- 1MEPHI, APHM, IRD 198, Aix Marseille University, IHU-Méditerranée Infection, Marseille, France
- 2Aix Marseille Université, Université de Toulon, CNRS, IRD, MIO UM 110, Marseille, France
The discovery of Acanthamoeba polyphaga mimivirus in 2003 using the free-living amoeba Acanthamoeba polyphaga caused a paradigm shift in the virology field. Twelve years later, using another amoeba as a host, i.e., Vermamoeba vermiformis, novel isolates of giant viruses have been discovered. This amoeba–virus relationship led scientists to study the evolution of giant viruses and explore the origins of eukaryotes. The purpose of this article is to review all the giant viruses that have been isolated from Vermamoeba vermiformis, compare their genomic features, and report the influence of these viruses on the cell cycle of their amoebal host. To date, viruses putatively belonging to eight different viral taxa have been described: 7 are lytic and 1 is non-lytic. The comparison of giant viruses infecting Vermamoeba vermiformis has suggested three homogenous groups according to their size, the replication time inside the host cell, and the number of encoding tRNAs. This approach is an attempt at determining the evolutionary origins and trajectories of the virus; therefore, more giant viruses infecting Vermamoeba must be discovered and studied to create a comprehensive knowledge on these intriguing biological entities.
Introduction: The Rise of Amoebal Virology
Much of the history of virology has been devoted to the study of viruses infecting organisms of interest to humankind, including human viruses themselves (Mehle et al., 2018; Loeffelholz and Fenwick, 2019), but also crop and livestock viruses (Chen et al., 2021). However, the fact that emerging diseases may be triggered by viruses infecting wild hosts and the increasing societal awareness of the importance of environmental preservation has fueled the birth of environmental virology (Gollakner and Capua, 2020; Haider et al., 2020). This has broadened our understanding of the viral world, especially of viruses that infect microbes. It is now universally recognized that viruses are the most abundant biological entities on the Earth (Jian et al., 2021) and have contributed substantially to the evolution of life. In this general context, the scientific community began to take an interest in amoeba viruses in the early 2000s. This specific niche was only a marginal part of virology of interest to only a few specialists.
This discipline got off to a flying start with the accidental discovery of Acanthamoeba polyphaga mimivirus (La Scola et al., 2003) the first giant virus (GV) which dismantled some of the most historical dogmas in virology. For the first time, a virus was shown to have a particle size (450 nm) comparable to the size of a small bacterium and was observable using light microscopy, thus abolishing the old dogma that viruses are sub-microscopic in size (Claverie and Abergel, 2016). Following this discovery, over the next decade, a “race” between several research groups took place to isolate and study other giant viruses using Acanthamoeba strains as prey. As a result, Acanthamoeba spp. are now the protist organisms for which the most remarkable diversity of viruses have been characterized (Abergel et al., 2015; Brandes and Linial, 2019).
As of May 2021, 51 GenBank entries corresponded to either partially or fully sequenced genomes of viruses isolated from an Acanthamoeba host. All these viruses have a double-stranded DNA genome. Most Acanthamoeba viruses are recognized or possible members of the Nucleocytoviricota phylum [also known as the nucleocytoplasmic large DNA viruses (NCLDV)]. They are classified in at least seven groups of Nucleocytoviricota (including two official taxonomic families: Mimiviriridae and Marseilleviridae), and five genera are still awaiting official classification: “Pandoravirus” (Philippe et al., 2013), “Pithovirus” (Legendre et al., 2014), “Medusavirus” (Yoshikawa et al., 2019), “Pacmanvirus”(Andreani et al., 2017) Mollivirus (Legendre et al., 2015) and one genus of Lavidaviridae («Sputnikvirus»). Most Nucleocytoviricota members have genome sizes > 350 kbp and particle sizes > 250 nm, with pandoravirus salinus holding the current record for genome size among viruses of almost 2.5 Mb (Philippe et al., 2013) and pithovirus the record for particle size (1,500 nm) (Legendre et al., 2014). Although each giant virus encodes hundreds or thousands of distinct proteins, they share at the most only three universally conserved genes (Colson and Raoult, 2010). A recent phylogeny-centered study proposed that phylum Nucleocytoviricota may be portioned in six orders comprising 32 families embracing all large DNA viruses including giant viruses (Aylward et al., 2021).
At the other end of the scale, virophages (with a particle size of 50–75 nm) (Lavidaviridae family), which parasitize the viral factories of viruses in the Mimiviridae family replicating within Acanthamoeba hosts, have genome sizes of around 17–18 kbp and encode about 20 genes (Desnues et al., 2012; Gaia et al., 2014). In addition, yaravirus, isolated from A. castellanii, represents a divergent lineage of viruses with a 45 kbp genome that potentially lies outside the Nucleocytoviricota phylum (Boratto et al., 2020). The successful use of Acanthamoeba strains as bait for isolating original viruses has led some research groups to diversify the panel of potential prey in their isolation protocols, in order to continue exploring the diversity of protist viruses. Thus, a mimivirus-like giant virus, namely, Platanovirus saccamoebae, has been isolated from a natural strain of Saccamoeba lacustris (Amoebozoa, Tubulinea), itself previously isolated from the surface of a Platanus tree (Hauröder et al., 2018). The Microbe, Evolution, PHylogeny and Infection (MEPHI) group at the Institut Hospitalo-Universitaire (IHU)-Méditerannée Infection has tested alternative protist species, including the cosmopolitan amoeba Vermamoeba vermiformis, and has refined its virus isolation protocols to allow high throughput screening of environmental samples (Khalil et al., 2016). Like Acanthamoeba spp., Vermamoeba vermiformis has proven to be particularly effective in “fishing for viruses,” with 26 viral isolates characterized to date using this approach. This review aims to present the diversity and some of the peculiar characteristics of these viruses.
Vermamoeba Vermiformis as Host Cell Support: A Novel Interest
The preference for using the Acanthamoeba protist in the viral co-culture strategy is historically due to the possibility of growing it under axenic conditions (PYG medium). Moreover, this strategy includes a step of antibiotic addition to eliminate bacterial contamination in the culture. Further, virus isolation attempts were successful after altering the medium substance in the relative Acanthamoeba’ strains. One example was the substitution of gentamicin to amoxicillin in A. castellanii, which established another medium (PPYG/NaC) for growing amoeba (La Scola, 2014). This strategy was found to be highly prolific as Acanthamoeba’s strains successfully isolated many viruses. Other successful viral isolation approaches used their natural hosts, such as Cafeteria roenbergensis virus (Fischer and Hackl, 2016) and Bodo saltans virus (Deeg et al., 2018).
Since 2015, a new strategy for isolating viruses from amoeba co-culture was implemented using Vermamoeba, a protist that belongs to the same order as the genus Acanthamoeba, the Amoebida (Khan, 2006). This strategy included ameliorating the antibiotic and the antifungal composition, as Acanthamoeba strains are more biocides-resistant than Vermamoeba (Fouque et al., 2015b), eliminating the potential bacterial and fungal contaminants, replacing the Page’s amoeba saline solution used for Acanthamoeba with a novel one named “starvation medium adapted to Vermamoeba vermiformis,” and switching from daily microscopic observations to flow cytometry for faster processing and detection of the cytopathic effect in Vermamoeba.
This new approach opened the way for the isolation of new viral strains in an automatic way (Bou Khalil et al., 2016a). At the same time, the observation that Vermamoeba endoparasites can be located outside vacuoles after phagocytosis and that Vermamoeba’s rapid encystment process and its failure to survive in Page’s amoeba saline solution, resulted in Vermamoeba being used as an alternative amoeba host for viral capture (Bou Khalil et al., 2016b). It is worth mentioning that not all amoebas seem to be suitable hosts for the isolation of new viruses. This seems to be the case for the amoeba Willaertia magna (Boudjemaa et al., 2020).
The V. vermiformis was first described in 1967 by F.C. Page as Hartmanella vermiformis (Page, 1967). In 2011, this organism was reclassified and renamed under its current name due to the formation that differentiates it from other Hartmanella spp. (Smirnov et al., 2011). This dissimilarity is the cylindrical shape of Vermamoeba as well as the length/width ratio in comparison with Hartmanella (Park, 2016). The taxonomic change was later confirmed through 18S rRNA gene analysis of different protists (Delafont et al., 2018). V. vermiformis thrives in soil (Maisonneuve et al., 2016) but occurs more frequently in water (Dillon et al., 2014; Buse et al., 2017). In addition, Vermamoeba has been reported in snow, tap water (Javanmard et al., 2017), thermal waters (Reyes-Batlle et al., 2016), cooling towers, hospital, household sewages, industrial composts, and bioaerosols, in mammals, birds (Masangkay et al., 2018), and humans (Cabello-Vílchez et al., 2014; Masangkay et al., 2018). Regarding humans, V. vermiformis was reported to be associated with keratitis (Fouque et al., 2015a), an eye disease (Siddiqui et al., 2021), and indirectly to Legionnaires’ disease (Park, 2016), a disease that infects the human respiratory system (König et al., 2019). Likewise, Vermamoeba served as a dwelling for pathogenic bacteria (develop endosymbiotic attributes; Scheid, 2019) such as Stenotrophomonas maltophilia (Cateau et al., 2014), Candidatus Rubidus massiliensis (Pagnier et al., 2015; Bou Khalil et al., 2016b), Bacillus anthracis, Pseudomonas aeruginosa, Legionella spp., Neochlamydia hartmannellae, Waddlia as well as Chlamydia-like endosymbionts (Masangkay et al., 2018) and Mycobacterium chelonae (Cabello-Vílchez et al., 2014).
