
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 24 November 2022
Sec. Infectious Agents and Disease
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1062399
This article is part of the Research TopicInsights in Infectious Agents and Disease: 2022View all 41 articles
The extended concept of one health integrates biological, geological, and chemical (bio-geo-chemical) components. Anthropogenic antibiotics are constantly and increasingly released into the soil and water environments. The fate of these drugs in the thin Earth space (“critical zone”) where the biosphere is placed determines the effect of antimicrobial agents on the microbiosphere, which can potentially alter the composition of the ecosystem and lead to the selection of antibiotic-resistant microorganisms including animal and human pathogens. However, soil and water environments are highly heterogeneous in their local composition; thus the permanence and activity of antibiotics. This is a case of “molecular ecology”: antibiotic molecules are adsorbed and eventually inactivated by interacting with biotic and abiotic molecules that are present at different concentrations in different places. There are poorly explored aspects of the pharmacodynamics (PD, biological action) and pharmacokinetics (PK, rates of decay) of antibiotics in water and soil environments. In this review, we explore the various biotic and abiotic factors contributing to antibiotic detoxification in the environment. These factors range from spontaneous degradation to the detoxifying effects produced by clay minerals (forming geochemical platforms with degradative reactions influenced by light, metals, or pH), charcoal, natural organic matter (including cellulose and chitin), biodegradation by bacterial populations and complex bacterial consortia (including “bacterial subsistence”; in other words, microbes taking antibiotics as nutrients), by planktonic microalgae, fungi, plant removal and degradation, or sequestration by living and dead cells (necrobiome detoxification). Many of these processes occur in particulated material where bacteria from various origins (microbiota coalescence) might also attach (microbiotic particles), thereby determining the antibiotic environmental PK/PD and influencing the local selection of antibiotic resistant bacteria. The exploration of this complex field requires a multidisciplinary effort in developing the molecular ecology of antibiotics, but could result in a much more precise determination of the one health hazards of antibiotic production and release.
The concept of “molecular ecology” was proposed in 1976 by the biochemist Carlos Asensio as a field of investigation of the fate and the interactions among molecules in a local or global chemosphere (Asensio, 1976). In this review, we consider the interactions between antibiotics and inorganic and organic molecules in the environment, and the potential consequences of these interactions on the “critical zone” of human health and welfare: the Earth’s thin microbiosphere (Brantley et al., 2007). One of these consequences is the emergence, selection, or spread of antibiotic resistant bacteria (Grenni et al., 2018). This ecological approach could help to predict the environmental effects of antimicrobial agents on the inorganic and organic composition of specific environments in a variety of conditions.
It has been estimated that at least one-half of all antibiotics used in human and animal diseases (including antibiotic production industry) and in farming activities are released into the water and soil environments (Fukahori et al., 2011; Dutta and Mala, 2020). There, they interact with a complex bio and chemosphere under the influence of environmental factors, such as light, temperature, and pH (Zhi et al., 2019). The fate of antibiotic activity in the environment, and therefore the impact produced on the ecosystem (Cycoń et al., 2019), is a result of geochemical and biological interactions including geo-and bio-adsorption, accumulation, and degradation; most of which remain under-investigated. For further reading, comprehensive reviews on antibiotics in the environment are available (Baquero et al., 2008; Kümmerer, 2009a,b; Sodhi et al., 2021). The fate and effects of drugs, including antibiotics, in the environment should be understood in a changing context: the global annual growth rate of the pharmaceutical industry is estimated to be 6.5% due to factors as age, life span expectancy, economic growth, intensified livestock practices, and exacerbation of diseases due to climate change (OECD, 2019 Pharmaceutical residues in freshwater: hazards and policy responses. Global Chemicals Outlook. United Nations Environment Programme, 2019).
Antibiotics lose their effects at varying rates due to spontaneous molecular alterations. The estimated half-life of antibiotics differs in relation with its concentration and the type (abiotic and biotic composition) of soil, and, certainly, also by sampling-analytical procedures (Parthasarathy et al., 2018). As an indication, fluoroquinolones have a longer half-life (more than 5 years), followed by macrolides (2–3 years), tetracyclines (2 years), sulfonamides (2–3 months), and beta-lactams (days; Cycoń et al., 2019). Even the more persistent antibiotics, such as the quinolones or macrolides, are degraded and mineralized over a long period of time (Topp et al., 2016). For beta-lactams, the cleavage readiness of their beta-lactam bonds in aqueous solution is dependent on the pH and on the chemical structure of the drug (Yamana and Tsuji, 1976). Antibiotics with a higher h-Woodward-Fieser value as a measure of chemical reactivity are more prone to hydrolysis; for example, carbapenems and clavulanate are easily decomposed and monobactams are less susceptible (Turner et al., 2022). The final result of these processes is mineralization (the transformation of antibiotics into inorganic forms), or their transformation into smaller, simpler, and inactive organic compounds if mineralization is not complete (Bridgham and Ye, 2013; Adeyemi et al., 2021).
Antibiotics may adsorb many organic and inorganic surfaces due to electrostatic interactions, π-π bonding, weak Van der Waal forces, H-bonding, and surface complexation (Mangla et al., 2022). Antibiotics have variable adsorption coefficients (Kds) to soil materials. Biosphere proteins are ubiquitous in the soil (mostly originating from dead cells), and protein-binding of some groups of antibiotics can detoxify them. Geogenic organic carbons, also from anthropogenic origins (e.g., biochar and graphite), and also bentonite, humic and fulvic substances (the final break-down constituents of the natural decay of plant and animal materials) and clay minerals adsorb antibiotics. Consequently, these substances can be used to remove antibiotics from the environment (Ahmed et al., 2015; Wang et al., 2018; Zethof et al., 2019). However, given that antibiotics accumulate within these compounds but are not degraded, this accumulation might alter the structure of the associated microbiomes. These and other aspects will be discussed in the following sections.
Colloidal soil particles such as clay minerals are frequent in the environment and they mainly form fine-grained sediments and rocks. They are an important component of soils and sediments from rivers, lakes, estuarine, delta, and oceans, which cover most of the Earth’s surface. Clay minerals consist of particulated hydrous aluminum phyllosilicates, with a characteristic stratified structure formed by sheets with varying topologies, typically tetrahedral, and octahedral sheets with a size slightly larger than a bacterial cell. Such structures act as “chemically active geochemical platforms” that influence bacterial metabolism (Rong et al., 2007), where inorganic (e.g., metals) and organic molecules (e.g., antibiotics) adsorb and interact. The adsorption rate can be high, with maximum adsorption capacities over 100 mg/g (Hacıosmanoğlu et al., 2022; Yin et al., 2022). Clay platforms located where water and light are available serve to accelerate processes influencing chemical modifications, including photochemical transformations of antibiotics, which can triple (in the case of tetracycline) the rate of modifications in pure water without colloids (Liu et al., 2019).
The mechanism involved in antibiotic degradation and detoxification in wet mineral clays mostly depends on oxidation and super-oxidation processes, which are accelerated by light and metals, in a pH-dependent process (Ahmad et al., 2021). Photolysis (also known as photodecomposition, photodissociation, or photodegradation) is modulated by the presence of dissolved inorganic (e.g., nitrates) and organic matter (e.g., humic acids; Andreozzi et al., 2003; Zhan et al., 2006). The net result is oxidative modification and degradation of the antibiotic chemical structure, which attacks the double bonds, aromatic rings, and functional groups essential for antibiotic activity. A key process in this catalytic degradation are the Fenton/Fenton-like reactions associated with the iron redox cycle, in which the antibiotic plus an hydroxyl radical gives rise to a middle product and OH-and ultimately CO2 and H2O (Jiang et al., 2022). Iron-rich minerals in the environments (such as biotite, Fe-smectite, jarosite, magnetite, pyrite, hematite, amphibole, and goethite) contribute to the antibiotics’ and other organic compounds’ mineralization processes, producing simpler organic compounds if mineralization is not complete (Bridgham and Ye, 2013; Meyer et al., 2015). Also, antibiotics adsorption and degradation due to hydroxides/oxides of Cu2+ and by Cu+ atomic species probably occurs in nature (Oliveira et al., 2018). Natural and human-produced (present and past) vegetation fires lead to a considerable increase of charcoal into soils (González-Pérez et al., 2004). Most probably, part of this charcoal could be naturally activated into highly porous charcoal, very efficient in adsorbing and inactivating antibiotics (Liao et al., 2013; Zhang et al., 2016).
