- 1College of Life Science, Baicheng Normal University, Baicheng, China
- 2Key Laboratory of Forage Cultivation, Processing and High Efficient Utilization of Ministry of Agriculture, Key Laboratory of Grassland Resources, Ministry of Education, College of Grassland, Resources and Environment, Inner Mongolia Agricultural University, Hohhot, China
- 3Inner Mongolia Academy of Agricultural Science & Animal Husbandry, Hohhot, China
- 4Japan International Research Center for Agricultural Sciences (JIRCAS), Tsukuba, Japan
Introduction: To prepare high-quality silage, we studied the chemical composition, silage fermentation, characterization, and identification of lactic acid bacteria (LAB) associated with the silage fermentation of native grass on the Inner Mongolian Plateau.
Methods: LAB were isolated from fresh native grass and their silage, and silages were prepared using a small-scale fermentation system with 2–3 cm length in plastic bags.
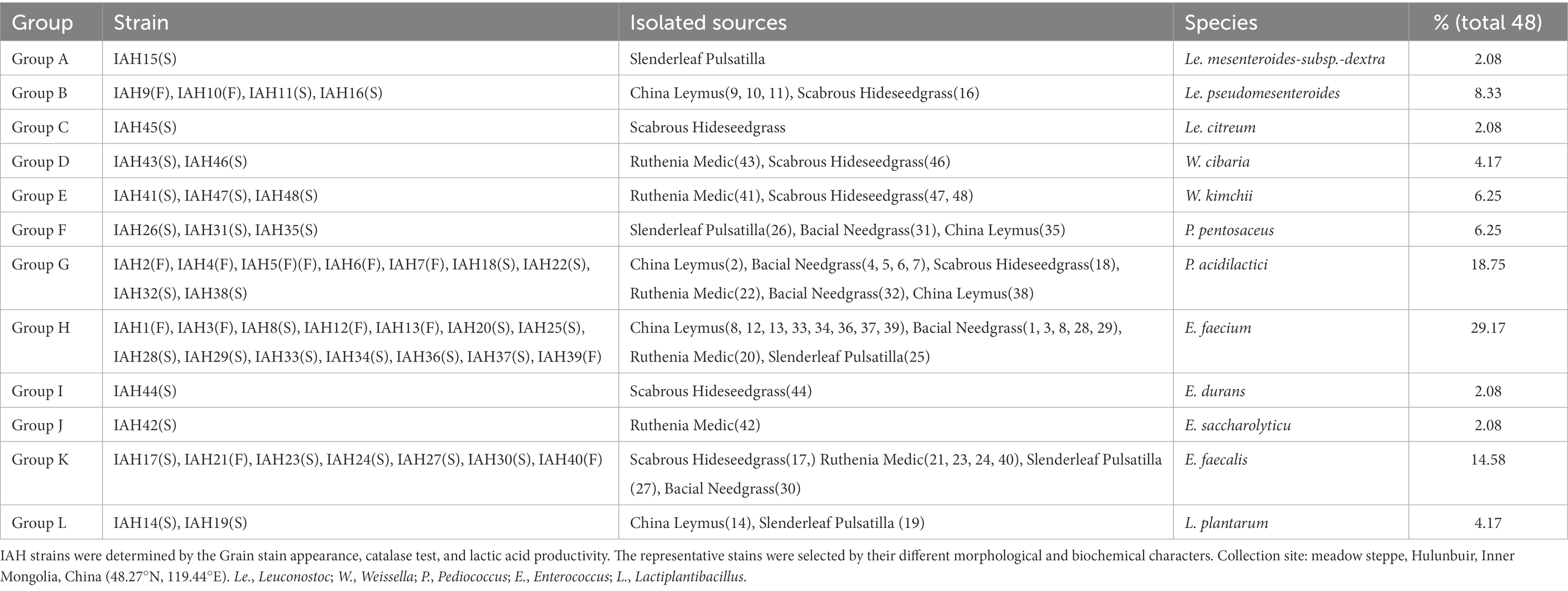
Results: The dominant species of native grasses used were Stipa baicalensis, Leymus chinensis, Cleistogenes squarrosa, Melissilus ruthenicus and Pulsatilla turczaninovii, which contained 47.83–59.43 % moisture, 55.12–67.74 % neutral detergent fiber (NDF), and 8.72–14.55 crude protein (CP), and these nutrients did not change greatly during ensiling. Good preservation with a relatively low pH (below 4.44) and high (p < 0.05) lactic acid content (>0.58) was obtained after ensiling. Based on the morphological and biochemical characteristics, these isolates were divided into 12 groups (A-L). All isolate strains were gram-positive and catalase-negative bacteria that produce lactic acid from glucose. Group A-K were cocci, while group L was rod-shaped. Group A-E formed D-lactic acid, but group H-K formed L-lactic acid, and other groups formed DL-lactic acid. Group A-E were heterofermentative, and Group F-L were homofermentative types of LAB. According to the 16S rRNA gene sequences analysis, strains were identified as genus Leuconostoc (A, B, and C), Weissellla (D, E), Pediococcus (F, G), Enterococcus (H, I, J and K), and Lactiplantibacillus (L). Enterococcus (E.) faecium (29.17%, percentage of total isolates) and Pediococcus (P.) acidilactici (18.75%) were the most frequently occurring dominant species.
Discussion: This study suggests that the native grasses contained abundant LAB species, and they can be used as good-quality silages in animal husbandry. In addition, the strains P. acidilactici and E. faecium were the most frequently isolated from native grass silages as dominant species which can be a potentially excellent inoculant for native grass silage.
1. Introduction
Alongside hay, native grass is regarded as an important resource for animal feed in the Mongolian Plateau, and a large quantity is produced annually in pastoral areas (Hou et al., 2017; Ge et al., 2018; You et al., 2021b). Grazing in native grasslands during summer/autumn and feeding hay during winter/early spring are traditional methods of animal rearing in this region, and these native grasses support the local livestock by providing roughage supplement throughout the year (Bu et al., 2021; Du et al., 2022). However, the harvest is always carried out in early autumn, and since weather is unpredictable, silage production has become a better way to preserve native grass than hay-making, which also allows for long-term storage of fresh grass, making it available throughout the year (Bu et al., 2021; Li Y. et al., 2022). Microorganisms including Lactobacilli, Bacilli, Clostridia, acetic acid bacteria, molds, and yeasts in preservation forages influence silage quality during ensiling. Studies have been conducted on microbe population and silage fermentation for ensiled corn, king grass, Italian ryegrass, and alfalfa (Pang et al., 2011; Shah et al., 2017; Yan et al., 2019; Wang et al., 2022). These showed that selected isolates from natural lactic acid bacteria (LAB) have the potential to improve silage quality and fermentation while inhibiting the growth of undesirable microbes under anaerobic conditions. Silage fermentation and their associated microbe communities have been studied using various microbiological techniques, such as 16S rRNA gene, DNA-DNA hybridization, and third-generation Pacific Biosciences (PacBio) single molecule real-time sequencing technology (SMRT), which generates long reads (Ennahar et al., 2003; Lee et al., 2018; Du et al., 2021). The phylogenetic relationships of LAB have been explored extensively in 16S rRNA sequence ribotyping and experiments, resulting in some new species, such as Lactiplantibacillus nasuensis sp. nov., Lactococcus uvarum sp. nov., Lactiplantibacillus terrae sp. nov., Lactiplantibacillus xiangfangensis sp. nov. isolated from silage, kimchi, milk, oil, and pickle being included in the microbe community.
High-quality silage from native grasses is generally difficult due to low water-soluble carbohydrate (WSC) content and higher buffering capacity (Hou et al., 2017). Certain natural LABs from native grasses play a vital role in fermentation (Ge et al., 2018). Further, the Mongolian Plateau might have special microorganisms regardless of water availability due to its frigid weather and dry climate (Zhang et al., 2015). Thus, collecting, screening, and identifying the LAB biochemically from native grass is important. In addition, some of the selected LAB might be potential well-sourced silage additions for forages.
2. Materials and methods
2.1. Silage preparation
Samples of native grass [Baical needlegrass (Stipa baicalensis), China leymus (Leymus chinensis), Scabrous hide seed grass (Cleistogenes squarrosa), Ruthenia medic (Melissilus ruthenicus), and Slenderleaf pulsatilla (Pulsatilla turczaninovii)] were collected from the meadow steppe of Hulunbuir (48.27°N, 119.44°E; Inner Mongolia, China) on 25 July 2019 (full-bloom stage). Before ensiling, the moisture of native grasses ranged from 48.34 to 59.43% FM. The grasses were cut into 3–5 cm lengths by using a chopper machine (DD-ZCD, Dedong Co., Ltd., Weifang, China), and the grass silages were prepared in the form of mini silos (polyethylene, 1L, Yijia, Beijing, China) without witling (density, 450–500 kg/m3). The silos were kept at ambient temperature (20–26°C). Five silos were used per treatment. After 30 days, the microbial community, chemical composition, and fermentation quality were analyzed.
2.2. Microbial population, chemical composition, and silage fermentation
Referring to Cai (1999), 10 g of each sample was blended with 90 ml saline solution (0.85% NaCl) and serially diluted from 10−1 to 10−6. The microbial community of the native grass was analyzed via plate cultivation. The numbers of LAB were measured using de Man, Rogosa, Sharp (MRS) agar after serial dilutions and incubated at 37°C for 48 h in an anaerobic box (Oxoid AnaeroJar, AG0025A, Thermo Fisher Scientific Inc., Waltham, MA, United States). Aerobic bacteria were counted on Blue Light Broth agar (Nissui-Seiyaku Ltd., Tokyo, Japan). Yeast and mold were counted on Potato Dextrose Agar (Oxoid, CM0139B, Thermo Fisher Scientific Inc., Waltham, MA, United States) and incubated at 37°C for 48 h. Yeasts were distinguished from mold and bacteria by colony appearance and observation of cell morphology. The coliform bacteria were counted on Blue Light Broth Agar (Nissui, Tokyo, Japan) at 37°C for 48 h. All microbial data were counted as viable numbers in colony-forming units (CFU) per gram on a fresh matter (FM) basis. For LAB isolation, 20 colonies of each sample were picked, 48 strains were collected and considered as LAB. The individual colonies were isolated and purified twice by steaking on MRS broth and Agar (Oxoid, CM1175/CM0003B, Thermo Fisher Scientific Inc., Waltham, MA, United States). Pure cultures were grown on MRS agar at 37°C for 24 h, resuspended in a solution of nutrient broth (Oxoid) containing 10% dimethyl sulfoxide, and stored at-80°C for further examination.
