
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 02 March 2023
Sec. Microbiotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1066133
This article is part of the Research Topic Trends of Microbial Technologies in Rehabilitation of Contaminated Environments View all 6 articles
 Rener De Jesus1†
Rener De Jesus1† Ruwaya Alkendi2†*
Ruwaya Alkendi2†*Accumulating plastics in the biosphere implicates adverse effects, raising serious concern among scientists worldwide. Plastic waste in nature disintegrates into microplastics. Because of their minute appearance, at a scale of <5 mm, microplastics easily penetrate different pristine water bodies and terrestrial niches, posing detrimental effects on flora and fauna. The potential bioremediative application of microbial enzymes is a sustainable solution for the degradation of microplastics. Studies have reported a plethora of bacterial and fungal species that can degrade synthetic plastics by excreting plastic-degrading enzymes. Identified microbial enzymes, such as IsPETase and IsMHETase from Ideonella sakaiensis 201-F6 and Thermobifida fusca cutinase (Tfc), are able to depolymerize plastic polymer chains producing ecologically harmless molecules like carbon dioxide and water. However, thermal stability and pH sensitivity are among the biochemical limitations of the plastic-degrading enzymes that affect their overall catalytic activities. The application of biotechnological approaches improves enzyme action and production. Protein-based engineering yields enzyme variants with higher enzymatic activity and temperature-stable properties, while site-directed mutagenesis using the Escherichia coli model system expresses mutant thermostable enzymes. Furthermore, microalgal chassis is a promising model system for “green” microplastic biodegradation. Hence, the bioremediative properties of microbial enzymes are genuinely encouraging for the biodegradation of synthetic microplastic polymers.
Plastics have a lot of good qualities, including weightless and stable physical properties, making them indispensable and highly resilient materials. The massive plastic production, which exponentially started in the 1950s, and the widespread usage of plastics have resulted in a large volume of post-consumer waste being dumped in landfills or marine environments (Jambeck et al., 2015; Geyer et al., 2017). In a recent United Nations Environment Programme (UNEP) report, around 400 million metric tons of plastic waste were produced annually (UNEP (United Nations Environment Programme), 2021). Experts believe that by 2060, global plastic waste production will be tripled, which half will end up in landfills, and the rest will be distributed in the environment (Organization for Economic Co-operation and Development, 2021). Thus, concrete environmental regulation and waste disposal management should be rationalized to control this impending environmental issue.
Plastic biodegradation is a natural process. Without human interference, the natural breakdown of plastic litter can occur via weathering, biodeterioration, and biofragmentation (Ojeda et al., 2011; Miles et al., 2017; Syranidou et al., 2019). However, this innate route is generally gradual. A plastic item can be degraded entirely after a hundred to thousand years (Delaney, 2013). Weathered or fragmented plastic items are significant sources of plastic particles called microplastics (MPs; Ter Halle et al., 2016; Kalogerakis et al., 2017). MPs are polymers with a size of <5 mm (about 0.2 in). MPs can be categorized into primary or secondary MPs (Arthur et al., 2009; Lehtiniemi et al., 2018). The primary MPs are product additives found in personal care and cosmetics (Habib et al., 2020), paint coatings, cleaning agents, and tire wear (Verschoor et al., 2016), to name a few. While secondary MPs originate from degraded plastic waste, such as water bottles (Winkler et al., 2019) and carry bags (Yurtsever and Yurtsever, 2018). Despite their different origins, both primary and secondary MPs are suspended in open waters (Han et al., 2020), in water columns (Choy et al., 2019), or embedded in the soil (Liu et al., 2018). MPs are chemically derived from various synthetic polymers, viz. polyethylene (PE), polypropylene (PP), polyvinyl chloride, polystyrene (PS), and polyamide (PA; Lise Nerland et al., 2014; Lin et al., 2018), that appear in different morphologies (i.e., fibers, fragments, beads), colors, and length. Because of their minute size, MPs cause detrimental effects on flora and fauna (Cao et al., 2017; Rodriguez-Seijo et al., 2017; de Souza Machado et al., 2019).
Bioprospecting is a process that explores biological products from plants, animals, and microorganisms (Ramesha et al., 2011). Bioprospecting offers a sustainable solution to many impending environmental issues like microplastic pollution. Many microorganisms can secrete enzymes with bioremediative potential against plastic particles (Table 1). These enzymes have shown remarkable biodegradation against various polymers and toxic compounds (Bhandari et al., 2021). The current waste disposal practices are inadequate in regulating litter quantities. As a result, there is a snowballing interest in exploiting efficient microbes to degrade many types of plastic. Therefore, this minireview paper focuses on the microbial enzymes involved in plastic polymer biodegradation, which offers a ‘bird’s-eye view’ of the bioremediative potential to assimilate microplastics.
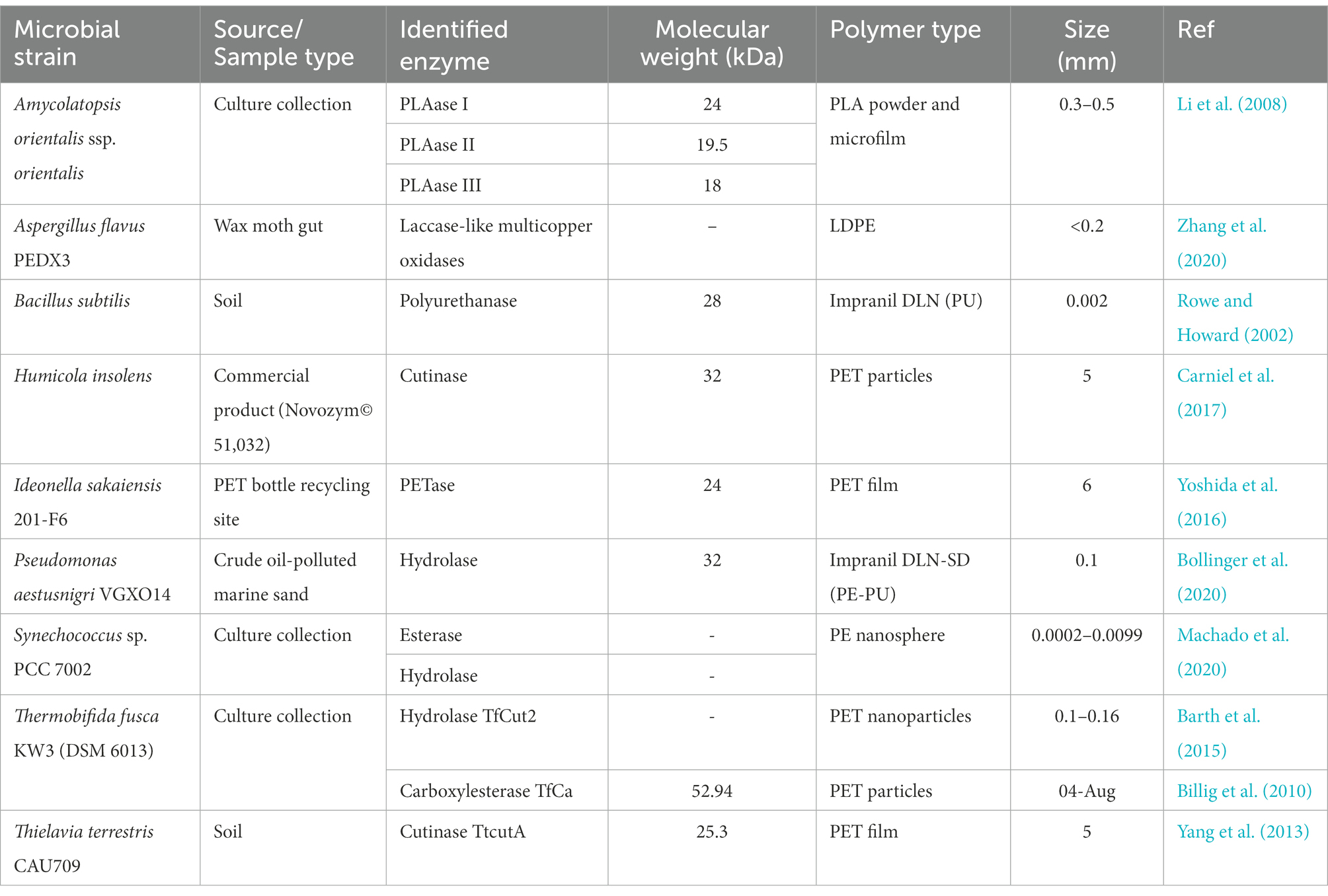
Table 1. List of some reported plastic-degrading enzymes from various microbial strains against various polymer types.