Organic matter (particulate or dissolved) from natural waters is photochemically reactive (Cottrell et al., 2013), being able to degrade antibiotics. Direct photodegradation occurs by sunlight absorption, and indirect photolysis involves reactions with reactive photo-induced species as singlet oxygen (1O2), hydroxyl radicals (HO●), and the triplet excited state of chromophoric dissolved organic matter (3CDOM*) formed in natural waters. Those photochemical effects have been detected in aminoglycosides (Li et al., 2016). This effect is complex; for example in tetracyclines indirect photolysis might be enhanced, but direct tetracycline photolysis (sunlight absorption) can be inhibited (Song et al., 2021). The effects are highly dependent on factors such as pH and water depth (Lastre-Acosta et al., 2019).
Antibiotics absorb to natural polymers ubiquitous in the soil and water. Cellulose and chitin are the most abundant biopolymer polysaccharides in the environment. Cellulose exposed hydroxyl and reduced and nonreduced end groups, facilitating reactivity with pollutants, is mostly found in plant cell walls, but bacteria and algae also biosynthesize cellulose (Sayen et al., 2018; Tao et al., 2020; Juela, 2021). Decontamination preparations using cellulose derivatives adsorb a variety of antibiotics, such as tetracyclines, quinolones, sulfonamides, chloramphenicol, beta-lactams, and macrolides (in order from higher to lower absorption; Yao et al., 2017). Chitin is present in variety of soil and water invertebrates, usually in the surface exoskeleton of arthropods such as crustaceans, and in the cuticle or extracellular matrix of insects, fungi, sponges, mollusks, and nematodes. Chitin is a good adsorber of some antibiotic agents (Tunç et al., 2020). The fact that soil animals constitute about one-quarter of all animals on Earth is frequently overlooked, but it suggests that the influence of soil invertebrates might play a significant, largely ignored role in the fate of antibiotics and, in general, in the ecosystem (Lavelle et al., 2006; Zhu et al., 2019). Chitosan (deacetylated chitin) is not a known natural compound in the environment, but it can be used in environmental antibiotic de-contamination processes (Abd El-Monaem et al., 2022).
One of the classic proposals regarding environmental effects and the natural degradation of antibiotics is the publication by Julian Davies, suggesting that antibiotic-producing microorganisms probably also contain mechanisms of antibiotic detoxification to avoid self-suicide of the population (Davies, 1994; Davies and Davies, 2010). Another possibility is that antibiotics could serve as weapons in “microbial wars,” essentially as defense mechanisms against competing organisms with antibiotic producers, to ensure permanence in their optimal niche. Antibiotic production is critical in sporulating microorganisms; their synthesis is triggered during the stationary phase of growth, which leads to spore formation. Ultimately, this energy-consuming process can require the degradation of mycelium (Streptomyces) or the mother cell (Bacillus; Yagüe et al., 2013; Roy et al., 2015). Since such self-nutrients should not be consumed by foreign microorganisms, such as bacteria and probably also Protozoa (Ahmetagic et al., 2011); the production of antibiotics against these competitors could prevent such consumption. If this hypothesis is true, a possible reaction of the potential invaders would be to biodegrade these inhibitory compounds. If antibiotic-producing microorganisms or widespread antibiotic resistant bacteria release a sufficient quantity of antibiotic-degrading molecules into the environment, this could impact the fate of antibiotics. However, the natural role of antibiotics in the environment could also be associated with cell-to cell communication; that is, “antibiotics as signaling agents” (Linares et al., 2006; Yim et al., 2006; Fajardo and Martínez, 2008; Aminov, 2009). By nature, signals should be ephemeral and should vanish after accomplishing their communication role. In fact, polymyxins (produced by Bacillaceae in relation with the sporulation process) are frequently hydrolyzed by Bacillus and Paenibacillus, but peptidases from Gram-negatives might also degrade these antibiotics (Yin et al., 2019).
The possibility of antibiotics serving as carbon or nitrogen nutrients cannot be ruled out. The term “antibiotic subsistence” was coined to refer to microbial organisms and communities subsisting on antibiotics (Dantas et al., 2008), a hypothesis suggested by Tony Medeiros in the 1990s (Medeiros, 1997). Several soil microorganisms, including Pseudomonas and Burkhordelia are able to grow on beta-lactams as a single carbon source (Jayaraman, 2009; Crofts et al., 2017). A later and broader study revealed that Burkholderiales, Pseudomonadales, Enterobacterales (mostly Serratia), Actinomycetales, Rhizobiales, and Sphingobacteriales from soil origin are able to subsist on antibiotics as a sole carbon source, and the spectrum of biodegraded antibiotics includes not only beta-lactams, but aminoglycosides, chloramphenicol, glycopeptides, quinolones and fluoroquinolones, sulphonamides, and trimethoprim (Dantas et al., 2008). These phenomena might also occur in the gut microbiota, and antibiotic-subsisting organisms from sewage communities might contribute to environmental antibiotic degradation, reducing selection for resistance (Perri et al., 2020; Deng et al., 2021; Lindell et al., 2022). A mechanism of extracellular molecular scavenging involving putrescine and lipocalins protects Burkholderia cenocepacia from bactericidal antibiotics, perhaps via an antioxidant effect (Naguib et al., 2022). All these functions imply that natural antibiotics should be present in the environment to fulfill ecological functions. However, as stated earlier, anthropogenic pollution with industrial antibiotics is a major source of antibiotics in the environment (Dantas et al., 2008).
At first glance, biotransformation of antibiotics with bacteria could represent a challenge due to the possible effect on biodegrading organisms (Olicón-Hernández et al., 2017). However, this effect is mitigated by non-microbial degradation and the typically low antibiotic concentrations in natural ecosystems (Bernier and Surette, 2013). In addition, antibiotic resistance could have evolved as a prior step to antibiotic catabolism with nutritional purposes, as can occur with aminoglycosides (de Bello González et al., 2015). On the other hand, we cannot rule out the possibility that biodegradative pathways with nutritional or signal-effacing purposes could be at the root of antibiotic resistance (Dantas et al., 2008; Lindell et al., 2022). Environmental microorganisms can degrade antibiotics in the environment by methyl-hydroxylation; aliphatic-aromatic rings hydroxylation; alcohols and amines oxidation; reduction of carboxyl groups; removal of methyl, carboxyl, fluoro, and cyano groups; addition of formyl, acetyl, nitrosyl, and cyclopentenone groups; opening aromatic rings, altering the loop structures, or removing functional chemical groups (Parshikov and Sutherland, 2012). For example, demethylations exerted by Klebsiella or Stenothrophomonas maltophilia can start the degradative process in tetracyclines (Leng et al., 2016; Ahmad et al., 2021). In the case of Pseudomonas and Burkhordelia, beta-lactam degradation occurs by co-expression of a β-lactamase, amidase, and up-regulation of phenylacetic acid catabolon (Crofts et al., 2018). Klebsiella pneumoniae and Proteus mirabilis could degrade ciprofloxacin in vitro by using mechanisms of hydroxylation, piperazine ring substitution and cleavage, and quinoline ring cleavage (Yang Y. et al., 2022). Labrys portucalensis, an alfa-Proteobacteia, also degrades fluoroquinolones (Amorim et al., 2014). Bacterial consortia could be more effective in antibiotic degradation; an ensemble of Acetobacterium, Desulfovibrio, Desulfobulbus, Peptococcaceae, Lentimicrobium, and Petrimonas might contribute to trimethoprim degradation in anaerobic conditions (Liang et al., 2019). In fact, consortia have been constructed on the bases of their high production of oxidases to increase biotransformation of antibiotics (Xu et al., 2022). Soil bacterial consortia efficiently degrade sulfonamides (Islas-Espinoza et al., 2012). Complex bacterial communities can be highly effective in antibiotic biodegradation, as has been described in the case of a consortium of Gamma, Beta-Proteobacteria, and Bacteroidetes degrading ciprofloxacin by deamination, hydroxylation, defluorination, and dealkylation (Liao et al., 2016). In this process, coupled with photocatalysis, Proteobacteria are particularly critical (Li et al., 2021).
Microalgae are prokaryotic and eukaryotic micro-organisms that can fix organic (autotrophic) and inorganic (heterotrophic) carbon. Cyanobacteria is probably the most common prokaryotic microalgae (Leng et al., 2020), and it significantly contributes to antibiotic removal via a process involving (as it was shown for tetracycline) biosorption and photodegradation (Pan et al., 2021; Wei et al., 2021). Eukaryotic microalgae include diatoms and green algae. Diatoms produce hydrogen peroxide (H2O2), which modifies and detoxifies complex organic molecules including antibiotics. A key mechanism in this process is the bio-Fenton reaction, which degrades hydrogen peroxide in the presence of iron particles, giving rise to the degradation of antibiotics, as has been shown with tetracycline (Pariyarath et al., 2021). Planktonic green algae can also degrade antibiotics. Early studies on antibiotics in the environment showed that green algae (genus Nitella) absorbed beta-lactams, phenicols, and aminoglycosides (Pramer, 1955). Scenedesmus obliquus is a frequent alga found in fresh and brackish water, particularly under conditions of anthropogenic pollution (Phinyo et al., 2017). It can degrade fluoroquinolones (such as levofloxacin) using a metabolic degrading pathway including cellular biocatalytic reactions including decarboxylation, demethylation, dihydroxylation, side chain breakdown, and ring cleavage (Xiong et al., 2017). The rate of antibiotic biodegradation (dissipation percentage) is variable among microalgae and various types of antibiotics. Selenastrum capricornutum and Chlorella vulgaris more efficiently degrade macrolides and fluoroquinolones than sulphonamides, which are better degraded by Scenedesmus quadricauda and Haematococcus pluvialis (Kiki et al., 2020). Microalgae communities with other microorganisms, such as filamentous fungi, could have synergistic effects in antibiotic detoxification (Leng et al., 2020).
