
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 01 December 2022
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1069109
 Tingting Chen1
Tingting Chen1 Zhuzhu Zhang1
Zhuzhu Zhang1 Wenzhi Li1
Wenzhi Li1 Jia Chen1
Jia Chen1 Xuetang Chen1
Xuetang Chen1 Bince Wang1
Bince Wang1 Jiling Ma1
Jiling Ma1 Yunyun Dai1
Yunyun Dai1 Haixia Ding1,2
Haixia Ding1,2 Weizhen Wang1
Weizhen Wang1 Youhua Long1,3*
Youhua Long1,3*Soft rot causes significant economic losses in the kiwifruit industry. This study isolated strain CTXW 7-6-2 from healthy kiwifruit tissue; this was a gram-positive bacterium that produced the red pigment pulcherrimin. The phylogenetic tree based on 16S ribosomal RNA, gyrA, rpoB, and purH gene sequences identified CTXW 7-6-2 as a strain of Bacillus subtilis. CTXW 7-6-2 inhibited hyphal growth of pathogenic fungi that cause kiwifruit soft rot, namely, Botryosphaeria dothidea, Phomopsis sp., and Alternaria alternata, by 81.76, 69.80, and 32.03%, respectively. CTXW 7-6-2 caused the hyphal surface to become swollen and deformed. Volatile compounds (VOC) produced by the strain inhibited the growth of A. alternata and Phomopsis sp. by 65.74 and 54.78%, respectively. Whole-genome sequencing revealed that CTXW 7-6-2 possessed a single circular chromosome of 4,221,676 bp that contained 4,428 protein-coding genes, with a guanine and cytosine (GC) content of 43.41%. Gene functions were annotated using the National Center for Biotechnology Information (NCBI) non-redundant protein, Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes, Clusters of Orthologous Groups of proteins, Gene Ontology, Pathogen–Host Interactions, Carbohydrate-Active enZYmes, and Rapid Annotations using Subsystem Technology databases, revealing non-ribosomal pathways associated with antifungal mechanisms, biofilm formation, chemotactic motility, VOC 3-hydroxy-2-butanone, cell wall-associated enzymes, and synthesis of various secondary metabolites. antiSMASH analysis predicted that CTXW 7-6-2 can produce the active substances bacillaene, bacillibactin, subtilosin A, bacilysin, and luminmide and has four gene clusters of unknown function. Quantitative real-time PCR (qRT-PCR) analysis verified that yvmC and cypX, key genes involved in the production of pulcherrimin, were highly expressed in CTXW 7-6-2. This study elucidates the mechanism by which B. subtilis strain CTXW 7-6-2 inhibits pathogenic fungi that cause kiwifruit soft rot, suggesting the benefit of further studying its antifungal active substances.
Kiwifruit is appreciated not only for its taste but also for its abundant nutrients and dietary fibre, which are beneficial to human health (He et al., 2019). Kiwifruit originated in China and is commonly grown in New Zealand, the USA, Chile, and some European countries, making it an economically important fruit with great potential for development. Owing to the industry’s rapid development, diseases have become increasingly prominent during the cultivation, harvesting, and storage of kiwifruit, including crown gall (He et al., 2021), bacterial canker (Song et al., 2019), blossom blight (Balestra et al., 2008), leaf spot disease (Chen et al., 2022a), and soft rot (Wang et al., 2021). Among them, soft rot is the most common disease during the storage and sale of kiwifruit. Soft rot outbreaks have occurred in New Zealand, China, South Korea, and other countries, causing significant economic losses and threatening the development of the global kiwifruit industry (Manning et al., 2016). Infected fruit are asymptomatic in the early storage period, gradually showing symptoms during the post-ripening period. During infection, the epidermis of the diseased site collapses and becomes soft, round or oval-shaped brown lesions appear, and the pulp turns yellow–green, resulting in rotten and inedible fruit. Several pathogenic fungi cause soft rot, including Botryosphaeria dothidea, Botrytis cinerea, Phomopsis sp., and Alternaria alternata.
Despite considerable progress in the management and control of soft rot, kiwifruit remain vulnerable to infection. Kiwifruit are usually soaked or sprayed with fungicides before harvesting, and bagging is used to reduce the spread of dangerous pathogens. However, these measures may increase chemical residue levels in the fruit or promote fungal resistance. Therefore, researchers are urgently seeking natural antifungal active substances as alternatives to traditional fungicides. For example, plant-derived methyl jasmonate was found to effectively inhibit the growth of B. dothidea hyphae to protect against kiwifruit soft rot (Pan et al., 2019). Natamycin, a natural antimicrobial, inhibited B. dothidea growth by causing the vacuolation and plasmolysis of hyphal cells, thereby reducing kiwifruit soft rot (Pan et al., 2022). In addition, Meng et al. (2022) reported that lemon essential oil-based nanoemulsions inhibited the growth of Phomopsis sp. to reduce postharvest decay of kiwifruit. In terms of microbial control, a culture filtrate of Bacillus amyloliquefaciens effectively inhibited B. dothidea growth and maintained kiwifruit quality during storage (Pang et al., 2020). Further, Bacillus subtilis was shown to antagonise pathogenic bacteria that cause kiwifruit soft rot (Kim et al., 2015) and promote the root growth of kiwifruit stem cuttings (Erturk et al., 2010). However, the mechanism by which B. subtilis counteracts soft rot-causing pathogens in kiwifruit remains unclear.
Bacillus subtilis can produce biologically active secondary metabolites and is widely used in medicine and agriculture (Caulier et al., 2019; Su et al., 2020), in the crucial role of controlling plant diseases. Bacillus spp. produce various antibacterial substances that can inhibit the normal growth of plant pathogens, including bacteriocins, lanthiopeptin, and polyketides (Sumi et al., 2015; Olishevska et al., 2019), and lipopeptide metabolites that can regulate plant hormones and inhibit pathogen proliferation (Cochrane and Vederas, 2016). Moreover, Bacillus spp. can form biofilms (Zeriouh et al., 2014), facilitating their survival and reproduction on plant surfaces and reducing pathogen abundance (Seneviratne et al., 2010). In addition, Bacillus spp. produce volatile compounds (VOC) that can control Fusarium oxysporum sp. spore germination (Wu et al., 2020), change the cell structure of Clavibacter michiganensis ssp. to prevent bacterial ring rot in potatoes (Rajer et al., 2017).
Researchers have applied omics techniques to determine the genetic components related to plant growth promotion, biosynthesis of secondary metabolites, and habitat adaptation of beneficial microorganisms (Zeng et al., 2018). The goal of the present study was to isolate a bacterial strain from healthy kiwifruit with biocontrol potential against soft rot disease and elucidate the underlying mechanism using phenotypic, phylogenetic, and comparative genomics, gene and protein sequence annotation analysis, and metabolic cluster prediction.
The tested pathogens were obtained from the Research Center for Engineering Technology of Kiwifruit at Guizhou University, including B. dothidea (OM802621), Phomopsis sp. (OP740383), Rhizoctonia solani (OK287282), Monilinia fructicola (MW281764), Fusarium fujikuroi (OL774567), A. alternata (MG189596.1), Colletotrichum spaethianum (OL347722), Pseudomonas syringae pv. actinidiae (NZ_CP032631.1), and Pantoea agglomerans (ON955285.1). The pathogenic fungi were cultured in potato dextrose agar (PDA: 250 g potato, 20 g glucose, and 15 g agar/L) for 5 days at 28°C in the dark. The pathogenic bacteria were cultured in beef extract peptone agar (NA: 5 g beef extract, 10 g peptone, 5 g NaCl, and 15 g agar/L) for 2 days at 28°C in the dark.
The kiwifruit were picked in an orchard in Xiuwen County, Guizhou Province (26°45′N, 106°65′E) in October 2021, packed in sterile sampling bags, and stored at 4°C. The kiwifruit surface was washed with sterile water to remove impurities, followed by soaking in 75% ethanol for 60 s, and subsequent washed with sterile water to remove the residual alcohol. Whole kiwifruit were crushed and 1 ml of crushed sample was diluted 1,000 times, spread on NA plates, and incubated at 28°C for 48 h. Subsequently, single colonies with uniform colour, glossiness, and other characteristics were selected and re-inoculated on new NA plates for 48 h. The dual-culture assay for assessing growth inhibition of pathogenic fungi was performed as described by Yan et al. (2020). Preliminarily, antagonistic bacteria that inhibited the growth of pathogenic fungi were screened out. The with obvious inhibition zones were selected for further study.
Antagonistic bacterial strains were preliminarily identified by referring to Buchanan and Gibbons (1984) and Dong and Cai (2001). Single bacterial colonies were picked using an inoculating needle and cultured in NA for 5 days at 28°C under 75% relative humidity. The morphological characteristics of the single colonies were observed with the naked eye and the results of Gram staining were observed using a DM500 binocular microscope (Leica Microsystems, Wetzlar, Germany). Simultaneously, single bacterial colonies were inoculated in NA and cultured at 28°C for 24 h with shaking at 180 rpm. The ultrastructure of the bacterial cells was observed using an SU8010 scanning electron microscope (SEM; Hitachi, Tokyo, Japan) with reference to the method previously reported by Hasan et al. (2014).
To further identify the taxonomy of the antagonistic bacterial strains, phylogenetic trees of the 16S ribosomal RNA, DNA gyrase subunit A (gyrA), RNA polymerase subunit B (rpoB) and phosphoribosylaminoimidazolecarboxamide formyltransferase (purH) genes were constructed using multilocus gene sequence analysis (Rooney et al., 2009; Kubo et al., 2011; Dhruw et al., 2020). DNA was extracted from bacterial cells using the TIANamp Bacteria DNA Kit (Tiangen Biotech Co., Ltd., Beijing, China) according to the manufacturer’s instructions. The PCR reaction was performed in a total volume of 20 μL containing 1 μL of 100 ng/μL DNA, 0.4 μL each of 10 μmol forward and reverse primers (Table 1), 10 μL of 2 × Taq Master Mix, and 8.2 μL ddH2O. The thermal cycling program included a denaturation step at 94°C for 30 s, an annealing step at 55 or 60°C for 30 s, and a final extension at 72°C for 10 min for 35 cycles. The obtained PCR products were separated on 1.0% w/v agarose gel at 150 V for 20 min to confirm the DNA amplification of a single product. The PCR products were sequenced by Sangon Biotech Co., Ltd., (Shanghai, China). BLAST was used to align the sequences with the National Center for Biotechnology Information (NCBI)1 nucleotide sequence database and the reference sequences were downloaded. Construction of multi-locus phylogenetic trees was performed using maximum likelihood, maximum parsimony, and Bayesian inference methods (Chen et al., 2022a). The sequencing data for 16S rRNA, gyrA, rpoB, and purH are deposited under GenBank accession numbers ON076886, ON086734, ON086735, and ON086736, respectively.
The in vitro inhibitory activity of the antagonistic bacterial strains against pathogenic bacteria or fungi was assessed with reference to the methods described by Yan et al. (2020) and Chen et al. (2022b). Briefly, a pathogen colony with a diameter of 6 mm was placed in the centre of a PDA plate. The bacterial strains were cultured in 50 ml of NA at 28°C for 48 h with shaking at 130 rpm. The bacterial suspension was adjusted to an OD600 reached 1.5–2 using a NanoPhotometer-N50 instrument (Implen, Munich, Germany). Subsequently, 100 μL aliquots of bacterial suspension were applied to four disc of filter paper (diameter of 6 mm), which were symmetrically placed 25 mm away from the pathogen colony. The control group contained only pathogen colonies without bacterial strains. All treatments were repeated three times and incubated at 28°C for 5 days.
