- 1Key Laboratory of Urban Environment and Health, Ningbo Urban Environment Observation and Research Station, Institute of Urban Environment, Chinese Academy of Sciences, Xiamen, China
- 2Zhejiang Key Laboratory of Urban Environmental Processes and Pollution Control, CAS Haixi Industrial Technology Innovation Center in Beilun, Ningbo, China
- 3Engineering Laboratory for AgroBiomass Recycling and Valorizing, College of Engineering, China Agricultural University, Beijing, China
- 4School of Water Resources and Environment, China University of Geosciences (Beijing), Beijing, China
The environmental accumulation and spread of antibiotic resistance pose a major threat to global health. Aerobic composting has become an important hotspot of combined pollution [e.g., antibiotic resistance genes (ARGs) and heavy metals (HMs)] in the process of centralized treatment and resource utilization of manure. However, the interaction mechanisms and environmental drivers of HMs resistome (MRGs), antibiotic resistance (genotype and phenotype), and microbiome during aerobic composting under the widely used amoxicillin (AMX) selection pressure are still poorly understood. Here, we investigated the dynamics of HMs bioavailability and their MRGs, AMX-resistant bacteria (ARB) and antibiotic resistome (ARGs and intI1), and bacterial community to decipher the impact mechanism of AMX by conducting aerobic composting experiments. We detected higher exchangeable HMs and MRGs in the AMX group than the control group, especially for the czrC gene, indicating that AMX exposure may inhibit HMs passivation and promote some MRGs. The presence of AMX significantly altered bacterial community composition and AMX-resistant and -sensitive bacterial structures, elevating antibiotic resistome and its potential transmission risks, in which the proportions of ARB and intI1 were greatly increased to 148- and 11.6-fold compared to the control group. Proteobacteria and Actinobacteria were significant biomarkers of AMX exposure and may be critical in promoting bacterial resistance development. S0134_terrestrial_group was significantly negatively correlated with blaTEM and czrC genes, which might play a role in the elimination of some ARGs and MRGs. Except for the basic physicochemical (MC, C/N, and pH) and nutritional indicators (NO3−-N, NH4+-N), Bio-Cu may be an important environmental driver regulating bacterial resistance during composting. These findings suggested the importance of the interaction mechanism of combined pollution and its synergistic treatment during aerobic composting need to be emphasized.
Introduction
With the overuse, misuse, and abuse of antibiotics in modern medicine and the livestock industry in the last decades (Yao L. et al., 2020; Zhang et al., 2020; Zou et al., 2020; Wang et al., 2021), antimicrobial resistance has developed into one of the most urgent public health crises in recent years (Zhu et al., 2017a; Vos et al., 2019; Joshi and Kim, 2020; Dai et al., 2022). According to statistics, the global defined daily doses of antibiotics reached 34.8 billion in 2015 and have increased by 65% since 2000 (Zhao R. et al., 2019). Amoxicillin (AMX) is currently the most important and frequently used broad-spectrum β-lactam antibiotic, accounting for 50–70% of total antibiotic consumption globally (Chen et al., 2017; Liu et al., 2018). Specifically, AMX is the most commonly used human and animal antibiotic among 36 different drug types in China (Liu et al., 2019), with doses up to 500 mg/kg used in pig feed (Liu et al., 2018). However, antibiotics usage in animal husbandry (Zhang et al., 2017a) can shift animal microbiome, enrich antibiotic resistance genes (ARGs), and subsequently transfer to livestock manure and its products (Qiu et al., 2022). Hence, livestock production is considered an important hotspot and key point-source of antibiotic resistant bacteria (ARB) and ARGs dissemination (Youngquist et al., 2016; Awasthi et al., 2019; Deng et al., 2020; Lu et al., 2020; Tang et al., 2020). Previous studies have shown that β-lactam resistance genes are the top three types of ARGs in manure samples from 17 dairy farms in Shanxi Province, China (Zhou et al., 2016; Liu et al., 2018). The Class I integrase gene (intI1), which gene cassette usually contains ARGs (Zhang et al., 2017a; Zhang R. et al., 2019), can mediate horizontal gene transfer (HGT) thereby facilitating ARGs spread among microorganisms (Yin et al., 2017; Zhang J. et al., 2019; Wang et al., 2023). Due to persistent environmental selection of antibiotic residue and fecal contamination containing antimicrobial resistant determinants, it is anticipated that about 300 million people will die prematurely by 2055 worldwide (Joshi and Kim, 2020).
It is noteworthy that heavy metals (HMs), such as copper (Cu) and zinc (Zn), were often used as important feed additives to promote the growth of livestock (Zhang J. et al., 2019; Zhang et al., 2020). However, due to the poor absorption and difficult degradation of HMs (Yin et al., 2017; Zhang et al., 2017b), they usually coexist with antibiotics and other harmful substances (Imran et al., 2019; Wang R.Z. et al., 2020; Yao N. et al., 2020) in breeding wastes (Awasthi et al., 2019; Zhang R. et al., 2019; Chen et al., 2020) and compost products (Guo et al., 2019; Deng et al., 2020; Zhang et al., 2020). These environmental pollutants (HMs, antibiotics, etc.) can release (co-)selection pressure, induce HMs resistome (MRGs) development and mobile genetic elements (including intI1) mediated HGT (Imran et al., 2019; Zhao Y. et al., 2019; Lu et al., 2020) and accelerate the spread of bacterial resistance (Zhao et al., 2017; Vos et al., 2019), thereby seriously threatening the environmental health (Guo et al., 2019; Imran et al., 2019; Yang et al., 2019). Worse still, even HMs of low concentration has high persistence (Yin et al., 2017; Vos et al., 2019) and strong selective pressure of antibiotic resistance in certain cases (Imran et al., 2019; Zhang J. et al., 2019), among which Cu was shown to be the strongest ability to promote ARGs conjugate transfer (Ji et al., 2012; Guo et al., 2019; Zhang R. et al., 2019).
Pig manure is a typical microenvironment contaminated with antibiotics and HMs (Zhang et al., 2017b; Zhang J. et al., 2019; Zhang R. et al., 2019), where the concentration of HMs is several times that of antibiotics (Awasthi et al., 2019; Imran et al., 2019). It has been reported that aerobic composting has the potential to reduce the bioavailability of HMs (Yin et al., 2017; Deng et al., 2020), antibiotic concentrations, and their drug resistance (Youngquist et al., 2016; Zhou et al., 2016; Awasthi et al., 2019), while utilizing livestock manure as resources (Liu et al., 2017a,b, 2018; Guo et al., 2019; Kumar Awasthi et al., 2019; Qiu et al., 2019; Tang et al., 2020). However, the removal effect of composting will largely vary depending on the composting environment, process conditions, and various pollutants (Zhang J. et al., 2019; Zhang et al., 2020). Up to date, many studies on the removal (Youngquist et al., 2016; Liu et al., 2018; Awasthi et al., 2019; Tang et al., 2020; Zhang et al., 2020) and effects (Meng et al., 2015; Qian et al., 2016; Yin et al., 2017; Liu et al., 2019; Deng et al., 2020) of various antibiotics or HMs during aerobic composting have been made. However, there is still a lack of research on the dynamics and interaction mechanisms of HMs resistome, AMX bacterial resistance (genotype and phenotype), and microbiome during aerobic composting under the widely used AMX selection pressure. Therefore, more studies are needed to decipher the impact mechanism of AMX on various complex pollutants in the process of fecal aerobic composting and the risk of bacterial resistance.
Thus, the objectives of our study were (1) to characterize the dynamics of HMs bioavailability and their resistome (MRGs) with or without AMX exposure, (2) to explore the effects of AMX selection pressure on AMX bacterial resistance (ARB) and antibiotic resistome (ARGs and intI1), and (3) to decipher the environmental drivers and interaction mechanisms of bacterial taxonomic and functional (ARGs, MRGs, intI1, etc.) community composition during composting. Our findings will provide insights into organic waste resource utilization and pollutant risk control mechanisms underpinning the cleaner production of intensive animal husbandry and better development of organic circular agriculture.