The dry matter (DM), organic matter (OM), crude protein (CP), and ether extract (EE) contents were measured according to the Association of Official Analytical Chemists (AOAC) methods (2005). The neutral (NDF) and acid (ADF) detergent fibers were obtained using the ANKOM A200i fiber analyzer (ANKOM Technology, Macedon, NY, United States) by Van Soest et al. (1991). The water-soluble carbohydrate (WSC) was determined by DNS colorimetric method (Sucrose/D-Glucose/D-Fructose UV-method, Roche Diagnostics, Tokyo, Japan) by Thomas (1997).
For silage fermentation analysis, another set of wet silage (10 g) samples was mixed with 90 ml of deionized water. The extracts were filtered through four layers of cheesecloth and filter paper (pore size 9 cm; Beimu, Hangzhou, China) and homogenized for 3 min. The pH was measured using a glass electrode pH meter (PHS-3E; LEICI, Shanghai, China). The NH3-N content was determined by the method of Broderick and Kang (1980) using steam distillation of the filtrates. The organic acid content was analyzed using the high-performance liquid chromatography (HPLC; GC-8A, Shimadzu, Kyoto, Japan) method (column: KC811, Shimadzu, Kyoto, Japan; detector: SPDM10AVP, Shimadzu, Kyoto, Japan; eluent: 3 mmol L−1 HCLO4, 1 ml min−1; temperature: 50°C) by Ennahar and Cai (2004).
2.3. Morphological, physiological, and biochemical tests
The Gram staining and observation of morphological characteristics of LAB were determined by MRS agar incubated at 37°C for 48 h (Zanoni et al., 1987). Catalase activity and glucose gas production were determined by the methods of Cai et al. (1999). Growth at different temperatures (5°C, 10°C, 45°C, and 50°C), different pH (2.5, 3.0, 3.5, 4.0, 7.0, and 9.0), and different salt solutions (3.0, 6.5% (w/v) NaCl) were determined on MRS agar at 30°C for 120 h. Carbohydrate fermentation of 49 different compounds was tested by Analytical Professorile Index (API 50 CH) strips (bioMerieux, Craponne, France) at 30°C for 24 h and verified for 48 h (Nomura et al., 1999).
2.4. Extraction of LAB genomic DNA and 16S rRNA gene sequence
Each strain was cultivated overnight in 5 ml MRS medium at 30°C, centrifuged at 9391 g for 5 min, washed twice with buffer (10 mmol L−1 Tris-HCl, 0.1 mmol L−1 EDTA, pH 8.0) in a clean 1.5 ml microcentrifuge tube, and recentrifuged. Genomic DNA was extracted through Plant Genomic DNA Kit (TIANGEN Biotech Co., Ltd., Beijing, China) according to the manufacturer’s instructions. The genomic DNA concentration of each strain was determined at 260 nm using a UV-VIS spectrophotometer (Shanghai Metash Instruments Co., Ltd., Shanghai, China; Zhang et al., 2016b). The extracted genomic DNA of each strain was stored at −20°C for future use.
For DNA extraction and purification, cells were grown at 37°C for 8 h (Akane et al., 1994), and the 16S ribosomal RNA (rRNA) gene sequences were amplified in a PCR Thermal Cycler (A48141, Thermo Fisher Scientific Inc., Waltham, MA, United States) by using the PCR and reagents from a Takara Taq PCR Kit (TIANGEN Biotech Co., Beijing, China). The sequences were determined directly with a sequencing kit (NOVA-5144, Bio Scientific Co., Ltd., Austin, United States) using the prokaryotic 16S rRNA universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) in combination with Applied Biosystems model 3500 (Applied Biosystems, Thermo Fisher Scientific Inc., Waltham, MA, United States) automated sequencing system (Cai, 1999). Sequence similarity searches were performed at the GenBank data library using the BLAST (Basic Local Alignment Search Tool) program.
2.5. Statistical analyses
The 16S rRNA sequences were constructed by the neighbor-joining method using Molecular Evolutionary Genetic Analysis (MEGA) 6.0 software (Saitou and Nei, 1987). Data on chemical composition and fermentation quality after 30 days were analyzed by one-way analysis of variance using the general linear model procedure of SAS version 9.3 (SAS Institute Inc., 2015). The differences between means were assessed using Turkey’s multiple comparison tests and were considered statistically significant at p < 0.05.
3. Results
3.1. Microbial parameters of native grass silages
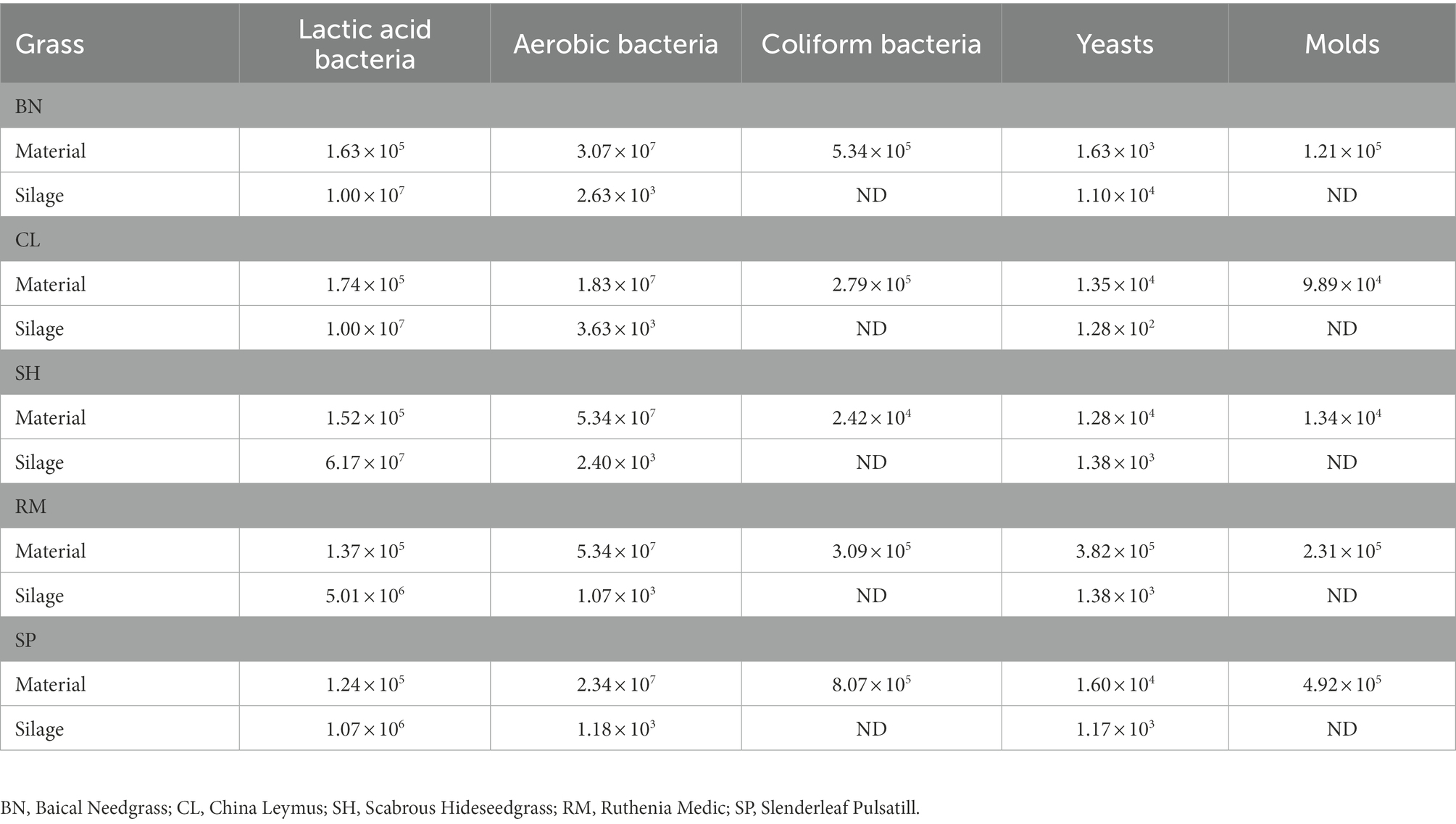
The counts of microorganisms of native grasses are shown in Table 1. Overall, 105 (cfu/g of FM) LAB, 107 (cfu/g of FM) aerobic bacteria, 104–105 (cfu/g of FM) coliform bacteria, 103–105 (cfu/g of FM) yeast, and 104–105 (cfu/g of FM) molds were found, and there was no big difference among the counts of bacteria from all silage samples. After 30 days of ensiling, 106–107 (cfu/g of FM) LAB, 103 (cfu/g of FM) aerobic bacteria, 102–104 (cfu/g of FM) yeasts were found in the native grasses. No coliform bacteria or mold was detected in any silage.
