
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 25 February 2022
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.821808
This article is part of the Research Topic Emergence and re-emergence of plant diseases caused by Xanthomonas species View all 5 articles
 Austin Herbert1
Austin Herbert1 C. Nathan Hancock2
C. Nathan Hancock2 Brodie Cox3
Brodie Cox3 Guido Schnabel3
Guido Schnabel3 Daniela Moreno1
Daniela Moreno1 Renato Carvalho4,5
Renato Carvalho4,5 Jeffrey Jones4
Jeffrey Jones4 Matthew Paret4,5
Matthew Paret4,5 Xueqing Geng6
Xueqing Geng6 Hehe Wang1*
Hehe Wang1*Xanthomonas arboricola pv. pruni (Xap) causes bacterial spot, a major worldwide disease of Prunus species. Very few chemical management options are available for this disease and frequent applications of oxytetracycline (OTC) in the United States peach orchards have raised concerns about resistance development. During 2017–2020, 430 Xap strains were collected from ten peach orchards in South Carolina. Seven OTC-resistant (OTCR) Xap strains were found in 2017 and 2020 from four orchards about 20–270 km apart. Interestingly, the seven strains were also resistant to streptomycin (STR). Six strains grew on media amended with ≤100 μg/mL OTC, while one strain, R1, grew on ≤250 μg/mL OTC. Genome sequence analysis of four representative OTCR strains revealed a 14–20 kb plasmid carrying tetC, tetR, and strAB in each strain. These three genes were transferable to Xanthomonas perforans via conjugation, and they were PCR confirmed in all seven OTCR Xap strains. When tetC and tetR were cloned and expressed together in a sensitive strain, the transconjugants showed resistance to ≤100 μg/mL OTC. When tetC was cloned and expressed alone in a sensitive strain, the transconjugants showed resistance to ≤250 μg/mL OTC. TetC and tetR expression was inducible by OTC in all six wild-type strains resistant to ≤100 μg/mL OTC. However, in the R1 strain resistant to ≤250 μg/mL OTC, tetR was not expressed, possibly due to the presence of Tn3 in the tetR gene, and in this case tetC was constitutively expressed. These data suggest that tetC confers OTC resistance in Xap strains, and tetR regulates the level of OTC resistance conferred by tetC. To our knowledge, this is the first report of OTC resistance in plant pathogenic xanthomonads.
Xanthomonas arboricola pv. pruni (Xap) causes bacterial spot, a major worldwide disease on stone fruits (e.g., peach, plum, nectarine, apricot, and cherry) and almond (Garita-Cambronero et al., 2018). Xap is an economically important pathogen for the peach industry worldwide and especially for producers in the southeastern United States. The disease produces substantial expenses for disease management throughout the growing season. Producers start spraying during “late dormancy” and continue the practice in 7–10-day intervals throughout the season. Despite these efforts, producers experience millions of dollars in yield loss at harvest in form of unmarketable fruit. Fruit infections result in damage that can affect majority of the fruit surface. Under favorable conditions, bacterial spot can affect 100% of the fruit in peach orchards (Stefani, 2010; Pothier et al., 2011a,b). Foliar infections may result in early defoliation, which leads to weakened trees that may impact winter hardiness. Twig infections lead to twig cankers (also called “spring cankers”), often observed at the beginning of the growing season, which are considered the major source of initial inoculum for leaf and fruit infection (Garita-Cambronero et al., 2018; Cox, 2020).
In United States, conventional management strategies for bacterial spot rely heavily on antibacterial sprays, primarily copper-based compounds and oxytetracycline (OTC) (Yang et al., 2013; Lamichhane, 2014). Cultural practices include optimizing fertilizer and water input, eliminating infected tissues, and intercropping; they are most effective when paired with antibacterial sprays (Yang et al., 2013; Lamichhane, 2014; Giovanardi et al., 2017; Garita-Cambronero et al., 2018). A few disease tolerant cultivars (i.e., Clayton and Candor) are available, however, these cultivars still develop bacterial spot symptoms under high disease pressure and they lack favorable fruit quality necessary for maximizing marketability (Yang et al., 2013; Aranzana et al., 2019).
Disease control with conventional bactericides can be a challenge. Even with weekly applications of copper and OTC, bacterial spot incidence can remain high in southeastern peach orchards under conducive environmental conditions (warm and rainy). Recently, Xap strains with different levels of copper tolerance were identified and they were found to reduce the efficacy of copper compounds used to control bacterial lesions on peach leaves; the copper rates applied at “shuck split” [599 μg/ml Metallic Copper Equivalent (MCE)] and “summer cover spray” (120 μg/ml MCE) were only able to reduce the bacterial spot incidence caused by the copper-sensitive strain, but not by the copper tolerant strains (Cox, 2020). OTC resistance in Xap has not been previously reported but the emergence of resistance is of grave concern since no other chemical control options are available.
Oxytetracycline is a tetracycline that exhibits a broad range of bacteriostatic activity on both Gram-negative and Gram-positive bacteria (Chopra and Roberts, 2001; Christiano et al., 2010). Once inside bacterial cells, OTC binds with high affinity to the 30S subunit and prevents the amino-acyl tRNA from binding to the A site of the ribosome, therefore preventing translation initiation and protein elongation (Chopra and Roberts, 2001). The efficacy of OTC for disease control is directly affected by the frequency and dosage of application, and environmental factors such as precipitation, sunlight (particularly UV light), and temperature (Christiano et al., 2010). Some peach orchards are treated almost weekly with OTC [150 ppm (μg/mL)] during the spring and summer months (Capasso, 2016). However, due to its broad-spectrum activity, the prolonged use of OTC spanning several decades may enrich bacterial communities with antibiotic resistant phenotypes in the phylloplane (Schnabel and Jones, 1999; Vidaver, 2002; Levy and Marshall, 2004).
Tetracyclines have been clinically relevant since the 1940s, and their long-term extensive clinic use for treating bacterial infections has contributed to the emergence and selection of tetracycline resistance genes among human and environmental bacteria (Roberts, 1996; Chopra and Roberts, 2001; Kyselková et al., 2015; Grossman, 2016). There are over 37 genes which are independently or cooperatively able to confer tetracycline resistance, and they are grouped by the biochemical pathways used by their proteins to provide resistance, including tetracycline efflux, ribosomal protection, modification of the tetracycline molecule, and modification of the tetracycline target site (Chopra and Roberts, 2001; Grossman, 2016; Sundin and Wang, 2018; Møller et al., 2020). Tetracycline efflux is the most common resistance mechanism; nearly 30 genes encode for tetracycline-specific efflux pumps, a family of membrane-bound pumps which readily sequester tetracyclines to the periplasm in exchange for a proton (Grossman, 2016; Møller et al., 2020). Several of these efflux genes (tetA, tetC, tetD, tetE, tetG, tetH, and tetI) are flanked upstream by a corresponding repressor tetR, which is oriented in the reverse direction and divergently transcribed (Chopra and Roberts, 2001). The tetR gene encodes a DNA binding protein, a repressor that inhibits transcription of the efflux gene in the absence of tetracycline (Hillen and Berens, 1994; Aleksandrov et al., 2009). Tetracycline efflux genes are commonly acquired through horizontal gene transfer (HGT), as they are often associated with mobile genetic elements with broad host range (Partridge et al., 2018). Furthermore, the tetracycline efflux genes are often co-inherited with other antibiotic resistance genes (e.g., strAB, sulI, and sulII) linked to the same mobile genetic element (Yamaguchi et al., 1990; Schnabel and Jones, 1999; Guardabassi et al., 2000; Shin et al., 2015).
Oxytetracycline was first registered as a pesticide in 1974 (EPA, 1993). Despite the long-term usage in agriculture, the previous reports of OTC resistance in phytopathogens have been limited to Pseudomonas syringae pv. syringae (resisting up to 500 μg/mL) and Agrobacterium tumefaciens (resisting >20 μg/mL), and the genetic determinants of the resistance were only characterized in A. tumefaciens as tetA and tetR (Spotts and Cervantes, 1995; Luo and Farrand, 1999; Hwang et al., 2005). Capasso (2016) surveyed for OTC resistance in Xap and epiphytic bacteria in the Pennsylvania peach orchards, and only found OTC resistance in epiphytic bacteria (minimum inhibitory concentration: 450 μg/mL), which included uncharacterized species under several genera (Pantoea, Rahnella, Pseudomonas, or Xanthomonas) that screened positive for one of the three tetracycline efflux genes (tetA, tetB, and tetC). These OTCR (OTC-resistant) epiphytic bacteria were isolated from peach leaves with or without OTC sprays, which indicates that bacterial epiphytes in the phylloplane of peach orchards could serve as reservoirs for tetracycline resistance genes regardless of selection pressure from OTC sprays.
We hypothesized that the prolonged usage of OTC may have led to the emergence of resistant Xap strains in peach orchards of southeastern United States. Therefore, in this study, we surveyed South Carolina peach orchards during 2017–2020 and identified the first OTCR Xap strains, which were also resistant to streptomycin (STRR). The genetic determinants for resistance to these antibiotics were identified and characterized via PCR, whole genome sequencing, and gene functional analysis.
During 2017–2020, symptomatic leaves and fruit were collected from a total of ten peach farms from six counties across South Carolina; symptomatic twig samples were also collected during 2018 and 2019 (Supplementary Table 1). Each sample was surface sterilized and Xap was isolated from the margins of bacterial spot lesions onto sucrose peptone agar (SPA, Willbrinks medium) (Schaad et al., 2001) and incubated at 25°C for 3–4 days; colonies with Xap morphology were then purified and confirmed with qPCR using Xap-specific primers (Palacio-Bielsa et al., 2011). All the strains were stored in cryogenic storage media (Burr, 1993) in −80°C. The suspensions (∼108 CFU/mL in 0.01% of Silwet® L-77) of the OTCR strains and a subset of the sensitive strains were sprayed onto the leaves of 1-year-old “CaroRed” trees in the greenhouse. Leaves sprayed with water served as control. Bacterial spot symptoms on inoculated peach leaves were evaluated weekly for up to 4 weeks after inoculation.