Abiotic-biotic factors have essential roles in the biodegradability of plastics. Abiotic factors, such as temperature, pH, light, and humidity, crucially influence biodegradation (Gewert et al., 2018; Oluwasina et al., 2019; Singh et al., 2019; Arisa-Tarazona et al., 2020). These factors enhance the hydrolysis of plastic polymers leading to chain scission. The scission allows biotic factors (i.e., microorganisms) to further polymer degradation. Temperature affects microbial diversity and activity (Zoungranan et al., 2020). Temperatures over 30°C decelerate plastic breakdown but increase microbial species abundance, which improves the biodegradation rate by 20% (Zoungranan et al., 2020). At the same time, pH promotes microbial growth and enzymatic activity that affects biodegradation. At 0°C with pH 3 and 11, MPs showed brittleness and fragmentations (Arisa-Tarazona et al., 2020). Furthermore, photolysis using UV light improves plastic degradation and applies as a pre-treatment method. Synthetic plastics exposed to UV for 12 months have produced fragments with decreasing sizes (Song et al., 2017). Humidity is a significant environmental factor influencing plastic biodegradability, as well. Humidity may negatively or positively stimulate microbial growth and activity. High moisture content would increase biodegradation, but excessive moisture content hinders biodegradation due to dilution effects (Oluwasina et al., 2019).
Moreover, the overall plastic biodegradation is also affected by the plastic’s surface area and polymer characteristics. High-molecular weight synthetic plastics (e.g., PE and PP) have reduced hydrophilicity because of their intact polymer chains and are thus more difficult to degrade than low-molecular weight plastics (Kawai, 1995). In addition, the absence of functional groups attribute to the durability of plastics. Some plastic additives have pro-oxidant functional groups with hydrophilic characteristics (Chiellini et al., 2006; Harshvardhan and Jha, 2013) and are receptive to attack by microbial enzymes, light, and water. Taken together, the abiotic-biotic factors determine the efficiency of microplastic biodegradation. However, the structural complexity of synthetic polymers affects the actions of these factors. Factors affecting the plastic biodegradation were discussed in many comprehensive review papers (Shah et al., 2008; Tokiwa et al., 2009; Yuan et al., 2020; Shilpa Basak and Meena, 2022).
Because microorganisms can produce enzymes that enable them to use plastic as a source of energy, microbes are ideal candidates for reducing plastic waste in the environment. Gambarini et al. (2021) identified a substantial number of putative microbial plastic degraders belonging to 12 different microbial phyla, of which just seven phyla have reported degraders to date. It indicates that bacterial and fungal phyla have a significant untapped potential for discovering enzymes that can degrade plastics. In fact, a broad family of microbial enzymes was already been isolated, such as hydrolases, laccases, peroxidases, and lipases that showed degradation of synthetic plastics. Though the differences between fungal and bacterial enzymes are not exclusively discussed in many literatures, their distinct physiologies are likely to differentially influence the rates of plastic biodegradation (Waring et al., 2013). Related studies found that most fungal enzymes have complete enzymatic systems for the depolymerization and mineralization of plastic (Blanchette, 1995; Nikhil et al., 2012; Zhu et al., 2016).
Since microbial enzymes are generally more stable than their plant and animal counterparts, microbes are gaining interest as a source of beneficial plastic-degrading enzymes. A notable example is the bacterial strain Ideonella sakaiensis 201-F6 that can degrade polyethylene terephthalate (PET). PET is one of the most widely used synthetic plastic, with an annual global output of over 50 million tons (Bornscheuer, 2016). The strain 201-F6 produced cutinase-like serine hydrolases named IsPETase and IsMHETase (Yoshida et al., 2016). The PET degradation process can be divided into two steps: the nick generation step and the terminal digestion step (Joo et al., 2018). In the nick generation step, the IsPETase cleaves one ester bond causing the formation of a nick in PET polymer chain, resulting in the generation of two PET chains with different terminals: terephthalic acid (TPA)-terminal and hydroxyethyl (HE)-terminal. Then, in the terminal digestion step, two PET chains having those termini are digested into bis-(2-hydroxyethyl)terephthalic acid (BHET) and mono-(2-hydroxyethyl)terephthalic acid (MHET) monomers (Joo et al., 2018). Subsequent digestion of these molecules, which IsMHETase breaks down MHET, produces ecologically harmless terephthalic acid (TPA) and ethylene glycol (EG) by-products (Carniel et al., 2017; Joo et al., 2018; Knott et al., 2020; Figure 1). Through assimilation and mineralization, TPA and EG are converted into carbon dioxide and water. However, one of the limitations of using IsPETase is its low thermal stability (Yoshida et al., 2016; da Costa et al., 2021). Nevertheless, because of impressive enzymatic activity against PET, IsPETase has been subjected to structural improvements using various biotechnological tools.
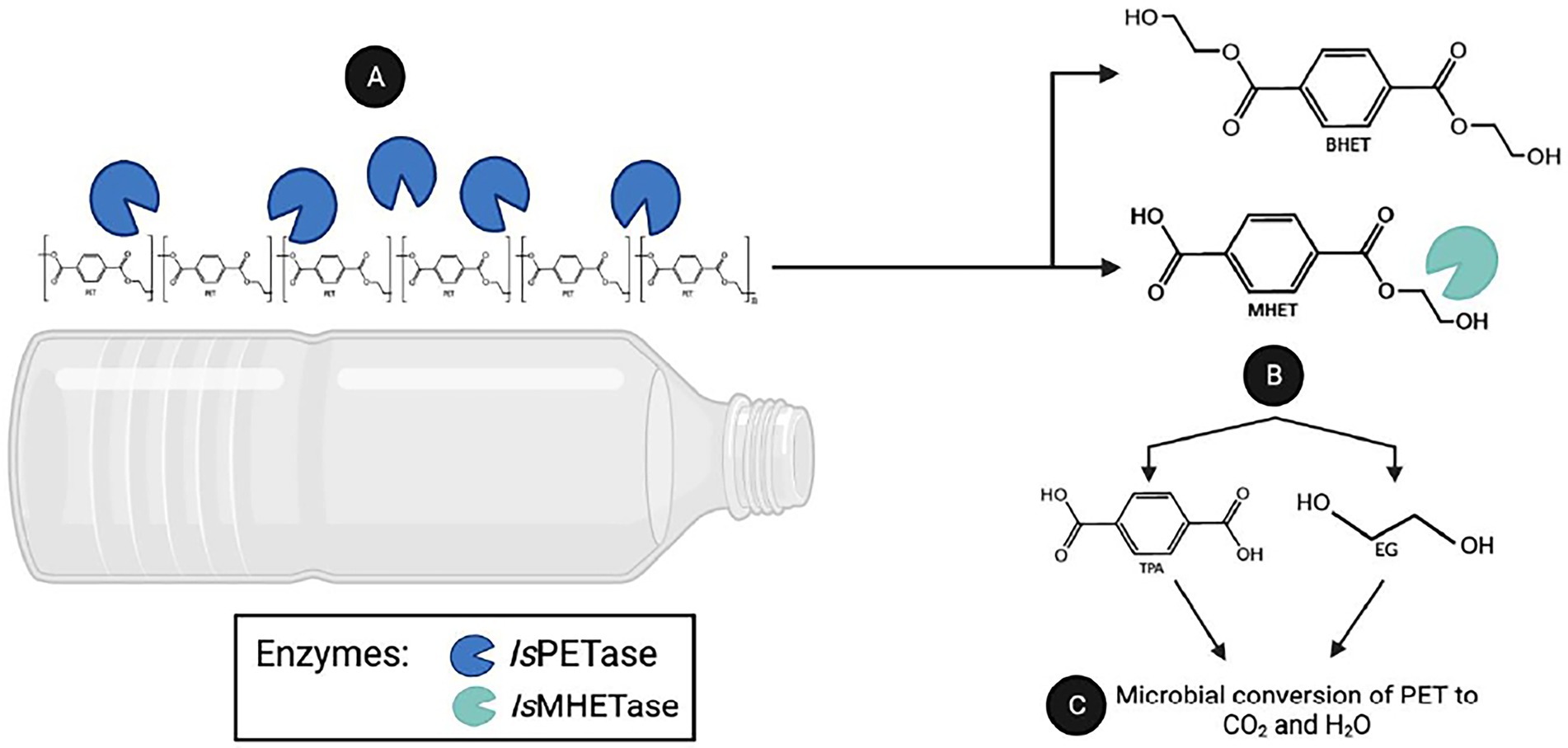
Figure 1. PET degradation by the enzymes PETase and MHETase. The Gram-negative bacterium I. sakaiensis 201-F6 is able to produce IsPETase and IsMHETase enzymes. (A) The IsPETase degrads polythylene chain producing bis(2-hydroxyethyl)terephthalate(BHET) or mono(2-hydroxyethyl) terepththalate(MHET). (B) While IsMHETase are converting MHET to non-toxic compounds: terephthalic acid and ethylene glycol. (C) By assimilation, these molecules converted to a carbon dioxide as byproduct of microbial conversion of PET.
