
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 28 December 2022
Sec. Evolutionary and Genomic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1078663
This article is part of the Research Topic Insights in Evolutionary & Genomic Microbiology: 2022 View all 11 articles
A correction has been applied to this article in:
Corrigendum: Out of the Qinghai-Tibetan plateau: origin, evolution and historical biogeography of Morchella (both Elata and Esculenta clades)
 Qing Meng1,2
Qing Meng1,2 Zhanling Xie1,2*
Zhanling Xie1,2* Hongyan Xu1,3
Hongyan Xu1,3 Jing Guo1,2
Jing Guo1,2 Yongpeng Tang4
Yongpeng Tang4 Ting Ma1
Ting Ma1 Qingqing Peng1
Qingqing Peng1 Bao Wang1,2
Bao Wang1,2 Yujing Mao1,2
Yujing Mao1,2 Shangjin Yan1
Shangjin Yan1 Jiabao Yang1,2
Jiabao Yang1,2 Deyu Dong1,2
Deyu Dong1,2 Yingzhu Duan4
Yingzhu Duan4 Fan Zhang5
Fan Zhang5 Taizhen Gao4
Taizhen Gao4Introduction: Morchella has become a research hotspot because of its wide distribution, delicious taste, and phenotypic plasticity. The Qinghai-Tibet Plateau subkingdoms (QTPs) are known as the cradle of Ice age biodiversity. However, the diversity of Morchella in the QTPs has been poorly investigated, especially in phylogenetic diversity, origin, and biogeography.
Methods: The genealogical concordance phylogenetic species recognition (GCPSR, based on Bayesian evolutionary analysis using sequences from the internal transcribed spacer (ITS), nuclear large subunit rDNA (nrLSU), translation elongation factor 1-α (EF1-α), and the largest and second largest subunits of RNA polymerase II (RPB1 and RPB2)), differentiation time estimation, and ancestral region reconstruction were used to infer Morchella’s phylogenetic relationships and historical biogeography in the QTPs.
Results: Firstly, a total of 18 Morchella phylogenetic species are recognized in the QTPs, including 10 Elata clades and 8 Esculenta clades of 216 individuals Secondly, the divergences of the 18 phylogenetic species were 50.24–4.20 Mya (Eocene-Pliocene), which was closely related to the geological activities in the QTPs. Furthermore, the ancestor of Morchella probably originated in the Northern regions (Qilian Shan, Elata cade) and southwestern regions (Shangri-La, Esculenta clade) of QTPs and might have migrated from North America (Rufobrunnea clade) via Beringian Land Bridge (BLB) and Long-Distance Dispersal (LDD) expansions during the Late Cretaceous. Moreover, as the cradle of species origin and diversity, the fungi species in the QTPs have spread out and diffused to Eurasia and South Africa starting in the Paleogene Period.
Conclusion: This is the first report that Esculenta and Elata clade of Morchella originated from the QTPs because of orogenic, and rapid differentiation of fungi is strongly linked to geological uplift movement and refuge in marginal areas of the QTPs. Our findings contribute to increasing the diversity of Morchella and offer more evidence for the origin theory of the QTPs.
As a famous edible mushroom, Morchella owns important ecological functions and has high commercial value around the world (Dissanayake et al., 2021; Wu H. et al., 2021; Yu et al., 2022). It was popular research in taxonomy, species diversity, distribution, ecological diversity, phylogeny, biogeography, and artificial cultivation of Morchella species (Annette et al., 1978; Dahlstrom et al., 2000; Hao et al., 2019; Liu et al., 2019; Ali et al., 2021; Cao et al., 2022; Deng et al., 2022). The distribution of Morchella exhibits a high level of cryptic speciation and provincialism due to phenotypic plasticity and unreliable morphological species recognition (O’Donnell et al., 2011; Du et al., 2012a, 2015, 2018; Richard et al., 2015;). There are 72 phylogenetically distinct species in the world that have been recognized in this genus based on GCPSR (Loizides et al., 2015, 2016; Yatsiuk et al., 2016; Baroni et al., 2018; Du et al., 2019). In China, which was known as the center of Morchella species diversification and rich floristic diversity, a total of 16 Elata clades and 27 Esculenta clades phylospecies have been recorded (Du et al., 2012a,b, 2019). The phylogenetic species diversity of the Morchella in the Qinghai-Tibet Plateau subkingdom (QTPs) is not yet known, though.
Great changes in crustal movement on earth occurred during the Phanerozoic Paleozoic (4.6 billion years ago), after occurring of biological explosions and forming of thick sedimentary limestone (Mittermeier et al., 2011; Kate, 2022). The QTPs have been uplifted steadily and rhythmically since the end of the Early Tertiary period, which is famous as the “Third Pole” of the earth (Mao et al., 2021; Spicer et al., 2021; Xiong et al., 2022); and were known as the ecological barrier of China and even Asia based on complex topography, variable plateau climate, and rich ecosystem (Sun et al., 2012; Qin et al., 2015; Miehe et al., 2019; Liu et al., 2022). Several fossil records of QTPs illustrated the cradle of mammalian fauna and mountain flora in the Ice age (Deng et al., 2020; Mao et al., 2021; Wu Y. D. et al., 2021). For fungus in the QTPs, the opportunities for genetic variation and speciation were strongly increased via the isolation of high-altitude geographic and geological complexity. Furthermore, the QTPs are also called a refugium created by microclimatic variations that provided some protection, and situ speciation and relic persistence in the early originated lineages (Shrestha et al., 2010; Yuan, 2015; Yan et al., 2017; Phonepaserd et al., 2019; Mao et al., 2021). The rapidly radiational differentiation of Morchella species was reported in North America, Asia, and Europe. M. rufobrunnea (Rufobrunnea clade), as the oldest taxon of the genus Morchella and might diverge into the basal lineage, originated in western North America in the late Jurassic (O’Donnell et al., 2011; Du et al., 2012a, 2015; Loizides et al., 2016, 2021). During the emergence of the Mid-Continental Seaway and the subsequent uplift of the Rocky Mountains, the ancestors of the Esculenta and Elata clades spread to eastern North America from western North America in the early Cretaceous (Sanmartín et al., 2001; Donoghue, 2008; Du et al., 2015). After that, Morchella experienced widespread extinction due to the new folding of the Rocky Mountains and the uplift of the Sierra Madre Oriental Range in central North America. It is also speculated that the Morchella species spread to Europe and Asia from North America via the Thulean North Atlantic Land Bridge and the Beringian Land Bridge (Du et al., 2015). During the middle Miocene to the Pleistocene, the Morchella species in East Asia and Europe rapidly evolved under the gradually cooling climate and environmental heterogeneity caused by the rise of the Qinghai-Tibetan Plateau (O’Donnell et al., 2011; Du et al., 2012a, 2015). However, further information regarding the differentiation, speciation, origination, and evolution of Morchella in the QTPs is still unclear.
In this study, ITS rDNA sequences of 174 Morchella individuals collected from QTPs were generated for the aims of (i) investigating phylogenetic species diversity and geographic distribution of Morchella in the QTPs by using genealogical concordance phylogenetic species recognition (GCPSR); (ii) estimating divergence times of Morchella species lineages in the QTPs; (iii) defining the geographic distributions of ancestor lineages of the Morchella in the QTPs; and (iv) estimating divergence times and reconstructing ancestral regions for world-widely distributed species of Morchella.
A total of 216 individuals were collected from three parts, among which 174 individuals were from the QTPs and 30 individuals were from Xinjiang and 12 individuals were from Jilin University. Total 7 natural localities approximately extending the whole distribution range of QTPs, ranging from 26.07°N–44.01°N to 81.60°E–105.05°E during the harvesting seasons (April–June). We divided the sampling sites into the higher latitude (35°N–40°N), the lower latitude (25°N–30°N), and the middle latitude (30°N–35°N) regions according to the north latitude lines (Figure 1). Of these, there are 56, 64, and 92 individuals were collected from high, low, and middle latitudes of the northeastern, central-eastern, and southwestern in the QTPs, respectively. Micromorphological data were obtained from the dried specimens and observed under a light microscope following Baran and Boroń (2017). Voucher specimens were deposited in Extreme Environment Microbiology Laboratory, Qinghai University, Xining, China. The codes, locations, sampling year, and sample numbers of Morchella from the QTPs are shown in Supplementary Table S1.
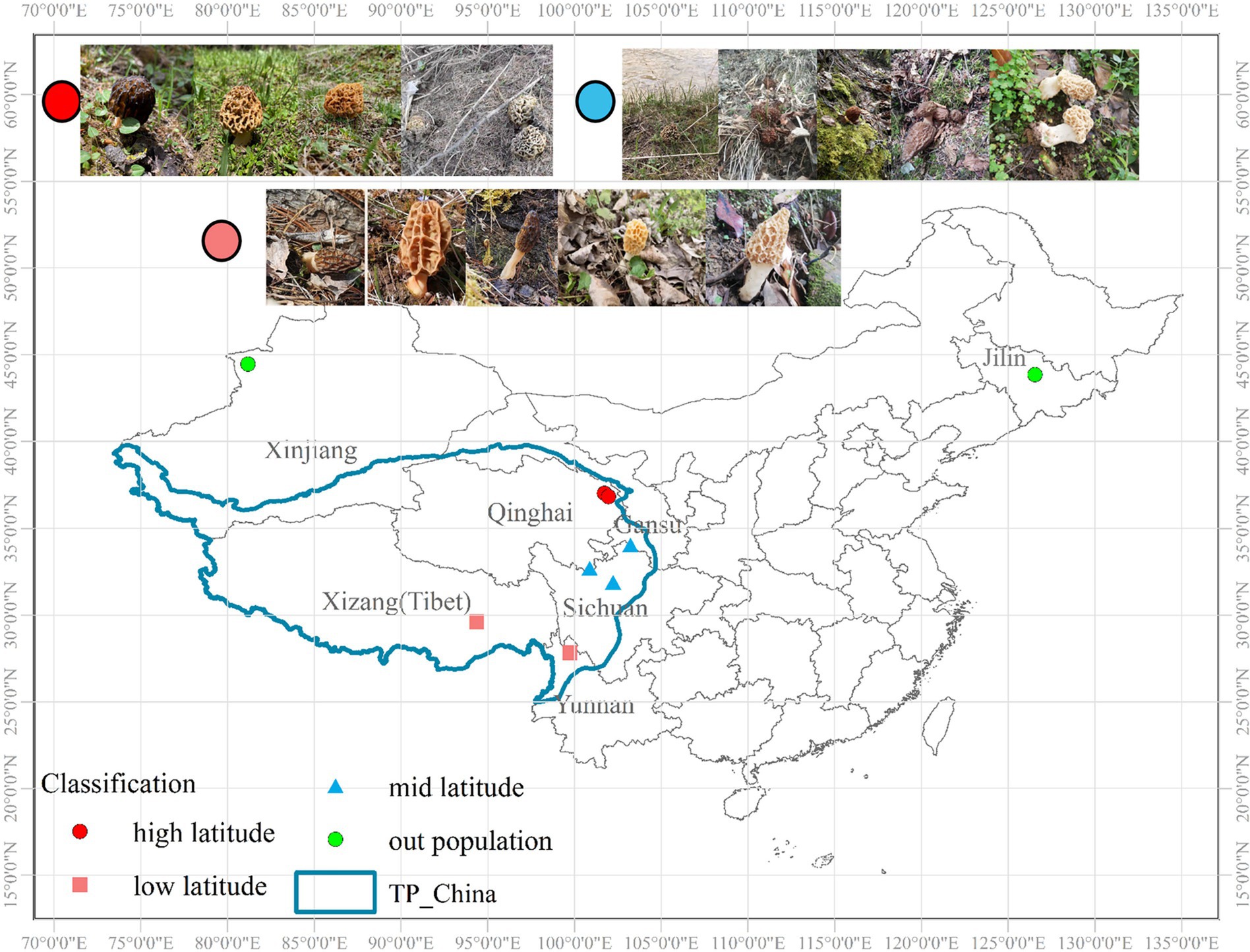
Figure 1. Geographical location and sampling site in Qinghai-Tibet Plateau subkingdom (QTPs) generated by ArcGIS v10.7. The red, pink, and blue sampling points are within the QTPs, where red is defined as the high-latitude region, blue is the middle-latitude region, and pink is the low-latitude region of the QTPs. The green sampling points are outside the QTPs.
