- 1Emergency Department of Hubei Third People's Hospital Affiliated to Jianghan University, Wuhan, Hubei Province, China
- 2School of Resources and Environmental Engineering, Wuhan University of Technology, Wuhan, China
- 3Research Center for Ecology, College of Science, Tibet University, Lhasa, China
- 4Wuchang District Shouyilu Street Community Health Service Center, Wuhan, Hubei Province, China
Gastrodia elata f.glauca (G. elata) is a commonly used Chinese Medicinal Materials with great medicinal value. The medicinal plant and its endophytic bacteria are a symbiotic whole, and the endophytic bacteria are rich in species, and their metabolites are a treasure trove of natural compounds. However, there is a relative lack of analysis on the diversity, flora composition and network interactions of the endophytic bacteria of G. elata. In this study, high-throughput sequencing technology based on the Illumina Miseq platform was used to reveal the core microbiota by examining the diversity and community structures of tuber endophytic bacteria in G. elata grown under different regions and exploring the effect of region on its endophytic bacteria. Here, 1,265 endophytic ASVs were found to coexist with G. elata tuber in Guizhou and Hubei. At the phylum level, the dominant phyla were Proteobacteria, Actinobacteria and Acdobacteriota. At the family level, the dominant family were Comamonadaceae, Nocardicaece, Xanthobacteraceae, and Burkholderiaceae. At the genus level, Delftia and Rhodococcus were represented the core microbiota in G. elata tuber, which served as the dominant genera that coexisted in all samples tested. Moreover, we found that the beta diversity of endophytic bacteria in G. elata tuber was higher level in the Guizhou region than Hubei region. Overall, this study results to provide a reference for screening active strains and interaction between plants and endophytic bacteria.
1. Introduction
Plant Endophytic bacteria are important microbial-plant symbionts and microbial resource that generally exists in healthy plant tissues, which is good for plants (Wang et al., 2019, 2021; Wu et al., 2020). Some studies have found that there are a large number of endophytic bacteria that can settle in the internal tissues of host plants and form a series of mutually beneficial symbiotic relationships (Hassani et al., 2018; Aswani et al., 2020; Li et al., 2020). Plant endophytes not only promote plant growth, but also have potential application prospects in medicine, agriculture, industry and other fields (Aswani et al., 2020; Wang et al., 2021). And the establishment of plant endophyte diversity and community structure is closely related to plant varieties, genotypes, growth environment, geographical location and other factors (Rangjaroen et al., 2019). The plant endophyte is seemed to be an important determinant of plant health and productivity, and has attracted extensive attention worldwide in recent years as a subject of scientific and commercial interest (Gaiero et al., 2013; Xia et al., 2016; Wang et al., 2021). The tuber of G. elata, as an important part of human nutrition and drug, which are rich in microbial resources (Steinbrecher and Leubner-Metzger, 2016).
Gastrodia elata f.glauca is commonly called “Tian ma” in Chinese and mainly distributed in the mountainous areas of eastern Asia (Howes et al., 2017), which is a perennial parasitic herb belonging to the Orchidaceae family, to treat headache, migraine, dizziness, epilepsy, infantile convulsion, tetany and other diseases, and it is widely used in clinical practice (Xu et al., 1998; Tsai et al., 2016; Hu et al., 2019). Up to now, there is no report on the composition and diversity of endophytic bacteria community in G. elata. Several recent studies have shown that the endophytic microbial community associated with wild plant species may play an important role in disease resistance (Pérez-Jaramillo et al., 2016; Tian et al., 2017, 2020; Wang et al., 2019). Some studies have shown that the predominant phylum of endophytic bacteria in rice seeds were Proteobacteria, Actinobacteria and Firmicutes phyla (Zhang et al., 2018; Liu et al., 2019), the Aspergillus, Thicket and Cysticercus was dominant in different varieties of cassava (Manihot esculenta Crantz; Li et al., 2020), is the same true in G. elata? Exploring these problems is of great significance for further development and utilization of G. elata.
However, the composition and diversity of endophytic bacteria community in G. elata has not yet been studied. The core microbiota of the G. elata tuber and their diversity level is not clear. Here, we studied the diversity and composition of the endophytic bacteria community in six G. elata tuber samples under two regions and aimed to discover the core microbiota of the G. elata tubers and the changes of the community composition with the different regions. In this study, the community structure, core microbiota and network interaction of endophytic bacteria in G. elata in Hubei and Guizhou regions were analyzed to provide a reference for screening active strains and interaction between plants and endophytes.
2. Materials and methods
2.1. Sample collection and treatment
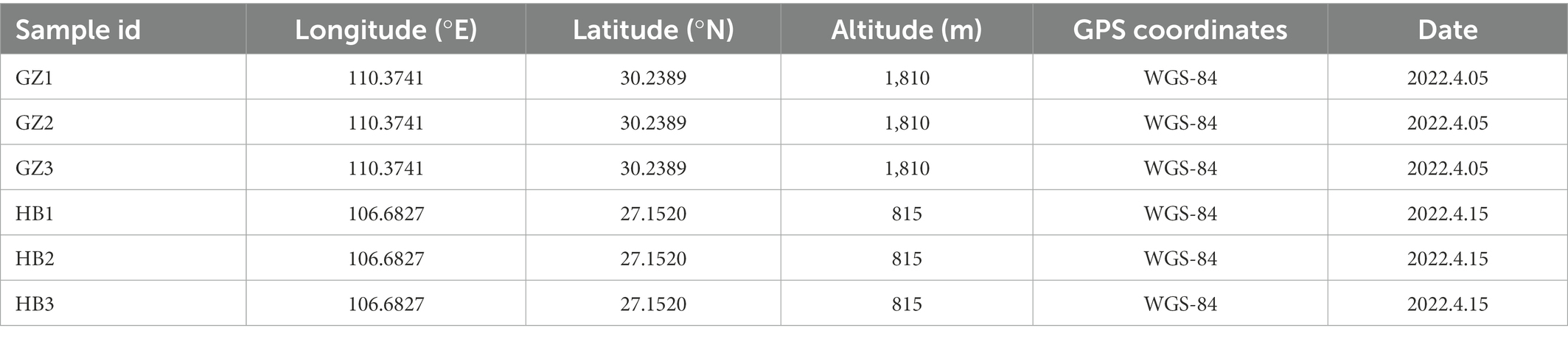
In total, six samples of G. elata tubers were collected from Wu Feng in Hu Bei and Da Fang in Gui zhou (Table 1) in 2022, and G. elata was identified by using the Flora Reipublicae Popularis Sinicae (Flora of China). During sampling, 3 healthy tubers of G. elata were selected and collected. Subsequently, the samples were loaded into sterile sampling bags, marked, placed in the car refrigerator at 4°C, and processed within 24 h. The samples latitude and longitude coordinates were used the World Geodetic System, 1984 (WGS-84) and were recorded using a hand-held GPS unit (Etrex 221x, Garmin, CH.).
