- 1Shandong Province Key Laboratory of Agricultural Microbiology, Department of Plant Pathology, College of Plant Protection, Shandong Agricultural University, Tai’an, China
- 2State Key Laboratory of Crop Biology, Shandong Agricultural University, Tai’an, China
Phytophthora sojae is a well-known destructive oomycete pathogen, which causes soybean stem and root rot and poses a serious threat to global food security. Growing soybean cultivars with the appropriate resistance to P. sojae (Rps) genes are the primary management strategy to reduce losses. In most Phytophthora pathosystems, host resistance protein encoded by a specific R gene in the plant recognizes corresponding RxLR effector protein, encoded by an avirulence gene. This gene-for-gene relationship has been exploited to help breeders and agronomists deploy soybean cultivars. To date, 6 Rps genes have been incorporated into commercial soybean germplasm and trigger plant immunity in response to 8 P. sojae avirulence effectors. The incorporation of Rps genes in the soybean population creates selection pressure in favor of novel pathotypes of P. sojae. The 8 avirulence genes evolved to evade the host immune system, driven by genetic selection pressures. Understanding the evading strategies has important reference value for the prevention and control of Phytophthora stem and root rot. This investigation primarily highlights the research on the strategies of P. sojae avirulence effector evasion of host recognition, looking forward to creating durable resistance genes and thereby enabling successful disease management.
1. Introduction
Phytophthora stem and root rot (PSRR) caused by Phytophthora sojae, is a devastating disease of soybean worldwide, causing US$ 1–2 billion in economic losses worldwide each year (Tyler, 2007). Reducing losses to PSRR primarily relies on growing soybean cultivars with predominant gene(s) that govern resistance to P. sojae (Rps genes). In PSRR-Soybean pathosystems, specific Rps genes that encode nucleotide binding site-leucine rich repeat (NBS-LRR) type of proteins, which recognize effector proteins of the pathogens to induce a defense response (Kang et al., 2012), which is known as the gene-for-gene interaction (Flor and Flor, 1955). Simply put, when P. sojae infects a soybean plant, it secretes an effector protein, encoded by an avirulence (Avr) gene. If the Rps protein is not contained in the soybean cultivars or the pathogen does not secrete the effector, no recognition is occurring, and disease will develop.
Prior to 2000, P. sojae is characterized into 55 races (now known as pathotypes) based upon its virulence on a standard differential set of 8 soybean genotypes that each contain a different Rps gene, and a universal susceptible genotype (Grau et al., 2004; Cerritos Garcia et al., 2022). Knowledge of P. sojae pathotypes provides important data on the utilization of the resistance genes in a region. In the United States and Canada, Rps1a, Rps1b, Rps1c, Rps1k, Rps3a, and Rps6 have been deployed in many commercial soybean cultivars, providing protection to soybean against PSRR in the field promptly (Grau et al., 2004; Dorrance, 2018; Van et al., 2021). Nevertheless, this imposes a directed selection pressure on the pathogen, and the populations of P. sojae are becoming more diverse and complex, which evolves to become virulent on the deployed Rps genes. For example, a survey conducted in Iowa in 2001–2003 identified 17 pathotypes (Niu, 2004), whereas 37 pathotypes were identified in 2012–2013 (Dorrance et al., 2016). The survey also showed that soybean cultivars with Rps1a, Rps1c, Rps1k, and Rps3a genes were mostly overcome by isolates of P. sojae in South Dakota (Dorrance et al., 2016). It is interesting that another survey reported a low level of genotypic diversity, and relatively a high level of pathotype diversity was found among the populations between states and within fields (Stewart et al., 2016; Grijalba et al., 2020). That means P. sojae avirulence effectors escape from host recognition under a low level of genotypic diversity. It is important to understand the changes in the virulence allele and in turn to help breeders and agronomists develop and deploy soybean cultivars.
So far, 9 P. sojae effectors have been shown to be avirulence proteins that were recognized by soybean Rps genes Avr1a (cognate Rps gene: Rps1a) (Qutob et al., 2009), Avr1b (Rps1b) (Shan et al., 2004), Avr1c (Rps1c) (Na et al., 2014), Avr1d (Rps1d) (Yin et al., 2013), Avr1k (Rps1k) (Song et al., 2013), Avr3a/5 (Rps3a, Rps5) (Qutob et al., 2009), Avr3b (Rps3b) (Dong et al., 2011a), Avr3c (Rps3c) (Dong et al., 2009), and Avr4/6 (Rps4, Rps6) (Dou et al., 2010).