Materials and methods
Materials and chemicals
Pig manure was obtained from a large-scale pig farm in Shunyi District (Beijing, China), with prior assurance that no antibiotics were applied to the sampled pigs during the breeding process. Wheat straw was collected from Shangzhuang Experimental Station of China Agricultural University and cut into 3–5 cm segments (Liu et al., 2017a,b, 2019). AMX (98%) was gained from Huamaike Biological Technology Company, Ltd. (Beijing, China). The chemicals used in HM extraction were of analytical grade.
Experimental design
In this study, two groups of fresh pig manure (about 5.2 kg) were fully mixed with 100 mg/kg (dry weight) AMX aqueous solution or an equivalent amount of ultrapure water (control), and then a certain amount of wheat straw and ultrapure water were mixed uniformly, according to the ratio of total carbon and total nitrogen (C/N) of 20:1 and moisture content (MC) of 65% (Liu et al., 2019). After being completely mixed and equilibrated for about 6 h (Qian et al., 2016; Liu et al., 2019), the two groups of initial compost mixture were placed in two parallel sets of in-laboratory aerobic composting reactor systems (Supplementary Figure S1), as the experimental group (AMX group) and the control group without AMX (CK group). Both groups of reactors were continuously ventilated by 0.2 L/(kg-VS-min). The basic physicochemical properties of initial composting materials have been determined and listed in Supplementary Table S1. The above settings of composting conditions and AMX concentration were based on the existing reports and our previous studies (Kotzerke et al., 2011; Liu et al., 2018, 2019).
According to the aerobic composting periods (Liu et al., 2018), about 120 g of the compost mixture was sampled on days 0, 2, 4, 6, 9, 15, and 21, and the basic physicochemical, biological, and heavy metal-related indicators were determined. Furthermore, the viable counts of AMX-resistant bacteria and total culturable bacteria (TCB) were performed immediately on the day of sampling. The presence of AMX resistance genes (blaTEM, blaVIM), Cu resistance gene (copA), Zn resistance gene (czrC), and intI1 were determined by real-time fluorescence quantitative PCR (qPCR) for day 0, 2, 6, 15, and 21 samples. Combined with the existing reports and our previous studies, TEM- and VIM-type β-lactam resistance genes, as the most common and highly abundant ARGs in animal breeding and clinical environments, can significantly enhance the zoonotic potential (Pomba et al., 2004). Together with copA- and czrC-type MRGs, they are often used as indicator genes of bacterial resistance contamination (Narciso-da-Rocha et al., 2014). Moreover, intI1 is widely used as an important universal marker for human pollutants (Gillings et al., 2015; Yin et al., 2017). The bacterial community of compost on days 2, 6, and 15 were analyzed according to different representative composting stages based on 16S rRNA gene sequencing. Samples used for molecular biological determinations were first lyophilized with a vacuum freeze dryer (Martin Christ Alpha 1–2 LD plus, Germany) for the same low MC, then ground with a 1-mm sieve and frozen at −80°C for DNA extraction. Other samples were kept at −20°C for physicochemical parameters analysis. The compost piles were fully mixed manually before and after each sampling.
Measurement and analysis methods
Basic physicochemical and biological indicators
Temperature sensor systems were used to monitor the temperature of the piles and ambient temperature in real-time during composting. Organic matter (OM), MC, C/N, pH, NH4+-N, and NO3−-N were determined concerning the existing methods (Liu et al., 2017a,b, 2019). Since the level of mobility is an important indicator to measure the potential risk of HMs in the environment (Nemati et al., 2011), exchangeable HMs have also attracted attention as the most active and bioavailable high-risk metal forms. Therefore, the most bioavailable and risky exchangeable copper (bio-Cu) and exchangeable zinc (bio-Zn) were detected according to the improved BCR sequential extraction method (Nemati et al., 2011).
Viable counts of ARB and TCB
According to the maximum value of the bacterial Minimum Inhibition Concentration, AMX-resistant bacteria and TCB were counted on R2A medium with and without 32 mg/L AMX by referring to previous experience (Liu et al., 2020). The colony formation unit (CFU) per gram was calculated by the formula (1). All measurements in this study were performed in duplicate.
DNA extraction and qPCR
According to the manufacturer’s instructions, a Soil Genomic DNA Extraction Kit (Kangwei Century, China) was used to extract DNA from composting samples. After the design of related primers (Supplementary Table S2), the presence of two β-lactam resistance genes (blaTEM, blaVIM), two MRGs (copA, czrC), and intI1 were determined by standard PCR (Liu et al., 2019). The PCR products were examined by 1% (w/v) agarose gel electrophoresis after the reaction, and the detected genes were quantified using the ABI 7500 Real-Time PCR System (Applied Biosystems, United States). Absolute abundances of genes were expressed as copy number per gram of dry compost.
Bacterial 16S rRNA gene high-throughput sequencing
16S rRNA gene sequencing was conducted with the Illumina MiSeq platform, and the 16S V3-V4 region was amplified using primers U341F (ACTCCTACGGGAGGCAGCAG) and U806R (GGACTACHVGGGTWTCTAAT). USEARCH was used for quality control of raw data, and UPARSE was used for clustering of qualified sequences with 97% similarity to form operational classification unit (OTU).
Statistical analysis
Repeated measurements were expressed as mean ± standard deviation (SD, indicated by the error bar in the figures). Excel 2016 (Microsoft, United States), SPSS 25 (IBM, United States), and OriginProV.8.5 SR1 (OriginLab Corp., United States) were used for basic statistical analysis. Shannon-Wiener curves, principal coordinates analysis (PCoA), heatmap, correlation analysis, and genus-level phylogenetic tree were performed by R3.4.1 software. Redundancy analysis (RDA) was carried out based on CANOCO 5.0. Correlation network analysis (Spearman, r > 0.6, p < 0.05) was conducted by R3.4.1 software and Gephi0.9.3 software.
Results and discussion
Changes of exchangeable HMs and their resistome (MRGs) during composting
As important indicators of environmental HMs potential risks, exchangeable HMs (bio-Cu and bio-Zn) can sensitively reflect the dynamic changes of HM morphology and toxicity during composting. The concentrations of bio-Cu and bio-Zn in the two groups leveled off after first decreasing during composting, presenting a relatively consistent change trend (Figures 1A,B). The results reflected that aerobic composting treatment will reduce the migration rate and bioavailability of Cu and Zn to a certain extent, thereby effectively reducing the toxicity and potential risks of HMs in the products. Comparatively, the concentrations of bio-Cu and bio-Zn in the AMX group were slightly higher than those in the CK group during the early stage of composting. This difference indicated that the addition of AMX likely hindered the passivation process of HMs and, thus, increased the risks of HMs contamination and the co-selection pressure of inducing bacterial resistance, which was also confirmed by previous reports (Song et al., 2017; Vos et al., 2019).
