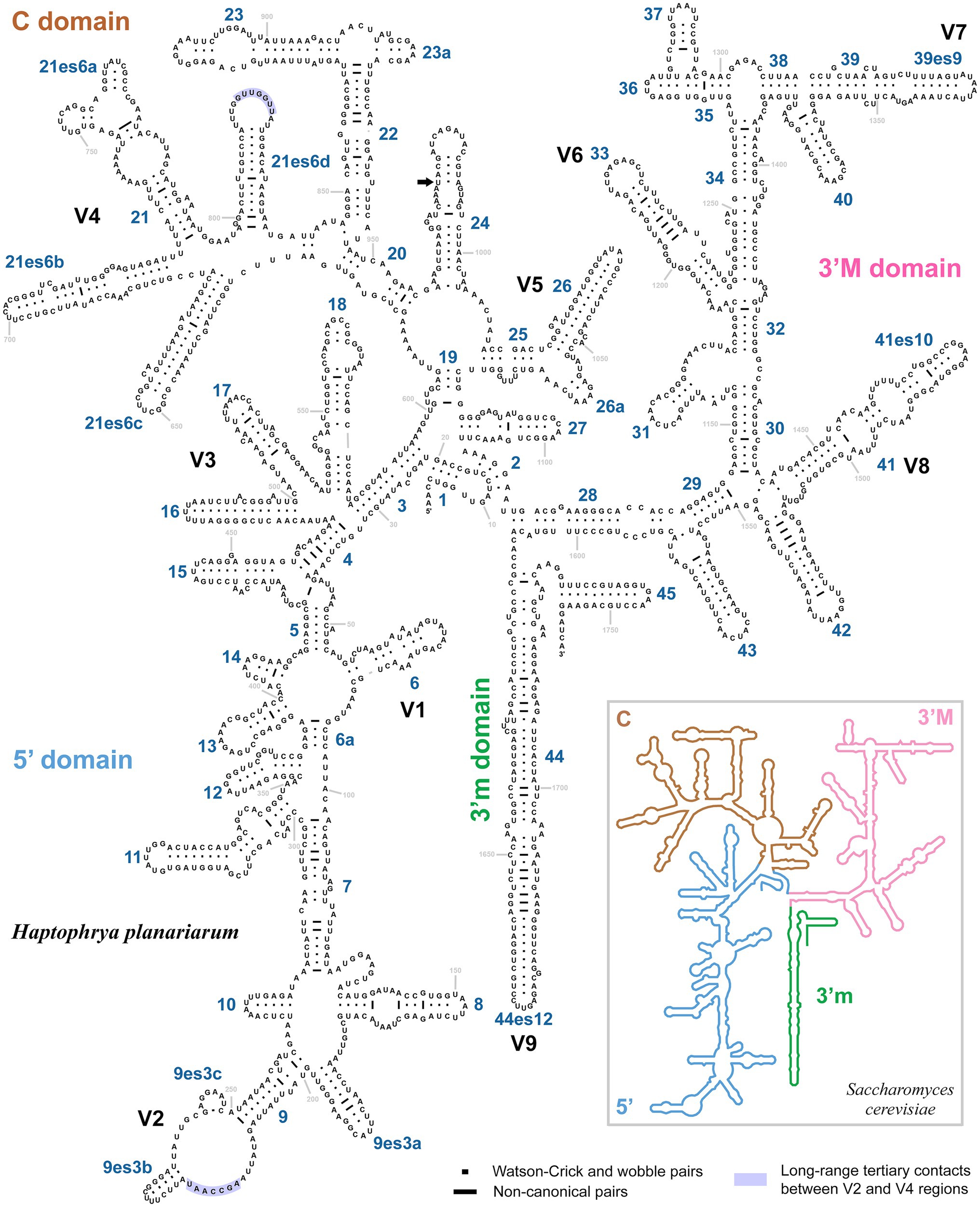
Figure 2. Secondary structure of the 18S rRNA molecule of Haptophrya planariarum, based on models taking into account 3D structures. 18S secondary structure map of Saccharomyces cerevisiae (inset) is from http://apollo.chemistry.gatech.edu/RibosomeGallery (Petrov et al., 2014). Arrow marks the ancestrally polymorphic position 975 in helix 24, where is either uracil or guanine. Both nucleotide states retain the RNA helical structure.
The amplified region of the mitochondrial 16S rRNA gene covers the C, 3’M, and 3’m domains and is 1,170 nt long in H. planariarum, 1,172 nt long in H. dugesiarum, and 1,167 nt long in H. schmidtearum. The GC content is 37.86% in H. planariarum, 37.27–37.37% in H. dugesiarum, and 39.33% in H. schmidtearum. According to the network analyses, there is a single ribotype in H. planariarum and H. schmidtearum, while two ribotypes in H. dugesiarum. The two Haptophrya species isolated from D. gonocephala differ by as many as 115–118 mutational steps (Figure 1B), which corresponds to a p-distance of 11.03–11.21%. Haptophrya schmidtearum is separated from their common ancestor by as many as 132 mutational steps (Figure 1B). This deep divergence is reflected also by high p-distances, which range from 16.87 to 17.03% between H. schmidtearum and the two other species. The 16S secondary structure models of the three Haptophrya species are shown in Figure 3 and Supplementary Figures S4, S5. The fraction of Watson–Crick pairs (AU and CG) ranges from 70.89% in H. planariarum to 72.72% in H. dugesiarum, wobble pairs (GU or UG) from 14.50% in H. dugesiarum to 18.73% in H. planariarum, and non-canonical pairs from 10.38% in H. planariarum to 12.77% in H. dugesiarum. Species-specific mutations tend to accumulate in helix 21 of the C domain (Figures 4A–C), helix 33 (Figures 4D–F), helices 39 and 40 (Supplementary Figures S6A–C) of the 3’M domain, as well as in helix 44 of the 3’m domain (Supplementary Figures S7A–C). The distinctness of all three Haptophrya species is also strengthened by the presence of CBCs in the 16S rRNA molecule. Specifically, H. schmidtearum is separated from H. planariarum and H. dugesiarum by 9 and 12 CBCs, respectively. The two latter species are distinguished by 10 CBCs.
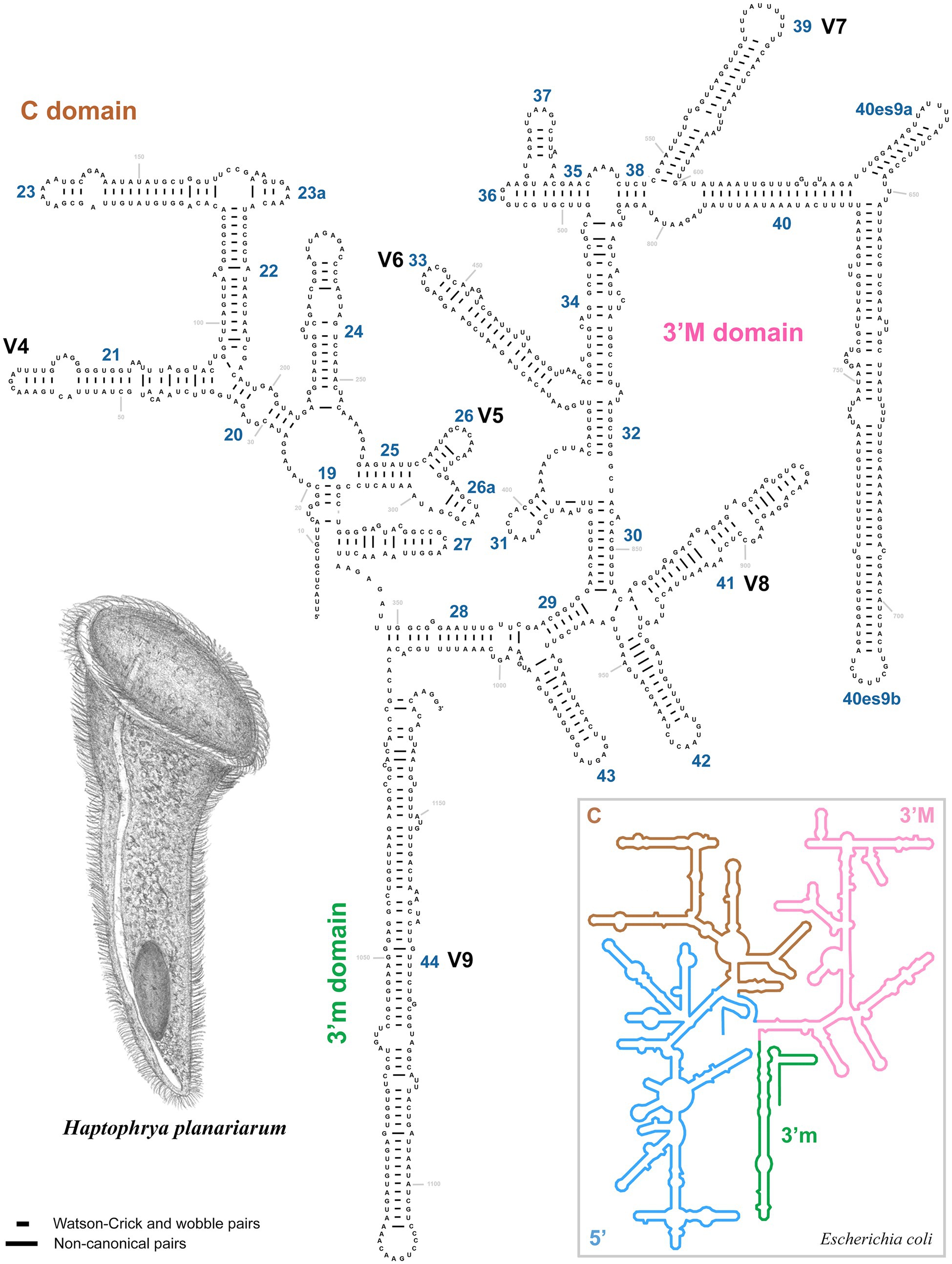
Figure 3. Secondary structure of the 16S rRNA molecule of Haptophrya planariarum, based on models taking into account 3D structures. 16S secondary structure map of Escherichia coli (inset) is from http://apollo.chemistry.gatech.edu/RibosomeGallery (Petrov et al., 2014).
