- 1State Key Laboratory of Infectious Disease Prevention and Control, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China
- 2OIE Reference Lab for Swine Streptococcosis, MOE Joint International Research Laboratory of Animal Health and Food Safety, College of Veterinary Medicine, Nanjing Agricultural University, Nanjing, China
- 3Nanxishan Hospital of Guangxi Zhuang Autonomous Region, Guilin, China
Streptococcus suis, an emerging zoonotic pathogen, is important reservoirs of antibiotic resistance genes that play critical roles in the horizontal transfer of corresponding resistances. In the present study, 656 antibiotic resistance (AR) genes were detected in 154 of 155 genomes of S. suis strains isolated from the nasopharynx of slaughtered pigs and the lungs of diseased pigs in China. The AR genes were clustered into 11 categories, consisting of tetracycline, macrolides, lincosamide, streptogramin, aminoglycoside, trimethoprim, amphenicols, nucleoside, quinupristin/dalfopristin, glycopeptide, and oxazolidinones resistance genes. In order to investigate the transmission patterns of the AR genes, AR genes-associated the mobile genetic elements (MGEs) were extracted and investigated. Twenty ICEs, one defective ICE, one tandem ICE, and ten prophages were found, which mainly carried tetracycline, macrolides/lincosamides/streptogramin (MLS), and aminoglycosides resistance genes. Three types of DNA cargo with AR genes were integrated into specific sites of ICEs: integrative mobilizable elements (IMEs), cis-IMEs (CIMEs), and transposon Tn916. Obvious differences in AR gene categories were found among the three cargo types. IMEs mainly harbored tetracycline and MLS resistance genes. CIMEs mainly carried aminoglycoside resistance genes, while transposon Tn916 carried only the tet (M) gene. Nearly all AR genes in ICEs were carried by IMEs and CIMEs. IMEs were prevalent and were also detected in additional 29 S. suis genomes. The horizontal transfer of IMEs and CIMEs may play critical role in ICE evolution and AR gene transmission in the S. suis population. Our findings provide novel insights into the transmission patterns of AR genes and the evolutionary mechanisms of ICEs in S. suis.
1. Introduction
Streptococcus suis is a major swine pathogen that causes septicemia, meningitis, and a variety of other diseases. It is emerging as a significant zoonotic agent responsible for the largest outbreak in China and increasing human meningitis worldwide (Ye et al., 2006; Gottschalk et al., 2007). High rates of resistance to tetracycline, macrolide, lincosamide, and aminoglycoside have been reported in both human and pig isolates of S. suis since 2000 (Wisselink et al., 2006; Ye et al., 2008; Zhang et al., 2008; Princivalli et al., 2009; Palmieri et al., 2011b). Furthermore, S. suis is likely an important reservoir of antibiotic resistance (AR) genes that may promote the interspecies horizontal transfer of these resistances (Palmieri et al., 2011b). A better understanding of the dissemination mechanism of AR genes is essential for monitoring and controlling drug resistance. Previous studies have revealed that mobile genetic elements (MGEs) play significant roles in the horizontal transfer of AR genes in Streptococcus (Puopolo et al., 2007; Chancey et al., 2015; Davies et al., 2015; Kambarev et al., 2018). Integrative conjugative elements (ICEs), prophages and tandem ICE-prophages located at different insertion sites were common MGEs described in S. suis (Uruen et al., 2022). Integrative and conjugative elements (ICEs) comprise the majority of MGEs and are critical contributors to the acquisition and distribution of AR genes in S. suis (Huang et al., 2016a), in which harbored tetracycline, macrolide, and lincomycin resistance genes (Li et al., 2011; Palmieri et al., 2011b; Marini et al., 2015; Athey et al., 2016; Huang et al., 2016a,b; Zheng et al., 2018; Libante et al., 2019; Pan et al., 2019; Shang et al., 2019; Zhu et al., 2019; Dechene-Tempier et al., 2021; Lunha et al., 2022; Yang et al., 2022). On the contrary, tetracycline, macrolide, lincomycin, nucleoside, and aminoglycoside resistance genes associated tandem ICE-prophages and prophage were often reported in S. suis clinical strains (Palmieri et al., 2011b; Huang et al., 2016a; Wang et al., 2019). In previous studies, tetracycline, macrolide, lincomycin, amphenicols, oxazolidinones, and aminoglycoside resistance genes associated prophages were common in S. suis strains (Wang et al., 2021; Uruen et al., 2022). Recently, the optrA-carrying prophage ΦSC181 was also reported in S. suis strain (Shang et al., 2019). In addition, IMEs mainly carrying tetracycline and macrolide resistance genes appeared to be more widespread than ICEs in S. suis and became major vehicles of AR genes in S. suis (Libante et al., 2019; Liang et al., 2021).
In order to thoroughly investigate the acquisition and dissemination mechanisms of AR genes, we genetically characterized AR genes-associated ICEs extracted from 155 genomes of S. suis strains isolated between 2006 and 2016 in China. Considering the fact that integrative mobilizable elements (IMEs) were widely spread in S. suis (Libante et al., 2019; Liang et al., 2021), the prevalence of IMEs with AR gens in the 155 genomes was also investigated.
2. Materials and methods
2.1. Bacterial strains, chromosomal DNA preparation, sequencing, and bioinformatic analysis
A total of 155 draft genomes of S. suis strains isolated from the nasopharynx of slaughtered pigs and the lungs of diseased pigs were included in this study (Supplementary Table S1). All strains were confirmed to belong to S. suis using 16S rRNA sequencing. They were sequenced on the Illumina platform as described in our previous studies (Chen et al., 2013; Qiu et al., 2016; Zheng et al., 2017). Sequences were assembled using SOAPdenovo (release 1.04), genes were predicted using Glimmer, and gene orthologs were determined using OrthoMCL.
The serotype of the tested strains was determined by capsular gene typing (Qiu et al., 2016; Zheng et al., 2017) and agglutination testing (serum provided by Statens Serum Institute, Copenhagen, Denmark; Supplementary Table S1). Multilocus sequence typing (MLST) were performed as described previously (King et al., 2002; Chen et al., 2013; Supplementary Table S1). The rplL gene, rum gene, mutT gene, luciferase-like monooxygenase gene, and ADP ribose pyrophosphatase gene are major insertion hot spots for the integration of MGEs in S. suis (Huang et al., 2016a; Libante et al., 2019). IMEs were usually inserted in snf2 and ppi loci. Downstream sequences of these genes were therefore analyzed. Inserted sequences with no gap were further annotated using the bioinformatic methods described in a previous study (Zheng et al., 2018). Genes were annotated with the Artemis program. The Clusters of Orthologous Groups (COG) and Pfam protein motif databases were used to search for conserved protein domains. For ICE identification, signature proteins integrase, relaxase, and VirB4 were typed using the database from a previous study (Huang et al., 2016a). The clade to which each protein belonged was determined based on protein identification and coverage of greater than 90%. If all three proteins were present, the element was considered an ICE. Elements that lacked any of the three proteins were considered defective ICEs (DICEs). Search strategies and the definitions of IMEs and CIMEs have been described in a previous study (Coluzzi et al., 2017). Antibiotic resistance analysis was performed by searching the Antibiotic Resistance Genes Database (ARDB), the Comprehensive Antibiotic Resistance Database (CARD), and ResFinder of Center for Genomic Epidemiology1. A gene was only regarded as a resistance gene homolog if its protein showed at least 80% amino acid identity to a known resistance protein across 80% of the protein’s length (Hu et al., 2013; Table 1). Sequence comparisons were performed using the BLASTn program (E-value cutoff 1e−10) and visualized using an in-house perl script (available on line:2).
2.2. Conjugation assay
The presence of the circular extrachromosomal form of ICEs in corresponding strains were firstly detected using specific primers facing outward from 5′ and 3′ side of each ICE (Supplementary Table S2). PCR amplification cycling parameters were 94°C for 5 min, then 30 cycles of 94°C for 30s, 48°C for 30s, 72°C for 60s. This was followed by a final elongation step at 72°C for 10 min. In mating experiment, S. suis derivative strain P1/7RIF (tetracycline-susceptible but rifampin resistant) was used as recipient and S. suis strains carrying circular ICEs (tetracycline-resistant but rifampin-susceptible) were utilized as donors. Donor and recipient strains were grown separately in THB broth containing the appropriate antibiotics at 5% CO2 37°C until OD600 nm value reaching 0.6 in the logarithmic growth phase, and then 0.2 ml of donor culture was mixed with 1 ml of recipient culture. Strains were collected by centrifugation and were resuspended in 100 μl THB supplemented with 10 mM MgCl2, 2 μg/ml BSA, and 100 U Dnase I, then spread onto a sterile nitrocellulose membrane overlaid on a THB agar plate and incubated 6 h. Strains were removed by washing the filters in 1 ml of THB broth, then 100 μl of the solution was spread onto THB agar plate containing tetracycline (8 μg/ml) and THB agar plate containing both tetracycline (8 μg/ml) and rifampin (8 μg/ml), respectively. Transconjugants grown on the THB containing both tetracycline and rifampin were confirmed by PCR and sequencing. Conjugative transfer frequencies were calculated as the number of transconjugants/the number of strains grown on the THB containing tetracycline.
2.3. Nucleotide sequence accession numbers
The sequences of DNA elements obtained in this study were deposited in GenBank under accession numbers MK211778, MK211782–MK211826, MN394765, MN420460, and MK473142 (Supplementary Table S1).