A variety of cutinases have been identified to degrade PET as well. Cutinases have been found in fungi and bacteria, such as Fusarium solani pisi (Stavila et al., 2013) and Thermobifida fusca (Chen et al., 2010). The cutinases from both groups belong to the α/ß-hydrolase superfamily with similar spatial structures, catalytic characteristics, and substrate specificities. Despite the similarities, fungal and bacterial cutinases lack sequence homology. Thus, cutinases can be classified into prokaryotic and eukaryotic cutinase subfamilies (Chen et al., 2008). The T. fusca cutinase (Tfc) was reported to improve PET degradation with microbial pre-treatment. Microbial pre-treatment with Stenotrophomonas pavanii JWG-G1 reduced PET surface hydrophobicity, causing an easy binding for Tfc (Huang et al., 2022). Microbial pre-treatment could be a novel approach for microplastic biodegradation and to increase the degradation rate. The synergistic action of JWG-G1 and Tfc initially starts with surface binding and biofilm development of JWG-G1, which leads to the formation of functional groups from breaking ester bonds to yield PET oligomers. Like IsPETase and IsMHETase, Tfc hydrolyzes PET oligomers (Huang et al., 2022) to produce carbon dioxide and water molecules. Furthermore, several microbial strains were reported of producing cutinases with beneficial bioremediative applications. Examples are Aspergillus sp. RL2Ct (Kumari et al., 2016), Pseudomonas cepacia NRRL B-2320 (Dutta et al., 2013), and Aspergillus nidulans that produced thermo-alkaline cutinase called ANCUT2 (Bermúdez-Garcia et al., 2017). Cutinases also degrade other polymers, like poly(butylene succinate) (PBS; Hu et al., 2016) and polyester (Baker et al., 2012).
Another family of plastic-degrading enzymes are laccases that primarily described in fungal lignin biodegradation. However, laccases have been identified in both bacterial and fungal species. Laccases are copper-dependent enzymes that perform oxidation reactions of an oxygen molecule to water (Mayer and Staples, 2002; Martínez et al., 2005; Munk et al., 2015). These enzymes showed degradation of PA, PE, and PP (Fujisawa et al., 2001; Sheik et al., 2015). The degradation steps are perhaps similar to the lignin decomposition, which proceeds by oxidative reactions that breaks carbon-to-carbon bonds or ether linkages to liberate functional groups (Asina et al., 2016). Fungal species like Cochliobolus sp. (Sumathi et al., 2016), Phlebia spp. (Arora and Rampal, 2002), Podospora anserina (Xie et al., 2014), and Yarrowia lipolytica (Lee et al., 2012) were reported of laccase production and involved in break down of lignin. Bacterial laccases, on the other hand, are more stable at varying conditions, like pH and temperature, than fungal laccases (Chauhan et al., 2017), which indicates an encouraging application in microplastic bioremediation. Soil bacterium Azospirillum lipoferum produced laccase-like polyphenol oxidase that was thermostable up to 70°C with optimal pH of 6.0 (Diamantidis et al., 2000). Other strains, such as Bacillus subtilis MTCC 2414 (Muthukumarasamy et al., 2015), Microbulbifer hydrolyticus IRE-3 (Li et al., 2020), Pseudomonas extremorientalis BU118 (Neifar et al., 2016), and Serratia marcescens MTCC 4822 (Kaira et al., 2015), were reported of producing laccases with broad deterioration activities against pollutants, including plastics. Nevertheless, the industrial application of laccases is restricted due to some limitations, like low yield and high-cost production (Akpinar and Ozturk Urek, 2017; Chenthamarakshan et al., 2017).
Peroxidases are a large family of oxidoreductases known to catalyze the oxidation of many inorganic and organic substrates by using hydrogen peroxide (Adewale and Adekunle, 2018; Twala et al., 2020). Most of the peroxidases were reported from various fungal species and involved in lignin degradation with laccases. The addition of manganese peroxidase showed increased PE degradation by lignin-degrading fungi (Iiyoshi et al., 1998), which is like the copper-induced laccase activity of IRE-3 (Li et al., 2020), improves biodegradation rates. Trace elements, such as manganese and copper, protect cells from oxidative stress resulting (Bonnarme and Jeffries, 1990; Levin et al., 2002) in the retention of polymer-degrading activities. Marine fungus Alternaria alternata FB1 efficiently degraded PE polymers by producing 153 exoenzymes, including peroxidase and laccase, and caused a 95% reduction in the polymer’s molecular weight (Gao et al., 2022). Compared to fungal peroxidase, studies about bacterial peroxidases are limited. Future biodegradation studies of plastics using bacterial peroxidase could open new avenues for breaking down many synthetic plastics.
Together with cutinase and hydrolase, lipase is one of the common enzymes associated with plastic degradation. Lipases have been produced in many bacterial and fungal strains. As discussed earlier, increasing molecular weight hinders the biodegradation rate, and specific fungal lipases can break down high-molecular weight polymers. Lipase is one of the best biocatalysts for PET degradation. Lipase B (CALB) from the yeast Candida antarctica, formerly Pseudozyma antarctica, is known for its high selectivity and catalytic activity. The action of CALB is similar to IsPETase and IsMHETase activities (Boneta et al., 2021). CALB demonstrated high-efficiency hydrolysis steps and polymer scission that led to the accumulation of TPA (Carniel et al., 2017; de Castro and Carniel, 2017; de Castro et al., 2017). Moreover, CALB and Humicola insolens cutinase resulted in complete PET depolymerization with a mole fraction of up to 0.88 and a 7.7-fold increase in PET to yield TPA (Carniel et al., 2017). Another, a purified lipase (CLE) from the Cryptococcus sp. strain S-2, effectively hydrolyzed high-molecular weight plastic polymers like poly (lactic acid) and other bio-based polymers, such as polybutylene succinate, poly (ɛ-caprolactone), and poly (3-hydroxybutyrate), at a concentration of 0.8 μg/ml in 88 h (Masaki et al., 2005). It was found that the hydrolytic action involves the activation of a catalytic triad following the formation of tetrahedral intermediates that stabilizes the enzyme structure (Yoshitake et al., 2009). Hydrolysis produced volatile fatty acids and glycerols that eventually assimilated by the microorganisms to yield lipid acyl chains for cell membrane maintenance (Poddar et al., 2020). Bacterial lipases are also recognized for breaking down plastic polymers like polyurethane (Gautam et al., 2007) and PET oligomers (Swiderek et al., 2022). Various bacterial species were reported to secrete novel lipases, including those from thermophilic and psychrophilic strains (Mobarak-Qamsari et al., 2011; Rabbani et al., 2013; Maiangwa et al., 2014). Members of mesophilic Bacillus spp. and Pseudomonas spp. have been described to produce lipases (Elwan et al., 1983; Khannous et al., 2014; Jaiswal et al., 2017) with potential microplastic biodegradation. However, the utilization of these enzymes in microplastic biodegradation has not been extensively explored. Further studies should be performed on the possible polymer degradation and assimilation of degradation by-products.
