- 1Key Laboratory of Animal Epidemic Disease Diagnostic Laboratory of Molecular Biology in Xianyang City, Institute of Animal Husbandry and Veterinary Medicine, Xianyang Vocational Technical College, Xianyang, Shaanxi, China
- 2Tianjin Institute of Animal Husbandry and Veterinary Medicine, Tianjin Academy of Agricultural Sciences, Tianjin, China
- 3College of Veterinary Medicine, Northwest A&F University, Yangling, Shaanxi, China
- 4Institute of Hemu Biotechnology, Beijing Hemu Biotechnology Co. Ltd., Beijing, China
- 5Liangshan County Animal Husbandry and Veterinary Development Center, Liangshan County Animal Husbandry Bureau, Jining, China
Porcine epidemic diarrhea virus (PEDV) in the Coronavirus family is a highly contagious enteric pathogen in the swine industry, which has evolved mechanisms to evade host innate immune responses. The PEDV-mediated inhibition of interferons (IFNs) has been linked to the nuclear factor-kappa B (NF-κB) pathway. MicroRNAs (miRNAs) are involved in virus–host interactions and IFN-I regulation. However, the mechanism by which the PEDV regulates IFN during PEDV infection has not yet been investigated in its natural target cells. We here report a novel mechanism of viral immune escape involving miR-615, which was screened from a high-throughput sequencing library of porcine intestinal epithelial cells (IECs) infected with PEDV. PEDV infection altered the profiles of miRNAs and the activities of several pathways involved in innate immunity. Overexpression of miR-615 increased PEDV replication, inhibited IFN expression, downregulated the NF-κB pathway, and blocked p65 nuclear translocation. In contrast, knockdown of miR-615 enhanced IFN expression, suppressed PEDV replication, and activated the NF-κB pathway. We further determined that IRAK1 is the target gene of miR-615 in IECs. Our findings show that miR-615 suppresses activation of the NF-κB pathway by suppressing the IRAK1 protein and reducing the generation of IFN-IIIs, which in turn facilitates PEDV infection in IECs. Moreover, miR-615 inhibited PEDV replication and NF-κB pathway activation in both IECs and MARC-145 cells. These findings support an important role for miR-615 in the innate immune regulation of PEDV infections and provide a novel perspective for developing new treatments.
Introduction
The emerging and re-emerging Coronavirus family member porcine epidemic diarrhea virus (PEDV) attacks neonatal piglets and causedto cause acute watery diarrhea, with a substantial mortality rate. The global swine industry has sustained tremendous financial losses as a result of PEDV infections (Akira et al., 2006; Li et al., 2012; Dong and Soong, 2021). The target cells for PEDV are porcine intestinal epithelial cells (IECs), and PEDV strain CV777 could successfully infect an immortalized IEC line (Cao et al., 2015b; Lin et al., 2015; Wang et al., 2016). Previous studies have shown that type-I interferons (IFN-Is), which are produced by the host innate immune response, are crucial in limiting PEDV replication by eliciting an innate antiviral response (Zhang et al., 2016; Jegaskanda et al., 2018). IFN-Is also promote adaptive immunity during influenza virus infections by enhancing natural killer cell function (Hoffmann et al., 2015).
Type-III interferons (IFN-IIIs) are believed to employ the same antiviral mechanism as IFN-Is and induce IFN-stimulated gene expression, with IFN-III receptors being distributed primarily in gastrointestinal and respiratory epithelial cells (Li et al., 2016). Studies on PEDV-infected IECs (IPEC-J2), as well as several other reports, have shown that IFN-IIIs play an important role in inhibiting PEDV infection in IECs (Li et al., 2016; Lui et al., 2016). IFN-IIIs also exhibit strong antiviral activity in Vero cells (Ma et al., 2018; Xue et al., 2018). Therefore, IFN-IIIs may be key factors regulating PEDV infection.
Viruses generally develop diverse mechanisms to evade the host innate immune response, such as by antagonizing IFN production (Li et al., 2017a; Zhang et al., 2018b). IFN antagonism and production have also been implicated in PEDV mechanisms for evading innate immunity (Annamalai et al., 2015; Li et al., 2017b). Therefore, identification of the antiviral factors of the innate immune system is crucial for the control of PEDV infection. IRF3, nuclear factor kappa B (NF-κB), and IRF7 activation are crucial for the release of IFN-IIIs. IFN-λ1, −λ4, and -λ3 have been identified in IPEC-J2 cells (Li et al., 2017b; Zhang et al., 2018b). In small intestinal epithelial cells (IPEC-J2), PEDV inhibited IFN-III secretion through interfering with IRF and NF-κB (Zhang et al., 2018b). However, PEDV regulates small intestinal epithelial cells of different origins in different ways. In IECs, PEDV could induce NF-κB activation through the Toll-like receptor (TLR)2, TLR3, and TLR9 pathways in porcine intestinal epithelial cells at 24 h.
The NF-κB pathway has been reported to affect the secretion of IFN-IIIs more potently than IFN-I pathways (Pu et al., 2017). Additionally, the majority of TLRs use myeloid differentiation primary response 88 (MyD88), which participates in the recruitment of interleukin-1 receptor-associated kinase (IRAK)1 and 4 (Fisher et al., 2021). Tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF-6) is triggered by IRAK1 phosphorylation, which activates NF-κB and mitogen-activated protein kinase (MAPK; Akira et al., 2006). Furthermore, PEDV-infected cells were found to alter the activity of the NF-κB pathway (Wang et al., 2016).
MicroRNAs (miRNAs) are small RNAs that are 18–23-nucleotides in length and exhibit various effects on cell proliferation, differentiation, and apoptosis, and in viral infections (Huang et al., 2018). Viral infections result in the dysregulated expression of miRNAs, and these changes in miRNA abundance can in turn affect viral infection and cellular physiological processes by regulating innate immunity. Several reports have shown that miRNAs inhibit PEDV infection by downregulating different target genes that are required for innate immunity (Wu et al., 2013; Zheng et al., 2018; Qi et al., 2021). In particular, miR-221-5p was found to decrease the rate of PEDV replication in MARC-145 cells by boosting activation of the NF-κB pathway (Zheng et al., 2018). In addition, miR-129-3p, which was identified in the process of studying porcine circovirus 2-infected cells, inhibited PEDV replication by targeting the NF-κB pathway in IPEC-J2 cells (Annamalai et al., 2015). By suppressing the expression of the proteins acting downstream of the NF-κB pathway in porcine kidney (PK) cells, miR-30c-5p reduces the expression of IFN-IIIs (Zhao et al., 2012; Buggele and Horvath, 2013; Song et al., 2015). However, the role and underlying mechanism of the miRNA-mediated regulation of the NF-κB pathway in PEDV infection in IECs have not yet been elucidated.
In this study, to ascertain the function of miRNAs in the innate immune response to PEDV infection, we performed high-throughput sequencing on PEDV-infected IECs, revealing a change in miRNA expression and innate immunity pathways under infectious conditions. Among the screened miRNAs, miR-615 was predicted to function in the NF-κB pathway and facilitate PEDV replication. We further found that miR-615 inhibits IFN-III expression and NF-κB pathway activation by targeting IRAK1. Conversely, miR-615 induced PEDV replication and inhibited the NF-κB pathway in two different types of cells (IECs and MARC-145). These data imply that miR-615 is a crucial PEDV target, and thus may be a potential target for PEDV treatment and prevention strategies.
Materials and methods
Cells and viruses
MARC-145 cells, which are kidney cells from an African green monkey, were grown in Dulbecco’s modified Eagle medium (DMEM; Hyclone, Logan, UT, USA) with 10% heat-inactivated fetal bovine serum (FBS; PAN-Biotech), and 100-times diluted penicillin and streptomycin (Hyclone). MARC-145 cells were used to validate the effect of miR-615 during PEDV infection. The IECs were cultivated in DMEM-F12 (Hyclone) supplemented with 10% FBS, penicillin, and streptomycin at the same concentrations as indicated above. All cells were kept in an incubator with 5% CO2 at 37°C as previously described (Wang et al., 2014). PEDV strains CV777 (GenBank accession number KT323979.1) and NW-17 (GenBank accession number MF782686.1) were provided by Nuoweilihua Biotechnology Co., which have an S gene from the epidemic strain group II. Strain NW-17 could infect IECs without trypsin and was therefore selected as an optimal strain for IEC infection. Vero cells were used to prepared the PEDV stock by three cycles of freezing and thawing, and were stored at −80°C.
Immunofluorescence assays (IFAs)
After 20 min of fixation at a 4:1 ratio of cold acetone and methanol, the cell samples were washed three times in phosphate-buffered saline (PBS). The cells were incubated with a monoclonal antibody (mAb) against the PEDV N or NF-κB p65 protein for 2 h. The cells were then rinsed three times with PBS and incubated for 1 h with a fluorescein isothiocyanate-conjugated AffiniPure Goat Anti-rabbit/Mouse secondary antibody (Sungene Biotech, Tianjin, China). Hoechst 33258 was utilized to stain the nucleus, and the cell samples were analyzed with a laser-scanning confocal microscope after three PBS rinses (Olympus). The experiment was performed at room temperature.
Mirna microarray and predicting the mRNA targets of differentially expressed miRNAs (DEMiRs)
Deep sequencing was carried out at Novogene (Beijing, China). IECs were infected in triplicates for 24 h with CV777, NW-17, or a mock infection at a multiplicity of infection (MOI) of 1. The mock group was treated with PBS. Total RNA was isolated using Trizol reagent (Invitrogen). Nine small RNA libraries (triplicate samples of the control mock-, CV777-, and NW-17-infected groups) were generated for Illumina sequencing. The microarray assay was conducted as previously described (Hallman et al., 2013). The prediction of target genes of DEMiRs was performed using miRanda (John et al., 2004; version 3.3a), PITA1, and RNAhybrid.2 Gene Ontology enrichment analysis was performed to screen the potential functions of the significantly enriched target genes. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was applied to identify the potential pathways associated with the target genes of the DEMiRs (Kanehisa et al., 2008). p-values were corrected using the Benjamini–Hochberg strategy (Benjamini and Hochberg, 1995). Statistical significance was defined as a corrected p-value <0.05.
