- 1Key Laboratory of Marine Drugs of Ministry of Education, Shandong Provincial Key Laboratory of Glycoscience and Glycotechnology, School of Medicine and Pharmacy, Ocean University of China, Qingdao, China
- 2Laboratory for Marine Drugs and Bioproducts, Pilot National Laboratory for Marine Science and Technology, Qingdao, China
Bacteroides thetaiotaomicron, one of the most eminent representative gut commensal Bacteroides species, is able to use the L-fucose in host-derived and dietary polysaccharides to modify its capsular polysaccharides and glycoproteins through a mammalian-like salvage metabolic pathway. This process is essential for the colonization of the bacteria and for symbiosis with the host. However, despite the importance of fucosylated proteins (FGPs) in B. thetaiotaomicron, their types, distribution, and functions remain unclear. In this study, the effects of different polysaccharide (corn starch, mucin, and fucoidan) nutrition conditions on newly synthesized FGPs expressions and fucosylation are investigated using a chemical biological method based on metabolic labeling and bioorthogonal reaction. According to the results of label-free quantification, 559 FGPs (205 downregulated and 354 upregulated) are affected by the dietary conditions. Of these differentially expressed proteins, 65 proteins show extremely sensitive to polysaccharide nutrition conditions (FGPs fold change/global protein fold change ≥2.0 or ≤0.5). Specifically, the fucosylation of the chondroitin sulfate ABC enzyme, Sus proteins, and cationic efflux system proteins varies significantly upon the addition of mucin, corn starch, or fucoidan. Moreover, these polysaccharides can trigger an appreciable increase in the fucosylation level of the two-component system and ammonium transport proteins. These results highlight the efficiency of the combined metabolic glycan labeling and bio-orthogonal reaction in enriching the intestinal Bacteroides glycoproteins. Moreover, it emphasizes the sensitivity of Bacteroides fucosylation to polysaccharide nutrition conditions, which allows for the regulation of bacterial growth.
Introduction
Bacteroides are the most abundant Gram-negative obligate anaerobic bacteria in the distal small intestine and colon of mammals (Moore and Holdeman, 1974; Ushijima et al., 1983; Oren et al., 2016). In fact, each gram of human feces contains 1010–1011 Bacteroides cells (Flint et al., 2008). Considering the essential role of Bacteroides in maintaining human health and in gut microbe–host symbiosis, these species have attracted considerable attention. Specifically, Bacteroides thetaiotaomicron, a representative gut commensal Bacteroides frequently detected in the feces of healthy adults, has been thoroughly investigated (Xu et al., 2003). Despite being an opportunistic pathogen, B. thetaiotaomicron normally forms a harmonious and mutually beneficial symbiotic relationship with the host during its long-term evolution (Brook et al., 2013; D’Argenio and Salvatore, 2015). In particular, B. thetaiotaomicron helps the host degrade complex carbohydrates (Donaldson et al., 2016), suppress obesity (Ley et al., 2006), ameliorate colon inflammation (Delday et al., 2019), regulate colonic neuronal innervation and neurogenic function (Aktar et al., 2020), and even resist the invasion of pathogens (Xu and Gordon, 2003).
As an important component of cell surface glycoconjugates in gut microbes and host intestinal mucosa, L-fucose plays an important role in Bacteroides colonization and in gut microbe-host symbiosis (Pickard and Chervonsky, 2015; Garber et al., 2021) and it mainly exists in the form of an O-glycosylation protein (Martens et al., 2008; Fletcher et al., 2009; González-Morelo et al., 2020). There are two pathways in Bacteroides for biosynthesizing GDP-fucose, the donor for fucosyltransferase-catalyzed incorporation of fucose into bacterial capsular polysaccharides and fucosylated proteins (FGPs). One is the novo pathway, which is characterized by conversion of GDP-mannose to GDP-fucose by GDP-mannose dehydratase and GDP-fucose synthetase. The other one is the salvage pathway, in which exogenous fucose also can be transformed to GDP-fucose by Fkp, a bifunctional enzyme with both fucokinase and pyrophosphorylase activity (Coyne et al., 2005). Among different gut Bacteroides, B. thetaiotaomicron can stimulate the host to increase the α-1,2 fucosylation level through a series of cascade reactions, which inhibits the growth of harmful bacteria and benefits the growth of symbiotic bacteria when the host is in sickness (Goto et al., 2014; Pickard et al., 2014; Kostopoulos et al., 2021). Therefore, fucosylation is an important factor for the harmonious symbiosis between Bacteroides and the host. As the main product of fucosylation, FGPs may play critical roles in this process.
Compared to other Bacteroides species, B. thetaiotaomicron can more efficiently degrade various types of polysaccharides that are ultimately used to supply carbon and energy to the bacteria. The type strain VPI-5482 has 88 polysaccharide utilization loci (PULs) which cover 18% of its genome, and it encodes the proteins involved in sensing, importing, degrading, and regulating dietary carbohydrates in the colonic ecosystem (Xu et al., 2003; Martens et al., 2008, 2009; Rogers et al., 2013). Interestingly, PULs are often implicated with the ECF-type sigma factor in an environment response mechanism that co-regulates the ability of the bacteria to utilize nutrition (Donaldson et al., 2016). For example, starch is easily degraded by B. thetaiotaomicron via the starch utilization system (Sus) that includes homologues of SusC, SusD, and various carbohydrate-active enzymes (CAZymes) (Foley et al., 2018; Stevenson et al., 2021). In addition to starch, these homologs and enzymes can break down a variety of polysaccharides that are supplied by the diet or generated by the host (El Kaoutari et al., 2013). Similarly, fucoidan, a complex sulfated polysaccharide, cannot be digested by the human intestine without the help of intestinal microbiota (Morya et al., 2012). Previous studies have shown that fucoidan in Ascophyllum nodosum, Laminaria japonica, and sea cucumbers increases the content of intestinal Bacteroides (Shang et al., 2018; Hu et al., 2019; Li et al., 2019). Moreover, when dietary carbohydrates are deficient (e.g., during fasting or in the case of low-fiber diets), B. thetaiotaomicron changes its transcriptional profile in order to produce multiple fucosidases that cleave fucose from the intestinal mucin of the host, which increases the level of fucose in the gut lumen (Koropatkin et al., 2009). The cleaved fucose is then utilized by gut microorganisms. For example, B. thetaiotaomicron transforms fucose into glycoproteins, which promotes the adaptation of the bacterial species to the intestinal environment (Sonnenburg et al., 2005). The ability of B. thetaiotaomicron to synthesize its own fucose glycoconjugates using host-derived and dietary polysaccharides raises questions regarding the response of FGPs to nutrient variations and the functional roles of these FGPs in mediating a symbiotic relationship with the host.
The traditional strategies for glycoprotein enrichment, including the methods based on lectins and antibodies, have been successfully used to characterize prokaryotic glycoproteomes (Gonzalez-Zamnorano et al., 2009; Fredriksen et al., 2013; Leon et al., 2015). For example, Comstock and co-workers (Fletcher et al., 2009, 2011) determined 20 proteins in B. fragilis that were fucosylated using Aleuria aurantia lectin (AAL). The confirmed glycoproteins have putative roles in crucial cellular processes including cell division, chromosomal segregation, signaling processes, chaperone functions, protein-protein interactions, and peptidase/protease reactions (Fletcher et al., 2011). However, despite the efficiency of lectin- and antibody-based methods, they are not free of limitations. Most importantly, these methods are limited by the inability to detect newly glycosylated proteins in response to external stimulation (Longwell and Dube, 2013; Parker and Pratt, 2020). This is due to the fact that lectins label all of the existing glycoproteins that bear their glycan ligands. In the recent years, a powerful strategy for discovering new glycoproteins in mammals and bacteria has been proposed (Hong et al., 2019; Inoue et al., 2019; Zhou et al., 2019; Pedowitz et al., 2020). This strategy is based on the combination of metabolic oligosaccharide engineering (MOE) (Dube and Bertozzi, 2003) and bioorthogonal reactions, such as Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC) (Kolb et al., 2001; Sletten and Bertozzi, 2009; Neumann et al., 2020). Briefly, the strategy entails the incorporation of an unnatural monosaccharide with an active group into the specific glycan via the biological salvage metabolic pathway. The incorporated monosaccharide is then reacted with a probe comprising complementary and reporter groups through a bio-orthogonal reaction. This allows for the labeling of specific glycoproteins that can thus, be selectively analyzed using a variety of techniques, such as imaging, gel-based assays, and mass spectrometry (MS) (Riley et al., 2021). This strategy has been successfully applied in the identification and quantification of newly synthesized glycoproteins in many systems, including hippocampal synapses, cardiac hypertrophy of live mice, secretome of activated Jurkat cells, and even gut bacteria (Liu et al., 2009; Champasa et al., 2013; Rong et al., 2014; Schanzenbächer et al., 2016; Witzke et al., 2017). MOE and bioorthogonal reaction have also been used to label glycoconjugates in various symbiotic anaerobic bacteria, including B. fragilis, to describe the distribution and colonization of these bacteria along the intestine (Besanceney-Webler et al., 2011; Geva-Zatorsky et al., 2015).
To the best of our knowledge, the FGPs in B. thetaiotaomicron have not yet been identified, and their types, distribution, and functions remain unknown.
In this study, we utilized MOE and CuAAC to selectively label, enrich and identify the FGPs in B. thetaiotaomicron for the first time. The effects of different nutritional conditions on FGPs expression patterns are also determined by coupling metabolic glycan labeling and label-free quantification. The results could help to preliminarily clarify the underlying molecular biological mechanisms of FGPs in promoting intestinal colonization and mediating the symbiotic relationship with the host.
Materials and Methods
Metabolic Labeling of Fucosylated Glycan in Bacteroides thetaiotaomicron
Bacteroides thetaiotaomicron VPI-5482 (purchased from China General Microbiological Culture Collection Center) was incubated under anaerobic conditions at 37°C for 24 h in modified anaerobic minimal medium (AMM) (Varel and Bryant, 1974; Baughn and Malamy, 2002) with or without 200 μM alkynyl-fucose (FucAl) [synthesized as previously described (Schneider et al., 2018)], then the cells were collected for flow cytometry or FGPs enrichment. For the comparative proteomic analysis of FGPs under different carbohydrate nutritional conditions, B. thetaiotaomicron VPI-5482 was incubated in AMM containing 0.5% glucose (Sigama, United States) in anaerobic conditions at 37 °C for about 24 h and grew to the middle logarithmic growth phase (OD600 nm = 0.5). The bacterial culture medium was divided into four groups: G (glucose), M (mucin), S (starch), and F (fucoidan). G group is the control, and no additional carbohydrate was added to this group. Mucin from porcine stomach (type II) (Sigma, St. Louis, MO, United States), corn starch (Solarbio, China) and fucoidan from L. japonica (Shandong Jiejing, China) were added to the other three corresponding groups to a final concentration of 0.25% (w/v), respectively. 200 μM FucAl was added at the same time. Cells were collected after culturing under anaerobic conditions at 37°C for 3 h. All groups mentioned above were done in three replicates.
Labeling of Fucosylated Proteins With Biotinylated Probes via Cu(I)-Catalyzed Azide-Alkyne Cycloaddition in Cell Lysates
Bacteroides thetaiotaomicron cells were harvested by centrifugation (5,000g, 10 min), washed two times with PBS and resuspended in lysis buffer (1% NP-40, 150 mM NaCl, Roche protease inhibitor, 100 mM sodium phosphate pH 7.5). Cells were intermittently vortexed and sonicated for 20 min at 4°C. The supernatant was obtained by centrifugation (12,000g, 10 min, 4°C) and the protein concentration was determined by BCA protein assay kit (Thermo Fisher Scientific, Waltham, MA, United States). Then cell lysates were diluted to 2 mg/mL and reacted with 100 μM biotin-picolyl-azide (Click Chemistry Tools, Scottsdale, AZ, United States) in the reaction solution containing premixed BTTAA (Click Chemistry Tools, Scottsdale, AZ, United States)-CuSO4 complex ([BTTAA]: [CuSO4] = 1 mM: 0.5 mM) and 2.5 mM freshly prepared sodium ascorbate for 1.5 h with agitation (25 °C, 1200 rpm).
Streptavidin Enrichment of Biotinylated Proteins
The biotin-labeled lysates were precipitated by 8 volumes of methanol at -40°C overnight. The precipitated proteins were collected by centrifugation at 5,000 g for 15 min at -10°C, washed with cold methanol for three times and resolubilized with 1.2% SDS/PBS. The solubilized proteins were incubated with streptavidin agarose resin (100 μL of slurry, Thermo Fisher Scientific, Waltham, MA, United States) in 0.2% SDS/PBS at RT for 3 h with rotation. After washing the beads with 0.2% SDS/PBS for one time, PBS for three times, and ddH2O for three times sequentially, biotinylated proteins were eluted from the beads by treating with 2 × SDS PAGE loading buffer in the presence of 2 mg/mL biotin for 10 min at 95°C to avoid streptavidin contamination (Cheah and Yamada, 2017). Excess biotin was removed by ultrafiltration.