In order to overcome the possible limitations of microbial enzymes in the biodegradation of MPs, new strategies need to be created. Researchers have recently demonstrated that biotechnological strategies improve enzyme structure and stability. One of the biotechnological tools widely used in protein engineering—the structural-based modeling, yields enzyme variants with higher enzymatic activity and temperature-stable properties. Son et al. (2020) successfully created IsPETaseS121E/D186H/S242T/N246D variant with enhanced substrate binding affinity and thermo-stable characteristics (Ma et al., 2018; Son et al., 2020; Meng et al., 2021). These enzyme variants exhibited 58-fold greater activity than the wild-type IsPETase (Son et al., 2020). Furthermore, site-directed mutagenesis (SDM) has efficiently been applied in degradation studies. Usually, an Escherichia coli strain is used to carry plasmid-encoding mutant and to express desirable enzymes. Furukawa et al. (2019) showed that the hydrolysis activity of mutant thermostable cutinase from T. fusca (TfCut2) expressed in the E. coli model system was 12.7 times higher than the wild-type TfCut2. Hence, the application of SDM will find bioremediative potential against MPs.
Microalgae have been extensively studied for biotechnological applications, mainly to make biofuels. However, several studies have reported the bioremediative potential of microalgae as microbial chassis. In synthetic biology, a chassis is an organism that shelters and sustains genetic components by supplying resources needed for cellular functions (Chi et al., 2019). Numerous functional expression studies were conducted using a green alga (Kim et al., 2020) and a diatom (Moog et al., 2019). Therefore, using eukaryotic microalgae instead of bacteria as model systems provide a viable and eco-friendly method for the bioremediation of microplastic-polluted water.
Plastic production has been increasing for the past decades due to the high demands of different sectors. Anthropogenic activities and improper waste disposal are the leading causes of rampant plastic pollution in the environment. Since the COVID-19 pandemic occurred in 2020, global plastic pollution has increased (Ammendolia et al., 2021), which could escalate the number of MPs (Liang et al., 2022). Thus, microplastic distribution is an emerging environmental issue that needs a long-term and sustainable solution. Bioremediation is a sustainable method to mitigate quantities of plastic contaminants, including MPs. Numerous studies stated the promising application of microbe-enzyme systems for the bioremediation of pollutants. Some of the prospective microbes are yeast (Tkavc et al., 2018), algae (Rehman et al., 2006), fungi (García-Delgado et al., 2015), and bacteria (Fu et al., 2021). This minireview paper concluded that the various enzymes of microbial origin have promising bioremediative applications in degrading synthetic microplastic particles.
Since enzymes could be produced extracellularly or intracellularly, studies focusing on intracellular enzymes are minimal. Nevertheless, a recent work published by Mohanan et al. (2022) demonstrated cloning and expression of intracellular lipases, which effectively hydrolyzed short- and medium-chain length plastics suggesting a potential bioremediative approach in microplastic biodegradation. Other innovative approaches, such as nanotechnology and enzyme immobilization, have started gaining attention for future applications to degrade MPs. The site-directed immobilization strategy for PETase on magnetic nanoparticles revealed a promising strategy for microplastic reduction (Schwaminger et al., 2021). Additionally, the application of microbial consortia in microplastic bioremediation is exceedingly encouraging. The synergistic actions of microbial consortia and enzymatic activities from various microbial networks should be thoroughly investigated. Enzyme cocktails also showed enhanced degrading action against complex polymers (Mekasha et al., 2016; Contreras et al., 2020), which can be considered an alternative option for microplastic bioremediation, especially for recalcitrant MPs. Given the seemingly endless potential of microorganisms and their constant adaptation to the changing environment, it is expected that further research in this area will soon lead to realistic biodegradation procedures that can be applied on a commercial scale.
RD wrote the original draft, revised original draft, collected references, and constructed illustrations. RA edited the original draft, revised original draft, and supervised. All authors contributed to the article and approved the submitted version.
This work was funded by the United Arab Emirates University Program for Advanced Research (2020-2022).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adewale, I. O., and Adekunle, A. T. (2018). Biochemical properties of peroxidase from white and red cultivars of kolanut (Cola nitida). Biocatal. Agric. Biotechnol. 14, 1–9. doi: 10.1016/j.bcab.2018.01.013
Akpinar, M., and Ozturk Urek, R. (2017). Induction of fungal laccase production under solid state bioprocessing of new agroindustrial waste and its application on dye decolorization. 3 Biotech. 7:98. doi: 10.1007/s13205-017-0742-5
Ammendolia, J., Saturno, J., Brooks, A. L., Jacobs, S., and Jambeck, J. R. (2021). An emerging source of plastic pollution: environmental presence of plastic personal protective equipment (PPE) debris related to COVID-19 in a metropolitan city. Environ. Pollut. 269:116160. doi: 10.1016/j.envpol.2020.116160
Arisa-Tarazona, M. C., Villareal-Chiu, J. F., Hernández-López, J. M., De la Rosa, J. R., Barbieri, V., Siligardi, C., et al. (2020). Microplastic pollution reduction by a carbon and nitrogen-doped TiO2: effect of pH and temperature in the photocatalytic degradation process. J. Hazard. Mater. 395:122632:122632. doi: 10.1016/j.jhazmat.2020.122632
Arora, D. S., and Rampal, P. (2002). Laccase production by some Phlebia species. J. Basic Microbiol. 42, 295–301. doi: 10.1002/1521-4028(200210)42:5<295::AID-JOBM295>3.0.CO;2-#
Arthur, C., Baker, J., and Bamford, H.. (2009). Proceedings of the international research workshop on the occurrence, effects, and fate of microplastic marine debris. Available at: www.MarineDebris.noaa.gov (Accessed November 10, 2022).
