- 1School of Pharmaceutical Sciences, South-Central Minzu University, Wuhan, China
- 2Graduate School of Pharmaceutical Sciences, The University of Tokyo, Bunkyo-ku, Japan
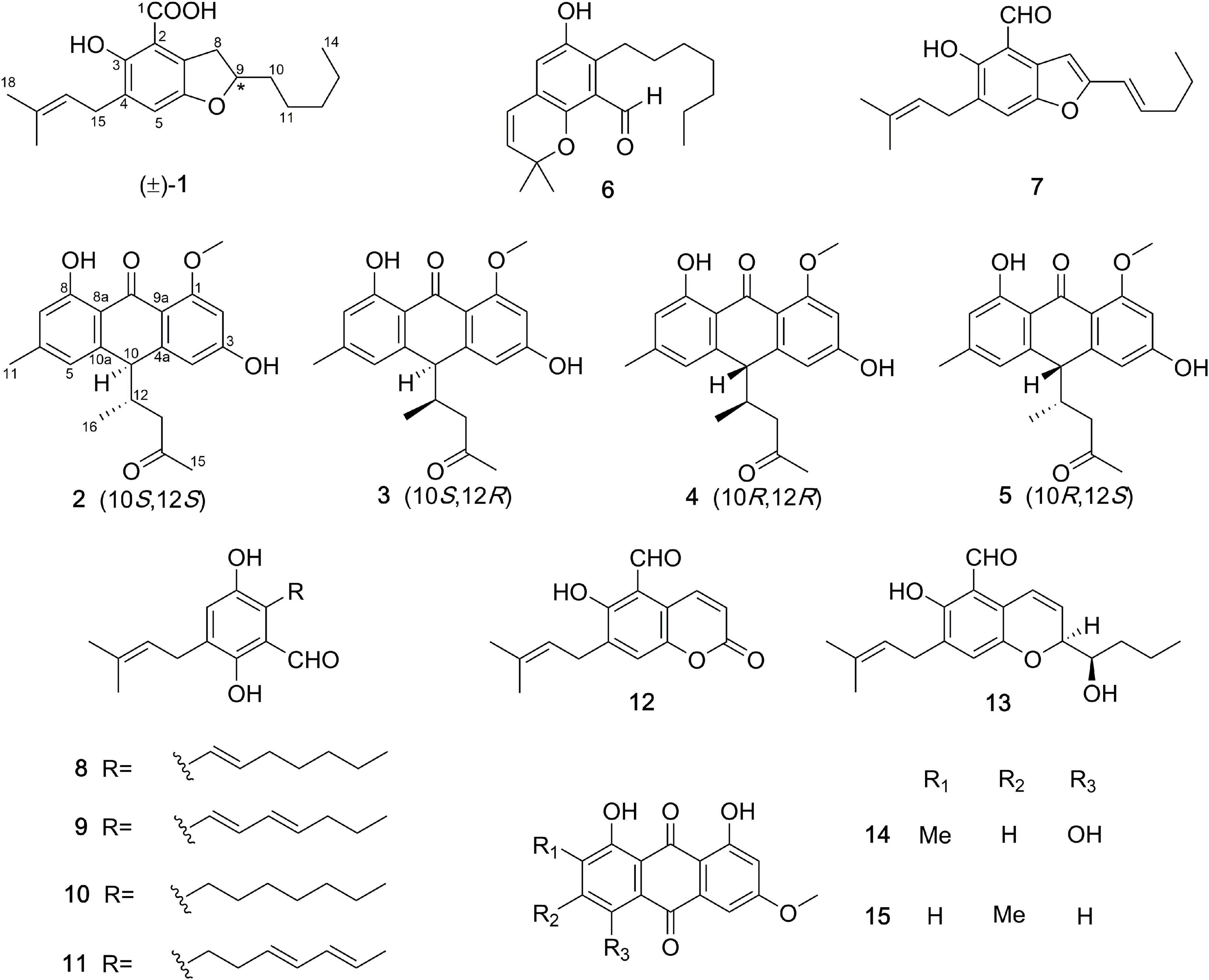
One new prenylated benzenoid, (±)-chevalieric acid (1), and four new anthraquinone derivatives, (10S,12S)-, (10S,12R)-, (10R,12S)-, and (10R,12R)-chevalierone (2–5), together with ten previously described compounds (6–15), were isolated from the fungus Aspergillus chevalieri (L. Mangin) Thom and Church. The structures of new compounds were elucidated by extensive 1D and 2D nuclear magnetic resonance (NMR), and HRESIMS spectroscopic analysis. The absolute configurations of 2–5 were determined by experimental and calculated electronic circular dichroism (ECD) and DP4+ analysis. Compound 10 showed weak cytotoxicity against human lung cancer cell line A549 with IC50 39.68 μM. Compounds 2–5 exhibited antibacterial activities against the methicillin-resistant Staphylococcus aureus (MRSA) and opportunistic pathogenic bacterium Pseudomonas aeruginosa. The MIC value for compound 6 against MRSA is 44.02 μM. Additionally, Compounds 8, 10, 11 showed weak to moderate inhibitory activities against the β-secretase (BACE1), with IC50 values of 36.1, 40.9, 34.9 μM, respectively.
Introduction
Dementia is a syndrome in which there is deterioration in cognitive function beyond the usual consequences of biological aging (World Health Organization, 2022). Dementia ranks the seventh leading cause of death and one of the major causes of disability among older people. Currently, more than 55 million people live with dementia worldwide, and a 10 million new cases increase every year. Alzheimer’s disease (AD) is the most common form of dementia which contributes to 60–70% of the cases. Current research indicate that amyloid β peptide (Aβ) aggregation is pathophysiological result of AD (Busche and Hyman, 2020; Piller, 2022). The Aβ protein is produced by cleavage of Aβ protein precursor by the β-secretase (BACE1) (Vassar and Kandalepas, 2011). However, in the field of AD drug development, more than 99% of clinical trials failed (Cummings et al., 2019). One of the main reasons is that lacking a potential lead compound reservoir which could feed the drug development process continuously. Additionally, human health is threatened by the emerging of “superbugs,” which show resistance to the antibiotics currently used (Rossiter et al., 2017). The pace for antibiotic drug development in recent years has slowed down due to the already widely used antibiotics with different mechanisms and lacking promising lead compounds. The rapid evolution of bacterial resistance to antibiotics urged us to keep the pace to find new types of lead compounds.
Fungal secondary metabolites are a natural reservoir which have contributed and still contributing to the development of drugs (Schueffler and Anke, 2014). Aspergillus chevalieri is a fungus belonging to the family Aspergillaceae. Fungi from this genus are widely distributed in the Dead Sea, desert, highlands, mangroves, or somewhere with high salt. Previous research on Aspergillus spp. led to the discovery of many diketopiperazine indole alkaloids (Zhong W.-M. et al., 2018), benzaldehyde derivatives (Zhong W.-M. et al., 2018; Zhong et al., 2020; Zhong W. et al., 2021), anthraquinone derivatives (Zhang et al., 2017; Zhao D. et al., 2018) and cochlioquinones (Koyama et al., 2005; Yamazaki et al., 2008). Most of the benzaldehyde derivatives displayed cytotoxic activity (Gao et al., 2012) and antioxidant activity (Miyake et al., 2009). In a follow-up program for mining bioactive natural products from fungi, the secondary metabolites of the fungus A. chevalieri were studied, which resulted in the isolation of five new and ten known compounds. We herein report the isolation, structural elucidation, and biological activities of the isolates in terms of cytotoxicity, antibacterial activity, and BACE1 inhibitory activity.
Materials and methods
General experimental procedures
UV spectra were recorded on a Hitachi UH5300 UV-Vis spectrophotometer (Hitachi, Co., Ltd., Tokyo, Japan). IR spectra were obtained on a Tracer-100 Fourier transform infrared spectrometer (Shimadzu Corporation, Tokyo, Japan) using KBr pellet. All nuclear magnetic resonance (NMR) spectra were recorded on a Bruker Ascend 600 MHz spectrometer (Bruker Corporation, Karlsruhe, Germany). HRESIMS were measured on a Q Exactive HF mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). Column chromatography (CC) was performed on silica gel (Qingdao Haiyang Chemical Co., Ltd., Qingdao, China) and Sephadex LH-20 (Amersham Biosciences, Uppsala, Sweden). Medium pressure liquid chromatography (MPLC) was performed on a Biotage SP1 equipment, and column was packed with C18 silica gel. Preparative high performance liquid chromatography (prep-HPLC) was performed on an Agilent 1260 liquid chromatography system equipped with Agilent Zorbax SB-C18 columns (particle size, 5 μm, i.d. 9.4 mm × 150 mm or 21.2 mm × 150 mm) and a diode array detector (DAD) (Agilent Technologies, Santa Clara, CA, US). Fractions were monitored by DAD detector, or thin layer chromatography (TLC) (GF254, Qingdao Haiyang Chemical Co., Ltd., Qingdao, China). The chiral phase HPLC analysis and preparation were performed on Agilent 1200 liquid chromatography system with Daicel CHIRALPAK AD-H and CHIRALPAK AS-H columns (particle size 5 μm, i.d. 4.6 mm × 250 mm, flow rate 1 mL/min) (Daicel Corporation, Tokyo, Japan). 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and LY2811376 were purchased from Sigma-Aldrich (St. Louis, MO, USA). Penicillin G sodium salt and ceftazidime were purchased from Solarbio (Beijing, China). Dulbecco’s modified Eagle’s medium (DMEM) and fetal bovine serum were obtained from HyClone (Logan, UT, USA). BACE1 FRET assay kit, red (P2985) was bought from Thermo Fisher Scientific (MA, USA).
