
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 24 May 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.815091
This article is part of the Research TopicAdvances in the discovery of Natural Molecules and their Analogues Against Microbial Infection-Related BiofilmsView all 14 articles
Candida biofilm-related infections cause increased morbidity and mortality in patients with a reduced immune response. Traditional antifungal therapies have proven to be insufficient as the biofilm matrix acts as a perfusion barrier. Thus, novel methods are required to improve drug delivery and kill Candida within the biofilm. In this study, chitosan nanoparticles (CSNPs) loaded with Amphotericin B (AMB), which were functionalized with β-1,3-glucanase (Gls), were fabricated (CSNPs-AMB-Gls), and their antibiofilm activity against Candida albicans biofilm was evaluated in vitro. Scanning electron microscopy (SEM) and confocal laser scanning microscopy (CLSM) were employed to examine biofilm architecture and cell viability. CSNPs-AMB-Gls inhibited planktonic cell growth and biofilm formation effectively and exhibited the highest efficacy on the removal of a mature biofilm than free AMB or CSNPs-AMB. The created nanoparticles (NPs) were found to penetrate the biofilm so as to directly interfere with the cells inside and disassemble the biofilm matrix. CSNPs-AMB-Gls could also eradicate biofilms from clinical isolates. These results suggest the potential applicability of CSNPs-AMB-Gls for the treatment of Candida biofilm-related infections.
Candida species are typical pathogenic microorganisms, which have been identified as the fourth most common cause of bloodstream infections in the United States and are associated with high morbidity and mortality (Gottardo et al., 2019; Nani et al., 2019). In addition to public health consequences, Candida is also related to food contamination, which reduces the nutritional value of products and leads to foodborne intoxications (Piccinelli et al., 2016; Rajkowska and Kunicka-Styczyńska, 2018). The predominant nosocomial fungal pathogen is Candida albicans, which is one of the leading causes of infections known to have a great biofilm-forming ability (Rajendran et al., 2016; Wall et al., 2019). Candida cells within biofilms, communities of cells embedded in a matrix, exhibit lower susceptibility to antimicrobials due to an extracellular matrix (Mitchell et al., 2016; Lohse et al., 2017). Even high therapeutic concentrations of existing antifungal drugs prove to be less efficient in inhibiting growth or removing biofilms while the risk of causing serious side effects, such as kidney or liver damage, is increased (Lohse et al., 2017).
Amphotericin B (AMB), a broad-spectrum antifungal drug, is recommended as first-line therapy for fungal-related infections (Benincasa et al., 2011; Vikelouda et al., 2017). However, C. albicans in the biofilm shows high resistance to AMB, which can be 1,000-fold less susceptible than planktonic ones. Several lipid-based drug delivery systems have been employed to enhance the therapeutic effect and decrease the toxicity of AMB (Weiler et al., 2008; Nieto et al., 2018). Unfortunately, the widespread use of these new formulations is impeded due to the high production cost (Weiler et al., 2009; Steimbach et al., 2017). Therefore, there is an urgent need to precise the delivery to enhance the efficiency and improve the inhibitory effect of AMB on ergosterol in fungal membranes.
Nanobiotechnology has gained attention in the pharmaceutical and medical fields, and the nano-sizing of antimicrobial agents seems to be a promising treatment for biofilm-related infections. Because nanoparticles (NPs) can more easily penetrate the biofilm matrix due to their size and surface charge (Baelo et al., 2015). It has been reported that both positively and negatively charged NPs could bind to biofilms. Negatively charged NPs could bind to microbial cells with hydrophobic interactions, while positively charged NPs could diffuse into the biofilm through binding to negatively charged biofilm polymers, such as eDNA (Nafee et al., 2014). Therefore, they can interfere with cells inside the biofilm directly (Jamil et al., 2017; Benoit et al., 2019). Moreover, the NP structure can enclose and protect antimicrobials from endogenous and exogenous factors of microbial cells, leading to enhanced bioactivity at the proximity of target cells (Jamil et al., 2016). Chitosan (CS) is a cost-effective biopolymer suitable for the preparation of biocompatible, biodegradable, and non-cytotoxic NPs with inherent antimicrobial and antibiofilm activity (Martinez et al., 2010; Klinger-Strobel et al., 2016). In our previous work, CS nanoparticle (CSNP) has been proven to be a kind of ideal carrier for biofilm treatment (Tan et al., 2018). Furthermore, CSNP is a promising carrier to overcome the poor aqueous solubility of hydrophobic drugs (Trapani et al., 2009; Quiñones et al., 2018).
Increased drug resistance of C. albicans in biofilms compared to its planktonic form is mainly due to the self-produced extrapolymeric substance (EPS) matrix (Limoli et al., 2015; Flemming et al., 2016). Moreover, the inactive structure of the biofilm still promotes the adhesion and regeneration of other microorganisms (MacDonald et al., 2000). Therefore, the biofilm matrix itself should also be a target in addition to fungal cells. Recently, EPS-degrading enzymes have been proposed as a new strategy to remove the biofilm and to enhance drug efficacy (Okshevsky and Meyer, 2015; Kimura and Urata, 2016; Klinger-Strobel et al., 2016). As one of the major polysaccharides in the C. albicans biofilm matrix, β-1,3-glucan plays an important role in the biofilm structure and cell protection. The enzyme β-1,3-glucanase (Gls) can disrupt the Candida biofilm matrix by degrading β-1,3-glucan (Nett et al., 2007). Our previous work also showed that Gls can enhance the efficacy of antimicrobial drugs (De Brucker et al., 2015; Tan et al., 2018). However, to our knowledge, there are no reports showing nanosystems that combine the enzyme specifically degrading the Candida biofilm matrix and the antifungal that kills Candida within the biofilm.
Therefore, the combination of CSNPs with antimicrobials and enzymes as novel nanoantimicrobials seems to be a plausible approach for the treatment of Candida biofilms. In this study, CSNPs were functionalized with Gls and loaded with AMB (CSNPs-AMB-Gls) to combine enhanced antibiofilm activity with biofilm matrix disruption ability.
Candida albicans DAY 185 was used in the experiments. Three clinical isolates of C. albicans, named C. albicans BF1, BF2, and BF3, were obtained from the Medical University of Vienna. Each experiment was repeated three times. All of them are able to form strong biofilms, which have been confirmed by the crystal violet method. The strains were cultured in a Yeast Peptone Dextrose (YPD) medium (Sigma-Aldrich, Austria) at 30°C. RPMI 1640 medium (Thermo Fisher Scientific, Waltham, MA, United States) was used for biofilm growth. Gls (lyticase from Arthrobacter luteus, ≥2,000 units/mg protein), CS (low molecular weight, degree of deacetylation 75–85%), AMB, and other reagents were purchased from Sigma-Aldrich.
Chitosan nanoparticles were prepared as described by the ion gelation method with polyanionic sodium triphosphate (TPP) in our previous work (Pu et al., 2014). CSNP-Gls was prepared with mixing CSNPs [1 mg/ml polybutylene succinate (PBS)] and Gls (100 μg/ml PBS) while stirring overnight at 4°C. NP suspensions were centrifuged (14,000 rpm, 30 min) and freeze-dried eventually. For the preparation of CSNPs-AMB-Gls, CSNPs-Gls suspension (1 mg/ml) was mixed with AMB (100 μg/ml DMSO) and stirred for 24 h. The mixture was centrifuged and lyophilized.
