- 1Heilongjiang Key Laboratory for Animal Disease Control and Pharmaceutical Development, Northeast Agricultural University, Harbin, China
- 2Department of Basic Veterinary Science, College of Veterinary Medicine, Northeast Agricultural University, Harbin, China
Colistin is the last line of defense for the treatment of multidrug-resistant gram-negative bacterial infections. However, colistin resistance is gradually increasing worldwide, with resistance commonly regulated by two-component system and mcr gene. Thus, this study aimed to investigate molecular epidemiology and colistin-resistant mechanism of mcr-positive and mcr-negative Escherichia coli isolates from animal in Sichuan Province, China. In this study, a total of 101 colistin-resistant E. coli strains were isolated from 300 fecal samples in six farms in Sichuan Province. PCR was used to detect mcr gene (mcr-1 to mcr-9). The prevalence of mcr-1 in colistin-resistant E. coli was 53.47% (54/101), and the prevalence of mcr-3 in colistin-resistant E. coli was 10.89% (11/101). The colistin-resistant E. coli and mcr-1–positive E. coli showed extensive antimicrobial resistance profiles. For follow-up experiments, we used 30 mcr-negative and 30 mcr-1–positive colistin-resistant E. coli isolates and E. coli K-12 MG1655 model strain. Multi-locus sequence typing (MLST) of 30 strains carrying mcr-1 as detected by PCR identified revealed six strains (20%) of ST10 and three strains (10%) of each ST206, ST48, and ST155 and either two (for ST542 and 2539) or just one for all other types. The conjugation experiment and plasmid replicon type analysis suggest that mcr-1 was more likely to be horizontally transferred and primarily localized on IncX4-type and IncI2-type plasmid. The ST diversity of the mcr-1 indicated a scattered and non-clonal spreading in mcr-1–positive E. coli. Twenty-eight mcr-negative colistin-resistant E. coli isolates carried diverse amino acid alterations in PmrA, PmrB, PhoP, PhoQ, and MgrB, whereas no mutation was found in the remaining isolates. The finding showed the high prevalence of colistin resistance in livestock farm environments in Sichuan Province, China. Our study demonstrates that colistin resistance is related to chromosomal point mutations including the two-component systems PhoP/PhoQ, PmrA/PmrB, and their regulators MgrB. These point mutations may confer colistin resistance in mcr-negative E. coli. These findings help in gaining insight of chromosomal-encoded colistin resistance in E. coli.
Introduction
Colistin is the last line of defense for the treatment of multidrug-resistant (MDR) gram-negative bacteria (Paterson et al., 2020). The rate of colistin resistance has risen to 30% of Carbapenem-resistant Enterobacteriaceae (CRE) isolates in Italy, Spain, and Greece over the past decade (Stefaniuk and Tyski, 2019). Colistin resistance is a growing public health concern worldwide. Colistin is a cyclic polycationic peptide that interacts with anionic lipopolysaccharide (LPS) molecules (Olaitan et al., 2014). However, overuse and misuse of colistin accelerate propagation of antibiotic resistance genes. The exact mechanism of colistin resistance is not well understood. It has been shown that an altered outer membrane, a loss of lipid A, and increase in drug efflux pumps are associated with colistin resistance (Raetz et al., 2007; Moffatt et al., 2010; Padilla et al., 2010).
The plasmid-mediated colistin resistance was not demonstrated until 2015. Liu et al. (2016) first systematically reported mcr-1 gene. Over the past few years, mcr-1 became common all over the world. The mcr-1 has the potential to spread rapidly by horizontal transfer and may pose a significant public health risk (Rapoport et al., 2016; Rolain et al., 2016; Sonnevend et al., 2016; Borowiak et al., 2017). In the past few years, novel plasmid-encoded colistin resistance genes have also been identified. So far, novel colistin resistance genes (mcr-2 to mcr-10) have been reported (Xavier et al., 2016; Carattoli et al., 2017; Yin et al., 2017; AbuOun et al., 2018; Yang et al., 2018; Wang et al., 2019, 2020). The mcr-2 to mcr-9 genes encode the proteins MCR-2 to MCR-9, which share 81, 32.5, 34, 36, 83, 35, 31, and 36% amino acid sequence identity with MCR-1, respectively (Carroll et al., 2019; Nang et al., 2019). MCR-10 is encoded by mcr-10, which contains 82.93% amino acids identical to that in MCR-9 (Wang et al., 2020). The mcr-1 has spread to more than 60 countries (Cheng et al., 2021). It has been reported that prevalence of mcr-1–positive bacteria ranged from 0.35 to 36.00% in pigs and 2.40–30.00% in poultry (Xiaomin et al., 2020). To date, mcr-1 was identified in many different plasmid types, including IncI2, IncHI2, IncX4, IncP, IncY, and IncF (Shen et al., 2018).
Before the discovery of mcr gene, clinical colistin resistance was associated with mutation of chromosomal gene (Kim et al., 2019; Mendes Oliveira et al., 2019). Chromosome-mediated colistin resistance had been linked to LPS modifications, which was associated with PmrAB or PhoPQ two-component systems (Poirel et al., 2017). MgrB is a negative regulator of the PhoPQ system. The inactivation of MgrB leads to over expression of the phoPQ operon (Cheng et al., 2010; Cannatelli et al., 2013). Moreover, amino acid substitutions of PmrA and/or PmrB have been reported in clinical isolates of Acinetobacter baumanni and Klebsiella pneumoniae (Poirel et al., 2017). First, amino acid substitutions may affect protein function. In addition, amino acid substitutions in MgrB, PmrA/B, and PhoP/Q are a common mechanism of colistin resistance among K. pneumoniae in clinical settings (Luo et al., 2017). However, amino acid substitutions in MgrB, PmrA/B, and PhoP/Q are rarely reported among mcr-negative colistin-resistant Escherichia coli isolates. A recent study showed that missense mutations may be responsible for colistin resistance (Olaitan et al., 2014).
This study aimed to investigate molecular epidemiology and colistin-resistant mechanism of mcr-positive and mcr-negative E. coli isolated from animals in Sichuan Province, China.
Materials and Methods
Sample Collection
From January 2016 to March 2018, a total of 300 fecal swabs were collected from six different farms in Sichuan Province. The samples were kept on ice and were immediately brought to laboratory. The samples were cultured on MacConkey agar at 37°C for 18–24 h, and then, five colonies with typical E. coli morphology were selected. The E. coli were identified using biochemical methods and confirmed by PCR amplification of 16S rRNA and sequencing. The primer of 16S rRNA is listed in Supplementary Table 1. The protocols used during this study were approved by the Northeast Agricultural University Institutional Animal Care and Use Committee, and all the animal care and treatment methods complied with the standards described in the Laboratory Animal Management Regulations (revised 2016) of Heilongjiang Province, China.
Detection of Colistin-Resistant Escherichia coli Isolates and mcr Gene
To screen colistin-resistant E. coli, E. coli were cultured on MacConkey agar at 37°C for 18–24 h. To determine the colistin minimum inhibitory concentration (MIC), E. coli isolates (>4μg/ml) were served as colistin-resistant E. coli. A DNA extraction kit (TIANGEN, Beijing, China) was used to extract genomic DNA of colistin-resistant E. coli isolates. The mcr-harboring isolates were screened by PCR amplification and were validated by sequencing. The primers of mcr gene are listed in Supplementary Table 1.
Antimicrobial Susceptibility Testing
The susceptibility of colistin-resistance E. coli isolates to 23 antibiotics, namely, Nitrofurantoin, Ciprofloxacin, Levofloxacin, Kanamycin, Amikacin, Amoxicillin, Tigecycline, Cefepime, Chloramphenicol, Fosfomycin, Aztreonam, Ampicillin, Ampicillin/sulbactam, Cefoxitin, Doxycycline, Streptomycin, Ceftriaxone, Florfenicol, Cefuroxime, Sulfamethoxazole, Gentamicin, and Tetracycline, was determined by the standard disk diffusion method in accordance with the Clinical and Laboratory Standards Institute (CLSI).1 Escherichia coli ATCC 25922 was served as a quality control strain for susceptibility testing.
Detection of Antimicrobial Resistance Genes
The presence of the β-lactamase genes (blaCTX–M, blaTEM, and blaSHV) (Dallenne et al., 2010), aminoglycoside resistance genes (strA, strB, aacC2, and aacC4) (Kozak et al., 2009), tetracycline resistance genes (tetA, tetB, and tetC) (Ji et al., 2020), fluoroquinolone resistance genes [qnrS, oqxA, oqxB, qepA, and aac(6′)-Ib-cr] (Ciesielczuk et al., 2013), florfenicol resistance gene (floR) (Lu et al., 2018), and sulfonamide resistance genes (sul1, sul2, and sul3) (Hammerum et al., 2006) were examined by PCR amplification and were validated by sequencing. The primers of resistance genes are listed in Supplementary Table 2. The obtained DNA sequences were analyzed using ChromasPro software and were compared with published sequences by BLAST.