Validation of DEMiRs using reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from IECs 24 h after infection at an MOI of 1. The Real-time Quantitative PCR Detection System and SYBR PrimeScript™ miRNA RT-PCR Kit were used to perform RT-qPCR. One microliter of each primer, 2.0 μl of diluted cDNA, and 12.5 μl of SYBR Green Premix Ex Taq II were included in each 25 μl reaction mixture. The thermocycling conditions comprised 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 20 s. Each sample was processed in triplicates. Fifteen miRNAs were selected for testing. The expression of the U6 small RNA was used as the reference for data normalization. Table 1 contains the primer sequences for each miRNA and gene.
Transfection of miRNA mimics, miRNA inhibitors, and siRNAs
A miRNA mimics is a chemically synthesized double-stranded RNA identical to a mature miRNA sequence. A miRNA inhibitor is a chemically modified single-stranded RNA that is complementary to the mature miRNA sequenc. After 12 h of culture, MARC-145 cells and IECs were transfected for 24 h using Lipofectamine 3000 (Invitrogen) with mimic control (MC), miR-615 mimics, miR-615 inhibitor (miR-615 inhi; 100 nM), inhibitor control (IC; 100 nM), small interfering RNA (siRNAs; si-IRAK1), or siRNA control (SC; 50 nM), which were synthesized at RiboBio (Guangzhou, China). Subsequently, the cells were infected with PEDV at an MOI of 1.0. After 24 h of infection, the cells were examined for indirect immunofluorescent labeling, or collected for RNA quantification or western blotting.
Plasmids
pCDNA3.1+ plasmid was used to clone the Flag-IRAK1 plasmid at Genecreate using the BamHI and EcoRI restriction sites (Wuhan, China). The miRNA reporter plasmids, WT-pmirGLO-IRAK1 (wild-type, WT), and MuTpmirGLO-IRAK1 (mutant type; MuT) were subcloned into pmirGLO using the Nhel and SalI sites at Genecreate. pNiFty-luc plasmids is composed of a minimal Promoter, five NF-κB repeated transcription factor binding sites and a Luc (Luciferase) reporter gene of mammal.
Dual-luciferase reporter assays
The potential miR-615 target genes in the 3′-untranslated region (3′-UTR) of IRAK1 were identified using the luciferase vector pmirGLO (Promega, Madison, WI, USA). The wild-type (WT-pmirGLO-IRAK1) or mutant (MuTpmirGLO-IRAK1) 3′-UTR sections of IRAK1 were subcloned into the pmirGLO vector and co-transfected into 293 T cells with miR-615. NF-κB activity was detected using pNiFty-luc and pRL-TK plasmids with the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer instructions.
Western blotting
The cells were lysed using radioimmunoprecipitation assay buffer (Beyotime, Shanghai, China) and centrifuged at 4°C for 12,000 rpm. The concentration of the lysate was evaluated using a bicinchoninic acid (Thermo Scientific) protein assay kit. Each sample was diluted with 5Хloading buffer, boiled for 10 min, resolved on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel, and then deposited onto polyvinylidene fluoride membranes at equal amounts. The membranes were blocked with 5% skim milk for 1 h and incubated overnight at 4°C with the primary antibodies against β-actin (1:1,000; 5,057; Cell Signaling Technology, Danvers, MA, USA), phospho-NF-κB p65 antibody (1:1,000; 3,033; Cell Signaling Technology), NF-κB p65 (1:1,000; 6,956; Cell Signaling Technology), and MyD88 (1:1,000; NB100-5698SS; RDSC). Proteins were detected using enhanced chemiluminescence detection reagents after being treated with secondary antibodies, HRP-conjugated goat anti-mouse IgG or goat anti-rabbit IgG (Beyotime). All samples were incubated along with β-actin as an internal standard. PEDV anti-nucleocapsid (N) protein antibody (1:1,000) was gifted by Prof. Tong Guangzhi, Shanghai Veterinary Research Institute.
Statistical analyses
All data were statistically analyzed using Student’s t-test in Graphpad Prism 9.3.1 for analysis of variance (ANOVA).
Results
Dysregulated expression of miRNAs in circulating and vaccine strain-infected IECs
Villous epithelial cells are the primary target cells of PEDV infection; therefore, IECs are a suitable model for studying virus–cell interactions in the intestinal epithelia (Wang et al., 2014). IECs have been found to be susceptible to certain PEDV strains such as CV777, independent of high concentrations of trypsin (Cao et al., 2015b).
The IFA results of the IECs at 24 h post-inoculation (hpi) with CV777 and NW-17 (MOI = 1) suggested significantly higher fluorescence in the CV777- and NW-17-infected groups than that in the mock-infected group (Figure 1A). This observation confirmed that CV777 and NW-17 could infect IECs effectively.
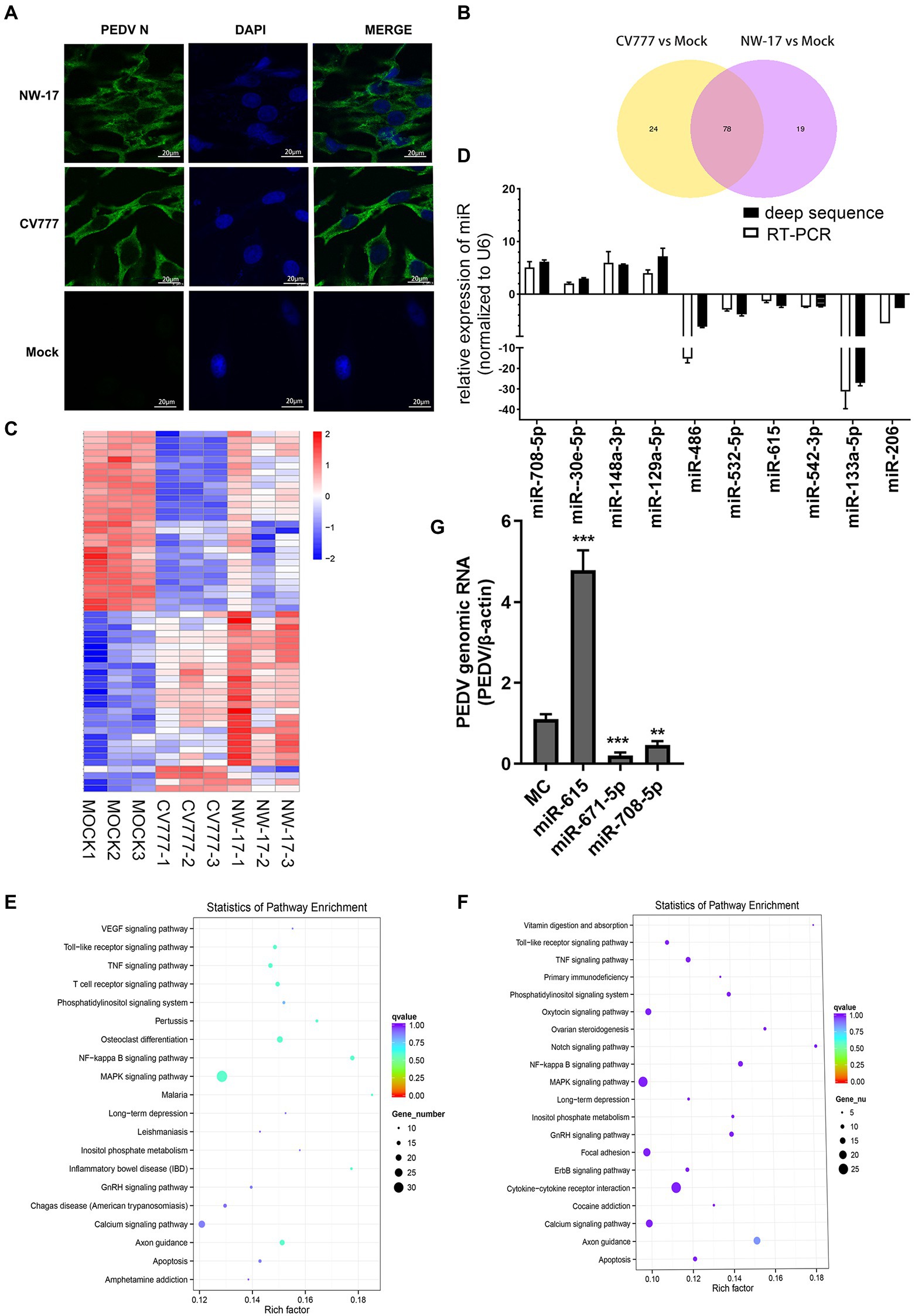
Figure 1. PEDV infection changes cellular microRNA profiles. (A) Immunofluorescence showing PEDV N protein expression in IECs (green). IECs were infected with the CV777 and NW-17 strains at a multiplicity of infection (MOI) of 1. Cells were fixed and stained with a mouse anti-N monoclonal antibody (mAb). Scale bar = 20 μm. (B) Venn diagrams showing the differentially expressed microRNAs (DEMiRs) among CV777-infected vs. mock-infected cells and NW-17-infected vs. mock-infected cells. (C) Heatmap showing the high abundance of DEMiRs. (D) Verification of the miRNA microarray assay using RT-qPCR. Data from RT-qPCR are shown as the mean ± SD of three independent experiments. U6 mRNA was detected as a control. (E and F) KEGG pathway enrichment analysis of the main DEMiRs for the CV777- and NW-17-infected groups, respectively. (G) Transfection of miRNAs inhibited or enhanced viral replication. IECs were transfected with the miR-671-5p, miR-708-5p, miR-615, or MC mimics at 50 nmol, followed by infection with PEDV (MOI = 1). Cells were collected for RT-qPCR at 24 h post-infection (hpi). PEDV genomic RNA was determined by RT-qPCR. Asterisks indicate statistical significance. *p < 0.05; **p < 0.01; ***p < 0.001.
