
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 07 April 2022
Sec. Terrestrial Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.825660
This article is part of the Research Topic Microbial Communities and Functions Contribute to Plant Performance under Various Stresses View all 25 articles
 Yihao Liu1
Yihao Liu1 Jinyang Yu1
Jinyang Yu1 Zuyu Wang1
Zuyu Wang1 Petri Penttinen1
Petri Penttinen1 Xiumei Yu1
Xiumei Yu1 Ke Zhao1
Ke Zhao1 Menggen Ma1
Menggen Ma1 Quanju Xiang1
Quanju Xiang1 Yunfu Gu1
Yunfu Gu1 Hanjun Liu1,2
Hanjun Liu1,2 Xiaoping Zhang1*
Xiaoping Zhang1* Qiang Chen1*
Qiang Chen1*
The barrenness of large mine tailing sand reservoirs increases the risks for landslides and erosion that may be accompanied with transfer of contaminants into the surrounding environment. The tailing sand is poor in nutrients, which effectively complicates the vegetation process. We investigated direct planting of Pennisetum giganteum into tailing sand using two pit planting methods: the plants were transplanted either directly into pits filled with soil or into soil-filled bio-matrix pots made of organic material. After growing P. giganteum in iron tailing sand for 360 days, the dry weight of the plants grown in the bio-matrix pot (T2) was approximately twofold higher than that of the plants grown in soil placed directly into the sand (T1). At 360 days, the organic matter (OM) content in the soil below the pit was the lowest in the not-planted treatment (T0) and the highest in T2, the available N (AN) contents were higher in T1 and T2 than in T0, and the available P and K contents were the highest in T2. At 360 days, the Shannon diversity of the soil microbial communities was higher in T1 and T2 than in T0, and the community compositions were clearly separated from each other. The profiles of predicted C cycle catabolism functions and N fixation-related functions in T1 and T2 at 360 days were different from those in the other communities. The results showed that P. giganteum grew well in the iron tailing sand, especially in the bio-matrix pot treatment, and the increased nutrient contents and changes in microbial communities indicated that using the bio-matrix pot in planting had potential to improve the vegetation process in iron tailing sands effectively.
The waste residue released from ore in mines after refining in the beneficiation plant is referred to as tailing sand (Dudeney et al., 2012; Kossoff et al., 2014; Lei et al., 2016). More than 10 billion tons of tailings are produced each year in the world, and approximately five billion tons of tailing sand are piled in China, with an annual increase of almost 500 million tons (Wang et al., 2017). The large mine tailing sand reservoirs are barren with no plants. The barrenness increases the risks for landslides and erosion that may be accompanied with transfer of contaminants into the surrounding environment. Tailing sand pond accidents, such as dam breaks and landslides, have been frequent and resulted in serious mudslides and collapses in recent years (Sun et al., 2018).
Several methods have been used to treat tailing sands, including refurbishment and reuse as building or chemical materials. However, the techniques are commonly of high cost and complicated or have resulted in secondary pollution (Alfonso et al., 2018; Galvão et al., 2018; Wu et al., 2018). Alarmingly, the wind-eroded dust from barren tailing sand reservoirs is a potent source of environmental contamination. In Fe-Pb-Zn mining area tailings in Murcia, Spain, the heavy metal concentrations in wind-eroded particulate matter were high, even though the concentrations in tailings were below hazardous levels (Khademi et al., 2018). Thus, the tailings should be stabilized. The tailings may be stabilized using chemical, for example, cementation, or biological methods, i.e., by vegetating the tailings. Tailing sand is poor in nutrients, which effectively complicates the vegetation process. Ensuring the growth of plants may require covering the tailings with soil, making the process laborious.
Pennisetum giganteum is a perennial gramineous plant that grows in the tropical, subtropical, and temperate area, with the merits of easy cultivation, drought and salinity tolerance, and the ability to thrive in barren soils (Hayat et al., 2020; Li et al., 2020). Since P. giganteum cultivation increased soil fertility and organic matter (OM) content in barren slopes or sandy decertified soils, it was often chosen as a pioneer plant for comprehensive ecological management in a variety of ecologically fragile areas (Hayat et al., 2020; Li et al., 2020).
In a preliminary study, a corn-based mini bio-matrix pot increased the growth of crop roots, yield, and microbial diversity in rhizosphere (unpublished). Thus, we hypothesized that in vegetating tailing sands, the bio-matrix pot would promote the growth of the plants and increase the OM and nutrient contents and microbial diversity in the sand. To test this, we applied two pit planting methods: the plants were transplanted either directly into pits filled with soil or into soil-filled bio-matrix pots; plant growth was measured, and soil properties and microbial communities as proxies of cultivability were analyzed.
Iron tailing sand was kindly provided by Chengdu Leejun Industrial Co., Ltd., Sichuan, China. The pH of the iron tailing sand was 7.90, and the sand contained OM 3.45 g kg–1, total nitrogen 0.12 g kg–1, total phosphorus 8.92 g kg–1, total potassium 7.96 g kg–1, alkaline nitrogen (AN) 16.64 mg kg–1, available phosphorus (AP) 1.78 mg kg–1, and available potassium (AK) 50.13 mg kg–1. The contents of available iron and exchangeable magnesium were 0.19 and 47.20 mg kg–1, respectively. The contents of chromium, copper, cadmium, lead, manganese, and zinc were 12.3, 52.4, 0.12, 4, 807, and 109 mg kg–1, respectively, all below the National Standard for Iron tailing sands in China (GB/T 31288-2014).
Soil for planting was from Jiange County (E105°28′52″, N32°0′52″), Sichuan, China. The soil was dried at room temperature, ground, and sieved through 20 mesh. The soil pH was 7.21; OM content was 11.70 g kg–1; and AN, AP, and AK contents were 35.98, 7.96, and 128.63 mg kg–1, respectively.
Bio-matrix pots were made by mixing corn cob powder (55%) and corn stalk powder (45%), adjusting water content to 55–60%, followed by autoclaving and cooling to room temperature. The mixture was inoculated with Ganoderma lucidum SZ-01, placed into a flowerpot mold, and incubated at 25°C for 10 days. During the incubation, the fungal mycelia bound the material firmly. After the incubation, the pots were dried at 80°C. The pots were 30- and 25-cm wide at the top and bottom, respectively, 20-cm high, and 2-cm thick. The OM, AN, AP, and AK contents of the pots were 642, 12.7, 21.7, and 52.8 g kg–1, respectively.
Two-to-three leaved, 15–20-cm high P. giganteum seedlings were purchased from the local growers.
The experiment, started in February 2016, was carried out in a greenhouse in 100 × 50 × 45 cm plastic containers. Three treatments were set up: iron tailing sand + soil (T0), iron tailing sand + soil + P. giganteum (T1), and iron tailing sand + soil + bio-matrix pot + P. giganteum (T2). Approximately 54 kg of iron tailing sand was placed into the container, and a hole the size of the bio-matrix pot was carved in the middle at the top (Figure 1). In T0 and T1, 6.5 kg of soil was placed into the hole; in T2, the soil was placed into the bio-matrix pot that was inserted into the hole. In T1 and T2, one P. giganteum seedling was transplanted into the soil. The containers were irrigated with distilled water.
