- State Key Laboratory of Conservation and Utilization of Subtropical Agro-Bioresources, Guangdong Laboratory for Lingnan Modern Agriculture, Guangdong Key Laboratory for Innovative Development and Utilization of Forest Plant Germplasm, College of Forestry and Landscape Architecture, South China Agricultural University, Guangzhou, China
Introduction: Phosphorus (P) is one of the most important nutrient elements for plant growth and development. Under P starvation, arbuscular mycorrhizal (AM) fungi can promote phosphate (Pi) uptake and homeostasis within host plants. However, the underlying mechanisms by which AM fungal symbiont regulates the AM symbiotic Pi acquisition from soil under P starvation are largely unknown. Here, we identify a HLH domain containing transcription factor RiPho4 from Rhizophagus irregularis.
Methods: To investigate the biological functions of the RiPho4, we combined the subcellular localization and Yeast One-Hybrid (Y1H) experiments in yeasts with gene expression and virus-induced gene silencing approach during AM symbiosis.
Results: The approach during AM symbiosis. The results indicated that RiPho4 encodes a conserved transcription factor among different fungi and is induced during the in planta phase. The transcription of RiPho4 is significantly up-regulated by P starvation. The subcellular localization analysis revealed that RiPho4 is located in the nuclei of yeast cells during P starvation. Moreover, knock-down of RiPho4 inhibits the arbuscule development and mycorrhizal Pi uptake under low Pi conditions. Importantly, RiPho4 can positively regulate the downstream components of the phosphate (PHO) pathway in R. irregularis.
Discussion: In summary, these new findings reveal that RiPho4 acts as a transcriptional activator in AM fungus to maintain arbuscule development and regulate Pi uptake and homeostasis in the AM symbiosis during Pi starvation.
Introduction
Arbuscular mycorrhizal fungi (AMF) belong to the Glomeromycotina in the Mucoromycota, and are a kind of the obligate soilborne fungi which can form the AM symbioses with more than 70% of land plants (Brundrett and Tedersoo, 2019; Bonfante and Venice, 2020; Genre et al., 2020; Rich et al., 2021). AMF have been shown to benefit plant productivity and they can absorb water and mineral nutrients such as phosphorus (P), nitrogen (N), iron, sulfur and zinc from soils, then transfer them to the host plants via the symbiotic interfaces (Watts-Williams and Cavagnaro, 2018; Kobae, 2019; Rahman et al., 2020; Ma et al., 2022). In return, host plants can transport fatty acids and sugars to AMF as the carbon and energy sources (Campos-Soriano and Segundo, 2011; Jiang et al., 2017; Luginbuehl and Oldroyd, 2017; An et al., 2019). These bidirectional processes effectively regulate the nutrient balance between the host plants and their AM fungal symbionts, and thus these associations are capable of promoting plant development and fitness (Pozo and Azcón-Aguilar, 2007; Hajong et al., 2013; Fellbaum et al., 2014).
Soil available P can be acquired at the root periphery and utilized by plants in the form of inorganic orthophosphate (Pi), however, Pi is always insufficient in the fields due to its low solubility and relative immobilization in soils (Vance et al., 2003; Hirsch et al., 2006; Nagy et al., 2009). The formation of AM symbiosis is an effective strategy for land plants to cope with low Pi availability (Cibichakravarthy et al., 2015; Dierks et al., 2021). During colonization, the branch hyphae of spores produce swellings called appressoria on the surface of the root epidermal cells after the perception of host plant-derived strigolactones (Giovannetti et al., 1993; Akiyama et al., 2005). Subsequently, the appressoria penetrate the epidermal cells to grow the intraradical hyphae assembled within the prepenetration apparatus (Genre et al., 2005, 2008, 2012; Russo et al., 2019); the developing intraradical mycelium (IRM) then run across the root cortical cells and form the tree-like structures called arbuscules in these cortical cells, where the nutrient transport and unloading (such as Pi and N) occurs (Parniske, 2008; Gutjahr and Parniske, 2013; Luginbuehl and Oldroyd, 2017; Hui et al., 2022). Meanwhile, arbuscules are surrounded by the extension of plant plasma membrane called the periarbuscular membrane (PAM) (Harrison et al., 2002; Pumplin et al., 2012). It is also considered to be the main nutrient exchange site of AM symbiosis (Balestrini and Bonfante, 2014; Ivanov and Harrison, 2014; Roth et al., 2019). During AM symbiosis, the extraradical mycelium (ERM) of AMF can reach up to 100 times length of root hairs (Javot et al., 2007) and form the large external hyphal networks to expand more area for Pi absorption beyond the rhizospheres, and also increase the phosphatase activities at the rhizospheres (Wang et al., 2016; Hu et al., 2019). Therefore, the AM symbioses can enhance plant Pi uptake and utilization during P starvation (Smith, 2009; Cibichakravarthy et al., 2015; Dierks et al., 2021).
Earlier radiotracer studies have demonstrated that Pi travels from soils through the AM fungal hyphae to the host plants (Pearson and Jakobsen, 1993; Smith et al., 2003). In past two decades, the high-affinity transporter genes belonging to the PHT1 (PHOSPHATE TRANSPORTER 1) family that are expressed in the ERM and IRM have been identified and characterized from some AMF species, for example, GmosPT, GigmPT, GvPT, and RiPT from Glomus mosseae, Gigaspora margarita, Glomus versiforme (currently Diversispora epigaea), and Rhizophagus irregularis (Harrison and van Buuren, 1995; Maldonado-Mendoza et al., 2001; Benedetto et al., 2005; Fiorilli et al., 2013; Xie et al., 2016; Sun et al., 2019; Venice et al., 2020). AMF can absorb Pi from soil by ERM, and polymerize Pi into polyphosphate (Poly-P) by vacuolar transporter chaperone (VTC) complex; the Poly-P was accumulated in the vacuoles, and then transferred to the IRM associating with water transport process (Kikuchi et al., 2016). The Poly-P phosphatases Ppn1 and Ppx1 in IRM can hydrolyze the Poly-P into Pi, and export it from vacuoles to the cytoplasm through the unknown P transporters located in the vacuole membrane (Solaiman et al., 1999; Ezawa and Saito, 2018; Xie et al., 2022).
It has been shown that there is a specialized Pi export system in the arbuscules, where free Pi is transported and unloaded into the periarbuscular space (PAS) (Ezawa and Saito, 2018; Zhou et al., 2021; Xie et al., 2022). After the Poly-P hydrolyzation in the IRM and arbuscules, the Pi transporters containing SPX (SYG1/Pho81/XPR1) domains participate in the Pi export process at the symbiotic interface, releasing Pi into the PAS (Ezawa and Saito, 2018; Plassard et al., 2019; Xie et al., 2022). Pi in the PAS cross the PAM to root cortical cells relies on the mycorrhiza-induced phosphate transporters belonging to the plant PHT1 gene family (Javot et al., 2007; Yang et al., 2012; Xie et al., 2013; Volpe et al., 2016). On the other hand, it has been found that AMF also possesses the low-affinity Pi transport system (such as Pho87/90/91) and phosphatases (Tisserant et al., 2012; Lin et al., 2014; Venice et al., 2020). The Pi transport systems in AMF is very similar to that of Saccharomyces cerevisiae, which is well-known to contain the high-affinity system Pho84p and Pho89p and the low-affinity system Pho87p, Pho90p, and Pho91p (Auesukaree et al., 2003). This suggests that there exists a conserved PHOSPHATE (PHO) signaling pathway between AMF and yeasts (Aono et al., 2004; Olsson et al., 2006; Kikuchi et al., 2014; Zhou et al., 2021).
Transcription factors (TFs) play crucial roles in the regulation of gene expression in fungal cells and determine the functions of eukaryotic cells (Shelest, 2008). Recent advances have been made in identifying several hub TFs in mycorrhizal plants functioning in the control of AM symbiosis nutrient uptake and exchange (Pimprikar and Gutjahr, 2018; Shi et al., 2021; Das et al., 2022; Ho-Plágaro and García-Garrido, 2022). By contrast, the studies on AM fungal TFs are very limited. In two previous studies, only a few TFs, such as RiMsn2 from R. irregularis and GintSTE from Glomus intraradices (currently R. irregularis) are preliminarily investigated (Tollot et al., 2009; Sun et al., 2018), while the key transcriptional regulators engaged in Pi absorption and homeostasis have not been explored in AMF. Some recent studies have shown that there exists the bHLH domain-containing transcription factors encoding genes involved in the PHO pathway in response to low Pi conditions in G. margarita, Gigaspora rosea and R. irregularis (Tang et al., 2016; Xie et al., 2016, 2022; Zhou et al., 2021). It is well-known that in S. cerevisiae, the AMF bHLH ortholog ScPho4 is the transcription factor regulating the PHO pathway to control Pi absorption and homeostasis (Lenburg and O’Shea, 1996; Auesukaree et al., 2004; Wykoff et al., 2007). Although several studies have revealed that some important genes are involved in the Pi signaling and metabolism pathways in AM fungal symbionts (Balestrini et al., 2007; Xie et al., 2016, 2022), the underlying molecular mechanisms on the regulation of Pi uptake and homeostasis in AMF during symbiosis remain elusive.
Eucalyptus species is the most valuable and widely planted hardwood in the world (Qin et al., 2021). It has many advantages such as fast growth and strong adaptability to drought, fire, insect pest, soil acidity and low fertility (Rockwood et al., 2008). Eucalyptus wood can be used as an important raw material for industrial pulp and paper making, fuel and charcoal production because of its high-density property (Rockwood et al., 2008; Kato and Hibino, 2009). Because of its economic and ecological values, it is important to enhance the productivity of Eucalyptus limited by environmental factors such as P and N (Santos et al., 2016; Yao et al., 2021; Che et al., 2022). AM symbiosis is an environmentally friendly strategy to promote the Eucalyptus plants nutrient absorption when compared with fertilizer excessive use (Smethurst, 2010). Recently, there are many researches on physiological roles of AMF and ectomycorrhizal fungi on the Eucalyptus plants (Pagano and Scotti, 2008; Santos et al., 2021), but little studies focus on the regulatory mechanisms of Pi uptake and exchange processes in AM fungal symbiont during AM fungus-Eucalyptus symbiosis.
After the investigation of Pi uptake and transport processes during AMF and plant interaction, to further understand the regulatory mechanisms of the Pi uptake and homeostasis at the symbiotic interface, we start to search the regulators (TFs) in R. irregularis expressed during the in planta phase. Here, we show a new transcription factor from R. irregularis (RiPho4), which contains a C-terminal bHLH domain, and provide experimental evidence for roles in the regulation of Pi uptake and homeostasis during AMF-Eucalyptus symbiosis. Moreover, our findings offer new insights into the control of Pi uptake and metabolism in the AM fungal symbionts at the symbiotic interfaces.
Materials and methods
AM fungus and plant materials and growth conditions
AM fungus used in this study was R. irregularis DAOM 197198, which was propagated in the pot cultures with maize (Zea mays). Spores of R. irregularis were collected from Z. mays root segments. The plant material used in this study was Eucalyptus grandis (The seeds was from the Research Institute of Tropical Forestry, Chinese Academy of Forestry). The surface-sterilization of E. grandis seeds was performed as the described previously (Plasencia et al., 2016). The seedlings germinated were transferred from solid medium to the pots, and then inoculated with R. irregularis (about 200 spores per plant). After inoculation, E. grandis plants are cultivated in a growth chamber under 16 h: 8 h, 24°C: 19°C, light: dark conditions (light intensity, 100–200 Wm−2; relative humidity, 55%).
Phosphate treatment
Eucalyptus grandis plants were cultivated in pots under AMF inoculation (AM) and uninoculated control (NM) treatments. Each treatment was carried out with three Pi concentrations including 30, 300 and 1,000 μM K2HPO4 (Sugimura and Saito, 2017; Fan et al., 2020). The plants were fertilized once a week using the modified Long-Ashton (mLA) solutions (Hewitt, 1966) containing the indicated Pi concentrations. After 45 days treatments, NM and AM plants were collected and stored at-80°C for subsequent experiments. And plant roots colonized with AMF in the pot experiment were collected to extract DNA and RNA from R. irregularis.
For RiPho4 subcellular localization analysis, yeast cells were cultured in YNB medium without uracil (Ura) for 24 h. And the yeast cells were cultured in YNB/-Ura medium for 12 h, supplemented with different Pi concentrations (the final Pi concentrations in each were 600 μM, 1 mM, 10 mM KH2PO4, respectively) (O’Neill et al., 1996; Komeili and O’Shea, 1999; Zhou and O’Shea, 2011).
Genomic and RNA-seq data analysis
To identify the candidate genes of the PHO pathway in R. irregularis, the genes of PHO signaling pathway in S. cerevisiae was used as the queries to search for the homologues genes in the genome of R. irregularis DAOM 197198 (Supplementary Table S2). Through genome BLAST in NCBI and GEO databases, tBLASTn and BLASTp searches were carried out to search the target genes in R. irregularis, and the best matching sequence ranking first from BLASTp results is used for subsequent analysis. The homologous amino acid sequences were collected from the NCBI database.
To analyze the expression profiles of the target genes in different fungal tissues of R. irregularis, the original RNA-seq sequences of non-symbiotic tissues (germinating spores) and symbiotic tissues (mycorrhizal roots) of R. irregularis download from DDBJ database. The accession numbers of RNA-seq reads are as follows: germinating spores harvested at a week after inoculation (DRA002591), germinating spores collected at 7 days after induction (GSE67913), laser microdissected cells contain IRM and arbuscules collected from Medicago truncatula mycorrhizal roots (GSE99655), mycorrhizal roots of M. truncatula (GSE99655, GSE67926), ERM collected from carrot root culture (GSE99655) (Zeng et al., 2018, 2020).
Gene expression analysis
The extraction of the genomic DNA from R. irregularis was referred to the method of Zézé et al. (1994) for amplification of gene fragments containing non-coding regions. Besides, total RNA was extracted by Trizol (Invitrogen) method, and the concentration and purity of total RNA were detected by NanoDrop 2000 (Thermo Scientific, United States). First-strand cDNA was produced from total RNA by a Hiscript III reverse transcriptase kit with gDNA wiper (Vazyme Biotech, Nanjing, China) following the manufacturer’s instructions. The qRT-PCR experiments were performed in a 96-well Real time PCR system instrument (BioRed, Hercules, CA, United States) (Xie et al., 2022). RiEF1α gene from R. irregularis was used as an internal control for qRT-PCR analysis. Relative expression levels were calculated using 2−ΔΔCt method. The list of gene-specific primers used for qRT-PCR analysis is given in Supplementary Table S3.
Yeast manipulations
The full-length of RiPho4 was amplified by gene-specific primers containing the BamHI site (Primer sequences are listed in Supplementary Table S4). One step cloning Kit (Vazyme Biotech, Nanjing, China) was used to recombine the RiPho4 cDNA into the pUG36 vector, and the resulting plasmid pUG36-GFP-RiPho4 was transformed into the yeast EY57 strain using the LiOAc/PEG-based method described previously (Gietz and Schiestl, 2007). Positive transformants were grown in YNB liquid medium lacking Ura for oscillation culture at 28°C for 24 h.
The ORF of RiPho4 was cloned from cDNA of R. irregularis using the primers ADRiPho4-F/R, and then cloned into the pGADT7 (Chien et al., 1991). To construct bait-specific pAbAi vector, cis-acting elements with their flanking nucleotides from the promoters of target genes (Tomar and Sinha, 2014) (listed in Supplementary Table S4) were synthesized and cloned into the SalI site of the pAbAi vector (Clontech Laboratories, United States). Then the constructed vectors were transformed into the Y1HGold yeast strain, and grown on YNB/-Ura solid medium with Aureobasidin A (AbA) concentration (100 mM) for testing the minimal inhibitory concentration of AbA for bait-specific pAbAi plasmids. We transformed the bait-specific pAbAi fragments and pGADT7-RiPho4 were co-transformed into yeast cells, and screened using the SD medium lacking Leucine. The yeast cells (OD600 = 0.2) containing both RiPho4-pGADT7 and promoter fragments were inoculated on SD medium lacking Leucine with different AbA concentrations (100–800 ng/ml). The yeast cells carrying pAbAi-p53 and pGADT7-SV40 were used as the positive control, whereas the yeast cells containing pGADT7-RiPho4 and promoter fragments lacking the cis-element served as the negative control. In addition, the inhibitory effect was adjusted based on the growth of yeast (Zhan et al., 2017; Sun et al., 2018; Yang et al., 2019).
Virus-induced gene silencing
Tobacco (Nicotiana benthamiana) is used for this virus-induced gene silencing (VIGS) experiment (Zhang and Liu, 2014; Kikuchi et al., 2016; Xie et al., 2022). Two specific cDNA fragments of RiPho4 from the-9 to +226 regions (RiPho4-RNAi-1) and + 1,200 to +1,429 regions (RiPho4-RNAi-2) relative to the start codon ATG (Supplementary Figure S5) were amplified by PCR following the method described by Senthil-Kumar and Mysore (2014). The cloned gene fragments (the primers VigsRiPho4-F1/R1 and VigsRiPho4-F2/R2 were listed in Supplementary Table S4) were ligated to the pTRV2 vector. And the resulting plasmids pTRV2-RiPho4-1 and pTRV2-RiPho4-2 were separately transformed into Agrobacterium tumefaciens GV3101 (Grønlund et al., 2013; Zhang and Liu, 2014). A. tumefaciens culture (OD600 = 1.0) with pTRV1 and that with pTRV2 or pTRV2-RiPho4 were mixed together and activated by adding 10 mM Acetosyringone, then stood in darkness for 2 h. The inoculums were injected into the leaves of N. benthamiana, whose roots had been inoculated with R. irregularis for 4 weeks as described by Kikuchi et al. (2016). Tobacco plants treated with three Pi concentrations (30 μM, 100 μM, 300 μM) were cultured in a small chamber for 2 weeks.
