
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 12 January 2023
Sec. Infectious Agents and Disease
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1099312
This article is part of the Research Topic Reviews in Microbial Pathogenesis View all 17 articles
Peptidoglycan, found within the cell wall of bacteria, is a structure critical for maintaining cell morphology and providing a protective barrier in diverse environments. Peptidoglycan is a remarkably dynamic structure that is constantly remodeled during cell growth and division by various peptidoglycan enzymes. Numerous peptidoglycan enzymes have been characterized from diverse bacteria and are highly sought after as targets for therapeutics. However, very little is known about these enzymes within the biothreat agent Francisella tularensis. As the causative agent of tularemia, F. tularensis is classified as a category A biothreat pathogen, in part due to its low infectious dose and lack of FDA-approved vaccine. Many bacterial species encode multiple peptidoglycan enzymes with redundant functions that allow for compensation if one of the enzymes are inactivated. In contrast, F. tularensis appears to lack this redundancy, indicating peptidoglycan enzymes may be completely essential for growth and could be exploited as targets for medical countermeasures. Indeed, several peptidoglycan enzymes in F. tularensis have been shown to play important roles in cell division, cell morphology, virulence, and modulation of host response. The aim of this review is to summarize findings from the current literature on peptidoglycan enzymes present in Francisella and discuss areas where future research efforts might be directed. We conclude that Francisella harbors a distinct set of peptidoglycan enzymes important for cell growth and virulence and represent potentially valuable targets for the development of novel therapeutics.
The vast majority of bacteria contain a cell wall consisting of peptidoglycan (PG), a structure critical for maintaining the cell morphology and membrane integrity, and providing a protective barrier against osmotic pressure, antibacterial compounds, and other environmental stresses. PG is composed of alternating N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) residues connected by β-(1–4) linkages. These glycan strands are further crosslinked by peptide chains, a feature which contributes to the cell wall stability and resistance to changes in osmotic pressure. In Gram-negative bacteria, these peptide chains are typically composed of L-Ala-γ-D-Glu-diaminopimelate (meso-DAP)-D-ala-D-ala, with the L-ala residue attaching the peptide chain to the C3 position of the MurNac residue (Vollmer et al., 2008; Sobhanifar et al., 2013). While these features are characteristic of most Gram-negative bacteria, some level of variation in the glycan strands or peptide chains exists across bacterial species (Schleifer and Kandler, 1972). Glycan strands are then further linked by short peptide chains to form a thin meshwork that surrounds the inner membrane. The number of peptidoglycan layers has been estimated to be between 1 and 3 in Escherichia coli, depending on the strain and growth conditions (Yao et al., 1999; Matias et al., 2003).
The bacterial cell wall is a dynamic structure that undergoes constant remodeling to accommodate cell division, response to environmental signals, and incorporation of membrane proteins. While the PG layer offers structural stability to the cell wall, it must also be flexible to accomplish these tasks. PG remodeling relies on the coordinated activities of a series of modifying enzymes, which target the glycan strands and the peptide cross-links. Extensive knowledge has been obtained from studying the array of PG remodeling proteins in E. coli and other bacteria. However, very few studies have investigated the PG remodeling enzymes in Francisella spp., and no characterization of the PG structure or composition in this organism has been performed. Francisella tularensis, the causative agent of tularemia, is a Gram-negative facultative intracellular bacterium. Within F. tularensis, there are two subspecies that cause disease in humans, subsp. tularensis and subsp. holarctica. A third subspecies, F. novicida, is commonly used as a laboratory surrogate for the fully virulent strains. F. tularensis poses a significant biothreat risk due its low infectious dose, ease of aerosolization, and lack of approved vaccine (McLendon et al., 2006). Moreover, the potential for antibiotic resistant strains makes finding novel antibiotics and therapeutics critical (Gestin et al., 2010; Sutera et al., 2014; Jaing et al., 2016; Biot et al., 2020). As a facultative intracellular pathogen, F. tularensis is able to replicate within multiple host cells, primarily macrophages, which is necessary for pathogenesis. Once taken up by a macrophage, F. tularensis resides within a phagosome. The phagosomal membrane is then disrupted, and the bacterium released to replicate freely within the cytosol. More detailed reviews on the intracellular life cycle of F. tularensis are available (Clemens and Horwitz, 2007; Chong and Celli, 2010; Santic et al., 2010). Exploring the PG enzymes of Francisella is an exciting area of study, given the importance of PG enzymes in cell division, virulence, antibiotic resistance, and their potential as therapeutic targets. The focus of this review is to summarize the current knowledge of PG enzymes in Francisella and highlight gaps in our understanding that require further investigation. We hope this work becomes part of the larger effort to reveal potential targets to combat tularemia.
