
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 16 February 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.820048
This article is part of the Research TopicMetal Resistance in MicroorganismsView all 12 articles
Recently, green silver nanoparticles (G-AgNPs) have gained much attention in medical science due to their extraordinary effects against multidrug-resistant microorganisms. The strong antimicrobial nature of G-AgNPs corresponds to their unique physicochemical properties such as size, shape, surface charge, and active surface groups available to interact with the pathogens. The current study demonstrates a simple, environmentally friendly, and economical method to produce G-AgNPs from an environmental isolate of Viridibacillus sp. The produced G-AgNPs were characterized by various analytical methods, including UV-Vis spectroscopy, single-particle inductively coupled plasma-mass spectrometry (sp-ICP-MS), scanning electron microscopy (SEM), energy dispersive x-ray spectroscopy (EDX), elemental mapping, transmission electron microscopy (TEM), dynamic light scattering (DLS), Fourier-transform infrared spectroscopy (FTIR), and Thermogravimetric analysis (TGA). The reduction of Ag+ to Ag° was observed by UV-Vis spectroscopy, which demonstrated the formation of stable G-AgNPs with a Surface Plasmon Resonance (SPR) band at the maximum of 430 nm. TEM analysis demonstrated that the G-AgNPs were spherical with a 5–30 nm size range. The produced G-AgNPs were stable for more than 1 year in an aqueous solution at 4°C. Importantly, G-AgNPs showed remarkable antimicrobial activity against Gram-negative pathogens- E. coli and P. aeruginosa with MIC values of 0.1 and 4 μg/mL and MBC values of 1 and 8 μg/mL, respectively. This level of antimicrobial activity is superior to other AgNPs reported in the literature.
Silver nanoparticles (AgNPs) are widely known for their industrial applications in the field of medicine, pharmacology, food, agriculture, cosmetics, and textiles due to their unique antimicrobial properties, which further depend on nanoparticles (NPs) structure (Gherasim et al., 2020). The most common methods applied for AgNPs production are physiochemical methods such as laser irradiation, thermal decomposition, electrochemical synthesis, chemical reduction, etc. (Zhang et al., 2018). However, these methodologies also bring many limitations, for instance, the use of toxic materials and volatiles organic solvents, demand for high energy consumption by using high temperature and pressure, the release of harmful byproducts, and toxic waste, which causes potential environmental damage (Garg et al., 2020). The most important limitation is the absorbance of unwanted toxic materials on the surface of produced nanoparticles, which further provide human and environmental toxicity, thus limiting the clinical use of NPs. These limitations motivate the development of green alternative methodologies, which leads to the formation of uniform and stable NPs with a biocompatible layer (called the corona) around them (Singh et al., 2016a; Heinemann et al., 2021). These green nanoparticles have a range of unlimited pharmaceutical applications, including drugs delivery, gene delivery, as a sensor for pathogens detection, and tissue engineering. Various green approaches to produce nanoparticles by using living entities have been reported, such as plant extracts, fungi, yeast, actinomycetes, algae, bacteria, and viruses (Zhang et al., 2020). One such popular approach is using bacteria as a cell factory to produce the AgNPs extracellularly. In addition, to producing biocompatible NPs, bacteria-mediated synthesis is low-cost, environmentally friendly, safe, and simple (Xu et al., 2020; Ssekatawa et al., 2021).
Developing resistance mechanisms in pathogenic microorganisms against current and developing drugs has become a prime concern in the medical field. Understanding the developing resistance mechanisms in these pathogens is important, and designing novel and strong antimicrobial agents that can overcome or circumvent the resistance is equally important (Liu et al., 2021). Indeed, with exposure to novel antimicrobial agents, there are always opportunities for microbes to become unresponsive or resistant. Pathogenic bacteria exert resistance by four different mechanisms: (a) by modification of target proteins, (b) enzymatic degradation or inactivation of drug, (c) decreased membrane permeability which blocks drugs intake, and (d) increased efflux of the drug (Kumar et al., 2021). In this context, G-AgNPs display a broad spectrum of antimicrobial activities and are therefore likely to escape the common mechanisms of resistance development (Prasher et al., 2018). G-AgNPs have been reported as effective treatments against many drug-resistant microorganisms, individually or with traditional/modern antibiotics (Burdusel et al., 2018; Deshmukh S.P. et al., 2019). AgNPs exert killing against multidrug-resistance bacteria by various mechanisms, including membrane damage/leakage, DNA damage, ROS generation, inactivation of intracellular proteins/enzymes, etc. (Figure 1). Recently Ssekatawa et al. (2021) showed the green synthesis of AgNPs from the extract of medicinal plants–Camellia sinensis and Prunus Africana. The current study deals with the extracellular synthesis of G-AgNPs from an environmental isolate without any additives for the reduction or stabilization process. The extracellular constituents of cells act as reducing and stabilizing agents. In addition to extensive characterization, we explored the synthesized G-AgNPs against E. coli, and P. aeruginosa, to study their antibacterial property.
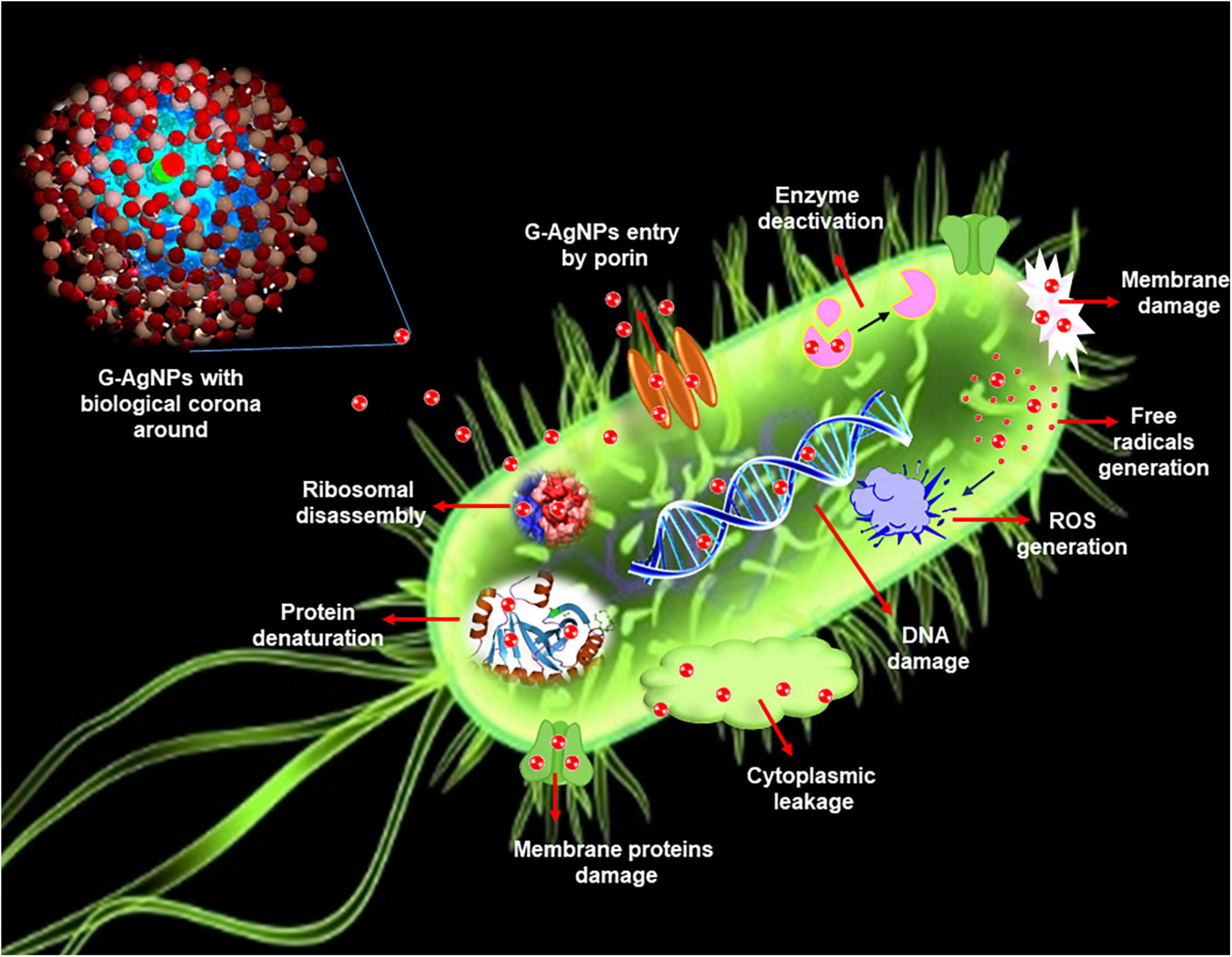
Figure 1. Schematic representation of silver nanoparticles (AgNPs) antimicrobial mechanisms in Escherichia coli.
Silver nitrate (AgNO3), tryptic soya agar (TSA), tryptic soya broth (TSB), and Luria broth (LB) were purchased from Sigma-Aldrich Chemicals, St. Louis, MO, United States.
A soil sample was collected in sterile poly bags from the Technical University of Denmark (DTU) field, Lyngby, Denmark. Single colonies were isolated by using the serial dilution technique on TSA plates. All the isolates were tested for primary AgNPs production, and the strongest strain was chosen for further studies. Molecular identification of the isolated potential strain was performed using 16S rDNA amplification and sequencing. Genomic DNA was isolated using the DNeasy Blood and Tissue Kit (Qiagen) and used as a template for PCR with the universal primers 27 F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492 R (3′-TACGGYTACCTTGTTACGACTT-5′) (Singh et al., 2017). Eurofins Genomics (Ebensburg, Germany) sequenced the PCR product, and the sequence was analyzed using the NCBI BLAST homepage against the reference sequence database.