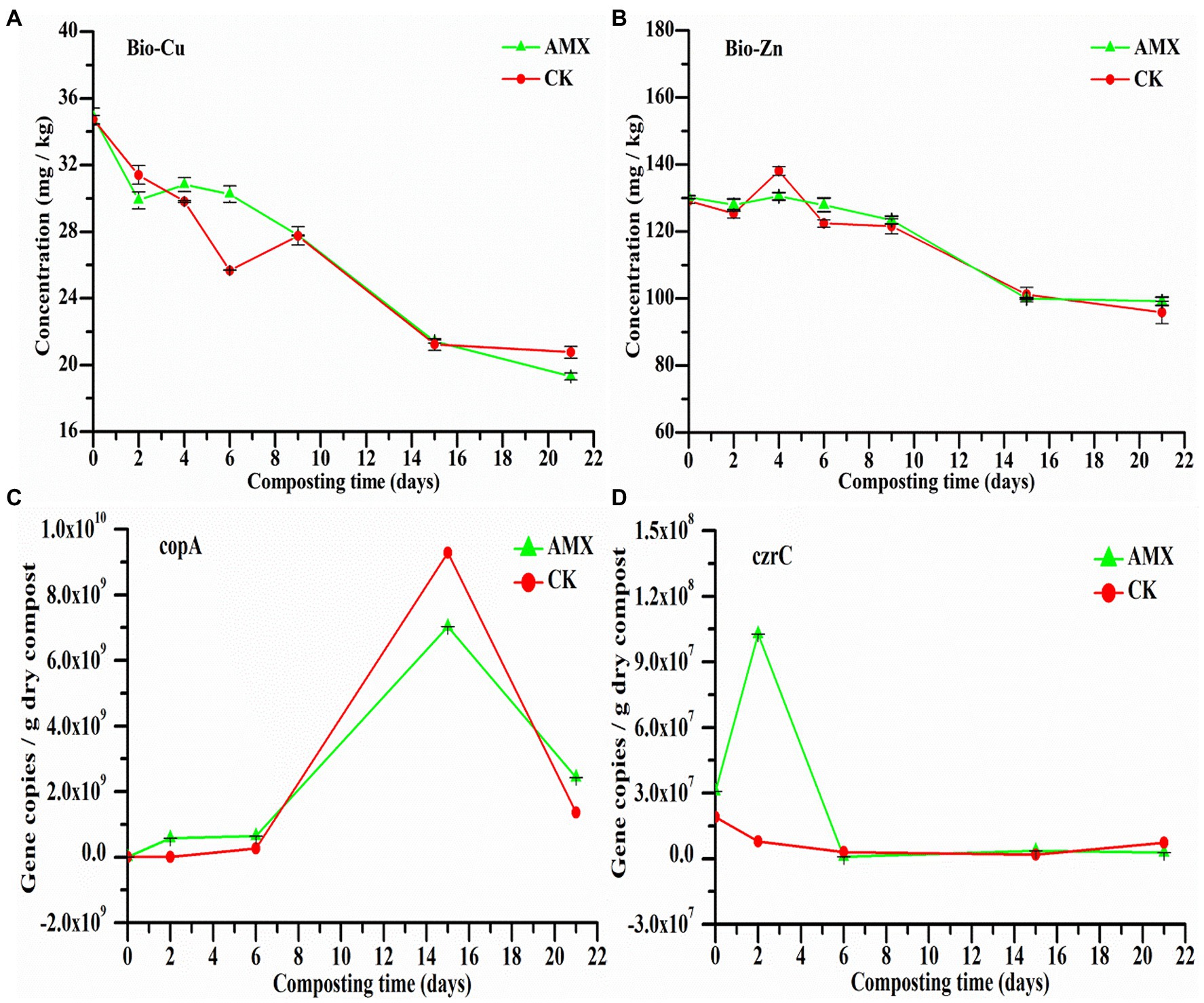
Figure 1. Dynamic changes of (A) exchangeable Cu (bio-Cu), (B) exchangeable Zn (bio-Zn), (C) Cu resistance gene (copA), and (D) Zn resistance gene (czrC) during composting.
We found that the abundance of copA and czrC in the AMX group was higher at the initial stage of composting (Figures 1C,D), especially the difference between the two groups of czrC was the most significant (p < 0.05, t-test), further illustrating the vital role of AMX selective pressure on the promotion of HM resistance and adaptation cost of MRGs-carrying bacteria (Imran et al., 2019; Zhang et al., 2020). Notably, AMX may have a stronger promotion effect on some Zn resistome. In the late stage of composting, the abundance of copA in the two groups increased rapidly then fell, while czrC of the two groups was very low and stable, probably because copA-carrying bacteria mainly dominated the composting maturity stage and were strongly affected by microbial succession at this stage. According to Figure 1, AMX selective pressure may significantly inhibit the passivation of HMs while promoting the development of some MRGs, thereby raising the potential risk of HM toxicity and bacterial resistance in the environment.
Changes of AMX bacterial resistance (phenotype and genotype) during composting
AMX-resistant bacteria and their proportion of resistance
As shown in Figure 2A, the number of AMX-resistant bacteria in the two groups fell sharply with the rapid increase in compost temperature (Supplementary Figure S2A) on days 0–2, which also restricted TCB (Supplementary Figure S2B). The temperature of compost piles on days 3–9 decreased slowly, with a significant recovery in the middle period, which resulted in a slight increase in the number of AMX-resistant bacteria and TCB in the two groups. As the compost temperature gradually decreased toward ambient temperature (days 10–21), the composting microenvironment became more suitable for the growth and reproduction of microorganisms. Therefore, a sharp increase in the number of AMX-resistant bacteria and TCB was observed, followed by a decrease due to nutrient deficiency.
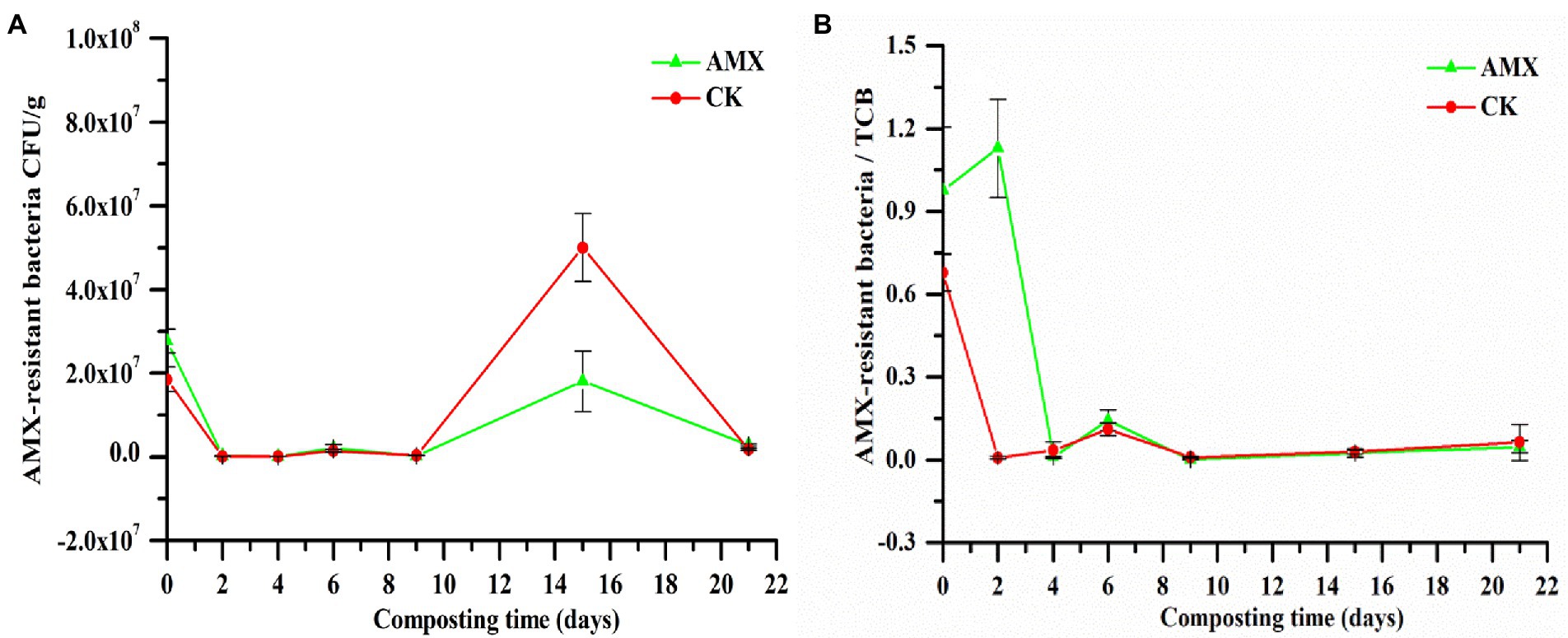
Figure 2. Dynamic changes of (A) AMX-resistant bacteria and (B) AMX-resistant bacteria / total culturable bacteria (TCB) during composting.
