
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 06 April 2022
Sec. Microbiotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.815366
This article is part of the Research TopicTrends in Applications and Improved Production of Biologically Active Metabolites using Microbial FermentationsView all 12 articles
Microbial cells utilizing electricity to produce high-value fuels and chemicals are the foundation of the biocathodic bioelectrochemical system. However, molecular mechanisms of electron transfer and utilization have not been elucidated. In this work, Escherichia coli engineered by introducing the Mtr pathway from Shewanella oneidensis exhibited stronger electrochemical activity than control and could utilize exogenous electrons to stimulate metabolite profiles and boost succinate production in the bioelectrochemical system. Proteomic analysis and real-time PCR were performed to investigate the effect of exogenous electrons on electroactive E. coli. Bioinformatics analysis suggested that the proteins of molecular function associated with oxidoreductase activity, 4 iron, 4 sulfur([4Fe-4S]) cluster binding, iron-sulfur cluster binding, and metal cluster binding were positively affected by exogenous electrons. Moreover, mapping to the Kyoto Encyclopedia of Genes and Genomes pathway database showed that the up-regulated proteins were mainly involved in metabolic pathways of tricarboxylic acid cycle, pyruvate metabolism, and nitrogen metabolism pathway, providing support for the metabolic balance of microbial cells shifting toward reduced end-products due to electron utilization. Using a biochemical method, the ompF-overexpressed strain was employed to investigate the function of the channel protein. These findings provided a theoretical basis for further improving electron transfer and utilization efficiency, and contributed to the potential applications of the bioelectrochemical system.
In a bioelectrochemical system (BES), microorganisms exchange electrons directly or indirectly with solid electrodes via extracellular electron transfer to manufacture or utilize renewable resources (Kracke et al., 2015; Sun et al., 2020). Biocathodic BESs, e.g., those using microbial electrosynthesis (MES) or cathodic electro-fermentation (CEF), are considered to be a green, sustainable, and attractive novel technology. In such BESs, high-value biofuels and chemicals are converted by microbial catalysis from CO2 or other substrates with electricity as the energy source, thereby achieving renewable energy storage in chemical bonds (Kracke et al., 2015; Patil et al., 2015;
Jiang and Zeng, 2018; Kracke et al., 2018). One advantage of these BESs is that low-cost carbon sources or wastes can be utilized by microorganisms as substrates. Another main advantage is the ability of microorganisms to utilize clean electricity to boost biofuel and chemical generation.
Microorganisms are the catalytic center of biocathodic BESs, whose parameters, activities, and functions are related to chemical production, reactor performance, and efficiency (Kracke et al., 2015; Kokko et al., 2016; Kracke et al., 2018). Electrons are transferred from cathodes into microbial cells, and then participate as essential elements in intracellular redox reactions and microbial metabolism (Sun et al., 2020). This process is applied to break through intracellular redox or energy limitations and shift metabolite profiles toward increasing terminal products of interest (Kracke et al., 2015; Kracke et al., 2018; Wu et al., 2019). Therefore, the ideal biocathodic BES must require microbial cells to take up electrons and efficiently incorporate them into a target cellular metabolic pathway (Kracke et al., 2018). Increasing efforts have focused on creating or promoting electron transfer (Tefft and TerAvest, 2019; Wu et al., 2019; Zhao et al., 2020). However, the molecular mechanism of electron transfer and effects on microbial cells are not clear (Korth and Harnisch, 2019; Sun et al., 2020). Existing research data, especially of biocathodes, are insufficient to conduct a comprehensive analysis, hindering further engineering of microorganisms to use electrons for efficiently generating target products, thereby limiting practical applications of biocathodic BES.
Proteomics technology is the most common method for conducting large-scale analysis on proteomic profiles and screening different target proteins (Zhang et al., 2020). A complete map of expression states derived from quantitative protein information provides insights into biological processes and helps reveal biological mechanisms (Schmidt et al., 2016; Zhang et al., 2020). The proteomes of many typical model microbes have been supported by numerous resources and databases. The Escherichia coli proteome is by far the most well defined bacterial proteome and is thus extensively used for studying general features of the prokaryotic proteome (Soufi et al., 2015; Fortuin et al., 2020).
Exploiting E. coli to produce electrosynthetic chemicals by engineering the electron transfer pathway is an important research area in BESs, which could make up for some deficiencies of electrochemically active bacteria and broaden the application prospects of biocathodic BES (Kracke et al., 2015; Mayr et al., 2019; Wu et al., 2019). The Mtr pathway from Shewanella oneidensis MR-1 is one of the most extensively studied electron transfer pathways and has been successfully constructed in E. coli (Jensen et al., 2010; Ross et al., 2011; Wu et al., 2019). The major components of this pathway are MtrCAB complexes (encoded by the mtrCAB cluster) containing MtrC, MtrB, and MtrA proteins (Jensen et al., 2010). We previously introduced the Mtr pathway into E. coli cells by expressing mtrCAB from S. oneidensis and the engineered E. coli could use the Mtr pathway to transfer electrons from the cathode into E. coli cells (Feng et al., 2020a). However, the molecular mechanism of electron utilization on E. coli cells has not been elucidated.
Based on these previous findings, this work first compared the metabolite profiles and electrochemical activity of engineered E. coli and the control strain without the Mtr pathway in the BES driven by electricity. The engineered E. coli exhibited better electrochemical activity and could use exogenous electrons to stimulate metabolite profiles, increasing succinate production by 18%. Subsequently, real-time PCR (RT-PCR) and proteomics methods were used to comprehensively analyze the specific gene expression changes and the differentially expressed proteins (DEPs) of E. coli. Moreover, the channel protein OmpF was found to be up-regulated in the proteomic data of subcellular location, and thus identified by further gene expression, showing the increased redox reactions between the electrode and the E. coli cells.
Bacterial strains and plasmids are listed in Table 1, and primers are in Supplementary Material. The recombinant strains used in this work were formed by transforming the corresponding recombinant plasmids (Table 1). The recombinant plasmid pCWJ-ompF was constructed by ligating ompF gene fragment into pCWJ between the EcoRI and SacI sites. The ompF gene fragment was amplified by PCR using ompF-1 and ompF-2 as primers and the E. coli BA102 genome as the template.
A suspension of overnight-activated strains was inoculated into 100 ml of 2 × Yeast extract and Typeptone (2 × YT) medium in 500 ml flasks supplemented with the corresponding antibiotics and isopropyl-beta-D-thiogalactopyranoside (0.1 mM chloramphenicol for E. coli-control; 0.1 mM gentamicin for E. coli-mtr; and 0.1 mM chloramphenicol and 0.1 mM gentamicin for E. coli-MtrCBA and E. coli-mtr-ompF). The cultures were grown at 30°C with shaking at 200 rpm for 12–14 h. The suspended cells harvested by centrifugation were saved for further studies.
The harvested cells dispersed into the cathode chamber (OD600 = 1) of the dual chamber “H” bioelectrochemical reactors with an internal volume of 100 mL and fine carbon felts as the cathode and anode electrodes. The anode electrolyte and cathode medium were described in our previous work (Feng et al., 2020a). The anodic electrolyte contained 25 mM NaCl, 15.7 mM NaH2PO4⋅2H2O, 7.0 mM NaHPO4⋅12H2O, and 0.02% dithiothreitol (DTT) (filtered through a 0.22 nm syringe filter before use). The cathodic medium was composed of 47.8 mM NaH2PO4⋅2H2O, 88 mM NaHPO4⋅12H2O, 130 mM NaHCO3, and 5.6 g L–1 yeast extract. Prior to operation, D-glucose was added to the cathodic medium. Cathodes were purged with CO2, imposed at -0.8 V (vs. Ag/AgCl), and stirred using a magnetic stirrer at 200 rpm.
The cyclic voltammograms (CVs), electrochemical impedance spectroscopy (EIS), and the constant potential [−0.8 V (vs. Ag/AgCl)] were performed using an electrochemical instrument (PMC 1000/DC, AMETEK, Berwyn, PA, United States). The CV scan rate was 20 mV/s in the range from -800 to 600 mV (vs. Ag/AgCl). The EIS experiments (0.01–105 Hz) were carried out at 10 mV amplitude.
