- 1State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China
- 2University of Chinese Academy of Sciences, Beijing, China
Glycerol is a readily available and low-cost simple polyol compound, which can be used as a carbon source for microorganisms to produce various value-added products. Understanding the underlying regulatory mechanism in glycerol metabolism is critical for making better use of glycerol for diverse applications. In a few reported Streptomyces strains, the glycerol utilization gene cluster (glp operon) was shown to be regulated by the IclR family transcriptional regulator GylR. However, the molecular regulatory mechanism mediated by GylR has not been fully elucidated. In this study, we first analyzed the available Actinobacteria genomes in the NCBI Genome database, and found that the glp operon-like gene clusters are conserved in Streptomyces and several other genera of Actinobacteria. By taking Streptomyces clavuligerus NRRL 3585 as a model system, we identified that GylR represses the expressions of glp operon and gylR by directly binding to their promoter regions. Both glycerol-3-phosphate and dihydroxyacetone phosphate can induce the dissociation of GylR from its binding sequences. Furthermore, we identified a minimal essential operator site (a palindromic 18-bp sequence) of GylR-like regulators in Streptomyces. Our study for the first time reported the binding sequences and effector molecules of GylR-like proteins in Streptomyces. The molecular regulatory mechanism mediated by GylR presumably exists widely in Streptomyces. Our findings would facilitate the design of glycerol utilization pathways for producing valuable products. Moreover, our study provided new basic elements for the development of glycerol-inducible regulatory tools for synthetic biology research in the future.
Introduction
Glycerol is a simple polyol compound existing widely in biological systems (Klein et al., 2017; Doi, 2019). In addition, glycerol is the main by-product of biodiesel production (da Silva et al., 2009; Poblete-Castro et al., 2020). These facts make glycerol as a relatively cheap and readily available substance. Glycerol has a number of industrial applications. It has been widely used in cosmetics and food industry due to its hygroscopic and other properties (Doi, 2019; Poblete-Castro et al., 2020). Furthermore, glycerol is an important substrate in biological industries as it can be converted into various value-added products, such as pharmaceuticals, resins, detergents, plastics and tobacco (Pagliaro et al., 2007; Poblete-Castro et al., 2020). In bacteria, glycerol can be metabolized mainly through two routes (Lin, 1976; Doi, 2019; Poblete-Castro et al., 2020). The first route involves the phosphorylation of glycerol to form glycerol-3-phosphate (G3P), and the subsequent oxidation of G3P to generate dihydroxyacetone phosphate (DHAP). In the other route, glycerol is firstly oxidized to dihydroxyacetone, which is then phosphorylated to afford DHAP. DHAP can be further channeled to downstream metabolic pathways like the glycolysis pathway. Thus it can be seen that glycerol can serve as a common precursor to biosynthesize various products. Usually, the genes encoding the enzymes for conversion of glycerol to DHAP are tightly controlled by different types of transcriptional regulators (Lin, 1976; Hindle and Smith, 1994; Bong et al., 2019; Poblete-Castro et al., 2020). Understanding the molecular regulatory mechanism of glycerol metabolism is critical for better use of glycerol to produce valuable products.
Streptomycetes are Gram-positive filamentous soil bacteria and are well known as prolific producers of a wide variety of natural products, including many antibacterial, anticancer, and immunosuppressive drugs (Kormanec et al., 2019; Lee et al., 2021). Glycerol can be used as a carbon source by streptomycetes to produce those valuable products, such as the clinically used beta-lactamase inhibitor clavulanic acid produced by Streptomyces clavuligerus NRRL 3585 (Saudagar et al., 2008; Guo et al., 2013; Shin et al., 2021). Until now, studies on the glycerol metabolism in streptomycetes have only been reported for Streptomyces coelicolor A3(2) (Smith and Chater, 1988a,b; Hindle and Smith, 1994; Lee et al., 2017) and S. clavuligerus NRRL 3585 (Baños et al., 2009), both of which contain a glycerol utilization gene cluster. In S. coelicolor A3(2), the glycerol utilization gene cluster consists of gylR and gylCAB, in which gylCAB corresponds to the glp operon (glpF1K1D1) in S. clavuligerus NRRL 3585 (Smith and Chater, 1988a,b; Baños et al., 2009). The three key glycerol utilization genes (gylC/glpF1, gylA/glpK1, and gylB/glpD1) encode a glycerol transporter, a glycerol kinase, and a FAD-dependent glycerol-3-phosphate dehydrogenase, respectively (Figure 1). Specifically, the transmembrane protein GylC/GlpF1 facilitates the uptake of glycerol into the cells, and then intracellular glycerol is phosphorylated by the kinase GylA/GlpK1 to form G3P, which is subsequently oxidized by the dehydrogenase GylB/GlpD1 to afford DHAP (Figure 1; Baños et al., 2009). Studies in S. coelicolor A3(2) suggested that GylR negatively regulates the transcription of the glp operon. G3P was proposed to be a true effector molecule of GylR, but there is a lack of experimental evidence (Seno and Chater, 1983; Seno et al., 1984). In S. clavuligerus NRRL 3585, similar results were observed. The glp operon is negatively regulated by GylR. As a member of the IclR family transcriptional regulators, GylR can directly bind to the promoter region of glp operon (Figure 1; Baños et al., 2009). In vivo studies have revealed that the expression of glp operon can be induced by the addition of glycerol during fermentation (Baños et al., 2009; Guo et al., 2013). Studies in non-Streptomyces strains, such as Mycolicibacterium smegmatis (previously known as Mycobacterium smegmatis), indicated that G3P rather than glycerol might bind to GylR-like proteins, and, as a result, regulate the glp operon (Bong et al., 2019). Despite the aforementioned studies, the exact binding sequences and the small molecule effectors of GylR-like proteins remain elusive in Streptomyces.
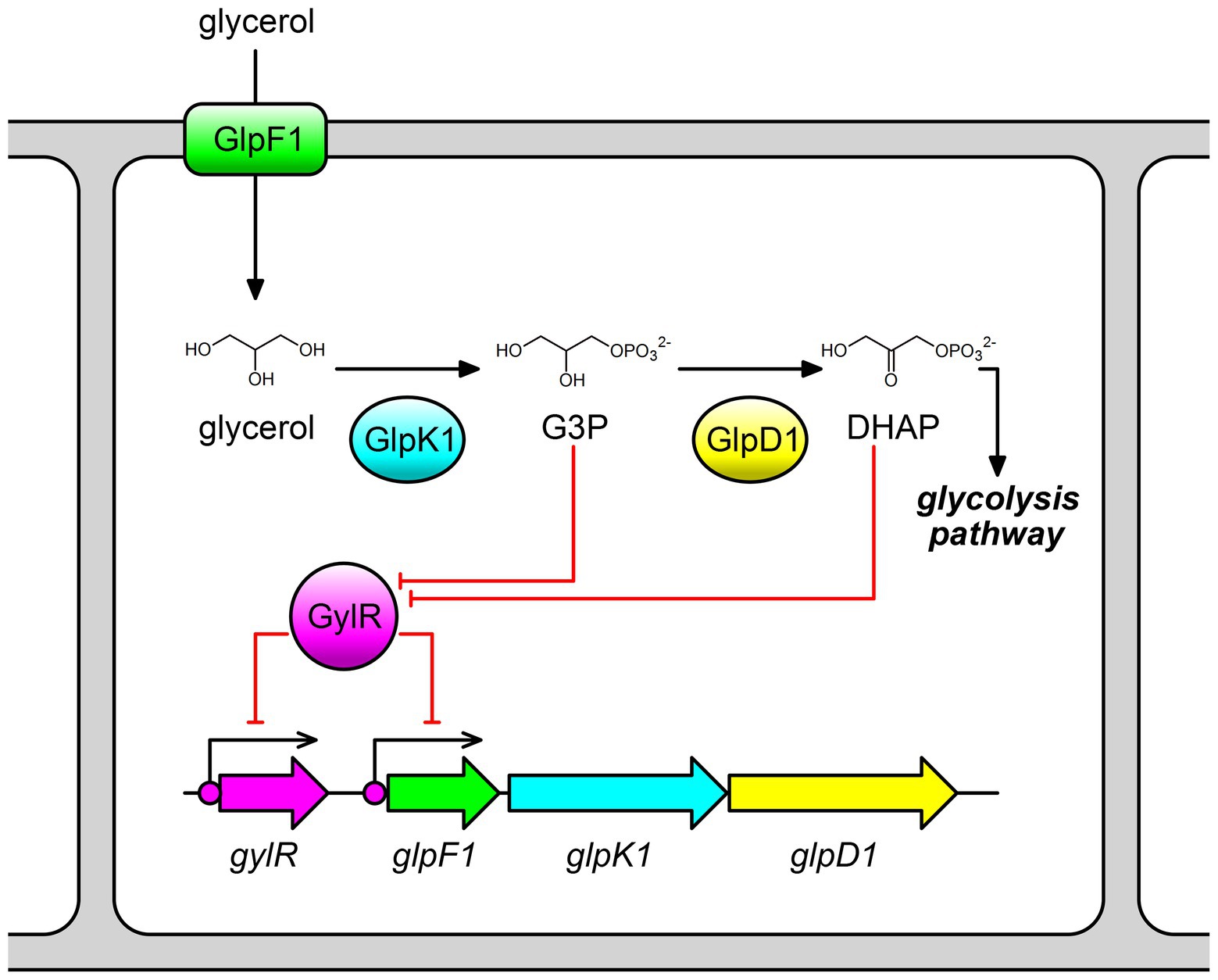
Figure 1. Schematic representation of GylR-mediated regulation on glycerol metabolism in Streptomyces clavuligerus NRRL 3585. The glp operon consisting of glpF1, glpK1, and glpD1, is negatively regulated by GylR. G3P and DHAP act as effector molecules that can bind to GylR and induce the dissociation of GylR from its binding sites. G3P, glycerol-3-phosphate; DHAP, dihydroxyacetone phosphate.