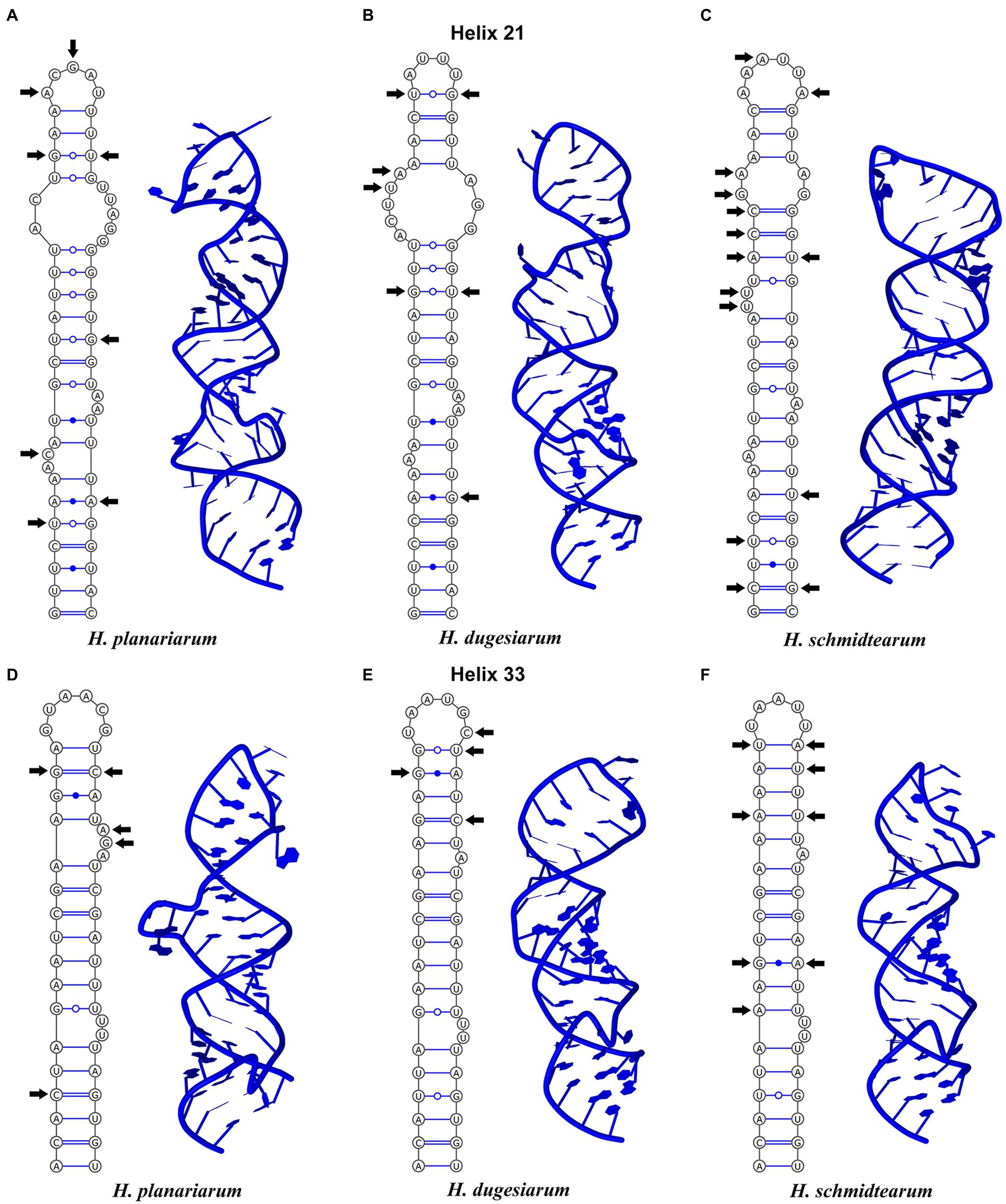
Figure 4. Secondary structure of the highly variable helices 21 (upper panel) and 33 (lower panel) from the C and 3’M domains of the 16S rRNA molecule of Haptophrya planariarum (A,D), Haptophrya dugesiarum (B,E), and Haptophrya schmidtearum (C,F). Arrows denote the molecular diagnostic characters. Note that each species can be unambiguously distinguished by both the primary and the secondary structure of helices 21 and 33. The 16S rRNA gene can be thus used as an optimal DNA barcode for the three Haptophrya species.
Phylogenetic Position of Haptophrya
Phylogenetic positions of mouthless ciliates, represented by Haptophrya, Clausilocola, and “core” astomes, within the class Oligohymenophorea were determined using the nuclear 18S and the mitochondrial 16S rRNA gene in a ML and BI framework. Masked and unmasked alignments constructed with the iterative G-INS-i and the progressive G-INS-1 method brought highly similar results. Phylogenetic trees inferred from the masked G-INS-1 datasets are shown in Figures 5, 6, as they received the highest ML bootstrap values and posterior probabilities. The phylogenetic position of Haptophrya slightly differed between the single-gene (18S) and the two-gene (16S + 18S) dataset, very likely due to the limited taxon sampling in the latter one. Despite that, Haptophrya never grouped with astomes isolated from annelids or with Clausilocola isolated from gastropods. In 18S rRNA gene phylogenies, Haptophrya very robustly clustered with free-living members of the scuticociliate genus Dexiotricha and representatives of the symbiotic genus Conchophthirus inhabiting the mantle cavity of bivalve mussels (100% ML, 1.00 BI). This heterogeneous clade was depicted as sister to the “core” scuticociliates (Pleuronematida + Philasterida) though with very poor statistical support (88% ML, 0.89 BI). Astomes isolated from annelids formed a monophylum (100% ML, 1.00 BI) that branched off before Haptophrya, while Clausilocola was nested within the subclass Hymenostomatia (100% ML, 1.00 BI). In trees inferred from 16S + 18S rRNA gene sequences, Haptophrya was placed in a sister position to a highly diverse clade comprising astomes and the “core” scuticociliates. Astomes grouped with the “core” scuticociliates with strong support (98% ML, 1.00 BI) in the two-gene phylogenies. On the other hand, the grouping of haptophryans with the “core” scuticociliates was only very weakly supported in the single-gene phylogenies (see above) and hence should be taken with caution.
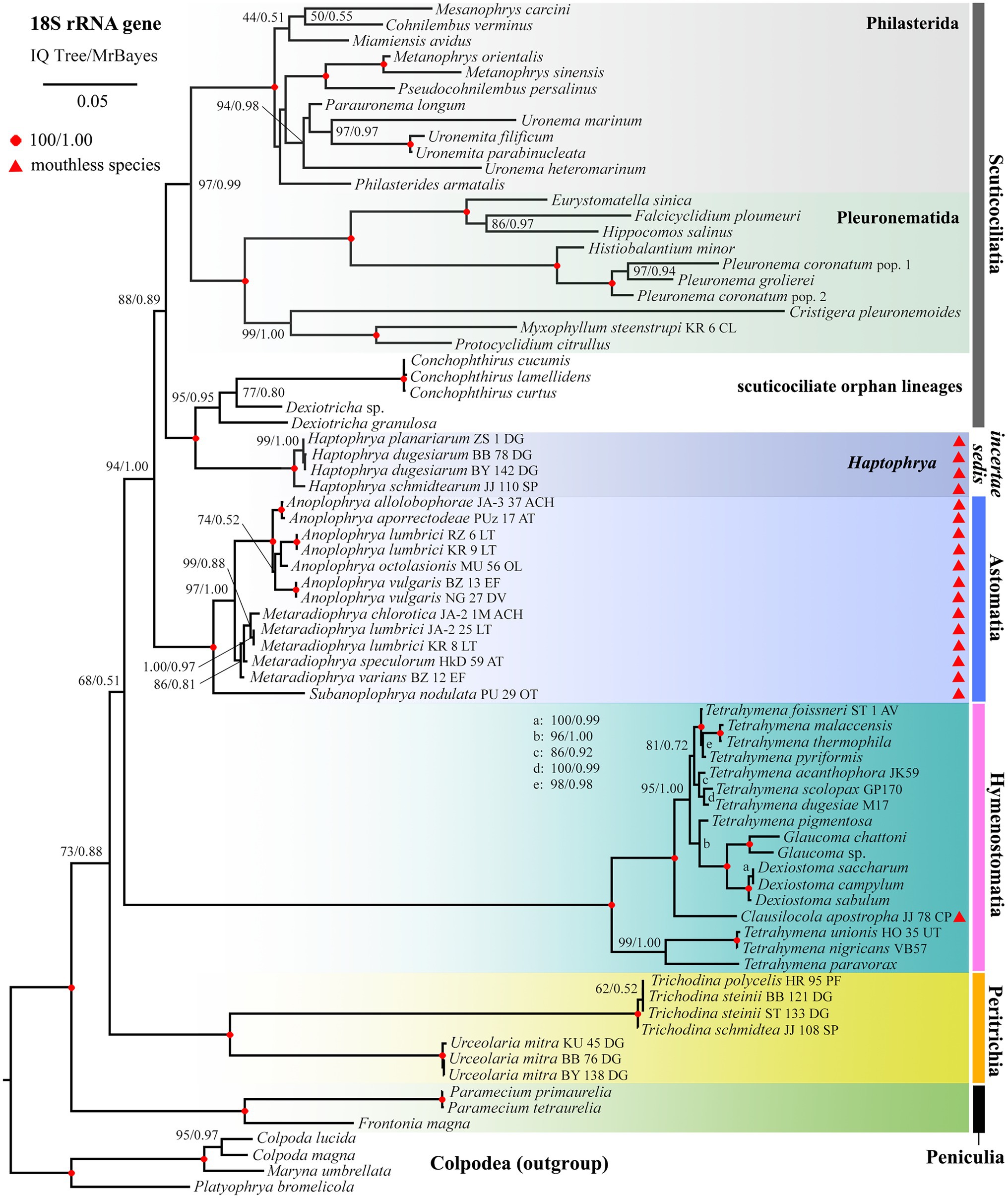
Figure 5. Phylogenetic tree based on the 18S rRNA gene, showing the systematic positions of Haptophrya and other mouthless species (marked by red triangles). Bootstrap values for maximum likelihood (ML) conducted in IQTrees as well as posterior probabilities for Bayesian inferences conducted in MrBayes were mapped onto the 50%-majority rule IQTree. Note that mouthless endosymbionts inhabiting planarians (Haptophrya), annelids (Astomatia), and mollusks (Clausilocola) do not cluster together. Haptophrya is most closely related to Conchophthirus and Dexiotricha, while Clausilocola is robustly nested within the subclass Hymenostomatia. The scale bar denotes five substitutions per one hundred nucleotide positions.