3. Results
3.1. The distribution of AR genes in 155 S. suis genomes
One of 155 genomes did not harbor any AR genes. Totally, 656 AR genes were present in the remaining 154 genomes. The AR genes were clustered into 11 categories, consisting of tetracycline, macrolides, lincosamide, streptogramin, aminoglycoside, trimethoprim, amphenicols, nucleoside, quinupristin/dalfopristin, glycopeptide, and oxazolidinones resistance genes (Supplementary Table S1). It is noteworthy that 145 genomes harbored at least two categories of AR genes. Among them, 14ND95 and YS39 harbored 11 and 10 AR genes, respectively.
3.1.1. The tetracyclines resistance genes
Totally, over 96% (150/155) of genomes carried 161 tetracycline-resistant genes, consisting of tet (O) (n = 102), tet (O/W/32/O) (n = 32), tet (M) (n = 12), tet (40) (n = 7), tet (W) (n = 4), tet (L) (n = 3), and tet (32) (n = 1) genes. Eleven of 150 genomes harbored two tetracycline-resistant genes simultaneously.
3.1.2. The MLS, macrolides/streptogramin, and lincosamide resistance genes
Totally, 111 genomes carried 122 MLS genes, consisting of erm (A) (n = 11) and erm (B) (n = 111) genes. Erm (B) gene was prevalent and present in 104 genomes, while erm (A) gene was present in 11 genomes. The 35 macrolides/streptogramin resistance gene msr (D) (n = 15) and mef (A) (n = 20) was present in 20 genomes. Fifty-one lincosamide resistance genes were present in 33 genomes, consisting of lnuB (n = 17), lnuC (n = 4), lnuD (n = 12), lsaE (n = 14), lsaC (n = 3), and lmrD (n = 1) genes.
3.1.3. The aminoglycosides resistance genes
In the present study, 60% (93/155) of carried 206 aminoglycosides resistance genes, consisting of ant (6)-Ia (n = 62), aac (6′)-Ie-aph (2″)-Ia (n = 50), spw (n = 39), aph (3′)-IIIa (n = 36), ant (4′)-Ib (n = 11), aad (6; n = 7), and ant (4)-Ia (n = 1) genes.
3.1.4. The trimethoprim and chloramphenicol resistance genes
Three genomes harbored trimethoprim resistance gene dfrF. Fourteen genomes harbored chloramphenicol resistance gene, consisting of cat (n = 10), cat-TC (n = 3), and cat-Q (n = 1) genes.
In addition, 18 and one genomes harbored nucleoside resistance gene sat-4 and quinupristin/dalfopristin-resistant gene vatE, respectively. It is noteworthy that YS191 harbored one glycopeptide resistance gene vanXYG.
3.2. Analysis of MGEs with ARs in 155 S. suis genomes
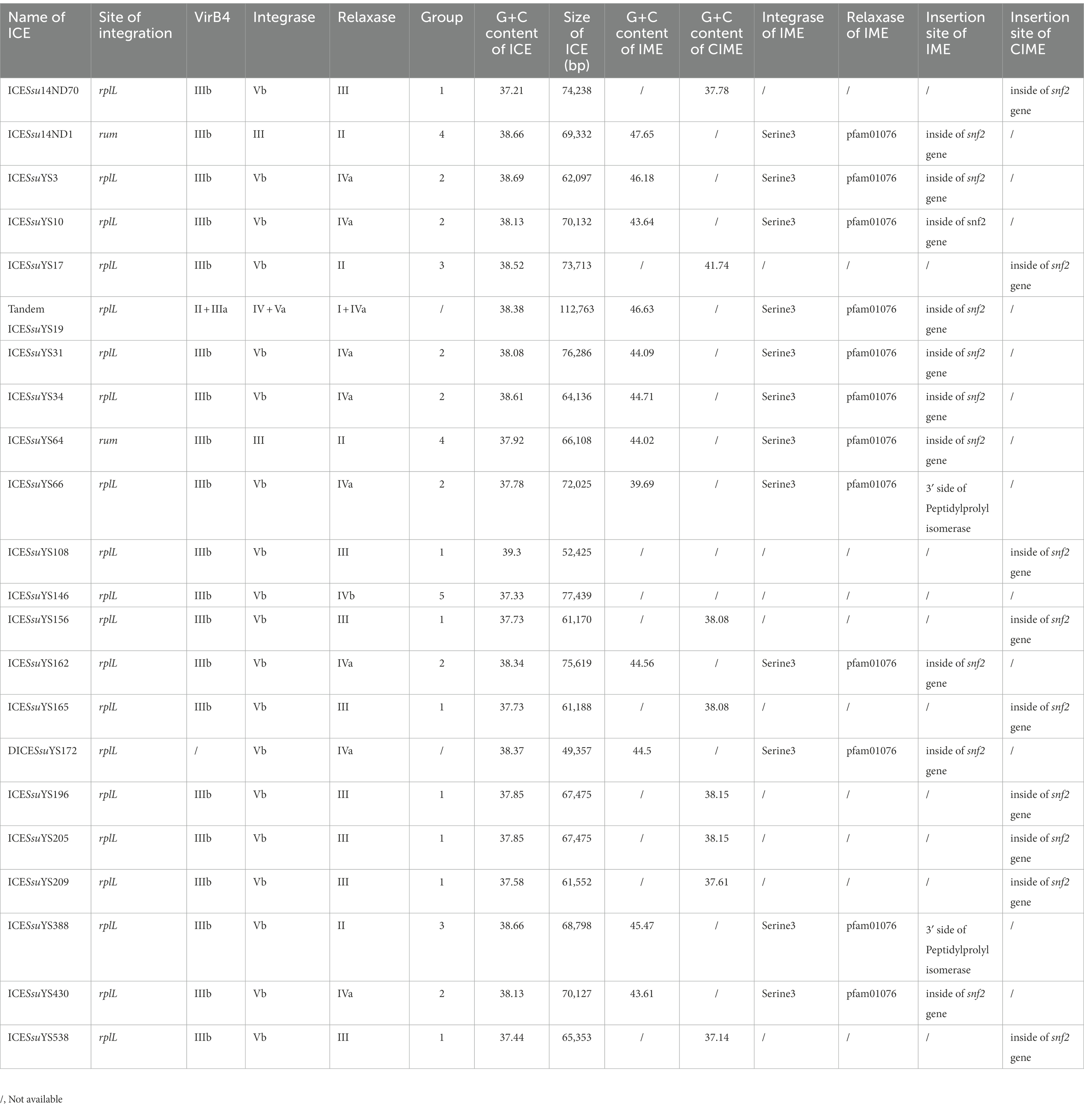
At rplL loci, 29 insertions were fully sequenced with no gap between the rplL and hdy genes. Nine of these 29 insertions lacked AR genes. Sequences of the remaining 20 insertions that contained AR genes were further analyzed. All of them harbored a 15-bp att sequence 5′-TTATTTAAGAGTAAC-3′ in the flanking region. The twenty insertions were composed of 18 ICEs, one DICESsuYS172 that lacked VirB4, and one tandem ICESsuYS19 (Table 1).
At rum loci, two intact ICEs with AR genes were identified and designated ICESsuYS64 and ICESsu14ND1 between the rum and glf genes (Table 1). ICESsu14ND1 harbored the 14-bp att sequences 5′-CACGTGGAGTGCGT-3′ and 5′-CACATAGAAGTTGT-3′ in the 5′ and 3′ side flanking regions, respectively. ICESsuYS64 harbored the 14-bp att sequences 5’-CACGTGGAGTGCGT-3′ and 5′ -CACGTTGAAGTTGT-3′ in the 5′ and 3′ side flanking regions, respectively. In the present study, ten intact prophages with AR genes were identified at rum loci (Supplementary Table S1). It is noteworthy that YS34 and YS162 harbored ICE and prophage simultaneously (Supplementary Table S1). Nine prophages with AR genes were integrated into downstream of rum gene and upstream of pgmA gene. One prophage (ΦSsuYS43) with AR genes was integrated into downstream of rum gene and upstream of glf gene.
