- 1Key Laboratory of Bovine and Ovine Germplasm and Straw Transfer into Feedstuff in Gansu Province, Institute of Livestock, Grass and Green Agriculture, Gansu Academy of Agricultural Sciences, Lanzhou, China
- 2College of Animal Science and Technology, Gansu Agricultural University, Lanzhou, China
Zanthoxylum bungeanum essential oil (EOZB) as an extract of Zanthoxylum bungeanum has a range of pharmacological effects such as antibacterial, anti-inflammatory, and antioxidant. However, there were no relevant studies on the regulation of gut microbes by EOZB in ruminants. In this study, the effects of different doses of EOZB on the structure and distribution of microorganisms in the small intestine of small-tailed Han sheep (STH) were investigated by 16s rRNA gene sequencing technique. We found that with the intervention of EOZB. The differential bacteria of duodenal at the phylum level were Firmicutes, Bacteroidetes, Tenericutes and Proteobacteria, and genus level differential bacteria were Prevotella 1, Ruminococcus 2 and Eubacterium coprostanoligenes group. The differential bacteria of jejunal at the phylum level were Firmicutes, Bacteroidetes, Tenericutes and Proteobacteria, and genus level differential bacteria were Prevotella 1, Rikenellaceae RC9 gut group, Christensenellaceae R-7 group, Ruminococcaceae UCG-014, Saccharofermentans, Ruminococcaceae NK4A214 group and Prevotellaceae UCG-001. The differential bacteria of ileal at the phylum level were Firmicutes, Bacteroidetes and Tenericutes, and genus level differential bacteria were Prevotella 1, Christensenellaceae R-7 group, Romboutsia and Ruminococcaceae UCG-014. In addition, at the same dose of EOZB, the five most abundant genera of bacteria varied in different regions of the small intestine. Among them, the abundance of Prevotella 1, Christensenellacea R-7 group and Ruminococcus 2 in ALW group was the highest in jejunum, duodenum and ileum, respectively. The abundance of Prevotella 1, Christensenellacea R-7 group and Rikenellacea RC9 gut group in BLW group was the highest in duodenum, jejunum and ileum, respectively. The abundance of Prevotella 1, Christensenellacea R-7 group and Ruminococcaeae NK4A214 group in CLW group was the highest in jejunum, duodenum and ileum, respectively. The abundance of Prevotella 1, Ruminococcus 2 and Ruminococcus NK4A214 groups in DLW group was the highest in jejunum, duodenum and ileum, respectively. Differential bacteria formed under the regulation of EOZB are associated with the digestion and absorption of nutrients and the state of intestinal health in the host. This study is the first to investigate the effect of EOZB on the distribution and structure of bacteria in the small intestine of STH. The results of the study enriched the structure and distribution of bacteria in the small intestine of ruminants and provided new insights into the future application of herbal medicine in ruminant production. Additionally, it provides a theoretical basis for the selection of probiotic bacteria for ruminants and the development and application of microecological preparations.
Introduction
Ruminants are essential for humans because they transform plant fibers into milk and meat (Eisler et al., 2014). In contrast to monogastric animals (such as pigs and poultry), ruminants have a compound stomach in the gastrointestinal tract (GIT) with four chambers: the rumen, reticulum, omasum, and abomasum. The numerous rumen microorganisms are essential in the processing of plant-based diets and produce precursors (such as volatile fatty acids) to provide ruminants with energy, as well as greenhouse gasses (such as methane; Russell and Rychlik, 2001; Moraïs and Mizrahi, 2019). Taxonomic profiles of rumen microorganisms and associated functions have been extensively investigated (Seshadri et al., 2018; Stewart et al., 2018, 2019; Li et al., 2020). The knowledge gained in previous studies has helped control rumen fermentation (Yáñez-Ruiz et al., 2015), increase feed effectiveness (Shabat et al., 2016), and reduce methane emissions (Shi et al., 2014), demonstrating that the GIT microbial community is crucial in rumen physiological processes (O'Hara et al., 2020). Additionally, the gut (GT) microbiota, commonly referred to as the “forgotten organ,” is a complex and varied community. The GT microbiota function as a bioinformatic hub that combines host metabolism, immunological functions, and genetic signals with outside inputs such as nutrition, medications, and probiotics (Thaiss et al., 2016). Anaerobic bacteria are prevalent in the mammalian distal gut where they break down nutrients such as polysaccharides (Backhed et al., 2005). The immune system and general health of hosts are substantially influenced by the large gut microbial community (Gillor et al., 2008; Schuijt et al., 2013). The ruminant digestive tract is divided into 10 compartments (the rumen, reticulum, omasum, abomasum, duodenum, jejunum, ileum, cecum, colon, and rectum), each compartment has a very diverse microbial composition and function. However, understanding of ruminant microbial processes is incomplete (Greening et al., 2019), even though the composition and function of microbial communities affect the ruminant digestive, immune, metabolic, and endocrine processes (Martinez-Guryn et al., 2019). Therefore, to completely comprehend the functions of the ruminant gastrointestinal microbiota, a detailed description of the bacteria in all gastrointestinal regions is required.
The plant Zanthoxylum bungeanum (Rutaceae) is currently widely distributed in most of China and some Southeast Asian countries. Zanthoxylum bungeanum is rich in flavonoids, primarily including rutin, quercetin, foeniculin, hyperin, and isoquercitrin (Li et al., 2001), and is currently frequently used as a flavoring agent and traditional Chinese herbal medicine because of its distinctive flavor and medicinal properties (Tezuka et al., 2001). The plant provides a range of pharmacological benefits, including anti-inflammatory (Zhang et al., 2017), antioxidant (Zhang et al., 2014), and antibacterial (Guleria et al., 2013) activities. The Z. bungeanum essential oil (EOZB) is a viscous liquid derived from plant husks with a distinct flavor that has pharmacological properties similar to those of Z. bungeanum (Zhang et al., 2022). Diarrhea is one of the common diseases in young ruminants and causes great problems to the development of animal husbandry. Diarrhea is usually accompanied by dysbiosis of the intestinal microbiota, damage to the intestinal mucosal barrier and inflammation (Karczewski et al., 2010), and EOZB has potential antibacterial, anti-inflammatory and antioxidant properties. However, most of the current ruminant studies have focused on the rumen and there are no reports on the regulation of EOZB on the intestinal bacteria in ruminants. Therefore, research in this area was needed for refinement.
Due to the incomplete study of the structure of ruminant gut bacterial composition, and whether EOZB affects the structure of the ruminant gut bacteria. We investigated the structure and distribution of bacteria communities in 60 STH small intestines (duodenum, jejunum, and ileum) samples under the EOZB intervention by 16s rRNA high-throughput sequencing. Aimed to supplement the effects of EOZB on ruminant small intestinal bacteria and the incomplete of ruminant gut bacteria.
Materials and methods
Experimental design and sampling
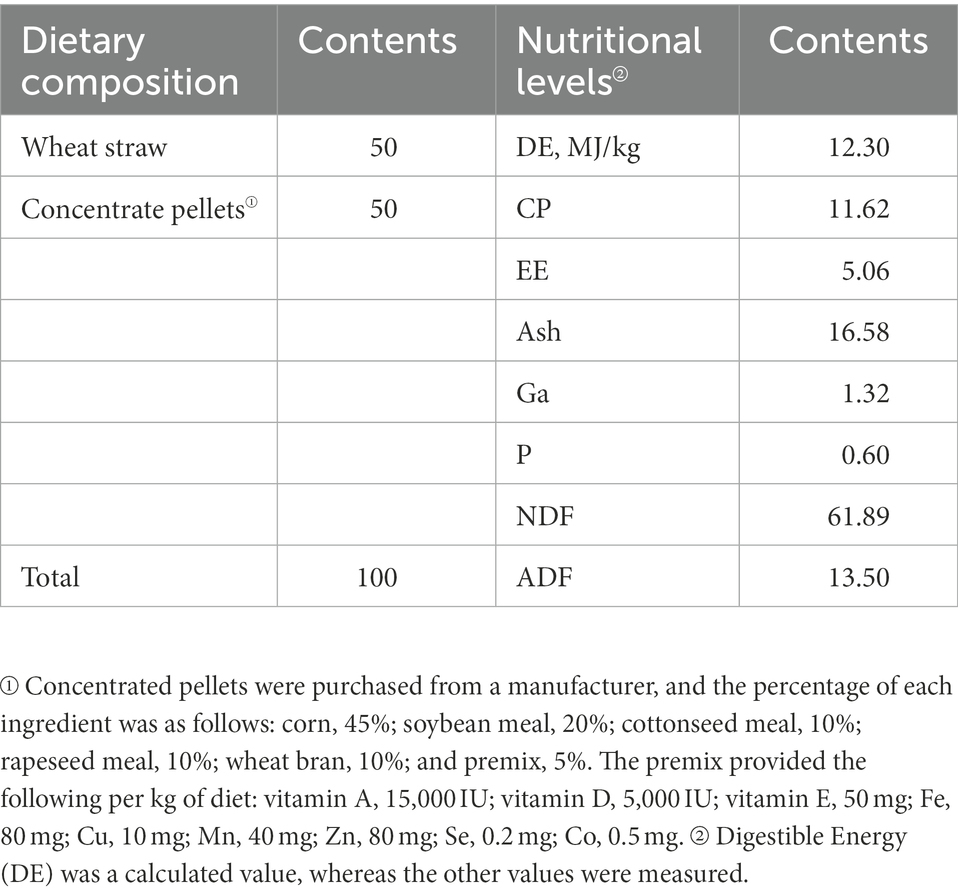
Twenty STH (male; 3-months-old; initial weight, 23.57 ± 4.61 kg, p > 0.05) were obtained from a farm in Dingxi, Gansu Province, China. Test sheep were housed in individual pens that were regularly disinfected and cleaned (once a week). The 20 experimental sheep were randomly divided into four groups and provided the diets shown in Table 1. The test groups were the following: ALW: basic diet (BD; without EOZB, nutritional level was basically the same as in other test groups); BLW: BD + EOZB at 5 ml/kg; CLW: BD + EOZB at 10 ml/kg; and DLW: BD + EOZB at 15 ml/kg. Different doses of EOZB were given daily to the different test groups (EOZB was added by spraying during feeding. The dose of EOZB was obtained with reference to Zhang et al. (2017) and adjusted to the actual situation). To distinguish samples from the small intestine sites of the duodenum, jejunum, and ileum, duodenal samples from ALW, BLW, CLW, and DLW groups were recorded as AD1-5, BD1-5, CD1-5, and DD1-5, respectively; jejunum samples were recorded as AJ1-5, BJ1-5, CJ1-5, and DJ1-5, respectively; and ileal samples were recorded as AI1-5, BI1-5, CI1-5, and DI1-5, respectively. The experiment was conducted for 52 days, and at the end, each group of test sheep was subjected to electric shock (head shock) and then slaughtered by means of neck bloodletting. Referring to Anatomy and Histology and Embryology of Domestic Animals ligation was performed at the division points of each part of the small intestinal tract, and the small intestinal tissue at each part was incised with a scalpel at the middle of the ligated segment. The contents of each part of the small intestine (duodenum, jejunum, and ileum) were collected in 1.5-ml sterile polypropylene tubes, immediately frozen in liquid nitrogen at −80°C, and stored for analysis of bacterial communities.