3.2. Chemical composition and fermentation quality of native grass silages
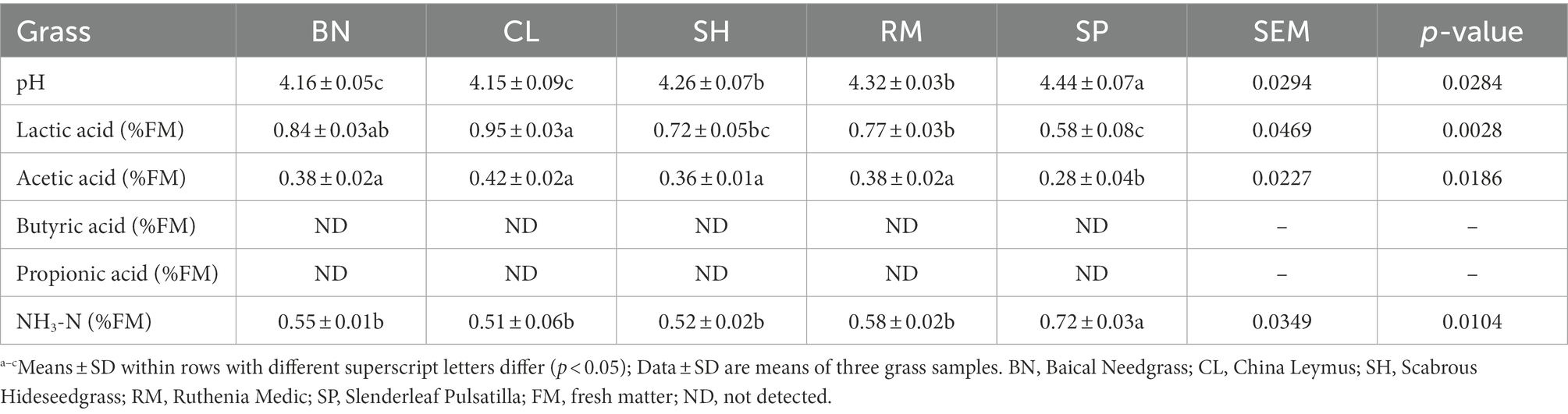
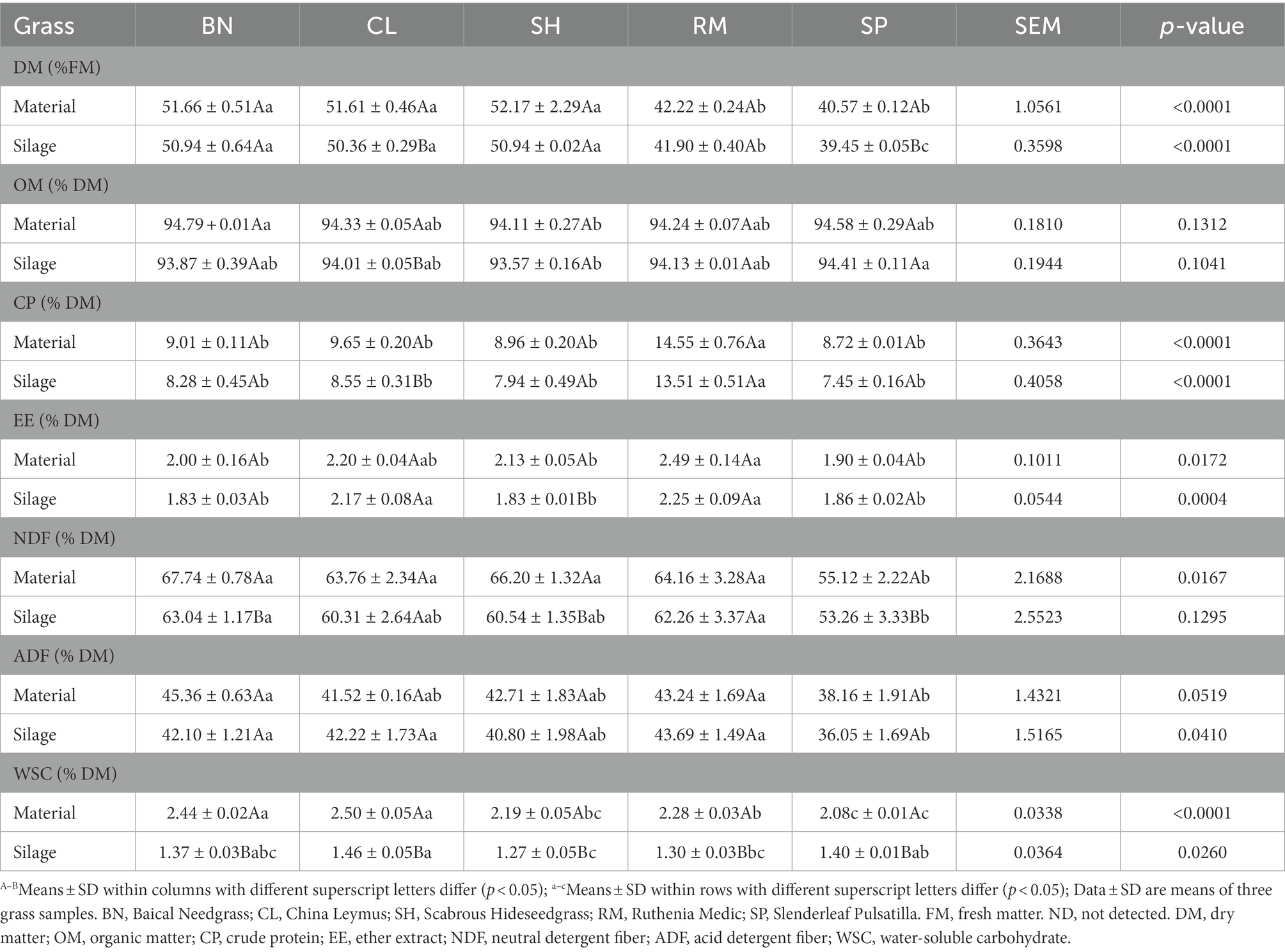
The chemical composition and fermentation quality of native grass silages are shown in Tables 2, 3. All silages had low moisture with DM contents of native grasses ranging from 40.57 to 52.17% on an FM basis. The DM contents of CL (50.36% DM) and SP (39.45% DM) silages were lower than those of other treatments (p < 0.05) after ensiling. The CP content of RM (fresh 13.51% DM, silage 14.55% DM) was higher than other treatments (p < 0.05). The NDF and ADF contents of the native grasses after ensiling were similar, and SP had the lowest NDF (53.26% DM) and ADF (36.05% DM) among the samples. Total WSC content of CL (1.46% DM), SP (1.40% DM) and BN (1.37% DM) silages were higher than RM (1.30% DM) and SH (1.27% DM; p < 0.05).
Native grasses significantly affected the pH, lactic acid, and NH3-N content (p < 0.05). The lowest pH (4.15), NH3-N (0.51% FM), and the highest lactic acid (0.95% FM) were observed in CL silage. No butyric acid or propionic acid was detected in any silage, indicating a good fermentation quality. Correlation coefficients (r) for the relationship between native grass chemical composition and silage fermentation are shown in Table 4. For pH, a significant negative relationship was observed with WSC content. The lactic acid content was significantly positive with WSC content. The NH3-N content was significantly negative with WSC and DM content (p < 0.05).
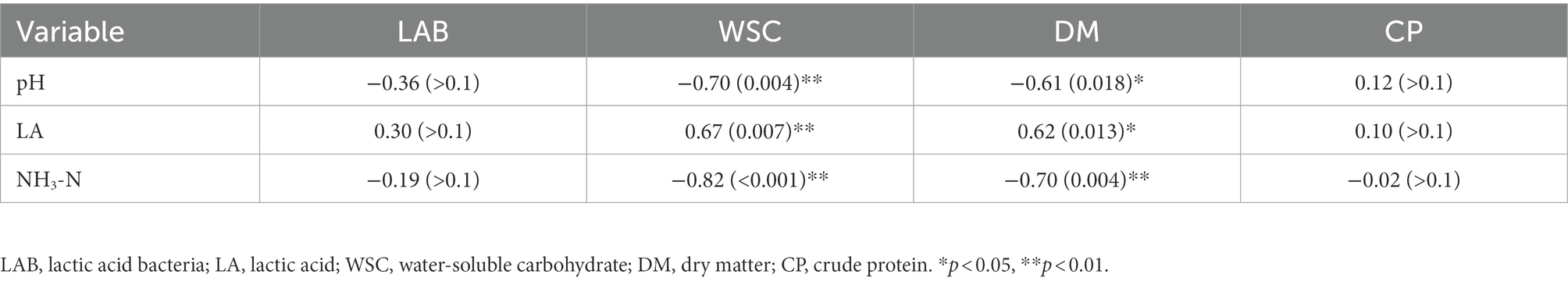
Table 4. Correlation coefficients (r) for relationships between chemical composition and fermentation silages after 60 days of ensiling.
3.3. Morphological, physiological, and biochemical
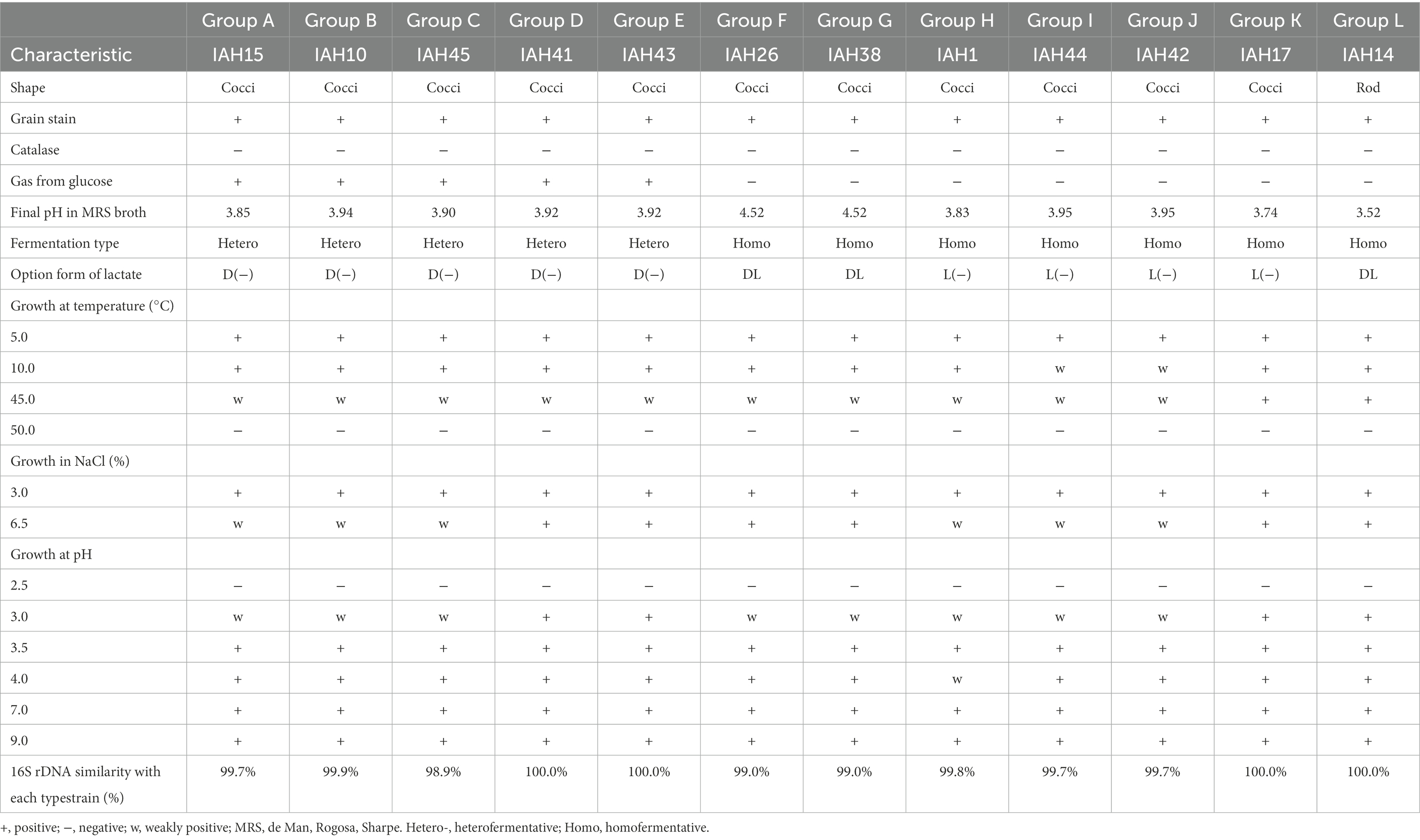
The morphological, physiological, and biochemical are shown in Table 5. A total of 54 isolates were collected, of which 48 were considered as LAB by Gram staining and catalase activity. According to the results of physiological and biochemical tests, these isolates were divided into 12 (A-L) groups, and 12 representative strains were used for phylogenetic analysis. Strains in groups A-K were cocci, while group L had rod-shaped bacteria. All isolates were gram-positive and catalase-negative bacteria, growing at temperatures of 5.0 and 10.0, at 3.0 and 6.5% (w/v) NaCl, and at pH 3.5 to 9.0, the 16S rRNA similarity of all isolates was higher than 98%.