Fungal material
Aspergillus chevalieri was isolated from the mud of the coast of Shenzhen Bay, China in May 2019, and was identified by ITS region sequencing. A voucher strain (No. HP-5) was stored at the School of Pharmaceutical Sciences, South-Central Minzu University, Wuhan, China. The fungus was fermented by cooked rice medium. The preparation of rice media was 100 g rice with 100 mL sea water each in 500-mL Erlenmeyer flasks. The fermentation was kept in dark environment for 40 days.
Extraction and isolation
The rice cultures of A. chevalieri (15 kg) were collected and extracted with methanol at room temperature (100 L, 5 days each). The methanol extract was evaporated to dry under reduced pressure. The residue (2.1 kg) was resuspended in H2O and partitioned against ethyl acetate (EtOAc) for three times to give an EtOAc extract (226 g). The EtOAc extract was subjected to CC eluting with the solvent mixture petroleum ether-acetone (v/v 20:1, 10:1, 5:1, 2:1, 0:1) to yield four fractions (A–D).
Fraction A was eluted by MPLC with a stepwise gradient of MeOH-H2O (v/v 0:100–100:0) to afford subfractions A1-A8. Fraction A4 was applied to silica gel CC (petroleum ether–acetone from v/v 10:1 to 0:1) to yield five subfractions A4a-A4e.
The subfraction A4b was separated by prep-HPLC (MeCN-H2O = v/v 46:54–56:44 in 25 min, 4 mL min–1) to yield 7 (4.1 mg, tR = 14.5 min) and 8 (2.8 mg, tR = 18.0 min).
Fraction A5 was purified to silica gel CC (petroleum ether–acetone from v/v 10:1 to 0:1) and was further isolated by prep-HPLC (MeCN-H2O = v/v 37:63–47:53 in 25 min, 4 mL min–1) to yield compound 14 (3.5 mg, tR = 13.6 min) and compound 15 (4.2 mg, tR = 16.5 min). Fraction A6 was subjected to prep-HPLC (MeCN-H2O = v/v 35:65–55:45 for 25 min, 4 mL min–1) to give compound 1 (2.8 mg, tR = 15.2 min).
Fraction B was purified by prep-HPLC (MeCN-H2O = v/v 47:53–57:43 for 25 min, 4 mL min–1) to yield the compound mixture 2–5 (3.6 mg, tR = 21.5 min). Then the compounds 2 and 3 were purified by semi-preparative HPLC with CHIRALPAK AS-H (n-hexane/2-propanol = v/v 97:3, isocratic, 1 mL min–1) to yield 2 (0.9 mg, tR = 20.0 min), 3 (0.7 mg, tR = 27.3 min) and compound mixture 4/5 (1.8 mg, tR = 17.0 min). The mixture 4/5 were further applied to semi-preparative HPLC with CHIRALPAK AD-H (n-hexane/2-propanol = v/v 94:6 isocratic, 1 mL min–1) to yield 4 (0.7 mg, tR = 13.2 min), 5 (0.6 mg, tR = 16.1 min).
Fraction D was fractionated by silica gel CC (CHCl3-MeOH from v/v 40:1, 30:1, 20:1, 10:1, 0:1) to give four subfractions D1–D3. Then subfraction D2 was isolated by prep-HPLC (MeCN-H2O = 49:51, 4 mL min–1, 25 min) to yield 9 (3.4 mg, tR = 17.5 min) and 10 (6.2 mg, tR = 18.6 min). Fraction D3 was subject to silica gel CC (petroleum ether–acetone from v/v 10: 1 to 0: 1) and further applied to prep-HPLC (MeCN-H2O = 43:57–63:37 in 25 min, 4 mL min–1) to yield 11 (6.2 mg, tR = 15.2 min) and 12 (4.3 mg, tR = 18.7 min).
Fraction E was separated by Sephadex LH-20 (MeOH), and further purified by prep-HPLC (MeCN-H2O = 53:47 in 25 min, 3 mL min–1) to yield 6 (2.4 mg, tR = 17.3 min), and 13 (2.6 mg, tR = 21.5 min).
Spectroscopic data
(±)-Chevalieric acid (1): yellow amorphous solid; UV (MeOH) λmax (log ε): 210 (3.01) nm; [α]D24 + 2.7 (c 0.05, MeOH); 1H and 13C NMR data, Table 1; HRESIMS m/z 319.19028 [M+H]+, calcd. for C19H27O4, 319.19093.
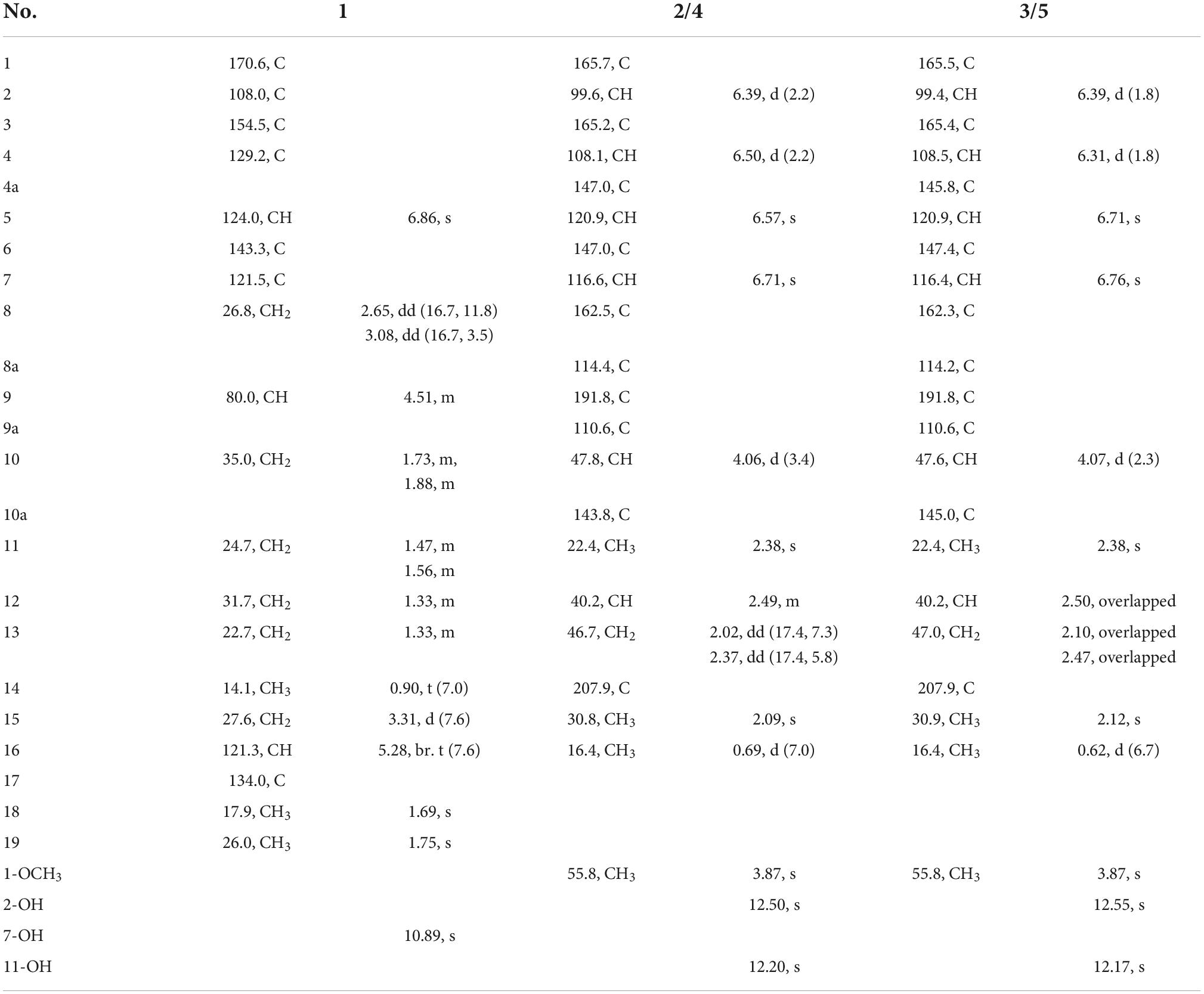
Table 1. 1H and 13C nuclear magnetic resonance (NMR) spectroscopic data of compounds 1–5 (600/150 MHz, CDCl3).