AM phenotypical analysis
The collected E. grandis roots were digested in 10% KOH solution at 90°C for 40 min for 3 times, and washed with distilled water, then neutralized in 2% HCI solution for 20 min. After washing with sterile water for 3 times, AM roots were stained with 5 μg/ml wheat germ agglutinin 488 (WGA488, Invitrogen) at 37°C for 30 min (Phillips and Hayman, 1970; Ivashuta et al., 2005). The enzyme activities of succinate dehydrogenase (SDH), alkaline phosphatase (ALP) and acid phosphatase (ACP) were performed as described previously (Zhao et al., 1997). Mycorrhizal colonization was estimated according to Trouvelot et al. (1986).
Microscopy
The fluorescent signals in yeast cells and AM roots were observed by a fluorescence microscope (Y-TV55; Nikon, Tokyo, Japan). The colonization levels of SDH, ACP and ALP enzyme activity staining was calculated under the light microscope (Y-TV55; Nikon, Tokyo, Japan).
Total P concentration analysis
Fresh samples of E. grandis were lyophilized for 6 h. To grind dried samples into powder, we added magnetic beads to them and grinded them with a grinder (35 Hz) for 2 min. 0.3 g of sample powders were digested by 6 M nitric acid under the water bath at 90°C for 1 h (Fan et al., 2020). The digested samples were filtered with filter membranes and diluted with 5% nitric acid to 10 ml. Total P concentrations in the E. grandis digests were measured by the inductively coupled plasma optical emission spectrometry (ICP-OES; Varian, United States). Total P concentrations of N. benthamiana were detected by the tissue total P content detection Kit (Cat. NO. BC2855, Solarbio, China) and measured with the Microplate Reader (Vaioskan, Thermo Scientific, United States).
Bioinformatics
The BLASTP1 was used to search homologs of RiPho4 protein in the fungi species. The characteristics of the secondary structure of RiPho4 were analyzed by the SMART program.2 The SWISS-MODEL website3 was employed to build the three-dimensional model of RiPho4. The conserved regions of RiPho4 and its homologous proteins were analyzed by Meme.4 The heat diagram for PHO pathway gene expression levels in different fungal tissues was made by the TBtools (Chen et al., 2020). The existence of cis-acting element by searching for 1.5 kb promoter sequences of the target gene coding regions through the NCBI database and PlantCARE software.5
Phylogenetic analysis
The unrooted phylogenetic tree of RiPho4 protein and other amino acid sequences in different fungi species were constructed with MEGA7.0 (Kumar et al., 2016) using the neighbor-joining method. Accession numbers of all the fungal proteins are shown in Supplementary Table S2.
Statistical analysis
Data are preliminarily accounted by Microsoft EXCEL 2016, and statistical significances between treatments were analyzed by analysis of variance (ANOVA) using SPSS software (Version 26.0, SPSS Inc., United States). The Duncan’s multiple range test were used for comparing more than two datasets. A value of p < 0.05 was considered to be statistically significant. The different letters indicate significant differences among phenotypes or treatments. Huang and Freiser (1993, Origin Lab, United States) to plot and illustrate diagram for changing curves of different treatments.
Accession numbers
Sequence data from this article can be found in the AM fungal genome and GenBank libraries under the following accession numbers: RiPho4 (XP_025175129.1), RdPho4 (RGB28534.1), RcPho4 (GBB98521.1), GcPho4 (RIA86088.1), GmPho4 (KAF0357978.1), GrPho4 (RIB17793.1), DePho4 (RHZ83467.1). Other accession numbers of fungal Pho4 proteins were provided in Supplementary Table S2.
Results
Effects of AMF on the growth and Pi uptake of Eucalyptus grandis in roots under Pi-deficient conditions
To study the effects of AMF inoculation on the growth and Pi uptake of E. grandis subjected to different Pi conditions, we treated mycorrhizal (AM) E. grandis with three phosphate concentrations when compared with non-mycorrhizal (NM) E. grandis. After 7 weeks inoculation, the growth performance of AM E. grandis was better than NM plants under medium and low phosphate conditions (30 and 300 μM) (Supplementary Figure S1). Overall growth (such as plant height, root length, and biomass) also showed such a trend (Figures 1A–D). Compared with NM plants, the total P concentrations of both roots and shoots showed significant increases in AM plants under low and medium phosphate concentrations (30 and 300 μM), while there were no significant differences between AM and NM plants under high Pi conditions (Figure 1E). In addition, it was found that the colonization levels of E. grandis exposed to low Pi (30 μM) were significantly higher than those grown under the medium and high Pi treatments (300 and 1,000 μM) (Figure 1F). Moreover, we also detected the activities of ACP, ALP and SDH involved in AM fungal function and mycorrhizal Pi utilization efficiency (Guillemin et al., 1995; Vivas et al., 2003; Kouas et al., 2009; Li et al., 2017). Activities of these three enzymes in roots exposed to 30 μM Pi were significantly higher than that subjected to 300 μM and 1,000 μM Pi (Figures 1G–I; Supplementary Table S1). Taken together, these results revealed that, during AM symbiosis, AM fungus R. irregularis can promote the E. grandis Pi uptake and utilization efficiency to improve the plant growth under Pi-deficient conditions.
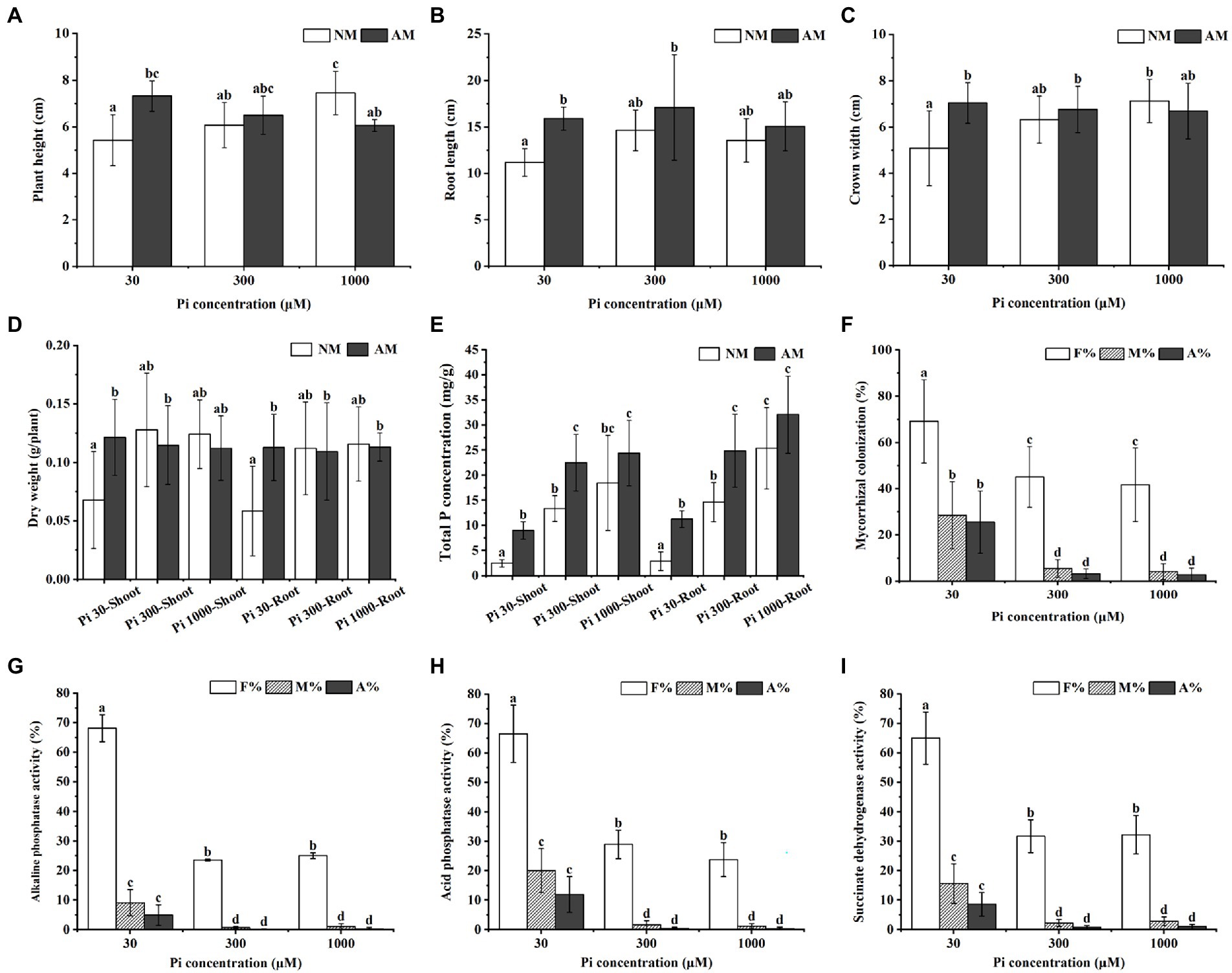
Figure 1. Impact of different Pi concentrations and AMF colonization on Eucalyptus grandis growth and enzyme activities within mycorrhizal roots. (A–D) The growth performances of NM and AM E. grandis plants under different phosphate concentrations. (E) Total P concentrations of shoots and roots of NM and AM E. grandis with R. irregularis grown at different Pi levels. (F) Mycorrhizal colonization levels among different phosphate concentrations were determined in roots after the WGA488 staining. (G–J) Impact of phosphate concentrations on three enzymes activities, including alkaline phosphatase (ALP), acid phosphatase (ACP) and succinate dehydrogenase (SDH), which are involved in AM fungal metabolism and Pi homeostasis during symbiosis. Data on the pictures are obtained by the ALP (G), ACP (H), and SDH (I) activities staining, respectively. (F–I) F%, the total colonization frequency; M%, the percentage of mycorrhizal intensity; A%, the percentage of arbuscule abundance. Error bars represent the mean of five biological replicates ± SD. Averages with the different letters on the top of the column mean significantly differences at p < 0.05 level, based on Duncan’s new multiple range test.
Transcription levels of PHO pathway genes in Rhizophagus irregularis are dependent on Pi availability
To investigate the effect of external phosphate concentrations on the transcription levels of AM fungal genes involved in Pi uptake and metabolism, we examined the expression profiles of 12 genes in the PHO pathway of R. irregularis in mycorrhizal E. grandis roots during different Pi conditions. As shown in Figure 2A, relative to medium and high P concentrations (300 and 1,000 μM) conditions, the transcript of RiPho2, which was predicted to be a cofactor for RiPho4 (Chen et al., 2018; Xie et al., 2022), was significantly higher in mycorrhizal roots under low Pi (30 μM) conditions. Similarly, the expression levels of RiPT1, RiPT2, RiPT3 and RiPT6, which encode the potential phosphate transporters responsible for Pi uptake, were much higher in mycorrhizal roots during low Pi concentration when compared with high Pi treatments (Figures 2B–E). Moreover, the RiACP1 and RiALP1 involved in Pi and Poly-P metabolisms were expressed in response to the low Pi application (Figures 2F,G). Besides, the expressions of key genes in response to Pi starvation signaling, such as RiPho81 and RiPho85 were also detected in mycorrhizal roots under such conditions. RiPho81 was significantly induced at low Pi concentration, while transcripts of RiPho85 were slightly but not significantly reduced at high Pi treatments (Figures 2H,I). On the other hand, the expressions of Poly-P accumulation-related genes RiVTC1 and RiVTC4 were significantly higher at 30 μM Pi than medium and high Pi concentrations (300 and 1,000 μM), whereas RiVTC2 expression was not significantly enhanced in roots exposed to low Pi concentration (Figures 2J–L). Overall, these gene expression profiles revealed that the Pi sensing and transport/metabolism genes are regulated in response to Pi starvation in R. irregularis during AM symbiosis.
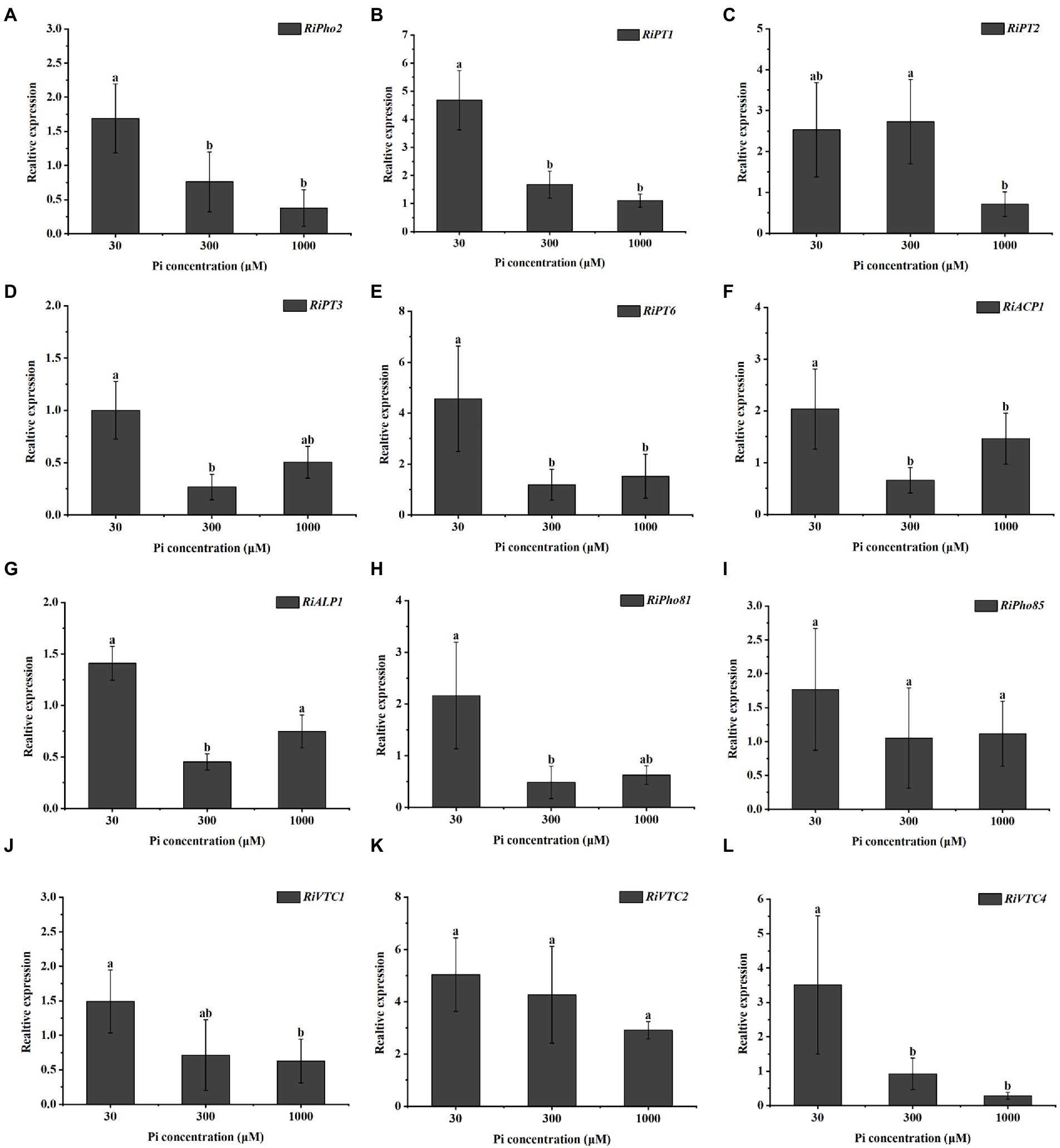
Figure 2. Expression profiles of the genes involved in the phosphate (PHO) signaling pathway from R. irregularis in mycorrhizal E. grandis roots at different phosphate concentrations. (A–L) Expression levels of genes of PHO pathway under different phosphate concentrations, including (A) transcription cofactor, (B–E) Pi transporters, (F,G) phosphatase related genes, (H,I) cyclin-protein genes and (J–L) vacuolar transporters, suggesting that the expressions of PHO pathway genes are affected by phosphate concentrations. AM fungal RiEF1α is set as the reference gene. The data represent the means of three biological replicates with standard deviations. Different letters indicate the Duncan’s multiple comparison results. Significance, p < 0.05.
Identification of RiPho4, which encodes a HLH domain-containing transcription factor
The above results (see Figure 2) indicate that the downstream genes of the PHO pathway in R. irregularis are transcriptionally dependent on the Pi availability. In order to identify the key transcription factor regulating the expression of downstream PHO genes in response to phosphate starvation, we searched for the sequences that correspond to the TFs in PHO pathway of S. cerevisiae using the genome sequencing projects of R. irregularis DAOM 197198 (Tisserant et al., 2013; Chen et al., 2018), and found a transcription factor called RiPho4. According to the GenBank annotation, RiPho4 contains 5 exons and 4 introns, a total length of 2052 bp with 1710 bp of ORF (Figure 3A). Using Smart program, it is predicted that RiPho4 encoding protein has 6 domains, 5 of which are low complex domains (LCDs), while the region from +436 to +515 is a HLH (helix loop helix) domain (Figure 3B), which is one of the specific domains of TFs (Ferré-D'Amaré et al., 1993; Kiribuchi et al., 2004; Carretero-Paulet et al., 2010; Zhu and Huq, 2011). Further the three-dimensional conformation of RiPho4 (Figure 3C) showed that RiPho4 is a typical HLH domain containing protein. Therefore, it is predicted that RiPho4 may serve as a key transcription factor containing a HLH domain in R. irregularis.
