- 1Engineering and Research Center for Southwest Bio-Pharmaceutical Resources of National Education Ministry of China, Guizhou University, Guiyang, China
- 2School of Pharmaceutical Engineering, Guizhou Institute of Technology, Guiyang, China
- 3Center of Excellence in Fungal Research, Mae Fah Luang University, Chiang Rai, Thailand
- 4Guizhou Key Laboratory of Agricultural Biotechnology, Guizhou Academy of Agricultural Sciences, Guiyang, China
Helicosporous hyphomycetes have the potential to produce a variety of bioactive compounds. However, the strain resources of this fungal group are relatively scarce, which limits their further exploitation and utilization. In this study, based on phylogenetic analyses of combined ITS, LSU, RPB2, and TEF1α sequence data and the morphology from 11 isolates, we introduce four new species of helicosporous hyphomycetes, viz. Helicoma wuzhishanense, Helicosporium hainanense, H. viridisporum, and Neohelicomyces hainanensis, as well as three new records, viz. Helicoma guttulatum, H. longisporum, and Helicosporium sexuale. Detailed morphological comparisons of the four new species that distinguish them are provided.
Introduction
The most remarkable feature that distinguishes helicosporous hyphomycetes from other fungal groups is that its conidia curve through at least 180° in one plane as they extend in length (Goos, 1986; Zhao et al., 2007; Luo et al., 2017; Lu et al., 2018a,b; Tian et al., 2022). They are distributed in the Dothideomycetes (Capnodiales, Microthyriales, Pleosporales, Tubeufiales, and Venturiales), Leotiomycetes (Helotiales), Orbiliomycetes (Orbiliales), Sordariomycetes (Hypocreales, Lulworthiales, Microascales, Torpedosporales), Agaricomycetes (Agaricales), Atractiellomycetes (Atractiellales), Exobasidiomycetes (Exobasidiales), Tremellomycetes (Tremellales), and Zoopagomycetes (Zoopagales) (Lu and Kang, 2020). Helicosporous fungi are widespread in tropical and temperate regions (Lu et al., 2018b). Most species in this group, which were published more than 10 years ago, were saprophytic on terrestrial woody substrates, and most of them were lacking in DNA molecular data (Goos, 1986; Zhao et al., 2007; Boonmee et al., 2014; Lu et al., 2018b). However, the species of this group discovered in the last decade mainly come from aquatic habitats (Lu et al., 2018b; Boonmee et al., 2021; Tian et al., 2022), and almost all newly published helicosporous species have molecular data. The latest comprehensive revision on helicosporous hyphomycetes was carried out by Lu et al. (2018b), who established nine new helicosporous genera based on morphology and phylogeny, viz. Dematiohelicoma, Dematiohelicomyces, Dematiohelicosporum, Helicoarctatus, Helicohyalinum, Helicotruncatum, Pleurohelicosporium, Pseudohelicomyces, and Pseudohelicoon, and reassessed the taxonomic system of the three earliest described helicosporous hyphomycete genera, viz. Helicomyces, Helicosporium, and Helicoma. For example, in the genus Helicosporium, Lu et al. (2018b) redefined its generic concept based on morphological and phylogenetic evidence, and accepted 13 species, including five new species, and excluded 25 species from this genus which were transferred to the genera Neohelicosporium and Helicoma. In addition, although Lu et al. (2018b) proposed some suggestions on how to classify and identify helicosporous fungi, there are still some species in this group that need more morphological and molecular data to solve their taxonomic status.
The focus of research on helicosporous fungi has been mainly in the field of taxonomy. However, these fungi are not only morphologically fascinating but also a potential source to produce a variety of bioactive secondary metabolites. For example, species of Helicomyces, Helicosporium, and Helicoma have been reported to produce natural products with antibacterial, anticancer, and anti-diabetic activities (Itazaki et al., 1990; Hanada et al., 1996; Ohtsu et al., 2003; Yoshimura et al., 2003; Zenkoh et al., 2003; Dong et al., 2004; Hu et al., 2006; Jiao et al., 2006; Jung et al., 2012; Lee et al., 2013). Furthermore, recent studies have revealed that other helicosporous fungi also show great potential for exploring new active natural products (Qian et al., 2022; Zeng et al., 2022; Zheng et al., 2022). Zheng et al. (2022) reported two novel compounds in Tubeufia rubra; one of which reverses multidrug resistance of tumor cell lines to Doxorubicin. Qian et al. (2022) also discovered another two new compounds in Tubeufia rubra, and one, namely, Rubrosin-D displayed significant multidrug resistance reversal effects. Zheng et al. (2022) discovered that some alkaloids in Neohelicomyces hyalosporus were cytotoxic against human cancer (A549, TCA, and RD) cells.
In order to solve the classification problems related to helicosporous hyphomycetes and enrich the species resources of the fungal group, we have recently collected a large number of specimens of this group from various terrestrial and aquatic environments. In this study, we report on 11 helicosporous hyphomycetes collected from decaying woody substrates from freshwater streams and terrestrial habitats in southern China. The taxa are characterized based on morphological features and phylogenetic analyses. The new species are morphologically and phylogenetically distinct. Detailed descriptions, illustrations, and phylogenetic analyses are provided.
Materials and methods
Sample collection and specimen examination
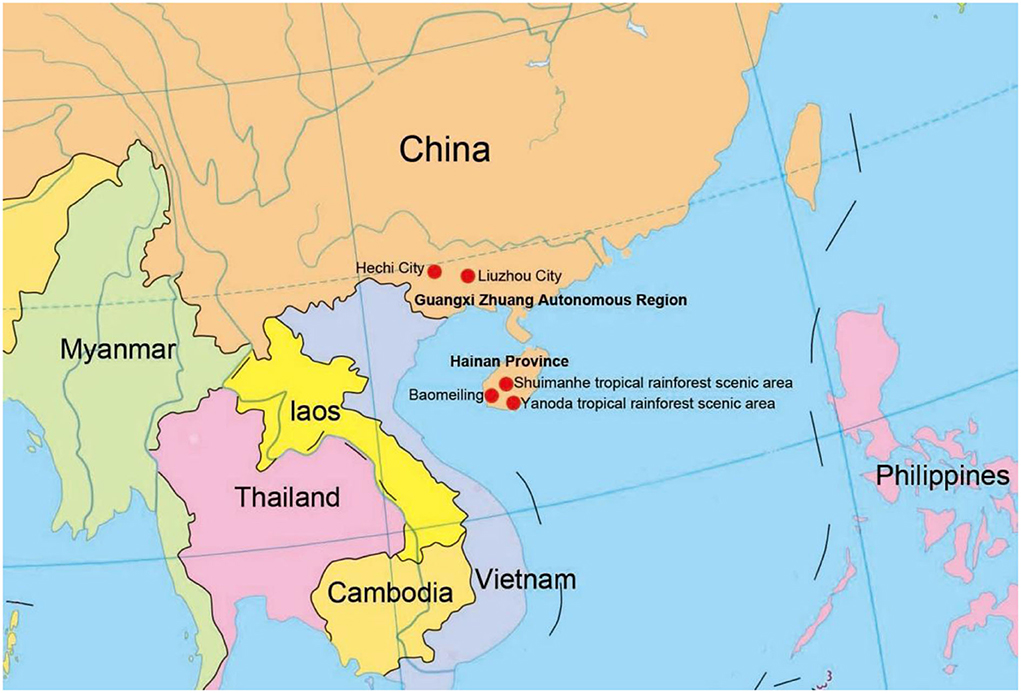
Submerged decaying wood samples were collected from various sites in freshwater streams and terrestrial environments in Guangxi Zhuang Autonomous Region and Hainan Provinces, China (Figure 1). Techniques in Senanayake et al. (2020) were followed for morphological study and single spore isolation. Morphological characteristics were examined with a stereomicroscope (SMZ 745 Nikon, Japan). Micro-morphological characters were photographed using a Nikon EOS 70D digital camera attached to an ECLIPSE Ni compound microscope (Nikon, Japan). Measurements were made with a Tarosoft (R) Image Frame Work program. Figures were processed and combined using Adobe Photoshop CS6 Extended version 10.0 software (Adobe Systems, USA).
