- 1Insect Vector Laboratory, Advanced Centre for Plant Virology, ICAR-Indian Agricultural Research Institute, New Delhi, India
- 2Division of Entomology, ICAR-Indian Agricultural Research Institute, New Delhi, India
- 3Center for Agricultural Bioinformatics, ICAR-Indian Agricultural Statistics Research Institute, New Delhi, India
- 4ICAR- National Research Centre for Integrated Pest Management, New Delhi, India
Thrips palmi (Thysanoptera: Thripidae) is the predominant tospovirus vector in Asia-Pacific region. It transmits economically damaging groundnut bud necrosis virus (GBNV, family Tospoviridae) in a persistent propagative manner. Thrips serve as the alternate host, and virus reservoirs making tospovirus management very challenging. Insecticides and host plant resistance remain ineffective in managing thrips–tospoviruses. Recent genomic approaches have led to understanding the molecular interactions of thrips–tospoviruses and identifying novel genetic targets. However, most of the studies are limited to Frankliniella species and tomato spotted wilt virus (TSWV). Amidst the limited information available on T. palmi–tospovirus relationships, the present study is the first report of the transcriptome-wide response of T. palmi associated with GBNV infection. The differential expression analyses of the triplicate transcriptome of viruliferous vs. nonviruliferous adult T. palmi identified a total of 2,363 (1,383 upregulated and 980 downregulated) significant transcripts. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses showed the abundance of differentially expressed genes (DEGs) involved in innate immune response, endocytosis, cuticle development, and receptor binding and signaling that mediate the virus invasion and multiplication in the vector system. Also, the gene regulatory network (GRN) of most significant DEGs showed the genes like ABC transporter, cytochrome P450, endocuticle structural glycoprotein, gamma-aminobutyric acid (GABA) receptor, heat shock protein 70, larval and pupal cuticle proteins, nephrin, proline-rich protein, sperm-associated antigen, UHRF1-binding protein, serpin, tyrosine–protein kinase receptor, etc., were enriched with higher degrees of interactions. Further, the expression of the candidate genes in response to GBNV infection was validated in reverse transcriptase-quantitative real-time PCR (RT-qPCR). This study leads to an understanding of molecular interactions between T. palmi and GBNV and suggests potential genetic targets for generic pest control.
Introduction
Thrips are minute, fringed winged insects that belong to the family Thripidae in the order Thysanoptera. Globally, thrips are considered as one of the most economically damaging pests of a wide range of food, feed, and fiber crops (Riley et al., 2011). Thrips rasp the soft plant tissues and suck plant sap from leaves, stems, flowers, and fruits. The infested plant parts develop distinctive silvery or bronze scarring. High incidence leads to curling, rolling, folding, distortion of leaves, and deformation of fruits. Besides, thrips are sole transmitters of tospoviruses (genus Orthotospovirus, family Tospoviridae, and order Bunyavirales) that cause significant economic losses to crop production across the globe (Ullman et al., 1992, 1997; Whitfield et al., 2005; Oliver and Whitfield, 2016). Annual losses due to tospovirus outbreaks are estimated to cost growers over US$ 1 billion worldwide (Pappu et al., 2009). To date, 16 thrips species are reported to vector 29 tospoviruses (Rotenberg et al., 2015; Ghosh et al., 2017). Tospoviruses propagate within thrips and thrips serve as effective alternate hosts and virus reservoirs which make tospovirus management very difficult (Ullman et al., 2002; Whitfield et al., 2005).
Melon thrips (Thrips palmi Karny) is a predominant thrips species in Asia and has spread to the Pacific, Florida, Hawaii, the Caribbean, South America, West Africa, and Australia (Bhatti, 1980; Johnson, 1986; Pantoja et al., 1988; Cooper, 1991; Palmer, 1992; Monteiro et al., 1995; Guyot, 1998; Kawai, 2001; Cannon et al., 2007; Ghosh et al., 2020). It infests over 200 plant species of family Asteraceae, Cucurbitaceae, Fabaceae, and Solanaceae (Walker, 1992; Cermeli and Montagne, 1993). T. palmi is known to transmit seven species of tospoviruses, such as calla lily chlorotic spot virus (CCSV), capsicum chlorosis virus (CaCV), groundnut bud necrosis virus (GBNV), melon yellow spot virus (MYSV), tomato necrotic ringspot virus (TNRV), watermelon bud necrosis virus (WBNV), and watermelon silver mottle virus (WSMoV; Ghosh et al., 2017, 2020). GBNV is the predominant tospovirus in Asia causing annual losses of approximately US$ 89 million. As much as 70%–90% of losses in groundnut were recorded due to infection of GBNV in India (Singh and Srivastava, 1995). Outbreaks of GBNV in tomatoes caused up to 100% disease incidence from 2003 to 2006 in India (Kunkalikar et al., 2011). Yield losses up to 29% have been reported in potatoes due to GBNV infection (Singh et al., 1997). Several management strategies have been adopted around the world, with insecticides and host plant resistance being the core components for thrips and tospovirus management, respectively. Farmers worldwide spend more than US$ 50 million annually to manage this vector. Insecticides are largely ineffective due to the emergence of resistant thrips population. Genetic host plant resistance is not available for all the crop species that are infected by thrips–tospoviruses. Understanding the interactions between thrips and tospovirus will be helpful to identify novel genetic targets to interrupt the interrelationship and restrict the epidemics of thrips–tospoviruses.
Various genomic approaches have been reported to understand the molecular interactions of thrips and tospoviruses. However, most of these studies are focused on tomato spotted wilt virus (TSWV) and Frankliniella species (Badillo-Vargas et al., 2012; Zhang et al., 2013; Stafford-Banks et al., 2014; Schneweis et al., 2017; Han and Rotenberg, 2021). Frankliniella occidentalis genes associated with host defense, insect cuticle structure and development, metabolism, and transport are regulated by TSWV infection (Schneweis et al., 2017). Genes associated with intracellular transport, development, and immune responses are regulated by TSWV infection in Frankliniella fusca (Shrestha et al., 2017). Transcriptome-wide responses of T. palmi associated with CaCV infection revealed upregulation of innate immune, digestion of proteins, and lipids-related genes, while the genes associated with the structural constituent of the cuticle were downregulated (Gamage et al., 2018). Tospovirus (WBNV) infects the epithelial cells of the anterior midgut of T. palmi and spreads to primary salivary glands via connecting ligaments and other parts of the alimentary canal (Ghosh et al., 2021). Tospovirus (WSMoV) propagates in the primary salivary glands and midgut of T. palmi (Mou et al., 2021). GBNV infection was found to negatively alter the survivability, adult longevity, and reproductive potential of T. palmi (Ghosh et al., 2019). Host plant also influences the biology and transmission of tospovirus (WBNV) by T. palmi (Dhall et al., 2021). Little is known about the differentially expressed genes (DEGs), pathways, and molecular interactions of T. palmi in response to GBNV infection. The present study reports the candidate genes of T. palmi responsive to GBNV infection through a comparative transcriptomic approach that would be novel targets for generic management of the thrips–tospovirus complex.
Materials and Methods
Establishing an Isofemale Population of Thrips palmi
An isofemale population of T. palmi maintained at Advanced Centre for Plant Virology, Indian Agricultural Research Institute (IARI), New Delhi, was used in the study. The identification of T. palmi was based on the morphological keys and mitochondrial cytochrome oxidase subunit I (mtCOI) sequences amplified in PCR using primer pairs, LCO 1490 and HCO 2198 (Folmer et al., 1994; Jangra et al., 2020). The population was established from a single adult female of T. palmi on eggplant (var. Navkiran, Mahyco). The population has been maintained at the thrips rearing facility under controlled environmental conditions at 28 ± 1°C temperature, 60 ± 10% relative humidity, and 16 h of light–8 h of the dark since 2018. Fresh virus-free healthy eggplants were supplied as and when required. The isofemale population has been used in replicate throughout the experiment.
Establishment of Pure Culture of GBNV
The initial inoculum of GBNV was collected from the pure culture maintained at Advanced Centre for Plant Virology, IARI. Healthy cowpea plants (var. Pusa Komal) were raised from seeds in a plant growth chamber (A2000, Conviron, Canada). The healthy cowpea plants at a two-leaf stage were sap-inoculated with GBNV as described by Ghosh et al. (2021). Briefly, the sap was extracted by grinding the inoculum in a pre-sterilized mortar on ice. Ice-cold sodium phosphate buffer (0.01 M, pH 7.0) was added at tissue to buffer ratio of 1:6 wt/vol. β-mercaptoethanol (0.2%) was added while grinding the tissue. A pinch of Celite was dusted on the cowpea plants. The sap was applied to the leaves by gently rubbing with gloved hands. The inoculated plants were washed with sterile distilled water after a while. The inoculated plants were maintained at 25 ± 1°C, 60 ± 10% relative humidity, and 16 h of light–8 h of dark under insect-proof conditions. Nutrient solution containing macro, meso, and trace elements at appropriate concentrations was provided to plants and regularly monitored for symptom appearance. All the inoculated plants were tested in reverse-transcriptase PCR (RT-PCR) using GBNV-specific primers, AG109F and AG110R (Supplementary Table 1) as described below.
Diagnosis of GBNV Infection by RT-PCR
The total RNA was isolated from cowpea plants using RNeasy Plant Mini Kit (Qiagen, Germantown, MD, United States). Complementary DNA (cDNA) was synthesized with random primers using FIREScript RT cDNA synthesis kit (Solis Biodyne, Estonia). The reaction mixture contained 1X RT reaction buffer, 1.0 μg template RNA, 5 μM random primers, 2 mM dNTP mix, 10-unit FIREScript RT, and 20-unit RiboGrip RNase inhibitor. The reverse transcription was carried out in a T100 thermocycler (Bio-Rad, United States) by primer annealing at 25°C for 10 min, reverse transcription at 50°C for 30 min and followed by enzyme inactivation at 85°C for 5 min. PCR was carried out in 25 μl reactions containing 2 μl cDNA, 2.5 μl 10X PCR buffer (Thermo Fischer Scientific), 0.4 μM each of forward and reverse primers, 260 μM dNTP mix (Thermo Fischer Scientific), and 2 U DreamTaq polymerase (Thermo Fisher Scientific). PCR was performed in a T100 Thermal Cycler with 95°C for 5 min followed by 30 cycles of 94°C for 30 s, 59°C for 45 s, and 72°C for 1 min followed by a final extension at 72°C for 7 min. RT-PCR products were resolved on 1% agarose gel stained with GoodView (BR Biochem, India) and observed in a gel documentation system (MasteroGen Inc., Taiwan). The purified product was sequenced bidirectional. The sequences were processed by BioEdit, and BLASTn was performed to check the species homology. The consensus sequence was submitted to GenBank.