The initial steps of PG biosynthesis occur in the cytosol via the Mur pathway that results in the formation of the lipid II PG precursor, consisting of a single disaccharide, pentapeptide unit attached to the lipid carrier (Egan et al., 2020). This precursor is then flipped across the inner membrane to allow for subsequent polymerization of the PG, which is carried out primarily by penicillin-binding proteins (PBPs). PBPs are a large group of enzymes broadly divided into two classes, high molecular mass (HMM) and low molecular mass (LMM) PBPs (Sauvage et al., 2008). The HMM PBPs are involved in the first steps of PG synthesis in the periplasm, catalyzing glycosyltransferase and transpeptidase reactions, involving transfer of the growing PG chain to the PG precursor and cross-linking of the glycan chains, respectively. The LMM PBPs are involved in cell separation and PG recycling (Sauvage et al., 2008). For the purpose of simplicity and therapeutic applications of the PG enzymes, this review will focus on the final steps of PG synthesis and recycling that occur in the periplasmic space.
The LMM PBPs consist of both carboxypeptidases, which cleave the terminal D-alanine from the stem pentapeptide creating a tetrapeptide, and endopeptidases, which cleave within the peptide crosslinks (Aguilera Rossi et al., 2016). Cell wall hydrolases, also referred to as autolysins, consist of three enzyme types, amidases, peptidases, and glycosidases, all of which are involved in cell wall remodeling during cell division. These enzymes are distinct from PBPs, though share overlapping functions (Vermassen et al., 2019). The amidases cleave the bond between the peptide stem and glycan chain, peptidases cleave the peptide stem, and glycosidases act upon the glycan chains. The lytic transglycosylases (LTs), structurally related to the glycosidases, are a unique family of enzymes that cleave the beta-glycosidic bond between the N-acetylglucosamine and N-acetylmuramic acid residues of the glycan strands, and catalyze the formation of 1,6-anhydro-N-acetyl-β-D-muramoyl residue (Dik et al., 2017). The schematic in Figure 1 shows where in the PG each type of enzyme cleaves, along with the known PG enzymes in Francisella spp. Currently, there are at least five carboxypeptidases and two lytic transglycosylases known to be present in Francisella, with LdcA harboring both carboxypeptidase and endopeptidase activity, and no known amidases or glucosaminidases reported in the literature. Known and putative peptidoglycan enzymes and their locus tags in three representative Francisella strains are listed in Table 1. Both LT enzymes from Francisella and two of the carboxypeptidases, DacD and LdcA, have been investigated and will be discussed herein.
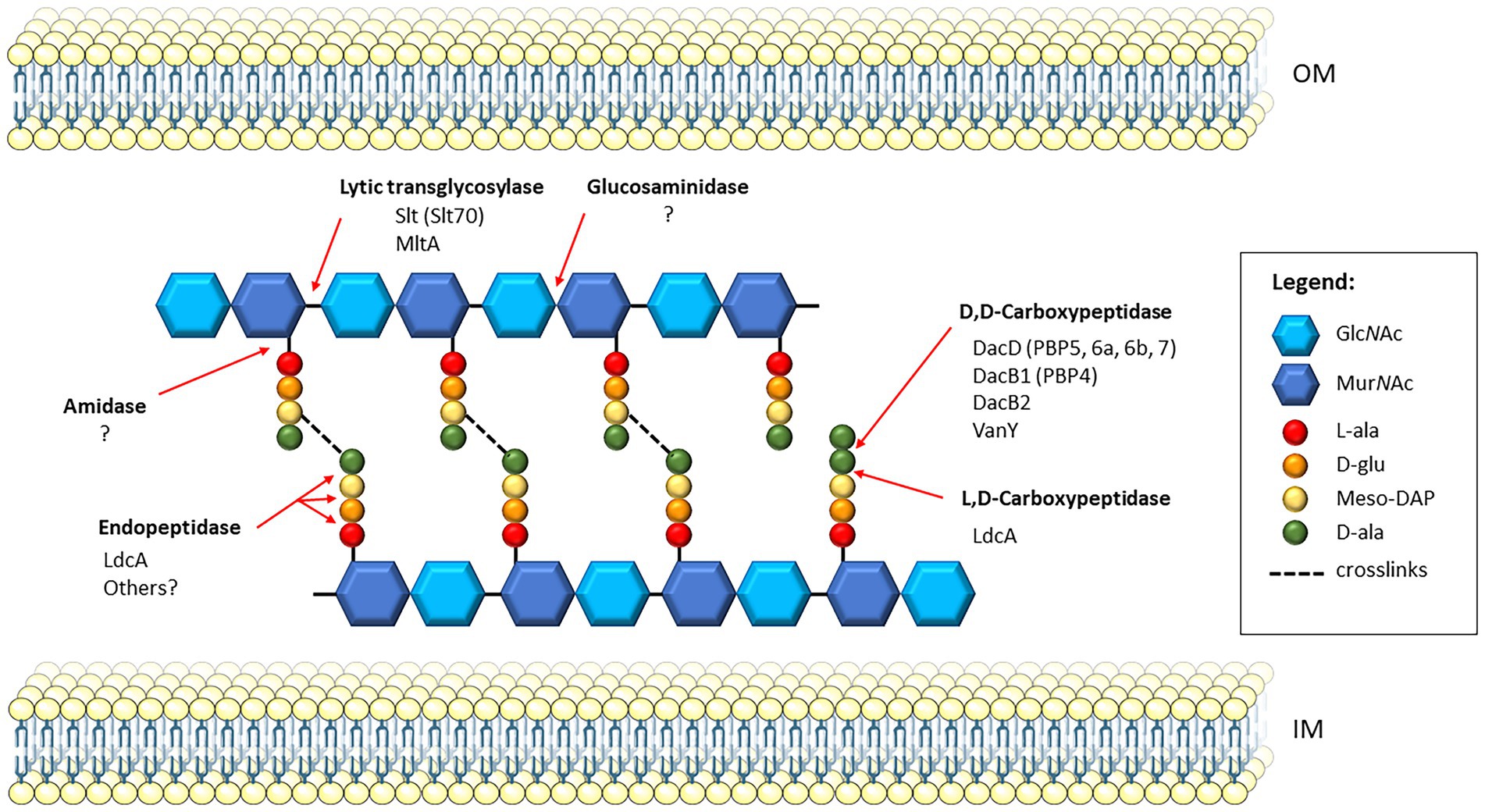
Figure 1. Schematic of PG and PG recycling enzymes in Francisella. A section of cross-linked PG is shown between the inner membrane (IM) and outer membrane (OM) of the bacterial cell. Since Francisella PG has not yet been characterized, this model shows the predicted PG composition based on that of Escherichia coli. The glycan strands composed of alternating disaccharide residues, N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc), are crosslinked via the stem peptide. The stem peptide is composed of L-Ala-γ-D-Glu-diaminopimelate (meso-DAP)-D-ala-D-ala. Transpeptidases catalyze the formation of crosslinks in a two-step process, first removing the