- 1Research Center of Avian Diseases, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 2Institute of Preventive Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 3Key Laboratory of Animal Disease and Human Health of Sichuan Province, Chengdu, China
The herpesvirus UL11 protein is encoded by the UL11 gene and is a membrane-anchored protein with multiple functions. In the last stage of viral replication, UL11 participates in the secondary envelopment process. It also plays a key role in primary envelopment, the transportation of newly assembled viral particles through cytoplasmic vesicles, and virion egress from the cell. UL11 is an important accessory protein and sometimes cooperates with other proteins that participate in virus-induced cell fusion. Cell fusion is necessary for cell-to-cell transmissions. This review summarizes the latest literature and discusses the roles of UL11 in viral assembly, primary and secondary envelopment, and cell-to-cell transmission to obtain a better understanding of the UL11 protein in the life cycle of herpesviruses and to serve as a reference for studying other viruses. Additionally, some recently discovered characteristics of UL11 are summarized.
Introduction
The Herpesviridae family is composed of Alphaherpesvirinae, Betaherpesvirinae, and Gammaherpesvirinae. The double-stranded DNA genome, capsid, tegument, and envelope form a mature herpesvirus virion (Guiping et al., 2007; Boštíková et al., 2014). The Aphaherpesvirinae subfamily includes many viruses, including human alphaherpesvirus-1/2 (HHV-1/2), also called herpes simplex virus-1/2 (HSV-1/2); pseudorabies virus (PRV); duck plague virus (DPV) (Qi et al., 2008; Zhao et al., 2008; Chang et al., 2009; Guo et al., 2009; Jia et al., 2009; Wu et al., 2012); human alphaherpesvirus-3 (HHV-3), also called varicella-zoster virus (VZV); equine herpesviruses (EHV); bovine herpesvirus (BoHV); infectious laryngotracheitis virus (ILTV); and Marek’s disease virus (MDV) (Kobty, 2015; Smith, 2017; You et al., 2017; Lefkowitz et al., 2018). The Betaherpesvirinae subfamily includes human alphaherpesvirus-5 (HCMV) and murine cytomegalovirus (MCMV). Human alphaherpesvirus-4 (HHV-4), also called Epstein–Barr virus (EBV), murine gammaherpesvirus-68 (MHV-68), and human alphaherpesvirus-8 (HHV-8) (Yuan et al., 2005), also called Kaposi’s sarcoma-associated herpesvirus (KSHV), are classified into the Gammaherpesvirinae subfamily (Foulon, 1992; Wu et al., 2020).
Tegument proteins are key components of herpesviruses (Newcomb et al., 2007). They are classified into two types, “inner” or “outer” tegument proteins, according to their preferential association with either the capsid or viral membrane during the viral lifecycle. When virions are lysed with non-ionic detergents, the inner or outer tegument proteins can be detected (Wolfstein et al., 2006; Mettenleiter et al., 2009; Radtke et al., 2010). Tegument proteins have multiple functions, including affecting viral replication by regulating gene transcription, destroying host innate immune responses, halting cell protein synthesis, and facilitating viral particle assembly by constructing a bridge that links viral capsids and envelope proteins (Nicoll et al., 2012; Owen et al., 2015), and evading innate immunity, as previously summarized (Yang et al., 2019).
Herpesviruses display two characteristic replication modes, namely, a rapid, productive replication cycle and a life-long quiescent infection (Zeev-Ben-Mordehai et al., 2014; You et al., 2017). To ensure lytic replication, after a virus has entered a cell, capsid DNA is released into the nucleus, and then viral DNA begins to replicate. After assembly and genome packaging, the intact capsid leaves the nucleus, and it immediately undergoes primary and de-envelopment at the nuclear envelope, followed by tegumentation and secondary envelopment in the cytoplasm. Ultimately, mature virions are released from infected cells by exocytosis (Yang et al., 2019).
The UL11 protein and its homologs are essential to the viral replication cycle, especially for cell-to-cell transmissions and, particularly, the secondary envelopment process (Yang et al., 2020a). In recent years, UL11 functions have been characterized (Kopp et al., 2004; Koshizuka et al., 2006; Yang et al., 2021, 2022). However, the roles played by herpesvirus UL11 in the viral replication cycle have not been fully elucidated. This article reviews the characteristics and roles of herpesvirus UL11 in the viral life cycle, especially its cell-to-cell transmission and cell fusion functions and UL11 interaction partners. On the basis of UL11 studies, we propose hypotheses to test and describe problems that remain to be solved to further characterize the importance of UL11 in future studies.
The UL11 Protein
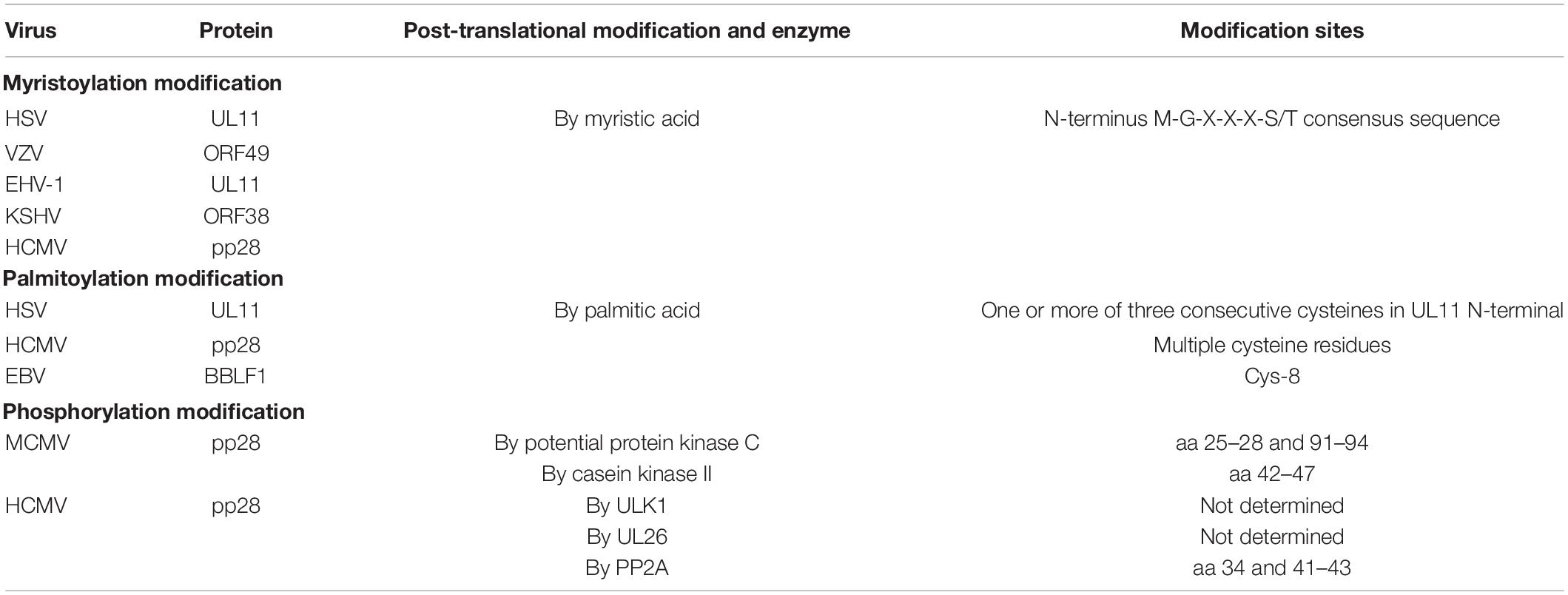
Approximately 700 copies of UL11 proteins per HSV-1 particle have been detected (Loomis et al., 2006). Using circular dichroism, limited proteolysis, small-angle X-ray scattering (SAXS), liquid–liquid phase separation (LLPS) analysis, and light microscopy, researchers identified that UL11 in vitro has an intrinsically disordered (ID) structure (Metrick et al., 2020). UL11 and its homologs in virions have been classified as tegument components (Johannsen et al., 2004; Tomtishen, 2012). The molecular size and localization of UL11 differ in herpesvirus species (Loomis et al., 2006). For example, the molecular size of UL11 is 15 kDa in HSV-1 (McGeoch et al., 1988), the molecular size of the UL11 homolog ORF51 in EHV-1 is 12 kDa (Badr et al., 2018), and the molecular size of UL11 in DPV (Yang et al., 2021) is approximately 15 kDa. In VZV, the ORF49 encoded protein is 13 kDa (Sadaoka et al., 2007). In β-herpesviruses, the molecular sizes of HCMV UL99 and MCMV UL99, each encoding pp28 protein, are 28 and 11.8 kDa, respectively (Cranmer et al., 1994; Hitomi et al., 1997). In γ-herpesviruses, the ORF38 protein in KSHV has been detected as 10 kDa (Wu et al., 2016). A 12-kDa protein has been found in MHV-68-infected cell lysate, which is the molecular size of ORF38 (Efstathiou et al., 1990). Interestingly, in all herpesviruses, UL11 expression partially overlaps with UL12 expression (Adam et al., 1995; Johannsen et al., 2004). Although HCMV pp28 has been shown to multimerize without other viral proteins, the researchers of this study did not rule out the possibility of an interaction with a cellular protein following pp28 multimerization (Seo and Britt, 2008). Table 1 shows detailed information about the sizes of the UL11 protein in various herpesviruses.