The size of NPs was analyzed by dynamic light scattering (DLS). Laser Doppler velocimetry assays were used to determine the zeta potential (ZetaSizer Nano ZS, Malvern Instruments, United Kingdom). Morphological characterization was confirmed by scanning electron microscopy (SEM, JSM 6310, JEOL Ltd., Akishima, Tokyo, Japan).
The CSNPs-Gls suspension (prepared as reported above) was evaluated using a suspension of yeast as the substrate according to the Sigma-Aldrich protocol for Gls activity assay. The loading capacity (LC) of AMB loaded on the NP was estimated by measuring the absorption at 405 nm spectrophotometrically (Spectroscopy, Persee, TU-1810, China) and calculated as follows:
Where tAMB, fAMB, and NPs, represent total AMB, free AMB, and NP amount, respectively.
Release kinetics in vitro was carried out as follows: 4 ml of CSNPs-AMB-Gls solution (1 mg/ml in a dialysis bag) was placed in 40 ml of PBS with stirring (100 rpm). At any given time point, 4 ml of PBS was taken and replaced with 4 ml of fresh PBS to maintain the sink volume. The amount of AMB in the solution was measured at 405 nm by UV spectrometry (Persee, TU-1810, China).
About 100 μl of Candida (1 × 106 CFU/ml), including C. albicans DAY 185 and three clinical isolates of C. albicans, was added to wells of 96-well microplates (Thermo Fisher Scientific, Waltham, MA, United States) with different concentrations of free AMB, CSNPs, and CSNPs-AMB-Gls (4, 2, 1, 0.5, 0.25, 0.125, 0.0625, or 0 μg/ml). The microplate was incubated at 37°C for 24 h at 150 rpm. Minimum inhibitory concentration (MIC) was defined as the lowest concentration of AMB at which no visible growth was detected.
Biofilms in a 96-well microplate and on silicone platelets were conducted and assayed as previously described (Tan et al., 2018). In brief, C. albicans was diluted to 1 × 106 CFU/ml with RPMI 1640 and 100 μl of fungal culture was pipetted into each well of a 96-well microplate. Biofilms were formed at 37°C for 24 h without shaking.
Chitosan nanoparticles were labeled with rhodamine B isothiocyanate (CSNPs-RBITC) as previously described (Pu et al., 2014). An autoclaved medical grade silicone platelet (3-mm-diameter, Websinger, Austria) was placed in each well of the 96-well microplate. C. albicans DAY 185 biofilms were formed on medical grade silicone platelets for 24 h as described above in the 96-well microplate and mixed with RBITC-CSNPs (100 μg/ml) for 2 h and washed with PBS. The penetration of CSNPs was determined by confocal laser scanning microscopy (CLSM).
Biofilms were formed in 96-well microplates as previously described. C. albicans DAY 185 cells were incubated with free AMB, a combination of AMB and Gls (AMB+Gls, 2 μg/ml), CSNPs-AMB, and CSNPs-AMB-Gls with different drug concentrations (0, 0.25, 0.5, and 1 μg/ml) for 24 h. Biofilms were washed with PBS and quantified with a cell counting kit-8 (CCK-8, Dojindo Molecular Technologies, Gaithersburg, MD, United States) reduction assay. The absorbance was examined at 450 nm.
Candida albicans DAY 185 biofilms were formed as described above for 24 h and added with fresh medium containing different concentrations (0, 1, 2, and 4 μg/ml) of free AMB, AMB+Gls, CSNPs-AMB, and CSNPs-AMB-Gls. After another 24 h, the biofilms were washed with PBS and quantified with the CCK-8 reduction assay.
Biofilms on silicone platelets were treated with free AMB, AMB+Gls, CSNPs-AMB, and CSNPs-AMB-Gls (4 μg/ml) as described above.
Biofilm architecture was investigated by SEM. Biofilms on silicone platelets were fixed in 3% glutaraldehyde (v/v) in PBS solution overnight at 4°C, and then subjected to serial dehydration with 25, 50, 75, and 100% ethanol for 10 min each. The biofilms were coated with gold and examined by SEM (JSM 6310, JEOL Ltd., Akishima, Tokyo, Japan).
The cells of biofilms were stained with a LIVE/DEAD® BacLight™ Bacterial Viability and Counting kit (L34856, Invitrogen, United States) following the manufacturer’s instructions. Cell viability was observed with CLSM.
The antibiofilm efficacy of free AMB, AMB+Gls, CSNPs-AMB, and CSNPs-AMB-Gls (2 μg/ml) was tested on three clinical isolates. Biofilms of clinical strains formed in the 96-well microplate were evaluated as described above.
Statistical analyses of data were determined with GraphPad Prism software program (GraphPad Software, San Diego, CA, United States). Values are given as mean ± standard deviation (SD) of the number of experiments (n = 3). Statistical significance was determined by the t-test analysis with p < 0.05.
Scanning electron microscopy images showed that CSNPs-AMB-Gls was round particles (Figure 1). The average size of CSNPs-AMB-Gls was 174.47 ± 5.12 nm (size PDI 0.17), and the surface zeta potential was +15.84 ± 1.41 mV. The loading capacity of AMB on CSNPs-AMB-Gls was 3.05% ± 0.13%. Gls loaded on the NP-retained Gls activity, the activity of which was 128.6 ± 4.54 U/mg NP.
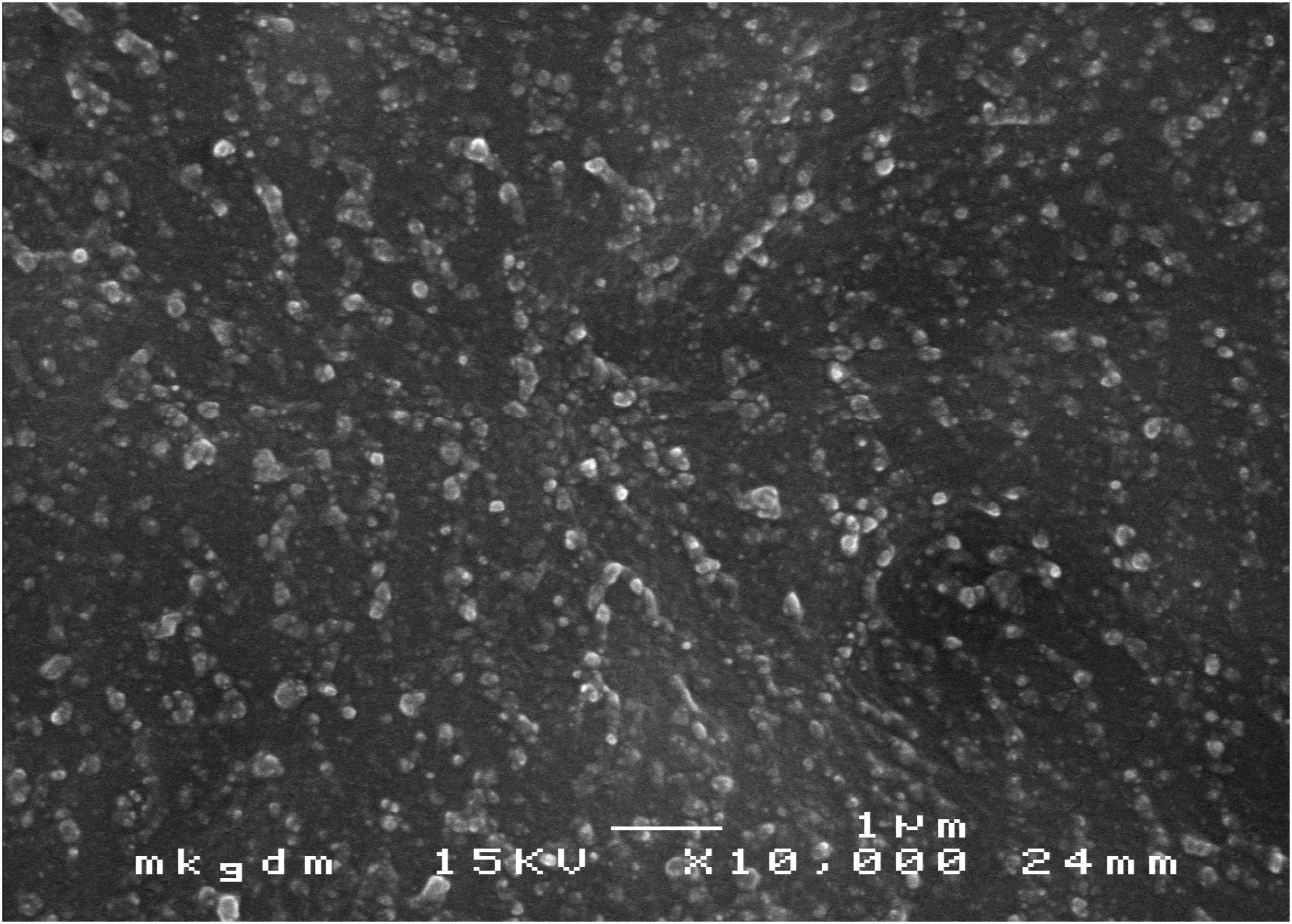
Figure 1. Scanning electron microscopy (SEM) image of chitosan nanoparticles (CSNPs) loaded with amphotericin B (AMB), which were functionalized with β-1,3-glucanase (Gls) (CSNPs-AMB-Gls). Magnification, ×10,000.