Organic acids were analyzed with high-performance liquid chromatography (HPLC) (Agilent 1290; Agilent Technologies, Santa Clara, CA, United States) via an HPX-87H column (300–7.8 mm, Bio-Rad, Hercules, CA, United States), equipped with an ultraviolet spectrophotometric detector (at 215 nm) and a refractive index detector. The mobile phase was 8 mM sulfuric acid and the flow rate was 0.5 mL/min.
RNA isolation was prepared using the RNAprep pure cell/bacteria kit (TIANGEN BIOTECH, Beijing, China). RNA concentration was determined using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, United States), and RNA purity was assessed based on the signals at 230, 260, and 280 nm. The integrity of extracted RNA was determined by agarose gel electrophoresis. RNA was transcribed into cDNA as an RT-PCR template using the PrimeScript RT Master Mix (Perfect Real Time) (Takara, Kusatsu, Shiga, Japan). RT-PCR was performed using SYBR Premix EX Taq (Takara, Kusatsu, Shiga, Japan) and a 7300 Plus Real-Time PCR System (Thermo Scientific, Waltham, MA, United States). rpoD, ihfB, and recA were used as housekeeping genes (Nitzschke and Bettenbrock, 2018), and gene expression was calculated by relative quantification applying the ΔΔCt method with three replicates. Reaction systems of RT-PCR analysis and gene-specific primers used for quantifying corresponding transcript levels are provided in Supplementary Material. The amplification conditions for RT-PCR analysis were: 95°C for 30 s, followed by 40 cycles each consisting of 95°C for 5 s, 60°C for 34 s, and 60°C for 1 min.
The bacterial sample was added to lysis buffer (8 M urea, 1% Protease Inhibitor Cocktail, 3 μM TSA, 50 mM NAM, and 2 mM EDTA) and sonicated on ice three times using a high intensity ultrasonic processor. After centrifugation (12,000 × g, 10 min, 4°C), the supernatant was collected into a new EP tube and the protein concentration was measured according to the manufacturer’s instructions for the BCA kit. For digestion, the protein solution was reduced, alkylated, diluted, and digested by trypsin twice according to a standard protocol reported in previous studies (Li et al., 2018; Zhang et al., 2020). The peptide obtained by trypsin digestion was desalted using a Strata X C18 SPE column (Phenomenex, Torrance, CA, United States) and dried by vacuum. The peptide was reconstituted in 0.5 M triethylammonium bicarbonate (TEAB) and processed with a Tandem Mass Tag (TMT) kit. TMT kits enable multiplex relative quantitation by mass spectrometry (MS). Briefly, one unit of TMT reagent was thawed and reconstituted in acetonitrile. Then TMT label reagent was added to differentially label the sample with six TMT tags (Control group 1–3: 126, 127, and 128 label; MtrCBA group 1–3: 129, 130, and 131 label). After incubation for 2 h at room temperature, the peptide mixtures were pooled, desalted, and dried by vacuum centrifugation.
The peptides were separated into fractions using high pH reverse-phase HPLC. Thermo Betasil C18 column (5 μm particles, 250 mm, and 10 mm i.d.) was used to perform the HPLC fractionation under a gradient of 8–32% acetonitrile (pH 9.0) within 60 min. Sixty fractions were collected and then combined to 10 fractions. After drying in a vacuum centrifuge, the fractions were dissolved in 0.1% formic acid for liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis.
For the collected fractions, peptides were analyzed (three repeats per component) using an EASY-nLC 1000 UPLC system coupled online with Q Exactive™ Plus (Thermo, United States). A home-made reversed-phase analytical column (15 cm length, 75 μm i.d.) was used to separate the peptides. Mobile phases A (2% acetonitrile in 0.1% formic acid) and B (0.1% formic acid in 90% acetonitrile) were employed at a constant flow rate of 500 nL/min by the following method: linear gradient from 9% mobile phase B to 26% over 23 min; linear gradient to 38% mobile phase B in 9 min; 38–80% in 4 min and holding at 80% for 4 min. The mass spectrometry parameters were set as follows: the electrospray voltage was set as 2.0 kV; the m/z scan range was set as 350 to 1,800; and intact peptides were detected at 70,000 resolution in the Orbitrap. Peptides were then selected for MS/MS using NCE setting as 28 and the fragments were detected at 17,500 resolution in the Orbitrap. A data-dependent process was conducted that alternated between one MS scan followed by 20 MS/MS scans with 15.0 s dynamic exclusion. The fixed first mass was 100 m/z.
The resulting MS/MS data were searched against the E. coli K12 database (4,446 sequences) using Maxquant search engine (v.1.5.2.8). The parameters were as follows: cleavage enzyme was trypsin/P; missing cleavages was set to 2; first search peptide tolerance and main search peptide tolerance were 20 and 5 ppm, respectively; the fragment mass tolerance was 0.02 Da; fixed modification was carbamidomethyl, variable modifications were acetylation modification and oxidation on Met. The false discovery rate (FDR) was adjusted to below 1%.
Gene Ontology (GO) proteins were annotated using InterProScan and derived from the UniProt-GOA database,1 which were classified according to three categories of GO annotation: biological process, cellular component, and molecular function. The protein pathway annotation was performed using Kyoto Encyclopedia of Genes and Genomes (KEGG) online tool KAAS (KEGG Automatic Annotation Server) based on the KEGG database. Subcellular localization predication software wolfpsort was used to predict subcellular localization. For functional enrichment, enrichment of GO analysis and pathway analysis were researched by employing a two-tailed Fisher’s exact test to test the enrichment of the DEPs against all identified proteins, and the corrected p-value < 0.05 is considered significant. Based on DEP functional classification, further hierarchical clustering was conducted, then visualized by a heat map using the “heatmap.2” function from the “gplots” R-package.
The biocathodic BES has been widely applied to convert electrons from electrodes into intracellular reducing power, promoting the production of target metabolites. In the succinate anaerobic fermentation pathway (see Supplementary Material for metabolic pathway), two molecules of NADH are produced from one molecule of glucose into two molecules of intermediate phosphoenolpyruvate (PEP), while four NADH molecules are needed to synthesize two molecules of succinate from two PEP molecules with CO2 supplementation. Therefore, insufficient reducing power is the critical limiting factor in succinate yield (Zhu and Tang, 2017; Wu et al., 2019). In our work, succinate production in the BES was investigated and the succinate-producing strain E. coli BA102 was used as a host strain. This host strain was constructed to reduce the main byproduct by deleting pflB and ldhA genes. In addition, the pool of available PEP was increased by disabling the ptsG gene encoding an enzyme for the PEP-dependent phosphotransferase system to preserve enhanced succinate production (see Supplementary Material for metabolic pathway) (Cao et al., 2011; Singh et al., 2011; Feng et al., 2020b).
MtrCAB-expressing E. coli strain E. coli-Mtr was constructed by introducing the Mtr pathway from S. oneidensis MR-1 and the engineered Mtr pathway allowed electron transfer from the cathode to E. coli cells, which realized the use of exogenous electrons (Feng et al., 2020a). To better understand the effect of exogenous electrons on metabolite profiles, E. coli-MtrCBA and E. coli-control were used in BES with glucose as the carbon source and CO2 supplementation. As shown in Figure 1, E. coli-MtrCBA had faster glucose consumption and succinate production rates, and the succinate concentration of E. coli-MtrCBA was 18% higher than that of E. coli-control after 44 h. Pyruvate was accumulated as a result of gene knockouts. Production of the main by-products pyruvate and acetate showed no significant difference between E. coli-MtrCBA and E. coli-control, while higher levels of succinate/pyruvate and succinate/acetate were shown in E. coli-MtrCBA (increased by 17 and 2%, respectively). The results indicated that the intracellular metabolite profiles were stimulated and changed to facilitate the production of reduced end-products (succinate) in the BES driven by electricity (Wu et al., 2019).