The effects of the VOCs produced by the antagonistic strains were evaluated using the double-plate pair-button method (You et al., 2021). Briefly, 150 μL of bacterial suspension (OD600 reached 1.5–2) was spread evenly on an NA plate, the pathogenic fungus plug (diameter = 6 mm) was placed in the centre of a PDA plate, and the two plates were sealed together. The pathogen colony diameter was measured using a ruler and hyphal growth inhibition was calculated as follows: A = (a1–a2)/a1 × 100, where A is the growth inhibition of the hyphae, a1 is the colony diameter of the control pathogen, and a2 is the colony diameter of the treated pathogen. The control group used 150 μL of sterile distilled water on the NA plate instead of 150 μL of bacterial suspension.
Hyphae at the edge of the antifungal zone were selected to evaluate the antagonistic effects of the isolated bacterial strains. Morphological changes were observed using a DM500 binocular microscope with magnification multiples of 400 (Leica) and SU8010 SEM (Hitachi) (Mo et al., 2021).
The genomic DNA of CTXW 7-6-2 was extracted using the Rapid Bacterial Genomic DNA Isolation Kit (Sangon Biotech Co., Ltd.) according to the manufacturer’s instructions. Whole-genome sequencing was performed on the Illumina HiSeq sequencing platform (San Diego, CA, USA) and established using the NEB Next® Ultra™ DNA Library Prep Kit for Illumina® (New England Biolabs, Ipswich, MA, USA). Trimmomatic v. 0.36 was used to filter the sequencing data to obtain high-quality data, and FastQC v. 0.11.2 was used to visually assess quality, reads, trimming, and de novo assembly. The next-generation sequencing data were assembled using SPAdes v. 3.5.0. GapFiller v. 1.11 was used to complement the contigs obtained by splicing, and PrInSeS-G v. 1.0.0 was used to correct editing errors and small insertions and deletions during the splicing process. The completed whole-genome sequence was submitted to NCBI under GenBank accession number JAMKCC000000000.1.
Prokka v. 1.10 was used to predict the gene elements of the assembled results, using Prodigal to predict coding genes, Aragorn to predict tRNAs, RNAmmer to predict rRNAs, and Infernal to predict miscRNAs. The Bacterial Pan Genome Analysis tool (BPGA) (Chaudhari et al., 2016) pipeline was used with default parameters, and a phylogenetic tree was constructed for the pan-/core-genome analysis of 25 representative Bacillus strains with sequenced genomes.
National Center for Biotechnology Information Blast+ v. 2.2.28 was used to align the protein sequences with those in the Pathogen-Host Interactions Database (PHI-base),2 along with those in the Clusters of Orthologous Groups of proteins (COG), Conserved Domain Database (CDD), NCBI non-redundant protein sequences (nr), and protein family (Pfam) databases. Gene Ontology (GO3) analysis was performed using the protein annotation results from the Swiss-Prot and TrEMBL databases and annotation information from the UniProt database. Pathway enrichment analysis was conducted using the Kyoto Encyclopedia of Genes and Genomes (KEGG) Automatic Annotation Server.4 HMMER3 v. 3.1b1 was used to align the gene set protein sequences with the Carbohydrate-Active enZYmes database (CAZy),5 thereby obtaining the corresponding carbohydrate-active enzyme annotation information (E-value < 10 ––5). The Rapid Annotations using Subsystem Technology (RAST)6 (Overbeek et al., 2013) approach was used to categorise the gene sets of proteins involved in specific biological processes.
The secondary metabolite synthesis gene clusters of B. subtilis CTXW 7-6-2 were predicted and analysed for potential bacteriostatic ability using antiSMASH v. 4.0 (Blin et al., 2017).7 B. subtilis NCIB 3610 (produces red pigment, GenBank: CP020102.1) and B. subtilis subsp. subtilis 168 (does not produce red pigment, Gen Bank: NC_000964.1) were used as control strains for analysis of secondary metabolite synthesis gene clusters.
The enzymes encoded by yvmC [cyclo(L-leucyl-L-leucyl) synthase] and cypX (pulcherriminic acid synthase) are known to be involved in the production of the red pigment pulcherriminic acid by Bacillus spp. (Li et al., 2017). Hence, the amino acid sequences of these two enzymes were selected from the whole gene sequence of the antagonistic bacteria. ProtParam8 was used to analyse the physicochemical properties of the proteins. Meanwhile, Predict Protein9 and SWISS-MODEL10 were used to predict the secondary and tertiary protein structures, respectively.
Quantitative real-time PCR (qRT-PCR) was used to determine the relative expression levels of yvmC and cypX in the antagonistic bacteria cultured at 28°C for 1 and 5 days with shaking at 180 rpm, respectively. B. subtilis GUMT323, which cannot produce red pigment, was used as the control strain (provided by Professor Ding Haixia of Guizhou University, CCTCC no. M 2018873). Sangon Biotech Co., Ltd., designed and synthesised the primers for yvmC and cypX, the sequences of which were submitted to their online platform (Table 1).11 The rpoB gene was used as the internal control (Jiang et al., 2021). Total RNA extraction and reverse transcription were performed using the RNAprep Pure Bacteria Kit (Tiangen Biotech) and First Strand cDNA Synthesis Kit (Tiangen Biotech), according to the manufacturer’s instructions. The cDNA was diluted to 200 ng/μL for the qPCR assay with each gene-specific primer. qRT-PCR was performed using Talent qRT-PCR SYBR Green PreMix (Sangon Biotech Co., Ltd.) on the Bio-Rad CFX96 qRT-PCR system (Bio-Rad, Hercules, CA, USA). Four biological replicates were analysed. Relative gene expression was calculated using the 2 –Δ Ct method.
Statistical analysis was conducted using SPSS Statistics v. 13.0 software (SPSS Inc., Chicago, IL, USA). All treatments were performed using three biological replicates. Data are expressed as the mean ± standard deviation (SD). Differences between treatments were determined using one-way ANOVA and Tukey’s honest significant difference test. A p-value < 0.05 was considered to be statistically significant.
Strain CTXW 7-6-2 was a rod-shaped gram-positive bacterium with dimensions of 0.62–0.75 μm × 1.5–2.0 μm (Figure 1A). After culturing for 5 days, the strain formed a single medium-sized, round, flat, and dry colony that was opaque, grey-white in colour, and later produced a light red pigment. Following PCR amplification and sequence alignment analyses of 16S rRNA, gyrA, rpoB, and purH, the constructed phylogenetic tree showed that strain CTXW 7-6-2 clustered with B. subtilis, with a self-sustaining rate of 100% maximum likelihood and 1.00 Bayesian inference. Bacillus cereus ATCC 14579 was shown to be an outgroup (Figure 1B).
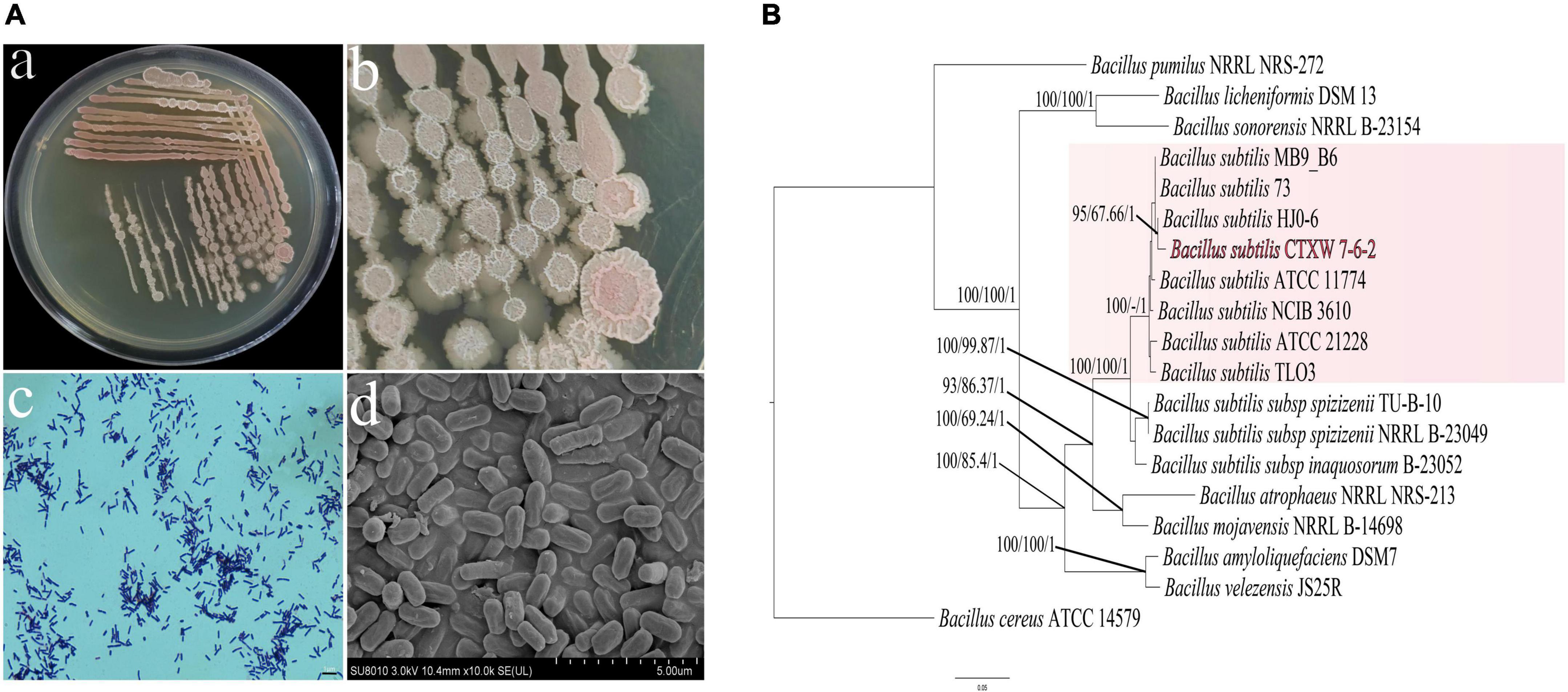
Figure 1. Morphological observations and analysis of housekeeping genes used to construct a phylogenetic tree. (Aa,b) Colony morphology in nutrient agar, (Ac) Gram staining. (Ad) Rod-shaped bacteria viewed under scanning electron microscopy. (B) A maximum likelihood tree was constructed using combined sequence analysis of the 16S ribosomal RNA, gyrA, rpoB, and purH genes. Bootstrap support values, maximum parsimony, and Bayesian inference posterior probabilities are given at each node based on 1000 bootstrap replicates.
A total of 25 bacterial strains were isolated, among which strain CTXW 7-6-2 displayed strong antagonistic effects using a double-layer double-culture assay. Table 2 lists the antagonistic effects of strain CTXW 7-6-2 against kiwifruit soft rot-causing pathogens, with growth of Phomopsis sp., A. alternata, and B. dothidea inhibited by 81.76 ± 3.554%, 69.80 ± 1.242%, and 32.03 ± 0.608%, respectively. Additionally, strain CTXW 7-6-2 inhibited the growth of F. fujikuroi, which causes leaf spot disease in kiwifruit, by 38.37 ± 2.265%. No inhibitory effect was observed against P. syringae pv. actinidiae, which causes bacterial canker in kiwifruit.
