- 1Microbiology Unit, Interdisciplinary Biosciences, Belgian Nuclear Research Centre, SCK CEN, Mol, Belgium
- 2Center for Microbial Ecology and Technology, Ghent University, Ghent, Belgium
- 3Centre Etudes Nucléaires de Bordeaux Gradignan (CENBG), Bordeaux, France
Environmental uranium pollution due to industries producing naturally occurring radioactive material or nuclear accidents and releases is a global concern. Uranium is hazardous for ecosystems as well as for humans when accumulated through the food chain, through contaminated groundwater and potable water sources, or through inhalation. In particular, uranium pollution pressures microbial communities, which are essential for healthy ecosystems. In turn, microorganisms can influence the mobility and toxicity of uranium through processes like biosorption, bioreduction, biomineralization, and bioaccumulation. These processes were characterized by studying the interaction of different bacteria with uranium. However, most studies unraveling the underlying molecular mechanisms originate from the last decade. Molecular mechanisms help to understand how bacteria interact with radionuclides in the environment. Furthermore, knowledge on these underlying mechanisms could be exploited to improve bioremediation technologies. Here, we review the current knowledge on bacterial uranium resistance and how this could be used for bioremediation applications.
Introduction
Controlled and accidental releases by nuclear industries, nuclear weapon tests and nuclear accidents have globally spread radionuclides in our environment, including synthetic radionuclides like 3H, 90Sr, 131I, 137Cs, and 241Am (UNSCEAR, 1988; Van der Stricht and Janssens, 2010; Taira et al., 2013; Prăvălie, 2014; International Atomic Energy Agency [IAEA], 2018). In addition, human activities can lead to an increased exposure to naturally occurring radioactive material (NORM), which primarily include 238U, 232Th, 40K and their decay products (International Atomic Energy Agency [IAEA], 2003). Examples of NORM industries are oil and gas production, coal mining and combustion, metal and uranium mining and processing, geothermal energy production, groundwater treatment, and phosphate mining for fertilizer production (International Atomic Energy Agency [IAEA], 2003; UNSCEAR, 2008). Environmental accumulation of natural and synthetic radionuclides can be hazardous for ecosystems. Transfer of radionuclides to vegetation mainly occurs through water bodies (Salbu et al., 2013), resulting in cytogenetic damage that decreases the reproductive ability (Geras’kin et al., 2013). Furthermore, contamination of water bodies, such as lakes, results in a substantial transfer of radionuclides to fish, herbivores and carnivores (Whicker, 1983; Salbu et al., 2013; Strømman et al., 2013). Humans can be exposed when radionuclides are accumulated through the food chain, through contaminated potable water sources, such as groundwater, or through inhalation (Tompson et al., 2002; Tykva, 2004; Zachara et al., 2013).
Uranium, atomic number 92, is a silvery-white metal belonging to the actinides that is naturally found in minerals such as pitchblende, uraninite, carnotite, autunite, uranophane, davidite and tobernite, but can also occur in phosphate rock, lignite and monazite sands (Lide, 2003). It is one of the principal contaminants of concern in NORM and nuclear industry (NEA/IAEA, 1999; International Atomic Energy Agency [IAEA], 2003). The most common natural isotopes are 238U (99.27%), 235U (0.72%) and 234U (<0.01%). Both 238U and 235U can be used as nuclear fuel, but 235U is more important as it is able to self-sustain a fission chain reaction. Therefore, 238U with slightly enriched 235U is used for the generation of electricity (Hammond, 2004). 238U, 235U, and 234U decay by emitting an alpha particle and have half-lives of 4.5 × 109 years, 700 × 106 years, and 246 × 103 years, respectively. 238U decays 14 times by alpha or beta emission before reaching stable lead-206 (206Pb).
As uranium accumulation could potentially be harmful for humans and ecosystems, strict control and monitoring is essential, and protection and remediation strategies are deployed. Various physical and chemical methods are available, some more advanced than others, but each with its own limitations and drawbacks such as high cost, high complexity and long time span (Godheja et al., 2016). Consequently, there is a need for more simple and ecofriendly alternatives, including biologically based methods. Microorganisms are often found in uranium-contaminated sites and can influence uranium mobility, toxicity and distribution (Choudhary and Sar, 2015). Processes such as biosorption, bioaccumulation, biomineralization and redox transformations are currently well known (Figure 1; Merroun and Selenska-Pobell, 2008). In turn, uranium exerts a permanent pressure on the prevailing microbial population, disrupting microbial communities and processes (Tapia-Rodríguez et al., 2012; Lopez-Fernandez et al., 2017; Sutcliffe et al., 2017). Consequently, fundamental understanding of the interaction between microorganisms and uranium is essential to assess the microbial impact in contaminated environments correctly. Moreover, knowledge on the underlying cellular response can be exploited to improve bioremediation technologies.
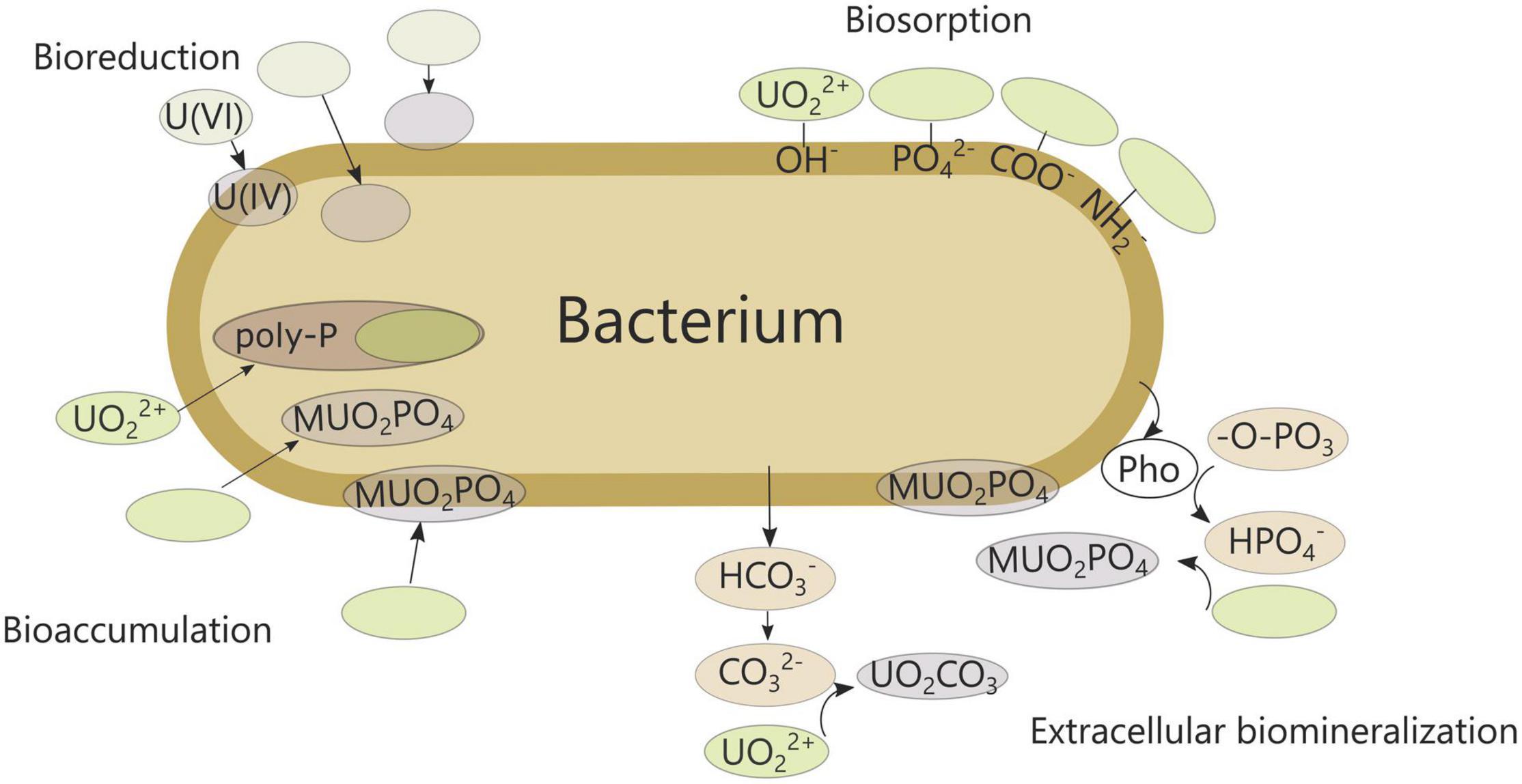
Figure 1. Bacterial interactions with uranium. Uranium minerals are presented as UO2CO3 or MUO2PO4 with M indicating a metal ion. Pho, phosphatase; poly-P, polyphosphate granules.
The most prevalent oxidation states of uranium in natural environments are U(VI) and U(IV), although it can also exist as U(III) and U(V). Uranium speciation and redox state are important to consider as they influence the mobility of the compound and the toxicity toward biological systems. A large number of factors, such as the aeration state, pH, organic matter, carbonates and phosphates, are able to influence its mobility complicating uranium chemistry (reviewed in Cumberland et al., 2016). In general, the aeration state determines the oxidation state of uranium. Uranyl (UO22+) is the main form in oxic systems. Uranyl ions are more mobile and more toxic compared to the reduced uraninite (UO2), which can be formed in anaerobic conditions and reoxidized with oxygen (Finch and Murakami, 1999; Markich, 2002; Liu et al., 2017). Also pH determines the solubility of U(VI) and U(IV) complexes. In general, the presence of carbonates has a positive effect on the solubility of U(VI) complexes, especially above pH 5.5. On the other hand, U(IV) is expected to be only slightly soluble in most environmental pH conditions, except at extreme low pH (pH < 3), which can be associated with anthropogenic environments such as acid mine drainage. It is also possible that U(IV) could be mobilized in a colloidal phase. Furthermore, the presence of other ions (e.g., PO43–, OH–, SO42–) and/or organic material can compete for binding to uranyl ions, thereby influencing the mobility of uranium and its sorption to mineral surfaces (Cumberland et al., 2016). Therefore, it is important to consider these factors when investigating microbial interactions with uranium since the toxicity, mobility and interaction strongly depend on the experimental setup.
Although the interaction of microorganisms with uranium is extensively studied, there is far less information about the cellular response of microorganisms to uranium exposure. Data on bacterial uranium resistance mechanisms is rather exploratory. Therefore, instead of discussing the outcome of bacterial interaction with uranium, this review focusses on the different active cellular mechanisms for uranium processing. Microbial reduction of soluble U(VI) to insoluble U(IV) is one of the best-studied mechanisms and many uranium-reducing bacteria have been identified. As such, different reduction mechanisms are discussed in a first section. In a following section, several types of phosphatases are reviewed as metal-phosphate complexation and metal-phosphate biomineralization are common mechanisms for metal detoxification. Afterward, the involvement of membrane proteins, metal efflux and regulatory systems in uranium resistance is reviewed. Finally, the potential application in bioremediation is discussed.
Uranium Reduction Mechanisms
Enzymatic uranium reduction can occur directly or indirectly in the cytoplasm, periplasm, at the outer membrane or extracellularly (Figure 2 and Table 1) (reviewed in You et al., 2021). It has been investigated particularly in Geobacter species, which often dominate in anaerobic uranium-reductive bioremediation setups (Yun-Juan et al., 2005; Shelobolina et al., 2008; Chandler et al., 2010). In general, cytochromes are imperative in the reduction process and were found to be increasingly expressed in Geobacter uraniireducens when growing in uranium-contaminated subsurface sediments (Holmes et al., 2009). Moreover, GscA (Geobacter subsurface c-type cytochrome A) of Geobacter sp. M18 was highly abundant during in situ uranium bioremediation (Yun et al., 2016). In addition, the diheme c-type cytochrome peroxidase MacA and the outer-surface c-type cytochrome OmcZ are essential for uranium reduction, and the periplasmic c7-type cytochrome PpcA is an important intermediate electron carrier (in the absence of hydrogen) in Geobacter sulfurreducens (Lloyd et al., 2003; Shelobolina et al., 2007; Orellana et al., 2013). On the other hand, two outer membrane cytochromes, OmcB and OmcC, showed no or less contribution to U(VI) reduction in G. sulfurreducens (Shelobolina et al., 2007). Functional c-type cytochromes were also shown to be essential for uranium reduction in other bacteria. For instance, a cytochrome c maturation deficient mutant of Shewanella oneidensis MR-1 was unable to reduce uranium. However, the precise electron transfer pathways involved in uranium reduction are not yet clear. It has been shown that the outer membrane c-type cytochrome MtrC (also known as OmcB), but not OmcA, can function as a terminal uranium reductase. In addition, deletion of both genes decreased the uranium reduction rate and changed the characteristics of the formed uranium nanoparticles (Marshall et al., 2006). A decreased uranium reduction rate was also observed for deletion mutants of mtrA (encoding a periplasmic decaheme cytochrome involved in metal reduction), mtrB (encoding an outer membrane protein involved in metal reduction) and menC (encoding a precursor of menaquinone), which are involved in the electron transfer. In addition, other Mtr-independent pathways can exist (Bencheikh-Latmani et al., 2005). Periplasmic uranium reduction in Desulfovibrio occurs mainly via cytochrome c3. Deletion of cycA, encoding cytochrome c3, in Desulfovibrio alaskensis G20 (formerly Desulfovibrio desulfuricans G20) reduced the uranium reduction rate in the presence of lactate and pyruvate, and almost completely inhibited it with hydrogen gas as electron donor in sulfate-reducing conditions. This indicates that cycA is responsible for uranium reduction with hydrogen gas as electron donor, but can be bypassed in the presence of other electron donors (Payne et al., 2002, 2004).