The proportion of AMX-resistant bacteria in the AMX group in the early stage of composting were significantly higher than those in the CK group (days 0–4; Figure 2B), implying the influence of AMX selective pressure on the enhancement of AMX bacterial resistance (Vos et al., 2019; Zou et al., 2020). However, in the later stage of composting, the number of AMX-resistant bacteria and TCB in the AMX group was significantly lower (p < 0.01, t-test) than that in the CK group (Figure 2A; Supplementary Figure S2B), while the proportion of AMX resistance (Figure 2B) was similar between the two groups, suggesting the inhibitory effect of AMX addition on microbial growth and reproduction (Liu et al., 2019).
ARGs and intI1 of the compost piles
According to Figures 3A,B, the blaTEM and blaVIM gene abundances in the AMX treatment were initially higher than those of the CK treatment. Both genes in the AMX group experienced a tendency to decline first, followed by an increase, and then a gradual decrease, which may be due to the dual effect of the high-temperature environment (Supplementary Figure S2A) and antibiotic selective pressure in the early stage of composting. With the piles’ temperature decrease in the late stage of composting and the long-term co-selection of AMX-HM pollution (Imran et al., 2019; Wang R.Z. et al., 2020; Yao N. et al., 2020), the abundance of the two β-lactam resistance genes in the AMX group obviously increased compared with the CK group.

Figure 3. Dynamic changes in the copy number of (A) blaTEM, (B) blaVIM, and (C) intI1 during composting.
As shown in Figure 3C, the absolute abundance of intI1 gene in the two groups changed similarly during composting, both demonstrating an initially decreasing trend followed by an increase. Comparatively, the abundance of intI1 in the AMX group was always 1.3–11.6 times greater than that in the CK group, even at the day 0, and maintained a rapid growth trend in the late stage of composting. Compared with blaTEM and blaVIM, intI1 was significantly increased during the equilibration phase before the start of composting (day 0), indicating that it would be greatly increased in a very short period of time and persistently affected by AMX exposure, which may be a non-negligible problem in the spread of bacterial resistance. Furthermore, while the high temperature of the thermophilic phase greatly reduced the number of bacteria (Supplementary Figure S2) and intI1 gene in both groups, the co-selection pressure of AMX and HMs still provided a strong selective advantage for their resistant bacteria (Figure 2) and resistome (Figures 1C,D, 3) in the AMX group. This further confirmed the crucial role of antibiotics in promoting the development of bacterial resistance (antibiotics, HMs, etc.) and its transmission risk (Imran et al., 2019).
Changes of bacterial communities during composting
A total of 397 operational taxonomic units (OTUs) were detected in compost samples. According to the Shannon-Wiener curves of the compost samples (Supplementary Figure S3), the sequencing data were sufficient to reflect the vast majority of microbial information in all samples. After leveling the minimum sequence number of the samples, alpha diversity analysis was performed on the two groups of compost samples (Supplementary Figure S4). The three diversity indices of the AMX group were higher than those of the CK group as a whole, confirming that the bacterial community of the AMX group was relatively rich and uniform with higher community diversity (Supplementary Figures S4A–C). Moreover, there was a greater difference in species lineage among samples in the CK group when considering species abundance and evolutionary distance (Supplementary Figure S4D). In summary, although AMX selective pressure reduced the absolute abundances of culturable bacteria (Supplementary Figure S2B), the richness and evenness of the bacterial community were relatively high (Supplementary Figure S4), which may indicate the effect of AMX on the alteration of microbial community structure during composting and the selective pressure on AMX-sensitive and -resistant bacteria. According to the PCoA, PC1 and PC2 accounted for 85.6% of the total variation of the bacterial community (Figure 4A). Principal coordinates analysis of the bacterial community revealed that the samples of different composting periods were clearly separated along the PCo1 axis, and some clustering of composting samples from the same period was also observed. The difference in bacterial community composition between the two groups decreased gradually as the number of composting days increased, illustrating that the impact of the composting periods and habitat on the bacterial community structure was particularly significant (Yin et al., 2017). Furthermore, AMX addition significantly affected the bacterial community structure in the early stage of composting, which is consistent with the above results (Supplementary Figure S4).
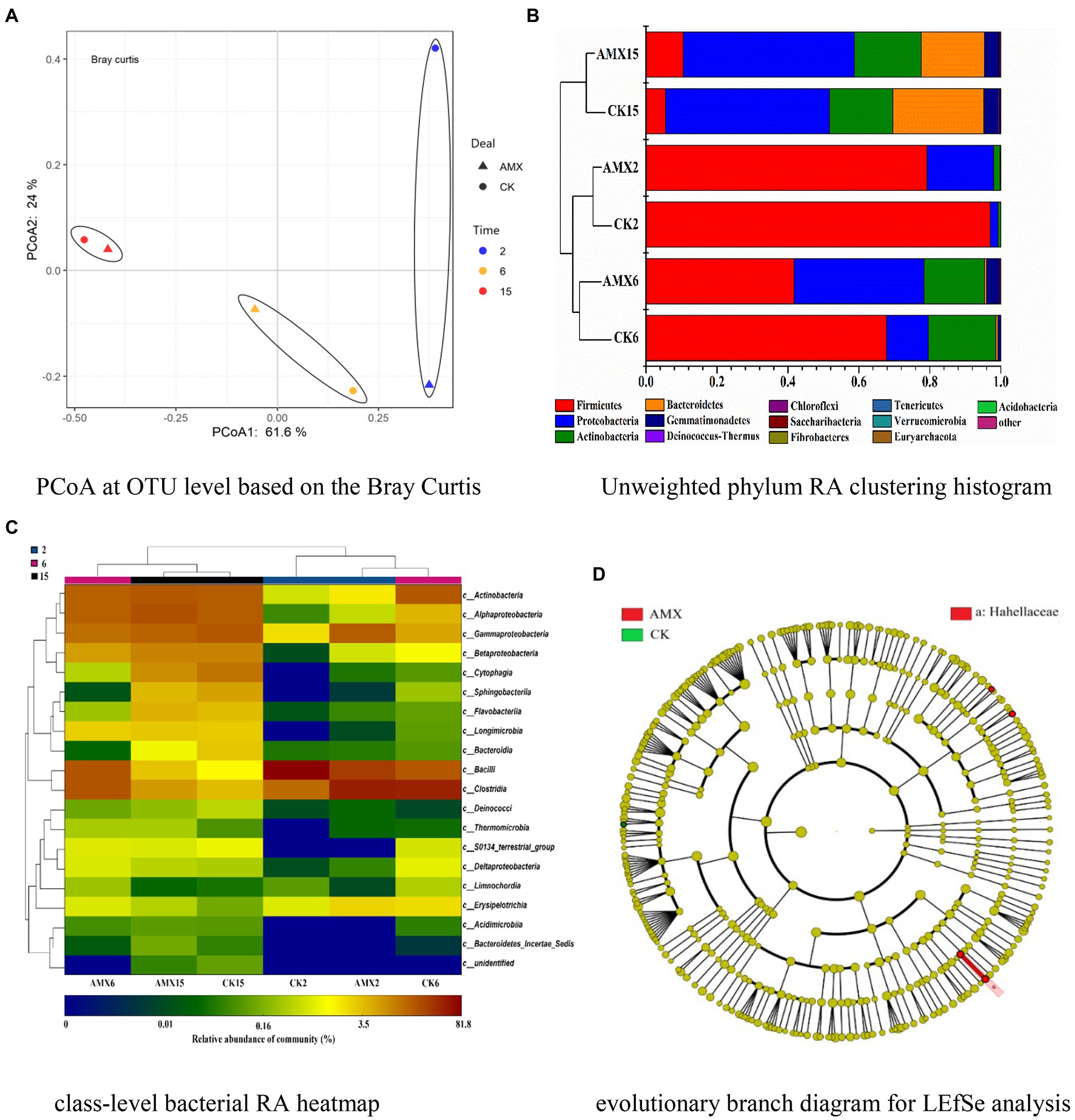
Figure 4. Features of bacterial community during composting. (A) PCoA at OTU level based on the Bray Curtis. (B) Unweighted phylum RA clustering histogram. (C) Class-level bacterial RA heatmap. (D) Evolutionary branch diagram for LEfSe analysis.