Take the sample of tubers of G. elata, rinse the sample with tap water, and place the sample in the ultra-clean table for surface sterilization. The specific method is as follows: Soak in 75% alcohol for 3 min, rinse with prepared sterile water for 3–5 times, soak in 5% NaClO for 3 min, rinse with prepared sterile water for 3–5 times, soak in 75% alcohol for 2 min, rinse with prepared sterile water for 3–5 times, take the last prepared sterile water to coat the plate, test the surface disinfection effect. The surface sterilized tubers of G. elata, respectively, cut into small pieces and put into sterile 2 ml centrifuge tubes and store at −80°C until used.
2.2. DNA extraction and high-throughput sequencing
The sample DNA was extracted from the filter membranes using a Power DNA Isolation Kit (Qiagen, Germantown, MD, United States) according to the manufacturer’s protocol, and DNA quality was checked using 1% agarose gel electrophoresis. DNA concentration and purity were determined with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, United States). Next, the 16S rDNA V3-V4 hypervariable region was PCR-amplified using the following primers: 799F (AACMGGATTAGATACCCKG) and 1392R (ACGGGCGGTGTGTRC). All of the thermocycling steps were as follows: 5 min at 95°C; 20 cycles of 45 s at 95°C, 30 s at 57°C, and 30 s at 72°C. The amplified products were purified and mixed in equivalent amounts PCR products were sequenced using the PE250 strategy on the Illumina Miseq2500 platform by Majorbio (Shanghai, China).
2.3. Bioinformatics and statistical analysis
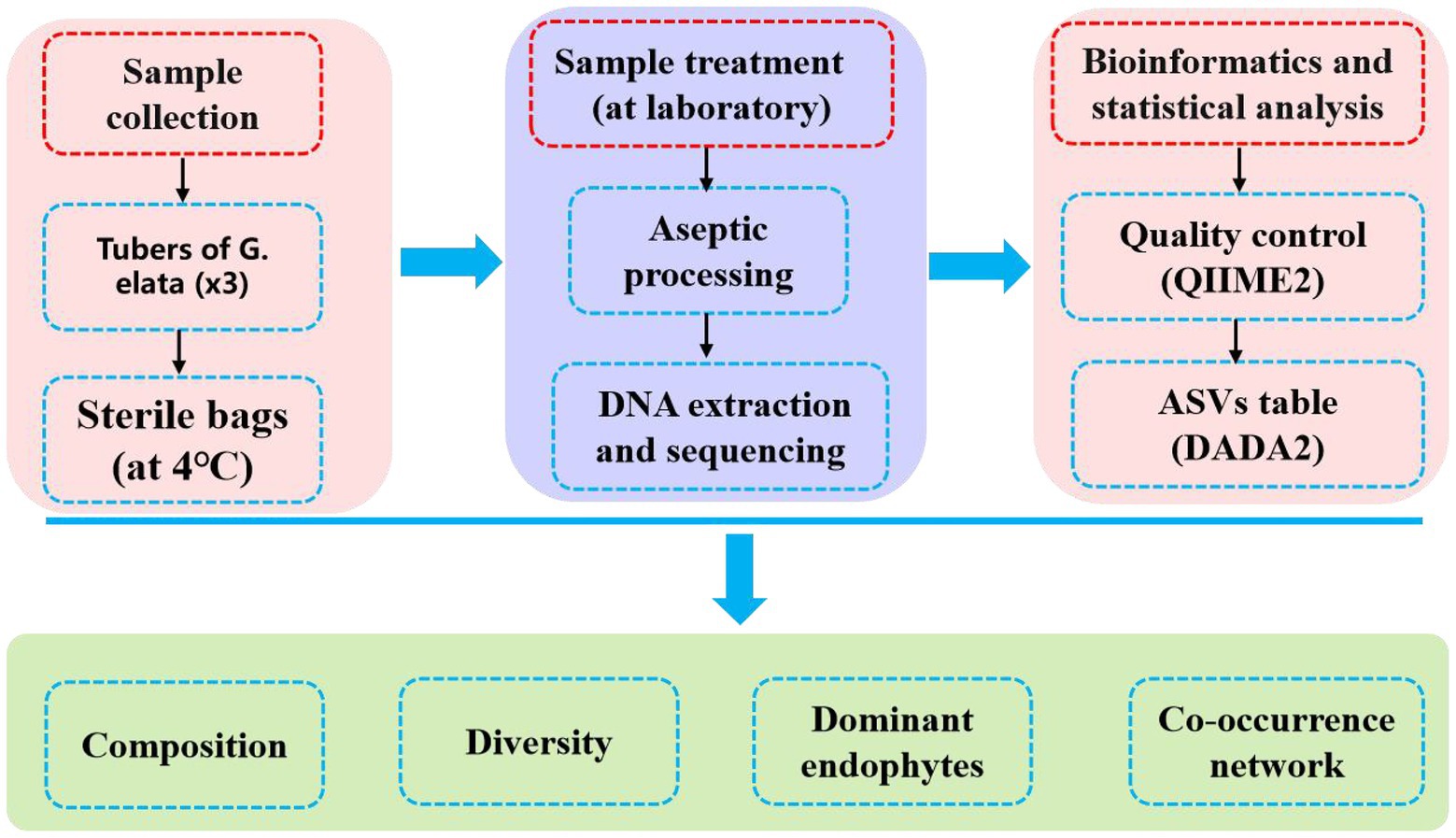
After quality filtering the raw data, high quality clean data was received for subsequent analysis. The clean data were demultiplexed separately by their unique barcodes. A standard denoising pipeline was used to obtain the amplicon sequence variants (ASVs) were obtained using the DADA2 plug-in in QIIME2 software (version 2022.8; Bokulich et al., 2018; Bolyen et al., 2019), after which an ASV abundance table was constructed. The ASVs were annotated using the SILVA database (version 138; Quast et al., 2013). Low-abundance ASVs (<10 reads) were removed. Three replicates were used to reduce the sampling bias. The ASV table was then rarefied to 40,000 reads per sample for downstream analysis. Alpha diversity indices of endophytic bacteria communities, including Richness index, Shannon-Wiener diversity index, Chao1 index, ACE index, Good’s coverage index and Simpson dominance index, were calculated using the “vegan” package in R software (version 4.1.1). Principal coordinate analysis (PCoA) and Multiple samples Non-metric multidimensional scaling (NMDS) were performed based on the Bray–Curtis distance using the “vegan” and “ggplot2” package in R software. Co-occurrence patterns of endophytic bacteria communities were constructed based on Spearman’s rank correlation coefficients. Co-occurrence events were identified as statistically robust correlations (|R| > 0.6, p < 0.05) and the co-occurrence network was visualized in Gephi (version 0.9.6). The basic skeleton diagram is shown in Figure 1.