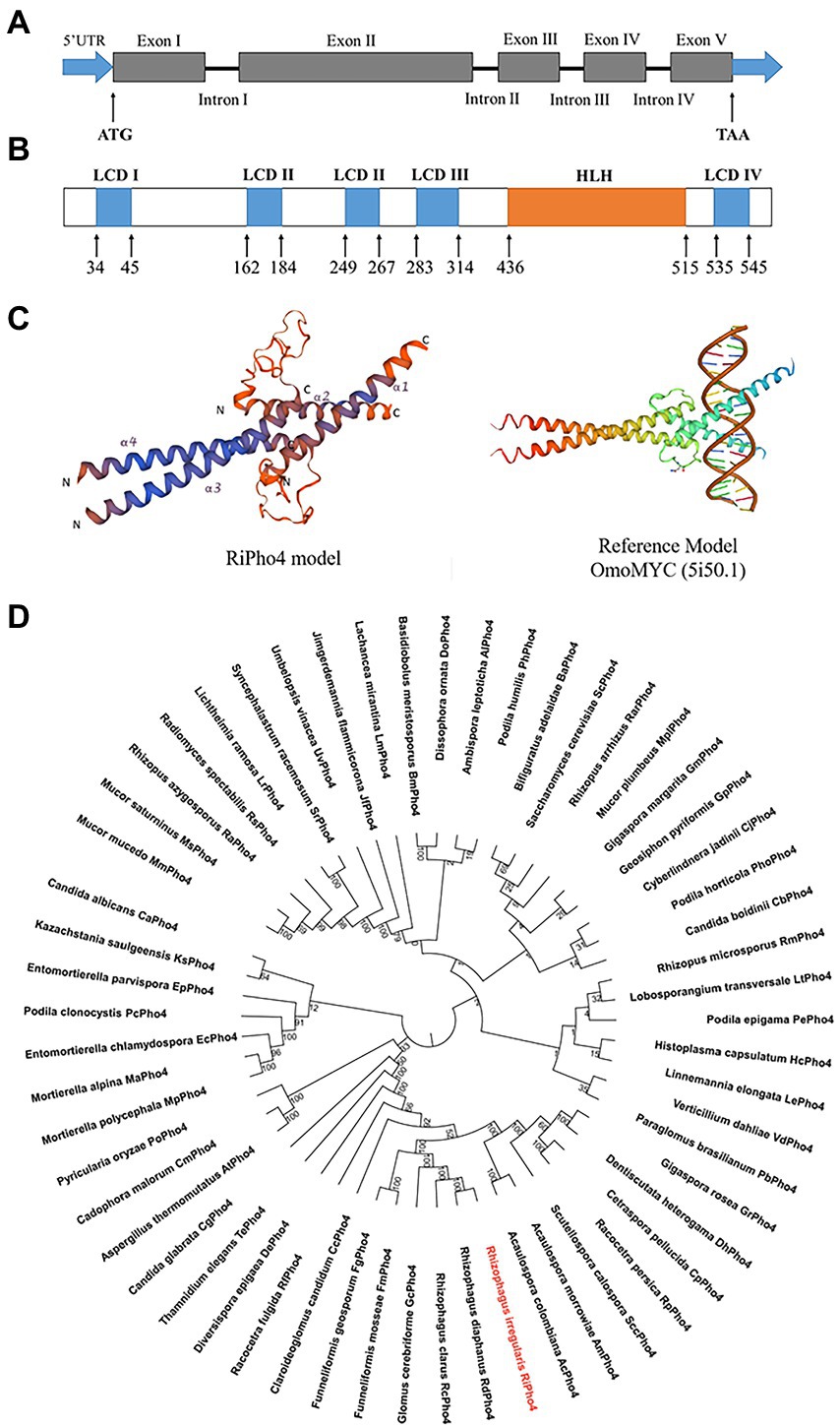
Figure 3. Gene and protein structures of RiPho4 in R. irregularis and the evolutionary relationships with Pho4 proteins in various fungi species. (A) The RiPho4 gene contains five exons and four introns, corresponding to gene ID 36869193. The untranslated regions (UTRs) are shown in the figure, ATG and TAA represent the translation start site and translation termination site, respectively. (B) Inferred protein domain of RiPho4. RiPho4 protein contains 570 amino acids and consists of one HLH domain and four low complex domains (LCDs) (http://smart.embl.de/). (C) Predicted three-dimensional structure of RiPho4 model by SWISS-MODEL. It was predicted that the 3D structure of RiPho4 is similar to that of OmoMYC (5i50.1), a protein encoded by MYC dominant-negative allele in Mus musculus (Jung et al., 2017). (D) The unrooted phylogenetic tree was constructed with the neighbor-joining method, based on multiple sequence alignments of RiPho4 (red color) and other Pho4 proteins from different fungi species using MEGA v7.0 software. Bootstrap tests were performed using 1,000 replicates. The accession numbers of all Pho4 proteins are provided in the Supplementary Table S2.
RiPho4 is conserved across fungi species
To determine the evolutionary relationships of Pho4 proteins between R. irregularis and other different fungi species, we performed the phylogenetic analysis and conserved motif identification. The result shows that RiPho4 is related to AM fungal TFs GmPho4, GcPho4, FgPho4 and FmPho4, and has closely relative to the RdPho4 and RcPho4 from Rhizophagus diaphanous and Rhizophagus clarus (Figure 3D). RiPho4 protein is >98% identical to Pho4 from R. diaphanous, 41% to LtPho4 from the filamentous fungi Lobosporangium transversale and 38% to ScPho4 of S. cerevisiae which has been reported to be a typical HLH-type transcription factor (Oshima, 1997; Tomar and Sinha, 2014). In addition, we identified RiPho4 functional orthologs with Pho4 from other AMF (Supplementary Figure S2). These in silico results suggest that R. irregularis RiPho4 is highly conserved across fungi species.
RiPho4 is induced in mycorrhizal roots
To determine the spatiotemporal expression patterns of RiPho4 from R. irregularis, the transcriptional analysis of RiPho4 and relevant PHO genes were performed in different tissues of R. irregularis using the RNA-sequencing data (Zeng et al., 2018, 2020). As shown in Figure 4A, the transcription of RiPho2, RiPT1, RiPho81, RiPho80, and RiPho91 (equally RiPT7, Xie et al., 2022) were up-regulated in mycorrhizal roots when compared with other fungal tissues, while the transcriptional levels of RiPho4 were induced in both mycorrhizal roots and arbuscules of R. irregularis, and RiPho85 were constitutively expressed in these six different fungal tissues. To confirm the transcriptomic data, we further conducted the qRT-PCR experiments with roots of E. grandis colonized with R. irregularis. The time-course analysis indicated that RiPho4 expression was still high in the later stage of mycorrhizal symbiosis, and its transcript reached the highest level at 56 days post inoculation (Figure 4B), this pattern was correlated with the colonization and development processes of R. irregularis within E. grandis roots (Supplementary Figure S3). Additionally, transcriptional patterns of the monosaccharide transporter gene RiMst2 and Pi transporter gene EgPT4, which are thought to be the AM marker genes (Helber et al., 2011; Che et al., 2022), were also similar to that of RiPho4 during symbiosis (Figures 4C,D). Therefore, these results indicated that RiPho4 is induced in the arbuscles of mycorrhizal roots.
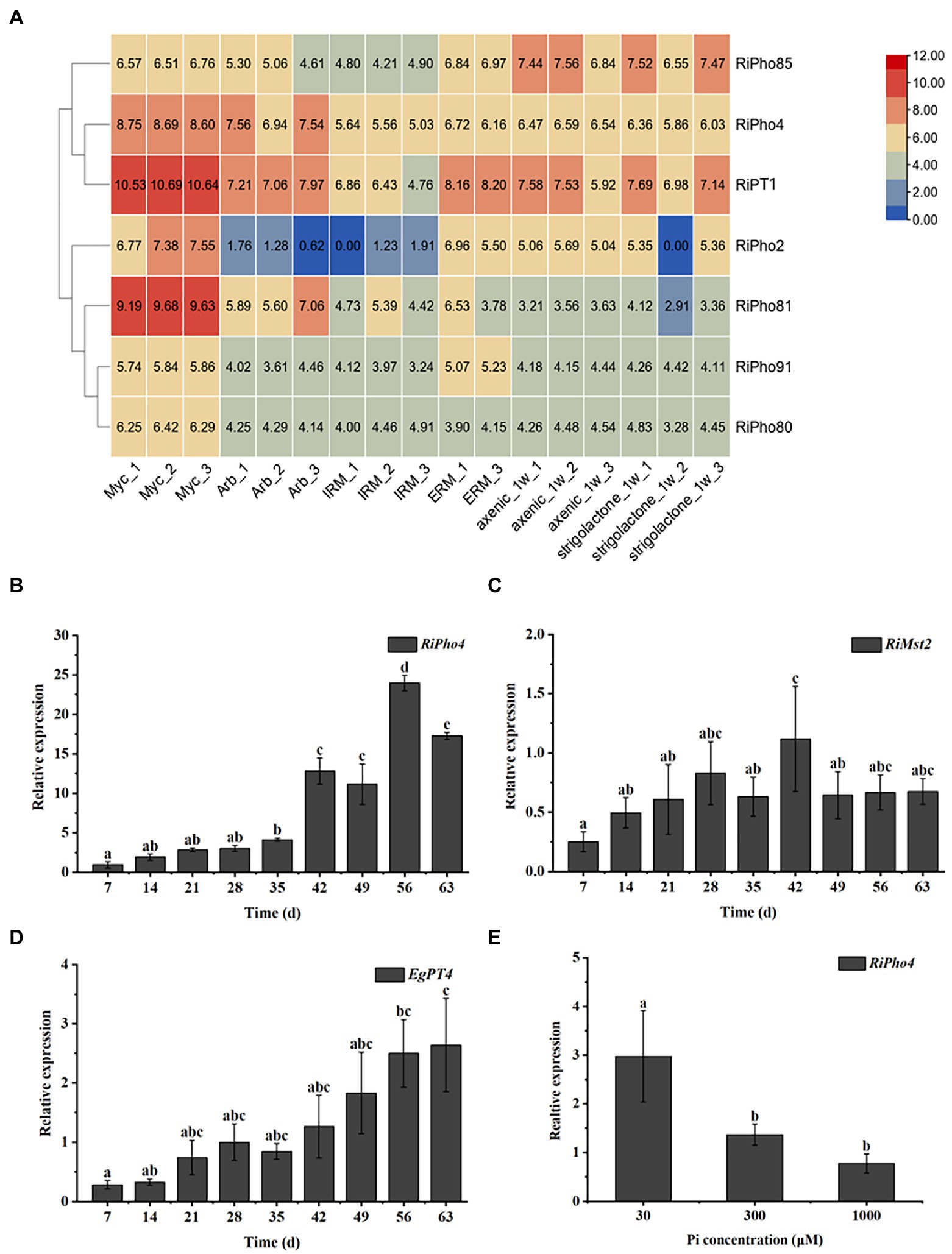
Figure 4. Spatiotemporal expression patterns of RiPho4 from the AM fungus R. irregularis. (A) Analysis of the heat diagram for transcriptional levels of RiPho4 and some PHO pathway genes in different fungal tissues: mycorrhizal roots of Medicago truncatula (Myc), arbuscules from M. truncatula mycorrhizal roots (Arb), intraradical mycelium (IRM), extraradical mycelium (ERM), axenic germinating spores harvested at a week after inoculation (axenic_1w) and germinating spores at 7 days after GR24 induction (strigolactone_1w). The heatmap of identities was visualized by the TBtools (Chen et al., 2020). (B–D) Time-course assay of the expression of RiPho4, RiMst2 and EgPT4 in E. grandis mycorrhizal roots after 7, 14, 21, 28, 35, 42, 49, 56 and 63 days (d) inoculation with R. irregularis. RiEF1α and EgUBI3 were used as the reference genes for R. irregularis and E. grandis gene normalization, respectively. (E) Expression analysis of RiPho4 in mycorrhizal roots of E. grandis grown under different phosphate concentrations. The error bars are the means with standard errors of three biological replications. Treatments with the same letters do not differ from others by the Duncan’s test at 5% probability.
RiPho4 is expressed in response to Pi starvation
To further determine the effect of external Pi concentrations on the transcription of RiPho4 during the in planta phase, E. grandis plants inoculated with R. irregularis were cultured in the pots supplemented with different phosphate concentrations (30, 300 or 1,000 μM). The qRT-PCR experiment was then performed on the mycorrhizal roots of E. grandis subjected to different Pi concentrations as mentioned above. As a result, the expression levels of RiPho4 were significantly inhibited in roots during high Pi concentrations (300, 1,000 μM) (Figure 4E). This result is consistent with the above findings on PHO gene expression profiles (see Figure 2), suggesting that RiPho4 is induced during mycorrhizal symbiosis in response to Pi starvation.
RiPho4 encodes a transcription factor and localizes in nucleus at low phosphate concentrations
Previous studies have found that ScPho4 protein in S. cerevisiae imports into the nucleus under low phosphorus supply, whereas the Pho4 exports into the cytoplasm nucleus of yeast cells exposed to high phosphorus conditions (Kaffman et al., 1994; Byrne et al., 2004; Urrialde et al., 2015). To investigate whether RiPho4 has a similar function to ScPho4 from S. cerevisiae, we carried out the subcellular localization experiments in yeast cells. As expected, GFP-RiPho4 fusion protein was localized in the nuclei of yeast strain EY57 cells during low Pi treatment (600 μM), while this fusion protein was found in the cytoplasm of yeast cells exposed to high P concentration (10 mM) (Figure 5A). To further verify the localization results as mentioned above, we used a labeling dye DAPI, which can bind to DNA to label the nuclei of yeast cells. The co-localization analysis indicated that the expression of GFP-RiPho4 fusion protein in yeast cells was confirmed to be correctly localized in the nuclei of yeast cells under Pi-limited (600 μM and 1 mM) conditions (Figure 5B). In conclusion, these results reveal that RiPho4 protein locates in the nucleus under low Pi conditions and may serve as a transcription factor in fungal cells.
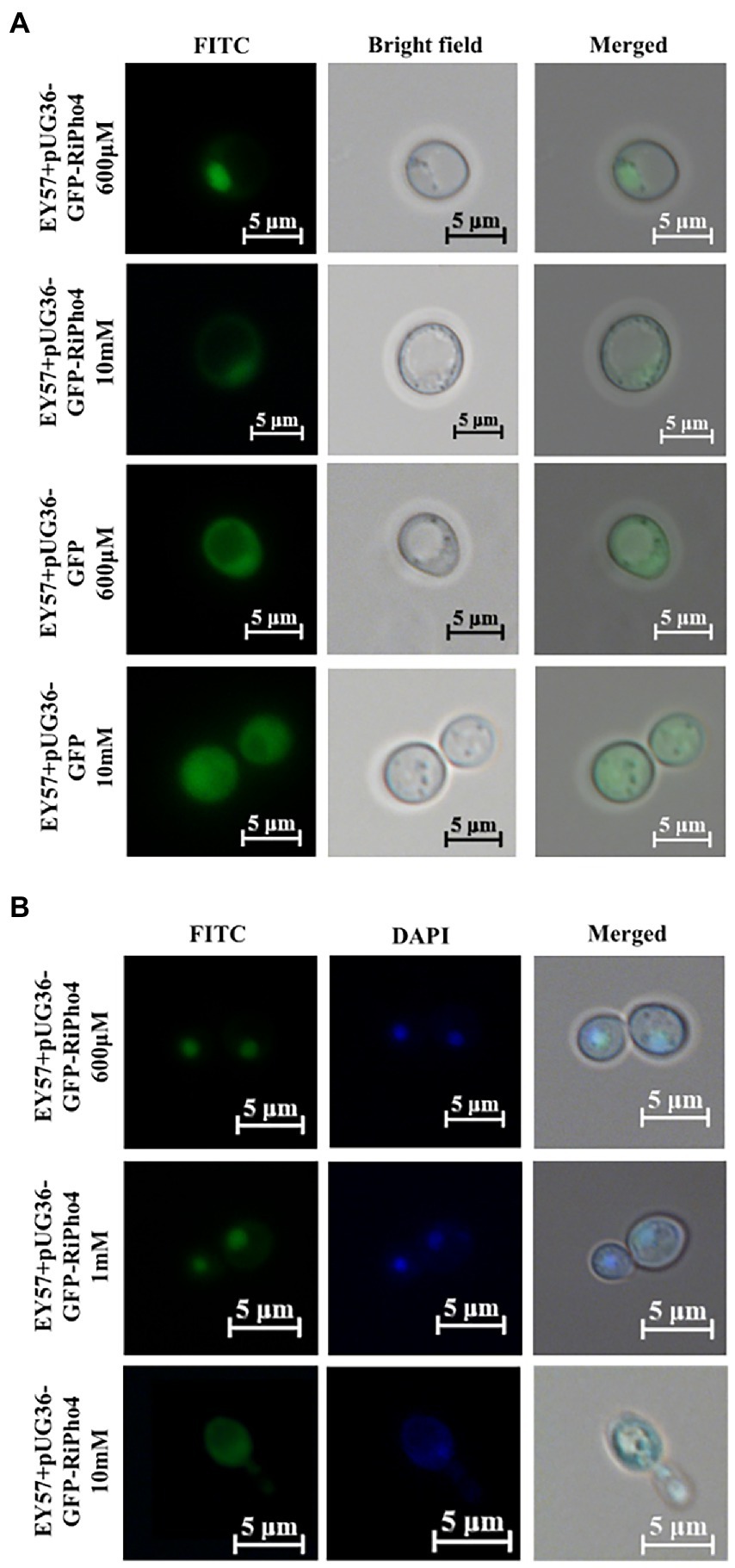
Figure 5. Subcellular localization of RiPho4 in Saccharomyces cerevisiae grown under different phosphate conditions. Fluorescence microscope images of S. cerevisiae EY57 cells expressing pUG36-GFP or pUG36-GFP-RiPho4. (A) Localization of RiPho4 was captured by fluorescence microscopy, the pUG36 containing the GFP reporter gene was used as the control, showing the fluorescence signal in the cytoplasm. The yeast cells were grown in YNB liquid medium with different Pi (600 μM, 10 mM KH2PO4) treatments for 12 h. (B) The transformed cells were grown in YNB medium under different Pi (600 μM, 1 mM, 10 mM K2HPO4) conditions for 12 h. FITC showed the green fluorescence channel; DAPI presented transformed yeast cells stained with DAPI dye. Scale bars, 5 μm.
