
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 09 January 2023
Sec. Food Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1061223
This article is part of the Research TopicAntimicrobial Resistant Pathogens in Sustainable Food Systems: How to Prevent, Detect or Control Contamination?View all 7 articles
 Agnieszka Kijewska1†
Agnieszka Kijewska1† Aleksandra Koroza2†
Aleksandra Koroza2† Katarzyna Grudlewska-Buda3†
Katarzyna Grudlewska-Buda3† Tomasz Kijewski2
Tomasz Kijewski2 Natalia Wiktorczyk-Kapischke3
Natalia Wiktorczyk-Kapischke3 Katarzyna Zorena1
Katarzyna Zorena1 Krzysztof Skowron3*
Krzysztof Skowron3*Bivalve shellfish consumption (ark shells, clams, cockles, and oysters) has increased over the last decades. Following this trend, infectious disease outbreaks associated with their consumption have been reported more frequently. Molluscs are a diverse group of organisms found wild and farmed. They are common on our tables, but unfortunately, despite their great taste, they can also pose a threat as a potential vector for numerous species of pathogenic microorganisms. Clams, in particular, might be filled with pathogens because of their filter-feeding diet. This specific way of feeding favors the accumulation of excessive amounts of pathogenic microorganisms like Vibrio spp., including Vibrio cholerae and V. parahaemolyticus, Pseudomonas aeruginosa, Escherichia coli, Arcobacter spp., and fecal coliforms, and intestinal enterococci. The problems of pathogen dissemination and disease outbreaks caused by exogenous bacteria in many geographical regions quickly became an unwanted effect of globalized food supply chains, global climate change, and natural pathogen transmission dynamics. Moreover, some pathogens like Shewanella spp., with high zoonotic potential, are spreading worldwide along with food transport. These bacteria, contained in food, are also responsible for the potential transmission of antibiotic-resistance genes to species belonging to the human microbiota. Finally, they end up in wastewater, thus colonizing new areas, which enables them to introduce new antibiotic-resistance genes (ARG) into the environment and extend the existing spectrum of ARGs already present in local biomes. Foodborne pathogens require modern methods of detection. Similarly, detecting ARGs is necessary to prevent resistance dissemination in new environments, thus preventing future outbreaks, which could threaten associated consumers and workers in the food processing industry.
Graphical Abstract. Bivalve aquacultures are frequently treated with antibiotics for the protection of farmed animals. Extensive antibiotic treatment, by the presence of antibiotics in the water, may generate bacterial strains carrying the antibiotic-resistance (AR) genes in Mobile Genetic Elements (MGE). MGEs transmitted among marine bacteria may also be passed to pathogens in farmed animals and cause foodborne outbreaks. New AR strains might spread in a different environment through water, infected animals and supply chains of seafood and consumers. One of the most known spreading mechanisms is passing AR strains through faeces removed to the water environment. The circulation mechanism of AR genes leads to the need to use new antibiotics, which will probably stop being effective after a certain period of use.
Various bivalve molluscs (phylum Mollusca, class Bivalvia) constitute a significant commodity in fisheries and aquacultures but are also crucial to preserving our ecosystem’s complexity and function. All over the world, bivalve production has increased significantly in recent years. In 2014, global aquaculture production reached 101.1 million tonnes (live weight); in 2018, global aquaculture production, including aquatic plants, was 114.5 million tonnes, with an estimated USD 263 billion (Food and Agriculture Organization [FAO], 2020). In 2022, FAO (Food and Agriculture Organization of United Nations) reported that between 1992 and 2022, world aquaculture production of molluscs increased by about 70 million tonnes (Food and Agriculture Organization [FAO], 2022).
Aquaculture worldwide experienced significant development from the beginning of the 1980s. Increased awareness of the consequences of the long-term, intensive fishing for the wild fish stocks raised an ecological concern which intensified the development of bivalve aquacultures, now widely spread along worldwide coasts. In consequence, the European Union (EU) report about aquaculture displays that EU aquaculture accounts for about 20% of fish and shellfish supply in the EU and directly employs about 70,000 people. In 2020, more than 45% of aquaculture production was shellfish (European Market Observatory for Fisheries and Aquaculture Products [EUMOFA], 2022). According to information published by European Market Observatory for Fisheries and Aquaculture Products [EUMOFA] (2022), the EU’s organic aquaculture production has increased by 60% in 5 years (2015–2020), mainly due to a rise in organic mussel production. The Netherlands, Italy, Germany, Ireland, Denmark, France, Spain, and Bulgaria noted a significant increase in organic mussel production. Organic mussel production accounted for 41,936 tonnes in 2020 (10% of EU mussel production), compared to 18,379 tonnes in 2015. Only production of exclusive oysters increased in France, the most significant European producer, from 900 tonnes in 2018 to 3,220 tonnes in 2020. These numbers represent the scale of production and growth of shellfish share in the European seafood market (European Market Observatory for Fisheries and Aquaculture Products [EUMOFA], 2022).
The bivalves are very attractive food rich in protein (Wright et al., 2018). Usually, shellfish are prepared, cooked, and marinated, but locally, they are also sold fresh. The microbiota in molluscs depends on regions and may change according to climatic changes, salinity, pollution, and other factors. Gut microorganisms play a significant role in the absorption of essential nutrients and produce the energy they need throughout the day, and without gut microbiomes, digesting and breaking down compound molecules from food cannot be accomplished (Ramamurthy et al., 2014; Diwan et al., 2022). Previously, phenotypic characterization, including culture media (also quantitative assessment) and biochemical tests, were used to identify different species that make up the microbiome.
Molluscs such as oysters, mussels, scallops, cockles, and clams might filter and concentrate environmental microorganisms in their tissues (Wright et al., 2018; Bighiu et al., 2019). These microorganisms may still be active when the molluscs are eaten raw. Bivalve molluscs, including blue mussels (Mytilus edulis), great scallops (Pecten maximus), flat oysters (Ostrea edulis), horse mussels (Modiolus modiolus), and carpet shells (Mya arenaria), are suspension feeders and actively filter and retain particles from their surrounding water, including free-living or particle-bound bacteria (Kueh and Chan, 1985; Romalde et al., 2014; Hsu and Lee, 2015; Grevskott et al., 2016, 2017; Zannella et al., 2017; Mudadu et al., 2021). Bivalves and the blue mussel (Mytilus edulis) are successfully used as indicator organisms for marine pollution monitoring. The general assumption is that the mussel appears to be an appropriate sentinel organism because of its global distribution of large and accessible populations, its large size and sedentary adulthood, its tolerance to diverse environmental conditions, the ventilation of large volumes of water for nutrition, respiration, and excretion (Krieger et al., 1981) and its ability to accumulate numerous contaminants (Moy and Walday, 1996).
Musella et al. (2020) noticed that alongside their ecological importance, mussels have a relevant economic value as a species of interest in aquaculture and, at the same time, have long been employed in the biomonitoring of environmental quality in coastal areas. That factor is one of the most interesting because any change in the environment, including the anthropogenic transformation, may result in changes in the microbiota (Lopez-Joven et al., 2018), also showing the increasing problem of antibiotic-resistance genes (ARG) presence in the marine environment and, in consequence, in marine animals.
Antibiotic-resistance (AR) is a serious and growing global health threat. One of the significant sources of AR is the massive use of the broad spectrum of antibiotics in livestock production. Antibiotics are used there for the following purposes: Therapeutic—to treat sick animals; prophylactic—administrated at sub-therapeutic exposure concentrations to prevent disease; metaphylactic—in mass medication to eliminate or minimize an expected outbreak, and as growth, promoters to improve the growth rate and the food conversion (Pepi and Focardi, 2021). According to Chen et al. (2020), only a few countries/international institutions systematically confine the latter purpose due to AR spreading in the environment.
This monitoring of the current status of AR genes in the marine environment is essential due to the possible role of marine bacteria as the reservoir of AR genes and because the spreading of AR genes can increase the number of potentially resistant human and animal pathogens (Ramamurthy et al., 2014; Chen et al., 2020).
Marine pollution is a global and complex problem. Chemical compounds (including antibiotics) and pathogens, mainly introduced to the environment through human activities, appear to be several risks to biota, human health and ecosystems (Weis, 2014; Kolm et al., 2018; Verga et al., 2020). Sewage contains diverse polluting agents that directly and indirectly affect ecosystems and organisms (Islam and Tanaka, 2004; Wright et al., 2018; Bagi and Skogerbø, 2022; Diwan et al., 2022). In many coastal areas, submarine sewage outfalls are used to dispose of partially or fully treated wastewater. In addition to microbial diversity, antibiotics released to sewage may cause significant changes in gene abundances, especially virulence and ARGs, which have been identified in anthropogenically impacted marine and freshwater environments (Bondarczuk and Piotrowska-Seget, 2019; Fresia et al., 2019). In particular, sewage discharges into shallow coastal and subtidal environments may favor eutrophication processes, leading to water and sediment quality deterioration and, finally, decrease or affect biodiversity, with related ecological problems (Grabowska et al., 2020; Figure 1). Water contamination can result in increased numbers of pathogenic bacteria in mussels (Martinez-Albores et al., 2020) because most of the bivalves feed on suspended phytoplankton caught from water by pumping water across the gills. Laboratory results show that bacteria concentration in mussels Dreissena polymorpha is 132 times higher than in surrounding water, persisting several days after peak exposition (Martinez-Albores et al., 2020). This way, the pathogenic bacteria’s level increases compared to contaminated water (Bighiu et al., 2019).
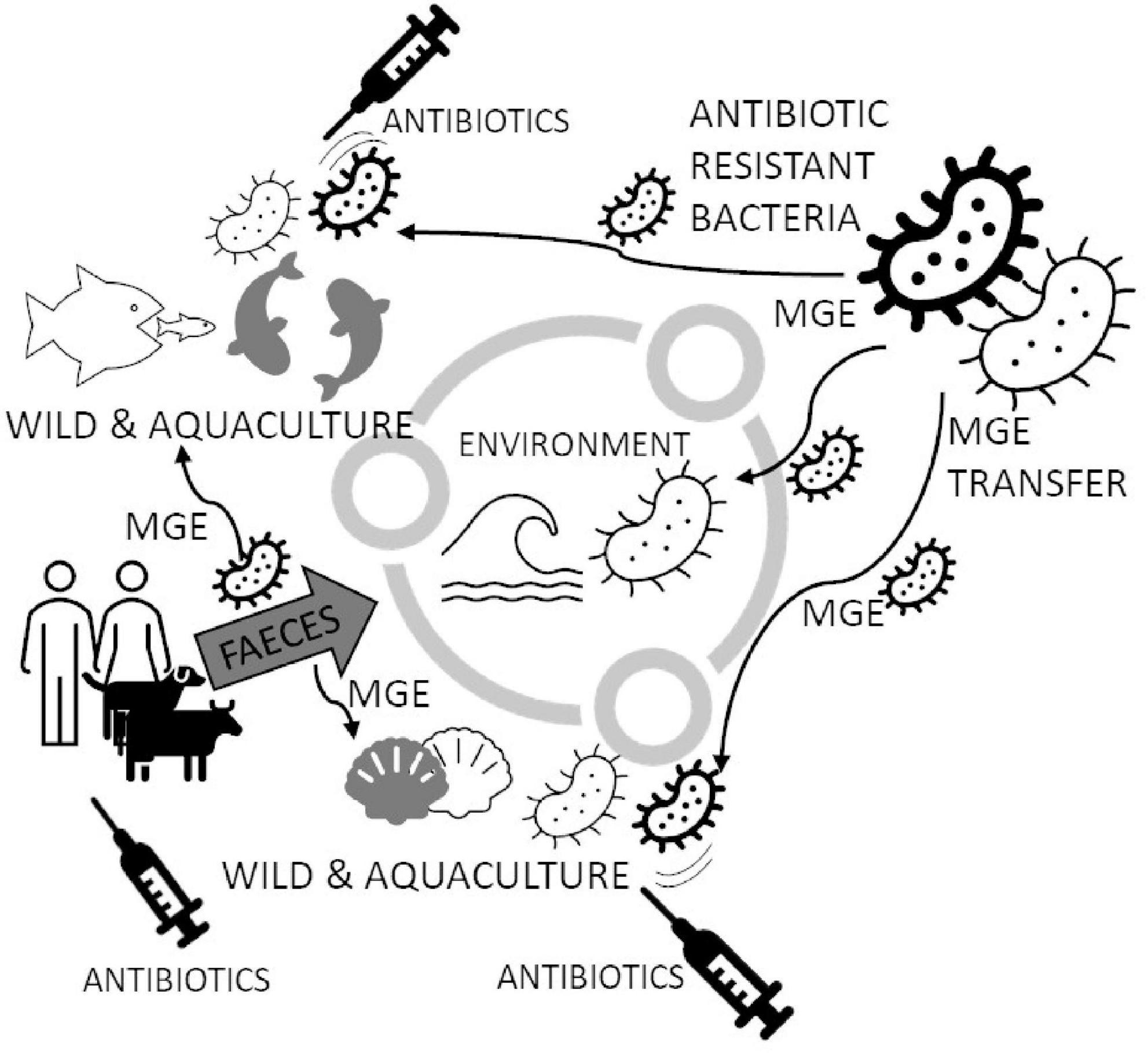
Figure 1. Circulating Mobile Genetic Elements (MGE) may increase Antibiotic Resistant (AR) marine bacterial species in the marine environment. Bacteria in the marine environment may spread resistance genes to other species, especially when antibiotics appear (like in fish aquacultures and bivalves’ larval hatcheries). The spread of MGE generates new strains of bacteria capable of successfully breaking out and becoming resistant to popular antibiotics. Antibiotics are delivered to water also with feces from coastlines, which are usually densely populated. The figure is prepared based on literature (Krieger et al., 1981; Kueh and Chan, 1985; Moy and Walday, 1996; Islam and Tanaka, 2004; Li et al., 2012; Weis, 2014; Hsu and Lee, 2015; Beaudry et al., 2016; Zannella et al., 2017; Chiesa et al., 2018; Kolm et al., 2018; Lopez-Joven et al., 2018; MacFadden et al., 2018; Bondarczuk and Piotrowska-Seget, 2019; Fresia et al., 2019; Hoegh-Guldberg et al., 2019; Pinsky et al., 2019; Chen et al., 2020; Grabowska et al., 2020; Martinez-Albores et al., 2020; Musella et al., 2020; Preena et al., 2020; Verga et al., 2020; Albano et al., 2021; Mudadu et al., 2021; Pepi and Focardi, 2021; Bagi and Skogerbø, 2022; Baker et al., 2022; Kamermans et al., 2022).