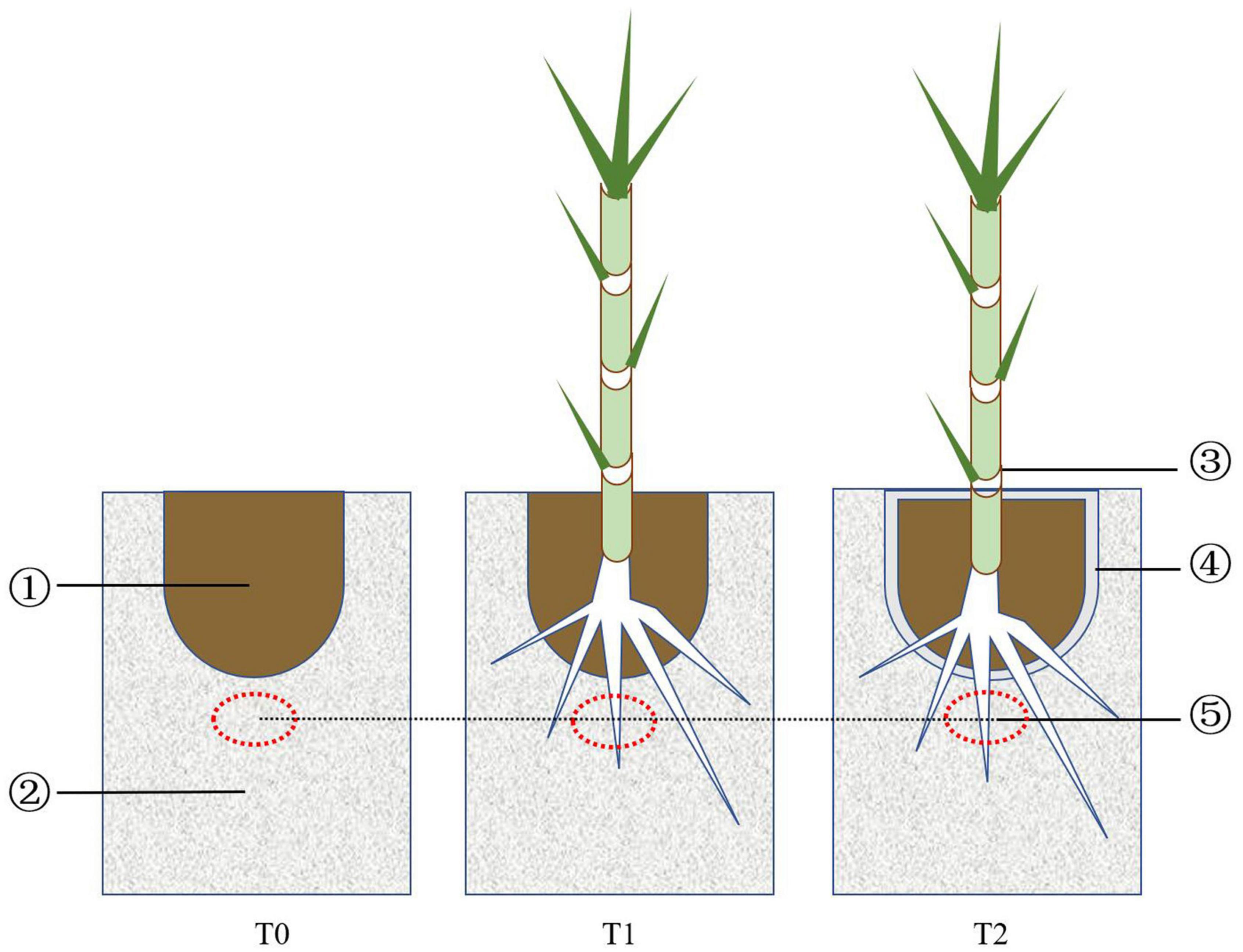
Figure 1. Design of the treatments: (1) soil, (2) iron tailing sand, (3) Pennisetum giganteum seedling, (4) bio-matrix pot, and (5) sampling spot.
Iron tailing sand samples from three containers per treatment were collected at 90, 180, and 360 days (Figure 1). The pH and OM, AN, AP, and AK contents were determined as described previously (Siddiqi and Glass, 1981). The plants were harvested at 360 days, and the shoot and root biomasses were measured after drying at 60°C to constant weight.
DNA was extracted from 0.5 g of sample using a Fast DNA ® SPIN kit for soil (MP Biomedicals, United States) according to the manufacturer’s instructions. The DNA concentration and quality were assessed by 0.8% agarose gel electrophoresis and UV spectrophotometer (Thermo Scientific NC2000). DNA extracts were stored at −20°C.
The universal primers 338F (5′-ACTC CTA CGG GAG GCA GCA-3′) and 806R (5′-GGA CTA CHV GGG TWT CTA AT-3′) were used to amplify the V3–V4 region of the 16S rRNA gene of sample DNA (Huse et al., 2008; Göransson et al., 2011). The amplification was done in 25-μl reactions containing 5× reaction buffer 5 μl, 5× GC buffer 5 μl, dNTP (2.5 mM) 2 μl, forward primer (10 mM) 1 μl, reverse primer (10 mM) 1 μl, DNA template 2 μl, ddH2O 8.75 μl, and Q5 DNA Polymerase 0.25 μl (Magoc and Salzberg, 2011). The thermal program included denaturation at 98°C for 2 min, 26 cycles with denaturation at 98°C for 15 s, annealing at 55°C for 30 s, extension at 72°C for 30 s, and a final extension at 72°C for 5 min. The amplified products were separated by 2% agarose gel electrophoresis, and the target fragments were recovered using AXYGEN’s gel recovery kit (AXYGEN, United States). PCR products were mixed in equimolar ratios and purified using the GeneJET PCR Purification Kit (Thermo Fisher). Amplicon libraries were prepared using the NEBNext ® Ultra™ DNA Library Prep Kit (New England Biolabs) prior to paired-end sequencing on the Illumina Miseq platform (Illumina, San Diego, CA, United States). The PCR products were sequenced at Shanghai Personal Medical Laboratory Co., Ltd. (Shanghai, China).
Illumina sequencing reads were paired and quality-filtered using the Quantitative Insights into Microbial Ecology (QIIME, v1.7.0) pipeline with default settings (Caporaso et al., 2010). Chimeric sequences were removed using UCHIME algorithm after alignment with the Gold database1 (Edgar, 2010). The sequences were assigned into operational taxonomic units (OTUs) at 97% similarity using UPARSE in the USEARCH pipeline v7.0.1090 (Edgar, 2010). The representative sequences of OTUs were assigned into taxa using SILVA rRNA database release 132 (Quast et al., 2013).
Rarefaction curves were created using R package vegan v2.5e2 in R studio (Oksanen et al., 2019). Chao1 and Shannon α-diversity indices at the OTU level were calculated in QIIME pipeline v1.7.0. The microbial communities were visualized at the genus level using cluster heatmaps based on 30 taxa with the highest relative abundances. The relative abundances of taxa were normalized using the z-score standardization method. The taxa were hierarchically clustered according to the Pearson correlation distance measure.
Microbial community composition at the genus level was visualized using principal component analysis (PCA) in R package vegan v2.5 in R studio (Oksanen et al., 2019). Partial least squares discriminant analysis (PLS-DA) at the OTU level was done using R packages BiocManager and mixOmics in R studio2 (Matthew and Rayens, 2003).
Differential abundance at the OTU level was analyzed pairwise at 90, 180, and 360 days using ALDEx2 (Fernandes et al., 2013). In the analysis, the underlying distributions were estimated using 128 Monte Carlo samples, the abundances were transformed using centered log-ratios, differences were tested using t-test, and effect sizes were measured as the relative fold difference. OTUs were considered differentially abundant when the effect size is >|1| and p < 0.1.
Carbon and nitrogen cycle-related functions were predicted using Functional Annotation of Prokaryotic Taxa (FAPROTAX) analysis in the Python-based FAPROTAX 1.1 package (Louca et al., 2016). Results were visualized as a clustered heatmap with a Pearson correlation dendrogram using the function “pheatmap” v1.0.12 in R studio.