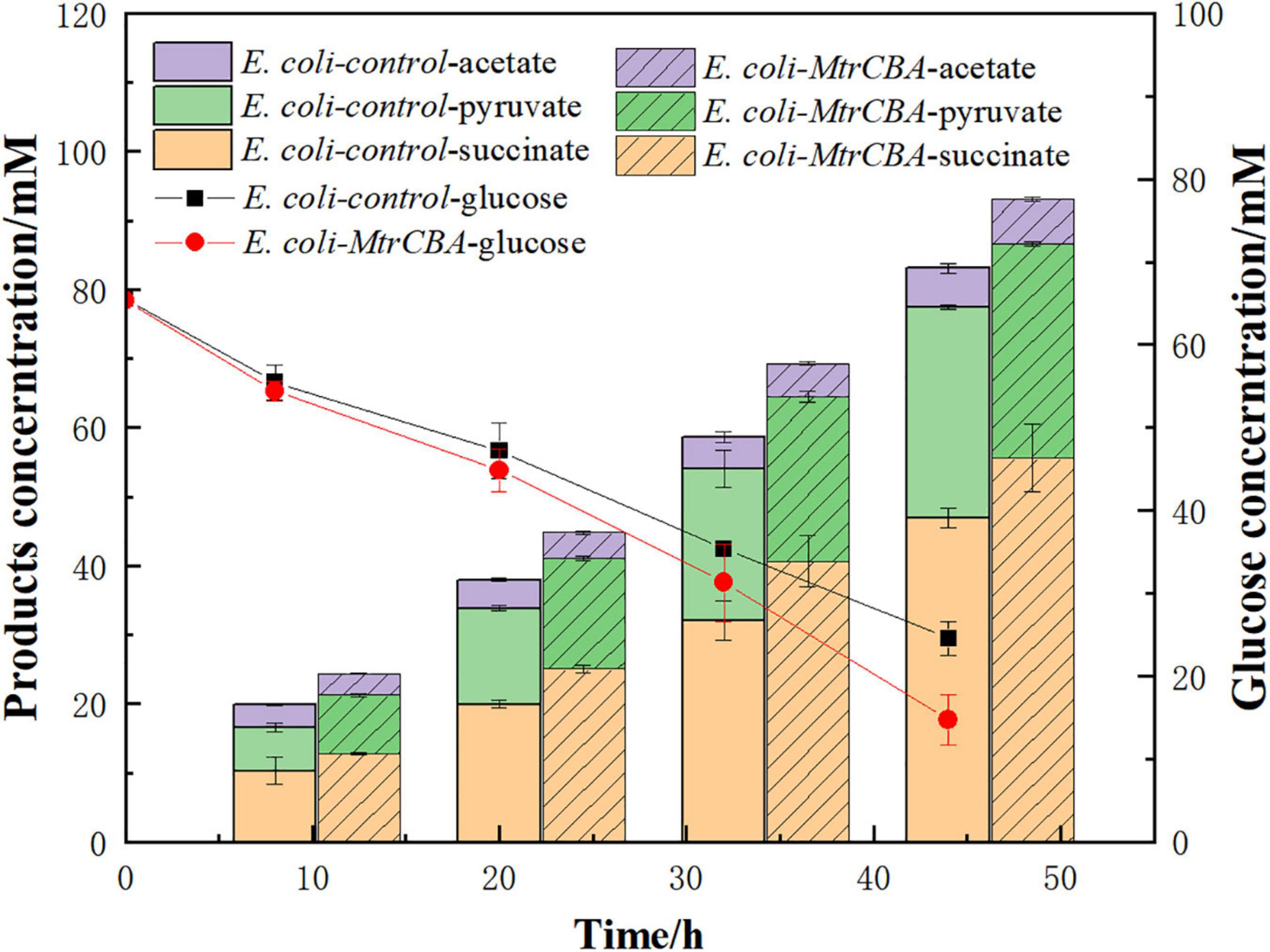
Figure 1. Effects of the exogenous electrons on major metabolite profiles. Bioelectrochemical reactors inoculated with E. coli-control or E. coli-MtrCBA (OD600 = 1) in cathode mediums were imposed at a constant potential [-0.8 V (vs. Ag/AgCl)].
To analyze the electrochemical performance of the BES, CV curves and nyquist plots of impedance were obtained from CV and EIS test to investigate the redox reaction characteristics between the electrode and the microbial cells. CVs of E. coli-MtrCBA and E. coli-control are presented in Figure 2A. Two oxidation peaks [ranging from +0.2 to +0.40 V (vs. Ag/AgCl) and −0.20 to 0.0 V (vs. Ag/AgCl)] and one reduction peak between −0.30 and −0.10 V (vs. Ag/AgCl) were observed, consistent with the broad potential of c-type cytochromes ranging from −0.45 to +0.20 V and the mediator molecules excreted by E. coli ranging from −0.45 to −0.3 V (Qiao et al., 2008; Ross et al., 2011; Kracke et al., 2015). This demonstrated that the redox peaks were caused by c-type cytochromes and endogenous redox species under the effect of exogenous electrons (Feng et al., 2020a). The higher peak area obtained from E. coli-MtrCBA compared with the control indicated that stronger redox reactions occurred in the BES inoculated with E. coli-MtrCBA. Moreover, the oxidation and reduction peaks of E. coli-MtrCBA shifted positively compared with the control group. Since proton transfer coupled with electron transfer is involved in the redox reaction, the CV result is affected by solution pH value. A previous report showed that the midpoint potential of the redox peak shifts positively with decreased pH (Qiao et al., 2008). More succinic acid produced by E. coli-MtrCBA led to a lower pH, resulting in a slightly positive shift in the redox peaks of E. coli-MtrCBA. In addition, the electron transfer efficiency at the cell-electrode interfaces was analyzed by EIS. EIS analysis reflected an improved electron transfer rate of E. coli-MtrCBA (Figure 2B) (Liu et al., 2012). These results confirmed that the engineered E. coli exhibited much better electrochemical activity and more effective biocatalytic behavior in BES compared to control.
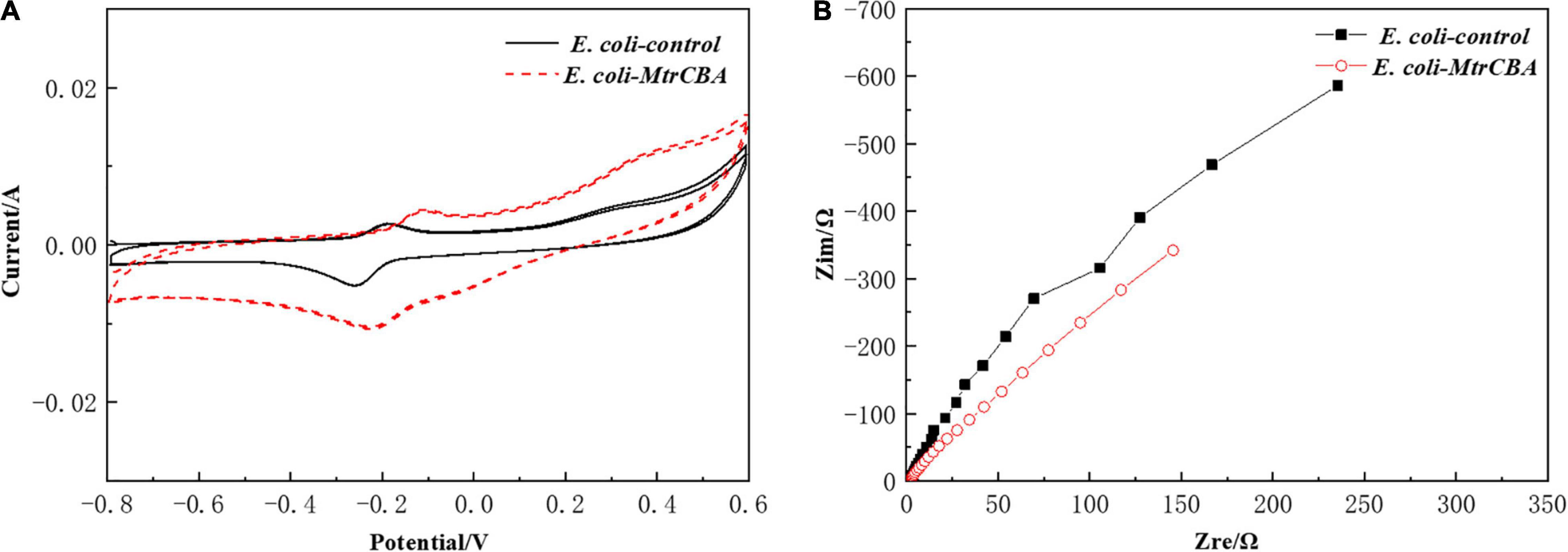
Figure 2. Effects of exogenous electrons on electrochemical activity. Cyclic voltammogram (CV) analysis scanned at 10 mV/s in the range of -800–600 mV (A). Electrochemical impedance spectroscopy scanned at 0.01−105 Hz with a 10 mV amplitude (B).
