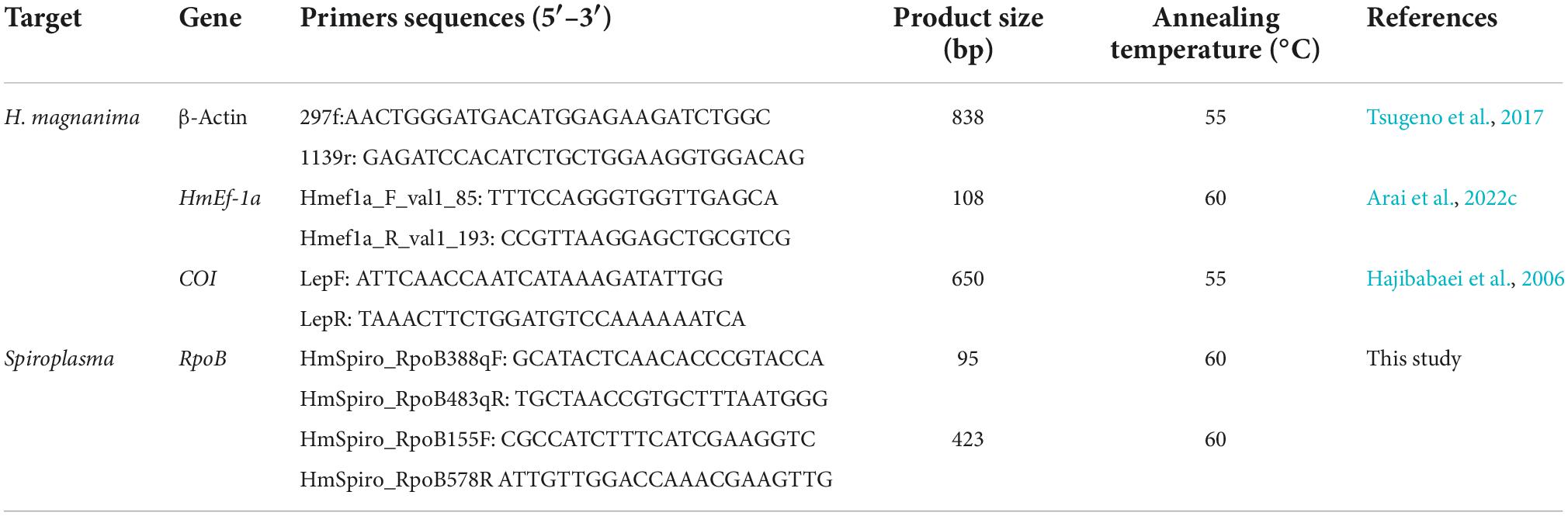
- 1United Graduate School of Agricultural Science, Tokyo University of Agriculture and Technology, Fuchu, Japan
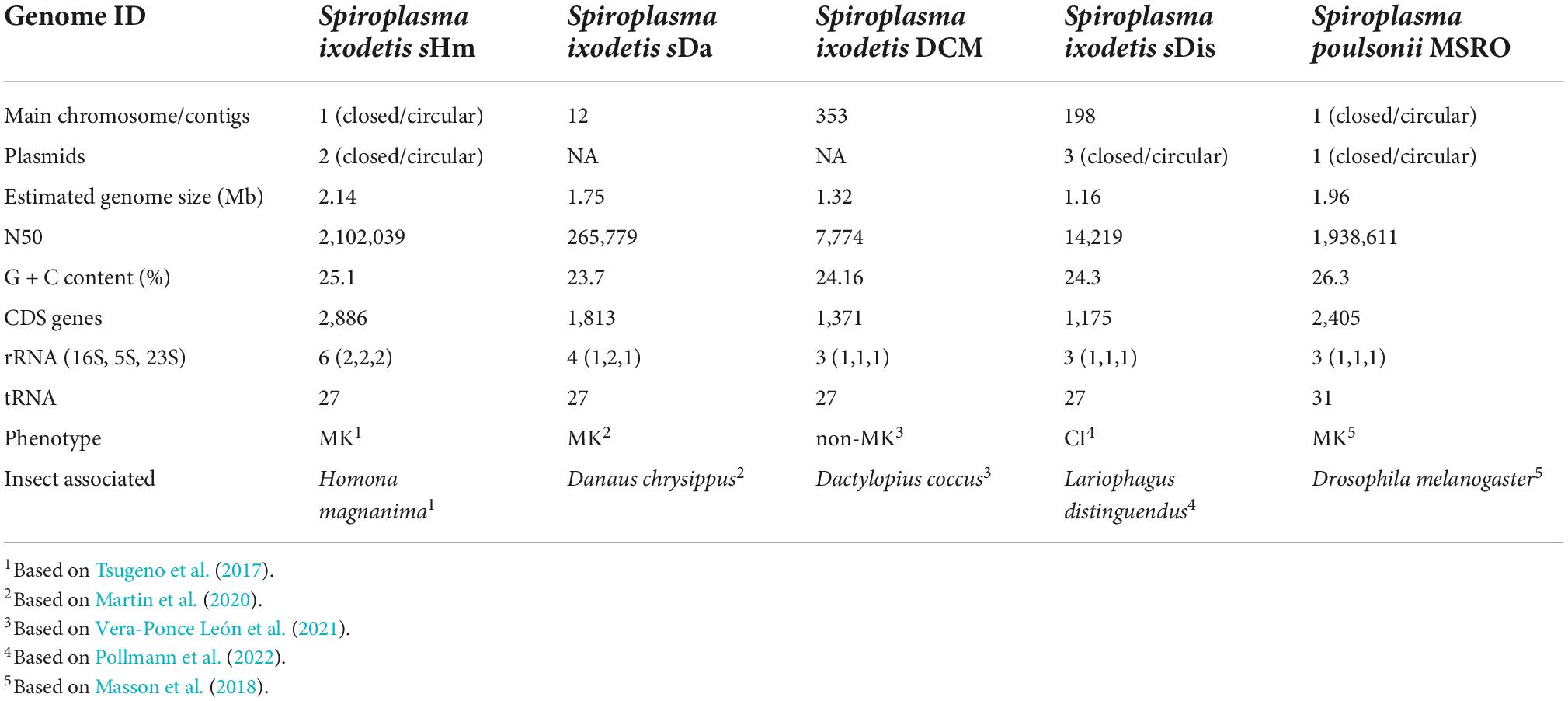
- 2Institute of Agrobiological Sciences, National Agriculture and Food Research Organization (NARO), Tsukuba, Japan
Male-killing, a male-specific death of arthropod hosts during development, is induced by Spiroplasma (Mollicutes) endosymbionts of the Citri–Poulsonii and the Ixodetis groups, which are phylogenetically distant groups. Spiroplasma poulsonii induces male-killing in Drosophila melanogaster (Diptera) using the Spaid toxin that harbors ankyrin repeats, whereas little is known about the origin and mechanisms of male-killing induced by Spiroplasma ixodetis. Here, we analyzed the genome and the biological characteristics of a male-killing S. ixodetis strain sHm in the moth Homona magnanima (Tortricidae, Lepidoptera). Strain sHm harbored a 2.1 Mb chromosome and two potential plasmids encoding Type IV effectors, putatively involved in virulence and host–symbiont interactions. Moreover, sHm did not harbor the spaid gene but harbored 10 ankyrin genes that were homologous to those in other S. ixodetis strains. In contrast to the predominant existence of S. poulsonii in hemolymph, our quantitative PCR assays revealed a systemic distribution of strain sHm in H. magnanima, with particularly high titers in Malpighian tubules but low titers in hemolymph. Furthermore, transinfection assays confirmed that strain sHm can infect cultured cells derived from distantly related insects, namely Aedes albopictus (Diptera) and Bombyx mori (Lepidoptera). These results suggest different origins and characteristics of S. ixodetis- and S. poulsonii-induced male-killing.
Introduction
In arthropods, maternally inherited endosymbiotic microbes frequently interact with the hosts in a mutualistic or a parasitic manner. Male-killing (MK), male-specific death in insects during development, is one of the reproductive manipulations induced by various intracellular bacteria, microsporidia, and viruses (Duron et al., 2008; Werren et al., 2008; Kageyama et al., 2012; Fujita et al., 2021). MK leads to the advantage of female siblings and is considered a selfish strategy of the intracellular microbes that promotes their spread and survival in nature (Hurst, 1991; Hurst and Jiggins, 2000; Hornett et al., 2006). The genus Spiroplasma (class: Mollicutes) are the most studied bacteria that induce MK in diverse insects (Anbutsu and Fukatsu, 2011; Lo et al., 2015; Harumoto and Lemaitre, 2018; Binetruy et al., 2019). Spiroplasma are small, helical, and motile bacteria that include commensal, pathogenic, and mutualistic species and have a diverse host range, including plants and animals (Regassa and Gasparich, 2006; Duperron et al., 2013; Viver et al., 2017; He et al., 2018). Phylogenetically, the MK Spiroplasma strains are clustered into the Citri–Poulsonii group (harbored by Drosophila flies and lacewings) (Williamson and Poulson, 1979; Hayashi et al., 2016) and the Ixodetis clade (harbored by ladybugs, butterflies, moths, and aphids) (Hurst et al., 1999; Simon et al., 2011; Tabata et al., 2011; Smith et al., 2016).
The molecular mechanisms underlying Spiroplasma-induced MK have been mostly investigated using S. poulsonii–Drosophila systems (Harumoto and Lemaitre, 2018). S. poulsonii strain MSRO induces MK in Drosophila melanogaster by a toxic protein androcidin (Spaid) harboring ankyrin repeats that damage the male X chromosome (Harumoto and Lemaitre, 2018). In contrast, information regarding the mechanism underlying MK induced by the members of the Ixodetis group is limited. The spaid gene is conserved among S. poulsonii strains (Harumoto and Lemaitre, 2018; Gerth et al., 2021), whereas whether the S. ixodetis group uses Spaid as an MK factor is unknown. The genus Spiroplasma exhibits high genomic flexibility and dynamic evolution of various toxin loci, such as Spaid and ribosome-inactivating protein (RIP) (Hamilton et al., 2016; Ballinger et al., 2019; Gerth et al., 2021; Massey and Newton, 2022; Pollmann et al., 2022). Gnomic analyses have revealed dynamic Spiroplasma evolution driven by bacteriophage lysogenization (Ye et al., 1996; Carle et al., 2010; Ku et al., 2013) and by horizontal gene transfer (Mouches et al., 1984; Joshi et al., 2005). Virulence-associated genes are frequently exchanged between microbes sharing the same niche (Kent and Bordenstein, 2010; Wiedenbeck and Cohan, 2011). Likewise, Spiroplasma may have acquired MK genes by horizontal gene transfer because they often coexist with other endosymbionts, such as Wolbachia and Rickettsia, in the same host (Hurst et al., 1999; Majerus et al., 2000; Watanabe et al., 2012; Hayashi et al., 2016; Takamatsu et al., 2021). However, the horizontal gene transfer to Spiroplasma may be constrained by the unusual codon usage by Spiroplasma compared with other bacteria (notably, the use of UGA as a tryptophan rather than a stop codon; Lo et al., 2015). Although genomic studies on MK S. poulsonii have been done, comparative genomic analyses of other MK Spiroplasma species, such as S. ixodetis, are essential to infer the origin and evolution of the MK machinery.
In this study, we sequenced the genome of S. ixodetis strain sHm that causes MK in the tea tortrix moth Homona magnanima (Tortricidae, Lepidoptera). Against the full-genome sequence of strain sHm, we searched for genes encoding Spaid and RIP toxin homologs, as well as putative MK genes of other MK endosymbionts such as Wolbachia (Arai et al., 2020, 2022a) and Partiti-like virus Osugoroshivirus (OGVs) (Fujita et al., 2021) in H. magnanima. We also examined the propagation characteristics and infectivity of strain sHm using quantification and transinfection assays. Finally, we argue that MK mechanisms and ecological characteristics are substantially different between Spiroplasma species.