The marine environment’s microbiological quality depends on many factors, but aquacultures might be considered one of the sources of pollutants. An exponential growth of aquaculture production in recent decades, which increased from less than 1 million tonnes in the 1950s to over 1225 million tonnes in 2020 (FAO), brought this problem into the Ocean (Preena et al., 2020). Bivalves are a significant part of aquaculture production, reaching nearly 18 million tonnes, including ornamental seashells and pearls farming. Aquaculture products’ microbiological quality depends on the water quality and other factors, including seafood preparation to consumption. These factors might represent human and animal sources of microorganisms, contaminating, among other, sedentary molluscs (Wright et al., 2018; Bighiu et al., 2019; Diwan et al., 2022). Also, as a consequence of the use of antibiotics in aquaculture, antibiotic-resistance is induced in the surrounding bacteria in the column water, sediment, and fish-associated bacterial strains (Pepi and Focardi, 2021).
Climate change may directly affect the molluscs’ aquacultures by increasing the water temperature above their optimal tolerance limits (Pinsky et al., 2019; Albano et al., 2021), supporting hypoxia and bacterial growth and their resistance to antimicrobials. A positive correlation between higher temperatures and raised antimicrobial resistance of Staphylococcus aureus, Escherichia coli, and Klebsiella pneumoniae was described by MacFadden et al. (2018). The climate anomalies also may lead to decreasing primary food production because of suboptimal living conditions led to low oxygen saturation and lowered immunocompetence, which increases the risk of infections (Beaudry et al., 2016; Kamermans et al., 2022). Also, climate change, including high air and water temperatures, rainfalls and droughts, may support the spread of food- and waterborne pathogens and, in effect, probably the increased number of outbreaks in the human population (Pepi and Focardi, 2021; Baker et al., 2022). Higher temperatures also favor spoilage processes and require earlier attempts to stop the degradation of the biochemical and microbiological processes of animal tissue, given the fact that for many regions, including the Mediterranean and central Europe, food availability is estimated to be reduced following a 2°C temperature increase (Hoegh-Guldberg et al., 2019).
The continuous presence of antibiotics in bivalves may affect animals and bacterial microbiota living in and on water animals (Figure 1). Chiesa et al. (2018), using an HPLC–MS/MS multiclass method, developed a method for the determination of 29 antibiotics belonging to eight different chemical classes (penicillin, quinolones, tetracyclines, sulphonamides, macrolides, lincosamides, cephalosporins, and amphenicols), in mussels and clams, both wild and farmed, collected from various geographic areas of the world and, particularly, Italy. In 100 samples of Meretrix lyrata (the lyrate Asiatic hard clam), M. meretrix (the Asiatic hard clam), Venerupis decussata (the cross-cut carpet shell), V. philippinarum (the Manila clam), Paphia textile (the carpet clam), and Chamelea gallina (the Venus clam), they found four representatives of tetracycline group (49.45 ng g–1 tetracycline, 125.03 ng g–1 oxytetracycline, 60.45 ng g–1 doxycycline, and 77.48 ng g–1 chlortetracycline) in one pool of farmed clams obtained from the Italian side of the North Adriatic Sea. Considering the annual Per capita consumption of 0.33 kg clams (European Commission, data), the daily consumption is 0.91 g; the result of the multiplication of this value by the sum of the concentrations of the four tetracyclines (312.41 ng g–1) found in the clam sample of North Adriatic Sea, is 0.29 μg per day. Molluscs from Bohai Sea (11 species of commercially available molluscs Crassostrea talienwhanensis, Chlamys farreri, Amussium, Scapharca subcrenata, Meretrix meretrix L., Mactra veneriformis, Mactra chinesis, Mya arenaria, Neverita didyma, Rapana venosa, and Mytilus edulis) were also analyzed for the presence of antibiotics in their flesh (Li et al., 2012). The authors found 22 antibiotics, including quinolones, sulfonamides and macrolides, in all samples of molluscs. The analysis of seawater from the Antarctic (Hernández et al., 2019) around South Shetland Island Fildes Peninsula, where are few scientific stations from several countries, showed the presence of antibiotics in seawater. The most prevalent antibiotics were two quinolones, ciprofloxacin, and norfloxacin, at average concentrations of 0.89 μg/L and 0.75 μg/L, respectively. Three macrolides were identified: Azithromycin and clarithromycin (average concentrations close to 0.4 μg/L), and erythromycin (average concentration 0.003 μg/L). González-Alonso et al. (2017) also found antibiotics in this area. Clarithromycin, ofloxacin, sulfadiazine, sulfamethazine, trimethoprim, and sulfamethoxazole were among the most consumed pharmaceuticals at all Antarctic stations nearby the sampling area. This datum could represent a risk mainly associated with the increase in antibiotic-resistance phenomenon. Continuous delivery of antibiotics to the marine environment strongly signals that anthropogenic activities are one of the primary sources of antibiotics in marine environments. The presence of antibiotics can lead to the conclusion that pharmaceuticals’ continuous pressure on microbial communities can lead to independent ways to develop resistance against the most concentrated or most effectively eliminating substances.
Even not targeted on environment purifying, bivalve farms perfectly fulfill their role in assessing AR spreading. In contrast to finfish aquaculture, bivalve aquaculture is fed with natural phytoplankton or detritus, and no additives such as antibiotics are added during their post-larval life (Wijsman et al., 2019). Moreover, the ecological importance of bivalve molluscs as filter feeders gives them a unique position in aquaculture systems as they are used to purify water from farming in multi-trophic aquaculture systems (MTA) (Sanz-Lazaro and Sanchez-Jerez, 2020). Thus they hold a particular position in AR distribution in marine environments (Figure 1). A field study comparing traditional and MTA aquaculture of fish Chanos chanos revealed that the latter system can prevent the shift in microbial communities and persistence of sulphonamide-resistant phenotypes (Ying et al., 2018). In the spatiotemporal study of bacterial resistance against florfenicol in the area of intense salmon farming, the microbiota of mussel M. chilensis revealed a striking influence depending on the distance to the next fish farm concerning water currents (Ramírez et al., 2022). Another study pinpoints AR against antibiotics used in aquaculture but against colistin–an antibiotic of last resort against multidrug-resistant Gram-negative bacteria in human medicine in Portuguese mussel farms (Salgueiro et al., 2021).
Similarly, a broad spectrum of AR genes was found in Ruditapes philippinarum clam (Dahanayake et al., 2019). Despite the importance of mussels in assessment and AR clearance, the products from mussels farms may become a public health threat for consumers. Selective pressures exerted by the overuse or misuse of antibiotics in primary food production, genetically modified crops with antibiotic-resistance marker genes, microorganisms added intentionally to the food chain (probiotic or technological) with potentially transferable antimicrobial resistance genes, food processing technologies used at sub-lethal doses (e.g., alternative non-thermal treatments), and the use of biocides are among the main driving forces behind the selection and spread of antimicrobial resistance throughout the food chains (Udoekong et al., 2021).
In the supply chain, molluscs usually are subjected to several stressors during capture, depuration, storage, and transportation, including prolonged periods of air exposure and temperature fluctuation. The stressors strongly decrease the bivalves’ quality, cause some changes in the bivalves’ metabolism, and may favor the growth of bacteria, an inadvisable component of meat. According to Pan et al. (2018), depuration with UV light decreases the number of bacteria significantly but lowered temperature during the depuration process extends the number of surviving clams and lowers the observed bacteria number compared to molluscs depurated at higher temperature (2,630 vs. 1,090 CFU/ml (colony forming units). The temperature can also affect the glycogen content and pH of bivalve tissues. Unlike crustaceans and finfish, molluscs’ flesh contains an appreciable amount of glycogen. Thus, the spoilage of molluscs is primarily glycolytic (1–5% glycogen content) rather than proteolytic, which may lead to a pH decrease resulting in a lower pH than in the case of other seafood. Lower pH at later stages of spoilage may favor the growth of bacterial species preferring lower pH, like enterococci, lactobacilli, and yeasts. For this reason, animals are usually transported to the point of sale, where they undergo further processing (Kontominas et al., 2021).
The analysis of bacterial communities on flash during the storage process of Manila clams (R. philippinarum) confirmed that each day of storage changes the proportions of bacterial taxons significantly. Authors showed it in the example of Proteobacteriales, including the class of Gammaproteobacteria with, among other, pathogenic genera like Vibrio, Escherichia, Pseudomonas, and Aeromonas. Proteobacteria are considered typical spoilage organisms for fresh seafood (Liu et al., 2020) and, like Aeromonas hydrophila, A. sobria or Shewanella putrefaciens, Vibrio spp. dominate the bacterial communities in many seafoods (Madigan et al., 2014; Chen et al., 2019; Zhuang et al., 2021).
Both fresh and cooked immediately after collection, bivalves may be at risk of colonization caused by pathogenic bacteria (Ekanem, 1998; Baris Bingol et al., 2008; Ates et al., 2011). These bacteria can originate from the natural environment or result from human, animal or abiotic activities, e.g., rain washing out the organic pollution from lands into the water (Alegbeleye and Sant’Ana, 2020), wave action and rainfall (Hernandez et al., 2020; Udoekong et al., 2021). The marine ecosystem can be considered an extensive reservoir of ARB (Antibiotic Resistant Bacteria) and ARGs mainly acquired through fecal contamination of human and/or animal origin (Figure 1). Publications report the bacterial microbiota in coastal and marine ecosystems, including E. coli, Clostridium perfringens, Bacillus cereus, S. aureus, Salmonella spp. and others (Islam and Tanaka, 2004; Moreno Roldán et al., 2011; Grabowska et al., 2020; Verga et al., 2020; Silva dos Santos et al., 2022). Although global seafood production is growing yearly, systematic surveillance of AMR in this sector is mainly lacking or has only been at a pilot study level. One main reason for this gap is the difficulties of investigating AMR phenotypes of marine bacteria, which often have specific growth requirements, and often lack standardized antimicrobial susceptibility testing methods and interpretation criteria (Delannoy et al., 2022). Among the most frequently found bacteria in bivalve molluscs are those belonging to the Proteobacteria phylum (Pierce and Ward, 2019). In this phylum, there are found commensal bacteria (e.g., Bacillus spp., Vibrio spp., and Aeromonas spp.) and non-commensal bacteria (e.g., Shewanella algae). Most bacterial species’ prevalence in bivalves corresponds to the beach’s pollution degree (Silva dos Santos et al., 2022) and general water bacteriological quality (Verga et al., 2020). However, there are much more bacteria than “most interesting” human and animal pathogens. We observe bacterial genera being narrow pathogens of animals like Francisella spp. (Duodu et al., 2012; Charles et al., 2021; Seong et al., 2021) and species like F. halioticida, which is a dangerous pathogen of giant abalone (Haliotis gigantea) and Yesso scallops (Mizuhopecten yessoensis) (Charles et al., 2021). This particular species is now known as endangering the mussel cultures (Cano et al., 2022), causing economic losses and a decline in mussels. Some bacteria may also break the barrier between the animal and human-animal species and cause disease symptoms and outbreaks like Vibrio fluvialis (Ramamurthy et al., 2014; Onohuean et al., 2022).
The magnitude of antimicrobial resistance in many parts of the world has reached alarming levels and suggests that a “post-antibiotic era” may be a real possibility for the 21st century (Vidovic and Vidovic, 2020). Monitoring the pathogenic bacteria in seafood still needs more detailed and accessible programs. Table 1 presents programs that directly or indirectly refer to microbiota monitoring in the food/seafood in European countries or USA, only sometimes mentioning worldwide regulations (Food and Agriculture Organization [FAO], and World Health Organization [WHO], 2020a). The fundamental problem of most of these programs is the limited time of operation or incomplete data with a relatively small geographical coverage. Apart from those programs, there were also found regulations and directions aiming to control the quality of the seafood that included regulations of safety checks of microbiota, e.g., Europe Commission implementing regulation regarding imports of live, chilled, frozen, or processed bivalve molluscs, echinoderms, tunicates, marine gastropods for human consumption from USA (last updated 2022). Pathogenic bacteria species that were more likely reported and/or required to be reported by all sorts of programs and conventions in different countries were: E. coli, V. vulnificus, V. parahaemolyticus, and V. cholerae.