Detection of pmrA/B, mgrB, and PhoP/Q Amino Acid Variants
The entire pmrAB, mgrB, and phoPQ genes were amplified of colistin-resistant isolates using the primers listed in Supplementary Table 3. The amplification products were validated with Sanger sequencing. Amino acid sequences of mcr-negative colistin-resistant E. coli isolates were compared with the reference strain E. coli K-12 MG1655. Missense mutations of PmrA/B, PhoP/Q, and MgrB identified in mcr-negative colistin-resistant E. coli were analyzed using the PROVEAN (Choi and Chan, 2015) prediction software. In addition, amino acid substitutions were considered “deleterious” if the PROVEAN score was ≤-2.5 and “neutral replacements” if the PROVEAN score was >-2.5 (Choo et al., 2016; Higuchi et al., 2016). We used PROVEAN bioinformatic tool2 to predict whether amino acid substitutions in MgrB, PmrA/B, and PhoP/Q affect protein function (Choi et al., 2012). SMART (Simple Modular Architecture Research Tool) is a web resource,3 providing simple identification and extensive annotation of protein domains and the exploration of protein domain architectures (Schultz et al., 1998; Letunic et al., 2015). SMART analysis was performed to determine the domain architectures of PmrA, PmrB, PhoP, PhoQ, and MgrB protein.
Conjugation Experiment and Plasmid Replicon Type Analysis
The transferability of mcr-1 was tested by conjugation experiment with 30 mcr-1–positive E. coli (MCRPEC) as donors and E. coli J53 as recipient strains. MacConkey agar plates containing rifampicin (256 μg/ml) and colistin (4 μg/ml) were used to select mcr-1–positive transconjugants. The mcr-1 gene of transconjugants was examined by PCR amplification and was validated by sequencing. The replicon types of the transconjugants were determined according to previous studies (Carattoli et al., 2005).
Multi-Locus Sequence Typing Analysis
Sequence type of 30 MCRPEC was determined according to the primers and protocol specified in E. coli multi-locus sequence typing (MLST) database website, which is based on the housekeeping genes adk, fumC, gyrB, icd, mdh, purA, and recA. The primers of housekeeping gene are listed in Supplementary Table 4. The obtained DNA sequence alignments were performed by using ChromasPro software. The phylogenetic tree was constructed by the neighbor-joining method.
Results and Discussion
Prevalence of mcr-Positive Escherichia coli Isolated From Animal
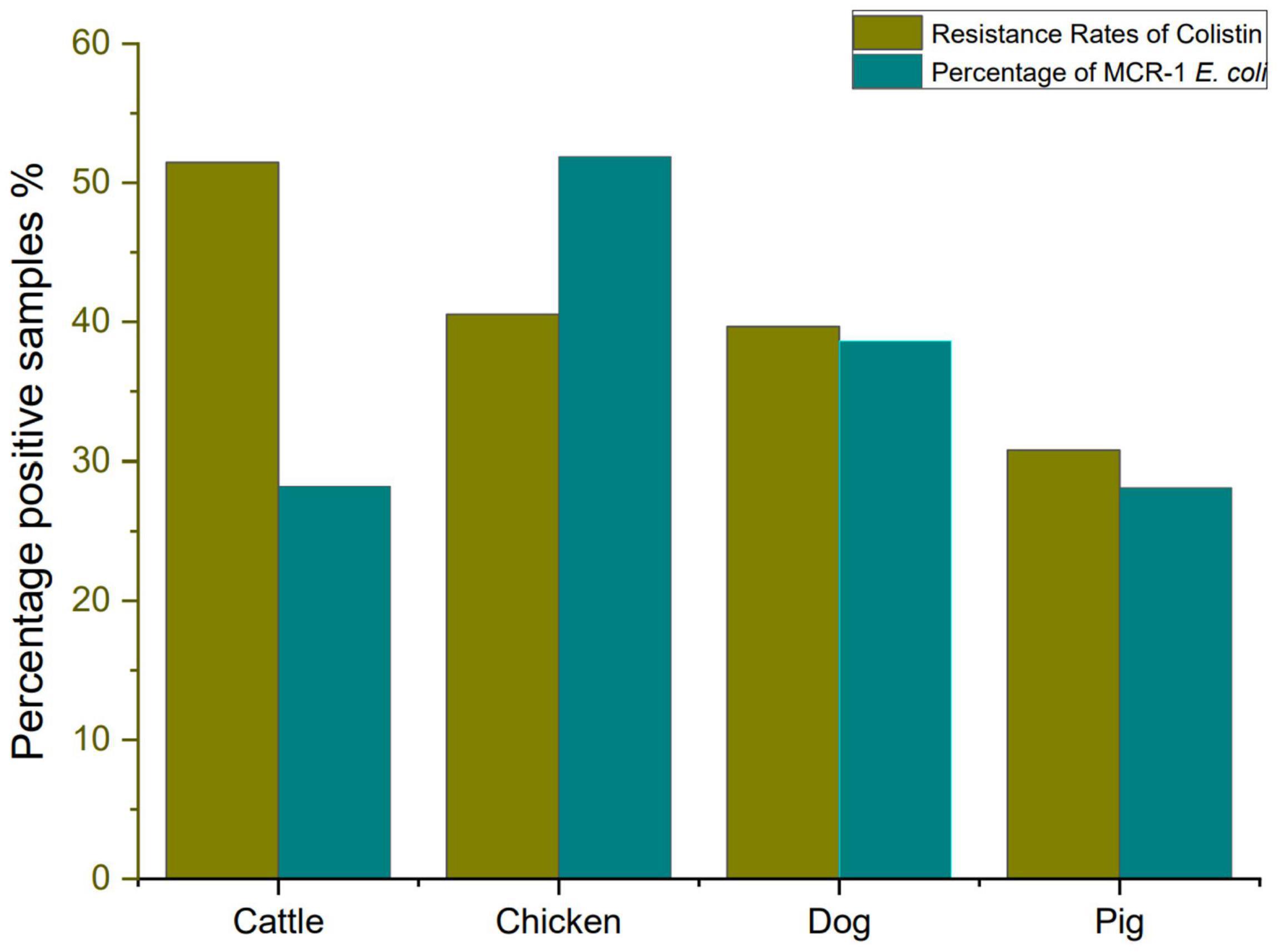
In this study, we investigate colistin resistance rate in E. coli isolated from six different farms in Sichuan Province during 2016–2018. A total of 254 E. coli strains were isolated from 300 fecal samples, including 37 chickens, 91 pigs, 68 cattle, and 58 dogs. The E. coli isolates from cattle showed a high resistance rate to colistin (51.47%, 35/68), followed by E. coli isolates from chicken (40.54%, 15/37), E. coli isolates from dog (39.66%, 23/58), and E. coli isolates from pig (30.77%, 28/91). However, it has been reported that the prevalence of colistin resistance in E. coli isolated from farms in different areas of China during 2013–2014, which revealed that colistin resistance rates in E. coli from pigs, chickens, and cattle were 26.5, 14.0, and 0.9%, respectively (Zhuge et al., 2019). Our data also showed that colistin resistance rate has risen significantly high. Colistin-resistant E. coli isolates (MIC of colistin ≥ 4 μg/ml) are listed in Supplementary Table 5. The resistance rate to colistin and percentage of MCRPEC are shown in Figure 1.
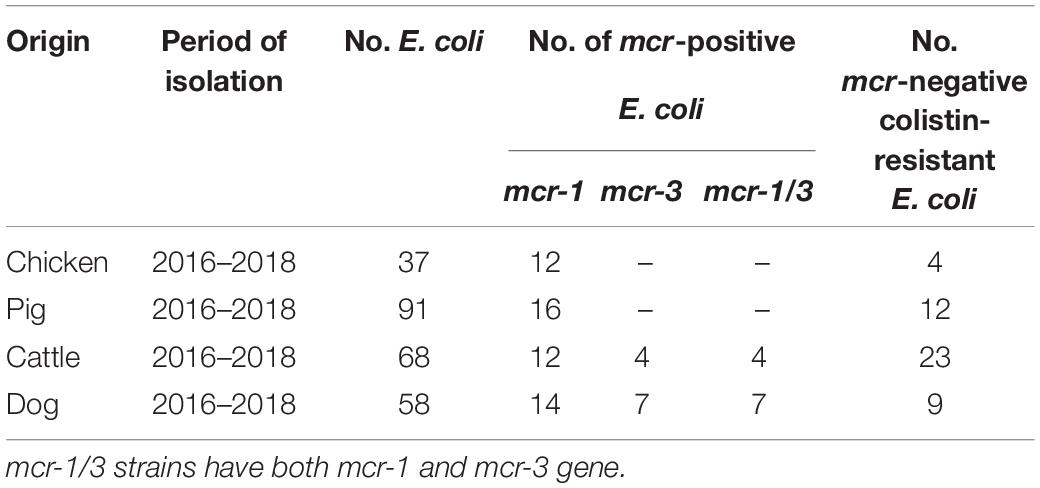
However, discoveries of plasmid-mediated colistin resistance gene in many countries have heralded a significant threat to public health worldwide (Kai and Wang, 2020). As shown in Table 1, mcr-1 gene was detected in 54 (53.47%) out of the selected 101 isolates, and mcr-3 gene was detected in 11 (10.89%) out of the selected 101 isolates. Eleven E. coli isolates have both mcr-1 and mcr-3 gene. No other mcr genes were found in this survey. Similar to our result, a surveillance of colistin resistance performed in Jiangsu Province revealed that the mcr-1 prevalence was 68.86% in pigs (Zhang et al., 2019). More detailed studies of mcr-1 have been performed in colistin-resistant isolates (Tong et al., 2018). The previous study showed that mcr-1 had been identified in approximately 60 countries across five different continents (Cao et al., 2018; Sun et al., 2018; Wang et al., 2018). The finding of mobilizable mcr-like genes became a global concern due to the possibility of horizontal transfer of the plasmid that often carry resistance determinants to beta-lactams and/or quinolones (Mendes Oliveira et al., 2019).