Fungi belonging to the Basidiomycota, Ascomycota, and Mucoromycotina (formerly Zygomicota) subphyla can remove and transform antibiotic molecules. Ciprofloxacin is detoxified by conjugation with formyl, vinyl, or acetyl groups, or by hydroxylation or polymerization (Olicón-Hernández et al., 2017). Aspergillus and Penicillium appear to decrease the amount of ciprofloxacin in soil, but the underlying mechanism has not been elucidated; Trichoderma produce ciprofloxacin-conjugated inactive compounds when incubated with fluoroquinolones. Mucoromycotina incertae sedis (formerly Zygomycota) are also able to detoxify fluoroquinolones in a process involving N-oxidation, N-dealkylation, and N-acetylation. White-rot fungi (as the Basidiomycota Pleurotus eryngii or Trametes versicolor), widespread in nature due to their capability to degrade ubiquitous lignin, induce the production of extracellular low-molecular-weight extracellular oxidants, including oxygen-free radicals, mainly hydroxyl radicals, and lipid peroxidation radicals activating O2 in the environment and removing pollutants (Gómez-Toribio et al., 2009), most probably also antibiotic molecules.
Independently from antibiotic adsorption to plant residues (Balarak et al., 2017), living plants may absorb a variety of antibiotics present in the soil, including anthropogenic quinolones (Eggen et al., 2011) This process is highly antibiotic dependent; for example, absorption is high for tetracyclines and low for macrolides (Kumar et al., 2005). Also, the type of plant determines absorption; oxytetracycline is accumulated in radish roots but not in lettuce leaves (Matamoros et al., 2022). Macrophytes such as duckweed and water fern absorb antibiotics by the roots and detoxify them by oxidation, conjugation, and storage in the plant (Maldonado et al., 2022). One example of such degradation is duckweed Spirodela polyrhiza, which degrades fluoroquinolones (Singh et al., 2019).
After cell death, cellular components can remain for extended periods of time in the soil or water. Many antibiotics, particularly macrolides, lincosamides, fluoroquinolones, tetracyclines, rifamycins, chloramphenicol, trimethoprim, and sulfonamides, and also beta-lactams to a lesser extent, enter eukaryotic cells where they are sequestered and inactivated. Interestingly, extracellular antibiotics have more activity than intracellular ones, although some of them accumulate intracellularly and reach high concentrations. One reason for the reduced activity of intracellular antibiotics is a presumed “impairment of the expression of antibiotic activity inside the cells” (van Bambeke et al., 2006). This field is important but poorly explored, and we know from human clinical trials that a renal dipeptidase, dehydropeptidase-I, can hydrolyze imipenem and other carbapenems (Birnbaum et al., 1985). Also, human and mammal liver microsomes (mimicking the activity of the endoplasmic reticulum) are able to biotransform fluoroquinolones, lincosamides, fluconazole, gentamicin, metronidazole, oxazolidinones, and even beta-lactams (Wynalda et al., 2000; Szultka et al., 2014; Szultka and Buszewski, 2016). Whether these results apply to other eukaryotic microsomes (including algae, fungi, plants, small animals, and protozoa) is not yet known. Nevertheless, an antioxidant defense mechanism is activated and glutathione S-transferase activity is significantly increased in aquatic plants such as Azolla caroliniana and Taxiphyllum barbieri exposed to tetracycline (Vilvert et al., 2017). Glutathione S-transferases, present in bacteria, fungi, plants, and animals inhibit beta-lactams sulfathiazole and tetracycline (Al-Mohaimeed et al., 2022).
Many antibiotics can be ultimately inactivated in matrixes constituted by massive amounts of dead bacteria (eventually killed by the antibiotics themselves; Hunt et al., 1987; Podlesek et al., 2016). This adsorption/detoxification of antibiotics by dead bacterial cells might be common in natural environments (Smakman and Hall, 2022). Envelopes of dead bacteria (such as lipopolysaccharide) and probably proteins (as in the case of “inoculum effect”) might bind to antibiotics (Peterson et al., 1985; Corona and Martínez, 2013). The same is possible with free DNA or RNA ribosomal fragments. Both aminoglycosides and beta-lactams can bind to the DNA helix via a minor groove binding model (Arya, 2005; Shahabadi and Hashempour, 2019).
This review is oriented toward antibiotic “natural detoxification” in wild environments. However, human activities increasingly contribute to the composition of the Earth global environment. Most clinically-used antibiotics are released in areas close to densely human-populated patches, where also farming, agricultural, and industrial activities polluting the natural environment takes place. As stated before, metals are important agents in the detoxification of antibiotics, mostly involving oxidation and super-oxidation processes. Heavy (significant) metals pollution, involving lead, cadmium, chromium, mercury, or arsenic, and also iron, copper, cobalt, and silver are released from metal processing and smelting, chemical and manufacturing activities, factory emissions, and sewage irrigation (Yang et al., 2018). Even if metals might detoxify antibiotics, they also have antibacterial activities, frequently synergistic with clinical antimicrobial agents, and antibiotic-resistance genes are frequently found in antibiotic resistant bacteria, contributing to the evolutionary biology of these organisms (Baquero et al., 2021). Thus detoxification might be compensated by an enhanced antimicrobial effect, resulting in a stronger selection. Oil–water interfaces might influence antibiotic degradation (Basáez and Vanýsek, 1999) but also contribute to bacterial aggregations, as marine bacteria in oil spill (Ahmadzadegan et al., 2019), so that selection for antibiotic resistance effectively occurs (Shen et al., 2020). Water chlorination (partially?) detoxifies some antibiotics as azithromycin or fluoroquinolones (Jaén-Gil et al., 2020) but eventually have additive or synergistic effects with these drugs. The result is a decrease the bacterial count, which not excludes increased selection of resistance; in any case, this field has been scarcely explored. Bacteria from minerally-fertilized soils and crops reduce their content in antimicrobial resistance genes (Sanz et al., 2022), suggesting that chemical compounds, such as ammonium, sodium, or potassium sulfates, or superphosphates might reduce the selective effect of environmental antibiotics. Organic fertilizers, as pig manure and sewage sludge contain bacterial consortia able to detoxify antibiotics (see above), but it is to note that this effect could be compensated by the heavier pollution with antibiotic molecules originated in abusive use of antibiotics in humans and animals (Dong et al., 2021). Industrial composting (organic matter recycling) removes antibiotics and alters the local microbial ecology (Chen et al., 2021). Pollution by anthropogenic microplastics, is another aspect of anthropogenic pollution. Microplastics, small (less than 5 mm in length) fragments of any type of plastic, which also adsorb/detoxify antibiotics but also adhere bacterial cells and therefore contribute to the selection of resistance (Peng et al., 2022; Wang et al., 2022).
Deactivation of antibiotics in the environment should be beneficial for reducing selection and the environmental reservoir of antibiotic resistant bacteria. However, many mineral and biotic biomolecules deactivating antimicrobial agents are also part of soil and water particles that attach bacterial cells and in some cases are of anthropogenic origin (Baquero et al., 2022; Cydzik-Kwiatkowska et al., 2022). The key question is whether the adhesion of antibiotic molecules to these particles concentrates the antimicrobial agent, so that even if deactivation takes place, there could be enough selective power to select antibiotic resistant co-adhered cells. For example, if clays are able to remove antibiotics from the environment, high antibiotic concentrations would be found on the surface of clay particles, thereby contributing to the local selection of antibiotic resistant microorganisms (Lv et al., 2019). What we really need is to know better the PK/PD of antibiotics on environmental surfaces where bacteria might attach; in particular, if attached bacteria are phenotypically resistant to antibiotics, and how much of this effect is due to the local enrichment in persistent cells (because of the superoxide’s action). We cannot discard an antibiotic action on attached cells, but that should depend on the rates of absorption and desorption of the different antibiotics (Figure 1) and the local bacterial growth rates. The local microecological conditions as light and water availability, temperature, osmolarity, and pH are expected to modify such kinetics.
