
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 10 January 2023
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1067284
This article is part of the Research TopicPrevalence, Transmission and Control of Clinically Important Antimicrobial-Resistant Bacteria/Genes within One Health FrameworkView all 22 articles
 Muhammad Shoaib1†
Muhammad Shoaib1† Amjad Islam Aqib2*†
Amjad Islam Aqib2*† Iqra Muzammil3
Iqra Muzammil3 Noreen Majeed4
Noreen Majeed4 Zeeshan Ahmad Bhutta5
Zeeshan Ahmad Bhutta5 Muhammad Fakhar-e-Alam Kulyar6
Muhammad Fakhar-e-Alam Kulyar6 Mahreen Fatima7
Mahreen Fatima7 C-Neen Fatima Zaheer8
C-Neen Fatima Zaheer8 Afshan Muneer9
Afshan Muneer9 Maheen Murtaza9
Maheen Murtaza9 Muhammad Kashif10
Muhammad Kashif10 Furqan Shafqat10
Furqan Shafqat10 Wanxia Pu1*
Wanxia Pu1*Staphylococcus aureus is recognized as commensal as well as opportunistic pathogen of humans and animals. Methicillin resistant strain of S. aureus (MRSA) has emerged as a major pathogen in hospitals, community and veterinary settings that compromises the public health and livestock production. MRSA basically emerged from MSSA after acquiring SCCmec element through gene transfer containing mecA gene responsible for encoding PBP-2α. This protein renders the MRSA resistant to most of the β-lactam antibiotics. Due to the continuous increasing prevalence and transmission of MRSA in hospitals, community and veterinary settings posing a major threat to public health. Furthermore, high pathogenicity of MRSA due to a number of virulence factors produced by S. aureus along with antibiotic resistance help to breach the immunity of host and responsible for causing severe infections in humans and animals. The clinical manifestations of MRSA consist of skin and soft tissues infection to bacteremia, septicemia, toxic shock, and scalded skin syndrome. Moreover, due to the increasing resistance of MRSA to number of antibiotics, there is need to approach alternatives ways to overcome economic as well as human losses. This review is going to discuss various aspects of MRSA starting from emergence, transmission, epidemiology, pathophysiology, disease patterns in hosts, novel treatment, and control strategies.
Currently, there are 81 species and numerous subspecies in the genus Staphylococcus. The majority of the genus’s species are opportunistic pathogens or commensals of mammals. Numerous species have veterinary as well as medical significance. Staphylococcus aureus (S. aureus) is among the most prodigious and important staphylococcal species for human pathogenicity (Haag et al., 2019). The name Staphylococcus is derived from two Greek words, “staphyle,” which means cluster or bunch, and “kokkos,” means grapes, and so-called as “bunch of grapes” upon observation under microscope. The term “golden staph” is derived from the phrase “Staphylococcus aureus” which means “Golden Cluster Seed” (Ogston, 1881). S. aureus is a coccus-shaped, gram-positive, non-motile, non-spore-forming, opportunistic bacteria with biochemical profile as catalase, nucleases, lipases, coagulase, catalase, proteases, collagenases, and β-lactamase are the enzymes produced by S. aureus. It produces colonies in a variety of colors on various culture media such as pink colonies on chromogenic agar, golden or grayish-white colonies on blood agar, and yellow colonies on mannitol salt agar (Carter et al., 1995). Under microscope, S. aureus appears as rounded seeds arranged in bunches, demonstrating its growth in various planes. S. aureus is the cause of a wide spectrum of illnesses whose symptoms range from superficial to fatal manifestations. It may colonize diverse sites on both human and animal body surfaces due to its commensal as well as opportunistic characteristics. S. aureus is a common inhabitant of the skin, mucosa, urinary tract, gastrointestinal tract, and, in particular, the anterior nares of the respiratory tract (Cuny et al., 2010). S. aureus has the ability to produce a wide variety of virulence substances such as various types of proteins, enzymes, toxins, and other substances responsible for high pathogenicity. S. aureus produces fibronectin-binding protein and protein A, both of which contribute to the bacterium’s ability to adhere to and colonize cell surfaces. The type of toxins produced by S. aureus are alpha, beta, gamma hemolysins, Panton-Valentine leukocidin (PVL) toxins, exotoxins, and enterotoxins. These all aid in the spread of S. aureus infection, which can cause severe blood stream and necrotizing infections in individuals (Gillet et al., 2002). This review article will cover the following aspects related to methicillin resistant strain of S. aureus; prevalence, transmission, pathogenesis, diseases, treatment, and prevention.
First time this bacteria was isolated from pus sample and given the name of “S. aureus” in 1881 (Ogston, 1881). Before the availability of penicillin, which was discovered by Radetsky (1996), an increased number of deaths were reported due to S. aureus infection that reached 90% case fatality rate which persisted up-to 19th century (Jevons, 1961). Later on production of β-lactamase enzyme by S. aureus makes the penicillin useless due to hydrolyzes of β-lactam ring of penicillin and S. aureus become resistant to penicillin soon after its discovery. Then, another antibiotic with the name of methicillin were discovered in 1950 which also found effective against S. aureus for long time. Unfortunately, the bacteria also acquired a significant resistance to this antibiotic and makes it ineffective anymore. The resistance to this antibiotic was reported at increased percentage which was name as methicillin resistant strain of S. aureus (MRSA). The molecular basis of MRSA was originated from a large mobile genetic element known as the staphylococcal cassette chromosome mec (SCC mec) genes such as mecA gene which are obtained by methicillin susceptible S. aureus (MSSA) through horizontal gene transfer among bacteria. The resistance was exhibited to all β-lactam antibiotics due to production of penicillin binding protein (PBP-2α) encoded by mecA gene (Vengust et al., 2006). In 1961, a study was conducted which noted among 50 staphylococci samples, 18 were found resistant to methicillin indicating high percentage (36.0%) of MRSA. These isolates were discovered to have the potential to keep both coagulase and hemolytic activity. MRSA detection was difficult in the early years of its discovery because methicillin resistance in S. aureus was diverse among different isolates and in a study, 5444 S. aureus samples were tested and only 3 isolates were diagnosed as MRSA (Barrett et al., 1968). As a result, heterogeneous strains mostly consist of bacterial cells that are both highly resistant and susceptible to methicillin. However, the addition of sugar or sodium chloride (NaCl) to the culture medium may promote the expression of phenotypic resistance in presence of β-lactam antibiotics (Datta et al., 2011).
From the long time, MRSA is considered as prototype of MDR and nosocomial pathogens which cause infection in hospitals and other healthcare settings. In the past three decades, the percentage of MRSA infections has significantly risen, and new strain of MRSA known as healthcare-associated (HA-MRSA) has spread and become endemic throughout industrialized nations as the leading cause of life-threatening infections like pneumonia, skin and bloodstream infections (Diekema and Pfaller, 2001). According to a US study, S. aureus infections are responsible for seven million hospitalizations of humans in the country, demonstrating the significant loss caused by hospital-acquired MRSA (HA-MRSA). The estimated yearly cost of these infections is $2.7 million, a considerable loss of 12,000 annual deaths, and sets the country’s economy at financial stress of over $9.5 billion (Noskin et al., 2005). First, MRSA was primarily associated with healthcare settings, and the risk factors that contributed to its spread were well-known (Chambers, 2001). A new type of MRSA rapidly emerged toward the end of the 1990s in the community and was known as community-acquired (CA-MRSA), with very high level of pathogenicity and the potential to spread, making it capable of infecting young as well as healthy individuals (DeLeo et al., 2010). Recently, MRSA also reported as frequent colonizer among animals due to extensive and frequent use of antibiotics in animal production. This new MRSA strain called as livestock associated MRSA (LA-MRSA) is primarily identified in food animals such as pigs, cattle, sheep, and goat with having extensive zoonotic potential (Pantosti, 2012). These cases of MRSA infection in humans and animals show how animals can serve as a source for the transmission of the disease, thus posing a serious threat to the population (Cefai et al., 1994). Rising public concern over MRSA has led to monitoring recommendations, such as information on the prevalence of the infection in healthy dogs, cats, and humans (Noskin et al., 2005).
The global emergence and transmission of MRSA is one the most important aspect in the epidemiology of MRSA. The spread of all types of MRSA have been reported from many countries as listed in Table 1. The spread of MRSA is known to occur by one of the two ways which are either spread of existing clones among humans, animals, from animals to humans or humans to animals and acquisition of SSCmec element through horizontal genes transfer (Lee et al., 2018). Currently, MRSA is known to be more endemic in hospital settings and is among the major nosocomial pathogens. According to the statement of CDC, MRSA is known to be a major threat to public health because of its increasing prevalence in hospitals, community and animals, transmission between humans and animals, infection rates, resistance, and therapeutic issues (Ferri et al., 2017). On an average, it was estimated the annual health cost due to MRSA infections accounts 3 billion dollars. CA-MRSA has been also emerging as a principle pathogen from the recent years. It is noted MRSA mostly causes skin and soft tissue infection leading to bacteremia which lead to higher mortality rates ranging from 15 to 60% (Lee et al., 2013).
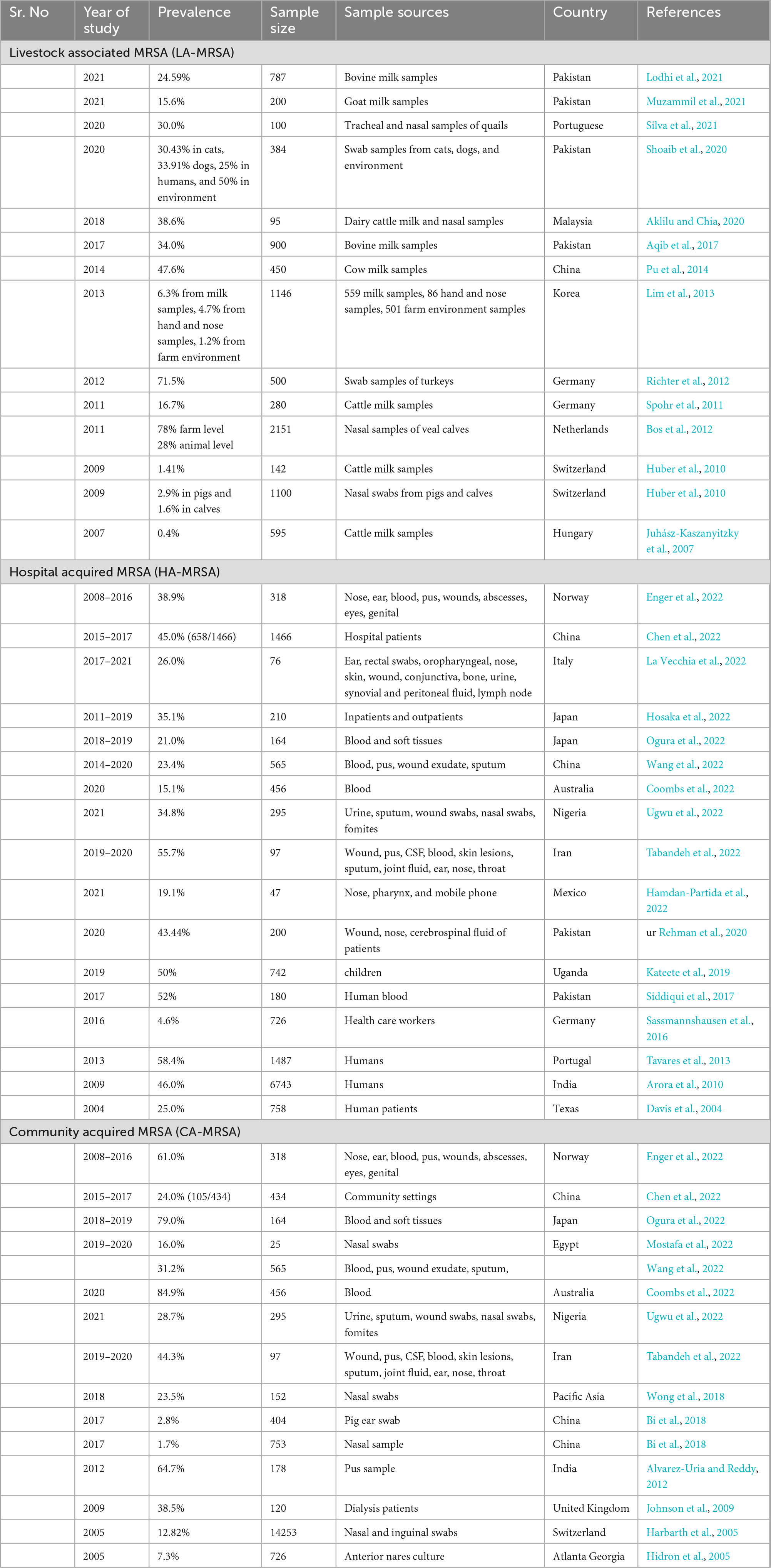
Table 1. Prevalence of livestock associated MRSA (LA-MRSA), healthcare-associated MRSA (HA-MRSA), and community-acquired MRSA (CA-MRSA) in different countries.
Recent trend in the prevalence of HA-MRSA was noted varying among the countries for example it was noted higher 58.4% from Portugal in 2013 (Tavares et al., 2013), 46% from India in 2009 (Arora et al., 2010), 52% from Pakistan in 2017 (Siddiqui et al., 2017), 45% from China from 2015 to 2017 (Chen et al., 2022), and 38.9% from Norway from 2008 to 2016 (Enger et al., 2022). However, with the increasing prevalence of HA-MRSA in different countries, MRSA prevalence also noted lower in many such as 4.6% from Germany (Sassmannshausen et al., 2016), 25% from Texas (Davis et al., 2004), 19.1% from Mexico (Hamdan-Partida et al., 2022), 15.1% from Australia (Coombs et al., 2022), and 26% from Italy (La Vecchia et al., 2022) are summarized in Table 1. Similar increasing and decreasing trend of MRSA prevalence from different countries also noted for CA-MRSA such as 79% from Japan (Ogura et al., 2022), 84.9% from Australia (Coombs et al., 2022), 64.7% from India (Alvarez-Uria and Reddy, 2012), 61% from Norway (Enger et al., 2022), and 44.3% from Iran (Tabandeh et al., 2022) with lower prevalence from Egypt (16%) (Mostafa et al., 2022), China [1.7% (Bi et al., 2018); 24% (Chen et al., 2022)], 7.3% from Gerorgia (Hidron et al., 2005), and 12.8% from Switzerland (Harbarth et al., 2005). This decline in the prevalence of HA-MRSA and CA-MRSA may be linked with better implementation national prevention measures. This increase and decline in the prevalence of HA−MRSA and CA-MRSA may be linked with increasing acquisition of LA-MRSA from animal reservoirs to humans especially from food and companion animals. The predominant LA-MRSA strain which is also identified from human MRSA isolates belong to CC398 as illustrated in Figure 1. However, person to person transmission of LA-MRSA found to be uncommon. The prevalence of LA-MRSA from different studies also noted higher from different countries as mentioned in Table 1. Recent studies conducted in Pakistan detected 15.6% LA-MRSA from goat milk (Muzammil et al., 2021), 24.5% from cow milk (Lodhi et al., 2021), 30.4% from cats (Shoaib et al., 2020), and 33.9% from dogs (Shoaib et al., 2020). Another study conducted in Malaysia in 2018 detected 38.6% MRSA from cow milk (Aklilu and Chia, 2020). Furthermore, two studies from Switzerland reported 1.41% from cow milk (Huber et al., 2010), 2.9% from pig nasal swabs (Huber et al., 2010), and 1.6% from calf nasal swabs (Huber et al., 2010) (summarized in Table 1).
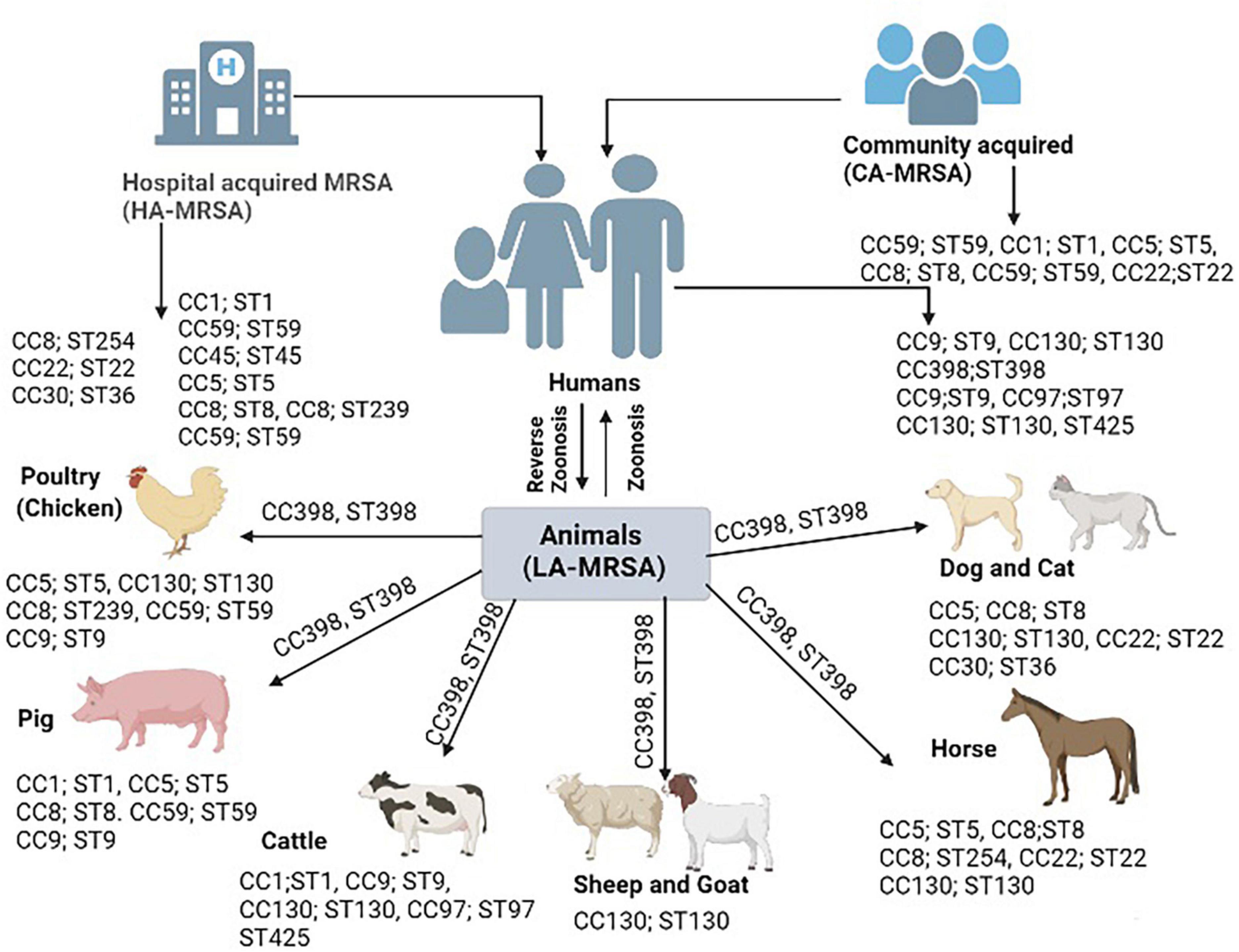
Figure 1. Bovine-adapted clonal complexes Staphylococcus aureus (S. aureus CCs) appear to have derived from human CCs and acquired bovine affinities through a series of spill-over events that resulted in the acquisition of various mobile genetic elements (MGEs). Several hosts have a high prevalence of the CC398 lineage. This lineage seems to have started in humans via reverse zoonosis, then it spread to pigs, then it returned to humans via pig zoonosis, and ultimately it spread to other species.
Transmission of MRSA among different hosts is primarily known to happen by physical contact with source. The capability of transfer of MRSA among different host species including humans and animals is the characteristic feature of MRSA lineages. HA-MRSA is primarily acquired from hospital settings such as contaminated instruments, bedding, doors, and equipment’s while CA-MRSA is primarily acquired by physical contact with infected or healthy person as S. aureus is a commensal bacterial in the nares of healthy individuals. LA-MRSA transmission to humans when the individual has physical contact with animal and environment (Pantosti, 2012). Firstly, LA-MRSA was restricted only to animals until 1961 before the Hungarian cow was reported to be the source of LA-MRSA transfer to its caretaker by testing throat swabs (Cefai et al., 1994). This was the first report of MRSA transmission from animal to human which proved the ability of MRSA horizontal transmission among animals and humans. Later on a number of reports were published by various authors from different regions of world from different animal’s species such as poultry, pigs, cattle, sheep and goat, equines, and companion animals. These reports noted number of clonal complexes (CCs) such as CC5, CC8, CC9, CC59, CC1, CC30, CC45, CC22, CC130, CC97, and CC398 with multi-locus sequence types (STs) were found similar among human isolated MRSA strains and animal isolated MRSA strains. On the other hand, various HA-MRSA and CA-MRSA strains are also found similar to other LA-MRSA strains which are elaborated in Figure 1. A human clone ST1 was found in animals and is responsible of causing mastitis in bovines (Grundmann et al., 2010). Similarly, animals clone CC398 and lineage ST398 found among humans’ which cause infections similar to HA-MRSA and CA-MRSA (Witte et al., 2007). Moreover, a worldwide clone of poultry ST5 also found among humans working at poultry farms (Lowder et al., 2009). Similarly, a clone of small ruminants CC130; ST 130 also recovered from humans (Guinane et al., 2010). The MRSA transmission from companion animals to humans is also well-documented in various studies for example a study conducted in USA and Canada documented 18% carriage rate of MRSA among the owners of companion animals (Faires et al., 2009). Another study in UK in nursing home documented similar strain in patient, hospital staff and nursing cat (Scott et al., 1988). Similarly, a study conducted at veterinary hospital detected transmission of MRSA lineage ST22 from infected dogs to veterinary staff (Baptiste et al., 2005). However, pets also found to be colonized with human PVL positive CA-MRSA strain by a household member in Netherland (Van Duijkeren et al., 2005). The risk of getting MRSA in animal caretakers (1.7%) was higher compared to those who were not exposed to animals (Wulf et al., 2006). For 13 months, eleven horse patients were hospitalized at the veterinary hospital for various diagnoses and surgical procedures. After procedures, a strain of MRSA was identified. MRSA strain was also isolated from 3 of 5 and with 1 person found to be colonized with 2 biotypes of MRSA. The isolates of human MRSA appeared to be identical to those of horses. The results showed that the isolates of horses and humans are members of a very close group and, apparently come from a common source. According to the pattern associated with the infection, a special mode of transmission was still unknown, but it was assumed that the main cause of the infection is the staff of the veterinary hospital (Seguin et al., 1999).
A number of risk factors also play a significant role in the spread of CA-MRSA and HA-MRSA infections. Important risk variables for cellulitis were overweight, the existence of abscesses, and head-and-neck sores relative to infections produced by other microorganisms according to an analysis of individuals with the condition. The presence of abscesses and obesity were additional important risk variables for MRSA dermatitis (Khawcharoenporn et al., 2010). Significant correlations between MRSA colonization, skin infection in the previous 3 months, sharing soap, and MRSA skin and soft tissue infection (SSTI) against no SSTI were found, college education, knowing about “staph” before, taking bath daily, and the previous contact with a health care worker (Haysom et al., 2018). Further research compared those with MRSA to those with MSSA to identify risk variables for MRSA colonization. According to research, there are a variety of meaningful risk factors for MRSA infection, such as the involvement of family members under the 7 years of age, a smoking habit, and the consumption of antibiotics the year before. Additional characteristics including age, sex, married status, chronically sick patients, education, taking a daily shower, and family income, however, were not meaningful risk factors for MRSA colonization (Wang et al., 2009).