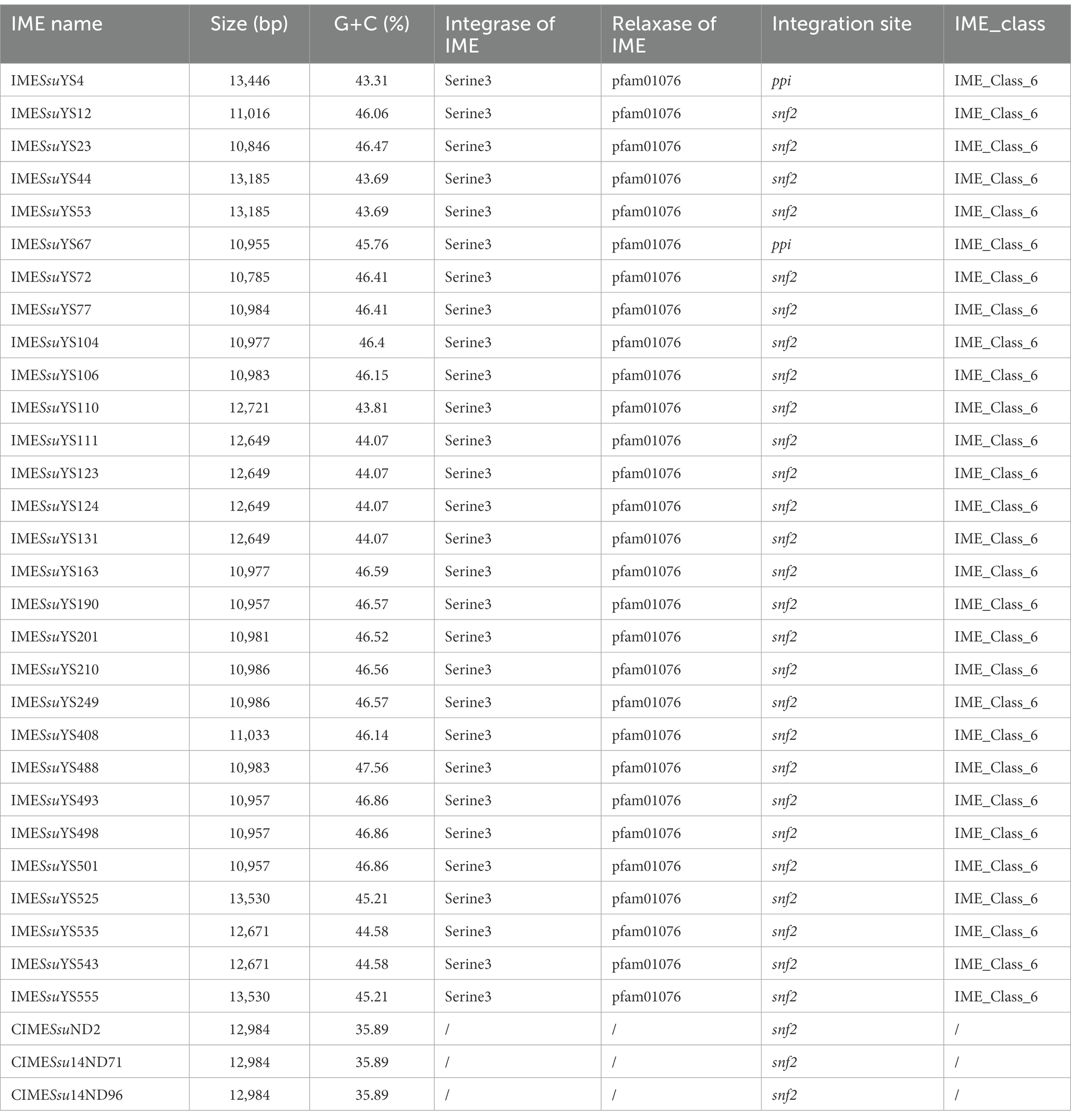
In addition to the aforementioned MGEs present in 30 genomes (18 genomes carrying ICE, eight genomes carrying prophage, two genomes carrying both ICE and prophage, one genome carrying DICE, one genome carrying tandem ICE), 29 IMEs and 3 identical CIMEs with AR genes were found in remaining 125 genomes. The majority of AR-containing IMEs (27 out of 29) were integrated into the snf2 gene (Table 2). The remaining two IMEs from genomes YS4 and YS67 were integrated into ppi loci (Table 2). It is noteworthy that 11 IMEs were integrated into the same contigs with either rplL or hdy genes, and the IME of genome YS12 was located in the same contig with the glf gene.
No insertions with ARs were identified at mutT, luciferase-like monooxygenase, and ADP ribose pyrophosphatase loci in the present study.
3.3. AR genes of MGEs
The 57 AR genes that were harbored in the 20 ICEs belonged to three categories: tetracycline, MLS, and aminoglycosides. Five ICEs carried all three categories of antibiotic resistance genes (Supplementary Table S1). The ICEs carried four types of tetracycline resistance genes, including tet (O/W/32/O), tet (O), tet (M), and tet (40) genes. The tet (O) gene was predominant and appeared in 11 ICEs. It is noteworthy that eight ICEs carried the tet (O/W/32/O) gene. The tet (40) and tet (M) genes were present in two and one ICEs, respectively (Supplementary Table S1). The erm (B) gene was found in 14 ICEs. Moreover, ICESsu14ND70 and ICESsuYS162 each carried three and two copies of the erm (B) gene, respectively (Supplementary Table S1). Eight ICEs that carried genes coding for aminoglycoside resistance, consisting of the ant (6)-Ia (n = 8), spw (n = 5), and aph (3′)-IIIa (n = 4) genes. Additionally, the sat-4 gene that encodes for streptothricin resistance was found in ICESsuYS17.
Tandem ICESsuYS19 only carried the tet (O) gene, and the DICESsuYS172 carried both the tet (O) and erm (B) genes (Supplementary Table S1).
The 43 AR genes present in 29 IMEs were mainly tet (O) and erm (B) genes. Among the 29 IMEs, 13 IMEs only carried the tet (O) gene. Ten IMEs carried both the tet (O) and erm (B) genes. Only two IMEs from genome YS525 and YS555 simultaneously carried tet (O), erm (B) and the aminoglycoside resistance gene ant (4′)-ia (Figure 1; Supplementary Table S1). Three identical CIMEs contained a combination of aph (3′)-IIIa, aac (6′)-Ie-aph (2″)-Ia, ant (6)-Ia, tet (O/W/32/O), erm (B), and sat-4 genes (Supplementary Table S1).
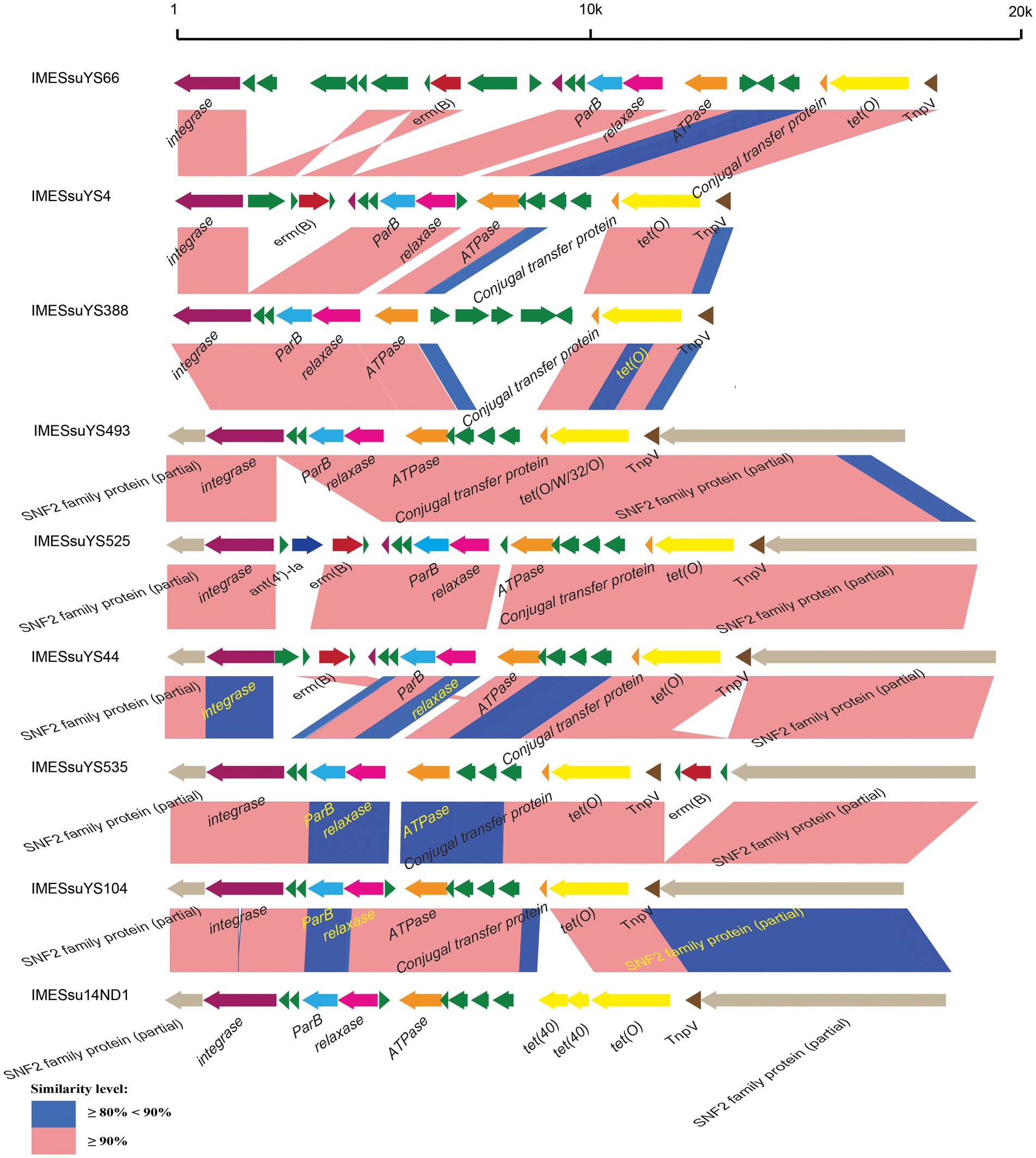
Figure 1. Schematic comparison of representative IMEs. The direction of the arrow indicates the direction of transcription. Regions of >90 and 80–90% identity are marked by pink and blue shading, respectively. The AR genes, integrase genes, ParB genes, relaxase genes, ATPase genes, conjugal transfer genes, TnpV genes, snf2 genes are indicated by different colors.
The 38 AR genes harbored in the ten prophages belonged to five categories: tetracycline, macrolides, streptogramin, lincosamide, and aminoglycosides. The tet (O) and erm (B) gene was present in seven and four prophages, respectively. The prophages also carried mef (A) (n = 7), msr (D) (n = 5), ant (6)-Ia (n = 5), aac (6′)-Ie-aph (2″)-Ia (n = 4), lnuD (n = 2), aph (3′)-IIIa (n = 1), lsaE (n = 1), lnuB (n = 1), and spw (n = 1) genes (Supplementary Table S1).