(10S,12S)-Chevalierone (2): yellow powder; UV (MeOH) λmax (log ε): 230 (3.40), 270.0 (3.17), 340.0 (3.46) nm; IR (KBr) νmax 3429, 2962, 2927, 1712, 1620, 1600, 1288, 1103, 817 cm–1. [α]D24 + 30.7 (c 0.07, MeOH); 1H and 13C NMR data, Table 1; HRESIMS m/z 355.15402 [M+H]+, calcd. for C21H23O5, 355.15455.
(10S,12R)-Chevalierone (3): yellow powder; UV (MeOH) λmax (log ε): 220.0 (3.29), 275.0 (2.89), 360.0 (3.22) nm; [α]D24 + 30.39 (c 0.05, MeOH); 1H and 13C NMR data, Table 1; HRESIMS m/z 355.15405 [M+H]+, calcd. for C21H23O5, 355.15455.
(10R,12R)-Chevalierone (4): yellow powder; UV (MeOH) λmax (log ε): 230.0 (3.38), 270.0 (3.13), 350.0 (3.45) nm; [α]D24 −33.10 (c 0.05, MeOH); 1H and 13C NMR data, Table 1; HRESIMS m/z 355.15402 [M+H]+, cacld. for C21H23O5, 355.15455.
(10R,12S)-Chevalierone (5): yellow powder; UV (MeOH) λmax (log ε): 230.0 (3.42) 275.0 (3.32) 340.0 (3.49) nm; [α]D24 −33.14 (c 0.06, MeOH); 1H and 13C NMR data, Table 1; HRESIMS m/z 355.15393 [M+H]+, calcd. for C21H23O5, 355.15455.
Electronic circular dichroism and nuclear magnetic resonance calculations
Conformational analyses of the candidate structures were performed using the MMFF94s force field. For ECD calculation, the conformers with a distribution higher than 1% were further optimized at B3LYP/6-31G(d) level of theory in the Gaussian 16 software package (Frisch et al., 2016). The conformers within 3 kcal/mol energy of global minimum were subjected to theoretical calculation of ECD using time-dependent density functional theory (TDDFT) at B3LYP/6-31G(d,p) level in methanol with IEFPCM model. The calculated and weighted ECD curves were all generated using SpecDis 1.71 (Bruhn et al., 2013, 2017), respectively, and plotted in Microsoft Office Excel Program. For NMR calculation, the conformers with a distribution higher than 1% were further optimized at B3LYP/6-311G(2d,p) level of theory in Gaussian 16 software package. The conformers within 3 kcal/mol energy of global minimum were subjected to theoretical calculation of ECD using TDDFT at B3LYP/6-311G(2d,p) level in chloroform with IEFPCM model. The calculated data were processed with an in-house Office Excel Workbook.
DP4+ calculation and analysis
The conformations of compounds 2–5 from ECD calculations were further optimized at the B3LYP/6-311G(2d,p) level. With these optimized conformers, chemical shift values at mPW1PW91/6-31 + G(d,p) for DP4+ for conformational analysis were computed in the solvent phase with chloroform. The averaged shielding tensors were used for probabilities calculation with the spreadsheet provided by the original publication (Grimblat et al., 2015).
Cytotoxicity assays
The human lung cancer cell line A549 used in this study was purchased from Conservation Genetics CAS Kunming Cell Bank. The cells were cultured in high-glucose DMEM with 10% fetal bovine serum, 100 mg/mL streptomycin, 100 U/mL penicillin, and kept in an incubator at 37°C under an atmosphere of 5% CO2. The cell viability was assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) method. The presence of NAD(P)H-dependent cellular oxidoreductases reflects the number of viable cells. These enzymes are capable of reducing MTT to its insoluble formazan, which has a purple color. Therefore, colorimetric measurements of the amount of insoluble formazan which produced in living cells based on the reduction of MTT can evaluate the cell viability. In brief, 100 μL of adherent cells was seeded to each well of a 96-well cell culture plate. The plate was kept for 12 h for adherence. The DMSO solution of test compounds were added to each well to give a final compound concentration of 40 μM. However, suspended cells were seeded with the same density of 1 × 105 cells/mL every 100 μL of culture medium before adding the test compounds. Then the plate was incubated for another 48 h. After 48 h of incubation, 100 μg of MTT was added to each well, another 4 h of incubation was continued at 37°C. After removal of 100-μL culture medium, the cells were lysed with 20% SDS-50% DMF (100 μL). The remained lysates were subjected to measure the optical density value at 595 nm with a 96-well microtiter plate reader (Tecan Spark 10M, Switzerland). Cisplatin was used as positive control. All the experiments were in triplicates. The IC50 value was calculated by a published method (Chen et al., 2018).
Antibacterial assays
In this study, the tested bacteria strains, Escherichia coli ATCC25922, Staphylococcus aureus subsp. aureus ATCC29213, Salmonella enterica subsp. enterica ATCC14028, Pseudomonas aeruginosa ATCC27853, methicillin-resistant S. aureus (MRSA) were purchased from China General Microbiological Culture Collection Center (CGMCC). All the strains were cultured in Mueller-Hinton broth (Oxoid, Thermo Fisher Scientific) at 37°C with shaking at 200 rpm overnight. Each culture was diluted 40-fold in fresh MHB broth and incubated at 37°C with shaking (200 rpm) for another 2–3 h. The mid-log phase cultures were diluted to a concentration of 5 × 105 CFU/mL by McFarland Standards, which was used as seeding solution for following antibacterial assays. Each well of the compound-containing 96-well plates was seeded with 100 μL of aforementioned seeding solution, giving a final compound concentration 200 μM. Plates were covered and incubated at 37°C for 24 h. The inhibitory rates were calculated by [(1-OD625sample)/OD625negative control] × 100%. Penicillin G sodium and ceftazidime were used as positive controls (inhibitory rates >99%) (Zhao Z.-Z. et al., 2018). Compounds were first tested for their potential against five bacterial strains at a single concentration of 200 μM. If the inhibitory rate above 50%, different concentrations of the compounds were further evaluated for the antibacterial ability.
β-amyloid precursor protein-cleaving enzyme 1 FRET assays
β-amyloid precursor protein-cleaving enzyme 1 (β-secretase, BACE1) assays were performed according to the literature (Qi et al., 2018; Yatsu et al., 2019). First, 10 μL of the test sample, 10 μL of the 722 nM BACE1 substrate (Rh-EVNNLDAEFK-Quencher in 50 mM ammonium bicarbonate), and 10 μL of the BACE1 enzyme (1 U/mL) were mixed in black 384-well plates and incubated for 3 h at room temperature. The fluorescence intensity was measured at 540 nm excitation and 590 nm emission using a microplate reader (Tecan Spark 10M, TECAN, Männedorf, Switzerland) at the time points of 1 h and 3 h. The BACE1 inhibition rate was calculated by the following equation: BACE1 inhibition rate (%) = {1–[(S–S0)–(B–B0)/(C–C0)–(B–B0)]} × 100, where C is the fluorescence of a DMSO control (enzyme, substrate, and assay buffer with DMSO) after 3 h of incubation, C0 is the fluorescence of the DMSO control at 1 h after incubation, B is the fluorescence of a non-enzyme control (substrate and assay buffer with DMSO) after 3 h of incubation, B0 is the fluorescence of the non-enzyme control at 1 h after incubation, S is the fluorescence of the tested samples (enzyme, sample solution, and substrate) after 3 h of incubation, and S0 is the fluorescence of the tested samples at 1 h after incubation. The IC50 values were calculated by GraphPad Prism software (v5.02, CA, USA).