The isolated strain was cultured overnight in 100 mL of TSB, at 37°C, 120 rpm. Next, the growth medium was centrifuged to separate the cells at 8,000 rpm for 10 min. The cell-free supernatant was supplemented with 1 mM AgNO3 and incubated in a shake flask incubator at 37°C, 200 rpm, and 24–48 h. The silver salt mix supernatant (reaction medium) was monitored continuously for AgNPs production, by visual inspection, and by recording the UV-Vis spectra of the reaction medium at definite time intervals. Once the G-AgNPs were formed, for purification, the reaction medium was centrifuged at 3,000 for 5 min to remove any big and unwanted components. Then the same medium was centrifuged at 14,000 rpm for 15 min (Singh et al., 2016b). The supernatant was decanted off to collect the pellets then washed several times with distilled water. This residue was suspended again into sterile water and used for all experiments.
The reduction of silver ions (Ag +) to G-AgNPs was initially monitored via visible inspection and then by scanning the reaction medium in UV-Vis spectroscopy at a specific interval. The UV-Vis spectrum was obtained using 6705 UV-Vis spectrophotometer, JENWAY, by scanning 1 mL of the reaction medium in the range of 300–700 nm. The optimization studies for G-AgNPs production were also conducted using visible and UV-Vis spectrum analysis.
To know the concentration of produced G-AgNPs, sp-ICP-MS (NexION 350D; PerkinElmer Inc., Waltham, MA, United States) was performed. The stability of G-AgNPs was examined by using the purified G-AgNPs and keeping them for a different time, temperatures, and indifferent bacteriological media such as TSB and LB. The results were observed by visible inspection of which pictures are taken, UV-Vis, and sp-ICP-MS analysis before and after the defined period (Singh et al., 2021).
Thermogravimetric analysis (TA Instruments, New Castle, DE, United States) was performed to check the temperature stability G-AgNPs. For analysis, G-AgNPs samples in dried and powdered form were placed in an alumina pan and heated from 20 to 700°C at a ramping time of 10°C/min.
Scanning electron microscopy examination with energy dispersive X-ray (EDX) and elemental mapping was performed to study the G-AgNPs morphology and elemental composition. EDX analysis setup was coupled with the SEM instrument. Sample preparation was done by dropping 5 μl of pure G-AgNPs (0.1 mg/mL) on carbon tape and air-dried at room temperature (RT) for 15 min. SEM Micrographs were recorded using a Quanta FEG 200 ESEM microscope (Quorum Technologies, Hitachi High-Tech Europe GmbH, Sweden).
Transmission electron microscopy study using FEI Tecnai T20 G2 was conducted to analyze the internal morphology, composition, and crystallographic information of G-AgNPs. The instrument was operated at an acceleration voltage of 200 kV. A sample of G-AgNPs was prepared by spotting a drop of pure NPs solution suspended in water on a carbon-coated copper grid. The sample-containing grid was completely dried before analysis.
Atomic force microscopy (Park NX20)1 measurements were carried out in intermittent contact mode using standard probes of single-crystal highly doped silicon with a radius of curvature of less than 30 nm (SuperSharpSiliconTM Non-contact AFM probes from Nanosensors). The standard uncertainty u(d) of the measured diameters is u(d) < 0.05 day (Singh et al., 2018c).
Dynamic light sattering measurements were performed to study the size distribution concerning intensity and zeta potential of pure G-AgNPs. Particle size measurement was executed using Zetasizer Nano ZS, Chuo-ku Kobe-shi, Japan. The autocorrelation functions of the samples were analyzed using the Contin algorithm through the Zetasizer 7.12 software. Samples were run in triplicates (Wypij et al., 2021).
Green silver nanoparticles were subjected to Fourier transform infrared (FTIR) analysis to determine the presence of biomolecules, functional groups responsible for the reduction and capping/stabilization. The FTIR measurements were carried out using Nicolet iS50 (Thermo Fisher Scientific, Waltham, MA, United States) by scanning the air-dried purified G-AgNPs and freeze-dried cell’s supernatant within the range of 500–4,000 cm–1. The recorded spectra recorded were plotted as transmittance (%) vs. wavenumber (cm–1).
The antimicrobial activity of G-AgNPs was evaluated against two Gram-negative pathogens: Escherichia coli UTI 89, and Pseudomonas aeruginosa PAO1. Both the strains were grown overnight in LB medium at 37°C for 24 h. The overnight grown cultures were diluted to approximately 1–2 × 105 colony-forming units (CFU)/mL using LB medium. Then, the G-AgNPs were added in concentrations ranging from 0.1 to 16 μg/mL. The LB medium containing respective pathogenic bacteria and G-AgNPs were further incubated in a shake flask incubator at 37°C, 150 rpm, for 24 h. After 24 h, the samples were analyzed by measuring the optical density (OD) at 550 nm. The MIC was defined as the lowest concentration of G-AgNPs, which inhibited the bacterial growth, measured as OD550. The MBC value was defined as the lowest concentration of G-AgNPs required to kill the respective bacterial strain. To measure MBC, 100 μL of the LB medium containing respective pathogenic bacteria and G-AgNPs were spread on agar plates and incubated at 37°C overnight, followed by a CFU count.
To visualize the viable and dead cells, control cells and cells treated with G-AgNPs were stained for 20 min with a mixture of 6.0 μM SYTO 9 and 30 μM KI from Live/Dead BacLight Viability kit L13152 (Invitrogen, Molecular Probes, Inc., Eugene, OR, United States). Fluorescence microscopic imaging of the cells was performed using a LEICA DM 4000 B (Leica Microsystems, Copenhagen, Denmark).
To evaluate the drastic effects of G-AgNPs on individual cells, SEM was carried out. SEM was performed by fixing the control and treated cells with 3% of glutaraldehyde overnight at 4°C. The next day, the samples were dehydrated with graded series of ethanol concentrations (40, 50, 60, 70, 80, and 90%) for 15 min and with absolute ethanol for 20 min. The dehydrated samples were placed on SEM carbon tape and left to dry at RT. The samples were coated with gold before SEM imaging. EDX and elemental mapping of G-AgNPs treated cells was also performed to check that the killing effects are due to the action of G-AgNPs only.
The rRNA sequencing of isolated bacterial strain indicated 99.05% identity with Viridibacillus arvi strain LMG 22165. The isolated strain sequence number is submitted to NCBI with GenBank ID: SUB10641455. Viridibacillus arvi is reported to be Gram-positive, aerobic, spore-forming, rod-shaped bacteria (Albert et al., 2007). Based on 16rRNA sequence similarity, the isolated strain was referred to as Viridibacillus sp.
The supernatant of a culture of Viridibacillius sp. grown for 24 h was used as a reaction medium to reduce silver salt and provide capping/stabilizing components to the formed G-AgNPs. The culture supernatant was supplemented with 1 mM AgNO3 and incubated further in a shake flask incubator at 37°C, 200 rpm, for 24–48 h. After the incubation period, the supernatant showed a visible color change from pale yellow to deep brown, which is attributed to the surface plasmon resonance (SPR) property of G-AgNPs (Ronavari et al., 2021). The observation was further confirmed by simultaneously recording the SPR band via UV-Vis at a specified time interval. After incubation, the reaction medium was first directly scanned, and then the purified G-AgNPs samples were also scanned to confirm the peaks intensity and overlapping range (Figures 2A,B). Purified G-AgNPs showed a strong and sharp SPR peak at 430 nm. The kinetics of G-AgNPs formation was monitored by recording the spectrum of the reaction medium at different temperatures, times, and various salt concentrations. For temperature optimization studies, the highest and clear peak in UV-Vis spectra was observed at 37°C (Figure 2C). According to the time-resolved UV-Vis spectra, the SPR absorbance band increased with the reaction time for up to 48 h (Figure 2D). For the salt concentration optimization, as the salt concentration increased, the intensity of the SPR band also increased. This trend continued to up to 3.5 mM (Figures 2E–H); any further increase in salt concentration led to broadening the peak and accumulation of NPs in the reaction medium. Thus the optimal conditions for G-AgNPs production using cell-free supernatant of Viridibacillus sp., were AgNO3 concentration of 3.5 mM, the temperature of 37°C, and the incubation period of 48 h. Any deviations from these key parameters resulted in disturbance of the UV-Vis peaks, signifying particle agglomeration.

Figure 2. Visible and UV-Vis spectral examination of optimization studies for green silver nanoparticles (G-AgNPs) production. (A) UV-Vis peak of unpurified G-AgNPs after 48 h of synthesis, (B), purified G-AgNPs. (C) Visible and UV-Vis peaks for temperature optimization, (D) time optimization, (E–H) salt optimization for G-AgNPs production at 24 and 48 h.
To check the yield of G-AgNPs, sp-ICP-MS was used. The result showed that the measured total mass concentration of G-AgNPs was 0.078 μg/μl, with a negligible dissolved fraction (<0.1 ppb). We then examined the stability of G-AgNPs over short and long periods. The measurements were made after 24, 48, 86 h, and 1 year (Figures 3A–D). Our study found no significant differences in particle size recorded using sp-ICP-MS, resulting in mean diameter of 15–60 nm, thus indicating the long-term stability of produced G-AgNPs. Based on visible observation, no dissolution or agglomeration of G-AgNPs occurred even after 1 year of storage in an aqueous solution. In addition, UV-Vis observations of G-AgNPs showed a sharp and overlapping peak before and after 1 year, thus confirming their aqueous stability (Figure 3E). A bacterial growth medium, such as TSB and LB, and water were tested for G-AgNPs stability (Figure 3F). The results showed that the G-AgNPs are completely stable in water as well as in the growth media. For thermal stability, the TGA measurement was taken (Figure 3G). According to the obtained spectra, there were two stages of weight loss, first at 150°C and second at 400°C. The physical adsorption of water molecules caused the initial loss of weight up to 150°C on the surfaces of the G-AgNPs. At 400°C, most of the weight loss was due to the biomolecules decomposing and evaporating from the surface of the G-AgNPs (Deshmukh A.R. et al., 2019). A further increase in temperature led to the complete degradation of G-AgNPs.