Materials and methods
Rearing and sexing of Homona magnanima
To construct S. ixodetis sHm genome, we used the laboratory-maintained Spiroplasma-positive MK-inducing line (S+ line) of H. magnanima (Tsugeno et al., 2017). In the present study, we accidentally obtained a Spiroplasma-positive 1:1 sex ratio line (S+M+ line) as a subline of the S+ line. For every generation, the male moths picked up from the 1:1 sex ratio line, which had been confirmed negative for Spiroplasma, Wolbachia, and OGV (NSR line) (Takamatsu et al., 2021), were crossed with the female moths of the S+ and S+M+ lines as described by Arai et al. (2022a). The obtained larvae were reared using artificial diet SilkMate 2S (Nosan Co., Yokohama, Japan) at 25°C under a long photoperiod (16L:8D), i.e., till pupation. To eliminate Spiroplasma from the S+ line, the first instar larvae were reared with SilkMate 2S supplemented with 0.05% tetracycline (w/w) as described by Arai et al. (2019). Adult moths were sexed based on their morphology, and the hatched larvae and the unhatched pharate larvae (mature embryo) were sexed based on the presence or absence of the female-specific sex chromatin body (a condensed W chromosome), which was detected via lactic-acetic orcein staining (Arai et al., 2022a).
Spiroplasma detection and quantification in Homona magnanima
Total DNA was extracted from the abdomen of female adults (0-day post eclosion), the whole body of larvae and pupae (0-day post molting), and dissected tissues of H. magnanima larvae (0-day post molting) using cell lysis buffer, as described by Arai et al. (2019). To detect Spiroplasma, a pair of Spiroplasma-specific primers was used to amplify RNA polymerase β subunit gene (RpoB), which is a single copy conserved gene in Spiroplasma spp., from the extracted DNA (adjusted to 50–100 ng/reaction) with EmeraldAmp MAX PCR Master Mix (TaKaRa Bio, Shiga, Japan); the primer sets are listed in Table 1. The PCR conditions were as follows: 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, followed by 72°C for 7 min. β-Actin gene of H. magnanima was used as the control. To quantify Spiroplasma density, qPCR was performed using the extracted DNA, which was diluted to a concentration of 10 ng/μL with MilliQ water, Spiroplasma RpoB primers (Table 1), and KOD SYBR® qPCR Mix (Toyobo, Osaka, Japan) in a LightCycler® 96 system (Roche, Basel, Switzerland). The PCR consisted of 45 cycles of 98°C for 10 s, 60°C for 10 s, and 68°C for 30 s. Relative abundance of the gene was calculated using the expression of elongation factor 1a gene (ef1a) of H. magnanima as the control. Spiroplasma density (RpoB copies) and relative abundance (RpoB/ef1a) were calculated as described in Arai et al. (2019, 2022c).
Genome sequence of the Spiroplasma sHm strain
For genome sequencing of strain sHm, high molecular weight DNA was extracted from the egg masses of S+ line moths using Nanobind Tissue Big DNA Kit (Circulomics Inc., MD, USA) and used for library construction using Ultra-Long DNA Sequencing Kit (Oxford Nanopore Technologies, Oxford, UK) following the manufacture’s protocol. The constructed libraries were sequenced using ONT MinION flow cell (R 9.4.1) (Oxford Nanopore Technologies). The obtained reads were mapped to the H. magnanima reference genome (Jouraku et al., in preparation) with minimap2 (Li, 2018), and the non-mapped reads containing Spiroplasma reads were extracted with SAMtools v.1.9 (Li et al., 2009) and assembled using Canu 1.6 (Koren et al., 2017). The draft Spiroplasma genome (a circular main chromosome and plasmids) was annotated via BLASTn (NCBI nr database). The extracted DNA was also subjected to Illumina paired-end 150 bp sequencing (PE-150) at Novogene (Beijing, China). The Illumina data were used to polish the draft genome using minimap2 (Li, 2018) and Pilon v. 1.23 (Walker et al., 2014). Since no sequence changes were observed after the second polishing, the polished genome was considered as the complete genome of strain sHm. The circularity of the sHm genome was confirmed by BLASTn search, followed by manual deletion of overlapping sequence.
Resequencing of the sHm strain in the S+ and S+M+ moth lines
S+ (MK line) and S+M+ H. magnanima lines (non-MK line) were used for DNA extraction as described by Arai et al. (2022a). The DNA extracted from Spiroplasma cells was amplified using whole genome amplification (WGA) by REPLI-g Mini Kit (Qiagen, Hilden, Germany), following the manufacture’s protocol. The WGA products, purified using AMPure XP beads (Beckman Coulter, Inc., CA, USA) and dissolved into TE buffer, were sequenced on Illumina platform (PE-150). The Illumina data assembled with unicycler (Wick et al., 2017) and Illumina raw read data were mapped to the sHm reference genome using minimap2 (Li, 2018) to detect the genomic changes in the genome of sHm in the S + M + line.
Genome annotations and homology surveys
The constructed sHm genome was annotated via DFAST (Tanizawa et al., 2018). Effector genes were further annotated using EffectiveDB (Eichinger et al., 2016). Functional analysis of proteins (i.e., domain predictions and Gene ontology annotations) was conducted using InterPro.1 Phage WO infections were annotated using PHASTER (Arndt et al., 2016). Protein homology between different Spiroplasma strains was analyzed using S. apis B31 (CP006682.1), S. citri strain BLH-MB (CP047437.1–CP047446.1), S. syrphidicola strain EA-1 (NC_021284.1), D. melanogaster endosymbiont S. poulsonii MSRO (CM020866.1–CM020867.1) (sMel, MK strain, Masson et al., 2018), Danaus chrysippus (Nymphalidae) endosymbiont S. ixodetis (NZ_CADDIL010000001.1–NZ_CADDIL010000012.1) (sDa, MK strain, Martin et al., 2020), Lariophagus distinguendus (Pteromalidae) endosymbiont S. ixodetis (NZ_JALMUW010000001.1–NZ_JALMUW010000198.1) [sDis, cytoplasmic incompatibility (CI) strain, Pollmann et al., 2022], and Dactylopius coccus (Dactylopiidae) endosymbiont S. ixodetis (JACSER010000001.1–JACSER010000358.1) (sCoc, non-MK strain, Vera-Ponce León et al., 2021) with OrthoVenn2.2 Homology of sHm genes and proteins with spaid from strain sMel (Harumoto and Lemaitre, 2018), ankyrin genes from S. ixodetis (Yeoman et al., 2019; Martin et al., 2020; Vera-Ponce León et al., 2021), and the Wolbachia MK candidate factor responsible for WO-mediated killing (Wmk, presumed helix-turn-helix transcriptional regulator, Perlmutter et al., 2019; Arai et al., 2022b) was evaluated using both BLASTn and BLASTp. Moreover, to verify whether MK microbes of H. magnanima carried conserved genes, the genes on the MK-associated prophage region WOwHm-t76 of MK Wolbachia wHm-t (Arai et al., 2022b) and those of the Partiti-like virus OGVs (Fujita et al., 2021) were compared to the sHm genes using both BLASTn and BLASTp. Unique genomic features of the sHm strain in the MK S+ and non-MK S+M+ H. magnanima lines were analyzed using GView3 and BV-BRC variation analysis service.4 Metabolic pathways of S. poulsonii sMel and S. ixodetis sHm were compared by using BV-BRC comparative analysis service (see text footnote 4). Phylogenetic trees of 16S rRNA gene and ankyrin genes of Spiroplasma strains were constructed by maximum likelihood with bootstrap re-sampling of 1,000 replicates using MEGA7 (Kumar et al., 2016). Mycoplasma genitalium G-37 (NR074611.1) was used as an outgroup.
Transinfection assays
A fifth instar female larva was sterilized in 50% bleach (ca. 3% sodium hypochlorite) for 10 min, in 70% ethanol for 10 min, and dissected in IPL-41 Insect Medium (Gibco, Carlsbad, CA, USA) supplemented with 10% (v/v) fetal bovine serum (FBS). Malpighian tubules of the dissected larva were transferred to flasks containing either the Bombyx mori NIAS-Bm-aff3 (aff3) cell line (Takahashi et al., 2006) or the Aedes albopictus NIAS-AeAl-2 (AeAl2) cell line (Mitsuhashi, 1981) maintained in IPL-41 Insect Medium (Gibco) with 10% (v/v) of FBS. The cells and Malpighian tubules were co-cultured at 23°C. Fresh medium was supplied to the flask every 10 d. Purified cells centrifuged at 1,000 g for 2 min were used to analyze infections and titers of the transinfected strain sHm in the cells. DNA extraction, PCR, and qPCR assays were performed as mentioned in section “Spiroplasma detection and quantification in Homona magnanima.”
Statistical analysis
Sex ratio bias was assessed using Fisher’s exact test. Spiroplasma densities, male ratio in hatched larvae, and male ratio in unhatched pharate larvae were analyzed using either the Wilcoxon test or the Steel–Dwass test. All analyses were performed using R software v4.05.
Results and discussion
Spiroplasma ixodetis strain sHm induced embryonic male death in Homona magnanima
The S+ line moths harboring sHm exhibited lower egg-hatching rates than the NSR line (Figure 1A), which is consistent with the results from previous studies that Spiroplasma infection halved the egg hatching rates of H. magnanima (Tsugeno et al., 2017; Takamatsu et al., 2021). Cytogenetic sexing based on the presence or absence of a sex chromatin body (W chromosome) revealed that the sex ratio of hatched larvae was strongly biased toward females in the S+ line moths but not in the NSR line moths (P < 0.01, Figure 1B). In contrast, the sex ratio of unhatched pharate larvae (late-stage embryos) were male-biased in the sHm-infected line (S+) (Fisher’s exact test, P < 0.01), confirming that sHm killed male H. magnanima during embryogenesis. Moreover, the elimination of Spiroplasma by tetracycline treatment resulted in non-biased sex ratios in the subsequent generation (P < 0.01, Figure 1C).
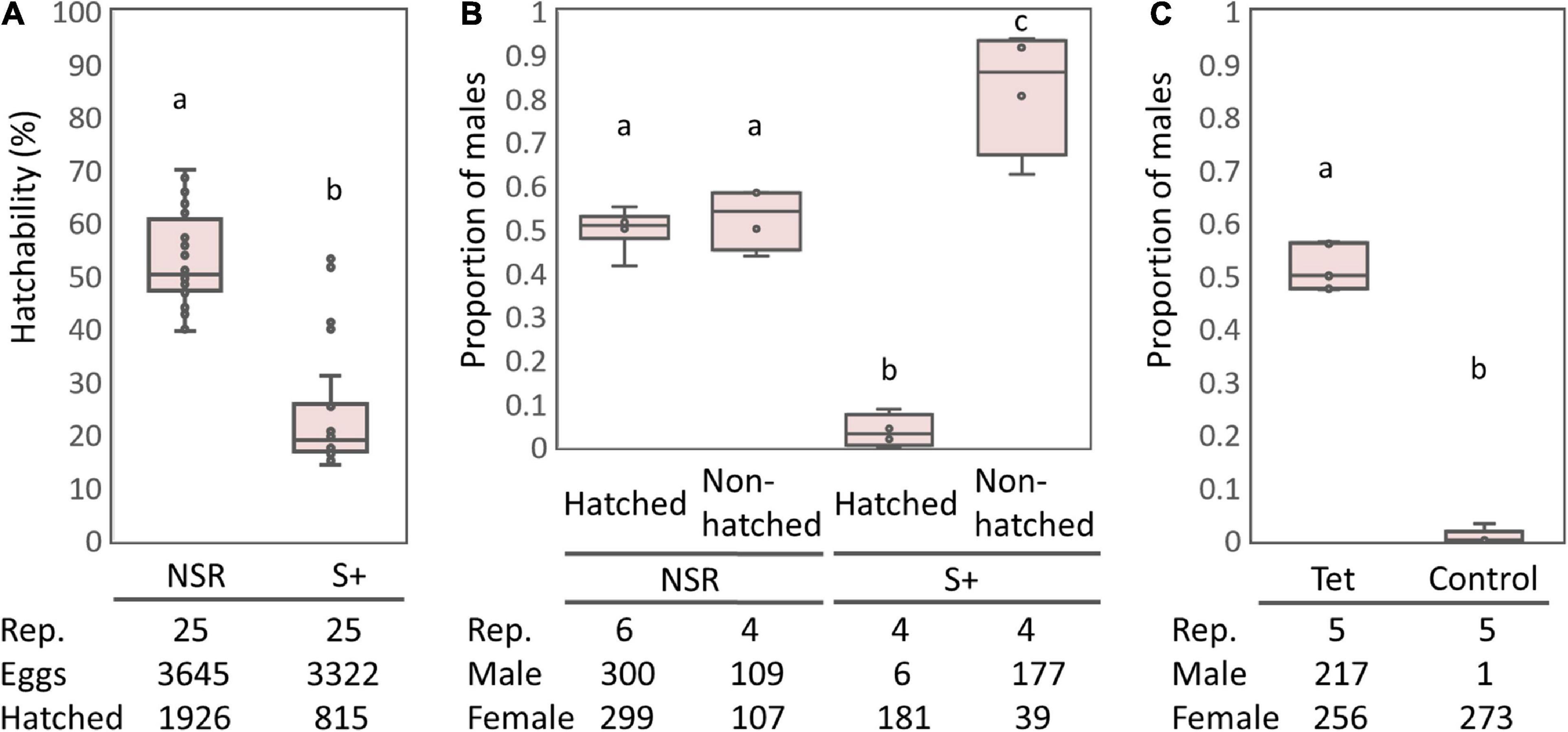
Figure 1. Egg hatching rates and sex ratios of the S+ line of Homona magnanima. (A) Egg hatching rates of 25 egg masses for each of the NSR and S+ lines (total 3,645 and 3,322 eggs, respectively). (B) Proportion of males among the hatched larvae and the unhatched pharate larvae. (C) Proportion of males of the subsequent generation after tetracycline treatment. Tet, tetracycline treatment; Control, non-treated control. The horizontal bar within the box represents the median. The upper and lower hinges of the box indicate upper quartile and lower quartile, respectively. Sample sizes are indicated below the panels. Different letters indicate significant differences between groups (Steel–Dwass test, P < 0.05).