Post-translational Modification of the UL11 Protein
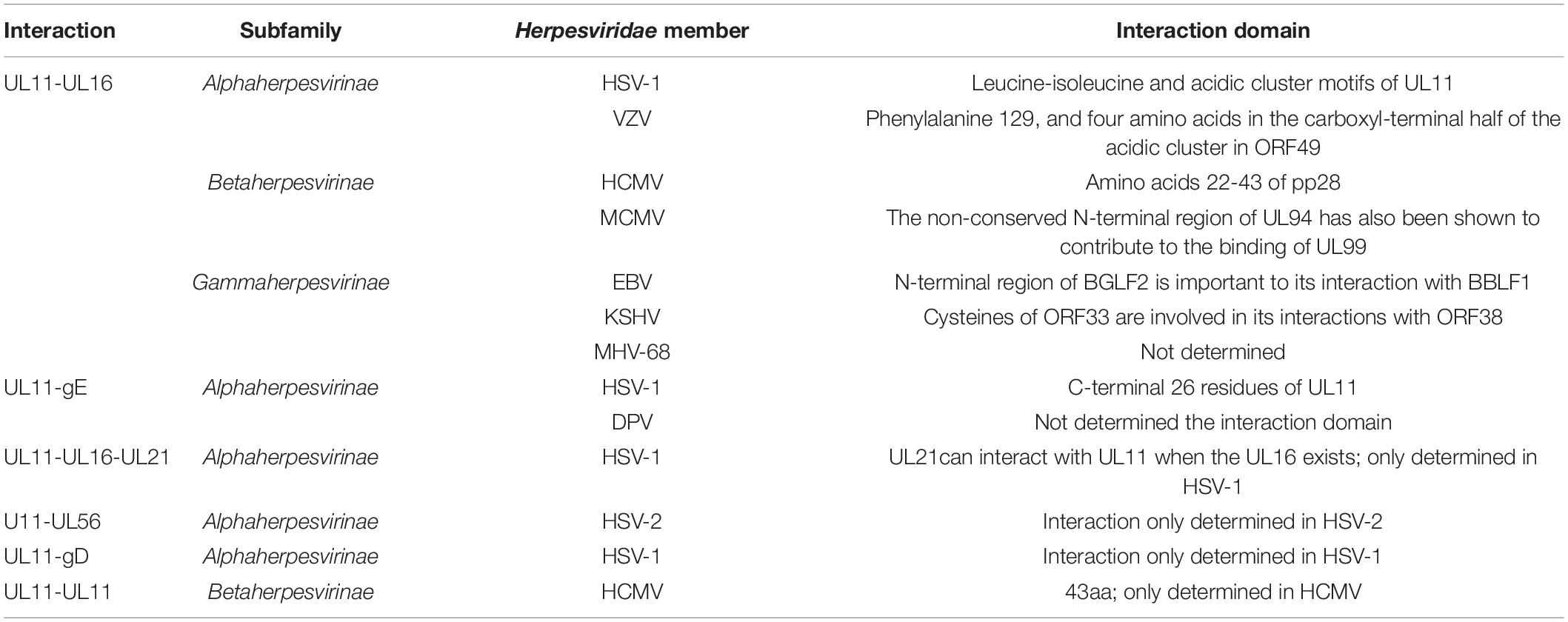
The UL11 protein undergoes multiple forms of post-translational modification, namely, myristoylation, and palmitoylation. Covalent modification with a fatty acid is an established characteristic in several cell types and viral polypeptides (Sefton and Buss, 1987), and palmitoylation and myristoylation are two fatty acid modifications. Studies on α-herpesviruses have shown that HSV UL11 and VZV ORF49 are myristoylated with myristic acid (MacLean et al., 1989; Harper and Kangro, 1990; Sadaoka et al., 2007). Additionally, EHV-1 UL11 has a conserved myristylation consensus sequence (M-G-X-X-X-S/T) (Resh, 1999). In β-herpesviruses, HCMV UL99 encodes a tegument protein that undergoes myristoylation (Chee et al., 1990). In KSHV, ORF38 is also myristoylated (Sanchez et al., 2000). In addition to myristoylation, HSV-1 UL11 is palmitoylated. The palmitoylated modification sites in UL11 contain one or more of three consecutive cysteines in the UL11 amino terminus (N-terminus) (Loomis et al., 2001). UL11 homologs of HSV-1, HCMV, EBV, and likely HSV-2 are also palmitoylated (Baird et al., 2008; Seo and Britt, 2008; Chiu et al., 2012; Jones and Lee, 2004).
In addition, UL11 undergoes phosphorylation. The HSV-1 UL11 homolog—HCMV UL99 phosphoprotein—has been identified strictly as a late kinetic reaction product (Mocarski and Courcelle, 2001). Studies have suggested that HCMV pp28 phosphorylation contributes to its intracellular trafficking and efficient viral assembly and incorporation. For example, when protein phosphatase 2 (PP2A) modified pp28, the mobility of pp28 was altered in SDS–PAGE, indicating that pp28 was indeed phosphorylated, potentially on several residues (Seo et al., 2020). Moreover, phosphorylation of pp28 tyrosine 34 and serines 41–43 was found to be important for transfected pp28 trafficking to the endoplasmic reticulum-Golgi-intermediate compartment (ERGIC) (Seo and Britt, 2006; Seo et al., 2020). Another study reported that deletion of HCMV UL26 caused pp28 hypophosphorylation, suggesting that the UL26 protein affected the normal phosphorylation of pp28 in virions and possibly additional tegument proteins (Munger et al., 2006). In MCMV, pp28 has been found to be phosphorylated, potentially by protein kinase C and casein kinase II (Cranmer et al., 1994), with amino acids (aa) 25–28 and 91–94 of pp28 being the protein kinase C targets and aa 42-47 being the casein kinase II targets (Cranmer et al., 1994). For HCMV, pp28 is the autophagy-initiating protein kinase ULK1 phosphorylation target (König et al., 2021). Table 2 contains information on these post-translational modifications.
Localization of the UL11 Protein
The localization of the UL11 protein is dynamic. In HSV-1, UL11 has been shown to be located at membranes, including nuclear and Golgi-derived membranes, and especially at the trans-Golgi network (TGN) (Baines et al., 1995; Loomis et al., 2001). In EHV-1, UL11 was predominantly localized at the TGN in infected cells, while in transfected cells, UL11 was localized at the plasma membrane, as determined through confocal laser scanning microscopy (Schimmer and Neubauer, 2003). Similar to UL11 in EHV-1, the virion component VZV ORF49 localized predominantly at the TGN in infected cells (Sadaoka et al., 2007). In DPV, UL11 is localized at the perinuclear area in infected cells and membranes, especially at the TGN membranes, in transfected cells (Yang et al., 2021). HCMV pp28 is localized in the ERGIC, which is a dynamic compartment in the secretory pathway that interfaces with both the ER and the Golgi apparatus (Sanchez et al., 2000). Another study identified MHV-68 ORF38 as a tegument protein that localized to cytoplasmic compartments (CT) during transient transfection and viral infection (Shen et al., 2014). Furthermore, EBV BBLF1 is localized to the TGN and perinuclear areas (Chiu et al., 2012). Hence, in most herpesviruses, UL11 is localized in the Golgi apparatus.
Gene Type of UL11
When a target cell is infected by herpesviruses, viral genomic DNA is circularized, and a temporal cascade of viral gene transcription is initiated; this cascade consists of three stages: immediate-early (IE), also called the α gene; early (E), also called the β gene; and late (L), also called the γ gene, indicating the expression period and dependence on other gene products (Mahmoudian et al., 2012; Yang et al., 2020b). ILTV UL11 has been identified as an L gene in alphaherpesviruses. Similarly, UL11 is also an L gene in HSV-1 and BoHV (Baines and Roizman, 1992; Desloges and Simard, 2001). In DPV and VZV, UL11 is also an L gene, as determined by its late-stage expression (Sadaoka et al., 2007; Yang et al., 2021). In EHV-1, UL11 is an early-late gene (Schimmer and Neubauer, 2003). Researchers characterized the HCMV pp28 upstream (pp28US) promoter with two regulatory components, one dependent on the onset of viral DNA synthesis and the other replication-independent, responding to viral trans-acting factors (Wu et al., 2001). Kohler et al. (1994) found that the pp28US promoter was sufficient to establish UL99 late kinetics (Kerry et al., 1997). In contrast, Jones and Lee showed that UL99 was expressed as an early-late protein, not an L protein, similar to UL99 in EHV-1 (Jones and Lee, 2004). MCMV UL99 has also been classified as an L gene based on its gene expression kinetics (Cranmer et al., 1994). Homologs of HSV-1 UL11 gene products in the herpesvirus family are nearly all conserved tegument proteins (Baer et al., 1984; Davison and Scott, 1986; Chee et al., 1990; Telford et al., 1992; Dijkstra et al., 1997; Osterrieder, 1999; Shen et al., 2014). Surprisingly, in contrast to other herpesvirus genes, MHV-68 ORF38 has been designated an IE gene after treatment with cycloheximide (Ebrahimi et al., 2003). Detailed information can be found in Table 1.
Functions of UL11 in the Viral Life Cycle
Viral Replication of the UL11 Null Mutant
The effect of the UL11 deletion on viral replication differs by virus species. In HSV-1 and PRV, the absence of UL11 resulted in a 10-fold reduction in viral titer (Baines et al., 1995; Kopp et al., 2003; Fulmer et al., 2007), and neither myristoylation nor palmitoylation modification of HSV-1 UL11 was found to be necessary for viral replication (MacLean et al., 1992; Baird et al., 2010). In EHV-1, deletion of UL11 in the neuropathogenic strain Ab4p resulted in abrogated progeny production. That is, in these mutant viruses, UL11 is replicated only in complementing cells; hence, UL11 is considered to be essential for EHV-1 (Badr et al., 2018). In addition, the absence of UL11 in the EHV-1 RacH and RacL22 strains, which exhibited one-step growth kinetics and were used to infect RK13 cells, resulted in an approximate 10- to 20-fold reduction in intracellular and extracellular virus titers (Schimmer and Neubauer, 2003). The reasons for the differences in these results were discussed in the article (Badr et al., 2018): First, the Ab4p UL11 sequence is the same as that in the RacH strain (data not shown); however, the restriction patterns of UL11 expression in the RacH and RacL22 strains differ from that in the Ab4 strain, which has a shorter left UL terminus and lacks a BamHI site (Hubert et al., 1996). Second, 0.85 kbp sequences in the inverted repeat regions are deleted in both copies of RacH (Hubert et al., 1996). Moreover, other viral proteins may compensate for the reduction in UL11 function in the Rac strain but not in Ab4p. Additionally, there may be experimental shortcomings in one of the studies. Similarly, HCMV UL99 is essential for the production of infectious viruses (Silva et al., 2003; Seo and Britt, 2007). No infectious progeny viruses were detected in fibroblasts infected with an HCMV pp28-deficient mutant, which also showed impaired viral replication due to the lack of enveloped virus particles. Additionally, although ORF49 is not essential for viral replication (Sadaoka et al., 2007), the VZV titer of the cell-free virus was 3 to 5% of that of the control virus (Sadaoka et al., 2014). In contrast to the effect of deletion of the essential gene UL99 in HCMV, deletion of BBLF1 only partially impaired EBV lytic replication (Chiu et al., 2012), reducing the production of EBV particles by approximately 54%. In another γ-herpesvirus, KSHV, ORF38 has not been considered essential for virus replication; however, the viral yield of the ORF38-null mutant was reduced 10-fold (Wu et al., 2016).
UL11 Influences Primary and Secondary Viral Envelopment
When the capsid is intact, it undergoes primary envelopment. The specific process of primary envelopment involves intranuclear capsid budding at the inner nuclear leaflet, and later, fusion with the outer leaflet of the nuclear membrane. Specifically, after primary envelopment, capsids undergo nuclear membrane fusion and de-envelopment, and then the nucleocapsids bud from the nucleus and are transported to the TGN, where the tegument is formed and where the nucleocapsid undergoes secondary envelopment (Mettenleiter, 2002).