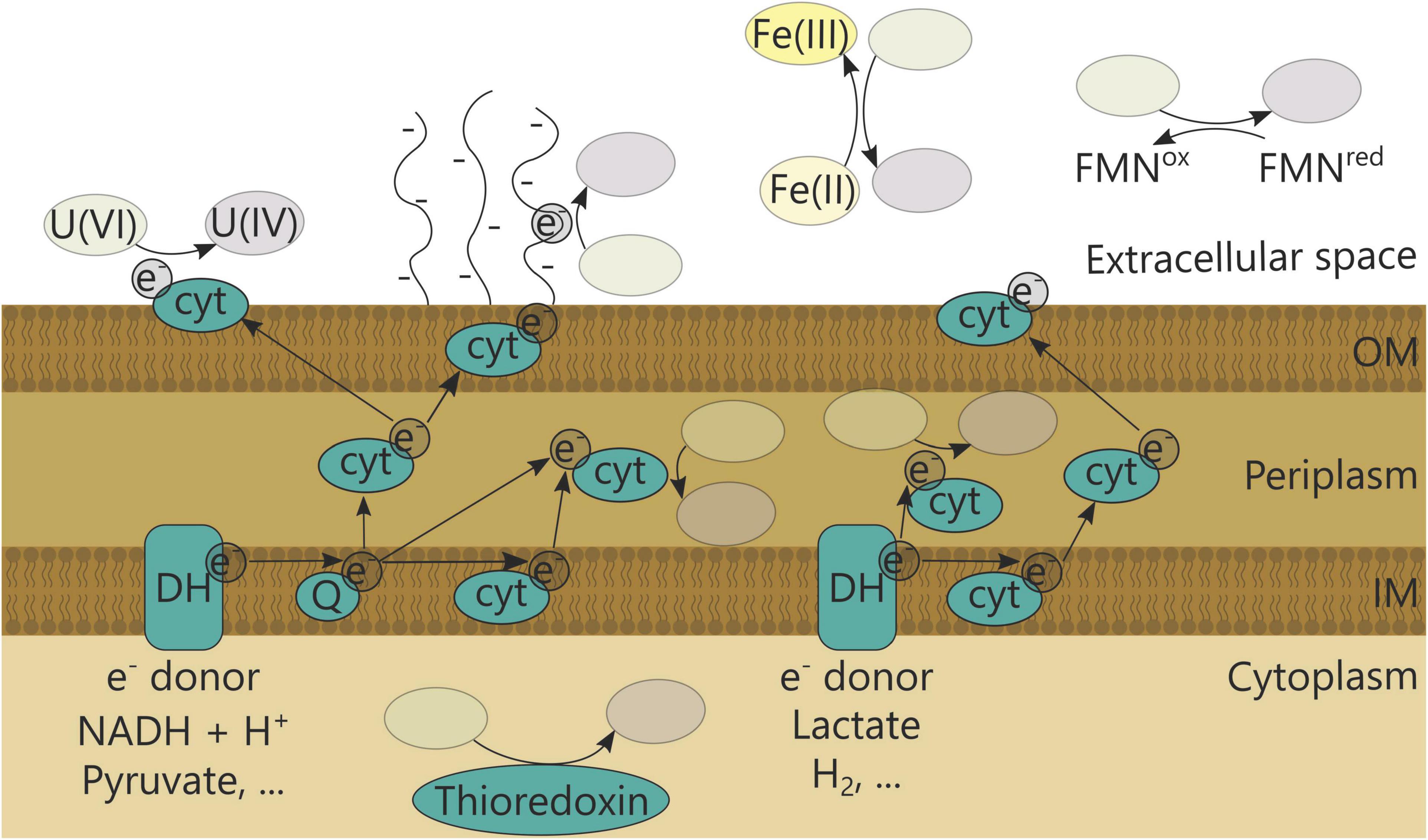
Figure 2. Bacterial uranium reduction mechanisms. DH, Dehydrogenase; Q, quinone; cyt, cytochrome; FMN, flavin mononucleotide.
Uranium can also be reduced abiotically by iron (Du et al., 2011). However, since cytochromes are often involved in iron reduction pathways (Weber et al., 2006), there could be a link between uranium reduction and iron metabolism. Indeed, siderophores have been shown to form stable complexes with metals and some radionuclides (Bouby et al., 1998; Rajkumar et al., 2010; Rashmi et al., 2013). Hydroxamate-type siderophores were shown to chelate uranium better when complexed with carbonate (Mo et al., 2016), and desferrioxamine-B increased dissolution of uraninite under reducing conditions and could therefore increase uranium mobility (Frazier et al., 2005). In addition, uranium stress induced siderophore production in the cyanobacterium Synechococcus elongatus BDU 130911 (Rashmi et al., 2013) as well as a large number of proteins related to iron metabolism in the Chernobyl isolate Microbacterium oleivorans A9. The latter include components of the siderophore iron uptake system such as ABC-transport type subunits and siderophore modification enzymes (Gallois et al., 2018). Moreover, uranium exposure evokes an iron starvation response, thereby enhancing the synthesis of iron uptake systems. This is in line with observations in Desulfotomaculum reducens MI-1, where several ferrous iron uptake and transport proteins, and a transcriptional regulator of the Fur family were upregulated in the presence of U(VI) (Junier et al., 2011). Although the actual link between iron metabolism and uranium resistance is currently unknown, these observations indicate that siderophores could protect cells from uranium stress through sequestration.
In Geobacter species, evidence has also emerged for U(VI) reduction farther from the cell via extracellular pili. G. sulfurreducens expressing pili increased the rate and extent of uranium reduction with carbon ligands outside of the cell, while pili-deficient strains precipitated U(IV) mainly in the periplasm (Cologgi et al., 2011). Furthermore, these conductive pili contributed more to uranium reduction than cytochrome OmcZ (Cologgi et al., 2014). This could indicate that the extracellular pili function as a protective mechanism to avoid excessive precipitation in the periplasm, thereby minimizing cytotoxic effects. Furthermore, the metal-chelating properties of rough lipopolysaccharides could complement the extracellular pili by preventing uranium crossing the outer membrane, thereby creating a barrier to maximize extracellular reduction (Clark et al., 2021). However, detailed knowledge about the mechanism is currently lacking. Another form of extracellular U(VI) reduction is through extracellular electron shuttle compounds. The electron shuttle is able to transfer electrons to an electron acceptor in the extracellular environment, such as U(VI), without the direct interaction of cellular compounds with the electron acceptor. This was shown for Shewanella species that secreted a flavin mononucleotide, which is able to mediate and accelerate reduction of U(VI) (Suzuki et al., 2010; Yamasaki et al., 2017).
Most studies demonstrated that reduced U(IV) was predominantly localized in the periplasm and at the outside of the cell. Nevertheless, cytoplasmic U(VI) reduction with thioredoxins as electron donor was also found. Transposon mutagenesis studies in D. alaskensis G20 showed that the mre operon, coding for a thioredoxin (MreD), thioredoxin reductase (MreE) and an additional oxidoreductase (MreG) was essential for uranium reduction (Li and Krumholz, 2009; Li et al., 2014).
Although much research has been performed to elucidate the precise uranium reduction pathway, there is currently no general model that completely explains the electron transport chain during uranium reduction. Moreover, different mechanisms seem to exist in different strains. More research is necessary to reveal the molecular pathways allowing uranium reduction.
Phosphatases
Metal-phosphate complexation and biomineralization are common mechanisms for limiting metal bioavailability and toxicity (Gudavalli et al., 2018; Zhang et al., 2021), including uranium (Table 1; Wufuer et al., 2017). Since phosphorus is an essential element (Smil, 2000) and soluble phosphate can be scarce in some environments like soil and water bodies, many bacteria use phosphatases to liberate phosphate ions from mineral or organic phosphorus. In fact, organic forms of phosphorus often constitute 30–50% of the total phosphorus (Ruttenberg, 2014). The liberated phosphate ions are able to interact with uranyl, facilitating complexation and precipitation of uranium. Phosphatases, which are either secreted outside the cell or membrane-bound, are broadly categorized based on the pH required for their optimum activity as acid or alkaline. Different sources of phosphate have been used to study phosphatase-mediated uranium-phosphate biomineralization, such as glycerol-2-phosphate (Appukuttan et al., 2006), glycerol-3-phosphate (Powers et al., 2002), phytate (Li et al., 2019) and polyphosphate (Renninger et al., 2004). An overview of the current knowledge of each of these mechanisms is discussed in the following subsections.
Acid Phosphatase
Enzymatic uranium phosphate precipitation was first observed in Serratia sp. N14 (Macaskie et al., 1992), originally classified as Citrobacter sp. N14 (Pattanapipitpaisal et al., 2002). Since then, uranium phosphate biomineralization has been shown in different Serratia spp. under diverse conditions, including anaerobic conditions and even in the presence of high doses of gamma irradiation (Newsome et al., 2015; Chandwadkar et al., 2018). The periplasmic acid phosphatase PhoN was responsible for uranium complexation as a phosphatase deficient mutant was unable to remove uranium from the growth medium (Macaskie et al., 1994; Jeong et al., 1997). Fragments of the purified PhoN were homologous to PhoN of Morganella morganii, Providencia stuartii, and Salmonella enterica subsp. enterica serovar Typhimurium (Macaskie et al., 1994; Jeong et al., 1997). However, uranium removal in those strains was negligible (Macaskie et al., 1994), indicating that additional strain-specific characteristics were needed, e.g., suitable sites for uranium nucleation (Macaskie et al., 2000). While the inner and outer membrane were initially identified as nucleation sites for biocrystallization, further research also proposed initial exocellular nucleation within the lipopolysaccharides aided by supposedly liposome-entrapped acid phosphatases that released Pi in close juxtaposition (Jeong et al., 1997; Macaskie et al., 2000). Moreover, the presence of phosphate-containing extracellular polymeric material putatively provided a protective function and enabled uranium removal (Jeong et al., 1997; Macaskie et al., 2000). Other studies on PhoN-type acid phosphatases of Serratia spp. showed that two isoenzymes exhibited a different pH optimum and glycerol 2-phosphate affinity (Jeong et al., 1998). Both isoenzymes were also sensitive to uranyl causing a reduction in their activity (Jeong and Macaskie, 1995).
To explore the role of phosphatases in uranium biomineralization more in detail, several recombinant strains expressing phosphatases have been studied (Basnakova et al., 1998). Although E. coli DH5α expressing either phoN from S. Typhi or the related phoC from M. morganii exhibited acid phosphatase activity comparable to Serratia sp. N14, different uranium removal capabilities were observed. E. coli expressing phoN exhibited increased removal compared to Serratia sp., and both were superior to E. coli expressing phoC (Basnakova et al., 1998), suggesting different in vivo properties of both phosphatases. Putatively, the tertiary structure of PhoN protects the sensitive sites of the enzyme for uranyl ions to access (Basnakova et al., 1998). In addition, heterologous expression of PhoN from a S. Typhi isolate in Deinococcus radiodurans R1 and in E. coli showed comparable uranium removal although a higher phosphatase activity was observed in E. coli (Appukuttan et al., 2006). Also, in M. oleivorans A9, the expression of a broad specificity phosphatase PhoE coincided with phosphate efflux and showed uranium-phosphate precipitation in different stadia (Gallois et al., 2018).
