- 1Key Laboratory of Coastal Biology and Biological Resource Conservation, Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences, Yantai, China
- 2College of Environmental Science and Engineering, Ocean University of China, Qingdao, China
- 3CAS Key Laboratory for Experimental Study Under Deep-Sea Extreme Conditions, Institute of Deep-Sea Science and Engineering, Chinese Academy of Sciences, Sanya, China
- 4University of Chinese Academy of Sciences, Beijing, China
- 5College of Marine Life Science, Ocean University of China, Qingdao, China
- 6Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao, China
Marine Synechococcus, a significant contributor to primary production, shows high phylogenetic diversity. However, studies on its phylogenetic composition in the Bohai Sea, the largest continental sea in China, are lacking. We sequenced rpoC1 (encodes the RNA polymerase β’ subunit protein) in samples from the Laizhou Bay (LZB) and Bohai Strait (BS) in June and November using high-throughput sequencing to reveal the phylogenetic composition of Synechococcus assemblages in the bay and strait areas of the Bohai Sea. In total, 12 lineages representing Synechococcus subclusters S5.1, S5.2, and S5.3 were identified. Spatially, clade I was obligately dominant in BS. In contrast, the Synechococcus assemblage in LZB was more diverse, with clades VI and III being highly abundant. In addition, we detected strong variation in Synechococcus structure between June and November in the Bohai Sea. Clades II, III, XX, and miyav were only detected in November. Vertically, variation in Synechococcus assemblage was not apparent among the water layers probably due to the shallow water depth with intense water mixing. Results of redundancy analysis (RDA) and random forest (RF) analysis together highlighted the key role of silicate in the Synechococcus assemblage. Our results suggested that the Bohai Sea provides various niches for different Synechococcus clades, resulting in a special phylogenetic composition of the Synechococcus assemblage, compared with that in the adjacent shelf sea and other continental seas in the world.
Highlights
–Synechococcus composition varied spatially between Laizhou Bay (LZB) and Bohai Strait (BS) in Bohai Sea.
–Synechococcus clades II, III, XX, and miyav were detected only in November.
–Vertical variation in Synechococcus assemblage was not apparent.
–Silicate played a key role in the Synechococcus assemblage in this continental sea.
Introduction
Marine Synechococcus, a widely distributed autotrophic picoplanktonic organism, plays a key role in the euphotic zone of global oceans in terms of primary production and carbon fluxes (Flombaum et al., 2013; Paulsen et al., 2016). The phylogenetic diversity of Synechococcus has been described using a range of conserved markers, such as 16S rRNA (Dufresne et al., 2008), ITS (16S-23S internally transcribed spacer; Ahlgren and Rocap, 2006; Huang et al., 2012), rpoC1 (subunit of RNA polymerase; Xia et al., 2017a), ntcA (a nitrogen regulatory gene; Penno et al., 2006), and petB (cytochrome b6) genes (Mazard et al., 2012). Among these gene markers, housekeeping gene of rpoC1 is single copy in all known Synechococcus genomes and has higher genetic resolution than the 16S rRNA gene (Mühling et al., 2006), so it has been widely used in studies involving the genetic diversity of Synechococcus (Xia et al., 2017a). So far, marine Synechococcus is classified into three subclusters: S5.1, S5.2, and S5.3 (Ahlgren and Rocap, 2012). In each subcluster, various clades can be further distinguished. S5.1 shows the highest phylogenetic diversity, consisting of at least 20 clades including clades I to XVI, CRD1/2, ENV-B/C, and so on (Choi et al., 2013a).
Studies targeting the global distribution of Synechococcus assemblages have revealed clear spatial partitioning for the main Synechococcus lineages (Farrant et al., 2016; Sohm et al., 2016; Xia et al., 2019; Li et al., 2021). The current notion regarding the biogeography of Synechococcus lineages indicates that S5.1 clades I and IV are most abundant in colder and eutrophic waters (Kent et al., 2019). S5.1 clades II, III, V, VI, and VII appear frequently in tropical and subtropical waters, while clades CRD1 and ENV-B/C dominate upwelling zones with low iron content (Zwirglmaier et al., 2008; Post et al., 2011; Sohm et al., 2016). In addition, the abundance and structure of Synechococcus assemblages also change with seasons (Tai and Palenik, 2009; Tai et al., 2011). In the coastal Pacific Ocean, for example, Synechococcus clade IV was prevalent throughout the year, while clades II and III appeared only in late summer and winter (Tai and Palenik, 2009).
The phylogenetic diversity of the Synechococcus community has been well studied in the South China Sea and East China Sea (Ma et al., 2004; Choi and Noh, 2009; Jing and Liu, 2012; Choi et al., 2013b; Chung et al., 2015). Furthermore, it has been studied in subtropical estuarine waters and coastal waters, such as Hong Kong estuary and coast, Pearl River estuary, and Changjiang River estuary (Jing et al., 2009b; Chung et al., 2015; Xia et al., 2015, 2017a). However, relevant studies in sea areas at higher latitudes in China are scarce (Li et al., 2021). In particular, little is known about the community composition of, and environment driven by, Synechococcus assemblages in the Bohai Sea, the largest continental sea of China. In fact, a few studies have focused on the population structure of Synechococcus in the continental seas in the world. So far, such studies have been conducted in limited continental seas, such as the Sea of Okhotsk (Jing et al., 2009a), Mediterranean Sea (Mella-Flores et al., 2011), and Red Sea (Fuller et al., 2005). The hydrologic characteristics of these regions are significantly influenced by continents, including the input of continental rivers, the human activity along the coasts, etc (Wang et al., 2018a; Temino-Boes et al., 2019). It is of great significance to investigate the phylogenetic composition and niche division of Synechococcus assemblages in these sea areas with anthropic environmental conditions.
Bohai Sea enjoys monsoon climate of medium latitudes with clear seasonal climate change. It is connected to the Yellow Sea only through the Bohai Strait. Serious anthropogenic interference and bad water quality exchange have resulted in poor water conditions in the Bohai Sea (Zhang et al., 2015). The bay areas, especially the Laizhou Bay, are polluted because of the discharge of large amounts of industrial wastewaters, domestic sewage, and wastewater from animal raising industries (Lü et al., 2015). The inflow of continental rivers, especially the Yellow River (the second largest river in China), also influences the biology in these areas. In contrast, BS has better water quality conditions owing to water exchange with the northern Yellow Sea. Synechococcus generally dominated the total cell abundance in most seasons in the above regions (Yan et al., 2020; Wang et al., 2021). However, the phylogenetic diversity of Synechococcus in this region has not been investigated. We collected samples from the Laizhou Bay and Bohai Strait of Bohai Sea in early summer and winter to investigate the population composition and effect of the environment on Synechococcus clades based on rpoC1 high-throughput sequencing, which is of great importance for understanding the niche division and population shift of Synechococcus assemblages seriously affected by human activities.
Materials and Methods
Sampling
Field sampling was conducted at stations LZB and BS in the Bohai Sea onboard the Research Vessel “Chuangxin I” in June and November 2019 (Table 1; Supplementary Figure S1). Station LZB is located in Laizhou Bay, which is seriously affected and polluted by human activities and the inflow of continental rivers. In comparison, station BS located in Bohai Strait has water exchanges with the North Yellow Sea, and the water quality is better. At each station, samples were collected from three or four standard depths according to the specifications for the oceanographic survey (National Standard of China, GB/T 12763-2007). Seawater was collected in Niskin bottles carried by a CTD rosette sampler (Sea-Bird 911Plus, Sea-Bird Electronics Inc., Bellevue, WA, United States). For DNA extraction, 1,000 ml of seawater was sequentially filtered through 48-μm nylon mesh (to remove microphytoplankton) and 0.22-μm polycarbonate filters (Millipore Co., Bedford, MA, United States). Then, the collected filters were flash frozen in liquid nitrogen and stored at −80°C until further analysis.
Measurement of Environmental Variables
Temperature, salinity, depth, pH, turbidity, dissolved oxygen, and concentration of chlorophyll a (chl. a) were measured in situ using the CTD (Sea-Bird 911Plus, Sea-Bird Electronics Inc., Bellevue, WA, United States). The nutrient concentrations, including nitrite (NO2−), nitrate (NO3−), ammonium (NH4+), phosphate (PO43−), and silicate (SiO44−) were measured using standard colorimetric methods with an AA3 segmented flow analyzer (Seal Analytical GmbH, Germany; Dafner, 2015).
DNA Extraction, PCR, and Sequencing
Each polycarbonate membrane filter was disrupted by bead-beating method and then taken for total genomic DNA extraction using the Fast DNA SPIN Kit (MP BIO, United States; Li et al., 2021). The DNA concentrations were determined using a Nano Drop 2000c spectrophotometer (Thermo Fisher, United States). PCR was performed in the thermocycler GeneAmp® PCR system 9700 (Perkin Elmer, United States) in triplicate (Li et al., 2019) with primers rpoC1-39F (5′-adaptor+barcode+GGNATYGTYTGYGAGCGYTG-3′) and rpoC1-462R (5′-adaptor+CGYAGRCGCTTGRTCAGCTT-3′; Mühling et al., 2006). All reactions were performed in 20 μl containing 4 μl of 5× FastPfu Buffer, 2 μl of 2.5 mM dNTPs, 0.8 μl of 5 μM forward and reverse primers, 0.4 μl of FastPfu Polymerase, 0.2 μl of BSA, 10 ng template DNA, and ddH2O. Thermal cycling protocol was as follows: following an initial denaturation step of 95°C for 3 min, 37 cycles at 95°C for 30 s, 53°C for 30 s, and 72°C for 45 s, followed by a final extension step at 72°C for 10 min. The triplicate amplified products were combined in one, and then it was gel-purified using the Qiaquick gel purification kit (Qiagen, Hilgen, Germany) as described by the manufacturer. The purified amplicon was roughly quantified with agarose gel imaging by QuantiFluor-ST Fluorometers (Promega, Madison, WI, United States). The amplicons obtained were sequenced by the Majorbio Co., Ltd. (Shanghai, China) using Illumina MiSeq sequencing platform (PE300).