Flow Cytometry
Bacteroides thetaiotaomicron cells were harvested and resuspended in cold PBS. For cells treated with CuAAC, 100 μL reactions were setup containing 5 × 107 cells, 100 μM biotin-picolyl-azide, 2.5 mM sodium ascorbate and BTTAA-CuSO4 complex ([BTTAA]:[CuSO4] = 360 μM: 60 μM). After 10 min reaction the cells were washed 3× with cold PBS and incubated in 1 μg/mL Streptavidin Alexa Fluor 488 conjugate (Thermo Fisher Scientific, Waltham, MA, United States) at 4°C for 30 min. Then the cells were washed 3× in cold PBS and applied to ACEA NovoCyte benchtop flow cytometer (Acea Biosciences, San Diego, CA, United States) using a 488 nm argon laser. Flow cytometry data were analyzed using NovoExpress.
Verification of Fucosylation of Candidate Proteins
Escherichia coli S17-1λpir chemically competent cells containing the Rp4-2 plasmid (Beijing Zoman Biotechnology, China) were used as the donor for mobilization of plasmids to Bacteroides strains. E. coli-Bacteroides shuttle expression plasmid pFD340 (Whitehead and Hespell, 1990) was a gift from Dr. Smith and Dr. Rocha (East Carolina University). A 3.9 kb fragment of Q8A818, containing the coding regions and 1500 bp of upstream and 18 bp of 6× His tag gene at the C terminus was amplified from the B. thetaiotaomicron genome, and ligated to pFD340. Similarly, A 4.3 kb fragment of Q8A0P4 was amplified and ligated to BamHI and NarI digested pFD340. All primers were designed using Primer Premier and shown below. Underlined nucleotides indicate engineered restriction sites, and lowercase letters represent homology arm sequences.
Q8A818
F: 5′-agaattgactctagaggatccTCGTTTTTGGGGACGTT TGT-3′
R1: 5′-TTAATGGTGATGGTGATGATGTTTCTTTATAAG ATTAGCAATAGAAGCA-3′
R2: 5′-gaaaataccgcatcaggcgccTTAATGGTGATGGTGATG ATGTTTC-3′
Q8A0P4
F: 5′-agaattgactctagaggatccTTCTTATCATGTTGCATAA ATGCG-3′
R: 5′-gaaaataccgcatcaggcgccTTAATGGTGATGGTGATGA TGGAAACGCATAGGTCTGC-3′
Subclones were obtained after transformation of E. coli S17-1λpir chemically competent cells (containing the Rp4-2 plasmid) with the ligation mixtures. The resulting plasmids, pFD340-Q8A818 and pFD340-Q8A0P4, were mobilized into B. thetaiotaomicron by E. coli-to-Bacteroides matings as described previously (Shoemaker et al., 1985; Strand et al., 2014; Liou et al., 2020). Recombinant B. thetaiotaomicron was incubated in brain–heart infusion medium (BHI) (Thompson and Malamy, 1990) with and without 200 μL FucAl (containing 10 μg/mL Ermr) under anaerobic conditions at 37°C to the logarithmic growth phase of bacteria. Cell lysates were prepared and reacted with biotin-picolyl-azide via CuAAC, followed by streptavidin enrichment as described above.
Western Blot Analysis
Proteins separated by SDS-PAGE were transferred to PVDF membrane. The membrane was incubated with blocking buffer [5% non-fat milk in TBST (Tris buffered saline with 0.1% Tween-20, pH 7.5)] for 1 h at room temperature (RT). The blocked membrane was incubated with HRP-anti-biotin antibody (Jackson ImmunoResearch, West Grove, PA, United States) (1:100,000) or an HRP-anti-His (27E8) antibody (Cell Signaling Technology, Danvers, MA, United States) (1:1000) in blocking buffer overnight at 4°C, washed three times with TBST and visualized with ECL Western blotting detection reagents (Thermo Fisher Scientific, Waltham, MA, United States). Odyssey FC imaging system (LI-COR, United States) was used to detect the chemiluminescence. For Aleuria Aurantia Lectin (AAL) blot, the membrane was blocked in 5% bovine serum albumin (BSA) (Sigma, St. Louis, MO, United States) for 1 h at RT and incubated with biotinylated-AAL (Vector Laboratories, Burlingame, CA, United States) (1:500) in PBS for 1 h at RT. After being washed for three times with TBST, the membrane was incubated with HRP-anti-biotin antibody and visualized.
Label-Free Comparative Proteomic Quantitative Analysis
The total protein or enriched FGPs of various groups were reduced with 5 mM TCEP for 30 min at 37°C, and alkylated with 15 mM IAA for 30 min in the dark at RT. Each sample was trypsinized by the filter aided proteome preparation (FASP) method. The peptides were desalted by C18 Cartridge, lyophilized and reconstituted by 40 μL of 0.1% formic acid solution.
Peptides were submitted to label-free differential proteomic analysis. Each sample was separated with a nanoliter flow rate HPLC liquid system Easy nLC. The mobile phases consisted of a 0.1% (v/v) formic acid aqueous solution (A) and 0.1% (v/v) formic acid in acetonitrile/water (84% acetonitrile) (B). The column was equilibrated with 95% solution A. Samples were loaded from the autosampler to the loading column (Thermo Scientific Acclaim PepMap100, 100 μm × 2 cm, nanoViper C18), and passed through the analytical column (Thermo scientific EASY column, 75 μm × 10 cm, C18-A2), with a flow rate of 300 nL/min. Samples were chromatographed and analyzed by Q-Exactive mass spectrometer. Detection parameters were used as follows: detection method (positive ion); scanning range of the precursor ion (300–1800 m/z); resolution of the first-order mass spectrum (70,000 at 200 m/z); automatic gain control target (1e6); maximum IT (50 ms); dynamic exclusion time (60.0 s). Mass-to-charge ratios of peptides and peptide fragments were collected according to the following methods: fragment maps taken after each full scan (20); MS2 activation type (HCD); isolation window (2 m/z); MS2 resolution (17,500 at 200 m/z); normalized collision energy (30 eV); underfill (0.1%).
MaxQuant software (version 1.5.3.17) was used to the database searching against UniProt -Bacteroides thetaiotaomicron (strain ATCC 29148/DSM 2079/NCTC 10582/E50/VPI-5482) and FASTA database (2019_07 release, 4783 reviewed entries). Searching parameters were used as follows: max missed cleavages (2); fixed modification, carbamidomethyl (C); variable modifications, oxidation (M); MS/MS tolerance (20 ppm); protein and peptide FDR (≤0.01); peptides used for protein quantification (razor and unique peptides); time window (2 min); protein quantification (LFQ/iBAQ). Groups M, S, and F were compared with group G, and the statistical analysis of T test and FDR (Benjamini-Hochberg) were performed. A fold change cut-off of ≥2 with p-value < 0.05 was used as threshold to define significantly changed proteins.
Bioinformatic Analysis of Protein Identifications
Blast2Go was used to annotate the GO functions of the FGPs. GO function enrichment analysis was performed by Fisher’s exact test method. KEGG was used to perform pathway-related analysis on the FGPs. The unit was KEGG pathway, and the total proteins identified were used as the background. Fisher’s exact test was used to analyze and calculate the significance level of protein enrichment in each pathway to determine the metabolic and signal transduction pathways that were significantly affected. In addition to the GO and KEGG annotation and enrichment analysis of the differential proteins according to the above methods, cluster analysis and protein-protein interaction of these proteins were also performed. The quantitative information of target proteins was normalized. The ComplexHeatmap R package (R Version 3.4) was used to classify the two dimensions of the sample and protein expression levels to generate a hierarchical clustering heatmap. The protein–protein interaction of differential proteins was conducted on the STRING database1 and Cytoscape software.
Results
Labeling and Enrichment of Fucosylated Proteins in Bacteroides thetaiotaomicron
The strategy adopted in this study for FGPs labeling in B. thetaiotaomicron is based on previous research (Besanceney-Webler et al., 2011). First, the fucose analog bearing a terminal alkynyl substituent at C-6 position, FucAl, was introduced into the culture media, and was metabolically incorporated into the glycoconjugates of B. thetaiotaomicron. Then, the alkyne-modified glycoproteins in the lysate solutions were chemically reacted with biotin-picolyl-azide via CuAAC, and then were enriched by streptavidin beads. This method allows for the assessment of the labeled FGPs using Western blot and flow cytometry analyses (Figure 1A). To evaluate the biocompatibility of FucAl, B. thetaiotaomicron was anaerobically cultured in minimal medium, in the presence or absence of 200 μM FucAl or fucose. The obtained growth curve shows that the addition of FucAl does not adversely affect the proliferation of B. thetaiotaomicron, even after 36 h, which suggests that it is biocompatible (Figure 1B). Moreover, based on the results of the Western blot and flow cytometry experiments, the added FucAl (200 μM) does not appreciably induce the hyperfucosylation of proteins and has no effect on the expression level of FGPs within 24 h. This confirms the biocompatibility of FucAl and indicates that it may be used to label FGPs (Supplementary Figure 1).
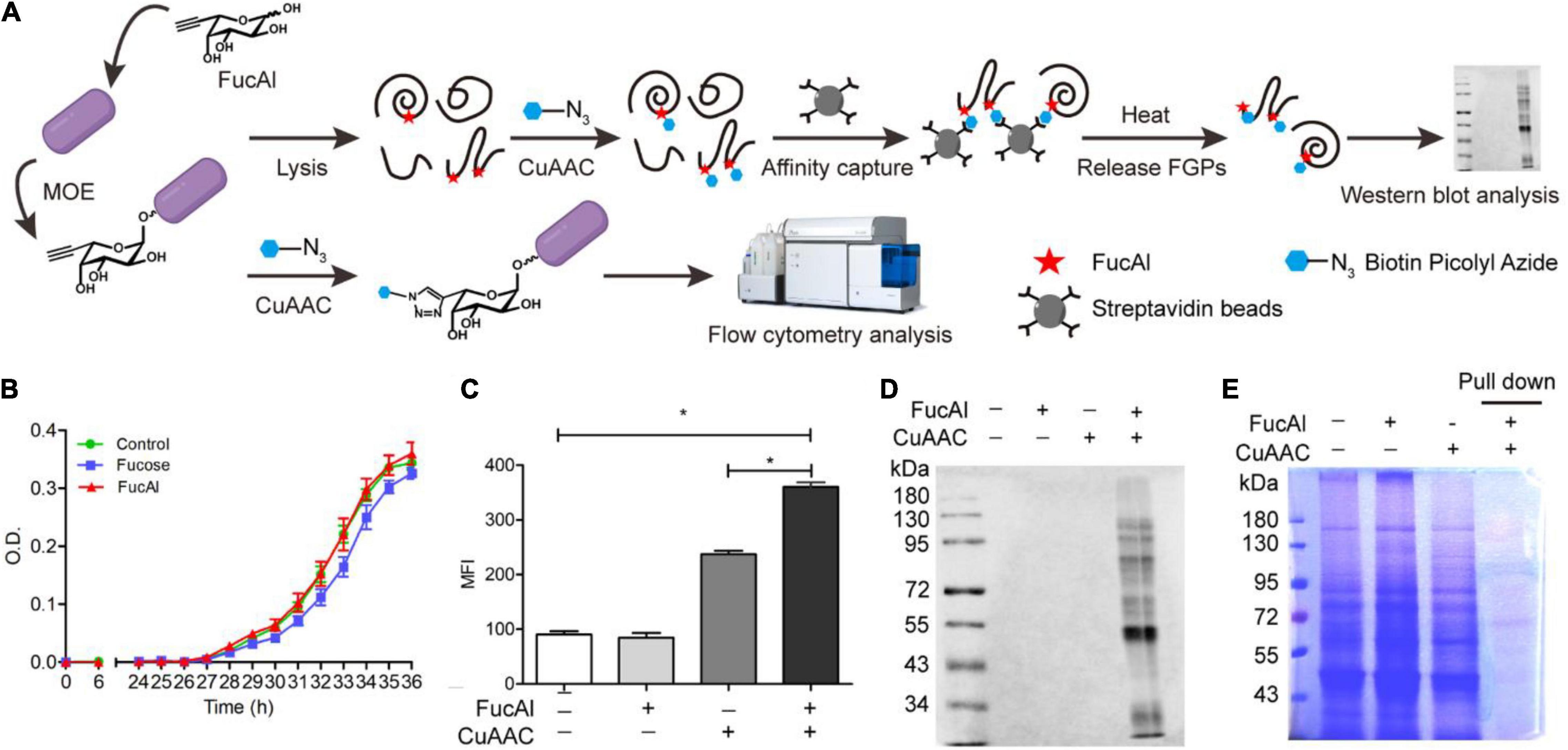
Figure 1. Detectionof B. thetaiotaomicron’s FGPs by metabolic labeling and CuAAC. (A) Workflow for labeling, enrichment and analysis of FGPs in B. thetaiotaomicron. (B) Evaluation of biocompatibility of FucAl in B. thetaiotaomicron by bacterial growth curve. (C) Flow cytometry analysis of labeling efficiency of FGPs on the cell surface of B. thetaiotaomicron. (D) Western blot analysis of labeling of B. thetaiotaomicron’s FGPs by HRP-anti-biotin antibody. (E) Coomassie brilliant blue staining of total proteins and the enriched FGPs. Errors were based on triplicate runs in one experiment. *P < 0.05.