3.4. Characterization of ICEs
We further investigated the characterizations of ICEs. In a previous study, the ICEs were classified into three groups (Tn5252, ICEsp1108, and TnGBS2) based on the presence of signature proteins VirB4, integrase, and relaxase (Huang et al., 2016a). The integrases of ICEs integrated into rplL and rum were tyrosine and serine integrase, respectively. The relaxases of all ICEs belonged to the MobP family. Moreover, all ICEs contained the conserved DNA processing unit composed of relaxase, MobC, and Tn5252 ORF10 (Figures 2, 3).
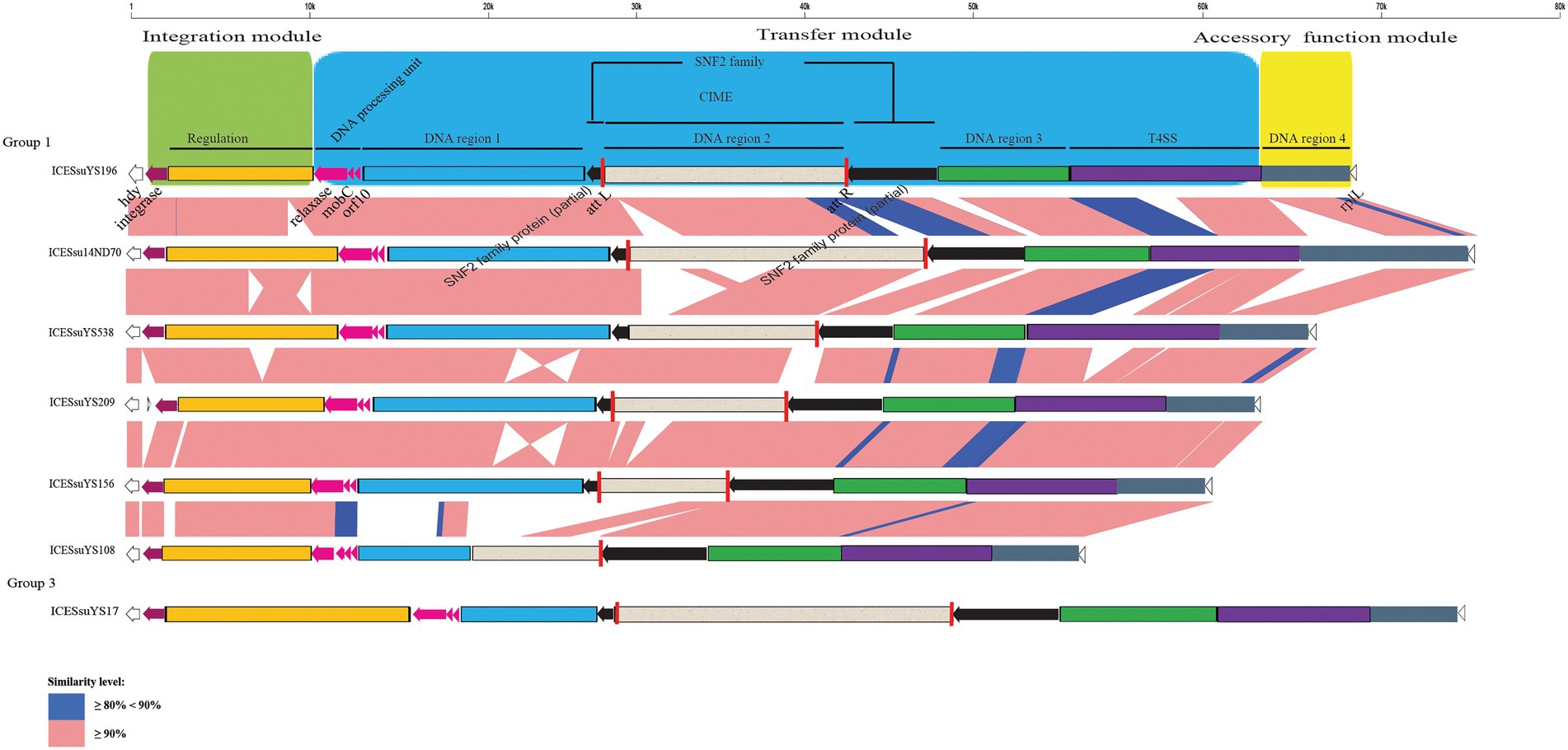
Figure 2. Schematic comparison of representative ICEs carrying CIMEs. The direction of the arrow indicates the direction of transcription. Regions of >90 and 80- 90% identity are marked by pink and blue shading, respectively. The DNA regions, T4SS, CIMEs, integrase gene, relaxase gene, mobC, orf10, snf2 gene, att site, integration module, transfer module, and accessory module of ICEs are indicated by different colors. The rplL and hdy genes are located in the flanking region of the ICEs.
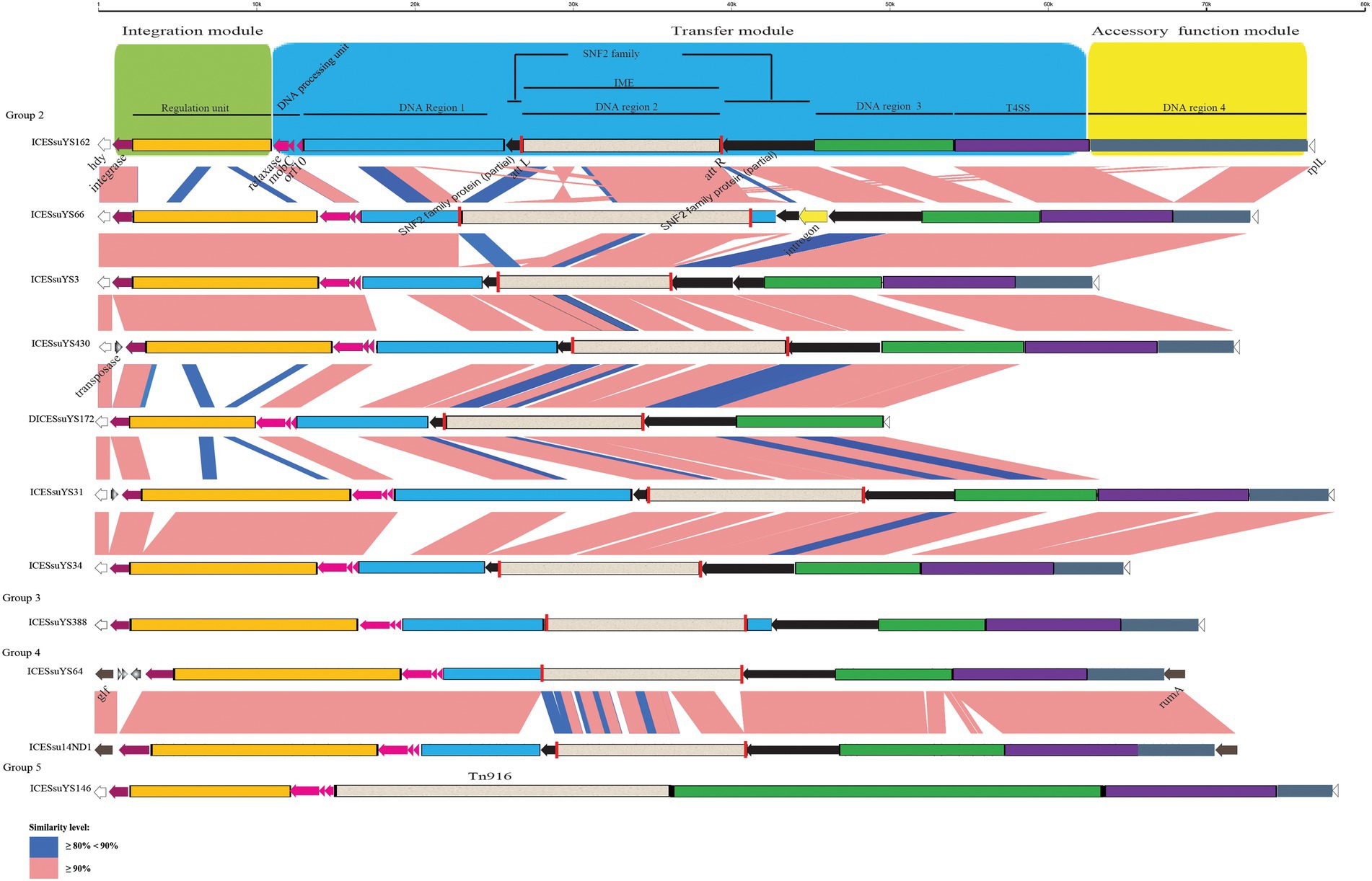
Figure 3. Schematic comparison of representative ICEs and DICEs carrying IMEs and transposon Tn916. The direction of the arrow indicates the direction of transcription. Regions of >90 and 80–90% identity are marked by pink and blue shading, respectively. The DNA regions, T4SS, IMEs, Tn916, integrase gene, relaxase gene, mobC, orf10, snf2 gene, att site, integration module, transfer module, and accessory module of ICEs and DICEs are indicated by different colors. The rplL and hdy genes or rumA and glf genes are located in the flanking region of the ICEs and DICE.