Herbarium specimens were deposited in the Herbarium of Guizhou Academy of Agriculture Sciences (Herb. GZAAS) and the Herbarium of Cryptogams Kunming Institute of Botany Academia Sinica (Herb. HKAS). Ex-type living cultures are deposited at Guizhou Culture Collection (GZCC). Facesoffungi database and Index Fungorum numbers are provided (Jayasiri et al., 2015).
DNA extraction, PCR amplification, and sequencing
Genomic DNA was extracted from at least 3-week-old living pure cultures grown on PDA at 28 °C using the Biospin Fungus Genomic DNA Extraction Kit (BioFlux, China), and following the manufacturer's protocol. The primer pairs of ITS5/ITS4, LR0R/LR5, fRPB2-5F/fRPB2-7cR, and EF1-983F/EF1-2218R were used to amplify the internal transcribed spacer (ITS) (White et al., 1990), the large subunit ribosomal DNA (LSU) (Vilgalys and Hester, 1990), the RNA polymerase II second largest subunit (RPB2) (Liu et al., 1999), and the translation elongation factor 1-alpha gene (TEF1α) (Rehner and Buckley, 2005) regions, respectively. The ITS, LSU, RPB2, and TEF1α amplification reactions were carried out using the method described by Lu et al. (2017b, 2018a). The PCR products were purified and sequenced with the same primers at Tsingke Biological Technology (Kunming) Co., China.
Phylogenetic analysis
DNASTAR Lasergene SeqMan Pro v. 7.1.0 (44.1) was used to edit ambiguous bases at both ends of the raw forward and reverse reads and to assemble them. The newly obtained sequences were used as queries to perform BLAST searches against the nr database to check for contamination, compare species, and create datasets. MAFFT v.7 was used to align the individual datasets (Katoh et al., 2019). Each alignment was trimmed using Trimal (Capella-Gutiérrez et al., 2009). BioEdit was used to check the alignment manually (Hall, 1999).
Four genetic markers, including ITS, LSU, RPB2, and TEF1α, were used for phylogenetic inferences (Table 1). The phylogeny tree was inferred using 147 taxa. IQ-Tree v.2 (Minh et al., 2020) was used to infer maximum likelihood trees (ML) according to the Bayesian information criterion (BIC). Partitioned analyses were carried out for the combined datasets, which were partitioned according to genetic markers. Branch support was estimated from 1,000 ultrafast bootstrap replicates. RAxML-HPC2 on XSEDE (8.2.12) (Stamatakis, 2014) in the CIPRES Science Gateway platform was also used. ModelTest, as implemented in MrMTgui (Nuin, 2007), was used to determine the best-fit evolution model for Bayesian inference analyses using the Akaike Information Criterion (AIC). Bootstrap support was estimated from 1,000 rapid bootstrap replicates. MrBayes v.3.1.2 (Ronquist et al., 2012) was utilized to evaluate the posterior probabilities (PP) by Markov Chain Monte Carlo sampling (MCMC). The number of generations was determined separately for each dataset and is noted in the individual tree legends. The first 25% of the trees were discarded, as they represented the burn-in phase of the analyses, while the remaining were used for calculating PP in the majority rule consensus tree. For all Bayesian inference trees, convergence was declared when the average standard deviation reached 0.01. The trees were figured in the FigTree v1.4.0 program (Rambaut and Drummond, 2008). The approximately unbiased (AU) test, implemented in CONSEL, was used to test the placement of the newly erected family (Shimodaira and Hasegawa, 2001). Topologies with AU test p-values < 0.05 were rejected.
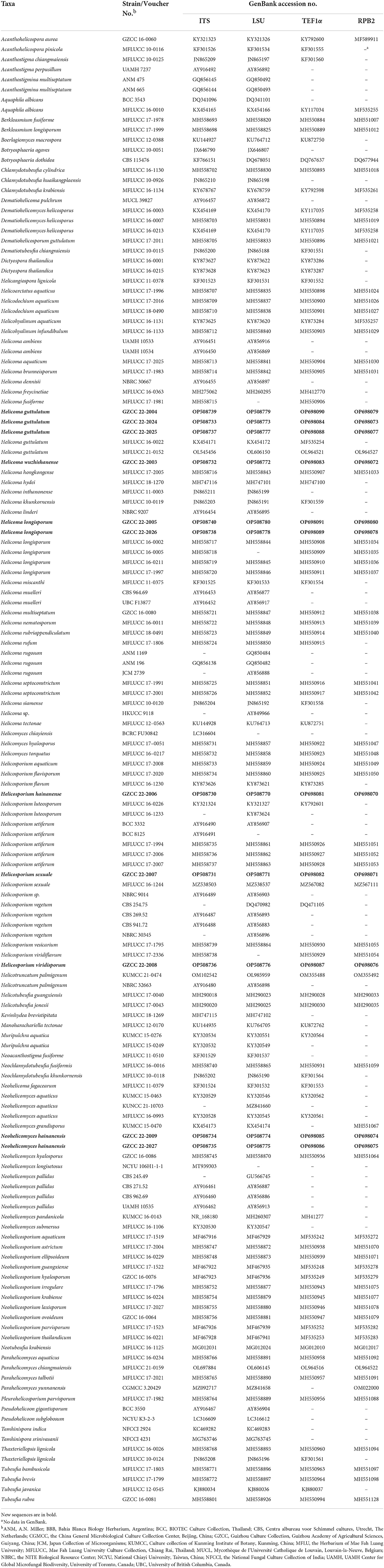
Table 1. Taxa used in this study and their GenBank accession numbers for ITS, LSU, RPB2, and TEF1α DNA sequence data.
Results
Phylogenetic analysis of combined ITS, LSU, RPB2, and TEF1α sequence data
The combined ITS, LSU, RPB2, and TEF1α datasets comprised 11 newly sequenced strains. Multiple genes were concatenated, which comprised 146 taxa and 3313 nucleotide characters, including gaps (ITS: 513 bp; LSU: 843 bp; RPB2: 1045 bp; TEF1α: 912 bp). The maximum likelihood and Bayesian analysis of the combined dataset resulted in phylogenetic reconstructions with largely similar topologies, and the IQ-Tree is shown in Figure 2.