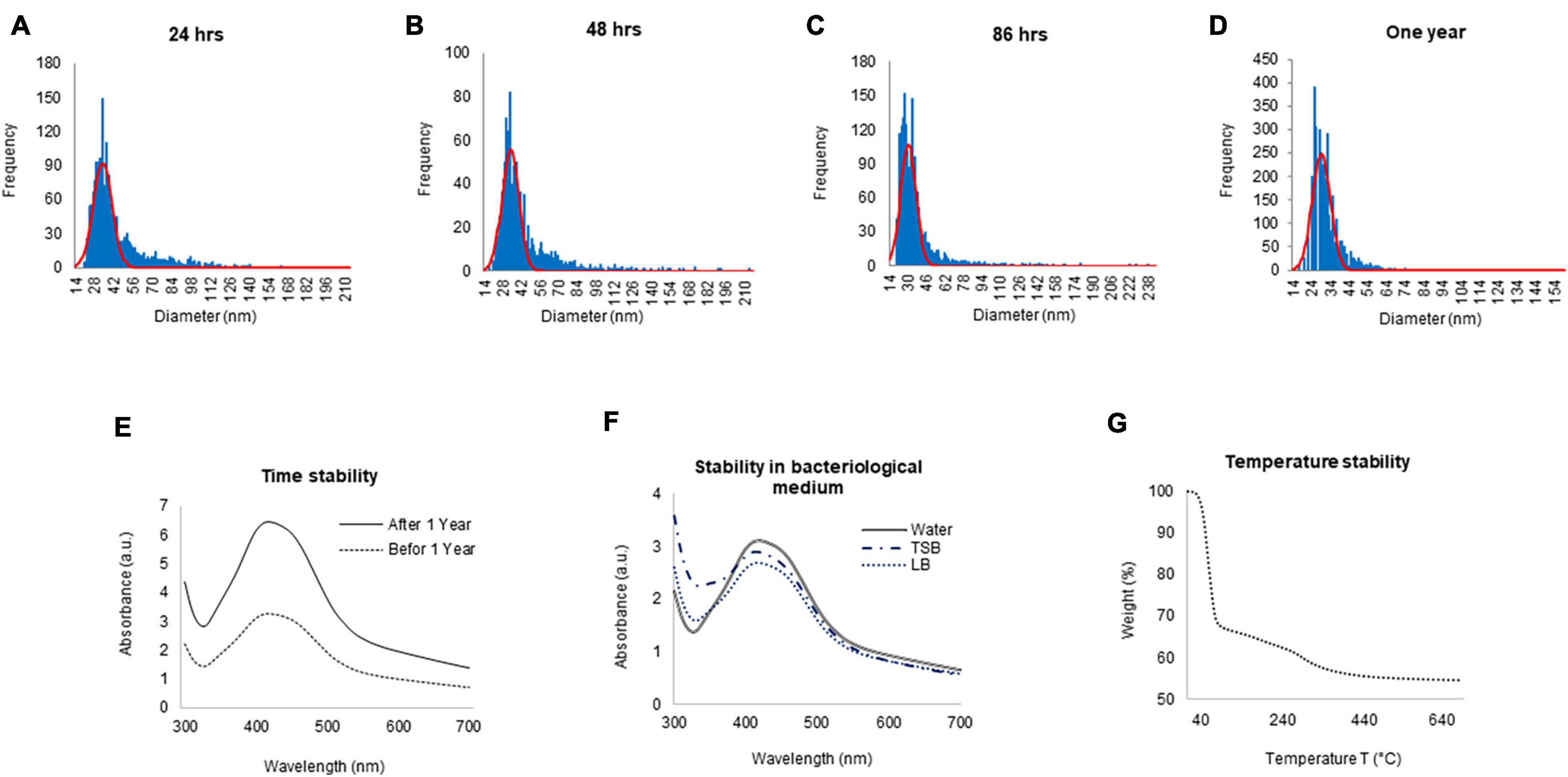
Figure 3. ICPMS and stability analysis of green silver nanoparticles (G-AgNPs). ICPMS histogram of G-AgNPs at different time intervals (A) 24 h, (B) 48 h, (C) 86 h, (D) 1 year. UV-Vis spectrum representing the stability analysis of G-AgNPs (E) before and after 1 year, (F) in a different medium, (G) at the temperature range from 20 to 700°C measured by the TGA instrument.
The purity and crystalline nature of G-AgNPs that were produced under optimized conditions were investigated by conducting SEM, EDX, elemental mapping, TEM, and Selected area (electron) diffraction (SAED) studies. SEM image examination showed the spherical structure of G-AgNPs (Figures 4A,B). The elemental mapping results demonstrated the selected scanned area of the G-AgNPs sample (Figures 4C–E) resembles the silver element (pink color) (Deshmukh A.R. et al., 2019). The elemental composition of G-AgNPs was determined via EDX spectroscopy, which reveals the presence of the strong elemental signal from silver at 3 keV (Figure 4F; Singh et al., 2018a). The core size and morphology of G-AgNPs were determined via TEM, which displayed the G-AgNPs are approximately spherical and uniformly distributed with an average particles size of 5–30 nm (Figure 4G). However, few polydispersity were found in hexagonal and truncated triangular form NPs. This morphology of nanoparticles depends on the experimental conditions and the constituents of the cellular supernatant. The SAED pattern of G-AgNPs indicated characteristic rings at 111, 200, 220, and 311 crystallographic planes, which corresponds to the face-centered cubic (fcc) of AgNPs (Figure 4H). These values are in accord with those reported in earlier studies (Sangaonkar and Pawar, 2018); they suggest the crystalline nature of AgNPs. Moreover, the AFM analysis also revealed similar size distribution, from 5 to 35 nm (Figure 4I). Hydrodynamic diameter and surface charge of the produced G-AgNPs were determined by DLS. The diameter and PDI were found to be 154.4 nm and 0.378, respectively (Figure 5A). The zeta potential value of the aqueous G-AgNPs solution at RT was found to be −25.7 mV (Figure 5B), which suggested that the G-AgNPs are negatively charged and quite stable at neutral conditions.
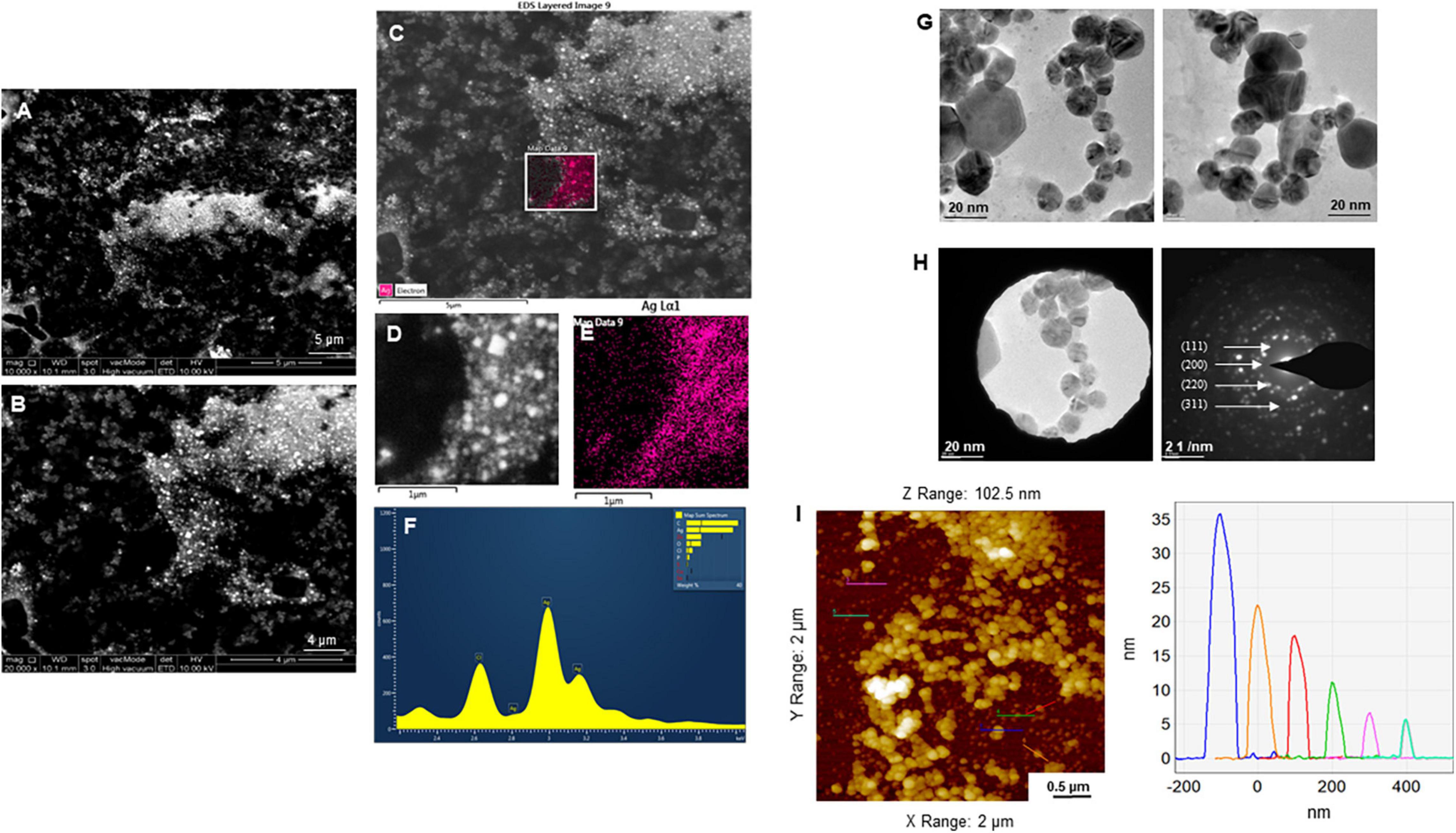
Figure 4. Structural analysis of green silver nanoparticles (G-AgNPs). (A,B) Scanning electron microscopy (SEM) images of nanoparticles, (C–E) elemental mapping of G-AgNPs showing scanned image of NPs and respective region with the individual silver nanoparticles (pink color), (F) energy dispersive x-ray spectroscopy (EDX) spectrum of the elemental mapped region is showing highest peak for silver element. (G) Transmission electron microscopy (TEM) image of G-AgNPs showing spherical nanoparticles, (H) selected area (electron) diffraction (SAED) pattern of G-AgNPs, and (I) AFM size analysis representation of G-AgNPs.