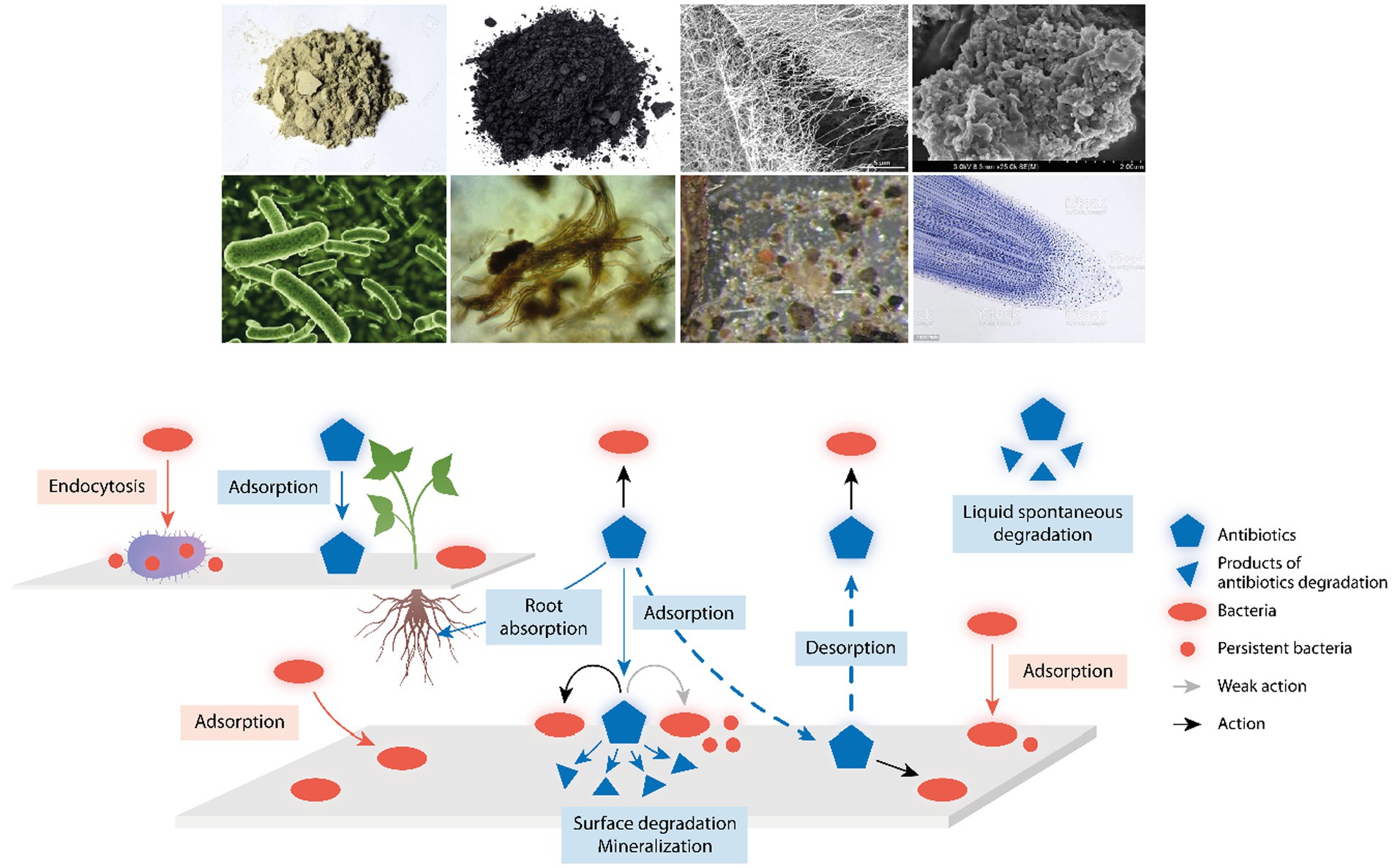
Figure 1. Environmental components influencing antibiotic action and detoxification. Up in the figure, examples of environmental components where antibiotics and bacterial cells might attach; from up left to down right, clay, charcoal, cellulose fibers, natural organic matter, bacteria, fungi, microfauna, and tip of plant roots. Down in the figure, a schema with a reactive surface from the above components where antibiotics (blue pentagons) and bacteria (red ovals) might attach; antibiotics can act (black arrows) or only weakly act (gray arrow) on sessile or attached bacteria; small blue triangles are the products of antibiotic degradation; and small red circles are persistent bacteria, phenotypically resistant to antibiotic action. The central broken arrows illustrate the key process of absorption/desorption of antibiotics in environmental surfaces, determining the PK/PD of these drugs, and therefore, the local selection of antibiotic resistant bacteria. Up left, a reactive surface illustrating the capture of bacteria by protozoa (as ciliates) favoring the release of bacterial pellets with antibiotic-persistent organisms, and the absorptive process of antibiotics by plant roots.
Absorption, photolysis, hydrolysis, cation-binding, adsorption, bioaccumulation, and biodegradation simultaneously contribute to the removal of antibiotics from the environment (Xiong et al., 2017). Most probably, the mechanisms of antibiotic removal that we have reviewed work in combination, perhaps in synergistic detoxification, as has been observed in pollution control studies. For example, the combination of microalgae with irradiation and oxidation treatment favors antibiotic degradation (Leng et al., 2020). The increasingly complex anthropogenic influence on the environment, which releases and removes both antibiotics and antibiotic-resistant bacteria, is certain to influence the entire kinetics of antimicrobial drugs in the microbiosphere. However, current information on the effects of antibiotic detoxification in the environment is still fragmentary and a global, and an integrated and ecological view on the elements contributing to this process is needed. For example, earthworms, which change the exposure of soil organisms to ciprofloxacin, result in a much higher mineralization rate of antibiotics and illustrate the complexity of predicting the antibiotic detoxification processes (Mougin et al., 2013). We are still lacking highly efficient and comprehensive analytic procedures to dissect and quantify the chemical and biological composition of specific soil or water environments that are exposed to intensive antibiotic pollution. Such integrated analyses could help measure the hazard of antibiotic release in particular places at defined time-periods. Among the required parameters, soil volumetric water content (Briciu-Burghina et al., 2022), the total organic matter (Yang C. et al., 2022), or the “amount of surface” in the soil (for instance, total surface of clay particles) can be calculated and expressed as “specific surface area,” the surface area/unit mass of the dry soil with units of m2/g (Cerrato and Lutenegger, 2002). Antibiotics with a high adsorption potential on clay or organic matter tend to accumulate and persist in this matrix, whereas those having a lower adsorption potential are easily transported to the aquatic environment. More study of the microbial ecology of antibiotic molecules is needed, given that there are potential gaps between the analytical results obtained in the lab and the in the environment (Polianciuc et al., 2020). Pedological sciences should approach microbiology to match soil classifications with local environmental pharmacokinetics and the pharmacodynamics of antimicrobial agents. Techniques able to measure the physical and chemical adsorption of antibiotic and bacterial molecules taking up by the different types of surfaces (with different energy distributions; Webb, 2003) should be developed to reach such a goal. Everything on Earth is intertwined, and the goal of One health (Hernando-Amado et al., 2019) is fully dependent on the geochemical and biological structure of the particular environments and requires an interdisciplinary effort (Brantley et al., 2007). We need to progress toward the definition of “local bio-geo-chemical reactive profiles,” so that we can understand the reactive transport (Carrera et al., 2022) of antibiotic molecules. That step will be indispensable in shaping appropriate environmental “One health” interventions to reduce microbial resistance to antimicrobial agents.
FB conceived the topic, discussed in depth with TC and J-LM, wrote the manuscript, and prepared the figure. All authors contributed to the article and approved the submitted version.