Microsoft Excel 2016 was used to calculate means and standard deviations. Differences in plant and soil characteristics were tested using one-way ANOVA and Tukey’s test in IBM SPSS Statistics 20.0 (SPSS Inc., Chicago, IL, United States). Differences were taken as statistically significant at p < 0.05.
After growing P. giganteum in iron tailing sand for 360 days, both the root dry weight (RDW) and shoot dry weight (SDW) of the plants grown in the bio-matrix pot (T2) were approximately twofold higher than those of the plants grown in soil placed directly into the sand (T1) (p < 0.05) (Figure 2), suggesting that the bio-matrix pot has promoted the growth of the plants.
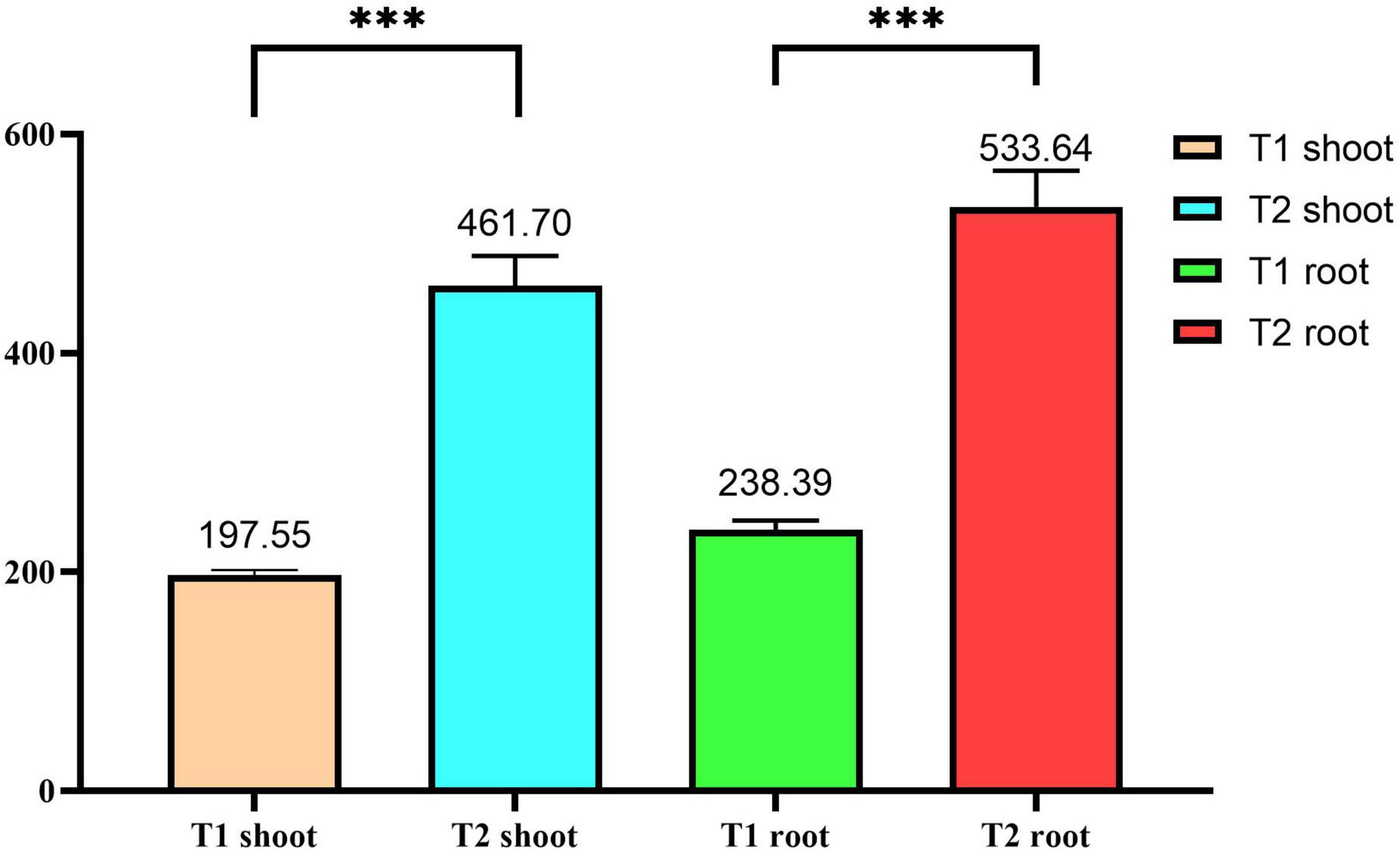
Figure 2. The shoot and root dry weights of Pennisetum giganteum grown in tailing sands for 360 days. The error bars indicate standard deviation (n = 3). ***Statistically significant difference (p < 0.05). T1, iron tailing sand + soil + Pennisetum giganteum; T2, iron tailing sand + soil + bio-matrix pot + Pennisetum giganteum.
The OM content of the iron tailing sand in T1 and T2 increased over time (Table 1). At 180 days, the OM content was the highest in T2, and at 360 days, the OM content was the lowest in the not-planted treatment (T0) and the highest in T2 (p < 0.05). The differences in pH values that ranged from 8.22 to 8.76 showed no clear trend. At 180 and 360 days, the AN contents were higher in T1 and T2 than in T0 (p < 0.05) (Table 1). The AP contents were the highest in T2 (p < 0.05). At 180 and 360 days, the AK contents were the highest in T2, and at 360 days, higher in T1 than in T0 (p < 0.05) (Table 1).
The 1,399,587 bacterial sequences from the 27 iron tailing sand samples were clustered into 1,436 OTUs at 97% similarity level. At 180 days, Chao1 index was higher in T1 than in T0 (p < 0.05) (Table 2). At 360 days, Shannon index was higher in T1 and T2 than in T0 (p < 0.05). Proteobacteria, Chloroflexi, Acidobacteria, and Actinobacteria were the most dominant phyla. The relative abundances of Proteobacteria accounted for 25.73–51.28% of the total abundance, Actinobacteria for 11.95–38.43%, Chloroflexi for 5.97–14.92%, and Acidobacteria for 4.92–8.76% (Figure 3 and Supplementary Table 1). At the genus level, the relative abundances of Pseudomonas, Sideroxydans, and Geobacter ranged from 0.53 to 9.41, 0.01 to 3.98, and 0.16 to 3.93%, respectively (Supplementary Table 2).