The carbohydrate fermentation patterns of LAB strains are shown in Table 6. All strains did not produce acid from Glycerol, Erythritol, D-Arabinose, Inositol, Fucose, and L-Arabitol. Group A-E were heterofermentative cocci that formed D (−) lactic acid, producing acid from D-Glucose, D-Fructose, and D-Mannose. Group F and G were homofermentative cocci that formed DL-lactic acid from D-Xylose, Galactose, D-Glucose, D-Fructose, D-Mannose, and Cellobiose. Group H-K were homofermentative cocci that formed L-lactic acid from L-Arabinose, Ribose, D-Xylose, D-Glucose, Esculin, and Cellobiose. Group L were homofermentative rods that formed DL lactic acid from Ribose, D-Xylose, D-Glucose, and Cellobiose.

Table 6. API 50 CH (bioMerieux, Tokyo, Japan) characteristics of representative strains isolated from natural grasses.
3.4. Phylogenetic trees of 16S rRNA gene sequence
The phylogenetic trees of 16S rRNA gene sequences are shown in Figures 1–3. Strains in Groups A-C were placed in the cluster of genera Leuconostoc (Le.), since they were grouped on the tree together with Le. mesenteroides-subsp.-dextra, Le. Pseudomesenteroides, and Le. citreum. Sktrains in Groups D-G were identified as Weissellla and Pediococcus. Strains in Group D and E were newly described as the newly species Weissellla cibaria and Weissellla kimchii with similarity exceeding 99.0%. Strains in Group F and G were assigned to Pediococcus pentosaceus and Pediococcus acidilactici. Strains in H to K were clearly identified as Enterococcus faecium, Enterococcus durans, Enterococcus saccharolyticus, and Enterococcus faecalis, Strains in Group L were placed in the cluster of genus Lactiplantibacillus on the phylogenetic tree and clearly identified as Lactiplantibacillus plantarum by their sequence similarity at 100%, which was only presented in BN and CL silages.
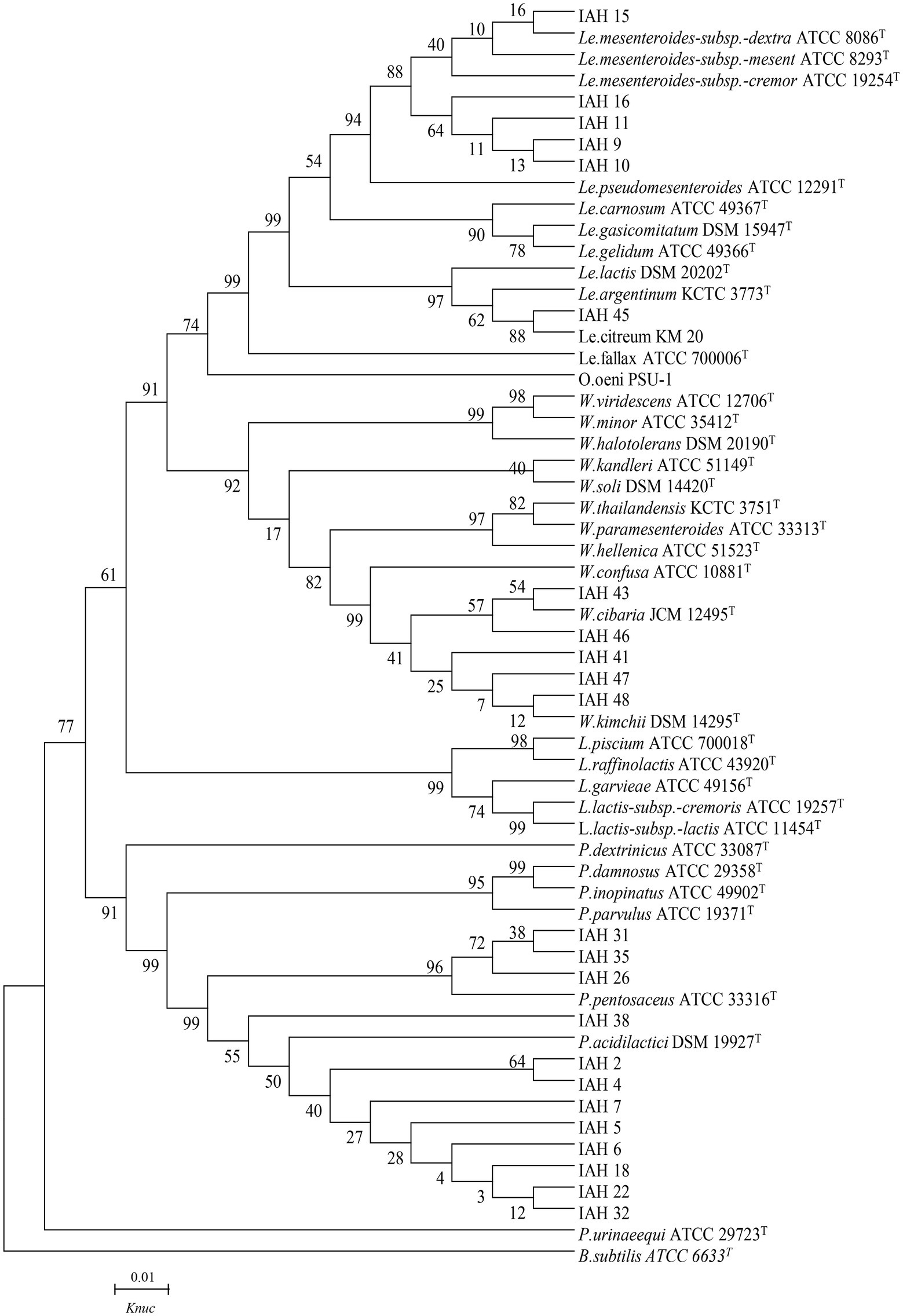
Figure 1. Phylogenetic tree showing the relative position of Leuconostoc, Weissella, Lactococcus, and Pediococcus species and representative strains isolated from natural grasses as inferred by the neighbor-joining method of complete 16S rDNA sequences with 1,000 replicates. Reference sequences of type strains from GenBank are used for comparison, Bacillus subtilis is used as the outgroup. The width is proportional to terminal branch lengths (1% sequence divergence, shown as a scale bar at lower right). Le., Leuconostoc; W., Weissella; L., Lactococcus; P., Pediococcus. Knuc, nucleotide substitution rates.
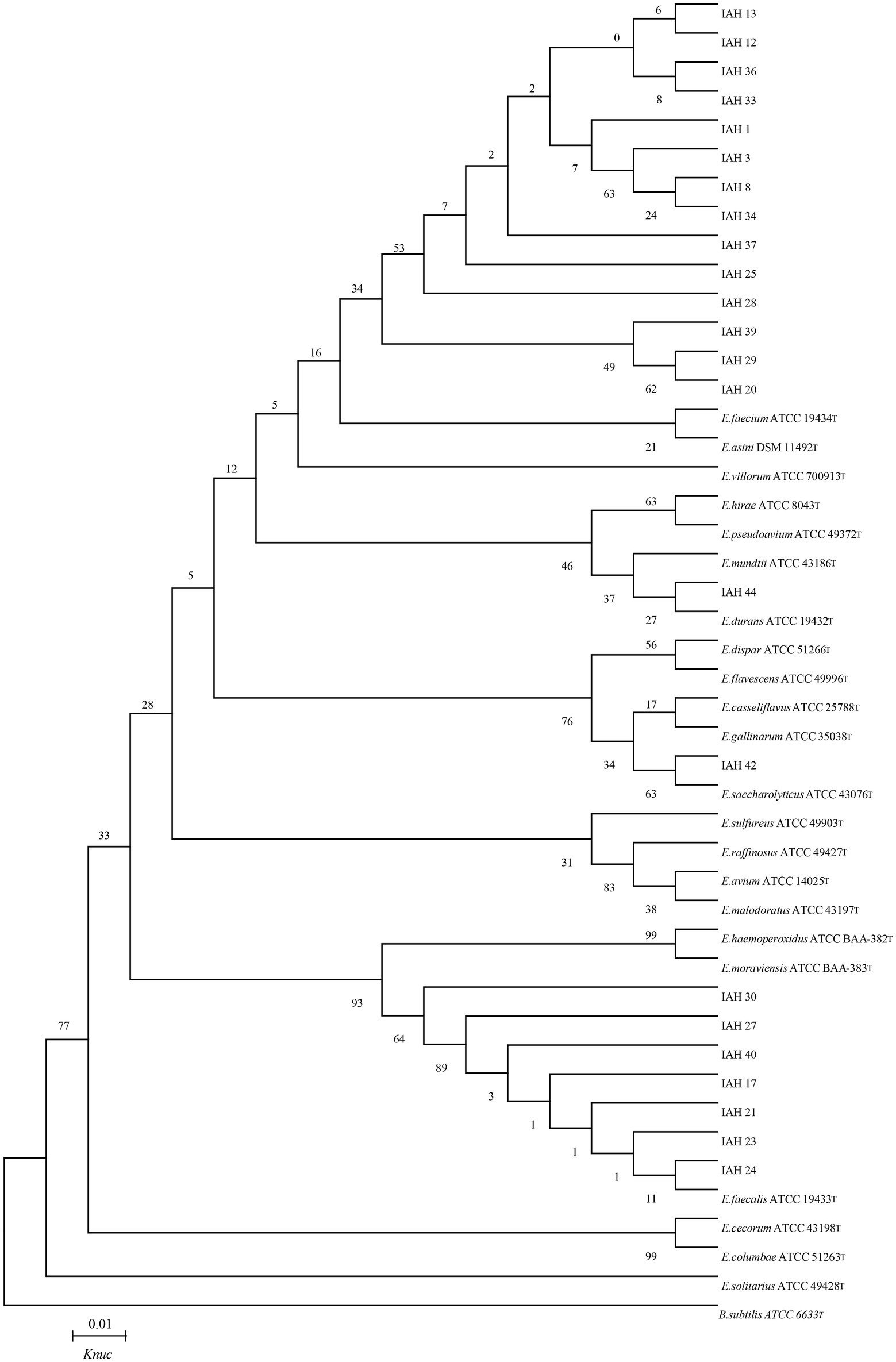
Figure 2. Phylogenetic tree showing the relative position of Enterococcus species and representative strains isolated from natural grasses as inferred by the neighbor-joining method of complete 16S rDNA sequences with 1,000 replicates. Reference sequences of type strains from GenBank are used for comparison, Bacillus subtilis is used as the outgroup. The width is proportional to terminal branch lengths (1% sequence divergence, shown as a scale bar at lower right). E., Enterococcus. Knuc, nucleotide substitution rates.