This work, developed in the Ecoevobiome lab of the Department of Microbiology of the Ramón y Cajal Hospital (IRYCIS; http://www.ecoevobiome.org/), was supported by the European Union—Next Generation EU (MISTAR Project—AC21_2/00041) and the Instituto de Salud Carlos III (ISCIII) of Spain, also funded by ISCIII through project PI21/02027, co-financed by the European Union, and by the Spanish Network for Research in Infectious Diseases, CIBERINFEC (CB21/13/00084), and CIBERESP (CB06/02/0053).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abd El-Monaem, E. M., Eltaweil, A. S., Elshishini, H. M., Hosny, M., Abou Alsoaud, M. M., Attia, N. F., et al. (2022). Sustainable adsorptive removal of antibiotic residues by chitosan composites: An insight into current developments and future recommendations. Arab. J. Chem. 15:103743. doi: 10.1016/j.arabjc.2022.103743
Adeyemi, J. O., Ajiboye, T., and Onwudiwe, D. C. (2021). Mineralization of antibiotics in wastewater via photocatalysis. Water Air Soil Pollut. 232, 1–28. doi: 10.1007/s11270-021-05167-3
Ahmad, F., Zhu, D., and Sun, J. (2021). Environmental fate of tetracycline antibiotics: degradation pathway mechanisms, challenges, and perspectives. Environ. Sci. Eur. 33, 1–17. doi: 10.1186/s12302-021-00505-y
Ahmadzadegan, A., Wang, S., Vlachos, P. P., and Ardekani, A. M. (2019). Hydrodynamic attraction of bacteria to gas and liquid interfaces. Phys. Rev. E 100:062605. doi: 10.1103/PhysRevE.100.062605
Ahmed, M. B., Zhou, J. L., Ngo, H. H., and Guo, W. (2015). Adsorptive removal of antibiotics from water and wastewater: Progress and challenges. Sci. Total Environ. 532, 112–126. doi: 10.1016/j.scitotenv.2015.05.130
Ahmetagic, A., Philip, D. S., Sarovich, D. S., Kluver, D. W., and Pemberton, J. M. (2011). Plasmid encoded antibiotics inhibit protozoan predation of Escherichia coli K12. Plasmid 66, 152–158. doi: 10.1016/j.plasmid.2011.07.006
Al-Mohaimeed, A. M., Abbasi, A. M., Ali, M. A., and Dhas, D. D. (2022). Reduction of multiple antibiotics from the waste water using coated glutathione S-transferase producing biocatalyst. Environ. Res. 206:112262. doi: 10.1016/j.envres.2021.112262
Aminov, R. I. (2009). The role of antibiotics and antibiotic resistance in nature. Environ. Microbiol. 11, 2970–2988. doi: 10.1111/j.1462-2920.2009.01972.x
Amorim, C. L., Moreira, L. S., Maia, A. S., Tiritan, M. E., and Castro, P. M. (2014). Biodegradation of ofloxacin, norfloxacin, and ciprofloxacin as single and mixed substrates by Labrys portucalensis. Appl. Microbiol. Biotechnol. 98, 3181–3190. doi: 10.1007/s00253-013-5333-8
Andreozzi, R., Raffaele, M., and Nicklas, P. (2003). Pharmaceuticals in STP effluents and their solar photodegradation in aquatic environment. Chemosphere 50, 1319–1330. doi: 10.1016/S0045-6535(02)00769-5
Arya, D. P. (2005). Aminoglycoside–nucleic acid interactions: the case for neomycin. Top. Curr. Chem. 253, 149–178. doi: 10.1007/b100446
Asensio, C. (1976). “Molecular ecology” in Reflections in Biochemistry. eds. A. Kornberg, B. L. Horecker, L. Cornudella, and J. Oro (Oxford: Pergamon Press), 235–240.
Balarak, D., Joghatayi, A., Mostafapour, F. K., and Azarpira, H. (2017). Biosorption of amoxicillin form contaminated water onto palm bark biomass. Int. J. Life Sci. Pharm. Res. 7, 1–8.
Baquero, F., Coque, T. M., Guerra-Pinto, N., Galán, J. C., Jiménez-Lalana, D., Tamames, J., et al. (2022). The influence of coalescent microbiotic particles from water and soil on the evolution and spread of antimicrobial resistance. Front. Environ. Sci. 10:824963. doi: 10.3389/fenvs.2022.824963
Baquero, F., Martínez, J. L., and Cantón, R. (2008). Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 19, 260–265. doi: 10.1016/j.copbio.2008.05.006
Baquero, F., Martínez, J. L., Lanza, V. F., Rodríguez-Beltrán, J., Galán, J. C., San Millán, A., et al. (2021). Evolutionary pathways and trajectories in antibiotic resistance. Clin. Microbiol. Rev. 34, e00050–e00119. doi: 10.1128/CMR.00050-19
Basáez, L., and Vanýsek, P. (1999). Transport studies of β-lactam antibiotics and their degradation products across electrified water/oil interface. J. Pharm. Biomed. Anal. 19, 183–192. doi: 10.1016/S0731-7085(98)00147-2
Bernier, S. P., and Surette, M. G. (2013). Concentration-dependent activity of antibiotics in natural environments. Front. Microbiol. 4:20. doi: 10.3389/fmicb.2013.00020
Birnbaum, J., Kahan, F. M., Kropp, H., and Mac Donald, J. S. (1985). Carbapenems, a new class of beta-lactam antibiotics. Discovery and development of imipenem/cilastatin. Am. J. Med. 78, 3–21. doi: 10.1016/0002-9343(85)90097-X
Brantley, S. L., Goldhaber, M. B., and Ragnarsdottir, K. V. (2007). Crossing disciplines and scales to understand the critical zone. Elements 3, 307–314. doi: 10.2113/gselements.3.5.307
Briciu-Burghina, C., Zhou, J., Ali, M. I., and Regan, F. (2022). Demonstrating the potential of a low-cost soil moisture sensor etwork. Sensors 22:987. doi: 10.3390/s22030987
Bridgham, S. D., and Ye, R. (2013). Organic matter mineralization and decomposition. Meth. Biogeochem. Wetl. 10, 385–406. doi: 10.2136/sssabookser10.c20
Carrera, J., Saaltink, M. W., Soler-Sagarra, J., Wang, J., and Valhondo, C. (2022). Reactive transport: a review of basic concepts with emphasis on biochemical processes. Energies 15:925. doi: 10.3390/en15030925
Cerrato, A. B., and Lutenegger, A. J. (2002). Determination of surface area of fine-grained soils by the ethylene glycol monoethyl ether (EGME) method. Geotech. Test. J. 25, 315–321.
Chen, Z., Fu, Q., Cao, Y., Wen, Q., and Wu, Y. (2021). Effects of lime amendment on the organic substances changes, antibiotics removal, and heavy metals speciation transformation during swine manure composting. Chemosphere 262:128342. doi: 10.1016/j.chemosphere.2020.128342
Corona, F., and Martínez, J. L. (2013). Phenotypic resistance to antibiotics. Antibiotics 2, 237–255. doi: 10.3390/antibiotics2020237
Cottrell, B. A., Timko, S. A., Devera, L., Robinson, A. K., Gonsior, M., Vizenor, A. E., et al. (2013). Photochemistry of excited-state species in natural waters: a role for particulate organic matter. Water Res. 47, 5189–5199. doi: 10.1016/j.watres.2013.05.059
Crofts, T. S., Wang, B., Spivak, A., Gianoulis, T. A., Forsberg, K. J., Gibson, M. K., et al. (2017). Draft genome sequences of three β-lactam-catabolizing soil Proteobacteria. Genome Announc. 5, e00653–e00717. doi: 10.1128/genomeA.00653-17
Crofts, T. S., Wang, B., Spivak, A., Gianoulis, T. A., Forsberg, K. J., Gibson, M. K., et al. (2018). Shared strategies for β-lactam catabolism in the soil microbiome. Nat. Chem. Biol. 14, 556–564. doi: 10.1038/s41589-018-0052-1
Cycoń, M., Mrozik, A., and Piotrowska-Seget, Z. (2019). Antibiotics in the soil environment—degradation and their impact on microbial activity and diversity. Front. Microbiol. 10:338. doi: 10.3389/fmicb.2019.00338
Cydzik-Kwiatkowska, A., De Jonge, N., Poulsen, J. S., and Nielsen, J. L. (2022). Unraveling gradient layers of microbial communities, proteins, and chemical structure in aerobic granules. Sci. Total Environ. 829:154253. doi: 10.1016/j.scitotenv.2022.154253
Dantas, G., Sommer, M. O., Oluwasegun, R. D., and Church, G. M. (2008). Bacteria subsisting on antibiotics. Science 320, 100–103. doi: 10.1126/science.1155157
Davies, J. (1994). Inactivation of antibiotics and the dissemination of resistance genes. Science 264, 375–382. doi: 10.1126/science.8153624
Davies, J., and Davies, D. (2010). Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433. doi: 10.1128/MMBR.00016-10
de Bello González, T. J., Zuidema, T., Bor, G., Smidt, H., and van Passel, M. W. J. (2015). Study of the aminoglycoside subsistence phenotype of bacteria residing in the gut of humans and zoo animals. Front. Microbiol. 6:1550. doi: 10.3389/fmicb.2015.01550
Deng, Y., Huang, Y., Che, Y., Yang, Y., Yin, X., Yan, A., et al. (2021). Microbiome assembly for sulfonamide subsistence and the transfer of genetic determinants. ISME J. 15, 2817–2829. doi: 10.1038/s41396-021-00969-z
Dong, B., Li, W., and Xu, W. (2021). Effects of partial organic substitution for chemical fertilizer on antibiotic residues in peri-urban agricultural soil in China. Antibiotics 10:1173. doi: 10.3390/antibiotics10101173
Dutta, J., and Mala, A. A. (2020). Removal of antibiotic from the water environment by the adsorption technologies: a review. Water Sci. Technol. 82, 401–426. doi: 10.2166/wst.2020.335
Eggen, T., Asp, T. N., Grave, K., and Hormazabal, V. (2011). Uptake and translocation of metformin, ciprofloxacin and narasin in forage-and crop plants. Chemosphere 85, 26–33. doi: 10.1016/j.chemosphere.2011.06.041