It can be seen from Figures 4B,C that the bacterial community of the two groups continued to change during composting, while the differences in community composition gradually decreased. Among the communities, Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetes were the dominant phylum bacteria during the whole composting process (Figure 4B), accounting for 95.2–99.9% of the bacterial 16S rRNA gene sequences (Supplementary Figure S5). As shown in Figure 4B, the relative abundance (RA) of Firmicutes was the highest in the early stage of composting and the RA in the AMX group was lower than that in the CK group, but decreased rapidly as the compost matured. This may be since Firmicutes, beneficial bacteria for promoting cellulose degradation, widely exist in animal intestines, compost, and soil environment, and dominate the heating and thermophilic phases of composting (Yin et al., 2017; Liu et al., 2019). Moreover, high concentrations of antibiotics will significantly inhibit their growth and reproduction (Qian et al., 2016; Guo et al., 2017; Liu et al., 2019), resulting in a significant reduction of Firmicutes RA in the AMX group. According to Figure 4B, RA of Bacteroides increased significantly in the mature composting stage of the two groups and was relatively higher in the CK group. In Figure 4C, it can be seen that Sphingobacteriia, Flavobacteriia, and Bacteroidia (which belong to Bacteroides) also displayed the same regularities and cluster. Bacteroides are a kind of bacteria attributed to the degradation of high-molecular-weight compounds and the growth of supporting materials, which mainly grow in the middle and late stages of composting (Zhu et al., 2017b; Liu et al., 2019). The RA of Proteobacteria and Actinobacteria (most notably in Proteobacteria) in the AMX group increased with composting maturity and was significantly higher than that in the CK group during the early stage of composting (Figure 4B). This large difference may be attributed to the dominance of both bacteria in the cooling and mature stages of composting, where the abundance of Actinobacteria is mostly used to mark the degree of maturity (Liu et al., 2019; Zhang et al., 2020). Previous studies have shown that both bacteria have a significant positive correlation with ARGs (Yin et al., 2017), and their RA will also be significantly increased in the presence of high concentrations of antibiotics (Guo et al., 2017). Besides, the driving force of the increased resistance in the compost was mainly attributed to Proteobacteria (Meng et al., 2015), in which γ-Proteobacteria has a strong resistance potential to high-concentration antibiotics (Zhu et al., 2017b). Moreover, β-Proteobacteria is considered to be the original source of intI1, which has become the core of antibiotic resistance due to its ability to capture and express multiple ARGs (Guo et al., 2017; Zhu et al., 2017b). The above conclusions were also further confirmed in Figure 4C, where the variation rules of α, β, γ-Proteobacteria, and Actinobacteria were similar and clustered with each other. Therefore, the high concentration of AMX added to the experimental group can selectively inhibit the growth and reproduction of sensitive bacteria, such as Firmicutes. This provided a more favorable living environment for drug-resistant Proteobacteria and Actinobacteria, thus affecting and changing the bacterial community structure and drug resistance level during composting.
LEfSe analysis (threshold set to 2) was used to further reveal the differences between the two groups of bacterial community during composting (Figure 4D). Compared with the CK group, the species with significantly different abundance in the AMX group were Hahellaceae, Hahella, Enteractinococcus, and Marmoricola (Supplementary Figure S6), where the former two species belong to γ-Proteobacteria and the latter two to Actinobacteria. Proteobacteria is the main host of ARGs and has a high tolerance to antibiotics, while Actinobacteria is the main producer of antibiotics and usually carries a variety of ARGs (Qian et al., 2016; Zhang et al., 2020). Therefore, this finding was consistent with the relevant conclusions in Figures 4B,C and previous literature reports, further confirming the significant role of AMX in reshaping the structure of environmental microbial communities and promoting bacterial resistance. Studies have shown that Enteractinococcus is a type of Micrococcaceae commonly found in animal feces (Cao et al., 2012), while Marmoricola is the gram-positive aerobic bacteria belonging to the Nocardiaceae (Habib et al., 2020). Both of them have been reported to exist in the aerobic composting environment of livestock feces as the main Actinobacteria (Chen et al., 2015).
Correlation analysis of the compost piles
Redundancy analysis was carried out to further reveal the correlation between the bacterial community structure and its function and environmental factors during composting, which can explain 86.8% of the total species variation (Figure 5A). Differences in the bacterial community structure gradually reduced with the extension of composting time, which intuitively reflected the severe succession and stable formation of the bacterial community structure during composting. A noteworthy correlation was observed between the thermophilic period of composting and the dynamic changes in the basic physiochemical indicators, exchangeable HMs, β-lactam resistance genes, czrC gene, and Firmicutes. Proteobacteria, Actinobacteria, and Bacteroidetes were mainly distributed in the composting maturation stage and were directly related to the dynamic changes of copA gene and intI1 gene. This correlation demonstrated the significant impact of the composting stages (especially the thermophilic period) and bacterial community structure on the composting microenvironment, HM forms, and resistance genes. Furthermore, it can also be suggested that Proteobacteria, Actinobacteria, and Bacteroidetes were likely the main factors directly affecting the development and spread of HM resistance and antibiotic resistance during composting.