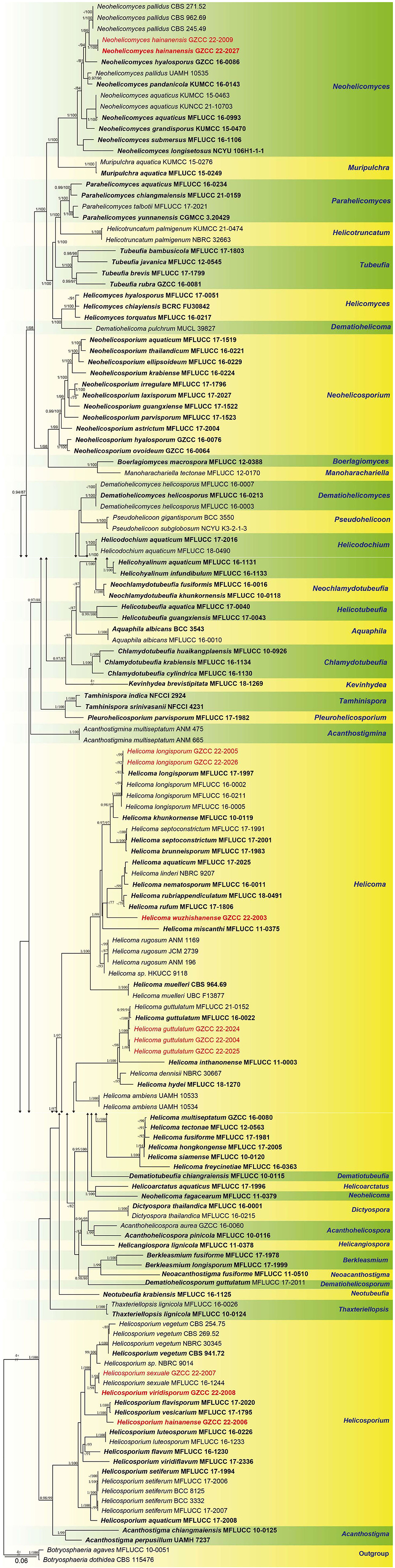
Figure 2. Phylogenetic tree generated from a maximum likelihood analysis based on a concatenated alignment of ITS, LSU, RPB2, and TEF1α sequence data. Bootstrap support values of maximum likelihood (ML) ≥75% and Bayesian posterior probabilities (PP) ≥0.95 are given near the nodes as PP/MLBS. The tree is rooted with Botryosphaeria agaves (MFLUCC 10-0051) and B. dothidea (CBS 115,476). Newly generated sequences are in red. Ex-type strains are in bold.
Representatives of the sequenced genera (with molecular data) of helicosporous hyphomycetes (Boonmee et al., 2011, 2014; Rajeshkumar and Sharma, 2013; Brahamanage et al., 2017; Doilom et al., 2017; Lu et al., 2017a, 2018a,b; Luo et al., 2017; Phookamsak et al., 2017; Liu et al., 2019; Tian et al., 2022) are included in our phylogenetic analysis (Figure 2). Thirty-six genera are represented by at least one species in Tubeufiaceae. Our 11 isolates are recognized as four new species, viz. Helicoma wuzhishanense, Helicosporium hainanense, H. viridisporum, and Neohelicomyces hainanensis, and three new records, viz. Helicoma guttulatum, H. longisporum, and Helicosporium sexuale.
Taxonomy
Helicoma guttulatum Y.Z. Lu, Boonmee & K.D. Hyde, Fungal Diversity 80: 125 (2016), Figure 3.

Figure 3. Helicoma guttulatum (GZAAS 22-2004). (a) Colony on decaying wood. (b–d) Conidiophores and conidia. (e–g) Conidiogenous cells. (i) Germinating conidium. (h,j–l) Conidia. (m,n) Colonies on PDA observed from above and below. Scale bars: (b–d) = 20 μm, (e–j,i–l) = 10 μm, and (h) = 5 μm.
Index Fungorum number: IF 552218; Facesoffungi number: FoF 02358.
Saprobic on submerged decaying wood in a freshwater stream. Sexual morph Undetermined. Asexual morph Hyphomycetous, helicosporous. Colonies superficial, effuse, gregarious, brown to dark brown. Mycelium mostly immersed, composed of branched, septate, brown hyphae. Conidiophores 120–202 × 4–6.5 μm ( = 169 × 5.5 μm, n = 20), macronematous, mononematous, cylindrical, erect, septate, unbranched, pale brown to brown at the apex, dark brown at the base, smooth-walled. Conidiogenous cells 18–37 × 4.5–6 μm ( = 24 × 5 μm, n = 20), holoblastic, mono- to polyblastic, integrated, terminal, cylindrical, brown, and smooth-walled. Conidia 20–26.5 μm ( = 22 μm, n = 25) in diam., and conidial filament 7.5–9.5 μm ( = 8.5 μm, n = 25) wide and 43–57 μm long ( = 51.5 μm, n = 25), solitary, acrogenous, helicoid, tightly coiled 1–11/2 times, guttulate, do not become loose in water, 7–8-septate, straight constricted at the septa, subhyaline to pale brown, tapering toward the flat end, rounded at the apex, conico-truncate at the base, smooth-walled.
Culture characteristics: Conidia germinating on PDA within 12 h; Colonies growing on PDA, reaching 9 mm in 2 weeks at 25°C, circular, with a flat surface, edge undulate, and pale brown to brown in the PDA medium.
Material examined: CHINA, Hainan Province, Yanoda Tropical rainforest scenic area, on submerged decaying wood in a freshwater stream, 23 October 2021, Jian Ma, Y16.2 (GZAAS 22-2004), living culture, GZCC 22-2004; Ibid., Y4 (GZAAS 22-2025), living culture, GZCC 22-2025; Hainan Province, Wuzhishan City, Shuimanhe tropical rainforest scenic area in Wuzhishan, on submerged decaying wood in a freshwater stream, 15 August 2021, Jian Ma, WZS34 (GZAAS 22-2024), living culture, GZCC 22-2024.
GenBank accession numbers: GZCC 22-2004: OP508739 (ITS), OP508779 (LSU), OP698079 (RPB2), and OP698090 (TEF1α); GZCC 22-2025: OP508737 (ITS), OP508777 (LSU), OP698077 (RPB2), and OP698088 (TEF1α); GZCC 22-2024: OP508733 (ITS), OP508773 (LSU), OP698073 (RPB2), and OP698084 (TEF1α).
Notes: Helicoma guttulatum was introduced by Hyde et al. (2016) with morphological and phylogenetic evidence. Tian et al. (2022) reported a new collection from Thailand. In this study, three newly obtained isolates clustered with two known strains of H. guttulatum (MFLUCC 16-0022 and MFLUCC 21-0152) with high statistical support (100% ML/1.00 PP, Figure 2). We note that there are two isolates (GZCC 22-2004 and GZCC 22-2025) clustered together with high statistical support and were phylogenetically different from the other isolates. However, there are only 5 bp and 12 bp differences in ITS and RPB2 between them and the ex-type strain of H. guttulatum (MFLUCC 16-0022), and their LSU and TEF1α data are identical. Moreover, we could not identify any morphological character differences to separate them, and these few gene base pair changes are within the accepted range of variation for a species; thus, we identify the newly obtained isolates as H. guttulatum. This species has only been previously reported in Thailand. It is the first record of H. guttulatum in China and in a terrestrial habitat.
Helicoma longisporum Y.Z. Lu, J.K. Liu & K.D. Hyde, Fungal Diversity 92: 178 (2018), Figure 4.