Genome sequence and genetic characteristics of male-killing Spiroplasma ixodetis strain sHm
Both Illumina (816.3 Mb, 5,442,459 reads, and 150 bp average length) and Nanopore data (93.5 Mb, 25,820 reads, and 3,624 bp average length) were used to reconstruct a complete genome consisting of a circular main chromosome (2, 102,039 bp in length) and two circular potential plasmids [20,119 bp (pSHM_1) and 16,408 bp (pSHM_2)]. Previously, Tsugeno et al. (2017) reported two 16S rRNA gene variants cloned from Spiroplasma-infected H. magnanima, but they did not elucidate whether the two sequences were interoperonic polymorphs of a single isolate or they were derived from two different strains. The present study confirmed that H. magnanima was infected with the MK S. ixodetis strain sHm that harbored two distinct 16S rRNA gene sequences in its genome (Figure 2). Moreover, Spiroplasma strains often encode multiple ribosomal RNA gene sets in their genome (Chang et al., 2014; Tsai et al., 2018; Vera-Ponce León et al., 2021).
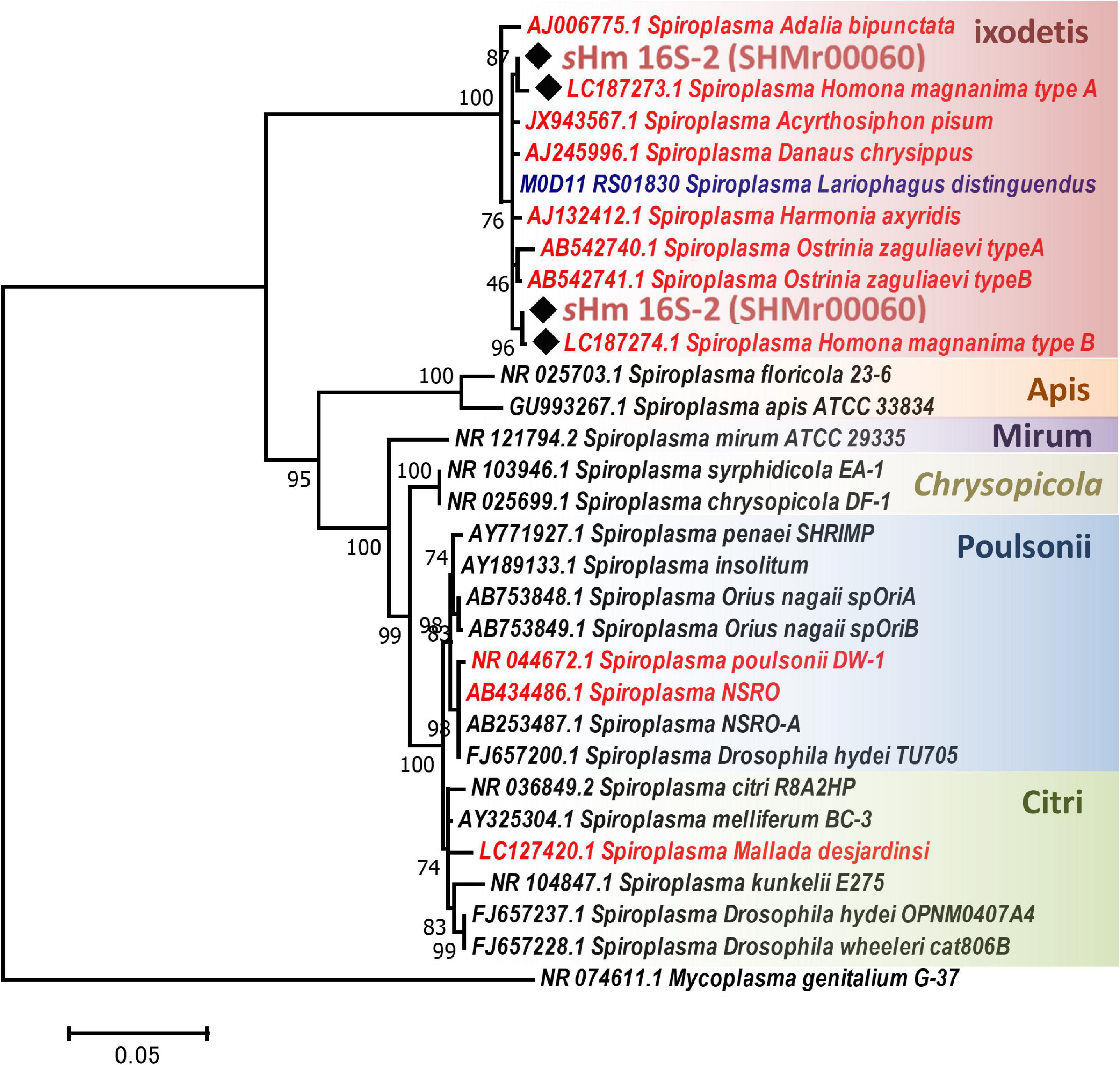
Figure 2. Phylogenetic tree of Spiroplasma strains based on 16S rRNA gene sequences. Phylogenetic tree based on 16S rRNA gene sequences of strain sHm and other Spiroplasma strains (retrieved from NCBI database) using maximum likelihood method based on the Tamura–Nei model with 1,000 bootstrap replicates. Accession numbers are shown along with the operational taxonomic units (OTUs). Samples highlighted with red and blue color fonts are MK and CI strains, respectively. Black diamonds indicate 16S rRNA sequences of the sHm. The classification of Spiroplasma is based on the study by Paredes et al. (2015). Mycoplasma genitalium was used as the outgroup.
Strain sHm harbored a higher number of coding sequences (CDS; 2,886 CDS) than strains sMel (1.9 Mb in genome size; 2,405 CDS; Masson et al., 2018) and sDa (1.7 Mb in genome size; 1,813 CDS; Martin et al., 2020; Table 2). Although plasmids often contain key accessory genes such as spaid of sMel (Harumoto and Lemaitre, 2018), genes on pSHM_1 (n = 24) and pSHM_2 (n = 20) mostly encoded hypothetical or uncharacterized proteins (Supplementary Table 1). In addition, strain sHm harbored 12 prophage regions (size: 6.6–14.8 kb) in its genome, which is consistent with previous reports that several phage sequences are found in Spiroplasma genomes (Bai and Hogenhout, 2002; Lo et al., 2013; Ramirez et al., 2021). Bacteriophages frequently carry virulence-associated genes that encode toxins (Waldor and Mekalanos, 1996; Brüssow et al., 2004). Recently, the mechanistic bases of Wolbachia-induced cytoplasmic incompatibility (CI) and MK have been attributed to phages (Beckmann et al., 2017; LePage et al., 2017; Perlmutter et al., 2019; Arai et al., 2022b). Besides, phages have also been implicated in the defense phenotype exhibited by bacteria against parasitoids, such as Hamiltonella defensa (Brandt et al., 2017). Therefore, it is possible that the phages of sHm contribute to the manifestation of MK phenotype or confer fitness advantage on hosts by protecting the hosts from natural enemies.
sHm harbored putative virulence-associated factors but did not harbor sMel spaid toxin
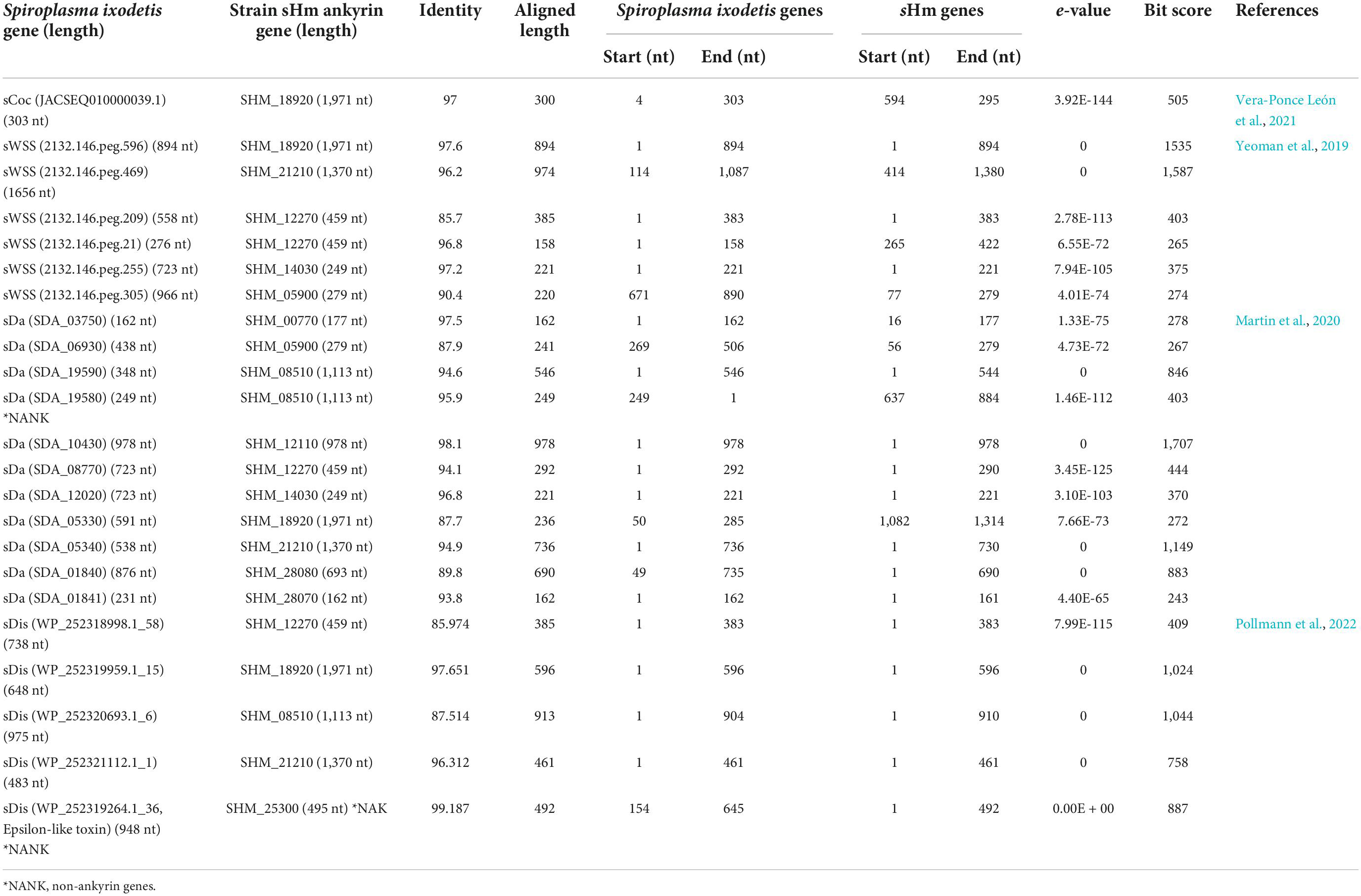
Recently, Yeoman et al. (2019) and Vera-Ponce León et al. (2021) reported that D. coccus-infecting S. ixodetis (sCoc) harbored a spaid homolog and Cephus cinctus (Cephidae)-infecting S. ixodetis harbored seven spaid homologs. Our BLAST searches confirmed that sHm did not harbor the spaid gene (Table 3), however, some of the ankyrin genes of sHm were homologous to the alleged gene sequences of sCoc and Cephidae-infecting S. ixodetis (Table 4). It is likely that S. ixodetis do not harbor the spaid gene. The superficial homology could be due to the presence of conserved ankyrin repeats (Table 3). Similarly, the amino acid sequences of ankyrin proteins of sHm (such as SHM_18920) showed partial homology to the ankyrin domain of Spaid from strain sMel (N-terminal 200 amino acids) as per BLASTp search, but the complete amino acid sequences of the proteins of these two strains were not homologous (Table 3). Moreover, we also confirmed the absence of Spaid homologs in a MK S. ixodetis strain sDa by using BLASTp search. Gerth et al. (2021) reported that the Spaid homologs are conserved among S. poulsonii strains regardless of the MK phenotype. Because the spaid gene is not likely to be possessed by S. ixodetis, MK mechanisms may differ between S. poulsonii and S. ixodetis (i.e., having different causative genes).