Figure 2 shows the release profiles of AMB from NPs. CSNPs-AMB-Gls presented an initial burst release phase in the first 1 h: 52.5% of the total AMB was released. Subsequently, the drug release was slowed down. Over 24 h, CSNPs-AMB-Gls showed a sustained release of 80.6% of the total AMB.
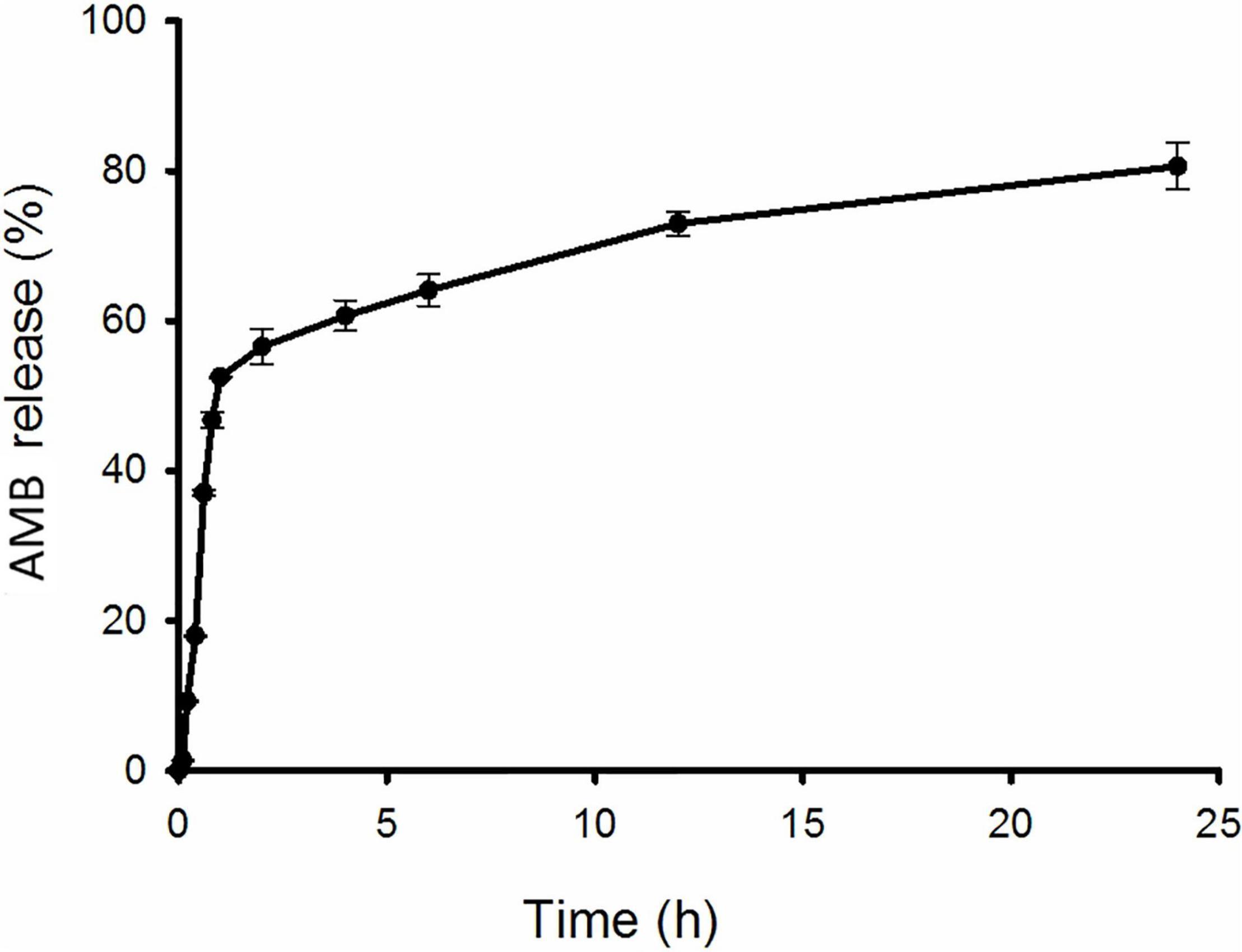
Figure 2. In vitro release kinetics of AMB from the nanoparticle (NP). CSNPs-AMB-Gls was placed in polybutylene succinate (PBS) with stirring. The amount of AMB in the solution was measured at 405 nm by UV spectrometry at any given time point.
Both free AMB and CSNPs-AMB-Gls were effective against C. albicans DAY 185 and the growth of clinical isolates, which showed the same MIC. MIC was 1 μg/ml against C. albicans DAY 185 and higher against clinical isolates (2, 2, and 4 μg/ml, respectively). CSNPs alone had no effect on planktonic Candida at the concentration used in this work (data not shown). This result suggested that AMB loading in CSNPs did not change the antifungal activity of free AMB to planktonic Candida.
After 2 h treatment with CSNPs, red color (RBITC-NPs) can be observed on the surface and inside the biofilm, suggesting that the penetration of CSNPs into the biofilm (Figure 3).

Figure 3. Penetration of CSNPs into biofilms. CSNPs were labeled with rhodamine B isothiocyanate (RBITC) and visualized by confocal laser scanning microscopy (CLSM). SYTO 9-labeled biofilms incubated with CSNPs (A). RBITC-NP penetration into biofilms (B). Overlap (C).