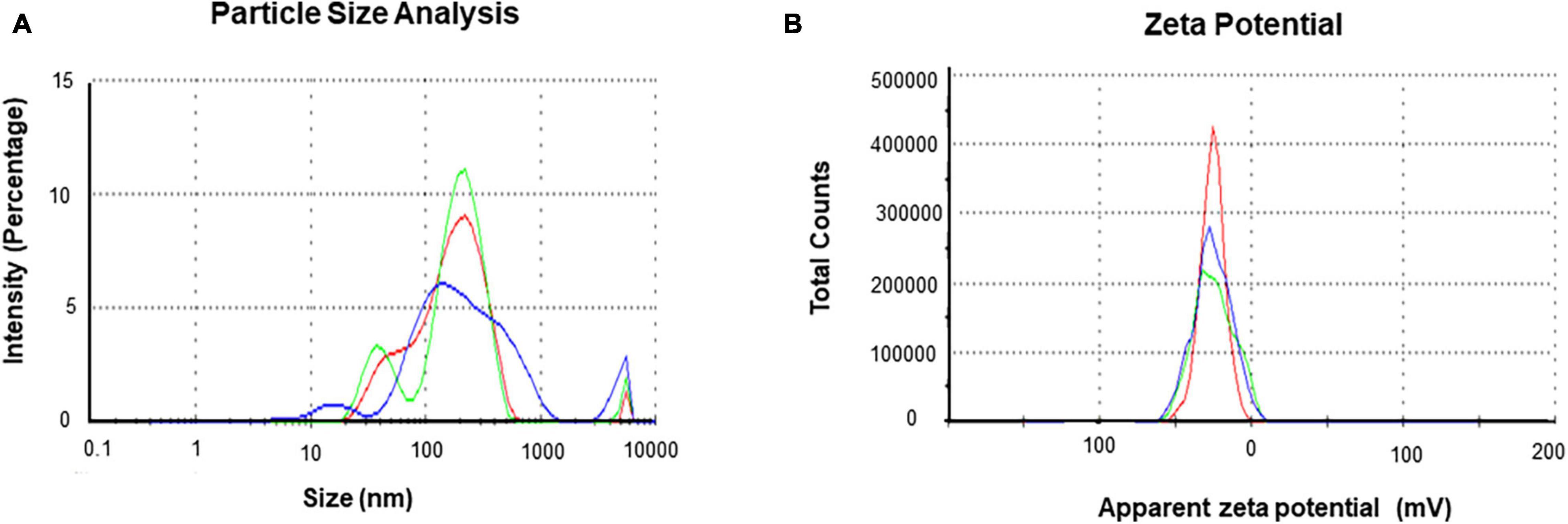
Figure 5. Dynamic light scattering (DLS) analysis of green silver nanoparticles (G-AgNPs) (A) nanoparticles distribution concerning size and intensity and (B) zeta potential of G-AgNPs representing highly negative surface charge.
Fourier-transform infrared spectroscopy measurements were conducted to identify the extracellular components released from isolated strain, present in the reaction medium, responsible for reducing, capping, and stabilizing G-AgNPs. The FTIR spectra of freeze-dried cell-free supernatant and purified G-AgNPs are shown in Figures 6A,B and Table 1. Comparing the FTIR spectrum of cellular supernatant with G-AgNPs, the high broad peaks for G-AgNPs appear at 2884.93 (asymmetric and symmetric C-H stretching, or secondary amines), 1635.21 [carboxyl groups (-C=O), and carbonyl group (-C=O)-stretching vibration of proteins], 1430.79 (C-H bending of COO– or carboxylate groups), 561.08 which are identical to the supernatant spectrum. The FTIR results indicate the presence of carboxyl groups (-C=O), and amine groups (−NH) which represents the presence of proteins, amino acids, and other biomolecules originating from the supernatant on the surface of the produced G-AgNPs, responsible for capping and stabilizing G-AgNPs (Abbai et al., 2016). The FTIR spectrum proved that the reaction medium contains reducing and stabilizing agents such as sugar, proteins, and amino acids responsible for the green synthesis of G-AgNPs.
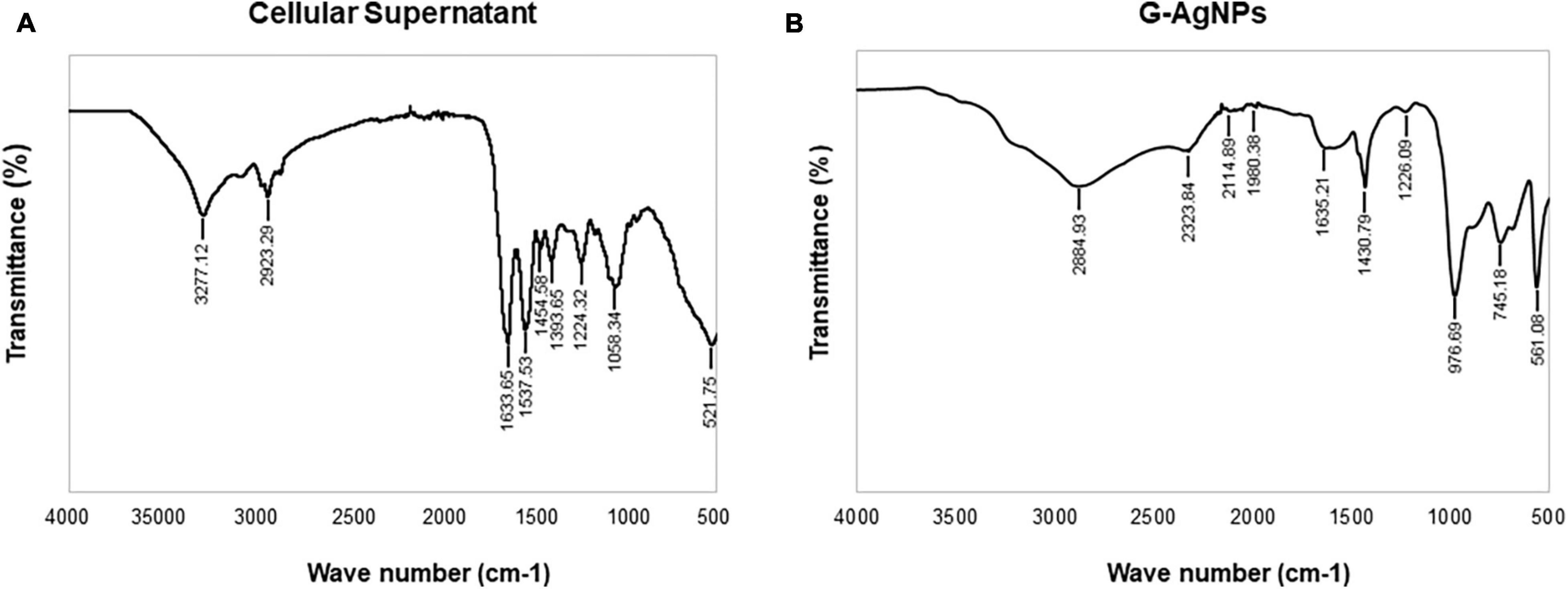
Figure 6. Fourier-transform infrared spectroscopy (FTIR) spectrum of panel (A) freeze-dried cellular supernatant and (B) green silver nanoparticles (G-AgNPs), which demonstrate the active surface groups for respective samples.
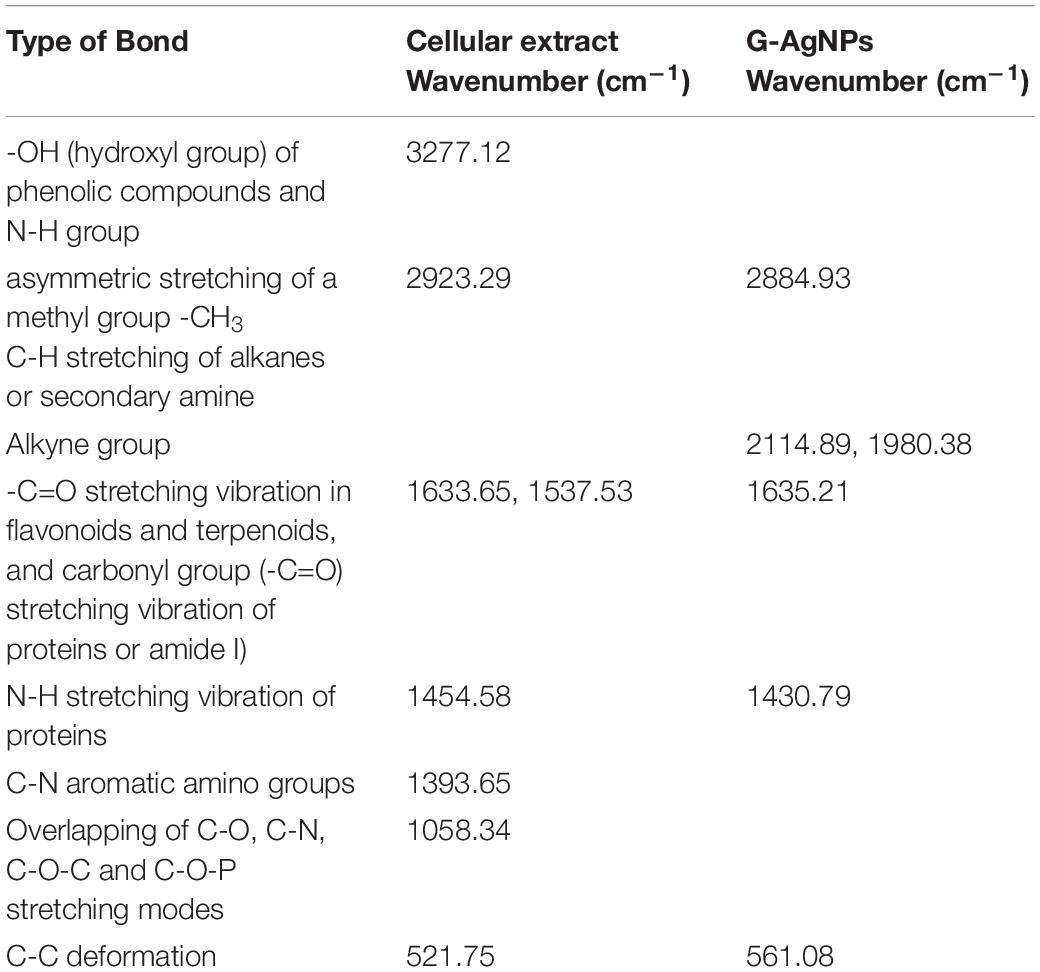
Table 1. Fourier transform-infrared spectroscopy (FT-IR) spectra of cellular extract and green silver nanoparticles (G-AgNPs).