Processing of High-Throughput Sequencing Data
QIIME2 was used to process raw sequence profiles as previously described with several modifications (Caporaso et al., 2010). First, the raw data were quality controlled to retain high-quality reads with criterions of Phred quality score > 20, no ambiguous bases, and consecutive high-quality bases >80% of total read length (Brown et al., 2017). USEARCH was used to filter chimeras (Edgar et al., 2011). Then operational taxonomic units (OTUs) were calculated at 95% similarity. OTU representative sequences were first identified using Basic Local Alignment Search Tool (BLAST) against the nt database, i.e., partially non-redundant nucleotide sequences National Center for Biotechnology Information (NCBI) database (Zhang et al., 2000), and those that do not belong to cyanobacteria are discarded. Representative sequences with relative abundance > 0.1% were extracted and aligned with the reference sequences. Modeltest and maximum likelihood phylogenetic tree construction were performed using Mega 7.014 with the model GTR + G + I and 200 bootstraps (Xia et al., 2017a). All sequences obtained from this study have been deposited in the NCBI Sequence Read Archive (SRA) under the accession number PRJNA753275.
Statistical Analysis and Visualization
Ocean Data View (ODV v. 5.1.7) was used to sketch the map of sampling stations (Schlitzer, 2002). The package corrplot in R Language v. 3.6.1 was used to calculate Spearman’s correlation (Tai and Palenik, 2009). A p-value less than 0.05 was considered statistically significant. The package pheatmap in R Language v. 3.6.1 was used to draw the heatmaps with sample clustering. The package vegan in R Language v. 3.6.1 was used to calculate alpha diversity indices, beta diversity distance, as well as redundancy analysis (RDA; Dixon, 2003). Statistical Product and Service Solutions (SPSS v. 17.0) was used to test the significance of differences in alpha diversity indices among samples at different stations and in different months through ANOVA with a post hoc test (Tukey test; George, 2011). A p-value less than 0.05 was considered statistically significant. The principal coordinate analysis (PCoA) ordination was conducted at the OTU level using the package vegan in R Language v. 3.6.1 (Dixon, 2003). Dissimilarity in community structure among samples at different stations and in different months was assessed with permutational multivariate ANOVA (PERMANOVA) using 999 permutations for significance testing (Anderson, 2017). Before RDA, environmental variables with variance inflation factor (VIF) more than 10 were filtered out. VIF is an indicator that tests the multicollinearity of regression analyses (Mansfield and Helms, 1982). VIF more than 10 represents that the corresponding variables provide little independent explanatory ability and need to be deleted in the calculation of regression analyses (Franke, 2010). The package randomForest in R Language v. 3.6.1 was used to conduct random forest (RF) analysis (Liaw and Wiener, 2002). The package psych in R Language v. 3.6.1 was used to characterize properties of co-occurrence networks (Revelle and Revelle, 2015). Then Gephi v0.9.2 was used to draw the network (Bastian et al., 2009). The hubba (key) score in each network was calculated using a plugin, CytoHubba, in the Betweenness method in software Cytoscape v3.8.0 (Chin et al., 2014). Finally, the package basicTrendline in R Language v. 3.6.1 was used to conduct linear ordinary least square (OLS) regression (Mei et al., 2018). OLS regression is a statistical method that estimates the relationship between the variables by minimizing the sum of squared differences between the observed and predicted values of the dependent variable (Hutcheson, 1999).
Results
Seawater Environmental Conditions
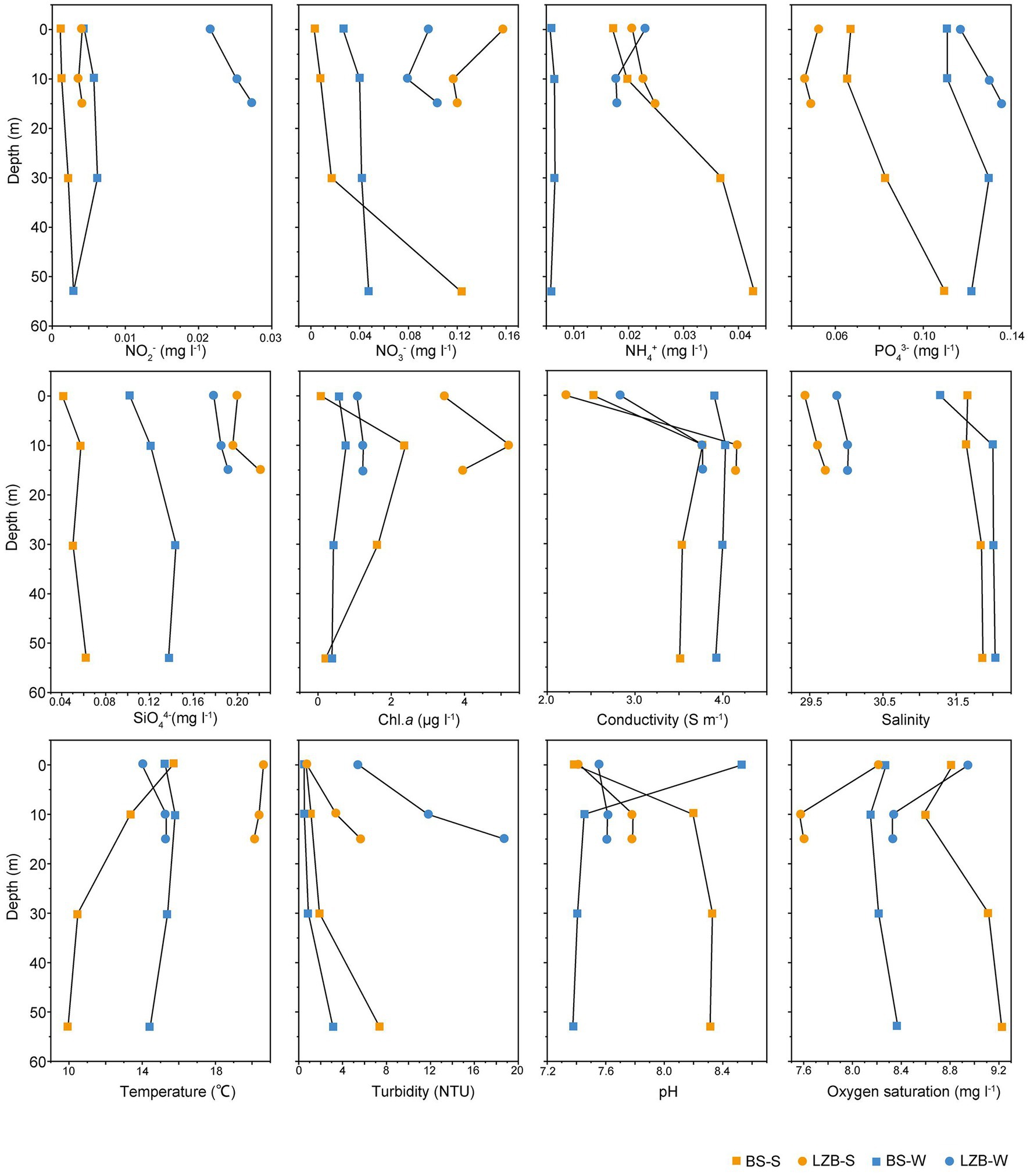
Environmental parameters of stations LZB and BS in June and November are shown in Figure 1. The highest mean values of nitrate (NO3−), silicate (SiO44−), chl. a, and temperature were detected in LZB-S, i.e., samples at station LZB in summer (June). In contrast, the highest mean values of nitrite (NO2−), phosphate, and (PO43−) concentrations, and turbidity appeared in LZB-W, i.e., samples at LZB in winter (November). The salinity was high in both June and November at the BS station. Hierarchical clustering of samples in the heatmap based on scaled environmental data showed that samples at station BS were separated from those at LZB (Supplementary Figure S2). Besides, the environmental condition between samples in June and November was usually different, no matter what station, BS or LZB. However, the environmental condition of samples in different water layers at the same stations and in the same month was very similar.
There was no significant correlation between environmental variables and water depth, indicating good water mixing among water layers (Supplementary Figure S3). However, multiple significant correlations were observed among the variables. In particular, the concentration of SiO44− significantly correlated with six variables, including the concentration of NO2− (ρ = 0.68, p < 0.01) and NO3− (ρ = 0.67, p < 0.01), temperature (ρ = 0.6, p < 0.05), turbidity (ρ = 0.55, p < 0.05), salinity (ρ = −0.79, p < 0.01), and oxygen saturation (ρ = −0.59, p < 0.01).
Community Characteristics of Synechococcus Assemblages
High-throughput sequencing of rpoC1 generated 431,129 quality sequences from 14 samples (Table 2). The Good’s coverage for the observed OTUs was >99%, indicating a near-complete sampling of the community. The Shannon diversity index ranged from 0.60 to 2.28, with an average of 1.81, and the Chao1 estimator ranged from 17 to 68, with an average of 41. On average, community diversity, represented by the Shannon diversity index and community richness represented by the Chao1 estimator of Synechococcus assemblage, were higher in winter than that in summer. Geographically, average community diversity and richness of Synechococcus assemblage were higher in LZB than in BS. Among all variables, only pH showed significant correlation with Shannon diversity index (ρ = −0.78, p < 0.01) and Chao1 estimator (ρ = −0.86, p < 0.01).