3. Results
3.1. High throughput sequencing statistics of endophytic bacteria communities in Gastrodia elata f.glauca
A total of 280,523 raw sequences were obtained by high-throughput sequencing. After quality control, 260,543 high-quality sequences were acquired from 6 samples of endophytic bacteria in G. elata, which were grouped into 1,265 ASVs were identified belonging to 23 phyla, 54 classes, 159 orders, 260 families, and 437 genera (Figure 2A). Rarefaction curves indicated that most of the diversity could be covered by the resampling depth of 40,000 reads (Figure 2B). Sequencing of 10–50 thousand V3-V4 rDNA reads from G. elata samples was sufficient to approach saturation in endophytic bacteria richness for all samples. For all samples combined, most ASVs were represented by 8–64 reads, and others had lower or greater abundance (Figure 2C). As displayed in the Venn diagram, 282 ASVs were common between GZ and HB group. The unique ASVs among all samples collected at different groups were 674, and 119, respectively, which were 62.7% in GZ group, and 11.1% in HB group (Figure 2D). The number of endophytic bacteria ASVs of HB group less than GZ group. In addition, only 282 ASVs were common between GZ and HB group, indicating a significant difference in the community structure of endophytic bacteria between GZ and HB group.
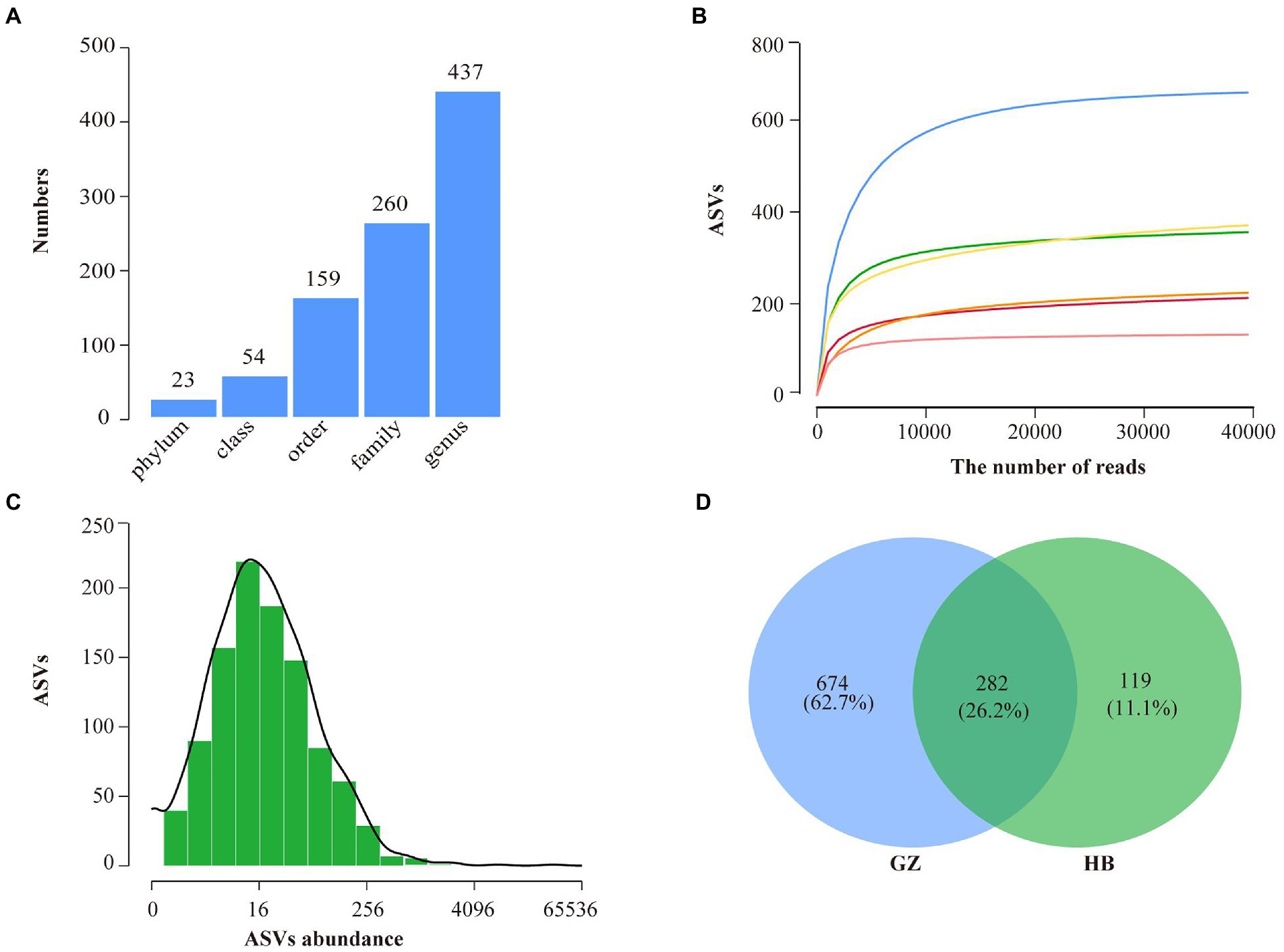
Figure 2. High throughput sequencing statistics and rank abundance curves. (A) ASV classification statistics. (B) ASV rarefaction curves. (C) Distribution of ASVs abundance of bacteria. (D) Venn diagram showing the number of shared and specific ASVs for each group.