Strain CTXW 7-6-2 also exhibited antifungal effects against pathogenic fungi that cause diseases in tobacco, plums, and Chinese ground orchids. The most significant antagonistic effects were observed against R. solani, which causes target spot disease in tobacco, with the colony diameter reduced to 0.36 ± 0.191 cm and growth inhibited by 93.36 ± 3.529%. The antagonistic effects of strain CTXW 7-6-2 were also observed against C. spaethianum, which causes anthracnose in Chinese ground orchids, and M. fructicola, which causes brown rot in plums, inhibiting growth by 72.38 ± 0.673% and 56.87 ± 3.801%, respectively. Strain CTXW 7-6-2 displayed no inhibitory effect against P. agglomerans, which causes bacterial shot-hole disease in plums.
The VOCs of strain CTXW 7-6-2 had differing inhibitory effects against soft rot-causing pathogens in kiwifruit. The VOCs displayed no inhibitory effect against B. dothidea growth (Figure 2), but the colony diameter of A. alternata was reduced to 1.46 ± 0.25 cm, growth was inhibited by 76.76 ± 3.93%, and no melanin was produced (Figure 2B). The VOCs of strain CTXW 7-6-2 inhibited the growth of Phomopsis sp. by 54.48 ± 7.96% and reduced the colony diameter to 3.10 ± 0.54 cm.
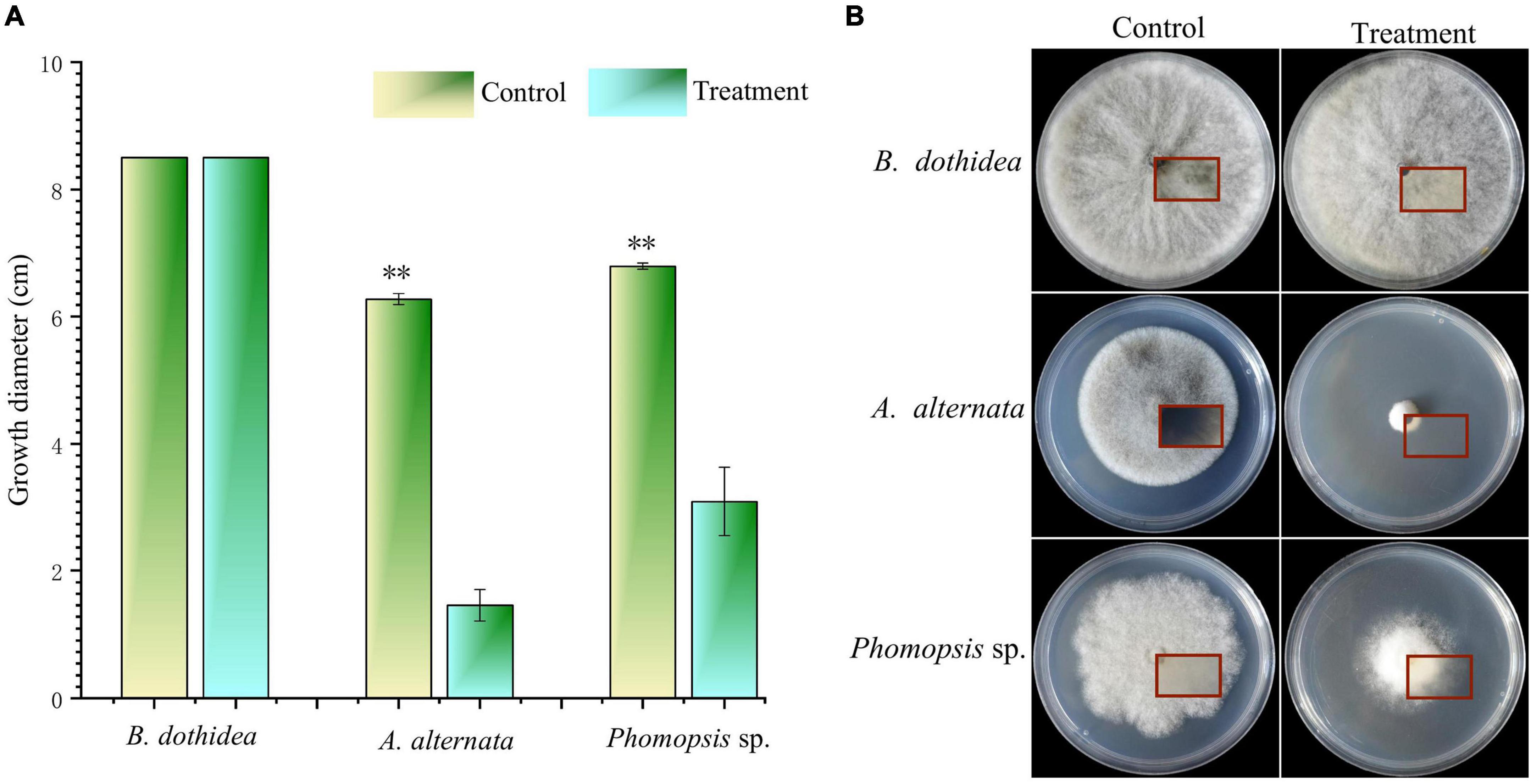
Figure 2. In vitro antagonism of the volatile compounds of strain CTXW 7-6-2 against kiwifruit soft rot-causing pathogenic fungi. Measurement of colony diameter (A) and colony morphology (B). The red square indicated the back of the colony. *A significant difference at p < 0.05. **A highly significant difference at p < 0.01.
Botryosphaeria dothidea, Phomopsis sp., and A. alternata displayed obvious growth inhibition zones near CTXW 7-6-2, with slow mycelial growth. The inhibited B. dothidea colonies were grey-black in colour, the tops and internodes of the hyphae were swollen, and their growth was hindered. The inhibited Phomopsis sp. colonies had neat edges, distorted hyphae, and showed dissolution. The inhibited A. alternata colonies were dense and whitish in colour, and the hyphae were swollen and became bead-like at each internode (Figure 3).
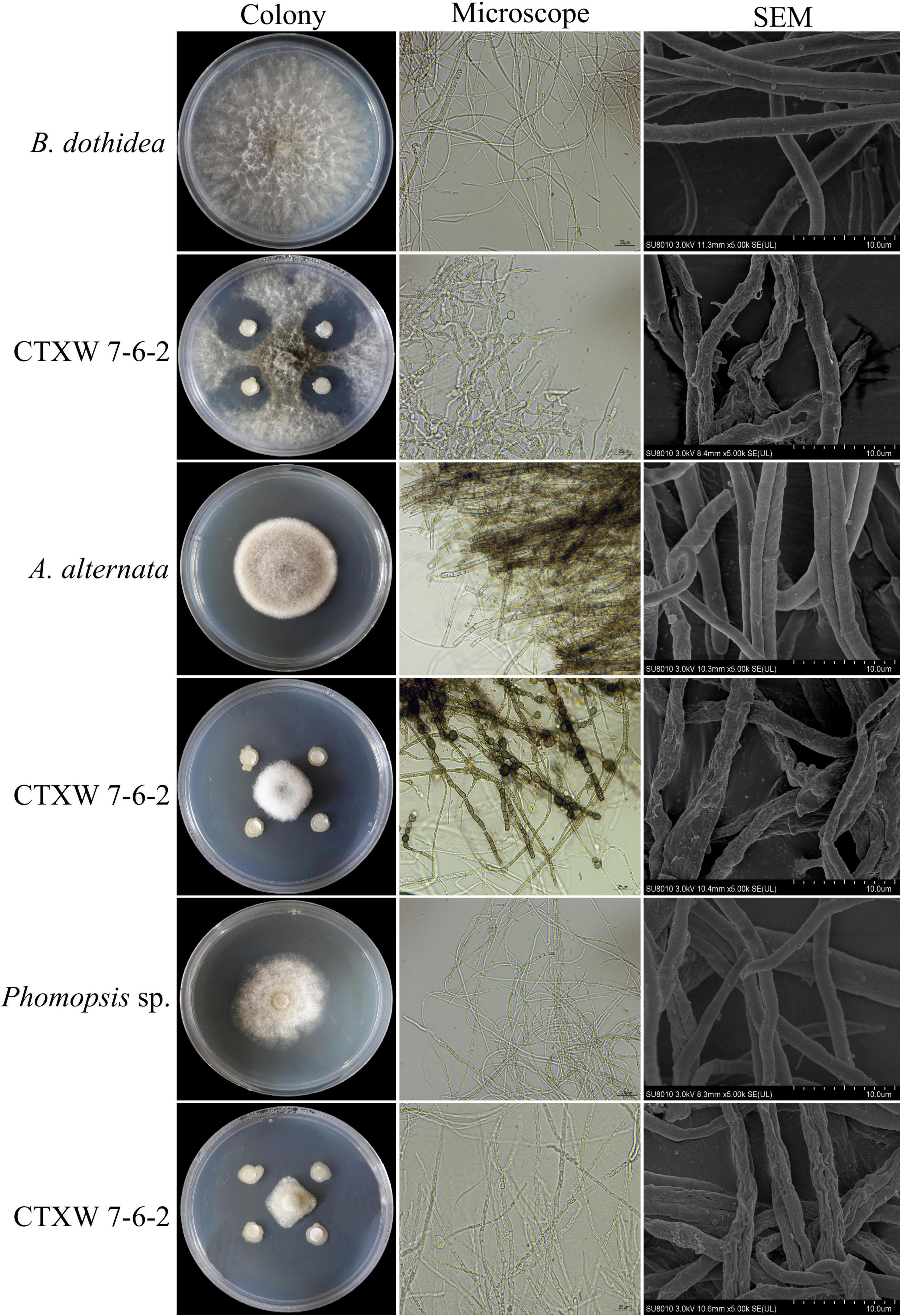
Figure 3. Comparison of colony and hyphal morphology of kiwifruit soft rot-causing pathogenic fungi before and after exposure to strain CTXW 7-6-2.
The SEM images revealed that the hyphae of the three pathogenic fungi were deformed to varying degrees. The untreated hyphae of each fungal species appeared smooth and plump. In comparison, the hyphae near CTXW 7-6-2 were rough, shrunken, twisted, and otherwise deformed, indicating severe damage (Figure 3).
The genomes of strain CTXW 7-6-2 and 25 representative Bacillus strains contained the same number of core genes and differing numbers of accessory, unique, and exclusively absent genes (Figure 4A). The phylogenetic analysis based on core and pan-genome data demonstrated that strain CTXW 7-6-2 was clearly clustered with B. subtilis and distinct from other Bacillus species (Figures 4B,C), with B. cereus ATCC 14579 as an outgroup. The B. subtilis CTXW 7-6-2 genome was finally assembled into 15 contigs with a total length of 4,221,676 bp (N50 contig size of approximately 1,046,756 bp), guanine and cytosine (GC) content of 43.41%, 4,428 protein-coding genes, 75 tRNAs, 10 rRNAs, and 45 unknown genes. A total of 3,079 annotations from the COG database were divided into 20 functional groups, mainly related to amino acid transport and metabolism, carbohydrate transport and metabolism, and transcription (Figure 4D).