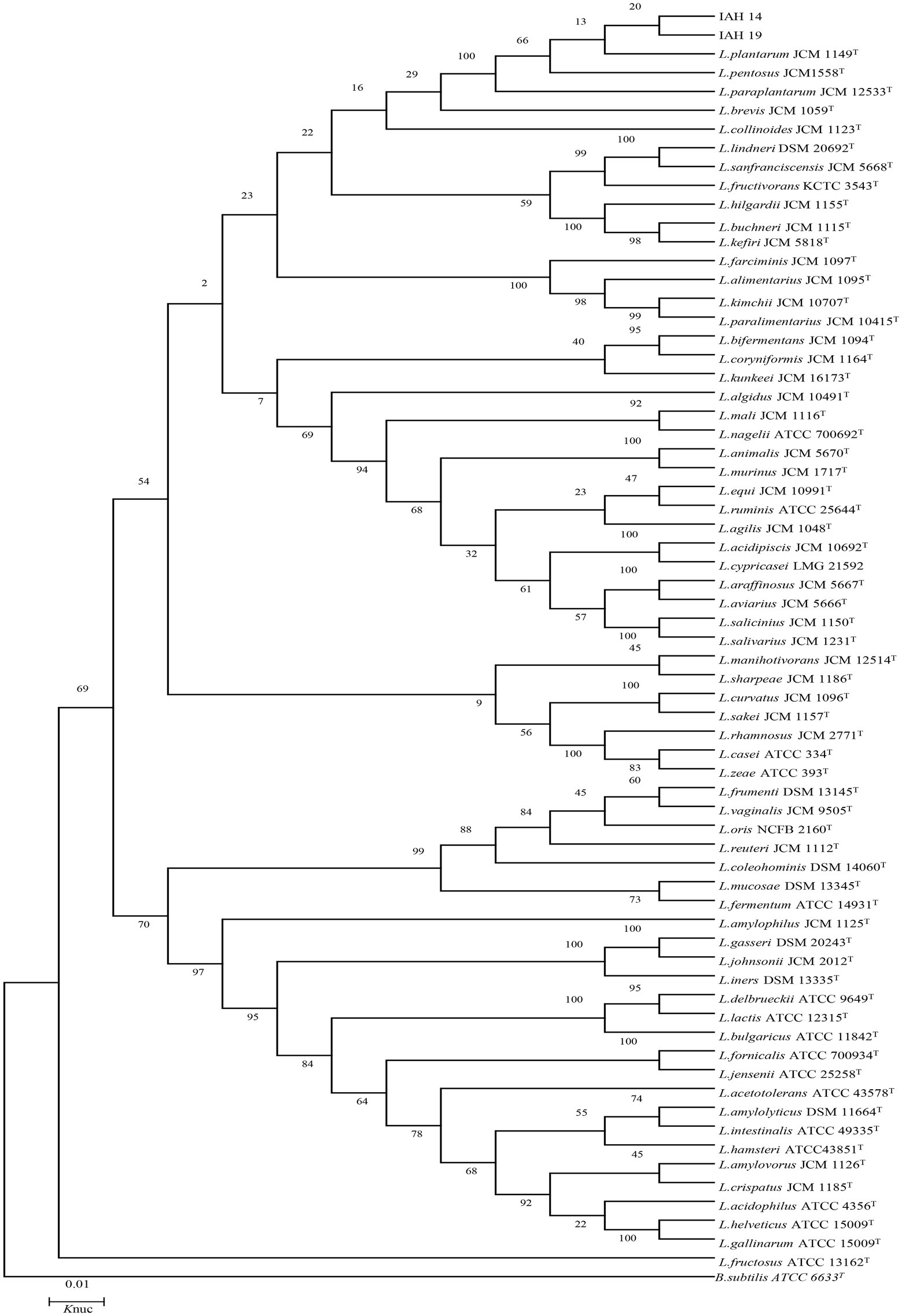
Figure 3. Phylogenetic tree showing the relative position of Lactiplantibacillus species and representative strains isolated from natural grasses as inferred by the neighbor-joining method of complete 16S rDNA sequences with 1,000 replicates. Reference sequences of type strains from GenBank are used for comparison, Bacillus subtilis is used as the outgroup. The width is proportional to terminal branch lengths (1% sequence divergence, shown as a scale bar at lower right). L., Lactiplantibacillus. Knuc, nucleotide substitution rates.
3.5. Microbial community of native grass silages
The LAB community of native grass and their silages are shown in Table 7 and Figure 4. The microbial diversity of LAB consisted of 12 species: Le. mesenteroides-subsp.-dextra (2.08%), Le. pseudomesenteroides (8.33%), Le. citreum (2.08%), W. cibaria (4.17%), W. kimchii (6.25%), P. pentosaceus (6.25%), P. acidilactici (18.75%), E. faecium (29.17%), E. durans (2.08%), E. saccharolyticu (2.08%), E. faecalis (14.58%), and L. plantarum (4.17%). Furthermore, most species were homofermentative and accounted for 76.92% of total fermentation. Pediococcus acidilactici, E. faecium, and E. faecalis were the predominant species in native grass. After 30 days of fermentation, the dominant LAB species were E. faecium (37.50 to 44.44%) in BN and CL silages, E. faecalis (28.57%) in RM silage, and W. kimchii (28.57%) in SH silage. The dominance of particular microbes was not detected in other silages.
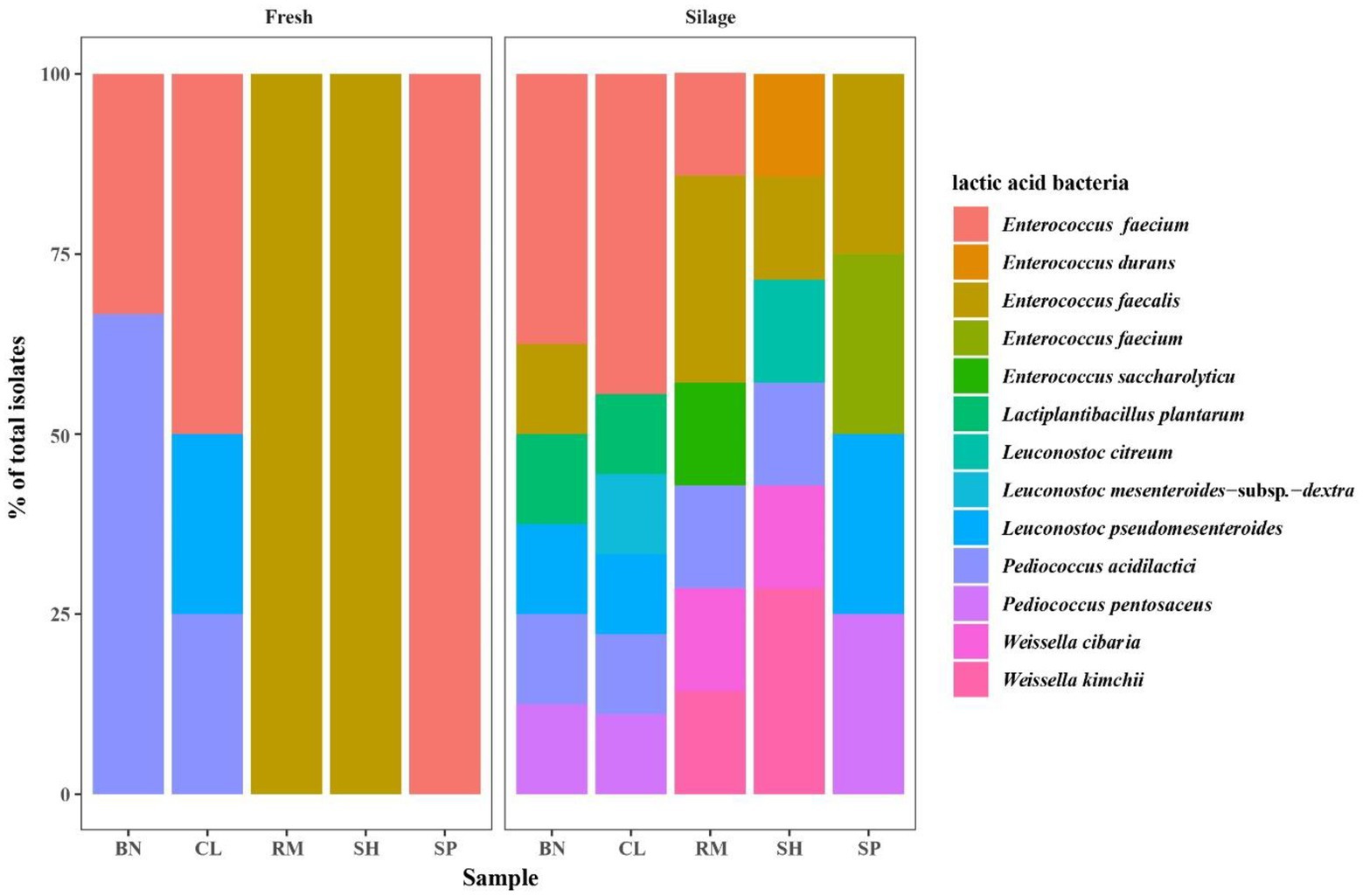
Figure 4. Community of lactic acid bacteria of native grass. BN, Baical Needgrass; CL, China Leymus; SH, Scabrous Hideseedgrass; RM, Ruthenia Medic; SP, Slenderleaf Pulsatilla.