Figure 4. Helicoma longisporum (GZAAS 22-2005). (a,b) Colony on decaying wood. (c,d) Conidiophores with attached conidia. (e,f,j) Conidiogenous cells. (g–i) Conidia. (k) Germinating conidium. (l,m) Colonies on PDA observed from above and below. Scale bars: (c–k) = 20 μm.
Index Fungorum number: IF 554840; Facesoffungi number: FoF 04715.
Saprobic on decaying wood in a freshwater stream. Sexual morph Undetermined. Asexual morph Hyphomycetous, helicosporous. Colonies on the substratum superficial, effuse, gregarious, light pink to brown. Mycelium partly immersed, pale brown to brown, septate, branched hyphae, with masses of crowded, glistening conidia. Conidiophores 114–281 × 6–10.5 μm ( = 197.5 × 7 μm, n = 20), macronematous, mononematous, cylindrical, straight, unbranched, septate, pale brown to brown, smooth-walled. Conidiogenous cells 11–21 × 6.5–10 μm ( = 13.5 × 7.5 μm, n = 20), holoblastic, monoblastic, integrated, intercalary, cylindrical, with denticles, rising laterally from the lower portion of conidiophores as tiny tooth-like protrusions (3–5.5 μm long, 3.5–4.5 μm wide), pale brown, smooth-walled. Conidia 51–70 μm in diam. and conidial filament 6.5–11 μm wide ( = 61 × 9 μm, n = 20), 325–508 μm long, solitary, pleurogenous, helicoid, coiled 2–3 times, becoming loosely coiled in water, rounded at tip, up to 34-septate, constricted at septa, pale brown to brown, smooth-walled.
Culture characteristics: Conidia germinating on PDA within 12 h. Colonies growing on PDA, reaching 10 mm in 2 weeks at 25°C, circular, with a flat surface, edge entire, and pale brown to brown in the PDA medium.
Material examined: CHINA, Hainan Province, Yanoda Tropical rainforest scenic area, on submerged decaying wood in a freshwater stream, 23 October 2021, Jian Ma, Y16.3 (GZAAS 22-2005), living culture, GZCC 22-2005; Ibid., Y5 (GZAAS 22-2026), living culture, GZCC 22-2026.
GenBank accession numbers: GZCC 22-2005: OP508740 (ITS), OP508780 (LSU), OP698080 (RPB2), and OP698091 (TEF1α); GZCC 22-2026: OP508738 (ITS), OP508778 (LSU), OP698078 (RPB2), and OP698089 (TEF1α).
Notes: Helicoma longisporum was introduced by Lu et al. (2018b) based on morphology and phylogeny. In this study, two newly obtained isolates are identified as H. longisporum based on their identical DNA molecular data, conidiophores, conidiogenous cells, and conidial characteristics (Lu et al., 2018b). This species has only been previously reported in Thailand (Lu et al., 2018b). It is the first record of H. longisporum in China.
Helicoma wuzhishanense Y.Z. Lu & J.C. Kang, sp. nov. Figure 5.

Figure 5. Helicoma wuzhishanense (GZAAS 22-2003, holotype). (a,b) Colony on decaying wood. (c–f) Conidiophores. (g,h) Conidiogenous cells with attached conidium. (i,j) Conidia. (k) Germinating conidium. (l,m) Colonies on PDA observed from above and below. Scale bars: (c–f,k) = 20 μm, (g–j) = 10 μm.
Index Fungorum number: IF 900032; Facesoffungi number: FoF 13100.
Holotype: GZAAS 22-2003.
Etymology: “wuzhishanense” referring to collecting site.
Saprobic on decaying wood in a freshwater stream. Sexual morph Undetermined. Asexual morph Hyphomycetous, helicosporous. Colonies on the substratum superficial, effuse, gregarious, brown to dark brown. Mycelium partly immersed, brown, septate, branched hyphae, with masses of crowded, glistening conidia. Conidiophores 90–130 μm long, 5.5–6.5 μm wide ( = 115 × 6 μm, n = 30), macronematous, mononematous, cylindrical, erect, straight to slightly bent, unbranched, septate, the lower part brown and the upper part pale brown, smooth-walled. Conidiogenous cells 10–13 × 5–6.5 μm ( = 11.5 × 5.5 μm, n = 20), holoblastic, mono- to polyblastic, integrated, intercalary, cylindrical, with denticles, rising laterally from the lower portion of conidiophores as tiny tooth-like protrusions (1.5–3 μm long, 1.5–2.5 μm wide), brown, smooth-walled. Conidia 34–58 μm diam., and conidial filament 2.5–5 μm wide ( = 45 × 4 μm, n = 20), 182–287 μm long, up to 34-septate, solitary, pleurogenous, helicoid, coiled 21/3-31/3 times, becoming loosely coiled in water, rounded at tip, guttulate, hyaline to pale brown, smooth-walled.
Culture characteristics: Conidia germinating on water agar and germ tubes produced from conidia within 12 h. Colonies growing on PDA, circular, with a flat surface, edge entire, reaching 29 mm in 4 weeks at 25°C, pale brown to yellowish in the PDA medium.
Material examined: CHINA, Hainan Province, Wuzhishan City, Shuimanhe tropical rainforest scenic area in Wuzhishan, on submerged decaying wood in a freshwater stream, 15 August 2021, Jian Ma, WZS23.2 (GZAAS 22-2003, holotype; HKAS 125862, isotype), ex-type living culture, GZCC 22-2003.
GenBank accession numbers: OP508732 (ITS), OP508772 (LSU), OP698072 (RPB2), and OP698083 (TEF1α).
Notes: Morphologically, Helicoma wuzhishanense resembles Helicoma rufum, having unbranched, straight to slightly bent, cylindrical conidiophores, and pleurogenous helicoid conidia. However, H. wuzhishanense can be distinguished from H. rufum by its smaller conidiophores (90–130 μm × 5.5–6.5 μm vs. 110–210 μm × 7–8.5 μm) and shorter conidial filament (182–287 μm vs. 240–410 μm) (Lu et al., 2018b). Furthermore, H. rufum produces a reddish brown pigment in the PDA medium in 7 days but H. wuzhishanense lacks this characteristic. Phylogenetically, H. wuzhishanense formed an independent lineage within the genus (Figure 2) and the phylogenetic analysis result supports it as a distinct species.
Helicosporium hainanense Y.Z. Lu & J.C. Kang, sp. nov. Figure 6.

Figure 6. Helicosporium hainanense (GZAAS 22-2006, holotype). (a,b) Colony on decaying wood. (c–f) Conidiophores and conidia. (g–i) Conidiogenous cells with attached conidia. (j) Germinating conidium. (k–m) Conidia. (n,o) Colonies on PDA observed from above and below. Scale bars: (c–f) = 20 μm, (g–j) = 10 μm, (k–m) = 5 μm.
Index Fungorum number: IF 900031; Facesoffungi number: FoF 13101.
Holotype: GZAAS 22-2006.
Etymology: “hainanense” referring to collecting site.