Total extracted DNAs of each sample were extracted using the modified 2× CTAB buffer method (Doyle and Doyle, 1987), checked by 1% agarose gel electrophoresis with ethidium bromide staining, and measured the concentration by spectrophotometer (Biospec-nano; Shimadzu). Five DNA gene fragments were analyzed, including those coding for RNA polymerase II largest subunit (RPB1) and second largest subunit (RPB2), translation elongation factor-1a (TEF1), along with two non-protein coding regions: internal transcribed spacer (ITS), nuclear large subunit rDNA (nrLSU). The PCR amplifications was performed according to Du et al. (2019), Baran and Boroń (2017), and Loizides et al. (2021). The primers used for PCR are listed in Supplementary Table S2. A total of 894 sequences of QTP Morchella were newly generated, including 216 ITS, 201 nrLSU, 161 EF1-α, 166 RPB1, and 125 RPB2. The ITS sequences generated in this study were combined with the representative 117 ITS sequences retrieved from GenBank (including 46 representatives of Morchella species recorded in NCBI) to identify the relationships between all of our individuals and the known related samples in GenBank. All newly generated sequences were submitted to GenBank (Supplementary Table S1).
For phylogenetic analyses, the ITS gene datasets were analyzed via Maximum likelihood (ML) and Bayesian inference (BI): a 110-taxon, 864 bp Esculenta Clade data set; and (ii) a 101-taxon, 612 bp Elata Clade dataset. Five-gene datasets were analyzed via Maximum parsimony (MP), ML, and BI: (i) a 14-taxon, 3,380 bp Esculenta Clade data set; and (ii) a 24-taxon, 2,517 bp Elata Clade dataset. The genes extracted and aligned were using the MAFFT (version 7; Katoh and Standley, 2013). The conservative region was selected in Gblocks 0.91b and the vacancy gap in the data were treated as missing data. We performed MP, ML, and BI based on the combined sequences of five genes to reconstruct the relationships of Morchella and related taxa were subsequently conducted on PAUP∗ version 4.0 beta 10 (Swofford, 2002) and PholySuite v1.2.2 (Zhang et al., 2020). For the MP analysis was performed in PAUP∗ version 4.0 beta 10 (Swofford, 2002). All characters were equally weighted, and gaps were treated as missing data. Trees were inferred using the heuristic search option with TBR branch swapping and 1,000 random sequence additions. The PholySuite v1.2.2 contains programs for sequence alignment and phylogenetic analysis, such as ModelFinder to find the best model. For the ML analyses, all parameters were kept at their default settings, the concatenated dataset was partitioned into five parts by sequence region, and 1,000 Ml searches under the K80 (K2P) model with all model parameters estimated using the ModelFinder program, IQ-TREE v1.6.8 web server1 to carry out the ML searches. The MrBayes v3.2.6 for BI phylogenetic analyses also used ModelFinder to generate the best model (Ronquist et al., 2012; Du et al., 2012a; Zhu et al., 2019; Zhao et al., 2021). The phylogenetic trees were modified using FigTree v1.4.4 and the iTOL website.2
we used BEAST v.2.6.6 (Chen et al., 2015; Zhu et al., 2019; Kim and Kim, 2022) to estimate the divergence times of Morchella phylospecies in the QTPs. In this study, we used Floccularia luteovirens, which is the endemic basidiomycete fungus in the QTPs, instead of P. devonicus as the calibration point 1 in Basidiomycota and Ascomycota (Taylor et al., 2004; Chen et al., 2015; Zhu et al., 2019; Guo et al., 2022). Normal distribution was applied by setting the mean and the standard deviation to 582.5 and 50.15, respectively. Calibration points 2 for analysis were obtained by including sequences of the following two species: Verpa and M. rufobrunnea (O’Donnell et al., 2011; Du et al., 2012a, 2015; Loizides et al., 2016, 2021). The origin time of Morchella was estimated in BEAST v.2.6.6 (Drummond and Rambaut, 2007) with the molecular clock and substitution models unlinked but with the trees linked for each gene partition. Two nuclear ribosomal RNA genes (ITS and nrLSU) and three protein-coding genes (EF1-α, RPB1, and RPB2), were concatenated for molecular dating. PholySuite v1.2.2 was also used to select the best models of evolution using the hierarchical likelihood ratio test. The GTR + I + G model was used for the EF1-α + RPB1 + RPB2 and the HKY + I + G model for the ITS + nrLSU data, based on the results from the PholySuite v1.2.2. The uncorrelated lognormal relaxed molecular clock and the Yule speciation prior set were used to estimate the divergence time and the corresponding credibility intervals by BEAUti 2. The Markov chain Monte Carlo (MCMC) analysis was 100 million generations, sampling parameters for every 1,000 generations. After discarding the first 10,000 (10%) trees as burn-in, the samples were summarized in a maximum clade credibility tree in TreeAnnotator v2.6.6 using a PP limit of 0.50 and summarizing the mean node heights. The means and 95% higher posterior densities (HPDs) of age estimates were obtained from the combined outputs using Tracer. FigTree v1.4.2 and iTOL website3 was used to visualize the resulting tree and to obtain the means and 95% HPD. A 95% HPD marks the shortest interval that contains 95% of the values sampled.
Ancestral area reconstruction and estimating spatial patterns of geographic diversification within Morchella in the QTPs were inferred using the Bayesian binary method (BBM) and statistical dispersal-vicariance analysis (S-DIVA) as implemented in Reconstruct Ancestral State in Phylogenies (RASP v3.1). The distribution range of the Elata clade in the QTPs was divided into five regions, consisting of A (Qinghai), B (Tibet), C (Gansu), D (Xinjiang), and E (Other). And the distribution range of the Esculenta clade in the QTPs was also divided into five regions, consisting of (A) Qinghai, (B) Gansu, (C) Sichuan, (D) Yunnan, and E (Other). For the BBM analysis, we used all post-burn-in trees obtained from the BEAST v2.6.6 analysis. The BBM was run using the fixed state frequencies model (Jukes-Cantor) with equal among-site rate variation for 50,000 generations, 10 chains each, and two parallel runs. In statistical dispersal-vicariance analysis (S-DIVA), the frequencies of an ancestral range at a node in ancestral reconstructions are averaged over all trees. In addition, the world-widely distribution of 8 Morchella species (Supplementary Table S2), M. spongiola, M. esculenta, M. crassipes, M. eohespera, M. eximia, M. costata, Mel-13, Mel-14, were downloaded in NCBI to estimate the differentiation time and reconstruction ancestral area using BEAST v2.6.6 and RASP v3.1, respectively. ArcGIS v10.7 was used to visualize the geographic distribution and possible dispersal routes of Morchella (Yu et al., 2015; Kim et al., 2019; Kim and Kim, 2022).
A total of 216 individuals of Morchella were classified into Esculenta and Elata clades, there is no Rufobrunnea clade. A total of 101 individuals clustered with 10 phylogenetic species, including Mel-14, M. deliciosa/Mel-13, M. norvegiensis = M. eohespera/Mel-19, Morchella eximia/Mel-5, Morchella costata, Morchella sextelata/Mel-6, Morchella septimelata/Mel-7, Morchella purpurascens/Mel-20, Mel-33, and Morchella pulchella/Mel-31 belongs to Elata clade (Figures 2A–C); and a total of 101 individuals clustered with 8 phylogenetic species, including M. vulgaris = M. spongiola/Mes-5, Mes-9, Mes-12, Mes-26, Morchella crassipes, Morchella esculenta/Mes-8, Mes-19, and Mes-6 belongs to Esculenta clade (Figures 3A–C). All Morchella species also exhibit extreme bradytelic morphological evolution as evidenced by the retention of the ancestral ascocarp body plan (Figures 2D, 3D). Overall, we identified 18 phylospecies that was widely distributed in the QTPs.
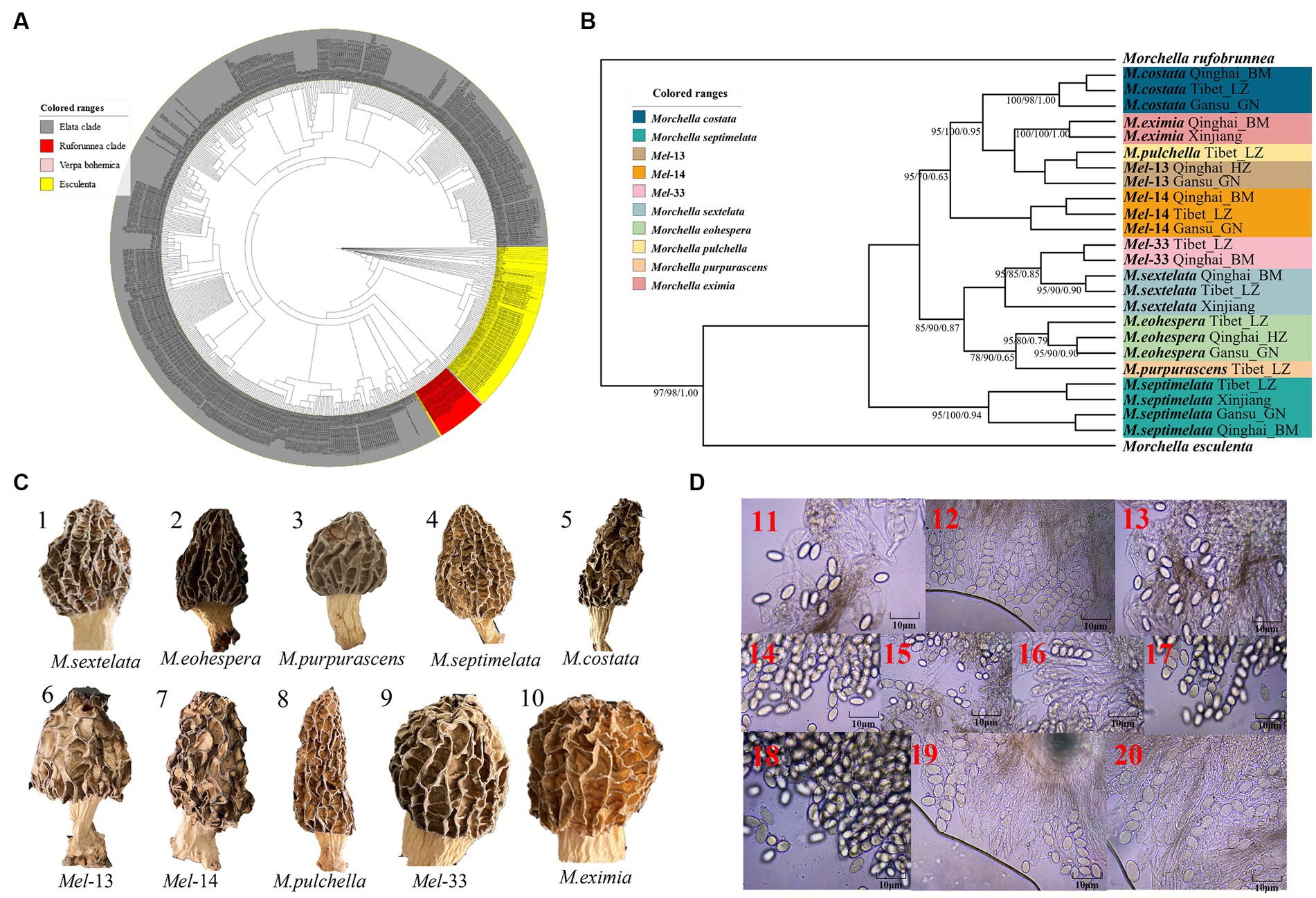
Figure 2. Species recognition of the Morchella in Elata clade from the QTPs. (A) Bayesian inference phylogenetic analyses of the Elata clade were inferred from 115 internal transcribed spacer (ITS) sequences representing a total of 10 phylospecies. (B) Phylogenetic analyses of the Elata clade were inferred from 120 (24*5) multi-genes (ITS+LSU + EF1-α + RPB1 + RPB2) sequences representing a total of ten phylospecies. Branches are labeled where MP/ML support is greater than 60% and collapsed below that support threshold. BPP is labeled were greater than 0.95. (C) Morphological diversity of the 10 Elata clades’ ascocarps from the QTPs: M. sextelata/Mel-6 (1), M. norvegiensis = M. eohespera/Mel-19 (2), M. purpurascens/Mel-20 (3), M. septimelata/Mel-7 (4), M. costata (5), M. deliciosa/Mel-13 (6), Mel-14 (7), M. pulchella/Mel-31 (8), Mel-33 (9), M. eximia/Mel-5 (10). (D) Micromorphological ascospores of the 10 Elata clades.