All Xap strains were screened for resistance to OTC. Isolates grown overnight on nutrient agar (NA, Thermo Fisher Scientific, Waltham, MA, United States) were suspended in 1x phosphate-buffered saline (PBS) to an OD600 value of 0.1 (∼1 × 108CFU/ml). Then, 10 μL of each suspension was dropped on to NA amended with 25 μg/mL of OTC using oxytetracycline hydrochloride (>99% purity, Sigma Aldrich; St. Louis, MO, United States) and incubated for 4 days at 28°C. The bacterial suspension dropped on NA without OTC was included as control. There were two experiments with two technical replicates for each strain. Compared to the growth on the NA control plates, the percentage of growth of each strain on OTC-amended NA was recorded at 3 and 4 days after incubation. Those strains with growth on plates amended with 25 μg/mL OTC were considered OTCR. The OTCR isolates were further screened on NA and NA amended with OTC at 25, 50, 75, 100, and 250 μg/mL and STR at 100, 200, and 300 μg/mL. There were two technical replicates per treatment and the whole experiment was independently replicated three times.
Oxytetracycline-resistant isolates were subjected to PCR screens for the potential resistance genes. One-to-two-day old culture was suspended in 1x PBS and used as DNA template for qPCR with tetC primers that target partial tetC sequence (Table 1; Fan et al., 2007) and SsoAdvanced Universal SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, United States) at 98°C for 3 min and 35 cycles of 98°C for 15 s, 60°C for 20 s and 72°C for 45 s, and a melt curve analysis from 65 to 95°C with an increment of 0.5°C every 5 s. tetR was amplified with the tetR-F and tetR-R primers (Table 1) using a GoTaq G2 Flexi DNA polymerase kit (Promega, Madison, WI, United States) with a temperature cycle of 95°C for 2 min, then 34 cycles of 95°C for 15 s, 57°C for 20 s and 72°C for 30 s, with a final extension at 72°C for 5 min. The strA gene was amplified with the strAP-F and strAT-R primers and strB with the strBP-F and strBT-R primers (Table 1), using the above mentioned GoTaq kit with a temperature cycle of 95°C for 5 min, then 34 cycles of 95°C for 15 s, 55°C (strA) or 56°C (strB) for 20 s, 72°C for 90 s, followed by a final extension of 72°C for 5 min.
The full-length tetC gene was amplified with the Phusion high fidelity enzyme (Thermo Fisher Scientific, Waltham, MA, United States) using the tetC-PF and tetC-TR primers (Table 1) with a temperature cycle of 98°C for 2 min followed by 35 cycles of 98°C for 10 s, 59°C for 30 s, 72°C for 1 min, with a final extension of 72°C for 10 min. PCR products were sent to Eton Biosciences (Research Triangle Park, NC) for sequencing. Gene sequences for tetA-E were retrieved from NCBI and used to build a phylogenetic tree with CLUSTAL (Sievers et al., 2011) based on sequence similarity.
Samples of one representative wild-type OTCR strain from each of the four orchards (F1, M1, R1, and T1) were grown overnight in nutrient broth (NB). DNA was extracted using the Wizard Genomic DNA purification kit (Promega, Madison, WI, United States) following the manufacturer’s instructions and sent for whole genome sequencing on the Illumina NextSeq 2000 system at the Microbial Genome Sequencing Center (MiGS, Pittsburg, PA, United States). DNA libraries were prepared with the Illumina DNA Prep, Tagmentation kit, with read lengths of 2 × 150 bp (Illumina Inc., San Diego, CA, United States), and bcl2fastq (v2.20.0.422)1 was used to demultiplex samples and trim the adapters. The raw reads were assembled with Unicycler v0.4.8 (Wick et al., 2017) utilizing its bold prediction strategy to achieve the highest contiguity.
From the Illumina sequencing, a multi-drug resistant (MDR) plasmid was found in each of the four OTCR strains, however, the plasmid in strain R1 was assembled ‘‘linear’’ instead of circular. To examine the structural variation of this plasmid in R1, we also sent R1 and T1 (as reference) for sequencing with Oxford Nanopore platform at MIGS with DNA extracted using Qiagen Blood and Tissue kit (Qiagen, Germantown, MD, United States). DNA libraries were prepared with the Genomic DNA by Ligation kit (Oxford Nanopore Technologies, New York, NY, United States) and the samples were loaded on R9 flow cells. Guppy (v 4.2.2)2 was used for high accuracy base calling.
The genomes of R1 and T1 were assembled to completion using the Nanopore long reads. Raw Nanopore reads for each genome were assembled with Flye v2.9 (Kolmogorov et al., 2019) utilizing customized options for a genome size of 5.2 M, one iteration of polishing, the plasmid rescue flag, and the meta flag for assembly of data with uneven coverage (Kolmogorov et al., 2020). Draft genomes were circularized using Circlator v1.5.5 (Hunt et al., 2015) with a merge minimum percent identity of 85 and a merge break length of 1000, and polished with Illumina short reads using Pilon v1.24 (Walker et al., 2014). Completeness and quality of the T1 and R1 genomes were assessed with BUSCO v5.2.2 (Simão et al., 2015) and Merqury v1.3 (Rhie et al., 2020). The k-mer databases in Merqury were built from the Illumina short reads of each strain with a k-mer size of 16.
Contig level assemblies were annotated with PROKKA v1.14.5 (Seemann, 2014), and gene clustering was performed with Panaroo v1.2.7 (Tonkin-Hill et al., 2020) using a percent identity threshold for gene clusters of 98% and Clustal (Sievers et al., 2011) to align the core genomes. Locations of the OTC and STR resistance genes were determined with a forward BLASTn of the tetC and strAB homologs from NCBI. Hypothetical proteins on the MDR plasmids were further annotated by BLASTp against the NCBI or UniProt databases. Potential structural variation among plasmids and transposons was first assessed through alignments with Clustal Omega (Sievers et al., 2011) and MAUVE (Darling et al., 2004), and later confirmed by read mapping using Hisat2 v2.2.1 (Kim et al., 2019). All formatting, sorting, and indexing of alignment files was conducted with Samtools v1.14 (Li et al., 2009).
Raw reads from each genome were aligned to each de novo assembled plasmid as stated above with Bowtie2 and variations were called with Freebayes. Read alignments were visualized in Geneious (Kearse et al., 2012) and universal primers were designed with Primer3 (Untergasser et al., 2012) to amplify the indel in strains T1, M1, F1, and R1. The structural variation and the proximity of tetR to tetC in strain R1 was assessed with two pairs of primers: tetR-F/strAT-R and tetR-All-F/tetR-R (Table 1). PCR reactions were performed with strains T1 and R1 using the Phusion DNA polymerase with a temperature program of 98°C for 2 min followed by 35 cycles of 98°C for 10 s, 65°C for 15 s, 72°C for 30 s, with a final extension of 72°C for 10 min. The primers 3708F/5276R (Table 1) flanking the insertion in T1 were used with the Phusion polymerase with the same PCR program described above. PCR products of 3708F/5276R were visualized on a 1% agarose gel and then sent to Eton Biosciences for Sanger sequencing.
Each of the four sequenced OTCR Xap strains were mated with X. perforans strain GEV1001 (Abrahamian et al., 2018) carrying a chromosomal mutation for rifampicin resistance. Xap and X. perforans were suspended in 1x PBS to an OD600 = 0.1, mixed at a ratio of 2:1 and grown on NA overnight at 28°C. The mixed culture was resuspended in 1x PBS (OD600 = 0.1) and plated on NA with 100 μg/mL of rifampicin and 25 μg/mL of OTC. The cell density (CFU/mL) of the recipient was calculated through plating dilutions of the combined culture on NA with 100 μg/mL rifampicin; the conjugation assay for each donor strain was independently performed twice. The species of transconjugants were verified with qPCR using the primers and probe (Table 1) specific for X. perforans (Strayer et al., 2016) or Xap (Palacio-Bielsa et al., 2011) using the SsoAdvanced Universal Probes Supermix at 98°C for 3 min, followed by 40 cycles of 98°C for 15 s and 69°C (for X. perforans) or 60°C (for Xap) for 60 s. Non-template control was included in each run. PCR amplification of tetC (primers: tetCP-F/tetCT-R), tetR (tetR-F/tetR-R), strA (strAP-F/strAT-R), strB (strBP-F/strBT-R) and another 4 kb fragment covering the mobA, mazEF, and repA genes (primers: pMDR_4KB_F/pMDR_4KB_R, Table 1) in the MDR plasmids were conducted with DNA from the transconjugants, Xap donors, and GEV1001. PCR reactions with the primers pMDR_4KB_F/pMDR_4KB_R were conducted with Phusion high fidelity enzyme with a temperature cycle of 98°C for 30 s, 35 cycles of 98°C for 10 s, 65°C for 20 s, 72°C for 3 min, and a final extension of 72°C for 10 min. PCR reactions with the other primers were as described above. Additionally, potential transfer of the ubiquitous plasmid pXap41 (Pothier et al., 2011b) from Xap to X. perforans was assessed with the pXap-41-F/pXap-41-R primers (Table 1) using the SsoAdvanced universal SYBR Green Supermix at 98°C for 3 min, and 39 cycles of 98°C for 15 s and 64°C for 30 s, and a melting curve analysis as described above.
