
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 01 March 2022
Sec. Infectious Agents and Disease
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.818398
 Kohei Ogura1
Kohei Ogura1 Hiroka Furuya2†
Hiroka Furuya2† Natsuki Takahashi1†
Natsuki Takahashi1† Kana Shibata1
Kana Shibata1 Maho Endo1
Maho Endo1 Shinya Watanabe3
Shinya Watanabe3 Longzhu Cui3
Longzhu Cui3 Tohru Miyoshi-Akiyama4
Tohru Miyoshi-Akiyama4 Shigefumi Okamoto1,2
Shigefumi Okamoto1,2 Kazuhiro Ogai5*
Kazuhiro Ogai5* Junko Sugama1,6*
Junko Sugama1,6*
Staphylococcus spp. colonize commensally on the human skin. Some commensal coagulase-negative staphylococci and Staphylococcus aureus are also involved in nosocomial infections. Bacteria were collected from skin healed from pressure injury (PI). After the collection time points, some patients suffered from recurrent PI (RPI). This study analyzed the characteristics of Staphylococcus spp. on healed skin before recurrence between healed skin that suffered from RPI within 6 weeks (RPI group) and healed skin that did not suffer within the duration (non-RPI group) by Staphylococcus spp.-specific sequencing. Of the seven patients in the RPI group, two were dominated by S. aureus and four by Staphylococcus caprae, coagulase-negative human commensal staphylococci in the RPI group. Using mouse models, both S. caprae and S. aureus, but not Staphylococcus epidermidis, colonized on skin healed from injury at significantly higher rates than normal skin. Although subcutaneous injection of S. caprae did not induce lesion formation, the bacterium exhibited high hemolytic activity on human red blood cells. Lesion formation by subcutaneous injection of S. aureus was significantly suppressed in the presence of S. caprae. The hemolytic activity of rabbit blood cells of S. aureus was suppressed by S. caprae, whereas the hemolytic activity of S. caprae was dramatically suppressed by S. aureus. Data indicated that each of the two Staphylococcus spp. suppresses the pathogenicity of the other and that the imbalance between the two is associated with RPI.
The skin is covered with numerous microorganisms, mainly bacteria (Kong, 2011). Bacteria reside on the epidermis and form commensal microbiota (Byrd et al., 2018). The skin microbiome is affected by skin microbiotopes, such as sebaceous, moist, and dry environments, and host factors, such as age and gender (Grice and Segre, 2011; Oh et al., 2014; Shibagaki et al., 2017). Although the skin’s microbial composition is not usually changed for a duration of years (Oh et al., 2016; Ogai et al., 2021a), in case of damage to the skin’s barrier function, the microbiome has an imbalance, followed by an increase of pathogenic bacteria, leading to cutaneous inflammatory disorders (Belkaid and Segre, 2014; Naik et al., 2015). For example, recent studies have shown that the skin microbiome is associated with diabetic foot ulcer, one of the major types of chronic wounds (Kalan and Brennan, 2019). On the diabetic foot ulcer, Staphylococcus spp. and Corynebacterium spp. tend to be highly prevalent, followed by mixed anaerobic communities (Loesche et al., 2017). Grice et al. (2010) found in mouse models that a longitudinal shift in the diabetic wound microbiota is correlated with prolonged skin defense response.
During aging, resident immune cells such as macrophages (Langerhans cells) and dendritic epidermal T cells become easily damaged, affecting the normal organization of the skin and its capacity for repair. Numerous mechanisms are involved in age-induced skin degradation; they also relate to non-healing or chronic wounds in the elderly (Farage et al., 2013; Blume-Peytavi et al., 2016; Russell-Goldman and Murphy, 2020). Pressure injuries (PIs) or pressure ulcers are clinical problems in bedridden people and develop by prolonged mechanical loading on the skin over a bony prominence, mostly at the sacral area (Schank, 2015). The mean incidence of PIs in Japanese long-term care hospitals is low (1.9%; Igarashi et al., 2013). In contrast to the low frequency of primary PI incidence, recurrent PIs (RPIs) are much higher, from 25.9% (Arisandi et al., 2020) to 26.7% (Shibata et al., 2021), in long-term care hospitals, although preventive care is constantly provided. Therefore, besides the occurrence of PIs, RPIs are deemed another clinical problem in a rapidly growing aging society.
In order to find out new factors associated with RPI, we have been focusing on skin microbiome. In a previous study, 16S rRNA-based sequencing of the healed site microbiome revealed significantly lower alpha diversity (Shannon index) and higher rates of Staphylococcus spp. in the RPI group than in the non-RPI group (Shibata et al., 2021). Since Staphylococcus spp. contain S. aureus, the dominant bacterial skin pathogen, we had suspected dominance of this bacterium on the healed sites which subsequently suffered from RPI. However, quantitative polymerase chain reaction (qPCR) showed that S. aureus did not show a significant difference between the groups, suggesting the involvement of other coagulase-negative staphylococci (CoNS). It is well known that CoNS could be involved in skin protections. For example, S. epidermidis and S. hominis produce antimicrobial peptides (AMPs) that synergize with the human cathelicidin AMP LL-37 and suppress colonization of S. aureus on human skins with atopic dermatitis (Nakatsuji et al., 2017). S. lugdunensis also inhibits S. aureus growth by producing AMP called lugdunin (Zipperer et al., 2016). In addition, recent reports showed that CoNS primes the skin immune system to limit the colonization of potential invaders (Parlet et al., 2019). In contrast, CoNS also could cause pathogenicity (Hatlen and Miller, 2021). Some S. epidermidis strains produce EspA proteases, which involve skin damages, while the amounts of the production vary among the strains (Cau et al., 2021). While S. lugdunensis inhibits S. aureus growth as mentioned above (Zipperer et al., 2016), this bacterium also possesses invasive pathogenic potential (Heilbronner and Foster, 2021).
These reports suggest that inter-species regulation of Staphylococcus spp. and mechanisms of pathogenicity are complicated. In order to analyze and clarify the relationships, identification of Staphylococcus spp. on the patients’ skins was required. Although 16S rRNA metataxonomics targeting the V3–V4 regions has been widely used for the determination of microbial populations (Wang et al., 2007; Claesson et al., 2010; Willis et al., 2019), this method lacks a species-level resolution; thus, the study could not identify which Staphylococcus spp. are related to RPIs. To determine Staphylococcus spp. associated with RPIs, this study conducted a sequencing method called single-locus sequence typing (SLST), recently developed by Scholz et al. for the strain-level identification of Propionibacterium acnes (Scholz et al., 2014; Scholz and Jensen, 2017) and utilized for Staphylococcus profiling on the skin (Smits et al., 2020). Based on the SLST profiling results of the healed site microbiome, Staphylococcus caprae, a coagulase-negative staphylococcus, and S. aureus reside on healed PIs. Although S. caprae has been regarded as a commensal bacterium in goat milk, the bacterium has been isolated from a patient with invasive infectious disease (Watanabe et al., 2018). Although it is known that hemolysin production of S. aureus is suppressed by AIP produced by goat milk-isolated S. caprae (Paharik et al., 2017), the pathogenicity of human-isolated S. caprae and cross-talk between the S. caprae and S. aureus have not been not fully clarified. This study presented the association of these two Staphylococcus spp. and the implications of these bacteria in the deterioration of skin integrity.
In this study, first we aimed to identify Staphylococcus spp. colonized on the skins healed from a PI. In order to analyze the pathogenicity of human-isolated S. caprae and cross-talk between the S. caprae and S. aureus, next we conducted some experiments such as mouse infection models and in vitro hemolysis assay.
A prospective cohort study was conducted in a long-term care hospital with 500 beds in Ishikawa Prefecture (Japan) from 2018 to 2019. Skin samples were collected from 30 patients who had healed from a PI within 1 month, although one sample was excluded because it was obtained immediately after bathing. After sample collection, whether the RPI (lesion or redness) was present at the sites where the samples were collected was assessed every week. Within 6 weeks, seven from a total of 29 patients suffered from RPI (Shibata et al., 2021). Written informed consent was obtained from the participants or her/his guardians before inclusion in the study. Patients who had a skin disease, hypertrophic scars, keloids, or other PIs were excluded. Further information on the participants was described in a previous report (Shibata et al., 2021).
DNA samples from the 29 patients were obtained by attaching an air-permeable medical tape with urethane glue to a healed PI site or a control site, according to a previous report (Ogai et al., 2021b). The control site was opposite the healed PI site when the healed PI was present on a trochanter or ∼5 cm superior to the healed PI site when present on the sacrum or coccyx (Arisandi et al., 2020). Sequence data of 16S rRNA genes (V3–V4 regions) were obtained in a previous report [DNA Data Bank of Japan (DDBJ) accession no. DRA009678 (Shibata et al., 2021)]. To determine Staphylococcus spp. associated with RPI, this study conducted SLST, a recently developed sequencing method that can type and profile the targeted bacterial family beyond the species level (Ederveen et al., 2019). This study selected a Staphylococcus-specific variable region in a gene encoding 30S ribosomal protein S11, which was chosen by a TaxPhlAn pipeline using 7,247 Staphylococcus genomes, for an SLST target. The regions were amplified by KAPA HiFi HS ReadyMix (Roche, Basel, Swiss) and a pair of SLST PCR primers (Supplementary Table 1), followed by purification by AMPure XP magnetic beads (Beckman Coulter, Brea, CA, United States). After the addition of index sequences using the Nextera XT Index Kit version 2 (Illumina, San Diego, CA, United States) and subsequent purification by AMPure, the equimolar mixtures of the products were applied for MiSeq sequencing with the MiSeq Reagent Kit version 3 (Illumina) and 15% of PhiX Control version 3 (Illumina). Sequence data were deposited to the DDBJ (accession no. DRA013063).