Asina, F., Brzonova, I., Voeller, K., Kozliak, E., Kubátova, A., Yao, B., et al. (2016). Biodegradation of lignin by fungi, bacteria and laccases. Bioresour. Technol. 220, 414–424. doi: 10.1016/j.biortech.2016.08.016
Baker, P. J., Poultney, C., Liu, Z., Gross, R., and Montclare, J. K. (2012). Identification and comparison of cutinases for synthetic polyester degradation. Appl. Microbiol. Biotechnol. 93, 229–240. doi: 10.1007/s00253-011-3402-4
Barth, M., Oeser, T., Wei, R., Then, J., Schmidt, J., and Zimmerman, W. (2015). Effect of hydrolysis products on the enzymatic degradation of polyethylene terephthlate nanoparticles by a polyester hydrolase from Thermobifida fusca. Biochem. Eng. J. 93, 222–228. doi: 10.1016/j.bej.2014.10.012
Bermúdez-Garcia, E., Peña-Montes, C., Castro-Rodríguez, J. A., González-Canto, A., Navarro-Ocaña, A., and Farrés, A. (2017). ANCUT2, a thermos-alkaline cutinase from aspergillus nidulans and its potential applications. Appl. Biochem. Biotechnol. 182, 1014–1036. doi: 10.1007/s12010-016-2378-z
Bhandari, S., Poudel, D. K., Marahatha, R., Dawadi, S., Khadayat, K., Phuyal, S., et al. (2021). Microbial enzymes used in bioremediation. J. Chem. 2021:8849512. doi: 10.1155/2021/8849512
Billig, S., Oeser, T., Birkemeyer, C., and Zimmerman, W. (2010). Hydrolysis of cyclic poly(ethylene terephthalate) trimers by a carboxylesterase from Thermobifida fusca KW3. Appl. Microbiol. Biotechnol. 87, 1753–1764. doi: 10.1007/s000253-010-2635-y
Blanchette, R. A. (1995). Degradation of the lignocellulose complex in wood. Can. J. Bot. 73, 999–1010. doi: 10.1139/b95-350
Bollinger, A., Thies, S., Knieps-Grünhagen, E., Gertzen, C., Kobus, S., Höppner, A., et al. (2020). A novel polyester hydrolase from the marine bacterium Pseudomonas aestusnigri – structural and functional insights. Front. Microbiol. 11:114. doi: 10.3389/fmicb.2020.00114
Boneta, S., Arafet, K., and Moliner, V. (2021). QM/MM study of the enzymatic biodegradation mechanism of polyethylene terephthalate. J. Chem. Inf. Model. 61, 3041–3051. doi: 10.1021/acs.jcim.1c00394
Bonnarme, P., and Jeffries, T. W. (1990). Mn(II) regulation of lignin peroxidases and manganese-dependent peroxidases from lignin-degrading white rot fungi. Appl. Environ. Microbiol. 56, 210–217. doi: 10.1128/aem.56.1.210-217.1990
Cao, D., Wang, X., Luo, X., Liu, G., and Zheng, H. (2017). Effects of polystyrene microplastics on the fitness of earthworms in an agricultural soil. IOP Conf. Ser.: Earth Environ. Sci. 61:012148. doi: 10.1088/1755-1315/61/1/012148
Carniel, A., Valoni, É., Nicomedes, J., Gomes, A. D. C., and De Castro, A. M. (2017). Lipase from Candida antarctica (CALB) and cutinase from Humicola insolens act synergistically for PET hydrolysis to terephthalic acid. Process Biochem. 59, 84–90. doi: 10.1016/j.procbio.2016.07.023
Chauhan, P. S., Goradia, B., and Saxena, A. (2017). Bacterial laccase: recent update on production, properties and industrial applications. 3 Biotech. 7, 7:323. doi: 10.1007/s13205-017-0955-7
Chen, S., Su, L., Billig, S., Zimmerman, W., Chen, J., and Wu, J. (2010). Biochemical characterization of the cutinases from Thermobifida fusca. J. Mol. Catal. 63, 121–127. doi: 10.1016/j.molcatb.2010.01.001
Chen, S., Tong, X., Woodard, R. W., Du, G., and Wu, J. (2008). Identification and characterization of bacterial cutinase. J. Biol. Chem. 283, 25854–25862. doi: 10.1074/jbc.M800848200
Chenthamarakshan, A., Parambayil, N., Miziriya, N., Soumya, P. S., Lakshmi, M. S. K., Ramgopal, A., et al. (2017). Optimization of laccase production from Marasmiellus palmivorus LA1 by Taguchi method of design of experiments. BMC Biotechnol. 17:12. doi: 10.1186/s12896-017-0333-x
Chi, H., Wang, X., Shao, Y., Qin, Y., Deng, Z., Wang, L., et al. (2019). Engineering and modification of microbial chassis for systems and synthetic biology. Synth. Syst. Biotechnol. 4, 25–33. doi: 10.1016/j.synbio.2018.12.001
Chiellini, E., Corti, A., D’Antone, S., and Baciu, R. (2006). Oxo-biodegradable carbon backbone polymers – oxidative degradation of polyethylene under accelerated test conditions. Polym. Degrad. Stab. 91, 2739–2747. doi: 10.1016/j.polymdegradstab.2006.03.022
Choy, C. A., Robison, B. H., Gagne, T. O., Erwin, B., Firl, E., Halden, R. U., et al. (2019). The vertical distribution and biological transport of marine microplastics across the epipelagic and mesopelagic water column. Sci. Rep. 9, 1–9. doi: 10.1038/s41598-019-44117-2
Contreras, F., Pramanik, S., Rozhkova, A. M., Zorov, I., Korotkova, O., Sinitsyn, A. P., et al. (2020). Engineering robust cellulases for tailored lignocellulosic degradation cocktails. Int. J. Mol. Sci. 21:1589. doi: 10.3390/ijms21051589
da Costa, C. H. S., dos Santos, A. M., Alves, C. N., Martí, S., Moliner, V., Santana, K., et al. (2021). Assess of the PETase conformational changes induced by poly(ethylene terephthalate) binding. Proteins 89, 1340–1352. doi: 10.1002/prot.26155
de Castro, A. M., and Carniel, A. (2017). A novel process for poly(ethylene terephthalate) depolymerization via enzyme-catalyzed glycolysis. Biochem. Eng. J. 124, 64–68. doi: 10.1016/j.bej.2017.04.011
de Castro, A. M., Carniel, A., Nicomedes, J., da Conceição Gomes, A., and Valoni, É. (2017). Screening of commercial enzymes for poly(ethylene terephthalate) (PET) hydrolysis and synergy studies on different substrate sources. J. Ind. Microbiol. Biotechnol. 44, 835–844. doi: 10.1007/S10295-017-1942-Z
de Souza Machado, A. A., Lau, C., Kloas, W., Bergmann, J., Bachelier, J., Faltin, E., et al. (2019). Microplastics can change soil properties and affect plant performance. Environ. Sci. Technol. 53, 6044–6052. doi: 10.1021/acs.est.9b01339
Delaney, P. (2013). How long it takes for some everyday items to decompose. In Eco Aware, General. Available at: www.down2earthmaterials.ie (Accessed August 21, 2022).
Diamantidis, G., Effosse, A., Potier, P., and Bally, R. (2000). Purification and characterization of the first bacterial laccase in the rhizospheric bacterium Azospirillum lipoferum. Soil Biol. Biochem. 32, 919–927. doi: 10.1016/s0038-0717(99)00221-7
Dutta, K., Dasu, V. V., and Hegde, K. (2013). Development of medium and kinetic modeling for enhanced production of cutinase from pseudomonas cepacian NRRL B-2320. Adv. Microbiol. 3, 479–489. doi: 10.4236/aim.2013.36064
Elwan, S. H., El-Hoseiny, M. M., Ammar, M. S., and Mostafa, S. A. (1983). Lipases production by Bacillus circulans under mesophilic and osmophilic conditions. Factors affecting lipases production. G. Batteriol. Virol. Immunol. 76, 187–199.