3.2. Overall alpha diversity of endophytic bacteria communities in Gastrodia elata f.glauca tubers
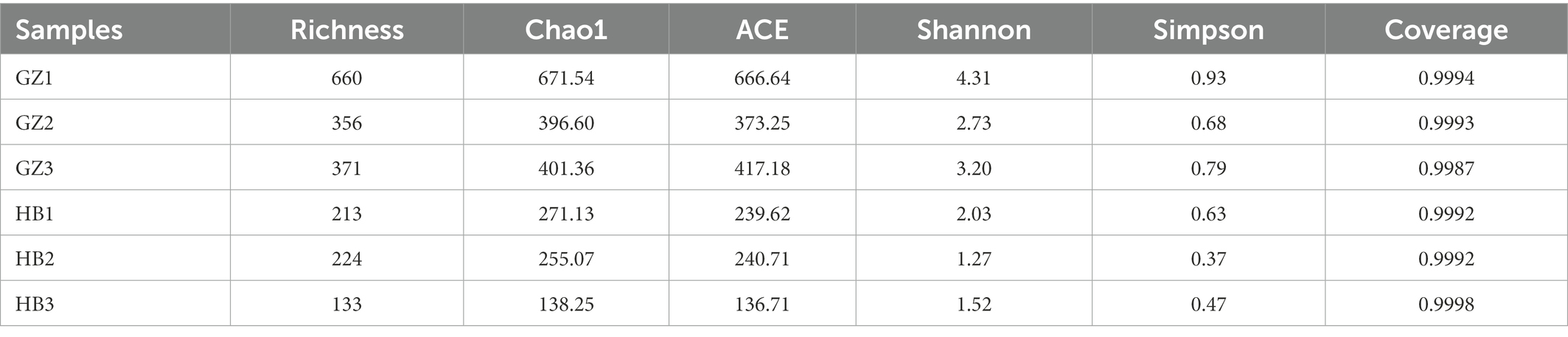
The Good’s coverage threshold of 16S sequences was obtained from endophytic bacteria in G. elata tubers were >0.9987, indicating that it represents the true endophytic bacteria populations in the microbial community of each sample (Table 2). Higher alpha diversity index (Shannon, Simpson, Richness, Chao1 and ACE index) indicate the high diversity of endophytic bacteria community in a sample. The alpha diversity of endophytic bacteria varied among different samples for the different indices: the ASV Richness index varied from 133 to 660, with the median value of 326.16. Shannon’s diversity index varied from 1.27 to 4.31, with the median value of 2.51. ACE index varied from 136.74 to 666.64, with the median value of 345.69. Chao1 index varied from 138.25 to 671.54, with the median value of 355.66. Simpson index varied from 0.37 to 93, with the median value of 0.65. We observed the highest alpha diversity of endophytic bacteria community in GZ1 sample followed by GZ2 sample, and the lowest alpha diversity in HB3 sample. These findings indicate that the alpha diversity of the endophytic bacteria of G. elata was quite difference in different regions (Table 2).
3.3. Beta diversity analysis of the endophytic bacteria communities in Gastrodia elata f.glauca tubers
To explore the Beta diversity, principal coordinate analysis (Figure 3A) and Non-metric multidimensional scaling (Figure 3B) analysis of samples of G. elata tubers were performed. The sum of PCoA1 and PCoA2 was 82.00% in PcoA and Stress <0.1 in NMDS, indicating a high diversity in the endophytic bacteria population structure in samples collected from G. elata tubers. The samples collected at HB1, HB2, and HB3 were clustered closely but were far from samples collected at GZ1, GZ2, and GZ3. These results proved the significant difference among endophytic bacteria communities in G. elata tubers in different regions.
