- 1College of Horticulture, Shenyang Agricultural University, Shenyang, China
- 2Institute of Grassland Research, Chinese Academy of Agricultural Sciences, Hohhot, China
- 3College of Grassland Science and Technology, China Agricultural University, Beijing, China
- 4Inner Mongolia Engineering Research Center of Development and Utilization of Microbial Resources in Silage, Inner Mongolia Academy of Agriculture and Animal Husbandry Science, Hohhot, China
- 5Inner Mongolia Key Laboratory of Microbial Ecology of Silage, Inner Mongolia Academy of Agriculture and Animal Husbandry Science, Hohhot, China
This study aims to investigate the effects of adding Lactobacillus buchneri (LB), Lactobacillus brevis (LBR) and Bacillus subtilis (BS) on the fermentation quality, nitrate degradation and bacterial community of sorghum-sudangrass silage. The results showed that the addition of LB significantly increased the pH and acetic acid content (p < 0.05), but high-quality silage was obtained. The addition of LBR and BS improved the fermentation quality of sorghum-sudangrass silage. The use of additives reduced the nitrate content in sorghum-sudangrass silage. The LB group increased the release of N2O at 3–7 days of ensiling (p < 0.05), and LBR and BS increased the release of N2O at 1–40 days of ensiling (p < 0.05). On the first day of ensiling, all silages were dominated by Weisslla, over 3 days of ensiling all silages were dominated by Lactobacillus. Acinetobacter, Serratia, Aquabacterium, and unclassified_f_enterobacteriaceae showed significant negative correlations with nitrate degradation during sorghum-sudangrass ensiling (p < 0.05). The BS and LBR groups increased the metabolic abundance of denitrification, dissimilatory nitrate reduction, and assimilatory nitrate reduction (p < 0.05). Overall, the additive ensures the fermentation quality of sorghum-sudangrass silage and promotes the degradation of nitrate by altering the bacterial community.
Introduction
With the development of the livestock industry and increasing consumer demand for meat and dairy products, there is a significant demand for the production of silage, and statistics show that China produces greater than 280 million tons of silage annually, especially in some arid areas where there is an increasing demand for high-yielding and water-saving fodder crops (Fijalkowska et al., 2020). Due to its excellent characteristics of high drought resistance, high water use efficiency, and high biological yield, sorghum-sudangrass (Sorghum bicolor×Sorghum sudanense) has become one of the high-quality modulated silage raw materials in arid areas (McCary et al., 2020). However, sorghum-sudangrass easily accumulates nitrate forage (Holman et al., 2019). Stress conditions such as drought and low temperature during its growth, and excessive use of nitrogen fertilizer in pursuit of high yield can affect the level of nitrate accumulation in sorghum-sudangrass (Crawford et al., 1961; Harada et al., 2000; Holman et al., 2019). Nitrate in silage can threaten animal health, the safety of milk, and other animal foods safety and even cause significant economic losses in severe cases (Vermunt and Visser, 2011; Schrenk et al., 2020). The high nitrate content in sorghum-sudangrass silage limits its use as animal feed. An urgent issue that needs to be resolved: is the reduction of nitrate content in sorghum-sudangrass silage.
Ensiling can reduce nitrate in sorghum-sudangrass to some extent content (Spoelstra, 1987; Bai C. et al., 2022). During the ensiling process, nitrate is degraded to produce the intermediate product nitrite, and the end products nitric oxide, ammonia, and nitrous oxide (Spoelstra, 1985). However, the nitrate content in silage may still threaten the health of ruminants by exceeding the safe limit of nitrate in forage (Spoelstra, 1983; Bai C. et al., 2022). In response, researchers have tried to use silage additives to regulate the nitrate content in silage. Takayoshi et al. (1980) used glucose as a silage additive to regulate the nitrate content in Italian ryegrass silage. Li et al. (1992) investigated the effect of ammonium hydroxide and calcium carbonate on the degradation of nitrate in corn silage. In the above studies, nitrates were degraded during ensiling after using chemical additives for silage preparation, but the quality of the silage was reduced. Today, producers often use microbial inoculants to regulate the silage fermentation process (Muck et al., 2018; Romero et al., 2018). Tian et al. (2021) demonstrated that Bacillus subtilis promoted fermentation quality and nutrient composition by altering bacterial communities and metabolic profiles. Xie et al. (2021) showed that the use of lactic acid bacteria to prepare sorghum silage improved fermentation quality. The ability of lactic acid bacteria and Bacillus subtilis to degrade nitrate has been demonstrated in studies on kimchi, whereas the use of those bacteria has rarely reported in research on silage fermentation (Sun et al., 2016; Zhang et al., 2022). Based on this, we hypothesized that inoculation lactic acid bacteria and Bacillus subtilis prior to sorghum-sudangrass silage would both enhance fermentation quality and promote nitrate degradation. However, experimental evidence is lacking to support this hypothesis.
Ensiling is a complex microbial fermentation process in which microorganisms interact with each other and with their environment during fermentation (Xu et al., 2019; Ding et al., 2020). Previous experimental cultivation studies have shown that microorganisms, such as Enterobacteriaceae and Lactobacillus, are involved in nitrate degradation (Spoelstra, 1985). Cultivation experiments are commonly applied in ensiling experiments, but microbial communities often contain a large number of bacteria that cannot be cultivated under laboratory conditions (Eikmeyer et al., 2013). Therefore, the complex microbial communities, microbial interactions, and their functions during nitrate degradation in silages have not been thoroughly studied in the previous experiments. In recent years, changes in silage fermentation parameters and their bacterial community dynamics during silage have been gradually analysed, with the application of molecular techniques in the field of silage (Xu Z. et al., 2017; You et al., 2021; Bai J. et al., 2022). This methodology provides the possibility to deeply explain the nitrate degradation during silage. For example, Bai C. et al. (2022) used high-throughput sequencing to illustrate the effects of Panobacter spp., Pseudomonas spp., and Enterobacter spp. on the nitrate content of silage sorghum-sudangrass. However, there are few studies on the effects of the use of Lactobacillus and Bacillus subtilis as additives for sorghum-sudangrass on nitrate content, nitrate degradation, and the correlation between nitrate content and silage fermentation quality and microbial community changes. Therefore, in this study, Lactobacillus buchneri, Lactobacillus brevis, and Bacillus subtilis were used as additives with the aim of investigating their role in silage with respect to fermentation quality and nitrate degradation in sorghum-sudangrass.
Materials and methods
Silage preparation
Sorghum-sudangrass (Jicao No.6) was grown at the experimental farm of Shenyang Agricultural University (123° 25′ E, 41° 46′ N) and harvested at the heading stage, leaving a stubble height of 15–20 cm. After harvest, the sorghum-sudangrass was chopped into 1–2 cm lengths using a grass shredding machine (Donghong No. 1, Donghong Mechanical Equipment Co., Ltd., China). Silage were treated with the following: sterile water control (CK), 2.0 × 105 colony-forming units (CFU)/g of Lactobacillus buchneri (LB, Gansu Pro-Bicon Biotech Co., Ltd., China), 1.0 × 105 colony-forming units (CFU)/g of Lactobacillus brevis (LBR, Laboratory of China Agricultural University) and 2.0 × 106 colony-forming units (CFU)/g of Bacillus subtilis (BS, Laboratory of Shenyang Agricultural University). The additives were dissolved in 5 ml of sterile water, and then each 5 ml of bacterial solution was sprayed onto 1 kg of fresh matter (FM). An equal amount of sterile water was added to the control group. After thoroughly mixing, 230 g of treated material was placed in anaerobic jars (300 ml; Saipu Instrument Technology Co., Ltd., Zhenjiang, China). A total of 60 jars (four treatments × five ensiling durations × three replicates) were prepared and stored at room temperature (25–30°C). Each treated silage was sampled after storage for 1,3, 7, 15, and 40 days for determining the fermentation quality, nutrient composition, microbial counts, and microbial community. In addition, the fermentation gas was collected at each sampling time point in a 500 ml aluminum foil gas sampling bag (Dalian Delin Co., Ltd., China) for N2O determination.
Analyses of silage fermentation quality and nutrient composition
To determine the fermentation traits of forage, a sample (10 g) of silage was mixed with 90 g of deionized water and kept in a refrigerator at 4°C for 24 h. The liquid extract was filtered through four layers of cheesecloth and filtered paper. Then, the filtrates were used to measure pH, ammonia nitrogen (NH3-N), and organic acids. The pH value of the extracts was determined using a pH meter (PB-10, Sartorius Group, Göttingen, Germany). Organic acids and nitrates were determined by high-performance liquid chromatography (Bai C. et al., 2022). The concentrations of N2O in the gas samples were analysed by gas chromatograph (Agilent 7890A, Agilent Technologies Limited Co., United States) within a week (Xu T. W. et al., 2017). A representative silage sample of 10 g was obtained, added to 90 ml of distilled water, shaken at 180 rpm for 30 min, and then the solution was diluted in a gradient. Microbial counts were analysed using the plate count method on Man Rogosa Sharpe agar, Violet Red Bile agar, and Rose Bengal agar (Beijing Aoboxing Bio-tech Co. Ltd., Beijing, China) as reported by Cai et al. (1999).
The dry matter (DM) of the fresh sorghum-sudangrass and silage was determined by oven drying at 65°C for 48 h. Colorimetry after reaction with anthrone reagent was used to determine the water-soluble carbohydrate (WSC) content (Murphy, 1958). The crude protein (CP = total N × 6.25) was determined using a Kjeldahl apparatus (Kjeltec 8,400; FOSS Co. Ltd., Hillerød, Denmark), according to previously published work (AOAC, 1990). The neutral detergent fiber (using heat-stable α-amylase, NDF) and acid detergent fiber (ADF) were measured using an Ankom 2000 fiber analyser (Ankom, Macedon, NY, United States), according to Van Soest’s procedures (Van Soest et al., 1991).
Bacterial community analysis
A 10 g fresh sample was placed into 90 ml of sterile distilled water. The mixture was placed in a low-temperature oscillator (THZ-98C, Shanghai Yiheng Scientific Instrument Co., Ltd., Shanghai, China) at 4°C, 180 rpm/min for 30 min and then subject to filtration through 4 layers of sterile gauze. The filtrate was centrifuged in a cryogenic centrifuge (ST 16R, Thermo Fisher Scientific, Inc., Waltham, United States) at 8000 rpm/min at 4°C for 15 min to enrich the sediment. Sediments were used for high-throughput sequencing (Wang et al., 2018).
Bacterial community genomic DNA was extracted from silage samples using the FastDNA® SPIN Kit and the FastPrep® Instrument (MP Biomedicals, Santa Ana, CA) according to the manufacturer’s instructions. The hypervariable region V3-V4 of the bacterial 16S rRNA gene was amplified with the primer pairs 341F (5′- CCTAYGGGRBGCASCAG-3′) and 806R(5’-GGACTACNNGGGTATCTAAT-3′) using an ABI GeneAmp® 9,700 PCR thermocycler (ABI, CA, United States). The PCR product was extracted from a 2% agarose gel, purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States) and quantified using a Quantus™ Fluorometer (Promega, United States). Purified amplicons were pooled in equimolar amounts and paired-end sequenced on an Illumina MiSeq PE300 platform (Illumina, San Diego, United States) by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). The raw 16S rRNA gene sequencing reads were demultiplexed, quality-filtered by fastp version 0.20.0 and merged by FLASH version 1.2.7 based on the following criteria. Operational taxonomic units (OTUs) with a 97% similarity cut-off were clustered using UPARSE version 7.1, and chimeric sequences were identified and removed. The taxonomy of each OTU representative sequence was analysed against the 16S rRNA database using a confidence threshold of 0.7 by RDP Classifier version 2.2. The microbial diversity within an individual sample was assessed using the following alpha diversity indices: Shannon diversity index, Simpson diversity index, Chao richness estimator, Ace richness estimator and Coverage. Beta diversity was analysed to assess the structural variation of the microbiota. Subsequently, principal component analysis (PCoA) was undertaken (Vazquez-Baeza et al., 2013). The heatmap function of R software2 and genus information for the silage were used to generate a heatmap. The data were analysed using the free online majorbio cloud platform. The raw sequence data have been uploaded to the sequence read archive (SRA) of the NCBI database (the accession number PRJNA882255).