Considering that B. thetaiotaomicron and B. fragilis share a common fucose acquisition system (Xu et al., 2003; Coyne et al., 2005), it was assumed that FucAl can also be applied in the labeling of B. thetaiotaomicron glycoproteins. To verify the labeling of B. thetaiotaomicron FGPs by FucAl and CuAAC, the biotinylated glycoconjugates on the bacterial surface were analyzed by flow cytometry using the streptavidin-Alexa Fluor 488. As expected, the FucAl- and CuAAC-treated B. thetaiotaomicron exhibit high intensity fluorescence, whereas the untreated cells show minor fluorescence (Figure 1C). These biotinylated FGPs were also detected in the lysates by Western blotting with HRP-anti-biotin (Figure 1D). After that, biotinylated FGPs were almost completely captured by streptavidin beads and eluted effectively (Supplementary Figure 2), and based on SDS-PAGE analysis of the eluent, the enriched FGPs account for only a small portion of total proteins (Figure 1E).
Bacteroides thetaiotaomicron Culturing and Fucosylated Proteins Profiling Under Different Polysaccharide Nutrition Conditions
Considering the importance of FGPs in maintaining the growth and survival of Bacteroides, it is important to assess the effects of different polysaccharide nutrition conditions on the expression of newly synthesized FGPs in B. thetaiotaomicron. For this purpose, we next sought to apply FucAl as the probe to investigate it. B. thetaiotaomicron was cultured in a normal glucose medium up to the mid-logarithmic phase with a relatively strong metabolic state. Then, FucAl and different types of polysaccharides, including those derived from dietary land plants (corn starch), dietary marine algea (fucoidan), and the host (porcine mucin-type O-glycans), were added to the cultures (Figure 2A). FucAl was used to label the newly fucosylated proteins in B. thetaiotaomicron within 3 h. Actually, very little of these added polysaccharides was used as the carbon sourse by the bacteria in the short period of time. Consequently, in this case, B. thetaiotaomicron was triggered by the newly added polysaccharides, and the changes of newly synthesized FGPs were due to the response to the changes of nutrient conditions, rather than the utilization of these polysaccharides. Finally, the total proteins and FGPs in these cultures were separately collected. As shown in Figure 2B, bacterial growth reaches a maximum at almost the same time, irrespective of the polysaccharide nutrition condition, and group M presents the highest amount of bacteria at the end of the exponential phase. Based on SDS-PAGE analysis, the expression patterns of total protein are similar in all groups (Figure 2C). In contrast, the expression patterns of FGPs were significantly different according to the Western blot result (Figure 2D). This stimulated our interest to further explore the expression differences of FGPs via comparative proteomics.
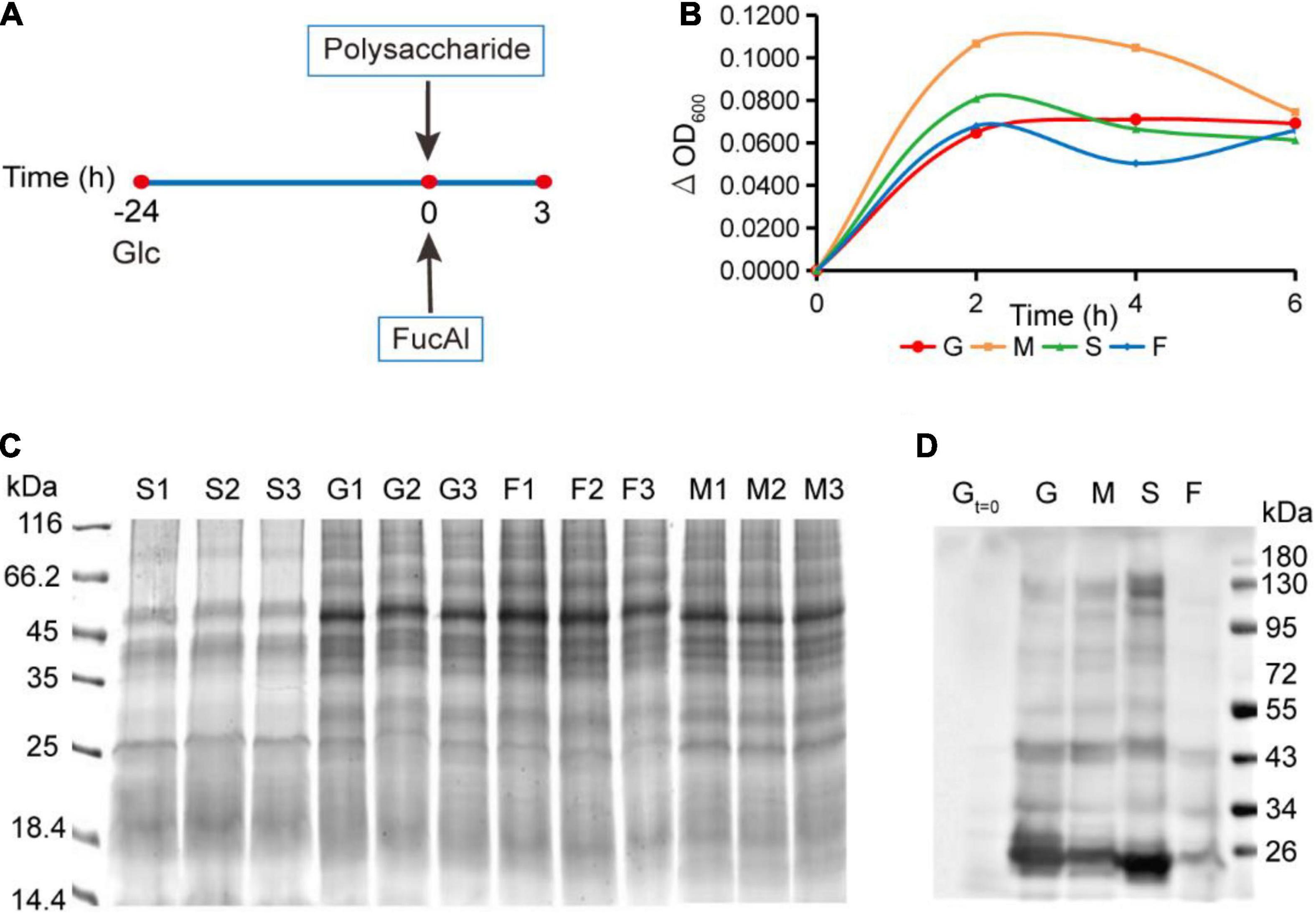
Figure 2. Culture of B. thetaiotaomicron and enrichment of FGPs in different polysaccharide nutrition conditions. (A) Scheme for culture of B. thetaiotaomicron. B. thetaiotaomicron was incubated in AMM (0.5% glucose) under anaerobic conditions at 37°C for 24 h and grew to the middle logarithmic growth phase. No additional carbohydrate was added to the G group. Mucin (M group), starch (S group), and fucoidan (F group) were added into the corresponding group to a final concentration of 0.25% (m/v), respectively. 200 μM FucAl was added at the same time. After 3 h, bacterial cultures were collected. (B) Growth curves of B. thetaiotaomicron under different polysaccharide nutrition conditions. G, glcucose; M, mucin; S, corn starch; F, fucoidan. (C) Gel imaging of global proteins in each experiment. (D) Western blot analysis of enriched FGPs in each group by anti-biotin antibody.
Fucosylated Proteins Identification by Label-Free Quantitative Mass Spectrometry Analysis
The above results indicate that the addition of exogenous polysaccharides has a greater impact on FGPs expression in B. thetaiotaomicron than on the level of total proteins. In order to identify the FGPs in response to different nutritional conditions, the methods of metabolic labeling and CuAAC with label-free comparative quantification were applied (Figure 3A). B. thetaiotaomicron was cultured as described in Figure 2A. At 3 h after polysaccharides treatment, bacterial cultures were collected, and the differences in expression patterns of total proteins and streptavidin enriched FGPs were both studied by label-free quantification (Figure 3A). The false discovery rate (FDR) for peptides and proteins was adjusted to less than 1%, and the mass deviations of all the identified peptides were mainly within 10 ppm. Each MS2 map presented an ideal Andromeda score, with 82.55% of the peptides scoring above 60 points. The median score of all the of the peptides was found to be 101.62 points. The peptide number distribution and peptide sequence coverage further confirm the high quality, accuracy, and reliability of the peptides identified herein (Supplementary Figure 3). The experiments were performed in triplicates, and the results indicate high reproducibility in terms of the identified FGPs (Figure 3B).
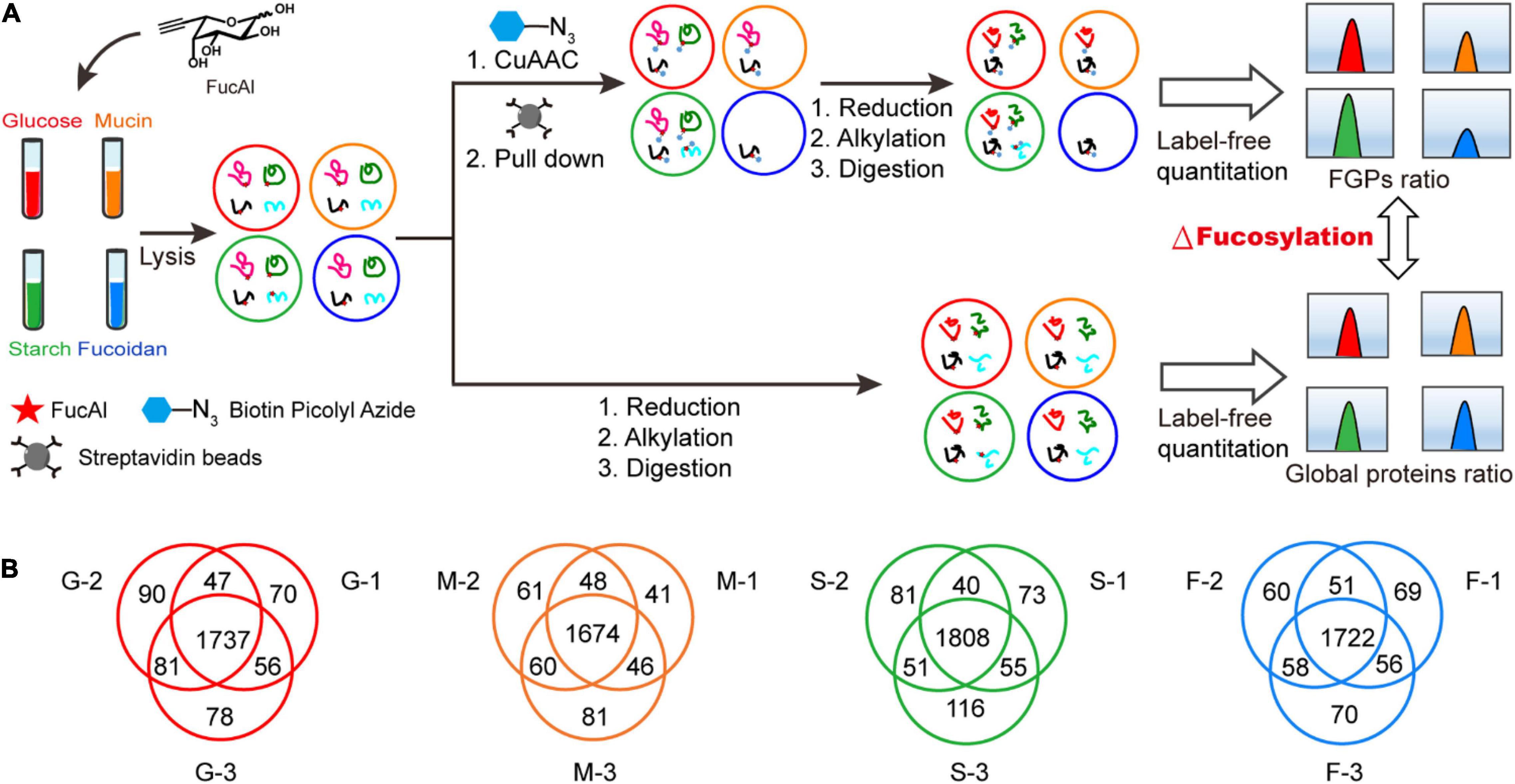
Figure 3. Label-free quantitative comprehensive proteomes of FGPs. (A) Workflow for label-free quantitative comprehensive proteomes of FGPs. (B) Venn diagrams of three repeated experiments in each group.
