
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 25 November 2022
Sec. Microbiological Chemistry and Geomicrobiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1070601
 Jinzhi Guo1†
Jinzhi Guo1† Jie Jiang1†
Jie Jiang1† Zhaofeng Peng1*
Zhaofeng Peng1* Yuhong Zhong1
Yuhong Zhong1 Yongguang Jiang1
Yongguang Jiang1 Zhou Jiang1,2
Zhou Jiang1,2 Yidan Hu1
Yidan Hu1 Yiran Dong1,2,3,4
Yiran Dong1,2,3,4 Liang Shi1,2,3,4*
Liang Shi1,2,3,4*The γ-proteobacterium Shewanella oneidensis MR-1 reduces iodate to iodide extracellularly. Both dmsEFAB and mtrCAB gene clusters are involved in extracellular reduction of iodate by S. oneidensis MR-1. DmsEFAB reduces iodate to hypoiodous acid and hydrogen peroxide (H2O2). Subsequently, H2O2 is reduced by MtrCAB to facilitate DmsEFAB-mediated extracellular reduction of iodate. To investigate the distribution of bacteria with the capability for extracellular reduction of iodate, bacterial genomes were systematically searched for both dmsEFAB and mtrCAB gene clusters. The dmsEFAB and mtrCAB gene clusters were found in three Ferrimonas and 26 Shewanella species. Coexistence of both dmsEFAB and mtrCAB gene clusters in these bacteria suggests their potentials for extracellular reduction of iodate. Further analyses demonstrated that these bacteria were isolated from a variety of ecosystems, including the lakes, rivers, and subsurface rocks in East and Southeast Asia, North Africa, and North America. Importantly, most of the bacteria with both dmsEFAB and mtrCAB gene clusters were found in different marine environments, which ranged from the Arctic Ocean to Antarctic coastal marine environments as well as from the Atlantic Ocean to the Indian and Pacific Oceans. Widespread distribution of the bacteria with capability for extracellular reduction of iodate around the world suggests their significant importance in global biogeochemical cycling of iodine. The genetic organization of dmsEFAB and mtrCAB gene clusters also varied substantially. The identified mtrCAB gene clusters often contained additional genes for multiheme c-type cytochromes. The numbers of dmsEFAB gene cluster detected in a given bacterial genome ranged from one to six. In latter, duplications of dmsEFAB gene clusters occurred. These results suggest different paths for these bacteria to acquire their capability for extracellular reduction of iodate.
Iodine (I) is a trace element of both health and environmental significance. As iodine is a crucial component of human thyroid hormones triiodothyronine and thyroxine, iodine deficiency disorders of humans (e.g., goiter and cretinism) are attributed to insufficient intake of iodine, such as drinking of the groundwater with low iodine level (Laurberg et al., 2001; Zimmermann, 2009; Zhang E. et al., 2013; Duan et al., 2016). Excessive intake of iodine, such as drinking of the groundwater with high iodine level, also results in thyroiditis and probably cancer (Laurberg et al., 2001; Andersen et al., 2009; Zhang E. et al., 2013; Duan et al., 2016). Thus, abnormal level of iodine in groundwater affects human health (Wen et al., 2013; Zhang E. et al., 2013; Duan et al., 2016). Furthermore, radioactive iodine-129 (129I) is an important risk driver of the Hanford and Savannah River Sites in Washington and South Carolina States, respectively, United States, where 129I level in groundwater is higher than that for the drinking water standard (Otosaka et al., 2011; Zhang S. et al., 2013; Kaplan et al., 2014). Finally, global cycling of iodine impacts air quality and climate (Carpenter et al., 2021).
In environment, iodate (IO3−) and iodide (I−) are the two dominant species of inorganic iodine. For example, IO3− is the major iodine species found in groundwater of the Hanford and Savannah River Sites (Otosaka et al., 2011; Zhang S. et al., 2013), while the dominant species found in the groundwater in Datong and Taiyuan Basins, China and North China Plains is I− (Li et al., 2013; Tang et al., 2013; Zhang E. et al., 2013). Both IO3−and I− are found in oceans where their combined concentration is 0.4–0.5 μM (Chance et al., 2014).
Microorganisms play crucial roles in redox transformation of IO3−and I− in environments. I−-oxidizing microorganisms oxidize I− to molecular iodine (I2), while IO3−-reducing microorganisms reduce IO3− to I− (Iino et al., 2016; Yamazaki et al., 2020; Reyes-Umana et al., 2022). The enzymes involved in microbial oxidation of I− to I2 include the extracellular multicopper oxidase LoxA (Suzuki et al., 2012; Shiroyama et al., 2015), while those involved in microbial reduction of IO3− to I− include two different types of enzymes (Yamazaki et al., 2020; Guo et al., 2022; Reyes-Umana et al., 2022; Shin et al., 2022).
IdrABP1P2 of the dissimilatory IO3−-reducing bacterium Pseudomonas sp. strain SCT is the first enzyme identified for IO3− reduction (Yamazaki et al., 2020). This enzyme consists of four subunits, in which IdrA is suggested to be the catalytic subunit, IdrB is the electron transfer subunit and IdrP1 and IdrP2 are the detoxification subunits. All of these subunits are believed to be localized in the periplasm. During IO3− reduction, IdrB is proposed to receive electrons from cytochrome c (Cyt-c) in the periplasm and then transfers the electrons to IdrA. IdrA is suggested to use the electrons to reduce IO3− to hypoiodous acid (HIO) and hydrogen peroxide (H2O2). The generated H2O2 is proposed to be reduced to H2O by IdrP1 and IdrP2. Cyt-c may also supply electrons to IdrP1 and IdrP2. HIO is suggested to be further reduced to I− probably by Cld (Yamazaki et al., 2020). Based on their polypeptide sequences, IdrA and IdrB are the homologs of dimethylsulfoxide (DMSO) reductase DmsA and DmsB, respectively. IdrP1 and IdrP2 are the c-type cytochromes (c-Cyt) that are peroxidases. The genes for IdrABP1P2 are clustered together in the genome of Pseudomonas sp. strain SCT (Yamazaki et al., 2020).
The idrABP1P2 gene cluster also exists in the genome of the dissimilatory IO3−-reducing bacterium Denitromonas sp. IR-12 (Reyes-Umana et al., 2022). Deletion of idrA gene impairs bacterial ability to grow with IO3− as the sole terminal electron acceptor. The proposed functions of IdrABP1P2 from Denitromonas sp. IR-12 in IO3− reduction are the same to those proposed for the IdrABP1P2 of Pseudomonas sp. strain SCT except that HIO is proposed to be disproportionate to IO3− and I−. The IO3− is further reduced by IdrAB (Reyes-Umana et al., 2022). Furthermore, microorganisms with idrABP1P2 gene cluster are widespread in oceans, suggesting their global significance in biogeochemical cycling of iodine (Reyes-Umana et al., 2022).
The dissimilatory metal-reducing bacterium Shewanella oneidensis MR-1 reduces IO3− extracellularly via DmsEFAB and MtrCAB (Mok et al., 2018; Toporek et al., 2019; Guo et al., 2022; Shin et al., 2022). The DmsEFAB is the first enzyme demonstrated experimentally for reducing IO3− to HIO and H2O2 (Guo et al., 2022). Correspondingly, MtrCAB is also confirmed experimentally to reduce H2O2 to H2O for facilitating DmsEFAB-mediated IO3− reduction. The Mtr extracellular electron transfer pathway is suggested to transfer electrons from the cytoplasmic membrane and across the periplasm to the DmsEF and MtrAB in the outer membrane. DmsEF and MtrAB transfer electrons across the outer membrane to DmsAB and MtrC, respectively. On bacterial surface, DmsAB and MtrC work collaboratively to reduce IO3− (Guo et al., 2022).
Unlike Pseudomonas sp. strain SCT and Denitromonas sp. IR-12, S. oneidensis MR-1 is not an IO3−-respiring bacterium (Toporek et al., 2019). In S. oneidensis MR-1, DmsEFAB and MtrCAB are for extracellular respiration of DMSO and ferric iron [Fe(III)]-containing minerals, respectively (Beliaev and Saffarini, 1998; Beliaev et al., 2001; Gralnick et al., 2006; Bretschger et al., 2007; Coursolle and Gralnick, 2010). It is believed that their extracellular localization enables DmsEFAB and MtrCAB to reduce IO3− collaboratively and extracellular reduction of IO3− by S. oneidensis MR-1 is a fortuitous function (Guo et al., 2022). The non-IO3− -respiring bacteria with the capability for extracellular reduction of IO3− are also believed to impact the fate, transport, and global biogeochemical cycling of iodine (Guo et al., 2022). However, to what extent the bacteria with the capability for extracellular reduction of IO3− distribute around the world has never been investigated before.