Statistical analyses
Factorial analysis of variance was performed to investigate the effects of additives, ensiling duration, and their interactions on the fermentation quality, microbial counts, bacterial community indices, nitrate, and N2O content of silage in the General Line Model of SPSS (SPSS 22.0 program, SPSS Inc., Chicago, Illinois, United States). Significant differences were compared using Tukey multiple range tests, and p < 0.05 indicated statistical significance.
Results
Characteristics of fresh sorghum-sudangrass
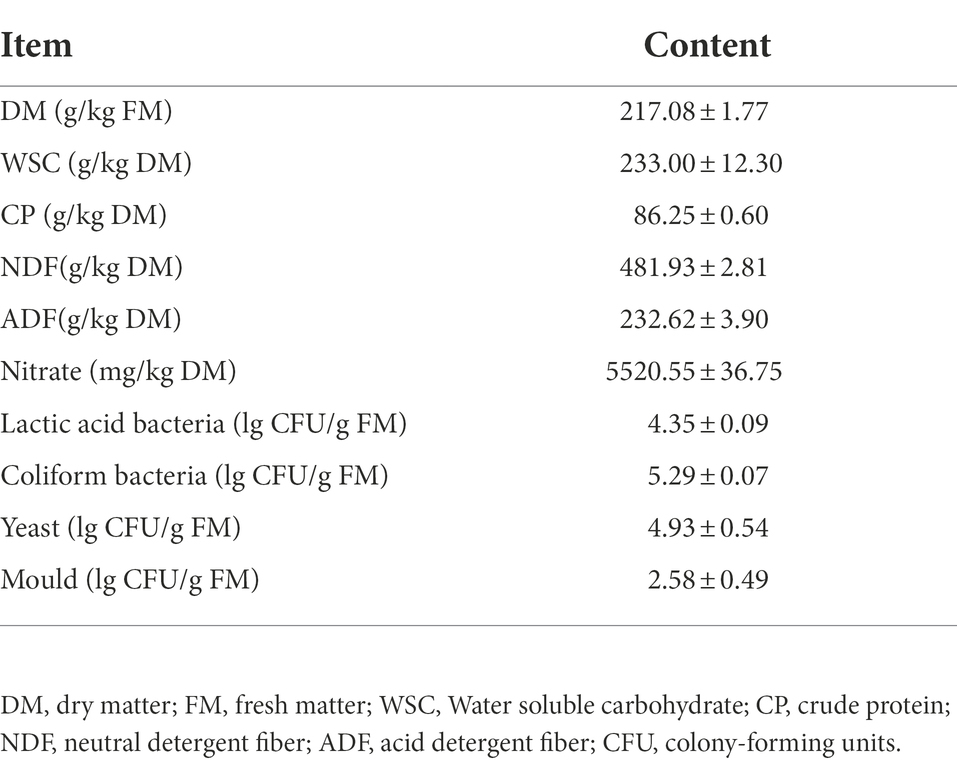
The chemical parameters and microbial counts of fresh sorghum-sudangrass are shown in Table 1. The concentrations of WSC, CP, NDF, and ADF in sorghum-sudangrass were 233.00, 86.25, 481.93, and 232.62 g/kg DM, respectively. The nitrate content was 5520.55 mg/kg DM. The counts of undesirable microbes containing coliform bacteria, yeast, and mould were 5.29, 4.93, and 2.58 lg CFU/g FM, respectively, whereas the lactic acid bacteria count was 4.35 lg CFU/g FM.
Fermentation quality and nutrient composition of silage
Additives significantly affected pH, lactic acid (LA), and acetic acid (AA) concentrations and lactic acid bacteria counts in sorghum-sudangrass silage (p < 0.05; Table 2). The pH of the LB group was significantly lower than that of CK at 3 days of ensiling; however, it was significantly higher than that of CK at 40 days of ensiling (p < 0.05). The AA content of the LB group increased rapidly during 15–40 days of ensiling and was significantly higher than that of CK at 40 days of ensiling (p < 0.05). The LBR group had a significantly lower pH than CK after 3 days of ensiling (p < 0.05). Compared to CK, the BS group had lower pH and higher LA content on days 3 and days 40 of ensiling (p < 0.05). The AA content of the LBR and BS group was significantly lower than that of CK at 40 days of ensiling (p < 0.05). Butyric acid was not detected during the fermentation process. After 40 days of ensiling, the NH3-N content of all silages was in the range of 4.88–5.31. The number of lactic acid bacteria reached the highest in all silages at 7 days (p < 0.05). At 40 days, the number of lactic acid bacteria in the LBR and BS groups was significantly lower than that of CK, and the number of lactic acid bacteria in the LB group was significantly higher than that in CK (p < 0.05). On the first day of ensiling, the numbers of coliform bacteria and yeast were significantly higher in the LBR and BS groups compared with CK (p < 0.05), and the number of coliform bacteria and yeast in the LB group was similar to that in CK (p > 0.05). However, as the number of ensiling days increased, coliform bacteria and yeast, which are harmful to silage, were not detected in all silages.
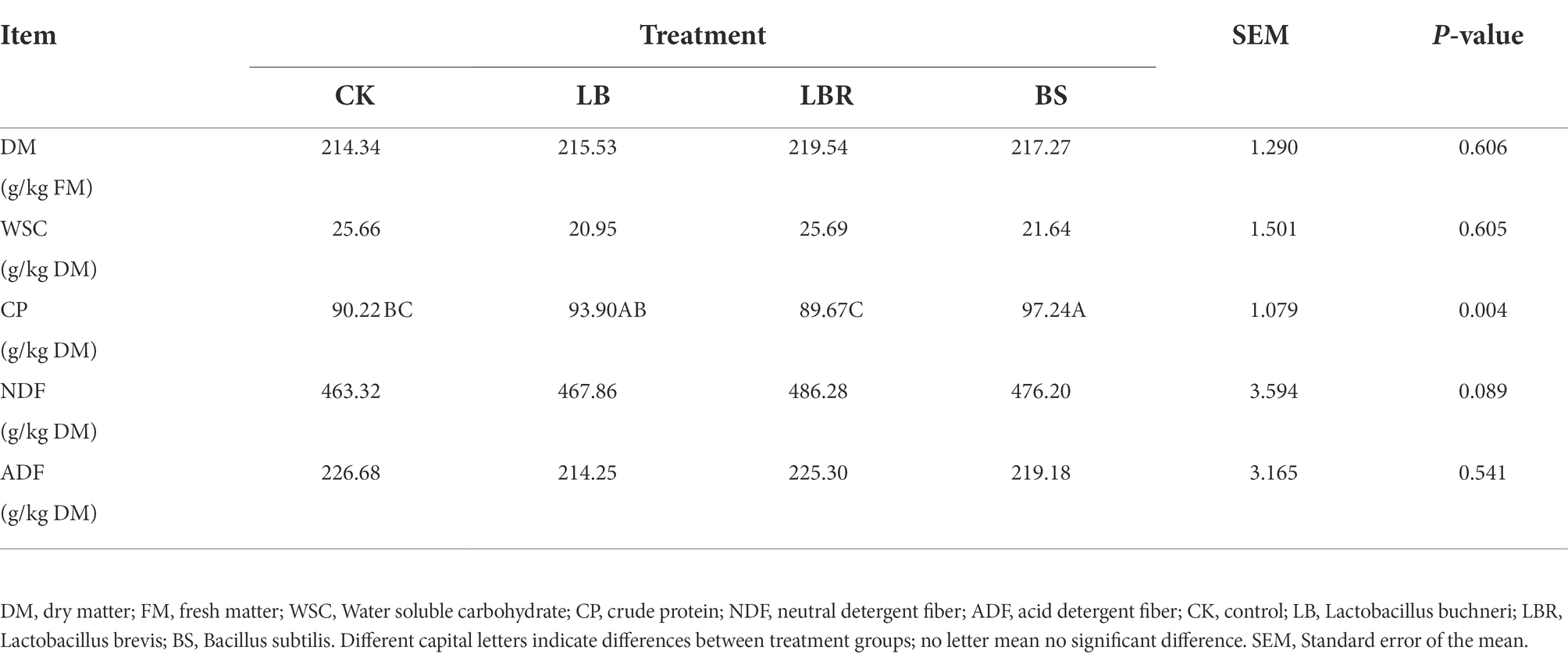
The nutrient composition of sorghum-sudangrass at 40 days of silage is listed in Table 3. The CP content of the BS group was significantly higher than that of CK after 40 days of ensiling (p < 0.05). Different additives did not affect the DM, WSC, NDF, and ADF contents of sorghum-sudangrass silage.
