
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 12 January 2023
Sec. Microbial Physiology and Metabolism
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1085918
 Neda Habibi Arejan1
Neda Habibi Arejan1 Delfina Ensinck2
Delfina Ensinck2 Lautaro Diacovich2
Lautaro Diacovich2 Parthvi Bharatkumar Patel1
Parthvi Bharatkumar Patel1 Samantha Y. Quintanilla1
Samantha Y. Quintanilla1 Arash Emami Saleh3
Arash Emami Saleh3 Hugo Gramajo2
Hugo Gramajo2 Cara C. Boutte1*
Cara C. Boutte1*Mycobacterial cell elongation occurs at the cell poles; however, it is not clear how cell wall insertion is restricted to the pole or how it is organized. Wag31 is a pole-localized cytoplasmic protein that is essential for polar growth, but its molecular function has not been described. In this study we used alanine scanning mutagenesis to identify Wag31 residues involved in cell morphogenesis. Our data show that Wag31 helps to control proper septation as well as new and old pole elongation. We have identified key amino acid residues involved in these essential functions. Enzyme assays revealed that Wag31 interacts with lipid metabolism by modulating acyl-CoA carboxylase (ACCase) activity. We show that Wag31 does not control polar growth by regulating the localization of cell wall precursor enzymes to the Intracellular Membrane Domain, and we also demonstrate that phosphorylation of Wag31 does not substantively regulate peptidoglycan metabolism. This work establishes new regulatory functions of Wag31 in the mycobacterial cell cycle and clarifies the need for new molecular models of Wag31 function.
In order to grow, bacteria need to expand the existing cell wall without disturbing its integrity. The peptidoglycan of the cell wall is critical for maintenance of cell shape in most bacteria, and is made of sugar chains crosslinked by small peptides that are assembled just outside of the cell membrane. Many rod shaped organisms expand the peptidoglycan network all along the lateral walls through the activity of membrane-anchored peptidoglycan synthases and their regulators (Zhao et al., 2017; Egan et al., 2020). Mycobacteria, as well as many other Actinomycetes and some Alphaproteobacteria, restrict peptidoglycan expansion to the cell poles (Thanky et al., 2007; Brown et al., 2012). In mycobacteria, the old pole elongates continuously, but there is a delay in initiation of elongation at the new pole – the one most recently created by cell division (Hannebelle et al., 2020) – which results in asymmetric cells (Joyce et al., 2012; Rego et al., 2017; Hannebelle et al., 2020).
The molecular mechanisms that control polar cell envelope expansion are not well described. In this paper we dissect some of the functions of Wag31 (AKA DivIVA, Rv2145c, MSMEG_4217), which is an essential protein for polar growth in mycobacteria (Kang et al., 2008; Melzer et al., 2018). Wag31 is a mycobacterial ortholog of DivIVA. DivIVA proteins are found in Firmicutes and Actinobacteria (Supplementary Figure S1) and they typically localize to the poles and/or septum. They function to recruit and regulate other factors involved in cell division, chromosome segregation and cell wall synthesis (Marston and Errington, 1999; Valbuena et al., 2007; van Baarle et al., 2013; Eswaramoorthy et al., 2014; Sieger and Bramkamp, 2015).
Wag31 is localized at the cell poles (Nguyen et al., 2007; Meniche et al., 2014) and at the septum just before division (Santi et al., 2013). In addition to its role in polar growth, it also helps with chromosome segregation by recruiting the segregation factor ParA to the poles (Pióro et al., 2018). Wag31 is not required for cell wall metabolism, but it is needed to direct this metabolism to the poles (Melzer et al., 2022), though it is not known how it performs this function.
Mycobacterium smegmatis (Msmeg) does not regulate polar cell wall synthesis by restricting its essential peptidoglycan transglycosylases to the poles, as some other pole-growing species do (Valbuena et al., 2007; Cameron et al., 2014; Sieger and Bramkamp, 2015; Sher et al., 2021), since both RodA and PBP1 are delocalized around the entire cell instead of restricted to the poles (Melzer et al., 2022). However, Msmeg does restrict synthesis of many cell wall precursor enzymes near the poles, through their association with the Intracellular Membrane Domain (IMD), a chemically distinct region of the plasma membrane found at the sub polar regions in growing mycobacterial cells (Hayashi et al., 2016; Melzer et al., 2022). Depletion of Wag31 delocalizes the IMD (García-Heredia et al., 2021), though it is not clear whether Wag31 regulates this membrane domain directly, or how the IMD contributes to polar elongation.
Wag31 has been shown to interact with AccA3, and FtsI, but the role of these interactions in regulating cell wall expansion is unknown (Mukherjee et al., 2009; Meniche et al., 2014; Xu et al., 2014). FtsI is a transpeptidase involved in peptidoglycan synthesis at the septum (Mukherjee et al., 2009; Boes et al., 2019), while AccA3 is a common subunit of the essential Acyl-CoA Carboxylase (ACCase) complexes involved in the first steps of lipid synthesis (Gago et al., 2018). The subunit composition of these ACCase complexes determines their preference for using either acetyl-CoA (ACC activity) or propionyl-CoA (PCC activity) as substrates, to yield either malonyl-or methylmalonyl-CoA, respectively, which are used as the elongation units for the synthesis of straight-chain or branched-chain fatty acids-containing lipids. The specificity constant of ACCase 5 (formed by the AccA3, AccD5 and AccE5 subunits) is five time higher for propionyl-CoA compared with acetyl-CoA (Gago et al., 2006) while ACCase 6 (constituted by the AccA3 and AccD6 subunits) has a high preference for acetyl-CoA as a substrate (Daniel et al., 2007).
Here, we obtained and characterized a series of point mutants of wag31 that exhibit diverse phenotypes, indicating that Wag31 has separate roles in regulating elongation at the new and old poles, and septation. We also show that while phosphorylation of Wag31 does moderately affect polar growth, it is not a critical regulator of this process. Furthermore, our data indicate that Wag31’s role in polar growth is not mediated through controlling the localization of IMD enzymes, or regulating FtsI via Wag31 residues NSD46-48. However, critical point mutants of Wag31 allowed us to demonstrate that it does have a role in regulating ACCase complex activity.
All Msmeg strains were grown in liquid culture in 7H9 (BD, Sparks, MD) medium supplemented with 0.2% glycerol, 0.05% Tween 80, and ADC (5 g/l albumin, 2 g/l dextrose, 0.85 g/l NaCl, 0.003 g/l catalase). For most experiments, Msmeg strains were grown on LB Lennox plates. DH5a, TOP10, and XL1-Blue E.coli cells were used for cloning. For E. coli, antibiotics concentrations were: kanamycin – 50 μg/ml; hygromycin – 100 μg/ml; nourseothricin – 40 μg/ml; Zeocin – 25 μg/ml. For Msmeg, antibiotic concentrations were: kanamycin – 25 μg/ml; hygromycin – 50 μg/ml; nourseothricin – 20 μg/ml; trimethoprim – 50 μg/ml; Zeocin – 20 μg/ml.
Strains were grown to logarithmic phase in 7H9 with appropriate antibiotics, then diluted to OD600 = 0.1 without antibiotics in a 96 well plate. A Synergy Neo2 linear Multi-Mode Reader was used to shake the plates continuously for 18 h at 37°C, and read OD every 15 min. To find the doubling time for each strain, the raw data were analyzed with exponential growth equation model using GraphPad Prism (version 9.1.2). Optical density reflects total cell material, not cell number (Schaechter et al., 1958), so it is a better predictor of differences in elongation rate than septation rate.
To build the allele replacement strains, first wag31Msm under its native promotor was cloned into a kanamycin-marked L5 integrating vector and transformed into Msmeg mc2155. Then, wag31 at its native locus was replaced with hygromycin resistance cassette by dsDNA recombineering (van Kessel and Hatfull, 2007). This cassette was oriented so the hygR promoter drives expression of downstream genes in the operon. Deletion of wag31 from the genome was confirmed with PCR checks. wag31Msm at the L5 site was swapped with wag31Mtb alleles in a nourseothricin-marked L5-integrating vector using L5-phage site allelic exchange as described (Pashley and Parish, 2003). The transformants were screened by antibiotic resistance in order to confirm allele swap vectors containing wag31Mtb mutants were created using PCR stitching. All vectors were made using Gibson cloning (Gibson et al., 2009). All strains, plasmids and primers used are listed in supplemental material part (Supplementary Tables S1–S3).
Msmeg strains carrying Mycobacterial protein fragment complementation (M-PFC) assay vectors were grown to logarithmic phase and struck on 7H11 agar (BD Difco™, Thermo Fisher Scientific) with or without trimethoprim at 50 μg/ml as described (Singh et al., 2006). Plates were incubated at 37°C for 4 days, and then photographed.
Msmeg strains carrying either M-PFC or swap allele vectors were grown to logarithmic phase (OD600 = 0.5–0.7), pelleted, and lysed by bead beating (Disruptor Beads 0.1 mm from Disruptor Genie®, Mini-Beadbeater-16, Biospect). Supernatants were separated by SDS-PAGE and transferred onto polyvinyl difluoride (PVDF) membranes (GE Healthcare) and blocked with BSA for 1 h. The blot was probed with a-C-terminal-DHFR rabbit antibody (1: 1000 Sigma-Aldrich, D0942), Wag31 antibody (1:10000 Thermo Fisher custom made peptide-induced antibody, epitope (KPPIGKRGYNEDEVDAFLD)), and goat anti-rabbit IgG HRP-conjugated secondary antibody (1:10,000, Thermo Fisher Scientific 31,460) in PBST. α-Rpob was used as loading control (10:10000, Thermo Fisher Scientific, MA1-25425). All bands were quantified using Fiji and normalized to the controls.
All microscopy was performed on three biological replicate cultures of each strain using a Nikon Ti-2 widefield epifluorescence microscope with a Photometrics Prime 95B camera and a Plan Apo 100x, 1.45-numerical-aperture (NA) lens objective. Cells were taken from logarithmic phase culture in 7H9 and immobilized on 1.5% agarose pads made with Hdb media. To detect GFPmut3 and Venus signal, a filter cube with a 470/40 nm excitation filter, a 525/50 nm emission filter and a 495 nm dichroic mirror was used. To detect mCherry2B signal, a filter cube with a 560/40 nm excitation filter, a 630/70 nm emission filter and a 585 nm dichroic mirror was used. To detect HADA, a filter cube with a 350/50 nm excitation filter, a 460/50 nm emission filter, and a 400 nm dichroic mirror was used. Image analysis was performed using MicrobeJ (Ducret et al., 2016) to make cell ROIs and extract fluorescence data from them. Fluorescence intensity data from MicrobeJ was further analyzed using bespoke MATLAB code (see supplement).
Cultures were stained with 1 μg/ml HADA (R&D systems) for 15 min with rolling and incubation at 37°C. Stained cultures were then pelleted and washed in 7H9 before imaging. For the elongation assay, cells after 15 min of HADA staining were resuspended into 7H9 for 1.5 h of rolling and incubation at 37°C, then stained with 1 μg/ml NADA (R&D systems) for 2 min at room temperature, washed and resuspended in 7H9 before imaging.