terminal D-ala of the stem peptide, then linking the D-ala of the resulting tetrapeptide on donor strand to the meso-DAP residue on the acceptor strand. Red arrows indicate the cleavage sites of specific PG enzymes, and those known to be present in Francisella; the homologous enzymes in E. coli, if applicable, are indicated in parentheses. Francisella harbors two known lytic transglycosylases, Slt and MltA, which cleave the β-1,4 glycosidic bond between MurNAc and GlcNAc residues. Currently, no glucosaminidases have been identified in Francisella, which also act upon glycan strands but cleave the non-reducing end of the GlcNAc residues. Five D, D-carboxypeptidases are present in Francisella, including DacD, DacB1, DacB2, and VanY. The LdcA enzyme is an L,D-carboxypeptidase that also exhibits activity as an endopeptidase, though no others have been yet identified. Lastly, amidases, which cleave the bond between the MurNac residues and the stem peptide, have not been identified in Francisella.
The D-alanyl-D-alanine carboxypeptidases, or DD-carboxypeptidases, are the most abundant PBPs within E. coli, though mutation of one or more genes does not cause lethality, owing to the highly redundant nature of these enzymes (Denome et al., 1999). The major DD-carboxypeptidase in Francisella, DacD, has been characterized in two separate studies. In the first study, dacD of the F. holarctica strain FSC200 was inactivated via insertional mutagenesis and shown to have a significant replication defect in murine bone-marrow derived macrophages (Spidlova et al., 2018). The dacD mutant was also attenuated in BALB/c mice after subcutaneous infection, with decreased spread to major organs in comparison to the wild-type strain. Transmission electron microscopy (TEM) revealed significantly increased cell size and breaks within the plasma membrane of the dacD mutant. The latter study generated an insertional dacD mutant in the fully virulent F. tularensis Schu S4 strain and showed a pH-dependent growth defect of the mutant relative to wild-type, that was restored by complementation and partially restored for growth at neutral pH (Kijek et al., 2019). Moreover, the dacD mutant had significant morphological defects and increased cell size as evidenced by scanning electron microscopy (SEM). Finally, during intranasal infection of BALB/c mice, the dacD mutant was significantly attenuated for virulence and protective against challenge with the parent Schu S4 strain, showing its potential as a live vaccine strain.
Recently, the L, D-carboxypeptidase, LdcA, of F. tularensis was described, which cleaves pentapeptide stems to tripeptide stems, as opposed to the D, D-carboxypeptidases which cleave the terminal D-alanine to form tetrapeptide stems (Zellner et al., 2021). Using a recombinant LdcA protein derived from the live vaccine strain (LVS), the authors demonstrated both L, D-carboxypeptidase and L, D-endopeptidase activities that were dependent on at least two residues of the Ser-Glu-His catalytic triad. Isogenic deletion of ldcA in LVS produced cells that were smaller and rounder than wild-type cells, with significantly thicker outer membranes and septum defects indicated by the presence of bacterial chains. The virulence of this mutant was completely attenuated during intranasal infection of C3H/HeN mice as compared to wild-type and complement strains, and further protected against challenge with the fully virulent Schu S4 strain.