DNA extraction and high-throughput sequencing
Total DNA was extracted from ruminal samples using an E.Z.N.A.1 Stool DNA Kit (Omega Bio-Tek, Norcross, GA, United States) according to the manufacturer’s protocol. The V4-V5 region of the bacterial 16S ribosomal RNA gene was amplified by PCR (95°C for 5 min, followed by 30 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s, and a final extension at 72°C for 5 min) using primers 515F (5′-GTGCCAGCMGCCGCGG-3′) and 907R (5′-CCGTCAATTCMTTTRAGTTT-3′), with the bar code a six-base sequence unique to each sample. The PCR reactions were performed in 30 μl of a mixture containing 15 μl of 2 × Phanta Master Mix, 1 μl of each primer (10 μM), and 20 ng of template DNA. Amplicons were extracted from 2% agarose gels and purified using an AxyPrep DNA Gel Extraction Kit (Axy-gen Biosciences, Union City, CA, United States) according to the manufacturer’s instructions. Purified PCR products were quantified by Qubit13.0 (Life Invitrogen, CA, United States), and every 20 amplicons with different bar codes were mixed equally. The pooled DNA product was used to construct an Illumina paired-end library following the Illumina genomic DNA library preparation procedure. The amplicon library was then pair-end sequenced (2 × 250) on an Illumina Novaseq 6000 platform (Nanjing GenePioneer Co. Ltd., Nanjing, China) according to the standard protocol.
Bioinformatics analysis
Raw data returned by the Illumina HiSeq sequencing platform were filtered using the software packages Pandaseq (Masella et al., 2012), PRINSEQ (Schmieder and Edwards, 2011), and Vsearch (Rognes et al., 2016; v2.15.0_linux_x86_64) to remove chimeras and obtain optimized sequences (tags). Operational taxonomic units (OTUs) were clustered using Vsearch, and the clustering similarity threshold was 97%. A self-developed Perl program was used for random rarefaction of the data of each sample (the number of rarefactions was the minimum number of sample sequences). To select the most abundant sequence of each OTU as the representative sequence, QIIME (v.1.9.1) was used. Then, the Uclust method was used to compare the representative sequence to the Silva rRNA database (release_132) and classify the OTU species. Based on the abundance and annotation information of OTUs, the proportion of sequences in each sample at different taxonomic levels was counted to assess sample species abundance and diversity. Alpha diversity was assessed by Observed species, Chao1, Shannon, and Simpson indices (Li et al., 2013) and by plotting the samples as species accumulation curves. Beta diversity analysis was used to compare differences in species diversity (microbial composition and structure) between different samples. Sample-level PCoA and Anosim analysis (Sakaki et al., 1994; Dubois et al., 2010; with grouping information) were conducted based on Bray–Curtis distance and Unweighted Unifrac distance, respectively. Analysis of significant differences between groups (LEfSe analysis) was used to identify biomarkers (Segata et al., 2011). To illustrate the composition of bacterial communities in all test groups, heat maps were constructed of genera in the duodenum, jejunum, and ileum of the small intestine.
Statistical data analysis
One-way ANOVA was performed using SPSS software (v21.0, SPSS Inc.). Analyzes were conducted on relative abundances and alpha diversity indices of bacteria in different parts of the small intestine at different levels. Multiple comparisons between groups were performed using Duncan’s method. Results are expressed as the mean and SEM, with p < 0.05 indicating significant differences.
Results
Effects of Zanthoxylum bungeanum essential oil on duodenal bacteria
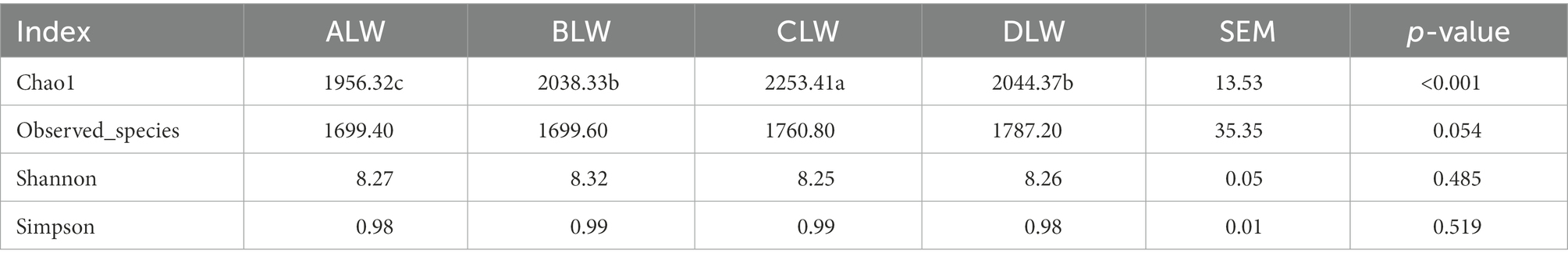
A total of 1,992,846 tags were obtained from 20 duodenum samples; 1,873,850 clean tags were obtained after filtering, optimization, and quality control; and 78,484 sequences were extracted from each sample for subsequent analysis based on the minimum value of detected sequences (Supplementary Table S1). Clustering was performed using Vsearch with 97% similarity to obtain the OTUs of each sample. A total of 4,617 OTUs were generated, and 3,877 were obtained after random rarefaction, with representative OTU sequences selected for species annotation. There were 2,617 shared OTUs in the four groups, whereas 81 OTUs were specific to ALW, 41 to BLW, 122 to CLW, and 63 to DLW (Figure 1A). Species accumulation curves, widely used to determine the adequacy of sample size and to estimate species richness, were used to describe the increase in species with increasing sample size. The curve began to plateau at the sample size of 10, indicating that the sample size was sufficient and that the sequencing coverage was saturated (Figure 1B).
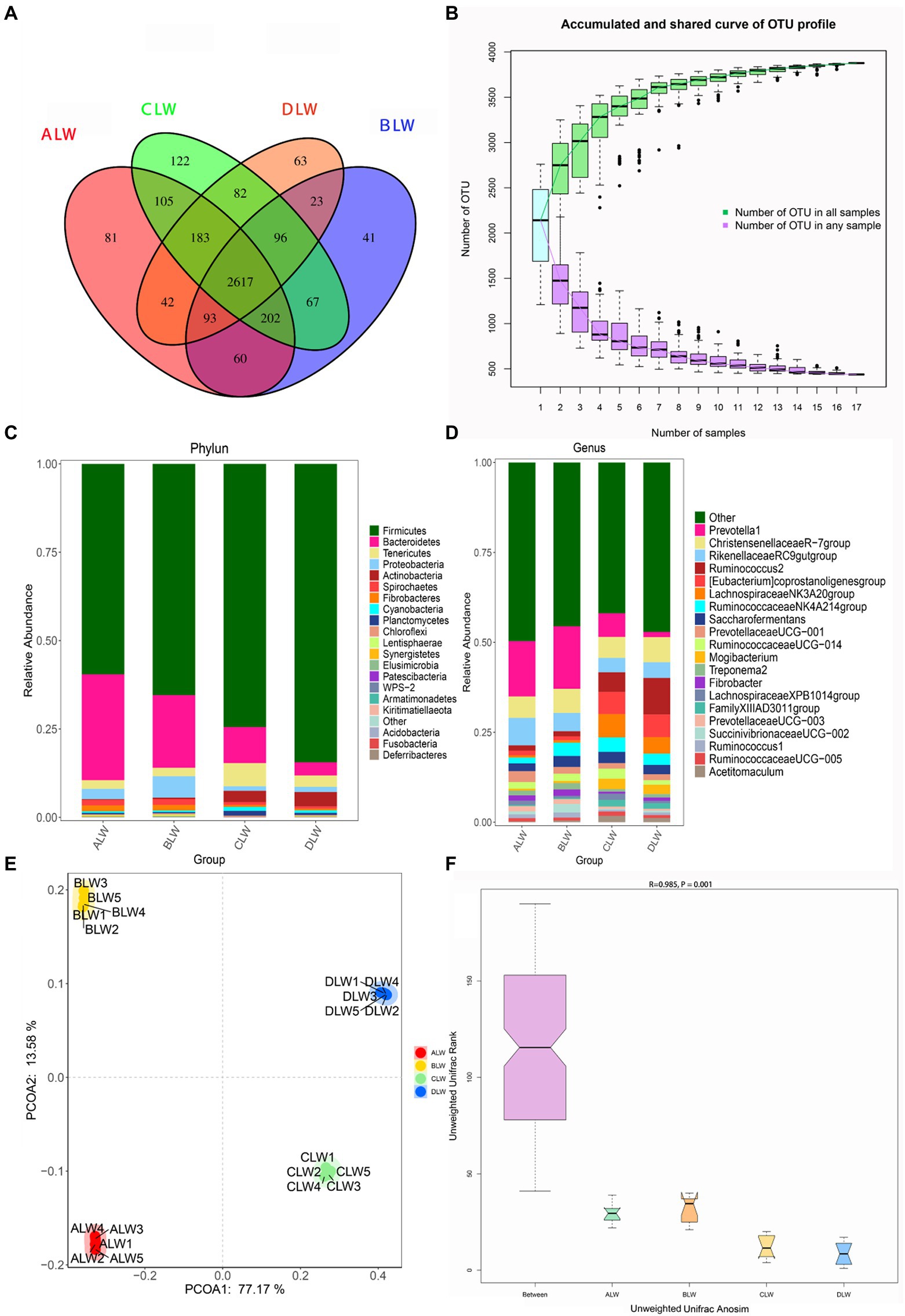
Figure 1. (A) OTU-Venn diagram (B) Species accumulation curves (C,D) Relative abundance of species at the phylum and genus level (E,F) PCoA and Anosim analysis.
The dominant phyla of bacteria were Firmicutes, Bacteroidetes, Tenericutes, and Proteobacteria, which had relative abundances >1% in all groups (Figure 1C). Relative abundance of Firmicutes was significantly higher in DLW than in the other groups (p < 0.001), whereas that of Bacteroidetes was significantly higher in ALW than in the other groups (p < 0.001). Relative abundance of Tenericutes was significantly higher in CLW than in the other groups (p = 0.034), whereas that of Proteobacteria was significantly higher in BLW than in the other groups (p = 0.008; Supplementary Table S2). The dominant genera were Prevotella 1, Christensenellaceae R-7 group, Ruminococcus 2, Rikenellaceae RC9 gut group, Eubacterium coprostanoligenes group, Ruminococcaceae NK4A214 group, Saccharofermentans, Ruminococcaceae UCG-014, and Prevotellaceae UCG-001, which had relative abundances >1% in each test group (Figure 1D). Relative abundance of Prevotella 1 was significantly higher in BLW than in the other groups (p < 0.001); whereas relative abundances of Ruminococcus 2 and Eubacterium coprostanoligenes group were significantly higher in DLW than in the other groups (p < 0.001 and p = 0.001, respectively; Supplementary Table S2).