ACC and PCC activities in cell-free extracts and in in vitro reconstituted complexes were measured by following the incorporation of radioactive HCO3 into acid non-volatile material (Hunaiti and Kolattukudy, 1982; Bramwell et al., 1996; Diacovich et al., 2002). The reaction mixture contained 100 mM potassium phosphate, pH 8.0, 300 μg of BSA, 3 mM ATP, 5 mM MgCl2, 50 mM NaH14CO3 (specific activity 200 μCi mmol−1 (740 kBq mmol−1)), 0.5 mM substrate (acetyl-CoA or propionyl-CoA), and 30 or 100 μg of cell-free protein extract in a total reaction volume of 100 μl. The reaction was initiated by the addition of NaH14CO3, allowed to proceed at 30°C for 15 min, and stopped with 200 μl of 6 M HCl. The contents of the tubes were then evaporated to dryness at 95°C. The residue was resuspended in 100 μl of water, 1 ml of Optiphase liquid scintillation medium (Wallac Oy) was added, and 14C radioactivity determined in a Beckman scintillation liquid counter. Nonspecific CO2 fixation by crude extracts was assayed in the absence of substrate. One unit of enzyme activity catalyzed the incorporation of 1 μmol of 14C into acid-stable products/min.
ACCase5 and 6 were reconstituted from purified components using equimolar amounts of the α (AccA3) and β (0.8 μM AccD5 or 1.6 μM AccD6) subunits. For the ACCase5 complex reconstitution AccE5 subunit was also included in a final concentration of 1.6 μM. After incubating the complexes for 15 min at room temperature, Wag31 was added in a molar ratio of 2:1 to AccA3. The activity was quantified using the ACC/PCC Assay Radioactive Method. The subunits of ACCases were purified as described (Gago et al., 2006). For Wag31 purification, the plasmid pET28-Wag31 was transformed into the E. coli strain BL21 (DE3) star, and the protein synthesis was induced by addition of 0.5 mM IPTG. H6-Wag31 was purified using a Ni Sepharose column (GE Healthcare) and aliquots containing Wag 31 were stored in presence of 0.2% Tween-20.
Depletion of Wag31 leads to arrest of polar elongation, loss of pole structure, (Kang et al., 2008; Jani et al., 2010) and delocalization of peptidoglycan and mycomembrane metabolism (Melzer et al., 2018). To dissect Wag31’s functional roles in these processes, we built Msmeg strains with wag31Mtb WT replaced by alanine mutants of wag31Mtb, using allele swapping with vectors that integrate at the L5 phage integration site (Pashley and Parish, 2003; Figure 1; Supplementary Figures S3–S6). We characterized phenotypes of allele swap mutants through growth curves, microscopy of fluorescent D-amino acid-stained cells, and a test of polar elongation. We tested the stability of the mutated Wag31 proteins by western blotting, and only characterized mutants with stable Wag31 (Figures 1C, 3C; Supplementary Figures S2, S3B). We were unable to swap some of the mutants, thus residues P2, T4, K15, and EQR 210–3 appear to be essential for Wag31 function (Supplementary Figure S4C).
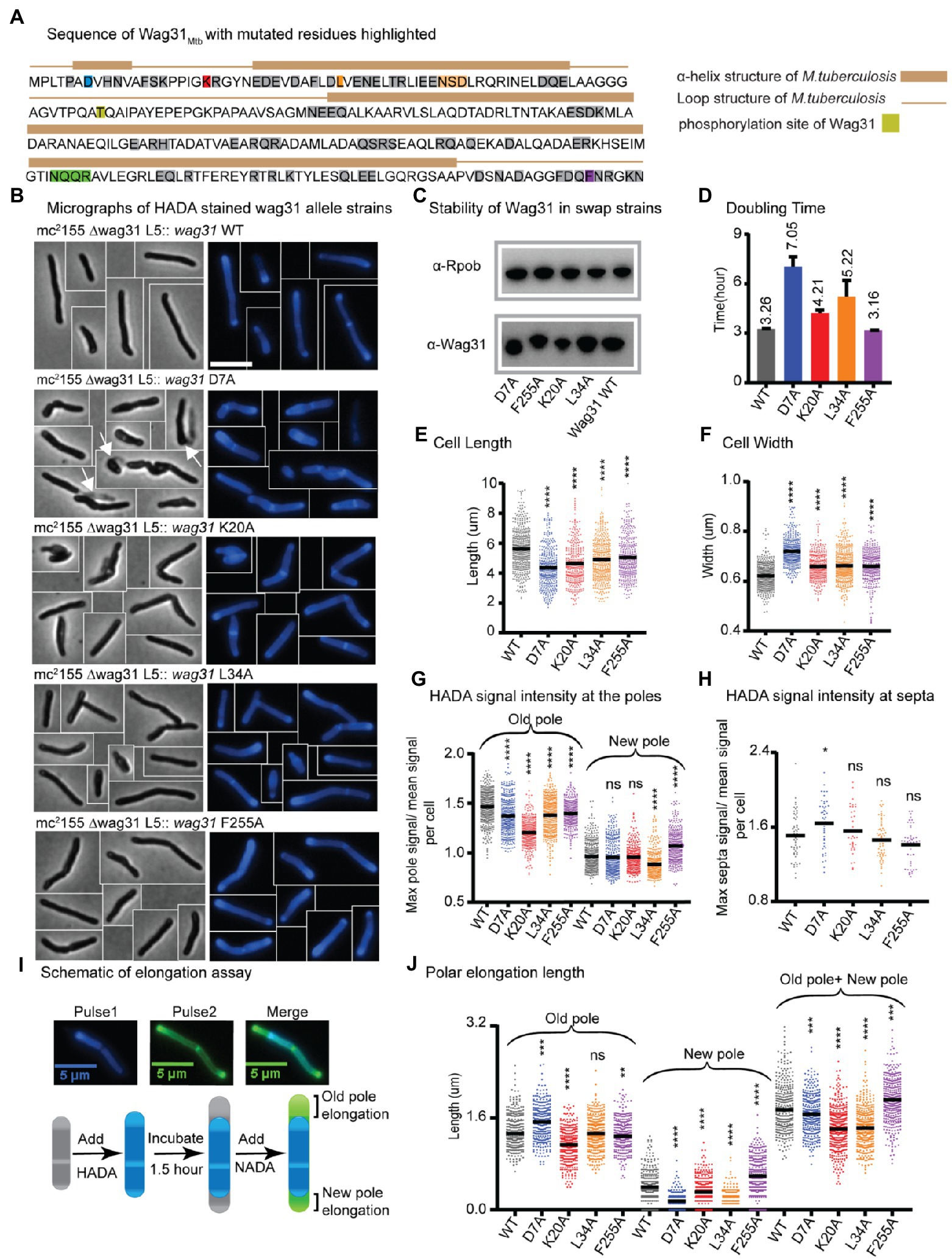
Figure 1. Wag31 has multiple distinct roles in both elongation and division. (A) Sequence of Wag31 from Mycobacterium tuberculosis. The residues mutated in the alanine scanning experiments are highlighted. (B) Phase (left) and fluorescence (right) images of Msmeg wag31 allele strains stained with HADA. The scale bar is 5 microns, and it applies to all images. Several cells from different images of the same strain are cropped together. White arrows indicate the ghosts in D7A (C) Western blots of Wag31 WT and mutant proteins in wag31 allele strains using α-Wag31 antibody. The epitope used to raise the antibodies is KPPIGKRGYNEDEVDAFLD, of which K is K20. Thus, the decreased signal seen in the K20A strain may be due to decreased association of the antibody against the mutant protein. RpoB serves as a loading control. (D) Doubling times of Msmeg cells expressing wag31 WT or alanine mutants. The means (on top of bars) are an average of three biological replicates. Error bars represent SD. (E) Cell lengths of the wag31 allele strains. Black bars are at the mean. (F) Cell widths of the wag31 allele strains. Black bars are at the mean. (G) Relative polar and septal (H) HADA intensity of wag31 allele strains. Equivalent to the maximum signal at one pole divided by the mean signal of that cell. (I) Schematic of elongation assay dual-FDAA staining method to measure elongation. The brackets indicate the measurements made. (J) Length of polar elongation in the wag31 allele strains, as measured by the elongation assay in I. Black bar is at the median. ns, p > 0.05, *p = < 0.05, **p = < 0.005, ***p = <0.0005, ****p = <0.0001. All p-values were calculated by one-way ANOVA, Dunnett’s multiple comparisons test, with comparisons of mutants to the wild-type in each case. All microscopy experiments were performed on at least 100 cells from each of three independent replicate strains.
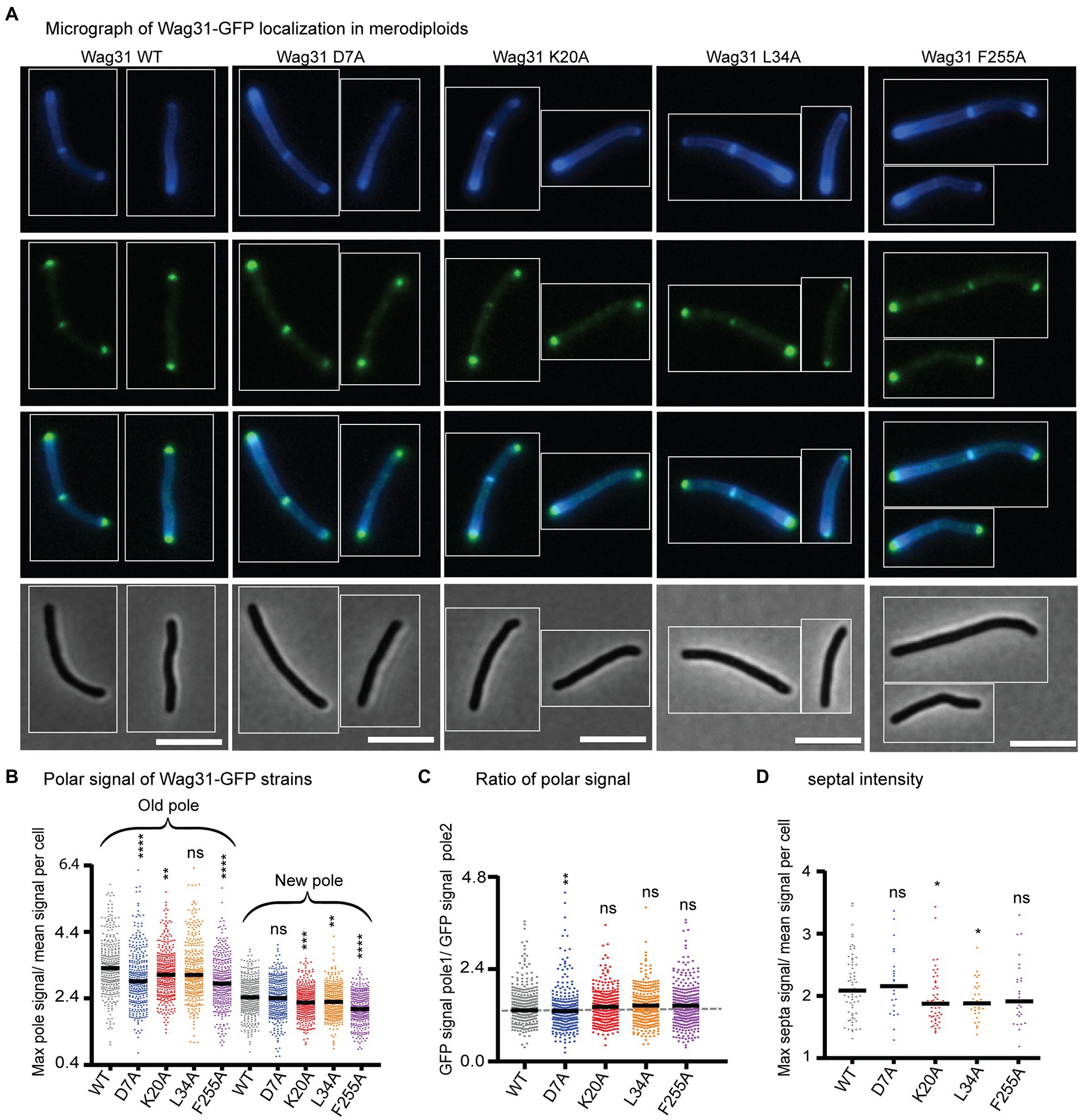
Figure 2. Wag31 mutant proteins localize normally in merodiploid strains. (A) Micrographs of WT Msmeg expressing Wag31-GFPmut3 constructs with the indicated mutations. Top: HADA; second: GFP; third: merged; bottom: phase. The scale bar is 5 microns, and it applies to all images. (B) Relative polar intensity of GFPmut3 signal from cells in A. (C) Ratio of normalized signal at the old pole over normalized signal at the new pole, from B. (D) Relative septal intensity of GFPmut3 signal from cells in A. ns, p > 0.05, *p = <0.05, **p = <0.005, ***p = <0.0005, ****p = <0.0001. All p-values (B), (C) were calculated by one-way ANOVA, Dunnett’s multiple comparisons test and the p-values (D) were calculated by the Welch’s t-test.