Among of the avirulence effectors shared a conserved RxLR (Arginine-any amino acid-Leucine-Arginine) motif at the N-terminus that mediates effector transport to plant cells (Jiang et al., 2008; Jiang and Tyler, 2012). The RxLR family effectors are unique in oomycetes, including all known avirulence proteins in downy mildews and Phytophthora pathogens (Jiang et al., 2008; Chepsergon et al., 2021). The genome of P. sojae contains over 400 RxLR family effectors (Tyler et al., 2006). RxLR effectors play an important role in the interaction between P. sojae and the host (Wang et al., 2022). The C-terminal of each RxLR effector mediates its specific function (Jiang et al., 2008; He et al., 2019; Zhang P. et al., 2019). Effector proteins are also the potential target for the plant immune system to recognize the invasion. The type of recognition is known as effector-triggered immunity (ETI), a part of the plant immune system (Jones and Dangl, 2006). ETI frequently undergo hypersensitive response (HR) to protect plant healthy from pathogen invasion, forming a “barrier wall” to block pathogen nutrient intake, resulting in rapid local programmed cell death (PCD; Ramírez-Zavaleta et al., 2022). However, pathogens, on the other hand, have developed strategies to suppress or evade ETI (Wang et al., 2011). Due to the frequent emergence of new pathotypes that can overcome resistance genes with time, the durability of a single Rps gene is estimated to be 8–20 years (Dorrance et al., 2003; Grau et al., 2004). So far, all of the Rps genes for commercial used have been “conquered” in the fields (Tremblay et al., 2021). Therefore, understanding the molecular mechanism of how avirulence effectors evade recognition is an urgent necessity to find out the failure of resistant varieties.
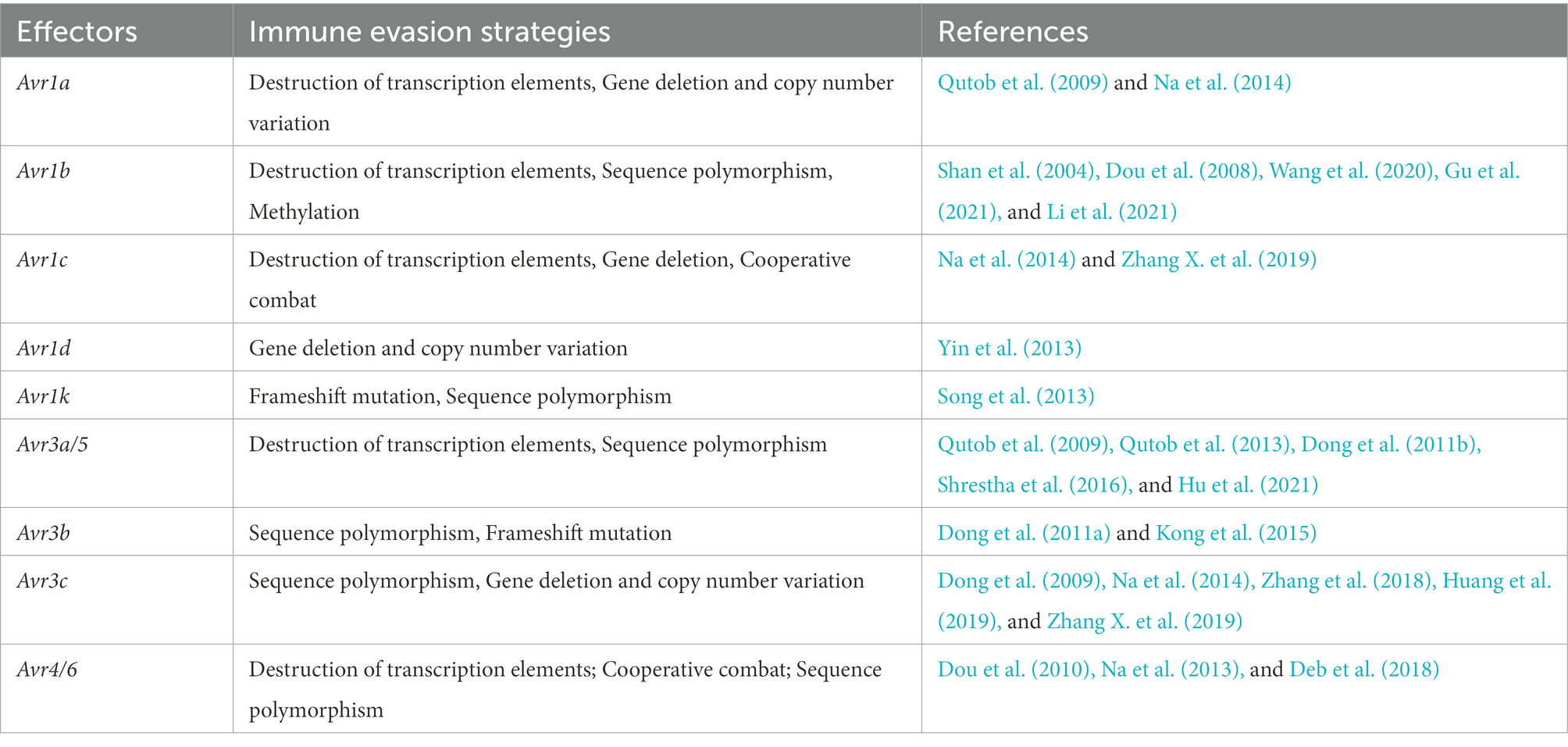
Here, we summarize RxLR avirulence effectors have evolved several distinct mechanisms to evade host recognition, such as by partial or complete deletions, transcriptional variations, transposon insertions, or point mutations. The strategies of evading host recognition reflect the competitive evolution of P. sojae and soybean. Both pathogens and their plant hosts continue to compete in this co-evolutionary dynamic.