The life cycle of V. vermiformis comprises two stages (Baron, 1996) including the trophozoites stage and the cyst stage (Fouque et al., 2015a; Masangkay et al., 2018). The trophozoite consists of an elongated cylindrical motile form, which uses pseudopods to move (Fouque et al., 2015b), and allows them to feed and replicate (Dobrowsky et al., 2016). The V. vermiformis cell is mononuclear with a central dense and homogenous karyoplasm and several mitochondria with tubular cristae spread over cytoplasm (Fouque et al., 2015b). The cyst is spherical in shape, with a two-layer cell wall containing proteins, a small amount of glucose polymers, and an absence of cellulose and ostioles (Delafont et al., 2018). The encystment process lasts 9 h (Fouque et al., 2015a), and involves the locomotive form which is implicated in the cyst clustering (Delafont et al., 2018). Encystment occurs in a hostile environment such as one which is depleted of nutrients and under osmotic pressure (Fouque et al., 2015a). This situation leads to the formation of the cyst wall (between the sixth and the ninth hour of the encystment procedure) with a two-layer composition (Fouque et al., 2015b). This composition is responsible for the spherical form of Vermamoeba vermiformis (Fouque et al., 2015b) and the extreme conditions resilience (nutrient starvation, heat, cold, desiccation, and biocidal treatments) (Dobrowsky et al., 2016). Its form consists of monopodial long slug-like cells, with some granular materials in the cytoplasm with a slight anterior hyaline zone, and are influenced by temperature, pH, and osmotic pressure (Fouque et al., 2014). These cells can change to bi- or multipodial when they change their directions (Delafont et al., 2018). Studies showed that at 4°C, the formation is spherical and immobilized, while at 50°C, the trophozoite is lysed (Fouque et al., 2014). However, the cell lysis is slower than that of Acanthamoeba (Andreani et al., 2018).
To date, many Vermamoeba strains have been characterized from various parts of the world such as Iranian mineral springs (Feiz Haddad et al., 2020), Myanmar (Arab-Mazar et al., 2019), Spain (Canary Islands) (Reyes-Batlle et al., 2016), central and northern Italy, France, South Korea, the United States, Pakistan, England, and China (Montalbano Di Filippo et al., 2019). Only one draft genome sequence has been released, namely, the one from strain CDC-19. The reported genome sequence is 59,550,895 bp in cumulated length and encodes 22,483 genes with 41.7% G+C. Its analyses revealed the number of ORFans (i.e., putative ORFs without match in public sequence databases) estimated at 7,220, plus 2,829 genes with an unknown function. Phylogenomic analysis revealed that ten of them had the best homologous match in the Candidate Phyla Radiation bacterial group (Chelkha et al., 2020). This group includes more than 70 different bacteria phyla (Danczak et al., 2017). In addition, 185 genes had best matches in viruses of the Nucleocytoviricota phylum, with the most extensive relationships with klosneuviruses and Bodo saltans virus, 101 and 69 genes, respectively (Chelkha et al., 2020). Furthermore, 1,680 genes were putatively involved in signal transduction, 1,208 genes in post-translational modification processes, protein turnover, and chaperones, and 622 genes in intracellular trafficking, secretion, and vesicular transport. Finally, 154 genes were identified as related to defense mechanisms. Regarding the possible origin of the genes, 55.9% had best matches in eukaryotes (19.8% of those eukaryotes were amoebas), 10.2% in bacteria (including proteobacteria, which was most represented, followed by bacteroidetes and cyanobacteria); 0.6% in archaea and 0.8% in viruses. Finally, two genes matched with Ralstonia phage phiRSL1, and the last one matched with Synechococcus phage S-SKS1. Another observation about V. vermiformis genome is that this amoeba encodes about 3.5 introns per gene (Chelkha et al., 2020).
Vermamoeba-Infecting Viruses From the Phylum Nucleocytoviricota
A total of 26 viruses infecting Vermamoeba (25 lytic viruses and one non-lytic virus) have been reported from multiple countries around the world, including France, Senegal, Algeria, Brazil, Saudi Arabia, and Lebanon. The most thoroughly studied virus is faustovirus, as 17 viral strains have thus far been described. The remaining viruses consist of a single strain or two strains. Reflecting on future perspectives, as more giant viruses will be subsequently described, the amount of information available will proportionally increase.
Faustoviruses
The development of a high-throughput characterization strategy in 2015 was fueled by the rapid isolation of eight faustovirus strains (Benamar et al., 2016), viruses that are close in phylogeny terms to the African swine fever virus (ASFV) (Reteno et al., 2015). The ASFV is responsible for the homonymous disease (Rolland et al., 2019), resulting in the death of domestic pigs (Galindo et al., 2015; Mazur-Panasiuk et al., 2020; Urbano and Ferreira, 2020). Transmission takes place through blood-sucking arthropods such as ticks (Guinat et al., 2016); although there is currently no evidence that mosquitoes may participate in ASFV transmission (Bonnet et al., 2020).
In the same context, it is believed that faustoviruses may possibly be transmitted by hematophagous arthropods (mainly biting midges), which suck blood from rodents, cattle, and humans. This suggestion is confirmed by FV traces isolated on sera from cattle and humans and various organs removed from rodents such as kidneys, tissue, and brains (Temmam et al., 2015). Notably, as most faustoviruses (FVs) have been isolated from wastewater samples, it could be assumed that their presence is an indirect indicator of fecal contamination (Bou Khalil et al., 2016a; Colson et al., 2017).
The FV-E12 isolate, one of the first to be described, was reported to have a 466,265 bp genome with a G + C% content of 36% and 451 predicted genes. In total, 164 of them (1/3 of all predicted proteins) were detected by nano-2D-LC-MS/MS to be present in its virion, and most of them were shared with proteins encoded by other viruses in the Nucleocytoviricota (Reteno et al., 2015). In contrast, ASFV members typically encode about 150 (Dixon et al., 2013; Jia et al., 2017; Shen et al., 2021) to 167 genes (Wang et al., 2021), which is only one-third of FVs. Furthermore, even though that FVs and ASFV are phylogenetically related, only 12% of FV genes appear to be homologous with ASFV (Aherfi et al., 2016). At the same time, FVs’ genome are larger than ASFV’s one (Koonin et al., 2020).
The sample containing FV-E12 was collected from a sewage environment in Marseille, France. After E12 stain, using the same capture technique in V. vermiformis, additional 16 FV strains have been sequenced and annotated so far. In general, the FV genomes are reported to be between 455,803 and 491,024 in length, encoding up to 506 genes (Geballa-Koukoulas et al., 2020). None of the FVs was found to contain tRNA genes. Finally, like most viruses in the Nucleocytoviricota, FVs contain a high number of ORFans.
An important structural characteristic concerns its virion. Indeed, the FV capsid has a double protein shell. The outer shell is made up of a double jelly roll protein, but the inner shell has a structure that is distinct from other “classic” capsid proteins. Such a structure is described as unique among DNA viruses (Klose et al., 2016). The Major Capsid Protein (MCP) gene spans over a 17,000 bp region and contains a surprisingly high number of type-I introns (Colson et al., 2017).
The capsid is about 2,400 Å (mature particles) in diameter and is fibril-free with an icosahedral shape (Klose et al., 2016). In addition, the reconstruction based on cryo-electron microscopy (Cryo-EM) observations revealed a double layer membrane between the external and the internal protein shells, allowing the MCP shells’ to touch the internal shell at several points. This formation consists of a double protein shell that seems to be in contrast with all other dsDNA viruses (Cherif Louazani et al., 2018), as some genomic materials are located between the internal and the external protein shells. Furthermore, it is believed that this agility found in the FVs structure is possibly created by either the presence of small lipids (mass spectrometry detection) or by an extra membrane that is not detectable by Cryo-EM and which encloses and protects the viral genome (Klose et al., 2016).
Using the transcriptomic sequencing results of the FV-E12 along with a splice-aware mapper and a double-round alignment strategy, 26 splice junctions were identified. These splice junctions are nested in the region that encodes the MCP gene (Cherif Louazani et al., 2018). In addition, this region is in the central part of the genome. Several group I introns that formed in this region were also recorded. In our previous study, introns were confirmed in the MCP gene of all FV isolates (varying in number between 13 and 18 per isolate), while some of them encoded several intron-homing endonucleases. Based on this observation, we hypothesized that there might be a link between the intron-generation mechanism (the endonucleases serve as a mediator before FV radiation in V. vermiformis) and MCP size. Genomic analysis revealed that all FV genomes contain high levels of gene colinearity, an amassing of indels, and local rearrangements in the MCP gene (close to the central region of the genome) along with their extremities which exhibit a faster evolutionary rate in that genomic region. Currently, known isolates belong to three different lineages among faustoviruses. The E9 and M/L clades consist of six isolates, and the D includes four isolates (Geballa-Koukoulas et al., 2020).