The tetC gene in the T1 strain was amplified using the tetCP-F and tetCT-R primers. The combined tetC, tetR, and their cis-regulatory elements (tetCR) in T1 were amplified using the tetR-xbal and tetR-kpn1 primers (Table 1). The high-fidelity PCR reactions used Phusion polymerase with a temperature program of 98°C for 2 min, 98°C for 10 s, 68°C for 10 s, and 72°C for 35 s. The PCR product was cloned into pGEM-T Easy (Promega, Madison, WI, United States) after adding poly-A tails using 2x APEX master mix (Genesee Scientific, San Diego, CA, United States). Plasmids were extracted with a ZymoPURE Plasmid Miniprep Kit (Zymo Research, Irvine, CA, United States) and the tetC and tetCR inserts were subcloned into pBBR1-MCS-2 (Bonomi et al., 2016) and transformed into Stellar competent E. coli cells (Takara Bio USA, Mountain View, CA, United States). The donor Stellar E. coli lines with the pBBR1-MCS2-tetC and pBBR1-MCS-2-tetCR plasmids were mated to the OTC- and STR-sensitive recipients X. perforans GEV1001 and Xap strain 2WF9 (a wild-type strain that was induced for rifampicin resistance on NA amended with 200 μg/mL rifampicin) (Table 1) with a chromosomal marker for rifampicin resistance. The matings were facilitated by a helper strain pRK2013 (Figurski and Helinski, 1979). All three strains were grown separately overnight in LB or NA with their corresponding antibiotics. Cells were spun down and resuspended in 1x PBS, mixed at a ratio of 2 (recipient) to 1 to 1, spot mixed on NA for overnight growth at 28°C before streaking on NA with 100 μg/mL of rifampicin and 25 μg/mL of OTC. All strains used in this experiment were identified to the species level using the qPCR assays specific for X. perforans (Strayer et al., 2016) or Xap (Palacio-Bielsa et al., 2011) as described above.
Strains T1, M1, F1, and R1 were grown for 24 h in NB and NB amended with 25 μg/mL OTC; OD600 values of each strain were standardized to 0.1 with NB before RNA extraction. RNA was extracted with a Zymo Quick RNA kit (Zymo Research, Irvine, CA, United States), and reverse transcription was conducted with the iScript gDNA Clear cDNA Synthesis kit (Bio-Rad Laboratories, Hercules, CA, United States). The cDNA samples were amplified for tetC, tetR, and gyrA (reference gene) with primers tetC-F/tetC-R, tetR-F/tetR-R, and gyrA-F/gyrA-R, respectively (Table 1). qPCR was conducted with the SsoAdvanced universal SYBR Green Supermix with 98°C for 3 min, and 39 cycles of 98°C for 15 s, 61°C for 20 s, and 72°C for 45 s, followed by a melt curve analysis described above. Standard curves for the PCR efficiency (E) of each primer set were calculated as (1 + E) = 10(–1/slope) (Pfaffl, 2001) and transcript abundance was calculated with the equation (1 + E)(–Δ Ct), where ΔCt equals the Ct of the reference gene subtracted from the Ct of the target gene. The change in expression of tetC and tetR in response to OTC treatment was represented by the transcript fold differences in OTC-amended (O) samples relative to controls (C), which was calculated from the equation (1 + Egene)^(CTc – CTo)/(1 + Eref)^(CTc – CTo). The experiment was conducted three times. The fold changes of tetC and tetR expression in response to OTC treatment were analyzed with a generalized linear mixed model in MiniTab v. 18, with strain as the fixed effect and replication as the random effect.
During 2017–2020, 430 Xap isolates were collected from symptomatic leaf, fruit, or twig tissues in the commercial peach orchards in 10 farms, located in six counties in South Carolina (Supplementary Table 1). Seven OTCRSTRR strains (11.6% of the 2017 collection and 2.3% of the 2020 collection) with two resistant phenotypes were identified from symptomatic leaves in four peach orchards 20–270 km apart (Table 2). These OTCRSTRR strains caused similar bacterial spot symptoms on inoculated peach leaves in the greenhouse compared to the sensitive Xap strains (Supplementary Figure 1). They account for 22.2, 1.8, 3.2, and 75% of Xap strains collected from Farms 1–4, respectively (Table 2 and Supplementary Table 1). Six of the seven strains exhibited the first resistant phenotype of growth on media containing OTC at concentrations up to 100 μg/mL and STR up to 300 μg/mL, while the other strain, R1, exhibited the second resistant phenotype of growth on media containing ≤250 μg/mL OTC and ≤100 μg/mL STR (Figure 1). The correlation between OTCR and STRR in Xap strains suggested that the genes controlling these traits are linked. All resistant strains were isolated from orchards that had been regularly treated with OTC but had never been exposed to STR.
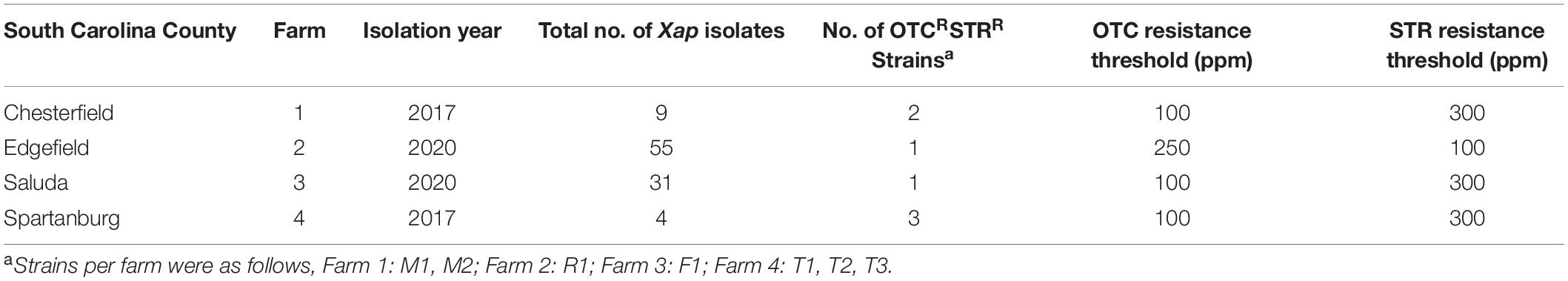
Table 2. Oxytetracycline- (OTC) and streptomycin (STR)-resistant Xanthomonas arboricola pv. pruni (Xap) strains used in this study.
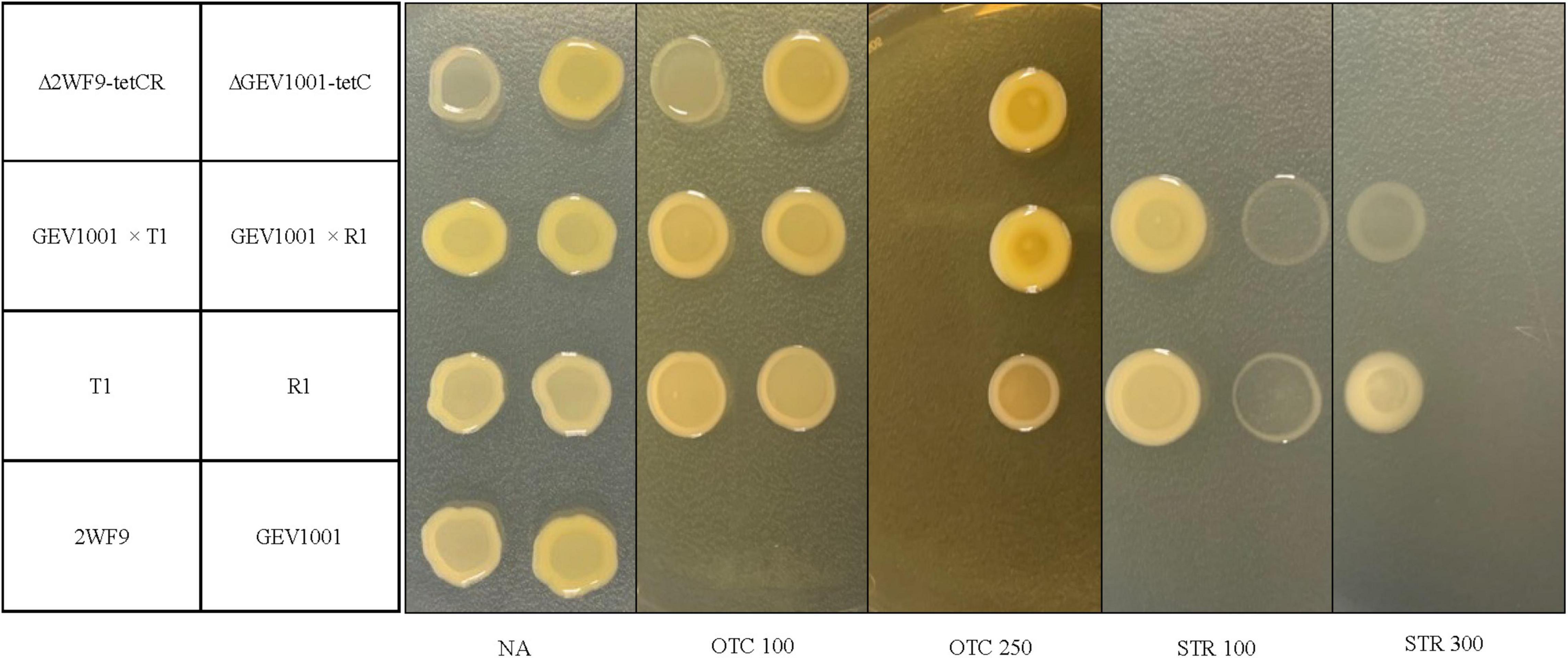
Figure 1. Growth of bacterial strains at 4 days after incubation on nutrient agar (NA), NA amended with 100 or 250 μg/mL of oxytetracycline (OTC 100 and OTC 250) and NA amended with 100 or 300 μg/mL of streptomycin (STR 100 and STR 300). Bacterial strains include: 2WF9 [an OTC- and STR-sensitive, rifampicin-resistant strain, Xanthomonas arboricola pv. pruni (Xap)], GEV1001 (an OTC- and STR-sensitive, rifampicin-resistant strain, Xanthomonas perforans), T1 and R1 (OTC- and STR-resistant wild-type strains, Xap), transconjugants with GEV1001 as recipient and T1 or R1 as donors (GEV1001 × pT1 and GEV1001 × pR1), the mutant of 2WF9 with insertion of tetCR genes cloned from T1 (Δ2WF9-tetCR), and the mutant of GEV1001 with the insertion of tetC gene cloned from T1 (ΔGEV1001-tetC). The strains were identified to the species levels using the Xap- and X. perforans-specific qPCR assays (Palacio-Bielsa et al., 2011; Strayer et al., 2016).