Fajardo, A., and Martínez, J. L. (2008). Antibiotics as signals that trigger specific bacterial responses. Curr. Opin. Microbiol. 11, 161–167. doi: 10.1016/j.mib.2008.02.006
Fukahori, S., Fujiwara, T., Ito, R., and Funamizu, N. (2011). pH-dependent adsorption of sulfa drugs on high silica zeolite: modeling and kinetic study. Desalination 275, 237–242. doi: 10.1016/j.desal.2011.03.006
Gómez-Toribio, V., García-Martín, A. B., Martínez, M. J., Martínez, Á. T., and Guillén, F. (2009). Induction of extracellular hydroxyl radical production by white-rot fungi through quinone redox cycling. Appl. Environ. Microbiol. 75, 3944–3953. doi: 10.1128/AEM.02137-08
González-Pérez, J. A., González-Vila, F. J., Almendros, G., and Knicker, H. (2004). The effect of fire on soil organic matter: a review. Environ. Int. 30, 855–870. doi: 10.1016/j.envint.2004.02.003
Grenni, P., Ancona, V., and Caracciolo, A. B. (2018). Ecological effects of antibiotics on natural ecosystems: a review. Microchem. J. 136, 25–39. doi: 10.1016/j.microc.2017.02.006
Hacıosmanoğlu, G. G., Mejías, C., Martín, J., Santos, J. L., Aparicio, I., and Alonso, E. (2022). Antibiotic adsorption by natural and modified clay minerals as designer adsorbents for wastewater treatment: a comprehensive review. J. Environ. Manag. 317:115397. doi: 10.1016/j.jenvman.2022.115397
Hernando-Amado, S., Coque, T. M., Baquero, F., and Martínez, J. L. (2019). Defining and combating antibiotic resistance from one health and Global Health perspectives. Nat. Microbiol. 4, 1432–1442. doi: 10.1038/s41564-019-0503-9
Hunt, G. H., Oberholzer, V. G., and Price, E. H. (1987). In-vitro antibiotic inactivation by mammalian cell and killed bacterial preparations. J. Antimicrob. Chemother. 19, 637–646. doi: 10.1093/jac/19.5.637
Islas-Espinoza, M., Reid, B. J., Wexler, M., and Bond, P. L. (2012). Soil bacterial consortia and previous exposure enhance the biodegradation of sulfonamides from pig manure. Microb. Ecol. 64, 140–151. doi: 10.1007/s00248-012-0010-5
Jaén-Gil, A., Farré, M. J., Sànchez-Melsió, A., Serra-Compte, A., Barceló, D., and Rodríguez-Mozaz, S. (2020). Effect-based identification of hazardous antibiotic transformation products after water chlorination. Environ. Sci. Technol. 54, 9062–9073. doi: 10.1021/acs.est.0c00944
Jayaraman, R. (2009). Antibiotic resistance: an overview of mechanisms and a paradigm shift. Curr. Sci. 96, 1475–1484.
Jiang, Y., Ran, J., Mao, K., Yang, X., Zhong, L., Yang, C., et al. (2022). Recent progress in Fenton/Fenton-like reactions for the removal of antibiotics in aqueous environments. Ecotoxicol. Environ. Saf. 236:113464. doi: 10.1016/j.ecoenv.2022.113464
Juela, D. M. (2021). Promising adsorptive materials derived from agricultural and industrial wastes for antibiotic removal: a comprehensive review. Sep. Purif. Technol. 284:120286. doi: 10.1016/j.seppur.2021.120286
Kiki, C., Rashid, A., Wang, Y., Li, Y., Zeng, Q., Yu, C. P., et al. (2020). Dissipation of antibiotics by microalgae: kinetics, identification of transformation products and pathways. J. Hazard. Mater. 387:121985. doi: 10.1016/j.jhazmat.2019.121985
Kumar, K., Gupta, S. C., Baidoo, S. K., Chander, Y., and Rosen, C. J. (2005). Antibiotic uptake by plants from soil fertilized with animal manure. J. Environ. Qual. 34, 2082–2085. doi: 10.2134/jeq2005.0026
Kümmerer, K. (2009a). Antibiotics in the aquatic environment–a review–part I. Chemosphere 75, 417–434. doi: 10.1016/j.chemosphere.2008.11.086
Kümmerer, K. (2009b). Antibiotics in the aquatic environment: a review–part II. Chemosphere 75, 435–441. doi: 10.1016/j.chemosphere.2008.12.006
Lastre-Acosta, A. M., Barberato, B., Parizi, M. P. S., and Teixeira, A. C. S. C. (2019). Direct and indirect photolysis of the antibiotic enoxacin: kinetics of oxidation by reactive photo-induced species and simulations. Environ. Sci. Pollut. Res. 26, 4337–4347. doi: 10.1007/s11356-018-2555-4
Lavelle, P., Decaëns, T., Aubert, M., Barot, S., Blouin, M., Bureau, F., et al. (2006). Soil invertebrates and ecosystem services. Eur. J. Soil Biol. 42, S3–S15. doi: 10.1016/j.ejsobi.2006.10.002
Leng, Y., Bao, J., Chang, G., Zheng, H., Li, X., Du, J., et al. (2016). Biotransformation of tetracycline by a novel bacterial strain Stenotrophomonas maltophilia DT1. J. Hazard. Mater. 318, 125–133. doi: 10.1016/j.jhazmat.2016.06.053
Leng, L., Wei, L., Xiong, Q., Xu, S., Li, W., Lv, S., et al. (2020). Use of microalgae based technology for the removal of antibiotics from wastewater: a review. Chemosphere 238:124680. doi: 10.1016/j.chemosphere.2019.124680
Li, Y., Chen, L., Tian, X., Lin, L., Ding, R., Yan, W., et al. (2021). Functional role of mixed-culture microbe in photocatalysis coupled with biodegradation: total organic carbon removal of ciprofloxacin. Sci. Total Environ. 784:147049. doi: 10.1016/j.scitotenv.2021.147049
Li, R., Zhao, C., Yao, B., Li, D., Yan, S., O’Shea, K. E., et al. (2016). Photochemical transformation of aminoglycoside antibiotics in simulated natural waters. Environ. Sci. Technol. 50, 2921–2930. doi: 10.1021/acs.est.5b05234
Liang, B., Kong, D., Qi, M., Yun, H., Li, Z., Shi, K., et al. (2019). Anaerobic biodegradation of trimethoprim with sulfate as an electron acceptor. Front. Environ. Sci. Eng. 13, 1–10. doi: 10.1007/s11783-019-1168-6
Liao, X., Li, B., Zou, R., Dai, Y., Xie, S., and Yuan, B. (2016). Biodegradation of antibiotic ciprofloxacin: pathways, influential factors, and bacterial community structure. Environ. Sci. Pollut. Res. 23, 7911–7918. doi: 10.1007/s11356-016-6054-1
Liao, P., Zhan, Z., Dai, J., Wu, X., Zhang, W., Wang, K., et al. (2013). Adsorption of tetracycline and chloramphenicol in aqueous solutions by bamboo charcoal: a batch and fixed-bed column study. Chem. Eng. J. 228, 496–505. doi: 10.1016/j.cej.2013.04.118
Linares, J. F., Gustafsson, I., Baquero, F., and Martinez, J. L. (2006). Antibiotics as intermicrobial signaling agents instead of weapons. Proc. Natl. Acad. Sci. U. S. A. 103, 19484–19489. doi: 10.1073/pnas.0608949103
Lindell, A. E., Zimmermann-Kogadeeva, M., and Patil, K. R. (2022). Multimodal interactions of drugs, natural compounds and pollutants with the gut microbiota. Nat. Rev. Microbiol. 20, 431–443. doi: 10.1038/s41579-022-00681-5
Liu, F., Liu, X., Zhao, S., Wang, J., Qian, X., Cui, B., et al. (2019). Photochemical transformations of tetracycline antibiotics influenced by natural colloidal particles: kinetics, factor effects and mechanisms. Chemosphere 235, 867–875. doi: 10.1016/j.chemosphere.2019.06.201
Lv, G., Li, Z., Elliott, L., Schmidt, M. J., MacWilliams, M. P., and Zhang, B. (2019). Impact of tetracycline-clay interactions on bacterial growth. J. Hazard. Mater. 370, 91–97. doi: 10.1016/j.jhazmat.2017.09.029
Maldonado, I., Moreno, E. G., and Vilca, F. Z. (2022). Application of duckweed (Lemna sp.) and water fern (Azolla sp.) in the removal of pharmaceutical residues in water: state of art focus on antibiotics. Sci. Total Environ. 838:156565. doi: 10.1016/j.scitotenv.2022.156565
Mangla, D., Sharma, A., and Ikram, S. (2022). Critical review on adsorptive removal of antibiotics: present situation, challenges and future perspective. J. Hazard. Mater. 425:127946. doi: 10.1016/j.jhazmat.2021.127946
Matamoros, V., Casas, M. E., Mansilla, S., Tadić, Đ., Cañameras, N., Carazo, N., et al. (2022). Occurrence of antibiotics in lettuce (Lactuca sativa L.) and radish (Raphanus sativus L.) following organic soil fertilization under plot-scale conditions: crop and human health implications. J. Hazard. Mater. 436:1290. doi: 10.1016/j.jhazmat.2022.129044
Medeiros, A. A. (1997). Evolution and dissemination of beta-lactamases accelerated by generations of beta-lactam antibiotics. Clin. Infect. Dis. 24, S19–S45. doi: 10.1093/clinids/24.Supplement_1.S19
Meyer, T. J., Ramlall, J., Thu, P., and Gadura, N. (2015). Antimicrobial properties of copper in gram-negative and gram-positive bacteria. Int J Pharm Pharm Sci 9, 274–278. doi: 10.5281/zenodo.1099692
Mougin, C., Cheviron, N., Repincay, C., Hedde, M., and Hernandez-Raquet, G. (2013). Earthworms highly increase ciprofloxacin mineralization in soils. Environ. Chem. Lett. 11, 127–133. doi: 10.1007/s10311-012-0385-z
Naguib, M., Feldman, N., Zarodkiewicz, P., Shropshire, H., Biamis, C., El-Halfawy, O. M., et al. (2022). An evolutionary conserved detoxification system for membrane lipid–derived peroxyl radicals in gram-negative bacteria. PLoS Biol. 20:e3001610. doi: 10.1371/journal.pbio.3001610
OECD (2019). Available at: https://www.oecd.org/publications/pharmaceutical-residues-in-freshwater-c936f42d-en.htm.