2. Destruction of transcription elements
PsAvr1b is the first cloned Phytophthora avirulence gene (Shan et al., 2004). Over-expression of PsAvr1b made the P. sojae transformants more virulent on susceptible soybean, indicating that Avr1b contributes to virulence (Dou et al., 2008). Avr1b interacts with the soybean U-box protein, GmPUB1-1, which is required for some R gene mediated resistance (Li et al., 2021). The sequence of the PsAvr1b allele from isolate Race2 (P6497), which has a virulent phenotype on Rps1b cultivars, has no mutations in the coding region, compared with the allele from avirulent isolate Race1 (P6954) (Shan et al., 2004). However, PsAvr1b has a high accumulation of mRNA in the avirulent strains but loses the transcript in Race2 (P6497) (Shan et al., 2004). Research found in virulent strain P6497, enhanced methylation of histone H3 lysine 27 were observed and resulting PsAvr1b silenced, allowing the pathogen to evade soybean Rps1b resistance gene-mediated defense surveillance (Wang et al., 2020). This indicates the presence of a genetic mechanism, gene silencing due to the destruction of transcription elements, by which avirulence is lost. The naturally occurring Avr1b-silenced also suggests that Avr1b is not an essential effectors, and the gene knockout assay also demonstrated that PsAvr1b is not required for full virulence of P. sojae (Gu et al., 2021).
On the other hand, the transcriptional profiling differences help researcher rapidly identify candidate Avr genes, in combination with genetic mapping and whole genome sequence information. Qutob used microarrays to find transcriptional polymorphisms associated with PsAvr3a allelic differences (Qutob et al., 2009). PsAvr3a is highly expressed in avirulent strains, but it is silenced in virulent strains. The silence is due to an insertion of a transposon-like fragment in its promoter region (Qutob et al., 2009). Promoter and 5′-UTR sequence analysis revealed eight unique mutations in the promoter region of PsAvr3a, providing new insights into the escape mechanisms of P. sojae (Hu et al., 2021). Further studies revealed that the virulence of PsAvr3a is also associated with the accumulation of small RNA. PsAvr3a small RNA expression could be detected in the virulent strain ACR10, but not in the avirulent strain Race19 (P7076) (Qutob et al., 2013). The presence of small RNAs generated from the Avr3a gene sequence in the parental strain and hybrid offspring was shown to be necessary but not sufficient for gene silencing (Shrestha et al., 2016).
PsAvr1c occurs in the PsAvr1a gene cluster. Many P. sojae strains were found to carry copies of PsAvr1a and PsAvr1c, but no transcript was detected, indicating that gain of virulence may also result from gene silencing (Na et al., 2014). Also, there are nucleotide substitutions and deletions in the 5′-noncoding region of the gene in the PsAvr4/6 virulent strain, but no changes in the coding region. Compared to avirulent isolates, virulent isolates Race6 (P7063) and Race30 (PT2004C2.S1) have two nucleotide substitutions (-2C-to-T, -33A-to-G) in the 5′Untranslated Region (UTR), and virulent isolate Race17 (P7074) has one nucleotide substitution (A to G at-33), and deletion of CAGTATCGGG at-170 to -161.The changes make PsAvr4/6 avoid the recognition of the Rps4 and Rps6 (Dou et al., 2010). That indicates the transcriptional profiling differences of P. sojae Avr genes represent mechanisms for evasion of Rps gene mediated immunity.
It happens that there is a similar case in other oomycetes pathogens, Phytophthora infestans Avr2 is also not transcribed in virulent strains, preventing resistance genes recognition (Gilroy et al., 2011; Yang et al., 2020).
3. Sequence polymorphism
Four sequenced isolates, Race2 (P6497), Race7 (P7064), Race17 (P7074), and Race19 (P7076), comprise the four major genotypes of P. sojae (Tyler et al., 2006; Wang et al., 2011). Of the 378 RxLR effectors predicted to be encoded in four P. sojae genomes (Jiang et al., 2008), 147 had small numbers (1–5) of nucleotide substitutions among any of the four P. sojae isolates, while 50 genes showed large numbers of substitutions (6 or more; Wang et al., 2011). Avirulence effectors that are encoded by Avr genes, often exhibit a high level of polymorphism as a result of Rps gene mediated selection.
By the way, PsAvr1b has a variety of sequence polymorphisms in virulent strains. For example, there are 21 amino acid mutations in the C-terminal of virulent strain P7076. PsAvr1bP7076 successfully evades recognition by resistant proteins due to such drastic sequence mutations (Dou et al., 2008). The substitution of the 174th glycine in PsAvr3c by serine greatly reduces its binding affinity to host target Ser/Lys/Arg-rich proteins, thus evading Rps3c-mediated soybean immunity (Dong et al., 2009; Huang et al., 2019).
There are also two distinct amino acid sequences of PsAvr3b in the virulent and avirulent strains of P. sojae. Compared with the avirulent strain P6497, the virulent strain has two amino acid deletions and 46 amino acid substitutions. The virulent strain encodes the same protein as the P7076 strain, but only 230 amino acids are successfully expressed due to a premature termination codon (Dong et al., 2011a). Through sequence polymorphism, PsAvr3b successfully evades recognition of resistance genes, thereby enhancing virulence.