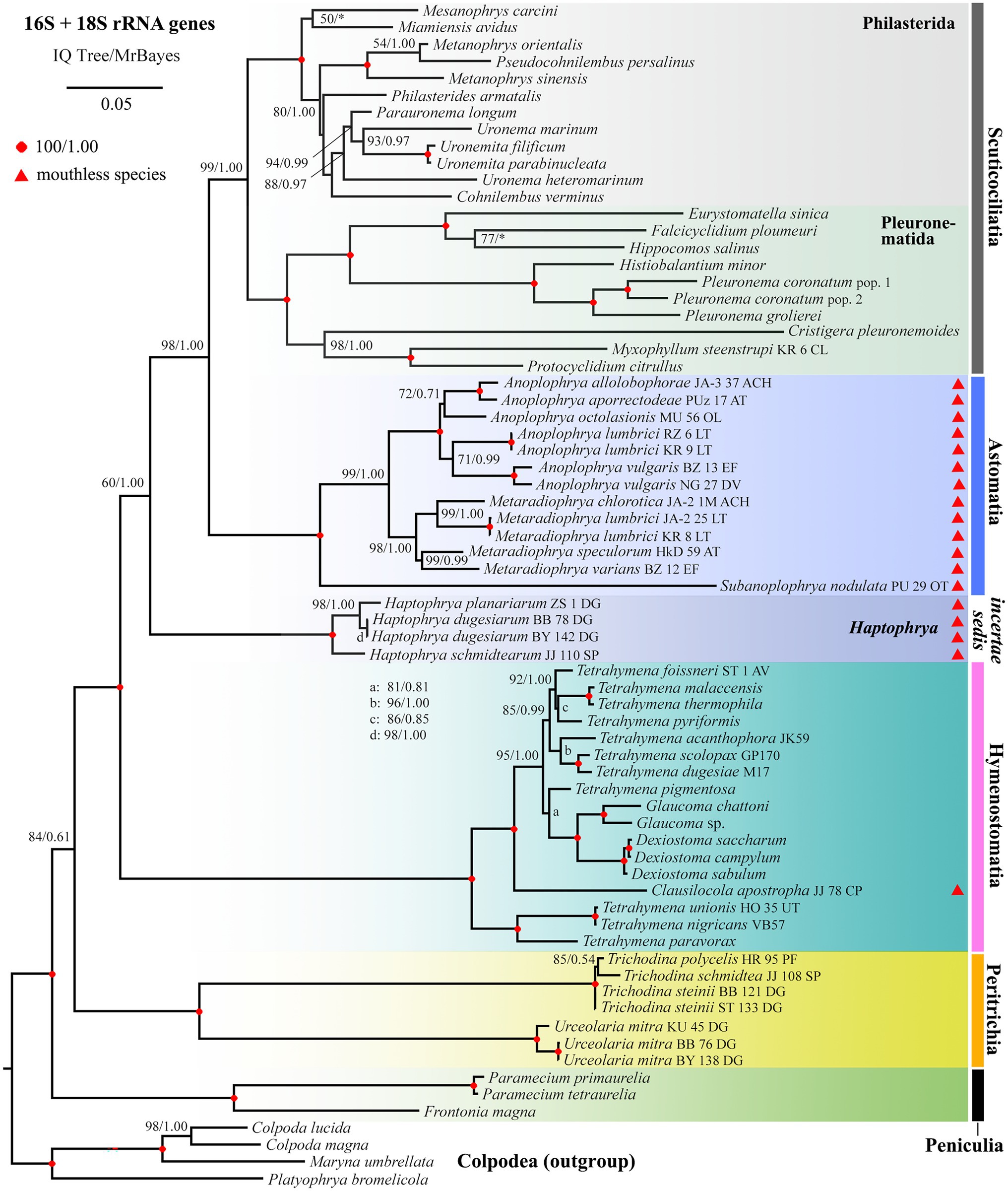
Figure 6. Phylogenetic tree based on the 16S and 18S rRNA genes, showing the systematic positions of Haptophrya and other mouthless species (marked by red triangles). Bootstrap values for ML conducted in IQTrees as well as posterior probabilities for Bayesian inferences conducted in MrBayes were mapped onto the 50%-majority rule IQTree. Note that mouthless endosymbionts inhabiting planarians (Haptophrya), annelids (Astomatia), and mollusks (Clausilocola) do not cluster together. Haptophrya represents an orphan lineage in the two-gene trees. Astomes group with relatives of the scuticociliate orders Philasterida and Pleuronematida, while Clausilocola is robustly nested within the subclass Hymenostomatia. The scale bar denotes five substitutions per one hundred nucleotide positions.
To summarize, mouthless endosymbionts inhabiting planarians (Haptophrya), annelids (“core” astomes), and mollusks (Clausilocola) never clustered together. Haptophrya was revealed to be most closely related to two scuticociliate orphan genera (Conchophthirus and Dexiotricha), “core” astomes are very likely relatives of the scuticociliate orders Philasterida and Pleuronematida, and Clausilocola is robustly nested within the hymenostome family Tetrahymenidae.
Morphological Characterization and Identification of Haptophryans
Haptophryans were isolated from two planarian hosts (D. gonocephala and S. polychroa) collected at 12 localities. According to molecular data, they belong to three species (H. planariarum, H. dugesiarum, and H. schmidtearum; Figures 1A,B). Nonetheless, they share all taxonomically important qualitative morphological characters (Figures 7A–E, 8A–C, 9A–E, 10A,B, and 11A–H): (i) a campanulate to truncate claviform body differentiated into a conspicuous anterior, shallow sucker, a more or less distinct neck-like constriction, and a cone-like trunk; (ii) the broadly to narrowly ellipsoidal macronucleus is situated in the rear body end in living cells and is accompanied by 1–3 globular micronuclei; (iii) the contractile canal extends along the whole dorsal cell margin; (iv) somatic kineties are built from monokinetids throughout; (v) ciliary rows are meridional and very narrowly spaced, about half of them extends onto the anterior sucker; (vi) the horseshoe-shaped suture runs along the dorsal and lateral borders of the sucker (shown only for H. planariarum in Figure 8A, opposed arrowheads); and (vii) two inconspicuous secant systems at lateral ends of the horseshoe-shaped suture (left suture shown only for H. schmidtearum in Figure 10A, opposed arrowheads). The general body organization as well as the ciliary pattern of Haptophrya is illustrated in detail in our previous study (Rataj and Vďačný, 2018) as well as in Lom (1959) and Corliss et al. (1965). The seeming differences between kinetid structures (Figures 7E, 9E) are caused by different bleaching and impregnation intensities of kinetodesmal fibers and postciliary microtubules. Thus, very deeply impregnated kinetodesmal fibers might cause the somatic monokinetid to appear as dikinetids especially when postciliary microtubules are only weakly impregnated.
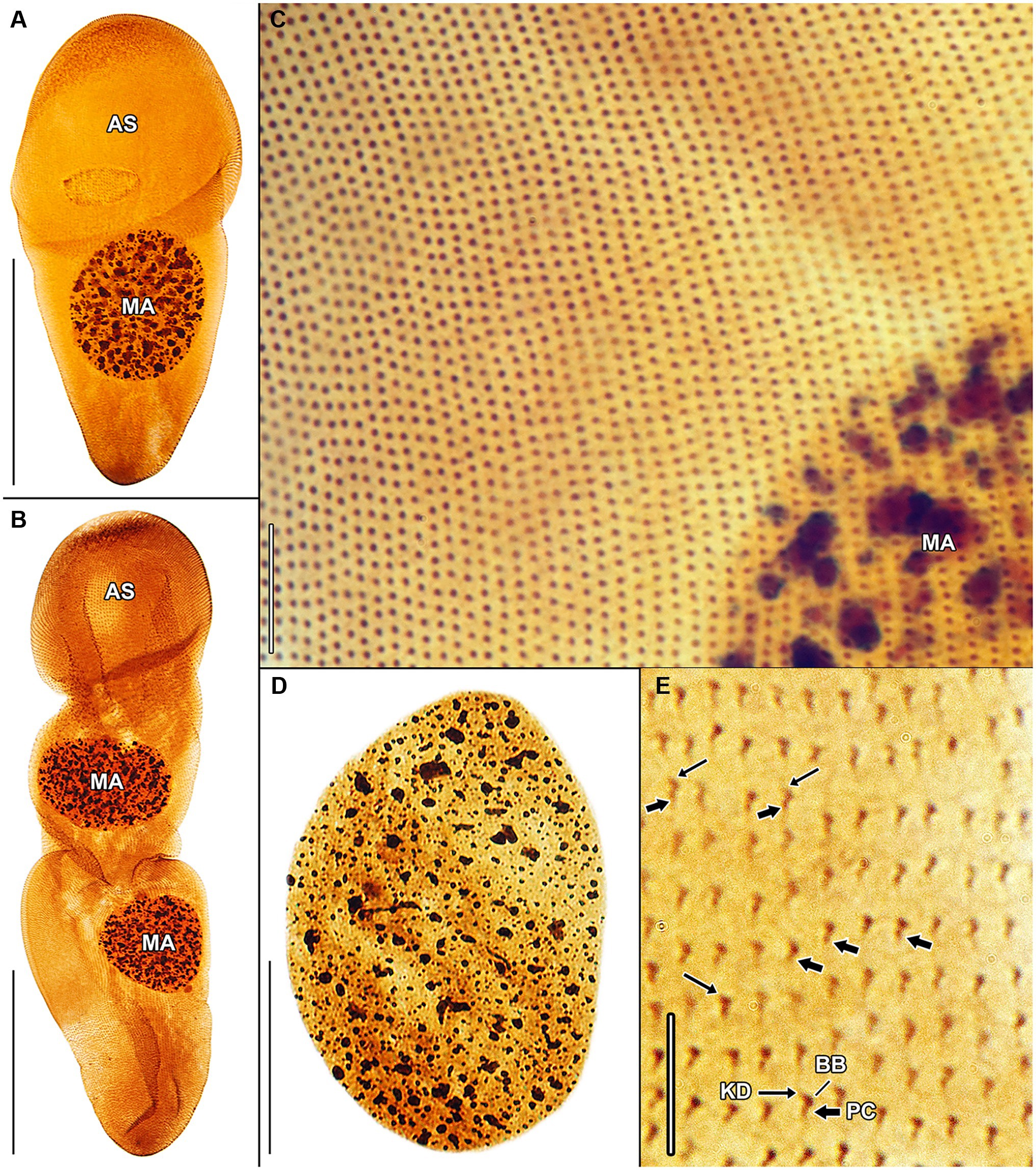
Figure 7. Haptophrya planariarum, protargol-impregnated specimens isolated from Dugesia gonocephala collected from the Malá Vydrica stream at the locality Železná studnička, Bratislava, Slovakia. (A) Ventral overview of a representative specimen. The body is campanulate and differentiated into a conspicuous anterior sucker, a more or less distinct neck-like constriction, and a cone-like trunk. (B) Ventral overview of a very late divider. Division occurs in freely motile conditions and is monotomic, i.e., yields two daughter cells. Axes of both daughter cells have the same orientation. (C) Ventral view, showing the very narrowly spaced somatic kineties composed of monokinetids. (D) The macronucleus is ellipsoidal and studded with globular to irregular nucleoli. (E) Detail of somatic kineties, showing the fibrillar associates of basal bodies. AS, adhesive sucker; BB, basal body; KD, kinetodesmal fiber (thin arrow); MA, macronucleus; and PC, postciliary microtubules (thick arrow). Scale bars = 5 μm (E), 10 μm (C), 30 μm (D), and 100 μm (A,B).