To identify whether miRNAs play a role in virus–cell interactions, we performed high-throughput sequencing to obtain the miRNA profiles of mock-infected or PEDV (CV777 and NW-17)-infected IECs at an MOI of 1 at 24 hpi. In comparison to the mock-infected group, 102 known miRNAs were found to be differentially expressed in the CV777-infected group and 98 known miRNAs were differentially expressed in the NW-17-infected group. Among them, 78 miRNAs were commonly differentially expressed in both infection groups (Figure 1B).
A total of 55 high-abundance miRNAs were selected by setting the read count to >250 (p < 0.05; Figure 1C). Of these miRNAs, 28 were upregulated and 27 were downregulated. To validate the high-throughput results of the DEMiRs, RT-qPCR was used to analyze the expression of 13 DEMiRs that were common to both groups (Figure 1D).
Using the KEGG pathway database, the roles of the DEMiRs in response to the PEDV strains were predicted, showing enrichment in several pathways, including the NF-κB, apoptosis, MAPK, TNF, and TLR signaling pathways, which participate in antiviral activities (Figures 1E,F). The high enrichment scores for the NF-κB and TLR signaling pathways indicated that innate immunity plays an important role in PEDV infection.
The DEMiRs enriched in the NF-κB pathway, miR-615, miR-708, miR-221-5p, and miR-671-5p, were selected for further investigation. Three miRNAs (miR-708, miR-671-5p, and miR-221-5p) were found to exert inhibition on PEDV replication by RT-qPCR. In particular, miR-615 overexpression significantly boosted viral replication compared to that in the MC group (p < 0.01; Figure 1G).
Mir-615 promoted PEDV infection and replication in IECs and MARC-145 cells
To further evaluate the interaction between miR-615 and PEDV in IECs and MARC-145 cells during PEDV infection, PEDV genomic and miR-615 RNA were detected using RT-qPCR. In IECs, the expression of miR-615 showed a downward trend (p < 0.01; Figure 2A). In MARC-145 cells, miR-615 expression was up-regulated at 12 h and was down-regulated at 24 h (p < 0.001; Figure 2B). The overexpression of miR-615 promoted viral replication in IECs (p < 0.01; Figure 2C) and MARC-145 cells (p < 0.001; Figure 2E). Conversely, the downregulation of miR-615 expression inhibited viral replication compared to that in the IC group in the IECs (p < 0.001; Figure 2D) and MARC-145 cells (p < 0.001; Figure 2F).
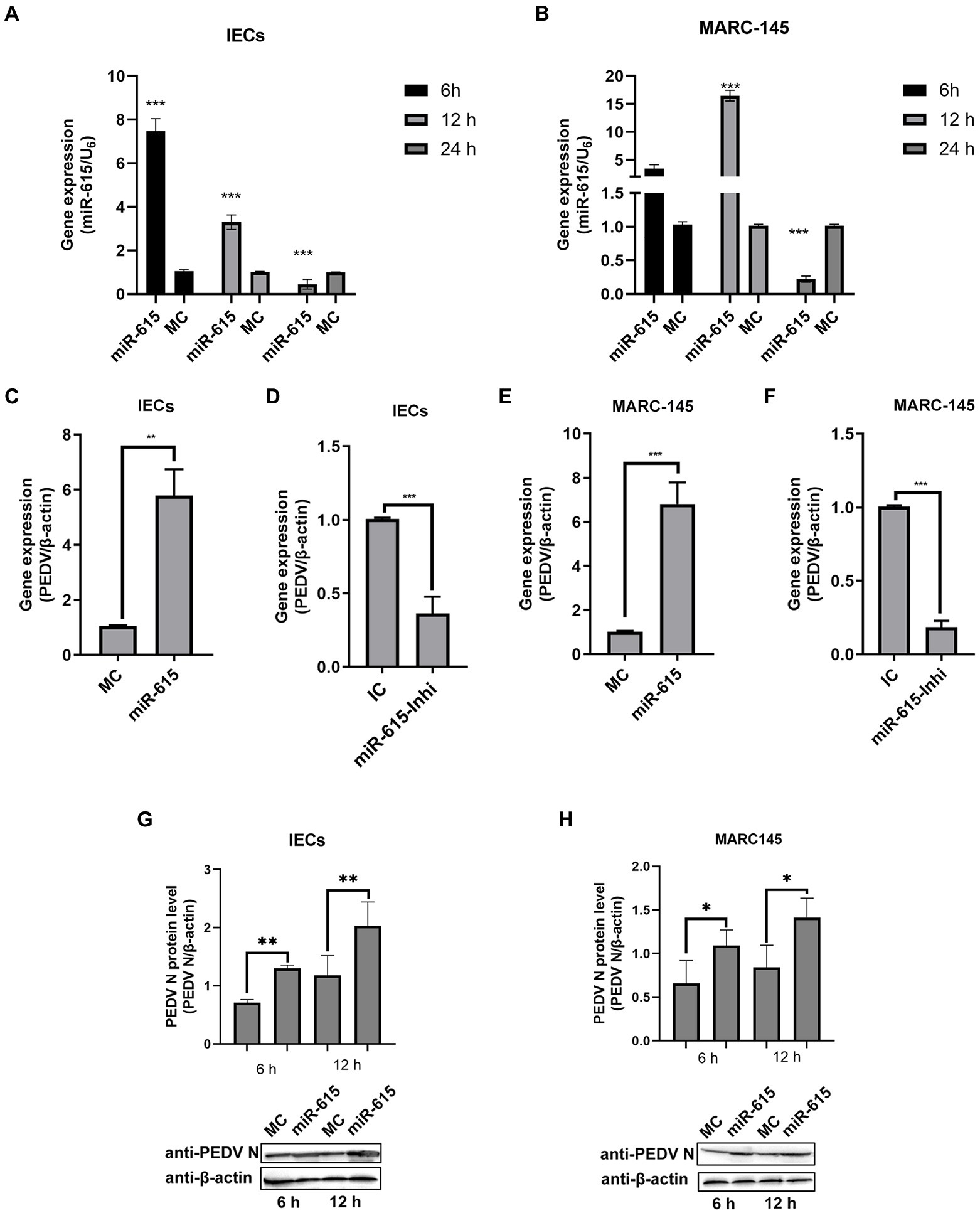
Figure 2. miR-615 facilitates PEDV infection in MARC-145 cells and IECs. (A and B) PEDV regulated miR-615 expression in IECs and MARC-145 cells. IECs (A) and MARC-145 cells (B) were infected with PEDV at a multiplicity of infection (MOI) of 1 and samples were collected at 6, 12, and 24 h post-infection. RT-qPCR was used to assess miR-615 expression. (C–F) IECs and MARC-145 cells were transfected with the MC, miR-615 mimic, IC, or miR-615 inhibitor (100 nmol). After 24 h, the cells were infected with PEDV at an MOI of 1. After 18 h, the PEDV genomic RNA of the MC, miR-615 mimic, IC, and miR-615 inhibitor in the IECs (A,B) and transfected MARC-145 cells (C,D) were assessed using RT-qPCR. PEDV N protein level was measured using western blotting following transfection with the MC or miR-615 mimic for 6 and 12 h in the IECs (G) and MARC-145 cells (H). The intensity represents PEDV N protein levels normalized against that of β-actin across three independent experiments in IECs and MARC-145 cells, respectively. The data are presented as the mean ± SD of three independent experiments, performed with technical duplicates. *p<0.05, **p < 0.01, ***p < 0.001.
Western blotting showed that the level of PEDV N protein expression increased in IECs (Figure 2G) and MARC-145 cells (Figure 2H) after transfection with the miR-615 mimic for 6 and 12 h.
Mir-615 inhibited the expression of IFN-IIIs in IECs and IFN-Is in MARC-145 cells
Recent studies suggested that IFN-III has an important effect on the antiviral activity of small IECs (Zhang et al., 2018b). Consistently, we found that PEDV replication could be inhibited by IFN-λ3 (Figure 3A). Therefore, we hypothesized that miR-615 affects the expression of IFN-IIIs or IFN-Is, which in turn enhances viral replication. To test this hypothesis, we assessed the expression of IFN-IIIs in PEDV-infected IECs and of IFN-I in MARC-145 cells.