The southern experiment revealed three different patterns of PsAvr3a, namely PsAvr3a-1, PsAvr3a-2, and PsAvr3a-3. The two amino acid differences between PsAvr3a-1 and PsAvr3a-3, are derived from avirulent strains. PsAvr3a-1 copies are also present in the two virulent strains, but no detectable transcripts. Both of them could be recognized by soybeans carrying Rps3a and causing cell death. PsAvr3a-2 is derived solely from virulent strains. Compared with PsAvr3a-1 and PsAvr3a-3, there are a large number of amino acid deletions and mutations, so PsAvr3a-2 evades soybean Rps3a recognition successfully (Qutob et al., 2009; Dong et al., 2011b).
The sequence polymorphism of RxLR effector-encoding genes revealed evidence for positive selection in groups of genes with seven or more mutations, such as in avirulence genes, elicitor genes, and highly expressed effector genes (Wang et al., 2011). PsAvh238 is a RxLR effector, which was induced during infection nearly 120-fold, could inhibit INF1-triggered PCD but triggers ETI itself. It is reported PsAvh238 evolved to evade host recognition by mutating a nucleotide site while retaining activity to suppress plant immunity to enhance P. sojae virulence. This single-point mutation helped PsAvh238 evade plant recognition (Yang et al., 2017). Plant immunity has also been circumvented by amino acid substitutions in the Avr3a allele domains of P. infestans (Armstrong et al., 2005; Bos et al., 2006). The avirulence genes ATR1, ATR13, and ATR5 of Peronospora parasitica also evade the recognition of host disease resistance genes through amino acid sequence variation (Allen et al., 2004; Rehmany et al., 2005; Bailey et al., 2011).
4. Gene deletion and copy number variation
The results of virulence test and gene amplification indicated that 11 P. sojae strains carry a copy of Avh6, which were avirulent in the host carrying Rps1d. Five strains lacked Avh6 and were virulent in the host carrying Rps1d.The predicted RxLR effector gene Avh6 precisely cosegregates with the Avr1d phenotype via genetic mapping (Na et al., 2013). Avh6 could suppress plant immunity induction by RxLR effectors, including Avh238, and Avh241, and that associated with PiAvr3a and R3a (Wang et al., 2011). The latest report showed that Avr1d physically binds to the E3 ligase GmPUB13, a susceptibility factor, and stabilized GmPUB13 by suppressing the self-ubiquitination (Lin et al., 2021).
Sequencing of PsAvr1d genes in 12 P. sojae strains revealed two alleles with significant sequence polymorphisms, indicating positive selection is observed for the avirulence gene. Although PsAvr1d undergoes drastic mutations, these mutated sites are not differentially recognized by Rps1d (Yin et al., 2013). By comparing the genomes of avirulent strains and virulent strains, the PsAvr1d gene is absent in virulent isolates, the gene deletion formed genotypic diversity and overcome Rps1d mediated resistance.
PsAvr1a shows copy number variation between strains. There are two copies of PsAvr1a in the avirulent strain Race2, while in the virulent strain, such as Race7(P7064), two copies of PsAvr1a are deleted, resulting in virulence changes. PsAvr1a is also deleted in Race6, Race8, Race9, and Race21, resulting in virulence in soybean carrying Rps1a (Qutob et al., 2009).
Recently, the whole genome re-sequencing of P. sojae reveals several novel variants that lead to the evading of host resistance, including a complete deletion in Avr3c and Avr1c (Na et al., 2014; Zhang X. et al., 2019). It is worth mentioning that Avr3c family genes are present in some other Phytophthora species, and both PsAvr3c paralog from P. sojae and ProbiAvh89 orthologs from P. cinnamomi var. robiniae interact with a novel soybean spliceosomal complex protein, GmSKRPs, and reprogram host pre-mRNA alternative splicing to promote infection (Zhang et al., 2018).
A similar situation occurs in other oomycete effectors. Avr2 deletion was found in some virulent strains of Phytophthora infestans, thus avoiding the recognition of resistance gene R2(Gilroy et al., 2011).
5. Frameshift mutation
The P.sojae avirulence gene PsAvr3b encodes a secreted NADH and ADP-ribose pyrophosphorylase, and Avr3b-like proteins are conserved in Phytophthora species (Dong et al., 2011a). Thus Avr3b might be delivered into plant cells as a Nudix hydrolase to impair host immunity. The PsAvr3b gene shows transcriptional polymorphisms between virulent and avirulent strains, indicating that it may try to evade host recognition by decreasing expression levels. However, transcripts of the Avr3b virulence allele remain detectable in both virulent and avirulent strains. By examining PsAvr3b polymorphisms among 20 P. sojae field isolates from China, 16 avirulent strains encoded a 315 amino acid protein while 4 virulent isolates encoded a protein that is truncated to 230 amino acids by a premature stop codon. The premature termination of the translation resulted in the deletion of about 100 amino acids, which precisely evades avirulence protein recognition (Dong et al., 2011a; Kong et al., 2015).