Olicón-Hernández, D. R., González-López, J., and Aranda, E. (2017). Overview on the biochemical potential of filamentous fungi to degrade pharmaceutical compounds. Front. Microbiol. 8:1792. doi: 10.3389/fmicb.2017.01792
Oliveira, L. M., Nascimento, M. A., Guimarães, Y. M., Oliveira, A. F., Silva, A. A., and Lopes, R. P. (2018). Removal of beta-lactams antibiotics through zero-valent copper nanoparticles. J. Braz. Chem. Soc. 29, 1630–1637. doi: 10.21577/0103-5053.20180034
Pan, M., Lyu, T., Zhan, L., Matamoros, V., Angelidaki, I., Cooper, M., et al. (2021). Mitigating antibiotic pollution using cyanobacteria: removal efficiency, pathways and metabolism. Water Res. 190:116735. doi: 10.1016/j.watres.2020.116735
Pariyarath, R. V., Inagati, Y., and Sakakibara, Y. (2021). Phycoremediation of tetracycline via bio-Fenton process using diatoms. J. Water Proc. Eng. 40:101851. doi: 10.1016/j.jwpe.2020.101851
Parshikov, I. A., and Sutherland, J. B. (2012). Microbial transformations of antimicrobial quinolones and related drugs. J. Ind. Microbiol. Biotechnol. 39, 1731–1740. doi: 10.1007/s10295-012-1194-x
Parthasarathy, R., Monette, C. E., Bracero, S. S., and Saha, M. (2018). Methods for field measurement of antibiotic concentrations: limitations and outlook. FEMS Microbiol. Ecol. 94:fiy105. doi: 10.1093/femsec/fiy105
Peng, C., Zhang, X., Zhang, X., Liu, C., Chen, Z., Sun, H., et al. (2022). Bacterial community under the influence of microplastics in indoor environment and the health hazards associated with antibiotic resistance genes. Environ. Sci. Technol. 56, 422–432. doi: 10.1021/acs.est.1c04520
Perri, R., Kolvenbach, B. A., and Corvini, P. F. (2020). Subsistence and complexity of antimicrobial resistance on a community-wide level. Environ. Microbiol. 22, 2463–2468. doi: 10.1111/1462-2920.15018
Peterson, A. A., Hancock, R. E., and McGroarty, E. J. (1985). Binding of polycationic antibiotics and polyamines to lipopolysaccharides of Pseudomonas aeruginosa. J. Bacteriol. 164, 1256–1261. doi: 10.1128/jb.164.3.1256-1261.1985
Phinyo, K., Pekkoh, J., and Peerapornpisal, Y. (2017). Distribution and ecological habitat of Scenedesmus and related genera in some freshwater resources of northern and north-eastern Thailand. J. Biol. Div. 18, 1092–1099. doi: 10.13057/biodiv/d180329
Podlesek, Z., Butala, M., Šakanović, A., and Žgur-Bertok, D. (2016). Antibiotic induced bacterial lysis provides a reservoir of persisters. Antonie Van Leeuwenhoek 109, 523–528. doi: 10.1007/s10482-016-0657-x
Polianciuc, S. I., Gurzău, A. E., Kiss, B., Ştefan, M. G., and Loghin, F. (2020). Antibiotics in the environment: causes and consequences. Med. Pharm. Rep. 93, 231–240. doi: 10.15386/mpr-1742
Pramer, D. (1955). Absorption of antibiotics by plant cells. Science 121, 507–508. doi: 10.1126/science.121.3145.507
Rong, X., Huang, Q., and Chen, W. (2007). Microcalorimetric investigation on the metabolic activity of bacillus thuringiensis as influenced by kaolinite, montmorillonite and goethite. Appl. Clay Sci. 38, 97–103. doi: 10.1016/j.clay.2007.01.015
Roy, S. V., Hajare, S. N., Gautam, S., Deobagkar, D., and Sharma, A. (2015). Sporulation-associated Mother Cell Lysis in Bacillus Displays Markers of Programmed Cell Death. Curr. Sci. 109, 1283–1292. doi: 10.18520/cs/v109/i7/1283-1292
Sanz, C., Casado, M., Navarro-Martin, L., Cañameras, N., Carazo, N., Matamoros, V., et al. (2022). Implications of the use of organic fertilizers for antibiotic resistance gene distribution in agricultural soils and fresh food products. Sci. Total Environ. 815:151973. doi: 10.1016/j.scitotenv.2021.151973
Sayen, S., Ortenbach-Lopez, M., and Guillon, E. (2018). Sorptive removal of enrofloxacin antibiotic from aqueous solution using a ligno-cellulosic substrate from wheat bran. J. Environ. Chem. Eng. 6, 5820–5829. doi: 10.1016/j.jece.2018.08.012
Shahabadi, N., and Hashempour, S. (2019). DNA binding studies of antibiotic drug cephalexin using spectroscopic and molecular docking techniques. Nucleosides Nucleotides Nucleic Acids 38, 428–447. doi: 10.1080/15257770.2018.1562071
Shen, S., Wu, W., Grimes, D. J., Saillant, E. A., and Griffitt, R. J. (2020). Community composition and antibiotic resistance of bacteria in bottlenose dolphins Tursiops truncatus–potential impact of 2010 BP oil spill. Sci. Total Environ. 732:139125. doi: 10.1016/j.scitotenv.2020.139125
Singh, V., Pandey, B., and Suthar, S. (2019). Phytotoxicity and degradation of antibiotic ofloxacin in duckweed (Spirodela polyrhiza) system. Ecotox. Environ. Safety 179, 88–95. doi: 10.1016/j.ecoenv.2019.04.018
Smakman, F., and Hall, A. R. (2022). Exposure to lysed bacteria can promote or inhibit growth of neighboring live bacteria depending on local abiotic conditions. FEMS Microbiol. Ecol. 98:fiac011. doi: 10.1093/femsec/fiac011
Sodhi, K. K., Kumar, M., Balan, B., Dhaulaniya, A. S., Shree, P., Sharma, N., et al. (2021). Perspectives on the antibiotic contamination, resistance, metabolomics, and systemic remediation. SN Appl. Sci. 3, 1–25. doi: 10.1007/s42452-020-04003-3
Song, C., Zhang, K. X., Wang, X. J., Zhao, S., and Wang, S. G. (2021). Effects of natural organic matter on the photolysis of tetracycline in aquatic environment: kinetics and mechanism. Chemosphere 263:128338. doi: 10.1016/j.chemosphere.2020.128338
Szultka, M., and Buszewski, B. (2016). Study of in-vitro metabolism of selected antibiotic drugs in human liver microsomes by liquid chromatography coupled with tandem mass spectrometry. Anal. Bioanal. Chem. 408, 8273–8287. doi: 10.1007/s00216-016-9929-6
Szultka, M., Krzeminski, R., Jackowski, M., and Buszewski, B. (2014). Identification of in vitro metabolites of amoxicillin in human liver microsomes by LC-ESI/MS. Chromatographia 77, 1027–1035. doi: 10.1007/s10337-014-2648-2
Tao, J., Yang, J., Ma, C., Li, J., Du, K., Wei, Z., et al. (2020). Cellulose nanocrystals/graphene oxide composite for the adsorption and removal of levofloxacin hydrochloride antibiotic from aqueous solution. R. Soc. Open Sci. 7:200857. doi: 10.1098/rsos.200857
Topp, E., Renaud, J., Sumarah, M., and Sabourin, L. (2016). Reduced persistence of the macrolide antibiotics erythromycin, clarithromycin and azithromycin in agricultural soil following several years of exposure in the field. Sci. Total Environ. 562, 136–144. doi: 10.1016/j.scitotenv.2016.03.210
Tunç, M. S., Hanay, Ö., and Yıldız, B. (2020). Adsorption of chlortetracycline from aqueous solution by chitin. Chem. Eng. Commun. 207, 1138–1147. doi: 10.1080/00986445.2019.1677628
Turner, J., Muraoka, A., Bedenbaugh, M., Childress, B., Pernot, L., Wiencek, M., et al. (2022). The chemical relationship among beta-lactam antibiotics and potential impacts on reactivity and decomposition. Front. Microbiol. 13:807955. doi: 10.3389/fmicb.2022.807955
United Nations Environment Programme (2019). Available at: https://www.unep.org/explore-topics/chemicals-waste/what-we-do/policy-and-governance/global-chemicals-outlook
van Bambeke, F., Barcia-Macay, M., Lemaire, S., and Tulkens, P. M. (2006). Cellular pharmacodynamics and pharmacokinetics of antibiotics: current views and perspectives. Curr. Opin. Drug Discov. Dev. 9, 218–230.
