- 1State Key Laboratory of Animal Nutrition, Institute of Animal Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Laboratory of Quality and Safety Risk Assessment for Dairy Products of Ministry of Agriculture and Rural Affairs, Institute of Animal Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
- 3Key Laboratory of Quality and Safety Control for Milk and Dairy Products of Ministry of Agriculture and Rural Affairs, Institute of Animal Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
- 4State Key Laboratory of Membrane Biology, Tsinghua University-Peking University Joint Center for Life Sciences, School of Life Sciences, Tsinghua University, Beijing, China
The presence of pathogenic bacteria and antibiotic resistance genes (ARGs) in milk are among the most important issues related to the safety of dairy products and the health of consumers. However, despite that dairy cow are housed for long periods of time on different beddings, the effect of different bedding materials on the microbiota and presence of ARGs is unclear. In this study, the composition of microorganisms, and the presence of mastitis pathogens and 33 ARGs targeting seven antibiotics in raw milk produced from farms using sand bedding, rice husk bedding, and recycled manure solids (RMS) bedding were compared by amplicon sequencing and real-time quantitative PCR. The results showed that the microbial composition of milk was related to the microbiota of bedding. None of the mastitis pathogens were detected in milk from cows housed on sand bedding (S-M). The proportion of ARGs was highest in the S-M group and lowest in the milk from cows housed on RMS bedding (RMS-M) group. In general, the content of ARGs in RMS-M was the lowest, however, the RMS bedding may pose a threat to the breast health of dairy cows.
Introduction
Milk is rich in nutrients, which not only supports human survival, but is also a good culture medium for microorganisms (Akindolire et al., 2018; Regasa et al., 2019). In addition to pathogenic bacteria, there may also be drug-resistant microorganisms in raw milk (Munsch-Alatossava and Alatossava, 2007; Caudell et al., 2018). Livestock feces can be used as an important potential source of pollution of pathogenic bacteria and antibiotic resistance genes (ARGs) in the environment. The use of antibiotics and epidemic prevention in intensive farming environments cause antibiotic pollution through feces, which further promotes the accumulation and spread of pathogenic bacteria and ARGs in the environment (Zhu et al., 2013; Wang et al., 2017). Multiple drug resistance genes were detected in manure samples, including tetracycline, sulfonamide, tylosin, macrolides, aminoglycosides, vancomycin, and macrolide lincomycin streptomycin B (Hölzel et al., 2010; Zhang et al., 2020).
Previous studies found that aerobic composting was a promising method to control pathogens and antibiotic contamination in feces (Du and Liu, 2012). However, although composting is an effective method to reduce the content of antibiotics and ARGs in animal feces, the risk of environmental exposure to ARGs in composted feces still exists (Selvam et al., 2012; Tien et al., 2017). In addition, sand and rice husk are often used as bedding materials for dairy cows. There is a small amount of microbiota in the sand bedding materials, and it has soft and comfortable performance, which can effectively maintain the breast health of dairy cows (Rowbotham and Ruegg, 2016b). And Rice husk bedding and other organic materials are easier to make pathogenic bacteria survive and grow (Hogan and Smith, 2012). However, the effect of bacteria in bedding on the bacteria and ARGs in raw milk remains unclear.
With increases in the number of dairy farms, there are an increasing number of large-scale pastures and dairy farming communities, and thus, the generation of fecal pollution has also increased greatly (Wang et al., 2018; Li et al., 2019). Fecal pollution not only occurs on the farm, where it provides a reservoir for mosquitoes and flies to breed and transmit disease, but also pollutes the air, soil, and water sources. Fecal pollution also restricts ranch production, pollutes the surrounding environment, endangers residents’ lives, and imparts great pressures on ranch managers.
Reasonable and effective treatment of cow manure and environmental protection has become an important topic in animal husbandry research. Since the 1970s, the United States and European Union countries have started the process of using recycled manure solids (RMS) as bedding material for dairy cows (Timms, 2008; Husfeldt et al., 2012). However, due to the existence of a large number of microorganisms in cow feces, the effect of RMS bedding on microorganisms in raw milk is unclear. The lack of knowledge surrounding the effect of RMS bedding on the microorganisms in raw milk restricts the popularization and application of RMS cushion-processing technologies. Therefore, it is urgent to study the impact of RMS on the microbiota and safety of raw milk compared to traditional bedding materials.
RMS cushions can be placed deep in the cow bed, like sand cushions, to increase the cow bedtime and reduce the occurrence of cow walking and joint injury (Tucker and Weary, 2004). RMS can effectively reduce the cost of pasture bedding and reduce the amount of pollution from cow dung in the environment. Manure-based bedding materials may cause harm to animals and human health, mainly due to the presence of a large number of microorganisms in the manure itself (Jahne et al., 2015). In previous studies, it was found that the processing of feces, such as extrusion, dehydration, fermentation, and drying, the microbial diversity in feces changed significantly, and the microbiota in the final material was significantly different from the original fecal microbiota (Wu et al., 2020). Most of the pathogenic bacteria in feces that cause mastitis are killed during the RMS processing and fermentation process, but some still remain (Wu et al., 2020). Some gram-negative bacteria, spore formers, heat-resistant bacteria, and food spoilage-related microorganisms, such as Pseudomonas, Bacillaceae, and Moraxellaceae, can remain in milk (Sorter et al., 2014; Leach et al., 2015; Rowbotham and Ruegg, 2016b; Wu et al., 2020). Although some studies have pointed out that the type of bedding will not affect the total bacterial count (TBC) in raw milk (Bradley et al., 2018). The effect of the bedding microbiota on milk microbiota is unclear.
The application of RMS bedding can reduce environmental pollution and the overall purchase cost of bedding; however, the impact of bedding on the safety of raw milk is not clear. The purpose of this study was to analyze the effects of different bedding materials on the microbial diversity of milk, as well as the pathogens and ARGs that raw milk contains.
Materials and Methods
Sample Collection
Bedding samples were collected from the same area as described in previous studies; from farms in Hebei, Heilongjiang, and Tianjin [Hebei: sand bedding (S-B); Heilongjiang: rice husk bedding (RH-B); Tianjin: RMS bedding (RMS-B) in healthy cowsheds] in October 2019 (Wu et al., 2020). The number of dairy cows in the three farms is about 5,000. Except for the types of bedding, the feed management and pasture facilities are similar. All pastures use milking parlor equipment to milk cows, and the milking process and method are the same. Bedding samples were collected from the center and four corners of the farms for 3 consecutive days, and five samples collected at the same time were mixed together. Raw milk samples were collected from the milk tanks in farms using rice husk beddings (RH-M), RMS bedding (RMS-M), and sand bedding (S-M) in the morning, noon, and evening after milking for 3 consecutive days. The three samples collected on the same day were mixed together. Samples were collected and stored directly on ice and transported to the laboratory. The samples were stored at −20°C until tested.
DNA Extraction, Amplification, and Sequencing
DNA extraction was performed as described previously (Wu et al., 2020). In brief, a total of 200 mg of samples were mixed with 70% alcohol and PBS and centrifuged to remove impurities. The DNA was extracted using the E.Z.N.ATM Mag-Bind Soil DNA Kit (M5635-02, OMEGA). After DNA quantification using the qubit 2.0 DNA kit, PCR was performed using v3-v4 universal primers 341f (5'-cctacggcgwgcag-3') and 805r (5'-gatachvggggatctatcc-3'). DNA was amplified using a Bio-Rad T 100TM thermal cycler (California, United States). The amplified products were evaluated by 1.5% agar gel electrophoresis. The SanPrep DNA Gel Extraction Kit (SANGON Biotechnology, Shanghai, China) was used to extract and purify the amplified DNA. The Qubit2.0 DNA kit was used to accurately quantify the recovered DNA, and the final sequencing concentration was 20 pmol. The purified DNA products were sequenced on the Illumina MiSeq pe300 sequencing platform (California, United States) at SANGON Biotechnology Co., Ltd. (Shanghai, China).