Rps1k has been the most widely used resistant gene to control PSRR, which responds to two distinct but closely linked Avr genes, PsAvr1b and PsAvr1k (Song et al., 2013). By sequencing the PsAvr1k from 8 isolates, 3 isolates (P6954, P6497, and P7360) contained identical sequences and were avirulent on varieties carrying Rpslk. The study also found that the virulent strain has a common 8 nucleotide (TGCTACTT) insertion, leading to an early stop at the front of the region encoding for RxLR motif, resulting in a frameshift and premature termination of translation (Song et al., 2013). The PsAvr1k gene shows sequence polymorphisms between virulent strains, indicating that it may try to evade host recognition by nucleotide substitutions, but ultimately chose the strategy of frameshift mutation.
6. Cooperative combat
Some effectors, such as PsAvr1d, PsAvr3a, and Avr1k, collaborate to escape the host recognition monitoring, by inhibiting cell death to cover plant immune response induced by other effectors (Wang et al., 2011). To achieve a protective effect, the effector PsAvh73 of P. sojae inhibits the death of soybean cells induced by the effector Avr4/6 and enhances virulence (Deb et al., 2018). The ACR9 strain, which expresses PsAvr1c, however, is virulent to RpFs1c plants. Maybe because there are other effectors in the strain ACR9 that inhibit ETI caused by Avr1c-Rps1c interaction, thus escaping the monitoring and recognition of plant immunity (Na et al., 2014). Phytophthora sojae effector PsCRN127 can inhibit RxLR effector Avh241-induced plant cell death in N. benthamiana, as well as improve plant resistance to Phytophthora (Zhang et al., 2015).
7. Conclusion and future prospects
Phytophthora sojae poses a serious threat to global food security, and RxLR-family effectors play an important role in the P. sojae-soybean interaction system. A large number of experimental evidence suggests that effectors interfere with host immunity by changing host protein stability, destroying protein complexes, repositioning targets, and other ways, thereby promoting pathogen infection. Simultaneously, the plant immune system also recognizes effectors to trigger plant immunity and inhibit pathogen colonization and expansion (He et al., 2020).
Avirulent effectors have evolved a series of strategies to evade the recognition and defense of host plants. They try to evade the surveillance and defense of host plants by the destruction of transcription elements, sequence polymorphisms, gene deletions, frameshift mutations, and cooperative combat (Table 1). As you see, sequence polymorphism is the most common strategy utilized to evade host recognition by these Avr genes. To some extent, the sequence polymorphism reflects the strategies and evolutionary process of P. sojae in avoiding recognition by the plant immune system (Ye et al., 2016). Though sometimes nucleotide substitutions were processed, such as Avr1d and Avr1k, the alleles remain avirulence, and thus gene deletion or frameshift mutation happens to help these effectors evade the host recognition. That suggests nucleotide substitutions the first choice for P.sojae evolved to evade the host immune system. The research on Avh238 showed that it evades host recognition by mutating a single nucleotide site while retaining activity to suppress plant immunity. That is the biggest challenge to crop breeding (Figure 1).
The in-depth analysis of RxLR effector escape mechanisms can provide new insights into the molecular mechanisms underlying the virulence functions of effectors, laying the groundwork for developing novel and practical approaches for sustainable, effective disease prevention and control strategies. Given that many escape mechanisms of P. sojae effectors have yet to be investigated, the main challenge will be to develop new system-level methods to identify effector escape strategies, accurately target effectors to reduce virulence and provide new theoretical strategies for improving future soybean disease resistance.
Author contributions
QW and XH initiated the idea and drafted the manuscript. XH, ZC, and HL performed the literature search and conducted data analysis. ZH and XT critically revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Outstanding Youth Foundation of Shandong Province (ZR2021YQ20) and the National Natural Science Foundation of China (31972249 and 32172387).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Allen, R. L., Bittner-Eddy, P. D., Grenville-Briggs, L. J., Meitz, J. C., Rehmany, A. P., Rose, L. E., et al. (2004). Host-parasite coevolutionary conflict between Arabidopsis and downy mildew. Science 306, 1957–1960. doi: 10.1126/science.1104022