Generation of Viruliferous and Nonviruliferous Thrips palmi Population
The freshly emerged first instar larvae (L1, less than 24 h old) of T. palmi were collected using a Camel hairbrush from the isofemale population on eggplants and used for GBNV acquisition in three replicates to generate the viruliferous population. The virus acquisition setup was established as described by Ghosh et al. (2021). About 50 T. palmi L1s in each replicate were placed on a GBNV-infected leaf and allowed to feed for 24 h at 28 ± 1°C and 60 ± 10% relative humidity. The setup was continuously monitored to confirm the feeding of larvae on the leaf. After the acquisition access period, the larvae were transferred on a detached healthy cowpea leaf. The petiole of the detached leaf was inserted in a slant of 0.8% dextrose agar within an insect breeding dish (10 cm diameter, 4 cm height, SPL Life Sciences, Korea). The GBNV-exposed T. palmi were reared up to the adult stage on cowpea leaves at 28 ± 1°C, 60 ± 10% relative humidity, and 16 h of light–8 h of dark. The breeding dishes were regularly monitored and supplied with fresh leaves as and when required. To generate nonviruliferous T. palmi adults, the L1s were released on the healthy cowpea leaves placed on the agar slant within the insect breeding dishes in three replicates as described above. Adults were collected immediately after their emergence and used in the study. Three sets of viruliferous (designated as TpTrI1, TpTrI2, and TpTrI3) and nonviruliferous (TpTrH1, TpTrH2, and TpTrH3) T. palmi adult populations were developed. Each set was divided into two parts. One part was used for transcriptome sequencing and another part was preserved at −80°C for gene expression analysis in reverse transcriptase-quantitative real-time PCR (RT-qPCR).
Infection of GBNV in T. palmi was confirmed by randomly collecting five individual adults from each of the viruliferous and nonviruliferous insect populations and testing in RT-PCR. Total RNA from adults of T. palmi was isolated using NucleoSpin RNA XS (Macherey-Nagel, Germany), and RT-PCR was performed as described above.
RNA Extraction, Library Preparation, and Sequencing
Three sets of viruliferous (TpTrI1, TpTrI2, and TpTrI3) and nonviruliferous (TpTrH1, TpTrH2, and TpTrH3) adults of T. palmi were subjected to total RNA isolation using TRIzol (Invitrogen, United States). Each set comprised 10 adult individuals that included both females and males (1:1).
RNA quality was checked using RNA 6000 Nano Kit (Agilent Technologies, United States) on 2100 Bioanalyzer (Agilent Technologies) with a minimum RNA Integrity Number (RIN) value of 7. RNA concentrations were determined with a NanoDrop ND-8000 spectrophotometer (Thermo Fischer Scientific). RNA-Seq libraries for all samples were prepared using NEBNext UltraII RNA library preparation kit for Illumina (New England Biolabs, United States), and sequencing was done in a single HiSEQ 4,000 (Illumina, Inc. United States) lane using 150 bp paired-end chemistry. The library preparation and sequencing were done by commercial service providers (NxGen Bio Life Sciences, India). Briefly, total RNA was used to purify poly(A) messenger RNA (mRNA) using oligo-dT beads. Magnetic beads were used for two rounds of purification. During the second elution of the poly-A RNA, the RNA was also fragmented into 200–500 bp pieces in the presence of divalent cations at 94°C for 5 min using an ultrasonicator. The cleaved RNA fragments were copied into first-strand cDNA using SuperScript-II Reverse Transcriptase (Thermo Fischer Scientific) and random primers. After second-strand cDNA synthesis, fragments were end-repaired and A-tailed, and indexed adapters were ligated. The products were purified and enriched with PCR to create the final cDNA library. The tagged cDNA libraries were pooled in equal ratios and used for 2 × 150 bp paired-end sequencing on a single lane of the Illumina HiSeq4000. Illumina clusters were generated and were loaded onto Illumina Flow Cell on Illumina HiSeq 4000 instrument and sequencing was carried out. After sequencing, the samples were demultiplexed and the indexed adapter sequences were trimmed using the CASAVA v1.8.2 software (Illumina, Inc.).
Data Preprocessing, Assembly, and Differential Gene Expression
The obtained low-quality raw reads were preprocessed to remove the adaptor contamination using Trim Galore v0.4.11 with the quality score <20%, and trimming ambiguous “N” nucleotides with a ratio of “N” > 5%. A reference genome index was established using BWAv0.7.5 (Li and Durbin, 2009). The processed clean paired-end reads were used to map the reference genome of T. palmi (Guo et al., 2020) by BWA-MEM (Li, 2013). Read numbers mapped to every gene were counted using Samtools v0.1.19 software. Differential expression between the viruliferous and nonviruliferous T. palmi populations was analyzed using the DESeq R package.2 An absolute value of log2 fold change >2 and value of p < 0.05 were used as the thresholds to judge the significance of gene expression differences.
GO and KEGG Pathway Enrichment of Differentially Expressed Genes
Gene annotations and functional enrichment analysis including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) biological pathways were performed to identify the DEGs that were significantly enriched in GO terms or biological pathways post-GBNV infection. GO enrichment analysis for the identified GO terms was performed using the Fisher’s exact test available in the Blast2Go program (Conesa and Götz, 2008). Gene annotations against the Uniprot GO database3 were performed by aligning DEGs to the NR database using Blast2Go v5.2.5 software. GO terms were also assigned using the QuickGO online search tool.4 KEGG pathway enrichment analysis of DEGs was performed using the KEGG database resource5 to identify the pathways that were differentially regulated between viruliferous and nonviruliferous T. palmi with corrected value of p < 0.05. KEGG pathway enrichment was performed by calculating the enrichment factor for all the identified KEGG pathways. The top 20 highly enriched pathways were plotted based on the rich factor.
Gene Regulatory Network Analysis
The top 20 genes each from up- and downregulated DEGs were selected for the construction of gene regulatory networks (GRNs). These DEGs were filtered based on the log fold change values. The network was analyzed and visualized using Cytoscape v3.7.2, where gene correlation was computed using the Pearson’s correlation coefficient of the normalized expression values (Shannon et al., 2003). Network Analyzer plug-in was used to estimate the network centrality and topology.
Validation of RNA-Seq Expression With Real-Time RT-qPCR
To validate the differential gene expression data by RNA-Seq analysis, a few highly up- and downregulated DEGs of T. palmi were selected to assess the gene expression in RT-qPCR. A part of the preserved viruliferous and nonviruliferous T. palmi populations was used in RT-qPCR analysis. Initially, three sets of primers were designed for each of the target genes, and finally, one pair of primers for each gene was selected after validation. β-tubulin was considered as an endogenous control. All the primer sets were validated and optimized in a gradient PCR. Following thermal cycling conditions were followed for validation and optimization of target genes and endogenous control primers: initial denaturation at 95°C for 5 min, then 35 cycles of 95°C for 40 s, 50°C–60°C (depending upon the Tm of primer pairs) for 40 s, and 72°C for 40 s followed by a final extension at 72° C for 10 min. The primer pairs that produced a single specific sharp band at the same PCR conditions for β-tubulin primers were further selected for RT-qPCR experiment. The list of primers that were validated and used in RT-qPCR assay is included in Supplementary Table 1.
The relative expressions of target genes were estimated by RT-qPCR following 2−ΔΔCT method (Livak and Schmittgen, 2001). Firstly, total RNA was isolated from viruliferous (TpTrI1, TpTrI2, and TpTrI3) and nonviruliferous (TpTrH1, TpTrH2, and TpTrH3) thrips samples stored at −80°C using NucleoSpin RNA XS. cDNA was synthesized with oligo (dT) primer using FIREScript RT cDNA synthesis kit. The reaction mixture contained 1X RT reaction buffer, 1.0 μg template RNA, 5 μM oligo dT primer, 2 mM dNTP mix, 10-unit FIREScript RT, and 20-unit RiboGrip RNase inhibitor. The reverse transcription was carried out in a T100 thermocycler at 50°C for 30 min followed by enzyme inactivation at 85°C for 5 min. RT-qPCR was performed in an Insta Q 48M (Himedia, India) using the DyNAmo ColorFlash SYBR Green qPCR Kit (Thermo Fisher Scientific). A 20 μl of RT-qPCR reaction mixture consisted of 10 μl of 1X DyNAmo ColorFlash SYBR Green Master Mix, 0.25 μM ROX passive reference dye, 0.2 μM each forward and reverse primer, and 2 μl template cDNA. Thermal cycling was performed as initial denaturation at 95°C for 5 min, 35 cycles of 95°C for 40 s, annealing at 53–59°C (depending upon the Tm of primer pairs) for 40 s, and 72°C for 40 s. Since SYBR Green I dye binds nonspecifically to any double-stranded DNA, a dissociation or melting stage was added after every reaction to determine the specificity of the amplicons based on the melting curves. The RT-qPCR was performed with three biological and two technical replicates. Log2 fold change value was calculated and relative expression of mRNA was determined by normalized log2 2−ΔΔCT (Livak and Schmittgen, 2001) values of viruliferous (TpTrI) in comparison to nonviruliferous (TpTrH) T. palmi. The ΔCT was calculated as CT of targeted gene—CT of endogenous control. ΔΔCT value was calculated as average ΔCT of nonviruliferous replicates—ΔCT of each replicate. The average 2-(ΔΔCT) was calculated for both the viruliferous and nonviruliferous populations and transformed to log2 2-(ΔΔCT). The log2-fold change of the target gene expression in viruliferous T. palmi population was estimated by normalizing the fold change values of the nonviruliferous population. The relative expressions of target genes post-GBNV exposure in RT-qPCR assay were compared with log2-fold changes obtained through RNA-Seq analysis, and Pearson’s correlation coefficient was calculated using CORREL function in MS Excel.
Results
Thrips palmi Population
The isofemale T. palmi population generated from a single adult female was confirmed on the basis of morphological characters. The adults of T. palmi were yellow in color measuring about 0.8 mm–1 mm in length. Three brick red ocelli were visible in a triangular fashion between the compound eyes. A pair of interocellar setae originated outside the ocellar triangle. The head was quadrangular in shape with seven segmented antennae. Female adults were comparatively larger than males. Female adults had sharp ovipositor at the apex of the abdomen while males were with rounded apex. Further, the nucleotide sequence of mtCOI confirmed the identity as T. palmi. A 657 bp fragment of mtCOI was amplified using primer pairs LCO 1490 and HCO 2198 (Folmer et al., 1994). The nucleotide sequence of the amplified product showed 100% homology to other T. palmi isolates (MN594549, NC_039437, and KX622379) available in GenBank. The sequence can be retrieved from the GenBank with the Accession number MW020349.
GBNV Infection of T. palmi and Cowpea Plant
The initial inoculum of GBNV was collected from the pure culture (tomato isolate) maintained at Advanced Centre for Plant Virology, IARI. Healthy cowpea plants (Vigna unguiculata var. Pusa Komal) were sap-inoculated with GBNV. Characteristic symptoms of GBNV were recorded on sap-inoculated cowpea plants 10–14 days post-inoculation (dpi). Local lesions appeared on inoculated leaves and yellow necrotic spots with multiple rings-like structures were visible on systemic leaves. RT-PCR with GBNV-specific primers produced an amplicon of 1.8 kb for all symptomatic samples that confirmed the GBNV infection in inoculated plants. No amplification was observed in RT-PCR for healthy plant samples. Further, the nucleotide sequence of the amplified product showed more than 97% identity to GBNV. The sequence can be retrieved from the GenBank with Accession No. MN566913.
The isofemale T. palmi adults in three replicates (TpTrI1, TpTrI2, and TpTrI3) exposed to GBNV-infected cowpea plants during the L1 stage were tested in RT-PCR. A 1.8 kb product was visualized on agarose gel that confirmed the infection of GBNV in T. palmi adults. T. palmi populations (TpTrH1, TpTrH2, and TpTrH3) that were exposed to healthy cowpea plants did not produce any GBNV-specific amplification in RT-PCR.