Results and discussion
Structural elucidation
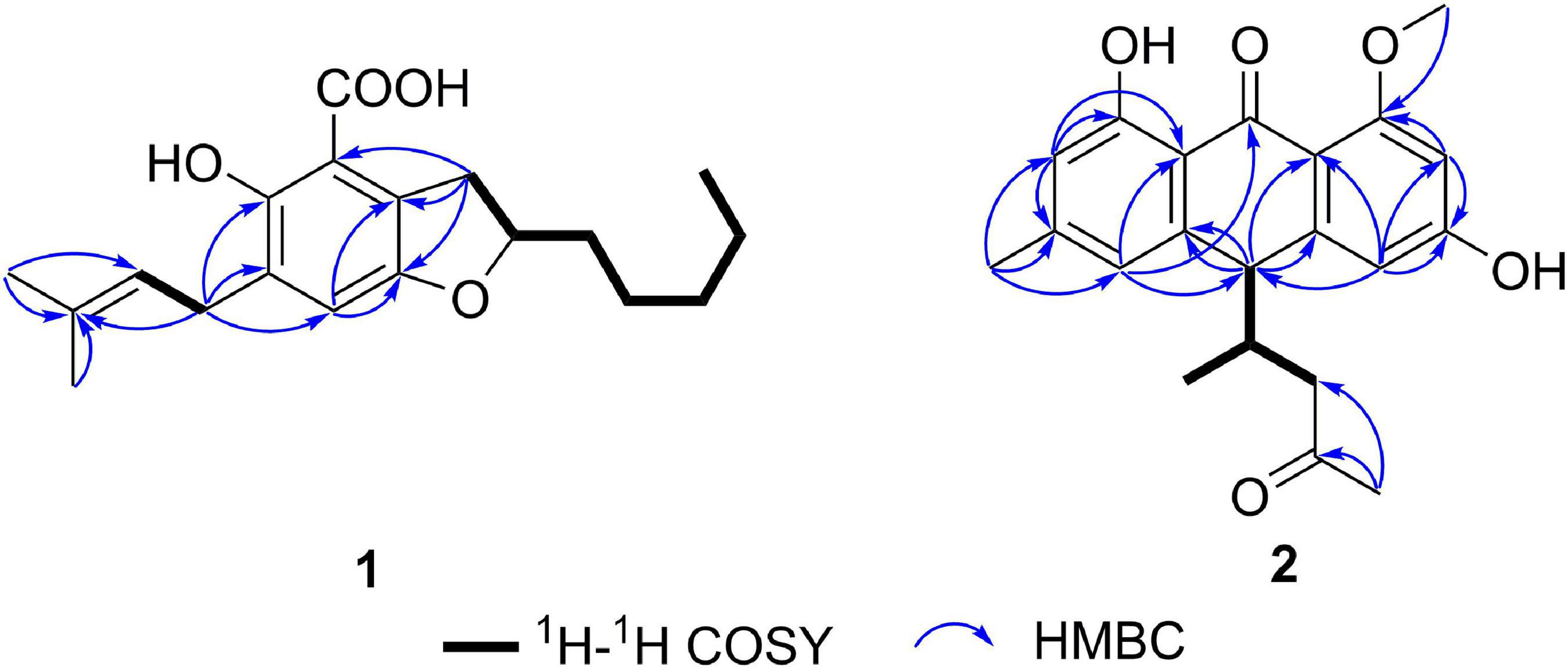
Compound 1 (Figure 1) was obtained as a racemate which was confirmed by chiral phase HPLC analysis. The HRESIMS spectroscopic analysis at m/z 319.19028 [M+H]+ (calcd. for C19H27O4, 319.19093) revealed the molecular formula is C19H26O4. According to the 1D NMR spectroscopic data of 1 (Table 1), two olefinic protons at δH 6.86 (1H, s, H-5) and at δH 5.28 (1H, br. t, J = 7.6 Hz, H-16), one methine at δH 4.51 (1H, m, H-9), three methyl groups at δH 0.90 (3H, t, J = 7.0 Hz, H3-14), 1.69 (3H, s, H3-18), 1.75 (3H, s, H3-19) were identified. The 13C NMR and HSQC spectra revealed one carbonyl carbon at δC 170.6 (C-1), six non-protonated carbons at δC 108.0 (C-2), 121.5 (C-7), 143.3 (C-6), 129.2 (C-4), 154.5 (C-3), 134.0 (C-17), three methines at δC 124.0 (C-5), 80.0 (C-9), 121.3 (C-16), six methylene carbons at δC 26.8 (C-8), 35.0 (C-10), 24.7 (C-11), 31.7 (C-12), 22.7 (C-13), 27.6 (C-15) and three methyl carbons at δC 14.1 (C-14), 17.9 (C-18), 26.0 (C-19). The data showed high similarity to those of 2-(2′,3-epoxy-1′,3′-heptadienyl)-6-hydroxy-5-(3-methyl-2-butenyl)benzaldehyde (7) (Wang et al., 2006), a previously described compound which was also been encountered in this study. The 1H-1H COSY correlations of H2-8, H-9, H2-10, H2-11, and the HMBC correlation from H2-8 to C-2, C-6, C-7, and from H-9 to C-6, together with the 13C chemical shifts of C-6 and C-9 suggested that the absence of conjugated double bonds at C-8, C-9, C-10, and C-11 in 1 compared to those of 7 (Figure 2).
Additionally, the 13C chemical shifts of C-1 (δC 170.6), as well as the absence of the aldehyde proton, indicated that C-1 in 1 was a carboxylic group instead of being an aldehyde carbonyl in 7. This change was also in accordance with the HRESIMS data. Considering that this isolate only has one chiral carbon, the question about whether compound 1 was optically pure was raised (Chen et al., 2016). Although the chiral phase HPLC analysis of this sample on two kinds of chiral phase columns, the CHIRALPAK AS-H and AD-H showed only one peak, suggesting either the sample was optically pure or the chiral phase columns used were unable to separate the antipodes. However, the small specific optical rotation value (+2.7) and the reported analog in the literature (Zhong W.-M. et al., 2018; Zhong W. et al., 2021) indicated that this sample is a racemic mixture. Hence, compound 1 was named chevalieric acid.
In this experiment, a chromatograph peak by reverse phase HPLC analysis on the C18 column was obtained, of which the purity met the requirement for further NMR analysis. However, the 1H NMR spectrum exhibited two sets of proton signals with a ratio 1:1 (Supplementary Figure 16), indicating the presence of two stereoisomers of this sample. Chiral phase HPLC analysis (column: Daicel CHIRALPAK AS-H) of this sample gave three peaks (Figure 3A), which were further subjected to measuring the 1H NMR spectra. The results suggested that the latter two peaks which were designated as compounds 2 and 3 were optically pure; however, the first peak was still a mixture. Further chiral phase HPLC analysis (column Daicel CHIRALPAK AD-H) of this mixture presented two isolated peaks which were designated as compounds 4 and 5 (Figure 3B). The clean NMR spectra of the optically pure isolates helped to unambiguously determine their structures by NMR spectroscopic analysis, specifical optical rotation measurements, and computational methods.
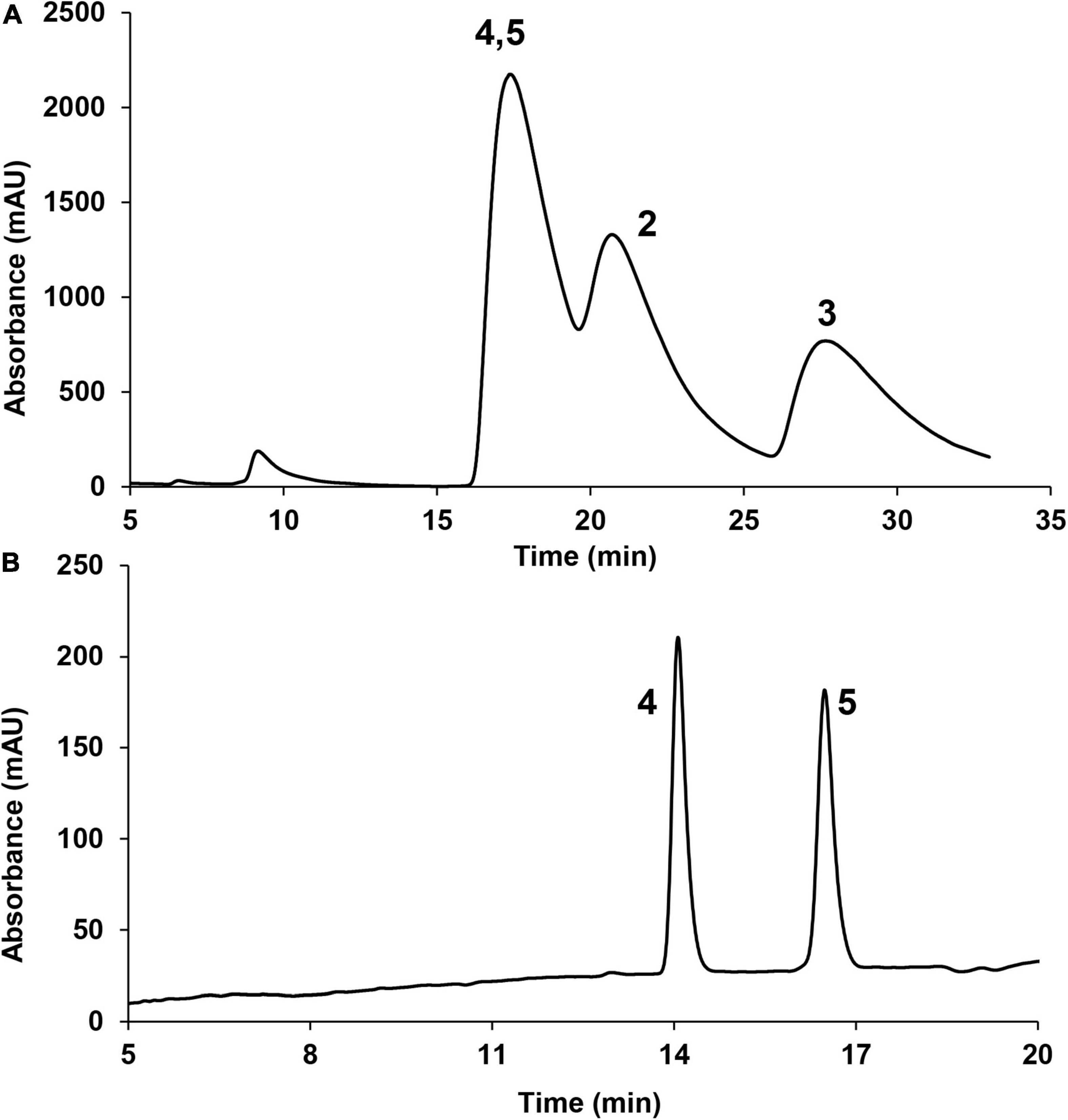
Figure 3. Chiral phase HPLC analysis of compounds 2–5 (220 nm). (A) CHIRALPAK AS-H column; (B) CHIRALPAK AD-H column.