Fu, X., Qiao, Y., Xue, J., Cheng, D., Chen, C., Bai, Y., et al. (2021). Analyses of community structure and role of immobilized bacteria system in the bioremediation process of diesel pollution seawater. Sci. Total Environ. 799:149439. doi: 10.1016/j.scitotenv.2021.149439
Fujisawa, M., Hirai, H., and Nishida, T. (2001). Degradation of polyethylene and nylon-66 by the laccase-mediator system. J. Polym. Environ. 9, 103–1008. doi: 10.1023/A:1020472426516
Furukawa, M., Kawakami, N., Tomizawa, A., and Miyamoto, K. (2019). Efficient degradation of poly(ethylene terephthalate) with Thermobifida fusca cutinase exhibiting improved catalytic activity generated using mutagenesis and additive-based approaches. Sci. Rep. 9:16038. doi: 10.1038/s41598-019-52379-z
Gambarini, V., Pantos, O., Kingsbury, J. M., Weaver, L., Handley, K. M., and Lear, G. (2021). Phylogenetic distribution of plastic-degrading microorganisms. MSystems 6:e01112-20. doi: 10.1128/msystems.01112-20
Gao, R., Liu, R., and Sun, C. (2022). A marine fungus Alternaria alternata FB1 efficiently degrades polyethylene. J. Hazard. Mater. 431:128617. doi: 10.1016/j.jhazmat.2022.128617
García-Delgado, C., Yunta, F., and Eymar, E. (2015). Bioremediation of multi-polluted soil by spent mushroom (Agaricus bisporus) substrate: polycyclic aromatic hydrocarbons degradation and Pb availability. J. Hazard. Mater. 300, 281–288. doi: 10.1016/j.jhazmat.2015.07.008
Gautam, R., Bassi, A., and Yanful, E. (2007). Candida rugosa lipase-catalyzed polyurethane degradation in aqueous medium. Biotechnol. Lett. 29, 1081–1086. doi: 10.1007/s10529-007-9354-1
Gewert, B., Plassman, M., Sandblom, O., and MacLeod, M. (2018). Identification of chain scission products released to water by plastic exposed to ultraviolet light. Environ. Sci. Technol. Lett. 5, 272–276. doi: 10.1021/acs.estlett.8b00119
Geyer, R., Jambeck, J., and Law, K. L. (2017). Production, use, and fate of all plastics ever made. Sci. Adv. 3:e1700782. doi: 10.1126/sciadv.1700782
Habib, R. Z., Salim Abdoon, M. M., Al Meqbaali, R. M., Ghebremedhin, F., Elkashlan, M., Kittaneh, W. F., et al. (2020). Analysis of microbeads in cosmetic products in the United Arab Emirates. Environ. Pollut. 258:113831. doi: 10.1016/j.envpol.2019.113831
Han, M., Niu, X., Tang, M., Zhang, B. T., Wang, G., Yue, W., et al. (2020). Distribution of microplastics in surface water of the lower Yellow River near estuary. Sci. Total Environ. 707:135601. doi: 10.1016/j.scitotenv.2019.135601
Harshvardhan, K., and Jha, B. (2013). Biodegradation of low-density polyethylene by marine bacteria from pelagic waters, Arabian Sea, India. Mar. Pollut. Bull. 77, 100–106. doi: 10.1016/j.marpolbul.2013.10.025
Hu, X., Gao, Z., Wang, Z., Su, T., Yang, L., and Li, P. (2016). Enzymatic degradation of poly(butylene succinate) by cutinase cloned from fusarium solani. Polym. Degrad. Stab. 134, 211–219. doi: 10.1016/j.polymdegradstab.2016.10.012
Huang, Q. S., Yan, Z. F., Chen, X. Q., Du, Y. Y., Li, J., Liu, Z. Z., et al. (2022). Accelerated biodegradation of polyethylene terephthalate by Thermobifida fusca cutinase mediated by Stenotrophomonas pavanii. Sci. Total Environ. 808:152107. doi: 10.1016/j.scitotenv.2021.152107
Iiyoshi, Y., Tsutsumi, Y., and Nishida, T. (1998). Polyethylene degradation by lignin-degrading fungi and manganese peroxidase. J. Wood Sci. 44, 222–229. doi: 10.1007/BF00521967
Jaiswal, A., Preet, M., and Tripti, B. (2017). Production and optimization of lipase enzyme from mesophiles and thermophiles. J. Microb. Biochem. Technol. 9:3. doi: 10.4172/1948-5948.1000355
Jambeck, J., Geyer, R., Wilcox, C., Siegler, T., Perryman, M., Andrady, A., et al. (2015). Plastic waste inputs from land into the ocean. Science 347, 768–771. doi: 10.1126/science.1260352
Joo, S., Cho, I. J., Seo, H., Son, H. F., Sagong, H.-H., Shin, T. J., et al. (2018). Structural insight into molecular mechanism of poly(ethylene terephthalate) degradation. Nat. Commun. 9:382. doi: 10.1038/s41467-018-02881-1
Kaira, G. S., Dhakar, K., and Pandey, A. (2015). A psychrotolerant strain of Serratia marcescens (MTCC 4822) produces laccase at wide temperature and pH range. AMB Express 5:1. doi: 10.1186/s13568-014-0092-1
Kalogerakis, N., Karkanorachaki, K., Kalogerakis, G. C., Triantafyllidi, E. I., Gotsis, A. D., Partsinevelos, P., et al. (2017). Microplastics generation: onset of fragmentation of polyethylene films in marine environment mesocosms. Front. Mar. Sci. 4, 1–15. doi: 10.3389/fmars.2017.00084
Kawai, F. (1995). Breakdown of plastics and polymers by microorganisms. Adv. Biochem. Eng. Biotechnol. 52, 151–194. doi: 10.1007/BFb0102319
Khannous, L., Jrad, M., Dammak, M., Miladi, R., Chaaben, N., Khemakhem, B., et al. (2014). Isolation of a novel amylase and lipase-producing Pseudomonas luteola strain: study of amylase production conditions. Lipids Health Dis. 13, 1–9. doi: 10.1186/1476-511x-13-9
Kim, J. W., Park, S. B., Tran, Q. G., Cho, D. H., Choi, D. Y., Lee, Y. J., et al. (2020). Functional expression of polyethylene terephthalate-degrading enzyme (PETase) in green microalgae. Microb. Cell Factories 19, 97–99. doi: 10.1186/s12934-020-01355-8
Knott, B. C., Erickson, E., Allen, M. D., Gado, J. E., Graham, R., Kearns, F. L., et al. (2020). Characterization and engineering of a two-enzyme system for plastics depolymerization. Proc. Natl. Acad. Sci. U. S. A. 117, 25476–25485. doi: 10.1073/pnas.2006753117
Kumari, V., Kumar, V., Chauhan, R., Asif, M., and Bhalla, T. C. (2016). Optimization of medium parameters by response surface methodology (RSM) for enhanced production of cutinase from Aspergillus sp. RL2Ct. 3 Biotech 6:149. doi: 10.1007/s13205-016-0460-4
Lee, K.-M., Kalyani, D., Tiwari, M., Kim, T.-S., and Dhiman, S. S. (2012). Enhanced enzymatic hydrolysis of rice straw by removal of phenolic compounds using a novel laccase from yeast Yarrowia lipolytica. Bioresour. Technol. 123, 636–645. doi: 10.1016/j.biortech.2012.07.066
Lehtiniemi, M., Hartikainen, S., Näkki, P., Engstròm-Öst, J., Koistinen, A., and Setälä, O. (2018). Size matters more than shape: ingestion of primary and secondary microplastics by small predators. Food Webs 16:e00097. doi: 10.1016/j.fooweb.2018
Levin, L., Forchiassin, F., and Ramos, A. M. (2002). Copper induction of lignin-modifying enzymes in the white-rot fungus Trametes trogii. Mycologia 94, 377–383. doi: 10.1080/15572536.2003.11833202
Li, F., Wang, S., Liu, W., and Chen, G. (2008). Purification and characterization of poly(L-lactic acid)-degrading enzymes from Amycolatopsis orientalis ssp. orientalis. FEMS Microbiol. Lett. 282, 52–58. doi: 10.1111/j.1574-6968.2008.01109.x
Li, Z., Wei, R., Gao, M., Ren, Y., Yu, B., Nie, K., et al. (2020). Biodegradation of low-density polyethylene by Microbulbifer hydrolyticus IRE-31. J. Environ. Manag. 263:110402. doi: 10.1016/j.jenvman.2020.110402
Liang, H., Ji, Y., Ge, W., Wu, J., Song, N., Yin, Z., et al. (2022). Release kinetics of microplastics from disposable face masks into the aqueous environment. Sci. Total Environ. 816:151650. doi: 10.1016/j.scitotenv.2021.151650
Lin, L., Zuo, L. Z., Peng, J. P., Cai, L. Q., Fok, L., Yan, Y., et al. (2018). Occurrence and distribution of microplastics in an urban river: a case study in the Pearl River along Guangzhou City, China. Sci. Total Environ. 644, 375–381. doi: 10.1016/j.scitotenv.2018.06.327
Lise Nerland, I., Halsband, C., Allan, I., Thomas, K. V., and Naes, K. (2014). Microplastics in marine environments: occurrence, distribution and effects (Issue 47). Report SNo. 6754-2014. Available at: www.miljodirektoratet.no (Accessed September 5, 2022).