In this study, we first carried out a systematic bioinformatic analysis, which revealed that the glp operon is highly conserved in Streptomyces genomes, and the glp operon-like glycerol utilization gene clusters (minimally containing one set of clustered glpF, glpK, and glpD homologous genes) are present in many genera of the phylum Actinobacteria. The results implied a common metabolic pathway of glycerol in Streptomyces. Then, we took S. clavuligerus NRRL 3585 as a model system to further investigate the molecular mechanism of GylR-mediated regulation of glycerol metabolism. Our results showed that GylR negatively regulates the transcriptions of gylR and glp operon by directly binding to their promoter regions, and the exact binding sequences were identified to contain a conserved 18-bp sequence motif. In addition to G3P, DHAP was found to be another effector molecule that could cause the dissociation of GylR from its binding sequences. Our study for the first time identified the binding sequences and effector molecules of GylR-like proteins in Streptomyces, which further elucidated the molecular mechanism of GylR-mediated regulation of glycerol metabolism. The results would facilitate the design of glycerol utilization pathways, and provide valuable regulatory elements for synthetic biology research.
Materials and methods
Strains, plasmids, primers, and growth conditions
Bacterial strains and plasmids used in this study are listed in Supplementary Table 1 in the Supplementary material. Primers in this study are listed in Supplementary Table 2. Escherichia coli DH5α was used as a general host for propagating plasmids. E. coli ET12567 (pUZ8002) (Kieser et al., 2000) was used as a host for transferring DNA from E. coli to Streptomyces in intergeneric conjugation experiments. E. coli Rosetta (DE3) was used as a host for production of GylR protein. S. clavuligerus NRRL 3585 and its derived strains were grown on yeast extract-dextrose (YD) agar medium at 28°C (Xiang et al., 2009). The SA medium used for fermentation and RNA extraction is composed of 10 g/l soluble starch, 2 g/l L-asparagine, 21 g/l MOPS, 0.6 g/l MgSO4·7H2O, and 0.1% (v/v) trace element solution (FeSO4·7H2O, 1 mg/ml; MnCl2·H2O, 1 mg/ml; ZnSO4·7H2O, 1 mg/ml; CaCl2·2H2O, 1.3 mg/ml; Guo et al., 2013). Antibiotics were used at the following concentrations unless otherwise specified: apramycin, 100 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 12.5 μg/ml; nalidixic acid, 25 μg/ml. General procedures for E. coli and Streptomyces manipulations were performed according to the standard protocols (Kieser et al., 2000).
Bioinformatic analyses
The completed genomes of the phylum Actinobacteria were downloaded from the NCBI Genome database (as of September 2022). The glpF, glpK, glpD, and gylR homologs were identified using standalone BLASTP program (Camacho et al., 2009) with the previously reported glycerol utilization genes in S. clavuligerus NRRL 3585 as query sequences (Baños et al., 2009). Protein sequence comparison was performed using EMBOSS 6.5.0 (Rice et al., 2000). DNA motif search and the data analysis were preformed using Python scripts. The conserved sequence motif logo was generated using MEME Suite 5.5.0 (Bailey et al., 2006). The information of all the identified homologs, including their GenBank accession numbers and sequences, was provided in Supplementary Sheet 1. The information of all the clustered homologs was provided in Supplementary Sheet 2. The information of all the DNA sequences corresponding to the conserved 18-bp sequence motif was provided in Supplementary Sheet 3.
Construction of the mutant strain Streptomyces clavuligerus ΔgylR
To construct a plasmid for inactivation of gylR, a 1,461-bp upstream DNA fragment was amplified with primers gylR-U-F and gylR-U-R, and a 1,469-bp downstream DNA fragment was amplified with primers gylR-D-F and gylR-D-R, using S. clavuligerus NRRL 3585 genomic DNA as the template. After digestion with appropriate enzymes (EcoRI and BamHI for upstream fragment, BamHI and HindIII for downstream fragment), the two fragments were inserted into EcoRI/HindIII-digested pKC1139 to generate pKC1139-gylR, which was verified by DNA sequencing. The plasmid pKC1139-gylR was transformed into E. coli ET12567 (pUZ8002) and then introduced into S. clavuligerus NRRL 3585 by intergeneric conjugation. After several rounds of passaging the exconjugants on solid YD medium, apramycin sensitive colonies were screened by PCR using primers gylR-del-F and gylR-del-R to obtain the double-crossover mutants ΔgylR (Supplementary Figure 1).
RNA extraction and quantitative reverse transcription PCR
The transcriptional levels of the analyzed genes in S. clavuligerus NRRL 3585 and ΔgylR mutant strains were evaluated by quantitative reverse transcription PCR (RT-qPCR). Total RNAs were isolated from the cultures grown in SA media without or with 163 mM glycerol at 24 h as in previous studies (Baños et al., 2009; Guo et al., 2013). The RNA samples were treated with RQ1 RNase-free DNase (Promega) to remove genomic DNA. Synthesis of cDNA and subsequent RT-qPCR were performed according to the standard methods (Li et al., 2015), and transcription of the hrdB gene was used as the internal control.
Promoter activity assay
The green fluorescent protein (GFP) gene was amplified by PCR from pIJ-Potr using primer pair GFP-F and GFP-R (Wang et al., 2016). The PCR product was digested with SpeI and EcoRI, and then inserted into the corresponding sites of the integrative plasmid pSET152 to generate pSET152-GFP. The promoter region of glp operon (Pglp) was amplified with primer pair Pglp-F and Pglp-R using S. clavuligerus NRRL 3585 genomic DNA as the template. The PCR product was digested with BamHI and SpeI, and then inserted into pSET152-GFP. The resultant plasmid pSET152-Pglp-GFP was introduced into S. clavuligerus NRRL 3585 and ΔgylR mutant strains through conjugation to obtain WT-Pglp-GFP and ΔgylR-Pglp-GFP, respectively. As a control, the plasmid pSET152-GFP was transformed into S. clavuligerus NRRL 3585 to generate the recombinant strain WT-GFP. The above recombinant strains were cultivated in SA medium for 12 h. Then, 200 μl of cell culture was washed twice with pure water, and transferred into 96-well plates. The fluorescence intensities were recorded using a Synergy™ HT Multi-Mode Microplate Reader with an excitation at 489 nm and an emission at 512 nm for analysis. Pure water was used as a blank control. The biomass of S. clavuligerus strains were detected by the simplified diphenylamine colorimetric method (Zhao et al., 2013).
Overproduction and purification of GylR
The coding region of gylR was amplified by PCR from S. clavuligerus NRRL 3585 genomic DNA using primer pair gylR-F and gylR-R. The PCR product was digested with NdeI and XhoI and then inserted into the corresponding sites of pET-30a. The resultant plasmid pET30a-gylR was verified by DNA sequencing and subsequently introduced into E. coli Rosetta (DE3) for production of GylR. The E. coli Rosetta (DE3) harboring pET30a-gylR was cultured in LB supplemented with 50 μg/ml kanamycin at 37°C and 250 rpm until an OD600 of 0.6 was reached, at which time point 0.1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) was added, and the strain culture was allowed to incubate at 30°C for 4 h. The cells were harvested, and resuspended in lysis buffer (20 mM Tris–HCl, pH 8.0, 500 mM NaCl, 5 mM imidazole, and 10% sorbitol).