RiPho4 is required for arbuscule development
In order to further study the roles of RiPho4 in AM symbiosis, we knocked down RiPho4 expression by the VIGS method. We designed two RNAi regions to target the corresponding specific regions of RiPho4 that was very divergent from the other genes containing bHLH domains in R. irregularis to avoid off-target silencing (see Supplementary Figure S5). First, we detected the transcriptional levels of RiPho4 in mycorrhizal roots of N. benthamiana under three different Pi concentrations (30, 100, or 300 μM). The results showed that the transcriptional levels of RiPho4 in the VIGS-RiPho4 roots were significantly reduced by more than 60%, when compared with the control roots, indicating that expression of RiPho4 was effectively knocked down (Figure 6A). Expressions of the monosaccharide transporter gene RiMst2, AM fungal reference gene RiEF1α and Pi transporter gene of N. benthamiana NbPT4, which are considered as symbiotic marker genes (Helber et al., 2011; Kikuchi et al., 2016; Xie et al., 2022), were significantly decreased in RiPho4-RNAi roots under different Pi concentrations (Figures 6B–D).
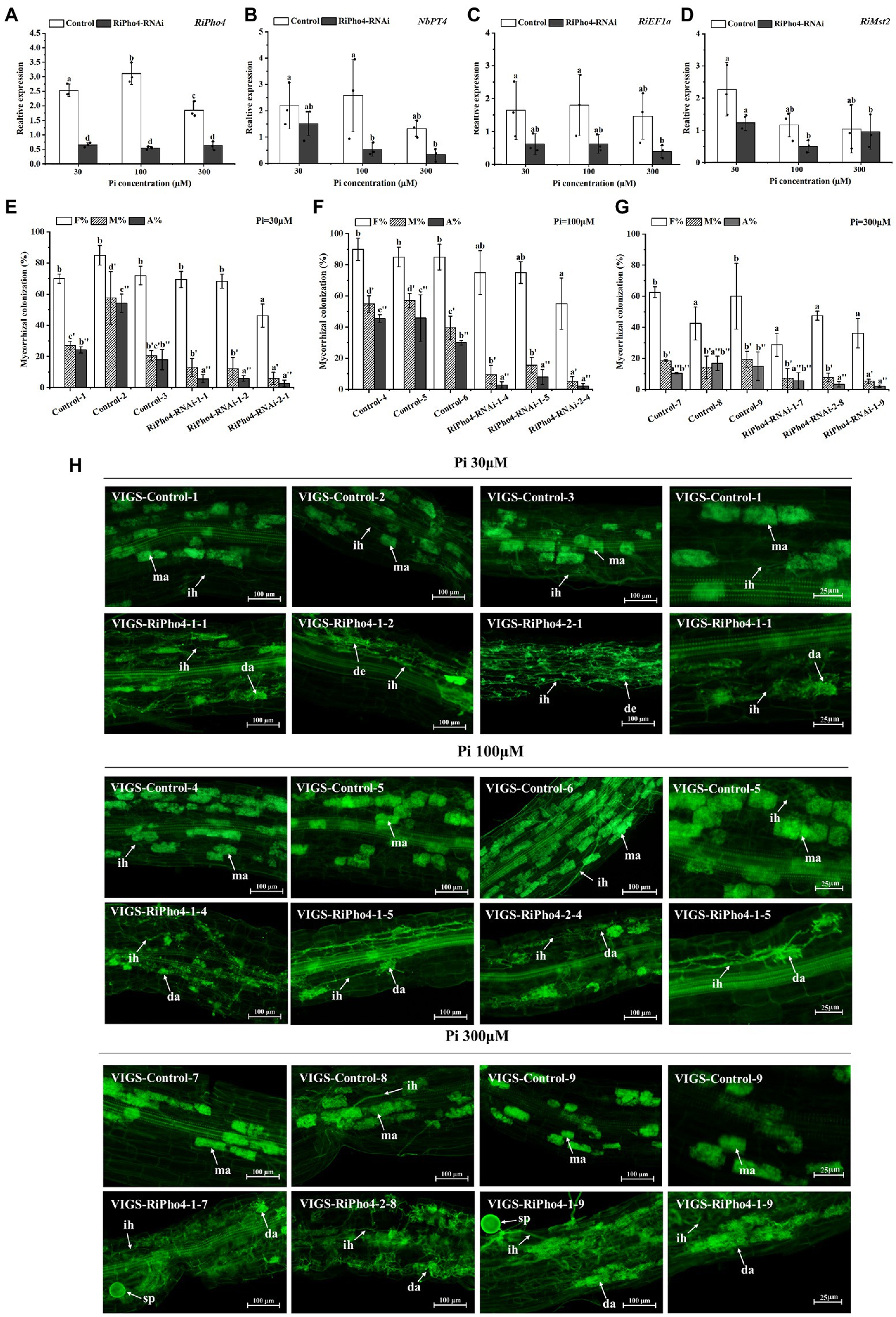
Figure 6. Molecular and arbuscular mycorrhizal phenotypes of virus-induced gene silencing (VIGS) of RiPho4 in tobacco (Nicotiana benthamiana) roots colonized by R. irregularis grown under different Pi conditions. (A–D) Expression levels of RiPho4, NbPT4, RiEF1α and RiMst2 in mycorrhizal tobacco RiPho4-RNAi roots under different phosphate conditions, estimated by quantitative RT-PCR. R. irregularis RiEF1a and N. benthamiana NbTEF1a were used as the endogenous genes for normalization of RiPho4, RiMst2, NbPT4 and RiEF1α expression, respectively. The data represented the means of three biological replicates. Bars indicated the standard errors of means. Means designated with the same letters are not significantly different (p ≥ 0.05) according to Duncan’s multiple range test. (E–G) Mycorrhizal colonization levels between the control and RiPho4-RNAi roots exposed to different Pi (30 μM, 100 μM and 300 μM K2HPO4) concentrations were determined after the WGA488 staining. F%, the total colonization frequency; M%, the percentage of mycorrhizal intensity; A%, the percentage of arbuscule abundance. (H) Fluorescence microscopic images of R. irregularis arbuscules in control and RiPho4-RNAi roots grown under different Pi conditions. The arrows and letters in the figure represent different structures of R. irregularis: ma, mature arbuscules; da, degraded arbuscules; de, dead arbuscules; ih, intraradical hyphae; sp., spores. Scale bars, 100 μm (1–3 columns), 25 μm (the 4th column).
Further observation of the mycorrhizal colonization uncovered the distinguishable AM phenotype between the RiPho4-RNAi and control roots during AM symbiosis under different Pi concentrations. Relative to the controls, the total AMF colonization in most of RiPho4-RNAi roots showed a slightly but not significantly decrease, while silencing of RiPho4 obviously decreased the mycorrhizal intensity and arbuscule abundance in roots (Figures 6E–G). Moreover, more arbuscules in the RiPho4-RNAi roots were abnormal, or degenerating under different Pi conditions (Figure 6H). Collectively, these findings reveal that RiPho4 is essential for arbuscule development.
RiPho4 plays an important role in regulating the symbiotic Pi absorption during AM symbiosis
To determine whether RiPho4 regulates the symbiotic Pi transport in AM symbiosis of N. benthamiana, we examined the total P concentration of AM tobacco shoots of control and RiPho4-RNAi plants grown under three different Pi (30, 100, or 300 μM) conditions. As a result, the P concentration in AM plants of RiPho4-silenced lines were significantly reduced compared with control plants (Figure 7). This result indicated that RiPho4 may regulate Pi transport at the symbiotic interface of arbuscular mycorrhizas.
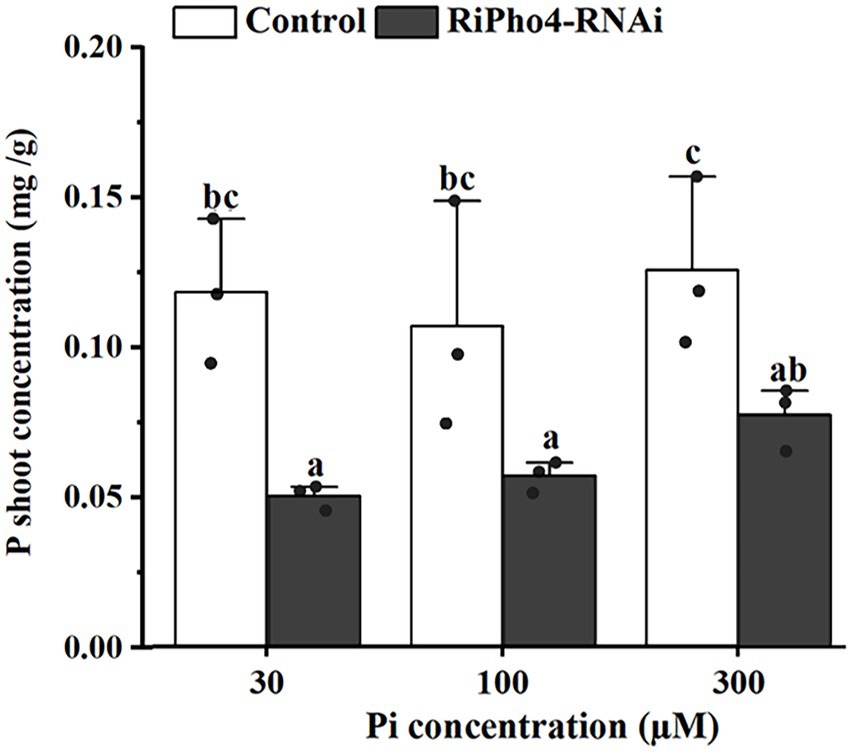
Figure 7. Phosphate concentrations in the tobacco (N. benthamiana) shoots of the control and VIGS of RiPho4 treatments under different Pi conditions. The data represented the means with SE. Different letters indicated the significant differences which statistically analyzed by Duncan’s multiple range test at the alpha = 0.05 level.
Knock-down of RiPho4 affects the expression of PHO pathway genes in Rhizophagus irregularis during AM symbiosis
To confirm the function of RiPho4 in the PHO pathway of R. irregularis, qRT-PCR was used to explore the effect of RiPho4 knock-down on the PHO pathway genes in mycorrhizal N. benthamiana roots exposed to different Pi concentrations. The results showed that RiPho2, RiPT1, RiPT2 and RiPT3 were significantly decreased in RiPho4-silenced roots under low Pi conditions (30–100 μM) when compared with control roots (Figures 8A–D). Furthermore, the expression levels of genes involved in Pi and Poly-P metabolism in R. irregularis, such as RiALP1, RiACP1, RiPPX1 and RiPPN1 (Xie et al., 2022), were also inhibited in the RiPho4-RNAi roots when compared to the controls (Figures 8E–H), suggesting that loss of RiPho4 function results in a reduction of Pi and Poly-P metabolisms under Pi-limited conditions. Therefore, these results indicate that RiPho4 may positively regulate the transcriptional levels of the downstream genes of PHO pathway in R. irregularis during AM symbiosis under Pi-deficient conditions.
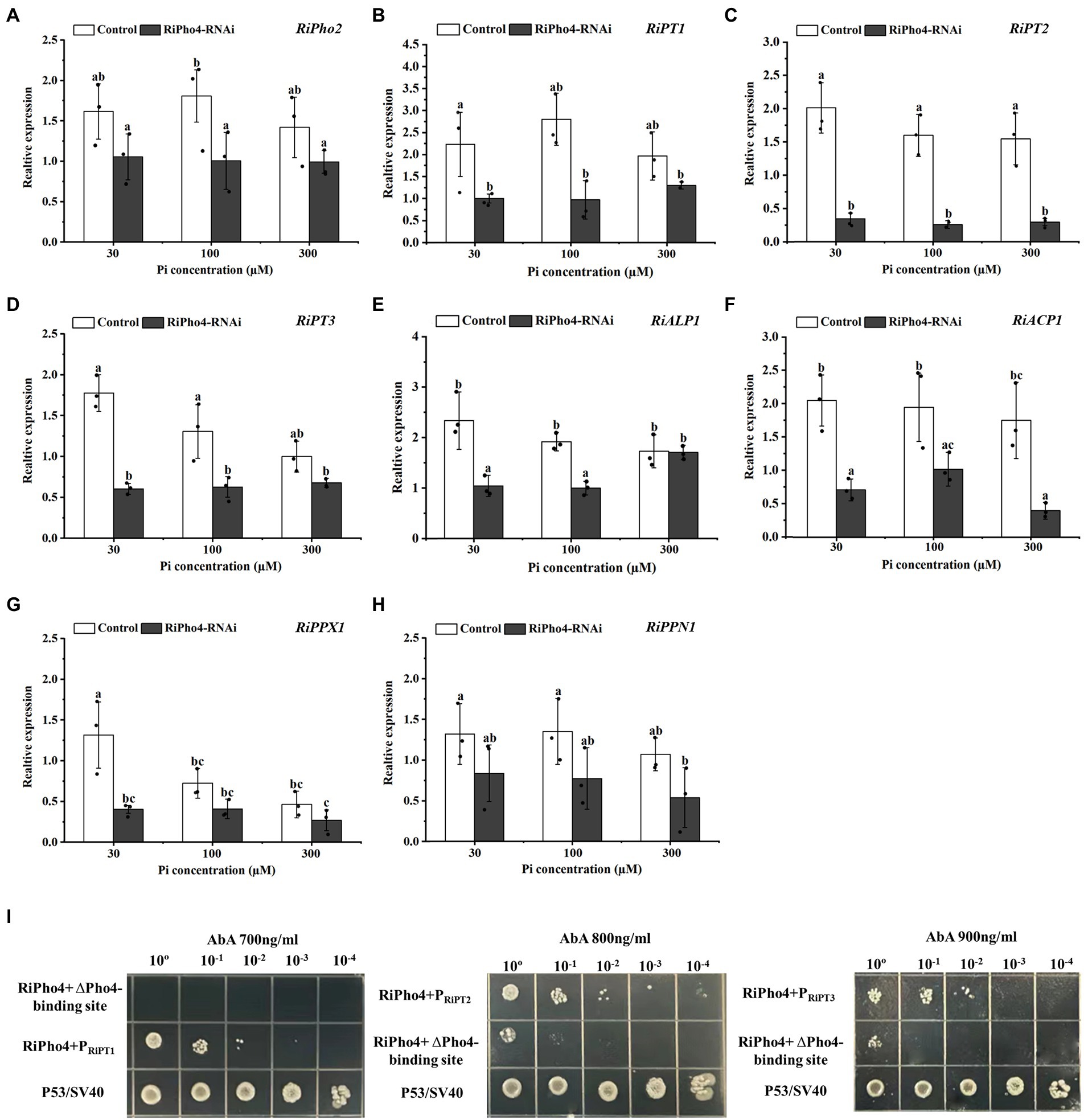
Figure 8. RiPho4 regulates the expression of downstream genes involved in Pi transport and metabolism processes in R. irregularis. (A–H) Expression analysis of R. irregularis genes engaged in PHO pathway between RiPho4-RNAi and control roots under different phosphate concentrations. Expression levels of RiPho2 (A), Pi transport genes RiPT1, RiPT2, RiPT3 (B–D), phosphatase related genes RiALP1, RiACP1 (E-F), RiPPX1 and RiPPN1 related to Poly-P metabolism (G,H) in response to different phosphate concentrations were estimated by real-time qRT-PCR analysis. Different letters indicated significant differences between control and RNAi lines. The differences between samples were analyzed using Duncan’s new multi-range test at the alpha = 0.05 level. (I) Yeast one-hybrid analysis of the interaction between RiPho4 with promoters of RiPT1, RiPT2, and RiPT3. The promoter regions of RiPT1, RiPT2 and RiPT3 containing the cis-acting elements CACGTG/T named PRiPT1, PRiPT2, and PRiPT3. The identification of interaction between RiPho4 and the promoter of RiPT1, RiPT2, or RiPT3 under the screened even higher AbA inhibition concentration indicated in Fig. S4. Yeast cells carrying both pGBKT7-P53 and pGADT7-SV40 grown on SD/−Leu were used as the positive control, PRiPT1, PRiPT2, PRiPT3-ΔPho4-binding sites with pGADT7-RiPho4 were used as the negative controls. ΔPho4-binding site indicates the deletion of the Pho4 binding site (CACGTG/T) elements located in PRiPT1, PRiPT2, PRiPT3 promoters. 10-fold serial dilutions of yeast cells were spotted on plates, and the initial yeast concentration was OD600 = 0.2.