Most of the strains with mutations at the N-terminus of Wag31 exhibit slowed growth, while the majority of strains with mutations in the middle and C-terminus of the protein grow as fast as the strain expressing WT Wag31 (Figure 1D; Supplementary Figure S4A). Microscopy experiments show that many of the mutant cells are short and/or wide (Figures 1E,F; Supplementary Figure S4A). Most of the wag31Mtb mutant strains have higher signal than the WT after staining with the fluorescent D-alanine HADA (Kuru et al., 2012; Supplementary Figures S5A, S6A, S8, S9), which may result from de-repressed peptidoglycan metabolism or increased cell wall permeability. To compare the localization of peptidoglycan metabolism between strains, we calculated relative HADA intensity at the poles and septa which is the ratio of the maximum intensity at each region to the mean intensity of the cell. Because the older pole almost always stains more brightly by HADA (Baranowski et al., 2018); throughout we assume that the brighter pole is the old pole.
Most mutants had lower relative HADA signal than the WT at the old pole, and none have higher signal (Figure 1G; Supplementary Figure S5B). At the new pole, most mutants stain like the WT, but some are brighter and others dimmer (Figure 1H; Supplementary Figure S5C). These data indicate that mutants of Wag31 can have different impacts on the peptidoglycan metabolism at the two cell poles. Relative septal HADA intensity is increased in a few, while it is decreased in others (Supplementary Figure S6B), suggesting that Wag31 affects peptidoglycan synthesis at the septa as well. We also observe that the septal placement is shifted toward one pole or the other in some mutants (Supplementary Figure S6C).
Previous work showed cell bending when Wag31-eGFP replaces native Wag31 (Meniche et al., 2014). Here, we observed a similar curved cell morphology in the wag31 NQQR199-202AAAA mutant (Supplementary Figures S3A,C,D). Although the wag31 NQQR199-202AAAA mutant has slightly decreased relative HADA intensity at the old pole (Supplementary Figure S3F), it elongates like the Wag31 WT at both poles (Supplementary Figure S3H). We surmise that the Wag31-eGFP C-terminal fusion and these mutations may disorder the Wag31 homo-oligomer in a way that could cause cell wall insertion to be uneven around the circumference of the cell, leading to slight polar bulging and curved cells (Supplementary Figure S3).
A variety of different phenotypes indicative of assorted functional roles for Wag31 were apparent across the collection of mutants. We chose for further study four strains with mutations in conserved amino acid residues (Supplementary Figure S1), and which represent a range of phenotypes: D7A, K20A, L34A, and F255A.
The wag31 D7A strain grows slowly, and the cells are short and wide (Figures 1B, D–F). There is a slight decrease in relative HADA intensity, compared to WT, at both poles (Figure 1G), but relative HADA intensity is significantly increased at the septa (Figure 1H). We performed an elongation assay with two colors of fluorescent D-amino acids to determine if the dimmer polar HADA staining and short cell length was due to decreased elongation compared to WT. We first stained the cells with blue fluorescent HADA, then outgrew them without stain for 1.5 h, then re-stained them with the green fluorescent D-alanine NADA, and measured the length of the poles that are green and not blue (Figure 1I). Our data (Figure 1J) show that the net elongation defect (7%) does not account for the substantially decreased cell length (22%; Figure 1E), indicating that division must be initiated at a shorter cell length in this strain. The relative septal HADA staining suggests that there is more peptidoglycan metabolism at the septum than in the WT cells (Figure 1H; Supplementary Figure S6B) and that septal staining occurs in shorter cells (Supplementary Figure S8). This cell division may occur earlier, which could lead to the unusual number of ghosts in the wag31 D7A phase images (Figure 1B). These data suggest that the wag31 D7A mutation causes septation to occur in an uncontrolled manner. We therefore conclude that Wag31 has a role in controlling septation.
We localized Wag31 D7A-GFPmut3 in a merodiploid strain – because the allele swaps were not viable – and find that it localizes like the Wag31 WT-GFPmut3 at the poles and septum (Figure 2), which allows us to infer that Wag31 D7A can associate normally with WT Wag31 at the polar focus.
The N-terminus of Wag31 is highly conserved and has been structurally characterized (Oliva et al., 2010; Choukate and Chaudhuri, 2020), while the C-terminus of Wag31 is less well conserved and characterized. In DivIVA from Bacillus subtilis, the amino acid F17 is involved in interaction with the membrane at the cell pole (Oliva et al., 2010). In Msmeg, K20 is in a comparable position as F17 in DivIVABsub in the structure (Choukate and Chaudhuri, 2020).
We found that mutations in two N-terminal residues, K20 and L34, and a C-terminal residue F255 of Wag31 lead to mis-regulation of polar growth (Figure 1). Point mutations in K20A and L34A cause defects in growth rate, while F255A does not (Figure 1D). All three mutants have short cell lengths (Figures 1B,E).
To determine whether the short cells result from decreased elongation or activated septation, we performed elongation assays (Figures 1I,J). We found that the defect in the wag31 K20A elongation (22% less than WT) is similar to the defect in cell length (18%), indicating that this mutant is defective in elongation, especially at the old pole (Figure 1J). The wag31 K20A mutant also has decreased HADA staining at the old pole, but equivalent staining at the new pole, compared to WT (Figure 1G), which corroborates the elongation data indicating that this mutant is specifically defective in old-pole elongation. The wag31 K20A strain has similar relative septal HADA intensity as the WT strain (Figure 1H); however, the septal location is shifted slightly toward the old pole (Supplementary Figure S6C), likely because the old pole is elongating more slowly.
The old pole of the wag31 L34A strain elongates the same as the WT strain, but the new pole has ~76% less elongation (Figure 1J). In the population of wag31 L34A mutants that we characterized, ~65% had no observable elongation at the new pole, while only 11% of cells in the wag31 WT population had no new pole elongation in the 1.5 h assay. These assays are performed in unsychronized populations of cells, so the new poles observed are at different stages in their development. Thus, it appears that the Wag31 L34 residue contributes to both initiation and extension of the new pole. The short cell length and slow growth rate (Figures 1B, D,E) are likely due to this defect in new pole elongation, as we did not observe defects in septal HADA staining (Figure 1H; Supplementary Figure S6C).
In the wag31 F255A mutant, there is an increased relative intensity of HADA at the new pole, and less at the old pole (Figure 1G). The elongation test shows that the wag31 F255A strain elongates ~47% more than the WT at the new pole, while the old pole elongates 6% less compared to the WT strain (Figure 1J). The HADA relative septal intensity is similar in the wag31 F255A mutant compared to the WT (Figure 1H), while septal location is shifted toward the old pole, likely due to the increased elongation at the new pole (Supplementary Figure S6C). The increased elongation must be accompanied by septation at a shorter cell length to cause the short cells observed in this mutant (Figure 1E). Our data indicate that the wag31 F255A mutant is defective in inhibition of new pole synthesis, suggesting that Wag31 plays a role in this function. The data from the K20A, L34A and F255A mutants combined indicates that Wag31 has distinct roles at the two cell poles.
We localized the Wag31 K20A-GFPmut3, Wag31 L34A-GFPmut3, and Wag31 F255A-GFPmut3 in merodiploid strains. Localization of the three mutant proteins is similar to the Wag31 WT at the cell poles (Figure 2C). Localization of the K20A and L34A mutant proteins are slightly dimmer than the WT at the septum (Figure 2D) where new poles are formed during division (Santi et al., 2013). These data indicate that the Wag31 K20A and L34A mutant proteins may be slightly impaired in their ability to adhere to the polar membranes.
Polar cell wall elongation is highly regulated and is known to decrease under stress (Wu et al., 2016; Melzer et al., 2022). In other species, DivIVA homologs involved in cell morphogenesis have been shown to be regulated by phosphorylation (Flärdh, 2003; Kang, 2005; Hempel et al., 2008, 2012; Saalbach et al., 2013; Pompeo et al., 2015; Junker et al., 2018; Passot et al., 2021). Phosphorylation of Wag31 at T73 by PknA (Kang, 2005) has been proposed as a possible means of controlling polar elongation (Jani et al., 2010), as expression of the wag31 T73E (phospho-mimetic) was observed to increase vancomycin staining of Msmeg, while expression of wag31 T73A (phospho-ablative) decreased staining (Jani et al., 2010).
To further test this hypothesis, we built the wag31 T73A and wag31 T73E strains through allele swapping with the native promoter, in order to control for protein levels, (Figure 3). We found that the Wag31 phospho-mutant proteins are both as stable as the WT (Figure 3C). The mutant strains have similar growth rates compared with the WT (Figure 3D) which indicates that phospho-site T73 does not have a significant role in regulating cell growth. However, we do observe some very subtle effects of these mutations on cell wall metabolism. Both mutants are a little longer (Figure 3E) and the wag31 T73E strain has slightly more HADA staining at the old pole, while both strains have similar relative HADA signal to the WT at the new pole (Figure 3G). The elongation assay showed that the wag31 T73E strain elongates ~9% more and the wag31 T73A strain elongates ~8% less than the WT at the old pole. We did not observe significant differences in elongation at the new pole (Figure 3H). The increased cell length (Figure 3E) and slightly decreased in old pole elongation of the wag31 T73A strain (Figure 3H) suggest that this mutant also activates division at longer cell lengths.