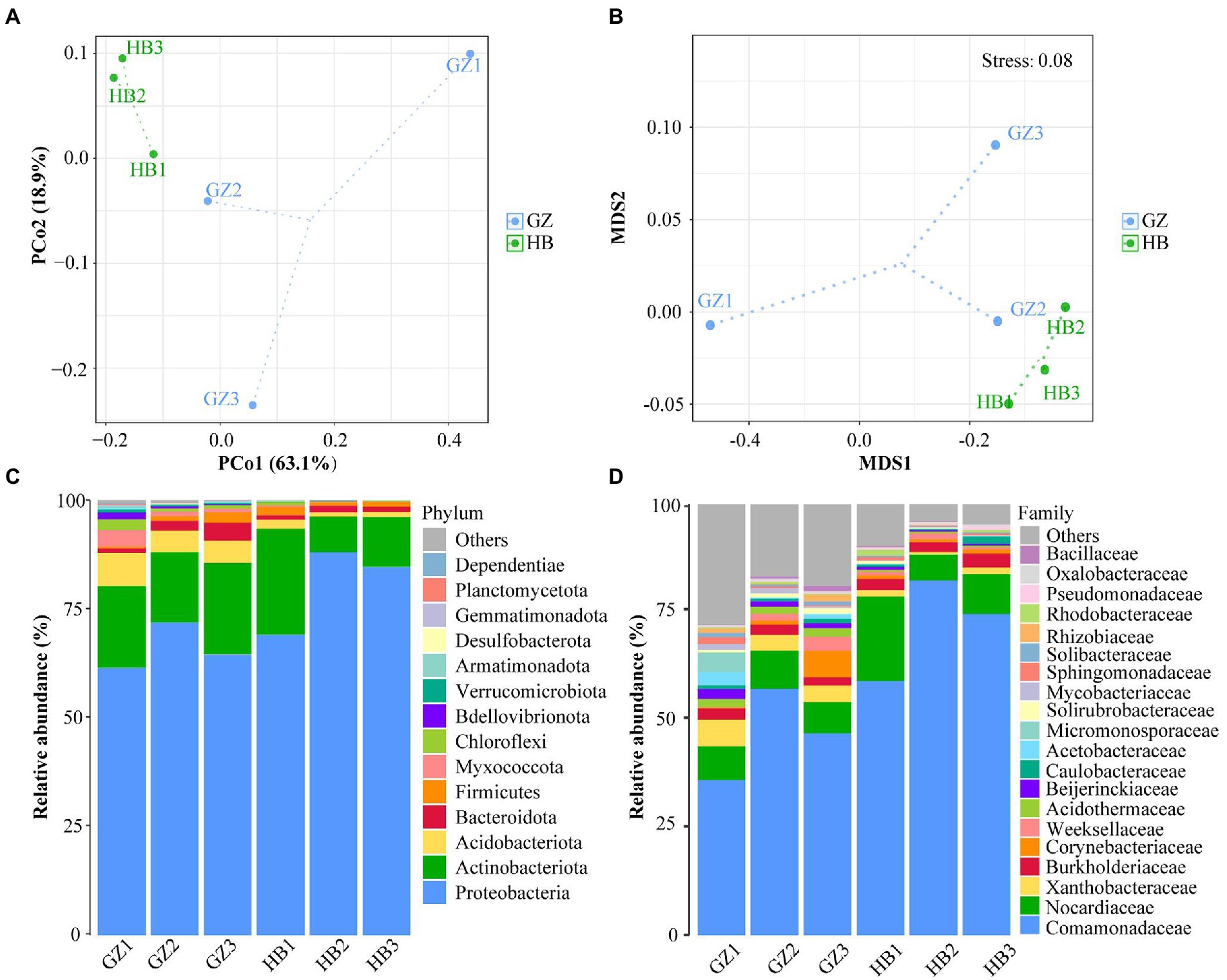
Figure 3. Composition of endophytic bacteria in Gastrodia elata f.glauca tuber samples. (A) Multiple sample principal coordinate analysis (PCoA) of the ASV level. (B) Multiple sample Non-metric multidimensional scaling (NMDS) of the ASV level. (C) Relative abundance of phyla in each sample. (D) Relative abundance of family in each sample.
The endophytic bacterial community composition of G. elata tubers in different regions is shown in Figure 3C at the phylum level. The results indicated that Proteobacteria (61.31%–87.95%), Actinobacteria (8.20%–18.84%) and Acdobacteriota (0.95%–7.64%) were the dominant endophytes that coexisted, in different proportions in G. elata tubers. Proteobacteria was the most dominant phylum in all tuber samples, with abundance ranging from 83.90%–99.87%. At the family level, every G. elata samples also had dominant endophytes, including mainly Comamonadaceae (35.71%–81.65%), Nocardicaece (7.24%–19.45%), Xanthobacteraceae (0.59%–6.10%) and Burkholderiaceae (1.87%–3.21%; Figure 3D).
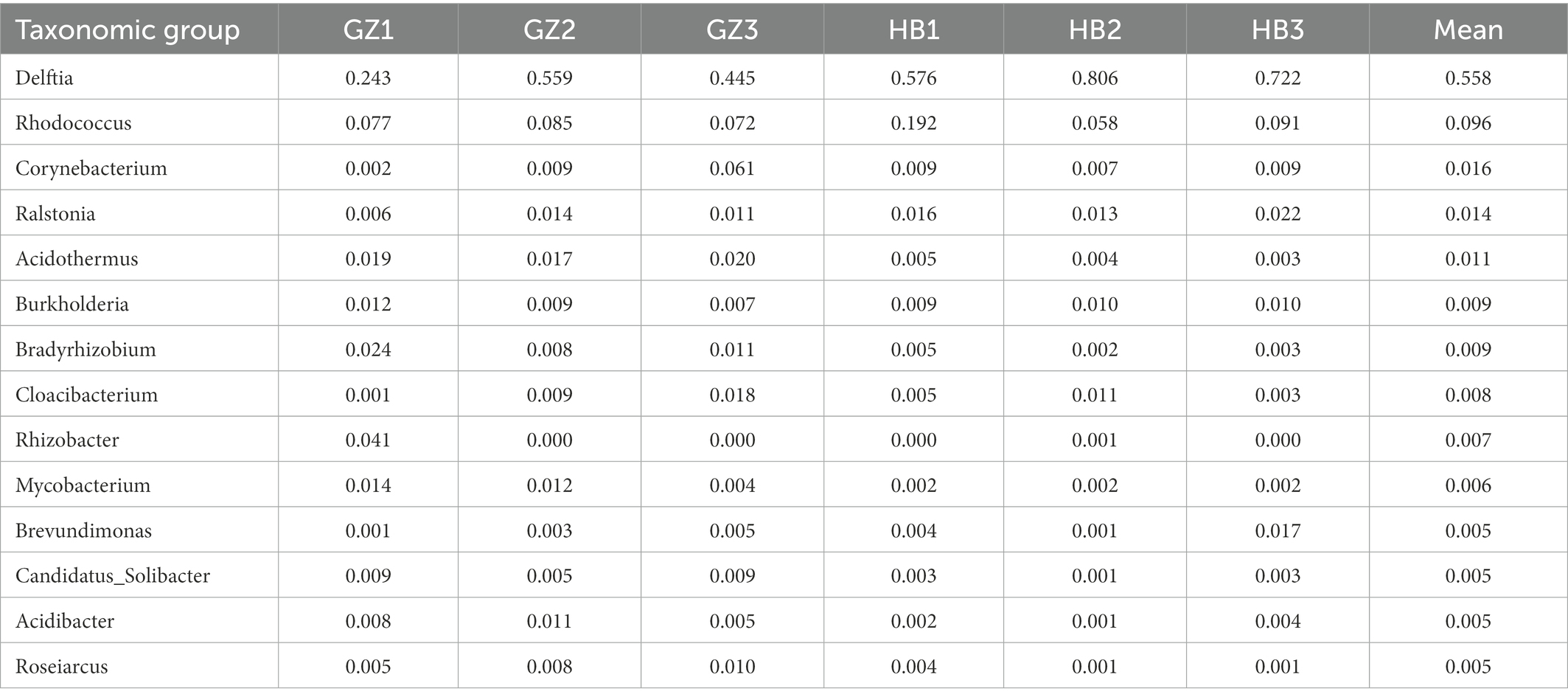
Analysis of the two regions showed that the predominant genus (mean relative abundance ≥0.5%) of endophytic bacteria in G. elata tuber was Delftia (55.8%), followed by Rhodococcus (9.6%), Corynebacterium (1.6%), Ralstonia (1.4%), Acidothermus (1.1%), Burkholderia (0.9%), Bradyrhizobium (1.9%), Cloacibacterium (0.8%), Rhizobacter (0.7%), Mycobacterium (0.6%), Brevundimonas (0.5%), Candidatus_Solibacter (0.5%), Acidibacter (0.5%), Roseiarcus (0.5%). We found Delftia was the most dominant endophytic bacterial genus (55.8%) and Rhodococcus (9.6%) was the second most abundant.
3.4. Co-occurrence network of endophytic bacteria communities in Gastrodia elata f.glauca
Co-occurrence network analysis was performed to explore potential relationships between endophytic bacteria communities in G. elata tuber. Modularity coefficients for all co-occurrence networks were > 0.4, indicating clear modularity. In the network, the predominant phylum (the ratio > 5%) of endophytic bacteria communities was Proteobacteria (37.43%), followed by Acidobacteriota (14.86%), Actinobacteriota (13.71%), Myxococcota (7.14%), Firmicutes (6.86%), Bacteroidota (5.71%), and Chloroflexi (5.43%; Figure 4A). In addition, there were five main modules (the ratio > 4%) in endophytic bacteria communities’ network of G. elata (Figure 4B).

Figure 4. Co-occurrence network pattern. (A) Co-occurrence networks of endophytic bacteria communities in G. elata tuber; Phyla of the network as shown in different colors. (B) Co-occurrence networks of endophytic bacteria communities in G. elata; Modules of the network as shown in different colors. (C) Co-occurrence relationships within endophytic bacteria communities in G. elata tuber; phyla as shown in different colors. (D) Co-occurrence relationships between phyla and modules.