Acid phosphatase activity and its involvement in uranium biomineralization was also observed in multiple isolates from uranium-contaminated environments. In Jaduguda (India), nine out of twelve strains isolated from uranium mine wastes possessed phosphatase activity (Choudhary et al., 2012). Furthermore, four out of eight isolates from uranium mill tailings pore waters in the region of Limousin (France) possessed acid phosphatase activity, while only one, Microbacterium oxydans Br5, possessed alkaline and acid phosphatase activity (Sanchez-Castro et al., 2017). Caulobacter sp. OR37, Rahnella sp. strain Y9602 and Bacillus sp. strain Y9-2 isolated from the Oak Ridge Field Research Center (ORFRC) were phosphatase-positive and removed uranium from growth medium supplemented with organic phosphate in different pH conditions (Beazley et al., 2007; Martinez et al., 2007; Morrison et al., 2021). Moreover, Rahnella sp. Y9602 was able to induce uranium phosphate complexation in nitrate-reducing conditions at pH 5.5, albeit with some differences in initial precipitation rates. The same uranium mineral was formed in both aerobic and anaerobic conditions and this was similar to uranium precipitated with free orthophosphate, suggesting that the precipitation is purely chemical through the liberation of Pi from organophosphate by phosphatases (Beazley et al., 2009). This further supports the hypothesis that cells govern nucleation sites for uranium phosphate complexation. Indeed, Morrison et al. (2021) indicated that abiotic precipitation does not occur at uranium concentrations below 1 μM with 500 μM Pi and below pH 5. However, introducing Caulobacter sp. strain OR37 resulted in uranium precipitation and the formation of intracellular polyphosphate granules. Presumably, the cells concentrated uranium at the membrane by sorption, which lowered the activation energy required for nucleation and mineralization that prevented abiotic uranium-phosphate mineralization. While most studies investigate uranium biomineralization at uranium concentrations higher than 20 μM, where uranium-phosphate precipitates will inevitable form if Pi is released, many contaminated sites have lower uranium concentrations (Morrison et al., 2021). Moreover, limitations for uranium in drinking water are often below 1 μM (Nolan and Weber, 2015). Therefore, studies investigating microbial interactions with low uranium concentrations are essential.
Results discussed above are mainly from acidic or near neutral environments. However, different observations could be made in alkaline conditions. Serratia sp. strain OT II 7, isolated from the acidic sub-surface soil of a uranium ore deposit, exhibited much higher phosphatase activity at pH 5 compared to pH 7 and 9 in the absence of uranium. However, this strain removed uranium much faster at pH 9 and pH 7, than at pH 5 (Chandwadkar et al., 2018). Moreover, for Serratia sp. strain OT II 7 as well as Chryseobacterium PMSPZI uranium precipitates were formed at a different cellular location depending on the pH. At pH 9, uranyl precipitated always extracellular. At pH 7, extracellular and cell-bound uranium precipitates were formed. Whereas, at pH 5, uranyl precipitates were mainly cell surface-associated or intracellular, which also decreased phosphatase activity and negatively impacted cell viability (Chandwadkar et al., 2018; Khare et al., 2020). It is clear that acid phosphatases play a prominent role in the precipitation of uranium and enable bacteria to withstand high uranium concentrations. Furthermore, uranium-contaminated environments are often acidic, also evidenced by mainly acidic phosphatase activity of environmental isolates.
Alkaline Phosphatases
Industrial processes can also lead to alkaline uranium waste (Seidel, 1981), for example by using carbonate-based reagents to recover uranium from historical mine waste (Santos and Ladeira, 2011) via soluble and stable uranium-carbonate complexes (Duff et al., 2004). In those environments, uranium phosphate mineralization is not expected, which indicates the need to study the physicochemical conditions to determine possible uranium-phosphate complexation. Uranium precipitation is possible at pH 9 in the presence of excess carbonate if log(HPO42–/HCO3–) > −3 (Zheng et al., 2006). Indeed, the secreted alkaline phosphatase PhoK from Sphingomonas sp. BSAR-1 precipitated the supplemented uranyl-carbonate at pH 9 as uranium-phosphate through glycerol-2-phosphate cleavage. The precipitation was even more rapidly at higher uranium concentrations when PhoK was over-expressed in Escherichia coli BL21 (Nilgiriwala et al., 2008). This PhoK has also been used to create the recombinant Deinococcus radiodurans Deino-PhoK strain, resulting in efficient extracellular uranium-phosphate precipitation in planktonic and alginate-encapsulated state. Precipitation occurred also during high doses of ionizing radiation and in the presence of cesium and strontium, which are often present in intermediate and low level liquid radioactive waste (Kulkarni et al., 2013). The advantages of using D. radiodurans in treating radioactive waste have recently been reviewed in Li et al. (2021). Uranium precipitation with PhoK-expressing recombinant E. coli DH5α and D. radiodurans was also examined in a carbonate-deficient condition at pH 6.8 (GC 1) versus a carbonate-abundant condition at pH 9 (GC 2). Uranium toxicity was clearly higher in GC 1 coinciding with more uranium adsorption to the biomass. Consequently, uranium precipitation was cell-associated in GC 1 for both E. coli and D. radiodurans, whereas precipitates were located more distant from cells in GC 2 (Kulkarni et al., 2016). These observations corroborate that uranium speciation and toxicity depend on the environmental conditions.
C. crescentus NA1000 forms uranium-phosphate precipitates extracellularly or on the cell surface by releasing Pi in modified M5G medium. The periplasmic alkaline phosphatase PhoY, related to the secreted PhoK from Sphingomonas sp. BSAR-1 (39% amino acid identity and 51% similarity), was identified as essential in this process and consequently also for uranium resistance (Yung and Jiao, 2014). Furthermore, heterologous expression of the E. coli alkaline periplasmic phosphatase PhoA in three Pseudomonas subsurface isolates released sufficient Pi in sterilized soil slurries to remove uranium from the cell-free supernatant (up to 69% of 20 μM uranyl acetate) for P. rhodesiae R1.2 and P. veronii V1.2, but not P. fluorescens F1.2 (Powers et al., 2002). In Stenotrophomonas bentonitica BII-R7, isolated from Spanish bentonite clay formations, four phosphatases were upregulated in the presences of uranium and facilitated the formation of extracellular uranium-phosphate precipitates (Pinel-Cabello et al., 2021). Overall, bacterial alkaline phosphatases are present in a wide range of species and could result in the precipitation of uranium. In the studies presented here, organophosphate was often provided as glycerol-2- or glycerol-3-phosphate. However, in natural environments, other forms of organophosphate can be present such as phytate, which requires specialized phosphatases to release inorganic phosphate.
Phytases
Phytate, or inositol hexaphosphate, is a naturally occurring organic phosphate that can be abundant in soils and is the main form of phosphorous storage in plants (Turner, 2006). Depending on the pH, on the amount of phytate present and on the available carbonates, abiotic uranium precipitation is not expected and thus phosphate needs to be released for uranium immobilization (Langmuir, 1978; Oh et al., 2004; Salome et al., 2017; Li et al., 2019). Phytases are a special class of phosphatases that catalyze the sequential hydrolysis of phytate to less phosphorylated myo-inositol derivatives and inorganic phosphate (Wyss et al., 1999). Although distinct phytate types exist, histidine acid phytases are mostly identified in microorganisms. Acidic, but not alkaline phytase activity was shown to be present in microbial communities from the uranium contaminated ORFRC. Experiments with sediment slurries at pH 5.5 indicated that uranium enhanced phytase activity but also resulted in the production of intermediate inositol phosphate species, probably due to the inactivation of other phosphatases, with a decrease in uranium solubility (Salome et al., 2017). C. crescentus CB15N is able to form calcium-uranium-phosphate precipitates in oligotrophic medium in the presence of inorganic phosphate. Interestingly, a putative phytase was the most highly upregulated protein in response to uranium in these conditions (Hu et al., 2005). The phytase is not essential for uranium resistance in M2G medium nor in rich PYE medium but does seem to enhance survival in the presence of uranium when phytate is provided as sole phosphate source. Furthermore, since the phytase-deficient mutant already showed reduced growth in the absence of uranium, it seems essential for growth but not necessarily for uranium biomineralization (Yung et al., 2014).
Phosphate Release From Cellular Phosphate Sources
Whereas most studies investigated bacterial uranium-phosphate precipitation by supplementing organic or inorganic phosphate, the possibility of phosphate release from innate phosphate sources for uranium biomineralization is also being scrutinized. In P. aeruginosa, overexpression of a polyphosphate kinase resulted in 100 times more accumulated polyphosphate. In the presence of uranyl nitrate, it was shown that uranyl adsorbed initially to the cells and was consecutively precipitated as uranyl phosphate mediated by the release of phosphate from polyphosphate (Renninger et al., 2004). In addition, D. radiodurans was shown to precipitate uranium in non-growth conditions at pH 4 without any supplemented phosphate source. Therefore, phosphate had to come from cellular material, such as polyphosphate, which was released during cell lysis (Suzuki and Banfield, 2004). Acharya et al. (2017) also hypothesized that in the cyanobacterium Anabaena torulosa alkaline phosphatases liberate Pi from organophosphate substrates released during the decomposition or degradation of cells by uranium. For Paenibacillus sp. JG-TB8, uranium was bound by organic phosphate at pH 2 and pH 3 independent on aeration conditions (Reitz et al., 2014). However, uranium seemed to precipitate more as meta-autunite-like uranyl phosphate at higher pH and in oxic conditions, while under anaerobic conditions no mineralization was observed due to decreased Pi release. Nonetheless, the only phosphate sources during the experiment were organic substrates from damaged cells (Reitz et al., 2014).
Membrane Proteins
The outer surface of many bacteria and archaea is covered by a proteinaceous surface layer (S-layer) that serves multiple functions, including survival in specific niches. Although its precise role in many organisms has not yet been identified (Fagan and Fairweather, 2014), its involvement in uranium biosorption has been shown in multiple bacteria (Table 1; Pollmann et al., 2006; Yung et al., 2015). This is mostly considered to be a passive process, but evidence emerged that bacteria might modulate their cell envelope to become more resistant. In C. crescentus NA1000, transposon mutagenesis revealed that the rsaFa and rsaFb genes encoding outer membrane transporters conferred uranium tolerance. While RsaFa and RsaFb are known for exporting the highly abundant S-layer protein RsaA, a role for the S-layer itself and other S-layer transport systems in uranium resistance was excluded. However, RsaF was found to be homologous to TolC and mutation resulted in a decreased resistance to cadmium and tetracycline, suggesting that resistance could be governed by interacting with other translocases/pumps. Furthermore, contrary to deletion of rsaFb and rsaA, deletion of rsaFa and rsaFaFbrsaA increased uranium accumulation. However, a role for RsaF in outer membrane integrity, which could have increased uranium accumulation, could not be excluded. Nevertheless, RsaF plays an important role in uranium resistance either via uranium efflux or via protection of the outer membrane integrity (Yung et al., 2015). The SlfB S-layer protein of B. sphaericus JG-A12, isolated from a uranium mining waste pile is much more effective in uranium binding than the SlfA S-layer protein of the reference strain B. sphaericus NCTC 9602. The different affinity for uranium could be explained by a distinct C-terminal region of both proteins. The C-terminal region of SlfB harbors significantly more serine and threonine residues, which are potential phosphorylation sites. Notably, analysis of the downstream region of slfA and slfB and comparison with S-layer proteins from other B. sphaericus strains indicated the involvement of horizontal gene transfer and genomic rearrangements (Pollmann et al., 2005). Interestingly, S-layer proteins can also be utilized to increase uranium removal. A protein fusion of the S-layer protein Hpi of D. radiodurans with PhoN displayed efficient uranium removal (Misra et al., 2021). Furthermore, a uranyl specific biosensor based on the S-layer proteins of B. sphaericus JG-A12 was developed (Conroy et al., 2010).
The involvement of other membrane proteins in uranium binding and resistance has also been shown recently. Comparison of four Microbacterium species revealed that protein UipA was only present in uranium-tolerant strains and was the most upregulated protein after uranium induction. Moreover, the C-terminal part of this single-pass transmembrane protein has a high uranyl binding affinity. The crystal structure of UipA displayed a tandem of PepSY domains in a swapped dimer with a negatively charged face, responsible for uranium binding (Gallois et al., 2021). In addition, the production of carboxymethyl cellulose modified iron sulfide complex (CMC-FeS) by sulfate reducing bacteria was shown to have increased U(VI) removal capacity compared to chemically produced CMC-FeS because of the presence of extracellular polymeric substances (EPS) containing tryptophan and tyrosine residues (He et al., 2021). Finally, increased expression of CreD, an inner membrane protein from S. bentonitica BII-R7, decreased membrane permeability and prevented uranium from entering the cytoplasm, thereby increasing uranium resistance during the lag phase (Pinel-Cabello et al., 2021).
Metal Efflux Systems
As evidenced in the previous sections, it is clear that bacteria are able to detoxify uranium via different mechanisms. Nevertheless, in some cases the actual process is not directly deducible. For instance, Arthrobacter sp. X34, isolated from the ORFRC, did not exhibit phosphatase activity and did not precipitate any uranium, but was equally and even more resistant to uranium than the uranium-biomineralizing Bacillus sp. strain Y9-2 and Rahnella sp. strain Y9602, respectively (Beazley et al., 2007; Martinez et al., 2007). Another example is the association of uranium-phosphate minerals with polyhydroxybutyrate in Cupriavidus metallidurans NA4 (Rogiers et al., 2021b), which is resistant to uranium independent of the presence of phaC1, encoding the poly(3-hydroxyalkanoate) polymerase subunit PhaC (Rogiers, 2022). Therefore, other mechanisms could mediate resistance, including metal efflux systems as uranium toxicity is mainly exerted through its chemical metal-related properties.