Liu, M., Lu, S., Song, Y., Lei, L., Hu, J., Zhou, W., et al. (2018). Microplastic and mesoplastic pollution in farmland soils in suburbs of Shanghai, China. Environ. Pollut. 242, 855–862. doi: 10.1016/j.envpol.2018.07.051
Ma, Y., Yao, M., Li, B., Ding, M., He, B., Chen, S., et al. (2018). Enhanced poly(ethylene terephthalate) hydrolase activity by protein engineering. Engineering 4, 888–893. doi: 10.1016/j.eng.2018.09.007
Machado, M., Vimbela, G., Silva-Oliveira, T., Bose, A., and Tripathi, A. (2020). The response of Synechococcus sp. PCC 7002 to micro−/nano polyethylene particles – investigation of a key anthropogenic stressor. PLoS One 15:e0232745. doi: 10.1371/journal.pone.0232745
Maiangwa, J., Shukuri, M., Ali, M., Bakar, A., Raja, S., Raja, N. Z., et al. (2014). Adaptational properties and applications of cold-active lipases from psychrophilic bacteria. Extremophiles 19, 235–247. doi: 10.1007/s00792-014-0710-5
Martínez, Á., Speranza, M., Ruiz-Dueñas, F., Ferreira, P., Camarero, S., Guillén, F., et al. (2005). Biodegradation of lignocellulosics: microbial, chemical, and enzymatic aspects of the fungal attack of lignin lignocellulosic materials. Int. Microbiol. 8, 195–204.
Masaki, K., Kamini, N. R., Ikeda, H., and Iefuji, H. (2005). Cutinase-like enzyme from the yeast Cryptococcus sp. strain S-2 hydrolyzes polylactic acid and other biodegradable plastics. Appl. Environ. Microbiol. 71, 7548–7550. doi: 10.1128/aem.71.11.7548-7550.2005
Mayer, A. M., and Staples, R. C. (2002). Laccase: new functions for an old enzyme. Phytochemistry 60, 551–565. doi: 10.1016/s0031-9422(02)00171-1
Mekasha, S., Toupalová, H., Linggadjaja, E., Tolani, H. A., Anděra, L., Arntzen, M., et al. (2016). A novel analytical method for D-glucosamine quantification and its application in the analysis of chitosan degradation by a minimal enzyme cocktail. Carbohydr. Res. 433, 18–24. doi: 10.1016/j.carres.2016.07.003
Meng, X., Yang, L., Liu, H., Li, Q., Xu, G., Zhang, Y., et al. (2021). Protein engineering of stable IsPETase for PET plastic degradation by premuse. Int. J. Biol. Macromol. 180, 667–676. doi: 10.1016/j.ijbiomac.2021.03.058
Miles, C., DeVetter, L., Ghimire, S., and Hayes, D. G. (2017). Suitability of biodegradable plastic mulches for organic and sustainable agricultural production systems. HortScience 52, 10–15. doi: 10.21273/hortsci11249-16
Mobarak-Qamsari, E., Kasra-Kermanshahi, R., and Moosavi-Nejad, Z. (2011). Isolation and identification of a novel, lipase-producing bacterium, Pseudomonas aeruginosa KM110. Iran. J. Microbiol. 3, 92–98.
Mohanan, N., Wong, C. H., Budisa, N., and Levin, D. B. (2022). Characterization of polymer degrading lipases, LIP1 and LIP2 from Pseudomonas chlororaphis PA23. Front. Bioeng. Biotechnol. 10:555. doi: 10.3389/fbioe.2022.854298
Moog, D., Schmitt, J., Senger, J., Zarzycki, J., Rexer, K. H., Linne, U., et al. (2019). Using a marine microalga as a chassis for polyethylene terephthalate (PET) degradation. Microb. Cell Factories 18, 171–115. doi: 10.1186/s12934-019-1220-z
Munk, L., Sitarz, A., Kalyani, D., Mikkelsen, D., and Meyer, A. (2015). Can laccases catalyze bond cleavage in lignin? Biotechnol. Adv. 33, 13–24. doi: 10.1016/j.biotechadv.2014.12.008
Muthukumarasamy, N., Jackson, B., Raj, A. J., and Sevanan, M. (2015). Production of extracellular laccase from Bacillus subtilis MTCC 2424 using agroresidues as a potential substrate. Biochem. Res. Int. 2015:765190. doi: 10.1155/2015/765190
Neifar, M., Chouchane, H., Mahjoubi, M., Jaouani, A., and Cherif, A. (2016). Pseudomonas extremorientalis BU118: a new salt-tolerant laccase-secreting bacterium with biotechnological potential in textile azo dye decolourization. 3 Biotech 6:107. doi: 10.1007/s13205-016-0425-7
Nikhil, B., Adhyaru, D., and Thakor, P. (2012). Production of xylanase by aspergillus flavus FPDN1 on pearl millet bran: optimization of culture conditions and applications in bioethanol production. Int. J. Res. Chem. Environ. 2, 204–210.
Ojeda, T., Freitas, A., Birck, K., Dalmolin, E., Jacques, R., Bento, F., et al. (2011). Degradability of linear polyolefins under natural weathering. Polym. Degrad. Stab. 96, 703–707. doi: 10.1016/j.polymdegradstab.2010.12.004
Oluwasina, O., Olaleyem, F., Olusegun, S., Oluwasina, O., and Mohallen, N. (2019). Influence of oxidized starch on physicomechanical, thermanl properties, and atomic force micrographs of cassava starch bioplastic film. Int. J. Biol. Macromol. 135, 282–293. doi: 10.1016/j.ijbiomac.2019.05.150
Organization for Economic Co-operation and Development (2021). Global plastics outlook policy scenarios to 2060. Available at: https://read.oecd-ilibrary.org/view/?ref=1143_1143481-88j1bxuktr&title=Global-Plastics-Outlook-Policy-Scenarios-to-2060-Policy-Highlights (Accessed November 10, 2022).
Poddar, N., Elahee Doomun, S., Callahan, D., Kowalski, G., and Martin, G. (2020). The assimilation of glycerol into lipid acyl chains and associated carbon backbones of Nannochloropsis Salina varies under nitrogen replete and deplete conditions. Biotechnol. Bioeng. 117, 3299–3309. doi: 10.1002/bit.27498
Rabbani, M., Bagherinejad, M. R., Sadeghi, H. M. M., Shariat, Z. S., Etemadifar, Z., Moazen, F., et al. (2013). Isolation and characterization of novel thermophilic lipase-secreting bacteria. Brazilian J. Microbiol. 44, 1113–1119. doi: 10.1590/S1517-83822013000400013
Ramesha, B. T., Gertsch, J., Ravikanth, G., Priti, V., Ganeshaiah, K. N., and Shaanker, U. R. (2011). Biodiversity and chemodiversity: future perspectives in bioprospecting. Curr. Drug Targets 12, 1515–1530. doi: 10.2174/138945011798109473
Rehman, A., Shakoori, F. R., and Shakoori, A. R. (2006). Heavy metal resistant Distigma proteus (Euglenophyta) isolated from industrial effluents and its possible role in bioremediation of contaminated wastewaters. World J. Microbiol. Biotechnol. 23, 753–758. doi: 10.1007/s11274-006-9291-5
Rodriguez-Seijo, A., Lourenço, J., Rocha-Santos, T. A. P., da Costa, J., Duarte, A. C., Vala, H., et al. (2017). Histopathological and molecular effects of microplastics in Eisenia andrei Bouché. Environ. Pollut. 220, 495–503. doi: 10.1016/j.envpol.2016.09.092
Rowe, L., and Howard, G. T. (2002). Growth of Bacillus subtilis on polyurethane and the purification and characterization of a polyurethanase-lipase enzyme. Int. Biodeterior. Biodegrad. 50, 33–40. doi: 10.1016/S0964-8305(02)00047-1
Schwaminger, S. P., Fehn, S., Steegmüller, T., Rauwolf, S., Löwe, H., Pflüger-Grau, K., et al. (2021). Immobilization of PETase enzymes on magnetic iron oxide nanoparticles for the decomposition of microplastic PET. Nanoscale Adv. 3, 4395–4399. doi: 10.1039/d1na00243k
Sheik, S., Chandrashekar, K. R., Swaroop, K., and Somashekarappa, H. M. (2015). Biodegradation of gamma irradiated low density polyethylene and polypropylene by endophytic fungi. Int. Biodeterior. Biodegradation 105, 21–29. doi: 10.1016/j.ibiod.2015.08.006