The NMR spectra between compounds 2 and 4, compounds 3 and 5 are identical, indicating that 2/4, 3/5 are two pairs of enantiomers. The HRESIMS spectroscopic analysis of 2/4 gave the protonated ion peak at m/z 355.15402 [M+H]+, revealing the molecular formula of C21H22O5 (calcd. for C21H23O5, 355.15455). The 1H NMR spectroscopic data (Table 1) of 2/4 showed four aromatic protons at δH 6.39 (d, J = 2.2 Hz, H-2), 6.50 (d, J = 2.2 Hz, H-4), 6.57 (s, H-5), 6.71 (s, H-7), two methine protons at δH 4.06 (d, J = 3.4 Hz, H-10), 2.49 (m, H-12), one methylene group at δH 2.02 (dd, J = 17.4, 7.3 Hz, H-13a), 2.37 (dd, J = 17.4, 5.8 Hz, H-13b), four methyl groups at δH 2.38 (s, H-11), 2.09 (s, H-15), 0.69 (d, J = 7.0 Hz, H-16), 3.87 (s, 1-OCH3). The 13C NMR data (Table 1) and HSQC spectra indicated a total of 21 carbon signals, including two carbonyl carbons at δC 191.8 (C-9), 207.9 (C-14), twelve aromatic carbons at δC 165.7 (C-1), 99.6 (C-2), 165.2 (C-3), 108.1 (C-4), 147.0 (C-4a), 120.9 (C-5), 147.0 (C-6), 116.6 (C-7), 162.5 (C-8), 114.4 (C-8a), 110.6 (C-9a), 143.8 (C-10a), two methine carbons at δC 47.8 (C-10), 40.2 (C-12), one methylene carbon at δC 46.7 (C-13), four methyl carbons at δC 22.4 (C-11), 30.8 (C-15), 16.4 (C-16), 55.8 (1-OCH3). The HMBC correlations from H-2 to C-1, C-3, C-9a, C-4, from H-4 to C-2, C-3, C-4a, C-9a, C-10, from H-5 to C-6, C-10, C-11, C-7, C-8a, and from H-7 to C-5, C-8, C-8a, C-11 (Figure 2) suggested the presence of an anthraquinone skeleton in compounds 2/4. The 1H-1H COSY correlations of H-10/H-12/H2-13, H-12/H3-16 in combination with the HMBC correlation from H3-15 to the carbonyl at δC 207.9 (C-14), and C-13 indicated that an additional chain with five carbons attached to the anthraquinone skeleton at the position C-10. The HMBC correlation from δH 3.87 to C-1 showed that C-1 was substituted by a methoxy group. According to the analysis above, the planar structure of 2/4 was determined as shown in Figure 2. Structural elucidation of the NMR spectra of 3/5 showed that the two compounds shared the same planar structures with compounds 2 and 4.
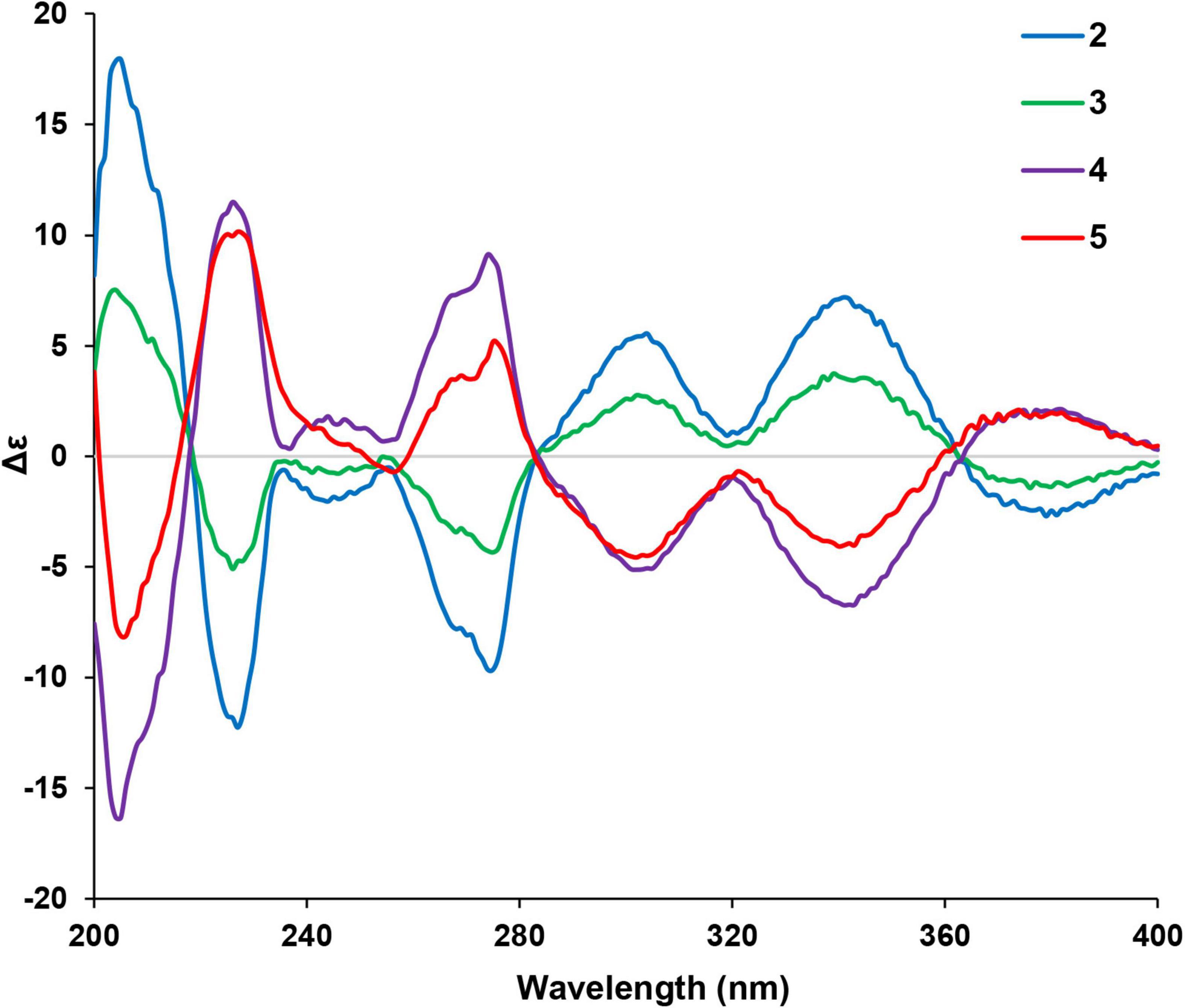
Given that there are two chiral centers in the planar structures of compounds 2–5 which correspond to four stereoisomers, including two pairs of enantiomers, the 10S,12S, the 10R,12R, the 10S,12R, and the 10R,12S. The absolute configurations of the four congeners were determined by computational methods. The experimental circular dichroism (CD) spectra of the four compounds were obviously classified into two groups which mirrors to each other, i.e., compounds 2 and 3 mirror with compounds 4 and 5, respectively, which was in accordance with the NMR data. Therefore, to simplify the calculation work, only compounds 2 and 3 were discussed hereinafter. First of all, the absolute configurations of 2 and 3 were unable to be determined by ECD calculations because of the remarkable similarity between the experimental CD spectra of these two compounds (Figure 4).