Like idrABP1P2 gene cluster, the genes for DmsEFAB and MtrCAB are also clustered together, respectively (Gralnick et al., 2006; Fredrickson et al., 2008; Wang et al., 2008). In this investigation, we searched bacterial genomes for dmsEFAB and mtrCAB gene clusters and found that the bacteria with both dmsEFAB and mtrCAB gene clusters were widespread, showing global distribution of the bacteria possessing capability for extracellular reduction of IO3−. Worldwide occurrence of the bacteria with capability for extracellular reduction of IO3− suggests the importance of these bacteria in global biogeochemical cycling of iodine.
DmsE and DmsF are homologous to MtrA and MtrB, respectively (Gralnick et al., 2006). Thus, we searched microbial genomes for DmsE/MtrA and DmsF/MtrB homologs by the approach that was described before (Shi et al., 2012a, 2014; Zhong and Shi, 2018). The DmsE, DmsF, MtrA, and MtrB of S. oneidensis MR-1served as templates for identifying microbial open reading frames (ORFs) whose protein sequences shared similarity to the templates by BLAST programs of the National Center for Biotechnology Information (NCBI) and of the Universal Protein Resource (UniProt; E < 0.01; Altschul et al., 1990), in which scoring matrix = BLOSUM62, gapopen = 0, gapextend = 0 and databases = non-redundant protein sequences database (nr) and UniprotKB database. The in-house Perl scripts and a hidden Markov model-based PRED-TMBB software were used to verify identified homologs with the CX2CH motifs and the trans-outer membrane motifs, respectively (Bagos et al., 2004;Shi et al., 2012b; Shi et al., 2014; Zhong and Shi, 2018). After verification, the identified homologs served as the templates for next round of genome search. The polypeptide sequences from the genes immediately upstream and downstream of the identified dmsEF/mtrAB gene clusters were further compared with previously identified DmsA, DmsB, and MtrC. The identified dmsEFAB gene clusters and the mtrCAB gene clusters co-existed with dmsEFAB gene clusters in the same bacterial genome were subjected to the additional analyses.
Clustal W (version 2.1) was used to align the polypeptide sequences identified. The parameters used were Gap Opening Penalty = 10; Gap Extension Penalty = 0.2; Protein matrix = BLOSUM series (Larkin et al., 2007). MEGA7 was used to analyze the aligned sequences of DmsA, DmsB, DmsE/MtrA, DmsF/MtrB, or MtrC homologs. Phylogenetic trees were constructed with Maximum Likelihood at a confidence level determined by 1,000 bootstrap replications (Kumar et al., 2016). The results of phylogenetic reconstruction were displayed with Evolview v2 (He et al., 2016). The genes for c-Cyts on the upstream and downstream of the mtrCAB gene clusters were also identified by the method described above (Shi et al., 2012b, 2014; Zhong and Shi, 2018). The map of distribution for the identified bacteria was constructed similarly to that described previously (Baker et al., 2022).
DmsE/MtrA is a decaheme c-Cyt that is inserted into the outer membrane porin protein DmsF/MtrB. The function of DmsF/MtrB is to insulate DmsE/MtrA from the outer membrane, which permits rapid electron transfer across the outer membrane by DmsE/MtrA (Hartshorne et al., 2009; White et al., 2013; Edwards et al., 2020). Thus, the identified microorganisms that possessed dmsEFAB and/or mtrCAB gene clusters were all the Gram-negative bacteria. Supplementary Table S1 listed the bacteria identified with both dmsEFAB and mtrCAB gene clusters from this investigation. These included three Ferrimonas and 26 Shewanella species. It should be noted that in S. oneidensis MR-1, dmsEFAB and mtrCAB gene clusters are for extracellular respiration of DMSO and Fe(III)-containing minerals, respectively (Beliaev and Saffarini, 1998; Beliaev et al., 2001; Gralnick et al., 2006; Bretschger et al., 2007; Coursolle et al., 2010). Involvement of dmsEFAB and mtrCAB gene clusters in extracellular reduction of IO3− is fortuitous (Guo et al., 2022). Restrictive distribution of the bacteria with capability for extracellular reduction of IO3− in Ferrimonas and Shewanella species may be attributed to this fortuitous function.
Possession of both dmsEFAB and mtrCAB gene clusters suggests that these bacteria are capable of reducing IO3− extracellularly. This is consistent with the previous observations that in addition to S. oneidensis MR-1, other Shewanella species, such as S. putrefaciens, reduced IO3− (Councell et al., 1997; Farrenkorf et al., 1997; Mok et al., 2018; Toporek et al., 2019; Guo et al., 2022; Shin et al., 2022).
The original habitats for this group of bacteria with both dmsEFAB and mtrCAB gene clusters varied substantially (Figure 1; Supplementary Table S1). Some of them were isolated from the sediments of the lakes located in China, India, and the United States (Myers and Nealson, 1988; Li et al., 2014; Rathour et al., 2021); city drainage in Vietnam (Dao et al., 2022); subsurface rock in United States (Fredrickson et al., 1998), river water in Tunisia and rainbow trout in South Korea (Figure 1; Supplementary Table S1). Notably, >82% of the identified bacteria with dmsEFAB and mtrCAB gene clusters were isolated from a variety of marine environments around the world. These included the costal sediments in Mallorca, Spain; Xiamen, China, and Nova Scotia, Canada (Rossello-Mora et al., 1995; Zhao et al., 2005; Huang et al., 2010); sediments in Ross Sea, South China Sea, and Arctic Ocean (Ivanova et al., 2003; Hwang et al., 2019; Li et al., 2021); a cold seep field in South China Sea (Figure 1; Supplementary Table S1) and deep-sea sediments in Southwest Indian Ocean and West Pacific Ocean (Wang et al., 2004, 2021; Yu et al., 2021). Some were also found in seawater in East Sea, Korea; North Sea, United Kingdom; Troitsa Bay, Russia; Alboran Sea and Black Sea (Makemson et al., 1997; Reid and Gordon, 1999; Venkateswaran et al., 1999; Kim et al., 2017; Bae et al., 2021) as well as sea ice floe close to Point Barrow, Alaska, United States (Figure 1; Supplementary Table S1); the gastric cavity of galaxy coral in the coastal area near Hainan Island, China (Tang et al., 2020), and the intestines of sea animals in Japan (Satomi et al., 2003). Thus, bacteria with both dmsEFAB and mtrCAB gene clusters occur globally. Distribution of the bacteria with dmsEFAB and mtrCAB gene clusters is also comparable to the distribution of bacteria with porin-cytochrome gene clusters (Baker et al., 2022). Widespread occurrence of these bacteria around the world, especially their distribution in a variety of marine environments, suggests significant importance of the bacteria with capability for extracellular reduction of IO3− in global biogeochemical cycling of iodine.