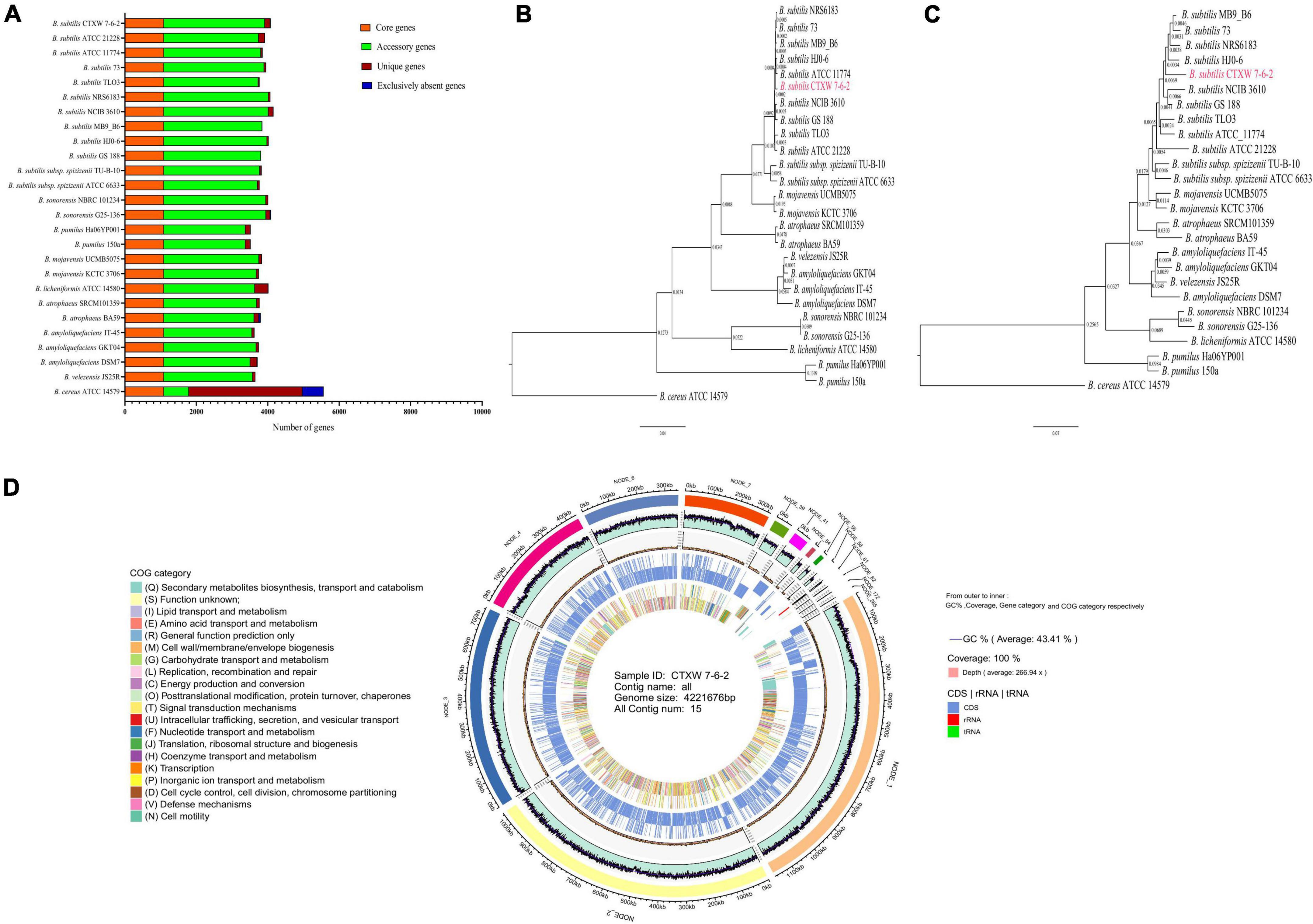
Figure 4. Comparison of genomes and circular genome map of strain CTXW 7-6-2. (A) Genome-wide proportions of core, accessory, unique, and exclusively absent genes were analysed among 26 Bacillus species. (B) Phylogenetic trees constructed based on core genomes. (C) Construction of phylogenetic trees based on pan-genomic analysis. (D) Displayed in order from outside to inside: scale bar, GC content (GC: ratio of guanine and cytosine), sequencing depth, gene element display, and clusters of orthologous group functional display.
The whole-genome of B. subtilis CTXW 7-6-2 was functionally annotated using eight gene annotation databases (Figure 5A). A total of 4,240 annotations were found in the nr database, accounting for 99.72% of all annotations. Additionally, 4,006 (94.21%), 3,583 (84.27%), 3,449 (81.11%), and 4,228 (99.44%) annotations were found in the Swiss-Prot, CDD, Pfam, and TrEMBL databases, respectively. A total of 1,775 KEGG annotations were associated with 34 metabolic pathways (Figure 5B). The genome of B. subtilis CTXW 7-6-2 had the highest number of genes associated with metabolism (2,104); carbohydrate, amino acid and cofactors, vitamin metabolism, and environmental information processing (412); genetic information processing (253); and cell processes (84). A total of 222 annotations were found in the CAZy database, including glycoside hydrolases (67), glycosyl transferases (61), carbohydrate esterases (46), auxiliary activities (11), carbohydrate-binding modules (27), and polysaccharide lyases (10) (Figure 5C). Glycosyl transferases, glycoside hydrolases, and carbohydrate esterase-related enzymes are involved in the synthesis of secondary metabolites via non-ribosomal pathways. Furthermore, B. subtilis CTXW 7-6-2 genome had 4 count proteins chitinase (GH18), 1 count proteins chitosanase (GH46), 13 count proteins endoglucanase (GH 5,51,16,26,11), and 8 count proteins lysozyme (GH23,73) encoding for possible antifungal CAZymes. AlsD (Alpha-acetolactate decarboxylase) and alsS (acetolactate synthase) genes encoding proteins are involved in the production of VOC 3-hydroxy-2-butanone. GO analysis revealed the main molecular functions of B. subtilis CTXW 7-6-2 were related to catalytic activity and binding function, while the main cellular components were related to cell and membrane parts (Figure 5D). In addition, 92 annotations were found in the PHI-base, including 10 proteins associated with loss of pathogenicity and 50 proteins associated with reduced virulence.
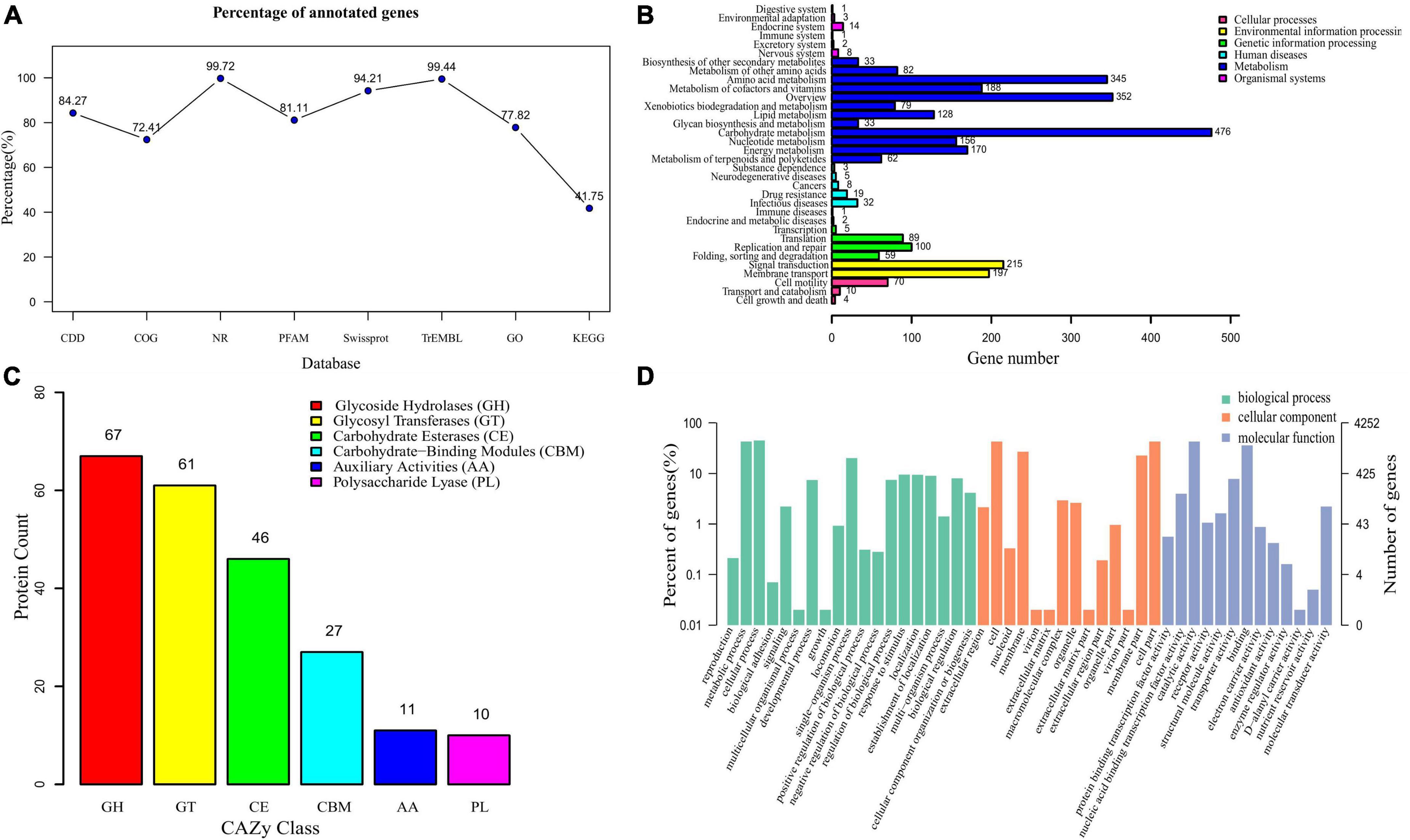
Figure 5. Functional analysis of gene and protein sequence annotations from B. subtilis CTXW 7-6-2. (A) Proportions of genes annotated in eight databases. (B) Metabolic pathways annotated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. (C) Proportions of enzymes annotated using the Carbohydrate-Active enZYmes database. (D) Biological process, cellular component, and molecular function terms annotated using the Gene Ontology database.
Rapid annotations using subsystem technology (RAST) subsystem analysis (Figure 6) revealed that the genome of B. subtilis CTXW 7-6-2 contained 245 genes involved in the metabolism of amino acids and their derivatives, 201 genes involved in carbohydrates, 161 genes involved in protein metabolism, 55 genes involved in RNA metabolism, and 132 genes involved in cofactors, prosthetic groups, vitamins, and pigment synthesis. The above annotation results indicated that various material and energy metabolism pathways provided the necessary nutrients and energy supply for the growth of B. subtilis CTXW 7-6-2. In addition, 27 genes were involved in iron acquisition and metabolism, 39 genes were involved in the stress response, 38 genes were involved in membrane transport, 40 genes were involved in motility and chemotaxis, 60 genes were involved in the cell wall and capsule, 6 genes were involved in secondary metabolism, 52 genes were involved in fatty acids, lipids, and isoprenoids, suggesting beneficial traits for potential biocontrol capabilities.