Antimicrobial Susceptibility of Colistin-Resistance and mcr-1–Positive Escherichia coli
The susceptibility of 101 colistin-resistant E. coli isolates to 23 antimicrobials was determined by the standard disk diffusion method. Figure 2 shows a significant difference in resistance rate of the E. coli isolates to 23 antibiotics. Among the colistin-resistant E. coli isolates, all isolates were susceptible to amikacin, tigecycline, and nitrofurantoin. Antibiotic sensitivity tests revealed that colistin-resistant E. coli isolates have a highly resistance to sulfisoxazole; a moderate rate of resistance to ampicillin, streptomycin, and tetracycline; and a low rate of resistance to cefuroxime, ceftazidime, cefepime, ceftriaxone, cefoxitin, aztreonam, ampicillin-sulbactam, amoxicillin-clavulanic acid, gentamicin, kanamycin, ciprofloxacin, and levofloxacin. The colistin-resistant E. coli isolates displayed high resistance rates to antibiotics that are commonly used in veterinary medicine in Sichuan Province (Ma et al., 2017; Zhang et al., 2021), including sulfisoxazole, ampicillin, streptomycin, tetracycline, and chloramphenicol. The antimicrobial resistance rates have reached to 30%, which may be due to the breadth of our sample or because of the generally high use of antibiotics in Sichuan Province. Three studies reported the rate of resistance to streptomycin was within the range 0–7% (Routman et al., 1985; Sayah et al., 2005; Tong et al., 2018). However, the rate of resistance to streptomycin has reached 30% in this study. This difference may reflect difference in antimicrobial use in different livestock.
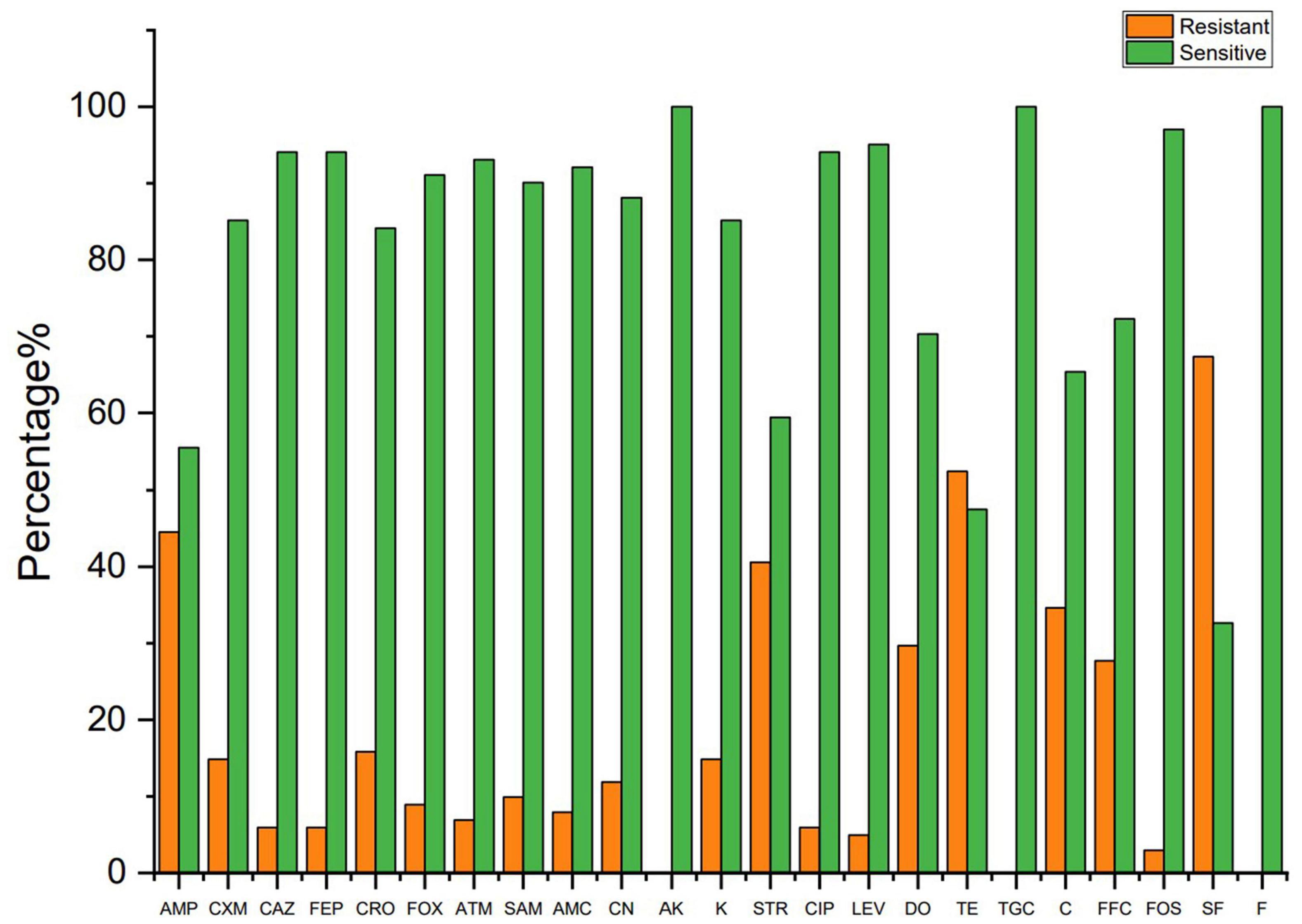
Figure 2. The resistant rate of colistin resistance E. coli to other antibiotics. AMP, ampicillin; CXM, cefuroxime; CAZ, ceftazidime; FEP, cefepime; CRO, ceftriaxone; FOX, cefoxitin; ATM, aztreonam; SAM, ampicillin-sulbactam; AMC, amoxicillin–clavulanic acid; CN, gentamicin; AK, amikacin; K, kanamycin; STR, streptomycin; CIP, ciprofloxacin; LEV, levofloxacin; TE, tetracycline; DO, doxycycline; TGC, tigecycline; C, chloramphenicol; FFC, florfenicol; FOS, fosfomycin; SF, sulfisoxazole; F, nitrofurantoin.
Figure 3 shows the resistance rate of the MCRPEC isolates to 23 antibiotics. MCRPEC isolate have a moderate rate of resistance (20–60%) to ampicillin, streptomycin, doxycycline, tetracycline, and sulfisoxazole; and a low rate of resistance (<20%) to cefuroxime, ceftazidime, cefepime, ceftriaxone, cefoxitin, aztreonam, ampicillin-sulbactam, amoxicillin-clavulanic acid, gentamicin, kanamycin, ciprofloxacin, levofloxacin, and fosfomycin. All the MCRPEC isolate were susceptible to amikacin, tigecycline, and nitrofurantoin. The resistance phenotype of mcr-1 positive E. coli is shown in Supplementary Table 6.