Bioinformatics
The forward and reverse sequences from the 16S rRNA amplicon gene were introduced into the qiime2-2021.04 system (Hall and Beiko, 2018). After confirming the data quality, dada2 plugin was used to remove the primer and chimeric sequences. After removing low-quality sequences, the sequence was spliced with each sequences. The spliced sequences were matched with 99% certainty to hits in the Greengene database to obtain bacterial taxonomic names. After adjusting all sample feature tables to 14,000 sequences, diversity analyses, including Shannon Diversity Index, Simpson Diversity Index, Chao1, and observed features were performed.
qPCR Analysis of Pathogenic Bacteria and Antimicrobial Resistance Genes
In order to improve the sensitivity of qPCR and improve the quality of the DNA extraction, DNA extraction was performed by referring to previous methods (Mertens et al., 2014). After centrifugation of 1.5 ml of milk at 15,000 rpm for 10 min, the supernatant was removed. Cell degradation and DNA extraction were mediated by protease K and cetyltrimethylammonium bromide for 30 min at 60°C. The mixed solution was extracted and purified following to the manufacturer’s instructions for the DNeasy Blood and Tissue Kit (Qiagen, Germantown, MD, United States; Munsch-Alatossava and Alatossava, 2007). The amount and purity of the extracted DNA were determined by NanoDrop (Thermo Fisher, United States).
The detection of mastitis pathogens in dairy cows was performed as described in the Bovine Mastitis Pathogenic Bacteria Nucleic Acid Typing Detection Kit (Shenzhen Bioeasy Biotechnologies, Co., Ltd.; Wu et al., 2020). Real-time PCR based on TaqMan probes was used to detect the common contact infectious pathogens (Staphylococcus aureus, Streptococcus agalactiae, Mycoplasma bovis, Corynebacterium bovis, Mycoplasma bovis, Mycobacterium bovis, and Mycoplasma spp.), environmental pathogens (i.e., Staphylococcus spp., Escherichia coli, Klebsiella spp., Prototheca spp., Streptococcus dysgalactiae, Streptococcus uberis, Trueperella pyogenes, Serratia marcescens, and yeasts), and β-lactamase resistance genes in milk. Four fluorescence channels, fam, hex, Rox, and Cy5 were used to collect quantitative data.
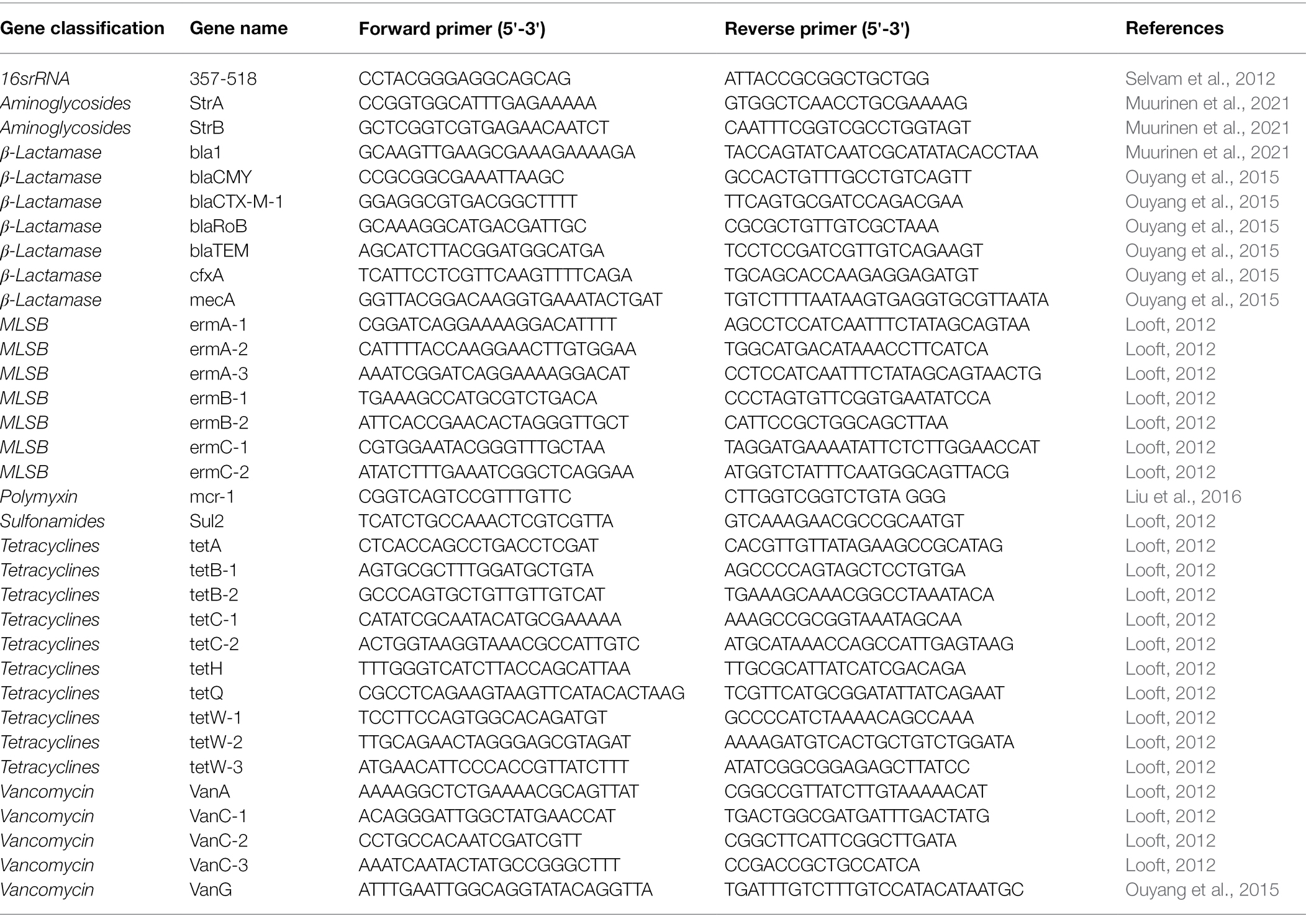
qPCR experiments were performed for ARGs in milk samples using an ABI 7900 HT system. The final reaction volume was 20 μl, including 2 μl of DNA template and 10 μl of 2× RealStar Green Fast Mixture (GeneStar, Beijing, China). The amplification conditions were as follows: after denaturation at 95°C for 3 min, 40 cycles of 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s were performed. The specific sequences of drug resistance genes were amplified and detected. Thirty-three antimicrobial resistance genes, including aminoglycosides (StrA and StrB), beta-lactamases (bla1, blaCMY, blaCTX-M-1, blaRoB, blaTEM, cfxA, and mecA), MLSB (ermA-1, ermA-2, ermA-3, ermB-1, ermB-2, ermC-1, and ermC-2), polymyxin (mcr-1), sulfonamides (Sul2), tetracyclines (tetA, tetB-1, tetB-2, tetC-1, tetC-2, tetH, tetQ, tetW-1, tetW-2, and tetW-3), and vancomycin (VanC-1, VanC-2, VanC-3, and VanG) were detected. The primers used to amplify the antimicrobial resistance genes were the same as those detailed previously (Table 1; Looft, 2012; Ouyang et al., 2015; Muurinen et al., 2021). The resistance genes were standardized to the 16S rRNA gene and quantified by amplification with 16S rRNA primers 357f (5'-cctacggaggcagg-3') and 517r (5'-attaccgcggctggg-3'; Selvam et al., 2012). After amplification, melt curves for all real-time PCR reactions were analyzed to determine the accuracy of the amplified target gene.