The 300-bp pair-end reads (fastq data) were filtered (QC > 20) and merged by the fastp tool (Chen et al., 2018), followed by conversion into FASTA-format sequence data by the seqtk tool.1 Sequence data were applied to the TaxPhlAn pipeline (Ederveen et al., 2019). The alleles were built with a Shannon diversity index threshold of 0.6. Unassigned alleles were manually extracted and applied to nucleotide BLAST in the National Center for Biotechnology Information website and regarded as those derived from species with the highest scores.
A 16S rRNA metataxonomic analysis was previously conducted for identical samples, and the abundance of Staphylococcus spp. in the microbiota was determined (Shibata et al., 2021). 16S rRNA and SLST data were assembled by adjusting the abundance of each Staphylococcus spp., which were obtained by SLST, to the abundance of Staphylococcus spp. in the whole microbiota obtained by 16S rRNA metataxonomics (Supplementary Figure 2).
Staphylococcus caprae ATCC 35538 [strain designation: 143.22 (CCM 3573)] (Devriese et al., 1983), derived from goat milk, and Staphylococcus epidermidis ATCC 14990 [strain designation: Fussel (NCTC 11047, R. Hugh 2466)] were obtained from the American Type Culture Collection. S. caprae JMUB145 was isolated from the blood culture of a woman with fever during chemotherapy treatment for uterine cancer at Jichi Medical University Hospital in Japan (Watanabe et al., 2018). S. aureus N315 (agr type II) was kindly provided by Dr. Keiichi Hiramatsu (Kuroda et al., 2001).
After incubation in TE buffer containing lysostaphin and lysozyme at 37°C for 30 min, genomic DNA was extracted by the QIAamp DNA mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
To analyze the allele types of the agrD gene in the clinical samples, qPCR was conducted using QuantiFast Probe PCR kits (Qiagen), primers, and probes described (Supplementary Table 1) in the AriaMx Real-time PCR System (Agilent Technologies, Santa Clara, CA, United States). Before the analysis, amplification specificity was validated using the indicated copy numbers of ATCC35538 and JMUB145 genomes that possess the agrD_YSTCSYYF and agrD_YRTCNTYF genes, respectively. To prepare standards, genomic DNA was extracted using QIAamp DNA mini kit according to the manufacturer’s instruction. Double-stranded DNA (dsDNA) concentrations were measured using the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA, United States).
Five-week-old female BALB/c mice (Sankyo Laboratory Service Corporation, Tokyo, Japan) were anesthetized through intraperitoneal administration of a mixture of Dormicum (4 mg/kg; Astellas Pharma, Tokyo, Japan), Vetorphale (5 mg/kg; Meiji Seika Pharma Co., Ltd., Tokyo, Japan), and Domitor (0.3 mg/kg; Nippon Zenyaku Kogyo Co., Ltd., Fukushima, Japan), followed by disinfection of mouse backs by ethanol. A circular full-thickness wound (4 mm in diameter), including the panniculus carnosus muscle, was made using a sterile biopsy punch (Kai Industries Co., Ltd., Gifu, Japan). Seven days after wound formation, the backs of control (no surgery) and healed mice were shaved under anesthesia by the Dormicum/Vetorphale/Domitor mixture. A 10 μL aliquot containing no bacteria [negative control for counting colony-forming units (CFUs) of commensal bacteria], 2.0 × 107 CFUs of S. aureus N315, 3.2 × 107 CFUs of S. caprae JMUB145, or 3.5 × 107 CFUs of S. epidermidis ATCC 14990 [strain designation: Fussel (NCTC 11047, R. Hugh 2466)] in phosphate-buffered saline (PBS) was inoculated directly onto the scabs or control sites by a micropipette. The droplets were confirmed to be dried under anesthesia. On Day 2 post-infection, mice were euthanized with sevoflurane (Maruishi Pharmaceutical Co., Ltd., Japan). The skin (∼1 × 1 cm) was collected by sterile scissors, suspended in sterile PBS, and mixed vigorously by a vortex mixer.
Eight-week-old female BALB/c mice were anesthetized through intraperitoneal administration of the Dormicum/Vetorphale/Domitor mixture. Mouse backs were shaved and disinfected by ethanol. A 100-μl bacterial solution in PBS was injected to the subcutis in two separate experiments in triplicate for each group. The S. aureus strain N315 (1.3 × 108–1.9 × 108 CFU/mouse), S. caprae strain JMUB145 (1.3 × 108–1.9 × 108 CFU/mouse), S. caprae strain ATCC 35538 (1.1 × 108–1.5 × 108 CFU/mouse), S. epidermidis ATCC strain 14490 (1.9 × 108–2.7 × 108 CFU/mouse), and coinjection of S. aureus strain N315 and S. caprae strain JMUB145 were tested. The size and color intensity of lesions were calculated by ImageJ. To quantify the intensity, image files (pictures) were converted to grayscale, the lesions were selected by drawing/selection tools, and the area-integrated intensity was calculated.
Using normal skin with the same size (1 cm apart from the selected area) as the control, the intensity was calculated as (intensity of the lesion)/(intensity of the normal skin).
Staphylococcus aureus N315, S. caprae JMUB145, and S. caprae ATCC 35538 were cultured in tryptic soy broth (TSB; Becton Dickinson, United State) overnight at 37°C. After centrifugation at 6,000 × g at 10 min, spent medium (supernatant) was collected. Cultured bacteria were suspended in fresh TSB or TSB containing spent medium (final 10%) at a density of OD600nm = 0.001. After overnight culture and centrifugation at 6,000 × g at 10 min, spent media were utilized for analysis.
Human blood was obtained from 22 to 38-year-old healthy volunteers. Human whole blood was collected in EDTA-containing tubes and centrifuged at 1,500 × g at 20 min. After washing with PBS thrice, cells were suspended at 20% (v/v) in PBS containing 0.5% bovine serum albumin (BSA; FujiFilm Wako Pure Chemical Corporation, Osaka, Japan). Rabbit whole blood in Alserver’s solution (Kohjin-Bio, Saitama, Japan) was centrifuged at 1,200 × g at 20 min. Rabbit blood cells were washed with PBS containing 0.1% BSA four times and suspended at 20% (v/v) in PBS containing 0.5% BSA.
The human or rabbit red blood cell (RBC) solution (200 μL) was incubated with 20 μL spent medium or fresh TSB (for blank control) for 6 h (human) or 18 h (rabbit). After centrifugation at 1,500 × g at 20 min, the absorbance of the supernatants was measured at a wavelength of 492 nm.
The spent medium was obtained after overnight culture, centrifuged at 6,000 × g at 10 min, and filtered. S. aureus N315 and S. caprae JMUB145 were suspended in fresh TSB or TSB containing spent medium (final 10%) at a density of OD600nm = 0.001 and cultured at 37°C with shaking for 10 h.
The clinical study (Kanazawa University no. 44) and animal experiments (Kanazawa University no. AP-214236) were approved by the Medical Ethics Committee of Kanazawa University and conducted in accordance with the Declaration of Helsinki and the Microorganism Safety Management Regulations of Kanazawa University.
We focused on skin microbiome associated with RPI. In a previous report, 16S rRNA metataxonomics revealed that the abundance of Staphylococcus spp. was significantly higher in RPI-healed sites than non-RPI-healed sites (p = 0.002, Mann–Whitney U test) (Shibata et al., 2021) and that the abundance was also significantly in the healed sites (including RPI and non-RPI) than the control sites (p < 0.01, Welch’s t-test), indicating composition of staphylococci is involved in RPI. In this study, Staphylococcus spp. were identified using a recently developed sequencing method named SLST. A Staphylococcus-specific region in a gene encoding 30S ribosomal protein S11 was amplified by PCR and sequenced by the MiSeq system. After merging pair-end reads, removing low-quality reads, and converting read data to multi-FASTA format, data were applied to the TaxPhlAn pipeline (Ederveen et al., 2019). The pipeline gave numbers of sequences derived from each Staphylococcus. The abundance of each Staphylococcus was adjusted to that of Staphylococcus spp. obtained by 16S rRNA sequencing in the previous report (Figure 1 and Supplementary Figures 2, 3). Staphylococcus spp. were mainly dominated by S. caprae and S. aureus, whereas the abundance of S. epidermidis was low. The abundance of S. caprae was significantly higher in healed sites than control sites (p < 0.01, Welch’s t-test; Figure 2A). The abundance of S. caprae was also significantly higher RPI-healed sites than non-RPI-healed sites (p = 0.01, Kruskal–Wallis test and Bonferroni correction; Figure 2B). S. aureus was high in healed sites (p = 0.07, Welch’s t-test; Figure 2C). The multiple comparison procedure (Kruskal–Wallis test) did not detect significance among the four groups (Figure 2D). There was no significant difference in the abundance s of Staphylococcus haemolyticus or S. epidermidis (Supplementary Figure 4). As shown in our previous report, there was no significant difference in the abundance of Corynebacterium spp. between RPI-healed sites and non-RPI-healed sites (Shibata et al., 2021) but RPI-control sites and RPI-healed sites (p = 0.02, Kruskal–Wallis test and Bonferroni correction) (Supplementary Figure 4).