Except these risk factors in humans, a study also highlighted the risk factors associated with LA-MRSA mammary infection in dairy animals. Among those risk factors are animal parity number, age, feeding status, body condition score, udder hygiene, hand or machine hygiene while milking were important factors associated with this infection while milking frequency was found non-significant risk factor for LA-MRSA infection (Aqib et al., 2017). Another study conducted by Shoaib et al. (2020) highlighted the risk factors associated with transmission of LA-MRSA from companion animals. Among those risk factors, animal health status, infection on body, long term antibiotic therapy, veterinarians, pet access to bedroom were found significant risk factors in transmitting MRSA to humans while the size of dog, owner’s sex, and sample site were found to be non-significant risk factors associated with MRSA transmission. Another study conducted by Mulders et al. (2010) highlighted the possible risk factors of MRSA transmission from poultry to humans are farm workers, individuals having contact with live birds at slaughterhouse, type of slaughtering method, and slaughtering environment are significantly correlated with higher carriage of MRSA among humans.
Staphylococcus aureus is a pathogenic and commensal bacterium that normally lives in the anterior nares of both humans and animals as well as in axillae, groin, and gastrointestinal tract are sites where it can also colonize. The major steps in pathogenesis of infection are colonization, virulence, initiation of infection, abscess formation, systemic infection, regulation and adaptation with the help of number of virulence factors. Colonization enhances the risk of bacterial infection when the host’s defenses are compromised either by physical disruption or other diseases (Wertheim et al., 2005). As MRSA is the methicillin-resistant strain of S. aureus, the S. aureus by self-contain a number of potential virulence factors which includes several surface proteins called “microbial surface components recognizing adhesive matrix molecules” (MSCRAMMs), which bind to fibrinogen, fibronectin, and collagen fibers of host cells to attack the host tissues. These factors may lead to infections of prosthetics, bones, joints, and endovascular system (Menzies, 2003). The ability of S. aureus to produce biofilm on both prosthetic surfaces and the host allows it to adhere to those surfaces by evading the effect of antimicrobials and host immune system. S. aureus also have the ability of produce small colony variants (SCVs) which have ability to cause persistent and recurrent infections. S. aureus also contain the anti-phagocytic microcapsule (type 5 or 8) which act as a primary defense mechanism. Moreover, due to internaction of Zwitter ionic capsule with Fc region of an immunoglobulin, the MSCRAMM protein A facilitate the protection of S. aureus from opsonization (Gordon and Lowy, 2008). Except these S. aureus contain a number of virulence factors such as adhesion proteins, chemotaxis inhibitory proteins, various enzymes such as proteases, lipases, hyaluronidase, staphylokinase, catalase, nucleases, lipases, coagulase, catalase, proteases, collagenases, β-lactamases, and elastases which help the S. aureus in causing infection in host. Except these virulence factors, MRSA also contain different mobile genetic elements (MGEs) in different animals’ species as listed in Table 2 that also aid and enhance its pathogenicity. Besides these, S. aureus produces various type of toxins such as exotoxins, enterotoxins, TSST-1, hemolysin toxins and PVL toxins as illustrated in Figure 2. Additionally, certain strains of S. aureus release superantigens that can also cause infections such as food poisoning and toxic shock syndrome (TSS) (Dings et al., 2000). Normal expression of S. aureus virulence factors plays a significant role in pathogenesis. Virulence factors express only according to the requirement of the bacterium to decrease unnecessary metabolic demands. Although secreted proteins like toxins are generated during the stationary phase, MSCRAMMs typically express during logarithmic growth phase. Early MSCRAMM protein expression helps in the initial colonization of tissue sites, while late toxin production helps in the dissemination of infection into the bloodstream. Mainly the S. aureus pathogenicity is regulated by the quorum-sensing accessory gene regulator (AGR) (Gordon and Lowy, 2008). S. aureus, in short, has a wide range of ways to cause disease and evade host defenses. However, the existence of some virulence factors is independent of the genomic structure and are related to clonal type (Peacock et al., 2002).

Table 2. Methicillin resistant strain of Staphylococcus aureus (MRSA) mobile genetic elements (MGEs) associated host determinants in different species.
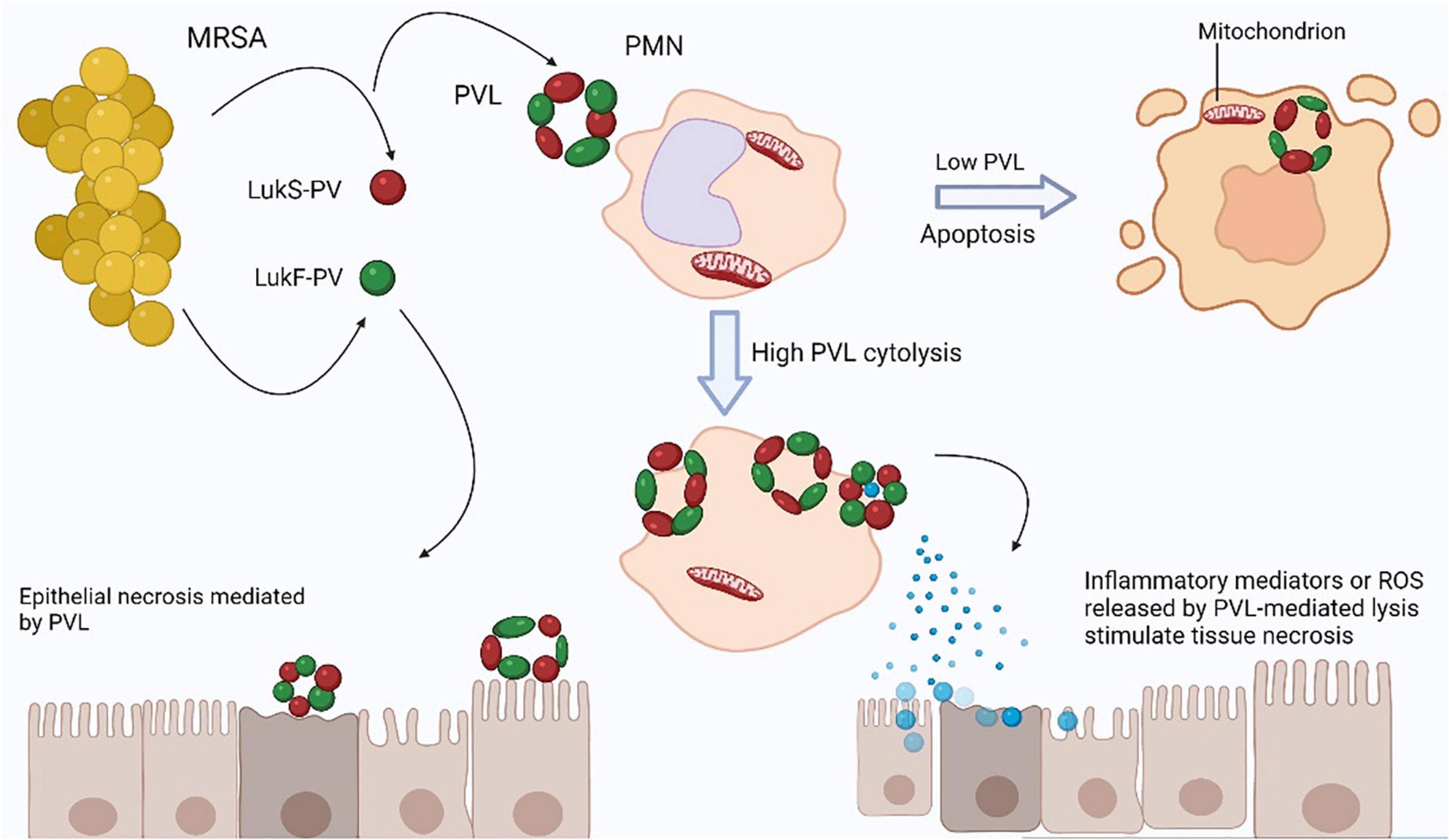
Figure 2. Tissue necrosis is caused by Panton-Valentine leukocidin (PVL). The two PVL components secreted by Staphylococcus aureus, LukS-PV, and LukF-PV, collectively form a pore-forming heptamer on the membranes of polymorphonuclear leukocytes (PMNs). Low PVL concentrations cause polymorphonuclear leukocytes (PMN) apoptosis through direct binding to mitochondrial membranes, whereas high PVL concentrations cause PMN lysis (Genestier et al., 2005). From lysed PMNs, reactive oxygen species (ROS) can cause tissue necrosis. Furthermore, the release of granules from PMNs that have been lysed may cause an inflammatory response that leads to tissue necrosis. PVL is unlikely to cause direct necrosis of epithelial cells.
Hospital acquired methicillin-sensitive S. aureus (MSSA) is less dangerous pathogen than HA-MRSA, which increases the pathogenicity and mortality. Nonetheless, the specific pathogenicity mechanism is unknown. However, it is thought that the PBP2-α protein, which is associated with β-lactam antibiotic resistance and expressed by the mecA gene, directly contributes to immunopathology during MRSA infection. Poor peptidoglycan cross-linking with β-lactam antibiotic caused by PBP2-α results in increased survival of MRSA strains as compared to MSSA (Yao et al., 2010). Improved immune system evasion and S. aureus-exclusive toxin synthesis all contribute to CA-MRSA strains’ enhanced virulence. Researchers have found that S. aureus’s PVL protein has dermonecrotic and leukocyte-lysing properties that lead to increased pathogenicity of CA-MRSA strains (Chini et al., 2006). Further investigations are required, since studies claim that the relationship between PVL and CA-MRSA virulence is complex (Wardenburg et al., 2007). Another study conducted by Wang et al. (2007) also showed that phenol-soluble modulin proteins which promote inflammation and impair the function of neutrophils in bacteremia patient and mice models were more abundant in CA-MRSA strains than HA-MRSA strains. The emergence of LA-MRSA and its transmission to humans and reports of humans strains in animals further increases the pathogenicity of MRSA as now MRSA have diversity of host species which results in genomic modification and increases the antibiotic resistance. SCCmec cassettes, particularly SCCmec IVa and SCCmec V, are present in LA-MRSA while other cassettes, such as SCCmec type XI, which includes mecC, have also been reported (Voss et al., 2005). According to several studies, the LA-MRSA CC398 misplaced human-associated virulence factors like exfoliative toxins and acquired the antibiotic resistance genes like mecA, tetM, TSS toxin I, and PVL genes (Ballhausen et al., 2017). The staphylococcal protein A gene (spa) in CC398 are also currently reported which aid in MRSA pathogenicity (Peeters et al., 2015).
Methicillin resistant strain of S. aureus (MRSA) is an emerging pathogen that can cause mild to serious infections in both animals and humans. Among humans, it mostly causes mild to life deadly infections such as skin and soft tissue infections which are staphylococcal scalded skin syndrome (SSSS), pustules, impetigo contagiosa, abscesses, and papules while deadly infections include TSS, pneumonia, or newborn TSS-like exanthematous disease in humans (Takahashi et al., 1998). Among the 100,000 cases of MRSA infections per year, 20% of the patients died (Klevens et al., 2007). Previously, CA-MRSA and HA-MRSA were two major types of developing infections in humans. HA-MRSA infections are seen in athletes, children, and in hospitalized individuals while CA-MRSA most offenly causes various types of SSTIs which range from mild such as fruncles, impetigo to deadly such as necrotizing fascilitis, and pneumonia infections. CA-MRSA causes less severe infections in animals and humans as compared to HA-MRSA (David and Daum, 2010).
Significant risk factors associated with HA-MRSA infections are aged and immunosuppressive patients due to extensive usage of broad-spectrum antibiotics for a long time (Kurkowski, 2007). HA-MRSA shows resistance to almost all β-lactam drugs which is the reason for its spread among hospital-acquired infections (Lindsay, 2013). Compared to HA-MRSA, CA-MRSA may be found in healthy people and its prevalence may be high among the people working at day care centers, prisons, players, and among military personnel because they are living in close contact with each other (Daum, 2007). HA-MRSA being a multidrug-resistant organism causes many healthcare-associated infections in children and adults. The infection rate is high in people that are immune compromised and in patients having cuts on the skin that may a source of spread of infection (Köck et al., 2010). HA-MRSA clones are dominant bacterial clones that are the major cause of these types of infections in humans and animals. These clones are distributed differently in their geographical areas. HA-MRSA epidemics start in the 1980s or 1990s and the major reason behind this was due to the development of novel clones of MRSA (Chambers and DeLeo, 2009). These clones circulate quickly among hospitals and lead to an increase in the death and morbidity rate. The important HA-MRSA clones include CC, spa type, sequence type (ST), PAGE type, and most simply by their lineage type. Among the CC includes CC30, CC5, CC45, CC8, and sequence type 239 (ST239) (Okuma et al., 2002; McDougal et al., 2003; Naimi et al., 2003; Klevens et al., 2007).
Community-acquired MRSA (CA-MRSA) causes infections on several body parts, most commonly skin and soft tissues, but also in lungs, bone, joints, bloodstream, surgical sites, and urinary tract (Dantes et al., 2013). Although, CA-MRSA is not limited to the skin and soft tissues but also the major cause of septicemia and necrotizing pneumonia. CA-MRSA also causes bacteremia that’s a complication that will lead to endocarditis and osteoarticular infections (Alzomor et al., 2017). CA-MRSA is becoming a global issue among infants, children, and adolescents (Stankovic and Mahajan, 2006). Researchers in Japan using techniques like agr typing, spa typing, coagulase typing, PCR assay for virulence genes, SCCmec typing, and multi-locus sequence typing (MLST) characterized the PVL-positive CA-MRSA in children in 2003 and similar strain was noted in athletes having cutaneous abscess (Takizawa et al., 2005). A rise in CA-MRSA infections was noted up-to 29.8%, and left 70.2% was due to unknown risk factors in Saudi Children Hospital. A study conducted in King Fahad Medical City, Riyadh, Saudi Arabia from 2005 to 2008 among outpatient children showed that 29.8% of cases were positive for CA-MRSA while the other 70.2% were due to unknown risk factors (Mermel et al., 2009). Initially, it was believed that CA-MRSA was a nosocomial strain that had been transmitted from hospitals to the population. However, unlike the HA-MRSA strains often identified in healthcare settings, CA-MRSA strains are paradoxically sensitive to non-β-lactam antimicrobials and showed clinical symptoms more resembling those of MSSA strains (Herold et al., 1998). The genetic lineage, genetic make-up of the methicillin resistance genes, and existence of PVL are the three main attributes that differentiate a CA-MRSA strain from a HA-MRSA strain. The current evidence indicates several distinct MSSA ancestral clones that are circulating in the world are incorporated by SCCmec especially SCCmec IV in CA-MRSA strains through gene transfer mechanism (Figure 3; Enright et al., 2002).
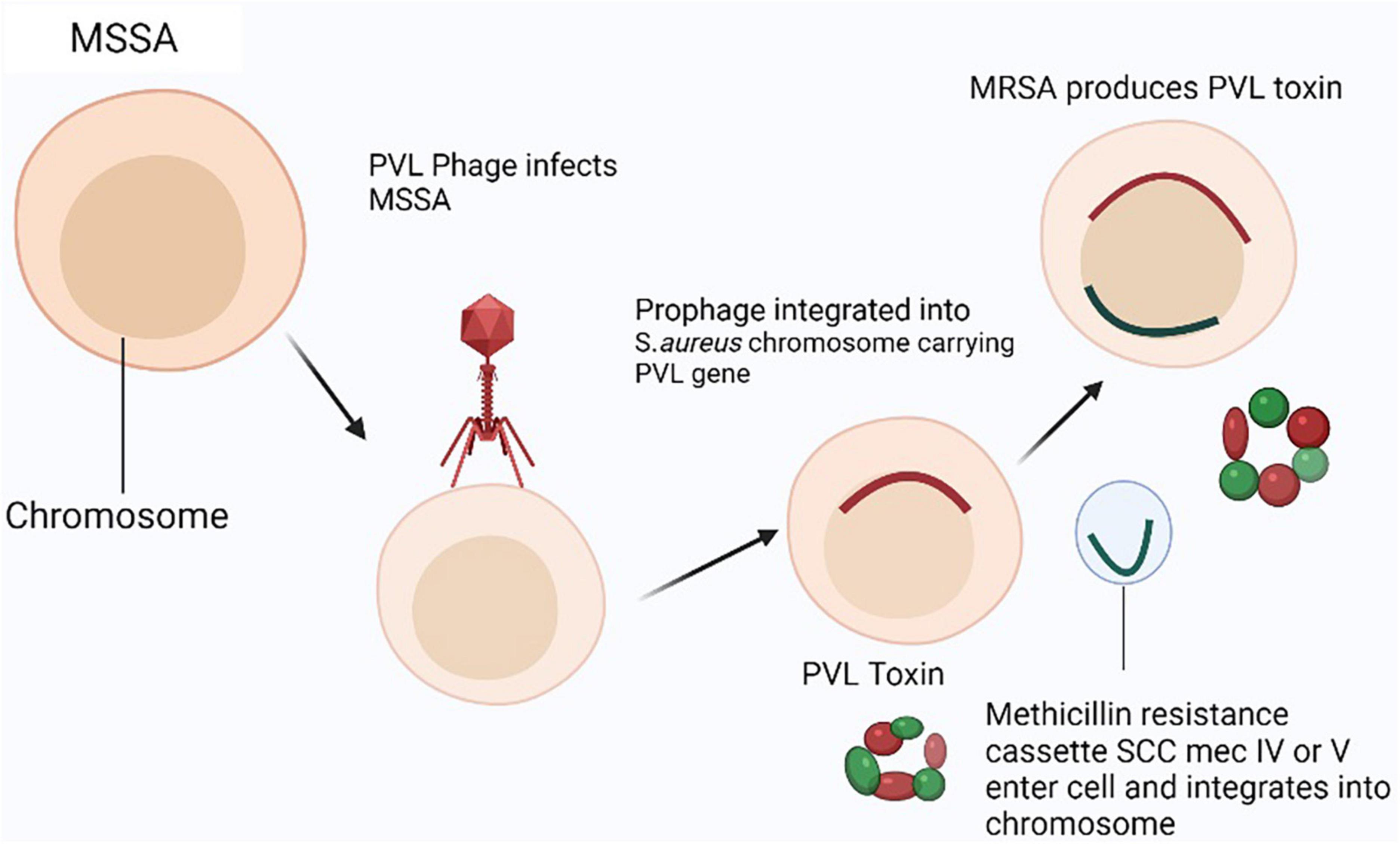
Figure 3. Panton-Valentine leukocidin (PVL) producing community-acquired–MRSA (CA-MRSA) model: In MSSA strain, two genes (pvl) encoding the methicillin-resistant phage virus (PVL) are infected and lysed by the phage (phiSLT) (Staphylococcal Leukocytolytic Toxin). Then, a horizontal transfer of a methicillin resistance cassette (SCCmec IV, V, or VT) carrying the mecA gene into the pvl-positive methicillin susceptible Staphylococcus aureus (MSSA) strain allows it to incorporate into the genome somewhere other than the phiSLT (Staphylococcal Leukocytolytic Toxin) integration site.
Although the spread of MRSA infections in food and companion animals was initially thought to be slower, it is now becoming a serious problem for food animals and food industries too. LA-MRSA is an important cause of mastitis in cows and buffaloes resulting in a decrease or no milk production (Javed et al., 2021). LA-MRSA also causes the infections in poultry such as comb necrosis, chondronecrosis, and septic conditions (Fluit, 2012). Almost all the companion animals like dogs, cats, and horses, are potential sources of LA-MRSA transmission to humans having direct or indirect contact with these animals. Besides mammals, LA-MRSA colonization in foxes, roes, rabbits, wild boars, and wild animals (e.g., pigeons, pheasants, ducks, buzzards, gulls, and rocks) has also been reported (Smith et al., 2009).
Mastitis is an important disease of dairy animals, which is linked to the maximum use of antibiotics responsible for huge economic losses. S. aureus is a chief pathogen of mastitis among all other causative agents throughout the world. LA-MRSA is also an important cause of pustular dermatitis in their milkers (Grinberg et al., 2004). Contrarily, all bovine MRSA clones are infrequently seen in dairy cows, responsible of subclinical mastitis in cattle (Aqib et al., 2018b; Abdeen et al., 2021). The first time MRSA was detected in Cattle in Belgium in 1972 in milk samples which thought to be spread from milker hands through contamination (Lee, 2003). MRSA infection of mammary gland in cattle leads to a decrease in milk production and may cause cessation of milk from mammary glands in severe cases which pose the dairy industry a big economic loss (Figure 4; Holden et al., 2013). Mammary gland inflammation results from cytotoxicity of lukMF9, a powerful virulence factor of LA-MRSA (Peton et al., 2014). lukMF9 has a high affinity for chemokine receptor CCR1 on bovine which results in significant inflammation and damage as a consequence of more accumulation of neutrophils in mammary gland (Fromageau et al., 2010; Vrieling et al., 2015). A tropical phage called wPV83, which is capable of hematogenous spread to S. aureus, carries the lukM and lukF genes (lukM-lukF-PV) which are closely associated with synthesis of lukMF9 (Yamada et al., 2005). A conformational change in the nucleotide of a gene produces a suppressor protein (rot) in S. aureus strains that block the activation of many toxin genes that exhibit large quantities of the lukMF9. Genetic studies have linked strains overexpressing lukMF9 to the lineage ST479 (Hoekstra et al., 2018). Superantigens (SAgs) are a class of bacterial toxins that stimulate the immune system and are released by staphylococcal species, particularly S. aureus. They have the potential to cause an unsustainable cytokine cascade (Tuffs et al., 2018). However, SAgs seem to perform a crucial function in bovine mastitis, and the majority of cattle S. aureus isolated have five or more genes encoding SAgs, although the precise function of SAgs in mastitis is yet unknown (Wilson et al., 2018). SAgs disrupt the human autoimmune reaction, which makes them potentially significant in chronic infections (Ferens and Bohach, 2000). By preventing immune-regulatory activation, the multiplication of T cells and the production of interleukins may stopped by SAgs that might reduce the efficiency of the immune system response (Tuffs et al., 2018). The cow susceptibility island SaPIbov is present in lineages that are related to cattle, including CC151 and CC133 (Wilson et al., 2018). Numerous toxins, including the toxic shock syndrome toxin 1 (TSST-1), the staphylococcal enterotoxin-like protein, and bovine staphylococcal enterotoxin C are also stimulated by SaPIbov (Fitzgerald et al., 2001).
Pigs in various studies exhibit higher than expected percentages of MRSA. Highly pathogenic strains are also rising in the pig population. In a study, 10% of samples were positive for the ST398 strain of MRSA, and the overall presence of MRSA among the three studied farms was considerably high. These 10% ST398 strains of MRSA were further found related to spa type, t1793 and t034 and they were exhibiting a high level of resistance to multiple antibiotics. This is another clue for the new emerging zoonotic strain of MRSA in Europe among the pig population (Hasman et al., 2010). Moreover, LA-MRSA ST398 is not only confined to pigs in Europe but it also spreads to Canada and USA (Khanna et al., 2008; Smith et al., 2009). Except ST398 strain, pigs also harbor other LA-MRSA strains such as ST1, ST97 and ST9 but at a lower frequency. A study conducted by Wagenaar et al. (2009) in China identified LA-MRSA ST9 lineage type colonization in pigs as well as workers. Another study was conducted by identified the human clone ST1 in pigs which indicating human to pig transmission of clone called as reverse zoonosis as depicted in Figure 1 (Monaco et al., 2010).
Methicillin resistant strain of S. aureus (MRSA) is a chief bacterium isolated from mastitis milk of goats which may infect individuals who are consuming contaminated milk. Contaminated milk may contain certain types of staphylococcal enterotoxins (SE), which is a major cause of food poisoning. All SE genes are capable of causing illnesses that are particular to their hosts, such as staphylococcal enterotoxin B causes infection in humans (Da Silva et al., 2005; Altaf et al., 2020). MRSA can also cause pyaemia which is a type of abscess which gain access to bloodstream and lead to sepsis (Webster and Mitchell, 1989). MRSA in sheep and goats is among the predominant cause of clinical and subclinical mastitis which leads to high somatic cell count in milk and make milk unfavorable to drink (Muzammil et al., 2021).