Nevertheless, some countries have developed programs to control seafood safety and quality (Table 1). The Food and Drug Administration (FDA) in the USA holds several large programs on microbiota presence and resistance in seafood directed explicitly at bivalves, e.g., NSSP (National Shellfish Sanitation Program run since 1925, available revisions from 2007-2019 every 2 years).1 This program aims to promote and improve molluscs and other shellfish sanitation. In Europe, the leading organization contributing to the collection and awareness of the need for the collection of microbiota data is European Center for Disease Prevention and Control (ECDC, run since 1988, changed its name to recent one in 2010). ECDC holds programs such as EARS-Net (collect, and compare AMR data, analyze temporal trends in Europe, and provide AMR data for policy decisions) and monitors the Vibrio spp. growth in the Baltic Sea during the summer seasons (since 2013) (Table 1).
Bacterial communities are selected and suppressed by antibiotics delivered from other natural producers—microorganisms and antibiotics produced by humans. These last ones may affect marine and freshwater bacteria and might be a selective factor for antibiotic-resistance developed by many strains individually or acquired by HGT (horizontal gene transfer), conjugation and transformation mechanisms. Previous studies have reported that ARGs might be horizontally transferred by the facilitation of class 1 integron-integrase gene (intI1) on urban beaches by conjugating bacterial plasmids (Jang et al., 2021).
Heavy use of antibiotics in aquaculture may highly contaminate the water bodies; hence, detecting antimicrobial residues in the water environment is also significant. Similarly, producing sewages may result in AR genes’ pollution and introduce new variants of these genes into the marine environment. The importance of this growing problem can be illustrated by the emergence of plasmid-mediated colistin-resistance among human, animal, and zoonotic pathogens (Vidovic and Vidovic, 2020). The presence of the mcr-1 gene responsible for the resistance to colistin was observed by Valdez et al. (2022) (Table 2) in undefined Vibrio species and coliforms (E. coli and Citrobacter freundii) isolated from clams (Ruditapes decussatus and R. philippinarum). Samples were collected in Óbidos Lagoon (Caldas da Rainha, Portugal) in 2019, and other R. decussatus samples were collected in Ria Formosa (Algarve, Portugal). Antimicrobial susceptibility tests were performed by the disk diffusion method, according to the EUCAST guidelines against the antimicrobial: Colistin (10 μg), cefotaxime (30 μg), imipenem (10 μg), ciprofloxacin (5 μg), and meropenem (10 μg). The colistin-resistance gene was carried in the Vibrio spp. chromosomes. The mcr-2 gene was previously found by Lei et al. (2019) in V. parahaemolyticus VP181, which was obtained from a shrimp sample collected from Hong Kong, China. The gene of colistin-resistance was carried on a plasmid. Cherak et al. (2021) expressed concern that antibiotic-resistance is widely distributed in the aquatic environment. The authors collected data concerning the colistin-resistance data and proved that mcr genes are widely distributed in many geographical regions, types of environments and many Gram-negative bacterial species. Khedher et al. (2020) published work that analyzed 64,628 genome sequences belonging to 1,050 bacterial species and had 6′651 significant positive hits indicating the presence of mcr genes. Among others, MCR-1, 2 and 6 reveal that the putative progenitors of these MCR variants would be bacteria belonging to Moraxella, Enhydrobacter, Dichelobacter, Psychrobacter, Methylophilaceae, Limnobacter, and Vibrio genera. It is interesting to note that all these bacteria originate from water and/or soil environments. Also, a considerable number of mcr-9-like sequences were identified with 3′056 hits. Most of these bacterial genera, including, Stenotrophomonas, Vibrio, Aeromonas, Shewanella, Moraxella, Buttiauxella, and Salmonella, are bacteria from environmental sources (Vidovic and Vidovic, 2020).
A work published in 2022 by Delannoy et al. (2022) concerned the presence of ARGs and mobile genetic elements (MGE) in the marine environment, including seafood. The team tested 268 seafood isolates of the bacterial microflora for the presence of 41 ARGs and 33 MGEs, including plasmid replicons, integrons, and insertion sequences in Gram-negative bacteria. They show the occurrence of sul-1, ant(3″)-Ia, aph(30)-Ia, strA, strB, dfrA1, qnrA, and blaCTX-M-9 genes in Pseudomonas spp., Providencia spp., Klebsiella spp., Proteus spp., and Shewanella spp. They found that the occurrence of MGE may be associated with the seafood type and the environmental, farming, and harvest conditions. Their results corroborate the hypothesis that the incidence of antimicrobial-resistant bacteria (ARB) and ARG decrease with increasing distance from potential sources of fecal contamination. By high throughput real-time PCR system and array designed for antimicrobial resistance markers, they confirmed the presence of some ARGs like blaCMX-M-9 and aph(30)-Ia in Klebsiella spp., qnrA in Shewanella spp., and sul1 and ant(3″)-Ia in Pseudomonas spp. In 2019, Fresia et al. (2019) analyzed microbial communities harboring ARG and their characteristics. They collected samples from 20 localities around Montevideo (Uruguay). They found that ARGs covering resistance to sulfonamides, beta-lactams, aminoglycosides, phenicols, macrolides, and streptogramins were more prevalent in plasmids than in bacterial chromosomes. At the same time, ARGs covering resistance to tetracyclines, lincosamides, fluoroquinolones, and elfamycins were more frequently encoded in chromosomes. In 61% of identified integrons, Fresia et al. (2019) found cassette ARGs with conserved attC sites. These cassette genes primarily coded for multi-drug efflux pumps, but authors also found genes coding for carbapenemases (OXA family), GES extended-spectrum beta-lactamases (ESBLs), and aadA1 aminoglycoside nucleotidyltransferases, chloramphenicol acetyltransferases, and aac6-Ib amikacin resistance. This fact may support a hypothesis that ARGs carried by bacteria mostly present in sewages are potentially one of the most significant anthropogenic problems supporting the spreading of microbial resistance into marine bacterial species. Paschoal et al. (2017) indirectly confirmed it by finding carbapenemase genes in Aeromonas, Pseudomonas, Enterococcus, Klebsiella, and other genera isolated from recreational coastal waters, gene variants such as blaGES-16, blaIMP-16, blaOXA-370, and blaOXA-143 which were so far reported only (or predominantly) in clinical isolates from Brazil.
In the Pacific Ocean and the Antarctic, similar research done by Jang et al. (2022) displayed the presence of ARGs in transects including the West Sea of Korea, the Philippine Sea, the Western Equatorial Pacific Ocean, the Tasman Sea, the New Zealand sector of the Southern Ocean, and the Ross Sea. They found tetracycline resistance genes (tetA, tetB, tetBP, tetD, and tetZ), sulfonamide-resistance gene (sul1), one macrolide resistance gene (ermB), one β-lactam resistance gene (blaTEM), one quinolone-resistance gene (qnrD), and one multi-antibiotic resistance gene (oqxA). Overall, ARG abundance in the Southern Ocean was lower than in the Western Pacific Ocean but increased toward Antarctica. This pattern may be affected by human activities in the research stations (e.g., residence for research purposes or tourism) and incomplete wastewater treatment in the Antarctic environment.
Probably the part of bacteria carrying the ARGs may be finally deposited in all filtering organisms living in local seawaters, but even most disturbing is the knowledge that the geographical distribution of these genes seems to be continuous and unlimited. This fact especially concerns human and animal pathogens and emerging species with the potential to outbreak in human populations. It must be noted that the occurrence of antimicrobial-resistant bacterial species is not always a signal of pollution. Many species exhibit natural antibiotic-resistance, which is also expected in the natural marine environment. Miranda et al. (2013, 2015) presented the role of scallops hatcheries as a reservoir of antibiotic-resistant bacterial species. According to the presented data, the resistance of bacteria occurred even when no antibiotics were used.
Gram-negative bacteria are the focus of the World Health Organization (WHO). In 2017, the World Health Organization (WHO) published a list of global priority pathogens (GPP)—12 species of bacteria with critical, high, and medium antibiotic-resistance (AR), where 9 out of the 12 pathogens were gram-negative (Asokan et al., 2019). The outer membrane of Gram-negative microorganisms may be a significant barrier that protects toxic compounds, which include antibiotics and host innate immune molecules such as cationic antimicrobial peptides. At the same time, antibiotic-resistance mechanisms in Gram-negative bacteria may involve the acquisition of enzymes that modify or destroy antibiotics. The mechanisms may also include acquiring enzymes that alter bacterial antibiotic targets, such as lipid A-modifying enzymes conferring resistance to colistin and acquiring mutations in bacterial targets, such as topoisomerases, ribosomes, and penicillin-binding proteins, and outer membrane porins that alter antibiotic efficacy or uptake (Miller, 2016).
Members of the genus Pseudomonas, which belongs to the γ-Proteobacteria, can colonize diverse habitats. The main fraction of Pseudomonas species described so far was assigned to terrestrial habitats (Manwar et al., 2004; Romanenko et al., 2005, 2008, 2015); however, there are also reports of species isolated from marine environments, e.g., P. glareae (Romanenko et al., 2015) or P. marincola (Romanenko et al., 2008), P. pachastrellae (Romanenko et al., 2005), and P. aeruginosa (Manwar et al., 2004). The species belonging to Pseudomonas, widespread in scallop hatcheries (Miranda et al., 2013, 2015), constitutes a significant group of antimicrobial-resistant species. Ramírez et al. (2022) published similar results for Mytilus species reared in different years and distances from salmon farms. Authors showed that Mytilus spp. microbiota differs depending on the time and proximity to salmon farms. They also showed that Pseudomonas spp. represent the majority of florfenicol-resistant isolates (89% in 2018 and 78% in 2019). The number of resistant isolates compared with several other species that acquired the resistance for florfenicol may suggest that Pseudomonas spp. are quick at obtaining the resistance to florfenicol and/or colonizing new hosts as members of the first wave of colonizing bacterial species. This hypothesis is supported by data in described publication showing the relative abundance of Pseudomonas spp. per location was <1% but represented the most resistant isolates. In the mentioned publication, along with Pseudomonas spp., only two more genera (Microbacterium spp. and Psychrobacter spp.) were represented by florfenicol-resistant isolates. Following the year, the number of florfenicol-resistant genera increased to four (Acinetobacter spp., Alcaligenes spp., Psychrobacter spp., and Serratia spp.).
Pseudomonas aeruginosa is the third most pathogenic Gram-negative bacteria capable of causing severe diseases (Nolan and Behrends, 2021). This species may be a serious issue in patients with ventilator-associated pneumonia, urinary/peritoneal dialysis catheter infections, bacterial keratitis, cystic fibrosis, and burn wounds (Nolan and Behrends, 2021). P. aeruginosa causes around 13–19% of hospital-acquired infections (HAIs) in the USA (Raman et al., 2018). This species is primarily a waterborne pathogen, thriving in moist habitats and water resources like outlets of sewage treatment plants, from where it can be further spread to the marine environment (Luczkiewicz et al., 2015). In 1975, Denis (1975) observed P. aeruginosa in over 70% of mussels from the Marennes–Oleron Basin (France). Maravić et al. (2018) analyzed Pseudomonas spp. by combining PCR detection of acquired genes and resistance-nodulation-cell division (RND) efflux, studying multifactorial resistant traits of 108 P. aeruginosa isolates recovered from wild-growing Mediterranean mussels (Mytilus galloprovincialis) in Croatia. The multi-drug resistance to at least three pharmaceuticals from three different classes was detected in 17 isolates (15.7%). The most common (8.3% of isolates) profile included resistance to piperacillin (PIP), piperacillin-tazobactam (TZP), ceftazidime (CAZ), cefotaxime (CTX), and aztreonam (ATM). These substances represented penicillins, β-lactam/β-lactamase inhibitor combinations, cephems, and monobactams. Generally, 23.14% of isolates were resistant to at least one pharmaceutical. Also, Udoekong et al. (2021) examined 117 clams, 88 oysters, and 136 periwinkles. The authors found P. aeruginosa strain JB2 in 7.4% of tested molluscs, P. aeruginosa strain N15-01092 in 2.2%. Other species of Pseudomonas found in examined samples were P. xiamenensis (3.7%) and P. anguillisceptica 4,029 (2.2%). Generally, 44 isolates of Pseudomonas species (100%) were resistant to ceftriaxone, cefpodoxime, ceftazidime, cefepime, imipenem, and chloramphenicol. Unfortunately, the authors did not check ARGs’ presence, but the appropriate genes like cmlA and integrons were undoubtedly present. The literature review did not provide enough data to compare the antibiotic-resistance of environmental isolates of P. aeruginosa and other species with terrestrial isolates, although the Pseudomonas clade probably has a water environment origin. Generally, Laborda et al. (2021) considered P. aeruginosa as the species that succeeded in colonizing multiple hosts by relying on its large genome, and the species’ virulence and resistance determinants evolved in natural ecosystems.