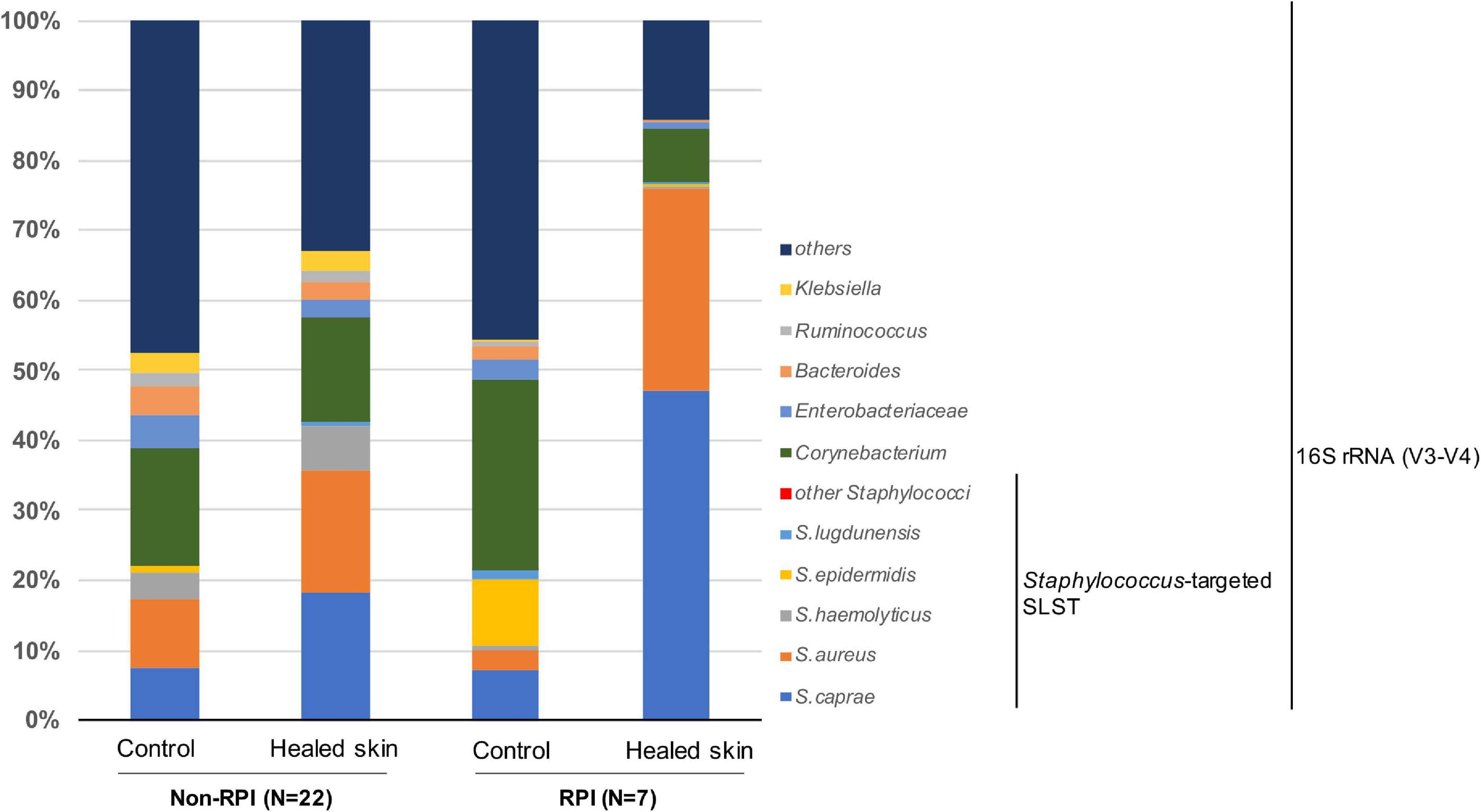
Figure 1. Composition of Staphylococcus spp. and the top five skin bacteria on the skin sites. Average abundance was shown for the control site of non-RPI patients (n = 22), healed skin sites of non-RPI patients (n = 22), control sites of RPI patients (n = 7), and healed skin sites of RPI patients (n = 7). The full sets of staphylococci are shown in Supplementary Figure 2. The composition of each sample is shown in Supplementary Figure 3.
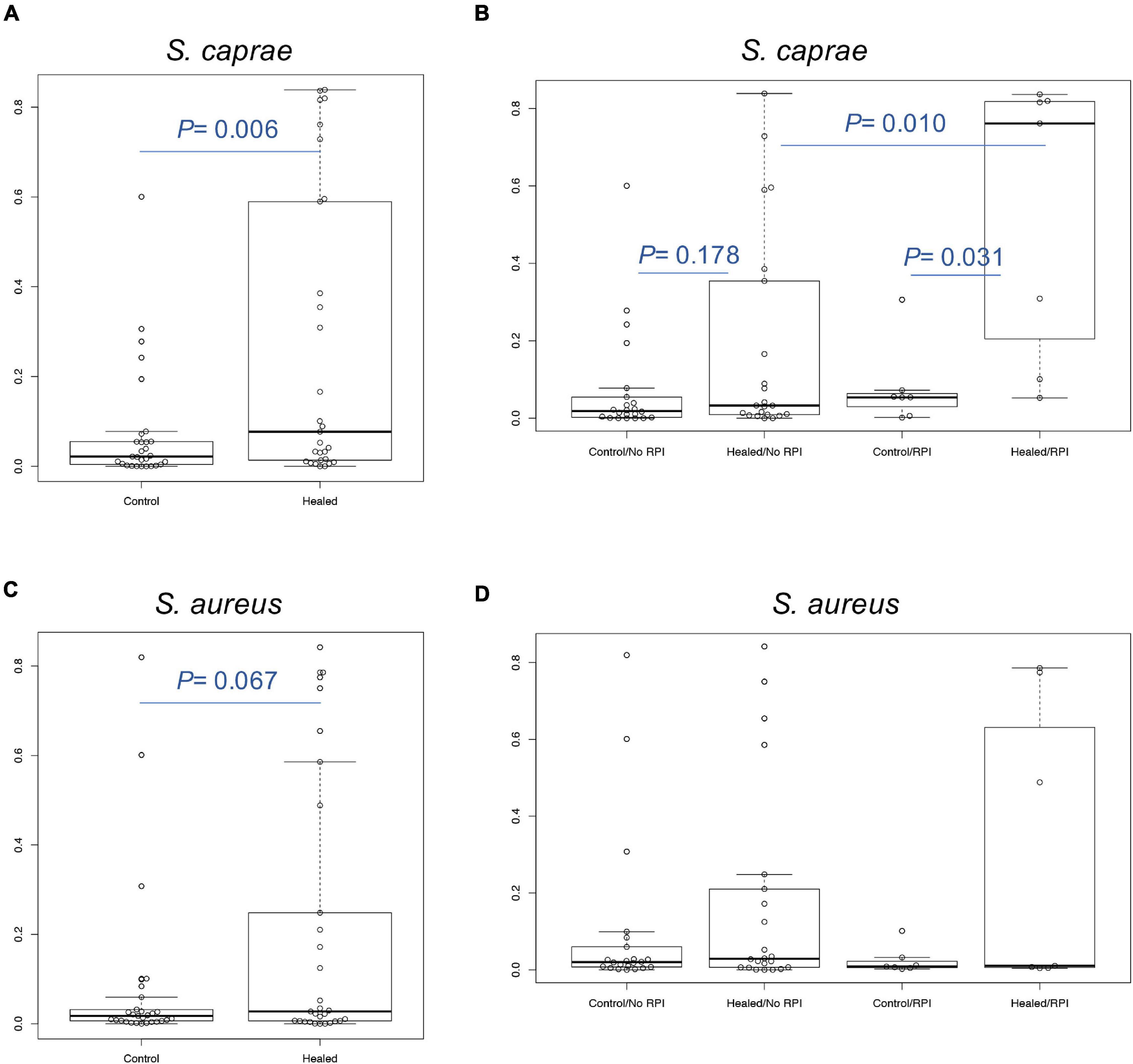
Figure 2. Relative abundance of S. caprae and S. aureus. Comparison of (A) S. caprae and (C) S. aureus abundance between control sites (including non-RPI and RPI) and healed sites. P-values were calculated by Welch’s t-test. (B) Comparison of S. caprae and the four groups. After the Kruskal–Wallis test (p = 0.006), Bonferroni correction was calculated for statistical significance. (D) Comparison of S. aureus and the four groups. No significance was detected by the Kruskal–Wallis test.
This study observed the Staphylococcus composition of the seven healed skins that suffered recurrence 1–6 weeks later. The dominance of S. caprae was found in four of the seven patients, whereas that of S. aureus was found in the other two samples (RS #6 and #7 in Figure 3). In one patient’s skin (RS #5) that suffered recurrence in 4 weeks, both S. caprae and S. aureus colonized. Patients who had the S. aureus-dominant microbiome suffered from RPI within 1 week, whereas those with the S. caprae-dominant one did after 2–6 weeks. There were no significant characteristics in the patient information.