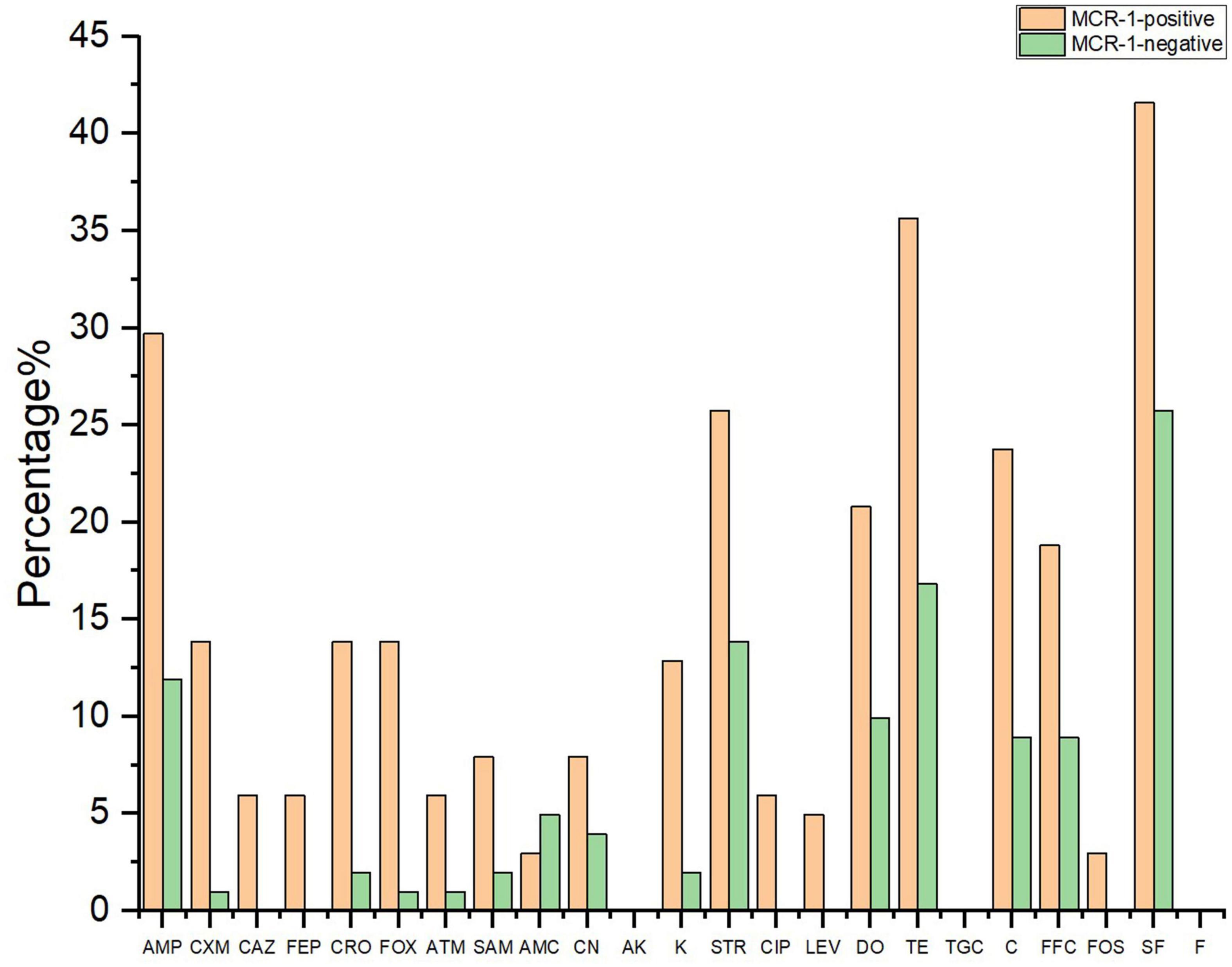
Figure 3. The resistant rate of mcr-1–positive and mcr-1–negative E. coli to 23 antibiotics. AMP, ampicillin; CXM, cefuroxime; CAZ, ceftazidime; FEP, cefepime; CRO, ceftriaxone; FOX, cefoxitin; ATM, aztreonam; SAM, ampicillin–sulbactam; AMC, amoxicillin–clavulanic acid; CN, gentamicin; AK, amikacin; K, kanamycin; STR, streptomycin; CIP, ciprofloxacin; LEV, levofloxacin; TE, tetracycline; DO, doxycycline; TGC, tigecycline; C, chloramphenicol; FFC, florfenicol; FOS, fosfomycin; SF, sulfisoxazole; F, nitrofurantoin.
Detection of Antimicrobial Resistance Genes of mcr-1–Positive Escherichia coli
The prevalence of additional antimicrobial resistance genes in 30 MCRPEC is shown in Figure 4. Overall, blaTEM (n = 29, 96.67%) and blaCTX–M (n = 29, 96.67%) were the most common ESBL genes, and blaSHV was not detected in this study. The aminoglycoside resistance genes with the highest detection rate were aacC2 (n = 24, 80.00%) and aacC4 (n = 25, 83.33%), followed by strA (n = 23, 76.67%), and strB were not detected. Among tetracycline resistance genes, tetA (n = 29, 96.67%), tetB (n = 29, 96.67%), and tetC (n = 29, 96.67%) were the most common tetracycline resistance genes. The fluoroquinolone resistance genes with the highest detection rate were aac(6′)-Ib-cr (n = 6, 20.00%), followed by qepA (n = 4, 13.33%) and qnrS (n = 4, 13.33%), and oqxA and oqxB were not detected in our study. The florfenicol resistance gene floR detection rate was (n = 30, 100.00%). The sulfonamide resistance genes with the highest detection rate were sul1 (n = 13, 43.33%), followed by sul2 (n = 7, 23.33%) and sul3 (n = 6, 20.00%). In our study, a high prevalence of ESBL genes (blaTEM and blaCTX–M), aminoglycoside resistance genes (aacC2, aacC4, and strA), tetracycline resistance genes (tetA, tetB, and tetC), and florfenicol resistance gene floR were found in 30 MCRPEC. This result may reflect that plasmid harboring mcr-1 usually carry other resistance genes (Rozwandowicz et al., 2018; Cheng et al., 2021).
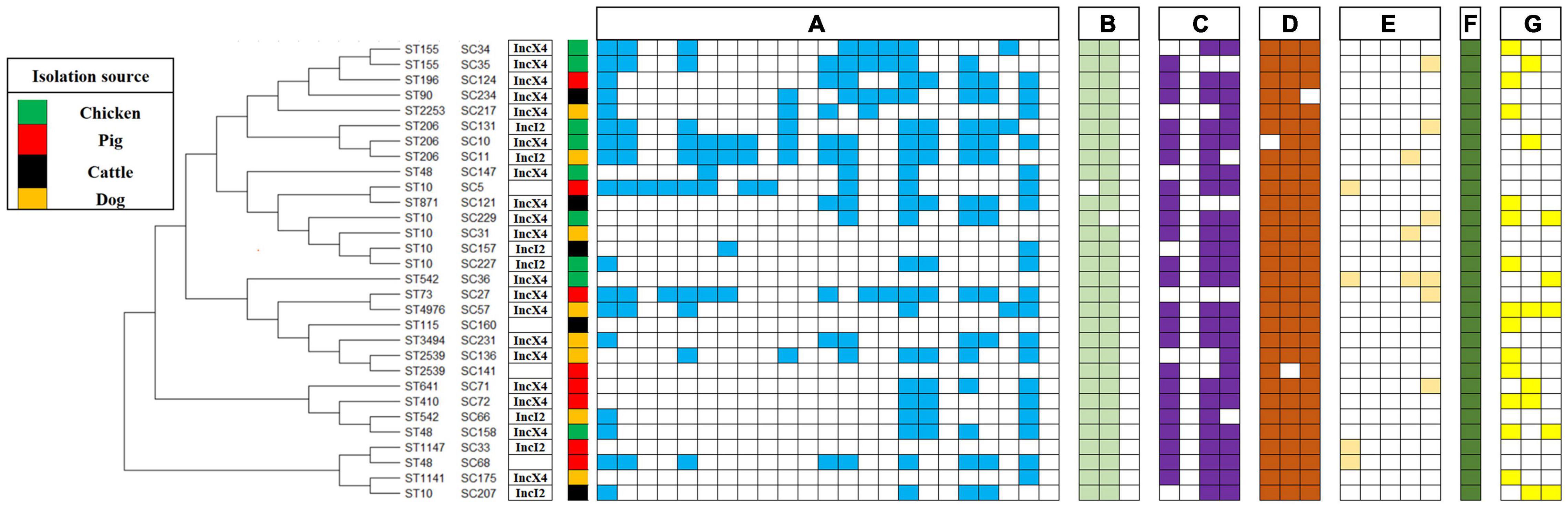
Figure 4. Phylogeny of mcr-1–positive E. coli (sequence types, strain number, plasmid type, source of isolation, and map of phenotypic resistance and resistance genes). Full and empty square mean presence and absence of antimicrobial resistance (AMR) gene, respectively, whereas empty space means non-defined. (A) Resistance profiles: ampicillin, cefuroxime, ceftazidime, cefepime, ceftriaxone, cefoxitin, aztreonam, ampicillin-sulbactam, amoxicillin-clavulanic acid, gentamicin, amikacin, kanamycin, streptomycin, ciprofloxacin, levofloxacin, tetracycline, doxycycline, tigecycline, chloramphenicol, florfenicol, fosfomycin, sulfisoxazole, and nitrofurantoin. (B) Beta-lactam resistance genes: blaCTX–M, blaTEM and blaSHV. (C) Aminoglycoside resistance genes: strA, strB, aacC2, and aacC4. (D) Tetracycline resistance genes: tetA, tetB, and tetC. (E) Fluoroquinolone resistance genes: qnrS, oqxA, oqxB, qepA, and aac(6′)-Ib-cr. (F) Florfenicol resistance gene: floR. (G) Sulfonamide resistance genes: sul1, sul2, and sul3.
Transferability and Plasmid Replicon Types of mcr-1–Positive Escherichia coli
As shown in Figure 4, the conjugation result showed that 27 isolates successfully conjugated among 30 representative MCRPEC. The success rate of conjugation was as high as 90%. mcr-1 has been observed in two types plasmids, including IncX4 (n = 19) and IncI2 (n = 7). In this study, the transferability of MCRPEC was assessed among 30 representative strains. The result was in line with previous study, which showed that majority of mcr-1–carrying plasmids were transferable (Cheng et al., 2021). Previous research has shown mcr-1–carrying plasmids belong to different replicon types, including IncI2 (Matamoros et al., 2017), IncHI1 (Zurfluh et al., 2016), IncHI2 (Matamoros et al., 2017), IncFIB (Wang et al., 2017; Khezri et al., 2020), IncFII (Xavier et al., 2016), IncP (Zhao et al., 2017), IncX4 (Gao et al., 2016), and IncY (Shen et al., 2018).