Bacteria belonging to the genus Aeromonas are ubiquitous in marine environments, and Aeromonas spp. has held the title “emerging foodborne pathogen” (Janda and Abbott, 2010; Chuang et al., 2011; Abdel-Latif and Khafaga, 2020; Fernández-Bravo and Figueras, 2020; de Silva et al., 2021). Aeromonas spp. is a Gram-negative, non-spore-forming rod-shaped, facultatively anaerobic bacteria easily found in aquatic environments. Members of the genus Aeromonas are potential pathogens to many hosts, such as fish, amphibians, reptiles, mammals, and humans (Janda and Abbott, 2010; Fernández-Bravo and Figueras, 2020). Aeromonas spp. may cause various human infections, such as gastroenteritis, soft tissue infection, septicemia, hepatobiliary tract infections and occasionally pleuropulmonary infections, indwelling device-related infections, meningitis, peritonitis, and haemolytic uremic syndrome (Chuang et al., 2011). Among Aeromonas species that cause fish diseases, A. hydrophila has been identified as one of the most dangerous pathogens causing mortality outbreaks in diverse cultured fish species (Abdel-Latif and Khafaga, 2020). Among pathogens associated with human diseases, A. caviae, A. veronii, and A. hydrophila biovar sobria were isolated from mussels, scallops, and oysters (de Silva et al., 2021). These species Aeromonas spp. can grow relatively uninhibited in food during refrigeration under a broad range of pH and NaCl concentrations and in various packaging atmospheres. Given its high prevalence in seafood included in many RTE (Ready-To-Eat) seafood meals, the significance of Aeromonas spp. as a potential foodborne pathogen and a food spoilage organism increases. Strains of several Aeromonas species have shown spoilage potential by producing spoilage-associated metabolites in various seafood products (Hoel et al., 2019). Another point is the presence of genes encoding exotoxins, such as haemolysin (hlyA), aerolysin (aerA), cytotoxic heat-labile enterotoxin (act), cytotonic heat-labile enterotoxin (alt), cytotonic heat-stable enterotoxin (ast), and Shiga-like toxins (stx). Mentioned exotoxins are well-known virulence factors related to clinical symptoms during Aeromonas infection. A problem is deepened by the simultaneous harboring of AR genes. Maravić et al. (2013) found isolates of Aeromonas spp. co-producing blaCTX-M-15, blaSHV-12, blaPER-1, and blaFOX-2 in Mediterranean mussels (Mytilus galloprovincialis) from Croatia (Table 2). Generally, A. caviae, and A. hydrophila displayed full phenotypic resistance to ampicillin; 86% were resistant to cefotaxime, 68% to ticarcillin, and 63% to aztreonam. Hossain and Heo (2022) found in Mytilus coruscus from South Korea, 33 Aeromonas species are carrying blaTEM (27.3%), blaSHV (24.2%), blaCTX-M (24.2%), tetB (18.2%), tetE (15.2%), and intI1 genes (27.3%). Their results suggest that mussels can serve as a reservoir of multi-drug resistant Aeromonas spp. Manila clams (R. philippinarum) purchased on the market in South Korea also contained A. hydrophila, A. media, A. veronii, A. allosaccharophila, and A. caviae resistant to various antibiotics (Sanz-Lazaro and Sanchez-Jerez, 2020). Isolates of Aeromonas spp. harbored seven resistance genes (blaTEM, blaSHV, blaCTX-M, tetB, tetE, qnrS, and StrA-strB) and class 1 integron [intl1, aac(6’)-Ib] in various combinations. The most universal was the β-lactamase-resistance complex of genes (bla) (Table 2) (Dahanayake et al., 2019).
Besides the prevalent pathogens from anthropogenic and animal pollution, marine bivalves can be colonized by Vibrio spp. This group of bacteria consists of species living in seawater and being pathogens both to bivalves and humans. In marine environments, metazoans constitute habitats for bacteria of the Vibrionaceae family (vibrios). These γ-proteobacteria are ubiquitous in marine and brackish environments representing one of the most abundant culturable fractions of the marine microbial community (Ceccarelli et al., 2019). The most known species are V. cholerae, V. vulnificus, V. parahaemolyticus, and V. alginolyticus, but other species are also recognized as associated with food consumption. V. vulnificus and V. parahaemolyticus are seasonally present in molluscs (Lorenzoni et al., 2021) V. fluvialis, V. mimicus, V. metschnikovii, V. metoecus, and V. furnissii are related to foodborne diseases (Ceccarelli et al., 2019). Among these vibrios, V. fluvialis is an emerging human pathogen (Ramamurthy et al., 2014) related to diarrhoeal outbreaks. Other pathogenic species are V. cincinnatensis and V. carchariae, whose significance as human pathogens remains determined as agents causing several types of vibriosis. About 13 species of Vibrionaceae have been reported to cause several human diseases (Hou et al., 2011; Ramamurthy et al., 2014; Cheng et al., 2015). At the same time, Vibrio spp. frequently coexists with Pseudomonas spp. and Aeromonas spp. species, which are another group of potential human pathogens. In Benin City (Nigeria), Igbinosa (2016) found isolates of V. fluvialis, V. vulnificus, V. parahaemolyticus and unidentified Vibrio spp. in four water ponds by collecting fish and water samples. These isolates were resistant to β-lactams, macrolides, quinolones, norfloxacin, sulphonamides, nitrofurantoin, aminoglycoside, tetracyclines, and phenicol derivatives. Isolates represented a broad spectrum of resistance, from 0% of isolates resistant to ciprofloxacin to 100% resistant to tetracycline. There is no reason to exclude the possibility that vibrios cannot infect the edible bivalves in the water. Many species of cultured or wild molluscs accompany fish aquacultures serving as potential filtration systems or just as free-living animals, settling the environment rich in food particles. According to Vu et al. (2018), the distribution of Vibrio spp. in different species (blue mussels, venus clams, razor shells, cockles, and white leg shrimps) from retail food found in Berlin included 40.8% of blue mussels, 100% of cockles, and 93.3% of venus clams and 27.3% of razor shells. Similar results were published by Roque et al. (2009) in the case of four species of cultured molluscs on the Spanish coast (Mytilus galloprovincialis, Crassostrea gigas, Ruditapes decussatus, and Ruditapes philippinarum). In 4.08% of samples, Obaidat et al. (2017) found V. parahaemolyticus isolates resistant to chloramphenicol and cefotaxime in seafood from Yemen, India and Egypt. Besides this, 100% of isolates were resistant to colistin sulfate, kanamycin, and neomycin. In hard-shelled mussel (Mytilus coruscus) and tiger shrimp (Penaeus monodon) purchased in South Korea, Ashrafudoulla et al. (2019) found isolates of V. parahaemolyticus resistant to erythromycin, vancomycin, kanamycin, ciprofloxacin, clindamycin, and other antibiotics. In Poland (Lopatek et al., 2015) found that 17.5% of 400 tested samples of mussels, clams, oysters, and scallops were positive for the presence of V. parahaemolyticus. Among positive isolates, the authors found resistance to β-lactams (56%), ciprofloxacin (1.6%), streptomycin (45%), and gentamicin (7%). The food represented by bivalves is not present in the Baltic Sea, and all specimens were imported from the Netherlands, Italy, Norway and France. Dubert et al. (2016) noted that chloramphenicol had been banned in food animals, including aquaculture, because it has been associated with aplastic anemia. However, the authors noticed that experimental hatchery aquacultures still use this antibiotic due to its broad antimicrobial spectra. They analyzed samples from The Centro de Investigacións Mariñas (Spain), where seven species of clams were produced, and found that among 121 isolates of Vibrio spp. representing V. hemicentroti, V. splendidus, V. kanaloae, V. neocaledonicus, and V. jasicida, of non-human-pathogenic bacteria, 19 were resistant to chloramphenicol (CHL), tetracycline (TET) and amoxicillin (AMX), and nine were resistant to chloramphenicol (CHL) and streptomycin (STR). In 28 isolates of AGR genes, they confirmed the presence of the gene responsible for the resistance to chloramphenicol. Moreover, they demonstrated that antibiotic-resistance genes could be transferred to E. coli and other marine bacteria from Vibrio spp. In light of information about the possibility of transferring the ARGs among bacterial clades (Ellabaan et al., 2021), it can be highly possible that marine vibrios can serve as a potential reservoir for antibiotic-resistance genes (Table 2) for many emerging bacterial species. Besides being the reservoir of AR, a secondary problem generated by this genus is the number of exotoxins secreted by vibrios. Even bacteria from this purely marine genus can produce exotoxins (ciliostatic factors and hemolysins), which cause deciliation and injure tissues. According to West (West, 1989), the increase of Vibrio spp. number (up to an infective dose) can occur as water temperatures rise seasonally, which facilitates the growth and rising concentration of bacteria on/in higher animals, such as chitinous plankton, or accumulation of the bacteria by shellfish and other seafood. Pathogenic Vibrio species must elaborate a series of virulence factors to elicit human disease. Activities predisposing to diarrhoeal and extraintestinal infections include ingesting seafood and occupational or recreational exposure to natural aquatic environments, especially warm ones with temperatures rising above 20°C. Iwamoto et al. (2010), analyzing V. parahaemolyticus and V. vulnificus outbreaks, noticed that cases of Vibrio spp. infections have a marked seasonal distribution. Most occur during summer and early fall, corresponding to the period of warmer temperatures. In Japan, V. parahaemolyticus is one of the leading causes of foodborne gastroenteritis; annually, 500–800 outbreaks affecting more than 10,000 people are reported (Food and Agriculture Organization [FAO], and World Health Organization [WHO], 2020b). According to FDA data, between 1973 and 1991, all occasional cholera cases related to raw or undercooked shellfish consumption were associated with a non-O1 biotype of V. cholerae (Kaysner et al., 2012).
Shewanella is the sole genus belonging to the family of marine bacteria Shewanellaceae. As far as four species of this genus are considered dangerous for humans: S. algae, S. putrefaciens, S. haliotis, and S. xiamenensis (Vignier et al., 2013; Cimmino et al., 2015; Tena et al., 2016; Huang et al., 2018). These species may cause bacteremia, Fournier’s gangrene, empyema, pneumonia, and intracranial infections (Yousfi et al., 2017). This family is represented by widespread species and extremophiles living in habitats with a broad range of temperatures and high pressures (Abboud et al., 2005; Lemaire et al., 2020). For example, S. loihica from thermal vents can grow at 4–42°C. The taxon includes species isolated from around all oceans, including Antarctic subglacial and arctic marine waters, tropical coastlines, deep oceanic trenches, and various fish and shellfish. Their survival in various ecological niches is due to their impressive physiological and respiratory versatility (Lemaire et al., 2020). Some strains have been isolated in particular geographic areas, suggesting a putative specificity, while other isolates were found in many distant zones. One of the most spread species, often related to seafood, is S. putrefaciens, the species responsible for food spoilage and disease outbreaks (Liu et al., 2006; To et al., 2010; Janda and Abbott, 2014). Many consumable products have yielded Shewanella species upon laboratory analysis. Among others, the analysis of oyster samples from Delaware Bay showed that concentrations of Shewanella (presumptive) in oyster meats ranged from non-detectable levels to ≥400 CFU/g per sample (Richards et al., 2008).
Members of this genus are generally susceptible to third- and fourth-generation cephalosporins, carbapenems, β-lactamase inhibitor combinations, aminoglycosides, chloramphenicol, erythromycin, aztreonam, and quinolones (Table 2) (Kang et al., 2013; Cimmino et al., 2015; Zago et al., 2020).
Zago et al. (2020) checked resistance against antibiotics of Shewanella algae. The authors collected water samples along the Veneto coastline of the Adriatic Sea, Lugi di Varano (Foggia) and the open sea. They found isolates of S. algae resistant to β-lactams (blaOXA-55-LIKE, ampC), tetracyclines (tetR, tet34, and tet35), fluoroquinolones (qnrA3, qnrA7, mepAB—multi-drug efflux pump, mfpA—qnr homolog, emrD- multi-drug efflux pump, and mtdN—multi-drug resistance protein), polymyxin (eptA), chloramphenicol (cat). Also, Delannoy et al. (2022) found Shewanella spp. (S. indica, S, haliotis, S. algae, and unidentified Shewanella spp.) isolates from shellfish resistant to quinolone and possessing the qnrA gene on a plasmid. In Asian hard clams (M. lusoria), Lee et al. (2019) found isolates identified as S. algae SYT4 harboring β-lactamase encoding blaOXA-55 responsible for carbapenem resistance. Authors suggested that environmental S. algae could be a potential class D β-lactamase gene source.
Besides blaOXA-55, authors also identified the dfrA3 gene associated with resistance to sulfonamide-trimethoprim and genes responsible for resistance to fluoroquinolone (mfd, qnrA3). Due to the high zoonotic potential and wide distribution, Shewanella spp. are found practically in most seafood and marine environments. The characteristics of their genomes (Nuñez et al., 2022) may suggest that this genus can intensively transfer genetic information, including AR genes. Nuñez et al. (2022) found 52% of analyzed genomes of Shewanella spp. with conservative integrase genes (intI). They showed that Shewanella spp. could acquire and disseminate integrase genes that may lead to the emergence of novel mobile integrons. The substantial diversity of gene cassettes found in the variable regions and their association with MGEs may imply the host’s constant evolution and adaptation. This relation responds to environmental niche changes and the composition of each microbial community. Furthermore, it must be considered that antimicrobial-resistance gene cassettes found in hospital settings can be recruited from environmental chromosomal integrons. The above characteristic may suggest the solid potential ability to adapt to new hosts and new environments, and the plasticity of that genus fully justifies treating this species as an emerging organism most dangerous to humans and animals.