For HSV-1, the incidence of UL11-deleted capsid juxtaposition with the inner lamellae of nuclear membranes has been shown to be greater than that of wild-type or repaired viruses. Specifically, approximately 3% of the virion particles in the nuclei of cells infected with wild-type or repaired viruses were in contact with the inner lamellae, whereas in cells infected with the mutant virus, 11% of the nuclear capsids were in contact with the inner lamellae. Moreover, the number of capsids partially enveloped by the inner lamellae of cells infected with the UL11-deleted virus was increased. In cells infected with wild-type, UL11-null or repaired viruses, enveloped particles, and, to a lesser extent, unenveloped particles accumulated in the space between the lamellae of the nuclear membrane 24 h after infection. The numbers of particles accumulating in this space were similar to the number of cells infected with each virus. This study suggested that UL11 gene deletion affected envelopment at the nuclear membrane by either slowing or inhibiting the process but had no effect on viral transit into the perinuclear space (Baines and Roizman, 1992). During the secondary envelopment stage, the HSV-1 UL11-null mutant produced fewer enveloped virions and more unenveloped CT capsids (Fulmer et al., 2007). In addition, deletion of PRV UL11 resulted in unenveloped capsid accumulation in the cytoplasm, which was inconsistently associated with recruited tegument proteins, similar to the effect of gE/gI and gM deletion mutants. Moreover, the absence of UL11 profoundly impaired the architecture of the Golgi-derived membrane, which is the typical site of secondary envelopment (Kopp et al., 2004). Similarly, when RK13 cells were infected with PRV UL11 and the UL51 double-deletion mutant, the morphology of the intracytoplasmic membranes was distorted, and nucleocapsids accumulated in the cytoplasm in association with aggregated tegument (Klupp et al., 2005b). The VZV ORF49-deletion mutant and the wild-type virus exerted the same effects (Sadaoka et al., 2007) in MRC-5 cells, as determined by ultrastructural analysis. This result suggests that VZV ORF49 did not affect primary or secondary envelopment in MRC-5 cells. In HCMV pp28-null mutant-infected cells, normal levels of viral DNA and L proteins were observed, but a large number of tegument-associated capsids accumulated in the cytoplasm and failed to undergo envelopment (Silva et al., 2003; Britt et al., 2004). Moreover, EBV BBLF1 has been hypothesized to be important for tegumented capsid budding into glycoprotein-embedded membranes during viral maturation (Chiu et al., 2012). MHV-68 ORF38 has also been reported to participate in secondary envelopment (Shen et al., 2014). As shown in a membrane flotation assay, KSHV ORF38 participated in the virion packaging process, promoting virion packaging into cytoplasmic vesicles critical for viral maturation and egress (Wu et al., 2016). Detailed information is presented in Table 3.
Effect of UL11 on Trafficking
Similar to that in another herpesvirus model, in an HSV-1 model, assembled nucleocapsid shuttling into the nucleus involved separate inner and outer nuclear membrane budding-fusion events; subsequently, these nucleocapsids moved through the cytoplasm as unenveloped capsids and underwent envelopment at TGN-derived vesicular membranes. As the nucleocapsids acquired this final lipid bilayer at the TGN, the tegument, which includes glycoproteins, was formed (Browne et al., 1996; Loomis et al., 2001). Notably, many other molecules are involved in the process of transporting non-enveloped viral particles such as microtubules.
The detected correlation between UL21 and microtubules (Takakuwa et al., 2001) has raised the possibility that UL21 participates in capsid transport to TGN-derived vesicles (Guo et al., 2010), where UL16 interacts with UL11, facilitating the budding process by linking capsids to the membrane (Loomis et al., 2003). A study showed that palmitoylation of HSV-1 UL11 was necessary for both Golgi targeting specificity and effective membrane binding. In addition, one-half of a conserved acidic sequence cluster in UL11 has been found to be important for tegument protein recycling from the plasma membrane to the Golgi apparatus (Loomis et al., 2001). HCMV UL99 has been shown to traffic to vacuole-like cytoplasmic structures, and the second glycine and a 44–57 acid sequence have been suggested to be necessary for envelopment (Jones and Lee, 2004). Further analyses of ORF33/ORF38 mutants revealed that the production of virion-containing vesicles was decreased, indicating that KSHV ORF38 facilitated the transport of newly assembled viral particles in cytoplasmic vesicles, a process that is essential for viral maturation and egress (Wu et al., 2016). When the first 100 amino acids in respiratory syncytial virus (RSV), Gag proteins were replaced by the entire UL11 sequence; the resulting chimeric protein was targeted in the Golgi apparatus rather than being localized to the plasma membrane. Studies have shown that a region consisting of the first 49 amino acids in UL11 is myristoylated, which is required for Golgi-specific targeting. In addition to the identification of a previously unknown modification site on UL11, these experiments showed that UL11 directed RSV Gag to internal cellular membranes (Bowzard et al., 2000). Taken together, these studies suggest that UL11 has highly dynamic membrane-trafficking properties.
Effect of UL11 on Cell Fusion and Cell-to-Cell Spread
Viruses spread to uninfected cells in two ways (Carmichael et al., 2018). Upon release from an infected cell and entry into the surrounding environment, a virus enters a new cell through cell-free spread. This modality describes both the viral infection of new hosts and transmission between cells within a host. However, some virions, including those of all herpesviruses, pass directly through cell junctions, enabling virion detection by neutralizing antibodies (Johnson and Huber, 2002; Mateo et al., 2015; Carmichael et al., 2018). In addition, virus-induced cell fusion, which facilitates transmission into adjacent cells, also protects virions from being exposed to neutralizing antibodies. This model of infectivity is observed in herpetic lesions (Roizman et al., 2007). This modality of transmission is important and worth studying because the mechanisms of cell-to-cell spread and cell fusion in herpesvirus infections remain unclear.
The mechanism of cell-to-cell spread is complex. In HSV-1, gE is crucial for cell-to-cell spread, as indicated by the inhibition of cell-to-cell spread in gE-deletion mutants treated with neutralizing antibodies in vitro and transmission between neurons in mouse models (Dingwell et al., 1994; Howard et al., 2014). Further research showed that the peripheral membrane-binding protein UL11, which is associated with tegument proteins UL16 and UL21, forms a complex on the cytoplasmic tail of gE. In cell culture, this complex appears to be critical for cell-to-cell spread (Han et al., 2012; Sarfo et al., 2017). The replication efficiency of the HSV-1 UL11-null mutant was nearly one logarithmic unit less than that of the wild-type virus, and the UL11-null virus formed plaques that were, on average, one-third the size of those created by the wild-type virus (Kim et al., 2014). Other HSV-1 proteins, including UL51, gI, and UL34, have also been shown to participate in cell-to-cell spread (Dingwell and Johnson, 1998; Haugo et al., 2011; Roller et al., 2014). When UL11 was deleted in EHV-1, the viral plaque decreased significantly—to approximately 20% of the parental virus (Schimmer and Neubauer, 2003). Moreover, the VZV UL11-null mutant showed diminished plaque compared to the wild-type strain, similar to its effect on HSV-1 infection of the human malignant melanoma cell line. However, no differences in plaque size or cell-to-cell spread were observed in MRC-5 human embryonic fibroblasts infected with the wild-type or mutant virus (Sadaoka et al., 2007).
Surprisingly, UL11 was also shown to be vital for the virus-induced cell fusion process. In the alphaherpesvirus HSV-1, when UL11 and gM were deleted, the virus entered cells more slowly than the wild-type virus, indicating that gM and UL11 both regulate the HSV-1 membrane fusion machinery during virus-induced cell fusion and virus entry (Kim et al., 2013). Cell fusion was induced when a 28 aa sequence in the carboxyl-terminus (C-terminus) of gB was deleted by the insertion of a stop codon and when the gK alanine residue at position 40 was replaced with valine. Among gB syncytial mutants, the gBΔ28 mutation is considered to exert the most profound inductive effect on the fusion of most cell types in tissue culture (Silverman et al., 2012). In addition, the gKsyn20 mutation caused extensive virus-induced cell fusion of most cell types (Chouljenko et al., 2009). Abrogation of UL11 led to severe impairment of gB△28 and gKsyn20-induced virus-induced cell fusion. In addition, UL11 affected gBA855V-induced cell fusion, confirming the aforementioned result that suggests that UL11 is required for gB△28 induced cell fusion (Silverman et al., 2012; Kim et al., 2013). In a study by Han, the UL11-null mutant in the presence of the UL11 interaction proteins UL16 and UL21 failed to form syncytia in HSV-1 (Han et al., 2012).
Effect of UL11 on Virion Egress
Upon maturation, viral particles are released from host cells. UL11 plays a key role in viral egress, as indicated by the profound delay in UL11-null mutant HSV-1 virus release from infected cells. Specifically, the wild-type virus showed an approximately 500-fold increase in virion release, while the UL11-null mutant showed only a 15-fold increase between 8 and 14 h post-infection (Baines and Roizman, 1992). Moreover, in the HSV-1 UL11 null mutant, Chouljenko et al. (2012) found that there were fewer extracellular virions than cytoplasmic virions. Interestingly, HCMV pp28 was phosphorylated by the autophagy-initiating protein kinase ULK1, which leads to efficient viral release (König et al., 2021). Thus, UL11 may facilitate the viral egress process. Figure 1 depicts a function map for the UL11 protein.