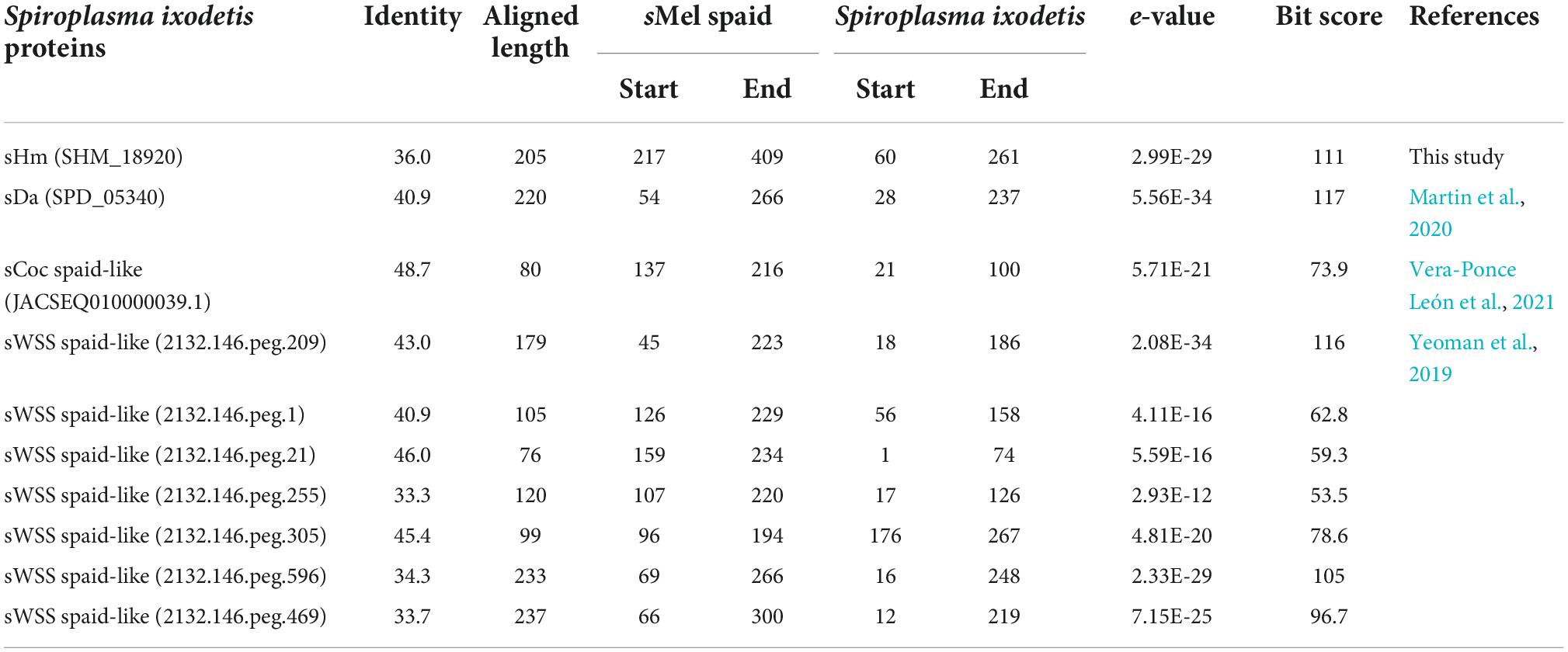
Table 3. Homology between Spaid [1,065 aa] of strain sMel and proteins of Spiroplasma ixodetis strains based on BLASTp search.
We then focused on genes conserved among Spiroplasma strains. Distantly related Spiroplasma species such as S. ixodetis (sHm), S. poulsonii (sMel), S. apis B31, S. citri BLH-MB, and S. syrphidicola EA-1 shared 345 protein clusters (Figure 3A). For S. ixodetis strains, two MK strains (sHm and sDa) and two non-MK strains (sCoc and sDis) shared 595 protein clusters (Figure 3A). In addition, MK strains sHm and sDa possessed additional 219 conserved protein clusters. sHm also harbored strain-specific 77 protein clusters (470 genes) associated with metabolism and transposition (Figure 3B and Supplementary Table 1) as well as many putative Type IV secretory system effector genes (n = 144, based on T4SEpre prediction at EffectiveDB, Supplementary Table 1), some of which were located in the prophage regions. In Spiroplasma, RIP toxin irreversibly inactivates eukaryotic cytosolic ribosomes (Hamilton et al., 2016; Ballinger et al., 2019; Garcia-Arraez et al., 2019). Based on our blast searches, RIP-4 encoded by Spiroplasma endosymbiont of Drosophila neotestacea (ASM46790.1) showed low homology to SHM_22560 (79–286 aa, e-value 6.7E-13, bit-score 60.8). Besides, SHM_22560 (hypothetical protein, 788 aa, Supplementary Table 1) was predicted to contain a RIP domain based on Interpro (hit: IPR016138, aligned length: 111–286 aa) and HHpred searches [hit: Sapolin (ID: 3HIQ), aligned length: 105–344 aa, e-value: 2.3E-29]. A homolog of an epsilon-like toxin (WP_252319264.1_36) encoded by CI-inducing sDis was detected in the sHm genome (SHM_25300, Table 4), while AbiEii abortive infection toxin (WP_252320055.1_19) and OTU-like cysteine protease (WP_252320277.1_1) were not detected.
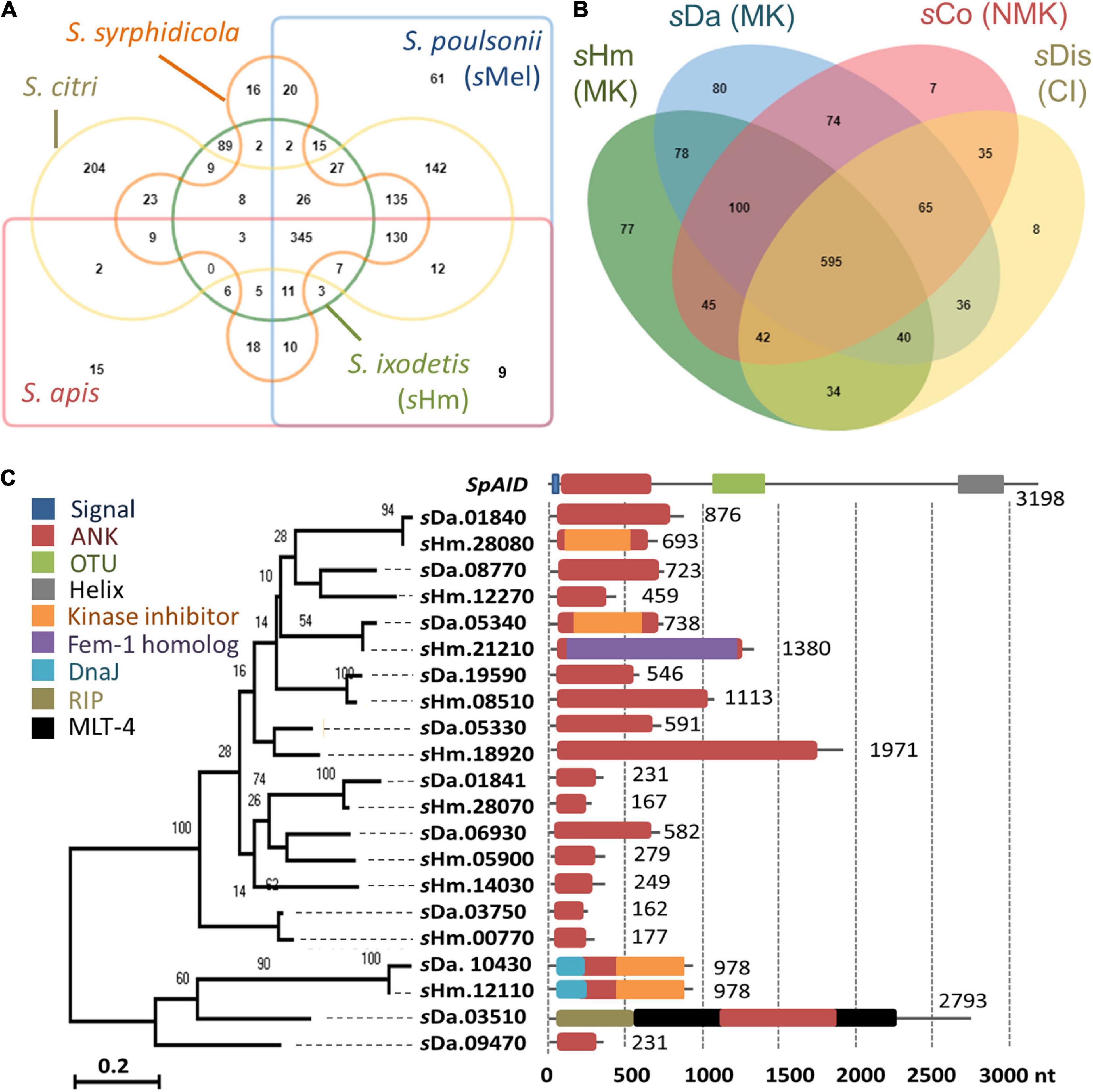
Figure 3. Comparison of Spiroplasma genomes and ankyrin genes. (A–B) Protein clusters conserved within Spiroplasma species (S. apis, S. citri, S. ixodetis, S. poulsonii, and S. syrphidicola) (A) and within S. ixodetis strains (sHm, sDa, sCoc, and sDis) (B). Venn diagrams were visualized using OrthoVenn2. The sHm-specific protein clusters and their functions are shown in Supplementary Table 1. MK, male-killing strain; NMK, non-male-killing strain; CI, cytoplasmic incompatibility strain. (C) Phylogeny of ankyrin genes in Spiroplasma strains sHm and sDa. Numbers indicates nucleotide length (nt). Other domains of the genes are highlighted in different colors. Red: ankyrin; Blue: signal peptide; Green: OTU; Gray: transmembrane helix; Orange: cyclin-dependent kinase inhibitor; Purple: ubiquitination associated FEM1A_DROME; Sky blue: DNA J; Khaki: Ribosome–inactivating protein (RIP); Black: molting protein (MLT-4).