Escherichia coli is one of the most popular and widespread bacteria in seawater, as it is a significant member of the bacterial microbiota community in the gut of many animals and humans (Michalska et al., 2021). There is a possibility of transmission of pathogenic E. coli strains to the seas from land animal feces and human feces, especially during periods of intense rainfall (Lunestad et al., 2016). Consumption of raw or inadequately cooked mussels poses a potential risk for consumers because of E. coli infections (Feng et al., 2020). According to the current EU regulations [Regulation (EU) 2017/625 of the European Parliament and of the Council, 2017], mussel sampling sites must be classified according to their suitability for microbiological and chemical water quality (De Witte et al., 2014). The fecal indicator organism (FIO) E. coli is frequently used as a general indicator of sewage contamination and for evaluating the success of mollusc cleaning (depuration) processes. Depending on the content of E. coli in mussels, all localities should be affiliated based on Most Probable Numbers (MPN) as Class A (<230 MPN E. coli/100 g mussel), B (<4,600 MPN E. coli/100 g mussel), or C (4,600–46,000 MPN E. coli/100 g mussel) areas (Grevskott et al., 2016). Most of the research on the effectiveness of depuration has been conducted using oysters, as these are usually eaten raw. In 2019, Martin et al. (2019) found Shiga-toxin-producing E. coli (STEC) in Norwegian bivalves (1.1%). This percentage in the work of other authors was higher in shellfish, ranging from 3.5% in France (Gourmelon et al., 2006) to 6.1% in Morocco (Bennani et al., 2011). The relatively low isolation rate of STEC strains does not exclude the presence of stx-carrying bacteriophages in the samples, which pose a threat by playing an essential role in the evolution of STEC strains but also can transfer stx genes from non-pathogenic strains E. coli (Krüger and Lucchesi, 2015).
The multi-drug resistance of E. coli has become a worrying problem (Table 2). E. coli is internally susceptible to almost all clinically relevant antimicrobials (the ability to accumulate resistance genes mainly through horizontal gene transfer). The most complicated mechanisms in E. coli are the acquisition of genes encoding broad spectrum β-lactamases (broad-spectrum cephalosporin resistance), carbapenemases (carbapenem resistance), resistance to (aminoglycosides), plasmid-mediated quinolone resistance (PMQR), and mcr genes(conferring resistance to polymyxins) (Poirel et al., 2018).
According to Zannella et al. (2017), only a few Gram-positive bacteria cause diseases in bivalves, and the main genus is Nocardia. Only single species were reported as potentially dangerous for humans, but bivalves can also host non-marine pathogens causing severe healthcare problems.
Of all Gram-positive bacteria characteristics for bivalves, one is definitively typical for the marine environment but may be pathogenic to humans. The Nocardia genus is a prevalent member of the molluscs microbiota. This genus can be responsible for a broad spectrum of human diseases. Species like Nocardia nova, N. farcinica, N. cyriacigeorgica, N. brasiliensis, and N. abscessus are responsible for such cases as invasive pulmonary infection, disseminated infection, or brain abscess; 20% present as cellulitis. From the marine environment, the most known species is N. crassostrea, responsible for Pacific oyster nocardiosis observed in North America from the Strait of Georgia, British Columbia, to California and Japan (Matsushima Bay). Carella et al. (2013) observed this disease in Mediterranean bivalves first time. In 2013 (Taj-Aldeen et al., 2013) and 2016 (Igbaseimokumo et al., 2016), two outbreaks of N. crassostrea were observed in immunocompromised patients independently. No comprehensive information about genes responsible for antibiotic-resistance or antibiogram analysis was available. Igbaseimokumo et al. (2016) added information about antibiogram results. N. crassostrea, in this analysis, was sensitive to beta-lactams, including carbapenems but was resistant to lincosamides (clindamycin), vancomycin, and teicoplanin. This observation suggests the constitutive mechanism of defense against mentioned groups of antibiotics if the environmental origin of N. crassostrea is considered. In the cases described by Taj-Aldeen et al. (2013), the authors also checked the antibiotics-resistance of N. crassostrea and found one isolate resistant to linezolid (oxazolidinones group) and moxifloxacin (fluoroquinolones group). Still, they found the tested strain’s sensitivity to beta-lactamases (including cefotaxime and imipenem) and aminoglycosides (amikacin).
Symptoms of botulism include fatigue, dizziness, double vision, progressive difficulties in speaking and swallowing, shortness of breath, and muscle weakness (Aberoumand, 2010). Most seafood-related botulism is associated with inadequate processing, temperatures or maintenance (National Advisory Committee on Microbiological Criteria for Foods [NACMCF], 2008; Hudson and Lake, 2012). The occurrence of another Clostridium sp. in shellfish, C. difficile, raises concern because spores can survive the cooking temperatures given that shellfish is often consumed poorly cooked or raw. For example, C. difficile has been isolated from edible bivalve molluscs in Spain and Italy (Agnoletti et al., 2019; Candel-Pérez et al., 2020). C. difficile collected from 702 molluscs harvested in the North Adriatic Sea (Agnoletti et al., 2019) were detected in 16.9% of all bivalves specimens. Isolates showed microbiological resistance against clindamycin (17%), erythromycin (23%), rifampicin (8.8%), and moxifloxacin (10.6%). All isolates were susceptible to metronidazole, and one showed MIC> ECOFF for vancomycin. C. difficile strains showed a wide variety in PCR-ribotypes, most already detected in other animals or known as highly virulent and epidemic in humans. In Spain, Candel-Pérez et al. (2020) analyzed 129 mollusc samples from different fishmongers and grocery stores in Murcia. C. difficile was isolated from 8.53% (11 specimens per total 129) of the investigated molluscs. Four C. difficile isolates harbored genes for producing toxins A and B. Examining mussels for antibiotic-resistant strains of C. difficile should be another essential aim of seafood monitoring.
Consumers’ growing interest in seafood, including mussels, should be determined by constantly monitoring microbiological hazards associated with these products. Such a network should include a system of bacterial monitoring in selected geographic locations dependent on seafood production centers. Another activity should supervise the distribution of genes encoding resistance to antibiotics independently on bacterial species. The sensitivity and selectivity of modern analysis methods should also be propagated to monitor seafood for the presence of the most emerging bacterial species, such as Pseudomonas spp. and Vibrio spp. The complete data concerning AR come from seawater genomic analyses (Paschoal et al., 2017; Lei et al., 2019; Khedher et al., 2020; Cherak et al., 2021), depicting the complete analysis of genes involved in AR (Table 2).
A unified global program of antibiotic treatment recommendations and monitoring of antibiotic use in aquacultures should be created to limit the spread of mobile genetic elements coding AR. Such a program could be helpful if synchronized with environmental and food monitoring.
On a global scale, disease outbreaks have increased over the last decades in the marine environment (Baker et al., 2022). Although the definitive causes for this increase are uncertain, ocean warming (Pinsky et al., 2019; Albano et al., 2021; Pepi and Focardi, 2021) and marine pathogen emergence, reemergence or evolution leading to the colonization of new niches, including humans, appear to be critical driving forces. For example, bacteria preferring subtropical and tropical seawater like V. vulnificus extended their distribution to new marine areas like the Baltic Sea (Kurpas et al., 2021).
The danger for humans and marine organisms includes harmful bacterial infections and the threat from toxins produced even by harmless marine bacteria due to massive contact with land-sourced bacteria and horizontal gene exchange. In the long-term, acquiring new AR genes from different bacterial species can be dangerous for humans and endangers seafood production by limiting the efficacy of antibiotics used to protect the aquacultures. Food quality should be monitored carefully to prevent the development of bacterial resistome in the marine environment and outbreaks of super-resistant bacteria. Some public information provided to clarify standards of molluscs’ freshness rating and procedures of seafood preparation should also be present.
AgK designed the form of the manuscript, wrote the text except for tables and subsections 3.1, 4.5, and 5.2, and prepared a graphic. AlK, TK, and KG-B wrote the text in the subsections mentioned above, and AlK and TK collected the data and synthesized them into tables. AgK, KG-B, and NW-K edited manuscript. AgK and NW-K prepared the literature in the present review and edited the manuscript. KS and KZ supervised the process of creation and edited the discussion. All authors contributed to the article and approved the submitted version.
The source of funds for publication was the statutory fund of the Medical University of Gdańsk (IDUB).
We would like to thank Dr. Monika Kurpas (Medical University of Gdańsk) for creating the first idea for the present manuscript and Dr. Marta Potrykus (Medical University of Gdańsk) for discussion and substantive comments.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abboud, R., Popa, R., Souza-Egipsy, V., Giometti, C., Tollaksen, S., Mosher, J., et al. (2005). Low-temperature growth of Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 71, 811–816. doi: 10.1128/AEM.71.2.811-816.2005
Abdel-Latif, H., and Khafaga, A. (2020). Natural co-infection of cultured Nile tilapia Oreochromis niloticus with Aeromonas hydrophila and Gyrodactylus cichlidarum experiencing high mortality during summer. Aquac. Res. 51, 1880–1892. doi: 10.1111/ARE.14538
Aberoumand, A. (2010). Occurrence of Clostridium botulinum in fish and fishery products in retail trade, a review article. World J. Fish Mar. Sci. 2, 246–250.
Agnoletti, F., Arcangeli, G., Barbanti, F., Barco, L., Brunetta, R., Cocchi, M., et al. (2019). Survey, characterization, antimicrobial susceptibility of Clostridium difficile from marine bivalve shellfish of North Adriatic Sea. Int. J. Food Microbiol. 298, 74–80. doi: 10.1016/j.ijfoodmicro.2019.03.003
Albano, P., Steger, J., Bošnjak, M., Dunne, B., Guifarro, Z., Turapova, E., et al. (2021). Native biodiversity collapse in the Eastern Mediterranean. Proc. R. Soc. B 288:20202469. doi: 10.1098/RSPB.2020.2469
Alegbeleye, O., and Sant’Ana, A. (2020). Manure-borne pathogens as an important source of water contamination: An update on the dynamics of pathogen survival/transport as well as practical risk mitigation strategies. Int. J. Hyg. Environ. Health 227:113524. doi: 10.1016/J.IJHEH.2020.113524
Ashrafudoulla, M., Mizan, M., Park, H., Byun, K., Lee, N., Park, S., et al. (2019). Genetic relationship, virulence factors, drug resistance profile and biofilm formation ability of Vibrio parahaemolyticus isolated from mussel. Front. Microbiol. 10:513. doi: 10.3389/fmicb.2019.00513
Asokan, G., Ramadhan, T., Ahmed, E., and Sanad, H. W. H. O. (2019). Global priority pathogens list: A bibliometric analysis of medline-pubmed for knowledge mobilization to infection prevention and control practices in Bahrain. Oman Med. J. 34, 184–193. doi: 10.5001/omj.2019.37
Ates, M., Ozkizilcik, A., and Tabakoglu, C. (2011). Microbiological analysis of stuffed mussels sold in the streets. Ind. J. Microbiol. 51:350. doi: 10.1007/S12088-011-0174-6
Bagi, A., and Skogerbø, G. (2022). Tracking bacterial pollution at a marine wastewater outfall site – A case study from Norway. Sci. Total Environ. 829:154257. doi: 10.1016/J.SCITOTENV.2022.154257
Baker, R. E., Mahmud, A. S., Miller, I. F., Rajeev, M., Rasambainarivo, F., Rice, B., et al. (2022). Infectious disease in an era of global change. Nat. Rev. Microbiol. 20, 193–205. doi: 10.1038/s41579-021-00639-z
Baris Bingol, E., Colak, H., Hampikyan, H., and Muratoglu, K. (2008). The microbiological quality of stuffed mussels (Midye Dolma) sold in Istanbul. Br. Food J. 110, 1079–1087. doi: 10.1108/00070700810917992
Beaudry, A., Fortier, M., Masson, S., Auffret, M., Brousseau, P., and Fournier, M. (2016). Effect of temperature on immunocompetence of the blue mussel (Mytilus edulis). J. Xenobiot. 6:5889. doi: 10.4081/XENO.2016.5889
Bennani, M., Badri, S., Baibai, T., Oubrim, N., Hassar, M., Cohen, N., et al. (2011). First detection of shiga toxin-producing Escherichia coli in shellfish and coastal environments of Morocco. Appl. Biochem. Biotechnol. 165, 290–299. doi: 10.1007/s12010-011-9251-x
Bighiu, M., Norman Haldén, A., Goedkoop, W., and Ottoson, J. (2019). Assessing microbial contamination and antibiotic resistant bacteria using Zebra mussels (Dreissena polymorpha). Sci. Total Environ. 650, 2141–2149. doi: 10.1016/j.scitotenv.2018.09.314
Bondarczuk, K., and Piotrowska-Seget, Z. (2019). Microbial diversity and antibiotic resistance in a final effluent-receiving lake. Sci. Total Environ. 650, 2951–2961. doi: 10.1016/J.SCITOTENV.2018.10.050