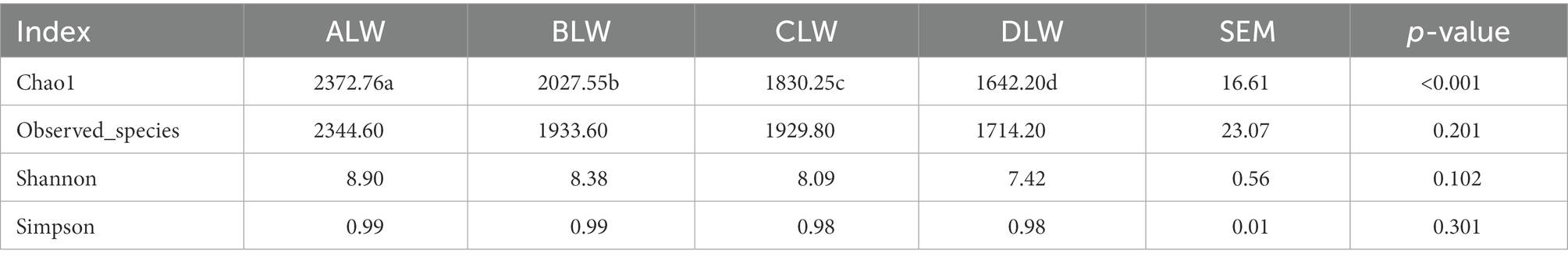
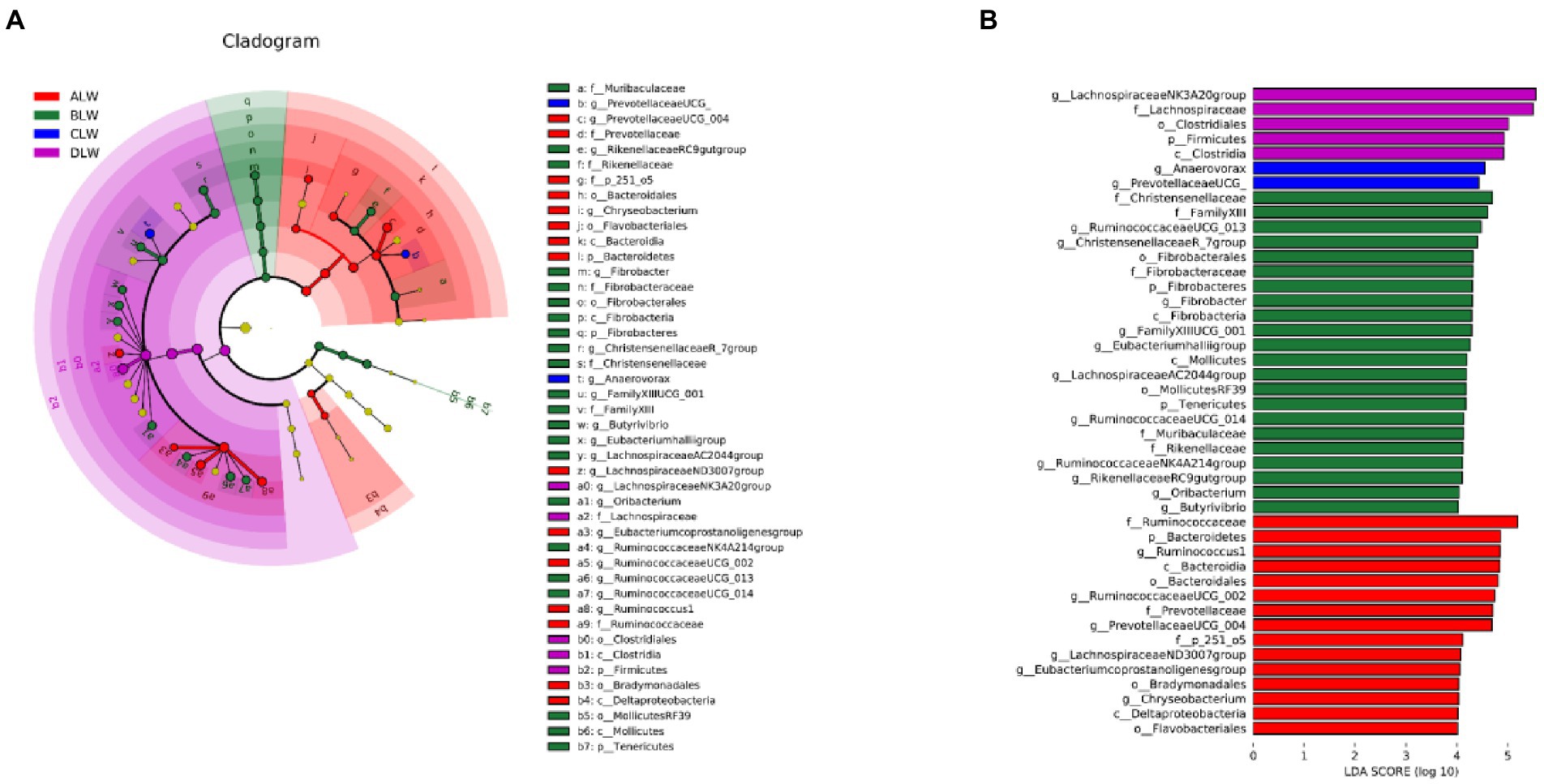
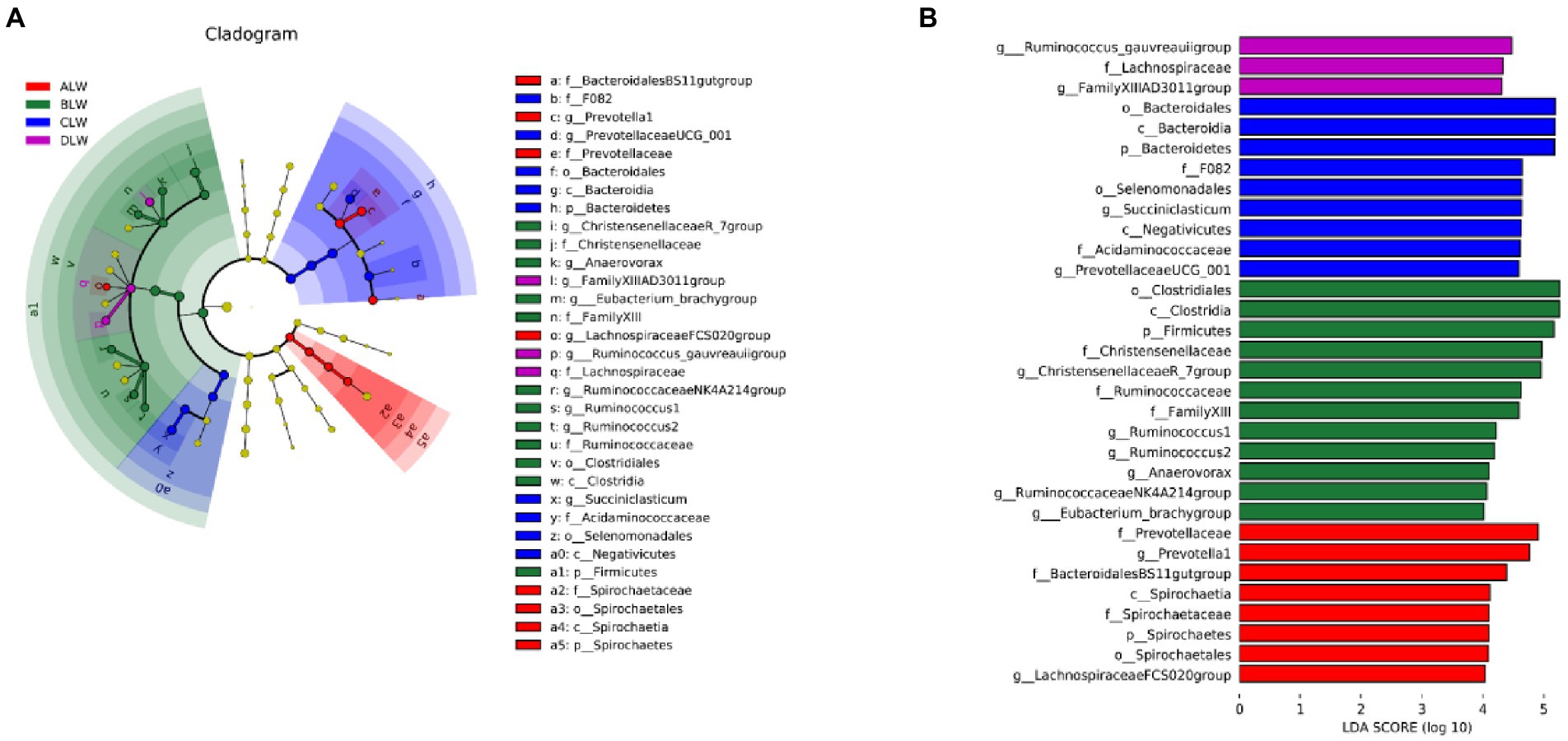
Principal coordinate analysis (PCoA) based on Bray–Curtis distance showed significant differences in bacterial taxa among the groups (Figure 1E). Anosim analysis based on Unweighted Unifrac distance further showed that the between-group differences were significantly greater than the within-group differences (Figure 1F). The Chao1 index, an indicator of community richness, was significantly higher in ALW than in the other groups (p < 0.001; Table 2). The LEfSe analysis showed that the addition of EOZB produced a total of 44 biomarkers (LDA score > 4; p < 0.05; Figures 2A,B).
Effect of Zanthoxylum bungeanum essential oil on jejunal bacteria
A total of 1,783,198 tags were obtained from 20 jejunum samples; 1,652,166 clean tags were obtained after filtering, optimization, and quality control; and 72,109 sequences were extracted from each sample for subsequent analysis based on the minimum value of detected sequences (Supplementary Table S3). Clustering was performed using Vsearch with 97% similarity to obtain the OTUs of each sample. A total of 3,776 OTUs were generated, and 3,038 were obtained after random rarefaction, with representative OTU sequences selected for species annotation. There were 2,038 shared OTUs in the four groups, whereas 38 OTUs were specific to ALW, 50 to BLW, 63 to CLW, and 106 to DLW (Figure 3A). The species accumulation curve began to plateau at the sample size of 10, indicating that the sample size was sufficient and that the sequencing coverage was saturated (Figure 3B).