Additional DD-carboxypeptidases, including DacB1 (FTL_1046), DacB2 (FTL_1509) and VanY (FTL_1004), have been annotated in Francisella spp. (Table 1), though no studies have yet been published on the characterization and confirmation of these putative enzymes. It is possible that additional PG enzymes are present in Francisella though not annotated; indeed, lack of consistent sequence annotation is a challenge when analyzing available genome data for Francisella. Homologs may be identified via BLAST search of known PG enzyme sequences, although Francisella harbors many hypothetical proteins and proteins which bear no homology to existing proteins, even if they exhibit similar functions. Interestingly, a previous study identified FTT_0924 as an essential gene in F. tularensis, encoding an unknown but highly conserved protein that was necessary for maintaining PG stability and replication in macrophages. This protein could represent one of the PG enzymes employed by Francisella spp., to maintain proper cell division (Brunton et al., 2015). Roughly 30% of the genes identified in Francisella are hypothetical proteins of unknown function (Titball and Petrosino, 2007).
Francisella tularensis harbors only two known lytic transglycosylases, the soluble lytic transglycosylase (Slt), and the murein lytic transglycosylase A (MltA). The role of the Slt enzyme in cell morphology and virulence was previously examined using a transposon mutant of F. novicida (Bachert et al., 2019). The slt mutant showed significant growth defects and loss of viability during growth in low pH conditions. SEM further showed morphological defects including aberrant cell shapes and sizes, and extensive clumping and fusion of cells which were restored via complementation. The F. novicida slt mutant was also significantly attenuated during intranasal infection of BALB/c mice, with limited dissemination to major organs, and showed decreased replication in J774 murine macrophage-like cells (Bachert et al., 2019). The slt gene was also identified in a cytotoxicity study, in which a transposon mutant of slt was found to be less cytotoxic to THP-1 human monocytes than the parental F. novicida strain (Nakamura et al., 2019). The authors further constructed an in-frame deletion of slt in F. novicida and showed reduced growth of the mutant in both THP-1 cells and J774 cells. Although the mutant was still able to escape the phagosome, mutant bacteria were degraded by autophagy during the later stages of infection. Furthermore, the mutant elicited high levels of pro-inflammatory cytokines TNF-α, IL-6, and IL-1β from the cells, indicating a role for Slt in immune suppression (Nakamura et al., 2019).
More recently, Nakamura et al. (2020) identified the related MltA enzyme as necessary for the survival of F. novicida in silkworms. Virulence in the silkworm model was significantly attenuated using an in-frame deletion mutant of mltA and restored by complementation. MltA was shown to suppress immune responses in this model as evidenced by increased induction of cecropin B expression in the mltA mutant. Lastly, F. novicida mltA failed to replicate in THP-1 cells and was less cytotoxic as assessed by LDH release (Nakamura et al., 2020).
Peptidoglycan modifying enzymes are reported to be highly redundant in a variety of bacteria. For example, E. coli harbors at least 35 PG hydrolases, including 7 carboxypeptidases and 8 lytic transglycosylases (van Heijenoort, 2011). Pseudomonas aeruginosa expresses 11 LTs (Lee et al., 2017). However, Francisella species appear to lack the redundancy of PG enzymes observed in many bacteria. A previous search for E. coli PBP homologs revealed only three PBPs in F. tularensis, in contrast to 12 PBPs in E. coli. The F. tularensis homologs are DacD, FtsI, and DacB1 with DacD representing the major DD-carboxypeptidase homologous to PBP5, 6a, 6b, and 7 from E. coli (Kijek et al., 2019). Furthermore, F. tularensis has been found to harbor only two LT enzymes, MltA (Nakamura et al., 2020) and Slt (Bachert et al., 2019; Nakamura et al., 2019). Moreover, dacD, dacB1, ftsI, slt, and mltA have all been identified as essential for in vitro growth of F. tularensis using a transposon library (Ireland et al., 2019). This finding does not rule out potential polar effects of a transposon insertion. However, multiple attempts by our laboratory to generate in-frame deletions of slt in LVS and the Schu S4 strain of F. tularensis were unsuccessful and suggest slt is essential for F. tularensis (Bachert et al., 2019). Such essential genes would be ideal targets for medical countermeasures against tularemia. The lack of gene redundancy observed is not unique to F. tularensis. It has been shown that host-evolved bacteria with reduced genomes, including Mycobacterium leprae and Rickettsia prowazekii, lost their genetic redundancy in favor of maintaining diverse protein families, in contrast to free living bacteria (Mendonça et al., 2011). Conversely, for pathogens with large genomes and frequent gene duplication, including duplication of PG enzymes, the effectiveness of potential PG enzymes inhibitors could be limited.