Bacterial community in silage
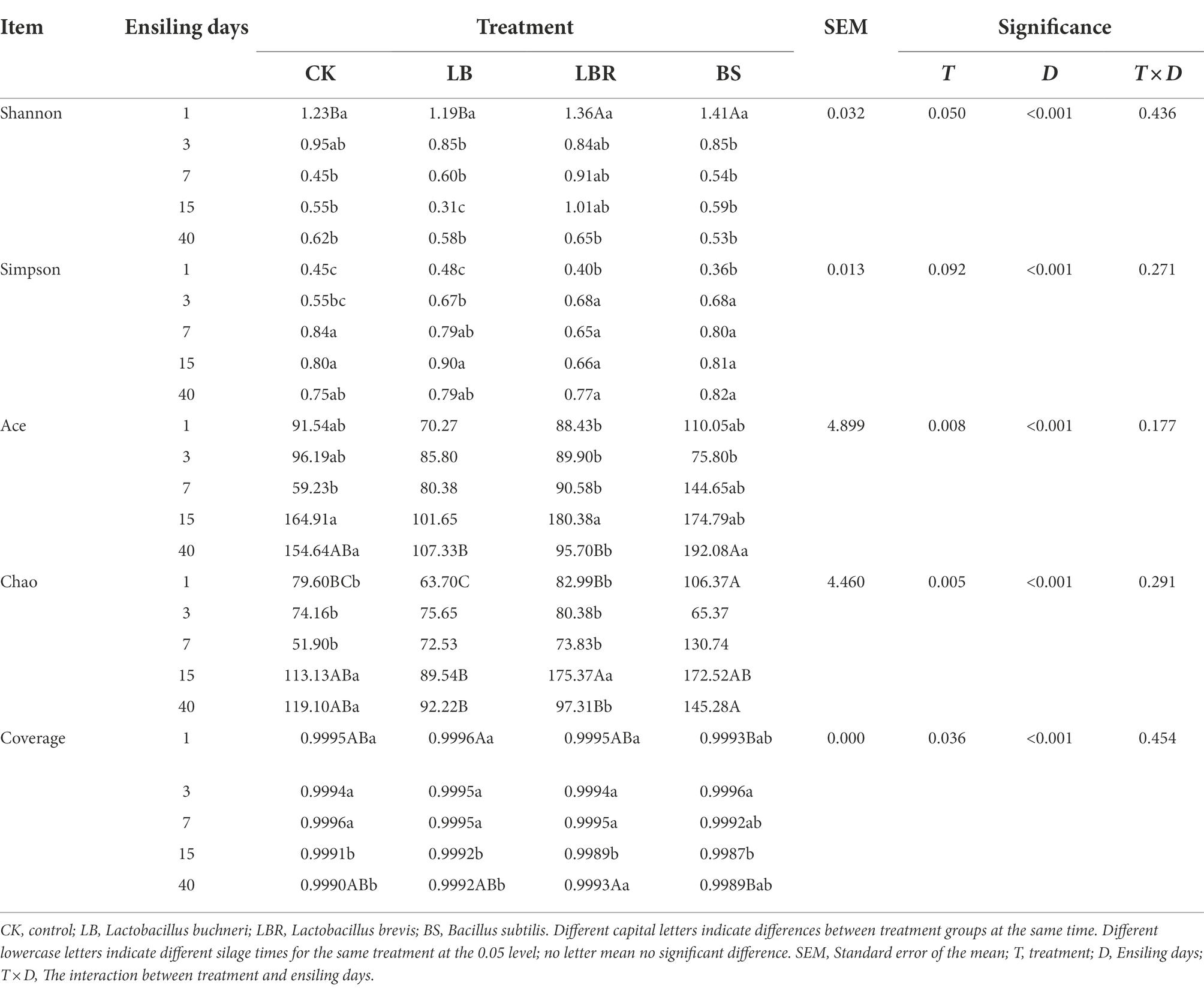
The bacterial alpha diversity of sorghum-sudangrass silage is shown in Table 4. The Coverage values of all treatment groups were greater than 0.99. Days of ensiling significantly affected the Shannon, Simpson, Ace, and Chao indices (p < 0.05), and additives had significant effects on the Ace and Chao values of sorghum-sudan grass silage (p < 0.05). The addition of LB had no significant effect on α-diversity during the ensiling process. The Shannon index of the LBR and BS groups was higher than that of the CK group on the first day of fermentation (p < 0.05). No significant differences in the Shannon, Simpson, Ace, and Chao indices of silages with additives and the CK were noted after 3 days of ensiling. As shown in Figure 1, principal component 1 (PCoA 1) and component 2 (PCoA 2) could explain 52.77 and 37.09% of the total variance, respectively. A clear separation of bacterial communities of sorghum-sudangrass silage between day 1 of ensiling and day 40 of ensiling could be observed. At 40 days of ensiling, the LBR and BS groups were significantly separated from CK.
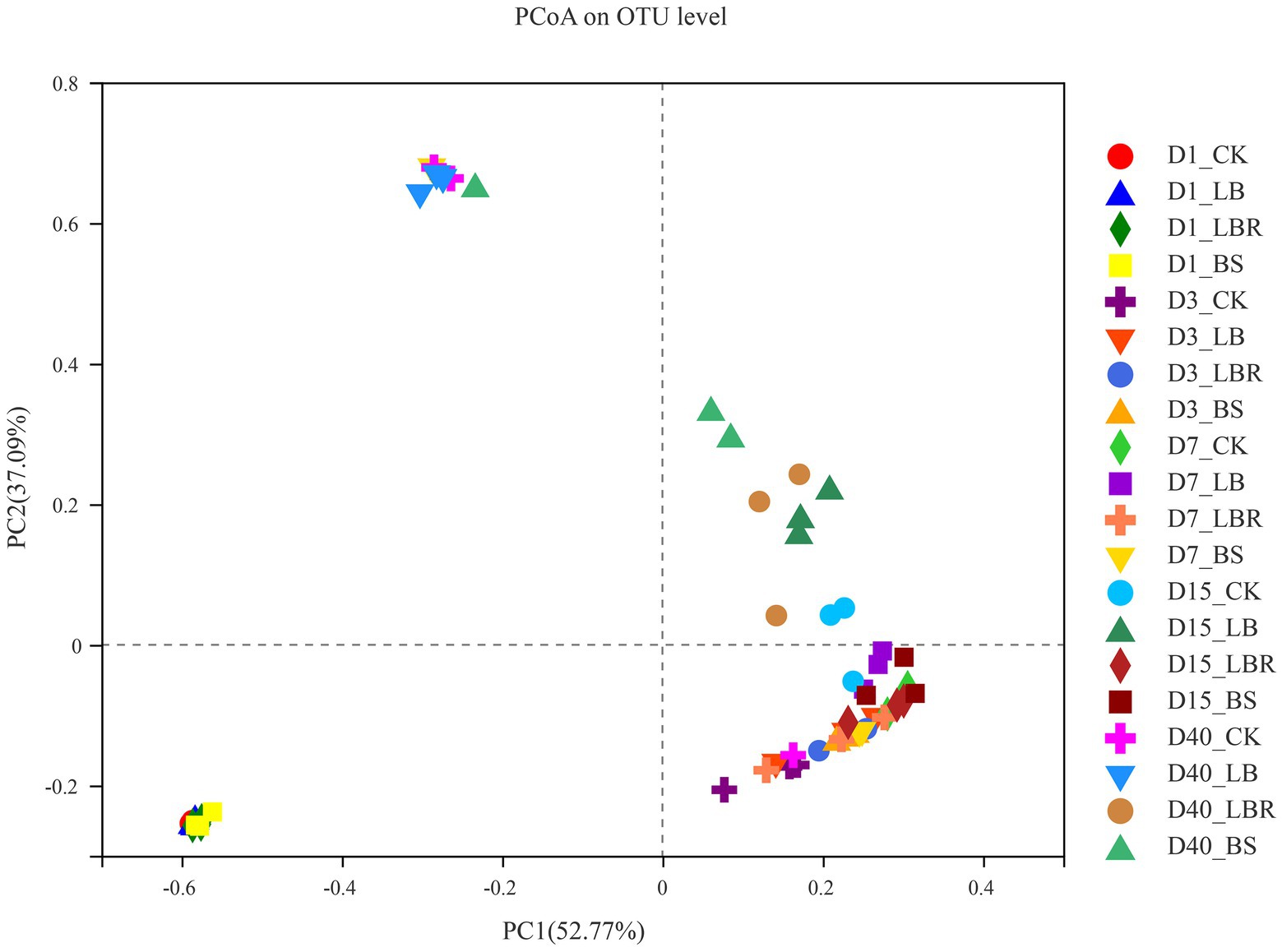
Figure 1. Principal component analysis (PCoA) of bacterial communities in sorghum-sudangrass silage after ensiling for 1, 3, 7, 15, and 40 days (CK, control; LB, Lactobacillus buchneri; LBR, Lactobacillus brevis; BS, Bacillus subtilis).
At the phylum level, the bacterial community in fresh sorghum-sudangrass was dominated by Proteobacteria, Firmicutes, Bacteroidota, and Actinobacteriota (Figure 2A). The dominant phylum in the fresh sorghum-sudangrass was proteobacteria (70.31%). After fermentation, the dominant bacteria changed to Firmicutes (92.02%). At the genus level, the dominant genus in fresh sorghum-sudangrass was Pantoea and the relative abundance of Pantoea decreased sharply after fermentation (< 5%; Figure 2B). The abundance of Weissella in all silages increased rapidly (> 60%) on the first day of ensiling, and the abundance of Lactobacillus in all silages increased rapidly (> 70%) on the third day of ensiling and remained high after 3 days of ensiling. Acinetobacter abundance was increased in the LB group compared with CK during 3–7 days of ensiling, and Serratia abundance was significantly increased in the LB group compared with CK during 7–15 days of ensiling (p < 0.05; Supplementary Figure S1). The LBR group exhibited increased Klebsiella abundance compared with CK on the first day of ensiling, significantly increased Acinetobacter abundance compared with CK during 3–7 days of ensiling, and significantly increased Serratia abundance compared with CK during 7–15 of ensiling (p < 0.05; Supplementary Figure S1). The BS group exhibited increased klebsiella than CK on the first day of ensiling (p < 0.05), significantly increased Acinetobater compared with CK during 3–7 days of ensiling (p < 0.05), and increased Serratia abundance compared with CK at 15 days of ensiling. Aquabacterium, Pelomonas, Brevundimonas, and Ralstonia levels were significantly increased in both the LBR and BS groups compared with CK at 40 days of ensiling (p < 0.05; Supplementary Figure S1).
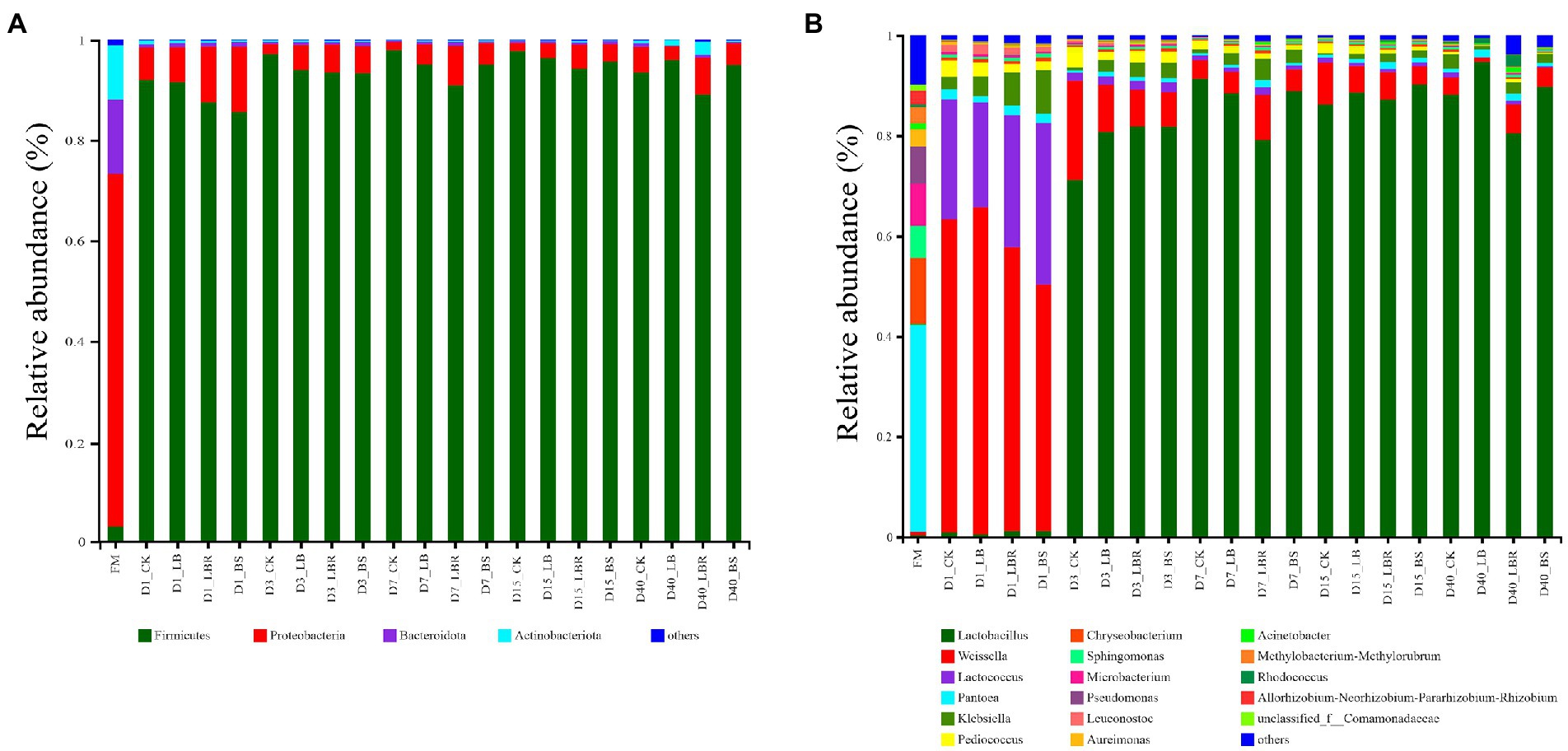
Figure 2. Relative abundance of bacterial communities at the phylum (A) and genus levels (B) in sorghum-sudangrass silage after ensiling for 1, 3, 7, 15, and 40 days (FM, fresh matter; CK, control; LB, Lactobacillus buchneri; LBR, Lactobacillus brevis; BS, Bacillus subtilis).