Molecular Genotyping of mcr-1–Positive Escherichia coli
As shown in Figure 4, 30 MCRPEC isolates were assigned to 17 STs. ST10 (n = 6) was the most numerous ST in this study, followed by ST206 (n = 3), ST48 (n = 3), ST155 (n = 3), ST542 (n = 2), and ST2539 (n = 2), and then by single ST type isolates, including ST196, ST90, ST2253, ST871, ST73, ST4976, ST3494, ST641, ST410, ST1147, and ST1141. Hence, colistin-resistant E. coli isolates comprised a variety of STs and were therefore genetically different, with the nosocomial transmission excluded. The ST diversity of the mcr-1–harboring E. coli isolates indicated a scattered and non-clonal prevalence. The discovery of the superbug MCRPEC has triggered a huge amount of innovative scientific inquiry (Yuan et al., 2021). Recently, a study analyzed 616 whole genomes of MCRPEC isolates from NCBI online database. Similar to our result, ST10 was the most common ST among the mcr-1–positive isolates (Zhuge et al., 2019).
Amino Acid Variations of PmrA/B, PhoP/Q, and MgrB in mcr-Negative Isolates
As shown before, colistin resistance was reported to be associated with chromosomal mutations. It is well-known that two-component PhoPQ and PmrAB were associated with LPS modification (Luo et al., 2017). LPS may play an important regulatory role in colistin-resistance isolates (Schurek et al., 2009; Kandehkar Ghahraman et al., 2021). To determine chromosome-mediated colistin-resistant mechanism in mcr-negative colistin-resistant isolates, we explored whether alterations in amino acid of PmrAB, PhoPQ, and MgrB affect protein function. We selected K-12 MG1655 E. coli as negative controls. The mutations are shown in Tables 2, 3, and we found that many variations were synonymous and non-synonymous mutations. The mgrB gene encodes a short 47–amino acid transmembrane protein. MgrB is a small transmembrane protein of 47 amino acids, which acts as negative feedback regulator of the PhoPQ two-component regulatory system (Lippa and Goulian, 2009). The multiple sequence alignment tool MEGA-X was used for multiple protein sequence alignment. Two mutations M1V (23 mcr-negative isolates) and V8A (one mcr-negative isolate) were detected. The mutation M1V [PROVEAN score = -4.670 (cutoff = -2.5)] and V8A [PROVEAN score = -2.808 (cutoff = -2.5)] were deleterious affecting protein function. We found similar mutations in MgrB (V8A) with previous studies (Delannoy et al., 2017; Luo et al., 2017). The mutation V8A may confer colistin resistance in mcr-negative E. coli.
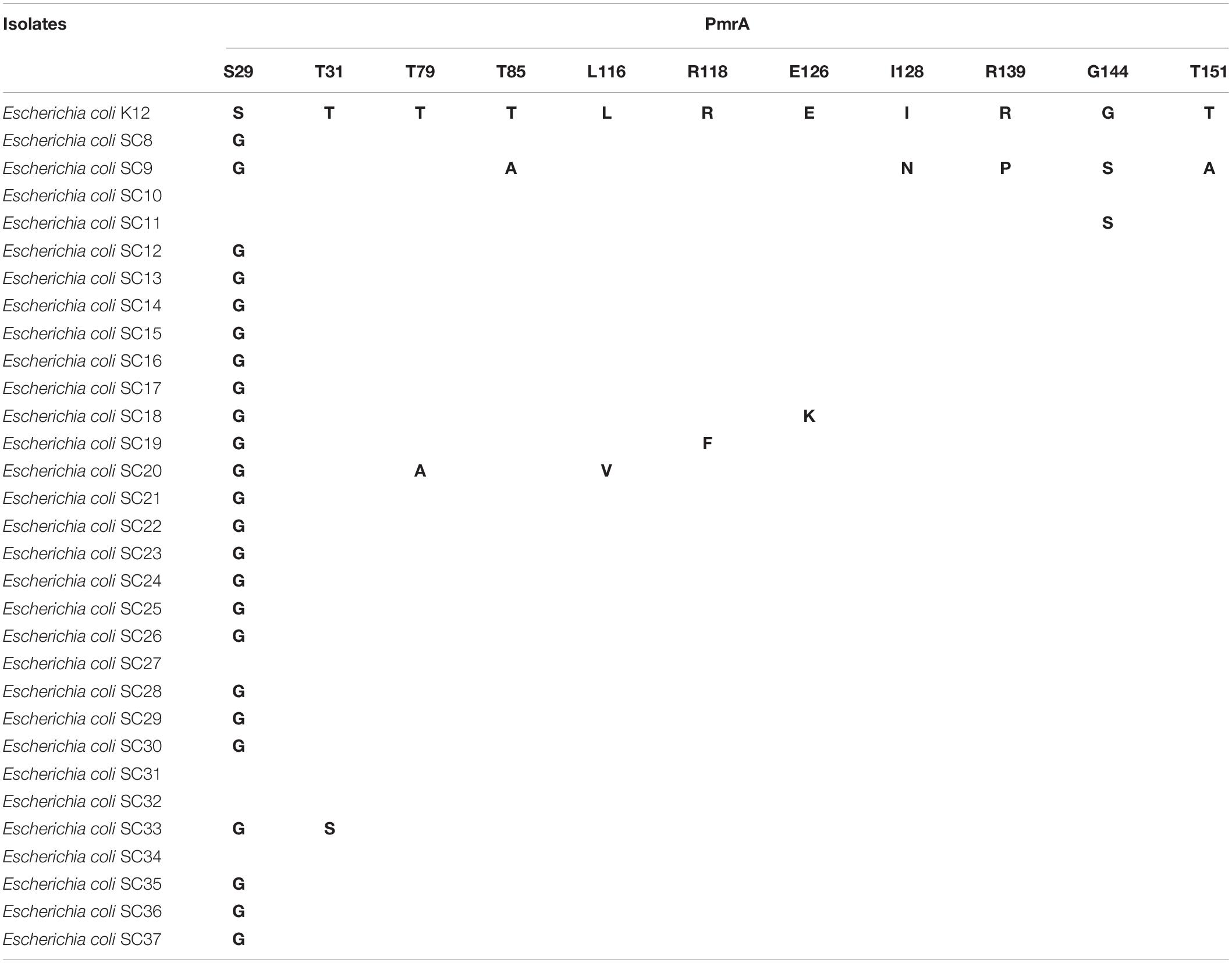
The pmrA gene encodes a short 222–amino acid transmembrane protein. The multiple sequence alignment tool MEGA-X was used for multiple protein sequence alignment. As shown in Table 2, 11 mutations S29G (24 mcr-negative isolates), T31S (one mcr-negative isolate), T79A (one mcr-negative isolate), T85A (one mcr-negative isolate), L116V (one mcr-negative isolate), R118F (one mcr-negative isolate), E126K (one mcr-negative isolate), I128N (one mcr-negative isolate), R139P (one mcr-negative isolate), G144S (two mcr-negative isolates), and T151A (one mcr-negative isolate) were detected. The mutations T79A [PROVEAN score = -4.586 (cutoff = -2.5)], R118F [PROVEAN score = -7.740 (cutoff = -2.5)], R139P [PROVEAN score = -3.456 (cutoff = -2.5)], and T151A [PROVEAN score = -2.790 (cutoff = -2.5)] were deleterious, affecting protein function. Figure 5 shows multiple sequence alignment of PmrA across seven mcr-1–negative colistin-resistant isolates. Mutations are shown with yellow boxes. The red boxes show that mutations were deleterious, affecting protein function. Amino acid deletion is shown with green box. In this study, 11 different mutations in PmrA (S29G, T31S, T79A, T85A, L116V, R118F, E126K, I128N, R139P, G144S, and T151A) were observed in 24 mcr-negative colistin-resistant isolates. Of them, four mutations, namely, T79A, R118F, R139P, and T151A, were deleterious, affecting protein function. In addition, we found similar mutations with previous study in PmrA (S29G, T31S, I128N, and G144S) (Luo et al., 2017). However, we also found many novel mutations in PmrA (T79A, T85A, L116V, R118F, E126K, R139P, and T151A). The seven mutations have not been reported in mcr-negative colistin-resistant E. coli. These mutations may confer colistin resistance in mcr-negative E. coli.
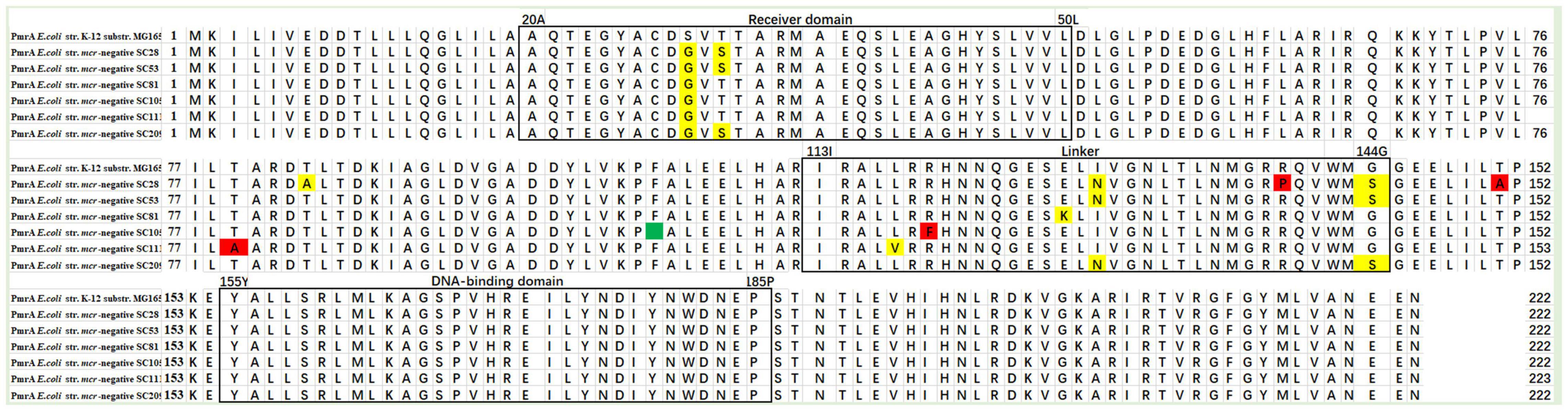
Figure 5. Multiple sequence alignment of PmrA across seven mcr-negative colistin-resistant E. coli. Mutations are shown with yellow boxes. The red boxes show that mutations were deleterious, affecting protein function. Amino acid deletion is shown with green box.