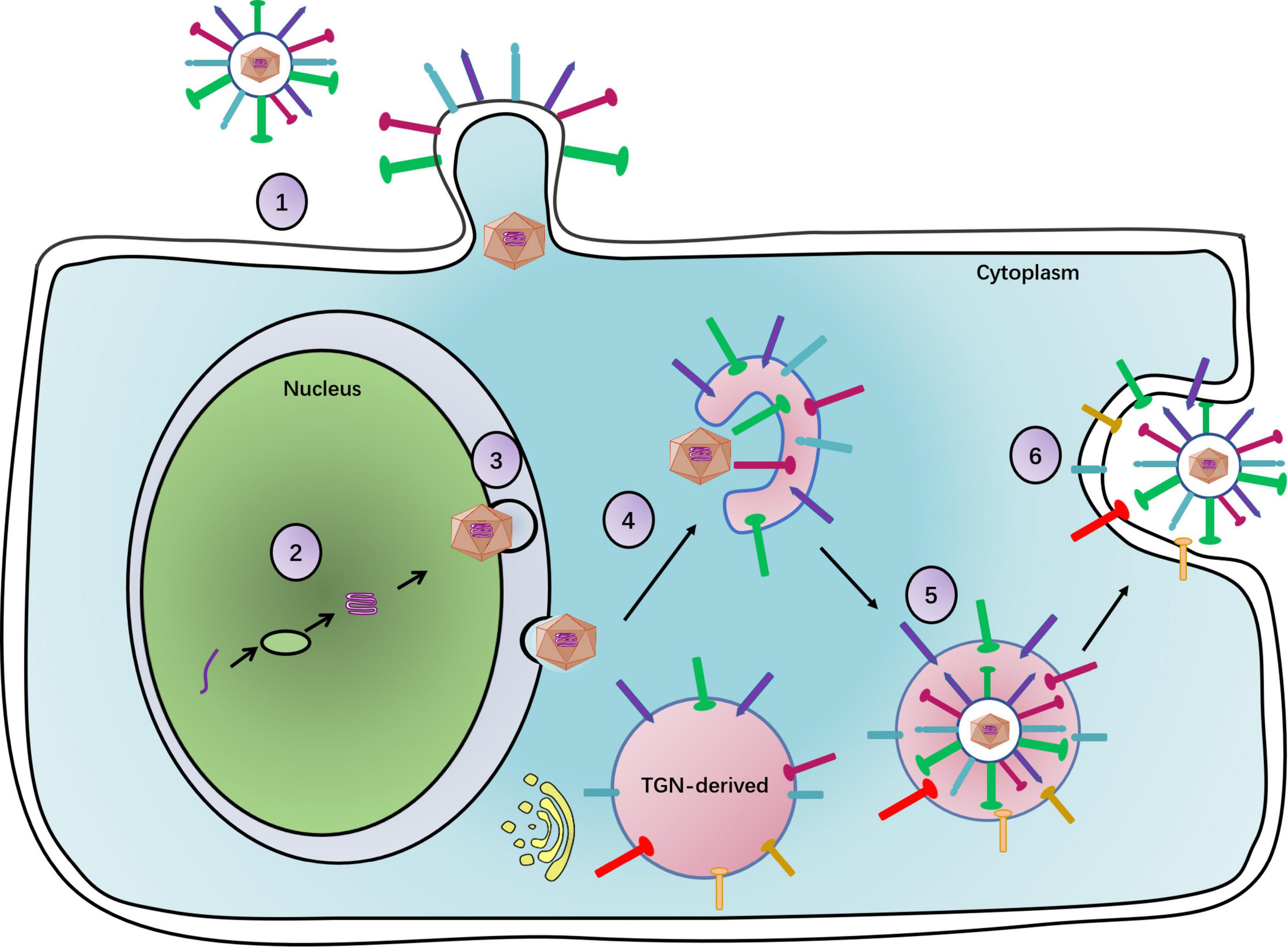
Figure 1. The complete process of the herpesvirus life cycle. (1) Viral particles enter a cell. With gM, UL11 promotes virus entry. (2) The viral DNA replication. (3) Primary envelopment of viral particles. UL11 participates in capsid primary envelopment. (4) Primary envelopment capsids arrive at the TGN to obtain secondary envelopment. UL11 can raise glycoproteins at the TGN to carry on secondary envelopment. (5) Secondary enveloped viral particles will be transported by the vesicle. UL11 promotes the association of the newly assembled viral particles with the vesicle. (6) Mature viral particles with the vesicle egress out of a cell. UL11 increases the number of viral particles released from the cell.
UL11 Interaction Partners
The gE-UL11-UL16-UL21 Complex
The classical interaction between UL11 and UL16 has been determined to be conserved in all members of the Herpesviridae family (Fossum et al., 2009; Diefenbach, 2015). Evidence has shown that the interaction between HSV-1 UL11 and UL16 may promote the correlation observed between nucleocapsids and the secondary envelopment process (Vittone et al., 2005; Meckes and Wills, 2007; Meckes et al., 2010; Johnson and Baines, 2011; Starkey et al., 2014). Because it is a nucleocapsid-associated protein, upon interaction with its partners, UL16 may recruit nucleocapsids to cytoplasmic membranes for envelopment within infected cells. In PRV, the interaction between UL11 and UL16 was identified in a glutathione S-transferase (GST) pull-down assay (Harper et al., 2010), and the complex promoted UL36 incorporation into mature virions (Michael et al., 2006), which was also observed in VZV (Sadaoka et al., 2014). Furthermore, binding with HCMV UL99 stabilized the UL16 homolog UL94 (Phillips et al., 2012), facilitating cytoplasmic nucleocapsid transport to membranes (Phillips and Bresnahan, 2012). In KSHV, ORF33 interacts with ORF38, and this interaction leads to the optimal production of infectious progeny viruses (Wu et al., 2016). The UL16-binding sites in homologs of HSV-1 and HCMV have been confirmed to be in the N-terminal half of the UL11 protein (Yeh et al., 2008; Liu et al., 2009): specifically, in HSV-1, they are in the acidic cluster sequence and leucine-isoleucine (LI) motifs within the UL11 N-terminus (Yeh et al., 2008). Similarly, in EBV, the interaction between BGLF2 and BBLF1 is required for the efficient production of infectious virus particles (Hung et al., 2020). These findings show that the interaction between the UL16 and UL11 proteins is conserved in the herpesvirus family.
Although both gE and pUL21 are unique to the Alphaherpesvirinae subfamily and have not yet been investigated, homologs of pUL11 and pUL16 may bind to the cytoplasmic tail of envelope proteins in the Beta and Gammaherpesvirinae subfamilies. Notably, a study reported that an HSV-1 UL11 acidic cluster motif is indispensable for gE packaging (Han et al., 2011). In DPV, research has shown that DPV gE and UL11 preferentially and exclusively interact. UL11 can interact with the CT and ET domains of gE, and the CT domain of gE is critical for UL11 incorporation into a viral particle (Yang et al., 2021). In HSV-1, UL11, and UL16 have been shown to interact with the cytoplasmic tail of gE, and UL11 has been shown to initiate the weak interaction observed between UL16 and gE (Farnsworth et al., 2007; Han et al., 2011, 2012; Yeh et al., 2011). In HSV, DPV, and PRV, UL21 can interact with UL16 (Klupp et al., 2005a; Harper et al., 2010; Gao et al., 2017; Yang et al., 2020b). In HSV, UL21 interacts with UL11 in the presence of UL16, and UL21 promotes the weak interaction between UL11 and UL16 (Han et al., 2012). Similar to the UL11-gE interaction, the UL11 and UL21 interaction has been found to be weak and has been identified only in the Alphaherpesvirinae subfamily (Diefenbach, 2015). UL21 and UL16 are capsid-associated proteins, and UL11-UL16-UL21 can form a complex. Therefore, UL11 may indirectly affect capsid function. In fact, UL11, UL16, UL21, and gE may have formed a quadruple complex in transfected Vero cells (Han et al., 2012). Table 4 contains additional information, and details of the relationship between UL11, UL16, UL21, and gE are presented in Figure 2.
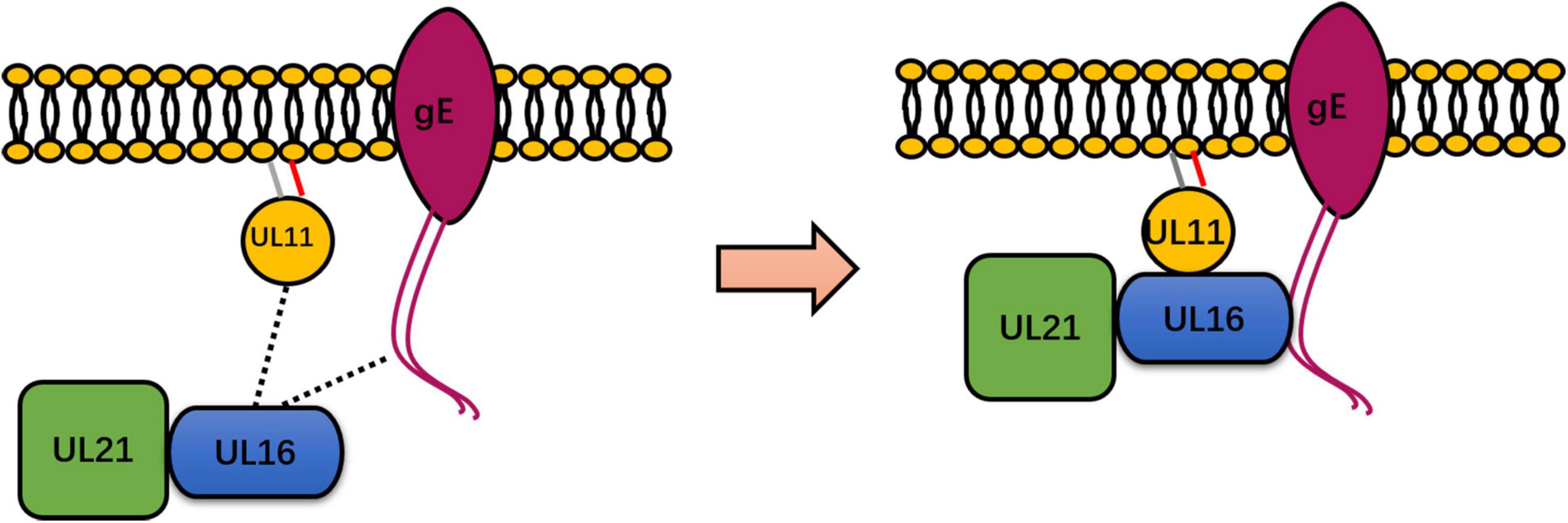
Figure 2. Summary of the known interactions between UL11 and UL16, UL21, and gE. UL11 binds to UL16, which is directly conserved in a representative member of Herpesviridae. However, UL11 formed a strong interaction with gE only in a study of HSV-1 and DPV. In HSV, DPV, and PRV, UL21 interacts directly with UL16. The dotted line represents the weak interaction between UL11 and UL16 and between UL16 and gE (left). UL16 may undergo enhanced binding with gE in the presence of UL11, UL16 may undergo enhanced binding with UL11, and UL16 may undergo enhanced binding with UL11 in the presence of UL21 in HSV-1 (right). Therefore, UL11, UL16, UL21, and gE form a complex. Some inspiration for this figure was obtained from previous articles (Han et al., 2012).
UL11 Can Form Homomultimers or Interact With Other Viral Proteins
Through GST pull-down assay and other immunoprecipitation experiments, Koshizuka et al. confirmed that UL11 interacted with UL56 in HSV-2-infected cells. UL56 is a tail-anchored type II membrane protein associated with the Golgi apparatus and cytoplasmic vesicles (Koshizuka et al., 2006). However, no reports have indicated that UL11 interacts with UL56 in other herpesviruses. On the basis of an immunoprecipitation assay, another study revealed that the CT domain of HSV-1 gD is bound to UL11 in HSV-1-infected cells (Farnsworth et al., 2007), but whether this interaction was direct or not was unclear, and it could not be induced in cells infected with other herpesviruses.
UL11 can undergo self-interaction to form homomultimers. A study found that the initial 43 amino acid sequences in HCMV pp28 contain a domain that is critical for the self-interaction of pp28 (Seo and Britt, 2008). Although this interaction was found to be independent of other viral proteins, the possibility of a pp28 interaction with cellular proteins was not ruled out. Additionally, multimerization of UL11 has only been reported in HCMV. Detailed information on the UL11 network is presented in Figure 3.