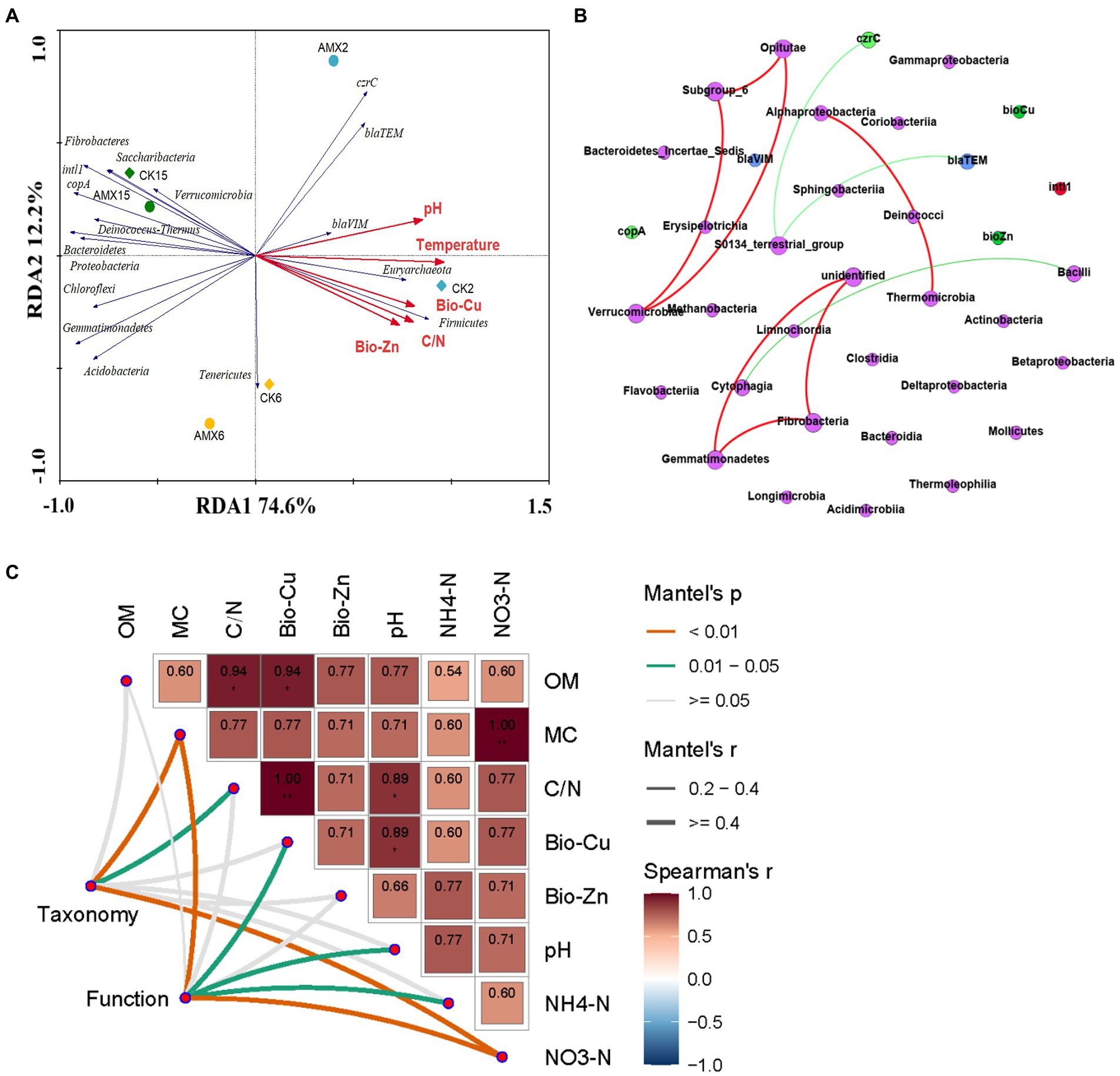
Figure 5. Environmental drivers of compost microbial community composition: (A) RDA of the relationships between the main bacterial phylum (more than 1% of the total bacterial abundance, blue arrows) and environmental factors (red arrows), (B) network analysis of the relationships among class-level bacteria (purple nodes), ARGs (blue nodes), MRGs (light green nodes), HMs (green nodes), and intI1 (red nodes), and (C) pairwise comparisons of environmental factors were shown, with color gradients representing Spearman’s correlation coefficients. Taxonomic (based on family-level bacteria) and functional (based on ARGs, MRGs, intI1, ARB, TCB, and ARB/TCB) community composition was related to each environmental factor by Mantel tests. Edge width denotes the Mantel’s r statistic for the corresponding distance correlations, and edge color represents the statistical significance.
To further verify the interaction pattern between the microbial communities in the two groups of different composting environments, the relationships among class-level bacteria, ARGs, MRGs, Bio-HMs, and intI1 were revealed by network analysis (Figure 5B). More positive edges (red, 7) than negative edges (green, 3) were detected in a network of the compost. Seven different class-level bacteria (except for unidentified) had a significant positive correlation with each other (p < 0.05), which was consistent with the results of the RDA analysis. Bacilli (belonging to Firmicutes) and Cytophagia (belonging to Bacteroidetes) were significantly negatively correlated (p < 0.05), which were the dominant bacterial communities in the early composting period and the late composting periods, respectively, (Figure 4B). S0134_terrestrial_group, which belongs to the Gemmatimonadetes, was significantly negatively correlated with blaTEM gene and czrC gene (p < 0.05). Combined with Figure 4B, Gemmatimonadetes were mainly detected in the middle and late stages of compost samples (AMX6, AMX15, and CK15), and probably play an important role in the reduction of blaTEM gene and czrC gene. Therefore, it is recommended in future studies to further explore the role of Gemmatimonadetes in removing other ARGs and MRGs during aerobic composting, especially β-lactams and zinc.
Moreover, to identify environmental drivers in our study (Sunagawa et al., 2015), we correlated distance-corrected dissimilarities of taxonomic and functional community composition with those of compost environmental factors (Figure 5C). Overall, MC and NO3−-N were the strongest correlates of both taxonomic and functional composition during composting, while no significant correlations were found for OM and Bio-Zn. Bio-Cu, pH, and NH4+-N were only weakly correlated with functional community, as well as C/N with taxonomic composition, and the other correlations were not statistically significant (Figure 5C). It can be seen that, except for the basic physicochemical (MC, C/N, and pH) and nutritional indicators (NO3−-N, NH4+-N), Bio-Cu may be an important environmental driver affecting taxonomic and functional community composition in composting environments, as well as an important potential factor regulating bacterial resistance.
Conclusion
Our study found that aerobic composting could effectively decrease HMs bioavailability, which may alleviate the combined pollutions and (co-)selection pressure of AMX and HMs. AMX selective pressure played an important role in inhibiting HMs passivation and bacterial growth, shifting bacterial community composition and AMX-sensitive and -resistant bacteria structures, and increasing some MRGs, AMX bacterial resistance, and their potential risks, especially for czrC and intI1. Proteobacteria and Actinobacteria, as significant biomarkers of AMX exposure, may be critical in promoting the development and spread of bacterial resistance. S0134_terrestrial_group was significantly negatively correlated with blaTEM and czrC gene, which may be critical in reducing some ARGs and MRGs. Except for the basic physicochemical (MC, C/N, and pH) and nutritional indicators (NO3−-N, NH4+-N) during composting, Bio-Cu may be an important environmental driver affecting taxonomic and functional community composition and regulating bacterial resistance. Therefore, it is necessary to further explore the efficient removal technology of combined pollutants and the interaction mechanisms between important microorganisms and contaminants in the process of composting, to promote the harmless treatment, resource recovery, and agricultural utilization of organic waste.
Data availability statement
The data presented in the study are deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) repository, accession number CRA009005 that are publicly accessible at: https://ngdc.cncb.ac.cn/gsa.