Statistical Analysis
Prism 8 was used to analyze the relative differences in ARGs between groups. One-way ANOVA was used to analyze the intergroup differences in the α-diversity index, ARGs, and bacterial communities. Canonical correspondence analysis (CCA) was performed using pass 4.02 software. Spearman correlation coefficients and heatmaps were performed between the bacterial communities and ARGs in milk, the microbiota in bedding and milk using Omicshare tools, a free online data analysis platform.1
Results
Milk Has Stable Bacterial α-Diversities
In the milk from cows housed on RMS bedding, rice husks, and sand bedding, an average of 19,233, 51,110, and 52,553 reads were obtained, respectively. Table 2 summarizes the results of the bacterial α-diversity indexes in bedding samples and milk samples. There was no significant difference in the Chao 1 index and observed features of milk bacteria under different bedding environments (p > 0.05). The Shannon index for microorganisms in the RMS-B group was significantly higher than that in RH-B and S-B (p < 0.05), while the Shannon index for milk bacteria was not different between the three bedding types (p > 0.05). In addition, the evenness index (Simpson index) of microbiota were no significant differences in the milk among the different bedding environments. These results shows that different bedding environments did not affect the diversity and evenness of the bacteria in milk.
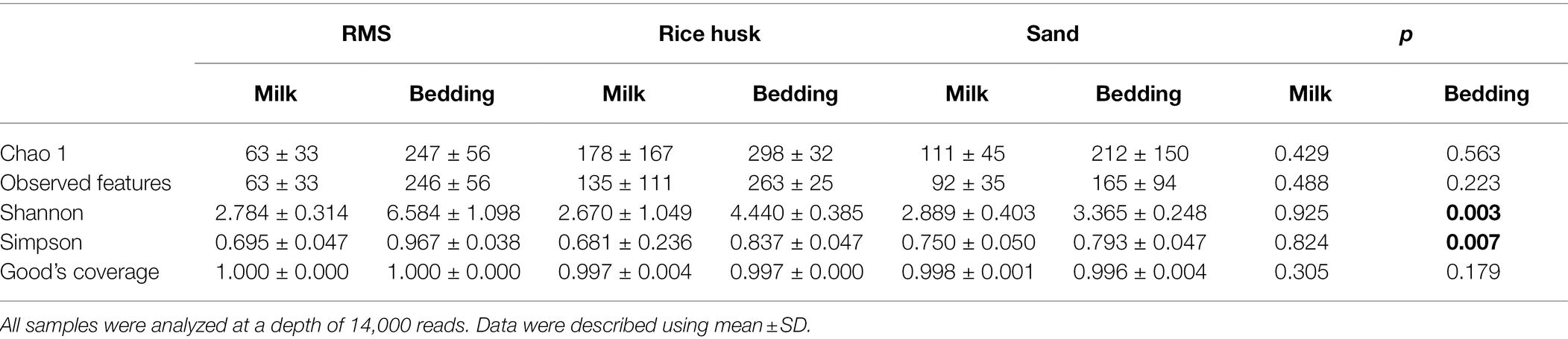
Table 2. One-way ANOVA of bacterial α-diversities of the microbiota in milk and bedding from different farms.
Microbiota Were Different in Milk From Cows Housed Under Different Bedding Conditions
The main bacterial phyla and families in milk samples and bedding samples were sorted and are summarized in Table 3 (family proportion >1%). There were significant differences in the microbiota of milk from farms using different bedding conditions. The proportion of Firmicutes in milk from the RMS-M group and S-M group was significantly higher than that of milk from the RH-M group (p = 0.001). Among those bacteria, Streptococcaceae and Staphylococcaceae were observed in a high proportion in the RMS-M and S-M groups (proportion >10%), and a high proportion of Enterococcaceae was found in the RMS-M group (proportion >10%). Conversely, the proportion of Proteobacteria in the RH-M group was the highest, and the proportion of Enterobacteriaceae was the highest (proportion >80%), being much higher than that of the RMS-M and S-M groups (p < 0.001). Although the value of p of One-way ANOVA analysis result was 0.052, the average proportion of Moraxellaceae in the RMS-M and S-M groups was as high as 35.0% and 34.2%, respectively, while the average proportion in the RH-M group was only 3.4%. The average content of Pseudomonadaceae in the S-M group was 16.0%, while that in the RH-M group was 4.9% and that in the RMS-M group was 1.0%.
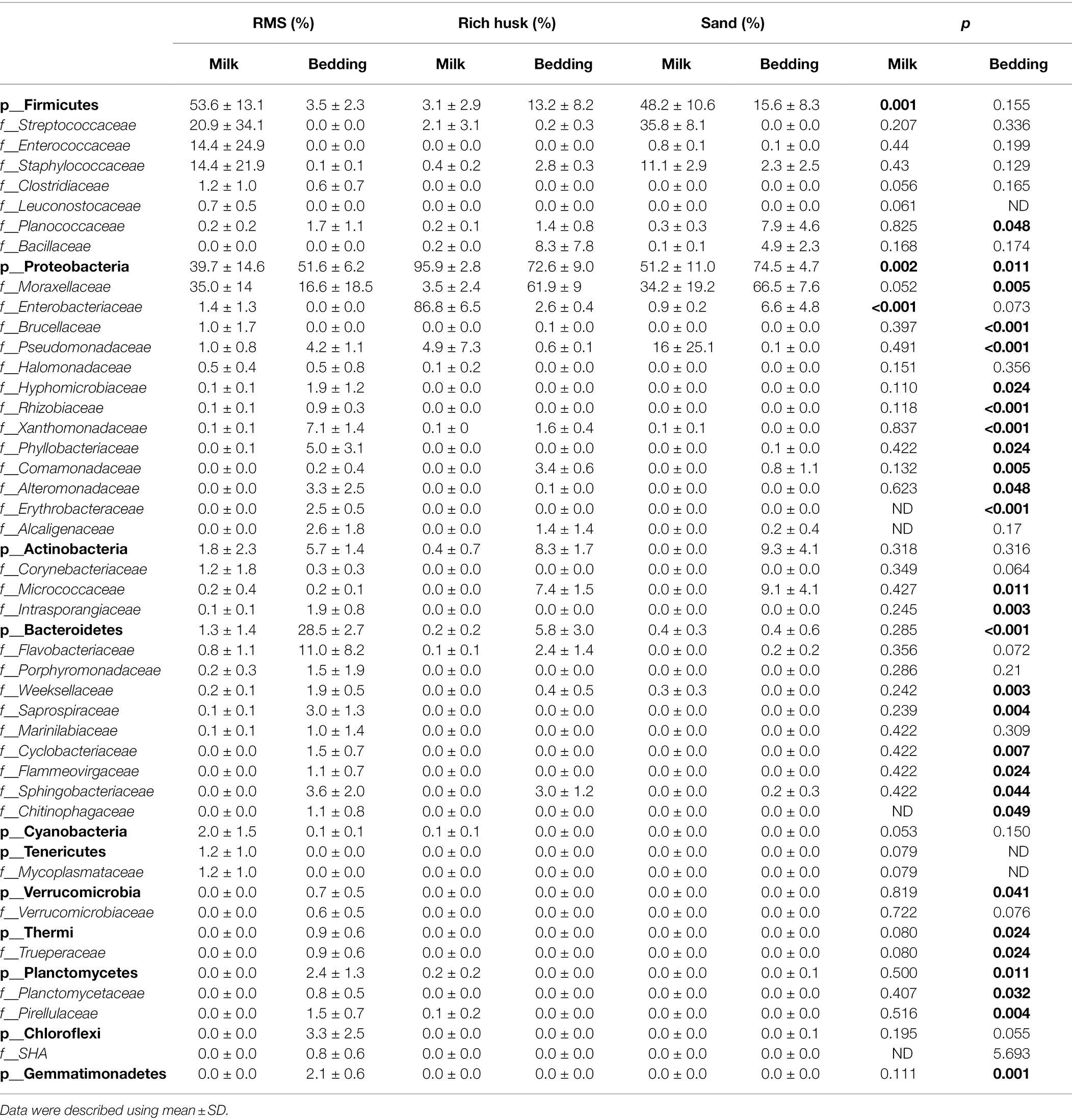
Table 3. Relative read abundance of different bacterial community structures at the phylum and family levels in different groups.