The two theoretical epimers with the configurations 10S,12S and 10S,12R were subjected to ECD calculations. As a result, the comparison between the calculated ECD and experimental CD failed to assign the absolute configuration of compounds 2 and 3 due to the close absorption patterns of the experimental CD of the two compounds (Figure 5).
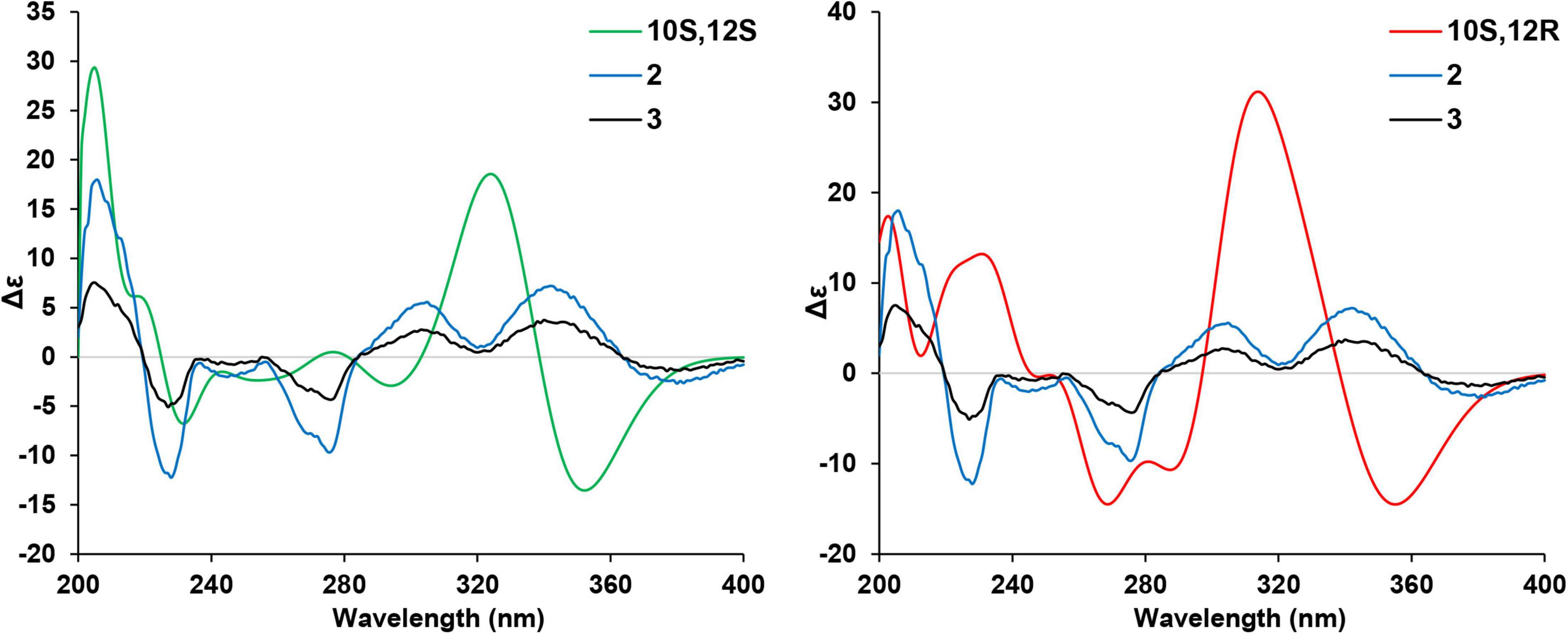
Figure 5. Comparison of the calculated electronic circular dichroism (ECD) of (10S,12S)- and (10S,12R)-isomers vs. compounds 2 and 3.
The two stereoisomers were further subjected to 13C NMR calculations at the B3LYP/6-311G(2d,p) level of theory with IEFPCM model in chloroform based on the optimized geometries at B3LYP/6-311G(2d,p) level of theory. As shown in Figure 6, all the linear regression analyses between the two theoretical epimers with compounds 2 and 3 gave good results since the close similarity of the 13C NMR between compounds 2 and 3. Therefore, this evidence was unable to assign the absolute configurations of compounds 2 and 3 unambiguously.
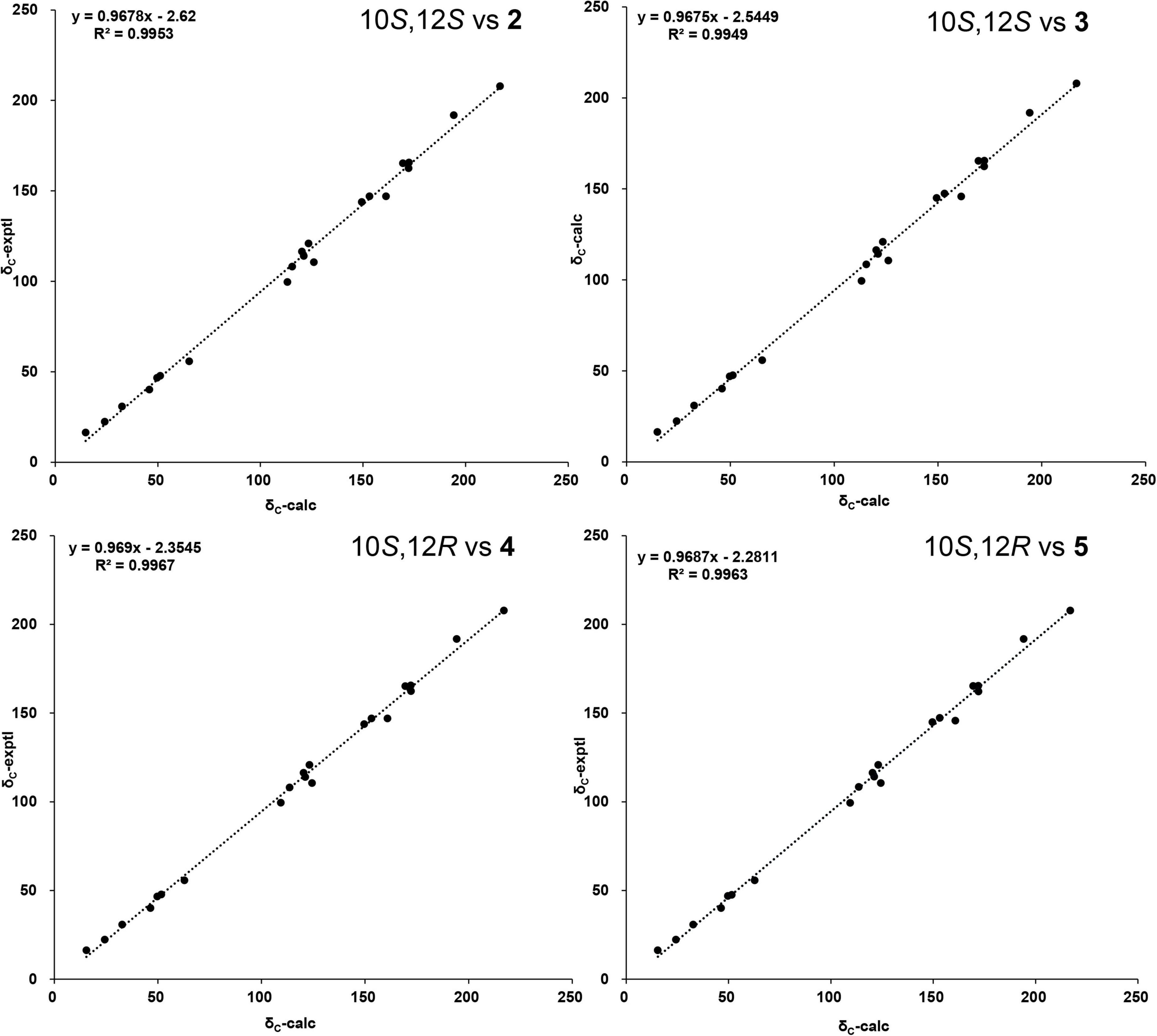
Figure 6. The linear regression analysis of calculated and experimental nuclear magnetic resonance (NMR) data of (10,12)-stereoisomers and compounds 2 and 3.
The DP4+ method is one of the leading strategies with reliable performance in structural assignment of compounds based on the Bayesian analysis of the calculated NMR data (Grimblat et al., 2015; Marcarino et al., 2021). The NMR shielding values of the (10,12)-stereoisomers were calculated with the recommended level of theory [mPW1PW91/6-31G + (d,p)//B3LYP/6-31G(d)], the calculated results were subjected to DP4+ analysis against the experimental chemical shifts of compounds 2 and 3, respectively. As shown in Table 2, the DP4+ analyses of all data (13C and 1H shielding values) suggested that compound 2 preferred the 10S,12S configuration instead of the 10S,12R configuration (98.01–1.99), and compound 3 preferred the 10S,12R configuration instead of the 10S,12S configuration (99.84–0.16), allowing the unambiguously assign the absolute configuration of compounds 2 and 3 as 10S,12S, and 10S,12R, respectively. Taking in consideration of the specific optical values and NMR results, the absolute configurations of compounds 4 and 5 were, thereby, determined as 10R,12R, and 10R,12S, respectively.