The most remarkable feature of the G-AgNPs synthesized in this study is their strong bacteriostatic and bactericidal activity against pathogenic E. coli and P. aeruginosa. The recorded MIC values against E. coli and P. aeruginosa were 4 and 0.1 μg/mL, respectively, while the respective MBC values were 8 and 1 μg/mL (Figures 7A,B). We used the live and dead staining technique to confirm the viability results. This technique allows one to distinguish the live cells (stained green) and dead cells (stained red) under a fluorescence microscope (Figures 8A–N). These results confirmed a dramatic onset of killing bacterial cells at G-AgNPs concentrations above 4 μg/mL for E. coli (Figure 8A–H) and 0.1 μg/mL for P. aeruginosa (Figures 8I–N). To investigate whether the killing effects involve drastic morphological changes in treated cells, we used SEM. A significant morphological alteration was observed in G-AgNPs treated cells E. coli cells (Figures 9A–F,K–P) and P. aeruginosa cells (Figures 10A–F,K–P). The severity of these effects was correlated to the concentration of applied G-AgNPs for both bacterial species. To confirm the damage that G-AgNPs provoked, we performed EDX and elemental mapping of individual cells. The results disclosed that the damaged cells generate a clear peak of the silver element in the EDX spectrum for E. coli (Figures 9G–J,Q–T) and P. aeruginosa cells (Figures 10G–J,Q–T). In addition, the mapping results also resemble the silver element in the scanned image of treated cells, indicating that G-AgNPs get internalized.
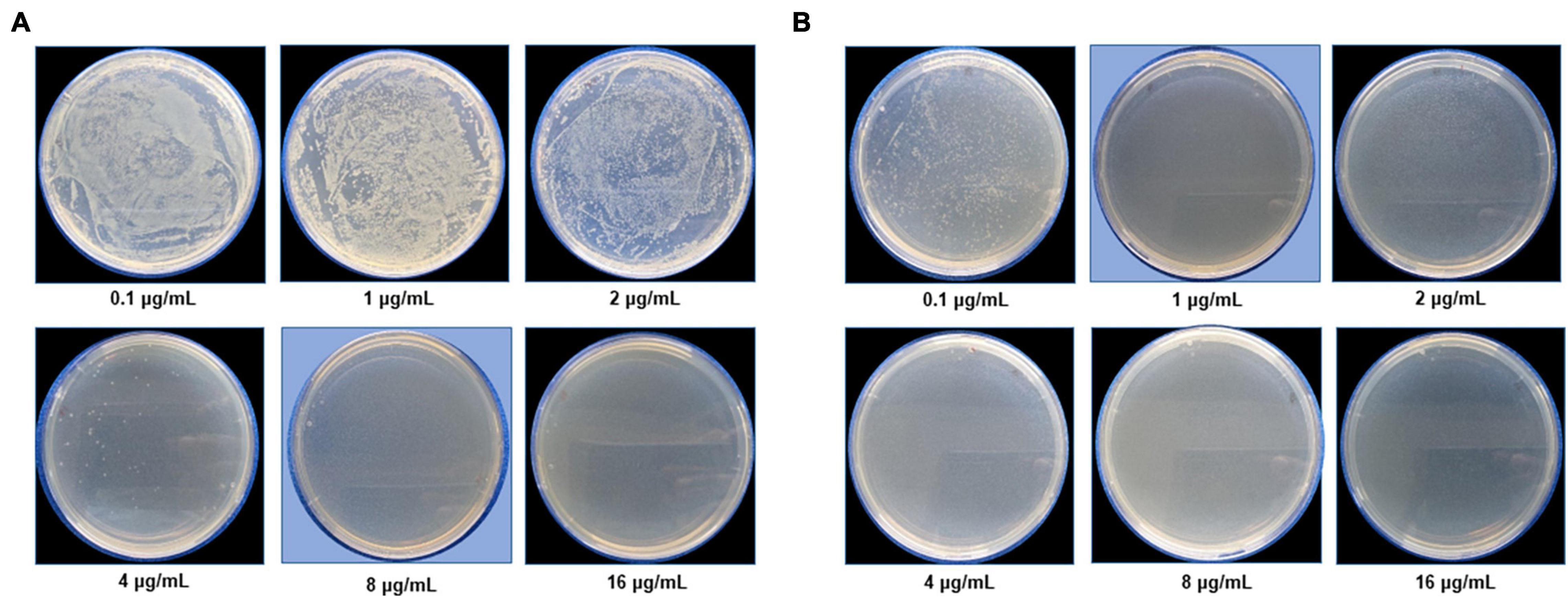
Figure 7. Cell viability test at a different concentration range from 0.1 to 16 μg/mL for (A) Escherichia coli and (B) Pseudomonas aeruginosa after green silver nanoparticles (G-AgNPs) treatment. The blue background shows the MBC values of G-AgNPs for respective pathogens.
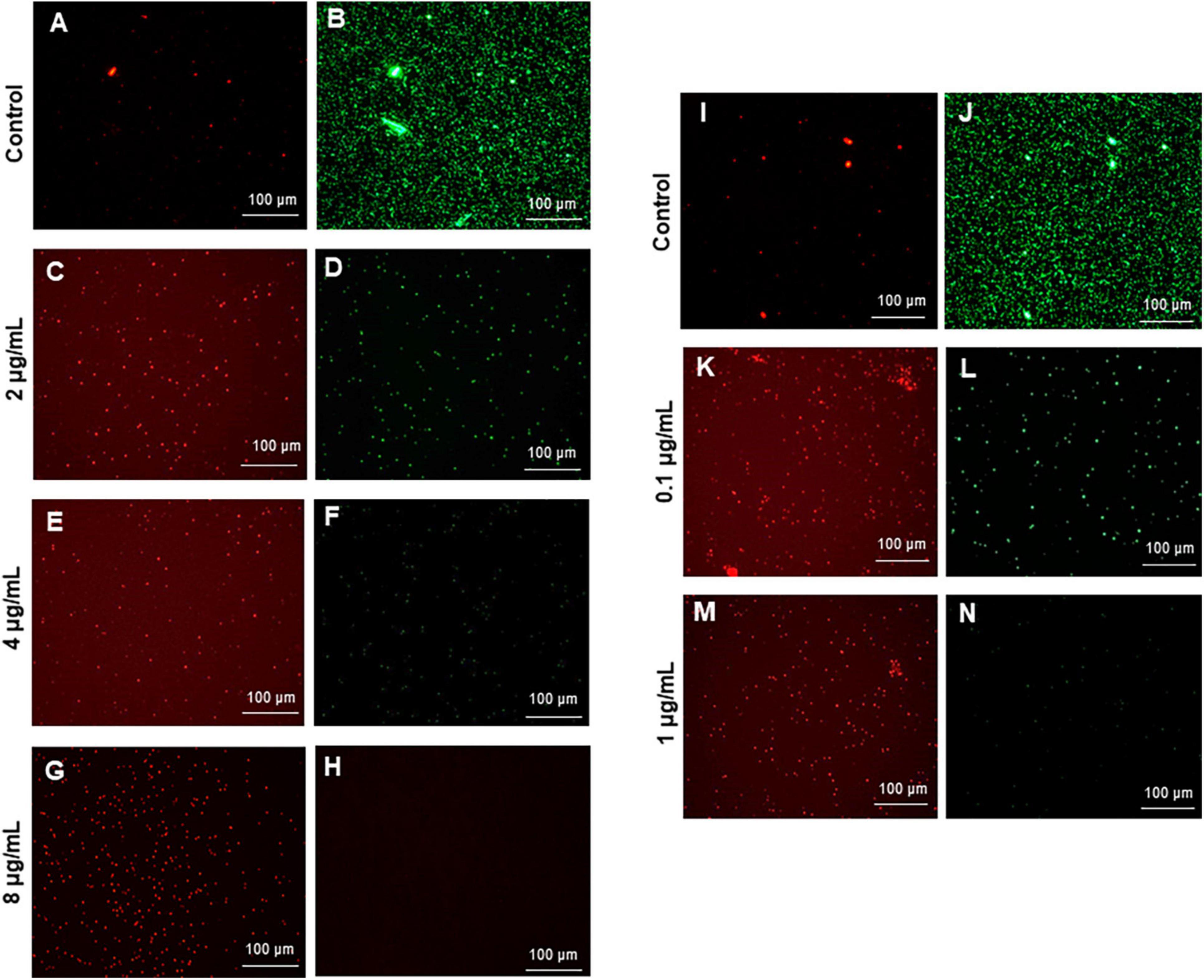
Figure 8. Live and dead staining of (A–H) Escherichia coli, and (I-N) Pseudomonas aeruginosa after treatment with green silver nanoparticles (G-AgNPs) at selected concentrations. E. coli cells: (A,B) control without G-AgNPs; (C,D) 2 μg/mL; (E,F) 4 μg/mL; (G,H) 8 μg/mL of G-AgNPs. P. aeruginosa cells: (I,J) control without G-AgNPs; (K,L) 0.1 μg/mL; (M,N) 1 μg/mL of G-AgNPs.