Bedding Bacteria Will Not Contribute to the Main Microbiota in Milk
Following Spearman correlation analyses, the proportion of microorganisms in milk and bedding was summarized and transformed into a heatmap (Figure 1). The Streptococcaceae, Staphylococcaceae, Enterococcaceae, Enterobacteriaceae, and Moraxellaceae in milk samples were higher (proportion > 0.05%), but there was no significant correlation with the contents of the same bacterial family in bedding samples. Cyanobacteria, Tenericutes, and Thermophiles, were only found in the RMS-M and RMS-B groups (proportion > 0.05%), had a positive correlation with the same phyla in bedding samples (p < 0.05). Clostridiaceae, Halomonadaceae, Intraporangiaceae, Porphyromonadaceae, Saprospiraceae, Marinilabiaceae, and Trueperaceae were only found in the RMS-M and RMS-B groups (proportion > 0.05%), and the proportion of these family in bedding samples were positively correlated (p < 0.05). In addition, Bacillaceae were only detected in a large proportion of the RH-B (8.3% ± 7.8%) and S-B groups (4.9% ± 2.3%) and were detected in milk samples from the same farm. Thus, there was a positive correlation between the presence of Bacillaceae in milk samples and bedding samples (p < 0.05).
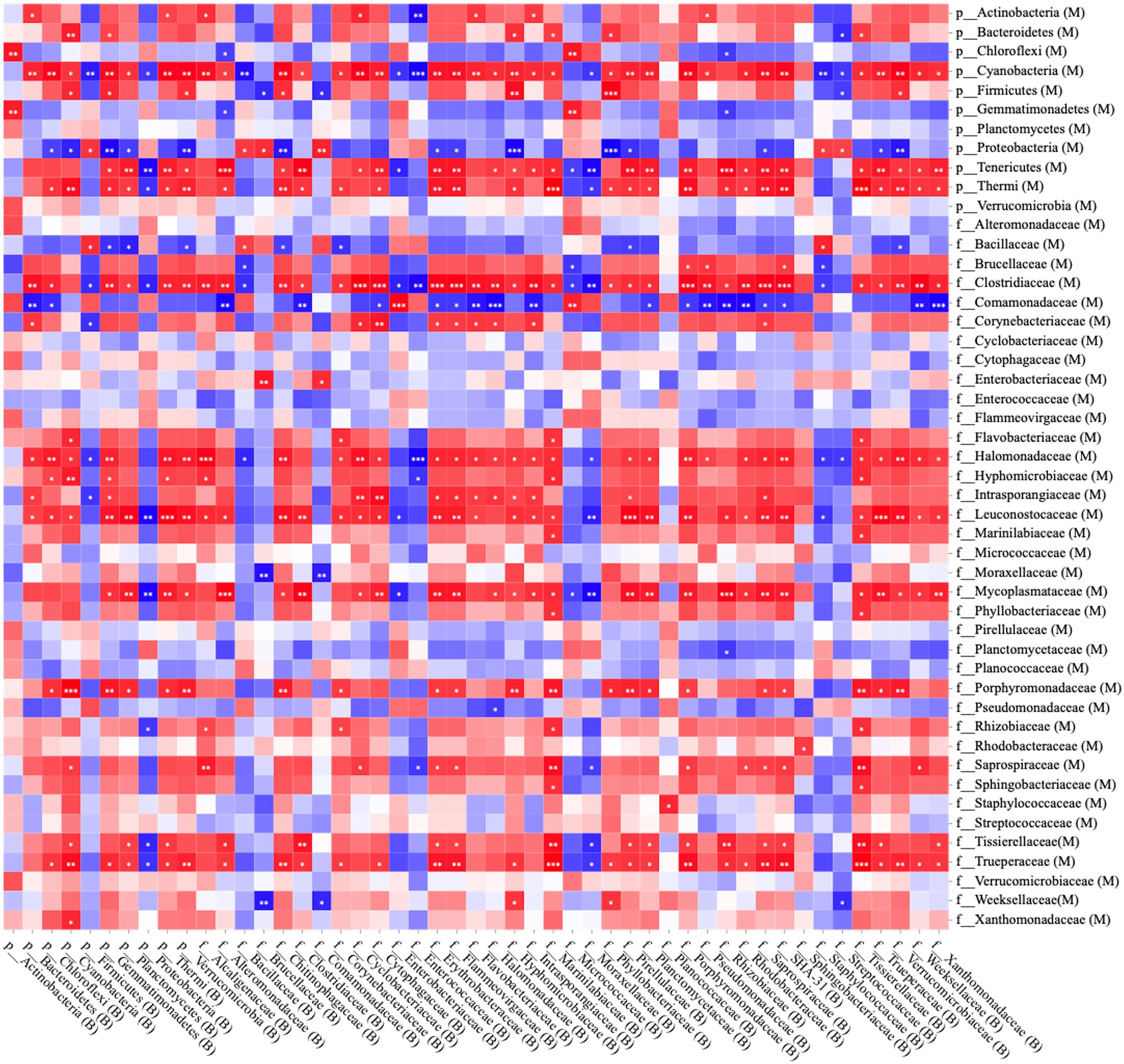
Figure 1. Spearman correlation analysis between bedding bacteria and milk bacteria at the phylum and family levels; red squares: positive correlation; blue squares: negative correlation; *p < 0.05; **p < 0.01; and ***p ≤ 0.001.
Detection of Pathogenic Bacteria in Milk
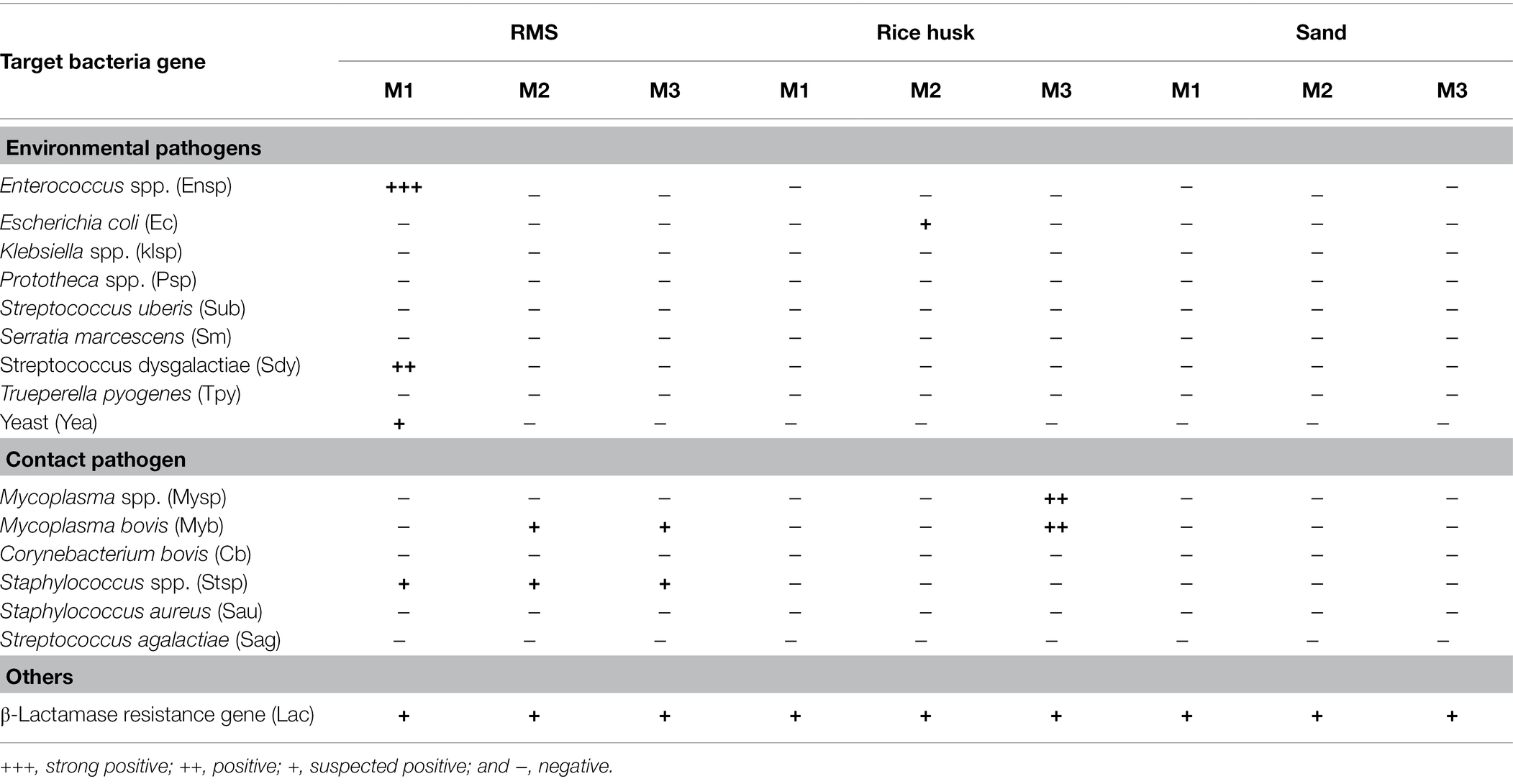
The qPCR results of pathogenic microorganisms in milk are summarized in Table 4. Raw milk in milk tanks after milking in the morning, noon, and evening in the farms where different bedding materials were used for 3 consecutive days. The raw milk of the same day was mixed together. Regarding environmental pathogens, it was found that 1/3 of the RMS-M samples had strong positive results of Enterococcus spp. (+++), positive results of Streptococcus dysgalatiae (++), and weak positive results of yeast (+). A weak positive result (+) for Escherichia coli was detected in 1/3 of RH-M samples. qPCR of contact pathogens revealed that Staphylococcus spp. were found in all RMS-M samples, while Mycoplasma bovis was weakly positive (+) in 2/3 RMS-M samples. Samples that were positive (+) for Mycoplasma spp. and Mycoplasma bovis were detected in 1/3 of RH-M samples. A weak positive (+) result was detected for β-lactamase genes in all milk samples. No environmental pathogens or contact pathogens were detected in S-M samples.