Livestock associated MRSA (LA-MRSA) has been also reported in cloaca and nares of healthy poultry birds. In poultry birds, it may cause pyoderma, omphalitis, urinary tract infection, arthritis, and otitis (Pickering et al., 2022). After performing different antimicrobial agents. MRSA was found spa type t011 and spa type t157. While ST398 is a new livestock-associated strain of MRSA that is also found in poultry (Nemati et al., 2008). Another investigation showed that all MRSA isolates of poultry origin were belonging to spa type t1456 (Persoons et al., 2009).
The companion animals mostly suffer skin and soft tissue infections predominately after surgical procedures. The United Kingdom observed 95/6519 MRSA-positive samples that were comprised of 24 cats, 69 dogs, 1 horse, and 1 rabbit (Boag et al., 2004). Molecular analysis of MRSA in dogs and cats using SCC mec typing and sequence typing of ccrAB gene (cassette chromosome recombinant gene AB), and established MRSA strains using MLST from cats and dogs similar to that of human SCC mec gene in staphylococci. A further cross-sectional study revealed the transmission of MRSA strains from humans to dogs especially MRSA strain ST239-III (Malik et al., 2006). A research finding at Irish Veterinary Hospital noted that 69.44% of canines, 22.22% of horses, and 2.78% of cats, rabbits, and seals were infected with MRSA (O’Mahony et al., 2005). Risk factors such as the site of infection, surgical history, medical history, intravenous catheterization, and use of previous antibiotics were inferred after 6 years of a controlled trial conducted in veterinary health care in the USA and Canada. The most prevalent sites of colonization were found skin and nares (Manian, 2003). MRSA also causes remarkable and life-threatening infections in horses and personals taking care of them. Horses may have septic arthritis, bacteremia, osteomyelitis, and inflammation of the skin and delicate tissues (Cuny et al., 2006), metritis, omphalitis, pneumonia, and infections related to catheters are a few examples of illnesses associated with implants. First case of MRSA in horses was noted in 1993 due to postoperative infections. Reverse zoonosis with CA-MRSA-5 (ST8) in horses has also been found with a wider range of antibiotic resistance, e.g., oxacillin, tetracycline, gentamycin, etc., (Weese et al., 2005).
Among individuals who have established MRSA infections, bacteremia that might result in mortality and could affect nearly 50% of the population (Nickerson et al., 2009). MRSA preventive measures and use of a recent antibiotic therapy alone and in combination may result in decrease MRSA infections (De Kraker et al., 2013). The ineffective disease management at the start of infection may be the reason of excessive death rate (Simor et al., 2016). Additionally, several virulence genes have been linked to increased mortality, such as acquisition of antibiotic resistance genes through horizontal gene transfer mechanisms are major cause of making antibiotics ineffective against MRSA infection (Albur et al., 2012). However, still there are antibiotics and futuristic approaches to treat MRSA infections which are discussed in further sections. The advent of several new antibiotics alone or in combination are available in the market, a hope for MRSA infected patients. Additionally, scientists now are switching from single-agent therapy to combination therapy, immunotherapy, and few latest alternative ways to combat MRSA infections such as phytochemicals, probiotics, nanoparticles, and bacteriophages as an antibiotic alternatives (Lee et al., 2016).
In comparison to MSSA infection, bacteremia caused by MRSA is more severe (Fowler et al., 2006). A prolonged duration of bacteremia may result in a more severe consequences (Fowler et al., 2003). A survey conducted in Australia noted that ceftaroline and cephalosporin resistance were found in 17% of MRSA cultures. Although several other new drugs have been approved for use, vancomycin remains the most effective (Abbott et al., 2015). Moreover, combining a β-lactam antibiotic with glycopeptide for example daptomycin to treat MRSA infections is recommended by the Spanish Society of Clinical Microbiology and Infectious Diseases (Gudiol et al., 2015). The mechanism of action of daptomycin somewhat different, it causes potassium and calcium ions to cross the plasma membrane, which causes apoptosis of the bacterial cell. Daptomycin inhibits the function of fem and aux genes, hence reducing the expression of the mecA gene. Daptomycin can reduce PBP-2α binding to its peptidoglycan moieties in the early phases of peptidoglycan synthesis (Rand and Houck, 2004). Daptomycin’s ability to bind to β-lactam antibiotics is improved when used in combination (Dhand et al., 2011). However, mutation at the gene level may makes the daptomycin a resistant drug against MRSA. The major genes involved in daptomycin resistance include multi-peptide resistance (mprF) and the regulatory gene walKR (also called yycGF). This is because of polymorphism in single nucleotide sequences (Jiang and Peleg, 2015). Another type of mutation e.g., point mutation in genes such as rpoB and rpoC also associated with the development of resistance against daptomycin (Peleg et al., 2012).
Vancomycin was once thought to be the most efficient antibiotic for treating MRSA-related severe infections. The suitability of vancomycin’s primary action is determined by gathering evidence of overall resistance, unachievable pharmacokinetic/pharmacodynamic (PK/PD) targets, and reduced effects (Holmes et al., 2012; Van Hal and Fowler, 2013). Vancomycin intermediate S. aureus (VISA) and hetero resistant (hVISA) are both commonly treated with glycopeptides. Vancomycin, therefore, begins to become resistant to them as well as losing sensitivity to glycopeptides, which causes the development of vancomycin-resistant S. aureus (VRSA). These isolates are more sensitive to other antibiotic classes, particularly β-lactam antibiotics, despite having the mecA gene. The “seesaw effect” refers to the sensitivity of MRSA isolates to anti-staphylococcal β-lactam antibiotics caused by higher minimum inhibitory concentrations (MICs) of daptomycin and vancomycin. Research on the synergistic effects between vancomycin and β-lactam antibiotics presents great potential (Werth et al., 2013). When the β-lactam seesaw effect and cross-resistance among glycopeptides, lipopeptides, and lipoglycopeptides against MRSA strains were examined, the important factors responsible for developing the cross-resistance to daptomycin, vancomycin, and dalbavancin is the change in membrane lipid composition such as fatty acyl and ultimately resulted in resistant strains. Abundance of long-chain fatty acyl peptidoglycans in membrane has a negative correlation with β-lactam sensitivity and a positive correlation with cross-resistance (Hines et al., 2020).
The combined effect of β-lactam antibiotics and vancomycin is found synergistic in in vitro studies when checked through antimicrobial sensitivity assays. The synergistic effect is highly linked with MIC of vancomycin. An in vivo trial was conducted on rabbits infected with VISA strain. The efficacy of two drugs, vancomycin and nafcillin, was checked to compare their effects. The results were ineffective with a single administration of drugs but give curative effects with combination therapy by reducing the magnitude of infection up to 4.52 log10 CFU/g. The extent of synergism was highly correlated with the MIC of vancomycin (Climo et al., 1999). Another in vitro study evaluated combinations of vancomycin and cephalosporin showed synergistic effects against MRSA isolates (Seibert et al., 1992). These isolates were also checked against the combination therapy of vancomycin and imipenem under the synergistic effect (Silva et al., 2011). Many more combinations have also been inquired such as rifampicin and gentamycin; daptomycin and rifampicin therapy against biofilm-producing MRSA isolates (Rosenberg Goldstein et al., 2012). These therapies proved better results by comparing the daptomycin and cloxacillin in a rat infected with MRSA and indicated better results against rifampicin-resistant MRSA infections.
The fifth generation cephalosporins are the most effective against MRSA infections than other antibiotic generations. The most widely utilized drugs in health care facilities among cephalosporins are ceftaroline and ceftobiprole. Ceftaroline is a drug that is licensed to treat skin associated infections and community-acquired pneumonia (CAP) (Purrello et al., 2016). To compare the efficacy of ceftobiprole, ceftriaxone, and linezolid, multi-center clinical studies were carried out in 2006–2007 among patients in hospitals who exposed to CAP. A multi-center FOCUS-1 study, conducted by a researcher in 2008–2009, investigated the effectiveness of ceftaroline and ceftriaxone against CAP. Nearly 168 locations from around the world were chosen. The modified intention to treat (mITT) and clinical evaluation (CE) rates of ceftaroline were found to be high, 86.6 and 83.3%, respectively while of ceftriaxone were 77.7% in mITT and 78.2% in CE (File et al., 2011). Many new approved and novel drugs are effective against MRSA infections, but further research is required to determine their efficacy at large scale. The next section discusses few of the most recent antimicrobials that are effective against MRSA infections (Thati et al., 2011).
A novel class of antibiotics known as oxazolidinones are effective against a variety of gram-positive bacteria, including vancomycin- and methicillin-resistant staphylococci. Oxazolidinone blocks protein synthesis by binding to the P-site of the 50S ribosome subunit. The development of oxazolidinone resistance with 23S rRNA does not influence oxazolidinone action when compared with the resistance of other protein synthesis blockers. Its application in surgical infections is made possible by its effective penetration in bone and infiltration into lungs, cerebrospinal fluid, and hematoma (Bozdogan and Appelbaum, 2004). Tedizolid, a brand-new drug among the oxazolidinones, has received approval for the standard 6-days treatment course for skin and soft tissue infections. Compared to linezolid, tedizolid is a drug that is more efficient and offers more benefits (Boucher et al., 2014; Flanagan et al., 2015). Tedizolid’s efficiency against chloramphenicol and florfenicol resistant (CFR) isolates harboring methyltransferase gene is also found (Flanagan et al., 2015). Novel oxazolidinone agent cadazolid is a potent agent against Clostridium difficile (Gerding et al., 2015), while radezolid is efficient against isolates of S. aureus that are resistant to linezolid (Lemaire et al., 2010).
This class of antibiotic inhibit the bacteria growth by inhibiting the protein synthesis. They attach to the 30S ribosomal subunit and prevent aminoacyl-tRNA from joining the translational mRNA complex, which inhibits the beginning of translation. In several researches, the drug tetracycline was found to bind to the rRNAs 16S and 23S (Chukwudi, 2016). The novel synthetic fluorocycline and eravacyline drugs are effective against infections caused by gram-positive and gram-negative bacteria, including MRSA. Eravacycline is four times more efficient than tigecycline for treating gram-positive bacteria (Zhanel et al., 2016). Omadacycline, an aminomethylcycline, is more efficient to treat CAP and acute bacterial skin and skin structural infections (ABSSSI) (Pfaller et al., 2017).
Quinolones are among the most common antibacterial drugs used worldwide to treat a range of bacterial diseases in both animals and humans. These drugs are structurally known as quinolones because they have a quinoline ring in their structure. Quinolones and fluoroquinolones prevent bacteria from multiplying by inhibiting their DNA replication pathway. These antibiotics basically damage the bacteria chromosome by targeting the enzymes gyrase and topoisomerase IV (Aldred et al., 2014). The fluoroquinolone antibiotic, delafloxacin is an drug that is effective against both gram-positive and gram-negative bacteria because of its unique electrochemical characteristics such as being anion at physiological pH and uncharged at acidic pH (Lemaire et al., 2011). In 2011, a research trial was carried out in the USA to evaluate delafloxacin effectiveness in comparison to vancomycin and linezolid. These three drugs were found in following order with highest rates of cure, delafloxacin, linezolid, and vancomycin (Kingsley et al., 2015). Zabofloxacin is another fluoroquinolone drug showing high activity against gram-positive microorganisms, especially against respiratory tract infections due to St. pneumoniae. MIC50 of zabofloxacin against MRSA was noted high as compared to delafloxacin i.e., 2 mg/ml in and 0.125 mg/ml respectively. Except for these characteristics of zabofloxacin, it is less efficient against gram-negative organisms. Thirdly, nemonoxacin was found to be similar to zabofloxacin in its pattern of activity and its effects against CAP have been also investigated. Fourthly, avarofloxacin efficacy against MRSA is also comparable to that of delafloxacin (Van Bambeke, 2014).
Fatty acid chains linked to glycopeptides, a family of antibiotics known as lipoglycopeptides, showed dose-dependent bactericidal action. They prevent the synthesis of cell wall and interfere with the bacterial cell membrane’s permeability function. Therefore, the terminal acyl-d-alanyl-d is where the glycopeptide core binds. The alanine chain in the cell wall interacts with hydrophobic filling and hydrogen bonds resulting in a high affinity. This inhibits the cell wall precursors from polymerizing and crosslinking (Damodaran and Madhan, 2011). Three novel drugs from the lipoglycopeptide family have been approved and launched in the market. A lipoglycopeptide called dalbavancin was approved by FDA and European Medicine Agency (EMA) for treating the ABSSSI in 2014 and 2015, respectively (Van Bambeke, 2015). Dalbavancin is a semi-synthetic lipoglycopeptide with a long half-life ∼10 days and prolonged duration pf action ∼7 days against MRSA with a single dosage of 500 mg (Chen et al., 2007). Dalbavancin is specifically used to treat complex infections in outpatients (Juul et al., 2016). A multi-center study was conducted to treat ABSSSI in 2011–2012 to compare efficacy of dalbavancin with vancomycin. Compared to vancomycin, dalbavancin showed great outcomes with fewer side effects (Boucher et al., 2014).
Ritavancin, a second lipoglycopeptide, authorized by FDA and EMA in 2014 and 2015, which is also a prolonged action lipopeptide used to treat ABSSSI infections (Takahashi and Igarashi, 2018). This drug act by supressing the transglycosylase and transpeptidase enzymes. This antibiotic also exhibits bactericidal activity against broad range of gram-positive pathogens such as vancomycin-resistant enterococci (VRE), VISA, and VRSA due to its enhanced capability of penetration through plasma membrane (Van Bambeke, 2014). Telavancin is another lipoglycopeptdie which is quite effective against hospital-acquired pneumonia (HAP) caused by MRSA (Sandrock and Shorr, 2015). The drug is approved by FDA in 2013 for the treatment of MRSA infections including HAP and VAP following approval for SSSS treatment (Wenzler and Rodvold, 2015). When other antibiotics become ineffective, the EMA has restricted its usage for the treatment of nosocomial pneumonia caused by MRSA (Masterton et al., 2015). Although the lipoglycopeptides are approved for a narrow range of treatment of infections, in the future they will play important role in the treatment of osteomyelitis, bacteremia, and infective endocarditis (Brade et al., 2016).
Other antimicrobial drugs may include doxycycline, clindamycin, and trimethoprim/sulphamethoxazole which are also found effective irrespective of the severity of the disease (Liu et al., 2011).
As stated by World Health Organization (WHO), among the largest threat to public health, is the rise in antibiotic-resistant microorganisms. Approximately 700, 000 fatalities are caused by antibiotic resistance bacteria each year globally, and by 2050, that number might reach 10 million (Tagliabue and Rappuoli, 2018). A post-antibiotic era, in which ordinary diseases and mild infections might kill, is a very real prospect for the twenty first century, according to a study from WHO (Streicher, 2021). Thus, the development of novel antibiotic-free strategies is urgently required for the management and treatment of antibiotic-resistant bacterial infections.
The use of antibiotic stimulators in association with antibiotics is one of the best methods for reducing antibiotic resistance and extending the life of current antibiotics. The most effective combination against MRSA was found β-lactam antibiotic and potassium clavulanate (De Araújo et al., 2013). As listed in Table 3, many studies shown that phytochemicals alone or in combination with antibiotic have a great organic potential to exhibit antibacterial activity as well as act as modulators of antibiotic resistance. The majority of these curative effects are attributed to the active secondary metabolites produced by plants (Lakshmi et al., 2013). Phytochemicals act as antibacterial agent by inhibiting efflux pumps, modification of active sites, increasing the permeability of plasma membrane, and alteration of bacterial enzymes as depicted in Figure 5 (Coutinho et al., 2009). Plant extract along with gentamicin and kanamycin have synergistic effects for example ethanol extracted of Turnera ulmifolia leaves may enhance antibacterial efficacy of antibiotic against MRSA strains. As a possible active efflux regulator, grapefruit oil was found to be beneficial against MRSA (Abulrob et al., 2004). In an interesting study, the traditional Korean medicine known as Sami Hyanglyum-Hwan, Aucklandiae radix, Coptidis rhizome, Rhei rhizome, and Arecae semen) restored the anti-microbial activity of ciprofloxacin when evaluated against multiple MRSA strains (Choi et al., 2015).
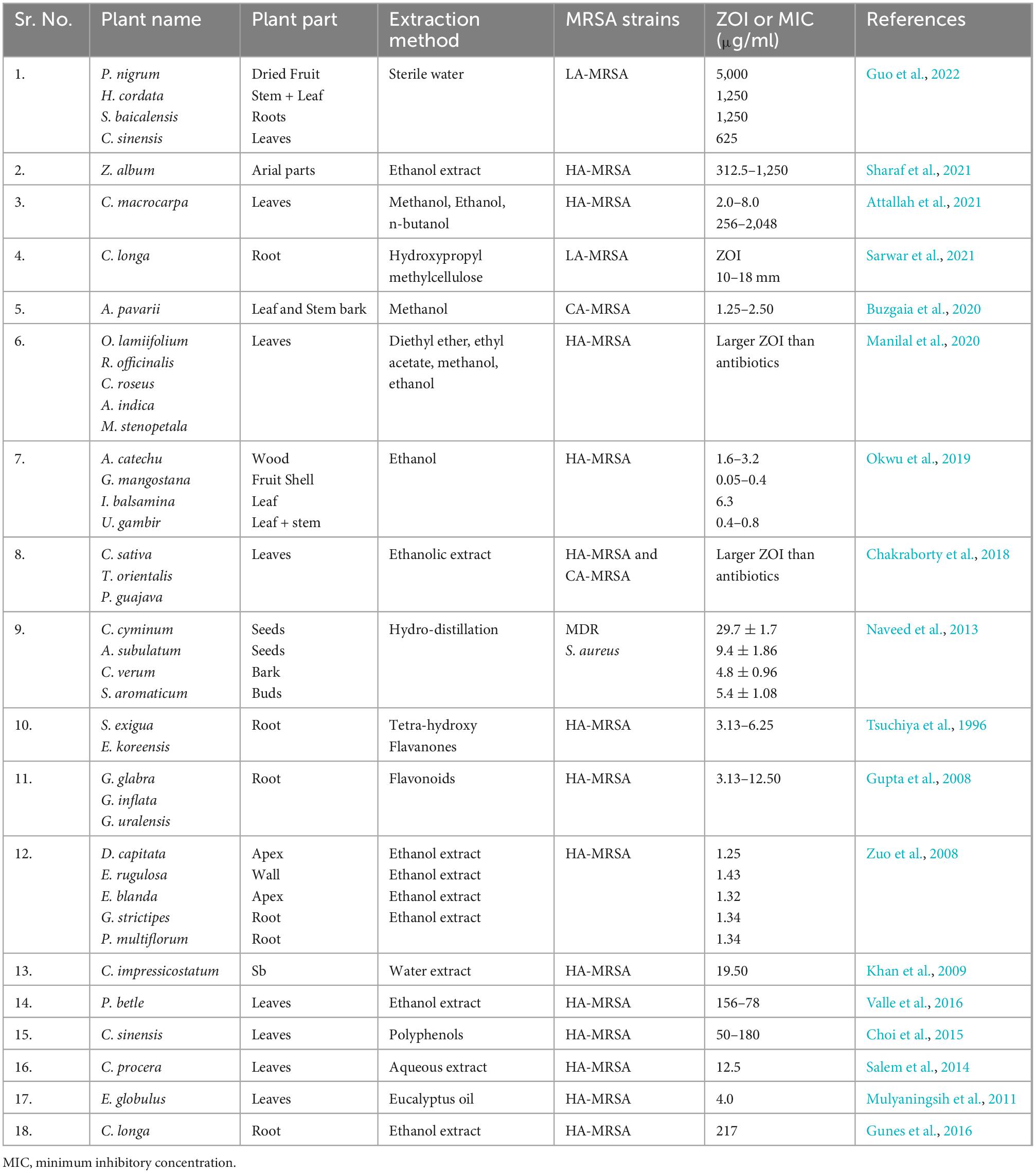
Table 3. Antibacterial activity of different herbal plants against different strains of methicillin-resistant Staphylococcus aureus (MRSA).
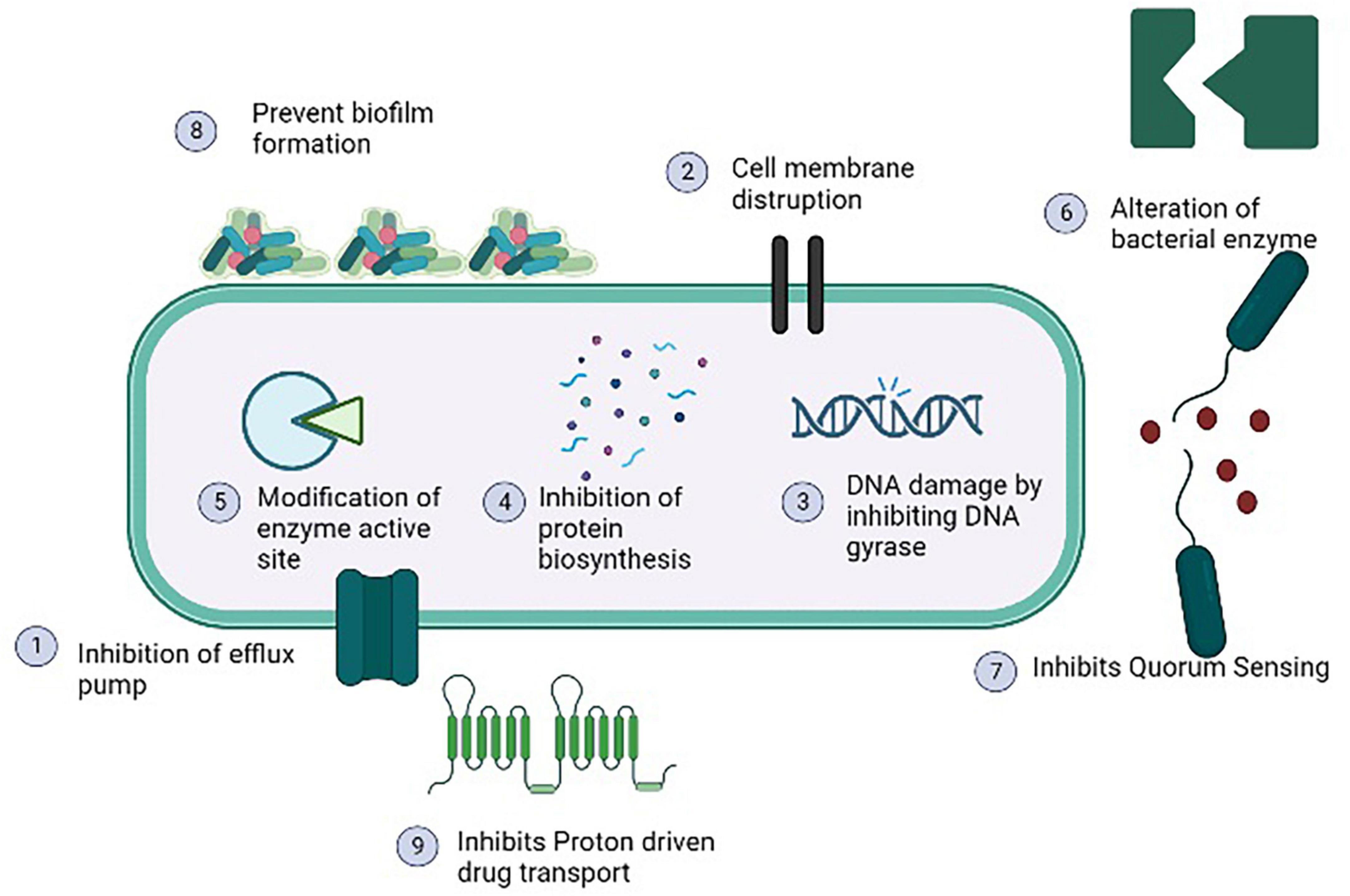
Figure 5. Antibacterial mechanisms of various phytochemicals against methicillin resistant strain of Staphylococcus aureus (MRSA).