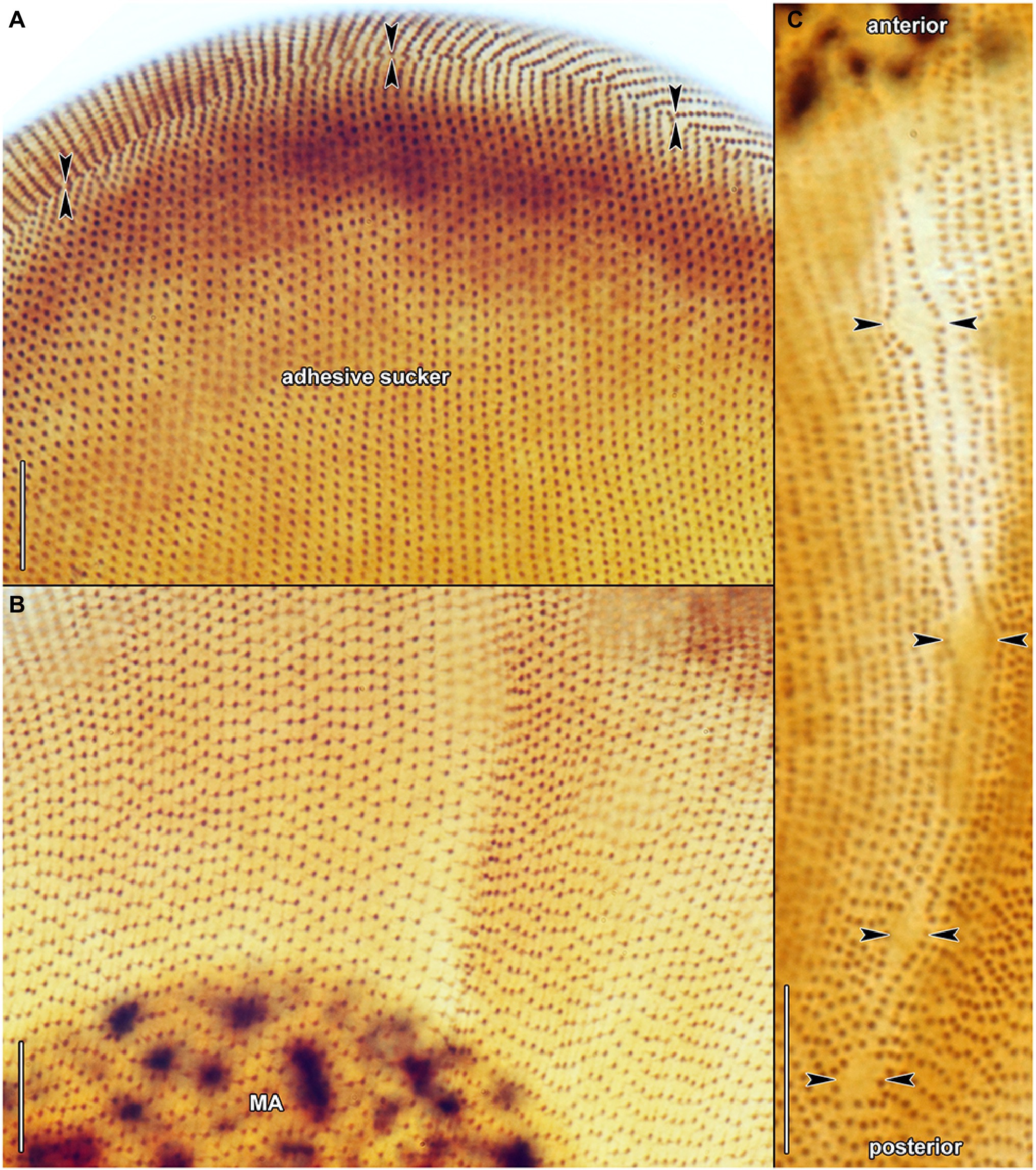
Figure 8. Haptophrya planariarum, protargol-impregnated specimens isolated from Dugesia gonocephala collected from the Malá Vydrica stream at the locality Železná studnička, Bratislava, Slovakia. (A) Ventral view of the adhesive sucker, showing the horseshoe-shaped suture (opposed arrowheads) that runs along the dorsal and lateral borders of the sucker. About 90 ventral kineties run onto the adhesive sucker to abut almost at right angle on the dorsal and lateral kineties along the whole horseshoe-shaped suture line. (B) Dorsal view of the adhesive sucker, showing the ciliary pattern. Somatic ciliature is holotrichous. Basal bodies are very densely spaced, i.e., the intrakinetidal distance is about 1.5 μm. Somatic kineties extend meridionally and they are also very narrowly spaced, i.e., the intrakinetal distance is approximately 1 μm. (C) Dorsal view of the trunk region, showing the ciliary pattern along the contractile canal. Opposed arrowheads denote the excretory pores of the canal. MA, macronucleus. Scale bars = 10 μm.
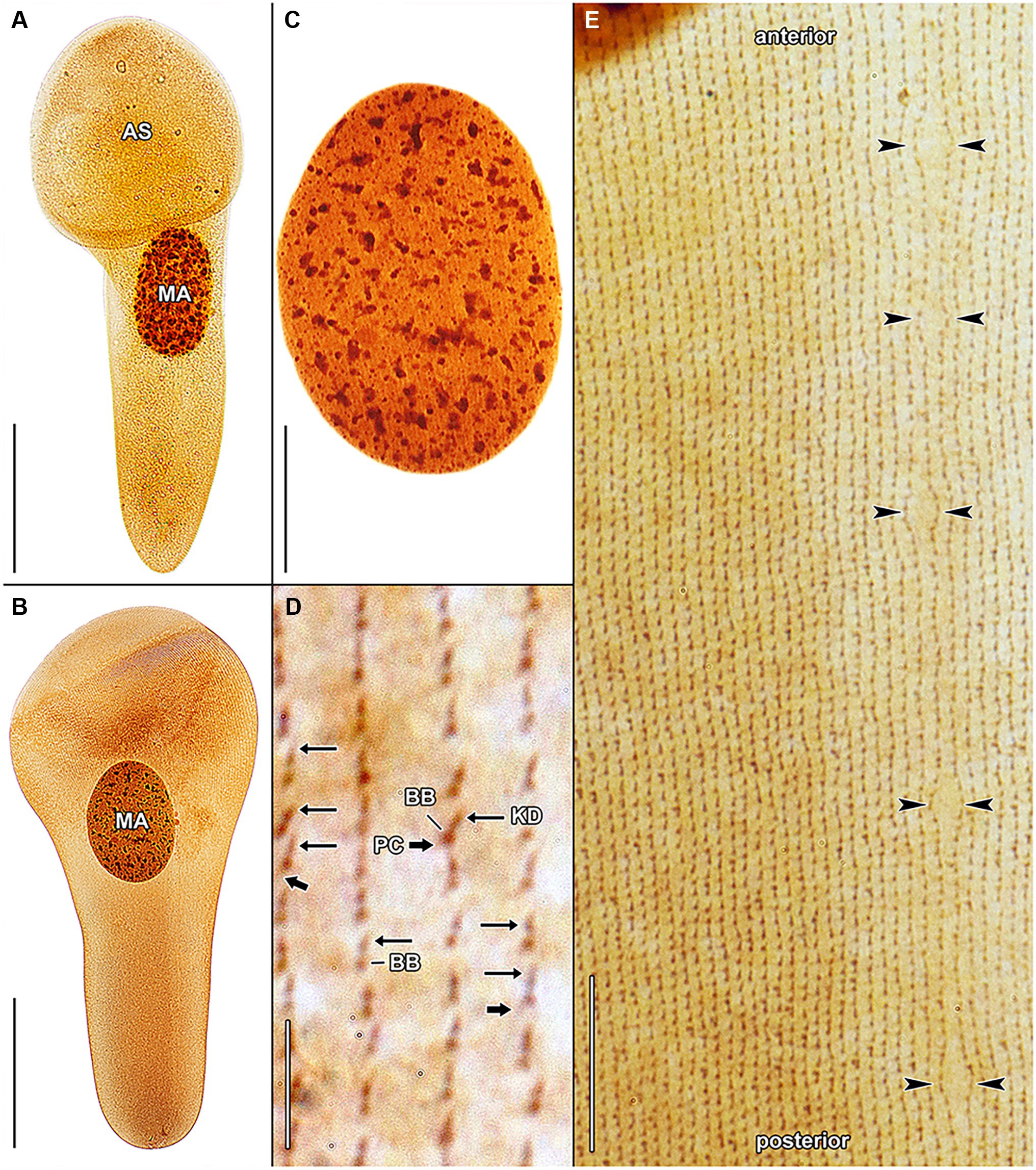
Figure 9. Haptophrya schmidtearum nov. spec., protargol-impregnated specimens isolated from Schmidtea polychroa collected from the Jurské jazierko pond in the district of the village of Svätý Jur, Malé Karpaty Mts., Slovakia. (A,B) Overviews of representative specimens. The body is campanulate to truncate claviform and differentiated into a conspicuous anterior sucker, a more or less distinct neck-like constriction, and a cone-like trunk. (C) The macronucleus is ellipsoidal and studded with globular to irregular nucleoli. (D) Detail of somatic kineties, showing the fibrillar associates of basal bodies. When kinetodesmal fibers are very deeply impregnated, somatic monokinetids might appear as dikinetids, especially when postciliary microtubules are only weakly impregnated. (E) Dorsal view of the trunk region, showing the ciliary pattern along the contractile canal. Opposed arrowheads denote the excretory pores of the canal. AS, adhesive sucker; BB, basal body; KD, kinetodesmal fiber (thin arrow); MA, macronucleus; and PC, postciliary microtubules (thick arrow). Scale bars = 5 μm (D), 10 μm (E), 30 μm (C), and 100 μm (A,B).
The quantitative characters significantly overlap and hence also do not allow discrimination of the three Haptophrya species. In mixed populations, the separation of H. dugesiarum from H. planariarum was impossible with morphological data and both species could be reliably discerned only after sequencing. Therefore, the following morphometric comparison is based only on a voucher population of H. planariarum (collected from the Malá Vydrica stream in the Kačínska dolina valley at the locality Železná studnička, Bratislava) and the type population of H. schmidtearum (collected from the Jurské jazierko pond in an urban oak-hornbeam forest, district of the village of Svätý Jur, Malé Karpaty Mts.). The “Malá Vydrica” population was selected to avoid mixing of H. planariarum and H. dugesiarum, as the latter species has been hitherto not detected in SW Slovakia. The body length in vivo spans a range of 205–335 μm in H. planariarum, while it is about 281 μm in H. schmidtearum. The maximum width of the adhesive sucker in vivo is 95 μm in H. planariarum, while the sucker is slightly wider (up to 131 μm) in H. schmidtearum. The maximum trunk width in living cells is 58 μm in H. planariarum, while H. schmidtearum is slightly broader (about 79 μm). The size of the macronucleus after protargol impregnation is 67–111 × 50–79 μm in H. planariarum and 81–104 × 53–63 μm in H. schmidtearum. Finally, the number of somatic kineties on one body side is 90–101 in H. planariarum and 98–108 in H. schmidtearum.