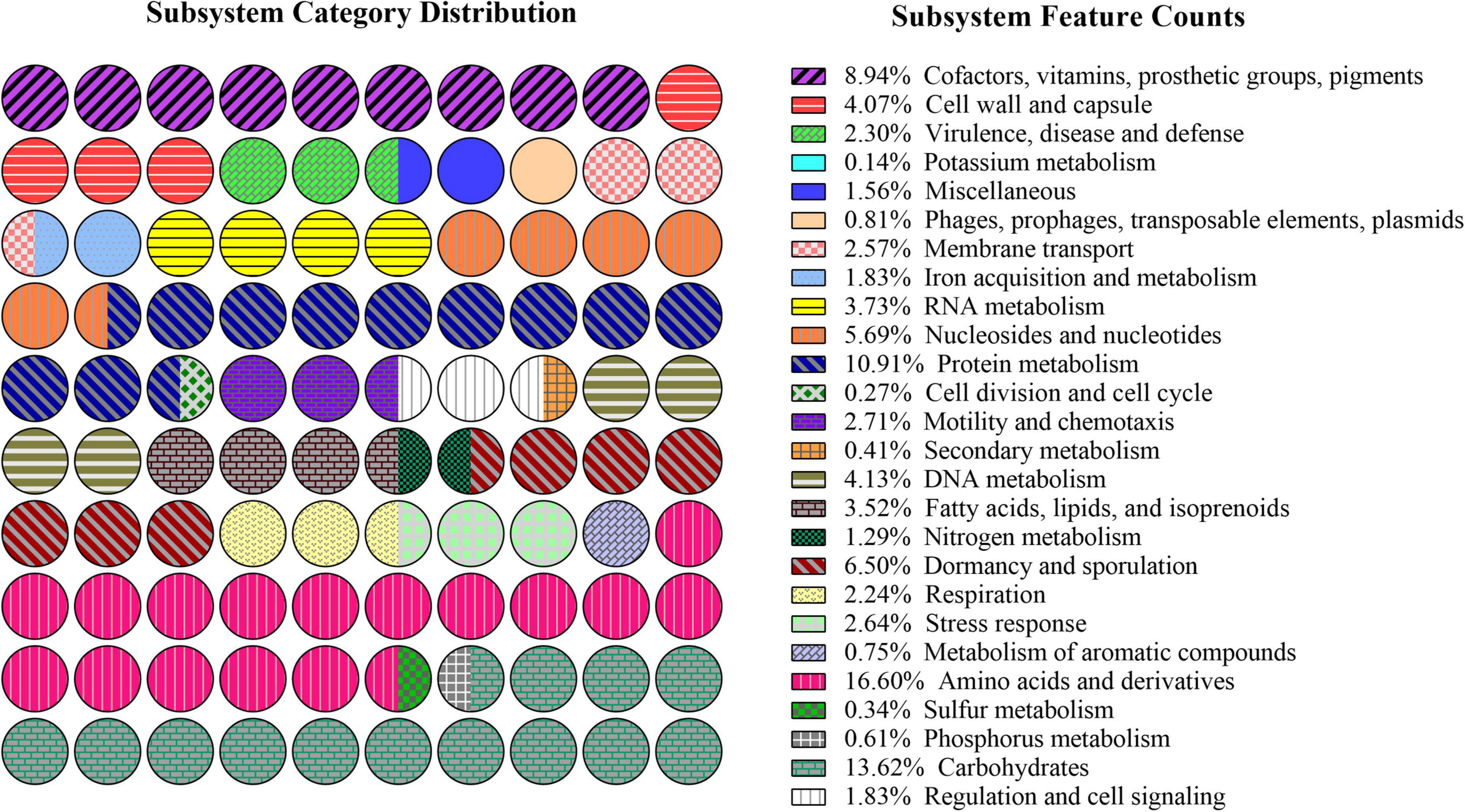
Figure 6. Subsystem distribution of the genome of B. subtilis CTXW 7-6-2 using the Rapid Annotations using Subsystem Technology (RAST) annotation server.
Predicting the metabolites produced by B. subtilis CTXW 7-6-2 is conducive to further analysis of its antifungal compounds. The antiSMASH analysis showed that B. subtilis CTXW 7-6-2 had 14 secondary metabolite biosynthetic gene clusters (BGCs), including five NRPS (non-ribosomal peptide synthetase) clusters, two putative NRPS + polyketide clusters, a polyketide cluster, and a thiopeptide cluster. The bioactive compounds with a predicted similarity of 100% were identified as bacillaene, bacillibactin, subtilosin A, bacilysin, and luminmide (Table 3). B. subtilis CTXW 7-6-2 also had four gene clusters with unknown functions, including one CDPS (tRNA-dependent cyclodipeptide synthase) cluster, one T3PKS (Type III PKS) cluster, and two terpene clusters (Figure 7), suggesting that some B. subtilis CTXW 7-6-2 gene clusters may synthesise new antibacterial substances. Schedule 1 and Table 3 show that CTXW 7-6-2 shared seven and six metabolic species classes with B. subtilis NCIB 3610 and 168, respectively. Luminmide, plipastatin, and basiliskamide A/B were unique to B. subtilis CTXW 7-6-2. Both B. subtilis CTXW 7-6-2 and NCIB 3610 had a predicted a gene cluster of unknown function containing yvmC as the core gene, in contrast to B. subtilis subsp. subtilis 168 (Schedule 1). Bacillus sp. that produce pulcherrimin are speculated to contain yvmC.
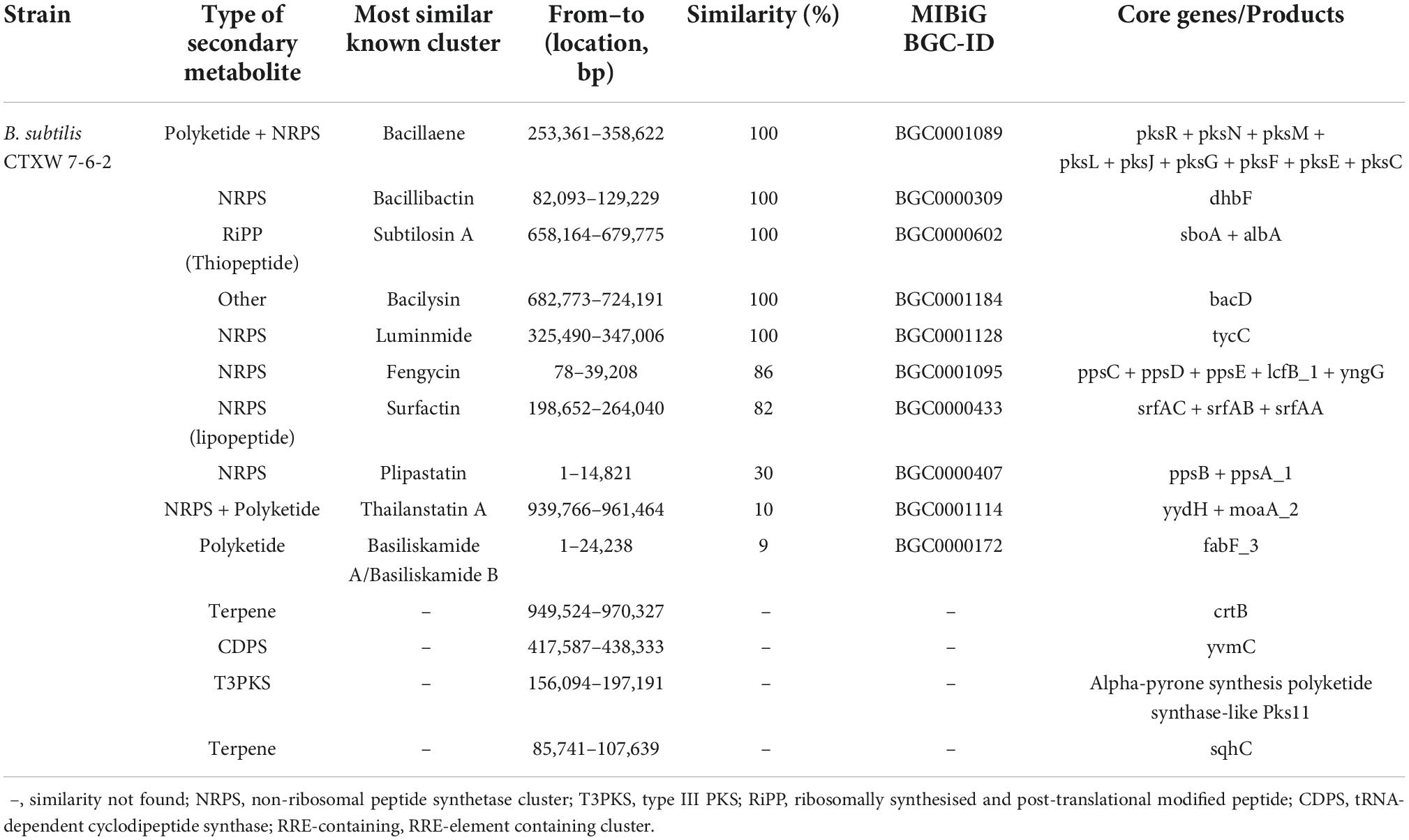
Table 3. Identification of secondary metabolite-related gene clusters in the genome of B. subtilis CTXW 7-6-2.
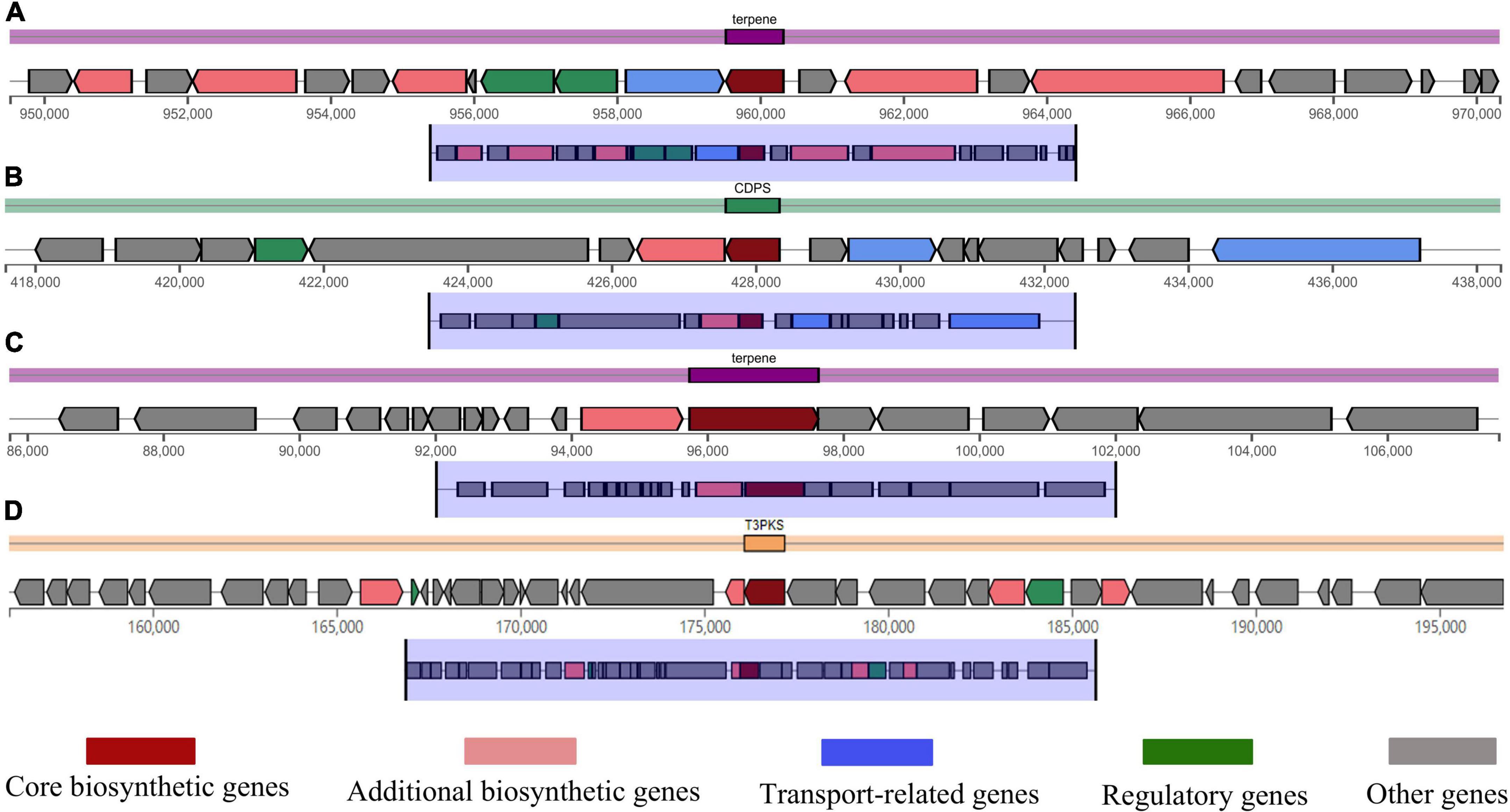
Figure 7. Domains of some gene clusters of B. subtilis CTXW 7-6-2. (A) Type terpene, crtB core gene. (B) Type CDPS, yvmC core gene. (C) Type T3PKS, alpha-pyrone synthesis polyketide synthase-like Pks11. (D) Type terpene, sqhC core gene.