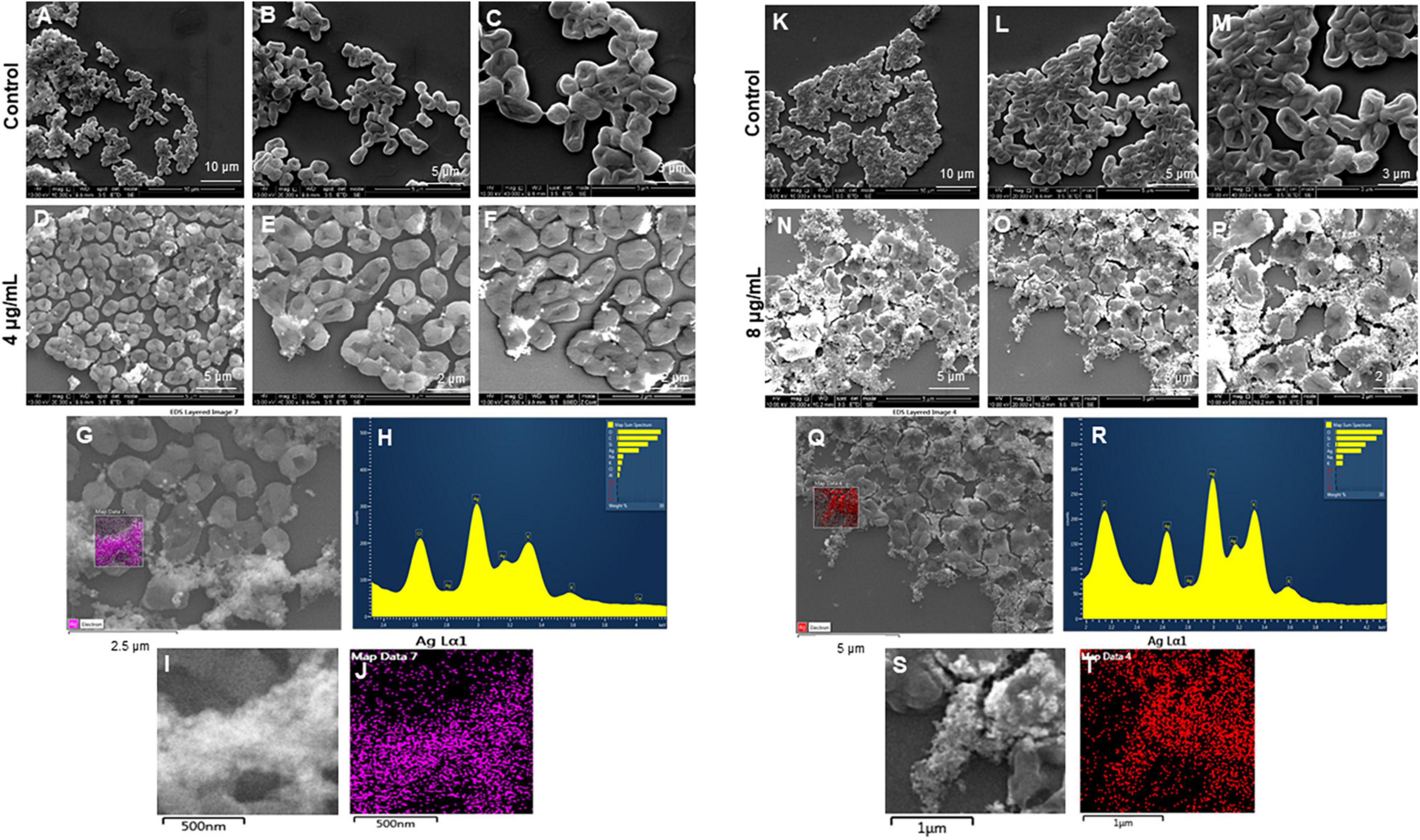
Figure 9. Scanning electron microscopy (SEM) analysis of Escherichia coli cells after treatment with green silver nanoparticles (G-AgNPs). (A–F) Control E. coli cells and G-AgNPs treated 4 μg/mL at different scales. (G) Scanned image of treated cells (H) energy dispersive x-ray spectroscopy (EDX) spectrum of choose area (I,J) elemental mapping of the selected area showing silver element in the treated cells, (K–P) control E. coli cells and G-AgNPs treated cells with 8 μg/mL at different scales. (Q) Scanned image of treated cells (R) energy dispersive x-ray spectroscopy (EDX) spectrum of choose area (S,T) elemental mapping of the selected area showing silver element in the treated cells.
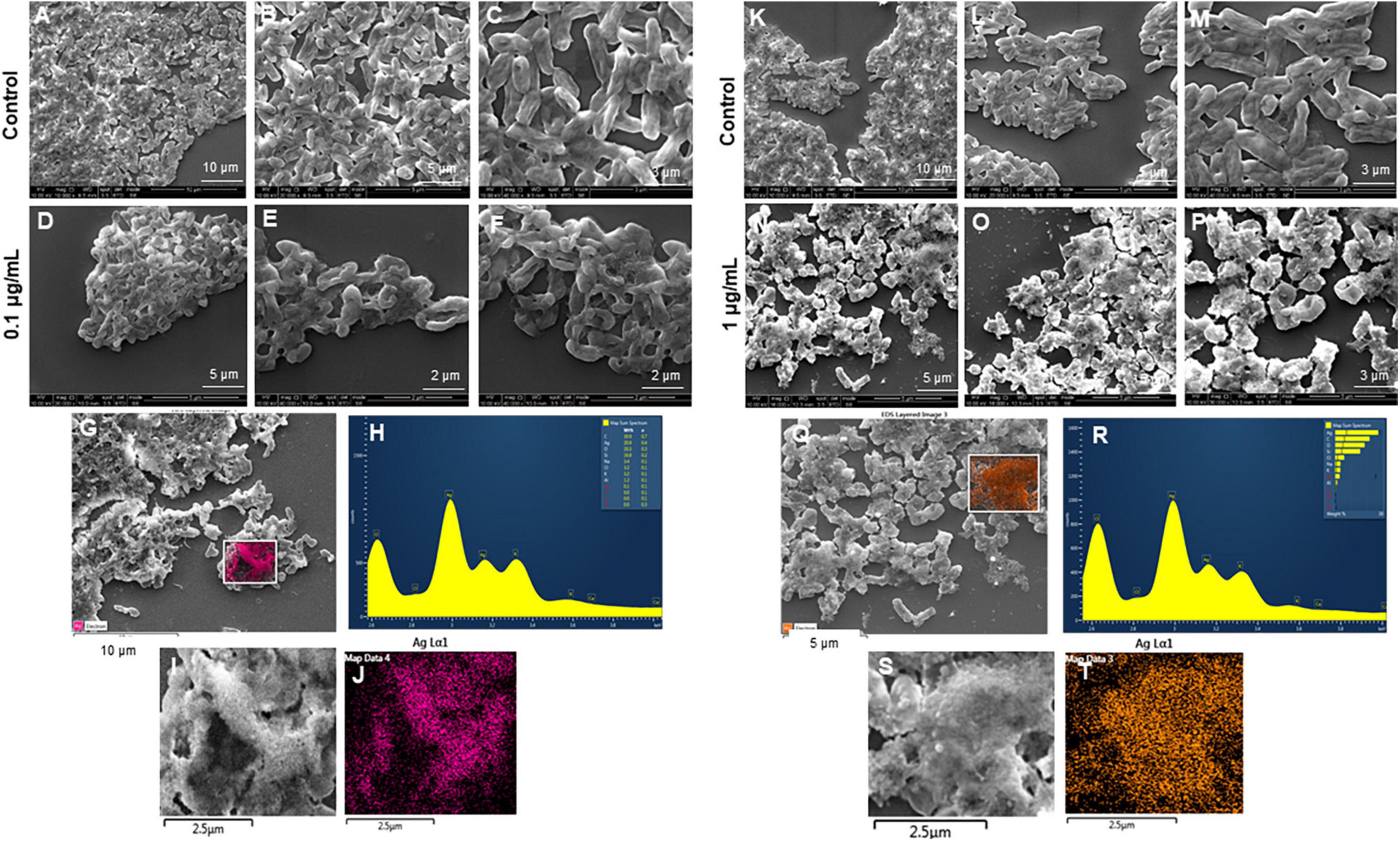
Figure 10. Scanning electron microscopy (SEM) analysis of Pseudomonas aeruginosa cells after treatment with green silver nanoparticles (G-AgNPs). (A–F) Control P. aeruginosa cells and G-AgNPs treated cells with 0.1 μg/mL at different scales. (G) Scanned image of treated cells (H) energy dispersive x-ray spectroscopy (EDX) spectrum of choose area (I,J) elemental mapping of the selected area showing silver element in the treated cells, (K–P) control P. aeruginosa cells and G-AgNPs treated cells with 1 μg/mL at different scales. (Q) Scanned image of treated cells (R) energy dispersive x-ray spectroscopy (EDX) spectrum of choose area (S,T) elemental mapping of the selected area showing silver element in the treated cells.