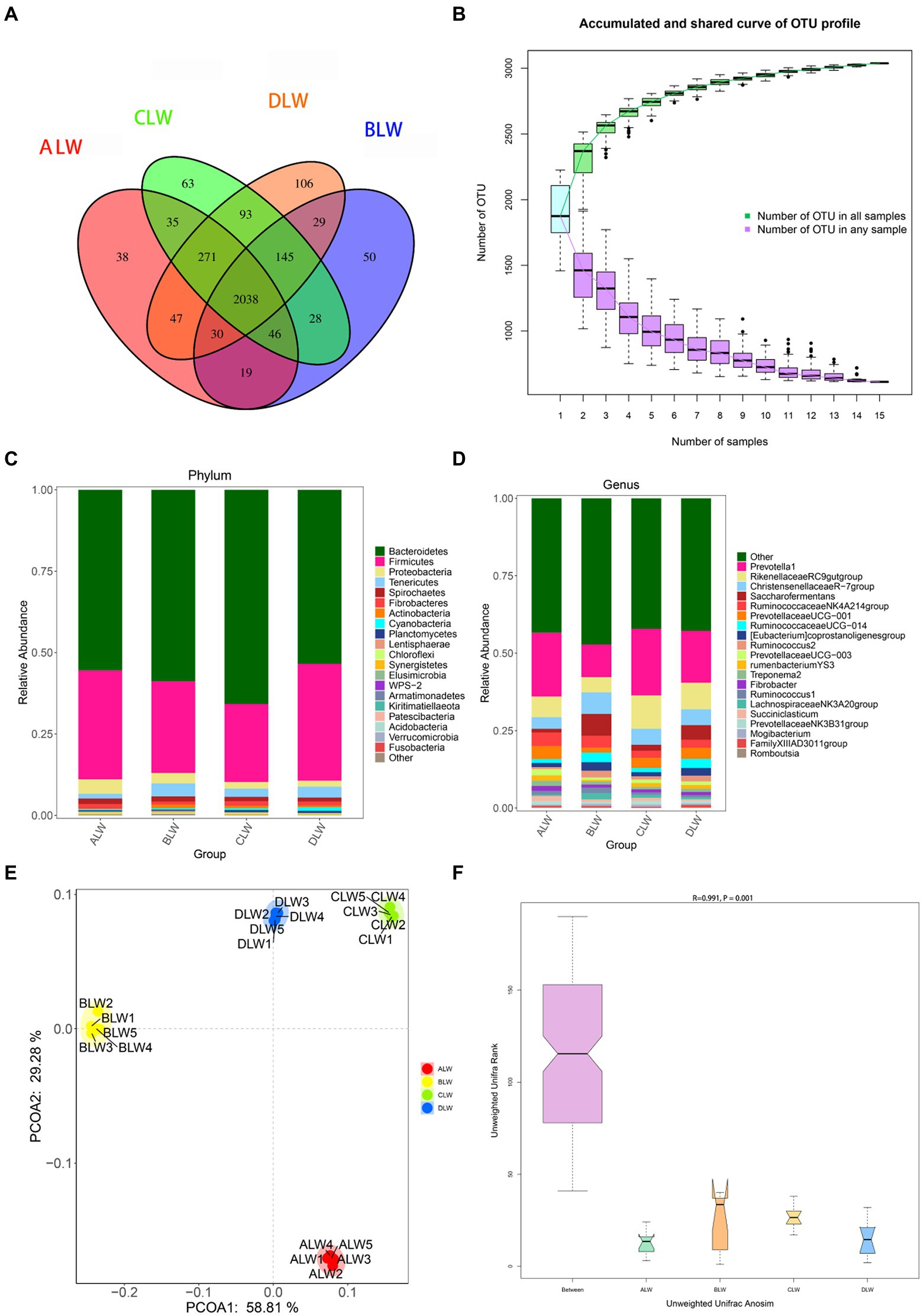
Figure 3. (A) OTU-Venn diagram (B) Species accumulation curves (C,D) Relative abundance of species at the phylum and genus level (E,F) PCoA and Anosim analysis.
The dominant phyla of bacteria were Firmicutes, Bacteroidetes, Tenericutes, Proteobacteria, and Spirochaetes, which had relative abundances >1% (Figure 3C). Compared with other groups, relative abundance of Firmicutes was significantly higher in DLW (p < 0.001); that of Bacteroidetes was significantly higher in CLW (p < 0.001); that of Tenericutes was significantly higher in BLW (p = 0.002); and that of Proteobacteria was significantly higher in ALW (p = 0.001; Supplementary Table S4). The dominant genera of bacteria were Prevotella 1, Rikenellaceae RC9 gut group, Christensenellaceae R-7 group, Saccharofermentans, Ruminococcaceae NK4A214 group, Prevotellaceae UCG-001, Eubacterium coprostanoligenes group, and Ruminococcaceae UCG-014, which had relative abundances >1% (Figure 3D). Compared with other groups, relative abundances of Prevotella 1 and Rikenellaceae RC9 gut group were significantly higher in CLW (both p < 0.001), those of Christensenellaceae R-7 group, Ruminococcaceae UCG-014, and Saccharofermentans were significantly higher in BLW (p = 0.006, p = 0.049, and p < 0.001, respectively), and those of Ruminococcaceae NK4A214 group and Prevotellaceae UCG-001 were significantly higher in ALW (p < 0.001 and p = 0.02, respectively; Supplementary Table S4).
PCoA analysis showed significant differences in bacterial taxa among the groups (Figure 3E). According to the Anosim analysis, between-group differences were significantly greater than within-group differences (Figure 3F). The Chao1 index of community richness was significantly higher in DLW than in the other groups (p < 0.001; Table 3). The LEfSe analysis showed that the addition of EOZB produced a total of 32 biomarkers (LDA score > 4; p < 0.05; Figures 4A,B).
Effect of Zanthoxylum bungeanum essential oil on Ileal bacteria
A total of 1,551,651 tags were obtained from 20 ileum samples; 1,470,809 clean tags were obtained after filtering, optimization, and quality control; and 62,344 sequences were extracted from each sample for subsequent analysis based on the minimum value of detected sequences (Supplementary Table S5). Clustering was performed with 97% similarity using Vsearch to obtain the OTUs of each sample. A total of 3,944 OTUs were generated, and 3,213 were obtained after random rarefaction, with representative OTU sequences selected for species annotation. There were 1,815 shared OTUs in the four groups, whereas 48 OTUs were specific to ALW, 40 to BLW, 371 to CLW, and 55 to DLW (Figure 5A). The species accumulation curve began to plateau at the sample size of 10, indicating that the sample size was sufficient and that the sequencing coverage was saturated (Figure 5B).
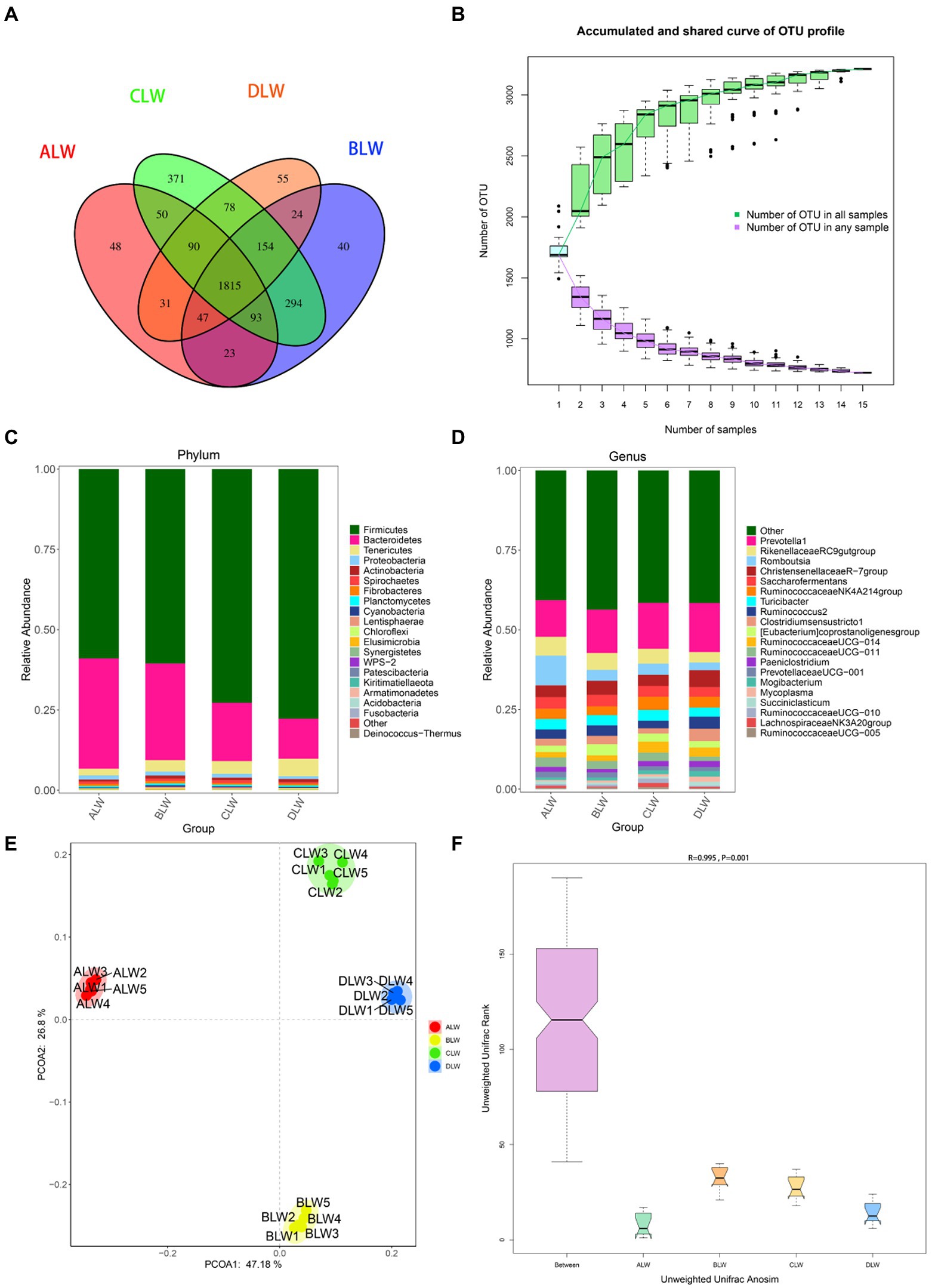
Figure 5. (A) OTU-Venn diagram (B) Species accumulation curves (C,D) Relative abundance of species at the phylum and genus level (E,F) PCoA and Anosim analysis.
The dominant phyla of bacteria were Firmicutes, Bacteroidetes, and Tenericutes, which had relative abundances >1% (Figure 5C). Compared with other groups, relative abundances of Firmicutes and Tenericutes were significantly higher in DLW (both p < 0.001) and that of Bacteroidetes was significantly higher in ALW (p < 0.001; Supplementary Table S6). At the genus level, Prevotella 1, Romboutsia, Christensenellaceae R-7 group, Rikenellaceae RC9 gut group, Turicibacter, Ruminococcaceae NK4A214 group, Saccharofermentans, Clostridium sensu stricto 1, Ruminococcus 2, Eubacterium coprostanoligenes group, Ruminococcaceae UCG-011, Ruminococcaceae UCG-014, Paeniclostridium, and Prevotellaceae UCG-001 were the dominant bacteria, which had relative abundances >1% (Figure 5D). Compared with other groups, relative abundances of Prevotella 1 and Christensenellaceae R-7 group were significantly higher in DLW (p < 0.001 and p = 0.001, respectively), that of Romboutsia was significantly higher in ALW (p < 0.001), and that of Ruminococcaceae UCG-014 was significantly higher in CLW (p = 0.039; Supplementary Table S6).