A spearman correlation heatmap of the lactic acid bacteria and fermentation products at the species level was created to better understand the relationship between fermentation and the microbial community (Figure 4). pH and NH3-N were negatively correlated with L. plantarum, E. faecium, and Le. mesenteroides-subsp.-dextra., while lactic acid showed the opposite patterns. During ensiling, E. faecium used the WSC as a substrate to produce lactic acid and preserve forages, while WSC was significantly positively correlated with E. faecium.
4. Discussion
The importance of native grass as a feed resource in animal husbandry across China has been increasing. Since the weather has become unpredictable, silage production has become a better method of preservation than hay-making in the Inner Mongolian Plateau for ensuring high quality and long-term storage of these resources (Bu et al., 2021; Li Y. et al., 2022). Therefore, studies on chemical composition, fermentation quality, microbial community, characterization, and identification of lactic acid bacteria related to silage fermentation of native grasses in the Mongolian Plateau are necessary to help create good quality silage and for long-term storage.
DM content is one of the key factors in achieving fermentation quality. In this study, the DM contents of all silages were lower than the fresh native grasses, which is consistent with other studies (Fu et al., 2022). The hydrolysis of organic acid during ensiling also reduced the DM content (Zhao et al., 2018). In this study, during the ensiling, WSC was transformed into organic acid, ethanol, and carbon dioxide by microorganisms, resulting in high DM content that inhibited the growth of Clostridium spp., which often produces butyric acid and ammonia nitrogen in silage material (McEniry et al., 2011; Muck et al., 2018; Wang et al., 2018). Overall, the lower CP and WSC and higher NDF and ADF agreed with our previous study in meadow steppes (Hou et al., 2017; Du et al., 2020; You et al., 2021a).
The epiphytic LAB used WSC to produce sufficient lactic acid to reduce pH and inhibit the growth of harmful bacteria (He et al., 2021). The LAB of all native grasses presented more than 105 cfu/g of FM, conforming with our previous study on native grasses (Hou et al., 2017; Ge et al., 2018; You et al., 2021a). The lower LAB in silages may have caused the acid-tolerant aerobic bacteria, assisted in some cases by yeasts, to use some lactic acid and nutrients, which is in agreement with the results of Ge et al. (2018). Coliform bacteria were not detected in all silages because acidification might have inhibited the growth of undesirable bacteria. Zhang et al. (2015, 2016a) reported that the activity of LAB caused rapid acidification by the increased lactic acid content and decreasing pH value, which might explain the suppression of the activity of some microorganisms like coliform bacteria on the L. chinensis silage.
Ensiling is a process of complex microbial fermentation, resulting in organic acid accumulation. In this study, the native grass silages were well preserved with lactic acid produced, exceeding 0.58% FM (Ding et al., 2020). pH below 4.20 could result in good fermentation quality and inhibit harmful bacteria (Wang, Y. et al., 2019). In our study, the pH in native grass after 30 days of fermentation was 4.15–4.44, similar to the results by You et al. (2021a). After 30 days of ensiling, the CP content of native grass was decreased, which might have been caused by the degradation of proteins by microorganisms (Du et al., 2022). The change of CP and NH3-N contents in SP was the highest out of all silages, showing that there was some undesirable microorganism and accumulation of NH3-N during the ensiling of SP, which is consistent with the findings of Ni et al. (2017) that the harmful bacteria Sphingomonas elevated NH3-N and protein breakdown. Liu et al. (2022) reported that Fungi such as Aspergillus and Monascus had significant positive correlations with CP and resulted in the decrease of nutrients in meadow steppe.
Fresh native grass had lactic acid bacteria, aerobic bacteria, coliform bacteria, yeasts, and molds, whereas silages contained no coliform bacteria or molds, and lactic acid bacteria was the most abundant microorganism. According to certain studies, prevalent LAB were the main microbial community in forage and grass silages [corn, paddy rice, oat, and Italian grass (Ennahar et al., 2003; Cai et al., 2020; Fu et al., 2022; Li H. et al., 2022)]. Cai (1999) found that epiphytic microbial flora isolated from forages and grasses were species of Lactiplantibacillus and Streptococci, Enterococci, Lactococci, Leuconostocs, Pediococci, and Weissella with the predominant LAB being homofermentative L (+) lactic acid-producing cocci. In the present study, a total of 54 isolates were screened and purified. According to the gram-stain and catalase tests, 48 isolates were considered as LAB. All strains were characterized by sugar fermentative assays and were divided into 12 groups, representing five genera. It is difficult to identify the isolates by adopting the phenotypic technique for classifying species (Ni et al., 2015). Hence, based on the 16S rRNA gene sequence, strains were identified as Lactiplantibacillus (4.17%), Weissella (10.42%), Leuconostoc (12.5%), Pediococcus (25%), and Enterococcus (47.92%).
Generally, the homofermentative LAB L. plantarum, Enterococcus faecium, and Pediococcus acidilactici are used as commercial inoculants (Ennahar et al., 2003). The epiphytic LAB cocci grew vigorously in the early stages of ensiling and created an aerobic environment, but lactobacilli played a more important role in long-term fermentation (Cai et al., 1998). It was previously reported that Lactiplantibacillus promoted silage quality, with L. plantarum being the most prevalent species (Kung et al., 2003; Chen et al., 2014; Da Silva et al., 2014; Xu et al., 2021). However, Lactiplantibacillus only accounted for 4.17% of the total LAB microflora in the native grass silages of our study. Zhang and Yu (2017), reported that the LAB species of L. chinensis also included Lactiplantibacillus brevis and L. casei, but we did not observe them. Lactiplantibacillus plantarum (IAH14) was only found in Stipa baicalensis and L. chinensis with lower pH and higher lactic acid than in other native grass silages. This could be because the L. chinensis in our study was native, but that in Zhang’s study was cultivated (Zhang et al., 2016a).
Group L was identified as L. plantarum or L. pentoses. Hammes et al. (2005) reported that these two species are genotypically closely related and show highly similar phenotypes in the same 16S rRNA phylogenetic group with only a 2 bp difference. Nevertheless, the only way to distinguish this was through phylogenetic analysis of sequences of the 16S-23S rRNA intergenic spacer region (ISR) or recA gene sequence comparison (Tannock et al., 1999; Torriani et al., 2001). In this study, all strains were tested for their sugar fermentation profiles and were unambiguously identified as L. plantarum by carbohydrate utilization pattern, as it produces acid from melezitose, D-raffinose, and α-methyl-D-mannoside (Spano et al., 2002; Cao et al., 2010), which was different from L. pentoses. L. pentoses can be distinguished from L. plantarum by growing D-xylose and glycerol (Zanoni et al., 1987). Hence, Group I (IAH14 and IAH19) were similar to L. plantarum. As expected, Group I could regularly grow at pH 3.5 and weakly at pH 3.0, which showed tolerance to acidic environments. Enterococcus faecium and Pediococcus acidilactici were dominant species in native grass silages, which is supported by a study where the Enterococcus and Pediococcus were the most frequently found on the surface of forage crops and food products (Cai, 1999; Fugaban et al., 2021). In fact, numerous studies found that Enterococcus and Pediococcus are active when pH > 5.0 and dominant in the early stages of ensiling, producing greater lactate concentration for further fermentation. In our study, the Pediococcus (Group F and G) and Enterococcus (Group H-K) could not grow in a low pH (<4.0) condition, which is in agreement with our previous study (Cai, 1999). The final pH of all silages was >4.0 after 30 days of ensiling, thus, some butyric acid fermentation might have occurred, but did not promote silage fermentation.
In Figure 5, we found L. plantarum was positively correlated with LA and negatively correlated with pH value, which is in accordance with Fang et al. (2022). The LA and the AA contents were negatively correlated with E. faecalis, in agreement with a previous study that illustrated the same (Fang et al., 2022). In addition, it was reported that W. paramesenteroides was the most abundant species of Weissela in forage silage (Libonatti et al., 2019; Wang, S. et al., 2019). However, Chen et al. (2010) found that W. cibara was the most abundant species in mulberry farms. This study did not find any W. paramesenteroides in native grass. Ennahar and Cai (2004) reported that W. cibaria and W. kimchii displayed very similar sugar-fermentation patterns and produced the D-form of lactic acid, which indicated that W. kimchii was a later heterotypic synonym for W. cibaria. Therefore, the strains IAH 43 and IAH 46 from our study, should belong to W. cibaria. In a previous study on fresh fruits and vegetables, W. cibara strain TM128 decreased infection levels of fungi by 50% (Ennahar and Cai, 2004). Similar work done on durum wheat showed that W. cibara strain C21-4 had the strongest antifungal activity, therefore, it is speculated that W. cibara may protect the native grass from fungi. This study is the first report of Weissella strain in native grass from the Mongolian Plateau. Further studies are necessary to test the effect of W. cibara as an additive to maintain native grass silage quality. Additionally, Weissella was another dominant genus found in silages throughout fermentation (Guan et al., 2018).