The pmrB gene encodes a short 363–amino acid transmembrane protein. Four mutations T235N (one mcr-negative isolate), D283G (11 mcr-negative isolates), V351I (one mcr-negative isolate), and Y358N (10 mcr-negative isolates) were detected. We found similar mutations in PmrB (D283G, V351I, and Y358N) with previous studies (Delannoy et al., 2017; Luo et al., 2017).
The phoP gene encodes a short 456–amino acid transmembrane protein. One mutation A416T (one mcr-1–negative isolate) was detected. The phoQ gene encodes a short 486–amino acid transmembrane protein. One mutation K46T (one mcr-negative isolate) was detected. The mutation K46T [PROVEAN score = -3.746 (cutoff = -2.5)] was deleterious, affecting protein function.
Domain Architectures of PmrAB, PhoPQ, and MgrB in mcr-Negative Isolates
SMART analysis was performed to determine the domain architectures of PmrA, PmrB, PhoP, PhoQ, and MgrB. As shown in Figure 6, we found that mutations have occurred in different domains of both PmrA and PmrB. In addition, pmrA appears to be the most commonly mutated gene in E. coli. The mutations T79A, R118F, R139P, and T151A were unique in PmrA. T91A is located in the cheY-homologous receiver domain. This domain contains a phosphoacceptor site that is phosphorylated by histidine kinase homologs. R118F, R139P, and T151A were located in transcriptional regulatory protein, C terminal. This domain is almost always found associated with the response regulator receiver domain. It may play a role in DNA binding. SMART analysis suggests that mutations T235N, V351I, and Y358N occurred in the HATPase_c domain in PmrB. The mutation V8A located in transmembrane region in MgrB, which starts at position 7 and ends at position 24. In summary, this study revealed diverse genetic mutations in two-component systems PmrAB and PhoPQ and their regulators MgrB in mcr-negative colistin-resistant E. coli isolates from Sichuan, China. However, mutated PmrB proteins do not contribute to colistin resistance (Wang et al., 2021). Although PmrAB, MgrB, and PhoPQ may be responsible for mcr-negative colistin-resistant E. coli, the mechanisms of colistin resistance appear to be highly diverse. These results suggest that the mechanisms underlying colistin resistance remain to be discovered in E. coli.
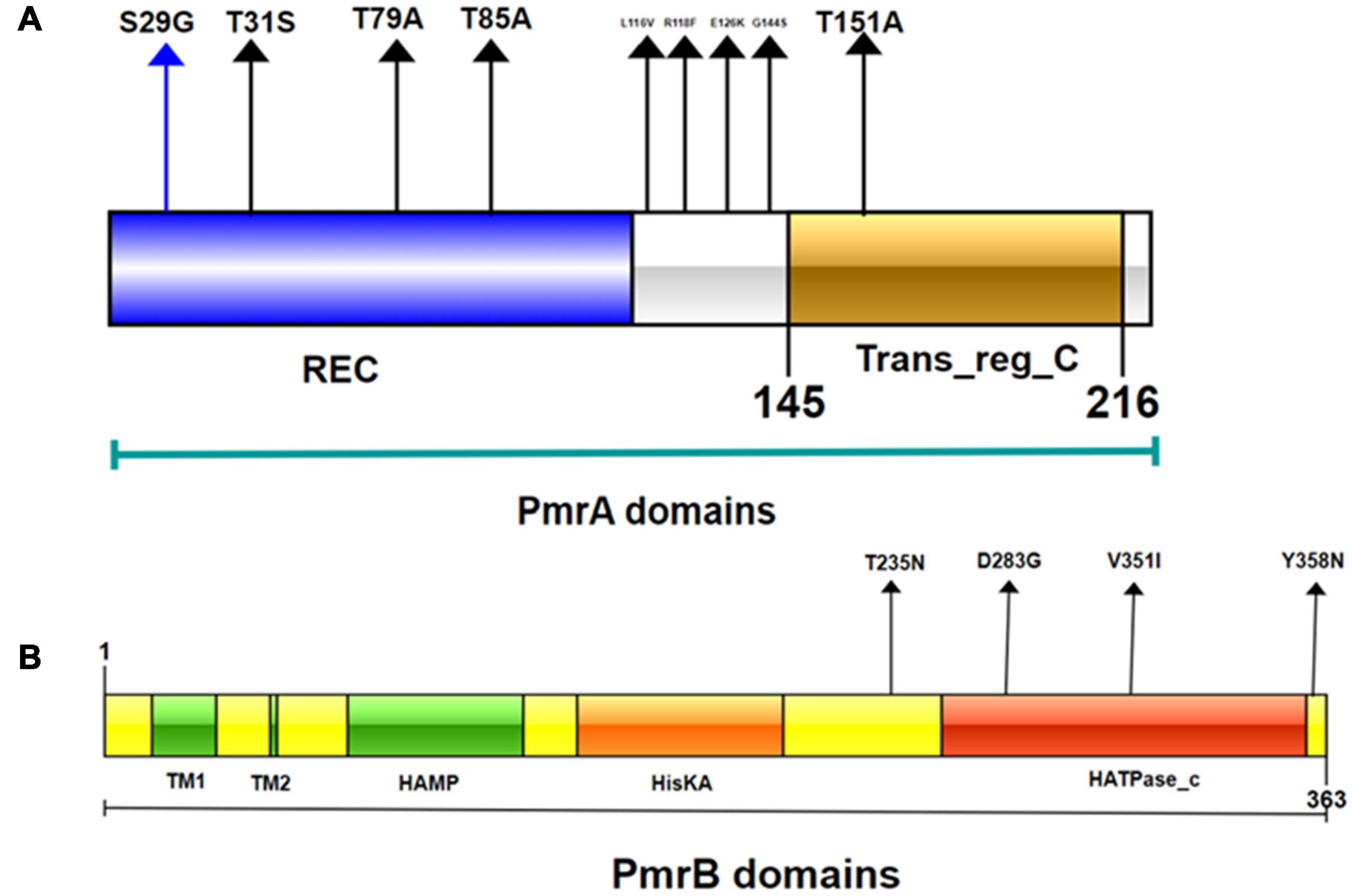
Figure 6. Domains of the PmrA/PmrB two-component system and positions of all mutations conferring colistin resistance to E. coli. (A) PmrA domains, cheY-homologous receiver domain [REC]; aa 1-112. Transcriptional regulatory protein, C-terminal domain [Trans_reg_C]; aa 145–216. S29G, T31S, T79A, T85A, L116V, R118F, E126K, I128N, G144S, and T151A represent mutation site. (B) PmrB domains, first transmembrane domain [TM1]; aa 15–34; second transmembrane domain [TM2]; aa 66–68. histidine kinases, adenylyl cyclases, methyl binding proteins, phosphatases [HAMP domain]; aa 89-141. Histidine kinase A (phosphoacceptor) domain [HisKA]; aa 142–202. Histidine kinase-like ATPases [HATPase_c]; aa 249-357. D283G, V351I, and Y358N represent smutation site.