Although spaid gene is the only ankyrin-coding gene identified in the genome of strain sMel (Harumoto and Lemaitre, 2018), sHm carried 10 ankyrin genes (Figure 3C). Some ankyrin genes harbored additional domains, such as those encoding for DnaJ (SHM_12110), cyclin-dependent kinase inhibitor (SHM_12110 and SHM_28080), and the protein ubiquitination associated FEM1A_DROME (SHM_21210); however, they did not encode for signal peptides, ovarian tumor-like deubiquitinase (OTU), or helix domains found in the spaid gene of strain sMe1 (Paredes et al., 2015; Harumoto and Lemaitre, 2018; Masson et al., 2018; Gerth et al., 2021). Wolbachia induces CI by the CI-inducing factors (Cif) harboring ankyrin repeats in insects (LePage et al., 2017). Pollmann et al. (2022) reported that CI-inducing sDis strain did not harbor the cif gene. Similarly, sHm-encoding 10 ankyrin genes has low homologies to those of other bacteria such as Wolbachia and Rickettsia and were not homologous to the cif as well as spaid genes. Intriguingly, MK sDa and CI sDis strains had 11 and 12 ankyrin genes, respectively. These findings suggest that S. ixodetis has similar characteristics to Wolbachia endosymbionts (Duplouy et al., 2013; Arai et al., 2022b) in terms of phenotypes (i.e., CI and MK) and genetic compositions (i.e., multiple ankyrin genes). Some ankyrin genes encoded by S. poulsonii and Wolbachia have been implicated in reproductive manipulation (LePage et al., 2017; Harumoto and Lemaitre, 2018), and the ankyrin genes found in the sHm genome may also be involved in MK mechanisms.
Male-killing genes of sHm are different from those of other male-killers in Homona magnanima
Homona magnanima harbors three different types of MK endosymbionts (i.e., Spiroplasma sHm, Partiti-like virus OGVs, and Wolbachia wHm-t strain), some of which can coinfect the same host (Arai et al., 2020; Takamatsu et al., 2021). Moreover, microbes sharing the same niche frequently exchange virulence-associated genes (Kent and Bordenstein, 2010; Wiedenbeck and Cohan, 2011). However, we found that strain sHm did not harbor any gene homologous to those of MK Partiti-like virus OGVs (Fujita et al., 2021). Moreover, strain sHm did not harbor wmk or effector genes (e.g., CifB-like) that are present on the MK-associated prophage WOwHm-t76 region of strain wHm-t (Arai et al., 2020, 2022b). The wmk gene, a candidate gene for Wolbachia-induced MK (Perlmutter et al., 2019, 2021; Arai et al., 2022b), possesses a helix-turn-helix (HTH) domain containing putative transcriptional regulator. Although no wmk homologs were identified, strain sHm harbored 87 HTH domain-encoding genes, namely putative transposase (classified into IS-30, IS-3, and IS-5 type transposase, n = 83), a type II toxin-antitoxin system antitoxin HipB (SHM_ 03650), an AAA family ATPase (SHM_24830), an XRE family transcriptional regulator (SHM_17560), and a helix-turn-helix transcriptional regulator (SHM_05440). Notably, a putative transposase SHM_03660, encoded by a gene adjacent to sHm-specific HipB-like SHM_3650, was homologous to the Wolbachia transcriptional regulator. Recently, Arai et al. (2022c) demonstrated that strains sHm, wHm-t, and OGVs affect H. magnanima males in different manners. Specifically, both strains sHm and wHm-t trigger abnormal apoptosis and interfere with sex determination in male embryos (manifested by the alteration of doublesex gene splicing), but only strain wHm-t impairs the dosage-compensation system of the host (manifested by the alteration of the global gene expression on sex chromosomes). In contrast, the OGVs do not affect sex-determination cascades or dosage-compensation systems. These findings and our current results support the view that phylogenetically distinct microbes have independently developed different MK machinery even for the same host, i.e., H. magnanima. Therefore, an unknown factor in the sHm genome may be responsible for the embryonic male death of H. magnanima.
sHm may require high infection density to kill Homona magnanima males
We observed that one of the sublines of the MK S+ line ceased to induce MK (Figure 4A). This subline, referred to as the S+M+ line, exhibited stable sHm infections for at least four generations. We simultaneously re-sequenced the genome of strain sHm from S+M+ and S+ lines at the second-generation stage since their divergence. We previously demonstrated from a genomic comparison of MK Wolbachia (wHm-t) and non-MK Wolbachia (wHm-c) that an MK-associated 76 kb prophage region was inserted only in wHm-t (Arai et al., 2022b). Similarly, we mapped the MK and non-MK sHm re-sequenced Illumina reads to the complete sHm genome (main chromosome and two plasmids) but did not detect any large-scale structural variation (insertions or deletions) as observed in wHm-t (Figure 4B). On the other hand, we found mutations specific to the non-MK sHm mutant (i.e., frameshifts or insertion of stop codons) in 21 genes encoding hypothetical proteins (n = 4), tyrosine-tRNA ligase (n = 1), and transposase (n = 16) (Supplementary Table 2). The 21 genes were found on the main chromosome, not in plasmids. Moreover, the sHm density in the S+M+ line was lower than that in the S+ line (Steel–Dwass test, P < 0.05, Figure 4C).
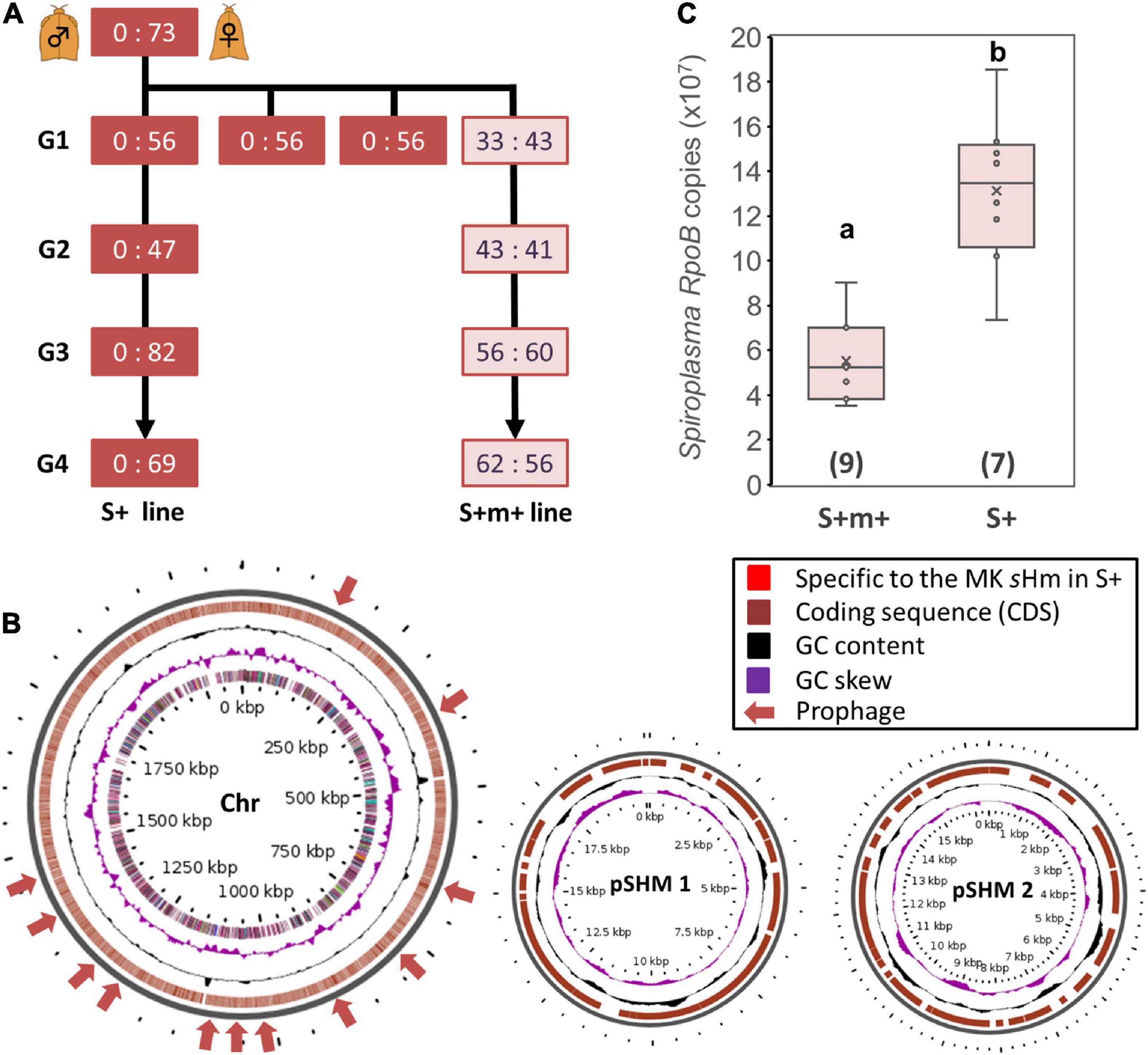
Figure 4. Comparison of Spiroplasma strain sHm in MK and non-MK Homona magnanima lines. (A) Spiroplasma-positive 1:1 sex ratio line (S+M+ line) was maintained for over four generations (G1–G4), parallelly with the all-female S+ line. (B) Structural variations of the genome of Spiroplasma strain sHm in the MK (S+) and non-MK (S+M+) H. magnanima lines were visualized via Gview software. No obvious structural variations (i.e., the red colored loci), specific to MK sHm strain in the S+ matriline, were identified. Chr: main chromosome (2.1 Mb); pSHM 1: sHm plasmid 1 (20 Kb); pSHM 2: sHm plasmid 2 (16 Kb). (C) Abundance of Spiroplasma (based on copy numbers of RpoB) in adult females (0-day post eclosion) of S+M+ line and S+ line. The horizontal bar within the box represents the median. The upper and lower hinges of the box indicate upper quartile and lower quartile, respectively. Sample sizes (numbers of examined individuals) are indicated in parentheses. Different letters indicate significant differences between groups (Steel–Dwass test, P < 0.05).
Rapid genetic evolution leading to resistance against MK Spiroplasma has been reported in various hosts, such as the lacewing (Hayashi et al., 2018) and the planthopper (Yoshida et al., 2021). However, we can exclude the host genetic changes from the possible cause of the observed loss of MK phenotype because females of the S+ and S+M+ lines were parallelly mated with the males of the genetically homogeneous NSR line that had been maintained via inbreeding in the laboratory for over 10 years (>120 generations).
Spiroplasma-induced phenotypic changes have been repeatedly observed in previous studies. For example, spontaneous loss of MK was found in S. poulsonii strains of Drosophila flies, wherein substitutions and deletions occurred in the MK gene spaid (Harumoto and Lemaitre, 2018). Moreover, the MK strain S. poulsonii NSRO and its non-MK variant NSRO-A exhibit difference in bacterial densities in D. melanogaster (Anbutsu and Fukatsu, 2003). Indeed, bacterial density is one of the crucial factors for Spiroplasma- and Wolbachia-induced MK phenotype (Hurst and Jiggins, 2000; Kageyama et al., 2007; Arai et al., 2020). Based on these results, we speculate that the loss of MK phenotype of sHm-infected H. magnanima was due to (i) reduced sHm density and/or (ii) mutations in sHm MK gene(s) or factors regulating MK gene expression levels. However, we still do not know how the small genomic rearrangements (i.e., inversions and insertions) detected in this study are involved in the phenotypic changes of sHm. Future de novo genome construction of sHm from S+M+ lines and gene function analysis would help in elucidating MK mechanisms.