Several studies have shown that NSAIDs exhibited antimicrobial properties, nevertheless, the exact mode of action is unclear. Except for mefenamic acid, it has been observed that diclofenac, aspirin, and ibuprofen have antibacterial effects at 5 mg/ml against certain gram-positive bacteria. Due to the presence of lipopolysaccharide in the cell wall of gram-negative bacteria, which is hydrophilic and inhibits most drugs metabolism, the only NSAID that is helpful against gram-negative bacteria is aspirin. The absence of lipopolysaccharide in cell wall of gram positive bacteria, makes it simple for antimicrobial drugs to enter the cells easily (Khalaf et al., 2015). In comparison to the typical therapeutic dosage in use for inflammatory, pain, or fever, NSAIDs have antimicrobial action at significantly lower concentrations (Ong et al., 2007). Opposite to diclofenac, aspirin and ibuprofen demonstrated bacteriostatic and bactericidal action against MRSA strains and may thus be used as antibiotic adjuvants to treat infections (Chan et al., 2017). The treatment of CA-MRSA infections involves the use of NSAIDs and antibiotics. Ineffective outcomes were seen when cefuroxime and chloramphenicol were taken alone to cure MRSA. Although aspirin and ibuprofen have bacteriostatic and bactericidal effects on MRSA strains, however their combination along with cefuroxime and chloramphenicol were examined. It was shown that the combination of ibuprofen/aspirin, chloramphenicol, and cefuroxime have increased antibacterial showing either synergistic or additive effect (Yin et al., 2014). MDR bacterial infections can be treated with NSAID and antibiotic combination (Chan et al., 2017). Another study conducted by Aqib et al. (2021) investigated the effect of antibiotics alone, in combination with NSAIDs, nanoparticles and plant extracts against MDR strains of S. aureus including MRSA. It was noted antibiotic in combination with NSAIDs such as meloxicam, diclofenac, aspirin and ibuprofen increases the ZOIs in in vitro studies against MRSA and MDR strains of S. aureus. The study concluded synergistic correlation between NSAIDs, antibiotics, nanoparticles and plant extracts by calculating fractional inhibitory concentration indices (FICIs).
Due to the special characteristics of metal nanoparticles (NPs), they are more ubiquitous and inexpensive manufacturing material that are getting much applications in today’s world. In 2016, it was noted that the nanometals market based on metal oxides reached USD 4.2 billion. A more rise in NPs manufacturing demand is found to be anticipated by 2025, which is due to increased use of metal based nanomaterials in biomedical research (Gudkov et al., 2022). Research studies going on to explore the potential of nanoparticles as biosensors (Singh et al., 2019), detection and treatment of oncological disorders (Vimala et al., 2019), and drug delivery have received a lot of interest (Spirescu et al., 2021). It is very interesting to employ metal oxide nanoparticle-based on nanomaterials to treat antibiotic resistant bacterial infections. Now a day, most commonly used metal oxide nanomaterials are silver nitrate, zinc oxide, platinum, aluminum oxide, titanium dioxide, gold, magnesium oxide, iron oxide and sodium alginate (Akhtar et al., 2019; Spirescu et al., 2021; Gudkov et al., 2022; Mendes et al., 2022).
According to the statement from WHO, antibiotic-resistant microorganisms are among the most remarkable barriers to public health and progress. The new paradigm to treat the diseases caused by resistant bacterial strains including MRSA is the use of metal oxide nanomaterials [95]. The NPs act on bacterial cell through various type specific mechanisms such as production of reactive oxygen species (ROS) to induce stress on cell, release of heavy metal ions, alter membrane permeability, DNA damage, protein damage, disrupt the function of efflux pump, act of cell wall and plasma membrane to cause damage and release cellular components outside of cell (Fernando et al., 2018) as illustrated in Figure 6. (Shameli et al., 2010) conducted study to evaluate the antibacterial activity of green silver nitrate nanoparticles against MRSA isolates and found that green silver nitrate nanoparticles exhibited the strong antibacterial activity by inhibiting the growth of MRSA in in vitro model. Another study was conducted by Lodhi et al. (2021) who evaluated the antibacterial activity of zinc oxide NPs alone and in combination with antibiotic and found increased antibacterial activity when NPs were used in combination with antibiotic. Another study conducted by Wichai et al. (2019) in Thailand who use a complex combination of nanomaterials with other materials (bacterial cellulose + sodium alginate NPs + chitosan + copper sulfate) and found that NPs exhibited a strong antibacterial activity against MRSA strains. A number of studies has been conducted by various researchers from different countries to check the antibacterial activity of various forms of nanoparticles against different strains of S. aureus including MRSA are listed in Table 4.
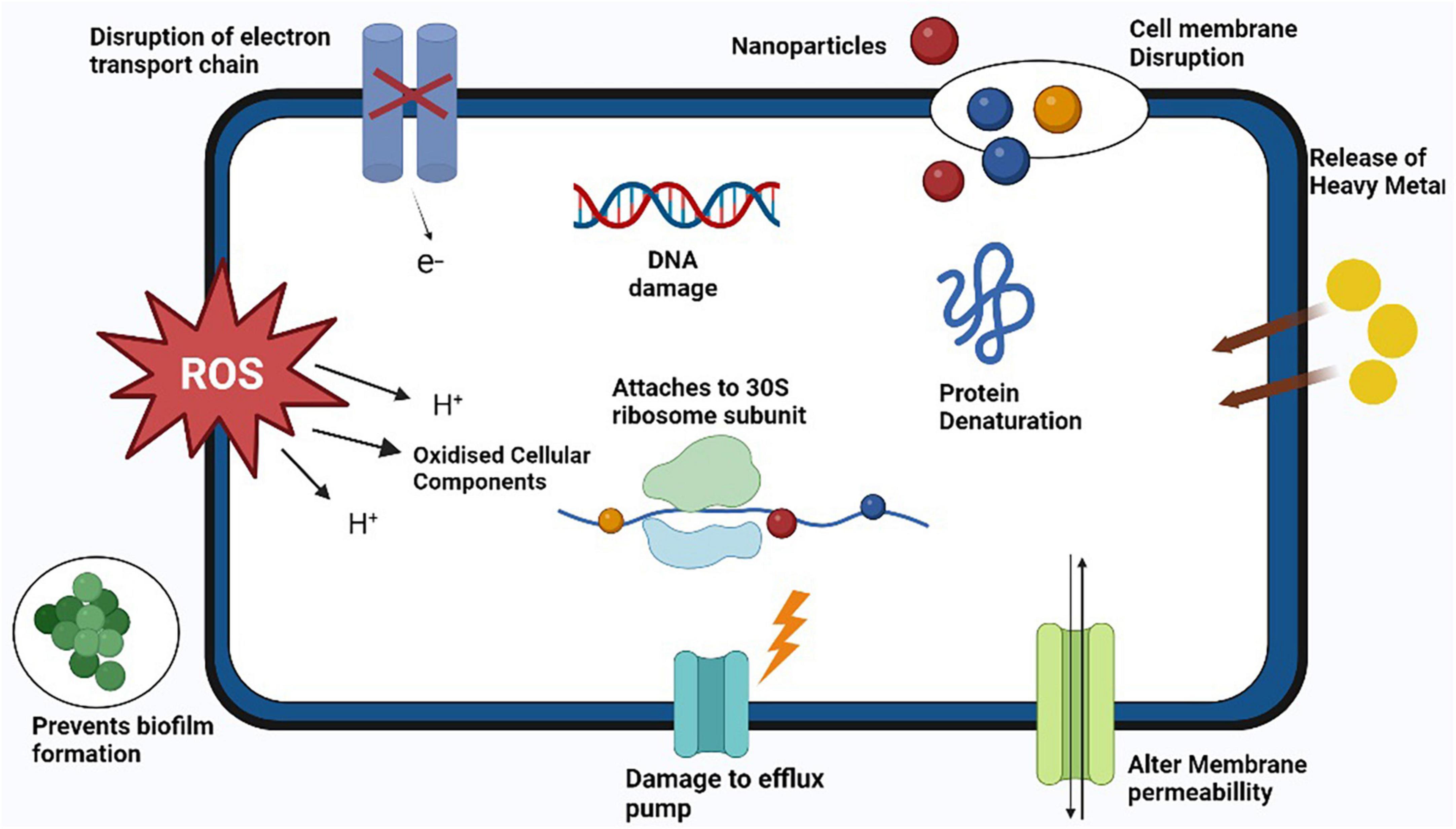
Figure 6. Antibacterial mechanisms of action of various nanoparticles against methicillin resistant strain of Staphylococcus aureus (MRSA).
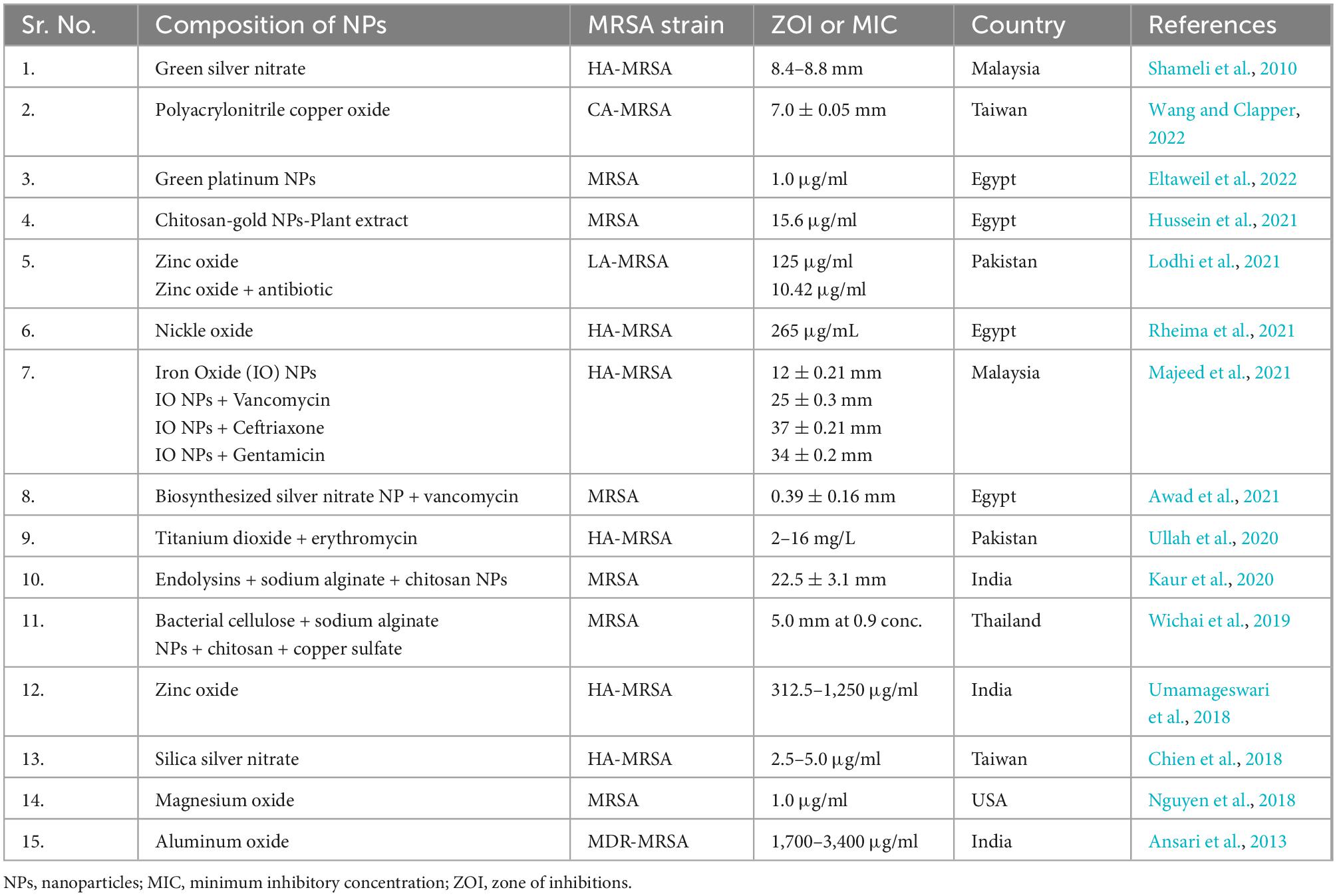
Table 4. In vitro studies on antibacterial activity of nanoparticles against methicillin resistant strain of Staphylococcus aureus (MRSA).
Bacteriophage therapy also known as phage therapy is a type of therapy that uses viruses to kill bacterial pathogens. Due to increasing resistance to antibiotics, phage therapy is considered as cost effective, highly specific and efficient in their mechanism of action against multiple MDR bacteria. Bacteriophages only cause damage to bacteria cells without causing any damage to human and animal cells (Tkhilaishvili et al., 2020). Phage therapy has been found as an effective treatment against multiple bacterial pathogens such as S. aureus, E. coli, P. aerogenosa, A. baumannii, S. pyogenes, S. suis, and B. cereus (Yang et al., 2017). This therapy is mostly used in Georgia and Russia for the treatment of those bacterial infections which don’t response to antibiotics. A study was conducted by Nandhini et al. (2022) who evaluated the lytic activity of kayvirus phages against MDR S. aurus. (Lubowska et al., 2019) also study the genomic, morphology, and lytic properties of three phages which are name as vB_SauM-A, vB_SauM-C, and vB_SauM-D and noted rapid adsorption with bacterial cell, short latent period, and increase lytic activity. Further genome study showed higher G + C content in these phages similar to phage K (Nandhini et al., 2022). As, bacteriophage act so specifically consist of following steps to destroy bacterial cell; (1) Adsorption which is attachment of virus with specific bacteria cell, (2) Penetration of phage DNA or RNA into the bacteria cell, (3) after penetration, phage uses host cell machinery to synthesis early viral proteins, (4) Replication of virus through using the early viral proteins, (5) Synthesis of late viral proteins to assemble again in new complete phage particles, (6) Lastly, new phage particles cause lysis of the bacterial cell and causes its death through release of cellular contents outside the cell and new phage particle starts infecting the bacterial cells (Sausset et al., 2020) as illustrated in Figure 7. Staphylococcal bacteriophage (Sb) and PYO bacteriophage are prepared by Eliava Institute, Georgia and available commercially as phage preparations for usage as replacement to antibiotic by humans (Fish et al., 2018). Bacteriophage Sb is a mono phage product and is effective against only one specific bacteria while PYO phage covers multiple bacterial species such as E. coli, S. aureus, Streptococcus spp., P. aeruginosa, and Proteus spp (Villarroel et al., 2017). The number of isolated phages against MRSA has significantly increased over the past 15 years, and several investigations have found that phages have effective and all-encompassing antibacterial effects as listed in Table 5.
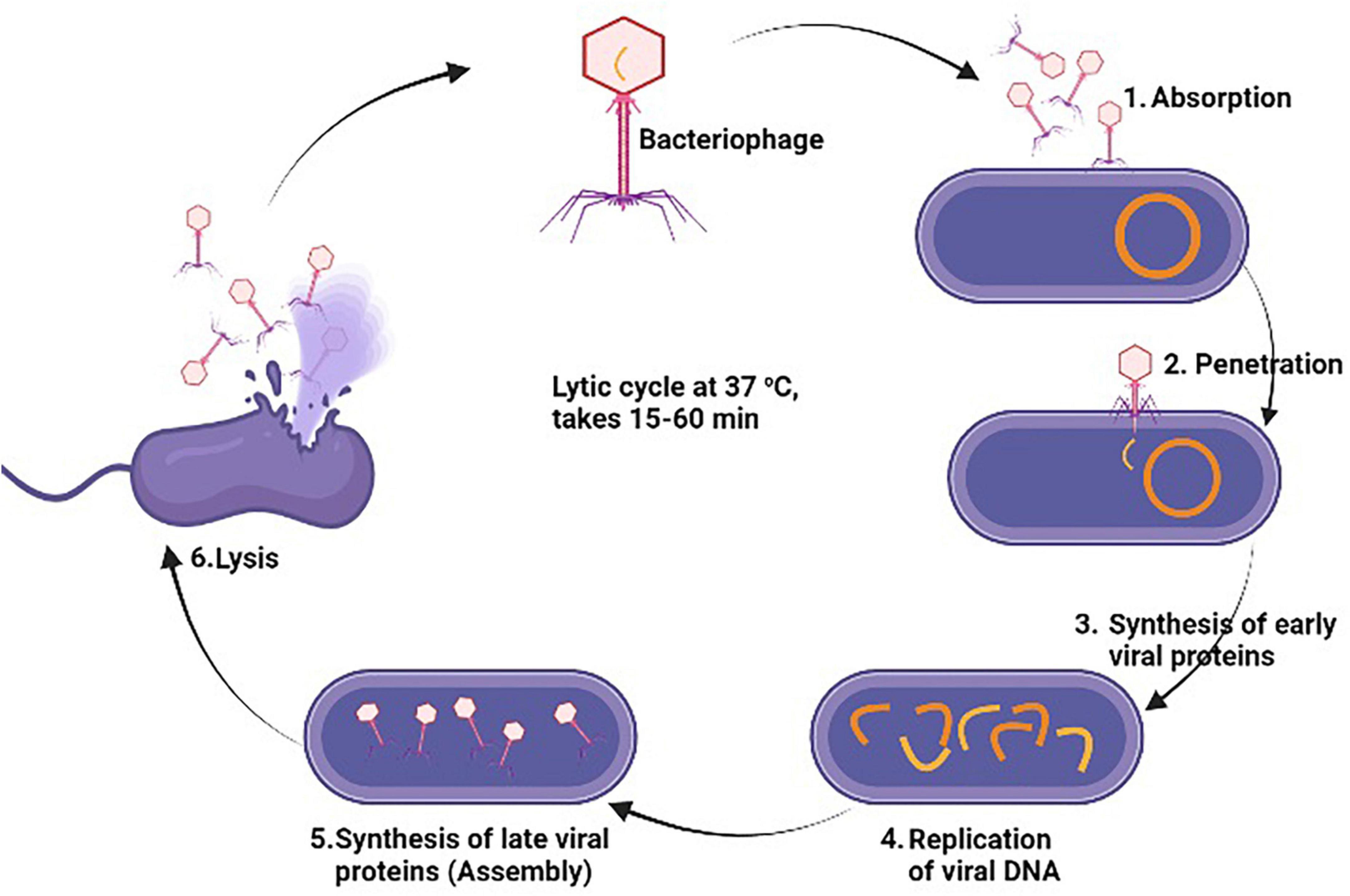
Figure 7. General antibacterial mechanism of bacteriophages against methicillin resistant strain of Staphylococcus aureus (MRSA).
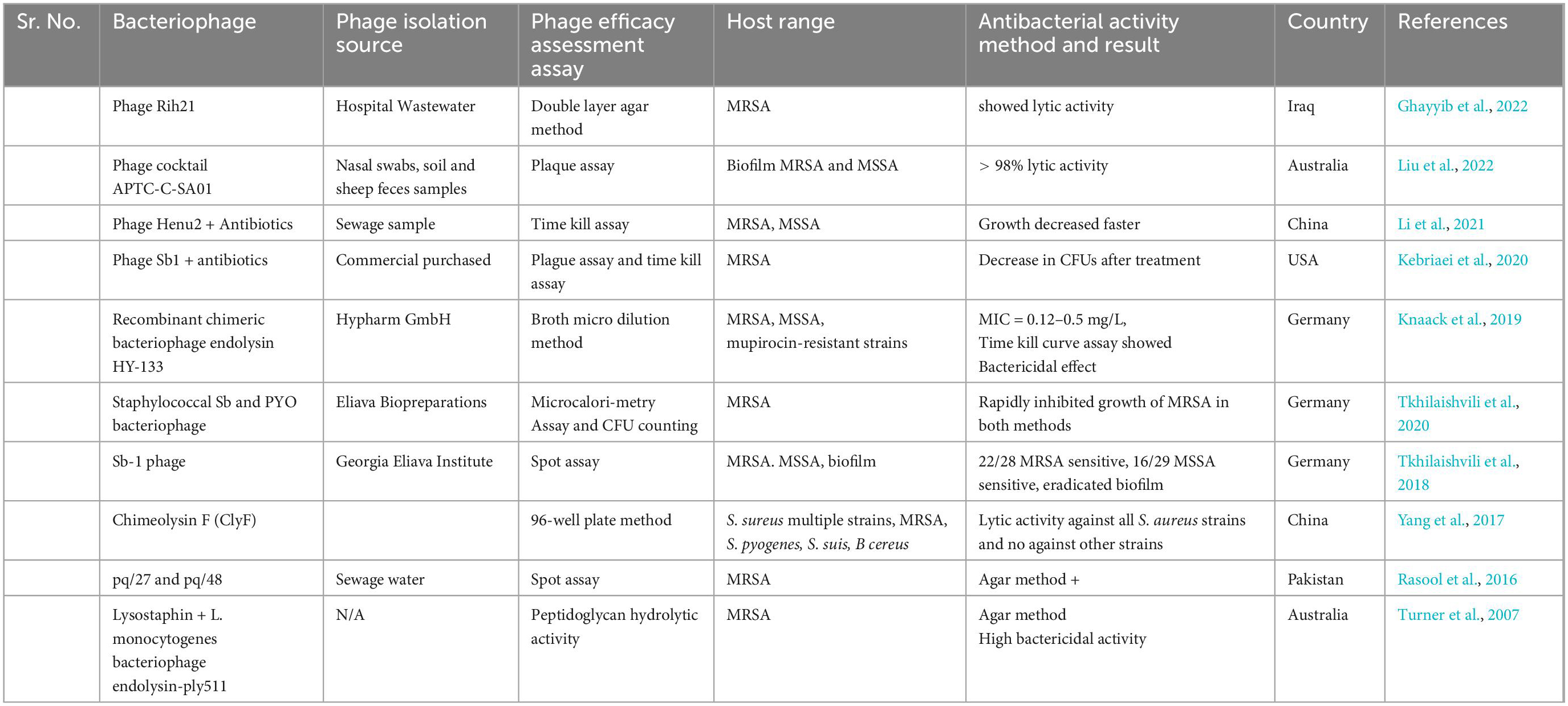
Table 5. In vitro studies on antibacterial activity of bacteriophages against methicillin resistant strain of Staphylococcus aureus (MRSA).
Utilizing probiotics is a prospective antibiotic substitute. Probiotics are live microorganism known to aid during infection or after antibiotic therapy which upsets the normal gut microbiota. Additionally, they could help to relieve certain other conditions such as irritable bowel syndrome (IBS) symptoms and antibiotic associated diarrhea (McFarland et al., 2021). Probiotic can improve the health of many other body tissues by replenishing their respective microbiota and releasing anti-pathogenic chemicals. So, the word probiotic is not only restricted to gut microbiota but they also provide various other health benefits such as enhances the host immunity by upregulating the immune cells, depression and anxiety disorders, tumor suppression, therapeutic role in COVID-19, overweight and obese patients (Herman, 2019; Shi et al., 2019; Shin et al., 2019; Ceccarelli et al., 2020; Daniali et al., 2020). Firstly, specific strains of Lactobacilli were used and evaluated for antimicrobial properties. Nowadays, many non-pathogenic species of bacterial as well as fungal genera are used as potential source of probiotic such as Streptococcus, Bifidobacterium, Bacillus, Escherichia, Enterococcus, and Saccharomyces from fungi. Various strains of Lactobacillus (L. rhamnosus, L. plantarum, L. animalis, L. reuteri, L. lactis, L. gasseri, L. curvattus, and L. lacis), Bacillus (B. subtilis, B. amyloliquefaciens), Bifidobacterium (B. lactis, B. bifidum, B. breve, B. dentium, B. longum, B. infantis, B. catenulatum, B. pseudo catenulatum), and Saccharomyces (S. cerevisiae) are all summarized in Table 6. All of the probiotic act by adopting one or more than one mechanism from the following; enhancement of epithelial barrier, increased adhesion to intestinal mucosa, synthesis of antimicrobial molecules, inhibition of pathogen adhesion to intestinal cells, competitive exclusion of pathogenic microorganisms, reducing the pH of gut lumen, and enhancement of immune response (Plaza-Diaz et al., 2019) as illustrated in Figure 8.