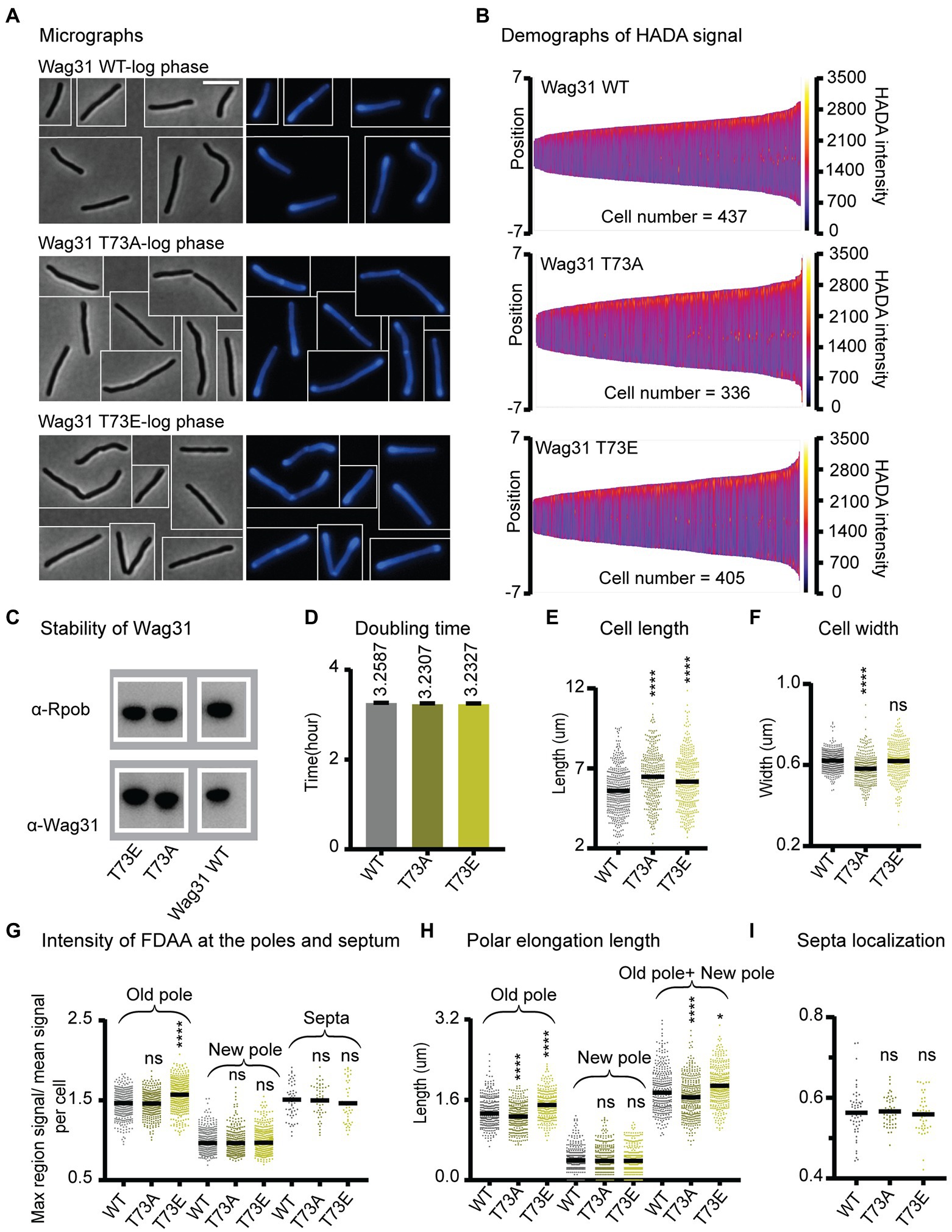
Figure 3. Phosphorylation of T73 on Wag31 subtly affects the cell wall metabolism, but does not affect growth rate. (A) Phase (left) and fluorescence (right) images of Msmeg wag31 allele strains stained with HADA. The scale bar is 5 microns, and it applies to all images. (B) Demographs of HADA intensity (color scale) across the length of the cell (Y axis) of the wag31 allele strains. The cells were sorted by length, with shortest cells on the left and longest on the right of each demograph. Cells were also pole-sorted according to HADA intensity, such that the brighter pole (presumed to be the old pole) was oriented to the top along the Y axis. At least 100 cells were analyzed from each of three independent biological replicates of each strain. (C) Western blots of Wag31 WT, T73A, and T73E proteins in wag31 allele strains using α-Wag31 antibody. RpoB serves as a loading control. (D) Doubling times of Msmeg cells expressing WT or wag31 mutants. The means (on top of bars) are an average of three biological replicates. Error bars represent SD. (E) Cell lengths of the wag31 allele strains. Black bars are at the mean. (F) Cell widths of the wag31 allele strains. Black bars are at the mean. (G) Relative polar and septal HADA intensity of wag31 allele strains. Relative intensity is equal to the maximum signal at a cell pole, divided by the mean signal of that cell. (H) Length of polar elongation in the wag31 allele strains, as measured by the elongation assay method diagrammed in Figure 1I. (I) Septal location in wag31 allele strains. Black bar is at the median. ns, p > 0.05, ****p = < 0.0001. All p-values were calculated by one-way ANOVA, Dunnett’s multiple comparisons test.
In order to determine whether phosphorylation of Wag31 might regulate cell wall metabolism under stress, we performed HADA staining and microscopy of the same strains in stationary phase (Supplementary Figure S7A). The results show that these phospho-mutants of Wag31 have the same cell length as the WT (Supplementary Figure S7C), indicating that phosphorylation on Wag31 is not required for regulating cell length and polar growth in stationary phase. We found slight differences in HADA staining at the old pole between the mutants and the WT, which are curiously in the opposite direction as in logarithmic phase, with the wag31 T73A phospho-ablative mutant staining more brightly and the phospho-mimetic mutant staining more dimly (Supplementary Figure S7F). This suggests that phosphorylation on Wag31 may have some minor role in regulating cell wall metabolism in stress, but that it is likely downstream of other regulators. The wag31 T73A mutant has a higher percentage of cells with active septa in stationary phase (Supplementary Figures S7E,F), indicating that the function of unphosphorylated Wag31 in slowing septation may continue into stationary phase, but that this regulation is not sufficient to affect cell length.
Recent work suggests that Wag31 is a favored substrate of the Serine Threonine Phosphatase PstP, which localizes to the septum during late division (Shamma et al., 2022). This suggests that Wag31 is dephosphorylated at the septum and may primarily be phosphorylated at the poles. It may be that the phosphorylation only accumulates sufficiently to affect peptidoglycan metabolism once the pole has matured into an old pole, since there are no differences in HADA staining between the WT and phospho-mutants at the new pole (Figures 3G,I). In summation, our data with the phospho-mutants indicates that phosphorylation on T73 of Wag31 has only very minor effects on polar growth at the old pole, while the unphosphorylated form may slightly inhibit both polar elongation and septation. However, as these effects are not enough to impact cell growth rates (Figure 3D), we conclude that phosphorylation on T73 is not an important regulator of cell wall metabolism under the conditions tested.
Wag31 has been shown to interact with AccA3 (Meniche et al., 2014; Xu et al., 2014), a member of the essential ACCase complexes that catalyze the first committed step of the synthesis of the precursors of all fatty acids, mycolic acids, and other methyl-branched lipids (Gago et al., 2011). We hypothesized that this interaction might allow Wag31 to localize and/or regulate the activity of the ACCase complexes, and might therefore affect polar elongation. To test this, we measured ACCase activity in the WT and two of the mutants with defects in polar elongation, wag31 K20A and wag31 L34A, as well as two mutants with little effect on polar elongation, the wag31 T73A and T73E mutants.
The ACCase activity was measured by the incorporation of radioactive sodium hydrogen carbonate into the non-volatile fraction of cell extracts with the addition of either acetyl-CoA (to assay ACC activity) or propionyl-CoA (to assay PCC activity) as the enzyme substrates. As shown in Figure 4A, lysates from the wag31 K20A and wag31 L34A mutants show a decrease in ACC activity compared with the WT strain, while PCC activity was only slightly modified in the K20A mutant (Figure 4B). As a metabolic control, we also assayed the NADPH dependent malic enzyme activity in the three strains, and found no differences in their levels (Figure 4C), suggesting that the differences in ACC activities was not due to a pleiotropic effect on cell metabolism resulting from the growth defect of the mutants.
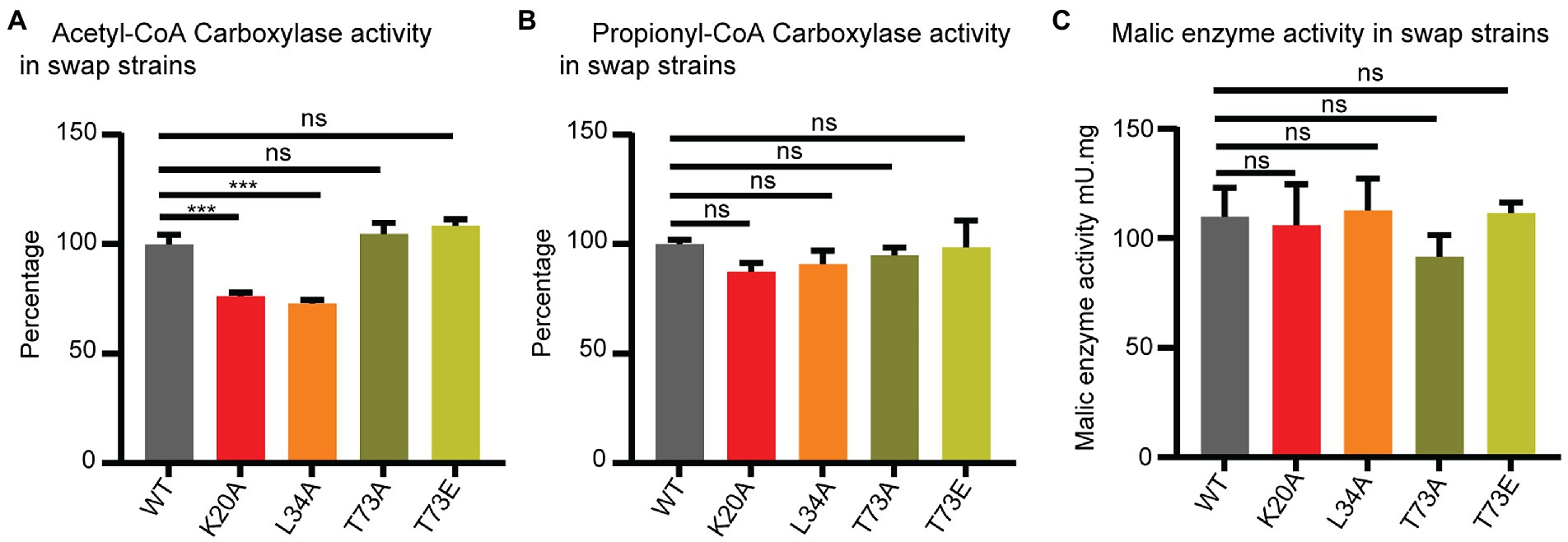
Figure 4. Wag31 modulates ACC activity. ACC (A), PCC (B) and malic enzyme (C) activity in cell-free extracts of Msmeg expressing WT or wag31 mutants. Results are the means of three independent experiments ± standard deviations (n = 3). ns, p > 0.05, ***p = <0.0005. All p-values were calculated by Tukey’s multiple comparison test.