Data Preprocessing, Assembly, and Differential Expression Analysis
T. palmi transcriptome data generated a total of 156,127,441 raw reads of 150 bp from three replicated cDNA libraries of each viruliferous and nonviruliferous T. palmi population. Around 46.84 Gb of the sequence was generated. The average reads per library were about 26,021,240 (Supplementary Table 2). After preprocessing, a total of 156,074,069 clean reads of 20–150 bp were filtered from all six libraries. The preprocessed reads ranged between 99.95 and 99.98%. After removing the low-quality reads from all the sets, the high-quality reads were used for further analyses. These were mapped with the T. palmi genome (Guo et al., 2020) yielding an average mapping percent of 97.31%.
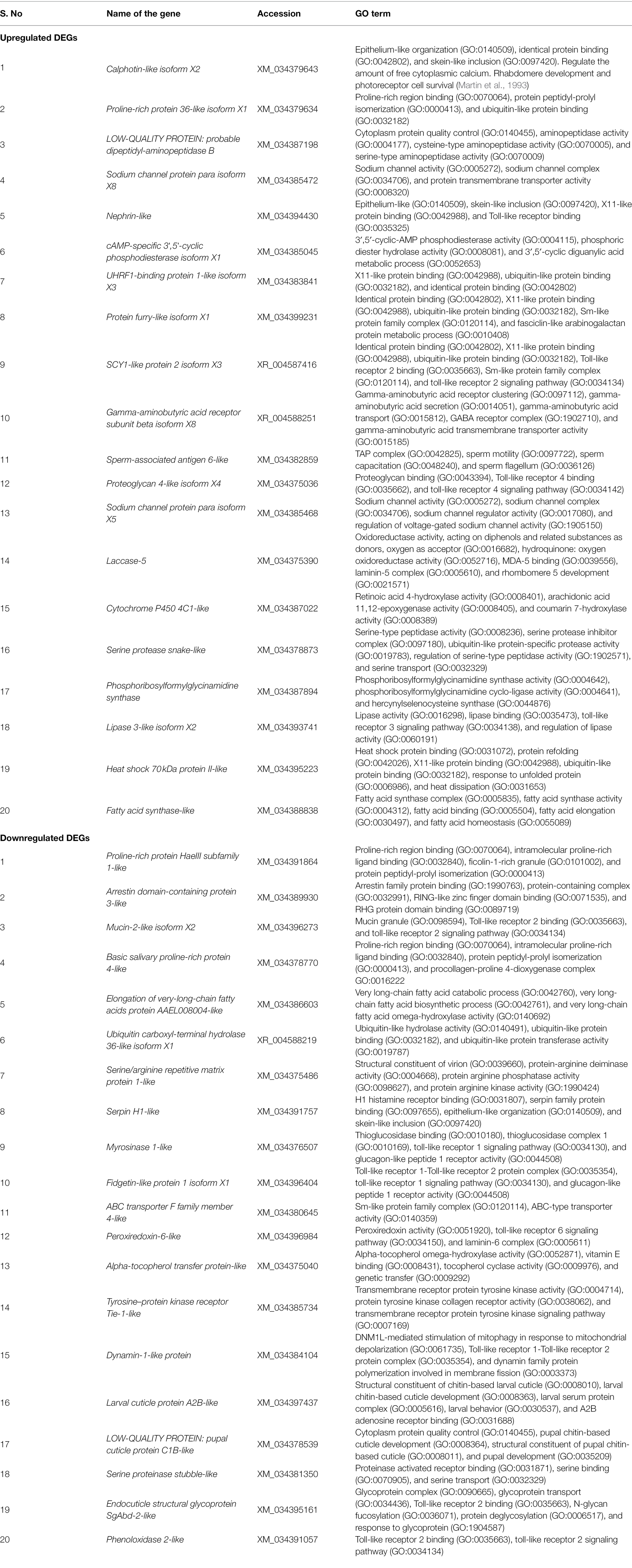
The differential expression analysis identified a total of 28,165 transcripts in the viruliferous vs. nonviruliferous T. palmi populations. Out of these, a total of 2,363 transcripts were significantly expressed in viruliferous T. palmi at stringent filtering parameters (log2 fold change > ± 2 and adjusted value of p < 0.05). A total of 1,383 differentially expressed transcripts were upregulated, while 980 transcripts were downregulated in viruliferous (pooled TpTrI) as compared to nonviruliferous (pooled TpTrH) populations (Figure 1). Table 1 represents the top 20 up- and downregulated genes of T. palmi under the stringent filtering criteria in response to GBNV-infection. The putative genes of T. palmi such as cytochrome P450 (CYP), gamma-aminobutyric acid (GABA) receptor, heat shock protein (hsp70), nephrin, sperm-associated antigen 6 (spag6)-like, and ubiquitin-like containing PHD and RING finger domains 1 (UHRF-1)-binding protein 1, etc. were found highly upregulated after exposure to GBNV. Significant downregulations of ABC transporter, arrestin domain-containing 3 (ARRDC3), elongation of very-long-chain fatty acid (ELOVL), dynamin-1-like, serpin H1-like, tyrosine–protein kinase receptor Tie-1 (TIE1), larval cuticle protein A2B-like, pupal cuticle protein C1B-like, and endocuticle structural glycoprotein SgAbd-2-like, etc., were also observed.
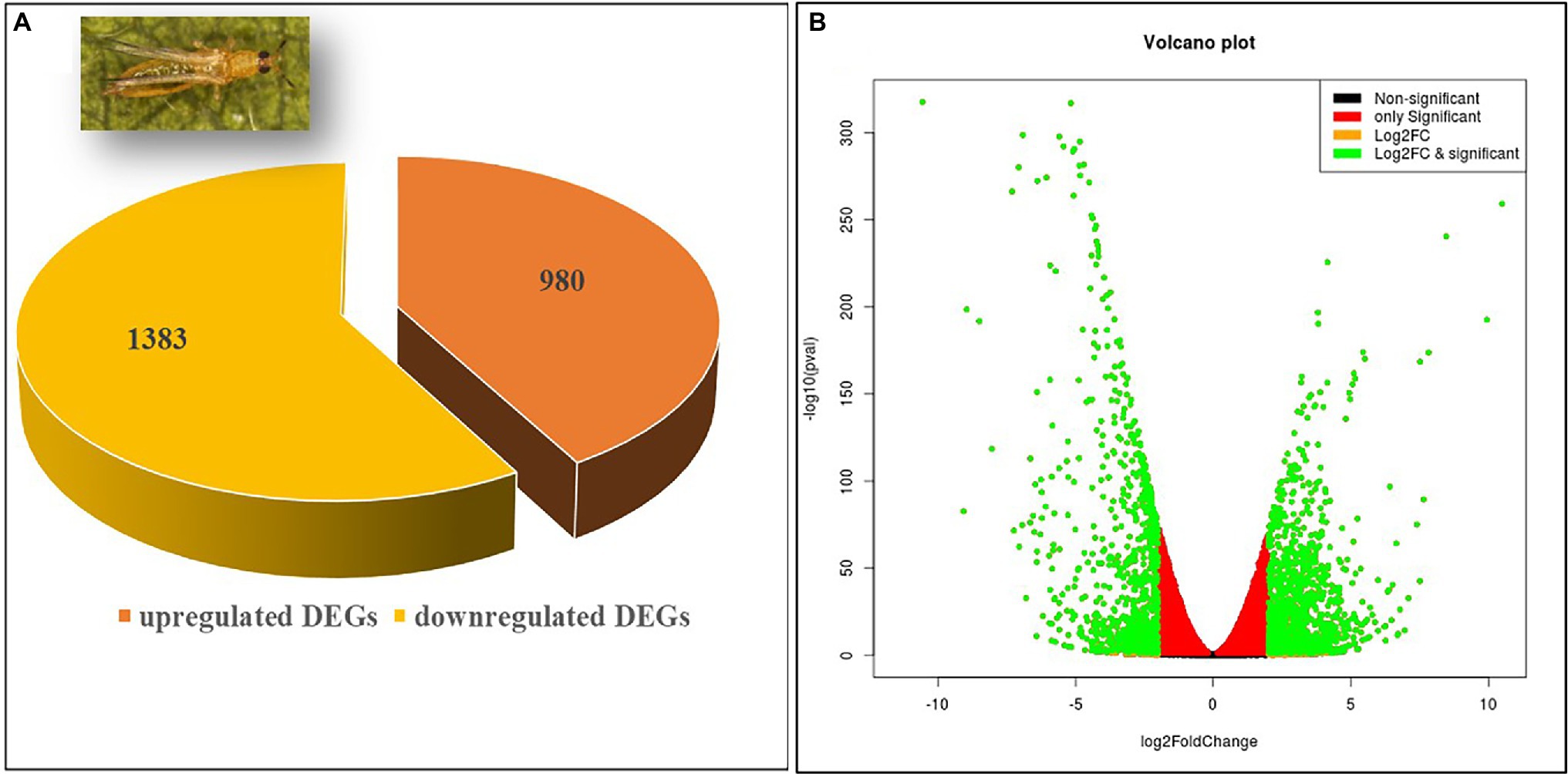
Figure 1. The proportion of differentially expressed genes (DEGs) in RNA-Seq analysis. (A) A total of 1,383 DEGs were found upregulated while 980 were downregulated. (B) Volcano plot of DEGs. The x axis shows the fold change in gene expression between different samples, and the y axis shows the statistical significance of the differences. Significantly up- and downregulated genes with log2 FC ≥ 2 are highlighted in green. The black dots represent insignificant differentially expressed genes.
GO and KEGG Pathway Enrichment Analyses of Differentially Expressed Genes
The 1,383 (58.52%) up- and 980 (41.48%) downregulated DEGs were categorized under the three major classes, namely, cellular components, biological processes, and molecular functions. The highest number of GO terms were categorized into molecular functions (70%) followed by cellular components (24%), and biological processes (6%; Figure 2A). In the molecular functions category, there was an abundance of genes associated with catalytic activity followed by binding, molecular transduction activity, antioxidant activity, and transporter activity. Similarly, genes associated with the membrane, membrane part, cell, cell part, protein-containing complex, organelle, synapse, synapse part were differentially enriched under the cellular components category. In the biological processes, genes involved in the cellular process, response to stimuli, signaling, regulation of the biological process, localization, metabolic process, biological regulation, and signaling were enriched. Within each of the functional groups, the top enrichment terms for both up- and downregulated DEGs identified as per Fisher’s exact test value of p are listed in Figures 2B,C, respectively.