Population dynamics and tissue tropism of sHm
Strain sHm was abundant at the late-developmental stages of H. magnanima (Figure 5A), and sHm densities drastically increased from pupal to adult stages of the insect (Figure 5B). In D. melanogaster, S. poulsonii copy numbers gradually increase as the host larval development proceeds and are generally higher in pupae than in larvae (Anbutsu and Fukatsu, 2003). In contrast to S. poulsonii, which is reported to be the most abundant in hemolymph (Anbutsu and Fukatsu, 2003, 2006), strain sHm exhibited low density in the hemolymph and high density in Malpighian tubules in the fifth instar larva stage (Figure 5C). High titers in Malpighian tubules are also a characteristic of Wolbachia; Wolbachia present in Malpighian tubules protects the host from RNA-virus infections and may constitute a secondary pool of vertically infected bacteria (Faria and Sucena, 2013; Pietri et al., 2016). The localization of strain sHm in somatic tissues may have contributed to the fitness of H. magnanima Although there have been no reports of S. ixodetis localization patterns in insects, our findings suggest that S. poulsonii and S. ixodetis have distinct proliferation strategies. The hemolymph is a nutrient-rich environment but is likely an extreme habitat for microorganisms because it is well-defended by the immune system of the host (Blow and Douglas, 2019). Indeed, only a few microbial taxa are known to persist in the hemolymph of insects for extended periods without causing insect morbidity and death (Blow and Douglas, 2019). Intriguingly, S. poulsonii sMel encoded more metabolic genes in its genome than S. ixodetis sHm (Supplementary Table 3). Hemolymph-inhabiting S. poulsonii may have developed specific adaptations for its habitat, which are distinct from those of S. ixodetis. Further characterization of genomic features and localization patterns of Spiroplasma strains will clarify the distinct proliferation strategies of the two species (e.g., nutrient requirements).
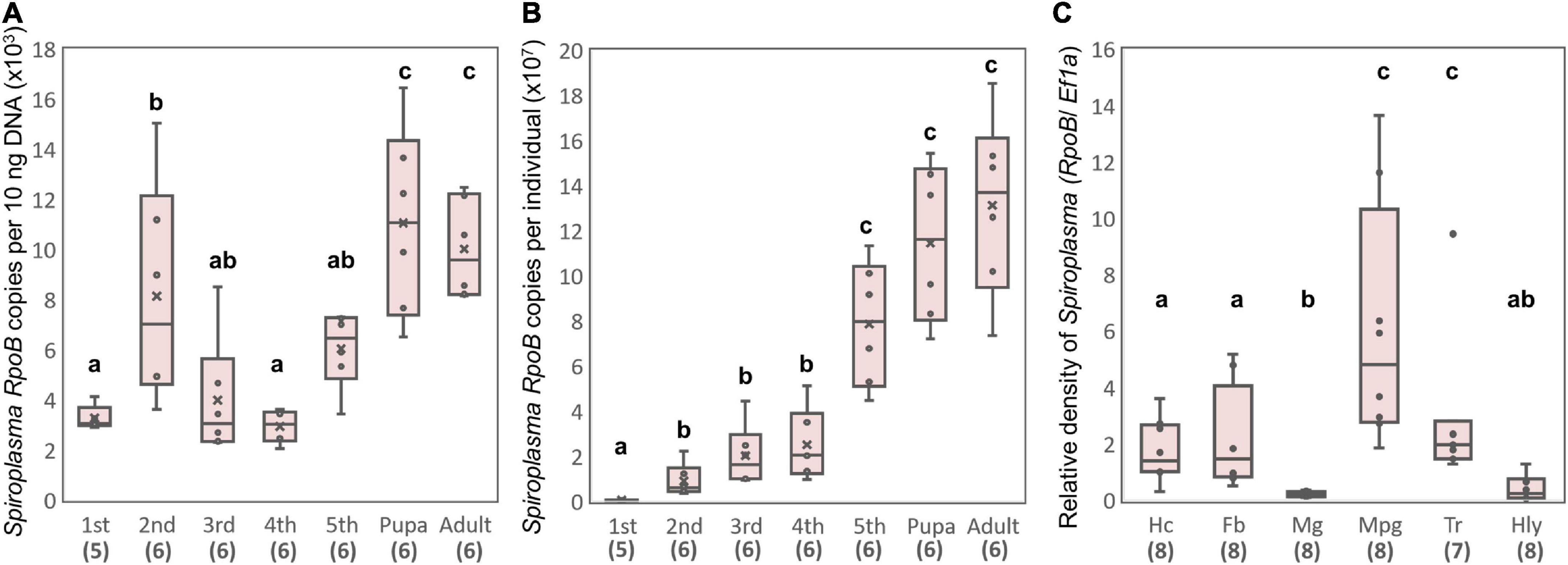
Figure 5. Propagation and localization of strain sHm in H. magnanima. Spiroplasma densities deduced from RpoB copy numbers per 10 ng DNA (A) and Spiroplasma densities deduced from RpoB copy numbers per individual (B) at each developmental stage. (C) Relative Spiroplasma density in each organ deduced from Spiroplasma RpoB copy numbers per host Ef1α copy. Hc, hemocyte; Fb, fat body; Mg, midgut; Mpg, Malpighian tubules; Tr, trachea; Hly, hemolymph. Different letters indicate significant differences between groups (Steel–Dwass test, P < 0.05).
Proliferation of sHm in insect cell culture
Tsugeno et al. (2017) reported that sHm is horizontally transmitted by inoculating non-infected H. magnanima with concentrated hemolymph collected from sHm-infected H. magnanima. Moreover, we revealed that S. ixodetis sHm exhibited Wolbachia-like genetic characteristics (i.e., multiple ankyrin genes) and localization patterns in somatic tissues. Wolbachia can infect and be maintained stably in insect cell lines derived from insect taxa that are distantly related to their native hosts (Fallon, 2021). To examine whether sHm can infect insect cells, we transinfected sHm to the cell lines of A. albopictus (AeAl2) and B. mori (aff3), which are known to be susceptible to Wolbachia. sHm proliferated successfully by placing a piece of fat bodies or Malpighian tubules derived from an S+ female larva into a flask containing the AeAl2 or aff3 cells (Figure 6). sHm was stably maintained in the cell lines for 12 weeks (Figure 6A) but not in cell-free medium IPL-41. qPCR revealed that sHm titers in AeAl2 cells were significantly higher at 12 weeks than at 4 weeks after the introduction of sHm (Figure 6B). This implies the potential of sHm to survive in a wide host range besides Homona (Tsugeno et al., 2017), such as other lepidopteran and dipteran insects. S. ixodetis strains isolated from Japanese ticks were also shown to be culturable in the A. albopictus cell line C6/36 (Thu et al., 2019). We hypothesize that S. ixodetis strains have a broad host range like that of Wolbachia. It is not clear whether S. poulsonii has a broad host range because there is no but one report by Hackett et al. (1986) that showed the infectivity of strain WSRO (derived from D. willistoni) in the Trichoplusia ni cell line IPLB-TN-R2. Several attempts to transinfect S. poulsonii (strain NSRO; derived from D. nebulosa) and MK Spiroplasma (derived from the lacewing Mallada desjardinsi) into AeAl2 and aff3 cells failed (personal observation by DK). It has been shown by hemolymph injection that S. poulsonii can infect drosophilid flies but not houseflies, suggesting its narrow host range (Williamson and Poulson, 1979).
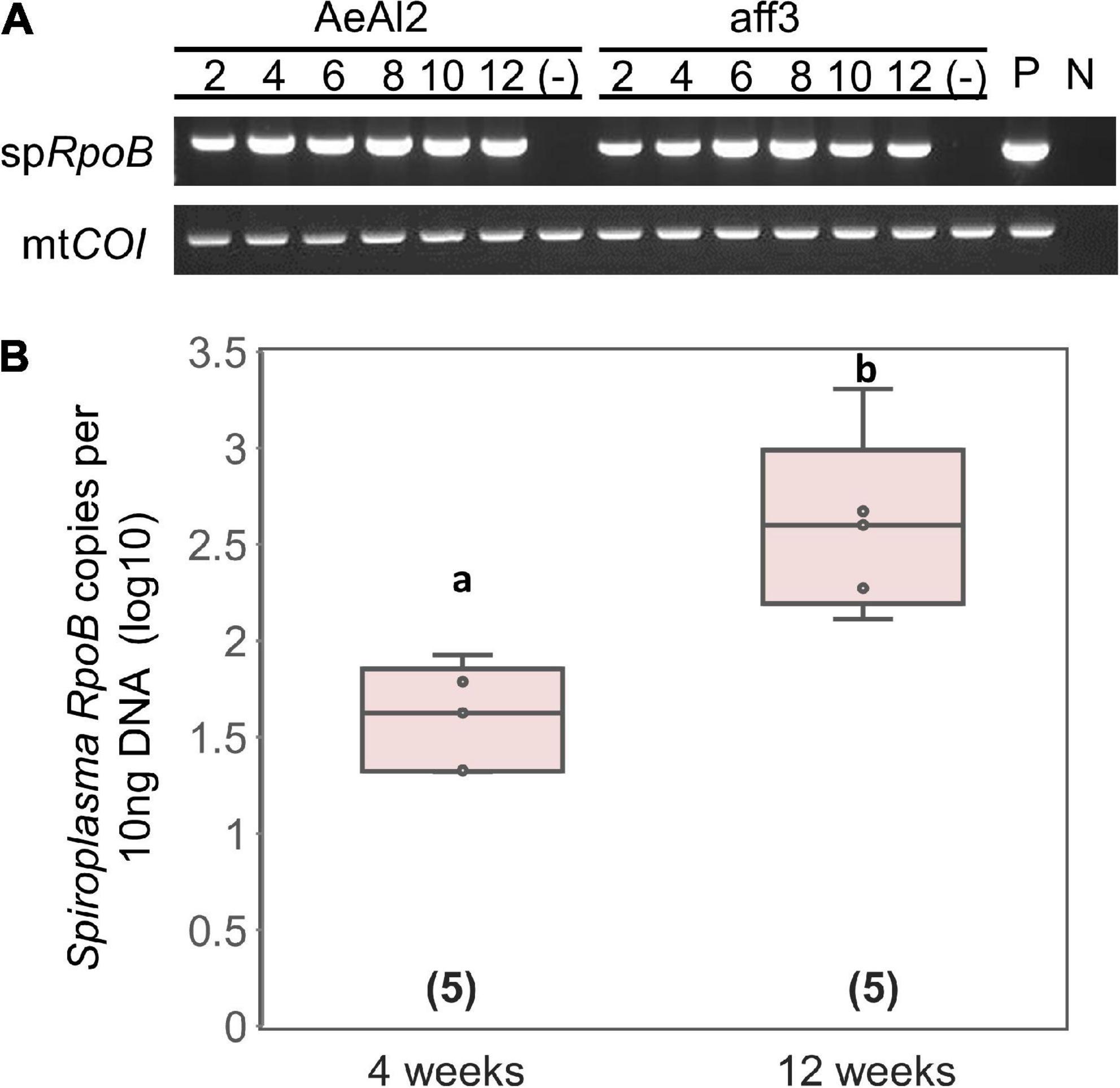
Figure 6. Transinfection of Spiroplasma strain sHm into mosquito and silkworm cell lines. (A) Detections of strain sHm in passaged cells. Numbers indicate periods (weeks) from the transinfection. P (positive control): S + female; N (negative control): NSR female. (B) Spiroplasma density in AeAl2 cells deduced from RpoB copy numbers per 10 ng DNA. Different letters indicate significant differences between groups (Steel–Dwass test, P < 0.05).
Summary and perspectives
In this study, we sequenced and analyzed the genome of an MK S. ixodetis strain sHm. S. poulsonii possesses the Spaid toxin as the MK factor, whereas our study revealed that MK S. ixodetis did not harbor spaid homologs. We speculate that MK S. ixodetis strains found in a diverse range of insects (Hurst et al., 1999; Jiggins et al., 2000; Simon et al., 2011; Tabata et al., 2011; Sanada-Morimura et al., 2013) harbor yet-unknown MK gene(s), other than spaid; thus, future studies should focus on the identification of these MK genes. Besides, high infection efficiencies of strain sHm in other insect cells led us to speculate that MK S. ixodetis has been horizontally transmitted among insect species, like Wolbachia, which has expanded its host range (Zhou et al., 1998; Baldo et al., 2006). Further studies would be required to understand whether closely related MK Spiroplasma strains (i.e., the S. ixodetis group) share common or different MK mechanisms, which will answer evolutionary questions such as how frequent novel MK genes arose, how MK genes moved between different Spiroplasma strains (if it did), and whether MK genes are associated with host sex determining systems.
Data availability statement
All sequence data are available at DRA under BioProject: PRJDB14468, Biosamples: SAMD00547685, SAMD00547900, and DRA014961. Spiroplasma genome data are available in the DDBJ database under the following accession numbers: AP026933–AP026935.