Fucosylation Verification of Candidate Fucosylated Proteins
To verify the fucosylation of the identified FGPs, two proteins (Q8A818 and Q8A0P4) with high scores and LFQ intensities were selected as candidate compounds. Q8A818 is a capsular polysaccharide transport protein that can transport polysaccharides to the capsule of B. thetaiotaomicron, resulting in the formation of loose mucus substances on the surface of Bacteroides cell walls. These substances are essential for the survival of the bacteria (Hsieh et al., 2020). Q8A0P4 is a TonB-dependent outer membrane receptor that is mainly responsible for the precipitation of ferritin in Gram-negative bacteria and for the intake of various nutrients, such as iron complexes, vitamin B12, and carbohydrates. Studies have shown that this type of receptor acts as an adhesion molecule that binds to the host fibronectin, and that it is related to the pathogenicity of Bacteroides (Koebnik, 2005; Miethke and Marahiel, 2007; Schauer et al., 2008; Pauer et al., 2009). Recombinant plasmids containing 6 × His-tagged recombinant proteins were constructed by homologous recombination and transferred to B. thetaiotaomicron by E. coli-to-Bacteroides mating. The total FGPs in the bacteria were then labeled as described above and subjected to pull-down with anti-biotin antibody. The western blot results illustrated in Figure 4 show that both the control and alkyne-labeled samples present His-tagged protein signals, which suggests that the candidate proteins had been successfully expressed in B. thetaiotaomicron, and that the addition of FucAl did not affect their expression. Compared with control, the enriched FGPs in alkyne-labeled samples show obvious signals of anti-biotin and anti-His, which suggests that the total FGPs contained Q8A818 and Q8A0P4. All the above results confirmed that the detected Q8A818 and Q8A0P4 proteins were indeed fucosylated.
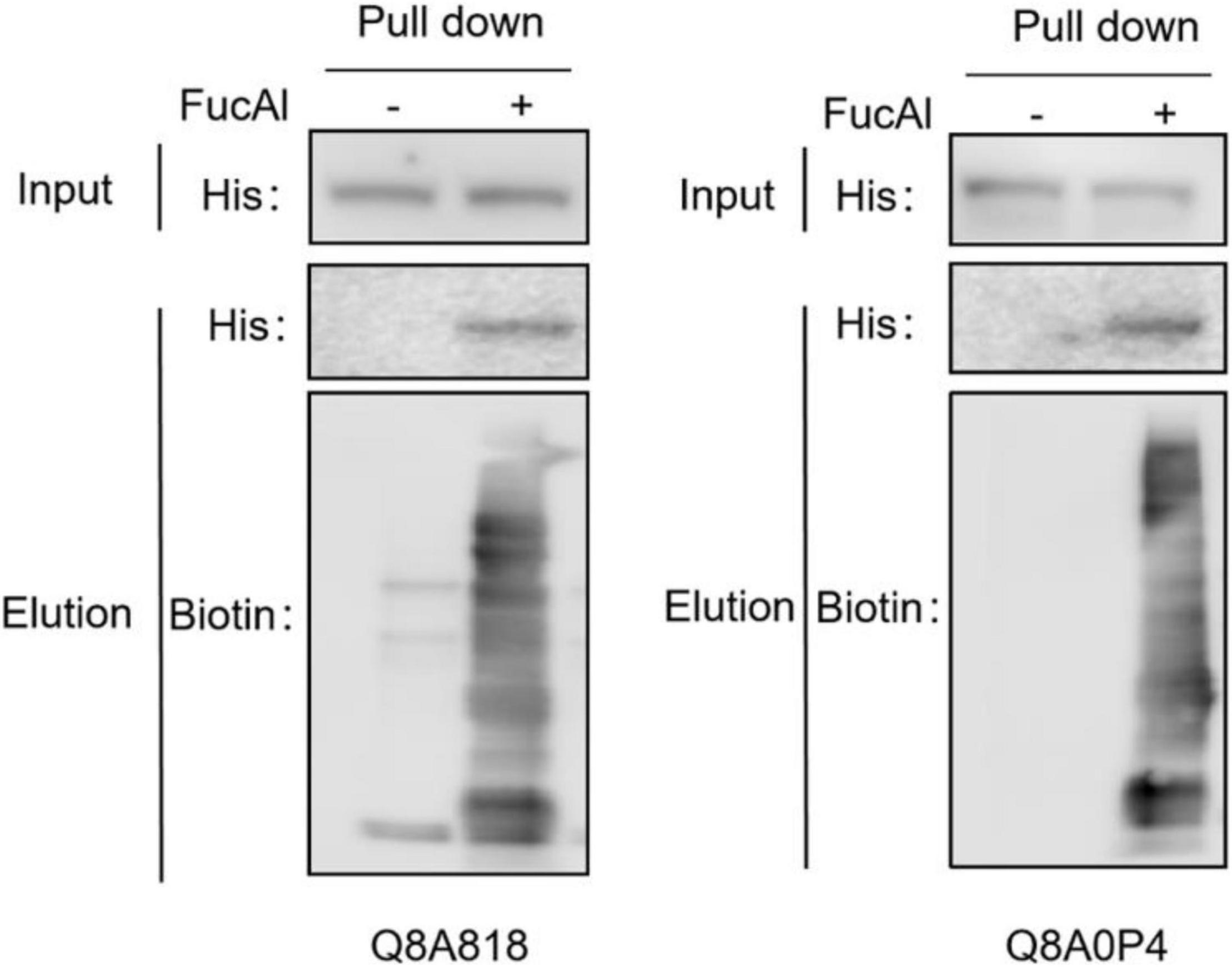
Figure 4. Fucosylation verification of two candidate FGPs. The cell lysates were prepared and reacted with biotin-picolyl-azide via CuAAC, followed by streptavidin beads pull-down. The enriched proteins were analyzed by Western blot with anti-His and anti-biotin antibody.
Screening and Annotation of Differentially Expressed Fucosylated Proteins
According to the threshold for differentially expressed proteins (p-values < 0.05, fold change ≥ 2), 285 (74 upregulated and 211 downregulated proteins), 120 (71 upregulated and 49 downregulated proteins), and 154 FGPs (60 upregulated and 94 downregulated proteins) are differentially expressed in the M, S, and F groups, respectively, compared to G group (Figure 5A). Moreover, some of these FGPs are exclusively detected in one group (Supplementary Data 1). Mostly, the differentially expressed FGPs in the M group are downregulated, whereas those in the S group are primarily upregulated. As for the F group, it has almost as many upregulated as downregulated differentially expressed FGPs. Overall, the volcano map and the normal distribution results illustrated in Figures 5B,C, respectively, indicated that the expression patterns of B. thetaiotaomicron FGPs varied significantly depending on the supplied polysaccharide diet. This is consistent with the Western blot results (Figure 2D), as well as with cluster analysis presented in Supplementary Figure 4.
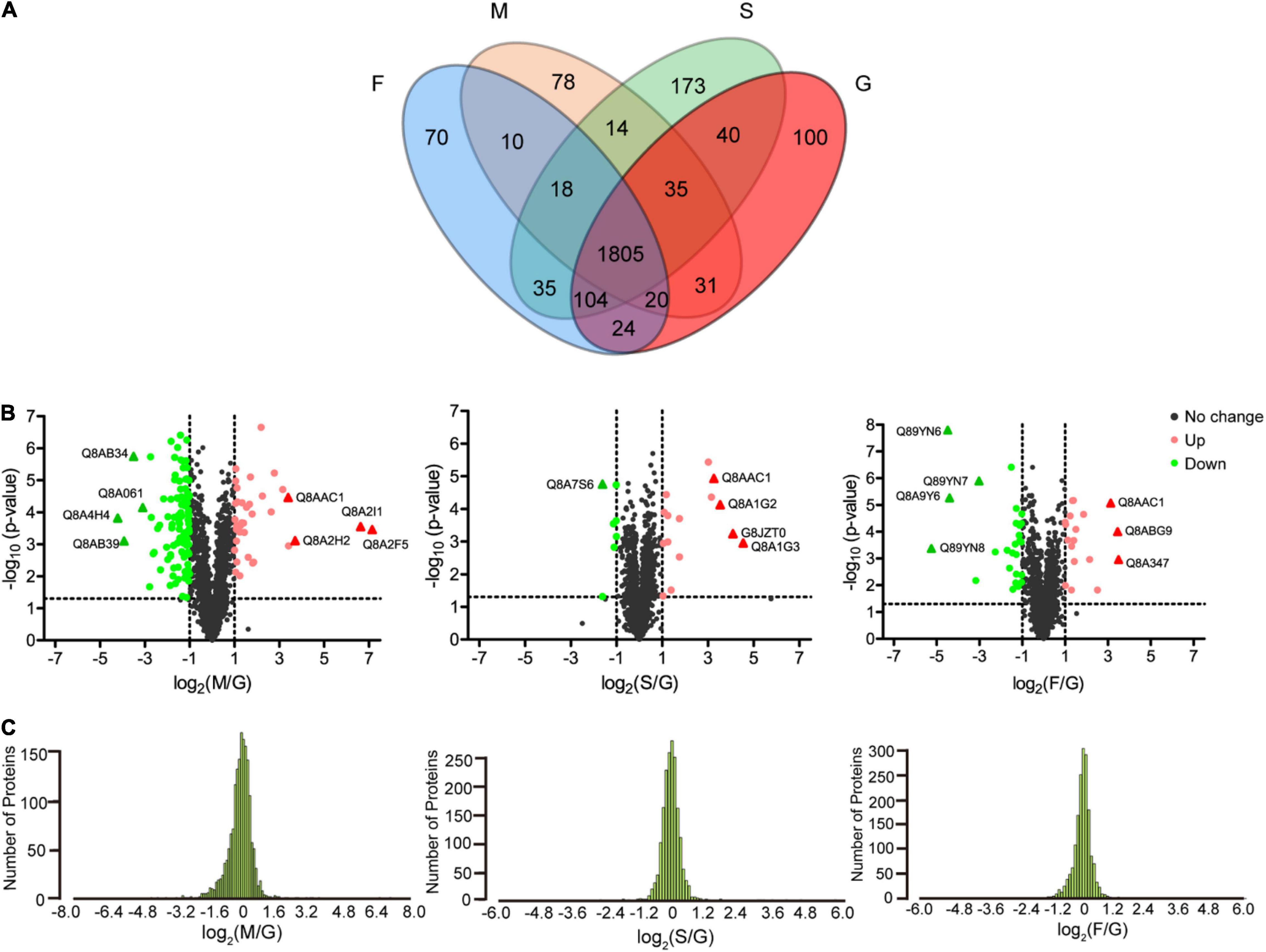
Figure 5. Analysis of differentially expressed FGPs in different comparisons. (A) Venn diagram of differentially expressed FGPs of B. thetaiotaomicron under four carbohydrate nutrition conditions. (B) Volcano plots depicting the enrichment (x axis) and significance (y axis) of the identified FGPs for mucin/glucose (left), starch/glucose (middle), and fucoidan/glucose (right). (C) Normal distribution of differentially expressed FGPs for mucin/glucose (left), starch/glucose (middle), and fucoidan/glucose (right).
The types and functions of the differentially expressed proteins in each group were analyzed by GO and KEGG enrichment analysis (Supplementary Data 2, 3). In group M, the FGPs are mostly downregulated, and the greatest change in expression is observed for Q8AB39 (SusC homolog) and Q8A4H4 (SusD homolog) proteins, both of which belong to the Sus protein family. Further, the most significantly upregulated FGPs in group M (more than 100 times) are Q8A2F5 (Chondroitin sulfate ABC lyase) and Q8A2I1 (Chondroitin sulfate ABC exolyase), both of which are chondroitin sulfate lyases (Supplementary Data 1). Based on the results of GO analysis (Figure 6A), the differentially expressed proteins are mainly outer membrane proteins that are involved in important metabolic biological processes and possess carbohydrate hydrolase activity. The KEGG analysis (Figure 6B) shows that galactose metabolism, starch and sucrose metabolism, butanoate metabolism and glycolysis are enhanced in the polysaccharide-treated samples, compared to the control.