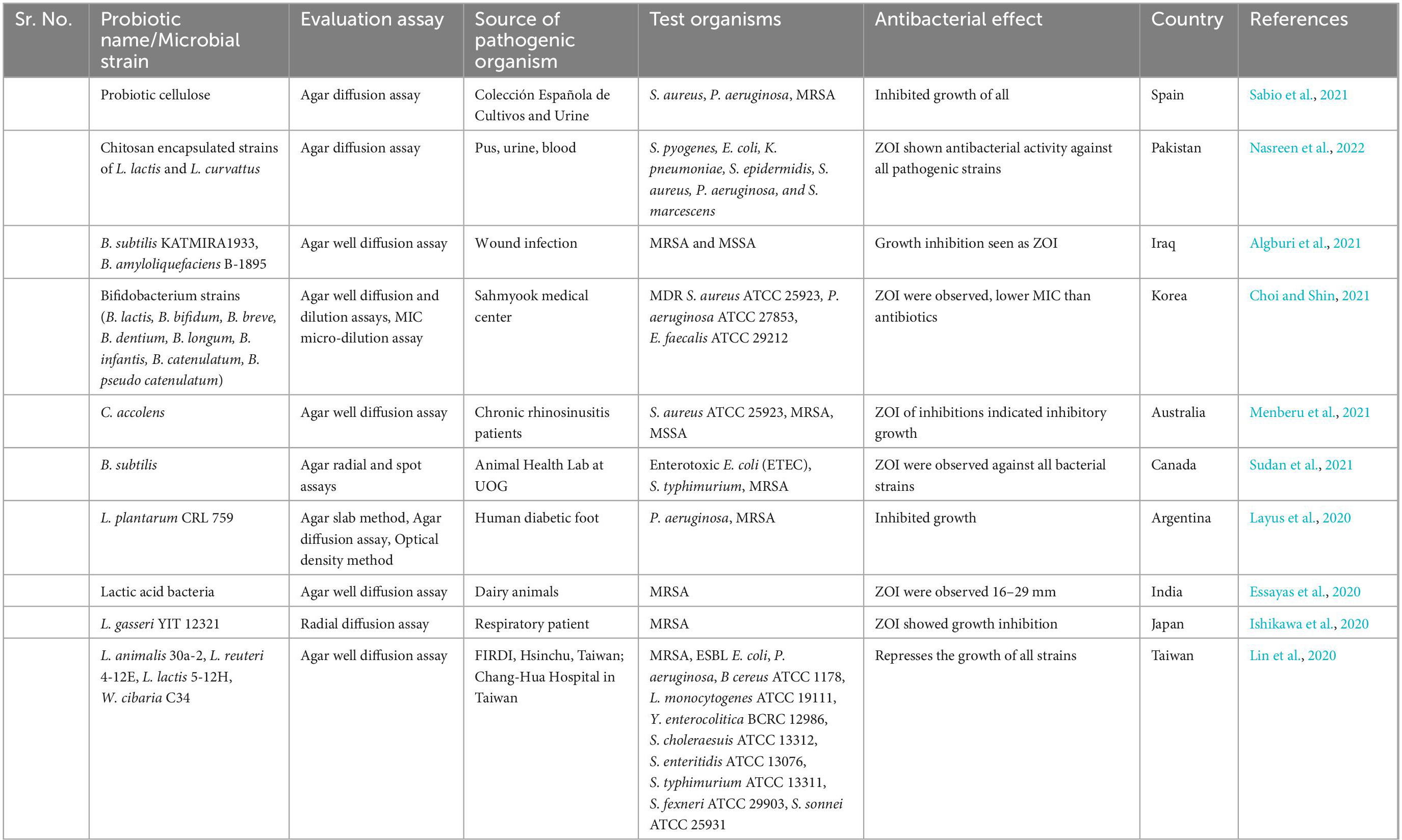
Table 6. In vitro studies on antibacterial activity of probiotics against methicillin resistant strain of Staphylococcus aureus (MRSA).
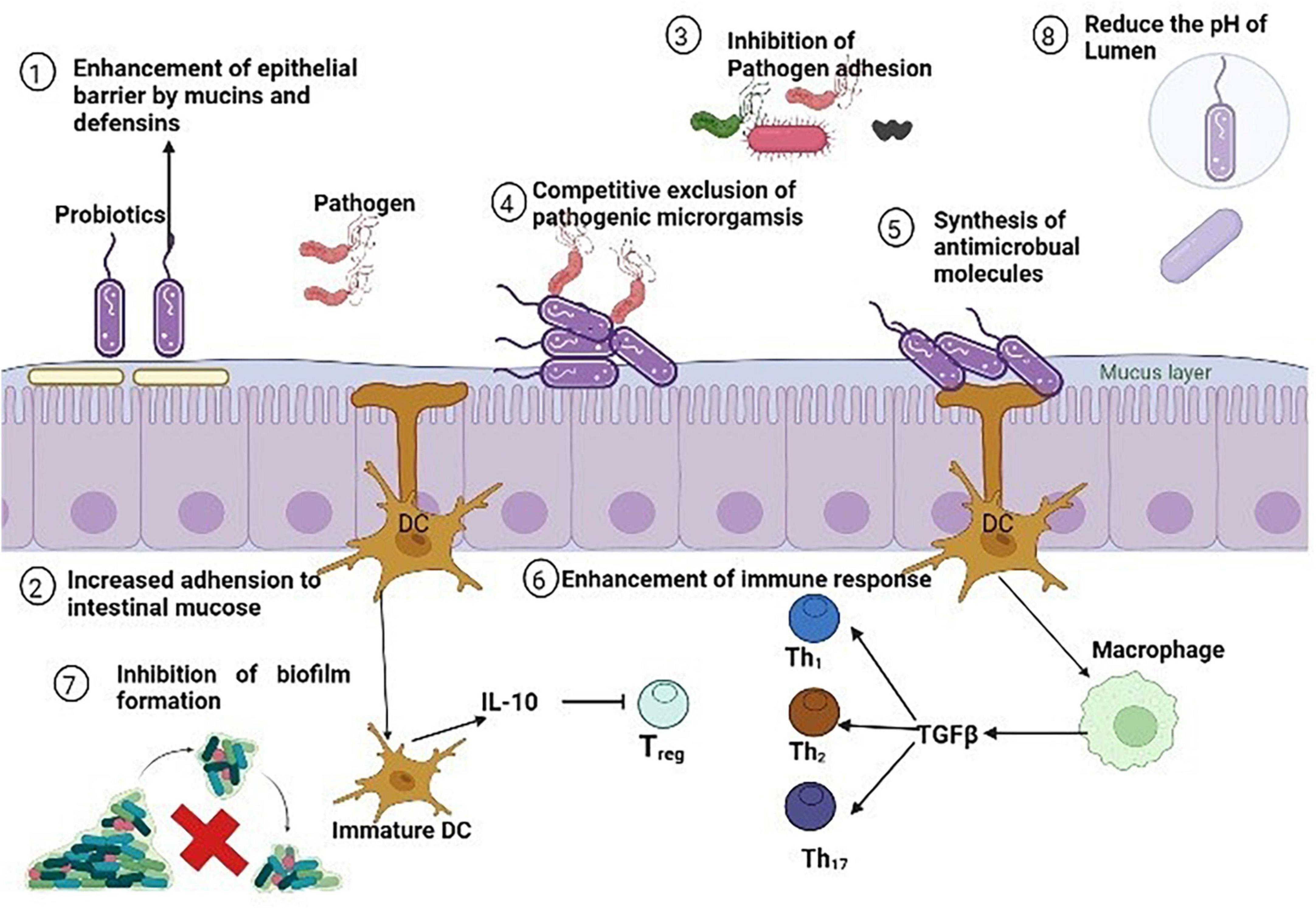
Figure 8. Antibacterial mechanisms of various probiotics against methicillin resistant strain of Staphylococcus aureus (MRSA).
Currently, many MRSA prevention and control interventions are carried out such as judicial use of antimicrobials, hand hygiene, controlling the interaction with S. aureus natural reservoirs, preventing transmission from infected patient, decolonization, isolation, disinfection of hospital environment, active surveillance, and many other (Lee et al., 2018). Now there is a need to develop most effective way to control MRSA at animal and human cadre i.e., vaccine production. The problem of antibiotic resistance including MRSA pointing to a notable concern and is under study by Center for Disease Control and Prevention and World Health Organization (Klevens et al., 2007). The development of novel ways to combat antibiotic resistance and production of vaccine very important in this era. MRSA is not well-known to be resistant to any known antibodies (Anderson et al., 2012). However, attempts to develop a potent vaccine against MRSA are under trial by various companies (Adhikari et al., 2012). To develop a potent vaccine against multiple MRSA strains, there is need to use multiple antigens to develop effective immunity against different strains (Aqib et al., 2018a). Additionally, an appropriate adjuvant needs to be added to the vaccine to improve the vaccine function as well as delivery into the host (Adamczyk-Poplawska et al., 2011). To create an effective multi-epitope component immunization against staphylococcal infections, three antigenic determinants are known to be much important which are clustered factor A (ClfA), alpha-enolase (Eno1), and iron regulated surface determinant protein B (IsdB). Eno1 is a polypeptide present in cytoplasm of cell and found in all S. aureus strains and has a very well-preserved lineage. Additionally, this protein aids in the mechanism of adhesion and contributes to the spread of infection, so this protein is a potential candidate for vaccine development against multiple S. aureus strains (Ghasemi et al., 2016). ClfA, another cell surface protein, aids in the pathogen’s adherence to the host. Previous investigations have shown that ClfA plays a key role in the development of staphylococcal illnesses (Garcia-Lara and Foster, 2009). Thus, to initiate a strong, active, and independent immune response to S. aureus, it is important to include this surface component as a vaccine agent (Brouillette et al., 2002). Another surface protein that aids in attachment to the cell membrane is IsdB, the third epitope marker (Zapotoczna et al., 2013).
A vaccine is an agent that prevent the infection before its onset and also disrupt the colonization of infection causing organism with host cell and thereby act as long lasting infection prevention agent and extensively reduces the use of antibiotics (Pozzi et al., 2017). A monoclonal antibody is developed by Medimmune, USA based company against α-haemolysin factors of S. aureus which may provide protective immunity administered alone or in combination with antibiotics. Another two monovalent trial based vaccines were prepared by USA based company and tested but failed to generate protective levels at the later stages of development. Then two more vaccines StaphVax and V710 were formulated by Nabi Pharmaceuticals, USA containing capsular polysaccharides CP5 plus CP8 and IsdB respectively as an antigenic component. Both of the vaccines provided immunity during animal model but failed in control phase III trials (Shinefield et al., 2002). The failure may be due to certain strains of S. aureus such as USA 300 does not contain CPs in their structure, lack of adjuvant in vaccine preparation, S. aureus immune evasion virulence factors such as IgG binding protein A have ability to compromise the function of antibodies and make them unable to provide effective immunity (Fowler and Proctor, 2014). However, the trials are underway to make a polyvalent vaccine containing number of antigens such as ClfA, CP5, CP8, secreted toxins (extracellular protein A and B, α-toxin, ESAT-6, lukS-PV), MntC, and Fhud2 (Pozzi et al., 2017). Another monoclonal vaccine containing WTA targeted antibodies plus rifampicin class antibiotic were tested and found protective effects in preclinical trials (Lehar et al., 2015). Moreover, further research is going on and hope soon a better vaccine will be able in market to treat multiple S. aureus strains including MRSA.
Methicillin resistant strain of S. aureus (MRSA) is found to be versatile and unpredictable pathogen with diversity of lineages common between humans and animals indicated its transmission between human and animals. The lineages found common between human and animal were CC398, CC9, CC130, CC97, CC398. Except this, few HA-MRSA and CA-MRSA lineages were identified in animals and LA-MRSA lineages were identified similar to HA-MRSA and CA-MRSA. Therefore, increasing prevalence and genetic adaptation of this pathogen at animal and human cadre exposing it a major threat for public health. This pathogen holds a diversity of host species ranging from humans to food and companion animals and many more. This pathogen has the ability to resist many antibiotics and to escape immune mechanisms through various virulence factors that unable this pathogen to cause mild to life threatening infections. Due to the increasing antibiotics resistance, this article highlighted the importance of alternative ways such as phytochemicals, bacteriophages, nanoparticles, and probiotics alone and in combination to use them as replacement of antibiotic. There is need to work more on these approaches to combat antibiotic resistance and making them available at commercial scale for public. One more approach is to work on successful vaccine production and immunization against MRSA.
MS and AA wrote the whole manuscript. WP structured and funded the project. IM and MK review the manuscript. ZB, AM, MF, and MM drew the illustrations. C-NZ, MK, FS, and NM added the references through endnote. All authors contributed to the article and approved the submitted version.
This project was funded by Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (25-LZIHPS-03).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbott, I., Jenney, A., Jeremiah, C., Mirèeta, M., Kandiah, J., Holt, D., et al. (2015). Reduced in-vitro activity of ceftaroline by etest among CC239 methicillin-resistant Staphylococcus aureus (MRSA) clinical strains from Australia. Antimicrob. Agents Chemother. 59, 7837–7841.
Abdeen, E. E., and Mousa, W. S. Abdel-Tawab, A. A. El-Faramawy, R. Abo-Shama, U. H. (2021). Phenotypic, genotypic and antibiogram among Staphylococcus aureus isolated from bovine subclinical mastitis. Pak. Vet. J. 41, 289–293.
Abulrob, A.-N., Suller, M. T., Gumbleton, M., Simons, C., and Russell, A. D. (2004). Identification and biological evaluation of grapefruit oil components as potential novel efflux pump modulators in methicillin-resistant Staphylococcus aureus bacterial strains. Phytochemistry 65, 3021–3027. doi: 10.1016/j.phytochem.2004.08.044
Adamczyk-Poplawska, M., Markowicz, S., and Jagusztyn-Krynicka, E. K. (2011). Proteomics for development of vaccine. J. Proteomics 74, 2596–2616.
Adhikari, R. P., Karauzum, H., Sarwar, J., Abaandou, L., Mahmoudieh, M., Boroun, A. R., et al. (2012). Novel structurally designed vaccine for S. aureus α-hemolysin: Protection against bacteremia and pneumonia. PLoS One 7:e38567. doi: 10.1371/journal.pone.0038567
Akhtar, S., Shahzad, K., Mushtaq, S., Ali, I., Rafe, M. H., and Fazal-ul-Karim, S. M. (2019). Antibacterial and antiviral potential of colloidal titanium dioxide (TiO2) nanoparticles suitable for biological applications. Mater. Res. Express 6:105409.
Aklilu, E., and Chia, H. Y. (2020). First mec C and mec A positive livestock-associated methicillin resistant Staphylococcus aureus (mec C MRSA/LA-MRSA) from dairy cattle in Malaysia. Microorganisms 8:147. doi: 10.3390/microorganisms8020147
Albur, M., Bowker, K., Weir, I., and MacGowan, A. (2012). Factors influencing the clinical outcome of methicillin-resistant Staphylococcus aureus bacteraemia. Eur. J. Clin. Microbiol. Infect. Dis. 31, 295–301.
Aldred, K. J., Kerns, R. J., and Osheroff, N. (2014). Mechanism of quinolone action and resistance. Biochemistry 53, 1565–1574.
Algburi, A., Al-Hasani, H. M., Ismael, T. K., Abdelhameed, A., Weeks, R., Ermakov, A. M., et al. (2021). Antimicrobial activity of Bacillus subtilis KATMIRA1933 and Bacillus amyloliquefaciens B-1895 against Staphylococcus aureus biofilms isolated from wound infection. Probiotics Antimicrob. Proteins 13, 125–134. doi: 10.1007/s12602-020-09673-4
Altaf, M., Ijaz, M., Iqbal, M. K., Rehman, A., Avais, M., Ghaffar, A., et al. (2020). Molecular characterization of methicillin-resistant Staphylococcus aureus (MRSA) and associated risk factors with the occurrence of goat mastitis. Pak. Vet. J. 40, 1–6.
Alvarez-Uria, G., and Reddy, R. (2012). Prevalence and antibiotic susceptibility of community-associated methicillin-resistant Staphylococcus aureus in a rural area of India: Is MRSA replacing methicillin-susceptible Staphylococcus aureus in the community? Int. Sch. Res. Notices 2012:248951. doi: 10.5402/2012/248951
Alzomor, O., Alfawaz, T., and Alshahrani, D. (2017). Invasive community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) infection in children: Case series and literature review. Int. J. Pediatr. Adolesc. Med. 4, 119–123. doi: 10.1016/j.ijpam.2017.07.001
Anderson, A. S., Miller, A. A., Donald, R. G., Scully, I. L., Nanra, J. S., Cooper, D., et al. (2012). Development of a multicomponent Staphylococcus aureus vaccine designed to counter multiple bacterial virulence factors. Hum. Vaccin. Immunother. 8, 1585–1594. doi: 10.4161/hv.21872
Ansari, M. A., Khan, H. M., Khan, A. A., Pal, R., and Cameotra, S. S. (2013). Antibacterial potential of Al2O3 nanoparticles against multidrug resistance strains of Staphylococcus aureus isolated from skin exudates. J. Nanopart. Res. 15:1970.
Aqib, A. I., Ijaz, M., Farooqi, S. H., Ahmed, R., Shoaib, M., Ali, M. M., et al. (2018b). Emerging discrepancies in conventional and molecular epidemiology of methicillin resistant Staphylococcus aureus isolated from bovine milk. Microb. Pathog. 116, 38–43. doi: 10.1016/j.micpath.2018.01.005
Aqib, A. I., Anjum, A. A., Ijaz, M., Hussain, R., Ahmed, R., Farooqi, S. H., et al. (2018a). Development and evaluation of vaccine against Staphylococcus aureus recovered from naturally occurring mastitis in she-camels. Microb. Pathog. 117, 341–347. doi: 10.1016/j.micpath.2018.03.003
Aqib, A. I., Ijaz, M., Anjum, A. A., Malik, M. A. R., Mehmood, K., Farooqi, S. H., et al. (2017). Antibiotic susceptibilities and prevalence of methicillin resistant Staphylococcus aureus (MRSA) isolated from bovine milk in Pakistan. Acta Trop. 176, 168–172. doi: 10.1016/j.actatropica.2017.08.008
Aqib, A. I., Saqib, M., Khan, S. R., Ahmad, T., Shah, S. A. R., Naseer, M. A., et al. (2021). Non-steroidal anti-inflammatory drugs, plant extracts, and characterized microparticles to modulate antimicrobial resistance of epidemic mecA positive S. aureus of dairy origin. Appl. Nanosci. 11, 553–563.
Arora, S., Devi, P., Arora, U., and Devi, B. (2010). Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in a tertiary care hospital in Northern India. J. Lab. Physicians 2, 078–081.
Attallah, N. G., Negm, W. A., Elekhnawy, E., Elmongy, E. I., Altwaijry, N., El-Haroun, H., et al. (2021). Elucidation of phytochemical content of Cupressus macrocarpa leaves: In vitro and in vivo antibacterial effect against methicillin-resistant Staphylococcus aureus clinical isolates. Antibiotics 10:890. doi: 10.3390/antibiotics10080890
Awad, M., Yosri, M., Abdel-Aziz, M. M., Younis, A. M., and Sidkey, N. M. (2021). Assessment of the antibacterial potential of biosynthesized silver nanoparticles combined with vancomycin against methicillin-resistant Staphylococcus aureus–induced infection in rats. Biol. Trace Elem. Res. 199, 4225–4236. doi: 10.1007/s12011-020-02561-6
Ballhausen, B., Kriegeskorte, A., van Alen, S., Jung, P., Köck, R., Peters, G., et al. (2017). The pathogenicity and host adaptation of livestock-associated MRSA CC398. Vet. Microbiol. 200, 39–45.
Baptiste, K. E., Williams, K., Willams, N. J., Wattret, A., Clegg, P. D., Dawson, S., et al. (2005). Methicillin-resistant staphylococci in companion animals. Emerg. Infect. Dis. 11:1942.
Bar-Gal, G. K., Blum, S., Hadas, L., Ehricht, R., Monecke, S., and Leitner, G. (2015). Host-specificity of Staphylococcus aureus causing intramammary infections in dairy animals assessed by genotyping and virulence genes. Vet. Microbiol. 176, 143–154. doi: 10.1016/j.vetmic.2015.01.007
Barrett, F. F., McGehee, R. F. Jr., and Finland, M. (1968). Methicillin-resistant Staphylococcus aureus at Boston City hospital: Bacteriologic and epidemiologic observations. N. Engl. J. Med. 279, 441–448. doi: 10.1056/NEJM196808292790901
Bi, Z., Sun, C., Börjesson, S., Chen, B., Ji, X., Berglund, B., et al. (2018). Identical genotypes of community-associated MRSA (ST 59) and livestock-associated MRSA (ST 9) in humans and pigs in rural China. Zoonoses Public Health 65, 367–371. doi: 10.1111/zph.12443
Boag, A., Loeffler, A., and Lloyd, D. (2004). Methicillin-resistant Staphylococcus aureus isolates from companion animals. Vet. Rec. 154, 411–411.
Bos, M. E., Graveland, H., Portengen, L., Wagenaar, J. A., and Heederik, D. J. (2012). Livestock-associated MRSA prevalence in veal calf production is associated with farm hygiene, use of antimicrobials, and age of the calves. Prev. Vet. Med. 105, 155–159. doi: 10.1016/j.prevetmed.2012.01.002
Boucher, H. W., Wilcox, M., Talbot, G. H., Puttagunta, S., Das, A. F., and Dunne, M. W. (2014). Once-weekly dalbavancin versus daily conventional therapy for skin infection. N. Engl. J. Med. 370, 2169–2179. doi: 10.1056/NEJMoa1310480
Bozdogan, B., and Appelbaum, P. C. (2004). Oxazolidinones: Activity, mode of action, and mechanism of resistance. Int. J. Antimicrob. Agents 23, 113–119.
Brade, K. D., Rybak, J. M., and Rybak, M. J. (2016). Oritavancin: A new lipoglycopeptide antibiotic in the treatment of Gram-positive infections. Infect. Dis. Ther. 5, 1–15.
Brouillette, E., Lacasse, P., Shkreta, L., Bélanger, J., Grondin, G., Diarra, M. S., et al. (2002). DNA immunization against the clumping factor A (ClfA) of Staphylococcus aureus. Vaccine 20, 2348–2357. doi: 10.1016/s0264-410x(02)00100-7
Buzgaia, N., Awin, T., Elabbar, F., Abdusalam, K., Lee, S. Y., Rukayadi, Y., et al. (2020). Antibacterial activity of Arbutus pavarii pamp against methicillin-resistant Staphylococcus aureus (MRSA) and UHPLC-MS/MS profile of the bioactive fraction. Plants 9:1539. doi: 10.3390/plants9111539
Carter, G. R., Chengappa, M., Roberts, A. W., Claus, G. W., and Rikihisa, Y. (1995). Essentials of veterinary microbiology. Hoboken, NJ: Wiley.
Ceccarelli, G., Scagnolari, C., Pugliese, F., Mastroianni, C. M., and d’Ettorre, G. (2020). Probiotics and COVID-19. Lancet Gastroenterol. Hepatol. 5, 721–722.
Cefai, C., Ashurst, S., and Owens, C. (1994). Human carriage of methicillin-resistant Staphylococcus aureus linked with pet dog. Lancet 344, 539–540. doi: 10.1016/s0140-6736(94)91926-7
Chakraborty, S., Afaq, N., Singh, N., and Majumdar, S. (2018). Antimicrobial activity of Cannabis sativa, Thuja orientalis and Psidium guajava leaf extracts against methicillin-resistant Staphylococcus aureus. J. Integr. Med. 16, 350–357. doi: 10.1016/j.joim.2018.07.005
Chambers, H. (2001). Methicillin-resistant Staphylococcus aureus. Mechanisms of resistance and implications for treatment. Postgrad. Med. 109(Suppl. 2) 43–50.