Figure 10. Haptophrya schmidtearum nov. spec., protargol-impregnated specimens isolated from Schmidtea polychroa collected from the Jurské jazierko pond in the district of the village of Svätý Jur, Malé Karpaty Mts., Slovakia. (A) Ventrolateral view, showing the secant system (opposed arrowheads) at the lateral margin of the adhesive sucker, whose ciliary rows abut on the anterior end of trunk ciliary rows. (B) Dorsal view of the adhesive sucker, showing the ciliary pattern. Somatic ciliature is holotrichous. Basal bodies are very densely spaced, i.e., the intrakinetidal distance is about 1.5 μm. Somatic kineties extend meridionally and they are also very narrowly spaced, i.e., the intrakinetal distance is approximately 1 μm. Note that kinetodesmal fibers are well-developed, directed anteriorly, and overlapping. They form kinetodesmal ribbons that extend close to the right of the somatic ciliary rows and appear as weakly impregnated lines, as typical of members of the class Oligohymenophorea. Scale bars = 10 μm.
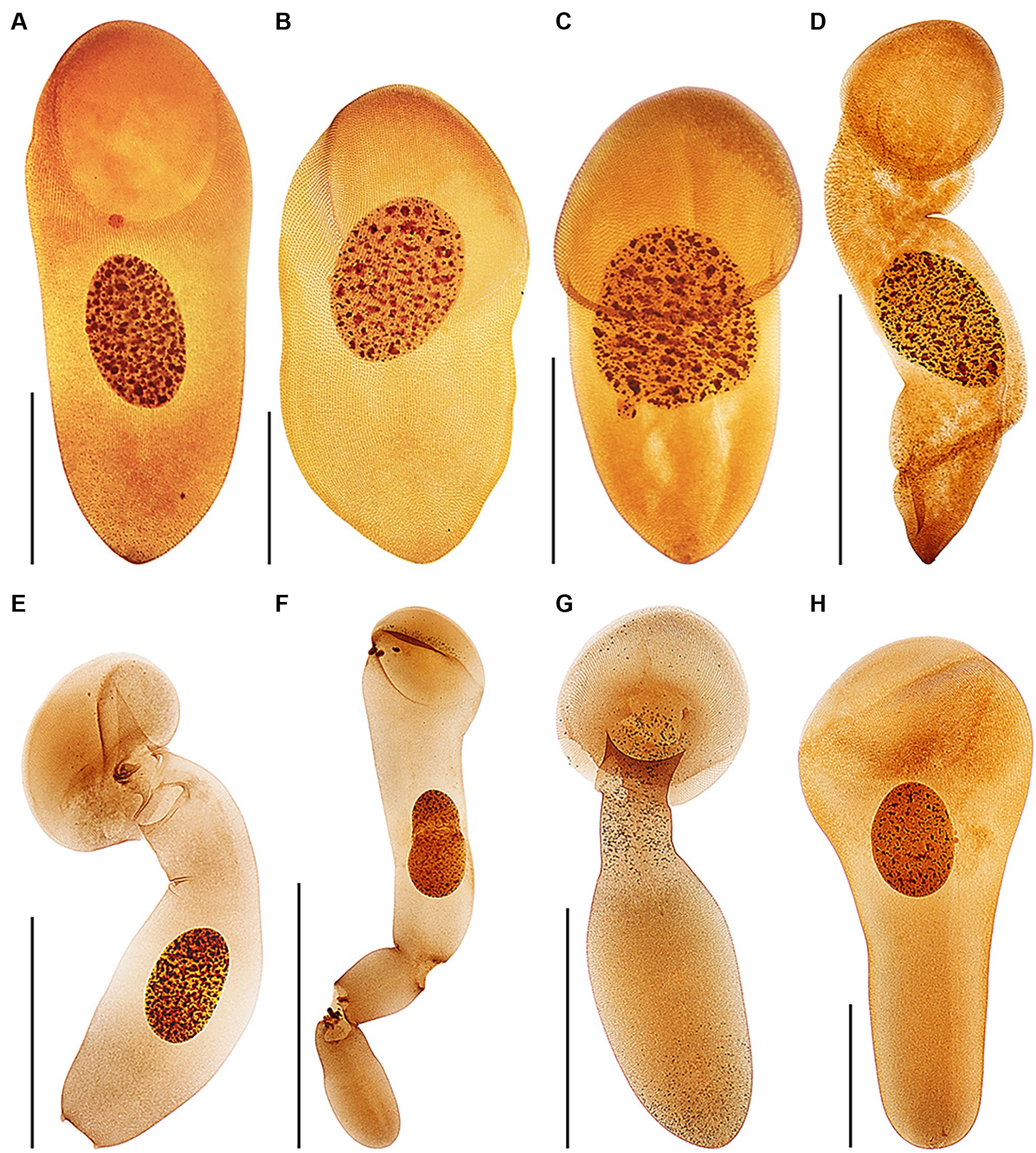
Figure 11. Protargol-impregnated specimens of Haptophrya planariarum (A–D) and Haptophrya schmidtearum nov. spec. (E–H). The typical body shape of Haptophrya species is campanulate to truncate claviform and the body is differentiated into a conspicuous anterior sucker, a more or less distinct neck-like constriction, and a cone-like trunk (A–C,H). The macronucleus is broadly to narrowly ellipsoidal and situated in the rear body end in living cells. Preparations artifacts and postmortal changes include elongation of the body, inflation of the anterior body third, and fragmentation of the posterior trunk region (D,E–G). Note that the macronucleus is gradually displaced from the rear end to the anterior body region in prepared cells. Already Schultze (1851) figured these morphological changes in H. planariarum specimens isolated from Planaria torva. Due to the dramatic changes in the body shape and size as well as in the localization of the nuclear apparatus in some protargol-impregnated specimens, fresh and living cells need to be investigated. Scale bars = 100 μm (A–C,H) and 200 μm (D–G).
Discussion
Cryptic Speciation in Haptophrya
Since the times of Ehrenberg (1838) and Stein (1859), it has been assumed that ciliate taxa should differ morphologically. However, there is no law in nature that would require phylogenetically distinct species to be also divergent in phenotype. Phenotypic divergence may be also delayed relative to phylogenetic divergence for multiple reasons: (i) species may have diverged too recently to have also diverged in phenotype, (ii) selective forces at work relative to a particular phenotype, and (iii) simple body plans offer only a little evidence for phenotypic divergence except for difficult to perceive ultrastructural or physiological features (Lücking et al., 2021). With the application of multiple molecular markers (especially the mitochondrial 16S rRNA and COI genes), cryptic or near-cryptic speciation (subtle, easily overlooked differences, or only statistically supported differences) has been detected in a variety of symbiotic ciliates. For instance, in clevelandellids associated with wood-feeding cockroaches (Pecina and Vďačný, 2020, 2021), in histophagous tetrahymenids parasitizing mollusks (Zhang and Vďačný, 2021), in astomes occupying the digestive tube of lumbricid earthworms (Obert and Vďačný, 2019, 2020; Obert et al., 2021), and in mobilids living on the surface of freshwater planarians (Rataj and Vďačný, 2021). Cryptic or near-cryptic speciation has apparently occurred also in haptophryans associated with freshwater planarians, as documented by the huge differences in the primary structure (interspecies p-distances range from 11.03 to 17.03%, while maximum intraspecies distances are only 0.26%), the secondary structure of the V4, V7, and V9 regions (Figure 4; Supplementary Figures S6, S7) as well as by 9–12 CBCs in the 16S rRNA molecule. Although no distinct qualitative or quantitative morphological differences were detected, the molecular differences among the phylogenetically delimited Haptophrya species are so huge that they cannot be interpreted as intra-species variability of H. planariarum. Haptophrya schmidtearum might be considered as a near-cryptic species, as it can be separated from H. planariarum and H. dugesiarum by the planarian host (the pond-dwelling S. polychroa vs. the stream- and river-dwelling D. gonocephala). Interestingly, the two latter Haptophrya species share the same host, which indicates a duplication event without host switching. This diversification mode was suggested also for astomes inhabiting megascolecid and glossoscolecid earthworms (Obert et al., 2021). In the absence of phenotypic divergence and diversification driven by duplication events with or without host switching, we prefer to employ molecular data to distinguish the three Haptophrya species. We anticipate that the current reluctance against the acceptance of phenotypically cryptic species (Warren et al., 2017) will diminish, as molecular tools have become more easily accessible and molecular delimitation of ciliate species has been successfully applied to multiple free-living (e.g., Doerder, 2019; Greczek-Stachura et al., 2021 and references cited therein) and symbiotic ciliates (e.g., Obert et al., 2021; Pecina and Vďačný, 2021). Finally, it is important to mention that the identity of H. planariarum populations reported from the Nearctic and Paleotropis is highly questionable, as the species was discovered in the Palearctic (for a review, see Rataj and Vďačný, 2018) and planarian species have biogeographies. Likewise, the conspecificity of populations isolated from various planarian hosts, belonging to the families Planariidae Stimpson, 1857 (Planaria Müller, 1776), Dendrocoelidae Hallez, 1892 (Dendrocoelum Ørsted, 1844), and Dugesiidae Ball, 1974 (Dugesia Girard, 1850 and Schmidtea Ball, 1974), is very doubtful in light of the present findings. Molecular data, especially 16S sequences, are indispensable to revealing their true identities.
Phylogenetic Relationships and Systematic Position of Haptophrya
Haptophrya was traditionally classified within the order Astomatida Schewiakoff, 1896 of the subclass Astomatia due to the complete lack of oral structures (Cépède, 1910; de Puytorac, 1957, 1963; Lom, 1959; Corliss et al., 1965). Annelophrya Lom, 1959, Cepedietta Kay, 1942, Lachmanella, Cépède, 1910, and Steinella Cépède, 1910 were considered the nearest relatives of Haptophrya and were assigned to the family Haptophryidae Cépède, 1923 in compendium of Lynn (2008). Since these genera differ by their attachment strategies (adhesive sucker vs. hooks), division mode (binary fission vs. chain formation), and host organisms (planarians vs. amphibians), they were divided into three subfamilies. The nominotypical subfamily Haptophryinae Cépède, 1923 (Haptophrya) occupies exclusively the digestive tract of freshwater planarians and is characterized by the absence of hooks and binary fission without chain formation. The subfamily Cepediettinae Corliss et al., 1965 unites endosymbionts of frogs and newts. They also attach to the host’s intestine employing an adhesive sucker but they form chains during reproduction. Finally, members of the subfamily Lachmannellinae Cépède, 1923 live in freshwater (Annelophrya) and marine (Lachmannella and Steinella) planarians. They attach to their digestive tract with a single hook (Lachmannella), two hooks of unequal size (Steinella), or with numerous, small spines surrounding the thigmotactic area (Annelophrya). No chain formation has been noted during the binary fission of lachmannellids. These profound morpho-ecological differences question the close relationship of cepediettids and lachmannellids as well as their assignment to the family Haptophryidae. Due to the lack of molecular data, their relatedness with Haptophrya and systematic positions remain unknown.