As shown in Figure 4, free AMB, AMB+Gls, CSNPs-AMB, and CSNPs-AMB-Gls showed concentration-dependent biofilm inhibition activity with all tested concentrations. At the concentration of 1 μg/ml, almost no biofilm formations were observed in the treatment with free AMB, AMB+Gls, CSNPs-AMB, and CSNPs-AMB-Gls. At other concentrations, CSNPs-AMB and CSNPs-AMB-Gls showed similar inhibition compared to free AMB.
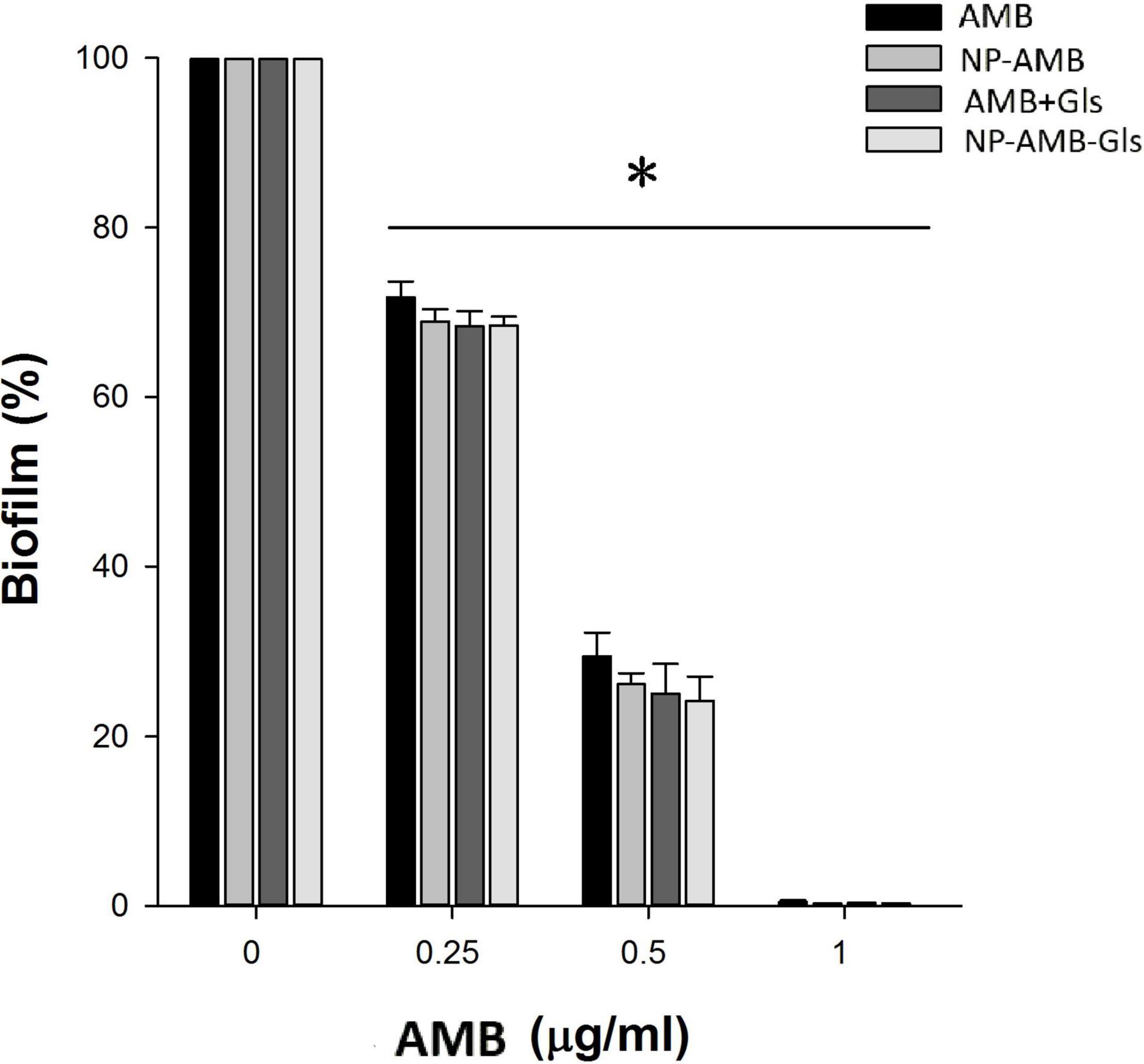
Figure 4. Inhibition efficacy on the biofilm in a 96-well microplate. Candida albicans DAY 185 cells were co-incubated with free AMB, AMB+Gls, CSNPs-AMB, and CSNPs-AMB-Gls with different drug concentrations (0, 0.25, 0.5, and 1 μg/ml) for 24 h. The results represent the means and standard deviations (SDs) (error bars), n = 3. Statistical significance was determined by the t-test analysis. *p < 0.05 for comparison between the untreated and treated groups.
Due to the high resistance of the biofilm to antimicrobial agents, we used 4 × MIC for evaluating the antibiofilm activity of free AMB, AMB+Gls, CSNPs-AMB, and CSNPs-AMB-Gls on the mature biofilm (Figure 5). As expected, C. albicans DAY 185 in the biofilm increases resistance to free AMB or CSNPs-AMB without Gls compared to planktonic forms. However, both free AMB with Gls (AMB+Gls) and CSNPs-AMB-Gls showed a good extent of biofilm eradication. Moreover, CSNPs-AMB-Gls exhibited the highest activity in the eradication of preformed biofilm at any of the concentrations tested. At a concentration of 4 μg/ml AMB, AMB+Gls caused a 83.1% biofilm reduction, but CSNPs-AMB-Gls reduced the biofilm by 90.9%.