PCR with primers that amplify known OTC and STR resistance genes were used to determine if these genes are present in the resistant strains (Table 1). We detected the tetC, tetR, and strA and strB genes in the OTCR and STRR strains, but not in the sensitive strains (Supplementary Figures 2, 3). Sanger sequencing of the tetC gene in all OTCR strains revealed 100% sequence identity among them.
Phylogenetic analysis confirmed close relationship of the Xap tetC with previously identified tetC sequences in the NCBI database (Supplementary Figure 4) and revealed 99.7–100% similarities to tetC sequences from Chitinibacter sp. 2T18, Candidatus saccharibacteria, Pseudomonas aeruginosa and many other species. Furthermore, the orientation and proximity of tetC to tetR in all strains except R1, similar to the configuration in other bacteria, was confirmed by PCR co-amplification of tetC and tetR using one set of primers (Figure 2; Roberts, 1996; Chopra and Roberts, 2001). These results indicated that there was genetic variation in the tetC or tetR region in the R1 strain.
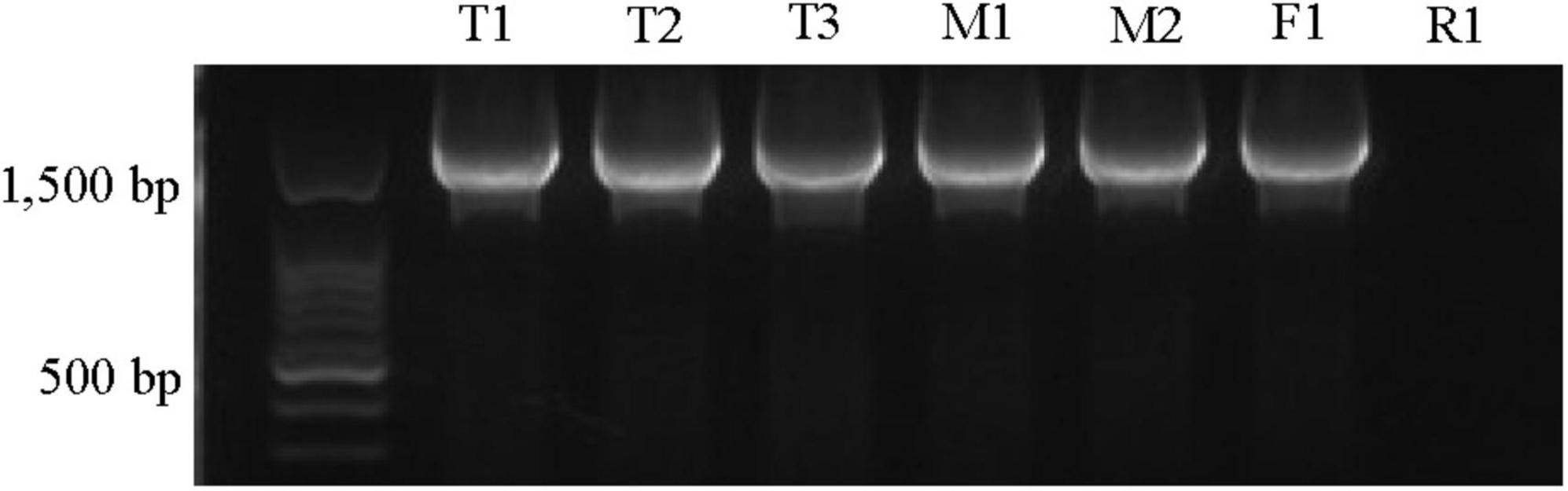
Figure 2. Amplification of tetC and tetR (1.9 kb amplicon) in oxytetracycline-resistant Xanthomonas arboricola pv. pruni strains. The primers tetR-Xbal/tetR-kpn1 were used. The strain IDs were labeled on top of the gel picture. The 100 bp ladder (Biotium, Fremont, CA, United States) was used. All of these strains tested positive for Xap using the probe-based qPCR assay (Palacio-Bielsa et al., 2011).
To gain further insight into the genetics of OTC and STR resistance, Illumina short-read sequencing was conducted for one representative OTCR strain from each peach orchard (Table 2). With these short sequencing reads, a MDR plasmid containing tetCR and strAB genes was identified in each of the four strains. This plasmid was circular in all strains except for R1 where the assembly of the plasmid was incomplete. Thus, we further sequenced strains R1 and T1 (as reference) with Nanopore technology. The Nanopore long reads helped complete the gapless genome assembly for R1 and T1. Each gapless assembly includes one circular chromosome (5,107,097 bp in R1 and 5,117,486 bp in T1), one universal circular plasmid pXap41 (41,093 bp in R1 and 41,084 bp in T1, Pothier et al., 2011b), and one circular MDR plasmid (20,174 bp in R1 and 15,112 bp in T1). Complete BUSCO scores of 99.5 and 99.6% were called for R1 and T1, respectively. The Merqury analysis generated a genome quality value of 43.9 and 42.0 with 98.1 and 99.1% of completeness for R1 and T1, respectively. The raw reads and genome assembly were deposited in Genbank under BioProject PRJNA762222, PRJNA762223, PRJNA762221, and PRJNA762226 for strain F1, M1, R1, and T1, respectively. The assembly data statistics for all four strains are shown in Table 3. Among the four strains, we observed a pan-genome of 4,606 genes and a shared core-genome of 4,146 genes, with each strain carrying the ubiquitous plasmid pXap41 (Pothier et al., 2011b). The complete assemblies of the circular MDR plasmids in these four strains were named pF1 (14,503 bp), pM1 (14,449 bp), pT1 (15,112 bp), and pR1 (20,174 bp) (Figure 3A and Supplementary Figure 5).
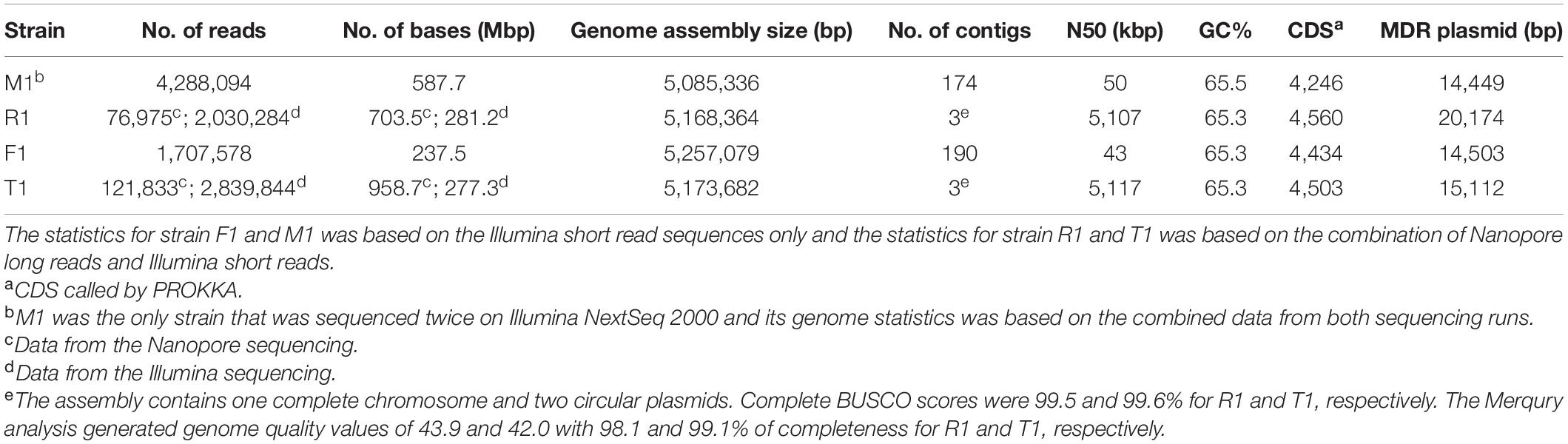
Table 3. Genome statistics of four oxytetracycline- and streptomycin-resistant Xanthomonas arboricola pv. pruni (Xap) strains.