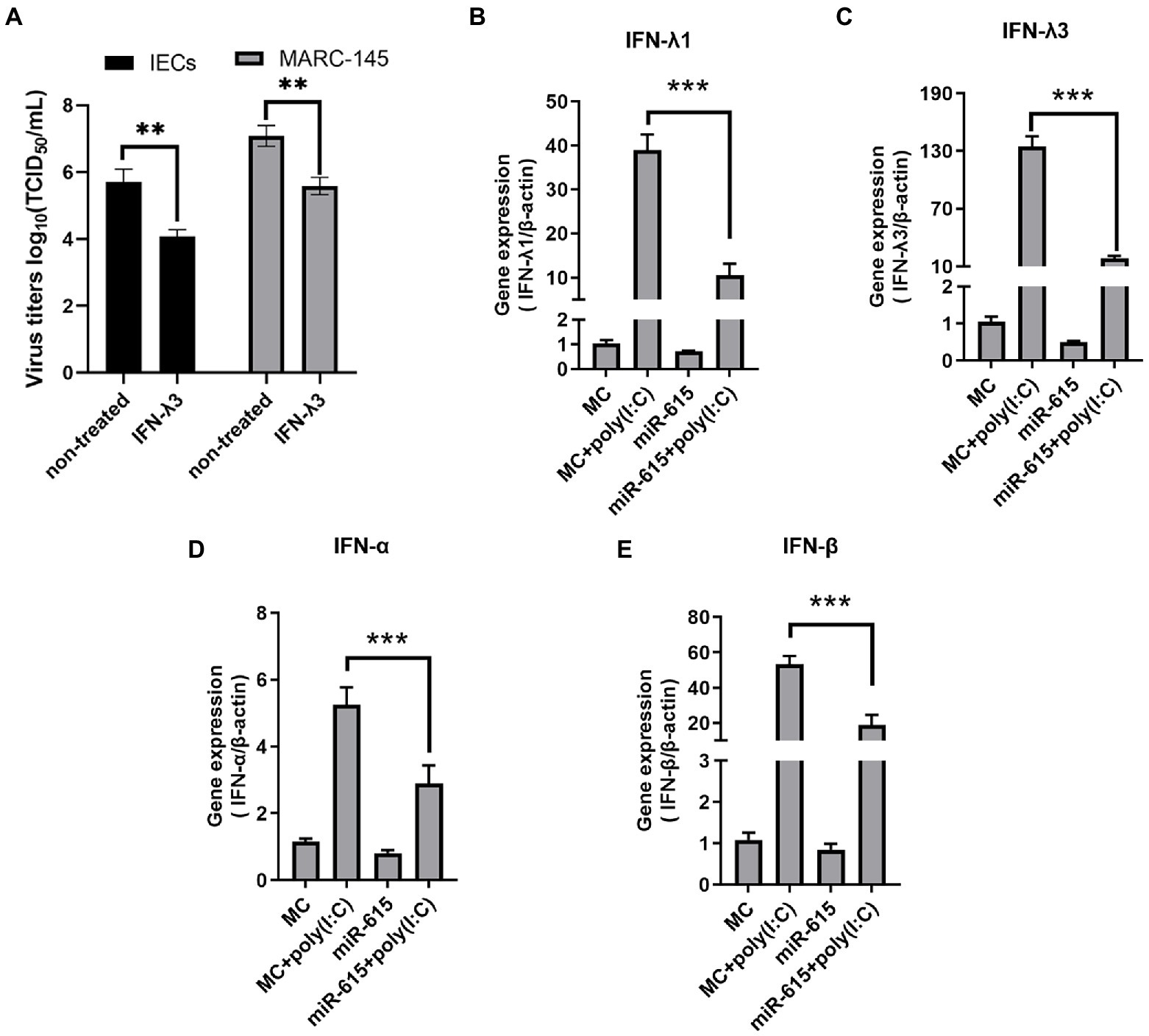
Figure 3. miR-615 downregulated the levels of IFN-Is in MARC-145 cells and of IFN-IIIs in IECs. (A) IFN-λ3 inhibited PEDV replication in IECs and MARC-145 cells. IECs and MARC-145 cells were seeded and treated with 100 ng/ml IFN-λ3 for 12 h, followed by infection with PEDV at a multiplicity of infection (MOI) of 1 with incubation for 2 h and replenished with fresh infection medium containing the IFN-λ3. Cell culture supernatants were collected at 12 and 24 h post-infection and titrated to determine the TCID50. MARC-145 and IECs were transfected with miR-615 for 24 h and then stimulated with poly (I:C; 10 μg/ml) for 24 h during PEDV infection. The cells were then harvested for RT-qPCR to determine the levels of (B) IFN-λ1 and (C) IFN-λ3 in the IECs, and of (D) IFN-α and (E) IFN-β in MARC-145 cells. The data are representative of three independent experiments (mean ± SD). ***p < 0.001, **p < 0.01.
miR-615 mimics were transfected into the cells and/or poly (I:C) was used to stimulate the cells for 24 h. RT-qPCR was used to examine the expression of the two IFN-III subtypes, IFN-λ1 and-λ3. IFN-λ1 and λ3 expression was upregulated after stimulation with poly (I:C), which served as a positive control; however, the expression levels of IFN-λ1 and -λ3 significantly decreased following miR-615 transfection in IECs (p < 0.01; Figures 3B,C).
We also evaluated IFN-β and -α levels in miR-615-transfected MARC-145 cells. Poly (I:C) stimulation upregulated IFN-β and -α expression, whereas miR-615 transfection significantly downregulated their expression (p < 0.01; Figures 3D,E). The downregulation of IFN-Is and -III expression suggests that miR-615 may affect the IFN pathway in these two types of cells when infected with PEDV.
Mir-615 inhibits NF-κB pathway activation in IECs and MARC-145 cells
Based on our preliminary findings, we hypothesized that miR-615 restricts the NF-κB pathway from being activated. The expression of IFNs is induced when NF-κB binds to the positive regulatory domain (PRD) II in the nucleus after translocation. Thus, we further examined NF-κB activity to explore whether the pathway was inhibited by miR-615 or miR-615 inhibitors. The NF-κB reporter luciferase plasmid (pNiFty-luc; containing five PRDII sites), thymidine kinase promoter-Renilla luciferase reporter plasmid (pRL-TK; an internal control plasmid), and miR-615 mimics or inhibitors were co-transfected into IECs, followed by poly (I:C) stimulation; thereafter, cell lysates were collected to detect luciferase activity at 24 hpi. The luciferase reporter assays showed that miR-615 markedly inhibited poly (I:C)-induced PRDII activity in IECs (p < 0.01; Figure 4A). Conversely, miR-615 inhibitors increased PRDII activity (p < 0.01; Figure 4B). Comparable results were observed in MARC-145 cells (Figures 4C,D). These results verified our hypothesis that miR-615 inhibits NF-κB pathway activity in both IECs and MARC-145 cells.
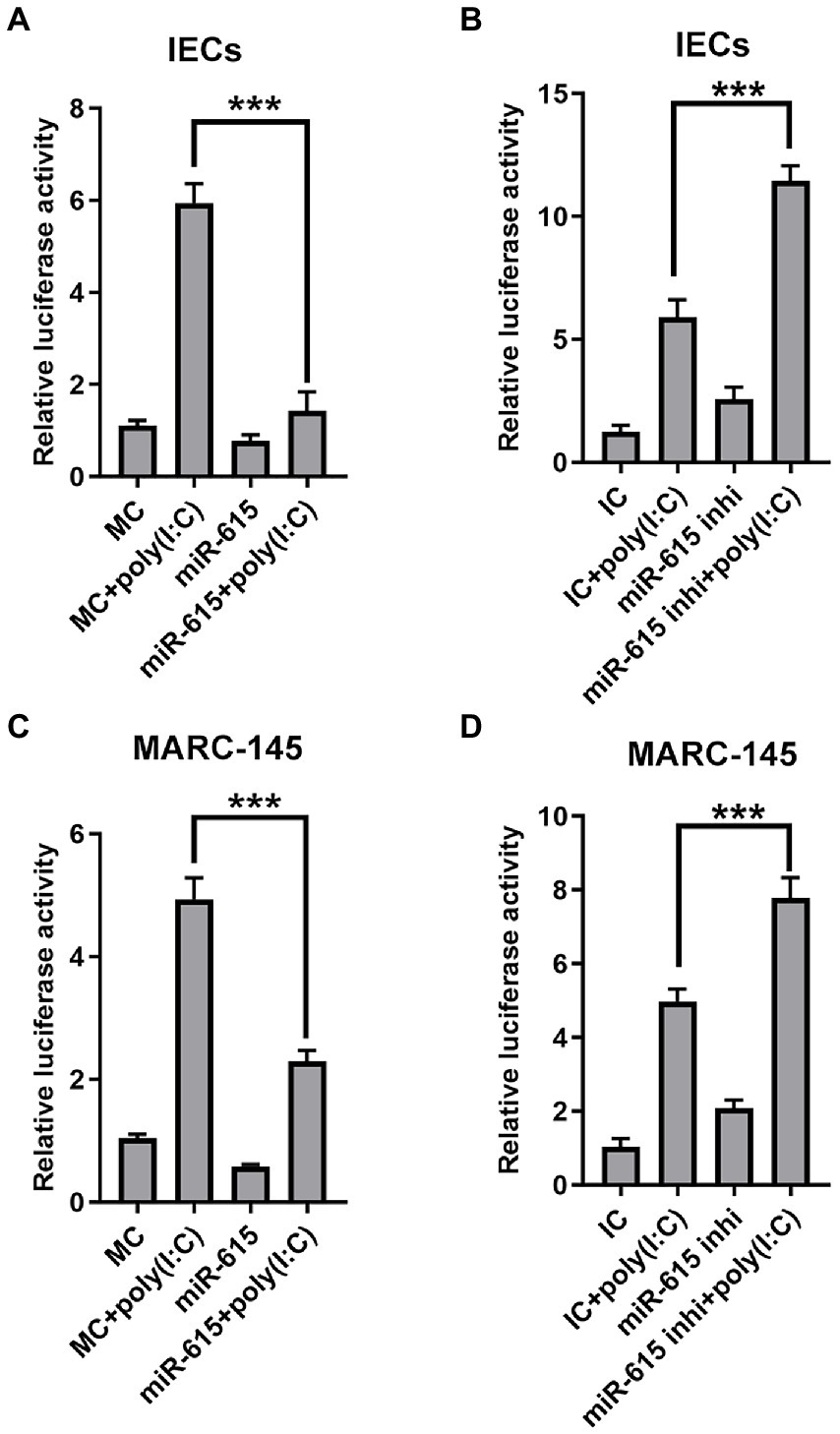
Figure 4. miR-615 inhibits NF-κB activation. Luciferase activity in IECs and MARC-145 cells. Cells were co-transfected using a dual-luciferase reporter system (pNiFty-luc and pRL-TK vectors, the activities of which indicate NF-κB-promoter activation) and miR-615 mimics or inhibitors for 24 h, and stimulated with poly (I:C) for 24 h during PEDV infection at a multiplicity of infection (MOI) of 1. Luciferase reporter activity of the miR-615 mimics in IECs (A) and MARC-145 cells (C). Luciferase reporter activity of the miR-615 inhibitors in IECs (B) and MARC-145 cells (D). The data are representative of three independent experiments (mean ± SD). ***p < 0.001.