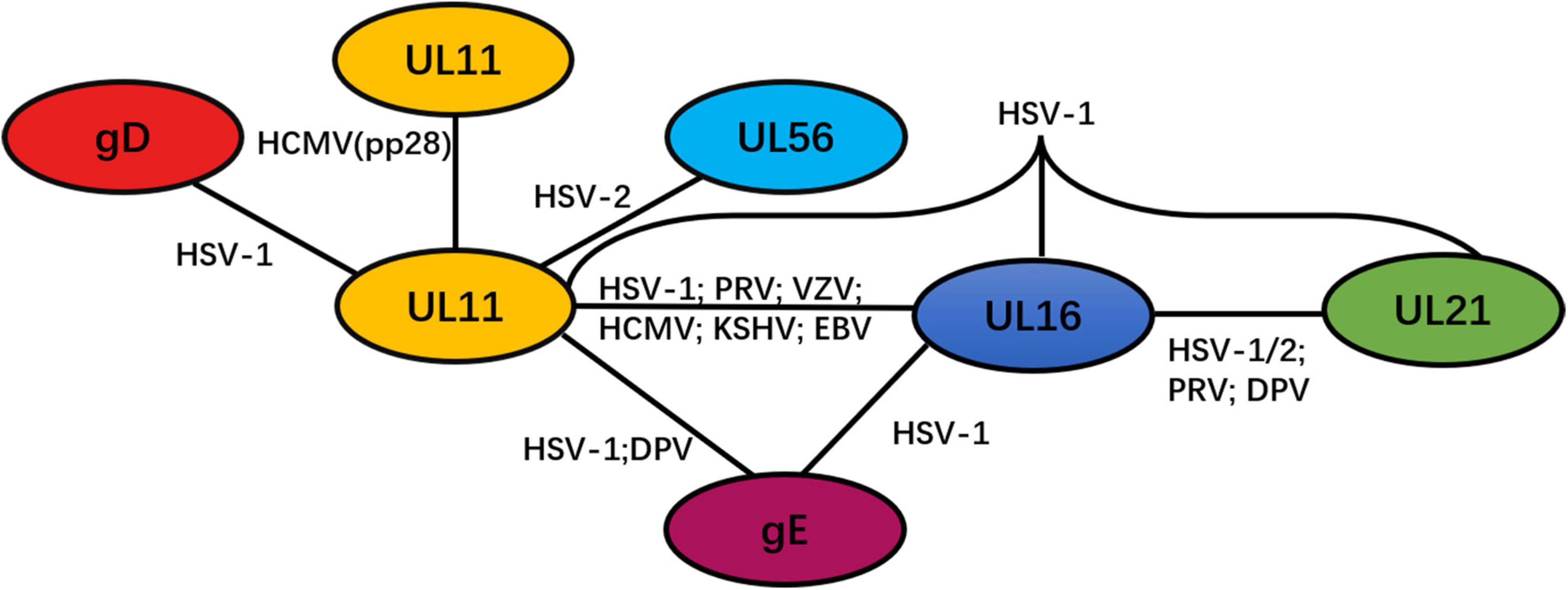
Figure 3. Network of protein–protein interactions between the tegument protein UL11 and other viral proteins. Black solid lines represent interactions.
UL11 Can Interact With Lipid Rafts
Lipid rafts, also called detergent-resistant membranes (DRMs), are microdomain structures that are rich in cholesterol and glycosphingolipids. Although lipid rafts are generally located on the plasma membrane, rafts are often assembled in the Golgi apparatus (Simons and Ikonen, 1997). Many proteins interacting with lipid rafts have important roles in trafficking, signal transduction, and pathogen entry and exit (Koshizuka et al., 2007). Researchers found that HSV-2 UL11 was associated with lipid rafts. In DPV research, UL11 has been shown to interact with lipid rafts, and this interaction depends on the second glycine of UL11 (Yang et al., 2022). The dual acylation of UL11 is required for lipid raft association because mutated myristoylation or palmitoylation sites block lipid rafts (Koshizuka et al., 2007; Baird et al., 2008). Through an immunoprecipitation assay, a study elucidated the DRMs associated with HSV-1 UL11 and showed that they depended on LI and acidic cluster motifs in UL11 (Baird et al., 2008).
Other Functions of UL11
Studying HSV-1, researchers found that the C-terminus of UL11 has a disordered structure, and further experiments indicated that this disordered structure fails to bind ribosomal RNA (rRNA) (Metrick et al., 2020). Additionally, this disordered structure was found not only in UL11 but also in all the tegument proteins of HSV-1. Moreover, this was the first report showing that UL11 is associated with RNA. Hence, the disordered tegument structure and its correlation with assembly mechanisms should be taken into account. The structural disorder in other tegument proteins needs to be further investigated because the details of these protein interactions are unclear.
For the study of HCMV, the UL99 transcript has been identified as a rapid marker of therapy effectiveness. Through a qRT–PCR assay analysis of 18 bronchoalveolar lavage samples from patients who were or were not subjected to antiviral treatment in vivo and vitro, UL99 has been developed as an antiviral control (Gambarino et al., 2014). In the past, it was difficult to study L gene promoters, as they were expressed in a disorderly fashion in transient assays (Kohler et al., 1994). However, a study of the entire viral genome showed that sequences from position –40 to +106 of the pp28 promoter are important for the L gene expression. The –6 to +46 sequences of the pp28 promoter repressed gene expression. The +46 to +88 sequence of the pp28 promoter was not particularly important to gene expression compared with the influences of the other sequences. These results indicate that translation of the L genes can be regulated by leader sequences in the pp28 gene (Kerry et al., 1997).
Conclusion
Among all α-, β-, and γ-herpesviruses, UL11 is a conserved gene. In the Herpesviridae family, the viral protein–protein interaction network is dynamic and necessary for the secondary envelopment process. Since it can associate with multiple proteins, UL11 is a key mediator in this interaction network. Moreover, the UL11 interaction network is important for the secondary envelopment process. Future studies on the structure of the conserved interactions, such as the UL11-UL16 interaction, will facilitate further research in herpesvirus assembly and tegument structure. The mechanism and function of the UL11, UL16, UL21, and gE quadruple complex also need to be investigated. Details on the function of the UL11 interaction with the two types of capsid-associated proteins need to be ascertained to determine whether they are related to the construction of a bridge between the capsid and the envelope. Studies of the interaction between the host protein and UL11 are rare, and elucidating the UL11-host cell protein interactions within virions may reveal the question regarding whether host cell proteins are simply passively acquired during viral assembly and trafficking or whether other mechanisms are involved is worth investigating. For research on β-herpesviruses, UL99 has been determined to be an important factor in antiviral therapy research, and it may be useful as an additional tool for overcoming drug resistance at the onset of infection.
In addition to the UL11 interaction partners that regulate the viral life cycle, other functions need to be investigated. Most tegument proteins, such as UL41, UL48, and UL49, have been reported to participate in innate immunity. However, because research into the effect of UL11 on innate immunity is rare, it remains unclear whether or to what extent UL11 is needed or functional. Investigating the role played by UL11 in immunity will lay a foundation for UL11 function research.
Author Contributions
LY conceived, designed, and drafted the manuscript. MW conceived and supervised the review. AC, QY, YW, JH, BT, RJ, ML, DZ, SC, XZ, SZ, XO, SM, QG, and DS approved the final manuscript for publication. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grants from the China Agriculture Research System of MOF and MARA and the Sichuan Veterinary Medicine and Drug Innovation Group of the China Agricultural Research System (SCCXTD-2020-18). The funding body played no role in the study’s design, collection, analysis, and interpretation of data or the writing of this manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adam, B. L., Jervey, T. Y., Kohler, C. P., Wright, G. L. Jr, Nelson, J. A., and Stenberg, R. M. (1995). The human cytomegalovirus UL98 gene transcription unit overlaps with the pp28 true late gene (UL99) and encodes a 58-kilodalton early protein. J. Virol. 69, 5304–5310. doi: 10.1128/JVI.69.9.5304-5310.1995
Badr, Y., Ayaka, O., Abo-Sakaya, R., Beshir, E., Ohya, K., and Fukushi, H. (2018). Equine herpesvirus type 1 ORF51 encoding UL11 as an essential gene for replication in cultured cells. Arch. Virol. 163, 599–607. doi: 10.1007/s00705-017-3650-4
Baer, R., Bankier, A. T., Biggin, M. D., Deininger, P. L., Farrell, P. J., Gibson, T. J., et al. (1984). DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310, 207–211. doi: 10.1038/310207a0
Baines, J. D., Jacob, R. J., Simmerman, L., and Roizman, B. (1995). The herpes simplex virus 1 UL11 protein is associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J. Virol. 69, 825–833. doi: 10.1128/jvi.69.2.825-833.1995
Baines, J. D., and Roizman, B. (1992). The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J. Virol. 66, 5168–5174. doi: 10.1128/JVI.66.8.5168-5174.1992
Baird, N. L., Starkey, J. L., Hughes, D. J., and Wills, J. W. (2010). Myristylation and palmitoylation of HSV-1 UL11 are not essential for its function. Virology 397, 80–88. doi: 10.1016/j.virol.2009.10.046
Baird, N. L., Yeh, P. C., Courtney, R. J., and Wills, J. W. (2008). Sequences in the UL11 tegument protein of the herpes simplex virus that controls association with detergent-resistant membranes. Virology 374, 315–321. doi: 10.1016/j.virol.2008.01.007
Boštíková, V., Salavec, M., Smetana, J., Sleha, R., and Boštík, P. (2014). Infections caused by human alpha herpes viruses. Epidemiol. Mikrobiol. Imunol. 63, 205–212.
