- 1Department of Crop and Soil Science, Oregon State University, Corvallis, OR, United States
- 2Department of Microbiology, Oregon State University, Corvallis, OR, United States
Microbially mediated nitrification plays an important role in the nitrogen (N) cycle, and rates of activity have been shown to change significantly with temperature. Despite this, the substrate affinities of nitrifying bacteria and archaea have not been comprehensively measured and are often assumed to be static in mathematical models of environmental systems. In this study, we measured the oxidation kinetics of ammonia- (NH3) oxidizing archaea (AOA), NH3-oxidizing bacteria (AOB), and two distinct groups of nitrite (NO2–)-oxidizing bacteria (NOB), of the genera Nitrobacter and Nitrospira, by measuring the maximum rates of apparent activity (Vmax(app)), the apparent half-saturation constant (Km(app)), and the overall catalytic efficiency (Vmax(app)/Km(app)) over a range of temperatures. Changes in Vmax(app) and Km(app) with temperature were different between groups, with Vmax(app) and catalytic efficiency increasing with temperature in AOA, while Vmax(app), Km(app), and catalytic efficiency increased in AOB. In Nitrobacter NOB, Vmax(app) and Km(app) increased, but catalytic efficiency decreased significantly with temperature. Nitrospira NOB were variable, but Vmax(app) increased while catalytic efficiency and Km(app) remained relatively unchanged. Michaelis–Menten (MM) and Haldane (H) kinetic models of NH3 oxidation and NO2– oxidation based on the collected data correctly predict nitrification potential in some soil incubation experiments, but not others. Despite previous observations of coupled nitrification in many natural systems, our results demonstrate significant differences in response to temperature strategies between the different groups of nitrifiers; and indicate the need to further investigate the response of nitrifiers to environmental changes.
Introduction
Anthropogenic activities have approximately doubled inorganic nitrogen (N) inputs to the global biosphere (Steffen et al., 2015), drastically altering nutrient cycling as evidenced by major changes in N utilization (Butterbach-Bahl et al., 2013; Liang et al., 2016) and carbon (C) sequestration (Davidson and Janssens, 2006). These changes have harmful effects on human and environmental health and severe economic impacts, including eutrophication of waterways (Shelton et al., 2018), significant changes in the sources and sinks of N oxide gas pollutants (Ni and Groffman, 2018), and the loss of biodiversity (Stevens, 2019). Nutrient cycling is microbially mediated by microorganisms that mineralize organic N to ammonium/ammonia (NH4+/NH3), oxidize NH3 to nitrite (NO2–) then nitrate (NO3–), and denitrify to N oxide and dinitrogen gases. Nitrification, the aerobic sequential oxidation of NH3 to NO2– and then to NO3–, is an important intermediate in the N cycle and contributes to N availability for plants, N oxide gas emissions, and NO2– and NO3– leaching that leads to eutrophication (Galloway et al., 2008; Ward, 2011).
While environmental change predictions and climate models have focused on plant effects and C sequestration, the evaluation of environmental changes on microbial physiology has lagged behind or simply been treated as a “black box” (Shade et al., 2009; McGuire and Treseder, 2010; Treseder et al., 2012; Bennett et al., 2019). Nitrifying microorganisms are an important group that controls the size and composition of inorganic N pools available to plants (Ward, 2011). There is a critical need to understand the physiological changes that N cycling microbes make in response to climate change, and whether their responses can sustain environmental functionality and resilience. Nitrification is carried out by diverse microorganisms, including bacteria and archaea. NH3-oxidizing bacteria and archaea (AOB and AOA, respectively) generally carry out NH3 oxidation in partnership with NO2–-oxidizing bacteria (NOB; Daims et al., 2011; Hidetoshi et al., 2011; Sayavedra-Soto and Arp, 2011; Starkenburg et al., 2011). There are several groups of NOB, but in this study we focused on two groups that are the most abundant in soils: the genus Nitrobacter and the genus Nitrospira. A subset of lineage II Nitrospira that has the capability to completely oxidize NH3 to NO3– (comammox) has not been included in this study (Ward, 2011; Daims et al., 2015; van Kessel et al., 2015).
Rates of biological enzymatic functions are temperature sensitive and there is evidence of differential responses of soil processes to temperature (Hobbs et al., 2013; Blagodatskaya et al., 2016; Taylor et al., 2017, 2019). Enzyme catalytic rates generally increase with temperature until an optimum is reached and, at higher temperatures, the reaction rate decreases due to thermal inactivation (Laidler and Peterman, 1979). Previous work by our research group and others suggests that AOB and AOA in soils respond differently to temperature (Pierre et al., 2017; Taylor et al., 2017; Duan et al., 2018), and it has recently shown that rates of activity and substrate affinity of soil NO2– oxidation are also temperature-sensitive (Taylor et al., 2019; Duan et al., 2020). Increasing temperature can also increase reaction rates which are associated with increased production of N oxide, nitric oxide (NO), and nitrous oxide (N2O) gases (Law et al., 2012; Schreiber et al., 2012; Bonnett et al., 2013; Mellbye et al., 2016). However, increasing temperature can also result in lower affinity for the substrate, which may offset higher reaction rates (Laidler and Peterman, 1979; Snider et al., 2000; Davidson and Janssens, 2006; Blagodatskaya et al., 2016). Different responses to temperature by associated NH3 oxidizers and NO2– oxidizers may affect nitrification coupling and potential NO2– accumulation (Schaefert and Hollibaugh, 2017; Taylor et al., 2019; Duan et al., 2020; Venterea et al., 2020), but significant biochemical data, particularly Vmax(app) and Km(app), to model the response of nitrifying microorganisms to temperature are lacking.
Most physiological data on nitrifying bacteria and archaea have been generated under optimal growth conditions. For example, Suzuki et al. (1974) showed the response of Nitrosomonas europaea Km(app) to changes in pH, but did not investigate temperature, and NH3 affinity has been measured in the AOBs Nitrosococcus oceanus and Nitrosospira briensis, but only at optimum temperature (Ward, 1987; Bollmann et al., 2005). The NO2– oxidation kinetics of strains of Nitrobacter and Nitrospira have been determined only at each strain’s optimal growth temperature, pH, and dO2 (Nowka et al., 2015; Ushiki et al., 2017). Exceptions include a study measuring whole-cell kinetics of Nitrosomonas sp. 4W30 at three temperatures, which showed significantly greater affinity for NH3 at lower temperatures (Jones and Morita, 1985), and the evaluation of the AOA “Ca. N. oleophilus” MY3 kinetic response where the apparent Km(app) increased ∼2-fold from 25 to 35°C (Jung et al., 2022).
Previous work demonstrated that the rates of nitrification changed with temperature (Kim et al., 2006; Schaefert and Hollibaugh, 2017; Taylor et al., 2017, 2019; Duan et al., 2018); however, it is unclear if changes in rates in response to temperature results from a shift in community structure or changes in microbial physiology (Blagodatskaya et al., 2016; Taylor et al., 2017; Duan et al., 2020). We hypothesized that individual nitrifiers respond differently to temperature, and that these responses are reflected in changes in substrate affinity and reaction rate. We chose pure culture representative members of AOA, AOB, Nitrobacter NOB, and Nitrospira NOB for evaluation and investigated changes in NH3 and NO2– affinity and oxidation rate over a large range of physiologically relevant temperatures. Finally, we applied Michaelis–Menten (MM) and Haldane (H) kinetic models, based on observations, to changes in nitrification rates observed in whole soils over a range of temperatures.
Materials and Methods
Nitrifying Strains and Growth Conditions
Nine nitrifying microorganisms were used in this study and cultivated as previously described (Table 1). We utilized the AOA Nitrososphaera viennensis (Tourna et al., 2011; Taylor et al., 2015), and the AOB N. europaea and Nitrosospira multiformis (Ensign et al., 1993; Chain et al., 2003; Norton et al., 2008; Taylor et al., 2013). We worked with a total of six NOB strains. Nitrobacter hamburgensis, Nitrobacter winogradskyi, Nitrobacter vulgaris, and Nitrospira moscoviensis were grown under the conditions previously used in our laboratories (Bock et al., 1983, 1990; Ehrich et al., 1995; Starkenburg et al., 2006, 2008; Koch et al., 2015; Mellbye et al., 2017a,b), and Nitrospira japonica NJ1 and Nitrospira ND1 were cultivated as previously described (Ushiki et al., 2017). Minimal media for all cultures were at pH 8. Cultures were grown at their optimal growth temperatures (30°C for all cultures except for N. viennensis, which was grown at 42°C and N. moscoviensis which was grown at 37°C, Table 1) to maximize growth yields and for the ease of comparison with other studies. Cultures were monitored at regular intervals to check cell density by optical density at 600 nm (OD600) and NO2– concentrations by the Griess assay.
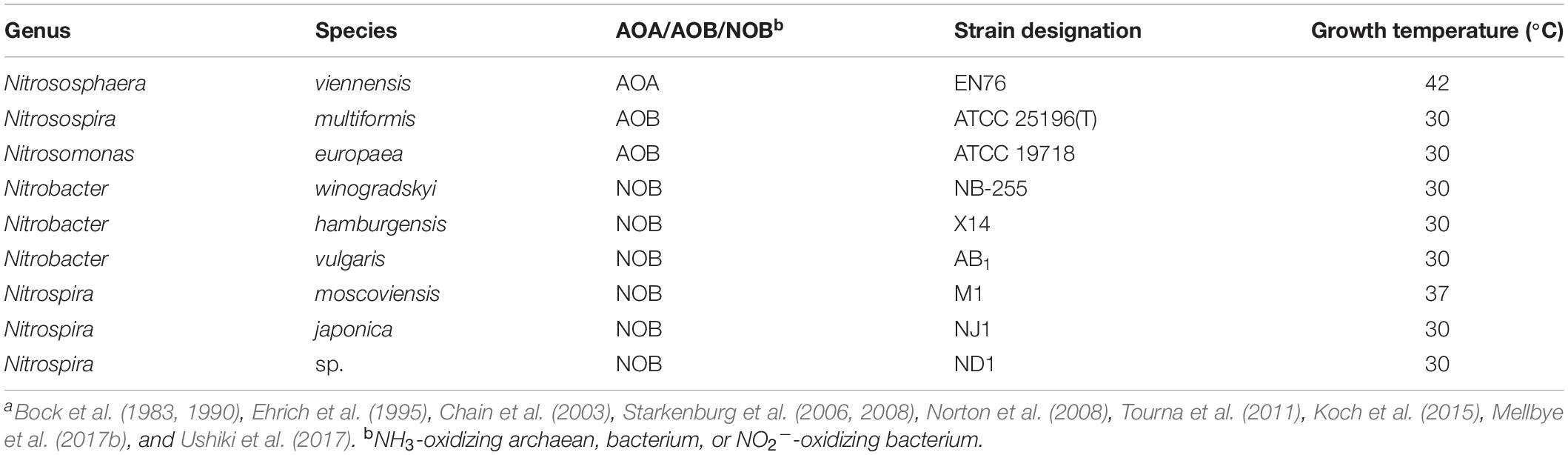
Table 1. Ammonia (NH3) and nitrite (NO2–) oxidizer strains for this study were grown under optimal growth conditions and temperaturesa.
Determination of Kinetic Parameters
Cultures were grown to early stationary phase and then harvested by centrifugation (10,000 × g, 15 min), washed with their respective substrate-free growth media to remove trace concentrations of NH3 and NO2–, and resuspended in ∼15-ml volumes of substrate-free growth media (pH 8) to yield a concentrated cell suspension. The cell density of the concentrated suspension was measured by OD600, and the protein content was measured with a Pierce bicinchoninic acid protein assay kit (Thermo Scientific, Rockford, IL, United States) to determine the volume of cells to be added to the incubation vials, yielding an average protein concentration of 2.6 ± 2.9 μg/μl for the NOB and 6.5 ± 1.7 μg/μl for NH3 oxidizers. For each experimental temperature, vials were prepared in triplicate with 5 ml portions of their respective growth media (pH 8) supplemented with a range of substrate concentrations (NH4+ for the AOA and AOB and NO2– for the NOB). The vials were closed with gray butyl stoppers and preincubated at the experimental temperatures. Experiments were initiated with the addition of aliquots of concentrated culture. Vials were sampled immediately and then at ∼30 min intervals to obtain NO2– concentration at five time points over 2 h, with a final time point obtained at ∼4 h after the initiation of the experiment. Rates of NH3 oxidation by the AOA and AOB were determined by the rate of NO2– accumulation in the incubation vials, and NO2– oxidation rates by its disappearance. The MM kinetic coefficients of substrate utilization at each temperature were determined by fitting the NO2– or NH3 oxidation rate (v) for a given substrate concentration (S) to the equation
to find the maximum reaction rate (Vmax) and the substrate concentration that yields half of this maximum rate (Km) using SigmaPlot (Systat Software, San Jose, CA, United States). Because we are working with whole cells, the calculated Vmax and Km should be considered to be the apparent Vmax (Vmax(app)) and apparent Km (Km(app)). Note that we calculated Vmax(app) as a relative kcat based on the maximum reaction rate per mg protein of whole cells (Snider et al., 2000). The effects of H inhibition on substrate utilization were determined by fitting the NO2– or NH3 oxidation rate for a given substrate concentration to the equation
to find the effects of substrate inhibition on Vmax(app) and Km(app), as well as to find the substrate concentration that inhibited Vmax(app) by 50% (Ki). Specific affinity for substrate (aos) and the affinity constant (KA) were determined as described by Button (1998). Temperature-dependent distribution of NH3/NH4+ was calculated as described by Groeneweg et al. (1994).