To further investigate co-occurrence relationships within endophytic bacteria communities, we analyzed ASVs interactions among the different phyla (Figure 4C). The top six pairs with co-occurrence relationships of endophytic bacteria communities were as follows: Proteobacteria to Proteobacteria > Myxococcota to Proteobacteria > Acidobacteriota to Proteobacteria > Proteobacteria to Chloroflexi > Proteobacteria to Actinobacteriota > Proteobacteria to Bdellovibrionota. In the co-occurrence network, a higher proportion of nodes of the Proteobacteria phylum interacted with ASVs in other phyla, indicating that Proteobacteria makes the greatest contribution to the network structure. Furthermore, we found that endophyte composition was significant differences in different modules (Figure 4D). The top 5 phyla with co-occurrence relationships for each module (M1 ~ M5) were as follows: In M1, Proteobacteria > Myxococcota > Chloroflexi > Acidobacteriota > Actinobacteriota; In M2, Proteobacteria > Firmicutes > Myxococcota > Acidobacteriota > Bacteroidota; In M3, Proteobacteria > Acidobacteriota > Chloroflexi > Actinobacteriota > Myxococcota; In M4, Proteobacteria > Acidobacteriota > Chloroflexi > Actinobacteriota > Myxococcota; and M5, Proteobacteria > Acidobacteriota > Myxococcota > Bdellovibrionota > Chloroflexi. In all modules, a higher proportion of nodes of the Proteobacteria interacted with ASVs in other phyla.
4. Discussion
Gastrodia elata f.glauca is an important Chinese medicinal materials and plays a pivotal role in the regional economy in Hubei, Guizhou and other places in China (Zhan et al., 2016). According to statistics, as of 2020, the dried production of G. elata in China was 30,000 tons, with a total output value of about 5 billion CNY. Carrying out scientific research on endophytes in G. elata is important to improve the potential yield of Chinese medicinal materials, identifying the quality of herbs, and to achieve healthy planting of G. elata. However, the present research on G. elata is mainly focused on breeding, planting, and medicine, but there is little research on its endophytic bacteria, especially tuber endophytic bacteria. Based high-throughput sequencing methods overcome the non-culturability problem of most microbes by analyzing DNA extracted from plant endophytes (Manter et al., 2010). Plant endophytes are conventionally defined as bacteria or fungi that reside internally in plant tissues, and are in healthy plant tissues and do not harm the plant (Gaiero et al., 2013; Wang et al., 2019; Li et al., 2020). High-throughput sequencing is an efficient way to reveal the diversity of plant endophytes (Gaiero et al., 2013; Rangjaroen et al., 2019; Aswani et al., 2020).