The L, D- and D, D-carboxypeptidases of Francisella appear to have important but very distinct roles in cell morphology, although they both act upon the peptide cross-links of the cell wall. For instance, inactivation of dacD resulted in a pH-dependent growth defect, but the ldcA mutant showed no replication defect in vitro (Kijek et al., 2019; Zellner et al., 2021). Moreover, some of the morphological defects observed for LVS ldcA were opposite those of the dacD mutant, including smaller rather than larger cell size and a thicker outer membrane compared to a discontinuous membrane (Spidlova et al., 2018; Zellner et al., 2021). These mutations have complex effects on the bacterial cell surface and its sensitivity to various stressors. For instance, ldcA mutant had increased sensitivity to cell-wall targeted antibiotics, such as vancomycin and ampicillin, but increased resistance to cytosol-targeted antibiotics such as gentamicin and ciprofloxacin (Zellner et al., 2021). Notably, increased sensitivity to sodium dodecyl sulfate was observed for both dacD and ldcA mutants (Spidlova et al., 2018; Kijek et al., 2019; Zellner et al., 2021). Interestingly, the ldcA mutant was reported to form chains of cells for ~10% of the bacteria (Zellner et al., 2021). We saw a similar phenotype with the slt mutant, although with clumping of cells rather than chains; Slt also acts upon the glycan strands rather than the cross-links (Bachert et al., 2019).
The overall phenotypes of slt and dacD mutants were highly similar, both showing increased cell size and altered membrane structure. Both mutants also exhibited pH-dependent phenotypes, with membrane defects apparent in low pH conditions that were partially restored by growth in neutral pH conditions (Bachert et al., 2019; Kijek et al., 2019). This pH-dependency could point to a role for Slt and DacD enzymes in maintaining bacterial cell morphology in the intracellular environment of the macrophage during host infection. The slt and dacD mutants were characterized using SEM, which allowed for the visualization of surface structures and observation of ruffled surface appearance, while the ldcA mutant was characterized using TEM; SEM analysis of the ldcA mutant could potentially reveal similar defects. It is also worth noting that mutation of the genes encoding for these enzymes may have different effects depending on the strain or subspecies. For example, mutation of dacD in the F. tularensis Schu S4 strain resulted in cells roughly two times larger than wild-type (Kijek et al., 2019), whereas mutation of dacD in the F. holarctica FSC200 strain resulted in cells 10 times larger than wild-type (Spidlova et al., 2018). No morphological defects have yet been reported for mltA mutants of Francisella, although the attenuated growth and virulence of F. novicida mltA would suggest such a phenotype.
Interestingly, both Slt and MltA were implicated in immunomodulation, although discovered in different models of infection, i.e., THP-1 cells for Slt and silkworm for MltA (Nakamura et al., 2019, 2020). It is possible that aberrations in the cell wall resulting from mutation of the genes encoding for the LT enzymes alters the recognition of the PG by the TLR2 receptor. It would be interesting to test Francisella mltA and slt mutants in parallel using a mouse model and obtaining cytokine profiles of the host challenged with the mutants compared to wild-type infected mice. Additionally, since the LT enzymes have been associated with assembly of the type VI secretion system and incorporation into the cell envelope of E. coli (Santin and Cascales, 2017), the immunosuppressive effects observed with some of these mutants may be due in part to alterations in the type VI secretion system. However, the contribution of these enzymes to type VI secretion system assembly or stability in Francisella spp., are currently unknown.
Modulation of the immune response by PG fragments released during cell wall recycling of bacteria has been reported in the literature. This phenomenon is especially well-characterized in Neisseria gonorrhoeae and Neisseria meningitidis, where toxic PG fragments, released from the activity of LTs during host infection, contribute to inflammation and cytotoxicity (Chan and Dillard, 2017). This is in contrast to the LTs of Francisella spp., which have been shown to dampen immune responses as described above. In general, Francisella is a stealth pathogen, replicating within macrophages without triggering host defenses or causing overt damage (Sjöstedt, 2006). The modified LPS structure expressed by Francisella is known to bypass typical TLR4 recognition, contributing to its ability to replicate unseen within the host (Okan and Kasper, 2013). However, the structure of the PG within Francisella has not yet been characterized and may harbor modifications that allow the bacterium to suppress or evade host immune responses.
NOD-like receptors within host cells are known to recognize the presence of intracellular PG in host cells and trigger the inflammasome, leading to caspase-1 activation and the production of IL-1B and IL-18 leading to cell death. Similar to other intracellular pathogens, Francisella has been shown to activate the inflammasome and caspase-1 responses (Weiss et al., 2007; Broz and Monack, 2011). However, the mechanisms described rely on AIM2 sensing of dsDNA and NLRP3 sensing of LPS (Man et al., 2015; Mitra et al., 2018). Whether Francisella PG within host cells triggers a similar immune response is not currently known.