Soil Incubations to Characterize NH3 Oxidation Response
Agricultural soils were collected from the western Oregon Coastal Plain in Tillamook County and from the eastern Oregon Columbia River basin in Morrow County (Waggoner et al., 2021). The western Oregon Coastal Plain soil is identified as a Quillamook series silt loam soil1. It receives a mean annual precipitation of 2,159 mm and has an isomesic temperature regime (temperatures averaging 6.5°C in winter and 14°C in summer) with a mean annual temperature of 10°C and has a pH of 5.8. The soil was cropped with a silage corn (Zea mays L.) and rye grass (Lolium spp.) rotation. The eastern Oregon Columbia River basin soil receives a mean annual precipitation of 178 mm and has a mesic temperature regime (temperatures averaging 2°C in winter to 22°C in summer) with a mean annual temperature of 12°C and has a pH of 8.5. It is identified as a Sagehill series sandy loam soil (see text footnote 1). It is cropped under a silage corn (Z. mays L.) and triticale (Triticosecale) rotation. Three field replicates were collected at each site, each of which was created using a composite of 3–5 samples from the surface 0–20 cm that were mixed in the field, and then brought to the laboratory where they were sieved at 2.36 mm and stored at 4°C until use.
Using a method previously adapted by DeAngelis et al. (2010), DNA extraction was performed using phenol:chloroform:isopropyl alcohol (25:24:1), followed by precipitation in PEG6000 solution [30% (w/v) polyethylene glycol 6000 in 1.6 M NaCl] (DeAngelis et al., 2010). Each soil sample was extracted two times per replicate, with three replicates. Extractions from each replicate were pooled. Quantitative polymerase chain reaction (qPCR) was then used to determine gene copy numbers of AOA amoA, AOB amoA, and NOB nxrA and nxrB using reagents, primers, and conditions as described in Supplementary Table 1. Standards for qPCR were prepared from N. viennensis (AOA amoA), N. multiformis (AOB amoA), N. winogradskyi (nxrA), and N. moscoviensis (nxrB).
Soil incubations were performed to evaluate the contributions of AOA and AOB to soil nitrification when NH4+ was supplied as NH4Cl. Addition of 1-octyne (4 μM) allows differentiation of AOA and AOB through inactivation of the AOB NH3 monooxygenase (AMO), and additions of acetylene (10 μM) were used to prevent all NH4+-mediated nitrification of both AOA and AOB, serving as a negative control (Taylor et al., 2013, 2015). To evaluate the effect of temperature on NH3 response, incubations were established at a range representative of temperatures during the growing season at each sampling site, plus one temperature 10°C higher than normal temperatures to evaluate the effect of temperature stress. The incubation temperatures for Coastal Plain soils were 10, 20, and 30°C, while Columbia Basin soils were incubated at 10, 20, 30, and 40°C. Prior to experimentation, three field replicates of each soil were leached to remove background NO3– to increase colorimetric assay sensitivity. Briefly, soil aliquots were placed in a funnel lined with a paper filter and covered with crushed ice, then held at room temperature while the ice melted and percolated through the soil. This leaching method allows the soil to retain most of its structure. Soils were then allowed to partially air dry to ∼30% gravimetric moisture content for the Coastal Plain and 5% for the Columbia Basin. Leached soils (2.5 g) were measured into 125-ml Wheaton bottles and preincubated at room temperature (23°C) for 12–24 h before experimentation.
To initiate the incubations, soils received 5 μmol NH4Cl g–1 soil in dH2O to bring the soil moisture content slightly above the field capacity for each soil (72% for the Coastal Plain and 22% for the Columbia Basin). A control that received dH2O, but no NH4+ source, was included for each treatment. Each bottle was capped using a lid with an n-butyl septum and alkyne treatments imposed as described above. Alkyne stocks were prepared and added in gaseous forms as previously described to the +octyne and +acetylene treatments (Giguere et al., 2015). The soils were then incubated in the dark for 24 h at 10, 20, 30, or 40°C.
At the end of the incubations, the accumulations of NO2– and NO2– + NO3– in soils were determined as previously described (Miranda et al., 2001; Hood-Nowotny et al., 2010). The difference in NO2– or NO2– + NO3– compared to incubations that included acetylene was considered to represent the rate of nitrification performed by the nitrifiers, or total AOA and AOB activity. The difference between +octyne treatments and +acetylene treatments represents only AOA activity. AOB activity was then calculated as the difference between total nitrification activity and AOA-only activity. NOB was considered to be equal to the total rate of soil nitrification minus any accumulated NO2–.
Modeling the Potential Soil Response of AOA, AOB, and NOB Cultures
Coastal Plain Soil
To determine whether MM kinetics and H (substrate-induced inhibition) parameters could be used to predict the rates of NH3 and NO2– oxidation activity in soil incubations, certain assumptions were made to apply the rates from cultures (μmol substrate mg protein–1 h–1) to soils in which population size was determined by qPCR of AMO and NO2– oxidoreductase (NXR) genes (Supplementary Table 1), and the rates were expressed as μmol substrate g soil–1 h–1. Because the qPCR primers did not discriminate between the strains of AOB or NOB, when evaluating the potential soil activity of each of the cultures we assumed that each strain made up the entire population. It was also assumed that all gene copy numbers determined through qPCR came from active cells, and this number was divided by the number of gene copies per genome as determined by the Basic Local Alignment Search Tool (BLAST; Supplementary Table 2). From Urakawa et al. (2011), the relationship between cell volume and cell protein content of Nitrosopumilus maritimus has been determined, and we extrapolated that information to estimate the protein content per cell of the cultures used in this study (Supplementary Table 3); and in turn estimate the amount of cell protein in each gram of soil (see Supplementary Material for further explanation on the conversion of μmol substrate g soil–1 h–1 to μmol substrate g soil–1 h–1).
It was also necessary to make assumptions about starting substrate concentrations in the soil. The Coastal Plain soil without NH4+ addition had 16 μmol g soil–1 KCl extractable NH3 + NH4+ and a soil water content of 72%, and the assumption was made that all extractable NH4+ would be in soil solution, yielding a concentration of 0.22 μM NH3 + NH4+. In the incubations with NH4+ addition (5 μmol g soil–1), it was assumed that all NH4+ would be in the soil solution yielding a concentration of 6 mM NH3 + NH. To evaluate how the rates of MM or H kinetics of NOB cultures would respond under soil conditions, it was necessary to estimate possible concentrations of NO2– in the soil. Our confident limit of detection of NO2– in soil is ≥2 μM NO2–. There was no detectable NO2– in Coastal Plain soils prior to, or during incubations, suggesting that the rate of soil NOB activity was equal to the rate of soil NH3 oxidation; but does not rule out the possibility that trace NO2– accumulated during active soil nitrification. For the Coastal Plain soil without NH4+ addition, there was 0.22 μM extractable NH3 + NH4+ and, if completely oxidized to NO2–, would yield 0.22 μM NO2–; this value was used as the substrate concentration. For Coastal Plain soils with NH4+ addition, we assumed a NO2– concentration just below our level of detection (2 μM NO2–).
The above assumptions were used with the kinetic parameters determined in this study (Supplementary Table 5) for each of the microbial strains to estimate the potential rate of activity in the soil using MM Eq. (1) and H Eq. (2) kinetics at each soil incubation temperature. The average value of population size, cell protein content, gene copy number, Vmax(app), Km(app), and Ki(app) were used in the calculations as there were an excessive number of potential combinations (26 or 64 combinations at each temperature) of these values if both extremes of the standard deviation (SD) were evaluated. We assumed no inhibition of NH3 oxidation in the incubations without NH4+ addition. Additionally, H kinetics were not determined for NOB in the treatments without NH4+ addition where NO2– was assumed to be 0.22 μM and several orders of magnitude lower than Ki determined in this study.
Columbia Basin Soil
The assumptions made on the population size were the same as for the Coastal Plain soil detailed above. The Columbia Basin soil without NH4+ addition had 0.01 μmol g soil–1 KCl extractable NH3 + NH4+ and a soil water content of 22%, and the assumption was made that all extractable NH4+ would be in the soil solution yielding a concentration of 0.05 μM NH3 + NH4+. In the incubations with NH4+ addition (5 μmol g soil–1), it was assumed that all NH4+ would be in the soil solution yielding a concentration of 22 mM NH3 + NH. There was no detectable NO2– in the Columbia Basin soil prior to incubation or in the incubations without NH4+ addition, suggesting that the rate of soil NOB activity was equal to the total rate of soil nitrification. In the Columbia Basin soil incubations without NH4+ addition, we assumed a NO2– concentration of 0.05 μM (all extractable soil NH4+ oxidized to NO2–). However, in the Columbia Basin soils with NH4+ addition, NO2– accumulated during incubations to concentrations of 2, 700, 1,900, and 400 μM at 10, 20, 30, and 40°C, respectively; and these values were used in the calculation of MM and H kinetic rates. In the treatments with NH4+ addition in which NO2– accumulated, the rate of NO2– oxidation was the NH3 oxidation rate minus the rate of accumulated NO2–.
Statistical Analysis
Analysis of variance (ANOVA) using the Holm–Sidak method in SigmaPlot (Systat Software, San Jose, CA, United States) was used to evaluate if there were significant differences in kinetic parameters across temperatures. Regression analyses in SigmaPlot compared the MM and H kinetic models with the rates of NH3 and NO2– oxidation measured in Coastal Plain and Columbia Basin soils.
Bioinformatic Analyses
The National Center for Biotechnology Information2, Kyoto Encyclopedia of Genes and Genomes3, and BLAST4 were used to identify AmoA and NxrA homologs in nitrifiers and other bacteria (Supplementary Datasets 1, 2; Boratyn et al., 2013; Kanehisa et al., 2017). MUSCLE was used for multiple alignment of amino acid sequences (Edgar, 2004), and phylogenetic analyses were conducted using MEGA11 (Kumar et al., 2016). The evolutionary history of AmoA and NxrA was inferred using the Maximum Likelihood method, based on the JTT matrix model (Jones et al., 1992). Phylogenetic trees were visualized using FigTree 1.4.45.
Results
Homology of AmoA and NrxA in Nitrifying Bacteria and Archaea
Enzyme kinetics are notably different between AOB and AOA, and between Nitrobacter NOB and Nitrospira NOB; perhaps due to different niche specialization and temperature optima (Martens-Habbena et al., 2009; Verhamme et al., 2011; Prosser and Nicol, 2012; Pester et al., 2014; Gruber-Dorninger et al., 2015; Nowka et al., 2015; Kits et al., 2017; Ushiki et al., 2017). We chose to analyze the oxidation rate and affinity of nine nitrifying microorganisms isolated from soils and wastewater systems, and we focused on microorganisms that could accumulate the biomass necessary for our experiments (Table 1). For ammonia oxidizers (AOA and AOB), we selected the AOA N. viennensis and the AOB N. multiformis and N. europaea. The evolutionary history of the putative substrate-binding subunits of AMO (putative substrate-binding subunit, AmoA) is shown in Figure 1A. For NOB, N. winogradskyi, N. hamburgensis, N. vulgaris, N. moscoviensis, N. japonica, and Ns. ND1 were selected. The evolutionary history of the putative substrate-binding subunits of NXR (putative substrate-binding subunit, NxrA) is shown in Figure 1B.