All 20 ICEs belonged to the Tn5252 group and ranged from 52,425 bp to 77,439 bp in size. Their G + C content ranged from 37.21 to 39.3%. Sequence alignment revealed high similarity between ICESsuYS10 and ICESsuYS430, ICESsuYS156 and ICESsuYS165, and ICESsuYS196 and ICESsuYS205, respectively. It is noteworthy that an intact type IV secretion system (T4SS) containing the VirB1, VirB4, VirB6, and VirD4 genes was identified in the 20 ICEs.
There were five VirB4, integrase, and relaxase combinations in the 20 ICEs, primarily distinguished by variations in relaxase. The combinations were classified into five groups: clade IIIb + clade Vb + clade III (group 1, n = 8), clade IIIb + clade Vb + clade IVa (group 2, n = 7), clade IIIb + clade Vb + clade II (group 3, n = 2), clade IIIb + clade III + clade II (group 4, n = 2) and clade IIIb + clade Vb + clade IVb (group 5, n = 1; Table 1).
The structures of all ICEs could be divided into an integration module, a transfer module, and an accessory function module. The integration module was composed of integrase and regulation unit, whereas the transfer module contained DNA processing unit, DNA region 1 to 3 and the T4SS. DNA region 4 constituted the accessory function module (Figures 2, 3).
Three modules were conserved within groups 1 and 4. Within groups 2 and 3, there were obvious variations in the integration and transfer modules. Variations were present mainly in regulation unit and DNA region 1 (Figures 2, 3).
The size and G + C content of DICESsuYS172 were 48,743 bp and 38.45%, respectively. The clades of integrase and relaxase in DICESsuYS172 were identical to those of group 2 ICEs (Table 1); However, an intact T4SS was absent. Compared with ICEs from other groups, the transfer module of DICESsuYS172 was most similar to that of group 2 ICEs (Figure 3).
The size and G + C content of the tandem ICESsuYS19 were 112,607 bp and 38.39%, respectively. Two clades of VirB4 (II + IIIa), integrase (IV + Va), and relaxase (I + IVa) were present in tandem ICESsuYS19, and it contained one copy of the T4SS (Table 1).
DNA region 2 of the ICEs contained various AR genes as DNA cargoes. In 15 ICEs, DICESsuYS172, and tandem ICESsuYS19, 17 cargoes were specifically integrated into the snf2 gene, which encodes an SNF2 family protein. These cargoes harbored an 11-bp inverted repeat 5’-TTTTGCGGACA-3′ in the flanking region. Moreover, they also carried a TnpV transposase gene. It is noteworthy that ten of them possessed integrase and relaxase genes with a pattern similar to that of IMEs (Coluzzi et al., 2017; Figure 3; Table 1; Supplementary Figure S1). The IMEs were identified in eight ICEs, DICESsuYS172, and tandem ICESsuYS19. The remaining seven DNA cargoes that had lost the integrase and relaxase genes were designated cis-IMEs (CIMEs; Coluzzi et al., 2017; Figure 2; Table 1; Supplementary Figure S2).
IMEs were also found in ICESsuYS66 and ICESsuYS388, where they were inserted downstream of genes encoding peptidylprolyl isomerase (PPI; Libante et al., 2019; Figure 3; Table 1). ICESsuYS388 harbored an intact snf2 gene, and a group II intron gene was integrated into the snf2 gene of ICESsuYS66 (Table 1; Supplementary Figure S1). The two IMEs found in ICESsuYS66 and ICESsuYS388 harbored an inverted repeat sequence in the flanking region that was identical to that of the aforementioned ten IMEs. The integrases of aforementioned 12 IMEs were identical and belonged to serine integrase type 3 (Coluzzi et al., 2017), regardless of their integration site. Meanwhile, the relaxases of the IMEs belonged to the PF01076/MOBV superfamily. Based on their integrase and relaxase types, all 12 IMEs belonged to IME_Class_6 (Coluzzi et al., 2017). Interestingly, the G + C content of the IMEs (from 39.69 to 47.65%) was clearly different from that of the corresponding ICEs (from 37.21 to 39.3%; Table 1).
CIMEs were also found in ICESsuYS108 and ICESsuYS538, in which the 3′ side of the snf2 gene (for ICESsuYS108) and the attL site (for ICESsuYS538 and ICESsuYS108) were absent (Figure 2; Supplementary Figure S2). On the contrary, the G + C content of the all CIMEs integrated into ICEs were similar to those of their host ICEs, except that of CIME harbored by ICESsuYS108 was unavailable. Interestingly, all ICEs from group 1 carried CIMEs (Table 1).
Transposon Tn916, prevalent in the epidemic strains that caused S. suis outbreak in China 2005 (Ye et al., 2008), was integrated into ICESsuYS146 (Figure 3; Supplementary Figure S2).
All AR genes in the ICEs were carried by integrated DNA cargos, except for an additional erm (B) gene that was carried in the accessory function module of ICESsuYS162 and ICESsu14ND70. Unlike IMEs and CIMEs, transposon Tn916 only harbored the tet (M) gene. Compared with IMEs that only carry the erm (B) gene and/or tetracycline resistance gene, aminoglycoside resistance genes were wildly present in nine CIMEs.
Moreover, additional 29 IMEs with AR genes present in remaining genomes also belonged to Class_6 IMEs. It is noteworthy that their G + C content ranged from 43.31 to 47.56% that were similar to those of IMEs integrated into ICEs (Table 2).
The size and G + C content of prophages ranged from 58,465 bp to 87,220 bp and from 38.83 to 39.61%, respectively. Among ten prophages, five prophages ΦSsuYS199, ΦSsuYS214, ΦSsuYS225, ΦSsuYS255 and ΦSsuYS262 were completely identical.
3.5. Transferability of ICEs
The presence of the circular extrachromosomal form of 18 ICEs at rpIL loci were detected in corresponding strains. The circularization of eight ICEs were detected in corresponding strains, consisting of ICESsuYS3, ICESsuYS17, ICESsuYS34, ICESsuYS66, ICESsuYS146, ICESsuYS162, ICESsuYS388, and ICESsuYS430. Notable, none of circular ICE carried CIMEs was detected in corresponding strain. In conjugation assay, eight S. suis strains carrying circular ICEs were used as donors and S. suis strain P1/7RIF were utilized as recipients. In mating experiments, we obtained clones of transconjugant carrying ICESsuYS3, ICESsuYS34, and ICESsuYS388 in S. suis P1/7RIF at a low frequency of ∼7.8 × 10−8 (3/3.84 × 107), 1.2 × 10−7 (4/3.26 × 107), and 7.1 × 10−8 (2/2.81 × 107), respectively.
4. Discussion
S. suis strains from swine are important reservoirs of antibiotic resistance genes in that tetracycline, lincosamide, macrolide, and aminoglicoside are extensively used for therapy and metaphylaxis in swine industry (Schwarz et al., 2007; Callens et al., 2012; Seitz et al., 2016; Xu et al., 2017). In the present study, 155 S. suis genomes harbored 656 AR genes which were mainly clustered into tetracycline, macrolide, lincosamide, and the aminoglycosides resistance genes. In the present study, 154 of 656 AR genes were present in MGEs, consisting of 20 ICEs, one tandem ICE, one DICE, 29 IMEs, three CIMEs, and ten prophages. Interestingly, the AR genes in MGEs were also responsible for the resistance to the aforementioned antibiotics in the present study. The MGEs played significant roles in the acquisition and dissemination of AR genes in S. suis. Interestingly, these resistance genes carried by MGEs also emerged and were prevalent in S. suis strains from patients in China (Wang et al., 2019), which is becoming emergent threat to local public health.
Consistent with previous report, ICEs and IMEs were prevalent vehicles of AR genes in S. suis (Huang et al., 2016a; Libante et al., 2019; Liang et al., 2021). The ICEs are able to excise from the chromosomes to form circular intermediates prior to conjugative transfer into recipient cells (Burrus et al., 2002). In the present study, eight of 20 ICEs did excise from the corresponding chromosome to produce a circular form, which represented an intermediate required for their transference. In addition, clones of transconjugant carrying three ICEs (ICESsuYS3, ICESsuYS34, and ICESsuYS388) were obtained in mating experiment. Our results indicated that the ICEs, as mobile genetic elements, may be crucial for the acquisition and dissemination of AR genes in S. suis.
Integrase, relaxase, and VirB4 as signature proteins of integration and transfer modules were used to classify the ICEs. The predominant clades of VirB4 and integrase in the Tn5252 group are Vb and IIIb (Huang et al., 2016a), consistent with what we have observed in the current study. Substantial diversity was found in relaxase, which was classified into four clades. Relaxase clade III and IVa were predominant in the present study, while clade IVb was prevalent in the Tn5252 group (Huang et al., 2016a). Five VirB4, integrase, and relaxase combinations were found in the 20 ICEs.
In the present study, three types of DNA cargo with AR genes were integrated into 20 ICEs, consisting of IMEs, CIMEs, and the transposon Tn916. It is noteworthy that nearly all AR genes in ICEs were carried by these three types of DNA cargoes. Therefore, it is reasonable to speculate that these cargoes play critical roles in the dissemination of AR genes. The three types of cargo contained different categories of AR genes. IMEs mainly harbored tetracycline and MLS resistance genes. CIMEs frequently contained genes coding for aminoglycoside resistance, while transposon Tn916 carried only the tet (M) gene. In contrary to IMEs mainly carrying the tet (O) gene, the predominant tetracycline resistance gene in CIMEs was tet (O/W/O/32).