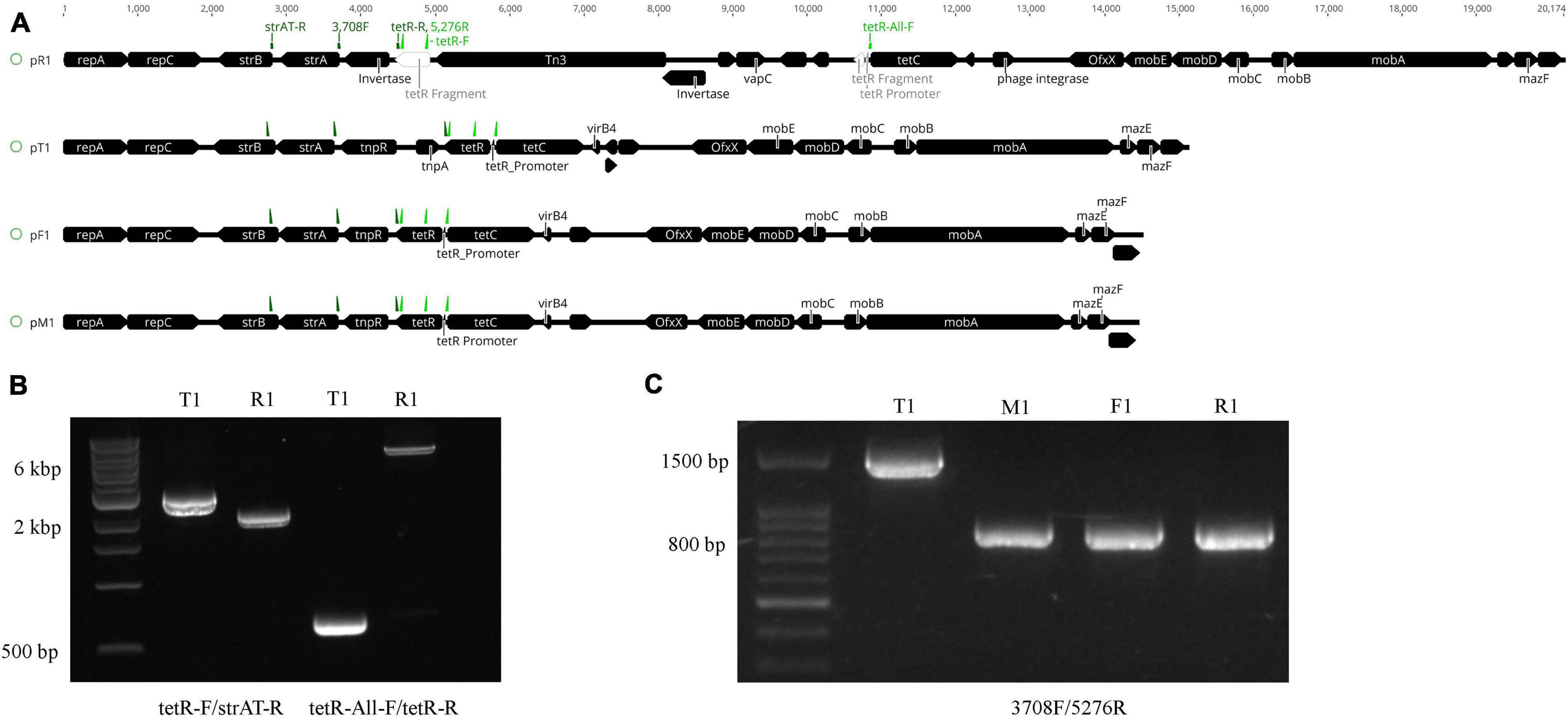
Figure 3. Structural variations of the plasmids harboring the oxytetracycline (OTC) and streptomycin (STR) resistance genes in Xanthomonas arboricola pv. pruni. (A) Linear plasmid map of the de novo assembled plasmids harboring the tetracycline efflux gene tetC, the OTC transcriptional repressor tetR, and the streptomycin resistance operon strAB. The black boxes indicate genes with the points denoting their directions. tetR in strain R1 was physically separated into two parts, bases 1–136 and 137–639, via the Tn3 insertion. On each plasmid, primers strAT-R, 3708F, and tetR-R were labeled as dark green arrows from left to right, and primers tetR-F, 5276R, and tetR-All-F were labeled as light green arrows from left to right. (B) PCR products of DNA from strains T1 and R1 amplified with the primers tetR-F/strAT-R and tetR-All-F/tetR-R. The amplicon sizes are 2,809, 2,109, 699, and 6,969 bp from left to right. The 1 kb Extend DNA ladder (New England Biolabs) was used in the gel. (C) PCR products of T1, M1, F1, and R1 with the 3708F/5276R primers. The amplicon size is 1,568 bp in T1 and 869 bp in the other three strains. The 100 bp ladder (Biotium) was used in the gel. All of the strains tested positive for Xap using the probe-based qPCR assay (Palacio-Bielsa et al., 2011).
Nanopore sequencing identified the Tn3 transposable element inserted into the tetR gene in pR1. The Tn3 element was found in the genome of the other three strains but was not present in their MDR plasmids. The ∼6 kb insertion including the Tn3 transposase and four other genes annotated as hypothetical proteins in pR1 was confirmed by PCR with primers flanking the region (Figures 3A,B). The Tn3 insertion separated the 5′ end of tetR (1–136 bp) and its promoter sequence (AGCTTTAAT, identified by BPROM) (Solovyev and Salamov, 2011) from the rest of the gene (137–503 bp) (Figure 3A and Supplementary Figure 5), which could impact tetR expression and the OTC resistance phenotype in strain R1. No variation was observed in the tetR promoter sequences between the four OTCR strains, but the C-terminus of tetR had sequence variation among all four strains (Supplementary Figure 6).
Aside from the Tn3 insertion on pR1, the other major site of sequence variation between the MDR plasmids was observed in the region between strA and tetC (Figure 3A). In pT1, this region has >68% nucleotide identity with the class II transposable element Tn5393, which contains the tnpA and tnpR genes (Figure 3A). In the other three plasmids, tnpA is missing and tnpR is truncated. The tnpR gene is 100% identical among strains R1, M1, and F1, but only shared 77.5% nucleotide identity with the tnpR gene from strain T1. PCR with the primer 5276R that flanks strA and 3708F that resides in the conserved region of tetR (Table 1) yielded a 1,568 bp amplicon for T1 and an 869 bp amplicon for the other three strains (Figure 3C), confirming the sequence variation identified from the genome sequencing. Sanger sequencing results of these amplicons matched 100% with the sequences in the genome assembly. Unlike pR1 and pT1, the pM1 and pF1 sequences lack structural variation and share a pairwise nucleotide identity of 99.6%.
Furthermore, BLASTn searches of pT1, pM1, and pF1 in the NCBI database revealed >11,000 identical nucleotides to plasmids from Edwardsiella tarda strain 150611-1, Citrobacter freundii strain RHBSTW-00697, Klebsiella sp. WP7-S18-ESBL-04, and Aeromonas salmonicida subsp. salmonicida strain SHY16-3432. The alignment of pT1 to A. salmonicida (Accession: NZ_CP038105.1) and E. tarda (Accession: NZ_MF925338.1) is shown in Supplementary Figure 7. Plasmid pR1 shared >9,000 bp with the above-mentioned bacteria. The main differences in gene content among these plasmids is the sequence variation in the tetracycline efflux genes and the presence of the strAB operon and tnpA and tnpR genes in Xap. The region containing the strAB operon in plasmid pT1 is annotated as a transposase in the Aeromonas and Edwardsiella plasmids and contributes to a significant portion of sequence variation among these species (Supplementary Figure 7).
To test the mobility of the OTC and STR resistance, we performed conjugation assays between the resistant Xap from all four geographic locations to a rifampicin-resistant (RifR) and OTC/STR-sensitive X. perforans strain GEV1001 (Abrahamian et al., 2018). OTCR and STRR GEV1001 transconjugants were produced with conjugation frequencies ranging from 3.53 × 10–7 to 6.81 × 10–6 per number of recipient cells (Table 4). All GEV1001 transconjugants were able to grow on solid media with OTC and STR concentrations matching those of the donor Xap strains (Figure 1). The transconjugants were confirmed to be X. perforans via species-specific PCR (Palacio-Bielsa et al., 2011; Strayer et al., 2016). The tetC, tetR, strAB, and another 4 kb fragment from the MDR plasmids were successfully amplified in all the X. perforans transconjugants and Xap donors, but not in GEV1001 (Supplementary Figures 2, 3, 8). Additionally, GEV1001 transconjugants were confirmed to not harbor the ubiquitous plasmid pXap41 specifically found in Xap with PCR (Pothier et al., 2011b). Together, this supports that the OTC and STR resistance genes are on a mobile plasmid in Xap which can be transferred to other bacterial species.
To confirm gene functions, tetC alone and tetCR (the resistance gene, its transcriptional repressor, and their cis-regulatory elements) were separately cloned from T1 into pBBR1-MCS-2, which were then individually transferred to GEV1001 via triparental mating. The GEV1001 mutants carrying tetCR grew on NA amended with OTC up to 100 μg/mL, the same resistance level as seen in six of the wild-type OTCR Xap strains, suggesting that tetC and tetR are the sole contributors to OTC resistance in these Xap strains (Figure 1). In contrast, when the tetR gene was not present, the GEV1001 mutants carrying tetC alone mimicked the growth patterns of strain R1 and were resistant to OTC concentrations up to 250 μg/mL (Figure 1), indicating that tetR is a crucial regulator and limits the level of OTC resistance in Xap.
In response to OTC treatment, tetC was upregulated in all four strains; tetR was also upregulated in all strains except in R1 where expression was not changed (Figure 4A). Without treating R1 with OTC, tetC was constitutively expressed (18.4–25.3-fold higher vs. the other three strains) (Figure 4B). Furthermore, R1 had significantly lower expression of tetR (3.1–75.5-fold) than the other three strains, which is likely responsible for the constitutive expression of tetC (Figure 4B).