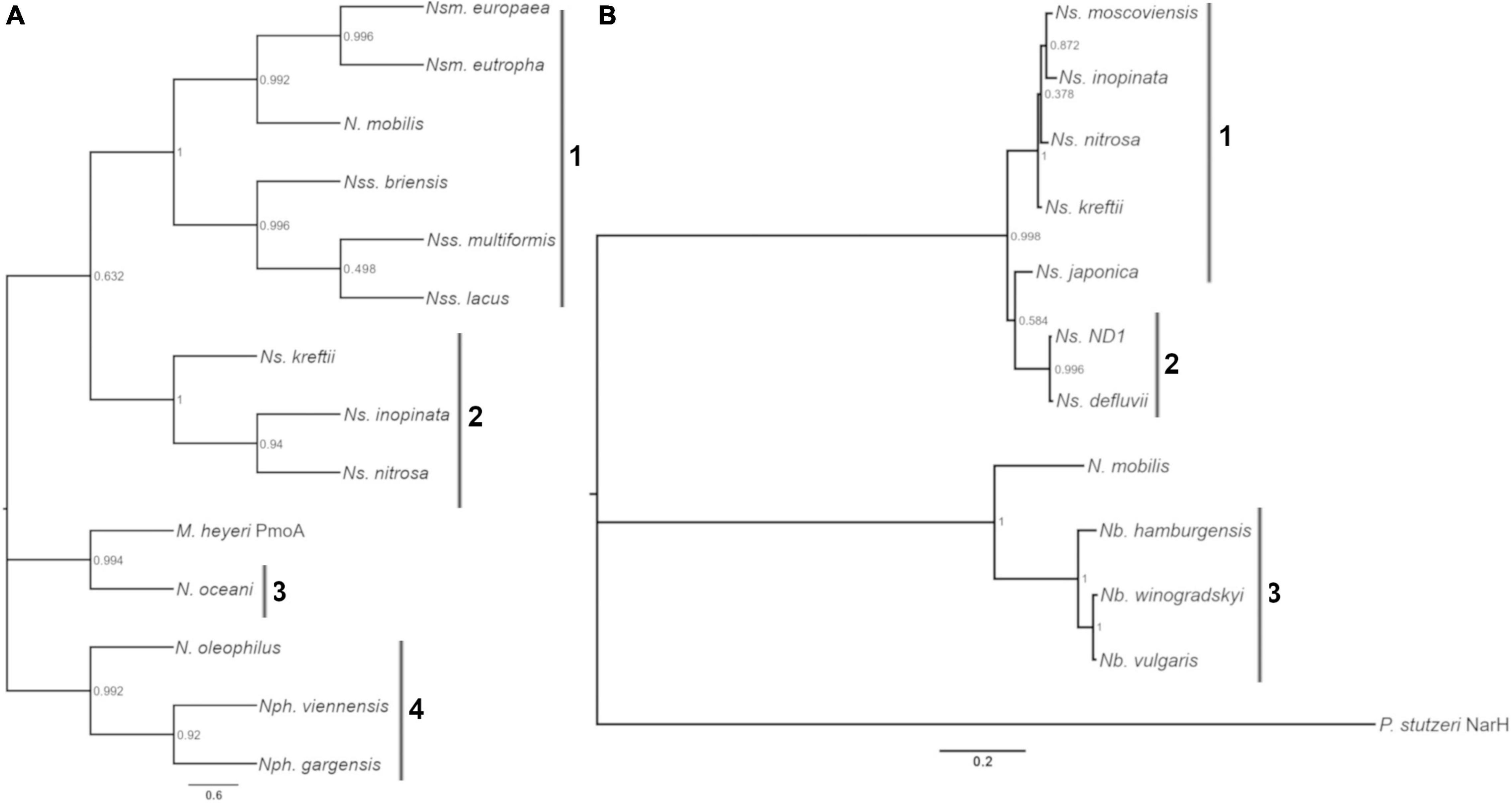
Figure 1. Evolutionary history of AmoA and NxrA. Phylogenetic trees of AmoA (A) and NxrA (B) homologs from nitrifiers and other bacteria. (A) Numbered bars indicate clades: 1, Nitrosomonas and Nitrosospira, 2, Comammox Nitrospira, 3, Nitrosococcus, and 4, AOA. (B) Numbered bars indicate clades: 1, Nitrospira lineage II, 2, Nitrospira Lineage I, and 3, Nitrobacter. The scale bar indicates the mean number of substitutions per site. Bootstrap values from 500 resamplings are rounded to two digits. The protein name of the homolog follows the genus and species, if not AmoA or NxrA. Strain designation and accession numbers can be found in Supplementary Datasets 1, 2.
Nitrifier Kinetics Change With Temperature
The oxidation rates of nitrifiers and their affinity in pure culture incubations were investigated. Both MM and H kinetic models were used to determine Vmax(app), Km(app), and catalytic efficiency (Vmax/Km) from pure culture data (Supplementary Table 5). We have presented these data showing changes in Vmax(app), Km(app), and catalytic efficiency in Figure 2. As expected, Vmax(app) increased with temperature in all microorganisms we investigated; however, there were significant differences in how enzyme affinity (Km(app)) changed with temperature between the different groups of nitrifiers.
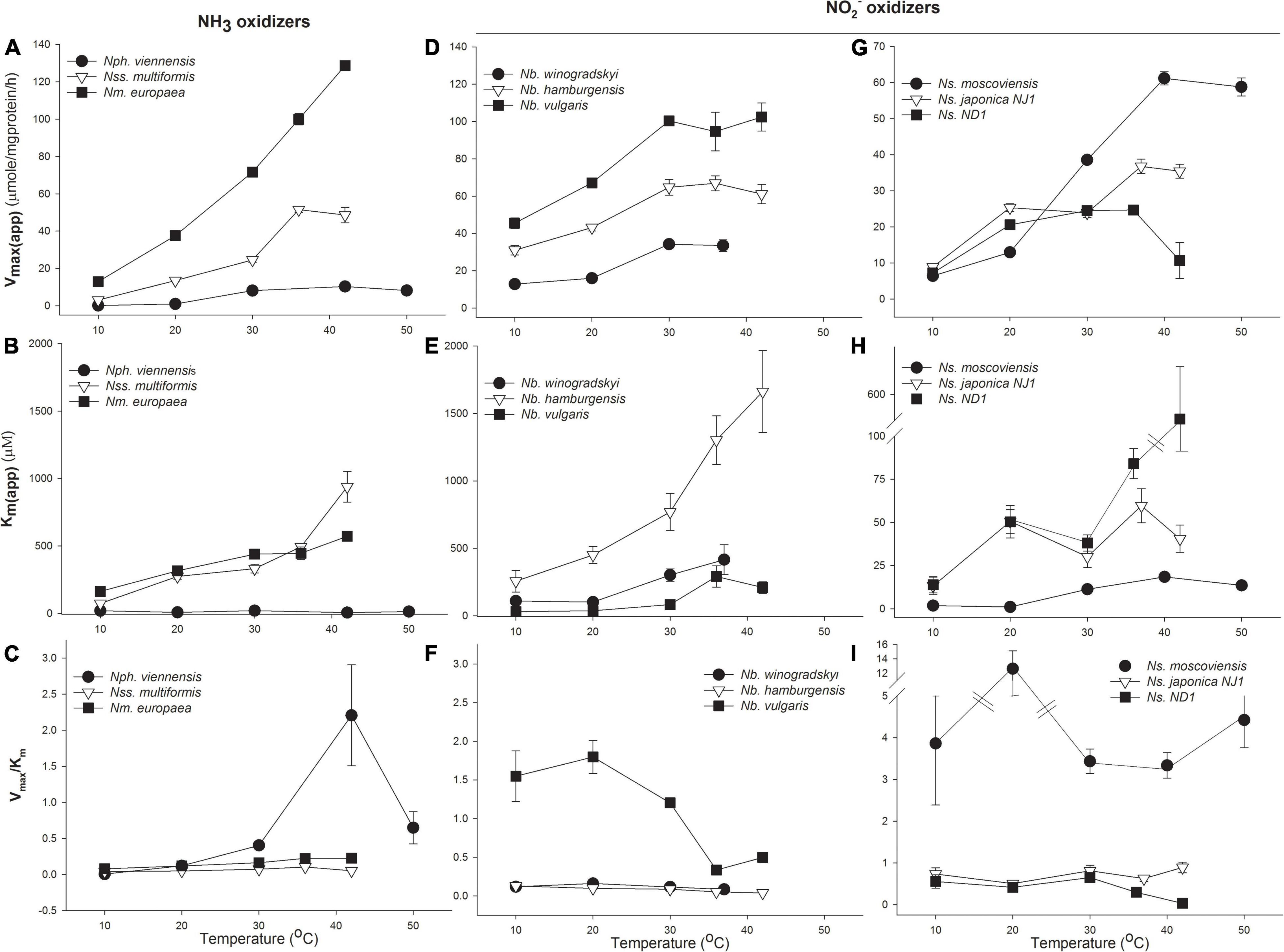
Figure 2. Response of ammonia (NH3) and nitrite (NO2–) oxidizers to temperature. Michaelis–Menten (MM) kinetic parameters for NH3-oxidizing bacteria and archaea (AOA, AOB; A–C), Nitrobacter NO2– oxidizing bacteria (NOB; D–F), and Nitrospira NOB (G–I). Cultures were grown at their optimal growth temperature, and Vmax(app) and Km(app) were determined at the indicated temperatures during short incubations. The response of NH3 oxidizers is expressed in terms of total NH3+NH4+; see Supplementary Table 6 for the response to NH3. Catalytic efficiency was calculated as Vmax/Km. Error bars represent the standard deviation (SD) of the average of three replicates.
Bacterial and archaeal NH3 oxidizers had significantly different changes in enzyme affinity with temperature. For the AOA representative, N. viennensis, Vmax(app) increased ∼100-fold from 10 to 42°C, with maximum rates of 10.3 μmol substrate mg protein–1 h–1. N. viennensis had no significant change in Km(app) with temperature, with values ranging from 6.1 to 20.3 μM NH3 + NH4+, suggesting that temperature generally does not significantly affect enzyme affinity but does increase catalytic efficiency (Figure 2A and Supplementary Tables 5, 6 compares the affinity of the NH3 oxidizers for total NH3 + NH4+ vs. NH3). There was evidence of temperature-independent H substrate-induced inhibition of N. viennensis at 0.3–2.4 mM NH3 + NH4+ (Supplementary Table 5). In contrast, the Km(app) of AOB significantly increased with temperature from 10 to 37°C, suggesting that increasing temperature decreases bacterial AMO affinity; potentially explaining previous observations of different temperature optima between AOA and AOB (Figure 2B). In addition, while Km(app) of NH3 oxidation for AOB increased 10- to 12.7-fold with temperature (from 73.7 to 938.6 μM NH3 + NH4+), the Vmax(app) of AOB also increased 10- to –17-fold (from 51.5 to 128.7 μmol NH3 + NH4+ mg protein–1 h–1), resulting in increased catalytic efficiency. In both N. viennensis and N. multiformis, Vmax(app) decreased when temperature increased past their temperature optima resulting in decreased catalytic efficiency (Figure 2C).
There were strikingly different kinetic responses of NO2– oxidation to temperature between Nitrobacter and Nitrospira and also between the different lineages (Figures 2D–I and Supplementary Table 5). In Nitrobacter, both Vmax(app) and Km(app) increased significantly as temperature increased from 20 to 37°C (Figures 2D,E). While Vmax(app) increased from ∼2- to 2.6-fold (from 33.6 to 102.4 μmol NO2– mg protein–1 h–1), fold increases in Km(app) were approximately two times that of Vmax(app) (3.8–6.9-fold) and ranged from 416.5 to 1662 μM NO2– as temperature increased; resulting in significantly decreased catalytic efficiency (Figure 2F and Supplementary Table 5).
There were interesting differences among the three Nitrospira strains tested. The lineage II Nitrospira, N. moscoviensis (growth temperature 37°C), and N. japonica NJ1 (growth temperature 30°C), behaved similarly, except for notable differences in temperature optima that resulted in maximum Vmax(app) at 37°C or 42°C for N. moscoviensis and N. japonica NJ1, respectively, (Figure 2G). There were also indications of H inhibition of N. moscoviensis that became most pronounced at 10°C (Supplementary Table 5). Vmax(app) increased from 1.5- to 4.1-fold, to 19.9 to 35.4 μmol NO2– mg protein–1 h–1 for lineage II Nitrospira while Km(app) varied between 6.6 and 36.8 μM NO2– with little relationship to temperature (Supplementary Table 5). Except for a ∼4-fold increase in catalytic efficiency for N. moscoviensis at 20°C, there was little difference in catalytic efficiency over the temperature ranges tested for the two evaluated lineage II Nitrospira (Figure 2I). The lineage I Nitrospira strain, Ns. ND1, had a 3.4-fold increase in Vmax(app) up to 24.5 μmol NO2– mg protein–1 h–1, and little change in Km(app) (13.9 to 50.4 μM NO2–), resulting in small changes in catalytic efficiency from 10 to 30°C. However, from 30 to 42°C, Vmax(app) stopped increasing and eventually decreased, and Km(app) drastically increased to 505.9 μM, and catalytic efficiency significantly decreased (Figure 2I and Supplementary Table 5).