The V. vermiformis infection begins about 30 min after the first contact with the FV, while the Vermamoeba’s phagosomes can detect FV’s particles between 2 and 4 h after the infection. The eclipse phase is completed between the fourth and sixth hours after the beginning of infection, while the cell’s burst is ready to begin from 16 to 18 h post-infection (h.p.i.). At 20 h after the initial infection, the Vermamoeba’s cell comes to its lysis (Reteno et al., 2015). On the other hand, FV-mariensis, a strain isolated from Pampulha Lagoon Brazil (Borges et al., 2019), seems to postpone the cell lysis by 4 h. This occurs possibly due to soluble factor, that was not able to impede the V. vermiformis’ encystment, post-infection, in the trophozoites stage, which remains conserved to cysts, and viral replication has not occurred. The same soluble factors were also described in A. castellanii, and consist of Mg2+ ions. In FV-mariensis, this soluble factor is enhanced by ethylenediaminetetraacetic acid (EDTA). It was also found that some intracellular bacteria can withstand and benefit from amoebal encystment suggesting FV-mariensis as a potential candidate antiviral model (Borges et al., 2019). The FVs genomes were initially hypothesized to have a circular conformation (Benamar et al., 2016). However, subsequent PCR assays and careful examination of the genome assemblies suggested instead that the typical FV genome is linear, like virtually in all other members of the Nucleocytoviricota (Geballa-Koukoulas et al., 2020).
Kaumoebaviruses
Kaumoebaviruses (KVs) is the second virus that has been isolated from Vermamoeba. The word “Kaumoeba” stands for King Abdulaziz University aMOEBA because the first member was collected from sewage waters in Jeddah, Saudi Arabia (Bajrai et al., 2016). This genus is currently not recognized by the ICTV (Koonin and Yutin, 2019), but phylogenetic reconstruction clusters it amidst ASFVs and FVs (Cherif Louazani et al., 2018; Rolland et al., 2019), suggesting that it is a genuine member of the Asfarviridae family (Geballa-Koukoulas et al., 2021a; Karki et al., 2021). This phylogenetic position is reflected in common features between those viruses (Rolland et al., 2019). More specifically, FVs and KV share the same morphology: an icosahedral (Colson et al., 2017) capsid of approximately 200 nm in diameter (Bajrai et al., 2016; Borges et al., 2019), the replication cycle lengths, and a spliced major capsid protein gene (Rolland et al., 2019), which encode a protein of about 500 AA in size (Geballa-Koukoulas et al., 2021b). Another significant aspect found is that the upstream regions of the core genes of ASFV, FVs, and KVs have TATATA and TATTA motifs (Oliveira et al., 2018), which resemble Asfarviridae promoter (Rolland et al., 2019) and support the hypothesis for a similar gene expression mechanism (Oliveira et al., 2019a; Rolland et al., 2019). In 2021, a second member of the KV group, strain “LCC10,” was isolated from La Ciotat in France. Their comparison revealed that the LCC10 genome is 11,030 bp larger and encodes 45 genes more than the original strain named “Sc.” Similar to other viruses in the Nucleocytoviricota, both strains of KVs encode numerous genes that have no detectable matches in any database. For KV-LCC10, these account for approximately 76% of their total genes, while for KV-Sc strain, ORFans are calculated to 67%. Finally, the number of unique genes is 98 for KV-LCC10 and 85 genes for KV-Sc. The GC content in both isolates is about 43%. Neither KVs encode any tRNAs (Geballa-Koukoulas et al., 2021b), a feature that is also found in all FVs (Geballa-Koukoulas et al., 2020). The MCP gene, like that of FV, is spliced by several type I introns, with some of them encoding homing endonucleases, which strengthen the hypothesis of a common ancestor between faustoviruses and kaumoebaviruses. KVs have two introns that encode homing endonucleases, and their replication cycle is complete after 20 h (Bajrai et al., 2016).
The KV genomes exhibited a significant gene strand bias over the two-thirds of genome length, over-represented in the positive strand, a unique feature among all members of the extended Asfarviridae. The mechanism supporting this phenomenon remains unknown, but there is no apparent link with the host cell (Geballa-Koukoulas et al., 2021b).
Orpheovirus
Orpheovirus (OV) was isolated from a rat stool sample collected in La Ciotat, France, in the same location where FV-LC9 was discovered (Andreani et al., 2018). The virus has an ovoid-shaped particle ranging between 900 and 1,100 nm in length and measuring 500 nm in diameter (Souza et al., 2019). In addition, it comprises an ostiole-like apex to the ovoid shape with a diameter ranging from 70 to 80 nm obstructed in a thick membrane, involving in the DNA delivery to the host cell (Andreani et al., 2018). The particle structure shared some morphological properties with cedratviruses, pithoviruses, and pandoraviruses (Souza et al., 2019). In addition, both phylogenetic and pan-genomic analyses reveal that cedratvirus and pithoviruses have a close relationship with OV. This double-stranded DNA virus genome size of 1.4 Mb is immense compared with FVs and KVs. That said, the GC% content is lower (about 25%), encodes 1,512 genes, of which more than half are annotated as ORFans (Andreani et al., 2018). The OV genome is estimated to be circular (Andreani et al., 2018).
Furthermore, a common feature of OV with KVs and FVs is the apparent lack of tRNA-coding genes (Souza et al., 2019). However, its MCP gene is different from KVs and FVs as it does not contain introns (Andreani et al., 2018). The replication cycle of OV in V. vermiformis is completed within 24–38 h, which seems to be longer compared to FV and KV. Remarkably, the virus enters to Vermamoeba cell by the process of phagocytosis. The eclipsed phase is observed during the fourth hour after the infection. The viral factories are detected at 14–16 h.p.i. The new virions fill the V. vermiformis cytoplasm after 20 h.p.i., and the cell bursts between 24 and 38 h.p.i, marking the end of the replication cycle (Souza et al., 2019).
Tupanviruses
Tupanviruses (TVs) clade consists of two isolated members from the Brazilian soda lake and soil sediments (Silva et al., 2019). Indicatively, tupanvirus soda lake (TV-SL) was isolated in 2014 from a soda lake in the Pantanal region with extreme high salinity and pH, and tupanvirus deep ocean (TV-DO) was isolated in 2016 from Campos dos Goytacazes sediment sample collected 3,000 m deep under the ocean (Rodrigues et al., 2019). Tupan stands for “God of Thunder,” an important mythological figure for indigenous south American tribes (Abrahão et al., 2018). Both viruses possess a linear genome and likely belong in the Mimiviridae family (Rodrigues et al., 2019). These viruses can both replicate in Acanthamoeba and Vermamoeba (Silva et al., 2019), and in other amoebas such as Dictyostelium discoideum and Willaertia magna (Rodrigues et al., 2019).
Tupanvirus strains were morphologically characterized and revealed an unusual structure (Silva et al., 2019), marked by the presence of a semi-icosahedral mimivirus-like capsid attached to an elongated cylindrical tail (de Miranda Boratto et al., 2019) of 550 nm in length (Rodrigues et al., 2019). The cylindrical tail may vary in size, so the overall length of some particles can exceed 2 μm. A mimivirus-like capsid has the stargate portal on one side, surrounded by fibrils (Silva et al., 2019). Even if the MCP structure is similar to the Mimiviridae members, their genome structure, ranking them as one of the largest (Oliveira et al., 2019a), distinguishes TVs from other mimivirids (Silva et al., 2019). With regards to their general features, TV genomes are much bigger in size compared with the rest of Vermamoeba viral isolates. Specifically, TV-SL genome is 1,439,508 bp, while TV-DO is 1,516,267 bp in size, and they encode 1,276 and 1,359 genes, respectively. In addition, TVs encode many tRNAs, with TV-SL encoding 67 and TV-DO three more. Tupanviruses encode a set of 20 aminoacyl-tRNA synthetases (aaRS) specific genes too. Their genomes also contain 11 translation-associated factors that are linked with tRNA/mRNA maturation, a ribosome modification protein, and a mannose-binding protein (Oliveira et al., 2019b). Curiously, TV-SL lacks the RNAse T2 gene, which plays an essential role in the RNA catabolism inside the cell (Rolland et al., 2020). Like many giant viruses, 26% of TV-DO genes (375 out of 1,276) and 28% of TV-SL ones (378 out of 1,359 genes) are annotated as ORFans (Abrahão et al., 2018). Until 2019 and the discovery of yasminevirus, TVs were the largest viruses infecting the Vermamoeba group (see section “Yasminevirus”).
A comparative genomic analysis between the TVs shows that, with exception of the terminal regions, in which the TV-SL shows local rearrangements and translocation compared to the TV-DO genome, the rest of their genomes are extremely conserved (Rodrigues et al., 2019). The cell replication starts almost instantly after the “adhesion” of the virus to the cell. This adhesion is believed to occur due to the mannose receptor gene expression. Also, it was reported that the expression of a mannose receptor gene participates in the pathogenesis of Acanthamoeba (Oliveira et al., 2019b). Tupanvirus replication time varies and depends on the eclipse phase time. For instance, for a 4-h post-infection eclipse phase, the replication cycle ends after 36 h.p.i., explaining why the final stage varies between 36 and 72 h (Silva et al., 2019).