The linear discriminant analysis effect size (LefSe) was applied to explore the relative richness (p < 0.05, LDA > 3.0) of silages (Figure 3). On the first day of ensiling, Weissella and Leuconostoc represent differential bacteria in the LB group. In the LBR group, the differential bacteria included Roseomonas, Serratia, and Pseudomonas. In the BS group, the differential bacteria were included Lactococcus, Klebsiella, Eschcrichia-Shigella, Chryseobacterium, Sphingomonas, Stenotrophomonas, and Enterococcus. At 40 days of ensiling, the differential bacteria in the LB group was Lactobacillus; the differential bacteria in the LBR group were Acinetobacter, Bacillus, Brevundimonas, and Paenibacillus; and the differential bacteria in BS was Aquabacterium.
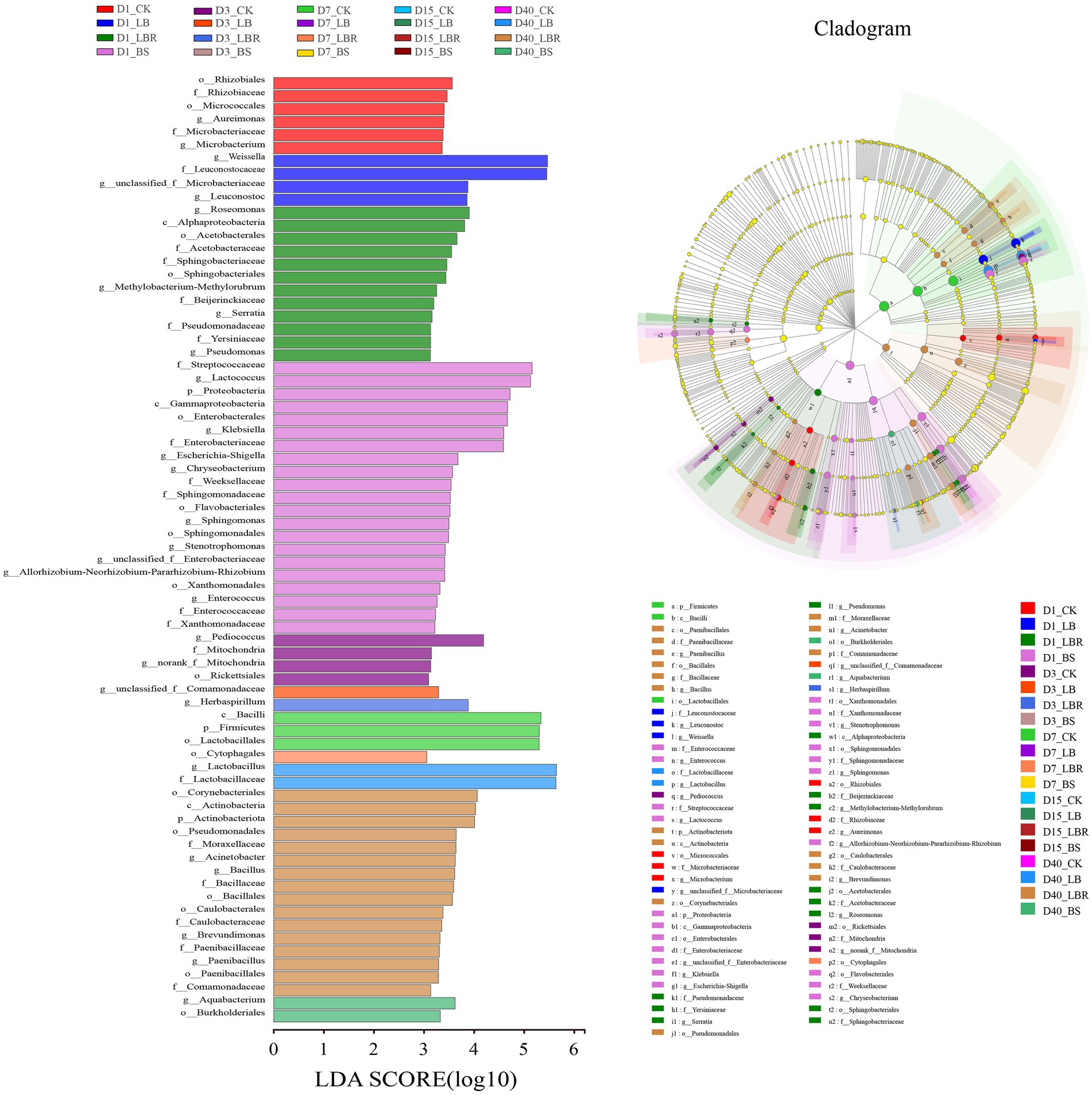
Figure 3. Comparison of microbial variations in sorghum-sudangrass after ensiling for 1, 3, 7, 15, and 40 days using the LEfSe online tool (CK, control; LB, Lactobacillus buchneri; LBR, Lactobacillus brevis; BS, Bacillus subtilis).
The relevance between fermentation quality and bacterial communities was assayed and is presented in Figure 4. The pH was positively associated with Weissella, Lactococcus, Kliebslella, Pediococcus, Chryseobacterium, Leuconostoc Aurelmonas, and Microbacterium (p < 0.05), and negatively related to Lactobacillus and Acinetobacter (p < 0.05). The LA content was positively correlated with Lactobacillus and Acinetobacter, and negatively correlated with Weissella Lactococcus, Kliebslella, Pediococcus, Chryseobacterium, Leuconostoc, Aurelmonas, and Microbacterium. The AA content was positively correlated with Lactobacillus (p < 0.05).
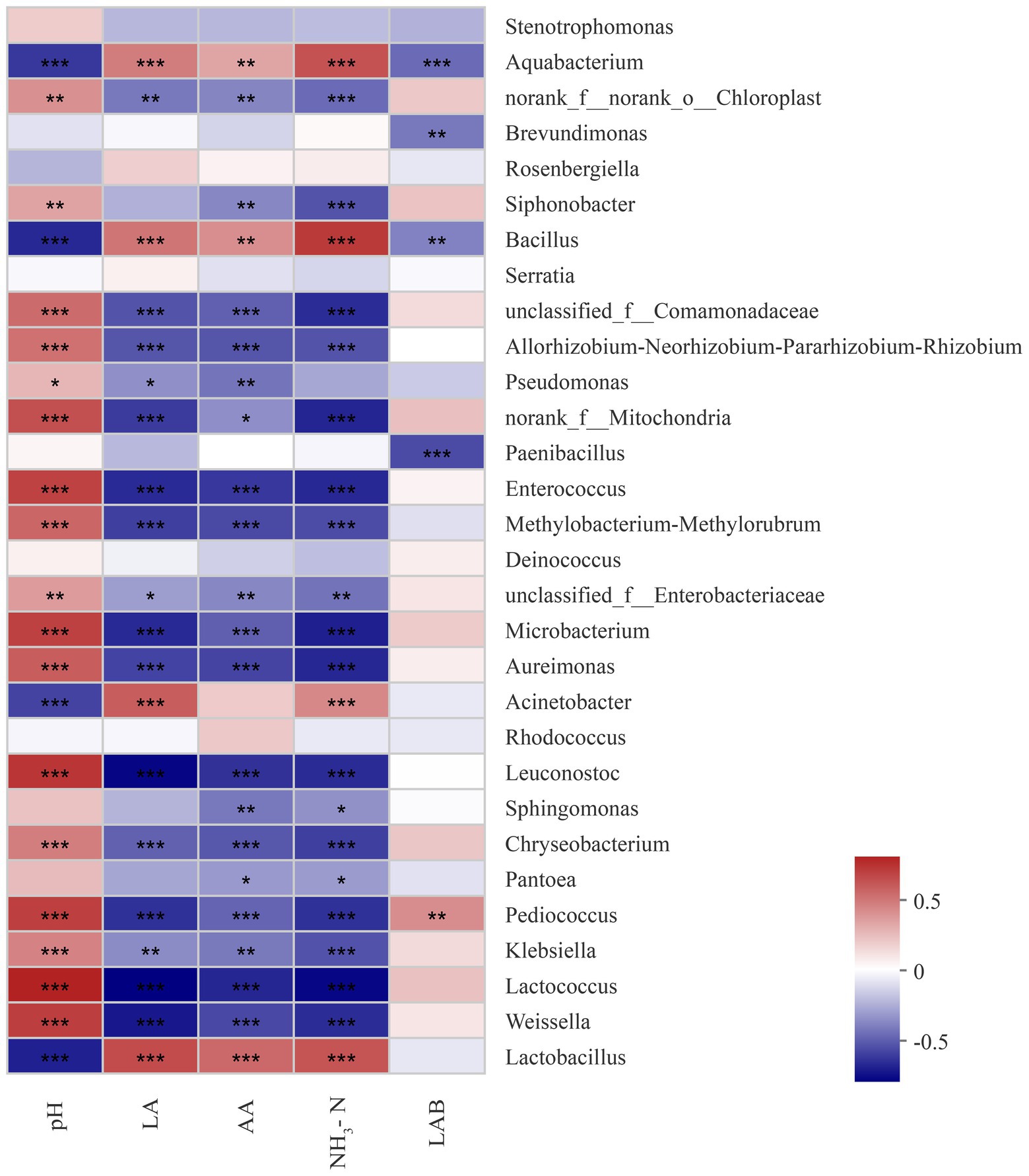
Figure 4. Correlation of the bacterial community and silage fermentation (LA, Lactic acid; AA, Acetic acid; NH3-N, ammonia nitrogen; LAB, Lactic acid bacteria; p-values are shown as *0.01 < P ≤ 0.05, **0.001 < p ≤ 0.01, ***p ≤ 0.001).
Nitrate, N2O and microbial function prediction of silage
The content of nitrate in sorghum-sudangrass was significantly influenced by additives and ensiling days (p < 0.05; Figure 5A). At 40 days, the nitrate content in the silages with additives was significantly lower than that in the CK group (p < 0.05). Additives and days of ensiling had a significant effect on the N2O content in sorghum-sudangrass silage (p < 0.05; Figures 5B). The LBR and BS groups had significantly higher N2O content than CK during ensiling (p < 0.05). The LB group had a significantly higher N2O content than CK from day 3 to day 7 of ensiling; however, the N2O content was not significantly different from CK after 15 days of ensiling (p < 0.05). The Nitrate content was positively correlated with Pediococcus and norank_f_mltochondria (p < 0.05), and negatively correlated with Acinetobacter and unclassified_f_enterobacteriaceae (p < 0.05; Figure 6). The N2O content was positively correlated with Acinetobacter and unclassified_f_enterobacteriaceae (p < 0.05), and negatively correlated with Pediococcus and norank_f_mltochondria (p < 0.05; Figure 6).