Comparison of the Nitrification Temperature Response Predicted by Michaelis–Menten and Haldane Kinetic Models to Soil Nitrification Activity
After determining kinetic values for NH3 and NO2– oxidation, we applied the MM and H kinetic models to the changes in nitrification rates observed in short-term incubations of whole soils with or without NH4+ addition and incubated at temperatures from 10 to 40°C (Figures 3, 4). We reasoned that this approach would yield insight into how well pure culture microorganisms used to make the model resemble naturally occurring nitrifying microorganisms. Agricultural soils from two climatic regions of Oregon that demonstrate different nitrification properties were used in this study. AOA outnumbered AOB in both soils (Supplementary Table 4). There was more Nitrobacter than Nitrospira NOB in Coastal Plain soils; however, there was no significant difference in these two populations in the Columbia Basin soil. Nitrification in Coastal Plain soils was coupled and NO2– did not accumulate, while NO2– accumulated in Columbia Basin soils despite having lower overall rates of NH3 oxidation (Figures 3, 4). H models include substrate inhibition and are expected to be important in AOA and Nitrospira NOB that prefer low concentrations of NH3 or NO2–, respectively.
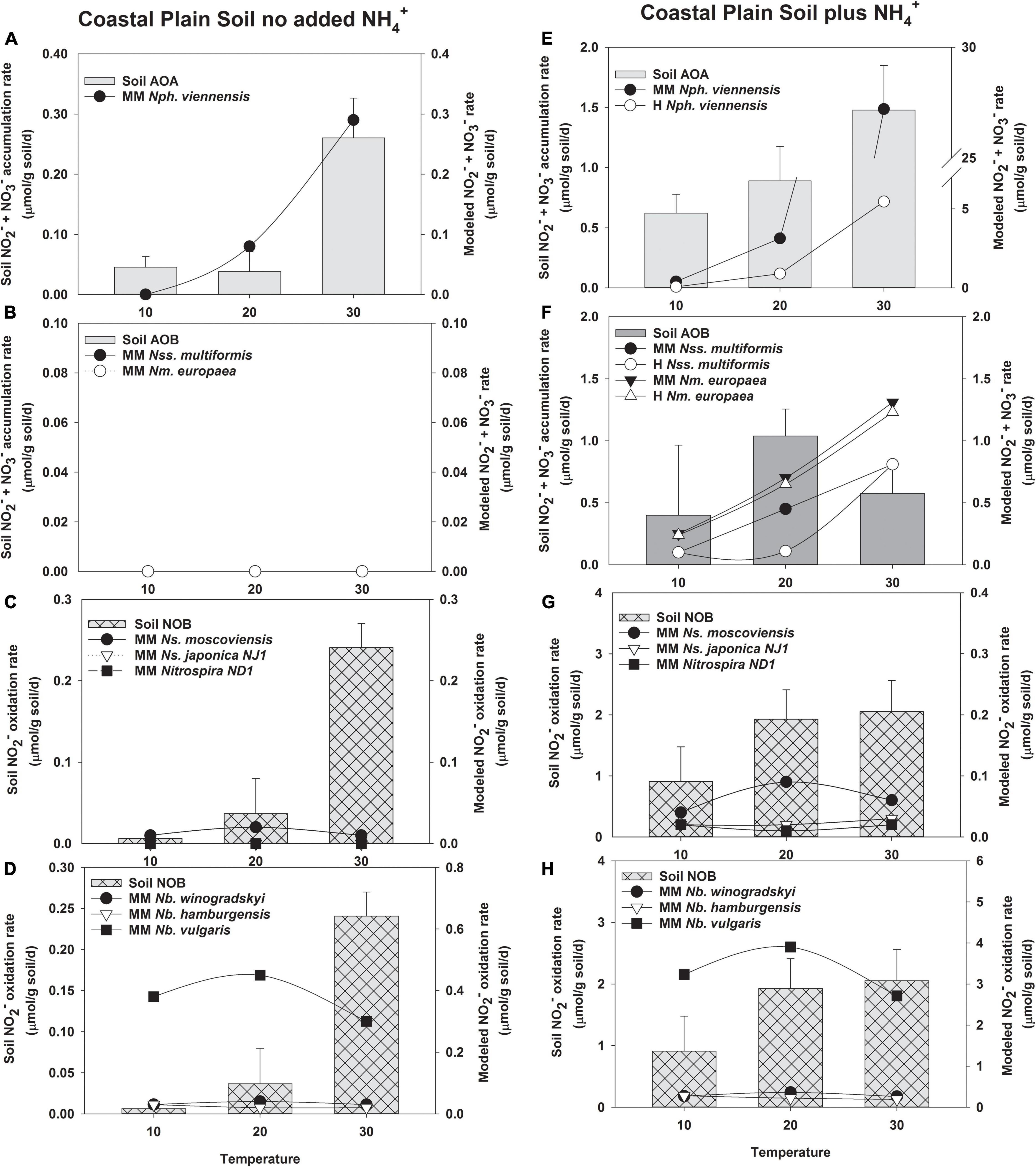
Figure 3. Comparison of the potential response of AOA, AOB, and NOB cultures evaluated with the response of soil nitrifiers in short-term (24 h) incubations of Coastal Plain soil without no NH4+ addition (A–D), and with NH4+ addition (5 μmol/g soil, E–H). The soil nitrifier response measured during the incubations is indicated by the bars corresponding to the left y-axis, and the response modeled using the kinetics of nitrifier cultures determined in this study (Supplementary Table 5) is indicated by the symbols corresponding to the right y-axis. Individual nitrifier names are preceded by either MM or Haldane (H) to indicate that MM or H parameters, respectively, were used to calculate the potential rates of activity.
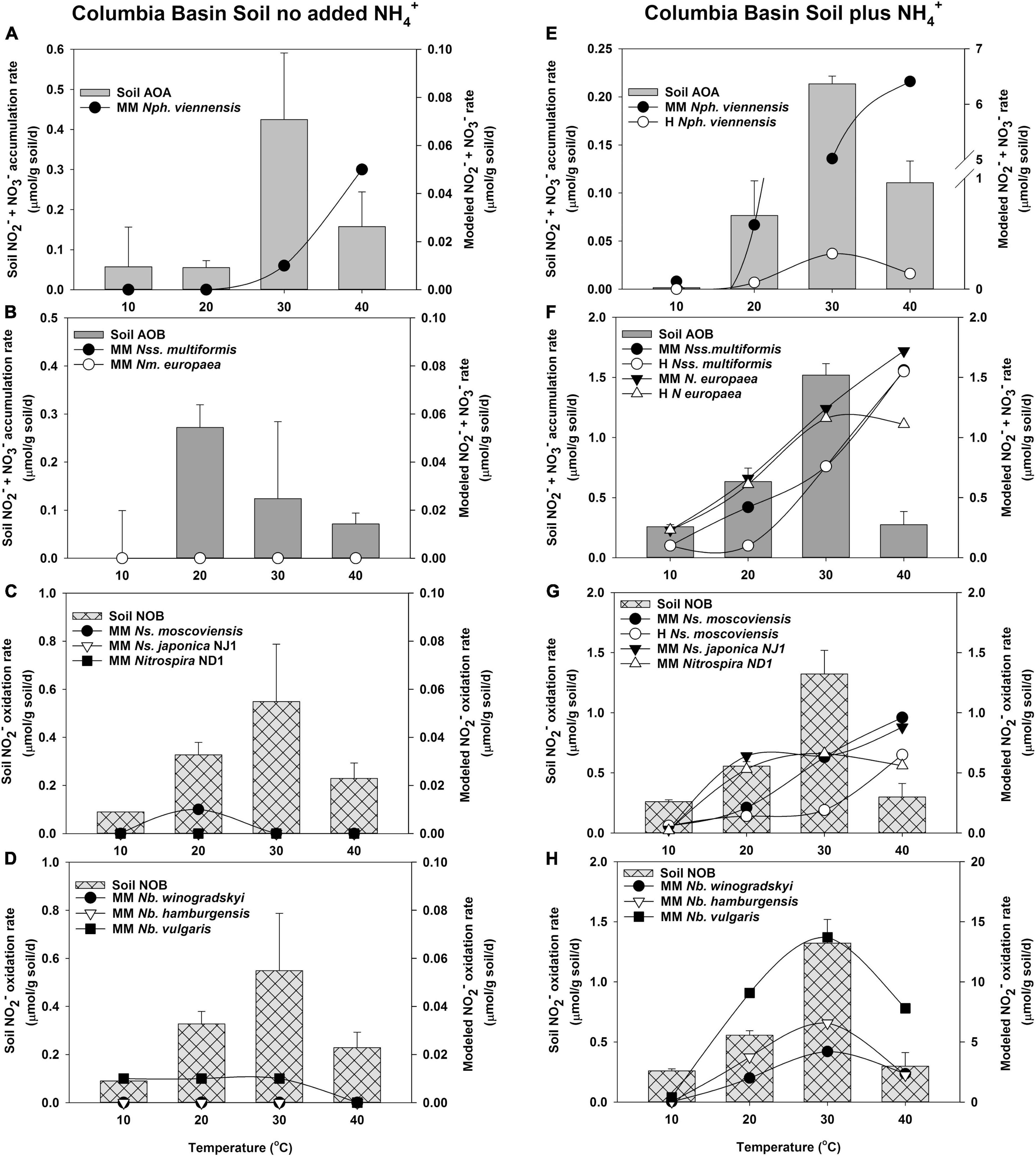
Figure 4. Comparison of the potential response of AOA, AOB, and NOB cultures evaluated with the response of soil nitrifiers in short-term (24 h) incubations of Columbia Basin soil with no NH4+ addition (A–D), and with NH4+ addition (5 μmol/g soil, E–H). The soil nitrifier response measured during incubations is indicated by the bars corresponding to the left y-axis, and the response modeled using the kinetics of nitrifier cultures determined in this study (Supplementary Table 5) is indicated by the symbols corresponding with the right y-axis. Individual nitrifier names are preceded by MM or H to indicate that MM or H parameters, respectively, were used to calculate the potential rates of activity.
Model Predictions Compared to Nitrification in Coastal Plain Soils
Soil AOA had activity in Coastal Plain soils without NH4+ addition, and the NH3 oxidation rate predicted by the MM model was a good fit for soil AOA activity (R2 = 0.815, p = 0.01, Supplementary Table 7, and Figure 3A). Unlike AOA, no activity of soil AOB was measured in the absence of NH4+ addition in Coastal Plain soils, and the AOB MM models also predicted no contribution to NH3 oxidation by soil AOB (Figure 3B). In the absence of NH4+ addition, the MM model did not predict any significant response by N. japonica NJ1 or Nitrospira ND1; however, the rate response of N. moscoviensis determined by the kinetic parameters of the MM model matched the data at 10 and 20°C, but not 30°C (Figure 3C). The rate response of N. vulgaris determined by the kinetic parameters of the MM model could potentially contribute to all the NO2– oxidation observed in this soil (R2 = 0.641, p = 0.01), but the MM models for the remaining NOB do not predict any contribution to NO2– oxidation (Figure 3D).
Addition of NH4+ to Coastal Plain soils increased the rates of NO2– + NO3– production by soil AOA ∼5–10-fold (Figure 3E). H substrate-induced inhibition of N. viennensis predicted a lower AOA contribution to NH3 oxidation than that observed in the soil at 10 and 20°C, although the potential response of AOA like N. viennensis exceeded the response observed in the soil at 30°C (R2 = 0.685, p = 0.006, Figure 3E). In the Coastal Plain soil with NH4+ addition, soil AOB contributed to nitrification at all three incubation temperatures, providing NH3 oxidation rates approximately equal to that of soil AOA at 10 and 20°C, and ∼25% of total NO2– + NO3– production at 30°C; however, neither the MM or H models for N. multiformis or N. europaea were a good fit to the data (Supplementary Table 7 and Figure 3F). The H model predicted no significant effects of substrate inhibition on N. europaea (data not shown) but predicted a reduction in N. multiformis contributions at 20°C. The MM models for N. europaea and N. multiformis showed increasing rates of NO2– + NO3– production as temperature increased from 10 to 30°C, whereas soil AOB rates were the highest at 20°C. The MM models predicted that AOB with kinetic characteristics like those of N. europaea and N. multiformis could contribute much of the NO2– + NO3– production observed at 10 and 20°C, but exceeded soil nitrification rates observed at 30°C. When NH4+ was added to increase NH3-oxidation activity in this soil, there was no accumulation of NO2– in the Coastal Plain soil above our limit of detection (2 μM NO2–), and under these conditions the MM models using kinetic parameters obtained for Nitrospira NOB in this study did not predict Nitrospira contributions to soil NO2– oxidation (R2 = 0.09–0.32, Figure 3G). The greatest MM model-predicted response for Nitrospira was N. moscoviensis, but it would only account for ∼3% of the total soil NOB activity at its highest activity rates. In contrast, the MM model for the Nitrobacter NOB N. vulgaris predicted rates of NO2– oxidation that met or exceeded the response observed by soil NOB, but the MM models did not predict that other Nitrobacter could contribute under these conditions with low concentrations of soil NO2– (Figure 3H).