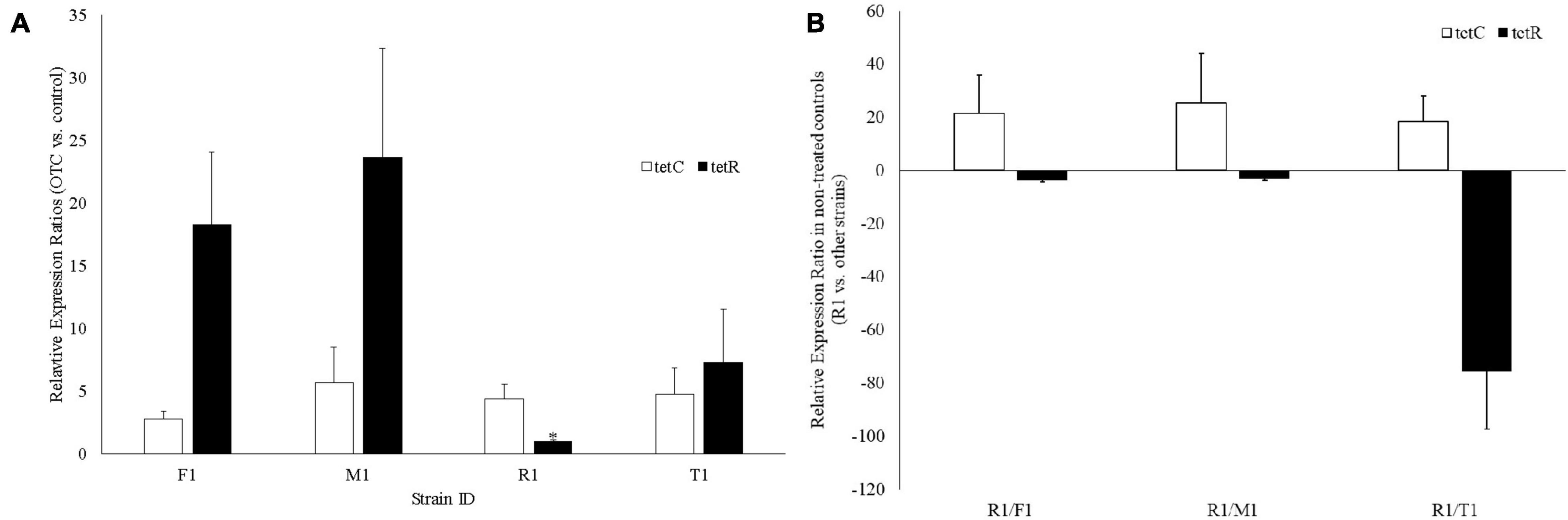
Figure 4. Gene expression analysis of the tetC and tetR genes in the four oxytetracycline-resistant Xanthomonas arboricola pv. pruni strains. (A) Fold expression changes of tetC and tetR after treatment with oxytetracycline (25 mg/L). *There was no change of tetR expression in R1 (relative expression ratio: 1.05, which was significantly lower than the tetR expression in the other three strains, p < 0.001); (B) Relative expression ratio of tetC and tetR in R1 vs. the other strains without oxytetracycline treatment.
This study reports and characterizes OTC resistance in a pathogenic Xanthomonas sp. for the first time. OTC was first registered as a pesticide in 1974 and has been used to control bacterial spot of peach since the 1980s (EPA, 1993). Despite decades of OTC application in agriculture, OTC resistance has only been reported in two plant pathogens (Spotts and Cervantes, 1995; Luo and Farrand, 1999; Hwang et al., 2005), P. syringae (resisting up to 500 μg/mL) and A. tumefaciens (resisting >20 μg/mL). In this study, Xap strains with two OTCR phenotypes were discovered in four orchards 20–270 km apart. In 2017, five strains with the first phenotype (22.2 and 75% of the total isolates in each orchard) were collected from two orchards. Such high percentages in a small sample size suggest that the resistant strains were prevalent or at least not rare in these two orchards at that time (Madden et al., 2017). This may have been a result of the increased selection pressure due to frequent application of OTC for bacterial spot control in peach orchards. Two different orchards were surveyed in 2020; only two resistant strains were found, one with the first phenotype and the other with the second phenotype (1.8 and 3.2% of the population within each orchard), suggesting that these strains may have recently emerged. Continued OTC application will likely maintain or even increase the prevalence of these resistant strains in commercial peach orchards (Schnabel and Jones, 1999; Chopra and Roberts, 2001; Mathers et al., 2011; Rysz et al., 2013; Kyselková et al., 2015; Abrahamian et al., 2018; Kang et al., 2018; Zhang et al., 2018). Given the limited control options available for bacterial spot, the spread of such resistant strains threatens to further increase the difficulty of bacterial spot management. This study provides a first report of the OTC and STR resistance in Xap. The data provided a snapshot of resistance prevalence, because each of these four orchards was only sampled once. Future studies will investigate the persistence and spread of these resistant strains in peach orchards of the southeastern United States.
The OTC resistance in our Xap strains was conferred by tetC interacting with its transcriptional repressor tetR on a 14–20 kb MDR plasmid that also carries strAB operon for STR resistance. The resistance genes on this MDR plasmid and their associated OTCRSTRR phenotype were found to be transferrable to X. perforans via conjugation (Figure 1 and Table 4). Mobility of these plasmids into a broad range of hosts through conjugation is supported by their gene content. Replication of these plasmids may be dependent on the presence of repA and repC (Figure 3A and Supplementary Figure 5), an atypical gene cassette associated with repABC operon reported in Rhizobium leguminosarum (Cevallos et al., 2008; Pérez-Segura et al., 2013). The repC gene encodes a replication initiator protein, and is known to be essential for plasmid replication, whereas repA encodes a plasmid partitioning protein that distributes ssDNA to segregating cells (Cevallos et al., 2008; Pinto et al., 2012; Pérez-Segura et al., 2013). The formation of the relaxosome and mobilization via conjugation is likely carried out by the mobABCDE genes on these plasmids (Figure 3A and Supplementary Figure 5). In this cluster, mobABC is necessary for the initiation and termination stages of conjugal transfer while mobD and mobE act as accessory genes by drastically increasing conjugation efficiency (Perwez and Meyer, 1996; Zhang and Meyer, 1997; Bravo-Angel et al., 1999; van Zyl et al., 2003). In addition, the type IV secretion system (T4SS) located on the chromosome of xanthomonads also contributes to conjugation (Wallden et al., 2010; Guglielmini et al., 2013; Cenens et al., 2020; Alvarez-Martinez et al., 2021). Therefore, mobilization of these small plasmids is likely the outcome from the interplay of the repAC and mobABCDE genes, T4SS genes, and selection pressure from OTC sprays (Guglielmini et al., 2013). The horizontal transfer of these MDR plasmids between bacterial species poses a threat to the well-being of the peach orchards and the efficacy of current spray programs.
Lineage-specific mutations on these newly identified MDR plasmids, like the acquisition of additional antibiotic resistance genes and indels are likely the outcome of the interplay of several contributing factors, including transposons, phages, or other mobile genetic elements. Characterization of the sequence and structure variation of these plasmids is important for reporting potential differences in antibiotic resistance phenotypes. Here, we report a unique example of how a transposon insertion on the plasmid pR1 directly affected the OTC resistance levels of its host strain. The Tn3 insertion in pR1 disrupts the tetR gene and helps to explain the lack of tetR expression, which is the most likely cause for the constitutive expression of tetC in strain R1, as no difference in the tetC promoter sequences was observed among the seven OTCR strains (Figures 3, 4). Although the tetC gene was upregulated upon OTC treatment in all four strains tested, the three strains without constitutive tetC expression may experience a shock from high OTC concentrations (>100 ppm), inhibiting their growth. In comparison, the constitutive expression of tetC may have protected the R1 strain from high OTC concentrations and allowed it to grow. Therefore, the observed gene expression patterns support the increased level of OTC resistance in strain R1 (same level of OTC resistance in the mutants expressing tetCR together) compared to the other six wild-type OTCR strains (same level of OTC resistance in the mutants expressing tetC alone) (Figure 1). All of these results together indicate tetR as the essential transcriptional regulator of tetC and that when tetR was suppressed or absent, tetC conferred much higher OTC resistance levels.
In addition to the structural variation in pR1, indels were also detected in the region between strA and tetC among the sequenced OTCRSTRR strains (Figure 3A). The presence of transposase and resolvase genes (tnpR and tnpA) suggests the involvement of transposons in the evolution of these plasmids. This region shares >68% nucleotide identity with Tn5393, a class II transposable element known for harboring tnpA and tnpR and disseminating the strAB cassette for STR resistance, including in xanthomonads (Sundin and Bender, 1995); the main sequence variations occur in the tnpR, tnpA, and tetCR genes. The tnpA and tnpR genes were both found in pT1, while tnpA was absent and tnpR was truncated in the other three plasmids. A double stranded break caused by faulty transposition of the tnpR and tnpA genes that has been recorded in Tn5393 (McManus et al., 2002) could have occurred and caused the observed deletion in pM1, pF1, and pR1. The proximity of tetR to this deletion site may be the cause for the sequence variation in its last nine C-terminal amino acids among our strains (Supplementary Figure 6).
The evolutionary history of these MDR plasmids is unknown, but we speculate that a potential transposon-mediated transfer of STR resistance into the tetracycline resistant plasmids could be the origin of these plasmids harboring OTC and STR resistance genes in the environmental bacteria, which was then passed on to Xap via conjugation. The MDR plasmids discovered in Xap have a similar backbone with over 73% nucleotide identity to the plasmids carrying tetracycline resistance genes in several different environmental bacteria, such as A. salmonicida and E. tarda (Supplementary Figure 7). They all carry a tetracycline efflux gene with tetR, and repAC and mobABCDE genes. Compared to these environmental bacteria, Xap strains have the additional region that contains strAB operon, tnpA and tnpR that resembles Tn5393 (as discussed above). Co-inheritance of tetracycline and streptomycin resistance genes on the same plasmid has also been reported in several bacterial species in the phylloplane of apple (Schnabel and Jones, 1999). Further studies characterizing the antibiotic resistome in the peach orchards and their water supply will help identify the underlying genetic reservoirs or sources of these plasmids.