Table 2. DP4+ analysis of the (10,12)-stereoisomers with compounds 2 and 3 (sDP4+: scaled chemical shifts; uDP4+: unscaled chemical shifts).
In this study, ten previously reported compounds were obtained, asperglaucin B (6), the 2-(2′,3-epoxy-1′,3′-heptadienyl)-6-hydroxy-5-(3-methyl-2-butenyl)benzalde-hyde (7) (Chen et al., 2016), tetrahydroauroglaucin (8) (Smetanina et al., 2007), dihydroauroglaucin (9) (Miyake et al., 2009), flavoglaucin (10) (Yoshihira et al., 1972), isodihydroauroglaucin (11) (Miyake et al., 2009), cristaldehyde A (12) (Zhang et al., 2019), (3′S,4′R)-6-(3′,5-epoxy-4′-hydroxy-1′-heptenyl)-2-hyd roxy-3-(3″-methyl-2″-butenyl)benzaldehyde (13) (Chen et al., 2017), 1,4,5-trihydroxy-7-methoxy-2-methyl-anthraquinone (14) (Awaad et al., 2014), physcion (15) (Yoshihira et al., 1972). All the isolated polyketides were classified into two groups, the salicylaldehyde derivatives with prenyl group modifications, and the anthraquinones. Compounds 2–5 are unusual anthraquinones with additional side chains, which worth to be studied in terms of biosynthetic origins.
Biological activity evaluation
All the benzaldehyde derivatives were subjected to screening the cytotoxicity against the human lung cancer line A549, only compound 10 showed weak cytotoxicity activity against the human lung cancer line A549 with the IC50 39.68 μM (positive control cisplatin, IC50 5.03 μM).
The Aβ protein, produced by the β-amyloid precursor protein-cleaving enzyme 1, is considered as the pathophysiology product of AD. We evaluated the BACE1 inhibitory of all benzaldehyde derivatives compounds isolated in this study. Compounds 8, 10, and 11 showed moderate to weak inhibition against the β-secretase with the IC50 values of 36.1, 40.9, 34.9 μM, respectively (Table 3). The IC50 value for the positive control LY2811376 was 0.6 μM. The results indicated the potential of salicylaldehydes serving as template structures for further chemical modifications in the field of anti-AD drug research and development.

Table 3. β-amyloid precursor protein-cleaving enzyme 1 (BACE1) inhibitory tests of compounds 1 and 6–13.
All the anthraquinone derivatives (2–5, 14–15) were evaluated for the antibacterial activity against five bacteria, the Escherichia coli ATCC25922, S. aureus subsp. aureus ATCC29213, S. enterica subsp. enterica ATCC14028, P. aeruginosa ATCC27853, and methicillin-resistant S. aureus (Table 4). Compounds 3–5 showed significant inhibition against the opportunistic pathogenic bacterium P. aeruginosa (inhibition rate: 81.0–91.5%) and methicillin-resistant S. aureus (inhibition rate: 74.0–88.5%) at the concentration of 200 μM, while the structural congener compound 2 only showed weak inhibition (inhibition rate: 38.2%) against the P. aeruginosa at 200 μM. Due to the shortage of samples, we only measured the inhibitory activity against MRSA of compound 5 with the MIC50 value of 44.02 μM.
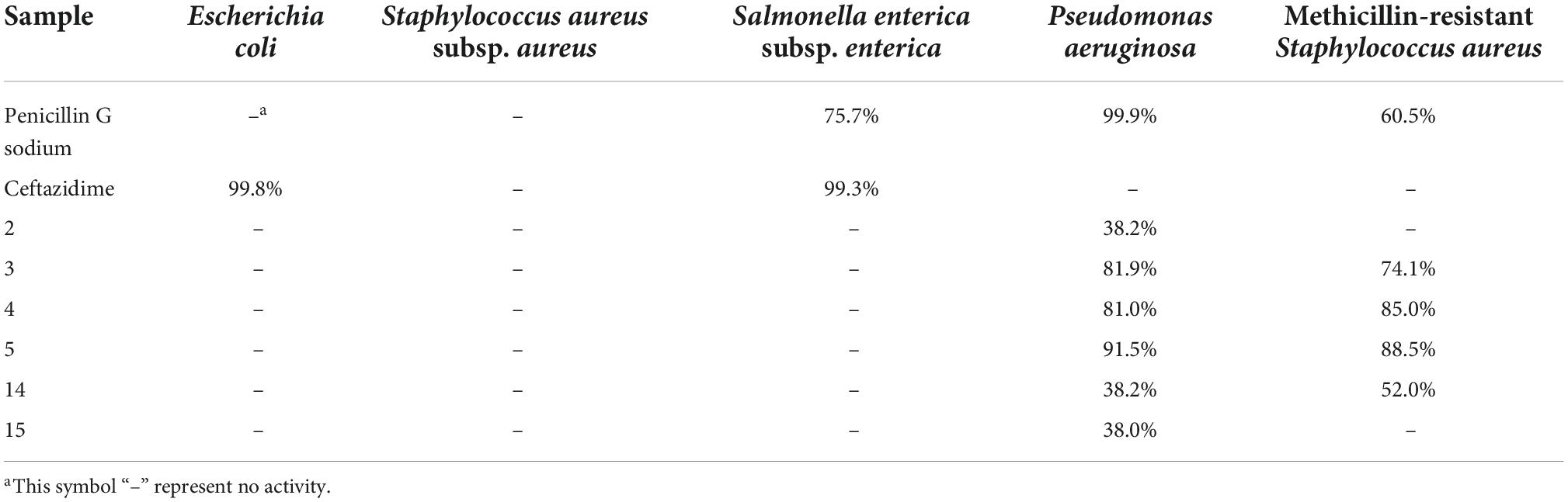
Table 4. Inhibition rates of anthraquinone derivatives against the bacteria at the concentration of 200 μM.
Conclusion
In summary, five previously undescribed polyketide derivatives with ten known compounds were isolated from the fungus A. chevalieri. Through comprehensive analysis of 1D, 2D NMR spectroscopic data, ECD calculations, 13C NMR calculations, and DP4+ analysis, the structures, including the absolute configurations of the new compounds, were unambiguously determined. In the bioassays, compound 10 showed weak cytotoxicity activity against the human lung cancer line A549 with the IC50 39.68 μM. Compounds 8, 10, and 11 exhibited moderate BACE1 inhibitory activities with IC50 values of 36.1, 40.9, 34.9 μM, respectively. Compounds 3, 4, and 5 showed significant antibacterial activities against both P. aeruginosa and MRSA. Notably, the four stereoisomers 2–5 displayed different antibacterial activities, raising the question about stereochemistry-bioactivity-relationship, which need to be addressed in the future. This study provides new horizons for follow-up drug development for anti-MRSA agents.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
XL and J-KL designed and supervised the project. Q-YW and H-PC performed the experiments. All authors analyzed data, discussed the results, wrote the manuscript, and read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (22177138 and 81903512).
Acknowledgments
We thank the Analytical and Measuring Center, School of Pharmaceutical Sciences, South-Central University for Nationalities for the spectra measurement.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1051281/full#supplementary-material
References
Awaad, A. S., Al-Zaylaee, H. M., Alqasoumi, S. I., Zain, M. E., Aloyan, E. M., Alafeefy, A. M., et al. (2014). Anti-leishmanial activities of extracts and isolated compounds from Drechslera rostrata and Eurotium tonpholium. Phytother. Res. 28, 774–780. doi: 10.1002/ptr.5096
Bruhn, T., Schaumlöffel, A., Hemberger, Y., and Bringmann, G. (2013). SpecDis: Quantifying the comparison of calculated and experimental electronic circular dichroism spectra. Chirality 25, 243–249. doi: 10.1002/chir.22138
Bruhn, T., Schaumlöffel, A., Hemberger, Y., and Pescitelli, G. (2017). SpecDis version 1.71, Berlin, Germany. Available online at: https://specdis-software.jimdofree.com (accessed November 12, 2022).