Model Predictions Compared to Columbia Basin Soils
In Columbia Basin soils with no NH4+ addition, soil AOA had much higher rates of activity than that predicted by the MM model of N. viennensis (Figure 4A). In the absence of addition of NH4+ to soil, AOB had activity at 20, 30, and 40°C, but neither of the MM models using the kinetic parameters from N. multiformis and N. europaea had high enough substrate affinity to predict the oxidation of NH3 at such low concentrations (Supplementary Table 7 and Figure 4B). NO2– did not accumulate in the Columbia Basin soil in the absence of the addition of NH4+; therefore, the rates of soil NOB activity matched that of NH3 oxidation. In the absence of accumulated NO2–, none of the Nitrospira NOB models predicted significant contributions under these low NO2– conditions (Figures 4C,D).
When NH4+ was added to the Columbia Basin soil, there was no significant change in the rates of NH3 oxidation by soil AOA over the incubations without NH4+ addition at 10, 20, and 40°C, but the rates at 30°C declined ∼2-fold, suggesting evidence of substrate inhibition (Figure 4E). While the MM model for N. viennensis predicted high rates of AOA NH3 oxidation in excess of those observed in the Columbia Basin soil, the H model accurately predicted the soil AOA activity (R2 = 0.918, p = 0.000, Figure 4E). With the addition of NH4+ to Columbia Basin soils, there were significant increases in the rates of activity by soil AOB with maximum rates of NH3 oxidation at 30°C (Figure 4F). Both N. europaea and N. multiformis MM models describe the NH3 oxidation response to NH4+ addition at 10, 20, and 30°C, but overestimated the AOB response at 40°C; resulting in the models that poorly fit the soil AOB activity (Supplementary Table 7). Although the rates of NO2– oxidation by soil NOB increased ∼2-fold in response to the higher rates of NH3 oxidation (Figures 4G,H), NO2– accumulated in the Columbia Basin soil when NH4+ was added (2, 700, 1,900, and 400 μM NO2– at 10, 20, 30, and 40°C, respectively), indicating that soil NOB did not have the capacity to immediately respond to higher rates of NH3 oxidation. However, the MM models of both Nitrospira and Nitrobacter NOB indicated a positive benefit in predicted rates of NO2– oxidation from the accumulated NO2–, and only N. moscoviensis showed substrate-induced inhibition at 30 and 40°C (Figure 4G). The MM model for Nitrospira NOB predicted rates equal to or greater than soil NOB at 20 and 40°C, but could not match the rate of soil NOB at 10 or 30°C. The MM and H models with accumulated NO2– for all three Nitrobacter NOB predicted the capacity to express a higher activity than the observed soil NOB activity but resulted in a significant fit of the soil data (p ≤ 0.004, Supplementary Table 7, and Figure 4H).
Discussion
The Effect of Temperature on Catalytic Efficiency
The Km(app) of all three Nitrobacter species increased substantially with temperature at a much greater rate than Vmax(app), resulting in a substantial decrease in catalytic efficiency at higher temperatures. In contrast, both Vmax(app) and Km(app) values of the Nitrospira species increased similarly with temperature, on a much smaller scale, and their overall catalytic efficiency generally did not change significantly. This difference between Nitrobacter and Nitrospira makes sense in view of the analysis of the phylogeny of NxrA suggesting that NO2– oxidation has arisen independently multiple times (Lücker et al., 2010; Sorokin et al., 2012), resulting in different biochemical or functional characteristics. For example, the NXR complex is located in the periplasm space of Nitrospira sp. and in the cytoplasm space of Nitrobacter sp. (Daims et al., 2016), which may contribute to the observed differences in substrate affinity between the groups (Nitrospira Km(app) = 9–24 μM, Nitrobacter Km(app) = 49–544 μM; Nowka et al., 2015; Daims et al., 2016; Ushiki et al., 2017). Additionally, while there was a 50–70% decline in catalytic efficiency with increasing temperature for each of the three Nitrobacter evaluated in this study, there was more variation in the response of Nitrospira; ranging from a ∼50% decline in catalytic efficiency over 10 to 37°C by Ns. ND1, to no change in catalytic efficiency by N. japonica NJ1, and a uniform response in catalytic efficiency across temperature by N. moscoviensis except at 20°C where there was a 4-fold increase. The phylogeny of NxrA indicates that there is more diversity within Nitrospira than in Nitrobacter (Daims 2016 Trends), and all Nitrobacter evaluated in this study have very similar NxrA (92–98% identity) compared to lineages I and II Nitrospira. As for N. japonica and N. moscoviensis, their NxrA have only ∼87% sequence identity and are clearly separated by phylogenetic analysis even though both NOB are in lineage II; the differences in their adaptation to different growth temperatures (30 and 37°C, respectively) may have contributed to their phylogenetic separation and differences in physiological response. N. japonica NJ1 is similarly divergent compared to Nitrospira ND1 (lineage I, 88% identity; Figure 1B).
Although the limited number of NH3 oxidizer cultures utilized in this study prevents definitive conclusions on the response of catalytic efficiency to temperature change, the results agree with previous studies and are in sharp contrast to that of the NOB. Numerous studies have demonstrated higher rates of NH3 oxidation in natural environments as temperatures increase (Myers, 1975; Groeneweg et al., 1994; Avrahami et al., 2003; Tourna et al., 2008; Gubry-Rangin et al., 2017; Ouyang et al., 2017; Taylor et al., 2017; Duan et al., 2018, 2020; Mukhtar et al., 2019; Zheng et al., 2020; Bello et al., 2021), but there is a dearth of information on temperature-dependent changes of substrate affinity and catalytic efficiency. In this study, AOB showed 3–7-fold increases in Km(app) with temperature (decrease in affinity), but greater increases in Vmax(app) with temperature (8–16-fold), which resulted in 2.5–2.8-fold increases in catalytic efficiency. This agrees with previous results with Nitrosomonas sp. 4W30 that had a 1.9–2.7-fold increase in catalytic efficiency as temperatures increased from 5 to 20°C (Jones and Morita, 1985). In the case of the AOA N. viennensis, there was no significant change in Km(app) but Vmax(app) increased nearly two orders of magnitude resulting in a significant increase of catalytic efficiency as temperatures increased. This increase in catalytic efficiency with temperature by N. viennensis was similar to the response of the AOA “Ca. N. oleophilus” MY3 over 25–35°C where there was ∼2-fold increase in catalytic efficiency (Jung et al., 2022).
To date, it is unknown if temperature differentially affects NH3 and NO2– oxidation in comammox Nitrospira. Previous evaluation of the kinetic parameters of NH4+ and NO2– oxidation in comammox cultures of Nitrospira inopinata and “Candidatus Nitrospira kreftii” at their optimal growth temperatures (37°C and room temperature, respectively) found that the catalytic efficiency of NH3 oxidation was one to two orders of magnitude greater than that of NO2– oxidation (Kits et al., 2017; Sakoula et al., 2020). The different catalytic efficiencies of NH3 oxidation (19.6) and NO2– oxidation (0.05) of N. inopinata may have caused the accumulation of NO2– until all NH4+ was consumed (Daims et al., 2015; Kits et al., 2017). In contrast, NO2– did not accumulate in “Candidatus Nitrospira kreftii” cultures where there were smaller differences between the catalytic efficiency of NH3 (33–42) and NO2– oxidation (4–8; van Kessel et al., 2015; Sakoula et al., 2020). Because the activity and catalytic efficiency of NH3 and NO2– oxidizers affect N availability and productivity in soil, marine, and surface waters, and engineered systems, future studies are needed to investigate the coupling of nitrification, including comammox in response to temperature.
A Few Observations on the Michaelis–Menten and Haldane Kinetic Parameters of NH3 and NO2– Oxidation
Ammonia and NO2– are poor energy sources to support an autolithotrophic lifestyle; therefore, it makes intuitive sense that the optimal growth temperature (Topt) of the NH3 oxidizers evaluated in this study was at, or near, the temperature where the response of MM kinetics resulted in near peak catalytic efficiency for NH3 oxidation (Figure 2). In contrast, the growth Topt of the six NOB evaluated in this study did not occur at the temperatures where their catalytic efficiency of NO2– oxidation was at its best. Under optimal growth conditions, it has been calculated that NOBs spend ∼26.7% of the energy generated from NO2– oxidation in cellular homeostasis, while those demands rise greater than 3-fold under non-growth conditions (Vadivelu et al., 2006; Ni et al., 2008). Perhaps, NOBs prioritize cell maintenance over growth. Nitrobacter NOBs have multiple energy storage cell inclusions, such as poly-β-hydroxybutyrate, glycogen, and polyphosphate granules (Watson et al., 1989; Spieck and Bock, 2005), and the energy generated during the oxidation of NO2– may be used to create or enrich these storage molecules until optimal growth conditions are met. We also observed that the H substrate inhibition of N. viennensis, N. europaea, N. moscoviensis, and N. winogradskyi, was temperature-sensitive (Supplementary Table 5), which may be another potential explanation for why Topt for growth and the temperature for optimal catalytic efficiency are not aligned, for it has been long known that substrate inhibition can occur during the isolation and culture of nitrifiers (Both et al., 1992; Bartosch et al., 2002; Spieck et al., 2006; Spieck and Lipski, 2011; Tourna et al., 2011). The H inhibition constant Ki was the highest (lowest sensitivity) at the optimal growth temperature (30°C) of N. winogradskyi and N. vulgaris, suggesting that substrate sensitivity may determine the Topt for growth rather than optimal catalytic efficiency. We also observed that N. moscoviensis, N. japonica strain NJ1, and N. vulgaris all had their highest sensitivity (Ki < 0.9 mM) to NO2– at 10°C, which is interesting because Venterea et al. (2020) reported NO2– accumulation in soil at temperatures of 5 and 10°C, whereas it was quickly consumed at 22 and 30°C. Intriguingly, we found that there was a trend for the AOA N. viennensis to be the most sensitive to NH4+ (Ki 0.3–0.5 mM) at its optimal growth temperature, which may help explain why it has been challenging to isolate AOA cultures.
Both Nitrobacter and Nitrospira have demonstrated the ability to utilize NO3– as a terminal electron acceptor in the presence of organic molecules, using NXR in the reverse direction to reduce NO3– to NO2– (Aleem and Sewell, 1981; Tanaka et al., 1983; Sundermeyerklinger et al., 1984; Freitag et al., 1987; Meincke et al., 1992; Lücker et al., 2010); and in the case of Nitrobacter, NXR possessed a higher affinity for NO3– (∼0.9 mM NO3–) than for NO2– (0.54–3.6 mM NO2–). Intriguingly, the highest rates of ATP production were observed in whole-cell incubation of N. winogradskyi when both oxygen and NO3– were provided as electron acceptors, suggesting the potential for NO2– oxidation and NO3– reduction to occur concurrently (Freitag and Bock, 1990). There are examples of temperature affecting the balance between the forward and reverse direction of enzymes in different ways. The equilibrium in favor of the forward reaction of glucose isomerase increased from 50 to 60°C (Dehkordi et al., 2009) while the affinity of the forward reaction of glycerol-2-phosphate dehydrogenase was higher at 5°C compared to 21°C or 37°C (Ruberto et al., 2019). It is unknown how temperature will affect the balance between the forward and reverse actions of NXR, or if NXR of Nitrobacter and Nitrospira will respond in different ways.
Protein Structure or Membrane Differences Between Groups
Bacterial and archaeal AMOs are integral membrane proteins (Norton et al., 2002; Urakawa et al., 2011), NXRs of Nitrobacter are membrane-bound proteins (Sundermeyerklinger et al., 1984; Meincke et al., 1992), and NXRs of Nitrospira are soluble proteins anchored to membranes (Mundinger et al., 2019); and it may be worth considering differences in membrane lipid composition between lineages to explain dissimilar temperature responses. This is based on (i) membrane dynamics and fluidity change in response to temperature, and the degree to which they change depends on lipid composition and lipid phase transition temperature (Mansilla et al., 2004), (ii) membrane protein activity and stability can be dependent on lipid composition and dynamics (Hong and Tamm, 2004; Reading et al., 2017; Sanders et al., 2018), (iii) lipid composition of Nitrobacter and Nitrospira is quite different (Lipski et al., 2001), indicating that their lipid membranes will respond differently to temperature, and (iv) AOA and other Thaumarchaeota have a high proportion of crenarchaeol, a core lipid that is unique in structure and potentially function from those of bacteria (Pitcher et al., 2010; Elling et al., 2014). It is well known that the archaeal and bacterial membrane lipid composition is dependent on growth temperature and is often used as a proxy for past temperatures (Elling et al., 2014; De Jonge et al., 2019), and that both bacteria and archaea actively modify lipid membrane composition in response to transient changes in temperature (Balogh et al., 2013; Siliakus et al., 2017). The cyclization of thaumarchaeal glycerol dialkyl glycerol tetraethers (GDGTs) membrane-spanning lipids is strongly correlated with sea surface temperature (Elling et al., 2014; Qin et al., 2015), and the degree of methylization and cyclization of branched bacterial glycerol dialkyl glycerol tetraethers (GDGTs) that are abundant in terrestrial and aquatic systems varies with the mean annual temperature (De Jonge et al., 2019). While NH3 and NO2– oxidizers may have the ability to modify the lipid composition of cellular and periplasmic membranes in response to transient temperature changes, the incubations in this study were likely not long enough for such changes to occur. However, the cultures in this study grown under optimal conditions would differ in lipid composition in ways that could influence lipid dynamics, and may be as important as variation in protein structure in governing the temperature response of individual nitrifying organisms.