Bowzard, J. B., Visalli, R. J., Wilson, C. B., Loomis, J. S., Callahan, E. M., Courtney, R. J., et al. (2000). Membrane targeting properties of a herpesvirus tegument protein-retrovirus Gag chimera. J. Virol. 74, 8692–8699. doi: 10.1128/jvi.74.18.8692-8699.2000.Z
Britt, W. J., Jarvis, M., Seo, J. Y., Drummond, D., and Nelson, J. (2004). Rapid genetic engineering of human cytomegalovirus using a lambda phage linear recombination system: demonstration that pp28 (UL99) is essential for the production of infectious virus. J. Virol. 78, 539–543. doi: 10.1128/jvi.78.1.539-543.2004
Browne, H., Bell, S., Minson, T., and Wilson, D. W. (1996). An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for redevelopment during egress. J. Virol. 70, 4311–4316. doi: 10.1128/JVI.70.7.4311-4316.1996
Carmichael, J. C., Yokota, H., Craven, R. C., Schmitt, A., and Wills, J. W. (2018). The HSV-1 mechanisms of cell-to-cell spread and fusion are critically dependent on host PTP1B. PLoS Pathog. 14:e1007054. doi: 10.1371/journal.ppat.1007054
Chang, H., Cheng, A., Wang, M., Guo, Y., Xie, W., and Lou, K. (2009). Complete nucleotide sequence of the duck plague virus gE gene. Arch. Virol. 154, 163–165. doi: 10.1007/s00705-008-0284-6
Chee, M. S., Bankier, A. T., Beck, S., Bohni, R., Brown, C. M., Cerny, R., et al. (1990). Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154, 125–169. doi: 10.1007/978-3-642-74980-3_6
Chiu, Y., Sugden, B., Chang, P., Chen, L., Lin, Y., Lan, Y., et al. (2012). Characterization and intracellular trafficking of Epstein-Barr virus BBLF1, a protein involved in virion maturation. J. Virol. 86, 9647–9655. doi: 10.1128/JVI.01126-12
Chouljenko, D. V., Kim, I. J., Chouljenko, V. N., Subramanian, R., Walker, J. D., and Kousoulas, K. G. (2012). Functional hierarchy of herpes simplex virus 1 viral glycoproteins in cytoplasmic virion envelopment and egress. J. Virol. 86, 4262–4270. doi: 10.1128/JVI.06766-11
Chouljenko, V. N., Iyer, A. V., Chowdhury, S., Chouljenko, D. V., and Kousoulas, K. G. (2009). The amino terminus of herpes simplex virus type 1 glycoprotein K (gK) modulates gB-mediated virus-induced cell fusion and virion egress. J. Virol. 83, 12301–12313. doi: 10.1128/JVI.01329-09
Cranmer, L. D., Clark, C., and Spector, D. H. (1994). Cloning, Characterization, and Expression of the Murine Cytomegalovirus Homologue of the Human Cytomegalovirus 28-kDa Matrix Phosphoprotein (UL99). Virology 205, 417–429. doi: 10.1006/viro.1994.1662
Davison, A. J., and Scott, J. E. (1986). The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67, 1759–1816. doi: 10.1099/0022-1317-67-9-1759
Desloges, N., and Simard, C. (2001). Expression kinetics of the late UL12 gene encoding the bovine herpesvirus 1 alkaline nuclease. Arch. Virol. 146, 1871–1884. doi: 10.1007/s007050170039
Diefenbach, R. J. (2015). Conserved tegument protein complexes: essential components in the assembly of herpesviruses. Virus Res. 2, 308–317. doi: 10.1016/j.virusres.2015.09.007
Dijkstra, J. M., Fuchs, W., Mettenleiter, T. C., and Klupp, B. G. (1997). Identification and transcriptional analysis of pseudorabies virus UL6 to UL12 genes. Arch. Virol. 142, 17–35. doi: 10.1007/s007050050056
Dingwell, K. S., Brunetti, C. R., Hendricks, R. L., Tang, Q., Tang, M., Rainbow, A. J., et al. (1994). Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J. Virol. 68, 834–845. doi: 10.1128/JVI.68.2.834-845.1994
Dingwell, K. S., and Johnson, D. C. (1998). The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. J. Virol. 72, 8933–8942. doi: 10.1128/JVI.72.11.8933-8942.1998
Ebrahimi, B., Dutia, B. M., Roberts, K. L., Garcia-Ramirez, J. J., Dickinson, P., Stewart, J. P., et al. (2003). Transcriptome profile of murine gammaherpesvirus-68 lytic infection. J. Gen. Virol. 84(Pt 1), 99–109. doi: 10.1099/vir.0.18639-0
Efstathiou, S., Ho, Y. M., Hall, S., Styles, C. J., Scott, S. D., and Gompels, U. A. (1990). Murine herpesvirus 68 is genetically related to the gammaherpesviruses Epstein-Barr virus and herpesvirus saimiri. J. Gen. Virol. 71(Pt 6), 1365–1372. doi: 10.1099/0022-1317-71-6-1365
Farnsworth, A., Wisner, T. W., and Johnson, D. C. (2007). Cytoplasmic residues of herpessimplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11 to gE and gD. J. Virol. 81, 319–331. doi: 10.1128/JVI.01842-06
Fossum, E., Friedel, C. C., Rajagopala, S. V., Titz, B., Baiker, A., Schmidt, T., et al. (2009). Evolutionarily conserved herpesviral protein interaction networks. PLoS Pathog. 5:e1000570. doi: 10.1371/journal.ppat.1000570
Foulon, T. (1992). Herpesviridae: classification and structure in 1991. Comp. Immunol. Microbiol. Infect. Dis. 15, 13–29. doi: 10.1016/0147-9571(92)90098-c
Fulmer, P. A., Melancon, J. M., Baines, J. D., and Kousoulas, K. G. (2007). UL20 protein functions precede and are required for the UL11 functions of herpes simplex virus type 1 cytoplasmic virion envelopment. J. Virol. 81, 3097–3108. doi: 10.1128/JVI.02201-06
Gambarino, S., Callea, S., Rizzo, G., Montanari, P., Loiacono, E., and Bergallo, M. (2014). Evaluation of UL99 transcript as a target for antiviral treatment efficacy. J. Virol. Methods 207, 104–109. doi: 10.1016/j.jviromet.2014.06.014
Gao, J., Hay, T. J. M., and Banfield, B. W. (2017). The Product of the Herpes Simplex Virus 2 UL16 Gene Is Critical for the Egress of Capsids from the Nuclei of Infected Cells. J. Virol. 91, e350–e317. doi: 10.1128/JVI.00350-17
Guiping, Y., Anchun, C., Mingshu, W., Xiaoying, H., Yi, Z., and Fei, L. (2007). Preliminary study on duck enteritis virus-induced lymphocyte apoptosis in vivo. Avian Dis. 51, 546–549. doi: 10.1637/0005-2086(2007)51[546:PSODEV]2.0.CO;2
Guo, H., Shen, S., Wang, L., and Deng, H. (2010). Role of tegument proteins in herpesvirus assembly and egress. Protein Cell 1, 987–998. doi: 10.1007/s13238-010-0120-0
Guo, H., Wang, L., Peng, L., Zhou, Z. H., and Deng, H. (2009). Open reading frame33 of a gammaherpesvirus encodes a tegument protein essential for virion morphogenesis and egress. J. Virol. 83, 10582–10595. doi: 10.1128/jvi.00497-09
Guo, Y., Cheng, A., Wang, M., and Zhou, Y. (2009). Purification of anatid herpesvirus 1 particles by tangential-flow ultrafiltration and sucrose gradient ultracentrifugation. J. Virol. Methods 161, 1–6. doi: 10.1016/j.jviromet.2008.12.017
Han, J., Chadha, P., Meckes, D. G. Jr., Baird, N. L., and Wills, J. W. (2011). Interaction and interdependent packaging of tegument protein UL11 and glycoprotein e of herpes simplex virus. J. Virol. 85, 9437–9446. doi: 10.1128/JVI.05207-11
Han, J., Chadha, P., Starkey, J. L., and Wills, J. W. (2012). Function of glycoprotein E of herpes simplex virus requires coordinated assembly of three tegument proteins on its cytoplasmic tail. Proc. Natl. Acad. Sci. U.S.A. 109, 19798–19803. doi: 10.1073/pnas.1212900109
Harper, A. L., Meckes, D. G. Jr., Marsh, J. A., Ward, M. D., Yeh, P. C., Baird, N. L., et al. (2010). Interaction domains of the UL16 and UL21 tegument proteins of herpes simplex virus. J. Virol. 84, 2963–2971. doi: 10.1128/JVI.02015-09
Harper, D. R., and Kangro, H. O. (1990). Lipoproteins of varicella-zoster virus. J. Gen. Virol. 71, 459–463. doi: 10.1099/0022-1317-71-2-459
Haugo, A. C., Szpara, M. L., Parsons, L., Enquist, L. W., and Roller, R. J. (2011). Herpes simplex virus 1 pUL34 plays a critical role in cell-to-cell spread of the virus in addition to its role in virus replication. J. Virol. 85, 7203–7215. doi: 10.1128/JVI.00262-11
Hitomi, S., Kozuka-Hata, H., Chen, Z., Sugano, S., Yamaguchi, N., and Watanabe, S. (1997). Human cytomegalovirus open reading frame UL11 encodes a highly polymorphic protein expressed on the infected cell surface. Arch. Virol. 142, 1407–1427. doi: 10.1007/s007050050169
Howard, P. W., Wright, C. C., Howard, T., and Johnson, D. C. (2014). Herpes simplex virus gE/gI extracellular domains promote axonal transport and spread from neurons to epithelial cells. J. Virol. 88, 11178–11186. doi: 10.1128/jvi.01627-14
Hubert, P. H., Birkenmaier, S., Rziha, H. J., and Osterrieder, N. (1996). Alterations in the equine herpesvirus type-1 (EHV-1) strain RacH during attenuation. J. Vet. Med. B 43, 1–14. doi: 10.1111/j.1439-0450.1996.tb00282.x
Hung, C. H., Chiu, Y. F., Wang, W. H., Chen, L. W., Chang, P. J., and Huang, T. Y. (2020). Interaction Between BGLF2 and BBLF1 Is Required for the Efficient Production of Infectious Epstein-Barr Virus Particles. Front. Microbiol. 24:3021. doi: 10.3389/fmicb.2019.03021
Jia, R., Cheng, A., Wang, M., Qi, X., Zhu, D., Ge, H., et al. (2009). Development and evaluation of an antigen-capture ELISA for detection of the UL24 antigen of the duck enteritis virus, based on a polyclonal antibody against the UL24 expression protein. J. Virol. Methods 161, 38–43. doi: 10.1016/j.jviromet.2009.05.011
Johannsen, E., Luftig, M., Chase, M. R., Weicksel, S., Cahir-McFarland, E., Illanes, D., et al. (2004). Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. U.S.A. 101, 16286–16291.