Saprobic on decaying woody substrate. Sexual morph Undetermined. Asexual morph Hyphomycetous, helicosporous. Colonies on the substratum superficial, effuse, gregarious, yellow green. Mycelium partly immersed, pale brown to brown, septate, branched hyphae, with masses of crowded, glistening conidia. Conidiophores 118–182 μm long, 2.5–4 μm wide ( = 155 × 3 μm, n = 30), macronematous, mononematous, cylindrical, unbranched, straight or slightly flexuous, septate, pale brown to dark brown, smooth-walled. Conidiogenous cells holoblastic, mono- to polyblastic, discrete, determinate, rising laterally from the lower portion of the conidiophores as tiny bladder-like protrusions, 2–8.5 μm long, 1.5–3.5 μm diam., each bearing 1–3 tiny conidiogenous loci, hyaline to pale brown, smooth-walled. Conidia 11–13 μm diam. and conidial filament 2–3 μm wide ( = 12 × 2.5 μm, n = 20), 55–60 μm long, solitary, pleurogenous, helicoid, tightly coiled 21/4-23/4 times, do not become loose in water, tapering toward the rounded ends, indistinctly multi-septate, guttulate, hyaline to yellowish, smooth-walled.
Culture characteristics: Conidia germinating on water agar and germ tubes produced from conidia within 12 h. Colonies growing on PDA, irregular, with a flat surface, edge undulate, reaching 19 mm in 5 weeks at 25°C, brown to dark brown in the PDA medium.
Material examined: CHINA, Hainan Province, Changjiang, Baomeiling, on decaying wood in a terrestrial habitat, 15 August 2021, Jian Ma, BM11 (GZAAS 22-2006, holotype; HKAS 125882, isotype), ex-type living culture, GZCC 22-2006.
GenBank accession numbers: OP508730 (ITS), OP508770 (LSU), OP698070 (RPB2), and OP698081 (TEF1α).
Notes: Phylogenetically, Helicosporium hainanense shares a sister relationship to H. flavisporum and H. vesicarium with high statistical support (100% ML/1.00 PP, Fig. 2), and can be considered as a distinct species. Morphologically, H. hainanense differs from H. flavisporum by its wider and shorter conidial filaments (2–3 μm wide, 55–60 μm long vs. 1–2 μm wide, 100–110 μm long), and from H. vesicarium by its longer conidiophores (118–182 μm vs. 65–120 μm) and smaller conidial diameter (11–13 μm vs. 13–18 μm) (Lu et al., 2018b).
Helicosporium sexuale Boonmee, Promputtha & K.D. Hyde, Fungal Diversity 111: 124 (2021), Figure 7.

Figure 7. Helicosporium sexuale (GZAAS 22-2007). (a,b) Colony on decaying wood. (c–h) Conidiophores. (i,j) Conidiogenous cells. (k) Germinating conidium. (l–o) Conidia. (p,q) Colonies on PDA observed from above and below. Scale bars: (c–h) = 20 μm, (i–o) = 10 μm.
Index Fungorum number: IF 558542; Facesoffungi number: FoF 09194.
Holotype: MFLU 21-0104.
Saprobic on decaying wood in a freshwater stream. Sexual morph see Boonmee et al. (2021). Asexual morph Hyphomycetous, helicosporous. Colonies on the substratum superficial, effuse, gregarious, yellow green. Mycelium partly immersed, partly superficial, brown to dark brown, septate, branched hyphae, with masses of crowded, glistening conidia. Conidiophores 60–129 μm long, 3.5–6 μm wide ( = 98 × 4.5 μm, n = 30), macronematous, mononematous, erect, setiferous, cylindrical, septate, brown to dark brown, smooth-walled. Conidiogenous cells holoblastic, monoblastic, discrete, determinate, denticulate, rising laterally from the lower parts of conidiophores as tiny tooth-like protrusions, hyaline to pale brown, smooth-walled. Conidia 11–20 μm diam. and conidial filament 1–2 μm wide ( = 14.5 × 1.5 μm, n = 20), 68–91 μm long, solitary, pleurogenous, helicoid, coiled 2–31/3 times, becoming loosely coiled in water, rounded at tip, guttulate, indistinctly multi-septate, hyaline to pale green, smooth-walled.
Culture characteristics: Conidia germinating on water agar and germ tubes produced from conidia within 12 h. Colonies growing on PDA, circular, with a flat surface, edge undulate, reaching 40 mm in 6 weeks at 25°C, brown to dark brown in the PDA medium.
Material examined: CHINA, Guangxi Zhuang Autonomous Region, Liuzhou City, Luzhai County, on submerged decaying wood in a freshwater stream, 4 May 2021, Jian Ma & Yongzhong Lu, LZ15 (GZAAS 22-2007 = HKAS 125866), living cultures, GZCC 22-2007.
GenBank accession numbers: OP508731 (ITS), OP508771 (LSU), OP698071 (RPB2), and OP698082 (TEF1α).
Notes: In this study, a new helicosporous hyphomycete (GZCC 22-2007) was phylogenetically grouped with Helicosporium sexuale (MFLUCC 16-1244) and did not show much divergence (Figure 2). We compared their DNA sequences and found that only 5 bp nucleotide differences between them in TEF1α sequence data, whereas their ITS, LSU, and RPB2 sequence data were identical. Therefore, we identify the new isolate GZCC 22-2007 as H. sexuale. Helicosporium sexuale was described as only a sexual morph (Boonmee et al., 2021). Its asexual morph is reported in this study for the first time. This is also the first record of H. sexuale in a freshwater habitat in China.
Helicosporium viridisporum Y.Z. Lu & J.C. Kang, sp. nov. Figure 8.

Figure 8. Helicosporium viridisporum (GZAAS 22-2008, holotype). (a,b) Colony on decaying wood. (c–e,g,i,j) Conidiophores and conidia. (f) Conidiogenous cells. (h) Germinating conidium. (k–n) Conidia. (o,p) Colonies on PDA observed from above and below. Scale bars: (c–f,i,j) = 20 μm, (g,h) = 10 μm, (k–n) = 5 μm.
Index Fungorum number: IF 900030; Facesoffungi number: FoF 13102.
Holotype: GZAAS 22-2008.
Etymology: “viridisporum” referring to the bright lime green conidia in a natural woody substrate.
Saprobic on decaying wood in a freshwater stream. Sexual morph Undetermined. Asexual morph Hyphomycetous, helicosporous. Colonies on the substratum superficial, effuse, gregarious, bright lime green. Mycelium partly immersed, brown to dark brown, septate, branched hyphae, with masses of crowded, glistening conidia. Conidiophores 80–206 μm long, 3–7 μm wide ( = 146 × 5 μm, n = 30), macronematous, mononematous, erect, setiferous, cylindrical, septate, brown to dark brown, smooth-walled. Conidiogenous cells holoblastic, polyblastic, discrete, determinate, denticulate, rising laterally from the lower parts of conidiophores as tiny tooth-like protrusions, hyaline to pale brown, smooth-walled. Conidia solitary, 12–14 μm diam. and conidial filament 1–2 μm wide ( = 13 × 1.5 μm, n = 30), 75–97 μm long, pleurogenous, helicoid, tightly coiled 2–31/3 times, becoming loosely coiled in water, rounded at tip, guttulate, indistinctly multi-septate, hyaline to pale green, smooth-walled.
Culture characteristics: Conidia germinating on water agar and germ tubes produced from conidia within 12 h. Colonies growing on PDA, circular, with a flat surface, edge undulate, reaching 40 mm in 5 weeks at 25°C, brown to dark brown in the PDA medium.
Material examined: CHINA, Guangxi Zhuang Autonomous Region, Hechi City, Xiayi Village, on submerged decaying wood in a freshwater stream, 3 May 2021, Jian Ma, XYC2 (GZAAS 22-2008, holotype; HKAS 125857, isotype), ex-type living culture, GZCC 22-2008.