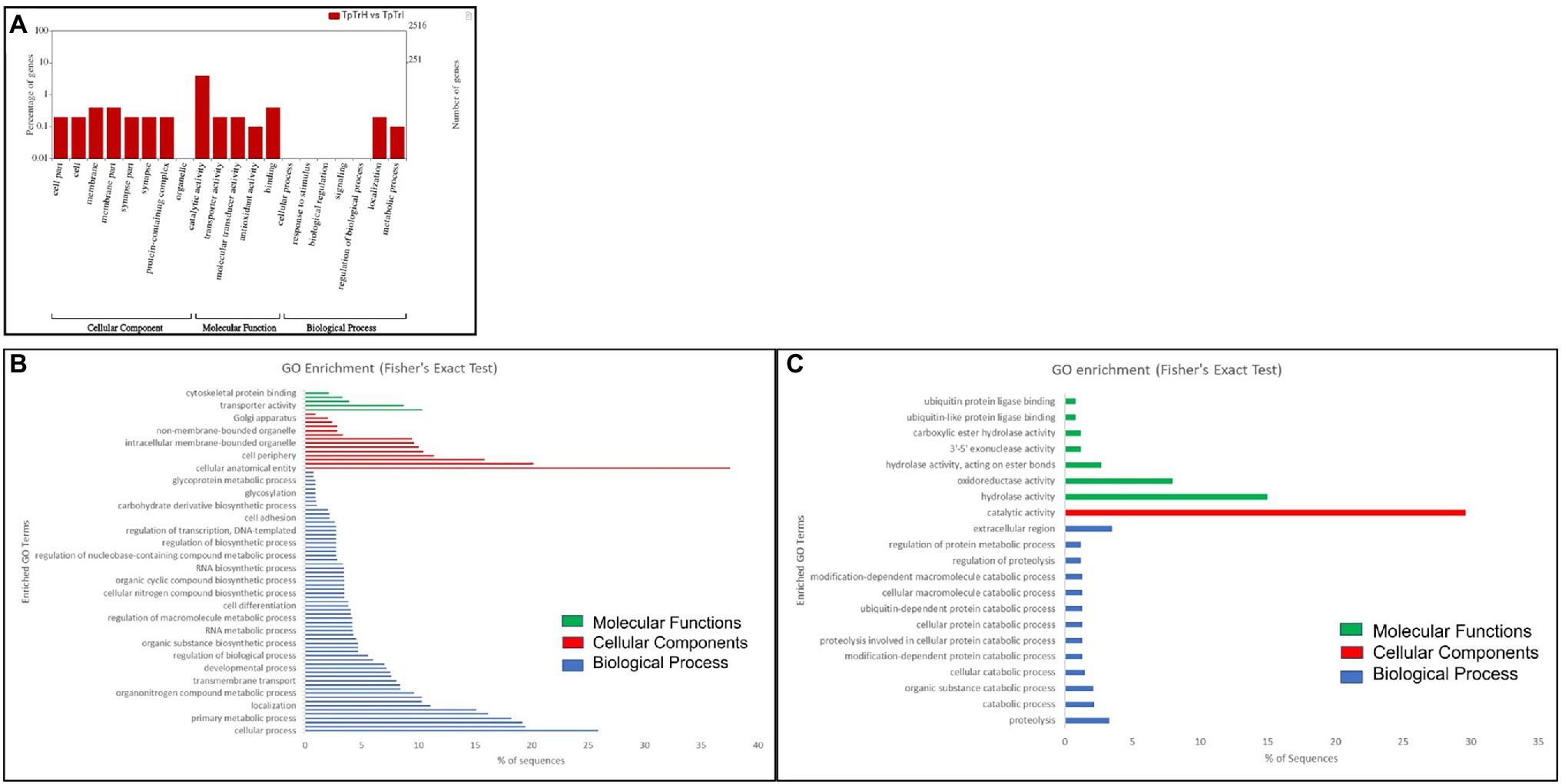
Figure 2. Gene ontology (GO) enrichment analysis of differentially expressed genes (DEGs). (A) DEGs were characterized under cellular components, molecular functions, and biological processes on the basis of GO analysis. GO of differential expressed (B) up- and (C) downregulated genes of viruliferous vs. nonviruliferous thrips populations. Green, red, and blue bars represent molecular functions, cellular components, and biological processes, respectively.
Kyoto Encyclopedia of Genes and Genomes pathway analysis of DEGs showed that the genes were involved in the functions like metabolic pathways, biosynthesis of secondary metabolites, cellular processes, endocytosis, organismal systems, and signaling pathways. Majorly affected metabolic pathways were glycerolipid, glycerophospholipid, purine, fructose and mannose, galactose, glutathione, and carbon metabolism. The differentially expressed signaling pathways were calcium, cyclic adenosine monophosphate (cAMP), glucagon, hypoxia-inducible factor 1 (HIF-1), mitogen-activated protein kinase (MAPK), neurotrophin signaling, phospholipase D, Ras-proximate-1 (Rap1) signaling, and Ras signaling pathways. The most affected cellular and organismal systems were dopaminergic synapses, endocytosis, glutamatergic synapse, lysine degradation, neuroactive ligand-receptor interaction, oocyte meiosis, and synaptic vesicle cycle. In KEGG pathway enrichment analysis, the highest number of DEGs was annotated in drug metabolism (map00982, 00983) under the class xenobiotics biodegradation and metabolism followed by glycerolipid metabolism (map00561), methane metabolism (map00680), glycerophospholipid metabolism (map00564), and fructose and mannose metabolism (map00051; Figure 3). The pathway enrichment of DEGs showed the DEGs were significantly enriched in ascorbate and aldarate metabolism (map00053), steroid hormone biosynthesis (map00140), and metabolism of xenobiotics by cytochrome P450 (map00980) pathways. The pathways identified in GO enrichment analysis were consistent with the findings of the KEGG pathway study.
Figure 3. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of DEGs of Thrips palmi in response to groundnut bud necrosis virus (GBNV) infection. The rich factor is defined as the ratio of the number of DEGs annotated in a pathway to the number of all genes annotated in this pathway. Y axis indicates the pathway name; x axis indicates the enriched factor in each of the pathways. The bubble size indicates the number of DEGs. The color bar indicates the corrected value of p, the red represents a higher value, and the green represents a lower value.
Gene Regulatory Network Analysis
A total of 40 nodes comprised the top up- and downregulated DEGs were considered in network analysis. The degree and betweenness centrality in the interaction network of the top DEGs were calculated. Interaction networks corresponding with the highly regulated DEGs demonstrated a total of 438 edges. The degree of certain nodes was markedly higher compared with the average degree in the networks (Figure 4). The upregulated DEGs with significantly higher degrees were calphotin-like isoform X2, proline-rich protein 36-like isoform X1, dipeptidyl-aminopeptidase B, sodium channel protein para isoform X8, nephrin-like, cAMP-specific 3′,5′-cyclic phosphodiesterase isoform X1, UHRF1-binding protein 1-like isoform X3, spag6-like, laccase-5, CYP 4C1-like, phosphoribosylformylglycinamidine synthase (PFAS), and hsp70 II-like. Among the downregulated nodes, a group of DEGs associated with cuticle development such as larval cuticle protein A2B-like, endocuticle structural glycoprotein SgAbd-2-like, and ABC transporter F family member 4-like were with higher degrees of interactions. Besides, proline-rich protein HaeIII subfamily 1-like, ARRDC3, serpin H1-like, TIE-1-like, myrosinase 1-like, alpha-tocopherol transfer protein (TTPA)-like, and dynamin-1-like protein showed significant interactions with higher degrees.
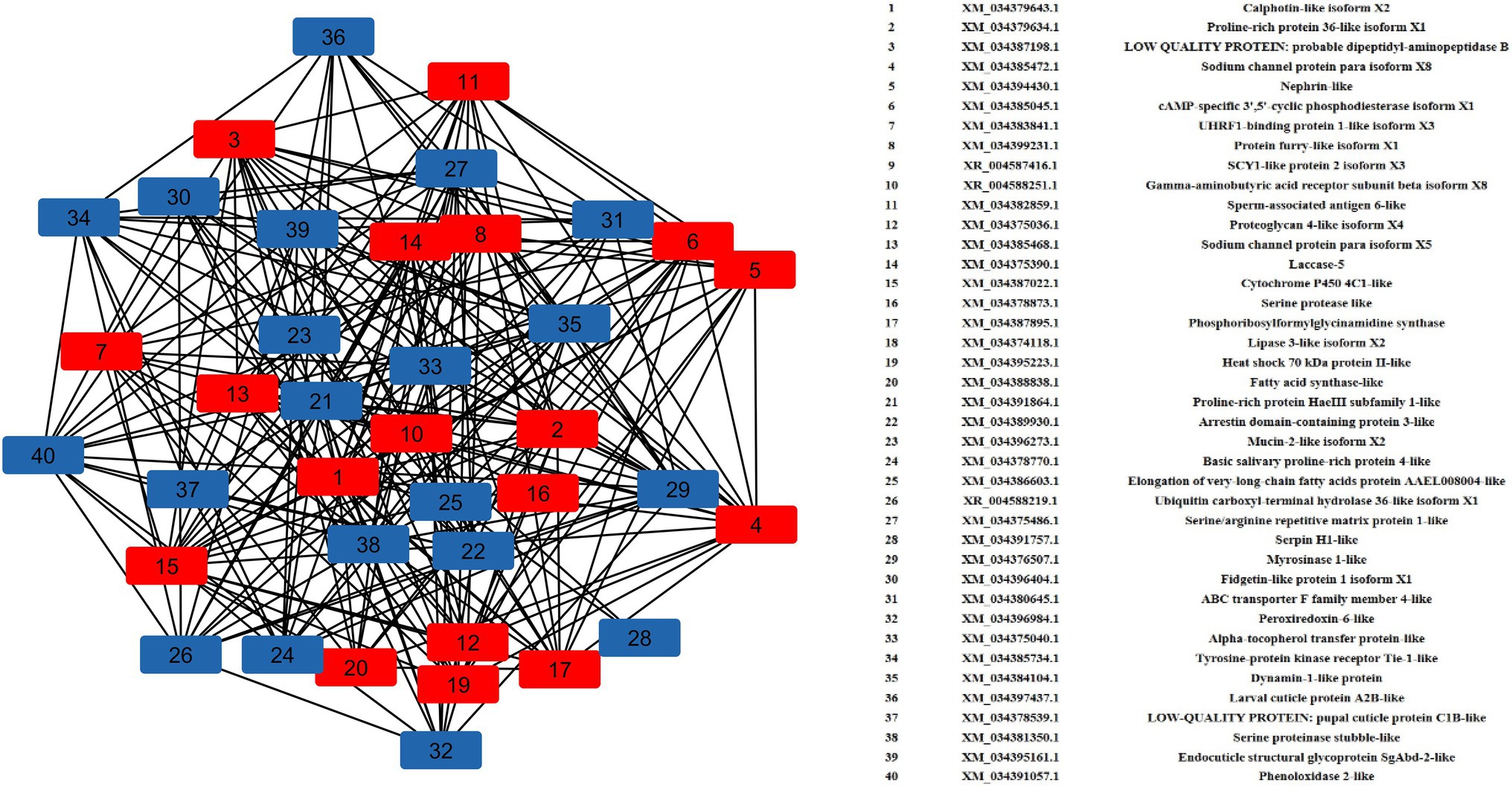
Figure 4. Gene regulatory network (GRN) of DEGs. Red squares represent upregulated DEGs, and blue squares represent downregulated DEGs.