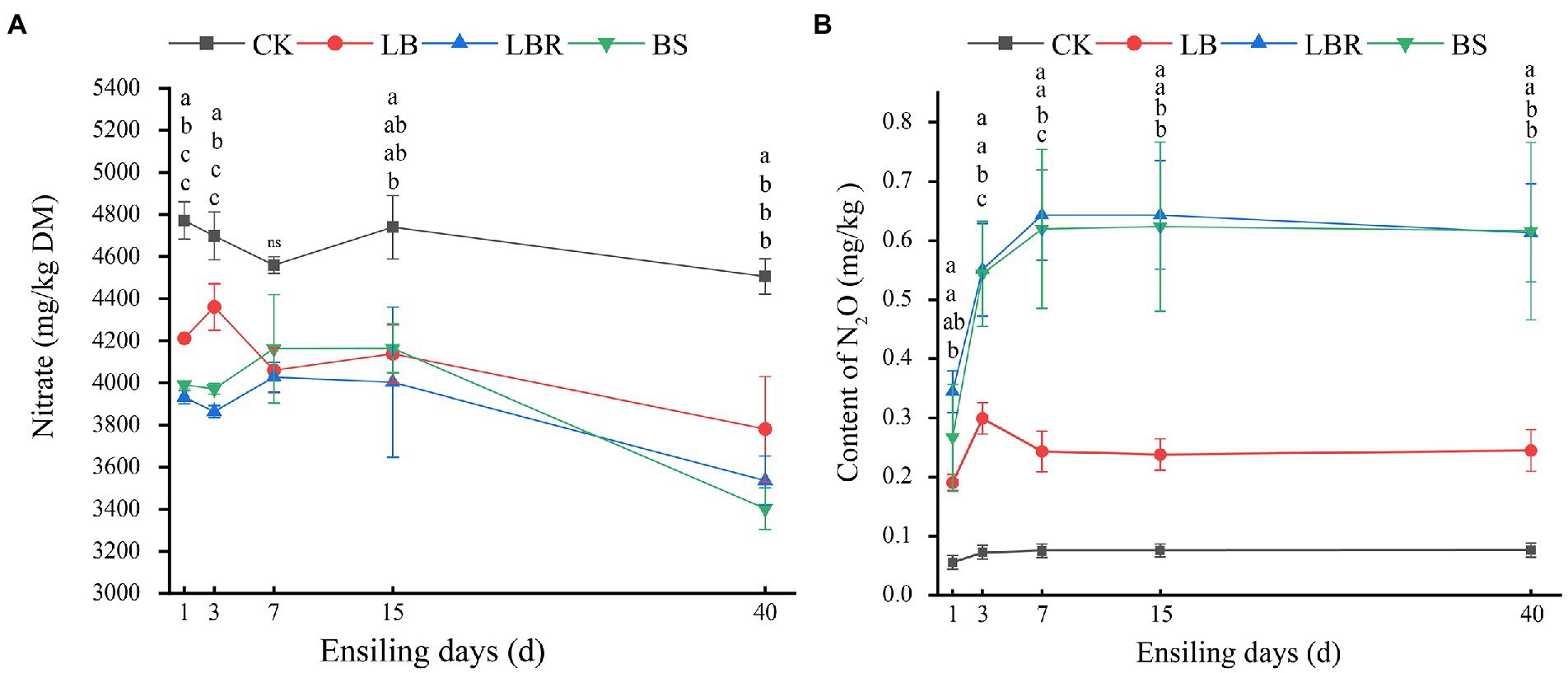
Figure 5. The nitrate content (A) and N2O (B) content in sorghum-sudangrass after ensiling for 1, 3, 7, 15, and 40 days (CK, control; LB, Lactobacillus buchneri; LBR, Lactobacillus brevis; BS, Bacillus subtilis; different lowercase indicate differences between treatment groups at the same time at the 0.05 level; ns, mean no significant difference).
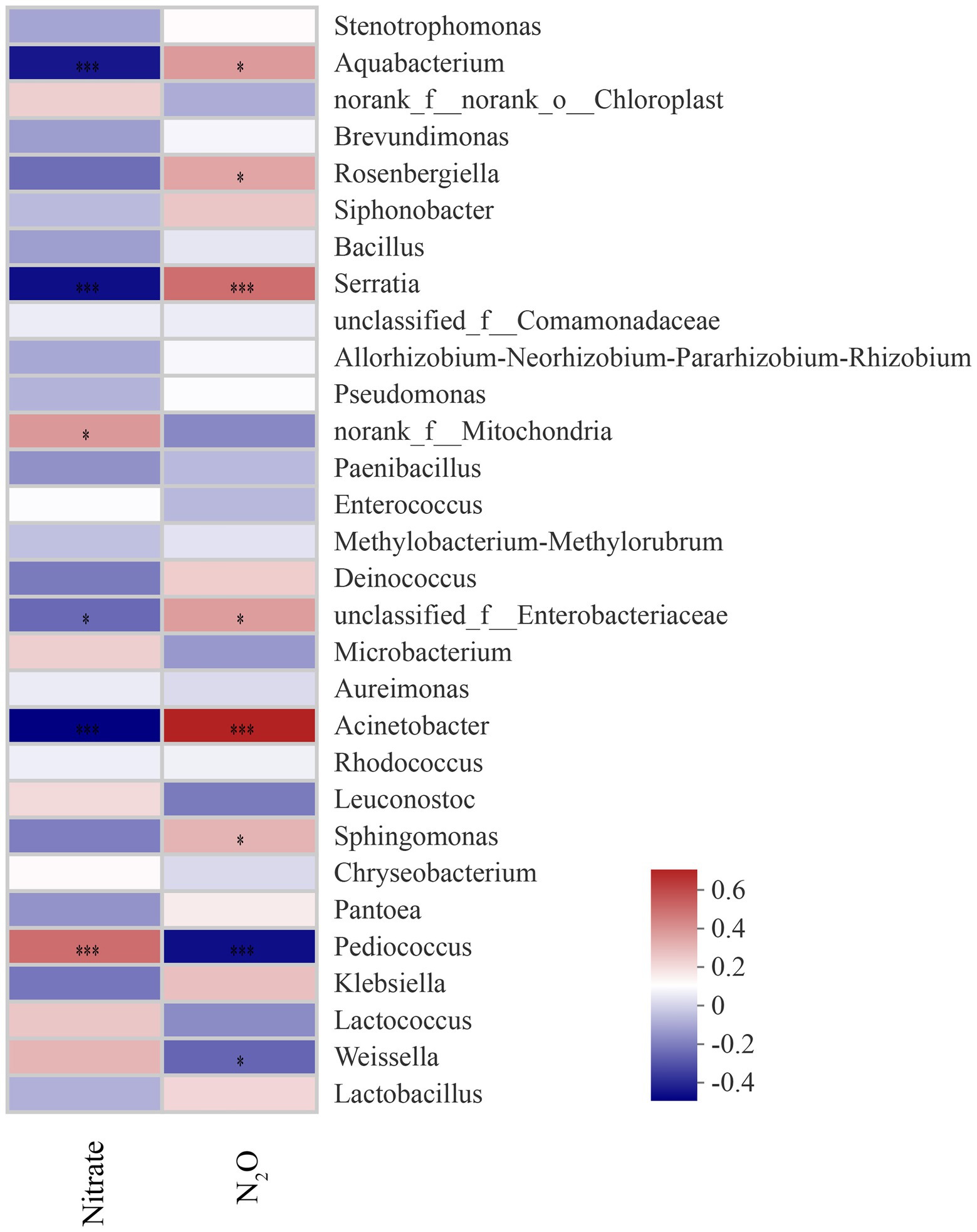
Figure 6. Correlation of bacterial communities with nitrate and N2O content (p-values are shown as *0.01 < p ≤ 0.05, ***p ≤ 0.001).
The Figure 7 predicts the effect of additives on the nitrate degradation function of bacteria during ensiling. Compared to CK, the metabolic abundance of denitrification increased significantly on the first day of ensiling in the LBR group, and the metabolic abundance of dissimilatory nitrate reduction and assimilatory nitrate reduction increased significantly on day 7 of ensiling (p < 0.05). Compared to CK, the BS group showed a higher metabolic abundance of silage denitrification, dissimilatory nitrate reduction, and assimilatory nitrate reduction on the first day of ensiling (p < 0.05).
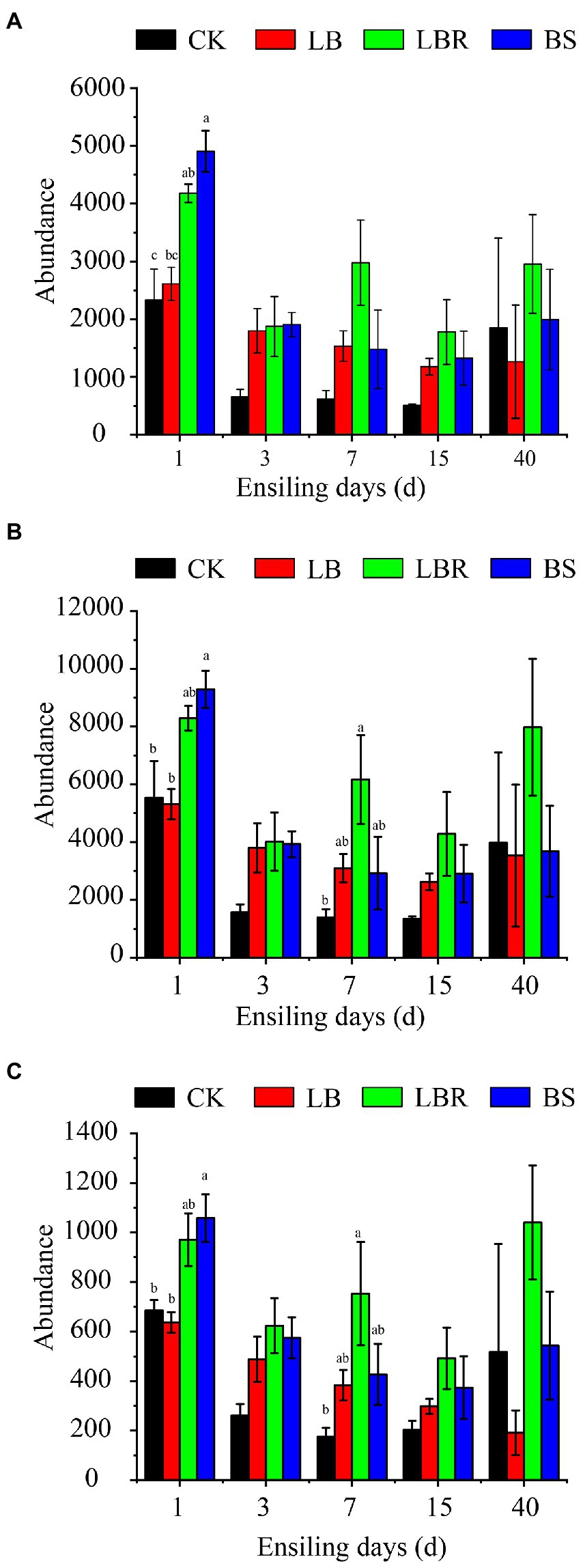
Figure 7. The denitrification (A), dissimilatory nitrate reduction (B), and assimilatory nitrate reduction (C) functional profiles of the microbial communities in sorghum-sudangrass after ensiling for 1, 3, 7, 15, and 40 days (CK, control; LB, Lactobacillus buchneri; LBR, Lactobacillus brevis; BS; Bacillus subtilis; different lowercase indicate differences between treatment groups at the same time at the 0.05 level).