Johnson, D. C., and Baines, J. D. (2011). Herpesviruses remodel host membranes for virus egress. Nat. Rev. Microbiol. 9, 382–394. doi: 10.1038/nrmicro2559
Johnson, D. C., and Huber, M. T. (2002). Directed egress of animal viruses promotes cell-to-cell spread. J. Virol. 76, 1–8. doi: 10.1128/JVI.76.1.1-8.2002
Jones, T. R., and Lee, S. W. (2004). An acidic cluster of human cytomegalovirus UL99 tegument protein is required for trafficking and function. J. Virol. 78, 1488–1502. doi: 10.1128/jvi.78.3.1488-1502.2004
Kerry, J. A., Priddy, M. A., Kohler, C. P., Staley, T. L., Weber, D., Jones, T. R., et al. (1997). Translational regulation of the human cytomegalovirus pp28 (UL99) late gene. J. Virol. 71, 981–987. doi: 10.1128/JVI.71.2.981-987.1997
Kim, I. J., Chouljenko, V. N., Walker, J. D., and Kousoulas, K. G. (2013). Herpes Simplex Virus 1 Glycoprotein M and the Membrane-Associated Protein UL11 Are Required for Virus-Induced Cell Fusion and Efficient Virus Entry. J. Virol. 87, 8029–8037. doi: 10.1128/JVI.01181-13
Kim, I. J., Saied, A. A., Chouljenko, V. N., Subramanian, R., and Kousoulas, K. G. (2014). Functional hierarchy of herpes simplex virus type-1 membrane proteins in corneal infection and virus transmission to ganglionic neurons. Curr. Eye Res. 39, 1169–1177. doi: 10.3109/02713683.2014.906626
Klupp, B. G., Granzow, H., Klopfleisch, R., Fuchs, W., Kopp, M., Lenk, M., et al. (2005b). Functional analysis of the pseudorabies virus UL51 protein. J. Virol. 79, 3831–3840. doi: 10.1128/JVI.79.6.3831-3840.2005
Klupp, B. G., Böttcher, S., Granzow, H., Kopp, M., and Mettenleiter, T. C. (2005a). Complex Formation between the UL16 and UL21 Tegument Proteins of Pseudorabies Virus. J. Virol. 79, 1510–1522. doi: 10.1128/JVI.79.3.1510-1522.2005
Kobty, M. (2015). Herpes simplex virus: beyond the basics. Neonatal. Netw. 34, 279–283. doi: 10.1891/0730-0832.34.5.279
Kohler, C. P., Kerry, J. A., Carter, M., Muzithras, V. P., Jones, T. R., and Stenberg, R. M. (1994). Use of recombinant virus to assess human cytomegalovirus early and late promoters in the context of the viral genome. J. Virol. 68, 6589–6597. doi: 10.1128/JVI.68.10.6589-6597.1994
König, P., Svrlanska, A., Read, C., Feichtinger, S., and Stamminger, T. (2021). The Autophagy-Initiating Protein Kinase ULK1 Phosphorylates Human Cytomegalovirus Tegument Protein pp28 and Regulates Efficient Virus Release. J. Virol. 95, e2346–e2320. doi: 10.1128/JVI.02346-20
Kopp, M., Granzow, H., Fuchs, W., Klupp, B., and Mettenleiter, T. C. (2004). Simultaneous deletion of pseudorabies virus tegument protein UL11 and glycoprotein M severely impairs secondary envelopment. J. Virol. 78, 3024–3034. doi: 10.1128/jvi.78.6.3024-3034.2004
Kopp, M., Granzow, H., Fuchs, W., Klupp, B. G., Mundt, E., Karger, A., et al. (2003). The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J. Virol. 77, 5339–5351. doi: 10.1128/jvi.77.9.5339-5351.2003
Koshizuka, T., Kawaguchi, Y., Goshima, F., Mori, I., and Nishiyama, Y. (2006). Association of two membrane proteins encoded by herpes simplex virus type 2, UL11 and UL56. Virus Genes 32, 153–163. doi: 10.1007/s11262-005-6871-7
Koshizuka, T., Kawaguchi, Y., Nozawa, N., Mori, I., and Nishiyama, Y. (2007). Herpes simplex virus protein UL11 but not UL51 is associated with lipid rafts. Virus Genes 5, 571–575. doi: 10.1007/s11262-007-0156-2
Lefkowitz, E. J., Dempsey, D. M., Hendrickson, R. C., Orton, R. J., Siddell, S. G., and Smith, D. B. (2018). Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 46, D708–D717. doi: 10.1093/nar/gkx932
Liu, Y., Cui, Z., Zhang, Z., Wei, H., Zhou, Y., Wang, M., et al. (2009). The tegument protein UL94 of human cytomegalovirus as a binding partner for tegument protein pp28 identified by intracellular imaging. Virology 388, 68–77. doi: 10.1016/j.virol.2009.03.007
Loomis, J. S., Bowzard, J. B., Courtney, R. J., and Wills, J. W. (2001). Intracellular Trafficking of the UL11 Tegument Protein of Herpes Simplex Virus Type 1. J. Virol. 75, 12209–12219. doi: 10.1128/JVI.75.24.12209-12219.2001
Loomis, J. S., Courtney, R. J., and Wills, J. W. (2003). Binding partners for the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 77, 11417–11424. doi: 10.1128/jvi.77.21.11417-11424.2003
Loomis, J. S., Courtney, R. J., and Wills, J. W. (2006). Packaging determinants in the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 80, 10534–10541. doi: 10.1128/JVI.01172-06
MacLean, C. A., Clark, B., and McGeoch, D. J. (1989). Gene UL11 of herpes simplex virus type 1 encodes a virion protein which is myristylated. J. Gen. Virol. 70(Pt 12), 3147–3157. doi: 10.1099/0022-1317-70-12-3147
MacLean, C. A., Dolan, A., Jamieson, F. E., and McGeoch, D. J. (1992). The myristylated virion proteins of herpes simplex virus type 1: investigation of their role in the virus life cycle. J. Gen. Virol. 73(Pt 3), 539–547. doi: 10.1099/0022-1317-73-3-539
Mahmoudian, A., Markham, P. F., Noormohammadi, A. H., and Browning, G. F. (2012). Kinetics of transcription of infectious laryngotracheitis virus genes. Comp. Immunol. Microbiol. Infect. Dis. 35, 103–115. doi: 10.1016/j.cimid.2011.11.001
Mateo, M., Generous, A., Sinn, P. L., and Cattaneo, R. (2015). Connections matter how viruses use cell to cell adhesion components. J. Cell Sci. 128, 431–439. doi: 10.1242/jcs.159400
McGeoch, D. J., Dalrymple, M. A., Davison, A. J., Dolan, A., Frame, M. C., McNab, D., et al. (1988). The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69(Pt 7), 1531–1574. doi: 10.1099/0022-1317-69-7-1531
Meckes, D. G. J. Jr., Marsh, J. A., and Wills, J. W. (2010). Complex mechanisms for the packaging of the UL16 tegument protein into herpes simplex virus. Virology 398, 208–213. doi: 10.1016/j.virol.2009.12.004
Meckes, J. D. G., and Wills, J. W. (2007). Dynamic interactions of the UL16 tegument protein with the capsid of herpes simplex virus. J. Virol. 81, 13028–13036. doi: 10.1128/JVI.01306-07
Metrick, C. M., Koenigsberg, A. L., and Heldweina, E. E. (2020). Conserved Outer Tegument Component UL11 from Herpes Simplex Virus 1 Is an Intrinsically Disordered, RNA-Binding Protein. mBio 11, e810–e820. doi: 10.1128/mBio.00810-20
Mettenleiter, T. C. (2002). Herpesvirus assembly and egress. J. Virol. 76, 1537–1547. doi: 10.1128/jvi.76.4.1537-1547.2002
Mettenleiter, T. C., Klupp, B. G., and Granzow, H. (2009). Herpesvirus assembly: an update. Virus Res. 143, 222–234. doi: 10.1016/j.virusres.2009.03.018
Michael, K., Böttcher, S., Klupp, B. G., Karger, A., and Mettenleiter, T. C. (2006). Pseudorabies virus particles lacking tegument proteins UL11 or pUL16 incorporate less full-length pUL36 than wild-type virus, but specifically accumulate a pUL36 N-terminal fragment. J. Gen. Virol. 87(Pt 12), 3503–3507. doi: 10.1099/vir.0.82168-0
Mocarski, E. S., and Courcelle, C. T. (2001). “Cytomegaloviruses and their replication,” in Fields Virology, 4th Edn, eds D. M. Knipe and P. M. Howley (Philadelphia, PA: Lippincott/The Williams & Wilkins Co.), 2629–2674.
Munger, J., Yu, D., and Shenk, T. (2006). UL26-deficient human cytomegalovirus produces virions with hypophosphorylated pp28 tegument protein that is unstable within newly infected cells. J. Virol. 80, 3541–3548. doi: 10.1128/JVI.80.7.3541-3548
Newcomb, W. W., Booy, F. P., and Brown, J. C. (2007). Uncoating the herpes simplex virus genome. J. Mol. Biol. 370, 633–642. doi: 10.1016/j.jmb.2007.05.023
Nicoll, M. P., Proença, J. T., and Efstathiou, S. (2012). The molecular basis of herpes simplex virus latency. FEMS Microbiol. Rev. 36, 684–705. doi: 10.1111/j.1574-6976.2011.00320.x
Osterrieder, N. (1999). Sequence and initial characterization of the UL10 (glycoprotein M) and UL11 homologous genes of serotype 1 Marek’s disease virus. Arch. Virol. 144, 1853–1863. doi: 10.1007/s007050050710
Owen, D. J., Crump, C. M., and Graham, S. C. (2015). Tegument assembly and secondary envelopment of alphaherpesviruses. Viruses 7, 5084–5114. doi: 10.3390/v7092861
Phillips, S. L., and Bresnahan, W. A. (2012). The human cytomegalovirus (HCMV) tegument protein UL94 is essential for secondary envelopment of HCMV virions. J. Virol. 86, 2523–2532. doi: 10.1128/JVI.06548-11
Phillips, S. L., Cygnar, D., Thomas, A., and Bresnahan, W. A. (2012). Interaction betweenthe human cytomegalovirus tegument proteins UL94 and UL99 is essential forvirus replication. J. Virol. 86, 9995–10005. doi: 10.1128/JVI.01078-12
Qi, X., Yang, X., Cheng, A., Wang, M., Zhu, D., and Jia, R. (2008). Quantitative analysis of virulent duck enteritis virus loads in experimentally infected ducklings. Avian. Dis. 52, 338–344. doi: 10.1637/8120-100207-ResNote.1
Radtke, K., Kieneke, D., Wolfstein, A., Michael, K., Steffen, W., Scholz, T., et al. (2010). Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog. 6:e1000991. doi: 10.1371/journal.ppat.1000991
Resh, M. D. (1999). Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451, 1–16. doi: 10.1016/s0167-4889(99)00075-0
Roizman, B., Knipe, D., and Whitley, R. (2007). “Herpes simplex viruses,” in Fields Virology, 5th Edn, eds D. Knipe and P. Howley (Philadelphia, PA: Lippincott Williams & Wilkins), 2501–2601.