To analyze the effect of exogenous electrons transported into cells by intracellular metabolic pathways, the expression changes of important genes related to terminal oxidases, respiratory chain, and the tricarboxylic acid (TCA) cycle were investigated via RT-PCR. The relative expression levels for selected genes are shown in Figure 3. As expected, genes connected to the TCA cycle including fum and mdh were upregulated in E. coli-MtrCBA. This is consistent with more succinate being produced by E. coli-MtrCBA through accepting cathodic electrons, hinting at a more active succinate synthesis pathway. There was an obvious change in expression level of intracellular redox regulation system protein gene arcB. The Arc two-component system comprising ArcA and ArcB is the key element in the global transcriptional regulatory network affecting the expression of numerous genes associated with substrate metabolism and energy metabolism to adapt to various conditions (van Beilen and Hellingwerf, 2016; Teran-Melo et al., 2018). Van Beilen and Hellingwerf reported that reduced quinone species could activate ArcB as a redox-sensitive, membrane-embedded kinase (van Beilen and Hellingwerf, 2016). These results suggested that exogenous electrons might affect a wide range of intracellular gene expression and expressed proteins and then biological reactions and pathways.
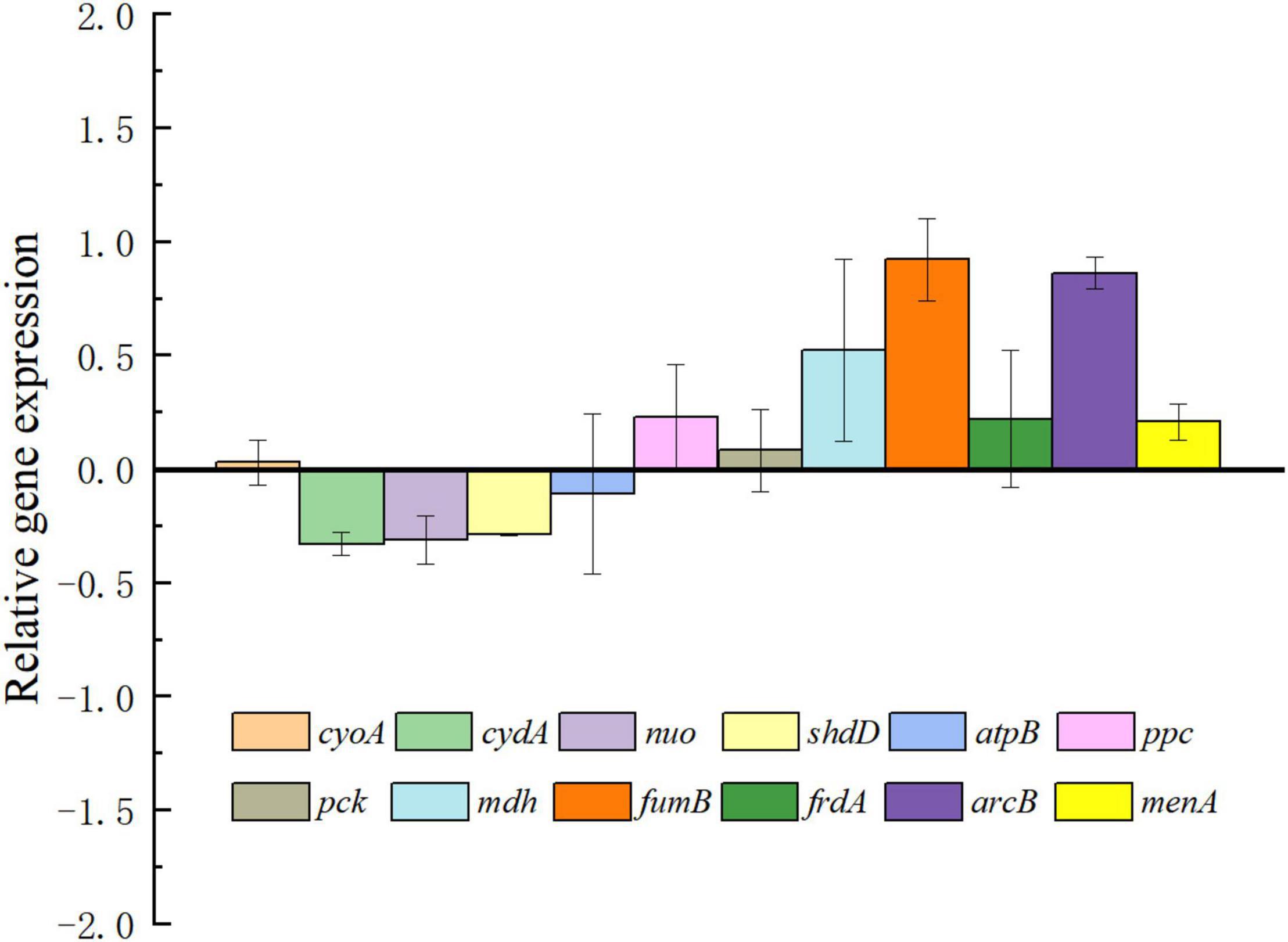
Figure 3. Results of RT-PCR analysis. The transcription pattern was normalized to the reference genes for the expression of E. coli-control. Genes encoding cytochrome bo3 (cyoA), cytochrome bd (cydA), NADH: ubiquinone oxidoreductase (nuo) and succinate dehydrogenase (sdhD), ATP synthase (atpB), phosphoenolpyruvate carboxylase (ppc), phosphoenolpyruvate carboxykinase (pck), malate dehydrogenase (mdh), fumarate hydratase (fumB), fumarate reductase (frdA), sensor histidine kinase (arcB), and 1,4-dihydroxy-2-naphthoate octaprenyltransferase (menA) were analyzed. The unchanged relative gene expression level was calculated as 0. The Y-axis was formatted in a logarithmic scale with base 10.
The mechanism of electron transfer and utilization is complex and not fully understood. To reveal key intracellular proteins or pathways responding to exogenous electrons and further comprehensively analyze the effect of electron transfer and utilization, systematic proteomic analysis research was conducted.
In total, 2,268 proteins were detected by TMT quantitative proteomics. Of these, 2,110 proteins were identified as quantifiable DEPs. Based on a change threshold of 1.2-fold, the results of protein expression levels are shown in the volcano plot in Figure 4A. Compared with the control, exogenous electrons from the cathode led to up-regulation of 152 proteins and down-regulation of 142 proteins in E. coli-MtrCBA.