Discussion
Characteristics of fresh sorghum-sudangrass
The WSC and epiphytic microorganisms in silage are key factors in determining the silage quality of the forage (Hu et al., 2009). The DM, WSC, and microbial counts of sorghum-sudangrass fulfill the theoretical requirements for well-fermented silage (Li et al., 2019). However, the nitrate content in the fresh sorghum-sudangrass in this experiment was as high as 5520.55 mg/kg DM, which can threaten the health of ruminants and pose a challenge to the safety of sorghum-sudangrass silage (Schrenk et al., 2020).
Fermentation quality and nutrient composition of silage
The pH of the sorghum-sudangrass silage decreased rapidly, and the LA content increased significantly during the initial 7 days of ensiling, indicating that extensive lactic acid fermentation occurred during the initial 7 days of ensiling. This phenomenon likely occurs because the forages are chopped into short parts to ensure the rapid release of plant juice, thus stimulating the growth of homofermentative lactic acid bacteria in the early period (Liu et al., 2019; Wang S. R. et al., 2022). Compared to CK, the LB group had a lower pH at 3 days of ensiling, whereas the pH was higher at 40 days of ensiling. The pH changes during ensiling were attributed to the accumulation of organic acids (Kung Jr. et al., 2018). In this study, the higher pH of the LB group at the end of ensiling was caused by the addition of LB to promote the accumulation of AA at the later stages of silage (Blajman et al., 2018). The pH was lower in the LBR group after 3 days of ensiling than in the CK. The LA content of the LBR group was significantly higher than that of CK on the third day of ensiling and during 15–40 days of ensiling. Therefore, the addition of LBR is more beneficial to obtain lower pH and higher LA content for sorghum-sudangrass silage. Similarly, lower pH and higher LA content were observed in silage with LBR in the study by Fijalkowska et al. (2020). Bai et al. (2020) reported that antimicrobial peptides produced by Bacillus subtilis could promote the fermentation of homozygous lactic acid bacteria. The BS group in our study had a higher LA concentration and lower pH at 40 days of fermentation. Butyric acid and NH3-N are both undesirable products in preserving forage (Kung Jr. et al., 2018; Li et al., 2018). In the present study, butyric acid was not detected throughout the sorghum-sudangrass silage, probably because the rapid decrease in pH during fermentation prevented Clostridium from fermenting soluble carbohydrates to butyric acid or converting lactic acid to butyric acid (Kung Jr. et al., 2018). The low NH3-N content in all groups in this study was due to the low number of undesirable microorganisms during the ensiling of sorghum-sudangrass (Wang et al., 2017). No butyric acid and lower content of NH3-N were detected which is beneficial for the quality of sorghum-sudangrass silage. Lactic acid bacteria can ferment a variety of substrates and rapidly produce large amounts of lactic acid for pH decline and microorganism inhibition. (Pang et al., 2012; Yang et al., 2021). The decrease in lactic acid bacteria numbers after 7 days of ensiling was related to insufficient silage substrate and the acidic fermentation environment of silage. At 40 days of ensiling, the higher pH in the LB group had a weaker inhibitory effect on lactic acid bacteria compared to the other group, so the LB group had a higher number of lactic acid bacteria. The number of yeast decreased rapidly after fermentation, which was due to the inhibition of their growth by AA produced after fermentation (Muck et al., 2018).
The nutrient composition of the sorghum-sudangrass silage after the addition of LB and LBR was similar to that of the CK, indicating that the addition of LB and LBR did not significantly alter the nutrient composition of sorghum-sudangrass. Similar findings were observed in the experiments performed by Alhaag et al. (2019) and Wang et al. (2020). Research has shown that the addition of BS inhibits the proteolysis in alfalfa silage and whole plant corn silage (Bai et al., 2020; Bai J. et al., 2022). In this study, the CP content of the BS group was significantly greater than that of the CK at 40 days of ensiling, indicating that the addition of BS to sorghum-sudangrass silage inhibited proteolysis.
Bacterial community in silage
The Coverage of all samples was 0.99, indicating that the depth of sequencing was sufficient to describe the dynamics of the bacterial community (He et al., 2020). The Shannon index was significantly lower in all treatment groups after 40 days of silage compared to the first day of silage and the Simpson index was significantly higher. As fermentation proceeded, the pH of the silage environment decreased, and the oxygen content gradually decreased to an anaerobic environment. This environment lead to a decrease in anaerobic microorganisms and acid-intolerant epiphytic microorganisms (Yan et al., 2019), resulting in a decrease in the diversity of the bacterial community of sorghum-sudangrass silage. In this study, the addition of LB did not affect α-diversity during the overall fermentation process. The Shannon index with LBR addition and the Shannon and Chao indices with BS addition were higher than those of the CK group on the first day of ensiling. The acidic environment had an inhibitory effect on the harmful microorganisms in silage fermentation (Ellis et al., 2016). On the first day of ensiling, the pH was slightly higher in the LBR and BS groups compared with CK, and both coliform bacteria and yeast counts were higher than in CK, which probably caused the differences in bacterial diversity between the LBR and BS groups and CK. In this study, there was no significant difference between the α-diversity of silages with additives and the CK during 3–40 days of ensiling. Although the dominant species changed for all silages with additives, the bacterial composition did not become simpler during the ensiling period from 3 to 40 days. (Wang et al., 2019; Xu et al., 2021). The plots of principal component analysis clearly reflected the changes of bacterial community during ensiling of sorghum-sudangrass. Storage time can affect the changes of bacterial communities in silage (Guo et al., 2021). The bacterial communities were significantly separated on day 1, days 3–15, and days 40, indicating that ensiling time affected the bacterial community of sorghum-sudangrass silage. Bacterial community changes during silage after the use of additives can indicate differences in the fermentation quality of silage (Ni et al., 2017a). In this study, the LBR and BS groups were significantly separated from the CK at 40 days of ensiling. We hypothesized that the addition of LBR and BS affected the quality of silage by changing the bacterial community structure at the end of ensiling.
In this study, Proteobacteria was the most abundant phylum in fresh sorghum-sudangrass, which is consistent with previous studies (Eikmeyer et al., 2013; Wang S. et al., 2022). At the beginning of ensiling (1 day), the bacterial community of all silages was dominated by the Firmicutes phylum because the anaerobic conditions and acidic environment formed during ensiling favoured the growth of the Firmicutes phylum (Keshri et al., 2018; Ali et al., 2020). Weissella is an important heterotypic lactic acid bacterium that initiates early fermentation and that the decrease in pH during silage inhibits its growth (Muck et al., 2018), after ensiling is performed by highly acid-tolerant homotypic fermenting lactic acid bacteria. In our study, Weissalla and Lactococcus were dominant in all silages on the first day of ensiling, and Lactobacillus was dominant in all silages after 3 days of ensiling. In addition, all silages had lower pH and higher lactic acid content, thus explaining the good fermentation quality of the silages (Guan et al., 2018). In the present study, the addition of LB increased the abundance of Serratia at 7–15 days of ensiling, and LBR significantly increased the abundance of Klebsiella at 1 day of ensiling and Serratia at 7–15 days of ensiling. Similarly, the addition of BS significantly increased the abundance of Klebsiella at 1 day of ensiling. Klebsiella and Serratia are usually considered as negative microorganisms for silage fermentation (Sun et al., 2021). However, in the present study, the LB, LBR, and BS were all dominated by Lactobacillus after 3 days of ensiling. In addition, the bacteria with the greatest contributions in the LB group included Weissella and Leuconostocaceae, and the bacteria with the greatest contributions in the BS group were Lactococcus and Streptococcaceae at 1 day of ensiling. Therefore, the addition of LBR and BS will not have a negative effect on the quality of sorghum-sudangrass silage.
Lactobacillus and Acinetbacter were significantly negatively correlated with the pH of the sorghum-sudangrass silage, while Lactococcus, Pediococcus, Leuconostoc, and Enterococcus were significantly positively correlated. The opposite trend was noted for fermentation acids, including LA and AA. Lactococcus starts lactic acid fermentation at the beginning of ensiling, but these species are sensitive to low pH and cannot survive in acidic environments (Ni et al., 2017b; Liu et al., 2019; Yang et al., 2019). Therefore, Lactobacillus plays a crucial role in lowering pH in the late silage stage.
Nitrate, N2O and microbial function prediction of silage
At the end of ensiling, the nitrate content in the additive group was significantly lower than that in CK, indicating that the addition of LB, LBR, and BS was beneficial for the degradation of nitrate in sorghum-sudangrass silage. Previous studies have shown that microorganisms, such as Enterobacteriaceae and Lactobacillus, in silage forage can degrade nitrate (Spoelstra, 1985). Bai C. et al. (2022) demonstrated that Panobacter spp., Pseudomonas spp., and Enterobacter spp. were negatively correlated with the nitrate content of silage sorghum-sudangrass. In addition, the Acinetobacter, Serratia, and Aquabacterium are also able to degrade nitrate in studies based on paddy soil and water process engineering (Zhang et al., 2016; Chen et al., 2021; Shelly et al., 2021). In the present study, nitrate content was negatively correlated with Acinetobacter, Serratia, Aquabacterium, and unclassified_f_enterobacteriaceae, indicating that they promoted the degradation of nitrate. In the LB group, Acinetobacter and Serratia were more abundant than in the CK group at 3–15 days of ensiling. The abundance of Klebsiella, Acinetobacter, Serratia, and Aquabacterium was greater in the LBR and BS groups compared with the CK group during ensiling. Moreover, Klebsiella was the differential bacteria in the LBR group, and Klebsiella and Aqubacterium were the differential bacteria in the BS group using LEfSe. This finding indicates that additives promoted nitrate degradation by affecting the bacterial community structure. N2O is one of the essential end products in the nitrate degradation process (Hill, 1999; Driehuis et al., 2018). The N2O content was significantly higher in the LBR and BS groups compared with CK at the end of silage, indicating that the addition of LBR, and BS contributed to the degradation of nitrate during ensiling of sorghum-sudangrass. This was also evidenced by the correlation between nitrate and N2O and the bacterial community of sorghum-sudangrass silage.
The silage process is mediated by microbial metabolic pathways to convert metabolites or degrade substrates (Xiang et al., 2022). We can estimate the impact of bacterial communities on changes in metabolic pathways during silage by predicting bacterial function. Therefore, this study used the KEGG pathway database PICRUSt2 to predict the function of bacterial communities in nitrate degradation in the silage of sorghum-sudangrass. Bacteria can degrade nitrate through denitrification, catabolism nitrate reduction, and assimilation nitrate reduction pathways (Shi et al., 2014). In this study, the addition of LB had a higher metabolic abundance of denitrification, dissimilatory nitrate reduction, and assimilatory nitrate reduction compared with CK, but no significant effects were noted at 3–15 days of ensiling. The addition of LBR significantly increased the metabolic abundance of denitrification on the first day of ensiling, and significantly increased the metabolic abundance of dissimilatory nitrate reduction and assimilatory nitrate reduction at 7 days of ensiling. The addition of BS significantly increased the metabolic abundance of denitrification, dissimilatory nitrate reduction, and assimilatory nitrate reduction on the first day of ensiling. This information explains why the addition of LBR and BS is more beneficial to the degradation of nitrate in sorghum-sudangrass silage.