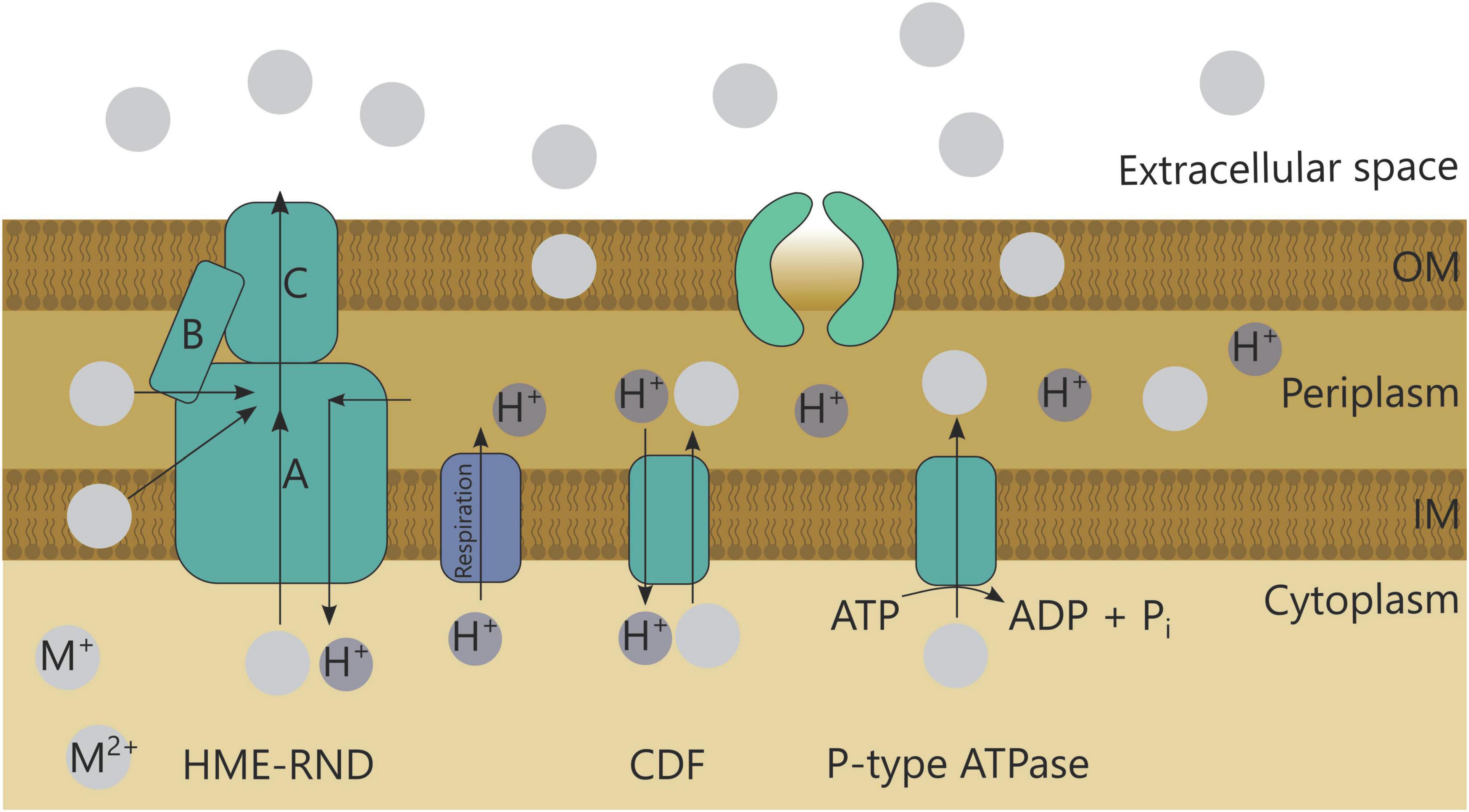
Metal efflux is a common detoxification strategy employed by bacteria. Although multiple systems exist, three systems are most common (Figure 3; Table 1; Nies, 2003). First, the resistance-nodulation-cell division (RND) superfamily includes seven protein families involved in several functions such as transport of hydrophobic compounds and nodulation factors, but also heavy metal efflux (HME-RND). The HME-RND protein (A) is usually combined with a membrane fusion protein (MFP, B) and an outer membrane factor (OMF, C) to form a protein efflux complex that can transport substrates from the cytoplasm, cytoplasmic membrane or periplasm to the outside (Nies, 2003). Typical examples are CzcCBA conferring resistance toward Cd2+, Zn2+, and Co2+ (Mergeay et al., 1985), and CusCBA and SilCBA providing resistance toward Cu+, Cu2+, and Ag+ ions (Munson et al., 2000; Mijnendonckx et al., 2013; Randall et al., 2014). A second export mechanism comprises efflux pumps driven by the proton motive force or potassium gradient known as cation diffusion facilitators (CDF) (Nies, 2003), such as CzcD mediating a small degree of Cd2+, Zn2+, and Co2+ resistance (Nies, 1992; Anton et al., 1999). Lastly, P-type ATPases are able to import and export cations through the hydrolysis of ATP (Nies, 2003). Import is important for essential metals, such as MgtA for Mg2+ (Snavely et al., 1989), but ATPases can also detoxify metals through export. Several examples are CadA for Cd2+ resistance (Nucifora et al., 1989), ZntA for Zn2+ resistance (Beard et al., 1997), and CzcP for Cd2+, Zn2+, and Co2+ resistance (Scherer and Nies, 2009).
Although efflux systems are often upregulated after uranium exposure, a designated efflux system for uranium has not yet been identified. For instance, in C. metallidurans NA4 almost all genes (from nine different clusters) involved in the response to and detoxification of silver and copper were upregulated after uranium exposure (Rogiers et al., 2021b). In Chryseobacterium sp. strain PMSZPI, isolated from a uranium-enriched environment and in S. bentonitica BII-R7 czcA and czcA/cusA genes were upregulated after uranium exposure, respectively (Nongkhlaw and Joshi, 2019; Pinel-Cabello et al., 2021). The expression of one of the two czc gene clusters present in C. metallidurans NA4 was also induced after exposure to uranium (Rogiers et al., 2021b). Three membrane fusion proteins and two outer membrane factors assisting HME-RND pumps were also upregulated in G. sulfurreducens after uranium induction, amongst which CzcC (Orellana et al., 2014). The expression of another czc gene, namely of the CDF encoding czcD, was found upregulated in both Chryseobacterium sp. strain PMSZPI and S. bentonitica BII-R7 (Nongkhlaw and Joshi, 2019; Pinel-Cabello et al., 2021). Furthermore, P-type ATPases are also often found upregulated after uranium exposure, such as cadA in Chryseobacterium sp. strain PMSZPI (Nongkhlaw and Joshi, 2019) and a cadmium- and copper-translocating P-type ATPAse in Desulfotomaculum reducens MI-1 (Junier et al., 2011). Altogether, these results suggest that uranium efflux, if active, occurs through systems conferring resistance to Cd2+, Zn2+, Co2+ or Cu2+, and Ag+. Other efflux-related genes are sometimes upregulated as well, such as the nickel and cobalt efflux regulator rcnB and antibiotic resistance efflux genes (mdtAB) in S. bentonitica BII-R7 (Pinel-Cabello et al., 2021).
In M. oleivorans A9, a fast initial biotic removal of uranium was followed by an active release of U(VI), which was accompanied with phosphate efflux for uranium-phosphate biomineralization. However, this release was only seen at a concentration of 10 μM but not at 50 μM uranyl nitrate. It was therefore hypothesized that higher concentrations of U(VI) could inhibit efflux-mediated resistance or that the influx of uranium into the cell would mask the efflux (Theodorakopoulos et al., 2015). In a follow-up proteomic study, the upregulation of several cation transporters (K+, Mn2+, Zn2+, Mg2+, and Co2+) and an ABC-type transport system co-occurred with uranium efflux (Gallois et al., 2018). A similar release of uranium after initial biosorption was observed for Halobacterium noricense (Bader et al., 2017). The studies on M. oleivorans A9 and H. noricense could indicate that uranium efflux is possible, but that uranium resistance is a complex process mediated by a combination of different mechanisms. This could explain why, although efflux systems are often upregulated, proof of essential efflux systems for uranium resistance is still lacking. Uranium efflux could contribute to limit uranium entry, but detoxification seems to be mediated by biomineralization, bioreduction or biosorption.
Regulatory Systems
To sense and respond to environmental changes bacteria deploy different regulatory systems, including two-component systems (TCSs) that are often involved in the response to metals (Chang and Stewart, 1998; Mergeay and Van Houdt, 2015). Typically, the sensor histidine kinase (HK) senses the metal ion, uses ATP to autophosphorylate a conserved histidine residue and transfers the phosphate group to a conserved aspartate residue on the corresponding response regulator (RR) (Chang and Stewart, 1998). On its turn, the phosphorylated RR regulates the expression of metal resistance genes, but often also autoregulates the expression of the TCS. In C. crescentus NA1000, two uranium-responsive TCSs have been identified by analyzing transcriptomic and proteomic data after exposure to non-toxic uranium concentrations (Hu et al., 2005; Yung et al., 2014; Park et al., 2017; Park and Taffet, 2019). UrpRS (uranium responsive phytase regulator and sensor, respectively, CCNA_01362 + 01363) was found to regulate a phytase gene (CCNA_01353) that confers uranium resistance when phytate is provided as the sole phosphate source (Yung et al., 2014; Park and Taffet, 2019). UzcRS (CCNA_02842 + 02845), also responsive toward zinc and copper, regulates the expression of urcA (uranium response in caulobacter, CC3302), the highest uranium-specific induced gene encoding a periplasmic protein with unknown function (Hu et al., 2005; Park et al., 2017). The promoter region of urcA contains two m_5 motives specific for uranium induction that are nearly identical to the 18-bp UzcR recognition motif (Hillson et al., 2007). This site contains the partially palindromic half sites 5′-CATTAC-N6-TTAA-3′ found in 44 of 57 UzcR binding regions determined by ChIP-seq (Park et al., 2017). Furthermore, deleting uzcR or uzcS prevented the Zn-, U- and Cu-dependent induction of the promoter region of urcA (PurcA). UzcR is thus presumably the regulator of the m_5 motif and could be involved in the direct recruitment of the RNA polymerase holoenzyme since the m_5 motif is located at a common binding site for transcriptional activators, 43 or 53 bp upstream of the transcription start site (Lee et al., 2012). In general, UzcR binds extensively throughout the genome mainly activating genes encoding proteins with a putative signal secretion signal and/or transmembrane domains (52 of 66 genes) such as metallopeptidases, multidrug-resistant efflux (MDR) pumps, TonB-dependent receptors and many proteins of unknown function (Park et al., 2017). The expression of uzcRS is modulated by auxiliary regulators and the TCS is part of a complex signaling network (Park et al., 2019). However, deletion mutants of uzcRS disputed an essential role in uranium resistance and the manner of uranium sensing is still unclear since UO22+, Zn2+, and Cu2+ displayed different coordination preferences (Haas and Franz, 2009; Park et al., 2017). It is hypothesized that expression of uzcRS and urpRS is induced indirectly by uranium. Nevertheless, the combination of both TCS systems has been used to develop a whole-cell biosensor for uranium that showed a highly improved selectivity toward uranium compared to the previously designed whole-cell biosensor based on the urcA promoter (Hillson et al., 2007; Park et al., 2019; Park and Taffet, 2019).
Upregulation of regulatory systems after uranium induction has also been observed in other bacteria. A negative regulator of stress-induced operons, ArsR (Silver and Phung, 2005), seemed to be less abundant when uranium is internally biomineralized in M. oleivorans A9 but more abundant as long as uranium remains extracellularly (Gallois et al., 2018). Interestingly, the TCS UipRS is located upstream the uranium binding protein UipA in multiple Microbacterium species, but a role in uranium sensing and resistance has not yet been shown (Gallois et al., 2021). In D. alaskensis G20, a cyclic AMP receptor protein (CRP) was found to possibly regulate expression of the mre operon for metal reduction, facilitating uranium reduction through thioredoxin (Li and Krumholz, 2009). Finally, ten TCSs are upregulated in C. metallidurans after uranium induction of which five are known to be involved in metal resistance (Rogiers et al., 2021b). While extensive work has only been done in C. crescentus, there is currently little known on how uranium is sensed and how this results in the transcription of target genes. Further research is therefore necessary, also in other bacteria, to unravel the underlying regulatory mechanisms. An overview of the current knowledge on the different molecular interaction mechanisms is presented in Table 1.