The Proportion of ARGs in the RMS-M Group Was the Lowest
In this study, 33 ARGs targeting seven drugs were detected and analyzed (Figure 2). The proportion of ARGs targeting aminoglycosides in all milk samples was lower than.5%, with no significant differences between groups (p > 0.05). The content of the mcr-1 gene targeting polymyxin in milk samples was also lower than 0.005, however, its proportion in the S-M group was higher than that in the RMS-M group (p < 0.05). Similarly, the results revealed that in the S-M group, the proportion of ARGs genes targeting β-lactamases, MLSBs, sulfonamides, tetracyclines, and vancomycin was higher than that in RMS-M group (p < 0.05). There were also ARGs in the S-M group, including those targeting β-lactamases (mecA and blaRoB), MLSBs (emrA-2 and emrB-1), sulfonamides (sul2), tetracyclines (tetW-1 and tetW-2), and vancomycin (vanC-2), there were more of them in S-M than RH-M (p < 0.05). The proportion of MLSBs (ermB-2) and tetracyclines (tetB-2 and tetW-1) in the RH-M group was also significantly higher than that in the RMS-M group (p < 0.05).
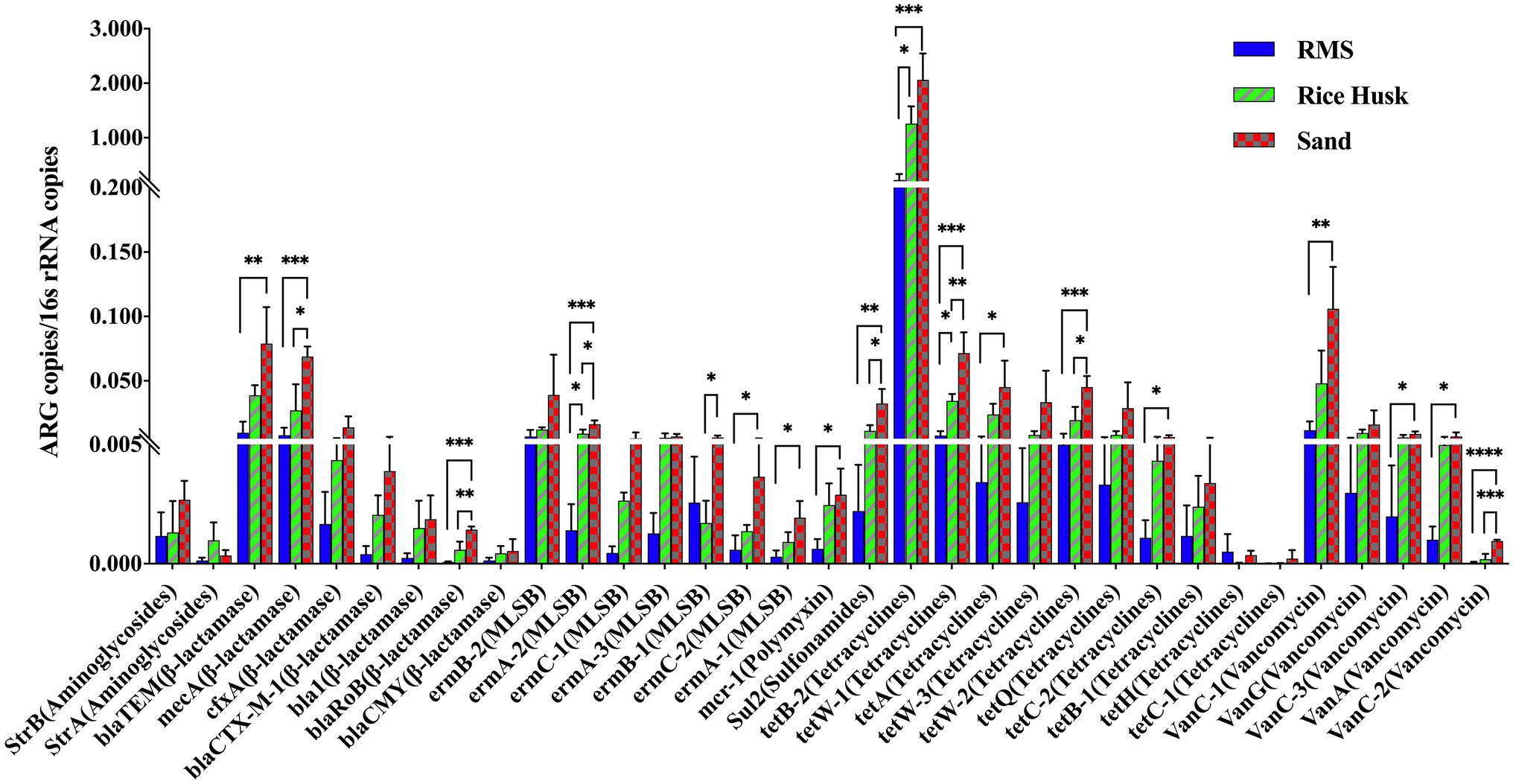
Figure 2. Detection proportion of ARGs detected in raw milk from different beddings. Data were described using mean ± SD. *p < 0.05; **p < 0.01; ***p ≤ 0.005; and ****p ≤ 0.001.
Relationship Between Milk Bacteria and ARGs
The clustering relationship between the main microbiota of milk using different bedding environments was analyzed by CCA. At the same time, the relationship between the main microbiota and ARGs content in milk was analyzed by Spearman correlation analysis (Figure 3). The CCA results showed that the microbiota in the RH-M group was low similarity from that of the other two groups due to the high proportion of Enterobacteriaceae. From the heatmap of the correlation analysis, it was observed that Enterococcaceae in milk samples was positively correlated with ARGs (ermb-1) targeting MLSBs (p < 0.05). Additionally, the proportion of Pseudomonadaceae was positively correlated with ARGs (cfxa) targeting lactamases and vancomycin (vanc-3; p < 0.05), and the presence of Staphylococcaceae was positively correlated with ARGs targeting tetracyclines (p < 0.01). Cyanobacteria and Tenericutes had a significant negative correlation with multiple ARGs. Cyanobacteria and Tenericutes were only detected in RMS bedding (proportion >1%), and the proportion of these phyla in bedding was positively correlated with that in milk (Figure 3).