The cell-free supernatant of environmental isolate Viridibacillus sp. acted as a reducing and stabilizing agent, which led to the formation of highly stable and monodisperse G-AgNPs. The reduction process does not require any additional reducing or stabilizing agents. Moreover, the synthesis took place in the cell-free medium, which means with the help of extracellularly released biomolecules from the isolated Viridibacillus sp. The extracellular components in the reaction medium form a biological corona around the nanoparticles, which helps long-term stabilization. This is an important feature of bacteria-mediated extracellular synthesis. Unlike intracellular synthesis, extracellular synthesis provides an opportunity to avoid additional steps in downstream processing, such as cell disruption by membrane lysis or sonication, removal of insoluble components, extraction of complete nanoparticles from intracellular organelles, etc. (Singh et al., 2018b). Thus, the proposed methodology is more economical once an appropriate bacterial strain is identified (Kapoor et al., 2021).
The G-AgNPs formation in our study was supported by visual observation and UV-Vis analysis. Based on kinetics and optimization studies, no significant change in the absorbance was observed beyond the optimized parameters, which suggested that the nucleation and growth process during this period supported the complete reduction of silver salt into G-AgNPs. Moreover, the color, ICPMS, and UV-Vis spectrum of G-AgNPs remained stable for more than 1 year and showed no aggregation (Singh et al., 2021). FTIR was used as a powerful tool to study the functional molecular vibrations. The spectrum of freeze-dried cell-free supernatant of Viridibacillus sp. showed that the medium contains proteins, reducing sugars, polysaccharides, various biomolecules, and amino acids, which help reduce and inhibit further agglomeration of produced G-AgNPs. This is the most important advantage of using green nano factories to produce AgNPs. It provides the additional biocompatible layer, which can keep nanoparticles stable for many years without any additives, thus enhancing colloidal stability (Belteky et al., 2019). In contrast, the physically or chemically produced NPs lack the additional biocompatible layer and require a surplus stabilizer. Most of the NPs produced by physical or chemical methods show complete agglomeration with time, even in the presence of stabilizing agents (Deshmukh A.R. et al., 2019).
Silver nanoparticles effects on pathogenic bacteria are well known (Figure 1; Liao et al., 2019; Crisan et al., 2021). The effects of AgNPs on Gram-negative and Gram-positive bacteria are mainly influenced by the thickness and composition of the cell wall, which means that Gram-negative bacteria are more susceptible, and Gram-positive bacteria can show resistance to some extent. Except for the thin cell membrane, the lipopolysaccharides (LPS) in the cell membrane promote the chemical interaction of the membrane with AgNPs (Loo et al., 2018). Although many studies have reported the antimicrobial activity and possible action mechanism of biological AgNPs, strong effects at a very low concentration of AgNPs are rare. The produced G-AgNPs were explored against two Gram-negative strains in the current study. Results showed extremely strong antimicrobial activity, i.e., total killing at 8 μg/mL for E. coli and 1 μg/mL for P. aeruginosa. The mentioned concentrations for total killing are very low compared to other reported green AgNPs. For instance, recently, Shah et al. (2021) showed that the LD50 dose (concentration of AgNPs causing 50% inhibition) obtained from Plantago lanceolate against E. coli was 45.54 mg/L, which is much higher than the G-AgNPs MBC value (kill 100% bacteria) against E. coli, i.e., 8 μg/mL, in our study. Similarly, Devanesan and AlSalhi, 2021 demonstrated the antimicrobial activity of AgNPs originated from flower extract of Abelmoschus esculentus. The authors showed the MBC value of AgNPs against E. coli and P. aeruginosa were 110 and 105 μg/mL (Devanesan and AlSalhi, 2021). Loo et al. (2018) described the MBC value of extremely small 4 nm AgNPs produced using pu-erh tea leaves against E. coli as 7.8 μg/mL. Ssekatawa et al. (2021) described the MBC value of two green AgNPs originating from Prunus africana and Camellia sinensis. The MIC and MBC value of 125 and 250 μg/mL against E. coli (Ssekatawa et al., 2021). Thus, our G-AgNPs showed MBC at 8 and 1 μg/mL against E. coli and P. aeruginosa, superior to all the cited examples. The possible reason for the strong antibacterial ability of G-AgNPs could be the biological corona, which provides a high negative surface charge, spherical shape of NPs, which allow them to interact with pathogens with the maximum surface area available.
We further confirmed the cells’ death by SEM. SEM has revealed that after contact with G-AgNPs, the cell membrane of E. coli and P. aeruginosa cells is completely ruptured. Thus, the G-AgNPs attach onto the negatively charged surface of the cell wall and membrane, which leads to the shrinkage of the cytoplasm and membrane detachment, finally leading to rupture of the cell wall. In addition, the interaction of G-AgNPs with the sulfur-containing proteins present in the cell wall could also affect the membrane permeability and cause cell leakage. It is reported that the porins on Gram-negative bacteria are also responsible for AgNPs uptake. Following penetration, G-AgNPs may interact with cellular components such as proteins, lipids, and DNA, corresponding to the damaging effects (Ullah Khan et al., 2018). AgNPs also cause DNA damage, mutations, inhibition of enzymes and proteins (Singh et al., 2020). It has been found that Ag (+) ions intercalate between the purine and pyrimidine base pairs, disrupt the H-bonds between base pairs of the anti-parallel DNA strands, and thereby disrupt the double-helical structure (Joshi et al., 2020). Another well-known mechanism of AgNPs action is their ability to produce ROS and free radical species and consequent increase in oxidative stress in cells and apoptosis (Loo et al., 2018; Ullah Khan et al., 2018). All these mechanisms were presented in Figure 1.
One of the most important physicochemical properties that affect antimicrobial activity is the size and shape of NPs (Karade et al., 2021). Typically, smaller NPs have the larger surface area available to interact and ascend intracellular penetration (Ginjupalli et al., 2018). We hypothesized that the bigger G-AgNPs >10 nm could cause membrane damage. In contrast, the smaller NPs (less than 10 nm) could enter the cells after adhesion and damage the intracellular structures, thus affecting vital cellular functioning. Thus, we believe that the size range of G-AgNPs from 5 to 30 nm offered strong interaction of nanoparticles on the surface (bigger NPs) and internal organelles (smaller NPs). In addition, sphere-shaped or quasi-spherical AgNPs are more susceptible to releasing Ag + ions; thus, G-AgNPs showed high antimicrobial effects. Moreover, as mentioned above, strong negative zeta potential and biological corona also play an important role in providing strong antimicrobial activity and stability in the biological environment so that NPs won’t degrade and act with their full potential. Thus, we strongly believe that the strong antimicrobial activity of produced G-AgNPs is due to the above-discussed mechanism. Exploring these nanoparticles further against more multidrug-resistant pathogens will answer the desire for strong antimicrobial agents in medical fields.
Green silver nanoparticles were successfully formed via a green synthetic method using cell-free supernatant of Viridibacillus sp., which acted as a reducing and capping agent. The G-AgNPs production was confirmed by the appearance of SPR band 430 nm and found to be highly stable, crystalline, and nearly spherical in size. The G-AgNPs showed excellent antimicrobial activity against two Gram-negative strains at very low concentrations. We propose that these nanoparticles constitute a promising opportunity for developing antimicrobial weapons with efficient antimicrobial properties, stability, and recyclability.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
PS designed and performed the experiments, analyzed the results, and prepared the manuscript and figures. IM supervised all experimental work and edited the manuscript. Both authors contributed to the article and approved the submitted version.
Lundbeckfonden to PS (R303-2018-3499), Novo Nordisk Foundation (NNF10CC1016517) and the NordForsk (project number 105121) to IM.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ICPMS was performed at DTU Environmental; TEM was performed at Center for electron microscopy; DTU and FTIR were conducted at Department of Chemical Engineering, DTU. Jørgen Garnaes helped with AFM analysis. Funds support AFMs contribution from The Danish Agency for Institutions and Educational Grants.
Abbai, R., Mathiyalagan, R., Markus, J., Kim, Y. J., Wang, C., Singh, P., et al. (2016). Green synthesis of multifunctional silver and gold nanoparticles from the oriental herbal adaptogen: siberian ginseng. Int. J. Nanomed. 11, 3131–3143. doi: 10.2147/IJN.S108549
Albert, R. A., Archambault, J., Lempa, M., Hurst, B., Richardson, C., Gruenloh, S., et al. (2007). Proposal of Viridibacillus gen. nov. and reclassification of Bacillus arvi, Bacillus arenosi and Bacillus neidei as Viridibacillus arvi gen. nov., comb. nov., Viridibacillus arenosi comb. nov. and Viridibacillus neidei comb. nov. Int. J. Syst. Evol. Microbiol. 57, 2729–2737. doi: 10.1099/ijs.0.65256-0
Belteky, P., Ronavari, A., Igaz, N., Szerencses, B., Toth, I. Y., Pfeiffer, I., et al. (2019). Silver nanoparticles: aggregation behavior in biorelevant conditions and its impact on biological activity. Int. J. Nanomed. 14, 667–687. doi: 10.2147/IJN.S185965
Burdusel, A. C., Gherasim, O., Grumezescu, A. M., Mogoanta, L., Ficai, A., and Andronescu, E. (2018). Biomedical applications of silver nanoparticles: an up-to-date overview. Nanomaterials 8:681. doi: 10.3390/nano8090681
Crisan, C. M., Mocan, T., Manolea, M., Lasca, L. I., Tǎbǎran, F.-A., and Mocan, L. (2021). Review on silver nanoparticles as a novel class of antibacterial solutions. Appl. Sci. 11:1120. doi: 10.3390/app11031120
Deshmukh, A. R., Gupta, A., and Kim, B. S. (2019). Ultrasound assisted green synthesis of silver and iron oxide nanoparticles using fenugreek seed extract and their enhanced antibacterial and antioxidant activities. Biomed. Res. Int. 2019:1714358. doi: 10.1155/2019/1714358
Deshmukh, S. P., Patil, S. M., Mullani, S. B., and Delekar, S. D. (2019). Silver nanoparticles as an effective disinfectant: a review. Mater Sci. Eng. C Mater. Biol. Appl. 97, 954–965. doi: 10.1016/j.msec.2018.12.102
Devanesan, S., and AlSalhi, M. S. (2021). Green synthesis of silver nanoparticles using the flower extract of abelmoschus esculentus for cytotoxicity and antimicrobial studies. Int. J. Nanomed. 16, 3343–3356. doi: 10.2147/IJN.S307676
Garg, D., Sarkar, A., Chand, P., Bansal, P., Gola, D., Sharma, S., et al. (2020). Synthesis of silver nanoparticles utilizing various biological systems: mechanisms and applications—a review. Prog. Biomater. 9, 81–95. doi: 10.1007/s40204-020-00135-2
Gherasim, O., Puiu, R. A., Birca, A. C., Burdusel, A. C., and Grumezescu, A. M. (2020). An updated review on silver nanoparticles in biomedicine. Nanomaterials 10:2318. doi: 10.3390/nano10112318
Ginjupalli, K., Shaw, T., Tellapragada, C., Alla, R., Gupta, L., and Perampalli, N. U. (2018). Does the size matter? Evaluation of effect of incorporation of silver nanoparticles of varying particle size on the antimicrobial activity and properties of irreversible hydrocolloid impression material. Dent Mater 34:e158–e165. doi: 10.1016/j.dental.2018.03.016
Heinemann, M. G., Rosa, C. H., Rosa, G. R., and Dias, D. (2021). Biogenic synthesis of gold and silver nanoparticles used in environmental applications: a review. Trends Environ. Anal. Chem. 30:e00129.