Implications of Changes in Catalytic Efficiency in Natural Environments
Based on these data, increasing temperature should result in an increase in the catalytic efficiency of NH3 oxidation, while Nitrospira NOBs are not predicted to increase catalytic efficiency with temperature, and the catalytic efficiency of Nitrobacter NOB declines significantly. This differential change in catalytic efficiency may help explain the accumulation of NO2– previously observed in other studies. For example, increasing temperature decoupled NO2– oxidation from NH3 oxidation in coastal waters (Heiss and Fulweiler, 2016; Schaefert and Hollibaugh, 2017), and in multiple soil types (Giguere et al., 2017; Taylor et al., 2019; Duan et al., 2020; Waggoner et al., 2021). We have previously observed that the accumulation of NO2– was required to stimulate the rates of NO2– oxidation to match those of NH3 oxidation in a range of soils (Giguere et al., 2018; Taylor et al., 2019); but accumulated NO2– also has the potential to inhibit NOB activity (Park and Bae, 2009; Zheng et al., 2021; Supplementary Table 5). Rates of NO2– oxidation were stimulated ∼2-fold across all temperatures when NO2– accumulated in NH4+-amended Columbia Basin soil incubations (Figure 3); however, these concentrations were greater than the temperature-dependent Ki of four of the NOB cultures evaluated in this study, and our modeling suggests temperature-dependent H inhibition, particularly of Nitrospira NOB.
Changes in enzyme catalytic efficiency with temperature are not specific to nitrification, and previous studies of some enzyme activities linked to processing of organic C, N, and P (cellobiohydrolase, β-glucosidase and xylanase, phosphatase, leucine-aminopeptidase, and tyrosine-aminopeptidase) in soil have observed changes in catalytic efficiency in response to temperature. Catalytic efficiency was positively correlated with temperature at sites ranging from boreal to tropical forests (German et al., 2012) and in long-term N addition plots (Stone et al., 2012). In other studies, catalytic efficiency increased with temperature or did not change depending on the enzyme evaluated (Blagodatskaya et al., 2016; Razavi et al., 2016). Curiously, none of these studies observed pronounced declines in catalytic efficiency as temperature increased as was observed in this study with Nitrobacter NOB.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
AT was responsible for experimental design, data analysis, and writing of the manuscript. BM was responsible for execution of experiments, phylogenetic analysis, and writing of the manuscript. Both authors contributed to the article and approved the submitted version.
Funding
This research was supported by USDA NIFA award 2012-67019-3028 and an Oregon Agricultural Research Foundation competitive grant.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Luis Sayavedra-Soto, Jenny Norton, Holger Daims, Christa Schelper, and Eva Spieck for providing pure cultures of AOB, NOB, and archaea, and Satoshi Tsuneda for providing pure cultures of N. japonica NJ1 and Nitrospira sp. ND1. We thank Peter Bottomley for his lab assistance and insights. We also thank Ashley Waggoner for the use of her soil incubation data set, and Rick Cooley for helpful discussions and insights into lipid composition.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.817986/full#supplementary-material
Footnotes
- ^ https://soilseries.sc.egov.usda.gov
- ^ https://www.ncbi.nlm.nih.gov/
- ^ http://www.genome.jp/kegg/
- ^ http://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ http://tree.bio.ed.ac.uk/software/figtree/
References
Aleem, M. I. H., and Sewell, D. L. (1981). Mechanism of nitrite oxidation and oxidoreductase systems in nitrobacter-agilis. Curr. Microbiol. 5, 267–272. doi: 10.1007/bf01567916
Avrahami, S., Liesack, W., and Conrad, R. (2003). Effects of temperature and fertilizer on activity and community structure of soil ammonia oxidizers. Environ. Microbiol. 5, 691–705. doi: 10.1046/j.1462-2920.2003.00457.x
Balogh, G., Peter, M., Glatz, A., Gombos, I., Torok, Z., Horvath, I., et al. (2013). Key role of lipids in heat stress management. FEBS Lett. 587, 1970–1980. doi: 10.1016/j.febslet.2013.05.016
Bartosch, S., Hartwig, C., Spieck, E., and Bock, E. (2002). Immunological detection of Nitrospira-like bacteria in various soils. Microb. Ecol. 43, 26–33. doi: 10.1007/s00248-001-0037-35
Bello, M. O., Aigle, A., Meng, Y. Y., Prosser, J. I., and Gubry-Rangin, C. (2021). Preferential temperature and ammonia concentration for in-situ growth of candidatus Nitrosocosmicus ammonia oxidising archaea. Soil Biol. Biochem. 162:108405. doi: 10.1016/j.soilbio.2021.108405
Bennett, A. E., Preedy, K., Golubski, A., Umbanhowar, J., Borrett, S. R., Byrne, L., et al. (2019). Beyond the black box: promoting mathematical collaborations for elucidating interactions in soil ecology. Ecosphere 10:e02799.
Blagodatskaya, E., Blagodatsky, S., Khomyakov, N., Myachina, O., and Kuzyakov, Y. (2016). Temperature sensitivity and enzymatic mechanisms of soil organic matter decomposition along an altitudinal gradient on Mount Kilimanjaro. Sci. Rep. 6:22240. doi: 10.1038/srep22240
Bock, E., Koops, H. P., Moller, U. C., and Rudert, M. (1990). A new facultatively nitrite oxidizing bacterium, Nitrobacter vulgaris Sp-Nov. Arch. Microbiol. 153, 105–110. doi: 10.1007/Bf00247805
Bock, E., Sundermeyerklinger, H., and Stackebrandt, E. (1983). New facultative lithoautotrophic nitrite-oxidizing bacteria. Arch. Microbiol. 136, 281–284. doi: 10.1007/Bf00425217
Bollmann, A., Schmidt, I., Saunders, A. M., and Nicolaisen, M. H. (2005). Influence of starvation on potential ammonia-oxidizing activity and amoA mRNA levels of Nitrosospira briensis. Appl. Environ. Microbiol. 71, 1276–1282. doi: 10.1128/AEM.71.3.1276-1282.2005
Bonnett, S. A. F., Blackwell, M. S. A., Leah, R., Cook, V., O’Connor, M., and Maltby, E. (2013). Temperature response of denitrification rate and greenhouse gas production in agricultural river marginal wetland soils. Geobiology 11, 252–267. doi: 10.1111/gbi.12032
Boratyn, G. M., Camacho, C., Cooper, P. S., Coulouris, G., Fong, A., Ma, N., et al. (2013). BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41, W29–W33. doi: 10.1093/nar/gkt282
Both, G. J., Gerards, S., and Laanbroek, H. J. (1992). Kinetics of nitrite oxidation in two nitrobacter species grown in nitrite-limited chemostats. Arch. Microbiol. 157, 436–441. doi: 10.1007/Bf00249101
Butterbach-Bahl, K., Baggs, E. M., Dannenmann, M., Kiese, R., and Zechmeister-Boltenstern, S. (2013). Nitrous oxide emissions from soils: how well do we understand the processes and their controls? Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20130122. doi: 10.1098/rstb.2013.0122
Button, D. K. (1998). Nutrient uptake by microorganisms according to kinetic parameters from theory as related to cytoarchitecture. Microbiol. Mol. Biol. Rev. 62, 636–645. doi: 10.1128/MMBR.62.3.636-645.1998
Chain, P., Lamerdin, J., Larimer, F., Regala, W., Lao, V., Land, M., et al. (2003). Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J. Bacteriol. 185, 6496–6496. doi: 10.1128/JB.185.9.2759-2773.2003
Daims, H., Lebedeva, E. V., Pjevac, P., Han, P., Herbold, C., Albertsen, M., et al. (2015). Complete nitrification by Nitrospira bacteria. Nature 528, 504–509. doi: 10.1038/nature16461
Daims, H., Lücker, S., and Wagner, M. (2016). A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. 24, 699–712. doi: 10.1016/j.tim.2016.05.004
Daims, H., Lucker, S., Paslier, D. L., and Wagner, M. (2011). “Diversity, environmental genomics, and ecophysiology of nitrite-oxidizing bacteria,” in Nitrification, eds B. B. Ward, D. J. Arp, and M. G. Klotz (Washington, DC: ASM Press), 295–322. doi: 10.1128/9781555817145.ch12
Davidson, E. A., and Janssens, I. A. (2006). Temperature sensitivity of soil carbon decomposition and feedbacks to climate change. Nature 440, 165–173. doi: 10.1038/nature04514
De Jonge, C., Radujkovic, D., Sigurdsson, B. D., Weedon, J. T., Janssens, I., and Peterse, F. (2019). Lipid biomarker temperature proxy responds to abrupt shift in the bacterial community composition in geothermally heated soils. Organ. Geochem. 137:103897. doi: 10.1016/j.orggeochem.2019.07.006
DeAngelis, K. M., Silver, W. L., Thompson, A. W., and Firestone, M. K. (2010). Microbial communities acclimate to recurring changes in soil redox potential status. Environ. Microbiol. 12, 3137–3149. doi: 10.1111/j.1462-2920.2010.02286.x
Dehkordi, A. M., Tehrany, M. S., and Safari, I. (2009). Kinetics of glucose isomerization to fructose by immobilized glucose isomerase (Sweetzyme IT). Industrial Eng. Chem. Res. 48, 3271–3278. doi: 10.1021/ie800400b
Duan, P. P., Wu, Z., Zhang, Q. Q., Fan, C. H., and Xiong, Z. Q. (2018). Thermodynamic responses of ammonia-oxidizing archaea and bacteria explain N2O production from greenhouse vegetable soils. Soil Biol. Biochem. 120, 37–47. doi: 10.1016/j.soilbio.2018.01.027
Duan, P. P., Zhang, Q. Q., and Xiong, Z. Q. (2020). Temperature decouples ammonia and nitrite oxidation in greenhouse vegetable soils. Sci. Total Environ. 733:139391. doi: 10.1016/j.scitotenv.2020.139391
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Ehrich, S., Behrens, D., Lebedeva, E., Ludwig, W., and Bock, E. (1995). A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira-moscoviensis Sp-Nov and its phylogenetic relationship. Arch. Microbiol. 164, 16–23. doi: 10.1007/bf02568729
Elling, F. J., Konneke, M., Lipp, J. S., Becker, K. W., Gagen, E. J., and Hinrichs, K. U. (2014). Effects of growth phase on the membrane lipid composition of the thaumarchaeon Nitrosopumilus maritimus and their implications for archaeal lipid distributions in the marine environment. Geochim. Cosmochim. Acta 141, 579–597. doi: 10.1016/j.gca.2014.07.005
Ensign, S. A., Hyman, M. R., and Arp, D. J. (1993). Invitro activation of ammonia monooxygenase from Nitrosomonas europaea by copper. J. Bacteriol. 175, 1971–1980. doi: 10.1128/jb.175.7.1971-1980.1993
Freitag, A., and Bock, E. (1990). Energy-conservation in nitrobacter. FEMS Microbiol. Lett. 66, 157–162. doi: 10.1111/j.1574-6968.1990.tb03989.x
Freitag, A., Rudert, M., and Bock, E. (1987). Growth of nitrobacter by dissimilatoric nitrate reduction. FEMS Microbiol. Lett. 48, 105–109. doi: 10.1111/j.1574-6968.1987.tb02524.x
Galloway, J. N., Townsend, A. R., Erisman, J. W., Bekunda, M., Cai, Z., Freney, J. R., et al. (2008). Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science 320, 889–892. doi: 10.1126/science.1136674
German, D. P., Marcelo, K. R. B., Stone, M. M., and Allison, S. D. (2012). The michaelis-menten kinetics of soil extracellular enzymes in response to temperature: a cross-latitudinal study. Global Change Biol. 18, 1468–1479. doi: 10.1111/j.1365-2486.2011.02615.x
Giguere, A. T., Taylor, A. E., Myrold, D. D., and Bottomley, P. J. (2015). Nitrification responses of soil ammonia-oxidizing archaea and bacteria to ammonium concentrations. Soil Sci. Soc. Am. J. 79, 1366–1374. doi: 10.2136/sssaj2015.03.0107
Giguere, A. T., Taylor, A. E., Myrold, D. D., Mellbye, B., Sayavedra-Soto, L., and Bottomley, P. J. (2018). Nitrite-oxidizing activity responds to nitrite accumulation in soil. FEMS Microbiol. Ecol. 94:fiy008. doi: 10.1093/femsec/fiy008
Giguere, A. T., Taylor, A. E., Suwa, Y., Myrold, D. D., and Bottomley, P. J. (2017). Uncoupling of ammonia oxidation from nitrite oxidation: impact upon nitrous oxide production in non-cropped Oregon soils. Soil Biol. Biochem. 104, 30–38. doi: 10.1016/j.soilbio.2016.10.011
Groeneweg, J., Sellner, B., and Tappe, W. (1994). Ammonia oxidation in nitrosomonas at NH3 concentrations near Km - effects of pH and temperature. Water Res. 28, 2561–2566. doi: 10.1016/0043-1354(94)90074-4
Gruber-Dorninger, C., Pester, M., Kitzinger, K., Savio, D. F., Loy, A., Rattei, T., et al. (2015). Functionally relevant diversity of closely related Nitrospira in activated sludge. ISME J. 9, 643–655. doi: 10.1038/ismej.2014.156
Gubry-Rangin, C., Novotnik, B., Mandic-Mulec, I., Nicol, G. W., and Prosser, J. I. (2017). Temperature responses of soil ammonia-oxidising archaea depend on pH. Soil Biol. Biochem. 106, 61–68. doi: 10.1016/j.soilbio.2016.12.007
Heiss, E. M., and Fulweiler, R. W. (2016). Coastal water column ammonium and nitrite oxidation are decoupled in summer. Estuarine Coastal Shelf Sci. 178, 110–119. doi: 10.1016/j.ecss.2016.06.002
Hidetoshi, U., Martens-Habbena, W., and Stahl, D. A. (2011). “Physiology and genomics of ammonia-oxidizing archaea,” in Nitrification, eds B. B. Ward, D. J. Arp, and M. G. Klotz (Washington, DC: ASM Press), 117–155.