The C-terminal His6-tagged GylR was purified using nickel-nitrilotriacetic acid (Ni-NTA) agarose chromatography according to the manufacturer’s protocol (Novagen) with glycerol in all the buffers being substituted by sorbitol. Of note, the elution buffer used for the purification of GylR contained 500 mM imidazole and 10% (w/v) sorbitol. As purified GylR was unstable and lost activity quickly, we used freshly purified GylR in all the assays in this study. The protein concentration was determined by using a bicinchoninic acid (BCA) protein assay kit (Novagen).
Size-exclusion chromatography assays
The polymerization state of GylR was analyzed by size-exclusion chromatography on a Shimadzu Prominence HPLC system using TSKgel G3000 SWxl column (7.8 × 300 mm, TOSOH Inc.) with an elution solvent of 0.1 M Na2SO4 (pH 6.7) and a flow rate of 0.5 ml/min as described previously (Pan et al., 2017). Bovine serum albumin, ovalbumin, and AlpJ were used as molecular weight standards.
Electrophoretic mobility shift assays
The electrophoretic mobility shift assays (EMSAs) were performed according to the protocol described previously (Wang et al., 2009). The DNA probes BS1 (upstream region of the coding sequence of gylR) and BS2 (upstream region of the coding sequence of glpF1) were amplified from S. clavuligerus NRRL 3585 genomic DNA with primer pairs BS1-F/BS1-R, and BS2-F/BS2-R, respectively. Other DNA probes were synthesized by Invitrogen. A typical 20 μl reaction mixture consisted of 10 ng/μl DNA probe, GylR (various concentrations), small molecules (glycerol or G3P or DHAP with different concentrations), 50 mM HEPES (pH 7.5), 1 mM dithiothreitol (DTT), 5 mM MgCl2, 0.2 mg/ml bovine serum albumin (BSA), 2 mM EDTA and 5% sorbitol. After incubation at 25°C for 25 min, protein-bound DNA and free DNA were separated by electrophoresis on nondenaturing 6.0% (w/v) polyacrylamide gels with a running buffer (pH 7.5) containing 200 mM HEPES and 4 mM EDTA. Gels were stained with SYBR Gold. The concentrations of DNA probes were determined with a NanoVue Plus spectrophotometer.
DNase I footprinting assay
In order to determine the binding sites of GylR, labeled BS1 and BS2 probes were amplified by PCR with primer pairs FAM-gylR/HEX-gylR and FAM-glpF1/HEX-glpF1, respectively. A typical 50 μl reaction mixture contained 200 ng FAM/HEX-labeled DNA probe, 0–40 ng/μl GylR, 100 mM KCl, 20 mM HEPES (pH 7.5), 2 mM DTT, 5 mM MgCl2, 0.5 mg/ml BSA, and 5% sorbitol. After incubation at 25°C for 25 min, 5.5 μl RQ1 RNase-free DNase I buffer and 0.375 U DNase I were added. The mixture was incubated at 25°C for an additional 80 s. Then, the reaction was stopped by addition of 10 μl stop solution (20 mM EGTA, pH 8.0) and 100 μl phenol-chloroform (1:1, v/v). Samples were analyzed by Genolab Biotech Co., Ltd.
Rapid amplification of 5′ cDNA ends (5′-RACE)
The transcription start points (tsp) of gylR, and glp operon were determined by using a FirstChoice RLM-RACE kit (Ambion) following the manufacturer’s instructions. Total RNAs from S. clavuligerus NRRL 3585 were prepared as described above (Guo et al., 2013). Total RNAs (10 ng) were successively treated with calf intestine alkaline phosphatase (CIP), tobacco acid pyrophosphatase (TAP) and ligated with a 45-base 5′-RACE adapter at the 5′-end using T4 RNA ligase. Reverse transcription was performed using SuperScript III reverse transcriptase with the supplement of random decamers. PCR was first performed using a 5’-RACE outer primer and gene-specific outer primer with Q5 DNA polymerase (New England Biolabs). This PCR product was used as the template of inner PCR using a 5′-RACE inner primer and a gene-specific inner primer. A 25 μl volume of each PCR reaction mixture was analyzed by 2% agarose gel electrophoresis. The product of inner PCR was sent for sequencing after gel purification.
Results
The distribution of glp operon-like glycerol utilization gene clusters in Actinobacteria
Previous studies strongly suggested that glp operon probably serves as a general pathway in glycerol metabolism in Streptomyces strains (Smith and Chater, 1988b; Hindle and Smith, 1994; Baños et al., 2009). Indeed, we found that the glp operon accompanied by gylR is highly conserved in the genus Streptomyces, as 441 out of 458 completed genomes of Streptomyces in the NCBI Genome database (as of Sep. 2022) each harbor a glp operon with an adjacent gylR homologous gene (Figure 2; Supplementary Sheets 1, 2). This encouraged us to further analyze the distribution of glp operon-like gene clusters (minimally containing one set of clustered glpF, glpK, and glpD homologous genes) in other genera of the phylum Actinobacteria. Therefore, we performed a systematic analysis of the remaining 3,112 completed genomes of Actinobacteria available in the NCBI Genome database (as of Sep. 2022). The glpF, glpK, glpD, and gylR homologous genes identified in each of the Actinobacteria genomes were shown in Supplementary Sheet 1. Overall, 1,517 out of 3,570 genomes each contain at least one set of glpF, glpK, and glpD homologous genes (not necessarily clustered). The bioinformatic analysis showed that the glp operon-like gene clusters often exist in Kitasatospora and Streptacidiphilus, the other two genera of the family Streptomycetaceae (Figure 2; Supplementary Sheet 2). In several genera of the family Pseudonocardiaceae, including Saccharopolyspora, Saccharothrix, Saccharopolyspora, Actinoalloteichus, and Amycolatopsis, almost every genome of them contains a gene cluster consisting of glpFK homologous genes, and an upstream glpD homolog (transcribed in the opposite direction). However, no gylR homolog was found around those gene clusters. Interestingly, over half of the Amycolatopsis genomes also contain another gene cluster. In each of these gene clusters, a gylR homolog and another glpK homolog are present upstream of the glpFKD homologous genes. Such a gene cluster is present in most of the genomes of Pseudonocardia, Actinoplanes, and Actinomadura. In the majority of Nocardiopsis genomes, a gylR homolog and glpKFD homologous genes were found clustered together. In most genomes of Brevibacterium, Micrococcus, Rothia, Kocuria, Brachybacterium, Cellulomonas, and Cellulosimicrobium, a gene cluster, in which a glpD homolog is located upstream of the glpFK homologous genes, was identified without gylR homologs nearby. In some other genera, such as Corynebacterium, Rhodococcus, Mycolicibacterium, and Nocardia, glp operon-like gene clusters were found in less than 50% analyzed genomes (Supplementary Sheet 2). Surprisingly, in the analyzed genomes of Mycobacterium, the glpF, glpK and glpD homologous genes were all found to be standalone proteins. It is worth pointing out again that the strain previously called Mycobacterium smegmatis (harboring a glp operon-like gene cluster) was renamed Mycolicibacterium smegmatis based on the updated taxonomy system of NCBI.