Chambers, H. F., and DeLeo, F. R. (2009). Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7:629.
Chan, E. W. L., Yee, Z. Y., Raja, I., and Yap, J. K. Y. (2017). Synergistic effect of non-steroidal anti-inflammatory drugs (NSAIDs) on antibacterial activity of cefuroxime and chloramphenicol against methicillin-resistant Staphylococcus aureus. J. Glob. Antimicrob. Resist. 10, 70–74. doi: 10.1016/j.jgar.2017.03.012
Chen, A., Zervos, M., and Vazquez, J. A. (2007). Dalbavancin: A novel antimicrobial. Int. J. Clin. Pract. 61, 853–863.
Chen, Y., Sun, L., Ba, X., Jiang, S., Zhuang, H., Zhu, F., et al. (2022). Epidemiology, evolution and cryptic susceptibility of methicillin-resistant Staphylococcus aureus in China: A whole-genome-based survey. Clin. Microbiol. Infect. 28, 85–92. doi: 10.1016/j.cmi.2021.05.024
Chien, C.-S., Lin, C.-J., Ko, C.-J., Tseng, S.-P., and Shih, C.-J. (2018). Antibacterial activity of silver nanoparticles (AgNP) confined to mesostructured silica against methicillin-resistant Staphylococcus aureus (MRSA). J. Alloys Compd. 747, 1–7. doi: 10.3390/nano10071264
Chini, V., Petinaki, E., Foka, A., Paratiras, S., Dimitracopoulos, G., and Spiliopoulou, I. (2006). Spread of Staphylococcus aureus clinical isolates carrying Panton–Valentine leukocidin genes during a 3-year period in Greece. Clin. Microbiol. Infect. 12, 29–34. doi: 10.1111/j.1469-0691.2005.01295.x
Choi, J.-G., Choi, J.-Y., Mun, S.-H., Kang, O.-H., Bharaj, P., Shin, D.-W., et al. (2015). Antimicrobial activity and synergism of Sami-Hyanglyun-Hwan with ciprofloxacin against methicillin-resistant Staphylococcus aureus. Asian Pac. J. Trop. Med. 8, 538–542. doi: 10.1016/j.apjtm.2015.06.010
Choi, Y. J., and Shin, H. S. (2021). Antibacterial effect of eight probiotic strains of bifidobacterium against pathogenic Staphylococcus aureus and Pseudomonas aeruginosa. J. Bacteriol. Virol. 51, 128–137.
Chukwudi, C. U. (2016). Ribosomal RNA binding sites and the molecular mechanism of action of the tetracyclines. Antimicrob. Agents Chemother. 60, 4433–4441.
Climo, M. W., Patron, R. L., and Archer, G. L. (1999). Combinations of vancomycin and β-lactams are synergistic against staphylococci with reduced susceptibilities to vancomycin. Antimicrob. Agents Chemother. 43, 1747–1753. doi: 10.1128/AAC.43.7.1747
Coombs, G. W., Daley, D. A., Yee, N. W., Shoby, P., and Mowlaboccus, S. (2022). Australian group on antimicrobial resistance (AGAR) Australian Staphylococcus aureus sepsis outcome programme (ASSOP) annual report 2020. Commun. Dis. Intell. 46, 1–17. doi: 10.33321/cdi.2020.44.71
Coutinho, H. D., Costa, J. G., Lima, E. O., Falcão-Silva, V. S., and Júnior, J. P. S. (2009). Herbal therapy associated with antibiotic therapy: Potentiation of the antibiotic activity against methicillin–resistant Staphylococcus aureus by Turnera ulmifolia L. BMC Complement. Altern. Med. 9:13. doi: 10.1186/1472-6882-9-13
Cuny, C., Friedrich, A., Kozytska, S., Layer, F., Nübel, U., Ohlsen, K., et al. (2010). Emergence of methicillin-resistant Staphylococcus aureus (MRSA) in different animal species. Int. J. Med. Microbiol. 300, 109–117.
Cuny, C., Kuemmerle, J., Stanek, C., Willey, B., Strommenger, B., and Witte, W. (2006). Emergence of MRSA infections in horses in a veterinary hospital: Strain characterisation and comparison with MRSA from humans. Euro Surveill. 11, 44–47.
Da Silva, E. R., do Carmo, L. S., and Da Silva, N. (2005). Detection of the enterotoxins A, B, and C genes in Staphylococcus aureus from goat and bovine mastitis in Brazilian dairy herds. Vet. Microbiol. 106, 103–107. doi: 10.1016/j.vetmic.2004.12.005
Damodaran, S., and Madhan, S. (2011). Telavancin: A novel lipoglycopeptide antibiotic. J. Pharmacol. Pharmacother. 2:135.
Daniali, M., Nikfar, S., and Abdollahi, M. (2020). A brief overview on the use of probiotics to treat overweight and obese patients. Expert Rev. Endocrinol. Metab. 15, 1–4.
Dantes, R., Mu, Y., Belflower, R., Aragon, D., Dumyati, G., Harrison, L. H., et al. (2013). National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA Intern. Med. 173, 1970–1978.
Datta, P., Gulati, N., Singla, N., Vasdeva, H. R., Bala, K., Chander, J., et al. (2011). Evaluation of various methods for the detection of meticillin-resistant Staphylococcus aureus strains and susceptibility patterns. J. Med. Microbiol. 60, 1613–1616. doi: 10.1099/jmm.0.032219-0
Daum, R. S. (2007). Skin and soft-tissue infections caused by methicillin-resistant Staphylococcus aureus. N. Engl. J. Med. 357, 380–390.
David, M. Z., and Daum, R. S. (2010). Community-associated methicillin-resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 23, 616–687. doi: 10.1128/CMR.00081-09
Davis, K. A., Stewart, J. J., Crouch, H. K., Florez, C. E., and Hospenthal, D. R. (2004). Methicillin-resistant Staphylococcus aureus (MRSA) nares colonization at hospital admission and its effect on subsequent MRSA infection. Clin. Infect. Dis. 39, 776–782. doi: 10.1086/422997
De Araújo, R. S., Guerra, F. Q., de, O., Lima, E., De Simone, C. A., Tavares, J. F., et al. (2013). Synthesis, structure-activity relationships (SAR) and in silico studies of coumarin derivatives with antifungal activity. Int. J. Mol. Sci. 14, 1293–1309. doi: 10.3390/ijms14011293
De Jong, N. W., Vrieling, M., Garcia, B. L., Koop, G., Brettmann, M., Aerts, P. C., et al. (2018). Identification of a staphylococcal complement inhibitor with broad host specificity in equid Staphylococcus aureus strains. J. Biol. Chem. 293, 4468–4477. doi: 10.1074/jbc.RA117.000599
De Kraker, M., Jarlier, V., Monen, J., Heuer, O., Van De Sande, N., and Grundmann, H. (2013). The changing epidemiology of bacteraemias in Europe: Trends from the European antimicrobial resistance surveillance system. Clin. Microbiol. Infect. 19, 860–868. doi: 10.1111/1469-0691.12028
DeLeo, F. R., Otto, M., Kreiswirth, B. N., and Chambers, H. F. (2010). Community-associated meticillin-resistant Staphylococcus aureus. Lancet 375, 1557–1568.
Dhand, A., Bayer, A. S., Pogliano, J., Yang, S.-J., Bolaris, M., Nizet, V., et al. (2011). Use of antistaphylococcal β-lactams to increase daptomycin activity in eradicating persistent bacteremia due to methicillin-resistant Staphylococcus aureus: Role of enhanced daptomycin binding. Clin. Infect. Dis. 53, 158–163. doi: 10.1093/cid/cir340
Diekema, D., and Pfaller, M. (2001). Schmitz FJet al. Survey of infections due to Staphylococcus species: Frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY antimicrobial surveillance program, 1997–1999. Clin. Infect. Dis. 32(Suppl. 2) S114–S132. doi: 10.1086/320184
Dings, M. M., Orwin, P. M., and Schlievert, P. (2000). Exotoxins of Staphyloccocus aureus. Clin. Microbiol. Rev. 13, 16–34.
Ehrlich, S. (1977). Replication and expression of plasmids from Staphylococcus aureus in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 74, 1680–1682.
Eltaweil, A. S., Fawzy, M., Hosny, M., Abd El-Monaem, E. M., Tamer, T. M., and Omer, A. M. (2022). Green synthesis of platinum nanoparticles using Atriplex halimus leaves for potential antimicrobial, antioxidant, and catalytic applications. Arab. J. Chem. 15:103517.
Enger, H., Larssen, K. W., Damås, E. S., Aamot, H. V., Blomfeldt, A., Elstrøm, P., et al. (2022). A tale of two STs: Molecular and clinical epidemiology of MRSA t304 in Norway 2008–2016. Eur. J. Clin. Microbiol. Infect. Dis. 41, 209–218. doi: 10.1007/s10096-021-04353-9
Enright, M. C., Robinson, D. A., Randle, G., Feil, E. J., Grundmann, H., and Spratt, B. G. (2002). The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. 99, 7687–7692.
Essayas, A., Pandit, S., and Taneja, P. (2020). Anti-microbial activity of potential probiotic lactic acid bacteria against methicillin-resistant Staphylococcus aureus (MRSA). bioRxiv [Preprint]. doi: 10.1101/2020.03.08.982512
Faires, M. C., Tater, K. C., and Weese, J. S. (2009). An investigation of methicillin-resistant Staphylococcus aureus colonization in people and pets in the same household with an infected person or infected pet. J. Am. Vet. Med. Assoc. 235, 540–543.
Ferens, W. A., and Bohach, G. A. (2000). Persistence of Staphylococcus aureus on mucosal membranes: Superantigens and internalization by host cells. J. Lab. Clin. Med. 135, 225–230. doi: 10.1067/mlc.2000.105179
Fernando, S., Gunasekara, T., and Holton, J. (2018). Antimicrobial nanoparticles: Applications and mechanisms of action. Sri Lankan J. Infect. Dis. 8, 2–11.
Ferri, M., Ranucci, E., Romagnoli, P., and Giaccone, V. (2017). Antimicrobial resistance: A global emerging threat to public health systems. Crit. Rev. Food Sci. Nutr. 57, 2857–2876.
File, T. M. Jr., Low, D. E., Eckburg, P. B., Talbot, G. H., Friedland, H. D., Lee, J., et al. (2011). FOCUS 1: A randomized, double-blinded, multicentre, phase III trial of the efficacy and safety of ceftaroline fosamil versus ceftriaxone in community-acquired pneumonia. J. Antimicrob. Chemother. 66(Suppl. 3) iii19–iii32.
Fish, R., Kutter, E., Wheat, G., Blasdel, B., Kutateladze, M., and Kuhl, S. (2018). Compassionate use of bacteriophage therapy for foot ulcer treatment as an effective step for moving toward clinical trials. Methods Mol. Biol. 1963, 159–170. doi: 10.1007/978-1-4939-7395-8_14
Fitzgerald, J. R., Monday, S. R., Foster, T. J., Bohach, G. A., Hartigan, P. J., Meaney, W. J., et al. (2001). Characterization of a putative pathogenicity island from bovine Staphylococcus aureus encoding multiple superantigens. J. Bacteriol. 183, 63–70. doi: 10.1128/JB.183.1.63-70.2001
Flanagan, S., McKee, E. E., Das, D., Tulkens, P. M., Hosako, H., Fiedler-Kelly, J., et al. (2015). Nonclinical and pharmacokinetic assessments to evaluate the potential of tedizolid and linezolid to affect mitochondrial function. Antimicrob. Agents Chemother. 59, 178–185. doi: 10.1128/AAC.03684-14
Foster, T. J. (2002). “Staphylococcus aureus,” in Molecular medical microbiology, ed. S. Baron (Galveston, TX: University of Texas Medical Branch at Galveston), 839–888.
Fowler, V. G. Jr., and Proctor, R. A. (2014). Where does a Staphylococcus aureus vaccine stand? Clin. Microbiol. Infect. 20, 66–75.
Fowler, V. G. Jr., Boucher, H. W., Corey, G. R., Abrutyn, E., Karchmer, A. W., Rupp, M. E., et al. (2006). Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N. Engl. J. Med. 355, 653–665. doi: 10.1056/NEJMoa053783
Fowler, V. G., Olsen, M. K., Corey, G. R., Woods, C. W., Cabell, C. H., Reller, L. B., et al. (2003). Clinical identifiers of complicated Staphylococcus aureus bacteremia. Arch. Intern. Med. 163, 2066–2072.
Fromageau, A., Gilbert, F. B., Prévost, G., and Rainard, P. (2010). Binding of the Staphylococcus aureus leucotoxin LukM to its leucocyte targets. Microb. Pathog. 49, 354–362. doi: 10.1016/j.micpath.2010.07.002
Garcia-Lara, J., and Foster, S. J. (2009). Anti-Staphylococcus aureus immunotherapy: Current status and prospects. Curr. Opin. Pharmacol. 9, 552–557.
Garzoni, C., and Kelley, W. L. (2011). Return of the Trojan horse: Intracellular phenotype switching and immune evasion by Staphylococcus aureus. EMBO Mol. Med. 3, 115–117. doi: 10.1002/emmm.201100123
Genestier, A.-L., Michallet, M.-C., Prévost, G., Bellot, G., Chalabreysse, L., Peyrol, S., et al. (2005). Staphylococcus aureus Panton-Valentine leukocidin directly targets mitochondria and induces Bax-independent apoptosis of human neutrophils. J. Clin. Invest. 115, 3117–3127. doi: 10.1172/JCI22684
Gerding, D., Hecht, D., Louie, T., Nord, C., Talbot, G., Cornely, O., et al. (2015). Susceptibility of Clostridium difficile isolates from a phase 2 clinical trial of cadazolid and vancomycin in C. difficile infection. J. Antimicrob. Chemother. 71, 213–219. doi: 10.1093/jac/dkv300
Ghasemi, Y., Hajighahramani, N., Dabbagh, F., Ghoshoon, M. B., Yarahmadi, E., Mobasher, M. A., et al. (2016). Cloning, expression, purification and expression condition optimization of alpha-enolase from Staphylococcus aureus in Escherichia coli. Minerva Biotecnol. 28, 33–38.
Ghayyib, A. A., Ahmed, I. A., and Ahmed, H. K. (2022). Isolation and characterization of Staphylococcus Phage Rih21 and evaluation of its antibacterial activity against methicillin-resistant Staphylococcus aureus clinical isolates. [Preprint]. doi: 10.20944/preprints202209.0004.v2
Gillet, Y., Issartel, B., Vanhems, P., Fournet, J.-C., Lina, G., Bes, M., et al. (2002). Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet 359, 753–759. doi: 10.1016/S0140-6736(02)07877-7
Gordon, R. J., and Lowy, F. D. (2008). Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clini. Infect. Dis. 46(Suppl. 5) S350–S359.
Grinberg, A., Hittman, A., Leyland, M., Rogers, L., and Le Quesne, B. (2004). Epidemiological and molecular evidence of a monophyletic infection with Staphylococcus aureus causing a purulent dermatitis in a dairy farmer and multiple cases of mastitis in his cows. Epidemiol. Infect. 132, 507–513. doi: 10.1017/s0950268804002079
Grundmann, H., Aanensen, D., van den Wijngaard, C. C., Spratt, B., Harmsen, D., and Friedrich, A. (2010). Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: A molecular-epidemiological analysis. PLoS Med. 7:e1000215. doi: 10.1371/journal.pmed.1000215
Gudiol, F., Aguado, J. M., Almirante, B., Bouza, E., Cercenado, E., and Domínguez, M. Á, et al. (2015). Executive summary of the diagnosis and treatment of bacteremia and endocarditis due to Staphylococcus aureus. A clinical guideline from the Spanish society of clinical microbiology and infectious diseases (SEIMC). Enferm. Infecc. Microbiol. Clin. 33, 626–632.
Gudkov, S. V., Burmistrov, D. E., Smirnova, V. V., Semenova, A. A., and Lisitsyn, A. B. (2022). A mini review of antibacterial properties of Al2O3 nanoparticles. Nanomaterials 12:2635. doi: 10.3390/nano12152635
Guinane, C. M., Ben Zakour, N. L., Tormo-Mas, M. A., Weinert, L. A., Lowder, B. V., Cartwright, R. A., et al. (2010). Evolutionary genomics of Staphylococcus aureus reveals insights into the origin and molecular basis of ruminant host adaptation. Genome Biol. Evol. 2, 454–466. doi: 10.1093/gbe/evq031
Gunes, H., Gulen, D., Mutlu, R., Gumus, A., Tas, T., and Topkaya, A. E. (2016). Antibacterial effects of curcumin: An in vitro minimum inhibitory concentration study. Toxicol. Ind. Health 32, 246–250. doi: 10.1177/0748233713498458
Guo, Q., Ren, C.-W., Cai, J.-H., Zhang, C.-Y., Li, Y. T., Xu, B., et al. (2022). The synergistic inhibition and mechanism of epicatechin gallate and chitosan against methicillin-resistant Staphylococcus aureus and the application in pork preservation. LWT 163:113575.
Gupta, V., Fatima, A., Faridi, U., Negi, A. S., Shanker, K., Kumar, J., et al. (2008). Antimicrobial potential of Glycyrrhiza glabra roots. J. Ethnopharmacol. 116, 377–380.
Haag, A. F., Fitzgerald, J. R., and Penadés, J. R. (2019). Staphylococcus aureus in animals. Microbiol. Spectr. 7, 731–746.
Hamdan-Partida, A., González-García, S., Martínez-Ruíz, F. J., Zavala-Sánchez, M. Á, Bustos-Hamdan, A., and Bustos-Martínez, J. (2022). Molecular characterization of Staphylococcus aureus strains isolated from mobile phones. Microorganisms 10:669.
Harbarth, S., François, P., Schrenzel, J., Fankhauser-Rodriguez, C., Hugonnet, S., Koessler, T., et al. (2005). Community-associated methicillin-resistant Staphylococcus aureus, Switzerland. Emerg. Infect. Dis. 11:962.
Hasman, H., Moodley, A., Guardabassi, L., Stegger, M., Skov, R., and Aarestrup, F. M. (2010). Spa type distribution in Staphylococcus aureus originating from pigs, cattle and poultry. Vet. Microbiol. 141, 326–331. doi: 10.1016/j.vetmic.2009.09.025
Haysom, L., Cross, M., Anastasas, R., Moore, E., and Hampton, S. (2018). Prevalence and risk factors for methicillin-resistant Staphylococcus aureus (MRSA) infections in custodial populations: A systematic review. J. Correct. Health Care 24, 197–213. doi: 10.1177/1078345818765271
Herman, A. (2019). Probiotics supplementation in prophylaxis and treatment of depressive and anxiety disorders-a review of current research. Psychiatr. Pol. 53, 459–473. doi: 10.12740/PP/92392
Herold, B. C., Immergluck, L. C., Maranan, M. C., Lauderdale, D. S., Gaskin, R. E., Boyle-Vavra, S., et al. (1998). Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279, 593–598. doi: 10.1001/jama.279.8.593
Hidron, A. I., Kourbatova, E. V., Halvosa, J. S., Terrell, B. J., McDougal, L. K., Tenover, F. C., et al. (2005). Risk factors for colonization with methicillin-resistant Staphylococcus aureus (MRSA) in patients admitted to an urban hospital: Emergence of community-associated MRSA nasal carriage. Clin. Infect. Dis. 41, 159–166. doi: 10.1086/430910
Hines, K. M., Shen, T., Ashford, N. K., Waalkes, A., Penewit, K., Holmes, E. A., et al. (2020). Occurrence of cross-resistance and β-lactam seesaw effect in glycopeptide-, lipopeptide-and lipoglycopeptide-resistant MRSA correlates with membrane phosphatidylglycerol levels. J. Antimicrob. Chemother. 75, 1182–1186. doi: 10.1093/jac/dkz562
Hoekstra, J., Rutten, V., Sommeling, L., Van Werven, T., Spaninks, M., Duim, B., et al. (2018). High production of LukMF’in Staphylococcus aureus field strains is associated with clinical bovine mastitis. Toxins 10:200. doi: 10.3390/toxins10050200
Holden, M. T., Hsu, L.-Y., Kurt, K., Weinert, L. A., Mather, A. E., Harris, S. R., et al. (2013). A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res. 23, 653–664. doi: 10.1101/gr.147710.112
Holmes, N. E., Johnson, P. D., and Howden, B. P. (2012). Relationship between vancomycin-resistant Staphylococcus aureus, vancomycin-intermediate S. aureus, high vancomycin MIC, and outcome in serious S. aureus infections. J. Clin. Microbiol. 50, 2548–2552.
Hosaka, Y., Yahara, K., Clark, A., Kitagawa, H., Hisatsune, J., Sugai, M., et al. (2022). Surveillance of multi-drug resistance phenotypes in Staphylococcus aureus in Japan and correlation with whole-genome sequence findings. J. Hosp. Infect. 123, 34–42. doi: 10.1016/j.jhin.2022.02.011
Huber, H., Koller, S., Giezendanner, N., Stephan, R., and Zweifel, C. (2010). Prevalence and characteristics of meticillin-resistant Staphylococcus aureus in humans in contact with farm animals, in livestock, and in food of animal origin, Switzerland, 2009. Eurosurveillance 15:19542.
Hussein, M. A. M., Grinholc, M., Dena, A. S. A., El-Sherbiny, I. M., and Megahed, M. (2021). Boosting the antibacterial activity of chitosan–gold nanoparticles against antibiotic–resistant bacteria by Punicagranatum L. extract. Carbohydr. Polym. 256:117498. doi: 10.1016/j.carbpol.2020.117498
Ishikawa, T., Imai, S., Nakano, T., Terai, T., Okumura, T., Hanada, N., et al. (2020). Antibacterial activity of the probiotic candidate Lactobacillus gasseri against methicillin-resistant Staphylococcus aureus. Asian Pac. J. Dent. 20, 1–8.
Javed, M. U., Ijaz, M., Fatima, Z., Anjum, A. A., Aqib, A. I., Ali, M. M., et al. (2021). Frequency and antimicrobial susceptibility of methicillin and vancomycin-resistant Staphylococcus aureus from bovine milk. Pak. Vet. J. 41, 463–468.
Jiang, J.-H., and Peleg, A. Y. (2015). Daptomycin-nonsusceptible Staphylococcus aureus: The role of combination therapy with daptomycin and gentamicin. Genes 6, 1256–1267. doi: 10.3390/genes6041256
Johnson, L. B., Jose, J., Yousif, F., Pawlak, J., and Saravolatz, L. D. (2009). Prevalence of colonization with community-associated methicillin-resistant Staphylococcus aureus among end-stage renal disease patients and healthcare workers. Infect. Control Hosp. Epidemiol. 30, 4–8. doi: 10.1086/592983
Juhász-Kaszanyitzky, É, Jánosi, S., Somogyi, P., Dán, Á, vanderGraaf van Bloois, L., Van Duijkeren, E., et al. (2007). MRSA transmission between cows and humans. Emerg. Infect. Dis. 13:630.
Juul, J. J., Mullins, C. F., Peppard, W. J., and Huang, A. M. (2016). New developments in the treatment of acute bacterial skin and skin structure infections: Considerations for the effective use of dalbavancin. Ther. Clin. Risk Manag. 12:225.
Kateete, D. P., Bwanga, F., Seni, J., Mayanja, R., Kigozi, E., Mujuni, B., et al. (2019). CA-MRSA and HA-MRSA coexist in community and hospital settings in Uganda. Antimicrob. Resist. Infect. Control 8, 1–9. doi: 10.1186/s13756-019-0551-1
Kaur, J., Kour, A., Panda, J. J., Harjai, K., and Chhibber, S. (2020). Exploring endolysin-loaded alginate-chitosan nanoparticles as future remedy for staphylococcal infections. AAPS PharmSciTech 21, 1–15. doi: 10.1208/s12249-020-01763-4
Kebriaei, R., Lev, K., Morrisette, T., Stamper, K. C., Abdul-Mutakabbir, J. C., Lehman, S. M., et al. (2020). Bacteriophage-antibiotic combination strategy: An alternative against methicillin-resistant phenotypes of Staphylococcus aureus. Antimicrob. Agents Chemother. 64, e00461–e00420. doi: 10.1128/AAC.00461-20
Khalaf, A., Kamal, M., Mokhtar, S., Mohamed, H., Salah, I., Abbas, R., et al. (2015). Antibacterial, anti-biofilm activity of some non-steroidal anti-inflammatory drugs and N-acetyl cysteine against some biofilm producing uropathogens. Am. J. Epidemiol. 3, 1–9.