Joshi, A. S., Singh, P., and Mijakovic, I. (2020). Interactions of gold and silver nanoparticles with bacterial biofilms: molecular interactions behind inhibition and resistance. Int. J. Mol. Sci. 21:7658. doi: 10.3390/ijms21207658
Kapoor, R. T., Salvadori, M. R., Rafatullah, M., Siddiqui, M. R., Khan, M. A., and Alshareef, S. A. (2021). Exploration of microbial factories for synthesis of nanoparticles - a sustainable approach for bioremediation of environmental contaminants. Front. Microbiol. 12:658294. doi: 10.3389/fmicb.2021.658294
Karade, V. C., Patil, R. B., Parit, S. B., Kim, J. H., Chougale, A. D., and Dawkar, V. V. (2021). Insights into shape-based silver nanoparticles: a weapon to cope with pathogenic attacks. ACS Sustain. Chem. Eng. 9, 12476–12507. doi: 10.1021/acssuschemeng.1c03797
Kumar, S., Basumatary, I. B., Sudhani, H. P. K., Bajpai, V. K., Chen, L., Shukla, S., et al. (2021). Plant extract mediated silver nanoparticles and their applications as antimicrobials and in sustainable food packaging: a state-of-the-art review. Trends Food Sci. Technol. 112, 651–666. doi: 10.1016/j.tifs.2021.04.031
Liao, C., Li, Y., and Tjong, S. C. (2019). Bactericidal and cytotoxic properties of silver nanoparticles. Int. J. Mol. Sci. 20:449.
Liu, X., Chen, J.-L., Yang, W.-Y., Qian, Y.-C., Pan, J.-Y., Zhu, C.-N., et al. (2021). Biosynthesis of silver nanoparticles with antimicrobial and anticancer properties using two novel yeasts. Scie. Rep. 11:15795. doi: 10.1038/s41598-021-95262-6
Loo, Y. Y., Rukayadi, Y., Nor-Khaizura, M.-A.-R., Kuan, C. H., Chieng, B. W., Nishibuchi, M., et al. (2018). In Vitro antimicrobial activity of green synthesized silver nanoparticles against selected gram-negative foodborne pathogens. Front. Microbiol. 9:1555. doi: 10.3389/fmicb.2018.01555
Prasher, P., Singh, M., and Mudila, H. (2018). Silver nanoparticles as antimicrobial therapeutics: current perspectives and future challenges. 3 Biotech 8:411. doi: 10.1007/s13205-018-1436-3
Ronavari, A., Igaz, N., Adamecz, D. I., Szerencses, B., Molnar, C., Konya, Z., et al. (2021). Green silver and gold nanoparticles: biological synthesis approaches and potentials for biomedical applications. Molecules 26:844. doi: 10.3390/molecules26040844
Sangaonkar, G. M., and Pawar, K. D. (2018). Garcinia indica mediated biogenic synthesis of silver nanoparticles with antibacterial and antioxidant activities. Colloids Surf. B Biointerfaces 164, 210–217. doi: 10.1016/j.colsurfb.2018.01.044
Shah, M. Z., Guan, Z.-H., Din, A. U., Ali, A., Rehman, A. U., Jan, K., et al. (2021). Synthesis of silver nanoparticles using Plantago lanceolata extract and assessing their antibacterial and antioxidant activities. Sci. Rep. 11:20754. doi: 10.1038/s41598-021-00296-5
Singh, P., Pandit, S., Garnaes, J., Tunjic, S., Mokkapati, V. R., Sultan, A., et al. (2018c). Green synthesis of gold and silver nanoparticles from Cannabis sativa (industrial hemp) and their capacity for biofilm inhibition. Int. J. Nanomed. 13, 3571–3591. doi: 10.2147/IJN.S157958
Singh, H., Du, J., Singh, P., and Yi, T. H. (2018a). Ecofriendly synthesis of silver and gold nanoparticles by Euphrasia officinalis leaf extract and its biomedical applications. Artif Cells Nanomed. Biotechnol. 46, 1163–1170. doi: 10.1080/21691401.2017.1362417
Singh, H., Du, J., Singh, P., and Yi, T. H. (2018b). Extracellular synthesis of silver nanoparticles by Pseudomonas sp. THG-LS1.4 and their antimicrobial application. J. Pharm. Anal. 8, 258–264. doi: 10.1016/j.jpha.2018.04.004
Singh, P., Kim, Y. J., Singh, H., Farh, M. E., and Yang, D. C. (2017). Achromobacter panacis sp. nov., isolated from rhizosphere of Panax ginseng. J. Microbiol. 55, 428–434. doi: 10.1007/s12275-017-6612-3
Singh, P., Kim, Y.-J., Zhang, D., and Yang, D.-C. (2016a). Biological synthesis of nanoparticles from plants and microorganisms. Trends Biotechnol. 34, 588–599. doi: 10.1016/j.tibtech.2016.02.006
Singh, P., Singh, H., Kim, Y. J., Mathiyalagan, R., Wang, C., and Yang, D. C. (2016b). Extracellular synthesis of silver and gold nanoparticles by Sporosarcina koreensis DC4 and their biological applications. Enzyme Microb. Technol. 86, 75–83. doi: 10.1016/j.enzmictec.2016.02.005
Singh, P., Pandit, S., Jers, C., Joshi, A. S., Garnaes, J., and Mijakovic, I. (2021). Silver nanoparticles produced from Cedecea sp. exhibit antibiofilm activity and remarkable stability. Sci. Rep. 11:12619. doi: 10.1038/s41598-021-92006-4
Singh, P., Pandit, S., Mokkapati, V., Garnaes, J., and Mijakovic, I. (2020). A sustainable approach for the green synthesis of silver nanoparticles from solibacillus isronensis sp. and their application in biofilm inhibition. Molecules 25:2783. doi: 10.3390/molecules25122783
Ssekatawa, K., Byarugaba, D. K., Kato, C. D., Wampande, E. M., Ejobi, F., Nakavuma, J. L., et al. (2021). Green strategy–based synthesis of silver nanoparticles for antibacterial applications. Fron. Nanotechnol. 3:697303. doi: 10.3389/fnano.2021.697303
Ullah Khan, S., Saleh, T. A., Wahab, A., Khan, M. H. U., Khan, D., Ullah Khan, W., et al. (2018). Nanosilver: new ageless and versatile biomedical therapeutic scaffold. Int. J. Nanomed. 13, 733–762. doi: 10.2147/IJN.S153167
Wypij, M., Jędrzejewski, T., Trzcińska-Wencel, J., Ostrowski, M., Rai, M., and Golińska, P. (2021). Green synthesized silver nanoparticles: antibacterial and anticancer activities, biocompatibility, and analyses of surface-attached proteins. Front. Microbiol. 12:632505. doi: 10.3389/fmicb.2021.632505
Xu, L., Wang, Y. Y., Huang, J., Chen, C. Y., Wang, Z. X., and Xie, H. (2020). Silver nanoparticles: Synthesis, medical applications and biosafety. Theranostics 10, 8996–9031. doi: 10.7150/thno.45413
Zhang, D., Ma, X.-L., Gu, Y., Huang, H., and Zhang, G.-W. (2020). Green synthesis of metallic nanoparticles and their potential applications to treat cancer. Front. Chem. 8:799. doi: 10.3389/fchem.2020.00799
Keywords: silver nanoparticles, green synthesis, strong antimicrobial activity, highly stable AgNPs, Gram-negative pathogenic microorganisms, environmental isolate
Citation: Singh P and Mijakovic I (2022) Strong Antimicrobial Activity of Silver Nanoparticles Obtained by the Green Synthesis in Viridibacillus sp. Extracts. Front. Microbiol. 13:820048. doi: 10.3389/fmicb.2022.820048
Received: 22 November 2021; Accepted: 25 January 2022;
Published: 16 February 2022.
Edited by:
Mirian A. F. Hayashi, Federal University of São Paulo, BrazilReviewed by:
Santosh Kumar, Central Institute of Technology, Kokrajhar, IndiaCopyright © 2022 Singh and Mijakovic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Priyanka Singh, cHJpc2luQGJpb3N1c3RhaW4uZHR1LmRr; Ivan Mijakovic, aXZhbi5taWpha292aWNAY2hhbG1lcnMuc2U=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.