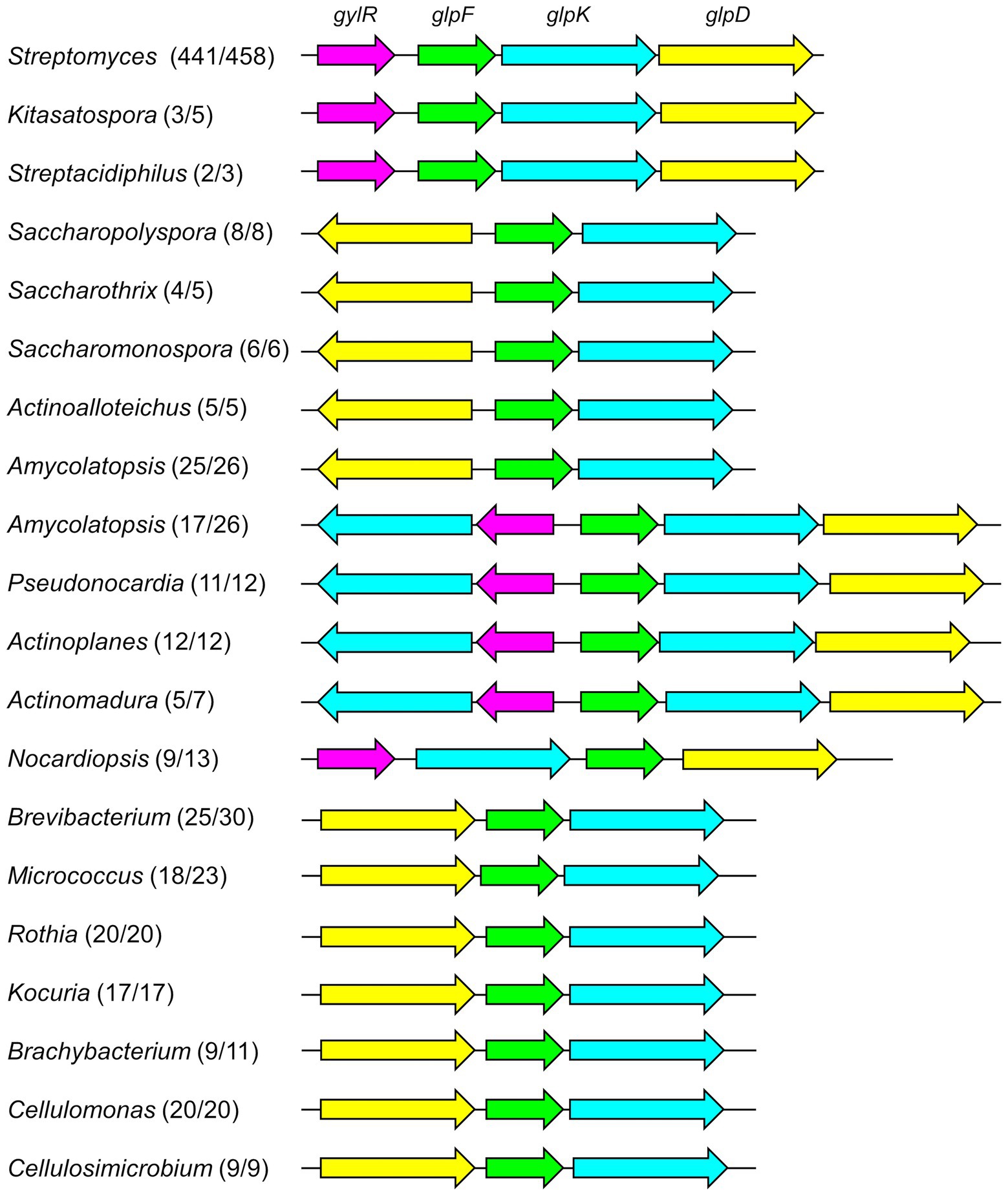
Figure 2. The distribution of glp operon-like gene clusters in analyzed genomes of the phylum Actinobacteria. The numbers after the name of each genus represented the frequency of glp operon-like gene clusters being identified, e.g., Streptomyces (441/458) denoted that 441 out 458 Streptomyces genomes each contains a glp operon-like gene cluster. The genera, in which less than 50% genomes harbor the glp operon-like gene clusters, were not included in this figure.
Overall, the results showed that glp operon-like gene clusters are present in many genera of Actinobacteria, and these clusters are likely to play important roles in the glycerol metabolism in Actinobacteria. It is obvious that Actinobacteria strains have evolved diverse glp operon-like gene clusters, which might be beneficial to different host strains for utilizing glycerol.
GylR represses the expression of glp operon and gylR itself
The above bioinformatic analysis further supported that glp operon together with gylR serves as an important general pathway in glycerol metabolism of Streptomyces. Here, we took S. clavuligerus NRRL 3585 as a model strain to further investigate the molecular regulatory mechanism of GylR in glycerol metabolism. To better understand the regulatory role of GylR, we generated the mutant strain S. clavuligerus ΔgylR with a truncation in gylR gene (Supplementary Figure 1). For the wild-type (WT) strain NRRL 3585, the transcriptions of glpF1, glpD1 and glpK1 were at low levels at the absence of glycerol, while the addition of 163 mM glycerol to the SA medium significantly augmented the transcriptions of glpF1, glpD1 and glpK1 (Figure 3A). However, for the mutant strain ΔgylR, the transcriptions of the three genes were all at high levels with or without the addition of glycerol. In addition, we found that the transcription of gylR was also induced by glycerol in the WT strain, and the transcription of truncated gylR in ΔgylR was at a similarly high level regardless of whether glycerol was added or not (Figure 3A). These results showed that GylR is a repressor regulating the transcriptions of the glp operon, as well as gylR itself in S. clavuligerus, which was consistent with the previous reports in S. clavuligerus (Baños et al., 2009) and S. coelicolor (Smith and Chater, 1988b).
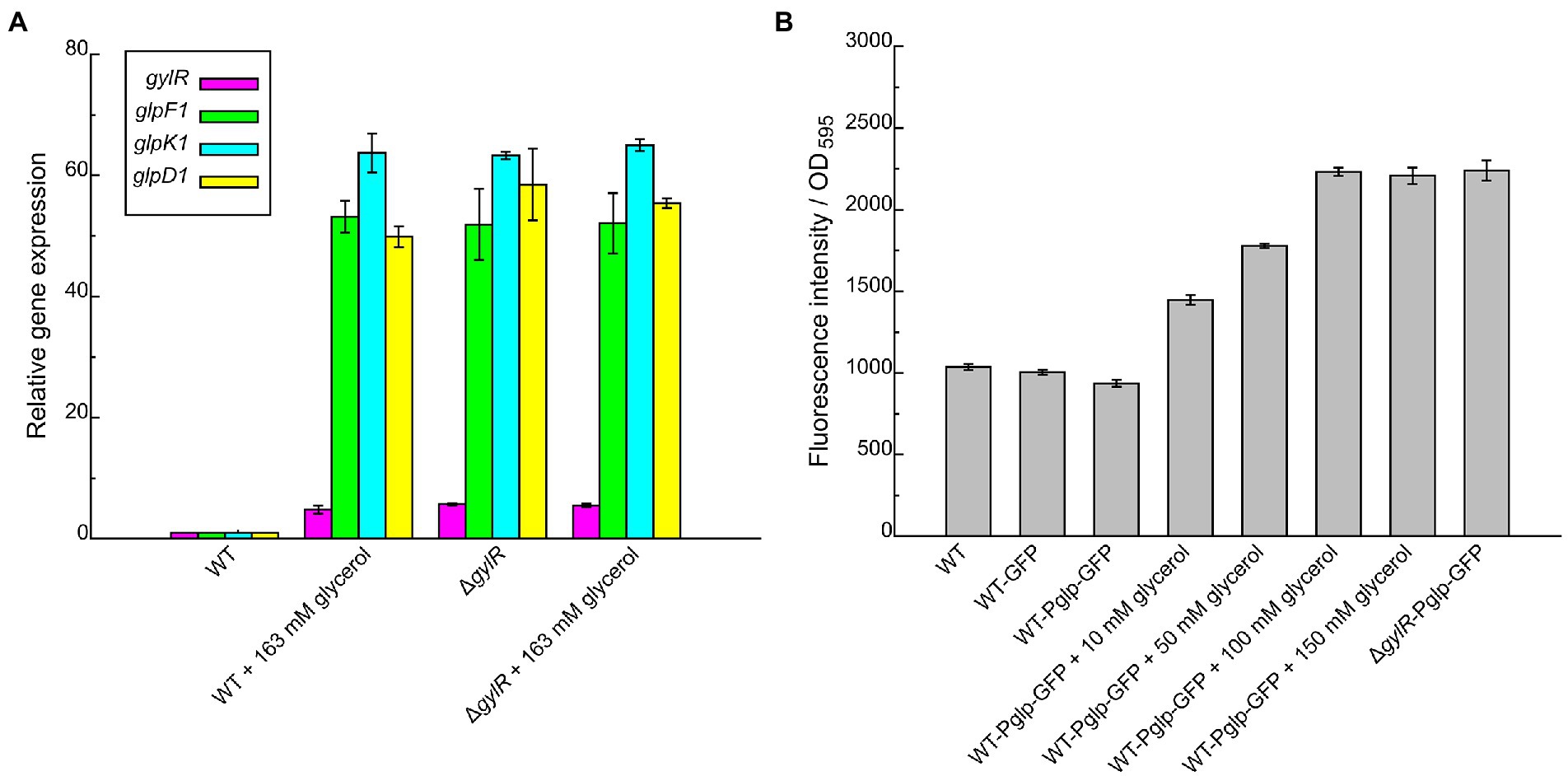
Figure 3. Transcriptional analysis of gylR and glp operon in Streptomyces clavuligerus NRRL 3585 and recombinant strains under different culture conditions. (A) RT-qPCR analysis of the transcriptional levels of gylR, glpF1, glpK1, and glpD1 in Streptomyces clavuligerus NRRL 3585 (WT) and ΔgylR cultivated in SA medium or SA medium supplemented with 163 mM glycerol. The hrdB gene (encoding the RNA polymerase principal sigma factor) was used as an internal control. The relative gene expression values in WT were arbitrarily assigned as 1. (B) Evaluation of the activities of Pglp (promoter region of glp operon) in WT and recombinant strains cultivated in SA medium or SA medium supplemented with different concentrations of glycerol. The fluorescence intensity of GFP and biomass (OD595) were analyzed after 12 h of cultivation. OD595 values were determined using the simplified diphenylamine colorimetric method (Zhao et al., 2013). Error bars represent standard deviations of three independent replicates.