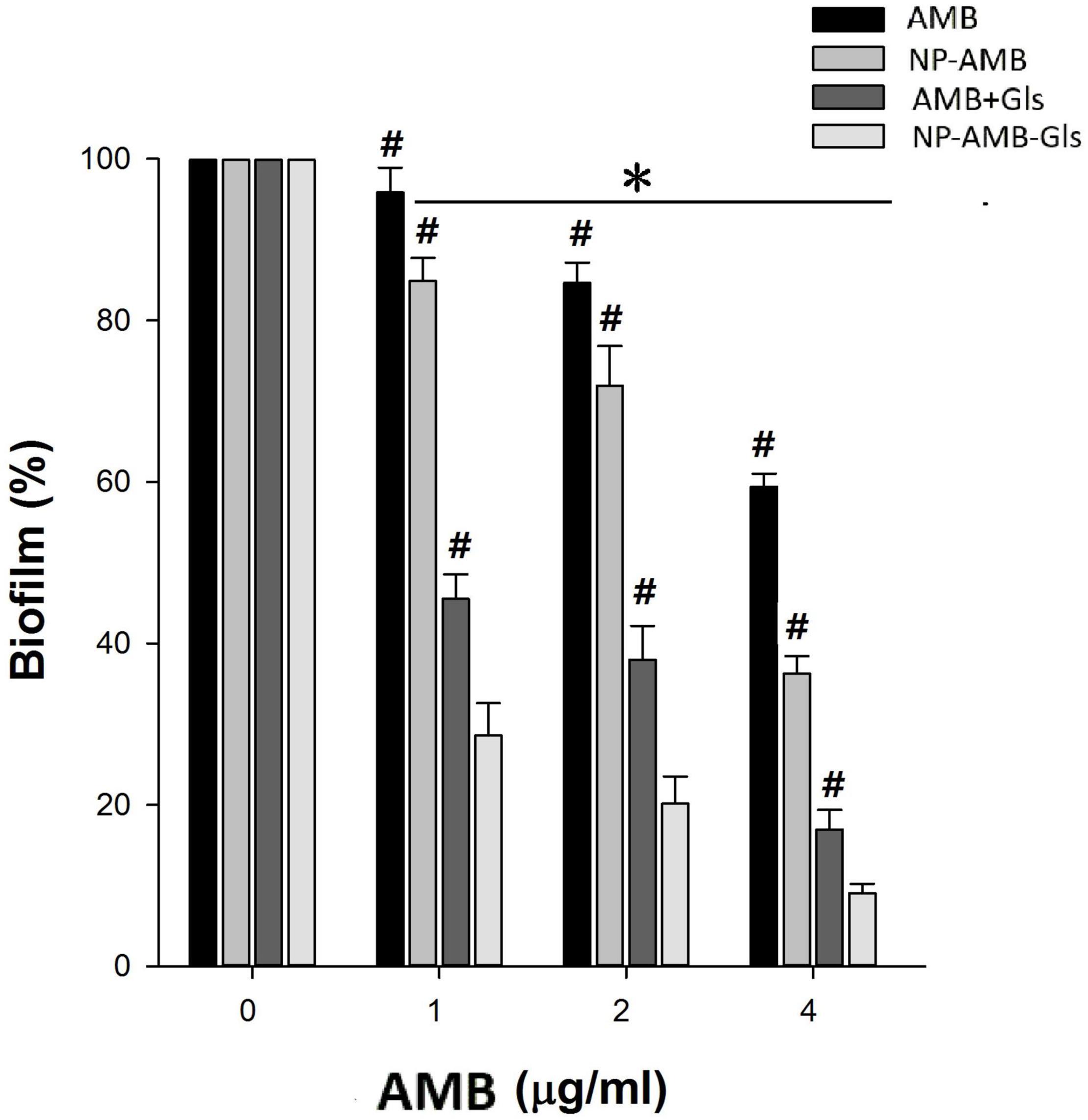
Figure 5. Eradication efficacy of NPs on the mature biofilm by C. albicans DAY 185 in a 96-well microplate. Mature biofilms were treated with different concentrations (0, 1, 2, and 4 μg/ml) of free AMB, AMB+Gls, CSNPs-AMB, and CSNPs-AMB-Gls for 24 h. The results represent the means and SDs (error bars), n = 3. Statistical significance was determined by the t-test analysis. *p < 0.05 for a comparison between the untreated and treated groups. #p < 0.05 for a comparison between CSNPs-AMB-Gls and other treated groups.
Biofilm architecture and LIVE/DEAD organisms within the biofilm on silicone surfaces were investigated. The cell viability of the biofilm on silicone platelets treated with or without drugs was observed with CLSM (Figure 6). A large number of green-colored cells (live cells) were observed without any treatment (Figure 6A), which indicated that the mature biofilm was mainly composed of active cells. Treated with free AMB and CSNPs-AMB, the green-colored cells (live cells) were drastically reduced (Figures 6B,C). With the combination of Gls, more red-colored cells (dead cells) and less biofilm thickness were shown compared to free AMB treatment (Figures 6D,E).
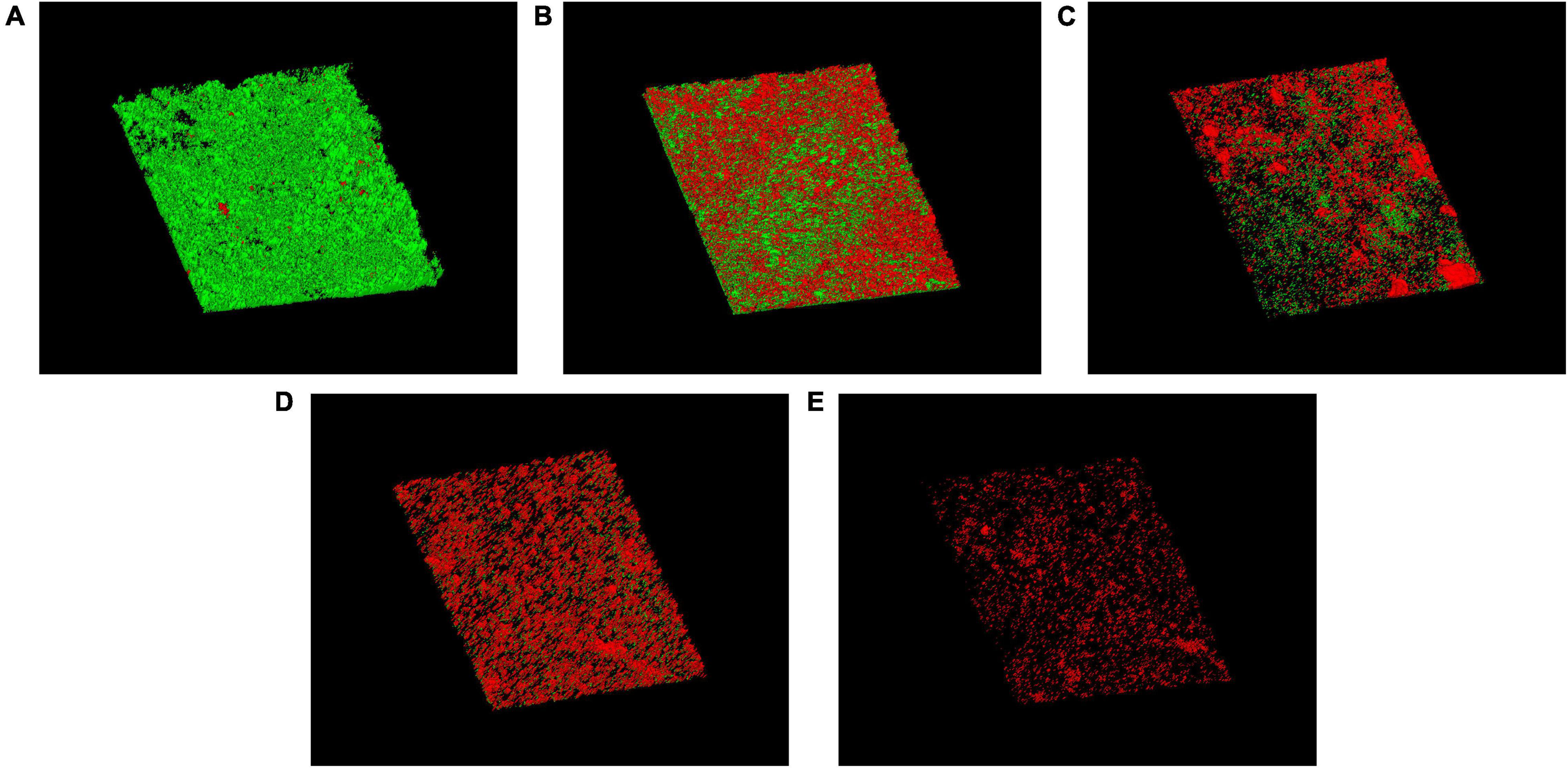
Figure 6. CLSM images of biofilms on untreated (A) or free AMB (B), CSNPs-AMB (C), AMB+Gls (D), and CSNPs-AMB-Gls (E) treated silicone surfaces. Biofilms were stained with the Live/Dead® BacLight™ Bacterial Viability and Counting kit. CLSM reconstructions show the three-dimensional staining pattern for live cells (SYTO-9, green) and dead cells (propidium iodide, red). Magnification, ×10.