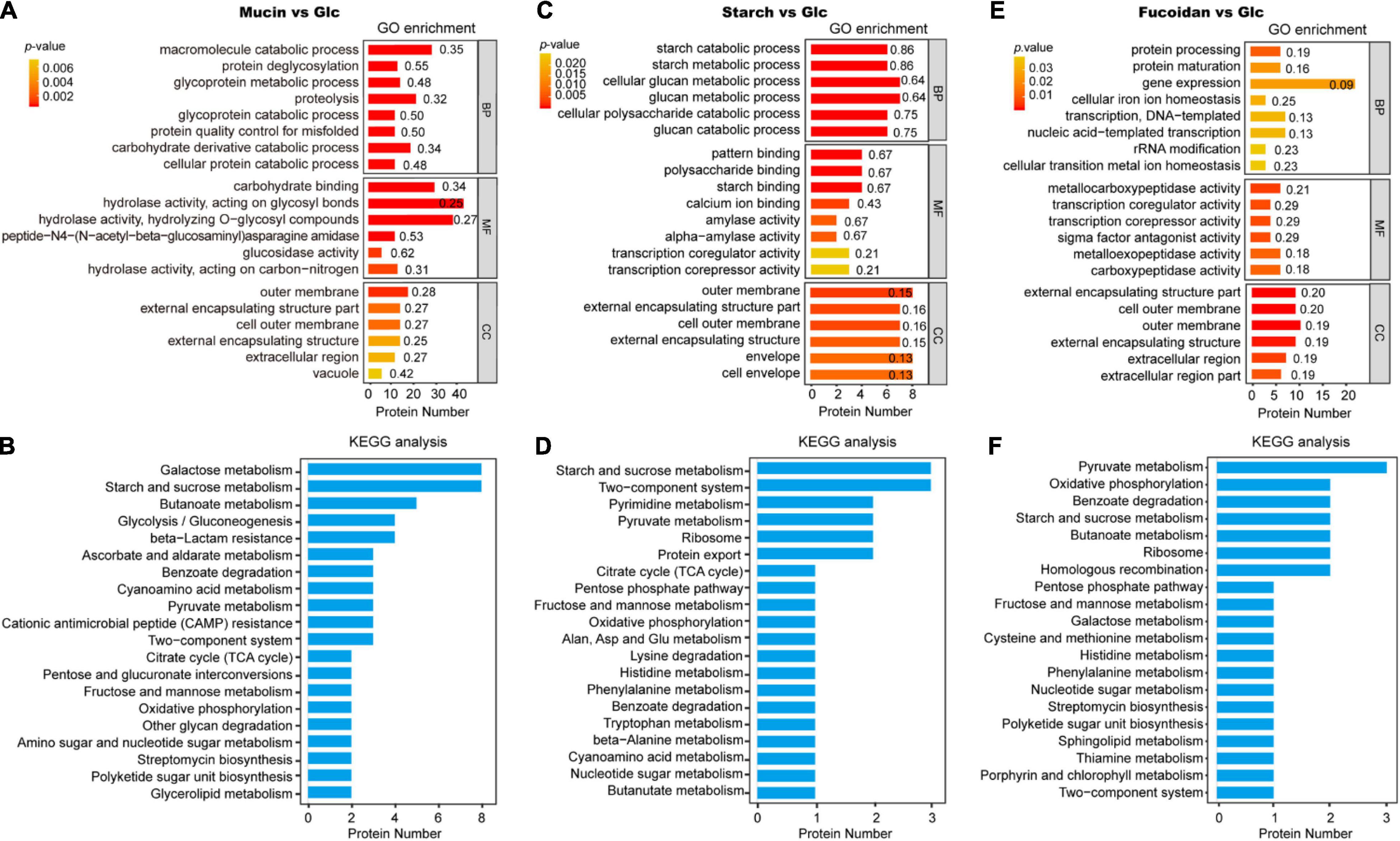
Figure 6. Gene ontology and Kyoto Encyclopedia of Genes and Genomes analysis of differentially expressed FGPs in different comparisons. GO and KEGG analysis of differentially expressed FGPs in mucin vs. glucose (A,B), starch vs. glucose (C,D), fucoidan vs. glucose (E,F). The p-values were obtained based on Fisher’s exact test. The smaller the p-value, and the higher the significance level of the corresponding functional category enrichment. Numbers beside the bars mean rich factors, indicating the ratio of the number of differentially expressed proteins annotated to the function to the number of all proteins identified to the function.
In the S group, the greatest FGPs upregulation is observed for Sus-like proteins Q8A1G3 (Alpha-amylase SusG) and G8JZT0 (Outer membrane protein SusE). The levels of proteins associated with quorum sensing and bacterial secretion (e.g., Q89YY8, Preprotein translocase subunit YajC) are also substantially increased in the S group. In general, these proteins are implicated in the regulation of multidrug resistance, biofilm formation, extracapsular polysaccharide synthesis, and host-gut bacteria symbiosis (Freitas et al., 2003; Antunes et al., 2005; Pumbwe et al., 2008). The most downregulated FGPs expressions detected in group S are Q8A7S6 (Biotin carboxyl carrier protein) and Q89ZW1 (Deoxycytidylate deaminase). The GO analysis results presented in Figure 6C show that most differentially expressed proteins in group S are involved in important biological processes, especially the polysaccharide catabolic process. These proteins also have essential molecular functions, such as amylase activity and the binding of patterns, polysaccharides, starch, and calcium ions. As for the KEGG analysis results, they emphasize the role of the differentially expressed FGPs in vital physiological metabolism and signaling pathways such as starch and sucrose metabolism, two-component systems, pyrimidine metabolism, pyruvate metabolism, and ribosome and protein export, among others (Figure 6D).
The addition of fucoidan to the culture medium increases the expression of some B. thetaiotaomicron FGPs while decreasing the level of others. Notably, the most downregulated proteins are all efflux system proteins, such as Q89YN8 and Q89YN6. These FGPs play conservative roles in bacterial cationic efflux systems, particularly in quorum sensing, drug resistance, and survival competition, which can protect bacteria from toxic chemical components (Pumbwe et al., 2008; Wang et al., 2016; Hibberd et al., 2017; Ghotaslou et al., 2018). As for the upregulated proteins, the top three are Q8A347 (putative beta-xylosidase), Q8ABG9 (SusD homolog), and Q8AAC1 (ammonium transporter). Based on GO analysis, the differentially expressed proteins in the F group are mainly involved in gene expression, protein processing, transcription, DNA template formation, gene transcription, and protein mature expression. These proteins also exhibit metallocarboxypeptidase activity, transcription coregulator activity, sigma factor antagonist activity, and metalloexopeptidase activity, among other transcription-related molecular functions (Figure 6E). Finally, the KEGG analysis results indicate that the differentially expressed proteins are implicated in various metabolic processes of biological macromolecules (Figure 6F).
For the most part, the FGPs present different variation trends, depending on the nutritional conditions. However, eight FGPs were found to be consistently upregulated (in all groups), and 13 were regularly downregulated (Supplementary Data 4). The upregulated proteins were involved in some important metabolic pathways such as ribosome binding, starch and sucrose metabolism, butanoate metabolism, pyruvate metabolism, and two-component systems. As for the downregulated FGPs, they mainly participated in metabolic pathways related to gene expression and carbohydrate metabolism. Although the string results showed that most of the consistently changed proteins didn’t exhibit obvious direct protein-protein interactions (data not shown), the regular variation in the expression levels of these proteins may be related to the regulation of their functions and to the adaptation of B. thetaiotaomicron to polysaccharide nutritional conditions.
Differentially Expressed Fucosylated Proteins With Nutrition-Sensitive Fucosylation Levels
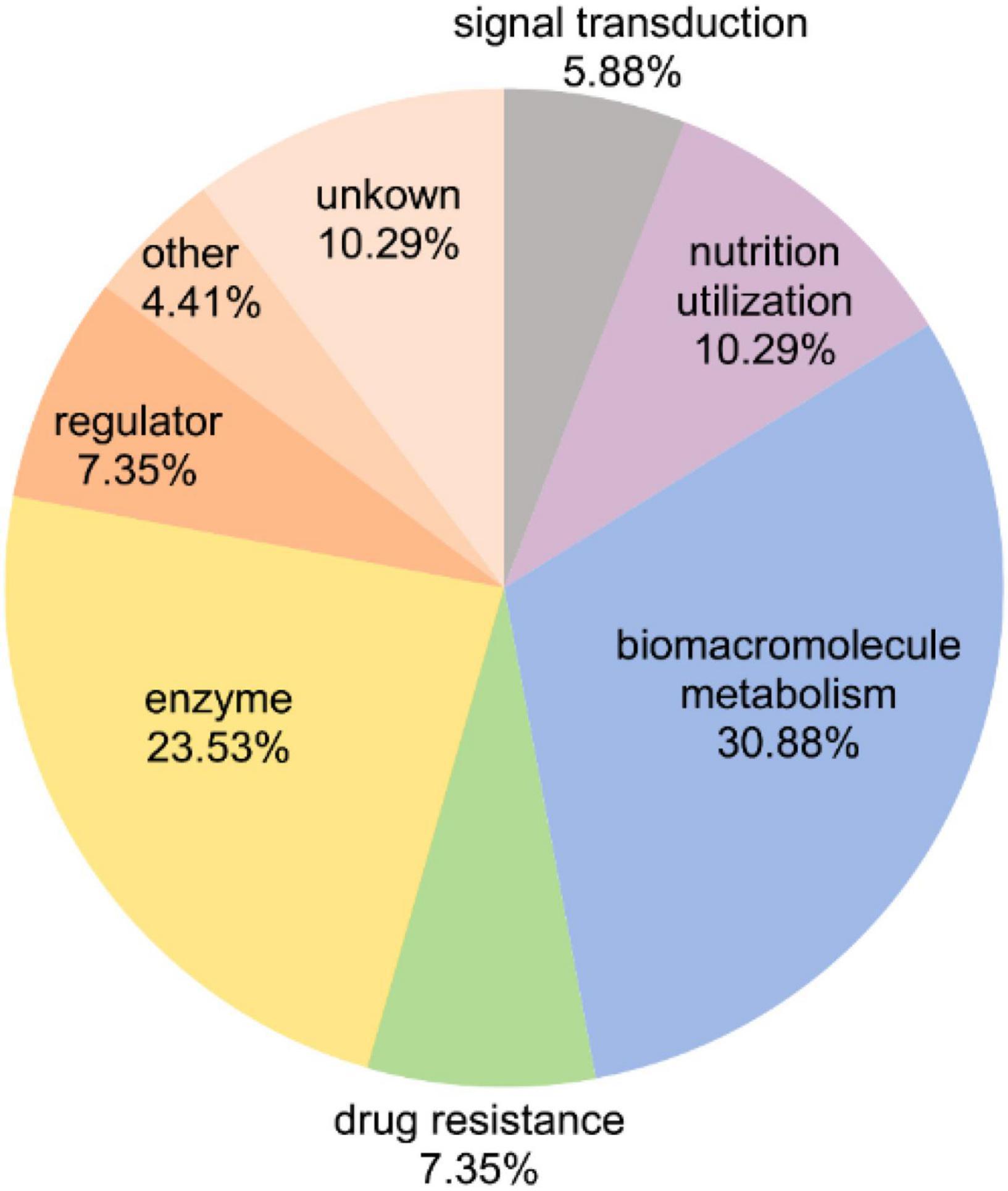
All above results showed the changes of the general FGPs expression levels. Notably, in addition to variations in protein expression levels, FGPs expressions were changed due to differences in the level of fucosylation. To identify the differentially expressed FGPs that were associated with altered fucosylation efficiency, quantitative global proteome analysis of B. thetaiotaomicron was also performed. Based on the obtained results (Supplementary Data 5), 65 key candidate FGPs were identified as being extremely sensitive to polysaccharide nutrition conditions (FGPs fold change/global protein fold change ≥2.0 or ≤0.5) (Table 1). They mainly involved in enzymatic activity, biomacromolecule metabolism, nutrition utilization, drug resistance and signal transduction (Figure 7).