GenBank accession numbers: OP508736 (ITS), OP508776 (LSU), OP698076 (RPB2), and OP698087 (TEF1α).
Notes: Helicosporium viridisporum is a typical Helicosporium species according to the redefined generic concept of Helicosporium by Lu et al. (2018b). Its colonies on natural woody substratum are bright lime green. H. viridisporum shares a sister relationship to H. sexuale and can be distinguished by its longer conidiophores (80–206 μm vs. 60–129 μm). The multi-gene phylogenetic analysis supports it as a new species.
Neohelicomyces hainanensis Y.Z. Lu & J.C. Kang, sp. nov. Figure 9.

Figure 9. Neohelicomyces hainanensis (GZAAS 22-2009, holotype). (a,b) Colony on decaying wood. (c–g) Conidiophores and conidia. (h–j) Conidiogenous cells. (k–n) Conidia. (o) Germinating conidium. (p,q) Colonies on PDA observed from above and below. Scale bars: (c–g) = 20 μm, (h,i,k–n) = 10 μm, (j) = 5 μm.
Index Fungorum number: IF 900029; Facesoffungi number: FoF 13103.
Holotype: GZAAS 22-2009.
Etymology: “hainanensis” referring to the collection site.
Saprobic on decaying wood. Sexual morph: Undetermined. Asexual morph Hyphomycetous, helicosporous. Colonies on the substratum superficial, effuse, gregarious, white to pink. Mycelium partly immersed, hyaline to pale brown, septate, with masses of crowded, glistening conidia. Conidiophores 137–197 μm long, 2.5–5 μm wide ( = 170 × 4 μm, n = 30), macronematous, mononematous, erect, septate, sparsely branched, pale brown, rising directly on the substrate, hyaline to pale brown, smooth-walled. Conidiogenous cells 11–17 × 3–4 μm ( = 14 × 3.5 μm, n = 30), holoblastic, mono- to polyblastic, integrated, cylindrical, with lateral minute denticles (1–2 μm long, 1–1.5 μm wide). Conidia 14–21 μm in diam., 1.5–3 μm wide ( = 17 × 2 μm, n = 30), conidial filament 82–136 μm long, solitary, acropleurogenous, helicoid, coiled 21/2-33/4 times, becoming loosely coiled in water, rounded at tip, guttulate, indistinctly multi-septate, hyaline to yellowish, smooth-walled.
Culture characteristics: Conidia germinating on water agar and germ tubes produced from conidia within 12 h. Colonies growing on PDA, circular, with umbonate surface, edge entire, reaching 29 mm in 5 weeks at 25°C, pale brown to brown.
Material examined: CHIAN, Hainan Province, Wuzhishan City, Shuimanhe tropical rainforest scenic area in Wuzhishan, on decaying wood in a terrestrial habitat, 24 August 2021, Jian Ma, WZS54 (GZAAS 22-2009, holotype; HKAS 125863, isotype), ex-type living culture, GZCC 22-2009; Ibid., WZS69 (GZAAS 22-2027, paratype), living culture, GZCC 22-2027.
GenBank accession numbers: GZCC 22-2009: OP508734 (ITS), OP508774 (LSU), OP698074 (RPB2), and OP698085 (TEF1α); GZCC 22-2027: OP508735 (ITS), OP508775 (LSU), OP698075 (RPB2), and OP698086 (TEF1α).
Notes: The conidiophores and conidial features of Neohelicomyces hainanensis are morphologically similar to those of N. hyalosporus but it can be distinguished from N. hyalosporus by its shorter conidiophores (137–197 μm vs. 210–290 μm) (Lu et al., 2018b). Its colonies change from white to pink on a natural woody substrate; a feature that other species of the genus do not have. Phylogenetically, N. hainanensis shares a sister relationship to N. pallidus with high statistical support (97 MLBS/0.99 PP), and the phylogenetic analysis results support it as a distinct species (Figure 2).
Discussion
The difficulty in the taxonomic study of helicosporous hyphomycete species is that their morphological characteristics are very similar; it is difficult to distinguish them only by morphological comparison (Linder, 1929; Pirozynski, 1972; Goos, 1985, 1986, 1989; Zhao et al., 2007; Kuo and Goh, 2018; Lu et al., 2018a,b; Hsieh et al., 2021; Tian et al., 2022). Therefore, polygenic phylogenetic analysis is required to accurately identify them. However, previous studies have mainly focused on the description of morphological characteristics; most of them without obtaining strains and DNA molecular data (Linder, 1929; Pirozynski, 1972; Goos, 1985, 1986, 1989; Zhao et al., 2007). What makes things more complicated is that standards for species identification are not uniform, which creates confusion in this taxonomic system. Some helicosporous fungi have been transferred several times. For example, Moore (1957) treated Drepanospora pannosa as Helicosporium pannosum; Matsushima (1975) classified Drepanospora pannosa, Helicosporium linderi, Helicosporium nematosporum, and Helicosporium serpentinum under Helicosporium pannosum; Goos (1989) treated them as Drepanospora pannosum; Zhao et al. (2007) treated all of them and Helicosporium gigasporum as Helicosporium pannosum. The reason the authors reassessed the taxonomic status of these species is that there were some differences in the morphological characteristics of the conidiophores, conidiogenous cells, and conidia; the authors used different taxonomic principles to identify these species (Moore, 1957; Matsushima, 1975; Goos, 1989; Zhao et al., 2007). In our previous study, we paid attention to the confusion regarding the classification of helicosporous hyphomycete, analyzed the existing problems, and proposed ideas to solve the problems (Lu et al., 2018b). Lu et al. (2018b) provided several examples to show that the morphological characteristics of conidiophores, conidiogenous cells, and conidia, including their color and size, are very important influencing factors that cannot be ignored in distinguishing helicosporous fungi. The key to solve this taxonomic system problem is to obtain more species resources such as molecular data and morphological characteristics, for both newly collected specimens and published specimens with incomplete morphological features. Specimens observed in previously published literature that have molecular data but lack morphological characteristics, and are well preserved, can be borrowed for further morphological research.
In addition, different fungal species with similar morphologies produced distinctly characteristic secondary metabolites. For example, the stromata and ascospores of Annulohypoxylon urceolatum were morphologically similar to those in A. leptascum. However, they could be distinguished by their unique stromatal HPLC profiles, in which A. urceolatum produced the sole main metabolite viz. urceoline, while A. leptascum produced large quantities of truncatone A and C (Kuhnert et al., 2017). Annulohypoxylon yungensis was morphologically similar to A. truncatum, but the former produced BNT (1,1′-binaphthalene-4,4′-5,5′-tetrol), whereas the latter produced truncaquenone A and B in large quantities as well as trace truncatone A (Surup et al., 2016; Kuhnert et al., 2017). Kuhnert et al. (2017) provided a good example, using chemotaxonomy to evaluate the taxonomic systems of fungi with similar morphologies. This may be a new way to solve the problem of the taxonomy of helicosporous hyphomycetes by using evidence from chemotaxonomic data together with phylogenetic and morphological data.