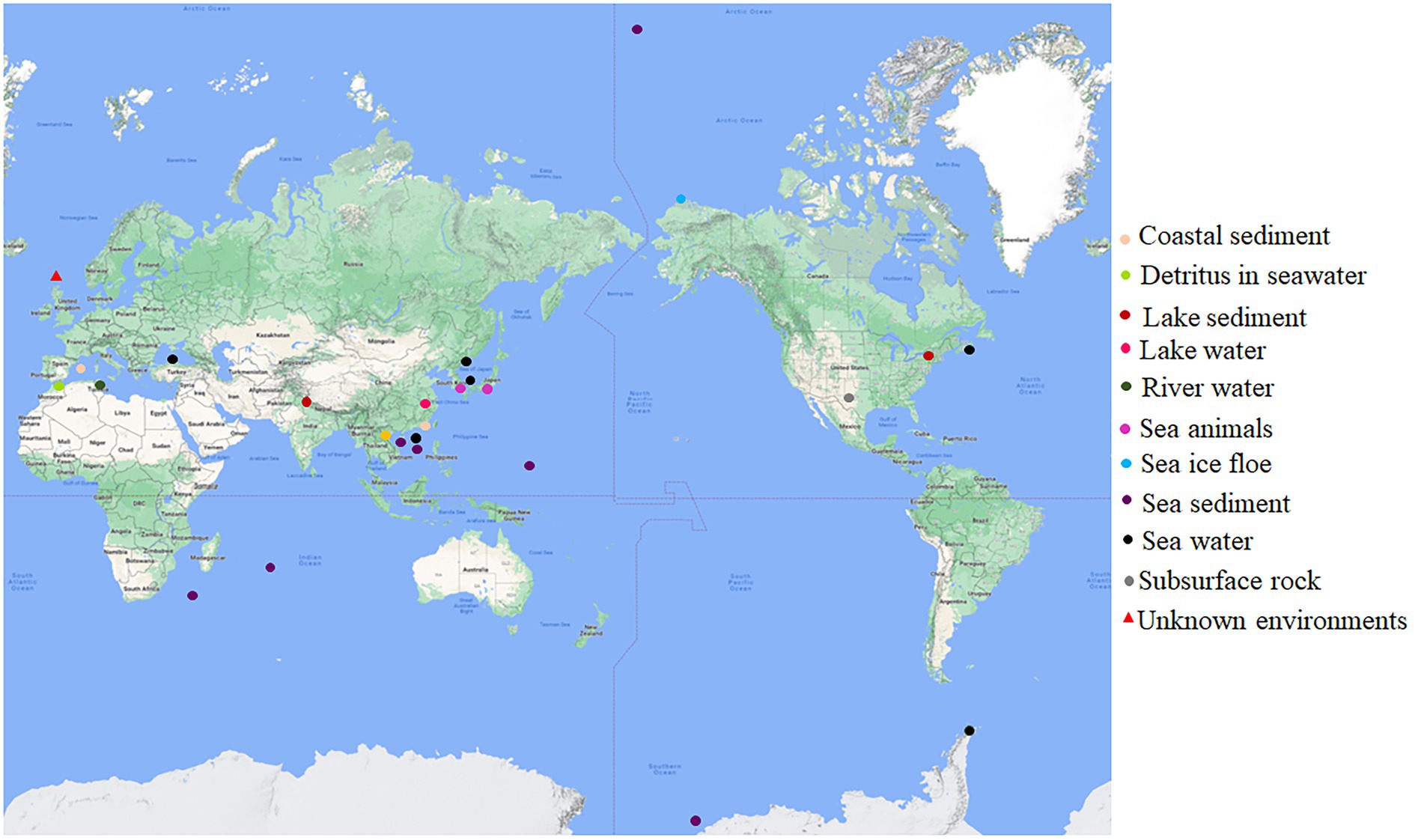
Figure 1. Global distribution of the bacteria with capability for extracellular reduction of iodate. Their original habits are indicated. See Supplementary Table S1 for details.
A total of 52 DmsA homologs were identified from the bacteria with dmsEFAB and mtrCAB gene clusters (Figure 2A; Supplementary Table S2). For comparison, IdrA of Pseudomonas sp. strain SCT and Denitromonas sp. IR-12 were included for phylogenetic analyses (Yamazaki et al., 2020; Reyes-Umana et al., 2022). Like DmsA and SO_4358 of S. oneidensis MR-1, all the identified DmsA homologs possessed the twin-arginine sequence at their N-termini for their secretion to the periplasm via the twin-arginine protein secretion system (Supplementary Figure S1; Gralnick et al., 2006). The identified DmsA homologs were 35–98% identical to DmsA of S. oneidensis MR-1. Notably, SO_4358 of S. oneidensis MR-1 was 36% identical to DmsA of S. oneidensis MR-1 (Supplementary Table S2). SO_4358 is not involved in extracellular reduction of iodate by S. oneidensis MR-1 (Guo et al., 2022). The identified DmsA homologs were distantly related to IdrA of Pseudomonas sp. strain SCT and Denitromonas sp. IR-12 (Figure 2A; Supplementary Table S2).
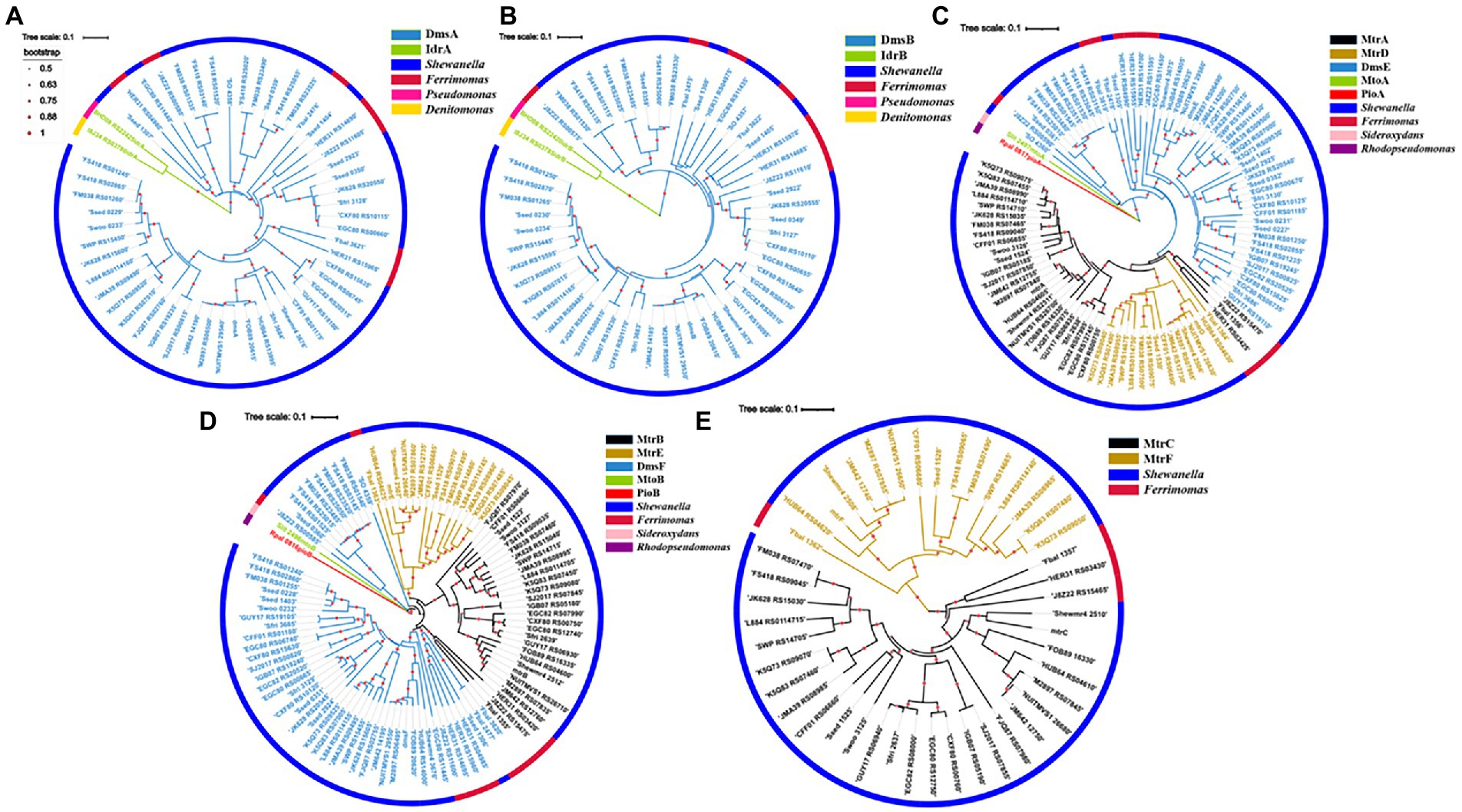
Figure 2. Phylogenetic analyses of identified DmsEFAB and MtrCAB homologs. (A) DmsA homologs, (B) DmsB homologs, (C) DmsE/MtrA/MtrD homologs, (D) DmsF/MtrB/MtrE homologs and (E). MtrC/MtrF homologs. Construction and graphical display of phylogenetic trees were described in text. The outer layer indicates bacterial genera with identified DmsEFAB and MtrCAB homologs. For comparison, IdrAB of Pseudomonas sp. strain SCT and Denitromonas sp. IR-12, MtoAB of Sideroxydans lithotrophicus ES-1, and PioAB of Rhodopseudomonas palustrisTIE-1 were included for the analyses.