Conclusion
The finding of this study showed the high prevalence of colistin in farms in Sichuan, China. The conjugation experiment and plasmid replicon type analysis suggest that the mcr-1 gene is more likely to be horizontally transferred. The ST diversity of the mcr-1 indicated a scattered and non-clonal. In addition, this study demonstrates diverse genetic mutations in two-component systems PmrAB and PhoPQ and their regulators MgrB of mcr-negative colistin-resistant E. coli isolates. In this study, we found several novel mutations, which have not been reported in mcr-negative colistin-resistant E. coli. These substitutions may confer colistin resistance in mcr-negative E. coli. To confirm our findings, further studies elucidating the resistance mechanism of mcr-negative E. coli to colistin are under way.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
FL conceived and designed the experiments and wrote the manuscript. PC, XL, RL, and HL provided assistance of the experiments and collected the samples. XZ supervised the experiments and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National 13th Five-Year Key R&D Program Special, Project under grant: 2018YFD0500306.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.818548/full#supplementary-material
Footnotes
References
AbuOun, M., Stubberfield, E. J., Duggett, N. A., Kirchner, M., Dormer, L., Nunez-Garcia, J., et al. (2018). mcr-1 and mcr-2 (mcr-6.1) variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J. Antimicrob. Chemother. 73:2904. doi: 10.1093/jac/dky272
Borowiak, M., Fischer, J., Hammerl, J. A., Hendriksen, R. S., Szabo, I., and Malorny, B. (2017). Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J. Antimicrob. Chemother. 72, 3317–3324. doi: 10.1093/jac/dkx327
Cannatelli, A., D’Andrea, M. M., Giani, T., Di Pilato, V., Arena, F., Ambretti, S., et al. (2013). In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob. Agents Chemother. 57, 5521–5526. doi: 10.1128/aac.01480-13
Cao, L., Li, X., Xu, Y., and Shen, J. (2018). Prevalence and molecular characteristics of mcr-1 colistin resistance in Escherichia coli: isolates of clinical infection from a Chinese University Hospital. Infect. Drug Resist. 11, 1597–1603. doi: 10.2147/idr.S166726
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Carattoli, A., Villa, L., Feudi, C., Curcio, L., Orsini, S., Luppi, A., et al. (2017). Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. 22:30589. doi: 10.2807/1560-7917.Es.2017.22.31.30589
Carroll, L. M., Gaballa, A., Guldimann, C., Sullivan, G., Henderson, L. O., and Wiedmann, M. (2019). Identification of Novel Mobilized Colistin Resistance Gene mcr-9 in a Multidrug-Resistant, Colistin-Susceptible Salmonella enterica Serotype Typhimurium Isolate. mBio 10:19. doi: 10.1128/mBio.00853-19
Cheng, H. Y., Chen, Y. F., and Peng, H. L. (2010). Molecular characterization of the PhoPQ-PmrD-PmrAB mediated pathway regulating polymyxin B resistance in Klebsiella pneumoniae CG43. J. Biomed. Sci. 17:60. doi: 10.1186/1423-0127-17-60
Cheng, P., Yang, Y., Cao, S., Liu, H., Li, X., Sun, J., et al. (2021). Prevalence and Characteristic of Swine-Origin mcr-1-Positive Escherichia coli in Northeastern China. Front. Microbiol. 12:712707. doi: 10.3389/fmicb.2021.712707
Choi, Y., and Chan, A. P. (2015). PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31, 2745–2747. doi: 10.1093/bioinformatics/btv195
Choi, Y., Sims, G. E., Murphy, S., Miller, J. R., and Chan, A. P. (2012). Predicting the functional effect of amino acid substitutions and indels. PLoS One 7:e46688. doi: 10.1371/journal.pone.0046688
Choo, S. W., Rayko, M., Tan, T. K., Hari, R., Komissarov, A., Wee, W. Y., et al. (2016). Pangolin genomes and the evolution of mammalian scales and immunity. Genome Res. 26, 1312–1322. doi: 10.1101/gr.203521.115
Ciesielczuk, H., Hornsey, M., Choi, V., Woodford, N., and Wareham, D. W. (2013). Development and evaluation of a multiplex PCR for eight plasmid-mediated quinolone-resistance determinants. J. Med. Microbiol. 62(Pt 12), 1823–1827. doi: 10.1099/jmm.0.064428-0
Dallenne, C., Da Costa, A., Decré, D., Favier, C., and Arlet, G. (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65, 490–495. doi: 10.1093/jac/dkp498
Delannoy, S., Le Devendec, L., Jouy, E., Fach, P., Drider, D., and Kempf, I. (2017). Characterization of Colistin-Resistant Escherichia coli Isolated from Diseased Pigs in France. Front. Microbiol. 8:2278. doi: 10.3389/fmicb.2017.02278
Gao, R., Hu, Y., Li, Z., Sun, J., Wang, Q., Lin, J., et al. (2016). Dissemination and Mechanism for the MCR-1 Colistin Resistance. PLoS Pathog. 12:e1005957. doi: 10.1371/journal.ppat.1005957
Hammerum, A. M., Sandvang, D., Andersen, S. R., Seyfarth, A. M., Porsbo, L. J., Frimodt-Møller, N., et al. (2006). Detection of sul1, sul2 and sul3 in sulphonamide resistant Escherichia coli isolates obtained from healthy humans, pork and pigs in Denmark. Int. J. Food Microbiol. 106, 235–237. doi: 10.1016/j.ijfoodmicro.2005.06.023
Higuchi, Y., Hashiguchi, A., Yuan, J., Yoshimura, A., Mitsui, J., Ishiura, H., et al. (2016). Mutations in MME cause an autosomal-recessive Charcot-Marie-Tooth disease type 2. Ann. Neurol. 79, 659–672. doi: 10.1002/ana.24612
Ji, K., Xu, Y., Sun, J., Huang, M., Jia, X., Jiang, C., et al. (2020). Harnessing efficient multiplex PCR methods to detect the expanding Tet(X) family of tigecycline resistance genes. Virulence 11, 49–56. doi: 10.1080/21505594.2019.1706913
Kai, J., and Wang, S. (2020). Recent progress on elucidating the molecular mechanism of plasmid-mediated colistin resistance and drug design. Int. Microbiol. 23, 355–366. doi: 10.1007/s10123-019-00112-1
Kandehkar Ghahraman, M. R., Hosseini-Nave, H., Azizi, O., Shakibaie, M. R., Mollaie, H. R., and Shakibaie, S. (2021). Stereochemical Trajectories of a Two-Component Regulatory System PmrA/B in a Colistin-Resistant Acinetobacter baumannii Clinical Isolate. Iran Biomed J. 25, 193–201. doi: 10.29252/ibj.25.3.157
Khezri, A., Avershina, E., and Ahmad, R. (2020). Plasmid Identification and Plasmid-Mediated Antimicrobial Gene Detection in Norwegian Isolates. Microorganisms 9:microorganisms9010052. doi: 10.3390/microorganisms9010052
Kim, S., Woo, J. H., Kim, N., Kim, M. H., Kim, S. Y., Son, J. H., et al. (2019). Characterization Of Chromosome-Mediated Colistin Resistance In Escherichia coli Isolates From Livestock In Korea. Infect. Drug Resist. 12, 3291–3299. doi: 10.2147/idr.S225383
Kozak, G. K., Boerlin, P., Janecko, N., Reid-Smith, R. J., and Jardine, C. (2009). Antimicrobial resistance in Escherichia coli isolates from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario, Canada. Appl. Environ. Microbiol. 75, 559–566. doi: 10.1128/aem.01821-08
Letunic, I., Doerks, T., and Bork, P. (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–D260. doi: 10.1093/nar/gku949
Lippa, A. M., and Goulian, M. (2009). Feedback inhibition in the PhoQ/PhoP signaling system by a membrane peptide. PLoS Genet. 5:e1000788. doi: 10.1371/journal.pgen.1000788
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/s1473-3099(15)00424-7
Lu, J., Zhang, J., Xu, L., Liu, Y., Li, P., Zhu, T., et al. (2018). Spread of the florfenicol resistance floR gene among clinical Klebsiella pneumoniae isolates in China. Antimicrob. Resist. Infect. Control 7:127. doi: 10.1186/s13756-018-0415-0
Luo, Q., Yu, W., Zhou, K., Guo, L., Shen, P., Lu, H., et al. (2017). Molecular Epidemiology and Colistin Resistant Mechanism of mcr-Positive and mcr-Negative Clinical Isolated Escherichia coli. Front. Microbiol. 8:2262. doi: 10.3389/fmicb.2017.02262
Ma, S., Lei, C., Kong, L., Jiang, W., Liu, B., Men, S., et al. (2017). Prevalence, Antimicrobial Resistance, and Relatedness of Salmonella Isolated from Chickens and Pigs on Farms, Abattoirs, and Markets in Sichuan Province, China. Foodborne Pathog. Dis. 14, 667–677. doi: 10.1089/fpd.2016.2264
Matamoros, S., van Hattem, J. M., Arcilla, M. S., Willemse, N., Melles, D. C., Penders, J., et al. (2017). Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci. Rep. 7:15364. doi: 10.1038/s41598-017-15539-7
Mendes Oliveira, V. R., Paiva, M. C., and Lima, W. G. (2019). Plasmid-mediated colistin resistance in Latin America and Caribbean: A systematic review. Travel Med. Infect. Dis. 31:101459. doi: 10.1016/j.tmaid.2019.07.015
Moffatt, J. H., Harper, M., Harrison, P., Hale, J. D., Vinogradov, E., Seemann, T., et al. (2010). Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 54, 4971–4977. doi: 10.1128/aac.00834-10