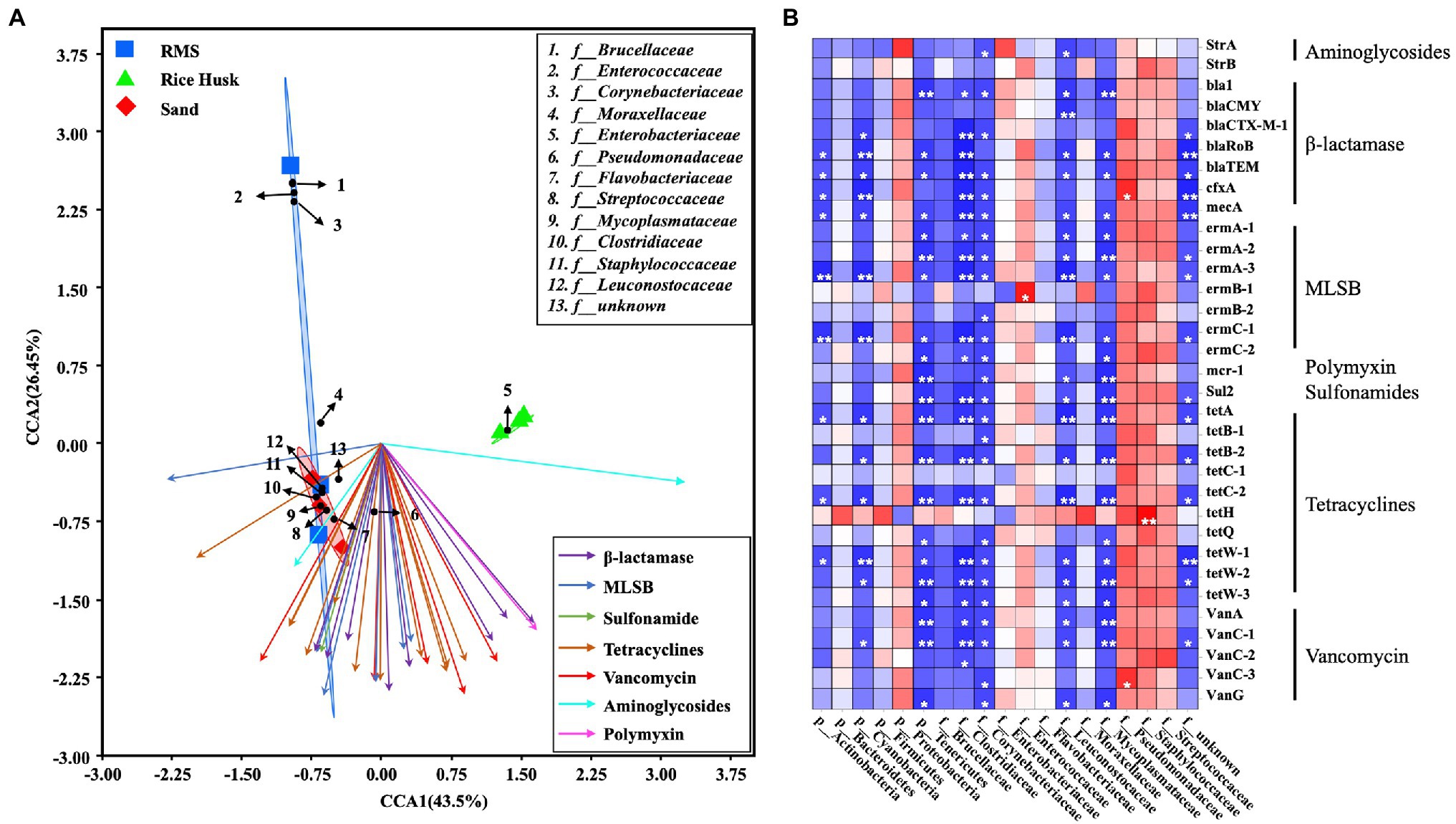
Figure 3. Combined analysis of ARGs and bacterial communities in milk. (A) Canonical correspondence analysis (CCA) of ARGs and bacterial family communities in milk. (B) Spearman correlation analysis between ARGs and milk bacteria at the phylum and family levels. Red squares: positive correlation; blue squares: negative correlation; *p < 0.05; **p < 0.01; and ***p ≤ 0.001.
Discussion
In this study, the contents of microbiota, pathogenic microorganisms, and ARGs in milk and RMS, rice husk, and sand bedding were collected and compared. As the surface where cow breasts contact for substantial periods of time, bedding materials and microorganisms in bedding, have a far-reaching impact on cow health and milk quality (Robles et al., 2020). Previous studies have confirmed that increases in the TBC in bedding are related to the TBC in the dairy cows udder skin (Rowbotham and Ruegg, 2016b). Microorganisms in beddings can pollute raw milk, thus, affecting its quality (Murphy et al., 2019). Some studies also pointed out that there was no significant difference in TBC content in milk under different bedding environments (Bradley et al., 2018). However, the effect of bedding microbiota on the milk microbiota is unclear.
In this study, it was found that there were a large number of Enterobacteriaceae (proportion >1%) in milk (RH-M) and bedding (RH-B) from farms using rice husk bedding. The use of sawdust bedding may be related to the animal epidemiology of coliform mastitis (Carroll and Jasper, 1980). In addition, a large number of Streptococcaceae, Staphylococcaceae, and Enterococcaceae were found in RMS-M group samples (proportion >10%). Previous studies also found that RMS bedding may increase the prevalence of Streptococcus thermophilus and Enterococcus in raw milk (Rowbotham and Ruegg, 2016a; Gagnon et al., 2020). Based on correlation analyses between bedding and milk microbiota, the presence of Bacillaceae in milk may be from the bedding (Figure 1). Those findings were also consistent with those found in previous studies (Driehuis et al., 2013; Skeie et al., 2019). It should be noted that the proportion of Moraxellaceae in all bedding and milk samples was greater than 1%. Previous studies also found that environmental Moraxellaceae may contaminate milk. Moraxellaceae can reproduce at low temperatures, secreting proteases, and lipases to alter the taste and odor of milk protein, thus, shortening the shelf life of milk and reducing its quality (Wu et al., 2019).
Sand cushion is considered to be the most ideal bedding material (Rowbotham and Ruegg, 2016a). The results of this study also revealed that no pathogenic bacteria were detected in the milk (S-M) produced from cows housed on sand bedding. Conversely, some environmental pathogens and contact pathogens were detected in RMS-M group, however.
Raw milk that has not been sterilized is not allowed to be sold in many countries. After the sterilization of raw milk in factories, pathogenic bacteria are killed. Of course, this does not mean that TBC in raw milk has no effect on the product. Compared with the threat of pathogenic bacteria to consumers, the drug resistance genes remaining in milk after processing may be more threatening. The prevalence of antibiotic resistance (AMR) continues to pose a global threat to human health (Ferroni et al., 2020). A recent study found that the direct transmission of microorganisms in raw milk is the main factor influencing the prevalence of AMR (Caudell et al., 2018), and raw milk plays an important role in maintaining the repository and spread of ARGs (Liu et al., 2020). In this study, S-M contained a higher proportion of ARGs than RMS-M (p < 0.05). The ARGs detected in the S-M group, including those targeting β-lactamases, MLSBs, sulfonamides, tetracyclines, and vancomycin, were significantly higher than those in RMS-M group samples. Some high level of ARGs may be caused by the antibiotic drugs used on farms, but it is unlikely that so many antibiotic drugs are used at the same time.