Busche, M. A., and Hyman, B. T. (2020). Synergy between amyloid-β and tau in Alzheimer’s disease. Nat. Neurosci. 23, 1183–1193. doi: 10.1038/s41593-020-0687-6
Chen, H.-P., Zhao, Z.-Z., Li, Z.-H., Huang, Y., Zhang, S.-B., Tang, Y., et al. (2018). Anti-proliferative and anti-inflammatory lanostane triterpenoids from the polish edible mushroom Macrolepiota procera. J. Agric. Food Chem. 66, 3146–3154. doi: 10.1021/acs.jafc.8b00287
Chen, H.-P., Zhao, Z.-Z., Zhang, Y., Bai, X., Zhang, L., and Liu, J.-K. (2016). (+)- and (–)-Ganodilactone, a pair of meroterpenoid dimers with pancreatic lipase inhibitory activities from the macromycete Ganoderma leucocontextum. RSC Adv. 6, 64469–64473. doi: 10.1039/C6RA10638B
Chen, M., Zhao, Q., Hao, J.-D., and Wang, C.-Y. (2017). Two benzaldehyde derivatives and their artefacts from a gorgonian-derived Eurotium sp. fungus. Nat. Prod. Res. 31, 268–274. doi: 10.1080/14786419.2016.1230116
Cummings, J., Feldman, H. H., and Scheltens, P. (2019). The “rights” of precision drug development for Alzheimer’s disease. Alz. Res. Ther. 11:76. doi: 10.1186/s13195-019-0529-5
Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., et al. (2016). Gaussian 16, Revision C.01. Wallingford CT: Gaussian, Inc.
Gao, J., Radwan, M. M., León, F., Wang, X., Jacob, M. R., Tekwani, B. L., et al. (2012). Antimicrobial and antiprotozoal activities of secondary metabolites from the fungus Eurotium repens. Med. Chem. Res. 21, 3080–3086. doi: 10.1007/s00044-011-9798-7
Grimblat, N., Zanardi, M. M., and Sarotti, A. M. (2015). Beyond DP4: An improved probability for the stereochemical assignment of isomeric compounds using quantum chemical calculations of NMR shifts. J. Org. Chem. 80, 12526–12534. doi: 10.1021/acs.joc.5b02396
Koyama, N., Nagahiro, T., Yamaguchi, Y., Masuma, R., Tomoda, H., and Ōmura, S. (2005). Stemphones, novel potentiators of imipenem activity against methicillin-resistant Staphylococcus aureus, Produced by Aspergillus sp. FKI-2136. J. Antibiot. 58, 695–703. doi: 10.1038/ja.2005.95
Marcarino, M. O., Cicetti, S., Zanardi, M. M., and Sarotti, A. M. (2021). A critical review on the use of DP4+ in the structural elucidation of natural products: The good, the bad and the ugly. A practical guide. Nat. Prod. Rep. 39, 58–76. doi: 10.1039/D1NP00030F
Miyake, Y., Ito, C., Itoigawa, M., and Osawa, T. (2009). Antioxidants produced by Eurotium herbariorum of filamentous fungi used for the manufacture of karebushi, dried bonito (katsuobushi). Biosci. Biotechnol. Biochem. 73, 1323–1327. doi: 10.1271/bbb.80887
Qi, C., Liu, M., Zhou, Q., Gao, W., Chen, C., Lai, Y., et al. (2018). BACE1 Inhibitory meroterpenoids from Aspergillus terreus. J. Nat. Prod. 81, 1937–1945. doi: 10.1021/acs.jnatprod.7b01050
Rossiter, S. E., Fletcher, M. H., and Wuest, W. M. (2017). Natural products as platforms to overcome antibiotic resistance. Chem. Rev. 117, 12415–12474. doi: 10.1021/acs.chemrev.7b00283
Schueffler, A., and Anke, T. (2014). Fungal natural products in research and development. Nat. Prod. Rep. 31, 1425–1448. doi: 10.1039/C4NP00060A
Smetanina, O. F., Kalinovskii, A. I., Khudyakova, Y. V., Slinkina, N. N., Pivkin, M. V., and Kuznetsova, T. A. (2007). Metabolites from the marine fungus Eurotium repens. Chem. Nat. Compd. 43, 395–398. doi: 10.1007/s10600-007-0147-5
Vassar, R., and Kandalepas, P. C. (2011). The β-secretase enzyme BACE1 as a therapeutic target for Alzheimer’s disease. Alz. Res. Ther. 3:20. doi: 10.1186/alzrt82
Wang, S., Li, X.-M., Teuscher, F., Li, D.-L., Diesel, A., Ebel, R., Proksch, P., et al. (2006). Chaetopyranin, a benzaldehyde derivative, and other related metabolites from Chaetomium globosum, an endophytic fungus derived from the marine red alga Polysiphonia urceolata. J. Nat. Prod. 69, 1622–1625. doi: 10.1021/np060248n
World Health Organization (2022). Dementia. Available online at: https://www.who.int/news-room/fact-sheets/detail/dementia (accessed September 21, 2022).
Yamazaki, H., Koyama, N., Ōmura, S., and Tomoda, H. (2008). Structure-activity relationships of stemphones, potentiators of imipenem activity against methicillin-resistant Staphylococcus aureus. J. Antibiot. 61, 426–441. doi: 10.1038/ja.2008.59
Yatsu, G., Kino, Y., Sasaki, H., Satoh, J.-I., Kinoshita, K., and Koyama, K. (2019). Meroterpenoids with BACE1 Inhibitory activity from the fruiting body of Boletinus asiaticus. J. Nat. Prod. 82, 1797–1801. doi: 10.1021/acs.jnatprod.8b01092
Yoshihira, K., Takahashi, C., Sekita, S., and Natori, S. (1972). Tetrahydroauroglaucin from Penicillium charlesii. Chem. Pharm. Bull. 20, 2727–2728. doi: 10.1248/cpb.20.2727
Zhang, P., Jia, C., Deng, Y., Chen, S., Chen, B., Yan, S., et al. (2019). Anti-inflammatory prenylbenzaldehyde derivatives isolated from Eurotium cristatum. Phytochemistry 158, 120–125. doi: 10.1016/j.phytochem.2018.11.017
Zhang, Y., Jia, A., Chen, H., Wang, M., Ding, G., Sun, L., et al. (2017). Anthraquinones from the saline-alkali plant endophytic fungus Eurotium rubrum. J. Antibiot. 70, 1138–1141. doi: 10.1038/ja.2017.121
Zhao, D., Cao, F., Guo, X.-J., Zhang, Y.-R., Kang, Z., and Zhu, H.-J. (2018). Antibacterial indole alkaloids and anthraquinones from a sewage-derived fungus Eurotium sp. Chem. Nat. Compd. 54, 399–401. doi: 10.1007/s10600-018-2361-8
Zhao, Z.-Z., Zhao, K., Chen, H.-P., Bai, X., Zhang, L., and Liu, J.-K. (2018). Terpenoids from the mushroom-associated fungus Montagnula donacina. Phytochemistry 147, 21–29. doi: 10.1016/j.phytochem.2017.12.015
Zhong, W., Chen, Y., Mai, Z., Wei, X., Wang, J., Zeng, Q., et al. (2020). Euroticins A and B, two pairs of highly constructed salicylaldehyde derivative enantiomers from a marine-derived fungus Eurotium sp. SCSIO F452. J. Org. Chem. 85, 12754–12759. doi: 10.1021/acs.joc.0c01407
Zhong, W., Chen, Y., Wei, X., Wang, J., Zhang, W., Wang, F., et al. (2021). Salicylaldehyde derivatives from a marine-derived fungus Eurotium sp. SCSIO F452. J. Antibiot. 74, 273–279. doi: 10.1038/s41429-020-00395-x
Zhong, W.-M., Wang, J.-F., Shi, X.-F., Wei, X.-Y., Chen, Y.-C., Zeng, Q., et al. (2018). Eurotiumins A–E, five new alkaloids from the marine-derived fungus Eurotium sp. SCSIO F452. Mar. Drugs 16:136. doi: 10.3390/md16040136
Keywords: Aspergillus chevalieri, polyketide, DP4+, antiproliferative activity, antibacterial activity, BACE1 inhibition
Citation: Wang Q-Y, Chen H-P, Wu K-Y, Li X and Liu J-K (2022) Antibacterial and β-amyloid precursor protein-cleaving enzyme 1 inhibitory polyketides from the fungus Aspergillus chevalieri. Front. Microbiol. 13:1051281. doi: 10.3389/fmicb.2022.1051281
Received: 22 September 2022; Accepted: 07 November 2022;
Published: 22 November 2022.
Edited by:
Dipesh Dhakal, University of Florida, United StatesReviewed by:
Wen-Bo Han, Northwest A&F University, ChinaFuhang Song, Beijing Technology and Business University, China
Man-Cheng Tang, Shanghai Jiao Tong University, China
Copyright © 2022 Wang, Chen, Wu, Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinyang Li, xinyangli131@hotmail.com; Ji-Kai Liu, liujikai@mail.scuec.edu.cn
†These authors have contributed equally to this work
 Qing-Yuan Wang1†
Qing-Yuan Wang1† He-Ping Chen
He-Ping Chen Xinyang Li
Xinyang Li