RiPho4 directly regulates the Pi transporter genes of PHO pathway from Rhizophagus irregularis
To confirm that RiPho4 encodes a transcription factor to regulate PHO genes, we tested its ability to interact with Pi transporters of R. irregularis by the yeast one-hybrid (Y1H) assay. It is known that the common binding motif of Pho4 transcription factor is CACGTG/T (Secco et al., 2012; Tomar and Sinha, 2014), we therefore selected the same CREs (cis-regulatory elements) in the promoters of three Pi transporter genes RiPT1, RiPT2, and RiPT3 for further studies (see Supplementary Table S5).
As a result, the Y1HGold yeast cells containing pAbAi vector with PRiPT1, PRiPT2, or PRiPT3 were significantly inhibited at 400, 600, and 800 ng/ml AbA, respectively (see Supplementary Figure S4). In the Y1H assay, the yeast cells containing AD-RiPho4 plasmid and wild-type promoter PRiPT1, PRiPT2, or PRiPT3 still grew well on medium supplemented with 700 ng/ml, 800 ng/ml, or 900 ng/ml AbA, while the yeasts containing mutant promoters without Pho4-binding sites (CACGTG/T) were strongly inhibited under such conditions (Figure 8I). These data showed that RiPho4 protein interacts with the promoters of RiPT1, RiPT2, and RiPT3 through the Pho4-binding sites (CACGTG/T). Therefore, the transcription factor RiPho4 can directly regulate the Pi transporter genes RiPT1, RiPT2 and RiPT3 in PHO pathway. Collectively, the Y1H results implicate that RiPho4 may act as a transcriptional activator in the PHO pathway in R. irregularis.
Discussion
Recent years, more researchers have reported the physiological responses of Eucalyptus species to low Pi stress (Wu et al., 2014; Niu et al., 2015; Bahar et al., 2018), and focused on the effects of different P levels on plant biomass and P content (Xu et al., 2001; Standish et al., 2007; Bichara et al., 2021), but little study on the molecular mechanisms of the interaction between AMF and Eucalyptus plants. Although the PHO pathway responsible for Pi absorption, transport and metabolism has been described in several fungi species (Kerwin and Wykoff, 2009; Zheng et al., 2020; Ahmed et al., 2022), the regulatory mechanisms of Pi nutrient exchange between AMF and host plants through PHO pathway is still partially understood so far (Xie et al., 2016, 2022). In this study, we focus on the AM fungus R. irregularis mediating Pi uptake and homeostasis in AM symbiosis of E. grandis by investigating the expression, localization and function of RiPho4, a transcription factor of the PHO pathway.
The PHO pathway of AM fungus plays a Key role in phosphate absorption during AM symbiosis
It has been reported that AMF can significantly promote Pi uptake and Pi stress adaptation abilities of host plants in both field and laboratory conditions (Abdel-Fattah, 2001; Kobae, 2019; Wang et al., 2019). Accordingly, a large number of studies have shown that the growth indices of AM plants, such as plant height, stem diameter, leaf area, root volume, shoot, root dry weight and P content, were significantly higher than those of NM plants under phosphorus limitation (Pumplin and Harrison, 2009; Frosi et al., 2016; Tian et al., 2017; Carballar-Hernandez et al., 2018; Wang et al., 2019). Especially at low Pi concentration, the host plants have high dependence to AMF (Chu et al., 2013). The results and data of the overall growth and physiological status of R. irregularis-E. grandis interaction in this study (Supplementary Figure S1; Figures 1A–E) are consistent with the previous results. AMF are sensitive to P supply and a low to moderate supply is required (Ezawa et al., 2002), the intensities of fungal ALP, ACP and SDH activities reflecting the metabolic activity and function of AMF also decreased with the increasing P input (Hamel et al., 1990; Saito, 1995; Vivas et al., 2003; Li et al., 2017; Wang et al., 2017). Similarly, high Pi supply strongly suppressed the AMF colonization and arbuscule formation (see Figure 1F) as well as the intensities of AM fungal ALP, ACP, and SDH activities (see Figures 1G–I; Supplementary Table S1) in this study. Because ACP and ALP are involved in the hydrolysis of Poly-P in the AMF (Ezawa et al., 2002), it can be considered that AMF greatly improved P nutrient of host plants by Poly-P hydrolysis in the IRM and apoplast when phosphate concentration was limited. The previous studies have found that the Pi uptake of mycorrhizal plants includes the direct uptake pathway and mycorrhizal uptake pathway (Smith and Smith, 2011). When Pi concentration was limited, Pi absorption of symbiotic plants is mainly through the mycorrhizal pathway (Zhang et al., 2021). However, when the phosphorus supply is sufficient, the direct pathway is activated within host plants to uptake from root surface, and the AMF has few Pi contribution (Chu et al., 2020). This is consistent with the results that the AMF colonization decreased and the phosphatase activities were decreased in mycorrhizal roots when Pi concentration was high (see Figure 1). In conclusion, plants absorb Pi mainly via the mycorrhizal pathway under low P environments, and AMF plays an important role in the growth of E. grandis.
Until now, molecular mechanisms by which AMF regulate Pi efflux from the IRM to symbiotic interface are partially understood (Wang et al., 2017; Nguyen and Saito, 2021; Xie et al., 2022). In yeast, several studies have implicated that transcription of PHO pathway genes are closely related to the Poly-P metabolism and cytosolic Pi transportation (Vardi et al., 2014; Desfougeres et al., 2016). The PHO pathway has been extensively characterized in yeast but less in AMF (Tisserant et al., 2012; Xie et al., 2016, 2022). In the case of yeasts and AMF, the homeobox transcription factor Pho2, Pi transporter genes, VTC complex VTC1/2/4, cyclin Pho80, CDK inhibitor Pho81, Cyclin-dependent kinase Pho85 are all controlled in the PHO pathway and in response to Pi deficiency (Lenburg and O’Shea, 1996; Ezawa et al., 2002; Tomar and Sinha, 2014; Xie et al., 2022). Similar identification was conducted on these homologous genes in Neurospora crassa (Gras et al., 2013). Through qRT-PCR analysis, it was found that the PHO pathway genes in R. irregularis were generally more active at low Pi levels than at medium and high Pi levels (see Figure 2). Besides, the transcriptional levels of RiALP1 and RiACP1 were influenced by high Pi supply (see Figures 2F,G) were consistent with the ALP and ACP staining results (see Figures 1G–I; Supplementary Table S1). Therefore, it is predicted that the PHO pathway genes of R. irregularis may play important roles in promoting plant Pi absorption during AM symbiosis under low Pi conditions, and the regulon which control the transcription of PHO pathway genes during Pi starvation is worthy to be further investigated.
RiPho4 acts as a key transcription factor of the PHO pathway in Rhizophagus irregularis
Very recently, it has been found that the core components of PHO pathway are evolutionarily conserved among AMF and yeast species (Zhou et al., 2021). In S. cerevisiae, Pho4 is known as a helical loop–helix (HLH) transcription factor to activate the expression of PHO downstream genes in response to Pi limitation (Vogel et al., 1989; Komeili and O’Shea, 1999; Urrialde et al., 2015). ScPho4 is a Pi-sensitive core regulation factor (Tomar and Sinha, 2014). Here, RiPho4, the homologous protein of ScPho4 in R. irregularis, is highly conserved in fungi species containing the HLH domain to bind to DNA (see Figure 3; Supplementary Figure S2). Therefore, RiPho4 can be considered as an important transcription factor of the PHO pathway in R. irregularis.
From expression patterns of RiPho4 (see Figure 4), it is more active in mycorrhizal roots and arbuscules during symbiosis, indicating that RiPho4 may function in the Pi nutrient exchange at the symbiotic interface, especially in arbuscules (Smith and Smith, 1997; Karandashov et al., 2004; Harrison, 2012; Luginbuehl and Oldroyd, 2017). Moreover, the transcription of RiPho4 is dependent on the Pi availability, this is similar to Pho4 in filamentous fungi (Peleg et al., 1996; Gras et al., 2013; Tomar and Sinha, 2014). The RiPho4 protein location in the nuclei of yeast cells is also dependent on Pi availability. When facing to the high phosphate concentrations, RiPho4 can be moved to the cytoplasm (see Figure 5). This finding is similar to Pho4 location patterns of yeasts and filamentous fungi (Peleg et al., 1996; Byrne et al., 2004; Urrialde et al., 2015). It is well-known that one of the mechanisms regulating the activation of TFs is cytoplasmic retention and subsequent translocation into the nucleus due to external stimuli (Reich and Liu, 2006; Hao and O’Shea, 2012). And for RiPho4, the transcription factor in the cytoplasm subsequently translocates into the nucleus in response to low Pi stimulus, like ScPho4. Therefore, our findings reveal that the transcription factor RiPho4 in R. irregularis is induced during P starvation, and may play a key role in the regulation of Pi uptake and homeostasis during AM symbiosis.
RiPho4 regulates arbuscule development and Pi concentration of mycorrhizal plants through modulating the PHO genes in Rhizophagus irregularis
Up to date, few Pi transporter genes of PHO pathway in AMF, such as GigmPT and RiPT7, have been functionally described by gene silencing, and knock-down of GigmPT or RiPT7 leads to fungal growth arrest and impaired arbuscule development (Xie et al., 2016, 2022). Correspondingly, in our study, RiPho4 silencing also results in the obvious phenotype of arbuscule degradation (see Figure 6). The previous studies have shown that the process of AMF Pi delivery to plant cells serve as a signal to maintain the arbuscule development (Javot et al., 2007; Xie et al., 2016, 2022). However, Pi levels of mycorrhizal tobacco were significantly reduced in the RiPho4-RNAi lines when compared with the control lines (see Figure 7), suggesting that the loss of RiPho4 function results in the hinder of Pi transportation from the arbuscules to host plants. Therefore, RiPho4 functions in maintaining the arbuscule development, resulting from its role in promoting Pi exchange at the symbiotic interface of mycorrhizas. Indeed, this regulatory roles of fungal Pho4 proteins in Pi uptake and homeostasis have been demonstrated. Previous studies showed that NUC-1 (Pho4 homologous gene) from N. crassa is considered to be a factor to activate the transcription of Pi transporter genes (Kang and Metzenberg, 1990; Gras et al., 2013), and in AMF, Pho4 is predicted to have a regulatory role on the PHO responsive genes (Xie et al., 2016; Zhou et al., 2021). As expected, in R. irregularis, the knock-down of RiPho4 by VIGS resulted in the significant decrease in expression levels of Pi transporter genes, Pi and Poly-P metabolism genes under Pi limited conditions (see Figures 8A–H). From this result, it is suggested that RiPho4 regulates Pi transport and homeostasis at the symbiotic interface through controlling the PHO gene expression.
Next, question is how RiPho4 regulates the transcription of PHO genes in R. irregularis. We used the Y1H assay to preliminarily address this issue. As shown in Figure 8I, the RiPho4 protein interacted with the promoters of Pi transporter genes RiPT1, RiPT2 and RiPT3 in R. irregularis through binding to the CACGTG/T sites. On the basis of the results, it is concluded that RiPho4 is able to directly regulate Pi transporter genes of PHO pathway. Since other downstream genes of PHO pathway, such as RiVTC1, RiVTC2 and RiALP1, also contain the Pho4-binding sites (CACGTG/T), it is predicted that RiPho4 may also have regulatory functions on these Pi and Poly-P metabolism genes (see Supplementary Table S5). In filamentous fungi, Pi responsive genes containing the Pho4-binding sites, including phosphate permeases and repressible alkaline phosphatase genes, help the cell to survive in the prevailing low Pi environment (Lenburg and O’Shea, 1996; Gras et al., 2007; Leal et al., 2007; Tomar and Sinha, 2014). These evidences indicate that RiPho4 can directly regulate the downstream genes of PHO pathway to control Pi uptake and homeostasis during AM symbiosis. However, whether RiPho4 binds to a large number of Pi responsive genes involved in the PHO pathway of R. irregularis need to be further identified in future. Taken together, it is proposed that RiPho4 as a transcription factor regulates arbuscule development and symbiotic Pi homeostasis through controlling the downstream genes in the PHO Pathway of R. irregularis.
A model of the key regulon RiPho4, which functions in the control of PHO genes in AM fungus during symbiosis
According to our results and previous studies (Oshima, 1997; Tomar and Sinha, 2014; Zhou et al., 2016; Ahmed et al., 2022), we proposed a working model in which RiPho4 acts as a transcription factor controlling the PHO genes to regulate Pi transport and homeostasis at the symbiotic interface (Figure 9). In this model, under low Pi conditions (Figure 9A), in arbuscules, the level of inositol heptakisphosphate (IP7), an evolutionary conserved metabolite (Lee et al., 2007), may increase. This induces the expression increasement of CDK inhibitor RiPho81 function (Lee et al., 2008; York and Lew, 2008), thereby preventing the formation of RiPho85 and RiPho80 complex (Schneider et al., 1994; Huang et al., 2007; Lee et al., 2008). The non-formation of the RiPho85 and RiPho80 complex results in hypo-phosphorylate of RiPho4, which is accumulated in the nucleus and activated with cofactor RiPho2 (Urrialde et al., 2015). Subsequently, RiPho4 binds to the CACGTG/T sites to activate the transcriptions of PHO pathway genes in arbuscules, such as the Pi transporter genes RiPT1/2/3 and Pi metabolism genes RiALP1 and RiACP1. Moreover, RiPPN1 and RiPPX1 are also induced to function in the Poly-P metabolism in vacuoles (Xie et al., 2016; Ezawa and Saito, 2018; Zhou et al., 2021). Therefore, during Pi deficiency, RiPho4 regulates the PHO-related genes to enhance the hydrolyzation of Poly-P in arbuscules, and these free Pi are transported into the PAS via the Pi transporters (Xie et al., 2022), then the PAM-located PT4 carriers transport Pi to the plant cells (Harrison et al., 2002; Javot et al., 2007; Che et al., 2022). Conversely, under Pi-sufficient conditions (Figure 9B), the IP7 may be lacking in arbuscules and prevent RiPho81 from inhibiting the kinase activity of RiPho85-RiPho80, thus enabling the phosphorylation of RiPho4 (Jeffery et al., 2001; Huang et al., 2007). However, the phosphorylated RiPho4 is exported from the nucleus into the cytoplasm where it cannot activate the transcription of PHO pathway genes (Oshima, 1997; Kaffman et al., 1998; Ghillebert et al., 2011). Under such conditions, the SPX domain-containing transporter RiPT7 can export Pi into the PAS, and AM-specific PT4 can acquire Pi from PAS (Tomar and Sinha, 2014; Che et al., 2022; Xie et al., 2022).