In this study, 260,543 high-quality sequences were acquired from 6 samples of endophytic bacteria in G. elata and 1,265 ASVs were identified belonging to 23 phyla, 54 classes, 159 orders, 260 families, and 437 genera, which could reveal the community composition of the endophytes in G. elata samples. Moreover, we found that the alpha and beta diversity of the endophytic bacteria of G. elata was the difference in different regions. The result demonstrated that many factors, including the level of oxygen, moisture and other environmental factors, could affect diversity levels of the endophytic bacteria community (Xia et al., 2016). This found is consistent with previous studies on endophytic bacteria communities in Camellia sinensis and saline-alkali tolerant rice, which also found that some key factors (e.g., salt, catechins, gallic acid and anthocyanidin etc.) determine the composition of endophytic bacteria (Wu et al., 2020; Wang et al., 2021). This also explains the significant differences in the diversity and composition of endophytic bacteria of G. elata in different geographic regions.
In addition, we found that Proteobacteria was the most dominant phylum in all tuber samples, with abundance ranging from 83.90% to 99.87%. This found is consistent with previous studies on endophytic bacteria communities in saline-alkali tolerant rice (Wang et al., 2021). The co-occurrence networks revealed that positive correlations were the most common interactions between species, to some extent reflecting co-aggregation, cross-feeding, and co-colonization (Faust and Raes, 2012; Zhang et al., 2022). The co-occurrence network of the endophytic bacteria of G. elata was mainly composed of positive correlations. The results of co-occurrence network analysis also showed that a higher proportion of nodes of the Proteobacteria phylum interacted with ASVs in other clades, indicating that Proteobacteria makes the greatest contribution to the endophytic network structure (Figure 4). Overall, the main endophytic bacterial groups and community structure of different G. elata tuber samples collected in two regions were similar at phylum level, but the abundance was different (Figure 3C). The mainly genera are Delftia, Rhodococcus, Corynebacterium, Ralstonia, Acidothermus, Burkholderia, Bradyrhizobium, Cloacibacterium, Rhizobacter, Mycobacterium, Brevundimonas, Candidatus_Solibacter, Acidibacter and Roseiarcus (Table 3). At the same time, we found that Delftia and Rhodococcus genus were the core bacterial genus observed (55.8, 9.6%, respectively). Shockingly, Delftia and Rhodococcus were first found in G. elata. Recent research shows that Delftia is also the dominant genus of probiotic endophytes in bananas (Beltran-Garcia et al., 2021). Delftia can also induce resistance of whole plant to bacterial pathogens (Kurokawa et al., 2021). The role of Delftia and Rhodococcus in G. elata and their relationship with asparagine synthesis deserve further investigation.
5. Conclusion
This study first investigated the endophytes of the G. elata tubers in the Guizhou and Hubei region using the high-throughput sequencing methodology. The results show that endophytes communities exhibited significant geographic variations. The alpha and beta diversity of endophytes in G. elata was higher in Guizhou and declined in Hubei. Moreover, At the phylum level, the dominant phyla were Proteobacteria, Actinobacteria and Acdobacteriota. At the family level, the dominant family were Comamonadaceae, Nocardicaece, Xanthobacteraceae and Burkholderiaceae. At the genus level, Delftia and Rhodococcus were represented the core microbiota in G. elata tuber. The coexistence of core bacteriome and tubers, indicates that should be beneficial to the growth and health of G. elata plants. The physiological and ecological functions of Delftia and Rhodococcus and their relationship with active components of G. elata are worthy of further study.
Data availability statement
The data presented in the study are deposited in the National Genomics Data Center repository, accession number CRA009458.
Author contributions
HZ, PZ, JD, and JG designed and participated in all experimental procedures, performed data analysis, and drafted the manuscript. JD participated in the samples collection and preparation. HZ, PZ, JQ, JD, and JG supervised the study and critically revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Hubei Provincial Administration of Traditional Chinese Medicine Research Project of Traditional Chinese Medicine (Project No. ZY2023F030).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
G. elata, Gastrodia elata Bl. form. Glauca S. Chow; ASVs, amplicon sequence variants; PCoA, principal coordinate analysis; NMDS, non-metric multidimensional scaling.
References
Aswani, R., Jishma, P., and Radhakrishnan, E. K. (2020). “2-endophytic bacteria from the medicinal plants and their potential applications,” in Microbial Endophytes, Woodhead Publishing Series in Food Science, Technology and Nutrition. eds. A. Kumar and V. K. Singh (Woodhead Publishing), 15–36.
Beltran-Garcia, M. J., Martinez-Rodriguez, A., Olmos-Arriaga, I., Valdez-Salas, B., Chavez-Castrillon, Y. Y., Di Mascio, P., et al. (2021). Probiotic endophytes for more sustainable Banana production. Microorganisms 9:1805. doi: 10.3390/microorganisms9091805
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Faust, K., and Raes, J. (2012). Microbial interactions: from networks to models. Nat. Rev. Microbiol. 10, 538–550. doi: 10.1038/nrmicro2832
Gaiero, J. R., McCall, C. A., Thompson, K. A., Day, N. J., Best, A. S., and Dunfield, K. E. (2013). Inside the root microbiome: bacterial root endophytes and plant growth promotion. Am. J. Bot. 100, 1738–1750. doi: 10.3732/ajb.1200572
Hassani, M. A., Durán, P., and Hacquard, S. (2018). Microbial interactions within the plant holobiont. Microbiome 6:58. doi: 10.1186/s40168-018-0445-0
Howes, M.-J. R., Fang, R., and Houghton, P. J. (2017). “Chapter two-effect of Chinese herbal medicine on Alzheimer’s disease,” in International Review of Neurobiology, Neurobiology of Chinese Herb Medicine. eds. B.-Y. Zeng and K. Zhao (Academic Press), 29–56.