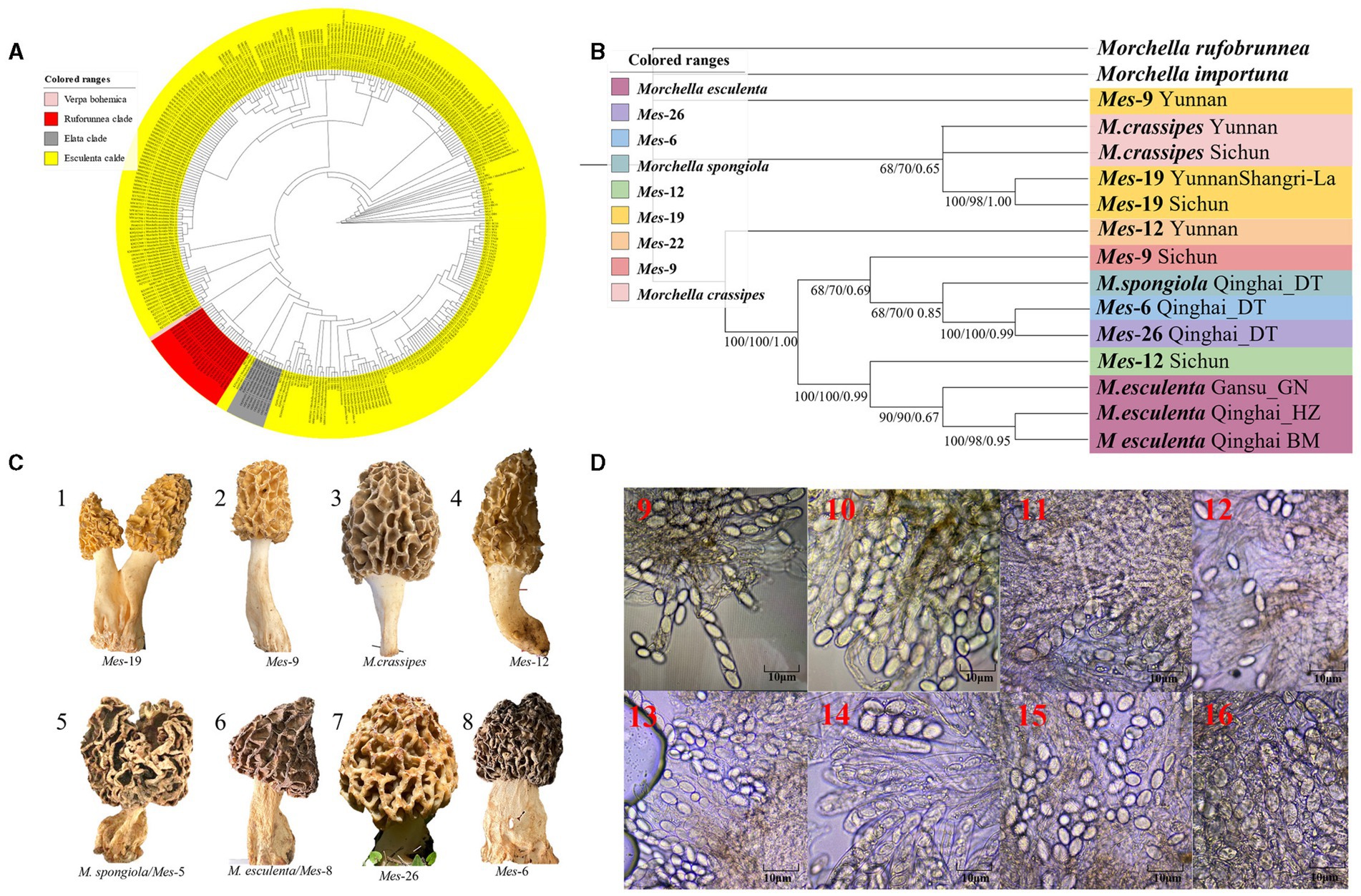
Figure 3. Species recognition of the Morchella in Esculenta clade from the QTPs. (A) Bayesian inference phylogenetic analyses of the Elata clade were inferred from 101 ITS sequences representing a total of 8 phylospecies. (B) Phylogenetic analyses of the Esculenta clade were inferred from 70 (14*5) multi-genes (ITS+LSU + EF1-α + RPB1 + RPB2) sequences representing a total of 8 phylospecies. Branches are labeled where MP/ML support is greater than 60% and collapsed below that support threshold. BPP is labeled were greater than 0.95. (C) Morphological diversity of the 8 Esculenta clades’ ascocarps from the QTPs: Mes-19 (1); Mes-9 (2), M. crassipes (3), Mes-12 (4), M. vulgaris = M. spongiola/Mes-5 (5), M. esculenta/Mes-8 (6), Mes-26 (7), Mes-6 (8). (D) Micromorphological ascospores of the 8 Escuenlta clades.
The divergence time of 18 Morchella phylospecies in the QTPs ranged from 50.24 to 4.20 Mya (Figure 4; Table 1). The earliest diverging branch of Morchella in the QTPs was represented by the monotypic M. rufobrunnea (originated in North America) lineage with an estimated divergence time of 154.15 Mya (95% HPD interval: 152.14–156.08); the second diverging branch of Esculenta and Elata clades at 98.63 Mya (95% HPD interval: 97.30–100.0); the third evolutionary diversification of the Elata Clade was dated at 57.94–68.97 Mya (95% HPD interval: 30.09–69.04; 40.17–98.45) and the Esculenta Clade at 20.41–20.83 Mya (95% HPD interval: 6.18–45.22; 2.62–52.67). The phylospecies of M. norvegiensis = M. eohespera/Mel-19, M. deliciosa/Mel-13, Mel-14, Morchella eximia, M. costata, M. esculenta/Mes-8, M. crassipes, and Mes-19, were estimated at 50.24 Mya (95% HPD interval: 40.17–98.45), 40.63 Mya (95% HPD interval: 18.95–63.83), 36.41 Mya (95% HPD interval: 2.83–66.27), 25.87 Mya (95% HPD interval: 4.77–48.44), 14.24 Mya (95% HPD interval: 6.18–45.22), 4.20 (95% HPD interval: 0.02–19.54), respectively. In summary, Morchella phylospecies in QTPs has maintained a very diversified evolutionary history during the Eocene and Pliocene, when the historical geological uplift and geological tectonic movement were experienced in the QTPs (Figure 5).
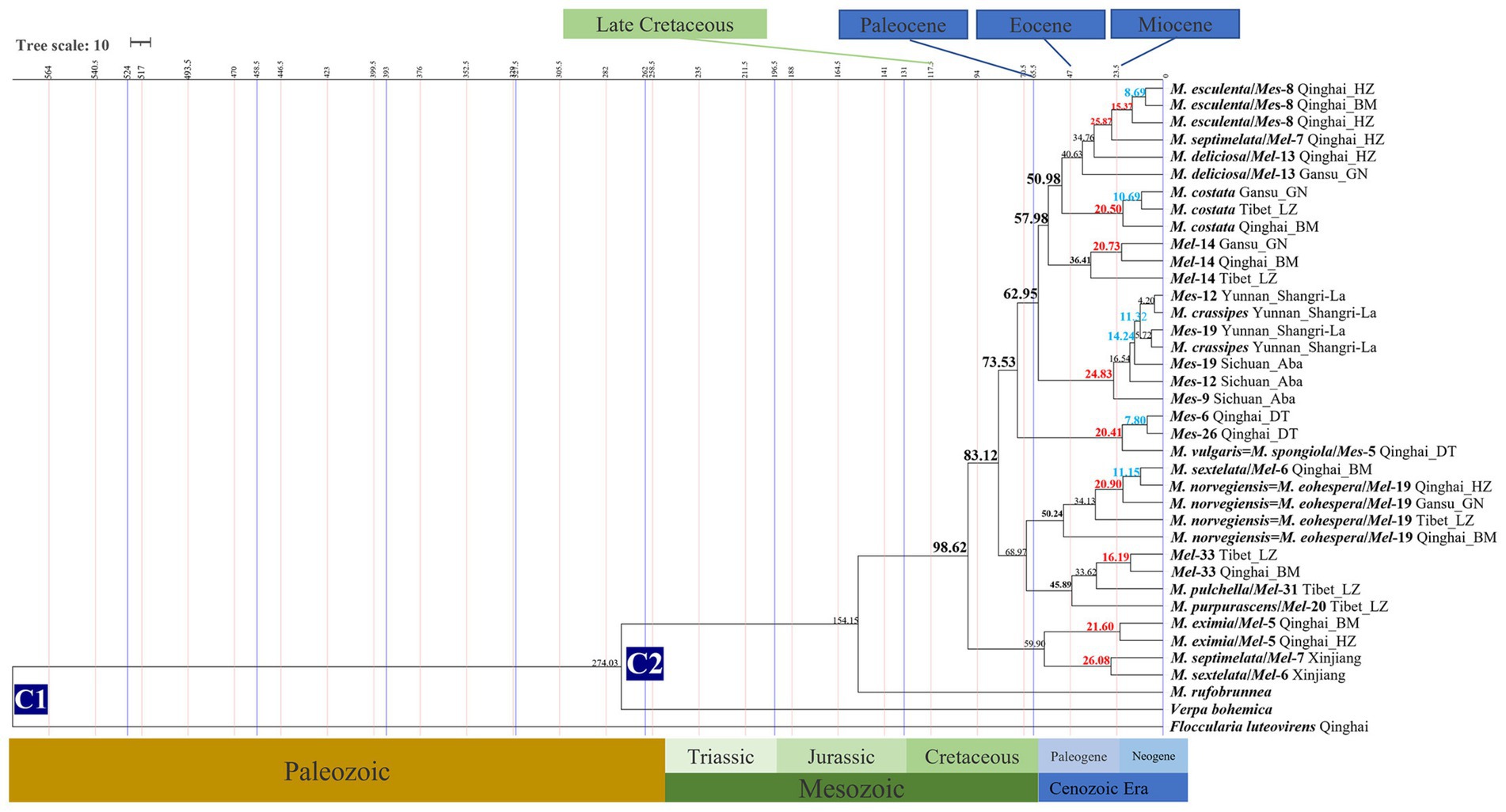
Figure 4. Chronogram and estimated divergence times of Morchella in QTPs generated by molecular clock analysis using the two concatenated datasets (ITS + LSU and EF1-α + RPB1 + RPB2) dataset. The chronogram was obtained using the Ascomycota-Basidiomycota divergence time of 582.08 Mya as the calibration point 1. The Morchella-Verpa bohenica divergence time of 274.06 Mya as the calibration point 2. The calibration point and objects of this study are marked in the chronogram. The geological time scale is millions of years ago (Mya). The red font is defined as the first uplift of the QTPs, and the blue font is defined as the second uplift of the QTPs.