The IMEs could excise and integrate but carry only some of the sequences or genes necessary for their conjugative transfer. Instead, they employ the conjugative elements of co-resident ICEs to affect transfer. In our study, several lines of evidence indicated that IMEs were exogenous and transferable, even though IMEs were unable to excise in laboratory experiment (Libante et al., 2019, (1) their G + C content was obviously different from that of corresponding ICEs, (2) they had an insertion hot spot, (3) they had an 11-bp inverted repeat sequence in the flanking region as att site, (4) spontaneous excision may occur through their common site-specific integrase gene, (5) they carried a TnpV transposase gene, which has been widely reported to be involved in conjugative element integration (Wang et al., 2006; Huang et al., 2016b), and (6) they were widely distributed in strains from different origins. In the present study, IMEs were distributed in ten ICEs, one DICE, one tandem ICE, and 29 additional genomes. Similar IMEs were also integrated into snf2 gene of ICESsuZJ20091101-1 (KX077882.1), ICESsuLP081102 (KX077885.1), ICESsuJH1301 (KX077887.1; Huang et al., 2016a) and ICESsD9 (Huang et al., 2016b). Through site-specific recombination mechanisms, IMEs integrated into ICEs/genomes and conferred antibiotic resistance to host ICEs/genomes. It is likely that the exchange, acquisition, and deletion of the IME module contributed to the evolution of ICEs.
IMEs encode tyrosine integrase, serine integrase, or DDE transposase. Tyrosine integrases were detected in more than 89% of Streptococcus IMEs (Coluzzi et al., 2017). In our study, all IMEs harbored the same serine integrase, Ser_3, which confers high specificity of IME integration. IME relaxases are classified into nine superfamilies, and the relaxases in this study belonged to the Rel_PF01076/MobV superfamily. In contrast to data from a previous study in which IMEs from classes 1, 2, 3, 4, 7, and 8 were present in 120 of 144 Streptococcus species (Coluzzi et al., 2017), all IMEs we analyzed in the present study were IME_class_6. IMEs were reported to be more widespread than ICEs in S. suis (Libante et al., 2019; Liang et al., 2021). In the present study, IMEs were also found to be highly prevalent in tested S. suis genomes. We propose that IMEs may play a critical role in the spread of tetracycline, macrolide, and lincosamide resistance genes and that their activity may explain the high rate of resistance to these antibiotics in the S. suis population.
CIMEs are decayed IMEs, which are cis-mobilizable elements that had lost their integration and relaxase genes but retained their attL and attR sites. CIMEs were found in nine ICEs and three additional genomes. A similar CIME was also inserted into the same integration site of ICESsuBSB6 (Supplementary Figure S2; Huang et al., 2018). The structure of the CIMEs was homologous to that of the mobile 15 K element of ICESsu32457 obtained from an Italian strain, although the latter had 1.3 kb att sequences in the flanking region and was integrated between the intact snf2 gene and the DNA primase gene (Palmieri et al., 2012; Supplementary Figure S1). Despite the lack of a recombinase gene, the 15 K element may be capable of spontaneous excision via its 1.3 kb att sequences in the flanking region (Palmieri et al., 2012). Unlike IMEs, CIMEs frequently contained genes coding for resistance to aminoglycosides. We suggest that CIMEs may play an important role in the dissemination of aminoglycoside resistance. Further study is needed to investigate whether CIMEs can be excised by their own 11-bp att sequences in the flanking region and whether the transfer can be mediated by the TnpV gene.
In previous study, most of the IMEs carrying AR genes were integrated into snf2 or ppi loci (Libante et al., 2019; Liang et al., 2021). In the present study, IMEs and CIMEs were predominantly inserted into snf2 site. Moreover, a group II intron was site-specifically integrated into the snf2 gene of ICESsuYS66. It is unclear why the snf2 gene was the preferred integration site for these cargoes. The snf2 gene was extensively distributed in the 155 S. suis genomes, only eight of which carried an intact snf2 gene (data not shown). We proposed that a variety of cargos specifically integrated into the snf2 gene may confer significant selective advantages to their host.
ICESsuYS146 harbored the transposon Tn916, which contributes to the spread of the tet (M) gene. It is noteworthy that tet (M) is a prevalent tetracycline resistance gene in S. suis strains from meningitis patients in Vietnam and outbreak in China, where it is also associated with the presence of transposon Tn916 (Ye et al., 2008; Hoa et al., 2011). The capacity of transposon Tn916 with the tet (M) gene to horizontal transfer in S. suis strains was revealed and played important roles in the evolution of the epidemic S. suis ST7 clone in our previous study (Ye et al., 2008).
The mobility of several ICEs was also evaluated in the present study. In S. suis, the ICEs carrying AR genes transferred in intra- and interspecies with frequency from 10−3 to 10−8 (Li et al., 2011; Marini et al., 2015; Huang et al., 2016a,b; Libante et al., 2019; Pan et al., 2019; Shang et al., 2019; Yang et al., 2022). The formation of circular intermediates between the attL and attR sites with the aid of the integrase was a requisite for the conjugative transfer of the ICEs (Flannery et al., 2011; Li et al., 2011). In the present study, the circular intermediates of eight ICEs were detected in corresponding strains, which were from group 2, 3, and 5. Notable, ICEs from group 1 did not appear to excise, even though they carried same clade of integrase to those of ICEs from group 2, 3, and 5. It is noteworthy that CIMEs mainly harbored aminoglycoside resistance genes were integrated into all ICEs from group 1. We proposed that multiple mutations in integrase genes and the integration of CIMEs may impair the excision and transferability capacity of ICEs from group 1.
In conjugation assay, the effective conjugation was not observed in five of excised ICEs in vitro. Similar observations were also reported in ICESsuHN105 (Zhu et al., 2019) and ICESsuNSU1086 (Libante et al., 2019). Moreover, clones of transconjugant carrying ICESsuYS3, ICESsuYS34, and ICESsuYS388 were obtained at a low frequency. Relaxase and T4SS components played critical roles in the transferability of ICEs (Li et al., 2011). Truncated virB4 and relaxase gene was present in ICESsuYS146 and ICESsuYS162, respectively. The mutations and truncation of relaxase and T4SS components genes may contribute to the transfer deficiencies of corresponding ICEs. It is possible that the conjugative frequency of these ICEs below the detection limit or their self-transmission occurred only in specific conditions not met in vitro. Previous study reported that IME was not mobile anymore as a single element but was transferred passively by the ICE harbored it (Libante et al., 2019). Further studies are needed to investigation the excision and conjugation of the IMEs.
In the present study, ten prophage carrying AR genes were also detected at rum loci. ΦSsuYS34 and ΦSsuYS771 closely resembled ΦSsUD.1 (Accession No. FN677480) of S. suis (Palmieri et al., 2011a). ΦSsuYS199, ΦSsuYS214, ΦSsuYS225, ΦSsuYS255 and ΦSsuYS262 were identical and closely resembled Φ46.1 (Accession No. FM864213) of S. pyogenes (Brenciani et al., 2010). ΦSsuYS43 closely resembled ΦSsuHCJ3 of S. suis (Accession No. MN270269). The prophage was reported to be the vehicle of tetracycline, macrolide, lincomycin, amphenicols, oxazolidinones, and aminoglycoside resistance genes in S. suis strains (Wang et al., 2021; Uruen et al., 2022). The transfer of prophage is mainly through transduction by three mechanisms, called generalized, specialized, and lateral transduction. There is in vitro evidence of transfer of prophage Φm46.1 between S. suis and S. pyogenes in both directions with different transfer rate (from S. suis to S. pyogenes with 8.5 × 10−4 and from S. pyogenes to S. suis with 2.3 × 10−9; Giovanetti et al., 2014). Further studies are needed to investigate the transferability of the prophage identified in the present study.
Most of clinical S. suis strains remain sensitive to beta-lactams antibiotic, including penicillin and cephalosporin (Hoa et al., 2011; Wang et al., 2019; Bamphensin et al., 2021). Resistance to penicillin in S. suis strains isolated from pigs is gradually increasing (Zhang et al., 2008; Lunha et al., 2022). In the present study, S. suis strains YS162 and YS388 were resistance to penicillin with the minimum inhibitory concentration (MIC) value 6 μg/ml for each strain, meanwhile S. suis strain YS34 was resistant to amoxicillin with MIC value 64 μg/ml. Moreover, S. suis strains 14ND70, YS34, and YS388 were resistant to cefaclor with MIC value 32 μg/ml, 6 μg/ml, and 8 μg/ml, respectively. In addition, S. suis strain YS34 was also resistant to cefepime, cefotaxime, ceftriaxone, and cefuroxime with MIC value 12 μg/ml, 16 μg/ml, 24 μg/ml and 24 μg/ml, respectively. In streptococci, variations of penicillin-binding proteins (PBP) were essential for the beta-lactam resistance (Sauvage et al., 2008; Li et al., 2016). Five pbp genes have been identified in S. suis strains, pbp1a, pbp1b, pbp2a, pbp2b, and pbp2x. It is noteworthy that none of pbp genes were present in ICEs, IME, and prophage identified in the present study. Compared to those of S. suis P1/7 (penicillin-susceptible isolate; pbp1a: SSU0370, pbp1b: SSU0121, pbp2a: SSU1777, pbp2b: SSU1186, and pbp2x: SSU1548,), multiple nucleotide substitutions were present in pbp genes of aforementioned beta-lactams resistant strains (data were not shown). Further studies are needed to corelated the pbp gene mutations to the reduction of drug affinity.