Clandestinovirus
Clandestinovirus (CLV), the first non-lytic isolate on V. vermiformis, was recently described (Rolland et al., 2021). This virus was separated by a single cell microaspiration strategy (Sahmi-Bounsiar et al., 2019) from a mixed co-culture associated with another giant virus (clandestinovirus ST1 and FV-ST1) collected from an environmental sample. Clandestinovirus has an icosahedral capsid devoid of fibrils and has a virion size between 175 and 202 nm in diameter. It possesses a linear genome of 581,987 bp (43.5% G + C content) assembled in two scaffolds which encodes 617 open reading frames, corresponding to a coding density of 86% (Rolland et al., 2021). In addition, it has one serine- and one histidine-like tRNAs. The clandestinovirus histidine-like (His-like) tRNA is also found in medusavirus. Indeed, according to the results of several analyses, these two viruses are phylogenetically related (Zhang et al., 2021).
The high-throughput automated technique detected the clandestinovirus in the V. vermifomis cytoplasm in which the fluorescence signal augmented after 24 h post-infection and remained up to 5 days confirming the non-lytic nature of viral infection (Francis et al., 2019). Specifically, after the viral entry into the Vermamoeba cell, the viral particles transposition from the cytoplasm to the nucleus to begin replication. This step was also observed during the replication of Herpersviridae (Jean Beltran and Cristea, 2014). Viral factories are observed 7–12 h.p.i, and at 16 h.p.i. The newly formed virions are discharged by exocytosis, which completes the replication cycle (Rolland et al., 2021).
Klosneuvirus-Like Viruses
The “klosneuvirus clade” consists of two viral isolates that infect V. vermiformis. yasminevirus and the fadolivirus, which is the newest member described. The main feature of these genomes is their large size, indeed the biggest among all the known Vermamoeba-infecting viruses.
Yasminevirus
One of the most recent GV isolates from V. vermiformis is the yasminevirus (YV). Like KV, YV was isolated from sewage water in Jeddah, Saudi Arabia (Bajrai et al., 2016). The structure and morphology of the mature YV particles are similar to the Bodo saltans virus, and its virion core is enclosed by putative thin membranes, which are surrounded by an icosahedral capsid of 330 nm in diameter. This icosahedral capsid structure is also found in most members of the phylum Nucleocytoviricota infecting Vermamoeba, such as KVs and FVs.
The capsid is surrounded by a layer of thin fibrils that mediate the start of the infection. Its mosaic genome structure is the biggest among all Vermamoeba-infecting viruses from the phylum Nucleocytoviricota, extending to 2,126,343 bp. The number of genes is predicted to be 1,541, which is the highest among Vermamoeba-infecting viruses belonging to Nucleocytoviricota. Comparative genomic analyses demonstrated that 34% of the genes have the best matches with those encoded by other viruses, 13.2% with those in Eukaryota, 7.2% in bacteria, and less than 1% in Archaea. The ORFans comprise about 44% of the total number of genes. Despite the YV genome being bigger than the TVs’, it encodes similar number of tRNAs (70 tRNAs in total) and contains 20 aminoacyl-tRNA synthetases (aaRSs) along with several translation and elongation factors. The GC content of YV was calculated to be 40.2%.
The replication cycle begins 30 min after infection initiation, particles can be observed in the host cytoplasm and, after a further 30–90 min, the cytoplasm is prepared for the eclipse phase during which the virus particles have vanished. This stage was further distinguished by the modification of the host cell nucleus and the development of intranuclear cleared regions. After 12 h.p.i., viral factories can be detected in the host cell, and the final cell lysis is completed after 24 h.p.i. (Bajrai et al., 2019).
Fadolivirus
Fadolivirus (FL) is the latest Vermamoeba-infecting isolate, described in 2020 from sewage water in Sidi bel Abbes in Algeria (Boudjemaa et al., 2020). This is a lytic virus with an icosahedral structure. The viral particles measured about 300 nm from which short fibrils extended on its capsid. Phylogenetically, this virus is clustered in a clade along with klosneuvirus, barrevirus, and indivirus.
Alike clandestinovirus, this virus has a linear genome and was assembled in two scaffolds totaling 1,595 Mbp (Andreani et al., 2021). It encodes 1,452 genes of which 38% have a detectable homology with other amoebal viruses, as well as 66 tRNAs and 23 aaRSs, a number that correlates with the yasminevirus that encodes 20. Moreover, 20 genes are shared with several related viruses of the “klosneuvirus clade” (core genes). The replication cycle of FL in V. vermiformis starts with phagocytosis. Up to 12 h.p.i, the eclipse phase is fully recognized. In total, 4–8 h after the eclipse phase, the viral particles started in the host cytoplasm, resulting in its final lysis after 36 h.p.i. (Andreani et al., 2021).
Comparison of the Vermamoeba-Infecting Viruses From the Phylum Nucleocytoviricota
As many giant viruses have been described, different replication stages are grouped into three phases for a precise description and comparison. The first phase comprises the early stage of infection after phagocytosis. The second one includes the eclipse phase and the stage when the viral factories become distinguished in the host cell, and finally, the last phase comprises the advanced stage and the final stage, in which an entire replication cycle is completed.
Sorting genomes in order of genomic size, the largest one is YV with 2,126,343 bp, followed by FL, TV-DO, OV, TV-SL, and FVs with the shortest genomes in KVs. In the same line, YV seems to encode more genes while KVs encode the least number. Regarding the GC content, orpheovirus has the smallest percentage, and the KVs have the highest percentage. About the structure of the viral isolates, all viruses exhibit an icosahedral capsid except orpheovirus with an ovoid shape and tupanviruses with a more elaborate shape comprising a ∼550 nm long cylindrical tail covered with fibrils. Regarding the infection time in V. vermiformis, all viruses start to visibly affect Vermamoeba at around 4 h, except FL which starts after 12 h; however, FL is a non-lytic virus and thus has a different infection behavior. The complete replication cycle appears to be the slowest in TVs, while in KVs and FVs, it replicates the most quickly among all the viruses.
For the tRNAs, CLV, YV, TVs, and the non-lytic virus of FL are the only viruses that contain tRNAs. One important observation is that these viruses could have common ancestry, as they are significantly related on the phylogenetic level. Of course, additional elements are required to validate this hypothesis. The origin of tRNAs is highly connected with the replication time (Maizels and Weiner, 1994). Furthermore, it is reported that the tRNA gene is associated with the DNA replication time (Müller and Nieduszynski, 2017) as they impede the replication process at the S phase (Molla-Herman et al., 2015). Unfortunately, based on the current bibliography, the role of viral tRNAs in cell life cannot be determined.
Considering the aforementioned information and the phylogenetic relationship (Geballa-Koukoulas et al., 2021a; Figure 1) between those viruses, we propose that viruses that infect V. vermiformis could be categorized into three groups. FVs and KV could comprise the first, OV the second, and the remaining viruses could form the latest group. Unfortunately, to make our estimations more accurate, new data or, even better, new isolates need to be investigated. Finally, all Nucleocytoviricota infecting Vermamoeba isolates contain a considerable number of ORFans proteins. Table 1 summarizes the main features for Vermamoeba-infecting giant viruses (such as the genome length size, the G + C content, the number of predicted genes, the number of tRNAs along with the three phases of the replication cycle described earlier).
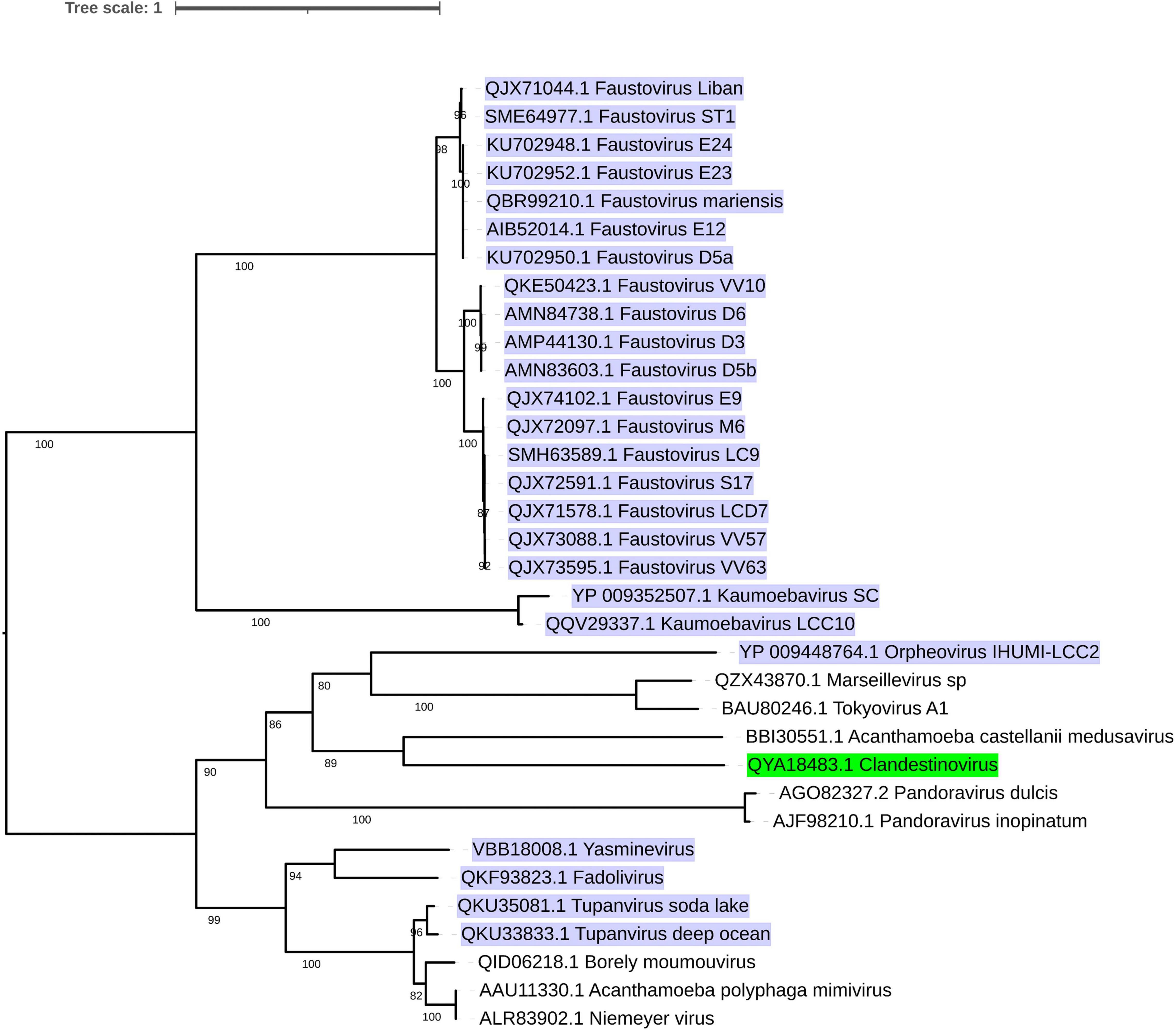
Figure 1. Phylogenetic tree of currently recognized and additional putative members of the phylum Nucleocytoviricota infecting Vermamoeba sp. based on DNA polymerase protein sequences. Amino acid sequences were aligned with MAFFT (Katoh and Standley, 2013) (with AUTO as parameter). The most conserved regions of the aligned sequences were selected by TrimAl (Capella-Gutierrez et al., 2009; v1.4.rev22 build (2015-05-21) with strictplus as a parameter]. IQ-TREE software (Nguyen et al., 2015; version 1.6.12 using -bb 1,000 and -m TEST as options) was applied for the tree reconstruction. Finally, ITOL (Letunic and Bork, 2021; version 10.0.5) was used for the visualization of the tree. Lytic viruses are highlighted in purple, while the non-lytic virus is marked in green.