Validation of DEGs in RT-qPCR
To validate the differential gene expression data, thirteen highly expressed genes of T. palmi were selected and mRNA expression levels were quantified in RT-qPCR in response to GBNV infection. A part of the samples used for RNA-Seq was preserved and used in RT-qPCR assay. Primer pairs designed for this study were initially optimized in a gradient PCR (Supplementary Table 1). Primer pairs of target genes that produced a single sharp amplicon at the same PCR conditions for endogenous control (β-tubulin) primers were selected for RT-qPCR assay. The primer pair specific to β-tubulin produced sharp bands at all annealing temperatures between 53 and 59°C. The annealing temperature of primer pairs for each target gene was standardized within the same temperature range. One best primer pair for each of the target genes was optimized for RT-qPCR assay. The annealing temperature, amplicon size, and melting temperature of each amplicon in RT-qPCR are included in Supplementary Table 1. The primer pairs for target and endogenous control genes produced single specific peaks without any secondary amplification in the RT-qPCR melting curve analysis that indicated the specificity of the reactions (Supplementary Figure 1).
The relative expression of target genes was estimated by a 2−ΔΔCT method. The variation among the biological replicates was normalized with respect to the CT value of endogenous control, β-tubulin. Among the selected highly expressed genes, the expression of UHRF1-binding protein 1 of T. palmi was upregulated by log2 5.5-fold in RT-qPCR in response to GBNV infection (Figure 5). Likewise, mRNA expression of nephrin was upregulated by log2 4.8-fold in viruliferous T. palmi adults in comparison to nonviruliferous adults. mRNA levels of spag6-like, hsp70, and GABA receptor were upregulated by log2 3.37, 0.98, and 0.80-fold, respectively, post-GBNV exposure. In viruliferous T. palmi, the expression of serpin was downregulated by log2 8.5-fold. Similarly, log2 4.3-fold downregulation of tyrosin kinase was recorded in RT-qPCR in response to GBNV infection. Expression of ABC transporter F family member 4-like gene was also downregulated by log2 3.57-fold, while it was log2 1.65-fold downregulations for ARRDC3 in viruliferous T. palmi. Some other genes such as elongation of fatty acid chain, dynamin-1-like, myrosinase 1-like showed log2 1.36, 0.66, and 2.57-fold, respectively, lower expression in viruliferous T. palmi as compared to nonviruliferous.
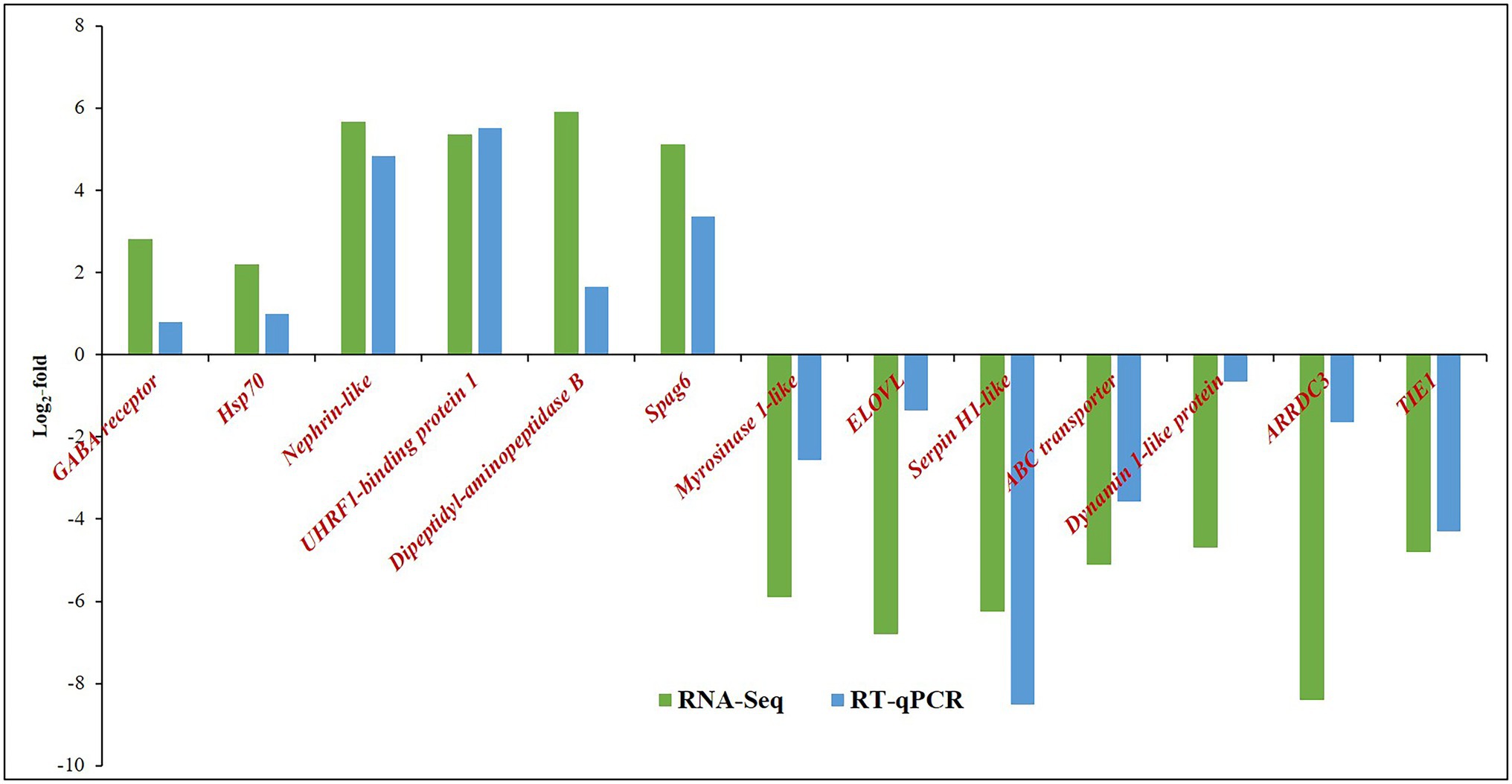
Figure 5. Expression of Thrips palmi putative genes in response to GBNV infection in RNA-Seq and reverse transcriptase-quantitative real-time PCR (RT-qPCR). The values of log2-fold changes calculated in RNA-Seq analysis were in accordance with the RT-qPCR fold change values. Pearson’s correlation coefficient value was 0.81 as calculated using the CORREL function in MS Excel.
The fold change values estimated from RNA-Seq were plotted against fold change values obtained in RT-qPCR assays of the selected genes (Figure 5). The Pearson’s correlation coefficient of 0.81 indicated the acceptability of DEGs through RNA-Seq (Supplementary Table 3).
Discussion
Multiplication of GBNV in T. palmi suggests a potential alteration in the physiological process of T. palmi. Pathogenic effects of GBNV are evident by reduced survival and adult longevity of T. palmi (Ghosh et al., 2019). The fecundity of T. palmi decreases sharply after GBNV exposure. GBNV infection in T. palmi also induces more females in progenies (Ghosh et al., 2019). We observed the same trend in the biological traits of T. palmi post-GBNV exposure in the current study. In thrips–tospovirus relationships, adults of thrips are vector competent only if the virus is acquired during the larval stages (Wetering et al., 1996; Mou et al., 2021). Only during the early larval stage, the virus is allowed to infect the midgut (Ullman et al., 1992; Ohnishi et al., 2001). The virus infects the epithelial cells of the anterior midgut of T. palmi and then progresses to the primary salivary glands through a pair of connecting ligaments (Ghosh et al., 2021). The tospovirus infection in adults of T. palmi is retained in visceral and longitudinal muscles of the midgut. Tospovirus may not be able to infect the epithelial cells of adult thrips (Ghosh et al., 2021). A 50-kDa protein receptor present on the epithelial cells of thrips is abundant during the larval stage only (Bandla et al., 1998). The tospovirus propagates in the midgut and salivary glands of T. palmi (Ghosh et al., 2021; Mou et al., 2021). In thrips, a transovarial infection barrier has been discovered, which prevents the tospovirus from transmitting vertically (Wijkamp et al., 1996; Mou et al., 2021). The next generation of thrips again needs to acquire the virus during their early larval stage.
Manipulation of insect vectors by plant viruses is common in thrips–tospovirus systems (Deangelis et al., 1993; Wijkamp et al., 1996; Belliure et al., 2008; Ingwell et al., 2012; Shrestha et al., 2012; Ghosh et al., 2019). The central hypothesis of the present study tells that the successful GBNV transmission by viruliferous T. palmi perturbs the expression of candidate genes associated with innate immunity, cuticle development, cellular receptor binding, and signaling of its vector. The differential transcriptomic response of T. palmi induced by GBNV infection was distinguished by RNA-Seq. KEGG analyses further explained several molecular pathways involved in T. palmi-GBNV interactions. Cluster analysis of expressed genes from viruliferous and nonviruliferous T. palmi revealed that 1,383 and 980 genes of T. palmi were up- and downregulated, respectively in response to GBNV infection. TSWV infection regulated expression of 127 transcripts in F. occidentalis adults. Expression of genes like arylphorin, chitinase, hemocyanin, and hexamerin was modulated in F. occidentalis adults in response to TSWV infection (Schneweis et al., 2017). A total of 562 contigs were differentially regulated by TSWV in adults of F. fusca. Reproduction, embryo development, and growth-related pathways were identified with upregulated contigs in virus-exposed adults (Shrestha et al., 2017). In the present study, candidate genes, such as CYP, GABA receptor, hsp70, laccase 5, nephrin, spag6, PFAS, proteoglycan, and UHRF1-binding protein 1 associated with innate immune response, receptor binding, signaling, endocytosis, viral replication, and apoptosis (Qin et al., 2009; González-Santoyo and Córdoba-Aguilar, 2012; Götz et al., 2012; Cooley et al., 2016; Zhang et al., 2016; Zhu et al., 2017; Lu et al., 2019) were upregulated in T. palmi adults in response to GBNV infection.
A group of genes associated with host innate immune and stress response, such as hsp70, UHRF1-binding protein 1, spag6, laccase-5, and PFAS was upregulated in viruliferous T. palmi (Zhao and Jones, 2012; Cooley et al., 2016; Zhang et al., 2016; Chen et al., 2020). Hsp belongs to a class of functionally related proteins involved in the folding and unfolding of other proteins. This group of proteins is elevated in insect cells in response to various biotic and abiotic stresses (Zhao and Jones, 2012). Besides, hsp70 is an active regulator of tomato yellow leaf curl virus (TYLCV) infection in its vector, Bemisia tabaci (Götz et al., 2012). In infected host cells, hsp70 interacts with TYLCV coat protein (CP) and is involved in the translocation of TYLCV CP from the cytoplasm to nucleus (Götz et al., 2012; Gorovits and Czosnek, 2013, 2017). Upregulations of T. palmi hsp70 were also recorded in response to CaCV infection (Gamage et al., 2018). However, interaction of hsp70 with GBNV surface proteins needs further experimental evidence. UHRF1-binding protein 1 is best known for its function in maintaining DNA methylation and acting as an epigenetic regulator. When the host is attacked by pathogens, gene expression levels are regulated by a DNA methylation pattern that leads to activation or suppression of several signaling pathways and triggers a series of immune response events against viral invasion (Zhang et al., 2016). Thus, upregulation of UHRF1-binding protein 1 in viruliferous T. palmi adults suggests its role in innate immunity. Upregulation of spag6 in the present study might also be due to the immune response of T. palmi to GBNV infection. The role of mouse spag6 in immune response was described by Cooley et al. (2016). In insects, laccase is abundant in the cuticle and has been involved in cuticle sclerotization (Hattori et al., 2010). Laccase 5 was significantly upregulated in viruliferous T. palmi. Laccases include phenol oxidases (Janusz et al., 2020) that are activated in response to viral infection and oxidative stress (González-Santoyo and Córdoba-Aguilar, 2012; Chen et al., 2020). A similar response of T. palmi laccase post-CaCV infection (Gamage et al., 2018) suggests its association in maintaining oxidative stress due to tospovirus infection in T. palmi. In consistent with the previous report of CaCV-exposed T. palmi (Gamage et al., 2018), PFAS was also upregulated in T. palmi in response to GBNV infection. PFAS was also found to be upregulated in Culex quinquefasciatus after infection with West Nile virus (Girard et al., 2010). PFAS not only plays a role in the immune escape process of the virus but also has a crucial impact on other pathological processes (Lu et al., 2019). Pathological effects such as reduced survival and adult longevity of T. palmi were also recorded post-GBNV exposure in the present study. The role of PFAS in triggering such pathological effects in T. palmi may be worth for further investigation.