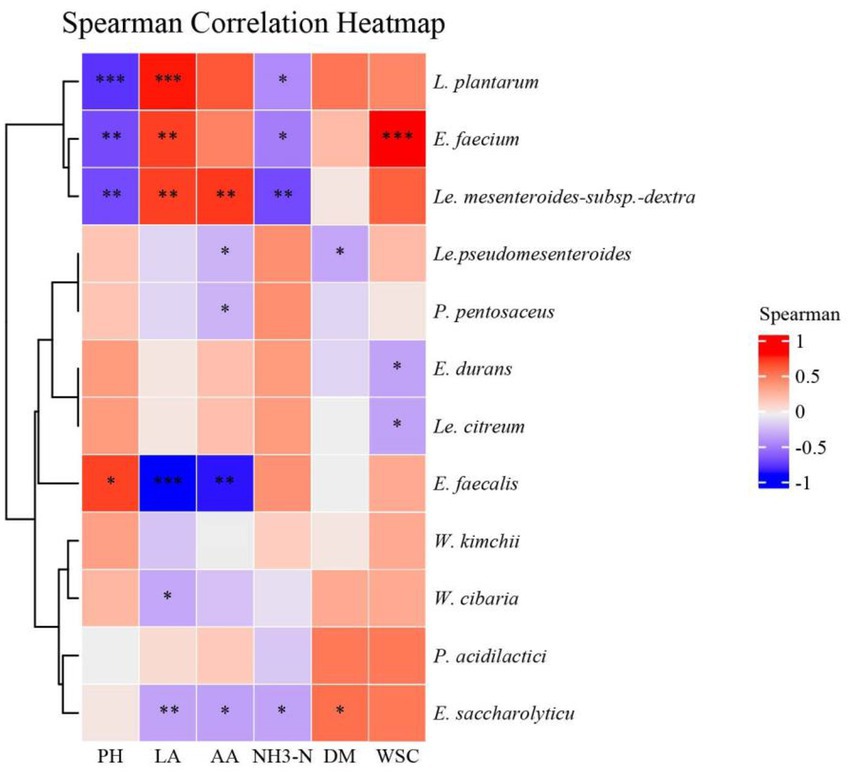
Figure 5. Spearman correlation heatmap of the lactic acid bacteria and fermentation products at species level. NH3-N, ammonia nitrogen; DM, dry matter; WSC, water-soluble carbohydrate; LA, lactic acid; AA, acetic acid. E., Enterococcus; L., Lactiplantibacillus. Le., Leuconostoc; W., Weissella; L., Lactococcus; P., Pediococcus. *p < 0.05, **p < 0.01, ***p < 0.001.
In Inner Mongolia, the studies on native grass silages have been increasing rapidly over the past 5 years (Hou et al., 2017; Ge et al., 2018; Du et al., 2020; You et al., 2021a; Li H. et al., 2022). The present study focused on L. plantarum and the silage fermentation of native grass. Future research must take into account this forage’s digestibility for sheep and cows, and how it affects the quality of mutton produced.
5. Conclusion
This study revealed that native grass has abundant LAB species and can be well preserved through silage. The strains P. acidilactici and E. faecium were the most frequently isolated from native grass silages as dominant species. Further studies about the LAB isolated from native grass and their use as additives in silage are needed for developing the potential inoculants for forge silages. Based on the chemical composition and silage fermentation, the Inner Mongolian meadow steppe produced good silage, which can help alleviate the burden of feed shortage in animal husbandry in China.
Data availability statement
The datasets generated for this study can be found in the NCBI under accession number PRJNA545378.
Author contributions
MH contributed to methodology, visualization, validation, and data curation and wrote the original draft. ZW contributed toward performing the experiments and funding acquisition. LS contributed significantly to the analysis and manuscript preparation. SW performed the data analyses and wrote the manuscript. YJ and YC contributed to conceptualization and discussions. All authors have read and agreed to publish this version of the manuscript.
Funding
This work was supported by the Key Laboratory of Forage Cultivation, the Processing and Highly Efficient Utilization of the Ministry of Agriculture, and the Key Laboratory of Grassland Resources of the Ministry of Education. It was funded by the National Key R&D Program of China “Construction of novel additives for total mixed ration and their application” (Grant No. 2022YFE0111000).
Acknowledgments
We would like to thank Editage (www.editage.cn) for the English language editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Akane, A., Matsubara, K., Nakamura, H., Takahashi, S., and Kimura, K. (1994). Identification of the heme compound copurified with deoxyribonucleic acid (DNA) from bloodstains, a major inhibitor of polymerase chain reaction (PCR) amplification. J. Forensic Sci. 39, 362–372.
Broderick, G. A., and Kang, J. H. (1980). Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media. J. Dairy Sci. 63, 64–75. doi: 10.3168/jds.S0022-0302(80)82888-8
Bu, Z., Ge, G., Jia, Y., and Du, S. (2021). Effect of hay with or without concentrate or pellets on growth performance and meat quality of Ujimqin lambs on the inner Mongolian Plateau. Anim. Sci. J. 92:e13553. doi: 10.1111/asj.13553
Cai, Y. (1999). Identification and characterization of enterococcus species isolated from forage crops and their influence on silage fermentation. J. Dairy Sci. 82, 2466–2471. doi: 10.3168/jds.S0022-0302(99)75498-6
Cai, Y., Benno, Y., Ogama, M., Ohmomo, S., Kumai, S., and Nakase, T. (1998). Influence of lactobacillus spp. from an inoculant and of Weissella and Leuconostoc spp. from forage crops on silage fermentation. Appl. Environ. Microbiol. 64, 2982–2987. doi: 10.1128/AEM.64.8.2982-2987.1998
Cai, Y., Benno, Y., Ogawa, M., and Kumai, S. (1999). Effect of applying lactic acid bacteria isolated from forage crops on fermentation characteristics and aerobic deterioration of silage. J. Dairy. Sci. 82, 520–526.
Cai, Y., Du, Z., Yamasaki, S., Nguluve, D., Tinga, B., Macome, F., et al. (2020). Community of natural lactic acid bacteria and silage fermentation of corn Stover and sugarcane tops in Africa. Asian Australas. J. Anim. Sci. 33, 1252–1264. doi: 10.5713/ajas.19.0348
Cao, Y., Takahashi, T., Horiguchi, K.-I., Yoshida, N., and Cai, Y. (2010). Methane emissions from sheep fed fermented or non-fermented total mixed ration containing whole-crop rice and rice bran. Anim. Feed Sci. Technol. 157, 72–78. doi: 10.1016/j.anifeedsci.2010.02.004
Chen, L., Guo, G., Yuan, X., Shimojo, M., Yu, C., and Shao, T. (2014). Effect of applying molasses and propionic acid on fermentation quality and aerobic stability of total mixed ration silage prepared with whole-plant corn in Tibet. Asian Australas. J. Anim. Sci. 27, 349–356. doi: 10.5713/ajas.2013.13378
Chen, Y. S., Wu, H. C., and Yanagida, F. (2010). Isolation and characteristics of lactic acid bacteria isolated from ripe mulberries in Taiwan. Braz. J. Microbiol. 41, 916–921. doi: 10.1590/S1517-838220100004000010
Da Silva, N. C., Dos Santos, J. P., Ávila, C. L. S., Evangelista, A. R., Casagrande, D. R., and Bernardes, T. F. (2014). Evaluation of the effects of two lactobacillus buchneri strains and sodium benzoate on the characteristics of corn silage in a hot-climate environment. Grassl. Sci. 60, 169–177. doi: 10.1111/grs.12053
Ding, Z., Bai, J., Xu, D., Li, F., Zhang, Y., and Guo, X. (2020). Microbial community dynamics and natural fermentation profiles of ensiled alpine grass Elymus nutans prepared from different regions of the Qinghai-Tibetan plateau. Front. Microbiol. 11:855. doi: 10.3389/fmicb.2020.00855
Du, Z., Risu, N., Gentu, G., Jia, Y., and Cai, Y. (2020). Dynamic changes and characterization of the protein and carbohydrate fractions of native grass grown in Inner Mongolia during ensiling and the aerobic stage. Asian Australas. J. Anim. Sci. 33, 556–567. doi: 10.5713/ajas.19.0212
Du, Z., Sun, L., Lin, Y., Yang, F., and Cai, Y. (2021). The use of PacBio SMRT technology to explore the microbial network and fermentation characteristics of woody silage prepared with exogenous carbohydrate additives. J. Appl. Microbiol. 131, 2193–2211. doi: 10.1111/jam.15124
Du, S., You, S., Sun, L., Wang, X., Jia, Y., and Zhou, Y. (2022). Effects of replacing alfalfa hay with native grass hay in pelleted total mixed ration on physicochemical parameters, fatty acid profile, and rumen microbiota in lamb. Front. Microbiol. 13:861025. doi: 10.3389/fmicb.2022.861025
Ennahar, S., and Cai, Y. M. (2004). Genetic evidence that Weissella kimchii Choi et al. 2002 is a later heterotypic synonym of Weissella cibaria Björkroth et al. 2002. Int. J. Syst. Evol. Microbiol. 54, 463–465. doi: 10.1099/ijs.0.02783-0
Ennahar, S., Cai, Y., and Fujita, Y. (2003). Phylogenetic diversity of lactic acid bacteria associated with paddy rice silage as determined by 16S ribosomal DNA analysis. Appl. Environ. Microbiol. 69, 444–451. doi: 10.1128/AEM.69.1.444-451.2003
Fang, D., Dong, Z., Wang, D., Li, B., Shi, P., Yan, J., et al. (2022). Evaluating the fermentation quality and bacterial community of high-moisture whole-plant quinoa silage ensiled with different additives. J. Appl. Microbiol. 132, 3578–3589. doi: 10.1111/jam.15506
Fu, Z., Sun, L., Hou, M., Hao, J., Lu, Q., Liu, T., et al. (2022). Effects of different harvest frequencies on microbial community and metabolomic properties of annual ryegrass silage. Front. Microbiol. 13:971449. doi: 10.3389/fmicb.2022.971449
Fugaban, J. I. I., Bucheli, J. E. V., Park, Y. J., Suh, D. H., Jung, E. S., Franco, B. D. G. D., et al. (2021). Antimicrobial properties of Pediococcus acidilactici and Pediococcus pentosaceus isolated from silage. J. Appl. Microbiol. 73, 725–734. doi: 10.1111/jam.15205
Ge, G. T., Hou, M., Liu, T., Jia, Y., and Cai, Y. (2018). Microbial population, chemical composition and silage fermentation of native grasses growing on the inner Mongolian plateau. Grassl. Sci. 64, 226–233. doi: 10.1111/grs.12207
Guan, H., Yan, Y., Li, X., Li, X., Shuai, Y., Feng, G., et al. (2018). Microbial communities and natural fermentation of corn silages prepared with farm bunker-silo in Southwest China. Bioresour. Technol. 265, 282–290. doi: 10.1016/j.biortech.2018.06.018
Hammes, W. P., Brandt, M. J., Francis, K. L., Rosenheim, J., Seitter, M. F. H., and Vogelmann, S. A. (2005). Microbial ecology of cereal fermentations. Trends Food Sci. Technol. 16, 4–11. doi: 10.1016/j.tifs.2004.02.010
He, L., Li, S., Wang, C., Chen, X., and Zhang, Q. (2021). Effects of vanillic acid on dynamic fermentation parameter, nitrogen distribution, bacterial community, and enzymatic hydrolysis of stylo silage. Front. Microbiol. 12:690801. doi: 10.3389/fmicb.2021.690801