An obvious drawback to the study was the use of draft genomes, which may have caused some limitations. Specifically, some ICEs and IMEs containing AR genes may not have been found. In addition, different classes of IMEs were not comprehensively identified. As a result, the prevalence, diversity, and contribution of ICEs and IMEs to the dissemination of AR genes may have been underestimated in the S. suis population.
In conclusion, AR-associated ICEs, DICEs, and a tandem ICE in S. suis strains from 155 genomes were identified and characterized. A particularly interesting finding was that the most common IMEs and CIMEs integrated into the ICEs and genomes in a site-specific manner. Our findings provide novel insights into the transmission patterns of AR genes and the evolutionary mechanisms of ICEs in S. suis.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
HZ, ZW, and JX designed the project. HZ drafted the manuscript. JW and XB carried out the experiments and generated the data. KQ, WK, PL, and HZ analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Priority Project on Infectious Disease Control and Prevention from the Ministry of Science and Technology of the People’s Republic of China (grant no. 2017ZX10303405-002, 2018ZX10734404) and the National Natural Science Foundation of China (grant no. 81572044).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1074844/full#supplementary-material
SUPPLEMENTARY FIGURE 1 | Schematic comparison of ICESsu32457 carrying 15K fragment and representative ICEs carrying IMEs. The direction of the arrow indicates the direction of transcription. Regions of >90% and 80%-90% identity are marked by pink and blue shading, respectively. The integrase genes, relaxase genes, snf2 genes, ppi genes, AR genes, and T4SS of ICEs are indicated by different colors. IMEs and the 15K fragment are highlighted in a red box. The AR genes and Tnpv genes of IMEs and the 15K fragment are indicated by different colors. The site-specific integrase, AR genes and Tnpv genes of IMEs are indicated by different colors. The rplL and hdy genes are located in the flanking regions of the ICEs.
SUPPLEMENTARY FIGURE 2 | Schematic comparison of ICESsuBSB6 carrying 15K fragment and representative ICEs carrying CIMEs or Tn916. The direction of the arrow indicates the direction of transcription. Regions of >90% and 80%-90% identity are marked by pink and blue shading, respectively. The integrase gene, relaxase gene, snf2 gene, AR genes, and T4SS of ICEs are indicated by different colors. CIMEs, Tn916, and the 15K fragment are highlighted in a red box. The AR genes and Tnpv genes of CIMEs and the 15K fragment are indicated by different colors. The rplL and hdy genes are located in the flanking regions of the ICEs.
SUPPLEMENTARY TABLE 1 | The information of the genomes and AR genes carried in the present study.
SUPPLEMENTARY TABLE 2 | The primers used in the present study for the circular ICEs analyses.
Footnotes
References
Athey, T. B., Teatero, S., Takamatsu, D., Wasserscheid, J., Dewar, K., Gottschalk, M., et al. (2016). Population structure and antimicrobial resistance profiles of Streptococcus suis serotype 2 sequence type 25 strains. PLoS One 11:e0150908. doi: 10.1371/journal.pone.0150908
Bamphensin, N., Chopjitt, P., Hatrongjit, R., Boueroy, P., Fittipaldi, N., Gottschalk, M., et al. (2021). Non-penicillin-susceptible Streptococcus suis isolated from humans. Pathogens 10:1178. doi: 10.3390/pathogens10091178
Brenciani, A., Bacciaglia, A., Vignaroli, C., Pugnaloni, A., Varaldo, P. E., and Giovanetti, E. (2010). Phim 46.1, the main streptococcus pyogenes element carrying mef (a) and tet (O) genes. Antimicrob. Agents Chemother. 54, 221–229. doi: 10.1128/AAC.00499-09
Burrus, V., Pavlovic, G., Decaris, B., and Guedon, G. (2002). Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46, 601–610. doi: 10.1046/j.1365-2958.2002.03191.x
Callens, B., Persoons, D., Maes, D., Laanen, M., Postma, M., Boyen, F., et al. (2012). Prophylactic and metaphylactic antimicrobial use in Belgian fattening pig herds. Prev. Vet. Med. 106, 53–62. doi: 10.1016/j.prevetmed.2012.03.001
Chancey, S. T., Agrawal, S., Schroeder, M. R., Farley, M. M., Tettelin, H., and Stephens, D. S. (2015). Composite mobile genetic elements disseminating macrolide resistance in Streptococcus pneumoniae. Front. Microbiol. 6:26. doi: 10.3389/fmicb.2015.00026
Chen, C., Zhang, W., Zheng, H., Lan, R., Wang, H., Du, P., et al. (2013). Minimum core genome sequence typing of bacterial pathogens: a unified approach for clinical and public health microbiology. J. Clin. Microbiol. 51, 2582–2591. doi: 10.1128/JCM.00535-13
Coluzzi, C., Guedon, G., Devignes, M. D., Ambroset, C., Loux, V., Lacroix, T., et al. (2017). A glimpse into the world of integrative and mobilizable elements in streptococci reveals an unexpected diversity and novel families of mobilization proteins. Front. Microbiol. 8:443. doi: 10.3389/fmicb.2017.00443
Davies, M. R., Holden, M. T., Coupland, P., Chen, J. H., Venturini, C., Barnett, T. C., et al. (2015). Emergence of scarlet fever streptococcus pyogenes emm 12 clones in Hong Kong is associated with toxin acquisition and multidrug resistance. Nat. Genet. 47, 84–87. doi: 10.1038/ng.3147
Dechene-Tempier, M., Marois-Crehan, C., Libante, V., Jouy, E., Leblond-Bourget, N., and Payot, S. (2021). Update on the mechanisms of antibiotic resistance and the Mobile Resistome in the emerging zoonotic pathogen Streptococcus suis. Microorganisms 9:1765. doi: 10.3390/microorganisms9081765
Flannery, E. L., Antczak, S. M., and Mobley, H. L. (2011). Self-transmissibility of the integrative and conjugative element ICEPm1 between clinical isolates requires a functional integrase, relaxase, and type IV secretion system. J. Bacteriol. 193, 4104–4112. doi: 10.1128/JB.05119-11
Giovanetti, E., Brenciani, A., Morroni, G., Tiberi, E., Pasquaroli, S., Mingoia, M., et al. (2014). Transduction of the streptococcus pyogenes bacteriophage Phim 46.1, carrying resistance genes mef (a) and tet (O), to other streptococcus species. Front. Microbiol. 5:746. doi: 10.3389/fmicb.2014.00746
Gottschalk, M., Segura, M., and Xu, J. (2007). Streptococcus suis infections in humans: the Chinese experience and the situation in North America. Anim. Health Res. Rev. 8, 29–45. doi: 10.1017/S1466252307001247
Hoa, N. T., Chieu, T. T., Nghia, H. D., Mai, N. T., Anh, P. H., Wolbers, M., et al. (2011). The antimicrobial resistance patterns and associated determinants in Streptococcus suis isolated from humans in southern Vietnam, 1997-2008. BMC Infect. Dis. 11:6. doi: 10.1186/1471-2334-11-6
Hu, Y., Yang, X., Qin, J., Lu, N., Cheng, G., Wu, N., et al. (2013). Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat. Commun. 4:2151. doi: 10.1038/ncomms3151
Huang, J., Chen, L., Li, D., Wang, M., Du, F., Gao, Y., et al. (2018). Emergence of a van G-carrying and multidrug resistant ICE in zoonotic pathogen Streptococccus suis. Vet. Microbiol. 222, 109–113. doi: 10.1016/j.vetmic.2018.07.008
Huang, J., Ma, J., Shang, K., Hu, X., Liang, Y., Li, D., et al. (2016a). Evolution and diversity of the antimicrobial resistance associated Mobilome in Streptococcus suis: a probable Mobile genetic elements reservoir for other streptococci. Front. Cell. Infect. Microbiol. 6:118. doi: 10.3389/fcimb.2016.00118
Huang, K., Song, Y., Zhang, Q., Zhang, A., and Jin, M. (2016b). Characterisation of a novel integrative and conjugative element ICESsD9 carrying erm (B) and tet (O) resistance determinants in Streptococcus suis, and the distribution of ICESsD9-like elements in clinical isolates. J Glob Antimicrob Resist 7, 13–18. doi: 10.1016/j.jgar.2016.05.008
Kambarev, S., Pecorari, F., and Corvec, S. (2018). Novel Tn916-like elements confer aminoglycoside/macrolide co-resistance in clinical isolates of Streptococcus gallolyticus ssp. gallolyticus. J. Antimicrob. Chemother. 73, 1201–1205. doi: 10.1093/jac/dky016
King, S. J., Leigh, J. A., Heath, P. J., Luque, I., Tarradas, C., Dowson, C. G., et al. (2002). Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. J. Clin. Microbiol. 40, 3671–3680. doi: 10.1128/JCM.40.10.3671-3680.2002