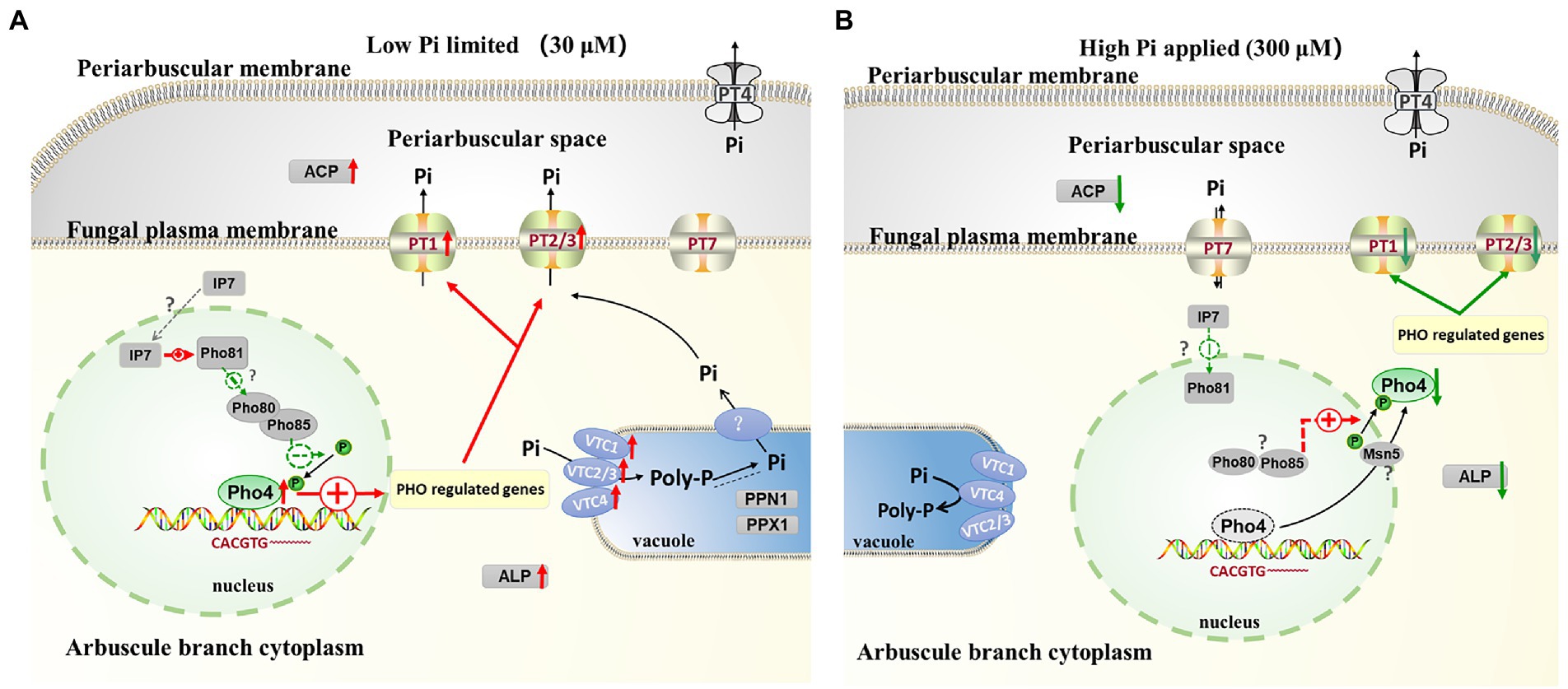
Figure 9. Schematic representation of the function of RiPho4 protein regulating phosphate transport and homeostasis in R. irregularis during AM symbiosis. (A) Under Pi limiting conditions, it is predicted that RiPho4 may be hypo-phosphorylated by RiPho80-RiPho85 complex (Jeffery et al., 2001), and Pho4 locates in nucleus to activate the transcription of downstream genes involved in the PHO pathway, such as PT1/2/3, ACP, ALP, PPN1 and PPX1. (B) Under high Pi conditions, the SPX domain-containing Pi transporter RiPT7 is responsible for Pi transport at the symbiotic interface (Xie et al., 2022), whereas the IP7 signal is predicted to be inhibited and the Pho85-Pho80 complex may phosphorylate Pho4 protein (Lee et al., 2008), which is exported from nucleus to cytoplasm, then switches off the PHO pathway. Negative and positive regulatory effects were indicated by green and red arrows, respectively. The relationships between the well-known genes and those not yet determined were shown by the solid and dashed heads, respectively. IP7, inositol heptakisphosphate; Poly-P, polyphosphate; Pi, inorganic phosphate. The green up and red down arrows next to genes indicated that transcript levels of genes were increased and decreased, respectively.
Conclusion
In conclusion, this study presented the expression, localization and functions of RiPho4 from R. irregularis. RiPho4 is a transcription factor containing a HLH domain, and is located in the nucleus of yeast cells under low Pi conditions. Further studies revealed that RiPho4 is a key regulatory factor in AM fungus to maintain arbuscule development through regulating the expression levels of the PHO pathway downstream genes in order to handle Pi transport and homeostasis at the symbiotic interface. Our findings provide new insights into the underlying mechanisms by which AMF control phosphate uptake and homeostasis during symbiosis.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
Author contributions
XX and MT designed the experiments and managed the projects. SZ and XF performed the experiments. YN and WW performed data analysis. XF and XX provided advice and guidance on the idea of bioinformatics analysis. SZ, XX, and MT wrote the manuscript. HC assisted with the interpretation of the results. All authors contributed to the manuscript read, edited, and approved the current version.
Funding
This research was funded by the National Natural Science Foundation of China (32170116 and 32071639), the Key Projects of Guangzhou of Science and Technology Plan (grant no. 201904020022), and the Laboratory of Lingnan Modern Agriculture Project (NZ2021025), the Guangdong Basic and Applied Basic Research Foundation (grant no. 2022A1515012013).
Acknowledgments
We are grateful to Jianyong An (Huazhong Agricultural University, Wuhan, China) for kindly providing for kindly providing the spores of R. irregularis DAOM 197198 for this study. We would like to thank Nianwu Tang (UMR Interactions Arbres/Microorganismes, Centre INRA-Grand Est-Nancy, Champenoux, France) for his help in analysis of transcriptional levels of PHO pathway genes in different fungal tissues. We are grateful to Honghui Zhu (South China Agricultural University, Guangzhou, China) for kindly providing the pAbAi vector for yeast one-hybrid experiment.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1114089/full#supplementary-material
Footnotes
1. ^http://blast.ncbi.nlm.nih.gov/Blast.cgi
3. ^https://swissmodel.expasy.org/
4. ^https://meme-suite.org/meme/tools/meme
5. ^http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
References
Abdel-Fattah, G. M. (2001). Measurement of the viability of arbuscular-mycorrhizal fungi using three different stains; relation to growth and metabolic activities of soybean plants. Microbiol. Res. 156, 359–367. doi: 10.1078/0944-5013-00121
Ahmed, Y., Ikeh, M. A. C., MacCallum, D. M., Day, A. M., Waldron, K., and Quinn, J. (2022). Blocking polyphosphate mobilization inhibits Pho4 activation and virulence in the pathogen Candida albicans. MBio 13:e0034222. doi: 10.1128/mbio.00342-22
Akiyama, K., Matsuzaki, K., and Hayashi, H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827. doi: 10.1038/nature03608
An, J. Y., Zeng, T., Ji, C. Y., de Graaf, S., Zheng, Z. J., Xiao, T. T., et al. (2019). A Medicago truncatula SWEET transporter implicated in arbuscule maintenance during arbuscular mycorrhizal symbiosis. New Phytol. 224, 396–408. doi: 10.1111/nph.15975
Aono, T., Maldonado-Mendoza, I. E., Dewbre, G. R., and Saito, H. M. (2004). Expression of alkaline phosphatase genes in arbuscular mycorrhizas. New Phytol. 162, 525–534. doi: 10.1111/j.1469-8137.2004.01041.x
Auesukaree, C., Homma, T., Kaneko, Y., and Harashima, S. (2003). Transcriptional regulation of phosphate-responsive genes in low-affinity phosphate-transporter-defective mutants in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 306, 843–850. doi: 10.1016/S0006-291X(03)01068-4
Auesukaree, C., Homma, T., Tochio, H., Shirakawa, M., Kaneko, Y., and Harashima, S. (2004). Intracellular phosphate serves as a signal for the regulation of the PHO pathway in Saccharomyces cerevisiae. J. Biol. Chem. 279, 17289–17294. doi: 10.1074/jbc.M312202200
Bahar, N. H. A., Gauthier, P. P. G., O'Sullivan, O. S., Brereton, T., Evans, J. R., and Atkin, O. K. (2018). Phosphorus deficiency alters scaling relationships between leaf gas exchange and associated traits in a wide range of contrasting eucalyptus species. Funct. Plant Biol. 45, 813–826. doi: 10.1071/fp17134
Balestrini, R., and Bonfante, P. (2014). Cell wall remodeling in mycorrhizal symbiosis: a way towards biotrophism. Front. Plant Sci. 5:237. doi: 10.3389/fpls.2014.00237
Balestrini, R., Gómez-Ariza, J., Lanfranco, L., and Bonfante, P. (2007). Laser microdissection reveals that transcripts for five plant and one fungal phosphate transporter genes are contemporaneously present in arbusculated cells. Mol. Plant-Microbe Interact. 20, 1055–1062. doi: 10.1094/mpmi-20-9-1055
Benedetto, A., Magurno, F., Bonfante, P., and Lanfranco, L. (2005). Expression profiles of a phosphate transporter gene (GmosPT) from the endomycorrhizal fungus glomus mosseae. Mycorrhiza 15, 620–627. doi: 10.1007/s00572-005-0006-9
Bichara, S., Mazzafera, P., and de Andrade, S. A. L. (2021). Root morphological changes in response to low phosphorus concentration in eucalypt species. Trees 35, 1933–1943. doi: 10.1007/s00468-021-02161-4
Bonfante, P., and Venice, F. (2020). Mucoromycota: going to the roots of plant-interacting fungi. Fungal Biol. Rev. 34, 100–113. doi: 10.1016/j.fbr.2019.12.003
Brundrett, M., and Tedersoo, L. (2019). Misdiagnosis of mycorrhizas and inappropriate recycling of data can lead to false conclusions. New Phytol. 221, 18–24. doi: 10.1111/nph.15440
Byrne, M., Miller, N., Springer, M., and O’Shea, E. K. (2004). A distal, high-affinity binding site on the cyclin-CDK substrate Pho4 is important for its phosphorylation and regulation. J. Mol. Biol. 335, 57–70. doi: 10.1016/j.jmb.2003.10.035
Campos-Soriano, L., and Segundo, B. S. (2011). New insights into the signaling pathways controlling defense gene expression in rice roots during the arbuscular mycorrhizal symbiosis. Plant Signal. Behav. 6, 553–557. doi: 10.4161/psb.6.4.14914
Carballar-Hernandez, S., Hernandez-Cuevas, L. V., Montano, N. M., Ferrera-Cerrato, R., and Alarcon, A. (2018). Species composition of native arbuscular mycorrhizal fungal consortia influences growth and nutrition of poblano pepper plants (Capsicum annuum L.). Appl. Soil Ecol. 130, 50–58. doi: 10.1016/j.apsoil.2018.05.022
Carretero-Paulet, L., Galstyan, A., Roig-Villanova, I., Martínez-García, J. F., Bilbao-Castro, J. R., and Robertson, D. L. (2010). Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153, 1398–1412. doi: 10.1104/pp.110.153593
Che, X. R., Lai, W. Z., Wang, S. J., Wang, X. Y., Hu, W. T., Chen, H., et al. (2022). Multiple PHT1 family phosphate transporters are recruited for mycorrhizal symbiosis in Eucalyptus grandis and conserved PHT1; 4 is a requirement for the arbuscular mycorrhizal symbiosis. Tree Physiol. 42, 2020–2039. doi: 10.1093/treephys/tpac050
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, E. C. H., Morin, E., Beaudet, D., Noel, J., Yildirir, G., Ndikumana, S., et al. (2018). High intraspecific genome diversity in the model arbuscular mycorrhizal symbiont Rhizophagus irregularis. New Phytol. 220, 1161–1171. doi: 10.1111/nph.14989
Chien, C. T., Bartel, P. L., Sternglanz, R., and Fields, S. (1991). The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl. Acad. Sci. U. S. A. 88, 9578–9582. doi: 10.1073/pnas.88.21.9578
Chu, Q., Wang, X., Yang, Y., Chen, F., Zhang, F., and Feng, G. (2013). Mycorrhizal responsiveness of maize (Zea mays L.) genotypes as related to releasing date and available P content in soil. Mycorrhiza 23, 497–505. doi: 10.1007/s00572-013-0492-0
Chu, Q., Zhang, L., Zhou, J. W., Yuan, L. X., Chen, F. J., Zhang, F. S., et al. (2020). Soil plant-available phosphorus levels and maize genotypes determine the phosphorus acquisition efficiency and contribution of mycorrhizal pathway. Plant Soil 449, 357–371. doi: 10.1007/s11104-020-04494-4
Cibichakravarthy, B., Kumutha, K., and Balachandar, D. (2015). Arbuscular mycorrhizal fungal diversity in phosphorus-deficient Alfisols of a dry north-western agro-ecosystem of Tamil Nadu, India. Ann. Microbiol. 65, 143–153. doi: 10.1007/s13213-014-0845-8
Das, D., Paries, M., Hobecker, K., Gigl, M., Dawid, C., Lam, H. M., et al. (2022). Phosphate starvation response transcription factors enable arbuscular mycorrhiza symbiosis. Nat. Commun. 13:477. doi: 10.1038/s41467-022-27976-8
Desfougeres, Y., Gerasimaite, R., Jessen, H. J., and Mayer, A. (2016). Vtc5, a novel subunit of the vacuolar transporter chaperone complex, regulates polyphosphate synthesis and phosphate homeostasis in yeast. J. Biol. Chem. 291, 22262–22275. doi: 10.1074/jbc.M116.746784
Dierks, J., Blaser-Hart, W. J., Gamper, H. A., Nyoka, I. B., and Six, J. (2021). Trees enhance abundance of arbuscular mycorrhizal fungi, soil structure, and nutrient retention in low-input maize cropping systems. Agric. Ecosyst. Environ. 318:107487. doi: 10.1016/j.agee.2021.107487
Ezawa, T., and Saito, K. (2018). How do arbuscular mycorrhizal fungi handle phosphate? New insight into fine-tuning of phosphate metabolism. New Phytol. 220, 1116–1121. doi: 10.1111/nph.15187
Ezawa, T., Smith, S. E., and Smith, F. A. (2002). P metabolism and transport in AM fungi. Plant Soil 244, 221–230. doi: 10.1023/A:1020258325010
Fan, X., Che, X., Lai, W., Wang, S., Hu, W., Chen, H., et al. (2020). The auxin-inducible phosphate transporter AsPT5 mediates phosphate transport and is indispensable for arbuscule formation in Chinese milk vetch at moderately high phosphate supply. Environ. Microbiol. 22, 2053–2079. doi: 10.1111/1462-2920.14952
Fellbaum, C. R., Mensah, J. A., Cloos, A. J., Strahan, G. E., Pfeffer, P. E., Kiers, E. T., et al. (2014). Fungal nutrient allocation in common mycorrhizal networks is regulated by the carbon source strength of individual host plants. New Phytol. 203, 646–656. doi: 10.1111/nph.12827
Ferré-D'Amaré, A. R., Prendergast, G. C., Ziff, E. B., and Burley, S. K. (1993). Recognition by max of its cognate DNA through a dimeric b/HLH/Z domain. Nature 363, 38–45. doi: 10.1038/363038a0
Fiorilli, V., Lanfranco, L., and Bonfante, P. (2013). The expression of GintPT, the phosphate transporter of Rhizophagus irregularis, depends on the symbiotic status and phosphate availability. Planta 237, 1267–1277. doi: 10.1007/s00425-013-1842-z
Frosi, G., Barros, V. A., Oliveira, M. T., Cavalcante, U. M. T., Maia, L. C., and Santos, M. G. (2016). Increase in biomass of two woody species from a seasonal dry tropical forest in association with AMF with different phosphorus levels. Appl. Soil Ecol. 102, 46–52. doi: 10.1016/j.apsoil.2016.02.009
Genre, A., Chabaud, M., Faccio, A., Barker, D. G., and Bonfante, P. (2008). Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell 20, 1407–1420. doi: 10.1105/tpc.108.059014
Genre, A., Chabaud, M., Timmers, T., Bonfante, P., and Barker, D. G. (2005). Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell 17, 3489–3499. doi: 10.1105/tpc.105.035410
Genre, A., Ivanov, S., Fendrych, M., Faccio, A., Zársky, V., Bisseling, T., et al. (2012). Multiple exocytotic markers accumulate at the sites of perifungal membrane biogenesis in arbuscular mycorrhizas. Plant Cell Physiol. 53, 244–255. doi: 10.1093/pcp/pcr170
Genre, A., Lanfranco, L., Perotto, S., and Bonfante, P. (2020). Unique and common traits in mycorrhizal symbioses. Nat. Rev. Microbiol. 18, 649–660. doi: 10.1038/s41579-020-0402-3
Ghillebert, R., Swinnen, E., De Snijder, P., Smets, B., and Winderickx, J. (2011). Differential roles for the low-affinity phosphate transporters Pho87 and Pho90 in Saccharomyces cerevisiae. Biochem. J. 434, 243–251. doi: 10.1042/bj20101118
Gietz, R. D., and Schiestl, R. H. (2007). High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34. doi: 10.1038/nprot.2007.13
Giovannetti, M., Sbrana, C., Avio, L., Citernesi, A. S., and Logi, C. (1993). Differential hyphal morphogenesis in arbuscular mycorrhizal fungi during pre-infection stages. New Phytol. 125, 587–593. doi: 10.1111/j.1469-8137.1993.tb03907.x
Gras, D. E., Persinoti, G. F., Peres, N. T. A., Martinez-Rossi, N. M., Tahira, A. C., Reis, E. M., et al. (2013). Transcriptional profiling of Neurospora crassa Delta mak-2 reveals that mitogen-activated protein kinase MAK-2 participates in the phosphate signaling pathway. Fungal Genet. Biol. 60, 140–149. doi: 10.1016/j.fgb.2013.05.007
Gras, D. E., Silveira, H. C., Martinez-Rossi, N. M., and Rossi, A. (2007). Identification of genes displaying differential expression in the nuc-2 mutant strain of the mold Neurospora crassa grown under phosphate starvation. FEMS Microbiol. Lett. 269, 196–200. doi: 10.1111/j.1574-6968.2006.00613.x
Grønlund, M., Albrechtsen, M., Johansen, I. E., Hammer, E. C., Nielsen, T. H., and Jakobsen, I. (2013). The interplay between P uptake pathways in mycorrhizal peas: a combined physiological and gene-silencing approach. Physiol. Plant. 149, 234–248. doi: 10.1111/ppl.12030
Guillemin, J. P., Orozco, M. O., Gianinazzi-Pearson, V., and Gianinazzi, S. (1995). Influence of phosphate fertilization on fungal alkaline phosphatase and succinate dehydrogenase activities in arbuscular mycorrhiza of soybean and pineapple. Agric. Ecosyst. Environ. 53, 63–69. doi: 10.1016/0167-8809(94)00555-S
Gutjahr, C., and Parniske, M. (2013). Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu. Rev. Cell Dev. Biol. 29, 593–617. doi: 10.1146/annurev-cellbio-101512-122413
Hajong, S., Kumaria, S., and Tandon, P. (2013). Comparative study of key phosphorus and nitrogen metabolizing enzymes in mycorrhizal and non-mycorrhizal plants of Dendrobium chrysanthum wall. Ex Lindl. Acta Physiol. Plant. 35, 2311–2322. doi: 10.1007/s11738-013-1268-z
Hamel, C., Fyles, H., and Smith, D. L. (1990). Measurement of development of endomycorrhizal mycelium using three different vital stains. New Phytol. 115, 297–302. doi: 10.1111/j.1469-8137.1990.tb00455.x
Hao, N., and O’Shea, E. K. (2012). Signal-dependent dynamics of transcription factor translocation controls gene expression. Nat. Struct. Mol. Biol. 19, 31–39. doi: 10.1038/nsmb.2192
Harrison, M. J. (2012). Cellular programs for arbuscular mycorrhizal symbiosis. Curr. Opin. Plant Biol. 15, 691–698. doi: 10.1016/j.pbi.2012.08.010
Harrison, M. J., Dewbre, G. R., and Liu, J. (2002). A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14, 2413–2429. doi: 10.1105/tpc.004861
Harrison, M. J., and van Buuren, M. L. (1995). A phosphate transporter from the mycorrhizal fungus glomus versiforme. Nature 378, 626–629. doi: 10.1038/378626a0
Helber, N., Wippel, K., Sauer, N., Schaarschmidt, S., Hause, B., and Requena, N. (2011). A versatile monosaccharide transporter that operates in the arbuscular mycorrhizal fungus glomus sp is crucial for the symbiotic relationship with plants. Plant Cell 23, 3812–3823. doi: 10.1105/tpc.111.089813
Hewitt, E. J. (1966). Sand and Water Culture Methods used in the Study of Plant Nutrition. Farnham Royal, UK: Commonwealth Agricultural Bureaux.