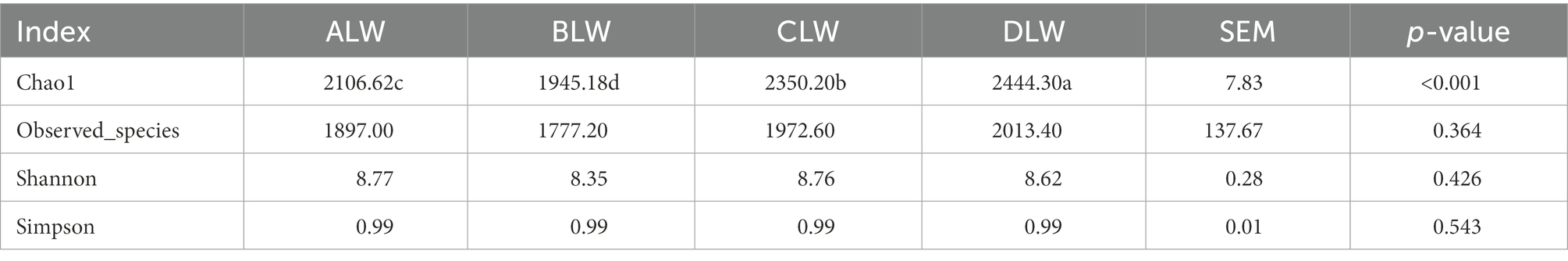
PCoA analysis showed significant differences in bacterial taxa among the test groups (Figure 5E). According to the Anosim analysis, between-group differences were significantly greater than within-group differences (Figure 5F). The Chao1 index of community richness was significantly higher in CLW than in the other groups (p < 0.001; Table 4). The LEfSe analysis showed that the addition of EOZB produced a total of 31 biomarkers (LDA score > 4; p < 0.05; Figures 6A,B).
Effect of Zanthoxylum bungeanum essential oil on bacteria in different sites of the small intestine
To further understand effects of EOZB on bacteria in the small intestine, a heat map was constructed for all genera (at the same dose of EOZB) in different sites of the small intestine.
The five most abundant genera in the duodenum of the ALW group were Prevotella 1, Rikenellaceae RC9 gut group, Christensenellaceae R-7 group, Prevotellaceae UCG-001, and Saccharofermentans. In the jejunum, the five most abundant genera were Prevotella 1, Rikenellaceae RC9 gut group, Ruminococcaceae NK4A214 group, Prevotellaceae UCG-001, and Christensenellaceae R-7 group. In the ileum, the five most abundant genera were Prevotella 1, Romboutsia, Rikenellaceae RC9 gut group, Christensenellaceae R-7 group, and Saccharofermentans. In addition, the same genus of bacteria was distributed differently in different parts of the small intestine. The highest abundance of Prevotella 1 was in the jejunum, that of Christensenellaceae R-7 group was in the duodenum, and that of Ruminococcus 2 was in the ileum (Figure 7A).
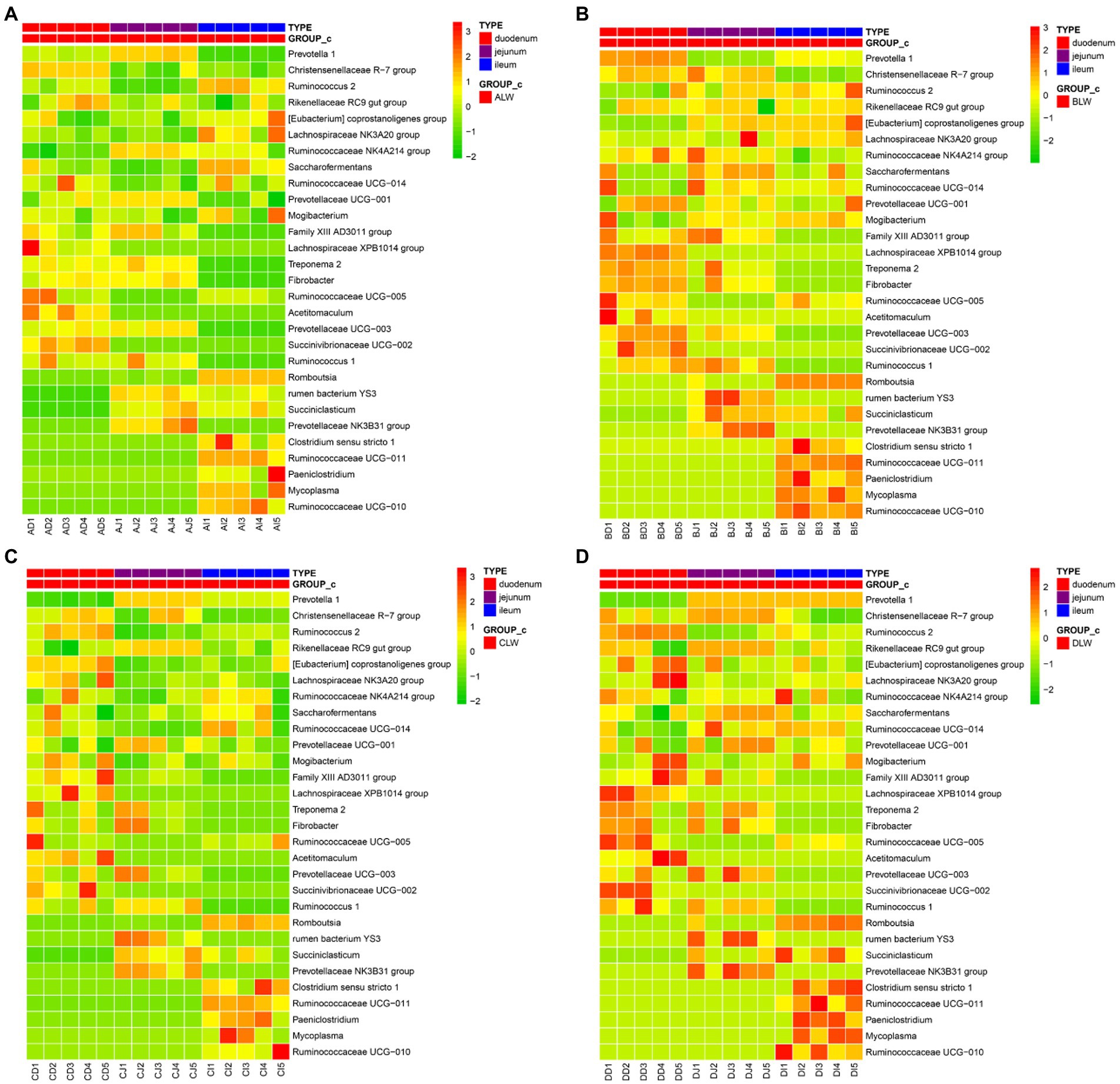
Figure 7. (A) Heat map of microbial distribution at the genus level in the ALW group (B) Heat map of microbial distribution at the genus level in the BLW group (C) Heat map of microbial distribution at the genus level in the CLW group (D) Heat map of microbial distribution at the genus level in the DLW group.
The five most abundant genera of bacteria in the duodenum of the BLW group were Prevotella 1, Christensenellaceae R-7 group, Rikenellaceae RC9 gut group, Ruminococcaceae NK4A214 group, and Saccharofermentans. In the jejunum, the five most abundant genera were Prevotella 1, Saccharofermentans, Christensenellaceae R-7 group, Rikenellaceae RC9 gut group, and Ruminococcaceae NK4A214 group. In the ileum, the five most abundant genera were Prevotella 1, Rikenellaceae RC9 gut group, Christensenellaceae R-7 group, Saccharofermentans, and Eubacterium coprostanoligenes group. In addition, the same genus of bacteria was distributed differently in different parts of the small intestine. The highest abundance of Prevotella 1 was in the duodenum, that of Christensenellaceae R-7 group was in the jejunum, and that of Rikenellaceae RC9 gut group was in the ileum (Figure 7B).
The five most abundant genera in the duodenum of the CLW group were Prevotella 1, Lachnospiraceae NK3A20 group, Eubacterium coprostanoligenes group, Christensenellaceae R-7 group, and Ruminococcus 2. In the jejunum, the five most abundant genera were Prevotella 1, Rikenellaceae RC9 gut group, Christensenellaceae R-7 group, Prevotellaceae UCG-001, and Ruminococcaceae NK4A214 group. In the ileum, the five most abundant genera were Prevotella 1, Rikenellaceae RC9 gut group, Christensenellaceae R-7 group, Saccharofermentans, and Ruminococcaceae UCG-014. In addition, the same genus of bacteria was distributed differently in different parts of the small intestine. The highest abundance of Prevotella 1 was in the jejunum, that of Christensenellaceae R-7 group was in the duodenum, and that of Ruminococcaceae NK4A214 group was in the ileum (Figure 7C).
The five most abundant genera in the duodenum of the DLW group were Ruminococcus 2, Christensenellaceae R-7 group, Eubacterium coprostanoligenes group, Lachnospiraceae NK3A20 group, and Rikenellaceae RC9 gut group. The five most abundant genera in the jejunum were Prevotella 1, Christensenellaceae R-7 group, Rikenellaceae RC9 gut group, Saccharofermentans, and Prevotellaceae UCG-001. The five most abundant genera in the ileum were Prevotella 1, Christensenellaceae R-7 group, Clostridium sensu stricto 1, Ruminococcus 2, and Rikenellaceae RC9 gut group. In addition, the same genus was distributed differently in different parts of the small intestine. The highest abundance of Prevotella 1 was in the jejunum, that of Ruminococcus 2 was in the duodenum, and that of Ruminococcaceae NK4A214 group was in the ileum (Figure 7D).
Discussion
In this study, Firmicutes, Bacteroidetes, Tenericutes, and Proteobacteria were the dominant phyla of bacteria in the duodenum of STH. Compared with other trial groups, Firmicutes, Tenericutes, and Proteobacteria increased significantly in the DLW, CLW, and BLW groups (p < 0.001, p = 0.034, and p = 0.008). Interestingly, Bacteroidetes decreased significantly in the DLW groups (p < 0.001). Firmicutes primarily include many gram-positive bacteria, including Lactococcus, Listeria, Bacillus, and Lactobacillus (Garneau et al., 2008). Lactobacillus, Listeria, and Lactococcus are considered beneficial bacteria that were important in maintaining the balance of intestinal microbiota and preventing pathogenic invasion (Wang et al., 2018; Li et al., 2019). Furthermore, Firmicutes have key roles in the digestion of proteins and carbohydrates (Spence et al., 2006). The main function of Bacteroidetes was to increase the utilization of carbohydrates (Jami et al., 2013). In addition, a high F/B (Firmicutes/Bacteroides) ratio helps a host to absorb energy and maintain microbiota homeostasis (Fernando et al., 2010; Murphy et al., 2010). In this study, the addition of EOZB increased significantly the abundance of Firmicutes and decreased significantly the abundance of Bacteroidetes in the DLW group, which caused an increase in the F/B ratio. This may be due to the antimicrobial properties of EOZB, where many microorganisms competed with each other, and a decrease in the number of one part of the microorganisms in competition may lead to an increase in the number of another part. It also indicated that EOZB may have a role in improving energy absorption, maintaining microbiota homeostasis, maintaining intestinal microorganism balance, and improving protein and carbohydrate digestion. Tenericutes were associated with fat deposition in Angus bull muscle (Zhan et al., 2017; Krause et al., 2020), and Proteobacteria were associated with energy accumulation (Amato et al., 2014; Chevalier et al., 2015). We found that the addition of EOZB increased significantly the abundance of Tenericutes in the CLW group and increased significantly the abundance of Proteobacteria in the BLW group. This may be due to the dose of EOZB, and different microorganisms may have different sensitivity to the dose of EOZB. It also indicated that EOZB may have a role in regulating energy accumulation and fat deposition. The dominant genera in the duodenum of STH were Prevotella 1, Christensenellaceae R-7 group, Ruminococcus 2, Rikenellaceae RC9 gut group, Eubacterium coprostanoligenes group, Ruminococcaceae NK4A214 group, Saccharofermentans, Ruminococcaceae UCG-014, and Prevotellaceae UCG-001. Relative abundances of Prevotella 1, Ruminococcus 2, and Eubacterium coprostanoligenes group were significantly higher in BLW, DLW, and DLW than in the other group (p < 0.001, p < 0.001, and p = 0.001, respectively). Prevotella 1 can degrade fiber-derived feeds such as hemicellulose or xylan (Emerson and Weimer, 2017). In addition, Prevotella can use starch, monosaccharides, and other noncellulosic polysaccharides (Purushe et al., 2010). Ruminococcus 2 belongs to Ruminococcus, which primarily inhabits the rumen and hindgut of ruminants and contributes to the degradation of cellulose and starch (Zhao et al., 2018). Ruminococcus also produces acetic acid, formic acid, and a small amount of lactic acid (Berg Miller et al., 2009). Short-chain fatty acids, including formic acid and acetate, have key roles in regulating gut microbiota balance and maintaining the morphology and function of intestinal epithelial cells (Tan et al., 2014; Li et al., 2019). The Eubacterium coprostanoligenes group functions in converting cholesterol to coproitol, which can further affect the fat metabolism of a host (Freier et al., 1994). We found that supplementation of EOZB increased significantly the abundance of Prevotella 1 in the BLW group, Ruminococcus 2 and Eubacterium coprostanoligenes group in the DLW group. This may be due to the dose and antimicrobial properties of EOZB, with different microorganisms responding differently to different doses of EOZB. It also suggests that EOZB may have a role in maintaining intestinal microorganism balance and enhancing carbohydrate digestion and absorption with lipid metabolism. This was similar to the function of differential microbiota at the phylum level.