Author contributions
HA conducted all experiments, data analysis, and wrote the original manuscripts. MI assisted insect rearing, experiments, and discussions. DK managed the experiments and revised the original manuscript. All authors approved the final version of the manuscript.
Funding
This study was supported by the Japan Society for the Promotion of Science (JSPS) Research Fellowships for Young Scientists (grant numbers: 19J13123 and 21J00895), JSPS Grant-in-Aid for Scientific Research (grant number: 22K14902), and Cabinet Office, Government of Japan, Cross-ministerial Moonshot Agriculture, Forestry and Fisheries Research and Development Program (grant number: JPJ009237).
Acknowledgments
We thank Dr. Akiya Jouraku [National Agriculture and Food Research Organization (NARO), 1-2 Owashi, Tsukuba, Ibaraki, Japan] for providing H. magnanima genome data.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1075199/full#supplementary-material
Abbreviations
CI, cytoplasmic incompatibility; FBS, fetal bovine serum; HTH, helix-turn-helix; JSPS, Japan Society for the Promotion of Science; MK, male-killing; OGV, Osugoroshivirus; OTU, operational taxonomic unit; RIP, ribosome-inactivating protein; WGA, whole genome amplification.
Footnotes
- ^ https://www.ebi.ac.uk/interpro/
- ^ https://orthovenn2.bioinfotoolkits.net/home
- ^ https://server.gview.ca
- ^ https://www.bv-brc.org
- ^ https://www.r-project.org/
References
Anbutsu, H., and Fukatsu, T. (2003). Population dynamics of male-killing and non-male-killing Spiroplasmas in Drosophila melanogaster. Appl. Environ. Microbiol. 69, 1428–1434. doi: 10.1128/AEM.69.3.1428-1434.2003
Anbutsu, H., and Fukatsu, T. (2006). Tissue-specific infection dynamics of male-killing and nonmale-killing Spiroplasmas in Drosophila melanogaster. FEMS Microbiol. Ecol. 57, 40–46. doi: 10.1111/j.1574-6941.2006.00087.x
Anbutsu, H., and Fukatsu, T. (2011). Spiroplasma as a model insect endosymbiont. Environ. Microbiol. Rep. 3, 144–153. doi: 10.1111/j.1758-2229.2010.00240.x
Arai, H., Ishitsubo, Y., Nakai, M., and Inoue, M. N. (2022a). Mass-rearing and molecular studies in Tortricidae Pest Insects. J. Vis. Exp. 181, doi: 10.3791/63737
Arai, H., Takamatsu, T., Lin, S. R., Mizutani, T., Omatsu, T., Katayama, Y., et al. (2022c). Distinct effects of the male-killing bacteria Wolbachia and Spiroplasma and a partiti-like virus in the tea pest moth, Homona magnanima. Biorxiv [Preprint]. doi: 10.1101/2022.04.29.490121
Arai, H., Anbutsu, H., Nishikawa, Y., Kogawa, M., Ishii, K., Hosokawa, M., et al. (2022b). Male-killing-associated bacteriophage WO identified from comparisons of Wolbachia endosymbionts of Homona magnanima. Biorxiv [Preprint]. doi: 10.1101/2022.06.12.495854v2
Arai, H., Hirano, T., Akizuki, N., Abe, A., Nakai, M., Kunimi, Y., et al. (2019). Multiple infection and reproductive manipulations of Wolbachia in Homona magnanima (Lepidoptera: Tortricidae). Microb. Ecol. 77, 257–266. doi: 10.1007/s00248-018-1210-4
Arai, H., Lin, S. R., Nakai, M., Kunimi, Y., and Inoue, M. N. (2020). Closely related male-killing and nonmale-killing Wolbachia strains in the Oriental tea tortrix Homona magnanima. Microb. Ecol. 79, 1011–1020. doi: 10.1007/s00248-019-01469-6
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Bai, X., and Hogenhout, S. A. (2002). A genome sequence survey of the mollicute corn stunt Spiroplasma Spiroplasma kunkelii. FEMS Microbiol. Lett. 210, 7–17. doi: 10.1111/j.1574-6968.2002.tb11153.x
Baldo, L., Dunning Hotopp, J. C., Jolley, K. A., Bordenstein, S. R., Biber, S. A., Choudhury, R. R., et al. (2006). Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl. Environ. Microbiol. 72, 7098–7110. doi: 10.1128/AEM.00731-06
Ballinger, M. J., Gawryluk, R. M., and Perlman, S. J. (2019). Toxin and genome evolution in a Drosophila defensive symbiosis. Genome Biol. Evol. 11, 253–262. doi: 10.1093/gbe/evy272
Beckmann, J. F., Ronau, J. A., and Hochstrasser, M. (2017). A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat. Microbiol. 2, 1–7. doi: 10.1038/nmicrobiol.2017.7
Binetruy, F., Bailly, X., Chevillon, C., Martin, O. Y., Bernasconi, M. V., and Duron, O. (2019). Phylogenetics of the Spiroplasma ixodetis endosymbiont reveals past transfers between ticks and other arthropods. Ticks Tick Borne Dis. 10, 575–584. doi: 10.1016/j.ttbdis.2019.02.001
Blow, F., and Douglas, A. E. (2019). The hemolymph microbiome of insects. J. Insect Physiol. 115, 33–39. doi: 10.1016/j.jinsphys.2019.04.002
Brandt, J. W., Chevignon, G., Oliver, K. M., and Strand, M. R. (2017). Culture of an aphid heritable symbiont demonstrates its direct role in defence against parasitoids. Proc. R. Soc. Lond. B Biol. Soc. 284:20171925. doi: 10.1098/rspb.2017.1925
Brüssow, H., Canchaya, C., and Hardt, W. D. (2004). Phages and the evolution of bacterial pathogens: From genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68, 560–602. doi: 10.1128/MMBR.68.3.560-602.2004
Carle, P., Saillard, C., Carrère, N., Carrère, S., Duret, S., Eveillard, S., et al. (2010). Partial chromosome sequence of Spiroplasma citri reveals extensive viral invasion and important gene decay. Appl. Environ. Microbiol. 76, 3420–3426. doi: 10.1128/AEM.02954-09
Chang, T. H., Lo, W. S., Ku, C., Chen, L. L., and Kuo, C. H. (2014). Molecular evolution of the substrate utilization strategies and putative virulence factors in mosquito-associated Spiroplasma species. Genome Biol. Evol. 6, 500–509. doi: 10.1093/gbe/evu033
Duperron, S., Pottier, M. A., Léger, N., Gaudron, S. M., Puillandre, N., Le Prieur, S. L., et al. (2013). A tale of two chitons: Is habitat specialisation linked to distinct associated bacterial communities? FEMS Microbiol. Ecol. 83, 552–567. doi: 10.1111/1574-6941.12014
Duplouy, A., Iturbe-Ormaetxe, I., Beatson, S. A., Szubert, J. M., Brownlie, J. C., McMeniman, C. J., et al. (2013). Draft genome sequence of the male-killing Wolbachia strain wBol1 reveals recent horizontal gene transfers from diverse sources. BMC Genom. 14:20. doi: 10.1186/1471-2164-14-20
Duron, O., Bouchon, D., Boutin, S., Bellamy, L., Zhou, L., Engelstädter, J., et al. (2008). The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol. 6:27. doi: 10.1186/1741-7007-6-27
Eichinger, V., Nussbaumer, T., Platzer, A., Jehl, M. A., Arnold, R., and Rattei, T. (2016). EffectiveDB—updates and novel features for a better annotation of bacterial secreted proteins and Type III, IV, VI secretion systems. Nucleic Acids Res. 44, D669–D674. doi: 10.1093/nar/gkv1269
Fallon, A. M. (2021). Growth and maintenance of Wolbachia in insect cell lines. Insects 12:706. doi: 10.3390/insects12080706
Faria, V. G., and Sucena, E. (2013). Wolbachia in the Malpighian tubules: Evolutionary dead-end or adaptation? J. Exp. Zool. B Mol. Dev. Evol. 320, 195–199. doi: 10.1002/jez.b.22498
Fujita, R., Inoue, M. N., Takamatsu, T., Arai, H., Nishino, M., Abe, N., et al. (2021). Late male-killing viruses in Homona magnanima identified as Osugoroshi viruses, novel members of Partitiviridae. Front. Microbiol. 11:620623. doi: 10.3389/fmicb.2020.620623
Garcia-Arraez, M. G., Masson, F., Escobar, J. C. P., and Lemaitre, B. (2019). Functional analysis of RIP toxins from the Drosophila endosymbiont Spiroplasma poulsonii. BMC Microbiol. 19:46. doi: 10.1186/s12866-019-1410-1
Gerth, M., Martinez-Montoya, H., Ramirez, P., Masson, F., Griffin, J. S., Aramayo, R., et al. (2021). Rapid molecular evolution of Spiroplasma symbionts of Drosophila. Microb. Genom. 7:000503. doi: 10.1099/mgen.0.000503
Hackett, K. J., Lynn, D. E., Williamson, D. L., Ginsberg, A. S., and Whitcomb, R. F. (1986). Cultivation of the Drosophila sex-ratio Spiroplasma. Science 232, 1253–1255. doi: 10.1126/science.232.4755.1253
Hajibabaei, M., Janzen, D. H., Burns, J. M., Hallwachs, W., and Hebert, P. D. (2006). DNA barcodes distinguish species of tropical Lepidoptera. Proc. Natl. Acad. Sci. U. S. A. 103, 968–971. doi: 10.1073/pnas.051046610
Hamilton, P. T., Peng, F., Boulanger, M. J., and Perlman, S. J. (2016). A ribosome-inactivating protein in a Drosophila defensive symbiont. Proc. Natl. Acad. Sci. U. S. A. 113, 350–355. doi: 10.1073/pnas.1518648113
Harumoto, T., and Lemaitre, B. (2018). Male-killing toxin in a bacterial symbiont of Drosophila. Nature 557, 252–255. doi: 10.1038/s41586-018-0086-2
Hayashi, M., Nomura, M., and Kageyama, D. (2018). Rapid comeback of males: Evolution of male-killer suppression in a green lacewing population. Proc. R. Soc. Lond. B Biol. Sci. 285:20180369. doi: 10.1098/rspb.2018.0369
Hayashi, M., Watanabe, M., Yukuhiro, F., Nomura, M., and Kageyama, D. (2016). A nightmare for males? A maternally transmitted male-killing bacterium and strong female bias in a green lacewing population. PLoS One 11:e0155794. doi: 10.1371/journal.pone.0155794
He, L. S., Zhang, P. W., Huang, J. M., Zhu, F. C., Danchin, A., and Wang, Y. (2018). The enigmatic genome of an obligate ancient Spiroplasma symbiont in a Hadal Holothurian. Appl. Environ. Microbiol. 84, e01965–17. doi: 10.1128/AEM.01965-17
Hornett, E. A., Charlat, S., Duplouy, A. M. R., Davies, N., Roderick, G. K., Wedell, N., et al. (2006). Evolution of male-killer suppression in a natural population. PLoS Biol. 4:e283. doi: 10.1371/journal.pbio.0040283
Hurst, G. D. D., Graf von der Schulenburg, J. H., Majerus, T. M. O., Bertrand, D., Zakharov, I. A., Baungaard, J., et al. (1999). Invasion of one insect species, Adalia bipunctata, by two different male-killing bacteria. Insect Mol. Biol. 8, 133–139. doi: 10.1046/j.1365-2583.1999.810133.x
Hurst, G. D., and Jiggins, F. M. (2000). Male-killing bacteria in insects: Mechanisms, incidence, and implications. Emerg. Infect. Dis. 6, 329–336. doi: 10.3201/eid0604.000402
Hurst, L. D. (1991). The incidences and evolution of cytoplasmic male killers. Proc. R. Lond. Soc. B Biol. Soc. 244, 91–99. doi: 10.1098/rspb.1991.0056
Jiggins, F. M., Hurst, G. D., Jiggins, C. D., v d Schulenburg, J. H., and Majerus, M. E. (2000). The butterfly Danaus chrysippus is infected by a male-killing Spiroplasma bacterium. Parasitology 120, 439–446. doi: 10.1017/s0031182099005867
Joshi, B. D., Berg, M., Rogers, J., Fletcher, J., and Melcher, U. (2005). Sequence comparisons of plasmids pBJS-O of Spiroplasma citri and pSKU146 of S. kunkelii: Implications for plasmid evolution. BMC Genom. 6:175. doi: 10.1186/1471-2164-6-175