Mir-615 inhibits NF-κB activation by suppressing p65 nuclear translocation
Next, we explored the mechanism of NF-κB inhibition. The IFA results revealed a significant reduction in the nuclear abundance of p65 protein in the miR-615 transfection group as compared to that in the MC-transfected group in both IECs (Figure 5A) and MARC-145 cells (Figure 5C). The abundance of p-p65 was found to decrease following transfection with miR-615 mimics in both the poly (I:C)-induced and non-poly (I:C)-induced groups (Figure 5B). We further examined p-p65 proteins upstream of MyD88 to investigate the mechanism underlying NF-κB downregulation. The results showed that p-p65 expression was downregulated, whereas MyD88 expression was not changed with the transfection of miR-615 in IECs (Figure 5B) and MARC-145 cells (Figure 5D). These results suggested that miR-615 inhibits the NF-κB pathway by repressing p65 nuclear translocation.
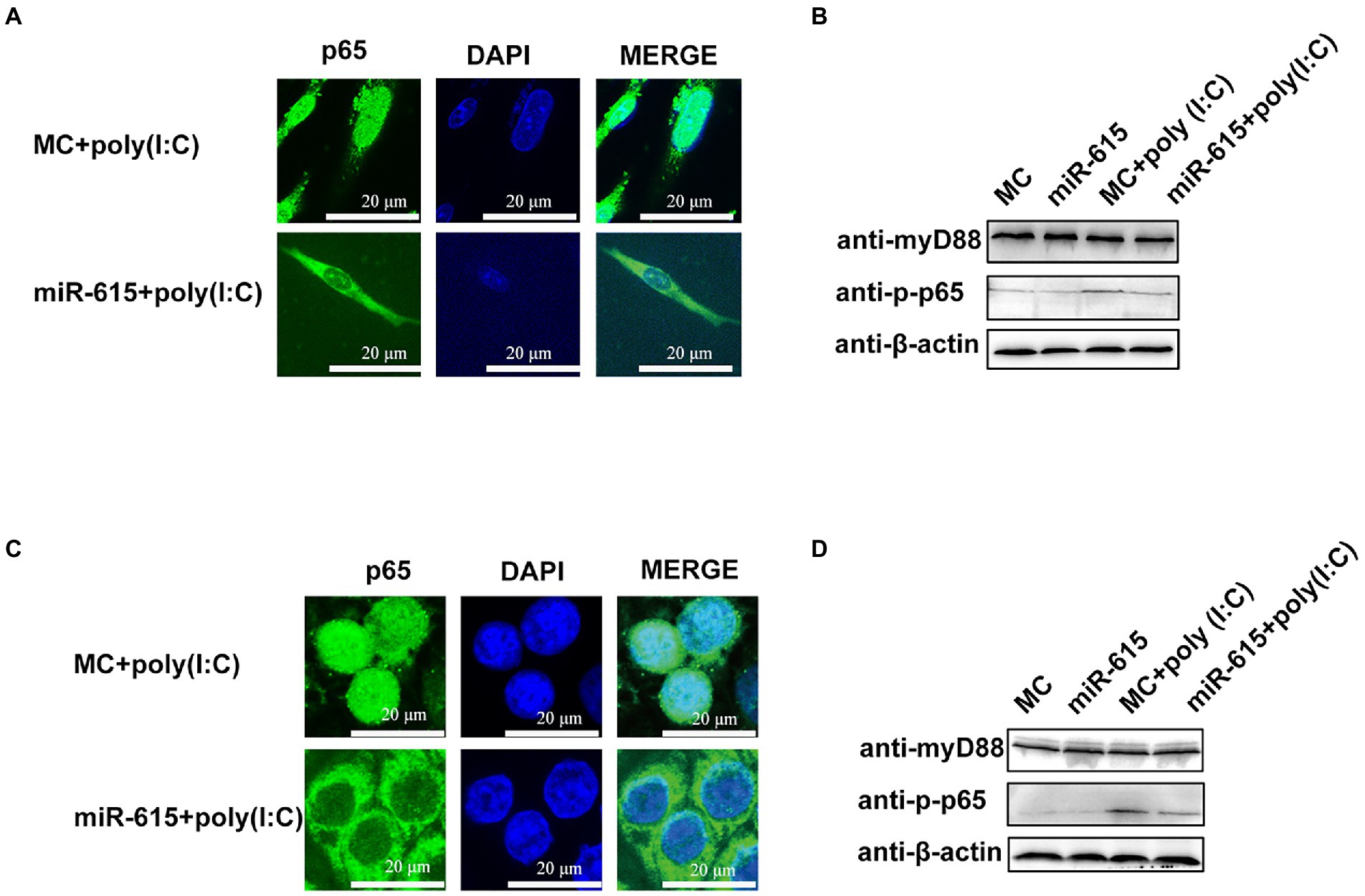
Figure 5. miR-615 inhibits the nuclear translocation and downregulates the phosphorylation of p65. (A,C) Immunofluorescence analysis of the nuclear translocation of p65 protein after transfection of miR-615 mimics during PEDV infection in IECs and MARC-145 cells. IECs and MARC-145 cells were transfected with miR-615. Poly (I:C) stimulation was added for 24 h during PEDV infection. Cells were fixed and stained with a rabbit anti-phosphorylated (p)-p65 and MyD88 monoclonal antibodies. Scale bar = 15 μm. (B,D) Western blot analysis for detection of the (p)-p65 and MyD88 proteins.
Identification of IRAK1 As a target gene of miR-615 in IECs
To explore the mechanism whereby miR-615 inhibits NF-κB activation, target gene prediction was conducted using miRanda3, PITA4, and RNAhybrid5, which identified 167 target genes (Supplementary Table S1). The most interesting candidate target gene was IRAK1 as it had the highest prediction score (174) and was enriched in the NF-κB pathway. Therefore, we hypothesized that IRAK1 is a target gene of miR-615.
Two reporter gene plasmids were constructed containing WT and seed region-Mut target sites (MuT) with matching or mutated target gene seed sites of miR-615 in the 3′-UTR of IRAK1 (Figure 6A). miR-615 inhibited the luciferase activity of the WT reporter plasmid when compared with that in the MC-transfected group; however, miR-615 did not repress the activity of the mutated dual-luciferase reporter gene plasmid (Figure 6B). Moreover, transfection with miR-615 mimics downregulated IRAK1 protein expression, whereas transfection with miR-615 inhibitors upregulated its expression (Figure 6C).
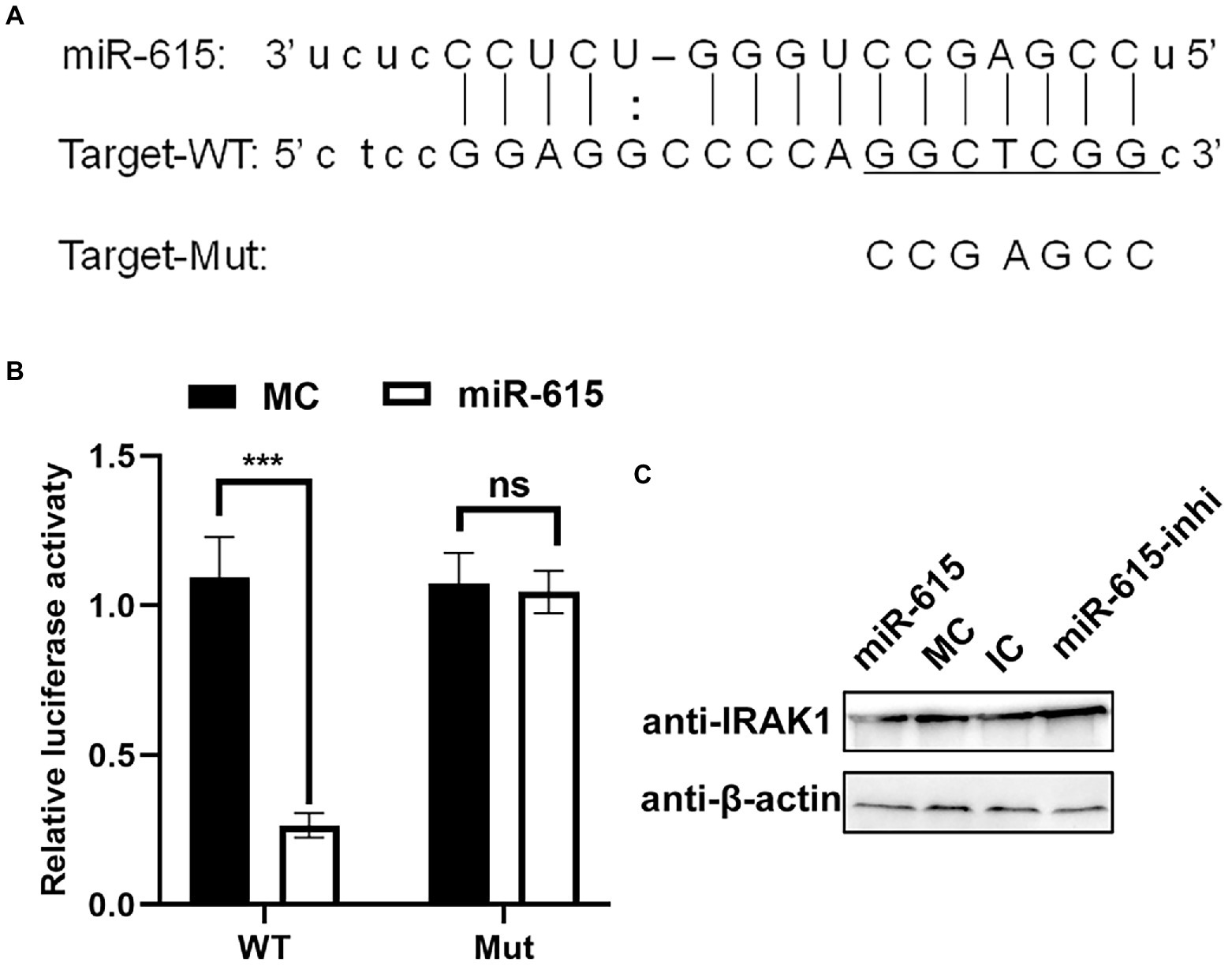
Figure 6. miR-615 targets IRAK1. (A) Bioinformatic prediction of the interactions between miR-615 and the 3′-UTR of swine IRAK1. For each schematic, the upper sequence is the sequence of mature miR-615, the middle sequence is the sequence in the binding site of miR-615 in the 3′-UTR of swine IRAK1, and the lower sequence is the mutated sequence of the IRAK1 3′-UTR. The seed sequence is underlined. (B) Luciferase activity in 293 T cells co-transfected with miR-615 mimics (or MC) and luciferase reporter gene plasmids containing the WT and Mut 3′-UTRs of IRAK1 for 48 h. Data are normalized against firefly luciferase activity. Comparisons between groups were determined using Student’s t-tests. ***p < 0.001, **p < 0.01. (C) The level of the IRAK1 protein during transfection with miR-615 mimics or inhibitors in IECs detected using western blotting during PEDV infection. Western blotting was conducted using anti-IRAK1 antibody at 24 h post-infection (hpi).