Firmicutes, Bacteroidetes, Tenericutes, Proteobacteria, and Spirochaetes were the dominant phyla of bacteria in the jejunum of STH. Relative abundances of Firmicutes, Bacteroidetes, Tenericutes, and Proteobacteria increased significantly in DLW, CLW, BLW, and ALW groups than in the other groups (p < 0.001, p < 0.001, p = 0.002, and p = 0.001, respectively). Spirochaetes were closely associated with fat metabolism pathways (Yang et al., 2022) and can degrade pectin and xylan (Liu et al., 2014; Tokuda et al., 2018). Spirochaetes was the dominant bacteria in this study, but the difference in relative abundance between groups was not significant. However, Firmicutes, Bacteroidetes, Tenericutes and Proteobacteria in the dominant bacteria increased significantly in the DLW, CLW, BLW and ALW groups than in the other trial groups. This may be due to jejunal function and the dose of EOZB, the jejunum of ruminants is dominated by chemical digestion and the main function of microorganisms is to secrete various digestive enzymes. Combined with the functions of the dominant bacteria mentioned earlier, this may indicate that EOZB has a role in regulating the microorganisms that secrete various digestive enzymes and thus improve carbohydrate and lipid absorption. At the genus level, Prevotella 1, Rikenellaceae RC9 gut group, Christensenellaceae R-7 group, Saccharofermentans, Ruminococcaceae NK4A214 group, Prevotellaceae UCG-001, Eubacterium coprostanoligenes group, and Ruminococcaceae UCG-014 were the dominant bacteria in the jejunum of STH. Relative abundances of Prevotella 1, Rikenellaceae RC9 gut group, Christensenellaceae R-7 group, Ruminococcaceae UCG-014, Saccharofermentans, Ruminococcaceae NK4A214 group, and Prevotellaceae UCG-001 were significantly higher in CLW, CLW, BLW, BLW, BLW, ALW, and ALW, respectively, than in the other groups (p < 0.001, p < 0.001, p = 0.006, p = 0.049, p < 0.001, p < 0.001, and p = 0.02, respectively). Rikenellaceae RC9 gut group belongs to Rikenellaceae and most can ferment unabsorbed polysaccharides in the host gut to produce short-chain fatty acids such as acetate, propionate, and butyrate (Su et al., 2014). Saccharofermentans are anaerobic gram-negative bacteria that convert glucose to acetate, lactate, and fumarate in vivo (Chen et al., 2010). We found that supplementation with EOZB significantly increased the relative abundance of Saccharofermentans in the BLW group and the Rikenellaceae RC9 gut group in the CLW group. This may be related to the dose of EOZB, with different microorganisms responding to different doses of EOZB. It also suggests that EOZB may have a role in improving polysaccharide absorption and VFA (Volatile Fatty Acid) production. Christensenellaceae R-7 group is in the Firmicutes (Waters and Ley, 2019) and has a key role in the decomposition of fibrous substances (Evans et al., 2011). Ruminococcaceae UCG-014 and Ruminococcaceae NK4A214 group are in the family Ruminococcaceae and have crucial roles in fiber degradation and biohydrogenation (Gagen et al., 2015; Opdahl et al., 2018). In addition, the relative abundance of Ruminococcaceae increases in beef cattle with high feed efficiency (Li and Guan, 2017). We found that EOZB supplementation significantly increased the abundance of Christensenellaceae R-7 group and Ruminococcaceae UCG-014 and significantly decreased the abundance of the Ruminococcaceae NK4A214 group in the BLW group. This may be due to the dose and antimicrobial characteristics of EOZB, with different microorganisms responding to different doses of EOZB. It also suggests that EOZB may have a role in improving fiber degradation and feed efficiency. Prevotellaceae UCG-001 is a Prevotella strain that has an important role in the degradation of cellulose and xylan. Prevotellaceae UCG-001 was significantly enriched after feeding mice with inulin and thereby alleviated glucose and lipid metabolism disorders (Song et al., 2019). In addition, the proportion of Prevotellaceae UCG-001 increases in the azomethane-induced mouse colon cancer model (Ibrahim et al., 2019). We found that Prevotellaceae UCG-001 was significantly increased in the ALW group than in the other trail groups. This may be due to the anti-inflammatory characteristics of EOZB. EOZB has an anti-inflammatory effect, which in turn inhibits the growth of microorganisms associated with pro-inflammatory responses. It also suggests that EOZB may have a role in regulating the catabolism of fibrous substances, glycolipid metabolism and improving the inflammatory process.
Firmicutes, Bacteroidetes, and Tenericutes were the dominant phyla of bacteria in the ileum of STH. Relative abundances of Firmicutes, Bacteroidetes, and Tenericutes were significantly higher in DLW, ALW, and DLW, respectively than in the other groups (all p < 0.001). The functions of those phyla of bacteria are discussed in the section on the duodenum. At the genus level, Prevotella 1, Romboutsia, Christensenellaceae R-7 group, Rikenellaceae RC9 gut group, Turicibacter, Ruminococcaceae NK4A214 group, Saccharofermentans, Clostridium sensu stricto 1, Ruminococcus 2, Eubacterium coprostanoligenes group, Ruminococcaceae UCG-011, Ruminococcaceae UCG-014, Paeniclostridium, and Prevotellaceae UCG-001 were the dominant bacteria in the ileum of STH. Relative abundances of Prevotella 1, Christensenellaceae R-7 group, Romboutsia, and Ruminococcaceae UCG-014 were significantly higher in DLW, DLW, ALW, and CLW, respectively, than in the other groups (p < 0.001, p = 0.001, p < 0.001, and p = 0.039). Romboutsia is significantly positively correlated with total carbohydrate digestibility (Li et al., 2022). In addition, Romboutsia has effects on the inflammatory process and is inversely related to inflammatory bowel disease (Schirmer et al., 2019). We found that Romboutsia was significantly increased in the ALW group than the other trial groups. This may be due to the anti-inflammatory characteristics of EOZB, which reduces the relative abundance of Romboutsia by inhibiting the inflammatory process and thus. It also suggests that EOZB may have a role in regulating carbohydrate digestion and inflammatory processes.
At the same dose of EOZB, the composition of the five most abundant genera of bacteria differed in different sites of the small intestine. In the ALW group, the Ruminococcaceae NK4A214 group was a jejunum-specific genus of bacteria, whereas Romboutsia was an ileum-specific genus of bacteria. The Ruminococcaceae NK4A214 group is in the family Ruminococcaceae and is highly associated with biohydrogenation in ruminants, in addition to its role in fiber degradation, which in turn affects lipid metabolism (Wang et al., 2019). The genus Romboutsia is more abundant in healthy guts than in polyp-associated gut tissue, suggesting that Romboutsia is important in maintaining gut health (Mangifesta et al., 2018). This may be due to the function of different small intestinal sites, which have different functions and therefore different compositions of microorganisms that maintain these functions. It also suggests that lipid metabolism may be one of the functions of the jejunum and that the function of the ileum may be related to intestinal health. Among the most abundant genera of bacteria in the BLW group, Eubacterium coprostanoligenes group was the ileum-specific genus of bacteria. Compared with patients with irritable bowel syndrome, the Eubacterium coprostanoligenes group is the dominant bacteria in healthy controls (Liu et al., 2021). This may be due to the function of different parts of the small intestine and the anti-inflammatory characteristics of EOZB, with differences in function resulting in different microbial compositions. It also suggests that the function of the ileum may be related to the regulation of inflammation and that EOZB may have a role in regulating the inflammatory process. Among the most abundant genera of bacteria in the CLW group, Lachnospiraceae NK3A20 group was the duodenum-specific genus of bacteria. Lachnospiraceae NK3A20 group is in the family Lachnospiraceae, which is one of the main components of ruminant gastrointestinal microbiota (Kittelmann et al., 2013) and is closely associated with butyrate production (Vital et al., 2014; Haas and Blanchard, 2017). Butyrate contributes to the development of gastrointestinal epithelial cells in ruminants (Xu et al., 2001). This may be related to the anti-inflammatory characteristics of EOZB, where intestinal epithelial cells are part of non-specific immunity and the proliferation of epithelial cells can reduce the permeability of intestinal epithelial tissue to some extent, and the reduced permeability leads to a decrease in the passage of pro-inflammatory factors and pathogenic microorganisms, which in turn regulates the inflammatory process. It also suggests that the anti-inflammatory effect of EOZB may have a relationship with an intestinal barrier function. Among the most abundant genera of bacteria in the DLW group, Ruminococcus 2 and Lachnospiraceae NK3A20 group were duodenal endemic bacteria, and Clostridium sensu stricto 1 was the ileal endemic bacteria. Clostridium sensu stricto 1 is a butyrate-producing genus of clostridia bacteria, and butyrate is essential for the development of gastrointestinal epithelial cells and as the final metabolite of unabsorbed polysaccharides by the gut microbiota, for host energy supply (Pryde et al., 2002). The results were similar to those of the CLW group, which may be related to the anti-inflammatory properties of EOZB. It also suggests that EOZB may have some connection with the production of butyrate. In addition, our other study also showed that EOZB significantly increased final weight, average daily gain, dry matter intake and feed conversion ratio in STH. This suggests that these bacteria that were altered by the intervention of EOZB may have a positive effect on the productive performance of STH.