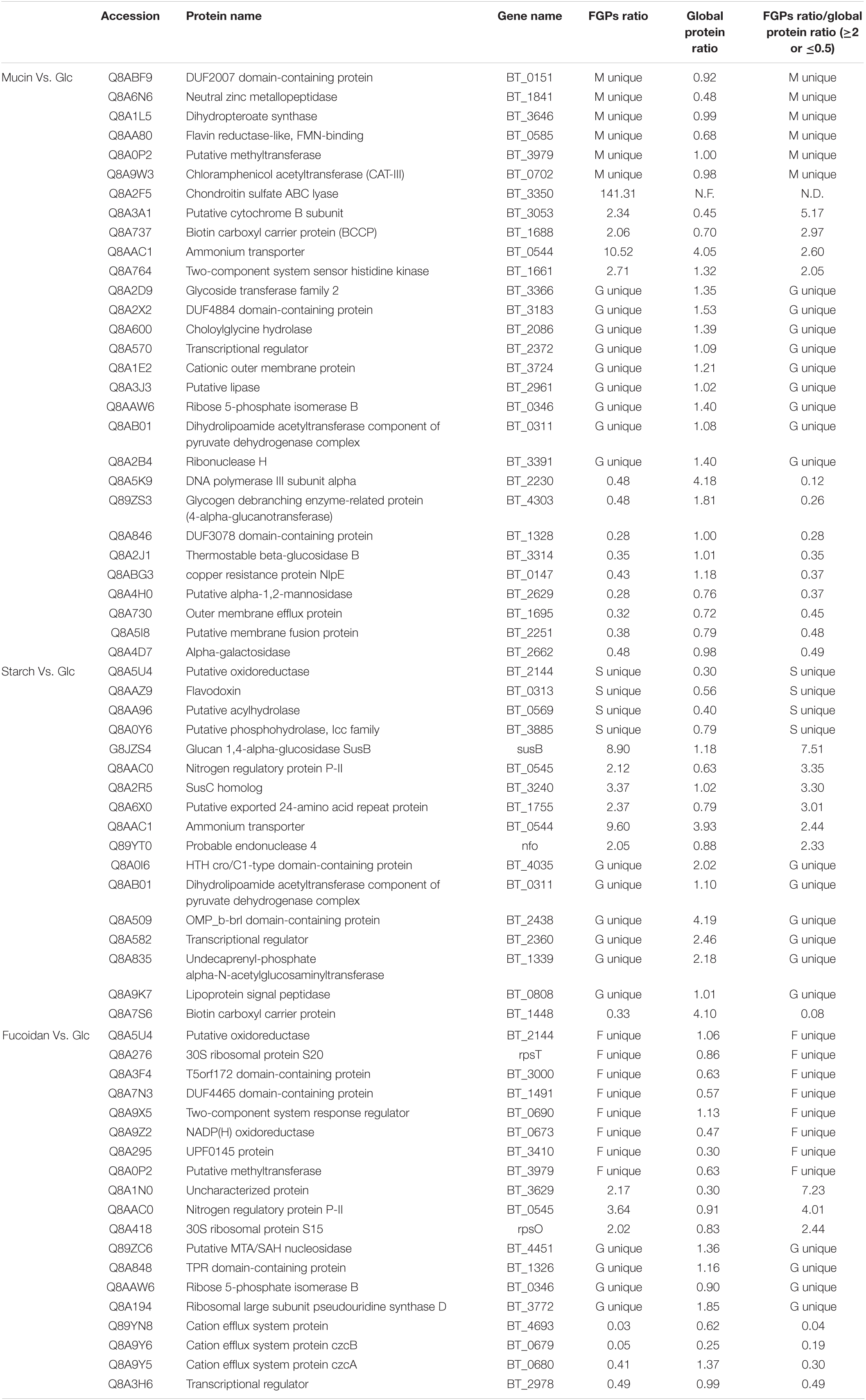
Table 1. Key FGPs with significantly altered fucosylation level under different polysaccharide nutritional conditions.
Overall, the addition of the three polysaccharides triggered a significant increase in the fucosylation levels of two-component system proteins and ammonium transporter proteins. In B. thetaiotaomicron, two-component systems (TCRS) are used to sense and degrade complex carbohydrates in the gut (Townsend, Raghavan, 2013). They are often co-localized with the genes encoding CAZymes and outer membrane sugar transporters so that they can co-regulate the utilization of polysaccharides (Xu et al., 2003; Raghavan and Groisman, 2010; Lynch and Sonnenburg, 2012; ,, and GroismanSchwalm, Townsend, and Groisman2017). However, the ammonium transporter proteins in B. thetaiotaomicron bacteria are involved in the absorption of nitrogen sources such as ammonia or ammonium (Luzhkov et al., 2006). A significant increase in the fucosylation levels of these two types of proteins is likely to affect their functions.
The FGPs related to chondroitin sulfate also show significant changes in fucosylation level under varying polysaccharide nutrition conditions. Particularly, the fucosylation levels of these FGPs are substantially increased (more than 100 times) upon the addition of mucin, which is consistent with the results presented above. Given that mucin cannot be degraded by the chondroitin sulfate ABC enzyme, it may significantly enhance the expression of this enzyme by acting as a signaling molecule that triggers a certain pathway in Bacteroides. Starch increases the fucosylation levels of Sus system proteins, which is possible to affect their ability to transport and utilize complex carbohydrates. In contrast, the fucosylation levels of efflux system proteins are substantially reduced by fucoidan. Other B. thetaiotaomicron FGPs showing changes in fucosylation levels upon the addition of different polysaccharides include those involved in polysaccharide metabolism, amino acid metabolism, short-chain fatty acid metabolism, folic acid biosynthesis, redox processes, RNA modification, and other important processes related to the growth and survival of Bacteroides. The above results show the fact that the fucosylation levels of proteins in Bacteroides are sensitive to various polysaccharide nutrition conditions, and the biological significance of this fact deserves to be further studied.
Discussion
In this study, the MOE and CuAAC methods were successfully applied to the labeling and enrichment of B. thetaiotaomicron FGPs. To understand the effect of the supplied diet on FGPs expression patterns, B. thetaiotaomicron were cultured under different polysaccharide nutrition conditions, and then the levels of newly synthesized FGPs and global proteins were determined by label-free proteome analysis. As expected, corn starch, mucin, and fucoidan present different effects on the expression levels of the newly synthesized FGPs in B. thetaiotaomicron. In addition, the fucosylation levels of different FGPs also vary depending on the polysaccharide nutrition conditions. As a result, 65 key FGPs showing significant differences in fucosylation levels were screened. The biological functions of these proteins, including nutrition utilization, environmental response, and multidrug resistance, are also expected to be influenced by exogenous nutritional conditions. Ultimately, the altered biological functions will affect the colonization, survival, and growth of B. thetaiotaomicron. Herein, we determine that the proteins whose fucosylation levels are the most sensitive to nutrition conditions are chondroitin sulfate ABC enzyme, Sus proteins, and cationic efflux system proteins. Moreover, we show that corn starch, mucin, and fucoidan can significantly promote the fucosylation of the two-component system and ammonium transport proteins. To the best of our knowledge, these data have never been previously reported.
It has been proved that the surface glycoproteins of B. fragilis and P. distasonis can be labeled by FucAl and CuAAC (Besanceney-Webler et al., 2011). Here, B. thetaiotaomicron, a Bacteroides species that shares a common fucose acquisition system with all the other Bacteroides except for P. distasonis, was also successfully labeled by FucAl (Xu et al., 2003; Coyne et al., 2005). Based on the obtained results, the functions of differentially expressed FGPs in B. thetaiotaomicron are similar to those of B. fragilis glycoproteins with many proteins in the former bacterial species having additional important functions, such as carbohydrate metabolism (Fletcher et al., 2009, 2011). This indicates that FGPs are essential for different Bacteroides species. Overall, the experimental method applied herein was found to be sensitive, accurate, and robust.
Compared to other labeling relative quantification technologies, such as iTRAQ, TMT, and SILAC, label-free quantification is simpler. By quantitatively comparing the results of a large number of samples, the latter method ensures the authenticity and accuracy of the reported data. The method is also more suitable for the analysis of differentially expressed proteins in microorganisms kept under strict culture conditions (Nie et al., 2014; Anand et al., 2017; Velasquez et al., 2017; Wu et al., 2019). Over the years, the instruments and algorithms of label-free quantification have been significantly improved. Today, the accuracy of this method is comparable to that of SILAC (Cox et al., 2014; Valikangas et al., 2018). Thus, label-free quantification was used to investigate the effects of polysaccharide nutrition on the expression pattern of FGPs in B. thetaiotaomicron.
Recently, studies have shown that dietary fibers can be used to regulate the intestinal microbiome and to guide the production of short-chain fatty acids. Depending on their structure, these fibers exhibit different and highly specific effects on the microbiome composition that is related to selective bacterial adhesion and substrate utilization (Deehan et al., 2020). Therefore, it is hypothesized that polysaccharides with different structures and sources would affect the abundance and metabolism of Bacteroides differently. Our results support this hypothesis. As key factors closely related to the growth and symbiosis of Bacteroides, as well as to substrate adhesion and nutrient utilization, FGPs and their fucosylation levels were found to be strongly influenced by the supplied polysaccharide diet. Moreover, most of the differentially expressed FGPs identified herein (especially CAZymes) are involved in macromolecular metabolic functions, such as polysaccharide and amino acid metabolism. These results indicate that different diets trigger different pathways of FGPs expression in B. thetaiotaomicron so that the bacteria may adapt to the environmental change. Compared with the results of global proteome, 65 key FGPs were selected and emphasized, which mainly involved in enzymatic activity, biomacromolecule metabolism, nutrition utilization, drug resistance and signal transduction. Changes of these proteins expression levels mainly came from the change of fucosylation levels, and reflected high sensitivity of their fucosylation to exogenous polysaccharide conditions. In contrast, some FGPs (such as Q8AAC1, Q8AB34) with significantly different expression levels were not selected, because their changes of protein expression level and changes of fucosylation level were similar or they were not found in global proteome. Therefore, sensitivity of fucosylation level to changes in exogenous nutritional conditions may play important roles in the growth, survival and colonization of Bacteroides.
Bacteroides thetaiotaomicron has an outstanding capability to degrade a wide range of polysaccharides including rhamnogalacturonan-II, the most structurally complex glycan known (Ndeh et al., 2017). Although the PULs for fucoidan has not been identified in B. thetaiotaomicron, this microbe encodes fucose cleavage and metabolism genes such as α-fucosidases (Sonnenburg et al., 2005) and several sulfatases genes (Ye et al., 2021), indicating its genetic potential for fucoidan degradation. So it’s reasonable to assume that during the 3-h incubation with B. thetaiotaomicron, the added polysaccharides were partially degraded and the released degradation products impacted cell metabolism. As a result, the proteome and protein fucosylation changed in B. thetaiotaomicron. Notably, it was possible that the fucose degraded from fucoidan or mucin could compete with FucAl for incorporation into newly synthesized proteins, which would lead to an underestimate of detectable protein fucosylation. To clarify to what extent the added polysaccharides are degraded, some analytical methods for sensitive determination and quantitation such as HPLC and multi angle light scattering (MALS) will be employed to monitor the degradation process in the follow-up work.
In this research, we just choose a relatively reasonable time point to study how the newly synthesized FGPs changes in response to the newly added polysaccharides. In the follow-up work, we will also explore the expression patterns of FGPs in Bacteroides at different time points after adding polysaccharide. Moreover, when the polysaccharide is the only carbon source, what changes of FGPs would make by the Bacteroides is an interesting question. To understand the reasons behind the changes in fucosylation levels and to assess the mechanisms of these changes, further biological experiments must be performed, including gene knockout and in vivo experiments. Given that the glycoproteins of most Bacteroides species have not yet been identified, the MOE and CuAAC methods must be applied to other Bacteroides. Overall, the results reported herein indicate that the combination of MOE and CuAAC is a suitable and effective strategy for the study of intestinal Bacteroides glycoproteomes, and that protein fucosylation is a potential key factor that may play a significant role in regulating multiple biological functions when B. thetaiotaomicron is subjected to changes in the nutritional environment.
Data Availability Statement
The data presented in the study are deposited in the ProteomeXchange repository, accession number PXD023756.