Khan, R., Islam, B., Akram, M., Shakil, S., Ahmad, A., Ali, S. M., et al. (2009). Antimicrobial activity of five herbal extracts against multi drug resistant (MDR) strains of bacteria and fungus of clinical origin. Molecules 14, 586–597.
Khanna, T., Friendship, R., Dewey, C., and Weese, J. (2008). Methicillin resistant Staphylococcus aureus colonization in pigs and pig farmers. Vet. Microbiol. 128, 298–303. doi: 10.1016/j.vetmic.2007.10.006
Khawcharoenporn, T., Tice, A. D., Grandinetti, A., and Chow, D. (2010). Risk factors for community-associated methicillin-resistant Staphylococcus aureus cellulitis-and the value of recognition. Hawaii Med. J. 69:232.
Kingsley, J., Mehra, P., Lawrence, L. E., Henry, E., Duffy, E., Cammarata, S. K., et al. (2015). A randomized, double-blind, Phase 2 study to evaluate subjective and objective outcomes in patients with acute bacterial skin and skin structure infections treated with delafloxacin, linezolid or vancomycin. J. Antimicrob. Chemother. 71, 821–829.
Klevens, R. M., Morrison, M. A., Nadle, J., Petit, S., Gershman, K., Ray, S., et al. (2007). Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298, 1763–1771.
Knaack, D., Idelevich, E. A., Schleimer, N., Molinaro, S., Kriegeskorte, A., Peters, G., et al. (2019). Bactericidal activity of bacteriophage endolysin HY-133 against Staphylococcus aureus in comparison to other antibiotics as determined by minimum bactericidal concentrations and time-kill analysis. Diagn. Microbiol. Infect. Dis. 93, 362–368. doi: 10.1016/j.diagmicrobio.2018.11.005
Köck, R., Becker, K., Cookson, B., van Gemert-Pijnen, J., Harbarth, S., Kluytmans, J., et al. (2010). Methicillin-resistant Staphylococcus aureus (MRSA): Burden of disease and control challenges in Europe. Euro Surveill. 15:19688. doi: 10.2807/ese.15.41.19688-en
Koop, G., Vrieling, M., Storisteanu, D. M., Lok, L. S., Monie, T., Van Wigcheren, G., et al. (2017). Identification of LukPQ, a novel, equid-adapted leukocidin of Staphylococcus aureus. Sci. Rep. 7, 1–10. doi: 10.1038/srep40660
La Vecchia, A., Ippolito, G., Taccani, V., Gatti, E., Bono, P., Bettocchi, S., et al. (2022). Epidemiology and antimicrobial susceptibility of Staphylococcus aureus in children in a tertiary care pediatric hospital in Milan, Italy, 2017–2021. Ital. J. Pediatr. 48, 1–8.
Lakshmi, A. V., Harasreeramulu, S., and Raju, D. S. (2013). Assessment of antibacterial potential of selected medicinal plants and their interactions with antibiotics on MRSA in the health care workers of Visakhapatnam hospitals. J. Pharm. Res. 6, 589–592.
Layus, B. I., Gerez, C. L., and Rodriguez, A. V. (2020). Antibacterial activity of Lactobacillus plantarum CRL 759 against methicillin-resistant Staphylococcus aureus and Pseudomonas aeruginosa. Arab. J. Sci. Eng. 45, 4503–4510.
Lee, A. S., De Lencastre, H., Garau, J., Kluytmans, J., Malhotra-Kumar, S., Peschel, A., et al. (2018). Methicillin-resistant Staphylococcus aureus. Nat. Rev. Dis. Primers 4, 1–23.
Lee, B. Y., Singh, A., David, M. Z., Bartsch, S. M., Slayton, R. B., Huang, S. S., et al. (2013). The economic burden of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA). Clin. Microbiol. Infect. 19, 528–536. doi: 10.1111/j.1469-0691.2012.03914.x
Lee, J. H. (2003). Methicillin (oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl. Environ. Microbiol. 69, 6489–6494. doi: 10.1128/AEM.69.11.6489-6494.2003
Lee, S. H., Wang, H., Labroli, M., Koseoglu, S., Zuck, P., Mayhood, T., et al. (2016). TarO-specific inhibitors of wall teichoic acid biosynthesis restore β-lactam efficacy against methicillin-resistant staphylococci. Sci. Transl. Med. 8:329ra32. doi: 10.1126/scitranslmed.aad7364
Lehar, S. M., Pillow, T., Xu, M., Staben, L., Kajihara, K. K., Vandlen, R., et al. (2015). Novel antibody–antibiotic conjugate eliminates intracellular S. aureus. Nature 527, 323–328. doi: 10.1038/nature16057
Lemaire, S., Tulkens, P. M., and Van Bambeke, F. (2010). Cellular pharmacokinetics of the novel biaryloxazolidinone radezolid in phagocytic cells: Studies with macrophages and polymorphonuclear neutrophils. Antimicrob. Agents Chemother. 54, 2540–2548. doi: 10.1128/AAC.01723-09
Lemaire, S., Tulkens, P. M., and Van Bambeke, F. (2011). Contrasting effects of acidic pH on the extracellular and intracellular activities of the anti-gram-positive fluoroquinolones moxifloxacin and delafloxacin against Staphylococcus aureus. Antimicrob. Agents Chemother. 55, 649–658. doi: 10.1128/AAC.01201-10
Li, X., Hu, T., Wei, J., He, Y., Abdalla, A. E., Wang, G., et al. (2021). Characterization of a novel bacteriophage Henu2 and evaluation of the synergistic antibacterial activity of phage-antibiotics. Antibiotics 10:174. doi: 10.3390/antibiotics10020174
Lim, S.-K., Nam, H.-M., Jang, G.-C., Lee, H.-S., Jung, S.-C., and Kim, T.-S. (2013). Transmission and persistence of methicillin-resistant Staphylococcus aureus in milk, environment, and workers in dairy cattle farms. Foodborne Pathog. Dis. 10, 731–736. doi: 10.1089/fpd.2012.1436
Lin, C.-F., Lin, M.-Y., Lin, C.-N., Chiou, M.-T., Chen, J.-W., Yang, K.-C., et al. (2020). Potential probiotic of Lactobacillus strains isolated from the intestinal tracts of pigs and feces of dogs with antibacterial activity against multidrug-resistant pathogenic bacteria. Arch. Microbiol. 202, 1849–1860. doi: 10.1007/s00203-020-01908-w
Lindsay, J. A. (2013). Hospital-associated MRSA and antibiotic resistance–what have we learned from genomics? Int. J. Med. Microbiol. 303, 318–323. doi: 10.1016/j.ijmm.2013.02.005
Liu, C., Bayer, A., Cosgrove, S. E., Daum, R. S., Fridkin, S. K., Gorwitz, R. J., et al. (2011). Clinical practice guidelines by the infectious diseases society of America for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children. Clin. Infect. Dis. 52, e18–e55.
Liu, S., Hon, K., Bouras, G. S., Psaltis, A. J., Shearwin, K., Wormald, P.-J., et al. (2022). APTC-C-SA01: A novel bacteriophage cocktail targeting Staphylococcus aureus and MRSA biofilms. Int. J. Mol. Sci. 23:6116. doi: 10.3390/ijms23116116
Lodhi, F. L., Saleem, M. I., Aqib, A. I., Rashid, I., Qureshi, Z. I., Anwar, M. A., et al. (2021). Bringing resistance modulation to epidemic methicillin resistant S. aureus of dairy through antibiotics coupled metallic oxide nanoparticles. Microb. Pathog. 159:105138. doi: 10.1016/j.micpath.2021.105138
Loeffler, A., McCarthy, A., Lloyd, D. H., Musilová, E., Pfeiffer, D. U., and Lindsay, J. A. (2013). Whole-genome comparison of meticillin-resistant Staphylococcus aureus CC22 SCCmecIV from people and their in-contact pets. Vet. Dermatol. 24, 538–e128. doi: 10.1111/vde.12062
Lubowska, N., Grygorcewicz, B. Kosznik-Kwasnicka, K. Zauszkiewicz-Pawlak, A., Wegrzyn, A., Dolegowska, B., et al. (2019). Characterization of the three new kayviruses and their lytic activity against multidrug-resistant Staphylococcus aureus. Microorganisms 7:471.
Lowder, B. V., Guinane, C. M., Ben Zakour, N. L., Weinert, L. A., Conway-Morris, A., Cartwright, R. A., et al. (2009). Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 106, 19545–19550. doi: 10.1073/pnas.0909285106
Majeed, S., Danish, M., Mohamad Ibrahim, M. N., Sekeri, S. H., Ansari, M. T., Nanda, A., et al. (2021). Bacteria mediated synthesis of iron oxide nanoparticles and their antibacterial, antioxidant, cytocompatibility properties. J. Clust. Sci. 32, 1083–1094.
Malik, S., Coombs, G. W., O’brien, F. G., Peng, H., and Barton, M. D. (2006). Molecular typing of methicillin-resistant staphylococci isolated from cats and dogs. J. Antimicrob. Chemother. 58, 428–431.
Manders, S. M. (1998). Toxin-mediated streptococcal and staphylococcal disease. J. Am. Acad. Dermatol. 39, 383–398.
Manian, F. A. (2003). Asymptomatic nasal carriage of mupirocin-resistant, methicillin-resistant Staphylococcus aureus (MRSA) in a pet dog associated with MRSA infection in household contacts. Clin. Infect. Dis. 36, e26–e28. doi: 10.1086/344772
Manilal, A., Sabu, K. R., Shewangizaw, M., Aklilu, A., Seid, M., Merdekios, B., et al. (2020). In vitro antibacterial activity of medicinal plants against biofilm-forming methicillin-resistant Staphylococcus aureus: Efficacy of Moringa stenopetala and Rosmarinus officinalis extracts. Heliyon 6:e03303. doi: 10.1016/j.heliyon.2020.e03303
Masterton, R., Cornaglia, G., Courvalin, P., Lode, H. M., Rello, J., and Torres, A. (2015). The clinical positioning of telavancin in Europe. Int. J. Antimicrob. Agents 45, 213–220.
McDougal, L. K., Steward, C. D., Killgore, G. E., Chaitram, J. M., McAllister, S. K., and Tenover, F. C. (2003). Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: Establishing a national database. J. Clin. Microbiol. 41, 5113–5120.
McFarland, L. V., Karakan, T., and Karatas, A. (2021). Strain-specific and outcome-specific efficacy of probiotics for the treatment of irritable bowel syndrome: A systematic review and meta-analysis. EClinicalMedicine 41:101154. doi: 10.1016/j.eclinm.2021.101154
Menberu, M. A., Liu, S., Cooksley, C., Hayes, A. J., Psaltis, A. J., Wormald, P.-J., et al. (2021). Corynebacterium accolens has antimicrobial activity against Staphylococcus aureus and methicillin-resistant S. aureus pathogens isolated from the sinonasal niche of chronic rhinosinusitis patients. Pathogens 10:207. doi: 10.3390/pathogens10020207
Mendes, C. R., Dilarri, G., Forsan, C. F., Sapata, V. M. R., Lopes, P. R. M., de Moraes, P. B., et al. (2022). Antibacterial action and target mechanisms of zinc oxide nanoparticles against bacterial pathogens. Sci. Rep. 12:2658.
Menzies, B. E. (2003). The role of fibronectin binding proteins in the pathogenesis of Staphylococcus aureus infections. Curr Opin. Infect. Dis. 16, 225–229.
Mermel, L. A., Allon, M., Bouza, E., Craven, D. E., Flynn, P., O’grady, N. P., et al. (2009). Clinical practice guidelines for the diagnosis and management of intravascular catheter-related infection: 2009 update by the infectious diseases society of America. Clin. Infect. Dis. 49, 1–45. doi: 10.1086/599376
Monaco, M., Sanchini, A., Grundmann, H., and Pantosti, A. (2010). Vancomycin-heteroresistant phenotype in invasive methicillin-resistant Staphylococcus aureus isolates belonging to spa type 041. Eur. J. Clin. Microbiol. Infect. Dis. 29, 771–777. doi: 10.1007/s10096-010-0922-2
Mostafa, G., Badr, M. F.-A., Zeid, M. S., and Eldegla, H. (2022). Nasal Carriage of community acquired and inducible dormant methicillin resistant Staphylococcus aureus among healthcare workers of Mansoura university children’s hospital. Egypt. J. Med. Microbiol. 31, 75–81.
Mulders, M., Haenen, A., Geenen, P., Vesseur, P., Poldervaart, E., Bosch, T., et al. (2010). Prevalence of livestock-associated MRSA in broiler flocks and risk factors for slaughterhouse personnel in The Netherlands. Epidemiol. Infect. 138, 743–755. doi: 10.1017/S0950268810000075
Mulyaningsih, S., Sporer, F., Reichling, J., and Wink, M. (2011). Antibacterial activity of essential oils from Eucalyptus and of selected components against multidrug-resistant bacterial pathogens. Pharm. Biol. 49, 893–899. doi: 10.3109/13880209.2011.553625
Murray, S., Pascoe, B., Meric, G., Mageiros, L., Yahara, K., Hitchings, M. D., et al. (2017). Recombination-mediated host adaptation by avian Staphylococcus aureus. Genome Biol. Evol. 9, 830–842. doi: 10.1093/gbe/evx037
Muzammil, I., Saleem, M. I, Aqib, A. I, Ashar, A., Mahfooz, S. A., Rahman, S., et al. (2021). Emergence of pathogenic strains of Staphylococcus aureus in goat milk and their comparative response to antibiotics. Pak. J. Zool. 53, 1659–1667.
Naimi, T. S., LeDell, K. H., Como-Sabetti, K., Borchardt, S. M., Boxrud, D. J., Etienne, J., et al. (2003). Comparison of community-and health care–associated methicillin-resistant Staphylococcus aureus infection. JAMA 290, 2976–2984.
Nandhini, P., Kumar, P., Mickymaray, S., Alothaim, A. S., Somasundaram, J., and Rajan, M. (2022). Recent developments in methicillin-resistant Staphylococcus aureus (MRSA) treatment: A review. Antibiotics 11:606. doi: 10.3390/antibiotics11050606
Nasreen, S., Andleeb, S., Ali, S., Imdad, K., Awan, U. A., Raja, S. A., et al. (2022). Screening of antibacterial efficacy of chitosan encapsulated probiotics (Lactococcus lactis and Lactobacillus curvattus) against clinical bacterial pathogens. J. Oleo Sci. 71, 1363–1374. doi: 10.5650/jos.ess22052
Naveed, R., Hussain, I., Tawab, A., Tariq, M., Rahman, M., Hameed, S., et al. (2013). Antimicrobial activity of the bioactive components of essential oils from Pakistani spices against Salmonella and other multi-drug resistant bacteria. BMC Complement. Altern. Med. 13:265. doi: 10.1186/1472-6882-13-265
Nemati, M., Hermans, K., Lipinska, U., Denis, O., Deplano, A., Struelens, M., et al. (2008). Antimicrobial resistance of old and recent Staphylococcus aureus isolates from poultry: First detection of livestock-associated methicillin-resistant strain ST398. Antimicrob. Agents Chemother. 52, 3817–3819. doi: 10.1128/AAC.00613-08
Nguyen, N.-Y. T., Grelling, N., Wetteland, C. L., Rosario, R., and Liu, H. (2018). Antimicrobial activities and mechanisms of magnesium oxide nanoparticles (nMgO) against pathogenic bacteria, yeasts, and biofilms. Sci. Rep. 8, 1–23. doi: 10.1038/s41598-018-34567-5
Nickerson, E. K., Hongsuwan, M., Limmathurotsakul, D., Wuthiekanun, V., Shah, K. R., Srisomang, P., et al. (2009). Staphylococcus aureus bacteraemia in a tropical setting: Patient outcome and impact of antibiotic resistance. PLoS One 4:e4308. doi: 10.1371/journal.pone.0004308
Noskin, G. A., Rubin, R. J., Schentag, J. J., Kluytmans, J., Hedblom, E. C., Smulders, M., et al. (2005). The burden of Staphylococcus aureus infections on hospitals in the United States: An analysis of the 2000 and 2001 nationwide inpatient sample database. Arch. Intern. Med. 165, 1756–1761. doi: 10.1001/archinte.165.15.1756
O’Mahony, R., Abbott, Y., Leonard, F., Markey, B., Quinn, P., Pollock, P., et al. (2005). Methicillin-resistant Staphylococcus aureus (MRSA) isolated from animals and veterinary personnel in Ireland. Vet. Microbiol. 109, 285–296. doi: 10.1016/j.vetmic.2005.06.003
Ogura, K., Kaji, D., Sasaki, M., Otsuka, Y., Takemoto, N., Miyoshi-Akiyama, T., et al. (2022). Predominance of ST8 and CC1/spa-t1784 methicillin-resistant Staphylococcus aureus isolates in Japan and their genomic characteristics. J. Glob. Antimicrob. Resist. 28, 195–202. doi: 10.1016/j.jgar.2022.01.011
Okuma, K., Iwakawa, K., Turnidge, J. D., Grubb, W. B., Bell, J. M., O’Brien, F. G., et al. (2002). Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40, 4289–4294.
Okwu, M. U., Olley, M., Akpoka, A. O., and Izevbuwa, O. E. (2019). Methicillin-resistant Staphylococcus aureus (MRSA) and anti-MRSA activities of extracts of some medicinal plants: A brief review. AIMS Microbiol. 5:117. doi: 10.3934/microbiol.2019.2.117
Ong, C., Lirk, P., Tan, C., and Seymour, R. (2007). An evidence-based update on nonsteroidal anti-inflammatory drugs. Clin. Med. Res. 5, 19–34.
Pantosti, A. (2012). Methicillin-resistant Staphylococcus aureus associated with animals and its relevance to human health. Front. Microbiol. 3:127. doi: 10.3389/fmicb.2012.00127
Peacock, S. J., Moore, C. E., Justice, A., Kantzanou, M., Story, L., Mackie, K., et al. (2002). Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect. Immun. 70, 4987–4996. doi: 10.1128/IAI.70.9.4987-4996.2002
Peeters, L. E., Argudín, M. A., Azadikhah, S., and Butaye, P. (2015). Antimicrobial resistance and population structure of Staphylococcus aureus recovered from pigs farms. Vet. Microbiol. 180, 151–156. doi: 10.1016/j.vetmic.2015.08.018
Peleg, A. Y., Miyakis, S., Ward, D. V., Earl, A. M., Rubio, A., Cameron, D. R., et al. (2012). Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. PLoS One 7:e28316. doi: 10.1371/journal.pone.0028316
Persoons, D., Van Hoorebeke, S., Hermans, K., Butaye, P., De Kruif, A., Haesebrouck, F., et al. (2009). Methicillin-resistant Staphylococcus aureus in poultry. Emerg. Infect. Dis. 15:452.
Peton, V., Bouchard, D. S., Almeida, S., Rault, L., Falentin, H., Jardin, J., et al. (2014). Fine-tuned characterization of Staphylococcus aureus Newbould 305, a strain associated with mild and chronic mastitis in bovines. Vet. Res. 45, 1–15. doi: 10.1186/s13567-014-0106-7
Pfaller, M. A., Rhomberg, P. R., Huband, M. D., and Flamm, R. K. (2017). Activities of omadacycline and comparator agents against Staphylococcus aureus isolates from a surveillance program conducted in North America and Europe. Antimicrob. Agents Chemother. 61, e02411–e02416. doi: 10.1128/AAC.02411-16
Pickering, A. C., Haag, A. F., Penades, J. R., and Fitzgerald, J. R. (2022). “Staphylococcus,” in Pathogenesis of bacterial infections in animals, eds J. F. Prescott, A. N. Rycroft, J. D. Boyce, J. I. MacInnes, F. Van Immerseel, and J. A. Vázquez-Boland (Hoboken, NJ: wiley), 543–564.
Plaza-Diaz, J., Ruiz-Ojeda, F. J., Gil-Campos, M., and Gil, A. (2019). Mechanisms of action of probiotics. Adv. Nutr. 10(Suppl. 1) S49–S66.
Pozzi, C., Olaniyi, R., Liljeroos, L., and Galgani, I. (2017). Vaccines for Staphylococcus aureus and target populations. Curr. Top. Microbiol. Immunol. 409, 491–528.
Pu, W., Su, Y., Li, J., Li, C., Yang, Z., Deng, H., et al. (2014). High incidence of oxacillin-susceptible mecA-positive Staphylococcus aureus (OS-MRSA) associated with bovine mastitis in China. PLoS One 9:e88134. doi: 10.1371/journal.pone.0088134
Purrello, S., Garau, J., Giamarellos, E., Mazzei, T., Pea, F., Soriano, A., et al. (2016). Methicillin-resistant Staphylococcus aureus infections: A review of the currently available treatment options. J. Glob. Antimicrob. Resist. 7, 178–186.
Radetsky, M. (1996). The discovery of penicillin. Pediatr. Infect. Dis. J. 15, 811–818. doi: 10.1097/00006454-199609000-00015
Rand, K. H., and Houck, H. J. (2004). Synergy of daptomycin with oxacillin and other beta-lactams against methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 48, 2871–2875. doi: 10.1128/aac.48.8.2871-2875.2004
Rasool, M. H., Yousaf, R., Siddique, A. B., Saqalein, M., and Khurshid, M. (2016). Isolation, characterization, and antibacterial activity of bacteriophages against methicillin-resistant Staphylococcus aureus in Pakistan. Jundishapur J. Microbiol. 9:e36135. doi: 10.5812/jjm.36135
Rehman, T., Aslam, R., Aqib, A. I, Mohsin, M., Manzoor, A., Shoaib, M., et al. (2020). Phylogeny of hospital acquired MRSA, and its comparative phenotypic clinico-epidemiology with vancomycin resistant S. aureus (VRSA). Microb. Pathog. 149:104537. doi: 10.1016/j.micpath.2020.104537
Resch, G., François, P., Morisset, D., Stojanov, M., Bonetti, E. J., Schrenzel, J., et al. (2013). Human-to-bovine jump of Staphylococcus aureus CC8 is associated with the loss of a β-hemolysin converting prophage and the acquisition of a new staphylococcal cassette chromosome. PLoS One 8:e58187. doi: 10.1371/journal.pone.0058187
Rheima, A. M., Al Marjani, M. F., Aboksour, M. F., and Mohammed, S. H. (2021). Evaluation of anti-biofilm formation effect of nickel oxide nanoparticles (NiO-NPs) against methicillin-resistant Staphylococcus aureus (MRSA). Int. J. Nanosci. Nanotechnol. 17, 221–230.
Richardson, E. J., Bacigalupe, R., Harrison, E. M., Weinert, L. A., Lycett, S., Vrieling, M., et al. (2018). Gene exchange drives the ecological success of a multi-host bacterial pathogen. Nat. Ecol. Evol. 2, 1468–1478. doi: 10.1038/s41559-018-0617-0
Richter, A., Sting, R., Popp, C., Rau, J., Tenhagen, B.-A., Guerra, B., et al. (2012). Prevalence of types of methicillin-resistant Staphylococcus aureus in turkey flocks and personnel attending the animals. Epidemiol. Infect. 140, 2223–2232. doi: 10.1017/S095026881200009X
Rosenberg Goldstein, R. E., Micallef, S. A., Gibbs, S. G., Davis, J. A., He, X., George, A., et al. (2012). Methicillin-resistant Staphylococcus aureus (MRSA) detected at four U.S. wastewater treatment plants. Environ. Health Perspect. 120, 1551–1558. doi: 10.1289/ehp.1205436
Sabio, L., González, A., Ramírez-Rodríguez, G. B., Gutiérrez-Fernández, J., Bañuelo, O., Olivares, M., et al. (2021). Probiotic cellulose: Antibiotic-free biomaterials with enhanced antibacterial activity. Acta Biomater. 124, 244–253.