Conclusion
The addition of LB increased the pH and acetic acid content, and the addition of LBR and BS increased the lactic acid content and decreased the pH. Sorghum-sudangrass silage has satisfactory fermentation quality. Additives had a significant effect on the bacterial community, increasing the abundance of microorganisms, such as Acinetobacter, Serratia, and Aquabacterium, but the fermentation process was still dominated by Lactobacillus. The addition of LB, LBR, and BS reduced the nitrate content in the silage of sorghum-sudangrass. Acinetobacter, Serratia, Aquabacterium, and unclassified_f_enterobacteriaceae were the main bacteria affecting nitrate degradation during sorghum-sudangrass ensiling. The LBR and BS could increase the metabolic abundance of denitrification, dissimilatory nitrate reduction, and assimilatory nitrate reduction. In conclusion, the use of LB, LBR, and BS additives in the silage of sorghum-sudangrass represents an effective strategy to degrade nitrate while guaranteeing its fermentation quality.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ebi.ac.uk/ena, PRJNA882255.
Author contributions
MZ, JS, ZY, CB, and YX designed the study and wrote the manuscript. MZ, HZ, GP, and HY performed the experiments. JS, ZY, CB, and YX reviewed and edited the manuscript. MZ, HZ, GP, and HY analyzed the data. ZY, CB, and YX funded and supervised the experiments. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the National Natural Science Foundation of China (nos. 31872422, 31772674, and 32160342), the Shenyang Agricultural University Graduate Student Innovation Cultivation Project (2022YCXS35), the Inner Mongolia Science and Technology Plan (2020GG0049), and the China Agriculture Research System of MOF and MARA.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1052837/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Differences in bacterial community on genus levels in sorghum-sudangrass silage after ensiling for 1 (A), 3 (B), 7 (C), 15 (D), and 40 (E) days.
References
Alhaag, H., Yuan, X. J., Mala, A., Bai, J. F., and Shao, T. (2019). Fermentation characteristics of Lactobacillus plantarum and pediococcus species isolated from sweet sorghum silage and their application as silage inoculants. Appl. Sci. 9:1247. doi: 10.3390/app9061247
Ali, N., Wang, S., Zhao, J., Dong, Z., Li, J., Nazar, M., et al. (2020). Microbial diversity and fermentation profile of red clover silage inoculated with reconstituted indigenous and exogenous epiphytic microbiota. Bioresour. Technol. 314:123606. doi: 10.1016/j.biortech.2020.123606
AOAC (1990). Official Methods of Analysis. 15th Edn. Arlington, VA: Association of Official Analytical Chemists.
Bai, J., Franco, M., Ding, Z., Hao, L., Ke, W., Wang, M., et al. (2022). Effect of bacillus amyloliquefaciens and Bacillus subtilis on fermentation, dynamics of bacterial community and their functional shifts of whole-plant corn silage. J. Anim. Sci. Biotechnol. 13:7. doi: 10.1186/s40104-021-00649-0
Bai, C., Pan, G., Leng, R., Ni, W., Yang, J., Sun, J., et al. (2022). Effect of ensiling density and storage temperature on fermentation quality, bacterial community, and nitrate concentration of sorghum-sudangrass silage. Front. Microbiol. 13:828320. doi: 10.3389/fmicb.2022.828320
Bai, J., Xu, D., Xie, D., Wang, M., Li, Z., and Guo, X. (2020). Effects of antibacterial peptide-producing Bacillus subtilis and lactobacillus buchneri on fermentation, aerobic stability, and microbial community of alfalfa silage. Bioresour. Technol. 315:123881. doi: 10.1016/j.biortech.2020.123881
Blajman, J. E., Paez, R. B., Vinderola, C. G., Lingua, M. S., and Signorini, M. L. (2018). A meta-analysis on the effectiveness of homofermentative and heterofermentative lactic acid bacteria for corn silage. J. Appl. Microbiol. 125, 1655–1669. doi: 10.1111/jam.14084
Cai, Y., Benno, Y., Ogawa, M., and Kumai, S. (1999). Effect of applying lactic acid bacteria isolated from forage crops on fermentation characteristics and aerobic deterioration of silage. J. Dairy Sci. 82, 520–526. doi: 10.3168/jds.S0022-0302(99)75263-X
Chen, X., Liu, Y., Liu, C. M., Zhang, W. Z., Qin, H. L., Sheng, R., et al. (2021). Functional differentiation of nitrate-reducing isolates regulated by long-term fertilization in a rice paddy soil. Can. J. Soil Sci. 101, 125–133. doi: 10.1139/cjss-2020-0038
Crawford, R. F., Kennedy, W. K., and Johnson, W. C. (1961). Some factors that affect nitrate accumulation in forages. Agron. J. 53, 159–162. doi: 10.2134/AGRONJ1961.00021962005300030010X
Ding, Z. T., Bai, J., Xu, D. M., Li, F. H., Zhang, Y. X., and Guo, X. S. (2020). Microbial community dynamics and natural fermentation profiles of ensiled alpine grass elymus nutans prepared from different regions of the Qinghai-Tibetan plateau. Front. Microbiol. 11:855. doi: 10.3389/fmicb.2020.00855
Driehuis, F., Wilkinson, J. M., Jiang, Y., Ogunade, I., and Adesogan, A. T. (2018). Silage review: animal and human health risks from silage. J. Dairy Sci. 101, 4093–4110. doi: 10.3168/jds.2017-13836
Eikmeyer, F. G., Kofinger, P., Poschenel, A., Junemann, S., Zakrzewski, M., Heinl, S., et al. (2013). Metagenome analyses reveal the influence of the inoculant lactobacillus buchneri CD034 on the microbial community involved in grass ensiling. J. Biotechnol. 167, 334–343. doi: 10.1016/j.jbiotec.2013.07.021
Ellis, J. L., Hindrichsen, I. K., Klop, G., Kinley, R. D., Milora, N., Bannink, A., et al. (2016). Effects of lactic acid bacteria silage inoculation on methane emission and productivity of Holstein Friesian dairy cattle. J. Dairy Sci. 99, 7159–7174. doi: 10.3168/jds.2015-10754
Fijalkowska, M., Przemieniecki, S. W., Purwin, C., Lipinski, K., Kurowski, T. P., and Karwowska, A. (2020). The effect of an additive containing three lactobacillus species on the fermentation pattern and microbiological status of silage. J. Sci. Food Agric. 100, 1174–1184. doi: 10.1002/jsfa.10126
Guan, H., Yan, Y. H., Li, X. L., Li, X. M., Shuai, Y., Feng, G. Y., et al. (2018). Microbial communities and natural fermentation of corn silages prepared with farm bunker-silo in Southwest China. Bioresour. Technol. 265, 282–290. doi: 10.1016/j.biortech.2018.06.018
Guo, L. N., Lu, Y. X., Li, P., Chen, L. Y., Gou, W. L., and Zhang, C. B. (2021). Effects of delayed harvest and additives on fermentation quality and bacterial community of corn stalk silage. Front. Microbiol. 12:687481. doi: 10.3389/fmicb.2021.687481
Harada, H., Yoshimura, Y., Sunaga, Y., and Hatanaka, T. (2000). Variations in nitrogen uptake and nitrate-nitrogen concentration among sorghum groups. Soil Sci. Plant Nutr. 46, 97–104. doi: 10.1080/00380768.2000.10408766
He, L. W., Lv, H. J., Xing, Y. Q., Chen, X. Y., and Zhang, Q. (2020). Intrinsic tannins affect ensiling characteristics and proteolysis of Neolamarckia cadamba leaf silage by largely altering bacterial community. Bioresour. Technol. 311:123496. doi: 10.1016/j.biortech.2020.123496
Hill, J. (1999). Nitrate in ensiled grass: a risk or benefit to livestock production. London, UK: Royal Society of Chemistry.