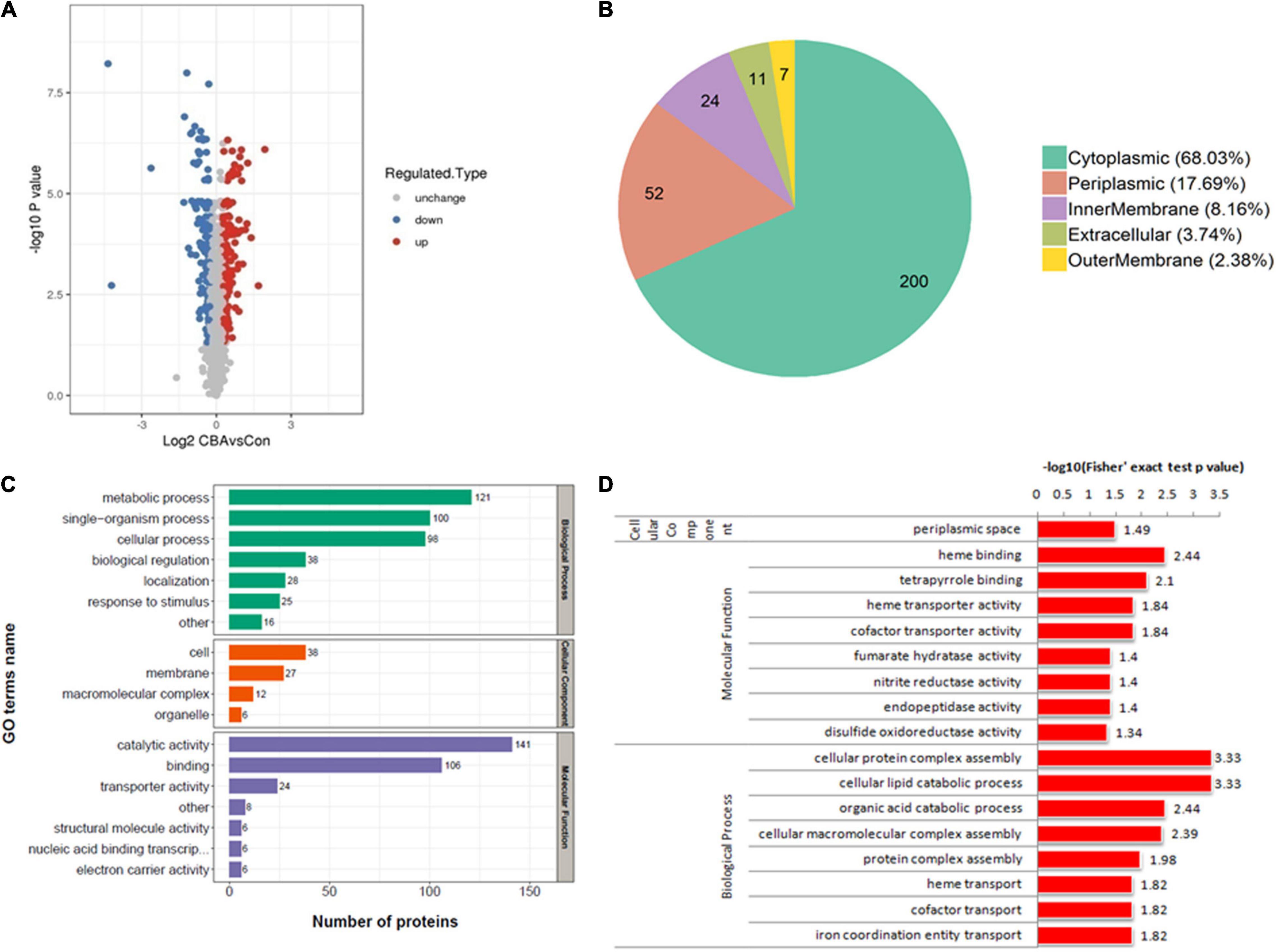
Figure 4. Results of proteomic analysis. The volcano plot shows the upregulated (red) or downregulated (blue) proteins between E. coli-MtrCBA/E. coli-control (A). Subcellular localization map of the differentially expressed proteins (B). The differentially expressed proteins classified according to Gene Ontology (GO) annotation (C). GO enrichment analysis of the differentially expressed proteins (D). –Log 10 P value represents the negative logarithm of p value calculated by Student’s t-test to base 10. Log 2 CBAvsCon represents the logarithm of the proteins’ difference multiple to the base 2. –Log10 (Fisher’s exact test p value) represents the negative logarithm of p value calculated by the Fisher’s exact test to base 10.
In order to acquire protein functional information, the DEPs were annotated and classified by GO annotation according to three categories (biological process, cellular component, and molecular function) and subcellular location (Figures 4B,C). Among them, the DEPs were respectively located in the cytoplasm (200 proteins, accounting for 68%), the periplasm (52 proteins, accounting for 18%), the innermembrane (24 proteins, accounting for 8%), the extracellular matrix (11 proteins, accounting for 4%), and the outermembrane (seven proteins, accounting for 2%).
Based on GO annotations and classification into three categories, GO enrichment analysis was performed using a two-tailed Fisher’s exact test to identify significantly enriched functional groups (p-value < 0.05) of the DEPs in the case of exogenous electron transfer into E. coli cells through the Mtr pathway (Figure 4D). In terms of cellular components, the DEPs were highly significantly enriched in the perplasmic space. MtrCAB complexes that span the outer membrane allow electron transfer from the decaheme cytochrome MtrC located on the extracellular face of the outer membrane to the periplasmic decaheme cytochrome MtrA, so proteins in the periplasmic space are directly affected by electrons (Jensen et al., 2010). DEPs were highly significantly categorized into eight biological processes related to the metabolism of protein, lipid, and organic acid, including “cellular protein complex assembly,” “cellular lipid catabolic process,” “cellular macromolecular complex assembly,” “protein complex assembly,” and “organic acid catabolic process,” and associated with GO terms about electrons, including “heme transport,” “cofactor transport,” and “iron coordination entity transport.” Among molecular function-associated GO terms, the main enriched molecular functions of the DEPs were related to binding and transport, including “heme binding,” “tetrapyrrole binding,” “heme transporter activity,” and “cofactor transporter activity.” MtrC and MtrA, the important parts of MtrCAB complexes, are decaheme cytochrome c, and their maturations are required to load heme (Jensen et al., 2010). Cofactors, such as NADH, MKH2, and FADH, are intracellular electron acceptors, which are used to store reducing power. In addition, “nitrite reductase activity” and “disulfide oxidoreductase activity,” which functionally catalyze the reactions commonly involved in electrons, were significantly enriched.
On the basis of enrichment analysis, further cluster analysis was carried out to find correlations or new insights into protein functions and pathways. According to differential expression multiple, the DEPs were divided into four parts: Q1, Q2, Q3, and Q4 (Figure 5A) and then clustered by hierarchical clustering. The results were visualized by heat maps.