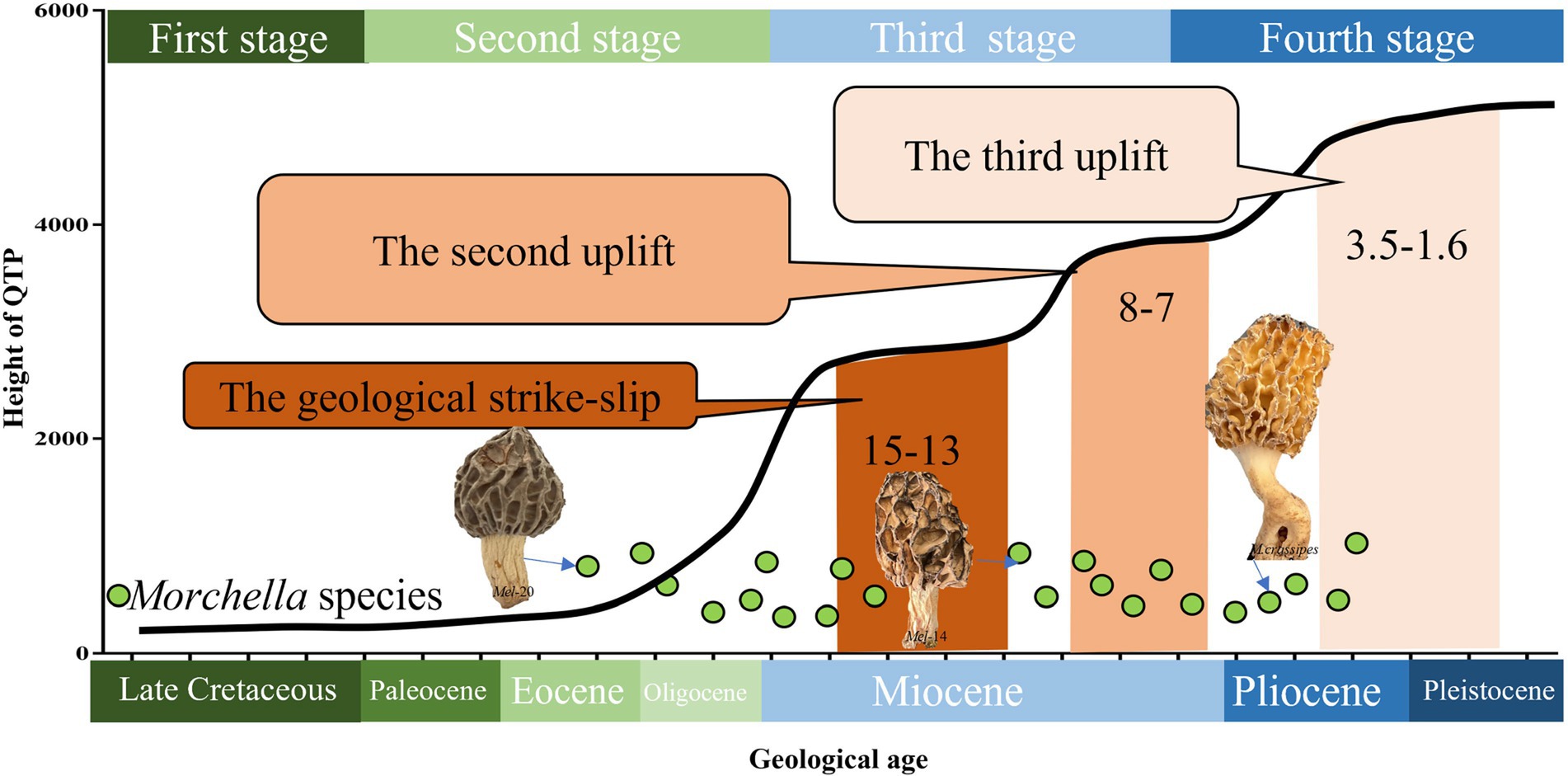
Figure 5. Species differentiation of Morchella related to the stage of plate movement and uplift on the QTPs from the Late Cretaceous to the present. The divergence time of Morchella phylospecies in QTPs has remained remarkably diverse during its long evolutionary history, ranging from 50.24 to 4.20 Mya (Eocene-Pliocene), when the historical geological uplift and geological tectonic movement were experienced in the QTPs. The plate activity and plateau uplift of QTP resulting from the collision are divided into four stages: (1) Late Cretaceous in the first stage marked as dark green; (2) Tertiary Period in the second stage marked as light green; (3) Neogene in the third stage marked as light blue; (4) Quaternary Period in the fourth stage marked as dark blue. The green dots represent the Morchella individual.
There were 19 dispersal events and 19 vicariance events that could explain the current distribution of the Morchella phylospecies in the QTPs. For the Elata clade, region A (Qilian Shan, Qinghai) located in the eastern part of QTPs, has the highest probability (50.08%) of being the ancestral area during the Eocene (Figure 6A). For the Esculenta clade, region D (Shangri-La, Yunnan), which was located in the southwestern part of QTPs, has the highest probability (77.48%) of being the ancestral area during the Miocene (Figure 6B). In general, based on the phylogeographic structures of the 8 subclades, 3 distribution patterns can be summarized: (i) the wide distribution around the QTPs, such as the M. deliciosa/Mel-13, Mel-14, M. norvegiensis = M. eohespera/Mel-19, suggests that not all of the Morchella species were narrowly distributed; (ii) the long-distance dispersal with latitude-based structure. (iii) Multi-origin: the two clades have different origins, the Easculenta clade originated from Shangri-La while the Esculenta clade originated from Qilian Shan (Figure 7).
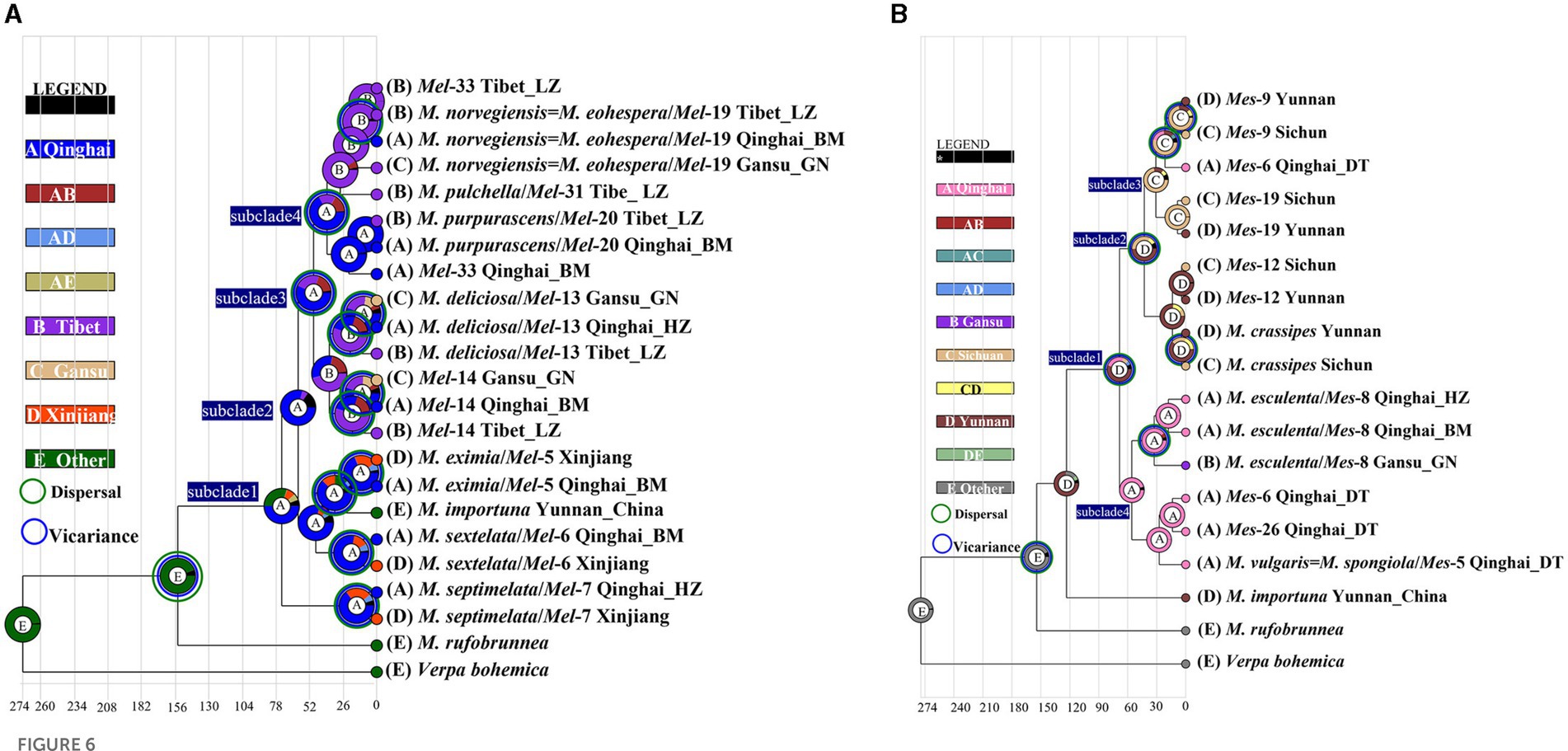
Figure 6. Ancestral area reconstruction of Morchella phylospecies in QTPs using the ITS dataset. The chronogram was obtained by molecular clock analysis using BEAST. The pie chart in each node indicates the possible ancestral distributions inferred from Bayesian Binary MCMC analysis (BBM) implemented in RASP. Bayesian credibility values (PP) over 0.85 are indicated near the pie chart of the tree. The green circle around the pie charts indicates possible dispersal events, the blue circle indicates possible vicariance events as suggested by BBM analysis. (A) Elata clade; (B) Esculenta clade.
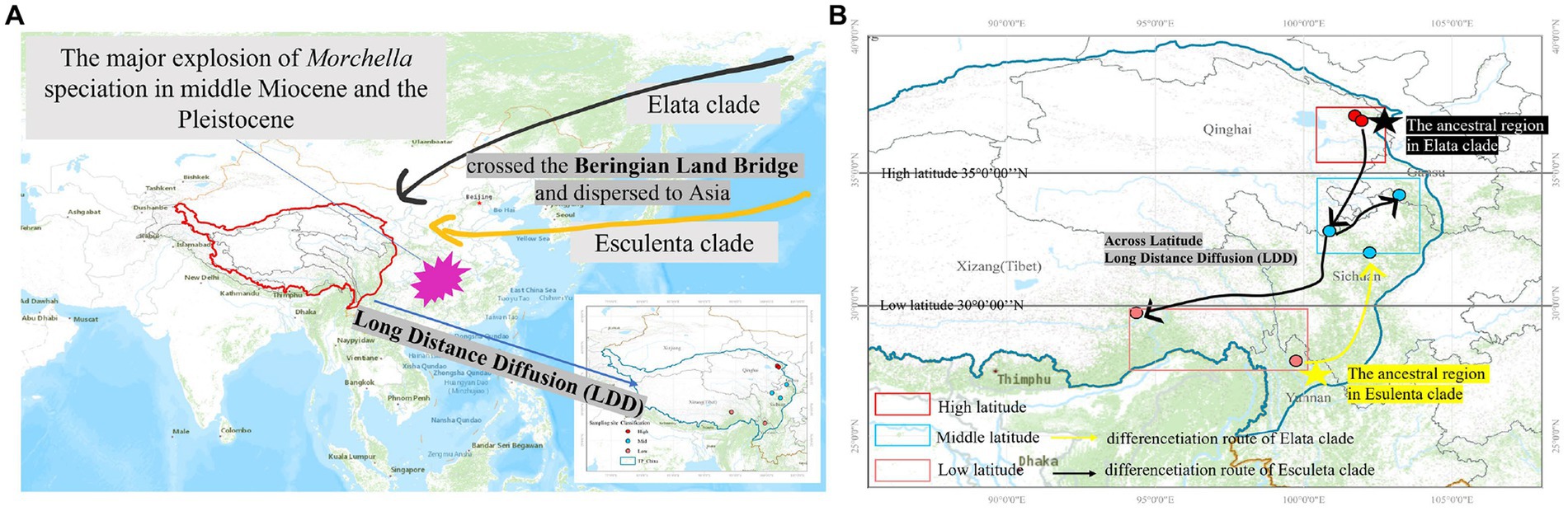
Figure 7. Map of the geographic distribution of Morchella and possible dispersal routes in QTPs generated by ArcGIS v10.1. (A) The major explosion of Morchella speciation in the middle Miocene and the Pleistocene crossed the Beringian Land Bridge and dispersed to Asia. (B) A hypothetical schematic depiction of the original locations, the migration routes the speciation of Morchella in QTPs.