To our knowledge, this study presents the first OTC and STR resistance in Xap and the first OTC resistance in a Xanthomonas pathogen. The OTC resistance is conferred by tetCR genes on a small mobilizable plasmid that also harbors strAB operons for STR resistance, and this MDR plasmid could potentially be transferred and maintained in other bacterial epiphytes of peach in addition to Xap. Moreover, these plasmids are prone to structural rearrangements and mutations that could alter the OTC resistance levels, as shown in one of our strains. Further studies are needed to assess the frequency of these mutation events in field populations. Although STR is not used for disease management in peaches, the coinheritance of OTCR and STRR genotypes and phenotypes suggests that STR cannot be used as an alternative to control OTCR strains. Finally, these results emphasize a need for antibiotics resistance monitoring in United States peach orchards and development of resistance management strategies for bacterial spot.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
AH, CH, GS, JJ, XG, and HW contributed to conception and design of the experiments. AH, BC, DM, and HW collected the Xap isolates and conducted chemical screening. AH conducted PCR, conjugation assay, gene expression experiments, and sequence data analysis. AH and CH conducted cloning and gene transfer. RC and HW prepared samples for genome sequencing. AH and HW conducted data analysis and wrote the manuscript. AH, CH, GS, RC, JJ, MP, XG, and HW contributed to manuscript revision. HW planned the study, secured the funding, and submitted the manuscript. All authors read the manuscript and approved the submitted version.
Funding for this work was supported by the USDA-NIFA project 2021-67013-33574 and SC-1700592, and South Carolina Peach Council.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We acknowledge Milan Panth for highly skillful technical assistance, Gary Vallad for providing us the GEV1001 strain and Clemson University for providing the Palmetto Cluster and the Secretariat High Performance computing cluster (supported by NIH grant P20GM139769) for our genome sequence analysis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.821808/full#supplementary-material
Abrahamian, P., Timilsina, S., Minsavage, G. V., Kc, S., Goss, E. M., Jones, J. B., et al. (2018). The Type III effector AvrBsT enhances Xanthomonas perforans fitness in field-grown tomato. Phytopathology 108, 1355–1362. doi: 10.1094/PHYTO-02-18-0052-R
Aleksandrov, A., Schuldt, L., Hinrichs, W., and Simonson, T. (2009). Tetracycline-tet repressor binding specificity: insights from experiments and simulations. Biophys. J. 97, 2829–2838. doi: 10.1016/j.bpj.2009.08.050
Alvarez-Martinez, C. E., Sgro, G. G., Araujo, G. G., Paiva, M. R. N., Matsuyama, B. Y., Guzzo, C. R., et al. (2021). Secrete or perish: the role of secretion systems in Xanthomonas biology. Comput. Struct. Biotechnol. J. 19, 279–302. doi: 10.1016/j.csbj.2020.12.020
Aranzana, M. J., Decroocq, V., Dirlewanger, E., Eduardo, I., Gao, Z. S., Gasic, K., et al. (2019). Prunus genetics and applications after de novo genome sequencing: achievements and prospects. Hortic. Res. 6:58. doi: 10.1038/s41438-019-0140-8
Bonomi, H. R., Toum, L., Sycz, G., Sieira, R., Toscani, A. M., Gudesblat, G. E., et al. (2016). Xanthomonas campestris attenuates virulence by sensing light through a bacteriophytochrome photoreceptor. EMBO Rep. 17, 1565–1577. doi: 10.15252/embr.201541691
Bravo-Angel, A. M., Gloeckler, V., Hohn, B., and Tinland, B. (1999). Bacterial conjugation protein MobA mediates integration of complex DNA structures into plant cells. J. Bacteriol. 181, 5758–5765. doi: 10.1128/JB.181.18.5758-5765.1999
Burr, T. J. (1993). Streptomycin-resistant bacteria associated with fire blight infections. Plant Dis. 77:63. doi: 10.1094/PD-77-0063
Capasso, S. (2016). Antibiotic Screens in Pennsylvania Stone Fruit Orchards. Available online at: https://etda.libraries.psu.edu/files/final_submissions/11693 (accessed May 15, 2021).
Cenens, W., Andrade, M. O., Llontop, E., Alvarez-Martinez, C. E., Sgro, G. G., and Farah, C. S. (2020). Bactericidal type IV secretion system homeostasis in Xanthomonas citri. PLoS Pathog. 16:e1008561. doi: 10.1371/journal.ppat.1008561
Cevallos, M. A., Cervantes-Rivera, R., and Gutiérrez-Ríos, R. M. (2008). The repABC plasmid family. Plasmid 60, 19–37. doi: 10.1016/j.plasmid.2008.03.001
Chopra, I., and Roberts, M. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. MMBR 65, 232–260. second page, table of contents. doi: 10.1128/MMBR.65.2.232-260.2001
Christiano, R. S. C., Reilly, C. C., Miller, W. P., and Scherm, H. (2010). Oxytetracycline dynamics on peach leaves in relation to temperature, sunlight, and simulated rain. Plant Dis. 94, 1213–1218. doi: 10.1094/PDIS-04-10-0282
Cox, B. (2020). Bacterial Spot of Peach: Epidemiology and Chemical Sensitivity of Xanthomonas Arboricola pv. Pruni in South Carolina Orchards. Available online at: https://tigerprints.clemson.edu/all_theses/3409/ (accessed May 15, 2021).
Darling, A. C. E., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
EPA (1993). Hydroxytetracycline Monohydrochloride and Oxytetracycline Calcium. Available online at: https://www3.epa.gov/pesticides/chem_search/reg_actions/reregistration/fs_PC-006308_1-Mar-93.pdf (accessed May 15, 2021).
Fan, W., Hamilton, T., Webster-Sesay, S., Nikolich, M. P., and Lindler, L. E. (2007). Multiplex real-time SYBR Green I PCR assay for detection of tetracycline efflux genes of Gram-negative bacteria. Mol. Cell. Probes 21, 245–256. doi: 10.1016/j.mcp.2006.12.005
Figurski, D. H., and Helinski, D. R. (1979). Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U.S.A. 76, 1648–1652. doi: 10.1073/pnas.76.4.1648
Garita-Cambronero, J., Palacio-Bielsa, A., and Cubero, J. (2018). Xanthomonas arboricola pv. pruni, causal agent of bacterial spot of stone fruits and almond: its genomic and phenotypic characteristics in the X. arboricola species context. Mol. Plant Pathol. 19, 2053–2065. doi: 10.1111/mpp.12679
Giovanardi, D., Dallai, D., and Stefani, E. (2017). Population features of Xanthomonas arboricola pv. pruni from Prunus spp. orchards in northern Italy. Eur. J. Plant Pathol. 147, 761–771. doi: 10.1007/s10658-016-1040-5
Grossman, T. H. (2016). Tetracycline antibiotics and resistance. Cold Spring Harb. Perspect. Med. 6:a025387. doi: 10.1101/cshperspect.a025387
Guardabassi, L., Dijkshoorn, L., Collard, J.-M., Olsen, J. E., and Dalsgaard, A. (2000). Distribution and in-vitro transfer of tetracycline resistance determinants in clinical and aquatic Acinetobacter strains. J. Med. Microbiol. 49, 929–936. doi: 10.1099/0022-1317-49-10-929
Guglielmini, J., de la Cruz, F., and Rocha, E. P. C. (2013). Evolution of conjugation and type IV secretion systems. Mol. Biol. Evol. 30, 315–331. doi: 10.1093/molbev/mss221
Hillen, W., and Berens, C. (1994). Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu. Rev. Microbiol. 48, 345–369. doi: 10.1146/annurev.mi.48.100194.002021
Hunt, M., Silva, N. D., Otto, T. D., Parkhill, J., Keane, J. A., and Harris, S. R. (2015). Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol. 16:294. doi: 10.1186/s13059-015-0849-0
Hwang, M. S. H., Morgan, R. L., Sarkar, S. F., Wang, P. W., and Guttman, D. S. (2005). Phylogenetic characterization of virulence and resistance phenotypes of Pseudomonas syringae. Appl. Environ. Microbiol. 71, 5182–5191. doi: 10.1128/AEM.71.9.5182-5191.2005
Kang, Y., Li, Q., Yin, Z., Shen, M., Zhao, H., Bai, Y., et al. (2018). High diversity and abundance of cultivable tetracycline-resistant bacteria in soil following pig manure application. Sci. Rep. 8:1489. doi: 10.1038/s41598-018-20050-8
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinform. Oxf. Engl. 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Kolmogorov, M., Bickhart, D. M., Behsaz, B., Gurevich, A., Rayko, M., Shin, S. B., et al. (2020). metaFlye: scalable long-read metagenome assembly using repeat graphs. Nat. Methods 17, 1103–1110. doi: 10.1038/s41592-020-00971-x
Kolmogorov, M., Yuan, J., Lin, Y., and Pevzner, P. A. (2019). Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546. doi: 10.1038/s41587-019-0072-8
Kyselková, M., Jirout, J., Vrchotova, N., Schmitt, H., and Elhottova, D. (2015). Spread of tetracycline resistance genes at a conventional dairy farm. Front. Microbiol. 6:536. doi: 10.3389/fmicb.2015.00536
Lamichhane, J. R. (2014). Xanthomonas arboricola diseases of stone fruit, almond, and walnut trees: progress toward understanding and management. Plant Dis. 98, 1600–1610. doi: 10.1094/PDIS-08-14-0831-FE
Levy, S. B., and Marshall, B. (2004). Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10, S122–S129. doi: 10.1038/nm1145
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence Alignment/Map format and SAMtools. Bioinform. Oxf. Engl. 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Luo, Z. Q., and Farrand, S. K. (1999). Cloning and characterization of a tetracycline resistance determinant present in Agrobacterium tumefaciens C58. J. Bacteriol. 181, 618–626. doi: 10.1128/JB.181.2.618-626.1999
Madden, L. V., Hughes, G., and van den Bosch, F. (2017). The Study of Plant Disease Epidemics. Saint Paul, MN: The American Phytopathological Society.