The GBNV is supposed to be internalized in T. palmi cells by clathrin-mediated endocytosis (CME; Jagdale and Ghosh, 2019). DEGs, such as nephrin, proteoglycan 4-like, and heparin sulfate proteoglycan (HSPG) associated with endocytosis and entry receptors (Qin et al., 2009; Christianson and Belting, 2014; Cagno et al., 2019) were upregulated upon GBNV infection. Nephrin is internalized during the signal transduction pathway of raft-mediated endocytosis and classical CME (Qin et al., 2009). The upregulation of nephrin in the present study indicated its involvement in CME of the GBNV proteins in T. palmi cells. Proteoglycan 4-like, upregulated in T. palmi post-GBNV infection, might be associated with receptor binding and entry of tospovirus in thrips. HSPG is a cell-surface endocytosis receptor and is involved in cellular interactions with several human viruses. The interaction with HSPG is used by viruses to increase their concentration at the cell surface and augment their chances of binding more specific entry receptors (Cagno et al., 2019). HSPG is an integral factor for the attachment of Arbovirus to the salivary gland duct in mosquitoes (Ciano et al., 2014).
In insects, GABA receptors are found throughout the central nervous system and are reported to be an important target site for several insecticides (Anthony et al., 1993). In mosquitoes, GABA receptor was associated with viral replication (Zhu et al., 2017). CYP imparts resistance against insecticides in F. occidentalis and T. tabaci (Zhang et al., 2013; Rosen et al., 2021). In the present study, upregulations of GABA receptor and CYP in viruliferous T. palmi were in accordance with the response of CaCV-exposed T. palmi (Gamage et al., 2018). CYP-mediated metabolism of xenobiotics pathway was also enriched in viruliferous T. palmi. An in-depth study of the mechanism involved would be necessary to shed further light on the role of GABA receptor and CYP in tospovirus replication in T. palmi.
A considerable number of T. palmi genes were under-regulated in response to GBNV infection. Downregulations of genes like ABC transporter, ARRDC3, cuticular proteins, dynamin-1-like protein, myrosinase-1-like, and serpin facilitate the circulation and multiplication of GBNV in T. palmi. Several cuticular protein genes were downregulated in transcriptomic studies of TSWV-infected F. occidentalis (Schneweis et al., 2017) and CaCV-infected T. palmi (Gamage et al., 2018). In the present study, downregulations of T. palmi larval cuticle protein A2B-like, pupal cuticle protein C1B-like, endocuticle structural glycoprotein SgAbd-2-like were recorded in response to GBNV infection. Endocuticle structural glycoprotein localizes in the midguts and salivary glands of F. occidentalis (Schneweis et al., 2017). ABC transporter has also been reported to be involved in molting and cuticle formation in insects (Broehan et al., 2013) and was downregulated in the present study. Virus infection suppresses the activities of cuticular proteins embedded in the peritrophic matrix, a structural barrier to pathogen attack (Kato et al., 2008; Levy et al., 2011). In consistent with the downregulation of genes associated with cuticle development, a delayed molting of the larval instars post-GBNV exposure was recorded in the present study. Delayed molting of viruliferous T. palmi was earlier reported by Ghosh et al. (2019). Under-expression of cuticular, endo-cuticular proteins, and ABC transporter of T. palmi gives an advantage of the delayed molting and membrane dysfunction for virus infection in the thrips gut. However, insect cuticle-related contigs were upregulated in TSWV-infected F. fusca (Shrestha et al., 2017). The innate differences among the thrips species, experimental setup, and virus isolates could be the reason behind the variation in response.
A few genes associated with innate immune responses like serpins and dynamin were also downregulated in T. palmi in response to GBNV infection. Insect innate immunity is regulated by serpins by inhibiting serine proteinase cascades that instigate innate immune responses. Serpins could also possess direct anti-pathogen activity upon infection (Levashina et al., 1999). Dynamin-1-like protein is close relative to dynamin and has a role in fission of mitochondrial and peroxisomal membranes. In mammals, mitochondrial dynamin affects disease resistance. Pathogens have evolved several strategies to interfere with mitochondrial dynamins to suppress the immune responses (Spier et al., 2019). Several pathogens directly target the dynamin-related protein (DRP1) to block the innate immune responses (Suzuki et al., 2014; Yu et al., 2015). Downregulation of dynamin-1-like protein and serpin H1-like genes in viruliferous T. palmi help in evading innate immune responses in T. palmi to favor the virus infection and multiplication in vector cells. Gene like ELOVL was downregulated in viruliferous T. palmi. Accumulation of ELOVL fatty acids in the cells leads to necroptosis (Parisi et al., 2017). Inhibition of ELOVL prevents the loss of plasma membrane integrity and cell death. Downregulation of ELOVL gene in T. palmi helps maintain the integrity of cells and supports viral growth.
A group of genes associated with cell surface receptor and cellular transport, such as TIE1, ARRDC3, TTPA-like, and potassium channel subfamily K member 18 (KCNK18)-like was regulated in T. palmi in response to GBNV infection. TIE1 is the major class of enzyme-coupled cell surface receptors activated in response to external signals such as growth factors, cytokines, and hormones. Host signaling through tyrosine receptor kinases has also been found to play a key role in virus replication (Kumar et al., 2011). Recently, it has been demonstrated that tyrosine kinases are required by viruses for intracellular entry (Stewart et al., 2021). ARRDC3 is a member of the family of α-arrestins in the superfamily of arrestin adaptor proteins (Patwari and Lee, 2012). The α-arrestin, ARRDC3 mediates ALIX ubiquitination and G protein-coupled receptor lysosomal sorting (Dores et al., 2015). Ubiquitination of ALIX has also been observed during the budding of viruses from the plasma membrane in mammals (Sette et al., 2010). TTPA-like and KCNK18-like that encode proteins associated with inter- and intracellular transport were downregulated in response to GBNV infection. Similar results were recorded by Gamage et al. (2018). TTPA isoform 1 and 2, and KCNK9 were also downregulated in response to TSWV infection in F. fusca (Shrestha et al., 2017). The requirement of potassium channels for infecting the host cell by bunyaviruses was reported by Hover et al. (2016). The antiviral activity of the potassium ion channel in honeybees was reported by O’Neal et al. (2017). However, the molecular mechanisms underlying the downregulation of these genes in response to GBNV infection are not known and need in-depth study.
The comparison of the DEGs reported in CaCV-exposed T. palmi (Gamage et al., 2018) with our work showed 103 functions in common (Supplementary Figure 2). However, we could find 2,260 genes uniquely expressed in the GBNV-exposed T. palmi, while 347 genes were unique to the CaCV-exposed T palmi. Among the DEGs, the responses of cuticular proteins, CYP, endocuticle structural glycoprotein, GABA receptor, hsp70, laccase 5, KCNK-like, and TTPA were conserved in both GBNV and CaCV infection but distinct to TSWV infection in F. occidentalis (Schneweis et al., 2017). These candidate genes would be potential targets for novel generic pest control.
Among the major pathways of T. palmi affected by GBNV, metabolic pathways, biosynthesis of secondary metabolites, endocytosis, and signaling pathways were important. Ascorbate and aldarate metabolisms, steroid hormone biosynthesis, cytochrome p 450, and other enzymes-mediated metabolism of xenobiotics, fructose, and mannose metabolism were highly enriched. GRN analysis showed that DEGs associated with innate immune response, cuticle development, receptor signaling, and endocytotic pathways are involved in possible manipulation in host invasion by GBNV.
In conclusion, we have assembled a whole-body transcriptome of adult T. palmi and reported the DEGs in response to GBNV infection. The majority of DEGs are involved in innate immunity, receptor binding and signaling, cuticle development, and endocytosis that facilitate the invasion, circulation, and multiplication of GBNV in T. palmi. The candidate genes of T. palmi that are highly regulated in response to GBNV infection would be potential targets for the effective management of tospoviruses. In addition, data generated in this study will enrich genomic information of thrips and will enable functional studies.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA758768.
Author Contributions
AG, VB, VK, and SC conceived and designed the research. DM prepared biological samples. DM, SJn, and Priti carried out the wet laboratory experiments and wrote the draft manuscript. Priti, MI, and SJi analyzed RNA-Seq data. AG and VB reviewed the results and edited the final manuscript. All authors read and approved the manuscript.
Funding
The scholarship of DM was supported by Indian Council of Agricultural Research.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This is a part of thesis work of DM submitted to Post Graduate School, IARI, New Delhi. The initial virus culture was supplied by Basavaraj YB (IARI). The authors are thankful to Heena (IARI) for her help in maintenance of virus and insect culture.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.773238/full#supplementary-material
Footnotes
1. ^https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
2. ^http://www.bioconductor.org/packages/release/bioc/html/DESeq.html
References
Anthony, N. M., Harrison, J. B., and Sattelle, D. B. (1993). GABA receptor molecules of insects, molecular neurobiology. EXS 63, 172–209. doi: 10.1007/978-3-0348-7265-2_8
Badillo-Vargas, I. E., Rotenberg, D., Schneweis, D. J., Hiromasa, Y., Tomich, J. M., and Whitfield, A. E. (2012). Proteomic analysis of Frankliniella occidentalis and differentially expressed proteins in response to tomato spotted wilt virus infection. J. Virol. 86, 8793–8809. doi: 10.1128/JVI.00285-12
Bandla, M. D., Campbell, L. R., Ullman, D. E., and Sherwood, J. L. (1998). Interaction of tomato spotted wilt tospovirus (TSWV) glycoproteins with a thrips midgut protein, a potential cellular receptor for TSWV. Phytopathology 88, 98–104. doi: 10.1094/PHYTO.1998.88.2.98
Belliure, B., Janssen, A., and Sabelis, M. W. (2008). Herbivore benefits from vectoring plant virus through reduction of period of vulnerability to predation. Oecologia 156, 797–806. doi: 10.1007/s00442-008-1027-9
Bhatti, J. S. (1980). Species of the genus Thrips from India (Thysanoptera). Syst. Entomol. 5, 109–166. doi: 10.1111/j.1365-3113.1980.tb00404.x
Broehan, G., Kroeger, T., Lorenzen, M., and Merzendorfer, H. (2013). Functional analysis of the ATP-binding cassette (ABC) transporter gene family of Tribolium castaneum. BMC Genomics 14:6. doi: 10.1186/1471-2164-14-6
Cagno, V., Tseligka, E. D., Jones, S. T., and Tapparel, C. (2019). Heparan sulfate proteoglycans, and viral attachment: true receptors or adaptation bias? Viruses 11:596. doi: 10.3390/v11070596
Cannon, R. J. C., Matthews, L., and Collins, D. W. (2007). A review of the pest status and control options for Thrips palmi. Crop Prot. 26, 1089–1098. doi: 10.1016/j.cropro.2006.10.023
Cermeli, M., and Montagne, A. (1993). Present situation of Thrips palmi Karny (Thysanoptera: Thripidae) in Venezuela. Manejo Integr. Plagas. 29, 22–23.