Mir-615 inhibits activation of the NF-κB pathway and promotes viral replication by targeting IRAK1
To determine whether repression of the NF-κB pathway by miR-615 is dependent on the regulation of IRAK1 expression, we first examined the effect of IRAK1 on the NF-κB pathway using a dual-luciferase reporter assay with overexpression and knockdown of IRAK1 in PEDV-infected IECs stimulated with poly (I:C). The NF-κB pathway was activated by IRAK1 overexpression (Figure 7A) and was downregulated by knockdown of IRAK1 (Figure 7B). In addition, IRAK1 overexpression upregulated IFN-λ1 and -λ3 expression during viral infection (Figures 7C,D).
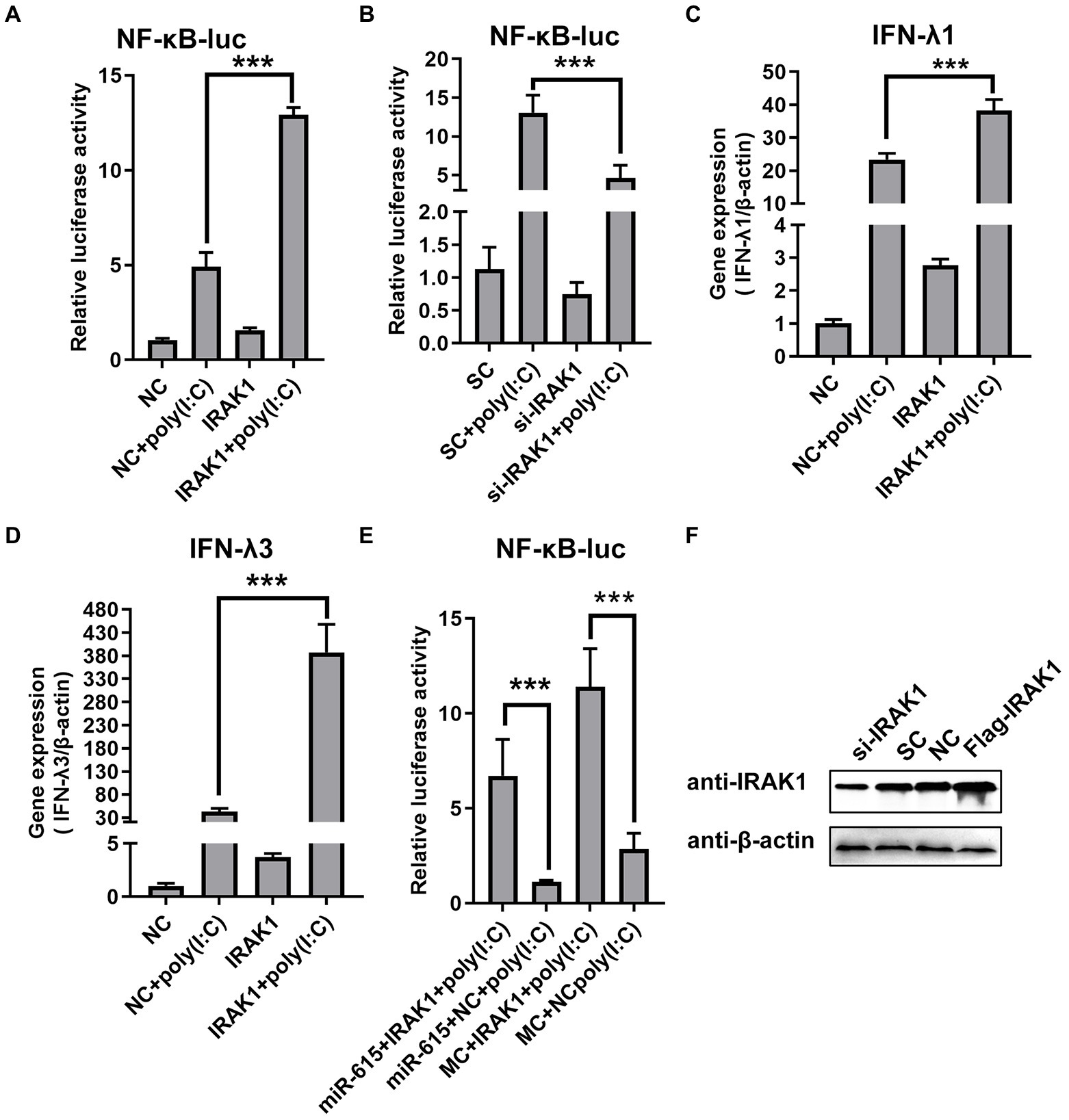
Figure 7. miR-615 inhibits NF-κB pathway activation by regulating IRAK1. (A) IRAK1-flag or negative control (NC) and (B) si-IRAK1 or siRNA control (SC) were co-transfected with pNiFty-luc and pRL-TK into IECs. Cells were then stimulated with poly (I:C) and infected with PEDV (MOI = 1) at 24 h post-infection (hpi) or left untreated (unstimulated). Cells were then collected for the dual-luciferase reporter assay. (C,D) IECs were transfected with IRAK1-flag (or NC) or si-IRAK1 (or SC) for 24 h. Poly (I:C) stimulation and viral infection were then induced. After 24 h, IECs were collected for RT-qPCR to determine (C) IFN-λ1 and (D) IFN-λ3 expression. (E) Rescue experiment. Luciferase activity was rescued by IRAK1 overexpression. IECs were co-transfected with miR-615 (or MC) and IRAK1-flag (or NC). At 24 hpi, poly (I:C) stimulation and viral infection were induced. After 24 h of stimulation, cells were collected and the luminescence activity was determined. (F) IRAK1 knockdown and overexpression were confirmed using western blotting. The data are representative of three independent experiments (mean ± SD). ***p < 0.001.
To further examine whether the effect of miR-615 on the NF-κB pathway was dependent on its regulation of IRAK1 expression, miR-615 and IRAK1 were overexpressed and the effects on the NF-κB pathway activity were examined. Porcine IECs were co-transfected with pNif-TK, pRL-TK, miR-615 mimics, or Flag-IRAK1, and poly (I:C) was used to induce NF-κB pathway activation. The rescue experiment showed that IRAK1 overexpression promoted NF-κB pathway activation and reversed the miR-615-dependent repression of the NF-κB pathway (Figure 7E). Collectively, these data indicate that miR-615 inhibits the NF-κB pathway by targeting IRAK1. The knockdown and overexpression of IRAK1 protein were further confirmed using western blotting (Figure 7F).
Discussion
PEDV infection has been reported to antagonize innate immunity (Annamalai et al., 2015; Cao et al., 2015a). The main factors involved in antiviral innate immunity are IFN-Is and -IIIs. Notably, IFN-IIIs are involved in innate immunity in IECs. Multiple PEDV-encoded proteins inhibit the production of IFNs (Ding et al., 2014; Zhang et al., 2017). miRNAs have been reported to play a pivotal role in the regulation of viral infections by targeting the viral genome or regulating host cytokines to modulate the cellular environment (Bartel, 2009). To further examine the mechanisms and interactions between miRNAs and PEDV in porcine IECs, deep-sequencing techniques were used to analyze miRNA expression profiles. We found that PEDV infection altered cellular miRNA expression profiles in IECs. Furthermore, the innate immunity pathway was confirmed to be involved in PEDV infection. miR-615 was enriched in this pathway, and its overexpression promoted viral replication by inhibiting IFN-III expression and targeting IRAK1 in the NF-κB pathway. To the best of our knowledge, this is the first report of an miRNA targeting IRAK1 during PEDV infection in IECs to function as a negative regulator of IFN-IIIs production.