Lom (1959) suggested a close kinship of Haptophrya and Clausilocola Lom, 1959 due to their similarities in body shape and the presence of anterior adhesive sucker. Their monophyletic origin was, however, rejected in the light of both detailed morphological and molecular analyses (Zhang and Vďačný, 2022). Specifically, Haptophrya displays a secant system at each lateral end of the horseshoe-shaped suture and does not have dikinetids and unciliated apical field (Corliss et al., 1965; Rataj and Vďačný, 2018; present study). By contrast, Clausilocola possesses a single subapical secant system in the midline of the ventral side, an extensive unciliated apical area, and numerous dikinetids at the top of all ciliary rows. Unrelatedness of Haptophrya and Clausilocola was recognized also by the previous (Zhang and Vďačný, 2022) and present (Figures 5, 6) phylogenetic analyses, as both genera were nested in different oligohymenophorean subclasses: Haptophrya in the Scuticociliatia Small, 1967 and Clausilocola in the Hymenostomatia Delage and Hérouard, 1896.
The classification of Haptophrya in the subclass Astomatia was challenged already by the first 18S rRNA gene analyses (Rataj and Vďačný, 2018). Haptophrya was placed in a sister position to the free-living scuticociliate genus Dexiotricha Stokes, 1885, and this relationship was corroborated also by two shared unique nucleotide positions. Later on, the orphan scuticociliate genus Conchophthirus Stein, 1861, which inhabits the mantle cavity of freshwater bivalves, was added to this clade (Antipa et al., 2020). Phylogenetic relationships among Haptophrya, Dexiotricha, and Conchophthirus remained, however, unresolved in 18S phylogenies (Antipa et al., 2020; Zhang and Vďačný, 2022). Despite this, Haptophrya has been never found as a close relative of astomes isolated from the digestive tube of annelids (Obert and Vďačný, 2019, 2021; Obert et al., 2021). The primary and secondary structures of the mitochondrial 16S rRNA molecule also do not support their close relationships. The amplified part of the 16S rRNA gene is 864–888 nt long in the “core” astomes (Obert et al., 2021), while 1,167–1,172 nt long in haptophryans (present study). This conspicuous length difference is caused by deletions/insertions, especially in helices 21 and 23 from the V4 region, helix 33 from the V6 region, helices 39 and 40 from the V7 region, and helix 44 from the V9 region. Especially, the structure of helix 40 is quite different among astomes, “core” scuticociliates, and haptophryans. Helix 40 of Haptophrya carries two extra hairpins (40es9a and 40es9b), which are absent in astomes (Obert et al., 2021), “core” scuticociliates (Zhang et al., 2019), peniculines and hymenostomes (Cannone et al., 2002). Whether these insertions are specific for Haptophrya or shared with Dexiotricha and Conchophthirus needs to be analyzed in the future when 16S sequences become available for the two latter genera. On the other hand, the length and structure of the astome 16S rRNA molecules correspond rather well to those of the “core” scuticociliates (Zhang et al., 2019), peniculines, and hymenostomes (Cannone et al., 2002). Based on the multi-gene phylogenies as well as the primary and the secondary structure of the 16S rRNA molecules, we proclaim that the “core” astomes are more closely related to the “core” scuticociliates than to Haptophrya. Very likely, Haptophrya evolved from an orphan scuticociliate lineage comprising also Dexiotricha and Conchophthirus by the loss of oral apparatus and by the transformation of the thigmotactic field into an adhesive sucker. Since astomy evolved at least three times independently (in Haptophrya, Clausilocola, and “core” astomes) within the class Oligohymenophorea (Zhang and Vďačný, 2022; present study), the loss of cell mouth cannot be used as a sole argument for the assignment of haptophryans to the subclass Astomatia anymore.
Taxonomic Summary
We use molecular data to diagnose the two new Haptophrya species because morphological data do not allow their unambiguous separation. We interpret the isolated DNA as type material of the new species, which is in accordance with Article 72.5.1 of the International Commission on Zoological Nomenclature (1999). The reference alignments are provided in Supplementary Material. The primary and secondary structures of the 16S and 18S molecules are shown in Figures 2–4 and Supplementary Figures S2–S7.
Zoobank Registration Number of Work
urn:lsid:zoobank.org:pub:3949D77D-194C-42A7-8BBE-687C52451603.
Phylum Ciliophora Doflein, 1901
Class Oligohymenophorea de Puytorac et al., 1974
Incertae sedis in Subclass Scuticociliatia Small, 1967
Family Haptophryidae Cépède, 1923
Genus Haptophrya Stein, 1867 (type species: Opalina planariarum von Siebold, 1839)
Haptophrya dugesiarum Nov. Spec.
Zoobank Registration Number of New Species
urn:lsid:zoobank.org:act:1CAD60AE-A806-4C2B-9119-7AFBC27EACE5.
Diagnosis. 16S rRNA gene: 55 G, 62 T, 63 A, 68 T, 73 G, 82 T, 92 G, 110 G, 118 G, 120 G, 167 A, 169 C, 180 C, 181 A, 183 A, 424 C, 426 T, 446 G, 453 C, 454 T, 457 C, 521 A, 535 G, 559 A, 568 A, 573 C, 581 C, 587 -, 597 C, 598 G, 617 G, 629 C, 630 C, 632 A, 637 -, 642 A, 643 A, 649 A, 651 T, 652 G, 663 T, 664 T, 665 A, 666 T, 667 T, 668 T, 673 G, 674 G, 683 T, 686 A, 688 T, 701 T, 702 A, 707 T, 711 A, 730 A, 731 T, 733 A, 738 A, 745 C, 746 C, 749 A, 751 A, 754 G, 755 G, 757 C, 762 T, 771 A, 772 A, 783 A, 790 A, 793 T, 796 C, 799 A, 800 A, 802 A, 805 C, 807 A, 808 A, 811 A, 817 G, 824 -, 825 -, 831 C, 837 G, 840 T, 925 A, 930 T, 1085 A, 1102 A, 1108 -, 1114 A, 1118 A, 1122 T, 1128 G, 1144 T, 1149 T, 1152 G, 1162 C, 1164 G, and 1175 A. 18S rRNA gene: 486 T, 1320 A.
Type Locality. An unnamed stream in the Fončorda residential area, Banská Bystrica, Zvolenská kotlina basin, Slovakia (48°43′21.4″N, 19°06′58.3″E).
Type Host. Dugesia gonocephala (Dugès, 1830) Girard, 1850.
Type MaterialA. DNA sample of holotype specimen has been deposited in Natural History Museum, Vajanského nábrežie 2, 810 06 Bratislava, Slovakia (ID Collection Code 01427586).
Gene Sequences. The 16S and 18S rRNA gene sequences of the holotype specimen have been deposited in GenBank under the following accession nos. OL752569 and OL752521, respectively.
Etymology. The specific epithet is a plural genitive case of the Neo-Latin noun Dugesi·a, ae [f] (generic name of freshwater planarians), meaning a Haptophrya from dugesians. The species-group name is to be treated as an adjective used as a substantive in the genitive case, because of its derivation from the host’s generic name (Article 11.9.1.4. of the International Commission on Zoological Nomenclature, 1999).
Haptophrya schmidtearum Nov. Spec.
Zoobank Registration Number of New Species
urn:lsid:zoobank.org:act:EF237EA8-DBC2-4E7C-94A7-00CCF191F284.
Diagnosis. 16S rRNA gene: 39 C, 41 T, 53 T, 54 T, 56 A, 57 C, 58 C, 59 G, 60 A, 61 -, 62 -, 68 A, 70 A, 73 A, 81 T, 92 -, 96 G, 106 A, 108 C, 111 A, 116 A, 152 C, 157 A, 170 G, 182 T, 189 G, 191 T, 278 T, 282 G, 428 C, 436 A, 438 G, 444 A, 447 T, 452 -, 454 A, 455 T, 457 T, 464 A, 515 C, 532 C, 547 C, 552 G, 554 G, 555 G, 558 C, 559 G, 564 A, 565 C, 567 G, 570 C, 574 T, 577 A, 578 C, 591 G, 597 -, 598 -, 601 T, 604 G, 606 A, 609 C, 612 C, 617 T, 620 A, 622 A, 628 -, 631 -, 637 A, 638 A, 639 A, 644 T, 645 T, 650 G, 651 G, 659 A, 660 C, 661 C, 664 -, 669 A, 671 C, 678 G, 680 T, 682 A, 683 G, 684 C, 685 G, 686 G, 692 A, 695 C, 697 C, 698 A, 700 A, 704 C, 705 A, 706 A, 708 A, 709 C, 711 G, 712 C, 713 C, 714 C, 715 C, 716 C, 719 C, 720 A, 722 T, 724 C, 725 C, 730 -, 733 C, 734 C, 735 T, 736 T, 737 G, 738 G, 739 T, 743 C, 744 C, 751 -, 754 -, 755 -, 756 -, 761 -, 773 A, 774 A, 775 A, 776 G, 777 G, 780 T, 781 T, 782 T, 784 G, 785 G, 788 T, 790 G, 791 C, 792 G, 795 G, 797 A, 799 T, 800 T, 805 G, 808 T, 812 T, 814 A, 816 C, 819 A, 824 T, 825 T, 827 G, 829 T, 837 -, 839 A, 844 G, 908 A, 920 G, 928 C, 931 A, 933 G, 934 T, 943 T, 957 T, 979 G, 1021 T, 1087 T, 1098 A, 1100 A, 1103 T, 1106 -, 1112 -, 1113 C, 1114 -, 1115 -, 1120 G, 1122 A, 1125 -, 1127 A, 1129 -, 1133 G, 1134 T, 1135 T, 1141 A, 1142 A, 1144 A, 1145 C, 1148 C, 1149 G, 1150 G, 1151 T, 1154 C, 1155 -, 1157 G, 1160 C, 1164 -, 1168 G, 1178 G, and 1180 A. 18S rRNA gene: 639 A, 640 T, 645 T, 661 G, 666 A, 680 T, 691 T, 704 A, 723 A, 725 C, 726 C, 766 A, 1336 -, 1349 T, 1370 T, 1463 A, and 1660 G.
Type Locality. Jurské jazierko pond in an urban oak-hornbeam forest, district of the village of Svätý Jur, Malé Karpaty Mts. (Little Carpathians), Slovakia (48°15′28.0″N, 17°09′14.6″E).