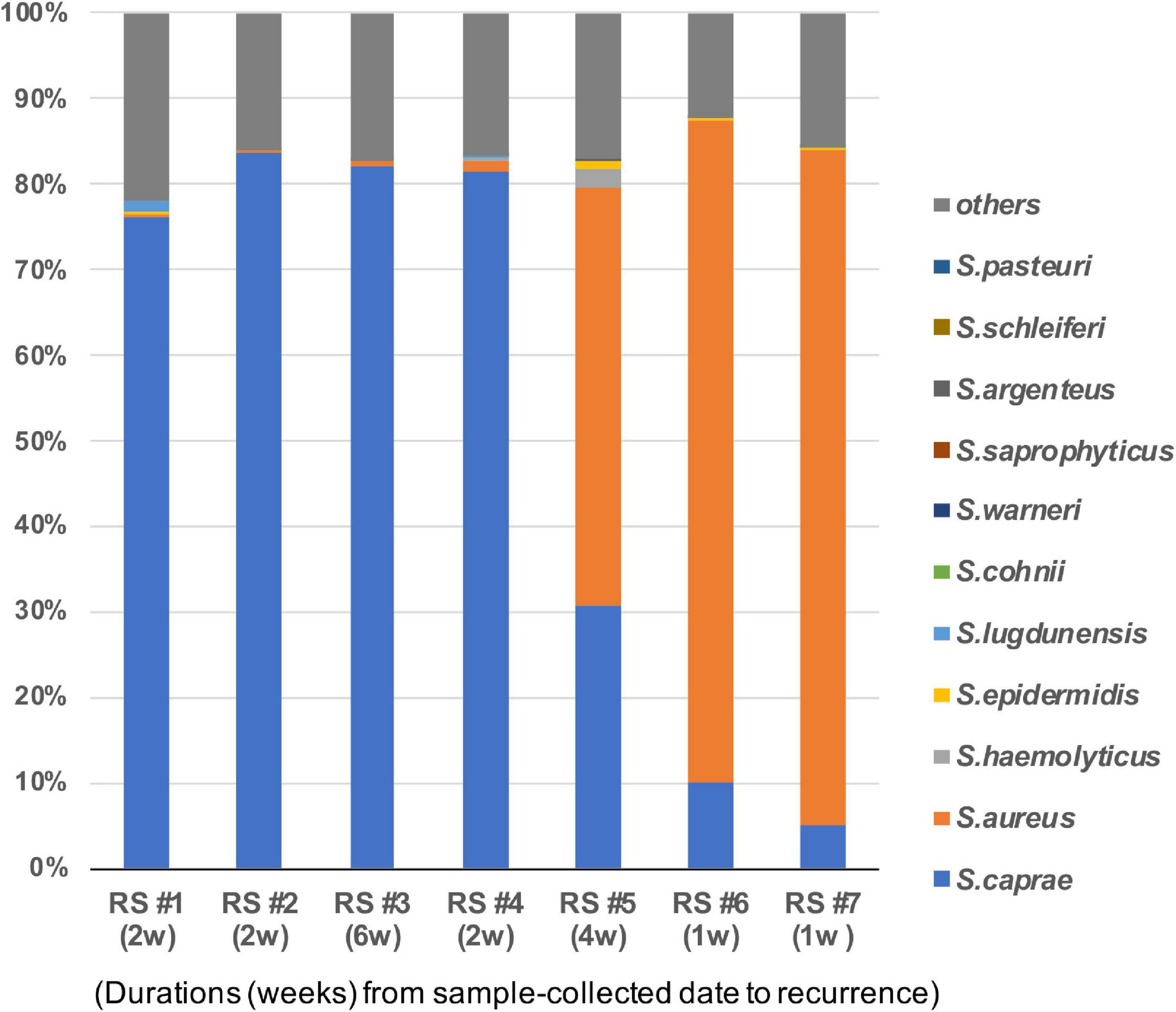
Figure 3. Staphylococcus caprae and S. aureus on healed skin of the seven patients who suffered from RPI within 1–6 weeks. Bacterial composition of the seven sites. Duration from the sample collection date to RPI is described in parentheses. The correlation of the abundance of S. aureus and S. caprae is shown in Supplementary Figure 4.
Staphylococcus caprae, CoNS, usually colonize goat skin and milk and occasionally cause goat mastitis (Devriese et al., 1983). Recent reports showed that S. caprae also reside as human commensal bacteria and occasionally cause nosocomial infections (Seng et al., 2014). Two S. caprae strains ATCC 35538 and JMUB145, isolated from goat and human, respectively, showed 98.5% average nucleotide identity (Yoon et al., 2017), indicating that their characteristics were slightly different. Some of staphylococci such as S. aureus is typed by an accessory-gene-regulator (agr) gene sequence (Dufour et al., 2002). Paharik et al. (2017) reported that S. caprae from goat milk possesses an agr gene encoding autoinducing peptide YSTCSYYF. Watanabe et al. (2018) reported that clinical isolate S. caprae possesses another allele type of the agrD gene encoding YRTCNTYF (Figure 4A), although mature structures of the peptides have not been confirmed yet. To determine which type of S. caprae was found in the healed ulcers, qPCR was conducted to count the copy numbers of agrD_YSTCSYYF and agrD_YRTCNTYF genes (Supplementary Figure 6). Most agrD genes in the samples were agrD_YRTCNTYF in healed sites (Figure 4B), indicating that genotypes of S. caprae strains in the clinical samples are similar to S. caprae JMUB145 rather than ATCC 35538. Thereafter, this study mainly utilized the human isolate S. caprae JMUB145 for analysis.

Figure 4. Staphylococcus caprae typing by AgrD genes in the clinical samples. (A) Different amino acid sequences of AgrD from ATCC 35538 (goat milk) and JMUB145 (clinical isolate), although mature structures of the peptides have not been confirmed yet. (B) Copy numbers were calculated by probe qPCR using the serial dilution of purified chromosomal DNAs of ATCC 35538 (agrD_YSTCSYYF) and JMUB145 (agrD_YRTCNTYF). Relative copy numbers were calculated as (agrD_YSTCSYYF or agrD_YRTCNTYF copy numbers)/[(agrD_YSTCSYYF copy numbers) + (agrD_YRTCNTYF copy numbers)].
This study indicated that S. caprae and S. aureus colonized on the healed PI site. To examine the ability of colonization on the skin after wound healing, a full-thickness wound on the back of mice was made, and 7 days were allowed to pass for the wound to heal (Figure 5A). Using this skin scab model, PBS containing S. caprae, S. aureus, and S. epidermidis were spotted (inoculated) onto the control (i.e., uninjured) and scab sites. The colonized bacteria were collected and counted on Day 2 postinoculation. S. epidermidis (strain ATCC 14990) colonized on the control skin at a significantly higher ratio than skin scab (p < 0.05, one-way analysis of variance) than S. aureus (agr type II strain N315) and S. caprae (strain JMUB145), although the ratio was not altered when this bacterium was inoculated on skin scab (Figure 5B). In contrast, S. aureus N315 and S. caprae JMUB145 colonized on skin scab at a significantly higher ratio than control skin (p < 0.05, Student’s t-test), suggesting that both S. aureus and S. caprae prefer colonization on healed skin rather than on the normal skin.
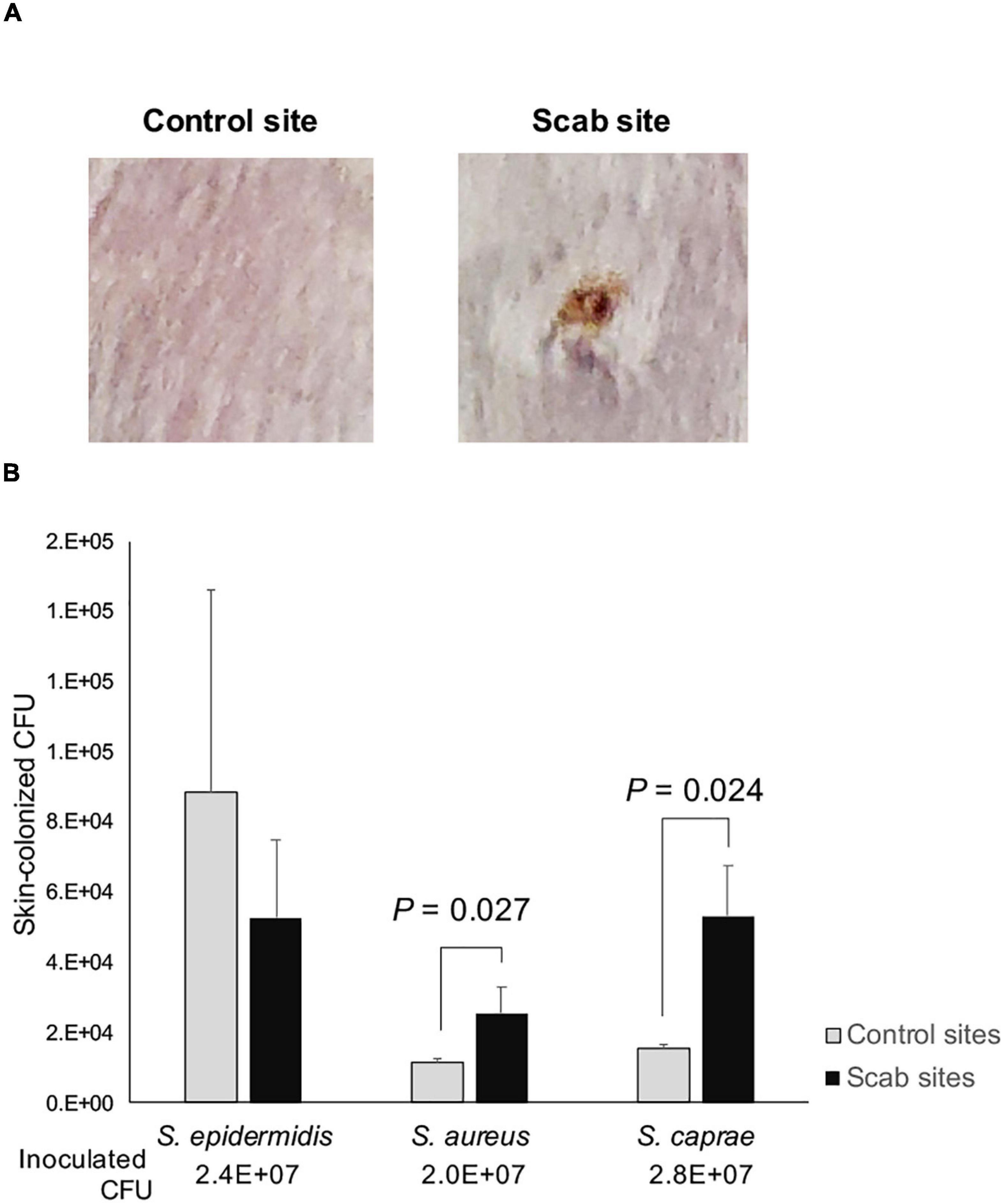
Figure 5. Skin colonization of staphylococci. (A) Representative images of control and healed skin. To prepare skin healed from wounds, mouse skin was punched out, and wounds were left for 1 week. (B) Colonized bacteria. On Day 2 post-infection, skin (1 × 1 cm) was suspended in PBS and vigorously mixed by vortexing. The relative CFU values were calculated as (CFUs of colonized bacteria)/(CFUs of infected bacteria). p-Values were calculated by Student’s t-test.