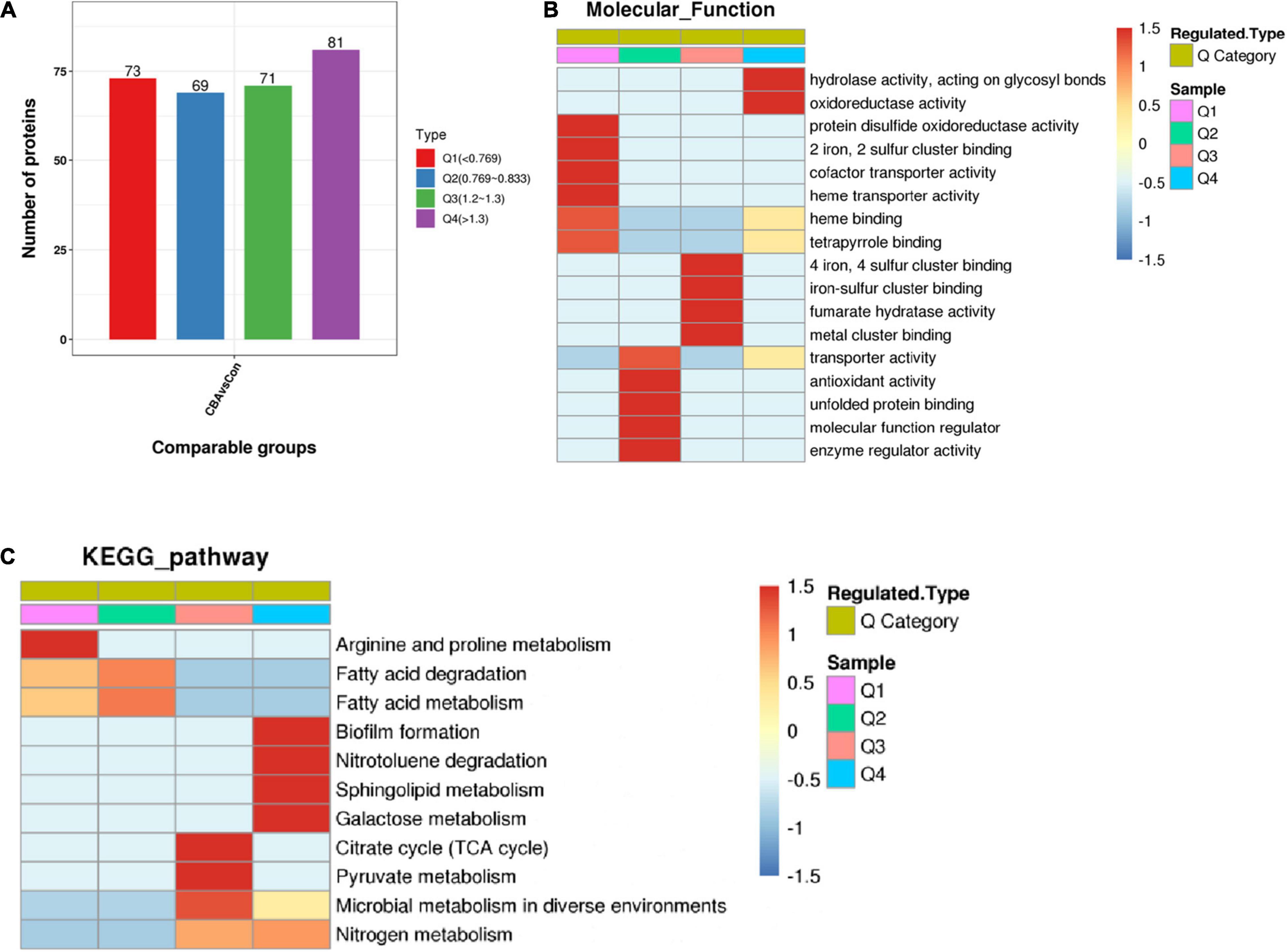
Figure 5. Enrichment-based clustering analysis. The differentially expressed proteins were divided into four parts: Q1 (<0.769), Q2 (0.769–0.833), Q3 (1.2–1.3), and Q4 (>1.3) according to differential expression multiple (A). Hierarchical clustering based on functional classification of differentially expressed proteins (Molecular Function) (B) and KEGG Pathway based on Escherichia coli K-12 MG1655 (C). Color from blue to red represents the degree of enrichment from weak to strong, respectively.
A cluster analysis heat map based on molecular function enrichment is shown in Figure 5B. “Hydrolase activity” and “oxidoreductase activity” (Q4 > 1.3) were significantly enhanced by exogenous electrons transferred into E. coli cells through the Mtr pathway, functionally catalyzing the reactions commonly accompanied with electrons or protons. DEPs of molecular function associated with “4 iron, 4 sulfur([4Fe-4S]) cluster binding,” “iron-sulfur (Fe-S) cluster binding,” and “metal cluster binding” (1.2 ≤ Q3 ≤ 1.3) were up-regulated, while those associated with “2 iron, 2 sulfur ([2Fe-2S]) cluster binding” (Q4 < 0.769) were down-regulated (Supplementary Material). Fe-S clusters are versatile and essential biological cofactors biosynthesized by the Isc and Suf pathways in E. coli (Mettert and Kiley, 2015). Reducing power such as in the presence of the reducing agent DTT is absolutely required for mature [4Fe-4S] proteins, due to the requirement of electrons to generate [4Fe-4S] from [2Fe-2S] clusters (Chandramouli et al., 2007; Beilschmidt et al., 2017). Electrons were transferred from the cathode into E. coli cells through the Mtr pathway, resulting in a reductive intracellular environment and providing electrons for [4Fe-4S] generation. This further promoted [4Fe-4S] cluster binding and [4Fe-4S] protein maturation.
Fe-S cluster-containing proteins are partly reductases and dehydrogenases, in which Fe-S clusters are the relay station or conduit participating in electron transfer. Fe-S clusters also serve as prosthetic groups acting as binding sites or participating in enzyme catalysis. In addition, the up-regulation of “fumarate hydratase activity” suggested that it promoted formation of fumarate, the precursor of succinate. Fumarate hydratase is also a [4Fe-4S] cluster-containing protein, in which the [4Fe-4S] cluster is used for substrate binding and readily oxidized to an inactive [3Fe-4S] cluster (Flint et al., 1992). A reductive intracellular environment caused by exogenous electrons could promote binding and stable activity of [4Fe-4S] clusters. Unexpectedly, the DEPs were significantly enriched in terms of negative regulation for “heme transporter activity,” “heme binding,” and “cofactor transporter activity.” Further analysis of the specific DEPs showed that CcmC and CcmF were the main DEPs in three terms related to the synthesis and maturation of the MtrCAB complexes. MtrCAB complexes spanning the outer membrane contain two cytochrome c MtrA and MtrC, whose maturation is required for cytochrome c maturation proteins CcmABCDEFGH (Jensen et al., 2010; Feng et al., 2020a). The polypeptides of MtrCAB and CcmABCDEFGH are secreted into the periplasm by the Sec secretion system, and then heme is translocated into the periplasm and loaded to form mature MtrC and MtrA by Ccm machinery. During synthesis and maturation of the MtrCAB complexes, there are multiple regulation points: excessive production or secretion of Mtr can inhibit and decrease Ccm, and inadequate heme production can lead to degradation of apocytochrome c or cytochrome c (Goldbeck et al., 2013). These might be the reasons why heme binding and transporter activity were down-regulated.
A cluster analysis heat map based on KEGG pathway enrichment is shown in Figure 5C. Among these metabolic pathways, DEPs are mainly connected with biofilm and cell membrane, such as the up-regulation of biofilm formation and sphingolipid metabolism (Q > 1.3), and down-regulation of fatty acid metabolism and fatty acid degradation (0.769 ≤ Q ≤ 0.833). These findings indicated that biofilm formation and cell membrane composition were significantly affected. The TCA cycle, pyruvate metabolism, and nitrogen metabolism pathway were promoted, which allowed the cells to produce more succinate (Feng et al., 2020a).
The results of electrochemical analysis confirmed that E. coli-MtrCBA exhibited much better bioelectrocatalytic activity and catalyzed stronger redox reactions compared with the control in the BES. The contributing factor was endogenous molecules under electrochemical stress (Feng et al., 2020a). Moreover, proteomic analysis indicated that the cell membranes were affected during the process of electron utilization in the BES. Combined with the data of subcellular location, seven of the significant DEPs were located in the outer membrane. Excluding uncharacterized proteins, the up-regulated outer membrane protein OmpF (E.coli-MtrCBA/E.coli-control Ratio = 1.653) was further identified. OmpF is a highly abundant primary porin in E. coli, which enables passive diffusion of hydrophilic molecules and influences outer membrane permeability (Duval et al., 2017).
To determine that the channel protein OmpF assisted with the redox reactions between the electrode and E. coli cells, ompF overexpressed strain E. coli-mtr-ompF and control E. coli-mtr were constructed and inserted into the BES to perform a CV test. As shown in Figure 6, significant redox peaks were observed from the BES inoculated with E. coli-mtr-ompF, indicating that OmpF might reduce transmission resistance of the cell membrane and improve outer membrane permeability to enhance the redox reactions. This was one of the reasons for the better bioelectrocatalytic activity exhibited by the engineered E. coli-MtrCBA.