Chen, Y.-H., Song, F., Miao, Y.-T., He, H.-H., Lian, Y.-Y., Li, X., et al. (2020). A novel Laccase gene from Litopenaeus vannamei is involved in the immune responses to pathogen infection and oxidative stress. Dev. Comp. Immunol. 105:103582. doi: 10.1016/j.dci.2019.103582
Christianson, H. C., and Belting, M. (2014). Heparan sulfate proteoglycan as a cell-surface endocytosis receptor. Matrix Biol. 35, 51–55. doi: 10.1016/j.matbio.2013.10.004
Ciano, K. A., Saredy, J. J., and Bowers, D. F. (2014). Heparan sulfate proteoglycan: An arbovirus attachment factor integral to mosquito salivary gland ducts. Viruses 6, 5182–5197. doi: 10.3390/v6125182
Conesa, A., and Götz, S. (2008). Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008:619832. doi: 10.1155/2008/619832
Cooley, L. F., El Shikh, M. E., Li, W., Keim, R. C., Zhang, Z., Strauss, J. F., et al. (2016). Impaired immunological synapse in sperm associated antigen 6 (SPAG6) deficient mice. Sci. Rep. 6:25840. doi: 10.1038/srep25840
Cooper, B. (1991). Infestation of the southern yellow thrips Thrips palmi, in vegetables. J. Agri. Soc. Trinidad Tobago 88, 37–38.
Deangelis, J. D., Sether, D. M., and Rossignol, P. A. (1993). Survival, development, and reproduction in western flower thrips (Thysanoptera: Thripidae) exposed to impatiens necrotic spot virus. Environ. Entomol. 22, 1308–1312. doi: 10.1093/ee/22.6.1308
Dhall, H., Jangra, S., Basavaraj, Y. B., and Ghosh, A. (2021). Hosts plant influences life cycle, reproduction, feeding, and vector competence of Thrips palmi (Thysanoptera: Thripidae), a vector of tospoviruses. Phytoparasitica 49, 501–512. doi: 10.1007/s12600-021-00893-0
Dores, M. R., Lin, H., Grimsey, N., Mendez, F., and Trejo, J. (2015). The α-arrestin ARRDC3 mediates ALIX ubiquitination and G protein-coupled receptor lysosomal sorting. Mol. Biol. Cell 26, 4660–4673. doi: 10.1091/mbc.E15-05-0284
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 5, 294–299.
Ghosh, A., Basavaraj, Y. B., Jangra, S., and Das, A. (2019). Exposure to watermelon bud necrosis virus and groundnut bud necrosis virus alters the life history traits of their vector, Thrips palmi (Thysanoptera: Thripidae). Arch. Virol. 164, 2799–2804. doi: 10.1007/s00705-019-04381-z
Ghosh, A., Dey, D., Mandal, B., and Jain, R. K. (2017). “Thrips as the vectors of tospoviruses in Indian agriculture,” in A Century of Plant Virology in India. eds. Mandal, B., Rao, G., Baranwal, V., and Jain, R. (Singapore: Springer), 537–561.
Ghosh, A., Jagdale, S. S., Basavaraj, Y. B., Dietzgen, R. G., and Jain, R. K. (2020). Genetics of Thrips palmi (Thysanoptera: Thripidae). J. Pest. Sci. 93, 27–39. doi: 10.1007/s10340-019-01160-2
Ghosh, A., Priti,, Mandal, B., and Dietzgen, R. G. (2021). Progression of watermelon bud necrosis virus infection in its vector, Thrips palmi. Cell 10:392. doi: 10.3390/cells10020392
Girard, Y. A., Mayhew, G. F., Fuchs, J. F., Li, H., Schneider, B. S., McGee, C. E., et al. (2010). Transcriptome changes in Culex quinquefasciatus (Diptera: Culicidae) salivary glands during West Nile virus infection. J. Med. Entomol. 47, 421–435. doi: 10.1093/jmedent/47.3.421
González-Santoyo, I., and Córdoba-Aguilar, A. (2012). Phenoloxidase: a key component of the insect immune system. Entomol. Exp. Appl. 142, 1–16. doi: 10.1111/j.1570-7458.2011.01187.x
Gorovits, R., and Czosnek, H. (2013). “Insect symbiotic bacterial GroEL (chaperonin 60) and plant virus transmission,” in Moonlighting Cell Stress Proteins in Microbial Infections. Heat Shock Proteins. Vol. 7. ed. B. Henderson (Dordrecht: Springer), 173–187.
Gorovits, R., and Czosnek, H. (2017). The involvement of heat shock proteins in the establishment of tomato yellow leaf curl virus infection. Front. Plant Sci. 8:355. doi: 10.3389/fpls.2017.00355
Götz, M., Popovski, S., Kollenberg, M., Gorovits, R., Brown, J. K., Cicero, J. M., et al. (2012). Implication of Bemisia tabaci heat shock protein 70 in Begomovirus-whitefly interactions. J. Virol. 86, 13241–13252. doi: 10.1128/JVI.00880-12
Guo, S. K., Cao, L. J., Song, W., Shi, P., Gao, Y. F., Gong, Y. J., et al. (2020). Chromosome-level assembly of the melon thrips genome yields insights into evolution of a sap-sucking lifestyle and pesticide resistance. Mol. Ecol. Resour. 20, 1110–1125. doi: 10.1111/1755-0998.13189
Guyot, J. (1998). Review and first observation in Guadeloupe on Thrips Palmi Karny. Agronomie 8, 565–575. doi: 10.1051/agro:19880701
Han, J., and Rotenberg, D. (2021). Integration of transcriptomics and network analysis reveals co-expressed genes in Frankliniella occidentalis larval guts that respond to tomato spotted wilt virus infection. BMC Genomics 22:810. doi: 10.1186/s12864-021-08100-4
Hattori, M., Tsuchihara, K., Noda, H., Konishi, H., Tamura, Y., Shinoda, T., et al. (2010). Molecular characterization and expression of laccase genes in the salivary glands of the green rice leafhopper, Nephotettix cincticeps (Hemiptera: Cicadellidae). Insect Biochem. Mol. Biol. 40, 331–338. doi: 10.1016/j.ibmb.2010.02.009
Hover, S., King, B., Hall, B., Loundras, E. A., Taqi, H., Daly, J., et al. (2016). Modulation of potassium channels inhibits bunyavirus infection. J. Biol. Chem. 291, 3411–3422. doi: 10.1074/jbc.M115.692673
Ingwell, L. L., Eigenbrode, S. D., and Bosque-Pérez, N. A. (2012). Plant viruses alter insect behavior to enhance their spread. Sci. Rep. 2:578. doi: 10.1038/srep00578
Jagdale, S. S., and Ghosh, A. (2019). In silico analyses of molecular interactions between groundnut bud necrosis virus and its vector, Thrips palmi. Virus Dis. 30, 245–251. doi: 10.1007/s13337-019-00521-w
Jangra, S., Mittal, A., Dhall, H., Jain, R. K., and Ghosh, A. (2020). A multiplex PCR assay for rapid identification of major tospovirus vectors reported in India. BMC Genomics 21:170. doi: 10.1186/s12864-020-6560-x
Janusz, G., Pawlik, A., Świderska-Burek, U., Polak, J., Sulej, J., Jarosz-Wilkołazka, A., et al. (2020). Laccase properties, physiological functions, and evolution. Int. J. Mol. Sci. 21:966. doi: 10.3390/ijms21030966
Johnson, M. W. (1986). Population trends of a newly introduced species, Thrips palmi (Thysanoptera: Thripidae), on commercial watermelon plantings in Hawaii. J. Econ. Entomol. 79, 718–720. doi: 10.1093/jee/79.3.718
Kato, N., Mueller, C. R., Fuchs, J. F., McElroy, K., Wessely, V., Higgs, S., et al. (2008). Evaluation of the function of a type I peritrophic matrix as a physical barrier for midgut epithelium invasion by mosquito-borne pathogens in Aedes aegypti. Vector Borne Zoonotic Dis. 8, 701–712. doi: 10.1089/vbz.2007.0270
Kawai, A. (2001). Population management of Thrips palmi Karny. Jpn. J. Appl. Entomol. Zool. 45, 39–59. doi: 10.1303/jjaez.2001.39
Kumar, N., Liang, Y., Parslow, T. G., and Liang, Y. (2011). Receptor tyrosine kinase inhibitors block multiple steps of influenza A virus replication. J. Virol. 85, 2818–2827. doi: 10.1128/JVI.01969-10
Kunkalikar, S. R., Poojari, S., Arun, B. M., Rajagopalan, P. A., Chen, T. C., Yeh, S. D., et al. (2011). Importance and genetic diversity of vegetable-infecting tospoviruses in India. Phytopathology 101, 367–376. doi: 10.1094/PHYTO-02-10-0046
Levashina, E. A., Langley, E., Green, C., Gubb, D., Ashburner, M., Hoffmann, J. A., et al. (1999). Constitutive activation of toll-mediated antifungal defense in serpin-deficient drosophila. Science 285, 1917–1919. doi: 10.1126/science.285.5435.1917
Levy, S. M., Falleiros, A. M. F., Moscardi, F., and Gregório, E. A. (2011). The role of peritrophic membrane in the resistance of Anticarsia Gemmatalis larvae (Lepidoptera: Noctuidae) during the infection by its nucleopolyhedrovirus (AgMNPV). Arthropod. Struct. Dev. 40, 429–434. doi: 10.1016/j.asd.2011.05.003
Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [Preprint]. doi: 10.6084/M9.FIGSHARE.963153.V1
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, A., Disoma, C., Zhou, Y., Chen, Z., Zhang, L., Shen, Y., et al. (2019). Protein interactome of the deamidase phosphoribosylformylglycinamidine synthetase (PFAS) by LC-MS/MS. Biochem. Biophys. Res. Commun. 513, 746–752. doi: 10.1016/j.bbrc.2019.04.039
Martin, J. H., Benzer, S., Rudnicka, M., and Miller, C. A. (1993). Calphotin: a Drosophila photoreceptor cell calcium-binding protein. Proc. Natl. Acad. Sci. 90, 1531–1535. doi: 10.1073/pnas.90.4.1531
Monteiro, R. C., Zucchi, R. A., and Mound, L. A. (1995). Record of Thrips palmi Karny, 1925 (Thysanoptera, Thripidae) in the state of São Paulo, Brazil. Revue de l’Agri. 70, 53–55.