The mouse model revealed that both S. caprae and S. aureus could colonize on healed skin (Figure 5). However, it remained unknown whether the dominance of these bacteria is directly associated with the deterioration of skin integrity, such as RPIs. In the colonization experiments (Figure 5), there was no significant inflammation or ulcer recurrence at the bacteria-inoculated healed skin, suggesting that another model is required to be developed to resemble RPI. Although several reports have shown how to mimic PI in animal models, it is still very difficult to mimic the RPI situation with animal models. Thus, this study analyzed the pathogenicity of S. caprae and S. aureus by a subcutaneous injection model. Subcutaneous injection of S. aureus strain N315 resulted in lesion formation with the largest size and highest color intensity on Day 4 (Figure 6 and Supplementary Figure 7). In contrast, injection of S. caprae JMUB145 alone or S. caprae ATCC 35538 alone did not result in lesion formation. When S. aureus N315 and S. caprae JMUB145 were co-injected, the resultant lesion sizes were significantly smaller than those caused by S. aureus alone (Figure 6).
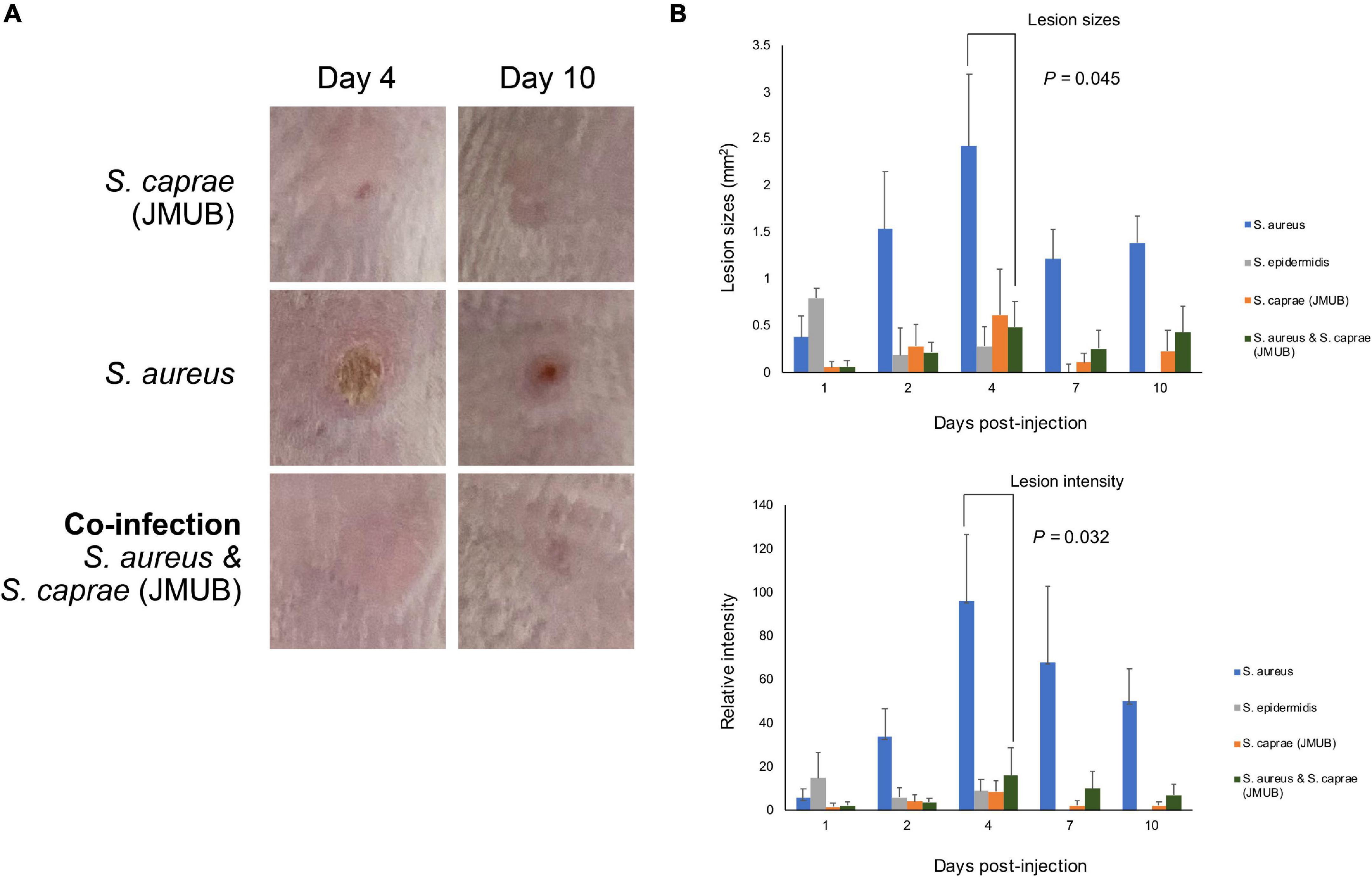
Figure 6. Subcutaneous injection. (A) Representative images of lesions after subcutaneous injection. Picture sizes are 1 × 1 cm. The other images are shown in Supplementary Figure 5. (B) Lesion sizes and intensity. The color intensity was calculated by ImageJ using normal skin (1 cm apart from injured sites) as control (=1). p-Values were calculated by Student’s t-test.
α-Hemolysin (Hla) plays a key virulence factor in the pathogenicity of S. aureus (Bayer et al., 1997). Regarding the suppression of Hla production, Paharik et al. (2017) reported that an autoinducer peptide composed of eight amino acids (YSTCSYYF) with a C-terminal five-membered thiolactone ring produced by S. caprae from goat milk could suppress Hla production and lesion formation of S. aureus on mouse skin. Their result was consistent with this study. Watanabe et al. (2018) reported the complete genome sequences of three S. caprae strains isolated from patients with nosocomial infections in Japan. Inconsistent with the report by Paharik et al. (2017) the three S. caprae strains isolated in Japan, including strain JMUB145, possessed agrD genes that encode different eight amino acids (YRTCNTYF; Figure 4A). To examine whether the two S. caprae agrD peptides (YSTCSYYF and YRTCNTYF) exhibit different characteristics to Hla production by S. aureus, this study tested the effects of AgrD peptides from S. caprae strains ATCC 35538 (agrD_YSTCSYYF) and JMUB145 (agrD_YRTCNTYF) on the hemolysis of rabbit RBCs by S. aureus. Spent media obtained from overnight TSB culture was utilized for hemolytic assay. Bacteria were also cultured in the presence of 10% spent medium. Hemolytic activity was tested using spent medium. As shown in Figure 7A, S. aureus N315 exhibited hemolytic activity on rabbit RBCs. The hemolytic activity was significantly enhanced when S. aureus N315 was cultured in the presence of 10% S. aureus N315 spent medium (P = 0.003, Student’s t-test). In contrast, the hemolytic activity using spent medium was partially suppressed by culture in the presence of 10% S. caprae JMUB145 (p < 0.001) but not ATCC 35538 (p = 0.898), indicating that ability of suppressing Hla production by S. aureus is different between the two S. caprae strains. In the previous report by Paharik et al. (2017) half maximal effective concentration (EC50) was shown, indicating that concentration might be required to examine the inhibitory effects of the ATCC 35538 supernatants.