Targeting the lytic transglycosylases, specifically in combination with beta-lactam antibiotics, may be useful for the design of novel therapeutics. Bulgecin A, an O-sulfonated glycopeptide, is an LT enzyme inhibitor that was first demonstrated to be active against the Slt70 enzyme of E. coli, and the Slt70-Bulgecin A complex was crystallized to determine the structure (Thunnissen et al., 1995). While Bulgecin A harbors no antibiotic activity itself, it works synergistically with beta-lactams to increase their efficacy. The combination treatment of Bulgecin A and beta-lactams has been demonstrated effective against multiple pathogens, including E. coli, P. aeruginosa, Helicobacter pylori, and Acinetobacter baumannii (Templin et al., 1992; Thunnissen et al., 1995; Bonis et al., 2012; Skalweit and Li, 2016). Bulgecin A has not yet been tested against Francisella, either alone or in combination with beta-lactams. Bulgecin A harbors great potential for improving efficacy of existing drugs when used as a combinational therapy.
Peptidoglycan enzymes have long been considered as potential targets for antibiotics, given that it is unique to bacteria and the active sites of these enzymes have been well characterized in common pathogens, such as E. coli. While very little is known about how PG remodeling occurs in Francisella and the specific enzymes involved, the few studies discussed above highlight a significant role of PG enzymes in the survival and virulence of Francisella. Importantly, the LT enzymes are essential for survival in fully virulent F. tularensis and when mutated in F. novicida, have a significant impact on cell morphology and virulence. This is in contrast to E. coli and other species, in which mutation of multiple PG enzymes result in no detected phenotype. These observations suggest antibiotics targeting the essential LT enzymes of Francisella may be especially potent against tularemia. Moreover, the threat of antibiotic resistance heightens the biothreat concern of Francisella, making the development of novel antibiotics and inhibitors essential. The active sites for LTs and carboxypeptidases seem to be conserved between Francisella and E. coli, so existing antibiotics may be applied or redesigned for use against Francisella. Combination therapies targeting both LT and PBP enzymes are currently being evaluated for other pathogens, such as P. aeruginosa, and could prove to be effective against F. tularensis (Dik et al., 2019). Characterization of the PG enzymes in Francisella and their functions is an underdeveloped (but very promising) area of study. Research efforts in this field will likely uncover strategies to develop more effective antibiotics and inhibitors against tularemia.
BB and JB contributed to the conceptualization, writing, and editing. All authors have read and agreed to the published version of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the US Army.
Aguilera Rossi, C. G., Gómez-Puertas, P., and Ayala Serrano, J. A. (2016). In vivo functional and molecular characterization of the penicillin-binding protein 4 (DacB) of Pseudomonas aeruginosa. BMC Microbiol. 16:234. doi: 10.1186/s12866-016-0853-x
Bachert, B. A., Biryukov, S. S., Chua, J., Rodriguez, S. A., Toothman, R. G. Jr., Cote, C. K., et al. (2019). A Francisella novicida mutant, lacking the soluble lytic transglycosylase Slt, exhibits defects in both growth and virulence. Front. Microbiol. 10:1343. doi: 10.3389/fmicb.2019.01343
Biot, F. V., Bachert, B. A., Mlynek, K. D., Toothman, R. G., Koroleva, G. I., Lovett, S. P., et al. (2020). Evolution of antibiotic resistance in surrogates of Francisella tularensis (LVS and Francisella novicida): effects on biofilm formation and fitness. Front. Microbiol. 11:593542. doi: 10.3389/fmicb.2020.593542
Bonis, M., Williams, A., Guadagnini, S., Werts, C., and Boneca, I. G. (2012). The effect of bulgecin a on peptidoglycan metabolism and physiology of helicobacter pylori. Microb. Drug Resist. 18, 230–239. doi: 10.1089/mdr.2011.0231
Broz, P., and Monack, D. M. (2011). Molecular mechanisms of inflammasome activation during microbial infections. Immunol. Rev. 243, 174–190. doi: 10.1111/j.1600-065X.2011.01041.x