For the Morchella genus, there is no Rufobrunnea clade on any continent except North America, and most of the species that are widely distributed in the continental region (besides Oceania) have the earliest divergence time in the QTPs, the ancestor of Morchella both the Esculenta and Elata probably originated in QTPs and initially covered Eurasian and South Africa in the Early Tertiary (Figure 8; Table 2).
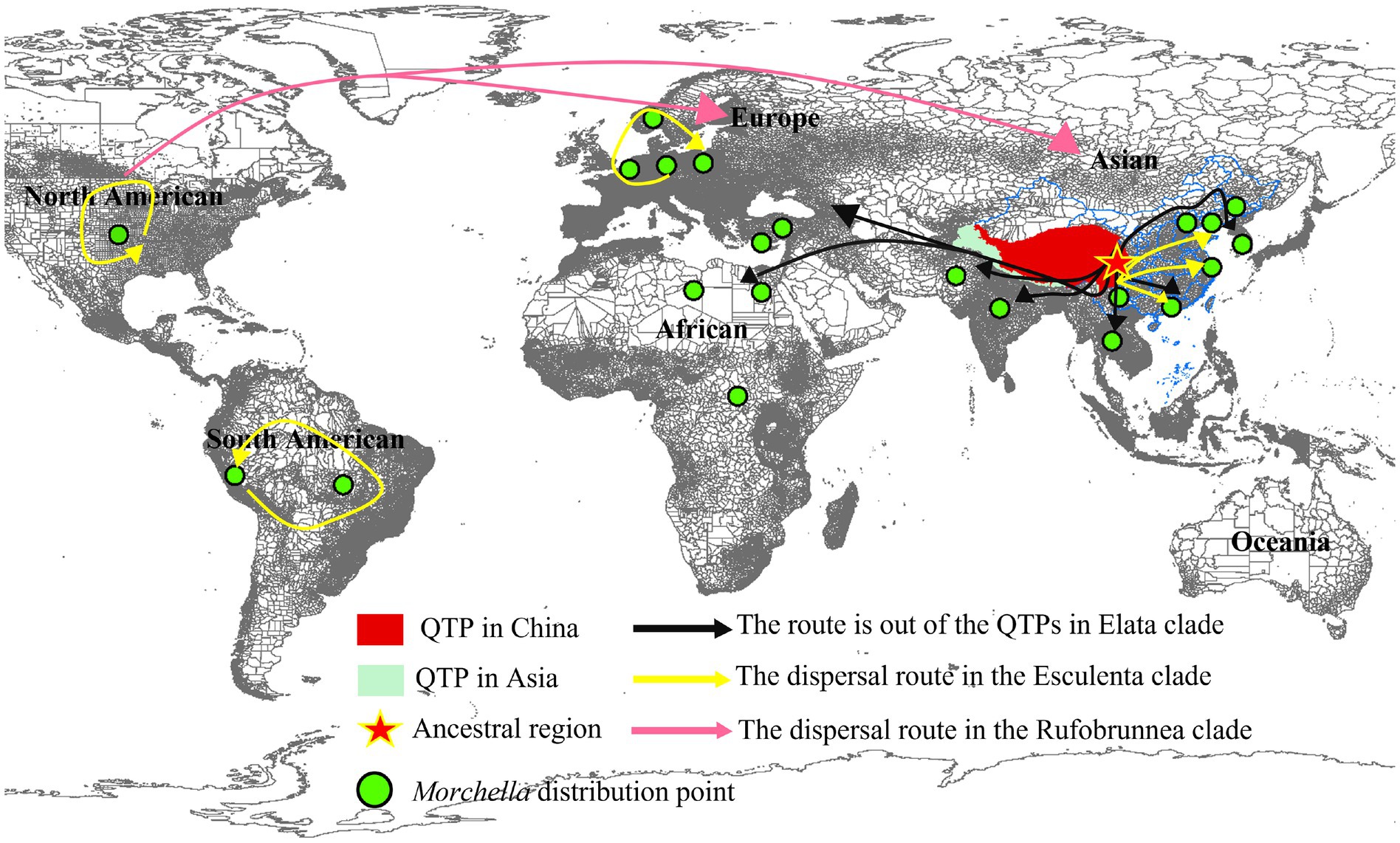
Figure 8. The origin of the QTPs and the route out of the QTPs in Morchella. Morchella originated from Qilian Shan and spread along the Himalayas on the edge of QTPs to surrounding Europe and South Africa in the Elata clade (Black route). Each continent has its ancestor in the Esculenta clade (Yellow route).
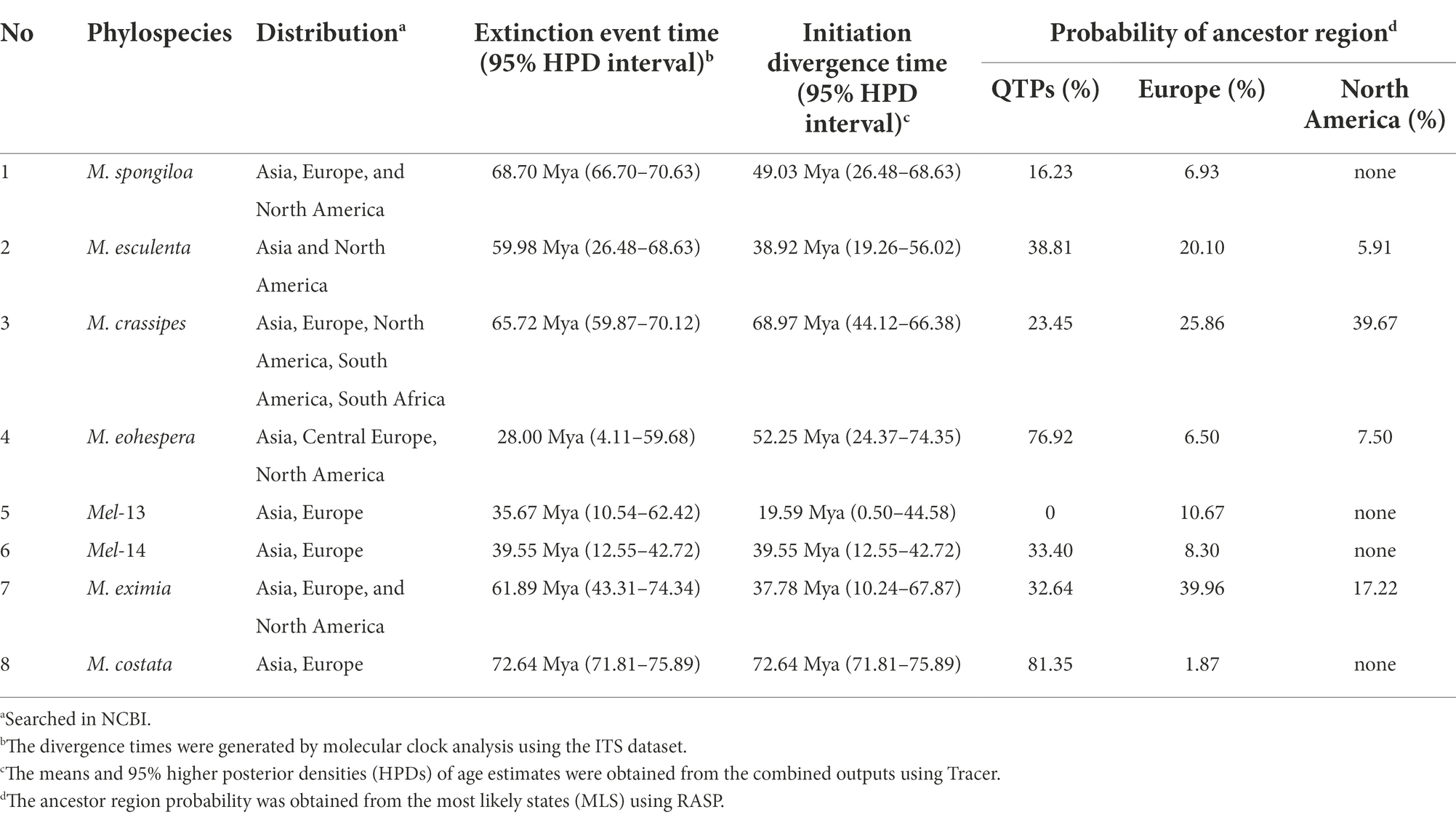
Table 2. Species initiation differentiation time and ancestor region reconstruction of worldwide Morchella species.
For the Esculenta clade: (1) M. vulgaris = M. spongiola/Mes-5 there were distributed in central Europe and Asia, and the initial differentiation of M. spongiloa in these two areas was 28.88 Mya (95% HPD interval: 4.11–59.68) and 49.03 Mya (95% HPD interval: 26.48–68.63), respectively. They went through an extinction event between Asia and Europe. QTPs of Asia and Germany of Europe were most likely ancestral regions. However, the Asian phylospecies diverged 11 Mya earlier than the European species, and the QTPs were the ancestral region of M. spongiloa (Table 2; Supplementary Figure S1). (2) M. esculenta/Mes-8 was widely distributed in Europe, North America, and Asia, among them, the earliest divergences of this species occurred in China at 38.92 Mya (95% HPD interval: 19.26–56.02). That phylospecies experienced one extinction event in the distribution area and the highest probability (38.81%) of being a putative ancestral region is the QTPs (Table 2; Supplementary Figure S2). (3) M. crassipes is a widely distributed species in Asia, Europe, North America, and South Africa. The earliest differentiation of this species is Shanghai during the 68.97 Mya (95% HPD interval: 67.04–70.93), but North America has the highest probability (39.67%) of being a putative ancestral region (Table 2; Supplementary Figure S3).
For Elata clade: (4) M. norvegiensis = M. eohespera/Mel-19 (Elata clade), were distributed in Central Europe, Asia North America. The initial divergence time in the QTPs is 52.25 Mya (95% HPD interval: 24.37–74.33), which is earlier than in North America and Central Europe [15.81Mya (95% HPD interval: 0.13–49.54), 9.79Mya (95% HPD interval: 0–36.55)]. The QTPs have the highest probability (76.92%) of being the ancestral area of M. eohespera and the dispersal events and extinction events also occurred in the continental range (Table 2; Supplementary Figure S4). (5) M. deliciosa/Mel-13 and Mel-14 are widely distributed in Eurasia, especially in the QTPs. M. deliciosa/Mel-13 and M. importuna diverged at 35.67 Mya (95% HPD interval: 21.92–72.25), while Mel-14 and M. importuna diverged at 39.55Mya (95% HPD interval: 12.05–72.82). Europe is the putative ancestral region of Mel-13 (10.67%) while QTPs have the highest probability of being an ancestral region of Mel-14 (33.40%), and they all experienced one extinction event (Table 2; Supplementary Figure S5). (6) M. eximia are widely distributed in America, Europe, and Asia, which were estimated at 16.56 Mya (95% HPD interval: 10.23–67.80) in QTPs (Supplementary Figure S6). (7) M. costata were widely distributed in Eurasia. The initial differentiation in QTPs was 40.53 Mya (95% HPD interval: 42.31–71.50), which has the highest probability (81.35%) of being an ancestral region of M. costata (Table 2; Supplementary Figure S7). Overall, six of the eight worldwide Morchella originated in the QTPs, the ancestral region of 60% (3/5) of the Elata clade and 66.66% (2/3) of the Esculenta clade were the QTPs; therefore, it is possible that QTPs was the center of origin for the current Morchella species diversity. Morchella originated from the QTPs and expanded out and spread to the other continents in Paleogene Period (Figure 8).