Furthermore, we analyzed the activity of Pglp (a DNA fragment containing the promoter of glp operon) using green fluorescent protein (GFP) as a reporter. The reporter plasmid pSET152-Pglp-GFP was constructed and transformed into the WT and ΔgylR strains, affording the recombinant strains WT-Pglp-GFP and ΔgylR-Pglp-GFP, respectively. A control plasmid pSET152-GFP was also constructed and transformed into WT to obtain the recombinant strain WT-GFP. We observed that, in comparison to that of WT-GFP, the transcription of glp operon in WT-Pglp-GFP was induced by glycerol in a concentration-dependent manner, and reached the maximum level when glycerol concentration was above 100 mM (Figure 3B). As expected, the transcription of glp operon in ΔgylR-Pglp-GFP was at a level comparable to the maximum level in the WT-Pglp-GFP strain induced by glycerol. These results further confirmed that GylR negatively regulates the expression of glp operon, and also implied that GylR serves as the main regulator of glp operon.
GylR directly binds to the promoter regions of glp operon and gylR
Although GylR was proposed to regulate the glp operon and gylR through binding to their promoter regions in S. clavuligerus, there is a lack of experimental evidences (Baños et al., 2009). We overproduced C-terminal His6-tagged GylR in E. coli Rosetta (DE3) and purified it to homogeneity by Ni-NTA affinity chromatography (Supplementary Figure 2A). Size-exclusion chromatography analysis revealed that GylR forms a dimer under the tested conditions (Supplementary Figure 2B), which is reasonable as IclR family transcription factors typically bind to their target sequences as dimers or tetramers (Suvorova and Gelfand, 2021). Then, the electrophoretic mobility shift assay (EMSA) was carried out to determine the direct interactions between GylR and the promoter regions of gylR and glp operon. Two DNA fragments (BS1 and BS2) containing the promoters of gylR and glp operon, respectively, were used as DNA probes in the EMSA studies. As shown in Figures 4A,B, GylR could directly bind to both BS1 and BS2 in a concentration dependent manner. Together, the above results strongly supported that GylR negatively regulates the expression of glp operon and gylR through directly binding to their promoter regions.
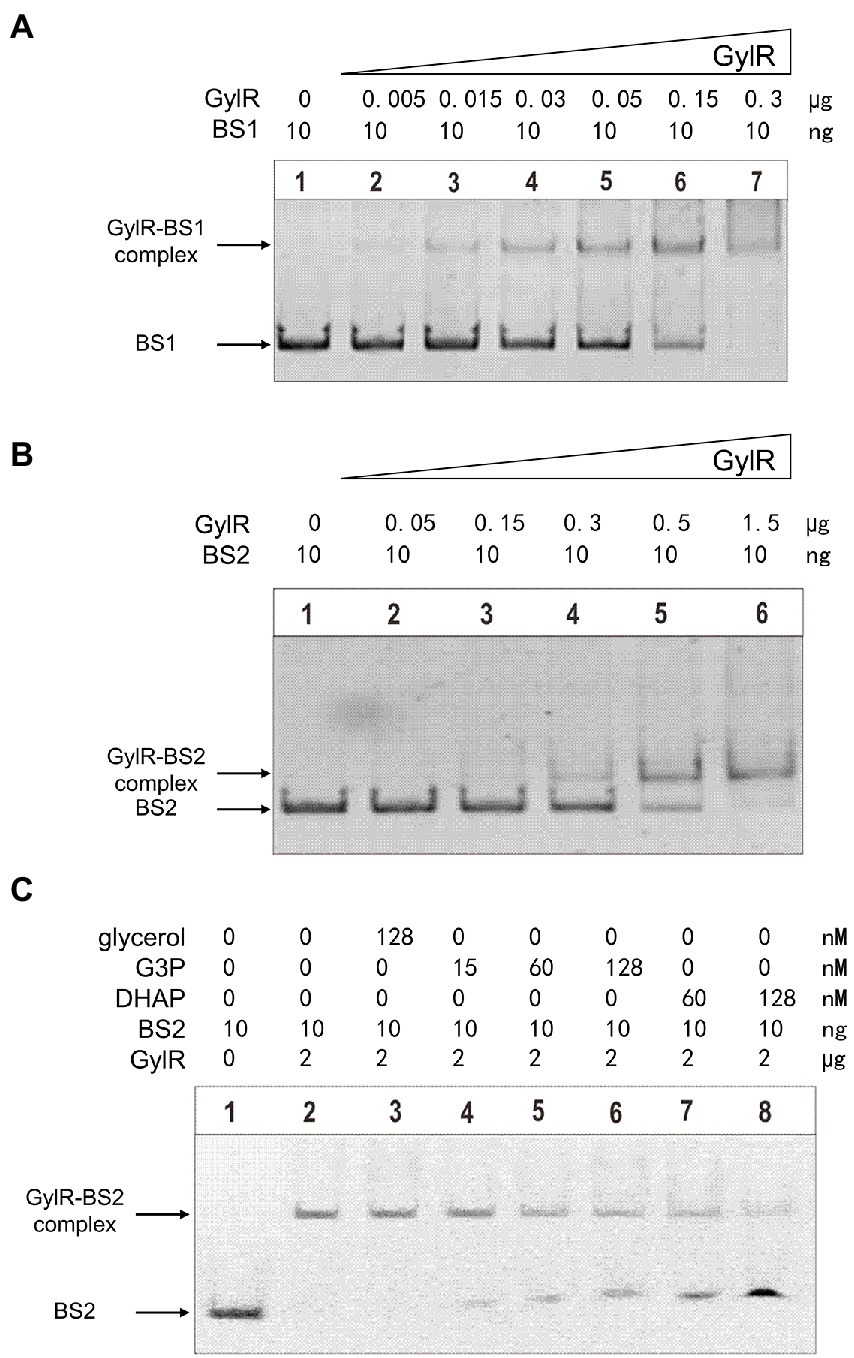
Figure 4. Binding of GylR to the promoter regions of gylR and glp operon, and identification of the effector molecules of GylR by EMSAs. (A) EMSAs of GylR binding to probe BS1 containing Pglp (promoter of glp operon). (B) EMSAs of GylR binding to probe BS2 containing Pgyl (promoter of gylR). (C) Effects of glycerol, G3P, and DHAP on GylR binding to BS2 by EMSAs.
Glycerol-3-phosphate and dihydroxyacetone phosphate are the effector molecules of GylR
To date, the effector molecules that could modulate the binding affinity of GylR to its target DNA regions have not been identified in streptomycetes. Previous preliminary studies in strains like S. coelicolor A3(2) and Mycolicibacterium smegmatis (Seno et al., 1984; Bong et al., 2019) inspired us to propose that the metabolites in the early stage of glycerol metabolic pathway might serve as the effector molecules of GylR in S. clavuligerus NRRL 3585. Therefore, we tested the effects of adding glycerol and its metabolites, G3P and DHAP, to the aforementioned EMSA reactions of GylR with the DNA probe BS2. The results showed that both G3P and DHAP were capable of inducing the dissociation of GylR from BS2, whereas glycerol could not induce the dissociation under tested conditions (Figure 4C). It is worth noting that DHAP was discovered for the first time as an effector molecule of GylR-like regulatory proteins.
Characterization of the binding sequences of GylR revealing a conserved sequence motif
To elucidate the exact binding sequences of GylR at the promoter regions of gylR and glp operon, DNase I footprinting experiments were carried out. The results revealed two protected regions including a region from position +11 to +57 (5′-TGGTCGACATCGACCAATGGTGTTCAGCATTGTCGAATCGCAGTGGG-3′) relative to the transcription start point (tsp) of gylR, and the other region from position −17 to −63 (5′-AAACTGCCGTTCATCGGTCGGCATTGTCGAACACCTACCGGAAATAC-3′) relative to the tsp. of the glp operon (Figure 5A). As shown in Figure 5B, the binding region upstream of gylR is located right after the −10 region of promoter Pgyl, which implied that GylR represses gylR through impeding the transcription elongation. On the other side, the binding region upstream glpF1 overlaps with the promoter Pglp, suggesting that GylR inhibits the transcription of glp operon by hindering the binding of RNA polymerase to Pglp.