Kageyama, D., Anbutsu, H., Shimada, M., and Fukatsu, T. (2007). Spiroplasma infection causes either early or late male killing in Drosophila, depending on maternal host age. Naturwissenschaften 94, 333–337. doi: 10.1007/s00114-006-0195-x
Kageyama, D., Narita, S., and Watanabe, M. (2012). Insect sex determination manipulated by their endosymbionts: Incidences, mechanisms and implications. Insects 3, 161–199. doi: 10.3390/insects3010161
Kent, B. N., and Bordenstein, S. R. (2010). Phage WO of Wolbachia: Lambda of the endosymbiont world. Trends Microbiol. 18, 173–181. doi: 10.1016/j.tim.2009.12.011
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Ku, C., Lo, W. S., Chen, L. L., and Kuo, C. H. (2013). Complete genomes of two dipteran-associated Spiroplasmas provided insights into the origin, dynamics, and impacts of viral invasion in Spiroplasma. Genome Biol. Evol. 5, 1151–1164. doi: 10.1093/gbe/evt084
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
LePage, D. P., Metcalf, J. A., Bordenstein, S. R., On, J., Perlmutter, J. I., Shropshire, J. D., et al. (2017). Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature 543, 243–247. doi: 10.1038/nature21391
Li, H. (2018). Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. doi: 10.1093/bioinformatics/bty191
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Lo, W. S., Chen, L. L., Chung, W. C., Gasparich, G. E., and Kuo, C. H. (2013). Comparative genome analysis of Spiroplasma melliferum IPMB4A, a honeybee-associated bacterium. BMC Genom. 14:22. doi: 10.1186/1471-2164-14-22
Lo, W. S., Gasparich, G. E., and Kuo, C. H. (2015). Found and lost: The fates of horizontally acquired genes in arthropod-symbiotic Spiroplasma. Genome Biol. Evol. 7, 2458–2472. doi: 10.1093/gbe/evv160
Majerus, M. E., Hinrich, J., Schulenburg, G. V. D., and Zakharov, I. A. (2000). Multiple causes of male-killing in a single sample of the two-spot ladybird, Adalia bipunctata (Coleoptera: Coccinellidae) from Moscow. Heredity 84, 605–609. doi: 10.1046/j.1365-2540.2000.00710.x
Martin, S. H., Singh, K. S., Gordon, I. J., Omufwoko, K. S., Collins, S., Warren, I. A., et al. (2020). Whole-chromosome hitchhiking driven by a male-killing endosymbiont. PLoS Biol. 18:e3000610. doi: 10.1371/journal.pbio.3000610
Massey, J. H., and Newton, I. L. (2022). Diversity and function of arthropod endosymbiont toxins. Trends Microbiol. 30, 185–198. doi: 10.1016/j.tim.2021.06.008
Masson, F., Calderon Copete, S., Schüpfer, F., Garcia-Arraez, G., and Lemaitre, B. (2018). In vitro culture of the insect endosymbiont Spiroplasma poulsonii highlights bacterial genes involved in host-symbiont interaction. Mbio 9, e00024–18. doi: 10.1128/mBio.00024-18
Mitsuhashi, J. (1981). A new continuous cell line from larvae of the mosquito Aedes albopictus (Diptera, Culicidae). Biomed. Res. 2, 599–606. doi: 10.2220/biomedres.2.599
Mouches, C., Barroso, G., Gadeau, A., and Bové, J. M. (1984). Characterization of two cryptic plasmids from Spiroplasma citri and occurrence of their DNA sequences among various Spiroplasmas. Ann. Inst. Pasteur Microbiol. 135A, 17–24. doi: 10.1016/s0769-2609(84)80054-x
Paredes, J. C., Herren, J. K., Schüpfer, F., Marin, R., Claverol, S., Kuo, C. H., et al. (2015). Genome sequence of the Drosophila melanogaster male-killing Spiroplasma strain MSRO endosymbiont. Mbio 6, e02437–14. doi: 10.1128/mBio.02437-14
Perlmutter, J. I., Bordenstein, S. R., Unckless, R. L., LePage, D. P., Metcalf, J. A., Hill, T., et al. (2019). The phage gene wmk is a candidate for male killing by a bacterial endosymbiont. PLoS Pathog. 15:e1007936. doi: 10.1371/journal.ppat.1007936
Perlmutter, J. I., Meyers, J. E., and Bordenstein, S. R. (2021). A single synonymous nucleotide change impacts the male-killing phenotype of prophage WO gene wmk. Elife 10:e67686. doi: 10.7554/eLife.67686
Pietri, J. E., DeBruhl, H., and Sullivan, W. (2016). The rich somatic life of Wolbachia. Microbiologyopen 5, 923–936. doi: 10.1002/mbo3.390
Pollmann, M., Moore, L. D., Krimmer, E., D’Alvise, P., Hasselmann, M., Perlman, S. J., et al. (2022). Highly transmissible cytoplasmic incompatibility by the extracellular insect symbiont Spiroplasma. Iscience 25:104335. doi: 10.1016/j.isci.2022.104335
Ramirez, P., Leavitt, J. C., Gill, J. J., and Mateos, M. (2021). Preliminary characterization of phage-like particles from the male-killing mollicute Spiroplasma poulsonii (an endosymbiont of Drosophila). Biorxiv [Preprint]. doi: 10.1101/2021.12.09.471767
Regassa, L. B., and Gasparich, G. E. (2006). Spiroplasmas: Evolutionary relationships and biodiversity. Front. Biosci. 11:2983–3002. doi: 10.2741/2027
Sanada-Morimura, S., Matsumura, M., and Noda, H. (2013). Male killing caused by a Spiroplasma symbiont in the small brown planthopper, Laodelphax striatellus. J. Hered. 104, 821–829. doi: 10.1093/jhered/est052
Simon, J. C., Boutin, S., Tsuchida, T., Koga, R., Le Gallic, J. F., Frantz, A., et al. (2011). Facultative symbiont infections affect aphid reproduction. PLoS One 6:e21831. doi: 10.1371/journal.pone.0021831
Smith, D. A., Gordon, I. J., Traut, W., Herren, J., Collins, S., Martins, D. J., et al. (2016). A neo-W chromosome in a tropical butterfly links colour pattern, male-killing, and speciation. Proc. R. Soc. Lond. B Biol. Sci. 283:20160821. doi: 10.1098/rspb.2016.0821
Tabata, J., Hattori, Y., Sakamoto, H., Yukuhiro, F., Fujii, T., Kugimiya, S., et al. (2011). Male killing and incomplete inheritance of a novel Spiroplasma in the moth Ostrinia zaguliaevi. Microb. Ecol. 61, 254–263. doi: 10.1007/s00248-010-9799-y
Takahashi, T., Murakami, H., Imanishi, S., Miyazaki, M., Kamiie, K., Suzuki, K., et al. (2006). Calreticulin is transiently induced after immunogen treatment in the fat body of the silkworm Bombyx mori. J. Insect Biotechnol. Sericol. 75, 79–84. doi: 10.11416/jibs.75.79
Takamatsu, T., Arai, H., Abe, N., Nakai, M., Kunimi, Y., and Inoue, M. N. (2021). Coexistence of two male-killers and their impact on the development of oriental tea tortrix Homona magnanima. Microb. Ecol. 81, 193–202. doi: 10.1007/s00248-020-01566-x
Tanizawa, Y., Fujisawa, T., and Nakamura, Y. (2018). DFAST: A flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 34, 1037–1039. doi: 10.1093/bioinformatics/btx713
Thu, M. J., Qiu, Y., Kataoka-Nakamura, C., Sugimoto, C., Katakura, K., Isoda, N., et al. (2019). Isolation of Rickettsia, Rickettsiella, and Spiroplasma from questing ticks in Japan using arthropod cells. Vector Borne Zoonotic Dis. 19, 474–485. doi: 10.1089/vbz.2018.2373
Tsai, Y. M., Chang, A., and Kuo, C. H. (2018). Horizontal gene acquisitions contributed to genome expansion in insect-symbiotic Spiroplasma clarkii. Genome Biol. Evol. 10, 1526–1532. doi: 10.1093/gbe/evy113
Tsugeno, Y., Koyama, H., Takamatsu, T., Nakai, M., Kunimi, Y., and Inoue, M. N. (2017). Identification of an early male-killing agent in the oriental tea tortrix, Homona magnanima. J. Hered. 108, 553–560. doi: 10.1093/jhered/esx049
Vera-Ponce León, A., Dominguez-Mirazo, M., Bustamante-Brito, R., Higareda-Alvear, V., Rosenblueth, M., and Martínez-Romero, E. (2021). Functional genomics of a Spiroplasma associated with the carmine cochineals Dactylopius coccus and Dactylopius opuntiae. BMC Genom. 22:240. doi: 10.1186/s12864-021-07540-2
Viver, T., Orellana, L. H., Hatt, J. K., Urdiain, M., Díaz, S., Richter, M., et al. (2017). The low diverse gastric microbiome of the jellyfish Cotylorhiza tuberculata is dominated by four novel taxa. Environ. Microbiol. 19, 3039–3058. doi: 10.1111/1462-2920.13763
Waldor, M. K., and Mekalanos, J. J. (1996). Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272, 1910–1914. doi: 10.1126/science.272.5270.1910
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Watanabe, M., Tagami, Y., Miura, K., Kageyama, D., and Stouthamer, R. (2012). Distribution patterns of Wolbachia endosymbionts in the closely related flower bugs of the genus Orius: Implications for coevolution and horizontal transfer. Microb. Ecol. 64, 537–545. doi: 10.1007/s00248-012-0042-x
Werren, J. H., Baldo, L., and Clark, M. E. (2008). Wolbachia: Master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751. doi: 10.1038/nrmicro1969
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comp. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wiedenbeck, J., and Cohan, F. M. (2011). Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol. Rev. 35, 957–976. doi: 10.1111/j.1574-6976.2011.00292.x
Williamson, D. L., and Poulson, D. F. (1979). “Plant and insect mycoplasmas,” in The Mycoplasmas, Volume III, eds R. F. Whitcomb and J. G. Tully (New York, NY: Academic Press), 175–208.
Ye, F., Melcher, U., Rascoe, J. E., and Fletcher, J. (1996). Extensive chromosome aberrations in Spiroplasma citri strain BR3. Biochem. Genet. 34, 269–286. doi: 10.1007/BF02399947
Yeoman, C. J., Brutscher, L. M., Esen, Ö. C., Ibaoglu, F., Fowler, C., Eren, A. M., et al. (2019). Genome-resolved insights into a novel Spiroplasma symbiont of the wheat stem sawfly (Cephus cinctus). Peerj 7:e7548. doi: 10.7717/peerj.7548
Yoshida, K., Sanada-Morimura, S., Huang, S. H., and Tokuda, M. (2021). Silence of the killers: Discovery of male-killing suppression in a rearing strain of the small brown planthopper, Laodelphax striatellus. Proc. R. Lond. B Biol. Sci. 288:20202125. doi: 10.1098/rspb.2020.2125
Keywords: Spiroplasma, male-killing, symbiosis, evolution, endosymbionts, Homona magnanima, spaid
Citation: Arai H, Inoue MN and Kageyama D (2022) Male-killing mechanisms vary between Spiroplasma species. Front. Microbiol. 13:1075199. doi: 10.3389/fmicb.2022.1075199
Received: 20 October 2022; Accepted: 08 November 2022;
Published: 28 November 2022.
Edited by:
Chih-Horng Kuo, Academia Sinica, TaiwanReviewed by:
Steve Perlman, University of Victoria, CanadaMatt Ballinger, Mississippi State University, United States
Copyright © 2022 Arai, Inoue and Kageyama. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hiroshi Arai, ZGF6YWkzOXBhcGlsaW9AZ21haWwuY29t; Daisuke Kageyama, a2FneW1hZEBhZmZyYy5nby5qcA==
 Hiroshi Arai
Hiroshi Arai Maki N. Inoue
Maki N. Inoue Daisuke Kageyama
Daisuke Kageyama