Scanning electron microscopy images confirmed the results mentioned above and revealed changes in the biofilm structures (Figure 7). In the control group, we can see the typical dense composition of biofilms (Figure 7A). However, treatment with free AMB and CSNPs-AMB significantly reduced colonization (Figures 7B,C). Furthermore, AMB+Gls and CSNPs-AMB-Gls resulted in only single cells or even free of cells on silicone platelets (Figures 7D,E), which meant that the biofilm structure was disrupted with the killing of Candida cells.
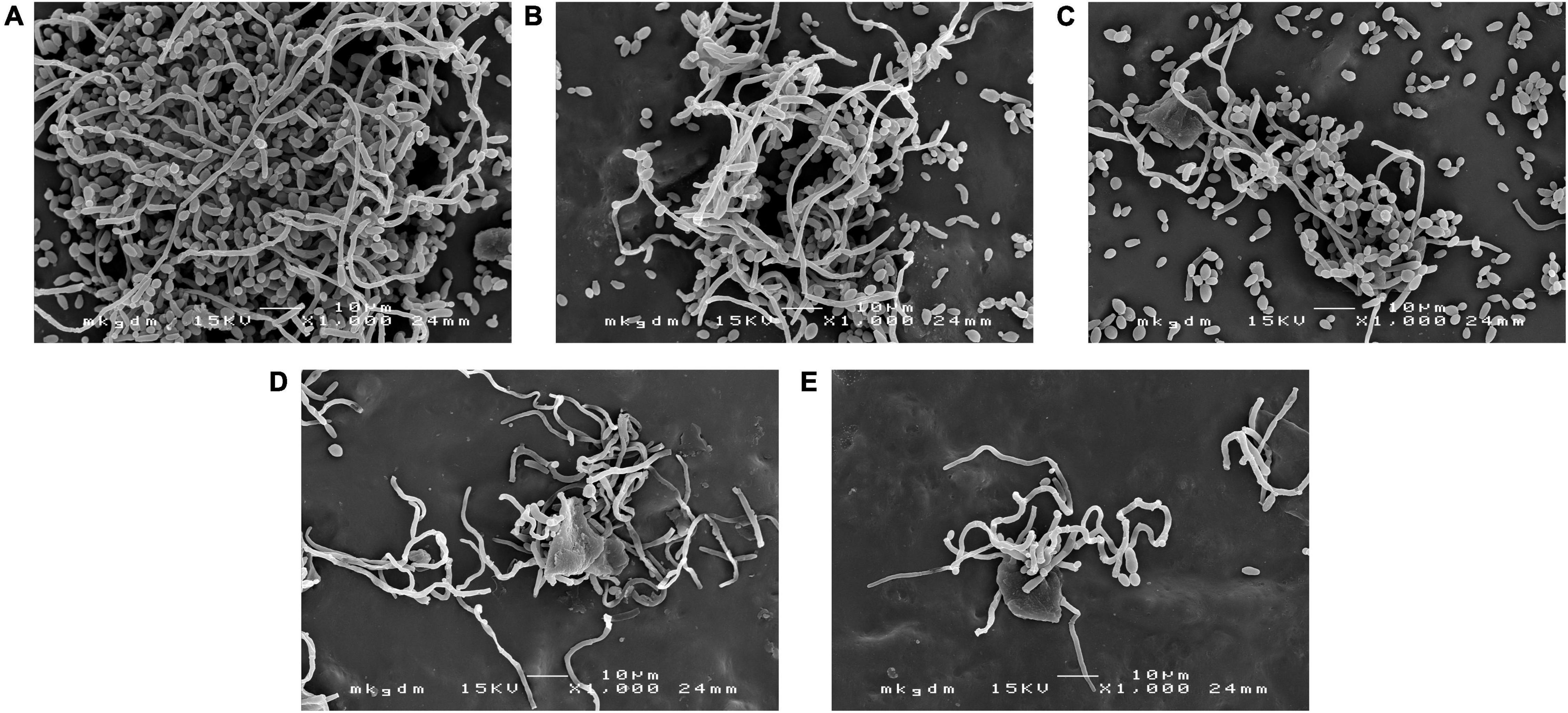
Figure 7. SEM images of the biofilm by C. albicans DAY 185 on untreated (A) or free AMB (B), CSNPs-AMB (C), AMB+Gls (D), and CSNPs-AMB-Gls (E) treated silicone surfaces.
As shown in Supplementary Figure 1, biofilm formations by all clinical isolates were reduced in the presence of free AMB, AMB+Gls, CSNPs-AMB, and CSNPs-AMB-Gls (4 × MIC). CSNPs-AMB-Gls exhibited the best antibiofilm activity, which reduced 74.5, 85.2, and 59.5% biofilms of the clinical isolates BF1, BF2, and BF3, respectively.
Recently, NPs have been employed to protect antibiofilm agents from drug sequestration and improve penetration into the matrix, so as to enhance the therapeutic effect for treating biofilms. Here, CSNPs were functionalized with Gls and loaded with AMB to develop a new antibiofilm strategy until now.
In this work, AMB from CSNPs-AMB-Gls showed an initial burst release phase followed by a sustained release phase. This release mode is more suitable for the removal of biofilms. Because the initial burst release means a higher initial drug dose, which will reduce microbial drug tolerance (Cheow et al., 2010; Forier et al., 2014).
It has been reported that positively charged NPs could bind to negatively charged bacterial biofilm matrix components (Nafee et al., 2014; Baelo et al., 2015). Moreover, CS has the ability of penetrating into biofilms (Lu et al., 2014; Pu et al., 2014). Here, the proven ability to penetrate biofilms facilitates CSNPs to be a good drug carrier, which can carry drugs to penetrate into the biofilm as well and kill the inner cells.
For biofilm inhibition efficacy, Gls alone exhibited no obvious inhibition and CSNPs-AMB-Gls was not able to enhance the inhibition efficacy of biofilm formation relative to CSNPs-AMB. In addition, AMB alone or whenever driven with CSNPs (at all concentrations) showed similar efficacy on biofilm formation and planktonic cells, because the drug could kill planktonic fungi directly before biofilm formation.
In the preformed biofilm, our results exhibited a better eradication activity of CSNPs-AMB and CSNPs-AMB-Gls than free AMB and AMB+Gls, which indicated the superiority of NPs for biofilm treatment. NPs can protect the loaded antimicrobial agents from drug sequestration by the biofilm matrix (Forier et al., 2013; Koo et al., 2017). Moreover, NPs can deliver the drug into the biofilm matrix and directly target the microbial cells in the biofilm so as to maximize the therapeutic benefit. AMB+Gls and CSNPs-AMB-Gls showed a better detachment activity than free AMB and CSNPs-AMB, which suggested the synergistic efficacy of Gls.