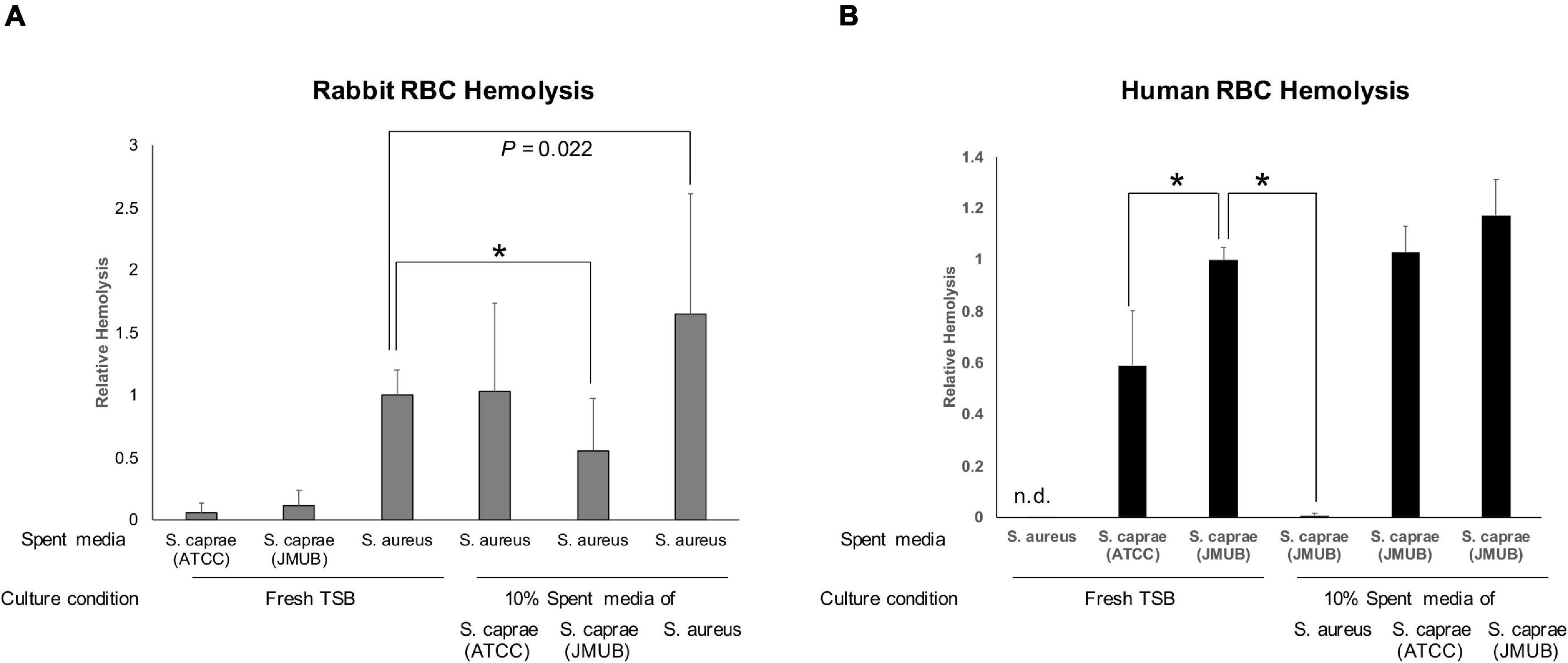
Figure 7. Hemolysis by spent medium. Autoinducer peptides are indicated by underlines. (A) Lysis of rabbit blood cells. Fresh or spent medium of S. aureus N315, S. caprae ATCC 35538, or S. caprae JMUB145 was added to the final 10%, followed by incubation of S. aureus N315 overnight. S. caprae ATCC 35538 and S. caprae JMUB145 were incubated in fresh medium. Spent media were utilized for the assay. (B) Human blood cells. Fresh or spent medium of S. aureus N315, S. caprae ATCC 35538, or S. caprae JMUB145 was added to the final 10%, followed by incubation of S. caprae JMUB145 overnight. S. aureus N315 and S. caprae ATCC 35538 were cultured in fresh medium. P-values were calculated by Student’s t-test. An asterisk indicates p < 0.001.
Hla exhibited hemolytic activity on rabbit RBCs but not human RBCs (Klainer et al., 1972). Consistently, the spent medium of S. aureus N315 did not exhibit hemolytic activity on human RBCs (Figure 7B). In contrast, S. caprae strains ATCC 35538 and JMUB145 possess several genes encoding β-hemolysin, δ-hemolysin, and putative hemolysins (Watanabe et al., 2018). The two S. caprae strains exhibited hemolytic activity on human RBCs (Figure 7B). Interestingly, the activity of S. caprae strain JMUB145 was significantly higher than the S. caprae strain ATCC 35538 (p < 0.001, Student’s t-test). Moreover, hemolysis was dramatically suppressed by culture in the presence of 10% S. aureus N315 spent medium (p < 0.001), showing that hemolysin production by S. caprae is suppressed by S. aureus. The spent medium did not affect the growth speed of S. aureus or S. caprae (Supplementary Figure 8).
Wound formation changes the skin microenvironment consisting of commensal bacteria and significantly changes the skin microbial community (Johnson et al., 2018). Staphylococcus spp. dominates chronic wounds (e.g., diabetic foot ulcers) (Wolcott et al., 2016; Gardiner et al., 2017). Gardner et al. (2013) revealed that the relative abundance of Staphylococcus spp. in chronic wounds is associated with wound duration. In this study, the microbiome of the skin healed from PI was analyzed and the composition of commensal Staphylococcus spp. was compared. The analysis showed the dominance of S. caprae on healed skins that suffered from RPI (four of the seven patients) within 1–6 weeks after healing (Supplementary Figure 1). Contrastingly, the relative abundance of S. epidermidis was extremely low in all skin sites (Figure 1). Therefore, S. caprae might have an inhibitory effect on S. aureus and/or S. epidermidis in the skin microenvironment during the healing process of RPI. Some CoNS species compete with S. aureus on specific skin microenvironments. S. epidermidis produces an AMP to eliminate S. aureus (Nakatsuji et al., 2017) and a phenol-soluble modulin to promote immune defense in the wound (Cogen et al., 2010). S. capitis was predominantly found in skin samples derived from patients with lesional atopic dermatitis compared with S. aureus and S. caprae (Smits et al., 2020). Similar to other CoNS, S. caprae might play an important role in the recovery process of skin damage from RPI.
It is well known that there is a crosstalk between S. aureus and CoNS via the agr quorum sensing system (Canovas et al., 2016), which contains agrACDB genes and a regulatory RNA (RNAIII) (Novick et al., 1995; Reynolds and Wigneshweraraj, 2011). The agr locus is ubiquitous in S. aureus and in CoNS. An autoinducing peptide (AIP), which is encoded and secreted by agrDB genes, binds to histidine kinase (AgrC), followed by promoting RNAIII expression via AgrC-dependent phosphorylation of the response regulator (AgrA) and upregulating specific virulence genes (Wang et al., 2014). In S. aureus, AIP accumulation leads to the upregulation of α-hemolysin (Queck et al., 2008), whereas this upregulation is suppressed by competitive binding of CoNS AIPs to S. aureus AgrC (Ji et al., 1997; Olson et al., 2014). AIPs have genetic and functional diversity, and this variety makes the intra/interspecies crosstalk via the agr systems complicated because sensitivity and specificity of AIP significantly vary depending on the combination of agr types. The agr of S. aureus strains is classified into four groups (I–IV) (Ji et al., 1997; Otto et al., 2001; Geisinger et al., 2012). It was also reported that the agr of other CoNS also has a sequence variety (Olson et al., 2014). Otto et al. (2001) reported that S. epidermidis AIP type I specifically inhibits S. aureus agr type III quorum sensing but not the other types. Regarding S. caprae, although AIP produced by a goat-isolated S. caprae strain inhibits all the four agr sensing systems, the effects were less toward type IV than the other three types (Paharik et al., 2017). In this study, it was found that S. caprae that colonized on the patients carried agr genes encoding different AIP from the goat-isolated one (Figure 4). Though all reported staphylococcal AIPs contained a five-membered thiolactone ring with an N-terminal extension (Thoendel and Horswill, 2013; Olson et al., 2014), the mature structure of the human-isolated S. caprae AIP remains unknown. Additionally, there are a few limitations in the present in vivo and in vitro studies (Figures 5–7). First, only the S. aureus N315 strain encoding a type II agr system was tested. Second, the suppression of α-toxins (Figure 7) might not be due to the agr system but another system such as saeR/S system in S. aureus (Voyich et al., 2009; Guerra et al., 2016). Thus, further analysis is necessary for the elucidation of agr-dependent crosstalk between human-isolated S. caprae and S. aureus such as testing other agr types of S. aureus strains, using synthetic AIPs of the human-isolated S. caprae, deleting the agrDBC genes, and/or exchanging the agr genes with those of the goat-isolated S. caprae.
As shown in Figure 3, the two patients with a high abundance of S. aureus on their healed ulcers suffered from RPI within 1 week, whereas the other four patients with a high abundance of S. caprae suffered from RPI between 1 and 6 weeks. These data showed that the balance of S. aureus and S. caprae could be related to RPIs; especially, the dominance of S. aureus can result in RPI at an earlier time point (i.e., within 1 week from healing). However, due to the small numbers of samples (only two samples of the S. aureus-dominant microbiome), it remained unclear whether such an imbalance of the two species is actually involved in RPI. S. caprae is considered part of a healthy human skin flora (Gowda et al., 2018). S. caprae, first isolated from goat milk, colonizes the skin and mammary glands of goats. This bacterium is also found in healthy human skin and can become pathogenic in humans (Seng et al., 2014; Mazur et al., 2017). The human-isolated S. caprae (strain JMUB145) displayed hemolytic activity on human RBCs (Figure 7A), indicating that this bacterium possesses pathogenicity to the skin of older people, although direct evidence was not obtained through the mouse models. Watanabe et al. (2018) reported that the three human-isolated S. caprae commonly possesses genes encoding phenol-soluble modulins (PSMs), including ∂-toxin, PSM-α, PSM-β, and PSM-δ, while the genes were also conserved in goat-isolated S. caprae strain ATCC 35538.