Armstrong, M. R., Whisson, S. C., Pritchard, L., Bos, J. I. B., Venter, E., Avrova, A. O., et al. (2005). An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl. Acad. Sci. 102, 7766–7771. doi: 10.1073/pnas.0500113102
Bailey, K., Çevik, V., Holton, N., Byrne-Richardson, J., Sohn, K. H., Coates, M., et al. (2011). Molecular cloning of ATR5Emoy2 from Hyaloperonospora arabidopsidis, an avirulence determinant that triggers RPP5-mediated defense in Arabidopsis. Mol. Plant-Microbe Interact. 24, 827–838. doi: 10.1094/mpmi-12-10-0278
Bos, J. I., Kanneganti, T. D., Young, C., Cakir, C., Huitema, E., Win, J., et al. (2006). The C-terminal half of Phytophthora infestans RxLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J. 48, 165–176. doi: 10.1111/j.1365-313X.2006.02866.x
Cerritos Garcia, D. G., Huang, S. Y., Kleczewski, N. M., and Mideros, S. (2022). Virulence, aggressiveness, and fungicide sensitivity of Phytophthora spp. associated with soybean in Illinois. Plant Dis. doi: 10.1094/pdis-07-22-1551-re [Epub ahead of print]
Chepsergon, J., Motaung, T. E., and Moleleki, L. N. (2021). "Core" RxLR effectors in phytopathogenic oomycetes: a promising way to breeding for durable resistance in plants? Virulence 12, 1921–1935. doi: 10.1080/21505594.2021.1948277
Deb, D., Anderson, R. G., How-Yew-Kin, T., Tyler, B. M., and McDowell, J. M. (2018). Conserved RxLR effectors from oomycetes Hyaloperonospora arabidopsidis and Phytophthora sojae suppress PAMP-and effector-triggered immunity in diverse plants. Mol. Plant-Microbe Interact. 31, 374–385. doi: 10.1094/mpmi-07-17-0169-fi
Dong, S., Qutob, D., Tedman-Jones, J., Kuflu, K., Wang, Y., Tyler, B. M., et al. (2009). The Phytophthora sojae avirulence locus Avr3c encodes a multi-copy RxLR effector with sequence polymorphisms among pathogen strains. PLoS One 4:e5556. doi: 10.1371/journal.pone.0005556
Dong, S., Yin, W., Kong, G., Yang, X., Qutob, D., Chen, Q., et al. (2011a). Phytophthora sojae avirulence effector Avr3b is a secreted NADH and ADP-ribose pyrophosphorylase that modulates plant immunity. PLoS Pathog. 7:e1002353. doi: 10.1371/journal.ppat.1002353
Dong, S., Yu, D., Cui, L., Qutob, D., Tedman-Jones, J., Kale, S. D., et al. (2011b). Sequence variants of the Phytophthora sojae RxLR effector Avr3a/5 are differentially recognized by Rps3a and Rps5 in soybean. PLoS One 6:e20172. doi: 10.1371/journal.pone.0020172
Dorrance, A. E. (2018). Management of Phytophthora sojae of soybean: a review and future perspectives. Can. J. Plant Pathol. 40, 210–219. doi: 10.1080/07060661.2018.1445127
Dorrance, A. E., Kurle, J., Robertson, A. E., Bradley, C. A., Giesler, L., Wise, K., et al. (2016). Pathotype diversity of Phytophthora sojae in eleven states in the United States. Plant Dis. 100, 1429–1437. doi: 10.1094/pdis-08-15-0879-re
Dorrance, A. E., McClure, S. A., and deSilva, A. (2003). Pathogenic diversity of Phytophthora sojae in Ohio soybean fields. Plant Dis. 87, 139–146. doi: 10.1094/pdis.2003.87.2.139
Dou, D., Kale, S. D., Liu, T., Tang, Q., Wang, X., Arredondo, F. D., et al. (2010). Different domains of Phytophthora sojae effector Avr4/6 are recognized by soybean resistance genes Rps4 and Rps6. Mol. Plant-Microbe Interact. 23, 425–435. doi: 10.1094/mpmi-23-4-0425
Dou, D., Kale, S. D., Wang, X., Chen, Y., Wang, Q., Wang, X., et al. (2008). Conserved C-terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. Plant Cell 20, 1118–1133. doi: 10.1105/tpc.107.057067
Flor, H. H., and Flor, H. H. (1955). Host-parasite interaction in flax rust–its genetics and other implications. Phytopath 45, 680–685.
Gilroy, E. M., Breen, S., Whisson, S. C., Squires, J., Hein, I., Kaczmarek, M., et al. (2011). Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2-like in Phytophthora infestans determine virulence on R2 plants. New Phytol. 191, 763–776. doi: 10.1111/j.1469-8137.2011.03736.x
Grau, C. R., Dorrance, A. E., Bond, J., and Russin, J. S. (2004). "Fungal diseases," in Soybeans: Improvement, Production, and Uses. eds. R. M. Shibles, J. E. Harper, R. F. Wilson, and R. C. Shoemaker (American Society of Agronomy, Inc.) 16, 679–763.