β-1,3-glucan in C. albicans biofilm is considered to contribute to the resistance of antifungal drugs. As one of the important matrix components, β-1,3-glucan can protect cells in the biofilm by sequestering drugs (Nett et al., 2007). Similar functions of β-1,3-glucan have also been found in other Candida species (Tan et al., 2017). Therefore, the degradation of β-1,3-glucan resulted in the disruption of the biofilm matrix, and then improved AMB efficiency. This result is also consistent with our previous work (Tan et al., 2017, 2018). It can be ascribed to that Gls could disassemble the biofilm structure, which facilitates the mobility of positively charged NPs and binds to negatively charged biofilm components, thus providing high local concentrations of AMB to enhance the antibiofilm activity. Similarly, it has also been reported that the antibiofilm effect of antibiotic-containing NPs can be enhanced if the biofilm matrix was degraded by enzymes, such as DNase (Pu et al., 2014; Baelo et al., 2015). This interpretation could be further strengthened by SEM and CLSM images. CSNPs-AMB-Gls was more effective in killing C. albicans DAY 185 cells and disassembling the biofilm structure, which indicated the CSNPs-AMB-Gls better disrupted the biofilm matrix and thus increased killing of fungal cells.
Standardized and idealized laboratory conditions might make the microbe to lose some important pathophysiological characteristics when they are subcultured for decades (Fux et al., 2005). Thus, in our work, clinically isolated specimens were used to mirror clinical efficacy. The results demonstrate that CSNPs-AMB-Gls can disrupt clinically isolated biofilms as well as standard strain biofilms.
In this study, we developed a biodegradable functional CSNP by loading AMB and Gls. CSNPs-AMB-Gls was homogeneously dispersed with a positive surface zeta potential. CSNPs-AMB-Gls was active in killing C. albicans cells and inhibiting biofilm formation, in addition to retaining the ability of Gls to disrupt the biofilm matrix. Excitingly, CSNPs-AMB-Gls exhibited the highest antibiofilm activity compared to free AMB and CSNPs-AMB in the mature biofilm. Although the assessment of biocompatibility and efficacy in vivo still needs to be evaluated, our studies pave the way for the application of CSNPs-AMB-Gls to treat C. albicans biofilm-related infections. Moreover, CSNPs-AMB-Gls can be employed as a platform to design more functions such as new drug delivery systems.
The original contributions and additional information presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
YT contributed to the conception of the study and finalized the manuscript. SM performed the experiment data and wrote the manuscript. JX contributed to analysis and manuscript preparation. RL helped to perform the analysis with constructive discussion. TD contributed to data analysis and manuscript editing. JL helped to edit the manuscript and evaluate the data. All authors contributed to the article and approved the submitted version.
This research was funded by the Young Taishan Scholars Program of Shandong Province (tsqn202103094).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.815091/full#supplementary-material
Supplementary Figure 1 | Antibiofilm activity against clinical isolates. The results represent the means and standard deviations (SDs; error bars) of three independent experiments. Statistical significance was determined by a t-test analysis. *p < 0.05 for a comparison between the untreated and treated groups. #p < 0.05 for a comparison between chitosan nanoparticle (CSNP) loaded with Amphotericin B (AMB), which were functionalized with β-1,3-glucanase (Gls) (CSNP-AMB-Gls) and other treated groups.
Baelo, A., Levato, R., Julian, E., Crespo, A., Astola, J., Gavalda, J., et al. (2015). Disassembling bacterial extracellular matrix with DNase-coated nanoparticles to enhance antibiotic delivery in biofilm infections. J. Control. Release 209, 150–158. doi: 10.1016/j.jconrel.2015.04.028
Benincasa, M., Pacor, S., Wu, W., Prato, M., Bianco, A., and Gennaro, R. (2011). Antifungal activity of Amphotericin B conjugated to carbon nanotubes. ACS Nano 5, 199–208. doi: 10.1021/nn1023522
Benoit, D., Sims, K. R., and Fraser, D. (2019). Nanoparticles for oral biofilm treatments. ACS Nano 13, 4869–4875. doi: 10.1021/acsnano.9b02816
Cheow, W. S., Chang, M. W., and Hadinoto, K. (2010). Antibacterial efficacy of inhalable levofloxacin-loaded polymeric nanoparticles against E. coli biofilm cells: the effect of antibiotic release profile. Pharm. Res. 27, 1597–1609. doi: 10.1007/s11095-010-0142-6
De Brucker, K., Tan, Y., Vints, K., De Cremer, K., Braem, A., Verstraeten, N., et al. (2015). Fungal β-1,3-glucan increases ofloxacin tolerance of Escherichia coli in a polymicrobial E. coli/Candida albicans biofilm. Antimicrob. Agents Chemother. 59, 3052–3058. doi: 10.1128/AAC.04650-14
Flemming, H.-C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., and Kjelleberg, S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–75. doi: 10.1038/nrmicro.2016.94
Forier, K., Messiaen, A.-S., Raemdonck, K., Deschout, H., Rejman, J., De Baets, F., et al. (2013). Transport of nanoparticles in cystic fibrosis sputum and bacterial biofilms by single-particle tracking microscopy. Nanomedicine 8, 935–949. doi: 10.2217/nnm.12.129
Forier, K., Raemdonck, K., De Smedt, S. C., Demeester, J., Coenye, T., and Braeckmans, K. (2014). Lipid and polymer nanoparticles for drug delivery to bacterial biofilms. J. Control. Release 190, 607–623. doi: 10.1016/j.jconrel.2014.03.055
Fux, C. A., Shirtliff, M., Stoodley, P., and Costerton, J. W. (2005). Can laboratory reference strains mirror ‘real-world’ pathogenesis? Trends Microbiol. 13, 58–63. doi: 10.1016/j.tim.2004.11.001
Gottardo, B., Lemes, T. H., Byzynski, G., Paziani, M. H., Von-Zeska-Kress, M. R., Almeida, M. D., et al. (2019). One-pot synthesis and antifungal activity of nontoxic silver-loaded hydroxyapatite nanocomposites against Candida species. ACS Appl. Nano Mater. 2, 2112–2120. doi: 10.1021/acsanm.9b00091
Jamil, B., Bokhari, H., and Imran, M. (2017). Mechanism of action: how nano-antimicrobials act? Curr. Drug Targets 18, 363–373. doi: 10.2174/1389450116666151019101826
Jamil, B., Habib, H., Abbasi, S. A., Ihsan, A., Nasir, H., and Imran, M. (2016). Development of cefotaxime impregnated chitosan as nano-antibiotics: de novo strategy to combat biofilm forming multi-drug resistant pathogens. Front. Microbiol. 7:330. doi: 10.3389/fmicb.2016.00330
Kimura, Y., and Urata, M. (2016). Characterization of a eukaryotic-like protein kinase, DspB, with an atypical catalytic loop motif from Myxococcus xanthus. Arch. Microbiol. 198, 219–226. doi: 10.1007/s00203-015-1181-5