Type Host. Schmidtea polychroa (Schmidt, 1861) Ball, 1974.
Type Material. A DNA sample of holotype specimen has been deposited in Natural History Museum, Vajanského nábrežie 2, 810 06 Bratislava, Slovakia (ID Collection Code 01427587).
Gene Sequences. The 16S and 18S rRNA gene sequences of the holotype specimen have been deposited in GenBank under the following accession nos. OL752574 and OL752526, respectively.
Etymology. The specific epithet is a plural genitive case of the Neo-Latin noun Schmidte·a, ae [f] (generic name of freshwater planarians), meaning a Haptophrya from schmidteans. The species-group name is to be treated as an adjective used as a substantive in the genitive case, because of its derivation from the host’s generic name (Article 11.9.1.4. of the International Commission on Zoological Nomenclature, 1999).
Haptophrya planariarum (von Siebold, 1839) Stein, 1867
Nomenclature and taxonomy of H. planariarum were reviewed by Rataj and Vďačný (2018). The species was neotypified by Corliss et al. (1965). However, at the present state of knowledge, the morphological data do not enable unambiguous identification of this species and gene sequences are not available from the neotype. To promote nomenclatural and taxonomic stability and correct usage of the name H. planariarum, we provide below a molecular diagnosis based on the barcoding 16S rRNA gene, voucher material, and gene sequences from the voucher specimen.
Molecular Diagnosis
16S rRNA gene: 36 T, 42 T, 46 C, 62 G, 66 A, 68 G, 73 T, 84 G, 92 A, 115 G, 129 T, 147 G, 153 A, 200 A, 279 G, 280 C, 286 -, 321 G, 433 C, 445 G, 454 C, 457 A, 458 G, 480 C, 485 T, 533 A, 559 -, 561 G, 563 G, 596 A, 597 T, 598 T, 599 T, 600 T, 605 A, 617 A, 625 G, 626 G, 627 T, 635 G, 637 T, 641 T, 647 G, 648 T, 651 A, 664 C, 681 G, 683 C, 686 -, 693 -, 699 -, 710 T, 711 -, 726 G, 727 G, 728 G, 730 C, 733 G, 738 T, 748 C, 751 G, 753 C, 754 T, 755 A, 759 G, 767 T, 768 T, 769 T, 770 T, 790 T, 794 G, 799 C, 800 G, 805 T, 808 G, 822 T, 824 C, 825 A, 834 T, 837 A, 841 G, 842 A, 923 A, 944 C, 960 T, 989 G, 1081 A, 1104 C, 1105 T, 1109 T, 1114 G, 1122 G, 1139 G, 1144 C, 1146 G, 1149 A, 1159 A, 1164 A, 1176 C, and 1190 G.
Voucher Material
A DNA sample of a voucher specimen has been deposited in Natural History Museum, Vajanského nábrežie 2, 810 06 Bratislava, Slovakia (ID Collection Code 01427588). The voucher originated from the Malá Vydrica stream in the Kačínska dolina valley at the locality Železná studnička, Bratislava (48°12′05.9″N, 17°04′34.7″E).
Gene Sequences
The 16S and 18S rRNA gene sequences of the voucher specimen have been deposited in GenBank under the following accession nos. OL752528 and OL752480, respectively.
Data Availability Statement
The data presented in the study are deposited in the GenBank database (https://www.ncbi.nlm.nih.gov/nucleotide/), accession numbers OL752480 to OL752575. Results of all analyses are included in this published article and Supplementary Material. GenBank accession numbers of sequences used in phylogenetic analyses can be found in the Supplementary Material.
Author Contributions
PV conceptualized the research. MR and TZ performed the laboratory work. PV and MR prepared and analyzed the data, created visualizations, and wrote the original draft of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Slovak Research and Development Agency under contract No. APVV-19-0076, by the Grant Agency of the Ministry of Education, Science, Research and Sport of the Slovak Republic and Slovak Academy of Sciences under the Grant VEGA 1/0013/21, as well as by the Comenius University in Bratislava under the Grants UK/28/2019 and UK/160/2020.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are grateful to Ján Kočišek, Erik Mravec, Ivan Rúrik, Martin Sečanský, Eva Tirjaková, and Michal Veselický, for their help with sampling, and to Ivan Rúrik for his help with some bioinformatics tools.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.830951/full#supplementary-material
Footnotes
1. ^http://apollo.chemistry.gatech.edu/RibosomeGallery
2. ^http://rna.urmc.rochester.edu/RNAstructureWeb
4. ^http://rnacomposer.cs.put.poznan.pl
5. ^http://rnapdbee.cs.put.poznan.pl
6. ^http://mafft.cbrc.jp/alignment/server/
7. ^http://molevol.cmima.csic.es/castresana/Gblocks_server.html
References
André, E. (1909). Sur un nouvel infusoire parasite des Dendrocoeles (Ophryoglena parasitica n. sp.). Rev. Suisse Zool. 17, 273–280. doi: 10.5962/bhl.part.3775
Antczak, M., Zok, T., Popenda, M., Lukasiak, P., Adamiak, R. W., Blazewicz, J., et al. (2014). RNApdbee—a webserver to derive secondary structures from pdb files of knotted and unknotted RNAs. Nucleic Acids Res. 42, W368–W372. doi: 10.1093/nar/gku330
Antipa, G. A., Strüder-Kypke, M. C., and Lynn, D. H. (2020). Molecular phylogeny, taxonomic relationships and north American distribution of Conchophthirus (Conchophthiridae, Scuticociliatia). Aquat. Ecosyst. Health Manag. 23, 58–68. doi: 10.1080/14634988.2020.1735919
Bignell, D. E. (2000). “Introduction to symbiosis,” in Termites: Evolution, Sociality, Symbioses, Ecology. eds T. Abe, D. E. Bignell, and M. Higashi (Dordrecht: Springer), 189–208.
Bowen, I. D., and Ryder, T. A. (1994). Urceolaria mitra (von Seib) epizoic on Polycelis tenuis (Ijima) an SEM study. Cell Biol. Int. 18, 881–888. doi: 10.1006/cbir.1994.1125
Cannone, J. J., Subramanian, S., Schnare, M. N., Collett, J. R., D’Souza, L. M., Du, Y., et al. (2002). The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:e2. doi: 10.1186/1471-2105-3-2
Caron, D. A. (2013). Towards a molecular taxonomy for protists: benefits, risks, and applications in plankton ecology. J. Eukaryot. Microbiol. 60, 407–413. doi: 10.1111/jeu.12044
Cépède, C. (1910). Recherches sur les infusoires astomes. Anatomie, biologie, ethologie parasitaire, systématique. Arch. Zool. Exp. Gén. 5, 341–609.
Clement, M., Posada, D., and Crandall, K. A. (2000). TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x
Corliss, J. O., de Puytorac, P., and Lom, J. (1965). Resolution of persistent taxonomic and nomenclatural problems involving ciliate protozoa assignable to the astome family Haptophryidae Cepede, 1923. J. Protozool. 12, 265–273. doi: 10.1111/j.1550-7408.1965.tb01849.x
Darty, K., Denise, A., and Ponty, Y. (2009). VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics 25, 1974–1975. doi: 10.1093/bioinformatics/btp250
de Puytorac, P. (1957). L’infraciliature de quelques ciliés Haptophryidae. Caparaison avec celle de certains thigmotriches. Compt. Rend. Hebd. Seanc. Acad. Sci. Paris 244, 1962–1965.
de Puytorac, P. (1963). Contribution à l’étude des ciliés astomes Haptophryidae Cépède, 1903 (cytologie, ultrastructure, taxinomie). Ann. Sci. Nat. Zool. Biol. Anim. 5, 173–210.
Dimijian, G. G. (2000a). Evolving together: the biology of symbiosis, part 1. Proc. (Baylor Univ. Med. Cent.) 13, 217–226. doi: 10.1080/08998280.2000.11927678
Dimijian, G. G. (2000b). Evolving together: the biology of symbiosis, part 2. Proc. (Baylor Univ. Med. Cent.) 13, 381–390. doi: 10.1080/08998280.2000.11927712
Doerder, F. P. (2019). Barcodes reveal 48 new species of Tetrahymena, Dexiostoma, and Glaucoma: phylogeny, ecology, and biogeography of new and established species. J. Eukaryot. Microbiol. 66, 182–208. doi: 10.1111/jeu.12642
Ehrenberg, C. G. (1838). Die Infusionsthierchen als vollkommene Organismen. Ein Blick in das tiefere organische Leben der Natur. Leipzig: Verlag von Leopold Voss.
Elias, R., and Hoksza, D. (2017). TRAVeLer: a tool for template-based RNA secondary structure visualization. BMC Bioinformatics 18:e487. doi: 10.1186/s12859-017-1885-4
Georgévitch, J. (1950). Nouvelles recherches sur les infusoires astomes des oligochètes et des triclades du Lac d’Ochrid. Bull. Acad. Serb. Sci. Math. Nat. 1, 63–73.
Greczek-Stachura, M., Rautian, M., and Tarcz, S. (2021). Paramecium bursaria—a complex of five cryptic species: mitochondrial DNA COI haplotype variation and biogeographic distribution. Diversity 13:e589. doi: 10.3390/d13110589
Haider, G. (1964). Monographie der Familie Urceolariidae (Ciliata, Peritricha, Mobilia) mit besonderer Berücksichtigung der in süddeutschen Raum vorkommenden Arten. Parasitol. Schriftenr. 17, 1–251.
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
International Commission on Zoological Nomenclature (1999). International Code of Zoological Nomenclature. 4th Edn. London: International Trust for Zoological Nomenclature.
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Lee, J. C., and Gutell, R. R. (2012). A comparison of the crystal structures of eukaryotic and bacterial SSU ribosomal RNAs reveals common structural features in the hypervariable regions. PLoS One 7:e38203. doi: 10.1371/journal.pone.0038203
Leigh, J. W., and Bryant, D. (2015). POPART: full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210X.12410
Lom, J. (1959). Beiträge zur Kenntnis der parasitischen Ciliaten aus Evertebraten IV. Neue Ciliaten aus der Familie Haptophryidae Cépède 1923, nebst einigen Bemerkungen zum heutigen Stand dieser Gruppe. Arch. Protistenkd. 104, 133–154.
Lücking, R., Leavitt, S. D., and Hawksworth, D. L. (2021). Species in lichen-forming fungi: balancing between conceptual and practical considerations, and between phenotype and phylogenomics. Fungal Divers. 109, 99–154. doi: 10.1007/s13225-021-00477-7
Lynn, D. H. (2008). The Ciliated Protozoa: Characterization, Classification, and Guide to the Literature. 3rd Edn. Dordrecht: Springer.