Hirsch, J., Marin, E., Floriani, M., Chiarenza, S., Richaud, P., Nussaume, L., et al. (2006). Phosphate deficiency promotes modification of iron distribution in Arabidopsis plants. Biochimie 88, 1767–1771. doi: 10.1016/j.biochi.2006.05.007
Ho-Plágaro, T., and García-Garrido, J. M. (2022). Multifarious and interactive roles of GRAS transcription factors during arbuscular mycorrhiza development. Front. Plant Sci. 13:836213. doi: 10.3389/fpls.2022.836213
Hu, J., Cui, X., Wang, J., and Lin, X. (2019). The non-simultaneous enhancement of phosphorus acquisition and mobilization respond to enhanced arbuscular mycorrhization on maize (Zea mays L.). Microorganisms 7:651. doi: 10.3390/microorganisms7120651
Huang, K., Ferrin-O’Connell, I., Zhang, W., Leonard, G. A., O’Shea, E. K., and Quiocho, F. A. (2007). Structure of the Pho85-Pho80 CDK-cyclin complex of the phosphate-responsive signal transduction pathway. Mol. Cell 28, 614–623. doi: 10.1016/j.molcel.2007.09.013
Huang, Y., and Freiser, B. S. (1993). Multiphoton infrared photoinduced ion-molecule reactions in the gas phase. J. Am. Chem. Soc. 115, 737–742. doi: 10.1021/ja00055a051
Hui, J., An, X., Li, Z., Neuhäuser, B., Ludewig, U., Wu, X., et al. (2022). The mycorrhiza-specific ammonium transporter ZmAMT3;1 mediates mycorrhiza-dependent nitrogen uptake in maize roots. Plant Cell 34, 4066–4087. doi: 10.1093/plcell/koac225
Ivanov, S., and Harrison, M. J. (2014). A set of fluorescent protein-based markers expressed from constitutive and arbuscular mycorrhiza-inducible promoters to label organelles, membranes and cytoskeletal elements in Medicago truncatula. Plant J. 80, 1151–1163. doi: 10.1111/tpj.12706
Ivashuta, S., Liu, J., Liu, J., Lohar, D. P., Haridas, S., Bucciarelli, B., et al. (2005). RNA interference identifies a calcium-dependent protein kinase involved in Medicago truncatula root development. Plant Cell 17, 2911–2921. doi: 10.1105/tpc.105.035394
Javot, H., Pumplin, N., and Harrison, M. J. (2007). Phosphate in the arbuscular mycorrhizal symbiosis: transport properties and regulatory roles. Plant Cell Environ. 30, 310–322. doi: 10.1111/j.1365-3040.2006.01617.x
Jeffery, D. A., Springer, M., King, D. S., and O’Shea, E. K. (2001). Multi-site phosphorylation of Pho4 by the cyclin-CDK Pho80-Pho85 is semi-processive with site preference. J. Mol. Biol. 306, 997–1010. doi: 10.1006/jmbi.2000.4417
Jiang, Y., Wang, W., Xie, Q., Liu, N., Liu, L., Wang, D., et al. (2017). Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356, 1172–1175. doi: 10.1126/science.aam9970
Jung, L. A., Gebhardt, A., Koelmel, W., Ade, C. P., Walz, S., Kuper, J., et al. (2017). OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors. Oncogene 36, 1911–1924. doi: 10.1038/onc.2016.354
Kaffman, A., Herskowitz, I., Tjian, R., and O’Shea, E. K. (1994). Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science 263, 1153–1156. doi: 10.1126/science.8108735
Kaffman, A., Rank, N. M., O’Neill, E. M., Huang, L. S., and O’Shea, E. K. (1998). The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature 396, 482–486. doi: 10.1038/24898
Kang, S., and Metzenberg, R. L. (1990). Molecular analysis of nuc-1+, a gene controlling phosphorus acquisition in Neurospora crassa. Mol. Cell. Biol. 10, 5839–5848. doi: 10.1128/mcb.10.11.5839-5848.1990
Karandashov, V., Nagy, R., Wegmuller, S., Amrhein, N., and Bucher, M. (2004). Evolutionary conservation of a phosphate transporter in the arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. U. S. A. 101, 6285–6290. doi: 10.1073/pnas.0306074101
Kato, T., and Hibino, T. (2009). Isolation and expression analysis of AGAMOUS-like genes from Eucalyptus grandis. Plant Biotechnol. 26, 121–124. doi: 10.5511/plantbiotechnology.26.121
Kerwin, C. L., and Wykoff, D. D. (2009). Candida glabrata PHO4 is necessary and sufficient for Pho2-independent transcription of phosphate starvation genes. Genetics 182, 471–479. doi: 10.1534/genetics.109.101063
Kikuchi, Y., Hijikata, N., Ohtomo, R., Handa, Y., Kawaguchi, M., Saito, K., et al. (2016). Aquaporin-mediated long-distance polyphosphate translocation directed towards the host in arbuscular mycorrhizal symbiosis: application of virus-induced gene silencing. New Phytol. 211, 1202–1208. doi: 10.1111/nph.14016
Kikuchi, Y., Hijikata, N., Yokoyama, K., Ohtomo, R., Handa, Y., Kawaguchi, M., et al. (2014). Polyphosphate accumulation is driven by transcriptome alterations that lead to near-synchronous and near-equivalent uptake of inorganic cations in an arbuscular mycorrhizal fungus. New Phytol. 204, 638–649. doi: 10.1111/nph.12937
Kiribuchi, K., Sugimori, M., Takeda, M., Otani, T., Okada, K., Onodera, H., et al. (2004). RERJ1, a jasmonic acid-responsive gene from rice, encodes a basic helix-loop-helix protein. Biochem. Biophys. Res. Commun. 325, 857–863. doi: 10.1016/j.bbrc.2004.10.126
Kobae, Y. (2019). Dynamic phosphate uptake in arbuscular mycorrhizal roots under field conditions. Front. Environ. Sci. 6:159. doi: 10.3389/fenvs.2018.00159
Komeili, A., and O’Shea, E. K. (1999). Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science 284, 977–980. doi: 10.1126/science.284.5416.977
Kouas, S., Debez, A., Slatni, T., Labidi, N., Drevon, J. J., and Abdelly, C. (2009). Root proliferation, proton efflux, and acid phosphatase activity in common bean (Phaseolus vulgaris) under phosphorus shortage. J. Plant Biol. 52, 395–402. doi: 10.1007/s12374-009-9050-x
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Leal, J., Squina, F. M., Martinez-Rossi, N. M., and Rossi, A. (2007). The transcription of the gene for iso-orotate decarboxylase (IDCase), an enzyme of the thymidine salvage pathway, is downregulated in the pregc mutant strain of Neurospora crassa grown under phosphate starvation. Can. J. Microbiol. 53, 1011–1015. doi: 10.1139/w07-064
Lee, Y. S., Huang, K. X., Quiocho, F. A., and O’Shea, E. K. (2008). Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat. Chem. Biol. 4, 25–32. doi: 10.1038/nchembio.2007.52
Lee, Y. S., Mulugu, S., York, J. D., and O’Shea, E. K. (2007). Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science 316, 109–112. doi: 10.1126/science.1139080
Lenburg, M. E., and O’Shea, E. K. (1996). Signaling phosphate starvation. Trends Biochem. Sci. 21, 383–387. doi: 10.1016/S0968-0004(96)10048-7
Li, C., Li, C., Zhang, H., Liao, H., and Wang, X. (2017). The purple acid phosphatase GmPAP21 enhances internal phosphorus utilization and possibly plays a role in symbiosis with rhizobia in soybean. Physiol. Plant. 159, 215–227. doi: 10.1111/ppl.12524
Lin, K., Limpens, E., Zhang, Z., Ivanov, S., Saunders, D. G., Mu, D., et al. (2014). Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus. PLoS Genet. 10:e1004078. doi: 10.1371/journal.pgen.1004078
Luginbuehl, L. H., and Oldroyd, G. E. D. (2017). Understanding the arbuscule at the heart of endomycorrhizal symbioses in plants. Curr. Biol. 27, R952–R963. doi: 10.1016/j.cub.2017.06.042
Ma, Q., Chadwick, D. R., Wu, L., and Jones, D. L. (2022). Arbuscular mycorrhiza fungi colonisation stimulates uptake of inorganic nitrogen and Sulphur but reduces utilisation of organic forms in tomato. Soil Biol. Biochem. 172:108719. doi: 10.1016/j.soilbio.2022.108719
Maldonado-Mendoza, I. E., Dewbre, G. R., and Harrison, M. J. (2001). A phosphate transporter gene from the extra-radical mycelium of an arbuscular mycorrhizal fungus glomus intraradices is regulated in response to phosphate in the environment. Mol. Plant-Microbe Interact. 14, 1140–1148. doi: 10.1094/mpmi.2001.14.10.1140
Nagy, R., Drissner, D., Amrhein, N., Jakobsen, I., and Bucher, M. (2009). Mycorrhizal phosphate uptake pathway in tomato is phosphorus-repressible and transcriptionally regulated. New Phytol. 181, 950–959. doi: 10.1111/j.1469-8137.2008.02721.x
Nguyen, C. T., and Saito, K. (2021). Role of cell wall polyphosphates in phosphorus transfer at the arbuscular interface in mycorrhizas. Front. Plant Sci. 12:725939. doi: 10.3389/fpls.2021.725939
Niu, F., Zhang, D., Li, Z., Iersel, M. V., and Alem, P. (2015). Morphological response of eucalypts seedlings to phosphorus supply through hydroponic system. Sci. Hortic. 194, 295–303. doi: 10.1016/j.scienta.2015.08.029
Olsson, P. A., Hansson, M. C., and Burleigh, S. H. (2006). Effect of P availability on temporal dynamics of carbon allocation and glomus intraradices high-affinity P transporter gene induction in arbuscular mycorrhiza. Appl. Environ. Microbiol. 72, 4115–4120. doi: 10.1128/aem.02154-05
O’Neill, E. M., Kaffman, A., Jolly, E. R., and O’Shea, E. K. (1996). Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science 271, 209–212. doi: 10.1126/science.271.5246.209
Oshima, Y. (1997). The phosphatase system in Saccharomyces cerevisiae. Genes Genet. Syst. 72, 323–334. doi: 10.1266/ggs.72.323
Pagano, M. C., and Scotti, M. R. (2008). Arbuscular and ectomycorrhizal colonization of two eucalyptus species in semiarid Brazil. Mycoscience 49, 379–384. doi: 10.1007/s10267-008-0435-3
Parniske, M. (2008). Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat. Rev. Microbiol. 6, 763–775. doi: 10.1038/nrmicro1987
Pearson, J. N., and Jakobsen, I. (1993). The relative contribution of hyphae and roots to phosphorus uptake by arbuscular mycorrhizal plants, measured by dual labelling with 32P and 33P. New Phytol. 124, 489–494. doi: 10.1111/j.1469-8137.1993.tb03840.x
Peleg, Y., Addison, R., Aramayo, R., and Metzenberg, R. L. (1996). Translocation of Neurospora crassa transcription factor NUC-1 into the nucleus is induced by phosphorus limitation. Fungal Genet. Biol. 20, 185–191. doi: 10.1006/fgbi.1996.0034
Phillips, J. M., and Hayman, D. S. (1970). Improved procedures for cleaning and staining parasitic and vesicular arbuscular mycorrhizal fungi for rapid assessment of infection. Trans. Br. Mycol. Soc. 55, 158–IN18. doi: 10.1016/S0007-1536(70)80110-3
Pimprikar, P., and Gutjahr, C. (2018). Transcriptional regulation of arbuscular mycorrhiza development. Plant Cell Physiol. 59, 678–695. doi: 10.1093/pcp/pcy024
Plasencia, A., Soler, M., Dupas, A., Ladouce, N., Silva-Martins, G., Martinez, Y., et al. (2016). Eucalyptus hairy roots, a fast, efficient and versatile tool to explore function and expression of genes involved in wood formation. Plant Biotechnol. J. 14, 1381–1393. doi: 10.1111/pbi.12502
Plassard, C., Becquer, A., and Garcia, K. (2019). Phosphorus transport in mycorrhiza: how far are we? Trends Plant Sci. 24, 794–801. doi: 10.1016/j.tplants.2019.06.004
Pozo, M. J., and Azcón-Aguilar, C. (2007). Unraveling mycorrhiza-induced resistance. Curr. Opin. Plant Biol. 10, 393–398. doi: 10.1016/j.pbi.2007.05.004
Pumplin, N., and Harrison, M. J. (2009). Live-cell imaging reveals periarbuscular membrane domains and organelle location in Medicago truncatula roots during arbuscular mycorrhizal symbiosis. Plant Physiol. 151, 809–819. doi: 10.1104/pp.109.141879
Pumplin, N., Zhang, X., Noar, R. D., and Harrison, M. J. (2012). Polar localization of a symbiosis-specific phosphate transporter is mediated by a transient reorientation of secretion. Proc. Natl. Acad. Sci. U. S. A. 109, E665–E672. doi: 10.1073/pnas.1110215109
Qin, Z., Li, J., Zhang, Y., Xiao, Y., Zhang, X., Zhong, L., et al. (2021). Genome-wide identification of microRNAs involved in the somatic embryogenesis of eucalyptus. G3-genes genomes. Genetics 11:jkab070. doi: 10.1093/g3journal/jkab070
Rahman, M. A., Parvin, M., Das, U., Ela, E. J., Lee, S. H., Lee, K. W., et al. (2020). Arbuscular mycorrhizal symbiosis mitigates iron (Fe)-deficiency retardation in alfalfa (Medicago sativa L.) through the enhancement of Fe accumulation and sulfur-assisted antioxidant defense. Int. J. Mol. Sci. 21:2219. doi: 10.3390/ijms21062219
Reich, N. C., and Liu, L. (2006). Tracking STAT nuclear traffic. Nat. Rev. Immunol. 6, 602–612. doi: 10.1038/nri1885
Rich, M. K., Vigneron, N., Libourel, C., Keller, J., Xue, L., Hajheidari, M., et al. (2021). Lipid exchanges drove the evolution of mutualism during plant terrestrialization. Science 372, 864–868. doi: 10.1126/science.abg0929
Rockwood, D. L., Rudie, A. W., Ralph, S. A., Zhu, J. Y., and Winandy, J. E. (2008). Energy product options for eucalyptus species grown as short rotation woody crops. Int. J. Mol. Sci. 9, 1361–1378. doi: 10.3390/ijms9081361
Roth, R., Hillmer, S., Funaya, C., Chiapello, M., Schumacher, K., Lo Presti, L., et al. (2019). Arbuscular cell invasion coincides with extracellular vesicles and membrane tubules. Nat. Plants 5, 204–211. doi: 10.1038/s41477-019-0365-4
Russo, G., Carotenuto, G., Fiorilli, V., Volpe, V., Faccio, A., Bonfante, P., et al. (2019). TPLATE recruitment reveals endocytic dynamics at sites of symbiotic interface assembly in arbuscular mycorrhizal interactions. Front. Plant Sci. 10:1628. doi: 10.3389/fpls.2019.01628
Saito, M. (1995). Enzyme activities of the internal hyphae and germinated spores of an arbuscular mycorrhizal fungus, Gigaspora margarita Becker & Hall. New Phytol. 129, 425–431. doi: 10.1111/j.1469-8137.1995.tb04313.x
Santos, F. M., Balieiro, F. D., Ataide, D. H. D., Diniz, A. R., and Chaer, G. M. (2016). Dynamics of aboveground biomass accumulation in monospecific and mixed-species plantations of eucalyptus and acacia on a Brazilian sandy soil. For. Ecol. Manag. 363, 86–97. doi: 10.1016/j.foreco.2015.12.028
Santos, L. H. D., Madi, J. P. S., Diaz, L., Ramirez, G. M., De Souza, E. C., Nunes, G. M., et al. (2021). Relationship between spectral variables with RapidEye images and dendrometric variables in teak plantations using principal component analysis. Sci. Forestalis. 49. doi: 10.18671/scifor.v49n132.09
Schneider, K. R., Smith, R. L., and O’Shea, E. K. (1994). Phosphate-regulated inactivation of the kinase PHO80-PHO85 by the CDK inhibitor PHO81. Science 266, 122–126. doi: 10.1126/science.7939631