The ProtParam analysis indicated that the molecular weights of the yvmC and cypX proteins of B. subtilis CTXW 7-6-2 were 28,506.74 and 45,499.08 Da and contained 248 and 405 amino acids, respectively. Additionally, the yvmC and cypX proteins of B. subtilis CTXW 7-6-2 had 33 and 41 positively charged residues (Arg + Lys) as well as 35 and 55 negatively charged residues (Asp + Glu), respectively. Moreover, their theoretical isoelectric points were 6.57 and 5.42 and their grand average of hydropathicity index values were −0.268 and −0.128, respectively. The calculated instability index values indicated that the yvmC (46.24) and cypX (50.78) proteins of B. subtilis CTXW 7-6-2 were negatively charged, unstable, and hydrophilic.
Predict Protein analysis of the secondary protein structure of yvmC showed that alpha helices, beta turns, extended strands, and random coils accounted for 48.39, 4.03, 12.90, and 34.68% of the total structure, respectively. Alpha helices, beta turns, extended strands, and random coils accounted for 50.37, 4.20, 10.86, and 34.57% of the secondary protein structure of cypX, respectively. The secondary structures of both proteins were dominated by alpha helix and random coil structures, followed by extended strands, and then beta turns (Figures 8A,B). The protein tertiary structure homology was predicted using SWISS-MODEL, as shown in Figures 8C,D.
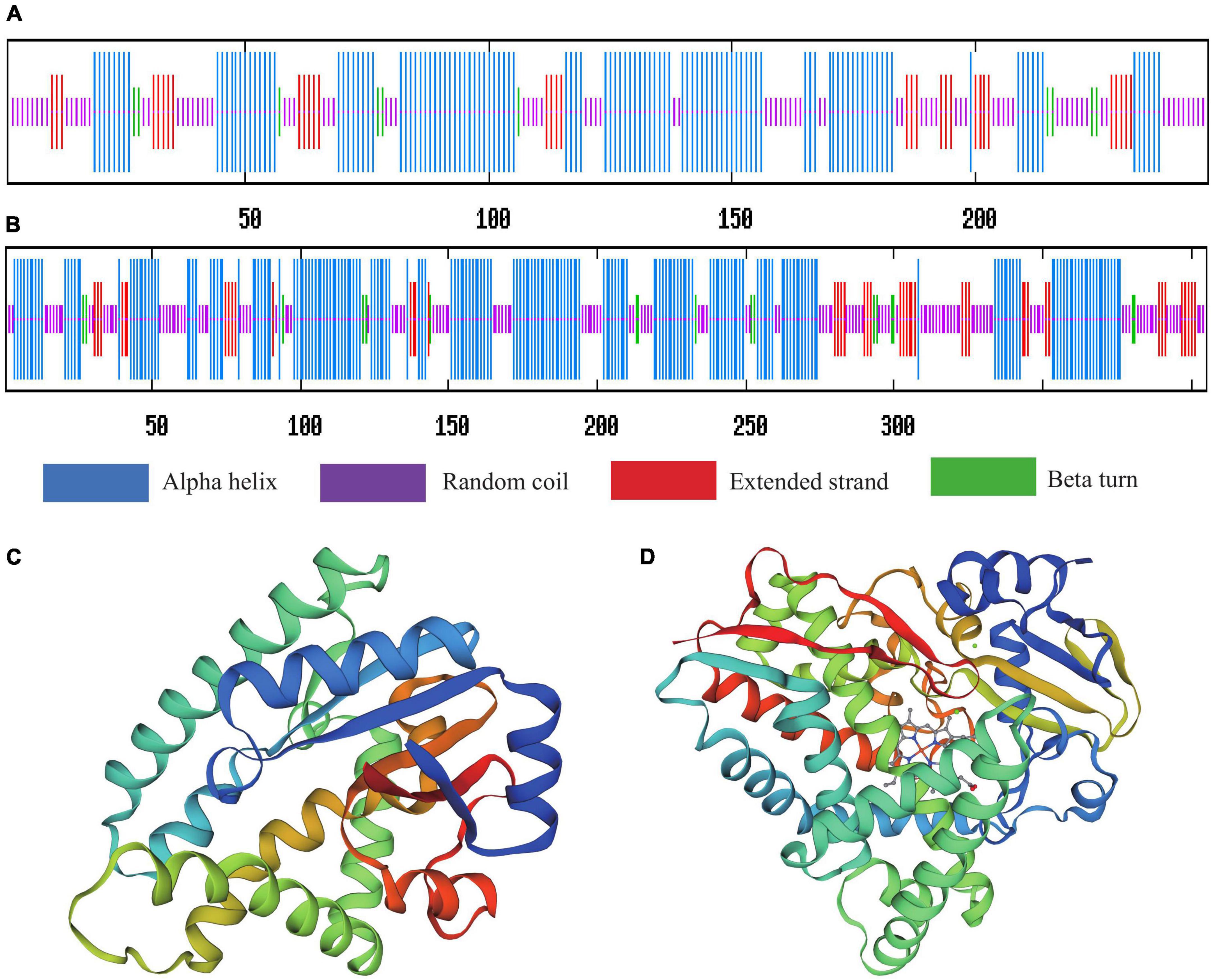
Figure 8. Secondary structure prediction and protein spatial conformation maps of (A,C) yvmC and (B,D) cypX of B. subtilis CTXW 7-6-2.
The relative expression levels of yvmC and cypX in B. subtilis CTXW 7-6-2 increased with culture time, increasing 6.6938 ± 0.517-fold and 0.983 ± 0.042-fold on day 5 compared to that on day 1, respectively (Figure 9). yvmC was more highly expressed than cypX in B. subtilis CTXW 7-6-2. In contrast, yvmC and cypX were relatively less expressed in B. subtilis GUMT323, with expression levels of lower than 0.06-fold.
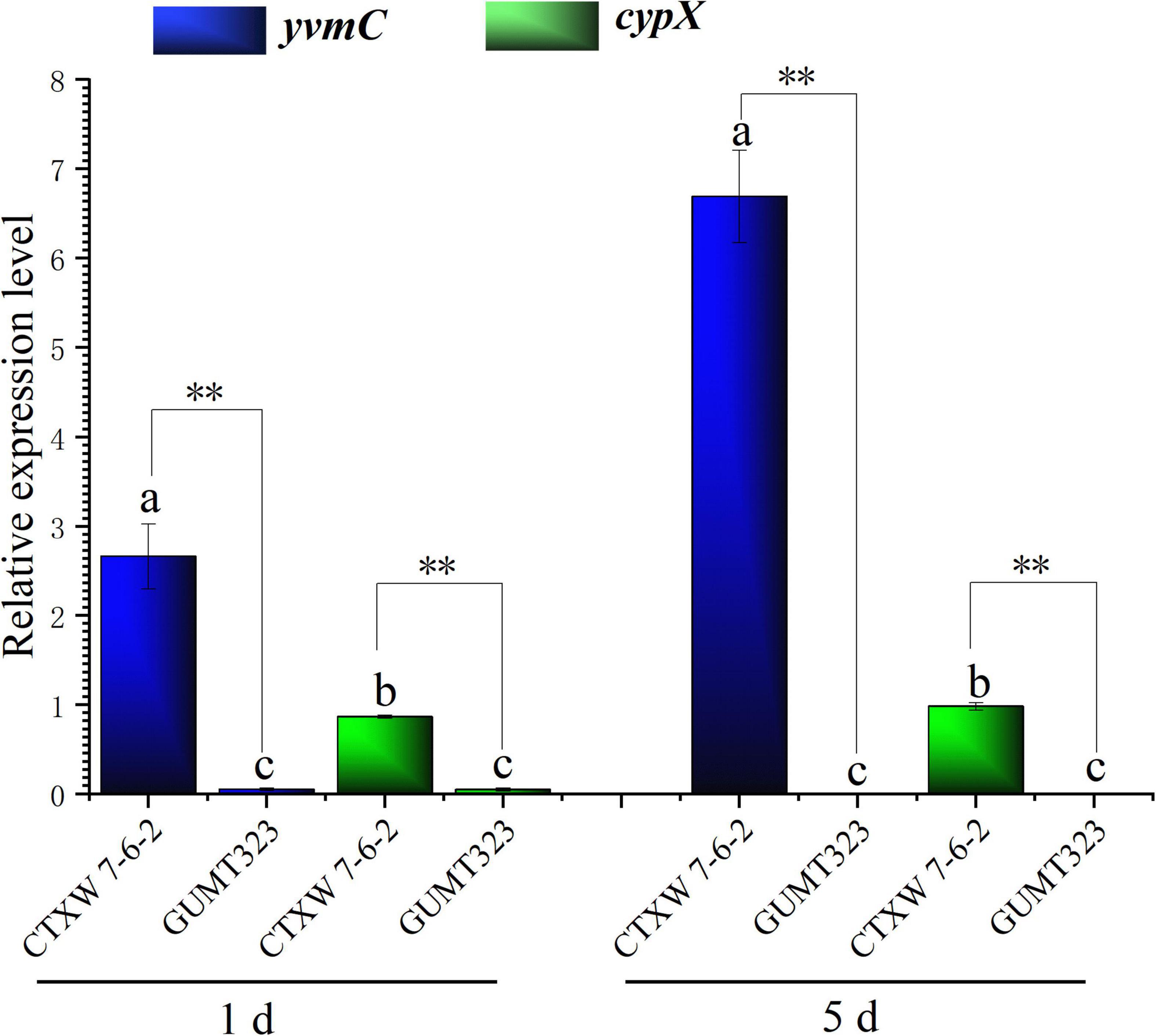
Figure 9. Quantitative real-time PCR analysis of yvmC and cypX gene expression in B. subtilis CTXW 7-6-2 and GUMT323, related to the production to pulcherriminic acid. Letters indicate significant differences between strains at p < 0.05, **indicates a significant difference at p < 0.01.