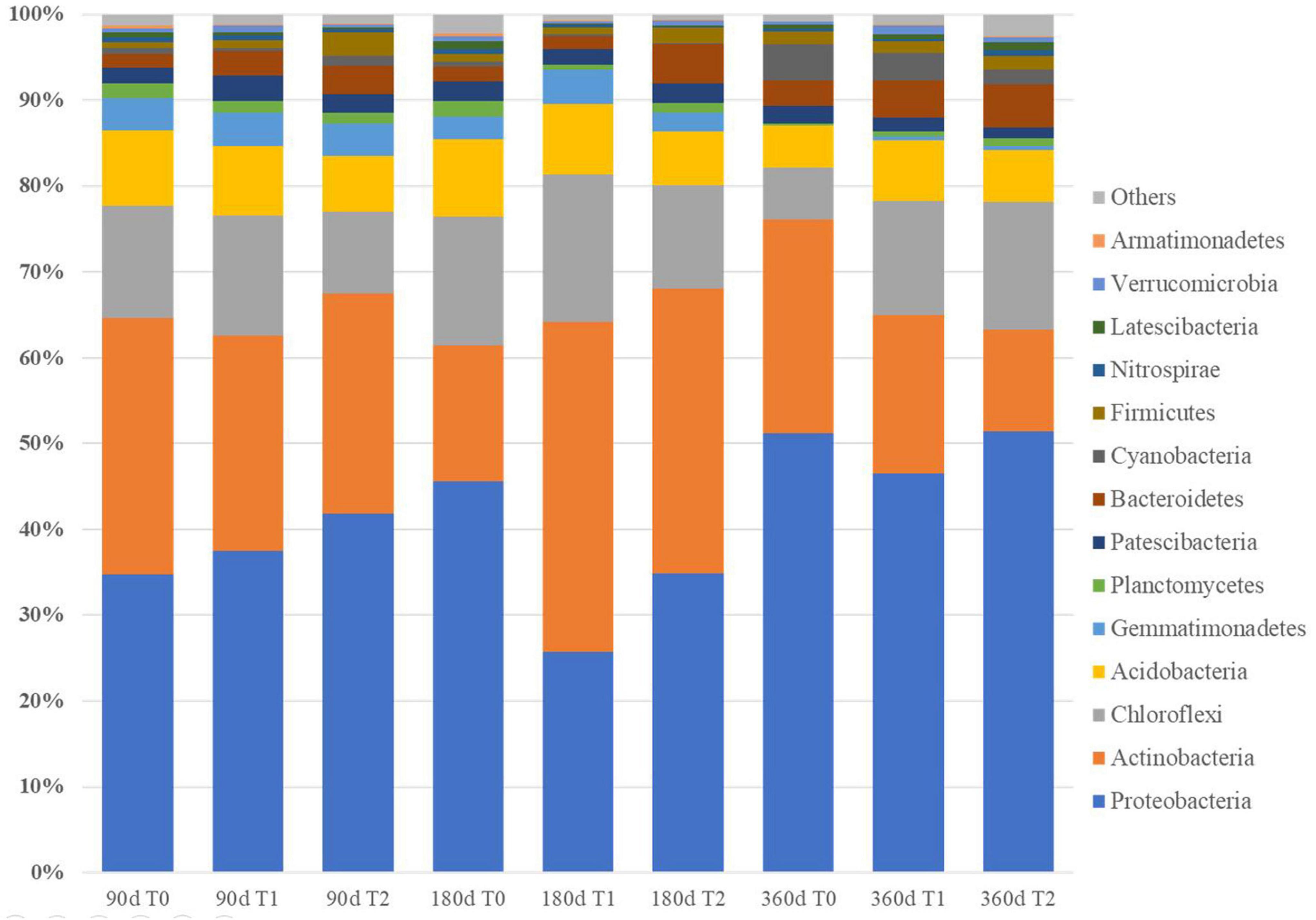
Figure 3. Bacterial community composition in the tailing sand at the phylum level. T0, iron tailing sand + soil; T1, iron tailing sand + soil + Pennisetum giganteum; T2, iron tailing sand + soil + bio-matrix pot + Pennisetum giganteum; 90 d, 180 d, and 360 d indicate the sampling time in days after the start of the experiment.
The microbial communities in T1 and T2 at 360 days were separated from the other communities in the hierarchical clustering (Figure 4). At 90 and 180 days, the communities in T0 and T1 were relatively similar in the PCA (Figure 5). The T2 community was separated from those in T0 and T1 at 180 days, and at 360 days, all the communities were clearly separated from each other (Figure 5). Based on the PLS-DA, the communities in T0 were different from those in T1 and T2 (Figure 6). At 180 days, the relative abundances of Anaerolineaceae (Chloroflexi) OTU7 and MND1 (Proteobacteria) OTU12 were lower in T2 than in T0 (effect size > |1|, p < 0.1) (Table 3). At 360 days, the relative abundances of Xanthobacteraceae (Proteobacteria) OTU24 and Marine Group II (Euryarchaeota) OTU59 were higher in T1 than in T0, and those of Micrococcaceae (Actinobacteria) OTU1, Solirubrobacteraceae (Actinobacteria) OTU30, Rubellimicrobium (Proteobacteria) OTU115, and Coleofasciculaceae (Cyanobacteria) OTU187 were lower (effect size > |1|, p < 0.1). The relative abundances of Sideroxydans (Proteobacteria) OTU31 and Marine Group II OTU59 were higher in T2 than in T0 at 360 days (effect size > |1|, p < 0.1). In the T1 treatment, the relative abundances of Aeromicrobium (Actinobacteria) OTU10 and Uncultured Gitt-GS-136 (Chloroflexi) OTU22 were higher at 180 and 360 days, respectively, than at 90 days (effect size > |1|, p < 0.1).
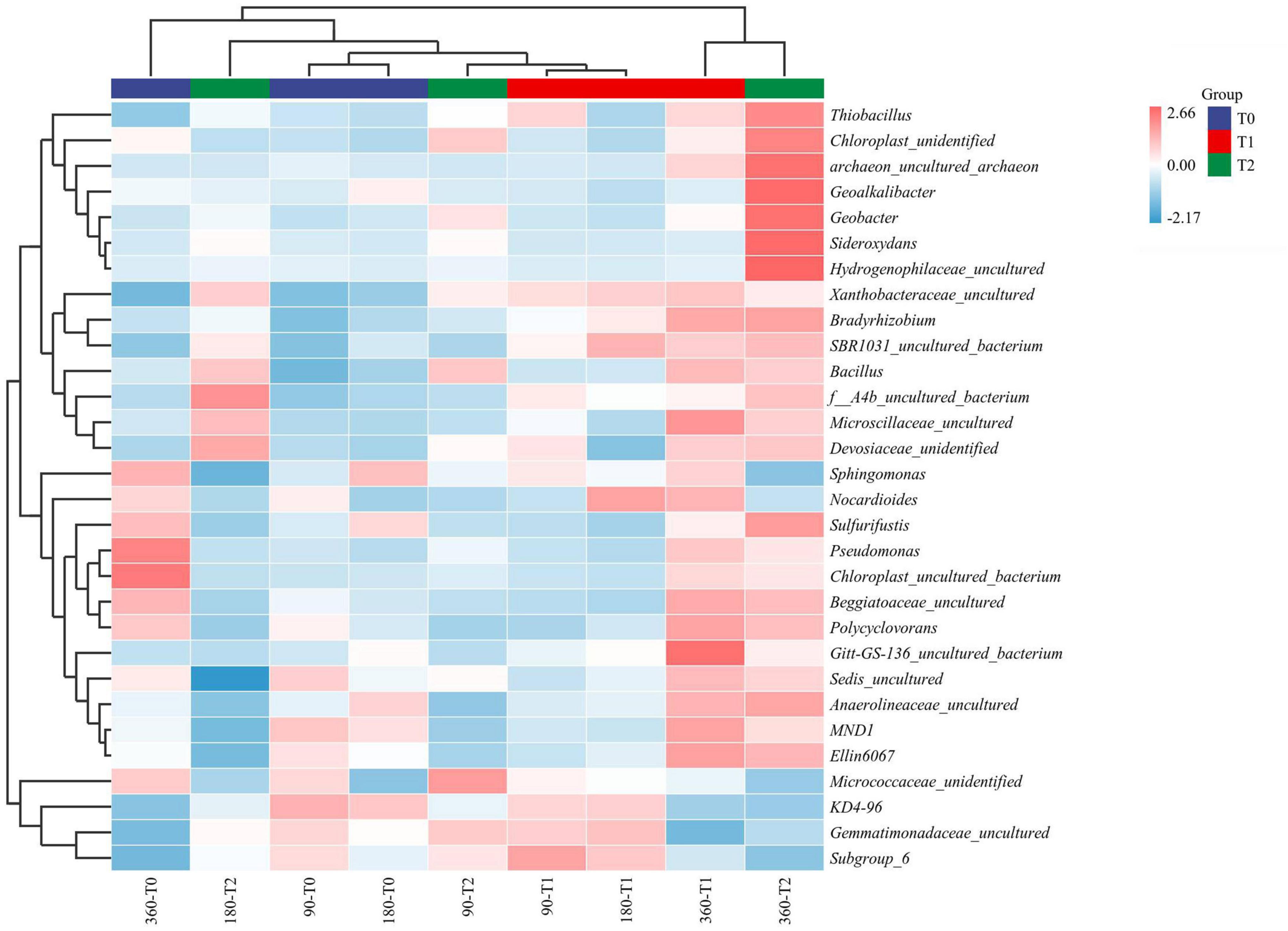
Figure 4. Bacterial community composition and their hierarchical clustering in the tailing sand at the genus level. T0, iron tailing sand + soil; T1, iron tailing sand + soil + Pennisetum giganteum; T2, iron tailing sand + soil + bio-matrix pot + Pennisetum giganteum; 90 d, 180 d, and 360 d indicate the sampling time in days after the start of the experiment.