The ACC and PCC activities in the cell lysates from the wag31 T73A and wag31 T73E strains did not show changes in either of the ACCases or malic enzyme activities.
From these results we conclude that Wag31 impacts the activity of these essential ACCases, either directly or indirectly. The phosphorylation state of Wag31 does not appear to affect the activity of the ACCase enzymes.
In order to assess whether native Wag31 has a direct effect on the activities of ACCases 5 and/or 6, we reconstituted the two enzyme complexes in vitro and assayed their activities with their preferred substrates, acetyl-CoA or propionyl-CoA, respectively, and in the presence and absence of Wag31. We found that Wag31 reduces ACCase6 activity ~20% (Supplementary Figure S10A). We also tested the ACCase5 complex, and found that Wag31 does not affect its PCCase activity (Supplementary Figure S10B). These results confirm that at least in vitro, native Wag31 modulates ACCase6 activity and suggests that the Wag31 K20A and Wag31 L34A mutants might have a stronger modulatory effect in vivo compared with the wild type protein.
Recent work suggests that localization of the enzymes that synthesize cell wall precursors to the subpolar Intracellular Membrane Domain (IMD) may be critical in restricting elongation to the pole (Hayashi et al., 2016; Melzer et al., 2022). Depletion of Wag31 has been shown to delocalize the IMD (García-Heredia et al., 2021), though it is not known if Wag31 regulates the IMD, or if the localization of IMD-associated enzymes is merely dependent on an intact cell pole.
To probe whether Wag31 regulates IMD structure, we localized both GlfT2-mcherry2B and MurG-Venus in a subset of wag31 alanine-mutant strains (Figure 5; Supplementary Figure S3). GlfT2 is the last cytoplasmic galactan enzyme in arabinogalactan synthesis (Beláňová et al., 2008), associates with the IMD, and localizes in the typical pattern of IMD-associated proteins (Hayashi et al., 2016) in the subpolar region and spottily along the lateral walls. MurG is the final cytoplasmic enzyme in peptidoglycan precursor synthesis, has a similar localization pattern as GlfT2, (Hayashi et al., 2016) and is found in both cytoplasmic and IMD membrane fractions (García-Heredia et al., 2021).
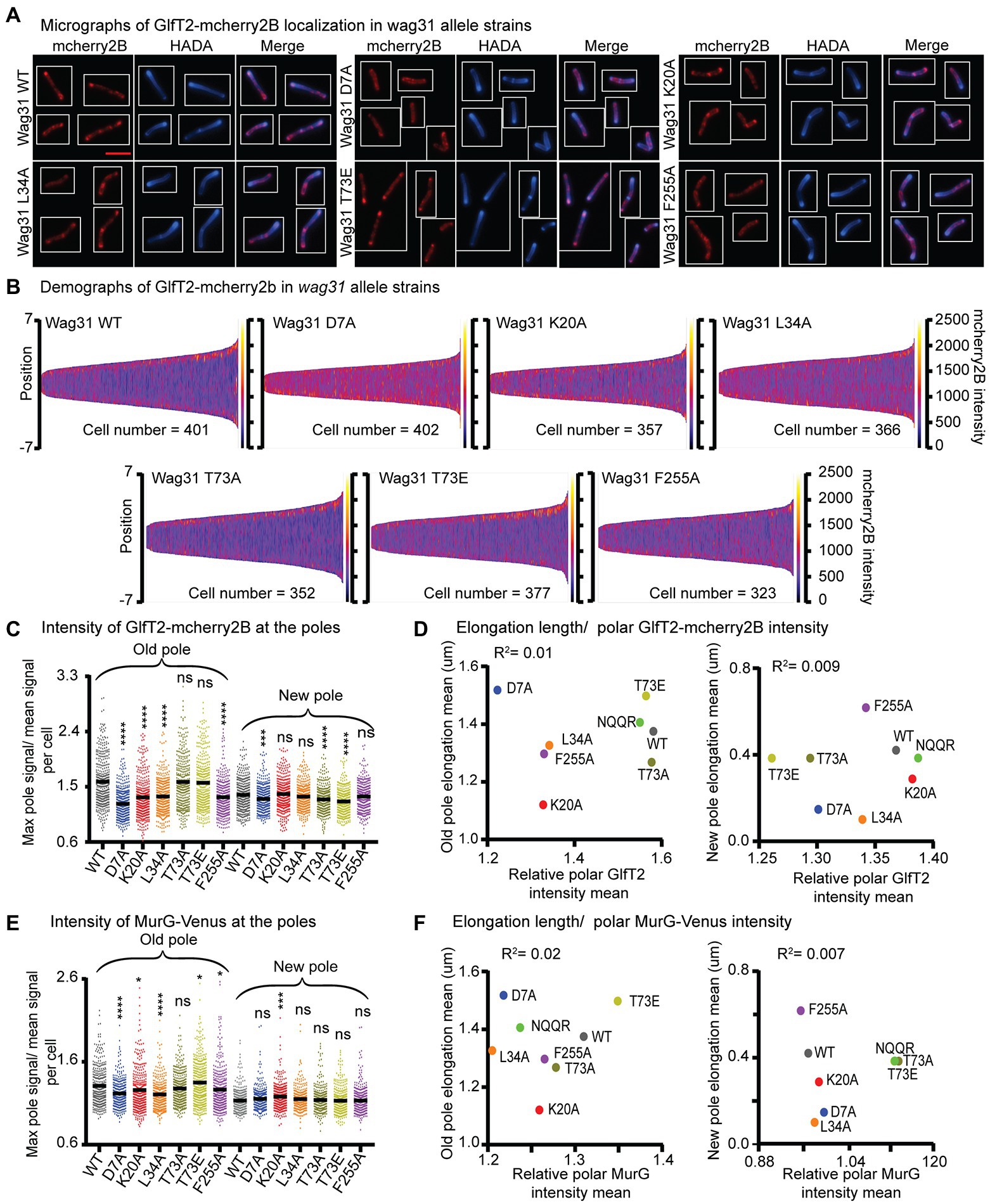
Figure 5. Localization of IMD proteins does not determine changes in polar growth. (A) Micrographs of wag31 allele strains expressing GlfT2-mcherry2B and stained with HADA. The scale bar is 5 microns, and it applies to all images. (B) Demographs of GlfT2-mcherry2B intensity across the length of each cell (Y axis of each plot, with intensity indicated by color – scale to the right of each plot), arranged by cell length (along the X axis). At least 100 cells were analyzed for each of three biological replicate cultures. (C) Relative polar intensity of GlfT2-mcherry2B in the Wag31 WT and Wag31 mutants at the old pole and the new pole. Relative intensity is equal to the maximum signal at a cell pole, divided by the mean signal of that cell. (D) Correlation between relative polar intensity of GlfT2-mcherry2B and elongation length (Figure 1J) at the poles. The eight mutants shown as colored dots. (E) Relative polar intensity of MurG-Venus in the Wag31 WT and Wag31 mutants at the old pole and the new pole. (F) Correlation between relative polar intensity of MurG-Venus and elongation length at the poles. ns, p > 0.05, *p = <0.05, ***p = <0.0005, ****p = <0.0001. p-values were calculated by one-way ANOVA, Dunnett’s multiple comparisons test.
Our microscopy data show that localization of GlfT2-mcherry2B at the old pole is significantly reduced in the Wag31 K20A, D7A, L34A and F255A mutants compared to the WT, while it was unchanged in the rest of the mutants (Figures 5A–C; Supplementary Figure S3I). At the new pole, localization of GlfT2-mcherry2B is largely unaffected, with modest defects in localization in the wag31 D7A, T73A, and T73E strains (Figures 5A–C; Supplementary Figure S3I). Localization of MurG-Venus is slightly decreased at the old pole in the Wag31 D7A and L34A mutants compared to the WT (Figure 5E; Supplementary Figure S3J), while it is unchanged in all mutants at the new pole (Figure 5E; Supplementary Figure S3J).
To test the hypothesis that the localization of IMD proteins helps determine polar growth (Mohammadi et al., 2007; Hayashi et al., 2018), we plotted the polar IMD signal of GlfT2 and MurG against the polar elongation metric for each wag31 allele strain. Across the mutants tested, polar elongation does not correlate either with GlfT2 (Figure 5D) or with MurG localization signals at the poles (Figure 5F). These data indicate that Wag31 may regulate the localization of IMD proteins, probably indirectly and largely at the old pole, but that this regulation does not directly control polar elongation.
Previous work showed that Wag31 interacts with the septal PBP3, FtsI under oxidative stress, and regulates FtsI stability through Wag31 residues 46–48 (NSD) (Mukherjee et al., 2009). FtsI is a peptidoglycan synthase with transpeptidation activity which is recruited to the septum by interacting with FtsW (Datta et al., 2006). We tested the interaction between Wag31Mtb and FtsIMtb with the Mycobacterial Protein Fragment Complementation two-hybrid assay (Singh et al., 2006). We used constructs with both the full length and cytoplasmic domain of FtsI. However, we were not able to confirm the Wag31-FtsI interaction in these conditions using this method (Figure 6A).