This is the first time report the phylospecies diversity of Morchella in the QTPs used GCSPR, and the results indicate that there is 18 phylospecies of Morchella in QTPs, which far exceeds the number of published taxa for these areas (Du et al., 2012b, 2016, 2019). Generally, both Esculenta and Elata clades have preferred habitats, known as phylogenetic niche conservation (PNC; Donoghue, 2008). We investigated the occurrence and preferred habitats of Morchella in the QTPs and found that occurred in primary forests with high vegetation coverage, deciduous forests, or mixed coniferous and broad-leaved forests, such as the primary forests of Qilian Shan, Belong River, Mote, and Shangri-la, all of which belong to the eastern or northwestern margin of the QTPs. In addition, about 90% of the species are found in temperate deciduous forests, while 80% of the species were found in coniferous forests (Figure 1). It can be inferred that the occurrence of Morchella in the QTPs was accompanied by the establishment of the temperate deciduous biome and coniferous biome in the late Cretaceous (O’Donnell et al., 2011; Du et al., 2012a, 2015; Chen et al., 2015).
The maximum crown age of Morchella in the QTPs was estimated to be around the Late Cretaceous (98.62 Mya). The majority of intracontinental range expansions within Morchella in the QTPs appear to have taken place relatively recently between the middle Oligocene and Miocene. During that time, the uplift of the southwest block (Yunnan-Guizhou Plateau) and the eastern margin of the QTP resulted in the distribution of plant diversity in this region. Previous studies have shown that the Beringian Land Bridge (BLB) plays a key role in the spread of Morchella species from North America to Asia in the early Cretaceous, which was a natural route linking Eurasia and North America (O’Donnell et al., 2011; Du et al., 2012a, 2015; Loizides et al., 2016, 2021). Palaeobotanical data indicate that the BLB route played a crucial role in plant dispersal for Populus, Lonicera, Leibnitzia, and Linnaea (Wang et al., 2009; Qiu et al., 2011; Wen et al., 2014; Du et al., 2015; Wang et al., 2015; Wu et al., 2017). The divergence events exhibited by the plant taxa (Pinus, Abies, and Picea) confirm the geological events associated with species diversification, and those indigenous florae provide native secluded habitats for ancestral species of Morchella in QTPs (O’Donnell et al., 2011; Loizides et al., 2021). The Beringian Land Bridge as a species expansion channel is a critical driver of Morchella migrating to the QTPs between the Cretaceous and Paleocene when plant diversity was already established on QTPs, which means that the QTPs had formed a variety of suitable habitats for living organisms before in the Paleocene.
The differentiation of the Morchella is strongly linked to the geological movements of the QTPs (Figure 5). Palaeogeographical evidence suggests that plate activity and plateau uplift of QTP resulting from the collision are divided into four stages (Shi et al., 1998; Ding et al., 2022). (i) Late Cretaceous, the ancient Mediterranean crust subducted to the Eurasian continental crust, blowing the prelude to the plateau uplift. At this stage, Morchella of western North America is diverging from its closest relatives in the early Cretaceous, which are divided into Rufobrunnea, Esculenta, and Elata clades, and palaeogeological events played a driving role in the dispersal of Morchella (O’Donnell et al., 2011). (ii) Tertiary Period (Including the Paleocene, Eocene, and Oligocene), the Indian plate collided with the Eurasian plate, opening the prelude to the plateau uplift. The Indian plate rapidly drifted northward, the Indo-Pak subcontinent and the subcontinent were getting closer, and the ancient Mediterranean crust gradually disappeared during the Oligocene. In our data, M. norvegiensis = M. eohespera/Mel-19 was differentiated at 50.24 Mya with the new uplift belts of Tengchong-Bango formatted and the uplift area of Songpan-Ganzi shrank to the east during Eocene (Wang et al., 2008, 2022). Eocene (55.8 ± 0.2) was a key period for Asian paleoenvironmental changes and was characterized by a warmer climate than any other interval in the Cenozoic (Shi et al., 1998; Zachos et al., 2001; Hoorn et al., 2012). M. norvegiensis = M. eohespera/Mel-19 in the middle latitudes region were differentiated at 34.24 Mya with the further uplifted of Kunlun-Algin-Qilian during the Oligocene, which has been called the beginning of the present ‘icehouse’ epoch (Ling et al., 2021; Dieter et al., 2022). (iii) Neogene, the Indian and Tarim plates compressed and subducted to the QTPs with greater stress, and the plateau was greatly uplifted, forming the QTPs and the Himalayas. Our results indicate that 9 of the 18 QTPs Morchella species lineages (i.e., 50%) diversified between the middle Miocene and the present (Figure 4). (iv) Quaternary Period, the plateau was greatly uplifted, forming the present towering QTP. The Esculenta clade of Morchella in the QTPs was undergoing differentiation at this time. Tectonic activity and climate change in geological periods can form geographical isolation barriers to promote species differentiation and increase diversity, and can also reduce biological diffusion barriers to expand species distribution areas and increase biological exchanges between different regions (Che et al., 2010; Antonelli et al., 2018; Rahbek et al., 2019; Ding et al., 2020). Generally speaking, the formation and evolution of Morchella phylospecies in the QTPs were affected by the tectonic uplift of the QTPs and the geological movements of QTPs were an important force for the differentiation of the Morchella species (Merckx et al., 2015; Dai et al., 2019). The early marginal geological movement of the QTPs caused strong habitat fragmentation and rapid expansion of dry and cold habitats, which further strengthened the monsoon climate in East Asia and promoted the differentiation of microorganisms in the QTPs (An et al., 2001; Yang, 2005; Wan et al., 2014; Liu et al., 2016).
At least three geographic distribution patterns were discovered to correspond to the following phylogeographic structures. (i) the wide distributions around the QTPs: geographic locations of 4 endemic species, such as Mel-13, Mel-14, Mel-19, and M. esculenta, cross over three regions (Qinghai, Gansu, and Tibet in high, middle, and low latitudes respectively) of the QTPs. (ii) provincialism in the QTPs: the specific local distributions of two species in the Elata clade (M. pulchella/Mel-31, M. purpurascens/Mel-20) were unique (only in the Tibet region). (iii) the long-distance dispersal with latitude-based distributions: the divergence of Mel-13 and Mel-14 experienced two dispersal and vicariance events in three regions of the QTPs (Figure 6A). Mel-13 diverged in the mid-latitudes at 40.63 Mya and subsequently differentiated to the higher-latitudes at 34.76 Mya, which was based on latitude long-distance dispersal. Mes-19 also experienced dispersal and vicariance events in mid-latitudes and lower-latitudes, although this species originated from low latitudes (Yunnan) of the southwestern QTP (Figure 6B). To sum up, long-distance dispersals based on latitudes may have contributed to the current disjunct distribution ranges of Morchella, which is supported by biogeographic studies of plants and animals on the QTPs (Figure 7). The spread and differentiated way of Morchella in the global range, including intercontinental, intracontinental, and putative transoceanic long-distance dispersals, was slightly different from our result in QTPs (Du et al., 2012; Quan et al., 2014).
The complex topography and a special eco-climate of QTPs provide multiple periglacial microrefugia for Morchella. The Qilian Shan, located in the northeastern of the QTP, was defined as the ancestral region of the Elata clade as early as the Eocene. The Qilian Shan is an intraplate orogenic belt that experienced multiple episodes of fold and thrust deformation throughout the Mesozoic and Cenozoic periods, which controlled the evolution of regional climatic conditions in a broad region in inland Asia since the Miocene (Jia et al., 2022). The Shangri-La region in the southeast of the QTPs, which was the ancestors’ region for the Esculenta clade of Morchella, not only served as an important glacial refugium but also as a center of diversification for a variety of plants and animals (Suzuki et al., 2003; Yang et al., 2008, 2009; Harrison and Noss, 2017). The QTPs contain several important biodiversity hotspots, particularly along its southeastern margin (the Henduan Mountains and the eastern Himalayas), which were proposed to be glacial refugia for alpine hepialid species that effectively avoided extinction during the Quaternary glacial period (Hewitt, 2000; Dai et al., 2019).
Multiple pieces of evidence suggest that the Elata clade originated from QTPs, whereas the origin of the Esculenta clade was diverse. Our results show that the worldwide distribution species was an initial differentiation time of 73.78 Mya–35.67 Mya earlier than Central Asia, Europe, and South Africa (Figure 8). During the Early Tertiary (65.5–23.03 Mya), the Indian plate collided with the Eurasian plate opening the prelude to the plateau uplift, which is the emergence time of modern organisms (Liu and Hu, 2003). For example, in M. norvegiensis = M. eohespera/Mel-19, the divergence time was estimated at 52.25 Mya, which was earlier than that of Europe (Supplementary Figure S4). The Morchella was out of QTPs probably because of Eurasia’s collision and plate migration. On the other hand, the probability of QTPs as the most likely ancestral region for these widely distributed species is greater than that of Europe, North America, and South Africa (Table 2). All world-widely phylospecies have experienced historical extinction events that may be caused by the palaeogeology events and tectonic movement of the QTPs during the Eocene, followed by the differentiation and expansion of the two clades of Morchella (Figure 8). For the Elata clade, Morchella originated from Qilian Shan and spread along the Himalayas on the edge of QTPs to surrounding Europe and South Africa. However, after the Esculenta clade originated from Shangri-La and spread to other regions of Asia, it was subsequently blocked by the geographical isolation formed in the Quaternary, which led to the diversification of the origin of the Esculenta clade. This result further enriches the theory of species origin in QTPs, except for the reported species such as Floccularia luteovirens, Ophiocordyceps sinensis, Saccharomyces pastorianus, etc. (Quan et al., 2014; Dai et al., 2019; Wu Y. et al., 2021; Bai, 2022; Guo et al., 2022).
In this study, a total of 216 individuals were identified 18 Morchella phylospecies of two clades (Elata and Escuelnta clade), and the results indicate that the Morchella’s phylogenetic diversity within the QTPs far exceeds the number of published taxa for these areas. The divergence time of the 18 Morchella phylospecies occurred in the Eocene-Pliocene period (50.24–4.2 Mya), it was strongly related to the geological strike-slip and the uplift movements of the QTPs, suggesting that the geographical movements had a large influence on the differentiation of the Morchella. Furthermore, the reconstructed ancestral areas of the Elata and Esculenta clades indicate that the northwestern and southeastern regions of QTPs are the likely ancestral area, which also has been suggested to coincide with the glacial refugia in the Quaternary. Moreover, we confirmed that Elata and Esculenta of Morchella originated from QTPs and spread out with plateau geological uplift, transoceanic or transcontinental long-distance dispersal, orogeny, or geographical movements during the Cenozoic Era. The origin of the Esculenta clade is not unique. These results offer strong evidence for the theory of the origin of species in the QTPs.
The data presented in the study are deposited in the NCBI repository, accession number of those data was list in Supplementary Table S1.
QM: conceptualization, methodology, and writing-original draft. ZX: supervision, validation, and writing review and editing. HX, JG, and QP and TM: writing-review and editing. YT, BW, YM, SY, JY, YD, FZ, and TG: investigation. All authors contributed to the article and approved the submitted version.
This work was supported by the Natural Science Planning Project of Qinghai Province (Grant no. 2021-HZ-802), and the first batch of central forestry and grassland ecological protection and restoration funds in 2021 (2021-87).