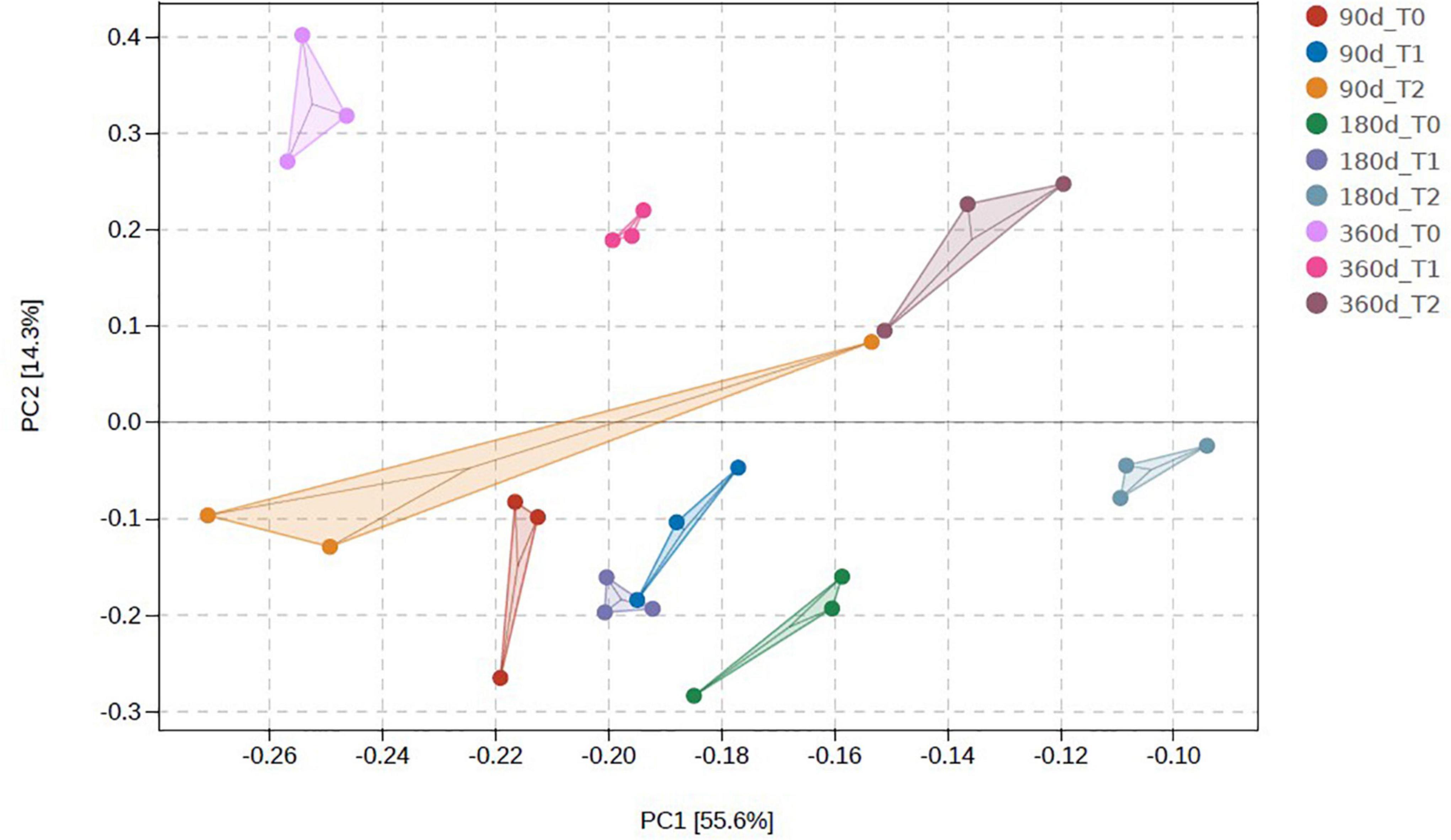
Figure 5. Principal component analysis of the microbial communities in the tailing sand at the operational taxonomic unit (OTU) level. T0, iron tailing sand + soil; T1, iron tailing sand + soil + Pennisetum giganteum; T2, iron tailing sand + soil + bio-matrix pot + Pennisetum giganteum; 90 d, 180 d, and 360 d indicate the sampling time in days after the start of the experiment.
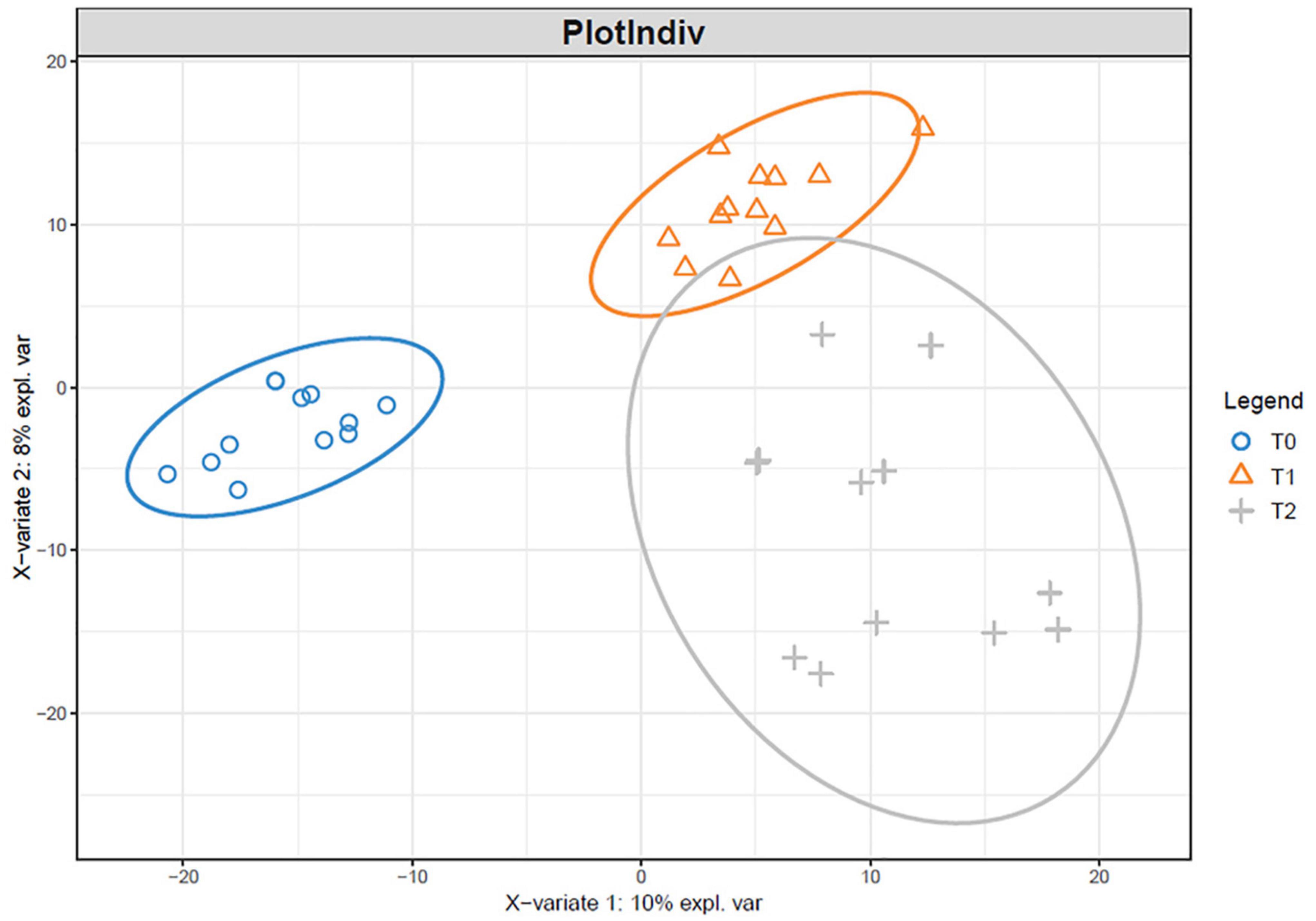
Figure 6. Partial least squares discriminant analysis of the microbial communities in the tailing sand at the OTU level. T0, iron tailing sand + soil; T1, iron tailing sand + soil + Pennisetum giganteum; T2, iron tailing sand + soil + bio-matrix pot + Pennisetum giganteum.