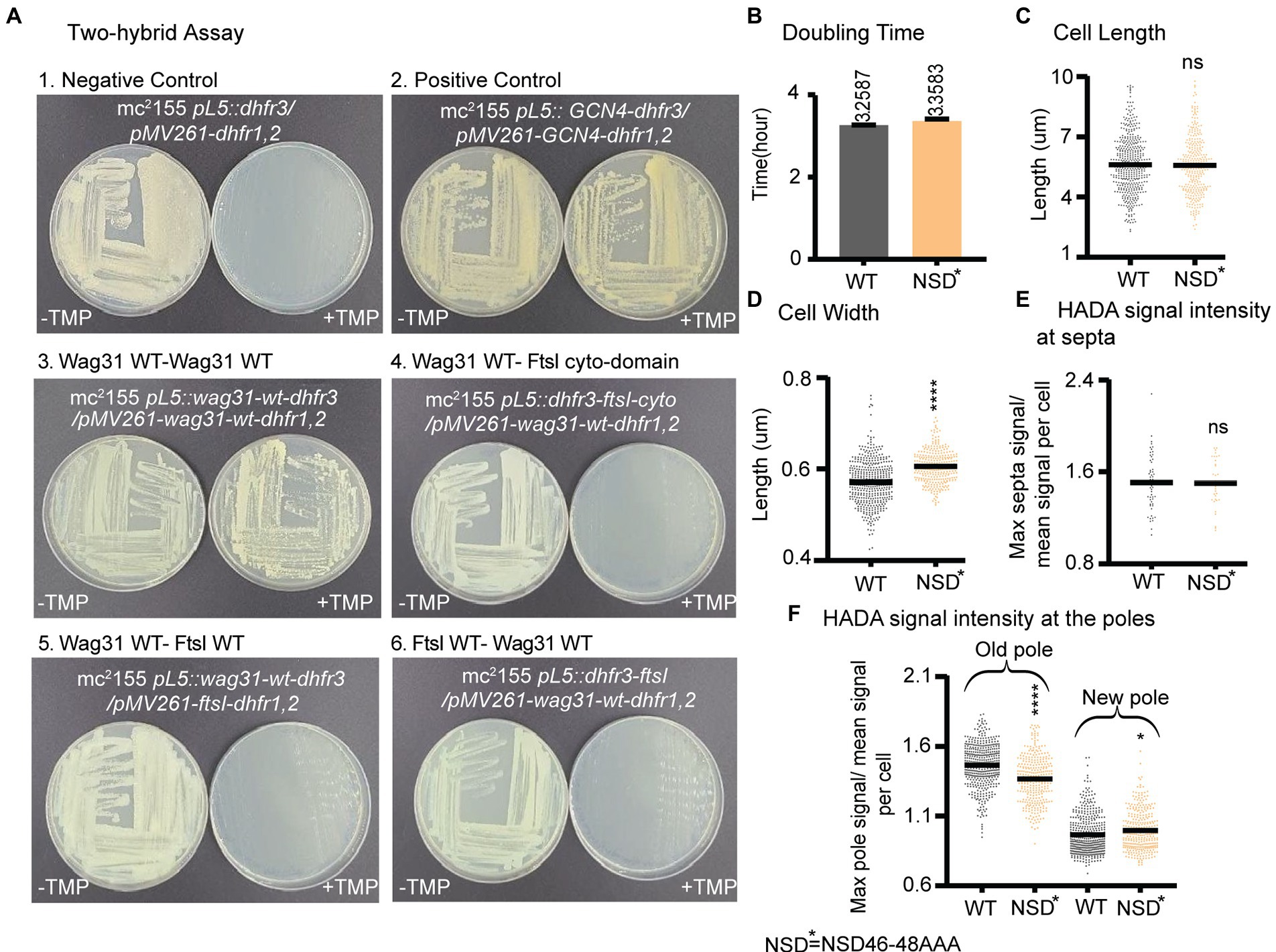
Figure 6. Wag31 does not regulate FtsI in logarithmic phase through residues NSD46-48. (A) Representative plates from the Mycobacterial Protein Fragment Complementation two-hybrid assay. Indicated strains were struck out on plates with and without trimethoprim (+TMP/ -TMP). (B) Doubling times of Msmeg cells expressing WT or wag31 NSD46-8AAA mutant. (C) Cell lengths and (D) Cell width of the wag31 NSD46-8AAA strain. Black bars are at the mean. (E) Relative septal and polar HADA (F) intensity of Wag31 WT and Wag31 NSD46-8AAA. ns, p > 0.05, *p = <0.05, ***p = <0.0005, ****p = <0.0001. p-values were calculated by unpaired t-test.
Next, we made an Msmeg allele swap strain with a wag31 ∆46–48 mutant, as per (Mukherjee et al., 2009) and found this mutant protein was unstable, while a wag31 NSD46-48AAA mutant was stable (Supplementary Figure S2). So, we used a wag31 NSD46-48AAA swap strain to test whether the Wag31-FtsI interaction, previously demonstrated in in oxidative stress (Mukherjee et al., 2009), has any role in regulating cell wall metabolism in logarithmic phase (Figure 6). We found that the wag31 NSD46-48AAA mutant has a normal cell length and growth rate (Figures 6B,C). We conclude that residues NSD46-48 of Wag31 do not affect FtsI activity in logarithmic phase (Figures 6E,F). Because we were not able to observe a protein interaction in the two-hybrid assay, we think it likely that Wag31 affects septation through another septal protein during logarithmic phase.
Wag31’s molecular function in controlling cell wall metabolism has remained poorly understood. Homology with other systems predicts that Wag31 orthologs might recruit and regulate cell wall enzymes at the pole (Marston and Errington, 1999; Sieger and Bramkamp, 2015). However, it is not clear how the hetero-oligomeric interactions that have been described for Wag31 – AccA3 (Mukherjee et al., 2009) and FtsI (Mukherjee et al., 2009) – connect to Wag31’s role in restricting peptidoglycan metabolism to the poles. In this work, we built and phenotyped a number of wag31 point mutant strains in an attempt to genetically separate Wag31’s functional roles.
We characterized a handful of wag31 point mutants which together indicate that Wag31 has distinct functions in polar elongation (Figures 7A,B). We found that the wag31 K20A mutant is specifically more defective in old pole elongation while the wag31 L34A mutant is specifically more defective in new pole elongation (Figure 1). These results indicate that Wag31 plays different roles at the new and old poles; it may have a distinct homo-oligomeric conformations at the two poles, or may form different hetero-oligomeric regulatory interactions. Both of these mutants exhibited slower growth rates, and decreased ACC activity compared to the WT (Figures 1D, 4A). The effect of Wag31 mutants on ACC activity suggests that the native conformation of Wag31 helps keep optimal levels of the ACCase complex working in the initial steps of lipid synthesis, contributing to preserve the integrity of the cell membranes (Gago et al., 2011). Modulation of the ACCase complex activities may occur through the Wag31-AccA3 interaction which has been described before (Meniche et al., 2014; Xu et al., 2014). Our data do not indicate whether Wag31 controls polar peptidoglycan synthesis by a separate pathway than that by which it regulates ACCase activity, or whether peptidoglycan synthesis is controlled in coordination with lipid synthesis. The transglycosylases that catalyze incorporation of new cell wall material are all strongly associated with the cytoplasmic membrane (Hayashi et al., 2016; Meeske et al., 2016; Arora et al., 2018), and the lipidII peptidoglycan precursor is made and transported through the membrane (Gee et al., 2012; Sham et al., 2014; Garcia-Heredia et al., 2019); thus, peptidoglycan synthesis could be regulated by the composition or protein-occupation status of the membrane, which could be regulated by Wag31’s effect on the ACCase complex.
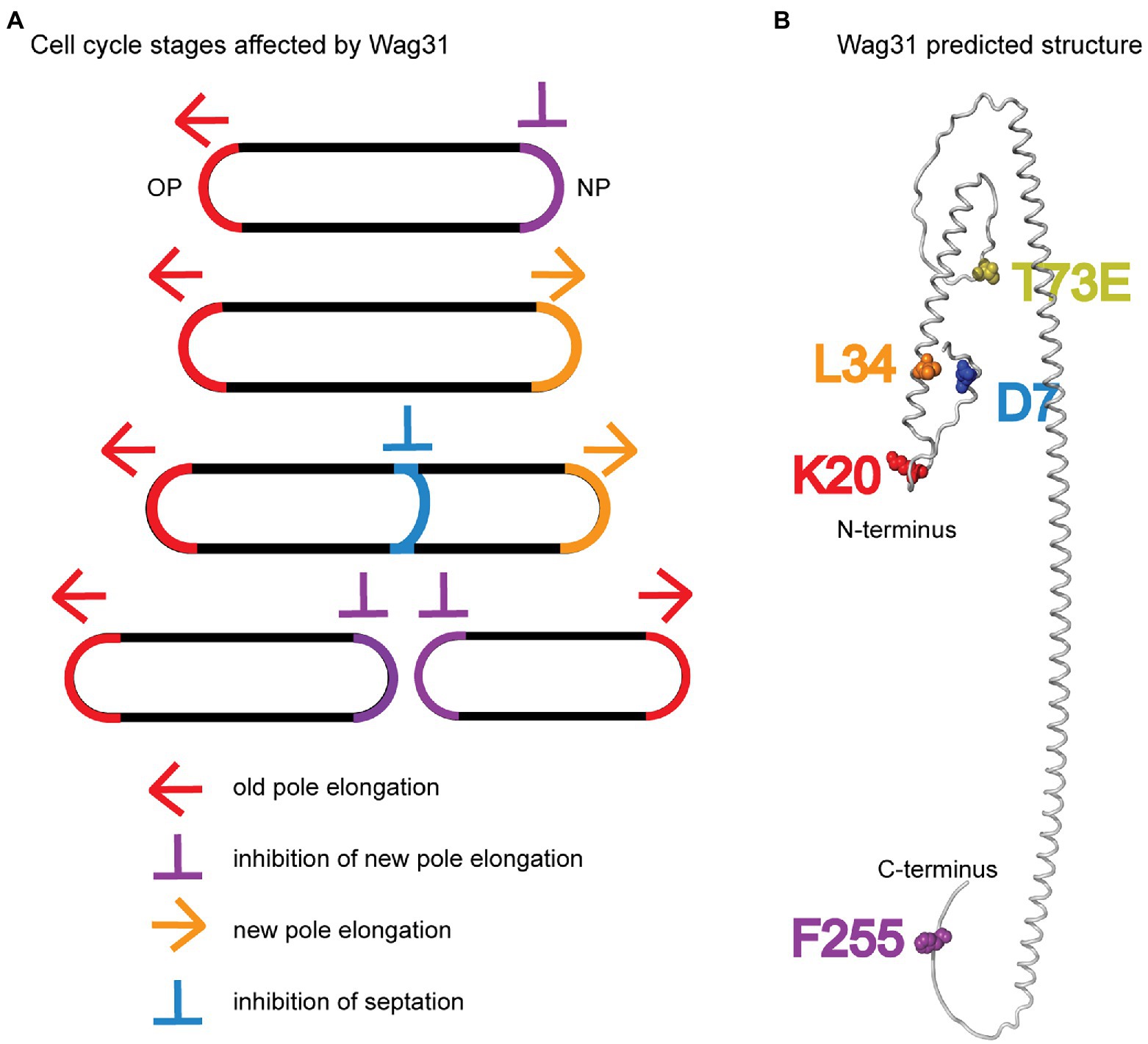
Figure 7. Models. (A) Multiple stages of the cell cycle affected by Wag31 are highlighted. (B) Alphafold2 predicted structure (Jumper et al., 2021) of a Wag31 monomer, with residues of interest highlighted.
We also found that the wag31 F255A mutant had increased polar elongation at the new pole (Figure 1). This suggests that Wag31 has a distinct role in regulating new pole elongation. LamA is a membrane protein that has previously been shown to inhibit new pole synthesis in Msmeg and which thereby contributes to cell asymmetry and heterogeneity (Rego et al., 2017). Wag31, through its C-terminus, may either work separately to inhibit new pole elongation, or may work in a pathway with LamA.