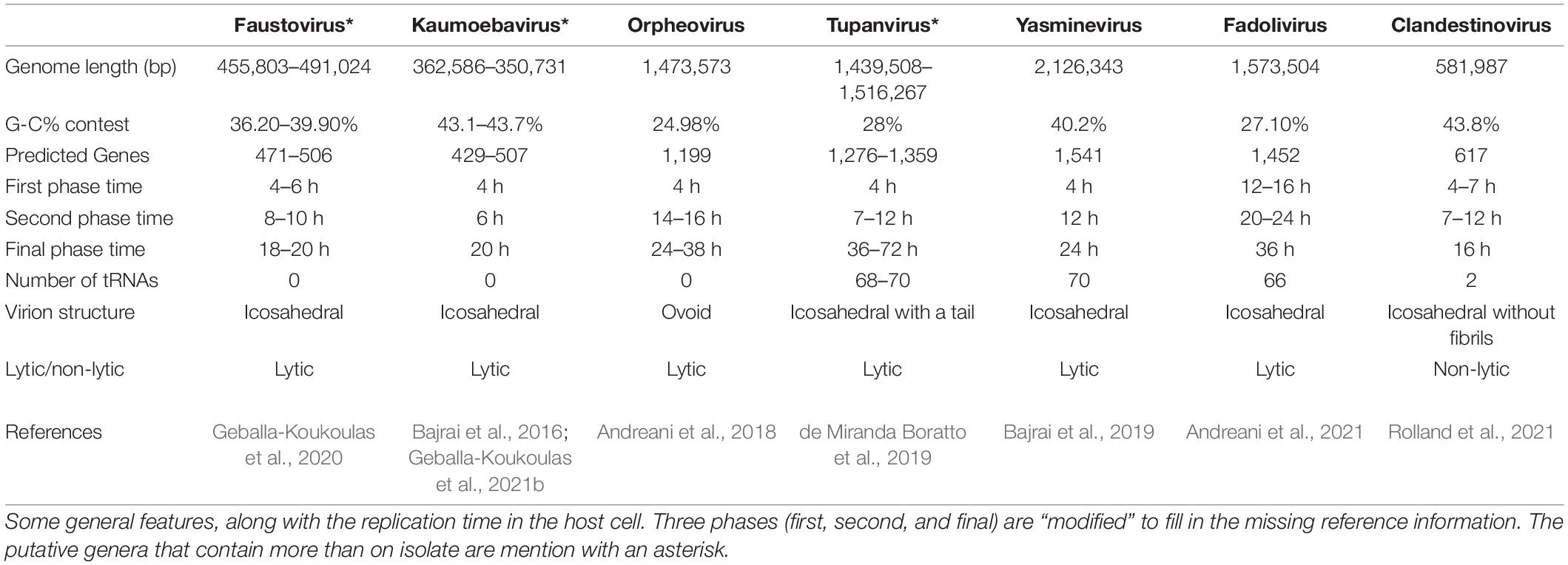
Table 1. General features of all Vermamoeba-infecting viruses, recognized and/or potential members of the phylum Nucleocytoviricota.
Conclusion
Giant viruses (GVs) are a relatively new and up-and-coming niche of virology linked with the entire world of eukaryotes. Scientists, using the free-living amoebas succeeded in isolating GVs. This technique was initially adopted in 2003 for the isolation of a first GV from Acanthamoeba polyphaga. By applying the same process, many new isolates have been characterized ever since. Acanthamoeba spp. were mainly used to capture GVs until the faustovirus (E12 strain) isolation in 2015, which pioneered V. vermiformis as a model organism, which blazed a trail for new GVs discoveries.
Currently, a total of 26 giant virus isolates are known to infect V. vermiformis. Here, for the first time, we present a systematic comparison of this group of viruses. A total of nineteen viral isolates likely belong to the extended Asfarviridae family (17 FVs and 2 KVs), two belong to “klosneuvirus clade” (YV and FL), two are comprised in the Mimiviridae family (2 TVs), and other isolates to be yet officially classified of OV and the non-lytic virus of CL. Regarding the replication cycle of those viruses in Vermamoeba, FVs and KVs replicate at a faster rate compared to the rest of Vermamoeba-infecting viruses from the phylum Nucleocytoviricota.
In summary, the 26 Vermamoeba-infecting viruses belonging to the phylum Nucleocytoviricota are here reviewed and informally classified into three groups based on their size, replication time, and the number of tRNAs. The first group consists of KV and FVs. The second is represented by OV, and the last group contains TVs and YV. We understand that this grouping may not stand the test of time as it is based upon limited data available at present time. We intend to provide an initial point for further discussions as the number of characterized isolates of GVs increases over time.
The world of viruses from the phylum Nucleocytoviricota infecting Vermamoeba remains shrouded in mystery, and the comparison with similar viruses from other aquatic microorganisms such as algae is a challenge. The discovery of endogenous giant virus-derived sequences from several green algae has also been reported recently (Moniruzzaman et al., 2020), which opens new possibilities in studying and understanding these viruses. However, using Vermamoeba spp., seven viral isolates have been described, and using Acanthamoeba spp., the number of isolates was much higher. Moreover, the amoeba co-cultivation technique allows isolation and studying different viruses in mixed infections and their separation as described by the single-cell microaspiration strategy (clandestinovirus ST1 and FV-ST1). To conclude, the amoeba-isolation technique is a relatively simple, reliable, and efficient method for discovering novel giant viruses. The use of new protists as potential host models could also facilitate novel discoveries.
Author Contributions
KG-K designed the figure and table, wrote the original draft, and edited the final version. BL, GB, and JA supervised this work and reviewed the manuscript. All authors read and approved this manuscript.
Funding
This work was supported by the French Government under the Investissements d’Avenir (Investments for the Future) program, managed by the Agence Nationale de la Recherche (National Agency for Research) (reference number Méditerranée Infection 10-IAHU-03).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Ariadni Geballa Koukoula for creating the cartoon drawings in Table 1.