We predicted C and N cycle-related functions to estimate the accompanying changes on gene level. In line with the OM content increase over time in T1 and T2, the predicted functions related to carbohydrate metabolism, C fixation in prokaryotes and photosynthetic organisms, and glycosyltransferases were on the same level at 90 days and high in T1 and T2 at 360 days (Figure 7A). The predicted C cycle biosynthesis and catabolism profiles were similar at 90 days (Figures 7B,C). The profiles of predicted biosynthesis functions in T1 and T2 at 360 days were relatively similar to those in T0 at 180 and 360 days (Figure 7B). The profiles of predicted C cycle catabolism functions in T1 and T2 at 360 days were different from those in the other communities (Figure 7C).
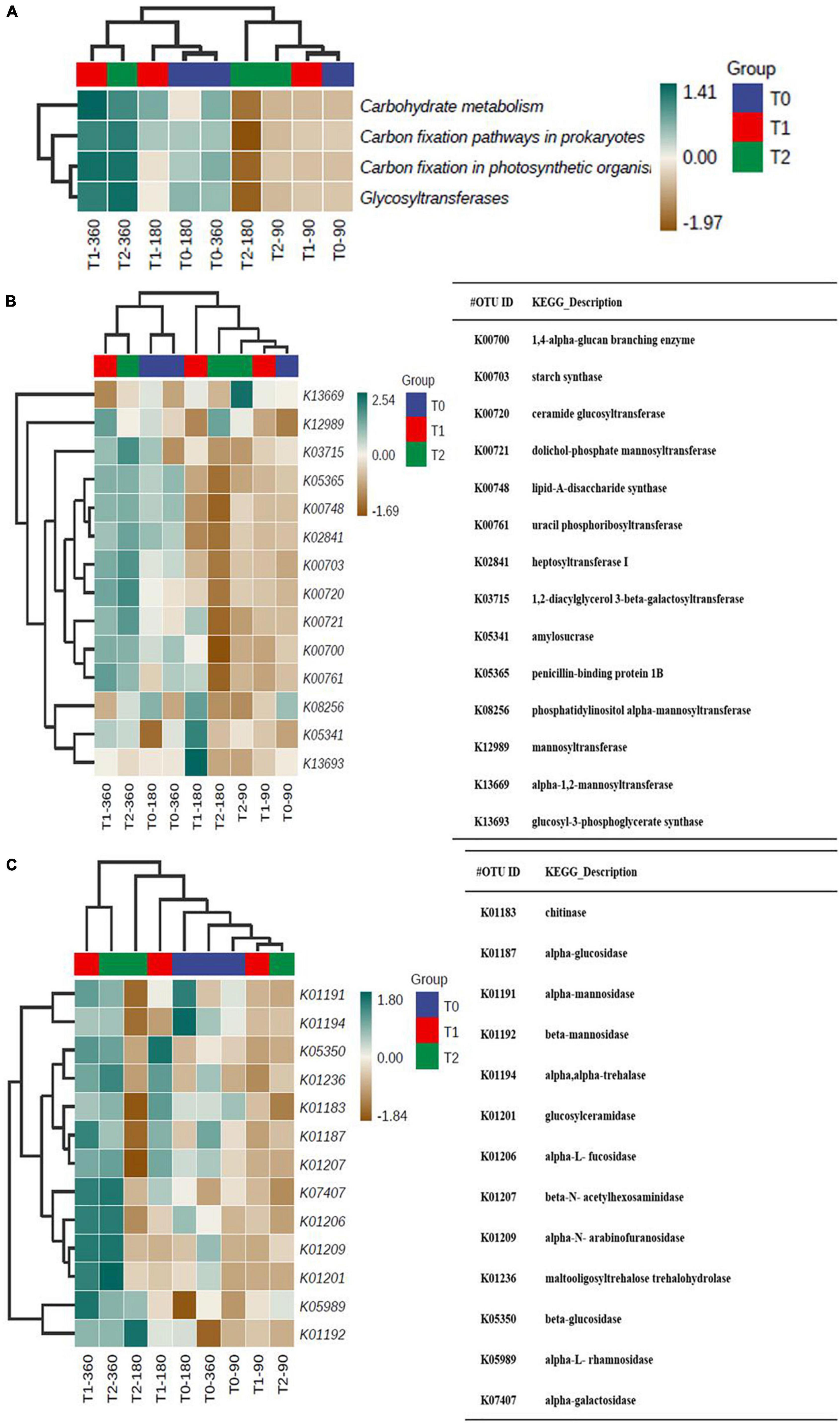
Figure 7. (A) The predicted C cycle-related functions in the microbial communities in the tailing sand samples and the predicted C cycle (B) biosynthesis and (C) catabolism profiles. T0, iron tailing sand + soil; T1, iron tailing sand + soil + Pennisetum giganteum; T2, iron tailing sand + soil + bio-matrix pot + Pennisetum giganteum. 90 d, 180 d, and 360 d indicate the sampling time in days after the start of the experiment.
The predicted N fixation-related functions were on approximately the same level at 90 and 180 days and high in T1 and T2 at 360 days (Figure 8A). The predicted denitrification-related functions were on approximately the same level at 360 days (Figure 8B).