The twin-arginine sequence was also detected in the DmsB homologs identified (Supplementary Figure S2; Gralnick et al., 2006). These DmsB homologs were 52%–99% identical to DmsB of S. oneidensis MR-1. Among them, SO_4359 of S. oneidensis MR-1 was 61% identical to DmsB of S. oneidensis MR-1 (Supplementary Table S3). The identified DmsB homologs were 11%–24% identical to IdrB of Pseudomonas sp. strain SCT and Denitromonas sp. IR-12 (Figure 2B; Supplementary Table S3).
In S. oneidensis MR-1, MtrDEF are homologous to MtrABC, respectively, and mtrDEF genes are clustered together and are part of mtrCAB gene cluster (Fredrickson et al., 2008). However, the mtrDEF gene cluster is not involved in extracellular reduction of IO3− by S. oneidensis MR-1 (Guo et al., 2022). Ninety-seven DmsE/MtrA/MtrD and DmsF/MtrB/MtrE homologs were identified, respectively, from the bacteria with dmsEFAB and mtrCAB gene cluster (Figures 2C,D; Supplementary Tables S4, S5). Identified DmsE/MtrA/MtrD homologs were 47%–98% identical to DmsE/MtrA/MtrD of S. oneidensis MR-1 (Supplementary Table S4), respectively; while the identified DmsF/MtrB/MtrE homologs were 24%–96% identical to DmsF/MtrB/MtrE of S. oneidensis MR-1, respectively (Supplementary Table S5). These DmsE/MtrA/MtrD and DmsF/MtrB/MtrE homologs were 32%–43% and 14%–23% identical to MtoA/PioA and MtoB/PioB of the Fe(II)-oxidizing bacteria Sideroxydans lithotrophicus ES-1 and Rhodopseudomonas palustrisTIE-1, respectively (Figures 2C,D; Supplementary Tables S4, S5; Jiao et al., 2005; Jiao and Newman, 2007; Liu et al., 2012; Shi et al., 2012b; Liu et al., 2013). Previous results showed that MtoA/PioA and MtoB/PioB of S. lithotrophicus ES-1 and R. palustrisTIE-1were homologous to MtrA and MtrB of S. oneidensis MR-1, respectively (Jiao and Newman, 2007; Shi et al., 2012b; Liu et al., 2014). Purified MtoA of S. lithotrophicus ES-1 was capable of oxidizing Fe(II), including the solid phase Fe(II) (Liu et al., 2012, 2013). PioA and PioB were also involved in extracellular oxidation of Fe(II) by R. palustrisTIE-1 (Jiao and Newman, 2007). Given that they are more homologous to DmsE/MtrA and DmsF/MtrB of S. oneidensis MR-1 than to MtoA/PioA and MtoB/PioB of S. lithotrophicus ES-1 and R. palustrisTIE-1, the identified DmsE/MtrA/MtrD and DmsF/MtrB/MtrE homologs are most likely to mediate electron transfer from inside cells to outside cells.
Forty-five MtrC/MtrF homologs were identified from the bacteria with dmsEFAB and mtrCAB gene clusters (Figure 2E; Supplementary Figure S3; Supplementary Table S6). These homologs were 29%–71% identical to MtrC of S. oneidensis MR-1 (Supplementary Table S6). MtrC of S. oneidensis MR-1is a lipoprotein with 10 c-type hemes and is located on the bacterial surface (Shi et al., 2006, 2008; Lower et al., 2009; Edwards et al., 2020). It contains a lipid-binding site in its N-terminus and replacement of this site renders MtrC unable to bind to the outer membrane (Edwards et al., 2015). Similar to MtrC of S. oneidensis MR-1, all identified homologs possessed this lipid-binding site in their N-termini (Supplementary Figure S3). Thus, these MtrC/MtrF homologs are most likely on the bacterial cell surface. All the identified MtrC/MtrF homologs also possessed 10 c-type heme-binding sites (Supplementary Figure S3). Similar to MtrC and other c-Cyts, these MtrC homologs should possess intrinsic peroxidase activity to degrade the H2O2 formed from extracellular reduction of IO3− (Thomas et al., 1976; Shi et al., 2006; Guo et al., 2022).
Further investigation revealed that the numbers of dmsEFAB gene cluster and genes associated with mtrCAB gene cluster varied among the genomes of identified bacteria. Because of these differences, the identified bacteria could be categorized into seven different groups (Supplementary Table S1; Figure 3).
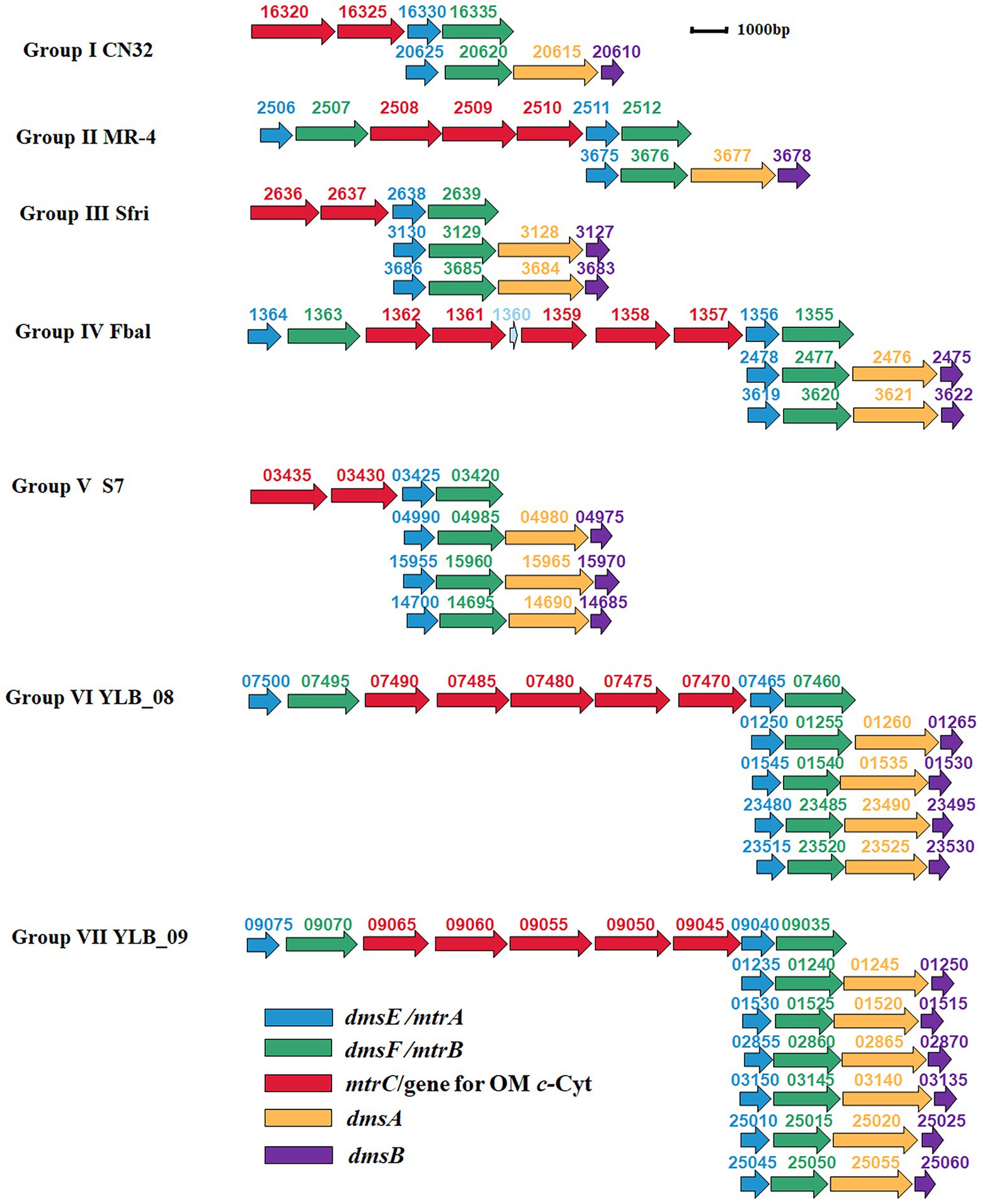
Figure 3. Genetic organization of identified mtrCAB and dmsEFAB gene clusters. The relative positions of genes identified within the complete genomes are shown. The identified genes are labeled with arrows. The arrow sizes indicate the relative lengths of identified genes. The arrow orientation indicates the presumed direction of gene transcription. The numbers above the identified genes are part of their locus tags. Group I: S. japonica KCTC 22435, S. livingstonensis LMG 19866, S. putrefaciens CN32 (CN32), S. woodyi ATCC 51908, Shewanella sp. ARC9_LZ, Shewanella sp. SUN WT4 and Shewanella sp. WPAGA9; Group II: S. fidelis ATCC-BAA-318, S. marisflavi EP1, S. piezotolerans WP3, S. schlegeliana JCM 11561, S. xiamenensis NUITM-VS1, Shewanella sp. 8A, Shewanella sp. ISTPL2, Shewanella sp. LZH-2, Shewanella sp. MBTL60-112-B1, Shewanella sp. MBTL60-112-B2 and Shewanella sp. MR-4 (MR-4); Group III: Ferrimonas sp. SCSIO 43195, S. frigidimarina NCIMB 400 (Sfri), Shewanella sp. Actino-trap-3 and Shewanella sp. KX20019; Group IV: Ferrimonas balerica DSM 9799 (Fbal) and S. oneidensis MR-1; Group V: F. lipolytica S7 (S7) and S. psychromarinicola M2; Group VI: S. eurypsychrophilus YLB-08 (YLB-08); and Group VII: S. sediminis HAW-EB3 and Shewanella sp. YLB-09 (YLB-09). OM c-Cyt: the outer membrane c-type cytochrome. See text for details.