The PCoA with the PERMANOVA test demonstrated that along the axis PC1, Synechococcus assemblages in LZB-S were significantly different from those in other samples (p < 0.01; Figure 2). Along the axis PC2, Synechococcus assemblage in BS-S, i.e., samples at station BS in summer (June), and BS-W, i.e., samples at station BS in winter (November), were significantly different from those in LZB-W (p < 0.01). In total, axes PC1 and PC2 explained 63% of the community variation of Synechococcus assemblage.
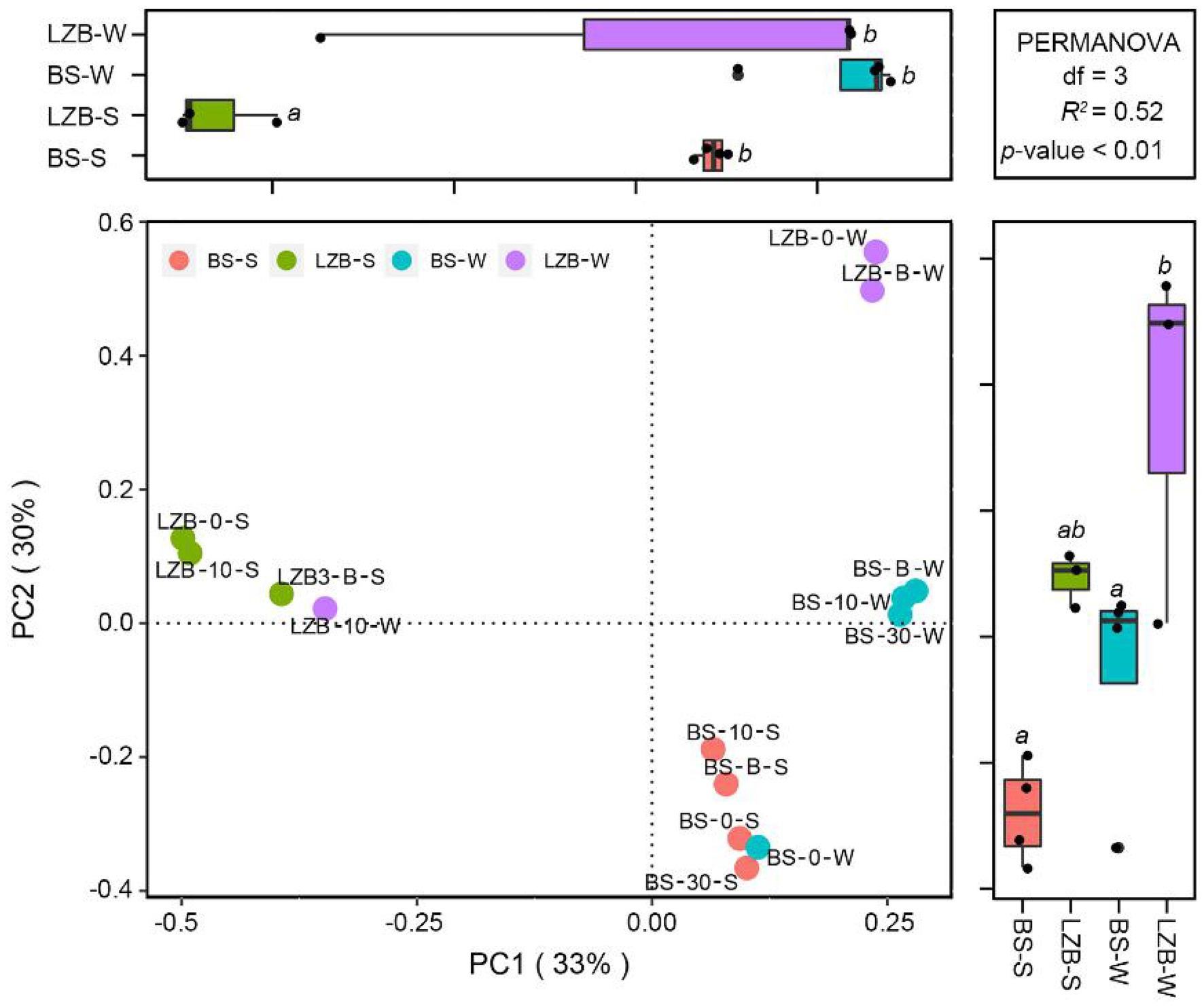
Figure 2. The principal coordinate analysis (PCoA) with permutational multivariate ANOVA (PERMANOVA) test showing the variation in Synechococcus assemblages at operational taxonomic units (OTU) level among samples at different stations and in different months. BS-S, samples at station Bohai Strait (BS) in summer (June); BS-W, samples at station BS in winter (November); LZB-S, samples at station Laizhou Bay (LZB) in summer (June); LZB-W, samples at station LZB in winter (November).
Community Composition of Synechococcus Assemblages
The maximum-likelihood (ML) tree constructed using rpoC1 sequences showed that all Synechococcus subclusters, S5.1, S5.2, and S5.3, were well identified, in which Synechococcus S5.1 was the dominant subcluster and could be further divided into 10 clades (Figure 3A). The percentage of Synechococcus S5.2 and S5.3 in the Synechococcus genotype of the studied area was negligible, accounting for only 0.10 and 1.21% on average, respectively. Among Synechococcus S5.1, clade I was the most preponderant clade, accounting for 60.98% of the clades in all samples. Synechococcus clade VI increased sharply in LZB-S and Synechococcus clade III was abundant in November, especially at depth of 0 m (LZB-0-W) and at the bottom of (LZB-B-W) layers of LZB-W. Synechococcus S5.1 clades I, IV, VI, VIII, IX, and XVI, S5.2, and S5.3 were detected in both months, while Synechococcus S5.1 clades II, III, XX, and miyav were only found in November (Figure 3B). In addition, in BS-S, a mass of Synechococcus clades, including clades II, III, VI, VIII, IX, XVI, XX, and miyav, were absent.
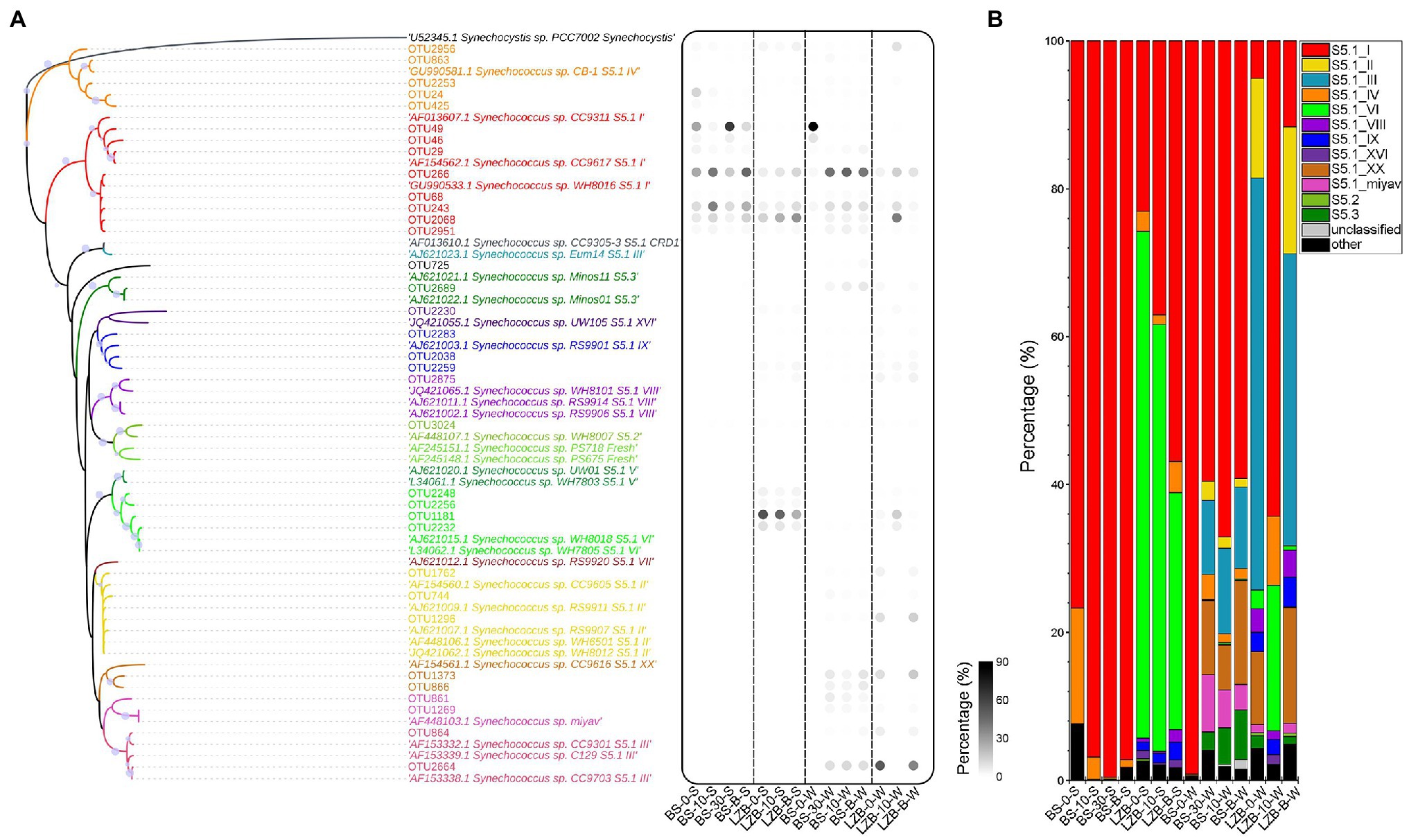
Figure 3. Genomic composition of Synechococcus assemblages. (A) The maximum-likelihood (ML) tree constructed using rpoC1 sequences with relative abundances of more than 0.1%. (B) The relative abundance of Synechococcus clades.