Hobbs, J. K., Jiao, W., Easter, A. D., Parker, E. J., Schipper, L. A., and Arcus, V. L. (2013). Change in heat capacity for enzyme catalysis determine temperature dependence of enzyme catalyzed rates. ACS Chem. Biol. 8, 2388–2393. doi: 10.1021/cb4005029
Hong, H. D., and Tamm, L. K. (2004). Elastic coupling of integral membrane protein stability to lipid bilayer forces. Proc. Natl. Acad. Sci. U S A. 101, 4065–4070. doi: 10.1073/pnas.0400358101
Hood-Nowotny, R., Hinko-Najera Umana, N., Inselbacher, E., Oswald-Lachouani, P., and Wanek, W. (2010). Alternative methods for measuring inorganic, organic, and total dissolved nitrogen in soil. Soil Sci. Soc. Am. J. 74, 1018–1027. doi: 10.2136/Sssaj2009.0389
Jones, D. T., Taylor, W. R., and Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282. doi: 10.1093/bioinformatics/8.3.275
Jones, R. D., and Morita, R. Y. (1985). Low-temperature growth and whole-cell kinetics of a marine ammonium oxidizer. Mar. Ecol. Prog. Series 21, 239–243. doi: 10.3354/meps021239
Jung, M. Y., Sedlacek, C. J., Kits, K. D., Mueller, A. J., Rhee, S. K., Hink, L., et al. (2022). Ammonia-oxidizing archaea possess a wide range of cellular ammonia affinities. ISME J. 16, 272–283. doi: 10.1038/s41396-021-01064-z
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y., and Morishima, K. (2017). KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361. doi: 10.1093/nar/gkw1092
Kim, D. J., Lee, D. I., and Keller, J. (2006). Effect of temperature and free ammonia on nitrification and nitrite accumulation in landfill leachate and analysis of its nitrifying bacterial community by FISH. Bioresour. Technol. 97, 459–468. doi: 10.1016/j.biortech.2005.03.032
Kits, K. D., Sedlacek, C. J., Lebedeva, E. V., Han, P., Bulaev, A., Pjevac, P., et al. (2017). Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature 549, 269–272. doi: 10.1038/nature23679
Koch, H., Lücker, S., Albertsen, M., Kitzinger, K., Herbold, C., Spieck, E., et al. (2015). Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc. Natl. Acad. Sci. U S A. 112, 11371–11376. doi: 10.1073/pnas.1506533112
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Laidler, K. J., and Peterman, B. F. (1979). Temperature effects in enzyme kinetics. Methods Enzymol. 63, 234–257. doi: 10.1016/0076-6879(79)63012-4
Law, Y., Ni, B. J., Lant, P., and Yuan, Z. (2012). N2O production rate of an enriched ammonia-oxidising bacteria culture exponentially correlates to its ammonia oxidation rate. Water Res. 46, 3409–3419. doi: 10.1016/j.watres.2012.03.043
Liang, J. Y., Qi, X., Souza, L., and Luo, Y. Q. (2016). Processes regulating progressive nitrogen limitation under elevated carbon dioxide: a meta-analysis. Biogeosciences 13, 2689–2699. doi: 10.5194/bg-13-2689-2016
Lipski, A., Spieck, E., Makolla, A., and Altendorf, K. (2001). Fatty acid profiles of nitrite-oxidizing bacteria reflect their phylogenetic heterogeneity. System. Appl. Microbiol. 24, 377–384. doi: 10.1078/0723-2020-2049
Lücker, S., Wagner, M., Maixner, F., Pelletier, E., Koch, H., Vacherie, B., et al. (2010). A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl. Acad. Sci. U S A. 107, 13479–13484. doi: 10.1073/pnas.1003860107
Mansilla, M. C., Cybulski, L. E., Albanesi, D., and de Mendoza, D. (2004). Control of membrane lipid fluidity by molecular thermosensors. J. Bacteriol. 186, 6681–6688. doi: 10.1128/Jb.186.20.6681-6688.2004
Martens-Habbena, W., Berube, P. M., Urakawa, H., de la Torre, J. R., and Stahl, D. A. (2009). Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461, 976–979. doi: 10.1038/nature08465
McGuire, K. L., and Treseder, K. K. (2010). Microbial communities and their relevance for ecosystem models: decomposition as a case study. Soil Biol. Biochem. 42, 529–535. doi: 10.1016/j.soilbio.2009.11.016
Meincke, M., Bock, E., Kastrau, D., and Kroneck, P. M. H. (1992). Nitrite oxidoreductase from Nitrobacter hamburgensis - redox centers and their catalytic role. Arch. Microbiol. 158, 127–131. doi: 10.1007/bf00245215
Mellbye, B. L., Giguere, A., Chaplen, F., Bottomley, P. J., and Sayavedra-Soto, L. A. (2016). Steady-state growth under inorganic carbon limitation conditions increases energy consumption for maintenance and enhances nitrous oxide production in Nitrosomonas europaea. Appl. Environ. Microbiol. 82, 3310–3318. doi: 10.1128/AEM.00294-216
Mellbye, B. L., Davis, E. W. II, Spieck, E., Chang, J. H., Bottomley, P. J., and Sayavedra-Soto, L. A. (2017a). Draft genome sequence of Nitrobacter vulgaris strain Ab1, a nitrite-oxidizing bacterium. Genome Announc. 5, 290–217. doi: 10.1128/genomeA.00290-217
Mellbye, B. L., Spieck, E., Bottomley, P. J., and Sayavedra-Soto, L. A. (2017b). Acyl-Homoserine lactone production in nitrifying bacteria of the genera Nitrosospira, Nitrobacter, and Nitrospira identified via a survey of putative quorum-sensing genes. Appl. Environ. Microbiol. 83:e01540-17. doi: 10.1128/AEM.01540-17
Miranda, K. M., Espey, M. G., and Wink, D. A. (2001). A rapid, simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide 5, 62–71. doi: 10.1006/niox.2000.0319
Mukhtar, H., Lin, Y. P., Lin, C. M., and Petway, J. R. (2019). Assessing thermodynamic parameter sensitivity for simulating temperature responses of soil nitrification. Environ. Science-Proc. Impacts 21, 1596–1608. doi: 10.1039/c9em00310j
Mundinger, A. B., Lawson, C. E., Jetten, M. S. M., Koch, H., and Lucker, S. (2019). Cultivation and transcriptional analysis of a canonical Nitrospira under stable growth conditions. Front. Microbiol. 10:1325. doi: 10.3389/fmicb.2019.01325
Myers, R. J. K. (1975). Temperature effects on ammonification and nitrification in a tropical soil. Soil Biol. Biochem. 7, 83–86. doi: 10.1016/0038-0717(75)90003-6
Ni, B. J., Fang, F., Xie, W. M., and Yu, H. Q. (2008). Growth, maintenance and product formation of autotrophs in activated sludge: taking the nitrite-oxidizing bacteria as an example. Water Res. 42, 4261–4270. doi: 10.1016/j.watres.2008.06.024
Ni, X., and Groffman, P. M. (2018). Declines in methane uptake in forest soils. PNAS 115, 8587–8590. doi: 10.1073/pnas.1807377115
Norton, J. M., Alzerreca, J. J., Suwa, Y., and Klotz, M. G. (2002). Diversity of ammonia monooxygenase operon in autotrophic ammonia-oxidizing bacteria. Arch. Microbiol. 177, 139–149. doi: 10.1007/s00203-001-0369-z
Norton, J. M., Klotz, M. G., Stein, L. Y., Arp, D. J., Bottomley, P. J., Chain, P. S., et al. (2008). Complete genome sequence of Nitrosospira multiformis, an ammonia-oxidizing bacterium from the soil environment. Appl. Environ. Microbiol. 74, 3559–3572. doi: 10.1128/AEM.02722-07
Nowka, B., Daims, H., and Spieck, E. (2015). Comparison of oxidation kinetics of nitrite-oxidizing Bacteria: nitrite availability as a key factor in niche differentiation. Appl. Environ. Microbiol. 81, 745–753. doi: 10.1128/AEM.02734-14
Ouyang, Y., Norton, J. M., and Stark, J. M. (2017). Ammonium availability and temperature control contributions of ammonia oxidizing bacteria and archaea to nitrification in an agricultural soil. Soil Biol. Biochem. 113, 161–172. doi: 10.1016/j.soilbio.2017.06.010
Park, S., and Bae, W. (2009). Modeling kinetics of ammonium oxidation and nitrite oxidation under simultaneous inhibition by free ammonia and free nitrous acid. Proc. Biochem. 44, 631–640. doi: 10.1016/j.procbio.2009.02.002
Pester, M., Maixner, F., Berry, D., Rattei, T., Koch, H., Lücker, S., et al. (2014). NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ. Microbiol. 16, 3055–3071. doi: 10.1111/1462-2920.12300
Pierre, S., Hewson, I., Sparks, J. P., Litton, C. M., Giardina, C., Groffman, P. M., et al. (2017). Ammonia oxidizer populations vary with nitrogen cycling across a tropical montane mean annual temperature gradient. Ecology 98, 1896–1907. doi: 10.1002/ecy.1863
Pitcher, A., Rychlik, N., Hopmans, E. C., Spieck, E., Rijpstra, W. I. C., Ossebaar, J., et al. (2010). Crenarchaeol dominates the membrane lipids of Candidatus Nitrososphaera gargensis, a thermophilic Group I. 1b Archaeon. ISME J. 4, 542–552. doi: 10.1038/ismej.2009.138
Prosser, J. I., and Nicol, G. W. (2012). Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol. 20, 523–531. doi: 10.1016/j.tim.2012.08.001
Qin, W., Carlson, L. T., Armbrust, E. V., Devol, A. H., Moffett, J. W., Stahl, D. A., et al. (2015). Confounding effects of oxygen and temperature on the TEX86 signature of marine Thaumarchaeota. Proc. Natl. Acad. Sci. U S A. 112, 10979–10984. doi: 10.1073/pnas.1501568112
Razavi, B. S., Blagodatskaya, E., and Kuzyakov, Y. (2016). Temperature selects for static soil enzyme systems to maintain high catalytic efficiency. Soil Biol. Biochem. 97, 15–22. doi: 10.1016/j.soilbio.2016.02.018
Reading, E., Hall, Z., Martens, C., Haghighi, T., Findlay, H., Ahdash, Z., et al. (2017). Interrogating membrane protein conformational dynamics within native lipid compositions. Angewandte Chemie-Int. Edn. 56, 15654–15657. doi: 10.1002/anie.201709657
Ruberto, A. A., Logan, S. M., and Storey, K. B. (2019). Temperature and serine phosphorylation regulate glycerol-3-phosphate dehydrogenase in skeletal muscle of hibernating Richardson’s ground squirrels. Biochem. Cell Biol. 97, 148–157. doi: 10.1139/bcb-2018-2198
Sakoula, D., Koch, H., Frank, J., Jetten, M. S. M., and van Kessel, M. A. H. J. (2020). Enrichment and physiological characterization of a novel comammox Nitrospira indicates ammonium inhibition of complete nitrification. ISME J. 15, 1010–1024. doi: 10.1038/s41396-020-00827-4
Sanders, M. R., Findlay, H. E., and Booth, P. J. (2018). Lipid bilayer composition modulates the unfolding free energy of a knotted alpha-helical membrane protein. Proc. Natl. Acad. Sci. U S A. 115, E1799–E1808. doi: 10.1073/pnas.1714668115
Sayavedra-Soto, L. A., and Arp, D. J. (2011). “Ammonia-oxidizing bacteria: their biochemistry and molecular biology,” in Nitrification, eds B. B. Ward, D. J. Arp, and M. G. Klotz (Washington, DC: ASM Press), 11–37. doi: 10.1155/2019/6957210
Schaefert, S. C., and Hollibaugh, J. T. (2017). Temperature decouples ammonium and nitrite oxidation in coastal waters. Environ. Sci. Technol. 51, 3157–3164. doi: 10.1021/acs.est.6b03483
Schreiber, F., Wunderlin, P., Udert, K. M., and Wells, G. F. (2012). Nitric oxide and nitrous oxide turnover in natural and engineered microbial communities: biological pathways, chemical reactions, and novel technologies. Front. Microbiol. 3:372. doi: 10.3389/fmicb.2012.00372
Shade, A., Carey, C. C., Kara, E., Bertilsson, S., McMahon, K. D., and SMith, M. C. (2009). Can the black box be cracked? the augmentation of microbial ecology to high-resolution, automated sensing technologies. ISME J. 3, 881–888. doi: 10.1038/ismej.2009.56
Shelton, R. E., Jacobsen, K. L., and McCulley, R. L. (2018). Cover crops and certilization alter nitrogen loss in organic and conventional conservation agriculture systems. Front. Plant Sci. 8:2260. doi: 10.3389/fpls.2017.02260
Siliakus, M. F., van der Oost, J., and Kengen, S. W. M. (2017). Adaptations of archaeal and bacterial membranes to variations in temperature, pH and pressure. Extremophiles 21, 651–670. doi: 10.1007/s00792-017-0939-x
Snider, M. J., Gaunitz, S., Ridgway, C., Short, S. A., and Wolfenden, R. (2000). Temperature effects on the catalytic efficiency, rate enhancement, and transition state affinity of cytidine deaminase, and the thermodynamic consequences for catalysis of removing a substrate “anchor”. Biochemistry 39, 9746–9753. doi: 10.1021/bi000914y
Sorokin, D. Y., Lücker, S., Vejmelkova, D., Kostrikina, N. A., Kleerebezem, R., Rijpstra, W. I. C., et al. (2012). Nitrification expanded: discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. ISME J. 6, 2245–2256. doi: 10.1038/ismej.2012.70
Spieck, E., and Bock, E. (2005). “The lithoautotrophic nitrite-oxidizing bacteria,” in Bergy’s Manual of Systematic Bacteriology, 2nd Edn, eds D. J. Brenner, N. R. Krieg, and J. T. Staley (New York, N.Y: Springer).