Vilvert, E., Contardo-Jara, V., Esterhuizen-Londt, M., and Pflugmacher, S. (2017). The effect of oxytetracycline on physiological and enzymatic defense responses in aquatic plant species Egeria densa, Azolla caroliniana, and Taxiphyllum barbieri. Toxicol. Environ. Chem. 99, 104–116. doi: 10.1080/02772248.2016.1165817
Wang, J., Peng, C., Dai, Y., Li, Y., Jiao, S., Ma, X., et al. (2022). Slower antibiotics degradation and higher resistance genes enrichment in plastisphere. Water Res. 222:118920. doi: 10.1016/j.watres.2022.118920
Wang, R., Yang, S., Fang, J., Wang, Z., Chen, Y., Zhang, D., et al. (2018). Characterizing the interaction between antibiotics and humic acid by fluorescence quenching method. Int. J. Environ. Res. Public Health 15:1458. doi: 10.3390/ijerph15071458
Webb, P. A. (2003). Introduction to chemical adsorption analytical techniques and their applications to catalysis. Micromerit. Instr. Corp. Tech. Pub., 1–12.
Wei, L., Li, H., and Lu, J. (2021). Algae-induced photodegradation of antibiotics: a review. Environ. Pollut. 272:115589. doi: 10.1016/j.envpol.2020.115589
Wynalda, M. A., Hauer, M. J., and Wienkers, L. C. (2000). Oxidation of the novel oxazolidinone antibiotic linezolid in human liver microsomes. Drug Metab. Dispos. 28, 1014–1017.
Xiong, J. Q., Kurade, M. B., Patil, D. V., Jang, M., Paeng, K. J., and Jeon, B. H. (2017). Biodegradation and metabolic fate of levofloxacin via a freshwater green alga, Scenedesmus obliquus in synthetic saline wastewater. Algal Res. 25, 54–61. doi: 10.1016/j.algal.2017.04.012
Xu, S. J., Chen, X. Y., Wang, X. F., Sun, H. Z., Hou, Z. J., Cheng, J. S., et al. (2022). Artificial microbial consortium producing oxidases enhanced the biotransformation efficiencies of multi-antibiotics. J. Hazard. Mater. 439:129674. doi: 10.1016/j.jhazmat.2022.129674
Yagüe, P., López-García, M. T., Rioseras, B., Sánchez, J., and Manteca, A. (2013). Pre-sporulation stages of Streptomyces differentiation: state-of-the-art and future perspectives. FEMS Microbiol. Lett. 342, 79–88. doi: 10.1111/1574-6968.12128
Yamana, T., and Tsuji, A. (1976). Comparative stability of cephalosporins in aqueous solution: kinetics and mechanisms of degradation. J. Pharm. Sci. 65, 1563–1574. doi: 10.1002/jps.2600651104
Yang, Y., Li, H., Deng, S., Yang, X., Wang, M., Tan, W., et al. (2022). Prediction and analysis of the soil organic matter distribution with the spatiotemporal kriging method. Earth Sci. Inf. 15, 1621–1633. doi: 10.1007/s12145-022-00815-6
Yang, Q., Li, Z., Lu, X., Duan, Q., Huang, L., and Bi, J. (2018). A review of soil heavy metal pollution from industrial and agricultural regions in China: pollution and risk assessment. Sci. Total Environ. 642, 690–700. doi: 10.1016/j.scitotenv.2018.06.068
Yang, C., Ma, S., Li, F., Zheng, L., Tomberlin, J. K., Yu, Z., et al. (2022). Characteristics and mechanisms of ciprofloxacin degradation by black soldier fly larvae combined with associated intestinal microorganisms. Sci. Total Environ. 811:151371. doi: 10.1016/j.scitotenv.2021.151371
Yao, Q., Fan, B., Xiong, Y., Jin, C., Sun, Q., and Sheng, C. (2017). 3D assembly based on 2D structure of cellulose nanofibril/graphene oxide hybrid aerogel for adsorptive removal of antibiotics in water. Sci. Rep. 7:45914. doi: 10.1038/srep45914
Yim, G., Wang, H. H., and Davies, J. (2006). The truth about antibiotics. Int. J. Med. Microbiol. 296, 163–170. doi: 10.1016/j.ijmm.2006.01.039
Yin, H., Cai, Y., Li, G., Wang, W., Wong, P. K., and An, T. (2022). Persistence and environmental geochemistry transformation of antibiotic-resistance bacteria/genes in water at the interface of natural minerals with light irradiation. Crit. Rev. Environ. Sci. Technol. 52, 2270–2301. doi: 10.1080/10643389.2021.1877062
Yin, J., Wang, G., Cheng, D., Fu, J., Qiu, J., and Yu, Z. (2019). Inactivation of polymyxin by hydrolytic mechanism. Antimicrob. Agents Chemother. 63, e02378–e02388. doi: 10.1128/AAC.02378-18
Zethof, J. H., Leue, M., Vogel, C., Stoner, S. W., and Kalbitz, K. (2019). Identifying and quantifying geogenic organic carbon in soils–the case of graphite. Soil 5, 383–398. doi: 10.5194/soil-5-383-2019
Zhan, M., Yang, X., Xian, Q., and Kong, L. (2006). Photosensitized degradation of bisphenol a involving reactive oxygen species in the presence of humic substances. Chemosphere 63, 378–386. doi: 10.1016/j.chemosphere.2005.08.046
Zhang, X., Guo, W., Ngo, H. H., Wen, H., Li, N., and Wu, W. (2016). Performance evaluation of powdered activated carbon for removing 28 types of antibiotics from water. J. Environ. Manag. 172, 193–200. doi: 10.1016/j.jenvman.2016.02.038
Zhi, D., Yang, D., Zheng, Y., Yang, Y., He, Y., Luo, L., et al. (2019). Current progress in the adsorption, transport and biodegradation of antibiotics in soil. J. Environ. Manag. 251:109598. doi: 10.1016/j.jenvman.2019.109598
Keywords: antibiotic detoxification, antibiotics in soil and water, antibiotic resistance in the environment, adsorption and desorption, particles in the environment
Citation: Baquero F, Coque TM and Martínez JL (2022) Natural detoxification of antibiotics in the environment: A one health perspective. Front. Microbiol. 13:1062399. doi: 10.3389/fmicb.2022.1062399
Received: 06 October 2022; Accepted: 03 November 2022;
Published: 24 November 2022.
Edited by:
Axel Cloeckaert, Institut National de recherche pour l’agriculture, l’alimentation et l’environnement, FranceReviewed by:
Magdalena Popowska, University of Warsaw, PolandCopyright © 2022 Baquero, Coque and Martínez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fernando Baquero, YmFxdWVyb0BiaXRtYWlsZXIubmV0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.