References
Abergel, C., Legendre, M., and Claverie, J.-M. (2015). The rapidly expanding universe of giant viruses: Mimivirus, Pandoravirus, Pithovirus and Mollivirus. FEMS Microbiol. Rev. 39, 779–796. doi: 10.1093/femsre/fuv037
Abrahão, J., Silva, L., Silva, L. S., Khalil, J. Y. B., Rodrigues, R., Arantes, T., et al. (2018). Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat. Commun. 9:749. doi: 10.1038/s41467-018-03168-3161
Aherfi, S., Colson, P., La Scola, B., and Raoult, D. (2016). Giant viruses of amoebas: an update. Front. Microbiol. 7:349. doi: 10.3389/fmicb.2016.00349
Andreani, J., Khalil, J. Y. B., Baptiste, E., Hasni, I., Michelle, C., Raoult, D., et al. (2018). Orpheovirus IHUMI-LCC2: a new virus among the giant viruses. Front. Microbiol. 8:2643. doi: 10.3389/fmicb.2017.02643
Andreani, J., Khalil, J. Y. B., Sevvana, M., Benamar, S., Di Pinto, F., Bitam, I., et al. (2017). Pacmanvirus, a new giant Icosahedral virus at the crossroads between asfarviridae and faustoviruses. J. Virol. 91:e00212-17. doi: 10.1128/JVI.00212-217
Andreani, J., Schulz, F., Di Pinto, F., Levasseur, A., Woyke, T., and La Scola, B. (2021). Morphological and genomic features of the new klosneuvirinae isolate Fadolivirus IHUMI-VV54. Front. Microbiol. 12:719703. doi: 10.3389/fmicb.2021.719703
Arab-Mazar, Z., Niyyati, M., Lasjerdi, Z., Spotin, A., Alavi Darzam, I., and Gachkar, L. (2019). Isolation, identification, and phylogenetic analysis of potentially pathogenic free-living amoebae isolated from nasal and oral mucosa of HIV/AIDS patients in Iran. Parasitol. Res. 118, 3061–3066. doi: 10.1007/s00436-019-06448-x
Aylward, F. O., Moniruzzaman, M., Ha, A. D., and Koonin, E. V. (2021). A phylogenomic framework for charting the diversity and evolution of giant viruses. PLoS Biol. 19:e3001430. doi: 10.1371/journal.pbio.3001430
Bajrai, L. H., Mougari, S., Andreani, J., Baptiste, E., Delerce, J., Raoult, D., et al. (2019). Isolation of yasminevirus, the first member of klosneuvirinae isolated in coculture with Vermamoeba vermiformis, demonstrates an extended arsenal of translational apparatus components. J. Virol. 94:e01534-19. doi: 10.1128/JVI.01534-19
Bajrai, L., Benamar, S., Azhar, E., Robert, C., Levasseur, A., Raoult, D., et al. (2016). Kaumoebavirus, a new virus that clusters with faustoviruses and asfarviridae. Viruses 8:278. doi: 10.3390/v8110278
Baron, S. (ed.) (1996). Medical Microbiology, 4th Edn. Galveston, Tex: University of Texas Medical Branch at Galveston.
Benamar, S., Reteno, D. G. I., Bandaly, V., Labas, N., Raoult, D., and La Scola, B. (2016). Faustoviruses: comparative genomics of new megavirales family members. Front. Microbiol. 7:3. doi: 10.3389/fmicb.2016.00003
Bonnet, S. I., Bouhsira, E., De Regge, N., Fite, J., Etoré, F., Garigliany, M.-M., et al. (2020). Putative role of arthropod vectors in african swine fever virus transmission in relation to their bio-ecological properties. Viruses 12:778. doi: 10.3390/v12070778
Boratto, P. V. M., Oliveira, G. P., Machado, T. B., Andrade, A. C. S. P., Baudoin, J.-P., Klose, T., et al. (2020). Yaravirus: a novel 80-nm virus infecting Acanthamoeba castellanii. Proc. Natl. Acad. Sci. U S A. 117, 16579–16586. doi: 10.1073/pnas.2001637117
Borges, I., Rodrigues, R. A. L., Dornas, F. P., Almeida, G., Aquino, I., Bonjardim, C. A., et al. (2019). Trapping the enemy: Vermamoeba vermiformis circumvents faustovirus mariensis dissemination by enclosing viral progeny inside cysts. J. Virol. 93:e00312-19. doi: 10.1128/JVI.00312-19
Bou Khalil, J. Y., Andreani, J., Raoult, D., and La Scola, B. (2016a). A rapid strategy for the isolation of new faustoviruses from environmental samples using Vermamoeba vermiformis. J. Vis. Exp. e54104. doi: 10.3791/54104
Bou Khalil, J. Y., Benamar, S., Baudoin, J.-P., Croce, O., Blanc-Tailleur, C., Pagnier, I., et al. (2016b). Developmental cycle and genome analysis of “rubidus massiliensis,” a new Vermamoeba vermiformis pathogen. Front. Cell. Infection Microbiol. 6:31. doi: 10.3389/fcimb.2016.00031
Boudjemaa, H., Andreani, J., Bitam, I., and La Scola, B. (2020). Diversity of amoeba-associated giant viruses isolated in algeria. Diversity 12:215. doi: 10.3390/d12060215
Brandes, N., and Linial, M. (2019). Giant viruses—big surprises. Viruses 11:404. doi: 10.3390/v11050404
Buse, H. Y., Ji, P., Gomez-Alvarez, V., Pruden, A., Edwards, M. A., and Ashbolt, N. J. (2017). Effect of temperature and colonization of Legionella pneumophila and Vermamoeba vermiformis on bacterial community composition of copper drinking water biofilms. Microbial Biotechnol. 10, 773–788. doi: 10.1111/1751-7915.12457
Cabello-Vílchez, A. M., Mena, R., Zuñiga, J., Cermeño, P., Martín-Navarro, C. M., González, A. C., et al. (2014). Endosymbiotic Mycobacterium chelonae in a Vermamoeba vermiformis strain isolated from the nasal mucosa of an HIV patient in Lima, Peru. Exp. Parasitol. 145, S127–S130. doi: 10.1016/j.exppara.2014.02.014
Capella-Gutierrez, S., Silla-Martinez, J. M., and Gabaldon, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Cateau, E., Maisonneuve, E., Peguilhan, S., Quellard, N., Hechard, Y., and Rodier, M.-H. (2014). Stenotrophomonas maltophilia and Vermamoeba vermiformis relationships: bacterial multiplication and protection in amoebal-derived structures. Res. Microbiol. 165, 847–851. doi: 10.1016/j.resmic.2014.10.004
Chelkha, N., Hasni, I., Louazani, A. C., Levasseur, A., La Scola, B., and Colson, P. (2020). Vermamoeba vermiformis CDC-19 draft genome sequence reveals considerable gene trafficking including with candidate phyla radiation and giant viruses. Sci. Rep. 10:5928. doi: 10.1038/s41598-020-62836-62839
Chen, S., Zhang, L., Wang, L., Ouyang, H., and Ren, L. (2021). Viruses from poultry and livestock pose continuous threats to human beings. Proc. Natl. Acad. Sci. U S A. 118:e2022344118. doi: 10.1073/pnas.2022344118
Cherif Louazani, A., Baptiste, E., Levasseur, A., Colson, P., and La Scola, B. (2018). Faustovirus E12 transcriptome analysis reveals complex splicing in capsid gene. Front. Microbiol. 9:2534. doi: 10.3389/fmicb.2018.02534
Claverie, J.-M., and Abergel, C. (2016). Giant viruses: the difficult breaking of multiple epistemological barriers. Stud. History Philosophy Sci. 59, 89–99. doi: 10.1016/j.shpsc.2016.02.015
Colson, P., and Raoult, D. (2010). Gene repertoire of amoeba-associated giant viruses. Intervirology 53, 330–343. doi: 10.1159/000312918
Colson, P., La Scola, B., and Raoult, D. (2017). Giant viruses of amoebae: a journey through innovative research and paradigm changes. Ann. Rev. Virol. 4, 61–85.
Danczak, R. E., Johnston, M. D., Kenah, C., Slattery, M., Wrighton, K. C., and Wilkins, M. J. (2017). Members of the candidate phyla radiation are functionally differentiated by carbon- and nitrogen-cycling capabilities. Microbiome 5:112. doi: 10.1186/s40168-017-0331-331
de Miranda Boratto, P. V., dos Santos, Pereira Andrade, A. C., Araújo Lima, Rodrigues, R., La Scola, B., et al. (2019). The multiple origins of proteins present in tupanvirus particles. Curr. Opin. Virol. 36, 25–31. doi: 10.1016/j.coviro.2019.02.007
Deeg, C. M., Chow, C.-E. T., and Suttle, C. A. (2018). The kinetoplastid-infecting Bodo saltans virus (BsV), a window into the most abundant giant viruses in the sea. eLife 7:e33014. doi: 10.7554/eLife.33014
Delafont, V., Rodier, M.-H., Maisonneuve, E., and Cateau, E. (2018). Vermamoeba vermiformis: a free-living amoeba of interest. Microb. Ecol. 76, 991–1001. doi: 10.1007/s00248-018-1199-1198
Desnues, C., Boyer, M., and Raoult, D. (2012). Sputnik, a virophage infecting the viral domain of life. Adv. Virus Res. 82, 63–89. doi: 10.1016/B978-0-12-394621-8.00013-3
Dillon, A., Achilles-Day, U. E. H., Singhrao, S. K., Pearce, M., Morton, L. H. G., and Crean, S. (2014). Biocide sensitivity of Vermamoeba vermiformis isolated from dental-unit-waterline systems. Int. Biodeterioration Biodegradation 88, 97–105. doi: 10.1016/j.ibiod.2013.10.026
Dixon, L. K., Chapman, D. A. G., Netherton, C. L., and Upton, C. (2013). African swine fever virus replication and genomics. Virus Res. 173, 3–14. doi: 10.1016/j.virusres.2012.10.020
Dobrowsky, P. H., Khan, S., Cloete, T. E., and Khan, W. (2016). Molecular detection of Acanthamoeba spp., Naegleria fowleri and Vermamoeba (Hartmannella) vermiformis as vectors for Legionella spp. in untreated and solar pasteurized harvested rainwater. Parasites Vectors 9:539. doi: 10.1186/s13071-016-1829-1822
Feiz Haddad, M. H., Habibpour, H., and Mahmoudi, M. R. (2020). Isolation and molecular identification of free-living amoebae (Naegleria spp., Acanthamoeba spp. and Vermamoeba spp.) from mineral springs in Guilan Province, northern Iran. J. Water Health 18, 60–66. doi: 10.2166/wh.2020.191
Fischer, M. G., and Hackl, T. (2016). Host genome integration and giant virus-induced reactivation of the virophage mavirus. Nature 540, 288–291. doi: 10.1038/nature20593
Fouque, E., Trouilhé, M.-C., Thomas, V., Humeau, P., and Héchard, Y. (2014). Encystment of Vermamoeba (Hartmannella) vermiformis: effects of environmental conditions and cell concentration. Exp. Parasitol. 145, S62–S68. doi: 10.1016/j.exppara.2014.03.029
Fouque, E., Héchard, Y., Hartemann, P., Humeau, P., and Trouilhé, M.-C. (2015a). Sensitivity of Vermamoeba (Hartmannella) vermiformis cysts to conventional disinfectants and protease. J. Water Health 13, 302–310. doi: 10.2166/wh.2014.154
Fouque, E., Yefimova, M., Trouilhé, M.-C., Quellard, N., Fernandez, B., Rodier, M.-H., et al. (2015b). Morphological study of the encystment and excystment of Vermamoeba vermiformis revealed original traits. J. Eukaryotic Microbiol. 62, 327–337. doi: 10.1111/jeu.12185
Francis, R., Ominami, Y., Bou Khalil, J. Y., and La Scola, B. (2019). High-throughput isolation of giant viruses using high-content screening. Commun. Biol. 2:216. doi: 10.1038/s42003-019-0475-476
Gaia, M., Benamar, S., Boughalmi, M., Pagnier, I., Croce, O., Colson, P., et al. (2014). Zamilon, a novel virophage with mimiviridae host specificity. PLoS One 9:e94923. doi: 10.1371/journal.pone.0094923
Galindo, I., Cuesta-Geijo, M. A., Hlavova, K., Muñoz-Moreno, R., Barrado-Gil, L., Dominguez, J., et al. (2015). African swine fever virus infects macrophages, the natural host cells, via clathrin- and cholesterol-dependent endocytosis. Virus Res. 200, 45–55. doi: 10.1016/j.virusres.2015.01.022
Geballa-Koukoulas, K., Abdi, S., La Scola, B., Blanc, G., Andreani, J., et al. (2021a). Pacmanvirus S19: the second Pacmanvirus to be isolated from sewage waters in Oran, Algeria. Am. Soc. Microbiol. 10:e0069321.