Our data suggest that Wag31 may have a minor role in controlling the timing or rate of septation. Wag31 had previously been shown to localize to the cell division site after FtsZ has left (Gupta et al., 2022) but before septation is completed (Santi et al., 2013), and overexpression of Wag31-GFP was shown to inhibit and mis-localize septa (Nguyen et al., 2007). Previous work showed that the Wag31-FtsI interaction protects FtsI from intra-membrane proteases under oxidative stress through Wag31 residues 46–48 (NSD) (Mukherjee et al., 2009). Our data show that these residues do not affect cell division in exponential phase (Figure 6). However, we do find evidence of mis-regulation of septation in the wag31 D7A strain. So, the Wag31 D7 residue could possibly establish a different interaction with FtsI, or with other septal factors (Wu et al., 2018). The phospho-site T73 on Wag31 may also have a role in regulating septation: we observed that the wag31 T73A phospho-ablative mutant has slower elongation but longer cell length in logarithmic phase (Figures 3E,G,H), which suggests that it has delayed cell division. Delayed cell division of the wag31 T73A is also apparent in stationary phase stress, as there are more cells with active peptidoglycan metabolism at the septum than in the WT strain (Supplementary Figure S7E).
Most of the mutants we characterized show a defect in GlfT2-mcherry2B and MurG-Venus localization at the old pole, but not new pole. The localization of GlfT2 and MurG at the poles did not correlate with polar elongation defects (Figure 5; Supplementary Figure S3), indicating that polar localization of the IMD proteins is not a key determinant of polar growth. These data suggest that Wag31 is at best an indirect regulator of the IMD, and that regulation of the localization of IMD proteins is likely not Wag31’s essential function.
In this work, we define new roles for Wag31 in regulating both septation and the inhibition of the new pole, and identify residues that have dominant roles in Wag31’s different functions (Figure 7). We show that Wag31 can modulate the activity of the ACCase complex and therefore may have a role in maintaining lipid homeostasis in mycobacteria (Figure 4). We also show that phosphorylation of Wag31 at T73 has very little effect on cell wall metabolism (Figure 3; Supplementary Figure S7). In addition, we show that Wag31 does not affect FtsI in logarithmic phase through the same residues used to regulate FtsI in stress (Mukherjee et al., 2009).
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
NH and CB were responsible for conceptualization, validation, and writing the first draft of the manuscript. HG, LD, and DE wrote the sections of the manuscript related to the ACCases. CB supervised the work together with HG and LD. NH, CB, HG, and LD contributed to reviewing and editing the manuscript. AE aided in data processing. NH, DE, and PP were responsible for formal analysis of the data. NH, SQ, and DE conducted the experimental work of the investigation. NH carried out all the microscopical visualization and data presentation. NH was responsible for data curation and methodology. All authors contributed to the article and approved the submitted version.
This work was supported by National Institutes of Health (NIH) grant R01AI148917 to CB.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1085918/full#supplementary-material
Arora, D., Chawla, Y., Malakar, B., Singh, A., and Nandicoori, V. K. (2018). The transpeptidase PbpA and noncanonical transglycosylase RodA of Mycobacterium tuberculosis play important roles in regulating bacterial cell lengths. J. Biol. Chem. 293, 6497–6516. doi: 10.1074/jbc.M117.811190
Baranowski, C., Welsh, M. A., Sham, L.-T., Eskandarian, H. A., Lim, H. C., Kieser, K. J., et al. (2018). Maturing Mycobacterium smegmatis peptidoglycan requires non-canonical crosslinks to maintain shape. Elife 7:e37516. doi: 10.7554/eLife.375161
Beláňová, M., Dianišková, P., Brennan, P. J., Completo, G. C., Rose, N. L., Lowary, T. L., et al. (2008). Galactosyl transferases in mycobacterial cell wall synthesis. J. Bacteriol. 190, 1141–1145. doi: 10.1128/JB.01326-07
Boes, A., Olatunji, S., Breukink, E., and Terrak, M. (2019). Regulation of the peptidoglycan polymerase activity of PBP1b by antagonist actions of the core divisome proteins FtsBLQ and FtsN. mBio 10:e01912-18. doi: 10.1128/mBio.01912-18
Bramwell, H., Hunter, L. S., Coggins, J. R., and Nimmo, H. G. (1996). Propionyl-CoA carboxylase from Streptomyces coelicolor A3(2): cloning of the gene encoding the biotinlcontaining subunit. Microbiology 142, 649–655. doi: 10.1099/13500872-142-3-649
Brown, P. J. B., de Pedro, M. A., Kysela, D. T., Van der Henst, C., Kim, J., De Bolle, X., et al. (2012). Polar growth in the Alphaproteobacterial order Rhizobiales. Proc. Natl. Acad. Sci. 109, 1697–1701. doi: 10.1073/pnas.1114476109
Cameron, T. A., Anderson-Furgeson, J., Zupan, J. R., Zik, J. J., and Zambryski, P. C. (2014). Peptidoglycan synthesis machinery in Agrobacterium tumefaciens during unipolar growth and cell division. MBio 5, e01219–e01214. doi: 10.1128/mBio.01219-14
Choukate, K., and Chaudhuri, B. (2020). Structural basis of self-assembly in the lipid-binding domain of mycobacterial polar growth factor Wag31. IUCrJ 7, 767–776. doi: 10.1107/S2052252520006053
Daniel, J., Oh, T.-J., Lee, C.-M., and Kolattukudy, P. E. (2007). AccD6, a member of the Fas II locus, is a functional carboxyltransferase subunit of the acyl-coenzyme a carboxylase in Mycobacterium tuberculosis. J. Bacteriol. 189, 911–917. doi: 10.1128/JB.01019-06
Datta, P., Dasgupta, A., Singh, A. K., Mukherjee, P., Kundu, M., and Basu, J. (2006). Interaction between FtsW and penicillin-binding protein 3 (PBP3) directs PBP3 to mid-cell, controls cell septation and mediates the formation of a trimeric complex involving FtsZ, FtsW and PBP3 in mycobacteria. Mol. Microbiol. 62, 1655–1673. doi: 10.1111/j.1365-2958.2006.05491.x
Diacovich, L., Peirú, S., Kurth, D., Rodriguez, E., Podestá, F., Khosla, C., et al. (2002). Kinetic and structural analysis of a new Group of Acyl-CoA carboxylases found in Streptomyces coelicolor A3(2). J. Biol. Chem. 277, 31228–31236. doi: 10.1074/jbc.M203263200
Ducret, A., Quardokus, E. M., and Brun, Y. V. (2016). MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 1:16077. doi: 10.1038/nmicrobiol.2016.77
Egan, A. J. F., Errington, J., and Vollmer, W. (2020). Regulation of peptidoglycan synthesis and remodelling. Nat. Rev. Microbiol. 18, 446–460. doi: 10.1038/s41579-020-0366-3
Eswaramoorthy, P., Winter, P. W., Wawrzusin, P., York, A. G., Shroff, H., and Ramamurthi, K. S. (2014). Asymmetric division and differential gene expression during a bacterial developmental program requires DivIVA. PLoS Genet. 10:e1004526. doi: 10.1371/journal.pgen.1004526
Flärdh, K. (2003). Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2): DivIVA and tip growth in Streptomyces. Mol. Microbiol. 49, 1523–1536. doi: 10.1046/j.1365-2958.2003.03660.x
Gago, G., Arabolaza, A., Diacovich, L., and Gramajo, H. (2018). “Components and key regulatory steps of lipid biosynthesis in Actinomycetes” in Biogenesis of Fatty Acids, Lipids and Membranes. ed. O. Geiger (Cham: Springer International Publishing), 1–25.
Gago, G., Diacovich, L., Arabolaza, A., Tsai, S.-C., and Gramajo, H. (2011). Fatty acid biosynthesis in actinomycetes. FEMS Microbiol. Rev. 35, 475–497. doi: 10.1111/j.1574-6976.2010.00259.x
Gago, G., Kurth, D., Diacovich, L., Tsai, S.-C., and Gramajo, H. (2006). Biochemical and structural characterization of an essential acyl coenzyme a carboxylase from Mycobacterium tuberculosis. J. Bacteriol. 188, 477–486. doi: 10.1128/JB.188.2.477-486.2006
García-Heredia, A., Kado, T., Sein, C. E., Puffal, J., Osman, S. H., Judd, J., et al. (2021). Membrane-partitioned cell wall synthesis in mycobacteria. Elife 10:e60263. doi: 10.7554/eLife.60263
Garcia-Heredia, A., Puffal, J., Julius Judd, J., Gray, T. A., Morita, Y. S., and Siegrist, S. (2019). Membrane compartmentalization of mycobacterial peptidoglycan synthesis. bioRxiv. doi: 10.1101/544338
Gee, C. L., Papavinasasundaram, K. G., Blair, S. R., Baer, C. E., Falick, A. M., King, D. S., et al. (2012). A phosphorylated pseudokinase complex controls cell wall synthesis in mycobacteria. Sci. Signal. 5:ra7. doi: 10.1126/scisignal.2002525
Gibson, D. G., Young, L., Chuang, R.-Y., Venter, J. C., Hutchison, C. A., and Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. doi: 10.1038/nmeth.1318
Gupta, K. R., Gwin, C. M., Rahlwes, K. C., Biegas, K. J., Wang, C., Park, J. H., et al. (2022). An essential periplasmic protein coordinates lipid trafficking and is required for asymmetric polar growth in mycobacteria. Elife 11:e80395. doi: 10.7554/eLife.80395
Hannebelle, M. T. M., Ven, J. X. Y., Toniolo, C., Eskandarian, H. A., Vuaridel-Thurre, G., McKinney, J. D., et al. (2020). A biphasic growth model for cell pole elongation in mycobacteria. Nat. Commun. 11:452. doi: 10.1038/s41467-019-14088-z
Hayashi, J. M., Luo, C.-Y., Mayfield, J. A., Hsu, T., Fukuda, T., Walfield, A. L., et al. (2016). Spatially distinct and metabolically active membrane domain in mycobacteria. PNAS 113, 5400–5405. doi: 10.1073/pnas.1525165113
Hayashi, J. M., Richardson, K., Melzer, E. S., Sandler, S. J., Aldridge, B. B., Siegrist, M. S., et al. (2018). Stress-induced reorganization of the mycobacterial membrane domain. MBio 9, e01823–e01817. doi: 10.1128/mBio.01823-17