Hu, M., Yan, H., Fu, Y., Jiang, Y., Yao, W., Yu, S., et al. (2019). Optimal extraction study of Gastrodin-type components from Gastrodia elata tubers by response surface design with integrated phytochemical and bioactivity evaluation. Molecules 24:547. doi: 10.3390/molecules24030547
Kurokawa, M., Nakano, M., Kitahata, N., Kuchitsu, K., and Furuya, T. (2021). An efficient direct screening system for microorganisms that activate plant immune responses based on plant–microbe interactions using cultured plant cells. Sci. Rep. 11:7396. doi: 10.1038/s41598-021-86560-0
Li, H., Yan, C., Tang, Y., Ma, X., Chen, Y., Chen, S., et al. (2020). Endophytic bacterial and fungal microbiota in different cultivars of cassava (Manihot esculenta Crantz). J. Microbiol. 58, 614–623. doi: 10.1007/s12275-020-9565-x
Liu, Y., Xu, P. P., Yang, F. Z., Li, M., Yan, H., Li, N., et al. (2019). Composition and diversity of endophytic bacterial community in seeds of super hybrid rice ‘Shenliangyou 5814’ (Oryza sativa L.) and its parental lines. Plant Growth Regul. 87, 257–266.
Manter, D. K., Delgado, J. A., Holm, D. G., and Stong, R. A. (2010). Pyrosequencing reveals a highly diverse and cultivar-specific bacterial endophyte Community in Potato Roots. Microb. Ecol. 60, 157–166. doi: 10.1007/s00248-010-9658-x
Pérez-Jaramillo, J. E., Mendes, R., and Raaijmakers, J. M. (2016). Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol. Biol. 90, 635–644. doi: 10.1007/s11103-015-0337-7
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., and Yarza, P. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic AcidsRes. 41, D590–596.
Rangjaroen, C., Lumyong, S., Sloan, W. T., and Sungthong, R. (2019). Herbicide-tolerant endophytic bacteria of rice plants as the biopriming agents for fertility recovery and disease suppression of unhealthy rice seeds. BMC Plant Biol. 19:580. doi: 10.1186/s12870-019-2206-z
Steinbrecher, T., and Leubner-Metzger, G. (2016). The biomechanics of seed germination. EXBOTJ erw 428:erw428. doi: 10.1093/jxb/erw428
Tian, L., Lin, X., Tian, J., Ji, L., Chen, Y., Tran, L.-S. P., et al. (2020). Research advances of beneficial microbiota associated with crop plants. Int. J. Mol. Sci. 21:1792. doi: 10.3390/ijms21051792
Tian, L., Zhou, X., Ma, L., Xu, S., Nasir, F., and Tian, C. (2017). Root-associated bacterial diversities of Oryza rufipogon and Oryza sativa and their influencing environmental factors. Arch. Microbiol. 199, 563–571. doi: 10.1007/s00203-016-1325-2
Tsai, C.-C., Wu, K.-M., Chiang, T.-Y., Huang, C.-Y., Chou, C.-H., Li, S.-J., et al. (2016). Comparative transcriptome analysis of Gastrodia elata (Orchidaceae) in response to fungus symbiosis to identify gastrodin biosynthesis-related genes. BMC Genomics 17:212. doi: 10.1186/s12864-016-2508-6
Wang, S.-S., Liu, J.-M., Sun, J., Sun, Y.-F., Liu, J.-N., Jia, N., et al. (2019). Diversity of culture-independent bacteria and antimicrobial activity of culturable endophytic bacteria isolated from different dendrobium stems. Sci. Rep. 9:10389. doi: 10.1038/s41598-019-46863-9
Wang, Z., Zhu, Y., Li, N., Liu, H., Zheng, H., Wang, W., et al. (2021). High-throughput sequencing-based analysis of the composition and diversity of endophytic bacterial community in seeds of saline-alkali tolerant rice. Microbiol. Res. 250:126794. doi: 10.1016/j.micres.2021.126794
Wu, Z., Su, Q., Cui, Y., He, H., Wang, J., Zhang, Y., et al. (2020). Temporal and spatial pattern of endophytic fungi diversity of Camellia sinensis (cv. Shu cha Zao). BMC Microbiol. 20:270. doi: 10.1186/s12866-020-01941-1
Xia, F., Chen, X., Guo, M.-Y., Bai, X.-H., Liu, Y., Shen, G.-R., et al. (2016). High-throughput sequencing-based analysis of endogenetic fungal communities inhabiting the Chinese Cordyceps reveals unexpectedly high fungal diversity. Sci. Rep. 6:33437. doi: 10.1038/srep33437
Xu, Q., Liu, Y., Wang, X., Gu, H., and Chen, Z. (1998). Purification and characterization of a novel anti-fungal protein from Gastrodia elata. Plant Physiol. Biochem. 36, 899–905. doi: 10.1016/S0981-9428(99)80008-4
Zhan, H.-D., Zhou, H.-Y., Sui, Y.-P., Du, X.-L., Wang, W., Dai, L., et al. (2016). The rhizome of Gastrodia elata Blume–An ethnopharmacological review. J. Ethnopharmacol. 189, 361–385. doi: 10.1016/j.jep.2016.06.057
Zhang, P., Xiong, J., Qiao, N., An, R., Da, Z., Miao, W., et al. (2022). Spatiotemporal distribution of protists in the Yarlung Zangbo River. Tibetan Plateau. Water Biology and Security 1:100064. doi: 10.1016/j.watbs.2022.100064
Keywords: Gastrodia elata, high-throughput sequencing, endophytic bacteria, diversity, composition
Citation: Zheng H, Zhang P, Qin J, Guo J and Deng J (2023) High-throughput sequencing-based analysis of the composition and diversity of endophytic bacteria community in tubers of Gastrodia elata f.glauca. Front. Microbiol. 13:1092552. doi: 10.3389/fmicb.2022.1092552
Edited by:
Zhihui Xu, Nanjing Agricultural University, ChinaReviewed by:
Laith Khalil Tawfeeq Al-Ani, Universiti Sains Malaysia, MalaysiaSahil Mehta, University of Delhi, India
Copyright © 2023 Zheng, Zhang, Qin, Guo and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiani Guo,  Z3VvamlhbmkwODA1QDE2My5jb20=; Jun Deng,
Z3VvamlhbmkwODA1QDE2My5jb20=; Jun Deng,  c2FsbWFuNTlAc2luYS5jb20=
c2FsbWFuNTlAc2luYS5jb20=
†These authors have contributed equally to this work and share first authorship
 Heng Zheng1,2†
Heng Zheng1,2† Jiani Guo
Jiani Guo