The Co-occurrence Patterns of Synechococcus Genotypes
In the co-occurrence network (Figure 4), Synechococcus S5.1 clades I, II, and III were the most widely correlated, associating with six lineages. The relationships of clade I with other lineages were all negative, while relationships of clades II or III with other lineages were mostly positive. Hubba scores ranked using the Closeness method revealed that the score of these three lineages was the highest, reaching the value of 7, which further demonstrated their importance in Synechococcus assemblage. In contrast, although Synechococcus S5.1 clades XVI and VI correlated significantly with each other, they did not associate with any other lineage. Furthermore, Synechococcus S5.1 clade IV was separated from the network and was not associated with any lineage.
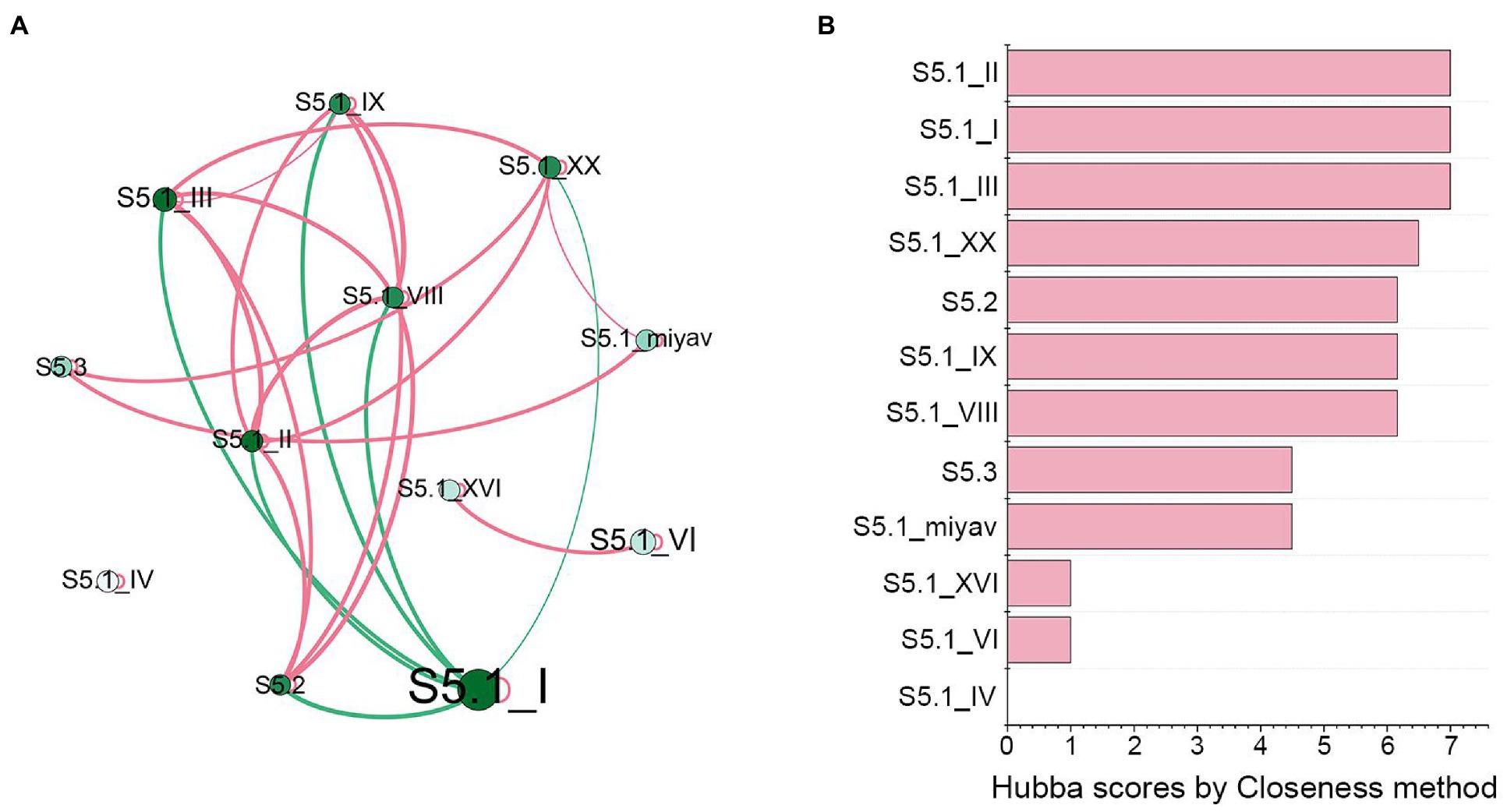
Figure 4. Co-occurrence patterns of Synechococcus genotypes. (A) The co-occurrence network of Synechococcus clades. The size of node represents its average percentage in all samples; the lightness of node represents its degree; the thickness of the edge represents its weight; the red edges represent positive correlations, and the green edges represent negative correlations. (B) Hubba scores ranked by the Closeness method represent the significance of each node.
Environmental Constraints of Synechococcus Assemblages
The analysis of VIF filtered out five environmental variables, including concentrations of NO2− and NO3−, salinity, temperature, and oxygen saturation. The remaining eight factors totally constrained 77.08% community variance of Synechococcus assemblage (p < 0.05; Figure 5). According to the permutation test, concentrations of SiO44−, PO43−, and chl. a were significant factors that correlated with the Synechococcus composition. Among them, SiO44− concentration was the key constraint variable (R2 = 0.80, p < 0.01).
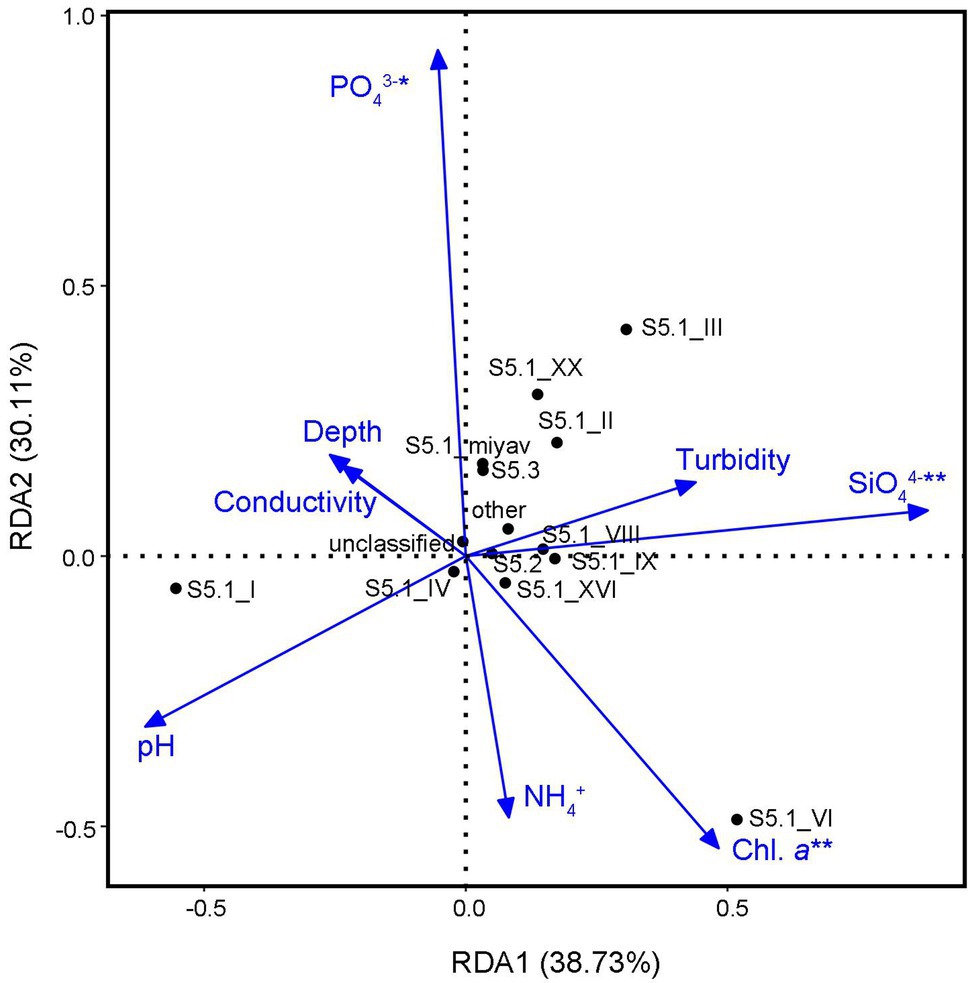
Figure 5. Redundancy analysis (RDA) with the permutation test between Synechococcus clades and environmental variables.
Random Forest Analysis Between Each Synechococcus Clade and Environmental Variables
Random forest analysis revealed that SiO44− concentration was the most significant variable affecting the distribution of six Synechococcus clades, covering S5.1 clade I, clade II, clade III, clade VIII, and clade IX, as well as S5.2 (Figure 6). Salinity was the most significant variable that affected the distribution of four Synechococcus clades, including S5.1 clade VI, clade XVI, and clade miyav, as well as S5.3. The temperature was the most significant variable that affected the distribution of Synechococcus S5.1 clade IV.