Group I bacteria contained a dmsEFAB gene cluster and a mtrCAB gene cluster. An additional gene for the outer membrane multiheme c-Cyt is also associated with the mtrCAB gene cluster (Group I, Figure 3). This group of bacteria included S. japonica KCTC 22435, S. livingstonensis LMG 19866, S. putrefaciens CN32, S. woodyi ATCC 51908, Shewanella sp. ARC9_LZ, Shewanella sp. SUN WT4, and Shewanella sp. WPAGA9 (Supplementary Table S1). Notably, the dmsEFAB and mtrCAB gene clusters of S. japonica KCTC 22435 were 99.5%–100% identical to those of Shewanella sp. WPAGA9, respectively (Figures 2A–E; Supplementary Tables S2–S6).
Like Group I bacteria, Group II bacteria also contained a dmsEFAB gene cluster and a mtrCAB gene cluster. However, the mtrCAB gene cluster of this group of bacteria also contained a mtrDEF gene cluster and the genes for other outer membrane multiheme c-Cyt (Group II, Figure 3). Group II bacteria included S. fidelis ATCC-BAA-318, S. marisflavi EP1, S. piezotolerans WP3, S. schlegeliana JCM 11561, S. xiamenensis NUITM-VS1, Shewanella sp. 8A, Shewanella sp. ISTPL2, Shewanella sp. LZH-2, Shewanella sp. MBTL60-112-B1, Shewanella sp. MBTL60-112-B2, and Shewanella sp. MR-4 (Supplementary Table S1). Among this group of bacteria, Shewanella sp. MBTL60-112-B1 and Shewanella sp. MBTL60-112-B2 shared identical dmsEFAB and mtrCAB gene clusters (Figures 2A–E; Supplementary Tables S2–S6). These results suggest that they are very closely related. Results also showed that dmsEFAB and mtrCAB gene clusters of S. xiamenensis NUITM-VS1, Shewanella sp. 8A, and Shewanella sp. LZH-2 were 97.6%–100% identical, respectively (Figures 2A–E; Supplementary Tables S2–S6), which suggest that these Shewanella spp. acquire dmsEFAB and mtrCAB gene clusters from a common ancestor.
Both Group III and IV bacteria contained two dmsEFAB gene clusters and a mtrCAB gene cluster. The major difference between these two groups of bacteria was that the mtrCAB gene of Group IV bacteria also had a mtrDEF gene cluster and one to three genes for the outer membrane c-Cyts (Group III and IV, Figure 3). Group III bacteria included Ferrimonas sp. SCSIO 43195, S. frigidimarina NCIMB 400, Shewanella sp. Actino-trap-3 and Shewanella sp. KX20019, among which an additional gene for the outer membrane multiheme c-Cyt associated with the mtrCAB gene cluster of S. frigidimarina NCIMB 400, Shewanella sp. Actino-trap-3 and Shewanella sp. KX20019 (Group III, Figure 3; Supplementary Table S1). The identified Group IV bacteria were Ferrimonas balerica DSM 9799 and S. oneidensis MR-1 (Supplementary Table S1).
Group V bacteria all possessed three dmsEFAB gene clusters and a mtrCAB gene cluster with an additional gene for the outer membrane c-Cyt (Group V, Figure 3), which included F. lipolytica S7 and S. psychromarinicola M2 (Supplementary Table S1). One of the dmsEFAB gene clusters, the 06735–06750 gene cluster, of S. psychromarinicola M2 was 100% identical to the 15625–15640 gene cluster, one of the dmsEFAB gene clusters of Shewanella sp. Actino-trap-3 from Group III (Figures 2A–D; Supplementary Tables S2–S5). Similarly, mtrCAB gene cluster of S. psychromarinicola M2 was 97.1%–99.7% identical to that of Shewanella sp. Actino-trap-3 (Figures 2C–E; Supplementary Tables S4–S6).
Both Group VI and VII bacteria had a mtrCAB gene cluster that also included a mtrDEF gene cluster and three genes for the outer membrane c-Cyts (Group VI and VI, Figure 3). However, the Group VI bacterium S. eurypsychrophilus YLB-08 possessed four dmsEFAB gene clusters (Group VI, Figure 3; Supplementary Table S1), while the Group VII bacteria S. sediminis HAW-EB3 and Shewanella sp. YLB-09 contained six dmsEFAB gene clusters (Group VII, Figure 3; Supplementary Table S1). Notably, among the six dmsEFAB gene clusters of Shewanella sp. YLB-09, the 01235–01250 and 01530–01515 gene cluster were 100% identical to the 02855–02870 and 03150–03135 gene clusters, respectively, which demonstrates duplications of dmsEFAB gene clusters in Shewanella sp. YLB-09 (Figures 2A–D; Group VII, Figure 3; Supplementary Tables S2–S5). Furthermore, the mtrCAB gene cluster and its associated genes (07500–07460) of S. eurypsychrophilus YLB-08 were 100% identical to the mtrCAB gene cluster and its associated genes (09075–09035) of Shewanella sp. YLB-09 (Group VI and VII, Figure 3; Supplementary Tables S4–S6). Similarly, dmsEFAB gene clusters 01250–01265, 01545–01530, 23480–23495, and 23515–23530 of S. eurypsychrophilus YLB-08 were 100% identical to dmsEFAB gene clusters 01235–01250/02855–02870, 01530–01515/03150–03135, 25010–25025 and 25045–25060 of Shewanella sp. YLB-09, respectively (Group VI and VII, Figure 3; Supplementary Tables S2–S5). Thus, the dmsEFAB and mtrCAB gene clusters of S. eurypsychrophilus YLB-08 and Shewanella sp. YLB-09 must be acquired from the same ancestor.
To investigate to what extent the extracellular IO3−-reducing organisms were distributed around world, the bacteria with both dmsEFAB and mtrCAB gene clusters were systemically searched. A total of 29 bacteria were identified to possess both gene clusters. They belonged to the genus of Ferrimonas and Shewanella. Possession of both dmsEFAB and mtrCAB gene clusters suggests the ability to mediate extracellular reduction of IO3− by these bacteria. The identified bacteria with capability for extracellular reduction of IO3− were widespread around the world. Although some were found in freshwater lakes and rivers as well as the rocks of deep continental subsurface, most of them were derived from geographically distributed marine environments. The latter included those found in the Arctic, Atlantic, Indian, and Pacific oceans. Widespread occurrence of the bacteria with capability for extracellular reduction of IO3− suggests a crucial role of this group of bacteria in global biogeochemical cycling of iodine.