Mou, D. F., Chen, W. T., Li, W. H., Chen, T. C., Tseng, C. H., Huang, L. H., et al. (2021). Transmission mode of watermelon silver mottle virus by Thrips palmi. PLoS One 16:e0247500. doi: 10.1371/journal.pone.0247500
Ohnishi, J., Knight, L. M., Hosokawa, D., Fujisawa, I., and Tsuda, S. (2001). Replication of tomato spotted wilt virus after ingestion by adult Thrips setosus is restricted to midgut epithelial cells. Phytopathology 91, 1149–1155. doi: 10.1094/PHYTO.2001.91.12.1149
Oliver, J. E., and Whitfield, A. E. (2016). The genus Tospovirus: emerging bunyaviruses that threaten food security. Annu. Rev. Virol. 3, 101–124. doi: 10.1146/annurev-virology-100114-055036
O’Neal, S. T., Swale, D. R., and Anderson, T. D. (2017). ATP-sensitive inwardly rectifying potassium channel regulation of viral infections in honey bees. Sci. Rep. 7:8668. doi: 10.1038/s41598-017-09448-y
Palmer, J. M. (1992). Thrips (Thysanoptera) from Pakistan to the Pacific: a review. Bull. Br. Mus. Nat. Hist. Entomol. Ser. 61, 1–76.
Pantoja, A., Segarra, A., Ruiz, H., and Medina-Gaud, S. (1988). Thrips palmi (Thysanoptera: Thripidae): a new insect pest for Puerto Rico. J. Agric. Univ. P. R. 2, 327–329.
Pappu, H. R., Jones, R. A. C., and Jain, R. K. (2009). Global status of Tospovirus epidemics in diverse cropping systems: successes achieved and challenges ahead. Virus Res. 141, 219–236. doi: 10.1016/j.virusres.2009.01.009
Parisi, L. R., Li, N., and Atilla-Gokcumen, G. E. (2017). Very long-chain fatty acids are functionally involved in necroptosis. Cell Chem. Biol. 24, 1445.e8–1454.e8. doi: 10.1016/j.chembiol.2017.08.026
Patwari, P., and Lee, R. T. (2012). An expanded family of arrestins regulate metabolism. Trends Endocrinol. Metab. 23, 216–222. doi: 10.1016/j.tem.2012.03.003
Qin, X.-S., Tsukaguchi, H., Shono, A., Yamamoto, A., Kurihara, H., and Doi, T. (2009). Phosphorylation of Nephrin triggers its internalization by raft-mediated endocytosis. JASN 20, 2534–2545. doi: 10.1681/ASN.2009010011
Riley, D. G., Joseph, S. V., Srinivasan, R., and Diffie, S. (2011). Thrips vectors of tospoviruses. J. Integr. Pest Manag. 2, I1–I10. doi: 10.1603/IPM10020
Rosen, R., Lebedev, G., Kontsedalov, S., Ben-Yakir, D., and Ghanim, M. A. (2021). De novo transcriptomics approach reveals genes involved in Thrips Tabaci resistance to spinosad. Insects 12:67. doi: 10.3390/insects12010067
Rotenberg, D., Jacobson, A. L., Schneweis, D. J., and Whitfield, A. E. (2015). Thrips transmission of tospoviruses. Curr. Opin. Virol. 15, 80–89. doi: 10.1016/j.coviro.2015.08.003
Schneweis, D. J., Whitfield, A. E., and Rotenberg, D. (2017). Thrips developmental stage-specific transcriptome response to tomato spotted wilt virus during the virus infection cycle in Frankliniella occidentalis, the primary vector. Virology 500, 226–237. doi: 10.1016/j.virol.2016.10.009
Sette, P., Jadwin, J. A., Dussupt, V., Bello, N. F., and Bouamr, F. (2010). The ESCRT-associated protein Alix recruits the ubiquitin ligase Nedd4-1 to facilitate HIV-1 release through the LYPXnL L domain motif. J. Virol. 84, 8181–8192. doi: 10.1128/JVI.00634-10
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shrestha, A., Champagne, D. E., Culbreath, A. K., Rotenberg, D., Whitfield, A. E., and Srinivasan, R. (2017). Transcriptome changes associated with tomato spotted wilt virus infection in various life stages of its thrips vector, Frankliniella fusca (hinds). J. Gen. Virol. 98, 2156–2170. doi: 10.1099/jgv.0.000874
Shrestha, A., Srinivasan, R., Riley, D. G., and Culbreath, A. K. (2012). Direct and indirect effects of a thrips-transmitted Tospovirus on the preference and fitness of its vector, Frankliniella fusca. Entomol. Exp. Appl. 145, 260–271. doi: 10.1111/eea.12011
Singh, A. B., and Srivastava, S. K. (1995). “Status and control strategy of peanut bud necrosis disease in Uttar Pradesh.” in Recent Studies on Peanut Bud Necrosis Disease: Proceedings of a Meeting. March 20, 1995; Patancheru, Andhra Pradesh, India: International Crop Research Institute for Semi-Arid Tropics, 65–68.
Singh, R. B., Srivastava, K. K., Pandey, S. K., and Khurana, S. M. (1997). Assessment of yield losses due to potato stem necrosis disease. Indian J. Virol. 13, 135–137.
Spier, A., Stavru, F., and Cossart, P. (2019). Interaction between intracellular bacterial pathogens and host cell mitochondria. Microbiol. Spectr. 7:10. doi: 10.1128/microbiolspec.BAI-0016-2019
Stafford-Banks, C. A., Rotenberg, D., Johnson, B. R., Whitfield, A. E., and Ullman, D. E. (2014). Analysis of the salivary gland transcriptome of Frankliniella occidentalis. PLoS One 9:e94447. doi: 10.1371/journal.pone.0094447
Stewart, C. M., Phan, A., Bo, Y., LeBlond, N. D., Smith, T. K. T., Laroche, G., et al. (2021). Ebola virus triggers receptor tyrosine kinase-dependent signaling to promote the delivery of viral particles to entry-conducive intracellular compartments. PLoS Pathog. 17:e1009275. doi: 10.1371/journal.ppat.1009275
Suzuki, M., Danilchanka, O., and Mekalanos, J. J. (2014). Vibrio cholerae T3SS effector VopE modulates mitochondrial dynamics and innate immune signaling by targeting Miro GTPases. Cell Host Microbe 16, 581–591. doi: 10.1016/j.chom.2014.09.015
Ullman, D., Cho, J. J., Mau, R. F. L., Hunter, W. B., Westcot, D. M., and Custer, D. M. (1992). “Thrips-tomato spotted wilt virus interactions: morphological, behavioral and cellular components influencing thrips transmission,” in Advances in Disease Vector Research 9. ed. Harris, K. F. (New York: Springer), 195–240.
Ullman, D., Meideros, R., Campbell, L. R., Whitfield, A. E., Sherwood, J. L., and German, T. L. (2002). Thrips as vectors of tospoviruses. Adv. Bot. Res. 36, 113–140. doi: 10.1016/S0065-2296(02)36061-0
Ullman, D., Sherwood, J., and German, T. L. (1997). “Thrips as vectors of plant pathogens,” in Thrips as Crop Pests. ed. Lewis, T. (Wallingford, UK: CAB International), 539–565.
Walker, A. K. (1992). “Pest status,” in Thrips palmi: A Literature Survey, International Institue of Biological Control. ed. Girling, D. J. (Ascot, Berks, UK: International Institute of Biological Control), 3–6.
Wetering, F., Goldbach, R., and Peters, D. (1996). Tomato spotted wilt tospovirus ingestion by first instar larvae of Frankliniella occidentalis is a prerequisite for transmission. Phytopathology 86, 900–905. doi: 10.1094/Phyto-86-900
Whitfield, A. E., Ullman, D. E., and German, T. L. (2005). Tospovirus-thrips interactions. Annu. Rev. Phytopathol. 43, 459–489. doi: 10.1146/annurev.phyto.43.040204.140017
Gamage, S. M. K. W., Rotenberg, D., Schneweis, D. J., Tsai, C. W., and Dietzgen, R. G. (2018). Transcriptome-wide responses of adult melon thrips (Thrips palmi) associated with capsicum chlorosis virus infection. PLoS One 13:e0208538. doi: 10.1371/journal.pone.0208538
Wijkamp, I., Goldbach, R., and Peters, D. (1996). Propagation of tomato spotted wilt virus in Frankliniella occidentalis does neither result in pathological effects nor in transovarial passage of the virus. Entomol. Exp. Appl. 81, 285–292. doi: 10.1046/j.1570-7458.1996.00098.x
Yu, C. Y., Liang, J. J., Li, J. K., Lee, Y. L., Chang, B. L., Su, C. I., et al. (2015). Dengue virus impairs mitochondrial fusion by cleaving mitofusins. PLoS Pathog. 11:e1005350. doi: 10.1371/journal.ppat.1005350
Zhang, Y., Schöttker, B., Florath, I., Stock, C., Butterbach, K., Holleczek, B., et al. (2016). Smoking-associated DNA methylation biomarkers and their predictive value for all-cause and cardiovascular mortality. Environ. Health Perspect. 124, 67–74. doi: 10.1289/ehp.1409020
Zhang, Z., Zhang, P., Li, W., Zhang, J., Huang, F., Yang, J., et al. (2013). De novo transcriptome sequencing in Frankliniella occidentalis to identify genes involved in plant virus transmission and insecticide resistance. Genomics 101, 296–305. doi: 10.1016/j.ygeno.2013.02.005
Zhao, L., and Jones, W. A. (2012). Expression of heat shock protein genes in insect stress responses. Invertebr. Surviv. J. 9, 93–101.
Keywords: melon thrips, tospovirus, virus–vector relationship, RT-qPCR, transcriptome
Citation: Mahanta DK, Jangra S, Priti, Ghosh A, Sharma PK, Iquebal MA, Jaiswal S, Baranwal VK, Kalia VK and Chander S (2022) Groundnut Bud Necrosis Virus Modulates the Expression of Innate Immune, Endocytosis, and Cuticle Development-Associated Genes to Circulate and Propagate in Its Vector, Thrips palmi. Front. Microbiol. 13:773238. doi: 10.3389/fmicb.2022.773238
Edited by:
Luca Nerva, Council for Agricultural and Economics Research (CREA), ItalyReviewed by:
Aleksandra Obrępalska-Stęplowska, National Research Institute, PolandPunya Nachappa, Colorado State University, United States
Copyright © 2022 Mahanta, Jangra, Priti, Ghosh, Sharma, Iquebal, Jaiswal, Baranwal, Kalia and Chander. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amalendu Ghosh, YW1hbDRlbnRvQGdtYWlsLmNvbQ==; YW1hbGVuZHUuZ2hvc2hAaWNhci5nb3YuaW4= orcid.org/0000-0001-6634-5771
 Deepak Kumar Mahanta1,2
Deepak Kumar Mahanta1,2 Sumit Jangra
Sumit Jangra Priti
Priti Amalendu Ghosh
Amalendu Ghosh Parva Kumar Sharma
Parva Kumar Sharma Mir Asif Iquebal
Mir Asif Iquebal Sarika Jaiswal
Sarika Jaiswal Vinay Kumari Kalia
Vinay Kumari Kalia