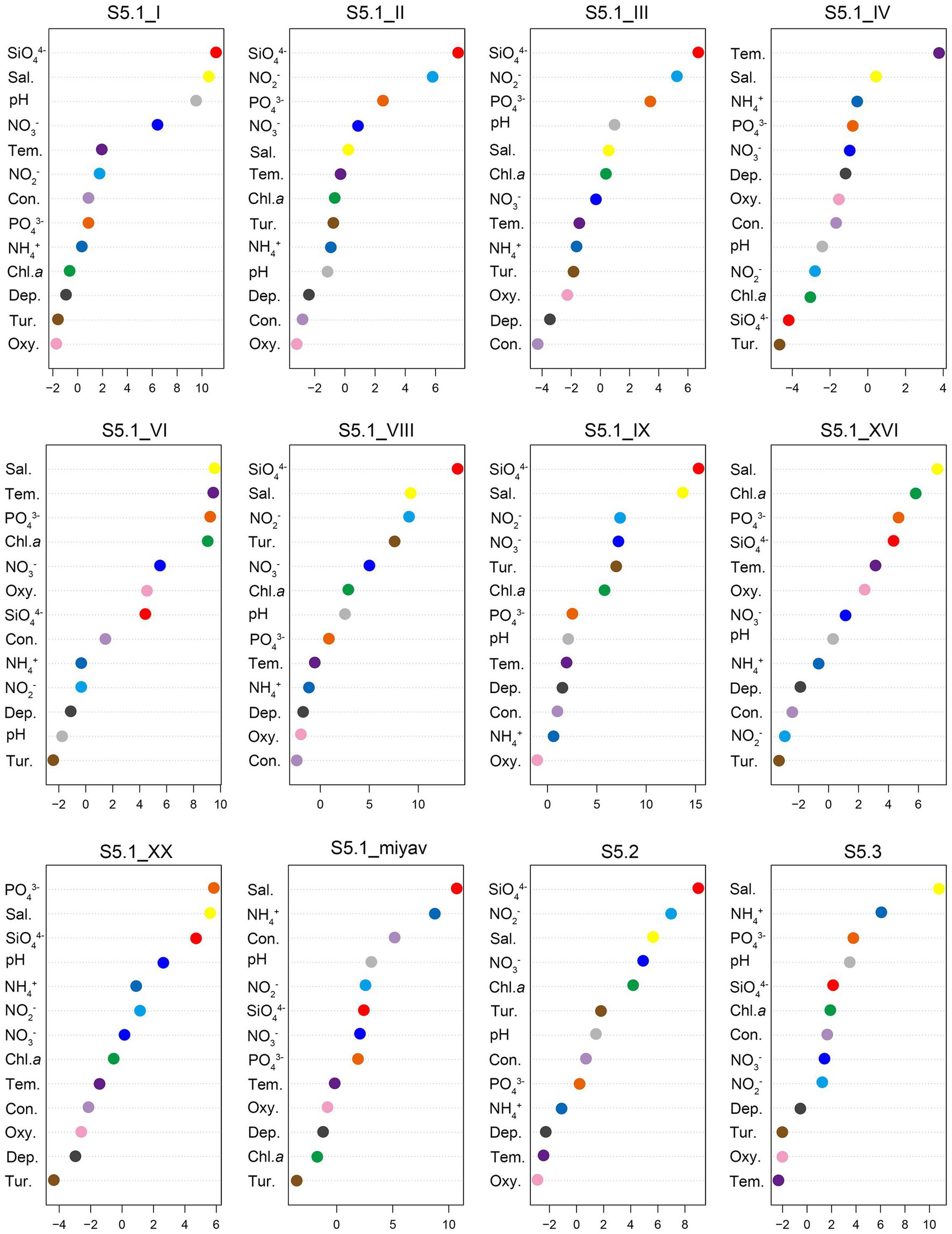
Figure 6. Random forest (RF) analysis between each Synechococcus clade and environmental variables. Sal, salinity; Tem, temperature; Con, conductivity; Dep, depth; Tur, turbidity; and Oxy, oxygen saturation. Abscissas are %ImcMSE evaluating the materiality of environmental variables to each Synechococcus clade in RF.
Linear Regression Analysis Between Synechococcus Genotypes and Environmental Variables
Linear OLS regression showed that except for clade IV, significant linear relationships were found between each Synechococcus clade and at least one environmental parameter (Table 3). Among them, clade VI related to the most environmental parameters with up to six. Clades I, IX, and S5.2 were next and correlated with five environmental parameters. Clades VIII, XVI, and II correlated with four, three, and two parameters, respectively, and the rest of the four clades correlated only with one parameter. Values of R2 greater than 0.6 were defined as a strong correlation (Supplementary Figure S4). Clade I correlated strongly to SiO44− concentration (negatively); clade VI correlated strongly with chl. a and temperature (both positively); clade VIII correlated strongly with NO2− concentration (positively). Clade IX correlated strongly with four parameters, namely, the concentrations of NO2− and SiO44−, salinity, and turbidity. Among them, the correlation between clade IX and salinity was negative, while that between clade IX and the other three parameters was positive.
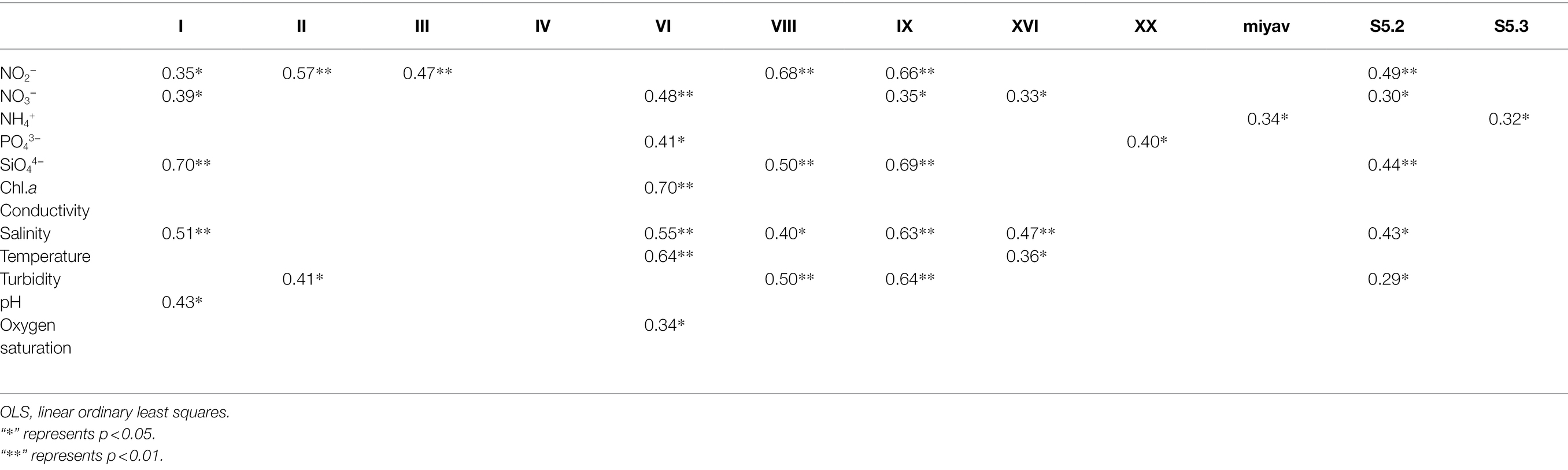
Table 3. Results of linear ordinary least square (OLS) regression for the relative abundances of Synechococcus genotypes and environmental variables.
Discussion
Shift in Hydrologic Conditions in Bohai Sea
The development of agriculture, aquaculture, and industry has increased nutrient inputs from land to sea water, which has changed the hydrological conditions in the Bohai Sea (Wang et al., 2018b). It is believed that a gradual change from nitrogen limitation to phosphate and silicate limitations has occurred here (Yu, 2000). We observed that the concentration of NO3− in 2019 is higher than that in 2014 (Wang et al., 2018b), which is consistent with the existing consensus that the dissolved inorganic nitrogen (DIN) concentration in the Bohai Sea increased continuously (Su et al., 2011; Wang et al., 2018b). PO44− and SiO44− concentrations were thought to be on a decline in the Bohai Sea (Ning et al., 2010). In comparison, we observed an increase in PO44− concentration but a decrease in SiO44− concentration compared with previous surveys (Wang et al., 2018b). In addition, we found that the ratio of SiO44−/DIN was lower than 1 in all samples except those from BS-W. Silicate limitation appears to have become a main hydrological characteristic in the Bohai Sea. According to our record in the Bohai Sea, the environmental conditions exhibited stronger variation between strait and bay areas than variation between June and November as well as different vertical water layers. Poor water exchange and continuous inflow of pollutants resulted in higher concentrations of various nutrients in the LZB than in BS (Su et al., 2011). Considering that environmental changes will stimulate the preferential growth of certain phytoplanktons under suitable conditions and lead to further changes in their communities, we expected the formation of a special phylogenetic composition of Synechococcus assemblage in the Bohai Sea, which was confirmed in this study (Ferreira et al., 2007; Yunev et al., 2007; Yan et al., 2020).
Diversity of Synechococcus Assemblages in the Bohai Sea
High-throughput sequencing of rpoC1 led to the detection of 12 clades representing Synechococcus S5.1, S5.2, and S5.3 (Figure 3). In comparison, previous studies found about 6 to 13 lineages in most of the sea areas of the world, such as open sea and coast of Pacific Ocean (Tai and Palenik, 2009), Gulf of Aqaba (Post et al., 2011), Chesapeake Bay (Chen et al., 2006), Sargasso Sea (Ahlgren and Rocap, 2006), and East China Sea (Choi et al., 2013b). More lineages were identified in estuarine waters, which included freshwater Synechococcus and Cyanobium species. For example, in total, 17 Synechococcus clades were reported in the estuarine and coastal waters of Hong Kong (Xia et al., 2015). Our results suggested that the phylogenetic diversity of Synechococcus lineages in the bay and strait areas of the Bohai Sea was similar to that in most worldwide seas, but is lower than that in estuarine waters.