The genetic organizations of identified dmsEFAB and mtrCAB gene clusters varied significantly. The mtrCAB gene clusters often associated with genes for the outer membrane c-Cyt of multiheme. Some of the mtrCAB gene clusters also contained a mtrDEF gene cluster. The numbers of dmsEFAB gene cluster detected in a given bacterial genome ranged from one to six. Duplications of the detected dmsEFAB gene clusters also occurred. Thus, this group of bacteria acquire their capability for extracellular reduction of iodate differently.
Collectively, the results from this investigation provide new insights into the distribution and evolution of as well as the role in global biogeochemical cycling of iodine by the bacteria with capability for extracellular reduction of iodate. Physiological characterization of the iodate-reducing capacity for the predicted strains and their ecological roles on iodine cycling in different ecosystems are in need of further investigation.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
LS designed the experiment and acquired funding. JG, JJ, and YZ performed the experiment. ZP, YJ, ZJ, YH, YD, and LS analyzed the data and prepared manuscript. All authors contributed to the article and approved the submitted version.
We would like to thank the National Key Research and Development Program of China (2018YFA0901303), National Natural Science Foundation of China (NSFC91851211, 42277065, 42272353), and the Fundamental Research Funds for the Universities of Chinese Central Government, China University of Geosciences-Wuhan (122-G1323522144) for their support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1070601/full#supplementary-material
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Andersen, S., Guan, H., Teng, W., and Laurberg, P. (2009). Speciation of iodine in high iodine groundwater in China associated with goitre and hypothyroidism. Biol. Trace Elem. Res. 128, 95–103. doi: 10.1007/s12011-008-8257-x
Bae, S. S., Jung, Y. H., Kwon, Y. M., Chung, D., and Baek, K. (2021). Ferrimonas lipolytica sp. nov., a facultatively anaerobic bacterium isolated from seawater. Int. J. Syst. Evol. Microbiol. 71:004665. doi: 10.1099/ijsem.0.004665
Bagos, P. G., Liakopoulos, T. D., Spyropoulos, I. C., and Hamodrakas, S. J. (2004). PRED-TMBB: a web server for predicting the topology of beta-barrel outer membrane proteins. Nucleic Acids Res. 32, W400–W404. doi: 10.1093/nar/gkh417
Baker, I. R., Conley, B. E., Gralnick, J. A., and Girguis, P. R. (2022). Evidence for horizontal and vertical transmission of Mtr-mediated extracellular electron transfer among the bacteria. MBio 13:e0290421. doi: 10.1128/mbio.02904-21
Beliaev, A. S., and Saffarini, D. A. (1998). Shewanella putrefaciens mtrB encodes an outer membrane protein required for Fe(III) and Mn(IV) reduction. J. Bacteriol. 180, 6292–6297. doi: 10.1128/JB.180.23.6292-6297.1998
Beliaev, A. S., Saffarini, D. A., Mclaughlin, J. L., and Hunnicutt, D. (2001). MtrC, an outer membrane decahaem c cytochrome required for metal reduction in Shewanella putrefaciens MR-1. Mol. Microbiol. 39, 722–730. doi: 10.1046/j.1365-2958.2001.02257.x
Bretschger, O., Obraztsova, A., Sturm, C. A., Chang, I. S., Gorby, Y. A., Reed, S. B., et al. (2007). Current production and metal oxide reduction by Shewanella oneidensis MR-1 wild type and mutants. Appl. Environ. Microbiol. 73, 7003–7012. doi: 10.1128/AEM.01087-07
Carpenter, L. J., Chance, R. J., Sherwen, T. J. A. T., Ball, S. M., Evans, M. J., Hepath, H., et al. (2021). Marine iodine emission in a changing world. Proc. R. Soc. U. S. A. 477:20200824. doi: 10.1098/rspa.2020.0824
Chance, R., Baker, A. R., Carpenter, L., and Jickells, T. D. (2014). The distribution of iodide at the sea surface. Environ Sci Process Impacts 16, 1841–1859. doi: 10.1039/C4EM00139G
Councell, T. B., Landa, E. C., and Lovley, D. R. (1997). Microbial reduction of iodate. Water Air Soil Pollut. 100, 99–106. doi: 10.1023/A:1018370423790
Coursolle, D., Baron, D. B., Bond, D. R., and Gralnick, J. A. (2010). The Mtr respiratory pathway is essential for reducing flavins and electrodes in Shewanella oneidensis. J. Bacteriol. 192, 467–474. doi: 10.1128/JB.00925-09
Coursolle, D., and Gralnick, J. A. (2010). Modularity of the Mtr respiratory pathway of Shewanella oneidensis strain MR-1. Mol. Microbiol. 77, 995–1008. doi: 10.1111/j.1365-2958.2010.07266.x
Dao, T. D., Kasuga, I., Hirabayashi, A., Nguyen, D. T., Tran, H. T., Vu, H., et al. (2022). Emergence of mobile tigecycline resistance gene tet(X4)-harbouring Shewanella xiamenensis in a water environment. J. Glob. Antimicrob. Resist. 28, 140–142. doi: 10.1016/j.jgar.2021.12.022
Duan, L., Wang, W., Sun, Y., and Zhang, C. (2016). Iodine in groundwater of the Guanzhong Basin, China: source and hydrogeochemical controls on its distribution. Environ. Earth Sci. 75:970. doi: 10.1007/s12665-016-5781-4
Edwards, M. J., White, G. F., Butt, J. N., Richardson, D. J., and Clarke, T. A. (2020). The crystal structure of a biological insulated transmembrane molecular wire. Cells 181:e610, 665–673.e10. doi: 10.1016/j.cell.2020.03.032
Edwards, M. J., White, G. F., Norman, M., Tome-Fernandez, A., Ainsworth, E., Shi, L., et al. (2015). Redox linked flavin sites in extracellular decaheme proteins involved in microbe-mineral electron transfer. Sci. Rep. 5:11677. doi: 10.1038/srep11677
Farrenkorf, A. M., Dollopf, M. E., Chadhain, S. N., Luther Iii, G. W., and Nealson, K. H. (1997). Reduction of iodate in seawater during Arabian Sea shipboard incubations and in laboratory culture of the marine bacterium Shewanella putrefaciens strain MR-4. Mar. Chem. 57, 347–354. doi: 10.1016/S0304-4203(97)00039-X
Fredrickson, J. K., Romine, M. F., Beliaev, A. S., Auchtung, J. M., Driscoll, M. E., Gardner, T. S., et al. (2008). Towards environmental systems biology of Shewanella. Nat. Rev. Microbiol. 6, 592–603. doi: 10.1038/nrmicro1947
Fredrickson, J., Zachara, J. N., Kennedy, D., Dong, H., Onstott, T. C., Hinman, N. W., et al. (1998). Biogenic iron mineralization accompnying the dissimilatory reduction of hydrous ferric oxide by a groundwater bacterium. Geochim. Cosmochim. Acta 62, 3239–3257. doi: 10.1016/S0016-7037(98)00243-9
Gralnick, J. A., Vali, H., Lies, D. P., and Newman, D. K. (2006). Extracellular respiration of dimethyl sulfoxide by Shewanella oneidensis strain MR-1. Proc. Natl. Acad. Sci. U. S. A. 103, 4669–4674. doi: 10.1073/pnas.0505959103
Guo, J., Jiang, Y., Hu, Y., Jiang, Z., Dong, Y., and Shi, L. (2022). The roles of DmsEFAB and MtrCAB in extracellular reduction of iodate by Shewanella oneidensis MR-1 with lactate as the sole electron donor. Environ. Microbiol. 24, 5039–5050. doi: 10.1111/1462-2920.16130
Hartshorne, R. S., Reardon, C. L., Ross, D., Nuester, J., Clarke, T. A., Gates, A. J., et al. (2009). Characterization of an electron conduit between bacteria and the extracellular environment. Proc. Natl. Acad. Sci. U. S. A. 106, 22169–22174. doi: 10.1073/pnas.0900086106