There were a few limitations for giving knowledge to clinical implementation through this study. Although this study attempted to investigate whether these Staphylococcus spp. are involved in RPI, a direct relationship between RPI and staphylococci remained unknown. It is challenging to develop RPI animal models. Although among the seven samples of the RPI groups, there was a significantly negative correlation between S. caprae and S. aureus (Figure 3 and Supplementary Figure 4), such negative correlation was on the basis of the relative abundance and did not directly elucidate the increase or decrease in amounts of colonized bacteria. This study could not count the absolute quantity using the present methods. Nevertheless, this study found new characteristics of human-isolated S. caprae, such as dominance on elderly skin healed from PI (Figure 2), the ability of colonization on mouse skin healed from injury (Figure 6), suppression of S. aureus-producing Hla, and production of hemolysin targeting human RBCs (Figure 7). In conclusion, this study showed that interspecies regulation between S. caprae and S. aureus on healed skin is associated with RPI.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by the Medical Ethics Committee of Kanazawa University. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the Medical Ethics Committee of Kanazawa University.
KoO designed the study, analyzed all data, and wrote the manuscript. HF conducted the animal experiments and hemolysis assay. NT collected the clinical samples and analyzed their sequence data. KS contributed to the sample collection. ME contributed to the animal experiments. SW revised the discussion and improved quality of the manuscript. SW and LC contributed to the experiments using S. caprae strains. TM-A analyzed the genomic data of S. caprae. SO supervised HF and ME and contributed to the launch of this project and its application for animal experiments. KaO conceived this study, collected sequence data, and revised the manuscript critically for content. JS launched this project, supervised NT and KS, and funded this study. All authors contributed to the article and approved the submitted version.
This work was partially supported by the Japan Society for the Promotion of Science KAKENHI (21H03220 to KaO and 17H04428 to JS).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank all the participants and their families for their cooperation in this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.818398/full#supplementary-material
PI, pressure injury; RPI, recurrent pressure injury; SLST, single-locus sequence typing; agr, accessory-gene-regulator; RBC, rabbit red blood cell.
Arisandi, D., Ogai, K., Urai, T., Aoki, M., Minematsu, T., Okamoto, S., et al. (2020). Development of recurrent pressure ulcers, risk factors in older patients: a prospective observational study. J. Wound Care 29, S14–S24. doi: 10.12968/jowc.2020.29.Sup4.S14
Bayer, A. S., Ramos, M. D., Menzies, B. E., Yeaman, M. R., Shen, A. J., and Cheung, A. L. (1997). Hyperproduction of alpha-toxin by Staphylococcus aureus results in paradoxically reduced virulence in experimental endocarditis: a host defense role for platelet microbicidal proteins. Infect. Immun. 65, 4652–4660. doi: 10.1128/iai.65.11.4652-4660.1997
Belkaid, Y., and Segre, J. A. (2014). Dialogue between skin microbiota and immunity. Science 346, 954–959. doi: 10.1126/science.1260144
Blume-Peytavi, U., Kottner, J., Sterry, W., Hodin, M. W., Griffiths, T. W., Watson, R. E. B., et al. (2016). Age-associated skin conditions and diseases: current perspectives and future options. Gerontologist 56, S230–S242. doi: 10.1093/geront/gnw003
Byrd, A. L., Belkaid, Y., and Segre, J. A. (2018). The human skin microbiome. Nat. Rev. Microbiol. 16, 143–155. doi: 10.1038/nrmicro.2017.157
Canovas, J., Baldry, M., Bojer, M. S., Andersen, P. S., Grzeskowiak, P. K., Stegger, M., et al. (2016). Cross-talk between Staphylococcus aureus and other Staphylococcal species via the agr quorum sensing system. Front. Microbiol. 7:1733. doi: 10.3389/fmicb.2016.01733
Cau, L., Williams, M. R., Butcher, A. M., Nakatsuji, T., Kavanaugh, J. S., Cheng, J. Y., et al. (2021). Staphylococcus epidermidis protease EcpA can be a deleterious component of the skin microbiome in atopic dermatitis. J. Allergy Clin. Immunol. 147, 955–966. doi: 10.1016/j.jaci.2020.06.024
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Claesson, M. J., Wang, Q., O’Sullivan, O., Greene-Diniz, R., Cole, J. R., Ross, R. P., et al. (2010). Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 38, e200. doi: 10.1093/nar/gkq873
Cogen, A. L., Yamasaki, K., Sanchez, K. M., Dorschner, R. A., Lai, Y., MacLeod, D. T., et al. (2010). Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J. Invest. Dermatol. 130, 192–200. doi: 10.1038/jid.2009.243
Devriese, L. A., Poutrel, B., Kilpper-Balz, R., and Schleifer, K. H. (1983). Staphylococcus gallinarum and Staphylococcus caprae, two new species from animals. Int. J. Syst. Bacteriol. 33, 480–486. doi: 10.1099/00207713-33-3-480
Dufour, P., Jarraud, S., Vandenesch, F., Greenland, T., Novick, R. P., Bes, M., et al. (2002). High genetic variability of the agr locus in Staphylococcus species. J. Bacteriol. 184, 1180–1186. doi: 10.1128/jb.184.4.1180-1186.2002
Ederveen, T. H. A., Smits, J. P. H., Hajo, K., van Schalkwijk, S., Kouwenhoven, T. A., Lukovac, S., et al. (2019). A generic workflow for single locus sequence typing (SLST) design and subspecies characterization of microbiota. Sci. Rep. 9:19834. doi: 10.1038/s41598-019-56065-y
Farage, M. A., Miller, K. W., Elsner, P., and Maibach, H. I. (2013). Characteristics of the aging skin. Adv. Wound Care 2, 5–10. doi: 10.1089/wound.2011.0356
Gardiner, M., Vicaretti, M., Sparks, J., Bansal, S., Bush, S., Liu, M., et al. (2017). A longitudinal study of the diabetic skin and wound microbiome. PeerJ 5:e3543. doi: 10.7717/peerj.3543
Gardner, S. E., Hillis, S. L., Heilmann, K., Segre, J. A., and Grice, E. A. (2013). The neuropathic diabetic foot ulcer microbiome is associated with clinical factors. Diabetes 62, 923–930. doi: 10.2337/db12-0771
Geisinger, E., Chen, J., and Novick, R. P. (2012). Allele-dependent differences in quorum-sensing dynamics result in variant expression of virulence genes in Staphylococcus aureus. J. Bacteriol. 194, 2854–2864. doi: 10.1128/JB.06685-11
Gowda, A., Pensiero, A. L., and Packer, C. D. (2018). Staphylococcus caprae: a skin commensal with pathogenic potential. Cureus 10:e3485. doi: 10.7759/cureus.3485
Grice, E. A., and Segre, J. A. (2011). The skin microbiome. Nat. Rev. Microbiol. 9, 244–253. doi: 10.1038/nrmicro2537
Grice, E. A., Snitkin, E. S., Yockey, L. J., Bermudez, D. M., Liechty, K. W., and Segre, J. A. (2010). Longitudinal shift in diabetic wound microbiota correlates with prolonged skin defense response. Proc. Natl. Acad. Sci. U.S.A. 107, 14799–14804. doi: 10.1073/pnas.1004204107
Guerra, F. E., Addison, C. B., de Jong, N. W. M., Azzolino, J., Pallister, K. B., van Strijp, J. A. G., et al. (2016). Staphylococcus aureus SaeR/S-regulated factors reduce human neutrophil reactive oxygen species production. J. Leukoc. Biol. 100, 1005–1010. doi: 10.1189/jlb.4vmab0316-100rr
Hatlen, T. J., and Miller, L. G. (2021). Staphylococcal skin and soft tissue infections. Infect. Dis. Clin. North Am. 35, 81–105. doi: 10.1016/j.idc.2020.10.003
Heilbronner, S., and Foster, T. J. (2021). Staphylococcus lugdunensis: a skin commensal with invasive pathogenic potential. Clin. Microbiol. Rev. 34:e00205-20. doi: 10.1128/CMR.00205-20
Igarashi, A., Yamamoto-Mitani, N., Gushiken, Y., Takai, Y., Tanaka, M., and Okamoto, Y. (2013). Prevalence and incidence of pressure ulcers in Japanese long-term-care hospitals. Arch. Gerontol. Geriatr. 56, 220–226. doi: 10.1016/j.archger.2012.08.011
Ji, G., Beavis, R., and Novick, R. P. (1997). Bacterial interference caused by autoinducing peptide variants. Science 276, 2027–2030. doi: 10.1126/science.276.5321.2027
Johnson, T. R., Gómez, B. I., McIntyre, M. K., Dubick, M. A., Christy, R. J., Nicholson, S. E., et al. (2018). The cutaneous microbiome and wounds: new molecular targets to promote wound healing. Int. J. Mol. Sci. 19:2699. doi: 10.3390/ijms19092699
Kalan, L. R., and Brennan, M. B. (2019). The role of the microbiome in nonhealing diabetic wounds. Ann. N. Y. Acad. Sci. 1435, 79–92. doi: 10.1111/nyas.13926
Klainer, A. S., Chang, T. W., and Weinstein, L. (1972). Effects of purified staphylococcal alpha toxin on the ultrastructure of human and rabbit erythrocytes. Infect. Immun. 5, 808–813. doi: 10.1128/iai.5.5.808-813.1972