Candel-Pérez, C., Zapata-Galián, E., López-Nicolás, R., Ros-Berruezo, G., and Martínez-Graciá, C. (2020). Presence of toxigenic Clostridioides (Clostridium) difficile in edible bivalve mollusks in Spain. Food Sci. Technol. Int. 26, 413–419. doi: 10.1177/1082013219894092
Cano, I., Parker, A., Ward, G., Green, M., Ross, S., Bignell, J., et al. (2022). First detection of Francisella halioticida infecting a wild population of blue mussels Mytilus edulis in the United Kingdom. Pathogens 11:329. doi: 10.3390/pathogens11030329
Carella, F., Carrasco, N., Andree, K., Lacuesta, B., Furones, D., and de Vico, G. (2013). Nocardiosis in Mediterranean bivalves: First detection of Nocardia crassostreae in a new host Mytilus galloprovincialis and in Ostrea edulis from the Gulf of Naples (Italy). J. Invertebr. Pathol. 114, 324–328. doi: 10.1016/J.JIP.2013.10.001
Ceccarelli, D., Amaro, C., Romalde, J., Suffredini, E., and Vezzulli, L. (2019). “Vibrio species,” in Food microbiology, eds M. P. Doyle, F. Diez-Gonzalez, and C. Hill (Amsterdam, NL: Elsevier), 347–388. doi: 10.1128/9781555819972.ch13
Charles, M., Quesnelle, Y., Glais, G., Trancart, S., and Houssin, M. (2021). Development of a semi-quantitative PCR assay for the detection of Francisella halioticida and its application to field samples. J. Fish Dis. 44, 1169–1177. doi: 10.1111/JFD.13377
Chen, H., Wang, M., Yang, C., Wan, X., Ding, H., Shi, Y., et al. (2019). Bacterial spoilage profiles in the gills of Pacific oysters (Crassostrea gigas) and Eastern oysters (C. virginica) during refrigerated storage. Food Microbiol. 82, 209–217. doi: 10.1016/J.FM.2019.02.008
Chen, J., Sun, R., Pan, C., Sun, Y., Mai, B., and Li, Q. (2020). Antibiotics and food safety in aquaculture. J. Agric. Food Chem. 68, 11908–11919. doi: 10.1021/acs.jafc.0c03996
Cheng, W., Jan, I., Chen, J., Teng, S., Teng, L., Sheng, W., et al. (2015). Evaluation of the bruker biotyper matrix-assisted laser desorption ionisation-time of flight mass spectrometry system for identification of blood isolates of Vibrio species. J. Clin. Microbiol. 53, 1741–1744. doi: 10.1128/JCM.00105-15
Cherak, Z., Loucif, L., Moussi, A., and Rolain, J. (2021). Epidemiology of mobile colistin resistance (mcr) genes in aquatic environments. J. Glob. Antimicrob. Resist. 27, 51–62. doi: 10.1016/J.JGAR.2021.07.021
Chiesa, L., Nobile, M., Malandra, R., Panseri, S., and Arioli, F. (2018). Occurrence of antibiotics in mussels and clams from various FAO areas. Food Chem. 240, 16–23. doi: 10.1016/J.FOODCHEM.2017.07.072
Chuang, H., Ho, Y., Lay, C., Wang, L., and Tsai, Y. (2011). Different clinical characteristics among Aeromonas hydrophila, Aeromonas veronii biovar sobria and Aeromonas caviae monomicrobial bacteremia. J. Korean Med. Sci. 26, 1415–1420. doi: 10.3346/JKMS.2011.26.11.1415
Cimmino, T., Olaitan, A., and Rolain, J. (2015). Whole genome sequence to decipher the resistome of Shewanella algae, a multidrug-resistant bacterium responsible for pneumonia, Marseille, France. Expert Rev. Anti Infect. Ther. 14, 269–275. doi: 10.1586/14787210.2016.1106936
Dahanayake, P., Hossain, S., Wickramanayake, M., and Heo, G. (2019). Antibiotic and heavy metal resistance genes in Aeromonas spp. isolated from marketed Manila Clam (Ruditapes philippinarum) in Korea. J. Appl. Microbiol. 127, 941–952. doi: 10.1111/jam.14355
Dahanayake, P., Hossain, S., Wickramanayake, M., and Heo, G. (2020). Prevalence of virulence and extended-spectrum β-lactamase (ESBL) genes harbouring Vibrio spp. isolated from cockles (Tegillarca granosa) marketed in Korea. Lett. Appl. Microbiol. 71, 61–69. doi: 10.1111/lam.13232
de Silva, L., Wickramanayake, M., and Heo, G. (2021). Virulence and antimicrobial resistance potential of Aeromonas spp. associated with shellfish. Lett. Appl. Microbiol. 73, 176–186. doi: 10.1111/LAM.13489
De Witte, B., Devriese, L., Bekaert, K., Hoffman, S., Vandermeersch, G., Cooreman, K., et al. (2014). Quality assessment of the blue mussel (Mytilus edulis): Comparison between commercial and wild types. Mar. Pollut. Bull. 85, 146–155. doi: 10.1016/j.marpolbul.2014.06.006
Delannoy, S., Hoffer, C., Youf, R., Dauvergne, E., Webb, H., Brauge, T., et al. (2022). High throughput screening of antimicrobial resistance genes in gram-negative seafood bacteria. Microorganisms 10:1225. doi: 10.3390/microorganisms10061225
Denis, F. (1975). Contamination of shellfish with strains of Pseudomonas aeruginosa and specific bacteriophages. Can. J. Microbiol. 21, 1055–1057. doi: 10.1139/M75-156
Diwan, A., Harke, S., and Gopalkrishna Panche, A. (2022). Aquaculture industry prospective from gut microbiome of fish and shellfish: An overview. J. Anim. Physiol. Anim. Nutr. 106, 441–469. doi: 10.1111/JPN.13619
Dubert, J., Osorio, C., Prado, S., and Barja, J. (2016). Persistence of antibiotic resistant Vibrio spp. in shellfish hatchery environment. Microb. Ecol. 72, 851–860. doi: 10.1007/s00248-015-0705-5
Duodu, S., Larsson, P., Sjödin, A., Forsman, M., and Colquhoun, D. (2012). The distribution of Francisella-like bacteria associated with coastal waters in Norway. Microb. Ecol. 64, 370–377. doi: 10.1007/s00248-012-0023-0
Ekanem, E. (1998). The street food trade in Africa: Safety and socio-environmental issues. Food Control 9, 211–215. doi: 10.1016/S0956-7135(97)00085-6
Ellabaan, M., Munck, C., Porse, A., Imamovic, L., and Sommer, M. (2021). Forecasting the dissemination of antibiotic resistance genes across bacterial genomes. Nat. Commun. 12, 1–10. doi: 10.1038/s41467-021-22757-1
European Market Observatory for Fisheries and Aquaculture Products [EUMOFA] (2022). Organic aquaculture in the EU: Current situation, drivers, barriers, potential for growth. Luxembourg: European Market Observatory for Fisheries and Aquaculture Products. doi: 10.2771/327564
Feng, P., Weagant, S., Grant, M., and Burkhardt, W. (2020). Enumeration of Escherichia coli and the coliform bacteria. Washington, DC: Food & Drug Administration, Bacteriological Analytical Manual.
Fernández-Bravo, A., and Figueras, M. (2020). An update on the genus Aeromonas: Taxonomy, epidemiology, and pathogenicity. Microorganisms 8:129. doi: 10.3390/MICROORGANISMS8010129
Food and Agriculture Organization [FAO] (2020). The state of world fisheries and aquaculture 2020. Sustainability in action. Rome: Food and Agriculture Organization. doi: 10.4060/ca9229en
Food and Agriculture Organization [FAO] (2022). The state of world fisheries and aquaculture 2022. Towards blue transformation. Rome: Food and Agriculture Organization. doi: 10.4060/cc0461en
Food and Agriculture Organization [FAO], and World Health Organization [WHO] (2020a). Code of practice for fish and fishery products. Rome: Food and Agriculture Organization.
Food and Agriculture Organization [FAO], and World Health Organization [WHO] (2020b). Risk assessment of Vibrio parahaemolyticus in seafood: Interpretative summary and technical report. Microbiological risk assessment series no. 16. Rome: Food and Agriculture Organization.
Fresia, P., Antelo, V., Salazar, C., Giménez, M., D’Alessandro, B., Afshinnekoo, E., et al. (2019). Urban metagenomics uncover antibiotic resistance reservoirs in coastal beach and sewage waters. Microbiome 7:35. doi: 10.1186/s40168-019-0648-z
González-Alonso, S., Merino, L., Esteban, S., López de Alda, M., Barceló, D., Durán, J., et al. (2017). Occurrence of pharmaceutical, recreational and psychotropic drug residues in surface water on the northern Antarctic Peninsula region. Environ. Pollut. 229, 241–254. doi: 10.1016/J.ENVPOL.2017.05.060
Gourmelon, M., Montet, M., Lozach, S., Le Mennec, C., Pommepuy, M., Beutin, L., et al. (2006). First isolation of Shiga toxin 1d producing Escherichia coli variant strains in shellfish from coastal areas in France. J. Appl. Microbiol. 100, 85–97. doi: 10.1111/j.1365-2672.2005.02753.x
Grabowska, K., Bukowska, A., Kaliński, T., Kiersztyn, B., Siuda, W., and Chróst, R. (2020). Presence and identification of legionella and Aeromonas spp. In the great Masurian lakes system in the context of eutrophication. J. Limnol. 79, 30–42. doi: 10.4081/JLIMNOL.2019.1924
Grevskott, D., Svanevik, C., Sunde, M., Wester, A., and Lunestad, B. (2017). Marine bivalve mollusks as possible indicators of multidrug-resistant Escherichia coli and other species of the Enterobacteriaceae family. Front. Microbiol. 8:24. doi: 10.3389/FMICB.2017.00024
Grevskott, D., Svanevik, C., Wester, A., and Lunestad, B. (2016). The species accuracy of the most probable number (MPN) European union reference method for enumeration of Escherichia coli in marine bivalves. J. Microbiol. Methods 131, 73–77. doi: 10.1016/J.MIMET.2016.10.006
Hernández, F., Calısto-Ulloa, N., Gómez-Fuentes, C., Gómez, M., Ferrer, J., González-Rocha, G., et al. (2019). Occurrence of antibiotics and bacterial resistance in wastewater and sea water from the Antarctic. J. Hazard. Mater. 363, 447–456. doi: 10.1016/j.jhazmat.2018.07.027
Hernandez, R., Acedo, I., and Dillon, J. (2020). Impact of wave action and rainfall on incidence and antibiotic resistance of total coliforms in Southern California beaches. J. Water Health 18, 766–775. doi: 10.2166/WH.2020.100
Hoegh-Guldberg, O., Jacob, D., Taylor, M., Guillén Bolaños, T., Bindi, M., Brown, S., et al. (2019). The human imperative of stabilising global climate change at 1.5°C. Science 365:eaaw6974. doi: 10.1126/SCIENCE.AAW6974
Hoel, S., Vadstein, O., and Jakobsen, A. (2019). The significance of Mesophilic Aeromonas spp. in minimally processed ready-to-eat seafood. Microorganisms 7:91. doi: 10.3390/microorganisms7030091
Hossain, S., and Heo, G. (2022). Detection of antimicrobial and heavy-metal resistance genes in Aeromonas spp. isolated from hard-shelled mussel (Mytilus Coruscus). Microb. Drug Resist. 28, 127–135. doi: 10.1089/mdr.2020.0590
Hossain, S., Wickramanayake, M., Dahanayake, P., and Heo, G. (2020). Occurrence of virulence and extended-spectrum β-lactamase determinants in Vibrio spp. isolated from marketed hard-shelled mussel (Mytilus coruscus). Microb. Drug Resist. 26, 391–401. doi: 10.1089/mdr.2019.0131
Hou, C., Lai, C., Liu, W., Chao, C., Chiu, Y., and Hsueh, P. (2011). Clinical manifestation and prognostic factors of non-cholerae Vibrio infections. Eur. J. Clin. Microbiol. Infect. Dis. 30, 819–824. doi: 10.1007/S10096-011-1162-9
Hsu, T., and Lee, J. (2015). Global distribution and prevalence of Arcobacter in food and water. Zoonoses Public Health 62, 579–589. doi: 10.1111/ZPH.12215
Huang, Y., Tang, Y., Cheng, J., Wu, Z., Mao, Y., and Liu, P. (2018). Genome analysis of multidrug-resistant Shewanella algae isolated from human soft tissue sample. Front. Pharmacol. 9:419. doi: 10.3389/FPHAR.2018.00419/FULL
Hudson, A., and Lake, R. (2012). Risk profile: Clostridium botulinum in ready-to-eat smoked fish and shellfish in sealed packaging. Technical Paper No: 2012/27 prepared for ministry for primary industries. Wellington: Ministry for Primary Industries.
Igbaseimokumo, U., El Shafie, S., and Al Khal, A. (2016). First human infection of Nocardia Crassostreae in an immunocompetent patient. Chin. Med. J. 129:114. doi: 10.4103/0366-6999.172609
Igbinosa, E. (2016). Detection and antimicrobial resistance of Vibrio isolates in aquaculture environments: Implications for public health. Microb. Drug Resist. 22, 238–245. doi: 10.1089/mdr.2015.0169
Islam, M., and Tanaka, M. (2004). Impacts of pollution on coastal and marine ecosystems including coastal and marine fisheries and approach for management: A review and synthesis. Mar. Pollut. Bull. 48, 624–649. doi: 10.1016/J.MARPOLBUL.2003.12.004
Iwamoto, M., Ayers, T., Mahon, B., and Swerdlow, D. (2010). Epidemiology of seafood-associated infections in the United States. Clin. Microbiol. Rev. 23:399. doi: 10.1128/CMR.00059-09
Janda, J., and Abbott, S. (2010). The genus Aeromonas: Taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23, 35–73. doi: 10.1128/CMR.00039-09/ASSET/E78450C2-50F6-41E4-A7F9-80D2C865F5A3/ASSETS/GRAPHIC/ZCM0011023010003.JPEG
Janda, J., and Abbott, S. (2014). The genus Shewanella: From the briny depths below to human pathogen. Crit. Rev. Microbiol. 40, 293–312. doi: 10.3109/1040841X.2012.726209
Jang, J., Kim, M., Baek, S., Shin, J., Shin, J., Shin, S., et al. (2021). Hydrometeorological Influence on antibiotic-resistance genes (ARGs) and bacterial community at a recreational beach in Korea. J. Hazard. Mater. 403:123599. doi: 10.1016/j.jhazmat.2020.123599
Jang, J., Park, J., Hwang, C., Choi, J., Shin, J., Kim, Y., et al. (2022). Abundance and diversity of antibiotic resistance genes and bacterial communities in the western Pacific and Southern Oceans. Sci. Total Environ. 822:153360. doi: 10.1016/J.SCITOTENV.2022.153360
Kamermans, P., Saurel, C., Boudry, P., and Kamermans, P. (2022). Interacting climate change effects on mussels (Mytilus edulis and M. galloprovincialis) and oysters (Crassostrea gigas and Ostrea edulis): Experiments for bivalve individual growth models. Aquat. Living Resour. 35:1. doi: 10.1051/ALR/2022001
Kang, C., Shin, Y., Jeon, H., Choi, J., Jeong, S., and So, J. (2013). Antibiotic resistance of Shewanella putrefaciens isolated from shellfish collected from the West Sea in Korea. Mar. Pollut. Bull. 76, 85–88. doi: 10.1016/J.MARPOLBUL.2013.09.025
Kaysner, C., DePAola, A., and Jones, J. (2012). “Vibrio cholerae, V. parahaemolyticus, V. vulnificus, and Other Vibrio spp,” in Vibrio cholerae serogroup NonO1, ed. Association of Official Analytical Chemists (Washington, DC: Food & Drug Administration, Bacteriological Analytical Manual).