He, Z., Zhang, H., Gao, S., Lercher, M. J., Chen, W. H., and Hu, S. (2016). Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 44, W236–W241. doi: 10.1093/nar/gkw370
Huang, J., Sun, B., and Zhang, X. (2010). Electricity generation at high ionic strength in microbial fuel cell by a newly isolated Shewanella marisflavi EP1. Appl. Microbiol. Biotechnol. 85, 1141–1149. doi: 10.1007/s00253-009-2259-2
Hwang, Y. J., Jang, G. I., Cho, B. C., Lee, J. I., Hwang, C. Y., and Hwang, C. Y. (2019). Shewanella psychromarinicola sp. nov., a psychrophilic bacterium isolated from pelagic sediment of the Ross Sea (Antarctica), and reclassification of Shewanella arctica Kim et al. 2012 as a later heterotypic synonym of Shewanella frigidimarina Bowman et al. 1997. Int. J. Syst. Evol. Microbiol. 69, 2415–2423. doi: 10.1099/ijsem.0.003490
Iino, T., Ohkuma, M., Kamagata, Y., and Amachi, S. (2016). Iodidimonas muriae gen. Nov., sp. nov., an aerobic iodide-oxidizing bacterium isolated from brine of a natural gas and iodine recovery facility, and proposals of Iodidimonadaceae fam. Nov., Iodidimonadales Ord. Nov., Emcibacteraceae fam. Nov. and Emcibacterales Ord. Nov. Int. J. Syst. Evol. Microbiol. 66, 5016–5022. doi: 10.1099/ijsem.0.001462
Ivanova, E. P., Sawabe, T., Hayashi, K., Gorshkova, N. M., Zhukova, N. V., Nedashkovskaya, O. I., et al. (2003). Shewanella fidelis sp. nov., isolated from sediments and sea water. Int. J. Syst. Evol. Microbiol. 53, 577–582. doi: 10.1099/ijs.0.02198-0
Jiao, Y., Kappler, A., Croal, L. R., and Newman, D. K. (2005). Isolation and characterization of a genetically tractable photoautotrophic Fe(II)-oxidizing bacterium, Rhodopseudomonas palustris strain TIE-1. Appl. Environ. Microbiol. 71, 4487–4496. doi: 10.1128/AEM.71.8.4487-4496.2005
Jiao, Y., and Newman, D. K. (2007). The pio operon is essential for phototrophic Fe(II) oxidation in Rhodopseudomonas palustris TIE-1. J. Bacteriol. 189, 1765–1773. doi: 10.1128/JB.00776-06
Kaplan, D. I., Denham, M. E., Zhang, S., Yeager, C., Xu, C., Sxhwehr, K. A., et al. (2014). Radioiodine biogeochemistry and prevalence in groundwater. Crit. Rev. Environ. Sci. Technol. 44, 2287–2335. doi: 10.1080/10643389.2013.828273
Kim, K. M., Choe, H., Kim, B. K., and Nasir, A. (2017). Complete genome of a metabolically-diverse marine bacterium Shewanella japonica KCTC 22435(T). Mar. Genomics 35, 39–42. doi: 10.1016/j.margen.2017.05.004
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., Mcwilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Laurberg, P., Bulow Pedersen, I., Knudsen, N., Ovesen, L., and Andersen, S. (2001). Environmental iodine intake affects the type of nonmalignant thyroid disease. Thyroid 11, 457–469. doi: 10.1089/105072501300176417
Li, Z., Lin, S., Liu, X., Tan, J., Pan, J., and Yang, H. (2014). A freshwater bacterial strain, Shewanella sp. Lzh-2, isolated from Lake Taihu and its two algicidal active substances, hexahydropyrrolo[1,2-a]pyrazine-1,4-dione and 2, 3-indolinedione. Appl. Microbiol. Biotechnol. 98, 4737–4748. doi: 10.1007/s00253-014-5602-1
Li, S., Niu, Y., Chen, H., and He, P. (2021). Complete genome sequence of an Arctic Ocean bacterium Shewanella sp. Arc9-LZ with capacity of synthesizing silver nanoparticles in darkness. Mar. Genomics 56:100808. doi: 10.1016/j.margen.2020.100808
Li, J., Wang, Y., Xie, X., Zhang, L., and Guo, W. (2013). Hydrogeochemistry of high iodine groundwater: a case study at the Datong Basin, northern China. Environ Sci Process Impacts 15, 848–859. doi: 10.1039/c3em30841c
Liu, J., Pearce, C. I., Liu, C., Wang, Z., Shi, L., Arenholz, E., et al. (2013). Fe(3-x)Ti(x)O4 nanoparticles as tunable probes of microbial metal oxidation. J. Am. Chem. Soc. 135, 8896–8907. doi: 10.1021/ja4015343
Liu, J., Wang, Z., Belchik, S. M., Edwards, M. J., Liu, C., Kennedy, D. W., et al. (2012). Identification and characterization of MtoA: a decaheme c-type cytochrome of the neutrophilic Fe(II)-oxidizing bacterium Sideroxydans lithotrophicus ES-1. Front. Microbiol. 3:37. doi: 10.3389/fmicb.2012.00037
Liu, Y., Wang, Z., Liu, J., Levar, C., Edwards, M. J., Babauta, J. T., et al. (2014). A trans-outer membrane porin-cytochrome protein complex for extracellular electron transfer by Geobacter sulfurreducens PCA. Environ. Microbiol. Rep. 6, 776–785. doi: 10.1111/1758-2229.12204
Lower, B. H., Yongsunthon, R., Shi, L., Wildling, L., Gruber, H. J., Wigginton, N. S., et al. (2009). Antibody recognition force microscopy shows that outer membrane cytochromes OmcA and MtrC are expressed on the exterior surface of Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 75, 2931–2935. doi: 10.1128/AEM.02108-08
Makemson, J. C., Fulayfil, N. R., Landry, W., Van Ert, L. M., Wimpee, C. F., Widder, E. A., et al. (1997). Shewanella woodyi sp. nov., an exclusively respiratory luminous bacterium isolated from the Alboran Sea. Int. J. Syst. Bacteriol. 47, 1034–1039. doi: 10.1099/00207713-47-4-1034
Mok, J. K., Toporek, Y. J., Shin, H. D., Lee, B. D., Lee, M. H., and Dichristina, T. J. (2018). Iodate reduction by Shewanella oneidensis does not involved nitrate reductase. Geomicrobiol. J. 35, 570–579. doi: 10.1080/01490451.2018.1430189
Myers, C. R., and Nealson, K. H. (1988). Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240, 1319–1321. doi: 10.1126/science.240.4857.1319
Otosaka, S., Schwehr, K. A., Kaplan, D. I., Roberts, K. A., Zhang, S., Xu, C., et al. (2011). Factors controlling mobility of 127I and 129I species in an acidic groundwater plume at the Savannah River site. Sci. Total Environ. 409, 3857–3865. doi: 10.1016/j.scitotenv.2011.05.018
Rathour, R., Medhi, K., Gupta, J., and Thakur, I. S. (2021). Integrated approach of whole-genome analysis, toxicological evaluation and life cycle assessment for pyrene biodegradation by a psychrophilic strain, Shewanella sp. ISTPL2. Environ. Pollut. 269:116176. doi: 10.1016/j.envpol.2020.116176
Reid, G. A., and Gordon, E. H. (1999). Phylogeny of marine and freshwater Shewanella: reclassification of Shewanella putrefaciens NCIMB 400 as Shewanella frigidimarina. Int. J. Syst. Bacteriol. 49, 189–191. doi: 10.1099/00207713-49-1-189
Reyes-Umana, V., Henning, Z., Lee, K., Barnum, T. P., and Coates, J. D. (2022). Genetic and phylogenetic analysis of dissimilatory iodate-reducing bacteria identifies potential niches across the world's oceans. ISME J. 16, 38–49. doi: 10.1038/s41396-021-01034-5
Rossello-Mora, R. A., Ludwig, W., Kampfer, P., Amann, R., and Schleifer, K.-H. (1995). Ferrimonas balearica gen. Nov., spec.Nov., a new marine facultative Fe(III)-reducing bacterium. Syst. Appl. Microbiol. 18, 196–202. doi: 10.1016/S0723-2020(11)80390-5
Satomi, M., Oikawa, H., and Yano, Y. (2003). Shewanella marinintestina sp. nov., Shewanella schlegeliana sp. nov. and Shewanella sairae sp. nov., novel eicosapentaenoic-acid-producing marine bacteria isolated from sea-animal intestines. Int. J. Syst. Evol. Microbiol. 53, 491–499. doi: 10.1099/ijs.0.02392-0