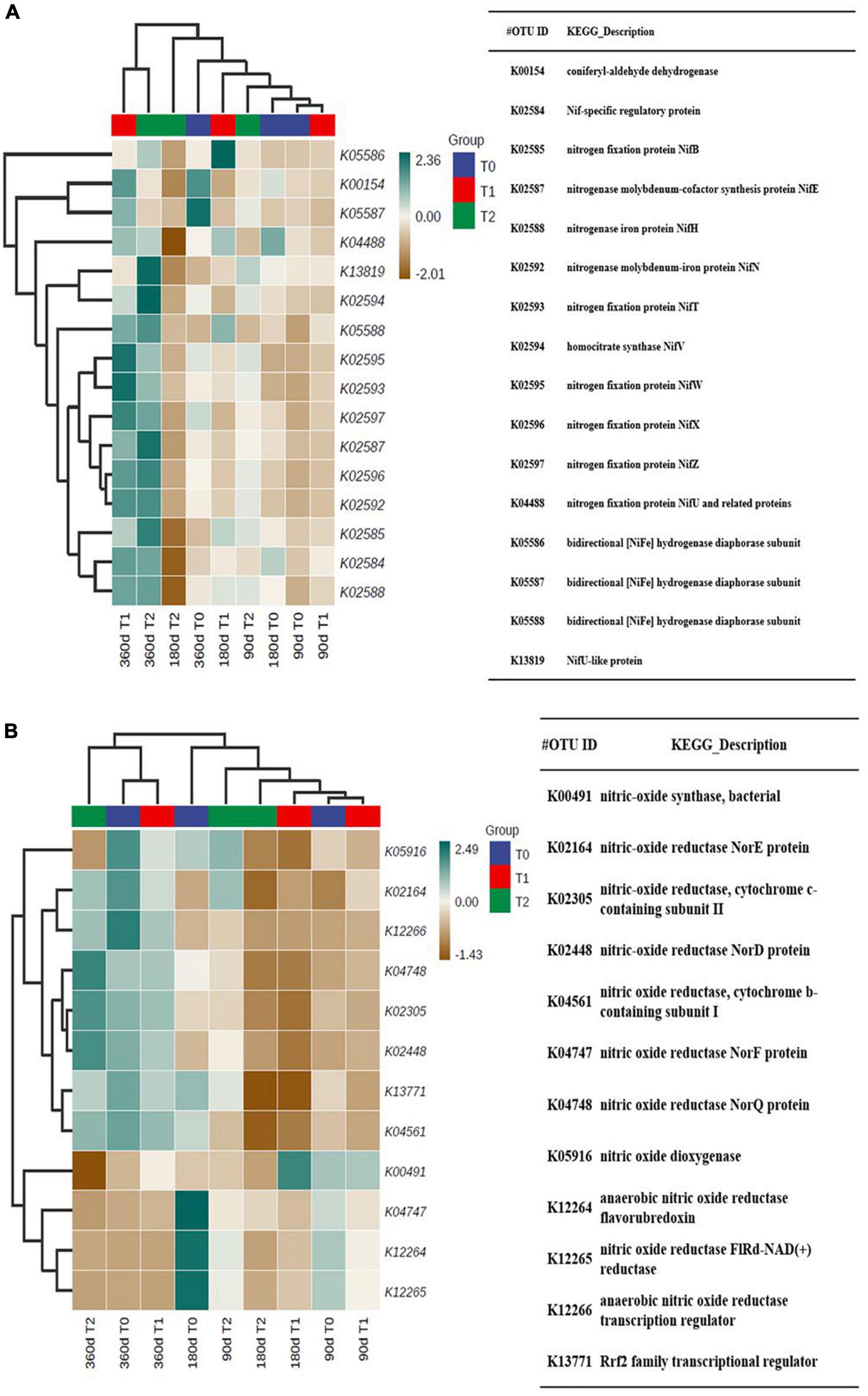
Figure 8. The predicted (A) N fixation-related functions and (B) denitrification-related functions in the microbial communities in the tailing sand samples. T0, iron tailing sand + soil; T1, iron tailing sand + soil + Pennisetum giganteum; T2, iron tailing sand + soil + bio-matrix pot + Pennisetum giganteum. 90 d, 180 d, and 360 d indicate the sampling time in days after the start of the experiment.
The large mine tailing sand reservoirs are barren with no plants, making them prone to landslides and erosion. Vegetating the tailings is recommended to prevent the transfer of contaminants into surrounding environment (Khademi et al., 2018), yet it is complicated due to the poor nutrient content of the tailing sand. We investigated direct planting into tailing sand using two pit planting methods: the plants were transplanted either directly into pits filled with soil or into soil-filled bio-matrix pots made of corn and G. lucidum.
The results showed that P. giganteum grew well in the iron tailing sand, especially in the bio-matrix pot treatment (T2). The higher biomass in T2 may have been due to the G. lucidum polysaccharides that are known to promote the growth of cotton (Zhang et al., 2019). The OM and AN contents increased in the planted iron tailing sand in the directly-into-soil-planted treatment (T1) and T2, whereas in the not-planted soil-only treatment (T0), the contents remained on the same level, indicating that the increases were due to the roots penetrating into the sand. At the end of the experiment, the AK had increased in the planted treatments and were the highest in T2. This, together with the over two times higher biomass of P. giganteum in T2 than in T1, indicated that using the bio-matrix pot in planting had potential to improve the vegetation process in iron tailing sands effectively.
The physicochemical properties of soil affect the diversity and activity of bacterial communities, and in natural tailing restoration, the diversity of soil microbial communities increases along the steps (Colin et al., 2019). In our study, at the end of the experiment, the diversity was higher in the planted treatments than in T0. As for the OM and AN content, the diversity increase was probably due to the plants and not only because of the soil in the pits. The community compositions were relatively similar at 90 days and in T0 and T1 even at 180 days, implying that the community composition changed at a faster rate in T2, possibly due to the quicker build-up of OM in T2. At the end of the experiment at 360 days, all the communities were clearly separated, implying that both the planting and the planting method had contributed to the differences in composition. Consistent with previous studies (Wang et al., 2017), Proteobacteria, Chloroflexi, Acidobacteria, and Actinobacteria were the dominant phyla, indicating the microbial communities have good adaptability. In barren gold mine tailings, Actinobacteria was the dominant phylum (Sibanda et al., 2019). In tailings undergoing restoration, Proteobacteria increased along the restoration process, and the abundance of actinobacteria decreased after remediation (Wu et al., 2018; Xu et al., 2018; Yu et al., 2019). In agreement, the differentially abundant OTUs were mostly assigned into Proteobacteria and Actinobacteria. Despite the community-level differences, the number of differentially abundant OTUs was small due to the large within-treatment variation.
The effect of the treatments on soil C and N cycle-related functions was estimated based on predicted functional profiles of microbial communities. At 360 days, functions related to C cycle catabolism and N fixation were estimated to be more abundant in the planted treatments, whereas the C cycle biosynthesis and denitrification-related functions seemed to change similarly in the treatments over time. Presumably, the increase in OM content with planting explained the increase in C cycle catabolism. In copper mine tailings, the diversity of nitrogen-fixing bacteria increased along natural restoration time together with development of pioneer plant communities (Zhan and Sun, 2012; Xiao et al., 2019). The increase in predicted N fixation functions implied that both planting methods had the potential to promote further development of vegetation, which is often limited by the availability of N (Göransson et al., 2011).
The results showed that P. giganteum grew well in the iron tailing sand, especially in the bio-matrix pot treatment, and increased the nutrient contents, indicating that using the bio-matrix pot in planting had the potential to improve the vegetation process in iron tailing sands effectively. At the end of the experiment, the diversity of the microbial communities in the tailing sands was higher in the planted treatments. The increase in predicted N fixation functions implied that both planting methods had the potential to promote further development of vegetation.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
QC, XZ, and YL designed the research. YL, JY, and ZW performed the research. YL, YG, MM, KZ, QX, and HL analyzed the data. YL, QC, XZ, XY, and PP analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This project was supported by the Key Research Project of Sichuan Province (No. 2019YFN0149) and the Subject Support Program of Sichuan Agricultural University.