Phylogeography of Synechococcus Assemblages in the Bohai Sea
Synechococcus clade I was the absolute dominant lineage in bay and strait areas of the Bohai Sea (Figure 3). Synechococcus clades VI and III were also highly abundant genotypes, while their average proportion in all samples was only about one-sixth of that of clade I. Synechococcus composition at station BS was similar to that in the adjacent north Yellow Sea (clade I was also found to be dominant), which is probably due to the water quality (Li et al., 2021). However, in sea areas that are farther away, such as south Yellow Sea, East China Sea, and South China Sea, the composition of Synechococcus varied highly (Jing and Liu, 2012; Chung et al., 2015; Xia et al., 2017b). In south Yellow Sea, Synechococcus clades VI, II, and III were relatively abundant (Xia et al., 2017b). In addition, Synechococcus clade II was dominant in the East China Sea and the South China Sea (Xia et al., 2017b).
Compared with other continental seas in the world, the phylogenetic composition of the Synechococcus assemblage in the bay and strait areas of the Bohai Sea resembled that in the Sea of Okhotsk (Jing et al., 2009a) and the northwestern waters of the Gulf of Lions (Mella-Flores et al., 2011). In the Sea of Okhotsk, the Synechococcus community was composed exclusively of S5.1 and dominated by clades I and IV (Jing et al., 2009a). In the Mediterranean Sea, the Synechococcus assemblage was dominated by clades I, III, and IV in the northwestern waters of the Gulf of Lions (Mella-Flores et al., 2011). In contrast, Synechococcus assemblage was dominated by a single clade, clade II, with the exception of a peak of clade III in June in the Gulf of Aqaba, Red Sea (Fuller et al., 2005). In the Mediterranean Sea, except for the northwestern waters of the Gulf of Lions, Synechococcus assemblage was dominated by groups genetically related to clades WPC1 and VI (Mella-Flores et al., 2011).
Geographical variation in Synechococcus composition between BS and LZB in the Bohai Sea was demonstrated (Figures 2, 3). In BS, the composition of Synechococcus genotype was similar to that found in open and high-latitude oceans, such as the North Atlantic, southern Bering, and northwestern Pacific Ocean (Huang et al., 2012; Sohm et al., 2016; Xia et al., 2017b). On the contrary, Synechococcus assemblage in LZB with higher phylogenetic diversity was mostly composed of clades VI and III, similar to that in coastal waters of Hong Kong and the Pearl River estuary (Xia et al., 2015, 2017a). This may be because of the relatively high nutritional condition of LZB caused by the discharge of exogenous substances from the mainland (Lü et al., 2015). However, the community structure of the Synechococcus assemblage did not vary among the vertical water layers in the Bohai Sea. This could be because the Bohai as a marginal sea of continental shelf possesses shallow water depth (the average depth is 18 m) with intense vertical water mixing. Overall, our study supports the conclusion that in addition to geographical proximity, Synechococcus phylogeography is driven by environmental conditions (Zwirglmaier et al., 2008).
Synechococcus clade VI was the third abundant clade in the bay and strait areas of the Bohai Sea (Figure 3). In particular, its proportion can reach 70% of Synechococcus assemblage in LZB-S. However, according to the co-occurrence network, its Hubba score was as low as 1 (Figure 4), representing a low contribution to Synechococcus community. This result might suggest that this clade showed low metabolic activity in the Bohai Sea. However, further studies at the transcriptional level are needed to confirm it.
Variations of Synechococcus Assemblages in the Bohai Sea Between June and November
The geographical features, as well as periodical discharges of pollution and freshwater from the continent, could together lead to the strong seasonal variation of Synechococcus in the Bohai Sea (Wang et al., 2021). Synechococcus S5.1 clades II, III, XX, and miyav appeared only in November. In the global oceans, clade II was the most ubiquitous clade dominant in the oligotrophic open ocean (Sohm et al., 2016). However, consistent with our results, with the increase in latitude, it was gradually replaced by clades I and IV above 37°N. Clade III appeared in the Mediterranean Sea and the Gulf of Mexico before (Farrant et al., 2016). As suggested by a year-round survey in the Red Sea, this clade is also found subject to seasonality, which is abundant during specific months and remains abundantly low over the rest of the year (Post et al., 2011). Clade miyav, represented by strain Synechococcus sp. strain miyav, was detected and defined in estuarine waters and coastal waters of Hong Kong (Xia et al., 2015). Similar to our results, this clade mainly occurred in winter. It should be noted that the seasonal variation of Synechococcus assemblage was only studied in June (representing summer) and in November (representing winter) due to the cruise limitation in this study. The characteristics of Synechococcus assemblage in spring and autumn need further study to clear.
Correlation Between Environmental Variables and Synechococcus Genotypes in the Bohai Sea
Environmental variables affect the community characteristics and structure of Synechococcus assemblage in specific environments (Xia et al., 2015). In wide global sea areas, the distribution of Synechococcus clades is decided by the oceanic condition defined by temperature, macronutrients, and iron (Sohm et al., 2016). The first axis was mainly a phosphate gradient, whereas the second axis was mainly nitrate concentration and biomass (chl. a) gradients in the RDA of the Gulf of Aqaba (Fuller et al., 2005). In contrast, Synechococcus assemblage in coastal and estuarine waters appeared to be influenced by physical marine environmental factors. In estuarine and coastal waters of Hong Kong, salinity and temperature significantly affected Synechococcus assemblage composition and accounted for 32.0 and 22.0% variance of the Synechococcus assemblages, respectively (Xia et al., 2015). However, concentrations of SiO44−, PO43−, and chl. a, were significant variables that correlated with the Synechococcus composition in the bay and strait areas of the Bohai Sea (Figure 5). This indicated that in addition to environmental pollution caused by human activities, other biological activities affect the Synechococcus assemblage in the environment.
In particular, each of the Synechococcus genotypes was linked to different environmental parameters and occupied special ecological niches. Clade I dominated the cold, high-latitude, and nutrient-rich waters; clade III was dominant in warm, oligotrophic, open-ocean habitats in the tropical and subtropical oceans; clade VI became apparent during the transition periods between mixing and stratification (Post et al., 2011; Sohm et al., 2016). According to the RF models, these dominant clades in the bay and strait areas of the Bohai Sea were considerably affected by SiO44− concentration and salinity (Figure 6). Baines et al. (2012) discovered the accumulation of silicon (Si) by Synechococcus; however, a clear role of Si in Synechococcus has not been revealed. Researchers have shown that Si uptake may be associated with the acquisition of phosphate and that Si accumulation by Synechococcus is inadvertent (Brzezinski et al., 2017). Our finding that the concentration of SiO44− was the most significant variable affecting the distribution of six Synechococcus clades contradicts the above conclusion of inadvertent accumulation of Si. It was also confirmed by the observation of Wei et al. (2019), which demonstrated that Synechococcus biomass including cellular sizes was favored by the dissolved Si (DSi) concentrations. Recent studies have demonstrated that picoplankton size populations, especially Synechococcus, contribute a measurable and significant proportion of total biogenic silica (bSi; Wei et al., 2021a). Meanwhile, it was found that atomically dense structures from Synechococcus cell lysis were enriched in Si element (Wei et al., 2021b). These signs all indicate the close relationship between Synechococcus and Si. In summary, our result demonstrated that Si may play a more important role in each Synechococcus genotype, which would add a new dimension to their nutrient physiology and to the suite of resources influencing Synechococcus abundance and community structure in nature, although further physiological and genomic studies are required to verify it.
Synechococcus can be considered a key environmental indicator. Rajaneesh et al. (2015) initially constructed the indicator relationship between two kinds of Synechococcus (phycoerythrin-rich Synechococcus and phycocyanin-rich Synechococcus), as well as between the trophic status index and various environmental parameters in the Cochin backwaters, west coast of India, and found the superiority of Synechococcus as an indicator organism. However, considering its high phylogenetic diversity and correlations with various environmental variables, their conclusion still underestimated the enormous potential of Synechococcus in indicating the marine environment. Here, we identified a rich linear correlation between Synechococcus clades and environmental parameters in the bay and strait areas of the Bohai Sea (Table 3; Supplementary Figure S4). A significant linear relationship existed between each of the 11 Synechococcus clades and at least one environmental parameter. In particular, clade VI correlated with the most parameters, reaching six. By expanding the number of samples and increasing the density of sampling sites, more stable equations regarding the relation between the Synechococcus clades and the environmental parameters can be achieved, which is our next research focus.
Conclusion
Continental seas, affected strongly by human activities, possess dynamic environmental conditions. In the Bohai Sea, the largest continental sea of China, silicate limitations appear to be a main hydrological characteristic. Because of this, a special phylogenetic composition pattern of Synechococcus assemblage was formed in this sea area. High-throughput sequencing of rpoC1 revealed that Synechococcus diversity in the bay and strait areas of the Bohai Sea was similar to that in most seas worldwide, but was lower than that in estuarine waters. Geographical variations in Synechococcus composition were observed between BS and LZB. Obligately dominated by clade I, the composition of Synechococcus genotype in BS was similar to that in open and high-latitude oceans. On the contrary, the Synechococcus assemblage in LZB, with higher phylogenetic diversity and highly abundant clades VI and III, was similar to that in coastal and estuarine waters. Synechococcus S5.1 clades II, III, XX, and miyav only appeared in winter. The community structure of the Synechococcus assemblage did not vary among the vertical water layers, which could be because of its shallow water depth with intense vertical water mixing. Silicate is a key environmental variable that affects the community characteristics and structure of the Synechococcus assemblage in the bay and strait areas of the Bohai Sea, probably because of silicon limitations. This may change the previous conclusion that silicate accumulation by Synechococcus is inadvertent, although further physiological and genomic studies are required for verifying this.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, SRR15404031—SRR15404044.