Shah, A. A., Hasan, F., Hameed, A., and Ahmed, S. (2008). Biological degradation of plastics: a comprehensive review. Biotechnol. Adv. 26, 246–265. doi: 10.1016/j.biotechadv.2007.12.005
Shilpa Basak, N., and Meena, S. S. (2022). Microbial biodegradation of plastics: challenges, opportunities, and a critical perspective. Front. Environ. Sci. Eng. 16:161. doi: 10.1007/s11783-022-1596-6
Singh, R. K., Ruj, B., Sadhukhan, A., and Gupta, P. (2019). Thermal degradation of waste plastics under non-sweeping atmosphere: part 1: effect of temperature, product optimization, and degradation mechanism. J. Environ. Manag. 239, 395–406. doi: 10.1016/j.jenvman.2019.03.067
Son, H. F., Joo, S., Seo, H., Sagong, H. Y., Lee, S. H., Hong, H., et al. (2020). Structural bioinformatics-based protein engineering of thermo-stable PETase from Ideonella sakaiensis. Enzym. Microb. Technol. 141:109656. doi: 10.1016/j.enzmictec.2020.109656
Song, Y. K., Hong, S. H., Jang, M., Han, G. M., Jung, S. W., and Shim, W. J. (2017). Combined effects of UV exposure duration and mechanical abrasion on microplastic fragmentation by polymer type. Environ. Sci. Technol. 51, 4368–4376. doi: 10.1021/acs.est.6b06155
Stavila, E., Arsyi, R. Z., Petrovic, D. M., and Loos, K. (2013). Fusarium solani pisi cutinase-catalyzed synthesis of polyamides. Eur. Polym. J. 49, 834–842. doi: 10.1016/j.europolymj.2012.12.010
Sumathi, T., Viswanath, B., Lakshmi, A. S., and SaiGopal, D. V. R. (2016). Production of laccase by Cochliobolus sp. isolated from plastic dumped soils and their ability to degrade low molecular weight PVC. Biochem. Res. Int. 2016:9519527. doi: 10.1155/2016/9519527
Swiderek, K., Velasco-Lozano, S., Galmés, M., Olazabal, I., Sardon, H., López-Gallego, F., et al. (2022). Mechanistic studies of a lipase unveil a dramatic effect of pH on the selectivity towards the hydrolysis of PET oligomers. Res. Square. doi: 10.21203/rs.3.rs-1930954/v1
Syranidou, E., Karkanorachaki, K., Amorotti, F., Avgeropoulos, A., Kolvenbach, B., Zhou, N. Y., et al. (2019). Biodegradation of mixture of plastic films by tailored marine consortia. J. Hazard. Mater. 375, 33–42. doi: 10.1016/j.jhazmat.2019.04.078
Ter Halle, A., Ladirat, L., Gendre, X., Goudouneche, D., Pusineri, C., Routaboul, C., et al. (2016). Understanding the fragmentation pattern of marine plastic debris. Environ. Sci. Technol. 50, 5668–5675. doi: 10.1021/acs.est.6b00594
Tkavc, R., Matrosova, V. Y., Grichenko, O. E., Gostincar, C., Volpe, R. P., Klimenkova, P., et al. (2018). Prospects for fungal bioremediation of acidic radioactive waste sites: characterization and genome sequence of Rhodotorula taiwanensis MD1149. Front. Microbiol. 8:2528. doi: 10.3389/fmicb.2017.02528
Tokiwa, Y., Calabia, B. P., Urwu, C. U., and Aiba, S. (2009). Biodegradability of plastics. Int. J. Mol. Sci. 10, 3722–3742. doi: 10.3390/ijms10093722
Twala, P. P., Mitema, A., Baburam, C., and Feto, N. A. (2020). Breakthroughs in the discovery and use of different peroxidase isoforms of microbial origin. AIMS Microbiol. 6, 330–349. doi: 10.3934/microbiol.2020020
UNEP (United Nations Environment Programme). (2021). Drowning in plastics – marine litter and plastic waste vital graphics. Available at: https://wedocs.unep.org/xmlui/bitstream/handle/20.500.11822/36964/VITGRAPH.pdf (Accessed November 10, 2022).
Verschoor, A., de Poorter, L., Dröge, R., Kuenen, J., and de Valk, E.. (2016). Emission of microplastics and potential mitigation measures abrasive cleaning agents, paints and Tyre wear. In RIVM Report. Available at: www.rivm.openrepository.com (Accessed July 28, 2022).
Waring, B. G., Averill, C., and Hawkes, C. V. (2013). Difference in fungal and bacterial physiology alter soil carbon and nitrogen cycling: insights from meta-analysis and theoretical models. Ecol. Lett. 16, 887–894. doi: 10.1111/ele.12125
Winkler, A., Santo, N., Ortenzi, M. A., Bolzoni, E., Bacchetta, R., and Tremolada, P. (2019). Does mechanical stress cause microplastic release from plastic water bottles? Water Res. 166:115082. doi: 10.1016/j.watres.2019.115082
Xie, N., Chapeland-Leclerc, F., Silar, P., and Ruprich-Robert, G. (2014). Systematic gene deletions evidences that laccases are involved in several stages of wood degradation in the filamentous fungus Podospora anserina. Environ. Microbiol. 16, 141–161. doi: 10.1111/1462-2920.12253
Yang, S., Xu, H., Yan, Q., Liu, Y., Zhou, P., and Jiang, Z. (2013). A low molecular mass cutinase of Thielavia terrestris efficiently hydrolyzes poly(esters). J. Ind. Microbiol. Biotechnol. 40, 217–226. doi: 10.1007/s10295-012-1222-x
Yoshida, S., Hiraga, K., Takehana, T., Taniguchi, I., Yamaji, H., Maeda, Y., et al. (2016). A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 351, 1196–1199. doi: 10.1126/science.aad6359
Yoshitake, S., Toshiaki, M., Misako, A., Hidemasa, K., Kazuo, M., and Haruyuki, I. (2009). ONIOM study of the mechanism of the enzymatic hydrolysis of biodegradable plastics. Bull. Chem. Soc. Jpn. 82, 338–346. doi: 10.1246/bcsj.82.338
Yuan, J., Ma, J., Sun, Y., Zhou, T., Zhao, Y., and Yu, F. (2020). Microbial degradation and other environmental aspects of microplastics/plastics. Sci. Total Environ. 715:136968. doi: 10.1016/j.scitotenv.2020.136968
Yurtsever, M., and Yurtsever, U. (2018). “Commonly Used Disposable Plastic Bags as a Source of Microplastic in Environment,” in Proceedings of the International Conference on Microplastic Pollution in the Mediterranean Sea. eds.
Zhang, J., Gao, D., Li, Q., Zhao, Y., Li, L., Lin, H., et al. (2020). Biodegradation of polyethylene microplastic particles by the fungus aspergillus flavus from the guts of wax moth galleria mellonella. Sci. Total Environ. 704:135931. doi: 10.1016/j.scitotenv.2019.135931
Zhu, N., Liu, J., Yang, J., Lin, Y., Yang, Y., Ji, L., et al. (2016). Comparative analysis of the secretomes of Schizophyllum commune and other wood-decay basidiomycetes during solid-state fermentation reveals its unique lignocellulose-degrading enzyme system. Biotechnol. Biofuels 9:42. doi: 10.1186/s13068-016-0461-x
Keywords: bioprospecting, bioremediation, plastic biodegradation, biotechnological tools, enzyme activity, microbial chassis
Citation: De Jesus R and Alkendi R (2023) A minireview on the bioremediative potential of microbial enzymes as solution to emerging microplastic pollution. Front. Microbiol. 13:1066133. doi: 10.3389/fmicb.2022.1066133
Received: 11 October 2022; Accepted: 29 November 2022;
Published: 02 March 2023.
Edited by:
Haibo Zhang, Qingdao Institute of Bioenergy and Bioprocess Technology (CAS), ChinaReviewed by:
Shivika Sharma, Sardar Swaran Singh National Institute of Renewable Energy, IndiaCopyright © 2023 De Jesus and Alkendi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruwaya Alkendi, cnV3YXlhYUB1YWV1LmFjLmFl
†ORCID: Rener De Jesus, https://orcid.org/0000-0002-5618-4000
Ruwaya Alkendi, https://orcid.org/0000-0003-2632-1163%0D
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.