Khedher, M. B., Baron, S., Riziki, T., Ruimy, R., Raoult, D., Diene, S., et al. (2020). Massive analysis of 64,628 bacterial genomes to decipher water reservoir and origin of mobile colistin resistance genes: Is there another role for these enzymes? Sci. Rep. 10:5970. doi: 10.1038/s41598-020-63167-5
Kolm, H., Gomes, K., Ishii, F., and Martins, C. (2018). An integrated appraisement of multiple faecal indicator bacteria and sterols in the detection of sewage contamination in subtropical tidal creeks. Int. J. Hyg. Environ. Health 221, 1032–1039. doi: 10.1016/J.IJHEH.2018.07.004
Kontominas, M., Badeka, A. V., Kosma, I., and Nathanailides, C. (2021). Recent developments in seafood packaging technologies. Foods 10:940. doi: 10.3390/foods10050940
Krieger, R., Gee, S., and Lim, L. (1981). Marine bivalves, particularly mussels, Mytilus sp., for assessment of environmental quality. Ecotoxicol. Environ. Saf. 5, 72–86. doi: 10.1016/0147-6513(81)90047-6
Krüger, A., and Lucchesi, P. (2015). Shiga toxins and stx phages: Highly diverse entities. Microbiology (Reading) 161(Pt 3), 451–462. doi: 10.1099/mic.0.000003
Kueh, C., and Chan, K. (1985). Bacteria in bivalve shellfish with special reference to the oyster. J. Appl. Bacteriol. 59, 41–47. doi: 10.1111/J.1365-2672.1985.TB01773.X
Kurpas, M., Michalska, M., Zakrzewski, A., and Zorena, K. (2021). First report of the presence of Vibrio vulnificus in the Gulf of Gdansk. Int. Marit. Health 72, 247–251. doi: 10.5603/IMH.2021.0048
Laborda, P., Sanz-García, F., Hernando-Amado, S., and Martínez, J. (2021). Pseudomonas aeruginosa: An antibiotic resilient pathogen with environmental origin. Curr. Opin. Microbiol. 64, 125–132. doi: 10.1016/J.MIB.2021.09.010
Lee, Y., Tung, K., Cheng, J., Wu, Z., Chen, S., Hong, Y., et al. (2019). Genomic characterisation of carbapenem-resistant Shewanella algae isolated from Asian hard clam (Meretrix lusoria). Aquaculture 500, 300–304. doi: 10.1016/J.AQUACULTURE.2018.10.028
Lei, T., Zhang, J., Jiang, F., He, M., Zeng, H., Chen, M., et al. (2019). First detection of the plasmid-mediated colistin resistance gene mcr-1 in virulent Vibrio parahaemolyticus. Int. J. Food Microbiol. 308:108290. doi: 10.1016/j.ijfoodmicro.2019.108290
Lemaire, O., Méjean, V., and Iobbi-Nivol, C. (2020). The Shewanella genus: Ubiquitous organisms sustaining and preserving aquatic ecosystems. FEMS Microbiol. Rev. 44, 155–170. doi: 10.1093/FEMSRE/FUZ031
Li, W., Shi, Y., Gao, L., Liu, J., and Cai, Y. (2012). Investigation of antibiotics in mollusks from coastal waters in the Bohai Sea of China. Environ. Pollut. 162, 56–62. doi: 10.1016/J.ENVPOL.2011.10.022
Liu, M., Gau, S., and Wu, H. (2006). Acute exudative tonsillitis caused by Shewanella algae in a healthy child. Scand. J. Infect. Dis. 38, 1104–1105. doi: 10.1080/00365540600664050
Liu, X., Teixeira, J., Ner, S., Ma, K. V., Petronella, N., Banerjee, S., et al. (2020). Exploring the potential of the microbiome as a marker of the geographic origin of fresh seafood. Front. Microbiol. 11:696. doi: 10.3389/FMICB.2020.00696/BIBTEX
Lopatek, M., Wieczorek, K., and Osek, J. (2015). Prevalence and antimicrobial resistance of Vibrio parahaemolyticus isolated from raw shellfish in Poland. J. Food Prot. 78, 1029–1033. doi: 10.4315/0362-028X.JFP-14-437
Lopez-Joven, C., Rolland, J., Haffner, P., Caro, A., Roques, C., Carré, C., et al. (2018). Oyster farming, temperature, and plankton influence the dynamics of pathogenic vibrios in the Thau Lagoon. Front. Microbiol. 9:2530. doi: 10.3389/FMICB.2018.02530/BIBTEX
Lorenzoni, G., Tedde, G., Mara, L., Bazzoni, A., Esposito, G., Salza, S., et al. (2021). Presence, seasonal distribution, and biomolecular characterization of Vibrio parahaemolyticus and Vibrio vulnificus in shellfish harvested and marketed in Sardinia (Italy) between 2017 and 2018. J. Food Prot. 84, 1549–1554. doi: 10.4315/JFP-21-059
Lozano-León, A., Rodríguez-Souto, R., González-Escalona, N., Llovo-Taboada, J., Iglesias-Canle, J., Álvarez-Castro, A., et al. (2021). Detection, molecular characterisation, and antimicrobial susceptibility, of Campylobacter spp. isolated from shellfish. Microb. Risk Anal. 18:100176. doi: 10.1016/j.mran.2021.100176
Luczkiewicz, A., Kotlarska, E., Artichowicz, W., Tarasewicz, K., and Fudala-Ksiazek, S. (2015). Antimicrobial resistance of Pseudomonas spp. isolated from wastewater and wastewater-impacted marine coastal zone. Environ. Sci. Pollut. Res. 22, 19823–19834. doi: 10.1007/s11356-015-5098-y
Lunestad, B., Frantzen, S., Svanevik, C., Roiha, I., and Duinker, A. (2016). Time trends in the prevalence of Escherichia coli and enterococci in bivalves harvested in Norway during 2007–2012. Food Control 60, 289–295. doi: 10.1016/j.foodcont.2015.08.001
MacFadden, D., McGough, S., Fisman, D., Santillana, M., and Brownstein, J. (2018). Antibiotic resistance increases with local temperature. Nat. Clim. Change 8, 510–514. doi: 10.1038/s41558-018-0161-6
Madigan, T., Bott, N., Torok, V., Percy, N., Carragher, J., de Barros Lopes, M., et al. (2014). A microbial spoilage profile of half shell Pacific oysters (Crassostrea gigas) and Sydney rock oysters (Saccostrea glomerata). Food Microbiol. 38, 219–227. doi: 10.1016/J.FM.2013.09.005
Manwar, A. V., Khandelwal, S., Chaudhari, B., Meyer, J., and Chincholkar, S. (2004). Siderophore production by a marine Pseudomonas aeruginosa and its antagonistic action against phytopathogenic fungi. Appl. Biochem. Biotechnol. 118, 243–251. doi: 10.1385/ABAB:118:1-3:243
Maravić, A., Šamanić, I., Šprung, M., Fredotović, Ž, Ilić, N., Dragičević, J., et al. (2018). Broad-spectrum resistance of Pseudomonas aeruginosa from shellfish: Infrequent acquisition of novel resistance mechanisms. Environ. Monit. Assess. 190:81. doi: 10.1007/s10661-018-6471-3
Maravić, A., Skočibušić, M., Šamanić, I., Fredotović, Ž, Cvjetan, S., Jutronić, M., et al. (2013). Aeromonas spp. simultaneously harbouring blaCTX-M-15, blaSHV-12, blaPER-1 and blaFOX-2, in wild-growing Mediterranean mussel (Mytilus galloprovincialis) from Adriatic Sea, Croatia. Int. J. Food Microbiol. 166, 301–308. doi: 10.1016/j.ijfoodmicro.2013.07.010
Martin, C., Svanevik, C., Lunestad, B., Sekse, C., and Johannessen, G. (2019). Isolation and characterisation of Shiga toxin-producing Escherichia coli from Norwegian bivalves. Food Microbiol. 84:103268. doi: 10.1016/j.fm.2019.103268
Martinez-Albores, A., Lopez-Santamarina, A., Rodriguez, J., Ibarra, I., del Carmen Mondragón, A., Miranda, J., et al. (2020). Complementary methods to improve the depuration of bivalves: A review. Foods 9:129. doi: 10.3390/FOODS9020129
Mechri, B., Monastiri, A., Medhioub, A., Medhioub, M., and Aouni, M. (2017). Molecular characterisation and phylogenetic analysis of highly pathogenic Vibrio alginolyticus strains isolated during mortality outbreaks in cultured Ruditapes decussatus juvenile. Microb. Pathog. 111, 487–496. doi: 10.1016/j.micpath.2017.09.020
Michalska, M., Zorena, K., Marks, R., and Wa̧ż, P. (2021). The emergency discharge of sewage to the Bay of Gdańsk as a source of bacterial enrichment in coastal air. Sci. Rep. 11:20959. doi: 10.1038/s41598-021-00390-8
Miller, S. (2016). Antibiotic resistance and regulation of the gram-negative bacterial outer membrane barrier by host innate immune molecules. mBio 7, e01541–16. doi: 10.1128/MBIO.01541-16/ASSET/0DE45D7F-4507-404C-919D-4BB32FC339C4/ASSETS/GRAPHIC/MBO0041629950001.JPEG
Miranda, C., Rojas, R., Garrido, M., Geisse, J., and González, G. (2013). Role of shellfish hatchery as a reservoir of antimicrobial resistant bacteria. Mar. Pollut. Bull. 74, 334–343. doi: 10.1016/J.MARPOLBUL.2013.06.032
Miranda, C., Rojas, R., Geisse, J., Romero, J., and González-Rocha, G. (2015). Scallop larvae hatcheries as source of bacteria carrying genes encoding for non-enzymatic phenicol resistance. Mar. Pollut. Bull. 95, 173–182. doi: 10.1016/J.MARPOLBUL.2015.04.026
Moreno Roldán, E., Rodríguez, E., Vicente, C., Navajas, M., and Abril, O. (2011). Microbial contamination of bivalve mollusks used for human consumption. J. Food Saf. 31, 257–261. doi: 10.1111/J.1745-4565.2010.00294.X
Moy, F., and Walday, M. (1996). Accumulation and depuration of organic micro-pollutants in marine hard bottom organisms. Mar. Pollut. Bull. 33, 56–63.