Secco, D., Wang, C., Shou, H., and Whelan, J. (2012). Phosphate homeostasis in the yeast Saccharomyces cerevisiae, the key role of the SPX domain-containing proteins. FEBS Lett. 586, 289–295. doi: 10.1016/j.febslet.2012.01.036
Senthil-Kumar, M., and Mysore, K. S. (2014). Tobacco rattle virus-based virus-induced gene silencing in Nicotiana benthamiana. Nat. Protoc. 9, 1549–1562. doi: 10.1038/nprot.2014.092
Shelest, E. (2008). Transcription factors in fungi. FEMS Microbiol. Lett. 286, 145–151. doi: 10.1111/j.1574-6968.2008.01293.x
Shi, J., Zhao, B., Zheng, S., Zhang, X., Wang, X., Dong, W., et al. (2021). A phosphate starvation response-centered network regulates mycorrhizal symbiosis. Cells 184, 5527–5540.e18. doi: 10.1016/j.cell.2021.09.030
Smethurst, P. J. (2010). Forest fertilization: trends in knowledge and practice compared to agriculture. Plant Soil 335, 83–100. doi: 10.1007/s11104-010-0316-3
Smith, J. (2009). Mycorrhizal symbiosis (third edition). Proc. Soil Sci. Soc. Am. 73. doi: 10.2136/sssaj2008.0015br
Smith, F. A., and Smith, S. E. (1997). Structural diversity in (vesicular)-arbuscular mycorrhizal symbioses. New Phytol. 137, 373–388. doi: 10.1046/j.1469-8137.1997.00848.x
Smith, S. E., and Smith, F. A. (2011). Roles of arbuscular mycorrhizas in plant nutrition and growth: new paradigms from cellular to ecosystem scales. Annu. Rev. Plant Biol. 62, 227–250. doi: 10.1146/annurev-arplant-042110-103846
Smith, S. E., Smith, F. A., and Jakobsen, I. (2003). Mycorrhizal fungi can dominate phosphate supply to plants irrespective of growth responses. Plant Physiol. 133, 16–20. doi: 10.1104/pp.103.024380
Solaiman, M. Z., Ezawa, T., Kojima, T., and Saito, M. (1999). Polyphosphates in Intraradical and Extraradical hyphae of an arbuscular mycorrhizal Fungus,Gigaspora margarita. Appl. Environ. Microbiol. 65, 5604–5606. doi: 10.1128/AEM.65.12.5604-5606.1999
Standish, R. J., Stokes, B. A., Tibbett, M., and Hobbs, R. J. (2007). Seedling response to phosphate addition and inoculation with arbuscular mycorrhizas and the implications for old-field restoration in Western Australia. Environ. Exp. Bot. 61, 58–65. doi: 10.1016/j.envexpbot.2007.03.004
Sugimura, Y., and Saito, K. (2017). Transcriptional profiling of arbuscular mycorrhizal roots exposed to high levels of phosphate reveals the repression of cell cycle-related genes and secreted protein genes in Rhizophagus irregularis. Mycorrhiza 27, 139–146. doi: 10.1007/s00572-016-0735-y
Sun, X., Chen, W., Ivanov, S., MacLean, A. M., Wight, H., Ramaraj, T., et al. (2019). Genome and evolution of the arbuscular mycorrhizal fungus Diversispora epigaea (formerly glomus versiforme) and its bacterial endosymbionts. New Phytol. 221, 1556–1573. doi: 10.1111/nph.15472
Sun, Z., Song, J., Xin, X., Xie, X., and Zhao, B. (2018). Arbuscular mycorrhizal fungal 14-3-3 proteins are involved in arbuscule formation and responses to abiotic stresses during AM symbiosis. Front. Microbiol. 9:91. doi: 10.3389/fmicb.2018.00091
Tang, N., San Clemente, H., Roy, S., Bécard, G., Zhao, B., and Roux, C. (2016). A survey of the gene repertoire of Gigaspora rosea unravels conserved features among Glomeromycota for obligate biotrophy. Front. Microbiol. 7:233. doi: 10.3389/fmicb.2016.00233
Tian, L., Nasrullah, H., and Wu, Q. S. (2017). Nitric oxide accelerates mycorrhizal effects on plant growth and root development of trifoliate orange. Sains Malaysiana 46, 1687–1691. doi: 10.17576/jsm-2017-4610-03
Tisserant, E., Kohler, A., Dozolme-Seddas, P., Balestrini, R., Benabdellah, K., Colard, A., et al. (2012). The transcriptome of the arbuscular mycorrhizal fungus glomus intraradices (DAOM 197198) reveals functional tradeoffs in an obligate symbiont. New Phytol. 193, 755–769. doi: 10.1111/j.1469-8137.2011.03948.x
Tisserant, E., Malbreil, M., Kuo, A., Kohler, A., Symeonidi, A., Balestrini, R., et al. (2013). Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc. Natl. Acad. Sci. U. S. A. 110, 20117–20122. doi: 10.1073/pnas.1313452110
Tollot, M., Wong Sak Hoi, J., van Tuinen, D., Arnould, C., Chatagnier, O., Dumas, B., et al. (2009). An STE12 gene identified in the mycorrhizal fungus glomus intraradices restores infectivity of a hemibiotrophic plant pathogen. New Phytol. 181, 693–707. doi: 10.1111/j.1469-8137.2008.02696.x
Tomar, P., and Sinha, H. (2014). Conservation of PHO pathway in ascomycetes and the role of Pho84. J. Biosci. 39, 525–536. doi: 10.1007/s12038-014-9435-y
Trouvelot, A., Kough, J. L., and Gianinazzi-Pearson, V. (1986). “Mesure du taux de mycorhization VA d'un systeme radiculaire. Recherche de methodes d'estimation ayant une signification fonctionnelle” in Physiological and Genetical Aspects of Mycorrhizae. Proceedings of the 1st European Symposium on Mycorrhizae. eds. V. Gianinazzi-Pearson and S. Gianinazzi (Paris: Institut National de la Recherche Agronomique), 217–221.
Urrialde, V., Prieto, D., Pla, J., and Alonso-Monge, R. (2015). The Pho4 transcription factor mediates the response to arsenate and arsenite in Candida albicans. Front. Microbiol. 6:118. doi: 10.3389/fmicb.2015.00118
Vance, C. P., Uhde-Stone, C., and Allan, D. L. (2003). Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol. 157, 423–447. doi: 10.1046/j.1469-8137.2003.00695.x
Vardi, N., Levy, S., Gurvich, Y., Polacheck, T., Carmi, M., Jaitin, D., et al. (2014). Sequential feedback induction stabilizes the phosphate starvation response in budding yeast. Cell Rep. 9, 1122–1134. doi: 10.1016/j.celrep.2014.10.002
Venice, F., Ghignone, S., Salvioli di Fossalunga, A., Amselem, J., Novero, M., Xianan, X., et al. (2020). At the nexus of three kingdoms: the genome of the mycorrhizal fungus Gigaspora margarita provides insights into plant, endobacterial and fungal interactions. Environ. Microbiol. 22, 122–141. doi: 10.1111/1462-2920.14827
Vivas, A., Marulanda, A., Gómez, M., Barea, J. M., and Azcón, R. (2003). Physiological characteristics (SDH and ALP activities) of arbuscular mycorrhizal colonization as affected by bacillus thuringiensis inoculation under two phosphorus levels. Soil Biol. Biochem. 35, 987–996. doi: 10.1016/S0038-0717(03)00161-5
Vogel, K., Hörz, W., and Hinnen, A. (1989). The two positively acting regulatory proteins PHO2 and PHO4 physically interact with PHO5 upstream activation regions. Mol. Cell. Biol. 9, 2050–2057. doi: 10.1128/mcb.9.5.2050-2057.1989
Volpe, V., Giovannetti, M., Sun, X. G., Fiorilli, V., and Bonfante, P. (2016). The phosphate transporters LjPT4 and MtPT4 mediate early root responses to phosphate status in non-mycorrhizal roots. Plant Cell Environ. 39, 660–671. doi: 10.1111/pce.12659
Wang, C., White, P. J., and Li, C. J. (2017). Colonization and community structure of arbuscular mycorrhizal fungi in maize roots at different depths in the soil profile respond differently to phosphorus inputs on a long-term experimental site. Mycorrhiza 27, 369–381. doi: 10.1007/s00572-016-0757-5
Wang, G. Z., Ye, C. C., Zhang, J. L., Koziol, L., Bever, J. D., and Li, X. L. (2019). Asymmetric facilitation induced by inoculation with arbuscular mycorrhizal fungi leads to overyielding in maize/faba bean intercropping. J. Plant Interact. 14, 10–20. doi: 10.1080/17429145.2018.1550218
Wang, X., Zhao, S., and Bücking, H. (2016). Arbuscular mycorrhizal growth responses are fungal specific but do not differ between soybean genotypes with different phosphate efficiency. Ann. Bot. 118, 11–21. doi: 10.1093/aob/mcw074
Wang, J. P., Zhong, H. N., Zhu, L. J., Yuan, Y. D., Xu, L. H., Wang, G. G., et al. (2019). Arbuscular mycorrhizal fungi effectively enhances the growth of Gleditsia sinensis Lam. Seedlings under greenhouse conditions. Forests 10:567. doi: 10.3390/f10070567
Watts-Williams, S. J., and Cavagnaro, T. R. (2018). Arbuscular mycorrhizal fungi increase grain zinc concentration and modify the expression of root ZIP transporter genes in a modern barley (Hordeum vulgare) cultivar. Plant Sci. 274, 163–170. doi: 10.1016/j.plantsci.2018.05.015
Wu, P. F., Ma, X. Q., Tigabu, M., Huang, Y., Zhou, L. L., Cai, L., et al. (2014). Comparative growth, dry matter accumulation and photosynthetic rate of seven species of eucalypt in response to phosphorus supply. J. For. Res. 25, 377–383. doi: 10.1007/s11676-014-0465-y
Wykoff, D. D., Rizvi, A. H., Raser, J. M., Margolin, B., and O’Shea, E. K. (2007). Positive feedback regulates switching of phosphate transporters in S. cerevisiae. Mol. Cell 27, 1005–1013. doi: 10.1016/j.molcel.2007.07.022
Xie, X., Huang, W., Liu, F., Tang, N., Liu, Y., Lin, H., et al. (2013). Functional analysis of the novel mycorrhiza-specific phosphate transporter AsPT1 and PHT1 family from Astragalus sinicus during the arbuscular mycorrhizal symbiosis. New Phytol. 198, 836–852. doi: 10.1111/nph.12188
Xie, X., Lai, W., Che, X., Wang, S., Ren, Y., Hu, W., et al. (2022). A SPX domain-containing phosphate transporter from Rhizophagus irregularis handles phosphate homeostasis at symbiotic interface of arbuscular mycorrhizas. New Phytol. 234, 650–671. doi: 10.1111/nph.17973
Xie, X., Lin, H., Peng, X., Xu, C., Sun, Z., Jiang, K., et al. (2016). Arbuscular mycorrhizal symbiosis requires a phosphate transceptor in the Gigaspora margarita fungal symbiont. Mol. Plant 9, 1583–1608. doi: 10.1016/j.molp.2016.08.011
Xu, D., Dell, B., Malajczuk, N., and Gong, M. (2001). Effects of P fertilisation and ectomycorrhizal fungal inoculation on early growth of eucalypt plantations in southern China. Plant Soil 233, 47–57. doi: 10.1023/A:1010355620452
Yang, G., Chao, D., Ming, Z., and Xia, J. (2019). A simple method to detect the inhibition of transcription factor-DNA binding due to protein-protein interactions in vivo. Genes 10:684. doi: 10.3390/genes10090684
Yang, S. Y., Grønlund, M., Jakobsen, I., Grotemeyer, M. S., Rentsch, D., Miyao, A., et al. (2012). Nonredundant regulation of rice arbuscular mycorrhizal symbiosis by two members of the phosphate transporter1 gene family. Plant Cell 24, 4236–4251. doi: 10.1105/tpc.112.104901
Yao, X. Y., Zhang, Q. C., Zhou, H. J., Nong, Z., Ye, S. M., and Deng, Q. (2021). Introduction of Dalbergia odorifera enhances nitrogen absorption on eucalyptus through stimulating microbially mediated soil nitrogen-cycling. For. Ecosyst. 8:59. doi: 10.1186/s40663-021-00339-3
York, J. D., and Lew, D. J. (2008). IP7 guards the CDK gate. Nat. Chem. Biol. 4, 16–17. doi: 10.1038/nchembio0108-16
Zeng, T., Holmer, R., Hontelez, J., Te Lintel-Hekkert, B., Marufu, L., de Zeeuw, T., et al. (2018). Host-and stage-dependent secretome of the arbuscular mycorrhizal fungus Rhizophagus irregularis. Plant J. 94, 411–425. doi: 10.1111/tpj.13908
Zeng, T., Rodriguez-Moreno, L., Mansurkhodzaev, A., Wang, P., van den Berg, W., Gasciolli, V., et al. (2020). A lysin motif effector subverts chitin-triggered immunity to facilitate arbuscular mycorrhizal symbiosis. New Phytol. 225, 448–460. doi: 10.1111/nph.16245
Zézé, A., Dulieu, H., and Gianinazzi-Pearson, V. (1994). DNA cloning and screening of a partial genomic library from an arbuscular mycorrhizal fungus, Scutellospora castanea. Mycorrhiza 4, 251–254. doi: 10.1007/BF00206773
Zhan, Y., Sun, X., Rong, G., Hou, C., Huang, Y., Jiang, D., et al. (2017). Identification of two transcription factors activating the expression of OsXIP in rice defence response. BMC Biotechnol. 17:26. doi: 10.1186/s12896-017-0344-7
Zhang, L., Chu, Q., Zhou, J. W., Rengel, Z., and Feng, G. (2021). Soil phosphorus availability determines the preference for direct or mycorrhizal phosphorus uptake pathway in maize. Geoderma 403:115261. doi: 10.1016/j.geoderma.2021.115261
Zhao, B., Trouvelot, A., Gianinazzi, S., and Gianinazzi-Pearson, V. (1997). Influence of two legume species on hyphal production and activity of two arbuscular mycorrhizal fungi. Mycorrhiza 7, 179–185. doi: 10.1007/s005720050179
Zheng, Q., Guan, G., Cao, C., Li, Q., and Huang, G. (2020). The PHO pathway regulates white-opaque switching and sexual mating in the human fungal pathogen Candida albicans. Curr. Genet. 66, 1155–1162. doi: 10.1007/s00294-020-01100-z
Zhou, X., Li, J., Tang, N., Xie, H., Fan, X., Chen, H., et al. (2021). Genome-wide analysis of nutrient signaling pathways conserved in arbuscular mycorrhizal fungi. Microorganisms 9:1557. doi: 10.3390/microorganisms9081557
Zhou, X., and O’Shea, E. K. (2011). Integrated approaches reveal determinants of genome-wide binding and function of the transcription factor Pho4. Mol. Cell 42, 826–836. doi: 10.1016/j.molcel.2011.05.025
Zhou, Y., Yuikawa, N., Nakatsuka, H., Maekawa, H., Harashima, S., Nakanishi, Y., et al. (2016). Core regulatory components of the PHO pathway are conserved in the methylotrophic yeast Hansenula polymorpha. Curr. Genet. 62, 595–605. doi: 10.1007/s00294-016-0565-7
Keywords: arbuscular mycorrhizal fungi, P starvation, P uptake, RiPho4, transcription factor, yeast one-hybrid, virus-induced gene silencing
Citation: Zhang S, Nie Y, Fan X, Wei W, Chen H, Xie X and Tang M (2023) A transcriptional activator from Rhizophagus irregularis regulates phosphate uptake and homeostasis in AM symbiosis during phosphorous starvation. Front. Microbiol. 13:1114089. doi: 10.3389/fmicb.2022.1114089
Edited by:
Kai Sun, Nanjing Normal University, ChinaReviewed by:
Zhiyong Pan, Huazhong Agricultural University, ChinaLin Zhang, China Agricultural University, China
Copyright © 2023 Zhang, Nie, Fan, Wei, Chen, Xie and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianan Xie, 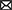 eGlleGlhbmFuODgzNDIwM0AxMjYuY29t; Ming Tang,
eGlleGlhbmFuODgzNDIwM0AxMjYuY29t; Ming Tang, 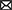 dGFuZ21pbmd5bEAxNjMuY29t
dGFuZ21pbmd5bEAxNjMuY29t
 Shuyuan Zhang
Shuyuan Zhang Yuying Nie
Yuying Nie Xiaoning Fan
Xiaoning Fan Wei Wei
Wei Wei Hui Chen
Hui Chen Xianan Xie
Xianan Xie Ming Tang
Ming Tang