Conclusion
In this study, the affection of EOZB on bacterial distribution in the small intestine of STH was characterized. There were significant differences in the horizontal distribution of bacteria in the same part of the small intestine at different doses of EOZB. With EOZB, in the duodenum, there were four different phyla of bacteria and three different genera, whereas in the jejunum, there were four different phyla of bacteria and seven different genera. In the ileum, there were three different phyla and four different genera. The composition (vertical) of the five most abundant genera of bacteria was different in different parts of the small intestine (duodenum, jejunum, and ileum) at the same dose of EOZB. Some genera were specific to different sites in the small intestine at the same dose of EOZB. The differential genera of bacteria are associated with the digestion and absorption of host proteins, carbohydrates, and lipids. Simultaneously, those bacteria can have positive effects on the development of host intestinal epithelial cells, maintenance of microbial and metabolic homeostasis, and control of inflammatory processes. Thus, although the evidence is indirect, it indicates that EOZB affects the above physiological processes. Our findings fill the gap of bacteria composition structure of small intestine in ruminants, and also provide some theoretical basis for the selection of probiotic bacteria and the development and application of EOZB.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The animal study was reviewed and approved by Experimental Animal Ethics Committee of Gansu Agricultural University (Approval No. GSAU-Eth-AST-2021-026). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
CW and XL conceived, designed, and supervised the project. HZ, XL, and YZ collected samples and performed experiments. HZ and XL carried out bioinformatic analyzes. HZ drafted the manuscript. HZ, XL, YZ, and CW revised the manuscript. XL, HZ, and CW, contributed ideas on the paper. All authors have read, edited, and approved the final manuscript.
Funding
This work was supported by the Project of Research Condition Construction and Achievement Transformation of Gansu Academy of Agricultural Sciences (Modern Biological Breeding, 2021GAAS01).
Acknowledgments
We are grateful to the many contributors to the sequence read achievement for making their data publicly available, and to colleagues at Genepioneer Biotechnologies (Jiangsu, China) for sequencing and bioinformatics analysis. We thank the staff at Sen Wuzhu Field and Pasture Farming Cooperative for helping to feed the test animals! We also thank the Gansu Agriculture University and the Gansu Academy of Agricultural Sciences for their help in collecting samples!
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1062077/full#supplementary-material
References
Amato, K. R., Leigh, S. R., Kent, A., Mackie, R. I., Yeoman, C. J., Stumpf, R. M., et al. (2014). The role of gut microbes in satisfying the nutritional demands of adult and juvenile wild, black howler monkeys (Alouatta pigra). Am. J. Phys. Anthropol. 155, 652–664. doi: 10.1002/ajpa.22621
Backhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A., and Gordon, J. I. (2005). Host-bacterial mutualism in the human intestine. Science 307, 1915–1920. doi: 10.1126/science.1104816
Berg Miller, M. E., Antonopoulos, D. A., Rincon, M. T., Band, M., Bari, A., Akraiko, T., et al. (2009). Diversity and strain specificity of plant cell wall degrading enzymes revealed by the draft genome of Ruminococcus flavefaciens FD-1. PLoS One 4:e6650. doi: 10.1371/journal.pone.0006650
Chen, S., Niu, L., and Zhang, Y. (2010). Saccharofermentans acetigenes gen. Nov., sp. nov., an anaerobic bacterium isolated from sludge treating brewery wastewater. Int. J. Syst. Evol. Microbiol. 60, 2735–2738. doi: 10.1099/ijs.0.017590-0
Chevalier, C., Stojanovic, O., Colin, D. J., Suarez-Zamorano, N., Tarallo, V., Veyrat-Durebex, C., et al. (2015). Gut microbiota orchestrates energy homeostasis during cold. Cells 163, 1360–1374. doi: 10.1016/j.cell.2015.11.004
Dubois, P. C., Trynka, G., Franke, L., Hunt, K. A., Romanos, J., Curtotti, A., et al. (2010). Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 42, 295–302. doi: 10.1038/ng.543
Eisler, M. C., Lee, M. R., Tarlton, J. F., Martin, G. B., Beddington, J., Dungait, J. A., et al. (2014). Agriculture: steps to sustainable livestock. Nature 507, 32–34. doi: 10.1038/507032a
Emerson, E. L., and Weimer, P. J. (2017). Fermentation of model hemicelluloses by Prevotella strains and Butyrivibrio fibrisolvens in pure culture and in ruminal enrichment cultures. Appl. Microbiol. Biotechnol. 101, 4269–4278. doi: 10.1007/s00253-017-8150-7
Evans, N. J., Brown, J. M., Murray, R. D., Getty, B., Birtles, R. J., Hart, C. A., et al. (2011). Characterization of novel bovine gastrointestinal tract Treponema isolates and comparison with bovine digital dermatitis treponemes. Appl. Environ. Microbiol. 77, 138–147. doi: 10.1128/AEM.00993-10
Fernando, S. C., Purvis, H. T., Najar, F. Z., Sukharnikov, L. O., Krehbiel, C. R., Nagaraja, T. G., et al. (2010). Rumen microbial population dynamics during adaptation to a high-grain diet. Appl. Environ. Microbiol. 76, 7482–7490. doi: 10.1128/AEM.00388-10
Freier, T. A., Beitz, D. C., Li, L., and Hartman, P. A. (1994). Characterization of Eubacterium coprostanoligenes sp. nov., a cholesterol-reducing anaerobe. Int. J. Syst. Bacteriol. 44, 137–142. doi: 10.1099/00207713-44-1-137
Gagen, E. J., Padmanabha, J., Denman, S. E., and McSweeney, C. S. (2015). Hydrogenotrophic culture enrichment reveals rumen Lachnospiraceae and Ruminococcaceae acetogens and hydrogen-responsive Bacteroidetes from pasture-fed cattle. FEMS Microbiol. Lett. 362:fnv104. doi: 10.1093/femsle/fnv104
Garneau, J. E., Tremblay, D. M., and Moineau, S. (2008). Characterization of 1706, a virulent phage from Lactococcus lactis with similarities to prophages from other Firmicutes. Virology 373, 298–309. doi: 10.1016/j.virol.2007.12.002
Gillor, O., Etzion, A., and Riley, M. (2008). The dual role of bacteriocins as anti-and probiotics. Appl. Microbiol. Biotechnol. 81, 591–606. doi: 10.1007/s00253-008-1726-5
Greening, C., Geier, R., Wang, C., Woods, L. C., Morales, S. E., McDonald, M. J., et al. (2019). Diverse hydrogen production and consumption pathways influence methane production in ruminants. ISME J. 13, 2617–2632. doi: 10.1038/s41396-019-0464-2
Guleria, S., Tiku, A. K., Koul, A., Gupta, S., Singh, G., and Razdan, V. K. (2013). Antioxidant and antimicrobial properties of the essential oil and extracts of Zanthoxylum alatum grown in north-western Himalaya. ScientificWorldJournal 2013:790580. doi: 10.1155/2013/790580
Haas, K. N., and Blanchard, J. L. (2017). Kineothrix alysoides, gen. Nov., sp. nov., a saccharolytic butyrate-producer within the family Lachnospiraceae. Int. J. Syst. Evol. Microbiol. 67, 402–410. doi: 10.1099/ijsem.0.001643
Ibrahim, A., Hugerth, L. W., Hases, L., Saxena, A., Seifert, M., Thomas, Q., et al. (2019). Colitis-induced colorectal cancer and intestinal epithelial estrogen receptor beta impact gut microbiota diversity. Int. J. Cancer 144, 3086–3098. doi: 10.1002/ijc.32037
Jami, E., Israel, A., Kotser, A., and Mizrahi, I. (2013). Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 7, 1069–1079. doi: 10.1038/ismej.2013.2
Karczewski, J., Troost, F. J., Konings, I., Dekker, J., Kleerebezem, M., Brummer, R. J., et al. (2010). Regulation of human epithelial tight junction proteins by Lactobacillus plantarum in vivo and protective effects on the epithelial barrier. Am. J. Physiol. Gastrointest. Liver Physiol. 298, G851–G859. doi: 10.1152/ajpgi.00327.2009
Kittelmann, S., Seedorf, H., Walters, W. A., Clemente, J. C., Knight, R., Gordon, J. I., et al. (2013). Simultaneous amplicon sequencing to explore co-occurrence patterns of bacterial, archaeal and eukaryotic microorganisms in rumen microbial communities. PLoS One 8:e47879. doi: 10.1371/journal.pone.0047879
Krause, T. R., Lourenco, J. M., Welch, C. B., Rothrock, M. J., Callaway, T. R., and Pringle, T. D. (2020). The relationship between the rumen microbiome and carcass merit in Angus steers. J. Anim. Sci. 98:skaa287. doi: 10.1093/jas/skaa287
Li, F., and Guan, L. L. (2017). Metatranscriptomic profiling reveals linkages between the active rumen microbiome and feed efficiency in beef cattle. Appl. Environ. Microbiol. 83, e00061–e00017. doi: 10.1128/AEM.00061-17
Li, Y., Han, Y., Zhao, Q., Tang, C., Zhang, J., and Qin, Y. (2022). Fermented soy and fish protein dietary sources shape Ileal and colonic microbiota, improving nutrient digestibility and host health in a piglet model. Front. Microbiol. 13:911500. doi: 10.3389/fmicb.2022.911500
Li, K., Nader, S. M., Zhang, X., Ray, B. C., Kim, C. Y., Das, A., et al. (2019). Novel lactate dehydrogenase inhibitors with in vivo efficacy against Cryptosporidium parvum. PLoS Pathog. 15:e1007953. doi: 10.1371/journal.ppat.1007953
Li, A., Wang, Y., Li, Z., Qamar, H., Mehmood, K., Zhang, L., et al. (2019). Probiotics isolated from yaks improves the growth performance, antioxidant activity, and cytokines related to immunity and inflammation in mice. Microb. Cell Fact. 18:112. doi: 10.1186/s12934-019-1161-6
Li, Y., Zeng, J., Liu, L., and Jin, X. (2001). GC-MS analysis of supercritical carbon dioxide extraction products from pericarp of Zanthoxylum bungeanum. Zhong Yao Cai 24, 572–573. (in Chinese). doi: 10.13863/j.issn1001-4454.2001.08.015
Li, B., Zhang, X., Guo, F., Wu, W., and Zhang, T. (2013). Characterization of tetracycline resistant bacterial community in saline activated sludge using batch stress incubation with high-throughput sequencing analysis. Water Res. 47, 4207–4216. doi: 10.1016/j.watres.2013.04.021
Li, J., Zhong, H., Ramayo-Caldas, Y., Terrapon, N., Lombard, V., Potocki-Veronese, G., et al. (2020). A catalog of microbial genes from the bovine rumen unveils a specialized and diverse biomass-degrading environment. Gigascience 9:giaa057. doi: 10.1093/gigascience/giaa057