General Implications for Technological Applications
The toxic characteristics of uranium urged researchers to investigate possible remediation strategies. This led to the discovery that bacteria could be used for uranium bioremediation, which is now one of the most promising bio-based approaches for the remediation of uranium-contaminated sites (Newsome et al., 2014). Uranium reduction is one of the most studied processes. Although the extensive research has already elucidated large parts of the uranium reduction pathway, it is not yet completely clarified. Further research on these pathways could be useful to understand the uranium reduction mechanism completely, but it could also enable full exploitation and modulation of the reduction process for bioremediation applications. However, one of the disadvantages of uranium reduction for bioremediation purposes is that it depends highly on environmental factors as it necessitates reducing conditions and often requires removal of soil compounds (e.g., nitrate) before it can be applied. In addition, one of the remaining problems for in situ bioremediation is that reduced uranium is more or less prone to reoxidation depending on the minerals formed. Moreover, the addition of cadmium, a known inhibitor of thioredoxin, showed complete inhibition of uranium reduction in some conditions, which is important to take into account for bioremediation purposes as uranium contaminated soils are very often co-contaminated with metals such as cadmium, zinc and copper (Li and Krumholz, 2009; Bigalke et al., 2017; Lu and Liu, 2018). Furthermore, zinc and copper were also found to completely inhibit uranium reduction when concentrations reached 25 and 15 mg/L, respectively, due to their toxic effects on sulfate-reducing bacteria (Yi et al., 2007). A more extensive screening of different metals can be useful to identify key inhibitors of uranium reduction. Besides metal ions, also uranyl speciation and concentration, pH, temperature, electron donors and acceptors can affect uranium reduction rates (recently reviewed by You et al., 2021). Nevertheless, uranium reduction can still be an asset in several conditions. For example, if in situ conditions are anaerobic, providing electron donors could quickly immobilize uranyl by forming uraninite. Also, if the recovery of uranium is necessary during ex situ remediation, reduction in column or batch setups could be more favorable, since U(IV) is easily remobilized.
To overcome a number of the limitations for long-term processes, the research focus shifted in recent years to other uranium interaction mechanisms, such as uranium phosphate biomineralization. For instance, if long-term immobilization is preferential or if the removal of uranium without recovery is the main goal, uranium-phosphate precipitation could be the best option. Uranium-phosphate minerals, such as autunite or meta-autunite, have generally low aqueous solubility (Lobeck et al., 2020), are stable over a wide temperature and pH range (Dzik et al., 2017; Wufuer et al., 2017; Gudavalli et al., 2018) and are not prone to remobilization through reoxidation (Williamson et al., 2014; Romanchuk et al., 2020). Abiotic remediation with Pi has been tested, but resulted rapidly in phosphate mineral precipitation not linked with uranium and clogged pore spaces that inhibited further diffusion, which was alleviated by using microbial activity to release Pi continuously from polyphosphates or phytate (Wellman et al., 2006). Phytate has been shown to be more recalcitrant to degradation than other organic phosphates, which may facilitate its migration in contaminated soils and can be advantages for bioremediation purposes (Wellman et al., 2006). However, also uranium phosphate biomineralization still depends on the environmental conditions. Partial protonation of inorganic phosphate starting below pH 4 could hamper abiotic uranyl phosphate mineralization (Hinsinger, 2001). Above circumneutral pH and in the presence of high (bi)carbonate concentrations, highly soluble uranium-carbonate complexes are formed, which can prevent uranyl phosphate precipitation or can solubilize autunite minerals (Pablo et al., 1999; Gudavalli et al., 2018). On the other hand, uranyl-hydroxide formation could allow precipitation (Chandwadkar et al., 2018). Furthermore, strong organic acids, such as oxalate and citrate, interact directly with uranyl and could affect uranium-phosphate biomineralization. However, organic ligands could also promote the conversion of colloidal particles UO2(OH)2 to free UO22+, which could facilitate uranyl phosphate biomineralization. Nonetheless, uranium-phosphate biomineralization was completely inhibited when organic ligands compete with biotic PO43– (Tu et al., 2019). Moreover, in anaerobic nitrate-reducing conditions, the combined toxicity of uranium and produced nitrite after nitrate reduction suppressed growth (Beazley et al., 2009). Even though uranium was still precipitated, presumably due to the early release of Pi before uranium addition or the continued activity of the phosphatases, nitrite accumulation might have implications on the sustainability of the process. Knowledge on compounds preventing uranium-phosphate biomineralization is currently limited to metals. Chromium was able to interfere with uranium biomineralization by PhoN-expressing D. radiodurans cells. Furthermore, introducing YieF, which is able to convert Cr(VI) to the less toxic Cr(III), alleviated this problem (Xu et al., 2018). Cd2+ and Hg2+ are known to affect soil acid phosphatase activity and Hg2+, Cu2+, and Cd2+ are able to inhibit E. coli alkaline phosphatase activity (Alnuaimi et al., 2012; Zheng et al., 2019). In general, since phosphatases are known to play a pivotal role, one can hypothesize that inhibition of phosphatase activity also inhibits uranium-phosphate biomineralization.
Overall, it is clear that the physico-chemical environment imposes restrictions on the applied method, especially for in situ processes. Since uranium-contaminated sites or often co-contaminated with toxic metals (Sitte et al., 2015; Boteva et al., 2016; Rogiers et al., 2021a), in situ bioremediation necessitates the presence of multiple metal resistance mechanisms, which are often present in the indigenous microbial communities thriving in such contaminated sites (Choudhary and Sar, 2015; Agarwal et al., 2020; Rogiers et al., 2021a).
Conclusion
We provided an overview of the state-of-the-art on active bacterial uranium detoxification mechanisms including uranium reduction, phosphatases, membrane proteins, efflux and regulatory systems. Although extensive work has been done, completely unraveling the molecular mechanistic insights behind uranium resistance and its regulation necessitates further research. Such mechanistic insights can augment bioremediation processes as evidenced throughout this review.
Author Contributions
TR, KM, and RV contributed to the conceptualization. TR wrote the original draft of the manuscript. KM and RV performed a critical revision of the manuscript. NL, AW, and NB contributed to the manuscript revision, read, and approved the submitted version. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Acharya, C., Chandwadkar, P., and Nayak, C. (2017). Unusual versatility of the filamentous, diazotrophic cyanobacterium Anabaena torulosa revealed for its survival during prolonged uranium exposure. Appl. Environ. Microbiol. 83, 1–17. doi: 10.1128/AEM.03356-16
Agarwal, M., Rathore, R. S., Black, A., Xu, X., Seaman, J., Chauhan, A., et al. (2020). Announcing the availability of a culture Collection of Uranium-Resistant Microbial Assemblages (CURMA) obtained from metalliferous soils of the Savannah River Site, USA. Microbiol. Resour. Announcements 9, e551–e520. doi: 10.1128/MRA.00551-20
Alnuaimi, M., Saeed, I., and Ashraf, S. (2012). Effect of Various Heavy Metals on the Enzymatic Activity of E. coli Alkaline Phosphatase. Asharij: United Arab Emirates University.
Anton, A., Große, C., Reißmann, J., Pribyl, T., and Nies, D. H. (1999). CzcD is a heavy metal ion transporter involved in regulation of heavy metal resistance in Ralstonia sp. strain CH34. J. Bacteriol. 181, 6876–6881. doi: 10.1128/jb.181.22.6876-6881.1999
Appukuttan, D., Rao, A. S., and Apte, S. K. (2006). Engineering of Deinococcus radiodurans R1 for bioprecipitation of uranium from dilute nuclear waste. Appl. Environ. Microbiol. 72, 7873–7878. doi: 10.1128/AEM.01362-06
Bader, M., Müller, K., Foerstendorf, H., Drobot, B., Schmidt, M., Musat, N., et al. (2017). Multistage bioassociation of uranium onto an extremely halophilic archaeon revealed by a unique combination of spectroscopic and microscopic techniques. J. Hazardous Mater. 327, 225–232. doi: 10.1016/j.jhazmat.2016.12.053
Basnakova, G., Stephens, E. R., Thaller, M. C., Rossolini, G. M., and Macaskie, L. E. (1998). The use of Escherichia coli bearing a phoN gene for the removal of uranium and nickel from aqueous flows. Appl. Microbiol. Biotechnol. 50, 266–272. doi: 10.1007/s002530051288
Beard, S. J., Hashim, R., Membrillo-Hernández, J., Hughes, M. N., and Poole, R. K. (1997). Zinc(II) tolerance in Escherichia coli K-12: evidence that the zntA gene (o732) encodes a cation transport ATPase. Mol. Microbiol. 25, 883–891. doi: 10.1111/j.1365-2958.1997.mmi518.x
Beazley, M. J., Martinez, R. J., Sobecky, P. A., Webb, S. M., and Taillefert, M. (2007). Uranium biomineralization as a result of bacterial phosphatase activity: insights from bacterial isolates from a contaminated subsurface. Environ. Sci. Technol. 41, 5701–5707. doi: 10.1021/es070567g
Beazley, M. J., Martinez, R. J., Sobecky, P. A., Webb, S. M., and Taillefert, M. (2009). Nonreductive biomineralization of uranium(VI) phosphate via microbial phosphatase activity in anaerobic conditions. Geomicrobiol. J. 26, 431–441. doi: 10.1080/01490450903060780
Bencheikh-Latmani, R., Williams, S. M., Haucke, L., Criddle, C. S., Wu, L., Zhou, J., et al. (2005). Global transcriptional profiling of Shewanella oneidensis MR-1 during Cr(VI) and U(VI) reduction. Appl. Environ. Microbiol. 71, 7453–7460. doi: 10.1128/AEM.71.11.7453-7460.2005
Bigalke, M., Ulrich, A., Rehmus, A., and Keller, A. (2017). Accumulation of cadmium and uranium in arable soils in Switzerland. Environ. Pollut. 221, 85–93. doi: 10.1016/j.envpol.2016.11.035
Boteva, S., Radeva, G., Traykov, I., and Kenarova, A. (2016). Effects of long-term radionuclide and heavy metal contamination on the activity of microbial communities, inhabiting uranium mining impacted soils. Environ. Sci. Pollut. Res. Int. 23, 5644–5653. doi: 10.1007/s11356-015-5788-5
Bouby, M., Billard, I., MacCordick, J., and Rossini, I. (1998). Complexation of Uranium (VI) with the Siderophore Pyoverdine. Radiochimica Acta 80, 95–100. doi: 10.1007/s10967-007-0931-5
Chandler, D. P., Kukhtin, A., Mokhiber, R., Knickerbocker, C., Ogles, D., Rudy, G., et al. (2010). Monitoring microbial community structure and dynamics during in situ U(VI) bioremediation with a field-portable microarray analysis system. Environ. Sci. Technol. 44, 5516–5522. doi: 10.1021/es1006498
Chandwadkar, P., Misra, H. S., and Acharya, C. (2018). Uranium biomineralization induced by a metal tolerant Serratia strain under acid, alkaline and irradiated conditions. Metallomics 10, 1078–1088. doi: 10.1039/c8mt00061a
Chang, C., and Stewart, R. C. (1998). The Two-component system. regulation of diverse signaling pathways in prokaryotes and eukaryotes. Plant Physiol. 117, 723–731. doi: 10.1104/pp.117.3.723
Choudhary, S., Islam, E., Kazy, S. K., and Sar, P. (2012). Uranium and other heavy metal resistance and accumulation in bacteria isolated from uranium mine wastes. J. Environ. Sci. Health Part a-Toxic/Hazardous Substances Environ. Eng. 47, 622–637. doi: 10.1080/10934529.2012.650584
Choudhary, S., and Sar, P. (2015). Interaction of uranium (VI) with bacteria: potential applications in bioremediation of U contaminated oxic environments. Rev. Environ. Sci. Bio-Technol. 14, 347–355. doi: 10.1007/s11157-015-9366-6
Clark, M. M., Paxhia, M. D., Young, J. M., Manzella, M. P., and Reguera, G. (2021). Adaptive synthesis of a rough lipopolysaccharide in geobacter sulfurreducens for metal reduction and detoxification. Appl. Environ. Microbiol. 87:e0096421. doi: 10.1128/AEM.00964-21
Cologgi, D. L., Lampa-Pastirk, S., Speers, A. M., Kelly, S. D., and Reguera, G. (2011). Extracellular reduction of uranium via Geobacter conductive pili as a protective cellular mechanism. Proc. Natl. Acad. Sci. U.S.A. 108, 15248–15252. doi: 10.1073/pnas.1108616108
Cologgi, D. L., Speers, A. M., Bullard, B. A., Kelly, S. D., and Reguera, G. (2014). Enhanced uranium immobilization and reduction by Geobacter sulfurreducens biofilms. Appl. Environ. Microbiol. 80, 6638–6646. doi: 10.1128/AEM.02289-14
Conroy, D. J. R., Millner, P. A., Stewart, D. I., and Pollmann, K. (2010). Biosensing for the environment and defence: aqueous Uranyl detection using bacterial surface layer proteins. Sensors 10, 4739–4755. doi: 10.3390/s100504739
Cumberland, S. A., Douglas, G., Grice, K., and Moreau, J. W. (2016). Uranium mobility in organic matter-rich sediments: a review of geological and geochemical processes. Earth-Sci. Rev. 159, 160–185. doi: 10.1016/j.earscirev.2016.05.010
Du, X., Boonchayaanant, B., Wu, W.-M., Fendorf, S., Bargar, J., and Criddle, C. S. (2011). Reduction of Uranium(VI) by Soluble Iron(II) conforms with thermodynamic predictions. Environ. Sci. Technol. 45, 4718–4725. doi: 10.1021/es2006012
Duff, M. C., Hunter, D. B., Hobbs, D. T., Fink, S. D., Dai, Z., and Bradley, J. P. (2004). Mechanisms of strontium and uranium removal from high-level radioactive waste simulant solutions by the sorbent monosodium titanate. Environ. Sci. Technol. 38, 5201–5207. doi: 10.1021/es035415+
Dzik, E. A., Lobeck, H. L., Zhang, L., and Burns, P. C. (2017). Thermodynamic properties of phosphate members of the meta-autunite group: a high-temperature calorimetric study. J. Chem. Thermodynamics 114, 165–171. doi: 10.1016/j.jct.2017.07.007
Fagan, R. P., and Fairweather, N. F. (2014). Biogenesis and functions of bacterial S-layers. Nat. Rev. Microbiol. 12, 211–222. doi: 10.1038/nrmicro3213
Finch, R., and Murakami, T. (1999). “Systematics and paragenesis of uranium minerals,” in Uranium: Mineralogy, Geochemistry, and the Environment, eds P. C. Burns and R. J. Finch (Berlin: De Gruyter), 91–179. doi: 10.1515/9781501509193-008
Frazier, S. W., Kretzschmar, R., and Kraemer, S. M. (2005). Bacterial siderophores promote dissolution of UO2 under reducing conditions. Environ. Sci. Technol. 39, 5709–5715. doi: 10.1021/es050270n
Gallois, N., Alpha-Bazin, B., Bremond, N., Ortet, P., Barakat, M., Piette, L., et al. (2021). Discovery and characterization of UipA, a uranium- and iron-binding PepSY protein involved in uranium tolerance by soil bacteria. ISME J. doi: 10.1038/s41396-021-01113-7
Gallois, N., Alpha-bazin, B., Ortet, P., Barakat, M., Piette, L., Long, J., et al. (2018). Proteogenomic insights into uranium tolerance of a Chernobyl’s Microbacterium bacterial isolate. J. Proteomics 177, 148–157. doi: 10.1016/j.jprot.2017.11.021
Geras’kin, S., Evseeva, T., and Oudalova, A. (2013). Effects of long-term chronic exposure to radionuclides in plant populations. J. Environ. Radioact. 121, 22–32. doi: 10.1016/j.jenvrad.2012.03.007
Godheja, J., Sk, S., Siddiqui, S. A., and Modi, D. R. (2016). Xenobiotic compounds present in soil and water: a review on remediation strategies. J. Environ. Analytical Toxicol. 6. doi: 10.4172/2161-0525.1000392
Gudavalli, R., Katsenovich, Y., and Wellman, D. (2018). Quantification of kinetic rate law parameters for the dissolution of natural autunite in the presence of aqueous bicarbonate ions at high concentrations. J. Environ. Radioact. 19, 1–9. doi: 10.1016/j.jenvrad.2018.04.007
Haas, K. L., and Franz, K. J. (2009). Application of metal coordination chemistry to explore and manipulate cell biology. Chem. Rev. 109, 4921–4960. doi: 10.1021/cr900134a
Hammond, C. R. (2004). The Elements, Handbook of Chemistry and Physics, 84th Edn. Boca Raton, FL: CRC Press.