Author contributions
NL: conceptualization, methodology, investigation, software, formal analysis, writing—original draft, visualization, and writing—review and editing. GL: investigation, methodology, and review and editing. YS: investigation and visualization. YZ: article revision and grammar correction. JM: methodology, review and editing, and supervision. GH: resources, writing—review and editing, supervision, and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the National Natural Science Foundation of China (31771684, 42090063, and 42107407), Ningbo Science and Technology Plan Project (2021S030) and the Project funded by China Postdoctoral Science Foundation (2022M713076).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1079114/full#supplementary-material
References
Awasthi, M. K., Chen, H., Awasthi, S. K., Duan, Y., Liu, T., Pandey, A., et al. (2019). Application of metagenomic analysis for detection of the reduction in the antibiotic resistance genes (ARGs) by the addition of clay during poultry manure composting. Chemosphere 220, 137–145. doi: 10.1016/j.chemosphere.2018.12.103
Cao, Y. R., Jiang, Y., Jin, R. X., Han, L., He, W. X., Li, Y. L., et al. (2012). Enteractinococcus coprophilus gen. Nov., sp. nov., of the family Micrococcaceae, isolated from Panthera tigris amoyensis faeces, and transfer of Yaniella fodinae Dhanjal et al. 2011 to the genus Enteractinococcus as Enteractinococcus fodinae comb. nov. Int. J. Syst. Evol. Microbiol. 62, 2710–2716. doi: 10.1099/ijs.0.034249-0
Chen, X., Li, G. D., Li, Q. Y., Hu, C. J., Qiu, S. M., Jiang, Y., et al. (2015). Enteractinococcus lamae sp. nov. and Enteractinococcus viverrae sp. nov., isolated from animal faeces. Antonie Van Leeuwenhoek 108, 1477–1483. doi: 10.1007/s10482-015-0603-3
Chen, J., Liu, Y. S., Zhang, J. N., Yang, Y. Q., Hu, L. X., Yang, Y. Y., et al. (2017). Removal of antibiotics from piggery wastewater by biological aerated filter system: treatment efficiency and biodegradation kinetics. Bioresour. Technol. 238, 70–77. doi: 10.1016/j.biortech.2017.04.023
Chen, J., Yu, X., Li, C., Tang, X., and Sun, Y. (2020). Removal of tetracycline via the synergistic effect of biochar adsorption and enhanced activation of persulfate. Chem. Eng. J. 382:122916. doi: 10.1016/j.cej.2019.122916
Dai, D., Brown, C., Burgmann, H., Larsson, D. G. J., Nambi, I., Zhang, T., et al. (2022). Long-read metagenomic sequencing reveals shifts in associations of antibiotic resistance genes with mobile genetic elements from sewage to activated sludge. Microbiome 10:20. doi: 10.1186/s40168-021-01216-5
Deng, W., Zhang, A., Chen, S., He, X., Jin, L., Yu, X., et al. (2020). Heavy metals, antibiotics and nutrients affect the bacterial community and resistance genes in chicken manure composting and fertilized soil. J. Environ. Manag. 257:109980. doi: 10.1016/j.jenvman.2019.109980
Gillings, M. R., Gaze, W. H., Pruden, A., Smalla, K., Tiedje, J. M., and Zhu, Y. G. (2015). Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 9, 1269–1279. doi: 10.1038/ismej.2014.226
Guo, H., Gu, J., Wang, X., Tuo, X., Yu, J., and Zhang, R. (2019). Key role of cyromazine in the distribution of antibiotic resistance genes and bacterial community variation in aerobic composting. Bioresour. Technol. 274, 418–424. doi: 10.1016/j.biortech.2018.12.005
Guo, A., Gu, J., Wang, X., Zhang, R., Yin, Y., Sun, W., et al. (2017). Effects of superabsorbent polymers on the abundances of antibiotic resistance genes, mobile genetic elements, and the bacterial community during swine manure composting. Bioresour. Technol. 244, 658–663. doi: 10.1016/j.biortech.2017.08.016
Habib, N., Khan, I. U., Xiao, M., Li, S., Saqib, M., Xian, W.-D., et al. (2020). Marmoricola caldifontis sp. nov., a novel actinobacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 70, 2053–2058. doi: 10.1099/ijsem.0.004016
Imran, M., Das, K. R., and Naik, M. M. (2019). Co-selection of multi-antibiotic resistance in bacterial pathogens in metal and microplastic contaminated environments: an emerging health threat. Chemosphere 215, 846–857. doi: 10.1016/j.chemosphere.2018.10.114
Ji, X., Shen, Q., Liu, F., Ma, J., Xu, G., Wang, Y., et al. (2012). Antibiotic resistance gene abundances associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in Shanghai; China. J. Hazard. Mater. 235-236, 178–185. doi: 10.1016/j.jhazmat.2012.07.040
Joshi, A., and Kim, K. H. (2020). Recent advances in nanomaterial-based electrochemical detection of antibiotics: challenges and future perspectives. Biosens. Bioelectron. 153:112046. doi: 10.1016/j.bios.2020.112046
Kotzerke, A., Fulle, M., Sharma, S., Kleineidam, K., Welzl, G., Lamshöft, M., et al. (2011). Alterations in total microbial activity and nitrification rates in soil due to amoxicillin-spiked pig manure. J. Plant Nutr. Soil Sci. 174, 56–64. doi: 10.1002/jpln.200900210
Kumar Awasthi, M., Chen, H., Duan, Y., Liu, T., Kumar Awasthi, S., Wang, Q., et al. (2019). An assessment of the persistence of pathogenic bacteria removal in chicken manure compost employing clay as additive via meta-genomic analysis. J. Hazard. Mater. 366, 184–191. doi: 10.1016/j.jhazmat.2018.11.108
Liu, N., Han, H., Yin, H., Han, L., and Huang, G. (2018). Variations in the fate and risk analysis of amoxicillin and its degradation products during pig manure aerobic composting. J. Hazard. Mater. 346, 234–241. doi: 10.1016/j.jhazmat.2017.11.050
Liu, N., Hou, T., Yin, H., Han, L., and Huang, G. (2019). Effects of amoxicillin on nitrogen transformation and bacterial community succession during aerobic composting. J. Hazard. Mater. 362, 258–265. doi: 10.1016/j.jhazmat.2018.09.028
Liu, N., Xu, L., Han, L., Huang, G., and Ciric, L. (2020). Microbiological safety and antibiotic resistance risks at a sustainable farm under large-scale open-air composting and composting toilet systems. J. Hazard. Mater. 401:123391. doi: 10.1016/j.jhazmat.2020.123391
Liu, N., Zhou, J., Han, L., and Huang, G. (2017a). Characterization of lignocellulosic compositions' degradation during chicken manure composting with added biochar by phospholipid fatty acid (PLFA) and correlation analysis. Sci. Total Environ. 586, 1003–1011. doi: 10.1016/j.scitotenv.2017.02.081
Liu, N., Zhou, J., Han, L., Ma, S., Sun, X., and Huang, G. (2017b). Role and multi-scale characterization of bamboo biochar during poultry manure aerobic composting. Bioresour. Technol. 241, 190–199. doi: 10.1016/j.biortech.2017.03.144
Lu, X. M., Lu, P. Z., and Liu, X. P. (2020). Fate and abundance of antibiotic resistance genes on microplastics in facility vegetable soil. Sci. Total Environ. 709:136276. doi: 10.1016/j.scitotenv.2019.136276
Meng, F., Gao, G., Yang, T.-T., Chen, X., Chao, Y., Na, G., et al. (2015). Effects of fluoroquinolone antibiotics on reactor performance and microbial community structure of a membrane bioreactor. Chem. Eng. J. 280, 448–458. doi: 10.1016/j.cej.2015.06.025
Narciso-da-Rocha, C., Varela, A. R., Schwartz, T., Nunes, O. C., and Manaia, C. M. (2014). blaTEM and vanA as indicator genes of antibiotic resistance contamination in a hospital-urban wastewater treatment plant system. J. Glob. Antimicrob. Resist. 2, 309–315. doi: 10.1016/j.jgar.2014.10.001
Nemati, K., Abu Bakar, N. K., Abas, M. R., and Sobhanzadeh, E. (2011). Speciation of heavy metals by modified BCR sequential extraction procedure in different depths of sediments from Sungai Buloh, Selangor, Malaysia. J. Hazard. Mater. 192, 402–410. doi: 10.1016/j.jhazmat.2011.05.039
Pomba, C. F., Correia, J. D., and Canica, M. M. (2004). Genetic relatedness between human and animal polymorphic blaTEM genes strengthen zoonotic potential. Infect. Genet. Evol. 4:285. doi: 10.1093/jac/dkh307