Hempel, A. M., Cantlay, S., Molle, V., Wang, S.-B., Naldrett, M. J., Parker, J. L., et al. (2012). The Ser/Thr protein kinase Afs K regulates polar growth and hyphal branching in the filamentous bacteria Streptomyces. Proc. Natl. Acad. Sci. 109, E2371–E2379. doi: 10.1073/pnas.1207409109
Hempel, A. M., Wang, S. B., Letek, M., Gil, J. A., and Flardh, K. (2008). Assemblies of DivIVA mark sites for Hyphal branching and can establish new zones of Cell Wall growth in Streptomyces coelicolor. J. Bacteriol. 190, 7579–7583. doi: 10.1128/JB.00839-08
Hunaiti, A. R., and Kolattukudy, P. E. (1982). Isolation and characterization of an acyl-coenzyme a carboxylase from an erythromycin-producing Streptomyces erythreus. Arch. Biochem. Biophys. 216, 362–371. doi: 10.1016/0003-9861(82)90222-3
Jani, C., Eoh, H., Lee, J., Hamasha, K., Sahana, M., Han, J.-S., et al. (2010). Regulation of polar peptidoglycan biosynthesis by Wag31 phosphorylation in mycobacteria. BMC Microbiol. 10:327. doi: 10.1186/1471-2180-10-327
Joyce, G., Williams, K. J., Robb, M., Noens, E., Tizzano, B., Shahrezaei, V., et al. (2012). Cell division site placement and asymmetric growth in mycobacteria. PLoS One 7:e44582. doi: 10.1371/journal.pone.0044582
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Junker, S., Maaβ, S., Otto, A., Michalik, S., Morgenroth, F., Gerth, U., et al. (2018). Spectral library based analysis of arginine phosphorylations in Staphylococcus aureus. Mol. Cell. Proteomics 17, 335–348. doi: 10.1074/mcp.RA117.000378
Kang, C.-M. (2005). The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 19, 1692–1704. doi: 10.1101/gad.1311105
Kang, C.-M., Nyayapathy, S., Lee, J.-Y., Suh, J.-W., and Husson, R. N. (2008). Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology 154, 725–735. doi: 10.1099/mic.0.2007/014076-0
Kuru, E., Hughes, H. V., Brown, P. J., Hall, E., Tekkam, S., Cava, F., et al. (2012). In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-AMINO acids. Angew. Chem. Int. Ed. 51, 12519–12523. doi: 10.1002/anie.201206749
Marston, A. L., and Errington, J. (1999). Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol. Microbiol. 33, 84–96. doi: 10.1046/j.1365-2958.1999.01450.x
Meeske, A. J., Riley, E. P., Robins, W. P., Uehara, T., Mekalanos, J. J., Kahne, D., et al. (2016). SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537, 634–638. doi: 10.1038/nature19331
Melzer, E. S., Kado, T., García-Heredia, A., Gupta, K. R., Meniche, X., Morita, Y. S., et al. (2022). Cell Wall damage reveals spatial flexibility in peptidoglycan synthesis and a nonredundant role for RodA in mycobacteria. J. Bacteriol. 204:e0054021. doi: 10.1128/jb.00540-21
Melzer, E. S., Sein, C. E., Chambers, J. J., and Sloan Siegrist, M. (2018). DivIVA concentrates mycobacterial cell envelope assembly for initiation and stabilization of polar growth. Cytoskeleton 75, 498–507. doi: 10.1002/cm.21490
Meniche, X., Otten, R., Siegrist, M. S., Baer, C. E., Murphy, K. C., Bertozzi, C. R., et al. (2014). Subpolar addition of new cell wall is directed by DivIVA in mycobacteria. Proc. Natl. Acad. Sci. 111, E3243–E3251. doi: 10.1073/pnas.1402158111
Mohammadi, T., Karczmarek, A., Crouvoisier, M., Bouhss, A., Mengin-Lecreulx, D., and den Blaauwen, T. (2007). The essential peptidoglycan glycosyltransferase MurG forms a complex with proteins involved in lateral envelope growth as well as with proteins involved in cell division in Escherichia coli. Mol. Microbiol. 65, 1106–1121. doi: 10.1111/j.1365-2958.2007.05851.x
Mukherjee, P., Sureka, K., Datta, P., Hossain, T., Barik, S., Das, K. P., et al. (2009). Novel role of Wag31 in protection of mycobacteria under oxidative stress. Mol. Microbiol. 73, 103–119. doi: 10.1111/j.1365-2958.2009.06750.x
Nguyen, L., Scherr, N., Gatfield, J., Walburger, A., Pieters, J., and Thompson, C. J. (2007). Antigen 84, an effector of Pleiomorphism in Mycobacterium smegmatis. J. Bacteriol. 189, 7896–7910. doi: 10.1128/JB.00726-07
Oliva, M. A., Halbedel, S., Freund, S. M., Dutow, P., Leonard, T. A., Veprintsev, D. B., et al. (2010). Features critical for membrane binding revealed by DivIVA crystal structure. EMBO J. 29, 1988–2001. doi: 10.1038/emboj.2010.99
Pashley, C. A., and Parish, T. (2003). Efficient switching of mycobacteriophage L5-based integrating plasmids in Mycobacterium tuberculosis. FEMS Microbiol. Lett. 229, 211–215. doi: 10.1016/S0378-1097(03)00823-1
Passot, F. M., Cantlay, S., and Flärdh, K. (2021). Protein phosphatase SppA regulates apical growth and dephosphorylates cell polarity determinant DivIVA in Streptomyces coelicolor. Mol. Microbiol. 117, 411–428. doi: 10.1111/mmi.14856
Pióro, M., Małecki, T., Portas, M., Magierowska, I., Trojanowski, D., Sherratt, D., et al. (2018). Competition between DivIVA and the nucleoid for ParA binding promotes segrosome separation and modulates mycobacterial cell elongation. Mol. Microbiol. 111, 204–220. doi: 10.1111/mmi.14149
Pompeo, F., Foulquier, E., Serrano, B., Grangeasse, C., and Galinier, A. (2015). Phosphorylation of the cell division protein GpsB regulates PrkC kinase activity through a negative feedback loop in Bacillus subtilis: regulation of PrkC activity by GpsB. Mol. Microbiol. 97, 139–150. doi: 10.1111/mmi.13015
Rego, E. H., Audette, R. E., and Rubin, E. J. (2017). Deletion of a mycobacterial divisome factor collapses single-cell phenotypic heterogeneity. Nature 546, 153–157. doi: 10.1038/nature22361
Saalbach, G., Hempel, A. M., Vigouroux, M., Flärdh, K., Buttner, M. J., and Naldrett, M. J. (2013). Determination of phosphorylation sites in the DivIVA cytoskeletal protein of Streptomyces coelicolor by targeted LC–MS/MS. J. Proteome Res. 12, 4187–4192. doi: 10.1021/pr400524d
Santi, I., Dhar, N., Bousbaine, D., Wakamoto, Y., and McKinney, J. D. (2013). Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria. Nat. Commun. 4:2470. doi: 10.1038/ncomms3470
Schaechter, M., MaalØe, O., and Kjeldgaard, N. O. (1958). Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella Typhimurium. Microbiology 19, 592–606. doi: 10.1099/00221287-19-3-592
Sham, L.-T., Butler, E. K., Lebar, M. D., Kahne, D., Bernhardt, T. G., and Ruiz, N. (2014). MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science 345, 220–222. doi: 10.1126/science.1254522
Shamma, F., Rego, E. H., and Boutte, C. C. (2022). Mycobacterial serine/threonine phosphatase PSTP is phosphoregulated and localized to mediate control of cell wall metabolism. Mol. Microbiol. 118, 47–60. doi: 10.1111/mmi.14951
Sher, J. W., Lim, H. C., and Bernhardt, T. G. (2021). Polar growth in Corynebacterium glutamicum has a flexible Cell Wall synthase requirement. MBio 12, e00682–e00621. doi: 10.1128/mBio.00682-21
Sieger, B., and Bramkamp, M. (2015). Interaction sites of DivIVA and RodA from Corynebacterium glutamicum. Front. Microbiol. 5:738. doi: 10.3389/fmicb.2014.00738
Singh, A., Mai, D., Kumar, A., and Steyn, A. J. C. (2006). Dissecting virulence pathways of Mycobacterium tuberculosis through protein-protein association. Proc. Natl. Acad. Sci. 103, 11346–11351. doi: 10.1073/pnas.0602817103
Thanky, N. R., Young, D. B., and Robertson, B. D. (2007). Unusual features of the cell cycle in mycobacteria: polar-restricted growth and the snapping-model of cell division. Tuberculosis 87, 231–236. doi: 10.1016/j.tube.2006.10.004
Valbuena, N., Letek, M., Ordóñez, E., Ayala, J., Daniel, R. A., Gil, J. A., et al. (2007). Characterization of HMW-PBPs from the rod-shaped actinomycete Corynebacterium glutamicum: peptidoglycan synthesis in cells lacking actin-like cytoskeletal structures. Mol. Microbiol. 66, 643–657. doi: 10.1111/j.1365-2958.2007.05943.x
van Baarle, S., Celik, I. N., Kaval, K. G., Bramkamp, M., Hamoen, L. W., and Halbedel, S. (2013). Protein-protein interaction domains of Bacillus subtilis DivIVA. J. Bacteriol. 195, 1012–1021. doi: 10.1128/JB.02171-12
van Kessel, J. C., and Hatfull, G. F. (2007). Recombineering in Mycobacterium tuberculosis. Nat. Methods 4, 147–152. doi: 10.1038/nmeth996
Wu, M.-L., Gengenbacher, M., and Dick, T. (2016). Mild nutrient starvation triggers the development of a small-cell survival Morphotype in mycobacteria. Front. Microbiol. 7:947. doi: 10.3389/fmicb.2016.00947
Wu, K. J., Zhang, J., Baranowski, C., Leung, V., Rego, E. H., Morita, Y. S., et al. (2018). Characterization of conserved and novel Septal factors in Mycobacterium smegmatis. J. Bacteriol. 200:e00649-17, /jb/200/6/e00649-17.atom. doi: 10.1128/JB.00649-17
Xu, W., Zhang, L., Mai, J., Peng, R., Yang, E., Peng, C., et al. (2014). The Wag31 protein interacts with AccA3 and coordinates cell wall lipid permeability and lipophilic drug resistance in Mycobacterium smegmatis. Biochem. Biophys. Res. Commun. 448, 255–260. doi: 10.1016/j.bbrc.2014.04.116
Keywords: Mycobacterium, polar growth regulation, petidoglycan synthesis, DivIVA, intracellular membrane domain (IMD), MurG, ACCase complex, FtsI
Citation: Habibi Arejan N, Ensinck D, Diacovich L, Patel PB, Quintanilla SY, Emami Saleh A, Gramajo H and Boutte CC (2023) Polar protein Wag31 both activates and inhibits cell wall metabolism at the poles and septum. Front. Microbiol. 13:1085918. doi: 10.3389/fmicb.2022.1085918
Received: 31 October 2022; Accepted: 20 December 2022;
Published: 12 January 2023.
Edited by:
Hari S. Misra, Bhabha Atomic Research Center (BARC), IndiaReviewed by:
Neeraj Dhar, International Vaccine Center (VIDO-InterVac), CanadaCopyright © 2023 Habibi Arejan, Ensinck, Diacovich, Patel, Quintanilla, Emami Saleh, Gramajo and Boutte. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cara C. Boutte,  Y2FyYS5ib3V0dGVAdXRhLmVkdQ==
Y2FyYS5ib3V0dGVAdXRhLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.