He, S., Hu, W., Liu, Y., Xie, Y., Zhou, H., Wang, X., et al. (2021). Mechanism of efficient remediation of U(VI) using biogenic CMC-FeS complex produced by sulfate-reducing bacteria. J. Hazardous Mater. 420:126645. doi: 10.1016/j.jhazmat.2021.126645
Hillson, N. J., Hu, P., Andersen, G. L., and Shapiro, L. (2007). Caulobacter crescentus as a whole-cell uranium biosensor. Appl. Environ. Microbiol. 73, 7615–7621. doi: 10.1128/AEM.01566-07
Hinsinger, P. (2001). Bioavailability of soil inorganic P in the rhizosphere as affected by root- induced chemical changes: a review. Plant Soil 237, 173–195. doi: 10.1023/A
Holmes, D. E., O’Neil, R. A., Chavan, M. A., N’Guessan, L. A., Vrionis, H. A., Perpetua, L. A., et al. (2009). Transcriptome of Geobacter uraniireducens growing in uranium- contaminated subsurface sediments. ISME J. 3, 216–230. doi: 10.1038/ismej.2008.89
Hu, P., Brodie, E. L., Suzuki, Y., McAdams, H. H., and Andersen, G. L. (2005). Whole-Genome transcriptional analysis of heavy metal stresses in Caulobacter crescentus. J. Bacteriol. 187, 8437–8449. doi: 10.1128/JB.187.24.8437
International Atomic Energy Agency [IAEA] (2003). Extent of Environmental Contamination by Naturally Occurring Radioactive Material (NORM) and Technological Options for Mitigation. IAEA Report 419. Vienna: IAEA
Jeong, B. C., Hawes, C., Bonthrone, K. M., and Macaskie, L. E. (1997). Localization of enzymically enhanced heavy metal accumulation by Citrobacter sp. and metal accumulation in vitro by liposomes containing entrapped enzyme. Microbiology 143, 2497–2507. doi: 10.1099/00221287-143-7-2497
Jeong, B. C., and Macaskie, L. E. (1995). PhoN-type acid phosphatases of a heavy metal-accumulating Citrobacter sp .: resistance to heavy metals and affinity towards phosphomonoester substrates. FEMS Microbiol. Lett. 130, 211–214. doi: 10.1111/j.1574-6968.1995.tb07722.x
Jeong, B. C., Poole, P. S., Willis, A. C., and Macaskie, L. E. (1998). Purification and chacterization of acid-type phosphatases from a heavy-metal-accumulating Citrobacter sp. Arch. Microbiol. 169, 166–173. doi: 10.1007/s002030050556
Junier, P., Vecchia, E. D., and Bernier-Latmani, R. (2011). The response of Desulfotomaculum reducens MI-1 to U(VI) exposure: a transcriptomic study. Geomicrobiol. J. 28, 483–496. doi: 10.1080/01490451.2010.512031
Khare, D., Kumar, R., and Acharya, C. (2020). Genomic and functional insights into the adaptation and survival of Chryseobacterium sp. strain PMSZPI in uranium enriched environment. Ecotoxicol. Environ. Saf. 191:110217. doi: 10.1016/j.ecoenv.2020.110217
Kulkarni, S., Ballal, A., and Apte, S. K. (2013). Bioprecipitation of uranium from alkaline waste solutions using recombinant Deinococcus radiodurans. J. Hazardous Mater. 262, 853–861. doi: 10.1016/j.jhazmat.2013.09.057
Kulkarni, S., Misra, C. S., Gupta, A., Ballal, A., and Apte, S. K. (2016). Interaction of uranium with bacterial cell surfaces: inferences from phosphatase-mediated uranium precipitation. Appl. Environ. Microbiol. 82, 4965–4974. doi: 10.1128/AEM.00728-16
Langmuir, D. (1978). Uranium solution-mineral equilibria at low temperatures with applications to sedimentary ore deposits. Geochimica Cosmochimica Acta 42, 547–569. doi: 10.1016/0016-7037(78)90001-7
Lee, D. J., Minchin, S. D., and Busby, S. J. W. (2012). Activating transcription in bacteria. Annu. Rev. Microbiol. 66, 125–152. doi: 10.1146/annurev-micro-092611-150012
Li, R., Ibeanusi, V., Hoyle-Gardner, J., Crandall, C., Jagoe, C., Seaman, J., et al. (2019). Bacterial-facilitated uranium transport in the presence of phytate at Savannah River Site. Chemosphere 223, 351–357. doi: 10.1016/j.chemosphere.2019.02.064
Li, S., Zhu, Q., Luo, J., Shu, Y., Guo, K., Xie, J., et al. (2021). Application progress of deinococcus radiodurans in biological treatment of radioactive uranium-containing wastewater. Ind. J. Microbiol. 61, 417–426. doi: 10.1007/s12088-021-00969-9
Li, X., and Krumholz, L. R. (2009). Thioredoxin is involved in U(VI) and Cr(VI) reduction in Desulfovibrio desulfuricans G20. J. Bacteriol. 191, 4924–4933. doi: 10.1128/JB.00197-09
Li, X., Zhang, H., Ma, Y., Liu, P., and Krumholz, L. R. (2014). Genes required for alleviation of uranium toxicity in sulfate reducing bacterium Desulfovibio alaskensis G20. Ecotoxicology 23, 726–733. doi: 10.1007/s10646-014-1201-2
Liu, W., Dai, X., Bai, Z., Wang, Y., Yang, Z., Zhang, L., et al. (2017). Highly sensitive and selective uranium detection in natural water systems using a luminescent mesoporous metal-organic framework equipped with abundant lewis basic sites: a combined batch, X-ray absorption spectroscopy, and first principles simulation Inv. Environ. Sci. Technol. 51, 3911–3921. doi: 10.1021/acs.est.6b06305
Lloyd, J. R., Leang, C., Hodges Myerson, A. L., Coppi, V. M., Cuifo, S., Methe, B., et al. (2003). Biochemical and genetic characterization of PpcA, a periplasmic c-type cytochrome in Geobacter sulfurreducens. Biochem. J. 369, 153–161. doi: 10.1042/BJ20020597
Lobeck, H. L., Balboni, E., Parker, C. J., Kohlgruber, T. A., Xu, M., Boukdad, S., et al. (2020). Dissolution of poorly soluble uranyl phosphate phases in the Metaautunite Subgroup under uranyl peroxide cage cluster forming conditions. Am. Mineralogist 105, 182–193. doi: 10.2138/am-2020-7106
Lopez-Fernandez, M., Vilchez-Vargas, R., Jroundi, F., Boon, N., Pieper, D., and Merroun, M. L. (2017). Microbial community changes induced by uranyl nitrate in bentonite clay microcosms. Appl. Clay Sci. 160, 206–216. doi: 10.1016/j.clay.2017.12.034
Lu, Z., and Liu, Z. (2018). Pollution characteristics and risk assessment of uranium and heavy metals of agricultural soil around the uranium tailing reservoir in Southern China. J. Radioanalytical Nuclear Chem. 318, 923–933. doi: 10.1007/s10967-018-6081-0
Macaskie, L. E., Bonthrone, K. M., and Rouch, D. A. (1994). Phosphatase-mediated heavy metal accumulation by a Citrobacter sp. and related enterobacteria. FEMS Microbiol. Lett. 121, 141–146. doi: 10.1111/j.1574-6968.1994.tb07090.x
Macaskie, L. E., Bonthrone, K. M., Yong, P., and Goddard, D. T. (2000). Enzymically mediated bioprecipitation of uranium by a Citrobacter sp.: a concerted role for exocellular lipopolysaccharide and associated phosphatase in biomineral formation. Microbiology 146, 1855–1867. doi: 10.1099/00221287-146-8-1855
Macaskie, L. E., Empson, R. M., Cheetham, A. K., Grey, C. P., and Skarnulis, A. J. (1992). Uranium bioaccumulation by a Citrobacter sp. as a result of enzymically mediated growth of polycrystalline HUO2PO4. Science 257, 782–785. doi: 10.1126/science.1496397
Markich, S. J. (2002). Uranium speciation and bioavailability in aquatic systems: an overview. ScientificWorldJournal 2, 707–729. doi: 10.1100/tsw.2002.130
Marshall, M. J., Beliaev, A. S., Dohnalkova, A. C., Kennedy, D. W., Shi, L., Wang, Z., et al. (2006). c-Type cytochrome-dependent formation of U(IV) nanoparticles by Shewanella oneidensis. PLoS Biol. 4:e268. doi: 10.1371/journal.pbio.0040268
Martinez, R. J., Beazley, M. J., Taillefert, M., Arakaki, A. K., Skolnick, J., and Sobecky, P. A. (2007). Aerobic uranium (VI) bioprecipitation by metal-resistant bacteria isolated from radionuclide- and metal-contaminated subsurface soils. Environ. Microbiol. 9, 3122–3133. doi: 10.1111/j.1462-2920.2007.01422.x
Mergeay, M., Nies, D. H., Schlegel, H. G., Gerits, J., Charles, P., and Van Gijsegem, F. (1985). Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J. Bacteriol. 162, 328–334. doi: 10.1128/jb.162.1.328-334.1985
Mergeay, M., and Van Houdt, R. (2015). Metal Response in Cupriavidus Metallidurans. Volume I, From Habitats to Genes and Proteins. Berlin: Springer.