PEDV infection has previously been shown to affect miRNA profiles and innate immunity (Huang et al., 2016; Zhang et al., 2018a; Qi et al., 2021). High-throughput sequencing of miRNAs in PEDV-infected IPEC-J2 cells revealed few identical differential miRNAs were expressed compared to our study. However, the KEGG pathway analysis showed that TLRs, Janus kinase-signal transducer and activator of transcription (JAK–STAT), retinoic acid-inducible gene I (RIG-I), and autophagy were involved in the response to PEDV infection. Our deep-sequencing KEGG analysis showed enrichment in the primary pathways involved in innate immunity (NF-κB and TLR signaling pathways; Zhang et al., 2018a). Another study using miRNA-mRNA high-throughput sequencing in PEDV-infected ST cells showed that innate immunity pathways were enriched following PEDV infection. Moreover, infection with different strains can induce the activation of different signaling pathways (Zhang et al., 2021). In PEDV-infected PK cells, high-throughput sequencing revealed few similar DEmiRNAs compared to those identified in other studies (Huang et al., 2016). However, PEDV-infected IECs (Cao et al., 2015b) showed activation of the NF-κB pathway. This may suggest that cells from the same source will have similar immune regulatory mechanisms after being infected by different PEDV strains. Our results thus provide insight into the mechanism of NF-κB pathway activation after PEDV infection of IECs. The inconsistency between our results and those of other studies may be attributed to different mechanisms of PEDV infection regulation in cells of different origins and the effects of different PEDV strains. Thus, our sequencing results provide insight on PEDV infection at the RNA level.
IFN-IIIs play a key role in antiviral innate immunity in the gut and at the mucosal surface. Compared with IFN-Is, IFN-IIIs preferentially inhibit PEDV infection in IECs (Li et al., 2017b). The robust activation of JAK–STAT signaling is induced to a greater degree by IFN-λ3 than by IFN-α. IFN-λ3 further plays a critical role in PEDV infection (Li et al., 2019). Similarly, IFN-λ1 exhibited strong anti-PEDV effects on IECs by activating the JAK–STAT signaling pathway. Furthermore, both IRF1 and NF-κB are related to PEDV-mediated IFN-IIIs suppression (Xue et al., 2018). In this study, we found that miR-615 downregulated IFN-λ1 and -λ3 expression by repressing the NF-κB pathway to facilitate PEDV replication. These results provide new insight into the mechanism by which IECs and PEDV interact through the NF-κB pathway. In addition, the transcription of IFN-III genes is more dependent on the NF-κB pathway than on the IRF system (Zhang et al., 2018b). This suggests that miR-615 is an important factor in cellular antiviral responses and an important anti-PEDV target. Notably, we did not identify changes in IFN-λ4 in IECs, which may be attributable to the low expression of IFN-λ4.
The inhibitory effect of miR-615 on the NF-κB pathway has been suggested in many other studies. In non-small cell lung cancer cells, miR-615-3p has been shown to be crucial in preventing cancer cell proliferation and metastasis by targeting insulin-like growth factor 2 (Liu et al., 2018). In breast cancer research, miR-615 was reported as a potential anti-onco-miR by targeting AKT serine/threonine kinase 2 expression (Bai et al., 2015). However, both the insulin-like growth factor 2 and AKT serine pathways could have an effect on the activity of the NF-κB pathway. In our study, miR-615 inhibited activation of the NF-κB pathway. This further suggests that miR-615 could exert its biological function by affecting the NF-κB pathway. It is worth noting that the NF-κB pathway is often activated in certain tumor cells, which further supports the role of miR-615 in affecting the NF-κB pathway. Finally, miR-615 promoted viral replication and inhibited the activation of the NF-κB pathway in both IECs and MARC-145 cells, suggesting the conserved role of miRNAs in different cells.
IRAK1 plays a critical role in TNF-α-induced NF-ĸB activation (Kim et al., 2012). The miRNAs miR-21, miR-146, miR-223, and miR-142a-3p have been reported to repress the NF-κB pathway by targeting IRAK1 (Chen et al., 2013; Hung et al., 2013; Xu et al., 2013). In this study, we strongly suggest that miR-615 inhibited activation of the NF-κB pathway by targeting IRAK1. This further demonstrated the key role of IRAK1 in NF-κB pathway activation and also supports that the same target gene can be regulated by multiple miRNAs.
Collectively, our study showed that PEDV infection affects the NF-κB pathway and other innate immune-related pathways by changing the miRNA profiles. Our data further revealed the mechanism by which PEDV infection inhibits the secretion of IFN-IIIs in IECs and provides a new perspective for understanding the function of miR-615. Furthermore, we provide novel information regarding the intricate interplay between PEDV and cellular innate immunity in IECs during PEDV infection, which may offer new targets for the development of effective therapies to control PEDV and other coronaviruses.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
Y-MZ and HH designed the study. H-QZ, X-FZ, W-XW, and CL performed the experiments and collected the data. B-YY, W-JZ, S-LF, and X-HY analyzed and interpreted the data. H-QZ and X-FZ wrote the draft of the manuscript. W-XW, S-LF, B-YY, and CL edited and revised the manuscript. Y-MZ and HH coordinated the whole project. All authors contributed to the article and approved the submitted version.
Funding
This research was funded in part by the Natural Science Basic Research Program of Shaanxi Province (grant number 2021JQ-900), Shaanxi Province Key Research and Development Project (grant number 2021NY-037), Research Fund Project of Xianyang Vocational and Technical College (2020KJA01 and 2018KYB01), Doctoral Research Foundation Project of Xianyang Vocational and Technical College (2021BK04 and 2019BK03), Key Research Projects of Xianyang Science, Technology Research and Development Program (grant number 2020 k02-63), and Innovative Research and Experimental Project of Young Scientific Researchers (grant number 2022012).
Acknowledgments
We would like to thank Editage (www.editage.cn) for English language editing.
Conflict of interest
Authors W-XW and HH were employed by Beijing Hemu Biotechnology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1071394/full#supplementary-material
Footnotes
2. ^https://bibiserv.cebitec.unibielefeld.de/rnahybrid/
3. ^http://www.microrna.org/microrna/hom.do
4. ^http://genie.weizmann.ac.il/pubs/mir07/mir07_dyn_data.html
References
Akira, S., Uematsu, S., and Takeuchi, O. (2006). Pathogen recognition and innate immunity. Cells 124, 783–801. doi: 10.1016/j.cell.2006.02.015
Annamalai, T., Saif, L. J., Lu, Z., and Jung, K. (2015). Age-dependent variation in innate immune responses to porcine epidemic diarrhea virus infection in suckling versus weaned pigs. Vet. Immunol. Immunopathol. 168, 193–202. doi: 10.1016/j.vetimm.2015.09.006
Bai, Y., Li, J., Li, J., Liu, Y., and Zhang, B. (2015). MiR-615 inhibited cell proliferation and cell cycle of human breast cancer cells by suppressing of AKT2 expression. Int. J. Clin. Exp. Med. 8, 3801–3808.
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cells 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Buggele, W. A., and Horvath, C. M. (2013). MicroRNA profiling of Sendai virus-infected A549 cells identifies miR-203 as an interferon-inducible regulator of IFIT1/ISG56. J. Virol. 87, 9260–9270. doi: 10.1128/jvi.01064-13
Cao, L., Ge, X., Gao, Y., Herrler, G., Ren, Y., Ren, X., et al. (2015a). Porcine epidemic diarrhea virus inhibits dsRNA-induced interferon-beta production in porcine intestinal epithelial cells by blockade of the RIG-I-mediated pathway. Virol. J. 12:127. doi: 10.1186/s12985-015-0345-x
Cao, L., Ge, X., Gao, Y., Ren, Y., Ren, X., and Li, G. (2015b). Porcine epidemic diarrhea virus infection induces NF-kappaB activation through the TLR2, TLR3 and TLR9 pathways in porcine intestinal epithelial cells. J. Gen. Virol. 96, 1757–1767. doi: 10.1099/vir.0.000133
Chen, Y., Chen, J., Wang, H., Shi, J., Wu, K., Liu, S., et al. (2013). HCV-induced miR-21 contributes to evasion of host immune system by targeting MyD88 and IRAK1. PLoS Pathog. 9:e1003248. doi: 10.1371/journal.ppat.1003248
Ding, Z., Fang, L., Jing, H., Zeng, S., Wang, D., Liu, L., et al. (2014). Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J. Virol. 88, 8936–8945. doi: 10.1128/jvi.00700-14
Dong, X., and Soong, L. (2021). Emerging and re-emerging zoonoses are major and global challenges for public health. Zoonoses 1:1. doi: 10.15212/ZOONOSES-2021-0001
Fisher, J. R., Chroust, Z. D., Onyoni, F., and Soong, L. (2021). Pattern recognition receptors in innate immunity to obligate intracellular bacteria. Zoonoses 1:10. doi: 10.15212/zoonoses-2021-0011
Hallman, J., Avesson, L., Reimegard, J., Kaller, M., and Soderbom, F. (2013). Identification and verification of microRNAs by high-throughput sequencing. Methods Mol. Biol. 983, 125–138. doi: 10.1007/978-1-62703-302-2_7
Hoffmann, H. H., Schneider, W. M., and Rice, C. M. (2015). Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol. 36, 124–138. doi: 10.1016/j.it.2015.01.004
Huang, J., Lang, Q., Li, X., Xu, Z., Zhu, L., and Zhou, Y. (2016). MicroRNA expression profiles of porcine kidney 15 cell line infected with porcine epidemic diahorrea virus. Bing Du Xue Bao 32, 465–471.