Grijalba, P. E., del Ridao, A. C., Guillin, E., and Steciow, M. (2020). Pathogenic diversity of Phytophthora sojae in the southeast of the province of Buenos Aires. Trop. Plant Pathol. 45, 397–401. doi: 10.1007/s40858-020-00364-7
Gu, B., Shao, G., Gao, W., Miao, J., Wang, Q., Liu, X., et al. (2021). Transcriptional variability associated with CRISPR-mediated gene replacements at the Phytophthora sojae Avr1b-1 locus. Front. Microbiol. 12:645331. doi: 10.3389/fmicb.2021.645331
He, Q., McLellan, H., Boevink, P. C., and Birch, P. R. J. (2020). All roads Lead to susceptibility: the many modes of action of fungal and oomycete intracellular effectors. Plant Commun. 1:100050. doi: 10.1016/j.xplc.2020.100050
He, J., Ye, W., Choi, D. S., Wu, B., Zhai, Y., Guo, B., et al. (2019). Structural analysis of Phytophthora suppressor of RNA silencing 2 (PSR2) reveals a conserved modular fold contributing to virulence. Proc. Natl. Acad. Sci. U. S. A. 116, 8054–8059. doi: 10.1073/pnas.1819481116
Hu, Y., He, Z., Kang, Y., and Cui, L. (2021). Mutations in the promoter and coding regions of Avr3a cause gain of virulence of Phytophthora sojae to Rps3a in soybean. Front. Microbiol. 12:759196. doi: 10.3389/fmicb.2021.759196
Huang, J., Chen, L., Lu, X., Peng, Q., Zhang, Y., Yang, J., et al. (2019). Natural allelic variations provide insights into host adaptation of Phytophthora avirulence effector PsAvr3c. New Phytol. 221, 1010–1022. doi: 10.1111/nph.15414
Jiang, R. H., Tripathy, S., Govers, F., and Tyler, B. M. (2008). RxLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc. Natl. Acad. Sci. U. S. A. 105, 4874–4879. doi: 10.1073/pnas.0709303105
Jiang, R. H., and Tyler, B. M. (2012). Mechanisms and evolution of virulence in oomycetes. Annu. Rev. Phytopathol. 50, 295–318. doi: 10.1146/annurev-phyto-081211-172912
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kang, Y. J., Kim, K. H., Shim, S., Yoon, M. Y., Sun, S., Kim, M. Y., et al. (2012). Genome-wide mapping of NBS-LRR genes and their association with disease resistance in soybean. BMC Plant Biol. 12:139. doi: 10.1186/1471-2229-12-139
Kong, G., Zhao, Y., Jing, M., Huang, J., Yang, J., Xia, Y., et al. (2015). The activation of Phytophthora effector Avr3b by plant cyclophilin is required for the nudix hydrolase activity of Avr3b. PLoS Pathog. 11:e1005139. doi: 10.1371/journal.ppat.1005139
Li, S., Hanlon, R., Wise, H., Pal, N., Brar, H., Liao, C., et al. (2021). Interaction of Phytophthora sojae effector Avr1b with E3 ubiquitin ligase GmPUB1 is required for recognition by soybeans carrying Phytophthora resistance Rps1-b and Rps1-k genes. Front. Plant Sci. 12:725571. doi: 10.3389/fpls.2021.725571
Lin, Y., Hu, Q., Zhou, J., Yin, W., Yao, D., Shao, Y., et al. (2021). Phytophthora sojae effector Avr1d functions as an E2 competitor and inhibits ubiquitination activity of GmPUB13 to facilitate infection. Proc. Natl. Acad. Sci. U. S. A. 118:e2018312118. doi: 10.1073/pnas.2018312118
Na, R., Yu, D., Chapman, B. P., Zhang, Y., Kuflu, K., Austin, R., et al. (2014). Genome re-sequencing and functional analysis places the Phytophthora sojae avirulence genes Avr1c and Avr1a in a tandem repeat at a single locus. PLoS One 9:e89738. doi: 10.1371/journal.pone.0089738
Na, R., Yu, D., Qutob, D., Zhao, J., and Gijzen, M. (2013). Deletion of the Phytophthora sojae avirulence gene Avr1d causes gain of virulence on Rps1d. Mol. Plant-Microbe Interact. 26, 969–976. doi: 10.1094/mpmi-02-13-0036-r
Niu, X. (2004). “Assessment of Phytophthora sojae Race Population and Fitness Components in Iowa ” St. Paul, MN APS Press.
Qutob, D., Chapman, B. P., and Gijzen, M. (2013). Transgenerational gene silencing causes gain of virulence in a plant pathogen. Nat. Commun. 4:1349. doi: 10.1038/ncomms2354
Qutob, D., Tedman-Jones, J., Dong, S., Kuflu, K., Pham, H., Wang, Y., et al. (2009). Copy number variation and transcriptional polymorphisms of Phytophthora sojae RxLR effector genes Avr1a and Avr3a. PLoS One 4:e5066. doi: 10.1371/journal.pone.0005066
Ramírez-Zavaleta, C. Y., García-Barrera, L. J., Rodríguez-Verástegui, L. L., Arrieta-Flores, D., and Gregorio-Jorge, J. (2022). An overview of PRR-and NLR-mediated immunities: conserved signaling components across the plant kingdom that communicate both pathways. Int. J. Mol. Sci. 23:12974. doi: 10.3390/ijms232112974
Rehmany, A. P., Gordon, A., Rose, L. E., Allen, R. L., Armstrong, M. R., Whisson, S. C., et al. (2005). Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell 17, 1839–1850. doi: 10.1105/tpc.105.031807
Shan, W., Cao, M., Leung, D., and Tyler, B. M. (2004). The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol. Plant-Microbe Interact. 17, 394–403. doi: 10.1094/mpmi.2004.17.4.394