Merroun, M. L., Raff, J., Rossberg, A., Hennig, C., Reich, T., and Selenska-Pobell, S. (2005). Complexation of uranium by cells and S-layer sheets of Bacillus sphaericus JG-A12. Appl. Environ. Microbiol. 71, 5532–5543. doi: 10.1128/AEM.71.9.5532-5543.2005
Merroun, M. L., and Selenska-Pobell, S. (2008). Bacterial interactions with uranium: an environmental perspective. J. Contaminant Hydrol. 102, 285–295. doi: 10.1016/j.jconhyd.2008.09.019
Mijnendonckx, K., Leys, N., Mahillon, J., Silver, S., and Van Houdt, R. (2013). Antimicrobial silver: uses, toxicity and potential for resistance. BioMetals 26, 609–621. doi: 10.1007/s10534-013-9645-z
Misra, C. S., Sounderajan, S., and Apte, S. K. (2021). Metal removal by metallothionein and an acid phosphatase PhoN, surface-displayed on the cells of the extremophile, Deinococcus radiodurans. J. Hazardous Mater. 419:126477. doi: 10.1016/j.jhazmat.2021.126477
Mo, K. F., Dai, Z., and Wunschel, D. S. (2016). Production and characterization of desmalonichrome relative binding affinity for Uranyl ions in relation to other siderophores. J. Nat. Prod. 79, 1492–1499. doi: 10.1021/acs.jnatprod.5b00933
Morrison, K. D., Zavarin, M., Kersting, A. B., Begg, J. D., Mason, H. E., Balboni, E., et al. (2021). Influence of Uranium concentration and pH on U-Phosphate biomineralization by Caulobacter OR37. Environ. Sci. Technol. 55, 1626–1636. doi: 10.1021/acs.est.0c05437
Munson, G. P., Lam, D. L., Outten, F. W., and O’Halloran, V. T. (2000). Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J. Bacteriol. 182, 5864–5871. doi: 10.1128/JB.182.20.5864-5871.2000
NEA/IAEA (1999). “Environmental activities in uranium mining and milling,” in Nuclear Development, ed. OECD (Vienna: IAEA).
Newsome, L., Morris, K., and Lloyd, J. R. (2014). The biogeochemistry and bioremediation of uranium and other priority radionuclides. Chem. Geol. 363, 164–184. doi: 10.1016/j.chemgeo.2013.10.034
Newsome, L., Morris, K., and Lloyd, J. R. (2015). Uranium biominerals precipitated by an environmental isolate of serratia under anaerobic conditions. PLoS One 10:e0132392. doi: 10.1371/journal.pone.0132392
Nies, D. H. (1992). CzcR and CzcD, gene products affecting regulation of resistance to cobalt, zinc, and cadmium (czc system) in Alcaligenes eutrophus. J. Bacteriol. 174, 8102–8110. doi: 10.1128/jb.174.24.8102-8110.1992
Nies, D. H. (2003). Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27, 313–339. doi: 10.1016/S0168-6445(03)00048-2
Nilgiriwala, K. S., Alahari, A., Rao, A. S., and Apte, S. K. (2008). Cloning and overexpression of alkaline phosphatase PhoK from Sphingomonas sp. strain BSAR-1 for bioprecipitation of uranium from alkaline solutions. Appl. Environ. Microbiol. 74, 5516–5523. doi: 10.1128/AEM.00107-08
Nolan, J., and Weber, K. A. (2015). Natural Uranium contamination in Major U.S. Aquifers linked to nitrate. Environ. Sci. Technol. Lett. 2, 215–220. doi: 10.1021/acs.estlett.5b00174
Nongkhlaw, M., and Joshi, S. R. (2019). Molecular insight into the expression of metal transporter genes in Chryseobacterium sp. PMSZPI isolated from uranium deposit. PLoS One 14:e0216995. doi: 10.1371/journal.pone.0216995
Nucifora, G., Chu, L., Misra, T. K., and Silver, S. (1989). Cadmium resistance from Staphylococcus aureus plasmid pI258 cadA gene results from a cadmium-efflux ATPase. Proc. Natl. Acad. Sci. U.S.A. 86, 3544–3548. doi: 10.1073/pnas.86.10.3544
Oh, B. C., Choi, W. C., Park, S., Kim, Y. O., and Oh, T. K. (2004). Biochemical properties and substrate specificities of alkaline and histidine acid phytases. Appl. Microbiol. Biotechnol. 63, 362–372. doi: 10.1007/s00253-003-1345-0
Orellana, R., Hixson, K. K., Murphy, S., Mester, T., Sharma, M. L., Lipton, M. S., et al. (2014). Proteome of Geobacter sulfurreducens in the presence of U(VI). Microbiology (United Kingdom) 160, 2607–2617. doi: 10.1099/mic.0.081398-0
Orellana, R., Leavitt, J. J., Comolli, L. R., Csencsits, R., Janot, N., Flanagan, K. A., et al. (2013). U(VI) reduction by diverse outer surface c-type cytochromes of Geobacter sulfurreducens. Appl. Environ. Microbiol. 79, 6369–6374. doi: 10.1128/AEM.02551-13
Pablo, D. J., Casas, I., Giménez, J., Molera, M., Rovira, M., Duro, L., et al. (1999). The oxidative dissolution mechanism of uranium dioxide. I. The effect of temperature in hydrogen carbonate medium. Geochimica Cosmochimica Acta 63, 3097–3103. doi: 10.1016/S0016-7037(99)00237-9
Park, D. M., Overton, K. W., and Jiao, Y. (2019). The UzcRS two-component system in Caulobacter crescentus integrates regulatory input from diverse auxiliary regulators. Mol. Microbiol. 111, 678–699. doi: 10.1111/mmi.14180
Park, D. M., Overton, K. W., Liou, M. J., and Jiao, Y. (2017). Identification of a U/Zn/Cu responsive global regulatory two-component system in Caulobacter crescentus. Mol. Microbiol. 104, 46–64. doi: 10.1111/mmi.13615
Park, D. M., and Taffet, M. J. (2019). Combinatorial sensor design in Caulobacter crescentus for selective environmental uranium detection. ACS Synthetic Biol. 8, 807–817. doi: 10.1021/acssynbio.8b00484
Pattanapipitpaisal, P., Mabbett, A. N., Finlay, J. A., Beswick, A. J., Paterson-Beedle, M., Essa, A., et al. (2002). Reduction of Cr(VI) and bioaccumulation of chromium by gram positive and gram negative microorganisms not previously exposed to Cr-stress. Environ. Technol. 23, 731–745. doi: 10.1080/09593332308618367
Payne, R. B., Casalot, L., Rivere, T., Terry, J. H., Larsen, L., Giles, B. J., et al. (2004). Interaction between uranium and the cytochrome c3 of Desulfovibrio desulfuricans strain G20. Arch. Microbiol. 181, 398–406. doi: 10.1007/s00203-004-0671-7
Payne, R. B., Gentry, D. M., Rapp-Giles, B. J., Casalot, L., and Wall, J. D. (2002). Uranium reduction by Desulfovibrio desulfuricans strain G20 and a cytochrome c3 mutant. Appl. Environ. Microbiol. 68, 3129–3132. doi: 10.1128/AEM.68.6.3129-3132.2002
Pinel-Cabello, M., Jroundi, F., López-Fernández, M., Geffers, R., Jarek, M., Jauregui, R., et al. (2021). Multisystem combined uranium resistance mechanisms and bioremediation potential of Stenotrophomonas bentonitica BII-R7: transcriptomics and microscopic study. J. Hazardous Mater. 403:123858. doi: 10.1016/j.jhazmat.2020.123858
Pollmann, K., Raff, J., Merroun, M., Fahmy, K., and Selenska-Pobell, S. (2006). Metal binding by bacteria from uranium mining waste piles and its technological applications. Biotechnol. Adv. 24, 58–68. doi: 10.1016/j.biotechadv.2005.06.002
Pollmann, K., Raff, J., Schnorpfeil, M., Radeva, G., and Selenska-Pobell, S. (2005). Novel surface layer protein genes in Bacillus sphaericus associated with unusual insertion elements. Microbiology 151, 2961–2973. doi: 10.1099/mic.0.28201-0
Powers, L. G., Mills, H. J., Palumbo, V. A., Zhang, C., Delaney, K., and Sobecky, P. A. (2002). Introduction of a plasmid-encoded phoA gene for constitutive overproduction of alkaline phosphatase in three subsurface Pseudomonas isolates. FEMS Microbiol. Ecol. 41, 115–123. doi: 10.1016/S0168-6496(02)00263-5
Prăvălie, R. (2014). Nuclear weapons tests and environmental consequences: a global perspective. Ambio 43, 729–744. doi: 10.1007/s13280-014-0491-1
Raff, J., Merroun, M., Roßberg, A., Hennig, C., and Selenska-Pobell, S. (2002). EXAFS Study of Uranium (VI) Complexes Formed by Native and Recrystallized S-layers of the Bacillus sphaericus StrainsJG-A12andNCTC9602, Vol. 36. Annual Report 2002 Institute of Radiochemistry. Warszawa.
Rajkumar, M., Ae, N., Prasad, M. N. V., and Freitas, H. (2010). Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol. 28, 142–149. doi: 10.1016/j.tibtech.2009.12.002
Randall, C. P., Gupta, A., Jackson, N., Busse, D., and O’Neill, A. J. (2014). Silver resistance in Gram-negative bacteria: a dissection of endogenous and exogenous mechanisms. J. Antimicrob. Chemother. 70, 1037–1046. doi: 10.1093/jac/dku523
Rashmi, V., ShylajaNaciyar, M., Rajalakshmi, R., D’Souza, S. F., Prabaharan, D., and Uma, L. (2013). Siderophore mediated uranium sequestration by marine cyanobacterium Synechococcus elongatus BDU 130911. Bioresour. Technol. 130, 204–210. doi: 10.1016/j.biortech.2012.12.016
Reitz, T., Rossberg, A., Barkleit, A., Selenska-Pobell, S., and Merroun, M. L. (2014). Decrease of U(VI) immobilization capability of the facultative anaerobic strain Paenibacillus sp. JG-TB8 under anoxic conditions due to strongly reduced phosphatase activity. PLoS One 9:e102447. doi: 10.1371/journal.pone.0102447
Renninger, N., Knopp, R., Nitsche, H., Clark, D. S., and Keasling, J. D. (2004). Uranyl precipitation by Pseudomonas aeruginosa via controlled polyphosphate metabolism. Appl. Environ. Microbiol. 70, 7404–7412. doi: 10.1128/AEM.70.12.7404-7412.2004
Rogiers, T. (2022). Uranium Resistance Mechanisms in Cupriavidus Metallidurans: From Environmental Relevance to Potential Applications. Ph.D. thesis. Ghent: Ghent University.