Author Contributions
SQ and JL: conceptualization and writing—review and editing. JL and TW: methodology and investigation. TW and XC: data curation and visualization. TW: writing—original draft preparation. SQ: supervision. JL: project administration and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This study was financially supported by the Key Deployment Project of Centre for Ocean Mega-Science, Chinese Academy of Sciences (COMS2020Q09), the National Key Research and Development Program of China (No. 2018YFD0901102), and the National Natural Science Foundation of China (No. 42176131).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The data and samples were collected by Research Vessel “Chuangxin I.” We acknowledge the assistance from the Operation and management center of large field observation equipment, Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences for organizing the research voyage and sharing open data. We extend our gratitude to the journal reviewers for their comments and suggestions, which helped in significantly improving the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.757896/full#supplementary-material
References
Ahlgren, N. A., and Rocap, G. (2006). Culture isolation and culture-independent clone libraries reveal new marine Synechococcus ecotypes with distinctive light and N physiologies. Appl. Environ. Microbiol. 72, 7193–7204. doi: 10.1128/AEM.00358-06
Ahlgren, N. A., and Rocap, G. (2012). Diversity and distribution of marine Synechococcus: multiple gene phylogenies for consensus classification and development of qPCR assays for sensitive measurement of clades in the ocean. Front. Microbiol. 3:213. doi: 10.3389/fmicb.2012.00213
Anderson, M. J. (2017). “Permutational multivariate analysis of variance (PERMANOVA),” in Wiley StatsRef: Statistics Reference Online. eds. N. Balakrishnan, T. Colton, B. Everitt, W. Piegorsch, F. Ruggeri, and J. L. Teugels 1–15.
Baines, S. B., Twining, B. S., Brzezinski, M. A., Krause, J. W., Vogt, S., Assael, D., et al. (2012). Significant silicon accumulation by marine picocyanobacteria. Nat. Geosci. 5, 886–891. doi: 10.1038/ngeo1641
Bastian, M., Heymann, S., and Jacomy, M. (2009). “Gephi: an open source software for exploring and manipulating networks.” in Third International AAAI Conference on Weblogs and Social Media; March 19, 2009.
Brown, J., Pirrung, M., and Mccue, L. A. (2017). FQC dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 33, 3137–3139. doi: 10.1093/bioinformatics/btx373
Brzezinski, M. A., Krause, J. W., Baines, S. B., Collier, J. L., Ohnemus, D. C., and Twining, B. S. (2017). Patterns and regulation of silicon accumulation in Synechococcus spp. J. Phycol. 53, 746–761. doi: 10.1111/jpy.12545
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chen, F., Wang, K., Kan, J., Suzuki, M. T., and Wommack, K. E. (2006). Diverse and unique picocyanobacteria in Chesapeake Bay, revealed by 16S-23S rRNA internal transcribed spacer sequences. Appl. Environ. Microbiol. 72, 2239–2243. doi: 10.1128/AEM.72.3.2239-2243.2006
Chin, C. H., Chen, S. H., Wu, H. H., Ho, C. W., Ko, M. T., and Lin, C. Y. (2014). cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 8:S11. doi: 10.1186/1752-0509-8-S4-S11
Choi, D. H., and Noh, J. H. (2009). Phylogenetic diversity of Synechococcus strains isolated from the East China Sea and the East Sea. FEMS Microbiol. Ecol. 69, 439–448. doi: 10.1111/j.1574-6941.2009.00729.x
Choi, D. H., Noh, J. H., and Lee, J. H. (2013a). Application of pyrosequencing method for investigating the diversity of Synechococcus subcluster 5.1 in open ocean. Microbes Environ. 29, 17–22. doi: 10.1264/jsme2.me13063
Choi, D. H., Noh, J. H., and Shim, J. (2013b). Seasonal changes in picocyanobacterial diversity as revealed by pyrosequencing in temperate waters of the East China Sea and the East Sea. Aquat. Microb. Ecol. 71, 75–90. doi: 10.3354/ame01669
Chung, C. C., Gong, G. C., Huang, C. Y., Lin, J. Y., and Lin, Y. C. (2015). Changes in the Synechococcus assemblage composition at the surface of the East China Sea due to flooding of the Changjiang river. Microb. Ecol. 70, 677–688. doi: 10.1007/s00248-015-0608-5
Dafner, E. V. (2015). Segmented continuous-flow analyses of nutrient in seawater: intralaboratory comparison of technicon autoanalyzer II and bran+ luebbe continuous flow autoanalyzer III. Limnol. Oceanogr. Methods 13, 511–520. doi: 10.1002/lom3.10035
Dixon, P. (2003). VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Dufresne, A., Ostrowski, M., Scanlan, D. J., Garczarek, L., Mazard, S., Palenik, B. P., et al. (2008). Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol. 9:R90. doi: 10.1186/gb-2008-9-5-r90
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Farrant, G. K., Doré, H., Cornejo-Castillo, F. M., Partensky, F., Ratin, M., Ostrowski, M., et al. (2016). Delineating ecologically significant taxonomic units from global patterns of marine picocyanobacteria. Proc. Natl. Acad. Sci. U. S. A. 113, E3365–E3374. doi: 10.1073/pnas.1524865113
Ferreira, J. G., Bricker, S. B., and Simas, T. C. (2007). Application and sensitivity testing of a eutrophication assessment method on coastal systems in the United States and European Union. J. Environ. Manag. 82, 433–445. doi: 10.1016/j.jenvman.2006.01.003
Flombaum, P., Gallegos, J. L., Gordillo, R. A., Rincón, J., and Martiny, A. C. (2013). Present and future global distributions of the marine cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. U. S. A. 110, 9824–9829. doi: 10.1073/pnas.1307701110
Franke, G. R. (2010). “Multicollinearity,” in Wiley International Encyclopedia of Marketing. eds. J. N. Sheth and N. K. Malhotra (New Jersey, USA: John Wiley & Sons Ltd.).
Fuller, N. J., West, N. J., Marie, D., Yallop, M., Rivlin, T., Post, A. F., et al. (2005). Dynamics of community structure and phosphate status of picocyanobacterial populations in the Gulf of Aqaba, Red Sea. Limnol. Oceanogr. 50, 363–375. doi: 10.4319/lo.2005.50.1.0363
George, D. (2011). SPSS for Windows Step by Step: A Simple Study Guide and Reference, 17.0 Update, 10/e. Chennai, India: Pearson Education India.
Huang, S., Wilhelm, S. W., Harvey, H. R., Taylor, K., Jiao, N., and Chen, F. (2012). Novel lineages of Prochlorococcus and Synechococcus in the global oceans. ISME J. 6, 285–297. doi: 10.1038/ismej.2011.106
Hutcheson, G. D. (1999). “Ordinary least-squares regression,” in The Multivariate Social Scientist. eds. L. Moutinho and G. D. Hutcheson (London, United Kingdom: SAGE Publications, Ltd.), 56–113.
Jing, H., and Liu, H. (2012). Phylogenetic composition of Prochlorococcus and Synechococcus in cold eddies of the South China Sea. Aquat. Microb. Ecol. 65, 207–219. doi: 10.3354/ame01546
Jing, H., Liu, H., and Suzuki, K. (2009a). Phylogenetic diversity of marine Synechococcus spp. in the Sea of Okhotsk. Aquat. Microb. Ecol. 56, 55–63. doi: 10.3354/ame01316
Jing, H., Zhang, R., Pointing, S. B., Liu, H., and Qian, P. (2009b). Genetic diversity and temporal variation of the marine Synechococcus community in the subtropical coastal waters of Hong Kong. Can. J. Microbiol. 55, 311–318. doi: 10.1139/W08-138
Kent, A. G., Baer, S. E., Mouginot, C., Huang, J. S., Larkin, A. A., Lomas, M. W., et al. (2019). Parallel phylogeography of Prochlorococcus and Synechococcus. ISME J. 13, 430–441. doi: 10.1038/s41396-018-0287-6
Li, G., Song, Q., Zheng, P., Zhang, X., Zou, S., Li, Y., et al. (2021). Dynamics and distribution of marine Synechococcus abundance and genotypes during seasonal hypoxia in a coastal marine ranch. J. Mar. Sci. Eng. 9:549. doi: 10.3390/jmse9050549
Li, J., Wang, T., Yu, S., Bai, J., and Qin, S. (2019). Community characteristics and ecological roles of bacterial biofilms associated with various algal settlements on coastal reefs. J. Environ. Manag. 250:109459. doi: 10.1016/j.jenvman.2019.109459
Lü, D., Zheng, B., Fang, Y., Shen, G., and Liu, H. (2015). Distribution and pollution assessment of trace metals in seawater and sediment in Laizhou Bay. J. Oceanol. Limnol. 33, 1053–1061. doi: 10.1007/s00343-015-4226-3
Ma, Y., Jiao, N., and Zeng, Y. (2004). Natural community structure of cyanobacteria in the South China Sea as revealed by rpoC1 gene sequence analysis. Lett. Appl. Microbiol. 39, 353–358. doi: 10.1111/j.1472-765X.2004.01588.x
Mazard, S., Ostrowski, M., Partensky, F., and Scanlan, D. J. (2012). Multi-locus sequence analysis, taxonomic resolution and biogeography of marine Synechococcus. Environ. Microbiol. 14, 372–386. doi: 10.1111/j.1462-2920.2011.02514.x
Mei, W., Yu, G., Lai, J., Rao, Q., and Umezawa, Y. (2018). basicTrendline: Add Trendline and Confidence Interval of Basic Regression Models to Plot. R Package Version 2.