Brunton, J., Steele, S., Miller, C., Lovullo, E., Taft-Benz, S., and Kawula, T. (2015). Identifying Francisella tularensis genes required for growth in host cells. Infect. Immun. 83, 3015–3025. doi: 10.1128/iai.00004-15
Chan, J. M., and Dillard, J. P. (2017). Attention seeker: production, modification, and release of inflammatory peptidoglycan fragments in Neisseria species. J. Bacteriol. 199:e00354-17. doi: 10.1128/jb.00354-17
Chong, A., and Celli, J. (2010). The Francisella intracellular life cycle: toward molecular mechanisms of intracellular survival and proliferation. Front. Microbiol. 1:138. doi: 10.3389/fmicb.2010.00138
Clemens, D. L., and Horwitz, M. A. (2007). Uptake and intracellular fate of Francisella tularensis in human macrophages. Ann. N. Y. Acad. Sci. 1105, 160–186. doi: 10.1196/annals.1409.001
Denome, S. A., Elf, P. K., Henderson, T. A., Nelson, D. E., and Young, K. D. (1999). Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J. Bacteriol. 181, 3981–3993. doi: 10.1128/jb.181.13.3981-3993.1999
Dik, D. A., Madukoma, C. S., Tomoshige, S., Kim, C., Lastochkin, E., Boggess, W. C., et al. (2019). Slt, MltD, and MltG of Pseudomonas aeruginosa as targets of bulgecin a in potentiation of β-lactam antibiotics. ACS Chem. Biol. 14, 296–303. doi: 10.1021/acschembio.8b01025
Dik, D. A., Marous, D. R., Fisher, J. F., and Mobashery, S. (2017). Lytic transglycosylases: concinnity in concision of the bacterial cell wall. Crit. Rev. Biochem. Mol. Biol. 52, 503–542. doi: 10.1080/10409238.2017.1337705
Egan, A. J. F., Errington, J., and Vollmer, W. (2020). Regulation of peptidoglycan synthesis and remodeling. Nat. Rev. Microbiol. 18, 446–460. doi: 10.1038/s41579-020-0366-3
Gestin, B., Valade, E., Thibault, F., Schneider, D., and Maurin, M. (2010). Phenotypic and genetic characterization of macrolide resistance in Francisella tularensis subsp. holarctica biovar I. J. Antimicrob. Chemother. 65, 2359–2367. doi: 10.1093/jac/dkq315
Ireland, P. M., Bullifent, H. L., Senior, N. J., Southern, S. J., Yang, Z. R., Ireland, R. E., et al. (2019). Global analysis of genes essential for Francisella tularensis Schu S4 growth in vitro and for fitness during competitive infection of Fischer 344 rats. J. Bacteriol. 201, e00630–e00618. doi: 10.1128/JB.00630-18
Jaing, C. J., McLoughlin, K. S., Thissen, J. B., Zemla, A., Gardner, S. N., Vergez, L. M., et al. (2016). Identification of genome-wide mutations in ciprofloxacin-resistant F. tularensis LVS usingwhole genome tiling arrays and next generation sequencing. PLoS One 11:e0163458. doi: 10.1371/journal.pone.0163458
Kijek, T. M., Mou, S., Bachert, B. A., Kuehl, K. A., Williams, J. A., Daye, S. P., et al. (2019). The D-alanyl-d-alanine carboxypeptidase enzyme is essential for virulence in the Schu S4 strain of Francisella tularensis and a dacD mutant is able to provide protection against a pneumonic challenge. Microb. Pathog. 137:103742. doi: 10.1016/j.micpath.2019.103742
Lee, M., Hesek, D., Dik, D. A., Fishovitz, J., Lastochkin, E., Boggess, B., et al. (2017). From genome to proteome to elucidation of reactions for all eleven known lytic transglycosylases from Pseudomonas aeruginosa. Angew. Chem. Int. Ed. Engl. 56, 2735–2739. doi: 10.1002/anie.201611279
Man, S. M., Karki, R., Malireddi, R. K. S., Neale, G., Vogel, P., Yamamoto, M., et al. (2015). The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat. Immunol. 16, 467–475.
Matias, V. R., Al-Amoudi, A., Dubochet, J., and Beveridge, T. J. (2003). Cryo-transmission electron microscopy of frozen-hydrated sections of Escherichia coli and Pseudomonas aeruginosa. J. Bacteriol. 185, 6112–6118. doi: 10.1128/jb.185.20.6112-6118.2003
McLendon, M. K., Apicella, M. A., and Allen, L. A. (2006). Francisella tularensis: taxonomy, genetics, and Immunopathogenesis of a potential agent of biowarfare. Annu. Rev. Microbiol. 60, 167–185. doi: 10.1146/annurev.micro.60.080805.142126
Mendonça, A. G., Alves, R. J., and Pereira-Leal, J. B. (2011). Loss of genetic redundancy in reductive genome evolution. PLoS Comput. Biol. 7:e1001082. doi: 10.1371/journal.pcbi.1001082
Mitra, S., Dolvin, E., Krishnamurthy, K., Wewers, M. D., and Sarkar, A. (2018). Francisella induced microparticulate caspase-1/gasdermin-D activation is regulated by NLRP3 independent of Pyrin. PLoS One 13:e0209931.