Rogiers, T., Claesen, J., Gompel, A. V., Vanhoudt, N., Mysara, M., Williamson, A., et al. (2021a). Soil microbial community structure and functionality changes in response to long-term metal and radionuclide pollution. Environ. Microbiol. 23, 1670–1683. doi: 10.1111/1462-2920.15394
Rogiers, T., Merroun, M. L., Williamson, A., Leys, N., Houdt, R. V., Boon, N., et al. (2021b). Cupriavidus metallidurans NA4 actively forms polyhydroxybutyrate-associated uranium-phosphate precipitates. J. Hazard Mater. 421:126737. doi: 10.1016/j.jhazmat.2021.126737
Romanchuk, A. Y., Vlasova, I. E., and Kalmykov, S. N. (2020). Speciation of uranium and plutonium from nuclear legacy sites to the environment: a mini review. Front. Chem. 8:630. doi: 10.3389/fchem.2020.00630
Ruttenberg, K. C. (2014). “10.13 - The global phosphorus cycle,” in Treatise on Geochemistry, 2nd Edn, eds H. D. Holland and K. K. Turekian (Oxford: Elsevier), 499–558. doi: 10.1016/b978-0-08-095975-7.00813-5
Salbu, B., Burkitbaev, M., Strømman, G., Shishkov, I., Kayukov, P., Uralbekov, B., et al. (2013). Environmental impact assessment of radionuclides and trace elements at the Kurday U mining site, Kazakhstan. J. Environ. Radioact. 123, 14–27. doi: 10.1016/j.jenvrad.2012.05.001
Salome, K. R., Beazley, M. J., Webb, S. M., Sobecky, P. A., and Taillefert, M. (2017). Biomineralization of U(VI) phosphate promoted by microbially-mediated phytate hydrolysis in contaminated soils. Geochimica Cosmochimica Acta 197, 27–42. doi: 10.1016/j.gca.2016.10.008
Sanchez-Castro, I., Amador-Garcia, A., Moreno-Romero, C., Lopez-Fernandez, M., Phrommavanh, V., Nos, J., et al. (2017). Screening of bacterial strains isolated from uranium mill tailings porewaters for bioremediation purposes. J. Environ. Radioact. 166(Pt 1), 130–141. doi: 10.1016/j.jenvrad.2016.03.016
Santos, E. A., and Ladeira, A. C. Q. (2011). Recovery of uranium from mine waste by leaching with carbonate-based reagents. Environ. Sci. Technol. 45, 3591–3597. doi: 10.1021/es2002056
Scherer, J., and Nies, D. H. (2009). CzcP is a novel efflux system contributing to transition metal resistance in Cupriavidus metallidurans CH34. Mol. Microbiol. 73, 601–621. doi: 10.1111/j.1365-2958.2009.06792.x
Shelobolina, E. S., Coppi, V. M., Korenevsky, A. A., Didonato, L. N., Sullivan, S. A., Konishi, H., et al. (2007). Importance of c-Type cytochromes for U(VI) reduction by Geobacter sulfurreducens. BMC Microbiol. 7:16. doi: 10.1186/1471-2180-7-16
Shelobolina, E. S., Vrionis, H. A., Findlay, R. H., and Lovley, D. R. (2008). Geobacter uraniireducens sp. nov., isolated from subsurface sediment undergoing uranium bioremediation. Int. J. Syst. Evol. Microbiol. 58, 1075–1078. doi: 10.1099/ijs.0.65377-0
Silver, S., and Phung, L. T. (2005). A bacterial view of the periodic table: genes and proteins for toxic inorganic ions. J. Industrial Microbiol. Biotechnol. 32, 587–605. doi: 10.1007/s10295-005-0019-6
Sitte, J., Loffler, S., Burkhardt, E. M., Goldfarb, K. C., Buchel, G., Hazen, T. C., et al. (2015). Metals other than uranium affected microbial community composition in a historical uranium-mining site. Environ. Sci. Pollut. Res. Int. 22, 19326–19341. doi: 10.1007/s11356-015-4791-1
Smil, V. (2000). Phosphorus in the environment: natural flows and human interferences. Annu. Rev. Energy Environ. 25, 53–88. doi: 10.1146/annurev.energy.25.1.53
Snavely, M. D., Florer, J. B., Miller, C. G., and Maguire, M. E. (1989). Magnesium transport in Salmonella typhimurium: 28Mg2+ transport by the CorA, MgtA, and MgtB systems. J. Bacteriol. 171, 4761–4766. doi: 10.1128/jb.171.9.4761-4766.1989
Strømman, G., Rosseland, B. O., Skipperud, L., Burkitbaev, L. M., Uralbekov, B., Heier, L. S., et al. (2013). Uranium activity ratio in water and fish from pit lakes in Kurday, Kazakhstan and Taboshar, Tajikistan. J. Environ. Radioact. 123, 71–81. doi: 10.1016/j.jenvrad.2012.05.014
Sutcliffe, B., Chariton, A. A., Harford, A. J., Hose, G. C., Greenfield, P., Elbourne, L. D. H., et al. (2017). Effects of uranium concentration on microbial community structure and functional potential. Environ. Microbiol. 19, 3323–3341. doi: 10.1111/1462-2920.13839
Suzuki, Y., and Banfield, J. F. (2004). Resistance to, and accumulation of, uranium by bacteria from a uranium-contaminated site. Geomicrobiol. J. 21, 113–121. doi: 10.1080/01490450490266361
Suzuki, Y., Kitatsuji, Y., Ohnuki, T., and Tsujimura, S. (2010). Flavin mononucleotide mediated electron pathway for microbial U(VI) reduction. Phys. Chem. Chem. Phys. 12, 10081–10087. doi: 10.1039/c0cp00339e
Taira, Y., Hayashida, N., Tsuchiya, R., Yamaguchi, H., Takahashi, J., Kazlovsky, A., et al. (2013). Vertical distribution and estimated doses from artificial radionuclides in soil samples around the chernobyl nuclear power plant and the semipalatinsk nuclear testing site. PLoS One 8:e57524. doi: 10.1371/journal.pone.0057524
Tapia-Rodríguez, A., Luna-Velasco, A., Field, J. A., and Sierra-Alvarez, R. (2012). Toxicity of uranium to microbial communities in anaerobic biofilms. Water Air Soil Pollut. 223, 3859–3868. doi: 10.1007/s11270-012-1154-0
Theodorakopoulos, N., Chapon, V., Coppin, F., Floriani, M., Vercouter, T., Sergeant, C., et al. (2015). Use of combined microscopic and spectroscopic techniques to reveal interactions between uranium and Microbacterium sp. A9, a strain isolated from the Chernobyl exclusion zone. J. Hazardous Mater. 285, 285–293. doi: 10.1016/j.jhazmat.2014.12.018
Tompson, A. F. B., Bruton, C. J., Pawloski, G. A., Smith, D. K., Bourcier, W. L., Shumaker, D. E., et al. (2002). On the evaluation of groundwater contamination from underground nuclear tests. Environ. Geol. 42, 235–247. doi: 10.1007/s00254-001-0493-8
Tu, H., Yuan, G., Zhao, C., Liu, J., Li, F., Yang, J., et al. (2019). U-phosphate biomineralization induced by Bacillus sp. dw-2 in the presence of organic acids. Nuclear Eng. Technol. 51, 1322–1332. doi: 10.1016/j.net.2019.03.002
Turner, B. L. (2006). “Inositol phosphates: linking agriculture and the environment,” in Inositol Phosphates in Soil: Amounts, Forms and Significance of the Phosphorylated Inositol Stereoisomers, eds B. L. Turner, A. E. Richardson, and E. J. Mullaney (Wallingford: CABI), 186–206. doi: 10.1079/9781845931520.0186
Tykva, R. (2004). Sources of environmental radionuclides and recent results in analyses of bioaccumulation. A review. Nukleonika 49(Suppl. 1), 3–7.
UNSCEAR (1988). Report to The General Assembly (Annex D— Exposures from Chernobyl Accident). New York, NY: UNSCEAR.
Van der Stricht, S., and Janssens, A. (2010). Radioactive Effluents from Nuclear Power Stations and Nuclear Fuel Reprocessing Plants in the European Union, 2004-08. Brussels: European Commission.
Weber, K. A., Achenbach, L. A., and Coates, J. D. (2006). Microorganisms pumping iron: anaerobic microbial iron oxidation and reduction. Nat. Rev. Microbiol. 4, 752–764. doi: 10.1038/nrmicro1490
Wellman, D. M., Icenhower, J. P., and Owen, A. T. (2006). Comparative analysis of soluble phosphate amendments for the remediation of heavy metal contaminants: effect on sediment hydraulic conductivity. Environ. Chem. 3, 219–224. doi: 10.1071/EN05023
Whicker, F. W. (1983). Radionuclide transport processes in terrestrial ecosystems. Radiation Res. 94, 135–150. doi: 10.2307/3575869
Williamson, A. J., Morris, K., Law, G. T. W., Rizoulis, A., Charnock, J. M., and Lloyd, J. R. (2014). Microbial reduction of U(VI) under alkaline conditions: implications for radioactive waste geodisposal. Environ. Sci. Technol. 48, 13549–13556. doi: 10.1021/es5017125
Wufuer, R., Wei, Y., Lin, Q., Wang, H., Song, W., Liu, W., et al. (2017). Uranium bioreduction and biomineralization. Adv. Appl. Microbiol. 101, 137–168. doi: 10.1016/bs.aambs.2017.01.003
Wyss, M., Brugger, R., Kronenberger, A., Re, R., Fimbel, R., Oesterhelt, G., et al. (1999). Biochemical characterization of fungal phytases (myo-Inositol Hexakisphosphate Phosphohydrolases): catalytic properties. Appl. Environ. Microbiol. 65, 367–373. doi: 10.1128/AEM.65.2.367-373.1999
Xu, R., Wu, K., Han, H., Ling, Z., Chen, Z., Liu, P., et al. (2018). Co-expression of YieF and PhoN in Deinococcus radiodurans R1 improves uranium bioprecipitation by reducing chromium interference. Chemosphere 211, 1156–1165. doi: 10.1016/j.chemosphere.2018.08.061
Yamasaki, S., Tanaka, K., Kozai, N., and Ohnuki, T. (2017). Effect of flavin compounds on uranium(VI) reduction- kinetic study using electrochemical methods with UV-vis spectroscopy. Appl. Geochem. 78, 279–286. doi: 10.1016/j.apgeochem.2017.01.014
Yi, Z.-J., Tan, K.-X., Tan, A.-L., Yu, Z.-X., and Wang, S.-Q. (2007). Influence of environmental factors on reductive bioprecipitation of uranium by sulfate reducing bacteria. Int. Biodeterioration Biodegradation 60, 258–266. doi: 10.1016/j.ibiod.2007.04.001
You, W., Peng, W., Tian, Z., and Zheng, M. (2021). Uranium bioremediation with U(VI)-reducing bacteria. Sci. Total Environ. 798:149107. doi: 10.1016/j.scitotenv.2021.149107
Yun, J., Malvankar, N. S., Ueki, T., and Lovley, D. R. (2016). Functional environmental proteomics: elucidating the role of a c-type cytochrome abundant during uranium bioremediation. ISME Journal 10, 310–320. doi: 10.1038/ismej.2015.113
Yung, M. C., and Jiao, Y. (2014). Biomineralization of uranium by PhoY phosphatase activity aids cell survival in Caulobacter crescentus. Appl. Environ. Microbiol. 80, 4795–4804. doi: 10.1128/AEM.01050-14
Yung, M. C., Ma, J., Salemi, M. R., Phinney, B. S., Bowman, G. R., and Jiao, Y. (2014). Shotgun proteomic analysis unveils survival and detoxification strategies by Caulobacter crescentus during exposure to uranium, chromium, and cadmium. J. Proteome Res. 13, 1833–1847. doi: 10.1021/pr400880s
Yung, M. C., Park, D. M., Overton, K. W., Blow, M. J., Hoover, C. A., Smit, J., et al. (2015). Transposon mutagenesis paired with deep sequencing of Caulobacter crescentus under Uranium stress reveals genes essential for detoxification and stress tolerance. J. Bacteriol. 197, 3160–3172. doi: 10.1128/jb.00382-15
Yun-Juan, C., Long, P. E., Geyer, R., Peacock, A. D., Resch, C. T., Sublette, K., et al. (2005). Microbial incorporation of 13C-labeled acetate at the field scale: detection of microbes responsible for reduction of U(VI). Environ. Sci. Technol. 39, 9039–9048. doi: 10.1021/es051218u
Zachara, J. M., Long, P. E., Bargar, J., Davis, J. A., Fox, P., Fredrickson, J. K., et al. (2013). Persistence of uranium groundwater plumes: contrasting mechanisms at two DOE sites in the groundwater-river interaction zone. J. Contaminant Hydrol. 147, 45–72. doi: 10.1016/j.jconhyd.2013.02.001
Zhang, K., Zhang, D., Wu, X., and Xue, Y. (2021). Continuous and efficient immobilization of heavy metals by phosphate-mineralized bacterial consortium. J. Hazardous Mater. 416:125800. doi: 10.1016/j.jhazmat.2021.125800
Zheng, L., Li, Y., Shang, W., Dong, X., Tang, Q., and Cheng, H. (2019). The inhibitory effect of cadmium and/or mercury on soil enzyme activity, basal respiration, and microbial community structure in coal mine–affected agricultural soil. Ann. Microbiol. 69, 849–859. doi: 10.1007/s13213-019-01478-3
Keywords: reduction, phosphatases, efflux systems, regulation, bioremediation
Citation: Rogiers T, Van Houdt R, Williamson A, Leys N, Boon N and Mijnendonckx K (2022) Molecular Mechanisms Underlying Bacterial Uranium Resistance. Front. Microbiol. 13:822197. doi: 10.3389/fmicb.2022.822197
Received: 25 November 2021; Accepted: 27 January 2022;
Published: 10 March 2022.
Edited by:
Daniela Ceccarelli, European Commission, BelgiumReviewed by:
Celin Acharya, Bhabha Atomic Research Centre (BARC), IndiaRaluca Maria Hlihor, Ion Ionescu de la Brad University of Agricultural Sciences and Veterinary Medicine of Iaşi, Romania
Alexey Vladimirovich Safonov, Frumkin Institute of Physical Chemistry and Electrochemistry RAS, Russia
Copyright © 2022 Rogiers, Van Houdt, Williamson, Leys, Boon and Mijnendonckx. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kristel Mijnendonckx, a21pam5lbmRAc2NrY2VuLmJl
 Tom Rogiers
Tom Rogiers Rob Van Houdt
Rob Van Houdt Adam Williamson
Adam Williamson Natalie Leys
Natalie Leys Nico Boon
Nico Boon Kristel Mijnendonckx
Kristel Mijnendonckx