In this study, we obtained 11 helicosporous fungal specimens and cultures and introduced four new species and three new records of helicosporous hyphomycetes based on morphological and phylogenetic evidence. We are also carrying out studies on the secondary metabolites of these fungi, and hope to find the characteristic compounds of each genus and solve the classification problem of helicosporous fungi with evidence from chemotaxonomic data in future.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
Y-ZL and JM conducted the experiments, analyzed the data, and wrote the article. J-CK planned the experiments. X-JX and Y-PX analyzed the data. JM and X-JX conducted the experiments. L-JZ and J-CK revised the article. Y-ZL and J-CK funded the experiments. All authors revised and agreed to the published version of the article.
Funding
This work was funded by the National Natural Science Foundation of China (NSFC 31900020, 32170019, and 31670027), the Science and Technology Foundation of Guizhou Province ([2020]1Y058), the China Post-doctoral Science Foundation Project (2020M683657XB), and the Guizhou Province high-level talent innovation and entrepreneurship merit funding project (No. 202104).
Acknowledgments
L-JZ would like to thank Mae Fah Luang University for granting a tuition scholarship for his Ph.D. studies.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Boonmee, S., Rossman, A. Y., Liu, J. K., Li, W. J., Dai, D. Q., Bhat, D. J., et al. (2014). Tubeufiales, ord. nov., integrating sexual and asexual generic names. Fungal Divers. 68, 239–298. doi: 10.1007/s13225-014-0304-7
Boonmee, S., Wanasinghe, D. N., Calabon, M. S., Huanraluek, N., Chandrasiri, S. K., Jones, G. E., et al. (2021). Fungal diversity notes 1387–1511: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 111, 1–335. doi: 10.1007/s13225-021-00489-3
Boonmee, S., Zhang, Y., Chomnunti, P., Chukeatirote, E., Tsui, C. K. M., Bahkali, A. H., et al. (2011). Revision of lignicolous Tubeufiaceae based on morphological reexamination and phylogenetic analysis. Fungal Divers. 51, 63–102. doi: 10.1007/s13225-011-0147-4
Brahamanage, R. S., Lu, Y. Z., Bhat, D. J., Wanasinghe, D. N., Yan, J. Y., Hyde, K. D., et al. (2017). Phylogenetic investigations on freshwater fungi in Tubeufiaceae (Tubeufiales) reveals the new genus Dictyospora and new species Chlamydotubeufia aquatica and Helicosporium flavum. Mycosphere 8, 917–933. doi: 10.5943/mycosphere/8/7/8
Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T. (2009). TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Doilom, M., Dissanayake, A. J., Wanasinghe, D. N., Boonmee, S., Liu, J. K., Bhat, D. J., et al. (2017). Microfungi on Tectona grandis (teak) in Northern Thailand. Fungal Divers. 82, 107–182. doi: 10.1007/s13225-016-0368-7
Dong, J. Y., Zhao, Z. X., Cai, L., Liu, S. Q., Zhang, H. R., Duan, M., et al. (2004). Nematicidal effect of freshwater fungal cultures against the pine-wood nematode, Bursaphelenchus xylophilus. Fungal Divers. 15, 125–135.
Goos, R. D. (1985). A review of the anamorph genus Helicomyces. Mycologia 77, 606–618. doi: 10.1080/00275514.1985.12025146
Goos, R. D. (1986). A review of the anamorph genus Helicoma. Mycologia 78, 744–761. doi: 10.1080/00275514.1986.12025318
Goos, R. D. (1989). On the anamorph genera Helicosporium and Drepanospora. Mycologia 81, 356–374. doi: 10.1080/00275514.1989.12025759
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic. Acids. Symp. Ser. 41, 95–98.
Hanada, T., Sato, T., Arioka, M., Uramoto, M., and Yamasaki, M. (1996). Purification and characterization of a 15 kDa Protein (p15) produced by Helicosporium that exhibits distinct effects on neurite outgrowth from cortical neurons and PC12 Cells. Biochem. Biophys. Res. Commun. 228, 209–215. doi: 10.1006/bbrc.1996.1641
Hsieh, S. Y., Goh, T. K., and Kuo, C. H. (2021). New species and records of Helicosporium sensu lato from Taiwan, with a reflection on current generic circumscription. Mycol. Prog. 20, 169–190. doi: 10.1007/s11557-020-01663-8
Hu, H., Guo, H., Li, E., Liu, X., Zhou, Y., and Che, Y. (2006). Decaspirones F-I, bioactive secondary metabolites from the saprophytic fungus Helicoma viridis. J. Nat. Prod. 69, 1672–1675. doi: 10.1021/np0603830
Hyde, K. D., Hongsanan, S., Jeewon, R., Bhat, D. J., McKenzie, E. H. C., Jones, E. B. G., et al. (2016). Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 80, 1–270. doi: 10.1007/s13225-016-0373-x
Itazaki, H., Nagashima, K., Sugita, K., Yoshida, H., Kawamura, Y., Yasuda, Y., et al. (1990). Solation and structural elucidation of new cyclotetrapeptides, trapoxins A and B, having detransformation activities as antitumor agents. J. Antibiot. 43, 1524–1532. doi: 10.7164/antibiotics.43.1524
Jayasiri, S. C., Hyde, K. D., Ariyawansa, H. A., Bhat, D. J., Buyck, B., Cai, L., et al. (2015). The Faces of Fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 74, 3–18. doi: 10.1007/s13225-015-0351-8
Jiao, P., Gloer, J. B., Campbell, J., and Shearer, C. A. (2006). Altenuene derivatives from an unidentified freshwater fungus in the family Tubeufiaceae. J. Nat. Prod. 69, 612–615. doi: 10.1021/np0504661
Jung, C. H., Lee, S. M., Kim, S. H., Kim, D. W., Choi, Y. W., and Joo, W. H. (2012). A novel Helicosporium isolate and its antimicrobial and cytotoxic pigment. J. Microbiol. Biotechnol. 22, 1214–1217. doi: 10.4014/jmb.1204.04063
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kuhnert, E., Esteban, B., Lambert, C., Hyde, K. D., Hladki, A. I., Romero, A. I., et al. (2017). Phylogenetic and chemotaxonomic resolution of the genus Annulohypoxylon (Xylariaceae) including four new species. Fungal Divers. 85, 1–43. doi: 10.1007/s13225-016-0377-6
Kuo, C. H., and Goh, T. K. (2018). Two new species of helicosporous hyphomycetes from Taiwan. Mycol. Prog. 17, 557–569. doi: 10.1007/s11557-018-1384-7
Lee, S. M., Kim, D. S., Lee, K. S., Lee, C. K., and Lee, D. W. (2013). Antibiotic Properties of Helicosporium sp. KCTC 0635BP to Rhizoctonia solani AG2-2 IV. Weed Turfgrass Sci. 2, 202–206. doi: 10.5660/WTS.2013.2.2.202
Linder, D. H. (1929). A monograph of the helicosporous fungi imperfecti. Ann. Mo. Bot. Gard. 16, 227–388. doi: 10.2307/2394038