Nang, S. C., Li, J., and Velkov, T. (2019). The rise and spread of mcr plasmid-mediated polymyxin resistance. Crit. Rev. Microbiol. 45, 131–161. doi: 10.1080/1040841x.2018.1492902
Olaitan, A. O., Morand, S., and Rolain, J. M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. doi: 10.3389/fmicb.2014.00643
Padilla, E., Llobet, E., Doménech-Sánchez, A., Martínez-Martínez, L., Bengoechea, J. A., and Albertí, S. (2010). Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 54, 177–183. doi: 10.1128/aac.00715-09
Paterson, D. L., Isler, B., and Stewart, A. (2020). New treatment options for multiresistant gram negatives. Curr. Opin. Infect. Dis. 33, 214–223. doi: 10.1097/qco.0000000000000627
Poirel, L., Jayol, A., and Nordmann, P. (2017). Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/cmr.00064-16
Raetz, C. R., Reynolds, C. M., Trent, M. S., and Bishop, R. E. (2007). Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 76, 295–329. doi: 10.1146/annurev.biochem.76.010307.145803
Rapoport, M., Faccone, D., Pasteran, F., Ceriana, P., Albornoz, E., Petroni, A., et al. (2016). First Description of mcr-1-Mediated Colistin Resistance in Human Infections Caused by Escherichia coli in Latin America. Antimicrob. Agents Chemother. 60, 4412–4413. doi: 10.1128/aac.00573-16
Rolain, J. M., Kempf, M., Leangapichart, T., Chabou, S., Olaitan, A. O., Le Page, S., et al. (2016). Plasmid-Mediated mcr-1 Gene in Colistin-Resistant Clinical Isolates of Klebsiella pneumoniae in France and Laos. Antimicrob. Agents Chemother. 60, 6994–6995. doi: 10.1128/aac.00960-16
Routman, E., Miller, R. D., Phillips-Conroy, J., and Hartl, D. L. (1985). Antibiotic resistance and population structure in Escherichia coli from free-ranging African yellow baboons. Appl. Environ. Microbiol. 50, 749–754. doi: 10.1128/aem.50.4.749-754.1985
Rozwandowicz, M., Brouwer, M. S. M., Fischer, J., Wagenaar, J. A., Gonzalez-Zorn, B., Guerra, B., et al. (2018). Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 73, 1121–1137. doi: 10.1093/jac/dkx488
Sayah, R. S., Kaneene, J. B., Johnson, Y., and Miller, R. (2005). Patterns of antimicrobial resistance observed in Escherichia coli isolates obtained from domestic- and wild-animal fecal samples, human septage, and surface water. Appl. Environ. Microbiol. 71, 1394–1404. doi: 10.1128/aem.71.3.1394-1404.2005
Schultz, J., Milpetz, F., Bork, P., and Ponting, C. P. (1998). SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U S A. 95, 5857–5864. doi: 10.1073/pnas.95.11.5857
Schurek, K. N., Sampaio, J. L., Kiffer, C. R., Sinto, S., Mendes, C. M., and Hancock, R. E. (2009). Involvement of pmrAB and phoPQ in polymyxin B adaptation and inducible resistance in non-cystic fibrosis clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53, 4345–4351. doi: 10.1128/aac.01267-08
Shen, Y., Zhou, H., Xu, J., Wang, Y., Zhang, Q., Walsh, T. R., et al. (2018). Anthropogenic and environmental factors associated with high incidence of mcr-1 carriage in humans across China. Nat. Microbiol. 3, 1054–1062. doi: 10.1038/s41564-018-0205-8
Sonnevend, Á, Ghazawi, A., Alqahtani, M., Shibl, A., Jamal, W., Hashmey, R., et al. (2016). Plasmid-mediated colistin resistance in Escherichia coli from the Arabian Peninsula. Int. J. Infect. Dis. 50, 85–90. doi: 10.1016/j.ijid.2016.07.007
Stefaniuk, E. M., and Tyski, S. (2019). Colistin Resistance in Enterobacterales Strains - A Current View. Pol. J. Microbiol. 68, 417–427. doi: 10.33073/pjm-2019-055
Sun, J., Zhang, H., Liu, Y. H., and Feng, Y. (2018). Towards Understanding MCR-like Colistin Resistance. Trends Microbiol. 26, 794–808. doi: 10.1016/j.tim.2018.02.006
Tong, H., Liu, J., Yao, X., Jia, H., Wei, J., Shao, D., et al. (2018). High carriage rate of mcr-1 and antimicrobial resistance profiles of mcr-1-positive Escherichia coli isolates in swine faecal samples collected from eighteen provinces in China. Vet. Microbiol. 225, 53–57. doi: 10.1016/j.vetmic.2018.09.018
Wang, C. H., Siu, L. K., Chang, F. Y., Chiu, S. K., and Lin, J. C. (2021). A Resistance Mechanism in Non-mcr Colistin-Resistant Escherichia coli in Taiwan: R81H Substitution in PmrA Is an Independent Factor Contributing to Colistin Resistance. Microbiol. Spectr. 9:e0002221. doi: 10.1128/Spectrum.00022-21
Wang, C., Feng, Y., Liu, L., Wei, L., Kang, M., and Zong, Z. (2020). Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 9, 508–516. doi: 10.1080/22221751.2020.1732231
Wang, Q., Sun, J., Li, J., Ding, Y., Li, X. P., Lin, J., et al. (2017). Expanding landscapes of the diversified mcr-1-bearing plasmid reservoirs. Microbiome 5:70. doi: 10.1186/s40168-017-0288-0
Wang, X., Wang, Y., Zhou, Y., Li, J., Yin, W., Wang, S., et al. (2018). Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microbes Infect. 7:122. doi: 10.1038/s41426-018-0124-z
Wang, X., Wang, Y., Zhou, Y., Wang, Z., Wang, Y., Zhang, S., et al. (2019). Emergence of Colistin Resistance Gene mcr-8 and Its Variant in Raoultella ornithinolytica. Front. Microbiol. 10:228. doi: 10.3389/fmicb.2019.00228
Xavier, B. B., Lammens, C., Butaye, P., Goossens, H., and Malhotra-Kumar, S. (2016). Complete sequence of an IncFII plasmid harbouring the colistin resistance gene mcr-1 isolated from Belgian pig farms. J. Antimicrob. Chemother. 71, 2342–2344. doi: 10.1093/jac/dkw191
Xiaomin, S., Yiming, L., Yuying, Y., Zhangqi, S., Yongning, W., and Shaolin, W. (2020). Global impact of mcr-1-positive Enterobacteriaceae bacteria on “one health”. Crit. Rev. Microbiol. 46, 565–577. doi: 10.1080/1040841x.2020.1812510
Yang, Y. Q., Li, Y. X., Lei, C. W., Zhang, A. Y., and Wang, H. N. (2018). Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 73, 1791–1795. doi: 10.1093/jac/dky111
Yin, W., Li, H., Shen, Y., Liu, Z., Wang, S., Shen, Z., et al. (2017). Novel Plasmid-Mediated Colistin Resistance Gene mcr-3 in Escherichia coli. mBio 8:17. doi: 10.1128/mBio.00543-17
Yuan, J., Wang, X., Shi, D., Ge, Q., Song, X., Hu, W., et al. (2021). Extensive antimicrobial resistance and plasmid-carrying resistance genes in mcr-1-positive E. coli sampled in swine, in Guangxi, South China. BMC Vet. Res. 17:86. doi: 10.1186/s12917-021-02758-4
Zhang, S., Chen, S., Abbas, M., Wang, M., Jia, R., Chen, S., et al. (2021). High incidence of multi-drug resistance and heterogeneity of mobile genetic elements in Escherichia coli isolates from diseased ducks in Sichuan province of China. Ecotoxicol. Environ. Saf. 222:112475. doi: 10.1016/j.ecoenv.2021.112475
Zhang, X., Zhang, B., Guo, Y., Wang, J., Zhao, P., Liu, J., et al. (2019). Colistin resistance prevalence in Escherichia coli from domestic animals in intensive breeding farms of Jiangsu Province. Int. J. Food Microbiol. 291, 87–90. doi: 10.1016/j.ijfoodmicro.2018.11.013
Zhao, F., Feng, Y., Lü, X., McNally, A., and Zong, Z. (2017). IncP Plasmid Carrying Colistin Resistance Gene mcr-1 in Klebsiella pneumoniae from Hospital Sewage. Antimicrob. Agents Chemother. 61:16. doi: 10.1128/aac.02229-16
Zhuge, X., Ji, Y., Tang, F., Sun, Y., Jiang, M., Hu, W., et al. (2019). Population structure and antimicrobial resistance traits of avian-origin mcr-1-positive Escherichia coli in Eastern China, 2015 to 2017. Transbound Emerg. Dis. 66, 1920–1929. doi: 10.1111/tbed.13222
Zurfluh, K., Klumpp, J., Nüesch-Inderbinen, M., and Stephan, R. (2016). Full-Length Nucleotide Sequences of mcr-1-Harboring Plasmids Isolated from Extended-Spectrum-β-Lactamase-Producing Escherichia coli Isolates of Different Origins. Antimicrob. Agents Chemother. 60, 5589–5591. doi: 10.1128/aac.00935-16
Keywords: colistin resistant mechanism, mcr, Escherichia coli, multidrug resistant, two-component systems
Citation: Li F, Cheng P, Li X, Liu R, Liu H and Zhang X (2022) Molecular Epidemiology and Colistin-Resistant Mechanism of mcr-Positive and mcr-Negative Escherichia coli Isolated From Animal in Sichuan Province, China. Front. Microbiol. 13:818548. doi: 10.3389/fmicb.2022.818548
Received: 19 November 2021; Accepted: 02 February 2022;
Published: 29 March 2022.
Edited by:
Ziad Daoud, Central Michigan University, United StatesReviewed by:
Thongpan Leangapichart, Norwegian Veterinary Institute (NVI), NorwayChristopher Morton Thomas, University of Birmingham, United Kingdom
Copyright © 2022 Li, Cheng, Li, Liu, Liu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiuying Zhang, emhhbmd4aXV5aW5nQG5lYXUuZWR1LmNu
 Fulei Li
Fulei Li Ping Cheng
Ping Cheng Xiaoting Li
Xiaoting Li Ruimeng Liu
Ruimeng Liu Haibin Liu
Haibin Liu Xiuying Zhang
Xiuying Zhang