Holman, J. D., Obour, A. K., and Mengel, D. B. (2019). Nitrogen application effects on forage sorghum production and nitrate concentration. J. Plant Nutr. 42, 2794–2804. doi: 10.1080/01904167.2019.1659321
Hu, W., Schmidt, R. J., McDonell, E. E., Klingerman, C. M., and Kung, L. (2009). The effect of Lactobacillus buchneri 40788 or Lactobacillus plantarum MTD-1 on the fermentation and aerobic stability of corn silages ensiled at two dry matter contents. J. Dairy Sci. 92, 3907–3914. doi: 10.3168/jds.2008-1788
Keshri, J., Chen, Y. R., Pinto, R., Kroupitski, Y., Weinberg, Z. G., and Sela, S. (2018). Microbiome dynamics during ensiling of corn with and without Lactobacillus plantarum inoculant. Appl. Microbiol. Biotechnol. 102, 4025–4037. doi: 10.1007/s00253-018-8903-y
Kung, L. Jr., Shaver, R. D., Grant, R. J., and Schmidt, R. J. (2018). Silage review: interpretation of chemical, microbial, and organoleptic components of silages. J. Dairy Sci. 101, 4020–4033. doi: 10.3168/jds.2017-13909
Li, X., Hansen, W. P., Otterby, D. E., Linn, J. G., and Kuehn, C. S. (1992). Effect of additives on fermentation of corn silage containing different amounts of added nitrate nitrogen. J. Dairy Sci. 75, 1555–1561. doi: 10.3168/jds.S0022-0302(92)77912-0
Li, X. J., Tian, J. P., Zhang, Q., Jiang, Y., Wu, Z., and Yu, Z. (2018). Effects of mixing red clover with alfalfa at different ratios on dynamics of proteolysis and protease activities during ensiling. J. Dairy Sci. 101, 8954–8964. doi: 10.3168/jds.2018-14763
Li, P., Zhang, Y., Gou, W. L., Cheng, Q. M., Bai, S. Q., and Cai, Y. M. (2019). Silage fermentation and bacterial community of bur clover, annual ryegrass and their mixtures prepared with microbial inoculant and chemical additive. Anim. Feed Sci. Technol. 247, 285–293. doi: 10.1016/j.anifeedsci.2018.11.009
Liu, B. Y., Huan, H. L., Gu, H. R., Xu, N. X., Shen, Q., and Ding, C. L. (2019). Dynamics of a microbial community during ensiling and upon aerobic exposure in lactic acid bacteria inoculation-treated and untreated barley silages. Bioresour. Technol. 273, 212–219. doi: 10.1016/j.biortech.2018.10.041
McCary, C. L., Vyas, D., Faciola, A. P., and Ferraretto, L. F. (2020). Graduate student literature review: current perspectives on whole-plant sorghum silage production and utilization by lactating dairy cows. J. Dairy Sci. 103, 5783–5790. doi: 10.3168/jds.2019-18122
Muck, R. E., Nadeau, E. M. G., McAllister, T. A., Contreras-Govea, F. E., Santos, M. C., and Kung, L. Jr. (2018). Silage review: recent advances and future uses of silage additives. J. Dairy Sci. 101, 3980–4000. doi: 10.3168/jds.2017-13839
Murphy, R. P. (1958). A method for the extraction of plant samples and the determination of total soluble carbohydrates. J. Sci. Food Agric. 9, 714–717. doi: 10.1002/jsfa.2740091104
Ni, K., Minh, T. T., Tu, T. T., Tsuruta, T., Pang, H., and Nishino, N. (2017a). Comparative microbiota assessment of wilted Italian ryegrass, whole crop corn, and wilted alfalfa silage using denaturing gradient gel electrophoresis and next-generation sequencing. Appl. Microbiol. Biotechnol. 101, 1385–1394. doi: 10.1007/s00253-016-7900-2
Ni, K., Wang, F., Zhu, B., Yang, J., Zhou, G., Pan, Y., et al. (2017b). Effects of lactic acid bacteria and molasses additives on the microbial community and fermentation quality of soybean silage. Bioresour. Technol. 238, 706–715. doi: 10.1016/j.biortech.2017.04.055
Pang, H. L., Tan, Z. F., Qin, G. Y., Wang, Y. P., Li, Z. W., Jin, Q. S., et al. (2012). Phenotypic and phylogenetic analysis of lactic acid bacteria isolated from forage crops and grasses in the Tibetan Plateau. J. Microbiol. 50, 63–71. doi: 10.1007/s12275-012-1284-5
Romero, J. J., Joo, Y., Park, J., Tiezzi, F., Gutierrez-Rodriguez, E., and Castillo, M. S. (2018). Bacterial and fungal communities, fermentation, and aerobic stability of conventional hybrids and brown midrib hybrids ensiled at low moisture with or without a homo- and heterofermentative inoculant. J. Dairy Sci. 101, 3057–3076. doi: 10.3168/jds.2017-13754
Schrenk, D., Bignami, M., Bodin, L., Chipman, J. K., del Mazo, J., and Grasl-Kraupp, B. (2020). Risk assessment of nitrate and nitrite in feed. EFSA J. 18:e06290. doi: 10.2903/j.efsa.2020.6290
Shelly, Y., Kuk, M., Menashe, O., Zeira, G., Azerrad, S., and Kurzbaum, E. (2021). Nitrate removal from a nitrate-rich reverse osmosis concentrate: superior efficiency using the bioaugmentation of an Acinetobacter biofilm. J. Water Process. Eng. 44:102425. doi: 10.1016/j.jwpe.2021.102425
Shi, W., Lu, W., Liu, Q., Zhi, Y., and Zhou, P. (2014). The identification of the nitrate assimilation related genes in the novel Bacillus megaterium NCT-2 accounts for its ability to use nitrate as its only source of nitrogen. Funct. Integr. Genomics 14, 219–227. doi: 10.1007/s10142-013-0339-y
Spoelstra, S. F. (1983). Inhibition of clostridial growth by nitrate during the early phase of silage fermentation. J. Sci. Food Agric. 34, 145–152. doi: 10.1002/JSFA.2740340206
Spoelstra, S. F. (1985). Nitrate in silage. Grass Forage Sci. 40, 1–11. doi: 10.1111/j.1365-2494.1985.tb01714.x
Spoelstra, S. F. (1987). Degradation of nitrate by enterobacteria during silage fermentation of grass. Neth. J. Agric. Sci. 35, 43–54. doi: 10.18174/njas.v35i1.16757
Sun, L., Bai, C., Xu, H., Na, N., Jiang, Y., Yin, G., et al. (2021). Succession of bacterial community during the initial aerobic, intense fermentation, and stable phases of whole-plant corn silages treated with lactic acid bacteria suspensions prepared from other silages. Front. Microbiol. 12:655095. doi: 10.3389/fmicb.2021.655095
Sun, Y., De Vos, P., and Heylen, K. (2016). Nitrous oxide emission by the non-denitrifying, nitrate ammonifier bacillus licheniformis. BMC Genomics 17:68. doi: 10.1186/s12864-016-2382-2
Takayoshi, M., Shinichi, W., Toshiyuki, S., and Awaya, K. (1980). Studies on the disappearance of nitrate in forage grops during ensilage: II. Relationship between disappearance of nitrate and fermentation products during ensilage. Jpn. J Grassland Sci. 26, 74–80. doi: 10.14941/grass.26.74
Tian, H., Zhu, Y., Dai, M., Li, T., Guo, Y., Deng, M., et al. (2021). Additives altered bacterial communities and metabolic profiles in silage hybrid pennisetum. Front. Microbiol. 12:770728. doi: 10.3389/fmicb.2021.770728
Van Soest, P. J., Robertson, J. B., and Lewis, B. A. (1991). Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74, 3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2
Vazquez-Baeza, Y., Pirrung, M., Gonzalez, A., and Knight, R. (2013). EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2:16. doi: 10.1186/2047-217X-2-16
Vermunt, J., and Visser, R. (2011). Nitrate toxicity in cattle. N. Z. Vet. J. 35, 136–137. doi: 10.1080/00480169.1987.35413
Wang, Y., Chen, X., Wang, C., He, L., Zhou, W., Yang, F., et al. (2019). The bacterial community and fermentation quality of mulberry (Morus alba) leaf silage with or without Lactobacillus casei and sucrose. Bioresour. Technol. 293:122059. doi: 10.1016/j.biortech.2019.122059
Wang, J., Chen, L., Yuan, X. J., Guo, G., Li, J. F., Bai, Y. F., et al. (2017). Effects of molasses on the fermentation characteristics of mixed silage prepared with rice straw, local vegetable by-products and alfalfa in Southeast China. J. Integr. Agric. 16, 664–670. doi: 10.1016/S2095-3119(16)61473-9
Wang, S., Li, J., Zhao, J., Dong, Z., and Shao, T. (2022). An investigation on fermentative profile, microbial numbers, bacterial community diversity and their predicted metabolic characteristics of Sudangrass (Sorghum sudanense Stapf.) silages. Anim. Biosci. 35, 1162–1173. doi: 10.5713/ab.21.0326
Wang, S. R., Shao, T., Li, J. F., Zhao, J., and Dong, Z. H. (2022). A survey of fermentation parameters, bacterial community compositions and their metabolic pathways during the ensiling of sorghum. J. Appl. Microbiol. 132, 3563–3577. doi: 10.1111/jam.15484
Wang, M., Xu, S., Wang, T., Jia, T., Xu, Z., Wang, X., et al. (2018). Effect of inoculants and storage temperature on the microbial, chemical and mycotoxin composition of corn silage. Asian Australas J. Anim. Sci 31, 1903–1912. doi: 10.5713/ajas.17.0801
Wang, S. R., Zhao, J., Dong, Z. H., Li, J. F., Kaka, N. A., and Shao, T. (2020). Sequencing and microbiota transplantation to determine the role of microbiota on the fermentation type of oat silage. Bioresour. Technol. 309:123371. doi: 10.1016/j.biortech.2020.123371
Xiang, Q., Zhang, J., Huang, X., Ma, M., Zhao, K., Yu, X., et al. (2022). Changes in the taxonomic and functional structures of microbial communities during vegetable waste mixed silage fermentation. Can. J. Microbiol. 68, 281–293. doi: 10.1139/cjm-2021-0059
Xie, Y., Guo, J., Li, W., Wu, Z., and Yu, Z. (2021). Effects of ferulic acid esterase-producing lactic acid bacteria and storage temperature on the fermentation quality, in vitro digestibility and phenolic acid extraction yields of Sorghum (Sorghum bicolor L.) Silage. Microorganisms 9:114. doi: 10.3390/microorganisms9010114
Xu, D. M., Ding, W. R., Ke, W. C., Li, F. H., Zhang, P., and Guo, X. S. (2019). Modulation of metabolome and bacterial community in whole crop corn silage by inoculating homofermentative Lactobacillus plantarum and heterofermentative Lactobacillus buchneri. Front. Microbiol. 9:3299. doi: 10.3389/fmicb.2018.03299
Xu, Z., He, H., Zhang, S., and Kong, J. (2017). Effects of inoculants Lactobacillus brevis and Lactobacillus parafarraginis on the fermentation characteristics and microbial communities of corn Stover silage. Sci. Rep. 7:13614. doi: 10.1038/s41598-017-14052-1
Xu, D., Wang, N., Rinne, M., Ke, W., Weinberg, Z. G., Da, M., et al. (2021). The bacterial community and metabolome dynamics and their interactions modulate fermentation process of whole crop corn silage prepared with or without inoculants. Microb. Biotechnol. 14, 561–576. doi: 10.1111/1751-7915.13623
Xu, T. W., Zhao, N., Hu, L. Y., Xu, S. X., Liu, H. J., Ma, L., et al. (2017). Characterizing CH4, CO2 and N2O emission from barn feeding Tibetan sheep in Tibetan alpine pastoral area in cold season. Atmos. Environ. 157, 84–90. doi: 10.1016/j.atmosenv.2017.03.023
Yan, Y., Li, X., Guan, H., Huang, L., Ma, X., Peng, Y., et al. (2019). Microbial community and fermentation characteristic of Italian ryegrass silage prepared with corn stover and lactic acid bacteria. Bioresour. Technol. 279, 166–173. doi: 10.1016/j.biortech.2019.01.107
Yang, L. L., Yuan, X. J., Li, J. F., Dong, Z. H., and Shao, T. (2019). Dynamics of microbial community and fermentation quality during ensiling of sterile and nonsterile alfalfa with or without Lactobacillus plantarum inoculant. Bioresour. Technol. 275, 280–287. doi: 10.1016/j.biortech.2018.12.067
Yang, F. Y., Zhao, S. S., Wang, Y., Fan, X. M., Wang, Y. P., and Feng, C. S. (2021). Assessment of bacterial community composition and dynamics in alfalfa silages with and without Lactobacillus plantarum inoculation using absolute quantification 16S rRNA sequencing. Front. Microbiol. 11:629894. doi: 10.3389/fmicb.2020.629894
You, S., Du, S., Ge, G., Wan, T., and Jia, Y. (2021). Selection of lactic acid bacteria from native grass silage and its effects as inoculant on silage fermentation. Agron. J. 113, 3169–3177. doi: 10.1002/agj2.20720
Zhang, H., Chen, X., Song, L., Liu, S., and Li, P. (2022). The mechanism by which Enteromorpha Linza polysaccharide promotes Bacillus subtilis growth and nitrate removal. Int. J. Biol. Macromol. 209, 840–849. doi: 10.1016/j.ijbiomac.2022.04.082
Keywords: sorghum-sudangrass silage, fermentation quality, nitrate, Lactobacillus buchneri, Lactobacillus brevis, Bacillus subtilis
Citation: Zhao M, Zhang H, Pan G, Yin H, Sun J, Yu Z, Bai C and Xue Y (2022) Effect of exogenous microorganisms on the fermentation quality, nitrate degradation and bacterial community of sorghum-sudangrass silage. Front. Microbiol. 13:1052837. doi: 10.3389/fmicb.2022.1052837
Edited by:
Siran Wang, Nanjing Agricultural University, ChinaCopyright © 2022 Zhao, Zhang, Pan, Yin, Sun, Yu, Bai and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunsheng Bai, YnNjQHN5YXUuZWR1LmNu; Yanlin Xue, eHVleWFubGluXzE5NzlAMTYzLmNvbQ==
 Meirong Zhao
Meirong Zhao Hongyu Zhang1
Hongyu Zhang1 Zhu Yu
Zhu Yu Chunsheng Bai
Chunsheng Bai Yanlin Xue
Yanlin Xue