Low moisture content in sand bedding material can reduce the diversity and concentration of microbial species. Previous studies found that microorganisms evolve faster under harsh living environment conditions (Li et al., 2014). In addition, studies have also shown that extreme living environment can increase microbial drug resistance (Patel and Amaresan, 2014). Therefore, we hypothesize that the use of sand bedding may increase the prevalence of drug-resistant bacteria and affect the prevalence of drug resistance genes in raw milk.
Correlation analyses between the microbial composition of milk and the presence of ARGs found that Pseudomonadaceae had a significant positive correlation with some ARGs targeting β-lactamases and vancomycin, while a positive correlation was also found between Enterococcaceae and MLSBs, and Staphylococcaceae and tetracyclines. No positive correlations were found between other families and ARGs. In other studies, it was also found that Pseudomonadaceae, Enterococcaceae, and Staphylococcaceae have significant drug resistance and may have adverse effects on consumers (Morandi et al., 2006; Quintieri et al., 2019; Machado et al., 2020). In this study, we found that the changes of these bacteria in milk may be related to the bedding material. Therefore, monitoring the content of individual microorganisms alone may not be sufficient to monitor ARGs in raw milk. This study also had some deficiencies. The joint analysis of influencing milk bacterial factors, such as bedding type with season and service time, need to be further studied.
Conclusion
In conclusion, this study compared the microbial composition of milk produced by cows housed on RMS bedding, sand bedding, and rice husk bedding. Microbial composition in cow milk under different bedding environments was significant differences. Pathogenic bacteria known to be involved in mastitis were found in the milk from cows housed on RMS bedding and rice husk bedding, but the ARGs in milk from cows housed on RMS bedding were significantly lower than those in milk from cows housed on sand bedding. These findings suggest that although RMS bedding may lead to the contamination of pathogenic microorganisms in milk, they may play a positive role in reducing the presence of ARGs in milk and, thus, protect the health of consumers.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author Contributions
HW and YW: conceptualization, methodology, investigation, and manuscript writing, review, and editing. HL and BD: formal analysis. LD, HH, and LM: resources. NZ: data curation. JW: supervision and project administration. JW and HW: funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the China Postdoctoral Science Foundation (grant number 2020TQ0357), the Scientific Research Project for Major Achievements of Agricultural Science and Technology Innovation Program (grant number CAAS-ZDXT2019004), the Agricultural Science and Technology Innovation Program (grant number ASTIP-IAS12), and the China Agriculture Research System of MOF and MARA.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Akindolire, M. A., Kumar, A., and Ateba, C. N. (2018). Genetic characterization of antibiotic-resistant Staphylococcus aureus from milk in the North-West Province, South Africa. Saudi J. Biol. Sci. 25, 1348–1355. doi: 10.1016/j.sjbs.2015.10.011
Bradley, A. J., Leach, K. A., Green, M. J., Gibbons, J., Ohnstad, I. C., Black, D. H., et al. (2018). The impact of dairy cows’ bedding material and its microbial content on the quality and safety of milk: a cross sectional study of UK farms. Int. J. Food Microbiol. 269, 36–45. doi: 10.1016/j.ijfoodmicro.2017.12.022
Carroll, E. J., and Jasper, D. E. (1980). “Coliform populations in bedding materials and coliform mastitis incidence.” in Annual Meeting, National Mastitis Council, Inc. February 18–20, 1980; 129–139.
Caudell, M. A., Mair, C., Subbiah, M., Matthews, L., Quinlan, R. J., Quinlan, M. B., et al. (2018). Identification of risk factors associated with carriage of resistant Escherichia coli in three culturally diverse ethnic groups in Tanzania: a biological and socioeconomic analysis. Lancet Planet. Health 2, e489–e497. doi: 10.1016/S2542-5196(18)30225-0
Driehuis, F., Lucas-van den Bos, E., and Wells-Bennik, M. H. J. (2013). Risico’s van het gebruik van gescheiden mest as beddinmateriaal voor de melkkwaliteit: sporen van Bacillus cereus en boterzuurbacteriën. (Risks of the use of cattle manure solids as bedding material for milk quality: Bacillus cereus and butyric acid bacteria). NIZORapport E2013/180 pp. 3.
Du, L., and Liu, W. (2012). Occurrence, fate, and ecotoxicity of antibiotics in agro-ecosystems. A review. Agron. Sustain. Dev. 32, 309–327. doi: 10.1007/s13593-011-0062-9
Ferroni, L., Lovito, C., Scoccia, E., Dalmonte, G., Sargenti, M., Pezzotti, G., et al. (2020). Antibiotic consumption on dairy and beef cattle farms of Central Italy based on paper registers. Antibiotics 9. doi: 10.3390/antibiotics9050273
Gagnon, M., Hamelin, L., Fréchette, A., Dufour, S., and Roy, D. (2020). Effect of recycled manure solids as bedding on bulk tank milk and implications for cheese microbiological quality. J. Dairy Sci. 103, 128–140. doi: 10.3168/jds.2019-16812
Hall, M., and Beiko, R. G. (2018). 16S rRNA gene analysis with QIIME2. Methods Mol. Biol. 1849, 113–129. doi: 10.1007/978-1-4939-8728-3_8
Hogan, J., and Smith, K. L. (2012). Managing environmental mastitis. Vet. Clin. Food Anim. Pract. 28, 217–224. doi: 10.1016/j.cvfa.2012.03.009
Hölzel, C. S., Schwaiger, K., Harms, K., Küchenhoff, H., Kunz, A., Meyer, K., et al. (2010). Sewage sludge and liquid pig manure as possible sources of antibiotic resistant bacteria. Environ. Res. 110, 318–326. doi: 10.1016/j.envres.2010.02.009
Husfeldt, A. W., Endres, M. I., Salfer, J. A., and Janni, K. A. (2012). Management and characteristics of recycled manure solids used for bedding in Midwest freestall dairy herds. J. Dairy Sci. 95, 2195–2203. doi: 10.3168/jds.2011-5105
Jahne, M. A., Rogers, S. W., Holsen, T. M., Grimberg, S. J., and Ramler, I. P. (2015). Emission and dispersion of bioaerosols from dairy manure application sites: human health risk assessment. Environ. Sci. Technol. 49, 9842–9849. doi: 10.1021/acs.est.5b01981
Leach, K. A., Archer, S. C., Breen, J. E., Green, M. J., Ohnstad, I. C., Tuer, S., et al. (2015). Recycling manure as cow bedding: potential benefits and risks for UK dairy farms. Vet. J. 206, 123–130. doi: 10.1016/j.tvjl.2015.08.013
Li, S. J., Hua, Z. S., Huang, L. N., Li, J., Shi, S. H., Chen, L. X., et al. (2014). Microbial communities evolve faster in extreme environments. Sci. Rep. 4, 1–9. doi: 10.1038/srep06205
Li, X., Shi, X. S., Lu, M. Y., Zhao, Y. Z., Li, X., Peng, H., et al. (2019). Succession of the bacterial community and functional characteristics during continuous thermophilic composting of dairy manure amended with recycled ceramsite. Bioresour. Technol. 294:122044. doi: 10.1016/j.biortech.2019.122044
Liu, J., Liu, J., Zhu, Y., Zhu, Y., Jay-Russell, M., Lemay, D. G., et al. (2020). Reservoirs of antimicrobial resistance genes in retail raw milk. Microbiome 8:99. doi: 10.1186/s40168-020-00861-6
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Looft, T. P. (2012). The Swine Intestinal Microbiota: Localized Adaptations and Responses to In-Feed Antibiotics. Dissertation/Thesis. ProQuest Dissertations Publishing, 194.