HL was employed by the company CNPC Chuanqing Drilling Engineering Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Mr. Fu Runqi for his assistance with data analysis and mapping.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.825660/full#supplementary-material
Alfonso, P., Tomasa, O., Garcia-Valles, M., Tarragó, M., Martínez, S., and Esteves, H. (2018). Potential of tungsten tailings as glass raw materials. Mater. Lett. 228, 456–458.
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of highthroughput community sequencing data. Nat. Methods 7, 335–336.
Colin, Y., Goberna, M., Verdu, M., and Navarro-Cano, J. A. (2019). Successional trajectories of soil bacterial communities in mine tailings: The role of plant functional traits. J. Environ. Manage. 241, 284–292. doi: 10.1016/j.jenvman.2019.04.023
Dudeney, A. W. L., Chan, B. K. C., Bouzalakos, S., and Huisman, J. L. (2012). Management of waste and wastewater from mineral industry processes, especially leaching of sulphide resources state of the art. Int. J. Min. Reclam. Environ. 27, 2–37.
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Fernandes, A. D., Macklaim, J. M., Linn, T. G., Reid, G., and Gloor, G. B. (2013). ANOVA-like differential expression (ALDEx) analysis for mixed population RNA-Seq. PLoS One 8:e67019. doi: 10.1371/journal.pone.0067019
Galvão, J. L. B., Andrade, H. D., Brigolini, G. J., Peixoto, R. A. F., and Mendes, J. C. (2018). Reuse of iron ore tailings from tailings dams as pigment for sustainable paints. J. Clean. Prod. 200, 412–422.
Göransson, H., Olde Venterink, H., and Bååth, E. (2011). Soil bacterial growth and nutrient limitation along a chronosequence from a glacier forefield. Soil Biol. Biochem. 43, 1333–1340.
Hayat, K., Zhou, Y., Menhas, S., Bundschuh, J., Hayat, S., Ullah, A., et al. (2020). Pennisetum giganteum: an emerging salt accumulating/tolerant non-conventional crop for sustainable saline agriculture and simultaneous phytoremediation. Environ. Pollut. 265:114876. doi: 10.1016/j.envpol.2020.114876
Huse, S. M., Dethlefsen, L., Huber, J. A., Mark Welch, D., Relman, D. A., and Sogin, M. L. (2008). Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 4:e1000255. doi: 10.1371/journal.pgen.1000255
Khademi, H., Abbaspour, A., Martinez-Martinez, S., Gabarron, M., Shahrokh, V., Faz, A., et al. (2018). Provenance and environmental risk of windblown materials from mine tailing ponds. Murcia. Spain. Environ. Pollut. 241, 432–440. doi: 10.1016/j.envpol.2018.05.084
Kossoff, D., Dubbin, W. E., Alfredsson, M., Edwards, S. J., Macklin, M. G., and Hudson-Edwards, K. A. (2014). Mine tailings dams: Characteristics, failure, environmental impacts, and remediation. Appl. Geochem. 51, 229–245.
Lei, H., Peng, Z., Yigang, H., and Yang, Z. (2016). Vegetation and soil restoration in refuse dumps from open pit coal mines. Ecol. Eng. 94, 638–646. doi: 10.3390/ijerph17061975
Li, Q., Xiang, C., Xu, L., Cui, J., Fu, S., Chen, B., et al. (2020). SMRT sequencing of a full-length transcriptome reveals transcript variants involved in C18 unsaturated fatty acid biosynthesis and metabolism pathways at chilling temperature in Pennisetum giganteum. BMC Genomics 21:52. doi: 10.1186/s12864-019-6441-3
Louca, S., Parfrey, L. W., and Doebeli, M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353, 1272–1277. doi: 10.1126/science.aaf4507
Magoc, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Matthew, B., and Rayens, W. (2003). Partial least squares for discrimination. J. Chemom. 17, 166–173. doi: 10.1016/j.nicl.2015.01.005
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’Hara, R. B., et al. (2019). Vegan: Community Ecology Package. R package version 2.5-6.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic. Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Sibanda, T., Selvarajan, R., Msagati, T., Venkatachalam, S., and Meddows-Taylor, S. (2019). Defunct gold mine tailings are natural reservoir for unique bacterial communities revealed by high-throughput sequencing analysis. Sci. Total Environ. 650, 2199–2209. doi: 10.1016/j.scitotenv.2018.09.380
Siddiqi, M. Y., and Glass, A. D. M. (1981). Utilization index A modified approach to the estimation and comparison of nutrient utilization efficiency in plants. J. Plant Nutr. 4, 289–302.
Sun, X., Zhou, Y., Tan, Y., Wu, Z., Lu, P., Zhang, G., et al. (2018). Restoration with pioneer plants changes soil properties and remodels the diversity and structure of bacterial communities in rhizosphere and bulk soil of copper mine tailings in Jiangxi Province. China. Environ. Sci. Pollut. Res. Int. 25, 22106–22119. doi: 10.1007/s11356-018-2244-3
Wang, L., Ji, B., Hu, Y., Liu, R., and Sun, W. (2017). A review on in situ phytoremediation of mine tailings. Chemosphere 184, 594–600. doi: 10.1016/j.chemosphere.2017.06.025
Wu, Z., Yu, F., Sun, X., Wu, S., Li, X., Liu, T., et al. (2018). Long term effects of Lespedeza bicolor revegetation on soil bacterial communities in Dexing copper mine tailings in Jiangxi Province. China. Appl. Soil Ecol. 125, 192–201.
Xiao, E., Ning, Z., Xiao, T., Sun, W., Qiu, Y., Zhang, Y., et al. (2019). Variation in rhizosphere microbiota correlates with edaphic factor in an abandoned antimony tailing dump. Environ. Pollut. 253, 141–151. doi: 10.1016/j.envpol.2019.06.097
Xu, L., Naylor, D., Dong, Z., Simmons, T., Pierroz, G., Hixson, K. K., et al. (2018). Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proc. Natl. Acad. Sci. U.S.A 115, E4284–E4293.
Yu, X., Kang, X., Li, Y., Cui, Y., Tu, W., Shen, T., et al. (2019). Rhizobia population was favoured during in situ phytoremediation of vanadium-titanium magnetite mine tailings dam using Pongamia pinnata. Environ Pollut 255:113167. doi: 10.1016/j.envpol.2019.113167
Zhan, J., and Sun, Q. (2012). Diversity of free-living nitrogen-fixing microorganisms in the rhizosphere and non-rhizosphere of pioneer plants growing on wastelands of copper mine tailings. Microbiol. Res. 167, 157–165. doi: 10.1016/j.micres.2011.05.006
Keywords: iron tailing sands, bio-matrix pot, microbial communities, C and N cycle, vegetation
Citation: Liu Y, Yu J, Wang Z, Penttinen P, Yu X, Zhao K, Ma M, Xiang Q, Gu Y, Liu H, Zhang X and Chen Q (2022) Bio-Matrix Pot Addition Enhanced the Vegetation Process of Iron Tailings by Pennisetum giganteum. Front. Microbiol. 13:825660. doi: 10.3389/fmicb.2022.825660
Received: 30 November 2021; Accepted: 24 February 2022;
Published: 07 April 2022.
Edited by:
Hai-Ming Zhao, Jinan University, ChinaReviewed by:
Aleksandr G. Bulaev, Federal Center Research Fundamentals of Biotechnology (RAS), RussiaCopyright © 2022 Liu, Yu, Wang, Penttinen, Yu, Zhao, Ma, Xiang, Gu, Liu, Zhang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoping Zhang, emhhbmd4aWFvcGluZ3BoZEAxMjYuY29t; Qiang Chen, Y3FpYW5nQHNpY2F1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.