Salem, W., Sayed, W., Hardy, M., and Hassan, N. (2014). Antibacterial activity of Calotropis procera and Ficus sycomorus extracts on some pathogenic microorganisms. Afr. J. Biotechnol. 13, 3271–3280.
Sandrock, C. E., and Shorr, A. F. (2015). The role of telavancin in hospital-acquired pneumonia and ventilator-associated pneumonia. Clin. Infect. Dis. 61(Suppl. 2) S79–S86.
Sarwar, I., Ashar, A., Mahfooz, A., Aqib, A. I., Saleem, M. I., Butt, A. A., et al. (2021). Evaluation of antibacterial potential of raw turmeric, nano-turmeric, and NSAIDs against multiple drug resistant Staphylococcus aureus and E. coli isolated from animal wounds. Pak. Vet. J. 41, 209–214.
Sassmannshausen, R., Deurenberg, R. H., Köck, R., Hendrix, R., Jurke, A., Rossen, J. W., et al. (2016). MRSA prevalence and associated risk factors among health-care workers in non-outbreak situations in the Dutch-German EUREGIO. Front. Microbiol. 7:1273. doi: 10.3389/fmicb.2016.01273
Sausset, R., Petit, M., Gaboriau-Routhiau, V., and De Paepe, M. (2020). New insights into intestinal phages. Mucosal Immunol. 13, 205–215.
Schijffelen, M. J., Boel, C., van Strijp, J. A., and Fluit, A. C. (2010). Whole genome analysis of a livestock-associated methicillin-resistant Staphylococcus aureus ST398 isolate from a case of human endocarditis. BMC Genomics 11:376. doi: 10.1186/1471-2164-11-376
Scott, G., Thomson, R., Malone-Lee, J., and Ridgway, G. (1988). Cross-infection between animals and man: Possible feline transmission of Staphylococcus aureus infection in humans? J. Hosp. Infect. 12, 29–34. doi: 10.1016/0195-6701(88)90119-3
Seguin, J. C., Walker, R. D., Caron, J. P., Kloos, W. E., George, C. G., Hollis, R. J., et al. (1999). Methicillin-resistant Staphylococcus aureus outbreak in a veterinary teaching hospital: Potential human-to-animal transmission. J. Clin. Microbiol. 37, 1459–1463. doi: 10.1128/JCM.37.5.1459-1463.1999
Seibert, G., Isert, D., Klesel, N., Limbert, M., Markus, A., and Schrinner, E. (1992). The in-vitro antibacterial activity of a combination of cefpirome or cefoperazone with vancomycin against enterococci and Staphylococcus aureus. J. Antimicrob. Chemother. 29(Suppl. A) 25–30.
Shameli, K., Ahmad, M. B., Yunus, W. M. Z. W., Rustaiyan, A., Ibrahim, N. A., Zargar, M., et al. (2010). Green synthesis of silver/montmorillonite/chitosan bionanocomposites using the UV irradiation method and evaluation of antibacterial activity. Int. J. Nanomed. 5, 875–887. doi: 10.2147/IJN.S13632
Sharaf, M. H., El-Sherbiny, G. M., Moghannem, S. A., Abdelmonem, M., Elsehemy, I. A., Metwaly, A. M., et al. (2021). New combination approaches to combat methicillin-resistant Staphylococcus aureus (MRSA). Sci. Rep. 11:4240. doi: 10.1038/s41598-021-82550-4
Shi, L., Sheng, J., Wang, M., Luo, H., Zhu, J., Zhang, B., et al. (2019). Combination therapy of TGF-β blockade and commensal-derived probiotics provides enhanced antitumor immune response and tumor suppression. Theranostics 9:4115. doi: 10.7150/thno.35131
Shin, D., Chang, S. Y., Bogere, P., Won, K., Choi, J. Y., Choi, Y. J., et al. (2019). Beneficial roles of probiotics on the modulation of gut microbiota and immune response in pigs. PLoS One 14:e0220843. doi: 10.1371/journal.pone.0220843
Shinefield, H., Black, S., Fattom, A., Horwith, G., Rasgon, S., Ordonez, J., et al. (2002). Use of a Staphylococcus aureus conjugate vaccine in patients receiving hemodialysis. N. Engl. J. Med. 346, 491–496.
Shoaib, M., Rahman, S. U., Aqib, A. I., Ashfaq, K., Naveed, A., Kulyar, M. F. A., et al. (2020). Diversified epidemiological pattern and antibiogram of mecA gene in Staphylococcus aureus isolates of pets, pet owners and environment. Pak. Vet. J. 40, 331–336. doi: 10.29261/pakvetj/2020.039
Siddiqui, T., Muhammad, I. N., Khan, M. N., Naz, S., Bashir, L., Sarosh, N., et al. (2017). MRSA: Prevalence and susceptibility pattern in health care setups of Karachi. Pak. J. Pharm. Sci. 30, 2417–2421.
Silva, L. V., Araújo, M. T., Santos, K. R., and Nunes, A. P. F. (2011). Evaluation of the synergistic potential of vancomycin combined with other antimicrobial agents against methicillin-resistant Staphylococcus aureus and coagulase-negative Staphylococcus spp strains. Mem. Inst. Oswaldo Cruz 106, 44–50. doi: 10.1590/s0074-02762011000100007
Silva, V., Vieira-Pinto, M., Saraiva, C., Manageiro, V., Reis, L., Ferreira, E., et al. (2021). Prevalence and characteristics of multidrug-resistant livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) CC398 isolated from quails (Coturnix Coturnix japonica) slaughtered for human consumption. Animals 11:2038. doi: 10.3390/ani11072038
Simor, A. E., Pelude, L., Golding, G., Fernandes, R., Bryce, E., Frenette, C., et al. (2016). Determinants of outcome in hospitalized patients with methicillin-resistant Staphylococcus aureus bloodstream infection: Results from national surveillance in Canada, 2008-2012. Infect. Control Hosp. Epidemiol. 37, 390–397. doi: 10.1017/ice.2015.323
Singh, S., Gill, A. A., Nlooto, M., and Karpoormath, R. (2019). Prostate cancer biomarkers detection using nanoparticles based electrochemical biosensors. Biosens. Bioelectron. 137, 213–221.
Smith, T. C., Male, M. J., Harper, A. L., Kroeger, J. S., Tinkler, G. P., Moritz, E. D., et al. (2009). Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 is present in midwestern US swine and swine workers. PLos One 4:e4258. doi: 10.1371/journal.pone.0004258
Spirescu, V. A., Chircov, C., Grumezescu, A. M., Vasile, B. Ş, and Andronescu, E. (2021). Inorganic nanoparticles and composite films for antimicrobial therapies. Int. J. Mol. Sci. 22:4595.
Spohr, M., Rau, J., Friedrich, A., Klittich, G., Fetsch, A., Guerra, B., et al. (2011). Methicillin-resistant Staphylococcus aureus (MRSA) in three dairy herds in southwest Germany. Zoonoses Public Health 58, 252–261. doi: 10.1111/j.1863-2378.2010.01344.x
Stankovic, C., and Mahajan, P. V. (2006). Healthy children with invasive community-acquired methicillin-resistant Staphylococcus aureus infections. Pediatr. Emerg. Care 22, 361–363. doi: 10.1097/01.pec.0000215652.27137.c7
Streicher, L. M. (2021). Exploring the future of infectious disease treatment in a post-antibiotic era: A comparative review of alternative therapeutics. J. Glob. Antimicrob. Resist. 24, 285–295. doi: 10.1016/j.jgar.2020.12.025
Sudan, S., Flick, R., Nong, L., and Li, J. (2021). Potential probiotic Bacillus subtilis isolated from a novel niche exhibits broad range antibacterial activity and causes virulence and metabolic dysregulation in enterotoxic E. coli. Microorganisms 9:1483. doi: 10.3390/microorganisms9071483
Tabandeh, M., Kaboosi, H., Taghizadeh Armaki, M., Pournajaf, A., and Peyravii Ghadikolaii, F. (2022). New update on molecular diversity of clinical Staphylococcus aureus isolates in Iran: Antimicrobial resistance, adhesion and virulence factors, biofilm formation and SCCmec typing. Mol. Biol. Rep. 49, 3099–3111. doi: 10.1007/s11033-022-07140-7
Tagliabue, A., and Rappuoli, R. (2018). Changing priorities in vaccinology: Antibiotic resistance moving to the top. Front. Immunol. 9:1068. doi: 10.3389/fimmu.2018.01068
Takahashi, N., Nishida, H., Kato, H., Sakata, Y., and Uchiyama, T. (1998). Exanthematous disease induced by toxic shock syndrome toxin 1 in the early neonatal period. Lancet 351, 1614–1619. doi: 10.1016/S0140-6736(97)11125-4
Takahashi, Y., and Igarashi, M. (2018). Destination of aminoglycoside antibiotics in the ‘post-antibiotic era’. J. Antibiot. 71:4. doi: 10.1038/ja.2017.117
Takizawa, Y., Taneike, I., Nakagawa, S., Oishi, T., Nitahara, Y., Iwakura, N., et al. (2005). A panton-valentine leucocidin (PVL)-positive community-acquired methicillin-resistant Staphylococcus aureus (MRSA) strain, another such strain carrying a multiple-drug resistance plasmid, and other more-typical PVL-negative MRSA strains found in Japan. J. Clin. Microbiol. 43, 3356–3363. doi: 10.1128/JCM.43.7.3356-3363.2005
Tavares, A., Miragaia, M., Rolo, J., Coelho, C., and De Lencastre, H. (2013). High prevalence of hospital-associated methicillin-resistant Staphylococcus aureus in the community in Portugal: Evidence for the blurring of community–hospital boundaries. Eur. J. Clin. Microbiol. Infect. Dis. 32, 1269–1283. doi: 10.1007/s10096-013-1872-2
Thati, V., Shivannavar, C. T., and Gaddad, S. M. (2011). Vancomycin resistance among methicillin resistant Staphylococcus aureus isolates from intensive care units of tertiary care hospitals in Hyderabad. Indian J. Med. Res. 134:704. doi: 10.4103/0971-5916.91001
Tkhilaishvili, T., Lombardi, L., Klatt, A.-B., Trampuz, A., and Di Luca, M. (2018). Bacteriophage Sb-1 enhances antibiotic activity against biofilm, degrades exopolysaccharide matrix and targets persisters of Staphylococcus aureus. Int. J. Antimicrob. Agents 52, 842–853. doi: 10.1016/j.ijantimicag.2018.09.006
Tkhilaishvili, T., Wang, L., Tavanti, A., Trampuz, A., and Di Luca, M. (2020). Antibacterial efficacy of two commercially available bacteriophage formulations, staphylococcal bacteriophage and PYO bacteriophage, against methicillin-resistant Staphylococcus aureus: Prevention and eradication of biofilm formation and control of a systemic infection of Galleria mellonella larvae. Front. Microbiol. 11:110. doi: 10.3389/fmicb.2020.00110
Tsuchiya, H., Sato, M., Miyazaki, T., Fujiwara, S., Tanigaki, S., Ohyama, M., et al. (1996). Comparative study on the antibacterial activity of phytochemical flavanones against methicillin-resistant Staphylococcus aureus. J. Ethnopharmacol. 50, 27–34. doi: 10.1016/0378-8741(96)85514-0
Tuffs, S. W., Haeryfar, S., and McCormick, J. K. (2018). Manipulation of innate and adaptive immunity by staphylococcal superantigens. Pathogens 7:53.
Turner, M. S., Waldherr, F., Loessner, M. J., and Giffard, P. M. (2007). Antimicrobial activity of lysostaphin and a Listeria monocytogenes bacteriophage endolysin produced and secreted by lactic acid bacteria. Syst. Appl. Microbiol. 30, 58–67. doi: 10.1016/j.syapm.2006.01.013
Ugwu, C. C., Ezeonu, I. M., Ike, A. C., and Nwaokorie, F. O. (2022). Clonality of methicillin-resistant Staphylococcus aureus (MRSA) in hospital and community samples in Nsukka, Nigeria. J. Sci. Technol. 7, 49–61.
Ullah, K., Khan, S. A., Mannan, A., Khan, R., Murtaza, G., and Yameen, M. A. (2020). Enhancing the antibacterial activity of erythromycin with titanium dioxide nanoparticles against MRSA. Curr. Pharm. Biotechnol. 21, 948–954. doi: 10.2174/1389201021666200128124142
Umamageswari, S., Manipriya, B., and Kalyani, M. (2018). Evaluation of antibacterial activity of zinc oxide nanoparticles against biofilm producing methicillin resistant Staphylococcus aureus (MRSA). Res. J. Pharm. Technol. 11, 1884–1888.
Valle, D. L. Jr., Cabrera, E. C., Puzon, J. J. M., and Rivera, W. L. (2016). Antimicrobial activities of methanol, ethanol and supercritical CO2 extracts of Philippine Piper betle L. On clinical isolates of Gram positive and Gram negative bacteria with transferable multiple drug resistance. PLoS One 11:e0146349. doi: 10.1371/journal.pone.0146349
Van Bambeke, F. (2014). Renaissance of antibiotics against difficult infections: Focus on oritavancin and new ketolides and quinolones. Ann. Med. 46, 512–529. doi: 10.3109/07853890.2014.935470
Van Bambeke, F. (2015). Lipoglycopeptide antibacterial agents in gram-positive infections: A comparative review. Drugs 75, 2073–2095.
Van Duijkeren, E., Wolfhagen, M., Heck, M., and Wannet, W. (2005). Transmission of a panton-valentine leucocidin-positive, methicillin-resistant Staphylococcus aureus strain between humans and a dog. J. Clin. Microbiol. 43, 6209–6211. doi: 10.1128/JCM.43.12.6209-6211.2005
Van Hal, S. J., and Fowler, V. G. Jr. (2013). Is it time to replace vancomycin in the treatment of methicillin-resistant Staphylococcus aureus infections? Clin. Infect. Dis. 56, 1779–1788. doi: 10.1093/cid/cit178
Vengust, M., Anderson, M., Rousseau, J., and Weese, J. (2006). Methicillin-resistant staphylococcal colonization in clinically normal dogs and horses in the community. Lett. Appl. Microbiol. 43, 602–606. doi: 10.1111/j.1472-765X.2006.02018.x
Viana, D., Blanco, J., Tormo-Más, M. Á, Selva, L., Guinane, C. M., Baselga, R., et al. (2010). Adaptation of Staphylococcus aureus to ruminant and equine hosts involves SaPI-carried variants of von Willebrand factor-binding protein. Mol. Microbiol. 77, 1583–1594. doi: 10.1111/j.1365-2958.2010.07312.x
Villarroel, J., Larsen, M. V., Kilstrup, M., and Nielsen, M. (2017). Metagenomic analysis of therapeutic PYO phage cocktails from 1997 to 2014. Viruses 9:328. doi: 10.3390/v9110328
Vimala, D. S., Murugesan, R., Francesco, M., Antara, B., Xiao, F. S., and Surajit, P. (2019). Comparative study on anti-proliferative potentials of zinc oxide and aluminium oxide nanoparticles in colon cancer cells. Acta Biomed. 90:241. doi: 10.23750/abm.v90i2.6939
Voss, A., Loeffen, F., Bakker, J., Klaassen, C., and Wulf, M. (2005). Methicillin-resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. 11:1965.
Vrieling, M., Koymans, K., Heesterbeek, D., Aerts, P., Rutten, V., De Haas, C., et al. (2015). Bovine Staphylococcus aureus secretes the leukocidin LukMF’ to kill migrating neutrophils through CCR1. mBio 6:e00335. doi: 10.1128/mBio.00335-15
Wagenaar, J. A., Yue, H., Pritchard, J., Broekhuizen-Stins, M., Huijsdens, X., Mevius, D. J., et al. (2009). Unexpected sequence types in livestock associated methicillin-resistant Staphylococcus aureus (MRSA): MRSA ST9 and a single locus variant of ST9 in pig farming in China. Vet. Microbiol. 139, 405–409. doi: 10.1016/j.vetmic.2009.06.014
Wang, B., Xu, Y., Zhao, H., Wang, X., Rao, L., Guo, Y., et al. (2022). Methicillin-resistant Staphylococcus aureus in China: A multicenter longitudinal study and whole-genome sequencing. Emerg. Microbes Infect. 11, 532–542.
Wang, P., Li, Z.-S., Liu, F., Ren, X., Lu, N.-H., Fan, Z.-N., et al. (2009). Risk factors for ERCP-related complications: A prospective multicenter study. Am. J. Gastroenterol. 104, 31–40.
Wang, R., Braughton, K. R., Kretschmer, D., Bach, T.-H. L., Queck, S. Y., Li, M., et al. (2007). Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 13:1510. doi: 10.1038/nm1656
Wang, W. B., and Clapper, J. C. (2022). Antibacterial activity of electrospun polyacrylonitrile copper nanoparticle nanofibers on antibiotic resistant pathogens and methicillin resistant Staphylococcus aureus (MRSA). Nanomaterials 12:2139. doi: 10.3390/nano12132139
Wardenburg, J. B., Bae, T., Otto, M., DeLeo, F. R., and Schneewind, O. (2007). Poring over pores: α-hemolysin and panton-valentine leukocidin in Staphylococcus aureus pneumonia. Nat. Med. 13:1405. doi: 10.1038/nm1207-1405
Webster, K., and Mitchell, G. (1989). Experimental production of tick pyaemia. Vet. Parasitol. 34, 129–133.
Weese, J. S., Rousseau, J., Traub-Dargatz, J. L., Willey, B. M., McGeer, A. J., and Low, D. E. (2005). Community-associated methicillin-resistant Staphylococcus aureus in horses and humans who work with horses. J. Am. Vet. Med. Assoc. 226, 580–583. doi: 10.2460/javma.2005.226.580
Wenzler, E., and Rodvold, K. A. (2015). Telavancin: The long and winding road from discovery to food and drug administration approvals and future directions. Clin. Infect. Dis. 61(Suppl. 2) S38–S47. doi: 10.1093/cid/civ522
Werth, B., Steed, M., Kaatz, G. W., and Rybak, M. J. (2013). Evaluation of ceftaroline (CPT) activity against heteroresistant vancomycin intermediate Staphylococcus aureus (hVISA) and VISA methicillin-resistant S. aureus (MRSA) strains in an in vitro pharmacokinetic/pharmacodynamic (PK/PD) model: Exploring the “Seesaw Effect”. Antimicrob. Agents Chemother. 57, 2664–2668.
Wertheim, H. F., Melles, D. C., Vos, M. C., van Leeuwen, W., van Belkum, A., Verbrugh, H. A., et al. (2005). The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 5, 751–762.
Wichai, S., Chuysinuan, P., Chaiarwut, S., Ekabutr, P., and Supaphol, P. (2019). Development of bacterial cellulose/alginate/chitosan composites incorporating copper (II) sulfate as an antibacterial wound dressing. J. Drug Deliv. Sci. Technol. 51, 662–671.
Wilson, G. J., Seo, K. S., Cartwright, R. A., Connelley, T., Chuang-Smith, O. N., Merriman, J. A., et al. (2011). A novel core genome-encoded superantigen contributes to lethality of community-associated MRSA necrotizing pneumonia. PLoS Pathog. 7:e1002271. doi: 10.1371/journal.ppat.1002271
Wilson, G. J., Tuffs, S. W., Wee, B. A., Seo, K. S., Park, N., Connelley, T., et al. (2018). Bovine Staphylococcus aureus superantigens stimulate the entire T cell repertoire of cattle. Infect. Immun. 86, e00505–e00518. doi: 10.1128/IAI.00505-18
Witte, W., Strommenger, B., Stanek, C., and Cuny, C. (2007). Methicillin-resistant Staphylococcus aureus ST398 in humans and animals, Central Europe. Emerg. Infect. Dis. 13:255.
Wong, J. W., Ip, M., Tang, A., Wei, V. W., Wong, S. Y., Riley, S., et al. (2018). Prevalence and risk factors of community-associated methicillin-resistant Staphylococcus aureus carriage in Asia-Pacific region from 2000 to 2016: A systematic review and meta-analysis. Clin. Epidemiol. 10:1489. doi: 10.2147/CLEP.S160595
Wulf, M., Van Nes, A., Eikelenboom-Boskamp, A., De Vries, J., Melchers, W., Klaassen, C., et al. (2006). Methicillin-resistant Staphylococcus aureus in veterinary doctors and students, the Netherlands. Emerg. Infect. Dis. 12:1939. doi: 10.3201/eid1212.060355
Yamada, T., Tochimaru, N., Nakasuji, S., Hata, E., Kobayashi, H., Eguchi, M., et al. (2005). Leukotoxin family genes in Staphylococcus aureus isolated from domestic animals and prevalence of lukM–lukF-PV genes by bacteriophages in bovine isolates. Vet. Microbiol. 110, 97–103. doi: 10.1016/j.vetmic.2005.07.006
Yang, H., Zhang, H., Wang, J., Yu, J., and Wei, H. (2017). A novel chimeric lysin with robust antibacterial activity against planktonic and biofilm methicillin-resistant Staphylococcus aureus. Sci. Rep. 7:40182. doi: 10.1038/srep40182
Yao, D., Yu, F. Y., Qin, Z. Q., Chen, C., He, S. S., Chen, Z. Q., et al. (2010). Molecular characterization of Staphylococcus aureus isolates causing skin and soft tissue infections (SSTIs). BMC Infect. Dis. 10:133. doi: 10.1186/1471-2334-10-133
Yin, Z., Wang, Y., Whittell, L. R., Jergic, S., Liu, M., Harry, E., et al. (2014). DNA replication is the target for the antibacterial effects of nonsteroidal anti-inflammatory drugs. Chem. Biol. 21, 481–487.
Zapotoczna, M., Jevnikar, Z., Miajlovic, H., Kos, J., and Foster, T. J. (2013). Iron-regulated surface determinant B (IsdB) promotes Staphylococcus aureus adherence to and internalization by non-phagocytic human cells. Cell. Microbiol. 15, 1026–1041. doi: 10.1111/cmi.12097
Zhanel, G. G., Cheung, D., Adam, H., Zelenitsky, S., Golden, A., Schweizer, F., et al. (2016). Review of eravacycline, a novel fluorocycline antibacterial agent. Drugs 76, 567–588.
Keywords: MRSA, epidemiology, pathophysiology, transmission, treatment, prevention, MRSA infections
Citation: Shoaib M, Aqib AI, Muzammil I, Majeed N, Bhutta ZA, Kulyar MF-e-A, Fatima M, Zaheer C-NF, Muneer A, Murtaza M, Kashif M, Shafqat F and Pu W (2023) MRSA compendium of epidemiology, transmission, pathophysiology, treatment, and prevention within one health framework. Front. Microbiol. 13:1067284. doi: 10.3389/fmicb.2022.1067284
Received: 17 October 2022; Accepted: 19 December 2022;
Published: 10 January 2023.
Edited by:
Ximin Zeng, The University of Tennessee, Knoxville, United StatesReviewed by:
İbrahim Hakkı CİĞERCİ, Afyon Kocatepe University, TurkeyCopyright © 2023 Shoaib, Aqib, Muzammil, Majeed, Bhutta, Kulyar, Fatima, Zaheer, Muneer, Murtaza, Kashif, Shafqat and Pu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amjad Islam Aqib,  YW1qYWRpc2xhbWFxaWJAY3V2YXMuZWR1LnBr; Wanxia Pu,
YW1qYWRpc2xhbWFxaWJAY3V2YXMuZWR1LnBr; Wanxia Pu,  cHV3YW54aWFAY2Fhcy5jbg==
cHV3YW54aWFAY2Fhcy5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.