Geballa-Koukoulas, K., Andreani, J., La Scola, B., and Blanc, G. (2021b). The kaumoebavirus LCC10 genome reveals a unique gene strand bias among “Extended Asfarviridae.”. Viruses 13:148. doi: 10.3390/v13020148
Geballa-Koukoulas, K., Boudjemaa, H., Andreani, J., La Scola, B., and Blanc, G. (2020). Comparative genomics unveils regionalized evolution of the faustovirus genomes. Viruses 12:577. doi: 10.3390/v12050577
Gollakner, R., and Capua, I. (2020). Is COVID-19 the first pandemic that evolves into a panzootic? Vet. Italiana 56, 7–8.
Guinat, C., Gogin, A., Blome, S., Keil, G., Pollin, R., Pfeiffer, D. U., et al. (2016). Transmission routes of African swine fever virus to domestic pigs: current knowledge and future research directions. Veterinary Record 178, 262–267. doi: 10.1136/vr.103593
Haider, N., Rothman-Ostrow, P., Osman, A. Y., Arruda, L. B., Macfarlane-Berry, L., Elton, L., et al. (2020). COVID-19—zoonosis or emerging infectious disease? Front. Public Health 8:596944. doi: 10.3389/fpubh.2020.596944
Hauröder, B., Junglas, L., Loch, S., Michel, R., Müller, K., and Wylezich, C. (2018). Experimental co-infection of Saccamoeba lacustris with Mimivirus-like Giant virus and a small Satellite virus. Endocytobiosis Cell Res. 29, 1–6.
Javanmard, E., Niyyati, M., Lorenzo-Morales, J., Lasjerdi, Z., Behniafar, H., and Mirjalali, H. (2017). Molecular identification of waterborne free living amoebae (Acanthamoeba, Naegleria and Vermamoeba) isolated from municipal drinking water and environmental sources, Semnan province, north half of Iran. Exp. Parasitol. 183, 240–244. doi: 10.1016/j.exppara.2017.09.016
Jean Beltran, P. M., and Cristea, I. M. (2014). The life cycle and pathogenesis of human cytomegalovirus infection: lessons from proteomics. Expert Rev. Proteomics 11, 697–711. doi: 10.1586/14789450.2014.971116
Jia, N., Ou, Y., Pejsak, Z., Zhang, Y., and Zhang, J. (2017). Roles of African swine fever virus structural proteins in viral infection. J. Veterinary Res. 61, 135–143. doi: 10.1515/jvetres-2017-2017
Jian, H., Yi, Y., Wang, J., Hao, Y., Zhang, M., Wang, S., et al. (2021). Diversity and distribution of viruses inhabiting the deepest ocean on Earth. ISME J. 15, 3094–3110. doi: 10.1038/s41396-021-00994-y
Karki, S., Moniruzzaman, M., and Aylward, F. O. (2021). Comparative genomics and environmental distribution of large dsDNA viruses in the family asfarviridae. Front. Microbiol. 12:657471. doi: 10.3389/fmicb.2021.657471
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software Version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Khalil, J. Y. B., Andreani, J., and La Scola, B. (2016). Updating strategies for isolating and discovering giant viruses. Curr. Opin. Microbiol. 31, 80–87. doi: 10.1016/j.mib.2016.03.004
Khan, N. A. (2006). Acanthamoeba : biology and increasing importance in human health. FEMS Microbiol. Rev. 30, 564–595. doi: 10.1111/j.1574-6976.2006.00023.x
Klose, T., Reteno, D. G., Benamar, S., Hollerbach, A., Colson, P., La Scola, B., et al. (2016). Structure of faustovirus, a large dsDNA virus. Proc. Natl. Acad. Sci. U S A. 113, 6206–6211. doi: 10.1073/pnas.1523999113
König, L., Wentrup, C., Schulz, F., Wascher, F., Escola, S., Swanson, M. S., et al. (2019). Symbiont-Mediated defense against Legionella pneumophila in amoebae. mBio 10:e00333-19. doi: 10.1128/mBio.00333-319
Koonin, E. V., and Yutin, N. (2019). Evolution of the large nucleocytoplasmic DNA viruses of eukaryotes and convergent origins of viral gigantism. Adv. Virus Res. 103, 167–202. doi: 10.1016/bs.aivir.2018.09.002
Koonin, E. V., Dolja, V. V., Krupovic, M., Varsani, A., Wolf, Y. I., Yutin, N., et al. (2020). Global organization and proposed megataxonomy of the virus world. Microbiol. Mol. Biol. Rev. 84:e00061-19. doi: 10.1128/MMBR.00061-19
La Scola, B. (2014). Looking at protists as a source of pathogenic viruses. Microb. Pathog. 77, 131–135. doi: 10.1016/j.micpath.2014.09.005
La Scola, B., Audic, S., Robert, C., Jungang, L., de Lamballerie, X., Drancourt, M., et al. (2003). A giant virus in amoebae. Science 299, 2033–2033. doi: 10.1126/science.1081867
Legendre, M., Bartoli, J., Shmakova, L., Jeudy, S., Labadie, K., Adrait, A., et al. (2014). Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc. Natl. Acad. Sci. U S A. 111, 4274–4279. doi: 10.1073/pnas.1320670111
Legendre, M., Lartigue, A., Bertaux, L., Jeudy, S., Bartoli, J., Lescot, M., et al. (2015). In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proc. Natl. Acad. Sci. U S A. 112, E5327–E5335. doi: 10.1073/pnas.1510795112
Letunic, I., and Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49:gkab301. doi: 10.1093/nar/gkab301
Loeffelholz, M. J., and Fenwick, B. W. (2019). Taxonomic changes for human and animal viruses, 2016 to 2018. J. Clin. Microbiol. 57:e01457–18. doi: 10.1128/JCM.01457-18
Maisonneuve, E., Cateau, E., Kaaki, S., and Rodier, M.-H. (2016). Vermamoeba vermiformis-Aspergillus fumigatus relationships and comparison with other phagocytic cells. Parasitol. Res. 115, 4097–4105. doi: 10.1007/s00436-016-5182-5183
Maizels, N., and Weiner, A. M. (1994). Phylogeny from function: evidence from the molecular fossil record that tRNA originated in replication, not translation. Proc. Natl. Acad. Sci. U S A. 91, 6729–6734. doi: 10.1073/pnas.91.15.6729
Masangkay, F., Milanez, G., Karanis, P., and Nissapatorn, V. (2018). “Vermamoeba vermiformis—global trend and future perspective,” in Encyclopedia of Environmental Health, ed. J. Nriagu (Amsterdam:Elsevier), 356–366. doi: 10.1016/B978-0-12-409548-9.11005-X
Mazur-Panasiuk, N., Walczak, M., Juszkiewicz, M., and Woźniakowski, G. (2020). The Spillover of African swine fever in Western Poland revealed its estimated origin on the basis of O174L, K145R, MGF 505-5R and IGR I73R/I329L Genomic Sequences. Viruses 12:1094. doi: 10.3390/v12101094
Mehle, N., Gutiérrez-Aguirre, I., Kutnjak, D., and Ravnikar, M. (2018). Water-Mediated transmission of plant, animal, and human viruses. Adv. Virus Res. 101, 85–128. doi: 10.1016/bs.aivir.2018.02.004
Molla-Herman, A., Vallés, A. M., Ganem-Elbaz, C., Antoniewski, C., and Huynh, J. (2015). tRNA processing defects induce replication stress and Chk2-dependent disruption of piRNA transcription. EMBO J. 34, 3009–3027. doi: 10.15252/embj.201591006
Moniruzzaman, M., Weinheimer, A. R., Martinez-Gutierrez, C. A., and Aylward, F. O. (2020). Widespread endogenization of giant viruses shapes genomes of green algae. Nature 588, 141–145. doi: 10.1038/s41586-020-2924-2922
Montalbano Di Filippo, M., Berrilli, F., Di Cave, D., and Novelletto, A. (2019). Novel data from Italian Vermamoeba vermiformis isolates from multiple sources add to genetic diversity within the genus. Parasitol. Res. 118, 1751–1759. doi: 10.1007/s00436-019-06294-x