Soft rot threatens the yield, quality, and economic value of kiwifruit. The current study conducted a preliminary exploration of the biological control exerted by strain CTXW 7-6-2 against kiwifruit soft rot. Strain CTXW 7-6-2 was identified as a B. subtilis strain via morphological observation and combined analysis of 16S rRNA, gyrA, rpoB, and purH. Additionally, strain CTXW 7-6-2 produced a light-red pigment, which was consistent with an unknown B. subtilis strain isolated from coconut tissue culture (Salo and Novero, 2020). Strain CTXW 7-6-2 inhibited pathogenic fungi associated with diseases that infect kiwifruit, tobacco, plums, and Chinese ground orchids, and adversely affected the hyphae of the pathogenic fungi B. dothidea, Phomopsis sp., and A. alternata, which cause soft rot. Although the biologically active substances were not extracted, secondary metabolites were predicted in the current study, thus providing a theoretical basis for the extraction of active substances in the future. Additionally, the volatile substances produced by strain CTXW 7-6-2 inhibited the growth of Phomopsis sp. and A. alternata. Volatile substances produced by Bacillus spp. reportedly inhibit phytopathogenic fungi (Li et al., 2020; De la Cruz-López et al., 2022; Wang et al., 2022), affect soil microbial richness (Yuan et al., 2017) and promote plant growth (Ghazala et al., 2022; Rojas-Sanchez et al., 2022). Nevertheless, the mechanisms by which the strain and its volatile substances affect pathogenic fungi in vivo require further study.
Whole-genome sequencing and comparative genomics have become increasingly useful for exploring the molecular mechanisms and antifungal genes of bacteria as potential biocontrol agents. The whole-genome sequencing results in the current study indicated that secondary metabolites were actively involved in the antifungal mechanisms of B. subtilis CTXW 7-6-2. Gene and protein sequence annotation analyses predicted that many carbon, amino acid, and energy metabolism pathways were involved in iron acquisition and metabolism, motility and chemotaxis, membrane transport, VOC 3-hydroxy-2-butanone, cell wall-associated enzymes, and other related favourable traits. Moreover, glycoside hydrolases, glycosyl transferases, and carbohydrate esterase enzymes accounted for most of the identified carbohydrate-active enzymes, which are beneficial for secondary metabolite synthesis via non-ribosomal pathways.
The predicted secondary metabolite gene clusters of B. subtilis CTXW 7-6-2 revealed hitherto undetected secondary metabolite gene clusters, as well as various predicted lipopeptide antibiotics or proteins, including bacillaene, bacillibactin, subtilosin A, bacilysin, and luminmide. Bacillaene, a polyketide composed of low-molecular weight fatty acids (Chen et al., 2006), exerts antibacterial activity by inhibiting pathogenic protein synthesis. Bacillibactin is an iron chelator that can competitively bind soluble iron ions necessary for the growth and activity of pathogens (Roy and Griffith, 2017). Subtilosin A is a ribosomally synthesised and post-translationally modified peptide (RiPP) with broad-spectrum antibacterial activity that can act on and interfere with the phospholipid bilayer of various bacteria (Thennarasu et al., 2005). Bacilysin mainly disrupts the integrity of pathogenic cell membranes and damages cell wall structure (Loffler et al., 2010).
Interestingly, yvmC was predicted to be present in a gene cluster with unknown function in both red pigment-producing B. subtilis CTXW 7-6-2 and NCBI 3160, but was not predicted to be present in B. subtilis 168, which does not produce pulcherrimin. The formation of cyclo(L-leucyl-L-leucyl) is mediated by cyclodipeptide synthase YvmC, which utilises charged leucyl tRNAs (Sauguet et al., 2011). cypX encodes a cytochrome P450 implicated in the transformation of cyclo(L-leucyl-L-leucyl) into pulcherriminic acid (Max et al., 2010). Pulcherriminic acid combines with extracellular ferric ions to form the red pigment pulcherrimin in a non-enzymatic reaction. Pulcherrimin has been shown to effectively inhibit the growth of yeast and fungi (Attila et al., 2015) and affect biofilm formation (Arnaouteli et al., 2019). yvmC and cypX were highly expressed in B. subtilis CTXW 7-6-2 and the relative expression of yvmC was higher than that of cypX. In contrast, yvmC and cypX were expressed at lower levels in non-red pigment-producing B. subtilis GUMT323. Additionally, the yvmC and cypX proteins in B. subtilis CTXW 7-6-2 were negatively charged, unstable, hydrophilic, and rich in alpha helix and random coil structures. The study findings suggest that pulcherrimin may be partly responsible for the antagonistic effects of B. subtilis CTXW 7-6-2 against soft rot-causing pathogenic fungi in kiwifruit, which warrants further investigation.
This study isolated B. subtilis strain CTXW 7-6-2 from healthy kiwifruit, which produced the red pigment pulcherrimin, inhibited the growth of soft rot-causing pathogenic fungi, and destroyed the hyphal structure of the tested fungi. In vitro experiments demonstrated that B. subtilis CTXW 7-6-2 possessed broad-spectrum antifungal activity. Whole-genome sequencing revealed the beneficial traits responsible for the antagonistic effects of B. subtilis CTXW 7-6-2 and predicted the production of active substances such as bacillaene, bacillibactin, subtilosin A, bacilysin, and luminmide. The key genes involved in pulcherrimin production, yvmC and cypX, were highly expressed in B. subtilis CTXW 7-6-2, which supports future in-depth studies of B. subtilis-derived pulcherrimin as an antifungal agent. The study findings may facilitate the development of natural prevention and control treatments against kiwifruit soft rot.
16S rRNA, gyrA, rpoB, and purH are deposited under GenBank accession numbers: ON076886, ON086734, ON086735, and ON086736, respectively. The completed whole-genome sequence was submitted to NCBI under GenBank accession number: JAMKCC000000000.1 (https://www.ncbi.nlm.nih.gov/nuccore/?term=Bacillus+subtilis+CTXW+7-6-2).
TC and YL designed the research and wrote the manuscript. TC, ZZ, WL, XC, JC, BW, JM, and HD performed all the experiments. TC and WW analysed the data. All authors reviewed the final manuscript.
This research was supported by the National Natural Science Foundation of China (No. 32260707), Agricultural Research Projects of the Science and Technology Department of Guizhou Province (Nos. 2019 2403 and 2021 YIBAN237), the China Agriculture Research System of MOF (CARS-26), and the Talent Introducing Scientific Research Fund of Guizhou University (No. X2021029).
We gratefully acknowledge YL for useful discussions about this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Arnaouteli, S., Matoz-Fernandez, D. A., Porter, M., Kalamara, M., Abbott, J., MacPhee, C. E., et al. (2019). Pulcherrimin formation controls growth arrest of the B. subtilis biofilm. Proc. Natl. Acad. Sci. U.S.A. 116, 13553–13562. doi: 10.1073/pnas.1903982116
Attila, K., Jana, H., Jana, P., Lukáš, H., and Miroslava, K. (2015). Antimicrobial activity of pulcherrimin pigment produced by Metschnikowia pulcherrima against various yeast species. J. Microbiol. Biotechnol. Food Sci. 5, 282–285. doi: 10.15414/jmbfs.2015/16.5.3.282-285
Balestra, G. M., Mazzaglia, A., and Rossetti, A. (2008). Outbreak of bacterial blossom blight caused by Pseudomonas viridiflava on Actinidia chinensis Kiwifruit Plants in Italy. Plant Dis. 92, 1707–1707. doi: 10.1094/PDIS-92-12-1707A
Blin, K., Wolf, T., Chevrette, M. G., Lu, X., Schwalen, C. J., Kautsar, S. A., et al. (2017). antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 45, W36–W41. doi: 10.1093/nar/gkx319
Buchanan, R. E., and Gibbons, N. E. (1984). Bergey’s Manual of Determinative Bacteriology, 8th Edn. Beijing: China Science Press, 729–759.
Caulier, S., Nannan, C., Gillis, A., Licciardi, F., Bragard, C., and Mahillon, J. (2019). Overview of the antimicrobial compounds produced by members of the B. subtilis group. Front. Microbiol. 10:302. doi: 10.3389/fmicb.2019.00302
Chaudhari, N. M., Gupta, V. K., and Dutta, C. (2016). BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 6:24373. doi: 10.1038/srep24373
Chen, T. T., Wu, X., Dai, Y. Y., Yin, X. H., Zhao, Z. B., Zhang, Z. Z., et al. (2022a). Sensitivity testing of natural antifungal agents on Fusarium fujikuroi to investigate the potential for sustainable control of kiwifruit leaf spot disease. J. Fungi. 8:239. doi: 10.3390/jof8030239
Chen, T. T., Ran, F., Shi, J. Q., Mo, F. X., Yin, X. H., and Zhao, Z. B. (2022b). First report of Bacillus velezensis JK-F2 for the biological control of crown gall in kiwifruit. J. Plant Dis. Protect. 129, 1153–1162. doi: 10.1007/s41348-022-00634-3
Chen, X. H., Vater, J., Piel, J., Franke, P., Scholz, R., Schneider, K., et al. (2006). Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB 42. J. Bacteriol. 188, 4024–4036. doi: 10.1128/JB.00052-06
Cochrane, S. A., and Vederas, J. C. (2016). Lipopeptides from Bacillus and Paenibacillus spp.: a gold mine of antibiotic candidates. Med. Res. Rev. 36, 4–31. doi: 10.1002/med.21321
De la Cruz-López, N., Cruz-López, L., Holguín-Meléndez, F., Guillén-Navarro, G. K., and Huerta-Palacios, G. (2022). Volatile organic compounds produced by cacao endophytic bacteria and their inhibitory activity on Moniliophthora roreri. Curr. Microbiol. 79:35. doi: 10.1007/s00284-021-02696-2
Dhruw, C., Husain, K., Kumar, V., and Sonawane, V. C. (2020). Novel xylanase producing Bacillus strain x2: molecular phylogenetic analysis and its application for production of xylooligosaccharides. 3 Biotech. 10, 1–15.
Dong, X. Z., and Cai, M. Y. (2001). Manual for Systematic Identification of Common Bacteria. Beijing: China Science Press, 353–398.
Erturk, Y., Ercisli, S., Haznedar, A., and Cakmakci, R. (2010). Effects of plant growth promoting rhizobacteria (PGPR) on rooting and root growth of kiwifruit (Actinidia deliciosa) stem cuttings. Biol. Res. 43, 91–98.
Ghazala, I., Chiab, N., Saidi, M. N., and Gargouri-Bouzid, R. (2022). Volatile organic compounds from Bacillus mojavensis I4 promote plant growth and inhibit phytopathogens. Physiol. Mol. Plant Pathol. 121:101887. doi: 10.1016/j.pmpp.2022.101887
Hasan, S., Singh, K., Danisuddin, M., Verma, P. K., and Khan, A. U. (2014). Inhibition of major virulence pathways of streptococcus mutans by quercitrin and deoxynojirimycin: a synergistic approach of infection control. PLoS One 9:e091736. doi: 10.1371/journal.pone.0091736
He, L. N., Shi, J. Q., Zhao, Z. B., Ran, F., Mo, F. X., Long, Y. H., et al. (2021). First Report of Crown Gall of Kiwifruit (Actinidia deliciosa) Caused by Agrobacterium fabacearum in China and the establishment of loop-mediated isothermal amplification technique. Int. J. Mol. Sci. 23:207. doi: 10.3390/ijms23010207
He, X. R., Fang, J. C., Chen, X. F., Zhao, Z. F., Li, Y. S., Meng, Y. B., et al. (2019). Actinidia chinensis planch.: a review of chemistry and pharmacology. Front. Pharmacol. 10:1236. doi: 10.3389/fphar.2019.01236
Jiang, Z. R., Wang, C., Wu, Z. X., Chen, K., Yang, W., Deng, H. X., et al. (2021). Enzymatic deamination of the epigenetic nucleoside N6-methyladenosine regulates gene expression. Nucleic Acids Res. 49, 12048–12068. doi: 10.1093/nar/gkab1124
Kim, G. H., Koh, Y. J., Jung, J. S., and Hur, J. S. (2015). Control of postharvest fruit rot diseases of kiwifruit by antagonistic bacterium B. subtilis. VIII Int. Symp. Kiwifruit 1096, 377–382.