Huang, Y., Wang, W., Xu, Z., Pan, J., Zhao, Z., and Ren, Q. (2018). Eriocheir sinensis microRNA-7 targets crab Myd88 to enhance white spot syndrome virus replication. Fish Shellfish Immunol. 79, 274–283. doi: 10.1016/j.fsi.2018.05.028
Hung, P. S., Liu, C. J., Chou, C. S., Kao, S. Y., Yang, C. C., Chang, K. W., et al. (2013). miR-146a enhances the oncogenicity of oral carcinoma by concomitant targeting of the IRAK1, TRAF6 and NUMB genes. PLoS One 8:e79926. doi: 10.1371/journal.pone.0079926
Jegaskanda, S., Vanderven, H. A., Tan, H. X., Alcantara, S., Wragg, K., Parsons, M. S., et al. (2018). Influenza infection enhances antibody-mediated NK cell functions via type I interferon dependent pathways. J. Virol. 93, e02090–e02018. doi: 10.1128/jvi.02090-18
John, B., Enright, A. J., Aravin, A., Tuschl, T., Sander, C., and Marks, D. S. (2004). Human MicroRNA targets. PLoS Biol. 2, e363–e1879. doi: 10.1371/journal.pbio.0020363
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. doi: 10.1093/nar/gkm882
Kim, J. M., Cho, H. H., Lee, S. Y., Hong, C. P., Yang, J., Kim, Y. S., et al. (2012). Role of IRAK1 on TNF-induced proliferation and NF-kB activation in human bone marrow mesenchymal stem cells. Cell. Physiol. Biochem. 30, 49–60. doi: 10.1159/000339045
Li, L., Fu, F., Xue, M., Chen, W., Liu, J., Shi, H., et al. (2017b). IFN-lambda preferably inhibits PEDV infection of porcine intestinal epithelial cells compared with IFN-alpha. Antivir. Res. 140, 76–82. doi: 10.1016/j.antiviral.2017.01.012
Li, W., Li, H., Liu, Y., Pan, Y., Deng, F., Song, Y., et al. (2012). New variants of porcine epidemic diarrhea virus, China, 2011. Emerg. Infect. Dis. 18, 1350–1353. doi: 10.3201/eid1808.120002
Li, S. W., Wang, C. Y., Jou, Y. J., Huang, S. H., Hsiao, L. H., Wan, L., et al. (2016). SARS coronavirus papain-like protease inhibits the TLR7 signaling pathway through removing Lys63-linked polyubiquitination of TRAF3 and TRAF6. Int. J. Mol. Sci. 17:678. doi: 10.3390/ijms17050678
Li, L., Xue, M., Fu, F., Yin, L., Feng, L., and Liu, P. (2019). IFN-lambda 3 mediates antiviral protection against porcine epidemic diarrhea virus by inducing a distinct antiviral transcript profile in porcine intestinal epithelia. Front. Immunol. 10:2394. doi: 10.3389/fimmu.2019.02394
Li, G., Zhang, W., Gong, L., and Huang, X. (2017a). MicroRNA-125a-5p inhibits cell proliferation and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma by downregulation of ErbB3. Oncol. Res. 27, 449–458. doi: 10.3727/096504017x15016337254623
Lin, C. M., Annamalai, T., Liu, X., Gao, X., Lu, Z., El-Tholoth, M., et al. (2015). Experimental infection of a US spike-insertion deletion porcine epidemic diarrhea virus in conventional nursing piglets and cross-protection to the original US PEDV infection. Vet. Res. 46:134. doi: 10.1186/s13567-015-0278-9
Liu, J., Jia, Y., Jia, L., Li, T., Yang, L., and Zhang, G. (2018). MicroRNA-615-3p inhibits the tumor growth and metastasis of NSCLC via inhibiting IGF2. Oncol. Res. 27, 269–279. doi: 10.3727/096504018x15215019227688
Lui, P. Y., Wong, L. Y., Fung, C. L., Siu, K. L., Yeung, M. L., Yuen, K. S., et al. (2016). Middle East respiratory syndrome coronavirus M protein suppresses type I interferon expression through the inhibition of TBK1-dependent phosphorylation of IRF3. Emerg. Microbes Infect. 5:e39, 1–9. doi: 10.1038/emi.2016.33
Ma, Y., Wang, C., Xue, M., Fu, F., Zhang, X., Li, L., et al. (2018). The coronavirus transmissible gastroenteritis virus evades the type I interferon response through IRE1alpha-mediated manipulation of the microRNA miR-30a-5p/SOCS1/3 axis. J. Virol. 92:e00728-18. doi: 10.1128/jvi.00728-18
Pu, J., Wu, S., Xie, H., Li, Y., Yang, Z., Wu, X., et al. (2017). miR-146a inhibits dengue-virus-induced autophagy by targeting TRAF6. Arch. Virol. 162, 3645–3659. doi: 10.1007/s00705-017-3516-9
Qi, X., Cao, Y., Wu, S., Wu, Z., and Bao, W. (2021). miR-129a-3p inhibits PEDV replication by targeting the EDA-mediated NF-κB pathway in IPEC-J2 cells. Int. J. Mol. Sci. 22:8133. doi: 10.3390/ijms22158133
Song, X., Zhao, X., Huang, Y., Xiang, H., Zhang, W., and Tong, D. (2015). Transmissible gastroenteritis virus (TGEV) infection alters the expression of cellular microRNA species that affect transcription of TGEV gene 7. Int. J. Biol. Sci. 11, 913–922. doi: 10.7150/ijbs.11585
Wang, D., Fang, L., Shi, Y., Zhang, H., Gao, L., Peng, G., et al. (2016). Porcine epidemic diarrhea virus 3C-like protease regulates its interferon antagonism by cleaving NEMO. J. Virol. 90, 2090–2101. doi: 10.1128/jvi.02514-15
Wang, J., Hu, G., Gao, W., Xu, L., Ning, P., and Zhang, Y. (2014). Immortalized porcine intestinal epithelial cell cultures susceptible to porcine rotavirus infection. J. Virol. Methods 202, 87–94. doi: 10.1016/j.jviromet.2014.03.007
Wu, S., He, L., Li, Y., Wang, T., Feng, L., Jiang, L., et al. (2013). miR-146a facilitates replication of dengue virus by dampening interferon induction by targeting TRAF6. J. Infect. 67, 329–341. doi: 10.1016/j.jinf.2013.05.003
Xu, G., Zhang, Z., Wei, J., Zhang, Y., Zhang, Y., Guo, L., et al. (2013). microR-142-3p down-regulates IRAK-1 in response to Mycobacterium bovis BCG infection in macrophages. Tuberculosis 93, 606–611. doi: 10.1016/j.tube.2013.08.006
Xue, M., Fu, F., Ma, Y., Zhang, X., Li, L., Feng, L., et al. (2018). The PERK arm of the unfolded protein response negatively regulates transmissible gastroenteritis virus replication by suppressing protein translation and promoting type I interferon production. J. Virol. 92, e00431–e00418. doi: 10.1128/jvi.00431-18
Zhang, C., Chen, J., Wen, J., and Liu, G. (2018a). Analysis and verification of miRNA expression profiles of PEDV infected IPEC-J2 and effect of novel-miR-877 and ssc-miR-219a on PEDV replication. Chin. J. Prevent Vet. Med. 40, 1095–1099.
Zhang, Q., Ke, H., Blikslager, A., Fujita, T., and Yoo, D. (2018b). Type III interferon restriction by porcine epidemic diarrhea virus and the role of viral protein nsp1 in IRF1 signaling. J. Virol. 92, e01677–e01617. doi: 10.1128/jvi.01677-17
Zhang, X., Li, C., Zhang, B., Li, Z., Zeng, W., Luo, R., et al. (2021). Differential expression and correlation analysis of miRNA-mRNA profiles in swine testicular cells infected with porcine epidemic diarrhea virus. Sci. Rep. 11:1868. doi: 10.1038/s41598-021-81189-5
Zhang, Q., Ma, J., and Yoo, D. (2017). Inhibition of NF-kappaB activity by the porcine epidemic diarrhea virus nonstructural protein 1 for innate immune evasion. Virology 510, 111–126. doi: 10.1016/j.virol.2017.07.009
Zhang, Q., Shi, K., and Yoo, D. (2016). Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology 489, 252–268. doi: 10.1016/j.virol.2015.12.010
Zhao, P., Zhao, L., Zhang, K., Feng, H., Wang, H., Wang, T., et al. (2012). Infection with street strain rabies virus induces modulation of the microRNA profile of the mouse brain. Virol. J. 9:159. doi: 10.1186/1743-422x-9-159
Keywords: miR-615, IFN, innate immunity, porcine epidemic diarrhea virus, intestinal epithelial cells, miRNA high-throughput, NF-kappa B, IRAK1
Citation: Zheng H-q, Li C, Zhu X-f, Wang W-X, Yin B-y, Zhang W-j, Feng S-l, Yin X-h, Huang H and Zhang Y-m (2022) miR-615 facilitates porcine epidemic diarrhea virus replication by targeting IRAK1 to inhibit type III interferon expression. Front. Microbiol. 13:1071394. doi: 10.3389/fmicb.2022.1071394
Edited by:
Jian Shang, Zhengzhou University, ChinaReviewed by:
Xin Yin, Harbin Veterinary Research Institute (CAAS), ChinaBenjie Chai, Tsinghua University, China
Weili Kong, Gladstone Institutes, United States
Copyright © 2022 Zheng, Li, Zhu, Wang, Yin, Zhang, Feng, Yin, Huang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: He Huang, aHVhbmdoZUBoZW11YmlvdGVjaC5jb20=; Yan-ming Zhang, emhhbmd5bUBud2FmdS5lZHUuY24=
†These authors have contributed equally to this work
 Hong-qing Zheng1,3†
Hong-qing Zheng1,3† Wei-Xiao Wang
Wei-Xiao Wang Yan-ming Zhang
Yan-ming Zhang