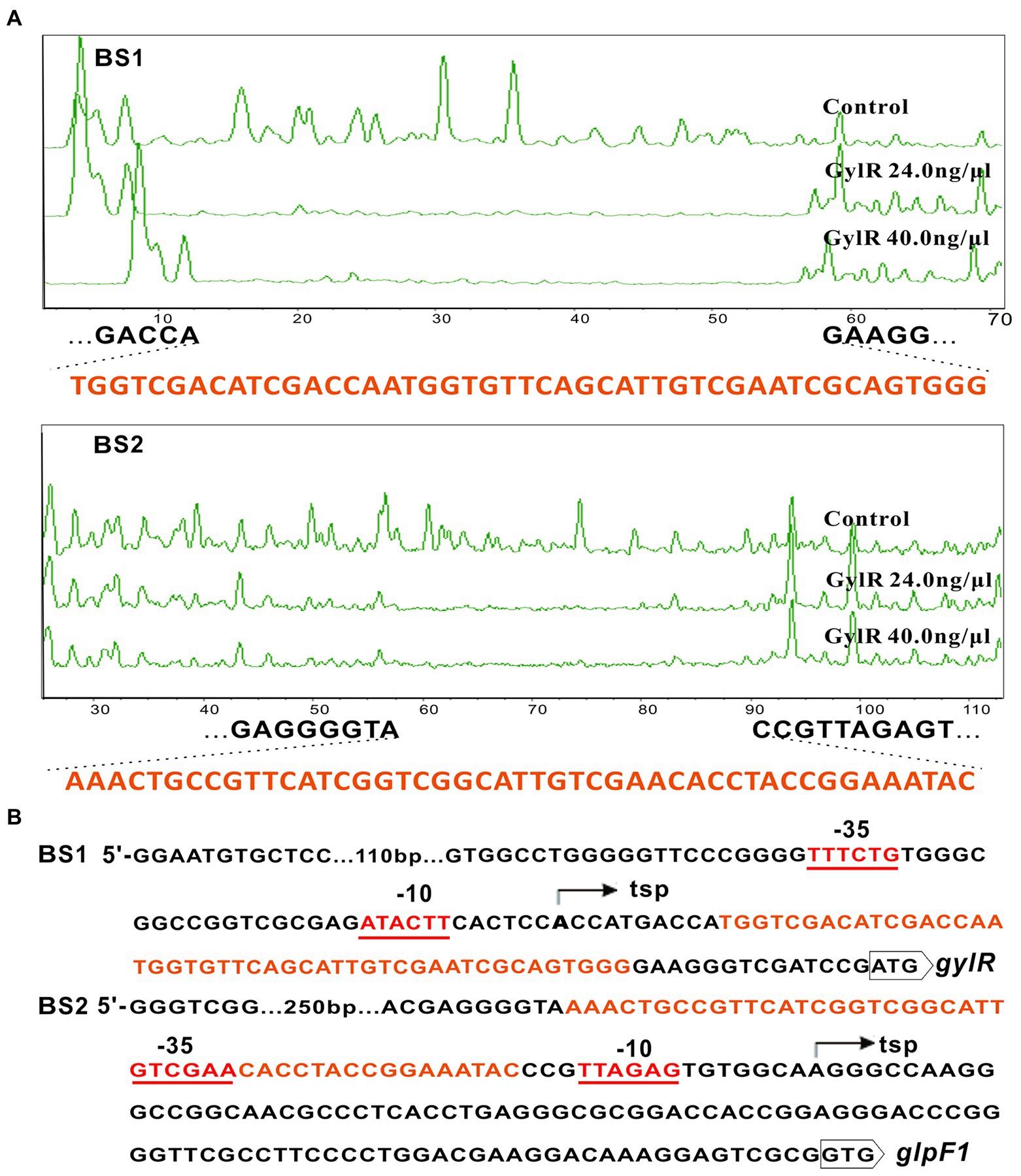
Figure 5. Identification of the binding sequences of GylR and the transcription start sites of gylR and glpF1. (A) DNase I footprinting assays of GylR with the probes BS1 and BS2. Footprints of GylR were shown between dashed lines. (B) Sequence analysis of BS1 and BS2. The identified binding sequences of GylR on BS1 and BS2 were shown in orange. The transcription start sites (tsp) of gylR and glpF1 were determined and marked by bent arrows. The putative − 10 and − 35 regions of both promoters Pgyl and Pglp were underlined.
Analysis of the two binding regions of GylR, and their homologous DNA regions upstream of gylR and glpF1 homologs in representative Streptomyces strains including S. coelicolor A3(2), Streptomyces albus DSM 41398, and Streptomyces avermitilis MA-4680, suggested a relatively conserved 21-bp sequence region (5′-YGKTCRRCATTGYCGAAYVVS-3′; Figure 6A). Then, the corresponding 21-bp DNA probes were synthesized and tested in the EMSAs. The results showed that GylR could bind to all these 21-bp DNA probes (Figure 6A). To identify the minimal core DNA sequences that interact with GylR to form a complex, the 21-bp probe F1 (derived from BS2) was trimmed from two ends separately to afford a set of shortened DNA probes, which were tested in the subsequent EMSAs (Figures 6B,C). The results allowed us to narrow down to a minimum 18-bp palindromic DNA sequence F1-c (5′-GGTCGGCATTGTCGAACA-3′), which could be bound by GylR (Figure 6D). Then, the aforementioned 21-bp DNA probes were all shortened to 18-bp DNA probes, and the EMSA results showed that GylR was able to bind all these 18-bp DNA sequences (Figure 6D). Furthermore, homologous 18-bp DNA sequences were identified in the upstream regions of gylR and glpF1 homologs in all the Streptomyces genomes harboring a glp operon (Supplementary Sheet 3). Analysis of these 18-bp DNA sequences led to the identification of an 18-bp sequence motif (Figure 6E). These results elucidated the detailed DNA binding sequences of GylR in S. clavuligerus NRRL 3585. Moreover, a core 18-bp sequence motif was identified to be bound by GylR-like regulators in Streptomyces based on experimental data and bioinformatic analysis. The molecular regulatory mechanism of GylR revealed by our study and previous reports presumably represents a general regulatory mechanism in glycerol metabolism in Streptomyces.
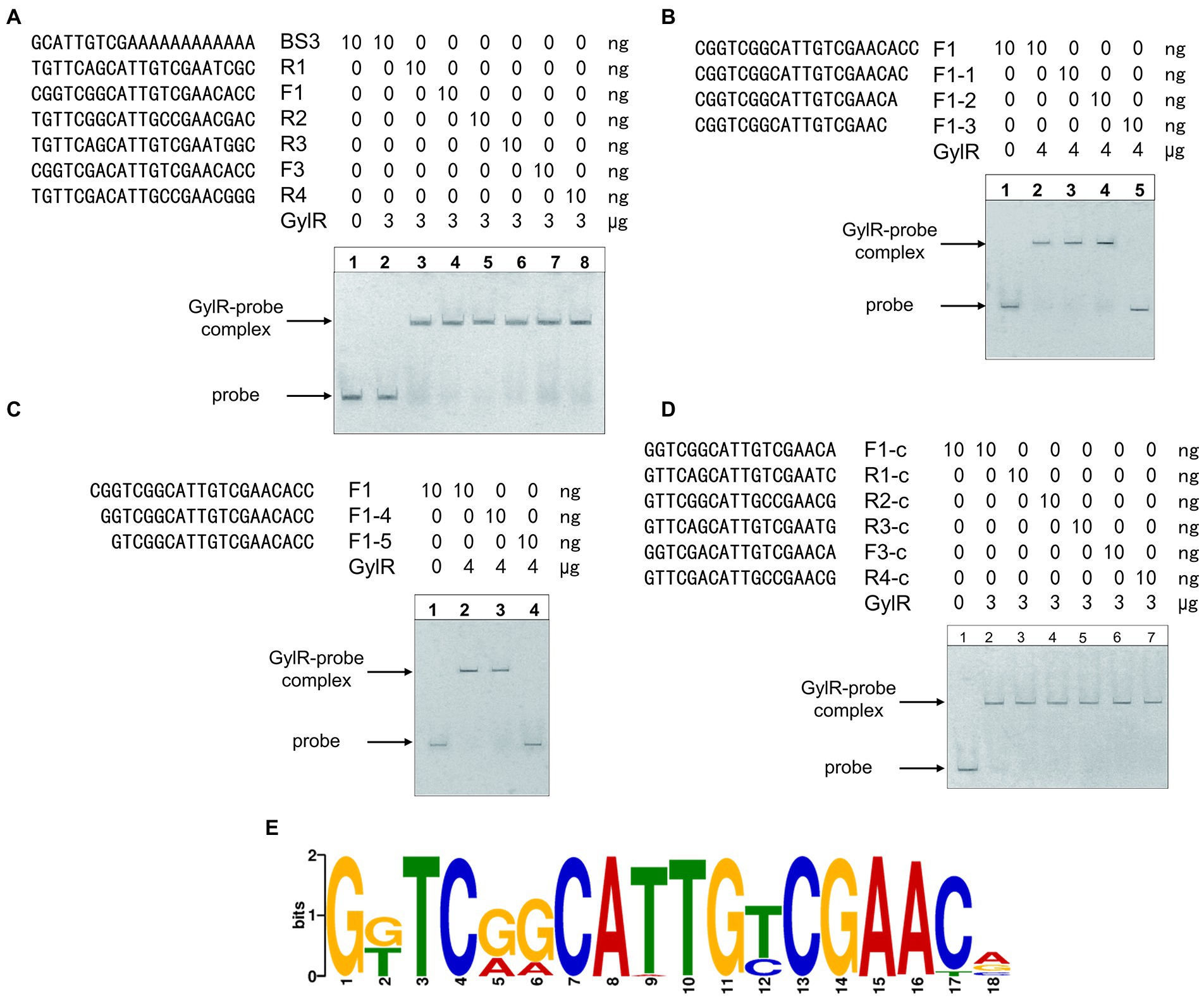
Figure 6. Identification of the core binding sequences of GylR revealing an 18-bp sequence motif. (A) EMSAs of GylR binding to the 21-bp DNA sequences from several representative Streptomyces strains. The DNA probes R1, R2, R3, and R4 referred to the corresponding 21-bp DNA sequences upstream of gylR homologs in S. clavuligerus NRRL 3585, S. coelicolor A3(2), S. albus DSM 41398, and S. avermitilis MA-4680, respectively. The DNA probes F1 and F3 referred to the corresponding 21-bp DNA sequences upstream of glpF1 homologs in S. clavuligerus NRRL 3585, and S. albus DSM 41398, respectively. The corresponding 21-bp DNA sequences upstream of glpF1 homologs in S. coelicolor A3(2) and S. avermitilis MA-4680 were identical to that of probe F1. The probe BS3 was used as a negative control. (B) EMSAs of GylR binding to trimmed F1 probes on the 3-prime end. (C) EMSAs of GylR binding to trimmed F1 probes on the 5-prime end. (D) EMSAs of GylR binding to the core 18-bp DNA sequences derived from the aforementioned representative 21-bp DNA sequences. F1-c, R1-c, R2-c, R3-c, F3-c, and R4-c referred to the core 18-bp DNA sequences derived from the probes F1, R1, R2, R3, F3, and R4, respectively. (E) The conserved 18-bp sequence motif bound by GylR-like proteins in Streptomyces strains. The 18-bp sequence motif was obtained by analyzing the putative binding sequences upstream of gylR homologs and glpF1 homologs in available Streptomyces genomes with MEME Suite 5.5.0 (Bailey et al., 2006).