Kubo, Y., Rooney, A. P., Tsukakoshi, Y., Nakagawa, R., and Kimura, K. (2011). Phylogenetic analysis of B. subtilis strains applicable to natto (fermented soybean) production. Appl. Environ. Microbiol. 77, 6463–6469. doi: 10.1128/AEM.00448-11
Li, X. X., Wang, X. H., Shi, X. Y., Wang, B. P., Li, M. P., Wang, Q., et al. (2020). Antifungal effect of volatile organic compounds from Bacillus velezensis CT32 against Verticillium dahliae and Fusarium oxysporum. Processes 8:1674. doi: 10.3390/pr8121674
Li, X. Y., Wang, D., Cai, D. B., Zhan, Y. Y., Wang, Q., and Chen, S. W. (2017). Identification and high-level production of pulcherrimin in Bacillus licheniformis DW2. Appl. Biochem. Biotechnol. 183, 1323–1335. doi: 10.1007/s12010-017-2500-x
Loffler, W., Tschen, J. S. M., Vanittanakom, N., Kugler, M., and Wu, T. G. (2010). Antifungal effects of bacilysin and fengymycin from B. subtilis F-29-3 A comparison with activities of other Bacillus antibiotics. J. Phytopathol. 115, 204–213. doi: 10.1111/j.1439-0434.1986.tb00878.x
Manning, M., Burdon, J., De Silva, N., Meier, X., Pidakala, P., and Punter, M. (2016). Maturity and postharvest temperature management affect rot expression in’Hort16A’kiwifruit. Postharvest. Biol. Technol. 113, 40–47. doi: 10.1016/j.postharvbio.2015.10.012
Max, J., CryleStephen, G., and BellIlme, S. (2010). Structural and biochemical characterization of the cytochrome P450 CypX (CYP134A1) from B. subtilis: A Cyclo-l-leucyl-l-leucyl Dipeptide Oxidase. Biochemistry 49, 7282–7296. doi: 10.1016/S0090-4295(99)00216-2
Meng, F. B., Gou, Z. Z., Li, Y. C., Zou, L. H., Chen, W. J., and Liu, D. Y. (2022). The efficiency of lemon essential oil-based nanoemulsions on the inhibition of Phomopsis sp. and reduction of postharvest decay of kiwifruit. Foods 11:1510. doi: 10.3390/foods11101510
Mo, F. X., Hu, X. F., Ding, Y., Li, R. Y., and Li, M. (2021). Naturally produced magnolol can significantly damage the plasma membrane of Rhizoctonia solani. Pesticide Biochem. Physiol. 178:104942. doi: 10.1016/j.pestbp.2021.104942
Olishevska, S., Nickzad, A., and Deziel, E. (2019). Bacillus and Paenibacillus secreted polyketides and peptides involved in controlling human and plant pathogens. Appl. Microbiol. Biotechnol. 103, 1189–1215. doi: 10.1007/s00253-018-9541-0
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2013). The seed and the rapid annotation of microbial genomes using subsystems technology (rast). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Pan, H., Zhong, C. H., Xia, L. G., Li, W. Y., Wang, Z. P., Deng, L., et al. (2022). Antifungal activity of natamycin against kiwifruit soft rot caused by B. dothidea and potential mechanisms. Sci. Horticult. 305:111344. doi: 10.1016/j.scienta.2022.111344
Pan, L. Y., Zhao, X. Y., Chen, M., Fu, Y. Q., Xiang, M. L., and Chen, J. Y. (2019). Effect of exogenous methyl jasmonate treatment on disease resistance of postharvest kiwifruit. Food Chem. 305:125483. doi: 10.1016/j.foodchem.2019.125483
Pang, L., Xia, B., Liu, X. B., Yi, Y. J., Jiang, L. W., Chen, C., et al. (2020). Improvement of antifungal activity of a culture filtrate of endophytic Bacillus amyloliquefaciens isolated from kiwifruit and its effect on postharvest quality of kiwifruit. J. Food Biochem. 45:e13551. doi: 10.1111/jfbc.13551
Rajer, F. U., Wu, H., Xie, Y., Xie, S., Raza, W., Has, T., et al. (2017). Volatile compounds produced by a soil-isolate, B. subtilis fa26 induce adverse ultra-structural changes to the cells of Clavibacter michiganensis ssp. sepedonicus, the causal agent of bacterial ring rot of potato. Microbiology-sgm 163:523. doi: 10.1099/mic.0.000451
Rojas-Sanchez, B., Santoyo, G., Delgado-Valerio, P., and Rocha-Granados, M. D. (2022). Endophytic Bacillus spp. differentially promotes growth of three blackberry varieties. Bioagro. 34, 99–110. doi: 10.51372/bioagro342.1
Rooney, A. P., Price, N. P. J., Ehrhardt, C., Swezey, J. L., and Bannan, J. D. (2009). Phylogeny and molecular taxonomy of the B. subtilis species complex and description of B. subtilis subsp. inaquosorum subsp. nov. Int. J. Syst. Evol. Microbiol. 59, 2429–2436. doi: 10.1099/ijs.0.009126-0
Roy, E. M., and Griffith, K. L. (2017). Characterization of a novel iron acquisition activity that coordinates the iron response with population density under iron-replete conditions in B. subtilis. J. Bacteriol. 199:e00487. doi: 10.1128/JB.00487-16
Salo, E. N., and Novero, A. (2020). Identification and characterisation of endophytic bacteria from coconut (Cocos nucifera) Tissue Culture. Trop. Life Sci. Res. 31, 57–68. doi: 10.21315/tlsr2020.31.1.4
Sauguet, L., Moutiez, M., Li, Y., Belin, P., Seguin, J., Le Du, M. H., et al. (2011). Cyclodipeptide synthases, a family of class-I aminoacyl-tRNA synthetase-like enzymes involved in non-ribosomal peptide synthesis. Nucleic Acids Res. 39, 4475–4489. doi: 10.1093/nar/gkr027
Seneviratne, G., Weerasekara, M., Seneviratne, K., Zavahir, J., Kecsk’es, M., and Kennedy, I. (2010). “Importance of biofilm formation in plant growth promoting rhizobacterial action,” in Plant Growth and Health Promoting Bacteria. Microbiology Monographs, Ed. D. Maheshwari (Berlin: Springer), 81–95. doi: 10.1007/978-3-642-13612-2_4
Song, Y. L., Sun, L. M., Lin, M. M., Chen, J. Y., Qi, X. J., Hu, C. G., et al. (2019). Comparative transcriptome analysis of resistant and susceptible kiwifruits in response to Pseudomonas syringae pv. actinidiae during early infection. PLoS One 14:e0211913. doi: 10.1371/journal.pone.0211913
Su, Y., Liu, C., Fang, H., and Zhang, D. W. (2020). B. subtilis: a universal cell factory for industry, agriculture, biomaterials and medicine. Microb. Cell Fact. 19:173. doi: 10.1186/s12934-020-01436-8
Sumi, C. D., Yang, B. W., Yeo, I. C., and Hahm, Y. T. (2015). Antimicrobial peptides of the genus Bacillus: a new era for antibiotics. Can. J. Microbiol. 61, 93–103. doi: 10.1139/cjm-2014-0613
Thennarasu, S., Lee, D. K., Poon, A., Kawulka, K. E., Vederas, J. C., and Ramamoorthy, A. (2005). Membrane permeabilization, orientation, and antimicrobial mechanism of subtilosin A. Chem. Phys. Lipids 137, 38–51.
Wang, D. K., Li, Y. C., Yuan, Y., Chu, D. P., Cao, J. M., Sun, G. J., et al. (2022). Identification of non-volatile and volatile organic compounds produced by Bacillus siamensis LZ88 and their antifungal activity against A. alternata. Biol. Control 169:104901. doi: 10.1016/j.biocontrol.2022.104901
Wang, X. J., Dong, H. J., Lan, J. B., Liu, Y. Y., Liang, K., Lu, Q., et al. (2021). High-quality genome resource of the pathogen of Diaporthe (Phomopsis) phragmitis causing kiwifruit soft rot. Mol. Plant-Microbe Interact. 34, 218–221. doi: 10.1094/MPMI-08-20-0236-A
Wu, J. J., Huang, J. W., and Deng, W. L. (2020). Phenylacetic acid and methylphenyl acetate from the biocontrol bacterium Bacillus mycoides BM02 suppress spore germination in Fusarium oxysporumf. sp. Lycopersici. Front. Microbiol. 11:569263. doi: 10.3389/fmicb.2020.569263
Yan, F., Li, C., Ye, X., Lian, Y., Wu, Y., and Wang, X. (2020). Antifungal activity of Lipopeptides from Bacillus amyloliquefaciens MG3 against Colletotrichum gloeosporioides in loquat fruits. Biol. Control 146, 104281. doi: 10.1016/j.biocontrol.2020.104281
You, W. J., Ge, C. H., Jiang, Z. C., Chen, M. M., Li, W., and Shao, Y. Z. (2021). Screening of a broad-spectrum antagonist-Bacillus siamensis, and its possible mechanisms to control postharvest disease in tropical fruits. Biol. Control 157:104584. doi: 10.1016/j.biocontrol.2021.104584
Yuan, J., Zhao, M., Li, R., Huang, Q., Raza, W., Rensing, C., et al. (2017). Microbial volatile compounds alter the soil microbial community. Environ. Sci. Pollut. Res. 24, 22485–22493. doi: 10.1007/s11356-017-9839-y
Zeng, Q., Xie, J., Li, Y., Gao, T., Xu, C., and Wang, Q. (2018). Comparative genomic and functional analyses of four sequenced Bacillus cereus genomes reveal conservation of genes relevant to plant-growth-promoting traits. Sci. Rep. 8:17009. doi: 10.1038/s41598-018-35300-y
Keywords: kiwifruit soft rot, Bacillus subtilis, biocontrol, pulcherrimin, antifungal
Citation: Chen T, Zhang Z, Li W, Chen J, Chen X, Wang B, Ma J, Dai Y, Ding H, Wang W and Long Y (2022) Biocontrol potential of Bacillus subtilis CTXW 7-6-2 against kiwifruit soft rot pathogens revealed by whole-genome sequencing and biochemical characterisation. Front. Microbiol. 13:1069109. doi: 10.3389/fmicb.2022.1069109
Received: 13 October 2022; Accepted: 14 November 2022;
Published: 01 December 2022.
Edited by:
Raza Waseem, Nanjing Agricultural University, ChinaReviewed by:
S. Nakkeeran, Tamil Nadu Agricultural University, IndiaCopyright © 2022 Chen, Zhang, Li, Chen, Chen, Wang, Ma, Dai, Ding, Wang and Long. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Youhua Long, Z3pseWgxMjZAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.