Nakamura, T., Shimizu, T., Inagaki, F., Okazaki, S., Saha, S. S., Uda, A., et al. (2020). Identification of membrane-bound lytic Murein Transglycosylase a (MltA) as a growth factor for Francisella novicida in a silkworm infection model. Front. Cell. Infect. Microbiol. 10:581864. doi: 10.3389/fcimb.2020.581864
Nakamura, T., Shimizu, T., Uda, A., Watanabe, K., and Watarai, M. (2019). Soluble lytic transglycosylase SLT of Francisella novicida is involved in intracellular growth and immune suppression. PLoS One 14:e0226778. doi: 10.1371/journal.pone.0226778
Okan, N. A., and Kasper, D. L. (2013). The atypical lipopolysaccharide of Francisella. Carbohydr. Res. 378, 79–83. doi: 10.1016/j.carres.2013.06.015
Santic, M., Al-Khodor, S., and Abu Kwaik, Y. (2010). Cell biology and molecular ecology of Francisella tularensis. Cell. Microbiol. 12, 129–139. doi: 10.1111/j.1462-5822.2009.01400.x
Santin, Y. G., and Cascales, E. (2017). Domestication of a housekeeping transglycosylase for assembly of a type VI secretion system. EMBO Rep. 18, 138–149. doi: 10.15252/embr.201643206
Sauvage, E., Kerff, F., Terrak, M., Ayala, J. A., and Charlier, P. (2008). The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32, 234–258. doi: 10.1111/j.1574-6976.2008.00105.x
Schleifer, K. H., and Kandler, O. (1972). Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36, 407–477. doi: 10.1128/br.36.4.407-477.1972
Sjöstedt, A. (2006). Intracellular survival mechanisms of Francisella tularensis, a stealth pathogen. Microbes Infect. 8, 561–567. doi: 10.1016/j.micinf.2005.08.001
Skalweit, M. J., and Li, M. (2016). Bulgecin a as a β-lactam enhancer for carbapenem-resistant Pseudomonas aeruginosa and carbapenem-resistant Acinetobacter baumannii clinical isolates containing various resistance mechanisms. Drug Des. Devel. Ther. 10, 3013–3020. doi: 10.2147/dddt.S110193
Sobhanifar, S., King, D. T., and Strynadka, N. C. (2013). Fortifying the wall: synthesis, regulation and degradation of bacterial peptidoglycan. Curr. Opin. Struct. Biol. 23, 695–703. doi: 10.1016/j.sbi.2013.07.008
Spidlova, P., Stojkova, P., Dankova, V., Senitkova, I., Santic, M., Pinkas, D., et al. (2018). Francisella tularensis D-ala D-ala carboxypeptidase DacD is involved in intracellular replication and it is necessary for bacterial cell wall integrity. Front. Cell. Infect. Microbiol. 8:111. doi: 10.3389/fcimb.2018.00111
Sutera, V., Levert, M., Burmeister, W. P., Schneider, D., and Maurin, M. (2014). Evolution toward high-level fluoroquinolone resistance in Francisella species. J. Antimicrob. Chemother. 69, 101–110. doi: 10.1093/jac/dkt321
Templin, M. F., Edwards, D. H., and Höltje, J. V. (1992). A murein hydrolase is the specific target of bulgecin in Escherichia coli. J. Biol. Chem. 267, 20039–20043. doi: 10.1016/S0021-9258(19)88662-3
Thunnissen, A. M., Rozeboom, H. J., Kalk, K. H., and Dijkstra, B. W. (1995). Structure of the 70-kDa soluble lytic transglycosylase complexed with bulgecin a implications for the enzymatic mechanism. Biochemistry 34, 12729–12737. doi: 10.1021/bi00039a032
Titball, R. W., and Petrosino, J. F. (2007). Francisella tularensis genomics and proteomics. Ann. N. Y. Acad. Sci. 1105, 98–121. doi: 10.1196/annals.1409.015
van Heijenoort, J. (2011). Peptidoglycan hydrolases of Escherichia coli. Microbiol. Mol. Biol. Rev. 75, 636–663. doi: 10.1128/mmbr.00022-11
Vermassen, A., Leroy, S., Talon, R., Provot, C., Popowska, M., and Desvaux, M. (2019). Cell wall hydrolases in bacteria: insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Front. Microbiol. 10:331. doi: 10.3389/fmicb.2019.00331
Vollmer, W., Blanot, D., and de Pedro, M. A. (2008). Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167. doi: 10.1111/j.1574-6976.2007.00094.x
Weiss, D. S., Henry, T., and Monack, D. M. (2007). Francisella tularensis: activation of the inflammasome. Ann. N. Y. Acad. Sci. 1105, 219–237. doi: 10.1196/annals.1409.005
Yao, X., Jericho, M., Pink, D., and Beveridge, T. (1999). Thickness and elasticity of gram-negative murein sacculi measured by atomic force microscopy. J. Bacteriol. 181, 6865–6875. doi: 10.1128/jb.181.22.6865-6875.1999
Keywords: Francisella, peptidoglycan, tularemia, antibiotics, therapeutics, lytic transglycosylase, carboxypeptidase, penicillin-binding protein
Citation: Bachert BA and Bozue JA (2023) Peptidoglycan enzymes of Francisella: Roles in cell morphology and pathogenesis, and potential as therapeutic targets. Front. Microbiol. 13:1099312. doi: 10.3389/fmicb.2022.1099312
Received: 15 November 2022; Accepted: 28 December 2022;
Published: 12 January 2023.
Edited by:
Svetlana Khaiboullina, University of Nevada, Reno, United StatesReviewed by:
Daniel Alford Powell, University of Arizona, United StatesCopyright © 2023 Bachert and Bozue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beth A. Bachert, ✉ YmV0aC5hLmJhY2hlcnQuY2l2QGhlYWx0aC5taWw=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.