Discussion
Glycerol can be obtained from multiple paths, especially the growing biodiesel industry that produces glycerol as a main byproduct. The expanding production and reducing cost have made glycerol readily available for versatile applications, one of which is to use it as a carbon source for microorganisms to produce value-added products (da Silva et al., 2009; Guo et al., 2013; Poblete-Castro et al., 2020). As the fertile producers of natural product drugs, streptomycetes can metabolize glycerol through the glp operon that is negatively regulated by GylR, as evident from previous studies in S. coelicolor A3(2) and S. clavuligerus NRRL 3585 (Smith and Chater, 1988b; Hindle and Smith, 1994; Baños et al., 2009). In this study, by analyzing all completed genomes of the phylum Actinobacteria available in the NCBI Genome database, we found that the glp operon-like glycerol utilization gene clusters are conserved in Streptomyces and several other genera of Actinobacteria, suggesting a common glycerol metabolic pathway in these microbes. Using S. clavuligerus NRRL 3585 as a model system, our in vivo data further confirmed that GylR represses the expression of glp operon and gylR, and the inhibition could be relieved upon addition of glycerol to the medium. Characterization of the effector molecules is important for fully understanding the mechanism of a regulatory protein. Prior to our research, there are only few studies on identifying the effectors of GylR-like regulators. In S. coelicolor A3(2), G3P was proposed to be an effector molecule of GylR based on in vivo study (Seno and Chater, 1983; Seno et al., 1984). In Mycolicibacterium smegmatis, in vitro data showed that G3P is an effector molecule of the GylR homolog, which positively regulates the glp operon-like gene cluster (Bong et al., 2019). In E. coli and Pseudomonas spp., the GlpR proteins regulating the glycerol utilization gene clusters show no significant sequence similarity to GylR, and G3P was suggested to be an effector molecule of these GlpR proteins (Lin, 1976; Poblete-Castro et al., 2020). However, there is a lack of direct evidences of identifying the effector molecules of GylR-like regulators in Streptomyces. Our study provided direct evidences showing that both G3P and DHAP are effector molecules of GylR in S. clavuligerus NRRL 3585, while glycerol is not. DHAP is for the first time discovered as an effector molecule of GylR-like regulatory proteins. Previous study in E. coli suggested that DHAP might have weak interaction with GlpR (Larson et al., 1987). These findings further supported the feedback activation mechanism in the regulation of glycerol metabolism (Lin, 1976; Baños et al., 2009). Specifically, due to the tight control of GylR, the glp operon expressed at a very low level initially, which enabled the slow conversion of glycerol to G3P and DHAP. Once these effector molecules reached to concentrations that could induce the dissociation of GylR from the binding of glp operon, the glycerol metabolic pathway was fully activated.
We further revealed the binding sequences of GylR at the promoter regions of glp operon and gylR, which was not known previously. The results suggested that GylR inhibits the expression of gylR and glp operon through impeding the transcription initiation and elongation, respectively. Based on experimental data and bioinformatic analysis, we were able to identify a palindromic 18-bp sequence motif important for the binding of GylR-like regulators in Streptomyces. It is worth noting that, the glp operon in Streptomyces could also be regulated by other proteins or pathways. In S. coelicolor A3(2), the glp operon is negatively regulated by the pleiotropic protein NdgR, the binding site of which does not overlap with the binding sequences of GylR (Lee et al., 2017). In addition, the glp operon in S. coelicolor A3(2) was reported to be subject to carbon catabolite repression (Smith and Chater, 1988a,b).
Glycerol-inducible regulatory elements have many applications in the metabolic engineering and synthetic biology research fields. The promoter of glp operon (Pglp) has been successfully used to coordinate the glycerol utilization with natural product biosynthesis, leading to 3.2-fold enhanced clavulanic acid production in S. clavuligerus NRRL 3585 (Guo et al., 2013). A glycerol-inducible expression system based on the antiterminator protein GlpP and its target promoter PglpD was applied in the heterologous production of aspartase and nattokinase in B. subtilis (Han et al., 2022). Replacing the promoters of zwf, pgi and pfk1 genes with the glycerol-inducible promoter pGUT1, resulting in significantly improved production of myo-inositol in Pichia pastoris (Zhang et al., 2022). Understanding the underlying molecular regulatory mechanisms in glycerol metabolism will lay a foundation for the design of efficient and simple glycerol-inducible regulatory tools.
In conclusion, by taking S. clavuligerus NRRL 3585 as a model system, we have identified the effector molecules and binding sequences of GylR, a key regulator in the glycerol metabolic pathway. Furthermore, we identified a core 18-bp sequence motif that is minimally required for the binding of GylR-like regulators in Streptomyces. These findings further revealed the molecular mechanism mediated by GylR in the regulation of glycerol metabolism. The information is of importance for the design of better glycerol utilization pathways that can improve utilization of glycerol to produce valuable products. Moreover, these findings would facilitate the development and application of glycerol-inducible regulatory elements for synthetic biology research in the future.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
YZ, KF, and GP conceived and designed the project. CZ and YZ performed the experiments. KF carried out the bioinformatics analyses. CZ, KF, and GP analyzed the data and wrote the manuscript with the input of all authors. YH, WW, and ZL provided key suggestions and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key R&D Program of China (2021YFC2100600 and 2020YFA0907700), and the National Natural Science Foundation of China (32070067).