Liu, Y., Li, W., Yang, H., Zhang, X., Wang, W., Jia, S., et al. (2021). Leveraging 16S rRNA microbiome sequencing data to identify bacterial signatures for irritable bowel syndrome. Front. Cell. Infect. Microbiol. 11:645951. doi: 10.3389/fcimb.2021.645951
Liu, J., Wang, J.-K., Zhu, W., Pu, Y.-Y., Guan, L.-L., and Liu, J.-X. (2014). Monitoring the rumen pectinolytic bacteria Treponema saccharophilum using real-time PCR. FEMS Microbiol. Ecol. 87, 576–585. doi: 10.1111/1574-6941.12246
Mangifesta, M., Mancabelli, L., Milani, C., Gaiani, F., Deangelis, N., Deangelis, G. L., et al. (2018). Mucosal microbiota of intestinal polyps reveals putative biomarkers of colorectal cancer. Sci. Rep. 8:13974. doi: 10.1038/s41598-018-32413-2
Martinez-Guryn, K., Leone, V., and Chang, E. B. (2019). Regional diversity of the gastrointestinal microbiome. Cell Host Microbe 26, 314–324. doi: 10.1016/j.chom.2019.08.011
Masella, A. P., Bartram, A. K., Truszkowski, J. M., Brown, D. G., and Neufeld, J. D. (2012). PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics 13:31. doi: 10.1186/1471-2105-13-31
Moraïs, S., and Mizrahi, I. (2019). The road not taken: the rumen microbiome, functional groups, and community states. Trends Microbiol. 27, 538–549. doi: 10.1016/j.tim.2018.12.011
Murphy, E. F., Cotter, P. D., Healy, S., Marques, T. M., O'Sullivan, O., Fouhy, F., et al. (2010). Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut 59, 1635–1642. doi: 10.1136/gut.2010.215665
O'Hara, E., Neves, A. L., Song, Y., and Guan, L. L. (2020). The role of the gut microbiome in cattle production and health: driver or passenger? Ann. Rev. Anim. Biosci. 8, 199–220. doi: 10.1146/annurev-animal-021419-083952
Opdahl, L. J., Gonda, M. G., and St-Pierre, B. (2018). Identification of uncultured bacterial species from Firmicutes, Bacteroidetes and CANDIDATUS Saccharibacteria as candidate cellulose utilizers from the rumen of beef cows. Microorganisms 6:17. doi: 10.3390/microorganisms6010017
Pryde, S. E., Duncan, S. H., Hold, G. L., Stewart, C. S., and Flint, H. J. (2002). The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 217, 133–139. doi: 10.1111/j.1574-6968.2002.tb11467.x
Purushe, J., Fouts, D. E., Morrison, M., White, B. A., Mackie, R. I., Coutinho, P. M., et al. (2010). Comparative genome analysis of Prevotella ruminicola and Prevotella bryantii: insights into their environmental niche. Microb. Ecol. 60, 721–729. doi: 10.1007/s00248-010-9692-8
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahe, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ. 4:e2584. doi: 10.7717/peerj.2584
Russell, J. B., and Rychlik, J. L. (2001). Factors that alter rumen microbial ecology. Science 292, 1119–1122. doi: 10.1126/science.1058830
Sakaki, T., Takeshima, T., Tominaga, M., Hashimoto, H., and Kawaguchi, S. (1994). Recurrence of ICA-PCoA aneurysms after neck clipping. J. Neurosurg. 80, 58–63. doi: 10.3171/jns.1994.80.1.0058
Schirmer, M., Garner, A., Vlamakis, H., and Xavier, R. J. (2019). Microbial genes and pathways in inflammatory bowel disease. Nat. Rev. Microbiol. 17, 497–511. doi: 10.1038/s41579-019-0213-6
Schmieder, R., and Edwards, R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864. doi: 10.1093/bioinformatics/btr026
Schuijt, T. J., van der Poll, T., de Vos, W. M., and Wiersinga, W. J. (2013). The intestinal microbiota and host immune interactions in the critically ill. Trends Microbiol. 21, 221–229. doi: 10.1016/j.tim.2013.02.001
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Seshadri, R., Leahy, S. C., Attwood, G. T., Teh, K. H., Lambie, S. C., Cookson, A. L., et al. (2018). Cultivation and sequencing of rumen microbiome members from the Hungate1000 collection. Nat. Biotechnol. 36, 359–367. doi: 10.1038/nbt.4110
Shabat, S. K., Sasson, G., Doron-Faigenboim, A., Durman, T., Yaacoby, S., Berg Miller, M. E., et al. (2016). Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 10, 2958–2972. doi: 10.1038/ismej.2016.62
Shi, W., Moon, C. D., Leahy, S. C., Kang, D., Froula, J., Kittelmann, S., et al. (2014). Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome Res. 24, 1517–1525. doi: 10.1101/gr.168245.113
Song, X., Zhong, L., Lyu, N., Liu, F., Li, B., Hao, Y., et al. (2019). Inulin can alleviate metabolism disorders in Ob/Ob mice by partially restoring leptin-related pathways mediated by gut microbiota. Genom. Proteom. Bioinformat. 17, 64–75. doi: 10.1016/j.gpb.2019.03.001
Spence, C., Wells, W. G., and Smith, C. J. (2006). Characterization of the primary starch utilization operon in the obligate anaerobe Bacteroides fragilis: regulation by carbon source and oxygen. J. Bacteriol. 188, 4663–4672. doi: 10.1128/JB.00125-06
Stewart, R. D., Auffret, M. D., Warr, A., Walker, A. W., Roehe, R., and Watson, M. (2019). Compendium of 4,941 rumen metagenome-assembled genomes for rumen microbiome biology and enzyme discovery. Nat. Biotechnol. 37, 953–961. doi: 10.1038/s41587-019-0202-3
Stewart, R. D., Auffret, M. D., Warr, A., Wiser, A. H., Press, M. O., Langford, K. W., et al. (2018). Assembly of 913 microbial genomes from metagenomic sequencing of the cow rumen. Nat. Commun. 9:870. doi: 10.1038/s41467-018-03317-6
Su, X. L., Tian, Q., Zhang, J., Yuan, X. Z., Shi, X. S., Guo, R. B., et al. (2014). Acetobacteroides hydrogenigenes gen. Nov., sp. nov., an anaerobic hydrogen-producing bacterium in the family Rikenellaceae isolated from a reed swamp. Int. J. Syst. Evol. Microbiol. 64, 2986–2991. doi: 10.1099/ijs.0.063917-0
Tan, J., McKenzie, C., Potamitis, M., Thorburn, A. N., Mackay, C. R., and Macia, L. (2014). The role of short-chain fatty acids in health and disease. Adv. Immunol. 121, 91–119. doi: 10.1016/B978-0-12-800100-4.00003-9
Tezuka, Y., Irikawa, S., Kaneko, T., Banskota, A. H., Nagaoka, T., Xiong, Q., et al. (2001). Screening of Chinese herbal drug extracts for inhibitory activity on nitric oxide production and identification of an active compound of Zanthoxylum bungeanum. J. Ethnopharmacol. 77, 209–217. doi: 10.1016/s0378-8741(01)00300-2
Thaiss, C. A., Zmora, N., Levy, M., and Elinav, E. (2016). The microbiome and innate immunity. Nature 535, 65–74. doi: 10.1038/nature18847
Tokuda, G., Mikaelyan, A., Fukui, C., Matsuura, Y., Watanabe, H., Fujishima, M., et al. (2018). Fiber-associated spirochetes are major agents of hemicellulose degradation in the hindgut of wood-feeding higher termites. Proc. Natl. Acad. Sci. U. S. A. 115, E11996–E12004. doi: 10.1073/pnas.1810550115
Vital, M., Howe, A. C., and Tiedje, J. M. (2014). Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio 5:e00889. doi: 10.1128/mBio.00889-14
Wang, Y., Li, A., Jiang, X., Zhang, H., Mehmood, K., Zhang, L., et al. (2018). Probiotic potential of Leuconostoc pseudomesenteroides and Lactobacillus strains isolated from yaks. Front. Microbiol. 9:2987. doi: 10.3389/fmicb.2018.02987
Wang, X., Martin, G. B., Wen, Q., Liu, S., Zhang, J., Yu, Y., et al. (2019). Linseed oil and heated linseed grain supplements have different effects on rumen bacterial community structures and fatty acid profiles in cashmere kids1. J. Anim. Sci. 97, 2099–2113. doi: 10.1093/jas/skz079
Waters, J. L., and Ley, R. E. (2019). The human gut bacteria Christensenellaceae are widespread, heritable, and associated with health. BMC Biol. 17:83. doi: 10.1186/s12915-019-0699-4
Xu, H., Collins, J. F., Bai, L., Kiela, P. R., Lynch, R. M., and Ghishan, F. K. (2001). Epidermal growth factor regulation of rat NHE2 gene expression. Am. J. Physiol. Cell Physiol. 281, C504–C513. doi: 10.1152/ajpcell.(2001).281.2.C504
Yáñez-Ruiz, D. R., Abecia, L., and Newbold, C. J. (2015). Manipulating rumen microbiome and fermentation through interventions during early life: a review. Front. Microbiol. 6:1133. doi: 10.3389/fmicb.2015.01133
Yang, T., Jiang, Y., Tang, J., Guobin, C., Zhao, W., Hou, S., et al. (2022). Comparison of cecal microbiota and performance indices between lean-type and fatty-type Pekin ducks. Front. Microbiol. 13:639. doi: 10.3389/fmicb.2022.820569
Zhan, J., Liu, M., Wu, C., Su, X., Zhan, K., and Zhao, G. Q. (2017). Effects of alfalfa flavonoids extract on the microbial flora of dairy cow rumen. Asian-Australas J. Anim Sci. 30, 1261–1269. doi: 10.5713/ajas.16.0839
Zhang, H., Lang, X., Li, X., Chen, G., and Wang, C. (2022). Effect of Zanthoxylum bungeanum essential oil on rumen enzyme activity, microbiome, and metabolites in lambs. PLoS One 17:e0272310. doi: 10.1371/journal.pone.0272310
Zhang, Z., Shen, P., Liu, J., Gu, C., Lu, X., Li, Y., et al. (2017). In vivo study of the efficacy of the essential oil of Zanthoxylum bungeanum pericarp in dextran sulfate sodium-induced murine experimental colitis. J. Agric. Food Chem. 65, 3311–3319. doi: 10.1021/acs.jafc.7b01323
Zhang, Z., Shen, P., Lu, X., Li, Y., Liu, J., Liu, B., et al. (2017). In vivo and in vitro study on the efficacy of Terpinen-4-ol in dextran sulfate sodium-induced mice experimental colitis. Front. Immunol. 8:558. doi: 10.3389/fimmu.2017.00558
Zhang, Y., Wang, D., Yang, L., Zhou, D., and Zhang, J. (2014). Purification and characterization of flavonoids from the leaves of Zanthoxylum bungeanum and correlation between their structure and antioxidant activity. PLoS One 9:e105725. doi: 10.1371/journal.pone.0105725
Keywords: ruminant, Zanthoxylum bungeanum essential oil, gut microbiota, 16s rRNA gene sequencing, LEfSe analysis
Citation: Zhang H, Lang X, Zhang Y and Wang C (2022) Distribution of bacteria in different regions of the small intestine with Zanthoxylum bungeanum essential oil supplement in small-tailed Han sheep. Front. Microbiol. 13:1062077. doi: 10.3389/fmicb.2022.1062077
Edited by:
Shengguo Zhao, Institute of Animal Sciences (CAAS), ChinaReviewed by:
Kaizhen Liu, Chinese Academy of Agricultural Sciences (CAAS), ChinaQingbiao Xu, Huazhong Agricultural University, China
Jianwei Zhou, Lanzhou University, China
Copyright © 2022 Zhang, Lang, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cailian Wang, d2FuZ2NsMTk3NEAxNjMuY29t
 Hailong Zhang
Hailong Zhang Xia Lang1
Xia Lang1