Roller, R. J., Haugo, A. C., Yang, K., and Baines, J. D. (2014). The herpes simplex virus 1 UL51 gene product has cell typespecific functions in cell-to-cell spread. J. Virol. 88, 4058–4068. doi: 10.1128/JVI.03707-13
Sadaoka, T., Serada, S., Kato, J., Hayashi, M., Gomi, Y., Naka, T., et al. (2014). Varicella-zoster virus ORF49 functions in the efficient production of progeny virus through its interaction with essential tegument protein ORF44. J. Virol. 88, 188–201. doi: 10.1128/JVI.02245-13
Sadaoka, T., Yoshii, H., Imazawa, T., Yamanishi, K., and Mori, Y. (2007). Deletion in open reading frame 49 of varicella-zoster virus reduces virus growth in human malignant melanoma cells but not in human embryonic fibroblasts. J. Virol. 81, 12654–12665. doi: 10.1128/JVI.01183-07
Sanchez, V., Sztul, E., and Britt, W. J. (2000). Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-golgi-intermediate compartment. J. Virol. 74, 3842–3851. doi: 10.1128/jvi.74.8.3842-3851
Sarfo, A., Starkey, J., Mellinger, E., Zhang, D., Chadha, P., Carmichael, J., et al. (2017). The UL21 Tegument Protein of Herpes Simplex Virus 1 Is Differentially Required for the Syncytial Phenotype. J. Virol. 91, e01161–e01117. doi: 10.1128/JVI.01161-17
Schimmer, C., and Neubauer, A. (2003). The equine herpesvirus 1 UL11 gene product localizes to the trans-Golgi network and is involved in cell-to-cell spread. Virology 308, 23–36. doi: 10.1016/s0042-6822(02)00060-0
Sefton, B. M., and Buss, J. E. (1987). The covalent modification of eukaryotic proteins with lipid. J. Cell Biol. 104, 1449–1453. doi: 10.1083/jcb.104.6.1449
Seo, J., and Britt, W. J. (2006). Sequence requirements for localization of human cytomegalovirus tegument protein pp28 to the virus assembly compartment and for assembly of infectious virus. J. Virol. 80, 5611–5626. doi: 10.1128/JVI.02630-05
Seo, J., Heo, J. A., and Britt, W. J. (2020). Phosphorylation of tegument protein pp28 contributes to trafficking to the assembly compartment in human cytomegalovirus infection. J. Microbiol. 58, 624–631. doi: 10.1007/s12275-020-0263-5
Seo, J. Y., and Britt, W. J. (2007). Cytoplasmic envelopment of human cytomegalovirus requires the postlocalization function of tegument protein pp28 within the assembly compartment. J. Virol. 81, 6536–6547. doi: 10.1128/JVI.02852-06
Seo, J. Y., and Britt, W. J. (2008). Multimerization of Tegument Protein pp28 within the Assembly Compartment Is Required for Cytoplasmic Envelopment of Human Cytomegalovirus. J. Virol. 82, 6272–6287. doi: 10.1128/JVI.02345-07
Shen, S., Guo, H., and Deng, H. (2014). Murine gammaherpesvirus-68 ORF38 encodes a tegument protein and is packaged into virions during secondary envelopment. Protein Cell 5, 141–150. doi: 10.1007/s13238-013-0005-0
Silva, M. C., Yu, Q. C., Enquist, L., and Shenk, T. (2003). Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 77, 10594–10605. doi: 10.1128/jvi.77.19.10594-10605.2003
Silverman, J. L., Greene, N. G., King, D. S., and Heldwein, E. E. (2012). Membrane requirement for folding of the herpes simplex virus 1 gB cytodomain suggests a unique mechanism of fusion regulation. J. Virol. 86, 8171–8184. doi: 10.1128/JVI.00932-12
Simons, K., and Ikonen, E. (1997). Functional rafts in cell membranes. Nature 387, 569–572. doi: 10.1038/42408
Smith, G. A. (2017). Assembly and Egress of an Alphaherpesvirus Clockwork. Adv. Anat. Embryol. Cell Biol. 223:171. doi: 10.1007/978-3-319-53168-7_8
Starkey, J. L., Han, J., Chadha, P., Marsh, J. A., and Wills, J. W. (2014). Elucidation of the block to herpes simplex virus egress in the absence of tegument protein UL16 reveals a novel interaction with VP22. J. Virol. 88, 110–119. doi: 10.1128/JVI.02555-13
Takakuwa, H., Goshima, F., Koshizuka, T., Murata, T., Daikoku, T., and Nishiyama, Y. (2001). Herpes simplex virus encodes a virion-associated protein which promotes long cellular processes in over expressing cells. Genes Cells 6, 955–996. doi: 10.1046/j.1365-2443.2001.00475.x
Telford, E. A., Watson, M. S., McBride, K., and Davison, A. J. (1992). The DNA sequence of equine herpesvirus-1. Virology 189, 304–316. doi: 10.1016/0042-6822(92)90706-u
Tomtishen, J. P. III (2012). Human cytomegalovirus tegument proteins (pp65, pp71, pp150, pp28). Virol. J. 17:22. doi: 10.1186/1743-422X-9-22
Vittone, V., Diefenbach, E., Triffett, D., Douglas, M. W., Cunningham, A. L., and Diefenbach, R. J. (2005). Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 79, 9566–9571. doi: 10.1128/JVI.79.15.9566-9571.2005
Wolfstein, A., Nagel, C. H., Radtke, K., Döhner, K., Allan, V. J., and Sodeik, B. (2006). The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic 7, 227–237. doi: 10.1111/j.1600-0854.2005.00379.x
Wu, J., Avey, D., Li, W., Gillen, J., Fu, B., Miley, W., et al. (2016). ORF33 and ORF38 of Kaposi’s Sarcoma-Associated Herpesvirus Interact and Are Required for Optimal Production of Infectious Progeny Viruses. J. Virol. 90, 1741–1756. doi: 10.1128/JVI.02738-15
Wu, J., Neill, J. O., and Barbosa, M. S. (2001). Late temporal gene expression from the human cytomegalovirus pp28US (UL99) promoter when integrated into the host cell chromosome. J. Gen. Virol. 82, 1147–1155. doi: 10.1099/0022-1317-82-5-1147
Wu, L., Cheng, A., Wang, M., Jia, R., Yang, Q., Wu, Y., et al. (2020). Alphaherpesvirus Major Tegument Protein VP22: its Precise Function in the Viral Life Cycle. Front. Microbiol. 7:1908. doi: 10.3389/fmicb.2020.01908
Wu, Y., Cheng, A., Wang, M., Yang, Q., Zhu, D., Jia, R., et al. (2012). Complete genomic sequence of Chinese virulent duck enteritis virus CHv strain. J. Virol. 86:5965. doi: 10.1128/JVI.00529-12
Yang, L., Shen, B., Wang, M., Cheng, A., Yang, Q., Wu, Y., et al. (2021). The intracellular domain of duck plague virus glycoprotein E affects UL11 protein incorporation into viral particles. Vet. Microbiol. 257:109078. doi: 10.1016/j.vetmic.2021.109078
Yang, L., Wang, M., and Cheng, A. (2020a). Research progress of herpesvirus UL11 gene and its coding protein. CHIN. J. Prevent. Vet. Med. 41, 868–873.
Yang, L., Wang, M., Zeng, C., Shi, Y., Cheng, A., Liu, M., et al. (2020b). Duck enteritis virus UL21 is a late gene encoding a protein that interacts with pUL16. BMC Vet. Res. 16:8. doi: 10.1186/s12917-019-2228-7
Yang, L., Wang, M., Cheng, A., Yang, Q., Wu, Y., Huang, J., et al. (2022). UL11 Protein Is a Key Participant of the Duck Plague Virus in Its Life Cycle. Front. Microbiol. 12:792361. doi: 10.3389/fmicb.2021.792361
Yang, L., Wang, M., Cheng, A., Yang, Q., Wu, Y., Jia, R., et al. (2019). Innate immune evasion of alphaherpesvirus tegument proteins. Front. Immunol. 10:2196. doi: 10.3389/fimmu.2019.02196
Yeh, P. C., Han, J., Chadha, P., Meckes, D. G. J. Jr., Ward, M. D., and Semmes, O. J. et al (2011). Direct and specific binding of the UL16 tegument protein of herpes simplex virus to the cytoplasmic tail of glycoprotein E. J. Virol. 85, 9425–9436. doi: 10.1128/JVI.05178-11
Yeh, P. C., Meckes, D. G. Jr., and Wills, J. W. (2008). Analysis of the interaction between the UL11 and UL16 tegument proteins of the herpes simplex virus. J. Virol. 82, 10693–10700. doi: 10.1128/jvi.01230-08
You, Y., Cheng, A., Wang, M., Jia, R., Sun, K., Yang, Q., et al. (2017). The suppression of apoptosis by α-herpesvirus. Cell Death Dis. 8:e2749. doi: 10.1038/cddis.2017.139
Yuan, G. P., Cheng, A. C., Wang, M. S., Liu, F., Han, X. Y., Liao, Y. H., et al. (2005). Electron microscopic studies of the morphogenesis of duck enteritis virus. Avian. Dis. 49, 50–55. doi: 10.1637/7237-071004r
Zeev-Ben-Mordehai, T., Hagen, C., and Grünewald, K. (2014). A cool hybrid approach to the herpesvirus “life” cycle. Curr. Opin. Virol. 5, 42–49. doi: 10.1016/j.coviro.2014.01.008
Keywords: UL11, interaction, herpesvirus, viral cycle, secondary envelopment
Citation: Yang L, Wang M, Cheng A, Yang Q, Wu Y, Huang J, Tian B, Jia R, Liu M, Zhu D, Chen S, Zhao X, Zhang S, Ou X, Mao S, Gao Q and Sun D (2022) Features and Functions of the Conserved Herpesvirus Tegument Protein UL11 and Its Binding Partners. Front. Microbiol. 13:829754. doi: 10.3389/fmicb.2022.829754
Received: 06 December 2021; Accepted: 26 April 2022;
Published: 03 June 2022.
Edited by:
Keith William Jarosinski, University of Illinois at Urbana-Champaign, United StatesReviewed by:
Hongyan Guo, Louisiana State University Health Science Center Shreveport, United StatesWalter Fuchs, Friedrich Loeffler Institute, Germany
Copyright © 2022 Yang, Wang, Cheng, Yang, Wu, Huang, Tian, Jia, Liu, Zhu, Chen, Zhao, Zhang, Ou, Mao, Gao and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anchun Cheng, Y2hlbmdhbmNodW5AdmlwLjE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Linjiang Yang
Linjiang Yang Mingshu Wang
Mingshu Wang Anchun Cheng
Anchun Cheng Qiao Yang
Qiao Yang Ying Wu
Ying Wu Juan Huang
Juan Huang Bin Tian
Bin Tian Renyong Jia
Renyong Jia Mafeng Liu
Mafeng Liu Dekang Zhu
Dekang Zhu Shun Chen
Shun Chen Xinxin Zhao
Xinxin Zhao Shaqiu Zhang
Shaqiu Zhang Xumin Ou1,2,3
Xumin Ou1,2,3 Sai Mao
Sai Mao Qun Gao
Qun Gao Di Sun
Di Sun