Kong, H. H. (2011). Skin microbiome: genomics-based insights into the diversity and role of skin microbes. Trends Mol. Med. 17, 320–328. doi: 10.1016/j.molmed.2011.01.013
Kuroda, M., Ohta, T., Uchiyama, I., Baba, T., Yuzawa, H., Kobayashi, I., et al. (2001). Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357, 1225–1240. doi: 10.1016/S0140-6736(00)04403-2
Loesche, M., Gardner, S. E., Kalan, L., Horwinski, J., Zheng, Q., Hodkinson, B. P., et al. (2017). Temporal stability in chronic wound microbiota is associated with poor healing. J. Invest. Dermatol. 137, 237–244. doi: 10.1016/j.jid.2016.08.009
Mazur, E., Żychowski, P., Juda, M., Korona-Głowniak, I., Niedzielska, G., Malm, A., et al. (2017). First report of a Staphylococcus caprae isolated from middle ear fluid of an infant with recurrent acute otitis media. Ann. Agric. Environ. Med. 24, 357–359. doi: 10.5604/12321966.1233553
Naik, S., Bouladoux, N., Linehan, J. L., Han, S. J., Harrison, O. J., Wilhelm, C., et al. (2015). Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature 520, 104–108. doi: 10.1038/nature14052
Nakatsuji, T., Chen, T. H., Narala, S., Chun, K. A., Two, A. M., Yun, T., et al. (2017). Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med 9:eaah4680. doi: 10.1126/scitranslmed.aah4680
Novick, R. P., Projan, S. J., Kornblum, J., Ross, H. F., Ji, G., Kreiswirth, B., et al. (1995). The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. MGG Mol. Gen. Genet. 248, 446–458. doi: 10.1007/BF02191645
Ogai, K., Ogura, K., Ohgi, N., Park, S., Aoki, M., Urai, T., et al. (2021a). Stability of skin microbiome at sacral regions of healthy young adults, ambulatory older adults, and bedridden older patients after 2 Years. Biol. Res. Nurs. 23, 82–90. doi: 10.1177/1099800420941151
Ogai, K., Shibata, K., Takahashi, N., Ogura, K., Okamoto, S., and Sugama, J. (2021b). Amplicon-based skin microbiome profiles collected by tape stripping with different adhesive film dressings: a comparative study. BMC Microbiol. 21:54. doi: 10.1186/s12866-021-02122-4
Oh, J., Byrd, A. L., Deming, C., Conlan, S., Kong, H. H., Segre, J. A., et al. (2014). Biogeography and individuality shape function in the human skin metagenome. Nature 514, 59–64. doi: 10.1038/nature13786
Oh, J., Byrd, A. L., Park, M., Kong, H. H., and Segre, J. A. (2016). Temporal stability of the human skin microbiome. Cell 165, 854–866. doi: 10.1016/j.cell.2016.04.008
Olson, M. E., Todd, D. A., Schaeffer, C. R., Paharik, A. E., Van Dyke, M. J., Buttner, H., et al. (2014). Staphylococcus epidermidis agr quorum-sensing system: signal identification, cross talk, and importance in colonization. J. Bacteriol. 196, 3482–3493. doi: 10.1128/JB.01882-14
Otto, M., Echner, H., Voelter, W., and Götz, F. (2001). Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 69, 1957–1960. doi: 10.1128/IAI.69.3.1957-1960.2001
Paharik, A. E., Parlet, C. P., Chung, N., Todd, D. A., Rodriguez, E. I., Van Dyke, M. J., et al. (2017). Coagulase-negative Staphylococcal strain prevents Staphylococcus aureus colonization and skin infection by blocking quorum sensing. Cell Host Microbe 22, 746–756. doi: 10.1016/j.chom.2017.11.001
Parlet, C. P., Brown, M. M., and Horswill, A. R. (2019). Commensal Staphylococci influence Staphylococcus aureus skin colonization and disease. Trends Microbiol. 27, 497–507. doi: 10.1016/j.tim.2019.01.008
Queck, S. Y., Jameson-Lee, M., Villaruz, A. E., Bach, T. H. L., Khan, B. A., Sturdevant, D. E., et al. (2008). RNAIII-Independent target gene control by the agr Quorum-Sensing System: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 32, 150–158. doi: 10.1016/j.molcel.2008.08.005
Reynolds, J., and Wigneshweraraj, S. (2011). Molecular insights into the control of transcription initiation at the Staphylococcus aureus agr operon. J. Mol. Biol. 412, 862–881. doi: 10.1016/j.jmb.2011.06.018
Russell-Goldman, E., and Murphy, G. F. (2020). The pathobiology of skin aging: new insights into an old dilemma. Am. J. Pathol. 190, 1356–1369. doi: 10.1016/j.ajpath.2020.03.007
Schank, J. E. (2015). The NPUAP meeting – this was no consensus conference. J. Am. Coll. Clin. Wound Spec. 7, 19–24. doi: 10.1016/j.jccw.2016.07.001
Scholz, C. F. P., and Jensen, A. (2017). Development of a single locus sequence typing (SLST) scheme for typing bacterial species directly from complex communities. Methods Mol. Biol. 1535, 97–107. doi: 10.1007/978-1-4939-6673-8_7
Scholz, C. F. P., Jensen, A., Lomholt, H. B., Brüggemann, H., and Kilian, M. (2014). A novel high-resolution single locus sequence typing scheme for mixed populations of Propionibacterium acnes in vivo. PLoS One 9:e104199. doi: 10.1371/journal.pone.0104199
Seng, P., Barbe, M., Pinelli, P. O., Gouriet, F., Drancourt, M., Minebois, A., et al. (2014). Staphylococcus caprae bone and joint infections: a re-emerging infection? Clin. Microbiol. Infect. 20, O1052–O1058. doi: 10.1111/1469-0691.12743
Shibagaki, N., Suda, W., Clavaud, C., Bastien, P., Takayasu, L., Iioka, E., et al. (2017). Aging-related changes in the diversity of women’s skin microbiomes associated with oral bacteria. Sci. Rep. 7:10567. doi: 10.1038/s41598-017-10834-9
Shibata, K., Ogai, K., Ogura, K., Urai, T., Aoki, M., Arisandi, D., et al. (2021). Skin physiology and its microbiome as factors associated with the recurrence of pressure injuries. Biol. Res. Nurs. 23, 75–81. doi: 10.1177/1099800420941100
Smits, J. P. H., Ederveen, T. H. A., Rikken, G., van den Brink, N. J. M., van Vlijmen-Willems, I. M. J. J., Boekhorst, J., et al. (2020). Targeting the cutaneous microbiota in atopic dermatitis by coal tar via AHR-dependent induction of antimicrobial peptides. J. Invest. Dermatol. 140, 415–424. doi: 10.1016/j.jid.2019.06.142
Thoendel, M., and Horswill, A. R. (2013). Random mutagenesis and topology analysis of the autoinducing peptide biosynthesis proteins in Staphylococcus aureus. Mol. Microbiol. 87, 318–337. doi: 10.1111/mmi.12100
Voyich, J. M., Vuong, C., DeWald, M., Nygaard, T. K., Kocianova, S., Griffith, S., et al. (2009). The SaeR/S gene regulatory system is essential for innate immune evasion by Staphylococcus aureus. J. Infect. Dis. 199, 1698–1706. doi: 10.1086/598967
Wang, B., Zhao, A., Novick, R. P., and Muir, T. W. (2014). Activation and inhibition of the receptor histidine kinase AgrC occurs through opposite helical transduction motions. Mol. Cell 53, 929–940. doi: 10.1016/j.molcel.2014.02.029
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Watanabe, S., Aiba, Y., Tan, X. E., Li, F. Y., Boonsiri, T., Thitiananpakorn, K., et al. (2018). Complete genome sequencing of three human clinical isolates of Staphylococcus caprae reveals virulence factors similar to those of Staphylococcus epidermidis and Staphylococcus capitis. BMC Genomics 19:810. doi: 10.1186/s12864-018-5185-9
Willis, C., Desai, D., and Laroche, J. (2019). Influence of 16S rRNA variable region on perceived diversity of marine microbial communities of the northern north atlantic. FEMS Microbiol. Lett. 366:fnz152. doi: 10.1093/femsle/fnz152
Wolcott, R. D., Hanson, J. D., Rees, E. J., Koenig, L. D., Phillips, C. D., Wolcott, R. A., et al. (2016). Analysis of the chronic wound microbiota of 2,963 patients by 16S rDNA pyrosequencing. Wound Repair Regen. 24, 163–174. doi: 10.1111/wrr.12370
Yoon, S.-H., Ha, S., Lim, J., Kwon, S., and Chun, J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1281–1286. doi: 10.1007/s10482-017-0844-4
Keywords: pressure injury, healed skin, recurrent injury, skin microbiome, single locus sequence typing, Staphylococcus aureus, Staphylococcus caprae
Citation: Ogura K, Furuya H, Takahashi N, Shibata K, Endo M, Watanabe S, Cui L, Miyoshi-Akiyama T, Okamoto S, Ogai K and Sugama J (2022) Interspecies Regulation Between Staphylococcus caprae and Staphylococcus aureus Colonized on Healed Skin After Injury. Front. Microbiol. 13:818398. doi: 10.3389/fmicb.2022.818398
Received: 19 November 2021; Accepted: 20 January 2022;
Published: 01 March 2022.
Edited by:
Mattias Collin, Lund University, SwedenReviewed by:
Alexander Horswill, University of Colorado Denver, United StatesCopyright © 2022 Ogura, Furuya, Takahashi, Shibata, Endo, Watanabe, Cui, Miyoshi-Akiyama, Okamoto, Ogai and Sugama. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kazuhiro Ogai, a2F6dWhpcm9Ac3RhZmYua2FuYXphd2EtdS5hYy5qcA==; Junko Sugama, anVua28uc3VnYW1hQGZ1aml0YS1odS5hYy5qcA==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.