Mella-Flores, D., Mazard, S., Humily, F., Partensky, F., Mahé, F., Bariat, L., et al. (2011). Is the distribution of Prochlorococcus and Synechococcus ecotypes in the Mediterranean Sea affected by global warming? Biogeosciences 8, 2785–2804. doi: 10.5194/bg-8-2785-2011
Mühling, M., Fuller, N. J., Somerfield, P. J., Post, A. F., Wilson, W. H., Scanlan, D. J., et al. (2006). High resolution genetic diversity studies of marine Synechococcus isolates using rpoC1-based restriction fragment length polymorphism. Aquat. Microb. Ecol. 45, 263–275. doi: 10.3354/ame045263
Ning, X., Lin, C., Su, J., Liu, C., Hao, Q., Le, F., et al. (2010). Long-term environmental changes and the responses of the ecosystems in the Bohai Sea during 1960–1996. Deep-Sea Res. Part II-Top. Stud. Oceanogr. 57, 1079–1091. doi: 10.1016/j.dsr2.2010.02.010
Paulsen, M. L., Doré, H., Garczarek, L., Seuthe, L., Müller, O., Sandaa, R. A., et al. (2016). Synechococcus in the Atlantic gateway to the Arctic Ocean. Front. Mar. Sci. 3:191. doi: 10.3389/fmars.2016.00191
Penno, S., Lindell, D., and Post, A. F. (2006). Diversity of Synechococcus and Prochlorococcus populations determined from DNA sequences of the N-regulatory gene ntcA. Environ. Microbiol. 8, 1200–1211. doi: 10.1111/j.1462-2920.2006.01010.x
Post, A. F., Penno, S., Zandbank, K., Paytan, A., Huse, S., and Mark Welch, D. (2011). Long term seasonal dynamics of Synechococcus population structure in the Gulf of Aqaba, Northern Red Sea. Front. Microbiol. 2:131. doi: 10.3389/fmicb.2011.00131
Rajaneesh, K. M., Mitbavkar, S., Anil, A. C., and Sawant, S. S. (2015). Synechococcus as an indicator of trophic status in the Cochin backwaters, west coast of India. Ecol. Indic. 55, 118–130. doi: 10.1016/j.ecolind.2015.02.033
Revelle, W., and Revelle, M. W. (2015). Package ‘Psych’. The Comprehensive R Archive Network. Available at: https://cran.rstudio.org/web/packages/psych/psych.pdf
Schlitzer, R. (2002). Interactive analysis and visualization of geoscience data with ocean data view. Comput. Geosci. 28, 1211–1218. doi: 10.1016/S0098-3004(02)00040-7
Sohm, J. A., Ahlgren, N. A., Thomson, Z. J., Williams, C., Moffett, J. W., Saito, M. A., et al. (2016). Co-occurring Synechococcus ecotypes occupy four major oceanic regimes defined by temperature, macronutrients and iron. ISME J. 10, 333–345. doi: 10.1038/ismej.2015.115
Su, M. L., Ling, W. L., and Zhang, Z. (2011). Inventory of nutrients in the Bohai. Cont. Shelf Res. 31, 1790–1797. doi: 10.1016/j.csr.2011.08.004
Tai, V., Burton, R. S., and Palenik, B. (2011). Temporal and spatial distributions of marine Synechococcus in the Southern California bight assessed by hybridization to bead-arrays. Mar. Ecol.-Prog. Ser. 426, 133–147. doi: 10.3354/meps09030
Tai, V., and Palenik, B. (2009). Temporal variation of Synechococcus clades at a coastal Pacific Ocean monitoring site. ISME J. 3, 903–915. doi: 10.1038/ismej.2009.35
Temino-Boes, R., Romero, I., Pachés, M., Martinez-Guijarro, R., and Romero-Lopez, R. (2019). Anthropogenic impact on nitrification dynamics in coastal waters of the Mediterranean Sea. Mar. Pollut. Bull. 145, 14–22. doi: 10.1016/j.marpolbul.2019.05.013
Wang, T., Chen, X., Li, J., and Qin, S. (2021). Distribution and phenogenetic diversity of Synechococcus in the Bohai Sea, China. J. Oceanol. Limnol. 39, 1–13. doi: 10.1007/s00343-021-1005-1
Wang, C., Lv, Y., and Li, Y. (2018a). Riverine input of organic carbon and nitrogen in water-sediment system from the Yellow River estuary reach to the coastal zone of Bohai Sea, China. Cont. Shelf Res. 157, 1–9. doi: 10.1016/j.csr.2018.02.004
Wang, J., Yu, Z., Wei, Q., and Yao, Q. (2018b). Long-term nutrient variations in the Bohai Sea over the past 40 years. J. Geophys. Res.-Oceans 124, 703–722. doi: 10.1029/2018JC014765
Wei, Y., Sun, J., Chen, Z., Zhang, Z., Zhang, G., and Liu, X. (2021a). Significant contribution of picoplankton size fraction to biogenic silica standing stocks in the Western Pacific Ocean. Prog. Oceanogr. 192:102516. doi: 10.1016/j.pocean.2021.102516
Wei, Y., Sun, J., Zhang, X., Wang, J., and Huang, K. (2019). Picophytoplankton size and biomass around equatorial eastern Indian Ocean. MicrobiologyOpen 8:e00629. doi: 10.1002/mbo3.629
Wei, Y., Wang, X., Gui, J., and Sun, J. (2021b). Significant pico- and nanoplankton contributions to biogenic silica standing stocks and production rates in the oligotrophic eastern Indian Ocean. Ecosystems 24, 1654–1669. doi: 10.1007/s10021-021-00608-w
Xia, X., Cheung, S., Endo, H., Suzuki, K., and Liu, H. (2019). Latitudinal and vertical variation of Synechococcus assemblage composition Along 170° W transect From the South Pacific to the Arctic Ocean. Microb. Ecol. 77, 333–342. doi: 10.1007/s00248-018-1308-8
Xia, X., Guo, W., Tan, S., and Liu, H. (2017a). Synechococcus assemblages across the salinity gradient in a salt wedge estuary. Front. Microbiol. 8:1254. doi: 10.3389/fmicb.2017.01254
Xia, X., Partensky, F., Garczarek, L., Suzuki, K., Guo, C., Yan Cheung, S., et al. (2017b). Phylogeography and pigment type diversity of Synechococcus cyanobacteria in surface waters of the northwestern Pacific Ocean. Environ. Microbiol. 19, 142–158. doi: 10.1111/1462-2920.13541
Xia, X., Vidyarathna, N. K., Palenik, B., Lee, P., and Liu, H. (2015). Comparison of the seasonal variations of Synechococcus assemblage structures in estuarine waters and coastal waters of Hong Kong. Appl. Environ. Microbiol. 81, 7644–7655. doi: 10.1128/AEM.01895-15
Yan, G., Jiang, T., Zhang, Y., Cui, Z., Qu, K., Zheng, Y., et al. (2020). Determining temporal and spatial distribution of autotrophic picoplankton community composition through HPLC-pigment method and flow cytometry in the central Bohai Sea (China). Mar. Pollut. Bull. 157:111261. doi: 10.1016/j.marpolbul.2020.111261
Yu, Z. (2000). Changes of the environmental parameters and their relationship in recent twenty years in the Bohai Sea. Mar. Environ. Sci. 19, 5–19.
Yunev, O. A., Carstensen, J., Moncheva, S., Khaliulin, A., Ærtebjerg, G., and Nixon, S. (2007). Nutrient and phytoplankton trends on the western Black Sea shelf in response to cultural eutrophication and climate changes. Estuar. Coast. Shelf Sci. 74, 63–76. doi: 10.1016/j.ecss.2007.03.030
Zhang, Y., Lu, X., Liu, H., Liu, Q., and Yu, D. (2015). Identifying the sources of organic matter in marine and riverine sediments of Bohai Bay and its catchment using carbon and nitrogen stable isotopes. J. Oceanol. Limnol. 33, 204–209. doi: 10.1007/s00343-015-4068-z
Zhang, Z., Schwartz, S., Wagner, L., and Miller, W. (2000). A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7, 203–214. doi: 10.1089/10665270050081478
Zwirglmaier, K., Jardillier, L., Ostrowski, M., Mazard, S., Garczarek, L., Vaulot, D., et al. (2008). Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ. Microbiol. 10, 147–161. doi: 10.1111/j.1462-2920.2007.01440.x
Keywords: picoplankton, cyanobacteria, genetic diversity, phylogeny, marginal sea, silicon accumulation, high-throughput sequencing
Citation: Wang T, Chen X, Li J and Qin S (2022) Phylogenetic Structure of Synechococcus Assemblages and Its Environmental Determinants in the Bay and Strait Areas of a Continental Sea. Front. Microbiol. 13:757896. doi: 10.3389/fmicb.2022.757896
Edited by:
Ondrej Prasil, Academy of Sciences of the Czech Republic (ASCR), CzechiaReviewed by:
Christina Pavloudi, George Washington University, United StatesWei Xie, Sun Yat-sen University, Zhuhai Campus, China
Copyright © 2022 Wang, Chen, Li and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jialin Li, amxsaUB5aWMuYWMuY24=
 Ting Wang
Ting Wang Xi Chen5
Xi Chen5 Jialin Li
Jialin Li Song Qin
Song Qin