Klinger-Strobel, M., Ernst, J., Lautenschläger, C., Pletz, M. W., Fischer, D., and Makarewicz, O. (2016). A blue fluorescent labeling technique utilizing micro-and nanoparticles for tracking in LIVE/DEAD® stained pathogenic biofilms of Staphylococcus aureus and Burkholderia cepacia. Int. J. Nanomedicine 11, 575–83. doi: 10.2147/IJN.S98401
Koo, H., Allan, R. N., Howlin, R. P., Stoodley, P., and Hall-Stoodley, L. (2017). Targeting microbial biofilms: current and prospective therapeutic strategies. Nat. Rev. Microbiol. 15, 740–755. doi: 10.1038/nrmicro.2017.99
Limoli, D. H., Jones, C. J., and Wozniak, D. J. (2015). Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol. Spectr. 3:2014. doi: 10.1128/microbiolspec.MB-0011-2014
Lohse, M. B., Gulati, M., Johnson, A. D., and Nobile, C. J. (2017). Development and regulation of single- and multi-species Candida albicans biofilms. Nat. Rev. Microbiol. 16, 19–31. doi: 10.1038/nrmicro.2017.107
Lu, Y., Slomberg, D. L., and Schoenfisch, M. H. (2014). Nitric oxide-releasing chitosan oligosaccharides as antibacterial agents. Biomaterials 35, 1716–1724. doi: 10.1016/j.biomaterials.2013.11.015
MacDonald, R., Santa, M., and Brozel, V. S. (2000). The response of a bacterial biofilm community in a simulated industrial cooling water system to treatment with an anionic dispersant. J. Appl. Microbiol. 89, 225–235. doi: 10.1046/j.1365-2672.2000.01099.x
Martinez, L. R., Mihu, M. R., Han, G., Frases, S., Cordero, R. J., Casadevall, A., et al. (2010). The use of chitosan to damage Cryptococcus neoformans biofilms. Biomaterials 31, 669–679. doi: 10.1016/j.biomaterials.2009.09.087
Mitchell, K. F., Zarnowski, R., and Andes, D. R. (2016). Fungal super glue: the biofilm matrix and its composition, assembly, and functions. PLoS Pathog. 12:e1005828. doi: 10.1371/journal.ppat.1005828
Nafee, N., Husari, A., Maurer, C. K., Lu, C., de Rossi, C., Steinbach, A., et al. (2014). Antibiotic-free nanotherapeutics: ultra-small, mucus-penetrating solid lipid nanoparticles enhance the pulmonary delivery and anti-virulence efficacy of novel quorum sensing inhibitors. J. Control. Release 192, 131–140. doi: 10.1016/j.jconrel.2014.06.055
Nani, B. D., Sardi, J., Lazarini, J. G., Silva, D. R., and Rosalen, P. L. (2019). Anti-inflammatory and anti-Candida effects of Brazilian organic propolis, a promising source of bioactive molecules and functional food. J. Agric. Food Chem. 68, 2861–2871. doi: 10.1021/acs.jafc.8b07304
Nett, J., Lincoln, L., Marchillo, K., Massey, R., Holoyda, K., Hoff, B., et al. (2007). Putative role of β-1,3 glucans in Candida albicans biofilm resistance. Antimicrob. Agents Chemother. 51, 510–520. doi: 10.1128/AAC.01056-06
Nieto, J., Alvar, J., Rodriguez, C., San Andres, M. I., San Andres, M. D., and Gonzalez, F. (2018). Comparison of conventional and lipid emulsion formulations of amphotericin B: pharmacokinetics and toxicokinetics in dogs. Res. Vet. Sci. 117, 125–132. doi: 10.1016/j.rvsc.2017.12.005
Okshevsky, M., and Meyer, R. L. (2015). The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit. Rev. Microbiol. 41, 341–352. doi: 10.3109/1040841X.2013.841639
Piccinelli, A. L., Pagano, I., Esposito, T., Mencherini, T., Porta, A., Petrone, A. M., et al. (2016). HRMS Profile of a hazelnut skin proanthocyanidin-rich fraction with antioxidant and anti-Candida albicans activities. J. Agric. Food Chem. 64, 585–595. doi: 10.1021/acs.jafc.5b05404
Pu, Y., Liu, A., Zheng, Y., and Ye, B. (2014). In vitro damage of Candida albicans biofilms by chitosan. Exp. Ther. Med. 8, 929–934. doi: 10.3892/etm.2014.1839
Quiñones, J. P., Peniche, H., and Peniche, C. (2018). Chitosan based self-assembled nanoparticles in drug delivery. Polymers (Basel) 10:235. doi: 10.3390/polym10030235
Rajendran, R., Sherry, L., Nile, C. J., Sherriff, A., Johnson, E., Hanson, M., et al. (2016). Biofilm formation is a risk factor for mortality in patients with Candida albicans bloodstream infection-Scotland, 2012-2013. Clin. Microbiol. Infect. 22, 87–93. doi: 10.1016/j.cmi.2015.09.018
Rajkowska, K., and Kunicka-Styczyńska, A. (2018). Typing and virulence factors of food-borne Candida spp. isolates. Int. J. Food Microbiol. 279, 57–63. doi: 10.1016/j.ijfoodmicro.2018.05.002
Steimbach, L. M., Tonin, F. S., Virtuoso, S., Borba, H. H., Sanches, A. C., Wiens, A., et al. (2017). Efficacy and safety of amphotericin B lipid-based formulations-a systematic review and meta-analysis. Mycoses 60, 146–154. doi: 10.1111/myc.12585
Tan, Y., Leonhard, M., Ma, S., Moser, D., and Schneider-Stickler, B. (2017). Dispersal of single and mixed non-albicans Candida species biofilms by β-1,3-glucanase in vitro. Microb. Pathog. 113, 342–347. doi: 10.1016/j.micpath.2017.10.057
Tan, Y., Ma, S., Leonhard, M., Moser, D., and Schneider-Stickler, B. (2018). β-1,3-glucanase disrupts biofilm formation and increases antifungal susceptibility of Candida albicans DAY185. Int. J. Biol. Macromol. 108, 942–946. doi: 10.1016/j.ijbiomac.2017.11.003
Trapani, A., Sitterberg, J., Bakowsky, U., and Kissel, T. (2009). The potential of glycol chitosan nanoparticles as carrier for low water soluble drugs. Int. J. Pharm. 375, 97–106. doi: 10.1016/j.ijpharm.2009.03.041
Vikelouda, A., Simitsopoulou, M., Kechagioglou, P., Papi, R., Kyriakidis, D., and Roilides, E. (2017). Antifungal activity of functionalized carbon nanotubes conjugated to Amphotericin-B against Candida albicans and Candida parapsilosis biofilms. Mycoses 60, 85–85. doi: 10.1021/nn1023522
Wall, G., Montelongo-Jauregui, D., Bonifacio, B. V., Lopez-Ribot, J. L., and Uppuluri, P. (2019). Candida albicans biofilm growth and dispersal: contributions to pathogenesis. Curr. Opin. Microbiol. 52, 1–6. doi: 10.1016/j.mib.2019.04.001
Weiler, S., Bellmann-Weiler, R., Dunzendorfer, S., Joannidis, M., and Bellmann, R. (2008). Levels of amphotericin B lipid formulations in ascites. J. Antimicrob. Chemother. 62, 1163–1164. doi: 10.1093/jac/dkn306
Keywords: antibiofilm, nanoparticle, chitosan, Candida, amphotericin B
Citation: Tan Y, Ma S, Ding T, Ludwig R, Lee J and Xu J (2022) Enhancing the Antibiofilm Activity of β-1,3-Glucanase-Functionalized Nanoparticles Loaded With Amphotericin B Against Candida albicans Biofilm. Front. Microbiol. 13:815091. doi: 10.3389/fmicb.2022.815091
Received: 15 November 2021; Accepted: 25 April 2022;
Published: 24 May 2022.
Edited by:
Laura Quintieri, Institute of Sciences of Food Production (CNR), ItalyReviewed by:
Leonardo Caputo, Institute of Sciences of Food Production (CNR), ItalyCopyright © 2022 Tan, Ma, Ding, Ludwig, Lee and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yulong Tan, dGFueXVsb25nQHFhdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.