The process by which microorganisms utilize electrons from a cathode to produce high-value fuels and chemicals is the basis of the biocathodic BES. However, major knowledge gaps on molecular mechanisms of electron transfer and the effect on microbial cells have limited further engineering of microorganisms and practical applications of biocathodic BES. In order to utilize the electrons of cathodes, E. coli engineered with the Mtr pathway was constructed by expressing mtrCAB from S. oneidensis MR-1. The engineered E. coli exhibited much better electrochemical activity and more effective biocatalytic behavior than control in BES based on CV and EIS analyses. The Mtr pathway in S. oneidensis MR-1 is a typical bidirectional electron transfer pathway, in which electrons flow from cells into anodes or from cathodes into cells (Ross et al., 2011; Feng et al., 2020a). Moreover, the Mtr pathway has been successfully constructed in E. coli (Jensen et al., 2010; Goldbeck et al., 2013; Wu et al., 2019). The electrochemical performance of E. coli-MtrCBA indicated that our engineered E. coli cells could transfer electrons from the electrodes to cells to utilize exogenous electrons. Compared with E. coli-control, the electroactive E. coli-MtrCBA could boost succinate production by 18% and displayed higher levels of succinate/pyruvate and succinate/acetate (enhanced by 17 and 2% respectively) in BES driven by electricity. The results suggested that E. coli-MtrCBA accepted electrons from the cathode through the Mtr pathway and utilized the electrons to obtain excess reducing power, which led to an imbalance of the intracellular redox state and thus metabolite profiles were stimulated and changed to facilitate the production of reduced products (Wu et al., 2019).
Subsequently, systematic proteomic analysis research and RT-PCR were performed to reveal key intracellular proteins or pathways in response to exogenous electrons and comprehensively analyze the effect of electron utilization on E. coli cells. Compared with the control, 152 up-regulated and 142 down-regulated proteins were identified in E. coli-MtrCBA, respectively. Bioinformatics analysis showed that the proteins of molecular function associated with hydrolase activity, oxidoreductase activity, [4Fe-4S] cluster binding, Fe-S cluster binding, metal cluster binding, and fumarate hydratase activity were positively affected by exogenous electrons. In addition to the proteins that functionally catalyze reactions involving electrons or protons, the Fe-S cluster was worth further study. Fe-S cluster-containing proteins are partly electron-accepting terminal oxidoreductases and dehydrogenases participating in multiple cellular processes, and Fe-S clusters are the relay station used to exchange electrons within the proteins (de Choudens and Barras, 2017; Feng et al., 2020a). Fe-S clusters can also function as prosthetic groups acting as the binding sites or participating in enzyme catalysis. Due to the requirement of electrons to generate [4Fe-4S] from [2Fe-2S] clusters, sufficient reducing power was absolutely required to generate mature [4Fe-4S] proteins and also stable activity of [4Fe-4S] clusters (Chandramouli et al., 2007; Beilschmidt et al., 2017).
Up-regulated proteins were mainly involved in the KEGG pathways of TCA, pyruvate metabolism, and nitrogen metabolism pathway, showing that the metabolic balance of microbial cells shifted toward reduced end-products due to electron utilization. Moreover, the DEPs enriched in sphingolipid metabolism and fatty acid metabolism indicated that cell membranes were affected during the process of electron utilization in BES. The ompF overexpressed strain was used to investigate the function of the channel protein OmpF located in the outer membrane. CV analysis showed that the up-regulated expression of ompF improved redox reactions between the electrode and the cells.
In conclusion, engineered E. coli transferred electrons from the cathode into the cells by the Mtr pathway and simultaneously up-regulated the channel protein to enhance redox reactions between the electrode and the cells to further improve electron transfer capacity. The electrons were transferred into E. coli cells, resulting in a reductive intracellular environment, which provided a basis to improve the maturation or stability of some proteins associated with “[4Fe-4S] cluster binding,” “Fe-S cluster binding,” and “metal cluster binding.” Sufficient reducing power provided the cofactor and electron donor for the oxidoreductases and dehydrogenases. The DEPs induced by exogenous electrons were mapped to the KEGG pathway database, which showed that the electrons disturbed metabolism in multiple ways by biofilm formation, sphingolipid metabolism, fatty acid metabolism, TCA, pyruvate metabolism, and nitrogen metabolism pathway.
In this work, E. coli engineered to contain the Mtr pathway exhibited better electrochemical activity and effective biocatalytic behavior. Based on electrochemical and metabolite profile analyses, the succinate yield increased by 18% showing the effect of exogenous electrons on E. coli cells. Subsequently, proteomics and RT-PCR were performed to investigate DEPs in E. coli in response to exogenous electrons. Based on bioinformatic analysis of the DEPs, combined with biochemical methods, potential proteins and metabolic pathways that might function in electron transfer and utilization of E. coli cells were identified. In future studies, more crucial gene targets for electron transfer and utilization need to be identified. Additionally, multi-level and large-scale connections should be explored. The detailed molecular mechanisms need much deeper research, as an in-depth understanding of molecular mechanisms is the basis for engineering microorganisms to improve electron utilization efficiency. This work revealed a global proteomic alteration of E. coli cells in response to exogenous electrons, which will aid in the understanding of molecular mechanisms on electron transfer and utilization, and provide a theoretical basis for improving electron transfer and utilization efficiency.
The datasets presented in this study can be found in online repositories. The names of the repository and accession number can be found below: http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD030650.
JiaoF: data curation, validation, investigation, writing–original draft, review, and editing, and funding acquisition. JiaF: data curation, formal analysis, and writing–review and editing. CL: data curation and writing–review and editing. SX: resources, project administration, funding acquisition, and writing–review and editing. XW and KC: resources and writing–review and editing. All authors contributed to the article and approved the submitted version.
This work was supported by the National Science Foundation of Young Scientists of China (Grant Nos. 22108123 and 21908099), the China Postdoctoral Science Foundation (2020M681569), and the Jiangsu Synergetic Innovation Center for Advanced Bio-Manufacture (XTD2208).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the PTM Biolabs, Inc. for technical assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.815366/full#supplementary-material
Beilschmidt, L. K., de Choudens, S. O., Fournier, M., Sanakis, I., Hograindleur, M. A., Clemancey, M., et al. (2017). ISCA1 is essential for mitochondrial Fe4S4 biogenesis in vivo. Nat. Commun. 8:15124. doi: 10.1038/Ncomms15124
Cao, Y. J., Cao, Y. G., and Lin, X. Z. (2011). Metabolically engineered Escherichia coli for biotechnological production of four-carbon 1,4-dicarboxylic acids. J. Ind. Microbiol. Biotechnol. 38, 649–656. doi: 10.1007/s10295-010-0913-4
Chandramouli, K., Unciuleac, M. C., Naik, S., Dean, D. R., Huynh, B. H., and Johnson, M. K. (2007). Formation and properties of [4Fe-4S] clusters on the IscU scaffold protein. Biochemistry 46, 6804–6811. doi: 10.1021/bi6026659