The authors appreciate Pan Tong (Chief Engineer of Qinghai Provincial Bureau of Geology and Mineral Exploration and Development) for the technical assistance and theoretical guidance in the Qinghai-Tibet Plateau uplift.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1078663/full#supplementary-material
Ali, S., Imran, A., Fiaz, M., Khalid, A. N., and Khan, S. M. (2021). Molecular identification of true morels (Morchella spp.) from the Hindu Kush temperate forests leads to three new records from Pakistan. Gene Rep. 23:101125. doi: 10.1016/j.genrep.2021.101125
An, Z. S., Kutzbach, J. E., Prell, W. L., and Porter, S. C. (2001). Evolution of Asian monsoons and phased uplift of the Himalaya-Tibetan plateau since late Miocene times. Nature 411, 62–66. doi: 10.1038/35075035
Annette, H., George, B., and Ina, L. (1978). Cultural studies of single Ascospore isolates of Morchella esculenta. Mycologia 70, 1269–1274. doi: 10.2307/3759331
Antonelli, A., Kissling, W. D., Flantua, S. G. A., Bermúdez, M. A., Mulch, A., Muellner-Riehl, A. N., et al. (2018). Geological and climatic influences on mountain biodiversity. Nat. Geosci. 11, 718–725. doi: 10.1038/s41561-018-0236-z
Bai, F. Y. (2022). Out of the Tibetan plateau: origin and evolution of lager yeast. Acta Microbiol. Sin. 62, 4188–4201. doi: 10.13343/j.cnki.wsxb.20220451
Baran, J., and Boroń, P. (2017). Two species of true morels (the genus Morchella, Ascomycota) recorded in the Ojców National Park (South Poland). Acta Mycol. 52:1094. doi: 10.5586/am.1094
Baroni, T. J., Beug, M. W., Cantrell, S. A., Clements, T. A., Iturriaga, T., Læssøe, T., et al. (2018). Four new species of 12 Morchella from the Americas. Mycologia 110, 1205–1221. doi: 10.1080/00275514.2018.1533772
Cao, Y. T., Lu, Z. P., Gao, X. Y., Liu, M. L., Sa, W., Liang, J., et al. (2022). Maximum entropy modeling the distribution area of Morchella dill. Ex Pers. species in China under changing climate. Biology 11:1027. doi: 10.3390/biology11071027
Che, J., Zhou, W. W., Hu, J. S., and Zhang, Y. P. (2010). Spiny frogs (Paini) illuminate the history of the Himalayan region and Southeast Asia. Proc. Natl. Acad. Sci. 107, 13765–13770. doi: 10.1073/pnas.1008415107
Chen, J. J., Bao, K. C., Zhou, L. W., Korhonen, K., and Dai, Y. C. (2015). Phylogeny, divergence time estimation, and biogeography of the genus Heterobasidion (Basidiomycota, Russulales). Fungal Divers. 71, 185–200. doi: 10.1007/s13225-014-0317-2
Dahlstrom, J. L., Smith, J. E., and Weber, N. S. (2000). Mycorrhiza-like interaction by Morchella with species of the Pinaceae in pure culture synthesis. Mycorrhiza 9, 279–285. doi: 10.1007/PL00009992
Dai, Y. D., Wu, C. K., Wang, Y. B., Wang, Y., Huang, L. D., Dang, X. J., et al. (2019). Phylogeographic structures of the host insects of Ophiocordyceps sinensis. Zoology 134, 27–37. doi: 10.1016/j.zool.2019.03.003
Deng, K., Lan, X., Chen, Y., Wang, T., Li, M., Xu, Y., et al. (2022). Integration of transcriptomics and metabolomics for understanding the different vegetative growth in Morchella sextElata. Front. Genet. 12:829379. doi: 10.3389/fgene.2021.829379
Deng, T., Wu, F., Zhou, Z., and Su, T. (2020). Tibetan plateau: An evolutionary junction for the history of modern biodiversity. Sci. China Earth Sci. 63, 172–187. doi: 10.1007/s11430-019-9507-5
Dieter, U., Rafael, S., Michael, W., Markus, J., and Poschmann, A. S. (2022). Wildfires during the Paleogene (late Eocene-late Oligocene) of the Neuwied Basin (W-Germany). Rev. Palaeobot. Palynol. 297:104565. doi: 10.1016/j.revpalbo.2021.104565
Ding, L., Kapp, P., Cai, F. C. N., Garzione, N., Xiong, Z. Y., Wang, H. Q., et al. (2022). Timing and mechanisms of Tibetan plateau uplift. Nat. Rev. Earth Environ. 3, 652–667. doi: 10.1038/s43017-022-00318-4
Ding, W., Ree, R. H., Spicer, R. A., and Xing, Y. W. (2020). Ancient orogenic and monsoon-driven assembly of the world’s richest temperate al⁃ pine flora. Science 369, 578–581. doi: 10.1126/science.abb4484
Dissanayake, A. A., Mills, G. L., Bonito, G., Rennick, B., and Nair, M. G. (2021). Chemical composition and anti-inflammatory and antioxidant activities of extracts from cultivated morel mushrooms, species of genus Morchella (Ascomycota). Int. J. Med. Mushrooms 23, 73–83. doi: 10.1615/IntJMedMushrooms.2021039297
Donoghue, M. J. (2008). A phylogenetic perspective on the distribution of plant diversity. Proc. Natl. Acad. Sci. U. S. A. 105, 11549–11555. doi: 10.1073/pnas.0801962105
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemistry 19, 11–15.
Drummond, A. J., and Rambaut, A. (2007). BEAST: bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. doi: 10.1186/1471-2148-7-214
Du, X. H., Wu, D. M., He, G. Q., Wei, W., Xu, N., and Li, T. L. (2019). Six new species and two new records of Morchella in China using phylogenetic and morphological analyses. Mycologia 111, 857–870. doi: 10.1080/00275514.2019.1640012
Du, X. H., Zhao, Q., O’Donnell, K., Rooney, A. P., and Yang, Z. L. (2012a). Multigene molecular phylogenetics reveals true morels (Morchella) are especially species-rich in China. Fungal Genet. Biol. 49, 455–469. doi: 10.1016/j.fgb.2012.03.006
Du, X. H., Zhao, Q., Xu, N., and Yang, Z. L. (2016). High inbreeding, limited recombination and divergent evolutionary patterns between two sympatric morel species in China. Sci. Rep. 6:22434. doi: 10.1038/srep22434
Du, X. H., Zhao, Q., Yang, Z. L., Hansen, K., Taşkın, H., Büyükalaca, S., et al. (2012b). How well do ITS rDNA sequences differentiate species of true morels (Morchella)? Mycologia 104, 1351–1368. doi: 10.3852/12-056
Du, X. H., Zhao, Q., and Zhu, L. Y. (2015). A review on research advances, issues, and perspectives of morels. Mycology 6, 78–85. doi: 10.1080/21501203.2015.1016561
Guo, J., Xie, Z., Jiang, H., Xu, H., Liu, B., Meng, Q., et al. (2022). The molecular mechanism of yellow mushroom (Floccularia luteovirens) response to strong ultraviolet radiation on the Qinghai-Tibet plateau. Front. Microbiol. 13:918491. doi: 10.3389/fmicb.2022.918491
Hao, H. B., Zhang, J. J., Wang, H., Wang, Q., Chen, M. J., Juan, J. X., et al. (2019). Comparative transcriptome analysis reveals potential fruiting body formation mechanisms in Morchella importuna. AMB Express 9:103. doi: 10.1186/s13568-019-0831-4
Harrison, S., and Noss, R. (2017). Endemism hotspots are linked to stable climatic refugia. Ann. Bot. 119, 207–214. doi: 10.1093/aob/mcw248
Hewitt, G. M. (2000). The genetic legacy of quaternary ice ages. Nature 405:907913. doi: 10.1038/35016000
Hoorn, C., Straathof, J., Abels, H. A., Xu, Y., Utescher, T., and Dupont-Nivet, G. (2012). A late Eocene palynological record of climate change and Tibetan plateau uplift (Xining Basin, China). Palaeogeogr. Palaeoclimatol. Palaeoecol. 344-345, 16–38. doi: 10.1016/j.palaeo.2012.05.011
Jia, J., Zheng, W., Zhang, Y., Wei, S., Liang, S., Feng, C., et al. (2022). Tectonic deformation of an intraplate Orogenic Belt: Mesozoic sedimentary basins in the northeastern Qilian Shan. China. Front. Earth Sci. 10:847921. doi: 10.3389/feart.2022.847921
Kate, T. (2022). Exceptional preservation of organs in Devonian placoderms from the Gogo lagerstätte. Science 337, 1311–1314. doi: 10.1126/science.abf3289Katoh
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kim, T. H., and Kim, J. H. (2022). Molecular phylogeny and historical biogeography of Goodyera R. Br. (Orchidaceae): a case of the vicariance between East Asia and North America. Front. Plant Sci. 10:396. doi: 10.3389/fpls.2022.850170
Kim, C., Kim, S. C., and Kim, J. H. (2019). Historical biogeography of melanthiaceae: a case of out-of-North America through the Bering land bridge. Front. Plant Sci. 10:396. doi: 10.3389/fpls.2019.00396
Ling, C. C., Ma, F. J., Dong, J. L., Zhou, G. H., Wang, Q. J., and Sun, B. N. (2021). A mid-latitude area in southwestern China experienced a humid subtropical climate with subtle monsoon signatures during the early Oligocene: evidence from the Ningming flora of Guangxi, Palaeogeography, Palaeoclimatology. Palaeoecology. 579:110601. doi: 10.1016/j.palaeo.2021.110601
Liu, W., Cai, Y. L., He, P. X., Chen, L. F., and Bian, Y. B. (2019). Comparative transcriptomics reveals potential genes involved in the vegetative growth of Morchella importuna. Biotech 3:81. doi: 10.1007/s13205-019-1614-y
Liu, Z. F., and Hu, X. M. (2003). Extreme climate events from the cretaceous and Paleogene. Adv. Earth Sci. 5, 681–690. doi: 1001-8166(2003)05-0681-10
Liu, H., Song, X., Wen, W., Jia, Q., and Zhu, D. (2022). Quantitative effects of climate change on vegetation dynamics in alpine grassland of Qinghai-Tibet plateau in a county. Atmos. 13:324. doi: 10.3390/atmos13020324
Liu, X. H., Xu, Q., and Ding, L. (2016). Differential surface uplift: Cenozoic paleoelevation history of the Tibetan plateau. Sci. China Earth Sci. 59, 2105–2120. doi: 10.1007/s11430-015-5486-y
Loizides, M., Alvarado, P., Clowez, P., Moreau, P. A., and Osa, L. R. D. L. (2015). Morchella tridentina, M. rufobrunnea, and M. kakiicolor: a study of three poorly known Mediterranean morels, with nomenclatural updates in section Distantes. Mycol. Prog. 14, 1–18. doi: 10.1007/s11557-015-1030-6
Loizides, M., Bellanger, J. M., Clowez, P., Richard, F., and Moreau, P. A. (2016). Combined phylogenetic and morphological studies of true morels (Pezizales, Ascomycota) in Cyprus reveal significant diversity, including Morchella arbutiphila and M.disparilis spp. Nov. Mycol. Progress. 15:39. doi: 10.1007/s11557-016-1180-1
Loizides, M., Gonou, Z. Z., Fransuas, G., Drakopoulos, P., Sammut, C., Martinis, A., et al. (2021). Extended phylogeography of the ancestral Morchella anatolica supports preglacial presence in Europe and the Mediterranean origin of morels. Mycologia 113, 559–573. doi: 10.1080/00275514.2020.1869489
Mao, K. S., Wang, Y., and Liu, J. Q. (2021). Evolutionary origin of species diversity on the Qinghai-Tibet plateau. J. Syst. Evol. 59, 1142–1158. doi: 10.1111/jse.12809
Merckx, V. S., Hendriks, K. P., Beentjes, K. K., Mennes, C. B., Becking, L. E., Peijnenburg, K. T., et al. (2015). Evolution of endemism on a young tropical mountain. Nature 524, 347–350. doi: 10.1038/nature14949
Miehe, G., Schleuss, P. M., Seeber, E., Babel, W., Biermann, T., Braendle, M., et al. (2019). The Kobresia pygmaea ecosystem of the Tibetan highlands-origin, functioning and degradation of the world's largest pastoral alpine ecosystem: Kobresia pastures of Tibet. Sci. Total Environ. 648, 754–771. doi: 10.1016/j.scitotenv.2018.08.164
Mittermeier, R. A., Turner, W. R., Larsen, F. W., Brooks, T. M., and Gascon, C. (2011). “Global biodiversity conservation: the critical role of hotspots” in Biodiversity Hotspots: Distribution and Protection of Conservation Priority Areas. eds. F. E. Zachos and J. C. Habel (Berlin: Springer), 3–22.