Author Contributions
HJ and GY conceived the project. XT, HF, and XW performed experiments. XT, HJ, and BC analyzed data. XT and HJ drafted the manuscript. XT, HJ, BC, and GY revised and approved the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by Natural Science Foundation of Shandong Province (ZR2017BC007 to HJ), National Natural Science Foundation of China (21807094 to HJ, 81991522, 31670811 to GY), National Science and Technology Major Project for Significant New Drugs Development (2018ZX09305-004 to GY), Shandong Provincial Major Science and Technology Innovation Project (2018SDKJ0404 to GY), and Taishan Scholar Climbing Project (TSPD20210304 to GY).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank C. Jeffrey Smith and Edson R. Rocha of East Carolina University for the gift of plasmid pFD340. We also thank Qingsen Shang of Ocean University of China for the insightful comments and suggestions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.826942/full#supplementary-material
Abbreviations
B. theaiotaomicron, Bacteroides theaiotaomicron; FGPs, fucosylated glycoproteins; PULs, polysaccharide utilization site; Sus, starch utilization system; CAZymes, carbohydrate-active enzymes; AAL, Aleuria aurantia lectin; MOE, metabolic oligosaccharide engineering; CuAAC, Cu(I)-catalyzed azide-alkyne cycloaddition; AMM, anaerobic minimal medium; LC-MS, liquid chromatography mass spectrometry; FucAl, alkynyl-fucose; FDR, false discovery rate; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; SILAC, stable isotope labeling with amino acids.
Footnotes
References
Aktar, R., Parkar, N., Stentz, R., Baumard, L., Parker, A., Goldson, A., et al. (2020). Human resident gut microbe Bacteroides thetaiotaomicron regulates colonic neuronal innervation and neurogenic function. Gut Microbes 11, 1745–1757. doi: 10.1080/19490976.2020.1766936
Anand, S., Samuel, M., Ang, C.-S., Keerthikumar, S., and Mathivanan, S. (2017). “Label-based and label-free strategies for protein quantitation,” in Proteome Bioinformatics, eds S. Keerthikumar and S. Mathivanan (Totowa, NJ: Humana Press:), 31–43. doi: 10.1007/978-1-4939-6740-7_4
Antunes, L. C. M., Ferreira, L. Q., Ferreira, E. O., Miranda, K. R., Avelar, K. E. S., Domingues, R., et al. (2005). Bacteroides species produce Vibrio harveyi autoinducer 2-related molecules. Anaerobe 11, 295–301. doi: 10.1016/j.anaerobe.2005.03.003
Baughn, A. D., and Malamy, M. H. (2002). A mitochondrial-like aconitase in the bacterium Bacteroides fragilis: imgplications for the evolution of the mitochondrial Krebs cycle. Proc. Natl. Acad. Sci. U.S.A. 99, 4662–4667. doi: 10.1073/pnas.052710199
Besanceney-Webler, C., Jiang, H., Wang, W., Baughn, A. D., and Wu, P. (2011). Metabolic labeling of fucosylated glycoproteins in Bacteroidales species. Bioorg. Med. Chem. Lett. 21, 4989–4992. doi: 10.1016/j.bmcl.2011.05.038
Brook, I., Wexler, H. M., and Goldstein, E. J. C. (2013). Antianaerobic antimicrobials: spectrum and susceptibility testing. Clin. Microbiol. Rev. 26, 526–546. doi: 10.1128/cmr.00086-12
Champasa, K., Longwell, S. A., Eldridge, A. M., Stemmler, E. A., and Dube, D. H. (2013). Targeted identification of glycosylated proteins in the gastric pathogen Helicobacter pylori (Hp). Mol. Cell. Proteomics 12, 2568–2586. doi: 10.1074/mcp.M113.029561
Cheah, J. S., and Yamada, S. (2017). A simple elution strategy for biotinylated proteins bound to streptavidin conjugated beads using excess biotin and heat. Biochem. Biophys. Res. Commun. 493, 1522–1527. doi: 10.1016/j.bbrc.2017.09.168
Cox, J., Hein, M. Y., Luber, C. A., Paron, I., Nagaraj, N., and Mann, M. (2014). Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526. doi: 10.1074/mcp.M113.031591
Coyne, M. J., Reinap, B., Lee, M. M., and Comstock, L. E. (2005). Human symbionts use a host-like pathway for surface fucosylation. Science 307, 1778–1781. doi: 10.1126/science.1106469
D’Argenio, V., and Salvatore, F. (2015). The role of the gut microbiome in the healthy adult status. Clin. Chim. Acta 451, 97–102. doi: 10.1016/j.cca.2015.01.003
Deehan, E. C., Yang, C., Perez-Muñoz, M. E., Nguyen, N. K., Cheng, C. C., Triador, L., et al. (2020). Precision microbiome modulation with discrete dietary fiber structures directs short-chain fatty acid production. Cell Host Microbe 27, 389–404.e386. doi: 10.1016/j.chom.2020.01.006
Delday, M., Mulder, I., Logan, E. T., and Grant, G. (2019). Bacteroides thetaiotaomicron ameliorates colon inflammation in preclinical models of crohn’s disease. Inflamm. Bowel Dis. 25, 85–96. doi: 10.1093/ibd/izy281
Donaldson, G. P., Lee, S. M., and Mazmanian, S. K. (2016). Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20–32. doi: 10.1038/nrmicro3552
Dube, D. H., and Bertozzi, C. R. (2003). Metabolic oligosaccharide engineering as a tool for glycobiology. Curr. Opin. Chem. Biol. 7, 616–625. doi: 10.1016/j.cbpa.2003.08.006
El Kaoutari, A., Armougom, F., Gordon, J. I., Raoult, D., and Henrissat, B. (2013). The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 11, 497–504. doi: 10.1038/nrmicro3050
Fletcher, C. M., Coyne, M. J., and Comstock, L. E. (2011). Theoretical and experimental characterization of the scope of protein o-glycosylation in Bacteroides fragilis. J. Biol. Chem. 286, 3219–3226. doi: 10.1074/jbc.M110.194506
Fletcher, C. M., Coyne, M. J., Villa, O. F., Chatzidaki-Livanis, M., and Comstock, L. E. (2009). A General O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell 137, 321–331. doi: 10.1016/j.cell.2009.02.041
Flint, H. J., Bayer, E. A., Rincon, M. T., Lamed, R., and White, B. A. (2008). Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat. Rev. Microbiol. 6, 121–131. doi: 10.1038/nrmicro1817
Foley, M. H., Martens, E. C., and Koropatkin, N. M. (2018). SusE facilitates starch uptake independent of starch binding in B-thetaiotaomicron. Mol. Microbiol. 108, 551–566. doi: 10.1111/mmi.13949
Fredriksen, L., Moen, A., Adzhubei, A. A., Mathiesen, G., Eijsink, V. G. H., and Egge-Jacobsen, W. (2013). Lactobacillus plantarum WCFS1 O-linked protein glycosylation: an extended spectrum of target proteins and modification sites detected by mass spectrometry. Glycobiology 23, 1439–1451. doi: 10.1093/glycob/cwt071
Freitas, M., Tavan, E., Cayuela, C., Diop, L., Sapin, C., and Trugnan, G. (2003). Host-pathogens cross-talk. Indigenous bacteria and probiotics also play the game. Biol. Cell 95, 503–506. doi: 10.1016/j.biolcel.2003.08.004
Garber, J. M., Hennet, T., and Szymanski, C. M. (2021). Significance of fucose in intestinal health and disease. Mol. Microbiol. 115, 1086–1093. doi: 10.1111/mmi.14681
Geva-Zatorsky, N., Alvarez, D., Hudak, J. E., Reading, N. C., Erturk-Hasdemir, D., Dasgupta, S., et al. (2015). In vivo imaging and tracking of host-microbiota interactions via metabolic labeling of gut anaerobic bacteria. Nat. Med. 21, 1091–1100. doi: 10.1038/nm.3929
Ghotaslou, R., Yekani, M., and Memar, M. Y. (2018). The role of efflux pumps in Bacteroides fragilis resistance to antibiotics. Microbiol Res. 210, 1–5. doi: 10.1016/j.micres.2018.02.007
González-Morelo, K. J., Vega-Sagardía, M., and Garrido, D. (2020). Molecular insights into o-linked glycan utilization by gut microbes. Front. Microbiol. 11:591568. doi: 10.3389/fmicb.2020.591568
Gonzalez-Zamnorano, M., Mendoza-Hernandez, G., Xolalpa, W., Parada, C., Vallecillo, A. J., Bigi, F., et al. (2009). Mycobacterium tuberculosis glycoproteomics based on cona-lectin affinity capture of mannosylated proteins. J. Proteome Res. 8, 721–733. doi: 10.1021/pr800756a
Goto, Y., Obata, T., Kunisawa, J., Sato, S., Ivanov, I. I., Lamichhane, A., et al. (2014). Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science 345:1254009. doi: 10.1126/science.1254009
Hibberd, M. C., Wu, M., Rodionov, D. A., Li, X., Cheng, J., Griffin, N. W., et al. (2017). The effects of micronutrient deficiencies on bacterial species from the human gut microbiota. Sci. Transl. Med. 9:eaal4069. doi: 10.1126/scitranslmed.aal4069
Hong, S., Sahai-Hernandez, P., Chapla, D. G., Moremen, K. W., Traver, D., and Wu, P. (2019). Direct visualization of live zebrafish glycans via single-step metabolic labeling with fluorophore-tagged nucleotide sugars. Angew. Chem. Int. Ed. 58, 14327–14333. doi: 10.1002/anie.201907410
Hsieh, S., Porter, N. T., Donermeyer, D. L., Horvath, S., Strout, G., Saunders, B. T., et al. (2020). Polysaccharide capsules equip the human symbiont Bacteroides thetaiotaomicron to modulate immune responses to a dominant antigen in the intestine. J. Immunol. 204, 1035–1046. doi: 10.4049/jimmunol.1901206
Hu, S., Wang, J., Wang, J., Yang, H., Yan, X., and Su, L. (2019). Fucoidan from Acaudina molpadioides improves insulin resistance by altering gut microbiota dysfunction. J. Funct. Foods 57, 59–67. doi: 10.1016/j.jff.2019.03.033
Inoue, N., Onoda, A., and Hayashi, T. (2019). Site-specific modification of proteins through n-terminal azide labeling and a chelation-assisted CuAAC reaction. Bioconjug. Chem. 30, 2427–2434. doi: 10.1021/acs.bioconjchem.9b00515
Koebnik, R. (2005). TonB-dependent trans-envelope signalling: the exception or the rule? Trends Microbiol. 13, 343–347. doi: 10.1016/j.tim.2005.06.005
Kolb, H. C., Finn, M. G., and Sharpless, K. B. (2001). Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. Engl. 40, 2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::aid-anie2004>3.3.co;2-x
Koropatkin, N., Martens, E. C., Gordon, J. I., and Smith, T. J. (2009). Structure of a SusD homologue, BT1043, involved in mucin o-glycan utilization in a prominent human gut symbiont. Biochemistry 48, 1532–1542. doi: 10.1021/bi801942a
Kostopoulos, I., Aalvink, S., Kovatcheva-Datchary, P., Nijsse, B., Bäckhed, F., Knol, J., et al. (2021). A continuous battle for host-derived glycans between a mucus specialist and a glycan generalist in vitro and in vivo. Front. Microbiol. 12:632454. doi: 10.3389/fmicb.2021.632454
Leon, D. R., Ytterberg, A. J., Boontheung, P., Kim, U., Loo, J. A., Gunsalus, R. P., et al. (2015). Mining proteonnic data to expose protein modifications in Methanosarcina mazei strain Go1. Front. Microbiol. 6:149. doi: 10.3389/fmicb.2015.00149
Ley, R. E., Turnbaugh, P. J., Klein, S., and Gordon, J. I. (2006). Microbial ecology – human gut microbes associated with obesity. Nature 444, 1022–1023. doi: 10.1038/4441022a
Li, S., Li, J., Mao, G., Yan, L., Hu, Y., Ye, X., et al. (2019). Effect of the sulfation pattern of sea cucumber-derived fucoidan oligosaccharides on modulating metabolic syndromes and gut microbiota dysbiosis caused by HFD in mice. J. Funct. Foods 55, 193–210. doi: 10.1016/j.jff.2019.02.001
Liou, C. S., Sirk, S. J., Diaz, C. A. C., Klein, A. P., Fischer, C. R., Higginbottom, S. K., et al. (2020). A metabolic pathway for activation of dietary glucosinolates by a human gut symbiont. Cell 180, 717–728.e719. doi: 10.1016/j.cell.2020.01.023