Machado, M. A. A., Ribeiro, W. A., Toledo, V. S., Ramos, G. L. P. A., Vigoder, H. C., and Nascimento, J. S. (2020). Antibiotic resistance and biofilm production in catalase-positive gram-positive cocci isolated from brazilian pasteurized milk. J. Food Qual. Hazards Control 7, 67–74. doi: 10.18502/JFQHC.7.2.2886
Mertens, K., Freund, L., Schmoock, G., Hänsel, C., Melzer, F., and Elschner, M. C. (2014). Comparative evaluation of eleven commercial DNA extraction kits for real-time PCR detection of Bacillus anthracis spores in spiked dairy samples. Int. J. Food Microbiol. 170, 29–37. doi: 10.1016/j.ijfoodmicro.2013.10.022
Morandi, S., Brasca, M., Andrighetto, C., Lombardi, A., and Lodi, R. (2006). Technological and molecular characterisation of enterococci isolated from north-west Italian dairy products. Int. Dairy J. 16, 867–875. doi: 10.1016/j.idairyj.2005.09.005
Munsch-Alatossava, P., and Alatossava, T. (2007). Antibiotic resistance of raw-milk-associated psychrotrophic bacteria. Microbiol. Res. 162, 115–123. doi: 10.1016/j.micres.2006.01.015
Murphy, S. I., Kent, D., Martin, N. H., Evanowski, R. L., Patel, K., Godden, S. M., et al. (2019). Bedding and bedding management practices are associated with mesophilic and thermophilic spore levels in bulk tank raw milk. J. Dairy Sci. 102, 6885–6900. doi: 10.3168/jds.2018-16022
Muurinen, J., Richert, J., Wickware, C. L., Richert, B., and Johnson, T. A. (2021). Swine growth promotion with antibiotics or alternatives can increase antibiotic resistance gene mobility potential. Sci. Rep. 11, 5485–5413. doi: 10.1038/s41598-021-84759-9
Ouyang, W.-Y., Huang, F.-Y., Zhao, Y., Li, H., and Su, J.-Q. (2015). Increased levels of antibiotic resistance in urban stream of Jiulongjiang River, China. Appl. Microbiol. Biotechnol. 99, 5697–5707. doi: 10.1007/s00253-015-6416-5
Patel, K., and Amaresan, N. (2014). Antimicrobials compounds from extreme environment rhizosphere organisms for plant growth. Int. J. Curr. Microbiol. App. Sci 3, 651–664.
Quintieri, L., Fanelli, F., and Caputo, L. (2019). Antibiotic resistant Pseudomonas spp. spoilers in fresh dairy products: an underestimated risk and the control strategies. Foods 8, 1–32. doi: 10.3390/foods8090372
Regasa, S., Mengistu, S., and Abraha, A. (2019). Milk safety assessment, isolation, and antimicrobial susceptibility profile of Staphylococcus aureus in selected dairy farms of Mukaturi and Sululta town, Oromia region, Ethiopia. Vet. Med. Int. 2019:3063185. doi: 10.1155/2019/3063185
Robles, I., Kelton, D. F., Barkema, H. W., Keefe, G. P., Roy, J. P., Von Keyserlingk, M. A. G., et al. (2020). Bacterial concentrations in bedding and their association with dairy cow hygiene and milk quality. Animal 14, 1052–1066. doi: 10.1017/S1751731119002787
Rowbotham, R. F., and Ruegg, P. L. (2016a). Associations of selected bedding types with incidence rates of subclinical and clinical mastitis in primiparous Holstein dairy cows. J. Dairy Sci. 99, 4707–4717. doi: 10.3168/jds.2015-10675
Rowbotham, R. F., and Ruegg, P. L. (2016b). Bacterial counts on teat skin and in new sand, recycled sand, and recycled manure solids used as bedding in freestalls. J. Dairy Sci. 99, 6594–6608. doi: 10.3168/jds.2015-10674
Selvam, A., Xu, D., Zhao, Z., and Wong, J. W. C. (2012). Fate of tetracycline, sulfonamide and fluoroquinolone resistance genes and the changes in bacterial diversity during composting of swine manure. Bioresour. Technol. 126, 383–390. doi: 10.1016/j.biortech.2012.03.045
Skeie, S. B., Håland, M., Thorsen, I. M., Narvhus, J., and Porcellato, D. (2019). Bulk tank raw milk microbiota differs within and between farms: a moving goalpost challenging quality control. J. Dairy Sci. 102, 1959–1971. doi: 10.3168/jds.2017-14083
Sorter, D. E., Kester, H. J., and Hogan, J. S. (2014). Short communication: bacterial counts in recycled manure solids bedding replaced daily or deep packed in freestalls. J. Dairy Sci. 97, 2965–2968. doi: 10.3168/jds.2013-7814
Tien, Y.-C., Li, B., Zhang, T., Scott, A., Murray, R., Sabourin, L., et al. (2017). Impact of dairy manure pre-application treatment on manure composition, soil dynamics of antibiotic resistance genes, and abundance of antibiotic-resistance genes on vegetables at harvest. Sci. Total Environ. 581-582, 32–39. doi: 10.1016/j.scitotenv.2016.12.138
Timms, L. (2008). Preliminary evaluation of separated manure solids characteristics at the new ISU dairy. Anim. Ind. Rep. 654, 10–12. doi: 10.31274/ans_air-180814-27
Tucker, C. B., and Weary, D. M. (2004). Bedding on geotextile mattresses: how much is needed to improve cow comfort? J. Dairy Sci. 87, 2889–2895. doi: 10.3168/jds.S0022-0302(04)73419-0
Wang, H., Sangwan, N., Li, H.-Y., Su, J.-Q., Oyang, W.-Y., Zhang, Z.-J., et al. (2017). The antibiotic resistome of swine manure is significantly altered by association with the Musca domestica larvae gut microbiome. ISME J. 11, 100–111. doi: 10.1038/ismej.2016.103
Wang, K., Yin, X., Mao, H., Chu, C., and Tian, Y. (2018). Changes in structure and function of fungal community in cow manure composting. Bioresour. Technol. 255, 123–130. doi: 10.1016/j.biortech.2018.01.064
Wu, H., Nguyen, Q. D., Tran, T. T. M., Tang, M. T., Tsuruta, T., and Nishino, N. (2019). Rumen fluid, feces, milk, water, feed, airborne dust, and bedding microbiota in dairy farms managed by automatic milking systems. Anim. Sci. J. 90, 445–452. doi: 10.1111/asj.13175
Wu, H., Wang, Y., Dong, L., Hu, H., Meng, L., Liu, H., et al. (2020). Microbial characteristics and safety of dairy manure ComPosting for reuse as dairy bedding. Biology 10:13. doi: 10.3390/biology10010013
Zhang, L., Li, L., Sha, G., Liu, C., Wang, Z., and Wang, L. (2020). Aerobic composting as an effective cow manure management strategy for reducing the dissemination of antibiotic resistance genes: an integrated meta-omics study. J. Hazard. Mater. 386:121895. doi: 10.1016/j.jhazmat.2019.121895
Keywords: tank milk, bedding material, Illumina MiSeq, bacterial diversity, ARGs
Citation: Wu H, Wang Y, Du B, Li H, Dong L, Hu H, Meng L, Zheng N and Wang J (2022) Influence of Dairy Cows Bedding Material on the Microbial Structure and Antibiotic Resistance Genes of Milk. Front. Microbiol. 13:830333. doi: 10.3389/fmicb.2022.830333
Edited by:
Buddhiman Tamang, Sikkim University, IndiaReviewed by:
Milena Brasca, National Research Council (CNR), ItalyYifan Zhong, Zhejiang University, China
Copyright © 2022 Wu, Wang, Du, Li, Dong, Hu, Meng, Zheng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nan Zheng, emhlbmduYW5AY2Fhcy5jbg==; Jiaqi Wang, amlhcWl3YW5nQHZpcC4xNjMuY29t
 Haoming Wu
Haoming Wu Yang Wang4
Yang Wang4 Bingyao Du
Bingyao Du Huiying Li
Huiying Li Haiyan Hu
Haiyan Hu Nan Zheng
Nan Zheng Jiaqi Wang
Jiaqi Wang