O’Donnell, K., Rooney, A. P., Mills, G. L., Kuo, M., Weber, N. S., and Rehner, S. A. (2011). Phylogeny and historical biogeography of true morels (Morchella) reveals an early cretaceous origin and high continental endemism and provincialism in the Holarctic. Fungal Genet. Biol. 48, 252–265. doi: 10.1016/j.fgb.2010.09.006
Phonepaserd, P., Yu, Z. D., and Li, T. (2019). High diversity of Morchella and a novel lineage of the Esculenta clade from the north Qinling Mountains revealed by GCPSR-based study. Sci. Rep. 9, 9–18. doi: 1001-8166(2003)05-0681-10
Qin, Y., Chen, J., and Yi, S. (2015). Plateau pikas burrowing activity accelerates ecosystem carbon emission from alpine grassland on the Qinghai-Tibetan plateau. Ecol. Eng. 84, 287–291. doi: 10.1016/j.ecoleng.2015.09.012
Qiu, Y. X., Fu, C. X., and Comes, H. P. (2011). Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of quaternary climate and environmental change in the world’s most diverse temperate flora. Mol. Phylogenet. Evol. 59, 225–244. doi: 10.1016/j.ympev.2011.01.012
Quan, Q. M., Chen, L. L., Wang, X., Li, S., Yang, X. L., Zhu, Y. G., et al. (2014). Genetic diversity and distribution patterns of host insects of Caterpillar fungus Ophiocordyceps sinensis in the Qinghai-Tibet plateau. PLoS One 9:92293. doi: 10.1371/journal.pone.0092293
Rahbek, C., Borregaard, M., Antonelli, A., Colwell, R., Holt, B., Nogués-Bravo, D., et al. (2019). Building mountain biodiversity: geological and evolutionary processes. Science 365, 1114–1119. doi: 10.1126/science.aax0151
Richard, F., Sauvé, M., Bellanger, J. M., Clowez, P., Hansen, K., O’Donnell, K., et al. (2015). True morels (Morchella, Pezizales) of Europe and North America: evolutionary relationships inferred from multilocus data and a unified taxonomy. Mycologia 107, 359–382. doi: 10.3852/14-166
Ronquist, F., Teslenko, M., Mark, P. V. D., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Sanmartín, I., Enghoff, H., and Ronquist, F. (2001). Patterns of animal dispersal, vicariance and diversification in the Holarctic. Biol. J. Linn. Soc. 73, 345–390. doi: 10.1006/bijl.2001.0542
Shi, Y. F., Li, J. J., and Li, B. Y. (1998). Uplift and Environmental Changes of Qinghai-Tibetan Plateau in the Late Cenozoic. Guangzhou: Guangdong Science and Technology Press.
Shrestha, B., Zhang, W. M., Zhang, Y. J., and Liu, X. Z. (2010). What is the Chinese caterpillar fungus Ophiocordyceps sinensis (Ophiocordy cipitaceae)? Mycology 1, 228–236. doi: 10.1080/21501203.2010.536791
Spicer, R. A., Su, T., Valdes, P. J., Farnsworth, A., Wu, F. X., Shi, G. l., et al. (2021). Why ‘the uplift of the Tibetan plateau’ is a myth. Natl. Sci. Rev. 8:nwaa091. doi: 10.1093/nsr/nwaa091
Sun, H., Zheng, D., Yao, T., and Zhang, Y. (2012). Protection and construction of the national ecological security shelter zone on Tibetan plateau. Acta Geograph. Sin. 67, 3–12. doi: 10.11821/xb201201001
Suzuki, H., Sato, J. J., Tsuchiya, K., Luo, J., Zhang, Y. P., Wang, Y. X., et al. (2003). Molecular phylogeny of wood mice (Apodemus, Muridae) in East Asia. Biol. J. Linn. Soc. 80, 469–481. doi: 10.1046/j.1095-8312.2003.00253.x
Swofford, D. L. (2002). PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods). Version 4.0b10. Sinauer Associates. doi: 10.1111/j.0014-3820.2002.tb00191.x
Taylor, T. N., Hass, H., Kerp, H., Krings, M., and Hanlin, R. T. (2004). Perithecial ascomycetes from the 400 million-year-old Rhynie chert: an example of ancestral polymorphism. Mycologia 96, 1403–1419. doi: 10.1080/15572536.2006.11832862
Wan, W., Xiao, P. F., Feng, X. Z., Ma, R. H., Duan, H. T., and Zhao, L. M. (2014). Monitoring lake changes of Qinghai-Tibetan plateau over the past 30 years using satellite remote sensing data. Chin. Sci. Bull. 59, 1021–1035. doi: 10.1007/s11434-014-0128-6
Wang, L. Y., Abbott, R. J., Zheng, W., Chen, P., Wang, Y. J., and Liu, J. Q. (2009). History and evolution of alpine plants endemic to the Qinghai-Tibetan plateau: Aconitum gymnandrum (Ranunculaceae). Mol. Ecol. 18, 709–721. doi: 10.1111/j.1365-294X.2008.04055.x
Wang, M. S., Li, Y., Peng, M. S., Zhong, L., Wang, Z. J., Li, Q. Y., et al. (2015). Genomic analyses reveal potential independent adaptation to high altitude in Tibetan chickens. Mol. Biol. Evol. 32, 1880–1889. doi: 10.1093/molbev/msv071
Wang, L., Yuan, Q., Shen, L., and Ding, L. (2022). Middle Eocene Paleoenvironmental reconstruction in the Gonjo Basin, Eastern Tibetan Plateau: evidence from palynological and evaporite records. Front. Earth Sci. 10, 1–14.doi: 10.3389/feart.2022.818418
Wang, C., Zhao, X., Liu, Z., Lippert, P. C., Graham, S. A., Coe, R. S., et al. (2008). Constraints on the early uplift history of the Tibetan plateau. PNAS 105, 4987–4992. doi: 10.1073/pnas.0703595105
Wen, J., Zhang, J. Q., Nie, Z. L., Zhong, Y., and Sun, H. (2014). Evolutionary diversification of plants on the Qinghai-Tibetan plateau. Front. Genet. 5:4. doi: 10.3389/fgene.2014.00004
Wu, H., Chen, J., Li, J., Liu, Y., Park, H. J., and Yang, L. (2021). Recent advances on bioactive ingredients of Morchella esculenta. Appl. Biochem. Biotechnol. 193, 4197–4213. doi: 10.1007/s12010-021-03670-1
Wu, Y. D., Li, L., Fan, Y. L., Ni, X. W., Ohiolei, J. A., Li, W. H., et al. (2021). Genetic evolution and implications of the mitochondrial genomes of two newly identified Taenia spp. in rodents from Qinghai-Tibet plateau. Front. Microbiol. 12:647119. doi: 10.3389/fmicb.2021.647119
Wu, F. X., Miao, D. S., Chang, M. M., Shi, G. L., and Wang, N. (2017). Fossil climbing perch and associated plant megafossils indicate a warm and wet Central Tibet during the late Oligocene. Sci. Rep. 7:878. doi: 10.1038/s41598-017-00928-9
Xiong, Z. Y., Liu, X. H., Ding, L., Alex, F., Robert, A. S., Xu, Q., et al. (2022). The rise and demise of the Paleogene Central Tibetan Valley. Sci. Adv. 8, 1–15. doi: 10.1126/sciadv.abj0944
Yan, Y. J., Li, Y., Wang, W. J., He, J. S., Yang, R. H., and Yao, Y. J. (2017). Range shifts in response to climate change of Ophiocordyceps sinensis, a fungus endemic to the Tibetan plateau. Biol. Conserv. 206, 143–150. doi: 10.1016/j.biocon.2016.12.023
Yang, P. (2005). The main environmental effect of Qinghai-Tibet plateau uplift. Qinghai Env. 3, 93–95. doi: 1007-2454(2005)03-0093-03
Yang, S. J., Dong, H. L., and Lei, F. M. (2009). Phylogeography of regional fauna on the Tibetan plateau: a review. Prog. Nat. Sci. 19, 789–799. doi: 10.1016/j.pnsc.2008.10.006
Yang, F. S., Li, Y. F., Ding, X., and Wang, X. Q. (2008). Extensive population expansion of Pedicularis longiflora (Orobanchaceae) on the Qinghai-Tibetan plateau and its correlation with the quaternary climate change. Mol. Ecol. 17, 5135–5145. doi: 10.1111/j.1365-294X.2008.03976.x
Yatsiuk, I., Saar, I., Kalamees, K., Sulaymonov, S., Gafforov, Y., and O’Donnell, K. (2016). Epitypification of Morchella steppicola (Morchellaceae, Pezizales), a morphologically, phylogenetically and biogeographically distinct member of the Esculenta clade from Central Eurasia. Phytotaxa. 284, 31–40. doi: 10.11646/phytotaxa.284.1.3
Yu, Y., Harris, A. J., Blair, C., and He, X. J. (2015). RASP (reconstruct ancestral state in phylogenies): a tool for historical biogeography. Mol. Phylogenet. Evol. 87, 46–49. doi: 10.1016/j.ympev.2015.03.008
Yu, F. M., Jayawardena, R. S., Thongklang, N., Lv, M. L., Zhu, X. T., and Zhao, Q. (2022). Morel production associated with soil nitrogen-fixing and nitrifying microorganisms. J. Fungi. 8:299. doi: 10.3390/jof8030299
Yuan, F. (2015). Study on Phylogeography and suitable distribution of Ophiocordyceps sinensis populations. Ph. D. dissertation. Yunnan: Yunnan University.
Zachos, J., Pagani, M., Sloan, L., Thomas, E., and Billups, K. (2001). Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 292, 686–693. doi: 10.1126/science.1059412
Zhang, D., Gao, F., Jakovlić, I., Zou, H., Zhang, J., Li, W. X., et al. (2020). PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20, 348–355. doi: 10.1111/1755-0998.13096
Zhao, H., Zhu, J., Zong, T., Liu, X., Ren, L., Lin, Q., et al. (2021). Two new species in the family Cunninghamellaceae from China. Mycobiology 49, 142–150. doi: 10.1080/12298093.2021.1904555
Keywords: Morchella, Qinghai-Tibet plateau subkingdoms, multigene phylogenetics, age estimation, phylogeographic structure
Citation: Meng Q, Xie Z, Xu H, Guo J, Tang Y, Ma T, Peng Q, Wang B, Mao Y, Yan S, Yang J, Dong D, Duan Y, Zhang F and Gao T (2022) Out of the Qinghai-Tibetan plateau: Origin, evolution and historical biogeography of Morchella (both Elata and Esculenta clades). Front. Microbiol. 13:1078663. doi: 10.3389/fmicb.2022.1078663
Received: 24 October 2022; Accepted: 30 November 2022;
Published: 28 December 2022.
Edited by:
Qiang Lin, University of Antwerp, BelgiumReviewed by:
Peng Zhao, Northwest University, ChinaCopyright © 2022 Meng, Xie, Xu, Guo, Tang, Ma, Peng, Wang, Mao, Yan, Yang, Dong, Duan, Zhang and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhanling Xie, eGllemhhbmxpbmcyMDEyQDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.