Müller, C. A., and Nieduszynski, C. A. (2017). DNA replication timing influences gene expression level. J. Cell Biol. 216, 1907–1914. doi: 10.1083/jcb.201701061
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Oliveira, G. P., de Aquino, I. L. M., Luiz, A. P. M. F., and Abrahão, J. S. (2018). Putative promoter motif analyses reinforce the evolutionary relationships among Faustoviruses, Kaumoebavirus, and Asfarvirus. Front. Microbiol. 9:1041. doi: 10.3389/fmicb.2018.01041
Oliveira, G., La Scola, B., and Abrahão, J. (2019a). Giant virus vs amoeba: fight for supremacy. Virology J. 16:126. doi: 10.1186/s12985-019-1244-1243
Oliveira, G., Silva, L., Leão, T., Mougari, S., da Fonseca, F. G., Kroon, E. G., et al. (2019b). Tupanvirus-infected amoebas are induced to aggregate with uninfected cells promoting viral dissemination. Sci. Rep. 9:183. doi: 10.1038/s41598-018-36552-36554
Page, F. C. (1967). Taxonomic criteria for limax amoebae, with descriptions of 3 new species of hartmannella and 3 of vahlkampfia. J. Protozool. 14, 499–521. doi: 10.1111/j.1550-7408.1967.tb02036.x
Pagnier, I., Valles, C., Raoult, D., and La Scola, B. (2015). Isolation of Vermamoeba vermiformis and associated bacteria in hospital water. Microb. Pathog. 80, 14–20. doi: 10.1016/j.micpath.2015.02.006
Park, J. S. (2016). First record of potentially pathogenic amoeba Vermamoeba vermiformis (Lobosea: Gymnamoebia) isolated from a freshwater of Dokdo Island in the East Sea, Korea. Animal Systematics Evol. Diversity 32, 1–8. doi: 10.5635/ASED.2016.32.1.001
Philippe, N., Legendre, M., Doutre, G., Couté, Y., Poirot, O., Lescot, M., et al. (2013). Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341, 281–286. doi: 10.1126/science.1239181
Reteno, D. G., Benamar, S., Khalil, J. B., Andreani, J., Armstrong, N., Klose, T., et al. (2015). Faustovirus, an asfarvirus-related new lineage of giant viruses infecting amoebae. J. Virol. 89, 6585–6594. doi: 10.1128/JVI.00115-115
Reyes-Batlle, M., Wagner, C., Zamora-Herrera, J., Vargas-Mesa, A., Sifaoui, I., González, A. C., et al. (2016). Isolation of thermotolerant Vermamoeba vermiformis strains from water sources in Lanzarote Island, Canary Islands, Spain. Acta Parasitol. 61, 650–653. doi: 10.1515/ap-2016-2088
Rodrigues, R. A. L., Mougari, S., Colson, P., La Scola, B., and Abrahão, J. S. (2019). “Tupanvirus”, a new genus in the family Mimiviridae. Arch. Virol. 164, 325–331. doi: 10.1007/s00705-018-4067-4064
Rolland, C., Andreani, J., Louazani, A., Aherfi, S., Francis, R., Rodrigues, R., et al. (2019). Discovery and further studies on giant viruses at the IHU mediterranee infection that modified the perception of the virosphere. Viruses 11:312. doi: 10.3390/v11040312
Rolland, C., Andreani, J., Sahmi-Bounsiar, D., Krupovic, M., La Scola, B., and Levasseur, A. (2021). Clandestinovirus: a giant virus with chromatin proteins and a potential to manipulate the cell cycle of its host Vermamoeba vermiformis. Front. Microbiol. 12:715608. doi: 10.3389/fmicb.2021.715608
Rolland, C., La Scola, B., and Levasseur, A. (2020). How tupanvirus degrades the ribosomal RNA of its amoebal host? the ribonuclease T2 track. Front. Microbiol. 11:1691. doi: 10.3389/fmicb.2020.01691
Sahmi-Bounsiar, D., Boratto, P. V., de, M., Oliveira, G. P., Bou Khalil, J. Y., La Scola, B., et al. (2019). Single cell micro-aspiration as an alternative strategy to fluorescence-activated cell sorting for giant virus mixture separation. JoVE 60148. doi: 10.3791/60148
Scheid, P. L. (2019). Vermamoeba vermiformis - a free-living amoeba with public health and environmental health significance. Open Parasitol. J. 7, 40–47. doi: 10.2174/1874421401907010040
Shen, Z., Chen, C., Yang, Y., Xie, Z., Ao, Q., Lv, L., et al. (2021). Novel function of African Swine Fever Virus pE66L in inhibition of host translation by the PKR/eIF2α Pathway. J. Virol. 95:e01872-20. doi: 10.1128/JVI.01872-1820
Siddiqui, R., Makhlouf, Z., and Khan, N. A. (2021). The Increasing Importance of Vermamoeba vermiformis. J. Eukaryotic Microbiol. 68:e12857. doi: 10.1111/jeu.12857
Silva, L. C. F., Rodrigues, R. A. L., Oliveira, G. P., Dornas, F. P., La Scola, B., Kroon, E. G., et al. (2019). Microscopic analysis of the tupanvirus cycle in Vermamoeba vermiformis. Front. Microbiol. 10:671. doi: 10.3389/fmicb.2019.00671
Smirnov, A. V., Chao, E., Nassonova, E. S., and Cavalier-Smith, T. (2011). A revised classification of naked lobose amoebae (Amoebozoa: Lobosa). Protist 162, 545–570. doi: 10.1016/j.protis.2011.04.004
Souza, F., Rodrigues, R., Reis, E., Lima, M., La Scola, B., and Abrahão, J. (2019). In-depth analysis of the replication cycle of Orpheovirus. Virol. J. 16:158. doi: 10.1186/s12985-019-1268-8
Temmam, S., Monteil-Bouchard, S., Sambou, M., Aubadie-Ladrix, M., Azza, S., Decloquement, P., et al. (2015). Faustovirus-Like asfarvirus in hematophagous biting midges and their vertebrate hosts. Front. Microbiol. 6:1406. doi: 10.3389/fmicb.2015.01406
Urbano, A. C., and Ferreira, F. (2020). Role of the DNA-Binding Protein pA104R in ASFV genome packaging and as a novel target for vaccine and drug development. Vaccines 8:585. doi: 10.3390/vaccines8040585
Wang, F., Zhang, H., Hou, L., Yang, C., and Wen, Y. (2021). Advance of African swine fever virus in recent years. Res. Veterinary Sci. 136, 535–539. doi: 10.1016/j.rvsc.2021.04.004
Yoshikawa, G., Blanc-Mathieu, R., Song, C., Kayama, Y., Mochizuki, T., Murata, K., et al. (2019). Medusavirus, a novel large DNA virus discovered from hot spring water. J. Virol. 93:e02130–18. doi: 10.1128/JVI.02130-18
Keywords: Vermamoeba vermiformis, kaumoebavirus, faustovirus, tupanvirus, yasminevirus, orpheovirus, fadolivirus, clandestinovirus
Citation: Geballa-Koukoulas K, La Scola B, Blanc G and Andreani J (2022) Diversity of Giant Viruses Infecting Vermamoeba vermiformis. Front. Microbiol. 13:808499. doi: 10.3389/fmicb.2022.808499
Received: 03 November 2021; Accepted: 18 March 2022;
Published: 22 April 2022.
Edited by:
Sead Sabanadzovic, Mississippi State University, United StatesReviewed by:
Kiran Kondabagil, Indian Institute of Technology Bombay, IndiaFrank O’Neill Aylward, Virginia Tech, United States
Copyright © 2022 Geballa-Koukoulas, La Scola, Blanc and Andreani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Khalil Geballa-Koukoulas, a2hhbGlsLmdlYmFsbGFAbWlvLm9zdXB5dGhlYXMuZnI=; Julien Andreani, SkFuZHJlYW5pQGNodS1ncmVub2JsZS5mcg==
†Present address: Julien Andreani, Univ. Grenoble Alpes, Laboratoire de Virologie, Centre Hospitalier Universitaire (CHU) Grenoble–Alpes, Grenoble, France
 Khalil Geballa-Koukoulas
Khalil Geballa-Koukoulas Bernard La Scola
Bernard La Scola Guillaume Blanc
Guillaume Blanc Julien Andreani
Julien Andreani