Shi, L., Chen, B., Wang, Z., Elias, D. A., Mayer, M. U., Gorby, Y. A., et al. (2006). Isolation of a high-affinity functional protein complex between OmcA and MtrC: two outer membrane decaheme c-type cytochromes of Shewanella oneidensis MR-1. J. Bacteriol. 188, 4705–4714. doi: 10.1128/JB.01966-05
Shi, L., Deng, S., Marshall, M. J., Wang, Z., Kennedy, D. W., Dohnalkova, A. C., et al. (2008). Direct involvement of type II secretion system in extracellular translocation of Shewanella oneidensis outer membrane cytochromes MtrC and OmcA. J. Bacteriol. 190, 5512–5516. doi: 10.1128/JB.00514-08
Shi, L., Fredrickson, J., and Zachara, J. (2014). Genomic analyses of bacterial porin-cytochrome gene clusters. Front. Microbiol. 5:657. doi: 10.3389/fmicb.2014.00657
Shi, L., Rosso, K. M., Clarke, T. A., Richardson, D. J., Zachara, J. M., and Fredrickson, J. K. (2012a). Molecular underpinnings of Fe(III) oxide reduction by Shewanella oneidensis MR-1. Front. Microbiol. 3:50. doi: 10.3389/fmicb.2012.00050
Shi, L., Rosso, K. M., Zachara, J. M., and Fredrickson, J. K. (2012b). Mtr extracellular electron-transfer pathways in Fe(III)-reducing or Fe(II)-oxidizing bacteria: a genomic perspective. Biochem. Soc. Trans. 40, 1261–1267. doi: 10.1042/BST20120098
Shin, H. D., Toporek, Y., Mok, J. K., Maekawa, R., Lee, B. D., Howard, M. H., et al. (2022). Iodate reduction by Shewanella oneidensis requires genes encoding an extracellular dimethylsulfoxide reductase. Front. Microbiol. 13:852942. doi: 10.3389/fmicb.2022.852942
Shiroyama, K., Kawasaki, Y., Unno, Y., and Amachi, S. (2015). A putative multicopper oxidase, IoxA, is involved in iodide oxidation by Roseovarius sp. strain A-2. Biosci. Biotechnol. Biochem. 79, 1898–1905. doi: 10.1080/09168451.2015.1052767
Suzuki, M., Eda, Y., Ohsawa, S., Kanesaki, Y., Yoshikawa, H., Tanaka, K., et al. (2012). Iodide oxidation by a novel multicopper oxidase from the alphaproteobacterium strain Q-1. Appl. Environ. Microbiol. 78, 3941–3949. doi: 10.1128/AEM.00084-12
Tang, Q., Xu, Q., Zhang, F., Huang, Y., Liu, J., Wang, X., et al. (2013). Geochemistry of iodine-rich groundwater in the Taiyuan Basin of Central Shanxi Province, North China. J. Geochem. Explor. 135, 117–123. doi: 10.1016/j.gexplo.2012.08.019
Tang, K., Zhan, W., Zhou, Y., Xu, T., Chen, X., Wang, W., et al. (2020). Antagonism between coral pathogen vibrio coralliilyticus and other bacteria in the gastric cavity of scleractinian coral Galzxea fascicularis. Sci. China Earth Sci. 63, 157–166. doi: 10.1007/s11430-019-9388-3
Thomas, P. E., Ryan, D., and Levin, W. (1976). An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75, 168–176. doi: 10.1016/0003-2697(76)90067-1
Toporek, Y. J., Mok, J. K., Shin, H. D., Lee, B. D., Lee, M. H., and Dichristina, T. J. (2019). Metal reduction and protein secretion genes required for iodate reduction by Shewanella oneidensis. Appl. Environ. Microbiol. 85, e02115–e02118. doi: 10.1128/AEM.02115-18
Venkateswaran, K., Moser, D. P., Dollhopf, M. E., Lies, D. P., Saffarini, D. A., Macgregor, B. J., et al. (1999). Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 49, 705–724.
Wang, F., Wang, P., Chen, M., and Xiao, X. (2004). Isolation of extremophiles with the detection and retrieval of Shewanella strains in deep-sea sediments from the West Pacific. Extremophiles 8, 165–168. doi: 10.1007/s00792-003-0365-0
Wang, F., Wang, J., Jian, H., Zhang, B., Li, S., Wang, F., et al. (2008). Environmental adaptation: genomic analysis of the piezotolerant and psychrotolerant deep-sea iron reducing bacterium Shewanella piezotolerans WP3. PLoS One 3:e1937. doi: 10.1371/journal.pone.0001937
Wang, D., Wang, J., Zeng, R., Wu, J., Michael, S. V., and Qu, W. (2021). The degradation activities for three seaweed polysaccharides of Shewanella sp. WPAGA9 isolated from deep-sea sediments. J. Basic Microbiol. 61, 406–418. doi: 10.1002/jobm.202000728
Wen, D., Zhang, F., Zhang, E., Wang, C., Han, S., and Zheng, Y. (2013). Arsenic, fluoride and iodine in groundwater of China. J. Geochem. Explor. 135, 1–21. doi: 10.1016/j.gexplo.2013.10.012
White, G. F., Shi, Z., Shi, L., Wang, Z., Dohnalkova, A. C., Marshall, M. J., et al. (2013). Rapid electron exchange between surface-exposed bacterial cytochromes and Fe(III) minerals. Proc. Natl. Acad. Sci. U. S. A. 110, 6346–6351. doi: 10.1073/pnas.1220074110
Yamazaki, C., Kashiwa, S., Horiuchi, A., Kasahara, Y., Yamamura, S., and Amachi, S. (2020). A novel dimethylsulfoxide reductase family of molybdenum enzyme, Idr, is involved in iodate respiration by pseudomonas sp. SCT Environ. Microbiol. 22, 2196–2212. doi: 10.1111/1462-2920.14988
Yu, L., Jian, H., Gai, Y., Yi, Z., Feng, Y., Qiu, X., et al. (2021). Characterization of two novel psychrophilic and piezotolerant strains, Shewanella psychropiezotolerans sp. nov. and Shewanella eurypsychrophilus sp. nov, adapted to an extreme deep-sea environment. Syst. Appl. Microbiol. 44:126266. doi: 10.1016/j.syapm.2021.126266
Zhang, E., Wang, Y., Qian, Y., Ma, T., Zhang, D., Zhan, H., et al. (2013). Iodine in groundwater of the North China Plains: spatial patterns and hydrogeochemical processes of enrichment. J. Geochem. Explor. 135, 40–53. doi: 10.1016/j.gexplo.2012.11.016
Zhang, S., Xu, C., Creeley, D., Ho, Y. F., Li, H. P., Grandbois, R., et al. (2013). Iodine-129 and iodine-127 speciation in groundwater at the Hanford site, US: iodate incorporation into calcite. Environ. Sci. Technol. 47, 9635–9642. doi: 10.1021/es401816e
Zhao, J. S., Manno, D., Beaulieu, C., Paquet, L., and Hawari, J. (2005). Shewanella sediminis sp. nov., a novel Na+−requiring and hexahydro-1,3,5-trinitro-1,3,5-triazine-degrading bacterium from marine sediment. Int. J. Syst. Evol. Microbiol. 55, 1511–1520. doi: 10.1099/ijs.0.63604-0
Zhong, Y., and Shi, L. (2018). Genomic analyses of the quinol oxidases and/or quinone reductases involved in bacterial extracellular electron transfer. Front. Microbiol. 9:3029. doi: 10.3389/fmicb.2018.03029
Keywords: DmsEFAB, MtrCAB, extracellular reduction of iodate, Ferrimonas, Shewanella, global biogeochemical cycling of iodine
Citation: Guo J, Jiang J, Peng Z, Zhong Y, Jiang Y, Jiang Z, Hu Y, Dong Y and Shi L (2022) Global occurrence of the bacteria with capability for extracellular reduction of iodate. Front. Microbiol. 13:1070601. doi: 10.3389/fmicb.2022.1070601
Received: 15 October 2022; Accepted: 11 November 2022;
Published: 25 November 2022.
Edited by:
Shungui Zhou, Fujian Agriculture and Forestry University, ChinaReviewed by:
Hyun-Dong Shin, Bereum Co. Ltd. Wonju, South KoreaCopyright © 2022 Guo, Jiang, Peng, Zhong, Jiang, Jiang, Hu, Dong and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaofeng Peng, cHpmLnNvZUAxMjYuY29t; Liang Shi, bGlhbmcuc2hpQGN1Zy5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.