Hou, M., Gentu, G., Liu, T., Jia, Y., and Cai, Y. (2017). Silage preparation and fermentation quality of natural grasses treated with lactic acid bacteria and cellulase in meadow steppe and typical steppe. Asian Australas. J. Anim. Sci. 30, 788–796. doi: 10.5713/ajas.16.0578
Kung, L., Taylor, C. C., Lynch, M. P., and Neylon, J. M. (2003). The effect of treating alfalfa with lactobacillus buchneri 40788 on silage fermentation, aerobic stability, and nutritive value for lactating dairy cows. J. Dairy Sci. 86, 336–343. doi: 10.3168/jds.S0022-0302(03)73611-X
Lee, Y., Cho, Y., Kim, E., Kim, H. J., and Kim, H. Y. (2018). Identification of lactic acid bacteria in Galchi-and Myeolchi-Jeotgal by 16S rRNA gene sequencing, MALDI-TOF mass spectrometry, and PCR-DGGE. J. Microbiol. Biotechnol. 28, 1112–1121. doi: 10.4014/jmb.1803.03034
Li, Y., Du, S., Sun, L., Cheng, Q., Hao, J., Lu, Q., et al. (2022). Effects of lactic acid bacteria and molasses additives on dynamic fermentation quality and microbial community of native grass silage. Front. Microbiol. 13:830121. doi: 10.3389/fmicb.2022.830121
Li, H., Zeng, T., Du, Z., Dong, X., Xin, Y., Wu, Y., et al. (2022). Assessment on the fermentation quality and bacterial community of mixed silage of faba bean with forage wheat or oat. Front. Microbiol. 13:875819. doi: 10.3389/fmicb.2022.875819
Libonatti, C., Agueria, D., Garcia, C., and Basualdo, M. (2019). Weissella paramesenteroides encapsulation and its application in the use of fish waste. Rev. Argent. Microbiol. 51, 81–83. doi: 10.1016/j.ram.2018.03.001
Liu, Y., Li, Y., Lu, Q., Sun, L., Du, S., Liu, T., et al. (2022). Effects of lactic acid bacteria additives on the quality, volatile chemicals and microbial community of Leymus chinensis silage during aerobic exposure. Front. Microbiol. 13:938153. doi: 10.3389/fmicb.2022.938153
McEniry, J., Forristal, P. D., and O'Kiely, P. (2011). Factors influencing the conservation characteristics of baled and precision-chop grass silages. Irish J. Agric. Food Res. 50, 175–188. doi: 10.2307/41549250
Muck, R. E., Nadeau, E. M. G., McAllister, T. A., Contreras-Govea, F. E., Santos, M. C., and Kung, L. Jr. (2018). Silage review: recent advances and future uses of silage additives. J. Dairy Sci. 101, 3980–4000. doi: 10.3168/jds.2017-13839
Ni, K. K., Wang, Y. P., Li, D. X., Cai, Y. M., and Pang, H. L. (2015). Characterization, identification and application of lactic acid bacteria isolated from forage paddy rice silage. PLoS One 10:e0121967. doi: 10.1371/journal.pone.0121967
Ni, K., Wang, F., Zhu, B., Yang, J., Zhou, G., Pan, Y., et al. (2017). Effects of lactic acid bacteria and molasses additives on the microbial community and fermentation quality of soybean silage. Bioresour. Technol. 238, 706–715. doi: 10.1016/j.biortech.2017.04.055
Nomura, M., Kimoto, H., Someya, Y., and Suzuki, I. (1999). Novel characteristic for distinguishing Lactococcus lactis subsp. lactis from subsp. crernoris. Int. J. Syst. Bacteriol. 49 Pt 1, 163–166. doi: 10.1099/00207713-49-1-163
Pang, H., Zhang, M., Qin, G., Tan, Z., Li, Z., Wang, Y., et al. (2011). Identification of lactic acid bacteria isolated from corn stovers. Anim. Sci. J. 82, 642–653. doi: 10.1111/j.1740-0929.2011.00894.x
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Shah, A. A., Xianjun, Y., Zhihao, D., Junfeng, L., and Shao, T. (2017). Isolation and molecular identification of lactic acid bacteria from king grass and their application to improve the fermentation quality of sweet sorghum. World J. Microbiol. Biotechnol. 34:4. doi: 10.1007/s11274-017-2387-2
Spano, G., Beneduce, L., Tarantino, D., Zapparoli, G., and Massa, S. (2002). Characterization of lactobacillus plantarum from wine must by PCR species-specific and RAPD-PCR. Lett. Appl. Microbiol. 35, 370–374. doi: 10.1046/j.1472-765X.2002.01200.x
Tannock, G. W., Tilsala, T. A., Rodtong, S., Ng, J., Munro, K., and Alatossava, T. (1999). Identification of lactobacillus isolates from the gastrointestinal tract, silage, and yoghurt by 16S-23S rRNA gene intergenic spacer region sequence comparisoons. Appl. Environ. 65, 4264–4267. doi: 10.1128/AEM.65.9.4264-4267.1999
Thomas, T. (1997). An automated procedure for the determination of soluble carbohydrates in herbage. J. Sci. Food. Agric. 28, 639–642. doi: 10.1002/jsfa.2740280711
Torriani, S., Clementi, F., Vancanneyt, M., Hoste, B., Dellaglio, F., and Kersters, K. (2001). Differentiation of Lactobacillus plantarum, L. pentosus and L. paraplantarum species by RAPD-PCR and AFLP. Syst. Appl. Microbiol. 24, 554–560. doi: 10.1078/0723-2020-00071
Van Soest, J. P., Robertson, J. B., and Lewis, B. A. (1991). Methods fro dietary fiber, neutral detergent fiber and non-starch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74, 3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2
Wang, S., Dong, Z., Li, J., Chen, L., and Shao, T. (2019). Pediococcus acidilactici strains as silage inoculants for improving the fermentation quality, nutritive value and in vitro ruminal digestibility in different forages. J. Appl. Microbiol. 126, 424–434. doi: 10.1111/jam.14146
Wang, Y., He, L., Xing, Y., Zhou, W., Pian, R., Yang, F., et al. (2019). Bacterial diversity and fermentation quality of Moringa oleifera leaves silage prepared with lactic acid bacteria inoculants and stored at different temperatures. Bioresour. Technol. 284, 349–358. doi: 10.1016/j.biortech.2019.03.139
Wang, S., Li, J., Dong, Z., Chen, L., Yuan, X., and Shao, T. (2018). The effects of lactic acid bacteria strains isolated from various substrates on the fermentation quality of common vetch (Vicia sativa L.) in Tibet. Grass Forage Sci. 73, 639–647. doi: 10.1111/gfs.12363
Wang, S., Li, J., Zhao, J., Dong, Z., Dong, D., and Shao, T. (2022). Dynamics of the bacterial communities and predicted functional profiles in wilted alfalfa silage. J. Appl. Microbiol. 132, 2613–2624. doi: 10.1111/jam.15417
Xu, H. W., Sun, L., Na, N., Wang, C., Yin, G. M., Liu, S. B., et al. (2021). Dynamics of bacterial community and fermentation quality in Leymus chinensis silage treated with lactic acid bacteria and/or water. Front. Microbiol. 12:717120. doi: 10.3389/fmicb.2021.717120
Yan, Y., Li, X., Guan, H., Huang, L., Ma, X., Peng, Y., et al. (2019). Microbial community and fermentation characteristic of Italian ryegrass silage prepared with corn stover and lactic acid bacteria. Bioresour. Technol. 279, 166–173. doi: 10.1016/j.biortech.2019.01.107
You, S., Du, S., Ge, G., Wan, T., and Jia, Y. (2021a). Microbial community and fermentation characteristics of native grass prepared without or with isolated lactic acid bacteria on the Mongolian plateau. Front. Microbiol. 12:731770. doi: 10.3389/fmicb.2021.731770
You, S., Du, S., Ge, G., Wan, T., and Jia, Y. (2021b). Selection of lactic acid bacteria from native grass silage and its effects as inoculant on silage fermentation. Agron. J. 113, 3169–3177. doi: 10.1002/agj2.20720
Zanoni, P., Farrow, J. A. E., and Phillips, B. A., and, Collins, M. D. (1987). Lactobacillus pentosus (Fred, Peterson, and Anderson) sp. nov., nom. rev. Int. J. Syst. Bacteriol. 37, 339–341, doi: 10.1099/00207713-37-4-339
Zhang, Q., Li, X., Zhao, M., and Yu, Z. (2016a). Lactic acid bacteria strains for enhancing the fermentation quality and aerobic stability of Leymus chinensis silage. Grass Forage Sci. 71, 472–481. doi: 10.1111/gfs.12190
Zhang, Q., Wu, B., Nishino, N., Wang, X., and Yu, Z. (2016b). Fermentation and microbial population dynamics during the ensiling of native grass and subsequent exposure to air. Anim. Sci. J. 87, 389–397. doi: 10.1111/asj.12427
Zhang, Q., and Yu, Z. (2017). Characterization, identification and application of lactic acid bacteria isolated from Leymus chinensis silage. Grassl. Sci. 63, 111–117. doi: 10.1111/grs.12156
Zhang, Q., Yu, Z., and Wang, X. (2015). Isolating and evaluating lactic acid bacteria strains with or without sucrose for effectiveness of silage fermentation. Grassl. Sci. 61, 167–176. doi: 10.1111/grs.12097
Keywords: 16S rRNA gene, native grass, lactic acid bacteria, silage fermentation, isolation and identification
Citation: Hou M, Wang Z, Sun L, Jia Y, Wang S and Cai Y (2023) Characteristics of lactic acid bacteria, microbial community and fermentation dynamics of native grass silage prepared in Inner Mongolian Plateau. Front. Microbiol. 13:1072140. doi: 10.3389/fmicb.2022.1072140
Edited by:
Annamalai Neelamegam, Dartmouth College, United StatesReviewed by:
Yang Cao, Heilongjiang Bayi Agricultural University, ChinaPing Li, Guizhou University, China
Copyright © 2023 Hou, Wang, Sun, Jia, Wang and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhijun Wang,  emhpanVud2FuZzMyMUAxMjYuY29t; Yimin Cai,
emhpanVud2FuZzMyMUAxMjYuY29t; Yimin Cai,  Y2FpQGFmZnJjLmdvLmpw
Y2FpQGFmZnJjLmdvLmpw
†These authors have contributed equally to this work
 Meiling Hou
Meiling Hou Zhijun Wang
Zhijun Wang Lin Sun
Lin Sun Yushan Jia
Yushan Jia Shicong Wang
Shicong Wang Yimin Cai
Yimin Cai