Lynn, D. H., Gómez-Gutiérrez, J., Strüder-Kypke, M. C., and Shaw, C. T. (2014). Ciliate species diversity and host-parasitoid codiversification in Pseudocollinia infecting krill, with description of Pseudocollinia similis sp. nov. Dis. Aquat. Org. 112, 89–102. doi: 10.3354/dao02796
Mathews, D. H. (2004). Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA 10, 1178–1190. doi: 10.1261/rna.7650904
Moon-van der Staay, S. Y., van der Staay, G. W., Michalowski, T., Jouany, J. P., Pristas, P., Javorský, P., et al. (2014). The symbiotic intestinal ciliates and the evolution of their hosts. Eur. J. Protistol. 50, 166–173. doi: 10.1016/j.ejop.2014.01.004
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Obert, T., Rurik, I., and Vďačný, P. (2021). Diversity and eco-evolutionary associations of endosymbiotic astome ciliates with their lumbricid earthworm hosts. Front. Microbiol. 12:e689987. doi: 10.3389/fmicb.2021.689987
Obert, T., and Vďačný, P. (2019). Integrative taxonomy of five astome ciliates (Ciliophora, Astomatia) isolated from earthworms in Central Europe. Eur. J. Taxon. 559, 1–37. doi: 10.5852/ejt.2019.559
Obert, T., and Vďačný, P. (2020). Delimitation of five astome ciliate species isolated from the digestive tube of three ecologically different groups of lumbricid earthworms, using the internal transcribed spacer region and the hypervariable D1/D2 region of the 28S rRNA gene. BMC Evol. Biol. 20:e37. doi: 10.1186/s12862-020-1601-2
Obert, T., and Vďačný, P. (2021). Multi-gene phylogeny of the subclass Astomatia (Protista: Ciliophora) refreshed with two rare astome ciliates from the digestive tube of endogeic earthworms. Org. Divers. Evol. 21, 59–77. doi: 10.1007/s13127-020-00469-6
Pecina, L., and Vďačný, P. (2020). Morphological versus molecular delimitation of ciliate species: a case study of the family Clevelandellidae (Protista, Ciliophora, Armophorea). Eur. J. Taxon. 697, 1–46. doi: 10.5852/ejt.2020.697
Pecina, L., and Vďačný, P. (2021). DNA barcoding and coalescent-based delimitation of endosymbiotic clevelandellid ciliates (Ciliophora: Clevelandellida): a shift to molecular taxonomy in the inventory of ciliate diversity in panesthiine cockroaches. Zool. J. Linn. Soc. doi: 10.1093/zoolinnean/zlab063 (in press).
Petrov, A. S., Bernier, C. R., Gulen, B., Waterbury, C. C., Hershkovits, E., Hsiao, C., et al. (2014). Secondary structures of rRNAs from all three domains of life. PLoS One 9:e88222. doi: 10.1371/journal.pone.0088222
Popenda, M., Szachniuk, M., Antczak, M., Purzycka, K. J., Lukasiak, P., Bartol, N., et al. (2012). Automated 3D structure composition for large RNAs. Nucleic Acids Res. 40:e112. doi: 10.1093/nar/gks339
Rataj, M., and Vďačný, P. (2018). Dawn of astome ciliates in light of morphology and time-calibrated phylogeny of Haptophrya planariarum, an obligate endosymbiont of freshwater turbellarians. Eur. J. Protistol. 64, 54–71. doi: 10.1016/j.ejop.2018.03.004
Rataj, M., and Vďačný, P. (2019). Living morphology and molecular phylogeny of oligohymenophorean ciliates associated with freshwater turbellarians. Dis. Aquat. Org. 134, 147–166. doi: 10.3354/dao03366
Rataj, M., and Vďačný, P. (2020). Multi-gene phylogeny of Tetrahymena refreshed with three new histophagous species invading freshwater planarians. Parasitol. Res. 119, 1523–1545. doi: 10.1007/s00436-020-06628-0
Rataj, M., and Vďačný, P. (2021). Cryptic host-driven speciation of mobilid ciliates epibiotic on freshwater planarians. Mol. Phylogenet. Evol. 161:e107174. doi: 10.1016/j.ympev.2021.107174
Reynoldson, T. B. (1947). Urceolarian epizoites of flatworms. Nature 160, 265–266. doi: 10.1038/160265a0
Reynoldson, T. B. (1950). Natural population fluctuations of Urceolaria mitra (Protozoa, Peritricha) epizoic on flatworms. J. Anim. Ecol. 19, 106–118. doi: 10.2307/1521
Reynoldson, T. B. (1951). The dispersal of Urceolaria mitra (Peritricha) epizoic on flatworms. J. Anim. Ecol. 20, 123–131. doi: 10.2307/1650
Reynoldson, T. B. (1955). Factors influencing population fluctuation of Urceolaria mitra (Peritricha) epizoic on freshwater triclads. J. Anim. Ecol. 24, 57–83. doi: 10.2307/1879
Reynoldson, T. B. (1956). The population dynamics of host specificity in Urceolaria mitra (Peritricha) epizoic on freshwater triclads. J. Anim. Ecol. 25, 127–143. doi: 10.2307/1854
Reynoldson, T. B. (1981). “The ecology of the turbellaria with special reference to the freshwater triclads,” in The Biology of the Turbellaria. eds. E. R. Schockaert and I. R. Ball (Dordrecht: Springer), 87–90.
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rossolimo, L. L. (1926). Parasitische Infusorien aus dem Baikal-See. Arch. Protistenkd. 54, 468–509.
Rybarczyk, A., Szostak, N., Antczak, M., Zok, T., Popenda, M., Adamiak, R., et al. (2015). New in silico approach to assessing RNA secondary structures with non-canonical base pairs. BMC Bioinformatics 16:e276. doi: 10.1186/s12859-015-0718-6
Sapp, J. (2004). The dynamics of symbiosis: an historical overview. Can. J. Bot. 82, 1046–1056. doi: 10.1139/b04-055
Schultze, M. S. (1851). Beiträge zur Naturgeschichte der Turbellarien. Greifswald: C. A. Koch’s Verlagshandlung.
Seibel, P. N., Müller, T., Dandekar, T., Schultz, J., and Wolf, M. (2006). 4SALE—a tool for synchronous RNA sequence and secondary structure alignment and editing. BMC Bioinformatics 7:e498. doi: 10.1186/1471-2105-7-498
Stein, F. (1859). Der Organismus der Infusionsthiere nach eigenen Forschungen in systematischer Reihenfolge bearbeitet. I. Abtheilung. Allgemeiner Theil und Naturgeschichte der hypotrichen Infusionsthiere. Leipzig: W. Engelmann.
Struck, T. H., Feder, J. L., Bendiksby, M., Birkeland, S., Cerca, J., Gusarov, V. I., et al. (2018). Finding evolutionary processes hidden in cryptic species. Trends Ecol. Evol. 33, 153–163. doi: 10.1016/j.tree.2017.11.007
Sweeney, B. A., Hoksza, D., Nawrocki, E. P., Ribas, C. E., Madeira, F., and Cannone, J. J., et al. (2021). R2DT is a framework for predicting and visualising RNA secondary structure using templates. Nat. Commun. 12:e3494. doi: 10.1038/s41467-021-23555-5
Talavera, G., and Castresana, J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577. doi: 10.1080/10635150701472164
Trifinopoulos, J., Nguyen, L. T., von Haeseler, A., and Minh, B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Vila-Farré, M., and Rink, J. C. (2018). “The ecology of freshwater planarians,” in Planarian Regeneration: Methods and Protocols, Methods in Molecular Biology. Vol. 1774. ed. J. C. Rink (New York, NY: Humana Press), 173–205.
von Siebold, C. T. (1839). Beiträge zur Naturgeschichte der Wirbellosen Thiere. N. Schrift. Naturf. Ges. Danzig 3, 56–71.
Warren, A., Patterson, D. J., Dunthorn, M., Clamp, J. C., Achilles-Day, U. E. M., Aescht, E., et al. (2017). Beyond the ‘Code’: a guide to the description and documentation of biodiversity in ciliate protists (Alveolata, Ciliophora). J. Eukaryot. Microbiol. 64, 539–554. doi: 10.1111/jeu.12391
Wilbert, N. (1975). Eine verbesserte Technik der Protargolimprägnation für Ciliaten. Mikrokosmos 6, 171–179.
Wolf, M., Friedrich, J., Dandekar, T., and Müller, T. (2005). CBCAnalyzer: inferring phylogenies based on compensatory base changes in RNA secondary structures. In Silico Biol. 5, 291–294.
Wright, J. F. (1981). Tetrahymena pyriformis (Ehrenberg) and T. corlissi Thompson parasitic in stream-dwelling triclads (Platyhelminthes: Turbellaria). J. Parasitol. 67, 131–133. doi: 10.2307/3280799
Zhang, T., Fan, X., Gao, F., Al-Farraj, S. A., El-Serehy, H. A., and Song, W. (2019). Further analyses on the phylogeny of the subclass Scuticociliatia (Protozoa, Ciliophora) based on both nuclear and mitochondrial data. Mol. Phylogenet. Evol. 139:e106565. doi: 10.1016/j.ympev.2019.106565
Zhang, T., and Vďačný, P. (2021). A discovery of two new Tetrahymena species parasitizing slugs and mussels: morphology and multi-gene phylogeny of T. foissneri sp. n. and T. unionis sp. n. Parasitol. Res. 120, 2595–2616. doi: 10.1007/s00436-021-07152-5
Zhang, T., and Vďačný, P. (2022). Multiple independent losses of cell mouth in phylogenetically distant endosymbiotic lineages of oligohymenophorean ciliates: a lesson from Clausilocola. Mol. Phylogenet. Evol. 166:e107310. doi: 10.1016/j.ympev.2021.107310
Zok, T., Antczak, M., Zurkowski, M., Popenda, M., Blazewicz, J., Adamiak, R. W., et al. (2018). RNApdbee 2.0: multifunctional tool for RNA structure annotation. Nucleic Acids Res. 46, W30–W35. doi: 10.1093/nar/gky314
Keywords: 16S and 18S rRNA genes, compensatory base changes, cryptic species, endosymbionts, Scuticociliatia, secondary structure
Citation: Rataj M, Zhang T and Vd’ačný P (2022) Nuclear and Mitochondrial SSU rRNA Genes Reveal Hidden Diversity of Haptophrya Endosymbionts in Freshwater Planarians and Challenge Their Traditional Classification in Astomatia. Front. Microbiol. 13:830951. doi: 10.3389/fmicb.2022.830951
Edited by:
Bettina Sonntag, University of Innsbruck, AustriaReviewed by:
Diogo Antonio Tschoeke, Federal University of Rio de Janeiro, BrazilMichaela Strueder-Kypke, University of Guelph, Canada
Copyright © 2022 Rataj, Zhang and Vďačný. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peter Vd’ačný, cGV0ZXIudmRhY255QHVuaWJhLnNr
 Matej Rataj
Matej Rataj Tengyue Zhang
Tengyue Zhang Peter Vd’ačný
Peter Vd’ačný