de Choudens, S. O., and Barras, F. (2017). Genetic, biochemical, and biophysical methods for studying Fe-S proteins and their assembly. Methods Enzymol. 595, 1–32. doi: 10.1016/bs.mie.2017.07.015
Duval, V., Foster, K., Brewster, J., and Levy, S. B. (2017). A novel regulatory cascade involving BluR, YcgZ, and Lon controls the expression of Escherichia coli OmpF porin. Front. Microbiol. 8:1148. doi: 10.3389/Fmicb.2017.01148
Feng, J., Jiang, M. J., Li, K., Lu, Q. H., Xu, S., Wang, X., et al. (2020a). Direct electron uptake from a cathode using the inward Mtr pathway in Escherichia coli. Bioelectrochemistry 134:107498. doi: 10.1016/j.bioelechem.2020.107498
Feng, J., Lu, Q., Li, K., Xu, S., Wang, X., Chen, K., et al. (2020b). Construction of an electron transfer mediator pathway for bioelectrosynthesis by Escherichia coli. Front. Bioeng. Biotechnol. 8:590667. doi: 10.3389/fbioe.2020.590667
Flint, D. H., Emptage, M. H., and Guest, J. R. (1992). Fumarase a from Escherichia coli: purification and characterization as an iron-sulfur cluster containing enzyme. Biochemistry 31, 10331–10337. doi: 10.1021/bi00157a022
Fortuin, S., Nel, A. J. M., Blackburn, J. M., and Soares, N. C. (2020). Comparison between the proteome of Escherichia coli single colony and during liquid culture. J. Proteomics 228:103929. doi: 10.1016/J.Jprot.2020.103929
Goldbeck, C. P., Jensen, H. M., TerAvest, M. A., Beedle, N., Appling, Y., Hepler, M., et al. (2013). Tuning promoter strengths for improved synthesis and function of electron conduits in Escherichia coli. ACS Synth. Biol. 2, 150–159. doi: 10.1021/sb300119v
Jensen, H. M., Albers, A. E., Malley, K. R., Londer, Y. Y., Cohen, B. E., Helms, B. A., et al. (2010). Engineering of a synthetic electron conduit in living cells. Proc. Natl. Acad. Sci. U.S.A. 107, 19213–19218. doi: 10.1073/pnas.1009645107
Jiang, Y., and Zeng, R. J. (2018). Expanding the product spectrum of value added chemicals in microbial electrosynthesis through integrated process design-A review. Bioresour. Technol. 269, 503–512. doi: 10.1016/j.biortech.2018.08.101
Kokko, M. E., Makinen, A. E., and Puhakka, J. A. (2016). “Anaerobes in bioelectrochemical systems,” in Anaerobes in Biotechnology, Advances in Biochemical Engineering/Biotechnology, eds R. Hatti-Kaul, G. Mamo, and B. Mattiasson (Berlin: Springer), 263–292. doi: 10.1007/10_2015_5001
Korth, B., and Harnisch, F. (2019). Modeling microbial electrosynthesis. Bioelectrosynthesis 167, 273–325. doi: 10.1007/10_2017_35
Kracke, F., Lai, B., Yu, S. Q., and Kromer, J. O. (2018). Balancing cellular redox metabolism in microbial electrosynthesis and electro fermentation - A chance for metabolic engineering. Metab. Eng. 45, 109–120. doi: 10.1016/j.ymben.2017.12.003
Kracke, F., Vassilev, I., and Kromer, J. O. (2015). Microbial electron transport and energy conservation - the foundation for optimizing bioelectrochemical systems. Front. Microbiol. 6:575. doi: 10.3389/fmicb.2015.00575
Li, F. D., Wang, Y. C., Li, Y., Yang, H. P., and Wang, H. M. (2018). Quantitative analysis of the global proteome in peripheral blood mononuclear cells from patients with new - onset psoriasis. Proteomics 18:e1800003. doi: 10.1002/Pmic.201800003
Liu, J., Yong, Y. C., Song, H., and Li, C. M. (2012). Activation enhancement of citric acid cycle to promote bioelectrocatalytic activity of arcA knockout Escherichia coli toward high-performance microbial fuel cell. ACS Catal. 2, 1749–1752. doi: 10.1021/cs3003808
Mayr, J. C., Grosch, J. H., Hartmann, L., Rosa, L. F. M., Spiess, A. C., and Harnisch, F. (2019). Resting Escherichia coli as chassis for microbial electrosynthesis: production of chiral alcohols. ChemSusChem 12, 1631–1634. doi: 10.1002/cssc.201900413
Mettert, E. L., and Kiley, P. J. (2015). How is Fe-S cluster formation regulated? Annu. Rev. Microbiol. 69, 505–526. doi: 10.1146/annurev-micro-091014-104457
Nitzschke, A., and Bettenbrock, K. (2018). All three quinone species play distinct roles in ensuring optimal growth under aerobic and fermentative conditions in E. coli K12. PLoS One 13:e0194699. doi: 10.1371/journal.pone.0194699
Patil, S. A., Gildemyn, S., Pant, D., Zengler, K., Logan, B. E., and Rabaey, K. (2015). A logical data representation framework for electricity-driven bioproduction processes. Biotechnol. Adv. 33, 736–744. doi: 10.1016/j.biotechadv.2015.03.002
Qiao, Y., Li, C. M., Bao, S. J., Lu, Z. S., and Hong, Y. H. (2008). Direct electrochemistry and electrocatalytic mechanism of evolved Escherichia coli cells in microbial fuel cells. Chem. Commun. 11, 1290–1292. doi: 10.1039/b719955d
Ross, D. E., Flynn, J. M., Baron, D. B., Gralnick, J. A., and Bond, D. R. (2011). Towards electrosynthesis in Shewanella: energetics of reversing the Mtr pathway for reductive metabolism. PLoS One 6:e16649. doi: 10.1371/journal.pone.0016649
Schmidt, A., Kochanowski, K., Vedelaar, S., Ahrne, E., Volkmer, B., Callipo, L., et al. (2016). The quantitative and condition-dependent Escherichia coli proteome. Nat. Biotechnol. 34, 104–110. doi: 10.1038/nbt.3418
Singh, A., Soh, K. C., Hatzimanikatis, V., and Gill, T. R. (2011). Manipulating redox and ATP balancing for improved production of succinate in E. coli. Metab. Eng. 13, 76–81. doi: 10.1016/j.ymben.2010.10.006
Soufi, B., Krug, K., Harst, A., and Macek, B. (2015). Characterization of the E. coli proteome and its modifications during growth and ethanol stress. Front. Microbiol. 6:103. doi: 10.3389/Fmicb.2015.00103
Sun, M., Zhai, L. F., Mu, Y., and Yu, H. Q. (2020). Bioelectrochemical element conversion reactions towards generation of energy and value-added chemicals. Prog. Energy Combust. Sci. 77:100814. doi: 10.1016/J.Pecs.2019.100814
Tefft, N. M., and TerAvest, M. A. (2019). Reversing an extracellular electron transfer pathway for electrode driven acetoin reduction. ACS Synth. Biol. 8, 1590–1600. doi: 10.1021/acssynbio.8b00498
Teran-Melo, J. L., Pena-Sandoval, G. R., Silva-Jimenez, H., Rodriguez, C., Alvarez, A. F., and Georgellis, D. (2018). Routes of phosphoryl group transfer during signal transmission and signal decay in the dimeric sensor histidine kinase ArcB. J. Biol. Chem. 293, 13214–13223. doi: 10.1074/jbc.RA118.003910
van Beilen, J. W. A., and Hellingwerf, K. J. (2016). All Three endogenous quinone species of Escherichia coli are involved in controlling the activity of the aerobic/anaerobic response regulator ArcA. Front. Microbiol. 7:1339. doi: 10.3389/Fmicb.2016.01339
Wu, Z. Q., Wang, J. S., Liu, J., Wang, Y., Bi, C., and Zhang, X. L. (2019). Engineering an electroactive Escherichia coli for the microbial electrosynthesis of succinate from glucose and CO2. Microb. Cell Fact. 18:15. doi: 10.1186/s12934-019-1067-3
Zhang, J., Li, Q. R., Liu, J. H., Lu, Y. H., Wang, Y., and Wang, Y. H. (2020). Astaxanthin overproduction and proteomic analysis of Phaffia rhodozyma under the oxidative stress induced by TiO2. Bioresour. Technol. 311: 123525. doi: 10.1016/j.biortech.2020.123525
Zhao, J., Li, F., Cao, Y., Zhang, X., Chen, T., Song, H., et al. (2020). Microbial extracellular electron transfer and strategies for engineering electroactive microorganisms. Biotechnol. Adv. 53:107682. doi: 10.1016/j.biotechadv.2020.107682
Keywords: Escherichia coli, exogenous electrons, proteomics, differentially expressed proteins, bioelectrochemical system
Citation: Feng J, Feng J, Li C, Xu S, Wang X and Chen K (2022) Proteomics Reveal the Effect of Exogenous Electrons on Electroactive Escherichia coli. Front. Microbiol. 13:815366. doi: 10.3389/fmicb.2022.815366
Received: 15 November 2021; Accepted: 17 February 2022;
Published: 06 April 2022.
Edited by:
Niranjan Koirala, University of Macau, ChinaReviewed by:
Wolfgang Buckel, University of Marburg, GermanyCopyright © 2022 Feng, Feng, Li, Xu, Wang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sheng Xu, aGVucnl4dUBuanRlY2guZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.