Li, Y., Metcalf, B. J., Chochua, S., Li, Z., Gertz, R. E. Jr., Walker, H., et al. (2016). Penicillin-binding protein Transpeptidase signatures for tracking and predicting beta-lactam resistance levels in Streptococcus pneumoniae. mBio 7:e00756-16. doi: 10.1128/mBio.00756-16
Li, M., Shen, X., Yan, J., Han, H., Zheng, B., Liu, D., et al. (2011). GI-type T4SS-mediated horizontal transfer of the 89K pathogenicity island in epidemic Streptococcus suis serotype 2. Mol. Microbiol. 79, 1670–1683. doi: 10.1111/j.1365-2958.2011.07553.x
Liang, P., Wang, M., Gottschalk, M., Vela, A. I., Estrada, A. A., Wang, J., et al. (2021). Genomic and pathogenic investigations of Streptococcus suis serotype 7 population derived from a human patient and pigs. Emerg Microbes Infect 10, 1960–1974. doi: 10.1080/22221751.2021.1988725
Libante, V., Nombre, Y., Coluzzi, C., Staub, J., Guedon, G., Gottschalk, M., et al. (2019). Chromosomal conjugative and Mobilizable elements in Streptococcus suis: major actors in the spreading of antimicrobial resistance and Bacteriocin synthesis genes. Pathogens 9:22. doi: 10.3390/pathogens9010022
Lunha, K., Chumpol, W., Samngamnim, S., Jiemsup, S., Assavacheep, P., and Yongkiettrakul, S. (2022). Antimicrobial susceptibility of Streptococcus suis isolated from diseased pigs in Thailand, 2018-2020. Antibiotics (Basel) 11:410. doi: 10.3390/antibiotics11030410
Marini, E., Palmieri, C., Magi, G., and Facinelli, B. (2015). Recombination between Streptococcus suis ICESsu32457 and Streptococcus agalactiae ICESa2603 yields a hybrid ICE transferable to streptococcus pyogenes. Vet. Microbiol. 178, 99–104. doi: 10.1016/j.vetmic.2015.04.013
Palmieri, C., Magi, G., Mingoia, M., Bagnarelli, P., Ripa, S., Varaldo, P. E., et al. (2012). Characterization of a Streptococcus suis tet (O/W/32/O)-carrying element transferable to major streptococcal pathogens. Antimicrob. Agents Chemother. 56, 4697–4702. doi: 10.1128/AAC.00629-12
Palmieri, C., Princivalli, M. S., Brenciani, A., Varaldo, P. E., and Facinelli, B. (2011a). Different genetic elements carrying the tet (W) gene in two human clinical isolates of Streptococcus suis. Antimicrob. Agents Chemother. 55, 631–636. doi: 10.1128/AAC.00965-10
Palmieri, C., Varaldo, P. E., and Facinelli, B. (2011b). Streptococcus suis, an emerging drug-resistant animal and human pathogen. Front. Microbiol. 2:235. doi: 10.3389/fmicb.2011.00235
Pan, Z., Liu, J., Zhang, Y., Chen, S., Ma, J., Dong, W., et al. (2019). A novel integrative conjugative element mediates transfer of multi-drug resistance between Streptococcus suis strains of different serotypes. Vet. Microbiol. 229, 110–116. doi: 10.1016/j.vetmic.2018.11.028
Princivalli, M. S., Palmieri, C., Magi, G., Vignaroli, C., Manzin, A., Camporese, A., et al. (2009). Genetic diversity of Streptococcus suis clinical isolates from pigs and humans in Italy (2003-2007). Euro Surveill. 14:19310. doi: 10.2807/ese.14.33.19310-en
Puopolo, K. M., Klinzing, D. C., Lin, M. P., Yesucevitz, D. L., and Cieslewicz, M. J. (2007). A composite transposon associated with erythromycin and clindamycin resistance in group B streptococcus. J. Med. Microbiol. 56, 947–955. doi: 10.1099/jmm.0.47131-0
Qiu, X., Bai, X., Lan, R., Zheng, H., and Xu, J. (2016). Novel capsular polysaccharide loci and new diagnostic tools for high-throughput capsular gene typing in Streptococcus suis. Appl. Environ. Microbiol. 82, 7102–7112. doi: 10.1128/AEM.02102-16
Sauvage, E., Kerff, F., Terrak, M., Ayala, J. A., and Charlier, P. (2008). The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32, 234–258. doi: 10.1111/j.1574-6976.2008.00105.x
Schwarz, S., Werckenthin, C., Alesik, E., Wieler, L. H., and Wallmann, J. (2007). Susceptibility of bacterial pathogens against lincomycin/spectinomycin (1/2), penicillin G/neomycin (1/1), and penicillin G/dihydrostreptomycin (1/1) as determined in the BfT-germ vet monitoring program 2004-2006. Berl. Munch. Tierarztl. Wochenschr. 120, 363–371.
Seitz, M., Valentin-Weigand, P., and Willenborg, J. (2016). Use of antibiotics and antimicrobial resistance in veterinary medicine as exemplified by the swine pathogen Streptococcus suis. Curr. Top. Microbiol. Immunol. 398, 103–121. doi: 10.1007/82_2016_506
Shang, Y., Li, D., Hao, W., Schwarz, S., Shan, X., Liu, B., et al. (2019). A prophage and two ICESa2603-family integrative and conjugative elements (ICEs) carrying optr a in Streptococcus suis. J. Antimicrob. Chemother. 74, 2876–2879. doi: 10.1093/jac/dkz309
Uruen, C., Garcia, C., Fraile, L., Tommassen, J., and Arenas, J. (2022). How Streptococcus suis escapes antibiotic treatments. Vet. Res. 53:91. doi: 10.1186/s13567-022-01111-3
Wang, M., Du, P., Wang, J., Lan, R., Huang, J., Luo, M., et al. (2019). Genomic epidemiology of Streptococcus suis sequence type 7 sporadic infections in the Guangxi Zhuang autonomous region of China. Pathogens 8:187. doi: 10.3390/pathogens8040187
Wang, H., Smith, M. C., and Mullany, P. (2006). The conjugative transposon Tn5397 has a strong preference for integration into its Clostridium difficile target site. J. Bacteriol. 188, 4871–4878. doi: 10.1128/JB.00210-06
Wang, X., Sun, J., Bian, C., Wang, J., Liang, Z., Shen, Y., et al. (2021). The population structure, antimicrobial resistance, and pathogenicity of Streptococcus suis cps 31. Vet. Microbiol. 259:109149. doi: 10.1016/j.vetmic.2021.109149
Wisselink, H. J., Veldman, K. T., Van den Eede, C., Salmon, S. A., and Mevius, D. J. (2006). Quantitative susceptibility of Streptococcus suis strains isolated from diseased pigs in seven European countries to antimicrobial agents licensed in veterinary medicine. Vet. Microbiol. 113, 73–82. doi: 10.1016/j.vetmic.2005.10.035
Xu, Z., Xie, J., Peters, B. M., Li, B., Li, L., Yu, G., et al. (2017). Longitudinal surveillance on antibiogram of important gram-positive pathogens in southern China, 2001 to 2015. Microb. Pathog. 103, 80–86. doi: 10.1016/j.micpath.2016.11.013
Yang, Y., Kuang, X., Han, R. J., Zhai, Y. J., He, D. D., Zhao, J. F., et al. (2022). Characterization of a novel linezolid resistance gene optr a and bacitracin resistance locus-carrying multiple antibiotic resistant integrative and conjugative element ICESsu1112S in Streptococccus Suis. Microbiol Spectr 10:e0196321. doi: 10.1128/spectrum.01963-21
Ye, C., Bai, X., Zhang, J., Jing, H., Zheng, H., Du, H., et al. (2008). Spread of Streptococcus suis sequence type 7, China. Emerg. Infect. Dis. 14, 787–791. doi: 10.3201/eid1405.070437
Ye, C., Zhu, X., Jing, H., Du, H., Segura, M., Zheng, H., et al. (2006). Streptococcus suis sequence type 7 outbreak, Sichuan, China. Emerg. Infect. Dis. 12, 1203–1208. doi: 10.3201/eid1208.060232
Zhang, C., Ning, Y., Zhang, Z., Song, L., Qiu, H., and Gao, H. (2008). In vitro antimicrobial susceptibility of Streptococcus suis strains isolated from clinically healthy sows in China. Vet. Microbiol. 131, 386–392. doi: 10.1016/j.vetmic.2008.04.005
Zheng, H., Du, P., Qiu, X., Kerdsin, A., Roy, D., Bai, X., et al. (2018). Genomic comparisons of Streptococcus suis serotype 9 strains recovered from diseased pigs in Spain and Canada. Vet. Res. 49:1. doi: 10.1186/s13567-017-0498-2
Zheng, H., Qiu, X., Roy, D., Segura, M., Du, P., Xu, J., et al. (2017). Genotyping and investigating capsular polysaccharide synthesis gene loci of non-serotypeable Streptococcus suis isolated from diseased pigs in Canada. Vet. Res. 48:10. doi: 10.1186/s13567-017-0417-6
Keywords: Streptococcus suis, antibiotic resistance, ICE, IME, snf2 gene
Citation: Wang J, Qi K, Bai X, Wu Z, Kang W, Liang P, Zheng H and Xu J (2022) Characterization of integrative and conjugative elements carrying antibiotic resistance genes of Streptococcus suis isolated in China. Front. Microbiol. 13:1074844. doi: 10.3389/fmicb.2022.1074844
Edited by:
Anusak Kerdsin, Kasetsart University Chalermphrakiat Sakon Nakhon Province Campus, ThailandReviewed by:
Zhang Wan Jiang, Chinese Academy of Agricultural Sciences (CAAS), ChinaSuganya Yongkiettrakul, National Center for Genetic Engineering and Biotechnology (BIOTEC), Thailand
Copyright © 2022 Wang, Qi, Bai, Wu, Kang, Liang, Zheng and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Han Zheng,  emhlbmdoYW5AaWNkYy5jbg==
emhlbmdoYW5AaWNkYy5jbg==
 Jianping Wang1
Jianping Wang1 Han Zheng
Han Zheng Jianguo Xu
Jianguo Xu