Mathers, J. J., Flick, S. C., and Cox, L. A. (2011). Longer-duration uses of tetracyclines and penicillins in U.S. food-producing animals: indications and microbiologic effects. Environ. Int. 37, 991–1004. doi: 10.1016/j.envint.2011.01.014
McManus, P. S., Stockwell, V. O., Sundin, G. W., and Jones, A. L. (2002). Antibiotic use in plant agriculture. Annu. Rev. Phytopathol. 40, 443–465. doi: 10.1146/annurev.phyto.40.120301.093927
Møller, T. S. B., Liu, G., Hartman, H. B., Rau, M. H., Mortensen, S., Thamsborg, K., et al. (2020). Global responses to oxytetracycline treatment in tetracycline-resistant Escherichia coli. Sci. Rep. 10:8438. doi: 10.1038/s41598-020-64995-1
Palacio-Bielsa, A., Cubero, J., Cambra, M. A., Collados, R., Berruete, I. M., and López, M. M. (2011). Development of an efficient real-time quantitative PCR protocol for detection of Xanthomonas arboricola pv. pruni in Prunus species. Appl. Environ. Microbiol. 77, 89–97. doi: 10.1128/AEM.01593-10
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31, e88–e17. doi: 10.1128/CMR.00088-17
Pérez-Segura, G., Pérez-Oseguera, A., and Cevallos, M. A. (2013). The repAC replication system of the Rhizobium leguminosarum pRL7 plasmid is functional: implications regarding the origin and evolution of repABC plasmids. Plasmid 69, 49–57. doi: 10.1016/j.plasmid.2012.08.003
Perwez, T., and Meyer, R. (1996). MobB protein stimulates nicking at the R1162 origin of transfer by increasing the proportion of complexed plasmid DNA. J. Bacteriol. 178, 5762–5767. doi: 10.1128/jb.178.19.5762-5767.1996
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Pinto, U. M., Pappas, K. M., and Winans, S. C. (2012). The ABCs of plasmid replication and segregation. Nat. Rev. Microbiol. 10, 755–765. doi: 10.1038/nrmicro2882
Pothier, J. F., Pagani, M. C., Pelludat, C., Ritchie, D. F., and Duffy, B. (2011a). A duplex-PCR method for species- and pathovar-level identification and detection of the quarantine plant pathogen Xanthomonas arboricola pv. pruni. J. Microbiol. Methods 86, 16–24. doi: 10.1016/j.mimet.2011.03.019
Pothier, J. F., Vorhölter, F.-J., Blom, J., Goesmann, A., Pühler, A., Smits, T. H. M., et al. (2011b). The ubiquitous plasmid pXap41 in the invasive phytopathogen Xanthomonas arboricola pv. pruni: complete sequence and comparative genomic analysis. FEMS Microbiol. Lett. 323, 52–60. doi: 10.1111/j.1574-6968.2011.02352.x
Rhie, A., Walenz, B. P., Koren, S., and Phillippy, A. M. (2020). Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 21:245. doi: 10.1186/s13059-020-02134-9
Roberts, M. C. (1996). Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol. Rev. 19, 1–24. doi: 10.1111/j.1574-6976.1996.tb00251.x
Rysz, M., Mansfield, W. R., Fortner, J. D., and Alvarez, P. J. J. (2013). Tetracycline resistance gene maintenance under varying bacterial growth rate, substrate and oxygen availability, and tetracycline concentration. Environ. Sci. Technol. 47, 6995–7001. doi: 10.1021/es3035329
Schaad, N. W., Jones, J. B., and Chun, W. (eds) (2001). Laboratory Guide for Identifiction of Plant Pathogenic Bacteria, 3rd Edn. St. Paul, MN: American Phytopathological Society.
Schnabel, E. L., and Jones, A. L. (1999). Distribution of tetracycline resistance genes and transposons among phylloplane bacteria in Michigan apple orchards. Appl. Environ. Microbiol. 65, 4898–4907. doi: 10.1128/AEM.65.11.4898-4907.1999
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinform. Oxf. Engl. 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shin, S. W., Shin, M. K., Jung, M., Belaynehe, K. M., and Yoo, H. S. (2015). Prevalence of antimicrobial resistance and transfer of tetracycline resistance genes in Escherichia coli isolates from beef cattle. Appl. Environ. Microbiol. 81, 5560–5566. doi: 10.1128/AEM.01511-15
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinform. Oxf. Engl. 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Solovyev, V., and Salamov, A. (2011). “Automatic annotation of microbial genomes and metagenomic sequences,” in Metagenomics and its Applications in Agriculture, Biomedicine and Environmental Studies, ed. R. W. Li (Hauppauge, NY: Nova Science Publishers), 61–78.
Spotts, R., and Cervantes, L. (1995). Copper, Oxytetracycline, and Streptomycin Resistance of Pseudomonas Syringae pv Syringae Strains from Pear Orchards in Oregon and Washington. Plant Disease. Available online at: https://www.apsnet.org/publications/plantdisease/backissues/Documents/1995Abstracts/PD_79_1132.htm (accessed May 15, 2021).
Stefani, E. (2010). Economic Significance and Control of Bacterial Spot/Canker of Stone Fruits by Xanthomonas Arboricola pv. Pruni. 92. Available online at: http://www.sipav.org/main/jpp/index.php/jpp/article/view/2511 (accessed May 15, 2021).
Strayer, A. L., Jeyaprakash, A., Minsavage, G. V., Timilsina, S., Vallad, G. E., Jones, J. B., et al. (2016). A multiplex real-time PCR assay differentiates four Xanthomonas species associated with bacterial spot of tomato. Plant Dis. 100, 1660–1668. doi: 10.1094/PDIS-09-15-1085-RE
Sundin, G. W., and Bender, C. L. (1995). Expression of the strA-strB streptomycin resistance genes in Pseudomonas syringae and Xanthomonas campestris and characterization of IS6100 in X. campestris. Appl. Environ. Microbiol. 61, 2891–2897. doi: 10.1128/AEM.61.8.2891-2897.1995
Sundin, G. W., and Wang, N. (2018). Antibiotic Resistance in Plant-Pathogenic Bacteria. Annu. Rev. Phytopathol. 56, 161–180. doi: 10.1146/annurev-phyto-080417-045946
Tonkin-Hill, G., MacAlasdair, N., Ruis, C., Weimann, A., Horesh, G., Lees, J. A., et al. (2020). Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 21:180. doi: 10.1186/s13059-020-02090-4
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3–new capabilities and interfaces. Nucleic Acids Res. 40:e115. doi: 10.1093/nar/gks596
van Zyl, L. J., Deane, S. M., and Rawlings, D. E. (2003). Analysis of the mobilization region of the broad-host-range IncQ-like plasmid pTC-F14 and its ability to interact with a related plasmid, pTF-FC2. J. Bacteriol. 185, 6104–6111. doi: 10.1128/jb.185.20.6104-6111.2003
Vidaver, A. K. (2002). Uses of antimicrobials in plant agriculture. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 34(Suppl. 3), S107–S110. doi: 10.1086/340247
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Wallden, K., Rivera-Calzada, A., and Waksman, G. (2010). Type IV secretion systems: versatility and diversity in function. Cell. Microbiol. 12, 1203–1212. doi: 10.1111/j.1462-5822.2010.01499.x
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Yamaguchi, A., Udagawa, T., and Sawai, T. (1990). Transport of divalent cations with tetracycline as mediated by the transposon Tn10-encoded tetracycline resistance protein. J. Biol. Chem. 265, 4809–4813. doi: 10.1016/S0021-9258(19)34044-X
Yang, N., Reighard, G., Ritchie, D., Okie, W., and Gasic, K. (2013). Mapping quantitative trait loci associated with resistance to bacterial spot (Xanthomonas arboricola pv. pruni) in peach. Tree Genet. Genomes 9, 573–586. doi: 10.1007/s11295-012-0580-x
Zhang, Q.-Q., Tian, G.-M., and Jin, R.-C. (2018). The occurrence, maintenance, and proliferation of antibiotic resistance genes (ARGs) in the environment: influencing factors, mechanisms, and elimination strategies. Appl. Microbiol. Biotechnol. 102, 8261–8274. doi: 10.1007/s00253-018-9235-7
Keywords: antibiotic resistance, plant disease, bacterial pathogen, stone fruits, horizontal gene transfer, transposon, plasmid
Citation: Herbert A, Hancock CN, Cox B, Schnabel G, Moreno D, Carvalho R, Jones J, Paret M, Geng X and Wang H (2022) Oxytetracycline and Streptomycin Resistance Genes in Xanthomonas arboricola pv. pruni, the Causal Agent of Bacterial Spot in Peach. Front. Microbiol. 13:821808. doi: 10.3389/fmicb.2022.821808
Received: 24 November 2021; Accepted: 19 January 2022;
Published: 25 February 2022.
Edited by:
Mohammad Arif, University of Hawai’i at Mānoa, United StatesReviewed by:
Xuefeng Wang, Citrus Research Institute (CAAS), ChinaCopyright © 2022 Herbert, Hancock, Cox, Schnabel, Moreno, Carvalho, Jones, Paret, Geng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hehe Wang, aGVoZXdAY2xlbXNvbi5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.