Qian, X., Sun, W., Gu, J., Wang, X. J., Sun, J. J., Yin, Y. N., et al. (2016). Variable effects of oxytetracycline on antibiotic resistance gene abundance and the bacterial community during aerobic composting of cow manure. J. Hazard. Mater. 315, 61–69. doi: 10.1016/j.jhazmat.2016.05.002
Qiu, T., Huo, L., Guo, Y., Gao, M., Wang, G., Hu, D., et al. (2022). Metagenomic assembly reveals hosts and mobility of common antibiotic resistome in animal manure and commercial compost. Environ. Microbiome 17:42. doi: 10.1186/s40793-022-00437-x
Qiu, X., Zhou, G., Zhang, J., and Wang, W. (2019). Microbial community responses to biochar addition when a green waste and manure mix are composted: a molecular ecological network analysis. Bioresour. Technol. 273, 666–671. doi: 10.1016/j.biortech.2018.12.001
Song, J., Rensing, C., Holm, P. E., Virta, M., and Brandt, K. K. (2017). Comparison of metals and tetracycline as selective agents for development of tetracycline resistant bacterial communities in agricultural soil. Environ. Sci. Technol. 51, 3040–3047. doi: 10.1021/acs.est.6b05342
Sunagawa, S., Coelho, L. P., Chaffron, S., Kultima, J. R., Labadie, K., Salazar, G., et al. (2015). Structure and function of the global ocean microbiome. Science 348:1261359. doi: 10.1126/science.1261359
Tang, Z., Xi, B., Huang, C., Tan, W., Li, W., Zhao, X., et al. (2020). Mobile genetic elements in potential host microorganisms are the key hindrance for the removal of antibiotic resistance genes in industrial-scale composting with municipal solid waste. Bioresour. Technol. 301:122723. doi: 10.1016/j.biortech.2019.122723
Vos, M., Sibleyras, L., Lo, L. K., Hesse, E., Gaze, W. H., and Klümper, U. (2019). Zinc can counteract selection for ciprofloxacin resistance. FEMS Microbiol. Lett. 367:fnaa038. doi: 10.1093/femsle/fnaa038
Wang, R. Z., Huang, D. L., Liu, Y. G., Zhang, C., Lai, C., Wang, X., et al. (2020). Synergistic removal of copper and tetracycline from aqueous solution by steam-activated bamboo-derived biochar. J. Hazard. Mater. 384:121470. doi: 10.1016/j.jhazmat.2019.121470
Wang, S., Zeng, D., Jin, B., Su, Y., and Zhang, Y. (2023). Deciphering the role of polyethylene microplastics on antibiotic resistance genes and mobile genetic elements fate in sludge thermophilic anaerobic digestion process. Chem. Eng. J. 452. doi: 10.1016/j.cej.2022.139520
Wang, L., Zheng, J., and Huang, X. (2021). Co-composting materials can further affect the attenuation of antibiotic resistome in soil application. Waste Manag. 135, 329–337. doi: 10.1016/j.wasman.2021.09.016
Yang, Y., Liu, G., Ye, C., and Liu, W. (2019). Bacterial community and climate change implication affected the diversity and abundance of antibiotic resistance genes in wetlands on the Qinghai-Tibetan plateau. J. Hazard. Mater. 361, 283–293. doi: 10.1016/j.jhazmat.2018.09.002
Yao, L., Li, Y., Li, Z., Shen, D., Feng, H., Zhou, H., et al. (2020). Prevalence of fluoroquinolone, macrolide and sulfonamide-related resistance genes in landfills from East China, mainly driven by MGEs. Ecotoxicol. Environ. Saf. 190:110131. doi: 10.1016/j.ecoenv.2019.110131
Yao, N., Li, C., Yu, J., Xu, Q., Wei, S., Tian, Z., et al. (2020). Insight into adsorption of combined antibiotic-heavy metal contaminants on graphene oxide in water. Sep. Purif. Technol. 236:116278. doi: 10.1016/j.seppur.2019.116278
Yin, Y., Gu, J., Wang, X., Song, W., Zhang, K., Sun, W., et al. (2017). Effects of copper addition on copper resistance, antibiotic resistance genes, and intl1 during swine manure composting. Front. Microbiol. 8:344. doi: 10.3389/fmicb.2017.00344
Youngquist, C. P., Mitchell, S. M., and Cogger, C. G. (2016). Fate of antibiotics and antibiotic resistance during digestion and composting: a review. J. Environ. Qual. 45, 537–545. doi: 10.2134/jeq2015.05.0256
Zhang, R., Gu, J., Wang, X., Li, Y., Liu, J., Lu, C., et al. (2019). Response of antibiotic resistance genes abundance by graphene oxide during the anaerobic digestion of swine manure with copper pollution. Sci. Total Environ. 654, 292–299. doi: 10.1016/j.scitotenv.2018.11.094
Zhang, R., Gu, J., Wang, X., Qian, X., Duan, M., Sun, W., et al. (2017a). Relationships between sulfachloropyridazine sodium, zinc, and sulfonamide resistance genes during the anaerobic digestion of swine manure. Bioresour. Technol. 225, 343–348. doi: 10.1016/j.biortech.2016.10.057
Zhang, M., He, L. Y., Liu, Y. S., Zhao, J. L., Zhang, J. N., Chen, J., et al. (2020). Variation of antibiotic resistome during commercial livestock manure composting. Environ. Int. 136:105458. doi: 10.1016/j.envint.2020.105458
Zhang, J., Lu, T., Chai, Y., Sui, Q., Shen, P., and Wei, Y. (2019). Which animal type contributes the most to the emission of antibiotic resistance genes in large-scale swine farms in China? Sci. Total Environ. 658, 152–159. doi: 10.1016/j.scitotenv.2018.12.175
Zhang, R., Wang, X., Gu, J., and Zhang, Y. (2017b). Influence of zinc on biogas production and antibiotic resistance gene profiles during anaerobic digestion of swine manure. Bioresour. Technol. 244, 63–70. doi: 10.1016/j.biortech.2017.07.032
Zhao, Y., Cocerva, T., Cox, S., Tardif, S., Su, J., Zhu, Y., et al. (2019). Evidence for co-selection of antibiotic resistance genes and mobile genetic elements in metal polluted urban soils. Sci. Total Environ. 656, 512–520. doi: 10.1016/j.scitotenv.2018.11.372
Zhao, R., Feng, J., Liu, J., Fu, W., Li, X., and Li, B. (2019). Deciphering of microbial community and antibiotic resistance genes in activated sludge reactors under high selective pressure of different antibiotics. Water Res. 151, 388–402. doi: 10.1016/j.watres.2018.12.034
Zhao, Z., Wang, J., Han, Y., Chen, J., Liu, G., Lu, H., et al. (2017). Nutrients, heavy metals and microbial communities co-driven distribution of antibiotic resistance genes in adjacent environment of mariculture. Environ. Pollut. 220, 909–918. doi: 10.1016/j.envpol.2016.10.075
Zhou, B., Wang, C., Zhao, Q., Wang, Y., Huo, M., Wang, J., et al. (2016). Prevalence and dissemination of antibiotic resistance genes and coselection of heavy metals in Chinese dairy farms. J. Hazard. Mater. 320, 10–17. doi: 10.1016/j.jhazmat.2016.08.007
Zhu, Y., Wang, Y., Jiang, X., Zhou, S., Wu, M., Pan, M., et al. (2017b). Microbial community compositional analysis for membrane bioreactor treating antibiotics containing wastewater. Chem. Eng. J. 325, 300–309. doi: 10.1016/j.cej.2017.05.073
Zhu, Y., Zhao, Y., Li, B., Huang, C., Zhang, S., Yu, S., et al. (2017a). Continental-scale pollution of estuaries with antibiotic resistance genes. Nat. Microbiol. 2:16270. doi: 10.1038/nmicrobiol.2016.270
Zou, Y., Xiao, Y., Wang, H., Fang, T., and Dong, P. (2020). New insight into fates of sulfonamide and tetracycline resistance genes and resistant bacteria during anaerobic digestion of manure at thermophilic and mesophilic temperatures. J. Hazard. Mater. 384:121433. doi: 10.1016/j.jhazmat.2019.121433
Keywords: beta-lactam resistance genes, antibiotic resistant bacteria, exchangeable heavy metals, heavy metal resistome, waste utilization
Citation: Liu N, Li G, Su Y, Zhao Y, Ma J and Huang G (2023) Environmental drivers and interaction mechanisms of heavy metal and antibiotic resistome exposed to amoxicillin during aerobic composting. Front. Microbiol. 13:1079114. doi: 10.3389/fmicb.2022.1079114
Edited by:
Hongbo Li, Nanjing University, ChinaCopyright © 2023 Liu, Li, Su, Zhao, Ma and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Ma, am1hQGl1ZS5hYy5jbg==; Guangqun Huang, aHVhbmdncUBjYXUuZWR1LmNu
 Ning Liu1,2
Ning Liu1,2 Gang Li
Gang Li Ya Su
Ya Su Yi Zhao
Yi Zhao Jun Ma
Jun Ma