Mudadu, A., Salza, S., Melillo, R., Mara, L., Piras, G., Spanu, C., et al. (2021). Prevalence and pathogenic potential of Arcobacter spp. isolated from edible bivalve molluscs in Sardinia. Food Control 127:108139. doi: 10.1016/J.FOODCONT.2021.108139
Musella, M., Wathsala, R., Tavella, T., Rampelli, S., Barone, M., Palladino, G., et al. (2020). Tissue-scale microbiota of the Mediterranean mussel (Mytilus galloprovincialis) and its relationship with the environment. Sci. Total Environ. 717:137209. doi: 10.1016/J.SCITOTENV.2020.137209
National Advisory Committee on Microbiological Criteria for Foods [NACMCF] (2008). Response to the questions posed by the food and drug administration and the national marine fisheries service regarding the determination of cooking parameters for safe seafood for consumers. J. Food Prot. 71, 1287–1308. doi: 10.4315/0362-028x-71.6.1287
Nolan, C., and Behrends, V. (2021). Sub-inhibitory antibiotic exposure and virulence in Pseudomonas aeruginosa. Antibiotics (Basel) 10:1393. doi: 10.3390/ANTIBIOTICS10111393
Nuñez, T., Cerbino, G., Rapisardi, M., Quiroga, C., and Centrón, D. (2022). Novel mobile integrons and strain-specific integrase genes within Shewanella spp. Unveil Multiple lateral genetic transfer events within the genus. Microorganisms 10:1102. doi: 10.3390/MICROORGANISMS10061102
Obaidat, M., Salman, A., and Roess, A. (2017). Virulence and antibiotic resistance of Vibrio parahaemolyticus isolates from seafood from three developing countries and of worldwide environmental, seafood, and clinical isolates from 2000 to 2017. J. Food Prot. 80, 2060–2067. doi: 10.4315/0362-028X.JFP-17-156
Onohuean, H., Agwu, E., and Nwodo, U. A. (2022). Global perspective of Vibrio species and associated diseases: Three-decade meta-synthesis of research advancement. Environ. Health Insights 16:117863022210994. doi: 10.1177/11786302221099406
Pan, L., Lin, C., Zhang, G., Mu, G., and Yang, X. (2018). The effect of pre-process and transport strategies on survival, microbiologic, and physiologic of Patinopecten yessoensis. Food Sci. Nutr. 6:1422. doi: 10.1002/FSN3.687
Paschoal, R., Campana, E., Corrêa, L., Montezzi, L., Barrueto, L., da Silva, I., et al. (2017). Concentration and variety of carbapenemase producers in recreational coastal waters showing distinct levels of pollution. Antimicrob. Agents Chemother. 61, e01963–17. doi: 10.1128/AAC.01963-17/ASSET/22CC8063-374B-4C8C-B360-3453BB2C8106/ASSETS/GRAPHIC/ZAC0121767210001.JPEG
Pepi, M., and Focardi, S. (2021). Antibiotic-resistant bacteria in aquaculture and climate change: A challenge for health in the Mediterranean area. Int. J. Environ. Res. Public Health 18:5723. doi: 10.3390/ijerph18115723
Pierce, M., and Ward, J. (2019). Gut microbiomes of the Eastern Oyster (Crassostrea virginica) and the blue mussel (Mytilus edulis): Temporal variation and the influence of marine aggregate-associated microbial communities. mSphere 4, e00730–19. doi: 10.1128/MSPHERE.00730-19
Pinsky, M., Eikeset, A., McCauley, D., Payne, J., and Sunday, J. (2019). Greater vulnerability to warming of marine versus terrestrial ectotherms. Nature 569, 108–111. doi: 10.1038/s41586-019-1132-4
Poirel, L., Madec, J., Lupo, A., Schink, A., Kieffer, N., Nordmann, P., et al. (2018). Antimicrobial resistance in Escherichia coli. Microbiol. Spectr. 6:14. doi: 10.1128/microbiolspec.ARBA-0026-2017
Preena, P., Swaminathan, T., Rejish Kumar, V., and Bright Singh, I. (2020). Unravelling the menace: Detection of antimicrobial resistance in aquaculture. Lett. Appl. Microbiol. 71, 26–38. doi: 10.1111/lam.13292
Ramamurthy, T., Chowdhury, G., Pazhani, G., and Shinoda, S. (2014). Vibrio fluvialis: An emerging human pathogen. Front. Microbiol. 5:91. doi: 10.3389/FMICB.2014.00091/BIBTEX
Raman, G., Avendano, E., Chan, J., Merchant, S., and Puzniak, L. (2018). Risk factors for hospitalised patients with resistant or multidrug-resistant Pseudomonas aeruginosa infections: A systematic review and meta-analysis. Antimicrob. Resist. Infect. Control 7, 1–14. doi: 10.1186/S13756-018-0370-9/TABLES/3
Ramírez, C., Gutiérrez, M., Venegas, L., Sapag, C., Araya, C., Caruffo, M., et al. (2022). Microbiota composition and susceptibility to florfenicol and oxytetracycline of bacterial isolates from mussels (Mytilus spp.) reared on different years and distance from salmon farms. Environ. Res. 204:112068. doi: 10.1016/J.ENVRES.2021.112068
Regulation (EU) 2017/625 of the European Parliament and of the Council (2017). Regulation (EU) 2017/625 of the European parliament and of the council of 15 March 2017 on official controls and other official activities performed to ensure the application of food and feed law, rules on animal health and welfare, plant health and plant protection products, amending regulations (EC) No 999/2001, (EC) No 396/2005, (EC) No 1069/2009, (EC) No 1107/2009, (EU) No 1151/2012, (EU) No 652/2014, (EU) 2016/429 and (EU) 2016/2031 of the European parliament and of the council, council regulations (EC) No 1/2005 and (EC) No 1099/2009 and council directives 98/58/EC, 1999/74/EC, 2007/43/EC, 2008/119/EC and 2008/120/EC, and repealing regulations (EC) No 854/2004 and (EC) No 882/2004 of the European parliament and of the council, council directives 89/608/EEC, 89/662/EEC, 90/425/EEC, 91/496/EEC, 96/23/EC, 96/93/EC and 97/78/EC and council decision 92/438/EEC (Official controls regulation). Washington, DC: Regulation (EU) 2017/625 of the European Parliament and of the Council.
Richards, G., Watson, M., Crane, E., Burt, I., and Bushek, D. (2008). Shewanella and Photobacterium spp. in oysters and seawater from the Delaware bay. Appl. Environ. Microbiol. 74, 3323–3327. doi: 10.1128/AEM.00060-08
Romalde, J., Diéguez, A., Lasa, A., and Balboa, S. (2014). New Vibrio species associated to molluscan microbiota: A review. Front. Microbiol. 4:413. doi: 10.3389/FMICB.2013.00413
Romanenko, L., Tanaka, N., Svetashev, V., and Mikhailov, V. (2015). Pseudomonas glareae sp. nov., a marine sediment-derived bacterium with antagonistic activity. Arch. Microbiol. 197, 693–699. doi: 10.1007/S00203-015-1103-6/TABLES/2
Romanenko, L., Uchino, M., Falsen, E., Frolova, G., Zhukova, N., and Mikhailov, V. (2005). Pseudomonas pachastrellae sp. nov., isolated from a marine sponge. Int. J. Syst. Evol. Microbiol. 55, 919–924. doi: 10.1099/IJS.0.63176-0
Romanenko, L., Uchino, M., Tebo, B., Tanaka, N., Frolova, G., and Mikhailov, V. (2008). Pseudomonas marincola sp. nov., isolated from marine environments. Int. J. Syst. Evol. Microbiol. 58, 706–710. doi: 10.1099/ijs.0.65406-0
Roque, A., Lopez-Joven, C., Lacuesta, B., Elandaloussi, L., Wagley, S., Furones, M., et al. (2009). Detection and identification of TDH-and TRH-positive Vibrio parahaemolyticus strains from four species of cultured bivalve molluscs on the Spanish Mediterranean Coast. Appl. Environ. Microbiol. 75, 7574–7577. doi: 10.3389/fmicb.2015.00736
Salgueiro, V., Reis, L., Ferreira, E., Botelho, M., Manageiro, V., and Caniça, M. (2021). Assessing the bacterial community composition of bivalve mollusks collected in aquaculture farms and respective susceptibility to antibiotics. Antibiotics 10:1135. doi: 10.3390/antibiotics10091135
Sanz-Lazaro, C., and Sanchez-Jerez, P. (2020). Regional integrated multi-trophic aquaculture (RIMTA): Spatially separated, ecologically linked. J. Environ. Manag. 271:110921. doi: 10.1016/J.JENVMAN.2020.110921
Seong, H., Kim, J., Kim, T., Ahn, S., Rho, M., and Sul, W. (2021). A case study on the distribution of the environmental resistome in Korean shrimp farms. Ecotoxicol. Environ. Saf. 227:112858. doi: 10.1016/J.ECOENV.2021.112858
Shahimi, S., Elias, A., Abd Mutalib, S., Salami, M., Fauzi, F., Mohd Zaini, N., et al. (2021). Antibiotic resistance and determination of resistant genes among cockle (Anadara granosa) isolates of Vibrio alginolyticus. Environ. Sci. Pollut. Res. 28, 44002–44013. doi: 10.1007/s11356-021-13665-4
Silva dos Santos, F., Krepsky, N., Teixeira, V., Martins, V., da Silva, P., and Neves, R. (2022). Fecal pollution increases susceptibility to diseases in brown mussel Perna perna from cultured and wild populations. Aquaculture 551:737922. doi: 10.1016/J.AQUACULTURE.2022.737922
Taj-Aldeen, S., Deshmukh, A., Doiphode, S., Wahab, A., Allangawi, M., Almuzrkchi, A., et al. (2013). Molecular identification and susceptibility pattern of clinical Nocardia species: Emergence of Nocardia crassostreae as an agent of invasive nocardiosis. Can. J. Infect. Dis. Med. Microbiol. 24:e33. doi: 10.1155/2013/256025
Tena, D., Losa, C., Carrasco, G., and Saez-Nieto, J. (2016). Surgical site infection caused by Shewanella putrefaciens case report and literature review. Infect. Dis. Clin. Pract. 24, 18–23. doi: 10.1097/IPC.0000000000000344
To, K., Wong, S., Cheng, V., Tang, B., Li, I., Chan, J., et al. (2010). Epidemiology and clinical features of Shewanella infection over an eight-year period. Scand. J. Infect. Dis. 42, 757–762. doi: 10.3109/00365548.2010.490562
Udoekong, N., Bassey, B., Asuquo, A., Akan, O., Ifeanyi, C., Udoekong, N., et al. (2021). Prevalence and antimicrobial resistance of gram-negative bacteria isolates in shellfish samples from two river estuaries in south-south Nigeria. Adv. Microbiol. 11, 428–443. doi: 10.4236/AIM.2021.119032
Valdez, C., Costa, C., Simões, M., de Carvalho, C., Baptista, T., and Campos, M. (2022). Detection of mcr-1 gene in undefined Vibrio species isolated from clams. Microorganisms 10:394. doi: 10.3390/MICROORGANISMS10020394
Verga, R., Tolosano, J., Cazzaniga, N., and Gil, D. (2020). Assessment of seawater quality and bacteriological pollution of rocky shores in the central coast of San Jorge Gulf (Patagonia, Argentina). Mar. Pollut. Bull. 150:110749. doi: 10.1016/j.marpolbul.2019.110749
Vidovic, N., and Vidovic, S. (2020). Antimicrobial resistance and food animals: Influence of livestock environment on the emergence and dissemination of antimicrobial resistance. Antibiotics 9:52. doi: 10.3390/ANTIBIOTICS9020052
Vignier, N., Barreau, M., Olive, C., Baubion, E., Theodose, R., Hochedez, P., et al. (2013). Human infection with Shewanella putrefaciens and S. algae: Report of 16 cases in Martinique and review of the literature. Am. J. Trop. Med. Hyg. 89, 151–156. doi: 10.4269/AJTMH.13-0055
Vu, T., Alter, T., and Huehn, S. (2018). Prevalence of Vibrio spp. In retail seafood in Berlin, Germany. J. Food Prot. 81, 593–597. doi: 10.4315/0362-028X.JFP-17-366
Weis, J. (2014). Physiological, developmental and behavioral effects of marine pollution, Vol. 1. Berlin: Springer Science & Business Media, 452. doi: 10.1007/978-94-007-6949-6/COVER
West, P. (1989). The human pathogenic vibrios–a public health update with environmental perspectives. Epidemiol. Infect. 103, 1–34. doi: 10.1017/s0950268800030326
Wijsman, J., Troost, K., Fang, J., and Roncarati, A. (2019). in Goods and services of marine bivalves, eds A. Smaal, J. Grant, Ø Strand, J. Ferreira, and J. Petersen (Cham: Springer Nature), 7–26. doi: 10.1007/978-3-319-96776-9
Wright, A., Fan, Y., and Baker, G. (2018). Nutritional value and food safety of bivalve Molluscan shellfish. J. Shellfish Res. 37, 695–708. doi: 10.2983/035.037.0403
Xu, D., Peng, X., Xie, L., and Chen, L. (2022). Survival and genome diversity of Vibrio parahaemolyticus isolated from edible aquatic animals. Diversity 14:350. doi: 10.3390/d14050350
Ying, C., Chang, M., Hu, C., Chang, Y., Chao, W., Yeh, S., et al. (2018). The effects of marine farm-scale sequentially integrated multi-trophic aquaculture systems on microbial community composition, prevalence of sulfonamide-resistant bacteria and sulfonamide resistance gene sul1. Sci. Total Environ. 643, 681–691. doi: 10.1016/J.SCITOTENV.2018.06.204
Yousfi, K., Bekal, S., Usongo, V., and Touati, A. (2017). Current trends of human infections and antibiotic resistance of the genus Shewanella. Eur. J. Clin. Microbiol. Infect. Dis. 36, 1353–1362. doi: 10.1007/S10096-017-2962-3/TABLES/3
Zago, V., Veschetti, L., Patuzzo, C., Malerba, G., and Lleo, M. (2020). Resistome, mobilome and virulome analysis of Shewanella algae and Vibrio spp. Strains isolated in Italian aquaculture centers. Microorganisms 8:572. doi: 10.3390/MICROORGANISMS8040572
Zannella, C., Mosca, F., Mariani, F., Franci, G., Folliero, V., Galdiero, M., et al. (2017). Microbial diseases of bivalve mollusks: Infections, immunology and antimicrobial defense. Mar. Drugs 15:182. doi: 10.3390/MD15060182
Keywords: molluscs, bivalves, bacteria, antibiotic-resistance, genes, foodborne diseases, zoonoses, aquaculture
Citation: Kijewska A, Koroza A, Grudlewska-Buda K, Kijewski T, Wiktorczyk-Kapischke N, Zorena K and Skowron K (2023) Molluscs—A ticking microbial bomb. Front. Microbiol. 13:1061223. doi: 10.3389/fmicb.2022.1061223
Received: 04 October 2022; Accepted: 20 December 2022;
Published: 09 January 2023.
Edited by:
Joseph D. Eifert, Virginia Tech, United StatesReviewed by:
Jessica L. Jones, United States Food and Drug Administration, United StatesCopyright © 2023 Kijewska, Koroza, Grudlewska-Buda, Kijewski, Wiktorczyk-Kapischke, Zorena and Skowron. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Krzysztof Skowron,  c2tvd3JvbjIzOEB3cC5wbA==
c2tvd3JvbjIzOEB3cC5wbA==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.