Liu, N. G., Lu, Y. Z., Bhat, D. J., McKenzie, E. H., Lumyong, S., Jumpathong, J., et al. (2019). Kevinhydea brevistipitata gen. et sp. nov. and Helicoma hydei sp. nov.,(Tubeufiaceae) from decaying wood habitats. Mycol. Prog. 18, 671–682. doi: 10.1007/s11557-019-01480-8
Liu, Y. J., Whelen, S., and Hall, B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092
Lu, Y. Z., Boonmee, S., Bhat, D. J., Hyde, K. D., and Kang, J. C. (2017a). Helicosporium luteosporum sp. nov. and Acanthohelicospora aurea (Tubeufiaceae, Tubeufiales) from terrestrial habitats. Phytotaxa 319, 24–253. doi: 10.11646/phytotaxa.319.3.3
Lu, Y. Z., Boonmee, S., Dai, D. Q., Liu, J. K., Hyde, K. D., Bhat, D. J., et al. (2017b). Four new species of Tubeufia (Tubeufiaceae, Tubeufiales) from Thailand. Mycol. Prog. 16, 403–417. doi: 10.1007/s11557-017-1280-6
Lu, Y. Z., Boonmee, S., Liu, J. K., Hyde, K. D., McKenzie, E. H. C, Eungwanichayapant, P. D., et al. (2018a). Multi-gene phylogenetic analyses reveals Neohelicosporium gen. nov. and five new species of helicosporous hyphomycetes from aquatic habitats. Mycol. Prog. 17, 631–646. doi: 10.1007/s11557-017-1366-1
Lu, Y. Z., and Kang, J. C. (2020). Research progress on helicosporous hyphomycetes. J. Fungal Res. 18, 304–314. doi: 10.13341/j.jfr.2020.8012
Lu, Y. Z., Liu, J. K., Hyde, K. D., Jeewon, R., Kang, J. C., Fan, C., et al. (2018b). A taxonomic reassessment of Tubeufiales based on multi-locus phylogeny and morphology. Fungal Divers. 92, 131–344. doi: 10.1007/s13225-018-0411-y
Luo, Z. L., Bhat, D. J., Jeewon, R., Boonmee, S., Bao, D. F., Zhao, Y. C., et al. (2017). Molecular phylogeny and morphological characterization of asexual fungi (Tubeufiaceae) from freshwater habitats in Yunnan, China. Cryptogam. Mycol. 38, 27–53. doi: 10.7872/crym/v38.iss1.2017.27
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Moore, R. T. (1957). Index to the Helicosporae: addenda. Mycologia 49, 580–587. doi: 10.1080/00275514.1957.12024670
Ohtsu, Y., Sasamura, H., Shibata, T., Nakajima, H., Hino, M., and Fujii, T. (2003). The novel gluconeogenesis inhibitors FR225659 and related compounds that originate from Helicomyces sp. No. 19353 II. Biological profiles. J. Antibiot. 56, 689–693. doi: 10.7164/antibiotics.56.689
Phookamsak, R., Lu, Y. Z., Hyde, K. D., Jeewon, R., Li, J. F., Doilom, M., et al. (2017). Phylogenetic characterization of two novel Kamalomyces species in Tubeufiaceae (Tubeufiales). Mycol. Prog. 17, 647–660. doi: 10.1007/s11557-017-1365-2
Pirozynski, K. A. (1972). Microfungi of Tanzania. I. Miscellaneous fungi on oil palm. II. New hyphomycetes. Mycol. Pap. 129, 1–29.
Qian, S.Y., Zeng, X.B., Qian, Y.X., Lu, Y.Z., He, Z.J., and Kang, J.C. (2022). A saprophytic fungus Tubeufia rubra Produces novel rubracin D and E reversing multidrug resistance in cancer cells. Appl. Microbiol. Biot. (Submitted).
Rajeshkumar, K. C., and Sharma, R. (2013). Tamhinispora a new genus belongs to family Tubeufiaceae from the Western Ghats, India based on morphology and phylogenetic analysis. Mycosphere 4, 165–175. doi: 10.5943/mycosphere/4/2/2
Rambaut, A., and Drummond, A. (2008). FigTree: Tree Figure Drawing Tool, Version 1.2.2. Edinburgh: Institute of Evolutionary Biology; The University of Edinburgh.
Rehner, S. A., and Buckley, E. (2005). A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97, 84–98. doi: 10.3852/mycologia.97.1.84
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Senanayake, I. C., Rathnayaka, A. R., Marasinghe, D. S., Calabon, M. S., Gentekaki, E., Lee, H. B., et al. (2020). Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. Mycosphere 11, 2678–2754. doi: 10.5943/mycosphere/11/1/20
Shimodaira, H., and Hasegawa, M. (2001). CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17, 1246–1247. doi: 10.1093/bioinformatics/17.12.1246
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Surup, F., Wiebach, V., Kuhnert, E., and Stadler, M. (2016). Truncaquinones A and B, asterriquinones from Annulohypoxylon truncatum. Tetrahedron Lett. 57, 2183–2185. doi: 10.1016/j.tetlet.2016.04.014
Tian, X., Karunarathna, S. C., Xu, R., Lu, Y., Suwannarach, N., Mapook, A., et al. (2022). Three new species, two new records and four new collections of Tubeufiaceae from Thailand and China. J. Fungi 8, 206. doi: 10.3390/jof8020206
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
White, T. J., Bruns, T., Lee, S., and Taylor, J. W. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols Guide Methods Appl. 18, 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Yoshimura, S., Zenkoh, T., Ohtsu, Y., Kanasaki, R., Shigematsu, N., Takase, S., et al. (2003). Isolation, structure determination and biological study of novel gluconeogenesis inhibitors, FR225659 family. Sympos. Chem. Nat. Prod. 45, 281–286.
Zeng, X., Qian, S., Lu, Y., Li, Y., Chen, L., Qian, Y., et al. (2022). A novel Nitrogen-containing Glyceride from fungal saprobe Tubeufia rubra reverses MDR of tumor cell lines to Doxorubicin. Rec. Nat. Prod. 16, 622–632. doi: 10.25135/rnp.320.2201.2334
Zenkoh, T., Ohtu, Y., Yoshimura, S., Shigematsu, N., Takase, S., and Hino, M. (2003). The novel gluconeogenesis inhibitors FR225659 and FR225656 from Helicomyces sp. No. 19353 III. Structure determination. J. Antibiot. 56, 694–699. doi: 10.7164/antibiotics.56.694
Zhao, G. Z., Liu, X., and Wu, W. (2007). Helicosporous hyphomycetes from China. Fungal Divers. 26, 313–524.
Keywords: freshwater fungi, taxonomy, Tubeufiales, woody substrates, saprophytic fungi
Citation: Lu Y-Z, Ma J, Xiao X-J, Zhang L-J, Xiao Y-P and Kang J-C (2022) Four new species and three new records of helicosporous hyphomycetes from China and their multi-gene phylogenies. Front. Microbiol. 13:1053849. doi: 10.3389/fmicb.2022.1053849
Received: 26 September 2022; Accepted: 31 October 2022;
Published: 25 November 2022.
Edited by:
Kezia Goldmann, Helmholtz Centre for Environmental Research (HZ), GermanyReviewed by:
Mingkwan Doilom, Zhongkai University of Agriculture and Engineering, ChinaNoelia Betiana Nuñez Otaño, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina
Danushka Sandaruwan Tennakoon, Chiang Mai University, Thailand
Copyright © 2022 Lu, Ma, Xiao, Zhang, Xiao and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ji-Chuan Kang, amNrYW5nQGd6dS5lZHUuY24=
 Yong-Zhong Lu
Yong-Zhong Lu Jian Ma2,3,4
Jian Ma2,3,4 Yuan-Pin Xiao
Yuan-Pin Xiao Ji-Chuan Kang
Ji-Chuan Kang