Spieck, E., and Lipski, A. (2011). Cultivation, growth physiology, and chemotaxonomy of nitrite-oxidizing bacteria. Methods Enzymol. Res. Nitrification Related Proc. 486, 109–130. doi: 10.1016/B978-0-12-381294-0.00005-5
Spieck, E., Hartwig, C., McCormack, I., Maixner, F., Wagner, M., Lipski, A., et al. (2006). Selective enrichment and molecular characterization of a previously uncultured Nitrospira-like bacterium from activated sludge. Environ. Microbiol. 8, 405–415. doi: 10.1111/j.1462-2920.2005.00905.x
Starkenburg, S. R., Chain, P. S. G., Sayavedra-Soto, L. A., Hauser, L., Land, M. L., Larimer, F. W., et al. (2006). Genome sequence of the chemolithoautotrophic nitrite-oxidizing bacterium Nitrobacter winogradskyi Nb-255. Appl. Environ. Microbiol. 72, 2050–2063. doi: 10.1128/Aem.72.3.2050-2063.2006
Starkenburg, S. R., Larimer, F. W., Stein, L. Y., Klotz, M. G., Chain, P. S. G., Sayavedra-Soto, L. A., et al. (2008). Complete genome sequence of Nitrobacter hamburgensis X14 and comparative genomic analysis of species within the genus Nitrobactei. Appl. Environ. Microbiol. 74, 2852–2863. doi: 10.1128/Aem.02311-2317
Starkenburg, S. R., Spieck, E., and Bottomley, P. J. (2011). “Metabolism and genomics of nitrite-oxidizing bacteria: emphasis on studies of pure cultures and of Nitrobacter species,” in Nitrification, eds B. B. Ward, D. J. Arp, and M. G. Klotz (Washington, DC: ASM Press), 267–293.
Steffen, W., Richardson, K., Rockstrom, J., Cornell, S. E., Fetzer, I., Bennett, E. M., et al. (2015). Sustainability. planetary boundaries: guiding human development on a changing planet. Science 347:1259855. doi: 10.1126/science.1259855
Stevens, C. J. (2019). Nitrogen in the environment. Science 363, 578–580. doi: 10.1126/science.aav8215
Stone, M. M., Weiss, M. S., Goodale, C. L., Adams, M. B., Fernandez, I. J., German, D. P., et al. (2012). Temperature sensitivity of soil enzyme kinetics under N-fertilization in two temperate forests. Global Change Biol. 18, 1173–1184. doi: 10.1111/j.1365-2486.2011.02545.x
Sundermeyerklinger, H., Meyer, W., Warninghoff, B., and Bock, E. (1984). Membrane-bound nitrite oxidoreductase of Nitrobacter - evidence for a nitrate reductase system. Arch. Microbiol. 140, 153–158. doi: 10.1007/bf00454918
Suzuki, I., Dular, U., and Kwok, S. C. (1974). Ammonia or ammonium ion as substrate for oxidation by Nitrosomonas europaea cells and extracts. J. Bacteriol. 120, 556–558. doi: 10.1128/jb.120.1.556-558.1974
Tanaka, Y., Fukumori, Y., and Yamanaka, T. (1983). Purification of cytochrome-a1c1 from Nitrobacter agilis and characterization of nitrite oxidation system of the bacterium. Arch. Microbiol. 135, 265–271. doi: 10.1007/bf00413479
Taylor, A. E., Giguere, A. T., Zoebelein, C. M., Myrold, D. D., and Bottomley, P. J. (2017). Modeling of soil nitrification responses to temperature reveals thermodynamic differences between ammonia-oxidizing activity of archaea and bacteria. ISME J. 11, 896–908. doi: 10.1038/ismej.2016.179
Taylor, A. E., Myrold, D. D., and Bottomley, P. J. (2019). Temperature affects the kinetics of nitrite oxidation and nitrification coupling in four agricultural soils. Soil Biol. Biochem. 136:107523. doi: 10.1016/j.soilbio.2019.107523
Taylor, A. E., Taylor, K., Tennigkeit, B., Palatinszky, M., Stieglmeier, M., Myrold, D. D., et al. (2015). Inhibitory effects of C2 to C10 1-alkynes on ammonia oxidation in two Nitrososphaera species. Appl. Environ. Microbiol. 81, 1942–1948. doi: 10.1128/AEM.03688-3614
Taylor, A. E., Vajrala, N., Giguere, A. T., Gitelman, A. I., Arp, D. J., Myrold, D. D., et al. (2013). Use of aliphatic n-alkynes to discriminate soil nitrification activities of ammonia-oxidizing Thaumarchaea and Bacteria. Appl. Environ. Microbiol. 79, 6544–6551. doi: 10.1128/AEM.01928-13
Tourna, M., Freitag, T. E., Nicol, G. W., and Prosser, J. I. (2008). Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ. Microbiol. 10, 1357–1364. doi: 10.1111/j.1462-2920.2007.01563.x
Tourna, M., Stieglmeier, M., Spang, A., Konneke, M., Schintlmeister, A., Urich, T., et al. (2011). Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc. Natl. Acad. Sci. U S A. 108, 8420–8425. doi: 10.1073/pnas.1013488108
Treseder, K. K., Balser, T. C., Bradford, M. A., Brodie, E. L., Dubinsky, E. A., Eviner, V. T., et al. (2012). Integrating microbial ecology into ecosystem models: challenges and priorities. Biogeochemistry 109, 7–18. doi: 10.1016/j.jenvman.2017.10.007
Urakawa, H., Martens-Habbena, W., and Stahl, D. A. (2011). “Physiology and genomics of ammonia-oxidizing Archaea,” in Nitrification, eds B. B. Ward, D. Arp, and M. Klotz (Washington D.C: ASM Press), 117–155.
Ushiki, N., Jinno, M., Fujitani, H., Suenaga, T., Terada, A., and Tsuneda, S. (2017). Nitrite oxidation kinetics of two Nitrospira strains: the quest for competition and ecological niche differentiation. J. Biosci. Bioeng. 123, 581–589. doi: 10.1016/j.jbiosc.2016.12.016
Vadivelu, V. M., Yuan, Z. G., Fux, C., and Keller, J. (2006). Stoichiometric and kinetic characterisation of Nitrobacter in mixed culture by decoupling the growth and energy generation processes. Biotechnol. Bioeng. 94, 1176–1188. doi: 10.1002/bit.20956
van Kessel, M. A. H. J., Speth, D. R., Albertsen, M., Nielsen, P. H., Op, den Camp, H. J. M., et al. (2015). Complete nitrification by a single microorganism. Nature 528, 555–559. doi: 10.1038/nature16459
Venterea, R. T., Coulter, J. A., and Clough, T. J. (2020). Nitrite accumulation and nitrogen gas production increase with decreasing temperature in urea-amended soils: experiments and modeling. Soil Biol. Biochem. 142:107727. doi: 10.1016/j.soilbio.2020.107727
Verhamme, D. T., Prosser, J. I., and Nicol, G. W. (2011). Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms. ISME J. 5, 1067–1071. doi: 10.1038/ismej.2010.191
Waggoner, A. L., Bottomley, P. J., Taylor, A. E., and Myrold, D. D. (2021). Soil nitrification response to dairy digestate and inorganic ammonium sources depends on soil pH and nitrifier abundances. Soil Sci. Soc. Am. J. 85, 1990–2006. doi: 10.1002/saj2.20325
Ward, B. B. (1987). Kinetic-studies on ammonia and methane oxidation by Nitrosococcus-Oceanus. Arch. Microbiol. 147, 126–133. doi: 10.1007/bf00415273
Ward, B. B. (2011). “Nitrification: an introduction and overview of the State of the Field,” in Nitrification, eds B. B. Ward, D. Arp, and M. Klotz (Washington D.C: ASM Press).
Watson, S. W., Bock, E., Harms, H., Koops, H. P., and Hooper, A. (1989). “Nitrifying bacteria,” in Bergy’s Manual of Systemic Bacteriology, eds J. T. Staley, M. P. Bryant, N. Pfennig, and J. G. Holt (Baltimore, MD: The Williams & Williams Co.), 1808–1834.
Zheng, M., Wang, Z. Y., Meng, J., Hu, Z. T., Liu, Y. C., Yuan, Z. G., et al. (2021). Inactivation kinetics of nitrite-oxidizing bacteria by free nitrous acid. Sci. Total Environ. 752. doi: 10.1016/j.scitotenv.2020.141876
Keywords: kinetics, substrate affinity, NH3 oxidation, NO2– oxidation, catalytic efficiency, nitrification
Citation: Taylor AE and Mellbye BL (2022) Differential Responses of the Catalytic Efficiency of Ammonia and Nitrite Oxidation to Changes in Temperature. Front. Microbiol. 13:817986. doi: 10.3389/fmicb.2022.817986
Received: 18 November 2021; Accepted: 08 April 2022;
Published: 10 May 2022.
Edited by:
Steffen Kolb, Leibniz Center for Agricultural Landscape Research (ZALF), GermanyReviewed by:
Petra Pjevac, University of Vienna, AustriaMan-Young Jung, Jeju National University, South Korea
Copyright © 2022 Taylor and Mellbye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anne E. Taylor, QW5uZS5UYXlsb3JAb3JlZ29uc3RhdGUuZWR1
†These authors have contributed equally to this work
 Anne E. Taylor
Anne E. Taylor Brett L. Mellbye
Brett L. Mellbye