Shrestha, S. D., Chapman, P., Zhang, Y., and Gijzen, M. (2016). Strain specific factors control effector gene silencing in Phytophthora sojae. PLoS One 11:e0150530. doi: 10.1371/journal.pone.0150530
Song, T., Kale, S. D., Arredondo, F. D., Shen, D., Su, L., Liu, L., et al. (2013). Two RxLR avirulence genes in Phytophthora sojae determine soybean Rps1k-mediated disease resistance. Mol. Plant-Microbe Interact. 26, 711–720. doi: 10.1094/mpmi-12-12-0289-r
Stewart, S., Robertson, A. E., Wickramasinghe, D., Draper, M. A., Michel, A., and Dorrance, A. E. (2016). Population structure among and within Iowa, Missouri, Ohio, and South Dakota populations of Phytophthora sojae. Plant Dis. 100, 367–379. doi: 10.1094/pdis-04-15-0437-re
Tremblay, V., McLaren, D. L., Kim, Y. M., Strelkov, S. E., Conner, R. L., Wally, O., et al. (2021). Molecular assessment of pathotype diversity of Phytophthora sojae in Canada highlights declining sources of resistance in soybean. Plant Dis. 105, 4006–4013. doi: 10.1094/pdis-04-21-0762-re
Tyler, B. M. (2007). Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol. Plant Pathol. 8, 1–8. doi: 10.1111/j.1364-3703.2006.00373.x
Tyler, B. M., Tripathy, S., Zhang, X., Dehal, P., Jiang, R. H., Aerts, A., et al. (2006). Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313, 1261–1266. doi: 10.1126/science.1128796
Van, K., Rolling, W., Biyashev, R. M., Matthiesen, R. L., Abeysekara, N. S., Robertson, A. E., et al. (2021). Mining germplasm panels and phenotypic datasets to identify loci for resistance to Phytophthora sojae in soybean. Plant Genome 14:e20063. doi: 10.1002/tpg2.20063
Wang, L., Chen, H., Li, J., Shu, H., Zhang, X., Wang, Y., et al. (2020). Effector gene silencing mediated by histone methylation underpins host adaptation in an oomycete plant pathogen. Nucleic Acids Res. 48, 1790–1799. doi: 10.1093/nar/gkz1160
Wang, Q., Han, C., Ferreira, A. O., Yu, X., Ye, W., Tripathy, S., et al. (2011). Transcriptional programming and functional interactions within the Phytophthora sojae RxLR effector repertoire. Plant Cell 23, 2064–2086. doi: 10.1105/tpc.111.086082
Wang, Y., Pruitt, R. N., Nürnberger, T., and Wang, Y. (2022). Evasion of plant immunity by microbial pathogens. Nat. Rev. Microbiol. 20, 449–464. doi: 10.1038/s41579-022-00710-3
Yang, L. N., Liu, H., Duan, G. H., Huang, Y. M., Liu, S., Fang, Z. G., et al. (2020). The Phytophthora infestans AVR2 effector escapes R2 recognition through effector disordering. Mol. Plant-Microbe Interact. 33, 921–931. doi: 10.1094/mpmi-07-19-0179-r
Yang, B., Wang, Q., Jing, M., Guo, B., Wu, J., Wang, H., et al. (2017). Distinct regions of the Phytophthora essential effector Avh238 determine its function in cell death activation and plant immunity suppression. New Phytol. 214, 361–375. doi: 10.1111/nph.14430
Ye, W., Wang, Y., Tyler, B. M., and Wang, Y. (2016). Comparative genomic analysis among four representative isolates of Phytophthora sojae reveals genes under evolutionary selection. Front. Microbiol. 7:1547. doi: 10.3389/fmicb.2016.01547
Yin, W., Dong, S., Zhai, L., Lin, Y., Zheng, X., and Wang, Y. (2013). The Phytophthora sojae Avr1d gene encodes an RxLR-dEER effector with presence and absence polymorphisms among pathogen strains. Mol. Plant-Microbe Interact. 26, 958–968. doi: 10.1094/mpmi-02-13-0035-r
Zhang, M., An, Y., Liu, T., Ru, Y., Li, W., and Dou, D. (2015). Transient expression of the PsCRN127 effector gene enhances Nicotiana benthamiana resistance to Phytophthora parasitica. J. Nanjing Agric. Univ. 38, 930–935. doi: 10.7685/j.issn.1000-2030.2015.06.009
Zhang, Y., Huang, J., Ochola, S. O., and Dong, S. (2018). Functional analysis of PsAvr3c effector family from Phytophthora provides probes to dissect SKRP mediated plant susceptibility. Front. Plant Sci. 9:1105. doi: 10.3389/fpls.2018.01105
Zhang, P., Jia, Y., Shi, J., Chen, C., Ye, W., Wang, Y., et al. (2019). The WY domain in the Phytophthora effector PSR1 is required for infection and RNA silencing suppression activity. New Phytol. 223, 839–852. doi: 10.1111/nph.15836
Keywords: Phytophthora sojae, RXLR effectors, host recognition, plant-microbe interactions, avirulence gene
Citation: Hou X, He Z, Che Z, Li H, Tan X and Wang Q (2023) Molecular mechanisms of Phytophthora sojae avirulence effectors escaping host recognition. Front. Microbiol. 13:1111774. doi: 10.3389/fmicb.2022.1111774
Edited by:
Jinliang Liu, Jilin University, ChinaReviewed by:
Jingtao Li, Qingdao Agricultural University, ChinaCopyright © 2023 Hou, He, Che, Li, Tan and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qunqing Wang,  d2FuZ3F1bnFpbmdAMTYzLmNvbQ==
d2FuZ3F1bnFpbmdAMTYzLmNvbQ==
 Xiaoyuan Hou
Xiaoyuan Hou Zheng He
Zheng He Zhengzheng Che
Zhengzheng Che Hengjing Li
Hengjing Li Xinwei Tan
Xinwei Tan Qunqing Wang
Qunqing Wang