Acknowledgments
We thank Yihua Chen (Institute of Microbiology, Chinese Academy of Sciences) for the insightful comments and suggestions on the preparation of the manuscript. This manuscript is dedicated to the memory of our beloved mentor Keqian Yang.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1078293/full#supplementary-material
References
Bailey, T. L., Williams, N., Misleh, C., and Li, W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. doi: 10.1093/nar/gkl198
Baños, S., Pérez-Redondo, R., Koekman, B., and Liras, P. (2009). Glycerol utilization gene cluster in Streptomyces clavuligerus. Appl. Environ. Microbiol. 75, 2991–2995. doi: 10.1128/AEM.00181-09
Bong, H.-J., Ko, E.-M., Song, S.-Y., Ko, I.-J., and Oh, J.-I. (2019). Tripartite regulation of the glpFKD operon involved in glycerol catabolism by GylR, Crp, and SigF in mycobacterium smegmatis. J. Bacteriol. 201:e00511. doi: 10.1128/JB.00511-19
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinform. 10:421. doi: 10.1186/1471-2105-10-421
Da Silva, G. P., Mack, M., and Contiero, J. (2009). Glycerol: a promising and abundant carbon source for industrial microbiology. Biotechnol. Adv. 27, 30–39. doi: 10.1016/J.BIOTECHADV.2008.07.006
Doi, Y. (2019). Glycerol metabolism and its regulation in lactic acid bacteria. Appl. Microbiol. Biotechnol. 103, 5079–5093. doi: 10.1007/S00253-019-09830-Y
Guo, D., Zhao, Y., and Yang, K. (2013). Coordination of glycerol utilization and clavulanic acid biosynthesis to improve clavulanic acid production in Streptomyces clavuligerus. Sci. China Life Sci. 56, 591–600. doi: 10.1007/s11427-013-4507-z
Han, L., Chen, Q., Luo, J., Cui, W., and Zhou, Z. (2022). Development of a glycerol-inducible expression system for high-yield heterologous protein production in Bacillus subtilis. Microbiol. Spectr. 10:e0132222. doi: 10.1128/SPECTRUM.01322-22
Hindle, Z., and Smith, C. P. (1994). Substrate induction and catabolite repression of the Streptomyces coelicolor glycerol operon are mediated through the GylR protein. Mol. Microbiol. 12, 737–745. doi: 10.1111/j.1365-2958.1994.tb01061.x
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F., and Hopwood, D. A. (2000). Practical Streptomyces Genetics. Norwich: The John Innes Foundation.
Klein, M., Swinnen, S., Thevelein, J. M., and Nevoigt, E. (2017). Glycerol metabolism and transport in yeast and fungi: established knowledge and ambiguities. Environ. Microbiol. 19, 878–893. doi: 10.1111/1462-2920.13617
Kormanec, J., Rezuchova, B., Homerova, D., Csolleiova, D., Sevcikova, B., Novakova, R., et al. (2019). Recent achievements in the generation of stable genome alterations/mutations in species of the genus Streptomyces. Appl. Microbiol. Biotechnol. 103, 5463–5482. doi: 10.1007/s00253-019-09901-0
Larson, T. J., Ye, S. Z., Weissenborn, D. L., Hoffmann, H. J., and Schweizer, H. (1987). Purification and characterization of the repressor for the sn-glycerol 3-phosphate regulon of Escherichia coli K12. J. Biol. Chem. 262, 15869–15874.
Lee, B. R., Bhatia, S. K., Song, H. S., Kim, J., Kim, W., Park, H., et al. (2017). The role of NdgR in glycerol metabolism in Streptomyces coelicolor. Bioprocess Biosyst. Eng. 40, 1573–1580. doi: 10.1007/S00449-017-1813-Z
Lee, N., Hwang, S., Kim, W., Lee, Y., Kim, J. H., Cho, S., et al. (2021). Systems and synthetic biology to elucidate secondary metabolite biosynthetic gene clusters encoded in Streptomyces genomes. Nat. Prod. Rep. 38, 1330–1361. doi: 10.1039/d0np00071j
Li, S., Wang, W., Li, X., Fan, K., and Yang, K. (2015). Genome-wide identification and characterization of reference genes with different transcript abundances for Streptomyces coelicolor. Sci. Rep. 5:15840. doi: 10.1038/srep15840
Lin, E. C. (1976). Glycerol dissimilation and its regulation in bacteria. Annu. Rev. Microbiol. 30, 535–578. doi: 10.1146/ANNUREV.MI.30.100176.002535
Pagliaro, M., Ciriminna, R., Kimura, H., Rossi, M., and della Pina, C. (2007). From glycerol to value-added products. Angew. Chem. Int. Ed. Engl. 46, 4434–4440. doi: 10.1002/ANIE.200604694
Pan, G., Gao, X., Fan, K., Liu, J., Meng, B., Gao, J., et al. (2017). Structure and function of a C-C bond cleaving oxygenase in atypical angucycline biosynthesis. ACS Chem. Biol. 12, 142–152. doi: 10.1021/acschembio.6b00621
Poblete-Castro, I., Wittmann, C., and Nikel, P. I. (2020). Biochemistry, genetics and biotechnology of glycerol utilization in pseudomonas species. Microb. Biotechnol. 13, 32–53. doi: 10.1111/1751-7915.13400
Rice, P., Longden, I., and Bleasby, A. (2000). EMBOSS: the European molecular biology open software suite. Trends Genet. 16, 276–277. doi: 10.1016/s0168-9525(00)02024-2
Saudagar, P. S., Survase, S. A., and Singhal, R. S. (2008). Clavulanic acid: a review. Biotechnol. Adv. 26, 335–351. doi: 10.1016/j.biotechadv.2008.03.002
Seno, E. T., Bruton, C. J., and Chater, K. F. (1984). The glycerol utilization operon of Streptomyces coelicolor: genetic mapping of gyl mutations and the analysis of cloned gylDNA. Mol. Gen. Genet. 193, 119–128. doi: 10.1007/BF00327424
Seno, E. T., and Chater, K. F. (1983). Glycerol catabolic enzymes and their regulation in wild-type and mutant strains of Streptomyces coelicolor A3(2). J. Gen. Microbiol. 129, 1403–1413. doi: 10.1099/00221287-129-5-1403
Shin, C. H., Cho, H. S., Won, H. J., Kwon, H. J., Kim, C. W., and Yoon, Y. J. (2021). Enhanced production of clavulanic acid by improving glycerol utilization using reporter-guided mutagenesis of an industrial Streptomyces clavuligerus strain. J. Ind. Microbiol. Biotechnol. 48:kuab004. doi: 10.1093/JIMB/KUAB004
Smith, C. P., and Chater, K. F. (1988a). Cloning and transcription analysis of the entire glycerol utilization (gylABX) operon of Streptomyces coelicolor A3(2) and identification of a closely associated transcription unit. Mol. Gen. Genet. 211, 129–137.
Smith, C. P., and Chater, K. F. (1988b). Structure and regulation of controlling sequences for the Streptomyces coelicolor glycerol operon. J. Mol. Biol. 204, 569–580.
Suvorova, I. A., and Gelfand, M. S. (2021). Comparative analysis of the IclR-family of bacterial transcription factors and their DNA-binding motifs: structure, positioning, co-evolution, regulon content. Front. Microbiol. 12:675815. doi: 10.3389/FMICB.2021.675815
Wang, L., Tian, X., Wang, J., Yang, H., Fan, K., Xu, G., et al. (2009). Autoregulation of antibiotic biosynthesis by binding of the end product to an atypical response regulator. Proc. Natl. Acad. Sci. U. S. A. 106, 8617–8622. doi: 10.1073/pnas.0900592106
Wang, W., Yang, T., Li, Y., Li, S., Yin, S., Styles, K., et al. (2016). Development of a synthetic oxytetracycline-inducible expression system for Streptomycetes using de novo characterized genetic parts. ACS Synth. Biol. 5, 765–773. doi: 10.1021/acssynbio.6b00087
Xiang, S., Li, J., Yin, H., Zheng, J., Yang, X., Wang, H.-B., et al. (2009). Application of a double-reporter-guided mutant selection method to improve clavulanic acid production in Streptomyces clavuligerus. Metab. Eng. 11, 310–318. doi: 10.1016/j.ymben.2009.06.003
Zhang, Q., Wang, X., Luo, H., Wang, Y., Wang, Y., Tu, T., et al. (2022). Metabolic engineering of Pichia pastoris for myo-inositol production by dynamic regulation of central metabolism. Microb. Cell Factories 21:112. doi: 10.1186/S12934-022-01837-X
Keywords: glycerol, metabolism, regulatory mechanism, sequence motif, Streptomyces, Actinobacteria
Citation: Zhang C, Zhao Y, Li Z, Wang W, Huang Y, Pan G and Fan K (2022) Molecular mechanism of GylR-mediated regulation of glycerol metabolism in Streptomyces clavuligerus NRRL 3585. Front. Microbiol. 13:1078293. doi: 10.3389/fmicb.2022.1078293
Edited by:
Dewu Zhang, Chinese Academy of Medical Sciences, ChinaReviewed by:
Guoqing Niu, Southwest University, ChinaChin-Yuan Chang, National Chiao Tung University, Taiwan
Copyright © 2022 Zhang, Zhao, Li, Wang, Huang, Pan and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Keqiang Fan, ZmFua3FAaW0uYWMuY24=
 Chaobo Zhang1,2
Chaobo Zhang1,2 Zilong Li
Zilong Li Weishan Wang
Weishan Wang Ying Huang
Ying Huang Guohui Pan
Guohui Pan Keqiang Fan
Keqiang Fan