Liu, F., Aubry, A. J., Schoenhofen, I. C., Logan, S. M., and Tanner, M. E. (2009). The engineering of bacteria bearing azido-pseudaminic acid-modified flagella. Chembiochem 10, 1317–1320. doi: 10.1002/cbic.200900018
Longwell, S. A., and Dube, D. H. (2013). Deciphering the bacterial glycocode: recent advances in bacterial glycoproteomics. Curr. Opin. Chem. Biol. 17, 41–48. doi: 10.1016/j.cbpa.2012.12.006
Luzhkov, V. B., Almlof, M., Nervall, M., and Aqvist, J. (2006). Computational study of the binding affinity and selectivity of the bacterial ammonium transporter AmtB. Biochemistry 45, 10807–10814. doi: 10.1021/bi0610799
Lynch, J. B., and Sonnenburg, J. L. (2012). Prioritization of a plant polysaccharide over a mucus carbohydrate is enforced by a Bacteroides hybrid two-component system. Mol. Microbiol. 85, 478–491. doi: 10.1111/j.1365-2958.2012.08123.x
Martens, E. C., Chiang, H. C., and Gordon, J. I. (2008). Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4, 447–457. doi: 10.1016/j.chom.2008.09.007
Martens, E. C., Koropatkin, N. M., Smith, T. J., and Gordon, J. I. (2009). Complex glycan catabolism by the human gut microbiota: the bacteroidetes sus-like paradigm. J. Biol. Chem. 284, 24673–24677. doi: 10.1074/jbc.R109.022848
Miethke, M., and Marahiel, M. A. (2007). Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71, 413–451. doi: 10.1128/mmbr.00012-07
Moore, W. E., and Holdeman, L. V. (1974). Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 27, 961–979. doi: 10.1128/aem.27.5.961-979.1974
Morya, V. K., Kim, J., and Kim, E.-K. (2012). Algal fucoidan: structural and size-dependent bioactivities and their perspectives. Appl. Microbiol. Biotechnol. 93, 71–82. doi: 10.1007/s00253-011-3666-8
Ndeh, D., Rogowski, A., Cartmell, A., Luis, A. S., Baslé, A., Gray, J., et al. (2017). Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature 544, 65–70. doi: 10.1038/nature21725
Neumann, S., Biewend, M., Rana, S., and Binder, W. H. (2020). The CuAAC: principles, homogeneous and heterogeneous catalysts, and novel developments and applications. Macromol. Rapid. Commun. 41:e1900359. doi: 10.1002/marc.201900359
Nie, S., Lo, A., Wu, J., Zhu, J., Tan, Z., Simeone, D. M., et al. (2014). Glycoprotein biomarker panel for pancreatic cancer discovered by quantitative proteomics analysis. J. Proteome Res. 13, 1873–1884. doi: 10.1021/pr400967x
Oren, A., Vandamme, P., and Schink, B. (2016). Notes on the use of Greek word roots in genus and species names of prokaryotes. Int. J. Syst. Evol. Microbiol. 66, 2129–2140. doi: 10.1099/ijsem.0.001063
Parker, C. G., and Pratt, M. R. (2020). Click chemistry in proteomic investigations. Cell 180, 605–632. doi: 10.1016/j.cell.2020.01.025
Pauer, H., Ferreira, E. D. O., dos Santos-Filho, J., Portela, M. B., Zingali, R. B., Araujo Soares, R. M., et al. (2009). A TonB-dependent outer membrane protein as a Bacteroides fragilis fibronectin-binding molecule. Fems Immunol. Med. Microbiol. 55, 388–395. doi: 10.1111/j.1574-695X.2009.00532.x
Pedowitz, N. J., Zaro, B. W., and Pratt, M. R. (2020). Metabolic labeling for the visualization and identification of potentially O-GlcNAc-modified proteins. Curr. Protocols chem. Biol. 12, e81–e81. doi: 10.1002/cpch.81
Pickard, J. M., and Chervonsky, A. V. (2015). Intestinal fucose as a mediator of host–microbe symbiosis. J. Immunol. 194, 5588–5593. doi: 10.4049/jimmunol.1500395
Pickard, J. M., Maurice, C. F., Kinnebrew, M. A., Abt, M. C., Schenten, D., Golovkina, T. V., et al. (2014). Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature 514, 638–641. doi: 10.1038/nature13823
Pumbwe, L., Skilbeck, C. A., and Wexler, H. M. (2008). Presence of quorum-sensing systems associated with multidrug resistance and biofilm formation in Bacteroides fragilis. Microb Ecol. 56, 412–419. doi: 10.1007/s00248-007-9358-3
Raghavan, V., and Groisman, E. A. (2010). Orphan and hybrid two-component system proteins in health and disease. Curr. Opin. Microbiology 13, 226–231. doi: 10.1016/j.mib.2009.12.010
Riley, N. M., Bertozzi, C. R., and Pitteri, S. J. (2021). A pragmatic guide to enrichment strategies for mass spectrometry-based glycoproteomics. Mol. Cell Proteomics 20:100029. doi: 10.1074/mcp.R120.002277
Rogers, T. E., Pudlo, N. A., Koropatkin, N. M., Bell, J. S. K., Balasch, M. M., Jasker, K., et al. (2013). Dynamic responses of Bacteroides thetaiotaomicron during growth on glycan mixtures. Mol. Microbiol. 88, 876–890. doi: 10.1111/mmi.12228
Rong, J., Han, J., Dong, L., Tan, Y., Yang, H., Feng, L., et al. (2014). Glycan imaging in intact rat hearts and glycoproteomic analysis reveal the upregulation of sialylation during cardiac hypertrophy. J. Am. Chem. Soc. 136, 17468–17476. doi: 10.1021/ja508484c
Schanzenbächer, C. T., Sambandan, S., Langer, J. D., and Schuman, E. M. (2016). Nascent proteome remodeling following homeostatic scaling at hippocampal synapses. Neuron 92, 358–371. doi: 10.1016/j.neuron.2016.09.058
Schauer, K., Rodionov, D. A., and de Reuse, H. (2008). New substrates for TonB-dependent transport: do we only see the ‘tip of the iceberg’? Trends Biochem. Sci. 33, 330–338. doi: 10.1016/j.tibs.2008.04.012
Schneider, M., Kumar, V., Nordstrom, L. U., Feng, L., Takeuchi, H., Hao, H., et al. (2018). Inhibition of delta-induced notch signaling using fucose analogs. Nat. Chem. Biol. 14, 65–71. doi: 10.1038/nchembio.2520
Schwalm, N. D. III, Townsend, G. E. II, and Groisman, E. A. (2017). Prioritization of polysaccharide utilization and control of regulator activation in Bacteroides thetaiotaomicron. Mol. Microbiol. 104, 32–45. doi: 10.1111/mmi.13609
Shang, Q., Shan, X., Cai, C., Hao, J., Li, G., and Yu, G. (2018). Dietary fucoidan modulates the gut microbiota in mice by increasing the abundance of Lactobacillus and Ruminococcaceae (vol 7, pg 3224, 2016). Food Funct. 9, 655–655. doi: 10.1039/c7fo90052j
Shoemaker, N. B., Guthrie, E. P., Salyers, A. A., and Gardner, J. F. (1985). Evidence that the clindamycin-erythromycin resistance gene of Bacteroides plasmid pBF4 is on a transposable element. J. Bacteriol. 162, 626–632. doi: 10.1128/jb.162.2.626-632.1985
Sletten, E. M., and Bertozzi, C. R. (2009). Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew. Chem. Int. Ed. 48, 6974–6998. doi: 10.1002/anie.200900942
Sonnenburg, J. L., Xu, J., Leip, D. D., Chen, C. H., Westover, B. P., Weatherford, J., et al. (2005). Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307, 1955–1959. doi: 10.1126/science.1109051
Stevenson, J., Ngo, M., Brandt, A., Weadge, J. T., and Suits, M. D. L. (2021). Analysis of two suse-like enzymes from bacteroides thetaiotaomicron reveals a potential degradative capacity for this protein family. Front. Microbiol. 12:645765. doi: 10.3389/fmicb.2021.645765
Strand, T. A., Lale, R., Degnes, K. F., Lando, M., and Valla, S. (2014). A new and improved host-independent plasmid system for RK2-based conjugal transfer. PLoS One 9:e90372. doi: 10.1371/journal.pone.0090372
Thompson, J. S., and Malamy, M. H. (1990). Sequencing the gene for an imipenem-cefoxitin-hydrolyzing enzyme (CfiA) from Bacteroides fragilis TAL2480 reveals strong similarity between CfiA and Bacillus cereus beta-lactamase II. J. Bacteriol. 172, 2584–2593. doi: 10.1128/jb.172.5.2584-2593.1990
Townsend, G. E. II, Raghavan, V., Zwir, I., and Groisman, E. A. (2013). Intramolecular arrangement of sensor and regulator overcomes relaxed specificity in hybrid two-component systems. Proc. Natl. Acad. Sci. U.S.A. 110, E161–E169. doi: 10.1073/pnas.1212102110
Ushijima, T., Takahashi, M., Tatewaki, K., and Ozaki, Y. (1983). A selective medium for isolation and presumptive identification of the Bacteriodes fragilis group. Microbiol. Immunol. 27, 985–993. doi: 10.1111/j.1348-0421.1983.tb02929.x
Valikangas, T., Suomi, T., and Elo, L. L. (2018). A systematic evaluation of normalization methods in quantitative label-free proteomics. Briefings Bioinformatics 19, 1–11. doi: 10.1093/bib/bbw095
Varel, V. H., and Bryant, M. P. (1974). Nutritional features of Bacteroides fragilis subsp. fragilis. Appl. Microbiol. 28, 251–257. doi: 10.1128/aem.28.2.251-257.1974
Velasquez, E., Nogueira, F. C. S., Velasquez, I., Schrnitt, A., Falkai, P., Domont, G. B., et al. (2017). Synaptosomal Proteome of the orbitofrontal cortex from schizophrenia patients using quantitative label-free and iTRAQ-based shotgun proteomics. J. Proteome Res. 16, 4481–4494. doi: 10.1021/acs.jproteome.7b00422
Wang, Y., Venter, H., and Ma, S. (2016). Efflux pump inhibitors: a novel approach to combat efflux-mediated drug resistance in bacteria. Curr. Drug Targets 17, 702–719. doi: 10.2174/1389450116666151001103948
Whitehead, T. R., and Hespell, R. B. (1990). Heterologous expression of the Bacteroides ruminicola xylanase gene in Bacteroides fragilis and Bacteroides uniformis. FEMS Microbiol. Lett. 54, 61–65. doi: 10.1016/0378-1097(90)90259-s
Witzke, K. E., Rosowski, K., Müller, C., Ahrens, M., Eisenacher, M., Megger, D. A., et al. (2017). Quantitative secretome analysis of activated jurkat cells using click chemistry-based enrichment of secreted glycoproteins. J. Proteome Res. 16, 137–146. doi: 10.1021/acs.jproteome.6b00575
Wu, Y., Mirzaei, M., Atwell, B. J., and Haynes, P. A. (2019). Label-free and isobaric tandem mass tag (TMT) multiplexed quantitative proteomic data of two contrasting rice cultivars exposed to drought stress and recovery. Data in Brief 22, 697–702. doi: 10.1016/j.dib.2018.12.041
Xu, J., and Gordon, J. I. (2003). Honor thy symbionts. Proc. Natl. Acad. Sci. U.S.A. 100, 10452–10459. doi: 10.1073/pnas.1734063100
Xu, J., Bjursell, M. K., Himrod, J., Deng, S., Carmichael, L. K., Chiang, H. C., et al. (2003). A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299, 2074–2076. doi: 10.1126/science.1080029
Ye, M., Yu, J., Shi, X., Zhu, J., Gao, X., and Liu, W. (2021). Polysaccharides catabolism by the human gut bacterium -Bacteroides thetaiotaomicron: advances and perspectives. Crit. Rev. Food Sci. Nutr. 61, 3569–3588. doi: 10.1080/10408398.2020.1803198
Keywords: Bacteroides thetaiotaomicron, fucosylation, proteomics, metabolic glycan labeling, polysaccharide
Citation: Tian X, Jiang H, Cai B, Feng H, Wang X and Yu G (2022) Comparative Proteomic Analysis of Fucosylated Glycoproteins Produced by Bacteroides thetaiotaomicron Under Different Polysaccharide Nutrition Conditions. Front. Microbiol. 13:826942. doi: 10.3389/fmicb.2022.826942
Received: 01 December 2021; Accepted: 31 January 2022;
Published: 04 March 2022.
Edited by:
Jennifer Angeline Gaddy, Vanderbilt University Medical Center, United StatesReviewed by:
David Leitsch, Medical University of Vienna, AustriaDarrell W. Cockburn, The Pennsylvania State University (PSU), United States
Copyright © 2022 Tian, Jiang, Cai, Feng, Wang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hao Jiang, aGFvamlhbmdAb3VjLmVkdS5jbg==; Guangli Yu, Z2x5dUBvdWMuZWR1LmNu
 Xiao Tian
Xiao Tian Hao Jiang
Hao Jiang Binbin Cai
Binbin Cai Huxin Feng
Huxin Feng Xuan Wang
Xuan Wang Guangli Yu
Guangli Yu